Abstract
Type 1 conventional dendritic cells (cDC1s) are necessary for cross-presentation (XP) of many viral and tumor antigens to CD8+ T cells. cDC1s can be identified in mice and humans by high expression of DNGR-1 (a.k.a. CLEC9A), a receptor that binds dead cell debris and facilitates XP of corpse-associated antigens. Here, we show that DNGR-1 is a dedicated XP receptor that signals upon ligand engagement to promote phagosomal rupture. This allows escape of phagosomal contents into the cytosol where they access the endogenous MHC class I antigen processing pathway. The activity of DNGR-1 maps to its signalling domain, which activates SYK and NADPH oxidase to cause phagosomal damage even when spliced into a heterologous receptor and expressed in heterologous cells. Our data reveal the existence of innate immune receptors that couple ligand binding to endocytic vesicle damage to permit MHC class I antigen presentation of exogenous antigens and regulate adaptive immunity.
Keywords: dendritic cells, cross-presentation, DNGR-1, CLEC9A, phagosome-to-cytosol
Introduction
The ability of antigen-presenting cells (APCs) to present exogenous antigens on major histocompatibility complex class I (MHC I), a process termed cross-presentation (XP), is essential to induce protective cytotoxic T cell (CTL) responses against tumors and many viruses1–6. Two basic XP models have emerged from studies in multiple cell types. In one, antigen processing and MHC I molecule loading occurs entirely within the phago-endosomal compartment of APCs (“vacuolar” pathway)7–9. In another, exogenous antigens somehow gain access to the APC cytosol (“cytosolic” or “phagosome-to-cytosol” (P2C) pathway) and are processed by the proteasome as for endogenous antigens7–9. P2C has been argued to involve transporters that selectively translocate polypeptide substrates across the phagosomal membrane (reviewed in 10). Alternatively, P2C could involve passive leakage of phagosomal contents, perhaps following membrane lipid damage by reactive oxygen species (ROS) produced by NADPH oxidase11,12. A version of this hypothesis, termed the “indigestion model”, suggests that P2C involves phagosomal rupture and wholesale release of internalised cargo into the cytosol13,14.
XP pathways may vary depending on cell type. Priming of CD8+ T cells against tumors and some viruses is often abrogated in Batf3 deficient (Batf3 –/–) mice lacking type 1 conventional dendritic cells (cDC1s)15, pointing to these cells as non-redundant cross-presenting APCs in those settings. cDC1s possess cell biological specialisations that favour XP, including adaptations in vesicular trafficking and a relative inability to degrade phagosomal cargo, preserving antigenic information5,6,16–20. Additionally, it is possible that cDC1 possess receptors that detect potentially antigenic cargo and specifically signal to induce XP. In this regard, cDC1s in mice and humans express high levels of DNGR-1 (a.k.a. CLEC9A), a type II transmembrane protein of the C-type lectin receptor superfamily21. DNGR-1 is used by cDC1 to detect dead cell debris, binding via its extracellular domain to exposed F-actin-myosin complexes on cell corpses that have adhered to or been internalised by cDC1s21,22. Within the short N-terminal intracellular domain of DNGR-1 is a single hemITAM motif that can recruit and activate the kinase SYK in response to DNGR-1 ligand engagement23. Notably, mice deficient in DNGR-1 or mice in which Syk has been ablated in CD11c+ cells (including cDC1) display a partial defect in XP of dead cell-associated antigens23, which results in impaired CD8+ T cell responses against cytopathic viruses3,24,25. As such, DNGR-1 and SYK contribute to the ability of cDC1 to translate dead cell recognition into adaptive immunity via XP. However, it is unclear whether this reflects a function of DNGR-1-SYK in routing dead cell cargo into specialised endocytic compartments that are permissive for XP (e.g., poorly degradative phagosomes) or a more active role of DNGR-1-SYK signalling in the process leading to XP. Here, we show that, distinct from any role in ligand uptake and phagosomal maturation, DNGR-1-SYK signalling actively promotes XP of ligand-associated antigens by inducing phagosomal membrane rupture. Our data indicate the existence of dedicated receptors that select cargo for P2C and promote XP.
Results
Phagosomal DNGR-1 engagement selectively regulates XP via the cytosolic pathway
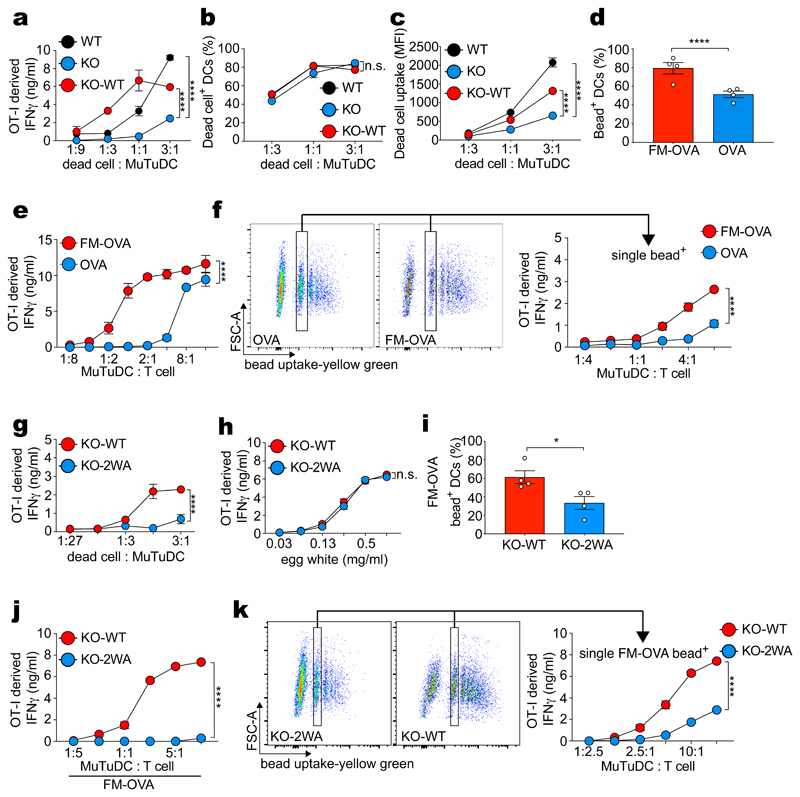
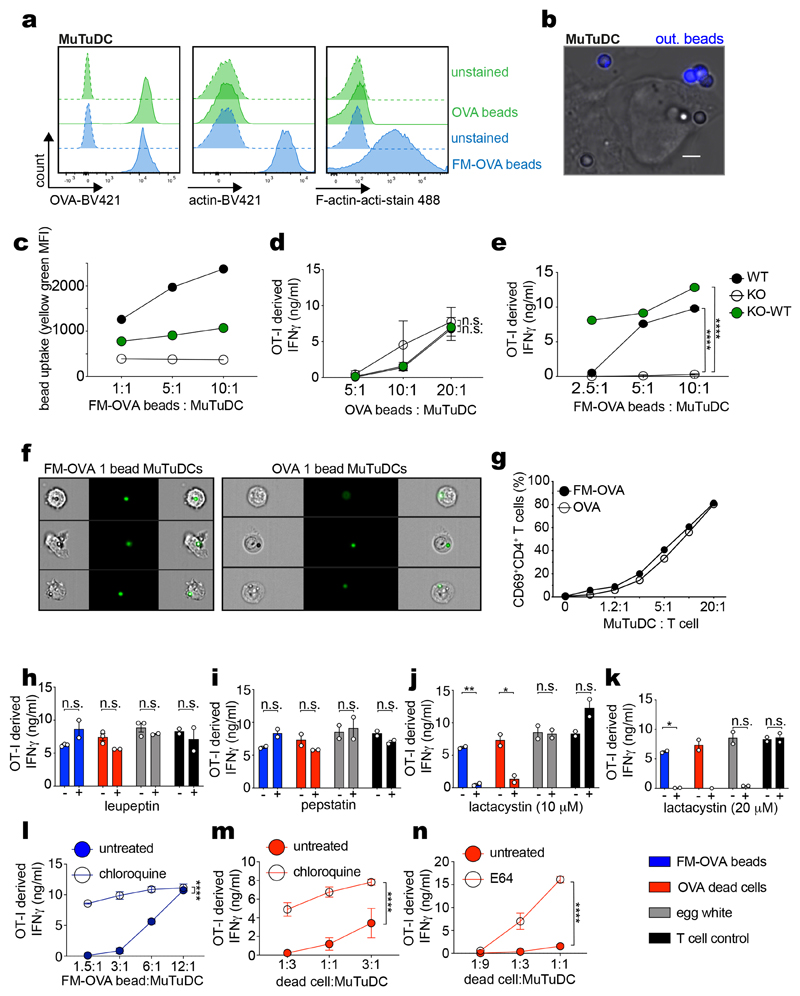
The cell line MuTuDC1940 (henceforth termed MuTuDCs) consists of transformed mouse spleen cDC1s that naturally express DNGR-1 and has been used to study XP 22,26–28. MuTuDCs were pulsed with UV-irradiated ovalbumin (OVA)-expressing H-2Kbm1 mouse embryonic fibroblasts (OVA dead cells) and then cultured with OVA peptide-H-2Kb-specific OT-I CD8+ effector T cells. As reported26, cultures with DNGR-1-deficient MuTuDCs (KO) accumulated lower levels of OT I-derived IFN-γ (an indirect measure of OVA XP) than cultures with wildtype (WT) MuTuDCs (Fig. 1a). This defect was corrected by ectopic re-expression of the receptor in DNGR-1-deficient MuTuDCs (KO-WT) (Fig. 1a). DNGR-1 expression did not markedly affect uptake of dead cell material (Fig. 1b-c), as reported23. In contrast, when measuring phagocytosis of OVA-coated latex beads (OVA beads; Extended Data Fig. 1a), we noticed that additional coating with DNGR-1 ligand (F-actin-myosin II - FM-OVA beads; Extended Data Fig. 1a), resulted in greater bead internalisation (Fig. 1d, Extended Data Fig. 1b) and enhanced OVA XP (Fig. 1e). Similarly, WT and KO-WT internalised FM-OVA beads with greater efficiency than KO MuTuDCs and this correlated with greater XP activity, an effect not seen when using OVA beads not coated with F-actin-myosin II (Extended Data Fig. 1c-e). Thus, DNGR-1 can play an important role in particle uptake when the target is relatively devoid of ligands for other phagocytic receptors. To separate the effect of DNGR-1 on XP from contribution to uptake, we sorted cells that had phagocytosed a single bead (Fig. 1f, Extended Data Fig. 1f). We found that MuTuDCs containing a single FM-OVA bead stimulated CD8+ OT-I T cells more efficiently than cells with single OVA beads (Fig. 1f). This was specific for XP as both sets of sorted MuTuDCs stimulated OVA-specific CD4+ T cells (OT-II) to the same extent (Extended Data Fig. 1g). We also used DNGR-1-deficient MuTuDCs complemented with either WT DNGR-1 or a mutant receptor that cannot bind F-actin (W155A-W250A; termed KO-2WA). Disrupting the binding capacity of DNGR-1 abrogated XP of dead cell-associated antigen (Fig. 1g and reference26) but did not impair XP of antigenic substrates lacking ligands for the receptor, such as soluble OVA antigen from hen egg white (Fig. 1h). KO-WA MuTuDC also displayed a markedly diminished ability to stimulate OT-I cells in response to FM-OVA beads, disproportionate to any reduction in bead uptake (Fig. 1i, j). Sorting single FM-OVA bead+ MuTuDCs (Fig. 1k, Extended Data Fig. 1f) formally confirmed a decrease in XP by cells bearing mutant DNGR-1 unable to engage its ligand (Fig. 1k). XP of soluble OVA, FM-OVA beads and cell-associated OVA was unaffected by inhibitors of lysosomal proteases or acidification (Extended Data Fig. 1h, l, m, n) but was blocked by the proteasome inhibitor lactacystin (Extended Data Fig. 1j-k). Altogether, these data indicate a specific effect of DNGR-1 engagement on promoting XP of ligand-associated antigen via the cytosolic pathway that is independent of receptor contribution to ligand uptake.
Figure 1. DNGR-1 has a dedicated role in XP of ligand-associated antigens.
a, WT MuTuDCs (WT), DNGR-1-deficient MuTUDCs (KO) and DNGR-1-deficient MuTuDCs complemented with WT DNGR-1 (KO-WT) were co-cultured with OVA-expressing dead cells for 4 hrs before effector OT-I T cells were added. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental duplicates and representative of three independent experiments (n = 3). b-c, CLARET-labelled dead cells were co-cultured with WT, KO and KO-WT MuTuDCs at the indicated ratios and uptake was analysed by flow cytometry. Frequency (b) and MFI (c) of CLARET+ MuTuDCs from one experiment is plotted. d, WT MuTuDCs were co-cultured with OVA or FM-OVA coated beads and bead internalisation was analysed by flow cytometry and microscopy after fixation and subsequent staining for OVA to exclude beads that were not fully internalised (“out. beads”). Data represent mean frequency (± s.d.) of bead+ MuTuDCs across 18-36 fields of view from one of three experiments (n = 3). P values were calculated using an unpaired nonparametric Kolmogorov-Smirnov test. e-f, WT MuTuDCs were pulsed with OVA or FM-OVA fluorescent beads for 4 hrs. Bead-pulsed bulk MuTuDCs (e) or FACS-sorted MuTuDCs containing a single bead (f) were co-cultured overnight with effector OT-I T cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental duplicates and representative of three independent experiments (n = 3). FACS profiles indicate gate used for sorting cells that internalised a single bead. g-i, DNGR-1 deficient MuTuDCs complemented with WT DNGR-1 (KO-WT) or F-actin binding deficient mutant DNGR-1 (W155A-W250A; KO-WA) were co-cultured with OVA-expressing dead cells (g), or hen egg white (h) for 4 hrs before effector OT-I T cells were added. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental duplicates and representative of three independent experiments (n = 3). i, KO-WT and KO-WA MuTuDCs were co-cultured with FM-OVA beads for 4 hrs and uptake was analysed by flow cytometry. Mean frequency (± s.d.) of bead+ MuTuDCs across four independent experiments is plotted. P values were calculated by Kolmogorov-Smirnov test. j-k, KO-WT or KO-WA MuTuDCs were pulsed with FM-OVA beads for 4 hrs. The bead pulsed bulk MuTuDCs or FACS sorted MuTuDCs containing a single bead were co-cultured overnight with effector OT-I T cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental duplicates and representative of three independent experiments (n = 3). P values for (a, b, e, f, g, h, j and k) were determined by two-way ANOVA. n.s., not significant; *P ≤ 0.05; ****P ≤ 0.00001.
DNGR-1 accumulates in poorly degradative phagosomes but is dispensable for their formation
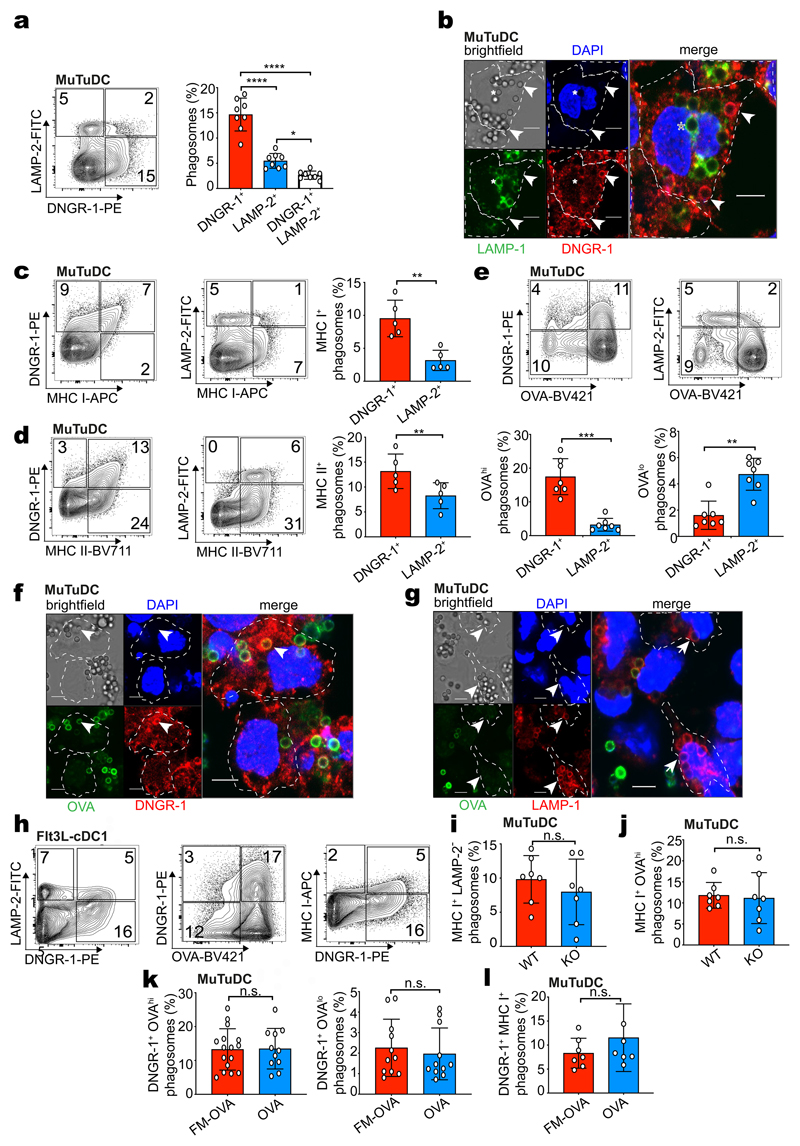
To investigate whether DNGR-1 affects the properties of phagosomes, we characterised FM-OVA bead phagosomes from MuTuDCs by PhagoFACS29 (Fig. 2a). DNGR-1 and the lysosomal marker LAMP-2 marked two mutually exclusive phagosome populations (Fig. 2a), which were found to co-exist in individual cells by microscopy (Fig. 2b). Further, DNGR-1+ phagosomes co-stained for MHC class I and II and expressed MHC I at higher levels than LAMP-2+ phagosomes (Fig. 2c, d). The two phagosome populations showed differential capacity to degrade antigen, as DNGR-1+ phagosomes displayed higher anti-OVA staining than LAMP-2+ phagosomes (Fig. 2e-g). After long chase periods, some DNGR-1+ MHC I+ OVAhi phagosomes lost DNGR-1 staining, acquired LAMP and degraded OVA, indicating that they were not fully arrested in maturation (Extended Data Fig. 2a-e). DNGR-1+ MHC I+ OVAhi phagosomes were not an aberrant compartment of MuTuDCs as they were also found in primary cDC1s obtained from bone marrow Flt3L cultures (Fig. 2h) or KID cells, a distinct cDC1 cell line30 (Extended Data Fig. 2f). Furthermore, ectopic expression of DNGR-1 in the macrophage cell line RAW264.7 also allowed for identification of DNGR-1+ OVAhi phagosomes distinct from LAMP-2+ OVAlow phagosomes (Extended Data Fig. 2g).
