Summary
Many microbial pathogens, including the malaria parasite Plasmodium falciparum, vary surface protein expression to evade host immune responses. P. falciparium antigenic variation is linked to var gene family-encoded clonally variant surface protein expression. Mututally exclusive var gene expression is partially controlled by spatial positioning; silent genes are retained at distinct perinuclear sites and relocated to transcriptionally active locations for monoallelic expression. We show that var introns can control this process and that var intron addition relocalizes episomes from a random to a perinuclear position. This var intron-regulated nuclear tethering and repositioning is linked to an 18 bp nuclear protein-binding element that recruits an actin protein complex. Pharmacologically induced F-actin formation, which is restricted to the nuclear periphery, repositions intron-carrying episomes and var genes and disrupts mutually exclusive var gene expression. Thus, actin polymerization relocates var genes from a repressive to an active perinuclear compartment, which is crucial for P. falciparium phenotypic variation and pathogenesis.
Introduction
Antigenic variation is a major survival strategy applied by the human malaria parasite Plasmodium falciparum to avoid destruction by the host’s immune system (reviewed in Scherf et al., 2008). This variation is mediated by the differential control of a family of surface adhesion molecules termed PfEMP1, which are encoded by ~60 var genes (Baruch et al., 1995; Smith et al., 1995; Su et al., 1995). The activation of var genes occurs in situ with no programmed DNA rearrangements, indicating that the underlying mechanism of mutually exclusive expression is mainly at the level of epigenetic control (Scherf et al., 1998).
Complete P. falciparum genome sequence analysis localized var gene members either to highly polymorphic chromosome ends or to central chromosome regions (Gardner et al., 2002). High rates of recombination in var genes, including gene conversion events, have been demonstrated (Freitas-Junior et al., 2000) and may account for the huge var gene repertoire diversity observed in clinical isolates (Barry et al., 2007). This chromosome region-specific genetic diversification process is probably promoted by the particular spatial organization of subtelomeres into perinuclear foci (four to seven) (Freitas-Junior et al., 2000). Surprisingly, this applies also to internal chromosome var genes, which also loop back to the perinuclear space (Lopez-Rubio et al., 2009; Ralph et al., 2005) by an as-yet-unknown mechanism.
In P. falciparum the spatial organization of chromosomes is also central to the expression of virulence gene families involved in immune evasion and pathogenesis (Scherf et al., 2008). The location at the nuclear periphery of var genes apparently is a precondition for their default silencing. This transcriptionally inactive state correlates to the presence of molecular markers for facultative heterochromatin (fHC) such as histone 3 lysine trimethylation (H3K9me3) (Chookajorn et al., 2007; Lopez-Rubio et al., 2007, 2009) and the recruitment of heterochromatin protein 1 (PfHP1) (Flueck et al., 2009; Perez-Toledo et al., 2009). Importantly, the enzymes involved in gene silencing, such as the H3K9 deacetylase (PfSir2) and H3K9 methyl transferase (PfKMT), are both recruited to the nuclear periphery and bind to subtelomeric chromatin (Freitas-Junior et al., 2005; Lopez-Rubio et al., 2009; Mancio-Silva et al., 2008). In P. falciparum, fHC is apparently restricted to genomic regions that are tethered to the nuclear periphery (Lopez-Rubio et al., 2009) and is virtually limited to clonally variant gene families involved in phenotypic variation and pathogenesis (Lopez-Rubio et al., 2009; Salcedo-Amaya et al., 2009). Activation of a single member of the var repertoire is linked to its relocation into a transcriptionally competent area (Duraisingh et al., 2005; Dzikowski et al., 2007; Ralph et al., 2005; Voss et al., 2006).
This expression site is still in the nuclear periphery but is distinct from telomere clusters, as shown in a recent RNA-DNA FISH analysis (Lopez-Rubio et al., 2009).
Valuable insight into the molecular process of antigenic variation came from two recent studies that demonstrated a key role of two genetic var elements, the var upstream sequence (ups) and the var intron, in mutually exclusive expression (Dzikowski et al., 2006; Voss et al., 2006). Importantly, var antigen production is dispensable in this process. Based on current data, taken mostly from reporter gene assays, silencing and monoallelic exclusion are regulated by two regions, one upstream of the coding region (5′ups) and the intron, which separates the two exons (Calderwood et al., 2003; Chookajorn et al., 2007; Deitsch et al., 2001). A molecular understanding of the role of the var intron in antigenic variation, however, remains elusive.
To address the molecular mechanism of the perinuclear anchoring and relocation-linked activation of var genes, we investigated the nuclear spatial location of an episome in the presence of different var gene DNA elements. We discovered that introns mediate episomal anchoring to the nuclear periphery. A nuclear actin-protein complex was identified that binds specifically to a single intron element of 18 bp. Actin-perturbing drugs demonstrate a role for perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of var genes. We provide important insight into the regulation of the major virulence gene family contributing to pathogenesis in malaria parasites through a previously unexplored mechanism.
