Abstract
Estrogen Receptor-α (ER) is the driving transcription factor in most breast cancers and its associated proteins can influence drug response, but direct methods for identifying interacting proteins have been limited. We purify endogenous ER using an approach termed RIME (Rapid Immunoprecipitation Mass Spec of Endogenous proteins) and discover the interactome under agonist and antagonist liganded conditions in breast cancer cells, revealing novel transcriptional networks in breast cancer. The most estrogen-enriched ER interactor is GREB1, a potential clinical biomarker, with no known function. GREB1 is shown to be a chromatin bound ER co-activator and is essential for ER function, where it stabilizes interactors between ER and additional co-factors. We show a GREB1-ER interaction in three xenograft tumors and using a directed protein-protein approach we find GREB1-ER interactions in half of ER+ primary breast cancers. This finding is supported by histological expression of GREB1, which shows that GREB1 is expressed in half of ER+ cancers and predicts good clinical outcome. These findings reveal an unexpected role for GREB1 as an estrogen-specific ER co-factor that is expressed in drug sensitive contexts.
Introduction
Estrogen Receptor-α (ER) is the key transcription factor that drives gene expression programs in ER+ luminal breast cancers (Ali and Coombes, 2002). ER+ breast cancers constitute the majority of breast tumors and these are generally treated with ER antagonists, although clinical response varies significantly (Ali and Coombes, 2002). ER-mediated transcription is determined by the associated co-factors and interacting transcription factors that form the ER complex (Shang et al., 2000). Increased levels of ER associated co-factors have been shown to correlate and contribute to drug resistance (Anzick et al., 1997; Torres-Arzayus et al., 2004), highlighting the importance of these co-factors in mediating ER transcriptional activity. Despite the fact that there are many known ER-associated factors (Metivier et al., 2003; Okada et al., 2008), it is clear that critical regulators are still being identified, as highlighted by the recent discovery of ER associated pioneer factors such as FoxA1, PBX-1 and AP-2γ (Carroll et al., 2005; Magnani et al., 2011; Tan et al., 2011), via enrichment of their DNA consensus sequence within ER binding domains. Direct experimental approaches for finding interacting proteins usually involve exogenous tagged methods or require very large-scale cell line culture (Malovannaya et al., 2011; Malovannaya et al., 2010; Selbach and Mann, 2006) and are non-existent in primary tissue.
We develop an endogenous protocol for systematic analysis of protein-protein interactions and protein-DNA binding events. We have combined several robust methods including formaldehyde cross-linking and on-bead digestion, permitting rapid and sensitive purification of endogenous interacting proteins. Cross-linking with formaldehyde is well established in chromatin immunoprecipitation and tissue fixation. Its size (~2Å) and ability to permeate membranes of intact living cells has two important implications: only amino acids in close proximity (2.3 to 2.7Å) will be cross-linked, and non-specific interactions by abundant proteins are minimized by the cells own architecture (Sutherland et al., 2008). This approach, termed RIME (Rapid Immunoprecipitation Mass Spec of Endogenous proteins) reveals novel ER associated transcriptional networks and identifies ligand specific interactions.
GREB1 is shown to be the most estrogen-specific ER interactor and endogenous association between ER, GREB1 and additional factors are verified both in vitro and in vivo. Very little is known about the function of GREB1, but GREB1 is one of the highest estrogen induced gene (Ghosh et al., 2000) and correlates well with changes in ER activity following breast cancer treatment (Dunbier et al., 2010). In addition it is known that GREB1 is required for growth of ER+ breast cancer cells (Rae et al., 2005) and overexpression can increase colony formation (Liu et al., 2012). However, it was not known what the mechanism is that allows GREB1 to mediate these effects. We now provide functional insight into how GREB1 mediates ER transcriptional activity and show that it is an independent predictor of clinical outcome in breast cancer patients.
