Abstract
Cadmium is one of the widely used heavy metals (HM) in commercial and industrial products and contributes to environmental contamination in an urban setting. In our previous studies, we established that An. gambiae sensu stricto, a vector of malaria, had adapted to HM pollutants in nature despite their proclivity for unpolluted aquatic habitats. We further demonstrated that heavy metal tolerance adaptation process impacts a biological cost to the fitness of the mosquito and potentially involves the induction of specific HM-responsive transcripts and proteins. Here we interrogated differential proteomic profiles of the cadmium tolerant vs. naïve strains of An. gambiae to shed light on proteomic processes that underpinned biological cost to fitness. We identified a total of 1067 larval proteins and observed significant down-regulation of proteins involved in larval immune responses, energy metabolism, antioxidant enzymes, protein synthesis, and proton transport. Our results suggest that mosquitoes can adjust their biological program through proteome changes to counter HM pollution. Since our study was done in controlled laboratory settings, we acknowledge this may not wholly represent the conditions HM polluted environments. Nevertheless, mosquitoes deploying this strategy have the potential of creating an urban enclave for breeding and thrive and become agents of sporadic malaria epidemics.
Keywords: Anopheles gambiae, Cadmium tolerance, Larvae, Proteomics, Immunity
1. Introduction
Anopheline mosquitos are the only known vectors of malaria including the most-deadly form caused by Plasmodium falciparum. Apart from malaria, some species of Anopheles contribute to the transmission of other tropical diseases including; lymphatic filariasis, canine heartworm and O’nyong’nyong fever (Cancrini et al., 2006; Nchoutpouen et al., 2019). Their success in disease transmission is primarily due to the capacity for mosquitoes to exploit different kinds of habitats created by humans, adaptive flexibility that has permitted the mosquito to exploit different larval habitats and colonize a variety of micro and macro environmental conditions throughout the Tropics (Lanzaro et al., 1998; Budiansky, 2002; Roberts et al., 2002; Rozendaal, 1992). Urbanization of Sub-Saharan Africa cities has favored adaptation of anopheline to various xenobiotics and expansion of their niche to habitats polluted by HMs and organic matter contaminants (Awolola et al., 2007; Djouaka et al., 2007; Mukhtar et al., 2003; Sattler et al., 2005; Sibomana, 2002; Mireji et al., 2008). This is contrary to the long-held dogma that An. gambiae s.l. exclusively thrive in clean water devoid of pollutants, a common scenario in rural settings (Mireji et al., 2010a). Behavioral plasticity and insecticide resistance have permitted mosquito to either resist insecticides or circumvent them through avoidance. Therefore, limiting the effectiveness of vector control tools such as long-lasting insecticide-treated nets (LLINs) or indoor residual sprays (IRS), thus leading to residual malaria transmission (Rydzanicz et al., 2009; Zhu et al., 2017). To combat malaria transmission especially in urban settings will require novel approaches to be included in the malaria control toolbox (Rydzanicz et al., 2009; Zhu et al., 2017). Understanding the biology of the vectors thriving in polluted urban environments forms the first steps towards designing more effective tools to match the changes in vector dynamics.
Previously, our work demonstrated through multigenerational chronic exposure of mosquito larvae to copper, lead and cadmium, that the adaptation, particularly to HM pollutants (Cancrini et al., 2006) is potentially accompanied by a biological cost that includes reduced egg viability, larval and pupal survivorship, adult emergence, fecundity, instantaneous birth, and reproductive rates, and increased population doubling times (Nchoutpouen et al., 2019). Besides, it was subsequently established that the adaptation was potentially mediated by alpha-tubulin and metallothionein (Lanzaro et al., 1998), a known HM responsive gene in insects (Budiansky, 2002), and spectrum of uncharacterized mosquito proteins revealed by 2D proteomic maps of differentially expressed peptides following the chronic exposures to the heavy metals (Roberts et al., 2002).
To further our understanding of mechanisms underlying the HM tolerance (hence shed light on the environmental adaptation of the mosquito to HMs), a one dimensional (1D) gel electrophoresis and high-throughput tandem mass spectrometry was performed to characterize the proteome of cadmium tolerant mosquitos (Mireji et al., 2010a) and identify molecular pathways functionally affected by the HM tolerance. Here we report that An. gambiae subjected to cadmium over several generations exhibited changes at the proteome level. The proteome changes further suggest the reduced capacity to detoxify reactive oxygen species, protein synthesis, and down-regulation of some aspects of mosquito immunity. These observations have identified a vital aspect of the mosquito physiological that can be deployed for adaptation to HM pollution. This implies that An. gambiae can adjust its biology and colonize HM polluted urban dwelling that provides sufficient access to water for mosquito breeding and a potential source for sporadic disease transmission.
