Abstract
Successful pregnancies rely on adaptations within the mother1, including marked changes within the immune system2. It has long been known that the thymus, the central lymphoid organ, changes markedly during pregnancy3. However, the molecular basis and importance of this process remain largely obscure. Here we show that the osteoclast differentiation receptor RANK4,5 couples female sex hormones to rewiring of the thymus during pregnancy. Genetic deletion of Rank (also known as Tnfrsf11a) in thymic epithelial cells results in impaired thymic involution and blunted expansion of natural regulatory T (Treg) cells in pregnant female mice. Sex hormones, in particular progesterone, drive the development of thymic Treg cells through RANK in a manner that depends on AIRE+ medullary thymic epithelial cells and depletion of Rank in the thymic epithelium results in reduced accumulation of natural Treg cells in the placenta, accompanied by an increased number of miscarriages. Thymic deletion of Rank also resulted in impaired accumulation of Treg cells in visceral adipose tissue, associated with enlarged adipocyte size, tissue inflammation, enhanced maternal glucose intolerance, fetal macrosomia, and a long-lasting transgenerational alteration in glucose homeostasis; key hallmarks of gestational diabetes. Transplantation of Treg cells rescued fetal loss, maternal glucose intolerance and fetal macrosomia. In human pregnancies, gestational diabetes also correlates with a reduced number of Treg cells in the placenta. Our findings show that RANK promotes the hormone-mediated development of thymic Treg cells during pregnancy and expand the functional role of maternal Treg cells to gestational diabetes and the transgenerational metabolic rewiring of glucose homeostasis.
During pregnancy, sex hormones help to coordinate marked anatomical and physiological adaptations in the pregnant mother in order to safeguard fetal development1. The receptor activator of NF-κB ligand (RANK), its ligand RANKL and the decoy osteoprotegerin regulate—under the control of female sex hormones—bone remodelling and the development of lactating mammary glands during pregnancy6,7. In the thymus, RANK signalling regulates the maturation of AIRE- and CD80-expressing medullary thymic epithelial cells (mTECs)8, which have been implicated in the development of natural thymic Treg cells9. We therefore hypothesized that RANK could mediate vital thymic adaptations during pregnancy.
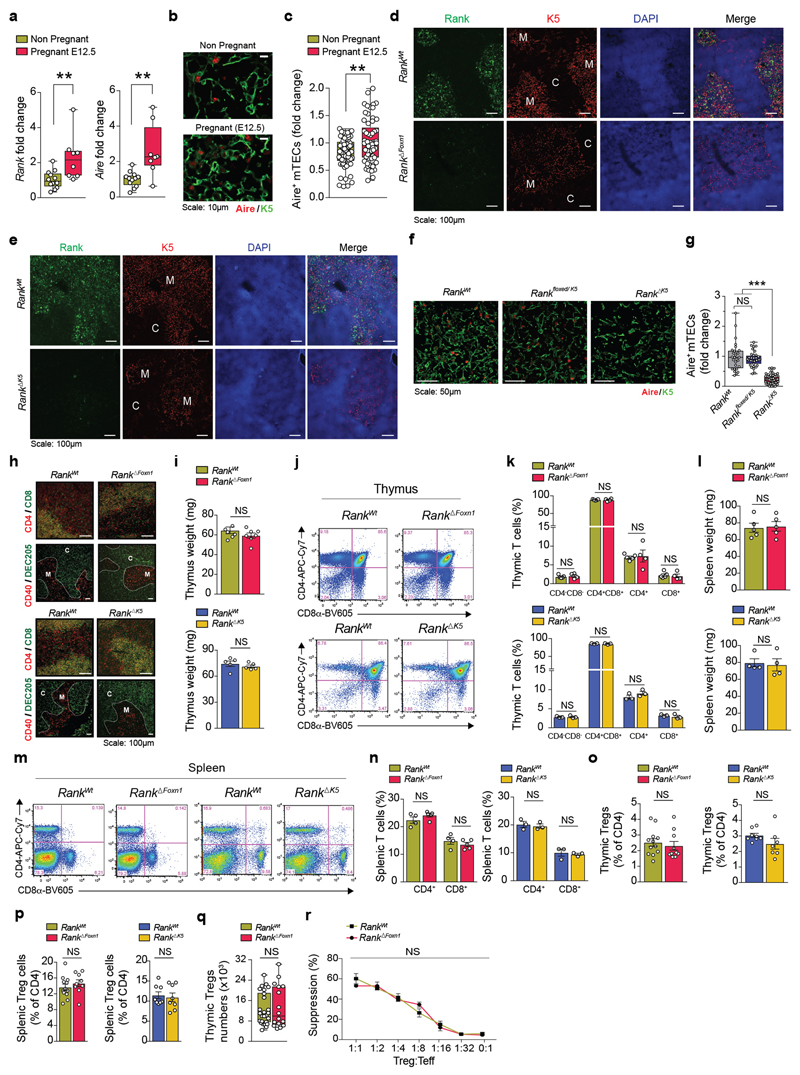
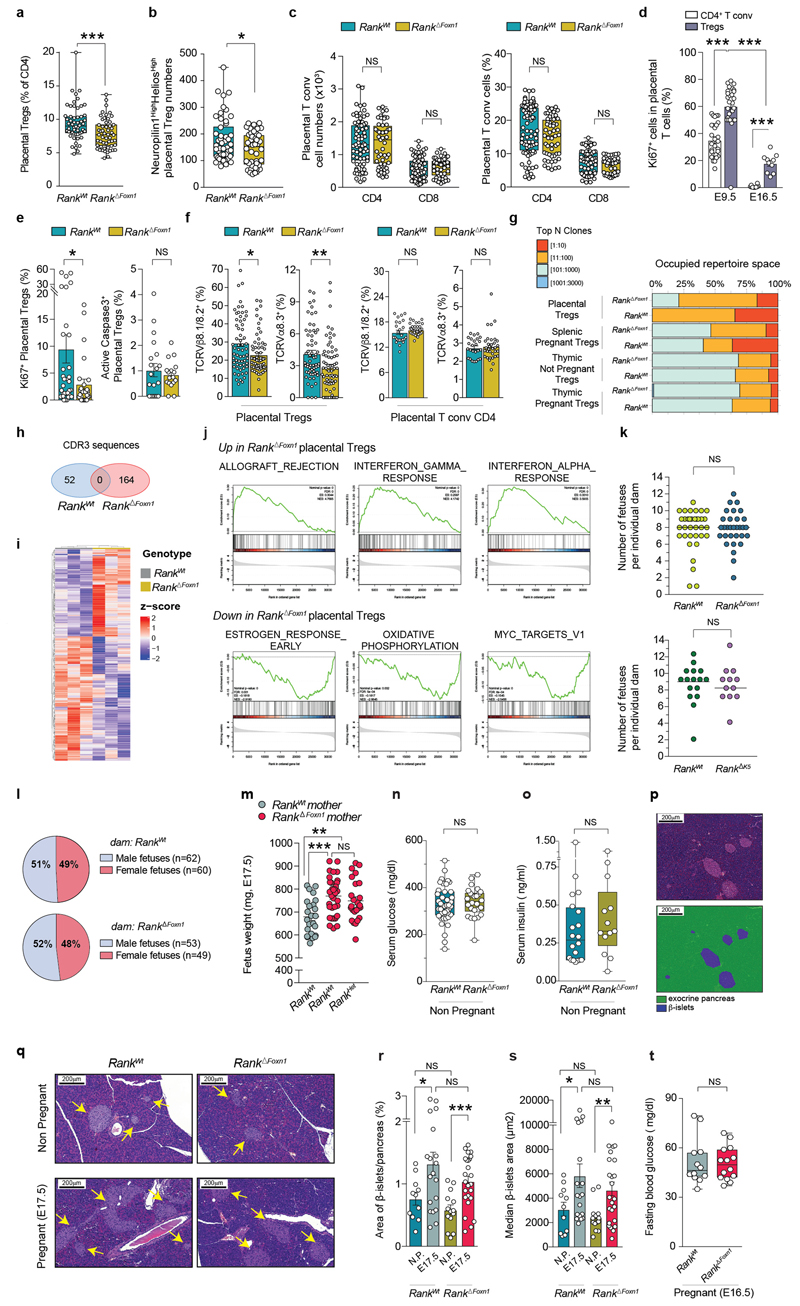
In the thymi of pregnant wild-type mice, Rank and Aire mRNA levels were upregulated during pregnancy, and pregnant female mice displayed increased numbers of AIRE+mTECs (Extended Data Fig. 1a–c). We next deleted Rank in thymic epithelial cells by crossing Rank floxed/floxed with Foxn1 cre/wt mice to generate Rank floxed/floxed Foxn1 wt/wt (hereafter, Rank WT) and Rank floxed/floxed Foxn1 Cre/wt animals (hereafter, Rank ΔFoxn1). We observed efficient deletion in the thymic medulla and reduced numbers of AIRE+mTECs in Rank ΔFoxn1 mice (Fig. 1a and Extended Data Fig. 1d). The findings in Rank ΔFoxn1 mice were corroborated by conditional Rank deletion in mTECs using K5 cre (K5 is also known as Krt5) mice (Rank ΔK5) (Extended Data Fig. 1e–g). Rank ΔFoxn1 and Rank ΔK5 females appeared overtly normal, exhibiting normal thymic size, thymic organization, thymocyte subsets, normal splenic T cell populations, percentages of thymic and splenic Treg cells, and suppressive activities of splenic Treg cells (Extended Data Fig. 1h–r). Thus, in non-pregnant female mice, the genetic inactivation of Rank in the thymic epithelium markedly reduces the numbers of mature mTECs, without any apparent effect on the thymic architecture, and the development of thymocytes or Treg cells.
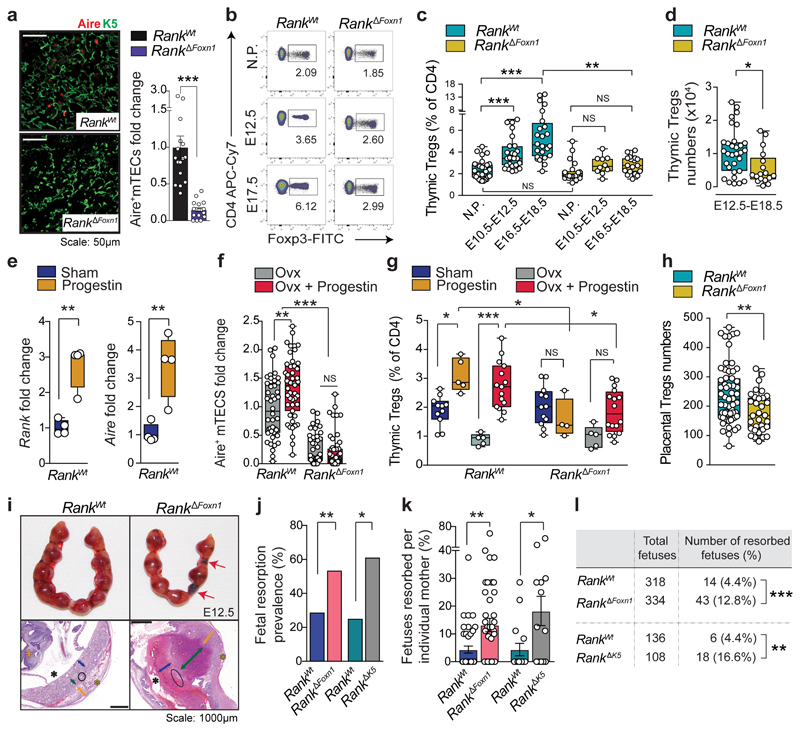
Fig. 1. RANK-expressing mTECs control thymic Treg cell expansion in pregnancy.
a, AIRE and K5 staining in thymic cross-sections and relative numbers of AIRE+mTECs in Rank WT and Rank ΔFoxn1 mice (n = 4, 4–6 fields per mouse). Scale bar, 50 μm. b, Fluorescence-activated cell sorting (FACS) plots for CD4+FOXP3+Treg cells in the thymus of non-pregnant (NP), and pregnant Rank WT and Rank ΔFoxn1 mice at E12.5 and E17.5 (the gating strategy is shown in Supplementary Data 1). c, Percentages of CD4+FOXP3+Treg cells in the thymus of non-pregnant, mid-pregnant (E10.5–E12.5) and late pregnant (E16.5–E18.5) Rank WT and Rank ΔFoxn1 females (n = 10–28). d, Numbers of CD4+FOXP3+Treg cells in the thymus of Rank WT and Rank ΔFoxn1 pregnant mice at E12.5–E18.5 (n = 31/17 for Rank WT/Rank ΔFoxn1 mice, respectively. e, Rank and Aire mRNA expression in the thymic stroma of sham- or progestin-treated Rank WT female mice (n = 4). f, AIRE+mTECs in ovariectomized (Ovx, n = 7/8), and ovariectomized and progestin-treated (n = 8/11) Rank WT and Rank ΔFoxn1 female mice (5–6 fields per mouse). g, Percentages of CD4+FOXP3+ Thymic Treg cells in sham-treated (n = 11/12), progestin-treated (n = 5/4), ovariectomized (n = 5/5), and ovariectomized and progestin-treated (n = 15/16) Rank WT and Rank ΔFoxn1 female mice. h, Numbers of CD4+FOXP3+Treg cells in E17.5 placentas of Rank WT and Rank ΔFoxn1 mice. n = 59/32 placentas from n = 11/8 dams. i, Top, uterine horns of Rank WT and Rank ΔFoxn1 dams displaying placental–fetus units. Images are representative of n = 10/9. Red arrows show fetal resorption. Bottom, congested and widened spongiotrophoblast (green arrows), necrotic labyrinth (blue arrows) and trophoblast giant cells (black circles) in placental cross-sections stained with haematoxylin and eosin (images are representative of n = 29/30 placentas). Orange arrows, decidua basalis; yellow asterisks, mesometrial lymphoid aggregate of pregnancy; black asterisks, amnion cavity; orange cross, embryo. Scale bars, 1,000 μm. j, k, Prevalence (percentage of mice with at least one fetal resorption) (j) and percentages (k) of fetal resorption in Rank WT and Rank ΔFoxn1 (n = 40/40), and Rank WT and Rank ΔK5 (n = 16/13) dams at E12.5–E18.5. l, Numbers and percentages of resorbed fetuses in Rank WT and Rank ΔFoxn1 (n = 41/42), and Rank WT and Rank ΔK5 (n = 16/13) dams at E12.5–E18.5. Data are shown as bar graphs (a, k; mean ± s.e.m.), and box-and-whisker plots (c–h, from minimal to maximal values. Dots represent individual data points. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Two-tailed Student’s t-test (a, e); Kruskal–Wallis test with Dunn’s post-test (c); two-tailed Mann–Whitney U-test (d, h, k); one-way analysis of variance (ANOVA) with Tukey’s post hoc test (f, g); two-tailed χ 2 test (j, l).
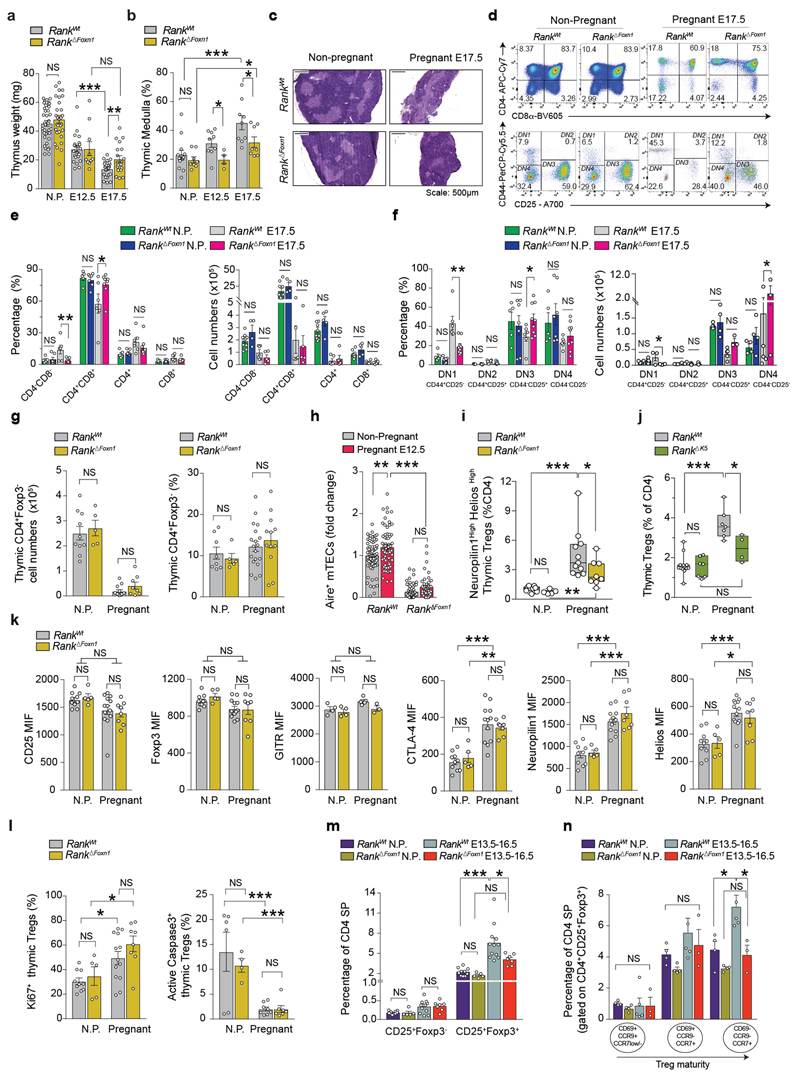
During pregnancy, the thymus undergoes marked changes, including partial involution, increased medullary-to-cortex ratios and altered thymocyte development; all of these thymic adaptions were altered in Rank ΔFoxn1 mice with syngeneic pregnancies (Extended Data Fig. 2a–g). Increased numbers of AIRE+mTECs and progressive thymic Treg cell expansion in pregnant female mice depended on Rank expression in the thymic epithelium (Fig. 1b–d and Extended Data Fig. 2h, i). Impaired thymic Treg cell expansion was confirmed in pregnant Rank ΔK5 female mice (Extended Data Fig. 2j). Whereas increased CTLA-4, neuropilin-1 and Helios expression as well as increased proliferation and improved survival of thymic Treg cells occurred in both pregnant Rank WT and Rank ΔFoxn1 female mice, Rank ΔFoxn1 mice exhibited impaired differentiation of CD69+CCR9+CCR7− to mature CD69−CCR9−CCR7+CD4+CD25+FOXP3+Treg cells in pregnancy (Extended Data Fig. 2k–n). Expansion of AIRE+mTECs and thymic Treg cells was confirmed in allogeneic pregnancies and was dependent on RANK expression in mTECs (Extended Data Fig. 3a–d). RNA-sequencing analysis showed that pregnancy is associated with marked changes in the molecular profiles of thymic Treg cells; Rank deletion in mTECs changed the transcriptome and T-cell-receptor (TCR) repertoire of Treg cells in pregnant as well as non-pregnant female mice (Extended Data Fig. 3e–g and Supplementary Table 1). Finally, RANK stimulation in fetal thymic organ cultures resulted in a marked increase in FOXP3+Treg cells, which could be reverted by the decoy osteoprotegerin (Extended Data Fig. 4a, b). Thus, loss of RANK expression in mTECs impairs thymic Treg cell expansion during pregnancy.
