Abstract
Objective
We hypothesized that genes within recently identified loci associated with waist–hip ratio (WHR) exhibit fat depot-specific mRNA expression, which correlates with obesity-related traits.
Methods
Adipose tissue (AT) mRNA expression of 6 genes (TBX15/WARS2, STAB1, PIGC, ZNRF3 and GRB14) within these loci showing coincident cis-expression quantitative trait loci was measured in 222 paired samples of human visceral (vis) and subcutaneous (sc) AT. The relationship of mRNA expression levels with obesity-related quantitative traits was assessed by Pearson's correlation analyses. Multivariate linear relationships were assessed by generalized linear regression models.
Results
Whereas only PIGC, ZNFR3 and STAB1 mRNA expression in sc AT correlated nominally with WHR (P < 0.05, adjusted for age and sex), mRNA expression of all studied genes in at least one of the fat depots correlated significantly with vis and/or sc fat area (P ranging from 0.05 to 4.0 x 106, adjusted for age and sex). Consistently, the transcript levels of WARS, PIGC and GRB14 were nominally associated with body mass index (BMI) (P ranging from 0.02 to 9.2 x 105, adjusted for age and sex). Moreover, independent of sex, obesity and diabetes status, differential expression between vis and sc AT was observed for all tested genes (P < 0.01). Finally, the rs10195252 T-allele was nominally associated with increased GRB14 sc mRNA expression (P = 0.025 after adjusting for age, sex and BMI).
Conclusions
Our data including the inter-depot variability of mRNA expression suggests that genes within the WHR-associated loci might be involved in the regulation of fat distribution.
Keywords: fat distribution, gene expression, waist–hip ratio, GWAS
Introduction
Body fat distribution (FD) is one of the main predictors of obesity-associated complications, such as type 2 diabetes, chronic inflammation, coronary heart disease as well as hepatic glucose production.1–3 There is good evidence that FD is controlled by genetic factors. Individual variation in waist–hip ratio (WHR), a measure of FD, is heritable with estimates ranging from 22-61%.4–7 However, the mechanisms and exact genetic variants causing adverse or visceral (vis) FD are still poorly understood.
Along with biological candidate genes, genome-wide association studies (GWAS) represent an important source of genes potentially controlling FD. Recently, 14 loci have been identified in a GWAS for WHR independent of BMI.8 Six out of the 14 loci did not only harbour polymorphisms associated with WHR but also showed coincident cis-expression quantitative trait loci (eQTLs) (expression quantitative trait loci) implicating that the observed associations with WHR are likely to be mediated by gene transcripts (TBX15, WARS2, PIGC, STAB1, GRB14 and ZNRF3) in various tissues/organs.
Although these data indicate a possible role of the six genes in the regulation of FD, more detailed analyses are inevitable to pinpoint causal transcripts underlying the observed association signals. Therefore, the aim of the study was to evaluate whether TBX15, WARS2, PIGC, STAB1, GRB14 and ZNRF3 exhibit fat depot-specific mRNA expression and whether their transcript levels correlate with obesity-related traits by analysing paired samples of human vis and subcutaneous (sc) AT from 222 metabolically well-characterized subjects. Further, we investigated the effects of WHR-associated genetic variants within/nearby these genes on AT gene expression.
Materials and Methods
Subjects
A total of 222 Caucasian men (N = 78) and women (N = 144), who underwent open abdominal surgery were included in the study, and paired samples of vis and sc AT were obtained.9 The subjects had a mean age of 51 ± 15 years, a mean BMI of 41.6 ± 13.7 kg m −2, a mean WHR of 0.94 ± 0.11 and a mean height of 1.69 ± 0.09 m. All subjects had a stable weight with no fluctuation > 2% of the body weight for at least 3 m before surgery. Patients with severe conditions including generalized inflammation or end-stage malignant diseases were excluded from the study. Samples of vis and sc AT were immediately frozen in liquid nitrogen after explantation.9 Among the 222 subjects, 42 were lean (mean age 63 ±12 years, mean BMI 22.0 ± 0.4 kg m −2), 21 were overweight (mean age 71 ± 10 years, mean BMI 27.1 ± 0.3 kg m −2) and 159 were obese (mean age 45 ± 13 years, mean BMI 48.6 ± 0.7 kg m −2). An oral glucose tolerance test was performed after an overnight fast with 75 g of standardized glucose solution (Glucodex Solution 75 g; Merieux, Montreal, Quebec, Canada). Fasting plasma insulin was measured with an enzyme immunometric assay for the IMMULITE automated analyzer (Diagnostic Products Corporation, Los Angeles, CA, USA). Insulin sensitivity was assessed with euglycemic-hyperinsulinemic clamps.10 In addition to above mentioned clinical parameters, abdominal vis and sc fat area were calculated using computed tomography scans at the level of L4-L5 and percentage body fat was measured by dual-energy x-ray absorptiometry.
