Abstract
In this chapter, we introduce core functionality of the Jalview interactive platform for the creation, analysis, and publication of multiple sequence alignments. A workflow is described based on Jalview’s core functions: from data import to figure generation, including import of alignment reliability scores from T-Coffee and use of Jalview from the command line. The accompanying notes provide background information on the underlying methods and discuss additional options for working with Jalview to perform multiple sequence alignment, functional site analysis, and publication of alignments on the web.
Keywords: Multiple sequence alignment visualization, Interactive analysis, Web application, Desktop application, Functional site inference, Web services
1. Introduction
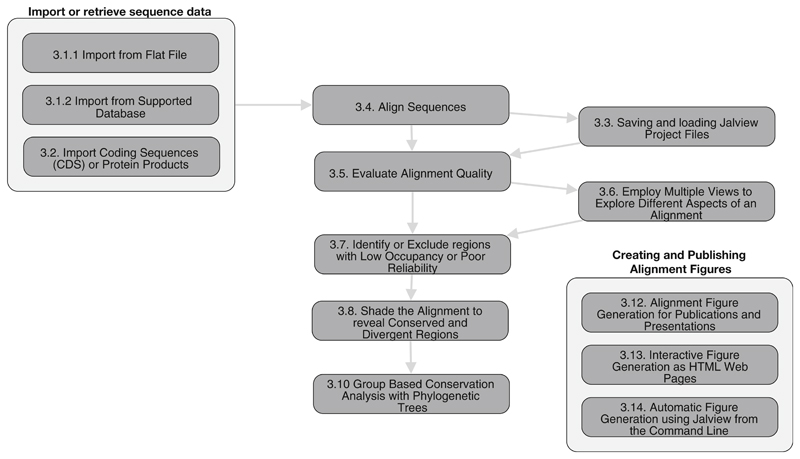
The Jalview [1] platform has many features for sequence analysis and visualization and is freely available both as a native “app” and single-page web application [2] from its web site [3]. A core function is to make it easy to run state-of-the-art methods for multiple sequence alignment (MSA). The resulting alignments can be visualized and integrated with other information to further interpret them and create figures for publication. In this chapter, we describe the steps involved in a typical Jalview sequence alignment workflow, as depicted in Fig. 1. Sequences for alignment may be retrieved from public databases or loaded via a variety of common file formats. Access to a range of alignment programs is achieved directly within Jalview through web services [4, 5], but Jalview also allows alignments generated by external programs to be imported. Built-in analysis routines calculate the consensus for each alignment column and for proteins, the amino acid physicochemical properties. These create column Annotations that are shown as histograms under the alignment. The alignment may be filtered to remove redundant sequences and to hide columns with low numbers of aligned residues. Aligned sequences may also be grouped either manually or by partitioning an associated tree which can be imported in a variety of formats or calculated using Jalview’s own tree algorithms. A range of shading and coloring schemes allow common patterns to be highlighted according to standard properties for amino acids (such as hydrophobicity), or whether nucleotides are purine or pyrimidine derivatives. Uniquely, coloring may be combined with conservation and consensus calculations to emphasize patterns of variation among conserved regions and to highlight different patterns of conservation among subgroups of sequences in the alignment.
Fig. 1.
Workflow for creating, analyzing, and generating figures for a multiple sequence alignment in Jalview. Numbers for each stage correspond to stages described in Sect. 3. An interactive version of this workflow can be viewed online at https:/tess.elixir-europe.org/workflows/multiple-sequence-alignment-analysis-and-figure-generation-with-jalview
2. Materials
Availability, download, and installation
The latest version of the Jalview Desktop application can be obtained from http://www.jalview.org/download. Installers are provided for ×86-based Windows, Linux, and OSX operating systems, and the minimum recommended physical memory is 1 GB. The installers provide the “Jalview Native Application”—which offers enhanced usability through features such as file-type associations, automatically managed Java installation, and “Over the Air Update” to ensure the latest and most secure version of Jalview is launched. An “executable Jar” file is also provided for users wishing to launch a specific version of Jalview or execute it on a platform for which a Jalview Native Application is not yet available. Jalview packages may also be obtained in Homebrew [6] and BioConda [7] and are primarily designed for users integrating Jalview into a command-line workflow rather than performing interactive analyses as described here.
Memory settings and working with larger datasets
The Jalview Native Application and Jalview Executable Jar will automatically request up to 90% of physical memory to be allocated to a Jalview session. The percentage of memory allocated can be modified via a command-line argument. Currently, no support is provided for working with alignments too big to load into memory.
The JalviewJS Web Application
An alternative to installing the Jalview Desktop is to access the JavaScript version of Jalview at https://www.jalview.org/jalview-js/JalviewJS.shtml. It is designed to work with modern web browsers such as Google Chrome or Mozilla Firefox, and provides the majority of functionality described below, except for access to public web services provided by JABAWS.
3. Methods
3.1. Import or Retrieve Sequence Data
A range of common bioinformatics sequence file formats (see Note 1) can be imported by “drag and drop” or via options provided on the “File” menu. Data from the system clipboard may also be pasted by right-clicking on the desktop background and selecting the “Paste to New Window” menu option that appears.
In addition, Jalview provides a “Sequence Fetcher” which allows import of sequences, alignments, and 3D structures from databases hosted by EMBL-EBI [8, 9] (see Note 2).
Once the import has completed, an alignment window containing the sequence data is displayed. More sequences may be added by the same methods (via the alignment window’s own “File” menu, by pasting from the clipboard, or by dragging files onto the window).
