Abstract
R-loops are three-stranded structures that harbour an RNA–DNA hybrid and frequently form during transcription. R-loop misregulation is associated with DNA damage, transcription elongation defects, hyper-recombination and genome instability. In contrast to such ‘unscheduled’ R-loops, evidence is mounting that cells harness the presence of RNA–DNA hybrids in scheduled, ’regulatory’ R-loops to promote DNA transactions including transcription termination and other steps of gene regulation, telomere stability and DNA repair. R-loops formed by cellular RNAs can regulate histone post-translational modification and may be recognized by dedicated reader proteins. The two-faced nature of R-loops implies that their formation, location and timely removal must be tightly regulated. In this review we discuss the cellular processes that regulatory R-loops modulate, the regulation of R-loops and the potential differences that may exist between regulatory R-loops and unscheduled R-loops.
Introduction
The past decade has revolutionized our thinking about regulatory RNAs. Primarily through using next-generation sequencing, it has been recognized that a large part of the genome is transcribed and that there are countless RNAs of different lengths with potential regulatory functions1. Although the biology of some non-coding RNAs (ncRNAs) is fairly well established, an understanding of the regulatory functions of other ncRNAs remains preliminary or elusive. One mode of how RNA may exert regulatory functions in the genome, in a sequence specific manner, is through the formation of RNA–DNA hybrids. The term RNA–DNA hybrid refers to base-pairing of RNA with DNA. When the formation of an RNA–DNA hybrid results in the displacement of single stranded DNA (ssDNA) it is referred to as an R-loop. RNA–DNA hybrids and R-loops have recently gained attention for having important roles in cellular processes such as gene regulation and DNA repair 2,3. Paradoxically, R-loops were long considered detrimental byproducts of transcription that interfere with transcription4 and contribute to genome instability. R-loop-induced genomic stress is directly linked with failure to remove the R-loops in a timely manner 5,6. Many of the negative consequences of faulty hybrid metabolism stem from replication stress owing to encounters of the DNA replication machinery with R-loops7-9, however R-loop-related defects in transcription elongation 10, double strand break (DSB) formation11, mutagenesis 8 and altered chromatin landscapes 12 can also occur in a replication-independent manner. We refer the reader to excellent Reviews covering the detrimental effects of misregulated R-loops 2-4,13-16 and the factors involved in their removal4,5,17,18.
In this review, we discuss how RNA–DNA hybrids and R-loops are used as positive regulators and signalling beacons of certain DNA transactions, with emphasis on transcription regulation and the maintenance of genomic integrity. We do not ignore, however, the detrimental aspects of R-loops that can affect the very same processes. We also discuss the factors that may determine the fate of R-loops and other hybrids and how they may influence whether hybrids are regulatory and productive or ’unscheduled’ and detrimental.
Roles of non-R-loop hybrids
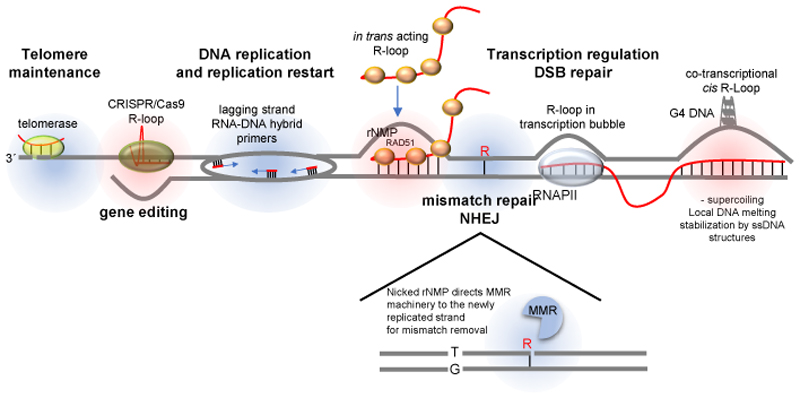
It has been known for decades that RNA–DNA hybrids have a key role in a variety of nuclear processes, most notably transcription and DNA replication (Fig. 1). During DNA synthesis, replication of the lagging strand offers a paramount example of RNA–DNA hybrid function, whereby the RNA primers, synthesized by DNA polymerase α, are used by DNA polymerase δ to polymerise mature Okazaki fragments 19–21. Another example of functional RNA–DNA hybridization occurs at telomeres, where the holoenzyme telomerase, which is made up of a ncRNA and a reverse transcriptase, specifically recognizes telomere sequences through RNA– DNA base pairing. The RNA moiety of telomerase hybridizes to the 3’ ssDNA extension that exists at chromosome ends and enables the extension of the ends to prevent telomere erosion (Fig. 1) 22. Frequently, RNA–DNA ’mini-hybrids’ are established when replicating DNA polymerases (most frequently DNA polymerase ε) misincorporate ribonucleotide triphosphates (rNTPs) into the newly synthesized DNA strand 23. Although misincorporated ribonucleoside monophosphates (rNMPs) can compromise genome integrity if not efficiently excised, they also have positive roles during DNA repair, as the mismatch repair (MMR) and non-homologous end-joining pathways utilize ribonucleotide insertions to promote genome integrity24–26. In the case of MMR, the rNMPs serve as a strand discrimination signal to direct the repair of a mismatched base on the newly synthesized, and not on the template strand (Fig. 1). The fact that rNMPs are so abundant (13,000 per cell cycle in yeast) is also suggestive of having a biological function 27. The positive and negative effects of rNMPs in the genome suggest they are transient and tightly regulated, to ensure optimal DNA repair while avoiding genome instability. The above hybrids are not considered R-loops as they do not result in DNA strand displacement (Fig. 1, blue circles).
Figure 1. Types of RNA–DNA hybrids.
RNA has the capacity to localize to genomic regions in a sequence specific manner and regulate downstream cellular processes (labelled in bold). R-loops are three-stranded structures harbouring an RNA–DNA hybrid and a displaced strand of DNA (highlighted in red). Non-R-loop forming RNA–DNA hybrids (highlighted in blue) are involved in different chromosomal transactions. At telomeres, the RNA moiety of the telomerase holoenzyme base-pairs with the 3´overhang at chromosome ends and provides a template for their extension. DNA replication, especially on the lagging strand, is dependent on the prior synthesis of small RNAs, which are used as polymerization primers by DNA polymerases. The cleavage of DNA-incorporated ribonucleoside monophosphates (rNMPs) on the 5´side serves as a signal for the DNA mismatch repair (MMR) machinery to distinguish the newly replicated strand from the template strand and ensure that the right mismatched base (here, ‘T’) is removed. R-loops are thought to form behind the RNA polymerase transcription machinery, where negative DNA supercoiling results in DNA unwinding, which provides an opportunity for the 5´end of the nascent transcript to base-pair with the template strand. Hybrid formation maybe be facilitated in the presence of G-rich sequences on the non-template strand, which may result in G4 structures. When RNA is produced from an exogenous source (such as a plasmid) or from a homologous chromosome or a distal homologous sequence, R-loops can also form in trans in a RAD51 recombinase-dependent manner. All RNA molecules are depicted in red, and DNA is depicted in blue. DSB, double strand break; G4, G-quadruplex secondary structure; NHEJ, non-homologous end joining; Pol II, RNA polymerase II.
R-loop formation and distribution
R-loops are three-stranded structures consisting of an RNA–DNA hybrid and the displaced strand of DNA (Fig. 1, red circles). R-loop features have been extensively discussed in several excellent reviews 2,5,6,17. Briefly, the three-stranded structures are typically, but not exclusively, linked to ongoing transcription, and until recently were assumed to be mere ’by-products’ of transcription that occur exclusively in cis, at the site of transcription. The negative supercoiling, and hence increased tendency for DNA melting (strand separation) that occurs behind the RNA polymerase provide an ideal opportunity for the nascent transcript to anneal to the complementary template strand and form an R-loop. Although RNA–DNA hybrids are continually formed within the transcription bubble of the RNA polymerase, it is unlikely that R-loops are simply an extension of this limited hybrid, but rather form through re-invasion of the 5’ end of the RNA (Fig. 1). This is supported by the structure of the RNA polymerase complex demonstrating that RNA and DNA are extruded from the complex at different exit channels, which would prevent an R-loop from simply being extended 28. Moreover, a recent study demonstrated that efficient formation of co-transcriptional R-loops requires a free RNA end and a GC skew (asymmetry in the distribution of guanines and cytosines between the strands; see below) 29. RNA–DNA hybrids can also form in trans when the RNA is produced at a spatially distinct site (Fig. 1) 30.