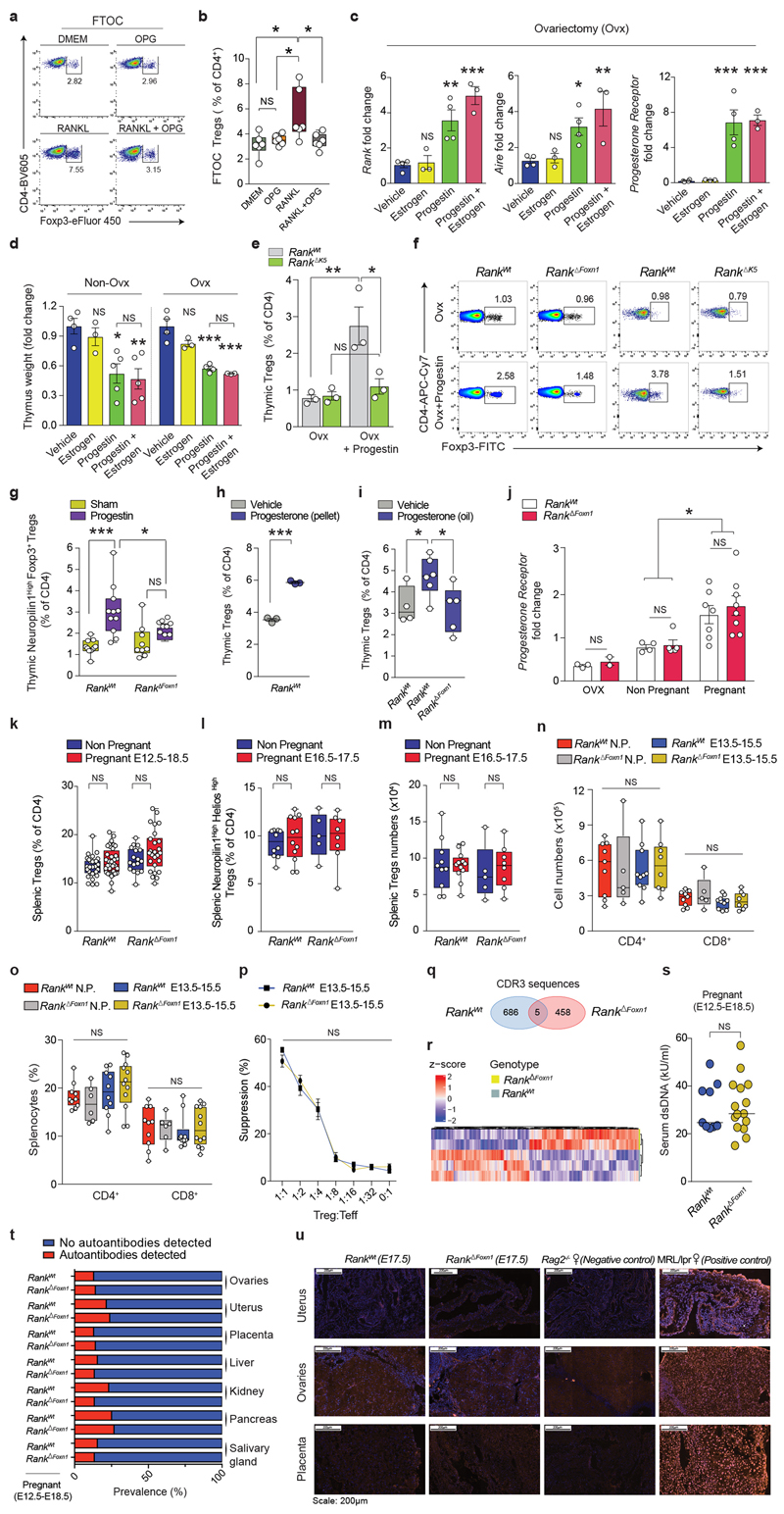
Female mice displayed thymic involution as well as an upregulation in the levels of Rank and Aire mRNA in the thymus in response to synthetic progestin, and to a lesser extent in response to oestrogen, regardless of whether the mice were ovariectomized (Fig. 1e and Extended Data Fig. 4c, d). Progestin increased AIRE+ mTEC and FOXP3+Thymic Treg cell numbers in ovariectomized Rank WT female mice, but not in ovariectomized Rank ΔFoxn1 littermates (Fig. 1f, g) nor in ovariectomized Rank ΔK5 female mice (Extended Data Fig. 4e, f). Progestin-expanded CD4+FOXP3+Treg cells expressed high levels of the thymic Treg cell marker neuropilin-110 (Extended Data Fig. 4g). Rank-dependent expansion of Treg cells was also induced by treatment with natural progesterone (Extended Data Fig. 4h, i). Thymic expression of the progesterone receptor was comparable among ovariectomized, non-pregnant and pregnant Rank WT and Rank ΔFoxn1 female mice (Extended Data Fig. 4j). Thus, progesterone induces thymic Treg cell development by acting on RANK+mTECs.
Next, we investigated the consequences of RANK-regulated thymic Treg cell expansion during pregnancy. We observed similar numbers and suppressive activities of Treg cells as well as CD4+ and CD8+T cells in the spleens of non-pregnant and mid-to-late stage pregnant Rank WT and Rank ΔFoxn1 female mice, and there were no overt signs of autoimmunity in pregnant Rank ΔFoxn1 female mice (Extended Data Fig. 4k–u and Supplementary Table 1). Whereas healthy pregnancies, in both humans and mice, correlate with the accumulation of Treg cells in the placenta11,12, placentas from Rank ΔFoxn1 dams displayed significantly lower numbers of Treg cells, in particular those of thymic origin, which were defined10,13 as neuropilin-1highHelioshigh (Fig. 1h and Extended Data Fig. 5a–c). We observed reduced proliferation as well as differential expression and a less-restricted TCR repertoire in placental Treg cells of Rank ΔFoxn1 mice, whereas they were similar among CD4+FOXP3− placental conventional T cells of both genotypes (Extended Data Fig. 5d–h and Supplementary Table 1). RNA-sequencing analysis revealed that placental Treg cells from Rank ΔFoxn1 dams displayed differential gene expression compared with Rank WT controls, with allograft rejection and inflammatory responses as the top overexpressed gene sets (Extended Data Fig. 5i, j and Supplementary Table 2). In line with the reduced numbers and molecular alterations in placental Treg cells, fetal resorptions rates were three times higher in pregnant Rank ΔFoxn1 and Rank ΔK5 mice compared with pregnant Rank WT controls (Fig. 1i–l). The total numbers of fetuses per mother as well as the male-to-female sex ratios were comparable (Extended Data Fig. 5k, l). Thus, the deletion of RANK in the thymic epithelium results in perturbed natural Treg cells in the placenta and increased fetal loss.
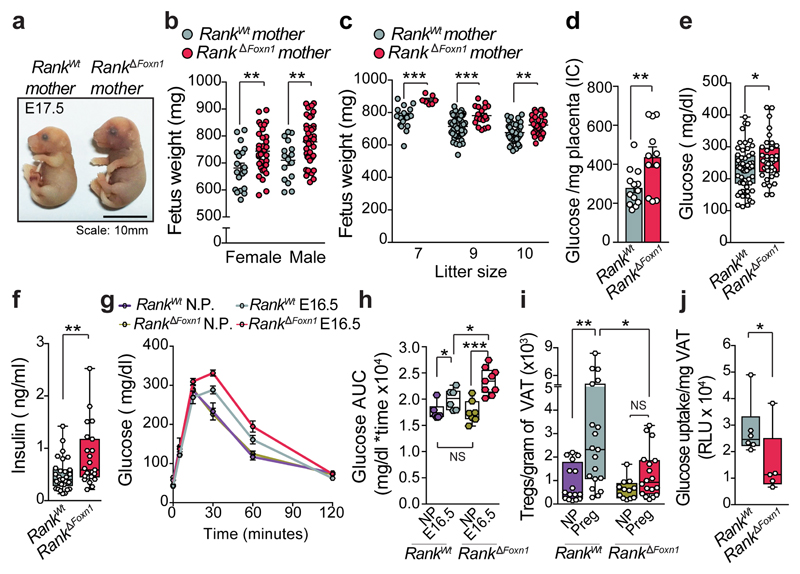
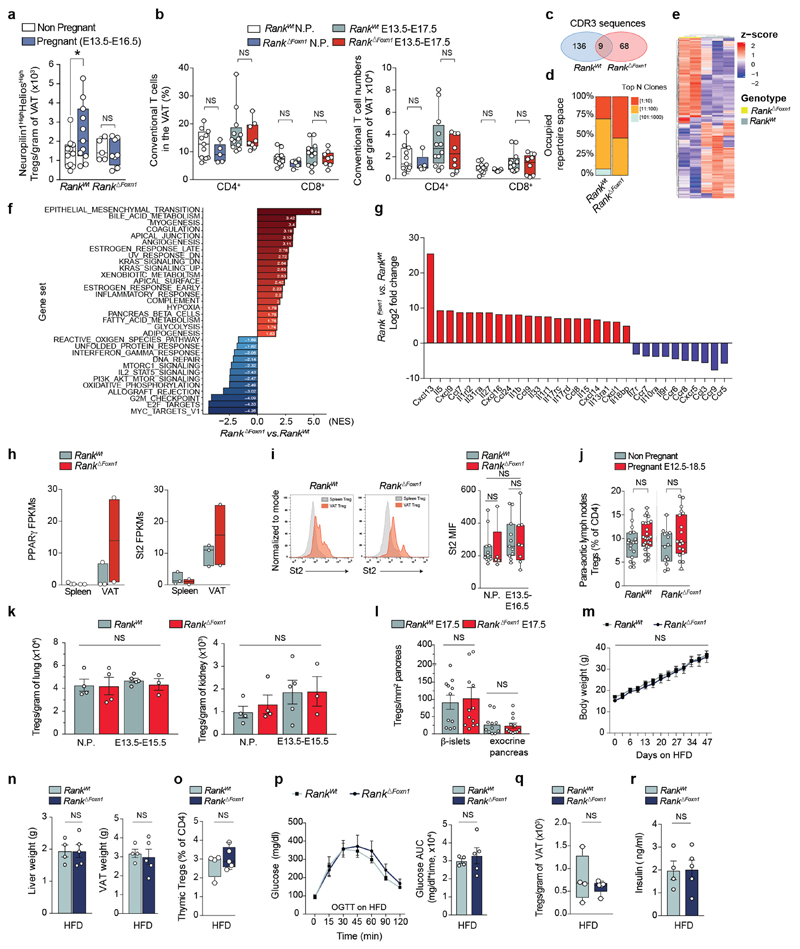
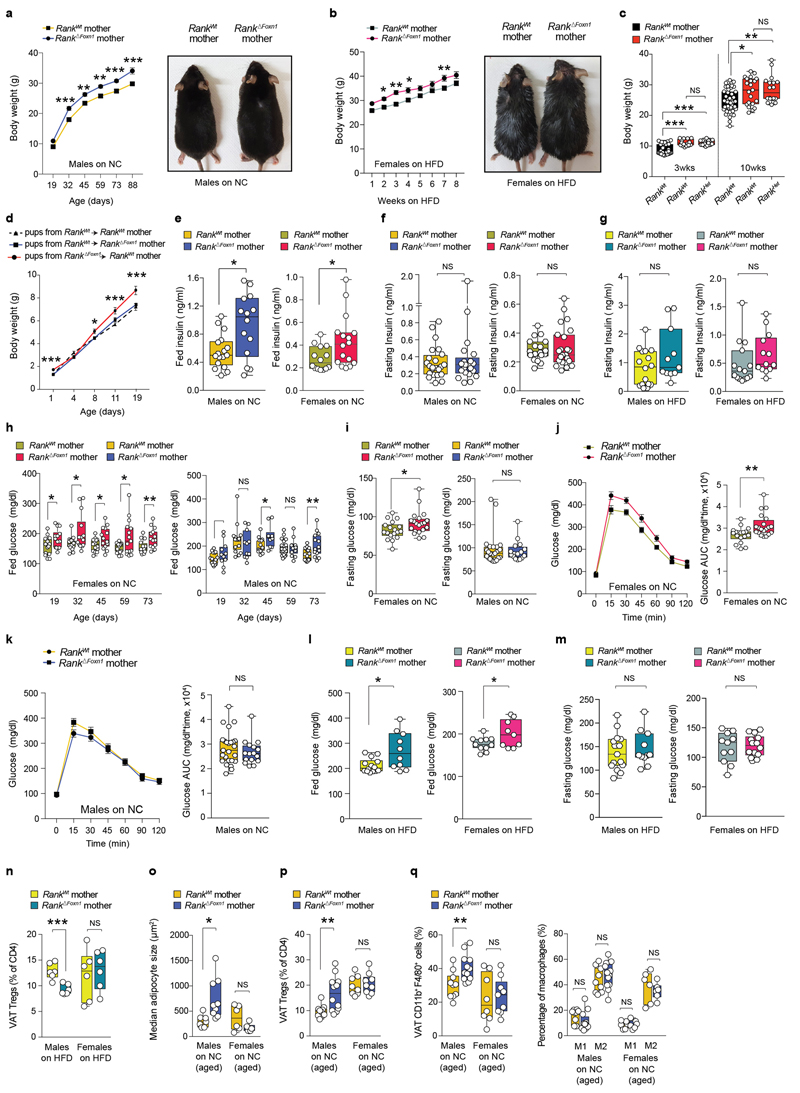
Surprisingly, viable fetuses of Rank ΔFoxn1 dams were significantly heavier at term regardless of sex, Rank heterozygosity or litter sizes (Fig. 2a–c and Extended Data Fig. 5m), a hallmark of gestational diabetes in humans14. Pregnant Rank ΔFoxn1 female mice displayed higher glucose levels in the placenta, and hyperglycaemia and hyperinsulinaemia in the serum (Fig. 2d–f); non-pregnant cohorts were indistinguishable (Extended Data Fig. 5n, o). Similarly to humans15, late pregnancy in mice was associated with decreased glucose tolerance; Rank ΔFoxn1 dams also showed significantly higher glucose response curves (Fig. 2g, h), which is indicative of glucose intolerance. There was no apparent difference in pregnancy-associated β-islet expansion16 and glucose levels were normalized in pregnant Rank ΔFoxn1 mice after fasting (Extended Data Fig. 5p–t), arguing against defects in insulin production and/or secretion. Previous studies in male mice have shown that Treg cells can alter glucose homeostasis by regulating inflammation in the visceral white adipose tissue (VAT)17–19. During pregnancy, we observed an accumulation of thymic Treg cells in the VAT of Rank WT, but not Rank ΔFoxn1, dams (Fig. 2i and Extended Data Fig. 6a, b). We also found molecular differences between VAT Treg cells from Rank WT and Rank ΔFoxn1 dams, including TCR expression, inflammation, metabolism, chemokine and cytokine signalling (Extended Data Fig. 6c–i and Supplementary Tables 1, 3). Concomitantly, the VAT of Rank ΔFoxn1 dams had a reduced glucose uptake capacity (Fig. 2j). Treg cell numbers in aortic lymph nodes, lungs, kidneys, exocrine pancreas and β-islets remained unchanged (Extended Data Fig. 6j–l). Body weight changes, thymic and VAT Treg cell numbers, glucose tolerance and insulin levels were comparable between non-pregnant Rank WT and Rank ΔFoxn1 female mice that were fed a high-fat diet (HFD) (Extended Data Fig. 6m–r). Thus, inactivation of Rank in the thymic epithelium results in impaired accumulation of Treg cells and reduced glucose uptake in the VAT of pregnant female mice, and is associated with increased gestational glucose intolerance and larger pups.
Fig. 2. Loss of Rank in mTECs augments gestational glucose intolerance.
a, Sizes of fetuses from Rank WT or Rank ΔFoxn1 mothers at E17.5. Scale bar, 10 mm. b, c, Macrosomia of E17.5 fetuses gestated in Rank ΔFoxn1 dams independent of sex (b) or litter size (c). n = 7–42 fetuses. d, Glucose levels in placentas of Rank WT and Rank ΔFoxn1 dams at E17.5 (n = 13/11 placentas from 4/5 dams). IC, ion current. e, f, Ad libitum-fed serum levels of glucose (e, n = 55/38) and insulin (f, n = 30/23) in pregnant (E12.5–E18.5) Rank WT and Rank ΔFoxn1 female mice. g, h, Oral glucose-tolerance test (mean ± s.e.m.) (g) and the corresponding area under the curve (AUC) (h) of non-pregnant (n = 6/7) and pregnant (E16.5) (n = 6/9) Rank WT and Rank ΔFoxn1 female mice. i, Total numbers of CD4+FOXP3+Treg cells in gonadal VAT of non-pregnant (n = 16/12) and pregnant (Preg, n = 19/18, E13.5–E17.5 pool) Rank WT and Rank ΔFoxn1 females. j, Uptake of 2-deoxyglucose in visceral adipose tissue of Rank WT and Rank ΔFoxn1 dams at E17.5. n = 6/5. RLU = relative luminescent units. Data are shown as bar charts, scatter dot plots (b-d, mean ± s.e.m.) and box-and-whisker plots (e,f,h-j, from minimal to maximal values). Dots represent individual data points. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed Student’s t-test for b-e; two-tailed Mann–Whitney U-test for f,h,j; One-way ANOVA, Tukey’s post hoc for i.
