Abstract
Using genome-wide CRISPR screens to understand endocrine drug resistance, we discovered ARID1A and other SWI/SNF complex components as the most critical factors required for response to two classes of Estrogen Receptor-alpha (ER) antagonists as these SWI/SNF-specific gene knockouts lead to drug resistance. Unexpectedly, ARID1A was also the top candidate for response to the BET inhibitor JQ1, but in the opposite direction, where loss of ARID1A sensitised breast cancer cells to BET inhibition. We show that ARID1A is a repressor which binds chromatin at ER cis-regulatory elements. However, ARID1A elicits repressive activity in an enhancer-specific, but FOXA1-dependent and active ER-independent manner. Deletion of ARID1A resulted in loss of Histone Deacetylase 1 (HDAC1) binding, increased histone 4 lysine acetylation and subsequent BRD4-driven transcription and growth. ARID1A mutations are more frequent in treatment-resistant disease and our findings provide mechanistic insight into this process whilst revealing rational treatment strategies for these patients.
Keywords: ARID1A, Breast cancer, Treatment resistance, CRISPR screens
Introduction
Three quarters of breast cancers are driven by Estrogen Receptor-alpha (ER) 1, which utilises a slew of associated proteins to access compacted chromatin (including Forkhead Box A1 (FOXA1) and GATA Binding Protein-3 (GATA3)) 2, 3. Drugs that target the ER pathway are effective treatments for a majority of women with ER+ disease 1, but a substantial fraction of women will present with de novo or acquired drug resistance. Mechanisms of resistance are varied and include changes in co-factor levels, growth factor activated transcription and mutations in ER and associated transcription factors and co-factors 4.
Significant effect has been invested in identifying associated protein complexes that influence ER transcriptional activity 5–7. A role for the ATP-dependent chromatin remodeling complex SWItch mating type/Sucrose Non-Fermenting chromatin remodeling complex (SWI/SNF), has been linked with nuclear receptor function 8,9, where this complex modulates chromatin accessibility. There are three ATPase complexes, BAF, P-BAF and a recently identified non-canonical BAF (ncBAF) and the BRG1 and BRM subunits are common between the three complexes. However, there are proteins that are specific to BAF (ARID1A, ARID1B, DPF1/2/3, SS18), P-BAF complex (ARID2, Polybromo (PBRM1), BRD7) and ncBAF (BRD9, GLTSCR1, GLTSCR1L) 10,11. Previous work has shown a physical association between the SWI/SNF component BRG1 and ER and a requirement for BRG1 for ER-mediated transcriptional activity 12,13. The recruitment of SWI/SNF to the ER complex, is mediated by shared co-factors 14 and BRG1 occupancy at ER regulatory elements, coincides with increased localised histone acetylation 15. On a locus-specific level, BRG1 can bind to ER regulatory elements independent of ER 12, suggesting that the SWI/SNF complex might contribute to chromatin preparation prior to ER recruitment.
The SWI/SNF complex is important for chromatin regulation and gene expression 16, it is mutated in ~20% of all human cancers 17 and has been linked with the transcriptional activity of numerous nuclear receptors 8,9,13,18. Wild type ARID1A expression is associated with better clinical outcome in ER+ breast cancer patients 19 and importantly ARID1A inactivating mutations are enriched in treatment-resistant tumours and metastases (in total 12% of cases) 20,21. In addition, ARID1A inactivation has been associated as a tumour promoting event in ER+ breast cancer 22.
To systematically identify genes involved in treatment response in breast cancer, we employed global Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) screening approaches, coupled with three different treatment modalities, which revealed a role for the SWI/SNF complex, as critical determinants of treatment response.
Results
A CRISPR screen reveals ARID1A as a gene involved in treatment response
We employed a CRISPR screening approach, which encompassed gRNAs that target a total of 18,009 human genes 23. We established Cas9-expressing MCF7 breast cancer cells (Supplementary Fig. 1) which were infected and grown for 20 days. All cell line experiments were conducted in asynchronous cells grown in estrogen-rich media. Three biological cell cultures with independent viral infections with CRISPR vectors were performed as described in the Online Methods section. Analysis of the depleted gRNAs at different post-infection time-points, revealed known ER interactors including Cyclin D1 (CCND1), FOXA1 and GATA3 (Fig. 1a and Supplementary Fig. 2, Supplementary Table 1), albeit with different essentiality kinetics (Fig. 1a and 1b). In addition, a number of gRNAs were enriched representing tumour suppressors or growth inhibitors (Fig. 1c). As expected, growth promoting genes required for cellular viability showed greater gRNA depletion with longer infection (Fig. 1d). After 9 days of infection, we subsequently treated cells for a total of 26 days with the Selective Estrogen Receptor Modulator (SERM) 4-hydroxytamoxifen (Tamoxifen) or the Selective Estrogen Receptor Degrader (SERD) Fulvestrant (ICI 182780). We also used the tool compound JQ1, which targets Bromodomain and Extraterminal Domain (BET)-containing proteins, since Bromodomain containing protein-4 (BRD4) is postulated to be a therapeutic target in ER+ breast cancers and BET inhibitors are currently being explored in clinical trials 24,25. Three independent infections were performed and the data was integrated as described in the methods sections (Complete data in Supplementary Table 2). When specifically assessing genes required for treatment response, we found that the Fulvestrant and Tamoxifen CRISPR screens looked largely similar (Fig. 1e). Despite the distinct mechanisms of growth suppression (Fulvestrant degrades ER, whereas Tamoxifen-bound ER is recruited to the chromatin as a repressive complex), 63.5% of the genes required for Fulvestrant’s antiproliferative effects were also required for Tamoxifen activity (Fig. 1e and Supplementary Fig. 2). One of the most significantly enriched gene was AT-Rich Interaction Domain 1A (ARID1A), a component of the BAF ATP-dependent chromatin remodeling complex. It was one of the most essential gene for both Tamoxifen and Fulvestrant activity and depletion of ARID1A (i.e. enrichment of gRNAs targeting ARID1A) resulted in drug resistance to both compounds. Unexpectedly, ARID1A was the highest ranked gene in the JQ1 treated cells (ranked 1 out of 18,009 genes), but in the opposite direction, where gRNAs were observed to be depleted in JQ1 treated conditions (Fig. 1f, 1g and Supplementary Fig. 2). Other BAF components, including ARID1B, SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily B, Member-1 (SMARCB1/BAF47/SNF5) and Synovial Sarcoma Translocation, Chromosome 18 (SS18) showed the same pattern (Fig. 1f, 1g and Extended Data 1), suggesting that the BAF complex is required for ER targeted drugs to work, but when lost, sensitises cells to BET inhibitors. The dependence on ARID1A for growth arrest mediated by ER-targeted agents was validated in MCF7 and ZR-75-1 cells using ARID1A siRNA (Extended Data Fig. 1, Source Data Fig. 1 and Supplementary Fig. 3).
Fig. 1. CRISPR screens reveal ARID1A and BAF components as essential genes for treatment response.
Log2 fold of gRNA counts change as a function of time per gene (red lines) and on average (black line) based on a sample of n=3 for three categories of genes: the ones showing a rapid growth depletion (a), the ones showing a longer-term growth depletion (b) and the ones showing increased proliferation (c). For each category, example genes are shown in red and ARID1A is shown in blue. d. Heatmap representing log2 fold change of significant genes (n=1915) in non-treated conditions (day 3 to day 20 of infection comparing to uninfected gRNA pool). Rows were ordered according to hierarchical clustering. e. Heatmap representing log2 fold change of genes after 26 days of treatment with Fulvestrant (Fulv, initiated with 300 nM and reduced to 100 nM gradually), 100 nM 4-hydroxytamoxifen (Tamox) or BETi (JQ1 – 1µM reduced to 250 nM) comparing to DMSO treatment (DMSO control after day 9 of infection). Rows were ordered according to hierarchical clustering. f. ARID1A and other BAF components were enriched, but in different directions depending on the specific drug treatment. The values show changes in gRNA levels for these genes, using a log2 fold change relative to DMSO control. g. Frequency of single gRNAs in log 2 scale against BAF complex subunits ARID1A, ARID1B, SMARCB1 and SS18, comparing 4-hydroxytamoxifen or JQ1 with non-treated conditions. h. Example of ARID1A ChIP-seq binding overlap with ER and FOXA1 from MCF7 cells grown in media containing 10% fetal bovine serum containing estrogen, from three independent biological ChIP-seq samples per group. i. Global overlap between ARID1A, ER and FOXA1 ChIP-seq data from MCF7 cells grown in media containing 10% fetal bovine serum containing estrogen (n=3 independent biological ChIP-seq samples per group). j. Heatmaps representing ARID1A binding in hormone-deprived cells treated with vehicle or 10 nM estrogen (n = 3) on the constant sites (n=24,754 sites) defined by DiffBind without any significant change with estrogen treatment and the DiffBind-defined significant sites (n=3,023) which show reduced ARID1A binding during estrogen treatment. Also shown are the relative ER and FOXA1 binding intensities at these regions.
Genomic characterisation of ARID1A function
We subsequently assessed the potential genomic interplay between ARID1A and ER. We performed three independent biological replicates of ChIP-seq for ARID1A in MCF7 and ZR-75-1 cells and peaks were called using MACS version 2 26, resulting in 21,226 ARID1A peaks in MCF7 and 56,966 peaks in ZR-75-1. ARID1A binding sites were found to commonly co-occur at ER and FOXA1 binding events (Fig. 1h and Extended Data 2) and global analysis revealed that more than 78% of all ARID1A binding events were shared with ER, FOXA1 or both proteins in MCF7 (Fig. 1i), implying a functional connection between ARID1A and the regulatory elements occupied by the ER/FOXA1 complex. Interestingly, ARID1A overlapped more with FOXA1 (78% ARID1A binding sites were co-bound by FOXA1) than with ER (66%) in ZR-75-1 cells (Extended Data 2). We assessed whether ARID1A binding to ER bound enhancers was dependent on ER, by hormone depriving cells, treating with vehicle (ethanol) or estrogen for 6hr and conducting ChIP-seq. ARID1A was able to bind to ER/FOXA1 binding events prior to ligand induced ER recruitment (Fig. 1j and Extended Data 2). These findings suggest that ARID1A is not a classic ER-associated co-factor and can bind to regulatory elements independent of active ER, likely in a repressive manner.
