Abstract
Fluorescence-based techniques have been used extensively in the malaria field to study the functional role of nuclear organization and gene positioning in blood stages of the human malaria parasite, Plasmodium falciparum. In this chapter, we present optimized protocols for bromouridine (BrUTP) incorporation into nascent RNA in live parasites and fluorescent in situ hybridization (FISH) in fixed parasites. Methodology to perform various combinations of the FISH assay, as well as a basic approach for quantitative analysis of nuclear position, is also described.
Keywords: Plasmodium falciparum, Blood stages, Nuclear organization, Gene positioning, Transcription sites, BrUTP incorporation, Nuclear transcripts, Nuclear periphery, Quantitative analysis, DNA FISH, RNA FISH, RNA/DNA FISH, Immunofluorescence, Immuno-FISH, Fluorescence microscopy
1. Introduction
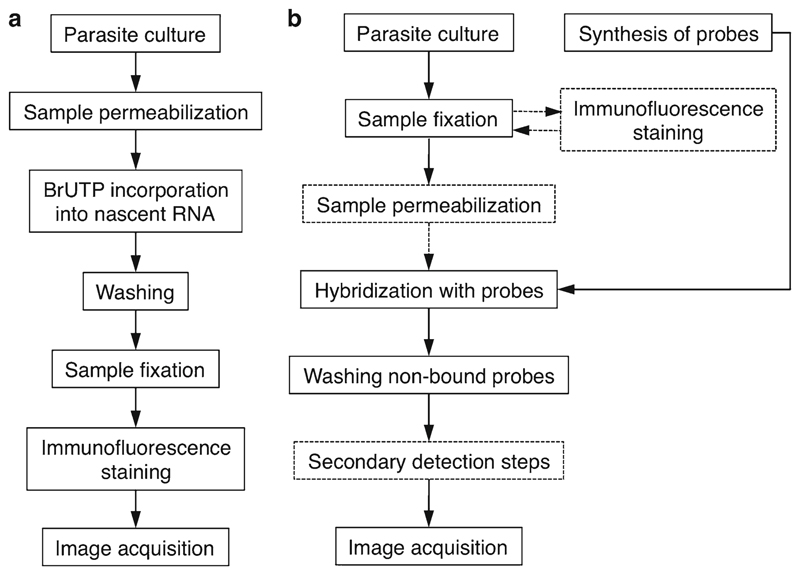
Fluorescence-based technology has been widely used to study the role of nuclear architecture in gene expression in malaria parasites (1–6). The current data point to a functional compartmentalization of the Plasmodium falciparum nucleus as observed in other eukaryotic cells (7, 8). The methods outlined in this review have been adapted for this parasite to allow the visualization of sites of active transcription by incorporation of bromouridine (Fig. 1a) and the localization of specific nuclear transcripts and genomic loci by in situ hybridization (Fig. 1b).
Fig. 1.
General outline of the BrUTP transcription and FISH assays. (a) The BrUTP incorporation and immunodetection by microscopy that has been adapted to P. falciparum parasites is a 2-day experiment. The parasites are first released from erythrocytes, lightly permeabilized, and incubated for 10–20 min in a transcriptional buffer containing BrUTP. After washing off the non-incorporated nucleotides, parasites are fixed and subject to immunofluorescence to detect BrRNA. The anti-BrdU antibodies are incubated overnight and the fluorophore-conjugated secondary antibodies for 1 h. This protocol preserves transcriptional activity and nuclear structure; it is performed entirely with parasites in suspension and is suited for simultaneous detection of proteins or nuclear substructures. (b) The traditional FISH is a 2-day experiment: on day 1 parasites are fixed, permeabilized, and hybridized overnight with labeled oligonucleotide probes, which bind to their complementary sequences. On day 2, the unbound probes are washed off and the slides are mounted and examined (dashed lines are optional or method-specific steps). The FISH probes are usually prepared in advance and can be directly coupled to biotin, digoxigenin, or fluorochromes. Nonfluorescent probes require a second step of detection (dashed box) using fluorophore-conjugated antibodies, which offers the advantage that signal can be amplified by using additional antibodies. The FISH protocol can be combined with immunofluorescence (immuno-FISH) allowing colocalization of protein, specific genes, and either genomic DNA or nuclear substructures. After immunofluorescence, parasites are fixed again and then subjected to FISH.
In situ fluorescence visualization of newly transcribed nuclear RNA has been used to discriminate sites of transcription by P. falciparum RNA Polymerase I and II (6). This non-isotopic and nondestructive method involves the incorporation of modified nucleotides (bromouridine triphosphate, BrUTP) into RNA of living cultured parasites by permeabilization followed by immunofluorescence detection (with anti-BrdU antibodies; Fig. 1a). The protocol allows the study of the dynamics of global transcription patterns in the parasite and can be combined with specific inhibitors of RNA synthesis and/or immunolocalization of proteins involved in different nuclear activities (Fig. 2a–d).