The methodology for the detection of RNA–DNA hybrids has been crucial for understanding how R-loops are regulated and for mapping where and when they form in the genome. Although the predominant tool for detecting RNA–DNA hybrids has been the monoclonal hybrid-specific S9.6 antibody 31, alternative reagents and techniques have emerged. Catalytic-dead versions of ribonuclease H1 (RNase H1; an enzyme that can degrade the RNA moiety of RNA–DNA hybrids), 29,32 or the RNase H1 hybrid binding domain fused to a fluorescent protein 33 can be used as hybrid sensors. Comprehensive summaries of these approaches, along with a discussion of their advantages and disadvantages, are available in refs.3,5,34. Although there is a general consensus on where hybrids accumulate, discrepancies between studies may be owing to different hybrid detection methods. One conserved feature of R-loops is their transient nature. This was first pointed out in a study using budding yeast, which demonstrated that only upon loss of both RNase H enzymes could a uniformly-distributed nuclear staining of RNA–DNA hybrids be detected 35. This indicated that hybrids frequently arise, but are rapidly removed, at least in part, by RNase H. Subsequently, in addition to RNase H, multiple helicases have been shown to contribute to hybrid resolution, reviewed in ref 3. The dispersed S9.6 staining throughout the nucleus also suggested that RNA–DNA hybrids were forming at multiple loci across the genome 35. These predictions were verified when genome-wide sequencing of hybrid-harboring loci in yeast revealed a widespread distribution, including a strong presence in retrotransposons, telomeres and highly expressed genes, such as the ribosomal RNA and tRNA loci and other structured ncRNAs (Fig. 2).
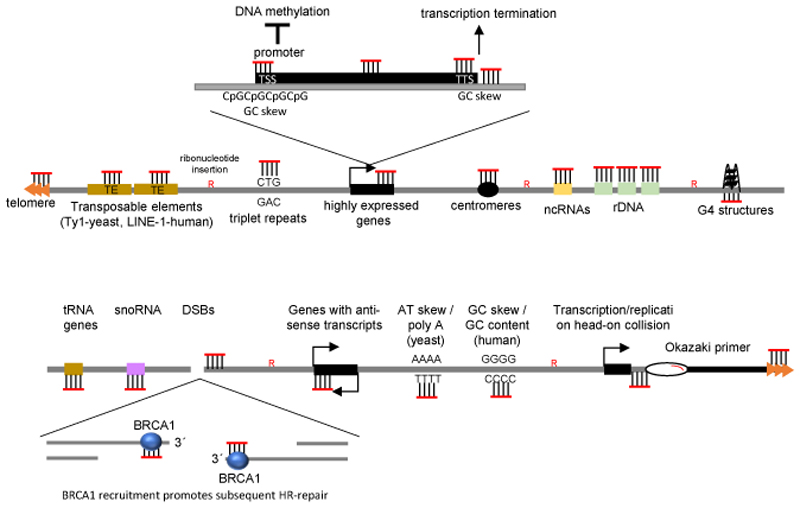
Figure 2. R-loops across the genome.
R-loops have been mapped genome-wide in a number of species. The most prevalent predictor of R-loop presence is high transcriptional activity; indeed, R-loops are found at promoter regions, where they promote transcription by inducing DNA demethylation, and at transcription termination regions, where they promote transcription termination. Other features of R-loop-rich areas include high GC content and GC skew, g-quadruplex (G4) structures, antisense transcription and regions where the replication and transcription machineries collide. RNA–DNA hybrids (and perhaps R-loops) also form at sites of DNA damage, particularly at double-stranded breaks (DSBs), where they promote homologous recombination (HR)-mediated repair through the recruitment of breast cancer susceptibility protein 1 (BRCA1). ncRNA, non-coding RNAs; snoRNAs, small nucleolar RNAs; rDNA, ribosomal DNA.
R-loops are considerably enriched at genes associated with anti-sense transcription and have recently been shown to directly promote anti-sense expression36–38(Fig. 2), suggesting they have a regulatory role in gene expression (see below). Although one study, which included treatment with S1 nuclease as part of the DRIP protocol to stabilize hybrids through removal of the displaced strand, indicated that AT skew and especially poly(A) tracts may be hotspots of R-loop formation37, the general consensus was that, in yeast, sequence per se is not a crucial determinant of R-loop accumulation, but rather the rate of transcription at the locus. Indeed, an R-loop-poor locus can be converted to an R-loop-rich locus, simply by boosting rates of transcription by changing promoters37. There is a tendency for GC-rich sequences to harbor more R-loops, however the same study revealed that GC-rich genes were typically more highly expressed in yeast 36. In mammalian cells, GC skew strongly favors R-loop formation (see below) (Fig. 2). However, yeast genomes do not display much GC skew, apart from at telomeres. In plants, both regions with GC skew or AT skew are enriched in R-loops 39. The link between high levels of expression and R-loops suggests either that high rates of transcription promote R-loop accumulation, or that R-loops promote high rates of transcription. One could also envision a positive feedback loop whereby transcription results in R-loop accumulation, which promotes further transcription. On the other hand, it is important to keep in mind that R-loops are known to be potent inhibitors of transcription and lead to polymerase stalling 10. The contradictory relationship between R-loops and transcription suggest that a tight control of R-loop persistence (half-life) is required to sustain high rates of transcription, or that a spatial separation exists between R-loops and open reading frames.
A study in yeast found little genomic overlap between R-loop hotspots and the localization of a subunit of RNA polymerase II (Pol II)36. This revealed that R-loop mapping by DRIP with the S9.6 antibody was not exclusively documenting ongoing transcription, but rather that some R-loops may persist, or be formed, post-transcriptionally. This was an early indication that R-loops may be more than mere transcription byproducts and suggested that hybrids may form independently of transcription, that is in trans, or influence transcription from a distance.
Similar to yeast, in human cells and in plants, R-loops accumulate at repetitive sequences such as transposable elements, ribosomal DNA, centromeres and telomeres32,39–41 (Fig. 2). Strikingly, in humans and plants they are also prominent at promoter regions that harbor CpG islands (CGIs) 29,32,39,42 (Fig. 2). CGIs are present in the promoters of approximately 60% of human genes. One characteristic of CGIs is the presence of a positive GC skew: enrichment of guanine over cytosine on the non-template strand, downstream of the transcription start site (TSS). A more precise R-loop localization within promoter regions revealed that R-loops are generally constrained between the TSS and the first intron–exon junction 43. In general, a strong GC skew correlates with high gene expression and with R-loop enrichment. Accumulation of R-loops in this sequence-specific manner may be due to the particularly strong thermodynamic binding properties of G-rich RNA to a complementary sequence 44,45. Moreover, the presence of DNA secondary structures such as G-quadruplexes (G4s) on the displaced DNA can contribute to R-loop stability46 (Fig. 1), either by preventing access of R-loop resolving proteins or by decreasing the re-annealing capacity of the DNA. A recent in vitro study using atomic force microscopy indicated that R-loops can attain secondary structures independently of the presence of G4 47. These secondary structures are formed at the displaced DNA strand, and include ‘blobs’, ‘spurs’ and ‘loops’. Such ‘R-loop objects’ impose local physical constraints on the surrounding DNA, for example by bending the DNA. A fascinating possibility is that R-loop objects could encode higher order regulatory information, for example by recruiting different chromatin modifiers. In the future, it would be important to confirm whether such R-loop objects exist in a chromatin context in vivo.
The strong presence of R-loops within gene promoters was a first indication that these structures may have important gene regulatory functions. In additions to promoter regions, genome-wide studies found considerable enrichment of R-loops at transcription termination sites, especially those with GC skew. This is in accordance with the proposed function of R-loops in promoting transcription termination 48,49. Finally, hybrids are produced at sites of DNA damage, including at dysfunctional telomeres and DSBs (Fig. 2), and have important functions in coordinating DNA repair (reviewed in 5 and discussed below).
R-loops have long been considered accidental by-products of transcription, detrimental to cellular physiology, and merely a source of genomic instability when not properly removed5,6. However, it has recently emerged that there is a class of beneficial, ‘regulatory’ R-loops, which have an essential role in a variety of biological processes. Regulatory R-loops harness the sequence specificity of RNA-DNA basepairing to either repel cofactors from, or target them to distinct chromosomal loci. A paramount example, from prokaryotes, is the CRISPR (Clusters of Regularly Interspaced Short Palindromic Repeats)–Cas9 system, which has evolved in bacteria as a natural defence mechanism to recognize and destroy foreign DNA elements of viral origin 50,51. Viral DNA that has been integrated into a CRISPR array in the bacterial genome is transcribed and associates with the Cas9 nuclease. The Cas9–RNA complex forms an R-loop with a matching sequence at the invading viral DNA and generates a DNA break, which eventually results in the destruction of the viral DNA. This module has now been harnessed as a highly precise and easily engineered gene editing tool using guide RNAs to target Cas9 to a specific sequence in the genome, a process which involves R-loop formation in trans 52,53.
