Abstract
Targeted protein degradation is a new therapeutic paradigm based on drugs that destabilize proteins by inducing their proximity to E3 ubiquitin ligases. Of particular interest are molecular glues that can degrade otherwise unligandable proteins by orchestrating direct interactions between target and ligase. However, their discovery has so far been serendipitous, thus hampering broad translational efforts. Here, we describe a scalable strategy towards glue degrader discovery that is based on chemical screening in hypo-neddylated cells coupled to a multi-omics target deconvolution campaign. This approach led us to identify compounds that induce ubiquitination and degradation of cyclin K by prompting an interaction of CDK12:cyclin K with a CRL4B ligase complex. Notably, this interaction is independent of a dedicated substrate receptor, thus functionally segregating this mechanism from all described degraders. Collectively, our data outline a versatile and broadly applicable strategy to identify degraders with non-obvious mechanisms and thus empower future drug discovery efforts.
A dependency on traditional antagonistic/agonistic pharmacologic concepts has limited the reach of small-molecule strategies. This in turn has contributed to a stagnation in therapeutic innovation caused by an inability to pursue some of the best-validated targets in life-threatening diseases such as cancer. A promising alternative to pharmacologic modulation of protein activity is to regulate the abundance of disease-causing proteins, for instance by chemically redirecting them to E3 ubiquitin ligases for degradation. A strong emphasis is currently placed on heterobifunctional degraders, often referred to as PROTACs (PROteolysis TArgeting Chimeras), which simultaneously bind target and ligase with dedicated warheads that are connected by a flexible linker1–6. While this modular design principle allows for a straightforward alteration of the degraded neosubstrate by switching the respective warhead, the overall proteomic space that is degradable via PROTACs is intrinsically limited to target proteins that can effectively be liganded via small organic molecules.
Conversely, molecular glue degraders were shown to degrade unligandable proteins. Mechanistic characterization of the clinically approved thalidomide analogs (IMiDs) as molecular glue degraders exemplify how this particular molecular pharmacology can expand the therapeutically actionable space in the human proteome7. Binding of IMiDs to the CRL4CRBN E3 ligase causes recruitment of selected zinc finger transcription factors (TFs), leading to their ubiquitination and subsequent proteasomal degradation8–11. Noteworthy, IMiDs have per se no measurable binding affinity to the degraded TFs. However, they orchestrate molecular recognition between ligase and TF by inducing several protein-protein interactions proximal to the binding interface. Mechanistically similar to IMiDs, certain aryl sulfonamides around the clinically tested compound indisulam have recently been shown to act as molecular glues between the CRL4DCAF15 ligase and the splicing factor RBM39, again causing the targeted degradation of the latter12–17.
In both cases, the molecular glue mechanism of action therefore enables the destabilization of target proteins otherwise considered unligandable and thus outside the reach of both traditional small-molecule inhibitors and likely also of heterobifunctional degraders. However, the discovery of molecular glue degraders has thus far unfortunately been serendipitous, and a lack of rational identification strategies has stymied its general applicability.
To address this limitation, we here describe a scalable strategy towards glue degrader discovery by comparative chemical screening in hypo-neddylated cellular models with broadly abrogated ligase activity. In a proof of concept, this led to the identification of a novel glue degrader that reprograms the CRL4DCAF15 ligase. Of note, this strategy also enabled the discovery of three small molecules that induce the destabilization of cyclin K. Integrating orthogonal functional genomics approaches with proteomics, drug-affinity chromatography and biochemical reconstitution experiments revealed an unprecedented mechanism of action that converges on a direct, drug-induced proximity between CDK12:cyclin K and DDB1 that causes ubiquitination and proteasomal degradation of cyclin K. Of note, drug-induced recruitment of CDK12:cyclin K is functionally independent of a dedicated substrate receptor, which functionally differentiates this mechanism from previously characterized glue degraders.
Results
Comparative drug profiling in hypo-neddylated cell lines
Currently known molecular glue degraders function by chemically redirecting cullin-RING ligases (CRLs). CRLs make up the largest family of E3 ubiquitin ligases and are diversified by modular assembly of substrate receptors (SRs) and adaptor proteins around different cullin backbones, leading to an overall complexity consisting of more than 250 CRLs. Small-molecule degraders typically function by rewiring CRL SRs such as CRBN or DCAF15. Intriguingly, CRL hijacking is a strategy of evolutionary dimensions, given that it is also employed by several viruses. In fact, certain viral proteins evolved to reprogram CRL activity independently of SRs by directly engaging other CRL components18. For instance, direct association of different viral proteins with the CRL4 adaptor protein DDB1 was shown to prompt the degradation of host proteins such as STAT transcription factors19–22.
Under physiological conditions, the activity of most CRLs is dependent on the reversible attachment of the ubiquitin-like protein NEDD8 to the respective cullin scaffold. NEDD8 conjugation is orchestrated by an E1 enzyme (NAE), two E2s (UBE2M and UBE2F) and several E3 enzymes. The neddylation E1 enzyme was identified in plants through a genetic screen of resistance to the molecular glue hormone auxin23. Similarly, we recently identified that CRISPR/Cas9-induced mutation of UBE2M causes resistance to known small-molecule degraders by inactivation of several CRLs including CRL4CRBN and CRL2VHL 24. Here, we set out to characterize the hypomorphic phenotype of the mutated UBE2M allele in the near-haploid KBM7 cells. Neddylation levels of most cullins were abrogated comparably to pharmacologic NAE inhibition with the exception of cullin 5, which is known to be predominantly neddylated by UBE2F25,26. No compensatory upregulation of UBE2F was detected, and levels of cullin neddylation could be restored upon genetic rescue via stable UBE2M cDNA expression (Fig. 1a). While pharmacologic NAE inhibition is toxic in KBM7 cells, UBE2Mmut cells did not show a significant proliferative defect despite the promiscuous impairment of CRL activity affecting the vast majority of the expressed ligases (Fig. 1b and Extended Data Fig. 1a-c). We hypothesized that comparative chemical profiling in hypo-neddylated versus neddylation-proficient cells would outline a strategy to screen for small molecules that are functionally linked to any of the >200 expressed CRLs. To this end, we screened a library of approximately 2,000 cytostatic/cytotoxic small molecules both in wildtype and UBE2Mmut KBM7 cells, which led us to identify several chemical scaffolds that appeared to be functionally dependent on uninterrupted neddylation levels (Fig. 1c and Supplementary Table 1). Dose-ranging validation that also extended to isogenic UBE2Mwt and UBE2Mmut HCT116 cells led us to prioritize four chemical scaffolds, dCeMM1 (1), dCeMM2 (2), dCeMM3 (3) and the closely related dCeMM3-1 (4), as well as dCeMM4 (5), for further mechanistic validation (Fig. 1d, e and Extended Data Fig. 1d-f). Of note, all four compounds were devoid of any prior functional annotation. To investigate the mechanism of action of dCeMM1, we conducted a focused CRISPR/Cas9 screen using a custom-designed sgRNA library covering all known CRLs and associated regulators (Supplementary Table 2 and Extended Data Fig. 1g). This confirmed the requirement on UBE2M, but most notably revealed a critical dependence of dCeMM1 efficacy on the CRL4DCAF15 ligase complex (Fig. 1f). This is in line with previous reports that certain serendipitously identified and structurally different sulfonamides can act as DCAF15 molecular glues capable of degrading the RRM containing proteins RBM39 and RBM2312,13,15–17. Coupling cellular dCeMM1 treatment with quantitative proteomics via isobaric tagging revealed that dCeMM1 exclusively destabilizes RBM39 (Fig. 1g and Supplementary Table 3). Further corroborating the mechanism as DCAF15 glue degrader, CRISPR-induced frameshift-mutations in DCAF15 rendered cells insensitive to dCeMM1 treatment and rescued RBM39 destabilization (Fig. 1h,i and Extended Data Fig. 1h). Collectively, this outlines that comparative drug profiling in hypo-neddylated cellular models enables the identification of novel molecular glue degraders that follow an established mechanism of action.
Fig.1. Screening in hypo-neddylated cells identifies novel molecular glue degraders rewiring the CRL4DCAF15 ligase.
a, Protein levels of UBE2M, UBE2F and neddylation status of different cullin backbones in WT, MLN4924-treated (1µM, 1h), UBE2M-deficient and UBE2M-reconstituted KBM7 cells. b, Substrate receptors expressed in KBM7 cells that associate with hypo-neddylated cullin scaffolds. c, Primary screening data comparing DMSO-normalized viability of WT and UBE2Mmut KBM7 cells treated with a (approximately) 2000 cytotoxic/cytostatic small-molecule library for 3 days. Doses tested were 10uM and 500nM. d, Chemical structure of the four prioritized chemical scaffolds (dCeMM1/2/3/4). dCeMM3-1 was structurally identical to dCeMM3 with a Br instead of the Cl. e, Dose-resolved, DMSO-normalized viability after 3-day dCeMM1/2/3/4 treatment in WT and UBE2Mmut KBM7 cells. Mean ± SEM; n=3 independent treatments. EC50s dCeMM1/2/3/4 (µM) WT=3;0,3;0.6;0.4 UBE2Mmut=8;4.2;10.7;7. f, CRL-focused CRISPR resistance screen for dCeMM1. Top: bubble plot displaying median sgRNA enrichment over DMSO, bubble size indicates significance. Bottom: sgRNAs enrichment targeting indicated genes, background indicates distribution of all sgRNAs. Results shown are the median of 2 independent screens. g, Expression proteomics after dCeMM1 treatment (25µM, 12h). h, DMSO-normalized viability after 3-day dCeMM1 treatment in WT and DCAF15mut KBM7 cells. Mean ± SEM; n=3 independent treatments. i, RBM39 levels in WT and DCAF15mut KBM7 cells after indisulam (IND) or dCeMM1 treatment.