Results
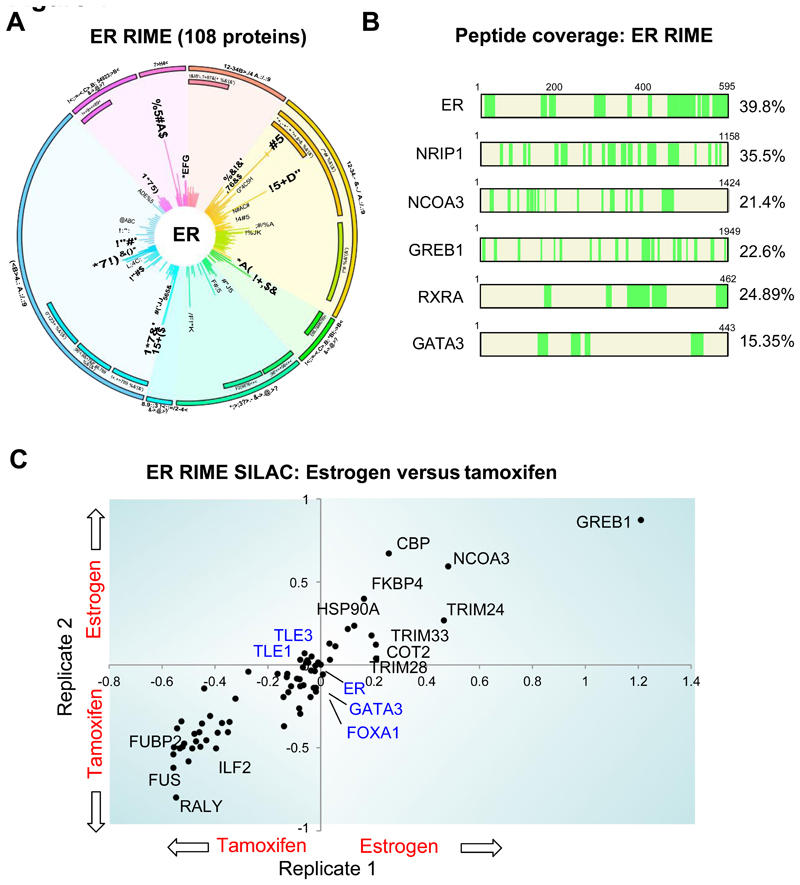
Endogenous ER was purified from 1 x 107 asynchronous MCF-7 breast cancer cells using RIME. The validation of the method is provided in Supplementary figure 1. In addition, RIME has been successfully utilized for purification of numerous other factors (an additional example (E2F4) is shown in Supplementary figure 2). We only considered ER associated proteins that occurred in three out of three independent replicates and excluded any protein that appeared in any one of five IgG control RIMEs. This resulted in 108 ER associated proteins (Figure 1A). The most confident protein identified from the ER RIME was ER. Also included within the ER interaction list are recently characterized ER pioneer factors FoxA1, TLE1, AP2-γ (Carroll et al., 2005; Holmes et al., 2011; Tan et al., 2011), putative pioneer factors such as GATA3 (Theodorou et al.), known co-factors including RIP140 (NRIP1), AIB1 (NCOA3), p300, CBP, CARM1, plus RAR family members which were recently implicated as ER associated protein (Hua et al., 2009; Ross-Innes et al., 2010). Peptide coverage of specific associated proteins is shown in Figure 1B. We also find a number of repressors, including N-CoR, SMRT and HDAC2, which are known to be recruited with estrogen-ER to repressed genes such as cyclin G2 (Stossi et al., 2006). A full list of ER interacting proteins is provided in Supplementary figure 3.
Figure 1.
ER RIME (Rapid Immunoprecipitation-Mass Spectrometry of Endogenous protein) was conducted in MCF-7 breast cancer cells. The graphical plot (termed MS-ARC) shows ER associated proteins. Non-specific interactions (identified from multiple IgG control replicates) have been removed. The ER associated proteins are clustered according to molecular function and the length of the line represents the mascot score. B. Peptide coverage (highlighted in green) of ER and additional identified interacting proteins. C. Hormone deprived MCF-7 cells were labeled with ‘heavy’ or ‘light’ SILAC isotopes, treated with estrogen or tamoxifen and ER RIME was conducted. Two replicates were conducted and shown are peptides found in both replicates. The axis represents log10 scale. The ER-DNA interacting complex, including putative pioneer factors are highlighted in blue text. GREB1 was the most estrogen-induced ER interacting protein based on SILAC ratio.