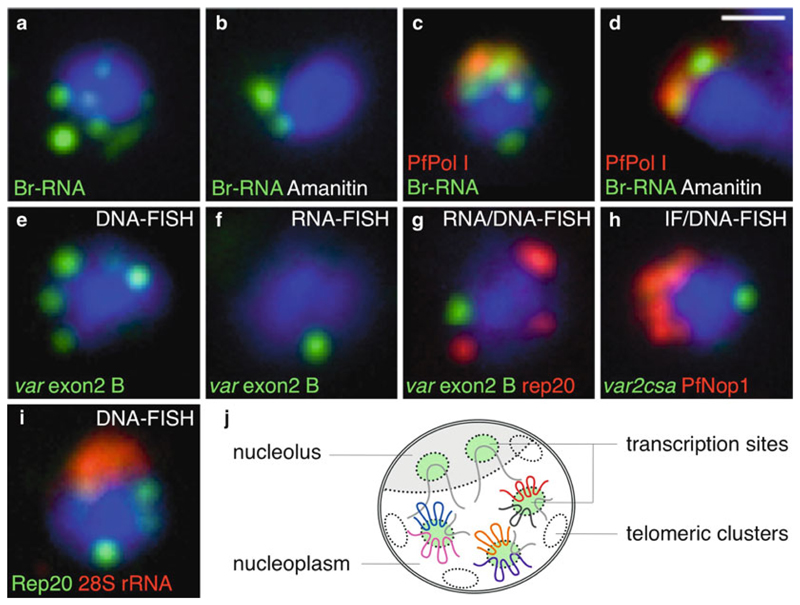
Fig. 2.
Representative images of pre-replicative blood-stage parasites in various combinations of BrUTP labeling and FISH techniques. (a–d) Br-UTP labeling of nascent RNA with and without α-amanitin, an inhibitor of Pol II and III. (c, d) Colocalization of BrRNA (green) with the nucleolus (red, PfPol I). (e) DNA-FISH analysis using the exon2 probe that cross-hybridizes to group B of the var multigene family. (f) RNA-FISH analysis of var transcripts from group B. Note that the same probe is used in (e, f) (only the hybridization and washing conditions were different for the DNA- and RNA-FISH). (g) RNA/DNA-FISH of var transcripts (green) and telomeric clusters (rep20, red). (h) Immunofluorescence using antibodies against the nucleolar protein PfNop1 (red) combined with DNA-FISH for an individual var gene (var2CSA, green). (i) Two-color DNA-FISH of telomeric clusters (rep20, green) and 28S rRNA (red). In all cases, parasites are in the ring stage and nuclear DNA stained with DAPI (blue). Representative examples are shown as merged images. Scale bar: 1 μm. (j) Current model of nuclear organization in ring-stage parasites. The nucleolus is represented in a half-moon shape at the nuclear periphery, opposing the telomeric clusters. These two DNA-based subnuclear compartments can be used as reference for colocalization studies. Other nuclear landmarks are the transcription sites or factories distributed in the nucleolus and nucleoplasm. It is speculated that the few transcription sites observed in the nucleoplasm do not correspond to single genes, thus implying nuclear clustering of multiple actively transcribing genes at each factory (6, 8).
Fluorescence in situ hybridization (FISH) was originally developed in P. falciparum for DNA analysis (DNA-FISH) to visualize the clustered chromosome ends (9). The basic technique involves the specific binding of fluorescently labeled oligonucleotide probes to their complementary sequences in fixed samples. The FISH protocol usually entails the following steps (Fig. 1b): parasite samples are fixed, permeabilized, hybridized with probes after DNA denaturation at high temperatures, and washed.
More recent studies have optimized FISH protocols for analysis of specific nuclear transcripts (RNA-FISH) (4, 5, 10, 11). RNAFISH methodology differs from the traditional DNA-FISH only by the fact that the FISH probes are allowed to hybridize without sample denaturation. If discrimination between sense and antisense transcripts is not necessary, the double-stranded DNA-FISH probes can also be used for RNA-FISH. In fact, if prepared in RNAse-free conditions, the same parasite fixed samples and probes can be employed on DNA- and RNA-FISH (Fig. 2e, f). RNA- and DNA-FISH can also be performed in a sequential manner (RNA/DNA-FISH), thus allowing to visualize DNA sites and nuclear transcripts at once in a single parasite (5) (Fig. 2g).
Another combination routinely used in Plasmodium parasites is that of FISH with immunofluorescence assays (immuno-FISH) (1, 6, 12) (Fig. 2h). Nonetheless, there are pitfalls in applying this methodology, as the optimal conditions for immunofluorescence are frequently poorly compatible with those for FISH. The nuclear architecture and the antibody reactivity should be preserved as much as possible, while allowing accessibility of the FISH probe to the nuclear chromatin. Usually, each pair of antibody/FISH probe requires specific optimized conditions. In our hands, most but not all antibodies against nuclear structures worked when combined with the FISH method.