The first examples of endogenous regulatory R-loops in eukaryotes came from studies on class-switch recombination at the Ig heavy chain locus 54. R-loops that form within the G-rich switch regions promote recombination-based deletions and drive antibody class diversity 55,56. A similar recombination-based switch occurring in the variant surface glycoprotein (VSG) locus during immune evasion in trypanosomes is also R-loop-regulated 57. The sequence specificity of RNA–DNA hybrids positions R-loops as prime candidate regulators of gene expression through precise targeting of promoter and termination sequences (Fig. 2).
Gene regulation by R-loops
When R-loop removal pathways are perturbed, the presence of R-loops in gene bodies or promoters can interfere with transcription initiation, elongation or termination 10,48,58. Although these consequences appear detrimental at first glance, cells exploit these very same features in a controlled manner to regulate gene expression.
Regulation of chromatin accessibility and transcription
Regulatory R-loops are implicated in chromatin and gene regulation, both by activating and by silencing gene expression. In Arabidopsis thaliana, the function of the promoter of the long non-coding RNA (lncRNA) COOLAIR is repressed by an R-loop, which is stabilized by the binding of the transcription factor ATNDX to the displaced ssDNA of the loop. ATNDX binding may impair the accessibility of R-loop resolving enzymes, thereby affecting transcription at the COOLAIR promoter 59. Also in A. thaliana, there is some evidence that circular RNAs may regulate exon-skipping by forming an R-loop with their cognate locus, although whether this happens in vivo and the underlying mechanism remain to be shown 60. In addition to promoter regions, certain genome-wide studies map R-loops to gene 3′ ends (Fig. 2). This correlates with a well-established role of R-loops of promoting transcription termination by stalling Pol II12,48, which we discuss below.
The chromatin signature (histone post-translational modifications) associated with R-loops resembles those associated with transcription at promoters and TSS, including mono-methylation and tri-methylation of histone H3 Lys 4 (H3K4me1 and H3K4me3, respectively) and histone H3 Lys 27 acetylation (H3K27ac), and with transcription elongation (H3K36me3) 12,32. Consistent with such activating histone modifications, R-loops can induce chromatin decondensation 61, but can also promote heterochromatin assembly 62–64 and chromatin compaction 65.
Regulatory R-loops are often generated by antisense lncRNAs, which recruit transcription regulators (Fig. 3). The gene RASSF1 and its antisense lncRNA RASSF1 antisense RNA 1 (ANRASSF1), for example, are transcribed in a convergent manner (head-to-head) from opposite strands of their locus; ANRASSF1 forms an RNA–DNA hybrid, which recruits Polycomb repressive complex 2 (PRC2) to silence the RASSF1A promoter 66 (Fig. 3a). A similar scenario, with a different outcome of expression, occurs when the head-to-head antisense lncRNA VIM-AS1 (VIM antisense RNA 1) activates the VIM gene, which encodes the intermediary filament vimentin. Transcription of VIM-AS1 initiates 709 bp downstream of the VIM transcription start site and forms an R-loop, which then promotes recruitment of transcription activators of the NF-κB pathway 67 (Fig. 3b). The same study also showed that expression of the gene HMGA2, which encodes a high-mobility group protein is induced by head-to-head transcription of the overlapping antisense lncRNA RPSAP52. In budding yeast, the galactose metabolism (GAL) gene cluster is tightly regulated by sugar availability in the environment. The GAL locus transcribes lncRNAs, which overlap with and positively regulate protein coding GAL genes by forming R-loops 68. These lncRNAs normally act in cis, but they also function in trans, when expressed from transgenes. The non-overlapping genes encoding GATA3-AS1 lncRNA and GATA3 are divergently transcribed. GATA3-AS1 forms an R-loop ~2kb upstream of the GATA3 TSS to upregulate GATA3 expression, likely by recruiting the H3K4 methyltransferase complex mixed-lineage leukaemia (MLL; also known as KMT2A) 69 (Fig. 3c). GATA3-AS1 expressed from plasmids can also activate GATA3, meaning the R-loop can also form in trans. Antisense RNAs are a widespread phenomenon and recent work showed that R-loops themselves facilitate antisense RNA transcription by acting as Pol II promoters 70.
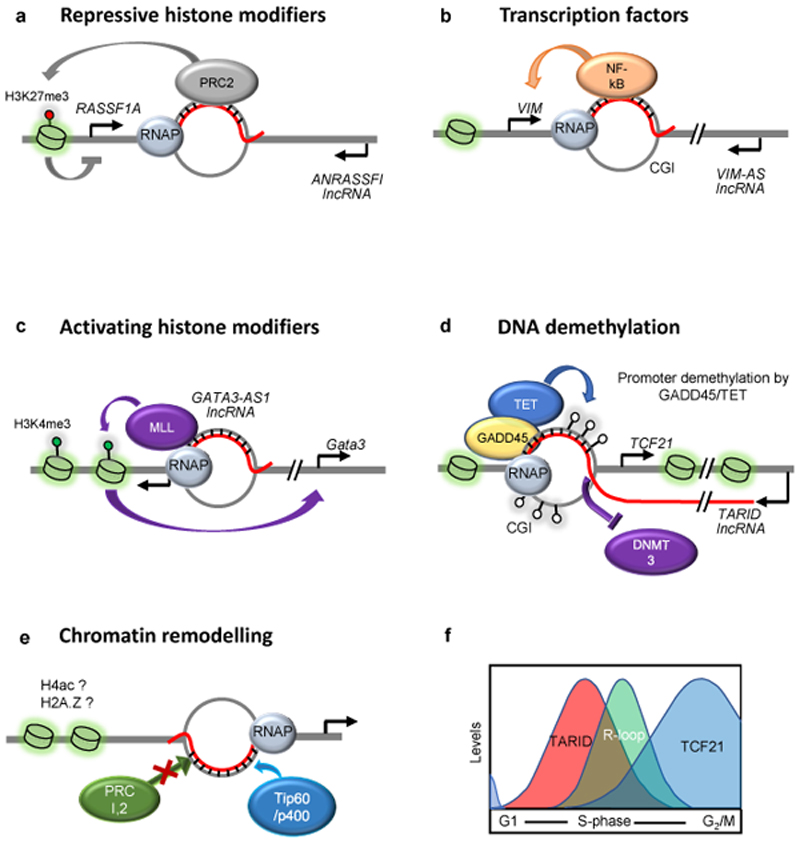
Figure 3. R-loops as regulators of gene expression.
a. At the RASSF1A locus, the anti-sense long non-coding RNA (lncRNA) RASSF1 antisense RNA 1 (ANRASSF1) forms an R-loop in the promoter region, which can serve as a recruitment platform for Polycomb repressive complex 2 (PRC2) to silence RASSF1A expression through histone H3 Lys27 tri-methylation (H3K27me3). b. R-loops can be recognised by transcription factors to promote gene expression. At the VIM locus, the anti-sense lncRNA VIM antisense RNA 1 (VIM-AS1) promotes NF-kB recruitment through the formation of an R-loop at the transcription start site c. Histone modifiers such a the methyltransferase mixed-lineage leukaemia (MLL), are recruited to R-loops in the promoter region of the GATA3 gene to facilitate transcription through H3K4me3 deposition. d. At a subset of CpG island (CGI)-containing promoters, growth arrest and DNA damage protein 45A (GADD45) serves as an R-loop ‘reader’ that recruits ten-eleven translocation (TET) DNA demethylases, which demethylate the promoter DNA and activate transcription. Shown here is the case of the antisense lncRNA TCF21 antisense RNA inducing demethylation (TARID) and its cis-target gene transcription factor 21 (TCF21). The effect of demethylation may be enhanced by the exclusion of DNA (cytosine-5)-methyltransferases (DNMTs) such as DNMT3 from R-loop-rich CGIs. e. During differentiation of embryonic stem cells, the binding of chromatin modifiers with opposing functions (PRC1 and PRC2 versus TIP60–p400) can be dictated by R-loop occupancy, potentially resulting in histone modifications, such as histone H4 acetylation (H4ac) or incorporation of histone variants such as H2A.Z. f. Transcription of the TARID lncRNA, R-loop formation and transcription of the TCF21 mRNA proceed sequentially during the cell cycle. Tight temporal restriction of the formation of regulatory R-loops may be essential to prevent their detrimental effects.