More than 25% of the 108 ER associated proteins were previously validated ER interactors (Supplementary figure 3). In addition, six previously uncharacterized ER associated proteins were selected for validation by standard Co-IP (no cross-linked) and ChIP-seq in MCF-7 cells (Supplementary figure 4).
ER RIME was repeated in asynchronous SILAC labeled cells following 3hr of estrogen treatment or 3hr of treatment with tamoxifen, the clinical ER antagonist. Two biological replicates were conducted. A number of proteins were predominantly bound with ER following treatment of either the agonist (estrogen) or the antagonist (tamoxifen) (Figure 1C). The core ER-DNA complex, composed of ER and the putative pioneer factors FoxA1, GATA3 and TLE1 are found under both ligand conditions (Figure 1C). GREB1 was the most markedly induced estrogen-ER interacting protein, based on SILAC ratio. Information on the spectra is provided in Supplementary figure 5. These data support the hypothesis that GREB1 is an ER associated protein only recruited under estrogenic conditions.
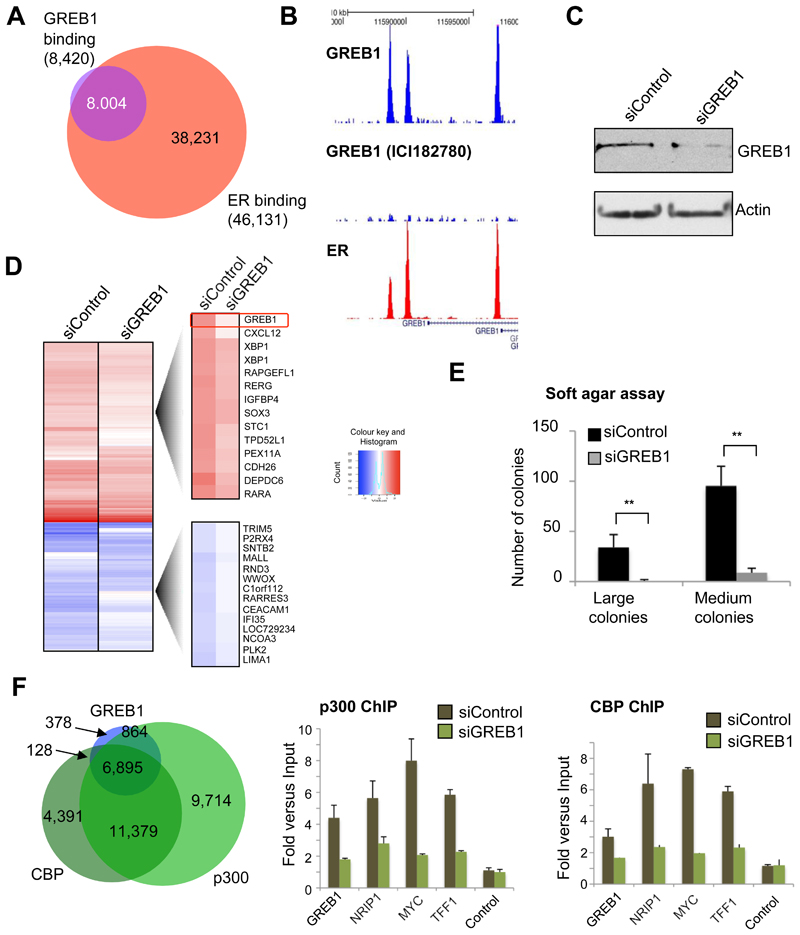
GREB1 is a highly estrogen induced gene (Ghosh et al., 2000) with almost no known function. ChIP-seq of GREB1 in estrogen-rich asynchronous MCF-7 cells revealed 8,420 GREB1 binding regions that occurred in two independent replicates (Figure 2A). Almost all (~95%) of the GREB1 binding events were shared by ER (Robinson et al., 2011) (Figure 2A). GREB1 binding was dependent on ER, since pre-treatment with ICI 182780 (an ER degrader) reduced GREB1 binding, as assessed by ChIP-seq (Figure 2B), despite GREB1 protein levels remaining the same at this short treatment time point (Supplementary figure 6). The GREB1 binding to ER cis-regulatory elements was validated by standard ChIP (Supplementary figure 6) and with an independent antibody (Supplementary figure 6). The interaction between ER and GREB1 was further validated by performing genome-wide ER and GREB1 Re-ChIP-seq, confirming global co-occupancy of these two proteins to the same genomic loci (data not shown).