The ethics committee at the Medical Faculty of the University of Leipzig specifically approved this study and all subjects gave written informed consent before taking part in the study.
Analysis of human mRNA expression
Total RNA was isolated from paired sc and vis AT samples using TRIzol (Life Technologies, Grand Island, NY, USA), and 1 μ g RNA was reverse transcribed with standard reagents (Life Technologies). Human gene expression was measured by quantitative real-time RT–PCR by using TaqMan methodology and fluorescence was detected on an ABI PRISM 7500 sequence detector (Applied Biosystems, Darmstadt, Germany) according to the manufacturer's instructions (Applied Biosystems; assay Hs00537087_m1 with ACCESSION AK096396 for TBX15; Hs00210571_m1 with ACCESSION AJ242739 for WARS2; Hs00267516_s1 with ACCESSION AK308201 for PIGC; Hs01109068_m1 with ACCESSION AB052956 for STAB1; Hs00610307_m1 with ACCESSION AK301961 for GRB14; Hs00393094_m1 with ACCESSION AB051436 and AB032959 for ZNRF3). Human mRNA expression was calculated relative to the mRNA expression of HPRT and 18S rRNA, determined by a premixed assay on demand (PE Biosystems, Darmstadt, Germany). The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis.
Genotyping of single-nucleotide polymorphisms
Genotyping of the single-nucleotide polymorphisms (SNPs) (rs984222 G > C, rs6784615 T > C, rs1011731 A > G, rs4823006 A > G and rs10195252 T > C was done using the TaqMan SNP Genotyping assays according to the manufacturer's protocol (Applied Biosystems, Inc., Foster City, CA, USA). To limit genotyping errors, according to recommendation by Pompanon et al., 11 a random ~5% selection of the sample was re-genotyped in all SNPs; all genotypes matched initial designated genotypes.
Statistical analyses
Prior to statistical analysis, non-normally distributed parameters were logarithmically transformed to approximate a normal distribution. Differences in mRNA expression between vis and sc AT were assessed using the paired Student's t-test. Multivariate linear relationships were assessed by generalized linear regression models. P-values were adjusted for age, sex and BMI (if appropriate) and the genetic analyses were done under the additive model of inheritance. Furthermore, SNP × sex interaction term has been included to test possible interaction effects of sex in linear regression models in the whole sample.
P-values ≤ 0.05 were considered to provide evidence for nominal association and are presented without correction for multiple hypothesis testing. Only two-sided P-values are provided. Pearson correlation coefficients (r) are provided for relationships of mRNA expression levels with obesity-related quantitative traits.
All statistical analyses were performed using SPSS version 20 (SPSS, Inc.; Chicago, IL, USA).
