Abstract
Phosphorylcholine (PC) is an epitope on oxidized low-density lipoprotein (oxLDL), apoptotic cells and several pathogens like Streptococcus pneumoniae. Immunoglobulin M against PC (IgM anti-PC) has the ability to inhibit uptake of oxLDL by macrophages and increase clearance of apoptotic cells. From our genome-wide association studies (GWASs) in four European-ancestry cohorts, six single nucleotide polymorphisms (SNPs) in 11q24.1 were discovered (in 3002 individuals) and replicated (in 646 individuals) to be associated with serum level of IgM anti-PC (the leading SNP rs35923643-G, combined β = 0.19, 95% confidence interval 0.13–0.24, P = 4.3 × 10−11). The haplotype tagged by rs35923643-G (or its proxy SNP rs735665-A) is also known as the top risk allele for chronic lymphocytic leukemia (CLL), and a main increasing allele for general IgM. By using summary GWAS results of IgM anti-PC and CLL in the polygenic risk score (PRS) analysis, PRS on the basis of IgM anti-PC risk alleles positively associated with CLL risk (explained 0.6% of CLL variance, P = 1.2 × 10−15). Functional prediction suggested that rs35923643-G might impede the binding of Runt-related transcription factor 3, a tumor suppressor playing a central role in the immune regulation of cancers. Contrary to the expectations from the shared genetics between IgM anti-PC and CLL, an inverse relationship at the phenotypic level was found in a nested case–control study (30 CLL cases with 90 age- and sex-matched controls), potentially reflecting reverse causation. The suggested function of the top variant as well as the phenotypic association between IgM anti-PC and CLL risk needs replication and motivates further studies.
Introduction
Phosphorylcholine (PC) is an epitope on oxidized low-density lipoprotein (oxLDL), apoptotic cells and several pathogens like Streptococcus pneumoniae (1,2). Pneumococcal vaccination has been shown to decrease atherosclerotic lesion formation in animal models, maybe because of this molecular mimicry (3). PC is reported to be important in oxLDL-induced immune activation (4). PC-targeting immunization can reduce atherosclerosis in apolipoprotein E knockout mice, in which the immunoglobulin M and G against PC (IgM and IgG anti-PC) are increased, although neither the general IgM nor IgG are increased (5). IgM anti-PC has the ability to inhibit uptake of oxLDL by macrophages and increase clearance of apoptotic cells (2). Therefore, IgM anti-PC has been put forward as a potential novel biomarker for several diseases such as autoimmune diseases, stroke and coronary heart disease (2,6–8). About 40% of IgM anti-PC variation can be attributed to genetics (9), but the associated genetic variants have not been identified so far.
PC also links to chronic lymphocytic leukemia (CLL), a B-cell malignancy usually occurring in older age (10). The antibodies derived from CLL cell lines have been reported to recognize limited target structures including oxLDL and PC (11). Among 10 types of respiratory tract infections, only pneumonia has been reported to be associated with increased risk of CLL (12). However, the phenotypic relationship between IgM anti-PC and CLL has not been statistically evaluated.
In this study, we aim to identify the genetic variants regulating serum level of IgM anti-PC by conducting genome-wide association studies (GWASs) in four Swedish cohorts; and to evaluate the association between IgM anti-PC and CLL risk in a nested case–control study.
Results
GWAS meta-analysis of IgM anti-PC
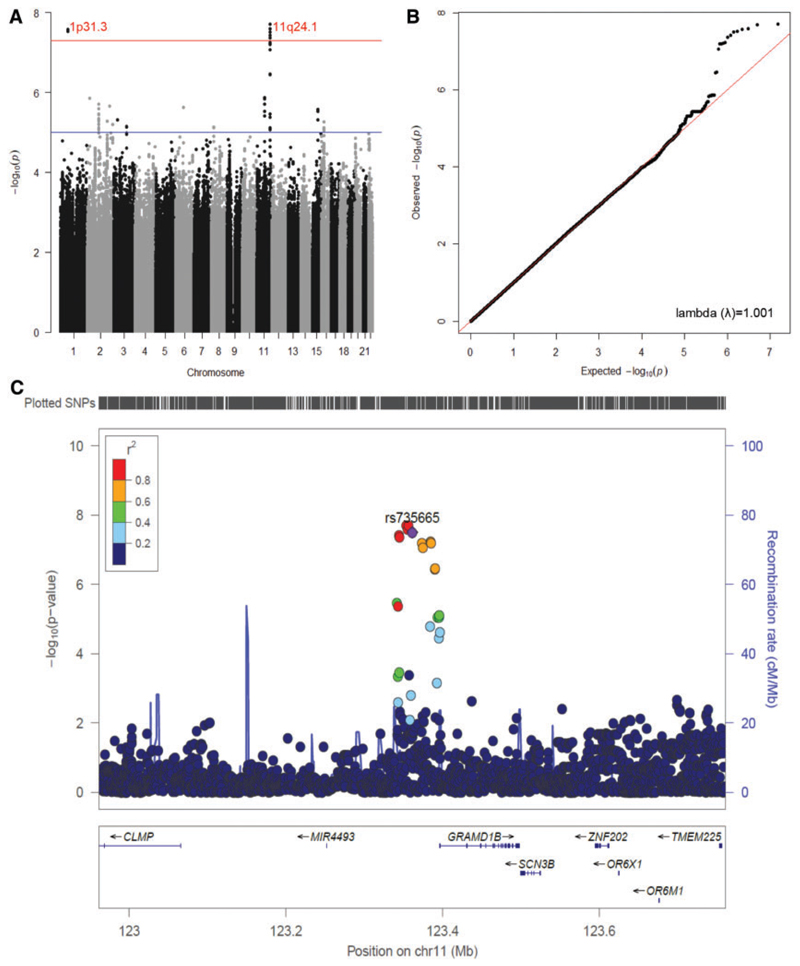