Figure 2.
GREB1 is an estrogen-mediated ER interacting protein required for ER function. A. Venn diagram showing overlap of GREB1 binding events and ER binding events, as determined by ChIP-seq. Almost all GREB1 binding events are shared with ER. B. ER ChIP-seq was conducted in asynchronous MCF-7 cells. GREB1 ChIP-seq was conducted in asynchronous cells treated with control or ICI 182780 (an ER down-regulator). An example of a binding region is shown. C. MCF-7 cells were transfected with siControl or siGREB1 and Western blotting confirmed effective knock-down of GREB1. D. Hormone deprived MCF-7 cells were transfected with siControl or siGREB1 (4 replicates) and treated with vehicle or estrogen for 6 hr. Microarray analysis was conducted and estrogen-mediated differential gene changes (in siControl conditions) were determined. The heatmap represents all genes selected as differentially expressed following estrogen treatment relative to vehicle, in either siControl or siGREB1 transfected cells. E. MCF-7 cells were transfected with siControl or siGREB1 and soft agar assays were conducted. ** denotes p < 0.001 and was determined using student t-test. F. Overlap of GREB1 binding with the co-activators p300 and CBP. MCF-7 cells were transfected with siControl or siGREB1 and p300 or CBP ChIP was performed followed by real time PCR of known ER regulatory regions. In the absence of GREB1, p300 and CBP co-activators cannot bind to enhancer elements.
Since GREB1 was found to be the most estrogen-enriched ER interactor, we investigated the functional role of this poorly understood protein. MCF-7 cells were hormone deprived, transfected with control siRNA or a siRNA to GREB1 (Figure 2C) and treated with estrogen or vehicle control for 6 hr for microarray analysis. Differential gene expression analysis revealed 739 genes that were estrogen regulated (p < 0.01) and almost half of these were no longer differentially expressed when GREB1 was specifically silenced (Figure 2D). In addition, silencing of GREB1 significantly impaired the ability of MCF7 breast cancer cells to form colonies in soft agar, suggesting that it is required for their transformed phenotype (Figure 2E).
We overlapped GREB1 binding events with p300 or CBP binding events previously mapped in MCF-7 cells (Zwart et al., 2011) and found that most GREB1 binding events were also co-occupied by the co-activators p300 and CBP (Figure 2F). GREB1 possesses two LXXLL motifs, used by co-factors to interact with nuclear receptors (Heery et al., 1997) and as such, we assessed whether GREB1 may be involved in modulating binding between p300/CBP and ER. GREB1 was silenced with siRNA and ChIP of ER, p300 and CBP were assessed at shared cis-regulatory elements. Specific silencing of GREB1 did not affect ER binding (data not shown), but p300 and CBP binding were decreased on all the tested regions (Figure 2F), suggesting that GREB1 may stabilize the interaction with the classic, enzymatically active co-activators at specific ER binding regions.
