Abstract
Inhibition of sclerostin is a therapeutic approach to lowering fracture risk in patients with osteoporosis. However, data from phase 3 randomized controlled trials (RCTs) of romosozumab, a first-in-class monoclonal antibody that inhibits sclerostin, suggest an imbalance of serious cardiovascular events, and regulatory agencies have issued marketing authorizations with warnings of cardiovascular disease. Here, we meta-analyze published and unpublished cardiovascular outcome trial data of romosozumab and investigate whether genetic variants that mimic therapeutic inhibition of sclerostin are associated with higher risk of cardiovascular disease. Meta-analysis of up to three RCTs indicated a probable higher risk of cardiovascular events with romosozumab. Scaled to the equivalent dose of romosozumab (210 milligrams per month; 0.09 grams per square centimeter of higher bone mineral density), the SOST genetic variants were associated with lower risk of fracture and osteoporosis (commensurate with the therapeutic effect of romosozumab) and with a higher risk of myocardial infarction and/or coronary revascularization and major adverse cardiovascular events. The same variants were also associated with increased risk of type 2 diabetes mellitus and higher systolic blood pressure and central adiposity. Together, our findings indicate that inhibition of sclerostin may elevate cardiovascular risk, warranting a rigorous evaluation of the cardiovascular safety of romosozumab and other sclerostin inhibitors.
Introduction
Osteoporosis, a disorder characterized by low bone mass, is a common condition associated with considerable morbidity and mortality, particularly among postmenopausal women and the elderly (1). A range of therapeutics for osteoporosis is currently available, but these agents are plagued by poor adherence [fewer than 40% of patients prescribed oral bisphosphonates continue to take these medications after 1 year (2)], occurrence of rare but serious adverse events such as osteonecrosis of the jaw, high cost, and uncertainty regarding long-term efficacy (3). There is therefore a substantial demand for effective, safe, and well-tolerated antiosteoporotic therapies (4).
Sclerostin, a glycoprotein encoded by the SOST gene, is a negative regulator of bone formation that is secreted by osteocytes. Sclerostin inhibits Wnt signaling, which leads to down-regulation of osteoblast development and function (5). Loss-of-function mutations in SOST lead to sclerosteosis, a rare autosomal recessive condition characterized by bone overgrowth (6). Similarly, van Buchem’s disease, another rare autosomal recessive condition with a phenotype similar to but generally milder than sclerosteosis, is caused by a deletion of a SOST-specific regulatory element (7). The discovery of functional variants in SOST as the underlying cause of these rare conditions of bone overgrowth nearly two decades ago led to the development of sclerostin inhibitors as a treatment for osteoporosis (8).
Three antisclerostin monoclonal antibodies have been or are currently in clinical development (5): romosozumab and blosozumab for osteoporosis and setrusumab—currently in phase 2b for osteogenesis imperfecta and previously also studied in adults with hypophosphatasia (9). Despite phase 2 results showing increases in bone mineral density (BMD) (10), a biomarker used to evaluate the effect of antiosteoporotic treatments, clinical development for blosozumab was halted in 2015, reportedly due to injection site reactions (11). Phase 2 and 3 randomized controlled trials (RCTs) have shown that romosozumab effectively increases BMD in both men and women while also reducing vertebral and nonvertebral fracture risks in women (12–15). However, adverse event data reported in the phase 3 BRIDGE (PlaceBo-ContRolled Study EvaluatIng The Efficacy AnD Safety Of Romosozumab In TreatinG MEn With Osteoporosis) and ARCH (Active-contRolled FraCture Study in Postmenopausal Women with Osteoporosis at High Risk of Fracture) trials (in men and postmenopausal women, respectively) have suggested that romosozumab may be associated with an excess risk of cardiovascular events (14, 15). Concerns about the cardiovascular safety of romosozumab, and sclerostin inhibition more generally, have previously been raised (8, 16–18) and, in 2017, the U.S. Food and Drug Administration (FDA) rejected an initial Biologics Licence Application for romosozumab due to concerns regarding cardiovascular adverse effects seen in the ARCH trial (19). Further licencing submissions were subsequently made to various regulatory agencies, including a resubmission to the U.S. FDA (20, 21). On 9 April 2019, the U.S. FDA approved romosozumab for the treatment of osteoporosis in postmenopausal women at high risk of fracture, with a boxed warning highlighting the risk of cardiovascular adverse events, and a postmarketing requirement to assess the cardiovascular safety of romosozumab (22). Further approvals have since been issued in Canada, Japan, and South Korea (including for both men and women in the latter two countries and with no specific warning label in Japan). The European Medicines Agency (EMA) issued a refusal of an application to marketing authorization for romosozumab in June 2019, citing “an increased risk of serious effects on the heart or circulatory system” (23). In October 2019, after a reexamination procedure, the EMA recommended granting a market authorization (24), leading to approval by the European Commission in December 2019, with a special warning relating to the risk of myocardial infarction (MI) and stroke (25).
A detailed appraisal of the cardiovascular effects of sclerostin inhibition by exploiting randomized data from orthogonal sources is therefore both timely and warranted, particularly given the discrepancy in opinions issued by regulatory agencies, and may assist in the evaluation of whether this class of drugs represents a rational and safe therapeutic strategy for the prevention of fracture. Naturally occurring human genetic variation can serve as a proxy for therapeutic stimulation or inhibition of a drug target, presenting a valuable opportunity to assess the likely consequences of modifying a therapeutic target on both the intended therapeutic effects and target-mediated adverse drug reactions (26, 27). The application of this approach in a Mendelian randomization (MR) framework was previously shown to recapitulate known clinical effects of drug target modulation (28–30). In this study, we used this genetic approach to examine the effect of BMD-increasing alleles in the SOST locus (as a proxy for sclerostin inhibition) on the risk of bone fracture, osteoporosis, and cardiovascular risk factors and disease (fig. S1) and complemented this with meta-analysis of cardiovascular data from RCTs of sclerostin inhibitors, to shed light on whether treatment with this class of drugs is likely to adversely affect cardiovascular disease.
Results
Risk of cardiovascular events in phase 3 RCTs of sclerostin inhibitors
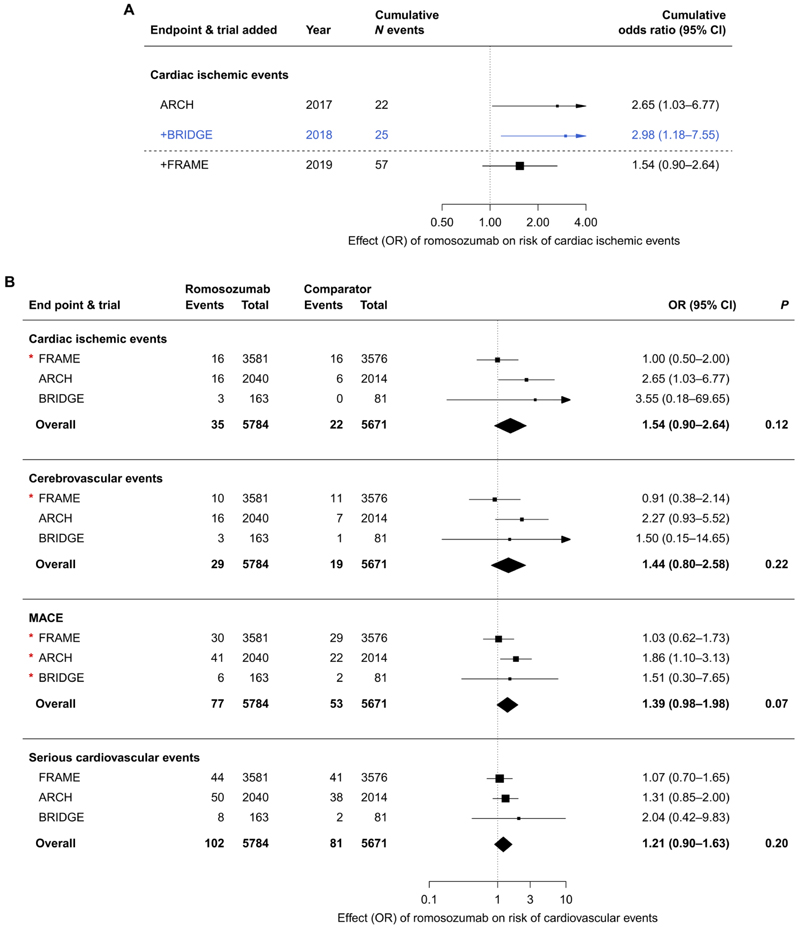
Romosozumab was the only sclerostin inhibitor with data from phase 3 RCTs. Four published phase 3 RCTs of romosozumab, including 11,954 individuals, were identified (table S1), of which three trials [BRIDGE (15), ARCH (14), and FRAME (FRActure study in post-menopausal woMen with ostEoporosis) (13)] reported cardiovascular adverse event data (table S2). Only the BRIDGE (15) and ARCH (14) trials reported data on cardiac ischemic events and cerebro-vascular events in their respective peer-reviewed publications. Unpublished data from the FRAME trial pertaining to these outcomes and data pertaining to major adverse cardiovascular events (MACE) (a composite of cardiovascular death, nonfatal MI, or nonfatal stroke) were extracted from U.S. FDA Drugs Advisory Committee briefing documentation (31).
Meta-analysis (using the Mantel-Haenszel method without continuity correction) of 25 events in 4298 individuals from two RCTs [BRIDGE (15) and ARCH (14)] identified that romosozumab (210 mg monthly) led to a higher risk of cardiac ischemic events as compared to the comparator [odds ratio (OR), 2.98; 95% confidence interval (CI), 1.18 to 7.55; P = 0.02; Fig. 1A]. Addition of the unpublished and non–peer-reviewed FRAME data made available to the U.S. FDA Drugs Advisory Committee brought the total number of cardiac ischemic events to 57 in 11,455 individuals and resulted in a pooled OR of 1.54 (95% CI, 0.90 to 2.64; P = 0.12; Fig. 1A). Corresponding estimates for cerebrovascular events were 27 events with OR of 2.15 (95% CI, 0.94 to 4.92; P = 0.07) before U.S. FDA meeting and 48 events with OR of 1.44 (95% CI, 0.80 to 2.58; P = 0.22) afterward. Estimates for MACE (130 events; OR, 1.39; 95%, 0.98 to 1.98; P = 0.07) and serious cardiovascular events (183 events; OR, 1.21; 95% CI, 0.90 to 1.63; P = 0.20) were directionally concordant with increased vascular risk arising from romosozumab treatment (Fig. 1B). Sensitivity analyses using the Peto method showed similar results (fig. S2A and table S3), as did inclusion of an alternative number of serious cardiovascular events in the FRAME trial, using U.S. FDA–sourced data (fig. S2B).
Fig. 1. Cumulative meta-analysis of cardiac ischemic events and meta-analysis of romosozumab and risk of cardiovascular events from phase 3 RCTs.
(A) Meta-cumulative plot of cardiac ischemic events, with RCTs ordered by data availability. Each line represents the addition of a further trial to the meta-analysis. The dashed horizontal line separates RCTs with data that were peer-reviewed and available before the U.S. FDA Drugs Advisory Committee meeting in January 2019 (ARCH and BRIDGE trials, with the meta-analyzed estimate highlighted in blue) from the FRAME trial. (B) Conventional meta-analysis of data from phase 3 RCTs of romosozumab, including unpublished data released for the U.S. FDA Drugs Advisory Committee meeting in January 2019 (indicated with a red asterisk). Events represent a number of events adjudicated per arm during initial 12-month double-blind period in each trial. The romosozumab group received 210 mg of romosozumab monthly in all trials; comparator group received placebo (FRAME and BRIDGE trials) or 70 mg of alendronate weekly (ARCH). Estimates were derived using the Mantel-Haenszel method. Boxes represent point estimates of effects. Lines represent 95% confidence intervals (CIs). OR, odds ratio.
Expression of SOST mRNA
We identified two conditionally independent genetic variants in the SOST locus associated with BMD, rs7209826 [A > G; G allele frequency in U.K. Biobank (UKBB), 40%] and rs188810925 (G > A; A allele frequency, 8%), from a recent large-scale genome-wide association study (GWAS) of estimated heel bone BMD (eBMD) (32). Expression of SOST across 53 human tissue types in the Genotype-Tissue Expression (GTEx) consortium was highest in arterial tissue (fig. S3; bone expression data not available). The minor alleles of rs7209826 (G allele) and rs188810925 (A allele) were both associated with lower expression of SOST mRNA in various human tissues, with the strongest associations with SOST expression for each variant identified in tibial artery (rs7209826, P = 1.4 × 10−8; 388 samples) and aorta (rs188810925, P = 7.6 × 10−6; 267 samples; fig. S4 and table S4). Colocalization analyses showed evidence of a shared causal variant driving the association with eBMD and SOST gene expression in tibial artery tissue (posterior probability of a shared causal variant of 74% in tibial artery for the rs7209826 signal, after adjusting for other independent signals in the region; table S5 and fig. S5), with other arterial tissues underpowered to provide a reliable probability of colocalization. No valid sclerostin protein quantitative trait loci (pQTLs) could be identified.
