Abstract
Metabolic reprogramming is a key hallmark of cancer, but less is known about metabolic plasticity of the same tumor at different sites. Here, we investigated the metabolic adaptation of leukemia in two different microenvironments, the bone marrow and the central nervous system (CNS). We identified a metabolic signature of fatty-acid synthesis in CNS leukemia, highlighting Stearoyl-CoA desaturase (SCD1) as a key player. In vivo SCD1 overexpression increases CNS disease, whilst genetic or pharmacological inhibition of SCD1 decreases CNS load. Overall, we demonstrated that leukemic cells dynamically rewire metabolic pathways to suit local conditions and that targeting these adaptations can be exploited therapeutically.
Keywords: metabolic reprogramming, acute lymphoblastic leukemia, central nervous system, SCD1, fatty acid synthesis
Introduction
Metabolic reprogramming is acknowledged as a hallmark of cancer, encompassing the idea that cancer cells adapt their metabolism to support cell proliferation and to survive in hypoxic and or nutrient-poor environments1–3. In hematologic malignancies, harnessing metabolic reprogramming of cancer cells has proven to be clinically useful in diagnostics such as Positron Emission Tomography and in identification of discrete subgroups in diffuse large B-cell lymphoma requiring specific therapeutic approaches4,5. Metabolic vulnerabilities of hematologic malignancies can be potentially exploited therapeutically6,7. In fact, successes in exploiting metabolic vulnerabilities as a therapeutic approach have been most notable in acute lymphoblastic leukemia (ALL) where the use of asparaginase is a cornerstone of most modern treatment protocols, targeting the asparagine-dependent leukemic cells8. Despite a recent explosion in cancer metabolism research, little is known about how metabolism dynamically changes in cancer cells in response to migration from one tissue compartment to another. Here we investigated how ALL cells metabolically-adapt to the CNS microenvironment upon migration from the bone marrow (BM) and tested whether this adaptation can inform novel targeted therapies for CNS leukemia.
ALL commonly infiltrates the CNS and successful eradication is essential for long-term remission9. Indeed, the largest advance in cure rates for ALL patients came with the introduction of universal CNS-directed therapy in the 1970s10. Although relatively effective, current CNS-directed therapy in the form of intrathecal and systemic high dose methotrexate, or less commonly craniospinal irradiation, is associated with significant neurotoxicity11,12. A lack of mechanistic understanding of CNS leukemia has hindered the development of targeted, less toxic, therapeutic approaches.
Early post-mortem histopathology of CNS leukemia identified that blast cells infiltrate the leptomeninges and generally stay confined within this compartment13. More recent work using animal models and patient-derived xenografts has identified a generic property of ALL cells that enables CNS colonization. Previous published work from our laboratory in mouse and human has established that there is no evidence of a selection for subclones within the CNS microenvironment although altered expression of certain genes, such as CCR7, can enhance the process of CNS-entry14–19. It is postulated that the CNS may act as a “sanctuary site” where leukemic cells are able to evade the immune response and be less exposed to systemic chemotherapy9,20. Successful adaptation to the CNS niche is an essential pre-requisite for long-term survival of leukemic cells, and for subsequent disease relapse in this extramedullary site.
Within the CNS niche, ALL cells are bathed in cerebrospinal fluid (CSF) within the leptomeningeal regions which is poorly vascularized and restricted by the blood-CSF barrier and therefore poor in nutrients and oxygen21–23. In contrast, sites of systemic engraftment, such as spleen or bone marrow, are well vascularized and constitute a rich niche with mesenchymal cells, cytokines and chemokines, favoring leukemic cell survival and proliferation24–26. Since leukemic blasts need energy and metabolic building blocks to survive and proliferate, and CSF nutrient supplies are scarce, we hypothesized that changes in metabolism would take place when ALL engrafts in the CNS. Using gene expression profiles and metabolic characterizations of leukemic cells in different niches, followed by gene ablation and pharmacologic manipulation of the cells, we identified an essential role for the fatty-acid synthesis gene, stearoyl co-A desaturase (SCD1), in CNS localization.
Results
CNS-derived ALL cells display a lipid oriented metabolic transcriptional signature
To investigate transcriptional adaptations of ALL to the CNS microenvironment two human leukemic cell lines, SEM and REH, were used. SEM cells have the t(4;11) KMT2A-AFF1 (MLL-AF4) chromosomal translocation and REH cells have the t(12;21) ETV6-RUNX1 (TEL-AML1) translocation, representing high-risk and standard-risk subtypes respectively. Cells were transplanted into immunodeficient mice. CNS tropism was demonstrated for both cell lines and they invaded the leptomeninges in a timeframe ranging from 4 to 5 weeks (Supplementary Figure 1). At experimental endpoint, blasts were retrieved from the CNS and periphery (spleen), and their gene expression profiles were compared using RNA-sequencing (experimental workflow shown in Supplementary Figure 2A).
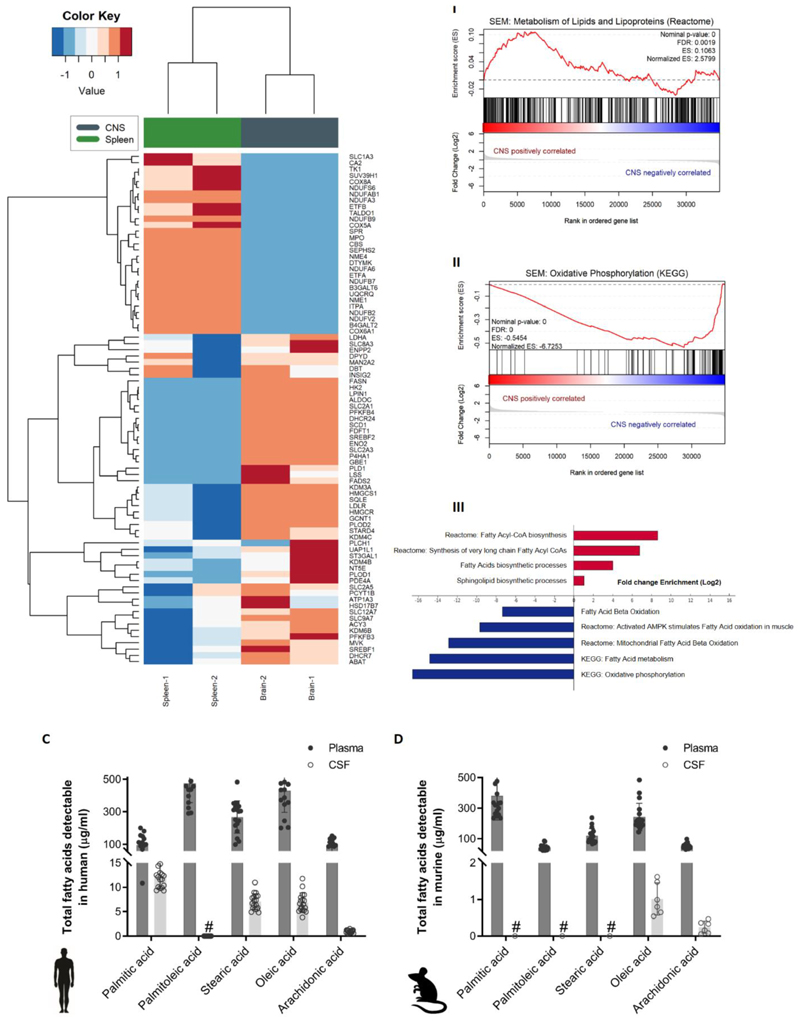
The human SEM cells’ gene expression profiles clustered according to site of colonization in the transplanted mice; CNS versus spleen (Figure 1A). Differentially expressed genes were ranked according to adjusted p-values and interestingly, genes associated with cellular metabolism or stress responses comprised 12 of the top 20 protein-coding genes (Supplementary Figure 2B-I). This was also the case with transplanted REH cells, where 14 of the top 20 genes where associated with metabolism or stress response (Supplementary Figure 2B-II), supporting our hypothesis that metabolic adaptation is a key feature of successful CNS engraftment. Further analysis using gene set enrichment algorithms (GSEA) identified positive correlation of lipid and lipoprotein metabolism signatures in CNS-derived cells, with particular propensity towards fatty acid synthesis (Figure 1B I and III). On the other hand, oxidative phosphorylation processes, linked to fatty acid degradation, were negatively correlated with CNS involvement and enriched in SEM cells from the spleen of transplanted mice (Figure 1B II-III). The same pattern was observed in mice transplanted with REH cells (Supplementary Figure 2C).
Figure 1. Fatty acid synthesis-related genes are upregulated in CNS-derived ALL cells in xenograft models.
(A) Heatmap representation of hierarchical clustering of genes differentially expressed between human SEM cells isolated from CNS and spleen of xenografted mice (n=2 independent groups of 5 mice). (B)I-II Enrichment plots of metabolism of lipids and lipoproteins (REACTOME) and oxidative phosphorylation (KEGG) for SEM cell line extracted from the CNS and spleen of xenografted mice. Profile of the running ES score & Positions of the GeneSet Members on the Rank Ordered List. III Statistically significant biological functions in SEM cells isolated from CNS and spleen of xenografted mice. Single sample GSEA scores were calculated for an array of metabolic gene signatures from MSIGDB, including KEGG, Reactome and GO term signatures. p-values for positive association with a signature (enrichment) were calculated by permutation test. Plotted are signatures with significant fold-changes in enrichment between the CNS versus spleen groups (log2 scale). Red bars indicate signatures with positive log fold-change (gain) in CNS versus spleen, blue bars indicate negative log fold-change (loss) in CNS versus spleen samples. (C) Total fatty acid levels measured by GC-MS in non-leukemic human plasma and CSF samples. n=18. Bars represent mean +/- SD. #: below detection levels. (D) Total fatty acid levels measured by GC-MS in non-leukemic murine plasma and paired CSF samples. n=9. Bars represent mean +/- SD. #: below detection levels.