3.2. Importing Coding Sequences (CDS) or Protein Products for CDS
CDS for proteins shown in an alignment view can be added simply by dragging and dropping the file containing CDS onto the protein alignment. Protein products for CDS are loaded in the same way. Providing each CDS has the same name as its corresponding protein product, and codons from the CDS exactly match the amino acid sequence (under one of Jalview’s supported translation tables), Jalview will offer the option of opening a “Linked Protein and CDS View” (see Note 3). This configuration allows proteins to be multiply-aligned according to their amino acid sequence, but analyzed at both the nucleotide and amino acid sequence levels.
3.3. Saving and Loading Project Files
A Jalview Project File (JVP) can be created via the Desktop’s File- > Save Project menu entry or an alignment window’s File- > Save As… menu entry. JVP files store data and visualization settings for alignment windows, including any associated views such as trees and 3D structures (e.g., the example file that is automatically loaded by default on startup—see Note 4). It is recommended that work is saved after each stage below to avoid data loss, and if the same filename is used, then Jalview will create versioned backups (see Note 5).
3.4. Align Sequences
A range of alignment programs is offered via the Alignment submenu of the Alignment Window’s “Web Services” menu. Jalview provides access to public services provided by the University of Dundee which are suitable for aligning up to 1000 sequences of at most 1000 residues each, and jobs are allowed to run for up to 1 h. Note 6 discusses how to perform alignments of larger sequence sets with Jalview.
For some alignment programs, Jalview provides a “Realignment” option. This allows sequences to be added to an existing alignment. See Note 7 for a discussion of its effective use.
If a selected region is defined in the alignment window (see Note 8), then only data in that region will be submitted for alignment. Similarly, hidden sequences will not be included in the alignment and hidden columns will force a series of local multiple alignments to be performed on just the visible regions (see Note 9).
Jalview must be left running for the duration of the alignment procedure. For current versions of Jalview (2.11), there is no way to “reconnect” to an alignment or analysis procedure performed via the “Web Services” menu, and Jalview projects do not preserve any information about Web Service jobs that have completed or are currently in progress.
3.5. Evaluating Alignment Quality
Jalview provides visual analytics that help judge the degree of similarity among sequences in an aligned region, but it does not on its own compute measures that indicate how reliable all or part of an alignment may be. Such calculations are provided by the T-Coffee suite via M-COFFEE [10] or Transitive Consistency Score (TCS) analysis [11] (http://www.tcoffee.org/Projects/tcs/) (see Note 10 and Chapter 6). Below we briefly outline a protocol for manually performing an alignment reliability calculation in T-Coffee.
Export the alignment to be assessed as a FASTA file via the File- > Save As… menu entry from the alignment window.
-
Submit the FASTA file for assessment with T-Coffee.
Either via the command line t_coffee -infile prot.aln -evaluate -output score_ascii. or, alternatively, via the T-Coffee web server’s TCS submission form at http://tcoffee.crg.cat/apps/tcoffee/do:core
T-Coffee’s TCS analysis produces a “score_ascii” file which can be read by Jalview. If the web service was employed, then this file should first be downloaded. To view the results, simply drag and drop the file onto the alignment window, or load it via the File- > Load Annotations option.
Jalview provides a “T-Coffee Score” color scheme in its Colour menu for viewing the reliability scores produced by TCS. This is enabled by default when the score_ascii file is loaded and mimics T-Coffee’s standard TCS coloring: where red indicates the most reliably aligned regions, transitioning to green, yellow, and blue for poor quality regions.
3.6. Employing Multiple Views to Explore Different Aspects of an Alignment
Alignment Views allow different regions of the alignment to be shown or hidden and sequences to be independently grouped and colored. Operations on the alignment, sequence, and annotation data affect all Views. When preparing an alignment figure for inclusion in a publication, it is often useful to create a series of Views corresponding to each panel of the figure.
A new View for the alignment is created with the New View option in the View menu. Its presence is indicated by a new tab appearing above the alignment ruler.
Views may be displayed simultaneously via the Expand option in the View menu. The Gather option returns all Views to the tab bar on the current View’s alignment window.
The current View is removed by pressing “Control” or “CMD” and “W” (or if the Views are expanded to their own window, by simply closing that View’s window).
Multiple Views allow specific features of an alignment to be highlighted. Accordingly, a View may be given a unique name via the dialog box opened by right-clicking the View’s tab.
3.7. Identification or Exclusion of Regions with Low Occupancy or Poor Reliability
Occupancy measures the number of sequences aligned at each position and is one of the dynamic Annotation rows automatically computed for an alignment (see Note 11).
Select the Select/Hide Columns dialog from the Alignment Window’s “Select” menu.
Choose “Occupancy” from the first drop-down menu as the annotation row to be queried.
Choose “Below” from the “Threshold Type” drop-down menu, and check the “Percentage” checkbox. Enter “10” to select columns in the alignment where less than 10% of sequences are aligned.
To hide the columns selected by the filter, then select the “Hide” option in the dialog rather than the “Select” option.
Press OK to close the dialog, or cancel to reset column visibility.
To exclude unreliable regions according to T-Coffee TCS scores, select “T-COFFEE” from the list of annotation rows in Sect. 3.5, step 2.
3.8. Shading the Alignment to Reveal Conserved and Divergent Regions
Jalview has a range of protein and nucleotide color schemes (see Note 12) which can be applied in combination with values from the Consensus (and for proteins, Conservation and Quality) dynamic Annotation rows (see Note 11) to highlight variation in columns that exhibit a high degree of amino acid or nucleotide conservation.
- Revealing conservation patterns with Colour by Conservation (Proteins only).