Association of rs7209826 and rs188810925 with BMD, osteoporosis, and fracture risk
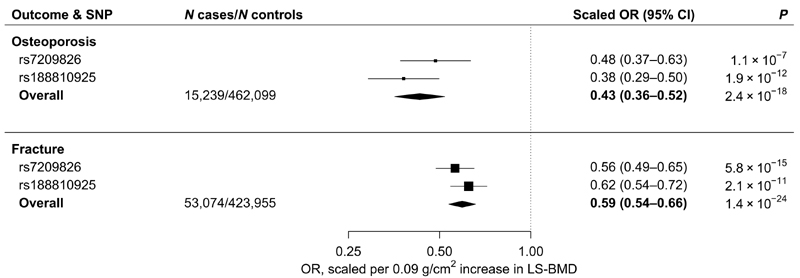
The minor alleles of both single-nucleotide polymorphisms (SNPs) were associated with higher eBMD in UKBB (32) [rs7209826, 0.04 g/cm2 [95% CI, 0.04 to 0.05; P = 2.3 × 10−36] per G allele; rs188810925, 0.07 g/cm2 (95% CI, 0.05 to 0.08; P = 1.3 × 10−26) per A allele; fig. S6] and with higher lumbar spine BMD (LS-BMD) (33) [rs7209826, 0.008 g/cm2 (95% CI, 0.005 to 0.01; P = 5.4 × 10−7) per G allele; rs188810925, 0.016 g/cm2 (95% CI, 0.01 to 0.022; P = 4.3 × 10−7) per A allele; fig. S6]. We subsequently scaled all further genetic estimates to the increase in LS-BMD seen with 12 months of romosozumab (210 mg monthly), equivalent to a higher BMD (0.09 g/cm2; see Materials and Methods for a detailed description of scaling) (34).
Scaled estimates from meta-analysis of rs7209826 and rs188810925 yielded a 57% lower risk of osteoporosis (OR, 0.43; 95% CI, 0.36 to 0.52; P = 2.4 × 10−18) and a 41% lower risk of sustaining a bone fracture (OR, 0.59; 95% CI, 0.54 to 0.66; P = 1.4 × 10−24; Fig. 2 and fig. S7). Associations were consistent across fracture sites (fig. S8).
Fig. 2. Scaled estimates and meta-analysis of BMD-increasing SOST variants with risk of osteoporosis and fracture.
Estimates are scaled to match the effect of romosozumab (210 mg monthly for 12 months) on LS-BMD (0.09 g/cm2; see Materials and Methods) and aligned to the BMD-increasing alleles. A total of 15,239 cases of osteoporosis and 53,074 cases of fracture were analyzed. See Materials and Methods for outcome definitions. Boxes represent point estimates of effects. Lines represent 95% CIs.
Association with cardiovascular events and cardiometabolic risk factors and diseases
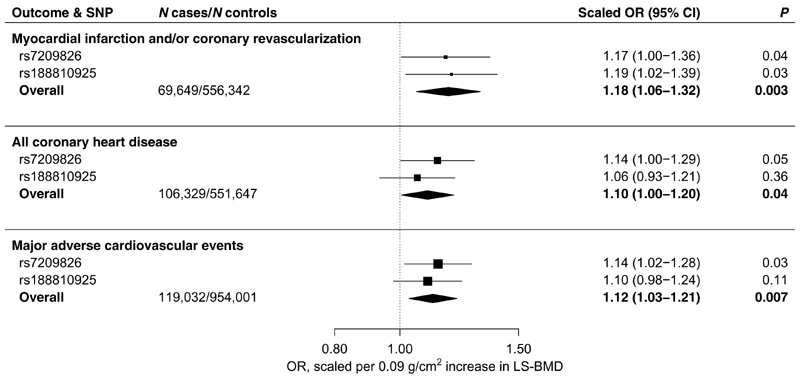
Next, we investigated the associations of the SOST variants with cardiovascular outcomes comparable to those used in the RCTs of romosozumab. In meta-analysis of scaled genetic estimates, the BMD-increasing SOST variants associated with an 18% higher risk of MI and/or coronary revascularization (69,649 cases; OR, 1.18; 95% CI, 1.06 to 1.32; P = 0.003) and a 12% higher risk of MACE (119,032 cases; OR 1.12, 95% CI 1.03 to 1.21, P = 0.007; Fig. 3 and fig. S9). Using a broader and less specific definition of coronary heart disease (CHD), which included self-reported angina and chronic stable ischemic heart disease (up to 106,329 cases), the SOST variants associated with a 10% increased risk of disease (OR, 1.10; 95% CI, 1.00 to 1.20; P = 0.04; Fig. 3 and fig. S9).
Fig. 3. Scaled estimates and meta-analysis of BMD-increasing SOST variants with risk of MI and/or coronary revascularization, CHD, and MACE.
Estimates are scaled to match the effect of romosozumab (210 mg monthly for 12 months) on LS-BMD (0.09 g/cm2; see Materials and Methods) and aligned to the BMD-increasing al-leles. MI and/or coronary revascularization includes codes pertaining to MI, coronary artery bypass graft surgery, and/or percutaneous transluminal coronary angioplasty; up to 69,649 cases were analyzed. CHD includes all codes pertaining to MI and/or coronary revascularization, as well as codes for angina and chronic stable ischemic heart disease; up to 106,329 cases were analyzed. MACE includes codes pertaining to a composite of MI and/or coronary revascularization, stroke, or death from either; up to 119,032 cases were analyzed. Boxes represent point estimates of effects. Lines represent 95% CIs.
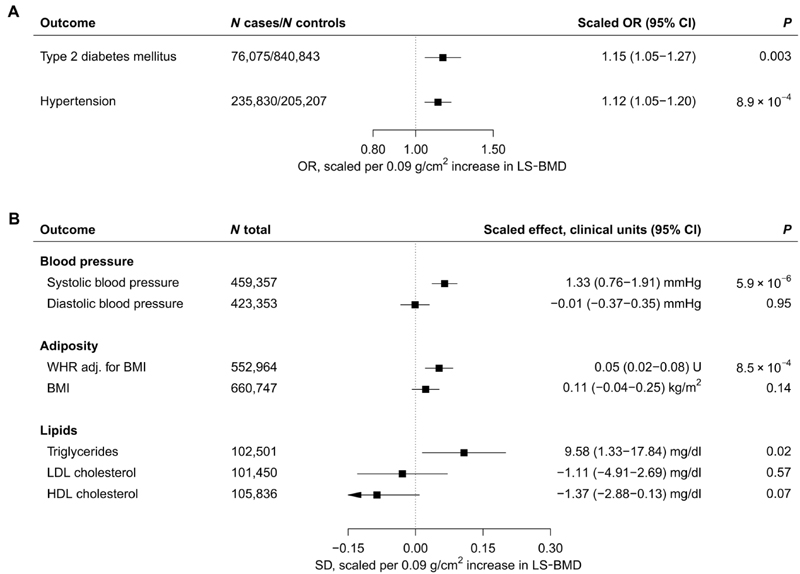
Evaluation of cardiometabolic risk factors and diseases revealed that the BMD-increasing SOST variants associated with higher risks of hypertension (OR, 1.12; 95% CI, 1.05 to 1.20; P = 8.9 × 10−4) and type 2 diabetes mellitus (T2D) (OR, 1.15; 95% CI, 1.05 to 1.27; P = 0.003; Fig. 4A).
Fig. 4. Meta-analysis of BMD-increasing SOST variants and cardiometabolic risk factors.
(A) Association with hypertension and T2D. (B) Association with quantitative traits plotted in SD units, with clinical units for each trait indicated in the column on the right. All estimates are scaled to match the effect of romosozumab (210 mg monthly for 12 months) on LS-BMD (0.09 g/cm2; see Materials and Methods) and aligned to the BMD-increasing alleles. The significance threshold was set at 0.004 (see Materials and Methods for details). Boxes represent point estimates of effects in OR (A) or SD (B) units. Lines represent 95% CIs. U, units.
Consistent with the effect on hypertension, we found the SOST variants to be associated with 1.3 mmHg higher systolic blood pressure (SBP) (95% CI, 0.76 to 1.91; P = 5.9 × 10−6) but observed no effect on diastolic blood pressure (DBP; Fig. 4B). In addition, the SOST variants associated with 0.05 SD units higher waist-hip ratio adjusted for body mass index (WHRadjBMI; 95% CI, 0.02 to 0.08; P = 8.5 × 10−4) and, nominally, with higher serum triglycerides (9.58 mg/dl higher; 95% CI, 1.33 to 17.84 mg/dl; P = 0.02). We found no further associations (figs. S10 to S13 and tables S6 and S7). Sex-stratified analyses revealed that, on average, the point estimates for cardiometabolic risk factors and disease appeared stronger in women than men; however, there was little statistical evidence of heterogeneity (table S8). Colocalization analyses of SBP and WHRadjBMI with eBMD were inconclusive because of low power (table S9), and a sensitivity analysis using conditionally independent SOST variants from a more recent GWAS of eBMD (35) yielded comparable results (figs. S14 and S15). An exploratory phenome-wide association analysis of 1074 binary outcomes in UKBB did not reveal any further suggestive associations (fig. S16).
Triangulation of RCTs and human genetics
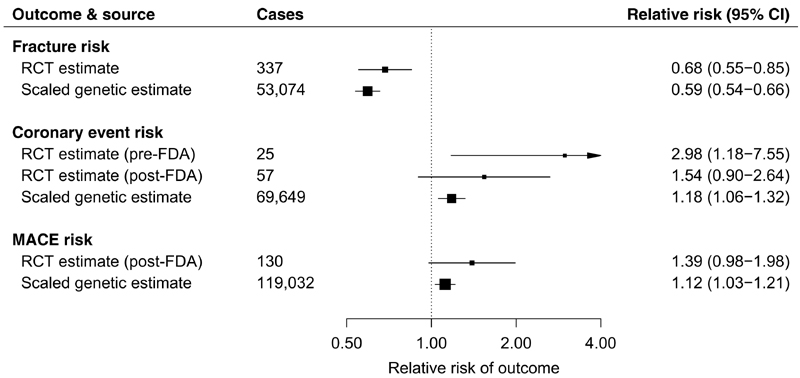
Triangulation refers to the integration of results from orthogonal approaches to study design, where each such approach has different sources of potential bias, to arrive at more reliable answers (36, 37). If the results from such approaches all support the same conclusion, then this bolsters our confidence in the finding. To this end, we first performed fixed-effect meta-analysis of 337 clinical fractures in 11,273 individuals from two RCTs with fracture outcome data [FRAME (13) and ARCH (14); table S10] and identified that romosozumab (210 mg monthly) led to a 32% lower risk of clinical fracture as compared to comparator (hazard ratio, 0.68; 95% CI, 0.55 to 0.85; P = 6.0 × 10−4; Fig. 5 and fig. S17) at 12 months. This compared well to the scaled genetic estimate, using the two SOST variants, of a 41% lower risk of fracture (Fig. 5). Evidence from treatment trials [up to three romosozumab phase 3 RCTs (14, 15)] and human genetics (two SOST variants) demonstrated an increased risk of coronary events (Fig. 5), indicating that this adverse effect is likely real and target mediated.
Fig. 5. Inhibition of sclerostin and risk of fracture and cardiovascular events derived from meta-analysis of phase 3 RCTs of romosozumab and human genetics.
Fracture risk RCT estimate represents inverse variance– weighted fixed-effect meta-analysis of estimates for hazard ratio of clinical fracture at 12 months (a composite of nonvertebral or symptomatic vertebral fracture) in the ARCH and FRAME trials. Coronary event risk RCT estimates represent fixed-effect meta-analysis (using the Mantel-Haenszel method) of estimates for OR of cardiac ischemic events in the ARCH and BRIDGE trials only [RCT estimate (pre-FDA)] and if including unpublished FRAME trial data released for U.S. FDA Drugs Advisory Committee meeting in January 2019 [RCT estimate (post-FDA)]. Data pertaining to the MACE risk RCT estimates were solely sourced from the U.S. FDA Drugs Advisory Committee meeting data. Scaled genetic estimates for fracture risk, coronary event risk, and MACE risk represent inverse variance–weighted fixed-effect meta-analyses of the scaled allelic estimates for the OR of fracture risk, MI and/or coronary revascularization, and MACE, respectively. All RCT estimates refer to the effect of romosozumab (210 mg monthly for 12 months) relative to comparator, and all genetic estimates are scaled to match the effect of romosozumab (210 mg monthly for 12 months) on LS-BMD (0.09 g/cm2 increase). Boxes represent point estimates of effects. Lines represent 95% CIs.
Last, we compared the estimates of our SOST-specific instrument with those of a recent MR study evaluating the causal effect of genetically predicted eBMD on risk of T2D and CHD (38). Scaling to the same difference in eBMD (an increase of 0.14 g/cm2 in eBMD), we found that estimates for the SOST-specific instrument were very similar to those of the eBMD instrument for both T2D and CHD (fig. S18). This suggests that the associations of our instrument with risk of T2D and CHD are consistent with the overall effect of BMD on these cardiometabolic outcomes and reduces the likelihood of the SOST associations with disease arising because of locus-specific pleiotropy.
Discussion
Using data from human genetics and randomized interventional trials, we have shown that sclerostin inhibition leads not only to higher BMD and lower risk of osteoporosis and fractures but also to a higher risk of CHD. Although prior phase 3 RCTs of sclerostin inhibition by romosozumab suggested an increased risk of adverse cardiac events (14, 15), it remained possible that the finding was due to chance owing to the low number of events. With the addition of data that had not undergone peer review, made available by the sponsors of romosozumab to the U.S. FDA in January 2019, the cumulative estimate for cardiac ischemic events attenuated. Thus, data from clinical trials support a probable but not unambiguous excess risk of cardiac disease arising from treatment with romosozumab. Our genetic analysis, which includes up to 106,329 CHD cases, shows that the excess risk of CHD from sclerostin inhibition is likely to be real and supports regulatory bodies’ addition of a warning for cardiovascular events to romosozumab labeling. Our findings also suggest that the excess risk of coronary events related to inhibition of sclerostin is driven, at least in part, by an increase in cardiovascular risk factors with an established etiological role in CHD, including adiposity, blood pressure, and T2D.