Results
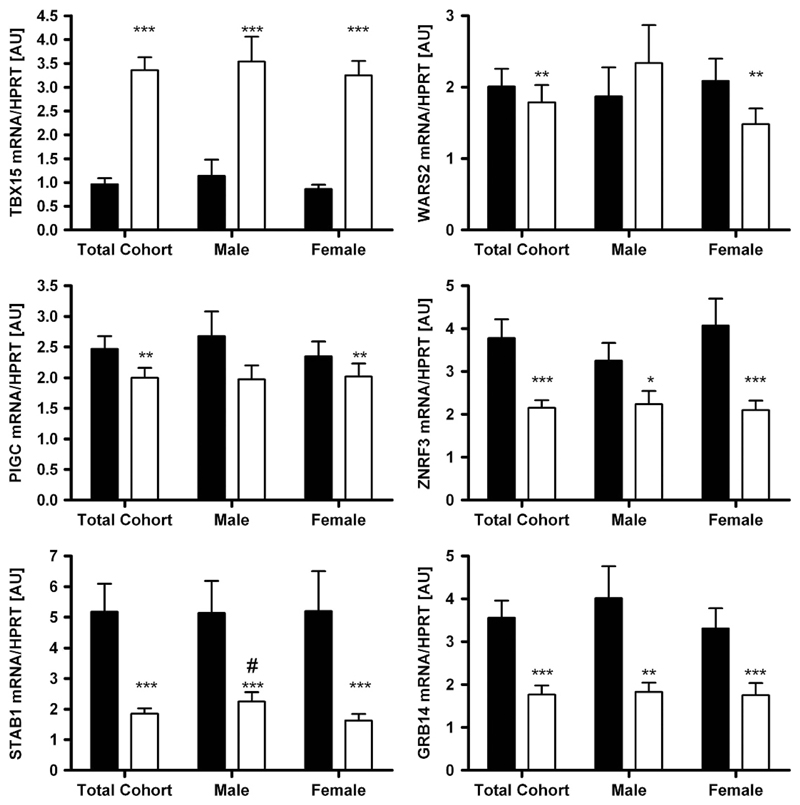
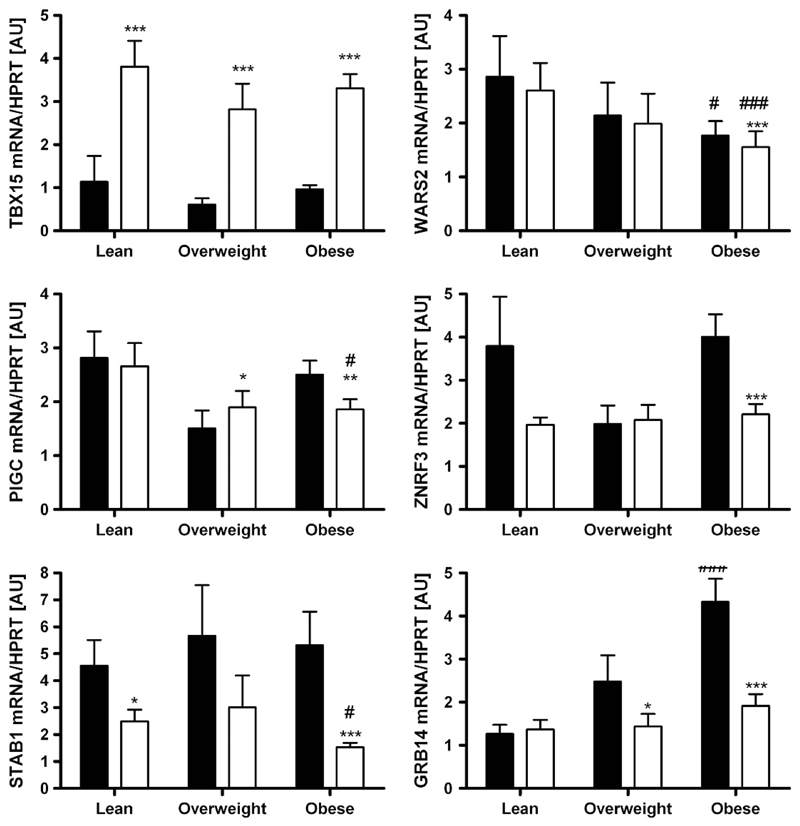
mRNA expression according to sex, obesity and diabetes status As sexual dimorphism has been reported for WHR and waist circumference in the initial GWAS,8 we analysed the expression data not only in the total cohort but also in men and women separately. Except for STAB1 with higher mRNA expression in sc AT in men when compared with women (P<0.05), there were no gender differences in gene expression in either tissue (Figure 1). Changes in expression activity of a gene/pathway in AT may be linked (either as a cause or a consequence) to a certain FD pattern or metabolically relevant phenotypes. Therefore, we compared the mRNA expression levels between lean (BMI < 25 kg m −2), overweight (BMI 25–30 kg m −2) and obese (BMI > 30kg m −2) subjects. WARS2, PIGC, STAB1 and GRB14 showed significantly different transcript levels between obese and lean subjects (Figure 2). Surprisingly, only PIGC, ZNFR3 and STAB1 mRNA expression in sc AT correlated significantly with WHR (P < 0.05 after adjusting for age and sex; Table 1). As WHR might be masked in severely obese conditions, we additionally included sc and vis fat area measurements as a more precise measure of FD. As summarized in Table 1, mRNA expression of all studied genes in at least one of the fat depots correlated with vis and/or sc fat area (P ranging from 0.05 to 4.0 × 106 after adjusting for age and sex; Supplementary Figure). Correlations of GRB14 (vis), WARS (vis) and PIGC (sc) AT mRNA expression with both vis and sc fat area were in accordance with correlations of their gene expression with BMI (Table 1).