Figure 2. DNGR-1 marks an early MHC I+ non-degradative phagosome population.
a, WT MuTuDCs were pulsed with FM-OVA beads and incubated for 2 hrs. Phagosomes were isolated and stained with anti-DNGR-1 and anti-LAMP-2 antibodies and analysed by flow cytometry. Frequency of DNGR-1+, LAMP-2+ and DNGR-1+LAMP-2+ phagosomes is plotted as mean (± s.d.) from eight independent experiments (n = 8). P values were calculated by one-way ANOVA. b, WT MuTuDCs were pulsed with FM-OVA beads (5:1 beads to DCs, 15 mins). DNGR-1 (red) and LAMP-2 (green) cells were imaged by confocal microscopy (bar scales 5 μm, nucleus stained with DAPI). The arrow marks a DNGR-1+ phagosome and the asterisk marks a LAMP-1+ phagosome. Individual cells are outlined with dotted lines. Images are representative of two independent experiments (n = 2). c-e, WT MuTuDCs were pulsed with FM-OVA beads and incubated for 2 hrs. Phagosomes were isolated and stained with anti-DNGR-1, anti-LAMP-2 and anti-MHC I or anti-MHC II antibodies or anti-OVA and analysed by flow cytometry. Frequency of phagosomes with the indicated phenotype is plotted as mean ± SD from 6 (c, d) or 7 (e) independent experiments. P values were calculated by two-tailed t-test, Welch’s test. f, g, WT MuTuDCs were pulsed with FM-OVA beads (5 : 1 beads to DCs, 15 mins). (f) DNGR-1 (red) and LAMP-2 (green) or (g) LAMP-1 (red) and anti-OVA (green) cells were imaged by confocal microscopy (bar scales 5 μm, nucleus stained with DAPI). The arrows mark DNGR-1+OVAhi phagosomes (f) or LAMP-1+OVAlow phagosomes (g) Images are representative of 2 independent experiments. h, cDC1s purified from Flt3L bone marrow cultures were pulsed with FM-OVA beads and incubated for 4 hrs. Phagosomes were isolated and stained with anti-DNGR-1, anti-LAMP-2, anti-OVA and anti-MHC I antibodies and analysed by flow cytometry. i, j, WT and DNGR-1-deficient (KO) MuTuDCs were pulsed with FM-OVA beads and incubated for 2 hrs. Phagosomes were isolated and stained with anti-LAMP-1 or anti-OVA and anti-MHC I antibodies and analysed by flow cytometry. Frequency of MHC I+LAMP-2– phagosomes or MHC I+OVAhi phagosomes is plotted from seven independent experiments. P values were calculated by an unpaired nonparametric test, Kolmogorov-Smirnov test. k, l, WT MuTuDCs were pulsed with FM-OVA or OVA beads and incubated for 2 hrs. Phagosomes were isolated and stained with anti-DNGR-1 and anti-OVA or anti MHC I antibodies and analysed by flow cytometry. Frequency of DNGR-1+ OVAhi and DNGR-1+OVAlow or DNGR-1+ MHC I+ phagosomes is plotted as mean (± s.d.) from seven independent experiments (n = 7). P values were calculated by an unpaired nonparametric test, Kolmogorov-Smirnov test. n.s., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.00001.
To ask if DNGR-1 was required for the formation of MHC I+ OVAhi phagosomal compartments, we compared FM-OVA bead phagosomes from WT and KO MuTuDCs. MHC I+ LAMP-2- and MHC I+ OVAhi phagosomes were identified at similar frequency in both cells (Fig. 2i-j) and matured at a similar rate (Extended Data Fig.2a-e). We further compared DNGR-1+ phagosomes from WT MuTuDCs that had been fed OVA beads vs. FM-OVA beads and found no differences with respect to OVA degradation (Fig. 2k) or MHC I recruitment (Fig. 2l). Finally, H-2Kb-expressing (see below) RAW264.7 macrophages fed particles that engage Fcγ phagocytic receptors also displayed MHC I+ phagosomes (Extended Data Fig. 2h). Taken together, these data suggest that DNGR-1 marks an MHC I+ phagosomal compartment in cDC1 (and, when ectopically expressed, in RAW264.7 macrophages) that has low degradative potential and the ability to preserve antigen, at least temporarily. However, neither the presence of DNGR-1 nor of its ligand is required for phagocytic cargo to access this compartment.
DNGR-1 ligand engagement induces phagosomal membrane damage
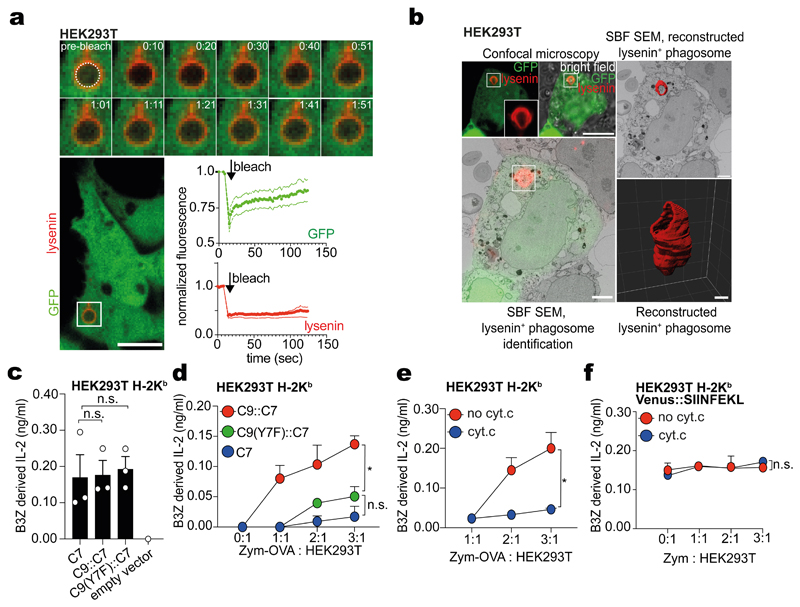
We therefore searched for subsequent steps in cargo handling that might be modulated by DNGR-1 engagement and explain induction of XP via the cytosolic pathway. To assess if DNGR-1+ phagosomes displayed signs of membrane damage, we measured recruitment of cytosolic galectin-3 or 8, which bind to sugar moieties attached to membrane proteins on the luminal side of phagosomes31. By PhagoFACS, we found that galectin-8 recruitment was higher for DNGR-1+ phagosomes than LAMP-2+ phagosomes in MuTuDCs (Extended Data Fig. 3a). By confocal microscopy, an mCherry::galectin-3 fusion protein could be found decorating phagosomes when MuTuDCs were fed FM-OVA beads, but much less frequently with OVA beads that do not bind DNGR-1 (Fig. 3a-b and Extended Data Fig. 3b). To test the role of the DNGR-1 hemITAM, we expressed mCherry::galectin-3 in DNGR-1-deficient MuTuDCs reconstituted with either WT (KO-WT) or hemITAM signalling-incompetent DNGR-1 (tyrosine to phenylalanine mutation at position 7: KO-Y7F). Upon feeding FM-OVA beads, mCherry::galectin-3 was recruited to phagosomes in cells expressing WT but not Y7F receptor (Fig. 3b). We also tested a different probe containing mCherry fused to a version of lysenin that binds sphingomyelin exposed to the cytosol upon endosomal damage32. Consistent with the galectin results, the mCherry::lysenin probe accumulated specifically on FM-OVA bead phagosomes in MuTuDCs expressing WT DNGR-1 but not the Y7F mutant (Fig. 3c). These data suggest that ligand-dependent DNGR-1 signalling via its hemITAM induces phagosomal membrane damage in MuTuDCs.
Figure 3. DNGR-1 signalling causes phagosomal damage resulting in rupture.
a-b, WT MuTuDCs (a and b left panel) or DNGR-1-deficient MuTuDCs complemented with either WT or mutant (Y7F) DNGR-1 (b, right panel) expressing mCherry::galectin-3 were incubated with either OVA or FM-OVA beads (b, left panel) or FM-OVA beads (b, right panel) (5:1 ratio beads to DCs) for 4 hrs. Uninternalised beads (“out. bead”) marked by anti-OVA staining and cells were imaged by confocal microscopy (scale bar = 5 μm). Galectin-3 phagosome+ cells were counted and are plotted as a ratio (index) of total bead+ cells. Each dot reflects one representative image out of 18 (FM-OVA), 36 (OVA), 25 (KO-WT) and 40 (KO-Y7F) images. P values were calculated by an unpaired, nonparametric Kolmogorov-Smirnov test. c, mCherry::lysenin-expressing DNGR-1-deficient MuTuDCs complemented with either WT (KO-WT, represented in image) or mutant (KO-Y7F) DNGR-1 were incubated with FM-OVA beads (5 : 1 ratio beads to DCs) for 4 hrs. Cells were imaged by confocal microscopy (scale bar = 5 μm). Lysenin phagosome+ cells were counted and plotted as a ratio (index) of total bead+ cells. Each dot reflects one image of 15 images (n = 15). P values were calculated by an unpaired, nonparametric Kolmogorov-Smirnov test. d, Left panel: Schematic of C7::C9 chimera receptors. HEK293T cells were transiently transfected with a plasmid encoding mCherry::lysenin and plasmids encoding cytosolic GFP with either C9::C7, C9(Y7F)::C7 or C7. Cells were then challenged with biotinylated zymosan A (ratio of zymosan particles to cell 10 : 1). After 4 hrs, biotinylated zymosan A particles were labelled with fluorescent streptavidin and the preparations were imaged by confocal microscopy (scale bar = 10 μm). A representative image of a cell expressing C9::C7 is shown. Image is representative of 3 independent experiments. e, The number of lysenin+ phagosomes per cell from panel d is plotted and is representative of three independent experiments (n = 3). Each dot represents a single image out of 10 images per condition (n = 10). P values we determined using an unpaired t-test. f, Phagosomal GFP influx was quantified by measuring the mean fluorescence intensity (MFI) in the lumen and expressed as a ratio to cytosolic GFP MFI. Each dot represents the ratio value from a single phagosome. Data are from 25 (lysenin+) and 60 (lysenin-) phagosomes per category and representative of three independent experiments (n = 3). P values were determined using an unpaired t-test. g, Confocal microscope images of HEK293T cells expressing either C9::C7 or C9(Y7F)::C7 receptors, treated with zymosan for 4 hrs. Red insets were magnified to show individual phagosomes (outlined by white circles). White arrow heads indicate individual phagosomes (scale bar = 10 μm). Image is representative of 3 independent experiments. h, The phagosome:cytosol GFP MFI ratios were binned and plotted against the number of phagosomes per bin to generate a histogram. A curve was fitted to the histogram to visualise to relative distribution of the phagosome:cytosol GFP MFI ratios. Quantification is from 3 independent experiments with n = 50 cells per condition. i, HEK293T cells expressing the indicated receptors were loaded with the FRET probe CCF-4 for 30 mins. Excess CCF-4 was washed away and the cells were then pulsed with zymosan pre-soaked with β-lactamase. The number of cells losing FRET was quantified after 1 hr by live cell confocal microscopy (scale bar = 5 μm). Data is one representative example of three independent experiments (n = 82 cells for C9(Y7F)::C7 and n = 114 cells for C9::C7). Each dot represents the average fluorescence emission at 460 nm from a single cell. P values were determined using an unpaired t-test. j, Confocal microscope images of HEK293T cells expressing either C7, C9::C7 or C9(Y7F)::C7 receptors, treated with zymosan or zymosan-cyt. c for 24 hrs. The total number of cells in each condition was counted using FIJI software and represented as a ratio of cells treated with zymosan-cyt. c to cells treated with zymosan alone. Top: representative images (scale bar = 5 μm, n = 3 experiments); bottom: each dot represents a single field of view out of 25 total from one representative experiment. Data (b, i) represented as mean (± s.d). Data (c, e, f, j) represented as mean (± s.e.m). P values were determined using an unpaired t-test. *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.00001.
Signalling by the DNGR-1 hemITAM is sufficient to induce phagosomal rupture in heterologous cells
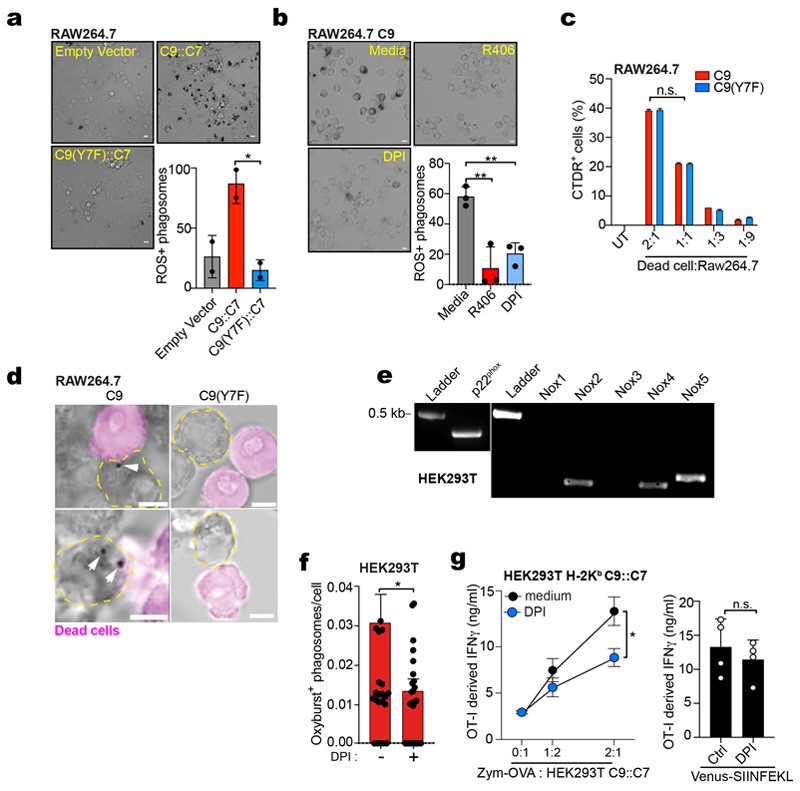
To distinguish effects of receptor signalling from phagosomal properties of cDC1, we asked if DNGR-1 hemITAM signalling could mediate phagosomal damage in a heterologous cell and receptor type. We chose the easily transfectable non-APC cell line HEK293T and ectopically expressed Dectin-1 (aka CLEC7A; a receptor structurally homologous to DNGR-1) or receptor chimeras comprising the extracellular domain and transmembrane region of Dectin-1 fused to variants of the cytoplasmic tail of DNGR-1 (Fig. 3d and Extended Data Fig. 3c). Dectin-1 binds to yeast β-glucans, allowing us to analyse uptake of zymosan (i.e., yeast cell walls) instead of latex beads. Dectin-1 (C7), Dectin-1 fused to WT (C9::C7) or hemITAM tyrosine-mutated cytoplasmic tail of DNGR-1 (C9(Y7F)::C7) conferred upon HEK293T the ability to phagocytose zymosan (Extended Data Fig. 3d). Notably, when we co-expressed the mCherry::lysenin probe, we observed a higher proportion of lysenin+ phagosomes in HEK293T cells expressing the C9::C7 chimera compared with cells expressing WT C7 or the signalling incompetent chimera C9(Y7F)::C7 (Fig. 3d-e).