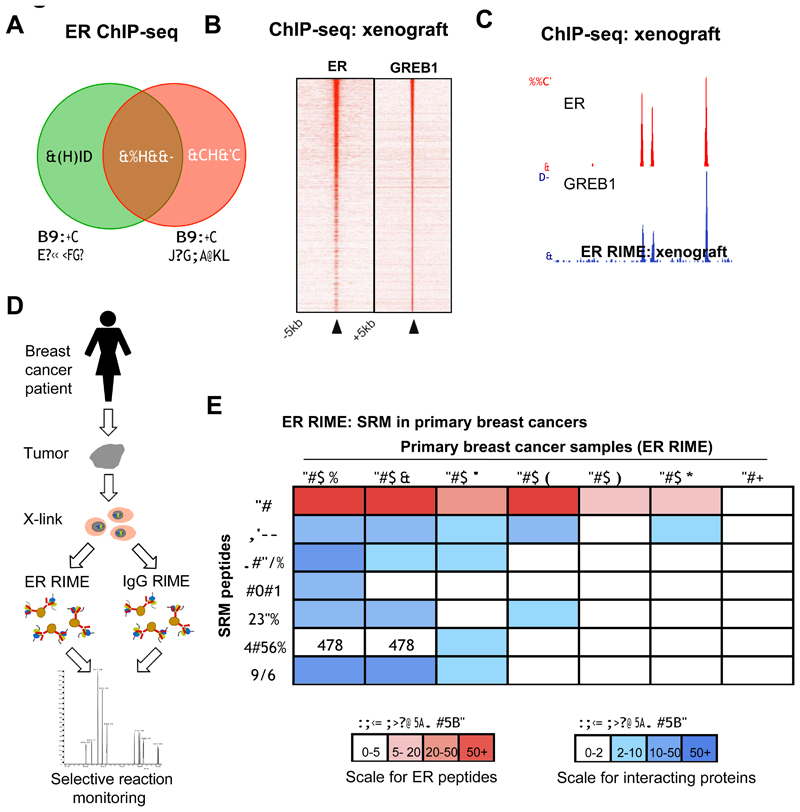
ER RIME was conducted from three independent MCF-7 xenograft tumors, implanted in immunocompromised mice (data not shown). In all three xenografts, ER-GREB1 interactions were observed (relative to matched IgG controls) (Supplementary figure 7) providing evidence that ER-GREB1 associations occur in solid tumor models. In addition to proteomic analysis, ER and GREB1 ChIP-seq was conducted on one of the MCF7 xenografts. Unexpectedly, MCF7 cells grown as a xenograft have a different ER binding profile compared to MCF7 cells growth in vitro (Figure 3A). In the xenografts, ER and GREB1 binding overlapped significantly, confirming that GREB1 binding paralleled the ER-DNA interactions observed in vivo (Figure 3B and 3C). We recently established a method for transcription factor mapping in primary human breast cancer samples (Ross-Innes et al., 2012), enabling genomic interrogation of ER binding properties. We sought to establish a method for protein-protein assessment into primary human tumors, to complement the genomic mapping approaches with proteomic analyses. Since these primary tumors were very small, potentially degraded and heterogeneous, we opted for a targeted approach by coupling RIME with Selective Reaction Monitoring (RIME-SRM), a method for sensitive and quantitative assessment of specific peptides of interest. A schematic of the approach taken is shown in Figure 3D. Seven primary frozen breast cancers were used, including six ER+ tumors and one ER- control tumor (tumor details are provided in Supplementary figure 7). Each tumor was cross-linked and split into ER or IgG RIME-SRM. We assessed multiple peptides that represent ER or some of its interactors identified from the cell line experiments, including GREB1, RXRα, TLE1, CBP, p300 and NRIP1. The data represents the average of all peptides for a specific protein (all peptides are provided in Supplementary figure 8) and the enrichment was normalized to the matched IgG control. We could successfully identify ER in all six ER+ tumors, but not the ER- tumor (Figure 3E and supplementary figure 7). Interestingly, we could also find several ER interacting proteins in some of the ER+ tumors (Figure 3E). We confirmed the identity of peptide precursors from tumor lysates by fragmentation (MS/MS) using a parallel platform (AB Sciex 5500MS) (data not shown). GREB1-ER interactions were not found in the ER negative tumor or in any of the IgG control and were observed in 3 out of the 6 ER RIME-SRMs from ER+ tumors (Figure 3E), suggesting that not all ER+ tumors have ER-GREB1 interactions.
Figure 3.
ER and GREB1 interactions in solid tumors. A. MCF7 cells were implanted into a immunocompromised mice and the xenografts were removed for ER and GREB1 ChIP-seq. Overlap in ER binding, as determined by ChIP-seq from the MCF7 cell line grown in vitro and as a xenograft. The growth of MCF7 cells as xenografts results in altered ER binding, when compared to MCF7 cells grown in vitro. B. Heatmap showing binding signal intensity of ER and GREB1 from a xenograft. C. Example of common ER and GREB1 binding events in a MCF7 xenograft. D. Establishment of RIME in clinical samples. Schematic representation of the ER RIME-SRM protocol used for assessing protein interactions in clinical samples. Frozen intact breast tumors were cross-linked and nuclei were collected. Each sample was split into an ER RIME or IgG RIME and subjected to selective reaction monitoring (SRM) to quantify specific peptides representing proteins of interest. E. ER interactions from six ER positive (ER+) and one ER negative (ER-) tumor. Each tumor was split and ER RIME or IgG RIME was conducted. Selective reaction monitoring (SRM) was used to identify the presence of specific peptides that were quantified relative to matched IgG control RIME. Where multiple peptides for a protein were identified, the average was taken. The peptide information can be found in Supplementary figure 8. All enrichments are shown relative to the matched IgG control. N/D = not determined.