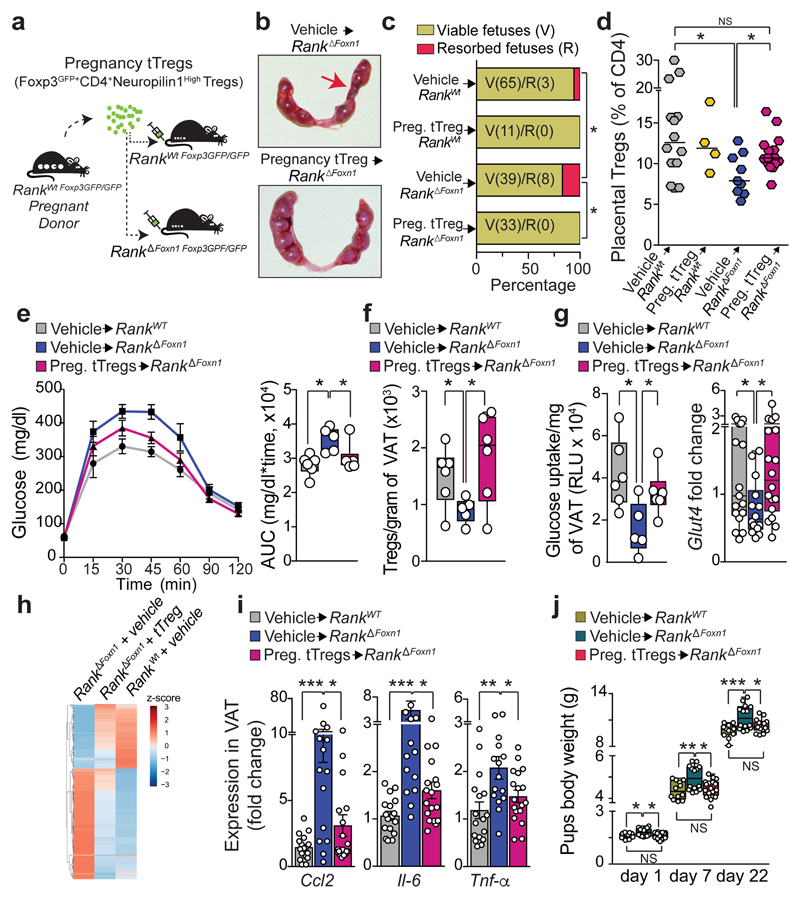
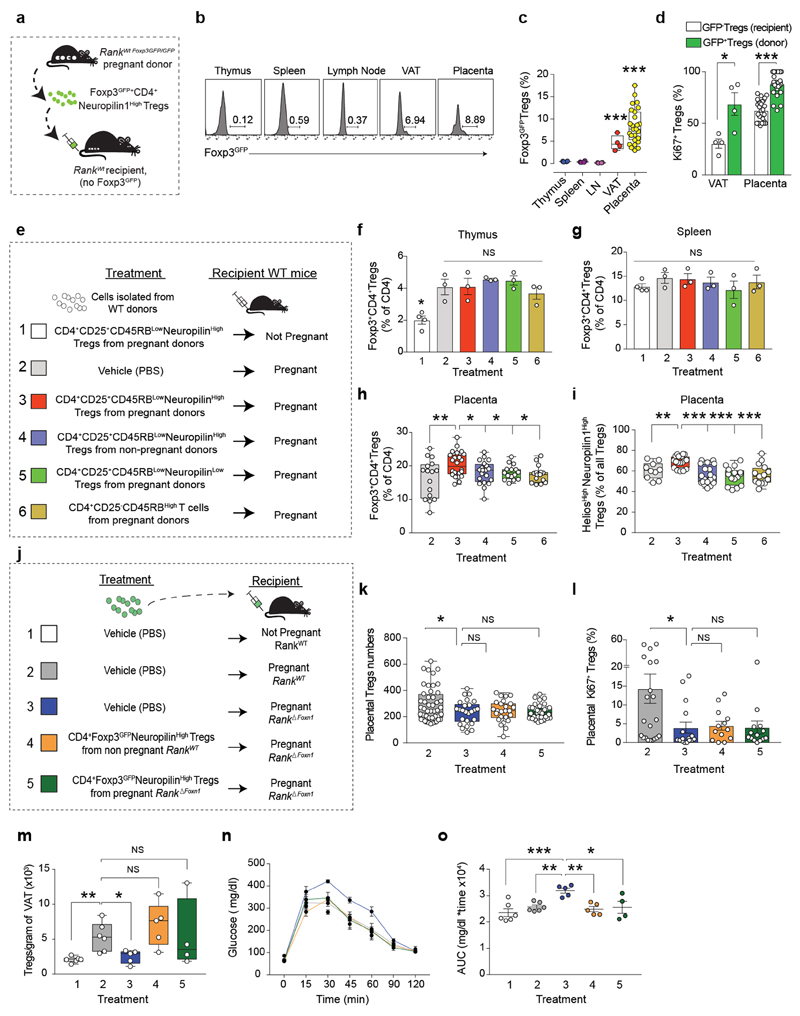
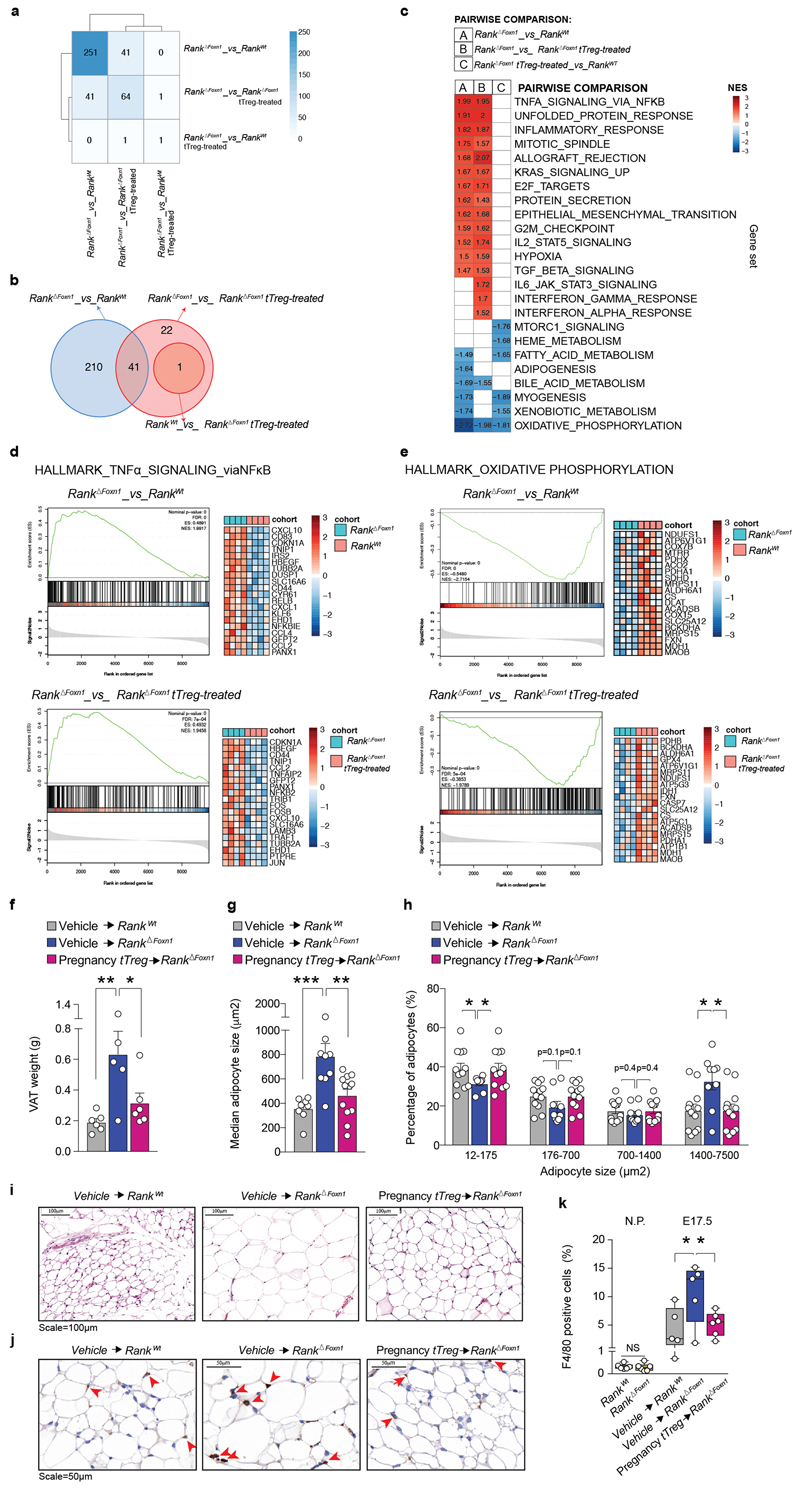
To demonstrate a causal role of thymic Treg cells in maternal metabolism and fetal loss, we established an adoptive Treg cell transfer model, showing that FOXP3–GFP+neuropilin-1high Treg cells from pregnant wild-type donors selectively home to the placenta; reconstitution of Rank ΔFoxn1 dams with regulatory T cells from pregnant Rank WT females restored the impaired placental Treg cell numbers and prevented miscarriages found in Rank ΔFoxn1 dams (Fig. 3a–d and Extended Data Fig. 7a–l). The transplantation of pregnancy-associated Treg cells also resulted in the homing of Treg cells to the VAT and restoration of the number of Treg cells, glucose tolerance, glucose uptake and expression of the glucose transporter Glut4 (also known as Slc2a4) in the VAT of Rank ΔFoxn1 pregnant mice (Fig. 3e–g and Extended Data Fig. 7a–d). In contrast to the placenta, transfer of Treg cells from non-pregnant Rank WT Foxp3 GFP/GFP and from pregnant Rank ΔFoxn1 Foxp3 GFP/GFP female mice could improve glucose tolerance and VAT Treg cell numbers in recipient Rank ΔFoxn1 dams (Extended Data Fig. 7m–o). RNA-sequencing analysis revealed significant differences in gene expression between the VAT of Rank ΔFoxn1and Rank WT dams, including TNF-dependent inflammation and oxidative phosphorylation (Fig. 3h and Extended Data Fig. 8a–e) —gene signature pathways linked to insulin resistance20,21. Treg cell transplantation restored gene expression in the VAT of Rank ΔFoxn1 dams (Fig. 3h, i and Extended Data Fig. 8a–e). The VAT of Rank ΔFoxn1 dams was significantly larger, showing enlarged adipocytes and marked macrophage infiltration; all of which was restored by Treg cell transplantation (Extended Data Fig. 8f–k). Finally, reconstitution of Treg cell numbers during pregnancy prevented macrosomia in the offspring of Rank ΔFoxn1 dams (Fig. 3j). Thus, reconstitution of Rank ΔFoxn1 dams with regulatory T cells from pregnant Rank WT female mice prevented miscarriages, VAT inflammation, gestational glucose intolerance and fetal macrosomia.
Fig. 3. Treg cells rescue fetal tolerance and maternal glucose metabolism.
a, Schematic of Treg cell adoptive transfer of pregnancy thymic Treg cells (Foxp3–GFP+CD4+neuropilin-1high Treg cells). For details, see Methods. b, Uterine horns of Rank ΔFoxn1 dams treated with vehicle (PBS) or pregnancy thymic Treg cells. Arrow shows fetal resorption. c, Percentages and total numbers of viable and resorpted fetuses gestated in Rank WT and Rank ΔFoxn1 pregnant mice, treated with vehicle or pregnancy thymic Treg cells. n = 8/2/6/4 dams. d, Percentage of CD4+FOXP3+Treg cells in E12.5 placentas of Rank WT and Rank ΔFoxn1 dams receiving vehicle or pregnancy thymic Treg cells. n = 14/4/9/15. e–g, Oral glucose-tolerance test (mean ± s.e.m.) and corresponding area under the curve (e), numbers of CD4+FOXP3+Treg cells in gonadal VAT (f) and insulin-stimulated 2-deoxyglucose uptake and relative expression of the glucose transporter Glut4 (g) in the VAT of vehicle-treated Rank WT (n = 6) and vehicle- and pregnancy thymic Treg cell-treated Rank ΔFoxn1 (n = 5/6) dams at E16.5 (e, f) and E17.5 (g, 2 VAT samples per mouse). RLU, relative luminescent units. h, Relative expression levels of differentially expressed genes in the pairwise comparisons (DESeq2 v.1.16.1, false-discovery rate (FDR) threshold of 0.05). n = 4. i, Ccl2, Il6 and Tnf qPCR expression in VAT of vehicle-treated Rank WT (n = 6) and vehicle- and pregnancy thymic Treg cell-treated Rank ΔFoxn1 (n = 5/6) dams at E17.5 (3 VAT samples per mouse). j, Body weights of pups, born to the indicated mothers, at perinatal and weaning ages (n = 12/14/17). Data are shown as scatter dot plots (d, lines are median), bar charts (i, mean ± s.e.m.) and box-and-whisker plots (e–g, j, from minimal to maximal values. Dots represent individual data points. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed χ 2 test (c); Student’s t-test (d, f, g); two-tailed Mann–Whitney U-test (i); Kruskal–Wallis test with Dunn’s post-test (e); one-way ANOVA with Tukey’s post hoc test (j).
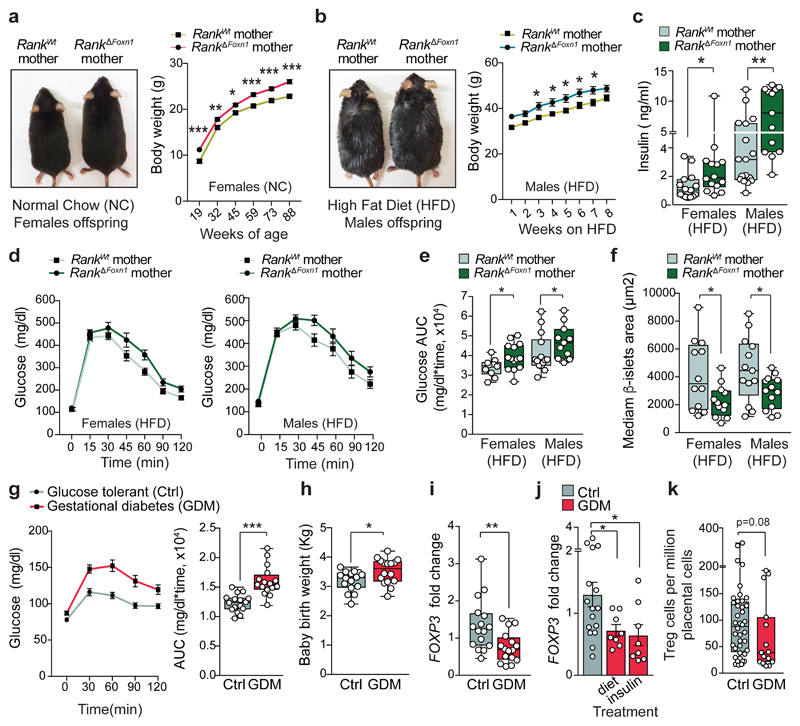
In humans, children born to women with gestational diabetes can have long-term health complications, such as proclivity to glucose intolerance and obesity22,23. We therefore followed the fate of macrosomic pups born to Rank ΔFoxn1 mothers, observing increased body weights throughout the entire timespan of follow-up, a phenotype observed in both male and female mice regardless of whether the mice were fed normal chow, HFD or nurtured by wild-type mothers (Fig. 4a, b and Extended Data Fig. 9a–d). We consistently observed hyperinsulinaemia in male and female offspring of Rank ΔFoxn1 mothers, again under both diet regiments; this effect was normalized after fasting (Fig. 4c and Extended Data Fig. 9e–g). When fed normal chow, female, but not male, offspring of Rank ΔFoxn1 mothers had higher glucose levels and impaired glucose tolerance (Extended Data Fig. 9h–k). When fed a HFD, both male and female offspring of Rank ΔFoxn1 mothers exhibited hyperglycaemia and marked glucose intolerance (Fig. 4d, e and Extended Data Fig. 9l, m). Similar to patients with type 1 and type 2 diabetes24,25, offspring of Rank ΔFoxn1 mothers have a reduced β-islet mass (Fig. 4f). Male offspring from Rank ΔFoxn1 mothers had reduced Treg cell frequencies when fed a HFD and, when fed normal chow, showed an increase in the size of adipocytes as well as altered frequencies of Treg cells and macrophages in the VAT (Extended Data Fig. 9n–q). Thus, ablation of Rank in the thymic epithelium results in glucose intolerance in pregnant female mice and transgenerationally in their offspring, with long-lasting effects.
Fig. 4. Transgenerational metabolic disorders in the offspring of Rank ΔFoxn1 mothers.
a, Body weights (mean ± s.e.m.) of the female offspring of Rank WT and Rank ΔFoxn1 dams (n = 15 and 14) fed normal chow. Image, 80-day-old mice. b, Photograph (6 weeks on HFD) and body weights (mean ± s.e.m.) of male offspring of Rank WT and Rank ΔFoxn1 dams (n = 16/11) fed an HFD. c–e, Ad libitum-fed serum insulin levels in female (n = 15/14) and male (n = 17/13) offspring of Rank WT and Rank ΔFoxn1dams (c), oral glucose-tolerance test (mean ± s.e.m.) (d) and area under the curve (e) for female (n = 14/10) and male (n = 14/11) offspring of Rank WT and Rank ΔFoxn1 dams fed a HFD for 9 weeks. f, Median β-islets area in female (n = 12/12) and male (n = 13/13) offspring of Rank WT and Rank ΔFoxn1 dams on HFD for 12 weeks. g, Oral glucose-tolerance test (mean ± s.e.m.) and area under the curve for pregnant women with gestational diabetes mellitus (GDM) and a glucose-tolerant pregnant control group (control). n = 16/16. h, Birth weights of babies born to mothers with gestational diabetes and control mothers (n = 16/16). i, FOXP3 transcripts in the placentas of control pregnant women and pregnant women with gestational diabetes (n = 16/16). j, FOXP3 transcripts in the placentas of control pregnant women and pregnant women with gestational diabetes treated with a controlled diet or by administration of insulin (mean ± s.e.m., n = 16/8/8). k, FOXP3+Treg cells in the placentas (maternal side) of control pregnant women and pregnant women with gestational diabetes (n = 17/8) detected by immunohistochemistry (2–3 sites per placenta). Data in c, e–i, k are shown as box-and-whisker plots (from minimal to maximal values). Dots represent individual data points. *P < 0.05; **P < 0.01; ***P < 0.001. Two-way ANOVA with Sidak’s multiple comparison (a, b); two-tailed Mann–Whitney U-test (c, e, g, i–k); two-tailed Student’s t-test (f, h).
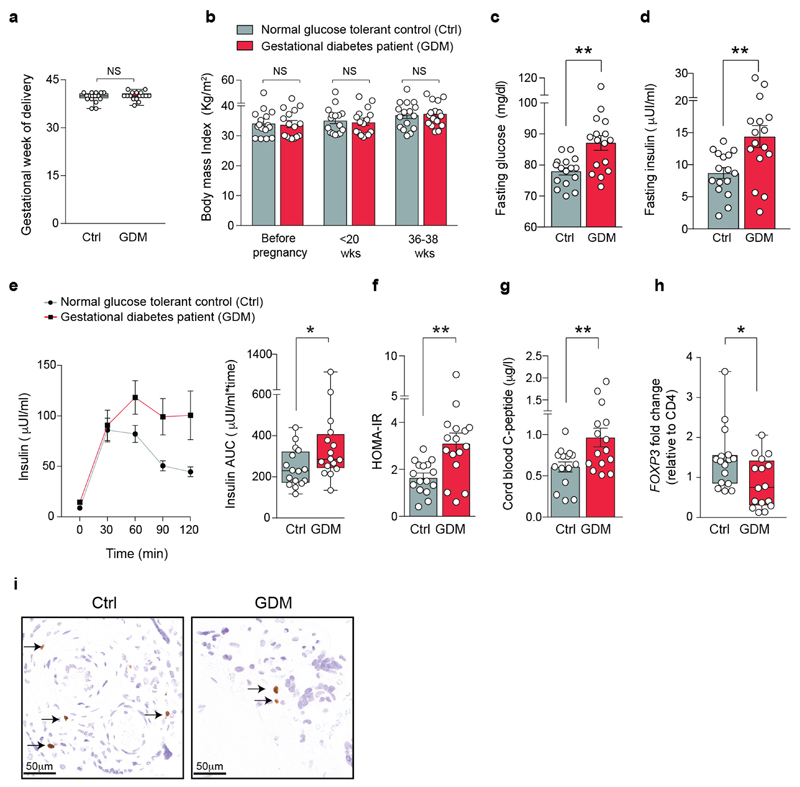
The estimated rates of gestational diabetes have reached up to 18–39% of human pregnancies26,27. We therefore assessed whether regulatory T cells are also altered in a clinical cohort study of women with gestational diabetes compared with women matched for body-mass index who had a normal glucose tolerance, and additionally matched for the sex of the babies, and week and mode of delivery (Fig. 4g and Extended Data Fig. 10a–f). As expected, babies born to mothers with gestational diabetes exhibited an increased birth weight and enhanced C-peptide levels, as a marker for insulin levels, in the cord blood (Fig. 4h and Extended Data Fig. 10g). Notably, we observed reduced Treg cell numbers in the placentas of women with gestational diabetes, regardless of diet or insulin treatments (Fig. 4i–k and Extended Data Fig. 10h, i). Therefore, in line with our studies in pregnant Rank ΔFoxn1 mice, reduced Treg cell numbers in the placenta correlate with gestational diabetes in human pregnancy.
How the immune system deals with the fetus during pregnancy remains unclear and is a central question of immunology and evolution. Various layers of immune regulation at the maternal–fetal interface have been linked to viable pregnancies2, including the presence of Treg cells11,12. Despite having a comparable suppression potency28,29, only pregnancy-associated Treg cells, but not Treg cells from non-pregnant female mice, can rescue miscarriages29, suggesting a dedicated development and/or function for pregnancy Treg cells. AIRE+mTECs preferentially express placental and lactating mammary gland genes30,31, providing further evidence for a role of these cells in pregnancy. In line with these studies, we now show that Treg cells from pregnant, but not from non-pregnant, female mice home to the placenta where they clonally expand and can prevent miscarriages; all of these mechanisms are dependent on RANK expression in mTECs.
Human pregnancy is associated with progressive insulin resistance that, if not compensated, can manifest as overt gestational diabetes32—a state that, although transitory, considerably increases the risk for both mother and child to develop glucose intolerance and diabetes33,34. The molecular and cellular bases of gestational diabetes remain largely unknown. Our data uncover a connection between the sex-hormone- and RANK-regulated rewiring of the thymic epithelial environment, thymic Treg cell expansion in pregnancy and miscarriages. Notably, RANK-regulated rewiring of the thymus is required to maintain, through VAT Treg cells, glucose homeostasis in the pregnant mother. We therefore provide molecular insights into gestational diabetes and show that sex hormones, RANK signalling in the thymic epithelium and natural Treg cells are a central mechanism for the integration of hormonal, immune and metabolic maternal adaptations in pregnancy.
Methods
Mice
Rank conditional mice (Rank flox) were generated in our laboratory and have been previously described35. We were given the following mouse strains: Foxn1 cre mice36 (G. A. Holländer), Foxp3 GFP reporter mice37 (A. Rudensky) and K5 cre mice38 (J. Takeda). Rag2 −/− mice were obtained from our in-house stock and MRL/lpr mice were purchased from Jackson Laboratories. For fetal thymic organ culture experiments and allogeneic crosses, BALB/cJ wild-type in-house mice were used. Except where stated otherwise, all other mouse experimental studies were performed on mice that were extensively backcrossed to the C57BL/6J background and only young (7–17-week-old) age-matched littermate female mice were used for experimentation. Mice were allocated to experimental groups based on their genotype and randomized within the given sex- and age-matched group. Given that our mice were inbred and matched for age and sex, we assumed similar variance between the different experimental groups. No statistical methods were used to predetermine sample size. We always used as many mice per group as possible in an attempt to minimize type-I and type-II errors. Critical experiments have high n numbers and were repeated multiple times. All sample sizes are indicated in the figure legends. Investigators were not blinded to allocation during experiments. For most studies (pregnancy studies and offspring studies) this was important, as we wanted to co-cage cohorts to reduce other variables (microbiota, matings to the same breeder, and so on). Genotyping was performed by PCR on genomic DNA and re-confirmed at the experimental end point, minimizing any potential bias during data collection. Housing conditions were: ambient temperature of 22 °C ± 1 °C; humidity of 55% +/-5% and a light/dark cycle of 14 h:10 h (light summertime, 07:00–21:00; light wintertime, 06:00–20:00). Images of mice shown in the figures were taken by M.P. in our animal facility. All animal experiments were performed following the principles of the 3Rs, in accordance with the Austrian Animal Experiments Acts and valid project licenses, which were approved and regularly controlled by the Austrian Federal Ministry of Education, Science and Research, and monitored by the institutional IMBA Ethics and Biosafety department.