2. Materials and methods
2.1. Mosquito propagation and tolerance experiments
In this study, proteomic analysis on larval proteins from cadmium tolerant (Cd) and naïve (control) (Ctrl) An. gambiae larvae was performed. All experiments were done using three biological replicates per group. Anopheles gambiae s. s. mosquitoes previously collected from Mbita field station (00.025’S, 34.013’E), Homa Bay County, Kenya were used. The mosquito colony were kept under environmental conditions (28 ± 2 °C, 75–80% RH and LD 12: 12h photoperiod) at the International Centre of Insect Physiology and Ecology (ICIPE), Nairobi, Kenya as previously described (Mireji et al., 2010b). Briefly, selection of cadmium tolerance in larval stages of An. gambiae s. s. was done in triplicates through chronic generational exposures to empirically determine Maximum Acceptable Toxicant Concentration (MATCs) of cadmium to the mosquito. Eggs (1500 per replicate) and subsequent emergent immature stages (larvae and pupae) were exposed to the metal solution at 0.36 μg/L of cadmium. The larvae were reared in 1500 mL of metal solutions in polypropylene cylindrical pans with a radius and height of 10.5 and 24.1 cm, respectively (Mireji et al.,2010b). At the time of this work, the colony was in the 90th filial generational selection for cadmium tolerance. We sourced and used cadmium chloride (CdCl2) 99.99% pure from Fisher Scientific LLC (Fair Lawn, NJ, U.S.A).
2.2. Extraction of An. gambiae s.s larvae proteome
Mosquito proteins were extracted from triplicate biological samples (each 50 mosquito L3 larvae) recovered after Trizol extraction (ZYMO Research Corporation, Irvine, CA, USA) according to the manufacturer’s instructions. Briefly, we performed RNA extraction step using Direct-zol kit (ZYMO Research Corporation, Irvine, CA, USA) and subsequently precipitated proteins from the flow through mixture by using 1 ml of isopropanol for every 750 μL sample incubating the solution for ten minutes at room temperature and centrifuging the solution at 12,000 × gat 4oC for ten minutes to recover the protein pellet. The recovered pellet was washed three times in 0.3M guanidine hydrochloride in 100% ethanol with 20 min incubation between each the washes and recovered the pellet after centrifugation at 12,000 × g for ten minutes. Pellet was then briefly air dried and re-suspended in the pellet in 100ul of SUBT buffer (4.5 M urea in 0.5% SDS, 25 mM Tris/HCl pH 7.5) (Bai and Laiho, 2012) supplemented with protease inhibitors after the final wash. The amount of recovered proteins was estimated through Bradford assay (Bradford, 1976).
2.3. Liquid chromatography and tandem mass spectrometry of An. gambiae s.s larvae proteome
LC-MS/MS analysis was performed following the methods and procedures of Njunge et al. (2017). Briefly, equal amounts of the larval proteins were diluted in Laemmle sample buffer (60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5%p-mercaptoethanol, 0.01% bromophenol blue), boiled the resultant solution and separated the inherent protein on a 12% acrylamide SDS PAGE gel (NuPAGE 4–12% Bis-Tris Gel, Life Technology) in triplicate fixed the gel in methanol/acetic acid, stained the gel with colloidal Coomassie (Sigma-Aldrich Laborchemikalien GmbH, Seelze, Germany) and destained the gel in methanol as previously described (Njunge et al., 2017). Each gel lane was excised to six pieces, and destained the gel pieces in 50 mM ammonium bicarbonate/50% CH3CN and digested the pieces by trypsin (Pierce MS Grade, Thermo Fisher Scientific) overnight at 37°C. This was followed by peptide extraction in 0.5% formic acid (FA)/50% CH3CN and drying in a SpeedVac (Thermo Fisher Scientific). Peptides were later re-suspended in 20 μl of 0.5% FA just before LC-MS/MS analysis. Peptides (5 μL) were loaded using a Dionex Ultimate 3000 nano-flow ultra-high-pressure liquid chromatography system (Thermo Scientific) on to a 75 μm × 2 cm C18 trap column (Thermo Scientific) and separated on a 75 μm × 25 cm C18 reverse-phase analytical column (Thermo Scientific). Elution was carried out with mobile phase B (80% acetonitrile with 0.1% formic acid) gradient (5-35%) over 120 min. Peptides were measured using an Exactive Orbitrap mass spectrometer (Thermo Scientific) coupled to the chromatography system via a nano-electrospray ion source (Thermo Scientific). The ms’1 settings were: Resolution, 70,000; AGC target, 3e6; scan range, 400–1800 m/z; while the ms’2 settings were: Resolution, 17,500; AGC, 5e4; isolation window, 1.6 m/z. The top 15 most intense ions were selected for ms’2, which were subsequently excluded for the next 30 s.
The false discovery rate was set at (FDR = 0.01) 1% for both the peptide-spectrum match level determined by the target-decoy approach and the protein FDR. The raw files were processed in MaxQuant (Version 1.5.3.30, www.MaxQuant.org), and searched against a combined database of Anopheles gambiae proteins (Agamp4; downloaded on 19th November 2014 from Vectorbase) (Giraldo-Calderon et al., 2015; Megy et al., 2012) and a contaminate database supplied by MaxQuant. The mass spectrometry proteomics data has been deposited in the ProteomeXchange Consortium via the PRIDE (Deutsch et al., 2017; Vizcaino et al., 2014) partner repository, with the dataset identifier PXD010707.