The constructs used to transfect HEK293T additionally encode GFP, which accumulates in the cytosol of transfected cells. Unlike latex beads, zymosan particles are porous, acting as a sponge for any probe that accesses the phagosomal lumen. We noticed that phagosomes in HEK293T cells expressing the C9::C7 chimera became positive for GFP (Fig. 3d-h). Intra-phagosomal GFP was largely absent from zymosan-containing phagosomes in HEK293T expressing C7 or C9(Y7F)::C7, indicating a specific requirement for DNGR-1 hemITAM signalling (Fig. 3d-h). Lysenin+ phagosomes showed a higher mean fluorescent intensity (MFI) for GFP compared to lysenin- phagosomes, suggesting that access of cytosolic GFP to phagosomes was coupled to membrane damage and permeability (Fig. 3d and f).
To ask whether permeability is bi-directional, we pulsed HEK293T expressing either C9::C7 or C9(Y7F)::C7 with zymosan soaked in sulforhodamine B (SRB). We detected a significant increase in SRB fluorescence in the cytosol of HEK293T expressing C9::C7 but not C9(Y7F)::C7 (Extended Data Fig. 3e). Using a previously-reported P2C assay33, we found that zymosan-adsorbed β-lactamase was released into the cytosol of HEK293T cells expressing C9::C7 to a greater extent than cells expressing C9(Y7F)::C7 (Fig. 3i). cDC1s can be selectively depleted in vivo by their ability to translocate internalised cytochrome c into the cytoplasm, triggering apoptosis34. We soaked zymosan in cytochrome c (zymosan-cyt. c) and added it to C7, C9::C7 and C9(Y7F)::C7 expressing HEK293T cells (Fig. 3j). Cells expressing C7 or C9(Y7F)::C7 internalised the particles and survived (Fig. 3j). In contrast, nearly all C9::C7-expressing cells died within a 24h period (Fig. 3j). Together, these data indicate that hemITAM signalling by DNGR-1 permeabilises phagosomes so as to allow efflux of luminal contents into the cytosol, a property that is intrinsic to the tail of the receptor and can function in heterologous cells.
To examine the nature of DNGR-1 hemITAM-induced phagosomal permeability, we first tested whether it was permanent. When we photobleached GFP within the lumen of lysenin+ phagosomes, signal was recovered within 2 minutes, indicating continuous GFP influx and irreversible phagosomal membrane damage. As a control, bleaching of the lysenin signal did not lead to fluorescence recovery (Fig. 4a). We then analysed the ultrastructure of GFP+lysenin+ zymosan phagosomes in C9::C7-expressing cells by 3D correlative light and electron microscopy (3D CLEM) using serial block face scanning electron microscopy. This revealed that the phagosomal membrane contained a large hole with a diameter of roughly 1 - 1.5 μm (Fig. 4b and Supplementary Video 1). Thus, DNGR-1 signalling can cause large scale rupture of phagosomes, allowing for even sizeable luminal contents to be released into the cytosol.
Figure 4. Phagosomal rupture results in P2C transfer and XP.
a, HEK293T cells expressing C9::C7, mCherry::lysenin and cytosolic GFP were pulsed with unlabelled zymosan A for 4 hrs (scale bar = 10 μm). Lysenin+ phagosomes were photobleached and the fluorescence recovery was monitored for both GFP and mCherry::lysenin for the indicated time by confocal microscopy. Data is representative of three independent experiments (n = 3). b, 3D CLEM of HEK293T cells expressing C9::C7 and GFP and mCherry::lysenin were challenged with unlabelled zymosan A for 4 hrs before confocal microscope imaging. Overlay of the confocal stack onto the serial block face scanning electron microscopy (SBF SEM) stack confirms the location of the lysenin+ phagosome of interest. Segmentation of the SBF SEM data and 3D reconstruction of the phagosomal membrane (red) shows the 3D structure of the phagosomal membrane. (Scale bar for confocal image = 10 μm, scale bar for CLEM = 2 μm, scale bar for 3D reconstruction = 0.5 μm. Image is representative of two independent experiments (n = 2). c-f, B3Z hybridoma cells were cultured with fixed c, HEK293T C7, C9::C7 or C9(Y7F)::C7 cells transfected with VENUS::SIINFEKL; d, HEK293T C7, C9::C7 or C9(Y7F)::C7 cells previously fed zymosan-OVA for 4 hrs; e, HEK293T C9::C7 cells fed with either zymosan-OVA or zymosan-OVA-cytochrome-c for 4 hrs before fixation with PFA (1 %); f, B3Z hybridoma cells were cultured with HEK293T C9::C7 cells transfected with VENUS::SIINFEKL fed with either zymosan or zymosan-cyt. c for 4 hrs before fixation with PFA (1 %). (c-f) IL-2 was assessed by ELISA, plotted as mean (± s.d) of experimental triplicates and representative of three independent experiments (n = 3). P values in (c) were determined using an unpaired t-test. P values in (d, e, and f) were determined by two-way ANOVA. n.s., not significant; *P ≤ 0.05.
Phagosomal rupture is necessary for DNGR-1 signalling-mediated XP
To link phagosomal rupture to XP, we generated HEK293T lines stably expressing murine H-2Kb and β2-microglobulin, which were further transfected to express C7, C9::C7 or C9(Y7F)::C7 chimeras and selected for equal H-2Kb and Dectin-1 extracellular domain expression levels (Extended Data Fig. 4a, b). When pulsed with OVA peptide (SIINFEKL), all cell lines showed equivalent capacity to elicit IL-2 production by B3Z (Extended Data Fig. 4c), an OVA-H-2Kb-specific T cell hybridoma that can be activated even by fixed APCs (used below). All cell lines were also equally competent at presenting endogenous Venus::SIINFEKL, a fusion protein mimic of OVA peptide35 (Fig. 4c). We then exposed the cell lines to zymosan pre-soaked in hen egg white (zymosan-OVA) and chemically fixed them before adding B3Z cells. Efficient XP, as measured by B3Z activation, was only observed with cells expressing the C9::C7 chimera (Fig. 4d) even though they internalised less zymosan than control C7 HEK293T cells (Extended Data Fig. 4d). We also fed HEK293T cells expressing C9::C7 with zymosan soaked in both hen egg white and cytochrome c, titrating the time of exposure so as to kill only a fraction of the cells. If phagosomal rupture and XP are linked, the killed fraction should contain all the cells able to stimulate B3Z. Indeed, as predicted, this treatment led to loss of XP activity when compared to feeding cells with zymosan particles soaked with OVA alone (Fig. 4e). Cytotoxicity of cytochrome c leaching into the culture was excluded by the fact that no decrease in B3Z activation was observed when zymosan-cyt. c particles were incubated with HEK293T cells expressing the C9::C7 chimera and Venus::SIINFEKL (Fig 4f). Finally, we generated RAW264.7 cells expressing H-2Kb and C9::C7 or C9(Y7F)::C7 chimeras. When fed zymosan-OVA, C9::C7- but not C9(Y7F)::C7-expressing RAW264.7-H-2Kb cells could stimulate OT-I cells (Extended Data Fig. 4e). As a control, XP activity in RAW264.7 cells expressing the C9(Y7F)::C7 chimera, could be restored by enforcing phagosomal rupture using zymosan co-soaked with OVA and the endosome-disrupting chemical L-leucyl-L-leucine methyl ester (LLOMe) (Extended Data Fig. 4e, f). Overall, these data indicate that phagosomal rupture induced by DNGR-1 hemITAM signalling promotes XP even in non-cDC1 cells.
DNGR-1 signalling-dependent phagosomal rupture requires SYK
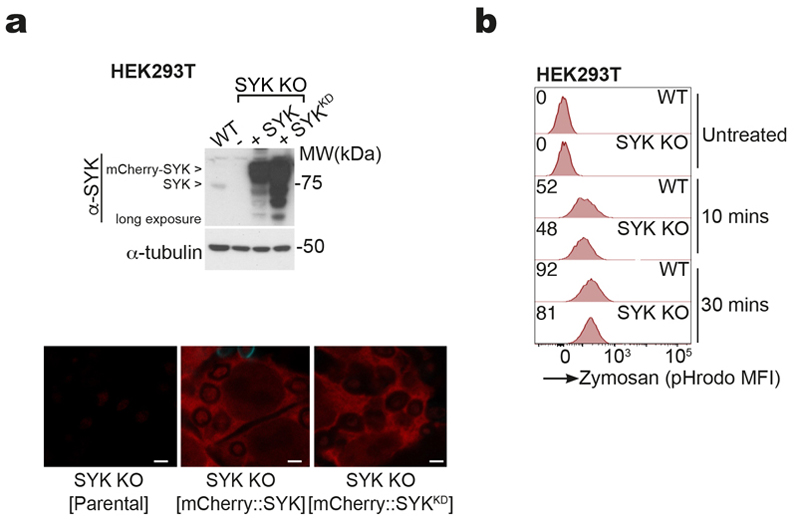
SYK phosphorylation at two distinct tyrosine residues was observed in MuTuDCs treated with anti-DNGR-1 crosslinking antibody, F-actin-myosin II complexes (DNGR-1 ligand; DNGR-1L) or the Dectin-1 agonist curdlan (Fig. 5a). C7 or C9::C7 but not C7(Y15F) (a hemITAM-mutated version of Dectin-1) or C9(Y7F)::C7 chimeras also induced phosphorylation of SYK in HEK293T in response to zymosan, which could be observed at the level of phagosomes (Fig. 5b). To determine whether this was upstream of phagosomal rupture, we generated SYK-deficient C9::C7-expressing HEK293T (SYK KO) (Extended Data Fig. 5a). SYK loss did not affect uptake of zymosan but completely blocked the influx of cytoplasmic GFP into phagosomes, which could then be restored by complementation with WT SYK (SYK WT) but not a catalytically-deficient mutant (K396R - kinase dead; SYK KD) (Fig. 5c and Extended Data Fig. 5b). Similarly, phagosomal GFP influx (Fig. 5d) was prevented when C9::C7-expressing HEK293T cells were treated with SYK inhibitor (R406 or SYK inhibitor IV).
Figure 5. SYK signalling promotes phagosomal rupture and XP.
a, Western blot images of phospho-SYK in lysates of MuTuDC incubated with isotype-matched irrelevant specificity control mAb (MAC49 clone, 0.5 μg), anti-DNGR-1 (7H11 clone, 0.5 μg), DNGR-1 ligand (DNGR-1-L, 100 nM) or Dectin-1 ligand (Curdlan 200 μg/ml) over the indicated timecourse. Data is representative of three independent experiments (n = 3). b, Confocal microscope images of phospho-SYK staining on phagosomes in HEK293T C9::C7 and C9(Y7F)::C7 cells expressing mCherry::SYK and incubated with zymosan for 20 and 30 min, respectively. White arrows indicate phagosomes (scale bars 5 μm). Quantification of fold enrichment of phospho-SYK staining on phagosomes (n > 50 phagosomes) in HEK293T C7, C7(Y15F), C9::C7 and C9(Y7F)::C7 cells. Data represented as mean (± s.d). P values were calculated by one-way ANOVA. Data are representative of two independent experiments (n = 2). c, Confocal microscope images of HEK293T C9::C7 SYK KO cells complemented with either an empty vector, or a plasmid encoding mCherry::SYK or kinase-dead (KD) mCherry::SYK K396R and incubated with zymosan for 5 hrs. Lower images are higher magnifications of areas of the upper images (insets indicated by white boxes). White arrows point to some of the phagosomes (scale bars 5 μm). Images are representative of two independent experiments (n = 2). d, HEK293T cells expressing C9::C7 and cytosolic GFP were preincubated in the presence or absence with R406 (1 μM) for 30 mins. The transfected cells were then pulsed with biotinylated zymosan A at a ratio of 10 : 1 particles per cell for 5 hrs. Images show GFP fluorescence (scale bar = 5 μm). The phagosome:cytosol GFP MFI ratios were binned and plotted against the number of phagosomes per bin to generate a histogram. A curve was fitted to the histogram to visualize to relative distribution of the phagosome:cytosol GFP MFI ratios. Data are from a single experiment (n = 50 phagosomes) representative of three independent experiments (n = 3). e, Confocal microscope images of mCherry::lysenin-positive phagosomes from MuTuDC incubated with anti-DNGR-1-coated 2 μm beads for 4 hrs in the presence or absence of SYK inhibitor IV (1 μM) (scale bar, 10 μm). Lysenin-positive phagosomes were quantified across 5 fields of view. Data represented as mean (± s.e.m). P values were calculated by unpaired two-tailed Mann-Whitney test and are representative of two independent experiments (n = 2). f, MuTuDC were pulsed with (left panel) FM-OVA beads or (right panel) SIINFEKL peptide (1 pM) for 30 mins, then in the presence or absence of SYK inhibitor IV (0.5 μM) or R406 (1 μM) for 4 hrs before adding of OT-I Rag1 –/– T-cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental triplicates and representative of three independent experiments (n = 3). P values were calculated by two-way ANOVA. n.s., not significant; *P ≤ 0.05; ****P ≤ 0.00001.
To confirm these findings in cDC1 expressing endogenous DNGR-1, we fed MuTuDCs with anti-DNGR-1-coated beads in the presence or absence of SYK inhibitor. SYK inhibition abrogated staining of phagosomes with the lysenin probe (Fig. 5e) and blocked XP of FM-OVA beads but not presentation of SIINFEKL peptide to OT-I (Fig. 5f). Together, these data suggest that SYK activation and kinase activity induced by DNGR-1 signalling are required for the induction of phagosomal rupture and XP.
DNGR-1 signalling-dependent phagosomal rupture involves NADPH oxidase
SYK can activate NADPH oxidase, leading to production of ROS that damage endosomal membranes11,12. NADPH oxidase is often studied in macrophages and in RAW264.7 macrophage-like cells ectopically expressing C9::C7, exposure to zymosan led to a potent oxidative burst at the level of individual phagosomes that was diminished in cells treated with the NADPH oxidase inhibitor, DPI, or expressing C7 or C9(Y7F)::C7 (Fig. 6a and Extended Data Fig. 6a). Similarly, in RAW264.7 cells expressing DNGR-1 (C9) and fed fixed and permeabilised sheep red blood cells (a phagocytic target bearing exposed F-actin recognised by DNGR-136), we observed an oxidative burst around phagosomes that was diminished by SYK inhibition or DPI treatment (Extended Data Fig. 6b). Further, RAW264.7 cells expressing C9 or C9(Y7F) took up dead cell debris equivalently but an oxidative burst around those debris was seen only with C9 (Extended Data Fig. 6c, d). Consistent with a role for ROS in membrane damage, inhibition of the NADPH oxidase by DPI blocked lysenin accumulation on phagosomes in RAW264.7 cells expressing C9::C7 to the same extent as the Y7F mutation (Fig. 6b). We then used siRNA targeting NOX2 (CYBB), the predominant catalytic subunit of the NADPH oxidase in myeloid cells, and observed decreased XP of zymosan-OVA by RAW264.7 expressing C9::C7 and H-2Kb but no effect on presentation of SIINFEKL peptide (Fig. 6c).
Figure 6. Phagosomal ROS promotes damage and rupture of phagosomes.
a, Confocal microscope images of ROS-producing phagosomes from RAW264.7 C7, C9::C7, or C9(Y7F)::C7 cells were challenged with zymosan-Oxyburst in the presence or absence of DPI (10 μM) for 30 mins (scale bar, 10 μm). Oxyburst+ positive phagosomes were quantified across 5 fields of view. b, Confocal images of RAW264.7 C9::C7 C9(Y7F)::C7 or C7 cells and mCherry::Lysenin and challenged with zymosan in the presence or absence of DPI (10 μM) for 30 mins (scale bar, 10 μm). Lysenin positive phagosomes quantified across 5 fields of view. Data represented as mean (± s.e.m). P values calculated using an unpaired t test. c, Left: Western blot images of RAW264.7 C9::C7 cells 48 hrs after transfection with non-sense (NS) or NOX2 siRNA. Right: siRNA treated RAW264.7 C9::C7 were pulsed with zymosan-OVA for 4 hrs before adding of OT-I Rag1 –/– T-cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental triplicates and representative of two independent experiments (n = 2). d, MuTuDC were pulsed with either FM-OVA beads or UV-irradiated bm1 T OVA MEFs for 30 mins, then in the presence or absence of DPI (10 μM) or α-tocopherol (5 μM) for 4 hrs before adding of OT-I Rag1 –/– T-cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of experimental duplicates and representative of two independent experiments (n = 2). P values were calculated by two-way ANOVA. n.s., not significant; **P ≤ 0.01; ****P ≤ 0.00001.