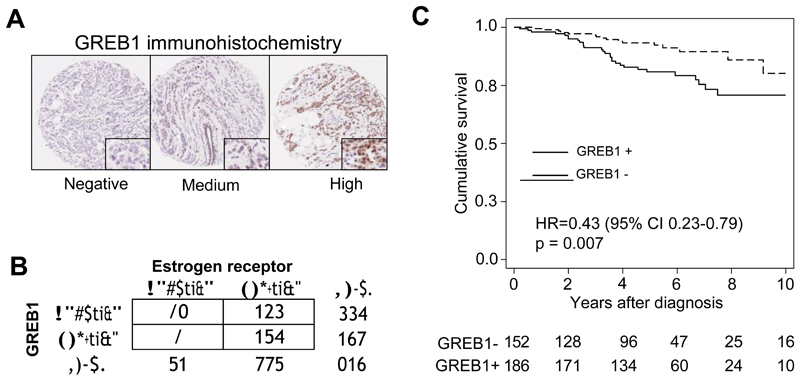
Given the observation that ER-GREB1 interactions could be detected in 3 out of 6 ER+ breast cancers by ER RIME-SRM (Figure 3E), we assessed GREB1 expression in a larger cohort (n = 419) of breast cancer patients by immunohistochemistry. An antibody for GREB1 was assessed in cell lines (Supplementary figure 9) and examples of GREB1 staining is shown in Figure 4A. This antibody has been previous validated for immunohistochemistry (Hnatyszyn et al., 2010). GREB1 expression was seen in 46.0% of all breast cancers, and of these 96.3% were ER+ breast cancers (Figure 4B), implying that GREB1 expression is mostly limited to ER+ disease. Of all ER+ breast cancers, 55% were GREB1 positive, slightly more than the 40% previously observed (Hnatyszyn et al., 2010). Although GREB1 is an estrogen-induced gene and GREB1 expression is limited to ER+ cancers, ER mRNA levels did not increase with increasing levels of GREB1 mRNA levels (Supplementary figure 9). Within ER+ breast cancers, GREB1 positivity (Allred > 2) correlated with a good clinical outcome (Figure 4C). Although GREB1 is estrogen induced, this correlation with good outcome may also reflect the functional role of GREB1 as a component of the ER complex. In support of this, GREB1 was found to be a prognostic factor independent of the Nottingham prognostic index (multivariate model GREB1 HR = 0.49 (0.26-0.90); p = 0.021; NPI HR = 2.3 (1.7-3.1); p < 0.001) (Supplementary figure 9) and in combination with the classic prognostic marker PR, GREB1 predicted an additive clinical benefit, when compared to PR alone (Supplementary figure 9).
Figure 4.
A. Example of GREB1 immunohistochemistry. B. Table representing the number of ER or GREB1 expressing breast cancers. C. Kaplan Meier curve representing clinical outcome relative to GREB1 expression in 338 ER+ cases (HR estimate from multivariate analysis). An Allred score of 2 was used as a cut point for GREB1 expression.
These findings place GREB1 as a key estrogen specific ER associated protein, where it is functionally linked with the transcriptional output of the ER complex. We observed that GREB1 is not expressed in a tamoxifen resistant (Tam-R) breast cancer model (Supplementary figure 10), but interestingly, re-expression of GREB1 in these Tam-R cells results in decreased cell growth in the presence of tamoxifen. As such, there may be pressure to inactivate or down-regulate GREB1 in endocrine resistant cells, potentially explaining why GREB1 expression in restricted to the 50% of ER+ breast cancer with a good clinical outcome.
