Abstract
Epigenetic regulators are the largest group of genes mutated in MDS patients. Most mutated genes belong to one of three groups of genes with normal functions in DNA methylation, in H3K27 methylation/acetylation or in H3K4 methylation. Mutations in the majority of epigenetic regulators disrupt their normal function and induce a loss-of-function phenotype. The transcriptional consequences are often failure to repress differentiation programs and upregulation of self-renewal pathways. However, the mechanisms how different epigenetic regulators result in similar transcriptional consequences are not well understood. Hypomethylating agents are active in higher risk MDS patients, but their efficacy does not correlate with mutations in epigenetic regulators and the median duration of hematologic response is limited to 10-13 months. Inhibitors of histone deacetylases (HDAC) yielded disappointing results so far, questioning this approach in MDS patients. We review the clinical relevance of epigenetic mutations in MDS, discuss their functional consequences and highlight the role of epigenetic therapies in this difficult to treat disease.
Keywords: MDS, epigenetics, ASXL1, DNMT3A, EP300, EZH2, IDH1, IDH2, KDM2B, KDM5A, KDM6A, MLL, MLL2, MLL3, MLL5, TET2, WT1, hypomethylating agent, HDAC inhibitor
Incidence and prognostic impact of mutations in epigenetic regulators
Our knowledge about the mutational landscape of MDS has markedly increased over the last 5-10 years due to advancement of sequencing technologies and comprehensive sequencing approaches by cooperative groups.1,2 Currently, while more than 100 recurrently mutated genes are known to occur in MDS, only about 5 are known to be mutated in more than 10% and only about 10-15 in more than 5% of patients.1,2 In this review we refer to MDS based on the WHO 2008 and 2016 classifications thus excluding patients with chronic myelomonocytic leukemia (CMML). Gene mutations in MDS can be classified according to their function. The following functional groups have been described in MDS:
-
-
DNA methylation and Chromatin modification = Epigenetic modifiers (DNA methylation: e.g. TET2, DNMT3A, IDH1/2; Chromatin modification: e.g. ASXL1, EZH2)
-
-
Splicing (e.g. SF3B1, SRSF2, U2AF1)
-
-
Transcription (e.g. RUNX, BCOR, BCORL1, ETV6)
-
-
Cohesin (e.g. STAG2, RAD21, SMC1A, SMC3)
-
-
Receptor/Kinases (e.g. JAK, MPL)
-
-
Ras Pathways (e.g. CBL, PTPN11)
-
-
DNA repair (e.g. BRCC3, ATM)
Gene mutations belonging to the group of epigenetic modifiers represent the largest group of mutated genes in MDS along with genes in the splicing machinery. The most frequently mutated genes in this group are Ten-eleven-translocation 2 (TET2)3, DNA (cytosine-5)-methyltransferase 3A (DNMT3A)4, Isocitrate dehydrogenase 1 and 2 (IDH1 and 2)5, Additional sex combs like 1 (ASXL1)6 and Enhancer of zeste homolog 2 (EZH2).7,8 These genes play a distinct role in epigenetic regulation.9 Thus, we can propose that mutations in epigenetic modifier genes have also a direct epigenetic impact. The exact functional impact of these different mutations on the epigenetic fingerprint of hematopoetic stem cells (HSC) is of key interest for understanding the pathogenesis of MDS.
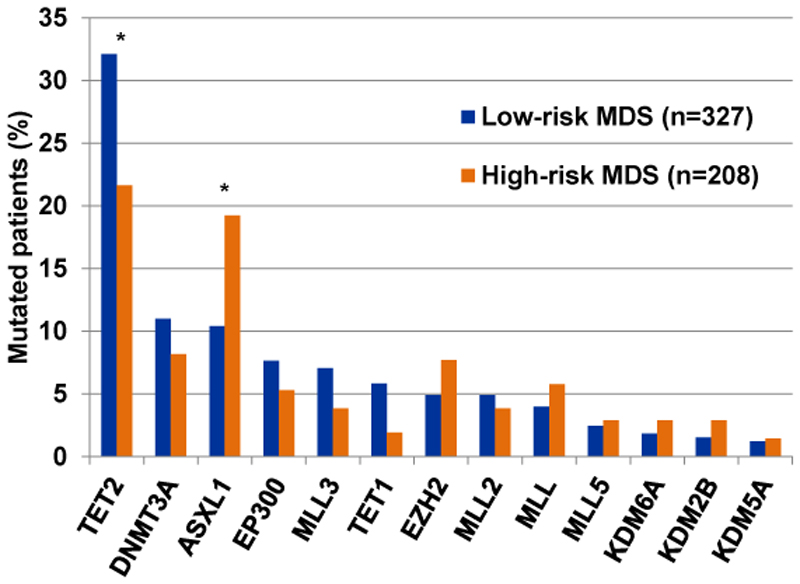
TET2 is an important epigenetic regulator that may lead to DNA demethylation. It encodes a protein that is known to convert methylcytosine, a modified DNA base, to 5-hydroxymethylcytosine (methylcytosine dioxygenase). We know that DNA methylation and hence also demethylation are key epigenetic mechanisms that impact cell differentiation and proliferation. Mutations in TET2 occur in 22-35% of patients with MDS1,2 and are more frequent in low-risk WHO categories compared to high-risk WHO-categories of MDS (Figure 1).2 Unlike mutations in SF3B1 they are not specifically associated with one morphologic subgroup, but can occur in all subgroups of MDS, which is also true for other gene mutations involving epigenetic modifiers.1,2 TET2 mutations in hematologic malignancies can be single nucleotide variants (SNV) as well as frameshift mutations in MDS. While TET2 mutations in the C-terminal catalytic region are mostly missense mutations, nonsense and frameshift mutations mostly occur in the N-terminal region. Thus, all of these mutations can lead to a premature truncation of the catalytic part of the enzymes, which disrupts the catalytic activity of TET2.10
Figure 1.
Mutation frequency in epigenetic regulators in low-risk (refractory anemia (RA), refractory anemia with multilineage dysplasia (RCMD), and RCMD with ringsideroblasts (RCMD-RS) and high-risk (refractory anemia with excess of blasts (RAEB-1 and RAEB2)) WHO categories of MDS patients including mutations of oncogenic, possibly oncogenic as well as of unknown oncogenic potential. The data is based on a publication by Papaemmanuil et al. using mutation information in Supplementary Table S2.2 * indicates P<0.01.
