Abstract
We integrated molecular data with available prognostic factors in patients undergoing allogeneic hematopoietic cell transplantation (alloHCT) for myelodysplastic syndrome (MDS) or secondary acute myeloid leukemia from MDS (sAML) to evaluate their impact on prognosis. 304 patients were sequenced for mutations in 54 genes. We used a Cox multivariate model and competing risk analysis with internal and cross validation to identify factors prognostic of overall survival (OS), cumulative incidence of relapse (CIR) and non-relapse mortality (NRM). In multivariate analysis, mutated NRAS, U2AF1, IDH2, TP53 and/or a complex karyotype were significant prognostic markers for OS besides age above 60 years, remission status, IPSS-R cytogenetic risk, HCT-CI > 2 and female donor sex. Mutated NRAS, IDH1, EZH2 and TP53 and/or a complex karyotype were genetic aberrations with prognostic impact on CIR. No molecular markers were associated with the risk of NRM. The inclusion of molecular information results in better risk prediction models for OS and CIR when assessed by the Akaike information criterion. Internal cross validation confirmed the robustness of our comprehensive risk model. In summary, we propose to combine molecular, cytogenetic, patient-and transplantation associated risk factors into a comprehensive risk modelto provide personalized predictions of outcome after alloHCT.
Introduction
Allogeneic hematopoietic cell transplantation (alloHCT) represents the only curative treatment approach for patients with MDS and sAML,1 but requires careful patient and donor selection. The European Leukemia Net recommends alloHCT in higher risk MDS patients, i.e. patients with IPSS intermediate 2 or high risk.2 This recommendation is based on statistical models in patients younger than 60 years and patients 60 to 70 years old suggesting that upfront alloHCT is beneficial for IPSS intermediate 2 and high risk patients, while delaying alloHCT is beneficial for IPSS low and intermediate 1 risk patients.3, 4 Comorbidities of the patient may put him at risk of non-relapse mortality (NRM). The most common scores for evaluating patients’ comorbidities are the HCT comorbidity index (HCT-CI)5 and the EBMT score. 6 Besides disease and patient associated risk factors the transplantation associated risk factors like conditioning regimen or donor sex also influence disease outcome, but these factors have not been integrated into current risk prediction tools. In de novo AML, molecular markers have important implications for prognosis7, 8 and selection of patients for allogeneic transplantation.9 However, less is known about the implications of molecular markers in the setting of alloSCT for MDS and sAML patients. One study included 87 MDS patients undergoing alloHCT and found that TP53, TET2 and DNMT3A mutations were each independently associated with shorter OS,10 while another study of 62 patients found no prognostic impact of gene mutations.11 In a recent study of 401 MDS and sAML patients ASXL1, RUNX1 and TP53 mutations were prognostic after alloHCT.12 To validate these findings and possibly clarify the discrepancies we evaluated molecular, cytogenetic, disease and transplant associated risk factors for their prognostic effect after alloHCT in MDS/sAML patients, and integrated the relevant risk factors for alloHCT into a model that allows personalized predictions of outcome for future MDS/sAML patients undergoing alloHCT.
Methods
Patients
304 patients with MDS or sAML after a prior diagnosis of MDS were included in this study who underwent allogeneic HCT at four German university medical centers (Dresden, Düsseldorf, Hamburg and Hannover) between 1996 and 2011 and for whom genomic DNA was available from an MDS or sAML sample before transplantation.13 77 patients did not have DNA available and were not included in the analysis. There were no differences regarding OS (P=0.54), cumulative incidence of relapse (CIR, P=0.24) and non-relapse mortality (NRM, P=0.85) between in-and excluded patients (data not shown). Written informed consent was obtained according to the Declaration of Helsinki, and the study was approved by the institutional review board of Hannover Medical School. Patients with chronic myelomonocytic leukemia and patients with haploidentical donor were excluded. DNA from 14 healthy blood donors was used for myeloid panel sequencing as a normal control.
Cytogenetic and molecular analysis
Pretransplant blood or bone marrow samples were studied centrally by G-and R-banding analysis. Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature.14 DNA was extracted as described before using the Allprep DNA/RNA purification kit (Qiagen, Hilden, Germany).15 DNA sequencing libraries were prepared with the TruSight Myeloid sequencing panel according to the manufacturer instructions (Illumina, San Diego, CA), which included 54 entire genes or hotspots recurrently found in leukemia. All samples received individual dual indexes and were pooled at equimolar concentrations. 80 samples per lane were sequenced on an Illumina HiSeq2500 sequencer using the HiSeq Rapid SBS Kit v2 (Illumina, San Diego, CA) for 250 cycles in both directions. The sequencing data was analysed as described before16 and as detailed in the supplement.
Statistical analysis
Median follow-up time for survival was calculated according to the method of Korn.17 Overall survival endpoints, measured from the date of alloHSCT, were death (failure) and alive at last follow-up (censored). The Kaplan-Meier method and log-rank tests were used to estimate the distribution of overall survival (OS),, and to compare differences between survival curves. The Gray test was used to compare and visually represent cumulative incidences of non-relapse mortality (NRM) and cumulative incidence of relapse (CIR) as competing risks using R package cmprsk.18 51 categorized variables were considered in univariate analysis for OS, CIR and NRM (Supplementary Table S1 for variables and Supplementary Table S2 for definition of variable categories). Variables were used for multivariate analysis, if they had a P value of ≤0.33 in univariate analysis and had distinct categorical values in at least 13 patients (>4% of all patients). We used the categories of the IPSS score (cytopenias, IPSS-R cytogenetic risk and bone marrow blasts) but not the IPSS itself, as it is not established for sAML patients. Individual cytogenetic aberrations were not included separately in multivariate analysis, as they are included in the IPSS-R cytogenetic risk score, except complex karyotype (KT), which had strong IPSS-R independent prognostic impact. Altogether twenty-two variables fulfilled these criteria and were used in multivariate analyses (16 variables for OS, 14 for CIR and 12 for NRM, see Supplementary Table S1). Missing values were imputed in SPSS by the fully conditional specification (FCS) method using Markov chain Monte Carlo (MCMC) sampling.19 Each variable was processed separately using OS, NRM status, age at transplantation and MDS vs sAML as independent variables known for each patient. For multivariate analysis, a Cox proportional hazards model was constructed for OS, CIR, and NRM adjusting for potential confounding covariates.20 Variables were selected by backward elimination. This process was initiated with all selected variables. At each step the variable with the worst fit (judged from individual P-values reported by the survival or the cmprsk packages of R) was removed from the model, until the model had a single variable. The model quality was monitored via the score of the model according to the loglikelihood ratio test and the Akaike information criterion (AIC).21 The two quality measures resulted in the same optimal variable set for OS. The optimal model was further tested by exclusion of each variable one by one showing that this did not further improve the model. This procedure was repeated with all molecular variables that had been removed during backward elimination.