To extend these observations to the other cell lines used in our study, we first examined whether DNGR-1 signalling also induced ROS production in HEK293T cells, which express NADPH oxidase (Extended Data Fig. 6e)37. Notably, a phagosomal oxidative burst was also observed in HEK293T cells expressing C9::C7 upon zymosan uptake (Extended Data Fig. 6f). DPI treatment decreased XP of zymosan-OVA by HEK293T cells expressing C9::C7 but did not diminish presentation of endogenous Venus-SIINFEKL antigen (Extended Data Fig. 6g). Finally, we assessed the role of ROS in DNGR-1-dependent XP by MuTuDCs. DPI, as well as the ROS scavenger alpha-tocopherol, greatly decreased XP of both FM-OVA beads and OVA-bearing dead cells without impacting presentation of SIINFEKL peptide (Fig. 6d). Thus, efficient DNGR-1-dependent XP across three distinct cell lines requires NADPH oxidase activity.
Phagosomal damage and cross-presentation of dead cell-associated antigens by cDC1 is decreased in absence of NOX2
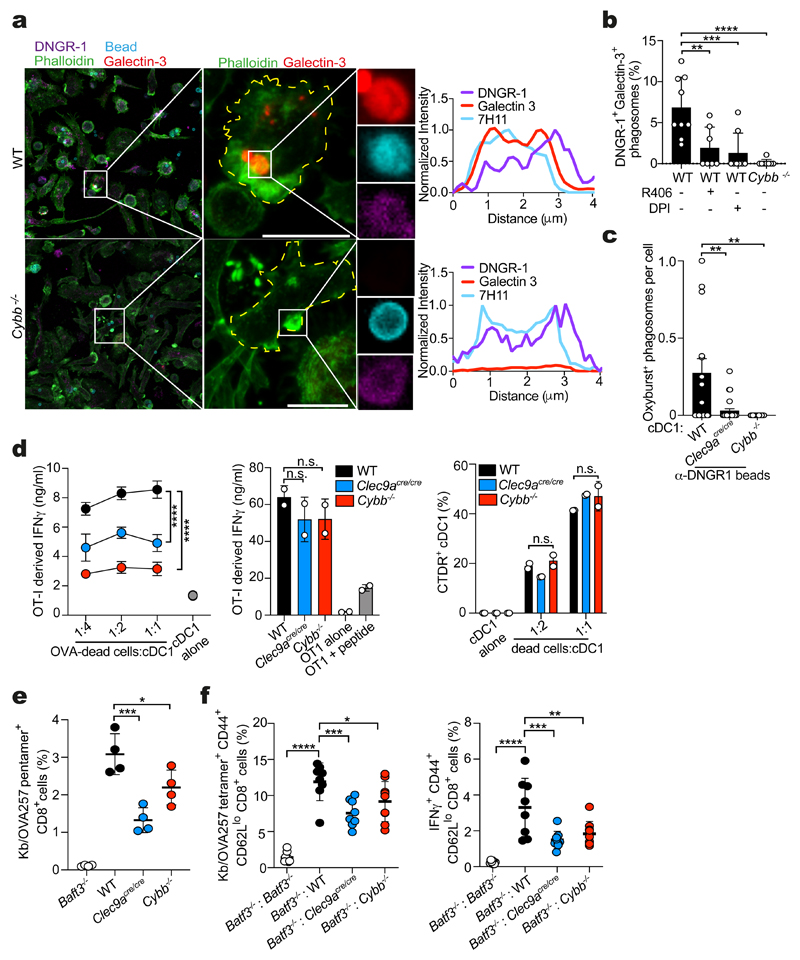
To extend these data to primary cDC1s, we established Flt3L cultures from bone marrow and exposed Flt3L-cDCs to anti-DNGR-1-coated beads. In DNGR-1+ cDC1, we observed numerous bead-containing phagosomes that were decorated with the receptor (Fig. 7a). Notably, in cells grown from WT but not NOX2-deficient (Cybb −/−) bone marrow, many of these phagosomes were galectin-3+, indicative of damage (Fig 7a, b). Similarly, the frequency of galectin-3+ phagosomes was decreased in WT cDC1 given DPI or R406 at the time of stimulation (Fig. 7b). Further, purified Cybb −/− Flt3L-cDC1 were defective in phagosomal ROS in response to DNGR-1-stimulating beads (Fig. 7c) and both Clec9a cre/cre and Cybb −/− cDC1s displayed a reduction in XP of OVA-bearing dead cells relative to WT cDC1s despite being equally effective at presenting SIINFEKL peptide and internalising dead cell material (Fig. 7d).
Figure 7. NOX2 in cDC1 is necessary for cross-presentation of dead-cell associated antigens.
a, Confocal microscope images of Flt3L bone marrow cultures from wild-type (WT) and Cybb -/– mice challenged with anti-DNGR-1 beads and stained for DNGR-1 (magenta), galectin-3 (red) and phalloidin (cyan) (scale bar, 10 μm). Fluorescence intensity profiles from a single line drawn through the phagosomes shown are plotted to the right. Image is representative of 10 similar fields of view. b, DNGR-1+ galectin-3+ phagosomes were quantified from wild-type (WT) and Cybb -/- Flt3L cultures challenged as in (a) and treated with R406 (1 μM) or DPI (10 μM) where indicated. DNGR-1+ galectin-3+ phagosomes were quantified across 10 fields of view. Data represented as mean (± s.e.m). c, ROS-producing phagosomes from wild-type (WT), Clec9a cre/cre or Cybb -/- cDC1s purified from Flt3L bone marrow cultures and challenged with anti-DNGR-1-Oxyburst beads (scale bar, 10 μm). Oxyburst+ positive phagosomes were quantified across 5 fields of view. Data represented as mean (± s.e.m). P values were calculated by two-tailed Welch’s t test. d, Wild-type (WT), Clec9a cre/cre or Cybb -/- cDC1s purified from Flt3L bone marrow cultures were incubated with either UVC-irradiated bm1 T OVA MEFs (left panel) or SIINFEKL peptide (middle panel) and incubated with OT-I Rag1 –/– T-cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.). of experimental duplicates and representative of two independent experiments (n = 2). P values were calculated by two-way ANOVA. Percentage of wild-type (WT), Clec9a cre/cre or Cybb -/- XCR1+ cDC1s purified from Flt3L bone marrow cultures that are positive for CTDR-labelled UVC-killed 5555 V600E cell after 2 hrs. Data representative of two independent experiments (n = 2). e, Batf3 -\-, wild-type (WT), Clec9a cre/cre and Cybb −/− were immunised i.v. with 4 × 107 FM-OVA latex beads and 50 μg poly (I:C). Six days later, the frequency of OVA-specific CD8+ T cells in the spleen was determined. Each dot is an individual mouse (Batf3 -/- n = 4, WT n = 4, Clec9a cre/cre n = 4, Cybb -/- n = 4). Data represented as mean (± s.d.). Data is representative of two independent experiments (n = 2). f, Mixed bone marrow chimeras were immunised i.v. with 1 × 106 UVC-killed 5555 BRAFV600E cells pulsed with OVA and poly(I:C). Six days later, the frequency of OVA-specific CD8+ T cells in the spleen was determined. Right panel: Intracellular IFN-γ production in response to SIINFEKL peptide (1 nM) restimulation ex vivo. Each dot is an individual mouse (Batf3 -/- : Batf3 -/- n =7, Batf3 -/- : WT n =8, Batf3 -/- : Clec9a cre/cre n = 8, Batf3 -/- : Cybb -/- n = 9). Data are pooled from two independent experiments (n = 2). Data represented as mean (± s.d.). P values in (e, f) were calculated by one-way ANOVA, Sidak’s multiple comparison test. n.s., not significant; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.00001.
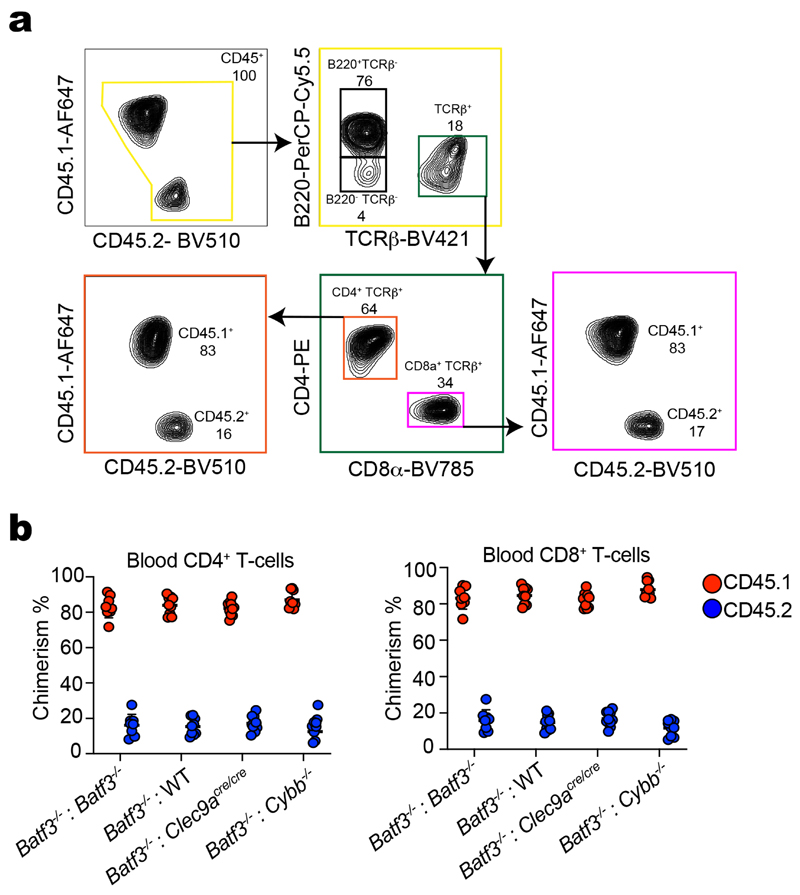
To assess the importance of NOX2 for DNGR-1-dependent XP in vivo, we first immunised WT, Clec9a cre/cre, Batf3 −/− or Cybb −/− mice with FM-OVA beads + poly I:C and measured OVA-specific CD8+ T cells responses by H-2Kb-OVA-pentamer staining (Fig. 7e). As reported22, WT mice mounted a robust response to FM-OVA beads + poly I:C that was significantly decreased in both Clec9a cre/cre and Batf3 −/− mice. Cybb −/− mice displayed a reduction in OVA-specific CD8+ T cell cross-priming comparable to that in Clec9a cre/cre animals (Fig. 7e). To confirm that this reflected an essential function for NOX2 in cDC1s and to extend to cross-priming to dead cell-associated antigens, we generated radiation chimeras using bone marrow from Batf3 −/− CD45.1 mice mixed at a ratio of 80:20 with bone marrow from either Batf3 −/−, WT, Clec9a cre/cre or Cybb −/− CD45.2 mice (Extended Data Fig. 7a, b). We used Batf3 −/− CD45.1 mice as recipients, further ensuring that the only cDC1 that develop after reconstitution arise from the CD45.2 donor bone marrow, and we analysed exclusively the response of CD45.1 T cells to exclude any cell-intrinsic effects of NOX2 deletion in lymphocytes. Following immunisation with OVA+poly I:C-pulsed dead cells, Batf3 −/−:Batf3 −/− chimeras failed to generate OVA-specific CD8+ T cells, as expected15 (Fig. 7f). In contrast, robust cross-priming to OVA was seen in Batf3 −/−:WT chimeras as measured by either H-2Kb-OVA-pentamer staining or by intracellular staining for IFN-γ upon ex vivo CD8+ T cell restimulation with SIINFEKL peptide (Fig. 7f). Consistent with previous observations in DNGR-1-deficient mice3,23–25, the OVA-specific CD8+ T cell response was significantly diminished but not abrogated in Batf3 −/−:Clec9a cre/cre chimeras (Fig. 7f). Notably, it was diminished to a similar extent in Batf3 −/−:Cybb −/− chimeras, indicating that cross-priming of CD8+ T cells in vivo to dead cell-associated antigen requires cDC1 expressing a NOX2-containing NADPH oxidase (Fig. 7f). Together with the fact that CD11cΔSyk mice display a defect in cross-priming to dead cell-associated antigens3, these data are consistent with the notion that DNGR-1-SYK-dependent activation of the NADPH oxidase and subsequent ROS production promotes phagosomal rupture and P2C in cDC1s in vivo.
Discussion
Macrophages, monocyte-derived dendritic cells and other myeloid cell types, as well as non-immune cells, have been used extensively to dissect some of the mechanisms involved in XP7–9. Together, they have led to the view that XP often involves the cytosolic pathway, as also noted in vivo 38, but have generally fallen short of explaining how P2C occurs, especially for complex substrates such as dead cells. cDC1s are non-redundant APCs in many instances of cross-priming in vivo and we focused on the possibility that they express receptors to promote XP of relevant antigenic substrates such as dead cells. We show that ligand-dependent DNGR-1 signalling at the level of phagosomes induces a local NADPH-dependent oxidative burst that destabilises the phagosomal membrane causing rupture and wholesale access of luminal contents to the cytoplasmic compartment where they can enter the endogenous MHC I processing pathway. Notably, the ability of DNGR-1 to signal for phagosomal rupture is intrinsic to its cytoplasmic signalling domain and can be grafted onto other receptors and other cell types, including non-APCs. Thus, machinery for ROS production in endosomes can be subverted by specialised XP receptors to deliberately provoke vacuolar membrane damage and P2C.
Two mechanisms for P2C transfer have been postulated8: the transporter39,40 and the membrane disruption or “indigestion”11,13,14 hypotheses. Our data clearly favour the latter and suggest that “indigestion” can be an active process whereby ingested material triggers cDC1 receptors that cause “ulcers” and permit P2C. It remains unclear whether the cargo released from ulcerating phagosomes is selectively targeted for degradation as in the case of invasive bacteria, where phagosomal damage detection by galectins rapidly recruits autophagy machinery31. Whether autophagy contributes to XP by cDC1s remains to be established, as does the fate of the phagosomal remnants that have disgorged their contents. A variation of the P2C model posits that phagosomes fuse with an ER-Golgi intermediate compartment that provides the relevant transporters and peptide-loading machinery. This fusion, in turn, allows proteasome-processed peptides to be imported back into the vacuolar space for local loading onto MHC I molecules (“phagosome-to-cytosol-to-phagosome” (P2C2P) pathway)41–43. Clearly, in DNGR-1-dependent XP, the P2C2P pathway is unlikely to involve the ruptured phagosomes but could still occur in phagosomes that have remained unbroken and have fused with ER-derived membranes.
Our observations do not exclude the possibility that cDC1s possess additional receptors that may engage distinct mechanisms of P2C40. A multitude of compensatory receptors and XP mechanisms is necessary for robustness, presumably explaining why loss of DNGR-13,23–25, Syk3 or NOX2 by cDC1 does not decrease XP as profoundly as ablation of the cells altogether. Our data also do not exclude the notion that cDC1s possess cell biological adaptations that favour XP6,16–18,27,34. Indeed, we identify in these cells slowly maturing phagosomal compartments that can retain undegraded cargo for long periods, known to bolster XP in other cell types44,45. The fact that DNGR-1 preferentially localises to these early phagosomes but does not impact their maturation is consistent with the notion that the main function of the receptor is to survey vacuolar compartments for the presence of exposed F-actin-myosin complexes indicative of putatively antigenic cargo that is relatively intact. Receptor engagement then leads to SYK-dependent localised production of ROS and membrane damage. Rupture of any given phagosome is likely to be a stochastic event partly determined by the extent of damage and, possibly, offset by membrane repair. It is nevertheless sufficiently frequent so as to occur at least once per cell as suggested by our zymosan-cyt. c experiments in which, essentially, all cells die in a period of 24h. Phagosomes that do not rupture can continue to mature and lose DNGR-1, generating the LAMP+ DNGR-1– degradative late phagosome pool that we detect in our assays. The limited nature of the rupture event, and the fact that it is circumscribed to early non-degradative endosomes, may contribute to preventing the toxicity that would be expected from wholesale introduction of proteolytic enzymes into the cytosol46. Notably, other receptors that can signal via SYK with varying efficiency (e.g., integrins) might also plug into the phagosomal damage pathway, which could explain instances of XP with ligands such as latex beads that can engage a Vav-Rac-NADPH oxidase-dependent XP pathway47. NADPH activation may further favour XP by increasing phagosomal pH and thereby preventing the activity of lysosomal proteases16 although this may occur independently of DNGR-1 as engagement of the latter did not noticeably alter cargo degradation.