CSF is a fatty acid-poor compartment compared to the plasma
CNS leukemic blasts are surrounded by the CSF. Since the strongest metabolic signature reported above indicated that de novo fatty acid and lipid synthesis is upregulated when ALL cells reach the CNS, we analyzed the fatty acid content of the CSF to understand this transcriptional adaptive response. The fatty acid content of normal CSF and plasma from both non-leukemic humans and mice was characterized by mass spectrometry (Figure 1C-D). In both species, fatty acids are scarce in the CSF compared to plasma. This suggests that ALL cells require to perform de novo synthesis of fatty acids rather than rely on exogenous supply, in order to support their survival and proliferation in the CNS. Mass spectrometric analysis of mouse CSF demonstrated that all metabolic precursors for fatty acid synthesis are present in comparable levels to those in plasma (Table 1), as observed in humans (Supplementary table 1). Furthermore, precursors for fatty acid synthesis were also present in the CSF even in the presence of leukemic infiltration (Table 1).
Table 1. Precursors for de novo fatty acid synthesis in mice.
| Metabolite | CSF: Plasma ratio (control) | CSF:Plasma ratio (leukemia) |
|---|---|---|
| Glucose | 0.22(0.13 – 0.33) | 0.24 (0.16 – 0.46) |
| Glutamine | 0.88(0.62 – 1.08) | 0.90 (0.62 – 1.38) |
| Lactate | 0.19(0.11 – 0.28) | 0.19(0.12 – 0.23) |
| Alanine | 0.20(0.14 – 0.24) | 0.18(0.10 – 0.26) |
| Pyruvate | 0.87(0.53 – 1.59) | 1.05(0.59 – 2.92) |
LC-MS quantification of key nutrients extracted from CSF and plasma of healthy (Control, n=8) and NSG mice and mice transplanted with the SEM cell line at clinical endpoint (n=6).
SCD1 is highly expressed in human CNS leukemic blasts
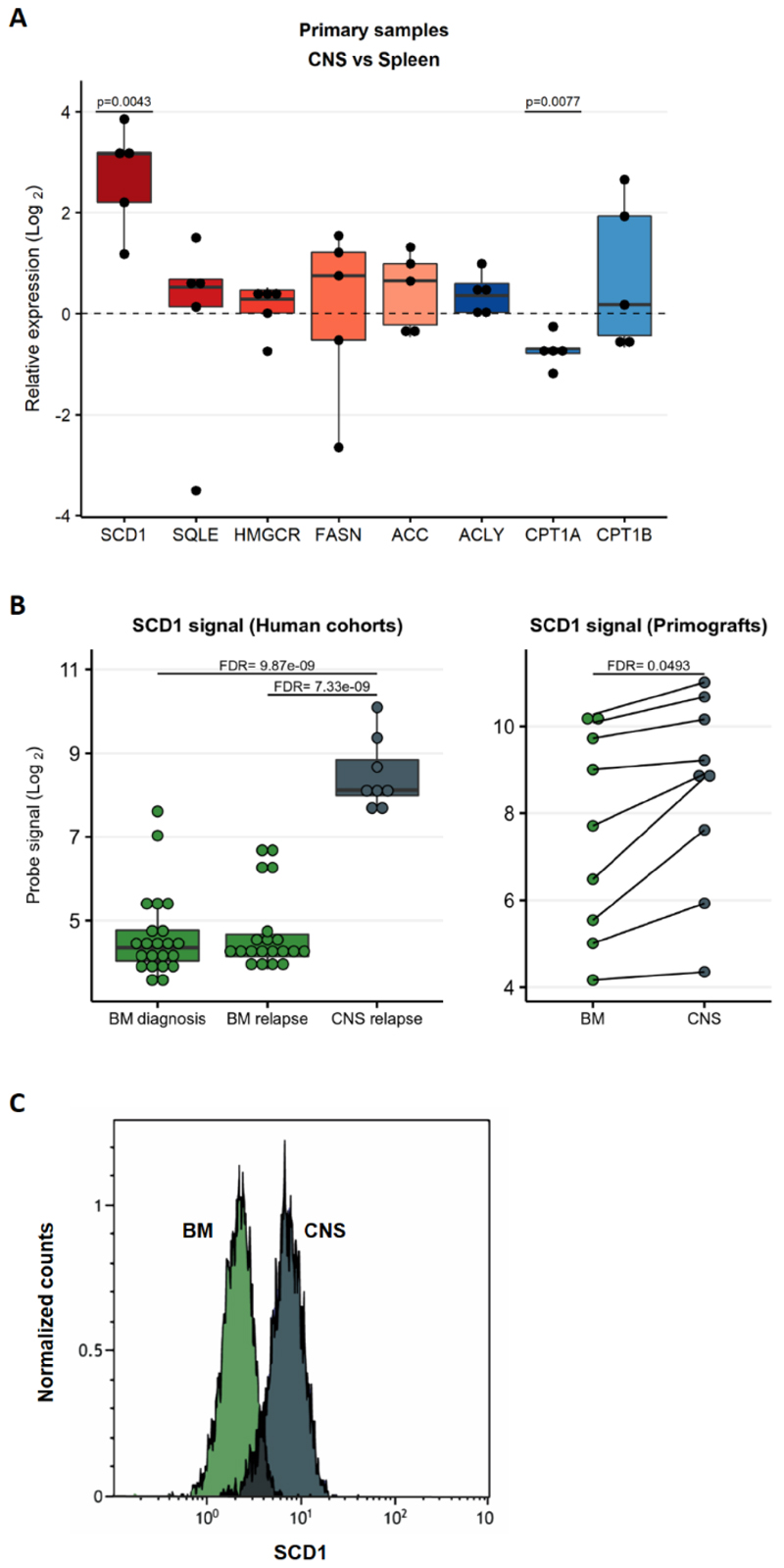
Lipid anabolism is a pivotal cellular process converting nutrients into building blocks for membrane biogenesis and for generating signaling molecules. The main lipid metabolic genes and pathways, for both fatty acid synthesis and degradation, are depicted in Supplementary Figure 3. To examine if upregulation of de novo fatty acid synthesis, was a generic adaptation to the CNS microenvironment in ALL, we expanded our investigation to primary patient samples and additional ALL cell lines. Expression levels of a panel of genes were measured by qPCR in blasts collected from CNS and spleen of mice transplanted with 5 ALL patient-derived samples (3 with t(12;21) ALL and 2 with t(4;11)) (Figure 2A). Further validation was obtained from an independent cohort of mice transplanted with REH and SEM cells (Supplementary Figure 4A) and with a third transplanted human cell line, 018z with known high CNS-tropism9,27 (Supplementary Figure 4B). Furthermore, analysis of two publicly available gene expression datasets from ALL patients14,15 confirmed a lipid biosynthetic signature in CNS ALL compared with bone marrow (Figure 2B, Supplementary Figure 5). The first dataset compared ALL cells from patients with CNS relapse to ALL cells from the bone marrow at diagnosis and after relapse (GSE60926)15.The second dataset compared primary samples xenografted in mice and collected from the CNS or bone marrow (GSE89710)14. Altogether, our results derived from cell lines and patients confirmed upregulation of genes involved in fatty acid synthesis and downregulation of oxidative phosphorylation genes in the CNS. In particular, SCD1 was consistently and strongly upregulated in all data sets (Figures 2A-B and Supplementary Figures 4A-B). Furthermore, SCD1 protein levels were high in 018z and SEM cells derived from the CNS compared to bone marrow derived cells (Figure 2C and Supplementary Figure 4C).
Figure 2. SCD1 is upregulated in primary patient samples in the central nervous system.
(A) Quantitative PCR validation of top ranked genes from RNA-Seq in n=5 patient derived xenografts with t(12;21) ETV6-RUNX1 (TEL-AML1) translocation or t(4;11) MLL-AFF1 (MLL-AF4)] translocation. Results are normalized to 36B4 human housekeeping gene and presented as Log2 fold change enrichment comparing CNS to spleen. p (two tailed) = One sample T and Wilcoxon test. (B) Gene expression of SCD1 from RNAseq data deposited in public databases. Left: Samples of BM of patients at diagnosis and relapse and CNS at relapse, unpaired analysis. Right: Patient-derived xenograft samples established by transplantation of patients’ ALL cells onto NSG mice. SCD1 expression in cells isolated from paired CNS and spleen is shown in the graph. FDR – False discovery rate. (C) Intracellular staining of SCD1 in cells from the BM (green) and CNS (grey) of a mouse transplanted with 018z cells. The peaks are relative to the percentage of human CD19+ cells normalized to mode.