Identification of structurally distinct cyclin K degraders
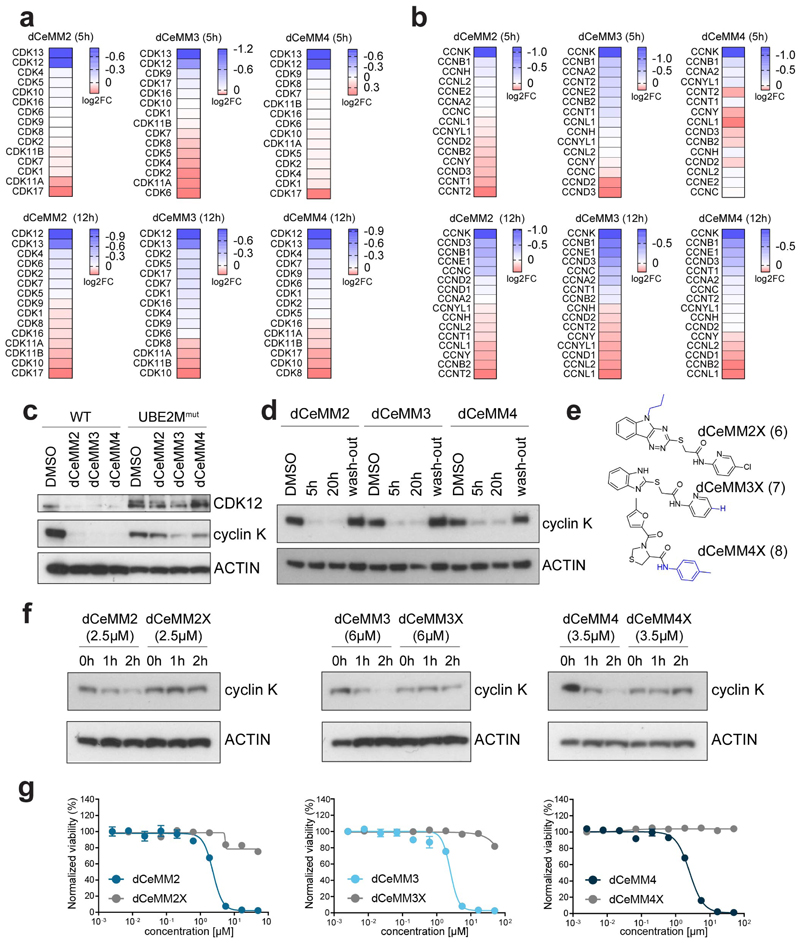
To characterize dCeMM2/3/4, we again coupled cellular treatment with quantitative expression proteomics. We found that all compounds lead to a pronounced destabilization of cyclin K alongside a milder destabilization of both associated kinases CDK12 and CDK13 (Fig. 2a and Supplementary Table 3). No other CDKs or cyclins were significantly destabilized (Extended Data Fig. 2a, b). We next validated cyclin K destabilization of all three scaffolds via time-resolved immunoblots. Near-complete cyclin K degradation was evident already after two hours (Fig. 2b) whereas levels of CDK12/13 appeared not acutely affected, but only destabilized upon prolonged drug incubation. Retained UBE2M dependency of CDK12/13 degradation however indicated that this delayed destabilization still occurs via a CRL-dependent mechanism that could be direct or indirect in nature (Extended Data Fig. 2c). Moreover, coupling cellular treatment with washout experiments revealed a reversible binding mode (Extended Data Fig. 2d) To develop a structure-activity relationship for the assayed scaffolds, we tested a total of 53 structurally related analogs in dose-ranging viability assays in KBM7 wildtype- and UBE2Mmut cells. This informed not only on sites amenable for derivatization of functionalized analogs, but also led us to identify structurally similar small molecules with no measurable activity that are thus ideally suited negative controls. Inactive analogs dCeMM2X (6), dCeMM3X (7) and dCeMM4X (8) fail to induce pronounced cyclin K destabilization and do not affect KBM7 cells in dose-ranging viability assays (Extended Data Fig. 2e-g). In line with a mechanism of targeted degradation, induced cyclin K destabilization was dependent on an active proteasome, could be rescued by pharmacologic NAE inhibition and by blocking the ubiquitin activating enzyme UBA1 (Fig. 2c).
Fig.2. dCeMM2/3/4 are novel and structurally different cyclin K degraders.
a, DMSO-normalized expression proteomics after 5h dCeMM2/3/4 treatment (2.5µM, 7µM, 3.5µM) in KBM7 cells. Limma statistical analysis was used. b Cyclin K levels upon dCeMM2/3/4 treatment in KBM7 cells. c, dCeMM2 (2.5µM, 5h) destabilizes cyclin K. 30min pretreatment with 1µM carfilzomib, 1µM MLN4924, 10µM TAK-243 or 1µM THZ531 rescues cyclin K destabilization. d, Recombinant kinase assays of dCeMM2/2X and THZ531 inhibition on enzymatic activity of CDK12/13/7. Mean ± SD n=2. e, dCeMM2/3/4 induce global transcriptional downregulation with phenotypic similarity to CDK12/13 inhibition by THZ531. Heatmap displays DMSO-normalized log2FC in gene expression for 27,051 transcripts, ranked by THZ531 log2FC. f, Gene set enrichment analysis of 984 genes (log2FC<-4 and adj. P-value<0.05) significantly downregulated upon THZ531 treatment in comparison to dCeMM2/3/4 (FDR<0.001). GSEA pre-ranked function was used (1000 permutations).
Of note, saturating the ATP binding pocket of CDK12/13 by treatment with the selective covalent CDK12/13 inhibitor THZ531 similarly rescued cyclin K destabilization27. This suggests that CDK12/13 active site engagement is required for the cyclin K destabilizing effect of the identified hit compounds (Fig. 2c). In line with this observation, recombinant kinase assays uncovered a measurable inhibitory activity of dCeMM2/3/4 on the enzymatic activity of CDK12/13 with a remarkable selectivity over CDK7. However, the 10-fold reduced potency compared to THZ531 likely doesn’t account for the strong cellular efficacy (Fig. 2d and Extended Data Fig. 3a). We next aimed to further corroborate CDK12/13:cyclin K as phenotypically relevant target of dCeMM2/3/4. To that end, we conducted mRNA-sequencing after five hours of cellular treatment with all identified small molecules and compared the associated, cell-count normalized transcriptional footprint with transcriptional profiles obtained after selective CDK12/13 inhibition with THZ531. Upon association with cyclin K, CDK12 and CDK13 are known to phosphorylate the carboxyterminal domain of RNA Pol II, licensing it for processive transcription elongation on a genome-wide scale28. In line with a functional impairment of CDK12/13:cyclin K complexes, we observed a global transcriptional downregulation following treatment with dCeMM2/3/4, as well as profound induction of apoptosis, without a phase-specific cell-cycle arrest (Fig. 2e, Extended Data Fig. 3b, c and Supplementary Table 4). Gene Set Enrichment Analysis further validated a profound phenotypic similarity between cellular treatment with dCeMM2/3/4 and selective pharmacologic inhibition of CDK12/13 (Fig. 2f and Extended Data Fig. 4a-c). Proteomics profiling at a later timepoint (12h) enabled functional enrichment analysis of the differentially expressed protein-protein interaction networks prompted by cyclin K degradation. In line with the proposed mechanism, “Regulation of cell cycle” and “RNA polymerase II CTD heptapeptide repeat kinase activity” were the major Gene Ontology biological processes/molecular functions affected (Extended Data Fig. 4d-e, and Supplementary Table 3). Collectively, intersection of quantitative proteomics and transcriptomics functionally validated cyclin K as a proteolytically destabilized target of dCeMM2/3/4. The convergence of all three scaffolds on the degradation of the same targets came unexpected given structural differences between dCeMM2/3 and dCeMM4.
Cyclin K degradation is mediated via CRL4B ligase complex
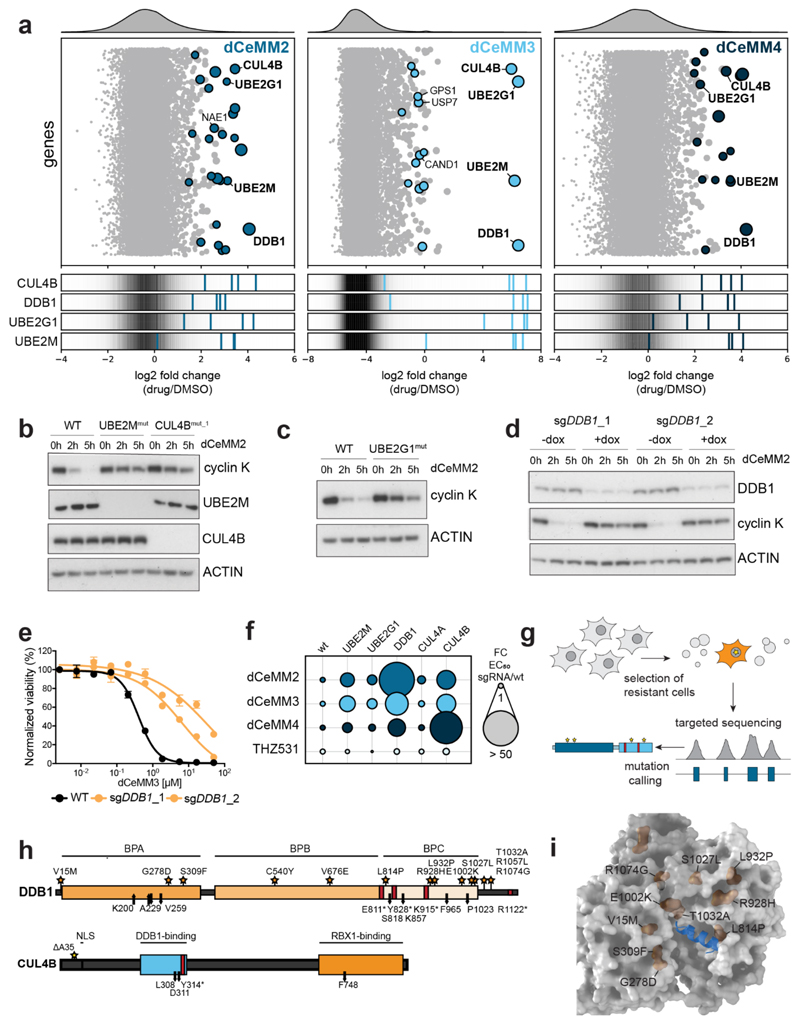
Next, we wanted to understand the molecular mechanism of drug-induced cyclin K destabilization and in particular identify the involved ubiquitin ligase complex. To that end, KBM7 cells were again subjected to CRISPR/Cas9-induced mutagenesis using the aforementioned CRL-focused sgRNA library (Extended Data Fig 5 and 6, Supplementary Table 2). Mutagenized cell pools were selected via continuous exposure to dCeMM2/3/4 to identify loss of function (LOF) mutations that allow clonal outgrowth and thus to identify genes that are functionally required for the anti-proliferative effects of the assayed compounds. In line with the initial chemical profiling in hypo-neddylated cells, we again identified UBE2M as hit in all tested conditions. Moreover, we identified mutations in three additional genes that constitute a partial CRL complex: the cullin scaffold CUL4B, the adaptor protein DDB1, and the E2 ubiquitin-conjugating enzyme UBE2G1. In addition to UBE2G1, known to extend ubiquitin chains, we also identified UBE2Z, an E2 conjugating enzyme known to prime ubiquitin chains, alongside the E1 ubiquitin-activating enzyme UBA6 (Extended Data Fig. 5a-c). Noteworthy, compared to previous profiling of the novel CRL4DCAF15 modulator dCeMM1 with the identical library, each of these screens failed to identify a consensus CRL SR. To exclude involvement of an orphan SR, we turned to genome-scale CRISPR/Cas9 screens using a previously reported library29. Strikingly, we again found a pronounced enrichment of the top four genes of the CRL-focused sgRNA library (DDB1, CUL4B, UBE2M and UBE2G1), which was particularly evident in the most stringently, dCeMM3-selected screen (Fig. 3a and Extended Data Fig. 5b, 6e). No similarly enriched genes were identified that would indicate the involvement of a hitherto orphan SR. Hence, these results point to a novel type of molecular degrader mechanism that is SR independent. Validating the pooled screening approach, targeted CRISPR-induced inactivation of CUL4B and UBE2G1 strongly abrogated drug-induced cyclin K degradation (Fig. 3b, c and Extended Data Fig. 7a). The essentiality of DDB1 prevented the isolation of individual LOF clones, but pooled DDB1 inactivation in KBM7 cells with doxycycline inducible Cas9 expression similarly abrogated the degradation of cyclin K (Fig. 3d). Inactivation of DDB1, UBE2G1 and UBE2M also strongly rescued cytotoxicity induced by dCeMM2/3/4, but not THZ531. This further corroborates a functional link between cyclin K degradation and the anti-proliferative consequences of the tested compounds and provides another layer of evidence that the cellular efficacy of dCeMM2/3/4 is independent of their weak inhibitory activity in CDK12 kinase assays (Fig. 3e, f, Fig. 1e and Extended Data Fig. 7b-e). Finally, despite sharing >80% sequence identity, dCeMM2/3/4 appear to be selectively dependent on CUL4B over CUL4A, suggesting a unique situation of non-redundancy of these cullin scaffolds (Extended Data Fig. 7f-h). Of note, dCeMM1/2/3/4 showed no direct effect on CUL4B abundance, or on levels of the consensus CUL4B substrate H2A-K119ub1 (Extended Data Fig. 7i, j).