Here, we describe the synthesis of FISH probes with fluorescently labeled or hapten-labeled nucleotides and the preparation of parasite samples for BrUTP incorporation, immunofluorescence, and FISH analysis. We then provide methodology to perform the various combinations of FISH protocols: (1) DNA-FISH, (2) RNA-FISH, (3) RNA/DNA-FISH, and (4) immuno-FISH. Finally, we give details on how to perform image acquisition, analysis, and quantitative measurements.
2. Materials
2.1. Br-UTP Incorporation
P. falciparum synchronized blood-stage cultures.
RPMI 1640.
1× PBS (DNAse- and RNAse-free).
Water (DNAse- and RNAse-free).
RNase A and RNAse inhibitors (Roche).
Anti-BrdU monoclonal antibodies (Santa Cruz Biotechnology).
Secondary Alexa-fluor antibodies (highly cross-adsorbed) (Invitrogen).
Transcriptional buffer stock solutions: 1 M DTT, 100 mM PMSF, 100 mM EGTA, 1 M HEPES, 1 M MgCl2, 2 M KCl, 100 mM individual nucleotide triphosphates (ATP, CTP, GTP; Roche).
BrUTP (Sigma): Prepare a 100 mM stock solution. Store aliquots at −20°C.
α-Amanitin (Sigma): Prepare a 1 mg/ml stock solution. Store aliquots at −20°C.
2% (w/v) saponin (in RNAse-free PBS), freshly prepared.
1.5- and 15-ml tubes (DNAse- and RNAse-free tubes).
10% (w/v) paraformaldehyde (PFA, Electron Microscopy Sciences).
Decontamination solution to clean pipettes, equipment, and surfaces (e.g., RNaseZap, Ambion).
Heating block with shaker (at 37°C).
Rotating wheel.
2.2. Fixation in Suspension
P. falciparum synchronized blood-stage cultures.
0.15% (w/v) saponin (in PBS). Store aliquots at −20°C.
10% (w/v) PFA (Electron Microscopy Sciences).
1× PBS (DNAse- and RNAse-free).
2.3. FISH Probes
Parasite gDNA or cDNA and appropriated primers.
Biotin-, digoxigenin (DIG), fluorescein-high prime labeling reaction kits (Roche).
Anti-DIG, streptavidin-conjugated with Alexa-fluor antibodies (Invitrogen).
4 M LiCl, ethanol 70 and 100%.
2.4. RNA- and DNA-FISH
Rubber in situ frames (25 μl, AbGene or Eppendorf) and plastic coverslips.
Microscope slides (1–3 wells, Teflon coated).
4% (w/v) BSA (in PBS). Store aliquots at −20°C.
0.1% (v/v) Triton X-100 (in PBS, freshly prepared).
Stock solutions: 20× SSC, 20× SSPE, 50% (w/v) dextran sulfate.
Hybridization solution: 50% (v/v) formamide, 10% (w/v) dextran sulfate, 2× SSPE, 250 μg/ml herring sperm DNA. Store at 4°C. Stable for 1–2 months.
Washing solution freshly prepared: 50% (v/v) formamide 2× SSC.
Washing solutions stable at room temperature (RT): 1× SSC, 2× SSC, 4× SSC, 0.5% (v/v) Tween20 (in PBS).
Forceps, humid chamber.
Thermal cycler with in situ adapter.
Heating block (at 100°C).
Water bath (at 37°C).
Hybridization oven (at 50°C).
Shaker.
2.5. DNA Counterstaining and Mounting
Antifade mounting medium with 4′,6-diamidino-2-phenylindole (DAPI).
Glass coverslips, nail polish.
3. Methods
3.1. BrUTP Incorporation into Live Parasites
Prepare 10 ml of fresh permeabilization solution (RPMI, RNAse inhibitor 5 U/ml, 0.02% saponin) and 5 ml of transcriptional buffer (50 mM HEPES, 100 mM KCl, 5 mM MgCl2, 0.5 mM EGTA in water). Warm the solutions at 37°C (water bath, see Note 1). Use appropriate precautions to avoid RNase contamination.
Collect the parasite culture by centrifugation (800 × g for 5 min in a 15-ml tube, see Note 2).
Wash the pellet in warmed RPMI (containing 5 U/ml RNAse Inhibitor). Centrifuge as above.
Gently resuspend the pellet in 10 ml of warmed permeabilization solution (see Note 3), mix by inversion, and incubate at 37°C (water bath) for up to 5 min or until the suspension becomes translucent (due to red blood cell (RBC) lysis).
Centrifuge (3200 × g for 5 min) and discard the supernatant (but not completely; to avoid parasite loss keep 0.5–1 ml of pellet and supernatant).