Increased SCD1 expression enhances CNS infiltration in ALL
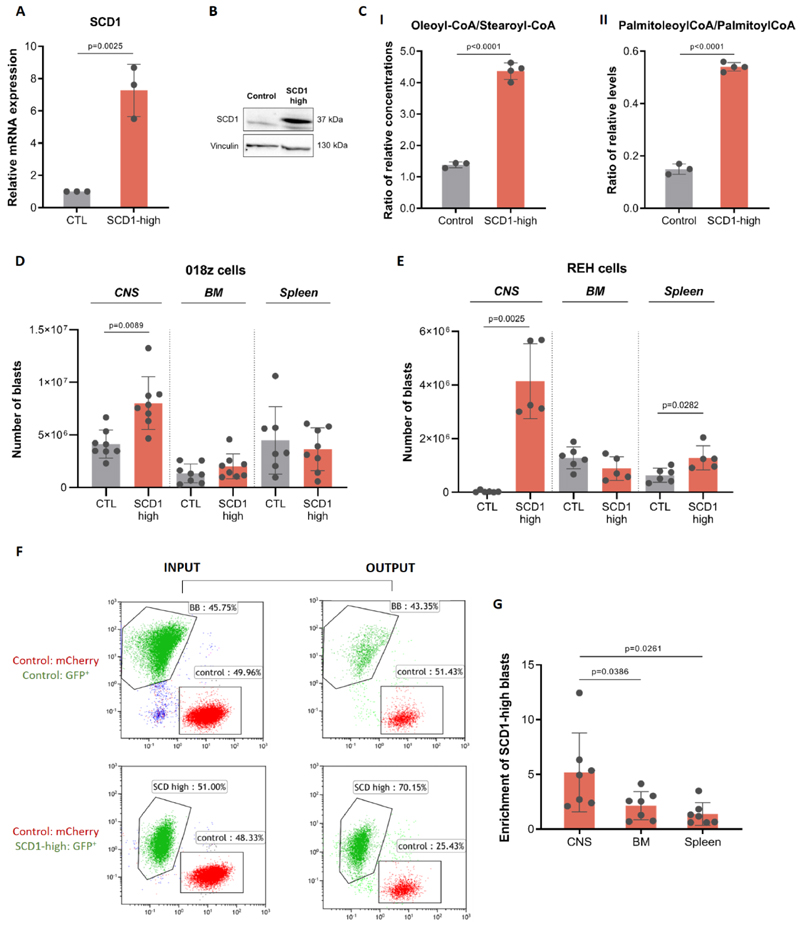
SCD1 is responsible for the generation of mono-unsaturated fatty acids via the addition of a double bond in the 9th position of extracellular-derived and de novo synthesized saturated fatty acyl-CoAs. This leads to sequential synthesis of different classes of fatty acids, eventually resulting in either triglyceride storage in lipid droplets or phospholipid production for biomass (Supplementary Figure 3). Interestingly, our analysis of the fatty acid composition of the CSF demonstrated that it is very low in fatty acids in general and extremely poor in oleic and palmitoleic acids, the main products of SCD1 (Figure 1C-D). Given the requirement to overcome a lipid poor microenvironment, we hypothesized that increased SCD1 expression would provide a relative growth or survival advantage to the cells in the CNS. To test whether upregulation of SCD1 supports CNS leukemia tropism, we overexpressed SCD1 in leukemic cells. SCD1 overexpression in 018z cells was confirmed by qPCR and immunoblotting (Figure 3A-B). Relative SCD1 activity in cells was assessed by the ratio of mono-unsaturated fatty acyl-CoAs to their saturated precursors, i.e. the ratio of oleoyl-CoA to stearoyl-CoA and the ratio of palmitoleoyl-CoA to palmitoloyl-CoA. The ratio between these mono-unsaturated to saturated fatty acyl-CoAs increased in the SCD1 overexpressing 018z cells, confirming increased SCD1 activity (Figure 3C I-II). The increased ratio of unsaturated/saturated fatty acyl-CoAs in SCD1 overexpressing cells mirrored the ratio of these fatty acids either as free fatty acids or as lipid esterified pools (Supplementary Figure 6A-B).
Figure 3. SCD1 overexpression confirms a competitive advantage for “SCD1-high” cells in the CNS microenvironment.
(A) SCD1 gene expression levels following SCD1 overexpression in 018z cells. (B) Western blot of SCD1 protein after overexpression in 018z cells. (C)I-II Ratio of relative levels of oleoyl-CoA/stearoyl-CoA and of palmitoleoyl-CoA/palmitoyl-CoA in control (CTL) and SCD1-high 018z cells. p=Student’s t-test. (D) Human leukemia burden in 018z xenograft model. Total amount of leukemic cells in CNS, BM and spleen of NSG mice xenografted with human 018z ALL cell line overexpressing SCD1 (SCD1-high) or control (CTL) GFP+ at the time of sacrifice. (E) Human leukemia burden in REH xenograft model. Total amount of leukemic cells in CNS, BM and spleen of NSG mice xenografted with human REH ALL cell lines overexpressing SCD1 (SCD1-high) or control (CTL) GFP+ at the time of sacrifice. p=paired Student’s t-test. (F) Competition assay in vivo: FACS plots of cells injected (INPUT) and cells isolated from the CNS of the mice at sacrifice (OUTPUT). Top: GFP+ control cells transduced with a GFP-carrying lentiviral backbone and mCherry+ control cells transduced with a mCherry carrying lentiviral backbone injected in a ratio of 1:1. Bottom: GFP+ SCD1 overexpressing (SCD1-high) cells and mCherry+ control cells transduced with a mCherry carrying lentiviral backbone injected in a ratio of 1:1. (G) Ratio of total number of SCD1-high 018z cells (GFP+) to control 018z cells (mCherry+) in CNS, BM and spleen of NSG mice transplanted with a mixture of the two cell types in a ratio of 1:1. The backbone vector for SCD1 overexpression was used as control vector. p=paired Student’s t-test.
In vivo, mice transplanted with SCD1-high 018z cells demonstrated significantly increased disease burden in the CNS but not in the bone marrow or spleen at experimental endpoint (Figure 3D). This correlated with a faster onset of CNS disease manifested by a clinical phenotype of earlier hind limb paralysis compared to controls (day 15 in SCD1-high cells as opposed to day 18-21 in control cells – Supplementary Video 1). Analysis of free fatty acids ratios in the CNS-engrafted cells demonstrated increased SCD1 activity in overexpressing vs. control 018z cells (Supplementary Figure 7A).
SCD1 overexpression in REH cells with generally lower propensity for CNS tropism (Supplementary Figure 7B) also induced rapid CNS engraftment at 3 weeks, a time at which control REH cells do not invade the CNS (Figure 3E). Mice transplanted with SCD1-high REH cells showed the same phenotype of earlier hind limb paralysis and accumulation of leukemic cells in the CNS seen with the more naturally CNS-tropic 018z cells. Control REH transplanted mice did not show any CNS engraftment at 3 weeks. SCD-1 high REH cells also showed a less pronounced but statistically significant increase in spleen tumor load and comparable tumor load in the bone marrow (Figure 3E).
To directly test if high SCD1 expression confers a competitive advantage to the formation of CNS ALL we labelled the SCD1-high 018z cells with GFP and the control 018z cells with either GFP or mCherry. While GFP expression in control cells did not provide any advantage in CNS colonization compared to control cells expressing the mCherry, the GFP-labelled SCD1-high cells showed a clear competitive advantage in the CNS but not in the periphery (Figure 3F-G).
SCD1 inhibition decreases CNS leukemia burden
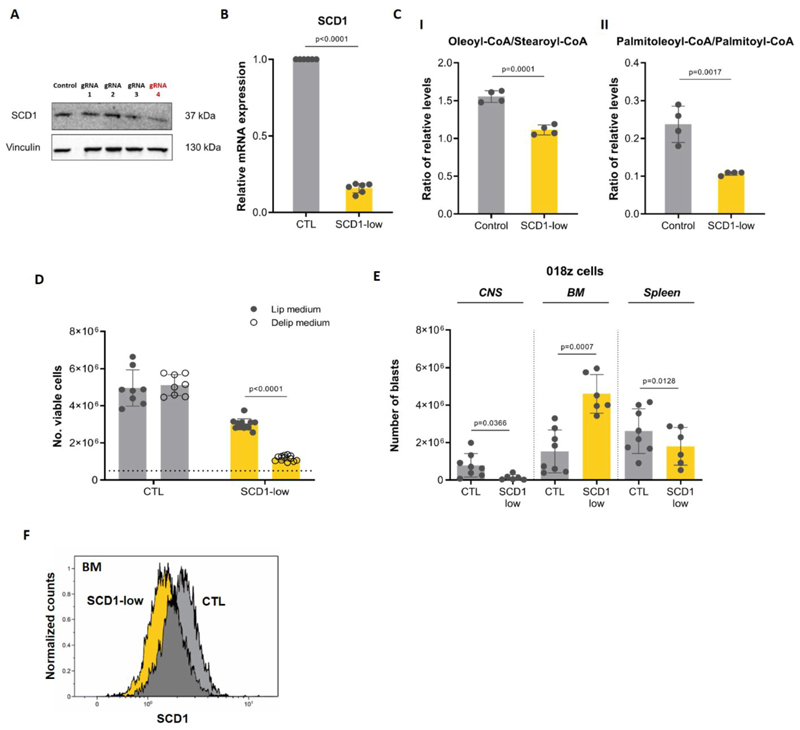
As SCD1 controls a key step in fatty acid synthesis, we hypothesized that blocking the activity of the enzyme would impair ALL survival and/or proliferation in the lipid-deprived CNS microenvironment. CRISPR/CAS-9 technology was used to ablate SCD1 in CNS-tropic 018z cells. Several guide RNAs were screened and gRNA4 resulted in the most pronounced decrease in SCD1 protein (Figure 4A) consistent with decreased mRNA transcripts (Figure 4B). To examine if SCD1 affects cell growth in lipid poor environments similar to the CSF, we cultured the cells in media containing delipidated serum. Delipidation itself did not affect the concentration of common nutrients in the serum (Supplementary Figure 8). SCD1-low cells cultured in medium supplemented with the delipidated serum showed decreased proliferation compared with cells cultured in full medium, while proliferation of control cells was not affected (Figure 4C). A significant decrease in the oleoyl-CoA to stearoyl-CoA ratio, and palmitoleoyl-CoA/palmitoyl-CoA ratio (Figure 4D I-II) indicated that SCD1 enzymatic activity was indeed decreased in SCD1-low cells. The decrease in the ratio of monounsaturated fatty acyl-CoAs to their saturated precursors reflected the decreased ratios of free and lipid esterified fatty acids following SCD1 downregulation (Supplementary Figure 6C-D).