Figure 1.
Expression of TBX15, WARS2, PIGC, ZNRF3, STAB1 and GRB14 in 222 paired human samples of visceral (vis) and subcutaneous (sc) adipose tissue (AT) in the total cohort and grouped by gender (78 males, 144 females). Mean±s.e.m. Black bars represent vis and white bars sc AT. *sc vs vis AT depot; #male vs female in the same AT depot. */#P < 0.05; **/##P < 0.01; ***/###P < 0.001.
Figure 2.
Expression of TBX15, WARS2, PIGC, ZNRF3, STAB1 and GRB14 in paired human samples of visceral (vis) and subcutaneous (sc) adipose tissue (AT) grouped by obesity status (lean N = 42, overweight N = 21, obese N = 159). Mean ±s.e.m. Black bars represent vis and white bars sc AT. *sc vs vis AT depot; #vs lean depot. */#P<0.05; **/##P < 0.01; ***/###P < 0.001.
Table 1. Correlations between gene expression and obesity-related parameters.
| mRNA expression | vis fat area (r; P-value) | sc fat area (r; P -value) | BMI (r; P-value) | WHR (r; P-value) |
|---|---|---|---|---|
| TBX15 | ||||
| vis | 0.02; 0.920 | 0.099; 0.146 | 0.125; 0.094 | − 0.008; 0.691 |
| sc | − 0.137; 0.054 | − 0.089; 0.189 | − 0.094; 0.133 | − 0.113; 0.092 |
| WARS | ||||
| vis | − 0.345; 8.3 × 10 − 5 | − 0.344; 2.8 × 10 − 4 | − 0.248; 0.001 | − 0.089; 0.106 |
| sc | − 0.403; 4.0 × 10 − 6 | − 0.414; 3.0 × 10 − 5 | 0.339; 9.2 × 10 − 5 | − 0.065; 0.056 |
| PIGC | ||||
| vis | − 0.230; 0.003 | − 0.148; 0.055 | − 0.072; 0.121 | − 0.118; 0.095 |
| sc | − 0.305; 0.001 | − 0.256; 0.035 | − 0.233; 0.021 | − 0.115; 0.021 |
| ZNFR3 | ||||
| vis | − 0.056; 0.371 | − 0.014; 0.719 | 0.043; 0.956 | − 0.071; 0.336 |
| sc | − 0.199; 0.009 | − 0.095; 0.248 | − 0.082; 0.293 | − 0.147; 0.020 |
| STAB1 | ||||
| vis | − 0.111; 0.238 | − 0.135; 0.240 | − 0.104; 0.243 | 0.015; 0.748 |
| sc | − 0.243; 0.015 | − 0.242; 0.077 | − 0.214; 0.168 | − 0.089; 0.014 |
| GRB14 | ||||
| vis | 0.197; 0.056 | 0.256; 0.005 | 0.284; 0.001 | 0.066; 0.489 |
| sc | 0.004; 0.516 | 0.006; 0.173 | 0.036; 0.071 | 0.042; 0.956 |
Abbreviations: AT, adipose tissue; sc, subcutaneous; vis, visceral adipose tissue. Correlations with P ≤ 0.05 are indicated in bold. All P-values adjusted for age and sex in linear regression models.