SYK can activate NADPH via Vav and Rac, all of which have been implicated in XP of particulate antigens by myeloid cells16,47,48. However, it appears that not all SYK-activating receptors engage this pathway and cause phagosomal damage to the same extent. As shown here, Dectin-1 only weakly induces P2C and XP and signals primarily for myeloid cell activation and inflammatory gene expression (which DNGR-1 does not24). How signal divergence downstream of each receptor occurs at the level of SYK will need investigation but it means that DNGR-1 acts primarily to decode the antigenicity of internalised cargo. Therefore, additional signals emanating from dead cells are required to activate cross-presenting cDC1s and render them competent to prime CD8+ T cells in anti-viral or anti-tumor immunity. Activation signals can also impact XP49,50 and understanding how they synergise with signals emanating from XP-promoting receptors such as DNGR-1 will be interesting to explore and may help with strategies to prime CD8+ T cells for cancer immunotherapy and vaccination.
Methods
Materials
| Product | Catalogue Number | Company |
|---|---|---|
| RPMI1640 | 31870-025 | Gibco |
| DMEM | 41966-029 | Gibco |
| IMDM | 12440-053 | Gibco |
| DPBS | 14190-144 | Gibco |
| Live cell Imaging Solution | A14291DJ | Invitrogen |
| Fetal Bovine Serum | A15-102 | PAA Cell Culture Company |
| β-mercaptoethanol | 31350-010 | Gibco |
| Pen Strep solution | 15070-063 | Gibco |
| Puromycin | A11138-03 | Gibco |
| Blasticidin | Ant-bl-1 | Invivogen |
| Hygromycin B Gold | Ant-hg-1 | Invivogen |
| Dimethyl sulfoxide (DMSO) | D2650-5X10ML | Sigma Aldrich |
| Paraformaldehyde (16% solution) | 15710 | Electron Microscopy Sciences |
| Sulforhodamine B | S1402 | Sigma Aldrich |
| In-Fusion HD EcoDry Cloning Kit | 639689 | Takara |
| β-lactamase loading solution | K1085 | Invitrogen |
| β-lactamase | P4524 | Sigma Aldrich |
| CCF4-AM | K1028 | ThermoFisher |
| LiveBLAzer Fret-B/G Loading Kit with CCF4-AM | K1030 | Thermo Fisher |
| Prolong diamond anti-fade mountant | P36961 | Invitrogen |
| EZ-Link Sulfo-NHS-LC-biotin | 21335 | ThermoFisher |
| Streptavidin, Alexa Fluor 488 | S32354 | Invitrogen |
| Streptavidin, Alexa Fluor 647 | S32357 | Invitrogen |
| Streptavidin, Alexa Fluor 594 | S32356 | Invitrogen |
| OxyBURST Green H2DCFDA, SE | D2935 | Invitrogen |
| Fluorescein Isothiocyanate (FITC) | F1906 | Invitrogen |
| Hen eggs | N/A | Marks and Spencer |
| Zymosan, depleted | tlrl-zyd | Invivogen |
| Fugene HD transfection reagent | E2311 | Promega |
| Bovine Cytochrome C | C3131 | Sigma Aldrich |
| DSB-X biotin protein labelling kit | D20655 | Invitrogen |
| Actin, skeletal muscle | AKL99 | Cytoskeleton Inc. |
| Myosin II, skeletal muscle | MY02-A | Cytoskeleton Inc. |
| Biotinylated actin, skeletal muscle | AB07-A | Cytoskeleton Inc. |
| Streptavidin coated beads/microspheres (2μm) | 24160-5 | Polysciences inc. |
| Streptavidin Fluoresbrite YG beads/microspheres (2μm) | 24159-5 | Polysciences Inc. |
| PKH26 red fluorescent cell linker kit | PKH26GL-1KT | Sigma Aldrich |
| CellVue Claret far red fluorescent cell linker kit | MIDCLARET-1KT | Sigma Aldrich |
| LIVE/DEAD fixable far red dead cell stain | L10120 | Invitrogen |
| LIVE/DEAD fixable blue dead cell stain | L34962 | Invitrogen |
| DAPI | D1306 | Invitrogen |
| Cell Tracker Deep Red Dye | C34565 | ThermoFischer Scientific |
| Coverslips (18mm) | 631-0153 | VWR |
| Glass Slides | 7101 | Sailing Boat |
| μ-dish (35mm) | 81156 | Ibidi |
| μ-dish gridded (35mm) | 81168 | Ibidi |
| GlutaMAX supplement | 35050061 | ThermoFisher |
| FIX&PERM cell fixation and permeabilisation kit (CE) | GAS-002 | Nordic MUBio |
| Ammonium chloride | 254134 | Sigma Aldrich |
| R406 | Inh-r406 | InvivoGen |
| Syk inhibitor IV | 574714 | Merck Millipore |
| Alpha-tocopherol | T3251 | Sigma Aldrich |
| DPI | D2926 | Sigma Aldrich |
| Lactacystin | L6785-.2MG | Sigma Aldrich |
| Nitroblue Tetrazolium (NBT) | N5514 | Sigma Aldrich |
| EasySep™ Release Mouse PE positive selection kit | 17656 | STEMCELL |
| Sheep red blood cells 100% washed pooled cells | R406-0050 | Rockland antibodies & assays |
| 2X Laemmli Sample Buffer | 1610737 | Bio-Rad |
| 4-20% Mini-PROTEAN® TGX™ Precast Protein gels, 15-well | 4561096 | Bio-Rad |
| GeneJET RNA purification kit | K0731 | Thermo Fisher |
| Qiagen OneStep RT-PCR kit | 210210 | Qiagen |
| Poly-L-lysine solution | P4707 | Sigma Aldrich |
| Glutaraldehyde solution | G5882 | Sigma Aldrich |
| Acti-stain 488 phalloidin | PHDG1 | Cytoskeleton Inc. |
Addgene plasmids
| Product | Catalogue Number | Company | Deposited by |
|---|---|---|---|
| pMSCV-mCherry-Syk | 50045 | Addgene | Hidde Ploegh |
| lentiCRISPR v2 | 52961 | Addgene | Feng Zhang |
| pSBbi-pur H-2Kb | 111623 | Addgene | Jon Yewdell |
| pISH-β2-microglobulin | 15883 | Addgene | Peter Mombaerts |
| pSBbi-GH | 60514 | Addgene | Eric Kowarz |
| pSBbi-Bla | 60526 | Addgene | Eric Kowarz |
| pCMV(CAT)T7-SB100 | 34879 | Addgene | Zsuzsanna Izsvak |
| pT2/SV-Venus-SIINFEKL | 111624 | Addgene | Jon Yewdell |
Antibodies
Unconjugated primary antibodies
Abbreviations: FC (flow cytometry), IF (immunofluorescence), CI (cell isolation)
| Target | Clone | Stock | Application | Company |
|---|---|---|---|---|
| DNGR-1 | polyclonal sheep | 0.2mg/ml | FC 1:200 IF 1:500 | R&D AF6776 |
| DNGR-1 | monoclonal rat (7H11) | 1.5mg/ml | Bead labeling: 1:100 | Generated in lab |
| Galectin-8 | polyclonal goat | 0.2mg/ml | FC 1:100 | R&D AF1305-SP |
| Galectin-3 | monoclonal mouse | 1mg/ml | IF 1:50 | Abcam Ab2785 |
| lnterferon-γ | R4-6A2 | 1mg/ml | ELISA 1:200 | BD Biosciences 551216 |
| IL-2 | JES6-1A12 | 0.5mg/ml | ELISA 1:200 | BD Biosciences 554424 |
| LAMP-1 (CD107a) | polyclonal rabbit | 1mg/ml | FC 1:200 IF 1:200 | abcam ab24170 |
| OVA | polyclonal rabbit | 2.3mg/ml | FC 1:500 | Sigma SAB4301164 |
| Dectin-1 | monoclonal rat (2A11) | 1mg/ml | IF 1:100 FC 1:100 | GeneTex GTX41467 |
| Syk | Monoclonal Rabbit (D3Z1E) | n/a | WB: 1:100 | Cell Signaling 13198S |
| Phospho-Syk Y352 | Monoclonal Rabbit (65E4) | n/a | WB: 1:1000 | Cell Signaling 2717S |
| Phospho-Syk Y525/526 | Monoclonal Rabbit (6C87C1) | n/a | IF: 1/50 | Cell Signaling 2710S |
| Anti-beta Actin | Monoclonal Rabbit (13E5) | n/a | WB: 1:1000 | Cell Signaling 4970L |
Conjugated primary antibodies
Abbreviations: FC (flow cytometry), CI (cell isolation)
| Target | Conjugate | Clone | Stock (mg/ml) | Application | Company |
|---|---|---|---|---|---|
| CD4 | PE | RM4-5 | 0.2 | CI 1:200 | BD Biosciences 553049 |
| CD8a | BV785 | 53-6.7 | 0.2 | FC: 1:200 | Biolegend 100750 |
| CD11b | PE | M1/70 | 0.2 | CI: 1:200 | BD Biosciences 553311 |
| CD11c | PE | HL3 | 0.2 | CI: 1:200 | BD Biosciences 553802 |
| CD16/32 | PE | 2.4G2 | 0.2 | CI: 1:200 | BD Biosciences 553145 |
| CD40 | PE | 3/23 | 0.2 | FC: 1:200 | BD Biosciences 553791 |
| CD45.1 | AF647 | A20 | 0.5 | FC: 1:200 | Biolegend 110720 |
| CD45.2 | BV510 | 104 | 0.2 | FC: 1:200 | Biolegend 109838 |
| CD44 | FITC | 1M7 | 0.5 | FC: 1:200 | BD Pharmingen 553133 |
| CD45R (B220) | PE | RA3-6B2 | 0.2 | CI: 1:200 | BD Biosciences 553090 |
| CD62L | PE-Cy7 | MEL-14 | 0.2 | FC 1:200 | Biolegend 104418 |
| TCRβ | BV421 | 104 | 0.2 | FC 1:200 | Biolegend 109230 |
| CD107b (LAMP-2) | FITC | M3/84 | 0.5 | FC 1:200 | Biolegend 108504 |
| CD107b (LAMP-2) | AF647 | M3/84 | 0.5 | FC 1:200 | BD Biosciences 564843 |
| DNGR-1 | PE | 1F6 | 0.2 | FC: 1:50 | Biolegend custom |
| DNGR-1 | APC | 1F6 | 0.2 | FC 1:50 | Biolegend custom |
| H-2Kb (MHC I) | APC | AF6-88.5 | 0.2 | FC 1:200 | Biolegend 116518 |
| H-2Kd (MHC I) | FITC | SF1-1.1 | 0.2 | FC 1:200 | BD Pharmingen 553565 |
| I-A/I-E (MHC II) | BV711 | M5/11 4.15.2 | 0.2 | FC 1:200 | BD Biosciences 563414 |
| Kb/OVA257 (SIINFEKL) tetramer | PE | - | 2 | FC 1:400 | Baylor College of Medicine |
| Pro5® MHC pentamer H-2Kb SIINFEKL | R-PE | - | - | FC 1:100 | ProImmune |
| Interferon-γ | biotin | XMG1.2 | 0.5 | ELISA 1:1,000 | BD Biosciences 554410 |
| Interferon-γ | PE | XMG1.2 | 0.2 | FC 1:100 | Biolegend 505808 |
| Ly6c/Ly6g (Gr-1) | PE | RB-8C5 | 0.2 | CI 1:200 | Biolegend 108408 |
| XCR-1 | PE | ZET | 0.2 | CI 1:50 | Biolegend 148204 |
| alpha-Tubulin | HRP | 11H10 | n/a | WB: 1000 | Cell Signaling 9099S |
Conjugated secondary antibodies
Abbreviations: FC (flow cytometry), IF (immunofluorescence)
| Target | Conjugate | Species | Stock (mg/ml) | Application | Company |
|---|---|---|---|---|---|
| biotin | alkaline phosphatase | Extravidin from egg white avidin | 1.5 | ELISA 1:5,000 | Sigma E2636 |
| Goat IgG | AF647 | donkey | 2 | FC 1:200 | Thermo Fisher A21447 |
| Rabbit IgG | AF555 | polyclonal donkey | 2 | IF 1:200 | Thermo Fisher A31572 |
| Rabbit IgG | AF647 | polyclonal goat | 2 | FC 1:200 IF 1:200 | Thermo Fisher A21245 |
| Rabbit IgG | BV421 | polyclonal donkey | 0.4 | FC 1:200 | Biolegend 406410 |
| Rat IgG | Cy5 | polyclonal goat | 2 | IF 1:200 | Thermo Fisher A10525 |
| Rabbit IgG | HRP | Goat polyclonal | 1 | WB:1:2000 | Jackson Immunoresearch 111035003 |
Cells
RPMI 1640 (Gibco) supplemented with glutamine, penicillin, streptomycin, 2-mercaptoethanol (all from Gibco) and 10 % heat-inactivated foetal calf serum (R10 medium) was used for culture of B3Z, RAW264.7, HEK293T, Flt3L differentiated bone marrow-derived dendritic cells (Flt3L-cDCs) and OT-1 Rag1 –/– T-cells. IMDM (Gibco) with the same additives (I10 medium) was used for culture of the MuTuDC 1940 cDC1 line51. MuTuDC Clec9a –/– sub-lines complemented with WT DNGR-1 cDNA or harbouring W155A, W250A (2WA) mutations were previously described26. B3Z cells containing a reporter plasmid for NFAT coupled to LacZ activity have been previously described52 and were a gift from N. Shastri (UC Berkeley). Mouse embryonic fibroblasts (MEFs) from H-2bm1 mice expressing a non-secreted form of OVA were previously described (bm1 T OVA MEFs)23. The BRAF V600E mutant 5555 mouse melanoma cell line has been previously described53.
Retroviral transduction of MuTuDCs
GP2-293 packaging cells were seeded in a 10cm dish. Upon reaching 70 % confluence, they were transfected with a mixture of 18μl GeneJuice (Novagen), 6 μg of pMDG.2 and 6μg of pFB-Neo plasmid coding for the protein of interest in 600 μl of Opti-MEM medium. On days 1, 2 and 3 post-transfection, the pseudotyped virus-containing culture medium was replaced with fresh one, filtered through 0.44 μm filter, supplemented with 8μg/ml polybrene (Sigma) and immediately applied onto Phoenix cells grown in a 6-well plates. The plate was centrifuged for 150 mins at 2500 xg at room temperature. The medium was exchanged for fresh complete RPMI1640 medium. On the third day, the Phoenix cells were re-plated into a 10 cm dish and expanded. For transduction of MuTuDC, ecotropic virus-containing supernatant from Phoenix cells was used for spinfection in presence of polybrene.
Transient Transfection of HEK293T cells and RAW264.7 cells
HEK293T or RAW264.7 cells were seeded onto glass coverslips (either 18 mm or 12 mm, thickness = #1.5 or 0.17 mm, VWR) and allowed to adhere overnight. Then, they were transfected with the indicated construct using FugeneHD transfection reagent (Promega) according to the provider’s manual. Cells were then allowed to take up and begin expressing the indicated constructs for an additional 18-24 hrs. When transfecting RAW264.7 cells, the medium was changed from RPMI1640 (Gibco) supplemented with 10 % FBS (PAA) to DMEM (Gibco) supplemented with 10% FBS immediately prior to transfection.
Generation of SYK-deficient HEK293T cell line
HEK293T were transfected using FugeneHD transfection reagent (Promega) according to the provider’s manual with lentiCRISPRV2 plasmid containing the SYK gRNA sequence - CACACCACTACACCATCGAG. After 48 hr, bulk population was selected based on Puromycin sensitivity (1 μg/ml). Then, selected cells were seeded as single clones (1 cell / well) in 96-well plates. After 3-4 weeks, clones were screened for SYK expression by Immunoblot. HEK293T SYK KO line was complemented with mouse SYK cloned into pMSCV-mCherry-SYK (from Addgene, deposited by Hidde Ploegh). A SYK kinase-dead version (pMSCV-mCherry-SYK K396R) was generated in-house by QuickChange Lightning kit (Agilent Technologies) using the primers (gtgaaaaccgtggctgtgCGaatcctgaagaacgaggcc; ggcctcgttcttcaggattCGcacagccacggttttcac) and introduced into the HEK293T SYK CRISPR line using FugeneHD transfection reagent (Promega). After 48 hrs, bulk population was selected based on mCherry expression and cell sorted using an Aria Fusion Sorter (BD Biosciences).