Results
Var Introns Direct Episomes to the Nuclear Periphery
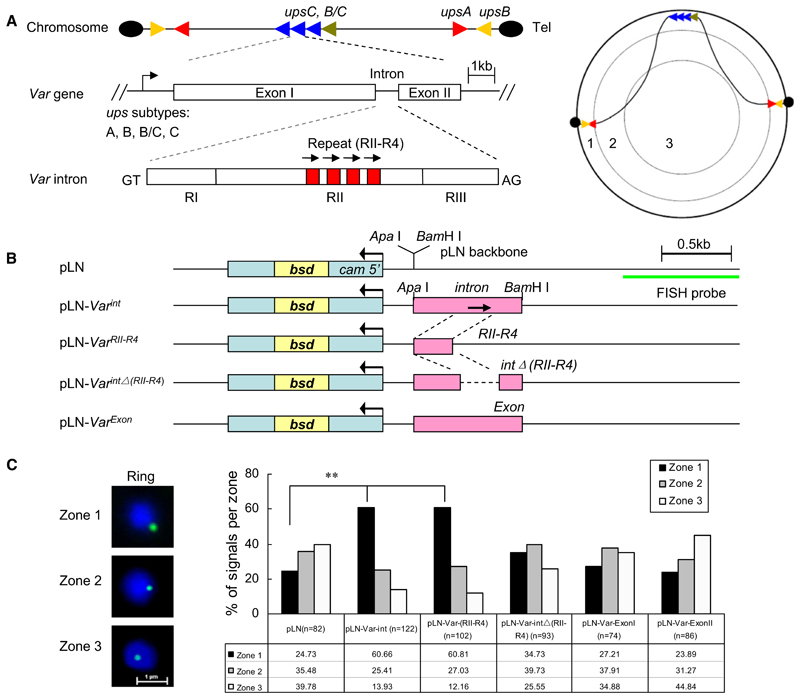
Given the apparent role of var gene positioning to the perinuclear space for monoallelic expression, we aimed to identify the genetic elements in the gene tethering process. We explored the capacity of the most conserved DNA region of var genes, the intron (Su et al., 1995), to alter the virtually random episomal location in transfected P. falciparum. Common structural and organizational features between members of the var gene family and their chromosomal spatial organization are shown in Figure 1A. We chose to clone the intron from a chromosome internal upsC var gene (PFD1000c) into the plasmid pLN (Nkrumah et al., 2006) containing a selectable marker gene blasticidin deaminase (bsd) (Figure 1B and see Figures S1A and S1B and Table S1 available online). Constructs were transfected into 3D7 strain, and all episomes were recovered after the transfection experiment to validate the integrity of the cloned inserts. To remove any possible influence of drug pressure on episome localization, the cultures were maintained in a drug-free medium for 2 days before 2D-FISH analysis on tightly synchronized ring stage parasites. Var introns shift episomes from a random (pLN episome) to a perinuclear position (pLN-Var int) (p < 0.001), as shown in Figure 1C. A pLN construct (pLN-Var RII-R4) with a 200 bp sequence of intron RII, which contains four repetitions of 25 bp separated by short AT-rich sequences (referred to as the RII-R4 domain, Figure S1B), shifted episomes in a manner similar to that of the full-length intron. Deleting this region from the intron virtually reversed the spatial preference of the episome (pLN-Var intΔ[RII-R4]) close to values observed for the intronless construct. Two DNA regions corresponding to coding regions of var exon 1 and 2 (pLN-Var Exon) did not change the spatial location of pLN. To exclude the potential localization effect of the pLN backbone sequence, we repeated the experiments with another vector, pARL, which contains a different selectable marker gene (human dhfr gene) (Crabb et al., 2004). FISH assay of parasites episomally transfected with pARL-Var int, pARL-Var intΔ(RII-R4), or pARL vector revealed pattern similar to that observed in pLN transfectans (Figures S1A-S1D). Moreover, another intron of upsC var gene (PF07_0048) containing a single copy of this repetition was also tested in the same strategy, and exhibited a similar result (Figures S1E–S1H). These data suggest that the var intron central repeat region is the major effector in this perinuclear tethering process. This region was previously noted to be critical for the var intron silencing and monoallelic expression of a drug-selectable marker under var promoter control (Calderwood et al., 2003; Gannoun-Zaki et al., 2005).
Figure 1. Var Introns Target Episomes to the Nuclear Periphery.
(A) Genomic organization, structure, and nuclear position of var genes. (Left panel) upsB or upsA corresponds to subtelomeric and upsC corresponds to chromosome internal var gene members. Each subtype var gene is labeled with a different color. Var introns contain up to four 18 bp repetitions in its region II (RII) domain. (Right panel) Schematic illustration of subnuclear localization of various subtype var genes on different chromosome sites. The area of the nuclear section was divided into three concentric zones with equal surfaces, and nuclear periphery corresponds to the zone 1.
(B) pLN vector constructs with 3D7 upsC-type full-length intron (PFD1000c), the intron repeat region (RII-R4), the intron without the RII-R4 region, two distinct regions from var exon I and exon II, and the vector backbone control. The drug-selectable marker gene (bsd) and the region used as FISH probe are shown, respectively.
(C) FISH signals of episomes (green) were localized into zones 1, 2, or 3 with equal surface (see A, right panel). The bar graph represents the position of FISH signals with respect to three concentric zones. DAPI (blue), nuclear DNA. n, number of nuclei analyzed from two independent ring stage samples. **p < 0.001 (χ2 test).
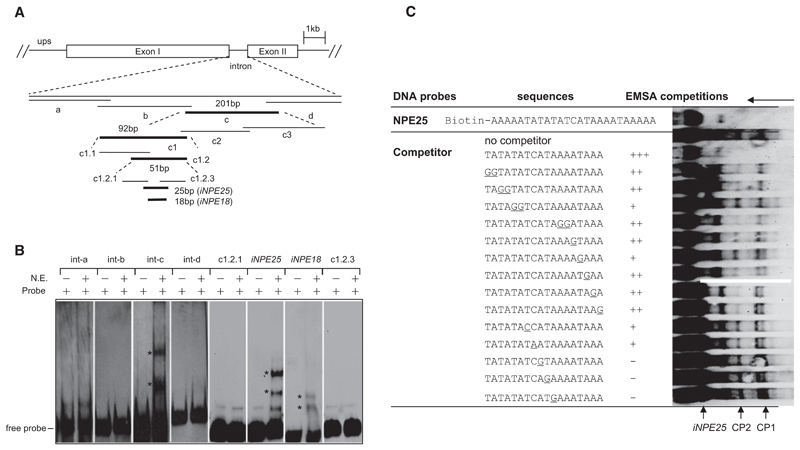
To explore this finding, we performed gel electrophoretic mobility shift assays (EMSA) with 20 bp overlapping intron sequences (int-a, -b, -c, and -d). upsC var introns are highly conserved except for variable numbers of central repetitions. We chose the single repetition containing var intron (PF07_0048) for our EMSA assays (Figure 2A, Figure S1F, and Table S3). Two shift bands (CP1 and CP2) were observed with the int-c probe and nuclear extracts of asynchronous blood stage parasites (Figure 2B). After several rounds of EMSA screening, we identified a 25 bp fragment that recruits similar nuclear protein complexes as the full intron. A shorter 18 bp version of this sequence produced similar shifted bands in EMSA, but with lower intensity (Figures 2A and 2B). We named this intron sequence the intron nuclear protein-binding element (iNPE). Importantly, the central var intron RII-R4 region that shifts the episome location to the nuclear periphery comprised the iNPE motif, which corresponds to one of the four repeat elements.
Figure 2. Identification of a Nuclear Protein-Binding Element in var Introns.
(A) EMSA screening strategy using biotin-labeled overlapping DNA fragments (bold sequences bound to nuclear proteins). The var intron (PF07_0048) containing a single repeat (RII-R1) was analyzed here. The sequences of the full-length intron and each probe are shown in Figure S1F and Table S4.
(B) A single intron region (iNPE25 or iNPE18) binds to nuclear proteins. The two iNPE-binding complexes (CP1 and CP2) were labeled with asterisks, respectively.
(C) Competition EMSA of biotin-labeled iNPE25 motif with various mutated nonlabeled iNPE18 sequences. Base pair exchanges in the mutated iNPE18 oligonucleotides are shown underlined. Competitor was used at a concentration of 100-fold excess of the labeled iNPE25 probe in each competition reaction. The decrease in intensity of CP1/CP2 bands was defined as strong (+++), medium (++), weak (+), and none (–), respectively, as shown in the table. Free probe, shifted bands CP1 and CP2, and running orientation of gel were labeled by arrows.
