Abstract
Polycystic Ovary Syndrome (PCOS) is the most common endocrine disorder amongst women of reproductive age, whose aetiology remains unclear. To improve our understanding of the molecular mechanisms underlying the disease, we conducted a genome-wide DNA methylation profiling in granulosa lutein cells collected from 16 women suffering from PCOS, in comparison to 16 healthy controls. Samples were collected by follicular aspiration during routine egg collection for IVF treatment. Study groups were matched for age and BMI, did not suffer from other disease and were not taking confounding medication.
Comparing women with polycystic versus normal ovarian morphology, after correcting for multiple comparisons, we identified 106 differentially methylated CpG sites with p-values < 5.8 ×10–8 that were associated with 88 genes, several of which are known to relate either to PCOS or to ovarian function. Replication and validation of the experiment was done using pyrosequencing to analyse six of the identified differentially methylated sites. Pathway analysis indicated potential disruption in canonical pathways and gene networks that are, amongst other, associated with cancer, cardiogenesis, Hedgehog signalling and immune response. In conclusion, these novel findings indicate that women with PCOS display epigenetic changes in ovarian granulosa cells that may be associated with the heterogeneity of the disorder.
Keywords: PCOS, EWAS, DNA methylation, Metabolic syndrome, Reproduction
1. Introduction
Polycystic Ovary Syndrome (PCOS) is a common, heterogeneous endocrine disorder with an estimated prevalence up to 15% amongst women of reproductive age, depending on the diagnostic criteria used (Azziz et al., 2004; Franks, 1995; March et al., 2010; Sirmans and Pate, 2013). It comprises metabolic and reproductive disturbances, whereas environmental influences, such as diet, are known to affect the phenotype. Its main biochemical characteristic is the hypersecretion of androgens, predominantly of ovarian origin that is associated with, and may be causally related to, infertility (anovulation, menstrual irregularities), hirsutism (hair excess), psychological distress (anxiety, depression) and metabolic defects (obesity, insulin resistance). Women with PCOS have an increased risk of developing type 2 diabetes and cardiovascular disease later in life (Diamanti-Kandarakis and Dunaif, 2012; Jayasena and Franks, 2014; Mani et al., 2013; Qu et al., 2012).
The aetiology of PCOS remains unclear. There is, however, evidence for genetic predisposition with familial clustering of cases as well as genetic variants of endocrine and metabolic markers (Barber and Franks, 2013; Chen et al., 2011; Day et al., 2015; Franks et al., 1997, 2008; Hayes et al., 2015; Shi et al., 2012; Vink et al., 2006). It has been proposed that PCOS originates in early (possibly fetal) life due to “programming” by exposure to excessive androgen production. (Abbott et al., 2002, 2005; Franks and Berga, 2012; Li and Huang, 2008; Xita and Tsatsoulis, 2006).
Results of protein and expression profiling experiments in adults support the view that there is an important contribution of androgen-dependent genes to the aetiology of PCOS (Adams et al., 2016; Coskun et al., 2013; Insenser and Escobar-Morreale, 2011; Li et al., 2016; Lv et al., 2017). There are however very few studies regarding the epigenetic changes associated with PCOS development, with only a handful of genome-wide studies that were conducted mainly on whole blood, or ovarian tissue (Table S1).
The rationale of our study is based on the hypothesis that PCOS can be explained by an integrated epigenetic model, whereby environmental factors modify the effect of susceptibility genes and therefore, influence the clinical and biochemical heterogeneity that is characteristic of the syndrome during adult life. In this investigation, we report results from a PCOS case-control, genome-wide methylation study using DNA from granulosa lutein cells (GLCs) that are known to be androgen responsive, with the aim to advance our understanding of the molecular mechanisms underlying the disease, to provide a novel insight into the role of epigenetic programming in PCOS and ultimately to improve its diagnosis and treatment.
2. Materials and Methods
2.1. Patient recruitment and sample collection
GLC samples from consecutive subjects with or without PCOS, were collected from the Hammersmith IVF clinic (Wolfson Fertility Centre, Hammersmith Hospital) by follicular aspiration of mature oocytes (> 14 mm diameter on the day of HCG administration), during routine egg collection.
All participants provided informed consent. PCOS was diagnosed according to the Rotterdam Consensus criteria (Rotterdam, 2004). Sample collection was approved by the National Research Ethics Service (NRES) (Hammersmith & Queen Charlotte's Chelsea: 08/H0707/152).
The array-based analysis cohort comprised 32 samples, 16 PCOS women with oligomenorrhea and 16 healthy women (controls), who received IVF treatment due to male infertility factors. All participants were of similar age and BMI, did not suffer either currently, or in the past from other disease and were not taking Metformin medication for insulin resistance (Table 1).
Table 1.
Clinical characteristics of PCOS patients and controls used for the array-based and the pyrosequencing analysis. The pyrosequencing analysis includes all samples used for the genome-wide study, extra samples for each group and an extra group comprising women suffering from PCOS, but with regular cycles. Group comparisons were performed using a two-tailed, unpaired t-test. Values are mean, ± standard error of means (SEM). N: number of patients, GnRHA: Gonadotropin-releasing hormone agonist, hCG: Human chorionic gonadotrophin, BMI: Body Mass Index, LH: Luteinizing Hormone, FSH: Follicle-Stimulating Hormone, Hep B, C: Hepatitis B, C, HIV: Human Immunodeficiency Virus. Number of * indicate level of significance.