Interestingly, while TET2 mutations are more frequent in MDS compared to AML, the reverse is true for IDH1 and IDH2 mutations.1,2,11 While both IDH1 and IDH2 mutations occur in nearly 10% of AML patients and thus affect nearly 20% of AML patients combined, IDH1 and IDH2 mutations occur in only 2-5% of MDS patients.1,2,12 In AML, there is a strong association between IDH mutations and normal karyotype, and NPM1 mutations and functional synergisms have been described for these two gene mutations.11,13 NPM1 mutations affect less than 2% of MDS patients and it is not surprising this association has not been described in MDS.1,2 IDH mutations are always a heterozygous point mutation affecting an arginine in codon R132 in IDH1 and codon R140 or R172 in IDH2. IDH1 and IDH2 are both enzymes in the citric acid cycle that catalyze the conversion of isocitrate to α-ketoglutarate. However, the mutated enzymes produce instead of α-ketoglutarate R-2-hydroxyglutarate (R-2HG).14 α-ketoglutarate is a cofactor for more than 80 enzymes like TET2, several histone demethylases, and prolyl hydroxylases.15 Several studies showed that R-2HG is a competitive inhibitor of α-ketoglutarate-dependent enzymes and thus IDH1/2 mutations have broad epigenetic consequences.16
DNMT3A is another important epigenetic regulator by encoding an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA.17 DNMT3A mutations were first described in AML where they present one of the most frequent mutations occurring in over 20% of AML patients.18,19 In MDS however, DNMT3A mutations are rarer occurring in not more than 10% of patients.1,2,4 About two thirds of mutations are missense mutations affecting arginine R882. HSCs with mutated DNMT3A appear to have a proliferative advantage comparted to wild-type HSCs and predispose HSCs to malignant transformaiton.20,21 DNMT3A is also the most commonly mutated gene identified in clonal hematopoiesis of indeterminate potential (CHIP).22 Additionally, there appears to be a direct link between DNMT3A and TET2 in hematopoietic cells. Both genes were shown to compete as well as cooperate in repressing lineage-specific transcription factors in HSCs.23
ASXL1 encodes for a chromatin-binding protein. The protein is thought to disrupt chromatin in localized areas which leads to enhanced transcription of some genes, while repressing the transcription of others.24 Importantly it belongs to the enhancer of trithorax and polycomb (ETP) genes that can both activate and repress HOX genes.24 Mutations in ASXL1 are after TET2 and SF3B1 mutations the most frequent mutations occurring in MDS with a frequency of 15-20%.1,2,6 ASXL1 mutations are more frequent in high-risk than in low-risk MDS patients (Figure 1).2 The mutations are associated with intermediate risk karyotype but not with other clinical parameters.6 Missense, nonsense and frameshift mutations have been described and all mutations affect exon 12, leading to truncation or changes in the plant homeodomain (PHD) of the gene, which is the main functional domain of the protein.6 It was shown that truncation of the C-Terminus involving the PHD domain induces MDS in vivo via inhibition of PRC2.25 In mice, the changes seen with C-terminal truncation of ASXL1 include multi-lineage myelodysplasia, pancytopenia, and occasional progression to overt leukemia.25,26 This functional finding is in line with the finding that frameshift and nonsense, but not missense mutations are responsible for the adverse prognostic impact of ASXL1, as frameshift and nonsense mutations have the strongest impact on C-terminal truncation.6
EZH2 mutations only affect 5-6% of patients with MDS and are rare in AML.1,2 Together with other proteins (EED, SUZ12 and RBBP4) EZH2 forms the polycomb repressive complex 2 (PRC2). Within the PRC2 complex EZH2 is of key functional significance as it forms the catalytic region of the complex. The PRC2 complex is a highly conserved histone H3 lysine 27 (H3K27) methyltransferase that contributes to epigenetic silencing of many genes.27 The functional activity of EZH2 can be disrupted by mutations as well as loss of chromosome 7q, where EZH2 is located. The latter is more frequently seen in MDS compared to AML patients. It was shown that premature chain termination of EZH2, as seen with EZH2 mutations and 7q loss found in MDS, causes direct abrogation of histone methyltransferase activity which again changes the epigenetic structure of cells.7 Another mechanism of EZH2 inhibition in MDS was found in SRSF2 mutated CMML and AML patients. Mutations in SRSF2 are frequent in MDS patients28 and disrupt regular splicing of many target genes.29 Interestingly, mutated SRSF2 also aberrantly splices EZH2, which was associated with lower EZH2 protein levels as well as lower global levels of histone H3K27 trimethylation.30 Frequent loss of EZH2 protein expression was found in chemotherapy resistant AML patients.31 Loss of EZH2 expression was dependent on phosphorylation by CDK1, stabilization by HSP90 and proteasomal degradation. However, whether this mechanism is relevant to MDS is yet unknown.
In the genomic study of MDS by Papaemmanuil et al. a significant proportion of patients displayed mutations in EP300, KMT2A/C/D/E (MLL family) genes, KDM6A/UTX, KDM5A and KDM2B (Figure 1).2 For many mutations it was unclear at the time whether these mutations are oncogenic or not. Nevertheless, a similar frequency of these mutations was found in AML patients in a more recent study and many of them were classified as oncogenic.11 KMT2A/C/D/E genes encode histone 3 lysine 4 (H3K4) methyltransferases, while KDM5A and KDM2B are demethylases at H3K4, and KDM6A is a demethylase at H3K27. KMT2A/C/D/E gene mutations were found in 17.6% of MDS patients (excluding del5q and CMML) and mutations in the demethylases were found in 5.6% of the patients.2 Thus, mutations in KMT2A/C/D/E genes and lysine demethylases may be more common than previously appreciated, but their prognostic and functional impact in MDS has not been defined.
The detection of mutations in MDS can not only help us to unravel the pathogenesis of the disease and find novel treatments, but it can also help us with prognostication of individual patients. While TET2 mutations appear to have no prognostic impact in MDS, ASXL1, EZH2, and DNMT3A mutations were shown to have an independently adverse prognostic impact in MDS patients.4,6–8,32 IDH1 mutations were initially found associated with poor prognosis,5 but a large study did not find a prognostic impact of IDH1/2 mutations on OS or AML progression in MDS patients.12 In patients undergoing allogeneic hematopoietic stem cell transplantation (alloHSCT) most epigenetic regulators lose their prognostic impact. In the largest study to date epigenetic regulators did not affect prognosis after alloHSCT, while a smaller study found an adverse prognostic effect for patients with ASXL1 mutations after alloHSCT.33,34 In our own study of 304 MDS and secondary AML patients we did not find a prognostic impact of epigenetic regulators after alloHSCT.35
Causes and consequences of epigenetic dysregulation in MDS
Modifiers of DNA methylation
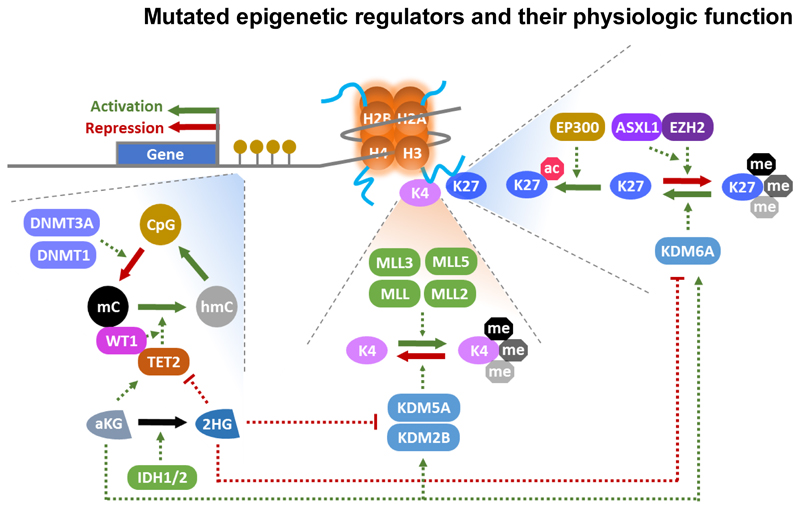
DNA hypermethylation has been recognized as a common feature of MDS and has been associated with poor prognosis in MDS patients.36 The efficacy of DNMT inhibitors like azacitidine and decitabine in MDS patients strongly suggested the pathophysiologic relevance of DNA methylation. However, the origin of DNA hypermethylation is currently being unraveled and causes and consequences have to be carefully distinguished. Besides mutated epigenetic regulators discussed in detail below, mutations in splicing factors may also dysregulate epigenetics through splicing of epigenetic enzymes or direct binding to nucleosomes in exonic DNA as shown for SF3B1.37 Figure 2 provides an overview on the physiologic function of epigenetic regulators that are mutated in MDS and Table 1 summarizes mutation frequency, molecular function, epigenetic target and target genes/pathways of these genes.