The specificity of the protein-DNA binding activities was verified by a competition assay with various DNA competitors (Figure S2A and Table S3). To identify nucleotides involved in the interaction between iNPE motif and nuclear proteins, we mutated individual nucleotides of iNPE18 to investigate their contribution to the specific interactions in competition gel shift assays. Figure 2C demonstrated that most mutations along the 18 bp motif reduce to different degrees the binding of CP1 and CP2. In a differnt EMSA experiment, larger blocks (five to seven bases) were mutated in the 18 bp motif (Figure S2B). Complete loss of the CP1 and CP2 complexes was observed for mutations in the 5′ and 3′ motif region (mut 1 and mut 3), whereas the mutations in the middle part (mut 2) significantly reduced binding. Taken together, our results sugggest that the entire 18 bp motif is important for the DNA protein binding complex.
Next we analyzed the distribution of the iNPE18 motif in the 3D7 genome (PlasmoDB) by sequence blast. Identical iNPE18 motifs were only observed in introns of the var gene family, including a few truncated pseudo vars. A total of 76 copies of iNPE18 with 100% identity were found in 39 of 60 var introns with variable numbers of (TA) repeats (three to twelve). One to four iNPE motifs were found in upsB, BC, and C type vars (Figure S2C). The average copy number of the intron motif increased from the upsB to upsC subgroups of var genes, and all iNPE motifs were exclusively distributed within the central intron region (Figure S2D). The iNPE element is absent in a small subset of introns belonging to the upsA type var genes. We searched the genomes of other malaria species and eukaryotic organisms including yeast, mouse, and human for this iNPE intron motif. In malaria parasites, the presence of multiple copies of this motif was restricted to var gene introns of P. falciparum (and the closely related P. reichenowi; see Table S2) and was virtually absent from other malaria species. In addition, several iNPE motifs were found in the genomes of higher eukaryotes (mainly in intergenic regions).
Actin Is a Component in the var Intron iNPE Motif-Protein Binding Complex
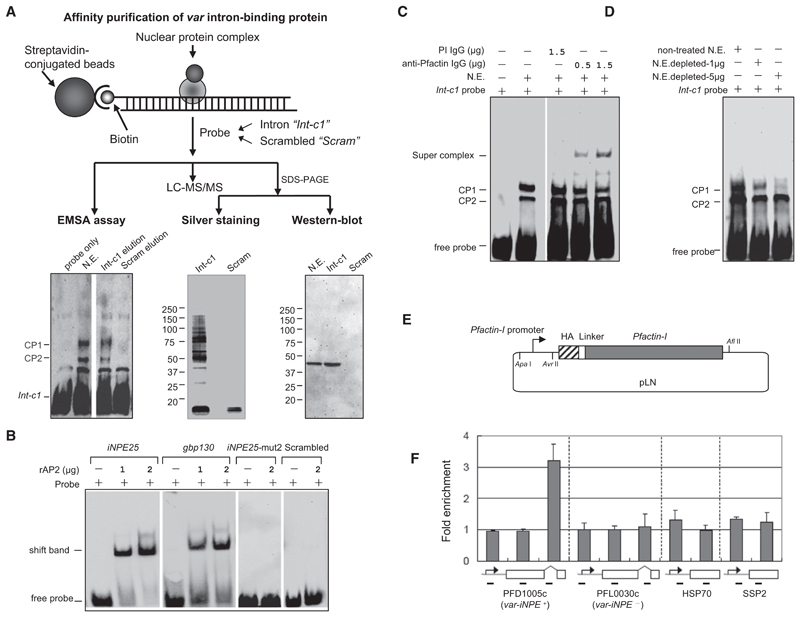
Affinity purification was performed to identify nuclear factors that bind to var introns. Before affinity purification, we tested the disassociation dynamics of the intron/protein binding activity under various KCl salt concentrations. Binding activities were completely disrupted by the addition of KCl at a final concentration of 800 mM (data not shown). Next, the biotinylated int-c1 DNA fragment was bound to streptavidin-coated beads and incubated with nuclear extracts from asynchronous parasites (Figure 3A, upper panel). As control, all purification steps were performed in parallel with a scrambled DNA probe with AT content comparable to that of int-c1 (see Table S3). Bound proteins were recovered with elution buffer containing 1 M KCl, and samples were analyzed by EMSA. Eluted fractions from int-c1 contained nuclear proteins able of forming a gel shift with int-c1 similar to the crude nuclear extracts (Figure 3A, lower panel, left). Equal volumes of purified fractions from int-c1 and control were analyzed by SDS-PAGE followed by silver stain. Int-c1 revealed a complex mix of proteins in the range of 10 to 150 KDa, whereas the control probe showed only a small protein complex in the range of 10 KDa (Figure 3A, lower panel, middle).
Figure 3. Actin Is a Component of the iNPE Protein Complex.
(A) (Upper panel) Schematic procedure of affinity purification steps. Equal amount of preadsorbed nuclear extract were incubated with int-c1 or scrambled control DNA-coupled beads. Bound components were eluted in equal volume. (Lower panels) Equal volumes of the two eluted fractions were analyzed by EMSA with crude nuclear extract (N.E.) as positive control (left). Another aliquot was analyzed by 4%–12% SDS-PAGE/silver stain (middle) and western blot with rabbit antibody against Pfactin-I (right). A third aliquot was used for LC-MS/MS analysis.
(B) Gel shift assay of recombinant AP2 domain (PF11_0091) with iNPE25, gbp130, iNPE25-mut2 (see Figure S2B), and scrambled probe, respectively.
(C) Super gel shift assay with int-c1 probe and rabbit anti-Pfactin-I antibody (affinity purified IgG) and preimmune (PI) rabbit IgG as a negative control. The two shifted bands CP1/CP2 and anti-actin antibody-induced supershift band are indicated.
(D) EMSA assay with int-c1 probe and nuclear extract depleted in actin (N.E. depleted) by incubation with 1 or 5 μg mouse anti-Pfactin-I antibody, respectively. The untreated crude nuclear extract (N.E.) was used as control.
(E) Schematic representation of the HA-tagged Pfactin-I episomally transfected into 3D7 strain. A detailed description of the construction of this vector is provided in the Supplemental Experimental Procedures.
(F) ChIP assay of HA-actin transfectant using monoclonal mouse anti-HA antibody. The enrichment folds of various gene loci were shown respectively. Regions used for PCR amplification are shown under the schematic gene structure. Data are represented as mean ± SEM of two independent experiments.
To obtain the sequence information, eluted proteins were digested with trypsin, and the resulting peptide mixtures were subjected to LC-MS/MS analysis using an Orbitrap velos. Bio-informatic analysis of the affinity purified nuclear proteins identified by mass spectrometry (for raw data see Table S4 and Table S6) showed the enrichment of 12 putative DNA/RNA-binding proteins not detected with scrambled control DNA probe (see Table 1). Similar sets of candidates were identified in two independent experiments. Among those we identified four Alba domain proteins. However, one Alba (PF10_0063) was present in both int-c1 and scrambled samples. Unpublished data from the Scherf laboratory showed that all four Albas bind to DNA in a non-sequence-specific manner (data not shown), suggesting that Albas do not bind specifically to var intron sequences. The only candidate with a demonstrated sequence-specific DNA-binding domain is a member of the P. falciparum AP2 protein family. We produced the recombinant AP2 domain (1486–1548 aa) of PF11_0091 in E.coli as previously published (Campbell et al., 2010). In gel shift assays this protein did bind to the iNPE sequence. The signal obtained was comparable to the gbp130 sequence motif that contained the binding motif identified by Llinas’s group (Figure 3C). No binding was observed with the TCATA box-mutated iNPE probe or scrambled sequence probe used in the affinity purification experiment. While the consensus binding sequence identified by the Llinas’s laboratory was AGCATAC, the putative binding site ATCATA in the iNPE motif is slightly different, indicating that this AP2 protein can also bind efficiently to related sequence motifs. Based on the results from Figure 2C and Figure S2B it is conceivable that the intron-binding AP2 protein needs flanking sequences in the iNPE motif for optimal DNA affinity.