Discussion
We have established a rapid protocol for the identification of endogenous interacting proteins from limited starting material, which enables assessment of endogenous protein interactions in tumor material. By applying RIME to ER, the major driving transcription factor in luminal breast cancer, we have revealed novel protein networks and functional components of this transcription complex. These include unexpected transcriptional associations between ER and COUP-TFII (COT2), KLF4, TLE3 and GREB1 (Supplementary figure 4). In addition to ER, we have successfully used this approach to identify associated complexes for numerous transcription factors. An additional example is shown in Supplementary figure 2, where E2F4 is purified and the components of the core E2F4 complex are found by RIME in two independent treatment conditions. Our approach provides a cost effective, sensitive and rapid method for discovery of endogenous interacting proteins. We exploit the size and permeability of formaldehyde to preserve bona-fide protein-protein interactions, avoiding non-specific interactions that would be anticipated in any post lysis procedure. Furthermore, the low affinity interactions between protein constituents of transcription factor complexes (interactions with rapid on-off rates) that would in any other protocol be missed with even moderate wash stringencies, are maintained using RIME. This is further controlled for by the use of parallel IgG controls in all experiments and quantitative SILAC approaches to confirm specificity. It is also possible that adjacent transcription complexes that occupy similar DNA binding domains may be cross-linked, yet digestion of DNA did not greatly affect the ER interactome identified (Supplementary figure 1). The sensitivity of RIME from limited material enables directed approaches for identification of protein-protein interactions in primary material, such as breast cancer tissue used in this study.
The most confident estrogen enriched ER-associated protein discovered using RIME, was the poorly characterized protein GREB1, a gene with little known function. The interaction between ER and GREB1 was observed in cell lines, xenograft tumors and in several primary human ER+ breast cancer samples. We show that GREB1 is recruited with ER to transcriptionally active cis-regulatory regions, where it functions as an essential factor, stabilizing associations with additional co-factors. This is of particular significance given the potential of GREB1 as a clinical biomarker (Dunbier et al., 2010). Our findings suggest that GREB1 expression functions as both a read-out of ER transcriptional activity and as a contributing factor to that transcriptional potential. Interestingly GREB1 has been shown to be important for growth of ER+ breast cancer cells (Rae et al., 2005) and AR+ prostate cancer cells (Rae et al., 2006), suggesting that it might also be involved in AR function in the prostate cancer.
Our observations suggest that GREB1 is expressed in half of ER+ tumors, where it correlates with positive clinical outcome (Figure 4C). In this subset of ER+ cancers, GREB1 is induced by the ER complex and is functionally linked with ER transcriptional activity. The absence of GREB1 in the remaining 50% of ER+ breast cancer (Figure 4B) may reflect a non-functional ER complex, where GREB1 is no longer ER induced. Our data would suggest (Supplementary figure 10) that GREB1 loss may be a contributing factor to the changes in hormonal responses and acquisition of antiestrogen resistance. This is supported by the observation that GREB1 expression is down-regulated in multiple endocrine resistant cell line models (McCune et al., 2010; Oyama et al., 2011; Shen et al., 2011) (and Supplementary figure 10) and that re-expression of GREB1 results in abrogation of the tamoxifen resistance (Supplementary figure 10). In drug sensitive cells, GREB1 functions as a contributor towards a functional ER complex, whereas in drug resistant cells, where tamoxifen-ER complex mediates cell growth, GREB1 is inhibitory. Given the findings that GREB1 is the least tamoxifen enriched ER associated protein (Figure 1C), tamoxifen liganded ER complex may acquire dependence on other up-regulated co-factors and may not be able to drive gene expression in the presence of GREB1. While it is currently unclear why GREB1 needs to be down-regulated for acquisition of endocrine resistance, our findings place GREB1 as one of the central estrogen-specific ER co-factors, where it functions to mediate interactions between ER and additional proteins.
Methods and materials
Detailed methods is available in Supplementary methods and materials.
Data deposition
All microrray data is deposited in GEO accession number GSE37386.
All ChIP-seq data is deposited in GEO accession number GSE41561.
All proteomic data is deposited with the PRIDE database with the accession number 27371-2738.