Timed pregnancy studies
To exclude the potential effects of Rank deletion in the fetus, Rank WT and Rank ΔFoxn1 female littermates were co-caged and crossed to the same wild-type syngeneic C57BL/6J male breeder, resulting in RANK-sufficient fetuses with a comparable genetic background. Syngeneic pregnancies allowed us to better assess hormone-regulated Treg cells in pregnancy. For comparison, an allogeneic pregnancy study was set up; in this case, Rank WT and Rank ΔFoxn1 female littermates were also co-caged and crossed to the same wild-type BALB/cJ male breeder. All breeding experiments were setup in the afternoon and the presence of a vaginal plug was checked daily. On the morning of plug detection—considered E0.5—females were separated from the male breeder. At the embryological day of the experimental end point, pregnant females were euthanized and all placenta–fetus units in the uterus were macroscopically evaluated for the presence of viable or resorbed fetuses. Prevalence of fetal loss is presented as a percentage of pregnant mice showing any (at least one) fetal resorption event (detected from E12.5 to E18.5); the statistical analysis of prevalence was performed with a two-tailed χ 2 test using the raw data (number of pregnant females with and without resorptions). To detect fetal macrosomia, fetuses were collected at E17.5 for measurement of their body weight using a sensitive scale. Sex genotyping of the fetuses was performed by PCR on genomic DNA using male-specific primers: Sry1, TCATGAGACTGCCAACCACAG; Sry2, CATGACCACCACCACCACCAA. Moreover, at each pregnancy end point the maternal blood, thymus, spleen, para-aortic lymph nodes, pancreas, gonadal (peri-uterine) visceral adipose tissue, lung, kidneys and placentas were collected for FACS, histological and ELISA analyses.
Flow cytometry
T cells from the thymus, spleen, lymph nodes and placenta were recovered by mechanical disruption of isolated tissues. For quantification of Treg cells and macrophages in the gonadal VAT (peri-uterine for females and epididymal for males) was dissected, weighed and disaggregated (up to 0.5 g) in a gentle MACS Dissociator (Miltenyi Biotec) using an adipose tissue dissociation kit optimized for a high yield of viable cells and preservation of cell-surface markers (Miltenyi Biotec). Dissociation of lungs and kidney tissues for FACS analysis was achieved by incubation in DMEM containing collagenase type IV and DNase I (both from Worthington Biochemical) in the gentle MACS Dissociator with heat. After digestion, samples were treated with red-blood-cell lysis buffer before staining. Single-cell suspensions were immunostained for 30 min at 4 °C in FACS buffer (PBS, 2% FCS, 2 mM EDTA). Before staining, Fc receptors were blocked with anti-CD16/32 antibodies (BD Pharmingen, 1:100) and dead cells were labelled using a fluorescently labelled and fixable viability dye (eBioscience, 1:1,000). For flow cytometry, directly labelled antibodies against CD45 (30-F11, 1:800), TCRβ (clone Η57-597, 1:200), CD4 (RM4-5, 1:400), CD8α (53-6.7, 1:250), CD25 (PC61, 1:200), CD44 (IM7, 1:200), KI-67 (16A, 1:100), CCR9 (CW-1.2, 1:150), CD69 (H1.2F3, 1:150), CD11b (M1/70, 1:250), ST2 (DIH9, 1:200), GITR (DTA-1, 1:150), F4/80 (BM8, 1:300), CD11c (N418, 1:250), CD301 (LOM-14, 1:100), Ly6G (1A8, 1:300), neuropilin-1 (3E12, 1:150), Helios (22F6, 1:100) (all from Biolegend), and CCR7 (4B12, 1:150), active CASP3 (C92-605, 20 μl per sample), CTLA-4 (UC10-4F10-11, 1:150), Vβ 8.1 and Vβ 8.2 TCRs (MR5-2, 1:150), Vα 8.3 TCRs (KT50, 1:150) (all from BD Biosciences), as well as FOXP3 (FJK-16s, eBioscience, 1:100) were used. A list of all antibodies, including clones, catalogue numbers, companies and dilutions used is provided in the Reporting Summary. Treg cells of thymic origin were defined as CD4+FOXP3+ cells that co-express high levels of neuropilin-110,39 and Helios13. Intracellular staining for FOXP3, Helios, KI-67 and active CASP3 were carried out using the FOXP3/Transcription Factor Staining kit (eBioscience). Data were acquired using a multi-colour flow cytometer (FACSCanto or LSR Fortessa, BD) equipped with FACSDiva software (BD). Sorting of cells was performed using a AriaIII cell sorter (BD Bioscience). Data analysis was performed using FlowJo v.10.0.0 software (TreeStar). For FACS analysis and sorting only viable cells (LIVE/DEAD Fixable dye-negative cells) were considered. Absolute splenocyte and thymocyte numbers were determined using a FACS LSR Fortessa flow cytometer equipped with a BD High Throughput Sampler for 96 wells, which allows counting of cells in a fix volume, and subsequent extrapolation for the total sample volume.
Ovariectomy and hormonal treatment
For hormonal studies in females, bilateral ovariectomy was performed under ketamine/xylazine anaesthesia through a small abdominal incision to open the peritoneal cavity40. Ovaries including the surrounding adipose tissue were excised. The sham surgery was only the abdominal incision. Mice were allowed to recover for 3–4 weeks before hormonal treatment. For hormonal treatment, anaesthetized female mice were implanted subcutaneously on the left flank—then moved to the interscapular region to avoid scratching and potential loss of the implant at the incision point—with slow-release pellets containing the progesterone derivative medroxyprogesterone acetate (MPA; 50 mg per pellet, 90-day release; Innovative Research of America). Oestrogen (17β-oestradiol, Sigma) was dissolved in sesame oil and was administrated subcutaneously (10 μg kg−1), either alone or in combination with MPA pellets (same dose and implantation as the single treatment). Progesterone was delivered subcutaneously by daily injections (450 μg per mouse, Sigma) or by implantation of a progesterone-releasing pellet (50 mg per pellet, 21-day release, Innovative Research of America). Control mice for pellet implantation were treated with sham surgery; controls for subcutaneous hormone injections were treated with only vehicle. We also tested placebo pellets as controls (15 mg per pellet, 21-day release, Innovative Research of America); neither the placebo pellet nor sham surgery had any overt effects on Treg cell numbers when compared to untreated mice. Hormonal treatment lasted for 5 or 7 days; thereafter mice were euthanized and analysed for qPCR, expansion of Treg cells and thymic alterations.
Adoptive Treg cell transfer
To isolate viable bona fide FOXP3+Treg cells, Rank WT and Rank ΔFoxn1 animals were crossed to Foxp3 GFP/GFP reporter mice to allow sorting of Treg cells based on GFP expression. Three adoptive Treg cell transfer models were set up with different aims, as described below.
Tracking transplanted Treg cells
To track transplanted Treg cells, recipient early pregnant (E3.5–E5.5) Rank WT females (no Foxp3 GFP transgene) were intravenously inoculated with 3 × 105 sorted purified CD4+FOXP3–GFP+neuropilin-1high Treg cells isolated and pooled from spleen, lymph nodes and thymus of E6.5–E8.5 Rank WT Foxp3 GFP/GFP pregnant donors. Recipients were euthanized 5 days after transfer to determine the relative percentage of FOXP3–GPF+ Transplanted Treg cells in different organs (thymus, spleen, lymph nodes, VAT and placentas) using FACS analysis.
Determination of the tissue homing capacities of different T cells
To determine which cell type migrates to the placenta, wild-type female mice were used as donors as well as recipients. As wild-type mice carried no Foxp3 GFP transgene, Treg cells were sorted as CD4+CD25+CD45RBlow and conventional CD4+T cells as CD4+CD25+CD45RBhigh cells. Treg cells from thymic origin (defined as neuropilin-1high), Treg cells from non-thymic origin (defined as neuropilin-1low) as well as conventional T cells were sorted and pooled from spleen, lymph nodes and thymus of non-pregnant and pregnant (E8.5–E9.5) donors and transferred (intravenous inoculation, 1 × 105) into early pregnant recipients (E0.5–E2.5). Thymic Treg cells (CD4+CD25+CD45RBlowneuropilin-1high) were also transplanted into non-pregnant wild-type female mice. Vehicle-control pregnant females received only PBS. Recipient mice were euthanized 15 days after transfer (E15.5–E17.5) for FACS analysis of Treg cell numbers and percentages in thymus, spleen and placentas.
Phenotypic rescue experiments
For phenotypic rescue experiments, non-pregnant Rank WT Foxp3 GFP/GFP as well as E7.5–E9.5 Rank WT Foxp3 GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP dams were used as Treg cell donors. Treg cells of thymic origin were FACS-sorted and pooled from the thymus, lymph nodes and spleen as CD4+FOXP3–GFP+neuropilin-1high T cells. Then, 1 × 105 sorted Treg cells were transferred intravenously into the tail veins of E0.5–E2.5 Rank WT Foxp3 GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP pregnant recipient females. Vehicle-control pregnant females received only PBS on days E0.5–E2.5. To assess rescue of fetal resorption, recipient pregnant mice were euthanized on day E12.5. To assess Treg cell numbers in the placenta, recipient mice were euthanized on days E12.5 and E16.5. To assess rescue of metabolic parameters, an oral glucose-tolerance test was performed on day E16.5. Rescue of fetal macrosomia, as well as inflammation, adipocyte size and Treg cell numbers in the visceral adipose tissue, was determined at E17.5. In all adoptive transfer experiments, pregnancies were from crosses to syngeneic C57BL/6J male mice and sorting of Treg cells and T cells was performed after exclusion of dead cells (viability dye-negative).
Histology and immunohistochemistry
For histological analysis of the cortico-medullary organization of the thymus, 2.5-μm paraffin-sections and 5-μm cryosections were stained with haematoxylin and eosin (H&E) or assayed by immunofluorescence for K5 expression (medullary marker; Poly19055, Biolegend) and counterstained with DAPI. Slides were then scanned on a Mirax Scanner (Zeiss) and the thymic medulla area/total thymus area (expressed as a percentage) was quantified using the Fiji software (ImageJ v.1.49b). For histological analysis of mouse placentas, dissected placenta–fetus units were fixed in 4% paraformaldehyde and embedded in paraffin after dehydration. Cross-sections (2–3 μm thick) were stained with H&E and histologically examined independently by two certified pathologists. For quantification of pancreatic β-islets and white adipocyte size during pregnancy, pancreata and gonadal (peri-uterine) white adipose tissue from pregnant and non-pregnant control littermate female mice were fixed in 4% paraformaldehyde, paraffin embedded and micro-sectioned (2–3 μm thick) at 2 distal sites. Individual adipocyte and β-islets sizes as well as total pancreas areas were quantified using an automated image analysis software (Definiens Tissue Studio v.4.4.3 and Definiens Developer v.2.7; both from Definiens). An algorithm was developed to identify, categorize and measure the area of the white adipocytes as well as β-islets and the exocrine pancreas, based on morphology and intensity of the H&E staining and under the guidance of a pathologist. The validity of the analysis was manually inspected for each slide and incorrect delimitations were manually adjusted if necessary (Extended Data Fig. 5p). Detection of macrophages in the adipose tissue of non-pregnant and E17.5 pregnant females was performed using the automated staining platform Leica BOND-III System (Leica Biosystems). In brief, 2-μm thick visceral (gonadal, peri-uterine) adipose tissue paraffin sections were deparaffinized by Leica Dewax solution (AR9222) and rehydrated before epitope retrieval using Leica epitope retrieval solution 1 (AR9961) for 20 min. Sections were blocked 1 h in PBS 1% BSA and 5% rabbit serum (R9133-5, Sigma Aldrich) and incubated for 1 h with anti-F4/80 primary antibody (MCA497G, Biorad, 1:100) diluted in Leica primary antibody diluent. Slides were then incubated with biotinylated secondary rabbit anti-rat antibodies (ab6733, Abcam), diluted 1:500 in Leica primary antibody diluent, for 30 min before detection using the Leica Bond Intense R Detection system (DS9263). Staining of FOXP3+Treg cells in mouse pancreas cross-sections was performed manually using anti-FOXP3 primary (ab54501, 1:1,000) and goat anti-rabbit IgG (E0432, 1:500, DAKO) antibodies, following routine immunohistochemistry procedures. All sections were counterstained with Harris haematoxylin (6765002, Thermo Fischer) and subsequently dehydrated in a Microm HMS 740 slide stainer. Quantification of F4/80+ and FOXP3+ cells was automatically performed using the Definiens Tissue Studio software v.4.4.3 (Definiens), under the guidance of a pathologist who validated the staining using positive and negative control slides. Every cell (detected by nuclear DAPI staining) in the section was classified as positively or negatively stained. The quantification of F4/80+ cells is presented as relative to other cells; that is, (number of positive nuclei/positive + negative nuclei) × 100; the quantification of FOXP3+ cells in the pancreas is presented relative to the tissue area (β-islets or exocrine pancreas), determined automatically by the software. All slides were scanned on a Mirax Scanner (Zeiss) and representative images of thymic, placental, gonadal white adipose tissue and pancreas cross-sections were acquired using the Panoramic Viewer Software v.1.15.4 (3DHistech Ltd).
Immunofluorescence of thymic cross-sections
Thymi were snap-frozen in OCT and 5 μm cryosections were prepared, air-dried and subsequently fixed in ice-cold acetone (−20 °C for 10 min). Slides were briefly blocked in 1% BSA in PBS and stained for 45 min at room temperature with the following antibodies: anti-CD4 AlexaFluor647 (clone RM4-5, Biolegend, 1:200), anti-CD8 biotin (53-6.7, eBioscience, 1:200), anti-DEC205 FITC (NLDC-145, Abcam, 1:100), anti-CD40 biotin (3/23, BD Bioscience, 1:100), anti-K5 (Poly19055, Biolegend, 1:2,000) and anti-AIRE AlexaFluor488 (5H12, eBioscience, 1:100). If primary antibodies were not directly conjugated to a fluorophore, AlexaFluor-conjugated streptavidin or secondary antibodies (Invitrogen, 1:1,000) were used for detection. DAPI was used for nuclear counterstaining. Confocal images were obtained using a Zeiss LSM 780 and Zeiss LSM 510 microscopes. For RANK staining, biotinylated anti-RANK antibodies (BAF692; R&D systems; 1:50, overnight incubation) and the TSA Fluorescence System (PerkinElmer) were used according to the manufacturer’s protocol. For quantification of AIRE+ medullary thymic epithelial cells (mTECs), 4–7 different medullary areas (identified by K5+ staining) were quantified per mouse. The total number of AIRE+ cells (manually annotated) was related to the thymic medullary area (automatically calculated using Fiji v.1.49b software). The amount of AIRE+ mTECs is presented in the figures as a fold change after normalization to the average value of the control mouse group for each experiment. We verified that Rank heterozygosity (Rank floxed/wt K5 cre/wt) did not alter mTEC development, excluding Cre-dependent effects or haploinsufficiency (Extended Data Fig. 1f, g).
Quantitative PCR
Total RNA was extracted from human placenta samples of pregnant women at delivery (central part, maternal side) as well as from the gonadal VAT, whole thymi or from the thymic stroma-enriched fraction of pregnant, non-pregnant and hormone-treated female mice. Thymic stroma enrichment was obtained by enzymatic disaggregation with 0.05% (w/v) of Liberase TH and 100 U ml−1 of DNase I as previously described41, followed by depletion of CD45+ haematopoietic cells using magnetic microbeads (Miltenyi Biotec). For a more-representative analysis of large tissues (that is, gonadal white adipose mouse tissue and human placenta samples), total RNA was independently extracted and analysed from 3 distal sites. RNA extraction was performed using the RNAeasy Mini Kit (Qiagen) and included DNase I digestion to avoid potential DNA contamination. Then, 1 μg of total RNA was reverse transcribed using iScript cDNA Synthesis Kit (BioRad). Gene-expression levels were quantified by real-time quantitative PCR (iCycler iQ BioRad) using the iQ SYBR Green Supermix from BioRad and normalized to Actb or Pum1 housekeeping genes. mRNA fold changes were calculated using the ΔΔC t method.
The following primers were used for mouse genes: Aire forward, CTCTGCTAGTCACGACCCTG; Aire reverse, AGAGAAGGGTGGTGTCTCGG. Rank forward, CCCAGGAGAGGCATTATGAG; Rank reverse, CACACACTGTCGGAGGTAGG. Actb forward, CGGTTCCGATGCCCTGAGGCTCTT; Actb reverse, CGTCACACTTCATGATGGAATTGA. Pum1 forward, TGTGGTCCAGAAGATGATCG; Pum1 reverse, GGATGTGCTTGCCATAGGTG. Il6 forward, CAAAGCCAGAGTCCTTCAGAG; Il6 reverse, AGCATTGGAAATTGGGGTAG. Tnf forward, TCTTCTCATTCCTGCTTGTGG; Tnf reverse, ATGAGAGGGAGGCCATTTG. Ccl2 forward, CAGCAGGTGTCCCAAAGAAG; Ccl2 reverse, TTCCGATCCAGGTTTTTAAT. Glut4 forward, CTGTCGCTGGTTTCTCCAAC; Glut4 reverse, CGGCAAATAGAAGGAAGACG. Pgr forward, ACAGCGCTTCTACCAACTCAC; Pgr reverse, CAACTGGGCAGCAATAACTTC.
The following primers were used for human genes: PUM1 forward, CGGTCGTCCTGAGGATAAAA; PUM1 reverse, CGTACGTGAGGCGTGAGTAA. CD4 forward, TTTTCATTGGGCTAGGCATC; CD4 reverse, ACTGGCAGGTCTTCTTCTCAC; FOXP3 forward, AAGCAGCGGACACTCAATG; FOXP3 reverse, TGTGCAGACTCAGGTTGTGG.
RNA sequencing
QuantSeq analysis of VAT
Using the RNAeasy Mini Kit (Qiagen), total RNA was extracted from the gonadal VAT of dams at E17.5 treated according to our Treg-cell adoptive transfer protocol. Tissues from four independent dams were analysed for each of the three pregnant mouse cohorts: vehicle-treated Rank WT Foxp3 GFP/GFP as well as vehicle-treated Rank ΔFoxn1 Foxp3 GFP/GFP dams and pregnancy thymic-Treg-cell-treated (hereafter, pregnancy tTreg-cell-treated) Rank ΔFoxn1 Foxp3 GFP/GFP dams. After RNA extraction, RNA quantification and quality control were performed with the Agilent RNA 6000 Nano Kit on the bioanalyser. Thereafter, 200 ng of total isolated RNA was used for library preparation using the Lexogen QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina. Preparations were performed according to the manufacturer’s recommendations. The quality controls of the final libraries were composed of a fragment analyser run to determine the average size, followed by qPCR to quantify library concentration. Libraries were pooled at an equimolar ratio and sequenced on an Illumina HiSeq 2500 instrument using the single-read 50-read mode. 3′ RNA-sequencing (Quantseq) reads were prepared for analysis by removing adaptor contamination, poly(A) read through and low-quality tails using bbmap v.36.92. Thereafter reads were aligned to the Mus musculus genome mm10 using TopHat v.2.1.1 (GTF annotation file mm10, RefSeq from UCSC, 2015/02, protein-coding genes). Reads in genes were counted with htseq-count v.0.6.1. Differential expression analysis was performed with DESeq2 v.1.16.1. Raw expression counts were converted to counts per million (CPM) using edgeR v3.18.1. For gene set enrichment analyses (GSEA), genes with very low expression levels were filtered out by setting an expression threshold of CPM > 1 in at least 2 samples. GSEA was performed using GSEA v.3.0 and gene signatures from MSigDB v.6.1.