Figure 2.
Molecular function of epigenetic regulators that are frequently mutated in MDS. A nucleosome is shown with histone tails. Enzymes regulating H3K4 and H3K27 are shown on the right side of the graph. Yellow circles represent CpG dinucleotides and their methylation is described on the left side of the graph; Me: methylation mark; green arrows: positive regulation, red arrows: negative regulation.
Table 1. Molecular function and transcriptional consequences of mutated epigenetic regulators in MDS.
| Mutated gene | Mutation frequency in MDS | Molecular function | Epigenetic targets | Regulated genes/pathways |
|---|---|---|---|---|
| ASXL1 | 15-20%1,2,6 | Accessory protein of PRC2 complex | Targets of the PRC2 complex | HOX (mainly HOXA5-9)26 |
| DNMT1 | <1%2 | Maintenance of DNA methylation | CpG methylation | Upregulation of Ezh2 targets, Klf4, Dusp6, Ccnd2, Tgfbi120 |
| DNMT3A | <10%1,2,4 | De novo DNA methylation | Cytocine methylation, is recruited to H3K36me3, can recruit HDAC | Upregulation of HSC genes (Gata3, Runx1, Pbx1, Cdkn1a, Vasni), downregulation of differentiation genes (Flk2, Ikaros, Sfpi1 (PU.1), Mef2c)41 |
| EP300 | ~7%2 | Histone acetyltransferas e (writer) | H3K122ac, H3K27ac | SIRT2, HDAC1, ALX1121 |
| EZH2/KM T6A | ~6% 1,2 | Histone methyltransfera se (writer) | H3K27me3 | Myc targets, cell cycle genes, PRC2 targets (Hox), Adhesion molecules ICAM, NCAM, E-cadherin and VE-cadherin, inflammatory cytokine response genes (Il6 and Tnfa pathways)61 |
| KDM2B (FBXL10) | ~2%2 | Histone demethylase (eraser) | H3K4me3 and H3K36me2 | Binds ribosomal RNA and inhibits its transcription, maybe also part of E3 ubiquitin ligase complex84,85 |
| KDM5A (JARID1A) | ~1%2 | Histone demethylase (eraser) | H3K4me2 and H3K4me3 | HOX, RB, CXCL12, nuclear receptors88 |
| KDM6A (UTX) | ~2%2 | Histone demethylase (eraser) | H3K27me2 and H3K27me3 | Loss of UTX induces down-regulation of GATA-1, LYL-1, SCL, KLF1, and IKAROS, as well as significant up-regulation of PU.1 |
| MLL/KMT2A | ~5%2 | Lysine methyltransferase (writer) | H3K4me | HOX (HOXA9)68 |
| MLL2/KMT2D | ~4%2 | Lysine methyltransferase (writer) | H3K4me | Estrogen receptor, beta globin79 |
| MLL3/KMT2C | ~6%2 | Lysine methyltransferase (writer) | H3K4me | Loss of MLL3 leads to inhibition of the following pathways: Antigen processing and presentation, Immune response, lineage differentiation77 |
| MLL5/KMT2E | ~3%2 | unknown | unknown | Cell cycle genes122 |
| TET1 | ~4%2 | Methylcytosine dioxygenase | 5 methylcytosine (hydroxylation), H4R3 | Indirectly represses B lymphoid transcription factors EBF1, Pax5 and IRF4123 |
| TET2 | 22-35%1,2 | Methylcytosine dioxygenase | 5 methylcytosine (hydroxylation) | Activates HSC specific genes (Meis1, Evi1), represses RBC signature genes (Klf1, Epor), myeloid transcription factors (Cebpa, Cebpδ, Mpo, and Csf1) and the IL-6 pathway (independent of DNA hydroxymethylation but through PRC2)124 |
| WT1 | ~1%2 | Transcription factor | Interaction parter of TET2 | TBL1X, BTRC, DACT1, LEF1, NLK125 |
| IDH1 | ~2%1,2,12 | No direct epigenetic activity | indirect inhibitor of TET2, KDM2A, KDM4A/C, and KDM5B | HOXA cluster, MAPK signaling, DNA damage response and DNA repair pathways through activating ATM expression (independent of Tet2) 51, 55 |
| IDH2 | ~4%1,2,12 | No direct epigenetic activity | indirect inhibitor of TET2, KDM2A, KDM4A/C, and KDM5B | TGFβ, WNT and HOX pathways16 |
DNMT3A
DNMT3A modifies DNA enzymatically by methylation of cytosines. The most common mutation of DNMT3A at amino acid 882 is a loss of function mutation, which also inhibits the wildtype allele in patients with heterozygous mutations and reduces the enzymatic activity of DNMT3A by approximately 80%.38 Functional loss of Dnmt3a in murine hematopoiesis resulted in an increase of hematopoietic stem cells upon serial transplantation.39 Transcriptional profiling confirmed that Dnmt3a null HSCs are characterized by increased HSC signature genes and concomitant decreased lineage specification genes.39 Earlier work had already suggested that DNMT1 acts as a tumor suppressor by restricting lineage fates of hematopoietic progenitor cells and more recent work confirmed a tumor suppressive role of DNMT3A.40,44 More recently it was shown that mutant DNMT3A causes focal DNA hypomethylation and restricts the degree of hypermethylation in AML cells.41,42 They showed that DNA hypermethylation is a consequence of cell proliferation that is independent of oncogenic signaling and can be also found in non-malignant cells upon cytokine stimulation.41 One might assume that DNMT3A mutant-associated hypomethylation results in failed repression of self-renewal pathways and lineage programs. However, hypomethylation had minimal effects on gene transcription, and additional mechanisms have to be evaluated to understand the mechanism of stem cell expansion in DNMT3A mutated cells. Translational studies evaluated genetic mutations as predictors of response to hypomethylating agents, but did not find mutated DNMT3A to correlate with response, which is expected for a loss-of-function mutation in DNMT3A and its associated DNA hypomethylation.
TET2
TET2 modifies DNA enzymatically by hydroxylation of methylcytosines.43 During replication the hydroxyl-methylcytosines become demethylated representing an important mechanism of DNA demethylation. TET2 mutations in MDS are loss-of-function mutations, which are associated with DNA hypermethylation.43 Specifically, a progressive and widespread DNA hypermethylation of active enhancer elements was shown in preleukemic hematopoietic cells, while CpG island and promoter methylation did not change upon Tet2 deletion.44 Tet2-dependent hypermethylation resulted in down regulation of putative tumor suppressor genes (e.g. Mtss1, Las2, Lxn, Ctdspl, Grap2) and upregulation of putative oncogenes (e.g. Aff3, Pim2, Nepn, Notch3, Igf1r).44 Functionally, it was shown that loss of Tet2 leads to increased stem cell self-renewal leading to enlargement of the hematopoietic stem cell compartment and repopulating activity.45 Concomitantly, upregulation of the self-renewal regulators (Meis1, Evi1) and downregulation of myeloid lineage transcription factors (Cebpa, Cebpδ, Mpo, Csf1) were observed upon Tet2 deletion.45 A modest differentiation block was noted in the myeloid, erythroid and lymphoid lineages upon Tet2 deletion and some mice developed a disease reminiscent of chronic myelomonocytic leukemia.46
IDH1 and IDH2
IDH1 and IDH2 mutations have been functionally linked to TET2 as the oncometabolite R-2-hydroxyglutatarate, which is produced by mutant IDH1/2, inhibits the enzymatic function of TET2.47 However, two observations suggest that TET2 is not the primary target of R-2HG: First, R-2HG is a much weaker inhibitor of TET2 than S-2HG, yet R-2HG but not S-2HG is leukemogenic (see below).48 Second, DNA hypermethylation occurs at much later passage than histone H3K9 methylation in cells overexpressing mutant IDH1,49 suggesting that DNA hypermethylation is rather a consequence than a cause of mutant IDH1/2. The H3K9 demethylases KDM4A and KDM4C are inhibited by R-2HG and S-2HG with equal potency and H3K9 hypermethylation is one of the first epigenetic changes to occur upon expression of mutant IDH1,49 suggesting that KDM4A/C are primary targets of mutant IDH1.