Table 1. Putative iNPE-Binding Protein Candidates Identified by LC-MS/MS.
| PlasmoDB Accession | Annotation | Protein Probability (%) | MW (kDa) | Unique Peptides | Sequence Coverage (%) | Experiment 1 | Experiment 2 |
|---|---|---|---|---|---|---|---|
| PF11_0091a | Transcription factor with AP2 domain(s), putative | 100 | 206.9 | 3 | 1.48 | + | + |
| MAL13P1.237 | Nucleic acid binding protein Alba, putative | 100 | 42.2 | 7 | 25 | + | + |
| PF08_0074 | DNA/RNA-binding protein Alba, putative | 100 | 27.2 | 6 | 29 | + | + |
| MAL13P1.233 | Nucleic acid binding protein Alba, putative | 100 | 25.0 | 4 | 22 | + | + |
| MAL8P1.40 | RNA binding protein, putative | 100 | 32.4 | 8 | 30 | - | + |
| PFI1025w | RNA binding protein, putative | 100 | 56.6 | 3 | 11 | - | + |
| PF10_0068 | RNA binding protein, putative | 100 | 29.5 | 4 | 29 | + | + |
| PF14_0096 | RNA binding protein, putative | 100 | 59.3 | 3 | 7 | - | + |
| PFI1435w | RNA binding protein, putative | 99.8 | 33.6 | 3 | 10 | + | + |
| PFE0435c | DNA binding protein, putative | 99.8 | 33.8 | 3 | 13 | + | + |
| PFL1170w | Polyadenylate-binding protein, putative | 100 | 97.2 | 14 | 20 | + | + |
| PFI0235w | Replication protein A | 100 | 56.2 | 16 | 40 | + | + |
| PFL2215wb | Pfactin-I | 100 | 41.9 | 8 | 52 | + | + |
Accession numbers (http://www.plasmodb.org/) of putative DNA/RNA binding proteins exclusively identified in the int-c1 elution.
Pfactin-I has been validated by ChIP as a member of the iNPE-binding protein complex.
An unexpected protein, P. falciparum actin-I gene (Pfactin-I; PlasmoDB accession number PFL2215w) was exclusively present in the int-c1 elution, showing a sequence coverage of ~52%. The P. falciparum genome encodes two actin genes: one is expressed in all life cycle stages (actin I), and the second one is expressed almost exlusively in sexual stages (actin II). Equal volumes of purified fractions from int-c1 and control probes were analyzed for actin enrichment. Western blot analysis showed the enrichment of actin in the int-c1 purified protein but no detectable band in the control purification (see Figure 3A, lower panel, right). This unexpected actin-intron interaction was further validated by supershift EMSA and ChIP experiments using antibodies raised against Pfactin-I (see schematic in Figure S3A) or HA-tagged actin (Figure S3D, upper panel). Actin antibodies resulted in a specific supershift band that had formed with the preincubation of nuclear extracts with anti-Pfactin-I antibodies. The CP1 complex showed a decreased intensity in the supershift reaction, while that of the CP2 band remained at a similar intensity (Figure 3C). Moreover, the intensity of the CP1 complex dramatically decreased with nuclear extracts depleted in actin, indicating that actin was one of the essential components of this complex (Figure 3D).
To investigate the actin/iNPE interaction in vivo, we performed chromatin immunoprecipitation (ChIP) using transfected 3D7 parasites expressing HA-tagged actin (Figure 3E and Figure S3D). ChIP experiments with ChIP grade anti-HA antibody demonstrated a >3-fold enrichment of actin on var introns (only with iNPE motif) compared to regions corresponding to coding or 5′ flanking sequences of var and two control genes (Figure 3F). Taken together, these data validate Pfactin-I as a prominent member of a protein complex (CP1) that binds to the var intron iNPE region. Our attempts to obtain a gel shift with purified bacterial recombinant full-length actin and int-c1 fragments failed (data not shown), indicating that a specific DNA-binding protein recruits actin.
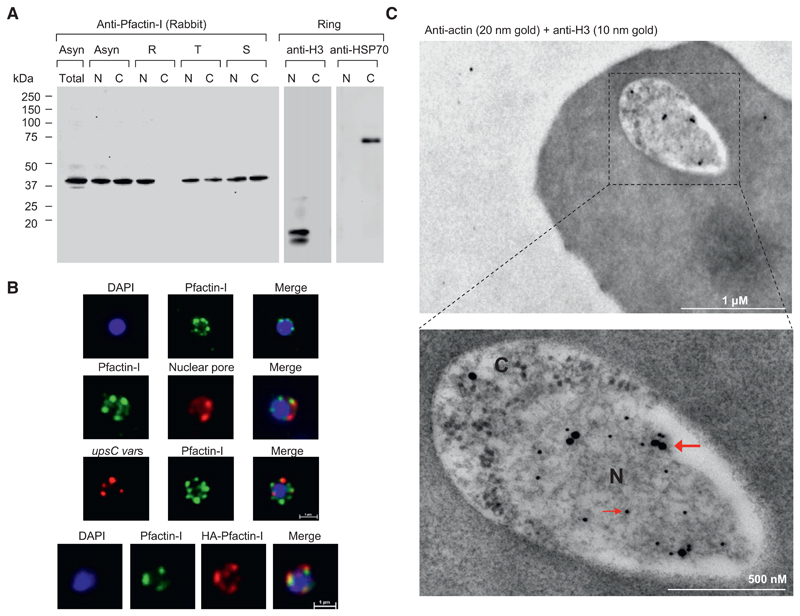
Our results point to the perinuclear enrichment of actin, a notion that had not been reported previously for P. falciparum and other organisms. We produced a panel of mouse and affinity purifed rabbit antibodies against Pfactin-I (see schematic in Figure S3A). All antibodies recognized a 42 kDa band by western blot (the predicted molecular weight for actin) in asexual blood stages (see Figure 4A and Figures S3B–S3D). The analysis of nuclear and cytoplasmic blood stage extracts revealed that actin is highly enriched in the nucleus in ring stage parasites (Figure 4A and Figure S3C). Control antibodies for nuclear extracts (anti-histone H3) and cytoplasmic extracts (anti-PfHSP70) confirmed the quality of the fractionation. IFA analysis with anti-Pfactin-I antibodies or with anti-HA antibody in HA-actin-transfected 3D7 parasites confirmed the restricted location of actin to the nucleus in ring stage parasites and demonstrated that it concentrated in foci at the nuclear periphery (Figure 4B and Figure S3E). Colocalization with a nuclear pore antibody (Lopez-Rubio et al., 2009) indicates that the staining is limited to the inner side of the nucleus (Figure 4B, second panel). This was further confirmed by performing immunoelectron microscopy on ring stages stained with Pfactin-I antibodies. Gold grains are virtually confined to the nuclear periphery (Figure 4C and Figure S4A). Since ring stage nuclear membranes are difficult to visualize, we used anti-histone H3 antibodies to delimit the nucleoplasm. Intranuclear actin was detected by immuno-EM during the entire blood stage cycle in trophozoite and schizont stage parasites (Figures S4B and S4C).