The head-to-head antisense lncRNA, TARID (TCF21 antisense RNA inducing demethylation), which overlaps with the promoter of the tumor suppressor transcription factor 21 (TCF21), forms an R-loop that induces local DNA demethylation and TCF21 expression 71 (Fig. 3d). Again, co-transcription of the sense and antisense transcripts from the same locus appears not to be essential, as exogenously-expressed lncRNA can form an R-loop and bring about gene activation. R-loop accumulation is a characteristic feature of unmethylated CGI-containing promoters that also show GC skew 42. Transcription through GC skew regions promotes R-loop accumulation because G-rich RNAs bound to C-rich DNA templates have a high thermodynamic stability 44,45. Genome-wide mapping showed that R-loops are enriched at loci with decreased DNA methylation and increased DNase hypersensitivity (chromatin accessibility) 41. R-loops can inhibit DNA methylation at promoters by preventing binding DNA (cytosine-5)-methyltransferases (DNMTs) 32,72; another mechanism whereby R-loops may favor DNA hypomethylation is by attracting ten-eleven translocation (TET) DNA demethylases 71 (Fig. 3d). In embryonic stem cells (ESCs), some 4% of the ~90,000 TET1 binding sites at CGI-containing promoters may be R-loop-dependent. Concordantly, the oxidized DNA demethylation intermediates 5-formyl cytosine and 5-carboxyl cytosine are enriched at R-loops and their levels increase upon RNase H1 depletion 71.
Regulatory R-loops and lncRNAs are also implicated in chromatin changes during differentiation of mouse embryonic stem cells. Two chromatin modifying complexes associated with these lncRNAs are Polycomb complex PRC2 (gene repression) and the histone acetyltransferase complex TIP60 (also known as KAT5)–p400 (gene activation) 73 (Fig. 3e). Promoter regions with strong tendency for R-loop formation are not bound by PRC2 or PRC; instead, they recruit TIP60–p400 and activate transcription. Indeed, the overexpression of RNase H1 in mouse ESCs decreases the localization of both TIP60 and p400 at most of their target genes and allows the recruitment of PRC2. In contrast, another study found that R-loops attract PRC to a subset of PRC-repressed target genes. Loss of R-loops leads to transcriptional activation of R-loop-positive PRC targets74. What distinguishes R-loops, which attract-from those that repel the PRC is unclear.
Taken together, these studies strongly indicate that regulatory R-loops can function as epigenetic marks, by altering chromatin and gene expression without affecting the underlying sequences. If R-loops function as epigenetic marks, what are their ‘readers’? Proteomics studies have documented many R-loop-associated proteins, including chromatin-modifying enzymes 75,76. However, whether these proteins bind to R-loops directly or indirectly, are associated with regulatory R-loops or with detrimental R-loops, and merely ‘co-associate’ with R-loops to the same chromatin environment, is unknown. The ssDNA binding proteins replication protein A (RPA) 77, RAD51 30 and ATNDX 59 may bind to the displaced DNA strand of R-loops to stabilize the hybrids and mediate transcription repression. In A. thaliana, ALBA DNA/RNA binding proteins recognize RNA–DNA hybrids in vitro and may protect R-loops from forming DNA breaks in vivo 78. Recently, the stress response protein growth arrest and DNA damage protein 45A (GADD45A) was identified as a reader of R-loops. GADD45A functions as adapter for the DNA demethylation enzymes TET and thymidine DNA glycosylase, and targets them for locus-specific DNA demethylation 79–81. GADD45A directly and selectively binds RNA–DNA hybrids in vitro, and in vivo it promotes DNA demethylation and gene expression through the regulatory R-loop formed by TARID at the TCF21 promoter 71 (Fig. 3d). A summary of the discussed regulatory R-loops and the processes they regulate is provided in Table 1.
Table 1. Selected examples of regulatory R-loops and other RNA–DNA hybrids and their effects.
| RNA in a regulatory R-loop or hybrid | Hybrid location | Effect |
|---|---|---|
| ANRASSF1A 66 | RASSF1A promoter | H3K27me3 deposition and RASSF1A repression |
| VIM-AS1 67 | VIM1 promoter | NFkB recruitment and VIM1 transcription |
| GATA3-AS1 69 | GATA3 promoter | H3K4me3 deposition and GATA3 transcription |
| TARID 71 | TCF21 promoter | DNA demethylation and TCF21 repression |
| TERRA 118 | Telomeres | Repair of short telomeres by homologous recombination |
| TIP60–p400-dependent genes73 | Subset of lncRNA in ESCs | Prevent PRC-mediated gene repression |
| Select PRC target genes 74 | Promoters | Promote PRC target gene repression |
| CEN-RNA 122 | Centromeres | Activation of Aurora B and centromere function |
| dilincRNA 110 and other RNAs at DSBs111 | Any DSB in a transcribed region | RAD51 recruitment and homologous recombination |
CEN-RNA, centromeric RNA; dilincRNA, damage induced long noncoding RNA; DSB, DNA double-stranded break; lncRNA, long non-coding RNA; PRC, Polycomb repressive complex; TARID, TCF21 antisense RNA inducing demethylation; TERRA, telomere repeat-containing RNA.
Roles of R-loops in transcription termination
Pol II transcription termination at gene 3′ ends is a complex process, failure of which can affect gene expression. For example, in tandemly-arranged genes, read-through RNA from an ill-terminated upstream gene can restrict the activity of a downstream gene through transcription interference 82. Genome-wide analyses showed that about 2000 genes with G-rich Pol II termination sites map to R-loop regions 42. Consistent with a role of R-loops in transcription termination, gene-dense regions, where read-through RNA could be most deleterious are enriched in genes with R-loops at their 3’-ends. R-loops can play a role in transcription termination of certain genes by facilitating Pol II pausing at the 3′ end. Stalling of elongating Pol II downstream of extended R-loops formed by the nascent mRNA is well documented and various mechanisms have been proposed to account for it. For example, an elongating Pol II complex was proposed to ’sense’ the conformation of the DNA in its vicinity and terminate transcription when this conformation is altered 83. Pol II is known to oscillate between elongation and backtracking, and extended R-loops were proposed to terminate backtracked Pol II (Fig. 4a). Furthermore, while the position of the R-loop remains fixed relative to the DNA axis, elongating Pol II rotates along the helical path during transcription. Hence, the nascent transcript produced downstream of the R-loop must wrap around the DNA and it was proposed that this unfavorable torsional stress could lead to transcription termination 84 (Fig. 4b).
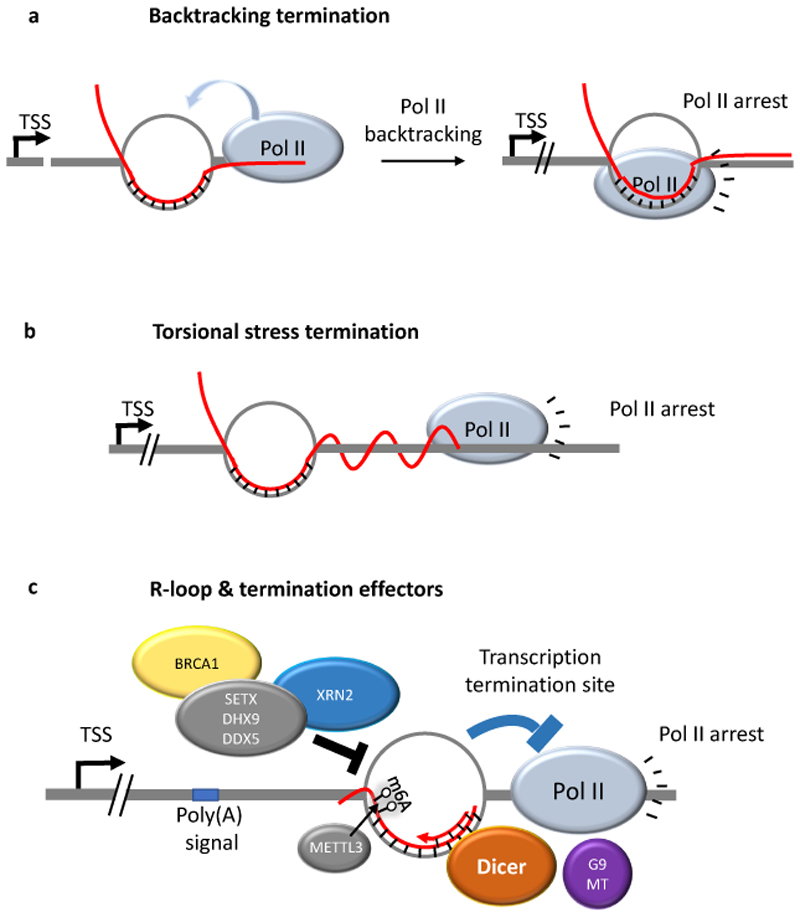
Figure 4. R-loops promote transcription termination.
Shown are mechanisms by which R-loops may lead to pausing of elongating RNA polymerase II (Pol II) and transcription termination. a. Backtracking may arrest Pol II over the R-loop. Pol II oscillates between transcription elongation and backtracking. The presence of an R-loop may terminate the function of backtracked Pol II. b. During transcription, the R-loop remains fixed relative to the DNA helix, while elongating Pol II continues rotating along the helical path. The nascent transcript produced downstream of the R-loop must therefore wrap around the DNA, which may lead to Pol II arrest due to buildup of torsional stress. c. R-loops can promote the production of double-stranded RNA that recruits the RNase Dicer and the histone methyltransferase G9a, which forms a repressive chromatin environment that promotes Pol II pausing. Conversely, R-loop recognition and dissolving enzymes such as BRCA1, the RNA helicases senataxin (SETX), DHX9 and DDX5, and 5'-3' exoribonuclease 2 (XRN2) are also required for proper transcription termination at certain genes.