-
(a)Select “Blosum 62” from the Colour menu, which colors each residue on a scale from white to blue according to the likelihood of mutation from the reference or consensus sequence for the alignment view.
-
(b)Enable “Colour by Conservation.” Columns that exhibit a high physicochemical property conservation score (see Note 11) will appear more strongly colored than those with fewer conserved properties.
-
(a)
- Revealing regions with high percentage identity.
-
(a)Select “Percent Identity.” This scheme shades nucleotides and amino acids according to their abundance at each column in the alignment.
-
(b)To shade the alignment according to some other property (e.g., Purines and Pyrimidines, or Taylor’s physicochemical property-based color scheme), first apply this scheme, and then select “Above Identity Threshold” from the “Colour” menu to only color symbols present in more than the specified minimum percentage of aligned sequences at each column in the alignment.
-
(a)
3.9. Shading the Alignment According to Conservation Scores from the AACon Web Service
Select the “Change AAcon settings…” option from the Conservation submenu of the Web Services drop-down menu in the alignment window.
Choose which scores to calculate from the dialog and press “Submit” to enable the calculation and display of AACon annotations. After a short delay, the additional scores will appear as histograms below the alignment.
Open the “Colour by Annotation” dialog from the Color menu. Choose one of the annotation rows from the dropdown menu in the dialog. By default, a linear shading will be applied to columns of the alignment according to values in the chosen row. To apply a threshold (similar to the “Percent Identity” threshold above), select the “Use Original Colours” option and select a threshold type from the drop-down menu.
3.10. Group-Based Conservation Analysis with Phylogenetic Trees
Shading and filtering according to column statistics do not always reveal regions of similarity or divergence not shared by all sequences in the alignment (see Note 13). Once sequences have been grouped, however, the shading schemes introduced in Sect. 3.8 will reveal the patterns of conservation and divergence unique to each group. Jalview is able to create groups interactively from selections and also to subdivide a selection to group sequences according to their identity. However, the most powerful sources of groups are phylogenetic trees (see Note 14). Jalview can import existing trees or calculate one for aligned sequences. The built-in tree viewer then offers a way to subdivide aligned sequences into groups according to their relatedness as defined by the tree.
A tree for all sequences or a selected region of a view can be calculated and displayed via the “Calculate” dialog (accessed via the Calculations menu). For best results when computing subgroups, particularly for a selected region of an alignment, we recommend selecting the UPGMA Average Distance Tree.
Use the “Sort alignment by tree” option in the Tree viewer’s submenu to reorder sequences in the alignment view according to the tree (Jalview’s preferences allow this action to be configured to be performed automatically).
Select a position between the root and leaves of the displayed tree to define a set of groups on the alignment. (Warning: previously defined groups will be removed.) In the alignment view, the names of sequences in the same group will have a similar background color. For each group, coloring based on Conservation and Percent Identity will employ values computed for just the grouped sequences: locally conserved regions will therefore be more strongly colored in comparison to regions of local divergence.
For alignments too large to fit on screen, the Alignment Overview (opened via the option in the View menu) allows differences between groups to be compared more easily.
When the overview is open, “Colour by Sequence ID” can be applied in combination with “Colour by Conservation” or “Above Identity Threshold” to more easily distinguish sequences in different groups.
3.11. Visualizing Group Conservation and Consensus
The “Autocalculated annotation” submenu in the Annotations menu provides options controlling the display of conservation and consensus rows for the currently selected group. To modify settings for all groups, enable the “Apply to all groups” option.
The consensus (and for proteins—conservation) annotation for groups on the alignment can be shown by enabling the “Group Consensus” and “Group Conservation” options under the Annotations menu’s Autocalculated Annotation submenu.
Clicking the label of a group’s annotation row will highlight the sequences in that group. Double clicking the label will select the whole group.
-
Display of sequence logos.
Amino acid and nucleotide distributions may be visualized for an alignment or sequence group as sequence logos [12]—where for each column, letters are stacked in order of increased frequency of observation, and their height also scaled accordingly.-
(a)Select the Annotation menu’s Show Logo option to display sequence logos on group or alignment consensus annotation rows.
-
(b)Distributions across different sites of the alignment can be more easily compared by enabling the “Normalise Logo” option. In this case, disabling the “Show Histogram” option allows the logo to be more clearly viewed.
-
(a)
3.12. Alignment Figure Generation for Presentations and Papers
Alignment views can be exported in a range of ways via the File menu’s “Export” submenu. Views can be exported as shown, rendered as a Portable Network Graphic (PNG) raster image—suitable for onscreen display and Scalable Vector Graphic (SVG) or Encapsulated PostScript (EPS) format vector graphic drawings which are recommended when preparing figures for publication. HTML pages can also be generated via the options in the menu—these interactive export options are discussed in the next step.
3.12.1. Preparing for Figure Export
When preparing to export views as static figures, it is recommended that a new View is created to allow layout, font size, colors, and data visibility to be configured, since no “Undo” functionality is provided to revert changes.
3.12.2. “Wrap mode”: Formatting Alignments to Fit Within the Margins of a Page
Enable the Wrap Mode option in the “Layout” menu to format the MSA as a series of fixed width blocks. The number of columns shown in each block is defined by the width of the alignment view. These are reflected by the numberings shown in the alignment ruler. The number of columns may be changed by:
-
(a)
Adjusting the width of the alignment view window. How this is done depends on which operating system (OS) is used: On Mac OSX, move the mouse pointer to the bottom right corner of the window and click-drag to adjust the size. Other OSes allow resizing by click-dragging any edge of the window.