After phenotype and genotype matching, individuals with both IgM anti-PC measurements and genotypes available were used in this study (Table 1). Among the three individual discovery GWAS of IgM anti-PC, one single nucleotide polymorphism (SNP) rs74420772 in 3p14.1 achieved genome-wide significance in TwinGene (P = 2.7 × 10−8), whereas no genome-wide significant SNPs were observed in the other two studies (Supplementary Material, Fig. S1). In the discovery phase meta-analysis of these three GWAS results (total n = 3002), two SNPs in 1p31.3 and six SNPs in 11q24.1 achieved genome-wide significance (P < 5.0 × 10™8, Fig. 1A; Supplementary Material, Table S1), with negligible inflation of the signal from population stratification or other sources, lambda (λ) = 1.001 (Fig. 1B). Validation of the eight associated SNPs from the discovery GWAS meta-analysis was sought in the fourth cohort (n = 646). The two SNPs in 1p31.3 were not significant (P ≥ 0.164), but all of the six SNPs in 11q24.1 were successfully replicated (P < 6.5×10−3, Supplementary Material, Table S2). On the basis of all four studies, the allelic effect size (β) of the leading SNP rs35923643-G in 11q24.1 was 0.19 [95% confidence interval (CI) 0.13–0.24, P =4.3 × 10−11] rank order normalized standard deviation (SD) of IgM anti-PC per allele (Table 2; Supplementary Material, Table S3). These six SNPs are in strong linkage disequilibrium (LD) within a 131 kb block that overlaps with the GRAM domain containing 1B (GRAMD1B) gene (Fig. 1C). The mean levels of IgM anti-PC in the genotype groups of rs35923643 (and its proxy SNP rs735665) are presented in Supplementary Material, Table S4.
Table 1. Subjects used in the genome-wide association meta-analysis of IgM anti-PC.
| Discovery phase | Replication | ||||
|---|---|---|---|---|---|
| TwinGene | PIVUS | MDC | PRACSIS | ||
| Numbers | |||||
| All | 1175 | 945 | 882 | 646 | |
| Male | 629 | 473 | 494 | 461 | |
| Female | 546 | 472 | 388 | 185 | |
| Age (years) | |||||
| All | 74.2 ± 5.6 | 70.2 ± 0.2 | 60.8 ± 5.0 | 64.5 ± 9.5 | |
| Male | 73.9 ± 5.4 | 70.1 ± 0.1 | 60.7 ± 4.9 | 63.7 ± 9.6 | |
| Female | 74.5 ± 5.7 | 70.2 ± 0.2 | 60.9 ± 5.2 | 66.4 ± 8.9 | |
| IgM anti-PC (U/ml) | |||||
| All | 42.7 (23.3−72.9) | 42.4 (27.0−73.4) | 48.0 (29.9−75.8) | 34.9 (21.2−60.4) | |
| Male | 42.6 (22.7−69.8) | 39.1 (24.5−65.3) | 48.0 (28.9−78.2) | 35.1 (22.2−58.7) | |
| Female | 42.8 (24.1−74.3) | 47.9 (29.7−79.9) | 48.0 (21.9−74.4) | 34.0 (20.3−67.4) | |
Distribution of age is described as mean±SD; the raw values of IgM anti-PC are skewed distributed and presented by median (interquartile range, the 25th percentile– the 75th percentile).
Figure 1.
Meta-analysis on three discovery GWASs of IgM anti-PC (total n = 3002). (A) Manhattan plot. It includes associations between 7 665 664 autosomal SNPs and serum level of IgM anti-PC, two SNPs in 1p31.3 (P = 2.6 × 10−8) and six SNPs in llq24.1 (P = 2.0 × 10−8) achieve genome-wide significance. (B) Quantile–quantile plot. It indicates negligible inflation of the signal from population stratification or other sources, genomic inflation factor lambda (λ) = 1.001. (C) Regional plot for the six genome-wide significant SNPs in 11q24.1 (highly correlated and rs735665 is directly genotyped). They locate in a 131 kb linkage disequilibrium block that overlaps with the GRAMD1B gene.
Table 2. Details of the lead SNP rs35923643 from the meta-analysis on four studies.
| SNP (Position) | Study | Info | β | SE | P-value | Phet |
|---|---|---|---|---|---|---|
| rs35923643 G/A | Meta | 0.189 | 0.029 | 4.34 × 10−11 | 0.159 | |
| (Chr11: 123355391) | TwinGene | 0.999 | 0.153 | 0.050 | 0.002 | |
| PIVUS | 0.980 | 0.263 | 0.054 | 1.18 × 10−6 | ||
| MDC | 0.987 | 0.101 | 0.061 | 0.097 | ||
| PRACSIS | 0.975 | 0.247 | 0.069 | 0.0004 |
SNP is presented with effect allele/alternative allele (chromosome number and position in human genome GRCh37/hg19). Info, imputation quality; β, effect size per SD of rank order normalized IgM anti-PC per allele; SE, standard error; Phet, P-value for heterogeneity.