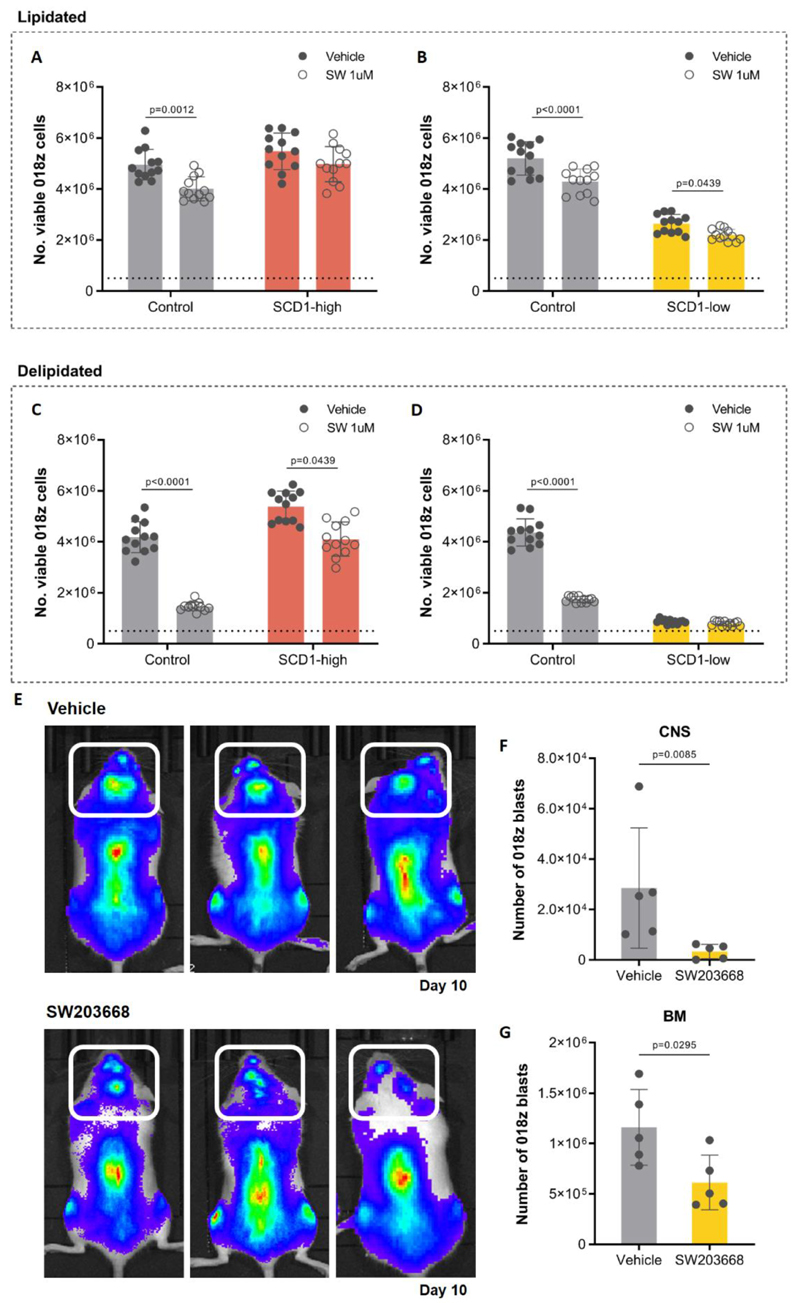
Figure 4. SCD1 ablation decreases 018z cells engraftment in the CNS.
(A) Western blot analysis of SCD1 after CRISPR-Cas9 gene ablation in 018z cells. Each lane represents a different gRNA used for the initial screening. gRNA4 decreased SCD1 protein expression while gRNA1-3 did not affect its levels. (B) Gene expression level of SCD1 after CRISPR-Cas9 knockout in 018z cells by gRNA4. (C)I-II Ratio of relative levels of oleoyl-CoA/stearoyl-CoA and of palmitoleoyl-CoA/palmitoyl-CoA in control (CTL) and SCD1-low 018z cells. p=Student’s t-test (D) In vitro proliferation of SCD1-low and control (CTL) 018z cells after 96 hours in in medium supplemented with 10% lipidated or delipidated FBS. The dotted line represents the initial number of cells plated at T0. p=two-way ANOVA. (E) Human leukemia burden in 018z xenograft model. Total amount of leukemic cells in CNS, BM and spleen of NSG mice xenografted with human 018z ALL cell lines SCD1-low or control GFP at the time of sacrifice. p=paired Student’s t-test. (F) Intracellular staining of SCD1 in human cells isolated from the BM of mice transplanted with SCD1-low and control (CTL) 018z cells. The scramble vector for SCD1 downregulation was used as control vector. The peaks are relative to the percentage of human CD19+ cells normalized to mode.
Importantly, the tumor load in the CNS was significantly decreased in mice engrafted with SCD1-low cells compared to control (Figure 4E). This was not associated with a general decline in biological fitness of the cells as decrease in cell number was not consistent among all tissues, as indicated by an elevated engraftment of SCD1-low cells in the bone marrow compared with the control. Clonal escape from SCD1 downregulation in the bone marrow was excluded by measuring SCD1 protein expression in leukemic cells engrafting the bone marrow, which confirmed ongoing suppression (Figure 4F).
To further confirm the dependency on SCD1 under lipid deprived conditions using a pharmacologic approach, 018z cells were cultured in lipid rich or lipid deprived conditions in the presence or absence of the SCD1 inhibitor SW20366828. While the inhibitor caused a slight decrease in proliferation of control cells in full serum, the effect of SCD1 inhibition was more pronounced when cells were cultured in delipidated serum (Figure 5A-D). This decrease in cell number was mitigated in SCD1 overexpressing cells (Figure 5A, 5C) and as expected, SCD1 inhibition showed minimal or no effect in SCD1-low cells (Figure 5B, 5D).
Figure 5. SCD1 pharmacological inhibition decreases 018z cells engraftment in the CNS.
In vitro proliferation of “SCD1-high”, “SCD1-low” and matching control (CTL) 018z cells after seeding 0.5x106 cells/well for 96 hours in medium supplemented with 10% lipidated (A-B) or delipidated (C-D) FBS treated with 1μM of the SCD1 inhibitor SW203668 or vehicle (DMSO).The dotted line represents the initial number of cells plated at T0. p=two-way ANOVA. (D) GFP+mCherry+FFLuc+ 018z cells were injected intravenously to NSG mice and treated from day 1 to day 10 with the SCD1 inhibitor SW203668 or vehicle (n=5 group). Representative bioluminescence of the tumor load at the time of sacrifice in three different pairs of mice, top: vehicle treated mice; bottom: drug treated mice. Decrease in the tumor load is clear in the spine and the skull area (CNS – marked with white box). Total amount of leukemic cells in CNS (E) and BM (F) of NSG mice xenografted with human GFP+mCherry+FFLuc+ 018z and treated with SW203668 for 10 days at the time of sacrifice. The backbone vector for SCD1 overexpression was used as control vector for SCD1-high and the scramble vector was used as control for SCD1-low. BM – bone marrow. p=Student’s t-test.
In order to explore whether SCD1 inhibitors could be a therapeutic option to prevent CNS relapse, we treated transplanted mice with the above mentioned SCD1 inhibitor. To better monitor the localization of engrafted cells, 018z cells were transduced with a lentivirus expressing the luciferase reporter gene. Following transplantation, mice were treated with SCD1 inhibitor for 10 days after which bioluminescent imaging was used to assess leukemic burden. This was followed by CNS and bone marrow blast counts as before. Both whole-body imaging and assessment of blasts in different tissue sites demonstrated a general impairment of 018z engraftment in transplanted mice (Figure 5E), decreasing the tumor load both in the bone marrow and CNS (Figure 5F-G). It should be noted however that the decrease in disease burden was greater in the CNS (88.31% decrease) than the bone marrow (47.21% decrease).
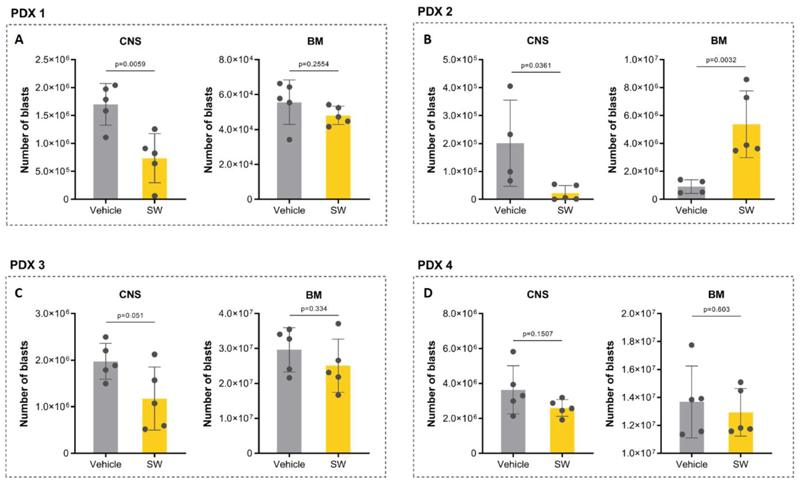
Moreover, we extended our experiments to primary ALL samples (patient-derived xenografts – PDXs). We selected 4 PDXs with variable but overall fast and aggressive CNS phenotype (PDXs features are summarized in Supplementary Table 2). Treatment with SW203668 (20 mg/kg-daily) started 1 week after the transplant. The treatment was not associated with systemic toxicity as confirmed by no changes in body weight of treated mice (Supplementary Figure 9). At sacrifice, treated mice from all PDX groups showed a consistent reduction in CNS tumor load compared with respective vehicle control groups, though not all reached statistical significance (Figure 6A-D). Importantly, in all PDX models, the first mouse to reach endpoint with signs of sickness was from the vehicle treated group. In 3 of the 4 PDX groups (PDXs 1, 3, 4) there was no significant effect on BM engraftment (Figure 6A, 6C-D), while PDX 2 showed increased BM tumor load upon SCD1 inhibitor treatment (Figure 6B), similar to the effect observed in 018z SCD1-low model (Figure 4E). Taken together, these results confirm that CNS leukemia is sensitive to SCD1 pharmacological inhibition pointing to SCD1 inhibition as a specific therapeutic strategy.
Figure 6. SCD1 pharmacological inhibition decreases patient-derived xenograft (PDXs) engraftment in the CNS.