Pair-wise comparisons of variables were performed using the Kolmogorov-Smirnovtest and Student’s t-test for continuous variables and the Chi-squared test forcategorical variables for exploratory purposes.
The two-sided level of significance was set at P <0.05. The statistical analyses wereperformed with the statistical software package SPSS 23.0 (IBM Corporation,Armonk, NY), statistical program R22 using packages “survival”, cmprsk”, “forestplot”;Microsoft excel 2010 (Microsoft Corporation, Redmond, WA) and custom linuxscripts. Additional details on statistical methods can be found in the supplement.
Results
Patient characteristics
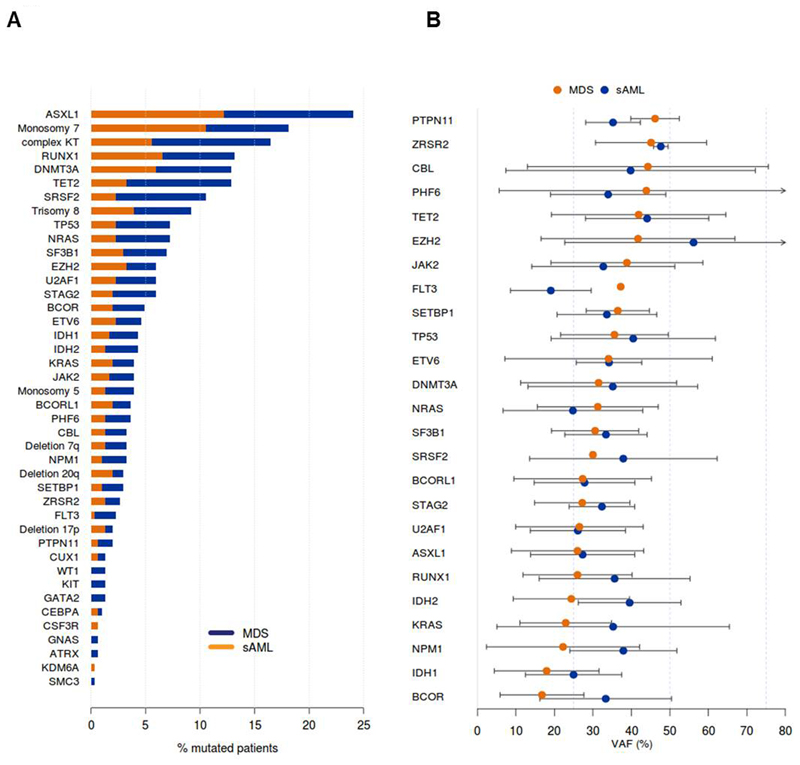
We included 304 patients with MDS (n=144) or sAML from MDS (n=160) with a median age of 58 years (range 19-75) who underwent alloHCT at one of four German transplant centers. As expected, more patients with MDS were untreated or in CR after treatment at the time of transplantation compared to sAML patients (67% vs 46%, see Table 1). Most other patient-and transplantation associated characteristics were similarly distributed between MDS and sAML patients (Table 1). The conditioning regimen was non-myeloablative in most patients (85%). 95% of patients received mobilized peripheral blood stem/progenitor cells. We sequenced all patients with a 54 gene myeloid panel in cell samples that were collected before transplantation at a time when the patient had active disease. 75% of the patients had at least one mutation and the mean number of mutations was 1.8 (Supplementary Table S3 for all mutations). The most frequently mutated gene was ASXL1 (24%), followed by RUNX1 (13%), DNMT3A (12%), TET2 (12%) and SRSF2 (10%, Figure 1A and Supplementary Table S4). Complex KT, TET2 and SRSF2 mutations were found more frequently in sAML than in MDS patients, while the other mutations were similarly distributed. Previously, mutations in signaling genes have been considered as late events in disease evolution in AML patients.23 However, in our MDS cohort the mutations in signaling genes like PTPN11, CBL, JAK2 and FLT3 had the highest variant allele frequency (VAF) besides ZRSR2 and epigenetic regulators TET2 and EZH2 (Figure 1B). In sAML patients EZH2, TET2, ZRSR2 and TP53 mutations had the highest VAF (Supplementary Figure S1). Overall, MDS and sAML patients had similar patient, transplant, cytogenetic and molecular characteristics and were combined for evaluation of prognostic markers.
Table 1. Clinical and transplant characteristics of all patients and comparison between MDS and sAML patients.