To validate the CRISPR screen, we specifically deleted ARID1A from MCF7 cells, resulting in two separate ARID1A knock-out clones (Clones 11 and 14). ARID1A deletion was confirmed by Sanger and amplicon-based next generation sequencing and Western blotting (Fig. 2a, Supplementary Fig. 4, Source Data Fig. 2) and potential off-target effects were assessed. In vitro growth of these clones and the wild type control (WT clone 219) validated the CRISPR screening results, showing that both clones had increased intrinsic proliferation and were resistant to Tamoxifen, but showed sensitivity to JQ1 (Fig. 2b and Supplementary Fig. 5) and two additional clinically relevant BET inhibitors, OTX015 (from OncoEthix/Merck) and IBET762 (from GlaxoSmithKline) (Supplementary Fig. 4).
Fig. 2. ARID1A knock-out clonal cells show loss of response to ER antagonists, but responsiveness to BET inhibitors.
a. ARID1A was knocked-out of MCF7 cells using CRISPR deletion. Western blots of ARID1A or ER confirm effective gene deletion in clones 11 and 14, with no change in total ER levels. This figure shows the data of one representative experiment (Source Data Fig. 2) out of the three independent experiments. b. Percentage confluence as a function treatment time, in an in-vitro proliferation assay using Incucyte conducted in asynchronous MCF7 cells treated with vehicle or 1 µM 4-hydroxytamoxifen. This figure shows the data of one representative experiment out of the four independent experiments. Each experiment considered n=3 replicated per group. Mean ± Standard error of the mean is shown in the graph. c. Xenograft tumour volume of MCF-7 (n=13 animals), ARID1A K.O clone 14 (n=8 animals), ARID1A K.O clone 11 (n=12 animals) as a function of time since day of enrolment. The dots and arrows respectively show the average tumour volume and corresponding 95% confidence intervals of mice at risk. Tumour size of animals at different time-points were fitted by means of a linear mixed model on the cubic root scale, with time and group as fixed effect and random intercepts and slopes for mice (Full details are provided in Supplementary Note). The colored curves and shaded areas correspond to the fitted growth curves for each group and 95% confidence intervals, and the p-values to the mixed model difference in growth rate tests. Test statistics in Fig. S5d. p-values were calculated by two-sided Wald test. d. RNA-seq was conducted on the ARID1A knockout cells treated with Vehicle, 10 nM Fulvestrant, 100 nM 4-hydroxytamoxifen or 250 µM JQ1 (n=4 independent biological samples). As controls, both parental MCF7 cells and three wild type clonal lines were included. The plot shows fold change of Fulvestrant-regulated genes (n=1094) (ordered by means of a hierarchical clustering) in wild type cells. Highlighted gene cluster (with a star) shows the maintained downregulated effect of JQ1 regardless of ARID1A status, but upregulation with Vehicle and 4-hydroxytamoxifen upon ARID1A loss. e. Survival rate as a function of time-to-event for 2 groups of ER+ cancer patients: patients showing up- (red) (n=104 for Vehicle and 72 for 4-hydroxytamoxifen) and down- (blue) (n=101 for Vehicle and 61 for 4-hydroxytamoxifen) regulation according to a gene signature defined by ARID1A targeted genes shown to be repressed by vehicle or 4-hydroxytamoxifen. p-values correspond to log-rank tests (two-sided) (estimated test statistics available in Supplementary Fig. 6) respectively comparing the survival distribution of patients with up and down - regulated genes. Total METABRIC cases: 1181.
We established xenograft tumours from the wild type or the two ARID1A knock-out clones in the presence of estrogen pellets to maintain ER+ tumour growth and subsequently treated cells with vehicle or 4-hydroxytamoxifen. Tumour growth at day 25 was increased in the two ARID1A knock-out clones in the presence of 4-hydroxytamoxifen, when compared to wild type mice (Supplementary Fig. 5 which includes details of the statistical tests), validating that ARID1A is required for antiestrogen efficacy. However, the greatest difference in growth rate was in ARID1A wild type versus knock-out contexts in non-treated conditions (Fig. 2c and Supplementary Fig. 5) and we postulated that the diminution in Tamoxifen efficacy in ARID1A-null tumours may simply be due to an increased overall intrinsic proliferative potential.
ARID1A regulates ER target genes and is part of the ER complex
To explore the mechanistic role of ARID1A in drug response, RNA-seq was conducted using four biological cell culture samples of the wild type or ARID1A knock-out lines, treated with vehicle, Fulvestrant, 4-hydroxytamoxifen or BETi (JQ1). Gene expression analysis of the ARID1A knock-out clones and controls revealed several findings. The control lines looked similar, regardless of whether they were parental cells or wild type clonal lines (Supplementary Fig. 8). Whilst Fulvestrant and Tamoxifen showed similar gene repression patterns, JQ1 treatment resulted in a substantially different gene expression profile (Fig. 2d and Supplementary Fig. 8). In the ARID1A knock-out clones, JQ1 treatment showed a more consistent expression pattern when compared to the wild type cells, whereas the majority of genes repressed by Fulvestrant/Tamoxifen, were up-regulated or not changed in the ARID1A knock-out cells (Fig. 2d and Supplementary Fig. 6). In total, 86% of the Fulvestrant and 85% of the Tamoxifen-repressed genes were no longer significantly repressed in the ARID1A knock-out cells and a cluster of them (highlighted in Fig. 2d) are significantly downregulated by JQ1 treatment, to the same degree as in wild type cells. ARID1A deletion therefore, resulted in induction of the Fulvestrant/Tamoxifen repressed genes, even in the absence of an ER antagonist, implying ARID1A-mediated basal repression of the ER target genes. We generated a gene signature from the RNA-seq data and could show in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort of ER+ breast cancer patients 27 that the ARID1A repressed genes in both vehicle and anti-estrogen conditions (those that were up-regulated in the ARID1A knock-out cell lines) were associated with poor clinical outcome when up-regulated in patients (Fig. 2e and Supplementary Fig. 6 which includes details of the statistical tests), again supporting the notion that ARID1A can repress genes linked with clinical outcome.
To understand the mechanism behind ARID1A regulation of innate proliferation, we used an unbiased proteomic approach called RIME (Rapid IP-Mass Spec of endogenous interactions) combined with a label-free quantification method 28 to identity interactors of ARID1A, BRG1 or ER, from asynchronous MCF-7 cells, using an IgG pulldown as a negative control (information is provided in supplemental material) (Supplementary Fig. 7). ARID1A and BRG1 purification revealed almost all the known BAF components, as well as ER and similarly, the ER RIME contained ARID1A and BRG1 in the complex (Fig. 3a, Extended Data 3 and Supplementary Table 3 and 4). The other ATP-ase complexes, P-BAF and ncBAF 11, are identified in BRG1 pulldown, but not in the ARID1A pulldown. BRG1 RIME identified all the BAF, P-BAF and ncBAF components, validating that BRG1 is common to these complexes 21. It also showed enrichment of GLTSCR1/GLTSCR1L (BICRA/BICRL) subunits. BRG1 RIME revealed BET proteins as interactors (data not shown). We extended on these observations by re-analysing our recently published ER quantitative multiplexed RIME (qPLEX-RIME) data from five ER+ primary tumour samples from different patients 29. We discovered ARID1A and several SWI/SNF components, including BRG1, BRM, BAF57, BAF170 and BAF155 as physical interactors of ER, even in surgical tumour tissue (Fig. 3b, Extended Data 3). Importantly, we also observed an interaction between ER and BRD4, a target of the BETi, in the patient tumour material (Fig. 3b), verifying physical associations between endogenous ER, the SWI/SNF complex and BRD4 in vivo. We re-analysed our previous proteomic data 29 to identify proteins that interact with Tamoxifen-bound ER 29. ARID1A, BRG1 and a number of additional SWI/SNF components were enriched with Tamoxifen-liganded ER complex after treatment with 4-hydroxytamoxifen for 6hr (Fig. 3c and Extended Data 3), confirming that the SWI/SNF-ER complex formation is repressive.
Fig. 3. The SWI/SNF complex interacts with ER and is recruited to chromatin following drug treatment.
a. ER, ARID1A or BRG1 RIME was conducted in asynchronous MCF7 cells. IgG was used as a negative control. ER, FOXA1 and HDAC1 were identified as interactors in the ARID1A and BRG1 pull downs and vice versa. Boxplots shows the enrichment of selected known interactors in the pulldown samples compared to IgG controls. Pull downs were performed in two biological cell culture samples and label free quantification was performed using Minora algorithm. The log2 intensities are normalised so that the median of IgGs is zero. Centre line shows the median. n=2 independent biological cell culture samples. b. Five ER+ PR+ primary tumour samples were split for ER or IgG pull downs and the enrichment of known co-factors in the ER compare to IgGs such as HDAC1 and BAF components are shown. Boxplots shows the enrichment of selected known ERα interactors in the ERα RIME samples compared to IgG controls in human breast cancer tissues. The log2 values are normalised so that the median of IgGs is zero. Centre line shows the median. c. ER qPLEX-RIME was conducted in asynchronous MCF7 cells treated with 100 nM 4-hydroxytamoxifen in a 4-point time course (n = 6 independent biological samples per group). Specific BAF proteins are highlighted and the enrichment of the BAF components in the ER complex upon 4-hydroxytamoxifen treatment is shown. Centre line shows the median. d. ChIP-seq of ARID1A, BRG1 or SNF5 (SMARCB1/BAF47) in asynchronous MCF7 cells treated with vehicle (ethanol) or 100 nM 4-hydroxytamoxifen (n = 3 independent biological ChIP-seq samples). The heatmaps represent the 39,214 ARID1A binding events observed after 4-hydroxytamoxifen treatment. Also included are H3K27Ac, ER and FOXA1 binding signal intensity at these regions.