-
(b)
Adjusting font size and column width. Either via the Layout’s Font dialog, or with a three-button mouse by clicking the middle button in the alignment view and moving it left or right to adjust width, and up or down to decrease or increase font size (respectively).
-
(c)
Adjusting the sequence ID margin. When the mouse pointer is moved to the right-hand side of the sequence ID panel, it will change to indicate that the margin can be adjusted by clickdragging to the left and right.
3.12.3. EPS Export as “Characters” or Line Art
When EPS export is selected, Jalview can either represent each sequence symbol as a character or render the shape of each symbol in the EPS file. The former allows EPS files that can be easily edited in a vector graphics program such as Illustrator—e.g., to modify sequence ID labels, but can result in EPS files that appear different to Jalview’s alignment view. The latter results in larger files but ensures all aspects of the MSA visualization (e.g., sequence logos, character alignment) are faithfully reproduced.
3.13. Interactive Figure Export in HTML Web Pages
Other options in the Export submenu of the Alignment View’s File menu allow HTML pages to be generated containing either an embedded SVG rendering of the view or a JavaScript visualization such as the BioJS Multiple Sequence Alignment Viewer (BioJS-msaviewer) [13]. In general, interactive figure export results in web pages that will look and behave differently to visualizations provided by the Jalview Desktop app or the JalviewJS web component (see Note 15).
3.13.1. Export as an Interactive HTML Figure
The “HTML” export option produces a web page containing two SVG figures—one for the ID panel and one for the columns of the MSA and Annotation rows. Buttons are provided to open the exported view in the Jalview application, and view the embedded data (stored as BioJSON [14]).
3.13.2. Export Alignment for Visualization with BioJS-msaviewer
-
(a)
The “BioJS” export option generates an HTML page containing embedded BioJSON and the JavaScript code necessary to display the alignment with BioJS-msaviewer.
HTML files generated by Jalview that contain BioJSON can be imported like any other alignment file (see step 3.1). However, Jalview Project files are recommended for long-term archiving of data and visualizations generated during multiple sequence alignment and analysis.
3.14. Automated Alignment Figure Generation in Batch Mode
Jalview’s command line allows figures to be generated without user intervention.
3.14.1. Prepare a Custom Jalview Properties File
When running in batch mode, a custom preferences file allows alignment layout parameters to be specified. To do this:
-
(a)
Make a backup of your existing “.jalview_properties” file.
-
(b)
Adjust the various Jalview User preferences provided in the Visual, Colour, Output, and Editing panels so that when an alignment is imported, it has the desired appearance.
-
(c)
Make a copy of your customized “.jalview_properties” file using a unique name such as “jalview_batch.properties.”
3.14.2. Running Jalview as a Command-Line Program
The precise way that Jalview is called from the command line depends on how it was installed. The procedure below assumes you have downloaded the Jalview executable JAR and that your system has an existing Java 8 installation.
-
(a)Execute the following command to generate a figure from the command line:
- Java -jar jalview-2.11.0.jar -headless -props jalview_batch.properties -open “http://www.jalview.org/examples/uniref50.fa” -png example_fig.png
-
(b)
Verify that the generated PNG file: “example_fig.png” exists and has the desired appearance.
4. Notes
-
Table 1 lists the file types currently supported by Jalview, the type of data provided, and whether Jalview can export as well as import in that format, along with any caveats regarding their use. For completeness, we also include here formats for annotation, 3D structure, phylogenetic trees, and the Jalview specific formats: Jalview Features Format, Jalview Annotations Format, Jalview Project, and BioJSON.
When importing sequence data, Jalview employs a series of rules to determine the format. If no format is found to match based on these rules or an error is encountered while parsing the data according to the determined format, then Jalview will default to the “Pfam” format reader. The Pfam format is also used by default when sequence data are copied directly from an alignment window to the system clipboard for other applications.
Table 2 details the databases that Jalview 2.11 is able to access and the kinds of data they provide. Jalview will present an interactive query dialog for databases that support free text search or a simple “Fetch IDs” dialog for databases for which no query client is available. The specific search capabilities provided when performing a free text search depend on the particular database being queried: all provide a range of fields that can be used to restrict a query (via a drop-down menu to the left of the search box) and also allow structured queries to be entered directly (please see Jalview documentation for details of these).
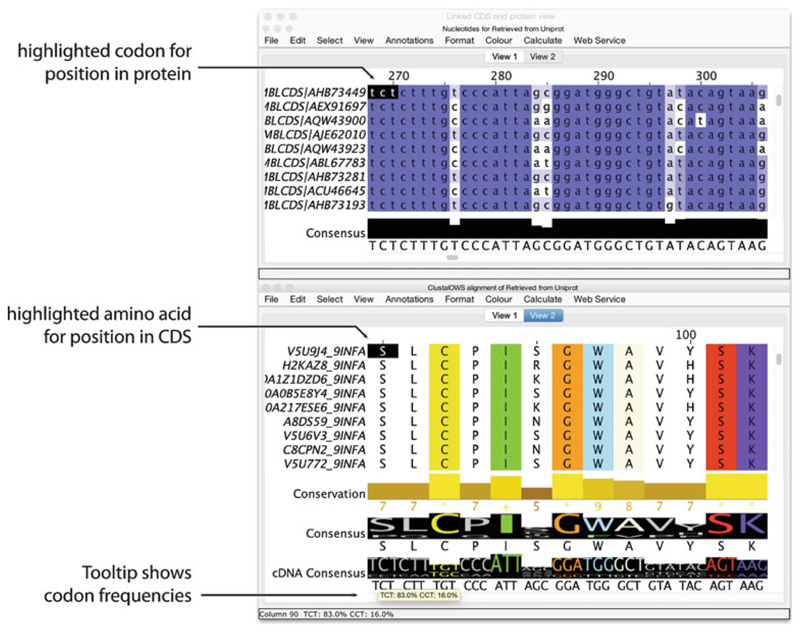
Jalview allows alignments of proteins and their CDS to be visualized and interactively analyzed as a pair of linked alignment views shown docked bottom to top (see Fig. 2). Operations on protein sequences are mirrored on the CDS, allowing proteins to be aligned using their amino acid sequence and the resultant CDS multiple sequence alignment analyzed to investigate the presence of bias that might indicate selection. All analysis steps described above can be performed on a linked CDS/Protein view, with the added benefits that (1) codon bias and diversity can also be visualized and used for filtering the alignment, (2) genome- and transcript-level sequence features can be visualized via the CDS alignment view, and (3) phylogenetic trees computed using the CDS alignment using either Jalview’s built-in score models or via an external program and loaded back onto the alignment can provide additional evolutionary insight when used to partition the protein alignment.