Fig.3. Induced cyclin K degradation is mediated via a CRL4B ligase complex in a SR-independent manner.
a, Genome-wide CRISPR dCeMM2/3/4 resistance screens. Top: bubble plot displaying median sgRNA enrichment over DMSO, bubble size indicates significance. Bottom: sgRNA enrichment targeting indicated genes, background indicates distribution of all sgRNAs. b-d, dCeMM2-induced cyclin K degradation (2.5µM) is rescued in UBE2M-, CUL4B- (b), UBE2G1- (c) and DDB1- (d) deficient cells. e, DMSO-normalized viability in WT and 3-day doxycycline (dox) pretreated sgDDB1_1 and sgDDB1_2 dox-inducible Cas9 KBM7 cells after 3-day dCeMM3 treatment. Mean ± SEM; n=3 independent treatments. f, WT-normalized fold change in EC50 based on dose-resolved 3-day viability experiments upon exposure to the indicated drugs in the indicated genetic backgrounds. g, Targeted hybrid-capture approach coupled to next generation sequencing to identify mutations in spontaneously dCeMM2/3/4-resistant cells. h, Depiction of DDB1 and CUL4B mutations identified by hybrid capture sequencing in drug-resistant cell pools. Stars: point mutations. Red bars: premature stop codons. Arrows: frameshift mutations. i, Structure of DDB1 (gray) in complex with SV5V peptide (aa 21-39, blue), PDB: 2HYE. DDB1 point mutations located within 20Å of the peptide are highlighted in orange.
To gain further confidence in the identified genes, we exposed various cell types to excess concentrations of the identified hit compounds to isolate drug-resistant clones. To increase the mutational depth, KBM7 and MV4-11 cells were additionally engineered to carry full LOF mutations in the DNA mismatch repair protein MLH1 (Extended Data Fig. 8a, b). Drug-resistant subclones were isolated in batches and subjected to targeted next generation sequencing of DDB1, CUL4A, CUL4B, UBE2M, UBE2G1, CDK12/13 and cyclin K via a hybrid-capture based approach coupled to next generation sequencing (Fig. 3g). Supporting the CRISPR screening data, we identified a profound enrichment of missense mutations or focal deletions particularly in the BPA and BPC domains of DDB1 (Fig. 3h and Supplementary Table 5). Intriguingly, most identified mutations cluster inside a pocket formed between BPA and BPC that has previously been shown to engulf N-terminal helices of viral proteins such as the SV5-V protein (Fig. 3i), thus highlighting it as a putative actionable site for induced protein-protein interactions.21,22
Isolation of an HCT116 clone harboring the DDB1 L932P mutation allowed us to test effects on dCeMM2/3/4-induced cyclin K degradation. Indeed, cyclin K degradation was strongly reduced in the mutant background that was also characterized by a subtle DDB1 destabilization (Extended Data Fig. 8c). Additional mutations in dCeMM2/3/4-selected populations were also identified in the DDB1-binding domain of CUL4B (Fig. 3h and Supplementary Table 5). Finally, we identified point mutations in CDK12 and CDK13, albeit outside of the kinase domain. Again, we managed to isolate a clone mutant for CDK13 (P1043H) with impaired cyclin K degradation following dCeMM2/3/4 treatment (Extended Data Fig. 8d-f and Supplementary Table 5). Collectively, orthogonal functional genomics profiling and targeted validation uncovered a critical involvement of a partial CRL4B complex in drug-induced cyclin K degradation. Again, the highly congruent chemogenomic profile of the three tested molecules confirms a unifying mechanism of action, despite their different chemical structures. A hotspot of spontaneously arising resistance mutations in a DDB1 pocket previously described as a critical interaction site with viral proteins establishes a rationale for an analogous, dCeMM2/3/4-mediated association between CDK12/13:cyclin K and DDB1 that is independent of a dedicated SR.
Drug-induced dimerization between DDB1 and CDK12:cyclin K
To validate if the drug-induced degradation of cyclin K via the CRL4B:DDB1 complex is mediated via direct drug engagement, we initially set out to perform drug affinity chromatography using tethered analogs of dCeMM3. To that end, we devised two complementary strategies. First, we synthesized dCeMM3-NH2 (9) by installing a free amine on dCeMM3. This allowed for immobilizing dCeMM3-NH2 on sepharose beads, and purification of interacting proteins out of whole cell lysates (Fig. 4a, b). Coupling dCeMM3-NH2 pulldowns with immunoblotting revealed both cyclin K and DDB1 as interacting proteins (Fig. 4c). Treating lysates with THZ531 and thus covalently blocking the active sites on CDK12/13 prevented DDB1 and cyclin K enrichment via the dCeMM3-NH2 resin. This is in line with the fact that (i) cellular co-treatment with THZ531 was able to abrogate drug-induced cyclin K degradation (Fig. 2c), as well as (ii) with a weak inhibitory activity of dCeMM2/3/4 in biochemical CDK12 activity assays (Fig. 2d). Next, we devised an alternative method for drug-target enrichment that is based on cellular treatment with dCeMM3-PAP (10), a functionalized analog of dCeMM3 containing a photoactive diazirine moiety and an alkyne handle (Fig. 4d). Following cellular treatment with dCeMM3-PAP P, cells were UV crosslinked, lysed and the alkyne handle in dCeMM3-PAP was biotinylated for subsequent immobilization on streptavidin beads and identification of drug-interacting proteins via immunoblots. Again, DDB1 and cyclin K were enriched in the eluates, indicating that target engagement also occurs in intact cells and that this interaction is independent of post-lysis artifacts (Fig. 4e, f). Also in this setup, covalently saturating the CDK12/13 binding site with THZ-531 competed with dCeMM3-PAP enrichment (Fig. 4e, f). While a certain bias of compound derivatization can’t be excluded for tethering approaches, both strategies suggest simultaneous drug engagement with the destabilized cyclin K and the CRL4 adaptor protein DDB1 in a CDK12/13-dependent manner.
Fig.4. dCeMM2/3/4 induce proximity between CUL4B:DDB1 and CDK12/13:cyclin K.
a, dCeMM3-NH2 chemical structure (dCeMM3 tethered analog). b, Drug affinity chromatography strategy based on probe-coupled agarose beads pulldowns after DMSO or THZ531 (competition) pretreatment in lysates. c, Cyclin K and DDB1 enrichment in dCeMM3-NH2 -based pulldowns. For quantification, eluted protein was normalized to protein available (input panels). THZ531-competed (100µM, 1h) ratios were set to 1. d, Chemical structure of dCeMM3-PAP (PAP: photo-affinity probe). e, Drug-target enrichment strategy based on cellular dCeMM3-PAP co-treatment with DMSO or THZ531 (100µM, competition) after Carfilzomib pretreatment (10µM, 30min). f, Cyclin K and DDB1 enrichment in dCeMM3-PAP -based pulldowns. For quantification, eluted protein was normalized to protein available (input panels). THZ531-competed (100µM, 1h) ratios were set to 1. g, Proximity labeling strategy to assess drug-induced dimerization in intact cells based on the biotin ligase miniTurbo (mTurbo). h, Biotin-labeled CDK12 enrichment following 1h DMSO or dCeMM2 treatment in the presence of carfilzomib (10µM) in HEKs transfected with DDB1-mTurbo fusion. i, Direct cyclin K immunoblotting of carfilzomib-pretreated KBM7 cells after 2h DMSO, dCeMM2 or THZ531+dCeMM2 (10µM) treatments. j, TR-FRET signal for CDK12-Alexa488cyclin K (0–5μM) titrated to terbiumDDB1 in DMSO or 10μM dCeMM2. “No DDB1” only contains streptavidin-terbium. Data are means ± SD (n=3). Kapparent (nM): DMSO=n.d., no DDB1 = n.d., dCeMM2 = 628.