Figure 4. Pfactin-I Concentrates at the Nuclear Periphery and Colocalizes with var Genes.
(A) Western blot assay of total protein extract, nuclear and cytoplasmic fractions of asynchronous or synchronous parasites at different intraerythrocytic stages with rabbit (#2) antibodies against Pfactin-I. The amount of nuclear and cytoplasmic fractions of a given stage corresponded to equal parasite numbers. Ring stage fractions were monitored using anti-histone H3 and anti-PfHSP70 antibodies. N, nuclear extract; C, cytoplasmic extract.
(B) (Upper panel) IFA visualization of subcellular actin in rings using an anti-Pfactin-I antibody (rabbit #2). (Second panel) Two-color IFA assay using anti-Pfactin-I (rabbit #2, green) and anti-nuclear pore (PF14_0706) antibody (rat, red). (Third panel) Combined FISH/IFA assay in rings showing partial colocalization of chromosome internal var genes (upsC-type) and actin. (Fourth panel) IFA analysis of HA-actin-transfected ring stage parasites using mouse anti-HA antibody (red) and anti-Pfactin-I antibody (rabbit #2, green).
(C) Immunoelectron microscopy of 3D7 rings. Double labeling with mouse anti-Pfactin-I (20 nm gold grain, bold arrow) and rabbit anti-histone H3 (10 nm gold grain, slim arrow). N, nucleus; C, cytoplasm.
A DNA FISH probe that hybridizes with upsC-type var genes colocalized with the actin signal (Figure 4B, third panel), supporting our EMSA data that actin is recruited to var introns in the nuclear periphery.
Nuclear Actin Polymerization Leads to the Spatial Repositioning and Derepression of var Gene Members
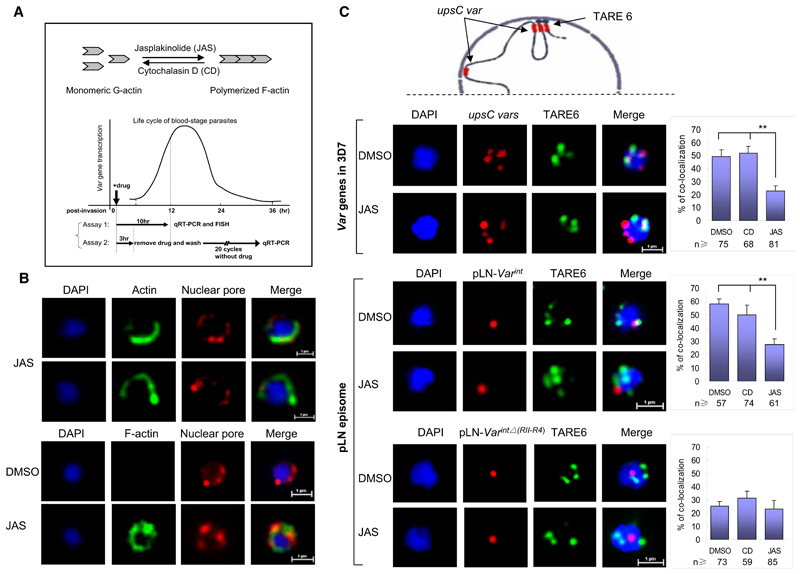
We used the actin-perturbing agents jasplakinolide (JAS) and cytochalasin D (CD) to investigate the role of nuclear actin in the regulation of var gene expression. CD promotes the dispersion of existing actin filaments, while JAS stabilizes actin filaments (see the schematic in Figure 5A). To explore the potential killing effect of CD and JAS on cultured parasites at 10 mM concentrations, we treated very early ring stage parasites (0–5 hr postinvasion) with drugs for 10 hr. Drug-treated parasites were washed twice in RPMI 1640 and put back into culture. Parasitemia was monitored by Giemsa staining until the second generation (Figure S5A). We observed that in the first blood stage cycle, almost all the parasites were able to develop into late-stage schizonts. We did not see a difference in the growth rate or morphology when compared to DMSO-treated parasites. However, the parasitemia was significantly lower compared to the control group during the second cycle, indicating that residual drug concentrations may interfere with merozoite invasion. Next, we investigated the nuclear distribution of actin in the presence of different actin inhibitors. Anti-Pfactin-I antibodies revealed a modification from a punctuate pattern in non- or DMSO-treated cells to a more horseshoe-like perinuclear staining in JAS-treated samples (Figure 5B, upper panel), whereas the CD treatment induced a spread of the actin signal toward nuclear interior regions (Figure S5B). In order to confirm that F-actin is formed, we stained the parasite with fluorescent-labeled phalloidin. In DMSO- or CD-treated parasites, we did not detect any IF staining, whereas JAS-treated parasites showed a perinuclear IF staining (Figure 5B, lower panel), similar to one observed with anti-Pfactin-I antibodies. In P. falciparum, it has been reported that polymerized actin forms extremely short filaments that cannot be visualized, and its actin dynamics differ fundamentally from the turnover of actin in mammalian cells (Schmitz et al., 2005). Our results are compatible with G-actin being the main component in the perinuclear space. Cytoplasmic actin was detected with our antibodies in mature blood stages (Figures S3C, S3E, S4B, and S4C) and is consistent with its role in hemoglobin uptake (Lazarus et al., 2008) and as a molecular motor in parasite invasion of erythrocytes (Baum et al., 2008; Schuler and Matuschewski, 2006a).
Figure 5. JAS Induces Spatial Repositioning of var int Episomes and var Loci.
(A) Schematic illustration of the opposite effect of actin inhibitors (CD and JAS) on dynamic equilibrium between G-actin and F-actin. (Lower section) var gene transcription profile during the 48 hr blood stage cycle and the protocol of two distinct assays.
(B) (Upper panel) IFA of nuclear actin after JAS treatment using an anti-Pfactin-I antibody (rabbit #2, green) and anti-nuclear pore antibody (rat, red). (Lower panel) IFA visualization of F-actin using fluorescence-labeled phalloidin (green). DMSO- or CD-treated parasites were used as control (see also Figure S5B).