Given the high short-term mortality associated with some types of fragility fracture—up to 25% mortality in the 12 months after a hip fracture (39)—a careful assessment of the potential risks versus benefits is warranted, weighing the merits of lower fracture and fracture-related morbidity and mortality against the potential harm from higher risk of metabolic and vascular disease. Clinical trials of romosozumab suggest that the reduction in relative risk of fracture (32%) is greater than the increased relative risk of coronary events (18%); however, such estimates relate to relative risks versus absolute risks, with absolute risks being more clinically meaningful, and furthermore, the reported relative risks do not take into account the differential impact that each disease has on an individual’s quality of life (as may be necessary to estimate the “benefit risk” of a therapeutic).The use of human genetics in this context may pose a translational challenge in estimating the expected effects of a therapy in both relative and absolute terms: For example, genotype approximates prolonged or lifetime exposure, whereas a clinical trial is typically of considerably shorter duration (40).
We did not observe statistical evidence of heterogeneity according to sex in our genetic analyses, a finding that is supported by romosozumab’s comparable effect on BMD in men and women in clinical trials (15). It is, however, notable that the genetic estimates for CHD, blood pressure, and T2D tended to be of greater magnitude in women than men. These findings emphasize that, despite limiting romosozumab’s label in the United States and Europe to only women (women being at relatively lower risk of vascular disease than men), treatment may still lead to an increase in vascular risk among women.
There are several lines of evidence that support the findings of our study. Sclerostin is expressed in cardiovascular tissues (41, 42), supporting a potential biological role for this protein in these tissue types. In observational studies, higher concentrations of circulating sclerostin are associated with a higher risk of cardiovascular disease together with increased cardiometabolic risk factors such as hypertension, T2D, and central adiposity (43–48). In contrast to our genetic findings, these data suggest that lowering of sclerostin may confer protection from cardiovascular disease; however, observational studies also show that osteoporosis and lower BMD are associated with lower sclerostin (49, 50), suggesting that increasing sclerostin would lead to higher BMD. We know this to be spurious given the well-established effects of therapeutic sclerostin inhibition on BMD. Furthermore, recent investigations of BMD show that the observational association with risk of CHD and the corresponding estimate from MR analyses are directionally opposite (38), making observational associations of bone density and vascular disease unreliable. More recent evidence has suggested that sclerostin may be up-regulated in the vasculature in response to vascular calcification, as part of a regulatory process aimed at counteracting such calcification (18), whereas murine studies have implicated sclerostin in adipocyte metabolism (51) and the development of atherosclerosis, aortic aneurysm, and hemopericardium (52, 53). Although these human and animal data serve as evidence for the biological plausibility of sclerostin inhibition altering cardiovascular risk, extrapolating findings from animal models of disease to humans is plagued by failures of translation (54), and observational studies of humans can be influenced by sources of error. This is why studies using a randomized design in humans provide more reliable evidence on causation (37).
Elucidation of whether adverse effects are on or off target is critical (8) because on-target effects would mean that any drug under development in the same class, such as a sclerostin inhibitor, would be expected to share a similar adverse effect profile, such as higher risk of vascular events. Our genetic data provide strong evidence in support of target-mediated adverse effects of sclerostin inhibition on coronary events. Sclerostin exerts its effects on bone as an inhibitor of the Wnt signaling pathway—a pathway also previously linked to vascular calcification (55–57)—by binding to the Wnt co-receptor low-density lipoprotein (LDL) receptor–related protein (LRP) 5 and 6 (58). Protein-coding mutations in both LRP5 and LRP6 have been linked to alterations in BMD and cardiometabolic risk profiles, including insulin resistance, dyslipidemia, hypertension, and CHD (59, 60), which supports our findings of altered cardiometabolic disease in carriers of SOST variants.
Concerns of cardiovascular safety have also plagued other antiosteoporotic agents. Odanacatib, a cathepsin K inhibitor developed for the treatment of osteoporosis, was not developed further after demonstrating reduced fracture risk in phase 3 trials because of an increased risk of stroke (61). We did not identify strong associations of SOST variants with risk of stroke subtypes in our analyses; however, the point estimates for both ischemic and hemorrhagic stroke had ORs of >1. With genetic lowering of sclerostin leading to elevations in SBP and WHRadjBMI, each of which plays a causal role in stroke (62, 63), it is plausible that additional stroke cases could identify such an association. More broadly, genetically elevated BMD may exert a causal effect on risk of T2D and CHD (38). Other BMD-raising therapies such as denosumab or bisphosphonates have not shown an association with increased risk of cardiovascular events in clinical trials (64–66). Some reports (31) using network meta-analysis have suggested that the increased cardiovascular risk seen in the romosozumab arm of the ARCH trial (14) may have been due to a cardioprotective effect of alendronate, a bisphosphonate used in the ARCH control arm. However, meta-analyses of RCTs of bisphosphonate s have shown that the effect of bisphosphonates on risk of cardiovascular events is null (65, 66), arguing against this hypothesis. In addition, this would not explain the imbalance seen in the placebo-controlled BRIDGE trial (15) nor would it account for our genetic findings.
The approach to exploiting human genetics to validate target-mediated effects is well established (28–30). In our study, we selected variants associated with reduced expression of SOST and increased BMD as proxies for the effect of sclerostin inhibition. We validated the effects of the SOST genetic variants on risk of osteoporosis and bone fracture (including fracture across several sites) and leveraged a large number of CHD cases—a more than 1800-fold increase over the number of cases reported in phase 3 trials of romosozumab (13–15). Our approach also facilitated identification of potential mechanisms and mediators that may drive this excess coronary risk. For example, the association of SOST variants with central adiposity, SBP, and T2D has not been previously reported, and if not measured in a clinical trial, such associations cannot be readily explored.
Our study is subject to several limitations. First, we examined lifelong effects of genetic variants, which are not necessarily representative of the effects seen with pharmacological perturbation of shorter duration later in life. Second, the common, noncoding variants that we use in this study have not been conclusively linked to altered sclerostin protein expression or function. A recent GWAS of circulating serum sclerostin (67) reported three trans-pQTLs but found that SNPs in the SOST locus were only nominally associated with serum sclerostin concentration. The authors speculated that this finding may suggest that SOST SNPs influence BMD and fracture risk by altering local sclerostin activity in the bone, independently of serum sclerostin. Further studies of larger sample size will be needed to explore the effect of genetic variants in the SOST locus on circulating sclerostin. We attempted to investigate the effects of the trans-pQTLs but found that they were unlikely to be reliable instruments. Nonetheless, the validity of our variants as suitable genetic instruments to mimic treatment with an inhibitor of sclerostin is supported by their association and colocalization with SOST mRNA expression across various tissue types and with relevant therapeutic outcomes including BMD, fracture risk, and osteoporosis. Third, although rare mutations in SOST have been linked to an extreme BMD phenotype indicating the presence of an allelic series of common and rare trait-associated alleles (68), we were not able to evaluate the effect of loss-of-function coding variants in SOST, primarily due to the relative scarcity of such variants in the general population. The most common predicted loss-of-function SOST variant in the Genome Aggregation Database (gnomAD) (69) has a minor allele frequency of 9.2 × 10−6, with no homozygotes reported. Increased cardiovascular risk has not been reported in individuals with Mendelian disorders of absent or low sclerostin expression such as van Buchem disease or sclerosteosis. Reasons for this may include a shortened life expectancy observed in some of these patients (for instance, due to raised intracranial pressure) (70) and having not been examined for cardiovascular disease specifically. Deep phenotyping, including comprehensive cardiovascular evaluation, of individuals with loss-of-function variants in SOST would likely be of value (71, 72). Fourth, in scaling the allelic estimates to match the effect of pharmacologic inhibition of sclerostin, we assumed that the effect of the variants on BMD and the outcomes of interest is linear in nature. Observational evidence suggests that the association between BMD and cardiovascular risk is inversely linear, with lower BMD and osteoporosis associated with higher cardiovascular risk (38, 73, 74); however, this is directionally opposite to the evidence from MR studies, which suggests that higher BMD is causally associated with higher cardiovascular risk (38) and therefore poses challenges in testing the assumption of linearity in the genetic associations. Nevertheless, the same approach to scaling allelic estimates has also been applied in studies similar to ours and is an established approach in MR (28, 29, 75). Last, we note that the effect estimates derived from the meta-analysis of RCTs of sclerostin inhibition provide only weak evidence of an adverse effect on cardiac disease. At the time of initiating this study, before the U.S. FDA meeting of January 2019, the unpublished data from the FRAME trial had not been made available. Our meta-cumulative plot shows the evolving nature of the vascular disease effects of sclerostin: Before the U.S. FDA hearing, the OR of cardiac ischemic events was 2.98 (95% CI, 1.18 to 7.55) with 25 events. Addition of non–peer-reviewed vascular disease events, which more than doubled the number of cases, led to a halving of the effect estimate (OR, 1.54) with the 95% CI including the null (95% CI, 0.90 to 2.64). Thus, the data from clinical trials are consistent with a potential elevated risk of vascular disease arising as an adverse consequence of treatment with sclerostin inhibition, but the trial evidence is not conclusive. Network meta-analysis may suggest that the excess risk from rosomozumab could have arisen because of a protective effect of alendronate (31); however, this is not supported by large-scale meta-analyses of bisphosphonates. Irrespectively, our genetic estimates are consistent with an excess risk of cardiac events being real and target mediated. Further, the pattern of associations of SOST variants not only with CHD but also with risk factors with an established causal role in CHD including adiposity, blood pressure, and T2D makes it implausible that these genetic findings have arisen by chance.
Our findings support the U.S. FDA and EMA’s addition of a warning for cardiovascular risk to romosozumab’s label (22, 25) and indicate that other therapies inhibiting sclerostin are also likely to exert similar cardiovascular effects. With the benefit of hindsight, our analyses could have identified this potential for an increased cardiovascular risk before initiating clinical development of romosozumab (and other sclerostin inhibitors). This information may have been incorporated into trial design, particularly for directly evaluating the effect of romosozumab on cardiovascular outcomes and risk factors in adequately powered RCTs. The postmarketing surveillance mandated by the U.S. FDA consisting initially of a 5-year observational feasibility study (22), although important, is subject to multiple sources of bias (76, 77). Our results therefore emphasize the utility of large-scale human genetic data in characterizing target-mediated effects to inform preclinical drug target validation (27, 68, 78).
In conclusion, our results warrant a rigorous assessment of the effect of romosozumab (and other sclerostin inhibitors in clinical development) on cardiovascular disease and cardiometabolic risk factors. This adds valuable information to whether pharmacological inhibition of sclerostin should be pursued as a therapeutic strategy for the prevention of fracture.
Materials and Methods
Study design
We performed two major analyses to investigate whether treatment with sclerostin inhibitors is likely to adversely affect on cardiovascular disease. First, we meta-analyzed cardiovascular outcomes data from RCTs of sclerostin inhibitors. Second, we examined the effect of BMD-increasing alleles in the SOST gene (encoding sclerostin) on the risk of therapeutic and cardiovascular outcomes. An overview of the genetic analysis is shown in fig. S1.
Study population
We examined individual-level genotypic and phenotypic data for 502,617 individuals in UKBB, a population-based cohort based in the United Kingdom. In addition, we included data from a further two European ancestry cohorts: Partners HealthCare Biobank (PHB; up to 19,132 individuals) and Estonian Biobank (EGCUT; up to 36,074 individuals). Trans-ethnic replication was attempted in China Kadoorie Biobank (CKB; up to 81,546 individuals). Further detail pertaining to each cohort is described below.
U.K. Biobank
UKBB is a prospective study of more than 500,000 British individuals recruited from 2006 to 2010 and aged between 45 and 69 (79). Phenotypic information available includes self-reported medical history as ascertained by verbal interview with a medical professional at enrolment, hospital-derived electronic health record (EHR) data, including International Classification of Diseases, ninth and tenth revision (ICD-9 and ICD-10) codes and Office of Population and Censuses Surveys Classification of Interventions and Procedures version 4 (OPCS-4) procedure codes, and an extensive set of physical measurements. Genotypic data (see below for detail) are available for 488,377 individuals, of whom 487,409 have imputed genotype data.
For the purposes of this study, we excluded all samples indicated to have poor-quality genotypes by UKBB (on the basis of high sample heterozygosity and missingness) and further excluded individuals that had reported non-white or mixed ethnicity at any point during follow-up, withdrawn their consent for participation, >10 third-degree relatives, putative sex chromosome aneuploidy, sex mismatches (comparing genetically determined versus self-reported sex and comparing between assessments), and ethnicity mismatches (mismatches between genetically determined and self-reported ethnicity for white British individuals and any ethnicity mismatches between assessments). We reviewed pairwise genetic relatedness between individuals and excluded one individual per pair of individuals with an estimated second degree or closer relatedness [equivalent to a kinship coefficient of greater than 0.0884 (80)]. After applying these filters, 423,766 individuals remained. Baseline characteristics of participants in UKBB are shown in table S11.