Polygenic risk score analysis
The allele rs35923643-G (or its proxy SNP rs735665-A, r 2 = 1, D′ = 1) has also been recognized as the top risk allele for CLL, as judged by the latest meta-analysis of six GWASs of CLL (13). In the latest GWAS of general immunoglobulin levels, rs735665-A was also found to be a main genetic variant specific for general IgM, rather than general IgA and general IgG (14). The shared genetic variants were investigated in polygenic risk score (PRS) and correlation analyses by using the summary GWAS results of IgM anti-PC (from our discovery meta-analysis), IgM [SNPs with association P < 1×10−6 from Jonsson et al. (14)] and CLL [from the InterLymph consortium (15)].
PRSs on the basis of general IgM increasing alleles were positively associated with IgM anti-PC, with β close to 0.5 and P < 5 × 10−6 across all tested P-value threshold quantiles (Fig. 2A). Eight of the nine genome-wide significant SNPs for general IgM were identified in our IgM anti-PC GWAS, in which rs2476601 (P = 0.007) and rs735665 (P = 3.1 × 10−8) achieved nominal significance (Fig. 2B).
Figure 2.
Shared genetics between general IgM, CLL and IgM anti-PC. (A) Standardized PRSs on the basis of the alleles increasing general IgM are used to predict IgM anti-PC. (B) Genome-wide significant SNPs for general IgM are identified in our IgM anti-PC GWAS result to investigate the correlations between their effect sizes. (C) Standardized PRSs on the basis of the alleles increasing IgM anti-PC are used to predict CLL risk. (D) Genome-wide significant SNPs for CLL are identified in our IgM anti-PC GWAS result to investigate the correlations between their effect sizes. The effect sizes of PRS or SNPs are presented as β for continuous IgM (anti-PC) levels and logOR for binary CLL. OR, odds ratio; N SNP, number of independent SNPs included in the P-threshold quantile; r 2, Nagelkerke r 2, the proportion of the target variation explained by independent SNPs in the P-threshold quantile; n SNP, genome-wide significant SNPs (for genral IgM and CLL) identified in our IgM anti-PC GWAS meta-analysis; n Y, smaple size of meta-analysis on discovery IgM anti-PC GWAS; SE, standard error; Phet, P-value for heterogeneity.
PRSs on the basis of IgM anti-PC increasing alleles were associated with higher risk of CLL. Across the 5000 quantiles including genome-wide SNPs with gradually increasing association P-value with IgM anti-PC, the quantile only including the SNP rs735665 was best-fitted and explained the largest variance of CLL (Nagelkerke r 2 = 0.006, P = 1.2×1015, Fig. 2C). All of the 33 independent genome-wide significant SNPs for CLL were identified in our IgM anti-PC GWAS, in which rs9392504 (P = 0.04) and rs735665 (P = 3.1 × 10−8) achieved nominal significance (Fig. 2D).
PRSs on the basis of increasing alleles for general IgM (across quantiles with gradually increasing threshold for association P-value, from P < 5.0 × 10−10 to P < 1.0 × 10−6) were associated with lower risk of CLL (Supplementary Material, Table S5). There were 43 independent SNPs (association P-values with general IgM was lower than 5.0 × 10−8) in the best-fitted quantile, explaining 0.003 of the variance of CLL risk with an odds ratio of 0.69 (95% CI 0.55–0.83, P = 4.2 × 10−8).
Functional prediction
The potential function of the variants in the top locus 11q24.1 shared between IgM anti-PC and CLL is not known. The LD block that rs35923643 located was scanned for regulatory marks in leukemia- or immune-cell lines (Supplementary Material, Fig. S2). Peaks of histone mark monomethylated histone H3 lysine 4 (H3K4Me1) (in GM12878) and deoxyribonuclease I (DNaseI) hypersensitivity (in GM12878, CD34+ and GM12865) were found at the position of rs35923643 (Supplementary Material, Fig. S3A and B).
In the chromatin immunoprecipitation-sequencing (ChIP-seq) experiments from Encyclopedia of DNA Elements (ENCODE), there were suggested binding sites for 22 different transcription factors (TFs) within the ~700bp region around rs35923643 (Supplementary Material, Fig. S3C). Among the matched sequences from Find Individual Motif Occurrences (FIMO) tool, the strongest signals were observed for Runt-related TF 3 (RUNX3) and SPI1 (Supplementary Material, Table S6). The RUNX3 binding motifs include the SNP rs35923643 (at the position of 391 bp), in which Haib_RUNX3_GM12878_Motif1_ fw_ic0 was from a B-cell line. Switching from the major allele T/A at rs35923643 to the minor C/G allele impeded the predicted binding affinity of RUNX3, making the P-value (defined as probability of match for a random sequence with the same length) to increase 10-fold (Supplementary Material, Fig. S3D).
The RegulomeDB database also gave support for rs35923643 to affect the binding of TFs in general (Supplementary Material, Fig. S4). Taken together, the results indicate that rs35923643 may be a more likely candidate to be the functional variant rather than rs735665 that has been frequently reported for CLL.
Phenotypic association between IgM anti-PC and CLL
In a small nested case–control study (7 prevalent CLL cases, 23 incident CLL cases, with 3 age- and sex-matched controls for each case), we found IgM anti-PC to be lower in prevalent CLL cases than in matched controls (P = 0.006); although it was not different between incident CLL cases and their matched controls (P = 0.227, Table 3). No association between IgM anti-PC and incident CLL risk estimated from the stratified Cox proportional hazards model (P = 0.354). Moreover, we tested whether the difference of IgM anti-PC levels between CLL cases and the matched controls was dependent on the time between sampling and diagnosis. There was a declining but not statistically significant trend (β = –0.08, P = 0.10, Fig. 3; Supplementary Material, Fig. S5).