(C) (Upper panel) Schematic of the perinuclear organization of subtelomeric and chromosome central var genes. (Middle and lower panels) Two-color FISH analysis of 3D7, pLN-Var int, and pLN-Var intΔ(RII-R4) transfectants treated with DMSO, CD, or JAS, respectively (assay 1). The upsC type var exon II and episomal probe were labeled with Alexa 568 (red); the subtelomeric DNA TARE6 probe was labeled with Alexa 488 (green). FISH signals that colocalize with TARE6 are shown in the right panels. n, counted number of nuclei from two independent samples. Data are represented as mean ± SEM of two independent experiments. **p < 0.001 (χ2 test).
See also Figure S5.
The monoallelic activation process has been linked to var locus repositioning at the perinuclear space. In a previous study (Lopez-Rubio et al., 2009), we had shown that var genes from central chromosome regions form several perinuclear foci (three to four foci per cell) and that about 50% of them are adjacent perinuclear telomere foci (see schematic of perinuclear var gene organization in Figure 5C, upper panel). To investigate JAS-induced changes in the spatial organization of var genes, we performed two-color FISH assays to visualize telomere foci (TARE6 probe) and chromosome central upsC-type var genes (Exon IIc probe) in ring stage parasites treated with DMSO, CD, and JAS for ~10 hr. Colocalization of telomere cluster and internal chromosomal var foci was considerably reduced in JAS-treated parasites (23%) (p < 0.001), whereas DMSO (49%) and CD (56%) incubation did not produce significant differences compared to previous results (Figure 5C, second panel). Regardless of the drug treatment, TARE6 and upsC var loci remained at the nuclear periphery (Figure S5C).
These data raise the possibility that introns may be directly involved in the actin-dependant spatial relocation process. To test this idea, we investigated if the pLN episome that carries a var intron (pLN-Var int) does change its spatial organization in JAS-treated parasites. By using the same experimental setup described in Figure 5C (second panel), we observed that the association of pLN-Var int with telomere cluster was drastically decreased from 58% to 28% colocalization when the transfectant parasites were treated with JAS. The episome carrying the intron which has deleted the iNPE region (pLN-Var intΔ(RII-R4)) showed the same low association with telomere cluster of around 23%–25% (see Figure 5C, middle and lower panels). In both transfectants, CD treatment did not show any significant effect on the colocalization of episomes and telomere clusters. These results link iNPE directly to JAS-induced repositioning of var genes.
Since JAS modulates perinuclear location of var genes, we investigated the influence of this drug on monoallelic expression. To this end, the var gene transcription profile was measured in the early ring stage, which coincides with the beginning of var gene transcription. We used 10 μM final drug concentrations and two distinct experimental setups. Assay 1 aimed to address the immediate effect of both drugs on var gene transcription and spatial organization after the incubation of ring stage parasites for 10 hr. Assay 2 aimed to investigate the role of actin perturbation in var gene switching. To this end, parasites were incubated for 3 hr during the ring stage, washed with medium to remove the drug, and cultured for 20 blood stage cycles. To study the effect of JAS on var gene regulation, we used a recently cloned 3D7 line (3D7G7) that expresses virtually only a single var gene (PFD0625c) at the ring and trophozoite stages (data not shown). Changes in the var gene transcription profile were measured by qRT-PCR using 3D7 var-specific primers. Assays 1 and 2 are schematically shown in Figure 5A.
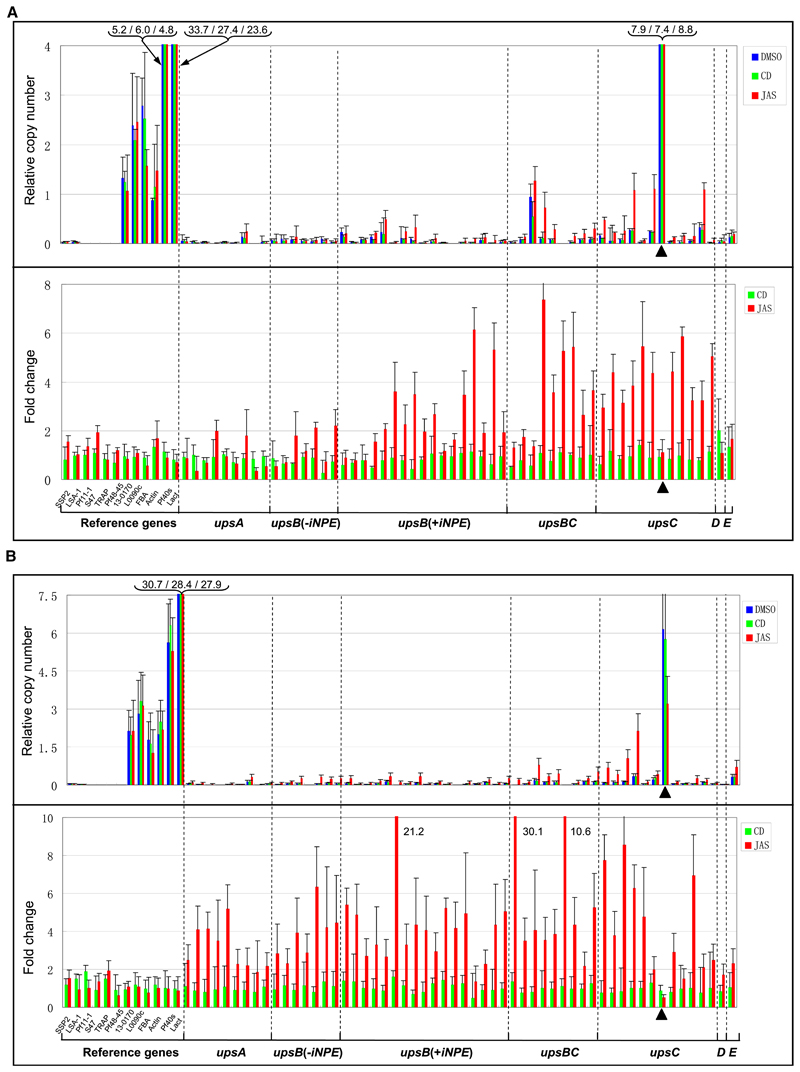
To begin, we tested the potential effect of drug treatment (JAS and CD) on RNA polymerase II-dependent gene transcription in P. falciparum. No significant changes in transcription levels (relative copy number and fold change) were observed for 12 reference genes (Figure 6A). FISH 2D location analysis showed that the majority of these genes show a random spatial location (Table S5). Six of the reference genes are expressed during asexual blood stage, and six are silent during this developmental stage. After that, we measured the drug effects on individual var genes (Salanti et al., 2003). Most silent var genes with an iNPE motif showed transcriptional upregulation >2-fold in the presence of JAS (25 out of 38), and eight of them were over 5-fold upregulated. CD did not induce significant transcriptional changes in var genes and, importantly, had no effect on the var gene dominantly expressed before adding the drugs (PFD0625c, indicated in Figure 6A). These data support a perinuclear scenario, in which filamentous actin (F-actin) supports var gene activation by moving genes into transcriptional competent regions.