Human leukemia burden in PDXs. Cells from 4 different PDXs were injected intravenously into NSG mice. Xenografted mice were treated daily from day 7 with the SCD1 inhibitor SW203668 or vehicle (n=5 group) at 20 mg/kg. The total amounts of leukemic cells in CNS and BM of NSG mice at the time of sacrifice are plotted in the graphs (A-D). p=Student’s t-test.
Discussion
Metabolic adaptation to environmental settings is a key hallmark of cancer, and therefore is potentially exploitable therapeutically. In this work, we used leukemic cells residing in the bone marrow and CNS to investigate metabolic changes occurring in a single cancer in two different compartments in vivo. These two regions are substantially different in their nutrient availability in general and fatty acid availability in particular.
Optimal management of CNS leukemia remains a clinical challenge with current treatments producing high levels of neurotoxic side effects and lacking efficacy in a subset of patients. An understanding of the mechanism by which leukemic blasts survive in the nutrient poor CNS is an important step to designing targeted therapies with improved toxicity profiles. Work to date using preclinical models has identified that CNS entry of leukemic blasts is relatively unrestricted and that survival in this hostile environment is likely to be the key determinant of relapse risk9,17,29. Therefore, we investigated adaptations of leukemic blasts to the CNS niche in order to identify potential vulnerabilities.
Our transcriptional data show that leukemic adaptation to the CNS is primarily metabolic with alterations in fatty acid synthetic pathways being particularly prominent. The SCD1 gene was identified as a consistently upregulated target in several complementary datasets. These findings are supported by the very low levels of fatty acids in CSF compared to plasma. Fatty acids are used by the cells catabolically for oxidation in order to generate energy, and anabolically, to produce triglycerides, phospholipids, and cholesterol esters, which serve as important sources of structural components and biomass. Monounsaturated fatty acids can either be taken up from the environment or produced de novo in a process involving SCD1. Interestingly, cancer cells under hypoxic conditions tend to bypass SCD1 and preferably import fatty acids30. While the CSF is relatively low in oxygen, it is also an extremely poor source of fatty acids, so cancer cells in the CSF may have a particular dependence on de novo synthetic pathways.
Upregulation of SCD1 expression has been reported in primary tumors in breast31, lung32, kidney33, gastrointestinal tract34 and prostate35 as well as chronic myeloid leukemia36 and B-cell lymphoma5,15,37. Our findings in ALL are strongly supported by a recent study investigating potential biomarkers for risk of CNS relapse15 which identified upregulated SCD1 and osteopontin expression in bone marrow blasts of patients who went on to experience a CNS relapse. Our work demonstrates a direct functional role for SCD1 in the CNS microenvironment mechanistically linked to the reduced availability of fatty acids in CSF compared to plasma.
SCD1 inhibitors have been developed by the pharmaceutical industry primarily for treatment of steatohepatitis and obesity. However, interest has increased recently in their use as anti-cancer agents38. Using a systems biology approach to predict selective drug targets on the basis of cancer metabolic networks, a specific dependence of cancer cells on lipid metabolism enzymes was identified39. This model predicted that targeting enzymes involved in these processes should selectively interfere with the growth of cancer cells without overt toxicity towards normal tissues. There is evidence of pre-clinical efficacy in a variety of tumor models, including Burkitt lymphoma and acute myeloid leukemia37. Although previous studies have reported side effects in mouse models40,41, including eye mucosal dysfunction, squinting and skin barrier dysfunction, they can be considered of less clinical severity compared to the wide spectrum of severe symptoms associated with commonly used chemotherapeutic agents such as heart failure and broader systemic toxicity. Of note, we did not detect any gross toxic effects in mice treated with SW203668. Therefore, the potential benefits of SCD1 anti-cancer properties may outweigh their risk of side effect.
Another possible concern is the potential adaptive metabolic rewiring of cancer cells to circumvent metabolic inhibitors. For example, a recent study identified activation of an alternative desaturation pathway from palmitate to sapienate in response to SCD1 inhibition in hepatocellular carcinoma cells42. There is also evidence that activation of autophagy may act as a resistance mechanism for SCD1 inhibition in colorectal cancer cell lines43. This suggests that multi-drug approaches may be needed to prevent compensatory “re-wiring” in the face of metabolic inhibitors.
Importantly, several lines of evidence from other works suggested that the metabolic adaptations we observed in the CNS are not “hardwired” as genetic mutations or selection of clones with pre-existing high levels of expression17,29. Rather, they are reversible adaptive changes to a novel microenvironment. This of course raises the possibility that cells may also be able to escape SCD1 inhibition by moving to a new microenvironment rich in exogenous lipids. Indeed, this may explain the variable effect of SCD1 inhibition on leukemic cells in the BM (Figure 6). Nevertheless, the bone marrow disease burden can be dealt with using other therapeutic approaches, while the SCD1 inhibition might be used to eliminate CNS involvement. The degree of dynamic movement in and out of the CNS by leukemic blasts is currently unknown. However, in childhood ALL, CNS relapse is rarely truly “isolated” and sub-microscopic bone marrow involvement is detected with sensitive molecular techniques in the majority of cases44.
The concept of metabolic heterogeneity in tumors until now has mainly focused on three areas. Firstly, the influence of different tumor-initiating genetic lesions in the same primary cancer type5,45. Secondly, differences between solid tumors originating in different tissues e.g. lung vs. breast45. Thirdly, the influence of local hypoxia or differences in nutrient availability as tumors enlarge in their primary sites46. All of these local effects can “prime” tumors to develop drug resistance or develop capacity to metastasize to distant sites47,48. In this paper, we highlight a fourth area – metabolic plasticity demonstrated by cancer cells when entering a new tissue microenvironment. This has important implications for therapeutic targeting of metabolism in the setting of metastatic disease or disseminated hematological malignancies and supports the use of multi-agent therapy. Moreover, it adds a further layer of complexity to the already recognized issue of intra-tumoral genetic heterogeneity49.
Methods
Cell culture
REH [t(12;21) ETV6-RUNX1 (TEL-AML1)] and SEM [t(4;11) KMT2A-AFF1 (MLL-AF4)] cell lines were bought from ATCC. 018z [(47, XY, +8, del(9)(p13))], were a kind gift of Dr. L.H. Meyer from Ulm University. 018z cells were maintained in RPMI supplemented with 20%FBS, L-glutamine 2mM, non-essential amino acids (0.1 mM glycine, L-alanine, L-asparagine, L-aspartic acid, L-glutamic acid, L-proline, L-serine) (Gibco-BRL), and sodium pyruvate 1 mM (Sigma). REH and SEM cells were maintained in Glutamax 25mM glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) FBS (Life Technologies), 1% penicillin/streptomycin solution. Cells were cultured at 37°C at 21% oxygen in a 5% CO2 incubator and were regularly monitored for mycoplasma contamination. Unless otherwise stated, all cell culture reagents were purchased from Thermo Fisher Scientific (Life Technologies).
SCD1 Knockout
CRISPR/Cas9 technology was used to knockout SCD1. Exon 3 of the SCD1 gene was targeted by designing guide RNAs (gRNA) using the publicly available software tool (crispr.mit.edu:8079/). Seven top-scored gRNAs were cloned into pLentiCRISPR.EFS.GFP (Addgene plasmid #57818, a gift from Benjamin Ebert, Division of Hematology, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston) according to published protocols50. Following cloning, plasmids were transformed into compatible stable bacteria. Single colonies were picked, and extracted plasmids were sequenced. For efficiency validation, a surveyor assay was performed using GUIDEIT validation kit (Clontech Laboratories) and the most-efficient gRNA was selected for virus production (Strand guide sequence: 5’-GGCCGAGCTTTGTAAGAGCGG-3’). 018z cells were then transduced and 3 days later were sorted for GFP and seeded for 7 days, with media changes every 72 hours. The scramble vector (pLentiCRISPR.EFS.GFP) was used as the control vector.
Verification of Endogenous SCD1 Gene Editing
Cells were transduced with the pLentiCRISPR.EFS.GFP construct targeting SCD1. Cells positive for GFP were sorted and cultured for 7 days. Genomic DNA was extracted from the sorted population and from wild type 018z cells with the QIAamp DNA Mini Kit (Quiagen), and CRISPR target regions amplified using appropriate locus-specific primers (FW: 5’-AGCCTGACGAAGACAGTTTCT-3’; REV: 5’-TGCTTTATGGACTTAAGGACTG-3’). Standard PCR conditions were used with Taq Ready mix (HyLabs) and 100 ng of genomic DNA per the manufacturer’s instructions for 35 cycles. The correct size of the PCR product was checked by agarose gel electrophoresis. PCR products were then purified with Wizard SV Gel and PCR Clean-Up System Protocol (Promega) and then Sanger-sequenced. RNA was extracted from 018z cells after sorting using Direct-zol RNA MiniPrep Kit (Zymo Research) and cDNA was synthesized using qScript cDNA Synthesis Kit. CRISPR target regions amplified using appropriate locus-specific primers were sequenced and compared using TIDE software.
SCD1 overexpression
Cloning of SCD1 gene was performed with standard cloning protocols. All fragments amplifications were carried out using Phusion High-Fidelity PCR Master Mix (Finnzymes, Espoo, Finland) in touchdown PCR program to account for different melting temperature of primers. Amplified fragments were purified directly from PCR reaction using the Wizard SV Gel and PCR Clean-up System (Promega). Fragments and vectors were digested using restriction enzymes according to manufacturer’s protocols (NEB Ipswich, MA) and purified after gel electrophoresis using the Wizard SV Gel and PCR Clean-up System (Promega). Quick ligation kit (NEB Ipswich, MA) was used to ligate the vectors and inserts. The ligations were transformed to NEB 5-alpha F’ Iq Competent E. coli suitable for toxic genes (NEB Ipswich, MA) according to the manufacturer’s protocol.
pCDH-EF1a-MCS-T2A-copGFP vector was purchased from SBI system biosciences (Mountain View, CA) to allow for efficient bi-cistronic expression51. SCD1 previously cloned from cDNA was amplified with primers carrying restriction enzymes sequences as following: NheI for 5′ amplification (5′-GCTAGCGCCACCATGCCGGCCCACTTGCTG-3′) and Bam HI for 3′ amplification (5′-GGATCCGCCACTCTTGTAGTTTCCATC-3′). Purified inserts and the pCDH-EF1a-MCS-T2A-copGFP vector were digested and with NheI and BamHI restriction enzymes and ligated to form pCDH-EF1a-SCD1-T2A-copGFP. The backbone of the vector (pCDH-EF1a-MCS-T2A-copGFP) was used as the control vector.