| EPIC array cohort | Pyrosequencing cohort | ||||||
|---|---|---|---|---|---|---|---|
| PCOS | Controls | P-value | PCOS | Controls | PCOSreg | P-value | |
| Menstrual cycle | Irregular | Regular | Irregular | Regular | |||
| Ovarian morphology | PCO | Normal | PCO | Normal | PCO | ||
| Mean age [y) | 30.19 ± 0.8, N=16 | 34.75 ± 0.9, N=16 | 0.0009*** | 30.2 ± 0.6, N=26 | 35.1 ± 0.6, N=43 | 32.9 ± 1.1, N=10 | 0.0001**** |
| Mean BMI (kg/m2) | 24.89 ± 0.8, N=16 | 24.38 ± 0.5, N=16 | 0.593 | 24.5 ± 0.6, N=26 | 24.4 ± 0.4, N=43 | 23.6 ± 1.1, N=10 | 0.714 |
| Protocol | Antagonist | Long agonist. Antagonist | Antagonist | Long agonist, Antagonist | Antagonist | ||
| Gonadotropin used | FSH (Gonal-f) | FSH (Gonal-f) | FSH (Gonal-f) | FSH (Gonal-f) | FSH (Gonal-f) | ||
| Maturation trigger | GnRHA, Kisspeptin | hCG | GnRHA, Kisspeptin | hCG | GnRHA, Kisspeptin | ||
| No. of antral follicles | 36 ± 3.4, N=16 | 14 ± 1.4, N=16 | <0.0001**** | 38.6 ± 2.9, N=26 | 12.2 ± 0.9, N=43 | 28.4 ± 2.1, N=10 | 0.0001**** |
| Baseline FSH (mlU/mL) | 5.2 ± 0.8, N=15 | 6.3 ± 0.6, N=12 | 0.2645 | 5.3 ± 0.5, N=24 | 6.5 ± 0.5, N=30 | 4.4 ± 0.7, N=9 | 0.0555 |
| Baseline LH (mlU/mL) | 5.3 ± 0.9, N=15 | 4.3 ± 0.4, N=10 | 0.4101 | 6.8 ± 0.9, N=23 | 5 ± 0.43 N=28 | 5.1 ± 0.9, N=9 | 0.1114 |
| Days of stimulation | 11.4 ± 0.8, N=16 | 11.1 ± 0.5, N=16 | 0.7815 | 11.1 ± 0.5, N=26 | 11.1 ± 0.3, N=43 | 11.3 ± 1.3, N=10 | 0.9674 |
| Cumulative FSH dose (iu) | 1578 ± 198, N=16 | 2986 ± 296, N=16 | 0.0004*** | 1466.3 ± 132, N=26 | 3087.2 ± 183, N=43 | 1551.5 ± 121.4, N=10 | 0.0001**** |
| No. of follicles >14mm | 15 ± 1.2, N=16 | 8 ± 0.7, N=16 | <0.0001**** | 15 ± 1.5, N=26 | 7.6 ± 0.6, N=43 | 9.8 ± 1.9, N=10 | <0.0001**** |
| No. of eggs collected | 20 ± 3.3, N=16 | 12 ± 1.3, N=16 | 0.0263* | 18.8 ± 2.3, N=26 | 10 ± 0.8, N=43 | 15.2 ± 2.8, N=10 | 0.0002*** |
| Smokers | 0 | 2 | 0 | 2 | 0 | ||
| Alcohol drinking | 1 | 4 | 2 | 6 | 0 | ||
| Hep B,C, HIV+ve | 1 | 0 | 1 | 0 | 0 | ||
| Family history | No | No | No | No | No | ||
| Medical history | No | Migraines (1), asthma (1), epilepsy (1), hypothyroidism (1), Raynaud’s syndrome (1) | No | Migraines (2), asthma (1), epilepsy (1), hypothyroidism (1), Raynaud’s syndrome (1) | No | ||
The validation cohort using pyrosequencing consisted of two PCOS subgroups, with (n = 27) and without (n=15) oligomenorrhea and healthy controls undergoing IVF treatment due to male infertility factor and/or physical blockage (n = 43, 2 of them apart from male infertility, also had salpingectomies). Samples were overlapping with the samples used for the genome-wide analysis. All extra samples were from subjects who had same clinical characteristics as those included in the array-based study (Table 1).
2.2. Genomic DNA isolation and quantitation
GLCs were isolated by density gradient centrifugation using Percoll (GE Healthcare Life Sciences, UK). Genomic DNA was extracted using the QIAGEN DNeasy Blood and tissue extraction kit according to the manufacturer’s instructions (QIAGEN Ltd).
Following extraction, DNA was quantified by measuring absorbance on a NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific) and fluorescence intensity using the Quant-iT PicoGreen dsDNA Assay (Life Technologies Limited) on a PHERAstar FS multi-mode reader (BMG LABTECH Ltd). Both quantitation methods were considered.
2.3. DNA methylation microarray
Genome-wide DNA methylation profiling was generated using the Infinium MethylationEPIC BeadChip array (Illumina, San Diego, CA, USA). 500 ng of DNA were bisulfite converted using the EZ-96 DNA Methylation™ Kit (Zymo Research Corporation, Irvine, CA), fragmented and hybridised on the BeadChip. Signal intensities were extracted using the Illumina iScan Reader (Illumina, San Diego, CA, USA).
Hybridization, scanning and raw data processing were performed by the Oxford Genomics Centre facilities (Wellcome Trust Centre for Human Genetics, Oxford), according to Illumina’s protocol (https://support.illumina.com).
2.4. Quality control and data pre-processing
Raw data files and genomic annotation from the EPIC beadchip array were provided by the Oxford Genomics Centre service. Raw intensities (.idat) were retrieved and pre-processed using the Bioconductor package minfi (version 1.18.6) in R (Fortin et al., 2017).
All samples passed the initial quality assessment, with an intensity detection p-value < 10−16 and a sample call rate > 98%. Intensity values were quantile normalised using limma (Smyth et al., 2005) and converted to β-values, reported as a score ranging from 0 (non-methylated) to 1 (completely methylated).
Global correlation patterns and sample relationships for the detection of biological clustering and outliers were assessed by Principal Component Analysis (PCA). We included all samples in the study and all markers that had no missing data (n = 850,514, prcomp function in R with default settings). PCA variance was 9.25% and 7.55% for PC1 and PC2 respectively. Linear models regressing the case-control status against the first two principal components indicated a separation between the two groups by PC1 (p = 0.04) and PC2 (p = 0.004).
PCA clustering indicated the presence of two potential outliers (1 PCOS; red colour and 1 control; black colour, Supplementary Fig. S1). However, comparing data of linear regression analysis with and without the two samples, we identified 62 overlapping hits within the cut-off threshold of p < 5.8 ×10−8, whereas 44 hits (out of 106) had p-values very close to the ones identified including all samples, ranging from 5.0×10−08 to 2.9× 10−07. The remaining 10 hits, identified when we excluded the two outliers, were not associated with any genes, and mainly had intergenic locations. Since neither sample deviated from the clinical phenotype that characterises the two study groups to which they belonged, they were considered not as biological, but rather as technical outliers, and were included in the analysis (Supplementary Table S2).
2.5. Statistical analysis
Downstream statistical analysis of the data was performed using the CPACOR (incorporating Control Probe Adjustment and reduction of global CORrelation) methylation analysis pipeline (Lehne et al., 2015) in R (http://cran.r-project.org, version 3.3). The analysis was corrected for multiple comparisons (Bonferroni correction for multiple testing; 0.05/853,307 = 5.8×10−8).
To improve data quality and adjust for technical variations introduced by the use of two probe types (I and II) with distinct differences, data were normalised as raw data (beta values) without preprocessing, and using Illumina Genome Studio (reverse engineered and implemented in minfi), Subset-quantile within array (SWAN), Quantile, Functional (FunNorm) and Noob normalisation methods (Fortin et al., 2017) (Supplementary Table S2). The highest correlations between paired methylation measurements were observed after quantile normalisation. Therefore, we based our analysis on a general linear regression model with quantile normalised data.
The distribution of p-values under the null hypothesis was determined by randomly re-assigning a case-control status to all 32 samples of the study and performing a linear regression analysis for each marker, using normalised data, with and without adjusting for the two control probes. 1000 permutations were performed each time, to obtain 2 x 1000 p-value sets under the assumption of no association.
For comparison of our data with the Xu et al. study (the only previously published study using granulosa-lutein cells), we considered a “hit” in the Xu et al. study as replicated if there was directional consistency between the effect sizes, with p < 0.05. We then compared how many hits were replicated versus the total number of published hits, in each category, using a one-sided binom. test () with unadjusted cut-off p = 0.05.
2.6. Pyrosequencing
Validation of the selected gene-associated CpG sites was performed using the Q96 MD Pyrosequencing platform (QIAGEN Ltd). Primers for the pyrosequencing assays were designed using the Pyromark Assay Design Software. The PCR and sequencing primers, PCR amplification conditions, and sequence that was analysed, are listed in Supplementary Table S3. Any CpGs overlapping common SNPs were excluded from the analysis as they can represent a source of discrepancy.