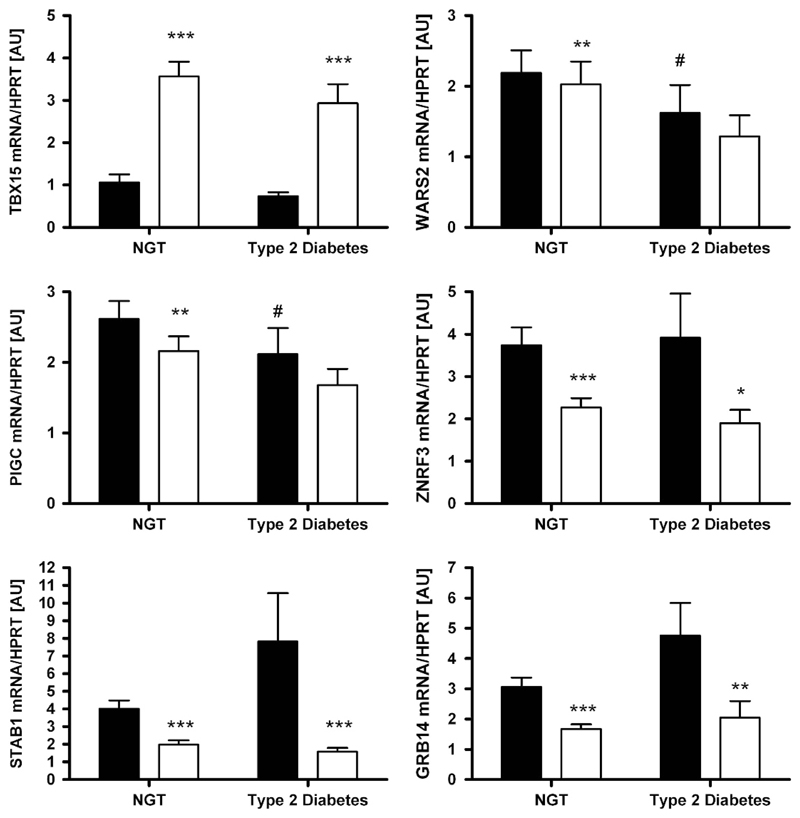
As glucose homeostasis status may eventually have an impact on expression of the transcripts, we tested this hypothesis as well. Compared with subjects with normal glucose tolerance, WARS2 and PIGC mRNA levels in vis AT were significantly lower in subjects with T2D (P <0.05; Figure 3). However, this appeared to be driven by obesity as no significant association was found after adjusting the analyses for BMI (all P> 0.05).
Figure 3.
Expression of TBX15, WARS2, PIGC, ZNRF3, STAB1 and GRB14 in paired human samples of visceral (vis) and subcutaneous (sc) adipose tissue (AT) grouped by type 2 diabetes status (NGT—subjects with normal glucose tolerance, N = 145; subjects with diabetes N = 71). Mean±s.e.m. Black bars represent vis and white bars sc AT. *vis vs sc AT depot; #vs NGT depot. *,#P < 0.05; **,##P < 0.01; ***/###P < 0.001.
Fat depot specific mRNA expression
The novel FD candidate genes TBX15, WARS2, PIGC, STAB1, GRB14 and ZNRF3 were differentially expressed between vis and sc AT. WARS2, PIGC, STAB1, GRB14 and ZNRF3 were predominantly expressed in vis fat, whereas TBX15 transcript levels were significantly higher in sc AT (P < 0.05;Figure 1). These differences seemed to be independent of sex, obesity or diabetes status, as they were observed in stratified analyses as well (Figures 1–3).
mRNA expression according to genotypes at WHR-associated loci
eQTL data can implicate regional transcripts that mediate trait associations8 and may help to sort out the expected direction. To elucidate the possible link between the initially described WHR-associated genetic variants8 and gene expression, we investigated the effects of rs984222 G > C (TBX15/WARS2), rs6784615 T > C (STAB1), rs1011731 A > G (PIGC), rs4823006 A > G (ZNRF3) and rs10195252 T > C (GRB14) within/nearby these genes on AT gene expression. All SNPs were in Hardy–Weinberg Equilibrium (all P > 0.05) and had following minor allele frequencies: rs984222— 38.8%, rs6784615—6.2%, rs1011731—40.7%, rs4823006—43% and rs10195252—41.1%.