When first launched, Jalview will automatically import and display an example alignment, tree, and 3D structure retrieved from the Jalview web site. The display of these examples is disabled by opening Jalview’s user preferences dialog (via the Preferences option of the “Tools” drop-down menu), un-ticking the “Open File” checkbox, and selecting “OK” to save the updated preferences. Jalview user preferences are stored in a “.jalview_properties” located in the user’s home directory. These are read every time Jalview is launched, and a customized properties file can also be provided when Jalview is run in batch mode from the command line to specify alignment and annotation layout for automatic generation of figures.
When saving alignment data, annotations, and projects, Jalview may not always ask you to confirm if you are about to overwrite an existing file—particularly if you employ the “Save File” shortcut key (either CTRL-S or CMD-S on Macs). In these situations, Jalview will by default automatically create a backup file (called, e.g., myfile.fa.bak001). Backup behavior can be changed in the “Backups” section of Jalview’s preferences panel: by default only the three most recent backups will be retained.
- Alignment of large sets of sequences is CPU intensive and can therefore take considerable time. Jalview’s public alignment services provided by the University of Dundee in Scotland, UK, do not permit execution of alignments of greater than 1000 sequences with up to 1000 amino acids each. To perform larger alignments, it is necessary to either (a) download and configure a local instance of a compatible Jalview web services system or (b) align sequences using an external program and then import the result.
-
(a)Downloading and configuring a local instance of Jalview Web Services. Jalview versions prior to 2.12 are able to access web services provided by the JABAWS system (http://www.compbio.dundee.ac.uk/jabaws), and instructions for local installation either as a virtual appliance or tomcat web application are provided (http://www.compbio.dundee.ac.uk/jabaws/docs/getting_started.html). It may then be necessary to compile binaries for your platform and modify the JABAWS execution limits to permit alignments to be performed of the size that you require (http://www.compbio.dundee.ac.uk/jabaws/docs/advanced.html#limiting-the-size-of-thejob-accepted-by-jabaws). Once configured, Jalview can be connected to your new JABAWS server by entering its URL in Jalview’s “Web Services” Preferences pane, and once validated, services will be accessible from the Web Services menu in the alignment view.A new web services system is currently being developed, and instructions will be made available via Jalview’s built-in help on how to download and deploy these new services once they are put into production.
-
(b)Exporting sequences for alignment and reimporting the result. Sequences may be exported via the File menu’s “Save as” option in the alignment window or for the current selection via the pop-up menu (opened by right-clicking the selected area). Once the alignment has been performed, it is straightforward to import the aligned result to a new alignment view, but there are potential problems:
-
•Alignment programs may have constraints on sequence name length, the range of characters permitted, and reject inputs containing duplicate sequence names.
-
•Import of the aligned sequences as a new alignment will not retain sequence metadata from the original Jalview alignment view such as CDS relationships, database cross-references, sequence features, and secondary structure annotation rows.
-
•Hidden columns in the original view will not be accounted for or included in the result of the alignment (see Note 9).
Problems with sequence names can be worked around through the use of custom scripts, but are outside the scope of this chapter. Assuming sequence identifiers have been preserved, then there are two ways to work around the loss of sequence metadata. Metadata originally retrieved by Jalview from an external database can be retrieved once again. The “Fetch DB Refs” option in the Web services menu provides options to retrieve records from either all standard databases or a specific one (e.g. UniProt for protein sequences), but this should be used with care since both options may take some time for large alignments. A second workaround is to manually export features and annotation from the original view and import the resultant Jalview features file (or GFF3 file) and Jalview annotations file to the newly imported aligned sequence set. -
•
-
(a)
- Jalview provides a Realignment option when performing alignments via the Clustal W and Clustal Omega web service programs. Normally, Jalview removes all gap characters from sequences passed to an alignment program, but for Realignment, gap characters will be preserved. The precise behavior depends upon which Clustal alignment program is used:
-
•Clustal W identifies aligned regions of the input as a range of sequences of equal length (including any gap characters) at the beginning of the input data. All other sequences in the input data are then aligned to that first block (with inserts into the block created as necessary). This process is quick, and preserves the original aligned region, provided that region was reordered to appear at the top of the alignment view when the alignment was submitted.
-
•Clustal Omega realigns sequences by performing a sequence-profile alignment. A profile is first constructed from the input sequences (including any gap characters present). Gaps are then removed from all sequences, and they are each aligned to the profile to generate the final multiple sequence alignment result. This method is more computationally expensive than ClustalW, and it is unlikely that relationships between aligned sequences will be preserved in regions of poor alignment reliability in the result.