Expression of mutant IDH1 from the endogenous locus in hematopoietic cells induced an expansion of hematopoietic progenitor cells, splenomegaly and anemia, while the mice did not develop leukocytosis or leukemia.50 It was shown that mutant IDH1 and IDH2 need a collaborating oncogene to induce leukemia in vivo.51,52 Coexpression of mutant IDH1 and HOXA9 accelerated the onset of monocytic leukemia in mice.51 When the potential oncogenic effects of R-2HG, S-2HG and αKG were evaluated in vivo, it became evident that only R-2HG is oncogenic in hematopoietic cells, but not S-2HG or αKG. Cytochrome c oxidase (COX), which represents complex IV of the mitochondrial electron transport chain, has been identified as a target of 2HG, which is specifically inhibited by R-2HG but not S-2HG.53 This inhibition was specific for complex IV of the mitochondrial electron transport chain, as complex I, II, III and V were not affected. Inhibition of COX lowered the apoptotic threshold and rendered the cells dependent on the activity of apoptosis inhibitor BCL2. Interestingly, IDH1 mutant AML patients were especially sensitive to the BCL2 inhibitor venetoclax underscoring the functional importance of COX inhibition by R-2HG.54 In addition, mutant IDH1 inhibits ATM expression and the DNA damage response pathway independently of TET2 through R-2HG-mediated inhibition of histone demethylases.55
Taken together, TET2 and DNMT3A mutations seem to result in opposite epigenetic changes at DNA level, but result in similar biologic effects like a gradual expansion of hematopoietic stem and progenitor cells. Double knockout of TET2 and DNMT3A revealed competitive and cooperative effects on gene expression with reduced expression of erythroid regulators Klf1 and Epor and upregulation of myeloid transcription factors Cebpα and Cebpε.23 TET2 and IDH1/2 mutations have been described as functionally redundant mutations, but recent observations suggest also TET2-independent mechanisms of these mutations.
Modifiers of H3K27 methylation and acetylation
Histone 3 lysine 27 (H3K27) is modified by several enzymes that are mutated in MDS patients. It is trimethylated by EZH2 as the catalytically active part of the PRC2 complex. ASXL1 interacts with and recruits the PRC2 complex to its target genes and therefore is also indirectly involved in H3K27 methylation. KDM6A/UTX demethylates H3K27 and thus is a functional opponent of EZH2 and ASXL1.56–58 EP300 acetylates the H3K27 residue besides other non-histone substrates.59 This acetylation mark is considered as an active enhancer mark and thus possibly also opposes the repressive function of EZH2 and ASXL1.
EZH2 and ASXL1
It is now well established that ASXL1 and EZH2 mutations are loss of function mutations in MDS.60,61 This loss of function prevents repression of PRC2 target genes like HOX genes, which are well known to promote self-renewal and leukemic progression.62 Loss of Ezh2 was sufficient to induce a MDS/MPN-like disease in mice,63 to promote myelofibrosis in mice expressing a constitutively active JAK2 (V617F),64 to collaborate with mutant RUNX1 thereby accelerating MDS in mice,61 and to induce chemoresistance and relapse in AML patients.31
ASXL1 interacts with BAP1 to form a polycomb deubiquitinase complex that removes monoubiquitin from histone H2AK119.65 Through interaction with the PRC2 complex loss of Asxl1 results in a genome-wide reduction in H3K27 trimethylation.26 Asxl1 deletion excludes Ezh2 and consequently diminishes H3K27me3 at the HoxA cluster and thereby activates HoxA gene expression.26 Constitutive loss of Asxl1 had no major impact on hematopoiesis and did not induce MDS in mice.24 However, conditional knockout of Asxl1 caused progressive multilineage cytopenias and dysplasia with increased numbers of hematopoietic stem and progenitor cells reminiscent of MDS25,66 likely mediated by HOXA9 and miR-125a activation and reduction of the miR-125a target gene CLEC5a.25
KDM6A/UTX
KDM6A/UTX demethylates H3K27me3 and thus is a functional antagonist to EZH2. Through H3K27 demethylation it suppresses PRC1-mediated repression of HOX genes, thus activating HOX gene expression.57 As EZH2 mutations are loss of function mutations in MDS, one may assume that KDM6A mutations should be gain of function mutations in MDS patients. However, it is not clear whether KDM6A acts as a tumor suppressor or oncogene and it is likely that these functions depend on cellular context.67 Conditional knockout of Kdm6a induced myelodysplasia and suppressed erythro- and megakaryopoiesis in the bone marrow,68 suggesting a tumor suppressor function of wildtype Kdm6a. Loss of Kdm6a is accompanied by significant down-regulation of GATA1, LYL1, SCL, and KLF1 as well as upregulation of Pu.1.68 However, in a model of T cell acute lymphoblastic leukemia KDM6A was required for maintaining a TAL1-positive leukemia, suggesting an oncogenic function.69 Analysis of MDS-specific mutations of KDM6A are required to clarify its pathophysiologic function.
EP300
The mutations in EP300 are not well characterized and it is not clear whether they are gain or loss of function mutations. Knockout of EP300 is embryonic lethal70 and EP300 deficiency impairs self-renewal and differentiation of hematopoietic stem cells in mice.71 Loss of EP300 positively contributed to myeloid leukemogenesis in cooperation with other oncogenes (NUP98-HOXD13, NUP98-HOXA9 and MOZ-TIF2).72,73 In RUNX1-RUNX1T1-driven AML cells it was shown that acetylation of RUNX1-RUNX1T1 by p300 was required for leukemogenesis.74 These studies suggest a gain-of-function mechanism for EP300 mutations, but additional proof is required.
From these data it is evident that modifying enzymes of H3K27 are critical for normal hematopoiesis and that their dysregulation may induce myelodysplasia. One of the best studied target genes affected by H3K27 dysregulation are HOX genes and failed repression of the HOX cluster is a powerful driver of leukemogenesis and is observed in many patients with MDS and AML.