All assay runs included standard DNA controls with 0%, 25%, 50%, 75% and 100% methylation status. Standards were prepared by diluting 100% methylated DNA (CpGenome Universal Methylated DNA, Millipore, UK) to 0% Whole Genome Amplified genomic DNA (Illustra GenomiPhi V2 DNA Amplification Kit, GE Healthcare Life Sciences). Whole genome amplified product was purified using the MinElute PCR Purification Kit (QIAGEN Ltd). 500 ng of the mixed standards were bisulfite converted using both the EZ methylation and EZ-Lightning methylation kits (Zymo Research Corporation, Irvine, CA), according to the manufacturer’s instructions. Each assay analysed two to six CpG sites. All the CpG sites within the assay, indicated the same directional effect on methylation levels.
Bisulfite treated DNA (1 μl) was amplified in 25 μl of PCR reaction mixture, containing 0.8 μM of primers and 1U of Taq (FastStart DNA Polymerase, Roche). DNA was amplified in a heated-lid thermocycler as follows; 95 °C for 5mins (x 1), 95 °C for 30sec, annealing for 30 s at temperature corresponding to each set of primers, 72 °C for 30sec (x 36), 72 °C for 5mins (x 1). Single PCR products corresponding to the expected product size were confirmed by 2% agarose gel electrophor-esis.
Sample preparation and pyrosequencing reactions were performed according to the manufacturer’s instructions. Methylation values were quantified as percentage of methylated cytosine over the sum of methylated and unmethylated cytosines applying formula (C/C + T) × 100, as determined by the Pyro Q-CpG™ software (QIAGEN).
Sample normality was checked using D’Agostino & Pearson omnibus K2 test; results with p < 0.05 were considered statistically significant and determined using unpaired, parametric t-test and one-way ANOVA analysis for samples with normal distribution and non-parametric Kruskal Wallis for skewed sample distributions. One-way ANOVA results were corrected for multiple testing using Bartlett’s test, assuming a Gaussian distribution with similar standard deviations between populations (run automatically by Prism). Graph analysis is presented as mean with 95% CI (confidence interval).
2.7. Data annotation and bioinformatics analysis
Annotations for the Infinium MethylationEPIC BeadChip were provided by Illumina (https://support.illumina.com/array/array_kits/infinium-methylationepic-beadchip-kit/downloads) and were based on GRCh37/hg19 build (Feb. 2009). Repeat Masker (Institute for Systems Biology, repeatmasker.org) was used to exclude repeat elements within primer sequences. Gene predictions were based on data from GENCODE v24 (currently hosted by the Ensembl Genome Browser), as well as RefSeq, Genbank, CCDS and Uniprot (Known Genes dataset).
Information on the 106 differentially methylated CpG sites and their associated genes was retrieved through mining the following databases; NCBI (www.ncbi.nlm.nih.gov), UCSC Genome Browser (genome.ucsc. edu, both GRCh37/hg19 and GRCh38/hg38 builds), meQTL database (http://www.mqtldb.org/) GeneCards suite (http://www.genecards.org/), EnSEMBL (www.ensembl.org), EMBL-EBI (www.ebi.ac.uk), The ReproGenomics viewer (rgv.genouest.org), Ovarian Kaleidoscope (okdb.appliedbioinfo.net), The Human Protein Atlas (www.proteinatlas.org) and the Polycystic Ovary Syndrome database (pcosdb.net). Gene predictions were based on data from GENCODE v24 (currently hosted by the Ensembl Genome Browser), as well as RefSeq, Genbank, CCDS and Uniprot (Known Genes dataset).
2.8. Pathway and gene ontology analyses
Pathway and gene ontology analysis were carried out using the Ingenuity Pathway Analysis (IPA) software (https://www.QIAGENbioinformatics.com/products/ingenuity-pathway-analysis, (Kramer et al., 2014)). A right-tailed Fisher’s exact test was used to estimate statistical significance (p < 0.05) for the network analysis.
3. Results
3.1. EWAS approach and classification of differentially methylated CpG sites
We performed a genome-wide, case-control epigenome profiling (EWAS), using the Illumina Infinium MethylationEPIC BeadChip that covers > 850,000 methylation sites (Moran et al., 2016) and DNA from GLCs, a known ovarian cell target for androgen action. Statistical analysis of the EPIC array was performed using the CPACOR (incorporating Control Probe Adjustment and reduction of global CORrelation) pipeline (Lehne et al., 2015), with several covariates.
Our EWAS sample population comprised a total of 32 participants (their clinical characteristics are presented in Table S2). Age, smoking and body mass index (BMI) have been known to affect global DNA methylation (Ashapkin et al., 2017; Bell, 2017; Sundar et al., 2017). However, our study population comprised women of reproductive age, who were predominantly non-smokers, and whose age and BMI were adjusted within a narrow range, with around 4 years mean age difference between patients and controls, and very similar normal BMI, in agreement with the eligibility criteria for receiving In Vitro Fertilisation (IVF) treatment (National Institute for Health and Care Excellence guidelines (NICE) https://www.nice.org.uk, last update-September 2017).
By using the presence or absence of PCOS ovarian morphology to compare datasets, we identified 106 differentially methylated CpG sites between PCOS and controls (p < 5.8×10−8). Correction of the analysis for one (CP1), or both (CP1 + CP2) internal control probes identified 76 and 11 CpGs, respectively, that were largely overlapping with the 106 CpGs originally identified (Supplementary Table S2). Since our study was based on a relatively small sample size (16 cases vs. 16 controls), we decided to consider all potential differentially methylated CpG sites.
Using the follicular maturation trigger as an independent predictor of outcome, we identified 6 CpG sites with altered methylation. The type of trigger typically differs between women with and without PCOS and so is a potential confounding variable, influenced by the PCOS status. Women with PCOS typically require lower doses of FSH for ovarian stimulation; thus, the cumulative dose of follicle-stimulating hormone (FSH) used during IVF treatment is another possible confounder (our analysis identified two CpG sites that could be FSH dose-dependent and therefore merit further investigation). Although the possible impact of age and BMI was minimised by the selection criteria for IVF treatment, we also adjusted for these, as well as for the total number of eggs collected, using all three covariates as negative controls. In all cases we found no effect on methylation status.
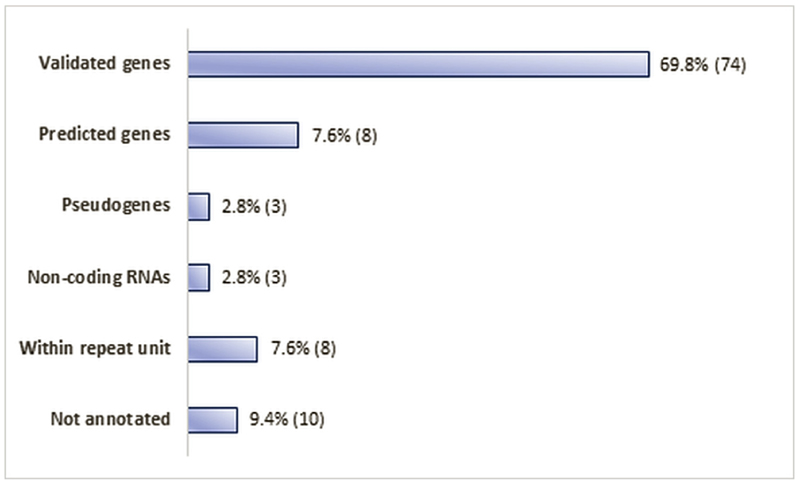
Overall, amongst the 106 identified hits, the percentage of hyperand hypo-methylated sites was similar, with 52% of them appearing to be hypermethylated and 48% hypomethylated (Supplementary Table S2). Apart from 18 CpGs that had intergenic locations (within either repetitive elements, or regions with no annotated genes), the remaining 88 were localised in the promoter regions and the body of validated and predicted genes, non-coding RNAs, and some pseudogenes (Figs. 1 and 2).