Next, we aimed to provide additional evidence of a direct, drug-induced association of CDK12:cyclin K and DDB1:CUL4B. Of note, under steady-state cellular conditions, cyclin K does not appear as a CUL4B substrate (Extended Data Fig. 9b). CRL decomposition after cellular lysis is known to challenge biochemical analysis but few approaches have been shown to overcome these problems.30,31 We opted to devise a cellular strategy based on enzyme-catalyzed proximity labeling via the efficient biotin ligase miniTurbo (mTurbo)32. This assay enables recording dynamic changes in molecular proximity of an mTurbo-tagged bait protein by streptavidin purification of covalently biotinylated, proximal proteins. We transiently expressed C-terminally tagged DDB1 mTurbo-fusions in HEK cells. Cells were treated for one hour with dCeMM2 or vehicle (DMSO) control, including a 30 minute biotin labeling pulse (Fig. 4g, h). In line with a drug-induced molecular proximity, CDK12 interactions with DDB1 were only identified in the presence of dCeMM2, but not after DMSO treatment. (Fig. 4h). Consistently, treatment with dCeMM2 induced polyubiquitination of cyclin K in a CDK12/13-dependent manner (Fig. 4i and Supplementary Fig. 9b). To provide further evidence for a direct, drug-induced binding of CDK12:cyclin K to DDB1, we reconstituted this interaction using recombinant proteins. Drug-induced proximity between CDK12:cyclin K and DDB1 was detected using time-resolved fluorescence energy transfer (TR-FRET). While weak binding between DDB1 and CDK12:cyclin K was observed in the presence of vehicle control (DMSO), treatment with dCeMM2 facilitated an interaction of DDB1 and CDK12:cyclin K with a Kapparent of 628 nM (Fig. 4j and Extended Data Fig. 9c). A similar affinity was measured for the dCeMM4-induced interaction between CDK12:cyclin K and DDB1, while inactive analogs of both hit compounds failed to strengthen the interaction in a comparable manner (Extended Data Fig. 9d).
Collectively, orthogonal experimental strategies allowed us to validate drug-dependent induction of proximity between CDK12:cyclin K and DDB1, which functionally leads to cyclin K ubiquitination and degradation. Phenotypically, cyclin K degradation by dCeMM2/3/4 validated a previously established hypersensitivity of T-cell malignancies for genetic cyclin K ablation (Extended Data Fig. 9e-g)33. Moreover, we observed a correlation between cyclin K degradation and sensitivity, as well as a synergistic potential when combining cyclin K degradation with DNA damage inducing agents (Extended Data Fig. 9e-g and Extended Data Fig. 10).
Discussion
Molecular glue degraders represent an intriguing strategy to deliver on the promise of chemically degrading disease-relevant proteins that have not been amenable to traditional, inhibitor-centric pharmacologic approaches. A major limitation to the systematic exploration and evaluation of molecular glue degraders as therapeutic strategies has thus far been their serendipitous identification and a lack of rational discovery principles. To address this bottleneck, we here introduce differential chemical profiling in hypo-neddylated cell lines that are characterized by a promiscuous defect in CRL activity. Lending proof of concept to this approach, we first identified dCeMM1, a glue degrader of RBM39 that functions by re-directing the activity of the CRL4DCAF15 ligase. Importantly, our strategy also led to the identification of three small molecules (dCeMM2/3/4) that appeared to be functionally independent of a dedicated substrate receptor, and that we mechanistically characterized via a subsequent multi-omics target deconvolution campaign.
To that end, we performed a comprehensive intersection of quantitative proteomics and functional genomic strategies, drug-affinity chromatography and cellular proximity assays to reveal an unprecedented and therapeutically yet unexplored mechanism for molecular glue-induced target degradation. Binding of dCeMM2/3/4 to CDK12:cyclin K prompts a dimerization with a DDB1:CUL4B E3 complex. Biochemical reconstitution experiments suggest a baseline affinity between DDB1 and CDK12:cyclin K that is strongly increased upon dCeMM2/3/4 addition. While future studies will address the underlying details from a structural perspective, this observation mechanistically differentiates the described cyclin K glue degraders from the well-studied thalidomide analogs where no baseline affinity between ligase and neo-substrate is measurable in the absence of the respective small molecule34. dCeMM2/3/4-induced dimerization between CDK12:cyclin K and DDB1:CUL4B leads to the positioning of cyclin K in the ubiquitination zone of the associated E2 ubiquitin-conjugating enzymes. UBE2Z and UBE2G1 function as the ubiquitin priming and extending E2s respectively, with UBA6 being the main E1 ubiquitin-activating enzyme. Consequently, cyclin K gets ubiquitinated and degraded by the proteasome via a mechanism that is independent of a dedicated CRL substrate receptor. Further research will explore the translational potential of targeted cyclin K degradation as single-agent therapy and in combination with DNA-damage inducing agents.
In summary, the presented data provides the first rational and scalable framework for the discovery of cellularly active molecular glue degraders with established, but also non-obvious mechanisms of action. Collectively, we expect this approach and derivatives that employ a different phenotypic readout to significantly increase the arsenal of molecular glue degraders. In contrast to the more commonly used heterobifunctional PROTACs, such molecules have the potential to induce the elimination of unligandable, disease-relevant proteins that are otherwise considered “undruggable”.
Online Methods
Cell lines and generation of mutants
KBM7 cells with the specified genetic backgrounds were grown in IMDM supplemented with 10% FBS and 1% penicillin/streptomycin (pen/strep). HCT116, and 293T cells were grown in DMEM 10% FBS and 1% pen/strep. MV4;11, MOLT4, Loucy, JURKAT, CCRFCEM, KOPTK1, DND41, CUTTL1, HSB2, HBPALL, MOLT16, SKW3 and SUPT11 were grown in RPMI 10% FBS and 1% pen/strep. KBM7 and HCT116 cells expressing Cas9 were generated using Lenti_Cas9_Blasti (Addgene 52962). LentiGuide-Puro (Addgene 52963) was used to express sgRNAs against UBE2M, DCAF15 and UBE2G1. LentiGuide-Puro-IRES-mCherry (modified from Addgene 52963) was used to express sgRNAs against CUL4A and DDB1, lentiCRISPRv2 (Addgene 52961) against CUL4B and lentiCRISPR2v2GFP (Addgene #82416) against MLH1. Lenti-PGK-Hygro-DEST-UBE2M was generated by Gateway cloning (UBE2M cDNA from pDEST17-UbE2M Addgene 15798 was cloned into pDONR221 by BP reaction, and then transferred to the destination vector Addgene 19066 by LR reaction), and used in UBE2Mmut cells to generate UBE2Mresc KBM7 cells. sgRNA sequences, plasmids and primers used to confirm single mutants are listed in Supplementary Table 6.
Reagents
Key reagents used were: dCeMM1 (Enamine Z2065717981, purity 97%), dCeMM2 (ChemDiv 8006-2691, purity 90%), dCeMM3 (ChemSpace PB54687009, purity 99%), dCeMM4 (Enamine Z1126858802, purity 91%) and THZ531 (MedChemExpress HY-103618, purity 99.49%).
Other reagents: Carfilzomib (Selleckchem, S2853), MLN4924 (Selleckchem, S7109), TAK243 (MedChemExpress, HY-100487), indisulam (Sigma-Aldrich, SML1225), doxycycline (VWR, A2951.0025).
Phenotypic screen based on differential viability
WT and UBE2Mmut KBM7 cells were treated with 2,000 cytostatic/cytotoxic small-molecules, mostly of unknown function/target, from the PLACEBO in-house collection. Briefly, the screening was divided in three parts 1) primary screening, 2) follow up and 3) validation. During the primary screening, cells were treated with 10μM and 500nM of every compound. DMSO was used as a negative control, YM155 1μM was used as a cytotoxic positive control, and the degrader ARV-771 (MedChemExpress, HY-100972) was used as a differential viability positive control at 50nM and 250nM (WT cells are sensitive and UBE2Mmut cells are resistant). After 3 days of treatment, cell viability was measured (CellTiter Glo, Promega). The screen was performed in 384-well plates (Corning 3712) with the compound dispensing system Echo 550 and for cell seeding the Liquid Handler Multidrop™ Combi. Control compounds were included in several wells, in all the plates. We considered as hits those small-molecules that inhibited the proliferation of WT cells, while sparing UBE2Mmut isogenic counterparts. >20 compounds fulfilled this criterion and were followed up by testing the dose-ranging differential effect. dCeMM1, dCeMM2, dCeMM3 and dCeMM4 were selected for further validation due to fold-change significance.
Cell viability assays
KBM7WT, mutant KBM7 clones (UBE2Mmut, UBE2Mresc, UBE2G1mut CUL4Amut, CUL4Bmut), and 3-day doxycycline pretreated KBM7iCas9_sgDDB1 cells were seeded at a cell density of 50,000 cells/mL in 96-well plates with DMSO or drug, in triplicates. Same conditions were used for viability assays with HCT116 and leukemia cell lines. Drugs used: dCeMM1, dCeMM2/2X, dCeMM3/3X, dCeMM4/4X or THZ531. Cells were treated for 3 days, after which cell viability was assessed according to manufacturer’s protocol (CellTiter Glo, Promega G7570). Survival curves and EC50 values were calculated by best-fit analysis of the log10 drug concentration to fold change of drug-treated cells over DMSO-treated cells. All survival assays included technical triplicates per sample, per experiment.
Western blot analysis
PBS-washed cell pellets were lysed in 50mM Tris pH 7.9, 8M Urea and 1% CHAPS and incubated with shaking at 4ºC for at least 30min. 20μg of supernatants were run and transferred for detection. Antibodies used: CUL1 (1:1000, Santa Cruz Biotechnology, sc-1276), CUL2 (1:500, Sigma-Aldrich, SAB2501565-100), CUL3 (1:1000, Cell Signaling Technology, 2759), CUL4A (1:1000, Cell Signaling Technology, 2699S), CUL4B (1:1000, Proteintech, 12916-1-AP), CUL5 (1:1000, Santa Cruz Biotechnology, sc-373822), UBE2M (1:1000, Santa Cruz Biotechnology, sc-390064), DDB1 (1:1000, Cell Signaling Technology, 5428S), cyclin K (1:5000, Bethyl, A301-939A), CDK12 (1:1000, Cell Signaling Technology, 11973S), CDK13 (1:1000, Bethyl, A301-458A), RBM39 (1:500, Santa Cruz Biotechnology sc-376531), V5 (1:1000 Cell Signaling Technology, 13202), Ubiquityl-Histone H2A (K119) (1:1000, Cell Signaling, 8240-20), MLH1 (1:1000, Cell Signaling Technology, 3515T). ACTIN (1:10000, Sigma-Aldrich, A5441), VINCULIN (1:1000, Santa Cruz Biotechnology, sc-25336). Secondary antibodies anti-mouse/rabbit/goat (1:10000, Jackson ImmunoResearch 115-035-003, 111-035-003 and 705-035-003) and anti-rabbit Alexa Fluor 488 (1:1000, Invitrogen, A21441).