Supplementary Material
Acknowledgements
The authors would like to thank the members of the proteomic and microscopy core facilities at Cancer Research UK. We would like to acknowledge the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. We thank Ian Ellis (Nottingham) for clinical samples and the people involved in METABRIC for their help with data analysis. The authors would like to thank Imperial College Healthcare NHS Trust, Human Biomaterials Resource Centre (Tissue Bank). Tumor samples from Cambridge were obtained with support from NIHR Biomedical Research Centre and the Experimental Cancer Medicine Centre. We thank AB SCIEX for help validating SRM peptides. We thank Claudia Kutter for help with figures and Wayne Tilley for reading the manuscript. Carlo Palmieri is funded by Cancer Research UK. Jason S. Carroll is supported by an ERC starting grant and an EMBO Young investigator award.
Footnotes
Conflict of interest
None of the authors have any conflicts of interest.
References
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nature reviews. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. Science. Vol. 277. New York, NY: 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer; pp. 965–968. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Dunbier AK, Anderson H, Ghazoui Z, Folkerd EJ, AHern R, Crowder RJ, Hoog J, Smith IE, Osin P, Nerurkar A, et al. Relationship between plasma estradiol levels and estrogen-responsive gene expression in estrogen receptor-positive breast cancer in postmenopausal women. J Clin Oncol. 2010;28:1161–1167. doi: 10.1200/JCO.2009.23.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer research. 2000;60:6367–6375. [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hnatyszyn HJ, Liu M, Hilger A, Herbert L, Gomez-Fernandez CR, Jorda M, Thomas D, Rae JM, El-Ashry D, Lippman ME. Correlation of GREB1 mRNA with protein expression in breast cancer: validation of a novel GREB1 monoclonal antibody. Breast cancer research and treatment. 2010;122:371–380. doi: 10.1007/s10549-009-0584-x. [DOI] [PubMed] [Google Scholar]
- Holmes KA, Hurtado A, Brown GD, Launchbury R, Ross-Innes CS, Hadfield J, Odom DT, Carroll JS. Breast Cancer Special Feature: Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc Natl Acad Sci; USA. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang G, Gomez-Fernandez CR, Guo S. GREB1 Functions as a Growth Promoter and Is Modulated by IL6/STAT3 in Breast Cancer. PLoS ONE. 2012;7:e46410. doi: 10.1371/journal.pone.0046410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O’Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci; USA. 2010. pp. 2431–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune K, Bhat-Nakshatri P, Thorat MA, Nephew KP, Badve S, Nakshatri H. Prognosis of hormone-dependent breast cancers: implications of the presence of dysfunctional transcriptional networks activated by insulin via the immune transcription factor T-bet. Cancer research. 2010;70:685–696. doi: 10.1158/0008-5472.CAN-09-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Okada M, Takezawa S, Mezaki Y, Yamaoka I, Takada I, Kitagawa H, Kato S. Switching of chromatin-remodelling complexes for oestrogen receptor-alpha. EMBO reports. 2008;9:563–568. doi: 10.1038/embor.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Oyama M, Nagashima T, Suzuki T, Kozuka-Hata H, Yumoto N, Shiraishi Y, Ikeda K, Kuroki Y, Gotoh N, Ishida T, et al. Integrated quantitative analysis of the phosphoproteome and transcriptome in tamoxifen-resistant breast cancer. The Journal of biological chemistry. 2011;286:818–829. doi: 10.1074/jbc.M110.156877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Cordero KE, Scheys JO, Larios JM, Gottardis MM, Pienta KJ, Lippman ME. GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate. 2006;66:886–894. doi: 10.1002/pros.20403. [DOI] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast cancer research and treatment. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011 doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes & development. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shen C, Huang Y, Liu Y, Wang G, Zhao Y, Wang Z, Teng M, Wang Y, Flockhart DA, Skaar TC, et al. A modulated empirical Bayes model for identifying topological and temporal estrogen receptor alpha regulatory networks in breast cancer. BMC Syst Biol. 2011;5:67. [Google Scholar]
- Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. The Journal of biological chemistry. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011 doi: 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Research In Press. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Arzayus MI, De Mora JF, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Zwart W, Theodorou V, Kok M, Canisius S, Linn S, Carroll JS. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 2011 doi: 10.1038/emboj.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.