Fig. 1.
Localisation of the 106 identified CpGs within the genome and their association (numbers and percentages) with types of genes (validated, predicted, pseudogenes, non-coding RNAs), repeat units and random non-annotated genomic locations.
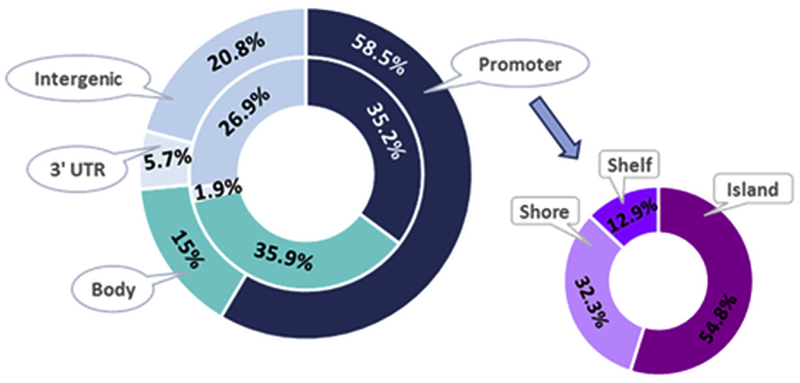
Fig. 2.
Distribution within the genome of the 106 CpG sites identified in our study (outside ring) and the published EPIC design coverage (Moran et al., 2016) (inside ring). Genomic locations are categorised as gene related (promoter, body, 3′UTR) and intergenic (no annotated genes in close vicinity). Promoter related sites are further sub-categorised depending on the CpG site location (island, shore, self).
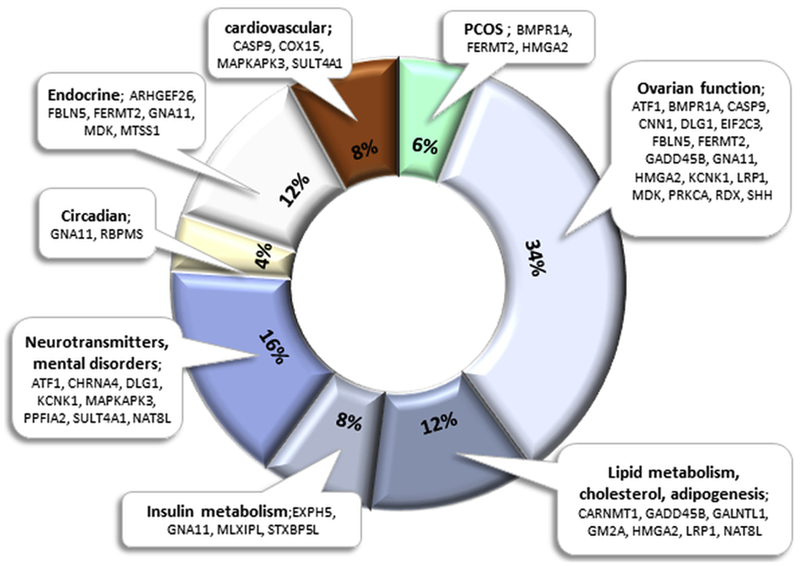
Database search indicated that these 88 genes were predominantly involved in gene regulation, endocrine and metabolic functions, immune response, cell signalling, cell death and survival and structural integrity (Supplementary Table S2). Of those, 36 genes are part of pathways relevant to PCOS phenotype, such as metabolic, cardiovascular, circadian, neurological and endocrine systems, or have an ovary related function (Fig. 3, Supplementary Table S2). Although there is a certain degree of complexity regarding the effect of methylation status on gene expression (Khalaf et al., 2013; Louwers et al., 2013; Yang et al., 2014), the identification of differential methylation in genes like BMPR1A, FERMT2 and HMGA2 that have been directly associated with the PCOS phenotype is very encouraging.
Fig. 3.
Graphical representation of the 36 gene-associated CpG sites identified in our analysis that are directly, or indirectly related to PCOS. Some genes are present in more than one category; hence percentage calculations are based on a total of 50 entries.
Of the106 identified CpGs, results from 6 were validated using pyrosequencing, with the addition of extra cases and control samples with similar clinical characteristics as those included in the microarray experiment (see Materials and Methods, Tables 1 and 2). These targeted selections were based on the degree of statistical significance (all target p-values were ≤1.66e−08), genomic location, relationship with ovarian functions and PCOS involvement.
Table 2.
Statistical analysis results and associated gene details on 10 differentially methylated CpG sites chosen for pyrosequencing validation. $ denotes closest gene to the CpG of interest, *N_ and S_ denote the upstream and downstream end of the island region. P-value corresponds to linear regression analysis of the EPIC array.
| Illumina ID | $Gene | chr | p-value | Comments | Disease | Ovary | PCOS | *CpG location |
|---|---|---|---|---|---|---|---|---|
| cg11683966 | FLJ40434 | 1p.32 | 2.09e−14 | Pseudogene | S_Shore | |||
| cg00112465 | CCDC48 | 3q.21 | 7.04e−13 | Predicted gene | Island | |||
| cg18364576 | FERMT2 | 14q.22 | 3.44e−10 | Scaffold protein related to PCOS androgen receptors | Hyper-androgenemia | Yes | Yes | N_Shore |
| cg10821050 | SHH | 7q36 | 1.40e−09 | Follicle and early embryo development, signal mediator btw granulosa and theca cells | Cancer, developmental disorders | Yes | N_Shore | |
| cg23044884 | RBPMS | 8p.12 | 1.39e−08 | Transcriptional regulator | S_Shelf | |||
| cg12976821 | BMPR1A | 10q22 | 1.66e−08 | TGF-b pathway downregulated by testosterone | Polyposis, Thyroid | Yes | Yes | 5′UTR |
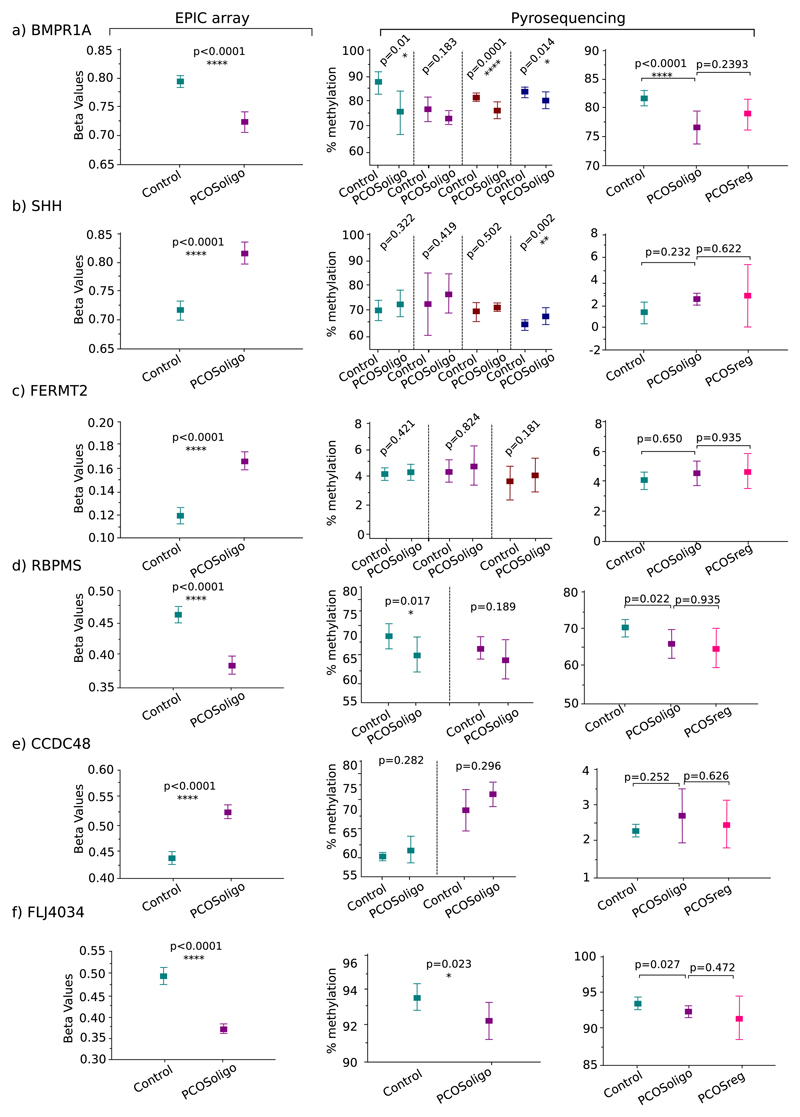
Pyrosequencing was also used to assess the methylation of a PCOS sub-group, comprising patients with regular cycles and compare the pattern with PCOS patients with oligomenorrhea, as well as healthy controls. There were no significant differences between the two PCOS groups, indicating that the two sub-phenotypes appear, at least for the examined CpGs, epigenetically similar (Fig. 4).