Modifiers of H3K4 methylation
Enzymes of the KMT2/MLL family methylate H3K4 to either mark distal regulatory enhancers (H3K4me1, e.g. by KMT2C/MLL3) or initiate transcription through recruitment of coactivators (H3K4me3, e.g. by KMT2A/MLL, KMT2D/MLL2, KMT2B/MLL4).75 While KMT2A is frequently involved in translocations or in partial tandem duplications in AML, these genetic aberrations are hardly found in MDS patients.2 KMT2 family members are frequently mutated in solid tumors and lymphoma,75 and it may not be surprising to find a high frequency of mutations also in MDS. Many KMT2 family gene mutations truncate the protein and are likely loss-of-function mutations.75 For KMT2C and KMT2D an inhibitory effect on cell growth has been described, and KMT2C was implicated as a tumor suppressor.76,77 Mice carrying the inactive form of Kmt2c showed a skewed hematopoiesis towards the myeloid lineage.78 Interestingly, knockdown of Kmt2c and neurofibromin1 (Nf1) in Tp53-null bone marrow cells induced AML in mice.77 In addition, suppression of Kmt2c significantly upregulated a differentiation-associated transcriptional signature.77 KMT2D also likely acts as a tumor suppressor, as its deletion in B lymphocytes promoted lymphoma development in mice.79
KMT2E/MLL5 does not exhibit methyltransferase activity and is therefore the most distant family member, which is more related to the SET3 gene family. However, its loss resulted in impaired neutrophil differentiation and increased infection susceptibility, mild impairment of erythropoiesis and reduced stem cell self-renewal, reminiscent of MDS.80 Loss of KMT2E resulted in accumulation of reactive oxygen species and DNA damage in hematopoietic stem and progenitor cells, which is mediated by type 1 interferon signaling and mitochondrial accumulation of Bid.81 As KMT2C and KMT2E are located on chromosome 7 and deletions of chromosome 7 are frequently observed in MDS patients, their reduced activity has been implicated in MDS pathogenesis.
KDM2B (also called JHDM1B or FBXL10) is a demethylase of H3K4me3 and of H3K36me2.82,83 Functional studies mostly implicated KDM2B as an oncogene. Its ectopic expression was sufficient to transform hematopoietic progenitor cells and induce leukemia through activation of Nsg2.84,85 Knockdown of KDM2B reduced cell proliferation in vitro and abrogated leukemogenesis in vivo in a humanized xenograft model.86 Loss of Kdm2b has been found to suppress Hoxa9/Meis1-induced leukemic transformation, supporting a potential oncogenic function.84
KDM5A (also called JARID1A and RBP2) is a demethylase of H3K4me2 and H3K4me3 and is recruited by PRC2 to repress its target genes in embryonic cells.87 Fusion of its c-terminal PHD-finger with NUP98 enforces the activation of lineage-specific transcription factors (Hox, Gata3, Meis1, Eya1 and Pbx1) by preventing loss of H3K4me3 at these loci in murine HSPC, leading to differentiation arrest and leukemia induction.88
In summary, missense and truncating mutations in modifiers of H3K4 methylation are frequently found in MDS patients and patients with solid tumors, but their contribution to disease pathogenesis is not well understood in stark contrast to KMT2A fusion genes, which are among the strongest oncogenes in hematopoiesis through activation of HOX genes and MEIS1.89 Gain- and loss of function studies and a detailed description of epigenetic consequences will help to define the role of these genes in MDS.
Epigenetic therapy in MDS
Hypomethylating agents
The therapy of MDS is risk adapted. The IPSS (International Prognostic Scoring System) and IPSS-R (revised-IPSS) are being used for the risk classification.90,91 The therapeutic recommendations by the European Leukemia Net (ELN) are based on the IPSS which considers blast percentage, karyotype and cytopenias and categorizes patients into low, intermediate-1 (int-1), intermediate-2 (int-2) and high risk.92 The two later groups, int-2 and high, are considered as high risk MDS while patients in the low and int-1 are considered to have low risk MDS. For patients with high risk MDS the recommended therapy is treatment with DNA hypomethylating agents (HMAs).92 Currently, HMAs are the only approved agents for higher risk MDS both by the FDA (decitabine93 and azacitidine94) and the EMA (azacitidine94) (Figure 3). Other therapeutic approaches are allogeneic stem cell transplantation for the relatively small percentage of younger and fit patients as well as solely supportive care for the very frail. The therapeutic benefit of HMA in high risk MDS can be understood when considering that recurrent methylation of tumor suppressor genes present a hallmark of high risk MDS. The hypermethylation of tumor suppressor genes leads to a reduction of their activity. Thus, the major therapeutic effect of HMA is believed to be due to a reactivation of tumor suppressor genes. The efficacy of HMA in high risk MDS is supported by the results of several studies showing that hypermethylation of the tumor suppressor gene p51INK4B is more pronounced in high risk MDS compared to lower risk MDS. Decitabine is the deoxynucleotide analog of azacitidine. Azacitidine is a pyrimidine nucleoside analogue of cytidine. It is mostly incorporated to RNA, and in part converted to deoxyazacitidine by ribonucleotide reductase, and thereafter incorporated in DNA. At low doses, azacitidine inhibits DNA methyltransferases (DNMTs) causing hypomethylation, and at high doses it induces a cytotoxic effect via effects on DNA and RNA.95
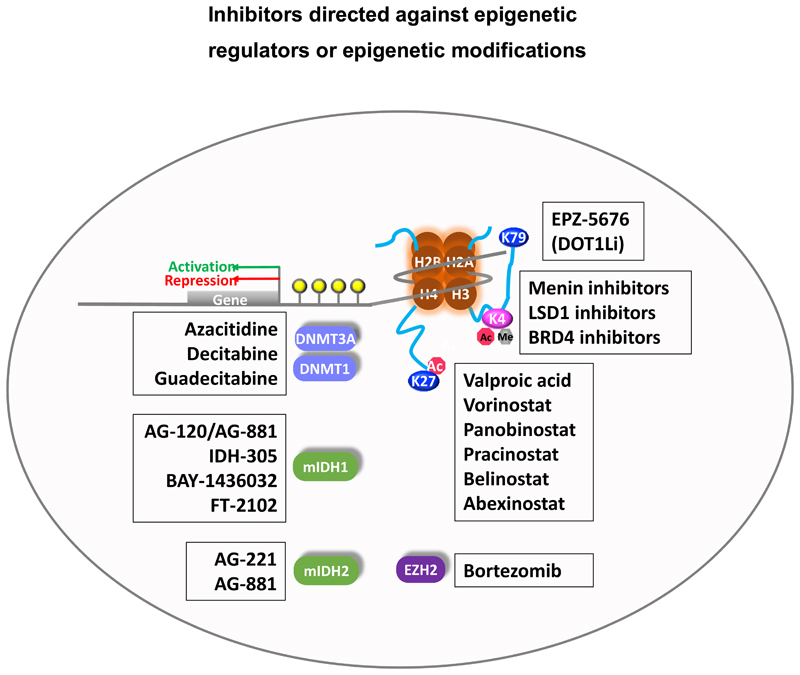
Figure 3.
Inhibitors in clinical development for MDS patients directed against epigenetic regulators or epigenetic modifications.
One of the first randomized trials with azacitidine was the CALGB trial which compared azacitidine with best supportive care. In this trial, 60 % of patients achieved a response to azacitidine. While only 7 % of patients achieved a complete remission, 16 % of patients obtained a partial remission and improvements in blood counts were seen in 37 % of patients.96 Although the observed complete remission rates were relatively low with azacitidine, these agents have been shown to lead to an overall survival benefit, improvements in hemoglobin, platelets and neutrophils and a better quality of life in later studies.94 Another important clinical point is that the response to these agents is generally not seen before the fourth or fifth cycle of treatment, which is different to many chemotherapy agents. As epigenetic changes take a longer time to evolve compared to cytotoxic therapy this timeline seems to be understandable. The median duration of response in patients with complete remission, partial remission or any hematological improvement was 13.6 months with azacitidine94 and 10.3 months with decitabine.93 Azacitidine is now standard of care for patients with intermediate-2 and high risk MDS. The typical regimen consists of a fix dose of 75 mg/m2/day subcutaneously for 7 days every 28 days. Treatment should be continued until patients loose clinical benefit.