Smart-Seq2 analysis of tissue Treg cells
For transcriptome analysis of pregnancy-associated Treg cells, placentas, thymi, spleen and VAT from Rank WT Foxp3 GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP pregnant females were isolated at E17.5. To recover thymocytes, thymi were mechanically disrupted. Isolated placentas and VAT (peri-uterine) were enzymatically digested in DMEM medium containing collagenase IV (1 mg ml−1) and DNase I (0.1 mg ml−1) (both from Worthington), shaking at 37 °C for 40 min at 100 rpm, or using an adipose tissue dissociation kit and a gentle MACS Dissociator (Miltenyi Biotec), respectively. After digestion, samples were put on ice and washed twice with DMEM containing 10 mM EDTA and 10% FCS to stop the enzymatic digestion and subsequently stained for FACS as described above. Cell sorting was performed on an AriaIII cell sorter (BD Bioscience) after excluding dead cells and doublets, and gating for low side scatter and CD45+ haematopoietic cells (gating strategy is shown in Supplementary Data 1). Equal numbers of Treg cells (280 cells) were sorted as CD45+CD8−CD4+FOXP3–GFP+neuropilin-1high cells from the thymus and placenta (both samples from the same female). To increase robustness, the 280 placental Treg cells were purified from 5 individual placentas per pregnant female (56 Treg cells per placenta). In a separate experiment with different pregnant mice than those used for thymus and placenta analysis, VAT Treg cells (20–30 cells) and splenic Treg cells (150 cells) were sorted (both samples from the same female). In all cases, cells were sorted directly into a cell lysis buffer consisting of a 0.2% (v/v) Triton X-100 solution and an RNase inhibitor (TaKaRa Bio Group). For the generation of full-length cDNA, the Smart-seq2 protocol42 was used. Subsequent library preparation from the amplified cDNA was performed using the Nextera XT DNA library prep kit (Illumina). Expression profiling libraries were sequenced on a HiSeq3000 instrument (Illumina) in 50-base-pair, single-end mode. Base calls, provided by the real-time analysis software (RTA v.2.7.3 and v.2.7.7, Illumina), were subsequently converted into multiplexed, unaligned BAM format before demultiplexing into sample-specific, unaligned BAM files. For raw data processing off the instruments, custom programs, based on Picard tools (v.2.19.2, https://broadinstitute.github.io/picard), were used. Next-generation sequencing (NGS) reads were mapped to the Genome Reference Consortium GRCm38 assembly using ‘Spliced Transcripts Alignment to a Reference’ (STAR, v.2.7.5a)43 using the ‘basic’ Ensembl transcript annotation from version e96 (April 2019) as reference transcriptome. As the mm10 assembly of the UCSC Genome Browser was preferred for downstream data processing with Bioconductor packages (v.3.11, https://bioconductor.org/packages), Ensembl transcript annotation had to be adjusted to UCSC Genome Browser sequence region names. STAR was run with options recommended by the ENCODE project. Aligned NGS reads overlapping Ensembl transcript features were counted with the summarizeOverlaps function of the Bioconductor GenomicAlignments package. Transcript-level counts were aggregated to gene-level counts and the Bioconductor DESeq244 package was used to test for differential expression based on a model using the negative binomial distribution. An initial exploratory analysis included principal component analysis, multi-dimensional scaling, sample distance and expression heat map plots, all annotated with variables used in the expression modelling (ggplot245, Bioconductor ComplexHeatmap), as well as volcano plots (Bioconductor EnhancedVolcano). On the basis of poor sample quality in principal component analysis plots and sample distance heat maps at the exploratory analysis stage, three low-input samples were excluded from the differential expression analysis of tissue Treg cells. The samples included one thymic Treg cell sample from a pregnant Rank WT mouse (GTT13_T_S13943) and two placental Treg cell samples, one from a Rank WT pregnant female (GTT31_P_S13938) and one from a Rank ΔFoxn1 pregnant female (GTT16_P_S13933) (all samples, including the excluded ones, have been deposited in the NCBI GEO). Resulting gene lists were annotated, filtered for significantly differentially up- and downregulated genes and independently subjected to GSEA (GSEAPreranked, using GSEA v.3.0 and gene signatures from MSigDB v.6.2). The TCR repertoire was estimated with the MiXCR46 software (v.3.0.13) and the immunarch (v.0.6.5) R package from GitHub (https://github.com/immunomind/immunarch). To obtain a sensible number of NGS reads from conventional, poly(A)-primed mRNA sequencing, reads of all replicate samples were pooled, aligned to immune genes and assembled into clones and clonotypes via MiXCR.
Fetal thymus organ culture
For fetal thymus organ cultures (FTOCs), day E16 thymic lobes from wild-type pregnant females were cultured for 7 days in DMEM, then cultured in the presence of 100 ng ml−1 soluble recombinant mouse RANKL (Peprotech) and/or 1,000 ng ml−1 osteoprotegerin (R&D systems) for an additional 4 days. Fetal thymic lobes were then mechanically disaggregated and stained with antibodies to detect CD4 (RM4-5, Biolegend), CD8 (53-6.7, eBioscience) and FOXP3 (FJK-16s, eBioscience, using the FOXP3 intracellular staining buffer). FACS data were analysed using a FACSFortessa BD.
Detection of autoantibodies
For the detection of autoantibodies, indirect immunofluorescence on Rag2 −/− tissue sections was performed as described previously47. In brief, 5-μm cryostat sections from several organs of Rag2 −/− mice were incubated with 1/40 dilutions of sera obtained from pregnant females, followed by detection with AlexaFluor555-labelled anti-mouse IgG secondary antibodies (Invitrogen, 1:1,000). Serum from antibody-deficient (Rag2 −/−) and autoimmune-prone (Mrl/lrp) females were used as positive and negative controls, respectively. DAPI was used for nuclear staining. Slides were examined and scored in a blinded fashion, and representative images were acquired using the Panoramic Viewer Software v.1.15.4 (3DHistech). Anti-dsDNA autoantibodies were detected by ELISA according to the manufacturer’s protocol (Alpha Diagnostic).
Treg cells in vitro suppression assays
For comparing the suppression capacity of Treg cells from spleen and lymph nodes, CD4+CD25+FOXP3GFP neuropilin-1high Treg cells were sorted from the freshly isolated spleen and lymph nodes of Rank WT Foxp3 GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP non-pregnant and E12.5–E14.5 pregnant female mice using an AriaIII cell sorter (BD Bioscience). To compare Rank WT Foxp3 GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP Treg cell populations against a common responder, purified FOXP3-GFP+Treg cells (2.5 × 104) were co-cultured at different ratios (from 1:1 to 1:32) with a pool of naive CD4+CD25−FOXP3–GFP− effector T cells that were similarly sorted from n = 3–4 Rank WT Foxp3 GFP/GFP donors and subsequently stained with a CellTrace Cell Proliferation Kit (violet, 5 μmol l−1, Life Technologies), according to the manufacturer’s instructions. Effector T cells from non-pregnant Rank WT Foxp3 GFP/GFP donors were used in co-cultures with Treg cells from non-pregnant Rank WT Foxp3 GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP mice. Similarly, effector T cells isolated from pregnant Rank WT Foxp3 GFP/GFP donors were used to test the suppression capacity of pregnancy-associated Rank WT Foxp 3GFP/GFP and Rank ΔFoxn1 Foxp3 GFP/GFP Treg cells. The proliferation of CD4+T effector cells was induced by incubation in 200 μl of IMDM containing 10% FCS, in a 96-round-well plate, in the presence of soluble anti-CD3 antibodies (0.25 μg ml−1) as well as CD11c+ dendritic cells (5 × 103) isolated from enzymatically digested spleens of wild-type C57BL/6J age-matched and non-pregnant female mice using MACS beads (Miltenyi Biotec). After 3 days of stimulation, the relative percentage of proliferating CD4+GFP− effector T cells was determined based on CellTrace Violet dilution data collected with a LSR II flow cytometer (BD Biosciences) and analysed using FlowJo v.10.0.0 software (Tree Star). Percentage of Treg cell suppression was calculated by comparing the proliferation of the effector T cells co-plated with Treg cells at each ratio to the proliferation of effector T cells plated without Treg cells (0:1 ratio).
Metabolic studies
Diet and husbandry
For metabolic studies during pregnancy, pregnant Rank WT and Rank ΔFoxn1 female mice as well their non-pregnant control littermates were fed normal chow. Rank WT and Rank ΔFoxn1 female mice were co-caged before and during pregnancy to minimize the effects of the microbiota on metabolic parameters. Their offspring were fed normal chow from weaning age until 80 days of age, after which they were fed a HFD (60% kcal% fat, Research Diets, D12492i) for up to 12 weeks. Offspring pups were weighted periodically starting from postnatal day 1 until weaning age. After weaning, tail blood glucose samples and body weight measurements of the offspring were obtained weekly, for mice fed normal chow or HFD. To discard milk or other mother-related effects, Rank WT and Rank ΔFoxn1 dams were interchanged to nurture each other litters, from postnatal day 1 until weaning age. After weaning, the offspring from Rank WT and Rank ΔFoxn1 were co-caged to minimize possible effects of the microbiota and/or cage. For analysis of long-term transgenerational effects on fat tissues, male and female offspring were kept on a normal chow diet for up to 17 months. For metabolic studies in non-pregnancy conditions, non-pregnant Rank WT and Rank ΔFoxn1 females were fed a HFD for 7 weeks, after which their glucose tolerance and adipose tissue were characterized.
Glucose and insulin measurements
All glucose-tolerance tests (GTT) were performed orally. For oral glucose-tolerance tests, mice were fasted for 16 h (15:00–07:00) and were then administrated 2 mg glucose per g body mass by oral gavage. Glucose values were measured using glucometers from blood samples taken by tail nick at 0, 5, 15, 30, 60 and 120 min after glucose ingestion. The area under the GTT curve was calculated for each mouse using GraphPad Prism v.7.0c (GraphPad Software). Fed and fasting glucose measurements were performed from blood tail samples or serum (OneTouch UltraEasy, according to the manufacturer’s instructions); insulin levels were measured from serum by ELISA (mouse ultrasensitive insulin ELISA, Alpco). For each litter of dams, offspring, as well as non-pregnant females, blood samples were taken the same day and at the same time for all genotypes of interest. The glucose uptake capacity of the adipose tissue was determined using 2-deoxyglucose and the bioluminescent Glu-Uptake Glo Assay (Promega). In brief, small pieces (around 4–9 mg) of VAT were incubated with 0.5 mM of 2-deoxyglucose for 15 min. For insulin-induced glucose uptake, tissues were pre-incubated for 20 min with 100 nM insulin, washed twice with PBS and then treated with 2-deoxyglucose (same time and dose). Glucose uptake values were measured using a luminometer as relative luminescence units and were normalized per mg adipose tissue.
Mass spectrometry measurements of glucose in the placental tissue
Metabolites were extracted from the E17.5 placentas of Rank WT and Rank ΔFoxn1 mice (around 30–40 mg) using a methanol:acetonitrile:water (2:2:1, v/v) ice-cold solvent mixture followed by subsequent rounds of snap-freezing, bead homogenization and sonication. Samples were then centrifuged at 4,000g for 10 min at 4 °C, the supernatants collected and transferred to another tube and evaporated to dryness in a vacuum concentrator. Extracted metabolites were resolubilized in a buffer containing 50% 20 mM ammonium acetate and 50% acetonitrile. Then, 1 μl of the sample was injected and separated using an Ulitimate U300 BioRSLC HPLC system (Dionex; Thermo Fisher Scientific), using a HILIC column (100 mm × 2.1 mm, 3.5 μm, 200 Å; Merck). Separation was carried out with a flow rate of 100 μl min−1 using a linear gradient of 13 min from 95% A (acetonitrile) to 45% B (10 mM ammonium acetate pH 7.5), followed by re-equilibration of the column. Eluting metabolites were analysed using a TSQ Quantiva mass spectrometer (Thermo Fisher Scientific) after electrospray ionization with single-reaction monitoring in negative ion mode using a spraying potential of 3,000 V. Glucose was quantified using the transitions m/z 239 to 179 (acetate adduct; quantifier) and m/z 179 to 119 (qualifier). Data were manually interpreted using the Xcalibur software (Thermo Fisher Scientific) and normalized per mg placental tissue.
Human data
For human studies, approval from the ethics committee of the Medical University of Vienna (EC number 1337/2016) as well as written informed consent from all participants was obtained. The study was performed in accordance with the principles of the Declaration of Helsinki. Pregnant women with gestational diabetes mellitus (GDM) and control glucose-tolerant pregnant women, older than 18 years (mean age, 34 years; with a 25th percentile of 30 years and 75th percentile of 36.25 years), carrying a singleton pregnancy, were both stratified as overweight or obese (body-mass index (BMI) ≥ 29.0 kg m−2). None of the women had any pre-existing diabetes, chronic medical condition or psychiatric disease. Participants underwent a standard 75 g oral glucose-tolerance test for which venous samples were taken at time 0 (fasting overnight) and at 30, 60, 90 and 120 min after glucose ingestion and measured for glucose and insulin levels in plasma by enzymatic methods at the General Hospital in Vienna (AKH Wien). Participants undertook a first oral glucose-tolerance test at early pregnancy (≤20 gestational weeks; Fig. 4g), which is advised for obese pregnant women48 and were retested at mid gestation (24–28 weeks) and late gestation (35–38 weeks), with similar results (data not shown). At each visit, anthropometric measurements for BMI calculations were also taken. GDM was diagnosed according to the IADPSG/WHO 2013 criteria49,50 of fasting plasma glucose ≥ 5.1mmol l−1, 1 h plasma glucose ≥ 10.0 mmol l−1 or 2 h plasma glucose ≥ 8.5 mmol l−1. Homeostatic model assessment of insulin resistance was calculated from fasting insulin and glucose values as follows: (fasting glucose (mmol l−1) × fasting insulin (pmol l−1))/135. The following data from the included patients were collected: age, height, weight (before pregnancy and at birth), BMI, gender of the baby, gestational week of delivery, method of delivery as well as complications in pregnancy, therapy of GDM, glucose control and glycaemic parameters. At delivery, samples from the cord blood (baby side) as well as samples from the centre of the placenta, at the maternal side (decidua) were collected. Cord blood samples were used to determine C-peptide levels using immunoassays. Birth weights were determined without diaper using a calibrated electronic scale. Preparations of placenta samples were performed at the same position in all cases. For FOXP3 qPCR studies, dissected placenta samples were immediately frozen and stored for later RNA analysis. Three distal placental slices (all on the maternal side) were analysed and averaged per woman. FOXP3 mRNA transcripts were normalized to a housekeeping gene (Fig. 4i, j), but similar results were observed if normalized to CD4 levels (Extended Data Fig. 10h). In a subsequent additional study for FOXP3 immunohistochemistry, the placenta samples were fixed in 10% neutral-buffered formalin, embedded in paraffin and micro-sectioned at 2–3 distal sites for detection of Treg cells using immunohistochemical analysis, using anti-FOXP3 (ab20034, 1:200) and goat anti-rabbit (DAKO, E0432, 1:500) antibodies in the automated staining platform Leica BOND-III System (Leica Biosystems). In total 2–3 distal placental slices (all on the maternal side) were analysed per woman. Slides were scanned on a Mirax Scanner (Zeiss) and the quantification of FOXP3+ cells was performed using the Definiens Tissue Studio software 4.4.3 (Definiens), under the guidance of a pathologist, who manually inspected the validity of the analysis, verifying nuclear staining and positive staining of human tonsil control slides. Every cell (nucleus) on the placenta section was quantified as negatively or positively stained. Images of the stained cross-sections were acquired using the Panoramic Viewer Software 1.15.4 (3DHistech). Of note, we had no access to fat tissue from the same female cohorts and hence could not determine Treg cell numbers in fat. For the analysis of the human data (except the immunohistochemistry study), all women with GDM were matched to control glucose-tolerant women for BMI (mean ± s.d. of 33.9 ± 4.8 for control and 34.3 ± 5.0 for GDM), gestational week of delivery (mean ± s.d. of 39.5 ± 1.6 for control and 40.0 ± 1.3 for GDM) and delivery method (vaginal or caesarean section; n = 7 and 9 for control and n = 6 and 10 for GDM, respectively). Of note, the reduction in FOXP3 transcripts in the placenta of pregnant women with GDM was observed, to a similar extent, in both vaginal and caesarean section deliveries; therefore, the Treg cell reduction appears to be independent of the mode of delivery of the baby (data not shown). Among pregnant women with GDM the distribution between those receiving insulin as treatment (maximum dose of 48 IE per day) and those who had sufficient metabolic control with dietary advice was also distributed equally (n = 8 and 8; diet and insulin treatment, respectively). Moreover, male and female sex of the babies were equally distributed in both cohorts (n = 8:8 male:female per group).
Statistical analyses
Unless otherwise stated, data are shown as individual data points or as mean ± s.e.m. The numbers of mice and individuals per group used in each experiment are annotated as ‘n’. Mouse experiments shown were reproduced 2–5 independent times. Figures and statistical analyses were generated using GraphPad Prism 7.0c (GraphPad Software). Normally distributed data were analysed using unpaired two-tailed Student’s t-tests for single comparisons, one-way ANOVAs for multiple comparisons or two-way ANOVAs for comparison of two groups over time. Ordinal and not normally distributed data (according to D’Agostino and Pearson omnibus normality tests) were analysed using unpaired two-tailed Mann–Whitney U-tests for single comparisons or Kruskal–Wallis tests for multiple comparisons. Post hoc tests used for multiple comparisons are indicated in each figure legend. The sex distribution of the offspring, as well as the prevalence of resorbed fetuses and autoimmunity, were analysed using a two-tailed χ 2 test. P ≤ 0.05 was considered to indicate statistical significance; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Extended Data
Extended Data Fig. 1. Characterization of non-pregnant Rank ΔFoxn1 and Rank ΔK5 conditional knockout mice.
a, qPCR analysis for relative Rank and Aire mRNA expression in the thymus of non-pregnant (N.P.; n = 12) and E12.5 pregnant (n = 8) Rank WT mice, normalized to non-pregnant values. Data are shown as box-and-whisker plots (from minimal to maximal values); dots represent individual mice.**P < 0.01. Two-tailed Student’s t-test. b, c, Immunofluorescence staining for AIRE (red) and K5 (K5, green) in thymus cross-sections (b, representative of n = 68/61 images) and relative quantification of AIRE+ mTECS in the thymic medullary regions (c) of non-pregnant and E12.5 pregnant Rank WT females (n = 11/12). Data are shown as box-and-whisker plots (from minimal to maximal values); dots represents individual data points (4–7 medullary fields per mouse), normalized to Rank WT non-pregnant values. **P < 0.01. Two-tailed Student’s t-test. d, e, Representative immunofluorescence staining for RANK (green), K5 (red), and DAPI (blue, to visualize nuclei) in thymic cross-sections of Rank WT and Rank ΔFoxn1 (d) and Rank WT and Rank ΔK5 (e) mice. Note that Rank expression is confined to the thymic medullary epithelium in Rank WT mice and is effectively ablated in Rank ΔFoxn1 as well as Rank ΔK5 mice. M, medulla; C, cortex. Images are representative of n = 5 mice per group. f, Immunofluorescence staining for AIRE (red) and K5 (green) in thymic cross-sections of Rank WT, Rank flox/K5, and Rank ΔK5 mice. Representative of n = 28/35/56 images. g, Relative numbers of AIRE+ mTECs in thymic medullary regions of Rank WT, Rank flox/K5 and Rank ΔK5 littermates, as determined by immunofluorescence. Note that Rank heterozygosity and Cre expression in the thymic epithelium does not affect AIRE+ mTECs numbers. Data were normalized to Rank WT values and are shown as box-and-whisker plots (from minimal to maximal values); dots represent individual data points. n = 5/5/9 (4–6 fields per mouse). ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. h, Organization of the thymic microenvironment. Representative immunofluorescence staining of thymic cross-sections of Rank WT and Rank ΔFoxn1 as well as Rank WT and Rank ΔK5 mice, to evaluate thymocyte distribution (top), defined by CD4 (red) and CD8 (green), as well as thymic epithelium organization (bottom), defined by CD40 (medulla, red) and DEC205 (cortex, green) expression. C, cortex; M, medulla. Representative of n = 5 mice per group. i, Thymus weights in Rank WT and Rank ΔFoxn1 (n = 7) as well as Rank WT and Rank ΔK5 (n = 5) female mice. Data are mean ± s.e.m. Two-tailed Student’s t-test. j, k, Representative FACS blots for CD4 and/or CD8α-expressing thymocytes (j) and their respective percentages (k, mean ± s.e.m.) in the thymus of Rank WT and Rank ΔFoxn1 (n = 5/4) and Rank WT and Rank ΔK5 (n = 3/3) female mice. Two-tailed Student’s t-test. l, Spleen weights of Rank WT and Rank ΔFoxn1 (n = 5/5) and Rank WT and Rank ΔK5 (n = 4/4) female mice. Data are mean ± s.e.m. Two-tailed Student’s t-test. m, n, Representative FACS blots for CD4 and/or CD8α expression (m) and percentages of CD4+ or CD8+T cells (n, mean ± s.e.m.) in the spleen of Rank WT and Rank ΔFoxn1 (n = 5/4) mice as well as Rank WT and Rank ΔK5 (n = 3/3) female mice. Two-tailed Student’s t-test. o, p, Percentages of CD4+FOXP3+Treg cells in the thymus (o) and spleen (p) of Rank WT and Rank ΔFoxn1 (n = 12/9) mice as well as Rank WT and Rank ΔK5 (n = 8/7) female mice. Data are mean ± s.e.m., determined by FACS. Two-tailed Mann–Whitney U-test. q, Total number of Treg cells in the thymus of Rank WT and Rank ΔFoxn1 (n = 26/16) female mice. These mice are the littermate controls of the pregnant cohorts shown in Fig. 1d. Data are shown as box-and-whisker plots (from minimal to maximal values) and dots represent individual mice. Two-tailed Student’s t-test. r, Percentage of suppression of CD4+T effector cells proliferation mediated by Treg cells, as determined by flow cytometry using CellTrace Violet dilution, for peripheral (pool of splenic and lymph node) Treg cells isolated from Rank WT (n = 4) and Rank ΔFoxn1 (n = 4) female mice and co-cultured with Rank WT effector T cells at the indicated ratios. Data are mean ± s.e.m. Two-way ANOVA, Sidak’s multiple comparisons test. All data in the figure are from young (4–12-week-old) and non-pregnant female mice.