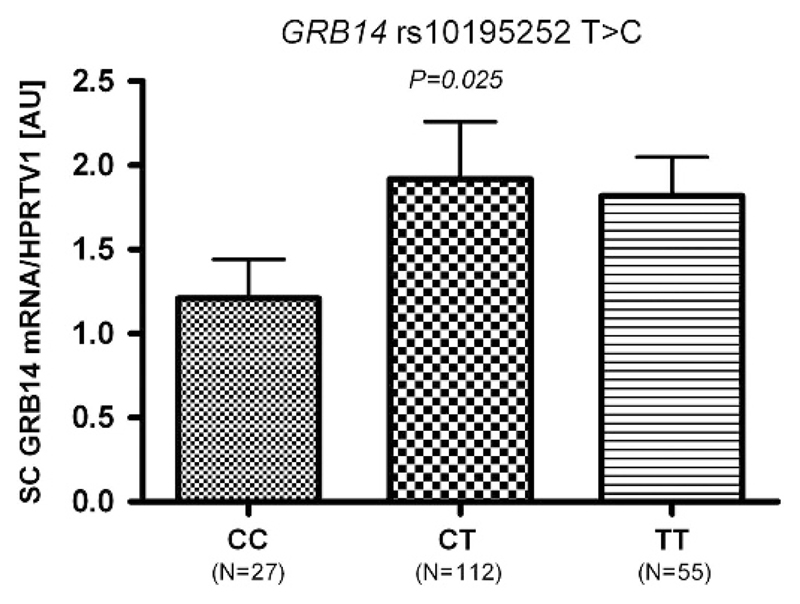
We found no sex-SNP interaction effects in the linear regression analyses in the whole sample and thus, subsequent analyses were performed without further sex stratifications. Except for rs10195252 in GRB14, none of the SNPs showed association with the respective gene transcript. The rs10195252 T-allele was nominally associated with increased GRB14 sc mRNA expression (P < 0.025 after adjusting for age, sex and BMI; Figure 4).
Figure 4.
Association of rs10195252 with GRB14 mRNA expression in subcutaneous (sc) adipose tissue. Data are given as arithmetic means±s.e.m. P < 0.05 in additive mode of inheritance; adjusted for age, sex and body mass index.
Discussion
In the present study, we measured the AT mRNA expression of 6 genes (TBX15/WARS2, STAB1, PIGC, ZNRF3 and GRB14) within the recently reported WHR loci8 in paired samples of human vis and sc AT. We observed differential expression between vis and sc AT for all tested genes. Furthermore, the mRNA expression of the studied genes correlated with measures of FD and obesity such as BMI, WHR and vis and sc fat area.
In general, mechanisms underlying differential gene expression between sc and vis AT are poorly understood. Nevertheless, many of these genes, including RBP4, LEP, PPARG, SERPINA12, AR and CB1R are not only differentially expressed in various fat depots, but their mRNA expression is also associated with traits related to obesity, such as insulin resistance or adipokine levels.3,12–14 Moreover, it has been shown that for some physiologically plausible candidate genes involved in the regulation of FD such as BMPRs, associations of genetic polymorphisms with anthropometric and metabolic measures might be mediated by changes in mRNA expression.15,16 Inter-depot specific differences have also been reported for genes involved in cytokine secretion or lipolysis but also for developmental genes involved in Wnt signaling.17 Also developmental genes such as TBX15, HOXA5 or GPC4 seem to play a relevant role in obesity and body FD.18 These genes did not only show strong inter-depot differences in mRNA expression in both mice and humans, but they also exhibited changes in expression that closely correlated with BMI and/or WHR.18 As postulated by Gesta et al.,18 these differences in gene expression are probably cell autonomous and independent of tissue microenvironment as indicated by their intrinsic nature and by persisting during in vitro culture and differentiation. One of the previously reported developmental genes, TBX15, was differentially expressed between vis and sc tissue also in the present study with significantly higher mRNA levels in sc AT. This direction is consistent with data obtained in mice studies, and data of an Australian child cohort,19 but opposite to human studies by Gesta et al. 18 One possible explanation might be the substantially smaller sample size of the previous study (53 vs 222) as well as the fact that only lean subjects have been investigated previously, whereas a wide range of BMI was included in the present work. Although the exact role in the regulation of either obesity or FD remains elusive, it is noteworthy that TBX15 encodes a transcription factor, which was shown to be involved in developmental processes as in dorsoventral patterning20 and skeletal development.21 Moreover, differential expression of TBX15 between fat depots is linked with differences in adipocyte differentiation, triglyceride accumulation and mitochondrial function.22