Fig. 4.
Box plot graphs of differential methylation levels of 6 differentially methylated CpG sites between two sub-groups of PCOS patients and healthy controls. Left hand graphs: microarray analysis adjusting for PCOS ovarian morphology; Y-axis: beta values. Middle graphs: pyrosequencing analysis using DNA from PCOS patients with oligomenorrhea; Y-axis: % methylation levels. Right hand graphs: pyrosequencing analysis using DNA from patients with regular cycles; Y-axis: % methylation levels. Expts 1, 2, 3, 4; number of pyrosequencing assays repeated for the same CpG methylation target.
3.2. Pathway and network analysis
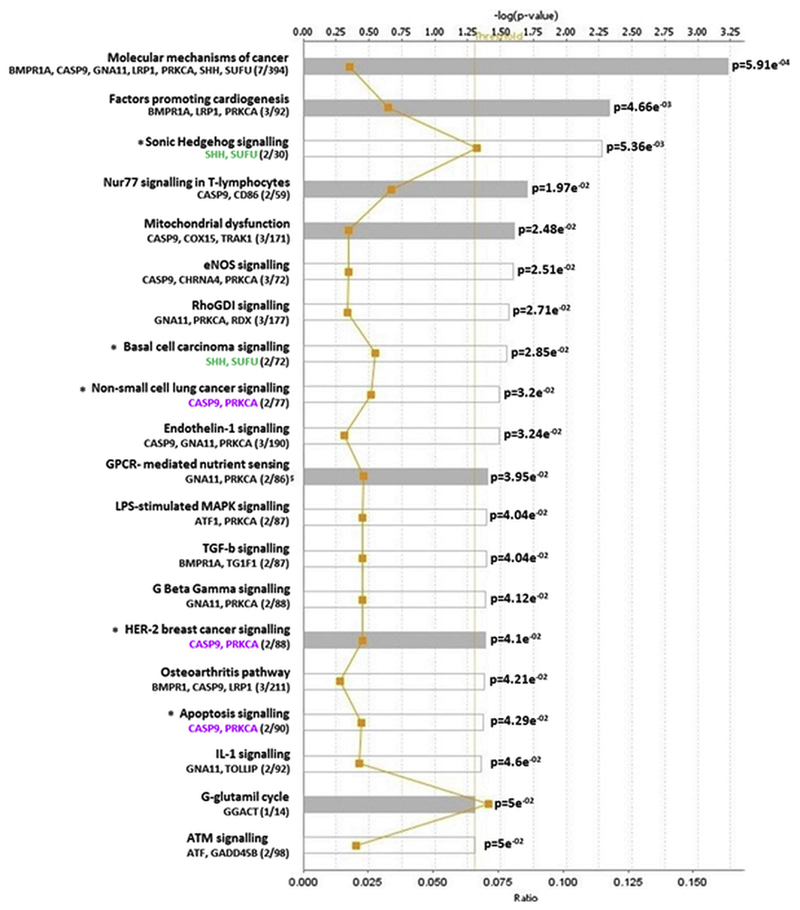
To assess the biological relevance and potential functional interactions of our findings, we carried out a core pathway analysis, using the Ingenuity Pathway Analysis software (IPA). A comparative review indicated 20 potentially disrupted canonical pathways with p-values ranging from 5.9 × 10−4 to 5.0 × 10−2. The most statistically significant pathways that were identified included molecular mechanisms of cancer, cardiogenesis, Sonic Hedgehog signalling, immune response and mitochondrial dysfunction, with some genes being present in more than one pathway (Fig. 5).
Fig. 5.
Pathway analysis of the 88 gene-associated differentially methylated CpGs. Y-axis; significant canonical pathways for the dataset. X-axis; -log (p value), calculated by Fisher’s right-tailed exact test. The ratio (orange line) is calculated as number of genes in a given pathway that meet cut-off criteria, divided by the total number of genes that make up that pathway. Threshold indicates the fraction of false positives among significant functions. Pathways that have a –log (p value) greater than the threshold of 1.3 (range 0–3.23) are displayed to the right-hand side of the graph. White bars; z-score at, or close to 0. Grey bars; pathways with no current prediction.
4. Discussion
PCOS is a polygenic disorder with a complex mode of inheritance. To date, the PCOS phenotype has been associated with 241 genes ((Joseph et al., 2016), http://pcoskb.bicnirrh.res.in) and 16 PCOS susceptibility loci, encompassing genes involved in neuroendocrine, metabolic and reproductive functions (Brower et al., 2015; Chen et al., 2011; Day et al., 2015, 2018; Hayes et al., 2015; Li et al., 2012; Louwers et al., 2013; Shi et al., 2012). Nevertheless, the complexity and heterogeneity by which the disease appears amongst the female population remains elusive.
To our knowledge, this PCOS epigenome wide association (EWAS) study is the first documented that uses GLCs, an ovary specific cell type, in combination with Illumina’s EPIC array that screens > 850,000 methylation sites, doubling the number from their previous version of 450 K. A similar study from Xu et al. (2016), also utilised GLCs for their analysis but on the 450 K array platform and without adjusting for multiple comparisons. In contrast, our study is based on data that have been thoroughly interrogated, corrected for genome-wide multiple comparisons and adjusted through the application of several normalisation methods and linear prediction models. Nevertheless, since both studies were performed using GLCs and overlapping Illumina arrays, we were able to directly compare the Xu et al. findings with our data (Supplementary Tables S3 and S4), identifying several CpGs in our study that were replicated in all three Xu et al. group analyses, as follows; 1) Controls versus PCOS obesity; 108/1472 replicated (p-value = 6.6 × 10−5), 2) Controls versus PCOS non-obesity; 155/2471 replicated (p-value = 0.003), 3) PCOS non-obesity vs. PCOS obesity; 45/1089 replicated (p-value = 0.9).