-
•
Many of Jalview’s operations apply either to the whole alignment or, when present, just the selected region. Selected regions in Jalview are highlighted with a red box. Columns are annotated with a red mark, and selected sequences highlighted in dark gray. Rows and columns on alignment view can be selected by clicking and dragging with the mouse on the sequence ID panel and on the alignment ruler. An area of the view can also be selected simply by clicking and dragging. These are summarized in the Jalview online video (http://www.jalview.org/videos/selectinginjalview) and FAQ “How Do I edit Sequences in Jalview” (http://www.jalview.org/faq#sequences). Functions in the Alignment window’s select menu allow columns to be selected on the basis of alignment annotation (with the Select/Hide By Annotation dialog) and when regions of the alignment are highlighted as the result of a find operation. Columns can also be selected according to the presence of sequence features.
Selected regions in a view can be hidden or shown simply by pressing “H” or one of the other key combinations shown under the View menu’s Hide and Show submenus. Columns containing gaps can also be hidden via the “Hide Inserts” function in the Selection and Sequence ID pop-up menu, opened by right-clicking in the sequence ID area. Hidden sequences and columns are excluded from alignment analysis (e.g., the conservation and consensus rows), tree calculation, principal component analysis, and secondary structure predictions (accessed via the submenu in the Web Services drop-down menu). Conversely, when multiple alignments are performed, hidden columns are “preserved” and not submitted to the server. Instead, the chosen program is executed several times, once for each contiguous region of the input set. Once all jobs are complete, Jalview concatenates the results and intervening hidden regions in order to construct the final alignment view.
Identification of reliably aligned regions in a multiple alignment is important for many applications. T-Coffee’s Transitive Consistency Score (TCS) provides one approach: it measures the average shift error between the sets of positions aligned in a multiple alignment and a library of pairwise alignments involving the same sequences. Optimally aligned regions will always be aligned in the same way, and an increasing shift error is more indicative of low reliability. T-Coffee also offers a consensus alignment tool, M-COFFEE, which computes multiple alignments for the input sequences with several different multiple alignment programs and then generates a final alignment from these different results. Here, TCS scores reflect shifts between the different multiple alignment results used to generate the consensus.
-
A range of MSA column statistics are automatically computed and can be displayed as alignment annotation rows below the alignment. These rows update automatically as sequences are added or removed from the view, or the MSA is otherwise adjusted via Jalview’s interactive MSA editing capabilities.
The “Visual” tab in Jalview’s user preferences dialog provides options for enabling or disabling Consensus, Occupancy, Conservation, and Alignment Quality Scores. The Consensus annotation row shows the modal residue in each column (or + if more than one residue is observed) and the proportion of sequences that contain that residue. Right-clicking on the Consensus row’s annotation label (on the left-hand side) opens a pop-up menu that allows the consensus sequence for the view to be copied to the clipboard (and so pasted to a new alignment). Options in the menu also allow gapped sites to be ignored when computing the height of the consensus histogram and a Sequence Logo to be overlaid or shown in place of the histogram. Occupancy simply reflects the number of sequences that are aligned at each column of the MSA. Alignment Quality and Conservation are only available for Protein MSAs. The Alignment Quality score reflects the total likelihood of observing mutations between amino acids aligned at the given column, based on the BLOSUM62 [15] substitution matrix. The Conservation score for a column is computed according to Zvelebil et al. [16] as implemented in the AMAS method [17] and reflects the number of physicochemical properties shared by all amino acids in a column. The tooltip for each column lists conserved properties with properties prefixed with an exclamation mark (!) to indicate the absence of that property among the aligned residues. Jalview also provides access to AACon [18] through the Alignment Conservation submenu of the Web Services menu, which permits a further 17 conservation scores to be computed.
Jalview’s built-in help provides a key and description for each of the color schemes available in the “Colour” menu. There are two classes of color scheme, symbol-based, such as Hydrophobic, or Taylor, and dynamic, such as ClustalX and Blosum62. When working with nucleotide alignments, the only dynamic scheme available is PID—which reflects abundance. The Purine and Pyrimidine color scheme, however, can be used to identify variation that may suggest differences in RNA secondary structure.
MSAs involving sequences that have diverse functions or complex evolutionary relationships such as duplications and domain expansions can be difficult to interpret for a number of reasons. The central problem is that while some columns are conserved, others are divergent as a result of evolutionary pressure; and at each site, the degree of divergence between any two sequences may not always be the same as for others. For instance, orthologous sequences will exhibit conservation across functionally relevant regions, but those same functional regions may not be conserved in paralogs. As a consequence, global statistics such as consensus and conservation are not always sufficient to identify regions of alignments that are important for the structure and function of a sequence family. A fully automated method for the identification of such regions solely on the basis of sequence remains a research problem, but hierarchical alignment analysis methods such as AMAS [17] (and when a 3D structure is available, Evolutionary Trace [19]) can be effective. These approaches reveal local patterns of conservation and divergence by subdividing aligned sequences into clusters according to their percentage identity, ideally with a tree computed from the alignment (see Note 14 below). Jalview enables alignments to be partitioned into groups in a similar manner, and its per-group conservation and consensus shading allow patterns of conservation to be revealed within each group. This approach does not on its own provide a way of quantifying the functional importance of a conserved region, but for proteins, Multi-Harmony [20] (via the Web Services’ Analysis submenu) can be applied to an MSA with subgroups defined in order to infer columns that exhibit functional variation.