2.4. Differential protein abundance analysis
To identify differentially expressed proteins, we performed labelfree quantification (LFQ) of proteins using MaxQuant software (Version 1.5.3.30, www.MaxQuant.org) according to the manufacturer’s protocols. LFQ values were normalized for all replicates (Kaur et al., 2017; Cox and Mann, 2008), and used for analysis. The mean protein expression values per group for each protein were obtained and used to determine differential protein abundance. The changes in protein abundance were computed from the difference in log2 values in (replicate–mean). Unpaired student t-test was performed on the mean protein abundance between the cadmium-tolerant and the control groups to identify proteins whose abundance were significantly altered. Benjamini Hochberg (BH) correction was performed on the p-values, proteins with adjusted pvalue p < 0.05 were considered significant for further downstream analyses. Patterns of variation among samples were analyzed by Principal Component Analysis (PCA). The global changes involving biological processes were investigated using STRING database (Szklarczyk et al., 2011) to identify known protein-protein interactions (PPIs) among the deferentially expressed peptides using orthologs of the An. gambiaeproteins in Drosophila melanogaster.
3. Results
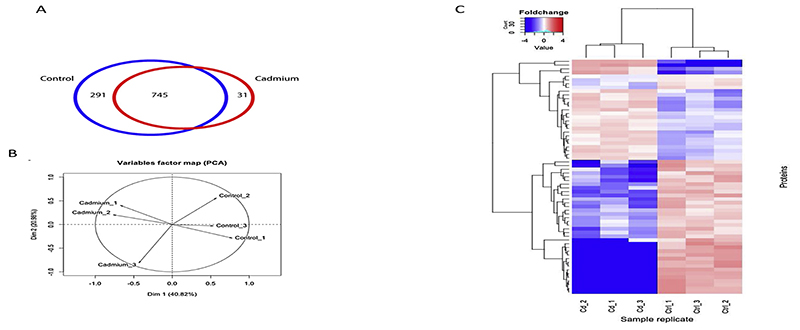
Our analysis identified 1067 larval proteins (FDR < 0.01) representing approximately 8% of the predicted proteins in the An. gambiae genome. Among these, 745 proteins were common to both groups while 322 were uniquely present in either the cadmium (31 proteins) or control groups (291 proteins) (Fig. 1A). Principal Component Analysis (PCA) analysis on the replicate samples distinguished the cadmium tolerant larvae from the rest (Fig. 1B). To determine protein signature associated with Cd tolerance we performed differential protein abundance analysis based on the LFQ values and revealed 63 proteins that were differentially expressed (Fig. 1C).
Fig. 1. The proteome of Anopheles gambiae larvae after multigenerational exposure to Cadmium metal.
Larvae proteins in Cd and control groups were extracted in triplicates, analyzed by 1D gel and tandem mass spectrometry. A) Venn plot shows the proportion of proteins found in both groups and those unique to each. B) principal component analysis of the replicate samples for Cd-tolerant vs. Control group C) Heatmap analysis of differentially expressed proteins. Log2 value of protein expression in (replicatemean) for each protein was calculated and 2tailed student t-test calculated in Cd-tolerant vs. Control. Protein with (BH corrected pvalue < 0.01) were used for generating heatmap in R Bioconductor heatmap.2 package.
3.1. Proteins suppressed by cadmium exposure in An. gambiae larvae
The down-regulated proteins by far were the largest group (n = 45). These were functionally associated with glucose and carbohydrate metabolism, metal binding and immunity. The most down-regulated was AGAP005163-PA a UDP-glucuronosyl/UDP-glucosyltransferase. From the immune group were proteins from the Class II-associated invariant chain peptide (CLIP) family (AGAP000315-PA, AGAP004719-PA, AGAP009216-PA and AGAP010731-PA, AGAP011783-PA), prophenoloxidase (AGAP010730-PA) and GNBP1 (AGAP004455-PA). Other proteins down-regulated were cuticular proteins (CPLCP13, CPTC1), UDP-galactose-4-epimerase, enzymes associated with redox pathway and free radical detoxification (AGAP004378 and AGAP005749), a protein involved with proteolysis (AGAP004015-PA), a ribosomal protein associated with oogenesis (RS3A_ANOGA), a cell growth regulator protein (AGAP006469-PA) and proton transport molecules (Tables 1 and S1).
Table 1.
Differentially expressed An. gambiae larval protein after multigenerational exposure with Cd.