More recent work revealed a molecular link between R-loops and termination-promoting heterochromatinization48,64 (Fig. 4c). The work provides evidence for a model where G-rich sequences promote R-loop formation and synthesis of antisense RNAs, which somehow forms double-stranded RNA over the termination region. Exactly how dsRNA is arranged within the R-loops is unknown, but it attracts the RNA interference RNase Dicer and the histone lysine methyltransferase G9a (also known as EHMT2) by unknown mechanisms48,64. The local deposition of H3K9me2 by G9 was proposed to promote the formation of a repressive chromatin environment (heterochromatin), which reinforces Pol II pausing and transcription termination. Inconsistent with this model is genome-wide R-loop mapping, which showed that R-loops at gene 3’ ends are in open chromatin regions enriched with gene activating histone modifications compared with matched gene 3’ ends devoid of R-loops 12. R-loops at gene termini may also recruit specific termination proteins such as the transcription elongation factor PAF1C, although the mechanism of recruitment is unknown. Recently, RNA N6-methyladenosine (m6A) was suggested to promote R-loop formation to facilitate transcription termination85. Produced by a methyltransferase complex containing the catalytic subunit METTL3, m6A is the most abundant reversible RNA modification. METTL3 depletion enhances read-through in m6A-containing protein-coding transcripts85. A recent report has demonstrated that the m6A modification by METTL3 is abundant at R-loops and demonstrated that its recognition by the m6A reader YTHDF2, crucial for RNA–DNA hybrid removal86. It will be important to reconcile how m6A may promote RNA–DNA hybrid formation, while at the same time enhance hybrid removal. Overall, the m6A modification may be important for defining regulatory R-loops and hence influencing transcription termination.
In contrast to the studies discussed above, deficiency in R-loop dissolving enzymes, including in the RNA helicases senataxin, DHX9 or DDX5 or in 5'-3' exoribonuclease 2 induces global R-loop accumulation and impairs transcription termination at certain genes 48,75,87,88 transcribed by Pol II or even by Pol III 89. Whether these enzymes directly bind and process the sites where their deficiency induces R-loops or whether enzyme deficiency acts indirectly remains to be demonstrated (Fig. 4c). It is also unclear what the consequences of accumulated R-loops are on transcription termination and read-through activity on a genome-wide level. These studies emphasize how crucial it is to strike the perfect R-loop balance, because both a failure to form and a failure to remove R-loops can affect a biological process in a similar manner.
Promoting genome integrity
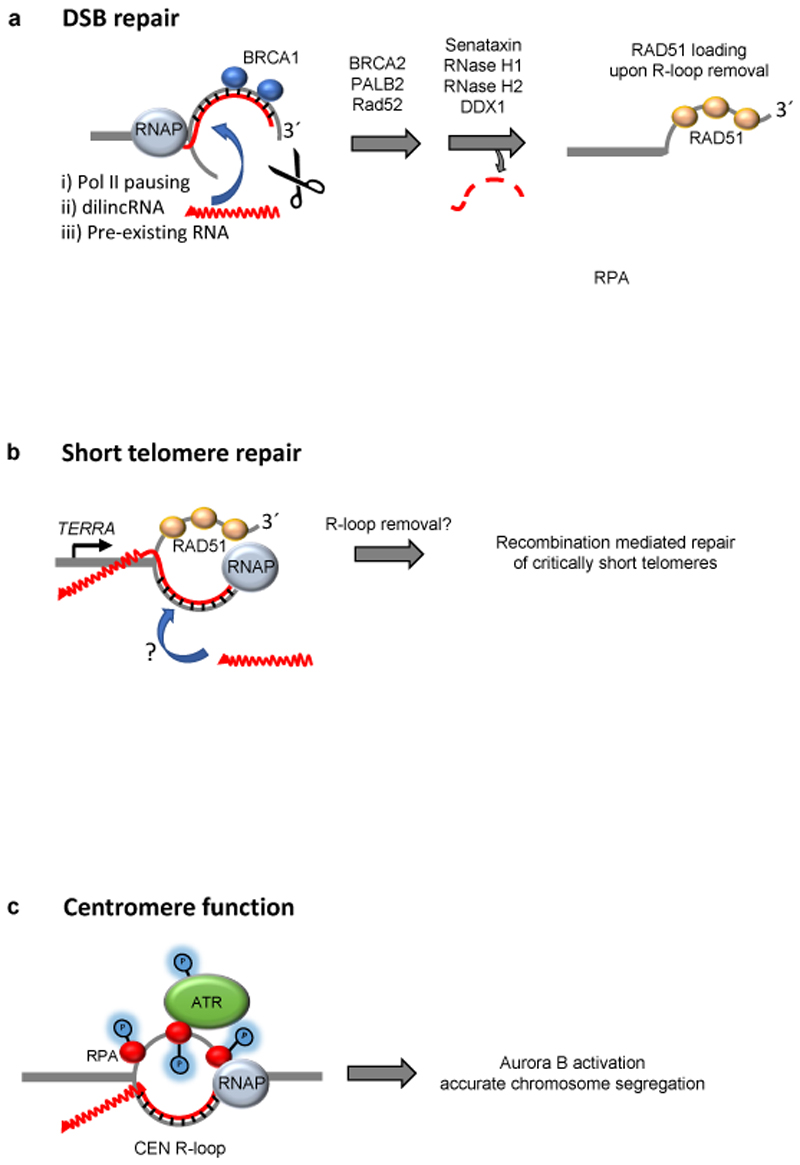
The initial perception of RNA–DNA hybrids was that of drivers of DNA damage and hence of genome instability10,35,90–93. This perception was based on observations made in conditions where hybrid prevention or removal was compromised. It was striking how the misregulation of RNA–DNA hybrids frequently leads to a considerable increase in the rates unscheduled recombination repair and in the number of cellular repair foci harbouring recombination factors. However, recent work has demonstrated that exactly these recombination supproting properties of R-loops can be channeled to promote DNA repair at breaks and at dysfunctional telomeres. R-loops have also been implicated in the establishment of centromere function to ensure accurate chromosome segregation, which is crucial for maintaining a stable genome. RNA– DNA hybrids at telomeres and centromeres form R-loops, however, for accuracy we will only refer to RNA–DNA hybrids at sites of DNA damage, as it is not clear that three-stranded R-loops are formed at these sites (Fig. 5).
Figure 5. RNA–DNA hybrids can promote genome stability.
a. RNA–DNA hybrids at DNA double-stranded breaks (DSBs) may form from transcription stalling within a gene (not shown); from de novo transcription from a free 3´end at the break site, as in the case of damage induced long noncoding RNA (dilncRNA); or alternatively a pre-existing transcript may associate with the newly resected strand in trans. The hybrid attracts BRCA1 and eventually other repair factors (BRCA2, PALB2, XPG (not shown)). The hybrid is then removed by either senataxin, RNase H1, RNase H2 or DDX1 or DDX1 to allow RAD51 loading and homologous recombination (HR). b. At short telomeres, the accumulation of telomere repeat-containing RNA (TERRA) R-loops promotes repair through RAD51-mediated homology-dependent repair. It is uncertain whether TERRA can form R-loops in trans at short telomeres, and the R-loop removal pathway has not been identified c. The post-replicative formation of R-loops at centromeres leads to replication protein A (RPA)-dependent recruitment of the kinase ATR. ATR catalyses multiple phosphorylation events and stimulates Aurora B activity, which promotes accurate microtubule attachment to kinetochores and chromosome segregation. Pol II, RNA polymerase II.
RNA–DNA hybrids can promote DNA repair in transcribed regions
When DNA single-stranded breaks (nicks) or double-stranded breaks (DSBs) are formed in regions of active transcription, multiple repair pathways converge to ensure that transcription in the immediate vicinity of the break is down-regulated. These include the checkpoint-mediated removal94 and pausing95,96 of Pol II; the latter can also result in R-loop stabilization. Additionally, the local chromatin structure is altered to a more repressive state around damage sites to ensure a decrease in transcription through a myriad of gene-repressive chromatin changes occur around break sites96–99. It is interesting that both PRC1 and PRC2, which localize to some promoters in an R-loop dependent manner (Fig. 3a), also localize to breaks and induce transcriptional silencing through H2AK119 ubiquitylation 100–102.