As some patients respond to demethylating therapy very well while others don’t, response prediction is an intriguing question. Is there a molecular signature that predicts response? Several trials have studied retrospectively the mutational profile of patients that responded in comparison to patients that did not. Specifically, the question was raised whether mutations in epigenetic modifiers correlate with response to hypomethylating agents. Response to HMAs was similar for patients with and without mutations in DNMT3A, IDH1, IDH2, ASXL1 and EZH2.12,97 When 213 MDS patients treated with HMA were analysed for commonly mutated genes in MDS, higher abundance of TET2 mutations were associated with increased response to hypomethylating agents, particularly when ASXL1 is not mutated.98 Similar results were shown in the study by Itzykson et al. looking at AML patients that were treated with demethylating agents. Here, mutations of TET2 detected by Sanger sequencing were found to predict a nearly twofold greater response rate with azacitidine.99 The mutations can remain detectable even if a good response has been observed.100 In the AZA-001 trial of azacitidine in AML patients ≥ 65 years of age an exploratory analysis of molecular predictors of response identified TP53 and NRAS mutations associated with improved overall survival.101 Patients with TP53 mutation and or an unfavourable cytogenetic risk profile were identified to show a clear clinical benefit with a 10-day course of decitabine in a study that looked at 116 patients with AML and MDS. The promising result in the TP53 mutated group is especially important as other therapies including stem cell transplantation usually fail in this high risk patient group.97 However, the current data does not allow tailoring hypomethylating agents to specific genetic subgroups withholding it in other patients, especially with the few alternatives currently available.
HDAC Inhibitors
Histone deacetylases (HDAC) remove acetyl groups from lysine in histones and thereby repress DNA transcription. HDAC inhibitors like the antiepileptic agent valproic acid can prevent DNA compaction to maintain active transcription and induce differentiation in leukemic cells.102 Preclinical and clinical data showed synergistic activity of HDAC inhibitors with hypomethylating agents.103 It was postulated that demethylation by HMAs and de-compaction of histones by HDAC inhibitors would synergistically activate repressed genes and thereby induce differentiation, apoptosis and cell cycle arrest. This led to the clinical evaluation of several HDAC inhibitors like valproic acid, vorinostat, panobinostat, pracinostat, belinostat, abexinostat, and others as single agents or in combination with HMAs (Figure 3).104–109 Single agent activity of HDAC inhibitors was very limited. Combination treatment with HMAs also has been disappointing so far. In a randomized phase II study with decitabine (20 mg/m2 days 1-5 with or without valproic acid 50 mg/m2 days 1-7) in 149 MDS and AML patients the median survival was 11.9 months with no difference between the treatment groups.110. The E1905 study treated 47 patients with therapy-related myeloid neoplasms with azacitidine (50 mg/m2 days 1-7) with or without entinostat (4 mg/m2 days 3-10). Median overall survival was 13 months in the azacitidine arm and 6 months in the combination arm.109 Pracinostat was evaluated in a randomized, placebo-controlled, double blind study of azacitidine (75 mg/m2 for 7days) with or without pracinostat (60 mg every other day for 3 days a week for the first 3 weeks of each cycle) in patients with higher-risk MDS (n=102). After 6 cycles the CR rate was 33% in the placebo compared to 18% in the pracinostat arms. Median overall survival was 19 and 16 months in the placebo and pracinostat arms (no significant difference). Adverse events and treatment discontinuation were more frequent in the pracinostat group.111 Thus, the addition of HDAC inhibitors to HMAs in the best case appears non-effective, but may add toxicity to the treatment with negative effects on survival probability of MDS and AML patients.
How can these clinical observations be explained? Two preclinical studies evaluated the effects of valproic acid on leukemic progenitor cells from AML patients. Valproic acid induced differentiation and partial leukemic regression, but spared leukemic progenitor cells and enhanced the colony-forming potential of primary AML cells.112,113 Thus, inhibition of HDACs may have opposing effects in bulk leukemic cells compared to leukemic stem and progenitor cells. The lack of clinical efficacy may be due to limited on-target activity of the inhibitors, or the most relevant HDAC enzymes may have not been targeted effectively yet. Alternatively, histone deacetylation and repression of associated target genes may be less relevant in the pathophysiology of MDS.
Novel treatment approaches
Various molecules targeting epigenetic regulators including IDH1, IDH2, EZH2, DOT1L (H3K79 methyltransferase, required by MLL fusion leukemias), and bromodomain proteins (epigenetic readers of lysine acetylation, e.g. BRD4) were developed for the treatment of specific hematological malignancies and their efficacy is currently under investigation (Figure 3). The results may provide promising treatment options and increase the pool of effective epigenetically targeted therapies in MDS patients. Development of new hypomethylating agents is ongoing with guadecitabine/SGI-110 being the most advanced drug. SGI-110 is a second-generation hypomethylating agent and a prodrug of decitabine and might even show benefit in patients that were heavily pretreated with hypomethylating agents.114 For the latter, combination of epigenetic therapy with immunotherapy is the focus of currently conducted trials. Treatment with azacitidine has been shown to increase the programmed death ligand-1 (PD-L1) expression of myeloblasts.115 The increase of PD-L1 is supposed to lead to higher response rates to immunotherapy. Thus, synergism between demethylating agents and immunotherapy is being postulated. Trials evaluating the combination of these two agents are ongoing.
It remains a challenge to restore the activity of mutated inactive epigenetic regulators. Two strategies are highlighted here for ASXL1 and EZH2. Although ASXL1 interacts with BAP1, loss of BAP1 had opposite effects than loss of ASXL1, as it resulted in increased H3K27 trimethylation, elevated expression of EZH2 and repression of PRC2 target genes.116 Thus, inhibition of BAP1 may be an interesting target in patients with mutated ASXL1 or repressed EZH2. Protein ubiquitination marks proteins for proteasomal degradation. In contrast, deubiquitination (e.g. by BAP1) or inhibition of the proteasome may increase the half-life of proteins. Along these lines the proteasome inhibitor bortezomib was used in chemotherapy-resistant AML cells, which had lost EZH2 expression during treatment. Bortezomib could restore EZH2 expression and resensitize the cells to chemotherapy, representing an interesting treatment approach for patients with low EZH2 expression.31 However, clinical trials with proteasome inhibitors in MDS as single agent or in combination with lenalidomide or low dose cytarabine showed only modest activity so far.117,118
In summary, epigenetic therapy is the standard treatment for high risk MDS patients leading to improved survival as well as improvements in blood counts. The effect is thought to be due reducing DNA hypermethylation and thus activation of tumor suppressor genes. The search for molecular response signatures, combination therapies and advancements of novel epigenetic drugs is ongoing.
Perspective
It has been a long-standing question whether epigenetic patterns are stably engraved in the genome to induce and maintain cell transformation and leukemogenesis. If this would be true, a treatment that reverses this epigenetic pattern would be sufficient to inhibit and cure leukemia. Alternatively, if aberrant epigenetic patterns are consequences of mutated and dysfunctional proteins that constantly maintain this epigenetic pattern, a treatment directed against the epigenetic changes would result only in temporary inhibition of leukemia.