| Protein_ID | Gene.name | description | l2fold | BH.corrected.Pvalue |
|---|---|---|---|---|
| AGAP005163-PA | Glucosyl/glucuronosyl transferases | < -7.9 | 2.34E-04 | |
| AGAP028204-PA | hypothetical protein | 2.56 | 4.69E-04 | |
| AGAP000315-PA | CLIPC6 | Clip-domain serine protease | −6.38 | 7.03E-04 |
| AGAP004877-PB | paramyosin | <-7.8 | 9.37E-04 | |
| AGAP010174-PA | oligosaccharyltransferase complex subuni | 2.37 | 1.17E-03 | |
| AGAP000399-PA | squid | −1.24 | 1.41E-03 | |
| AGAP013112-PA | mRNA binding protein | − 0.5 | 1.64E-03 | |
| AGAP004514-PA | nuclear GTP-binding protein | <− 7.7 | 1.87E-03 | |
| AGAP008499-PA | Mitochondrial transcription factor A | 1.74 | 2.11E-03 | |
| AGAP004395-PA | nucleophosmin 3 | <− 7.6 | 2.34E-03 | |
| AGAP013365-PA | hypothetical protein | − 0.88 | 2.58E-03 | |
| AGAP004895-PA | carbonic anhydrase | <− 7.5 | 2.81E-03 | |
| AGAP003216-PA | 26S proteasome regulatory subunit T2 | <− 7.4 | 3.05E-03 | |
| AGAP013036-PA | hypothetical protein | 1.58 | 3.28E-03 | |
| AGAP011802-PA | RpL39 | 60S ribosomal protein L39 | − 1.99 | 3.51E-03 |
| AGAP012317-PA | hypothetical protein | − 1.84 | 3.75E-03 | |
| AGAP007250-PA | hypothetical protein | − 2.85 | 3.98E-03 | |
| AGAP028178-PA | CPLCP13 | cuticular protein (putative) CPLCP13 | − 0.74 | 4.22E-03 |
| AGAP007507-PA | mRpL48 | Mitochondrial ribosomal protein L48 | − 7.35 | 4.45E-03 |
| AGAP005129-PA | 5-methylthioadenosine phosphorylase | <− 7.3 | 4.69E-03 | |
| AGAP011298-PA | RpL10a | 60S ribosomal protein L10a | − 0.18 | 4.92E-03 |
| AGAP004606-PA | HYPK | <− 7.2 | 5.15E-03 | |
| AGAP004719-PA | CLIPC9 | Clip-Domain Serine Protease | <− 7.1 | 5.39E-03 |
| AGAP011457-PA | ribose 5-phosphate isomerase A | 1.82 | 5.62E-03 | |
| AGAP007393-PC | protein disulfide isomerase family A, me | − 0.69 | 5.86E-03 | |
| AGAP010730-PA | Prophenoloxidase activating factor | − 0.7 | 6.09E-03 | |
| AGAP012504-PA | hypothetical protein | 0.36 | 6.33E-03 | |
| AGAP004618-PA | hypothetical protein | <− 7.0 | 6.56E-03 | |
| AGAP004015-PA | SP21408 | prolylcarboxypeptidase | <− 7.1 | 6.79E-03 |
| AGAP012407-PA | protein disulfide-isomerase A1 | 2.09 | 7.03E-03 | |
| AGAP011476-PA | hypothetical protein | − 2.7 | 7.26E-03 | |
| AGAP013347-PA | hypothetical protein | − 0.74 | 7.50E-03 | |
| AGAP007060-PA | hypothetical protein | − 3.32 | 7.73E-03 | |
| AGAP009216-PA | Clip-domain serine protease | − 2.52 | 7.97E-03 | |
| AGAP007249-PB | Flightin | Flightin | − 1.6 | 8.20E-03 |
| AGAP010392-PA | Calumenin | − 1.02 | 8.43E-03 | |
| AGAP011276-PA | hypothetical protein | − 0.17 | 8.67E-03 | |
| AGAP011477-PA | Eupolytin | 1.02 | 8.90E-03 | |
| AGAP007666-PA | Calcyphosin-like protein | − 1.21 | 9.14E-03 | |
| AGAP009652-PA | NADH dehydrogenase (ubiquinone) 1 alpha | − 2.13 | 9.37E-03 | |
| AGAP010181-PA | obelix | − 2.42 | 9.61E-03 | |
| AGAP004443-PB | glycogen synthase kinase 3 beta | < −7.2 | 9.84E-03 | |
| AGAP012990-PA | RpS23 | 40S ribosomal protein S23 | 0.22 | 1.01E-02 |
| AGAP008440-PA | urate oxidase | − 5.33 | 1.03E-02 | |
| AGAP010404-PB | GSTS1 | glutathione S-transferase | 1.54 | 1.05E-02 |
| AGAP009863-PA | Eukaryotic translation initiation factor | 0.99 | 1.08E-02 | |
| AGAP010613-PA | elongation factor 1-beta | − 1.16 | 1.12E-02 | |
| AGAP010100-PA | CPR84 | cuticular protein 84 RR-2 family | 1.06 | 1.15E-02 |
| AGAP009271-PA | Prosbeta1 | 20S proteasome subunit beta 1 | -3 | 1.17E-02 |
| AGAP012823-PA | NADH dehydrogenase 1 alpha subcomplex su | 1.8 | 1.19E-02 | |
| AGAP012056-PA | cofilin | 0.3 | 1.22E-02 | |
| AGAP006469-PA | cell growth-regulating nucleolar protein | − 2.51 | 1.24E-02 | |
| AGAP013185-PA | hypothetical protein | 1.62 | 1.27E-02 | |
| AGAP010477-PB | phosducin-like 3 | − 4.57 | 1.29E-02 | |
| AGAP005559-PA | 26S proteasome regulatory subunit N10 | 0.04 | 1.31E-02 | |
| AGAP012425-PA | hypothetical protein | 1.91 | 1.34E-02 | |
| AGAP012837-PA | mRpL43 | 39S ribosomal protein L43 | − 1.67 | 1.36E-02 |
| AGAP005696-PA | CPTC1 | cuticular protein from two-cysteine fami | − 7.04 | 1.38E-02 |
| AGAP007172-PB | translation initiation factor 4E | − 2.43 | 1.41E-02 | |
| AGAP010731-PA | CLIPA8 | Clip-Domain Serine Protease | − 1.15 | 1.43E-02 |
| AGAP003360-PA | dynein light chain roadblock-type | <− 7.3 | 1.45E-02 | |
| AGAP011098-PA | beta-aspartyl-peptidase (threonine type) | 2.15 | 1.48E-02 | |
| AGAP006117-PA | hypothetical protein | <− 7.4 | 1.50E-02 |
3.2. Proteins induced by cadmium exposure in An. gambiae larvae
We observed induction of nine proteins with catalytic activity in the cadmium tolerant strain (Fig. 1C and Table 1). Three each of these proteins were functionally associated with the redox (AGAP010404-PB, AGAP001325 and AGAP002170) and actin dynamics (AGAP012056-PA). We also detected induction of two signaling molecules of the small GTPase family (AGAP001902 and AGAP002219), and several hypothetical proteins including two most abundant proteins (AGAP028204-PA and AGAP028068-PA). We similarly saw the induction of transcription factors, ribosomal proteins, enzyme from GST family (AGAP010404-PB) and proteins that function in the degradation pathway (Table 1 and S1 table).