To explore the putative functional connection between SWI/SNF and the ER complex, we conducted a series of ChIP-seq experiments to map binding sites for ARID1A and two SWI/SNF common proteins, BRG1 and SNF5 (BAF47), in estrogen-rich asynchronous MCF7 cells treated with control or 4-hydroxytamoxifen for 6hr. Three independent biological replicates were conducted. Binding of all three proteins were increased globally following 4-hydroxytamoxifen treatment (Fig. 3d and Supplementary Fig. 11), supporting the hypothesis that they were involved in drug responsiveness. Both induced BRG1 and SNF5 sites overlapped with induced ARID1A sites, ER and FOXA1 (Fig. 3d and Supplementary Fig. 8). Unexpectedly, binding of these proteins were also increased following Fulvestrant treatment (Extended Data 4). The Fulvestrant-induced sites overlapped with both the Tamoxifen gained sites and estrogen lost sites from Fig. 1j (Extended Data 4), implying that these are the consistent hormone-regulated SWI/SNF binding regions. Altogether, our findings suggest that the recruitment of these factors, whilst able to associate with the ER complex, can bind to chromatin in an ER independent manner, in support of data showing basal repression of ER target genes by the BAF complex (Fig. 2d and 2e).
FOXA1 recruits ARID1A to chromatin
As Fulvestrant and Tamoxifen both increased BAF binding to chromatin, we speculated that the pioneer factor FOXA1, might modulate ARID1A and BRG1 recruitment to the chromatin, as supported by the data showing considerable overlap between ARID1A and FOXA1 binding (Fig. 1i and Extended Data 2). MCF7 and ZR-75-1 cells were hormone-deprived and transfected with FOXA1 or control siRNA and ChIP-seq of ARID1A or BRG1 was conducted. Both ARID1A and BRG1 binding was substantially reduced following FOXA1 silencing at enhancers, in both the cell lines assessed (Fig. 4a-e, Extended Data 5-6 and Supplementary Fig. 9-10), suggesting a degree of dependence on the pioneer factor FOXA1 for SWI/SNF recruitment. Importantly, the FOXA1-dependent ARID1A binding sites were the same regions where Tamoxifen induced ARID1A binding to the genome (Fig. 4f-g). To understand the importance of FOXA1 on ARID1A dependent genes, we identified the ER bound cis-regulatory elements close to ARID1A-repressed genes (those up-regulated in ARID1A knock-out cells), which we had previously shown to correlate with clinical outcome (Fig. 2e). We observed a modest change on ARID1A and BRG1 recruitment on these sites with FOXA1 loss (Fig. 4h). These findings show that the key ARID1A binding events are mediated by FOXA1 and not ER.
Fig. 4. FOXA1 promotes binding of ARID1A and BRG1 to a subset of potential enhancer elements.
a, b. Hormone-deprived MCF7 cells were transfected with control or FOXA1 siRNA and ChIP-seq was conducted for ARID1A (a) or BRG1 (b). n = 3 independent biological ChIP-seq samples. MA plots are shown with the average intensity of binding vs log2 fold change with FOXA1 siRNA comparing to control siRNA. c, d. Heatmaps (c) and boxplots (d) shown on ARID1A-BRG1 constant (n= 65563 sites) and ARID1A-BRG1 lost sites (n= 9355 sites) defined by DiffBind following FOXA1 silencing in MCF7 cells. ER and FOXA1 overlap are also shown on (c) these sites. n = 3 independent biological cell culture samples. p-values (d) were calculated by Welch’s t-test, two-sided. For boxplot, centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range). More statistical details are mentioned in Supplementary Table 5a. e. Scatterplot showing the association of decreased ARID1A and BRG1 binding following FOXA1 silencing. PCC – Pearson Correlation coefficient. p-values were calculated by Pearson Correlation test, two-sided. f-g. Scatterplot showing the association of ARID1A (f) and BRG1 (g) binding following FOXA1 silencing at tamoxifen-induced ARID1A (f) and BRG1 (g) binding sites from Fig. 3d. PCC – Pearson Correlation coefficient. p-values were calculated by Pearson Correlation test, two-sided. h. Boxplots illustrating the effect of siFOXA1 on ARID1A and BRG1 binding on the ER binding sites (n=2,746 sites) close to ARID1A repressed genes in Vehicle conditions. p-values were calculated by Welch’s t-test, two-sided. Window – 400 bp around center of the factor binding. Centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range). More statistical details are mentioned in Supplementary Table 5b.
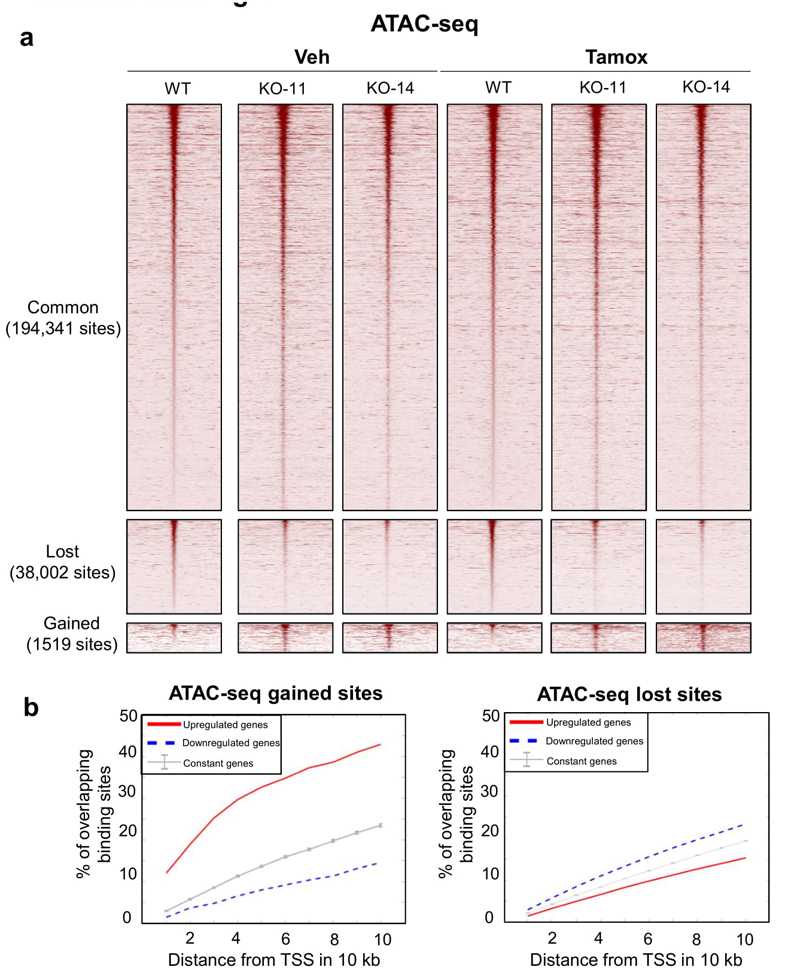
We sought to identify the molecular mechanism that dictated decreased drug responsiveness when SWI/SNF components were deleted (Fig. 1f and Extended Data 1). We performed Assay for Transposase-Accessible Chromatin (ATAC)-sequencing on MCF7 ARID1A knockout or wild type control cells, to assess if ARID1A was required for maintaining chromatin accessibility. Four independent cell culture samples were performed. We observed 233,862 total accessible regions in the genome, of which 83% (n=194,341) were not altered in ARID1A knock-out cells. Only 0.7% of sites showed a gain in accessibility in ARID1A knock-out cells and 16.3% of sites (n=38,002) sites had decreased accessibility in ARID1A knock-out cells (Extended Data 7). Integrative analyses of the chromatin accessibility and gene expression datasets showed that genes which are up-regulated in ARID1A knock-out cells are more associated with the ATAC-seq gained sites, implicating ARID1A in basal repression of these targets via inhibition of chromatin accessibility (Extended Data 7). However, there was no significant difference in accessibility at the regions co-bound by ARID1A and ER (data not shown), suggesting that loss of ARID1A is not altering chromatin accessibility at the regulatory regions bound by these protein complexes.
As previous work showed that a SWI/SNF subunit BRD9 inhibition results in a switch to P-BAF activity 18, we hypothesised that loss of ARID1A and BAF activity might result in a switch to a P-BAF-driven pathway. We therefore conducted ChIP-seq of ARID2 (a P-BAF-specific complex component) and BRG1 in wild type or ARID1A knock-out clonal cell lines and could show that ARID2 binding was not appreciably changed by Tamoxifen treatment and there was substantially less ARID2 binding in both ARID1A knock-out clones, regardless of the hormonal treatment conditions (Extended Data 8). This is a possible consequence of the decreased overall BRG1 binding in the ARID1A deleted cells (Fig. 5a and Extended Data 9). As such, loss of ARID1A does not result in recruitment of ARID2 and a switch to P-BAF dependency.