Jalview includes the algorithms “UPGMA” and Neighbor-Joining for the generation of dendrograms from distance matrices computed over a range of columns in an MSA. These functions are accessed from the “Calculate Tree or PCA…” dialog in the Calculations menu. A variety of score functions are provided [21] including protein substitution matrices such as BLOSUM62 and a Percent Identity score suitable for DNA. Trees calculated by external programs may also be imported as New Hampshire (Newick) and New Hampshire “Extended” format flat files—Jalview will attempt to automatically match leaves to sequences based on the displayed sequence IDs.
In addition to the Jalview Desktop application, Jalview is also available as a web-based application: JalviewJS. Launched in late 2019, JalviewJS is the Jalview application compiled to JavaScript [2] and adapted to run in-page either as the full-featured “Desktop” application or as interactive MSA visualization components designed for embedding in web pages. For more details, please see http://www.jalview.org/jalview-js/.
Table 1. File formats supported by Jalview.
| Format | Extension | Sequence | Features | Annotation | Import only | Notes |
|---|---|---|---|---|---|---|
| AMSA | .amsa | ✔ | ✔ | ✔ | ||
| FASTA | .fa, .fasta | ✔ | Jalview does not currently support “FASTQ” style quality data embedded in FASTA records | |||
| Block | .blc | ✔ | ✔ | Header lines are available via the “Alignment Properties” dialog. For BLC files produced by an iterative search program such as ScanPS, the first iteration is loaded by default. A specific iteration can be retrieved by appending the filename with “#2” (to retrieve iteration 2) | ||
| Clustal | .aln | ✔ | The “Clustal” version header in files written by Jalview is hard coded, which may cause compatibility problems with other programs that require a specific version header | |||
| MSF | .pileup, .aln | ✔ | ||||
| Protein Information Record | .pir | ✔ | ✔ | Jalview supports MODELLER [22] style PIR description lines [23] to specify the filename and chain in a PDB [24] or mmCIF [25] file that maps to a sequence | ||
| Phylip | ✔ | |||||
| Pfam | .pfam | ✔ | ||||
| Stockholm | .stk, .stockholm | ✔ | ✔ | ✔ | In addition to sequence, features, and annotation, Jalview also imports any provided database cross-references for a sequence (including PDB [24] database identifiers) and generic annotation tags associated with the alignment | |
| Jalview Features Format | .jvfeats | ✔ | ||||
| T-COFFEE Score file | .score_ascii | ✔ | Jalview will raise an error if the number of sequences and columns in the score file does not match the alignment | |||
| Jalview Annotations Format | .jvannot | ✔ | ||||
| Generic Features Format (V3) | .gff, .gff2, .gff3 | ✔ | ✔ | |||
| Variant Call Format (indexed) | .vcf, .vcf.tbi | ✔ | A protein or DNA contig with chromosomal coordinates must first be loaded before it can be annotated with variants from VCF files | |||
| BioJSON | .json, biojson, html | ✔ | ✔ | ✔ | A JSON hash containing elements structured according to BioJSON scheme can also store Groups, color schemes, Hidden columns, and sequences. Jalview will also search HTML files for embedded BioJSON | |
| Jalview Project | .jvp, .jar | ✔ | ✔ | ✔ | Projects preserve all aspects of a Jalview session, including the layout of windows showing alignments, trees, and 3D structures | |
| PDB | .pdb, .ent | ✔ | ✔ | ✔ | * | Only 3D structures imported as PDB [24] files can be exported as PDB files |
| mmCIF | .mmcif, .cif | ✔ | ✔ | ✔ | * | Only 3D structures imported as mmCIF files can be exported as mmCIF files |
| Newick | .tree, ,nw | * | Jalview supports both simple Newick [26] and Extended Newick [27] and will preserve distances and bootstrap values detected in the file. It only exports trees in simple Newick format |
Table 2. Sequence databases that Jalview can query and retrieve data.
| Database | Free text search | What is retrieved | |
|---|---|---|---|
| UniProt | Yes | Annotated protein sequences | Imported sequences include positional features and cross-references to other databases |
| Ensembl | Annotated genomic loci, transcripts, and protein products | Retrieved transcripts are shown aligned against their parent loci. Intronic regions are automatically hidden | |
| European Nucleotide Archive | Annotated contigs and protein products | Retrieved contigs that include CDS regions are shown along with retrieved protein products in a linked CDS/Protein alignment view | |
| PDBe | Yes | 3D structures (as mmCIF) | Sequences for all chains in the imported 3D structure file are extracted and shown in the alignment view. Sequence-associated Annotation rows and features provide information about secondary structure and mapping between sequence and 3D coordinate numbering schemes |
| Pfam | Annotated protein domain alignment (as Stockholm) | Alignments include an additional consensus line computed by the Pfam pipeline and a secondary structure annotation row when 3D structure data for the domain family is available. | |
| Rfam | Annotated RNA domain alignment (as Stockholm) | A consensus RNA secondary structure annotation for the Rfam family is also provided |
Fig. 2.