Generation of stable HEK293T and RAW264.7 cell lines
Stable HEK293T and RAW264.7 cell lines expressing chimeric receptors were generated using the optimized Sleeping Beauty transposon system. H-2Kb in the pSBbi-puromycin vector was obtained from Addgene (deposited by Jon Yewdell). The generation of the C7, C9::C7 and C9(Y7F)::C7 chimeric receptors in the pFB vector has been previously described24. The receptors were sub-cloned with the Takara In-Fusion Cloning system into the pSBbi-GH vector (Addgene) using the following primers:
| Primer Name | Primer Sequence |
|---|---|
| C7 forward | CCCCAAGCTTGGCCTTTACAGTTCCTTCTCACAGA |
| C7 reverse | ACCCCAAGCTGGCCTATGAAATATCACTCTCATATAGA |
| C9::C7 reverse | ACCCCAAGCTGGCCTATGCATGCGGAAGAAATATATACC |
| C9(Y7F)::C7 reverse | ACCCCAAGCTGGCCTATGCATGCGGAAGAAATATTTACC |
The murine β-2 microglobulin sequence in the pGEM-T Easy plasmid was obtained from Addgene (deposited by Peter Mombaerts) and sub-cloned with the Takara InFusion Cloning system into the pSBbi-bla plasmid (Addgene) using the following primers:
| Primer Name | Primer Sequence |
|---|---|
| β2M forward | CCCCAAGCTTGGCCTAAAGCAGAAGTAGCCACAGGG |
| β2M reverse | ACCCCAAGCTGGCCTTTTCAGTGGCTGCTACTCGG |
For the generation of stable lines. Cells were seeded into 6 well plates. They were then transfected with 1.9 μg of the various constructs described above along with 0.1 μg of a construct containing the SB100X transposase in the pCMV(CAT)T7 vector (Addgene). After 18-24 hrs, the appropriate selection antibiotic was added to the medium at the concentrations indicated below.
| Selection antibiotic | Concentration |
|---|---|
| Blasticidin | 10 μg/ml |
| Puromycin | 1 μg/ml |
| Hygromycin | 300 μg/ml |
Cells were grown with selection antibiotic for 5-7 days and then sorted for equal expression of the various receptors, and H-2Kb.
RT-PCR for NADPH oxidase subunits in HEK293t cells
Messenger RNA was isolated from HEK293T cells using the GeneJET RNA purification kit (Thermo Fisher) according to the manufacturer’s manual. The purified RNA was used as a template for complementary DNA generation and subsequent amplification using the OneStep RT-PCR kit from Qiagen. The forward and reverse primers used are listed below.
| Primer Name | Primer Sequence |
|---|---|
| hNOX1 forward | GTTTTACCGCTCCCAGCAGAA |
| hNOX1 reverse | GGATGCCATTCCAGGAGAGAG |
| hNOX2 forward | CCCAATCCCTCAGTTTGCT |
| hNOX2 reverse | CCTTCTGTTGAGATCGCCAA |
| hNOX3 forward | ACCTTCTGTAGAGACCGCTAT |
| hNOX3 reverse | TCACATGCATACAAGACCACA |
| hNOX4 forward | CTGTGGTGTTACTATCTGTATTTTCTC |
| hNOX4 reverse | CTTGCTGCATTCAGTTCAACA |
| hNOX5 forward | GCCAGTGCCTCAACTTCG |
| hNOX5 reverse | CCACTACCACGTAGCCCATA |
| hp22phox forward | ATGGGGCAGATCGAGTGGGCCATGT |
| hp22phox reverse | GTAGATGCCGCTCGCAATGGCCAG |
Preparation of egg white solution
Hen egg white can be used as a source of endotoxin-free soluble OVA54. The shell of an egg (Marks & Spencer) was sterilised with 70 % ethanol. Under a laminar flow cabinet, a 19 gauge needle was used to carefully draw 10-15 ml of egg white from the egg while avoiding the yolk. The egg white was transferred to a 50 ml tube and combined with 40 ml of PBS (Gibco). The resulting solution was sonicated for 10 mins at room temperature. An additional 50 ml of PBS was added to the solution and it was then filtered through a 0.2 micron filter tip syringe. The concentration of ovalbumin was calculated by measuring the OD at 280 nm and assuming that approximately 54 % of the egg white is ovalbumin54. The solution was aliquoted and stored at -80 °C.
Preparation of zymosan-OVA and zymosan-cytochrome C
Zymosan depleted of TLR agonists (Invivogen) was resuspended in PBS (Gibco) to a final concentration of 1 mg/ml and stored at -20 °C as a stock solution. 18-24 hrs prior to XP assays, the zymosan was thawed, centrifuged for 1 minute and the pellet was re-suspended in egg white solution and/or bovine cytochrome c (10 mg/ml) in PBS until immediately before the XP assay. The zymosan was then centriguged as above and washed 3X with 1 ml of PBS before re-suspending at 1 mg/ml in PBS. The resulting zymosan-OVA was used immediately and not stored for more than 2 hrs.
Preparation of biotinylated Zymosan
Zymosan (1 mg/ml) was incubated with EZ Link sulfo-NHS-LC-biotin (ThermoFisher) dissolved in 0.1 M sodium carbonate (pH 8.3) at a concentration of 1 mg/ml. To begin the reaction, 0.1 mg of EZ link was added to 1 mg of zymosan in 0.1 M sodium carbonate (pH 8.3). The reaction was allowed to proceed at room temperature for 1 hr. The zymosan was washed 5X-6X with fresh PBS, re-suspended to a concentration of 1 mg/ml and stored at -20 °C.
Preparation of zymosan-fluorescein and zymosan-OxyBurst
Zymosan (1mg/ml) was incubated with either Fluorescein isothiocyanate (FITC) (ThermoFisher) or the succinimidyl ester of OxyBurst Green (ThermoFisher) dissolved in DMSO (Merck). To begin the reaction, 0.1 mg of the isothiocyanate of succinimidyl ester was added to 1 mg of zymosan in 0.1 M sodium carbonate (pH 8.3 for succinimidyl esters and pH 9.0 for isothiocyanates). The reaction was allowed to proceed at room temperature for 1 hr. The zymosan was washed 5X-6X with fresh PBS, re-suspended to a concentration of 1 mg/ml and stored at -20 C.
Generation of F-actin-myosin
Lyophilised non-biotinylated G-actin and myosin II (Cytoskeleton) were reconstituted in sterile water at the final concentration of 10mg/ml and stored at -80 °C. Before use, the G-actin aliquots were diluted into G-actin buffer to final concentration 1 mg/ml and incubated for at least 30 mins on ice. To generate biotinylated F-actin, non-biotinylated G-actin was mixed in a 1:1 molar ratio with biotinylated G-actin (Cytoskeleton) which was always freshly reconstituted. 20 μg of each G-actin preparation was mixed with F-actin buffer and incubated for 1 hr at room temperature. To complex the biotinylated F-actin with myosin II, F-actin was mixed in a 1:1 molar ratio with myosin II and incubated for 1 hr at room temperature.
Preparation of FM-OVA and OVA beads
For the coating of microbeads, OVA was biotinylated using the DSB-X biotinylation kit (Thermo Fisher). The concentration of biotinylated OVA was adjusted to 2 mg/ml. Streptavidin coated microbeads with a diameter of 2.0 μm (Polysciences Inc.) were used in non-fluorescent or yellow-green fluorescent form and were labelled with biotinylated OVA (1:1000 unless stated otherwise) for 1 hr on ice. The OVA bead preparations were washed with 1 % BSA in PBS for 3 mins at 10,000 xg. The OVA beads were then split in two fractions, one of which was stored in 1%BSA containing PBS and the other was subjected to labelling with F-actin-myosin II. In vitro polymerised biotinylated F-actin-myosin II was added to OVA beads and incubated for at least 1 hr on ice. The coating of OVA and actin was monitored by flow cytometry.
Preparation of anti-DNGR1 (7H11) beads
25 μl of 2 μm streptavidin beads (Polysciences Inc.) were washed 3X with PBS supplemented with 1 % BSA. The beads were then re-suspended in 400 μl of PBS supplemented with 1 % BSA and 0.15 mg/ml of the biotinylated anti-DNGR1 monoclonal antibody 7H11 and incubated on ice for 30 mins. The beads were then washed with PBS supplemented with 1 % BSA 3X and then re-suspended in 800 μl of PBS.
Preparation of Cell Tracker Deep Red™ labelled, fixed and permeabilized sheep red blood cells (FP-sRBC) beads
In some experiments sRBCs were labelled with Cell Tracker Deep Red™ dye before use. Briefly, 1 ml of 100 % sRBCs in PBS were labelled with Cell Tracker Deep Red™ dye (1:1000) for 30 mins. sRBCs were centrifuged at 1000 xg for 5 mins and washed with 1 ml PBS 2X to remove excess dye. To fix sRBCs, 1ml of 100% sRBCs in PBS was incubated with 4 % PFA for 15 mins at room temperature. sRBCs were centrifuged at 1000 xg for 5 mins and washed with 1 ml PBS 2X. To permeabilise, sRBCs were resuspended in 1 ml PBS containing 0.05 % TX-100 for 10 mins. sRBCs were centrifuged at 1000 xg for 5 mins and washed with 1 ml PBS 2X and then resuspended in 200 μl PBS.
Phagocytosis assays
Flow cytometry: UV-irradiated BM1 T OVA MEFs were stained with the lipophilic dye PKH26 (Sigma Aldrich) or CLARET (Sigma Aldrich) according to the manufacturer’s instruction. Labelled dead cells were added to MuTuDCs at a 1:1 ratio in a 96-well plate and incubated for at least 1 hr. Yellow-green microbeads (PolySciences Inc.) were coated the day before analysis and applied to the cells at a 10:1 ratio. For analysis by flow cytometry, cells were resuspended in FACS buffer and data were acquired on a LSR Fortessa flow cytometer (BD Biosciences). To discriminate cells that had lost their membrane integrity, DAPI or live-dead far red dye (Thermo Fisher) were used. Flow cytometry data were analysed using FlowJo 10.3 software. Microscopy: MuTuDCs, RAW264.7 or HEK293T cells were plated onto glass coverslips (either 18 mm or 12 mm, thickness = # 1.5 or 0.17 mm, VWR) and allowed to adhere overnight. Cells were then challenged with the phagocytic target (zymosan, dead cells [labelled as above] or beads) at the indicated ratios. After 1-4 hrs the cells were fixed with a solution of 4 % paraformaldehyde in PBS. Outside particles were then labelled with either fluorescent streptavidin (0.002 mg/ml in PBS) or anti-OVA antibody. The cells were then washed 3X with fresh PBS and permeabilized using 0.1 % Triton X-100 in PBS. After permeabilization, total particles could be labelled with fluorescent streptavidin (0.002 mg/ml in PBS). Coverslips containing the cells were then mounted on slides using ProLong Diamond Antifade mounting medium (ThermoFisher) and imaged 18-24 hrs later. The images were then used to quantify the total number of particles internalized per cell, referred to as the phagocytic index.
PhagoFACS assay
MuTuDCs were resuspended in CO2-independent medium (Thermo Fisher) containing 2X of glutamax (Thermo Fisher) and were adjusted to 20×106 cells/ml. Microbeads were added in a 10:1 ratio of beads:DCs and the mixture was incubated for 25 mins at 16°C. After 5 mins incubation at 37 °C, cells were washed with ice-cold PBS at 450 xg for 4 mins. The cells were washed two more times with ice-cold PBS and were then resuspended in 1ml PBS which was applied on top of a 5ml FCS cushion. Cells were centrifuged for 4 mins at 150 xg and the supernatant was discarded. The cell pellet was resuspended in cell culture medium adjusting the cells to 20x106 cells/ml. The sample was then split into different time points using 5 x 106 cells per time point. The chase was performed for up to 360 mins and samples were collected after indicated time points by adding ice cold PBS and keeping them on ice. For outside bead labelling the cells were stained with anti-OVA for 30 mins on ice followed by anti-rabbit_BV421 for 30 mins on ice. Cells were then resuspended in homogenisation buffer (0.5ml per time point) and pushed 30 times through a 22 G needle. The post-nuclear supernatant was collected after centrifugation for 4 mins at 150 xg and washed with 1 % BSA in PBS, which was also used for the antibody staining. The crude phagosome isolations were washed stained in 1 % BSA in PBS with different antibodies overnight in a polypropylene V-bottom 96-well plate. The next day all secondary antibody staining were performed for 2 hrs before phagosomes were analysed by flow cytometry using a Fortessa flow cytometer (BD). The data were analysed using FlowJo 10.3 software.
Sorting of single bead containing MuTuDCs
MuTuDCs were pulsed with yellow-green FMO or OVA beads with a ratio of 10 beads: 1 cell for 25 mins at 16 °C. To allow for internalisation, cells were then incubated for 5 mins at 37 °C. Free beads were removed by 3X washes with ice cold PBS, before the pellet was resuspended in 1 ml PBS, which was then applied onto a 5 ml FCS cushion. The FCS cushion was centrifuged for 4 mins at 150 g and the pellet was resuspended to 20 × 106 cells/ml and incubated for 4 hrs at 37°C (chase period). Cells were washed and resuspended and filtered through a 70 μm strainer before sorting. The sorting of single and double bead containing MuTuDCs was performed using an Aria Fusion Sorter (BD Biosciences) by gating on live and single cells. The sorter was set-up with 100 μm nozzle size to allow good recovery of the cells and the sort mode was on 4-way purity precision. The flow rate was kept below 2.0 and the efficacies were usually kept between 70-95 %. To confirm that sorted cells had internalised a single bead, ImageStream analysis was performed. A small sample of the input sample was analysed in the Amnis ImageStream X analyser (Merck Milipore). Data was analysed using the Ideas software (Merck Milipore).
GFP influx assay
Phagocytosis assays were performed as above. After 4 hrs of phagocytosis, the cells were imaged live to avoid artefactual phagosomal rupture during the fixation process. To quantify luminal GFP, a Z-Stack was selected in the center of each phagosome. The intensity of GFP in the phagosomal lumen as well as in the cytosol was measured using FIJI. A ratio of luminal to cytosolic GFP was obtained and binned as shown in the figures.
MuTuDC stimulation with DNGR-1 agonists and curdlan
MuTuDC were seeded into 24 well plates (Corning) at a density of 4 × 105 cells per well. After 18-24 hrs, the cells were incubated with isotype-matched irrelevant specificity control mAb (MAC49 clone, 0.5 μg), anti-DNGR-1 (7H11 clone, 0.5 μg), F-actin-myosin-II (DNGR-1-L, 100 nM) or Dectin-1 ligand (Curdlan 200 μg/ml) over a 1 hr timecourse. For immunoblotting, cell lysates were prepared using laemmli sample buffer (Bio-Rad) and were electrophoresed on 4-20 % Mini-PROTEAN® TGX™ Precast Protein gels (Bio-Rad). Protein expression was examined by immunoblotting.
In vitro XP assay in HEK293T cells
HEK293T cells stably expressing H-2Kb plus murine β2m and the indicated receptors were seeded into 96 well U-bottom plates (Corning) at a density of 1 x 105 cells per well. After 18-24 hrs, the cells were incubated with Zymosan-OVA, SIINFEKL peptide, or egg white solution for 6-8 hrs. Cells were then fixed with 0.5 % paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS for 20 mins. Reactive PFA was quenched using 50 mM NH4Cl (ThermoFisher) in PBS for 10 mins. Cells were then washed 3X with fresh PBS and 1X with RPMI (Gibco) containing 10 % FBS and 50 μM β-mercaptoethanol (Gibco) (R10 medium). B3Z cells were added to the wells at a 2:1 (B3Z:HEK293T) ratio in R10 medium and incubated for 18-24 hrs. The plates were then frozen to lyse cells and the amount of IL-2 in the supernatants was quantified by ELISA.
In vitro XP assay in MuTuDCs
Microbeads and dead cells were prepared the day before the assay. MuTuDCs were seeded at a density of 0.1 × 106 cells per well in a U-bottom 96 well plate. In cases where inhibitors were titrated and a fixed antigen dose was applied, dead cells were administered in a 3:1 ratio and beads in a 20:1 ratio. All inhibitors were added 10-15 mins before adding antigens and centrifuging the plates for 3 mins at 200 xg before incubating at 37 °C for 4 hrs. 0.05 × 106 OT-I CD8+ T cells were added per well and co-cultured overnight. Next morning, the plate was freeze-thawed once, and the total amount of IFN-γ was determined by ELISA.
In vitro XP assay with FLT3L-culture cDC1
Microbeads and dead cells were prepared the day before the assay as above. Differentiated cDC1 were prepared from day 9 bone marrow cultures (Flt3L 150 ng/ml) using magnetic enrichment strategy with a XCR1-PE antibody (Clone: ZET – 2 μg/ml). Briefly, cells were incubated with an XCR1-PE antibody and cDC1 were isolated according to the instructions of the EasySep™ PE positive selection kit. Purified cDC1 (> 90% purity) were seeded at a density of 1 × 105 cells per well in a U-bottom 96-well plate. Titrations of different antigen sources were added to purified cDC1 and cultures were incubated at 37°C for 4 hrs. 5 × 104 OT-I CD8+ T cells were added per well and co-cultured overnight. Next morning, the plate was freeze-thawed once, and the total amount of IFN-γ in supernatant was determined by ELISA.
Mice
C57BL/6Jax, Batf3 −/−{Hildner:2008p29393}, Cybb −/− , Clec9a Cre/Cre (Cre knocked into Clec9a locus to disrupt gene function55), Batf3 −/−B6.SJL.CD45.1 , OT-I/Rag1 −/−, mice were bred at The Francis Crick Institute under specific pathogen-free conditions. Batf3 −/− mice were a gift from Dr. Kenneth Murphy. Mice were used at 6-8 weeks of age and littermates of the same sex were randomly assigned for bone marrow collection and chimera generation. All animal experiments were performed in accordance with national and institutional guidelines for animal care and were approved by the Francis Crick Institute Biological Resources Facility Strategic Oversight Committee (incorporating the Animal Welfare and Ethical Review Body) and by the Home Office, UK.