3.3. Functional enrichment and network analysis
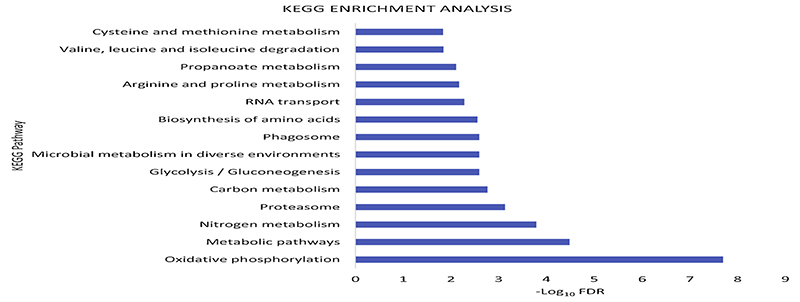
On performing functional enrichment using KEGG pathways, we identified 14 downregulated pathways, among these were pathways linked to oxidative phosphorylation, glycolysis, arginine and proline metabolism (Fig. 2). In addition, analysis based on protein families (Pfam) identified the CLIP protease, proteasome sub unit A, insect cuticle proteins downregulated (Table 2).
Fig. 2. Functional enrichment of differentially regulated proteins.
Protein enrichment, according to functional groups, was done by STRING online database using KEGG pathways. The x-axis shows biological pathways that were significantly altered based on the-log10 FDR values plotted in the y-axis. Enrichment was only observed in down-regulated proteins.
Table 2.
Biological pathways dysregulated in Anopheles gambiae larvae after multigenerational exposure to Cd metal.
| Pathway | Pathway ID | Pathway description | Observed gene count | False discovery rate | Regulation |
|---|---|---|---|---|---|
| Pfam | PF00089 | Trypsin | 30 | 1.89E-03 | down |
| Pfam | PF00227 | Proteasome subunit | 7 | 1.89E-03 | down |
| Pfam | PF10584 | Proteasome subunit A N-terminal signature | 5 | 3.37E-03 | down |
| Pfam | PF12032 | Regulatory CLIP domain of proteinases | 7 | 7.71E-03 | down |
| Pfam | PF00379 | Insect cuticle protein | 20 | 1.44E-02 | down |
| Pfam | PF01105 | emp24/gp25 L/p24 family/GOLD | 4 | 2.78E-02 | down |
| KEGG | 190 | Oxidative phosphorylation | 21 | 2.00E-08 | down |
| KEGG | 1100 | Metabolic pathways | 58 | 3.32E-05 | down |
| KEGG | 910 | Nitrogen metabolism | 6 | 1.63E-04 | down |
| KEGG | 3050 | Proteasome | 9 | 7.43E-04 | down |
| KEGG | 1200 | Carbon metabolism | 12 | 1.72E-03 | down |
| KEGG | 10 | Glycolysis / Gluconeogenesis | 7 | 2.57E-03 | down |
| KEGG | 1120 | Microbial metabolism in diverse environments | 14 | 2.57E-03 | down |
| KEGG | 4145 | Phagosome | 9 | 2.57E-03 | down |
| KEGG | 1230 | Biosynthesis of amino acids | 9 | 2.77E-03 | down |
| KEGG | 3013 | RNA transport | 14 | 5.22E-03 | down |
| KEGG | 330 | Arginine and proline metabolism | 7 | 6.79E-03 | down |
| KEGG | 640 | Propanoate metabolism | 5 | 7.76E-03 | down |
| KEGG | 280 | Valine, leucine and isoleucine degradation | 6 | 1.43E-02 | down |
| KEGG | 270 | Cysteine and methionine metabolism | 5 | 1.45E-02 | down |
4. Discussions
Zinc, copper, and cadmium are common environmental contaminants in urban and industrial settings. Zinc is required at trace levels for cellular and biological processes. However, cadmium has no physiological role. Multigenerational chronic exposure of cadmium in insects may be manifested in increased oxidative stress, blocking of signal transduction, and tissue necrosis (Zervas et al., 2010). Here we investigated the effects of cadmium exposure in mosquitoes as they expand their niche in the polluted urban environments in Kenya. Previously we showed that cadmium tolerance leads to increase expression of alpha-tubulin and metallothionein transcripts (Mireji et al., 2010b). We further identified over 30 proteins that were differentially expressed due to the multi-generational chronic exposure to cadmium (Mirejiet al., 2006). In our current work, we have expanded our understanding of cadmium tolerance in the mosquito by using an unbiased and more sensitive approach by combining the strength of 1D gel and a sensitive mass spectrometry analysis. We established that 63 showed significant changes, most of which were suppressed. The biological processes associated with these changes include immunity, signaling mechanism, oxidative response, and energy metabolism. The most significant change was observed for AGAP005163-PA, a UDP-glucuronosyl/UDP-glucosyltransferase that was consistently not identified in HM-tolerant mosquitos. This was not surprising given that UDP-glucuronosyl catalyzes the transfer of glucose from UDP-glucose to ecdysteroids and the vital role ecdysteroids play in regulating insect growth and transitions between stages. Low levels of ecdysteroids are linked to a disruption in mosquito development. HM-tolerance is known to induce delays in pupation and reduced adult size. Thus, we associate AGAP005163-PA abundance the fitness cost incurred for HM-adaptation. Molecules that regulate insect development have previously shown great potential as novel targets for vector control using system such as gene-drive to suppress vector populations. Based on our findings, AGAP005163-PA provides a potential target that can be exploited for vector control.