Genotyping in UKBB
Genotyping, quality control (QC), and imputation were performed centrally by UKBB, and details are fully described elsewhere (79). Briefly, genotype data are available for 488,377 individuals, 49,950 of whom were genotyped using the Applied Biosystems U.K. BiLEVE Axiom Array by Affymetrix [containing 807,411 markers (81)]. The remaining 438,427 individuals were genotyped using the Applied Biosystems U.K. Biobank Axiom Array by Affymetrix (containing 825,927 markers). Both arrays were specifically designed for use in the UKBB project and share ~95% of marker content. Phasing was performed using SHAPEIT3, and imputation was conducted using IMPUTE4. For imputation, the Haplotype Reference Consortium (HRC) panel (82) was used wherever possible, and for SNPs not in that reference panel, a merged UK10K + 1000 Genomes reference panel was used. SNPs were imputed from both panels, but the HRC imputation was preferentially used for SNPs present in both panels. Both SNPs selected as instruments in this study demonstrated high imputation quality in UKBB (info scores of 0.98 and 0.95 for rs7209826 and rs188810925, respectively).
Study outcomes and definitions in UKBB
Fracture status was based on an affirmative answer to the question “Have you fractured/broken any bones in the past 5 years?” at either baseline or at follow-up. Individuals were designated as missing if they answered “Do not know” or “Prefer not to answer” at baseline and at all follow-up occasions. All remaining individuals were designated as controls. An identical definition has been used in recent GWAS of BMD and fracture (32, 83), and the use of self-reported fracture has previously been validated (84). Individuals who reported having sustained a fracture were asked “Which bone(s) did you fracture/break?” and given a list of options to choose from (including the ankle, leg, hip, spine, wrist, arm, and other bones). For the purposes of our analysis, we grouped these into upper limb (wrist and arm), lower limb (ankle, leg, and hip), vertebral (spine), and other fractures.
Individual values for SBP and DBP and the definition of hypertension were derived using the same method as in a recent GWAS for blood pressure performed in UKBB (85). We derived the mean SBP and DBP for each individual from two blood pressure measurements performed at baseline. For individuals with only a single measurement available, we used this single value. We then adjusted these values for blood pressure–lowering medication use by addition of 10 and 15 mmHg to DBP and SBP, respectively, for individuals who had reported taking blood pressure medication at baseline (86). Individuals were coded as having hypertension if they had an adjusted SBP of 140 mmHg or higher, a DBP of 90 mmHg or higher, or reported use of antihypertensive medication.
To create phenotypes for BMI and WHRadjBMI, we followed steps consistent with those followed in the Genetic Investigation of Anthropometric Traits (GIANT) consortium (87–89). Baseline measures of BMI, waist circumference, and hip circumference were used. We first divided waist circumference by hip circumference to calculate the WHR and then regressed WHR on BMI, sex, age at time of assessment, and (age at time of assessment)2, whereas BMI was regressed on sex, age at time of assessment, and (age at time of assessment)2. Next, we performed rank inverse normalization on the resulting residuals from the regressions. These normalized residuals (for BMI and WHRadjBMI) were used for performing allelic association testing.
We included two definitions for CHD: an inclusive CHD phenotype including angina and other forms of chronic CHD and a more specific phenotype of MI and/or coronary revascularization only. Both of these definitions were derived from those used in a recent GWAS of CHD conducted in UKBB by the CARDIoGRAMplusC4D [Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) Genetics] consortium (90). Detailed definitions for all binary outcomes analyzed in UKBB (and not specifically defined above) are given in table S12.
Ethical considerations
The UKBB project was approved by the North West Multi-Centre Research Ethics Committee, and all participants provided written informed consent to participate. This research has been conducted under UKBB application number 11867.
Partners HealthCare Biobank
We identified patients with relevant phenotype information and genotype data from the PHB (91, 92), a biorepository of consented patient samples at Partners HealthCare hospitals in the Boston area of Massachusetts. The PHB maintains blood and DNA samples, clinical records, and genotype data from consented patients. Patients are recruited in the context of clinical care appointments and electronically. All patients who participate in the PHB are consented for their samples to be linked to their clinical information for the use in broad-based research.
A total of 19,136 patients of European ancestry were genome-wide genotyped in PHB, including 8868 males and 10,268 females with age at recruitment ranged from 19 to 102 (mean, 59.4; median, 62; SD, 16.61). For these patients, EHRs were available for extracting the following phenotypes: fracture, osteoporosis, CHD, MI and/or coronary revascularization, T2D, BMI, high-density lipoprotein (HDL) cholesterol, LDL cholesterol, triglycerides, and hypertension. We subset these patients according to the phenotype information availability for each genetic association analysis (see the “Study outcomes and definitions in PHB” section below).
Genotyping in PHB
DNA samples from whole blood and genome-wide genotyping were performed for 25,582 patients. Genotyping was performed with Illumina Multi-Ethnic Genotyping Array (first batch), Expanded Multi-Ethnic Genotyping Array (second batch), and Multi-Ethnic Global BeadChip (third batch), all of which were designed to capture the global diversity of genetic backgrounds (N of genotyped variants, 1,416,020 to 1,778,953).
Preimputation QC was performed on each genotyping batch separately as follows: We removed SNPs with a genotype missing rate of >0.05 before sample-based QC; excluded samples with a genotype missing rate of >0.02, an absolute value of heterozygosity of >0.2, or failed sex checks; and removed SNPs with a missing rate of >0.02 after sample-based QC. To merge genotyping batches for imputation and analyses, we performed batch QC by removing SNPs with significant batch association (P < 1.0 × 10−6 between different batches).
Because the PHB samples have diverse population backgrounds, we performed Hardy-Weinberg equilibrium (HWE) test (P < 1.0 × 10−6) for SNP-based QC after extracting samples of European ancestry (see below). We also performed relatedness tests by identifying pairs of samples with π > 0.2 and excluding one sample from each related sample pair (560 samples excluded). All QC were conducted using PLINK v1.9 and R software.
We extracted samples with European ancestry based on principal components analysis (PCA) with 1000 Genomes Project reference samples. To identify patients of European ancestry, we first performed PCA on linkage disequilibrium (LD)–pruned dataset merged with 1000 Genomes Project reference samples labeled with five distinct superpopulations [European (EUR), African (AFR), East Asian (EAS), admixed American (AMR), and South Asian (SAS)]. Then, we used the top four principal components to build a random forest classifier trained on 1000 Genomes Project reference samples with superpopulation labels. Last, we applied the trained random forest classifier to identify patients of European ancestry from PHB (with a predicted probability of European ancestry of >0.9).
Genotype imputation was performed on the quality-controlled patients of European ancestry with a two-step prephasing/imputation approach. We used Eagle2 for the prephasing and Minimac3 for imputation, with a reference panel from 1000 Genomes Project phase 3.
Study outcomes and definitions in PHB
We extracted relevant phenotype information from EHRs for the patients with imputed genome-wide genotype data. The phenotype definitions and number of samples are described in table S13.
Ethical considerations in PHB
The PHB maintains blood and DNA samples from consented patients seen at Partners HealthCare hospitals in the Boston area of Massachusetts. Patients are recruited in the context of clinical care appointments and also electronically. Biobank individuals provide consent for the use of their samples and data in broad-based research.
Estonian Biobank
The EGCUT is the population-based biobank containing longitudinal data and biological samples, including DNA, for 5% of the adult population of Estonia. The broad informed consent form signed by the participants of the biobank allows the Estonian Genome Center to continuously update their records through periodical linking to central EHR databases and registries. We studied the genotypic and phenotypic data of 51,881 individuals, and after removing relatives (PiHat, >0.2), 36,073 individuals (65% women and 35% men) with an average age of 45 years were included for further analysis. For the CHD analyses in EGCUT, we only included participants not previously included in the CARDIoGRAMplusC4D consortium.
Genotyping in EGCUT
Of all the studied biobank participants, 33,155 have been genotyped using the Global Screening Array, 8137 HumanOmniExpress BeadChip, 2640 HumanCNV370-Duo BeadChips, and 6861 Infinium CoreExome-24 BeadChips from Illumina. Furthermore, of 2056 individuals’ whole genomes have been sequenced at the Genomics Platform of the Broad Institute.
Sequenced reads were aligned against the GRCh37/hg19 version of the human genome reference using BWA-MEM1 v0.7.7; polymerase chain reaction duplicates were marked using Picard (http://broadinstitute.github.io/picard) v1.136, and the Genome Analysis Toolkit (GATK) v3.4-46 was applied for further processing of BAM files and genotype calling. All insertion-deletions (indels) in the Variant Call Format were normalized and multiallelic sites split using bcftools (https://samtools.github.io/bcftools/bcftools.html). The following genotypes were set to missing: a genotype quality of <20, a read depth of >200, and an allele balance of <0.2 or >0.8 for heterozygous calls. The GATK’s variant quality score recalibration metric was used to filter variants with a truth sensitivity of 99.8% for single nucleotide variants (SNVs) and of 99.9% for indels. Furthermore, variants with an inbreeding coefficient of <-0.3, a quality by depth of <2 for SNVs and <3 for indels, a call rate of <95%, or HWE P < 1 × 10−6 were excluded.
The genotype calling for the Illumina microarrays was performed using Illumina’s GenomeStudio V2010.3 software. The genotype calls for rare variants on the Global Screening Array (GSA) array were corrected using the zCall software (version 8 May 2012). After variant calling, the data were filtered using PLINK (v.1.90) by sample (call rate of >95%, no sex mismatches between phenotype and genotype data, heterozygosity < mean ± 3 SE) and marker-wise (HWE P >1 × 10−6 and call rate of >95% and for the GSA array additionally by Illumina GenomeStudio GenTrain score of >0.6 and cluster separation score of >0.4). Before the imputation, variants with a minor allele frequency (MAF) of <1% and C/G or T/A polymorphisms as well as indels were removed, as these genotype calls do not allow precise phasing and imputation. The genotype data obtained on all of the arrays were separately phased using Eagle2 (v. 2.3) and imputed using the BEAGLE (v. 4.1) software implementing a joint Estonian and Finnish reference panel (93).
Study outcomes and definitions in EGCUT
We tested the associations of rs7209826 and rs188810925 with fracture (ICD-10 codes S52.5, S82.6, S22.3, S42.2, S52.6, S22.4, S42.0, S82.8, 572.0, S71.1, S52.1, S32.0, S52.0, S82.4, S82.3, S72.1, S82.5, S22.0, S82.0, S82.7, S82.2, S82.0, S32.5, S32.2, S42.3, S52.2, S52.3, S42.4, S72.3, S52.8, S22.2, S52.4, S42.1, S72.2, S72.4, S32.1, S22.1, S12.2, S32.7, S32.8, S32.4, S82.9, S32.3, S52.9, S12.1, S42.8, S12.7, S72.8, S42.7, S72.9, S22.5, S72.7, S12.0, and S42.9), osteoporosis (ICD-10 codes M80 and M81), the prevalent coronary artery disease (ICD-10 codes I20, I21, I22, I23, I24, and I25), infarction (ICD-10 codes I21, I22, and I25.2), and SBP (measured at participant recruitment). For all of the outcome variables with an exception of cardiovascular disease, we considered prevalent case statuses reported at recruitment and individuals with records of diagnosis codes reported in the electronic registries before the recruitment. For the outcome of cardiovascular disease, we considered only prevalent cases reported at recruitment.
Ethical considerations in EGCUT
Analyses in EGCUT were approved by the Ethics Review Committee of the University of Tartu (243 T-12).
China Kadoorie Biobank
The CKB is a prospective cohort of 512,713 adults aged 30 to 79 years. Individuals were recruited between the years of 2004–2008 from five urban and five rural regions across China, as previously described (94). Baseline information was collected via detailed questionnaire (including demographic/lifestyle factors and medical history) and physical measurements (which included anthropometry, blood pressure, and spirometry). A nonfasting blood sample was taken and separated into plasma and buffy coat fractions for long-term storage. Longterm follow-up is through electronic linkage of each participant’s unique national identification number to the Chinese national health insurance system and established regional registries for death and disease. Health insurance reports include detailed information [e.g., disease description, ICD-10 code, and procedure or examination codes] about each hospital admission. Vascular disease events have been reviewed and standardized by clinicians.
Genotyping in CKB
A total of 102,783 CKB participants were genotyped using two custom-designed Affymetrix Axiom arrays including up to 800,000 variants, optimized for genome-wide coverage in Chinese populations. Stringent QC included an SNP call rate of >0.98, plate effect P > 10−6, batch effect P > 10−6, HWE P > 10−6 (combined 10 df χ2 test from 10 regions), biallelic, MAF difference from a 1000 Genomes Project (1KGP) EAS of <0.2, a sample call rate of >0.95, heterozygosity < mean + 3 SD, no chromosome XY aneuploidy, genetically determined sex concordant with database, and exclusion of recent immigrants to each study area as identified by region-specific PCA, resulting in genotypes for 532,415 variants present on both array versions in 94,592 individuals. Imputation into the 1000 Genomes phase 3 reference (EAS MAF, >0) using SHAPEIT3 and IMPUTE4 yielded genotypes for 10,276,634 variants with an MAF of >0.005 and an info of >0.3. rs188810925 was not imputed (1KGP EAS MAF, 0), and rs7209826 was imputed with an info of 0.986 and an MAF of 0.30.
Study outcomes and definitions in CKB
Outcomes, phenotypes, and transformations are defined in table S14. Disease end points are for hospitalizations, from electronic linkage to the national health insurance system. Phenotypes were measured either at baseline or in a subset of individuals at the second resurvey of ~5% of the CKB cohort.