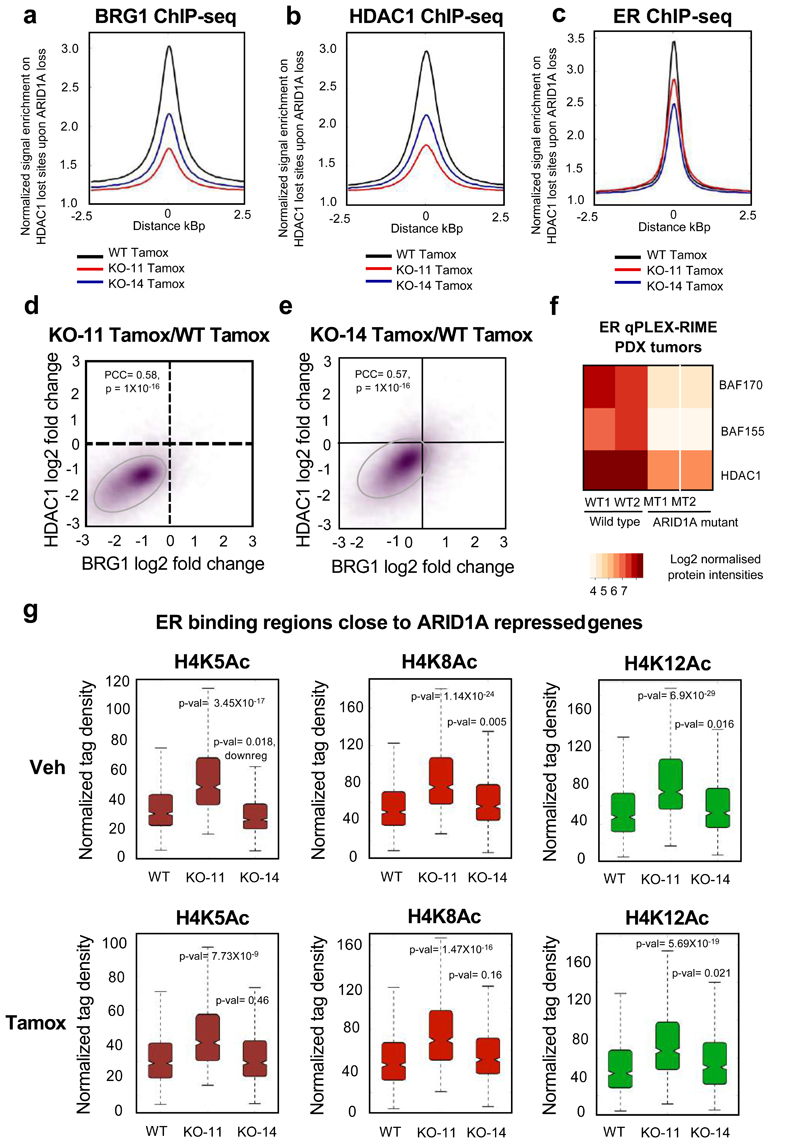
Fig. 5. Loss of ARID1A results in decreased BRG1 and HDAC1 recruitment and increased histone H4 acetylation.
a, b, c. Quantitative signal from BRG1 (a), HDAC1 (b) and ER (c) ChIP-seq within ARID1A knock-out cells (n=3 independent biological cell culture samples per group). ChIP-seq was conducted in the wild type cells or the two ARID1A knock-out clones, showing decreased binding of the factors in the absence of ARID1A. Average plots were shown on HDAC1 lost sites in the ARID1A knock-out cells. d, e. Scatterplots showing the association of decreased BRG1 and HDAC1 binding in ARID1A knockout clone 11 (d) and clone 14 (e) following 100 nM 4-hydroxytamoxifen treatment. n = 3 independent biological cell culture samples. PCC – Pearson Correlation coefficient. P-values were calculated by Pearson Correlation test, two-sided. f. ER qPLEX-RIME was conducted in four ER+ PDX tumours, two of which had loss of ARID1A via mutation (MT1/2) and two were wild type (WT1/2) for ARID1A. Heatmaps reveals decreased BAF and HDAC1 interactions with ER in ARID1A mutant tumours compare to the wild type tumours. g. We specifically identified ARID1A repressed genes in proximity to the ER-bound regulatory elements (n=686 sites) that display, according to PCA, more than 75% contribution to the variance in intensity of histone H4 acetylation. The data is shown as boxplots. ARID1A dependent genes acquired gained H4 acetylation, especially H4K8Ac and H4K12Ac at adjacent enhancers, coincident with increased gene expression. P-values were calculated by Welch’s t-test, two-sided. Window – 2 kb around center of the binding event. More statistical details are provided in Supplementary Table 5c.
ARID1A contributes to HDAC1 recruitment and mediating acetylation
To assess the mechanistic basis for the ARID1A repressive function, we performed H3K27Ac ChIP-seq and found that it was not affected in the ARID1A knock-out versus wild type cells (Extended Data 9). To identify other possibilities explaining the sustained gene expression in the presence of ER targeted drugs, when ARID1A was suppressed, we explored our RIME data and found that the histone deacetylase protein HDAC1 was an ARID1A interacting protein in non-treated conditions (Fig. 3a). In addition, in our qPLEX-RIME data, HDAC1 recruitment to the ER complex was enriched following Tamoxifen treatment, during active gene repression 29. Furthermore, HDAC1 was one of the most statistically enriched ER interactors in ER+ primary tumour samples (Extended Data 3) compared to IgG controls. We therefore conducted HDAC1 ChIP-seq and found a substantial decrease in HDAC1 recruitment, when ARID1A was specifically knocked-out (Fig. 5b and Extended Data 9). Only modest changes in ER binding were observed on HDAC1 lost sites (Fig. 5c, Extended Data 9 and Supplementary Fig. 11). We also observed a modest decrease in global FOXA1 binding as determined by ChIP-seq (data not shown). However, this was explained by a parallel decrease in FOXA1 expression, suggesting that ARID1A does not directly modulate FOXA1 recruitment to cis-regulatory elements and moderately influences FOXA1 binding by affecting total levels of this pioneer factor. BRG1 and HDAC1 binding was decreased at the same genomic regions in both the ARID1A knock-out clones (Fig. 5d and 5e), suggesting that both HDAC1 and BRG1 binding was dependent on ARID1A.
Additionally, we performed ER qPLEX-RIME on four ER+ Patient-Derived Xenograft (PDX) tumours 30, including two that had ARID1A loss via mutation and two ARID1A wild type control models (Extended Data 9 and Supplementary Fig. 12). We found a decrease in interactions between ER and HDAC1, BAF170 and BAF155 in the ARID1A mutant PDX models (Fig. 5f). As such, BRG1-associated SWI/SNF complex physically associates with HDAC1 in an ARID1A-dependent manner and the transcriptional repression elicited by HDAC1 requires functional ARID1A.
Given that HDAC proteins can actively remove the acetylation marks that are read by BET proteins 31, we speculated that changes in HDAC activity might explain the increased sensitivity to BET inhibition in the absence of a functional SWI/SNF complex. The histone acetylation marks that are read by BET proteins include Histone 4 lysine residues, including H4K5Ac, H4K8Ac and H4K12Ac 31. We assessed for increases in these histone marks in our ARID1A knock-out cells as a potential consequence of decreased HDAC1 recruitment. A distinct subset of histone H4 acetylated sites were increased under both non-treated and Tamoxifen treated conditions in the ARID1A knock-out cells, with the most prominent change observed in H4K8Ac (Supplementary Fig. 11). To understand the function of H4Ac upon ARID1A dependent genes, we examined the adjacent ER bound cis-regulatory elements on ARID1A target genes in ARID1A wild type versus knock-out cells. ARID1A was recruited to these enhancers in wild type cells and these sites showed substantial upregulation of the histone H4 acetylation, particularly H4K8/12Ac in both the clones (Fig. 5g and Extended Data 10). Given the decreased HDAC1 recruitment, the increase in H4K8/12Ac in ARID1A depleted cells and the increased responsiveness to BETi in ARID1A deleted contexts (Fig. 1f and 1g), we hypothesised that depletion of ARID1A would result in gained BRD4 binding and activity. BRD4 ChIP-seq in wild type and ARID1A knock-out cells, revealed a gain of 6,197 BRD4 binding sites in ARID1A depleted cells, confirming a significant increase in BRD4 chromatin binding. Analyses on ER binding sites close to ARID1A target genes showed increased BRD4 binding under ARID1A loss in both treatment conditions (Fig. 6a and 6c). The same regions showed a gain of histone H4 acetylation and BRD4 and decreased HDAC1 binding in ARID1A-deleted cells (Fig 6b and Supplementary Fig. 13-14). We integrated the gained BRD4 binding that was only observed in ARID1A-null cells, with the Fulvestrant/Tamoxifen-repressed genes and found a significant enrichment of BRD4 recruitment to the genes typically repressed by both ER antagonists (Supplementary Fig. 13). Mechanistically, our findings show that depletion of ARID1A results in decreased HDAC1 binding, a gain in histone 4 acetylation and coincident BRD4 recruitment at regulatory elements adjacent to genes normally repressed by ER targeted drugs in wild type contexts (Supplementary Fig. 14). This culminates in increased basal proliferation that occurs in a BET-dependent manner. In support of the intrinsic regulation of proliferation by ARID1A, we assessed breast cancer patients with ARID1A mutations, when compared to patients with wild type ARID1A 27, revealing a poorer clinical outcome in women with ARID1A-mutant tumours (Fig. 6d and Supplementary Fig. 14 with details of the statistical test). To explore the link between BET-driven growth in ARID1A null contexts and to assess other treatment options for women with ARID1A mutations, we established a tumour explant from an ARID1A mutant PDX tumour which has a frameshift mutation leading to ARID1A loss 30 (Supplementary Fig. 14). Tumour tissue was cultivated ex vivo and treated with vehicle or two different BETi for 48hr and we could show significant antiproliferative effects by assessing Ki67 expression, a surrogate marker for proliferation, following treatment (Figs. 6e and 6f), confirming the dependence on BET proteins in ARID1A mutant/deleted contexts similar to wild type contexts.