Jalview’s linked CDS and Protein views. Screenshot of the Jalview 2.11 Desktop showing a reconstructed coding sequence alignment for a Clustal Omega alignment of influenza (H5N1) neuraminidase protein sequences. Protein sequences were retrieved from UniProt via Jalview’s Sequence Fetcher and aligned with the Clustal Omega Web Service with default parameters. The coding sequence alignment was reconstructed by selecting “EMBLCDS” from the “Show Cross References” submenu of the “Calculate” menu, which triggered a retrieval of coding sequences from the European Nucleotide Archive. Two views have been created for the Linked CDS and Protein view (View 1 and View 2); the highlighted positions in CDS and Protein views are shown when the mouse is moved across the alignment area. Display of Sequence Logos have been enabled via the Consensus Annotation row’s pop-up menu in order to display logos for amino acid and cDNA codon frequencies
Acknowledgments
JBP and GJB wrote the manuscript. SLD created training videos based on exercises in the Jalview Manual developed by JBP and DMAM. All other authors contributed to Jalview development in the period 2009–2019. Development of the Jalview sequence analysis platform is coordinated at the University of Dundee with support from the UK’s Biotechnology and Biological Sciences Research Council (BB/L020742/1), the Scottish Funding Council, and the Wellcome Trust (101651/Z/13/Z). We also gratefully acknowledge contributions and feedback from the Jalview user community and student project support from the Google Summer of Code (2011 and 2012).
References
- 1.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson R, Barton GJ, Procter J, Carstairs G, Soares B. java2script/SwingJS for bio-informatics: reintroducing Jalview on the Web as JalviewJS. 2019 https://f1000research.com/posters/8-1578. [Google Scholar]
- 3.The Jalview Web Site. [Accessed 2nd Sept 2019];2019 http://www.jalview.org.
- 4.Troshin PV, Procter JB, Sherstnev A, Barton DL, Madeira F, Barton GJ. JABAWS 2.2 distributed web services for Bioinformatics: protein disorder, conservation and RNA secondary structure. Bioinformatics. 2018;34(11):1939–1940. doi: 10.1093/bioinformatics/bty045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troshin PV, Procter JB, Barton GJ. Java bioinformatics analysis web services for multiple sequence alignment—JABAWS:MSA. Bio-informatics. 2011;27(14):2001–2002. doi: 10.1093/bioinformatics/btr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moretti S. Jalview 2.11.0 package for Homebrew. 2019 https://formulae.brew.sh/cask/jalview. [Google Scholar]
- 7.Gruning B, Dale R, Sjodin A, Chapman BA, Rowe J, Tomkins-Tinch CH, Valieris R, Koster J, Bioconda T. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018;15(7):475–476. doi: 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeira F, Madhusoodanan N, Lee J, Tivey ARN, Lopez R. Using EMBL-EBI Services via web interface and programmatically via web services. Curr Protoc Bioinformatics. 2019;66(1):e74. doi: 10.1002/cpbi.74. [DOI] [PubMed] [Google Scholar]
- 9.Yates A, Beal K, Keenan S, McLaren W, Pignatelli M, Ritchie GR, Ruffier M, Taylor K, Vullo A, Flicek P. The Ensembl REST API: Ensembl data for any language. Bioinformatics. 2015;31(1):143–145. doi: 10.1093/bioinformatics/btu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace IM, O’Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34(6):1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang JM, Di Tommaso P, Notredame C. TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol Biol Evol. 2014;31(6):1625–1637. doi: 10.1093/molbev/msu117. [DOI] [PubMed] [Google Scholar]
- 12.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yachdav G, Wilzbach S, Rauscher B, Sheridan R, Sillitoe I, Procter J, Lewis SE, Rost B, Goldberg T. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics. 2016;32(22):3501–3503. doi: 10.1093/bioinformatics/btw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ofoegbu TP, Procter James B. BioJSON Version 1.0 Schema for representation and exchange of annotated Multiple Sequence Alignments with The Jalview Workbench. [Accessed 2nd Sept 2019];2015 https://jalview.github.io/biojson/v1.0/ [Google Scholar]
- 15.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zvelebil MJ, Barton GJ, Taylor WR, Sternberg MJ. Prediction of protein secondary structure and active sites using the alignment of homologous sequences. J Mol Biol. 1987;195(4):957–961. doi: 10.1016/0022-2836(87)90501-8. [DOI] [PubMed] [Google Scholar]
- 17.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9(6):745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 18.Golicz AT, Troshin PV, Madeira F, Martin DMA, Procter JB, Barton GJ. AACon: A Fast Amino Acid Conservation Calculation Service. [Accessed 2nd Sept 2019];2018 http://www.compbio.dundee.ac.uk/aacon/ [Google Scholar]
- 19.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257(2):342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 20.Brandt BW, Feenstra KA, Heringa J. Multi-harmony: detecting functional specificity from sequence alignment. Nucleic Acids Res. 2010;38:W35–W40. doi: 10.1093/nar/gkq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Calculate Tree or PCA Dialog. [Accessed 2nd Sept 2019];2019 http://www.jalview.org/help/html/calculations/tree.html.
- 22.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 23.Webb Bea. Alignment File (PIR)—from the Modeller Manual Online. [Accessed 2nd Sept 2019];2010 https://salilab.org/modeller/9v8/manual/node454.html.
- 24.Sussman JL, Lin D, Jiang J, Manning NO, Prilusky J, Ritter O, Abola EE. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 6 Pt 1):1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 25.Westbrook JD, Bourne PE. STAR/mmCIF: an ontology for macromolecular structure. Bioinformatics. 2000;16(2):159–168. doi: 10.1093/bioinformatics/16.2.159. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. The Newick Tree Format. [Accessed 2nd Sept 2019];1986 http://evolution.genetics.washington.edu/phylip/newicktree.html. [Google Scholar]
- 27.Cardona G, Rossello F, Valiente G. Extended Newick: it is time for a standard rep-resentation of phylogenetic networks. BMC Bioinformatics. 2008;9:532. doi: 10.1186/1471-2105-9-532. [DOI] [PMC free article] [PubMed] [Google Scholar]