As for any GWAS in general, one of the greatest challenges is to understand the biological consequences of the identified loci associated with the respective trait. Therefore, systematic strategies to elucidate how WHR-associated variants exert their effects will be inevitable. No doubt that fine-mapping efforts are crucial to understand the complete architecture of genetic variation in the associated regions and to narrow down the number of functionally interesting variants. However, for non-Mendelian traits, the majority of the variants identified in GWAS maps within non-coding regions and thus, it is likely that WHR-associated alleles exert their effects by affecting gene transcription. Therefore, identifying the variant associated with gene transcription in specific target tissues, often referred to as eQTLs appears to be one of the major steps to determine potential causality of the revealed associations. Since eQTLs might explain a greater proportion of phenotypic variance than usually observed for risk alleles and clinical traits, eQTL studies do not require as large sample sizes as mostly needed for clinical association studies.23 Still, even though the sample size in our study may be appropriate in the context of expression studies, we are aware that we may have been lacking adequate statistical power to detect significant associations between the SNPs and mRNA expression in AT. Indeed, only one of the SNPs (rs10195252) showed nominal association with the corresponding gene transcript (GRB14 mRNA levels in sc AT). However, by applying correction for multiple testing (for example, Bonferroni corrections would require a P < 0.01 considering five SNPs analysed and taken into account the strong correlations between clinical traits), none of the SNPs would be associated with the respective gene expression. Nevertheless, the present work provides correlations between mRNA expression and metabolic traits related to obesity and FD, which seem to be statistically robust and resisting corrections for multiple testing. In this context, our study provides novel data on mRNA expression of several WHR-associated genes in AT, suggesting their role in the regulation of FD and so strongly supports the previous GWAS for WHR.8 It is of note, however, that further mechanisms have to be taken into account when elucidating the causative mechanisms behind genetic associations. For instance, epigenetic regulation of gene expression including DNA-methylation, histone modifications and altered function of non-coding RNAs or miRNAs have to be considered as well. Ultimately, with sufficient evidence for a causal variant or a susceptibility gene, cell and tissue models as well as in vivo models of disease development may be employed.
In conclusion, besides the correlations of expression levels with BMI or WHR, the most striking feature of the expression of the six studied genes is the inter-depot variability as well as correlations with the vis and sc fat area. Even though the nature of our study does not allow clarifying causative chains underlying the observed genotype–phenotype and phenotype–phenotype correlations, it clearly supports the role of recently reported WHR-associated genes in the regulation of FD.
Supplementary Material
Acknowledgements
We thank all those who participated in the studies. We would like to acknowledge excellent technical assistance by Daniela Kox from the University of Leipzig and excellent clinical work by Edward Shang. This work was supported by grants from DFG (Deutsche Forschungsgemeinschaft—KO 3880/1-2, KL 2346, SFB-1052); German Diabetes Association (DDG; to J.B. and P.K.), LIFE (Leipzig Research Center for Civilization Diseases, Universität Leipzig. Leipzig Research Center for Civilization Diseases, Universität Leipzig), Federal Ministry of Education and Research (BMBF), Germany, FKZ: 01EO1001 (N.K.), German Obesity Biomaterial Bank (FKZ: 01GI1128 to MB) and Boehringer Ingelheim Foundation (to D.S. and P.K.).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Bonora E. Relationship between regional fat distribution and insulin resistance. Int J Obes Relat Metab Disord. 2000;24:S32–S35. doi: 10.1038/sj.ijo.0801274. [DOI] [PubMed] [Google Scholar]
- 2.Arner P. Regional differences in protein production by human adipose tissue. Biochem Soc Trans. 2001;29:72–75. doi: 10.1042/bst0290072. [DOI] [PubMed] [Google Scholar]
- 3.Wajchenberg BL. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr Rev. 2000;21:697–738. doi: 10.1210/edrv.21.6.0415. [DOI] [PubMed] [Google Scholar]
- 4.Mills GW, Avery PJ, McCarthy MI, Hattersley AT, Levy JC, Hitman GA, et al. Heritability estimates for beta cell function and features of the insulin resistance syndrome in UK families with an increased susceptibility to Type 2 diabetes. Diabetologia. 2004;47:732–738. doi: 10.1007/s00125-004-1338-2. [DOI] [PubMed] [Google Scholar]