Although there was general downregulation of oxidative phosphorylation pathway, we observed significant upregulation enzymes from GST family. GSTs function by catalyzing the conjugation of glutathione (GSH) (Xu et al., 2014) to various electrophilic compounds, including reactive oxygen species (ROS). The induction of these molecules is in response to oxidative stress due to the accumulation of ROS (Li et al., 2003). Deleterious effects of oxidative stress are not only associated with tissue, DNA, and protein damage, but reports indicate this may include insecticide resistance mechanisms in mosquitos (Jones et al., 2012; Oliver and Brooke, 2018; Ramirez and Gimenez, 2003). High levels of metabolic enzymes such as GSTs and cytochrome oxidases are strongly associated with this phenotype (Oliver and Brooke,2016). As insects become more accustomed to HM-polluted habitats, there is excellent potential for increased risk of high-level insecticide resistance and increased odds of disease transmission
The relationship between HM tolerance and mosquito immunity has not been well established. In our analysis, we observed downregulation of several immune molecules in exposed larvae. The importance of CLIPB9 and CLIPA8 for the activation of pro-phenol oxidase cascade and melanization of Plasmodium and bacteria has been demonstrated (An et al., 2011; Schnitger et al., 2007). GNBP1 (AGAP004455-PA) plays a vital role by acting as a pattern recognition receptor (PRR) with a broad repertoire for pathogen surveillance (Dimopoulos, 2003; Whitten et al., 2004). PRR activate Toll and IMD pathways that induce the expression of mosquito immunity genes. In the absence of effective immune surveillance, HM-tolerant mosquitos may be more prone to bacterial pathogens frequently found in aquatic habitats. Also, immune priming occurs at larval stages and shapes antiplasmodial responses in adult stages (Moreno-Garcia et al., 2015). However, we note that immune system development is a complex process and may be different in laboratory settings as compared to a natural environment where other abiotic factors may come in to play.
In conclusion, we performed proteomic analysis and identified specific proteins and biological processes associated with An. gambiaeadaptation to HM-tolerance. We note the limitations of our study being a laboratory setting, while the difference between this approach and field conditions have been demonstrated. Nevertheless, our data shows the genetic capacity in mosquitoes to adapt to harsh conditions and thrive. These opportunistic tendencies have been observed for both West and East African malaria mosquitos (Sattler et al., 2005; Kudom, 2015), and also in mosquitoes from Culex and Aedes species (Mukhtar et al., 2003) that can potentially become a source of disease transmission. Besides, Anopheles arabiensis, an efficient vector of human malaria, is currently replacing Anopheles gambiae s.s. as the dominant malaria vector in several regions in East Africa (Bayoh et al., 2010), interestingly Oliver and Brooke (2016) has shown that An. arabiensis adult mosquito emerging from larva exposed to heavy metals exhibit high levels of insecticide tolerance (Oliver and Brooke, 2018). Also, An. arabiensis is an opportunistic feeder that derives blood meals sources from a variety of vertebrate hosts and exhibits outdoor feeding and resting (Killeen et al., 2016; Charlwood et al., 2018). This behavior factors, together with the potential for HM selection for high insecticide tolerance, possess a significant threat and potential breakdown on vector control strategies that target adult mosquitoes using insecticides to control indoor resting and feeding mosquitoes.
Acknowledgements
We also acknowledge the support received from Kenya Medical Research Institute (KEMRI). This work is published with the permission of the director of KEMRI.
Grant information
Strategic Award B9ROUN0 (084538/Z/07/D) to SK, The German Academic Exchange Service (DAAD) fellowship to CM. MKR is supported by FLAIR fellowship (FLR\R1\190497).
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.actatropica.2019.05.024.
Author’s contribution
Conceived and designed the experiments POM, CM, MKR, performed the experiments CM, MKR, JN. Analyzed the data MKR, CM, JN. Drafting the manuscript MKR, CM, reviewed the manuscript POM, FK, RM, RO, SK, JM. All authors read approved the final version of the MS.
Competing interest
They have no competing interest.