Apoptosis induction: AnnexinV/propidium iodide (PI)
KBM7 cells were treated with DMSO, dCeMM2 (2.5µM), dCeMM3 (7µM) or dCeMM4 (3.5µM) for 4h, 8h and 12h. To asses apoptosis induction AnnexinV/PI staining was used (BD Bioscience 556547). 5x105 cells were collected, pelleted by centrifugation, washed with PBS and resuspended in 1X binding buffer at a concentration of 105 cells/mL, preparing a sufficient volume to have 100µL per sample. 5μl of staining solution was added per sample and incubated for 20min at room temperature in the dark. 400μl of 1X binding buffer was added and cells were analyzed (within 1h) by flow cytometry. Samples were analyzed on an LSR Fortessa (BD Biosciences) and data analysis was performed using FlowJov_10.6.1.
Cell cycle analysis by PI staining
2 million KBM7 cells were treated with DMSO, dCeMM2 (2.5µM), dCeMM3 (7µM) or dCeMM4 (3.5µM) for the indicated times and fixed in cold 70% ethanol for 30min at 4ºC. After addition of RNAse (100µg/mL) and PI (50µg/mL), cells were incubated for 1h at 37ºC before checking cell cycle profiles by flow cytometry. Samples were analyzed on an LSR Fortessa (BD Biosciences) and data analysis was performed using FlowJov_10.6.1.
Expression proteomics
We compared overall proteome-wide changes in KBM7WT cells treated with dCeMM1 (25µM), dCeMM2 (2.5µM), dCeMM3 (7µM) or dCeMM4 (3.5µM) for 5h and 12h, using quantitative proteomics based on isobaric tagging.
Sample preparation
50 million KBM7WT cells in duplicates per condition were collected, washed four times with ice-cold PBS, the supernatant aspirated and pellets snap-frozen in liquid nitrogen. Each washed cell pellet was lysed separately in 40μL lysis buffer as previously described24. Offline Fractionation via RP-HPLC at high pH and 2D-RP/RP Liquid Chromatography Mass Spectrometry were performed as previously described24.
Data Analysis
Acquired raw data files were processed using the Proteome Discoverer 2.2.0.388 platform, utilizing the Sequest HT database search engine and Percolator validation software node (V3.04) to remove false positives with a false discovery rate (FDR) of 1% on peptide and protein level under strict conditions. Searches were performed with full tryptic digestion against the human SwissProt database v2017.06 06 (20,456 sequences and appended known contaminants) with max two allowed miscleavage sites. Oxidation (+15.994Da) of methionine and deamidation (+0.984 Da) of glutamine and asparagine were set as variable modification, whilst carbamidomethylation (+57.021Da) of cysteine residues and TMT 10-plex labelling of peptide N-termini and lysine residues were set as fixed modifications. Data was searched with mass tolerances of ±10 ppm and ±0.025Da on the precursor and fragment ions, respectively. Results were filtered to include peptide spectrum matches (PSMs) with Sequest HT cross-correlation factor (Xcorr) scores of ≥1 and high peptide confidence assigned by Percolator. MS2 signal-to-noise values (S/N) values of TMT reporter ions were used to calculate peptide/protein abundance values. PSMs with precursor isolation interference values of ≥50% and average TMT-reporter ion S/N≤10 were excluded from quantitation. Only unique peptides were used for TMT quantitation. Isotopic impurity correction was applied and results exported for analysis for differential expression using limma R package35. Prior to performing limma analysis all proteins lacking reporter ion intensity values were removed and all protein TMT reporter channels normalized to equal median abundance. TMT ratios with P-values/Q-values (after FDR correction) lower than 0.01 were considered as significant. For the calling of destabilized proteins, a log2 fold change threshold (drug/DMSO) of -0.3 was applied.
CRISPR/Cas9 resistance screens (CLR-focused sgRNA library and genome-scale Brunello sgRNA library)
Design of the CLR-focused sgRNA library
CRL-focused library sgRNAs are listed in Supplementary Table 2.
Lentivirus production
293T cells seeded on 15cm culture plates 16h before were transfected with 5µg Brunello pooled library (Addgene 73178; 2-vector system), 2.5µg pMD2.G (Addgene 12259), and 3.75µg psPAX2 (Brunello sgRNA library, Addgene 12260) or CLR sgRNA library, using PEI (Polysciences, 24765-1). Viral supernatant was harvested 72h after transfection and concentrated using Lenti-X-concentrator (Takara). Concentrated viral supernatant was stored in aliquots at -80°C and titrated following a standard protocol29 to achieve a MOI of 0.2-0.3.
CLR sgRNA library screens
12 million KBM7Cas9 or KBM7iCas9 cells were transduced at MOI 0.3, yielding a calculated library representation of 635 cells/sgRNA (library representation = 2.5 million cells). For transduction, 100µL of concentrated viral supernatant was added to 3 million cells in 3 mL IMDM and 8µg/mL polybrene in a 12-well plate. Plate was centrifuged at 2000rpm for 1h at 30°C in a benchtop centrifuge and then incubated at 37°C overnight. Transduction efficiency was titrated following a standard protocol29. Pools were selected with 1µg/mL puromycin for 5 days (KBM7Cas9) or 500µg/mL neomycin for 8 days (iCas9 cells, followed by 5 days of cas9 expression by doxycycline 0.5µg/mL). Independent resistance screens were performed with both mutant libraries in duplicates, using drugs at starting concentrations of 4xEC50: dCeMM1 9µM, dCeMM2 0.9µM, dCeMM3 2.8µM and dCeMM4 1.25µM and a respective DMSO control. Every 4 days, cells were counted and re-seeded to 2.5 million cells in 5mL, applying fresh drug. Drug concentrations were dynamically adjusted to the growth curves to yield a consistent impact on cell proliferation. Drug resistant pools were harvested after 14 days of treatment, snap-frozen in liquid nitrogen and stored at -80°C.
Brunello pooled library screens
250 million KBM7-Cas9 cells or were transduced at MOI 0.23, yielding a calculated library representation of 668 cells/sgRNA (library representation = 50 million cells). For transduction, 20µL of concentrated viral supernatant was added to 5 million cells in 1.5 mL IMDM and 8µg/mL polybrene in 6-well plates. Plates were centrifuged at 2000rpm for 1h at 30°C in a benchtop centrifuge, 0.5mL IMDM were added and then incubated at 37°C overnight. The next day, transduced cells were pooled and diluted. Pools were selected with 1µg/mL puromycin for 5 days. Independent resistance screens were performed with the library using drugs at starting concentrations of 4xEC50: dCeMM2 0.9µM, dCeMM3 2.8µM and dCeMM4 1.25µM. Selective drug treatment was performed on 50 million cells/drug at a seeding density of 500,000 cells/mL. Every 5 days, cells were pooled, counted and re-seeded to 50 million cells in 100mL, applying fresh drug. Drug concentrations were dynamically adjusted to the growth curves to yield a consistent impact on cell proliferation. Drug resistant pools were harvested after 15 days of treatment, snap-frozen in liquid nitrogen and stored at -80°C.
Library preparation for next generation sequencing
Genomic DNA (gDNA) was extracted from cell-frozen pellets with the QIAamp DNA Mini Kit (Qiagen, 51304). PCR on the gDNA templates was done to amplify sgRNA sequences. Primer sequences used available in Supplementary Table 7.
For the screens performed with the CLR library: The isolated gDNA was processed in parallel to yield 5µg gDNA per 50µL reaction for the 1st PCR. One PCR reaction contained 0.75µL of ExTaq polymerase (Clontech), 2.5µL 10x buffer, 4µL of dNTP mix, 2.5µL of 10µM P7 forward primer mix, 2.5µL of 10µM condition-specific P5 barcoded primer, and water to reach 50µL. Target amplification was achieved by using: 1min at 95°C initial denaturation; 30s at 95°C, 30s at 53°C, 30s at 72°C, for 22-24 cycles; 10min at 72°C final elongation. Specific amplification of the 300bp target was confirmed by agarose gel electrophoresis. PCRs were purified using AMPure XP beads in a 1:1 ratio, following standard protocols. 10ng per condition were used for the 2nd PCR, using the same PCR conditions than for 1st PCR but using the primers Rev2_p5 and Fwd2_p7 primers, and 4 cycles.
For the Brunello screens, a published protocol was followed29. The isolated gDNA was processed in parallel to yield 10µg gDNA per 100µL reaction. One PCR reaction contained 1.5µL of ExTaq polymerase (Clontech), 10µL 10x buffer, 8µL of dNTP mix, 0.5µL of 100µM P5 forward primer mix, 10µL of 5µM condition-specific P7 barcoded primer, and water to reach 100µL. DNA oligo primers were ordered PAGE purified from Sigma Aldrich. Target amplification was achieved by using: 1min at 95°C initial denaturation; 30s at 95°C, 30s at 53°C, 30s at 72°C, for 26-27 cycles; 10min at 72°C final elongation. Specific amplification of the 360bp target was confirmed by agarose gel electrophoresis. All PCR reactions of a respective condition were pooled and 100µL were purified using AMPure XP beads in a 1:1 ratio, following standard protocols. Purified amplicon was eluted using 50µL TE buffer. Final sequencing libraries were pooled in equimolar amounts and sequenced on a HiSeq 3000/4000.
Next generation sequencing data analysis
Raw read files were converted to fastq format using the convert function from bamtools (v2.5.0)36. Sequencing adapters were trimmed using cutadapt (v2.7.12) with -g CGAAACACCG and --minimum-length = 10 (genome-wide screens only). The 20bp of spacer sequence were then extracted using fastx toolkit (v0.0.14) (http://hannonlab.cshl.edu/fastx_toolkit/) and aligned to the respective sgRNA index using bowtie2 (v2.2.4)37 allowing for one mismatch in the seed sequence. Spacers were counted using the bash command ‘cut -f 3 (0) | sort | uniq -c’ on the sorted SAM files. A count table with all conditions was then assembled, and the counts + 1 were converted to counts-per-million to normalize for sequencing depth. Log2-normalized fold changes compared to DMSO were calculated for each spacer. Statistical analysis was performed using the STARS algorithm v1.329. For this, spacers were rank-ordered based on log2 fold change and tested with the parameters --thr 10 --dir P against a null hypothesis of 10000 random permutations. Genes with q-value < 0.05 were called as hits.