| Characteristic | All | MDS | sAML | P |
|---|---|---|---|---|
| No. (%) | 304 | 144 (47) | 160 (53) | |
| Patient age, years | 1.0 | |||
| Median | 58 | 56 | 59 | |
| Range | 19-75 | 19-74 | 29-75 | |
| Patient sex | .74 | |||
| Male - no. (%) | 193 (63) | 90 (63) | 103 (64) | |
| Female - no. (%) | 111 (37) | 54 (37) | 57 (36) | |
| Disease status at transplant | .006 | |||
| Untreated or in CR- no. (%) | 170 (56) | 96 (67) | 74 (46) | |
| Treated but not in CR - no. (%) | 120 (39) | 48 (33) | 72 (45) | |
| Missing – no. (%) | 14 (5) | 0 (0) | 14 (9) | |
| Cytogenetic risk (according IPSS-R) | .53 | |||
| Very good - no. (%) | 2 (1) | 2 (1) | 0 (0 | |
| Good – no. (%) | 133 (44) | 64 (45) | 69 (43) | |
| Intermediate - no. (%) | 51 (17) | 21 (15) | 30 (19) | |
| Poor - no. (%) | 85 (28) | 42 (29) | 43 (27) | |
| Very poor – no. (%) | 11 (3) | 5 (3) | 6 (3) | |
| Missing - no. (%) | 22 (7) | 10 (7) | 12 (8) | |
| WBC count | 0.64 | |||
| Median (x109/µl) | 2.85 | 2.85 | 2.85 | |
| Range (x109/µl) | 0.2-293 | 0.5-293 | 0.2-261 | |
| Missing - no. (%) | 106 (35) | 52 (36) | 54 (34) | |
| Hemoglobin | 0.021 | |||
| Median (g/dl) | 9.3 | 8.9 | 9.9 | |
| Range (g/dl) | 2.6-15 | 4.6-13.7 | 2.6-15 | |
| Missing - no. (%) | 107 (35) | 52 (36) | 55 (34) | |
| Platelets | 0.073 | |||
| Median (x109/µl) | 16.2 | 23.2 | 13.6 | |
| Range (x109/µl) | 0.5-395 | 0.5-395 | 0.5-347 | |
| Missing - no. (%) | 105 (35) | 52 (36) | 53 (33) | |
| Cytopenias in peripheral blood | 0.75 | |||
| 0-1 cytopenia - no. (%) | 82 (27) | 37 (26) | 45 (28) | |
| 2-3 cytopenias - no. (%) | 116 (38) | 55 (38) | 61 (38) | |
| Missing - no. (%) | 106 (35) | 52 (36) | 54 (34) | |
| Bone marrow blasts before alloTx | <0.001 | |||
| Median (%) | 5 | 6 | 4 | |
| Range (%) | 0-80 | 0-19 | 0-80 | |
| Missing - no. (%) | 34 (11) | 19 (13) | 15 (9) | |
| WHO classification | n.a. | |||
| RA - no. (%) | 8 | 8 | ||
| RARS - no. (%) | 4 | 4 | ||
| RCMD - no. (%) | 35 | 35 | ||
| RAEB-1 - no. (%) | 30 | 30 | ||
| RAEB-2 - no. (%) | 59 | 59 | ||
| MDS-U - no. (%) | 1 | 1 | ||
| sAML - no. (%) | 160 | 160 | ||
| Missing - no. (%) | 7 | 7 | ||
| HCT-CI score | 0.91 | |||
| 0-2 - no. (%) | 192 (63) | 92 (64) | 100 (63) | |
| > 2 - no. (%) | 110 (36) | 52 (36) | 58 (36) | |
| Missing - no. (%) | 2 (1) | 0 (0) | 2 (1) | |
| Donor match | .87 | |||
| MRD - no. (%) | 71 (23) | 34 (24) | 37 (23) | |
| MUD - no. (%) | 171 (56) | 79 (55) | 92 (58) | |
| MMUD - no. (%) | 62 (21) | 31 (21) | 31 (19) | |
| Donor sex | .11 | |||
| Male - no. (%) | 208 (68) | 105 (73) | 103 (64) | |
| Female - no. (%) | 96 (32) | 39 (27) | 57 (36) | |
| Stem Cell source | .21 | |||
| PBSC - no. (%) | 288 (95) | 133 (93) | 155 (97) | |
| Bone marrow - no. (%) | 13 (4) | 9 (6) | 4 (2) | |
| Cord Blood - no (%) | 3 (1) | 2 (1) | 1 (1) | |
| CMV status | .44 | |||
| D pos/P pos - no. (%) | 108 (36) | 53 (37) | 55 (34) | |
| D pos/P neg - no. (%) | 45 (15) | 16 (11) | 29 (18) | |
| D neg/P neg - no. (%) | 94 (31) | 46 (32) | 48 (30) | |
| D neg/P pos - no. (%) | 50 (16) | 23 (16) | 27 (17) | |
| Missing - no. (%) | 7 (2) | 6 (4) | 1 (1) | |
| Conditioning | .93 | |||
| Myeloablative - no. (%) | 47 (15) | 22 (15) | 25 (16) | |
| Reduced intensity - no. (%) | 257 (85) | 122 (85) | 135 (84) | |
| Acute GvHD | .24 | |||
| Yes - no. (%) | 158 (52) | 80 (56) | 78 (49) | |
| No - no. (%) | 146 (48) | 64 (44) | 82 (51) | |
| Acute GvHD stage (n=158) | .16 | |||
| °I or °II - no. (%) | 107 (35) | 50 (35) | 57 (36) | |
| °III or °IV - no. (%) | 51 (17) | 30 (21) | 21 (13) | |
| Chronic GvHD | .23 | |||
| Yes - no. (%) | 118 (39) | 61 (42) | 57 (36) | |
| No - no. (%) | 186 (61) | 83 (58) | 103 (64) | |
| Chronic GvHD stage (n=118) | .016 | |||
| Limited - no. (%) | 52 (17) | 32 (22) | 20 (13) | |
| Extensive - no. (%) | 55 (18) | 21 (15) | 34 (21) | |
| Missing - no. (%) | 11 (4) | 8 (6) | 3 (2) | |
Abbreviations: MDS, myelodysplastic syndromes; sAML, secondary acute myeloid leukemia; no., number; CR, complete remission; IPSS-R, Revised International Prognostic Scoring System; WBC, white blood cell; alloTx, allogeneic stem cell transplantation; WHO, world health organization; RA, refractory anemia, RARS, refractory anemia with ring sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RAEB, refractory anemia with excess of blasts; HCT-CI, hematopoietic cell transplantation comorbidity index; MRD, matched related donor; MUD, matched unrelated donor; MMUD, mismatched unrelated donor; PBSC, peripheral blood stem cells; D, donor; P, patient; neg, negative; pos., positive; CMV, cytomegalovirus; GvHD, graft-versus-host-disease.
Figure 1. Genetic profile of MDS and sAML patients undergoing alloHCT.
(A) Frequency of cytogenetic and molecular aberrations in MDS and sAML patients undergoing alloHCT (for aberrations occurring in > 5 patients).
(B) Variant allele frequency of molecular aberrations in MDS and sAML patients (sorted in descending order according to frequency in MDS patients).