Table 3. Association between IgM anti-PC and CLL in the nested case–control study.
| n | Raw value (U/ml) | Normalized value | |
|---|---|---|---|
| All | 120 | 40.72 (25.33−83.45) | 0.00 ± 0.99 |
| t-test | |||
| CLL cases | 30 | 29.35 (18.68−69.73) | -0.38 ± 1.11 |
| Matched controls | 90 | 42.30 (30.32−87.06) | 0.13 ± 0.92 |
| P | 0.015 | ||
| CLL prevalent cases | 7 | 22.29 (9.29−29.53) | -1.12 ± 1.10 |
| Matched controls | 21 | 46.20 (38.04−74.36) | 0.12 ± 0.90 |
| P | 0.006 | ||
| CLL incident cases | 23 | 31.73 (18.86−88.51) | -0.15 ± 1.04 |
| Matched controls | 69 | 41.74 (27.67−91.26) | 0.13 ± 0.93 |
| P | 0.227 | ||
| CLL incident cases (>5 years)a | 11 | 29.77 (18.86−144.23) | -0.18 ± 1.09 |
| Matched controls | 33 | 38.72 (27.67−69.25) | 0.05 ± 0.84 |
| P | 0.471 | ||
| Stratified Cox proportional hazards model (CLL incident cases and matched controls) | |||
| Hazard ratio (95% CI) | 92 | 1.00 (0.99−1.01) | 0.75 (0.40-1.39) |
| P | 0.421 | 0.354 | |
Nested case–control study includes three age- and sex-matched controls for each CLL case. The distribution of raw IgM anti-PC value is skewed, so median (interquartile range, the 25th percentile–75th percentile) is used to describe the distribution. Mean6SD is used to describe the distribution of rank order normalized values of IgM anti-PC. Follow-up time is the years between dates of blood sampling and first CLL diagnosis.
Only cases with more than 5 years between sampling and diagnosis are included.
Figure 3.
The difference of IgM anti-PC across time between sampling and diagnosis. The difference (Δ) in rank order normalized IgM anti-PC level between each CLL case and the average of its three matched controls, each circle represents one matched case–control set.
Discussion
In summary, we show that the same top variant in 11q24.1 is shared between serum level of IgM anti-PC and CLL risk; and that rs35923643-G constitutes a more likely functional variant than rs735665 because it might impede the binding of tumor suppressor RUNX3. Even though PRS on the basis of increasing alleles of IgM anti-PC associated with higher risk of CLL, and the top variant contributes most to this, we saw an inverse relationship between IgM anti-PC and CLL at the phenotypic level.
The allele rs35923643-G shared between IgM anti-PC and CLL could reflect shared influences between susceptibility to initiate a B-cell malignant process and IgM anti-PC level, or it might be due to interactions with other factors such as infections. Serum level of IgM anti-PC is very low or undetectable in newborns (16), and it is lower in Swedish as compared with Kitavan people who are exposed to considerable more microorganisms (17). Among 10 types of respiratory tract infections, only pneumonia has been reported to be associated with increased CLL risk (12). Because PC is an epitope on S. pneumoniae, the production of IgM anti-PC might also be triggered by such infection.
The phenotypic correlation between general IgM and IgM anti-PC is not established in the literature, but our PRS and SNP effect size correlation analyses (Fig. 2A and B) indicate that they are expected to be markedly correlated. This is of course not surprising given that IgM anti-PC is a sub-fraction of general IgM. Because IgM anti-PC has been put forth as a promising bio-marker for autoimmune and cardiovascular diseases (2,6-8), it has been measured in several cohorts in Sweden. However, similar data for general IgM are not available in these materials, we were unable to investigate the genetic underpinnings of IgM anti-PC after adjustment for level of general IgM. We believe that such analysis is worth perusing because it might reveal other specific signals.
In the PRS analysis, the explained variance of CLL becomes reduced when including more SNPs with higher P-threshold in the quantiles (Fig. 2C), potentially because of the lack of power with the small sample size of our IgM anti-PC GWAS. However, the PRSs on the basis of general IgM increasing alleles are associated with lower risk of CLL throughout the tested P-thresholds (Supplementary Material, Table S5). The difference between these results may reflect that for general IgM, only the subset of 5934 SNPs with association P-value < 1.0 × 10−6 were available for analysis (the full summary statistics have not been published); or it might indicate actual differences in the associations to CLL between general IgM and IgM anti-PC. Therefore, the pleiotropic effects and shared genetics between general IgM, IgM anti-PC and CLL are worth to be further investigated by Mendelian randomization-Egger and LD score regression in studies with larger sample size.
The allele frequency of the proxy SNP rs735665-A is very different among ethnic groups (Supplementary Material, Fig. S6): 1%–2% in African and East Asian people, but 14%–22% in South Asian, European and American populations. We note that these differences coincide with CLL being one of the most common forms of leukemia in western countries, whereas rare in East Asia and Middle East (10,18), but the significance of these observations remains to be further investigated.
The GRAMD1B gene close to rs735665 and rs35923643 in 11q24.1 is highly conserved and mainly expressed in nervous-(brain) and immune-tissues (RNA-seq from GTEx Portal), but its function has not been well characterized. GRAM domain is an intracellular protein- or lipids-binding signaling domain, which exists in several membrane-associated proteins such as glucosyltransferase, myotubularin and GRAMD1B protein (19). Recently, GRAMD1B was reported to be involved in chemo-resistance and used as potential target to treat ovarian cancer (20).