Ethical considerations in CKB
Ethical approval for CKB was obtained jointly from the University of Oxford, the Chinese Center for Disease Control and Prevention (CCDC), and the regional CCDC from the 10 study areas.
Transethnic replication
We attempted transethnic replication in CKB. In 21,547 individuals with eBMD measurements available, the rs7209826 variant showed no evidence of association (β = 0.0016 g/cm2; SE = 0.001; P = 0.14) and strong evidence of heterogeneity with the corresponding eBMD estimate in UKBB (P heterogeneity = 1.35 × 10−28; fig. S19). A regional analysis showed no evidence of a suitable genetic instrument (fig. S20). Transethnic replication in a large East Asian biobank (CKB) could not be reliably performed because of a lack of association of SOST variants with BMD; however, there was a lack of association with CHD and other outcomes in CKB (table S15). This provides an opportune means to illustrate that the multiple cardiometabolic risk factor and disease associations identified in the European datasets, wherein genetic variants of SOST act as valid instruments for inhibition of sclerostin, did not arise as a consequence of horizontal pleiotropy (that is, the genetic associations did not arise through a mechanism other than that which operates through inhibition of sclerostin). The absence of suitable instruments in CKB does not imply that pharmacological inhibition of sclerostin will not be an effective therapeutic strategy for raising BMD in non-European populations.
GWAS consortia
We supplemented data from UKBB with summary-level data from nine GWAS consortia, including data for BMD [both ultrasound-derived eBMD (32) and dual-energy x-ray absorptiometry (DXA)–derived BMD measured at various anatomical sites (33)], CHD and MI (95), stroke (96), atrial fibrillation (97), T2D (98) and glycaemic traits (99, 100), serum lipid fractions (101), anthropometric traits (88, 89), and chronic kidney disease (102, 103). Where available, we selected data pertaining to analyses conducted in European ancestry individuals. Further details on each consortium are provided in table S16.
Genetic Factors for Osteoporosis (GEFOS) consortium
Detailed descriptions pertaining to measurement of eBMD and BMD are provided elsewhere (32, 33). Estimates for eBMD were provided in grams per square centimeter units (32). Estimates for DXA-derived BMD across three anatomical sites [LS-BMD, femoral neck BMD (FN-BMD), and forearm BMD (FA-BMD)] were provided in SD units (33). We converted these estimates to grams per square centimeter units using an estimate of the pooled population SD for each of these three measures (0.18, 0.14, and 0.07 g/cm2 for LS-BMD, FN-BMD, and FA-BMD, respectively) in the GEFOS consortium (33).
Genetic instrument selection and validation
A recent large-scale GWAS for eBMD, conducted in 142,487 individuals (32), identified two conditionally independent (r 2 = 0.13 among European ancestry individuals in UKBB) genetic variants in the SOST locus associated with eBMD: rs7209826 (A > G; G allele frequency in UKBB, 40%) and rs188810925 (G > A; A allele frequency, 8%). Both SNPs are located ~35-kb downstream from SOST and fall within or near a 52-kb area that contains the van Buchem disease deletion (fig. S21), a region previously shown to affect SOST expression in human bone (7). Recent functional evidence has also shown that SNPs in this area [one of which, rs7220711, is in high LD (r 2 = 0.99) with rs7209826] regulate SOST expression via differential transcription factor binding (104). Previous MR studies have also made use of noncoding variants with an effect on gene expression as proxies for pharmacologic modulation of the same target gene (29, 30, 105, 106).
We extracted estimates for these two SNPs (rs7209826 and rs188810925) from GWAS consortia listed in table S16. For GWAS datasets that did not include these variants, we selected proxy variants in high LD (r 2 > 0.9 in European ancestry individuals) with our selected SNPs using HaploReg (107). We identified 14 SNPs to be in high LD (r 2 = 0.99) with rs7209826. Of these 14, we selected rs7220711 as a proxy for rs7209826 based on high LD (r 2 = 0.99 in European ancestry populations), availability across most consortia, and prior functional evidence linking rs7220711 to SOST expression (104). In addition, we validated the effect of rs7220711 on various measures of BMD as being comparable to that of rs7209826 (fig. S22). There were no suitable proxies (r 2 > 0.9) for rs188810925.
We examined the associations of rs7209826 and rs188810925 (and their selected proxies) on DXA-derived measures of BMD measured at specific body sites (LS-BMD, FN-BMD, and FA-BMD), using data from the largest GWAS to date for these phenotypes (33). We next examined the effect of these variants on SOST mRNA expression in various human tissues in the GTEx project dataset (108).
Study outcomes
Specific definitions used in each cohort are specified above. We first tested the association of rs7209826 and rs188810925 with key efficacy outcomes, i.e., fracture risk and risk of osteoporosis (both defined as a combination of self-reported outcomes and ICD-9 and ICD-10 codes).
We selected coronary disease as the primary outcome of interest, given the association with cardiac ischemic events reported in a previous RCT of romosozumab (14). We examined the association of the BMD-increasing alleles of rs7209826 and rs188810925 with risk of MI and/or coronary revascularization (including self-reported and ICD-9/ICD-10 codes for MI, coronary artery bypass graft surgery, and/or percutaneous transluminal coronary angioplasty) and a broader composite of all CHD (including all codes for MI and/or coronary revascularization, as well as self-reported and ICD-9/ ICD-10 codes for angina and chronic stable ischemic heart disease; see tables S12 to S14 for specific codes included).
Next, we investigated the association of the SOST variants with further outcomes and traits in two tiers: First, we examined risk of MACE (a composite of MI and/or coronary revascularization, stroke, or death from either) and all stroke (analogous to the “MACE” and “cerebrovascular events” outcomes studied in RCTs of romosozumab) and association with cardiovascular risk factors previously shown to play a causal role in CHD including hypertension, T2D, SBP, DBP, BMI, WHRadjBMI, LDL cholesterol, triglycerides, fasting insulin, and glycated hemoglobin (HbA1c). Second, we evaluated an exploratory group of outcomes and traits to identify further putative associations including ischemic stroke, hemorrhagic stroke, peripheral vascular disease, atrial fibrillation, heart failure, chronic kidney disease, aortic aneurysm, aortic stenosis, HDL cholesterol, fasting glucose, and serum creatinine-estimated glomerular filtration rate (eGFR).
Statistical analyses
We derived cohort-specific SNP effect estimates from participants in the four prospective cohorts. Estimates [log(OR) and the SE of log(OR) for binary outcomes and β and the SE of β for quantitative traits] for the per-allele effect of these variants on disease outcomes and quantitative traits were aligned to the BMD-increasing alleles (as per the effect of therapeutic sclerostin inhibition) and scaled to an increase in BMD equivalent to that reported with romosozumab treatment (see below). Estimates in each cohort were derived as described below.
U.K. Biobank
The genotype-outcome association analyses in UKBB were performed using SNPTEST v2.5.4. We used an additive frequentist model (using -frequentist 1) and included sex, age at baseline, genotyping array (a binary variable), and the first 15 principal components as covariates in all analyses. We accounted for genotype uncertainty by using -method expected.
Partners HealthCare Biobank
Linear (for continuous phenotype) or logistic regression (for case-control binary phenotype) was used to test associations between SNPsrs188810925 and rs7209826 and phenotypes of interest using PLINK 1.9. For phenotypes without residual inverse rank normalization, we adjusted for sex, age at assessment, age squared, genotype batches, and 15 principal components in the regression model. For phenotypes with residual inverse rank normalization, we did not adjust for any covariates in the regression model.
Estonian Biobank
We calculated the effect estimates for the two SNPs using logistic regression and adjusting for age, sex, and 15 principal components. All associations were tested under an additive model using glm function with R software (3.3.2).
China Kadoorie Biobank
To avoid ascertainment biases, unless otherwise specified, controls for binary end points were restricted to 72,795 subsets who had been randomly selected for genotyping (~28,000 of genotyped samples were selected on the basis of incident cardiovascular disease or chronic obstructive pulmonary disease disease events). Covariates were as specified in table S14. All analyses used release version 15 of the CKB database and were performed using SAS software (version 9.3, SAS Institute Inc.).
Scaling of allelic estimates
Similar to other studies using genetic variants to estimate the effects of drug target modulation (28, 75), we scaled allelic estimates pertaining to risk of osteoporosis, fractures, cardiometabolic outcomes, and quantitative traits to an increase in BMD equivalent to that reported in a phase 2 RCT with 210 mg of romosozumab monthly for 12 months (34). This corresponds to the dose evaluated in phase 3 RCTs of romosozumab (i.e. the RCTs that reported cardiovascular outcomes) and represents an increase of 0.09 g/cm2 in LS-BMD in postmenopausal women (34). We selected LS-BMD as the reference phenotype because this was the only BMD phenotype for which we had genetic data (33) and clinical trial data (34) in the same units of measurements (grams per square centimeter). To do this, we derived the allelic effect estimates (in SD units) for the association of rs7209826 and rs188810925 with LS-BMD from a previous large-scale GWAS of LS-BMD (33) (0.046 and 0.09 SD units, respectively). Because these estimates were reported in SD units, we first converted these standardized estimates to grams per square centimeter units by using the pooled (median) SD of LS-BMD as reported in the same study (one SD, 0.18 g/cm2). We then multiplied the SD unit effect estimates by this value, to derive estimates in grams per square centimeter units (rs188810925, 0.016 g/cm2; rs7209826, 0.008 g/cm2). Next, we derived a scaling factor for each SNP by dividing the increase in LS-BMD arising from treatment with 210 mg of romosozumab monthly for 12 months (0.09 g/cm2) by the allelic effect estimate for LS-BMD (scaling factors: rs188810925, 5.52; rs7209826, 10.84). All per-allele effect estimates [log(OR) and the SE of log(OR) for binary outcomes and β and the SE of β for quantitative traits] were subsequently multiplied by these scaling factors as an approximation to estimating the effects expected with 12 months of romosozumab (210 mg monthly). We recognize that genetic instruments for exposures represent lifelong durations of exposure and the nature of the disease-exposure relationship (e.g. whether it is cumulative) makes direct comparisons of estimates derived from a genetic instrument to a treatment trial challenging.
Meta-analysis of scaled allelic estimates
We meta-analyzed scaled estimates from the prospective cohorts with (scaled) estimates from GWAS consortia for equivalent outcomes using inverse-variance weighted fixed-effect meta-analysis. We predefined a P value threshold of <0.05 for the association with CHD, given the association of sclerostin inhibition with cardiac ischemic events in prior RCTs (14, 15). For the first tier of outcomes and traits (MACE, all stroke, and cardiovascular risk factors previously shown to play a causal role in CHD including hypertension, T2D, SBP, DBP, BMI, WHRadjBMI, LDL cholesterol, triglycerides, fasting insulin, and HbA1c), we used a Bonferroni adjusted P value threshold of <0.004 (adjusting for a cumulative of 13 tests), and for the second tier [including ischemic stroke, hemorrhagic stroke, peripheral vascular disease, atrial fibrillation, heart failure, chronic kidney disease, aortic aneurysm, aortic stenosis, HDL cholesterol, fasting glucose, and serum creatinine-eGFR], an adjusted P value threshold of <0.002 (adjusting for a cumulative of 24 tests). The R package metafor was used for performing all meta-analyses of scaled allelic estimates (109). Fixed-effect meta-analyses were used in all instances. Cochran’s Q test and the I 2 statistic were used to evaluate heterogeneity for all meta-analyses performed (see table S7).
Sensitivity analyses
To evaluate the robustness of our instrument selection approach, we investigated the concordance of our findings with those of an alternative set of SOST instruments. We extracted conditionally independent SOST variants from a more recent GWAS of eBMD by Morris et al. (35), which included 426,824 individuals as compared to the 142,487 used in the GWAS by Kemp et al. (32), the latter being the primary data source for instrument derivation in this manuscript (the datasets were overlapping). Using Morris et al. (35), two alternative SNPs were identified as most strongly associating with eBMD, rs2741856 and rs7217502. Both variants were in high or moderately high LD with the two variants derived from Kemp et al. (32) used in the current manuscript (r 2 = 0.98 between rs7209826 and rs7217502 and r 2 = 0.60 between rs188810925 and rs2741856). We compared associations of a SOST instrument using the two SNPs identified from Morris et al. (35) to our instrument [derived from Kemp et al. (32)] with various measures of BMD (fig. S14) and with major outcomes of interest (osteoporosis, fracture, and MI and/or coronary revascularization; fig. S15). We used the same scaling and meta-analysis approach for the two variants identified from Morris et al. (35) as we used for Kemp et al. (32), described above, using the LS-BMD estimate for each variant (fig. S14) to scale the allelic estimates relating to the major outcomes of interest.
Sex-specific analyses
In light of the indication for romosozumab in the United States and Europe (in women only), we performed additional sex-specific sensitivity analyses for all associated outcomes from the combined sexes analysis. Sexual heterogeneity was evaluated using Cochran’s Q test and the I 2 statistic.