Gently resuspend the parasite pellet in warmed RPMI (containing 5 U/ml RNAse inhibitor), transfer to 1.5-ml tubes, and centrifuge (2000 × g for 1 min). Repeat the wash and keep the tubes at 37°C (block-heating shaker).
To 1–2 ml of the transcriptional buffer prepared in step 1, add to a final concentration 5 mM DTT, 0.5 mM PMSF, 100 U/ml RNase inhibitor, 2 mM ATP, 1 mM CTP, 1 mM GTP, and 0.5 mM BrUTP. Transcription inhibitors can be added as controls (see Note 4).
Gently resuspend the pellet in the final transcriptional buffer (500 μl/tube) and incubate at 37°C for 10–20 min with 300 rpm shaking (block-heating shaker, see Note 5).
Centrifuge for 2000 × g for 1 min. Resuspend the pellet in PBS (500 μl/tube) and centrifuge again.
Resuspend the pellet in ice-cold 4% PFA (500 μl/tube) and incubate on ice for 15–20 min.
Centrifuge for 2000 × g for 1 min. Wash the pellet in ice-cold PBS (500 μl/tube) and finally resuspend in 100–250 μl of icecold PBS (see Note 6).
Add anti-BrdU antibodies (1:50 in PBS, no BSA, see Note 7) to the samples and incubate overnight at 4°C (rotating wheel in a cold room).
Wash twice with PBS (200 μl/tube, centrifuge for 2000 × g for 1–2 min, see Note 8).
Resuspend the pellet in secondary antibodies conjugated with fluorophores (1:500 in PBS) and incubate at 37°C for 30–60 min (water bath).
Wash twice with PBS (200 μl/tube, centrifuge for 2000 × g for 1–2 min).
Resuspend the pellet in 15–50 μl PBS and deposit on microscope slides (apply 15–50 μl of parasite suspension, let the parasites adhere to the glass for 1–5 min, and return the suspension to the tube) (see Note 9).
Slightly air-dry the preparation at RT for ~15 min (keep in the dark) and mount the slide using antifade solution with DAPI (5–10 μl for each well).
Cover the slide with a coverslip (avoiding air bubbles and without pressure) and seal with nail polish (see Note 10).
3.2. FISH in Fixed Parasites
3.2.1. Preparing FISH Probes
FISH probes of 1–2 kb should be designed corresponding to highly specific regions of the locus of interest (if possible with high GC content) to minimize the risk of cross-hybridization to other genes or transcripts. The chosen genomic locus is then PCR-amplified and labeled by nick translation, random priming, or in vitro transcription in the presence of fluorescein, biotin, or digoxigenin (DIG). Fluorescent or hapten-tagged oligonucleotides can also be used. Although more costly, this is recommended for obtaining sense and antisense RNA probes. Usually, we label genomic DNA probes by random priming as follows:
Use ~300 ng of DNA template (PCR-amplified and purified) for a standard 20-μl reaction. Incubate the kit reaction mixture at 37°C for 20–24 h (see Note 11).
Precipitate the labeled probe by adding 2.5 μl 4 M LiCl and 75 μl prechilled 100% ethanol. Let the precipitate form overnight at −20°C.
Centrifuge 14000 × g for 30 min at 4°C, and discard the supernatant.
Wash the pellet with 500 μl cold 70% ethanol (centrifuge 14000 × g for 15 min at 4°C).
Discard the supernatant and let the pellet dry for at least 1 h at RT (see Note 12).
Resuspend the pellet in 30–50 μl of water and store at −20°C (see Note 13).
3.2.2. Preparing Parasites for Fixation in Suspension
Transfer the parasite culture (see Note 2) to a 15-ml tube and centrifuge (800 × g for 5 min).
Wash the pellet with warmed RPMI (centrifuge 800 × g for 5 min see Note 1).
Resuspend the pellet in 0.015% saponin (diluted in PBS or RPMI), mix by inversion, and incubate at 37°C (water bath) for up to 5 min or until the suspension becomes translucent (due to erythrocyte lysis).
Centrifuge for at 3200 × g for 5 min.
Carefully resuspend the pellet in warmed PBS or RPMI and transfer to 1.5-ml tubes.
Wash twice with warmed RPMI or PBS (1 ml/tube, centrifuge at 2000 × g for 1 min).
Resuspend the pellet in ice-cold 4% PFA (freshly diluted in PBS) and incubate on ice for 15–20 min.
Wash twice with ice-cold PBS (centrifuge at 2000 × g for 1 min).
Resuspend the pellet in ice-cold PBS (500 μl) and store at 4°C (see Note 14).
3.2.3. In Situ Hybridization
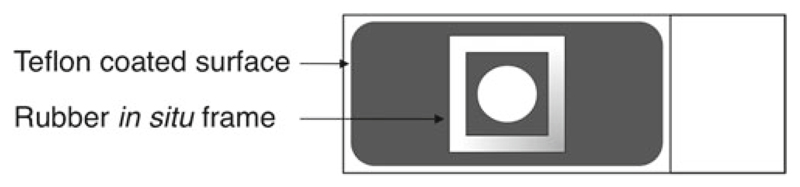
Deposit a monolayer of fixed parasites on the wells of the microscope slide (see Fig. 3) and air-dry at RT for 15–30 min (see Note 15).