Extended Data Fig. 2. Rank deletion in mTECs impairs thymic adaptations and thymic Treg cell expansion during pregnancy.
a, b, Thymus weights (a) and percentages (b) of the thymic medulla areas in non-pregnant as well as in E12.5 and E17.5 pregnant Rank WT and Rank ΔFoxn1 mice. Data are mean ± s.e.m., each dot represents an individual mouse. n = 37/22/22 and n = 29/10/19 in a; n = 13/9/9 and n = 8/4/8 in b; for the Rank WT and Rank ΔFoxn1 groups, respectively. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed Student’s t-test. Similar thymic involution defects were observed in Rank ΔK5 dams as compared to their Rank WT counterparts (data not shown). c, H&E-stained thymic cross-sections of non-pregnant and E17.5 pregnant Rank WT and Rank ΔFoxn1 mice. Representative of n = 13/8 and n = 9/8 (E17.5) mice. These sections as well as thymic cryo-sections stained for DAPI/K5 (data not shown) were used for the quantification of the thymic medulla expansion during pregnancy shown in b. d, Representative FACS blots for CD4 and CD8 expression (upper blots) and CD44 and CD25 expression (lower blots, gated on double negative -DN- CD4−CD8− cells) in thymocytes of non-pregnant and E17.5 pregnant Rank WT and Rank ΔFoxn1 females. Numbers indicate percentage of cells in each quadrant. DN1, CD44+CD25−; DN2, CD44+CD25+; DN3, CD44−CD25+; DN4, CD44−CD25−. e, f, Percentages and total cell numbers of thymocyte subpopulations expressing CD4 and/or CD8 (e) and early developing double-negative (DN) thymocytes (as detected by CD44 and CD25 staining on CD4−CD8− DN populations) (f) in the thymus of non-pregnant and E17.5 pregnant Rank WT and Rank ΔFoxn1 females. Data are mean ± s.e.m., dots represent individual mice. n = 3–10 animals per group *P < 0.05; **P < 0.01. One-way ANOVA. Similar percentage results were observed when comparing Rank WT and Rank ΔK5 dams at E18.5 (not shown). g, Total cell numbers and percentages of conventional single positive CD4+ thymocytes (FOXP3−) in the thymus of non-pregnant and E13.5–E16.5 pregnant Rank WT and Rank ΔFoxn1 females. Data are mean ± s.e.m., dots represent individual mice. n = 5–19 animals per group. Two-tailed Student’s t-test. h, Relative quantification of AIRE+ mTECS in the thymic medullary regions of non-pregnant and E12.5 pregnant Rank WT (n = 12/12) and Rank ΔFoxn1 (n = 13/10) mice, as determined by immunofluorescence (5–7 medullary fields per mouse). Data are mean ± s.e.m., normalized to Rank WT non-pregnant values. Each dot represents an individual data point. **P < 0.01; ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. i, Percentages of neuropilin-1highHelioshighFOXP3+Treg cells (of total CD4+ cells) in the thymus of non-pregnant (n = 9/5) and pregnant (pool from E15.5–E17.5, n = 11/8) Rank WT and Rank ΔFoxn1 females, as determined by FACS analysis. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed Mann–Whitney U-test. j, Percentages of CD4+FOXP3+Treg cells (of total CD4+ cells) in the thymus of non-pregnant and E18.5 pregnant Rank WT and Rank ΔK5 females. Data are shown as box-and-whisker plots (from minimal to maximal values); dots represent individual mice. n = 12/7 for Rank WT and n = 7/4 for Rank ΔK5 mice. *P < 0.05; ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. k, Mean fluorescence intensity (MIF) of CD25, FOXP3, GITR, CTLA-4, neuropilin-1 and Helios in thymic Treg cells of non-pregnant as well as in E13.5–E16.5 pregnant Rank WT and Rank ΔFoxn1 mice. Data are mean ± s.e.m., each dot represents an individual mouse with n = 4–13 animals per group. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. l, Percentage of proliferating (KI-67+) and apoptotic (active CASP3+) CD4+FOXP3+Treg cells in the thymus of non-pregnant as well as in E13.5–E16.5 pregnant Rank WT and Rank ΔFoxn1 mice. Data are mean ± s.e.m., each dot represents an individual mouse with n = 10/5/13/8 for KI-67 and n = 6/4/8/7 for CASP3. *P < 0.05; ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. m, Frequencies of Treg cell precursors (CD25+FOXP3−) and Treg cells (CD25+FOXP3+) in the thymus of non-pregnant (n = 10/5) and E13.5–E16.5 pregnant (n = 12/7) Rank WT and Rank ΔFoxn1 females. Data are mean ± s.e.m.; shown as percentage of CD4+ single-positive (SP) thymocytes. Dots represent individual mice. *P < 0.05; ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. n, Frequencies of the subsequent mature populations of thymic CD4+CD25+FOXP3+Treg cells, based on CCR7, CCR9, CD69 expression, in the thymus of non-pregnant (n = 4/4) and E13.5–E16.5 pregnant (n = 5/3) Rank WT and Rank ΔFoxn1 females. Data are mean ± s.e.m., each dot represents an individual mouse. *P < 0.05. One-way ANOVA, Tukey’s post hoc test. All data in this figure are from syngeneic C57BL/6J pregnancies.
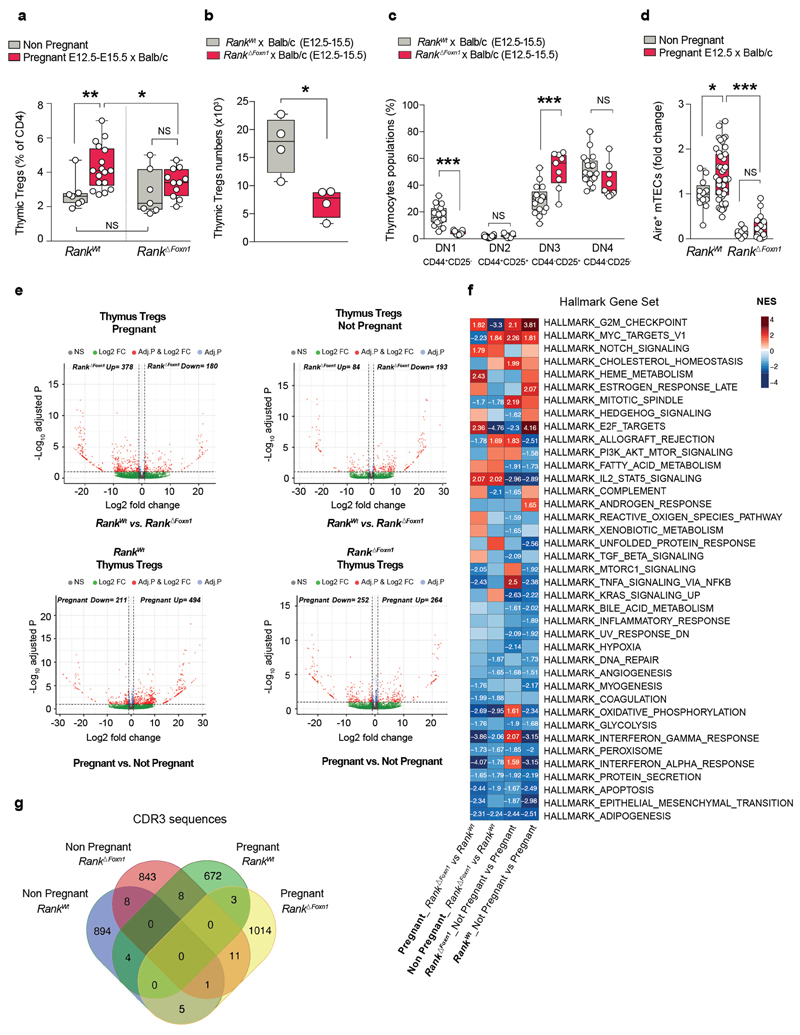
Extended Data Fig. 3. Rank ΔFoxn1 thymus in allogeneic pregnancies and transcriptome analysis of thymic Treg cells.
a, Percentages of CD4+FOXP3+Treg cells (of total CD4+ cells) in the thymus of individual non-pregnant (n = 7/7) and E12.5–E15.5 pregnant Rank WT and Rank ΔFoxn1 females (n = 17/11), crossed to allogeneic BALB/cJ males. *P < 0.05; **P < 0.01. Two-tailed Student’s t-test. b, Total numbers of CD4+FOXP3+Treg cells in the thymus of individual E12.5–E15.5 pregnant Rank WT and Rank ΔFoxn1 females (n = 4/4), crossed to allogeneic BALB/cJ males. *P < 0.05. Two-tailed Student’s t-test. c, Percentages of early developing double negative (DN) thymocytes (as detected by CD44 and CD25 staining on CD4−CD8− DN populations) in the thymus of E12.5–E15.5 pregnant Rank WT and Rank ΔFoxn1 females crossed to allogeneic BALB/cJ males. n = 18/9. ***P < 0.001. Two-tailed Student’s t-test. d, Relative quantification of AIRE+ mTECS in the thymic medullary regions of non-pregnant and E12.5 pregnant Rank WT (n = 3/7) and Rank ΔFoxn1 (n = 2/4) females, crossed to BALB/cJ males, as determined by immunofluorescence (3–6 medullary fields per mouse) and normalized to Rank WT non-pregnant values. *P < 0.05. One-way ANOVA, Tukey’s post hoc tes). a–d, The defects and Treg cell deficiencies in Rank ΔFoxn1 dams observed in syngeneic pregnancies are also present in allogeneic pregnancies. Data in a–d are box-and-whisker plots (from minimal to maximal values) and each dot represents an individual data point. e, Volcano plots for differentially expressed genes in Treg cells isolated from the thymus of non-pregnant as well as pregnant (E17.5) Rank WT (n = 4/3) and Rank ΔFoxn1 (n = 4/4) female mice. Dots represent genes, colored as: not significant (NS, dark grey); log2-transformed fold change above threshold (log2FC; abs(log2FC) > = 1.0, green); adjusted P value above threshold (Adj. P, adjusted P < = 0.1, light grey); and significant, with log2-transformed fold change and adjusted P value both above thresholds (red). The numbers of significantly up- and downregulated genes are indicated on each plot. P values are based on a two-tailed Wald test and adjusted via the Benjamini–Hochberg procedure. f, Summary of GSEA Hallmark gene sets enrichment analysis for the expression profiles of thymic Treg cells isolated from non-pregnant as well as E17.5 pregnant Rank WT (n = 4/3) and Rank ΔFoxn1 (n = 4/4) females. Numbers are normalized enrichment scores (NES) and are only shown for significantly enriched gene set (defined as NES > = 1.4 and FDR < = 10%). g, Venn diagram showing the number of unique CDR3 nucleotide sequences identified in the TCRs of thymic Treg cells isolated from non-pregnant as well as E17.5 pregnant Rank WT (n = 4/3) and Rank ΔFoxn1 (n = 4/4) females. Sequences were determined from the RNA-sequencing data using the MiXCR software. For primary data on TCR clones see Supplementary Table 1.
Extended Data Fig. 4. Progesterone-driven thymic Treg cell expansion as well as characterization of splenic Treg cells and autoimmunity in pregnant Rank ΔFoxn1 mice.
a, b, Representative FACS blots (a) and percentages (b) of CD4+FOXP3+ Treg cells (gated on CD4+ cells) in wild-type fetal thymic embryonic lobes (FTOCs) treated with DMEM, RANKL (100 ng ml−1) and/or osteoprotegerin (OPG;1,000 ng ml−1). Data in b is a pool of 3 independent experiments, total n = 6 per group. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual data point. *P < 0.05. One-way ANOVA, Tukey’s post hoc test. c, qPCR analysis for Rank, Aire and progesterone receptor (Pgr) mRNA expression in the thymus of ovariectomized wild-type C57BL/6J female mice (11 weeks old) that were vehicle-treated (n = 4), or subcutaneously-treated with oestrogen (10 μg kg−1 daily, n = 3), progestin (MPA; 90-day slow-release pellet of 50 mg, n = 4), or with both progestin and oestrogen (same doses as single treatments, n = 3) for 5 days. Data are mean ± s.e.m., each dot is an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA, Dunnett’s post hoc test versus the vehicle group. d, Thymus weights in non-ovariectomized (Non-Ovx) and ovariectomized wild-type C57BL/6J female mice (7 weeks and 11 weeks old, respectively) that were vehicle-treated, or subcutaneously-treated with oestrogen, progestin, or with progestin and oestrogen (same hormonal doses as in c) for 5 days. Data are mean ± s.e.m., each dot is an individual mouse (n = 3–5 per group). *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA, Dunnett’s post hoc test versus the vehicle group of each cohort. e, Percentage of thymic CD4+FOXP3+ Treg cells (of total CD4+ cells) in Rank WT and Rank ΔK5 ovariectomized females treated with sham surgery or subcutaneously with a progestin-releasing (MPA) pellet for 5 days. Data are mean ± s.e.m., each dot is an individual mouse (n = 3). *P < 0.05; **P < 0.01. One-way ANOVA, Tukey’s post hoc test. f, Representative FACS blots for thymic Treg cells (gated on single positive CD4+ cells, as shown in Supplementary Data 1) in Rank WT and Rank ΔFoxn1 as well as Rank WT and Rank ΔK5 ovariectomized females treated with sham surgery or subcutaneously with a progestin-releasing (MPA) pellet for 5 days. g, Percentages of neuropilin-1highFOXP3+ Treg cells (of total CD4+ cells) in the thymus of sham-treated (n = 11/9) and progestin-treated (n = 11/9) Rank WT and Rank ΔFoxn1 female mice. Data were determined by FACS analysis. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05; ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. h, Percentages of thymic Treg cells in Rank WT females treated with sham-surgery or treated subcutaneously with progesterone (50 mg/21 day-release pellets) for 6 days (n = 3, each dot is an independent mouse and horizontal lines are median, error bars are s.e.m.). ***P < 0.001. Two-tailed Student’s t-test. i, Percentages of thymic Treg cells in Rank WT mice treated with vehicle (oil, n = 4), and Rank WT (n = 6) and Rank ΔFoxn1 (n = 5) mice treated subcutaneously with progesterone (450 μg in oil per day, n = 6/5) for 7 days. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05. Two-tailed Student’s t-test. j, qPCR analysis for the relative progesterone receptor mRNA expression in the thymus of ovariectomized (n = 3/2), non-ovarectomized and non-pregnant (n = 4/5), and E12.5–E13.5 pregnant (n = 7/8) Rank WT and Rank ΔFoxn1females, which is an expected finding in line with previous studies showing that RANKL/RANK acts downstream of progesterone. Data are mean ± s.e.m. Each dot represents an individual animal. *P < 0.05. Two-tailed Student’s t-test. k, Percentages of CD4+FOXP3+ Treg cells in the spleen of non-pregnant and pregnant (pool from E12.5–E18.5) Rank WT (n = 29/36) and Rank ΔFoxn1 (n = 17/26) female mice. Two-tailed Student’s t-test. l, Percentages of neuropilin-1highHelioshigh Treg cells (of total CD4+) in the spleen of non-pregnant and E16.5–E17.5 pregnant Rank WT (n = 10/12) and Rank ΔFoxn1 (n = 5/8) female mice. Two-tailed Student’s t-test. m, Total numbers of CD4+FOXP3+ Treg cells in the spleen of non-pregnant and pregnant (pool from E16.5–E17.5) Rank WT (n = 10/12) and Rank ΔFoxn1 (n = 5/8) female mice. Two-tailed Student’s t-test. n, o, Total numbers (n) and percentages (o) of CD4+ and CD8+ splenocytes in individual non-pregnant (n = 10/5) and E13.5–E15.5 pregnant (n = 12/8) Rank WT and Rank ΔFoxn1 female mice. One-way ANOVA, Tukey’s post hoc test. k–o, Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. p, Percentage of suppression of conventional CD4+T cell proliferation mediated by Treg cells, as determined by flow cytometry using CellTrace Violet dilution, for peripheral (pool of splenic and lymph node) Treg cells isolated from E13.5–E15.5 pregnant Rank WT (n = 5) and Rank ΔFoxn1 (n = 3) female mice. Data are mean ± s.e.m. Two-way ANOVA, Sidak’s multiple comparisons test. q, Venn diagram showing the number of unique CDR3 nucleotide sequences identified in the TCRs of splenic Treg cells isolated from E17.5 pregnant Rank WT (n = 3) and Rank ΔFoxn1 (n = 2) females, as identified from the RNA-sequencing data using the MiXCR software. For a summary of clonal distributions see Extended Data Fig. 5g; for primary data on TCR clones see Supplementary Table 1. r, Clustered heat map showing the relative expression (z-score) of the 400 top-ranked genes of splenic Treg cells isolated from E17.5 pregnant Rank WT or Rank ΔFoxn1 mice. The initial ranking of genes is based on the maximum of the log2-transformed fold change, the absolute value and the statistical significance ranks. Rows indicate genes, columns samples (n = 3/2). s, Quantification of serum autoantibodies reactive against double-stranded DNA (dsDNA) in Rank WT (n = 9) and Rank ΔFoxn1 (n = 15) pregnant mice (from E12.5–E18.5), determined by ELISA. Values for individual mice (dots) are shown; horizontal lines are median values. Two-tailed Mann–Whitney U-test. t, Prevalence (shown as percentages) of Rank WT (n = 7–E14) and Rank ΔFoxn1 (n = 8–21) pregnant mice (from E12.5–E18.5) with detectable autoantibodies reactive towards different organs (red bars), as determined by indirect immunofluorescence shown in k. No statistically significant differences were observed between the cohorts of Rank WT and Rank ΔFoxn1 dams (two-tailed χ 2 test; data not shown). u, Images for the detection of autoantibodies (red) by indirect immunofluorescence staining on uterus, ovaries, and placenta sections of Rag2 −/− mice using serum obtained from pregnant Rank WT and Rank ΔFoxn1 mice. Red: autoantibody staining, blue: DAPI. Serum from antibody-deficient Rag2 −/− as well as autoimmune-prone MRL/lpr females were used as negative and positive controls, respectively. Images are representative of n = 7–14 (Rank WT) and n = 8–21 (Rank ΔFoxn1) mice per tissue. All pregnancies in this figure were C57BL/6J syngeneic.