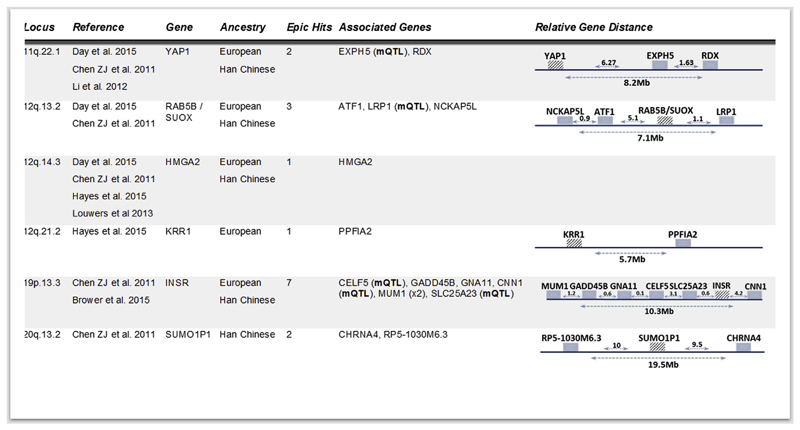
Overall, we identified 106 CpGs with differential methylation between PCOS patients and healthy controls. Of these, 88 were associated with genes, several of which are implicated in endocrine, metabolic and reproductive processes found to be associated with PCOS. In addition, 16 of the identified CpGs were mapped within 6 known PCOS susceptibility loci and of those, 5 were overlapping with known methylation quantitative trait loci (meQTLs) (Fig. 6, Supplementary Table S2) that may affect gene expression levels in a cis- or transacting fashion, strengthening the notion of a plausible environmental contribution to the genetic basis of PCOS (Schalkwyk et al., 2010; Shoemaker et al., 2010).
Fig. 6.
Mapping of 16 gene-associated CpGs within published PCOS susceptibility loci. Table lists information regarding the susceptibility loci (chromosomal region, publication, published gene/s and ancestry used for the GWAS study) and the number of CpGs from the analysis located within these regions, the name of the associated to the CpGs genes and a graphical representation of their localisation within the genome.
We validated our results using pyrosequencing targeted analysis of 6 specific CpG sites from the same patient-control cohort used in the genome wide approach, with the addition of extra samples from both patients and controls (see Methods and Materials, Table 1). Overall, the results from both experimental platforms were in agreement, having the same direction of effect, but with lower significance levels for the pyrosequencing experiment (Fig. 4). A possible explanation would be the use of two different experimental platforms and multiple batches of bisulfite treated DNA for each sample, both of which can add layers of technical bias and introduce a degree of variability in the replication of results.
In addition, using pyrosequencing, the CpG site associated with FLJ4034, the selected target with the most significant p-value, failed to replicate. FLJ4034 is a potential pseudogene and determination of its methylation status can be ambiguous due to the high methylation levels that pseudogenes usually exhibit in order to be silenced. Indeed, our analysis indicated > 95% methylation for FLJ4034 in both cases and controls.
A prominent feature of PCOS is its clinical heterogeneity that can easily result in a complex or inaccurate diagnosis. The main reason for such heterogeneity is that PCOS development and progression are controlled by several mechanisms and environmental factors, involving genes expressed at various degrees, in a multitude of ovarian cell types. Significantly, we found the methylation profile of PCOS patients with and without menstrual irregularities to be similar, suggesting a shared epigenetic and potentially genetic architecture between distinct sub-phenotypes (Fig. 4). This mirrors findings in a recent meta-analysis of GWAS studies in which there was lack of genetic heterogeneity between women with PCOS irrespective of the diagnostic criteria that were applied (Day et al., 2018).
Pathway analysis identified CpG-associated genes from our panel, as part of potential molecular synergies that could shed some light to the disease heterogeneity (Fig. 5, Table 3).
Table 3.
List of the top five Identified signalling pathways with CpG-associated genes from our EWAS analysis.
| IP A Pathway | Genes from our dataset | PCOS associated pathology or gene | References | |
|---|---|---|---|---|
| # 1 | Molecular mechanisms of cancer | BMPR1A, CASP9, GNA11, LRP1, PRKCA, SHH and SUFU | A well-recognised PCOS associated pathology is endometrial cancer | Shen et al. (2013) |
| #2 | Cardiogenesis | BMPR1A, LRP1 and PRKCA | BMPR1A is PCOS related and LRP1 is involved in lipid metabolism. Also, PCOS is frequently accompanied by an increased risk for cardiovascular disease, because of molecular interactions between obesity, testosterone and dyslipidemia Couto Alves et al., 2017 | |
| #3 | Sonic Hedgehog (SH) signalling | SHH and SUFU | SH is involved in ovarian follicular growth, regulating the steroidogenic capacity of endocrine cells like GLCs and signalling oocyte maturation, especially towards the final stages of antral follicle development. One of the key features of PCOS is the production of multiple prematurely arrested follicles leading to reduced fertility. Therefore, it is plausible that SH may be implicated in the development of anovulation in PCOS | Wijgerde et al., 2005; Spicer et al., 2009 Wang et al., 2014 |
| #4 | Nur77, T-lymphocytes signalling | CASP9 and CD86 | Nur77 is related to autoimmune response prevention. PCOS development and progression have been associated with immune response. It has been hypothesized that functional autoantibodies lead to a higher prevalence of autoimmune thyroiditis amongst PCOS patients. Also, several studies have indicated a link between increased androgen levels in PCOS patients, and inflammatory conditions, both systemic and GLC localised | Gleicher et al., 2007 Adams et al., 2016 Xu et al., 2016 |
| #5 | Mitochondrial dysfunction | CASP9, COX15 and TRAK1 | Mitochondrial biogenesis is particularly important during oocyte growth and its impairment has been associated with poor oocyte quality, diminished ovarian reserves and insulin resistance. Although patients selected for the study did not suffer from insulin resistance or hyperinsulinemia, these findings suggest a potential predisposition to insulin signalling disruption that may prove functionally significant given that methylation changes can precede clinical manifestation of the disease by several years | Wang et al., 2014 Wang and Moley, 2010 Brower et al., 2015 Ding et al., 2017 Moran et al., 2016 |
The top five identified signalling pathways include cardiogenesis, Sonic Hedgehog, Nur77, mitochondrial dysfunction and cancer, with pathologies like endometrial cancer (shown to be more common in women with PCOS), cardiovascular disease, oocyte growth and maturation, and immune response, all of which have been associated with the development of PCOS. It is not surprising that “Molecular mechanisms to cancer” was identified as the most important pathway, with the involvement of seven genes from our dataset. It is well known that epigenetic alterations can affect gene expression and create genetic instability, disrupting signalling cascades and predisposing not only to malignant phenotypes, but also to a wide spectrum of pathologies. Of significance is also the fact that a potential involvement of immune response to the PCOS development is - further to pathway analysis - supported by data mining of our dataset, where we identified a total of 16 CpG-associated genes involved in immune and inflammatory response (Supplementary Table S2). Given that the GLC purification procedure has removed any carry-over cells, including lymphocytes that could bias the results, our findings suggest that immune response may play an important role in the diversification of PCOS.