Chromatin structure and epigenetic programming play important roles in the maturation of B-cells and development of CLL (21). Our functional prediction suggests that the variant rs35923643 (in very high LD with rs735665) may constitute a functional variant that influence regulatory pathways in leukemia- or immune-cells. We found peaks of histone mark H3K4Me1 and DNaseI hypersensitivity at the position of rs35923643 in B-lymphocytes and hematopoietic stem cells, which indicate that chromatin at this position can be highly remodeled for transcriptional regulation. TF binding site screening by FIMO suggested that variation at rs35923643 could influence the binding of RUNX3.
RUNX3 is a TF with 429 amino acids that binds to the core site 5′-PYGPYGGT-3′, which presents in several enhancers and promoters. It acts as a tumor suppressor and may influence cancer development through its central role in immunity (22). The interaction network output from STRING database (23) indicates that functional partners of RUNX3 include core-binding factor β subunit, SMAD family 3/4 and ubiquitin C (Supplementary Material, Fig. S7), which are important components in networks and pathways of macromolecule metabolic processes, stem cell differentiation, cell cycle and protein phosphorylation (Supplementary Material, Fig. S8). The RUNX3 gene is a common site for somatic disruption and translocations in lymphoid malignancies, and it is usually expressed in proliferating B cells (24). Despite several indications, we realize that the mechanistic significance of our TF binding results admittedly is speculative. Validation of the involvement of RUNX3 binding affinity to the top genetic variant is needed from alternative analyses (e.g. other in silico approaches or allele specific ChIP-seq experiments).
The inverse phenotypic association between IgM anti-PC and CLL is opposite to what the identified shared genetic variant suggests (rs35923643-G is associated with increases in both IgM anti-PC and CLL risk). We note that the situation with alleles exerting opposite effects for different but related trait is a phenomenon increasingly seen in the immune-mediated diseases (25). With the limited sample size in the nested case–control study, we of course cannot rule out association because of chance. However, the low level of IgM anti-PC in cases with CLL may be due to a general disease-related hypoglobulinemia that presents some time before or after CLL manifests (26). In prevalent CLL cases, the affected B cells might have stopped producing immunoglobulin as a consequence of the CLL or treatment, i.e. reverse causation. We found the IgM anti-PC level to be lower also in incident CLL cases, even though the relationship was not statistically significant. Our nested case–control study was limited to serum samples from elderly subjects, which may accentuate the observed inverse phenotypic association. Other limitations are that we lack duplicate measurements, detailed clinical information about stage, therapy, immunoglobulin heavy chain and stereotyped B-cell status.
Because of the generally old age of onset, hypoglobulinemia and immunosuppressive treatments, 30%–50% of CLL patients die from infections. Therefore, immunoglobulin antimicrobial prophylaxis and different types of vaccines (e.g. influenza and pneumococcal vaccines) have been recommended to reduce mortality from infections in CLL patients (26). The potential predictive and preventive value of IgM anti-PC in CLL development also deserves more investigation in the future.
This is the first GWAS of IgM anti-PC, and more findings would be expected with increasing sample sizes in the future. Even so, we think that the robustness of the GWAS result combined with the overlap with genetic susceptibility to CLL indicates that the top locus in 11q24.1 likely harbors variants with joint effects on IgM anti-PC and CLL.
Materials and Methods
Phenotype and genotype in each cohort
Four Swedish cohorts (European-ancestry participants) were involved in this study: TwinGene, PIVUS (Prospective Investigation of the Vasculature in Uppsala Seniors), MDC (Malmö Diet and Cancer) and PRACSIS (Prognosis and Risk in Acute Coronary Syndromes in Sweden). These studies were approved by the local ethics committees, and all participants gave informed consent.
According to the same manufacturer’s protocol (CVDefine®, Athera Biotechnologies AB, Stockholm, Sweden), serum level of IgM anti-PC was measured by indirect non-competitive enzyme immunoassay in each cohort. IgM anti-PC was expressed as units per milliliter (U/ml), by using a six-point calibrators curve containing IgM anti-PC levels ranging from 0 to 100U/ml (27). Genomic DNA was extracted from the whole blood of participants in each cohort, DNA samples that passed sample quality control (QC) were sent to SNP genotyping. The 1000 Genome reference panel (GRCh 37/hg 19, Phase 1, version 3) was used for imputation in four cohorts, by using IMPUTE 2 in PIVUS and MDC, Mach 1.0 and Minimac in TwinGene and PRACSIS, respectively.
TwinGene is a population-based cohort of twins collected between 2004 and 2008, including 12 591 subjects born between 1911 and 1958 (28); in which 1018 complete twin pairs (2036 individuals) were randomly selected to measure IgM anti-PC and estimate the heritability (9). Genomic DNA from all available dizygotic twins and one member of each monozygotic twin pair were genotyped by using Illumina OmniExpress BeadChip. Genotyping QC exclusion criteria: genotypic or individual missingness > 0.03, minor allele frequency (MAF) < 0.01, Hardy-Weinberg equilibrium (HWE) P < 10−7, sex mismatch, heterozy-gosity (individuals with an F-statistic beyond ±5SD from the sample mean), or cryptic relatedness.
PIVUS cohort started in 2001 and includes a random sample of 1016 subjects who were 70 years old and living in the community of Uppsala (29). All participants were measured with IgM anti-PC and genotyped by Illumina OmniExpress BeadChip and Metabochip. Genotyping QC exclusion criteria: monomorphic SNPs, MAF < 0.01, genotypic missingness > 0.05 for SNPs with MAF > 0.05, or genotypic missingness > 0.01 for SNPs with MAF < 0.05, HWE P < 10−6, individual missingness > 0.01, sex mismatch, heterozygosity (beyond ±3SD from the mean), duplicated samples, identity-by-descent (IBD) match, ethnic outliers.