Prognostic factors for alloHCT in MDS and sAML patients
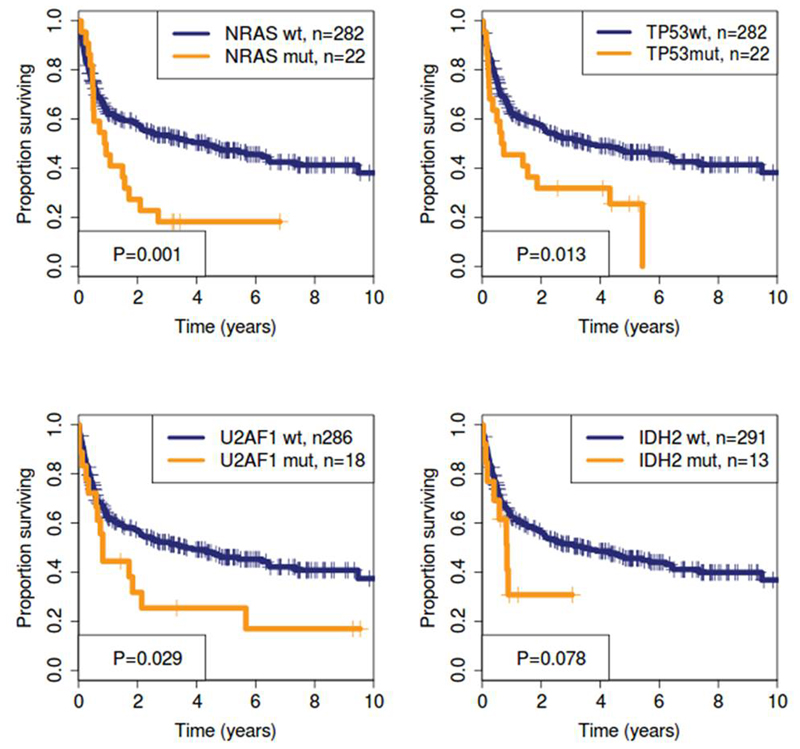
The median follow up for all patients was 5.03 years. Median OS was 3.1 years and 5-year OS was 45%. At 5 years CIR was 26% and NRM was 33% by competing risk analysis. By evaluating 51 potential risk factors by univariate analysis we found 10 significant risk factors for OS (Supplementary Table S1 and Supplementary Figures S2-S4 for Kaplan Meier curves and Supplementary Figures S5-S7 for forest plots). Three of those were molecular mutations in the genes NRAS, TP53 and U2AF1 (Figure 2). IDH2 mutated patients showed a trend for shorter OS (Figure 2). Other patient and transplant characteristics and especially other gene mutations including ASXL1 did not reach statistical significance. Complex KT and TP53 mutations frequently co-occured (14 of 50 (28%) patients with complex KT had a TP53 mutation, 14 of 22 (64%) of patients with TP53 mutation had a complex KT). As the prognostic effect of either a TP53 mutation or a complex KT or the combination of both had a similarly poor impact on OS, we used the presence of a complex KT and/or a TP53 mutation as a combined risk factor for multivariate analysis (Supplementary Figure S8).
Figure 2. Prognostic impact of significant molecular risk factors in univariate analysis for OS.
Next, we performed competing risk analysis for CIR and NRM using 51 potential risk factors. In univariate analysis 7 factors were significantly associated with CIR and 5 factors with NRM (Supplementary Table S1, Supplementary Figures S9 to S11 for Kaplan-Meier curves and Supplementary Figures S12-S17 for forest plots). Three of the risk factors for CIR were molecular mutations in one the genes NRAS, BCOR, and TP53, but no molecular mutations were significantly associated with NRM.
Comprehensive risk model integrating molecular, cytogenetic, patient-and transplantation-associated risk factors
To develop a comprehensive risk model for MDS/sAML patients undergoing alloHCT we selected all variables with P≤0.3 in univariate analysis and occurrence in at least 13 patients for multivariate analysis (Supplementary Table S1). By backward elimination we found 7 significant variables and two variables with trend which were predictive for OS (Table 2). Besides older age, remission status “treated but not in CR before transplantation”, HCT-CI ≥3, female donor sex and poor/very poor cytogenetic risk according to IPSS-R, we found four genetic risk factors that were associated with shorter OS (complex KT and/or TP53 mutated, mutations in NRAS, U2AF1 or IDH2).
Table 2. Multivariate analysis for OS, CIR and NRM.
| Overall survival multivariate analysis |
CIR Multivariate analysis |
NRM Multivariate analysis |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HR# | 95%CI | P | HR# | 95%CI | P | HR# | 95%CI | P | |
| IDH2 mutated | 2.14 | 1.13-4.05 | 0.02 | ||||||
| Complex karyotype and/or TP53 | 1.82 | 1.15-2.88 | 0.011 | 2.373 | 1.43-3.93 | 0.0008 | |||
| U2AF1 mutated | 1.82 | 1.10-3.02 | 0.021 | ||||||
| NRAS mutated | 1.7 | 1.07-2.72 | 0.026 | 2.221 | 1.14-4.32 | 0.019 | |||
| Remission status treated but not in CR | 1.58 | 1.16-2.15 | 0.004 | 1.56 | 0.99-2.46 | 0.056 | |||
| Donor Sex Female | 1.54 | 1.13-2.11 | 0.0068 | 1.58 | 1.06-2.35 | 0.024 | |||
| Cytogenetic risk (IPSS-R) | 1.46 | 0.95-2.26 | 0.085 | 1.56 | 1.05-2.31 | 0.029 | |||
| HCT-CI > 2 | 1.41 | 1.04-1.93 | 0.03 | 1.53 | 1.04-2.25 | 0.029 | |||
| Age > 60 | 1.32 | 0.97-1.81 | 0.081 | 1.57 | 1.05-2.33 | 0.027 | |||
| sAML | 1.89 | 1.14-3.15 | 0.014 | 0.68 | 0.45-1.01 | 0.054 | |||
| IDH1 mutated | 2.011 | 0.88-4.58 | 0.097 | ||||||
| Patient Sex Male | 1.746 | 1.04-2.94 | 0.036 | ||||||
| EZH2 mutated | 0.359 | 0.11-1.23 | 0.1 | ||||||
Hazard ratios greater than or less than 1 indicate an increased or decreased risk, respectively, of an event for the category listed.
Abbreviations: CIR, cumulative incidence of relapse; NRM, non-relapse mortality; HR, hazard ratio; CI, confidence interval; P, P value; CR, complete remission; IPSS-R, International Prognostic Scoring System revised; HCT-CI, hematopoietic cell transplantation comorbidity index; sAML, secondary acute myeloid leukemia.
By backward elminiation for CIR we found complex KT and/or TP53 mutation, mutated NRAS, sAML, male patient sex, remission status “treated but not in CR” before transplantation and mutated IDH1 associated with higher CIR, while mutated EZH2 was associated by trend with lower CIR (Table 2).
Finally, multivariate analysis identified female donor sex, IPSS-R cytogenetic risk, HCT-CI >2, and age >60 years associated with higher NRM, while sAML was associated with lower risk of NRM (Table 2). Thus, several molecular aberrations are associated with OS and CIR but not with NRM, which is in good accordance with a functional role of these mutations in disease recurrence.
Development and validation of an individual risk prediction tool
Based on the hazard ratios shown in Table 2 we developed a comprehensive risk model for OS, CIR and NRM to predict the individual patient risk after alloHCT. The baseline hazard function was multiplied by hazard ratios from Table 2 for each patient, when the risk variable value was not 0, and the respective survival curve was plotted. At each time point we can thereby predict the probability of death, relapse and NRM (the latter two were considered as competing events).