5. Conclusion
Over the past decade our view has shifted from the dogma of “genetic determinism” to a more flexible and interactive scenario of constant and dynamic relationship of our genome with the environment. It is therefore becoming increasingly evident that epigenetic modification is a crucial component of our genetic make-up and plays a critical role in key regulatory processes, genome stability and organismal adaptation to environmental exposure (Bjornsson, 2015).
These 106 differentially methylated CpGs present the opportunity, through further investigation, to be developed into diagnostic, as well as prognostic landmarks and risk indicators, since epigenetic changes can precede disease manifestation. Most importantly, epigenetic changes are environmentally dependent; hence, altering environmental exposure can reverse the effect, making them candidates for therapeutic agents (Cole et al., 2017; Kanwal et al., 2015; Nicoll et al., 2001; Teruel and Sawalha, 2017).
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mce.2019.110611.
Acknowledgements
We would like to thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for the generation of Methylation data.
Funding
E.M is funded by the Genesis Research Trust, as part of her Daphne Jackson Fellowship. C.M.L is supported by the Li Ka Shing Foundation, WT-SSI/John Fell funds and by the NIHR Biomedical Research Centre, Oxford, by Widenlife and NIH (CRR00070 CR00.01). The work was also supported by grants to S.F and K.H from MRC (G0802782 and MR/M012638/1).
Footnotes
Authors’ roles statement
Eleni Makrinou processed the samples, analysed and interpreted the array-based data, performed, analysed and interpreted the pyrosequencing generated data, prepared Figures and Tables, and wrote the manuscript;
Alexander W. Drong wrote the R scripts for the genome wide analysis, analysed and interpreted the array-based data; T.C. edited the manuscript;
Avigdor Lerner processed samples and offered advice;
George Christopoulos. and Stuart Lavery were involved in sample collection;
Stephen Franks conceived and supervised the study, edited the manuscript;
Kate Hardy conceived and supervised study;
Cecilia Lindgren supervised analysis of the array-based data.
References
- Abbott DH, Barnett DK, Bruns CM, Dumesic DA. Androgen excess fetal programming of female reproduction: a developmental aetiology for polycystic ovary syndrome? Hum Reprod Update. 2005;11:357–374. doi: 10.1093/humupd/dmi013. [DOI] [PubMed] [Google Scholar]
- Abbott DH, Dumesic DA, Franks S. Developmental origin of polycystic ovary syndrome - a hypothesis. J Endocrinol. 2002;174:1–5. doi: 10.1677/joe.0.1740001. [DOI] [PubMed] [Google Scholar]
- Adams J, Liu Z, Ren YA, Wun WS, Zhou W, Kenigsberg S, Librach C, Valdes C, Gibbons W, Richards J. Enhanced inflammatory transcriptome in the granulosa cells of women with polycystic ovarian syndrome. J Clin Endocrinol Metab. 2016;101:3459–3468. doi: 10.1210/jc.2015-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI, Vanyushin BF. Aging as an epigenetic phenomenon. Curr Genom. 2017;18:385–407. doi: 10.2174/1389202918666170412112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–2749. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- Barber TM, Franks S. Genetics of polycystic ovary syndrome. Front Horm Res. 2013;40:28–39. doi: 10.1159/000341682. [DOI] [PubMed] [Google Scholar]
- Bell CG. The epigenomic analysis of human obesity. Obesity. 2017;25:1471–1481. doi: 10.1002/oby.21909. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower MA, Jones MR, Rotter JI, Krauss RM, Legro RS, Azziz R, Goodarzi MO. Further investigation in europeans of susceptibility variants for polycystic ovary syndrome discovered in genome-wide association studies of Chinese individuals. J Clin Endocrinol Metab. 2015;100:E182–E186. doi: 10.1210/jc.2014-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- Cole E, Brown TA, Pinkerton KE, Postma B, Malany K, Yang M, Kim YJ, Hamilton RF, Jr, Holian A, Cho YH. Perinatal exposure to environmental tobacco smoke is associated with changes in DNA methylation that precede the adult onset of lung disease in a mouse model. Inhal Toxicol. 2017;29:435–442. doi: 10.1080/08958378.2017.1392655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun S, Otu HH, Awartani KA, Al-Alwan LA, Al-Hassan S, Al-Mayman H, Kaya N, Inan MS. Gene expression profiling of granulosa cells from PCOS patients following varying doses of human chorionic gonadotropin. J Assist Reprod Genet. 2013;30:341–352. doi: 10.1007/s10815-013-9935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day F, Karaderi T, Jones M, Meun C, He C, Drong A, Kraft P, Lin N, Huang H, Broer L, et al. Large-scale genome-wide meta analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnostic criteria. PLoS Genet. 2018 doi: 10.1371/journal.pgen.1007813. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto Alves A, Valcarcel B, Mäkinen VP, Morin-Papunen L, Sebert S, Kangas AJ, Soininen P, Das S, De Iorio M, Coin L, et al. Metabolic profiling of polycystic ovary syndrome reveals interactions with abdominal obesity. Int J Obes (Lond) 2017;41(9):1331–1340. doi: 10.1038/ijo.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, Bjonnes A, Broer L, Dunger DB, Halldorsson BV, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464. doi: 10.1038/ncomms9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev. 2012;33:981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Xia BH, Zhang CJ, Zhuo GC. Mutations in mitochondrial tRNA genes may be related to insulin resistance in women with polycystic ovary syndrome. Am J Transl Res Jun. 2017;9(6):2984–2996. [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Triche TJ, Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks S. Polycystic ovary syndrome. N Engl J Med. 1995;333:853–861. doi: 10.1056/NEJM199509283331307. [DOI] [PubMed] [Google Scholar]
- Franks S, Berga SL. Does PCOS have developmental origins? Fertil. Steril. 2012;97:2–6. doi: 10.1016/j.fertnstert.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks S, Gharani N, Waterworth D, Batty S, White D, Williamson R, McCarthy M. The genetic basis of polycystic ovary syndrome. Hum Reprod. 1997;12:2641–2648. doi: 10.1093/humrep/12.12.2641. [DOI] [PubMed] [Google Scholar]
- Franks S, Webber LJ, Goh M, Valentine A, White DM, Conway GS, Wiltshire S, McCarthy MI. Ovarian morphology is a marker of heriTable biochemical traits in sisters with polycystic ovaries. J Clin Endocrinol Metab. 2008;93:3396–3402. doi: 10.1210/jc.2008-0369. [DOI] [PubMed] [Google Scholar]
- Gleicher N, Barad D, Weghofer A. Functional autoantibodies, a new paradigm in autoimmunity? Autoimmun Rev Nov. 2007;7(1):42–45. doi: 10.1016/j.autrev.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Hayes MG, Urbanek M, Ehrmann DA, Armstrong LL, Lee JY, Sisk R, Karaderi T, Barber TM, McCarthy MI, Franks S, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. doi: 10.1038/ncomms8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insenser M, Escobar-Morreale HF. Application of proteomics to the study of polycystic ovary syndrome. J Endocrinol Investig. 2011;34:869–875. doi: 10.3275/8108. [DOI] [PubMed] [Google Scholar]