MDC study is a prospective cohort including about 30000 participants who live in Malmö city (30,31). IgM anti-PC was measured in 1042 individuals within a nested case–control sub-study for cardiovascular disease (32). SNPs were genotyped by Illumina OmniExpressExome BeadChip, with genotyping QC exclusions criteria as follows: genotypic or individual missingness > 0.05; IBD match; heterozygosity (absolute cryptic relatedness inbreeding coefficient > 0.20); sex mismatch; or population outliers.
PRACSIS cohort is a prospective risk stratification program, which including consecutive patients (18–79 years of age) with acute coronary syndrome but without other life threatening diseases from 1995 to 2001 (33). IgM anti-PC levels for 1185 patients were measured in blood samples collected within 24h of admission. Genotyping of 1268 patients was performed by using Illumina OmniExpressExome BeadChip. Genotyping QC exclusion criteria: sex mismatch, genotypic or individual missingness > 0.03, heterozygosity (beyond ±3SD from the mean), duplicated or related individuals (IBD > 0.185), divergent ancestry (principal component analysis with HapMap populations), MAF < 0.01 or HWE P < 10−7.
Raw values of IgM anti-PC were adjusted for age and sex in the linear regression model, after removing outliers (beyond ±4SD from the mean), residuals from the regression model were rank order normalized to achieve standard normal distribution. After phenotype and genotype matching, individuals with both IgM anti-PC measurements and SNPs available (1175 from TwinGene, 945 from PIVUS, 882 from MDC and 646 from PRACSIS) were used in our GWAS.
GWAS meta-analysis
Firstly, discovery GWAS was performed in three cohorts: TwinGene, PIVUS and MDC; by using PLINK in TwinGene, SNPTEST (version 2.5) in PIVUS and MDC. Analyses were restricted to autosomal SNPs with imputation quality (info or r 2) higher than 0.4. The first four principal components (first two for PIVUS because all individuals were of the same ethnicity and geographical area) were used as covariates in linear regression model to control population stratification. The “--within” option in PLINK was used to statistically adjust for relatedness (complete dizygotic twin pairs) in TwinGene. Manhattan and quantile–quantile plots were drawn by using qqman package in R 3.4.1, and the regional plot was generated from LocusZoom tool. Fixed-effect meta-analysis of three discovery IgM anti-PC GWAS results was performed by METAL (weighted by sample size). Genome-wide significant SNPs (P<5.0×10−8) from the discovery GWAS meta-analysis were validated in PRACSIS study (n = 646). The replicated SNPs were meta-analyzed by using the metaphor package in R 3.4.1.
PRS and effect sizes correlation analysis
PRS analysis was performed by using summary GWAS results of IgM anti-PC (from our meta-analysis of three discovery studies, total n = 3002), general IgM [public data including SNPs with P < 1 × 10−6 among ~19000 individuals from Jonsson et al. (14)] as the bases, and CLL (from the InterLymph consortium, including 3100 unrelated cases and 7677 controls) as the target (15). LD-linked SNPs in the base were clumped by using HapMap_ceu_all genotype as the reference (release 22, 60 individuals, 3.96 million SNPs), with the parameter settings recommended in PRSice (34); a clumping threshold of p1 = p2 = 0.5, an LD threshold of r 2 = 0.05 and a distance threshold of 300 kb. Independent SNPs were grouped into different quantiles with gradually increasing P thresholds (PT). The PT of the quantile that explained most variance of the target was defined as the best-fitted PT.
Independent genome-wide significant SNPs (P < 5 × 10−8 from each locus) for general IgM and CLL were identified in our discovery GWAS meta-analysis of IgM anti-PC. For each identified SNP, the effect sizes on general IgM, CLL versus IgM anti-PC were plotted in scatter plots by using GTX package in R 3.4.1.
Functional prediction
Correlated SNPs (r 2 > 0.10) within the LD block of the top finding were submitted to UCSC Genome Browser (GRCh37/hg19), to study marks of potential regulation (including transcription, histone modification, DNase hypersensitivity and TFs binding) determined by ENCODE on cell lines (35). Annotation database RegulomeDB with established and predicted regulatory elements in the intergenic regions of the Homo sapiens was used for searching evidence of TF binding (36). In order to identify the binding sites for potential TFs, 728bp of the DNA sequence surrounding the indicated functional variant was submitted to FIMO (37), a tool enabling searches of motifs derived from ChIP-seq and SELEX data (38,39).
Nested case–control study
The nested case–control study was designed to follow-up on the results of the GWAS meta-analysis indicating the top genetic variant shared between IgM anti-PC and CLL. We evaluate the phenotypic association between IgM anti-PC and CLL in the Swedish Twin Registry serum biobank, which contains serum samples of over 12 000 participants who have been linked to the Swedish National Patient Registry (40). By using international classification of diseases (ICDs) codes (ICD10: C91.1; ICD7/8/9: 204.1), all cases of CLL in the biobank were identified; 7 prevalent (onset before sampling) and 23 incident CLL cases (onset after blood sampling) were found, and three age- (birthdates were within 690 days and the same age at blood sampling) and sex-matched controls were also randomly selected for each case. For the 30 CLL cases and the 90 matched controls, serum samples were withdrawn and IgM anti-PC levels were measured according to the same manufacturer’s protocol (CVDefine® , Athera Biotechnologies AB) as used for the anti-PC GWAS. Measurements were calibrated by using a six-point calibrators curve containing IgM anti-PC levels ranging from 0 to 100U/ml (27). Raw values and rank order normalized values of IgM anti-PC were used in the t-test and stratified Cox proportional hazards models.