Fig. 6. Loss of ARID1A results in increased BRD4 recruitment and a gain in intrinsic proliferation.
a- c. BRD4 ChIP-seq was conducted in wild type or ARID1A knock-out cells (n=3 independent ChIP-sew samples per group). a. Boxplots were shown on ER bound regions close to ARID1A repressed genes (n=686) that display, according to PCA, more than 75% contribution to the variance in intensity of H4 acetylation. p-values were calculated by Welch’s t-test, two-sided. Window – 400 bp around center of the factor binding. For boxplot, centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range). More statistical details are mentioned in Supplementary Table 5d. b. Scatterplot showing the association of BRD4 and HDAC1 binding in the ARID1A knockout clone 11 cells. n = 3 independent biological cell culture samples. PCC – Pearson Correlation coefficient. P-values were calculated by Pearson Correlation test, two-sided. c. Heatmap shows the gained BRD4 occupancy and decreased HDAC1 binding on the ER-bound regulatory elements (n=2,746 sites) adjacent to ARID1A target genes. d. Overall patient survival was assessed based on ARID1A mutational status in a cohort of 1,824 breast cancer patients. p-value was calculated by log rank survival test, two-sided (estimated test statistics available in Supplementary Fig. 14). e. Ki67 IHC protein levels stained on an ARID1A mutant PDX cultivated ex vivo and treated with DMSO vehicle (n=10 explant chunks), 250 nM JQ1 (n=9 explant chunks) or 1µM IBET762 (n =10 explant chunks) for 48hr in a single experiment. Median values are shown with p-values calculated using Wilcoxon test, two-sided (Wilcoxon test Statistic W= 17 for both comparisons). f. Representative images of Ki67 IHC in BETi ex vivo tumour tissue, with each image representing a region of 100 μm in length. The explant chunks were treated with DMSO vehicle (n=10), 250 nM JQ1 (n=9) or 1µM IBET762 (n =10). g. Model of FOXA1-ARID1A-HDAC1-BRD4 axis in ARID1A wild type and mutant contexts.
Our study shows that the SWI/SNF complex is recruited to ER cis-regulatory elements prior to active ER binding, via the pioneer factor FOXA1. ARID1A exhibits transcriptional repression by recruiting HDAC1 and when ARID1A is functionally inactivated, HDAC1 binding is diminished, resulting in a gain in enhancer-specific acetylation, which is subsequently ‘read’ by BET proteins (Fig. 6g and Supplementary Fig. 15).
Discussion
Our unbiased genetic screening approach has revealed a critical role for the SWI/SNF complex in estrogen receptor-targeted treatment efficacy. Loss of ARID1A had profound effects on the gene expression program and growth phenotype, by affecting the chromatin environment. Tumour growth and clinical outcome were influenced by ARID1A status, independent of estrogen-bound ER activity, in support of previous work showed that BAF57 could be recruited to the ER target gene promoter, pS2 (TFF1) in an estrogen independent manner 12. In contrast, Glucocorticoid Receptor (GR) was shown to recruit the BAF complex to the MMTV chromatin template 8,32, implying that the mode of BAF-chromatin occupancy is nuclear receptor-specific. Our findings suggest that while ARID1A and SWI/SNF components can be recruited to ER cis-regulatory elements by ER antagonistic ligands, in particular Tamoxifen, this complex can associate with these enhancer elements independent of nuclear receptor activation. In this study, we identified that the pioneer factor FOXA1, which demarcates ER regulatory elements 2,33 and binds chromatin independently of hormonal status, is responsible for recruiting the BAF complex to the chromatin. FOXA1 can directly recruit the histone modifying methyltransferase that deposits the histone modification that is the hallmark of enhancer elements 34and previous work has shown that FOXA1 can open a compacted chromatin template, independently of other proteins 35,36, placing it upstream of all factors that subsequently get recruited to these enhancer elements.
Mutation of ARID1A occurs in ~5% of primary breast cancer, but the frequency increases to ~12% when looking in the metastatic context 20, implying a selection for tumour cells possessing loss-of-function ARID1A mutations 20,21. Our findings suggest that loss of ARID1A causes a shift in the H4 acetylation status, a result of decreased HDAC1 binding, which consequently results in BRD4 recruitment and BET-dependent growth (Fig. 6g). Since ARID1A (and other components of the BAF complex) is commonly mutated in many cancer types, a role for this complex in regulating general proliferation status may involve co-opting the key cell type-specific cis-regulatory elements. Recent studies highlighted the possibility of exploiting a synthetic lethality-based treatment strategy in ARID1A-mutant ovarian cancers, using inhibitors of BET proteins 37,38. BET inhibitors are proven to be effective in ER-dependent breast cancer cells 25 and our current work implies an increased dependency on epigenetic readers that drive cell division when the activity of the BAF complex is compromised. Given the frequency of BAF mutations in breast cancer, particularly drug resistant contexts, our findings would suggest exploring the potential of epigenetic inhibitors that target the BET proteins.
Online Methods
Preparation of Cas9-expressing clones
MCF7 cells were transduced with Cas9 lentiviral vector pKLV2-EF1aCas9T2ABsd-W with 8 ug/ml Polybrene in 2% serum containing media without antibiotics. Media was replaced after 24 hrs with 10% serum, grown for two more days and selected with 30 µg/ml Blasticidin for four days. These cells were single cell sorted using (BD FACSAria II) in one 96 well plate, seeded with very high suspension and diluted into two 15 cm dishes and grown in the presence of Blasticidin. After 10 days of growth, single cell clones were hand-picked and seeded and grown in two 96 well plates. After the clones were grown well, 48 clones were selected and assessed for Cas9 cutting efficiency using reporter assay in a 6 well plate. Cas9 clones were transduced separately with pKLV2-U6gRNA5(GFPg0)-PGKBFPGFP-W where the cells can express BFP and GFP (control) after 3 days of infection and pKLV2-U6gRNA5(GFPg5)-PGKBFPGFP-W which has a gRNA for GFP. Highly efficient clones were selected which shows ~95% BFP+ cells in the infected population sorted by BD Influx™ Cell Sorter (Supplementary Fig. 1). The average efficiency was calculated from 4 independent experiments. Cas9 cut efficiency is calculated as follows:
Cas9 efficiency (%) = 100 – (%. of GFP+BFP+cells/(%. of GFP-BFP+cells + %. of GFP+BFP+cells)*100) which means 100 – (%. of uncut cells/Total % of transfected cells*100)
1C3 clone was selected from FACS-sorted plate and showed 93.9% Cas9 cut efficiency. This had been used for initial essentiality screen. 3G1 clone sorted from highly diluted plate showed 94.62% Cas9 cut efficiency which was used for drug resistance screening. Both the clones were compared after infection with gRNA library after 9 days for their reproducibility.
Genome-wide CRISPR screening
Highly efficient Cas9-expressing cells were infected with the human gRNA pooled library version-1 with the vector backbone pKLV2-U6gRNA5(lib)-PGKpuroBFP-W23. Cells were seeded two days before in a 15 cm dish to ~30 million cells per replicate. Cells were infected with 30% transduction efficiency (Supplementary Fig. 1) so that only one gRNA gets integrated into the genome per cell. After 3 days (D3), the 30% transduction efficiency was verified by FACS sorting in Influx for BFP+ cells. 60 million cells were collected for next generation sequencing and antibiotic selection was performed on the remaining cells with 10 µg/ml Puromycin for 4 days. BFP+ cells were at least 95% after 4 days of antibiotic selection which was verified by FACS sorting in Influx. Consequently, 100 million cells were collected on different number of days (Day 7, 9, 12, 15 and 20). Genomic DNA was isolated from 20-50 million cells using Qiagen Blood & Cell Culture DNA Maxi Kit along with RNase treatment.
-
Drug resistance CRISPR screening
After 9 days of infection with gRNA library, cells were treated with 100 nM 4-hydroxy-tamoxifen. Fulvestrant and JQ1 were used at 300 nM and 1 µM, respectively, at the start of the assay and gradually reduced to 100 nM and 250 nM, respectively. All treatments were done for 26 days. DMSO was used as a control.
-
Library preparation for CRISPR screens
90 µg of genomic DNA from CRISPR library-infected cells which represents 10 million MCF7 cells (100X representation of gRNA library) were amplified as 5 ug per reaction (20 times) using primers with lentiviral and Illumina adapter sequences with Q5 Hot Start High-Fidelity 2X Master Mix (New England biolabs). Primers were noted in Supplementary Table 6. 15 ng of the whole-genome plasmid library per reaction was used corresponding to 1.7 × 1010 molecules of the plasmid DNA. PAGE-purified primers (Sigma) were used: PCR was done for 25 cycles as follows: Initial denaturation 98°C, 30sec; denaturation 98°C, 10sec: annealing 61°C, 15sec; extension 72°C, 20sec; final extension 72°C, 2min. The PCR reaction was verified using Agarose gel electrophoresis for the presence of 250 bp PCR product. 5 µl from each reaction was taken, pooled and purified using Qiagen PCR purification kit. Second amplification was performed on 100 ng of PCR-purified DNA using Illumina dual indices from Takara ThruPLEX DNA-seq 96D Kit R400407 and KAPA HiFi HotStart ReadyMix for 8 cycles as follows: initial denaturation 98°C, 30sec; denaturation 98°C, 10sec; annealing 66°C, 15sec and extension 72°C, 20sec; final extension 72°C, 5min. Final PCR product was purified using Beckman Coulter Agencourt Ampure XP beads with 0.7X ratio. Libraries were checked for size by Agilent Bioanalyser 2100 or Tapestation 4000 and quantified by qPCR using KAPA library quantification kit with ROX Low qPCR Master Mix or Qubit 3.0 Flurometer. These were pooled and sequenced using an Illumina HiSeq 4000 with 50 bp single end reads with 30% llumina PhiX Control spike-in version 3. 30 million reads per sample to sequence every gRNA from every independently infected cell.
-
CRISPR screening analyses
Short reads were depleted from low-quality sequences and aligned to human gRNA sequence library (GRC h37) using BLAT v. 34 39. Exact-matching reads were counted and treated as a measurement of gRNA abundance. Gene ranking was performed using MAGecK (Full data is available in the Supplementary Table 1 and 2) and log2 fold changes were calculated using DESeq. Heatmaps were generated using median log2 fold changes values from gRNAs specific to a gene. Plasmid library was used as the control for essential gene screening and DMSO for drug resistance screening. Time series clustering was performed using dtwclust R package on genes which showed significant enrichment or depletion. For the heat-map, Dynamic Time Warp algorithm from dtw R package was used for distance measurement followed by hierarchical clustering.