RNA extraction, synthetic RNA spike-in and RNA sequencing
10 million KBM7 cells were treated in triplicates for 5h with DMSO, dCeMM1 (25µM), dCeMM2 (2.5µM), dCeMM3 (7µM), dCeMM4 (3.5µM) or THZ531 (600nM). Total RNA was extracted with the RNeasy mini kit (Qiagen 74106), and to account for global differences in transcriptional output, we employed an exogenous spike-in approach, according to the manufacturer’s protocol (SIRV-Set3 Iso Mix E0/ERCC from Lexogen, 051). RNA amount was determined using Qubit RNA HS Kit (Thermo, Q32852). 1.5µg RNA per condition was subjected to Poly(A) enrichment (Lexogen #039). RNAseq library prep was performed according to the manufacturer’s protocol (Corall Total RNA-Seq Library Prep Kit, Lexogen 095). 2ng RNA were used as starting material. Before the endpoint PCR, a qPCR was done (PCR Add-on Kit for Illumina, Lexogen 020) to determine the optimal number of PCR cycles (16 cycles). Final sequencing libraries were analyzed on Agilent 2100 Bioanalyzer, pooled in equimolar amounts and sequenced on a HiSeq 3000/4000.
RNA sequencing data analysis
Next-generation sequencing libraries were sequenced by the Biomedical Sequencing Facility at CeMM using the Illumina HiSeq 3000 platform and 50bp single-end configuration. Raw reads were trimmed using Trimmomatic (v0.32)38. The following parameters were used: HEADCROP:13 ILLUMINACLIP:epignome_adapters_2_add.fa:2:10:4:1:true SLIDINGWINDOW:4:1 MAXINFO:16:0.40 MINLEN:18. Trimmed reads were mapped to the hg38/GRCh38 assembly of the human reference genome and to SIRV-Set 3 sequences using STAR aligner version 2.5.2b39 using the following parameters: --outFilterType BySJout --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverLmax 0.6 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --outSAMattributes NH HI NM MD --outSAMtype BAM SortedByCoordinate.
Genes were defined based on genome-build GRCh38.p7, genome-build-accession NCBI:GCA_000001405.22. Sequences and annotations in FASTA and GTF format for SIRV Set 3 (SIRV Isoform Mix E0 and ERCCs, Cat. No 051) included in SIRV_Set3_Sequences_170612a (ZIP) were downloaded from https://www.lexogen.com/sirvs/download/.
Read counts per gene were obtained from the aligned reads using the htseq-count command (v0.11.2) from the HTSeq framework40. The Bioconductor/R packages edgeR, affy and limma were used for normalization and differential gene expression analysis.
Gene set enrichment analysis of genes significantly downregulated upon THZ531 treatment (log2FC < -4 and adj. P-value < 0.05) in comparison to dCeMM2/3/4 treatments was performed using the GSEA v3.0. To build the gene rankings, genes were ranked based on their expression change (dCeMM2/3/4 treatment vs. DMSO), using the log2FC value from differential gene expression analysis. The following parameters were used: xtools.gsea.GseaPreranked -nperm 1000 -scoring_scheme weighted -norm meandiv.
The data analysis code is available at https://github.com/himrichova/MGs_RNAseq_analysis.
The RNA-seq data generated for this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE142405.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142405
Recombinant proteins and in vitro kinase assays
Human GST-CDK12 (714-1063)/GST-CyclinK (1-267), GST-CDK13 (694-1093)/GST-Cyclin K (1-267) and GST-Cdk7 (2-346)/ CyclinH (1-323)/Mat1 (1-309) proteins were recombinantly expressed in Sf9 insect cells using the MultiBacTurbo system41. For full activation GST-CDK12 (714-1063)/GST-Cyclin K (1-267) and GST-CDK13 (694-1093)/GST-Cyclin K (1-267) were co-expressed with CDK-Activating Kinase (CAK) from S. cerevisiae. After 72 hours cells were harvested by centrifugation and resuspended in lysis buffer (50 mM Hepes pH 7.6, 300 mM NaCl, 5% glycerol and 5 mM bMe), followed by sonication. The lysate was cleared by centrifugation in a Beckman Avanti J-26S XP centrifuge with a JA-25.50 rotor (20,000 rpm for 45 min at 4 °C) and applied to a GST Trap FF column (GE Healthcare) equilibrated with lysis buffer using an Äkta Prime chromatography system (GE Healthcare). After application of 10 column volumes (CV) of lysis buffer, protein was eluted in lysis buffer with 10 mM glutathione. After TEV protease cleavage overnight, kinase complexes were further purified by size exclusion chromatography (SEC) using a Superdex 200 PG column (GE Healthcare) equilibrated in SEC buffer (20 mM Hepes pH 7.4, 150 mM NaCl, 5% glycerol and 1 mM TCEP). Fractions containing a stoichiometric kinase complex were determined by SDS–PAGE, pooled and concentrated using Amicon filters (Millipore). Proteins were aliquoted, snap frozen in liquid nitrogen and stored at -80 °C.
Radioactive kinase assays were performed using 0.2 µM CDK/Cyclin K complex incubated with 0.2 mM ATP containing 0.45 mCi [32P]-g-ATP/mL (Perkin Elmer) and varying concentrations of inhibitory compound for 10 min prior to starting the kinase reaction by adding the substrate. Reactions were incubated for 20 min at 30º C and stopped by addition of EDTA to a final concentration of 50 mM. Samples were spotted onto Amersham Protran nitrocellulose membrane (GE Healthcare), followed by three washing steps for 5 min with 0.75% (v/v) phosphoric acid. Counts per minutes were estimated in a Beckman Liquid Scintillation Counter (Beckman-Coulter) for 1 min.
Acquired resistance and hybrid capture
MLH1 mutant lines
LentiCRISPR2v2GFP (Addgene #82416) was used to express sgRNAs against MLH1 in KBM7 and MV4;11 cells. Transduced pools of cells were sorted for GFP expression twice. Individual clones were expanded in 96-well format and treated with 500µM temozolomide (Sigma-Aldrich, T2577) in mirror plates to nominate MLH1 mutants. Knock-out was validated in resistant clones by WB (anti-MLH1, Cell Signaling Technology 3515T). Target sequences were amplified, A-tailing and TOPO-cloning was performed according to the manufacturer’s instructions (Thermo Scientific, K457502) and individual alleles were sequenced. Primers listed in Supplementary Table 6.
Generation of cells with acquired drug resistance and hybrid capture protocol
100 million KBM7 and MV4;11 cells (WT and MLH1mut) were treated with DMSO, 6µM dCeMM2, 11µM dCeMM3, or 12µM dCeMM4 in 50mL medium. 25d later, Ficoll-gradient centrifugation with Lymphocyte Separation Media (Corning, COR25-072-CV) was performed. The following day cells were counted, PBS-washed and pellets frozen. HCT116 cells (MLH1-/+) were seeded in 384-w plates at 30,000 cells/mL in the presence of 50µM dCeMM3 or 30µM dCeMM3.1. 9 individual resistant clones were isolated and acquired resistance was validated against dCeMM2, dCeMM3 and dCeMM4. They were sensitive to Paclitaxel. DMSO-treated HCT116 cells (control) and a pool of 500,000 cells from each of the 9 HCT116 clones were PBS-washed and pellets frozen. gDNA was extracted using QIAamp DNA Mini Kit (Qiagen 51304). DNA content was determined with Qubit HS Kit (Thermo Fisher Q32854). The pools of resistant cells were subjected to targeted next generation sequencing of DDB1, CUL4B, UBE2M, UBE2G1, CDK12/13 and cyclin K via a hybrid-capture based approach. For the library preparation, NEBnext Ultra II FS DNA Library Prep Kit for Illumina (NEB, E7805S) was used (protocol for inputs >100ng). 500ng of DNA per condition were used as starting material. Fragmentation and bead size selection was performed for insert size of 150-350bp. PCR enrichment of adaptor-ligated DNA was done with NEBnext Multiplex Oligos for Illumina (Set1 E7335 and Set2 E75000). 5 PCR cycles were performed. For the hybrid capture, 500ng of DNA were used. Hybridization and prep was performed according to the protocol xGen hybridization capture of DNA libraries from IDT. As xGen blocking oligos the xGen Universal Blocker-TS Mix (IDT 1075475) was used. Custom probes against UBE2M, UBE2G1, CUL4A, CUL4B, CDK12, CDK13 and cyclin K were used as xGen Lockdown Panel. Hybridization was performed for 16h. NEBNext High-Fidelity 2X PCR Master Mix (NEB, M0541S) was used for the post-capture PCR, using 16-25 PCR cycles. Final sequencing libraries were analyzed on Agilent 2100 Bioanalyzer, pooled according to resistance rate and sequenced on a MiSeq (paired-end sequencing).
Bioinformatics analysis of hybrid capture
Raw read files were converted to fastq format using the SamToFastq function from the picard tools package (v2.9.0) (http://broadinstitute.github.io/picard/) with standard settings. Sequencing adapters and low-quality reads were removed using trimmomatic (v0.36)38 in PE mode with standard settings. Data pre-processing was performed following the latest version of Best Practices of the GATK consortium42. Reads were first aligned to hg38 assembly using bwa mem (v0.7.8) (http://bio-bwa.sourceforge.net/bwa.shtml). Duplicate reads were then filtered using the picard tools MarkDuplicates function. The BaseRecalibrator function from the GenomeAnalysisTK package (v3.7)43 was applied supplying the hg38_dbsnp138 known sites of variation from the GATK resource bundle under the --known-sites argument. Quality score recalibration was then applied using the PrintReads function with the optional -BQSR argument. Variant calling was performed using the Mutect2 function (--output_mode EMIT_VARIANTS_ONLY). Variants were annotated utilizing the Ensembl Variant Effect Predictor webtool44 in its latest release (Ensembl v98). Variants that affected the coding sequence (inframe_deletion, inframe_insertion, frameshift_variant, missense_variant, protein_altering_variant, stop_gained) that had a greater than 5-fold enrichment in allele frequency (as determined by Mutect2) in the drug-resistant population compared to unselected cells were considered hits.
Flow cytometry analysis of CDK13 levels
WT and CDK13+/P1043H HCT116 cells were fixed with cold 70% ethanol for 30min on ice. After 15min 5% Triton X-100 permeabilization on ice, cells were incubated with CDK13 antibody (1:200, Bethyl, A301-458A) in 0.5% BSA PBS for 1h at RT. After washing twice with 0.5% BSA PBS, cells were incubated with anti-rabbit Alexa Fluor 488 secondary antibody (Invitrogen, A21441) for 30min and washed twice with PBS before analysis by flow cytometry.
Synthesis of dCeMM3-NH2 and pulldown
Detailed synthesis in Supplementary Information.
Coupling of dCeMM3-NH2 to NHS-sepharose beads
100µL NHS-Activated Sepharose 4 Fast Flow (GE Life Sciences, 17090601) per condition were washed with 500µL DMSO 3 times (3min 800rpm centrifugation at RT). Beads were resuspended in 50µL DMSO and 2.5µL dCeMM3 NH2 10mM and 0.75µL TEA (Sigma-Aldrich, T0886) were added. After 16-24h of incubation on roto-shaker at RT, the remaining free amino groups on beads were blocked by adding 2.5µL ethanolamine (Sigma-Aldrich, 11016-7) and incubating on a roto-shaker for at least 8h at RT. Beads were centrifuged and 500µL-DMSO washed twice before proceeding with drug pulldown.