To validate our model we split all patients into 3 risk groups (low, intermediate and high risk groups according to three equally large log hazard ratio intervals, Supplementary Figure S18). For internal (in-sample) validation we used our model to predict survival curves for these three groups and compared the predictions with actual Kaplan-Meier curves. The model was able to predict survival curves of each of the groups with high accuracy (Supplementary Figure S19). For internal cross (out-of-sample) validation we used different clinically defined patient groups as training and validation sets. First, we developed a comprehensive risk model using all MDS patients and predicted the OS of all sAML patients. The predicted and actual survival showed very good overlap and a crossvalidation Q2 value of 0.917 (Supplementary Figure S20A). Second, we used all sAML patients to predict the OS of all MDS patients. The predicted and actual survival showed excellent overlap with Q2 of 0.986 (Supplementary Figure S20B). Similarly, the survival of higher risk patients could be well predicted when the model was trained with lower risk patients and vice versa (Q2 values of 0.915 and 0.605, Supplementary Figures S20C, D).
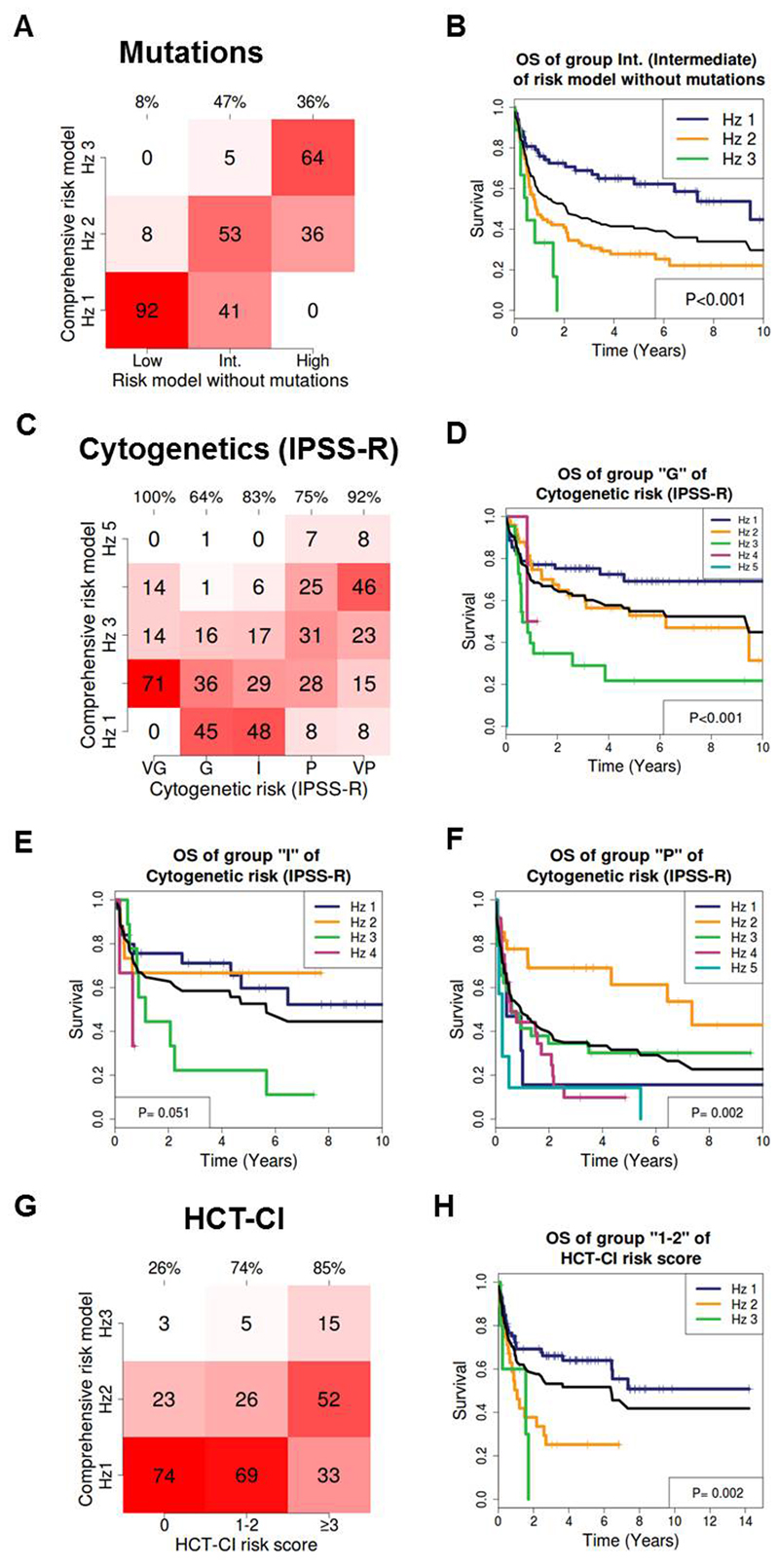
Next, we tested the clinical relevance of our model by (1) evaluating the contribution of molecular aberrations to risk stratification, (2) by comparison to the IPSS-R cytogenetic risk classifier, which has shown prognostic impact also in transplanted patients 24–26, and (3) by comparison to the HCT-CI score.5 First, we removed the molecular risk factors from our OS model (NRAS, TP53 mutation/complex KT, U2AF1, IDH2) and stratified the patients in 3 groups by increasing hazard ratios. We then added back the genetic markers and evaluated, how many patients were reclassified into a different risk group. Figure 3A shows for example that 41% of the patients in the intermediate risk group were reclassified to hazard group 1, and 5% of the intermediate risk patients were reclassified to hazard group 3 (47% in total) when molecular aberrations were added to the model. This reclassification resulted in a significant stratification of survival probabilities as shown by Kaplan-Meier survival curves in Figure 3B (the black line indicates the survival of group 2 according to the risk model without mutations). The addition of molecular risk factors to the model resulted in a reduction of the AIC value from 1656.2 to 1650.7, corresponding to a 16-fold reduced probability that prognostic information is lost, when using the model with vs. without molecular factors.
Figure 3. Validation of an individual risk prediction tool.
(A) Impact of mutations on risk prediction for OS: The risk model without mutations (x-axis) was compared to the comprehensive risk model with gene mutations (NRAS, TP53/complex KT, U2AF1 and IDH2, y-axis). Numbers in the graph represent percentage of patients within the respective risk group. Percentages on top of the graph indicate the percentage of patients that were reclassified by the comprehensive risk model. Hz, hazard risk group. The risk model without mutations is a recalculated multivariate model after mutation covariates are removed from the comprehensive risk model.
(B) OS of patients belonging to the intermediate (int.) risk group of the risk model without mutations, which were reclassified into 3 risk groups by the comprehensive risk model with mutations. The black line represents the survival of intermediate risk patients according to the risk model without mutations (baseline).