Fix the rubber in situ frames (see Fig. 3) around the wells containing the parasites.
Wash twice the wells with PBS for 5 min at RT (cover each well with 50–60 μl of PBS, remove the liquid carefully, and repeat the washing; from this step on, avoid air-drying the preparation).
Permeabilize nuclei using ice-cold 0.1% Triton X-100 for 5 min at RT. For DNA-FISH, this step is optional and may include RNaseA treatment. For RNA-FISH, the permeabilization step is absolutely necessary and the solution may contain an RNAse inhibitor (see Note 16).
Wash the wells three times with PBS at RT (5 min in total).
Prepare the probes: 1–2 μl of probe(s) with 23–24 μl of hybridization solution (in 1.5-ml tubes). The optimal concentration of the probes must be determined empirically (see Notes 13 and 17).
Denaturate the probes for 5 min at 100°C (heating block). Chill tubes on ice (for 5 min). For fluorescein-labeled probes, protect from light.
Apply the probes (25 μl for each well) (see Note 18).
Cover the rubber frame with the plastic coverslip avoiding air bubbles (make sure that the frame and coverslip are well fixed to the slide; otherwise the probe(s) will be lost during hybridization).
Place the slides on the Thermal cycler (use the in situ adapter). For DNA-FISH, perform 30 min at 80°C for sample denaturation, and then hold at 37°C for at least 16 h. For RNA-FISH, perform hybridization at 37°C for at least 16 h without denaturation (see Note 19).
Fig. 3.
Microscope slide setup for FISH analysis. The Teflon-coated surface delimitates and reduces the region to apply the parasite suspension and solutions or antibodies. The rubber in situ frame when covered with a coverslip (also in plastic) creates a small hybridization chamber that allows a reduction on the amount of probe required for each assay. This system also prevents against evaporation during the overnight hybridization. The frames are adhesive on both sides, and can be applied and removed easily from the microscope slide using forceps. We usually use microscope slides with three wells (and three frames), thus allowing for testing three different conditions per slide.
3.2.4. Washing DNA-FISH
Pre-warm the washing solutions (use ~40 ml of solution in 50-ml tubes) (see Note 20).
Remove the rubber frames and coverslips using forceps.
Wash the slides in 50% formamide-2× SSC for 30 min at 37°C (water bath), followed by 1× SSC, 2× SSC, and 4× SSC for 10 min each at 50°C (hybridization oven).
If using DIG or biotinylated probes, incubate the wells with anti-DIG or streptavidin-conjugated antibodies (diluted in 4% BSA, cover each well with 50–60 μl) at RT for 30 min in a humid chamber, protected from light. Wash three times in 0.5% Tween20 (10 min each with shaking, 50–70 rpm).
Final wash with PBS at RT for 5 min.
Dry the Teflon surfaces and bottom of the slides with a paper tissue.
Air-dry the wells at RT for ~15 min and mount the slide using antifade solution with DAPI (5–10 μl for each well).
Cover the slide with a coverslip (avoiding air bubbles and without pressure) and seal with nail polish (see Note 10).
3.2.5. Washing RNA-FISH
Pre-warm the 2× SSC solution at 37°C (water bath, see Notes 20 and 21).
Remove the rubber frames and coverslips using forceps.
Wash the slides three times in 2× SSC at 37°C for 10 min each.
If using nonfluorescent probes, proceed to step 4 in Subheading 3.2.4.
Wash with PBS at RT for 5 min.
To finalize, follow the steps 6–8 described in Subheading 3.2.4.
3.2.6. RNA-FISH Followed by DNA-FISH (RNA/DNA-FISH)
After RNA hybridization overnight at 37°C wash as described in Subheading 3.2.5, steps 1–5 (see Note 22).
Dry the Teflon surfaces and bottom of the slides with a paper tissue.
Fix new rubber in situ frames around the wells and prepare the new probes for DNA-FISH (see Subheading 3.2.3, step 6).
Apply the denaturated probes (25 μl for each well). Cover with a coverslip (avoiding air bubbles).
Place the slides on the Thermal cycler (using the adapter). Perform 30 min at 80°C for sample denaturation, and then hold at 37°C for at least 16 h.
Wash as described above for DNA-FISH (Subheading 3.2.4, steps 1–8).
3.2.7. Immunofluorescence Followed by DNA-FISH (Immuno-FISH)
Use 100–200 μl of the suspension of fixed parasites (prepared in Subheading 3.2.2).
Centrifuge at 2000 × g for 1 min. Carefully resuspend the pellet in the diluted primary antibodies and incubate at 37°C (water bath) for 30–60 min.