In vivo immunisation with F-actin- and myosin II-coated OVA-bearing latex beads
6-8 week old male and female mice were injected intravenous (i.v.) with 4 × 107 F-actin-myosin-OVA latex beads (2 μm) mixed with poly(I:C) (50 μg/mouse) in a total volume of 0.2 ml. Six days later, spleens were isolated and CD8+ T cell responses were measured by quantitating the number of H-2Kb-OVA-pentamer positive CD8+ T cells.
Bone marrow chimera experiments
6-8 week old Batf3 −/− B6.SJL.CD45.1 mice were lethally irradiated with 6.6 Gy twice (4 hr rest in between exposures), then rested for 1 day to recover. Mice were injected intravenous (i.v.) with 2 × 106 mixed bone marrow cells in a total volume of 0.2ml obtained from Batf3 −/− B6.SJL.CD45.1 mixed with either CD45.2+ Batf3 −/−, C57BL/6Jax, Cybb −/−, Clec9a Cre/Cre mice at an 80% to 20% ratio, respectively. After 8 weeks, chimerism was quantitated in the blood. To assess XP in these chimeras, we prepared dead cells by UVC-irradiation (250 mJ/cm2) of 5555V600E melanoma cells with and left overnight. Cell corpses were pulsed with OVA (10 mg/ml) and Poly(I:C) (250 μg/ml) for 1 hr at 37 °C. Dead cells were washed 3X in ice-cold PBS before being injected i.v. (1 × 106 cells/mouse). Six days later, spleens were isolated and CD8+ T cell responses were measured by quantitating the number of H-2Kb-OVA-tetramer positive CD8+ CD62Llo CD44+ T cells or by OVA peptide (SIINFEKL 1 nM for 16 hrs) re-stimulation and intracellular staining for IFN-γ (brefeldin A (10 μg/ml) added last 4 hrs).
Oxyburst assay
MuTuDCs, RAW264.7 or HEK293T cells were plated onto glass coverslips (either 18 mm or 12 mm, thickness = #1.5 or 0.17mm, VWR) and allowed to adhere overnight. Cells were then challenged with the phagocytic target (zymosan-OxyBurst or bead-OxyBurst) at the indicated ratio. After 1-4 hrs the cells were fixed with a solution of 4 % paraformaldehyde in PBS. Coverslips containing the cells were then mounted on slides using ProLong Gold Antifade mounting medium (ThermoFisher) and imaged 18-24 hrs later. The images were then used to quantify the total number of fluorescent particles internalised per cell referred to as OxyBurst+ phagosomes.
Nitroblue tetrazolium (NBT) assay
MuTuDCs, RAW264.7 or HEK293T cells were plated onto glass coverslips (either 18 mm or 12 mm, thickness = #1.5 or 0.17mm, VWR) and allowed to adhere overnight. Cells were then challenged with zymosan. NBT (Sigma) at 10 μg/ml was added to the medium, and phagocytosis was allowed to proceed for 1 hr. Cells were then fixed with 4 % PFA in PBS and imaged immediately by bright field microscopy.
Sulforhodamine B (SRB) release assay
HEK293T cells expressing the indicated receptors were plated in 35 mm μ-dishes (Ibidi) for direct visualization and microscopy. Cells were then incubated with zymosan to allow for the initiation of phagocytosis. SRB at 150 μg/ml was added to the medium to allow for its internalization, along with the phagocytic target, into the lumen of the phagosomes. After 5-10 mins of phagocytosis the SRB was washed 3X with fresh, ice-cold PBS and then the cells were bathed in culture medium and placed in a 37 °C incubator for 1-4 hrs. Cells were then imaged live to avoid artefactual rupture of the phagosomal membrane during fixation. The mean fluorescence intensity of SRB in the cytosol was then quantified using FIJI software and normalized to the intensity of SRB in the lumen of the phagosome.
β-lactamase release assay
HEK293T cells expressing the indicated receptors were plated in 35 mm μ-dishes (Ibidi) for direct visualization and microscopy. Cells were pre-incubated with the acetoxymethyl ester of CCF4 (CCF4-AM) (ThermoFisher) at 0.5 μg/ml in Live Cell Imaging Solution (ThermoFisher) containing Solution B and probenecid according to provider’s instructions (LiveBLAzer™ FRET B/G Loading Kit; ThermoFisher). Cells were then incubated with zymosan that had been pre-soaked in β-lactamase. After 5-10 mins of phagocytosis the excess zymosan was washed away 3X with fresh, ice-cold PBS and then the cells were bathed in culture medium and placed in a 37 °C incubator for 4 hrs. Cells were then imaged live (excitation : 405 nm, emission : 460-520 nm) to avoid artefactual rupture of the phagosomal membrane during fixation. The mean fluorescence intensity at 460 nm in the cytosol was then quantified using FIJI software.
Fluorescence recovery after photobleaching
HEK293T cells expressing the C9::C7 chimeric receptor were transiently transfected with mCherry-SBD-lysenin. After 18-24 hrs, the cells were incubated with zymosan for 4 hrs. Lysenin+ phagosomes in live cells were located by confocal microscopy. Regions of interest were drawn over the phagosome, over an area of cytosol, and over a Lysenin- phagosome within individual cells. The phagosomes were photo-bleached at 90-95 % laser power. Fluorescence recovery in both the GFP and mCherry channel was monitored at maximum interval speed for the indicated time-course.
Immunofluorescence staining
Cells were fixed with 4% PFA (Electron Microscopy Sciences) in PBS for 20 mins at room temperature. Cells were then washed 3X-4X with PBS (Gibco). Reactive PFA was quenched using 50 mM NH4Cl (Sigma-Aldrich) in PBS for 5-10 mins and then washed 3X-4X with PBS. Cells were then permeabilized with 0.1 % Triton X-100 (CalBioChem) in PBS for 15 mins at room temperature followed by 3-4 washes with PBS. The cells were then blocked with 2 % non-fat powdered milk (Marvel) in PBS (blocking buffer). The primary antibody incubation was performed by inverting the coverslips onto 80-100 μl droplets of blocking buffer containing a dilution of the primary antibody for 1 hr at room temperature. The coverslips were then flipped back into the tissue culture plate and washed 3X with blocking buffer. The secondary antibodies were then diluted in blocking buffer and added directly to the plate and incubated for 1 hr at room temperature. The cells were then washed 3X with blocking buffer and then 3X with PBS. Coverslips were mounted onto glass slides using ProLong Antifade Mounting Medium (Thermofisher).
Coating coverslips with α-MHC class II
Glass coverslips (18 mm) were coated with a 0.1 % solution of poly-L-lysine for 30 mins at room temperature. The excess poly-L-lysine was washed 3X with DPBS. The coverslips were then incubated in a 2.5 % solution of glutaraldehyde for 15 mins at room temperature. After washing three times with DPBS, coverslips were incubated in a solution of 1μg/mL α-MHC class II in DPBS for 30 mins at room temperature. Coverslips were washed again with DPBS and incubated overnight in 0.2 M glycine in PBS at 4°C. On the day of the experiment, the coverslips were washed three times with fresh DPBS.
Detection of Galectin-3 labelled phagosomes in Flt3L cultures
Flt3L cultures were plated at a density of 5 X 105 cells onto individual coverslips that were previously coated with α-MHC class II. Cells were allowed to attach for 1 hr at 37 °C. Cells were then challenged with beads coated with α-DNGR-1 (clone: 7H11) for 3 hrs. They were then fixed with 4% PFA and immunostained as above.
3D correlative light and electron microscopy (3D CLEM)
For serial block face scanning electron microscopy (SBF SEM), cells were grown on photoetched glass bottom dishes (Ibidi GmbH, Martinsried, Germany), imaged by confocal microscopy as described above and initially fixed with 4 % (v/v) formaldehyde (Taab Laboratory Equipment Ltd, Aldermaston, UK) in 0.1 M phosphate buffer (PB) pH 7.4 for 15 mins. The samples were then processed using a Pelco BioWave Pro+ microwave (Ted Pella Inc, Redding, USA) following a protocol adapted from the National Centre for Microscopy and Imaging Research protocol56. Each step was performed in the BioWave, except for the PB and water wash steps, which consisted of two washes on the bench followed by two washes in the BioWave without vacuum (at 250 W for 40 s). All the chemical incubations were performed in the BioWave for 14 mins under vacuum in 2 min cycles alternating with-without 100 W power. The SteadyTemp plate was set to 21 °C unless otherwise stated. In brief, the samples were fixed again in 2.5 % (v/v) gluteraldehyde (TAAB) / 4 % (v/v) formaldehyde in 0.1 M PB. The cells were then stained with 2 % (v/v) osmium tetroxide (TAAB) / 1.5 % (v/v) potassium ferricyanide (Sigma), incubated in 1 % (w/v) thiocarbohydrazide (Sigma) with SteadyTemp plate set to 40 °C, and further stained with 2 % osmium tetroxide in ddH2O (w/v). The cells were then incubated in 1 % aqueous uranyl acetate (Agar Scientific, Stansted, UK) with SteadyTemp plate set to 40 °C, and then washed in dH2O with SteadyTemp set to 40 °C. Samples were then stained with Walton's lead aspartate with SteadyTemp set to 50 °C, and dehydrated in a graded ethanol series (70 %, 90 %, and 100 %, twice each), at 250 W for 40 s without vacuum. Exchange into Durcupan ACM® resin (Sigma) was performed in 50 % resin in ethanol, followed by 4 pure Durcupan steps, at 250 W for 3 mins, with vacuum cycling (on-off at 30 sec intervals), before embedding at 60 °C for 48 hrs. Blocks were trimmed to a small trapezoid, excised from the resin block, and attached to an SBF SEM specimen holder using conductive epoxy resin. Prior to commencement of an SBF SEM imaging run, the sample were coated with a 2 nm layer of platinum to further enhance conductivity. SBF SEM data was collected using a 3View2XP (Gatan, Pleasanton, CA) attached to a Sigma VP SEM (Carl Zeiss Ltd, Cambridge, UK). Inverted backscattered electron images were acquired through the entire extent of the region of interest. For each of the 334 50nm slices needed to image the cell in its whole volume, a low resolution overview image (horizontal frame width 78 μm; pixel size of 52 nm; using a 2 μs dwell time) and a high resolution image of the cell of interest (horizontal frame width 37.85 μm; pixel size of 7.6 nm; using a 2 μs dwell time) were acquired. The SEM was operated in high vacuum with focal charge compensation on (80 %). The 30 μm aperture was used, at an accelerating voltage of 1.8 kV. Only minor adjustments in image alignment were needed, and were done with the TrakEM2 plug-in of the FIJI framework57.
For 3D light and electron microscopy data registration, image stacks from confocal microscopy and SBF SEM were manually aligned to each other using the BigWarp plugin of the FIJI framework58, with the SBF SEM stack set as ‘target’ and the confocal stack as ‘moving’ dataset. ‘Landmark mode’ was used to add 22 pairs of corresponding points within both datasets throughout the whole volume of the cell. Bright field and GFP channels were used to place landmarks in the confocal dataset. The SBF SEM dataset could be aligned to the plane of the platinum layer, which was coplanar with the coverslip in the confocal dataset. An affine transformation was applied to the confocal data and the transformed dataset was merged with the SBF SEM data to produce the final overlay. For segmentation, the SBF SEM stack was loaded in Microscopy Image Browser (MIB) 59.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software (GraphPad). Statistical significance between two samples was determined using an unpaired two-tailed Welch’s-Student’s t test, Kolmogorov-Smirnov test, or unpaired parametric Mann Whitney tests as indicated in figure legends. Statistical analyses for three or more groups was performed using one-way ANOVA. Statistical analyses for two independent variables was performed using two-way ANOVA. Data are shown as mean (± s.d.) or mean (± s.e.m.) as indicated in the figure legends.
Extended Data
Extended Data Fig. 1. Bead uptake, CD4+ T cell stimulation and DNGR-1 mediated XP by the cytosolic pathway.
a, OVA and FM-OVA beads stained with anti-ovalbumin, anti-actin or phalloidin analysed by flow cytometry. b, WT MuTuDCs were pulsed with OVA or FM-OVA beads, fixed, stained with anti-OVA to mark uninternalized beads (“out. beads”) and imaged by confocal microscopy (scale bar 5 μm). Image representative of one experiment. c, FM-OVA bead internalisation by WT, KO and KO-WT MuTuDCs analysed by flow cytometry. MFI from one of two experiments (n = 2). d-e, WT, KO and KO-WT MuTuDCs pulsed with OVA or FM-OVA beads for 4 hrs were co-cultured with OT-I cells. IFN-γ was assessed by ELISA. One of three experiments is shown. Plotted as mean (±s.d.) of experimental duplicates. P values determined using two-way ANOVA. f, ImageStream analysis of WT MuTuDCs pulsed with yellow-green FM-OVA or OVA beads for 4 hrs and sorted for single bead+ MuTuDCs. g, Single bead+ WT MuTuDCs were co-cultured with OT-II cells. CD69+CD4+ T cell frequency was determined by flow cytometry. One of three experiments is shown (n = 3). h-n, WT MuTuDCs were incubated with FM-OVA beads (blue, 10:1, beads:DCs), UV-irradiated bm1 OVA MEFs (red, 3:1, dead cells:DCs) or hen egg white (grey, 1 mg/ml) in the presence of 250μM leupeptin, 250 μM pepstatin, 10 μM lactacystin, 20 μM lactacystin, 50 μM chloroquine or 500 μM E64. After 4 hrs, OT-I cells were added overnight. OT-I cells alone were also incubated with SIINFEKL peptide (black, 1 nM) in the presence of the inhibitors. IFN-γ was assessed by ELISA, plotted as mean (±s.d.) of experimental duplicates and representative of two independent experiments (n =2). P values determined using an unpaired t test. n.s., not significant; *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.00001.
Extended Data Fig. 2. DNGR-1+ non-degradative phagosome in RAW264.7 macrophages ectopically expressing DNGR-1.
a-g, WT and KO MuTuDCs (a-e), RAW264.7 (f) or KID (g) cells were pulsed with FM-OVA beads for the indicated times. Phagosomes were isolated and stained with anti-DNGR-1, anti-LAMP-2, anti-OVA and anti-MHC I antibodies. Analysis was performed by flow cytometry. Data represented as mean (± s.d.) and from one of two experiments are shown (n = 2). h, Confocal microscopy image of RAW264.7 cells transiently expressing H-2Kb were pulsed with IgG-beads for 2 hrs (scale bar = 6 μm). Each dot represents a single image out of 10 images per condition (n = 10). Data from one representative experiment of two independent experiments (n = 2).
Extended Data Fig. 3. Recruitment of galectin-8 to DNGR-1+ phagosomes containing FM-OVA beads, phagocytosis by cells expressing chimeric receptors and sulforhodimine B release from phagosomes.
a, PhagoFACS of phagosomes isolated from WT MuTuDCs pulsed with FM-OVA beads for 4 hrs and stained with anti-DNGR-1, anti-LAMP-2 and anti-galectin-8 antibodies. Frequency of phagosomes is plotted as mean (± s.d.) for three independent experiments (n = 3). P values were determined by unpaired t test, Welch’s test. b, WT MuTuDCs expressing mCherry::galectin-3 were incubated with OVA or FM-OVA beads (5:1, beads to DCs) for 4 hrs. Cells were fixed, uninternalised beads (“out. bead”) marked by anti-OVA staining and cells were imaged by confocal microscopy (scale bar = 5 μm). Images are a representative image from 18 FM-OVA and 36 OVA images. c, HEK293T cells expressing cytosolic GFP and C7, C9::C7 or C9(Y7F)::C7 receptors and stained for Dectin-1. Images show a single representative image from 3 similar images. d, HEK293T cells expressing the indicated receptors were pulsed with biotinylated zymosan for 1 hr before fixation and labelling of uninternalised zymosan with fluorescent streptavidin. Internalised zymosan particles were enumerated from replicate confocal images (n = 3 images for empty vector and C7 tail-less, 15 images for C7, C9::C7 and C9(Y7F)::C7 and 5 images for C7 R406 and C9::C7 R406). P values were determined using an unpaired t test. e, HEK293T cells expressing the indicated receptors were challenged with zymosan pre-soaked in sulforhodamine B (SRB). SRB in the cytosol was quantified and normalized to phagosomal fluorescence. Data and images are representative of three independent repeats (n = 3) and n = 40 cells for C9::C7 and 28 cells for C9(Y7F)::C7. Data (d, e) represented as mean (± s.e.m.). P values were determined using an unpaired t test, Welch’s test. n.s., not significant; **P ≤ 0.01; ***P ≤ 0.001.