Transcription repression may, at first glance, appear incompatible with R-loop presence as high transcription rates are typically correlated with R-loops, however it is becoming clear that RNA–DNA hybrids do indeed accumulate at a subset of break sites, particularly in transcribed regions of the genome, where they actively contribute to DNA repair. The co-localization of hybrids to some breaks and sites of DNA damage has been demonstrated using different RNA– DNA hybrid detection techniques and multiple different sources of DNA damage 103–108. Although the pausing of Pol II at sites of damaged DNA may be responsible for the local accumulation of R-loops107,9,29,109, it has also been proposed that at sites if DNA damage, de novo transcription from the 3´end of the resected strand 105,108,110, which gives rise to damage induced long noncoding RNA (dilncRNA) 108, or the annealing of pre-existing transcripts from the same locus 30,106,107 may contribute to hybrid accumulation (Fig. 5a). The proposed sources of RNA–DNA hybrids at DNA breaks — stalled transcription, de novo transcription, and transcript re-annealing — are not mutually exclusive and the source of the hybrid may be context dependent, for example in terms of sequence, cell cycle and/or transcriptional activity. Although the source of hybrids at sites of DNA damage remains unresolved (reviewed in REF. 110), their transient presence is clearly important for repair111. It is important to emphasize that hybrids are not a universal feature of all DSBs and there are clear examples of site-specific breaks that are not associated with RNA–DNA hybrids 112.
R-loops appear to accumulate on both sides of a DSB, as assayed by DRIP-seq or DRIP-qPCR105,107,108. Breast cancer type 1 susceptibility protein (BRCA1), which is crucial for homologous recombination, is able to bind directly to RNA–DNA hybrids in vitro 108. Consistently, formation of BRCA1 foci following ionizing radiation (which generates DSBs) is strongly reduced when RNase H1 is overexpressed and RNA–DNA hybrids fail to form 108. BRCA1 then recruits BRCA2 and PALB2 (partner and localizer of BRCA2) to the DSB, which has the combined effect of attracting RNase H2 in order to remove the RNA from the hybrid, and stimulate the subsequent loading of RAD51 onto the newly exposed ssDNA to begin homology search (Fig. 5a). Although the factors involved may differ depending on the context of repair, the sequence of events seems to be consistently played out: following 5´-end resection at the DSB, an RNA–DNA hybrid forms at the exposed 3´end, which recruits repair factors such as RAD52103, XPG103, BRCA1108 and BRCA2108; the RNA is subsequently removed by either senataxin107, RNaseH1105,113, RNaseH2105,108 or DDX1106, thereby allowing RAD51107,108,106 loading and HR. The presence of RNA–DNA hybrids at DSBs exemplifies the importance of a transient hybrid that performs its function, but is subsequently evicted to avoid potential problems. It is interesting to speculate about why such a process would be implemented, when RAD51 could be just as easily loaded directly onto ssDNA, without the need for an RNA–DNA hybrid intermediate. It is possible that an ’R-loop-first approach’ at breaks allows the cell to distinguish between exposed ssDNA that occurs during DNA replication (where RAD51 and HR would be detrimental) and ssDNA generated at DSBs, where RAD51 is essential for repair. Alternatively, or additionally, an RNA–DNA hybrid may help to ensure that RPA does not outcompete RAD51 at a DSB.
Regulatory R-loops promote telomere repair
Another example of RNA–DNA hybrids driving DNA repair is found at telomeres. The telomere repeat-containing RNA (TERRA) has a strong tendency to form R-loops at chromosome ends 36,114–116. This is likely due to its repetitive, G-rich sequence, which may potentiate R-loops by the formation of G4 structures on the displaced DNA strand117. TERRA (and the R-loops at yeast telomeres) are unstable, being formed in the early S phase and degraded by the RNase H enzymes approximately at the time of telomere replication, in late S phase118,119. Therefore, similar to the bulk of R-loops, TERRA R-loops are also transient. They are stabilized, however, when telomeres become critically short through breakage or progressive shortening through replication, and RNase H enzymes no longer efficiently localize to the telomeres. In yeast, stabilized R-loops at a single shortened telomere promote recombination with the long telomeres and this has the outcome of preventing the onset of premature senescence (Fig. 5b). When short telomeres are experimentally prevented from R-loop stabilization — either through RNase H overexpression or TERRA ablation — recombination impairment ensues, which results in the accelerated onset of senescence114.
Telomeric R-loops also have a role in human cancer cells that exclusively rely on the recombination mechanism known as alternative lengthening of telomeres (ALT) to maintain their telomeres and achieve immortality40,115. Therefore, as is the case with DSB repair, the presence of R-loops at short telomeres drives recombination. One important difference, however, between telomeres and DSBs is the DNA strand that is engaged in the hybrid. Whereas at DSBs, the 3´overhang base-pairs with RNA (Fig. 5a), it is the 5´-end strand that hybridizes with TERRA at telomeres (Fig. 5b). In both cases, the presence of an R-loop facilitates RAD51 loading 107,108,118. Live cell imaging has revealed that TERRA has the ability to leave and subsequently return to the specific telomere were it was transcribed120. It will be interesting to determine whether short telomeres are better acceptors of TERRA acting in cis or in trans, whether they are inefficient resolvers of TERRA, or both. Similar to DSBs, it is presumed that the hybrid at short telomeres must eventually be removed to allow re-annealing of the newly extended strand. Indeed in yeast, the over-expression of the RNA annealing protein Yra1 stabilizes R-loops at short telomeres and leads to accelerated, rather than attenuated, replicative senescence (irreversible cell cycle arrest triggered by telomere attrition) 121. Whereas RNase H2 is responsible for R-loop removal at telomeres of normal length in a cell cycle dependent manner118, it remains to be determined how hybrids are eventually resolved at short telomeres that are subject to repair. In summary, telomeric R-loops are also subject to a Goldilocks-like effect, in which just the right amount of R-loops should be present in order to achieve a productive, and not destructive, outcome.
R-loops ensure centromere function
Centromeric loci are transcribed into centromeric RNA (CEN-RNA) and also harbour R-loops (CEN R-loops), which are regulated in a cell cycle dependent manner, a feature conserved from yeast to man122,123. In human cells, the displaced ssDNA at the CEN R-loop is bound by RPA and recruits the DNA damage response kinase ATR, leading to the activation of Aurora B, which promotes correct microtubule–kinetochore attachments (Fig. 5c). Accordingly, RNase H overexpression prevents ATR localization and hence full Aurora B activation, and leads to increase in the number of anaphases with lagging chromosomes, and. Importantly, in both yeast and human cells, CEN R-loops are formed only after DNA replication to prevent replication conflicts and are only present when required, that is, during mitosis. In both yeast and humans it has been shown that an increase in CEN R-loop levels leads to local chromatin compaction83,65,, and in the fission yeast Schizosaccharomyces pombe CEN R-loops stimulate the formation of RNAi-induced heterochromatin62. It is interesting to note that ATR and the related kinase ATM also transiently associate with telomeres in a cell-cycle regulated manner to ensure telomere functionality124. It will be interesting to determine whether R-loops provoke their recruitment as is the case at centromeres.
Regulating regulatory R-loops?
A key question is how the potentially detrimental effects of regulatory R-loops are constrained. For example, how can regulatory R-loops activate gene expression if they might stall Pol II? How can R-loops promote DNA repair, when they are themselves considered a source of DNA damage? We now understand that some detrimental repercussions of unscheduled RNA–DNA hybrids, that is hyper-recombination and transcription stalling, can be harnessed to promote essential cellular functions such as DNA repair and transcription termination, respectively. Therefore, RNA–DNA hybrid regulation is crucial as the hybrids should form at the right place and time to allow certain regulatory processes to occur without deleterious consequences.
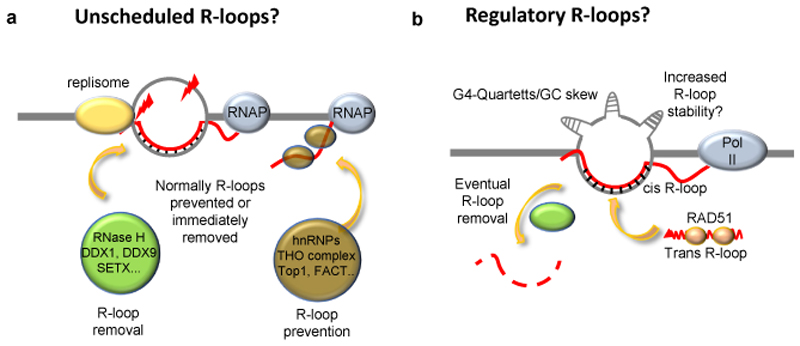
So how might such regulation be established? Firstly, R-loops that regulate gene expression could be spatially separated from the TSSs of the genes they regulate., for example R-loops regulating upstream genetic elements may avoid interference with transcription of the regulated genes. Accordingly, the GATA3-AS1 lncRNA forms a regulatory R-loop ~2kb upstream of the GATA3 TSS 69 (Fig. 3c). Second, when located at promoters and within gene bodies, the decoding (reading, interpreting and eventual removal) of regulatory R-loops could be temporally separated from transcription of the genes, for example during the cell cycle. R-loop levels change during the cell cycle65,125 and RNase H2 is tightly cell-cycle regulated126. Indeed, in the case of TCF21, transcription of its antisense lncRNA TARID, R-loop accumulation, DNA demethylation and TCF21 expression proceed sequentially during the cell cycle 71 (Fig. 3f). Such a sequence may avoid simultaneous R-loop formation at promoters, and at the coding strand by the regulatory antisense lncRNA, or at the non-coding strand by the transcribed mRNA, so that downstream processes could be initiated through R-loops, while the detrimental effects of R-loops would be avoided through their rapid removal. However, in postmitotic cells, temporal separation between the decoding of regulatory R-loops and gene transcription would require cell-cycle-independent modes of temporal separation. In such instances it would be beneficial if regulatory R-loops were only transient in nature. R-loops associated with DNA repair provide examples of such regulation. At DSBs and shortened telomeres, an R-loop is formed just long enough to recruit the repair machinery and is then removed. This regulation appears to be a key feature as both the hyper-stabilization and the absence of R-loops can hinder the DNA repair process 105,108,118,121. Therefore, similarly to unscheduled R-loops, regulatory R-loops are likely to be short-lived and tightly regulated by endonucleases and helicases to avoid having detrimental effects (Fig. 6).