Wash once or twice in 200 μl PBS (centrifuge at 2000 × g for 1–2 min).
Resuspend the pellet in the diluted secondary antibodies, incubate at 37°C (water bath) for 30–60 min, and wash as in step 3.
Resuspend the pellet in ice-cold 4% PFA and incubate on ice for 20–30 min (see Note 23).
Wash with 200 μl PBS (centrifuge at 2000 × g for 1–2 min).
Resuspend the pellet in 15–50 μl PBS and store at 4°C (see Note 24). Deposit the parasites on a slide and confirm on the fluorescence microscope the nuclei density and if immunofluorescence signals are as expected.
Deposit the immunolabeled parasites on a new slide and proceed for DNA-FISH as described in Subheading 3.2.3. For immuno-DNA-FISH, perform 10 min at 72°C for sample denaturation, and then hold at 37°C for at least 16 h (see Note 25).
3.3. Imaging Analysis and Quantitative Measurements
Plasmodium immunofluorescence and FISH preparations are generally examined using wide-field microscopes (1–6, 9–13), though confocal microscopy has also been applied with success (14). Since the parasite nucleus, in the asexual ring stage, is only 1.5–2 μm in diameter, the objective magnification recommended is 100×. Regardless of the instrumentation used for image acquisition, it is also recommended to correct the chromatic shift, by alignment of signals from beads that emit fluorescence at multiple wavelengths (e.g., 0.2 μm TetraSpeck, Invitrogen).
Because of the harsh FISH conditions, the diameter of the parasite nucleus (as determined by DAPI staining) should be monitored regularly to ensure that the nuclear structure has not been disrupted. Worth noting, the nuclear size and shape change as the parasite maturates in the 48-h blood-stage cycle. To discriminate large and mature parasite nuclei from disrupted nuclear structure, a bright-field or phase image should also be acquired. The presence and size of the hemozoin pigment can also serve as guide for determining the cell cycle stage.
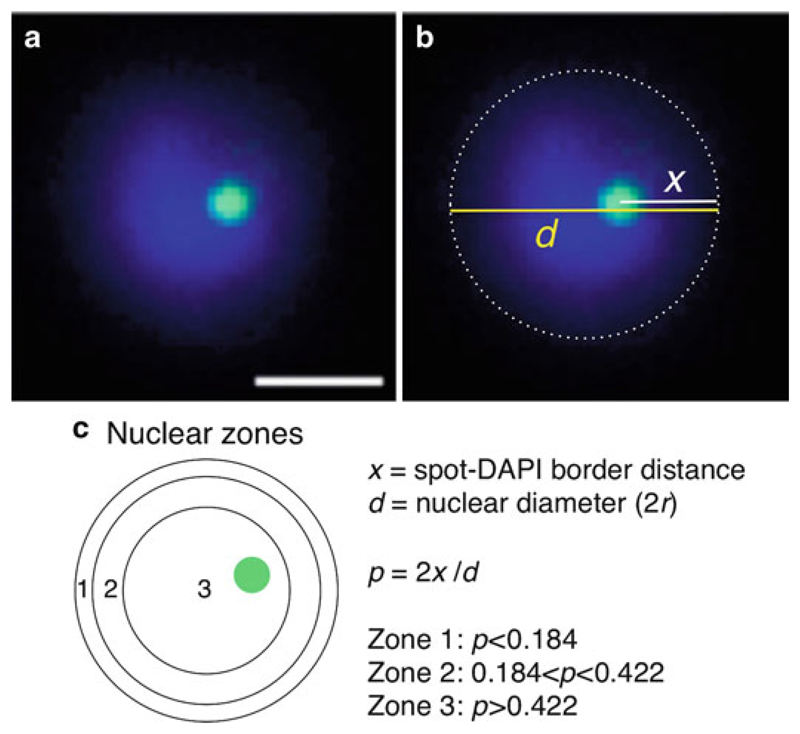
For quantitative 2D analysis of nuclear position, fluorescent signals are usually scored in relation to the parasite nuclear periphery using the zoning method (15). As described in Fig. 4, it consists of measuring the distance of the FISH signal to the extremity of the nucleus (defined as the DAPI border, see Note 26), as well as measuring the nuclear diameter. By dividing the first value by the half of the second (i.e., the radius), one can classify each spot falling into one of the three concentric zones of equal surface. All the measurements can be performed using the software for acquisition or the freely available ImageJ (http://rsbweb.nih.gov/ij/).
Fig. 4.