RNA-sequencing
RNA quality was checked using RNA Integrity Number (RIN) from Bioanalyser and 500 ng of RNA was used to prepare libraries using Illumina TruSeq stranded mRNA (HT) library preparation kit. Library size distribution was assessed using the Agilent Tapestation 4200 system. These were sequenced using HiSeq 4000 50 bp single end sequencing. 1% PhiX version 3 viral genome spike-in was introduced during sequencing. Fastaq single-end reads from multiple lanes were merged to make a single library per replicate. STAR40, version 2.5.1a, was used to align reads against hg38 reference genome. The read counting was performed using the intrinsic function of STAR. Differential gene expression analysis used the DESeq2 workflow. All p-values were corrected for multiplicity by means of the Benjamini and Hochberg FDR multiplicity correction.
ATAC-sequencing
Omni ATAC-sequencing was performed according to the established protocol from Corces et al 41. NX# TDE1, Tagment DNA enzyme and buffer from Illumina were used for the transposition reaction. Nextera dual indices were utilized for multiplexing. Sequencing was performed using HiSeq 4000 paired end 150 bp reads.
ChIP-seq and ATAC-seq analyses
Reads were mapped to hg38 genome using bowtie2 2.2.642. Aligned reads with the mapping quality less than 5 were filtered out. The read alignments from three cell culture samples were combined into single library and peaks were called with MACS2 version 2.0.10.2013121626 using sequences from MCF7 chromatin extracts as a background input control. The peaks yielded with MACS2 q value <= 1e-3 were selected for downstream analysis. Genrich (https://github.com/jsh58/Genrich) was used to verify the ATAC-seq peaks from MACS2. Meme version 4.9.143 was used to detect known and discover novel binding motifs amongst tag-enriched sequences.
Differential binding analysis (DiffBind) was performed as described previously 44. For visualizing tag density and signal distribution heatmap the read coverage in a window of +/- 2.5 or 5 kb region flanking the tag midpoint was generated using the bin size of 1/100 of the window length.
Gene signature analysis, KM plots
A set of genes that were evaluated as differentially-expressed in RNA-seq analysis and located in +/- 50kbp vicinity to the differentially-occupied sites evaluated in ChIP-seq analysis was qualified as a potential Gene Signature.
METABRIC27 gene-expression data was accessed via API available at Genomics Data Commons portal (https://gdc.cancer.gov/developers/gdc-application-programming-interface-api) ported to MATLAB. Kaplan-Meier plots and log-rank tests were respectively used to display the survival probabilities per group as a function of time and to test if the hazard functions of the groups of interest are different. Groups of clinical cases (n>=20) of BC ER+ cohorts were stratified by expression of group of genes established at a threshold corresponding to most significant difference in survival.
RIME and qPLEX-RIME
-
RIME on cell lines
Cells were double cross-linked with 2 mM DSG and 1% Formaldehyde as described in ChIP-sequencing in the Supplementary Note. The protocol was followed as in ChIP-seq with following modifications: beads were washed 10 times with RIPA and twice with 100mM ice-cold ammonium hydrogen carbonate. Antibodies used: ARID1A (HPA005456), BRG1 (ab215998), ERα (ab3575 and Merck Millipore 06-935 antibody mix) and negative control IgG (ab171870).
-
qPLEX-RIME on patient-derived xenografts
Frozen clinical tissues were cryosectioned at 30 micron sections and ~90 sections were double crosslinked with 2mM DSG for 25 mins and 1% formaldehyde in the same solution of DSG for 20 mins. Crosslinking was quenched with 0.25M Glycine. The pull down was performed with the ER antibody mix as mentioned in ChIP-seq and qPLEX-RIME sections.
-
Proteomic sample preparation, LC-MS analysis and Data processing
For sample preparation, trypsin at final concentration 15ng/ul (Pierce) was added to the beads followed by overnight incubation at 37°C. A second digestion step was performed the next day for 4h and peptides were cleaned with the Ultra-Micro C18 Spin Columns (Harvard Apparatus) according to manufacturer’s instructions. For the qPLEX-RIME experiment, samples were dried and labelled with the TMT-10plex reagents (Thermo Fisher) followed by fractionation using Reversed-Phase spin columns at high pH (Pierce #84868). For the qPLEX-RIME, peptide fractions were analysed on a Dionex Ultimate 3000 UHPLC system coupled with the nano-ESI Fusion Lumos (Thermo Scientific) mass spectrometer.. The full MS scans were performed in the Orbitrap in the range of 380-1500 m/z at 120K resolution. The MS2 scans were performed in the ion trap with CID collision energy 35%. Peptides were isolated in the quadrupole with isolation window 0.7Th. The top 10 most intense fragments were selected for Synchronous Precursor Selection (SPS) HCD-MS3 analysis with MS2 isolation window 2.0Th. The HCD collision energy was set at 65% and the detection was performed with Orbitrap resolution 50K. For RIME experiments, peptides were analysed on a Dionex Ultimate 3000 UHPLC system coupled with the Q-Exactive HF (Thermo Scientific) or the Q-Exactive mass spectrometers. The full MS scans were acquired in the Orbitrap within the range of 400-1600m/z at 60K or 70K resolution respectively. For MS2, the top 10 most intense precursor ions were selected with a 2.0Th window followed by HCD fragmentation with collision energy 28%. The collected CID and HCD tandem mass spectra were processed with the SequestHT search engine in Proteome Discoverer 2.1 and Proteome Discoverer 2.2 respectively. The SequestHT included the following parameters: Precursor Mass Tolerance 20ppm, Fragment Mass Tolerance 0.5Da for CID and 0.02Da for HCD, Dynamic Modifications were Oxidation of M (+15.995Da), Deamidation of N/Q (+0.984Da) and Static Modifications were TMT6plex at any N-Terminus/K (+229.163Da) for the qPLEX-RIME experiment only. The consensus workflow included calculation of TMT signal-to-noise and the confidence level for peptide identifications was estimated with the Percolator node with decoy database search. The peptide intensities for the qPLEX-RIME experiment were normalized and aggregated (by summing) to protein intensities. The differential protein expression was performed using limma45 implemented in the qPLEXanalyzer tool (10.18129/B9.bioc.qPLEXanalyzer). The Minora Feature Detector node implemented on Proteome Discoverer 2.2 was used for label-free quantification at Maximum ΔRT of Isotope Pattern Multiplets 0.2 min and minimum number of isotopes two peaks. The consensus workflow included Feature Mapper and Precursor Ions Quantifier for Precursor Abundance quantification based on intensity. Complete data is available in Supplementary Table 3 and 4.
Sample size calculation for in vivo MCF7 xenografts
The sample size of the study was defined so that, based on effect sizes defined on prior data and on nuisance parameters deduced from data of Mohammed et al 46, a global power of 0.8 would be achieve when testing a chosen set of differences in means of tumor volumes at the global 5% level for different time points by means of Welsh’s tests.
In vivo xenografts
All mouse experiments were carried out in Biological Resource Unit at CRUK Cambridge Institute. The experiments were in accordance with the UK Animals (Scientific Procedures) Act 1986, with approval from the CRUK Cambridge Institute Animal Ethical Review and Welfare Body. Age matched (8 weeks) NOD/SCID/IL2Rg−/− (NSG) female mice were purchased from Charles River. The animals were verified to be pathogen free and in excellent health. Subcutaneous xenografts of MCF-7 cells/ARID1A clones were conceived by implanting cells 105 in 50% growth media and 50% matrigel (BD Biosciences), in the right flank of 8 weeks old female NSG mice. The mice were also implanted subcutaneously with 90 day-slow release 17b-oestradiol (0.72 mg per pellet) hormone pellets (Innovative Research of America) into the left flank. After 4 weeks for the efficacy cohort, the tumors were randomized and enrolled to the study when the average tumor volume was 100-150 mm3 size. 8.8 mg/ml of Tamoxifen (Tocris Bioscience, 6342) was made in sterile filtered corn oil (Sigma, C8267). The mice were dosed at 20mg/kg, I.P, 6 days a week with Tamoxifen. Tumor sizes were monitored twice a week with Vernier caliper measurement.
As tumor volumes show linear growths on the cubic root scale, we used linear mixed models to compare the average tumor growth of the different groups as a function of time from enrolment on that scale. Linear mixed models allow to take both the within-mouse and time-dependence into account by means of random effects and auto-regressive parameters respectively. We considered here a random intercept and slope model with time since enrolment, groups and an interaction between time since enrolment and groups as fixed effects, and an autocorrelation structure of order 1 for the error term. Model checks suggested a good fit of the model to the data. Sensitivity analyses considering alternative modelling (like models including quadratic terms, other kind of time-dependence or other transformations of the tumor volumes) lead to similar conclusions. We used the program R (version 3.5.1) and the package nlme (version 3.1-137) to fit linear mixed models. Mean values drop for the clones especially at day 18 and 25 as the tumor volume exceeded the 1500 mm3 limit and were removed from the mice.
Test statistics were shown in Supplementary Fig. S5. In the table S5C, fixed effect estimates of the random intercept and slope model used to fit the growth curves in Figure 2C. The model intercept corresponds to the tumour size of the MCF7 WT group at day of enrolment on the cubic root scale. The coefficient related to the variable Days correspond to the daily increase in tumour size for the reference group (MCF7) on the cubic root scale. The two last parameters correspond to shift in daily growth of the KO 11 and KO 14 groups compared to MCF7.