Preparation of cell lysates
200 million KBM7 cells were resuspended in 2mL lysis buffer (50nM Tris pH 7.5, 0.2% NP-40, 5% glycerol, 1.5mM MgCl2, 100mM NaCl, 1mM EDTA) supplemented with protease inhibitors (Thermo Scientific, 78437) and and benzonase (Merck, US170746-370746) and incubated on ice for 30min. After centrifugation (full-speed, 4°C, 30min) supernatant was transferred to a new tube and extracted protein measured with BCA (Fisher Scientific, 23225). 3mg of protein lysate were pretreated with DMSO or THZ531 100µM for 1h on a roto-shaker at 4°C.
Protein affinity purification and elution
Drug-coupled beads were washed with 1x lysis buffer by centrifugation 3 times. Washed drug-coupled beads were gently resuspended in the pretreated cell lysates for 2h on a roto-shaker at 4°C. After incubation, beads were centrifuged, supernatant removed and washed with lysis buffer at 4°C. Beads were transferred to unplugged mini Bio-Spin Chromatography Columns (BioRad, 732-6207 in washing buffer (50mM pH8 HEPES, 150mM NaCl and 5mM EDTA) and washed 3 times with 1ml buffer II (all steps performed at 4°C). Columns were transferred to RT, plugged and beads incubated with 4% SDS buffer II for 15min. Unplugged columns were placed in 1.5mL Eppedendorf tubes and centrifuged for 1min at RT to let eluates enter into the tubes. Eluates were analyzed by WB. Quantification was performed with ImageJ.
Synthesis of dCeMM3-photoaffinity probe (dCeMM3-PAP) and pulldown
dCeMM3-NH2 was conjugated to a constant side chain consisting of a photosensitive diazirine group and an alkyne handle. Detailed synthesis in Supplementary Information.
20 million KBM7WT cells per condition were pre-treated with either DMSO or 100µM THZ531 (competition condition) for 1h in the presence of 10µM carfilzomib (Selleckchem, S2853) in serum-free IMDM medium (3ml/20million cells/10cm dish). Then, cells were treated with 10µM dCeMM2PAP for 1h. Target proteins were covalently linked to the probe via photo-crosslinking using an UV crosslinker at 4°C (365nm wavelength for 10min). Cell pellets were collected, PBS-washed and snap-frozen in liquid nitrogen. For protein extraction, thawed pellets were resuspended in 500μl lysis buffer (NP-40 0.8%, HEPES pH 7.5 50mM, Glycerol 5%, NaCl 150mM, Mg2Cl 1.5mM, SDS 1%, protease inhibitors and benzonase) and incubated on ice for 30min. Click reaction to conjugate azide-PEG3-biotin to the photoprobe was performed using 1000µg of protein per sample (1mL total volume): 20μl of 5mM Azide-PEG3-biotin (Sigma-Aldrich, 762024-25MG) was added to each sample followed by a mix of 60μl 1.7mM TBTA, 20μl 50mM CuSO4 and 20μl 50mM TCEP (=100μl per sample), and left at room temperature for 2h. SpinOUT™ G-600 columns (G-Biosciences, 786-1621) were used to purify protein samples after the click reaction, according to manufacturer’s protocol. 700μg per sample were used for the pulldowns. Enrichment of target proteins was done using Pierce™ High Capacity NeutrAvidin™ Agarose beads (Thermo Scientific, 29202). After the last washing step beads were resuspended in 100µl elution Buffer (HEPES pH8 50mM, NaCl 150mM, EDTA 5mM, SDS 4%), incubated at 75°C for 30min and eluted by centrifuging at full speed for 3min. Eluates were analyzed by WB. Quantification was performed with ImageJ.
Biotin proximity labeling with miniTurbo in live cells
Cloning of DDB1-miniTurbo-V5
attB1-MCS-mCherry-MCS-miniTurbo-MCS-V5-attB2 was synthesized as a gBlock (IDTDNA) and cloned into a Gateway-compatible donor vector (pDONR221) using BP clonase (Invitrogen, 11789-020) to generate pENTR221_MCS_mCherry_miniTurbo_V5 (miniTurbo sequence obtained from Addgene, 107170). DDB1 was cloned by Gibson assembly (NEB, E5510S) into NdeI+XbaI digested pENTR221_MCS_mCherry_miniTurbo_V5. Primers to amplify DDB1 with Gibson compatible overhangs are listed in Supplementary Table 6. pcDNA3-FLAG-DDB1 (Addgene, 19918) was used as a template. Resulting plasmid pENTR221_DDB1_miniTurbo_V5 was subsequently cloned into a Gateway compatible destination vector (Addgene, 19066) using LR clonase (Thermo Fisher Scientific, 11-791-020) to generate pLenti_DDB1_miniTurbo_V5.
Biotin labeling in live cells and pulldown
One 10-cm culture plate of 293T cells (70%–80% confluent) was transfected with 2μg of pLenti_DDB1_miniTurbo_V5 using Lipofectamine 2000 (Invitrogen, 11668019). On the next day, the plate was expanded to 2x 10-cm plates. 48h after transfection, cells were pretreated with 10μM Carfilzomib for 30min, then treated with either DMSO or 20μM dCeMM2 for 1.5h, adding 500μM biotin during the last 30min. Labeling was stopped by transferring the cells to ice and washing 5 times with ice-cold PBS32. Cells were collected and lysed in 600μL of lysis buffer (50mM Tris-HCl pH 7.5, 125mM NaCl, 5% glycerol, 0.2% NP-40, 1.5mM MgCl2 and protease inhibitors). After 15min incubation at RT, lysates were clarified by centrifuging at 15,000g for 10min. For streptavidin pull-down of the biotinylated proteins, 500μg of protein per condition were incubated with 50μL of lysis buffer-washed streptavidin magnetic beads (Thermo Fisher Scientific, 11205D) for 1h at RT on a rotator. Beads were pelleted using a magnetic rack and each bead sample was washed 3 times with lysis buffer. To elute biotinylated proteins, beads were resuspended in 65µl elution Buffer (HEPES pH8 50mM, NaCl 150mM, EDTA 5mM, SDS 4%) and incubated at 75°C for 30min. Beads were pelleted on a magnetic rack and eluate (60µl) was collected. Eluates were analyzed by WB.
Cyclin K ubiquitination in live cells
5 million KBM7 cells per condition were pretreated for 30min with Carfilzomib 1µM, then treated with either DMSO, dCeMM2 10µM or dCeMM2 10µM + THZ531 10µM for 2h, collected and processed for WB analysis. Cells were lysed in 200µl lysis buffer: 50mM Tris pH 7.5, 150mM NaCl, 0.1% NP40, 1mM EDTA, 5mM MgCl2, 5% Glycerol with protease inhibitors, phosphatase inhibitors and NEM 25mM (Sigma-Aldrich E3876). NEM was included in all washing steps, including PBS-washing after collection before lysis. For K-48 pulldown assays 20million cells per conditions were used and the protocol provided by manufacturer was followed (Nanotab biotechnologies N1810).
TR-FRET experiments
Protein purification
Human wild-type DDB1 (Uniprot entry Q16531), CDK12 (Q9NYV4) and cyclin K (O75909) were subcloned into pAC-derived vectors45 and recombinant proteins were expressed as N-terminal His6, His6-Spy, or StrepII-Avi fusions in Trichoplusia ni High Five insect cells using the baculovirus expression system (Invitrogen)46.
Wild-type full-length StrepII-Avi-DDB1 was purified as previously described for DDB1-DCAF complexes47. High Five insect cells co-expressing truncated versions of wild-type His6-CDK12 (713-1032) and His6-Spy-tagged cyclin K (aa 1-267) were lysed by sonication in 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 10% (v/v) glycerol, 10 mM MgCl2, 10 mM imidazole, 0.25 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1% (v/v) Triton X-100, 1 mM phenylmethylsulfonylfluoride (PMSF), and 1x protease inhibitor cocktail (Sigma). Following ultracentrifugation, the soluble fraction was passed over HIS-Select Ni2+ affinity resin (Sigma), washed with 50 mM Tris-HCl (pH 8.0), 1 M NaCl, 10% (v/v) glycerol, 0.25 mM TCEP, 10 mM imidazole and eluted in 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 10% (v/v) glycerol, 0.25 mM TCEP, 250 mM imidazole. The pH of the eluate was adjusted to 6.8 before ion exchange chromatography. For ion exchange chromatography, the pH of the eluate was adjusted to 6.8 and the affinity purified proteins were diluted in a 1:1 ratio with buffer A (50 mM Tris-HCl (pH 6.8), 10 mM NaCl, 2.5% (v/v) glycerol, 0.25 mM TCEP) and passed over an 8 mL Poros 50HQ column. The flow through was again diluted in a 1:1 ratio with buffer A and passed over an 8 mL Poros 50HS column. Bound proteins were eluted by a linear salt gradient mixing buffer A and buffer B (50 mM Tris-HCl (pH 6.8), 1 M NaCl, 2.5% (v/v) glycerol, 0.25 mM TCEP) over 15 column volumes to a final ratio of 80% buffer B. Poros 50HS peak fractions containing the CDK12:cyclin K complex were concentrated and subjected to size exclusion chromatography in 50 mM HEPES (pH 7.4), 200 mM NaCl, 2.5% (v/v) glycerol and 0.25 mM TCEP. The concentrated proteins were flash frozen in liquid nitrogen and stored at -80°C.
Biotinylation of DDB1
Purified full-length StrepII-Avi-DDB1 was biotinylated in vitro at a concentration of 8 μM by incubation with final concentrations of 2.5 μM BirA enzyme and 0.2 mM D-biotin in 50 mM HEPES (pH 7.4), 200 mM NaCl, 10 mM MgCl2, 0.25 mM TCEP and 20 mM ATP. The reaction was incubated for 1 hour at room temperature and stored at 4°C for 14-16 hours. Biotinylated DDB1 (biotinDDB1) was purified by gel filtration chromatography and stored at -80°C (~20 μM).