Figure 6. JAS Treatment Interferes with Mutually Exclusive var Gene Transcription.
(A) qRT-PCR analysis of the transcription level of individual var genes of 3D7 (G7 clone) after actin inhibitor treatment (Figure 5A, assay 1). The data are shown as relative copy numbers for all the three samples using the seryl-tRNA synthetase gene (PF07_0073) as endogenous control (upper panel) and the fold change of transcription level with respect to DMSO-treated sample (lower panel). Reference genes are the following: 1–6, transcriptionally silent in asexual blood stage; 7–12, transcriptionally active during asexual blood stage (see Tables S3 and S5). The dominantly expressed var gene before drug treatment (PFD0625c) in G7 clone is indicated with triangle.
(B) qRT-PCR analysis of transcription levels of individual var genes of 3D7 (G7 clone) treated with actin inhibitors for 3 hr and cultivated for 20 cycles in the absence of drugs (Figure 5A, assay 2). The data are treated and represented as in (A). Data of assays 1 and 2 are represented as mean ± SEM of two independent experiments.
Parasites treated for only 3 hr with the drugs (see Assay 2 in Figure 5A) were cultured in the absence of drugs for 20 life cycles. The total RNAs of these cultures were extracted to analyze the transcription profile of individual var genes and 12 reference genes as done in Assay 1. No significant change in the var transcription pattern was observed between the DMSO- and CD-treated cultures. JAS treatment induced the most significant upregulation in iNPE containing upsB, upsBC, and upsC subtypes. Meanwhile, a weak upregulation was also observed in a few iNPE-deficient upsA and upsB var genes (Figure 6B). Importantly, a significant downregulation of the dominantly transcribed var gene (PFD0625c) was observed (p < 0.05), indicating that the initial JAS treatment for 3 hr has resulted in var gene deregulation and subsequent expression of a heterogeneous population with regard to var gene transcription. None of the 12 reference genes showed any transcriptional up- or downregulation in JAS- and CD-treated parasites. These data further support the idea that actin polymerization overcomes the silencing state in our experimental conditions, which creates most likely the molecular basis leading to increased var switching rates once JAS-treated parasites resume to the monoallelic expression modus in subsequent cycles.
Discussion
The coordinated expression of a number of multigene families contributes in many key host-parasite interactions such as sequestration, invasion, and immune evasion. In this paper we demonstrate a critical role of introns in the perinuclear tethering process of the immune evasion var gene family, a function that has never been linked to introns in malaria parasites and in model eukaryote systems. The short repeated intron element (iNPE) is only present in the major virulence var gene family. Previous reports showed that the silencing of a var upstream promoter requires the presence of a var intron in the 3′ position of the luciferase reporter gene (Deitsch et al., 2001). Otherwise, the luciferase gene is expressed constitutively. In light of our data, we hypothesize that the silencing effect of the luciferase gene may result from the targeting of the plasmid to a repressive region in the nuclear periphery.
The spatial arrangement of virulence genes in other malaria species has not yet been experimentally addressed. A database search of several malaria genome sequences showed that iNPE motifs are restricted to var introns and are virtually absent in other malaria species (Table S2). It remains to be seen if var genes are the only driving force for the particular spatial organization of virulence genes or if alternative anchoring mechanisms exist. Although iNPE is completely absent in yeast, in this organism DNA motifs are able to mediate perinuclear localization through the interaction with inner nuclear membrane (INM) proteins involving noncoding regions (Ahmed et al., 2010). It is noteworthy that several iNPE copies were found in human, rat, and mouse genomes. It will be important to verify whether the described plasmodial perinuclear tethering process is conserved in higher eukaryotes.
Nuclear location experiments of episomes and EMSA experiments identified a short var intron repeat motif of 18 bp as having a key role in perinuclear anchoring and in recruiting an intron-specific protein complex. Surprisingly, plasmodial actin I was identified as a major component of the iNPE-binding protein complex. The P. falciparum genome encodes two actin genes: one is expressed in all life cycle stages (actin I), and the second one is expressed almost exlusively in sexual stages (actin II). Recent advances in the nuclear biology of eukaryotes have cast more light on the functions of nuclear actin and an evergrowing family of actin-associated proteins in eukaryotic gene transcription (reviewed in Hofmann, 2009). Although the presence of actin in the P. falciparum nucleus is somehow expected, its unusual concentration at the nuclear periphery and its association with introns has not been observed in other eukaryotes, and apparently is linked to a role in the antigenic variation of P. falciparum. Cytoplasmic staining appeared in more mature blood stages, which is compatible with actin’s function as a molecular motor in parasite motility and the invasion of different host cells (reviewed in Baum et al., 2008; Schuler and Matuschewski, 2006a) and in hemoglobin uptake (Lazarus et al., 2008).
Given that recombinant Pfactin-I does not directly bind iNPE, we assume that a specific intron-binding protein recruits actin. A number of putative actin-binding proteins have been acknowledged in the P. falciparum genome (Schuler and Matuschewski, 2006b), but none of these candidates have obvious DNA-binding domains. Our work has identified a number of candidate molecules. Among the nuclear proteins that were purified on the iNPE probe, 12 proteins have putative DNA-binding domains (see Table 1). Among those, only the member of the Ap2 family is predicted to bind to specific DNA motifs. We show that this protein binds specifically to the iNPE motif, making this Ap2 member a prime candidate for recruiting the actin protein complex at var introns. Further studies are needed to validate this hypothesis and to gain further insight into the tethering process and its implications regarding antigenic variation.
Intranuclear relocation of a var gene locus to an expression competent perinuclear region is a critical step in the process of mutually exclusive var expression. The underlying molecular mechanism, however, remained unidentified. In P. falciparum, polymerized actin forms extremely short filaments that cannot be visualized, and its actin dynamics differ fundamentally from the turnover of actin in mammalian cells (Schmitz et al., 2005). In this study, we were able to link distinct functions of G- and F-actin to antigenic variation using actin-perturbing drugs. JAS-induced actin polymerization did affect the nuclear spatial organization of internal chromosome var genes and iNPE-carried episomes that were significantly more dissociated from telomere clusters, strongly supporting a key role of var introns in actin-dependent nuclear gene repositioning. Moreover, JAS treatment induces transcriptional upregulation of many previously silent var genes in bulk culture. The parasite used in this experiment initially expresses a single var gene. After JAS treatment, we do not observe a reduction in the transcript level of this var gene, indicating that additional var genes are cotranscribed in the same cell (see a model in Figure S6). There is growing evidence for connecting transcription to nuclear pores in yeast and other eukaryotes (Dieppois and Stutz, 2010), and the molecular mechanisms responsible for chromosomal repositioning remain puzzling but are thought to involve actin and myosin (Dundr et al., 2007). Drugs favoring G-actin did not show a detectable effect on the spatial organization of var genes (or telomere cluster formation) or var gene activation. This result is compatible with G-actin being the main component in the perinuclear space possibly involved in the tethering of virulence genes to heterochromatin regions. Controlled actin polymerization may provide part of a molecular motor that allows the repositioning of virulence genes to an expression site. Importantly, our results show that increased switch rates of var genes are linked to nuclear filamentous actin. It is an important finding that a molecule involved in epigenetic switching of malaria parasites has been identified. It is tempting to speculate that by fine-tuning actin polymerization the parasite may adapt the expression of clonally variant surface antigens to the immune status of its host.