Colocalization analyses
We used coloc, a Bayesian modeling approach (110), to estimate the posterior probability of shared causal variant(s) driving associations in pairs of traits (colocalization). We assessed colocalization of eBMD and tissue-specific SOST mRNA expression between conditionally independent eBMD signals in the SOST locus (those pertaining to rs7209826 and rs188810925) by first applying conditional analysis using the GCTA-COJO statistical suite (111), using genotype data from UKBB as an LD reference panel. We conditioned all datasets on rs2523161 (an independent eBMD-associated variant ~600 kb from SOST), on either rs7209826 or rs188810925, and on any further independent expression quantitative trail loci (eQTLs) identified in specific tissues. We ran coloc on 400-kb and 2-Mb regions centered on each independent lead eBMD-associated SNP and used the default priors. We investigated colocalization between eBMD and SOST gene expression in various tissues, SBP, and WHRadjBMI. Colocalization analyses where PP3 (posterior probability of distinct causal variants) + PP4 (posterior probability of a shared causal variant) < 0.8 were considered to be underpowered to detect colocalization (112, 113). Visual assessment of colocalization was performed using the LocusCompareR R library (114). For adequately powered colocalization analyses, we considered a PP4 of >0.8 as being consistent with colocalization between the two traits, although lower thresholds have recently been proposed (115–118).
pQTL analysis
We sought to investigate whether lower circulating sclerostin protein concentration is also associated with an increased risk of cardiometabolic diseases. For this analysis, we used genetic variants associated with circulating sclerostin protein as reported in a recent GWAS of this trait (67). Three trans-pQTLs (and no cis-pQTLs) were reported [two (rs215226 and rs7241221) meeting genome-wide significance of P < 5 × 10−8 and one (rs1485303) suggestive at P = 7.70 × 10−8]. We first evaluated the effect of these variants on various measures of BMD to confirm that the variants had a robust and appropriate effect on these biomarkers, before investigating their effects on clinical outcomes of interest.
For one variant (rs1485303), we found that the sclerostin-lowering allele was associated with lower eBMD. This suggests that this variant is likely primarily lowering BMD through non–sclerostin-related mechanisms, leading to secondary lowering of sclerostin concentration in response to a reduced BMD. This is in keeping with the bidirectional relationship between circulating sclerostin and BMD as shown by Zheng et al. (67). The closest gene to rs1485303, TNFRS11B, encodes osteoprotegerin, a natural inhibitor of RANK (receptor activator of nuclear factor kappa-Β) ligand (RANKL). RANKL is also the therapeutic target of denosumab, a monoclonal antibody indicated for the treatment of osteoporosis. These observations suggest that rs1485303 is unlikely to be a valid proxy for therapeutic inhibition of sclerostin. For the remaining two variants, rs215226 (intronic to B4GALNT3) and rs7241221 (near GALNT1), a lack of robust association with LS-BMD (P = 0.06 and 0.45, respectively) precluded further evaluation. Trans-pQTLs are vulnerable to potential confounding by horizontal pleiotropy, thereby potentially limiting confidence that the phenotypic associations of such SNPs are solely attributable to their effect on the protein of interest (sclerostin in our case). This is a central tenet of instrumental variable analysis (the so-called exclusion restriction criterion).
Phenome-wide association analysis
We extracted estimates relating to the association of rs188810925 and rs7209826 with 1074 binary outcomes (those with more than 200 cases available) from publicly available summary statistics generated by the Lee Lab (www.leelabsg.org/resources). All estimates were scaled to approximate the effect of romosozumab (210 mg monthly for 12 months) on LS-BMD (0.09 g/cm2) and aligned to the BMD-increasing alleles.
Meta-analysis of RCTs of sclerostin inhibitors
We searched for all phase 3 RCTs performed for sclerostin inhibitors (romosozumab, blosozumab, and setrusumab). We collected a record of trials conducted for sclerostin inhibitors from the websites of the agents’ developers (www.amgentrials.com/amgen/study.aspx and www.mereobiopharma.com/pipeline/bps-804-setrusumab/) and supplemented this with further searches on clinical trials registries (ClinicalTrials.gov, European Union Clinical Trials Register and International Clinical Trials Registry Platform) and PubMed (1966 to present), using the keywords (“sclerostin” or “romosozumab” or “AMG-785” or “setrusumab” or “blosozumab”) and (“trial” or “randomized controlled trial” or “RCT” or “randomised controlled trial”) for PubMed. One of the authors (J.B.) screened the titles and abstracts of all identified studies for eligibility; where doubt arose as to eligibility, the full paper was evaluated. Potentially eligible studies were reviewed with two senior authors (C.M.L. and M.V.H.). All published studies with available cardiovascular outcome data pertaining to a double-blind, comparator-controlled phase 3 RCT were eligible for inclusion. Our search strategy and results are summarised in table S17, with last search performed on 8 November 2019. Seven phase 3 trials were identified (table S18), of which three trials met our criteria for inclusion in the meta-analysis. A PRISMA flow diagram is shown in fig. S23. Assessment of risk of bias for each eligible trial was completed using the Cochrane Risk of Bias tool (119) and is shown in fig. S24. We extracted each trial’s duration, blinding status, nature of the control intervention, and data pertaining to risk of “clinical fracture” (a composite of nonvertebral fracture and symptomatic vertebral fracture) and risk of four cardiovascular outcomes (“cardiac ischemic events,” cerebrovascular events, MACE, and “serious cardiovascular events”) from the peer-reviewed publications of the trials. Where trials reported data pertaining to multiple phases (e.g., an initial blinded phase, followed by an open-label phase), only data pertaining to the blinded phase were extracted. We also incorporated unpublished data pertaining to the cardiovascular outcomes newly released by the sponsors of romosozumab for the U.S. FDA Drugs Advisory Committee meeting in January 2019 (120), with the caveat that some of these data have not undergone peer review (albeit adjudicated by two independent bodies—the Duke Clinical Research Institute and the Thrombolysis in Myocardial Infarction Study Group). Inclusion of these data in our analyses is indicated where appropriate. Data extracted from trial publications were cross-checked with the U.S. FDA–reported data, where possible. If data for the same trial and outcome differed between the original publication and the U.S. FDA–reported data, then we included data from the original publication in our primary analysis and performed further sensitivity analyses with inclusion of the U.S. FDA–reported data. The risk of bias arising from selective publication of trials was deemed to be low—of the seven phase 3 trials of romosozumab we identified (table S18), three were unpublished. The largest unpublished trial included 294 participants (compared to the two largest, published phase 3 trials, ARCH and FRAME, totaling 4093 and 7180 participants, respectively).
We then performed meta-analyses of cardiovascular outcome data at 12 months (corresponding to the duration of the blinded phase) from phase 3 RCTs using 210 mg of romosozumab monthly for 12 months (because this was the only agent for which eligible RCTs were available and also the standard dose and duration used across the eligible phase 3 trials), with cardiac ischemic events as the primary outcome of interest, given the association with this outcome observed in a previous RCT of romosozumab (14). Further meta-analyses were performed for cerebrovascular events and two composite outcomes: serious cardiovascular events (including cardiac ischemic events, cerebrovascular events, heart failure, cardiovascular death, noncoronary revascularization, and peripheral vascular ischemic events not requiring revascularization) and MACE (a composite of MI, stroke, or cardiovascular death). Meta-analyses were performed by computing ORs and 95% CIs for each cardiovascular outcome, using fixed-effect models. Cochran’s Q test was used to evaluate heterogeneity between trials. We set a prespecified P value threshold of <0.05 for meta-analyses of the RCTs given the prior evidence for ischemic cardiovascular events seen in individual RCTs of romosozumab (14, 15).
All meta-analyses of RCT cardiovascular outcomes were performed according to the Mantel-Haenszel method without continuity correction. This method has been shown to perform relatively well when events are rare and is also the default fixed-effect meta-analysis method recommended by the Cochrane collaboration (121–123). We performed additional sensitivity analyses using the Peto method, a fixed-effect method shown to provide relatively unbiased results if within-trial intervention and control groups are of about equal size and if the effect size is modest (121, 122). Meta-analysis of fracture data was performed using inverse variance–weighted fixed-effect meta-analysis of fracture risk at 12 months in the FRAME (13) and ARCH (14) trials (table S10). All meta-analyses of trial data were performed using the R package metafor. We completed the PRISMA checklist (table S19) (124).
Supplementary Material
Acknowledgements
We wish to express our gratitude to the studies and consortia that provided summary association results and to the participants of the biobanks and research cohorts for valuable contributions to our research.
Funding
J.B. is supported by funding from the Rhodes Trust, Clarendon Fund, and the Medical Sciences Doctoral Training Centre, University of Oxford. J.C.C. is funded by the Oxford Medical Research Council Doctoral Training Partnership (Oxford MRC DTP) and the Nuffield Department of Clinical Medicine, University of Oxford. T.F. is supported by the NIHR Biomedical Research Centre, Oxford. S.L.P. has a Veni Fellowship (016.186.071; ZonMw) from the Dutch Organization for Scientific Research, Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO). S.L. is funded through a Novo Nordisk Postdoctoral Research Fellowship. L.M. is supported by funding from Estonian Research Council grants PRG184, IUT20-60, IUT34-4, IUT34-11, and IUT24-6. B.M.N. is supported by NIH project 5P50HD028138-27—Project 2: Common Complex Trait Genetics of Reproductive Phenotypes. R.M. is supported by funding from Estonian Research Council grants PRG184, IUT20-60, IUT34-4, IUT34-11, and IUT24-6. C.M.L. is supported by the Li Ka Shing Foundation, WT-SSI/John Fell funds, the NIHR Biomedical Research Centre, Oxford, Widenlife, and NIH (5P50HD028138-27). M.V.H. works in a unit that receives funding from the U.K. Medical Research Council and is supported by a British Heart Foundation Intermediate Clinical Research Fellowship (FS/18/23/33512) and the National Institute for Health Research Oxford Biomedical Research Centre. C.K.B. is supported by funding from Kadoorie Charitable Foundation in Hong Kong, the U.K. Wellcome Trust (grant numbers 202922/Z/16/Z, 088158/Z/09/Z, and 104085/Z/14/Z), Chinese Ministry of Science and Technology (grant number 2011BAI09B01), National Natural Science Foundation of China (grant numbers 81390540, 81390541, and 81390544), U.K. Medical Research Council (grant numbers MC_PC_13049 and MC_PC_14135), GlaxoSmithKline, BHF Centre of Research Excellence, Oxford (grant number RE/13/1/30181), British Heart Foundation, and Cancer Research UK. C.K.B. wishes to thank the CKB participants, CKB project staff based at Beijing, Oxford and the 10 regional centers, the China National CDC and its regional offices for assisting with the fieldwork, and the assistance of BGI (Shenzhen, China) for conducting DNA extraction and genotyping. G.D.S. is funded by the Medical Research Council (MRC) and the University of Bristol, which fund the MRC Integrative Epidemiology Unit (MC_UU_00011/1).
Data and materials availability
All data associated with this study are present in the paper or the Supplementary Materials. All allelic estimates generated during analyses are presented in this manuscript. GWAS summary statistics used in our analyses are publicly available.
Footnotes
Author contributions
J.B., M.V.H., and C.M.L. contributed to the conception and design of the study and coordinated the establishment of the consortium of prospective cohorts. All authors contributed to the data collection and analyses and interpretation of findings. J.B., M.V.H., and C.M.L. contributed to the first draft of the manuscript. All authors contributed to and approved the final version of the manuscript.
Competing interests
J.B. has served as a consultant to the Bill and Melinda Gates Foundation Strategic Investment Fund. B.M.N. is a member of the Scientific Advisory Board for Deep Genomics, a consultant for Camp4 Therapeutics Corporation, a consultant for Merck & Co., a consultant for Takeda Pharmaceutical, and a consultant for Avanir Pharmaceuticals Inc. C.M.L. has collaborated with Novo Nordisk and Bayer in research and, in accordance with a university agreement, did not accept any personal payment. M.V.H. has collaborated with Boehringer Ingelheim in research and, in accordance with the policy of the Clinical Trial Service Unit and Epidemiological Studies Unit (University of Oxford), did not accept any personal payment. Since October 2018, C.A.G. has been a full-time employee of BenevolentAI, United Kingdom. Since January 2019, C.Y.C. has been a full-time employee of Biogen, United States. Since January 2019, S.L.P. has been a full-time employee of Vertex Pharmaceuticals, United Kingdom. Contributions to this manuscript by C.A.G., C.Y.C., and S.L.P. were made before commencement of the stated employment. All other authors declare that they have no competing interests.