Patient samples
Primary patient material was obtained from approved biobanks. ALL live cells from diagnosis – Bloodwise Childhood Leukaemia Cell Bank, normal control CSF – Glasgow Neuroimmunology Biobank, ALL patient CSF – West of Scotland CSF Biobank, plasma – NHS Greater Glasgow & Clyde Bio-repository. Use of biobanked human samples for this specific project was approved by the West of Scotland Research Ethics Service (ref 09/S0703/77). All research was conducted in accordance with the Declaration of Helsinki.
Xenotransplants
JAX NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG; Charles River Laboratories, Harlow, United Kingdom) mice were kept in sterile isolators with autoclaved food, bedding, and water. Xenotransplants were performed in 6-10 weeks old mice, the experimental protocol varied according to the model, with details given in Supplementary Table 3. At the end of experiment, mice were sacrificed by overdose of pentobarbital (IP) and cells were collected from bone marrow, spleen and leptomeninges as previously described17. For RNA sequencing leukemic cells were purified using a Ficoll gradient (Lymphoprep, Stemcell) following the supplier instructions. Purity was determined using flow cytometry. Cells were dry pelleted and snap frozen for further use (see below). For SCD1 overexpression and inhibition experiments bone marrow, spleen and leptomeningeal cells were analyzed to check human engraftment by FACS using Absolute counting beads (Beckman Coulter). GFP was used to identify human cells. The tumor load was expressed as total amount of leukemic cells in each organ. Competition assays were performed by transplanting SCD1 overexpressing and control cells in the same mouse in a 1:1 ratio (1.25x106 cells/type, total 2.5x106 cells/mouse). Patient’s samples (PDXs) were expanded in NSG mice and experiments were performed upon secondary-tertiary passage in the mice. 1x106 cells from donor mice (90% human engraftment) were transplanted in recipients to perform the experiments.
All animal experiments were approved by Institutional Ethical Review Process Committees and were performed under UK Home Office license (PPL 60-4512), Israel Institutional Animal care and use committee approval (1074/16/ANIM) and Institutional Animal Care and use Committee at Memorial Sloan Kettering Cancer Center (protocol 11-10-025).
FBS delipidation using fumed silica
The protocol for delipidation of FBS was adapted from Agnese et al. (1983)52. Lipid depletion was achieved by adding two grams of fumed silica (Sigma Aldrich) to 100 mL of FBS, and the suspension was stirred for 2h30. Centrifugation at 5000 rpm for 35 minutes, at room temperature. The supernatant was filtered through a 0.45 μm filter at first, and then through a 0.22 μm filter.
In vitro treatment with the SCD1 inhibitor SW203668
At day 0, 0.5x106 cells were seeded in RPMI supplemented with lipidated or delipidated FBS. A concentration of 1 μM of the SCD1 inhibitor SW203668 or of the vehicle (DMSO) was added daily, over 96h.
In vivo treatment with the SCD1 inhibitor SW203668
Ten 6-week old NSG females were transplanted with 1.5x106 GFP+mCherry+FFLuc+ 018z cells. Treatment started 24 hours after transplantation with five mice receiving vehicle (10% DMSO+ 10% Kolliphor (Sigma Aldrich) + 80% lactic acid pH 5.5) and 5 other mice receiving the small molecule SCD1 inhibitor SW203668 (Cayman Chemical) at 20 mg/kg twice a day I.P. for 10 days. In vivo bioluminescence assay (IVIS Lumina LT, Series III, Caliper Life Sciences) was performed at day 10, immediately before sacrifice. Secondary and tertiary PDXs were transplanted into 6 weeks old NSG female mice at a dose of 1x106 cells. The treatment with 20 mg/kg SW203668 started 1 week after the transplant. Mice were sacrificed when they showed the first signs of sickness. Details on time of treatment for each PDX are displayed in Supplementary Table 3. Bone marrow, spleen and meninges were collected post-mortem and analysed to check human engraftment by FACS using absolute counting beads (Beckman Coulter). GFP expression was used to identify human cells. The tumor load was expressed as total amount of leukemic cells in each organ.
SCD1 intracellular staining
Cells isolated from bone marrow and CNS of mice transplanted with human leukemic cells were fixed with PFA 1.6% and permeabilized with ice-cold 90% methanol. GFP was used to gate on human cells. Rabbit anti human monoclonal SCD1 antibody (SCD1 (C12H5) Rabbit mAb #2794, Cell Signaling) was used at a dilution of 1:50 in PBS for 20 minutes at 4°C.
Western blot
SCD1-high and low cells were cultured in RPMI supplemented with 20% FBS and harvested in PBS, then pelleted and lysed in CelLytic™ MT Cell Lysis Reagent (Sigma Aldrich) supplemented with complete mini protease cocktail (11836153001 Roche) and phosphatase inhibitor cocktail (200-664-3 Sigma Aldrich). Cells were incubated on ice for 30 minutes before clarification. Standard procedures were used for western blotting. Fifty μg of total protein lysate were loaded on a 12% Polyacrylamide gel and incubated overnight with rabbit anti-human monoclonal SCD1 antibody 1:2000 (SCD1 (C12H5) Rabbit mAb #2794, Cell Signaling) and vinculin (Vinculin Antibody #4650, Cell Signaling) at 4°C. Proteins were detected using ECL Western Blotting Substrate (Promega).
CSF and plasma sampling from mice
CSF was collected from mice under terminal anesthesia with pentobarbital. A 25-gauge needle was percutaneously inserted into the cisterna magna and CSF collected by gravity. Blood was collected immediately post-mortem. Both CSF and blood were centrifuged at 2,000g for 15 min at 4°C. CSF supernatant and plasma were snap-frozen and stored at −80°C until analysis.
Gas Chromatography/Mass Spectrometry analyses
CSF or plasma samples were precipitated vol/vol with ice-cold glacial MetOH (15min, -20°C), followed by ice-cold glacial chloroform extraction (ratio 1:3). After centrifugation (13,000rpm, 5min), the chloroform fraction was extracted into GC-MS glass vials (Agilent). Samples were dried under nitrogen and stored at -20°C for subsequent use. On the day of analysis, samples were re-suspended in chloroform/MetPrepII solution to generate fatty acid methyl esters (FAMEs). Samples were analyzed using an Agilent 7890B GC system coupled with an Agilent 7000 Triple Quadrupole mass spectrometer. Phenomenex ZB-1701P columns (30 m x 0.25 mm x 0.25 μm) were used for FAME separation. Further details are given in Supplementary Table 4. Generated data files were pre-processed with Agilent Mass Hunter B.06.00 software and relative intensities of FAMEs were extracted using R-based script MetabQ53. Absolute quantities of fatty acids were calculated using the calibration curves.
Liquid Chromatography/Mass Spectrometry analyses
Plasma and CSF samples were diluted 50-fold with cold solvent, comprising 50% methanol, 30% acetonitrile, 20% water. Free fatty acids and fatty acid precursors were extracted from CNS and bone marrow cells, and from 1x106 cells from in vitro culture, by resuspending cells in 1mL of the same solvent. Samples were vortexed for 15min and then centrifuged at 16,100 x g for 10 minutes at 0-4°C. Supernatants were transferred to glass HPLC vials and kept at -80°C prior to LC-MS analysis.
Total fatty acids were extracted from 1x106 in vitro cultured cells. For lipid saponification, cells were resuspended in 1 mL of 90% methanol in water containing 0.3M of potassium hydroxide, followed by vortexing and incubation at 80°C for 60 minutes. Samples were then brought to room temperature and 100 μL of formic acid and 800 μL of hexane were added to each sample. After vortexing, the upper phase was transferred into a glass HPLC vial, and samples were re-extracted with 700 μL of hexane. The upper phase was again collected and pooled with previously collected volume. Samples were dried under nitrogen, reconstituted in 200 μL of methanol, and stored at -80°C until LC-MS analysis. Fatty acids were analyzed by a QExactive Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA, USA) together with a Thermo Ultimate 3000 HPLC system. The HPLC setup consisted of a HSS T3 column (150 x 2.1 mm, 1.8 μm, Waters Corporation, Milford, MA, USA), with a HSS T3 guard column (Waters, 1.8 μm) and an initial mobile phase of 80% 0.1% formic acid in water/ 20% 0.1% formic acid in acetonitrile. Samples (5 μl) were injected and metabolites were separated over a 7 minutes mobile phase gradient, increasing the organic mobile phase (0.1% formic acid in acetonitrile) to 95%, using a flow rate of 400 μL/min and a column temperature of 45°C. The total analysis time was 12 minutes.