References
- An C, Budd A, Kanost MR, Michel K. Characterization of a regulatory unit that controls melanization and affects longevity of mosquitoes. Cell Mol Life Sci. 2011;68:1929–1939. doi: 10.1007/s00018-010-0543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awolola TS, Oduola AO, Obansa JB, Chukwurar NJ, Unyimadu JP. Anopheles gambiae s.s. breeding in polluted water bodies in urban Lagos, southwestern Nigeria. J Vector Borne Dis. 2007;44:241–244. [PubMed] [Google Scholar]
- Bai B, Laiho M. Efficient sequential recovery of nucleolar macromolecular components. Proteomics. 2012;12:3044–3048. doi: 10.1002/pmic.201200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayoh MN, Mathias DK, Odiere MR, Mutuku FM, Kamau L, Gimnig JE, Vulule JM, Hawley WA, Hamel MJ, Walker ED. Anopheles gambiae: historical population decline associated with regional distribution of insecticide-treated bed nets in western Nyanza Province, Kenya. Malar J. 2010;9:62. doi: 10.1186/1475-2875-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Budiansky S. Creatures of our own making. Science. 2002;298:80–86. doi: 10.1126/science.298.5591.80. [DOI] [PubMed] [Google Scholar]
- Cancrini G, Magi M, Gabrielli S, Arispici M, Tolari F, Dell’Omodarme M, Prati MC. Natural vectors of dirofilariasis in rural and urban areas of the Tuscan region, central Italy. J Med Entomol. 2006;43:574–579. doi: 10.1603/0022-2585(2006)43[574:nvodir]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Charlwood JD, Kessy E, Yohannes K, Protopopoff N, Rowland M, LeClair C. Studies on the resting behaviour and host choice of Anopheles gambiae and An. arabiensis from Muleba, Tanzania. Med Vet Entomol. 2018;32:263–270. doi: 10.1111/mve.12299. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, in-dividualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G. Insect immunity and its implication in mosquito-malaria interactions. Cell Microbiol. 2003;5:3–14. doi: 10.1046/j.1462-5822.2003.00252.x. [DOI] [PubMed] [Google Scholar]
- Djouaka RF, Bakare AA, Bankole HS, Doannio JM, Coulibaly ON, Kossou H, Tamo M, Basene HI, Popoola OK, Akogbeto MC. Does the spillage of petroleum products in Anopheles breeding sites have an impact on the pyrethroid resistance? Malar J. 2007;6:159. doi: 10.1186/1475-2875-6-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, VectorBase C, Madey G, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Toe HK, Sanou A, Namountougou M, Hughes A, Diabate A, Dabire R, Simard F, Ranson H. Additional selection for insecticide resistance in urban malaria vectors: DDT resistance in Anopheles arabiensis from Bobo-Dioulasso, Burkina Faso. PLoS One. 2012;7:e45995. doi: 10.1371/journal.pone.0045995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Ali SA, Kumar S, Mohanty AK, Behare P. Label-free quantitative proteomic analysis of Lactobacillus fermentum NCDC 400 during bile salt exposure. J Proteomics. 2017;167:36–45. doi: 10.1016/j.jprot.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Killeen GF, Govella NJ, Lwetoijera DW, Okumu FO. Most outdoor malaria transmission by behaviourally-resistant Anopheles arabiensis is mediated by mosquitoes that have previously been inside houses. Malar J. 2016;15:225. doi: 10.1186/s12936-016-1280-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudom AA. Larval ecology of Anopheles coluzzii in Cape Coast, Ghana: water quality, nature of habitat and implication for larval control. Malar J. 2015;14:447. doi: 10.1186/s12936-015-0989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzaro GC, Toure YT, Carnahan J, Zheng L, Dolo G, Traore S, Petrarca V, Vernick KD, Taylor CE. Complexities in the genetic structure of Anopheles gambiae populations in west Africa as revealed by microsatellite DNA analysis. Proc Natl Acad Sci U S A. 1998;95:14260–14265. doi: 10.1073/pnas.95.24.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, Hauser MA, Scott WK, Small GW, Nance MA, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12:3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, et al. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40:D729–734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireji PO, Keating J, Kenya E, Mbogo C, Nyambaka H, Osir E, Githure J, Beier J. Differential induction of proteins in Anopheles gambiae sensu stricto (Diptera: cullicidae) larvae in response to heavy metal selection. Int J Trop Insect Sci. 2006;26:214–226. doi: 10.1017/S1742758406658955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireji PO, Keating J, Hassanali A, Mbogo CM, Nyambaka H, Kahindi S, Beier JC. Heavy metals in mosquito larval habitats in urban Kisumu and Malindi, Kenya, and their impact. Ecotoxicol Environ Saf. 2008;70:147–153. doi: 10.1016/j.ecoenv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireji PO, Keating J, Hassanali A, Mbogo CM, Muturi MN, Githure JI, Beier JC. Biological cost of tolerance to heavy metals in the mosquito Anopheles gambiae. Med Vet Entomol. 