Figure 6. Unscheduled vs. regulatory R-loops – a model.
a. We suggest that in normal conditions, potential sites of unscheduled-R-loop formation are free of RNA–DNA hybrids owing to either efficient hybrid prevention by RNA binding proteins, or to efficient R-loop removal. In the absence of R-loop prevention and removal, the dwell time of an R-loop increases, which can lead to replication stress, or to DNA damage on the, now vulnerable, displaced strand. Examples of proteins and protein complexes involved in R-loop removal or prevention are depicted. b. Regulatory R-loops may be associated with genomic features that allow R-loops to form more readily and with increased stability. Such features could include formation of stable G-quadruplexes (G4) on the non-template strand, GC skew, and a favourable chromatin environment. These features may also promote the formation of R-loops by trans-acting RNAs bound by RAD51, which promotes strand invasion at homologous sequences. Regulatory R-loops must also be resolved in order to prevent the very same problems that are associated with unscheduled R-loops. In the absence of R-loop removal factors, regulatory R-loops could be converted into unscheduled R-loops. DDX, DEAD-Box helicase; FACT, facilitates chromatin transcription; hnRNP, heterogeneous nuclear ribonucleoprotein; SETX, Senataxin; THO, suppressors of transcription defects of Hpr1 by overexpression, TOP1, Topoisomerase 1.
The prevailing model for how unscheduled R-loops form is by co-transcriptional invasion of the duplex DNA at the site of transcription by the nascent transcript in cis 6 (Fig. 1). The presence of unscheduled R-loops is likely related to defects in R-loop prevention or removal (Fig. 6). By contrast, a recurring theme of regulatory R-loops is the ability of lncRNAs to form in trans. Therefore hybrid formation may not require co-transcription from the same locus. An additional feature of regulatory R-loops is that the trans acting RNAs may contribute to the longevity of regulatory R-loops through continued de novo formation, which raises the question of how the lncRNA threads into the DNA duplex. Although RNA helicases may facilitate RNA threading, ATPase activity may not be required, as exemplified by the CRISPR–Cas system, which can cause DNA bending that leads to DNA unwinding and threading of the guide RNA 127. In vitro, RNA–DNA hybrids can form post-transcriptionally in trans with the aid of the ssDNA-binding protein RecA, which is the bacterial homologue of RAD51 128,129. In vivo, RAD51 is required for R-loop formation in trans 30 and may act analogously to CRISPR–Cas to facilitate hybrid formation 127 (Fig. 1). Thus, local unwinding of the DNA duplex may be sufficient to nucleate self-propagating R-loops in trans, notably in negatively supercoiled DNA, where hybrids relax this high-energy state 130. Certain DNA sequences or structures may also facilitate RNA threading at regulatory R-loops. One prominent DNA structure that may stabilise the unwound DNA is intramolecular G4s, which are prevalent in R-loops and, upon stabilization, increase R-loop levels 46,131. This suggests that G-rich lncRNAs may form R-loops in trans by threading into sites where G4 structures form and basepairing with the C-rich strand (Fig. 6).
Finally, it will be interesting to see how m6A contributes to unscheduled and to regulatory RNA–DNA hybrids. Although m6A was shown to promote hybrid formation85, it can also stimulated hybrid removal and prevent R-loop-mediated genotoxic stress86.
Conclusion
The fact that (at least some) regulatory R-loops function in trans, opens up the possibility that the expression of R-loop-regulated genes or the elongation of critically short telomeres, may be manipulated using RNA oligonucleotides. Transfection of R-loop-forming RNAs may be used to control gene expression, for example by targeting promoters exhibiting GC-skew. Conversely, RNA oligonucleotides complementary to endogenous R-loop RNAs could be used to prevent R-loop formation. Therefore, the regulation of regulatory R-loops could be potentially harnessed for therapeutic purposes.
Acknowledgements
We apologize to colleagues whose work could not be cited owing to space constraints. We thank Regina Otto for help with figures, Olga Vydzhak and Natalie Schindler for feedback on the manuscript. C.N. acknowledges support by an ERC Advanced Grant (HybReader) and B.L acknowledges support from the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) Heisenberg Program - LU 1709/2-1. Both laboratories are funded by the DFG, project number 393547839 – SFB 1361.
Footnotes
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Molecular Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
ToC
R-loops (three-stranded RNA–DNA structures) are often associated with transcription defects, DNA damage and genome instability, but ‘regulatory’ R-loops can promote gene regulation, telomere stability and DNA repair. This dual functionality of R-loops requires tight control of their formation, location and timely removal.
References
- 1.Morris KV, Mattick JS. The rise of regulatory RNA. Nature reviews Genetics. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nature reviews Genetics. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Muse T, Aguilera A. R Loops: From Physiological to Pathological Roles. Cell. 2019;179:604–618. doi: 10.1016/j.cell.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 4.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends in cell biology. 2015;25:514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Molecular cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Hamperl S, Cimprich KA. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang KS, et al. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell. 2017;170:787–799 e718. doi: 10.1016/j.cell.2017.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170:774–786 e719. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Cristini A, et al. Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks. Cell reports. 2019;28:3167–3181 e3166. doi: 10.1016/j.celrep.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz LA, et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Molecular cell. 2016;63:167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belotserkovskii BP, Tornaletti S, D'Souza AD, Hanawalt PC. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA repair. 2018;71:69–81. doi: 10.1016/j.dnarep.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freudenreich CH. R-loops: targets for nuclease cleavage and repeat instability. Current genetics. 2018;64:789–794. doi: 10.1007/s00294-018-0806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard P, Manley JL. R Loops and Links to Human Disease. Journal of molecular biology. 2017;429:3168–3180. doi: 10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes & development. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costantino L, Koshland D. The Yin and Yang of R-loop biology. Current opinion in cell biology. 2015;34:39–45. doi: 10.1016/j.ceb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilera A, Gomez-Gonzalez B. DNA-RNA hybrids: the risks of DNA breakage during transcription. Nature structural & molecular biology. 2017;24:439–443. doi: 10.1038/nsmb.3395. [DOI] [PubMed] [Google Scholar]
- 19.Stodola JL, Burgers PM. Mechanism of Lagging-Strand DNA Replication in Eukaryotes. Advances in experimental medicine and biology. 2017;1042:117–133. doi: 10.1007/978-981-10-6955-0_6. [DOI] [PubMed] [Google Scholar]
- 20.Burgers PM. Solution to the 50-year-old Okazaki-fragment problem. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:3358–3360. doi: 10.1073/pnas.1900372116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugino A, Hirose S, Okazaki R. RNA-linked nascent DNA fragments in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:1863–1867. doi: 10.1073/pnas.69.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 23.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA repair. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Molecular cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Molecular cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryor JM, et al. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science. 2018;361:1126–1129. doi: 10.1126/science.aat2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Molecular cell. 2017;68:745–757 e745. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boguslawski SJ, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. Journal of immunological methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 32.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Molecular cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatia V, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 34.Vanoosthuyse V. Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops. Noncoding RNA. 2018;4 doi: 10.3390/ncrna4020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Molecular cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan YA, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS genetics. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes & development. 2016;30:1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS genetics. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, et al. The R-loop is a common chromatin feature of the Arabidopsis genome. Nature plants. 2017;3:704–714. doi: 10.1038/s41477-017-0004-x. [DOI] [PubMed] [Google Scholar]
- 40.Arora R, et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nature communications. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadel J, et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics & chromatin. 2015;8:46. doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome research. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dumelie JG, Jaffrey SR. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. eLife. 2017;6 doi: 10.7554/eLife.28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratmeyer L, Vinayak R, Zhong YY, Zon G, Wilson WD. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 45.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 46.De Magis A, et al. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrasco-Salas Y, et al. The extruded non-template strand determines the architecture of R-loops. Nucleic acids research. 2019;47:6783–6795. doi: 10.1093/nar/gkz341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Molecular cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao DY, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529:48–53. doi: 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]