Web resources
PLINK, http://zzz.bwh.harvard.edu/plink/;
METAL, http://genome.sph.umich.edu/wiki/METAL;
qqman, https://cran.r-project.org/web/packages/qqman/;
LocusZoom, http://locuszoom.sph.umich.edu/locuszoom/;
1000 Genomes Project, http://www.1000genomes.org/;
InterLymph consortium, http://epi.grants.cancer.gov/InterLymph/;
Metaphor, http://www.metafor-project.org/doku.php;
PRSice: http://prsice.info/;
UCSC Genome Browser, https://genome.ucsc.edu/;
Encyclopedia of DNA Elements (ENCODE): https://www.encodeproject.org/;
Ensembl, http://www.ensembl.org/index.html;
RegulomeDB, http://regulome.stanford.edu/index;
Find Individual Motif Occurrences (FIMO), http://meme-suite.org/tools/fimo;
STRING database, http://string-db.org/;
GTX: Genetics ToolboX, https://cran.r-project.org/web/packages/gtx/.
All websites were last accessed on 22 March 2018.
Supplementary Material
Acknowledgements
The authors would like to thank the InterLymph consortium for sharing the summarized GWAS result of CLL.
Funding
Swedish Heart-Lung Foundation (20070481 to P.K.E.M.) and China Scholarship Council (201306210065 to X.C.). The Swedish Twin Registry is managed by Karolinska Institutet and receives funding through the Swedish Research Council (2017–00641).
Footnotes
Conflict of Interest statement. J.F. is named as inventor on patents of anti-PC. The remaining authors declare no other conflicts of interest.
References
- 1.Binder CJ, Chang MK, Shaw PX, Miller YI, Hartvigsen K, Dewan A, Witztum JL. Innate and acquired immunity in atherogenesis. Nat Med. 2002;8:1218–1226. doi: 10.1038/nm1102-1218. [DOI] [PubMed] [Google Scholar]
- 2.Frostegard J. Immunity, atherosclerosis and cardiovascular disease. BMC Med. 2013;11:117. doi: 10.1186/1741-7015-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder CJ, Horkko S, Dewan A, Chang MK, Kieu EP, Goodyear CS, Shaw PX, Palinski W, Witztum JL, Silverman GJ. Pneumococcal vaccination decreases atherosclerotic lesion formation: molecular mimicry between Streptococcus pneumoniae and oxidized LDL. Nat Med. 2003;9:736–743. doi: 10.1038/nm876. [DOI] [PubMed] [Google Scholar]
- 4.Frostegard J, Huang YH, Ronnelid J, Schafer-Elinder L. Platelet-activating factor and oxidized LDL induce immune activation by a common mechanism. Arterioscler Thromb Vasc Biol. 1997;17:963–968. doi: 10.1161/01.atv.17.5.963. [DOI] [PubMed] [Google Scholar]
- 5.Caligiuri G, Khallou-Laschet J, Vandaele M, Gaston AT, Delignat S, Mandet C, Kohler HV, Kaveri SV, Nicoletti A. Phosphorylcholine-targeting immunization reduces atherosclerosis. J Am Coll Cardiol. 2007;50:540–546. doi: 10.1016/j.jacc.2006.11.054. [DOI] [PubMed] [Google Scholar]
- 6.McMahon M, Skaggs B. Autoimmunity: do IgM antibodies protect against atherosclerosis in SLE? Nat Rev Rheumatol. 2016;12:442–444. doi: 10.1038/nrrheum.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverman GJ. Protective natural autoantibodies to apoptotic cells: evidence of convergent selection of recurrent innate-like clones. Ann N Y Acad Sci. 2015;1362:164–175. doi: 10.1111/nyas.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman M, Sing S, Golabkesh Z, Fiskesund R, Gustafsson T, Jogestrand T, Frostegard AG, Hafstrom I, Liu A, Frostegard J. IgM antibodies against malondialdehyde and phosphorylcholine are together strong protection markers for atherosclerosis in systemic lupus erythematosus: regulation and underlying mechanisms. Clin Immunol. 2016;166–167:27–37. doi: 10.1016/j.clim.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Rahman I, Atout R, Pedersen NL, de Faire U, Frostegard J, Ninio E, Bennet AM, Magnusson PK. Genetic and environmental regulation of inflammatory CVD bio-markers Lp-PLA2 and IgM anti-PC. Atherosclerosis. 2011;218:117–122. doi: 10.1016/j.atherosclerosis.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 10.Fabbri G, Dalla-Favera R. The molecular patho-genesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16:145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- 11.Lanemo Myhrinder A, Hellqvist E, Sidorova E, Soderberg A, Baxendale H, Dahle C, Willander K, Tobin G, Backman E, Soderberg O, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–3848. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- 12.Landgren O, Rapkin JS, Caporaso NE, Mellemkjaer L, Gridley G, Goldin LR, Engels EA. Respiratory tract infections and subsequent risk of chronic lymphocytic leukemia. Blood. 2007;109:2198–2201. doi: 10.1182/blood-2006-08-044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law PJ, Berndt SI, Speedy HE, Camp NJ, Sava GP, Skibola CF, Holroyd A, Joseph V, Sunter NJ, Nieters A, et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun. 2017;8:14175. doi: 10.1038/ncomms14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson S, Sveinbjornsson G, de Lapuente Portilla AL, Swaminathan B, Plomp R, Dekkers G, Ajore R, Ali M, Bentlage AEH, Elmer E, et al. Identification of sequence variants influencing immunoglobulin levels. Nat Genet. 2017;49:1182–1191. doi: 10.1038/ng.3897. [DOI] [PubMed] [Google Scholar]