Extended Data Fig. 4. Generation of HEK293T cell lines able to present OVA on H-2Kb.
a, Confocal microscope images of HEK293T cells stably expressing cytosolic GFP, H-2Kb and either C7, C9::C7 or C9(Y7F)::C7 receptors (anti-Dectin-1 staining). Nuclei were stained with DAPI. Images are representative of 3 similar images (scale bars 5 μm). b, As for (a) but using flow cytometry c, IL-2 from B3Z hybridoma cells cultured with the above H-2Kb-expressing HEK293T cells in the presence of the indicated concentrations of SIINFEKL peptide. Data are plotted as mean (± s.d.) of experimental triplicates from one of three independent experiments (n = 3). P values were determined by two-way ANOVA. d, Confocal images of HEK293T cells stably transfected with empty vector or plasmid encoding either C7, C9::C7 or C9(Y7F)::C7 receptors and pulsed with biotinylated zymosan for 1 hr before fixation and labelling of uninternalised zymosan with fluorescent streptavidin (scale bars 10 μm).. The number of internalised zymosan particles (right) was enumerated from the images (left). Data represented as mean (± s.e.m.) and each dot represents an independent experiment. e, RAW264.7 cells expressing H-2Kb and either C9::C7 or C9(Y7F)::C7 receptors were pulsed with zymosan-OVA for 4 hrs before fixation with 0.5 % PFA and incubated with OT-I cells. IFN-γ was assessed by ELISA. f, LLOMe (1 mM) was added during zymosan-OVA pulse of RAW264.7 cells expressing H-2Kb and C9(Y7F)::C7 receptor. All data in (e, f) are plotted as mean (± s.e.m.) of experimental triplicates. n.s., not significant; **P ≤ 0.01; ****P ≤ 0.00001.
Extended Data Fig. 5. SYK is required for GFP influx into phagosomes.
a, Western blot and confocal microscope images of SYK protein expression in HEK293T SYK KO cells, complemented with mouse mCherry::SYK or mCherry::SYK K396R kinase dead (KD). Bottom panel: mCherry expression of the same micrographs depicted as in Fig. 5c lower panel (scale bar 2 μm). b, Flow cytometry plots measuring phagocytosis of HEK293T WT and SYK KO cells incubated with zymosan-pHrodo-Red particles over 30 min timecourse. Data are representative of two independent experiments (n = 2).
Extended Data Fig. 6. DNGR signalling promotes phagosomal ROS production.
a-b, Confocal images of RAW264.7 cells transfected with empty vector or plasmid encoding C9::C7 or C9(Y7F)::C7 receptors and pulsed with zymosan (a) or dead sRBCs (b) in the presence of Nitroblue tetrazolium (NBT) (Scale bar 10 μm). Quantification of ROS+ phagosomes. Data represented as mean (± s.e.m.) (a) or (± s.d.) (b) and are representative of two independent determinations (n = 2). P values determined by one-way ANOVA. c, RAW264.7 stably expressing C9 or C9(Y7F) receptors were pulsed with CellTracker DeepRed (CTDR)-labelled FP-sRBCs for 2 hrs. Percentage of CTDR+ RAW264.7 cells was quantified by flow cytometry. Data represented as mean (± s.d.) and are representative of two independent experiments (n = 2). d, Confocal images of RAW264.7 stably expressing C9 or C9(Y7F) receptors pulsed with dead cells in the presence of NBT for 2 hrs (scale bars 10 μm). Image is a representative image of three similar images. e, RT-PCR of NADPH oxidase subunits in HEK293T. Representative of two experiments (n = 2). f, HEK293T cells stably expressing C9::C7 were challenged with zymosan-Oxyburst in the presence or absence of DPI for 1 hr. Oxyburst+ positive phagosomes were quantified across 5 fields of view (n > 100 phagosomes). Data represented as mean (± s.e.m.). P values were calculated by unpaired parametric test, Mann-Whitney and are representative of two independent experiments (n = 2). g, HEK293T C9::C7 cells were pulsed with zymosan-Ova (left) or transfected with plasmid encoding VENUS-SIINFEKL (right) in the presence or absence of DPI (10 μM) for 4 hrs before fixing and adding of OT-I Rag1 -/- T-cells. IFN-γ was assessed by ELISA, plotted as mean (± s.d.) of an experimental triplicate. n.s., not significant; *P ≤ 0.05; **P ≤ 0.01.
Extended Data Fig. 7. Generation of DNGR-1 and NOX2-deficient cDC1 chimeras.
a, Flow cytometry gating strategy to define and quantify chimeric T cell populations in blood. b, Quantification of percentage chimerism in the blood 8 weeks post adoptive transfer across the indicated genotypes. Each dot is an individual mouse sample (Batf3 -/- : Batf3 -/- n =7, Batf3 -/- : WT n =8, Batf3 -/- : Clec9a cre/cre n = 8, Batf3 -/- : Cybb -/- n = 9).
Supplementary Material
mCherry::lysenin and the C9::C7 receptor were incubated with zymosan for 4 hrs. The 3D reconstruction of a lysenin+ phagosome was carried out as described in Methods and the video was prepared using the IMARIS software.
Acknowledgements
We thank members of the Immunobiology Laboratory for helpful discussions and suggestions. We thank the Crick BRF and Flow Cytometry STPs for their support throughout this project. We are grateful to P. Faull and B. Snijders for helpful comments. This work was supported by The Francis Crick Institute, which receives core funding from Cancer Research UK (FC001136), the UK Medical Research Council (FC001136), and the Wellcome Trust (FC001136), by an ERC Advanced Investigator grant (AdG 268670), by a Wellcome Investigator Award (WT106973MA), and by a prize from the Louis-Jeantet Foundation. C.M. Henry was supported by a FEBS Long-Term Fellowship and a Marie Sklodowska-Curie Individual Fellowship under Horizon 2020 from the European Union. M.D. Buck was supported by an EMBO LTF (ALTF 1096-2018) and a Marie Sklodowska-Curie Individual Fellowship under Horizon 2020 from the European Union. F. Randow is supported by the MRC (U105170648) and the Wellcome Trust (WT104752MA).
Footnotes
Author contributions J.C., C.M.H., and H.B. performed most of the experiments. O.S. and H.R. helped with the sorting of single bead MuTuDCs. C.M.H and M.D.B performed in vivo experiments. O.S. performed initial experiments and provided key reagents. E.C. helped with confocal experiments. N.C.R. helped with mouse breeding and genotyping. H.R. helped with ImageStream analysis. S.Z. generated the DNGR-1::Dectin-1 chimeras. L.C. and M.C.D. performed electron microscopy. A.A. and S.A. provided advice and training for the PhagoFACS experiments. V.P. provided the Cybb −/− mouse strain. D.C.T. provided reagents that contributed to this study. C.J.E and F.R. provided the lysenin and galectin probes. H.B., J.C., C.M.H. and C.R.S. interpreted the data and wrote the manuscript. C.R.S. designed and supervised the study.
Competing interests The authors declare no competing interests
References
- 1.Huang AY, et al. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–965. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 2.Sigal LJ, Crotty S, Andino R, Rock KL. Cytotoxic T-cell immunity to virus-infected non-haematopoietic cells requires presentation of exogenous antigen. Nature. 1999;398:77–80. doi: 10.1038/18038. [DOI] [PubMed] [Google Scholar]
- 3.Iborra S, et al. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. J Clin Invest. 2012;122:1628–1643. doi: 10.1172/JCI60660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smed-Sörensen A, et al. Influenza A virus infection of human primary dendritic cells impairs their ability to cross-present antigen to CD8 T cells. PLoS Pathog. 2012;8:e1002572. doi: 10.1371/journal.ppat.1002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alloatti A, et al. Critical role for Sec22b-dependent antigen cross-presentation in antitumor immunity. Journal of Experimental Medicine. 2017 doi: 10.1084/jem.20170229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theisen DJ, et al. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science. 2018;362:694–699. doi: 10.1126/science.aat5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz FM, Colbert JD, Merino E, Kriegsman BA, Rock KL. The Biology and Underlying Mechanisms of Cross-Presentation of Exogenous Antigens on MHC-I Molecules. Annu Rev Immunol. 2017;35:149–176. doi: 10.1146/annurev-immunol-041015-055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gros M, Amigorena S. Regulation of Antigen Export to the Cytosol During Cross-Presentation. Front Immunol. 2019;10:1283–9. doi: 10.3389/fimmu.2019.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colbert JD, Cruz FM, Rock KL. Cross-presentation of exogenous antigens on MHC I molecules. Curr Opin Immunol. 2020;64:1–8. doi: 10.1016/j.coi.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grotzke JE, Cresswell P. Are ERAD components involved in cross-presentation? Mol Immunol. 2015;68:112–115. doi: 10.1016/j.molimm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Dingjan I, et al. Lipid peroxidation causes endosomal antigen release for cross-presentation. Sci Rep. 2016;6 doi: 10.1038/srep22064. 22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingjan I, et al. VAMP8-mediated NOX2 recruitment to endosomes is necessary for antigen release. Eur J Cell Biol. 2017 doi: 10.1016/j.ejcb.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reis e Sousa C, Germain RN. Major histocompatibility complex class I presentation of peptides derived from soluble exogenous antigen by a subset of cells engaged in phagocytosis. J Exp Med. 1995;182:841–851. doi: 10.1084/jem.182.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain RN, et al. Processing and presentation of endocytically acquired protein antigens by MHC class II and class I molecules. Immunol Rev. 1996;151:5–30. doi: 10.1111/j.1600-065x.1996.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 15.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savina A, et al. The Small GTPase Rac2 Controls Phagosomal Alkalinization and Antigen Crosspresentation Selectively in CD8+ Dendritic Cells. Immunity. 2009;30:544–555. doi: 10.1016/j.immuni.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Weimershaus M, et al. Conventional dendritic cells require IRAP-Rab14 endosomes for efficient cross-presentation. The Journal of Immunology. 2012;188:1840–1846. doi: 10.4049/jimmunol.1101504. [DOI] [PubMed] [Google Scholar]
- 18.Kretzer NM, et al. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. Journal of Experimental Medicine. 2016;213:2871–2883. doi: 10.1084/jem.20160597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han D, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou P, et al. Thioesterase PPT1 balances viral resistance and efficient T cell crosspriming in dendritic cells. J Exp Med. 2019;5 doi: 10.1084/jem.20190041. jem.20190041–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanc P, et al. In: C-Type Lectin Receptors in Immunity. Yamasaki S, editor. Springer; 2016. [DOI] [Google Scholar]
- 22.Schulz O, et al. Myosin II Synergizes with F-Actin to Promote DNGR-1-Dependent Cross-Presentation of Dead Cell-Associated Antigens. Cell Rep. 2018;24:419–428. doi: 10.1016/j.celrep.2018.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sancho D, et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelenay S, et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest. 2012;122:1615–1627. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iborra S, et al. Optimal Generation of Tissue-Resident but Not Circulating Memory T Cells during Viral Infection Requires Crosspriming by DNGR-1+ Dendritic Cells. Immunity. 2016;45:847–860. doi: 10.1016/j.immuni.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanc P, et al. A pH- and ionic strength-dependent conformational change in the neck region regulates DNGR-1 function in dendritic cells. EMBO J. 2016;35:2484–2497. doi: 10.15252/embj.201694695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grotzke JE, et al. Sec61 blockade by mycolactone inhibits antigen cross-presentation independently of endosome-to-cytosol export. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1705242114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozik P, et al. Small Molecule Enhancers of Endosome-to-Cytosol Import Augment Anti-tumor Immunity. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107905. 107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savina A, Vargas P, Guermonprez P, Lennon A-M, Amigorena S. Measuring pH, ROS production, maturation, and degradation in dendritic cell phagosomes using cytofluorometry-based assays. Methods Mol Biol. 2010;595:383–402. doi: 10.1007/978-1-60761-421-0_25. [DOI] [PubMed] [Google Scholar]
- 30.Böttcher JP, et al. Oncogenic Transformation of Dendritic Cells and Their Precursors Leads to Rapid Cancer Development in Mice. The Journal of Immunology. 2015;195:5066–5076. doi: 10.4049/jimmunol.1500889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thurston TLM, Muhlinen, von N, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison CJ, Kukulski W, Boyle KB, Munro S, Randow F. Transbilayer Movement of Sphingomyelin Precedes Catastrophic Breakage of Enterobacteria-Containing Vacuoles. Curr Biol. 2020;30:2974–2983.e6. doi: 10.1016/j.cub.2020.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cebrian I, et al. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell. 2011;147:1355–1368. doi: 10.1016/j.cell.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Lin ML, et al. Selective suicide of cross-presenting CD8+ dendritic cells by cytochrome c injection shows functional heterogeneity within this subset. Proc Natl Acad Sci USA. 2008;105:3029–3034. doi: 10.1073/pnas.0712394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dersh D, Yewdell JW, Wei J. Antigen Processing. Vol. 1988. Humana; New York, NY: 2019. pp. 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J-G, et al. The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments. Immunity. 2012;36:646–657. doi: 10.1016/j.immuni.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Lee SB, Bae IH, Bae YS, Um H-D. Link between mitochondria and NADPH oxidase 1 isozyme for the sustained production of reactive oxygen species and cell death. J Biol Chem. 2006;281:36228–36235. doi: 10.1074/jbc.M606702200. [DOI] [PubMed] [Google Scholar]
- 38.Palmowski MJ, et al. Role of immunoproteasomes in cross-presentation. J Immunol. 2006;177:983–990. doi: 10.4049/jimmunol.177.2.983. [DOI] [PubMed] [Google Scholar]
- 39.Ackerman AL, Giodini A, Cresswell P. A role for the endoplasmic reticulum protein retrotranslocation machinery during crosspresentation by dendritic cells. Immunity. 2006;25:607–617. doi: 10.1016/j.immuni.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Zehner M, et al. Mannose receptor polyubiquitination regulates endosomal recruitment of p97 and cytosolic antigen translocation for cross-presentation. Proc Natl Acad Sci USA. 2011;108:9933–9938. doi: 10.1073/pnas.1102397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackerman AL, Kyritsis C, Tampé R, Cresswell P. Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc Natl Acad Sci USA. 2003;100:12889–12894. doi: 10.1073/pnas.1735556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guermonprez P, et al. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature. 2003;425:397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- 43.Houde M, et al. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 44.Accapezzato D, et al. Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. J Exp Med. 2005;202:817–828. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belizaire R, Unanue ER. Targeting proteins to distinct subcellular compartments reveals unique requirements for MHC class I and II presentation. Proc Natl Acad Sci USA. 2009;106:17463–17468. doi: 10.1073/pnas.0908583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turk B, Turk V. Lysosomes as ‘suicide bags’ in cell death: myth or reality? J Biol Chem. 2009;284:21783–21787. doi: 10.1074/jbc.R109.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graham DB, et al. An ITAM-signaling pathway controls cross-presentation of particulate but not soluble antigens in dendritic cells. Journal of Experimental Medicine. 2007;204:2889–2897. doi: 10.1084/jem.20071283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savina A, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 49.Nair-Gupta P, et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell. 2014;158:506–521. doi: 10.1016/j.cell.2014.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alloatti A, et al. Toll-like Receptor 4 Engagement on Dendritic Cells Restrains Phago-Lysosome Fusion and Promotes Cross-Presentation of Antigens. Immunity. 2015;43:1087–1100. doi: 10.1016/j.immuni.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Fuertes Marraco SA, et al. Novel murine dendritic cell lines: a powerful auxiliary tool for dendritic cell research. Front Immunol. 2012;3:331. doi: 10.3389/fimmu.2012.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karttunen J, Sanderson S, Shastri N. Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proc Natl Acad Sci USA. 1992;89:6020–6024. doi: 10.1073/pnas.89.13.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhomen N, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 54.Boes M, et al. T cells induce extended class II MHC compartments in dendritic cells in a Toll-like receptor-dependent manner. J Immunol. 2003;171:4081–4088. doi: 10.4049/jimmunol.171.8.4081. [DOI] [PubMed] [Google Scholar]
- 55.Schraml BU, et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 56.Deerinck T, Bushong EA, Lev-Ram V, Shu X. Enhancing Serial Block-Face Scanning Electron Microscopy to Enable High Resolution 3-D Nanohistology of Cells and Tissues. Microscopy and Microanalysis. 2010;16:1138–1139. [Google Scholar]
- 57.Cardona A, et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogovich JA, Hanslovsky P, Wong A, Saalfeld S. Robust registration of calcium images by learned contrast synthesis. IEEE International Symposium on Biomedical Imaging; 2016. [Google Scholar]
- 59.Belevich I, Joensuu M, Kumar D, Vihinen H, Jokitalo E. Microscopy Image Browser: A Platform for Segmentation and Analysis of Multidimensional Datasets. PLoS Biol. 2016;14:e1002340. doi: 10.1371/journal.pbio.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mCherry::lysenin and the C9::C7 receptor were incubated with zymosan for 4 hrs. The 3D reconstruction of a lysenin+ phagosome was carried out as described in Methods and the video was prepared using the IMARIS software.