- Jayasena CN, Franks S. The management of patients with polycystic ovary syndrome. Nat Rev Endocrinol. 2014;10:624–636. doi: 10.1038/nrendo.2014.102. [DOI] [PubMed] [Google Scholar]
- Joseph S, Barai RS, Bhujbalrao R, Idicula-Thomas S. PCOSKB: a KnowledgeBase on genes, diseases, ontology terms and biochemical pathways associated with PolyCystic Ovary Syndrome. Nucleic Acids Res. 2016;44:D1032–D1035. doi: 10.1093/nar/gkv1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal R, Gupta K, Gupta S. Cancer epigenetics: an introduction. Methods Mol Biol. 2015;1238:3–25. doi: 10.1007/978-1-4939-1804-1_1. [DOI] [PubMed] [Google Scholar]
- Khalaf M, Morera J, Bourret A, Reznik Y, Denoual C, Herlicoviez M, Mittre H, Benhaim A. BMP system expression in GCs from polycystic ovary syndrome women and the in vitro effects of BMP4, BMP6, and BMP7 on GC steroidogenesis. Eur J Endocrinol. 2013;168:437–444. doi: 10.1530/EJE-12-0891. [DOI] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, Afzal U, Scott J, Jarvelin MR, Elliott P, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37. doi: 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang J, Deng Q, Li J, Li Z, Xiao Y, Hu S, Li T, Tan Q, Li X, et al. Proteomic profiling for identification of novel biomarkers differentially expressed in human ovaries from polycystic ovary syndrome patients. PLoS One. 2016;11:e0164538. doi: 10.1371/journal.pone.0164538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Zhao H, Zhao X, Zhang B, Cui L, Shi Y, Li G, Wang P, Chen ZJ. Identification of YAP1 as a novel susceptibility gene for polycystic ovary syndrome. J Med Genet. 2012;49:254–257. doi: 10.1136/jmedgenet-2011-100727. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang H. Epigenetic abnormality: a possible mechanism underlying the fetal origin of polycystic ovary syndrome. Med Hypotheses. 2008;70:638–642. doi: 10.1016/j.mehy.2006.09.076. [DOI] [PubMed] [Google Scholar]
- Louwers YV, Stolk L, Uitterlinden AG, Laven JS. Cross-ethnic meta-analysis of genetic variants for polycystic ovary syndrome. J Clin Endocrinol Metab. 2013;98:E2006–E2012. doi: 10.1210/jc.2013-2495. [DOI] [PubMed] [Google Scholar]
- Lv Y, Zhao SG, Lu G, Leung CK, Xiong ZQ, Su XW, Ma JL, Chan WY, Liu HB. Identification of reference genes for qRT-PCR in granulosa cells of healthy women and polycystic ovarian syndrome patients. Sci Rep. 2017;7:6961. doi: 10.1038/s41598-017-07346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani H, Levy MJ, Davies MJ, Morris DH, Gray LJ, Bankart J, Blackledge H, Khunti K, Howlett TA. Diabetes and cardiovascular events in women with polycystic ovary syndrome: a 20-year retrospective cohort study. Clin Endocrinol. 2013;78:926–934. doi: 10.1111/cen.12068. [DOI] [PubMed] [Google Scholar]
- March WA, Moore VM, Willson KJ, Phillips DI, Norman RJ, Davies MJ. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum Reprod. 2010;25:544–551. doi: 10.1093/humrep/dep399. [DOI] [PubMed] [Google Scholar]
- Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll G, Crichton DN, McDowell HE, Kernohan N, Hupp TR, Thompson AM. Expression of the Hypermethylated in Cancer gene (HIC-1) is associated with good outcome in human breast cancer. Br J Canc. 2001;85:1878–1882. doi: 10.1054/bjoc.2001.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Wang FF, Yin R, Ding GL, El-Prince M, Gao Q, Shi BW, Pan HH, Huang YT, Jin M, et al. A molecular mechanism underlying ovarian dysfunction of polycystic ovary syndrome: hyperandrogenism induces epigenetic alterations in the granulosa cells. J Mol Med (Berl.) 2012;90:911–923. doi: 10.1007/s00109-012-0881-4. [DOI] [PubMed] [Google Scholar]
- Rotterdam EA-SPCWG. Revised 2003 consensus on diagnostic criteria and longterm health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HR, Qiu LH, Zhang ZQ, Qin YY, Cao C, Di W. Genome-wide methylated DNA immunoprecipitation analysis of patients with polycystic ovary syndrome. PLoS One. 2013;8:e64801. doi: 10.1371/journal.pone.0064801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, Zhang B, Liang X, Li T, Chen J, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirmans SM, Pate KA. Epidemiology, diagnosis, and management of polycystic ovary syndrome. Clin Epidemiol. 2013;6:1–13. doi: 10.2147/CLEP.S37559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Spicer LJ, Sudo S, Aad PY, Wang LS, Chun SY, Ben-Shlomo I, Klein C, Hsueh AJ. The hedgehog-patched signaling pathway and function in the mammalian ovary: a novel role for hedgehog proteins in stimulating proliferation and steroidogenesis of theca cells. Reproduction Aug. 2009;138(2):329–339. doi: 10.1530/REP-08-0317. [DOI] [PubMed] [Google Scholar]
- Sundar IK, Yin Q, Baier BS, Yan L, Mazur W, Li D, Susiarjo M, Rahman I. DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD. Clin Epigenet. 2017;9:38. doi: 10.1186/s13148-017-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel M, Sawalha AH. Epigenetic variability in systemic lupus erythematosus: what we learned from genome-wide DNA methylation studies. Curr Rheumatol Rep. 2017;19:32. doi: 10.1007/s11926-017-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- Wang Q, Moley KH. Maternal diabetes and oocyte quality. Mitochondrion Aug. 2010;10(5):403–410. doi: 10.1016/j.mito.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XX, Wei JZ, Jiao J, Jiang SY, Yu DH, Li D. Genome-wide DNA methylation and gene expression patterns provide insight into polycystic ovary syndrome development. Oncotarget. 2014;5:6603–6610. doi: 10.18632/oncotarget.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M, Ooms M, Hoogerbrugge JW, Grootegoed JA. Hedgehog signaling in mouse ovary: Indian hedgehog and desert hedgehog from granulosa cells induce target gene expression in developing theca cells. Endocrinology Aug. 2005;146(8):3558–3566. doi: 10.1210/en.2005-0311. [DOI] [PubMed] [Google Scholar]
- Xita N, Tsatsoulis A. Review: fetal programming of polycystic ovary syndrome by androgen excess: evidence from experimental, clinical, and genetic association studies. J Clin Endocrinol Metab. 2006;91:1660–1666. doi: 10.1210/jc.2005-2757. [DOI] [PubMed] [Google Scholar]
- Xu J, Bao X, Peng Z, Wang L, Du L, Niu W, Sun Y. Comprehensive analysis of genome-wide DNA methylation across human polycystic ovary syndrome ovary granulosa cell. Oncotarget. 2016;7:27899–27909. doi: 10.18632/oncotarget.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Du J, Lu D, Ren C, Shen H, Qiao J, Chen X, Zhang H. Increased expression of kindlin 2 in luteinized granulosa cells correlates with androgen receptor level in patients with polycystic ovary syndrome having hyperandrogenemia. Reprod Sci. 2014;21:696–703. doi: 10.1177/1933719113512536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mce.2019.110611.