Explant culture
The ARID1A mutant Patient-Derived Xenograft AB555B was grown in dental sponges as previously described 47,48. Spongostan gelatine dental sponges were pre-soaked in explant culture media with or without inhibitors (250 nM JQ1 and 1 µM IBET762) and warmed in a 37°C incubator. One sponge per well was placed in a sterile 24-well tissue culture plate, along with 500 µl explant culture media RPMI 1640 (phenol red-free, L-glutamine-free) (Gibco, 32404-014) with 10% heat inactivated fetal bovine serum (Gibco A3840401), 2mM L-glutamine (Sigma G7513), 10 µg/ml Sigma hydrocortisone (Sigma H0888), 10 µg/ml human recombinant insulin (Sigma I9278)), 100 U penicillin, 100 µg streptomycin, 250 ng amphotericin B /ml (from 1x Sigma anti-biotic, anti-mycotic solution; #A5955). PDX material was cut into 9-12 smaller pieces and each piece was analysed as a replicate. Samples on the sponges were incubated with media with inhibitors for 2 days at 37°C with 5% CO2. These were collected from sponges and fixed in 10% neutral buffered formalin overnight at room temperature. Tissues were processed and embedded in paraffin for histological assessment. Slides were scanned on an Aperio AT2 (Leica) at 20X magnification (resolution 0.5um per pixel) and analysed using HALO software (Indica labs), with the multiplex IHC v2.1.1 module.
Statistical analyses
Two-sided tests were used for all the statistical analyses. Bar graphs were shown with average values and the box plots with median values. Standard deviation was used to denote the error bars in the bar graphs with average values except the proliferation data from Incucyte assays where standard error of the mean was used. For boxplots, centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range). More details about the boxplots on ChIP-seq data are mentioned in Supplementary Table 5.
Generation of Genome Edited ARID1A Knock Out clones
CRISPR guides (sgRNA) were designed against Exon 2 of ARID1A (NM_006015). Oligos (Sigma Aldrich) were cloned into pSpCas9(BB)-2A-GFP (PX458, Addgene # 48138) as previously described49. Guide cutting efficiency was determined in MCF7 and HEK293T cells using the T7 assay (New England Biolabs, following manufacturer’s instructions). To generate independent, non-sister clonal cell lines, MCF7 cells were transiently transfected (Lipofectamine 3000, Thermo Fisher Scientific) with PX458-empty (control), PX458-sgARID1A_2.1 and PX458-sgARID1A2.2, and single cell cloned in 5X 96 well plates per gRNA 96h post-transfection by FACS (BD FACSAria II). gDNA was extracted from each clone (Extracta DNA Prep, VWR, cat#95091-025) and Exon 2 of ARID1A was amplified by PCR (FastStart HF System (Sigma Aldrich, cat#3553361001), primers were noted in Supplementary Table 6 (universal Fluidigm tag in lower case). Amplicons were diluted 1:150 and re-amplified with Fluidigm barcoding primers (incorporating a unique sample barcode and Illumina P5 and P7 adapter sequences), pooled and subjected to sequencing (Illumina MiSeq platform). The AmpliconSeq analysis pipeline was used for data processing and variant calling. Briefly, reads were aligned against the reference genome (GRCh38) using BWA-MEM 50 and variants were called using two methods (VarDict51 and GATK HaplotypeCaller (https://doi.org/10.1101/201178)). Consensus variants and their effects on CRISPR clones were then calculated. All clones used in this paper were STR genotyped and confirmed as free from mycoplasma.
Assessment of off-target CRISPR effects
The top three predicted off targets (ACGGCTCCCTGTCCCCGCAG at chr1:205061276-205061299; AGAGGCCCCAGACCCCGCAG at chr7:1547994-1548017; CCGGCTCCCAGGCCCCGCAG at chr5:10555551-10555574) defined by Desktop Genetics with score 88 out of 10049 were verified for their absence of editing with Sanger sequencing by amplifying the regions with primers against 3 loci from the final knock-out and empty vector control clones (11, 14, 216, 219, 221). Primers are noted in Supplementary Table 6.
8. Extended Data
Extended Data Fig. 1. Enrichment of BAF and P-BAF components in the CRISPR screen.
a. Scatterplot of CRISPR screening data, showing enrichment of BAF components following 26 days of different drug treatment, relative to DMSO treated control cells. n= 3 independent viral infections. b. Log2 fold changes showing gRNA enrichment/depletion against all BAF, P-BAF and ncBAF components in the CRISPR screen. Treatment conditions are compared to DMSO control. More proliferative changes represent enriched gRNA after treatment, indicating genes that contribute to drug resistance. c, e. Validation of ARID1A perturbation effect on proliferation and drug response using ARID1A siRNA on MCF7 (c) and ZR-75-1 (e), representative experiments shown from 2 similar independent experiments each cell line. p-values calculated by One way ANOVA test. * denote p < 0.05, *** denotes p < 0.001. Sample size mentioned in S4. Measure of centre represents mean ± SEM (c) and mean ± SD (e). d. Western blot of ARID1A protein levels after siRNA transfection in MCF7 cells. A representative image is shown from 3 similar independent experiments. Unprocessed Western blot in Source Data Fig. 2.
Extended Data Fig. 2. ARID1A co-binds ER and FOXA1-bound regulatory elements, but is depleted with estrogen treatment.
a-c. Single gene profiles showing the binding of ER, FOXA1 and ARID1A on overlapping sites in MCF7 cells. ChIP-seq was performed using three independent biological cell cultures. d. Overlap of binding sites for ER, FOXA1 and ARID1A binding sites in ZR-75-1 cells. e. Boxplots showing the normalized ChIP-seq tag density around 400 bp window around the center of ARID1A binding on DiffBind-defined estrogen independent (constant) and dependent (reduced with estrogen) sites in MCF7. Both classes show reduced ARID1A binding upon estrogen. p-values were calculated by Welch’s t-test, two-sided. Centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range). Statistical test details are mentioned in Supplementary Table 5e.
Extended Data Fig. 3. Enrichment of SWI/SNF factors with ER and FOXA1 in RIME.
a. ARID1A and BRG1 RIME were conducted on asynchronous MCF7 cells on two biological cell cultures. Label free quantification was performed to show the log 2 scaled normalized intensities of the BAF, P-BAF, ncBAF and common subunits of SWI/SNF complex. Rabbit polyclonal IgG is used as the negative control. b. ER qPLEX-RIME was performed on five primary tumours from ER+ breast cancer patients and the ER interactors are shown as enrichment over IgG vs -log10 p-value, corrected by Benjamini and Hochberg multiplicity correction, two-sided. c, d. Boxplots illustrating the more enrichment of HDAC1 (c) and less enrichment of random factors (d) in ERα RIME in five patients compared to IgG negative control in human breast tumours. The values are scaled to the median of IgG and log2 transformed. e. Boxplots illustrating the enrichment of selected known ERα interactors from the RIME experiment in MCF7 cells at a representative timepoint (4-hydroxytamoxifen- 24 hrs) comparing to IgG negative control. The values are scaled to the median of IgG and log2 transformed. n = 5 independent biological cell cultures. For all boxplots, Centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range).
Extended Data Fig. 4. Enrichment of SWI/SNF factors during Tamoxifen and Fulvestrant in ChIP-seq experiments.
a-d. Asynchronous MCF7 cells were treated with vehicle or Fulvestrant, an ER degrader and ChIP-seq was conducted for ARID1A (b), BRG1 (c) or SNF5 (d). Triplicate independent cell cultures were conducted. d. Single gene profile showing the induction of SWI/SNF complex binding during Fulvestrant treatment. e. Overlap of ARID1A lost sites during estrogen treatment with gained sites during Tamoxifen and Fulvestrant from three independent biological cell cultures. f. Overlap of ARID1A gained sites during Tamoxifen treatment with Fulvestrant and Tamoxifen downregulated genes.
Extended Data Fig. 5. FOXA1 promotes the binding of ARID1A and BRG1.
Hormone-deprived ZR-75-1 cells were transfected with control or FOXA1 siRNA and ChIP-seq was conducted for ARID1A (a) and BRG1 (b). n = 3 independent biological cell cultures. MA plots are shown with the average intensity of binding vs log2 fold change with FOXA1 siRNA relative to control siRNA. c. Scatterplot showing the association of the loss of ARID1A and BRG1 binding upon FOXA1 knockdown. PCC – Pearson Correlation coefficient, two-sided. d. Heatmaps shown on ARID1A and BRG1 FOXA1 independent (common) and dependent (lost sites with FOXA1 knockdown) sites in ZR-75-1 cells. e. Boxplots showing the normalized ChIP-seq tag density around 400 bp window of ARID1A and BRG1 on FOXA1 independent (constant, n=70,429 sites) and dependent (lost sites with siFOXA1, n=17,357 sites) sites in ZR-75-1. p-value calculated by Welch’s test, two-sided. n = 3 independent biological cell culture samples. Centre line shows the median values with bounds of box corresponding to the first and third quartiles and the upper and lower whiskers extend to the largest or the smallest value no further than 1.5 × IQR (inter-quartile range). Statistical test details are mentioned in Supplementary Table 5f.
Extended Data Fig. 6. FOXA1 promotes the binding of ARID1A and BRG1.
Hormone-deprived MCF7 and ZR-75-1 cells were transfected with control or FOXA1 siRNA and ChIP-seq was conducted for ARID1A and BRG1. n = 3 independent biological cell cultures. (a-b) Single gene profiles of CCND1 (a) and CDH1 (b) showing the effect on SWI/SNF complex binding with FOXA1 knockdown on MCF7 and ZR-75-1 cells. ER and FOXA1 binding overlap is shown. (c-d) ChIP-qPCR analyses on specific sites (CCND1 and CDH1 ER binding sites) showing ARID1A and BRG1 binding with FOXA1 knockdown in hormone-deprived MCF7 and ZR-75-1 cells (c) or ARID1A binding following Tamoxifen treatment in asynchronous MCF7 cells (d). n = 3 independent biological cell cultures. * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001. Precise p-values are mentioned in Fig. S10. Mean is measured as centre shown with standard deviation. Details of the statistical tests are mentioned in Fig. S10.