(C) Refinement of IPSS-R cytogenetic risk classification (x-axis) by our comprehensive risk model with gene mutations (y-axis). Numbers in the graph represent percentage of patients within the respective risk group. Percentages on top of the graph indicate the percentage of patients that were reclassified by the comprehensive risk model. Hz, hazard risk group.
(D) OS of patients belonging to the IPSS-R cytogenetic good risk group, which was reclassified into 5 risk groups by the comprehensive risk model with mutations. The black line represents the survival of the IPSS-R cytogenetic good risk group (baseline).
(E) OS of patients belonging to the IPSS-R cytogenetic intermediate risk group, which was reclassified into 5 risk groups by the comprehensive risk model with mutations. The black line represents the survival of the IPSS-R cytogenetic intermediate risk group (baseline).
(F) OS of patients belonging to the IPSS-R cytogenetic poor risk group, which was reclassified into 5 risk groups by the comprehensive risk model with mutations. The black line represents the survival of the IPSS-R cytogenetic poor risk group (baseline).
(G) Refinement of the HCT-CI risk classification (x-axis) by our comprehensive risk model with gene mutations (y-axis). Numbers in the graph represent percentage of patients within the respective risk group. Percentages on top of the graph indicate the percentage of patients that were reclassified by the comprehensive risk model. Hz, hazard risk group.
(H) OS of patients with HCT-CI 1-2, who were reclassified into 3 risk groups by the comprehensive risk model with mutations. The black line represents the survival of patients with HCT-CI 1-2 (baseline).
The prediction of CIR was also better with molecular aberrations (Supplementary Figure S20), resulting in a reduction of the AIC value from 787.6 to 777.6 and a 148-fold reduced probability that prognostic information is lost. This suggests that these four gene mutations are associated with treatment resistance and relapse and represent the more aggressive biology of the disease.
We further reclassified the five cytogenetic IPSS-R risk groups by five risk groups of our comprehensive risk model (Figure 3C). As shown for patients with good, intermediate and poor risk cytogenetics, our model could stratify survival probabilities of these patient groups into significantly different risk groups (Figure 3D-3F). IPSS-R cytogenetics alone resulted in a model with an AIC of 1678.3, which is significantly inferior compared to our comprehensive risk model with an AIC of 1650.7, corresponding to a 106-fold reduced probability that information is lost). Similarly, a large proportion of patients with an HCT-CI score of 0, 1-2, or ≥ 3 was reclassified by our comprehensive risk model (Figure 3G) and OS was stratified into significantly different risk groups (Figure 3H). Using HCT-CI alone as a model resulted in an AIC value of 1687.9, which is significantly inferior to our comprehensive risk model (corresponding to a 108-fold reduced probability that information is lost). In summary, we have developed a comprehensive risk prediction model for OS, CIR and NRM of MDS and sAML patients after alloHCT, which can improve currently available risk prediction tools.
Application of the comprehensive risk model
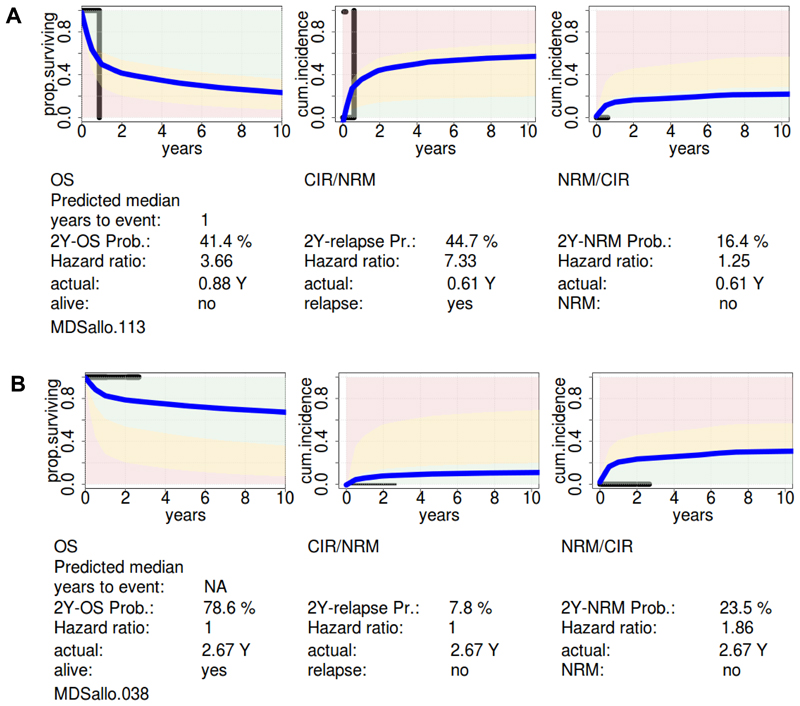
To further evaluate our comprehensive risk model we provide an online calculator, in which the probabilities of OS, CIR and NRM after defined time intervals are provided for an individual patient (https://webext.mh-hannover.de/mdsallo/). The survival probability in our online tool is compared to the survival probability of the normal population of the respective age and gender. An individualized prediction for a 53-year-old male with sAML with trisomy 11, mutated NRAS, IDH2 and DNMT3A and complete remission after double induction is shown in Figure 4A. The probability of CIR at 2 years was 45% and the patient relapsed after 0.61 years. The probability of OS at 2 years was 41% and the patient died after 0.88 years. Another prediction is shown for a 53-year-old female with MDS RAEB2 with normal karyotype and mutations in ASXL1, RUNX1, SF3B1, and ZRSR2, who was transplanted without preceding induction therapy (Figure 4B). The probabilities of CIR and NRM at 2 years were 8% and 23%, respectively, and the predicted 2-year OS was 79%. The patient was alive after a follow-up of 2.67 years with limited chronic GvHD.
Figure 4. Application of the comprehensive risk model.
(A) Individual risk prediction for a 53 year old male with secondary AML, normal cytogenetics, mutations in DNMT3A, IDH2 and NRAS, who achieved complete remission after two cycles of induction chemotherapy. The blue curve shows the predicted individual survival probability, and the black curve shows the actual survival (horizontal line) and the time point of the event (vertical line).
(B) Individual risk prediction for a 53 year old female with refractory anemia with excess of blasts 2 (RAEB2), IPSS risk score intermediate-2, normal cytogenetics, mutations in ASXL1, RUNX1, SF3B1 and ZRSR2. The blue curve shows the predicted individual survival probability, and the black curve shows the actual survival (horizontal line) and the time point of the event (vertical line).