- 5.Rose KM, Newman B, Mayer-Davis EJ, Selby JV. Genetic and behavioral determinants of waist–hip ratio and waist circumference in women twins. Obes Res. 1998;6:383–392. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 6.Selby JV, Newman B, Quesenberry CP, Fabsitz RR, Carmelli D, Meaney FJ, et al. Genetic and behavioral influences on body fat distribution. Int J Obes. 1990;14:593–602. [PubMed] [Google Scholar]
- 7.Souren NY, Paulussen ADC, Loos RJF, Gielen M, Beunen G, Fagard R, et al. Anthropometry, carbohydrate and lipid metabolism in the east flanders prospective twin survey: heritabilities. Diabetologia. 2007;50:2107–2116. doi: 10.1007/s00125-007-0784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist–hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt J, Klöting N, Kralisch S, Kovacs P, Fasshauer M, Schön MR, et al. Plasma visfatin concentrations and fat depot-specific mRNA expression in humans. Diabetes. 2005;54:2911–2916. doi: 10.2337/diabetes.54.10.2911. [DOI] [PubMed] [Google Scholar]
- 10.Blüher M, Unger R, Rassoul F, Richter V, Paschke R. Relation between glycaemic control, hyperinsulinaemia and plasma concentrations of soluble adhesion molecules in patients with impaired glucose tolerance or Type II diabetes. Diabetologia. 2002;45:210–216. doi: 10.1007/s00125-001-0723-3. [DOI] [PubMed] [Google Scholar]
- 11.Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat Rev/Genet. 2005;6:847–859. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- 12.Klöting N, Stumvoll M, Blüher M. The biology of visceral fat. Internist. 2007;48:126–133. doi: 10.1007/s00108-006-1781-x. [DOI] [PubMed] [Google Scholar]
- 13.Montague CT, Prins JB, Sanders L, Zhang JL, Sewter CP, Digby J, et al. Depot-related gene expression in human subcutaneous and omental adipocytes. Diabetes. 1998;47:1384–1391. doi: 10.2337/diabetes.47.9.1384. [DOI] [PubMed] [Google Scholar]
- 14.Lefebvre AM, Laville M, Vega N, Riou JP, van Gaal L, Auwerx J, et al. Depot-specific differences in adipose tissue gene expression in lean and obese subjects. Diabetes. 1998;47:98–103. doi: 10.2337/diab.47.1.98. [DOI] [PubMed] [Google Scholar]
- 15.Böttcher Y, Unbehauen H, Klöting N, Ruschke K, Körner A, Schleinitz D, et al. Adipose tissue expression and genetic variants of the bone morphogenetic protein receptor 1A gene (BMPR1A) are associated with human obesity. Diabetes. 2009;58:2119–2128. doi: 10.2337/db08-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleinitz D, Klöting N, Böttcher Y, Wolf S, Dietrich K, Tönjes A, et al. Genetic and evolutionary analyses of the human bone morphogenetic protein receptor 2 (BMPR2) in the pathophysiology of obesity. PLoS ONE. 2011;6:e16155. doi: 10.1371/journal.pone.0016155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vöhl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, et al. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- 18.Gesta S, Blüher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci USA. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam CS, Heilbronn LK, Henegar C, Wong MN, Cowell CT, Cowley MJ, et al. An early inflammatory gene profile in visceral adipose tissue in children. Int J Pediatr Obes. 2011;6:E360–E363. doi: 10.3109/17477166.2011.575152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candille SI, Van Raamsdonk CD, Chen CY, Kuijper S, Chen-Tsai Y, Russ A, et al. Dorsoventral patterning of the mouse coat by Tbx15. PLoS Biol. 2004;2:30–42. doi: 10.1371/journal.pbio.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev. 2005;122:131–144. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Gesta S, Bezy O, Mori MA, Macotela Y, Lee KY, Kahn CR. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc Natl Acad Sci USA. 2011;108:2771–2776. doi: 10.1073/pnas.1019704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman ML, Monteiro ANA, Gyther SA, Coetzee GA, Risch A, Plass C. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;46:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.