Extended Data Fig. 5. Characterization of the placental Treg cells, fetal macrosomia and glucose metabolism in pregnant Rank ΔFoxn1 mice.
a, Percentages of CD4+FOXP3+Treg cells (of total CD4+T cells) in E17.5 placentas of Rank WT and Rank ΔFoxn1 pregnant mice. Data were determined by FACS and are from individual placentas (n = 56/54) isolated from n = 11/13 dams, respectively. ***P < 0.001. Two-tailed Mann–Whitney U-test. b, c, Total numbers of thymic-derived Treg cells, as determined by neuropilin-1highHelioshigh expression (b), as well as total numbers and percentages of conventional CD4+ and CD8+T cells (c) in the E13.5–E16.5 placentas of Rank WT and Rank ΔFoxn1 pregnant mice. n = 39–76 placentas analysed from n = 8–17 dams per group. *P < 0.05. Two-tailed Mann–Whitney U-test. a–c, Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual data point. d, Percentage of KI-67+CD4+FOXP3+Treg cells as well as conventional CD4+T cells in the E9.5 and E16.5 placentas of Rank WT mice. Data are mean ± s.e.m., from n = 28/10 individual E9.5 and E16.5 placentas (each dot), respectively. ***P < 0.001. One-way ANOVA, Tukey’s post hoc test. e, Percentage of KI-67+ and active CASP3+Treg cells in the E13.5–E16.5 placentas of Rank WT and Rank ΔFoxn1 mice. Data are mean ± s.e.m., from n = 34/27 (for KI-67) and n = 20/17 (Caspase3) individual placentas (each dot) analysed from n = 12/8 (KI-67) and n = 8/7 (Caspase3) Rank WT and Rank ΔFoxn1 pregnant females, respectively. *P < 0.05. Two-tailed Mann–Whitney U-test. f, Frequency of CD4+FOXP3+Treg cells and conventional CD4+T cells in the E13.5–E17.5 placentas from Rank WT and Rank ΔFoxn1 pregnant females expressing TCRVβ8.1/8.2 and TCRVα8.3 receptors, as determined by FACS analysis. The non-significant differences observed in the conventional FOXP3−CD4+T cell population reinforce the thymic origin of the placental Treg cells, as opposed to a peripheral derivation from CD4+T cells. Data are mean ± s.e.m. and are from n = 61–68 (for Treg cells) and n = 36–25 (for conventional CD4+T cells) individual placentas (each dot) analysed from n = 17/17 (for Treg cells) and n = 7/11 (for conventional CD4+T cells) Rank WT and Rank ΔFoxn1 pregnant females, respectively. *P < 0.05; **P < 0.01. Two-tailed Mann–Whitney U-test. g, Distribution of TCR clonal proportions identified in thymic Treg cells isolated from non-pregnant Rank WT and Rank ΔFoxn1 females (n = 4/4), as well as in thymic (n = 3/4), splenic (n = 3/2) and placental (n = 3/3) Treg cells isolated from E17.5 pregnant Rank WT and Rank ΔFoxn1 mice. The proportions of the most abundant (top N) TCR clones by the number of next-generation sequencing reads are shown. The increased proportion of reads taken up by the top 1 to 10 (red) and 11 to 110 (orange) clones in placental Treg cells indicates a lower TCR clonal diversity compared to thymus and splenic Treg cells. h, Venn diagram showing the number of unique CDR3 nucleotide sequences identified in the TCRs of placental Treg cells isolated from E17.5 pregnant Rank WT (n = 3) and Rank ΔFoxn1 (n = 3) females. Data in g and h was determined from the RNA-sequencing data using the MiXCR software (for primary data on TCR clones see Supplementary Table 1). i, Clustered heat map showing the relative expression (z-score) of the 400 top-ranked genes of placental Treg cells isolated from E17.5 pregnant Rank WT and Rank ΔFoxn1 mice. The initial ranking of genes is based on the maximum of the log2-transformed fold change, the absolute value and the statistical significance ranks. Rows indicate genes, columns samples (n = 3/3). The genotypes, either Rank WT or Rank ΔFoxn1, are annotated for each sample at the top of the columns. j, GSEA enrichment plots of the top 3 positively as well as negatively enriched gene sets in the E17.5 placental Treg cells of Rank ΔFoxn1 versus Rank WT dams (n = 3 per group). A summary of the GSEA Hallmark gene sets enrichment analysis is shown in Supplementary Table 2. Data from g–j are from sort-purified neuropilin-1highCD4+FOXP3+Treg cells. k, Number of total fetuses (viable + resorbed) gestated per Rank WT (n = 35) and Rank ΔFoxn1 (n = 31), as well as Rank WT (n = 16) and Rank ΔK5 (n = 12) individual mothers (each dot). Horizontal lines are median values. Two-tailed Mann–Whitney U-test. l, Sex distribution of viable fetuses at term (E17.5) born to Rank WT and Rank ΔFoxn1 dams. Total numbers and percentage of fetuses per sex are shown for Rank WT (n = 14) and Rank ΔFoxn1 (n = 11) dams. No significant difference was detected in fetal sex distributions between both cohorts (two-tailed χ 2 test; data not shown). Sex was determined by PCR. m, Weight of E17.5 fetuses showing that the fetal macrosomia observed in fetuses gestated in Rank ΔFoxn1 dams is independent of the Rank ΔFoxn1 genotype of the offspring. All pregnancies were syngeneic to C57BL/6J males, thus the offspring of Rank ΔFoxn1 dams are either Rank WT wild-type (Rank flox/WT Foxn1 cre−) or Rank HET heterozygous (Rank flox/WT Foxn1 cre+) for RANK expression in the thymus; both groups are macrosomic compared to Rank WT (Rank flox/flox Foxn1 cre−) fetuses from Rank WT dams. Each dot represents an individual fetus, with n = 7–54 mice per group. Horizontal lines are mean, error bars are s.e.m. **P < 0.01; ***P < 0.001. Two-tailed Student’s t-test. n, o, Serum glucose (n; n = 42/26) and insulin (o; n = 20/13) levels in ad libitum-fed individual non-pregnant Rank WT and Rank ΔFoxn1 females. These are the littermate controls of the pregnant cohorts shown in Fig. 2e, f. n, o, Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. Two-tailed Student’s t-test (n) and two-tailed Mann–Whitney U-test (o). p, Images for the quantification of β-islet areas using the Definiens Tissue software to identify and categorize β-islets (blue) and the exocrine pancreas (green). Representative of n > 120 slides analysed. q, H&E-stained pancreas cross-sections for non–pregnant (upper panels) and E17.5 pregnant (lower panels) Rank WT and Rank ΔFoxn1 mice, depicting the expected enlargement of insulin-producing β-islets (arrows) during pregnancy, in both Rank WT and Rank ΔFoxn1 dams. Representative of n = 11–25 slides analysed per group. r, s, Relative total area of pancreatic β-islets (r) and median β-islets area (s) in non-pregnant and E17.5 pregnant Rank WT (n = 5/9) and Rank ΔFoxn1 (n = 7/13) mice, determined as shown in p (2 distal sections per mouse). *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed Student’s t-test. Data are mean ± s.e.m., each dot represents an individual data point. t, Fasting blood glucose levels in E16.5 pregnant Rank WT and Rank ΔFoxn1 mice. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual dam, n = 12/14. Two-tailed Student’s t-test. All pregnancies in this figure were from C57BL/6J syngeneic crosses.
Extended Data Fig. 6. Characterization of Treg cells in the VAT and other tissues in pregnant Rank ΔFoxn1 mice.
a, Total numbers of neuropilin-1highHelioshighCD4+FOXP3+ cells Treg cells (indicative of thymic origin), per gram of visceral white adipose tissue (gonadal VAT) in non-pregnant (n = 11/5) and E13.5–E16.5 pregnant (n = 11/8) Rank WT and Rank ΔFoxn1 females. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. Data were determined by FACS analysis. *P < 0.05. Two-tailed Student’s t-test. b, Percentages and total numbers of conventional CD4+ and CD8+T cells per gram of visceral white adipose tissue (gonadal VAT) in non-pregnant (n = 11/5) and E13.5–E17.5 pregnant (n = 12/8) Rank WT and Rank ΔFoxn1 females. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. Data were determined by FACS analysis. Two-tailed Mann–Whitney U-test. c, Venn diagram showing the number of unique CDR3 nucleotide sequences identified in the TCRs of VAT (gonadal) Treg cells isolated from E17.5 pregnant Rank WT (n = 3) and Rank ΔFoxn1 (n = 2) females. d, Distribution of TCR clonal proportions identified in VAT Treg cells isolated from E17.5 pregnant Rank WT (n = 3) and Rank ΔFoxn1 (n = 2) mice. The proportions of the most abundant (top N) TCR clones by the number of next-generation sequencing reads are shown. Data from c and d were determined using the MiXCR software (for primary data on TCR clones see Supplementary Table 1). e, Clustered heat maps showing the relative expression (z-score) of the 400 top-ranked genes of VAT Treg cells isolated from E17.5 pregnant Rank WT or Rank ΔFoxn1 mice. The initial ranking of genes is based on the maximum of the log2-transformed fold change, the absolute value and the statistical significance ranks. Rows indicate genes, columns samples. The genotypes, either Rank WT (n = 3) or Rank ΔFoxn1 (n = 2), are annotated for each sample at the top of the columns. f, Summary of GSEA Hallmark gene sets enrichment analysis for the expression profiles of VAT Treg cells isolated from E17.5 pregnant Rank ΔFoxn1 (n = 2) versus Rank WT (n = 3) females. Numbers are NES and all gene sets shown are significantly enriched (defined as NES > = 1.4 and false discovery rate FDR < = 10%). Red, positively enriched gene sets in Rank ΔFoxn1; blue, positively enriched gene sets in Rank WT (the full list of gene sets is shown in Supplementary Table 3). g, Bar plot showing log2-transformed fold change of expression for cytokines and chemokines genes in VAT Treg cells isolated from E17.5 pregnant Rank ΔFoxn1 (n = 2) versus Rank WT (n = 3) mice, as determined by RNA sequencing. Red and blue denote significantly up- and downregulated genes in Rank ΔFoxn1, respectively (defined as abs(log2FC) > = 1.0 and adjusted P < = 0.1). h, Expression levels (in Fragments per Kilobase Million -FPKM) of the signature VAT Treg cells markers PPARγ and ST2 (IL-33R) in splenic and VAT Treg cells from E17.5 pregnant Rank WT (n = 3) and Rank ΔFoxn1 females (n = 2); as determined by RNA sequencing. Floating bars run from minimal to maximal values; lines are median and dots are individual mouse. i, Left, Representatives FACS histograms for ST2 expression in Treg cells from the spleen and gonadal VAT of E13.5–E16.5 Rank WT and Rank ΔFoxn1 pregnant mice. Right, Mean fluorescence intensity (MIF) of ST2 in Treg cells from gonadal VAT of non-pregnant as well as E13.5–E16.5 Rank WT (n = 11/11) and Rank ΔFoxn1 (n = 5/8) pregnant mice. Data were determined by FACS and are shown as box-and-whisker plots (from minimal to maximal values) with each dot representing an individual mouse. Kruskal–Wallis test, Dunn’s post-test. j, Percentages of CD4+FOXP3+Treg cells (of total CD4+ cells) in the uterus-draining para-aortic lymph nodes of individual non-pregnant and pregnant (pool from E12.5–E18.5) Rank WT (n = 19/26) and Rank ΔFoxn1 (n = 14/19) female mice, as determined by FACS. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. Two-tailed Student’s t-test. k, Numbers of CD4+FOXP3+Treg cells per gram of lung and kidney of individual non-pregnant (n = 5/5) and E13.5–E15.5 pregnant (n = 5/3) Rank WT and Rank ΔFoxn1 female mice, as determined by FACS. Data are mean ± s.e.m., each dot represents an individual mouse. One-way ANOVA, Tukey’s post hoc test. l, Numbers of CD4+FOXP3+Treg cells per mm2 in β-islets and exocrine pancreas of E17.5 pregnant Rank WT (n = 11) and Rank ΔFoxn1 (n = 13) mice, as determined by immunohistochemistry. Data are mean ± s.e.m., each dot represents an individual mouse. Two-tailed Mann–Whitney U-test. m, Body weight increases for Rank WT or Rank ΔFoxn1 female mice (n = 4/5) fed a HFD for 47 days. Data are mean ± s.e.m. Two-way ANOVA, Sidak’s multiple comparison test. n, Liver and visceral white adipose tissue (gonadal VAT) weights in Rank WT and Rank ΔFoxn1 female mice (n = 4/5) fed a HFD for 7 weeks. Data are mean ± s.e.m., each dot represents an individual mouse. Two-tailed Student’s t-test. o, Percentages of CD4+FOXP3+Treg cells in the thymus of Rank WT (n = 4) and Rank ΔFoxn1 (n = 5) females fed a HFD for 7 weeks, as determined by FACS. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. Two-tailed Mann–Whitney U-test. p, Oral glucose-tolerance test (mean ± s.e.m.) and the corresponding area under the curve (AUC) calculations (data are mean ± s.e.m., values for individual mice are shown) for Rank WT and Rank ΔFoxn1 females fed a HFD for 7 weeks (n = 4/5). Two-tailed Student’s t-test. q, Total numbers of CD4+FOXP3+Treg cells per gram of VAT in Rank WT and Rank ΔFoxn1 females fed a HFD for 7 weeks. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse (n = 4/5). Data were determined by FACS analysis. Two-tailed Mann–Whitney U-test. r, Serum levels of insulin in Rank WT and Rank ΔFoxn1 females fed ad libitum a HFD for 7 weeks (n = 4/5). Data are mean ± s.e.m., each dot represents an individual mouse. Two-tailed Student’s t-test.
Extended Data Fig. 7. Treg cell transplantation studies.
a, Treg-cell adoptive-transfer experimental design to assess the migration of the transplanted pregnancy-associated Treg cells into different organs. Recipient early pregnant (E3.5–E5.5) Rank WT females (no Foxp3 GFP transgene) were inoculated (intravenous inoculation) with 3 × 105 sort-purified FOXP3–GFP+CD4+neuropilin-1high Treg cells isolated from E6.5–E8.5 Rank WT Foxp3 GFP/GFP pregnant donors. b, c, Representative histograms (b) and relative percentage (c) of FOXP3–GPF+ transplanted Treg cells (of total Treg cells) in the thymus, spleen, draining (peri-uterine) lymph nodes, gonadal VAT and placentas of recipient Rank WT wild-type pregnant females 5 days after transfer, as described in a. n = 4 recipient mice and n = 26 placentas were analysed. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual data point. ***P < 0.001 versus thymus, spleen or draining lymph nodes. One-way ANOVA, Dunnett’s post hoc test. d, Percentage of KI-67+GFP–(recipients’) and KI-67+GFP+ (donors’) Treg cells in the gonadal VAT and placentas of recipient Rank WT wild-type pregnant females 5 days after transfer, as described in a. Data are mean ± s.e.m., each dot represents an individual data point from n = 4 (for VAT) and n = 26 (for placenta). *P < 0.05; ***P < 0.001. Two-tailed Mann–Whitney U-test. e, Schematics of the adoptive Treg cell transfer study used to determine which cell type migrates to the placenta (only wild-type animals were used). Since mice did not express the Foxp3 GFP transgene, Treg cells were sorted as CD4+CD25+CD45RBlow cells and conventional T CD4 cells as CD4+CD25−CD45RBhigh. Treg cells of thymic origin (neuropilin-1high) were transplanted from pregnant donors into non-pregnant females (1). Vehicle-treated control pregnant recipients received only PBS (2). neuropilin-1high Treg cells isolated from pregnant (3) or non-pregnant donors (4) were transplanted into pregnant recipients. Finally, neuropilin-1low Treg cells (5) as well as conventional CD4+CD25−CD45RBhigh T cells (6) were transferred from pregnant donors to recipient dams. In all cases, cells were sorted and pooled from spleen, lymph nodes and thymus of donors and transferred (intravenous inoculation, 1 × 105) into early (E0.5–E2.5) pregnant mice. All recipients were euthanized 15 days post transfer (E15.5–E17.5) for FACS analysis of Treg cells in thymus, spleen and placentas shown in f–i. f, g, Percentages of CD4+FOXP3+Treg cells (of total CD4+) in the thymus (f) and spleen (g) of recipient mice treated with different T cells as described in e, at the experimental end point (n = 4–3 mice per group). Data are mean ± s.e.m., each dot represents an individual mouse. *P < 0.05. Two-tailed Student’s t-test. h, Percentages of CD4+FOXP3+Treg cells (of CD4+), as well as i, neuropilin-1highHelioshigh Treg cells (of all CD4+FOXP3+Treg cells) in the E15.5–E17.5 placentas of recipient mice treated as described in e. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual placenta. n = 10–24 individual placentas from n = 3 recipient mice were analysed per group. *P < 0.05; **P < 0.01. Two-tailed Student’s t-test versus group 3. j, Treg-cell adoptive-transfer scheme to assess whether Treg cells from non-pregnant Rank WT or from pregnant Rank ΔFoxn1 females can rescue placental Treg cell numbers and altered glucose metabolism in Rank ΔFoxn1 dams. As in the transplant study shown in Fig. 3a, thymic Treg cells were sorted and pooled from spleen, thymus and lymph nodes defined as CD4+FOXP3–GFP+neuropilin-1high Treg cells (thymic Treg cells). Both donor (E9.5) and recipient mice contained the FOXP3GFP transgene and 1 × 105 Treg cells were transplanted at E.0.5–E2.5. Vehicle was PBS. k, l, Total numbers of Treg cells (k) and percentages of KI-67+Treg cells (l) in the E16.5 placentas of vehicle-treated Rank WT (2) and Rank ΔFoxn1 (3) pregnant mice, as well as E16.5 placentas of Rank ΔFoxn1 pregnant mice treated with thymic Treg cells isolated from non-pregnant Rank WT females (4) or isolated from pregnant Rank ΔFoxn1 females (5) as described in j. Data were obtained by FACS analysis and are shown as box-and-whisker plots (k, from minimal to maximal values) and bar charts (l, mean ± s.e.m.) from n = 46/25/25/32 for k, and n = 21/16/13/14 for l, individual placentas (each dot) analysed from n = 6/5/5/4 mice for groups 2 to 5 respectively. *P < 0.05. One-Way ANOVA, Dunnett’s multiple comparison test versus group 3. m, Total numbers of CD4+FOXP3+Treg cells per gram of gonadal VAT in vehicle-treated non-pregnant Rank WT females (group 1), vehicle-treated Rank WT (2) and Rank ΔFoxn1 (3) E16.5 pregnant mice, as well as Rank ΔFoxn1 E16.5 pregnant mice treated with thymic Treg cells isolated from non-pregnant Rank WT females (4) or from pregnant Rank ΔFoxn1 females (5) as described in j. Data were obtained by FACS analysis and are from n = 6/6/5/5/4 mice for groups 1 to 5, respectively. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05; **P < 0.01. Two-tailed Student’s t-test versus group 2. n, o, Oral glucose-tolerant test (n; mean ± s.e.m.) and the corresponding area under the curve calculations (o) in vehicle-treated non-pregnant Rank WT females (group 1), vehicle-treated Rank WT (2) and Rank ΔFoxn1 (3) E15.5 pregnant mice, as well as Rank ΔFoxn1 E15.5 pregnant females treated with thymic Treg cells isolated from non-pregnant Rank WT females (4) or from pregnant Rank ΔFoxn1 females (5) as described in j. n = 6/6/5/5/4 mice for groups 1–5, respectively. Lines in scatter dot plots are mean, error bars are s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA, Dunnett’s multiple comparisons test versus group 3.
Extended Data Fig. 8. Gene expression profiling and characterization of the VAT in pregnant Rank ΔFoxn1 mice.
a, b, Pairwise intersection heat map showing the numbers of differentially expressed genes in the gonadal VAT (a) as well as overlap counts (b) resulting from all pairwise comparisons of the three experimental conditions: vehicle-treated E17.5 pregnant Rank WT, as well as E17.5 Rank ΔFoxn1 pregnant females treated with either vehicle- or with sort-purified FOXP3–GFP+CD4+neuropilin-1high Treg cells isolated from Rank WT Foxp3 GFP/GFP pregnant donors (pregnancy tTreg-cell-treated). DESeq2 v.1.16.1, FDR threshold of 0.05. c, Summary of GSEA Hallmark gene sets significantly enriched (NES > = 1.4 and FDR < = 10%) comparing the expression profiles (in CPM) of the visceral (gonadal) white adipose tissue of Rank ΔFoxn1, Rank WT, and Rank ΔFoxn1 tTreg-cell-treated dams at E17.5 (n = 4 per group). NES scores are only listed for the significant sets. d, e, GSEA enrichment plots of the top positively enriched gene set: (TNF signalling via NF-κB), as well as the top negatively enriched gene set (oxidative phosphorylation) in the VAT of Rank ΔFoxn1 versus Rank WT as well as Rank ΔFoxn1 versus Rank ΔFoxn1 dams at E17.5 treated with thymic Treg cells (n = 4 per group). GSEA-derived heat maps on the right show the relative level of gene expression (red, high; blue, low) in the leading edge subset (top 20 genes shown). All mice described in this figure were treated as described in Fig. 3a. Rank ΔFoxn1 versus Rank WT dams were treated with vehicle (PBS), which for simplification is not shown in the figure. f, Total adipose tissue weights and g, median adipocyte sizes in the gonadal VAT of vehicle-treated E17.5 pregnant Rank WT mice (n = 6) as well as Rank ΔFoxn1 recipient E17.5 pregnant females treated with either vehicle (n = 5) or with sort-purified FOXP3–GFP+CD4+neuropilin-1high Treg cells isolated from Rank WT Foxp3 GFP/GFP pregnant donors (n = 6; that is, pregnancy tTreg-cell-treated). Treg-cell adoptive transfer was performed as shown in Fig. 3a. Data are mean ± s.e.m., each dot represents an individual data point. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA, Dunnett’s post hoc test. h, Percentages of different adipocytes sizes in the VAT of vehicle-treated Rank WT dams (n = 6) as well as vehicle- and pregnancy tTreg-cell-treated Rank ΔFoxn1 dams at E17.5 (n = 5 and 6, respectively). *P < 0.05; other P values are shown. Two-tailed Student’s t-test. For g and h, two distal cross-sections were analysed per mouse. Data are mean ± s.e.m., each dot represents an individual data point. i, H&E cross-sections of the gonadal white adipose tissue of vehicle-treated Rank WT dams, as well as vehicle- and pregnancy tTreg-cell-treated Rank ΔFoxn1 dams at E17.5, showing markedly enlarged adipocytes in Rank ΔFoxn1 pregnant mice compared with Rank WT and pregnancy tTreg-cell-treated Rank ΔFoxn1 counterparts. Representative of n = 10–12 slides analysed from 5–6 mice per group. j, Immunohistochemistry detecting F4/80+ macrophages (arrows) in the VAT of vehicle-treated Rank WT, as well as of vehicle- and pregnancy tTreg-cell-treated Rank ΔFoxn1 dams at E17.5. Representative of n = 5–6 mice analysed. k, Percentages of F4/80+ macrophages in the VAT of non-pregnant Rank WT and Rank ΔFoxn1 females as well as vehicle-treated Rank WT and vehicle- and pregnancy tTreg-cell-treated Rank ΔFoxn1 dams at E17.5, as detected by immunohistochemistry and quantified using the Definiens Software (n = 5–6 per genotype). Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05. One-way ANOVA, Tukey’s post hoc test.