Time-resolved fluorescence resonance energy transfer (TR-FRET)
Increasing concentrations of Alexa488-SpyCatcher-labelled10 His6-Spy-cycK/His6-CDK12 (Alexa488cyclin K:CDK12) were added to a mixture of biotinylated DDB1 (biotinDDB1) at 50 nM, terbium-coupled streptavidin at 4 nM (Invitrogen) and kinase inhibitors at 10 µM (final concentrations) in 384-well microplates (Greiner, 784076) in a buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% pluronic acid and 0.5% DMSO (see also figure legends). Forward titrations were carried out by adding increasing concentrations of Alexa488cycK-CDK12 (0-5 μM) into premixed 10 µM compound, 50 nM biotinDDB1, and 4 nM terbium-coupled streptavidin. Before TR-FRET measurements, reactions were incubated for 15 minutes at room temperature. After excitation of terbium (Tb) fluorescence at 337 nm, emissions at 490 nm (Tb) and 520 nm (Alexa488) were measured with a 70 μs delay to reduce background fluorescence and the reactions were followed by recording 60 data points of each well over 1 hours using a PHERAstar FS microplate reader (BMG Labtech). The TR-FRET signal of each data point was extracted by calculating the 520:490 nm ratio. Data were analysed with Prism 7 (GraphPad) assuming equimolar binding of biotinDDB1 to Alexa488cyclin K:CDK12 using the equations described previously8. Three technical replicates were carried out per experiment.
Cyclin K degradation-sensitivity correlations and synergy experiments
Correlation of degradation and sensitivity (Supplementary Fig.9f and 10c) in a panel of cell lines was calculated using cyclin K levels by immunoblotting and EC50s by 3-day viability assays. CCNK levels were quantified with ImageJ, normalized to KBM7, and ploted against the EC50 of a particular cell line. For the two-way compound combinations of synergy experiments, compounds were added to 384-well plates in a 6x10 matrixed format (6-point titrations, including DMSO control of dCeMM2 and 10-point titrations od DNA damage drugs including DMSO control) were used. 1000 SKW3 cells per well in 50µL were used in 384-well plates. Cell viability was measured 72h later (CellTiter Glo, Promega G7570) and Bliss scores were calculated as previously described48.
Statistics and Reproducibility
Information regarding error bars, numbers of replicates or samples, and statistical analyses are described in the corresponding figure legends. Representative results of at least two independent experiments are shown unless otherwise indicated. No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
Supplementary Material
Acknowledgements
CeMM and the Winter laboratory are supported by the Austrian Academy of Sciences. This work has received funding the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 851478). This work was further enabled by funding from the Austrian Science Fund (FWF, project number P32125-B and P30271-B28). C. Mayor-Ruiz is supported by an individual Marie Skłodowska-Curie postdoctoral fellowship (grant agreement number: 796010). Z. Kozicka was supported by a European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement number: 765445. G. Petzold was supported by the Human Frontier Science Program (HFSP Long-Term Fellowship LT000210/2014) and the European Molecular Biology Organization (EMBO Advanced Fellowship ALTF 761-849 2016). We further acknowledge funding awarded from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program grant agreement no. 666068 and from the Novartis Research Foundation to N. H. Thomä. Sequencing was performed at the Biomedical Sequencing facility and proteomics at the Proteomics facility at CeMM. We thank Benjamin Ebert and Mikolai Slabicki for sharing and discussing results ahead of publication.
Footnotes
Author contribution
C.M.-R. planned and performed most of the experiments, analyzed data and made figures. S.B. provided technical help in many experiments and prepared RNAseq and hybrid capture sequencing libraries. M.B. analyzed CRISPR screens and hybrid capture experiments and made figures. Z.K. developed and conducted TR-FRET assays and performed purification and labelling of recombinant proteins. M.S. synthesized tethered analogs of dCeMM3. H.I. analyzed RNAseq data. I.K. performed recombinant kinase assays. E.H and K.R. provided technical help in acquired resistance and experiments with dCeMM3-PAP. A.K. provided technical help in the chemical screen. G.P. performed purification and labelling of recombinant proteins. M.F. cloned the CRL sgRNA library. C.B. provided input on bioinformatic analysis. A.C.M. supervised proteomics experiments. J.Z. supported sgRNA design for the CRL library and supervised the cloning strategy. M.G. supervised recombinant kinase assays. N.H.T. gave critical input to the manuscript and supervised TR-FRET experiments. S.K. supervised and helped to design chemical profiling screen. G.E.W. wrote the manuscript, planned and supervised the presented research and has overall project responsibility.
Competing Interests
C.M.-R. and G.E.W. are listed as inventors of a patent application for glue discovery in neddylation-deficient cellular systems. C.M.-R., S.K. and G.E.W. are listed as inventors of patent applications covering the chemical space of dCeMM2/3/4. M.B., S.K., G.E.W and CeMM are founders and equity holders of Proxygen.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Sequencing of sgRNA cassettes from all the CRISPR/Cas9 screens described in this study (Fig. 1h, Fig, 3a and Extended Data Fig. 5a and 6b-e) and hybrid capture experiments (Fig.3h and Extended Data Fig. 7h and 8d) have been deposited in the NCBI Sequence Read Archive (SRA) under the accession code: PRJNA599346. The analyzed data are provided in Supplementary Table 2 and Supplementary Table 5 respectively. Expression proteomics data (Fig. 1g, Fig. 2a and Extended Data Fig. 2a and 4d, e) are provided in Supplementary Table 3. Searches were performed with full tryptic digestion against the human SwissProt database v2017.06 06 (https://www.uniprot.org/statistics/Swiss-Prot%202017_06). Structure of DDB1 in complex with SV5V peptide (Fig. 3i) corresponds to PDB: 2HYE (https://www.rcsb.org/structure/2HYE). RNAseq data (Fig. 2e,f and Extended Data Fig.4a-c) have been deposited in Gene Expression Omnibus (GEO) under the accession code: GSE142405. The analyzed data are provided in Supplementary Table 4.
Code availability
A detailed description of the bioinformatics analysis used for CRISPR screens, hybrid capture and RNAseq is available in the Online Methods). RNAseq analysis code available at https://github.com/himrichova/MGs_RNAseq_analysis.
References
- 1.Winter GE, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–81. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–7. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zengerle M, Chan KH, Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol. 2015;10:1770–7. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–9. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneekloth JS, Jr, et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126:3748–54. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 6.Toure M, Crews CM. Small-Molecule PROTACS: New Approaches to Protein Degradation. Angew Chem Int Ed Engl. 2016;55:1966–73. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–50. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 8.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–9. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–5. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sievers QL, et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362 doi: 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br J Haematol. 2014;164:811–21. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han T, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017 doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- 13.Uehara T, et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat Chem Biol. 2017;13:675–680. doi: 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- 14.Bussiere DE, et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat Chem Biol. 2020;16:15–23. doi: 10.1038/s41589-019-0411-6. [DOI] [PubMed] [Google Scholar]
- 15.Ting TC, et al. Aryl Sulfonamides Degrade RBM39 and RBM23 by Recruitment to CRL4-DCAF15. Cell Rep. 2019;29:1499–1510.e6. doi: 10.1016/j.celrep.2019.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faust TB, et al. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat Chem Biol. 2020;16:7–14. doi: 10.1038/s41589-019-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du X, et al. Structural Basis and Kinetic Pathway of RBM39 Recruitment to DCAF15 by a Sulfonamide Molecular Glue E7820. Structure. 2019;27:1625–1633.e3. doi: 10.1016/j.str.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 19.Andrejeva J, Poole E, Young DF, Goodbourn S, Randall RE. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J Virol. 2002;76:11379–86. doi: 10.1128/JVI.76.22.11379-11386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin GY, Paterson RG, Richardson CD, Lamb RA. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology. 1998;249:189–200. doi: 10.1006/viro.1998.9317. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–17. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–11. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leyser HM, et al. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–4. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- 24.Mayor-Ruiz C, et al. Plasticity of the Cullin-RING Ligase Repertoire Shapes Sensitivity to Ligand-Induced Protein Degradation. Mol Cell. 2019;75:849–858.e8. doi: 10.1016/j.molcel.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Wei D, Sun Y. Small RING Finger Proteins RBX1 and RBX2 of SCF E3 Ubiquitin Ligases: The Role in Cancer and as Cancer Targets. Genes Cancer. 2010;1:700–7. doi: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DT, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–95. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol. 2016;12:876–84. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenleaf AL. Human CDK12 and CDK13, multi-tasking CTD kinases for the new millenium. Transcription. 2019;10:91–110. doi: 10.1080/21541264.2018.1535211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reitsma JM, et al. Composition and Regulation of the Cellular Repertoire of SCF Ubiquitin Ligases. Cell. 2017;171:1326–1339.e14. doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichermeier KM, et al. PIKES Analysis Reveals Response to Degraders and Key Regulatory Mechanisms of the CRL4 Network. Mol Cell. 2020;77:1092–1106.e9. doi: 10.1016/j.molcel.2019.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Branon TC, et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers RM, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chamberlain PP, Cathers BE. Cereblon modulators: Low molecular weight inducers of protein degradation. Drug Discov Today Technol. 2019;31:29–34. doi: 10.1016/j.ddtec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–2. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bieniossek C, Imasaki T, Takagi Y, Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37:49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 1–11 10 33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdulrahman W, et al. A set of baculovirus transfer vectors for screening of affinity tags and parallel expression strategies. Anal Biochem. 2009;385:383–5. doi: 10.1016/j.ab.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 46.Li T, Pavletich NP, Schulman BA, Zheng N. High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 2005;398:125–42. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- 47.Fischer ES, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winter GE, et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat Chem Biol. 2012;8:905–12. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Sequencing of sgRNA cassettes from all the CRISPR/Cas9 screens described in this study (Fig. 1h, Fig, 3a and Extended Data Fig. 5a and 6b-e) and hybrid capture experiments (Fig.3h and Extended Data Fig. 7h and 8d) have been deposited in the NCBI Sequence Read Archive (SRA) under the accession code: PRJNA599346. The analyzed data are provided in Supplementary Table 2 and Supplementary Table 5 respectively. Expression proteomics data (Fig. 1g, Fig. 2a and Extended Data Fig. 2a and 4d, e) are provided in Supplementary Table 3. Searches were performed with full tryptic digestion against the human SwissProt database v2017.06 06 (https://www.uniprot.org/statistics/Swiss-Prot%202017_06). Structure of DDB1 in complex with SV5V peptide (Fig. 3i) corresponds to PDB: 2HYE (https://www.rcsb.org/structure/2HYE). RNAseq data (Fig. 2e,f and Extended Data Fig.4a-c) have been deposited in Gene Expression Omnibus (GEO) under the accession code: GSE142405. The analyzed data are provided in Supplementary Table 4.
A detailed description of the bioinformatics analysis used for CRISPR screens, hybrid capture and RNAseq is available in the Online Methods). RNAseq analysis code available at https://github.com/himrichova/MGs_RNAseq_analysis.