Subnuclear localization relative to the nuclear periphery. (a) DNA-FISH analysis of a single locus gene (PFB0540w, green) in ring-stage parasites. Nuclear DNA is visualized with DAPI (blue). Scale bar: 1 μm. (b) Schematic representation of the measurements of nuclear diameter (d, yellow line) and the distance of the green fluorescent spot to the nearest periphery (x, white line). The extremity of DAPI staining (defined by the low-intensity staining, see Note 26) represents the perimeter of the nucleus (dashed line). (c) Illustration of the three concentric zones of equal surface (zones 1, 2, and 3). As described in (15), the most peripheral zone (zone 1) corresponds to a ring of width 0.184 × nuclear radius (r). Zone 2 lies between 0.184r and 0.442r from the periphery, and zone 3 is a central core of radius 0.578r. In the example shown in a, p = 0.53, which falls in zone 3. Analysis of position is generally performed on 100–200 parasite nuclei, and the percentage of measured distribution is compared to a random distribution (33.3% in the 3 zones) or to another condition, using a chi-square test.
To gain more insight into the function of nuclear position, fluorescent signals can also be localized with respect to parasite-specific nuclear landmarks, such as the telomeric clusters (2, 3, 9) (Fig. 2g), the nucleolus (6) (Fig. 2h), or the transcription sites (6) (Fig. 2a, b).
4. Notes
Until the fixation step, it is important to maintain the temperature at 37°C as much as possible to avoid alterations on gene expression and nuclear positioning induced by a temperature drop. As such, it is recommended to pre-warm the centrifuge and the solutions at 37°C. Also, it is convenient that the culture incubator, centrifuges, water bath, block-heating shaker, and bench work are close to each other to avoid temperature fluctuations. The block heating (at 37°C) should be used as tube stand.
For a synchronized culture on ring-stage parasites, start with ~5% parasitemia in 1 ml of packed blood cells. For parasite cultures of mature stages (trophozoites or schizonts), use 1–2% parasitemia in 500 μl of RBC pellet, as these forms are not so easily lost throughout the centrifugation steps.
The permeabilization is a critical step on the BrUTP transcription assay, as nucleotide triphosphates are taken up poorly by non-permeabilized cells. The saponin treatment here has a dual function of releasing the parasites from the erythrocytes and lightly permeabilizing the parasite membranes. Because saponin may change from batch to batch, it may be necessary to adjust the saponin concentration and duration of permeabilization.
To confirm that the observed pattern represents nascent RNA, RNaseA and/or transcription inhibitors (e.g., actinomycin D, α-amanitin (6, 16)) should be included in the transcription buffer (Fig. 2c, d).
To ensure that sites of RNA synthesis are labeled rather than processing sites, the duration of the BrUTP labeling should be optimized to be the minimum time required for producing a detectable signal. Longer incubations may also produce more intense signals, although less discrete.
These BrUTP-labeled and fixed parasites in suspension are ready to be used on immunodetection of BrRNA. Alternatively, they can be stored at 4°C (up to 1 week) and immunofluorescence performed later.
Antibodies to bromodeoxyuridine (BrdU) can be used to recognize bromouridine (BrUTP). However, the sensitivity of the commercially available monoclonal antibodies can vary and it may be necessary testing different antibodies. Overnight incubation of the anti-BrdU antibody is apparently better to obtain very discrete and bright fluorescence signals. For dual staining (Fig. 2c, d), if raised in different species, the antibodies can be mixed together with the anti-BrdU.
If the dark parasite pellet becomes very small during washing centrifugation, perform a single wash and/or increase the centrifugation to 4000 × g for 2 min instead.
The suspension of fluorescently BrUTP-labeled parasites can be stored at 4°C in the dark for a few weeks. The suspension is stable to be analyzed later. Cells need to be carefully resuspended before taking an aliquot. Alternatively, the BrUTPlabeled parasites may be used for subsequent FISH analysis (namely, RNA-FISH).
While mounting the preparation, no pressure on the coverslip should be applied to maintain the integrity of the nucleus of the parasite. The prepared slides can be immediately examined on the microscope or kept at 4°C in the dark for a few weeks.
For synthesis of FISH probes, labeling time is at least 1 h at 37°C; however, longer incubations increase the yield of labeled DNA. As an example, starting with 300 ng of template DNA is expected to produce 320 ng of fluorescein-labeled DNA after a 1-h incubation and 1,350 ng after a 20-h incubation.
Drying the pellet is important because small traces of residual ethanol will cause precipitation of the dextran sulfate present in the hybridization solution. Trace of ethanol can also lead to serious background problems.
The amount of newly synthesized labeled DNA may vary because of the template purity and sequence, incubation time, washing, etc. It is therefore advisable to test the probe prior to the final FISH experiment. Usually, we perform probe titrations by using 0.5, 1, and 2 μl of the newly labeled probe. If the labeling reaction was very efficient, dilution of the probe may be necessary in order to reduce background and increase signal specificity. Labeled FISH probes of this kind can be stored at −20°C for a few months.
The free-parasite fixation in suspension has been shown to improve conservation of the nuclear shape and size in Plasmodium parasites (14). Moreover, the suspension of fixed parasites can be stored at 4°C up to 2 months. This represents an important advantage of the protocol, as there is no need of fresh parasite culture to perform a new FISH experiment. Also, the same batch of fixed parasites can be used for testing multiple FISH probes or combinations.