References
- 1.Compston JE, McClung MR, Leslie WD. Osteoporosis. Lancet. 2019;393:364–376. doi: 10.1016/S0140-6736(18)32112-3. [DOI] [PubMed] [Google Scholar]
- 2.Black DM, Rosen CJ. Clinical practice. Postmenopausal osteoporosis. N Engl J Med. 2016;374:254–262. doi: 10.1056/NEJMcp1513724. [DOI] [PubMed] [Google Scholar]
- 3.Khosla S, Hofbauer LC. Osteoporosis treatment: Recent developments and ongoing challenges. Lancet Diabetes Endocrinol. 2017;5:898–907. doi: 10.1016/S2213-8587(17)30188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Villiers TJ. The quest for new drugs to prevent osteoporosis-related fractures. Climacteric. 2017;20:103–106. doi: 10.1080/13697137.2017.1289659. [DOI] [PubMed] [Google Scholar]
- 5.McClung MR. Clinical utility of anti-sclerostin antibodies. Bone. 2017;96:3–7. doi: 10.1016/j.bone.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y-H, Alisch RS, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot–containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen CJ. Romosozumab—Promising or practice changing? N Engl J Med. 2017;377:1479–1480. doi: 10.1056/NEJMe1711298. [DOI] [PubMed] [Google Scholar]
- 9.Seefried L, Baumann J, Hemsley S, Hofmann C, Kunstmann E, Kiese B, Huang Y, Chivers S, Valentin M-A, Borah B, Roubenoff R, et al. Efficacy of anti-sclerostin monoclonal antibody BPS804 in adult patients with hypophosphatasia. J Clin Invest. 2017;127:2148–2158. doi: 10.1172/JCI83731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recker RR, Benson CT, Matsumoto T, Bolognese MA, Robins DA, Alam J, Chiang AY, Hu L, Krege JH, Sowa H, Mitlak BH, et al. A randomized, double-blind phase 2 clinical trial of blosozumab, a sclerostin antibody, in postmenopausal women with low bone mineral density. J Bone Miner Res. 2015;30:216–224. doi: 10.1002/jbmr.2351. [DOI] [PubMed] [Google Scholar]
- 11.Reid IR. Targeting sclerostin in postmenopausal osteoporosis: Focus on romosozumab and blosozumab. BioDrugs. 2017;31:289–297. doi: 10.1007/s40259-017-0229-2. [DOI] [PubMed] [Google Scholar]
- 12.McClung MR, Grauer A, Boonen S, Bolognese MA, Brown JP, Diez-Perez A, Langdahl BL, Reginster J-Y, Zanchetta JR, Wasserman SM, Katz L, et al. Romosozumab in postmenopausal women with low bone mineral density. N Engl J Med. 2014;370:412–420. doi: 10.1056/NEJMoa1305224. [DOI] [PubMed] [Google Scholar]
- 13.Cosman F, Crittenden DB, Adachi JD, Binkley N, Czerwinski E, Ferrari S, Hofbauer LC, Lau E, Lewiecki EM, Miyauchi A, Zerbini CAF, et al. Romosozumab treatment in postmenopausal women with osteoporosis. N Engl J Med. 2016;375:1532–1543. doi: 10.1056/NEJMoa1607948. [DOI] [PubMed] [Google Scholar]
- 14.Saag KG, Petersen J, Brandi ML, Karaplis AC, Lorentzon M, Thomas T, Maddox J, Fan M, Meisner PD, Grauer A. Romosozumab or alendronate for fracture prevention in women with osteoporosis. N Engl J Med. 2017;377:1417–1427. doi: 10.1056/NEJMoa1708322. [DOI] [PubMed] [Google Scholar]
- 15.Lewiecki EM, Blicharski T, Goemaere S, Lippuner K, Meisner PD, Miller PD, Miyauchi A, Maddox J, Chen L, Horlait S. A phase III randomized placebo-controlled trial to evaluate efficacy and safety of romosozumab in men with osteoporosis. J Clin Endocrinol Metab. 2018;103:3183–3193. doi: 10.1210/jc.2017-02163. [DOI] [PubMed] [Google Scholar]
- 16.Evenepoel P, D’Haese P, Brandenburg V. Romosozumab in postmenopausal women with osteopenia. N Engl J Med. 2014;370:1664. doi: 10.1056/NEJMc1402396. [DOI] [PubMed] [Google Scholar]
- 17.Tsourdi E, Rachner TD, Hofbauer LC. Romosozumab versus alendronate and fracture risk in women with osteoporosis. N Engl J Med. 2018;378:195. doi: 10.1056/NEJMc1714810. [DOI] [PubMed] [Google Scholar]
- 18.Brandenburg VM, Verhulst A, Babler A, D’Haese PC, Evenepoel P, Kaesler N. Sclerostin in chronic kidney disease-mineral bone disorder think first before you block it! Nephrol Dial Transplant. 2018;34:408–414. doi: 10.1093/ndt/gfy129. [DOI] [PubMed] [Google Scholar]
- 19.Mullard A. FDA rejects first-in-class osteoporosis drug. Nat Rev Drug Discov. 2017;16:593. doi: 10.1038/nrd.2017.176. [DOI] [PubMed] [Google Scholar]
- 20.Amgen. European Medicines Agency accepts filing for EVENITY™ (romosozumab) 2018 www.amgen.com/en-au/media/news-releases/2018/01/european-medicines-agency-accepts-filing-for-evenity-romosozumab/
- 21.Amgen. Amgen and UCB resubmit biologics license application BLA for EVENITY™ (romosozumab) to the US FDA. 2018 www.amgen.com/media/news-releases/2018/07/amgen-and-ucb-resubmit-biologics-license-application-bla-for-evenity-romosozumab-to-the-us-fda/
- 22.Amgen. FDA approves EVENITY™ (romosozumab) for the treatment of osteoporosis in postmenopausal women at high risk for fracture. 2019 www.prnewswire.com/news-releases/fda-approves-evenity-romosozumab-aqqg-for-the-treatment-of-osteoporosis-in-postmenopausal-women-at-high-risk-for-fracture-300828376.html.
- 23.European Medicines Agency. Evenity: Pending EC decision. 2019 https://www.ema.europa.eu/en/medicines/human/EPAR/evenity.
- 24.UCB. EVENITY (romosozumab) receives positive CHMP opinion for the treatment of severe osteoporosis in postmenopausal women at high risk of fracture. 2019 www.ucb.com/stories-media/Press-Releases/article/EVENITY-romosozumab-Receives-Positive-CHMP-Opinion-for-the-Treatment-of-Severe-Osteoporosis-in-Postmenopausal-Women-at-High-Risk-of-Fracture.
- 25.Amgen. European Commission approves EVENITY® (romosozumab) for the treatment of severe osteoporosis in postmenopausal women at high risk of fracture. 2019 www.amgen.com/media/news-releases/2019/12/european-commission-approves-evenity-romosozumab-for-the-treatment-of-severe-osteoporosis-in-postmenopausal-women-at-high-risk-of-fracture/
- 26.Walker VM, Smith G Davey, Davies NM, Martin RM. Mendelian randomization: A novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol. 2017;46:2078–2089. doi: 10.1093/ije/dyx207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes MV. Human genetics and drug development. N Engl J Med. 2019;380:1076–1079. doi: 10.1056/NEJMe1901565. [DOI] [PubMed] [Google Scholar]
- 28.Scott RA, Freitag DF, Li L, Chu AY, Surendran P, Young R, Grarup N, Stancáková A, Chen Y, Varga TV, Yaghootkar H, et al. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aad3744. 341ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millwood IY, Bennett DA, Holmes MV, Boxall R, Guo Y, Bian Z, Yang L, Sansome S, Chen Y, Du H, Yu C, et al. Association of CETP gene variants with risk for vascular and nonvascular diseases among chinese adults. JAMA Cardiol. 2018;3:34–43. doi: 10.1001/jamacardio.2017.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes MV, Simon T, Exeter HJ, Folkersen L, Asselbergs FW, Guardiola M, Cooper JA, Palmen J, Hubacek JA, Carruthers KF, Horne BD, et al. Secretory phospholipase A2-IIA and cardiovascular disease: A Mendelian randomization study. J Am Coll Cardiol. 2013;62:1966–1976. doi: 10.1016/j.jacc.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FDA. FDA briefing document for Meeting of the Bone, Reproductive and Urologic Drugs Advisory Committee. 2019 www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/Drugs/ReproductiveHealthDrugsAdvisoryCommittee/UCM629456.pdf.
- 32.Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, Zheng J, Gregson CL, Grundberg E, Trajanoska K, Logan JG, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. 2017;49:1468–1475. doi: 10.1038/ng.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H-F, Forgetta V, Hsu Y-H, Estrada K, Rosello-Diez A, Leo PJ, Dahia CL, Park-Min KH, Tobias JH, Kooperberg C, Kleinman A, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishibashi H, Crittenden DB, Miyauchi A, Libanati C, Maddox J, Fan M, Chen L, Grauer A. Romosozumab increases bone mineral density in postmenopausal Japanese women with osteoporosis: A phase 2 study. Bone. 2017;103:209–215. doi: 10.1016/j.bone.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, Vulpescu NA, Forgetta V, Kleinman A, Mohanty ST, Sergio CM, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51:258–266. doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawlor DA, Tilling K, Davey Smith G. Triangulation in aetiological epidemiology. Int J Epidemiol. 2016;45:1866–1886. doi: 10.1093/ije/dyw314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. 2018;362 doi: 10.1136/bmj.k601. k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan W, Clarke RJ, Mahajan A, Kulohoma B, Kitajima H, Robertson NR, Rayner NW, Walters RG, Holmes MV, Chen Z, McCarthy MI. Bone mineral density and risk of type 2 diabetes and coronary heart disease: A Mendelian randomization study. Wellcome Open Res. 2017;2:68. doi: 10.12688/wellcomeopenres.12288.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyles KW, Colón-Emeric CS, Magaziner JS, Adachi JD, Pieper CF, Mautalen C, Hyldstrup L, Recknor C, Nordsletten L, Moore KA, Lavecchia C, et al. Zoledronic acid and clinical fractures and mortality after hip fracture. N Engl J Med. 2007;357:1799–1809. doi: 10.1056/NEJMoa074941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ference BA. How to use Mendelian randomization to anticipate the results of randomized trials. Eur Heart J. 2018;39:360–362. doi: 10.1093/eurheartj/ehx462. [DOI] [PubMed] [Google Scholar]
- 41.Zhu D, Mackenzie NCW, Millán JL, Farquharson C, MacRae VE. The appearance and modulation of osteocyte marker expression during calcification of vascular smooth muscle cells. PLOS ONE. 2011;6:e19595. doi: 10.1371/journal.pone.0019595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandenburg VM, Kramann R, Koos R, Krüger T, Schurgers L, Mühlenbruch G, Hübner S, Gladziwa U, Drechsler C, Ketteler M. Relationship between sclerostin and cardiovascular calcification in hemodialysis patients: A cross-sectional study. BMC Nephrol. 2013;14:219. doi: 10.1186/1471-2369-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claes KJ, Viaene L, Heye S, Meijers B, d’Haese P, Evenepoel P. Sclerostin: Another vascular calcification inhibitor? J Clin Endocrinol Metab. 2013;98:3221–3228. doi: 10.1210/jc.2013-1521. [DOI] [PubMed] [Google Scholar]
- 44.Novo-Rodríguez C, García-Fontana B, Luna-Del Castillo JDD, Andújar-Vera F, Ávila-Rubio V, García-Fontana C, Morales-Santana S, Rozas-Moreno P, Muñoz-Torres M. Circulating levels of sclerostin are associated with cardiovascular mortality. PLOS ONE. 2018;13:e0199504. doi: 10.1371/journal.pone.0199504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y-C, Hsu B-G, Liou H-H, Lee C-J, Wang J-H. Serum levels of sclerostin as a potential biomarker in central arterial stiffness among hypertensive patients. BMC Cardiovasc Disord. 2018;18:214. doi: 10.1186/s12872-018-0955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathold K, Wanby P, Brudin L, Von SP, Carlsson M. Alterations in bone turnover markers in patients with noncardio-embolic ischemic stroke. PLOS ONE. 2018;13:e0207348. doi: 10.1371/journal.pone.0207348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saadeldin MK, Elshaer SS, Emara IA, Maged M, Abdel-Aziz AK. Serum sclerostin and irisin as predictive markers for atherosclerosis in Egyptian type II diabetic female patients: A case control study. PLOS ONE. 2018;13:e0206761. doi: 10.1371/journal.pone.0206761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albassam RS, Sabico S, Alnaami AM, Khattak MNK, Lei KY, Al-Daghri NM, Reginster J-Y, Alokail MS. Bone metabolism markers are associated with neck circumference in adult Arab women. Osteoporos Int. 2019 doi: 10.1007/s00198-018-04830-6. [DOI] [PubMed] [Google Scholar]
- 49.Reppe S, Noer A, Grimholt RM, Halldórsson BV, Medina-Gomez C, Gautvik VT, Olstad OK, Berg JP, Datta H, Estrada K, Hofman A, et al. Methylation of bone SOST, its mRNA, and serum sclerostin levels correlate strongly with fracture risk in postmenopausal women. J Bone Miner Res. 2015;30:249–256. doi: 10.1002/jbmr.2342. [DOI] [PubMed] [Google Scholar]
- 50.Polyzos SA, Anastasilakis AD, Bratengeier C, Woloszczuk W, Papatheodorou A, Terpos E. Serum sclerostin levels positively correlate with lumbar spinal bone mineral density in postmenopausal women–the six-month effect of risedronate and teriparatide. Osteoporos Int. 2012;23:1171–1176. doi: 10.1007/s00198-010-1525-6. [DOI] [PubMed] [Google Scholar]
- 51.Kim SP, Frey JL, Li Z, Kushwaha P, Zoch ML, Tomlinson RE, Da H, Aja S, Noh HL, Kim JK, Hussain MA, et al. Sclerostin influences body composition by regulating catabolic and anabolic metabolism in adipocytes. Proc Natl Acad Sci USA. 2017;114:E11238–E11247. doi: 10.1073/pnas.1707876115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishna SM, Seto S-W, Jose RJ, Li J, Morton SK, Biros E, Wang Y, Nsengiyumva V, Lindeman JHN, Loots GG, Rush CM, et al. Wnt signaling pathway inhibitor sclerostin inhibits angiotensin II–induced aortic aneurysm and atherosclerosis. Arterioscler Thromb Vasc Biol. 2017;37:553–566. doi: 10.1161/ATVBAHA.116.308723. [DOI] [PubMed] [Google Scholar]
- 53.Javaheri B, Herbert E, Hopkinson M, Al-Jazzar A, Pitsillides AA. Sost haploinsufficiency provokes peracute lethal cardiac tamponade without rescuing the osteopenia in a mouse model of excess glucocorticoids. Am J Pathol. 2019;189:753–761. doi: 10.1016/j.ajpath.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pound P, Bracken MB. Is animal research sufficiently evidence based to be a cornerstone of biomedical research? BMJ. 2014;348 doi: 10.1136/bmj.g3387. g3387. [DOI] [PubMed] [Google Scholar]