In summary, our online tool allows convenient application of our risk model using a limited set of risk factors to predict probabilities of OS, CIR and NRM on an individual patient basis and to compare it to the survival of the age-and sex-matched normal population. This tool enables the comparison with other datasets and serves as a platform to improve risk prediction when larger datasets become available.
Discussion
We have developed a prediction tool for the outcome after alloHCT that integrates molecular, cytogenetic, patient-and transplantation-associated risk factors.
This prediction tool requires 12 parameters including the karyotype and the mutation status of NRAS, U2AF1, TP53, IDH2, IDH1 and EZH2. The mutation status of NRAS, U2AF1, TP53, and IDH2 is required to predict the OS probability and can be easily obtained by Sanger sequencing, thus making this model accessible also for patients who have not been sequenced with a gene panel.
Our model shows high reproducibility by internal cross (out-of-sample) validation, but will need further external validation before it can be applied in daily clinical practice. Our prediction tool shows the contribution of relapse and non-relapse mortality to the overall survival probability, compares the individual survival prediction with the survival of our transplantation cohort, and compares the predicted survival with the age-and sex-matched survival probability of the normal population. Thus, our prediction model provides additional and more detailed information for the individual patient than previous risk models.5, 27, 28 Our comprehensive risk model may also be used to define patient cohorts for specific trial interventions in future prospective trials and to establish comparability of patient populations between trials.
The risk model for OS includes nine variables that were selected by backward elimination. Five of these are disease-associated genetic variables like IPSS-R cytogenetic risk, complex KT/mutated TP53, NRAS, U2AF1 and IDH2. Complex KT was a strong predictor besides IPSS-R cytogenetic risk and therefore was included in addition to IPSS-R cytogenetics. Previous studies have already shown the strong prognostic impact of complex cytogenetics especially in the transplantation setting 10, 29. While Bejar et al. found a stronger effect of mutated TP53 compared to complex cytogenetics,10 in our analysis these two parameters alone or in combination had a similarly poor prognostic effect and therefore were combined into one risk factor. Complex KT/TP53 and mutated NRAS were also predictive for CIR, suggesting that these aberrations are drivers of relapse and may confer treatment resistance. IPSS-R cytogenetic risk and age were prognostic only by trend, but nevertheless were important risk factors in our model. Remission status before transplantation accounted for patients who received chemotherapy before alloHCT but did not achieve complete remission. In the future, this variable may be refined by minimal residual disease assessment, which has been identified as a strong prognostic marker in alloHCT patients with AML.30 The importance of disease burden before transplantation was recently underscored by Trottier and colleagues showing that patients with >10% blasts and/or 77-100% cytogenetically abnormal cells had significantly worse OS.31 In a study by Hamilton and colleagues mutations in U2AF1 and SRSF2 were predictive for outcome after alloHCT in AML patients but not MDS patients, though the patient number was small.32
The poor prognostic impact of mutated TP53 is consistent among the two previously published studies on molecular predictors of outcome after alloHCT and our analysis.10, 12 The negative prognostic impact of TP53 and RAS mutations found in our study was also described in a recent study of 1514 patients with MDS receiving alloHCT and supports the validity of our results.33 However, survival differences for TET2 and DNMT3A mutated and wildtype patients were only seen in the study by Bejar et al.10 The prognostic effect of ASXL1 and RUNX1 mutations in the study by Della Porta et al. and the effect of U2AF1 and IDH2 in our study was not replicated by the others. These discrepancies may be due to different patient populations: MDS/sAML patients were not included in the study by Bejar et al., constituted 32% of all patients in the study by Della Porta et al. and 53% in our study. The discrepancies may be also due to competing risk factors that were included in our model but not in the other models like comorbidities and transplantation associated risk factors.
Patients without any of our nine risk factors for OS have a base hazard of 27%, i.e. 27% of the mortality risk is not captured by any of our markers yet. When more patients become available, rare mutations will be found in large enough patient subgroups and may become prognostically relevant. In addition, immunologic parameters may be defined that explain some of this risk.34 Our model can then be easily adapted to larger patient numbers and will be updated accordingly.
In summary, we propose a comprehensive risk model integrating patient-related, transplantation-related, cytogenetic and molecular information into one prediction tool, which provides personalized predictions of outcome. After external validation our tool will help to inform physician and patient about the specific risk associated with alloHCT and may be helpful to stratify patients to address specific questions in prospective clinical trials.
Supplementary Material
Acknowledgements
We are indebted to all patients and contributing doctors. We thank Elke Dammann for documentation of clinical data.
This study was supported by the German Federal Ministry of Education and Research grant 01EO0802 (IFB-Tx), grants 110284, 110287, 110292 and 70111267 from Deutsche Krebshilfe; grant DJCLS R13/14 from the Deutsche José Carreras Leukämie-Stiftung e.V; DFG grant HE 5240/5-2 and HE 5240/6-1; an ERC grant under the European Union’s Horizon 2020 research and innovation programme (No. 638035), and by grants from Dieter-Schlag Stiftung.
Footnotes
Conflict of interest statement
The authors have no potential conflicts of interest.