For fatty acyl-CoAs extraction, 8x106 cells cultured in vitro were resuspended in 700 μL of cold methanol (-20°C). The samples were vigorously vortexed for 60 seconds and put at -20°C for 10 minutes. 500 μL of cold (-20°C) chloroform were added to the tube and the mixture was vigorously vortexed for 60 seconds. Then, 270 μL of water were added to the mixture, the samples were vortexed again and placed on ice for 10 min. Tubes were centrifuged at 4°C, 16,100 x g for 15 minutes. 500 μL of the upper layer were transferred into 1 mL glass vial and gently dried under nitrogen. The dried extracts were reconstituted in 50 μL of methanol:water (in a proportion of 1:1) and transferred to HPLC glass vials equipped with 50 μL inserts. Thermo Ultimate 3000 high-performance liquid chromatography (HPLC) system coupled to Q- Exactive Orbitrap Mass Spectrometer (Thermo Fisher Scientific) was used with a resolution of 70,000 at 200 mass/charge ratio (m/z), with electrospray ionization. Detection was at positive mode across a mass range of 600 to 1500 m/z. HPLC setup consisted HSS T3 column (150 x 2.1 mm, 1.8 μm, Waters). 5 μL of biological extracts were injected and the compounds were separated with mobile phase gradient of 12 min, starting at 90% aqueous (0.5% Ammonium hydroxide) and 10% organic (Acetonitrile with 0.5% Ammonium hydroxide) and terminated with 40% organic. Flow rate and column temperature were maintained at 0.3 mL/min and 30°C, respectively, for a total run time of 18 min. HESI source parameters: the sheath gas flow rate and the aux gas flow rate were set to 45 and 10 arbitrary units; the spray voltage was 4.5 Kv; The capillary temperature and the aux gas heater temperature were 300c and 350c respectively; the S-lens RF level was set at 50. All metabolites were detected using mass accuracy below 5 ppm. Thermo Xcalibur was used for data acquisition. TraceFinder 4.1 was used for analysis. Peak areas of metabolites were determined by using the exact mass of the singly charged ions.
Reverse transcription and quantitative Real Time Polymerase Chain Reaction (qPCR)
RNA was extracted from cells resuspended and lysed in TRIzol according to manufacturer’s instructions, and underwent DNase treatment using an RNase-free DNase kit (Qiagen). RNA quality was assessed with the RNA Nano Chip kit (Agilent) on an Agilent Bioanalyser 2100. 5 μg of total RNA from each sample was used to synthesize cDNA using the PrimeScript RT reagent kit (Promega) with oligo-dT primers, according to the manufacturer’s instructions. Quantitative PCR reactions were performed with specific primers listed in Supplementary Table 5 and the GoTaq qPCR master mix kit (Promega) using the Biorad system. Differential expressions of transcripts of interest were calculated in relation to the human 36B4 housekeeping transcript for SEM and REH cell line. For 018z cell line, quantitative PCR reactions were performed with the SYBR™ Green PCR Master Mix (Thermo Fisher) using the StepOnePlus™ Real-Time PCR System (Applied Biosystems). Human HPRT was used as a housekeeping gene.
For patient-derived xenograft samples cDNA was generated using Applied Biosystems High-Capacity RNA-to-cDNA™ Kit (Thermo Fisher, MA, #4387406). cDNA was amplified in a sequence-specific manner using a Multiplex PCR kit (Qiagen # 206143), then treated with Exonuclease I (4U/μL New England Biolabs, MA, #M0293L) to remove unincorporated primers. Samples and primers were loaded onto a 48.48 Dynamic Array™ chip. The chip was primed on a Biomark™ IFC controller MX then multiplex PCR performed using a Biomark™ system for Genetic Analysis (all Fluidigm Corporation, CA). All steps were performed as per manufacturer’s instructions. Data were analysed using the comparative CT method54.
RNA sequencing analyses
RNA samples were run on a Next Seq500 Sequencing system (Illumina). The raw fastq files containing paired-end 2 x 75bp reads were pre-processed to trim the 3’ end adapter with Cutadapt55 (version 1.5) and to trim the very low quality reads using Sickle (version 0.940) software (https://github.com/najoshi/sickle) with generous quality threshold -q 10. Only read pairs containing ≥54 bases were kept. Transcript expression quantification was performed using Kallisto (version 0.42.4) software56 against combined human and mouse transcriptomes, Ensembl GRCh38 and GRCm38, respectively, in order to remove the possible mouse RNA contamination. Read counts related to human transcripts were collected, rounded, and summarized into gene specific read counts. These were then processed with DESeq2 software57 to generate gene expression estimates after regularized log transformation which stabilized variance as a function of mean.
Metabolic enrichment
Preprocessed RNA expression data from RNA sequencing was analyzed using single sample gene set enrichment analysis (ssGSEA). Gene expression signatures for metabolic pathways were downloaded from the MSIGDB database (software.broadinstitute.org/gsea/msigdb) and included signatures from the KEGG and REACTOME pathway databases. For each signature S and each sample, a positive signature score was calculated together with a p-value pS indicating the significance of the score (permutation test, n=10,000). For a given signature S, we calculated the fold change in the expression of the signature between the two sample groups (CNS, Spleen) as the difference of the log-transformed p-values ES(sample)=-log2(pS(sample)), i.e. FC(S)=mean(ES(CNSsamples))-mean(ES(SPLEENsamples)). The fold changes were plotted in a bar plot. Student’s T-test was used to calculate significance of differential expression between the groups. Furthermore, a Pre-Ranked GSEA using fold change values was performed to display the enrichment profiles for several gene-sets (permutation test, n=1,000, classic enrichment statistic). All the regular log-transformed counts of selected genes were double-scaled. The biclustering was performed using Euclidean distance and complete linkage and the heatmap was made using the gplots package for R (v. 3.0.1)58.
For comparison, available human and primograft data was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), Refs. GSE60926 and GSE89710 respectively. Those datasets were analysed using the Affymetrix Transcriptome Analysis Console v4.0, employing Robust Multiarray Average summarization and the moderated differential expression analysis was carried out using the empirical Bayes method with FDR correction.
Histology
Murine heads and femurs were stripped of soft tissues, fixed in 10% neutral buffered formalin (CellPath, Powys, United Kingdom), and decalcified in Hilleman and Lee EDTA solution (5.5% EDTA in 10% formalin) for 2 to 3 weeks. Following paraffin embedding, hematoxylin and eosin staining (Sigma-Aldrich) was performed on 5-mm brain/femur sections. Photographs were taken using an Axio Observer Light microscope (Zeiss).
Statistical analysis
Parametric data were analyzed using two-sided Student unpaired or paired t-tests. Nonparametric data were analyzed using two-sided Mann-Whitney U tests. For wet lab experiments a p value of <0.05 was considered significant. In all figures, data are presented as Mean ± Standard Deviation (SD). Analysis was carried out using GraphPad Prism software (La Jolla, CA).
Supplementary Material
Acknowledgements
We thank the patients and their families who generously donated the samples used in this study to the NHS Greater Glasgow and Clyde Biorepository, Laboratory Medicine Building, Queen Elizabeth University Hospital (QEUH), the Bloodwise Childhood Leukaemia Cellbank, the Glasgow Neuroimmunology Biobank and the West of Scotland CSF biobank. In addition, we thank John Goodfellow, Hugh Willison, Saeeda Bhatti and Yasar Yousafzai for assistance with obtaining primary samples, Clare Orange and Lynn Stevenson for help with histology. Histology slides were scanned by University of Glasgow slide scanning and image analysis service at the QEUH. RNA sequencing was performed by the Glasgow Polyomics research facility at the University of Glasgow. We also thank Karen Keeshan and the Biological Services Unit, Cancer Research UK Beatson Institute of Cancer Research for animal assistance.
This work was supported by the Chief Scientist Office (O.O. and C.H. Grant ETM/374), Fondazione Italiana per la Ricerca sul Cancro *FIRC (A.M.S.), the William and Elizabeth Davies Foundation (A.C. – Clinical research fellowship), the Laura and Ike Perlmutter Fund (E.G. and I.A.), the German Israel Foundation (A.Z.) and the Israel Science Foundation 1775/12 (E.G., I.M.). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement META-CAN No 766214 (S.I.F., J.F-G., I.M. and E.G.).
Footnotes
Authors Contribution
A.M.S., S.I.F., O.O., E.G., C.H., P.H., S.I. designed the study. A.M.S., S.I.F., O.O., A.C., A.Z., S.B., I.G., L.F., Y.B., C.E., performed most of the experiments. A.M.S., S.I.F., O.O., A.C., P.H., E.K.M., J.G-F., C.H., I.M., E.G. analyzed and interpreted the data. S.T., I.A., J.J.K. and G.M. performed and analyzed the mass spectrometry experiments. A.M.S., S.I.F., O.O., C.H., E.G., I.M. and S.I. wrote the manuscript.
Competing interests
E.G. is a Board member and a Shareholder of Metabomed Ltd Israel, J.J.K. is an employee and shareholder of Rheos Medicines Inc. All other authors declare no potential conflicts of interest.