2010a;24:101–107. doi: 10.1111/j.1365-2915.2010.00863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireji PO, Keating J, Hassanali A, Impoinvil DE, Mbogo CM, Muturi MN, Nyambaka H, Kenya EU, Githure JI, Beier JC. Expression of metallothionein and alpha-tubulin in heavy metal-tolerant Anopheles gambiae sensu stricto (Diptera: Culicidae) Ecotoxicol Environ Saf. 2010b;73:46–50. doi: 10.1016/j.ecoenv.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Garcia M, Vargas V, Ramirez-Bello I, Hernandez-Martinez G, Lanz-Mendoza H. Bacterial exposure at the larval stage induced sexual immune dimorphism and priming in adult Aedes aegypti mosquitoes. PLoS One. 2015;10:e0133240. doi: 10.1371/journal.pone.0133240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M, Herrel N, Amerasinghe FP, Ensink J, van der Hoek W, Konradsen F. Role of wastewater irrigation in mosquito breeding in south Punjab, Pakistan. Southeast Asian J Trop Med Public Health. 2003;34:72–80. [PubMed] [Google Scholar]
- Nchoutpouen E, Talipouo A, Djiappi-Tchamen B, Djamouko-Djonkam L, Kopya E, Ngadjeu CS, Doumbe-Belisse P, Awono-Ambene P, Kekeunou S, Wondji CS, Antonio-Nkondjio C. Culex species diversity, susceptibility to insecticides and role as potential vector of Lymphatic filariasis in the city of Yaounde, Cameroon. PLoS Negl Trop Dis. 2019;13:e0007229. doi: 10.1371/journal.pntd.0007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njunge JM, Oyaro IN, Kibinge NK, Rono MK, Kariuki SM, Newton CR, Berkley JA, Gitau EN. Cerebrospinal fluid markers to distinguish bacterial meningitis from cerebral malaria in children. Wellcome Open Res. 2017;2:47. doi: 10.12688/wellcomeopenres.11958.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SV, Brooke BD. The role of oxidative stress in the longevity and insecticide resistance phenotype of the major malaria vectors Anopheles arabiensis and Anopheles funestus. PLoS One. 2016;11:e0151049. doi: 10.1371/journal.pone.0151049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SV, Brooke BD. The effect of metal pollution on the life history and insecticide resistance phenotype of the major malaria vector Anopheles arabiensis (Diptera: Culicidae) PLoS One. 2018;13:e0192551. doi: 10.1371/journal.pone.0192551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez DC, Gimenez MS. Induction of redox changes, inducible nitric oxide synthase and cyclooxygenase-2 by chronic cadmium exposure in mouse peritoneal macrophages. Toxicol Lett. 2003;145:121–132. doi: 10.1016/s0378-4274(03)00237-6. [DOI] [PubMed] [Google Scholar]
- Roberts DR, Manguin S, Rejmankova E, Andre R, Harbach RE, Vanzie E, Hakre S, Polanco J. Spatial distribution of adult Anopheles darlingi and Anopheles albimanus in relation to riparian habitats in Belize, Central America. J Vector Ecol. 2002;27:21–30. [PubMed] [Google Scholar]
- Rozendaal JA. Relations between Anopheles darlingi breeding habitats, rainfall, river level and malaria transmission rates in the rain forest of Suriname. Med Vet Entomol. 1992;6:16–22. doi: 10.1111/j.1365-2915.1992.tb00029.x. [DOI] [PubMed] [Google Scholar]
- Rydzanicz K, Lonc E, Becker N. Current procedures of the integrated urban vector-mosquito control as an example in Cotonou (Benin, West Africa) and Wroclaw area (Poland) Wiad Parazytol. 2009;55:335–340. [PubMed] [Google Scholar]
- Sattler MA, Mtasiwa D, Kiama M, Premji Z, Tanner M, Killeen GF, Lengeler C. Habitat characterization and spatial distribution of Anopheles sp. Mosquito larvae in Dar es Salaam (Tanzania) during an extended dry period. Malar J. 2005;4:4. doi: 10.1186/1475-2875-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitger AK, Kafatos FC, Osta MA. The melanization reaction is not required for survival of Anopheles gambiae mosquitoes after bacterial infections. J Biol Chem. 2007;282:21884–21888. doi: 10.1074/jbc.M701635200. [DOI] [PubMed] [Google Scholar]
- Sibomana I. Investigation into the Ecological Determinants of Distribution of Anopheles gambiae s.s. (Diptera: Culicidae) Larval Populations in Urban Accra, Ghana. University of Ghana; Legon: 2002. [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten MM, Tew IF, Lee BL, Ratcliffe NA. A novel role for an insect apolipoprotein (apolipophorin III) in beta-1,3-glucan pattern recognition and cellular encapsulation reactions. J Immunol. 2004;172:2177–2185. doi: 10.4049/jimmunol.172.4.2177. [DOI] [PubMed] [Google Scholar]
- Xu YT, Wang J, Yin R, Qiu MT, Xu L, Wang J, Xu L. Genetic poly-morphisms in Glutathione S-transferase Omega (GSTO) and cancer risk: a metaanalysis of 20 studies. Sci Rep. 2014;4 doi: 10.1038/srep06578. 6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas GPFSA, Pappas E, Zoidis K, Fegeros K. Cadmium Toxicity and the Antioxidant System. Nova Science; New York: 2010. [Google Scholar]
- Zhu L, Muller GC, Marshall JM, Arheart KL, Qualls WA, Hlaing WM, Schlein Y, Traore SF, Doumbia S, Beier JC. Is outdoor vector control needed for malaria elimination? An individual-based modelling study. Malar J. 2017;16:266. doi: 10.1186/s12936-017-1920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]