References
- 1.Komrokji RS. Current State of the Art: Management of Higher Risk Myelodysplastic Syndromes. Clin Lymphoma Myeloma Leuk. 2016;(16 Suppl):S39–43. doi: 10.1016/j.clml.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Malcovati L, Hellstrom-Lindberg E, Bowen D, Ades L, Cermak J, Del Canizo C, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood. 2013;122(17):2943–2964. doi: 10.1182/blood-2013-03-492884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutler CS, Lee SJ, Greenberg P, Deeg HJ, Perez WS, Anasetti C, et al. A decision analysis of allogeneic bone marrow transplantation for the myelodysplastic syndromes: delayed transplantation for low-risk myelodysplasia is associated with improved outcome. Blood. 2004;104(2):579–585. doi: 10.1182/blood-2004-01-0338. [DOI] [PubMed] [Google Scholar]
- 4.Koreth J, Pidala J, Perez WS, Deeg HJ, Garcia-Manero G, Malcovati L, et al. Role of reduced-intensity conditioning allogeneic hematopoietic stem-cell transplantation in older patients with de novo myelodysplastic syndromes: an international collaborative decision analysis. J Clin Oncol. 2013;31(21):2662–2670. doi: 10.1200/JCO.2012.46.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorror ML, Maris MB, Storb R, Baron F, Sandmaier BM, Maloney DG, et al. Hematopoietic cell transplantation (HCT)-specific comorbidity index: a new tool for risk assessment before allogeneic HCT. Blood. 2005;106(8):2912–2919. doi: 10.1182/blood-2005-05-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gratwohl A, Stern M, Brand R, Apperley J, Baldomero H, de Witte T, et al. Risk score for outcome after allogeneic hematopoietic stem cell transplantation: a retrospective analysis. Cancer. 2009;115(20):4715–4726. doi: 10.1002/cncr.24531. [DOI] [PubMed] [Google Scholar]
- 7.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellagatti A, Roy S, Di Genua C, Burns A, McGraw K, Valletta S, et al. Targeted resequencing analysis of 31 genes commonly mutated in myeloid disorders in serial samples from myelodysplastic syndrome patients showing disease progression. Leukemia. 2016;30(1):247–250. doi: 10.1038/leu.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 10.Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32(25):2691–2698. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christopeit M, Badbaran A, Alawi M, Zabelina T, Zeck G, Wolschke C, et al. Correlation of somatic mutations with outcome after FLAMSA-busulfan sequential conditioning and allogeneic stem cell transplantation in patients with myelodysplastic syndromes. Eur J Haematol. 2016;97(3):288–296. doi: 10.1111/ejh.12724. [DOI] [PubMed] [Google Scholar]
- 12.Della Porta MG, Galli A, Bacigalupo A, Zibellini S, Bernardi M, Rizzo E, et al. Clinical Effects of Driver Somatic Mutations on the Outcomes of Patients With Myelodysplastic Syndromes Treated With Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.67.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thol F, Kolking B, Hollink IH, Damm F, van den Heuvel-Eibrink MM, Michel Zwaan C, et al. Analysis of NUP98/NSD1 translocations in adult AML and MDS patients. Leukemia. 2012;27(3):750–754. doi: 10.1038/leu.2012.249. [DOI] [PubMed] [Google Scholar]
- 14.Mitelman F. ISCN (1995):an International System for Human Cytogenetic Nomenclature. Basel, Switzerland, SKarger. 1995 [Google Scholar]
- 15.Thol F, Suchanek KJ, Koenecke C, Stadler M, Platzbecker U, Thiede C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013;27(10):2072–2075. doi: 10.1038/leu.2013.145. [DOI] [PubMed] [Google Scholar]
- 16.Thol F, Klesse S, Kohler L, Gabdoulline R, Kloos A, Liebich A, et al. Acute myeloid leukemia derived from lympho-myeloid clonal hematopoiesis. Leukemia. 2017 doi: 10.1038/leu.2016.345. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korn EL. Censoring distributions as a measure of follow-up in survival analysis. Stat Med. 1986;5(3):255–260. doi: 10.1002/sim.4780050306. [DOI] [PubMed] [Google Scholar]
- 18.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Annals of Statistics. 1988;16:1141–1154. [Google Scholar]
- 19.Li KH, Raghunathan TE, Rubin DB. Large-Sample Significance Levels from Multiply Imputed Data Using Moment-Based Statistics and an F Reference Distribution. Journal of the American Statistical Association. 1991;86:1065–1073. [Google Scholar]
- 20.Cox DR. Regression models and life tables. J R Stat Soc B. 1972;34:187–202. [Google Scholar]
- 21.Burnham KP, Anderson DR. Multimodel Inference: Understanding AIC and BIC in Model Selection. Sociological methods and research. 2004;33:261–304. [Google Scholar]
- 22.Team RDC. R: A Language and Environment for Statistical Computing. 2008 [Google Scholar]
- 23.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koenecke C, Gohring G, de Wreede LC, van Biezen A, Scheid C, Volin L, et al. Impact of the revised International Prognostic Scoring System, cytogenetics and monosomal karyotype on outcome after allogeneic stem cell transplantation for myelodysplastic syndromes and secondary acute myeloid leukemia evolving from myelodysplastic syndromes: a retrospective multicenter study of the European Society of Blood and Marrow Transplantation. Haematologica. 2015;100(3):400–408. doi: 10.3324/haematol.2014.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deeg HJ, Scott BL, Fang M, Shulman HM, Gyurkocza B, Myerson D, et al. Five-group cytogenetic risk classification, monosomal karyotype, and outcome after hematopoietic cell transplantation for MDS or acute leukemia evolving from MDS. Blood. 2012;120(7):1398–1408. doi: 10.1182/blood-2012-04-423046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Della Porta MG, Alessandrino EP, Bacigalupo A, van Lint MT, Malcovati L, Pascutto C, et al. Predictive factors for the outcome of allogeneic transplantation in patients with MDS stratified according to the revised IPSS-R. Blood. 2014;123(15):2333–2342. doi: 10.1182/blood-2013-12-542720. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. [PubMed] [Google Scholar]
- 28.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schanz J, Tuchler H, Sole F, Mallo M, Luno E, Cervera J, et al. New comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J Clin Oncol. 2012;30(8):820–829. doi: 10.1200/JCO.2011.35.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buccisano F, Maurillo L, Spagnoli A, Del Principe MI, Fraboni D, Panetta P, et al. Cytogenetic and molecular diagnostic characterization combined to postconsolidation minimal residual disease assessment by flow cytometry improves risk stratification in adult acute myeloid leukemia. Blood. 2010;116(13):2295–2303. doi: 10.1182/blood-2009-12-258178. [DOI] [PubMed] [Google Scholar]
- 31.Trottier BJ, Sachs Z, DeFor TE, Shune L, Dolan M, Weisdorf DJ, et al. Novel disease burden assessment predicts allogeneic transplantation outcomes in myelodysplastic syndrome. Bone Marrow Transplant. 2016;51(2):199–204. doi: 10.1038/bmt.2015.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton BK, Visconte V, Jia X, Tabarroki A, Makishima H, Hasrouni E, et al. Impact of allogeneic hematopoietic cell transplant in patients with myeloid neoplasms carrying spliceosomal mutations. Am J Hematol. 2016;91(4):406–409. doi: 10.1002/ajh.24306. [DOI] [PubMed] [Google Scholar]
- 33.Lindsley RC, Saber W, Mar BG, Redd RA, Haagenson MD, Grauman PV, et al. Genetic Alterations Predict Outcomes in Patients with Myelodysplastic Syndrome Receiving Allogeneic Hematopoietic Stem Cell Transplantation. Blood. 2016 Abstrac #69. [Google Scholar]
- 34.Crucitti L, Crocchiolo R, Toffalori C, Mazzi B, Greco R, Signori A, et al. Incidence, risk factors and clinical outcome of leukemia relapses with loss of the mismatched HLA after partially incompatible hematopoietic stem cell transplantation. Leukemia. 2015;29(5):1143–1152. doi: 10.1038/leu.2014.314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.