Extended Data Fig. 7. ATAC-seq analyses shows a negligible regulation of ARID1A on transcription-associated chromatin opening.
a. Heatmap showing ATAC-seq analysis in ARID1A KO clones 11 and 14 following Tamoxifen treatment. Common, gained and lost sites defined by DiffBind analysis. n = 4 independent biological cell cultures. FDR ≤ 0.05 corrected by Benjamini-Hochberg multiplicity correction, two-sided. b. Association of ARID1A KO upregulated and downregulated genes with ATAC-seq gained and lost sites.
Extended Data Fig. 8. ARID1A perturbation regulates ARID2 binding.
a. ARID2 ChIP-seq was conducted in wild type cells or the two ARID1A knock-out clonal cell lines and heatmaps are shown on ARID2 binding sites after Tamoxifen treatment. Also included was ARID1A ChIP-seq from wild type cells treated with vehicle or Tamoxifen. ARID2 binding overlapped with ARID1A binding and was dependent on ARID1A. n = 3 independent biological cell cultures. b. Signal intensity plot showing changes in ARID2 binding in wild type control cells or ARID1A knock-out cells at ARID2 binding sites. n = 3 independent biological cell cultures.
Extended Data Fig. 9. ARID1A promotes BRG1 and HDAC1 binding without affecting ER and H3K27ac occupancy.
a, b. BRG1, H3K27Ac, HDAC1 and ER (b) ChIP-seq were conducted in asynchronous wild type cells treated with vehicle or tamoxifen or in the two ARID1A knock-out clones (Clones 11 and 14) following tamoxifen treatment. The binding is shown on regions where HDAC1 is lost in ARID1A knockout cells relative to wild type cells. n = 3 independent biological cell cultures. c, d. Scatterplot showing the correlation of ER (c) or H3K27Ac (d) and HDAC1 binding in ARID1A knockout clone 11 versus wild type cells. n = 3 independent biological cell cultures. PCC – Pearson Correlation coefficient. p-values were calculated by Pearson correlation test, two-sided. e. Principal Component Analysis (PCA) of normalised peptide intensities of PDX tumours after ER qPLEX-RIME. n= 2 PDX each group. f. Details of ARID1A mutations observed within ER+ PDX tumours used in ER qPLEX-RIME.
Extended Data Fig. 10. ARID1A regulates histone H4 acetylation.
Upregulation of histone H4 acetylation in ARID1A knock-out clone 11 and 14 in Vehicle (a) or Tamoxifen (b) treated cells comparing to wild type cells. Heatmap representing the changes in histone H4Ac marks upon ARID1A knockout with Vehicle or Tamoxifen treatment on ER binding sites close to ARID1A repressed genes. n =3. (c) Empirical cumulative probability distribution plots of H4K8Ac and H4K12Ac ChIP-seq signals showing upregulation in intensity (y-axis) with ARID1A knockouts clones 11 and 14. Plots were made on ER sites close to ARID1A repressed genes (n=686 sites) with more than 75% contribution to the variance in intensity. Window – 2 kb around the center of binding.
Supplementary Material
Acknowledgments
The authors would like to thank Genomics, Proteomics, Bioinformatics, Preclinical genome editing, Histopathology and the Flow cytometry core facilities at Cancer Research UK Cambridge Institute. In particular, we thank C. D’Santos from Proteomics, A. Smith from Preclinical genome editing and M. Clayton, G. Cronshaw, R. Ellis and all the staff in CRUK CI BRU. We acknowledge the support of the University of Cambridge and Cancer Research UK. pSpCas9(BB)-2A-GFP (PX458) used for making CRISPR knockout clones was a kind gift from F. Zhang, Broad Institute, Boston (Addgene # 48138) and the BRD4 antibody from Prof. Cheng Ming Chiang, UT Southwestern Medical Center, Texas. We acknowledge the suggestions from Z. Najafova for the mice experiments and A. Nicholson, M. Sen, X. Wang and S. A. Johnsen for reagents and antibodies. We thank R. Rony for his help in graphic design and models. J.S. Carroll is supported by an ERC Consolidator award (Project number 646876), a Susan G Komen leadership grant and CRUK core funding. S. Nagarajan is supported by Komen grant. S.V. Rao is funded by Innovation postdoc granted by the Norwegian Research Council. R. Siersbæk is funded by the Novo Nordisk Foundation (NNF15OC0014136).
Footnotes
Author contributions
S.N contributed on conceptualization, methodology, experimental work, formal analysis and figure assembly, writing of text, reviewing and advising on the manuscript. S.V.R contributed on experimental work, analysis and figure assembly of mice experiments and advising on the manuscript. J.S contributed on experimental work, analysis and figure assembly of mice experiments and advising on the manuscript. D.C contributed on experimental work and advising on the manuscript. S.D contributed on methodology and advising on the manuscript. E.K.P contributed on experimental work, formal analysis and figure assembly and advising on the manuscript. J-E.G.P contributed on experimental work and advising on the manuscript. D-L.C contributed on statistical analyses, figure assembly, reviewing and advising on the manuscript. S.K contributed on methodology and advising on the manuscript. K.K contributed on bioinformatic analyses, figure assembly, and advising on the manuscript. C.S.R.C contributed on bioinformatic analyses, figure assembly, and advising on the manuscript. S-E.G contributed on methodology and advising on the manuscript. E.A.G contributed on experimental work and advising on the manuscript. C.B contributed on histopathological analyses and advising on the manuscript. N.G performed ARID1A immunohistochemistry and advising on the manuscript. R.N performed ARID1A immunohistochemistry and advising on the manuscript. A.B contributed on methodology regarding PDX material and advising on the manuscript. C.C contributed on methodology regarding PDX material, funding acquisition and advising on the manuscript. A.R methodology and advising on the manuscript. R.S contributed on methodology, supervision and advising on the manuscript. K.Y contributed on methodology, funding acquisition and advising on the manuscript. I.C contributed on methodology, bioinformatic analyses and figure assembly, writing of text and advising on the manuscript. J.S.C contributed on conceptualization, supervision, funding acquisition, writing of text, reviewing and advising on the manuscript.
Declaration of Interests
Jason S. Carroll is the founder and CSO of Azeria Therapeutics.
Data Availability Statement
All CRISPR, ChIP-seq and RNA-seq data has been deposited at GEO and can be accessed at GSE123286. ATAC-seq data can be accessed at GSE134270. All proteomic data has been deposited at PRIDE and can be accessed at PXD011810.
References
- 1.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 2.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Eeckhoute J, et al. Positive cross-regulatory loop ties GATA-3 to Estrogen Receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–83. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 4.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–43. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 5.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 6.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–99. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammed H, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–9. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher TM, et al. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–63. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John S, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–24. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Michel BC, et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20:1410–1420. doi: 10.1038/s41556-018-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mashtalir N, et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell. 2018 doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. Embo J. 2002;21:4094–103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Pedrero JM, Kiskinis E, Parker MG, Belandia B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem. 2006;281:22656–64. doi: 10.1074/jbc.M602561200. [DOI] [PubMed] [Google Scholar]
- 14.Jeong KW, Lee YH, Stallcup MR. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J Biol Chem. 2009;284:29298–309. doi: 10.1074/jbc.M109.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiRenzo J, et al. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol. 2000;20:7541–9. doi: 10.1128/mcb.20.20.7541-7549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Z, et al. Vitamin D Switches BAF Complexes to Protect beta Cells. Cell. 2018;173:1135–1149 e15. doi: 10.1016/j.cell.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho HD, et al. Loss of Tumor Suppressor ARID1A Protein Expression Correlates with Poor Prognosis in Patients with Primary Breast Cancer. J Breast Cancer. 2015;18:339–46. doi: 10.4048/jbc.2015.18.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates LR, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell. 2017;32:169–184 e7. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67. doi: 10.1016/j.gde.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7 doi: 10.1038/ncomms11479. 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzelepis K, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–36. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagarajan S, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–9. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-9-r137. R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glont SE, et al. Identification of ChIP-seq and RIME grade antibodies for Estrogen Receptor alpha. PLoS One. 2019;14:e0215340. doi: 10.1371/journal.pone.0215340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papachristou EK, et al. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04619-5. 2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruna A, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167:260–274 e22. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol Biol Cell. 2008;19:3308–22. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30:3885–94. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell Rep. 2016;17:2715–2723. doi: 10.1016/j.celrep.2016.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–9. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 36.Cirillo LA, Zaret KS. Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J Mol Biol. 2007;366:720–4. doi: 10.1016/j.jmb.2006.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berns K, et al. ARID1A mutation sensitizes most ovarian clear cell carcinomas to BET inhibitors. Oncogene. 2018;37:4611–4625. doi: 10.1038/s41388-018-0300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caumanns JJ, Wisman GBA, Berns K, van der Zee AGJ, de Jong S. ARID1A mutant ovarian clear cell carcinoma: A clear target for synthetic lethal strategies. Biochim Biophys Acta Rev Cancer. 2018;1870:176–184. doi: 10.1016/j.bbcan.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark R, Brown GD. DiffBind: differential binding analysis of ChIP-Seq peak data. Bioconductor. http://www.http://http://bioconductor.org/packages/release/bioc/html/DiffBind.html. [Google Scholar]
- 45.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohammed H, et al. Progesterone receptor modulates ERalpha action in breast cancer. Nature. 2015;523:313–7. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10:483–7. doi: 10.1038/nrurol.2013.126. [DOI] [PubMed] [Google Scholar]
- 48.Centenera MM, et al. A patient-derived explant (PDE) model of hormone-dependent cancer. Mol Oncol. 2018;12:1608–1622. doi: 10.1002/1878-0261.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All CRISPR, ChIP-seq and RNA-seq data has been deposited at GEO and can be accessed at GSE123286. ATAC-seq data can be accessed at GSE134270. All proteomic data has been deposited at PRIDE and can be accessed at PXD011810.