A recent study eloquently addressed this question using AML cells: Leukemic cells were reprogrammed to induced pluripotent stem cells (iPSC) and were differentiated back to hematopoietic cells. While undifferentiated iPSCs did not induce leukemia in vivo, differentiated iPSCs induced AML with the same phenotype as the primary AML cells.119 When assessing absolute methylation differences, undifferentiated iPSC populations were globally hypermethylated compared to primary AML and differentiated AML-iPSCs. Differentiated AML-iPSCs, similar to primary AML cells, demonstrated relative hypermethylation of pluripotency gene sets, but showed hypomethylation of hematopoietic and leukemic gene sets. Interestingly, no residual epigenetic memory was found in differentiated AML-iPSCs that could contribute to re-acquisition of the leukemic phenotype. It was concluded that genetic mutations in AML-iPSCs reactivated leukemia target genes upon hematopoietic differentiation thus inducing the leukemic phenotype.119 This study suggests that efficient epigenetic targeting should be directed against the dysregulated epigenetic enzyme and not merely against the epigenetic pattern associated with this enzyme. The difficulty is that most of the mutations in epigenetic regulators are loss-of-function mutations and that no successful treatment strategies have been developed yet for loss-of-function mutations as exemplified for mutated p53.
Hypomethylating agents are undoubtedly helpful in the treatment of MDS patients. However, none of the patients treated with HMAs is cured with HMAs alone (median response duration is 10.3-13.6 months) and there is an urgent need to move beyond hypomethylating agents in MDS. A better understanding of the molecular mechanisms of mutated epigenetic regulators and their epigenetic consequences will eventually lead us to better treatments of our MDS patients.
Acknowledgement
This work was supported by an ERC grant under the European Union’s Horizon 2020 research and innovation programme (No. 638035), by grants 110284, 110287, 110292 and 111267 from Deutsche Krebshilfe; and DFG grants HE 5240/5-2 and HE 5240/6-1.
Footnotes
Conflict of interest
The authors have no conflicts of interest.
References
- 1.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 4.Thol F, Winschel C, Ludeking A, et al. Rare occurrence of DNMT3A mutations in myelodysplastic syndromes. Haematologica. 2011;96(12):1870–1873. doi: 10.3324/haematol.2011.045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thol F, Weissinger EM, Krauter J, et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica. 2010;95(10):1668–1674. doi: 10.3324/haematol.2010.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thol F, Friesen I, Damm F, et al. Prognostic Significance of ASXL1 Mutations in Patients With Myelodysplastic Syndromes. J Clin Oncol. 2011;29(18):2499–2506. doi: 10.1200/JCO.2010.33.4938. [DOI] [PubMed] [Google Scholar]
- 7.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 8.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejar R, Ebert BL. The genetic basis of myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24(2):295–315. doi: 10.1016/j.hoc.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9(3):193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiNardo CD, Jabbour E, Ravandi F, et al. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia. 2016;30(4):980–984. doi: 10.1038/leu.2015.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawara Y, Katsumoto T, Aikawa Y, et al. IDH2 and NPM1 Mutations Cooperate to Activate Hoxa9/Meis1 and Hypoxia Pathways in Acute Myeloid Leukemia. Cancer Res. 2015;75(10):2005–2016. doi: 10.1158/0008-5472.CAN-14-2200. [DOI] [PubMed] [Google Scholar]
- 14.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40(8):4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- 16.Heuser M, Araujo Cruz MM, Goparaju R, Chaturvedi A. Enigmas of IDH mutations in hematology/oncology. Exp Hematol. 2015;43(8):685–697. doi: 10.1016/j.exphem.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 18.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thol F, Damm F, Lüdeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayle A, Yang L, Rodriguez B, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125(4):629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Su J, Jeong M, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48(9):1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher CL, Pineault N, Brookes C, et al. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115(1):38–46. doi: 10.1182/blood-2009-07-230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue D, Kitaura J, Togami K, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest. 2013;123(11):4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdel-Wahab O, Adli M, LaFave LM, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1-2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1 and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 29.Qiu J, Zhou B, Thol F, et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA. 2016 doi: 10.1261/rna.056101.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E, Ilagan JO, Liang Y, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27(5):617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gollner S, Oellerich T, Agrawal-Singh S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23(1):69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Della Porta MG, Galli A, Bacigalupo A, et al. Clinical Effects of Driver Somatic Mutations on the Outcomes of Patients With Myelodysplastic Syndromes Treated With Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.67.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsley RC, Saber W, Mar BG, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heuser M, Koenecke C, Gabdoulline R, et al. Molecular Predictors of Outcome in Patients with MDS and AML Following MDS after Allogeneic Hematopoietic Stem Cell Transplantation. Blood. 2015;126(23):912–912. [Google Scholar]
- 36.Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113(6):1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kfir N, Lev-Maor G, Glaich O, et al. SF3B1 association with chromatin determines splicing outcomes. Cell Rep. 2015;11(4):618–629. doi: 10.1016/j.celrep.2015.03.048. [DOI] [PubMed] [Google Scholar]
- 38.Russler-Germain DA, Spencer DH, Young MA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009 doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 41.Spencer DH, Russler-Germain DA, Ketkar S, et al. CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell. 2017;168(5):801–816. doi: 10.1016/j.cell.2017.01.021. e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Rodriguez B, Mayle A, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;29(6):922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen KD, Jia G, Johansen JV, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29(9):910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quivoron C, Couronne L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaturvedi A, Araujo Cruz MM, Jyotsana N, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122(16):2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 52.Kats LM, Reschke M, Taulli R, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14(3):329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan SM, Thomas D, Corces-Zimmerman MR, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med. 2015;21(2):178–184. doi: 10.1038/nm.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konopleva M, Pollyea DA, Potluri J, et al. Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov. 2016;6(10):1106–1117. doi: 10.1158/2159-8290.CD-16-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue S, Li WY, Tseng A, et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell. 2016;30(2):337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan F, Bayliss PE, Rinn JL, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(7163):689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 57.Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 58.Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 59.Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95(3):745–755. [PubMed] [Google Scholar]
- 60.Wang J, Li Z, He Y, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123(4):541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sashida G, Harada H, Matsui H, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun. 2014;5:4177. doi: 10.1038/ncomms5177. [DOI] [PubMed] [Google Scholar]