- 50.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 51.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 52.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature immunology. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 55.Reaban ME, Griffin JA. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- 56.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 57.Briggs E, Crouch K, Lemgruber L, Lapsley C, McCulloch R. Ribonuclease H1-targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion. PLoS genetics. 2018;14:e1007729. doi: 10.1371/journal.pgen.1007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & development. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conn VM, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nature plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 61.Powell WT, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes to cells : devoted to molecular & cellular mechanisms. 2012;17:218–233. doi: 10.1111/j.1365-2443.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- 63.Groh M, Lufino MM, Wade-Martins R, Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS genetics. 2014;10:e1004318. doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castellano-Pozo M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Molecular cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Beckedorff FC, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS genetics. 2013;9:e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boque-Sastre R, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:5785–5790. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cloutier SC, et al. Regulated Formation of lncRNA-DNA Hybrids Enables Faster Transcriptional Induction and Environmental Adaptation. Molecular cell. 2016;61:393–404. doi: 10.1016/j.molcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibbons HR, et al. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Frontiers in immunology. 2018;9:2512. doi: 10.3389/fimmu.2018.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan-Wong SM, Dhir S, Proudfoot NJ. R-Loops Promote Antisense Transcription across the Mammalian Genome. Molecular cell. 2019 doi: 10.1016/j.molcel.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arab K, et al. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nature genetics. 2019;51:217–223. doi: 10.1038/s41588-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grunseich C, et al. Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Molecular cell. 2018;69:426–437.e427. doi: 10.1016/j.molcel.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nature structural & molecular biology. 2015;22:999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skourti-Stathaki K, et al. R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes. Molecular cell. 2019;73:930–945.e934. doi: 10.1016/j.molcel.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell reports. 2018;23:1891–1905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang IX, et al. Human proteins that interact with RNA/DNA hybrids. Genome research. 2018;28:1405–1414. doi: 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen HD, et al. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Molecular cell. 2017;65:832–847 e834. doi: 10.1016/j.molcel.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan W, et al. ALBA protein complex reads genic R-loops to maintain genome stability in Arabidopsis. Sci Adv. 2019;5:eaav9040. doi: 10.1126/sciadv.aav9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arab K, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Molecular cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 80.Kienhofer S, et al. GADD45a physically and functionally interacts with TET1. Differentiation; research in biological diversity. 2015;90:59–68. doi: 10.1016/j.diff.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z, et al. Gadd45a promotes DNA demethylation through TDG. Nucleic acids research. 2015;43:3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352:aad9926. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kireeva ML, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. Journal of molecular biology. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 84.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X, et al. m(6)A promotes R-loop formation to facilitate transcription termination. Cell research. 2019 doi: 10.1038/s41422-019-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abakir A, et al. N6-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nature genetics. doi: 10.1038/s41588-019-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morales JC, et al. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS genetics. 2016;12:e1006107. doi: 10.1371/journal.pgen.1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mersaoui SY, et al. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. The EMBO journal. 2019;38:e100986. doi: 10.15252/embj.2018100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rivosecchi J, et al. Senataxin homologue Sen1 is required for efficient termination of RNA polymerase III transcription. The EMBO journal. 2019:e101955. doi: 10.15252/embj.2019101955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 91.Stirling PC, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes & development. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Costantino L, Koshland D. Genome-wide Map of R-Loop-Induced Damage Reveals How a Subset of R-Loops Contributes to Genomic Instability. Molecular cell. 2018;71:487–497 e483. doi: 10.1016/j.molcel.2018.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mischo HE, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Molecular cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. S1097-2765(10)00963-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nature structural & molecular biology. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 95.Awwad SW, Abu-Zhayia ER, Guttmann-Raviv N, Ayoub N. NELF-E is recruited to DNA double-strand break sites to promote transcriptional repression and repair. EMBO reports. 2017;18:745–764. doi: 10.15252/embr.201643191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. The Journal of cell biology. 2017;216:1959–1974. doi: 10.1083/jcb.201611135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Savitsky P, et al. Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin. Cell reports. 2016;17:2724–2737. doi: 10.1016/j.celrep.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rona G, et al. PARP1-dependent recruitment of the FBXL10-RNF68-RNF2 ubiquitin ligase to sites of DNA damage controls H2A.Z loading. eLife. 2018;7 doi: 10.7554/eLife.38771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle. 2013;12:2675–2683. doi: 10.4161/cc.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ginjala V, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Molecular and cellular biology. 2011;31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. The Journal of cell biology. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yasuhara T, et al. Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell. 2018;175:558–570 e511. doi: 10.1016/j.cell.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 104.Teng Y, et al. ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nature communications. 2018;9:4115. doi: 10.1038/s41467-018-06586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohle C, et al. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell. 2016;167:1001–1013 e1007. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 106.Li L, et al. DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Molecular and cellular biology. 2016;36:2794–2810. doi: 10.1128/MCB.00415-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cohen S, et al. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nature communications. 2018;9:533. doi: 10.1038/s41467-018-02894-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.D'Alessandro G, et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nature communications. 2018;9:5376. doi: 10.1038/s41467-018-07799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Allison DF, Wang GG. R-loops: formation, function, and relevance to cell stress. Cell Stress. 2019;3:38–46. doi: 10.15698/cst2019.02.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Michelini F, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nature cell biology. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Puget N, Miller KM, Legube G. Non-canonical DNA/RNA structures during Transcription-Coupled Double-Strand Break Repair: Roadblocks or Bona fide repair intermediates? DNA repair. 2019:102661. doi: 10.1016/j.dnarep.2019.102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao H, Zhu M, Limbo O, Russell P. RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO reports. 2018;19 doi: 10.15252/embr.201745335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Britton S, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic acids research. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Balk B, et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nature structural & molecular biology. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 115.Arora R, Azzalin CM. Telomere elongation chooses TERRA ALTernatives. RNA biology. 2015 doi: 10.1080/15476286.2015.1065374. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. The EMBO journal. 2013;32:2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rippe K, Luke B. TERRA and the state of the telomere. Nature structural & molecular biology. 2015;22:853–858. doi: 10.1038/nsmb.3078. [DOI] [PubMed] [Google Scholar]
- 118.Graf M, et al. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell. 2017;170:72–85 e14. doi: 10.1016/j.cell.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 119.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Molecular and cellular biology. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. MCB.00460-10 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cusanelli E, Romero CA, Chartrand P. Telomeric Noncoding RNA TERRA Is Induced by Telomere Shortening to Nucleate Telomerase Molecules at Short Telomeres. Molecular cell. 2013;51:780–791. doi: 10.1016/j.molcel.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 121.Garcia-Rubio M, et al. Yra1-bound RNA-DNA hybrids cause orientation-independent transcription-replication collisions and telomere instability. Genes & development. 2018;32:965–977. doi: 10.1101/gad.311274.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kabeche L, Nguyen HD, Buisson R, Zou L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 2018;359:108–114. doi: 10.1126/science.aan6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen CF, Pohl TJ, Chan A, Slocum JS, Zakian VA. Saccharomyces cerevisiae Centromere RNA Is Negatively Regulated by Cbf1 and Its Unscheduled Synthesis Impacts CenH3 Binding. Genetics. 2019 doi: 10.1534/genetics.119.302528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 125.Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes & development. 2014;28:735–748. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lockhart A, et al. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell reports. 2019;29:2890–2900 e2895. doi: 10.1016/j.celrep.2019.10.108. [DOI] [PubMed] [Google Scholar]
- 127.Xiao Y, et al. Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System. Cell. 2017;170:48–60 e11. doi: 10.1016/j.cell.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kasahara M, Clikeman JA, Bates DB, Kogoma T. RecA protein-dependent R-loop formation in vitro. Genes & development. 2000;14:360–365. [PMC free article] [PubMed] [Google Scholar]
- 129.Zaitsev EN, Kowalczykowski SC. A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes & development. 2000;14:740–749. [PMC free article] [PubMed] [Google Scholar]
- 130.Stolz R, et al. Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:6260–6269. doi: 10.1073/pnas.1819476116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuznetsov VA, Bondarenko V, Wongsurawat T, Yenamandra SP, Jenjaroenpun P. Toward predictive R-loop computational biology: genome-scale prediction of R-loops reveals their association with complex promoter structures, G-quadruplexes and transcriptionally active enhancers. Nucleic acids research. 2018;46:7566–7585. doi: 10.1093/nar/gky554. [DOI] [PMC free article] [PubMed] [Google Scholar]