- 55.Wojczynski MK, Li M, Bielak LF, Kerr KF, Reiner AP, Wong ND, Yanek LR, Qu L, White CC, Lange LA, Ferguson JF, et al. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med Genet. 2013;14:75. doi: 10.1186/1471-2350-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boström KI, Rajamannan NM, Towler DA. The regulation of valvular and vascular sclerosis by osteogenic morphogens. Circ Res. 2011;109:564–577. doi: 10.1161/CIRCRESAHA.110.234278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Touw WA, Ueland T, Bollerslev J, Schousboe JT, Lim WH, Wong G, Thompson PL, Kiel DP, Prince RL, Rivadeneira F, Lewis JR. Association of circulating Wnt antagonists with severe abdominal aortic calcification in elderly women. J Endocr Soc. 2017;1:26–38. doi: 10.1210/js.2016-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 59.Mani A, Radhakrishnan J, Wang H, Mani A, Mani M-A, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saarinen A, Saukkonen T, Kivelä T, Lahtinen U, Laine C, Somer M, Toiviainen-Salo S, Cole WG, Lehesjoki A-E, Mäkitie O. Low density lipoprotein receptor-related protein 5 (LRP5) mutations and osteoporosis, impaired glucose metabolism and hypercholesterolaemia. Clin Endocrinol (Oxf) 2010;72:481–488. doi: 10.1111/j.1365-2265.2009.03680.x. [DOI] [PubMed] [Google Scholar]
- 61.Drake MT, Clarke BL, Oursler MJ, Khosla S. Cathepsin K inhibitors for osteoporosis: Biology, potential clinical utility, and lessons learned. Endocr Rev. 2017;38:325–350. doi: 10.1210/er.2015-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clarke R, Li L, Bennett D, Millwood IY, Walters RG, Guo Y, Bian Z, Peto R, Collins R, Parish S, Chen ZM. Abstract 16532: Impact of systolic blood pressure on cardiovascular disease in a chinese population: A Mendelian randomization study. Circulation. 2014;130 A16532. [Google Scholar]
- 63.Dale CE, Fatemifar G, Palmer TM, White J, Prieto-Merino D, Zabaneh D, Engmann JEL, Shah T, Wong A, Warren HR, McLachlan S, et al. Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes, and type 2 diabetes mellitus: A Mendelian randomization analysis. Circulation. 2017;135:2373–2388. doi: 10.1161/CIRCULATIONAHA.116.026560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Samelson EJ, Miller PD, Christiansen C, Daizadeh NS, Grazette L, Anthony MS, Egbuna O, Wang A, Siddhanti SR, Cheung AM, Franchimont N, et al. RANKL inhibition with denosumab does not influence 3-year progression of aortic calcification or incidence of adverse cardiovascular events in postmenopausal women with osteoporosis and high cardiovascular risk. J Bone Miner Res. 2014;29:450–457. doi: 10.1002/jbmr.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim DH, Rogers JR, Fulchino LA, Kim CA, Solomon DH, Kim SC. Bisphosphonates and risk of cardiovascular events: A meta-analysis. PLOS ONE. 2015;10:e0122646. doi: 10.1371/journal.pone.0122646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kranenburg G, Bartstra JW, Weijmans M, de Jong PA, Mali WP, Verhaar HJ, Visseren FLJ, Spiering W. Bisphosphonates for cardiovascular risk reduction: A systematic review and meta-analysis. Atherosclerosis. 2016;252:106–115. doi: 10.1016/j.atherosclerosis.2016.06.039. [DOI] [PubMed] [Google Scholar]
- 67.Zheng J, Maerz W, Gergei I, Kleber M, Drechsler C, Wanner C, Brandenburg V, Reppe S, Gautvik KM, Medina-Gomez C, Shevroja E, et al. Mendelian randomization analysis reveals a causal influence of circulating sclerostin levels on bone mineral density and fractures. J Bone Miner Res. 2019;34:1824–1836. doi: 10.1002/jbmr.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 69.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sebastian A, Loots GG. Genetics of Sost/SOST in sclerosteosis and van Buchem disease animal models. Metabolism. 2018;80:38–47. doi: 10.1016/j.metabol.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Corbin LJ, Tan VY, Hughes DA, Wade KH, Paul DS, Tansey KE, Butcher F, Dudbridge F, Howson JM, Jallow MW, John C, et al. Formalising recall by genotype as an efficient approach to detailed phenotyping and causal inference. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03109-y. 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGregor TL, Hunt KA, Nioi P, Mason D, Ticau S, Pelosi M, Perry L, Finer S, Griffiths C, MacArthur D, Trembath RC, et al. Deep phenotyping of a healthy human HAO1 knockout informs therapeutic development for primary hyperoxaluria type 1. bioRxiv. 2019 Jan 18; 524256 [Preprint. [Google Scholar]
- 73.Tankó LB, Christiansen C, Cox DA, Geiger MJ, McNabb MA, Cummings SR. Relationship between osteoporosis and cardiovascular disease in postmenopausal women. J Bone Miner Res. 2005;20:1912–1920. doi: 10.1359/JBMR.050711. [DOI] [PubMed] [Google Scholar]
- 74.Yesil Y, Ulger Z, Halil M, Halaçli B, Yavuz BB, Yeşil NK, Kuyumcu ME, Cankurtaran M, Ariogul S. Coexistence of osteoporosis (OP) and coronary artery disease (CAD) in the elderly: It is not just a by chance event. Arch Gerontol Geriatr. 2012;54:473–476. doi: 10.1016/j.archger.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 75.Schmidt AF, Swerdlow DI, Holmes MV, Patel RS, Fairhurst-Hunter Z, Lyall DM, Hartwig FP, Horta BL, Hyppönen E, Power C, Moldovan M, et al. PCSK9 genetic variants and risk of type 2 diabetes: A Mendelian randomisation study. Lancet Diabetes Endocrinol. 2017;5:97–105. doi: 10.1016/S2213-8587(16)30396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Resnik DB. Postmarketing Research and Surveillance: Issues and Challenges. Monitor. 2008;22:45–48. [PMC free article] [PubMed] [Google Scholar]
- 77.Spelsberg A, Prugger C, Doshi P, Ostrowski K, Witte T, Hüsgen D, Keil U, Working Group on Health and Working Group on Freedom of Information Transparency International Deutschland eV, Contribution of industry funded post-marketing studies to drug safety: Survey of notifications submitted to regulatory agencies. BMJ. 2017;356 doi: 10.1136/bmj.j337. j337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diogo D, Tian C, Franklin CS, Alanne-Kinnunen M, March M, Spencer CCA, Vangjeli C, Weale ME, Mattsson H, Kilpeläinen E, Sleiman PMA, et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06540-3. 4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS, Billington CK, Kheirallah AK, Allen R, Cook JP, Probert K, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): A genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trajanoska K, Morris JA, Oei L, Zheng H-F, Evans DM, Kiel DP, Ohlsson C, Richards JB, Rivadeneira F, GEFOS/GENOMOS Consortium and the 23andMe Research Team Assessment of the genetic and clinical determinants of fracture risk: Genome wide association and Mendelian randomisation study. BMJ. 2018;362 doi: 10.1136/bmj.k3225. k3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ismail AA, O’Neill TW, Cockerill W, Finn JD, Cannata JB, Hoszowski K, Johnell O, Matthis C, Raspe H, Raspe A, Reeve J, et al. Validity of self-report of fractures: Results from a prospective study in men and women across Europe. Osteoporos Int. 2000;11:248–254. doi: 10.1007/s001980050288. [DOI] [PubMed] [Google Scholar]
- 85.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, Ng FL, et al. Million Veteran Program, Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 87.Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, Yengo L, Ferreira T, Marouli E, Ji Y, Yang J, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694,649 individuals of European ancestry. Hum Mol Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, Workalemahu T, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 91.Smoller JW, Karlson EW, Green RC, Kathiresan S, MacArthur DG, Talkowski ME, Murphy SN, Weiss ST. An eMERGE clinical center at partners personalized medicine. J Pers Med. 2016;6:5. doi: 10.3390/jpm6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gainer VS, Cagan A, Castro VM, Duey S, Ghosh B, Goodson AP, Goryachev S, Metta R, Wang TD, Wattanasin N, Murphy SN. The biobank portal for partners personalized medicine: A query tool for working with consented biobank samples, genotypes, and phenotypes using i2b2. J Pers Med. 2016;6:11. doi: 10.3390/jpm6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mitt M, Kals M, Pärn K, Gabriel SB, Lander ES, Palotie A, Ripatti S, Morris AP, Metspalu A, Esko T, Mägi R, et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur J Hum Genet. 2017;25:869–876. doi: 10.1038/ejhg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Z, Chen J, Collins R, Guo Y, Peto R, Wu F, Li L, China Kadoorie Biobank (CKB) Collaborative Group China Kadoorie Biobank of 0.5 million people: Survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40:1652–1666. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.CARDIoGRAMplusC4D Consortium. A comprehensive 1000 genomes–based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, Anderson CD, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, Surakka I, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, Cook JP, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manning AK, Hivert M-F, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu C-T, Bielak LF, Prokopenko I, Amin N, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wheeler E, Leong A, Liu C-T, Hivert M-F, Strawbridge RJ, Podmore C, Li M, Yao J, Sim X, Hong J, Chu AY, et al. Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLOS Med. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, Taliun D, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7 doi: 10.1038/ncomms10023. 10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, Nolte IM, Cocca M, Taliun D, Gomez F, Li Y, et al. 1000 genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep. 2017;7 doi: 10.1038/srep45040. 45040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ye W, Wang Y, Mei B, Hou S, Liu X, Wu G, Qin L, Zhao K, Huang Q. Computational and functional characterization of four SNPs in the SOST locus associated with osteoporosis. Bone. 2018;108:132–144. doi: 10.1016/j.bone.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 105.Burgess S, Ference BA, Staley JR, Freitag DF, Mason AM, Nielsen SF, Willeit P, Young R, Surendran P, Karthikeyan S, Bolton TR, et al. Association of LPA variants with risk of coronary disease and the implications for lipoprotein(a)-lowering therapies: A Mendelian randomization analysis. JAMA Cardiol. 2018;3:619–627. doi: 10.1001/jamacardio.2018.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JEL, Shah T, Sofat R, Stender S, Johnson PCD, Scott RA, Leusink M, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: Evidence from genetic analysis and randomised trials. Lancet. 2015;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 110.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLOS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alasoo K, Rodrigues J, Mukhopadhyay S, Knights AJ, Mann AL, Kundu K, HIPSCI Consortium. Hale C, Dougan G, Gaffney DJ. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Çalışkan M, Manduchi E, Shanker Rao H, Segert JA, Beltrame MH, Trizzino M, Park Y, Baker SW, Chesi A, Johnson ME, Hodge KM, et al. Genetic and epigenetic fine mapping of complex trait associated loci in the human liver. bioRxiv. 2018 Oct 8; doi: 10.1016/j.ajhg.2019.05.010. 432823 [Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siewert KM, Voight BF. Bivariate genome-wide association scan identifies 6 novel loci associated with lipid levels and coronary artery disease. Circ Genom Precis Med. 2018;11:e002239. doi: 10.1161/CIRCGEN.118.002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardiñas AF, Rajagopal VM, Als TD, Nguyen HT, Girdhar K, Boocock J, et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019;51:659–674. doi: 10.1038/s41588-019-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sabik OL, Calabrese GM, Taleghani E, Ackert-Bicknell CL, Farber CR. Identification of a core module for bone mineral density through the integration of a co-expression network and GWAS data. bioRxiv. 2019 Oct 13; doi: 10.1016/j.celrep.2020.108145. 803197 [Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andaleon A, Mogil LS, Wheeler HE. Genetically regulated gene expression underlies lipid traits in Hispanic cohorts. PLOS ONE. 2019;14:e0220827. doi: 10.1371/journal.pone.0220827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sterne JAC, Savović J, Page MJ, Elbers RG, Blencowe NS, Boutron I, Cates CJ, Cheng H-Y, Corbett MS, Eldridge SM, Emberson JR, et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366 doi: 10.1136/bmj.l4898. 14898. [DOI] [PubMed] [Google Scholar]
- 120.FDA. January 16, 2019: Meeting of the Bone, Reproductive and Urologic Drugs Advisory Committee Meeting. 2019 www.fda.gov/advisory-committees/advisory-committee-calendar/january-16-2019-meeting-bone-reproductive-and-urologic-drugs-advisory-committee-meeting-announcement.
- 121.Bradburn MJ, Deeks JJ, Berlin JA, Russell Localio A. Much ado about nothing: A comparison of the performance of meta-analytical methods with rare events. Stat Med. 2007;26:53–77. doi: 10.1002/sim.2528. [DOI] [PubMed] [Google Scholar]
- 122.Sweeting MJ, Sutton AJ, Lambert PC. What to add to nothing? Use and avoidance of continuity corrections in meta-analysis of sparse data. Stat Med. 2004;23:1351–1375. doi: 10.1002/sim.1761. [DOI] [PubMed] [Google Scholar]
- 123.Efthimiou O. Practical guide to the meta-analysis of rare events. Evid Based Ment Health. 2018;21:72–76. doi: 10.1136/eb-2018-102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ. 2009;339 b2535. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. All allelic estimates generated during analyses are presented in this manuscript. GWAS summary statistics used in our analyses are publicly available.