Data availability
RNA-seq data supporting the findings of this study have been deposited in Gene Bank (accession numbers: GSE135115; GSE135113; GSE135114). The GSE135115 SuperSeries is entitled “Gene expression profiles of MLL-AF4 and TEL-AML1 acute lymphoblastic leukaemia blasts retrieved from central nervous system and spleen”. This SuperSeries contains two series related to SEM and REH experiments as follows: GSE135113 “Gene expression profiles of MLL-AF4 acute lymphoblastic leukaemia blasts retrieved from central nervous system and spleen”, GSE135114 “Gene expression profiles of TEL-AML1 acute lymphoblastic leukaemia blasts retrieved from central nervous system and spleen”. The source data associated with each figure is provided with the manuscript. All other data supporting the findings of this study is available from the corresponding author on reasonable request.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:1–18. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha J-Y, Lee H-J. Targeting Lipid Metabolic Reprogramming as Anticancer Therapeutics. J Cancer Prev. 2017;21:209–215. doi: 10.15430/JCP.2016.21.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gisselbrecht C. Positron emission tomography – Guided therapy of aggressive non-hodgkin lymphoma: Standard of care after the PETAL study? J Clin Oncol. 2018;36:3272–3273. doi: 10.1200/JCO.18.00498. [DOI] [PubMed] [Google Scholar]
- 5.Caro P, et al. Metabolic Signatures Uncover Distinct Targets in Molecular Subsets of Diffuse Large B Cell Lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuntz EM, et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med. 2017;23:1234–1240. doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachmias B, Schimmer AD. Metabolic Flexibility in Leukemia—Adapt or Die. Cancer Cell. 2018;34:695–696. doi: 10.1016/j.ccell.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Olivares O, Däbritz JHM, King A, Gottlieb E, Halsey C. Research into cancer metabolomics: Towards a clinical metamorphosis. Semin Cell Dev Biol. 2015;43:52–64. doi: 10.1016/j.semcdb.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Frishman-Levy L, Izraeli S. Advances in understanding the pathogenesis of CNS acute lymphoblastic leukaemia and potential for therapy. Br J Haematol. 2017;176:157–167. doi: 10.1111/bjh.14411. [DOI] [PubMed] [Google Scholar]
- 10.Pui CH, Howard SC. Current management and challenges of malignant disease in the CNS in paediatric leukaemia. Lancet Oncol. 2008;9:257–268. doi: 10.1016/S1470-2045(08)70070-6. [DOI] [PubMed] [Google Scholar]
- 11.Halsey C, et al. The impact of therapy for childhood acute lymphoblastic leukaemia on intelligence quotients; Results of the risk-stratified randomized central nervous system treatment trial MRC UKALL XI. J Hematol Oncol. 2011;4:1–12. doi: 10.1186/1756-8722-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer NS, Balsamo LM, Bracken MB, Kadan-Lottick NS. Chemotherapy-only treatment effects on long-term neurocognitive functioning in childhood ALL survivors: A review and meta-analysis. Blood. 2015;126:346–353. doi: 10.1182/blood-2015-02-627414. [DOI] [PubMed] [Google Scholar]
- 13.Price RA, Johnson WW. The central nervous system in childhood leukemia: I. The arachnoid. Cancer. 1973;31:520–533. doi: 10.1002/1097-0142(197303)31:3<520::aid-cncr2820310306>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Münch V, et al. Central nervous system involvement in acute lymphoblastic leukemia is mediated by vascular endothelial growth factor. Blood. 2017;130:643–654. doi: 10.1182/blood-2017-03-769315. [DOI] [PubMed] [Google Scholar]
- 15.van der Velden VHJ, et al. New cellular markers at diagnosis are associated with isolated central nervous system relapse in paediatric B-cell precursor acute lymphoblastic leukaemia. Br J Haematol. 2016;172:769–781. doi: 10.1111/bjh.13887. [DOI] [PubMed] [Google Scholar]
- 16.Krause S, et al. Mer tyrosine kinase promotes the survival of t(1;19)-positive acute lymphoblastic leukemia (ALL) in the central nervous system (CNS) Blood. 2015;125:820–830. doi: 10.1182/blood-2014-06-583062. [DOI] [PubMed] [Google Scholar]
- 17.Williams MTS, et al. The ability to cross the blood-cerebrospinal fluid barrier is a generic property of acute lymphoblastic leukemia blasts. Blood. 2016;127:1998–2006. doi: 10.1182/blood-2015-08-665034. [DOI] [PubMed] [Google Scholar]
- 18.Williams MTS, et al. Interleukin-15 enhances cellular proliferation and upregulates CNS homing molecules in pre-B acute lymphoblastic leukemia. Blood. 2014;123:3116–3127. doi: 10.1182/blood-2013-05-499970. [DOI] [PubMed] [Google Scholar]
- 19.Buonamici S, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frishman-Levy L, et al. Central nervous system acute lymphoblastic leukemia: Role of natural killer cells. Blood. 2015;125:3420–3431. doi: 10.1182/blood-2014-08-595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spector R, Snodgrass S Robert, Johanson CE. A balanced view of the cerebrospinal fluid composition and functions: Focus on adult humans. Exp Neurol. 2015;273:57–68. doi: 10.1016/j.expneurol.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 22.Hühmer AF, Biringer RG, Amato H, Fonteh AN, Harrington MG. Protein Analysis in Human Cerebrospinal Fluid: Physiological Aspects, Current Progress and Future Challenges. Dis Markers. 2006;22:3–26. doi: 10.1155/2006/158797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damkier HH, Brown PD, Praetorius J. Cerebrospinal Fluid Secretion by the Choroid Plexus. Physiol Rev. 2013;93:1847–1892. doi: 10.1152/physrev.00004.2013. [DOI] [PubMed] [Google Scholar]
- 24.Méndez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olechnowicz SWZ, Edwards CM. Contributions of the host microenvironment to cancer-induced bone disease. Cancer Res. 2014;74:1625–31. doi: 10.1158/0008-5472.CAN-13-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhoff EM, Queudeville M, Debatin K-M, Meyer LH. A novel B cell precursor ALL cell line (018Z) with prominent neurotropism and isolated CNS leukemia in a NOD/SCID/huALL xenotransplantation model. Blood. 2009;114:1630–1630. [Google Scholar]
- 28.Theodoropoulos PC, et al. Discovery of Tumor-Specific Irreversible Inhibitors of Stearoyl CoA Desaturase. Nat Chem Biol. 2016;12:218–225. doi: 10.1038/nchembio.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartram J, et al. High throughput sequencing in acute lymphoblastic leukemia reveals clonal architecture of central nervous system and bone marrow compartments. Haematologica. 2018;103:e110–e114. doi: 10.3324/haematol.2017.174987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelucci C, et al. Pivotal role of human stearoyl-CoA desaturases (SCD1 and 5) in breast cancer progression: oleic acid-based effect of SCD1 on cell migration and a novel pro-cell survival role for SCD5. Oncotarget. 2018;9:24364–24380. doi: 10.18632/oncotarget.25273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess D, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase activity blocks cell cycle progression and induces programmed cell death in lung cancer cells. PLoS One. 2010;5:e11394. doi: 10.1371/journal.pone.0011394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, et al. High expression of Stearoyl-CoA desaturase 1 Predicts Poor Prognosis in Patients with Clear-Cell Renal Cell Carcinoma. PLoS One. 2016;11:e0166231. doi: 10.1371/journal.pone.0166231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, et al. Stearoyl-CoA desaturase-1 mediated cell apoptosis in colorectal cancer by promoting ceramide synthesis. Sci Rep. 2016;6:1–11. doi: 10.1038/srep19665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SJ, Choi H, Park SS, Chang C, Kim E. Stearoyl CoA desaturase (SCD) facilitates proliferation of prostate cancer cells through enhancement of androgen receptor transactivation. Mol Cells. 2011;31:371–377. doi: 10.1007/s10059-011-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Li H, Ho N, Li D, Li S. Scd1 Plays a Tumor-Suppressive Role in Survival of Leukemia Stem Cells and the Development of Chronic Myeloid Leukemia. Mol Cell Biol. 2012;32:1776–1787. doi: 10.1128/MCB.05672-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southam AD, et al. Drug redeployment to kill leukemia and lymphoma cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids. Cancer Res. 2015;75:2530–2540. doi: 10.1158/0008-5472.CAN-15-0202. [DOI] [PubMed] [Google Scholar]
- 38.Imamura K, et al. Discovery of Novel and Potent Stearoyl Coenzyme A Desaturase 1 (SCD1) Inhibitors as Anticancer Agents. Bioorganic Med Chem. 2017;25:3768–3779. doi: 10.1016/j.bmc.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:1–10. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazaki M, Man WC, Ntambi JM. Targeted Disruption of Stearoyl-CoA Desaturase1 Gene in Mice Causes Atrophy of Sebaceous and Meibomian Glands and Depletion of Wax Esters in the Eyelid. J Nutr. 2001;131:2260–2268. doi: 10.1093/jn/131.9.2260. [DOI] [PubMed] [Google Scholar]
- 41.Brown JM, Rudel LL. Stearoyl-coenzyme A desaturase 1 inhibition and the metabolic syndrome: considerations for future drug discovery. Curr Opin Lipidol. 2010;21:192–197. doi: 10.1097/MOL.0b013e32833854ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vriens K, et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ono A, et al. Feedback activation of AMPK-mediated autophagy acceleration is a key resistance mechanism against SCD1 inhibitor-induced cell growth inhibition. PLoS One. 2017;12:e0181243. doi: 10.1371/journal.pone.0181243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagedorn N, et al. Submicroscopic bone marrow involvement in isolated extramedullary relapses in childhood acute lymphoblastic leukemia: a more precise definition of “isolated” and its possible clinical implications, a collaborative study of the Resistant Disease Committee. Blood. 2007;110:4022–4029. doi: 10.1182/blood-2007-04-082040. [DOI] [PubMed] [Google Scholar]
- 45.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sciacovelli M, Frezza C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 2017;284:3132–3144. doi: 10.1111/febs.14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 50.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ibrahimi A, et al. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther. 2009;20:845–860. doi: 10.1089/hum.2008.188. [DOI] [PubMed] [Google Scholar]
- 52.Agnese ST, Spierto FW, Hannon WH. Evaluation of four reagents for delipidation of serum. Clin Biochem. 1983;16:98–100. doi: 10.1016/s0009-9120(83)90676-8. [DOI] [PubMed] [Google Scholar]
- 53.Tumanov S, et al. Calibration curve-free GC–MS method for quantitation of amino and non-amino organic acids in biological samples. Metabolomics. 2016;12:1–13. [Google Scholar]
- 54.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 55.Martin M, N T. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011 [Google Scholar]
- 56.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warnes GR, et al. gplots: Various R Programming Tools for Plotting Data. R package version 3.0-1. 2015 http://CRAN.R-project.org/package=gplots.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data supporting the findings of this study have been deposited in Gene Bank (accession numbers: GSE135115; GSE135113; GSE135114). The GSE135115 SuperSeries is entitled “Gene expression profiles of MLL-AF4 and TEL-AML1 acute lymphoblastic leukaemia blasts retrieved from central nervous system and spleen”. This SuperSeries contains two series related to SEM and REH experiments as follows: GSE135113 “Gene expression profiles of MLL-AF4 acute lymphoblastic leukaemia blasts retrieved from central nervous system and spleen”, GSE135114 “Gene expression profiles of TEL-AML1 acute lymphoblastic leukaemia blasts retrieved from central nervous system and spleen”. The source data associated with each figure is provided with the manuscript. All other data supporting the findings of this study is available from the corresponding author on reasonable request.