- 15.Berndt SI, Camp NJ, Skibola CF, Vijai J, Wang Z, Gu J, Nieters A, Kelly RS, Smedby KE, Monnereau A, et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat Commun. 2016;7:10933. doi: 10.1038/ncomms10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frostegard AG, Sjoberg BG, Frostegard J, Norman M. IgM-antibodies against phosphorylcholine in mothers and normal or low birth weight term newborn infants. PLoS One. 2014;9:e106584. doi: 10.1371/journal.pone.0106584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frostegard J, Tao W, Rastam L, Lindblad U, Lindeberg S. Antibodies against phosphorylcholine among new guineans compared to swedes: an aspect of the hygiene/missing old friends hypothesis. Immunol Invest. 2017;46:59–69. doi: 10.1080/08820139.2016.1213279. [DOI] [PubMed] [Google Scholar]
- 18.Ruchlemer R, Polliack A. Geography, ethnicity and “roots” in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1142–1150. doi: 10.3109/10428194.2012.740670. [DOI] [PubMed] [Google Scholar]
- 19.Doerks T, Strauss M, Brendel M, Bork P. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem Sci. 2000;25:483–485. doi: 10.1016/s0968-0004(00)01664-9. [DOI] [PubMed] [Google Scholar]
- 20.Wu SY, Yang X, Gharpure KM, Hatakeyama H, Egli M, McGuire MH, Nagaraja AS, Miyake TM, Rupaimoole R, Pecot CV, et al. 2′-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat Commun. 2014;5:3459. doi: 10.1038/ncomms4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, Wang Q, Imbusch CD, Serva A, Koser SD, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48:253–264. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lotem J, Levanon D, Negreanu V, Bauer O, Hantisteanu S, Dicken J, Groner Y. Runx3 at the interface of immunity, inflammation and cancer. Biochim Biophys Acta. 2015;1855:131–143. doi: 10.1016/j.bbcan.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady G, Farrell PJ. RUNX3-mediated repression of RUNX1 in B cells. J Cell Physiol. 2009;221:283–287. doi: 10.1002/jcp.21880. [DOI] [PubMed] [Google Scholar]
- 25.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 26.Whitaker JA, Shanafelt TD, Poland GA, Kay NE. Room for improvement: immunizations for patients with monoclonal B-cell lymphocytosis or chronic lympho-cytic leukemia. Clin Adv Hematol Oncol. 2014;12:440–450. [PubMed] [Google Scholar]
- 27.de Faire U, Su J, Hua X, Frostegard A, Halldin M, Hellenius ML, Wikstrom M, Dahlbom I, Gronlund H, Frostegard J. Low levels of IgM antibodies to phos-phorylcholine predict cardiovascular disease in 60-year old men: effects on uptake of oxidized LDL in macrophages as a potential mechanism. J Autoimmun. 2010;34:73–79. doi: 10.1016/j.jaut.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Magnusson PK, Almqvist C, Rahman I, Ganna A, Viktorin A, Walum H, Halldner L, Lundstrom S, Ullen F, Langstrom N, et al. The Swedish Twin Registry: establishment of a biobank and other recent developments. Twin Res Hum Genet. 2013;16:317–329. doi: 10.1017/thg.2012.104. [DOI] [PubMed] [Google Scholar]
- 29.Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol. 2005;25:2368–2375. doi: 10.1161/01.ATV.0000184769.22061.da. [DOI] [PubMed] [Google Scholar]
- 30.Manjer J, Carlsson S, Elmstahl S, Gullberg B, Janzon L, Lindstrom M, Mattisson I, Berglund G. The Malmo Diet and Cancer Study: representativity, cancer incidence and mortality in participants and non-participants. Eur J Cancer Prev. 2001;10:489–499. doi: 10.1097/00008469-200112000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Hedblad B, Nilsson P, Janzon L, Berglund G. Relation between insulin resistance and carotid intima-media thickness and stenosis in non-diabetic subjects. Results from a cross-sectional study in Malmo. Sweden Diabet Med. 2000;17:299–307. doi: 10.1046/j.1464-5491.2000.00280.x. [DOI] [PubMed] [Google Scholar]
- 32.Sjoberg BG, Su J, Dahlbom I, Gronlund H, Wikstrom M, Hedblad B, Berglund G, de Faire U, Frostegard J. Low levels of IgM antibodies against phosphorylcholine—a potential risk marker for ischemic stroke in men. Atherosclerosis. 2009;203:528–532. doi: 10.1016/j.atherosclerosis.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Caidahl K, Hartford M, Karlsson T, Herlitz J, Pettersson K, de Faire U, Frostegard J. IgM-phosphorylcholine autoantibodies and outcome in acute coronary syndromes. Int J Cardiol. 2013;167:464–469. doi: 10.1016/j.ijcard.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Euesden J, Lewis CM, O’Reilly PF. PRSice: poly-genic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Ludvigsson JF, Andersson E, Ekbom A, Feychting M, Kim JL, Reuterwall C, Heurgren M, Olausson PO. External review and validation of the Swedish national inpatient register. BMC Public Health. 2011;11:450. doi: 10.1186/1471-2458-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.