Extended Data Fig. 9. Macrosomia, VAT and glucose metabolism in the offspring of Rank ΔFoxn1 dams.
a, Body weights (mean ± s.e.m.) and representative photograph (at 80 days of age) for the male offspring of Rank WT or Rank ΔFoxn1 dams (n = 16/11) fed a normal chow (NC). Data from female offspring are shown in Fig. 4a. **P < 0.01; ***P < 0.001. Two-way ANOVA, Sidak’s multiple comparison test. b, Body weights (mean ± s.e.m.) and representative photograph (taken after 6 weeks on a HFD) for the female offspring of Rank WT or Rank ΔFoxn1 dams (n = 14/12) fed a HFD. Male comparisons are shown in Fig. 4b. *P < 0.05; **P < 0.01. Two-way ANOVA, Sidak’s multiple comparison test. c, Body weights at 3 and 10 weeks of age for the different genotypes of the offspring of Rank ΔFoxn1 dams as compared to those from Rank WT dams. All pregnancies were syngeneic to C57BL/6J males, thus the offspring of Rank ΔFoxn1 dams are either Rank WT wild-type (Rank flox/WT Foxn1 cre−) or Rank HET heterozygous (Rank flox/WT Foxn1 cre+) for RANK expression in the thymus; both groups are macrosomic compared to Rank WT (Rank flox/flox Foxn1 Cre−) fetuses from Rank WT dams. Offspring of different genotypes were co-caged to minimize microbiota effects. Data are shown as box-and-whisker plots (from minimal to maximal values); dots represent individual mice, with n = 16–45 per group. *P < 0.05; **P < 0.01; ***P < 0.001. Kruskal–Wallis test, Dunn’s post-test. d, Body weights from postnatal day 1 until weaning (day 19) for pups born to Rank ΔFoxn1 dams but caged and breast-fed by Rank WT dams (n = 18), as well as the pups born to Rank WT dams but caged and breast-fed by Rank ΔFoxn1 dams (n = 21). As controls, pups born and breast-fed to Rank WT dams are shown (n = 11). Mothers were exchanged at day 1 after birth. Note the macrosomia is inherent to the pups and does not depend on mothers or breast milk-derived effects. Data are mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001. Two-way ANOVA, Dunnett’s multiple comparison test. e, f, Ad libitum-fed (e) and fasting (f) serum insulin levels in male and female offspring of Rank WT and Rank ΔFoxn1 dams fed a normal chow (NC) diet for 80 days. Data are shown as box-and-whisker plots (from minimal to maximal values). Dots represent individual mice, with n = 14–24 per group. *P < 0.05. Two-tailed Mann–Whitney U-test. g, Fasting serum insulin levels in male (n = 16/11) and female (n = 15/13) offspring of Rank WT and Rank ΔFoxn1 dams fed a HFD for 9 weeks. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. Two-tailed Mann–Whitney U-test. h, Ad libitum-fed blood glucose levels at different ages of females (n = 14–25) and males (n = 10–23) offspring of Rank WT or Rank ΔFoxn1 dams fed a normal chow (NC). Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05; **P < 0.01. Two-tailed Student’s t-test. i, Fasting blood glucose levels in female (n = 18/20) and male (n = 25/16) offspring of Rank WT or Rank ΔFoxn1 dams that were fed a normal chow for 80 days. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05. Two-tailed Mann–Whitney U-test. j, k, Oral glucose-tolerance test (mean ± s.e.m.) and the corresponding area under the curve calculations for the female (j, n = 18/20) and male offspring (k, n = 25/16) of Rank WT and Rank ΔFoxn1 dams. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse, fed 80 days a normal chow diet. **P < 0.01. Two-tailed Mann–Whitney U-test. l, m, Ad libitum-fed (l) and fasting (m) blood glucose levels in male and female offspring of Rank WT and Rank ΔFoxn1 dams fed a HFD for 9 weeks. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. n = 9–15. *P < 0.05. Two-tailed Student’s t-test. n, Percentages of CD4+FOXP3+ VAT Treg cells (of total CD4) in the gonadal white adipose tissue of the HFD-fed male and female offspring of Rank WT (n = 6/6) and Rank ΔFoxn1 (n = 6/6) dams. Mice were fed a HFD for 12 weeks. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. ***P < 0.001. Two-tailed Student’s t-test. o, Median adipocyte sizes in the gonadal VAT of aged male and female offspring of Rank WT (n = 7/6) and Rank ΔFoxn1 (n = 9/6) dams. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. *P < 0.05. Two-tailed Student’s t-test. p, Percentages of CD4+FOXP3+Treg cells (of total CD4) in the gonadal VAT of aged male and female offspring of Rank WT (n = 10/8) and Rank ΔFoxn1 (n = 12/9) dams. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse. **P < 0.01. Two-tailed Student’s t-test. q, Percentages of Cd11b+F4/80+ macrophages (left) as well as percentage of M1 (CD11c+CD301−) and M2 (CD11c−CD301+) macrophages (right, gated on Cd11b+F4/80+ macrophages) in the gonadal white adipose tissue of aged male and female offspring of Rank WT and Rank ΔFoxn1 dams. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual mouse, with n = 6–12 per group. **P < 0.01. Two-tailed Student’s t-test. Data in o–q are from individual 15–17-month-old mice, fed a normal chow since weaning.
Extended Data Fig. 10. Characterization of human patients with GDM.
a, Gestational week of delivery for control glucose-tolerant pregnant women (Ctrl) and pregnant women with GDM included in the study. All women were carrying singleton pregnancies with equal distribution of sex in each group (n = 8 carrying female and n = 8 carrying male babies). Placenta samples were collected at delivery and were also matched for gestational week and sex of the baby. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual woman, with n = 16/16 women per group. Two-tailed Mann–Whitney U-test. b, BMI before pregnancy, at early pregnancy (<20 weeks) and at late pregnancy (36–38 weeks) for control glucose-tolerant pregnant women and pregnant women with GDM, showing matched BMI between the cohorts. Data are mean ± s.e.m. and each dot represents a woman n = 16/16. Two-tailed Student’s t-test. c, d, Fasting glucose (c) and insulin (d) levels for pregnant women diagnosed with GDM as well as control glucose-tolerant pregnant women. n = 16 per group (data are mean ± s.e.m. and each dot represents an individual woman). **P < 0.01. Two-tailed Student’s t-test. e, Plasma insulin levels (mean ± s.e.m.) and the corresponding area under the curve calculations during the 2 h oral glucose test shown in Fig. 4g, for the same pregnant women with GDM and control pregnant women. Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual woman, with n = 16 per group. *P < 0.05. two-tailed Mann–Whitney U-test. f, Homeostasis model assessment of insulin resistance (HOMA-IR) for the pregnant women with GDM as well as control glucose-tolerant pregnant women. Data are mean ± s.e.m. and each dot represents an individual woman, n = 16/16. **P < 0.01. two-tailed Student’s t-test. Data in c–f were collected at early gestation (<20 weeks). Similar results were observed mid- (24–28 weeks) and late pregnancy (36–38 weeks) (data not shown). g, Birth levels of C-peptide (a by-product of insulin production in the β-islets) in the cord blood of babies born to GDM (n = 15) and control glucose-tolerant mothers (n = 15), revealing hyperinsulinaemia in the babies of mothers with GDM. Data are mean ± s.e.m. and each dot represents an individual sample. **P < 0.01. Two-tailed Student’s t-test. h, Relative expression of the Treg-cell-specific transcript FOXP3 normalized to CD4 transcript levels in the placentas of control glucose-tolerant pregnant women (n = 16) and in pregnant women with GDM (n = 16). Three distal placental tissue slices (all on the maternal side) were analysed for each woman and then averaged. Note that similar results were observed if normalized to a housekeeping gene (shown in Fig. 4i, j). Data are shown as box-and-whisker plots (from minimal to maximal values) and each dot represents an individual woman. *P < 0.05. Two-tailed Mann–Whitney U-test. i, Immunohistochemistry detecting FOXP3+Treg cells (arrows) in the placentas of control glucose-tolerant pregnant controls and women with GDM. Representative of n = 65/28 slides analysed per group. For relative quantification of the staining see Fig. 4k.
Supplementary Material
Acknowledgements
We thank all members of the Penninger and Paolino laboratories for discussions and technical support; Y. Redouane for help with RNA isolation; D. Hoffmann and L. Haas for assistance with oral gavage and intravenous injections; S. Gavali for proofreading the manuscript; G. Schmauss, G. Petri, M. Weninger and other members of the IMP-IMBA Biooptics service facility for assistance in cell sorting and image quantification; staff of the VBCF histopathology department and, in particular, M. Zeba, J. Klughofer and A. Kavirayani, as well as H. Schachner for histology support; and staff of the animal facility and all other IMP/IMBA service facilities. Mass spectrometry and QuantSeq were performed at the VBCF Metabolomics and Next Generation Sequencing units, funded by the City of Vienna. Smart-seq2 libraries were sequenced by the Biomedical Sequencing Facility at CeMM. M.P. is a Ragnar Söderberg Fellow in Medicine and is supported by IMBA, start-up grants from the Karolinska Institutet, the Swedish Research Council (Vetenskapsrådet NE 2016-04458) and the Ragnar Söderberg Foundation (M21/17). G.A.H. is supported by grants from the Swiss National Foundation (3100-68310.02 and 3100-122558) and The Wellcome Trust (105045/Z/14/Z). J.P.F. is funded by Agencia Nacional de Promoción Científica y Tecnológica (PICT-2017-2401), GlaxoSimthKline/Ministerio de Ciencia y Técnica (PCE-GSK-2017-0052) and Fundación para el Progreso de la Medicina (GF-N03/2017). G.A. is supported by a MRC Programme Grant (MR/N000919/1), A.J.W. is funded by a Wellcome Trust Seed Award (204375/Z/16/Z). L.K. is supported by the BM Fonds (15142), the Margaretha Hehberger Stiftung (15142), the COMET Competence Center CBmed (Center for Biomarker Research in Medicine) (FA791A0906.FFG), a FWF grant (P26011) and the Christian-Doppler Lab for Applied Metabolomics. A.K.-W. and J.H. are supported by the Austrian Science Fund No. I 4209 (GOING-FWD) and a Medical Scientific Fund of the Mayor of the City of Vienna (15205). J.M.P. is supported by the Austrian Federal Ministry of Education, Science and Research, the Austrian Academy of Sciences and the City of Vienna and grants from the Austrian Science Fund (FWF Wittgenstein award Z 271-B19), the T. von Zastrow foundation, an EU ERC Advanced Grant (European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 341036) and a Canada 150 Research Chairs Program (F18-01336).
Footnotes
Author contributions.
All experiments were performed by M.P. with the help of: R.K. in the maintenance of mouse colony and metabolic studies and E.R. for breeding and timed pregnancies; I.U. in qPCR analysis; S.J.F.C. in adoptive transfer experiments and Treg cell suppression assays; B.P. in mouse dissections and J.P.F. in confocal imaging. V.S. helped with sex-hormone in vivo experiments. A.J.W. performed FTOC cultures. L.K. and S.D. performed the pathology analysis of placentas and pancreas. J.H., D.B.-T. and A.K.-W. collected human samples and anthropometric data on pregnant women. T.P. and C.B. performed Smart-seq2 sequencing on Treg cells and M.S. performed bioinformatics analyses on the Smart-seq2 data. M.N. performed bioinformatics analyses on the QuantSeq data and GSEA analyses. G.A.H. provided the Foxn1 cre expressing mice. G.A. provided expertise in experimental design, training and data analysis of FTOCs and confocal microscopy, as well as key reagents. M.P. together with J.M.P. designed the experiments and wrote the manuscript.
Competing interests The authors declare no competing interests.
Data availability
RNA-sequencing data has been deposited in the NCBI Gene Expression Omnibus under the following accession numbers: QuantSeq data on VAT tissue: GSE117112; Smart-seq2 data on Treg cells: GSE97666.
References
- 1.Soma-Pillay P, Nelson-Piercy C, Tolppanen H, Mebazaa A. Physiological changes in pregnancy. Cardiovasc J Afr. 2016;27:89–94. doi: 10.5830/CVJA-2016-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19:548–556. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- 3.Persike EC., Jr Involution of the thymus during pregnancy in young mice. Proc Soc Exp Biol Med. 1940;45:315–317. [Google Scholar]
- 4.Dougall WC, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong Y-Y, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 6.Sigl V, Jones LP, Penninger JM. RANKL/RANK: from bone loss to the prevention of breast cancer. Open Biol. 2016;6 doi: 10.1098/rsob.160230. 160230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fata JE, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell. 2000;103:41–50. doi: 10.1016/s0092-8674(00)00103-3. [DOI] [PubMed] [Google Scholar]
- 8.Rossi SW, et al. RANK signals from CD4+3− inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowan JE, et al. The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+Thymocyte development. J Exp Med. 2013;210:675–681. doi: 10.1084/jem.20122070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilburgs T, et al. Evidence for a selective migration of fetus-specific CD4+CD25bright regulatory T cells from the peripheral blood to the decidua in human pregnancy. J Immunol. 2008;180:5737–5745. doi: 10.4049/jimmunol.180.8.5737. [DOI] [PubMed] [Google Scholar]
- 12.Aluvihare VR, Kallikourdis M, Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. 2004;5:266–271. doi: 10.1038/ni1037. [DOI] [PubMed] [Google Scholar]
- 13.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kc K, Shakya S, Zhang H. Gestational diabetes mellitus and macrosomia: a literature review. Ann Nutr Metab. 2015;66:14–20. doi: 10.1159/000371628. [DOI] [PubMed] [Google Scholar]
- 15.Kühl C. Glucose metabolism during and after pregnancy in normal and gestational diabetic women. 1. Influence of normal pregnancy on serum glucose and insulin concentration during basal fasting conditions and after a challenge with glucose. Acta Endocrinol (Copenh.) 1975;79:709–719. [PubMed] [Google Scholar]
- 16.Van Assche FA, Aerts L, De Prins F. A morphological study of the endocrine pancreas in human pregnancy. Br J Obstet Gynaecol. 1978;85:818–820. doi: 10.1111/j.1471-0528.1978.tb15835.x. [DOI] [PubMed] [Google Scholar]
- 17.Kolodin D, et al. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 2015;21:543–557. doi: 10.1016/j.cmet.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bapat SP, et al. Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature. 2015;528:137–141. doi: 10.1038/nature16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 21.Vernochet C, et al. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 2012;16:765–776. doi: 10.1016/j.cmet.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115:e290–e296. doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- 23.Vohr BR, McGarvey ST, Tucker R. Effects of maternal gestational diabetes on offspring adiposity at 4–7 years of age. Diabetes Care. 1999;22:1284–1291. doi: 10.2337/diacare.22.8.1284. [DOI] [PubMed] [Google Scholar]
- 24.Meier JJ, Bhushan A, Butler AE, Rizza RA, Butler PC. Sustained beta cell apoptosis in patients with long-standing type 1 diabetes: indirect evidence for islet regeneration? Diabetologia. 2005;48:2221–2228. doi: 10.1007/s00125-005-1949-2. [DOI] [PubMed] [Google Scholar]
- 25.Butler AE, et al. β-Cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 26.Sacks DA, et al. Frequency of gestational diabetes mellitus at collaborating centers based on IADPSG consensus panel-recommended criteria: the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study. Diabetes Care. 2012;35:526–528. doi: 10.2337/dc11-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egan AM, et al. Epidemiology of gestational diabetes mellitus according to IADPSG/WHO 2013 criteria among obese pregnant women in Europe. Diabetologia. 2017;60:1913–1921. doi: 10.1007/s00125-017-4353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somerset DA, Zheng Y, Kilby MD, Sansom DM, Drayson MT. Normal human pregnancy is associated with an elevation in the immune suppressive CD25+ CD4+ regulatory T-cell subset. Immunology. 2004;112:38–43. doi: 10.1111/j.1365-2567.2004.01869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zenclussen AC, et al. Abnormal T-cell reactivity against paternal antigens in spontaneous abortion: adoptive transfer of pregnancy-induced CD4+CD25+T regulatory cells prevents fetal rejection in a murine abortion model. Am J Pathol. 2005;166:811–822. doi: 10.1016/S0002-9440(10)62302-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansom SN, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014;24:1918–1931. doi: 10.1101/gr.171645.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J Immunol. 2015;195:498–506. doi: 10.4049/jimmunol.1500558. [DOI] [PubMed] [Google Scholar]
- 32.Butte NF. Carbohydrate and lipid metabolism in pregnancy: normal compared with gestational diabetes mellitus. Am J Clin Nutr. 2000;71:1256S–1261S. doi: 10.1093/ajcn/71.5.1256s. [DOI] [PubMed] [Google Scholar]
- 33.Bellamy L, Casas J-P, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;373:1773–1779. doi: 10.1016/S0140-6736(09)60731-5. [DOI] [PubMed] [Google Scholar]
- 34.Clausen TD, et al. High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes: the role of intrauterine hyperglycemia. Diabetes Care. 2008;31:340–346. doi: 10.2337/dc07-1596. [DOI] [PubMed] [Google Scholar]
- 35.Hanada R, et al. Central control of fever and female body temperature by RANKL/RANK. Nature. 2009;462:505–509. doi: 10.1038/nature08596. [DOI] [PubMed] [Google Scholar]
- 36.Zuklys S, et al. Stabilized β-catenin in thymic epithelial cells blocks thymus development and function. J Immunol. 2009;182:2997–3007. doi: 10.4049/jimmunol.0713723. [DOI] [PubMed] [Google Scholar]
- 37.Fontenot JD, et al. Regulatory T cell lineage specification by the Forkhead transcription factor Foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Tarutani M, et al. Tissue-specific knockout of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc Natl Acad Sci USA. 1997;94:7400–7405. doi: 10.1073/pnas.94.14.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+T reg cells. J Exp Med. 2012;209:1723–1742. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Idris AI. Ovariectomy/orchidectomy in rodents. Methods Mol Biol. 2012;816:545–551. doi: 10.1007/978-1-61779-415-5_34. [DOI] [PubMed] [Google Scholar]
- 41.Xing Y, Hogquist KA. Isolation, identification, and purification of murine thymic epithelial cells. J Vis Exp. 2014;90:e51780. doi: 10.3791/51780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2016. [Google Scholar]
- 46.Bolotin DA, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 47.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the Aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 48.Harreiter J, et al. IADPSG and WHO 2013 gestational diabetes mellitus criteria identify obese women with marked insulin resistance in early pregnancy. Diabetes Care. 2016;39:e90–e92. doi: 10.2337/dc16-0200. [DOI] [PubMed] [Google Scholar]
- 49.Metzger BE, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:e98. doi: 10.2337/dc09-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diagnostic criteria and classification of hyperglycaemia first detected in pregnancy: a World Health Organization Guideline. Diabetes Res Clin Pract. 2014;103:341–363. doi: 10.1016/j.diabres.2013.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data has been deposited in the NCBI Gene Expression Omnibus under the following accession numbers: QuantSeq data on VAT tissue: GSE117112; Smart-seq2 data on Treg cells: GSE97666.