- 62.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 63.Muto T, Sashida G, Oshima M, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210(12):2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sashida G, Wang C, Tomioka T, et al. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J Exp Med. 2016;213(8):1459–1477. doi: 10.1084/jem.20151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465(7295):243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdel-Wahab O, Gao J, Adli M, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210(12):2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van der Meulen J, Sanghvi V, Mavrakis K, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125(1):13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thieme S, Gyarfas T, Richter C, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121(13):2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 69.Benyoucef A, Palii CG, Wang C, et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30(5):508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao TP, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 71.Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci U S A. 2002;99(23):14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng G, Liu F, Asai T, et al. Loss of p300 accelerates MDS-associated leukemogenesis. Leukemia. 2017 doi: 10.1038/leu.2016.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giotopoulos G, Chan WI, Horton SJ, et al. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene. 2016;35(3):279–289. doi: 10.1038/onc.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, Gural A, Sun XJ, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011;333(6043):765–769. doi: 10.1126/science.1201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15(6):334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J, Kim DH, Lee S, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci U S A. 2009;106(21):8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen C, Liu Y, Rappaport AR, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25(5):652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arcipowski KM, Bulic M, Gurbuxani S, Licht JD. Loss of Mll3 Catalytic Function Promotes Aberrant Myelopoiesis. PLoS One. 2016;11(9):e0162515. doi: 10.1371/journal.pone.0162515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heuser M, Yap DB, Leung M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113(7):1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 81.Tasdogan A, Kumar S, Allies G, et al. DNA Damage-Induced HSPC Malfunction Depends on ROS Accumulation Downstream of IFN-1 Signaling and Bid Mobilization. Cell Stem Cell. 2016;19(6):752–767. doi: 10.1016/j.stem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 82.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450(7167):309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 83.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15(11):1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011;117(14):3869–3880. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ueda T, Nagamachi A, Takubo K, et al. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood. 2015;125(22):3437–3446. doi: 10.1182/blood-2014-03-562694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van den Boom V, Maat H, Geugien M, et al. Non-canonical PRC1.1 Targets Active Genes Independent of H3K27me3 and Is Essential for Leukemogenesis. Cell Rep. 2016;14(2):332–346. doi: 10.1016/j.celrep.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 87.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22(10):1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316(5824):600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 90.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. [PubMed] [Google Scholar]
- 91.Greenberg PL, Tuechler H, Schanz J, et al. Revised International Prognostic Scoring System (IPSS-R) for myelodysplastic syndromes. Blood. 2012 doi: 10.1182/blood-2012-03-420489. Advance Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malcovati L, Hellstrom-Lindberg E, Bowen D, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood. 2013;122(17):2943–2964. doi: 10.1182/blood-2013-03-492884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106(8):1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 94.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Navada SC, Steinmann J, Lubbert M, Silverman LR. Clinical development of demethylating agents in hematology. J Clin Invest. 2014;124(1):40–46. doi: 10.1172/JCI69739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20(10):2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 97.Welch JS, Petti AA, Miller CA, et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med. 2016;375(21):2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bejar R, Lord A, Stevenson K, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Itzykson R, Kosmider O, Cluzeau T, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25(7):1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 100.Uy GL, Duncavage EJ, Chang GS, et al. Dynamic changes in the clonal structure of MDS and AML in response to epigenetic therapy. Leukemia. 2016 doi: 10.1038/leu.2016.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang L, Dolnik A, MacBeth KJ, et al. Impact of Gene Mutations on Overall Survival in Older Patients with Acute Myeloid Leukemia (AML) Treated with Azacitidine (AZA) or Conventional Care Regimens (CCR) Blood. 2016;128:2859–2859. [Google Scholar]
- 102.Zapotocky M, Mejstrikova E, Smetana K, Stary J, Trka J, Starkova J. Valproic acid triggers differentiation and apoptosis in AML1/ETO-positive leukemic cells specifically. Cancer Lett. 2012;319(2):144–153. doi: 10.1016/j.canlet.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 103.Quintas-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011;25(2):226–235. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- 104.Cashen A, Juckett M, Jumonville A, et al. Phase II study of the histone deacetylase inhibitor belinostat (PXD101) for the treatment of myelodysplastic syndrome (MDS) Ann Hematol. 2012;91(1):33–38. doi: 10.1007/s00277-011-1240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kirschbaum M, Gojo I, Goldberg SL, et al. A phase 1 clinical trial of vorinostat in combination with decitabine in patients with acute myeloid leukaemia or myelodysplastic syndrome. Br J Haematol. 2014;167(2):185–193. doi: 10.1111/bjh.13016. [DOI] [PubMed] [Google Scholar]
- 106.Tan P, Wei A, Mithraprabhu S, et al. Dual epigenetic targeting with panobinostat and azacitidine in acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood Cancer J. 2014;4:e170. doi: 10.1038/bcj.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raffoux E, Cras A, Recher C, et al. Phase 2 clinical trial of 5-azacitidine, valproic acid, and all-trans retinoic acid in patients with high-risk acute myeloid leukemia or myelodysplastic syndrome. Oncotarget. 2010;1(1):34–42. doi: 10.18632/oncotarget.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vey N, Prebet T, Thalamas C, et al. Phase 1 dose-escalation study of oral abexinostat for the treatment of patients with relapsed/refractory higher-risk myelodysplastic syndromes, acute myeloid leukemia, or acute lymphoblastic leukemia. Leuk Lymphoma. 2017;58(8):1880–1886. doi: 10.1080/10428194.2016.1263843. [DOI] [PubMed] [Google Scholar]
- 109.Prebet T, Sun Z, Ketterling RP, et al. Azacitidine with or without Entinostat for the treatment of therapy-related myeloid neoplasm: further results of the E1905 North American Leukemia Intergroup study. Br J Haematol. 2016;172(3):384–391. doi: 10.1111/bjh.13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Issa JP, Garcia-Manero G, Huang X, et al. Results of phase 2 randomized study of low-dose decitabine with or without valproic acid in patients with myelodysplastic syndrome and acute myelogenous leukemia. Cancer. 2015;121(4):556–561. doi: 10.1002/cncr.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garcia-Manero G, Montalban-Bravo G, Berdeja JG, et al. Phase 2, randomized, double-blind study of pracinostat in combination with azacitidine in patients with untreated, higher-risk myelodysplastic syndromes. Cancer. 2017;123(6):994–1002. doi: 10.1002/cncr.30533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bug G, Schwarz K, Schoch C, et al. Effect of histone deacetylase inhibitor valproic acid on progenitor cells of acute myeloid leukemia. Haematologica. 2007;92(4):542–545. doi: 10.3324/haematol.10758. [DOI] [PubMed] [Google Scholar]
- 113.Leiva M, Moretti S, Soilihi H, et al. Valproic acid induces differentiation and transient tumor regression, but spares leukemia-initiating activity in mouse models of APL. Leukemia. 2012;26(7):1630–1637. doi: 10.1038/leu.2012.39. [DOI] [PubMed] [Google Scholar]
- 114.Griffiths EA, Choy G, Redkar S, Taverna P, Azab M, Karpf AR. SGI-110: DNA Methyltransferase Inhibitor Oncolytic. Drugs Future. 2013;38(8):535–543. [PMC free article] [PubMed] [Google Scholar]
- 115.Yang H, Bueso-Ramos C, DiNardo C, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2014;28(6):1280–1288. doi: 10.1038/leu.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.LaFave LM, Beguelin W, Koche R, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Daher M, Hidalgo Lopez JE, Randhawa JK, et al. An exploratory clinical trial of bortezomib in patients with lower risk myelodysplastic syndromes. Am J Hematol. 2017 doi: 10.1002/ajh.24746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Attar EC, Amrein PC, Fraser JW, et al. Phase I dose escalation study of bortezomib in combination with lenalidomide in patients with myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) Leuk Res. 2013;37(9):1016–1020. doi: 10.1016/j.leukres.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chao MP, Gentles AJ, Chatterjee S, et al. Human AML-iPSCs Reacquire Leukemic Properties after Differentiation and Model Clonal Variation of Disease. Cell Stem Cell. 2017;20(3):329–344.e327. doi: 10.1016/j.stem.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Trowbridge JJ, Sinha AU, Zhu N, Li M, Armstrong SA, Orkin SH. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 2012;26(4):344–349. doi: 10.1101/gad.184341.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nickerson ML, Das S, Im KM, et al. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene. 2017;36(15):2172–2183. doi: 10.1038/onc.2016.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng F, Liu J, Zhou SH, Wang XN, Chew JF, Deng LW. RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int J Biochem Cell Biol. 2008;40(11):2472–2481. doi: 10.1016/j.biocel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 123.Cimmino L, Dawlaty MM, Ndiaye-Lobry D, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Q, Zhao K, Shen Q, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525(7569):389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang Y, Xiao M, Chen X, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57(4):662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]