In conclusion, clonally variant gene families that express virulence factors involved in host parasite interactions adopt a particular spatial organization required for default silencing and monoallelic expression in malaria parasites. We show here that var introns contribute to this process and that F-actin is critical in the activation of epigenetically silenced var genes and in the process of var gene switching. These findings are highly relevant for organisms that depend on mutually exclusive gene expression such as many microbial pathogens and the mammalian olfactory system.
Experimental Procedures
Additional detailed methods are presented in the Supplemental Experimental Procedures available online.
Parasite Culture and Treatment with Actin Inhibitors
Plasmodium falciparum 3D7 parasites were cultivated and sorbitol synchronized as described previously (Lopez-Rubio et al., 2009). Ring stage parasites were incubated with 10 μM CD (Sigma), 10 μM JAS (Invitrogen), or DMSO (0.1%), respectively, for 3 or 10 hr.
Plasmid Constructs of Transfection
pLN-ENR-GFP (Nkrumah et al., 2006) was modified by deleting the whole pfenr-gfp cassette, then was used to clone various DNA fragments in Apa I/BamHI multicloning site. For pARL vector (Crabb et al., 2004), the crt promoter was exchanged with var intron sequences. pLN-ENR-GFP vector was also modified for pLN-HA-Pfactin-I construct. Three copies of HA tag (YPYDVPDYA) were ligated to the N terminus of Pfactin-I via a linker sequence (AAAAVDAAAA). The calmodulin promoter of pLN was replaed by 1.5 kb 5′UTR of Pfactin-I gene. The primers used in these constructs are listed in Table S1.
Quantitative Reverse Transcription PCR
Total RNA of synchronous parasite culture was extracted using Trizol reagent (Invitrogen) as described previously (Kyes et al., 2000). cDNA were prepared according to the manufacturer’s recommendations (Invitrogen). All qRT-PCR primers used in this study were shown in Table S1.
Antibodies and Western Blot Analysis
Rabbit and mouse anti-Pfactin-I antibodies were used in western blot studies. Total parasite extracts and nuclear and cytoplasmic extracts were separated on 4%–12% SDS-PAGE gel (Bio-Rad) and subjected to western blot analysis.
Gel Shift Assay
The preparation of nuclear and cytoplasmic extracts and EMSA analysis were described previously (Voss et al., 2003).
Identification of the Intron-Binding Proteins
The var intron binding proteins were isolated and identified as previously described (Flueck et al., 2010). Briefly, the biotinylated probe was bound to streptavidin-coupled beads and incubated with nuclear extracts according to the supplier’s protocol (Invitrogen). The eluted fraction was subjected to LC-MS/MS.
Immunofluorescence and FISH
Immunofluorescence assay and FISH were performed as previously described (Mancio-Silva et al., 2008).
Immunoelectron Microscopy
Immunolabeling for EMwas performed on ultrathin sections of glutaraldehydefixed ring stage parasites. The nucleoplasmic region was stained with rabbit anti-histone H3 antibodies and actin with mouse anti Pfactin-I antibodies.
Chromatin Immunoprecipitation
The ChIP assay was performed as described previously (Freitas-Junior et al., 2005). The mouse anti-HA antibody (Roche, 12CA5) and mouse IgG (Sigma, I5381) were used in this study.
Statistics
The nuclear distribution of FISH signals were compared using χ2 test; **p < 0.001 between tested group and control. For the qRT-PCR data, statistical significance was determined with the two-tailed Student’s t test; *p < 0.05; **p < 0.001.
Supplementary Material
Supplemental Information includes six figures, six tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:http://dx.doi.org/10.1016/j.chom.2011.09.013.
Acknowledgments
We would like to thank G. Wei and X. Xu for assistance in recombinant protein expression, M. Denise for anti-HSP70 antibody, and N. Siegel for the critical reading of this manuscript. This work was supported by the National Basic Research Program (973 Program) in China (2007CB513100), the National Natural Science Foundation of China (No. 30430610), an ERC Advanced Grant (PlasmoEscape 250320), and European Commission FP7 programs EVIMalaR and ANR Blanc International PARACTIN. Institutions 1 and 2 on the affiliation list contributed equally to this work.
References
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Leliwa-Sytek A, Tavul L, Imrie H, Migot-Nabias F, Brown SM, McVean GA, Day KP. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog. 2007;3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning of P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Baum J, Tonkin CJ, Paul AS, Rug M, Smith BJ, Gould SB, Richard D, Pollard TD, Cowman AF. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell Host Microbe. 2008;3:188–198. doi: 10.1016/j.chom.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinas M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb BS, Rug M, Gilberger TW, Thompson JK, Triglia T, Maier AG, Cowman AF. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol Biol. 2004;270:263–276. doi: 10.1385/1-59259-793-9:263. [DOI] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci. 2010;123:1989–1999. doi: 10.1242/jcs.053694. [DOI] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BT, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Gannoun-Zaki L, Jost A, Mu J, Deitsch KW, Wellems TE. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot. Cell. 2005;4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/S1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Lazarus MD, Schneider TG, Taraschi TF. A new model for hemoglobin ingestion and transport by the human malaria parasite Plasmodium falciparum. J Cell Sci. 2008;121:1937–1949. doi: 10.1242/jcs.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Mancio-Silva L, Rojas-Meza AP, Vargas M, Scherf A, Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J Cell Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, Martinez-Calvillo S, Scherf A, Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci USA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, van den Elzen AM, Cohen AM, Janssen-Megens EM, van de Vegte-Bolmer M, Selzer RR, Iniguez AL, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci USA. 2009;106:9655–9660. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- Schmitz S, Grainger M, Howell S, Calder LJ, Gaeb M, Pinder JC, Holder AA, Veigel C. Malaria parasite actin filaments are very short. J Mol Biol. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- Schuler H, Matuschewski K. Plasmodium motility: actin not actin’ like actin. Trends Parasitol. 2006a;22:146–147. doi: 10.1016/j.pt.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Schuler H, Matuschewski K. Regulation of apicomplexan microfilament dynamics by a minimal set of actin-binding proteins. Traffic. 2006b;7:1433–1439. doi: 10.1111/j.1600-0854.2006.00484.x. [DOI] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Voss TS, Kaestli M, Vogel D, Bopp S, Beck HP. Identification of nuclear proteins that interact differentially with Plasmodium falciparum var gene promoters. Mol Microbiol. 2003;48:1593–1607. doi: 10.1046/j.1365-2958.2003.03528.x. [DOI] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information includes six figures, six tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:http://dx.doi.org/10.1016/j.chom.2011.09.013.