Nucleus density of parasites fixed in suspension should be confirmed first on the fluorescence microscope. Apply 15–50 μl of the suspension to a glass slide, let the parasites adhere for 1–5 min, and recover the suspension to the initial tube. After air-dry, mount with DAPI and check on the microscope. Nonspecific signals increase when too many nuclei are present. Dilute or concentrate the samples if necessary.
RNaseA treatment of the samples is recommended for DNA-FISH protocol, particularly when the gene of interest is highly transcribed (e.g., ribosomal rRNA genes (6)) to reduce the background. It should also be performed as negative control for RNA-FISH. Treat for 30 min at 37°C in a humid chamber. The use of RNase inhibitor on RNA-FISH is most important when the nuclear transcripts of interest are expressed at very low level. For rRNA and var transcripts, the RNase inhibitor was not necessary (5, 6).
For a two-color DNA-FISH, the two differently labeled probes (e.g., fluorescein and biotin) can be mixed in the same hybridization solution and added to a single well (Fig. 2i).
Optional step: Before adding the probes, the parasite samples can be pre-hybridized or equilibrated with 50 μl of hybridization solution (denatured at 100°C for 5 min) for 30 min at 42°C (in a humid chamber). This blocking step may reduce background problems (14).
For DNA-FISH, the temperature and duration used for sample denaturation vary with the laboratory. Others have used 95°C for 2–5 min; we usually perform 80°C for 30 min, since lower temperatures appear to better conserve the nuclear structure. For RNA-FISH the denaturation step is not performed. However, the hybridization temperature can be modified from 37 to 42°C or 50°C in order to increase the specificity of probe binding.
For the washing steps, wrap the 50-ml tubes with aluminum foil to protect from light. It is possible to wash two slides at once by placing the two slides back to back. Avoid drying the slide while washing; make sure that the slides (wells) are immersed by the washing solutions. Transfer the slides from one tube to the other using forceps. To prevent background problems, do not reuse the washing solutions.
In case of high background on RNA-FISH, increase the temperature of the washing solution to 42°C or 50°C and/or include some shaking.
For sequential RNA/DNA-FISH, the combination that works best is first to perform the RNA-FISH using fluorescein-labeled probes and second the DNA-FISH using biotinylated probes (Fig. 2g). It is recommended to confirm that the RNA-FISH produced the expected signal on the first day. As such, one extra slide should be prepared and used as control. Wash the control slide as described in Subheading 3.2.5 and observe under the fluorescence microscope. If the RNA-FISH signals are fine in the control slide, then proceed for a second day of hybridization (DNA-FISH) on the other slide. In case of weak RNA-FISH signals, a fixation step with 4% PFA for 10–20 min can be done before proceeding for DNA-FISH (wash the wells three times with PBS). If RNA-FISH signals are barely seen or the background is extremely high, there is no point to carry on the DNA-FISH. The RNA-FISH should be restarted using different conditions for the probes and temperatures of hybridization or washing. Note that after all the procedures, part of the RNA-FISH signals is lost and the number of double-positive nuclei is usually no more than 30%.
In the immuno-FISH protocol, the second fixation with PFA prevents primary and secondary antibody dissociation under the harsh conditions used for the subsequent DNA-FISH.
The suspension of immunolabeled and post-fixed parasites can be stored at 4°C for a few weeks. This provides the possibility of stopping the experiment at this point and also of repeating the DNA-FISH later or to perform other combinations with DNA probes without the need to carry on a new IF. The combination that works best for immuno-DNA-FISH is to perform IF with red fluorescent secondary antibodies and DNA-FISH using fluorescein-labeled probes (Fig. 2h).
For a combined immuno-DNA-FISH protocol, be aware that denaturating temperatures higher than 72°C will revert the cross-link by the PFA and so the antibody staining will be lost.
In order to precisely define the nuclear boundaries, we have used an antibody against a putative nuclear pore protein and observed that the nuclear diameter delineated by the immunofluorescence signals was larger than that of the most intense DAPI staining (2.06 ± 0.30 vs. 1.67 ± 0.23 μm) (5). In contrast to the budding yeast model, inclusion of nuclear pore staining on a regular basis on Plasmodium FISH experiments is unworkable. Thus, to define the nuclear periphery for FISH quantitative analysis, we take into consideration the less intense extremity of the DAPI staining. This border is better visualized using a black/white image.
Acknowledgments
This work was supported by the French Agency for Research (ANR Blanc 0274-01) and European Research Council Executive Agency Advanced Grant (PlasmoEscape 250320).
References
- 1.Freitas-Junior LH, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 2.Ralph SA, et al. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci USA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voss TS, et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 4.Dzikowski R, et al. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Rubio JJ, et al. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Mancio-Silva L, et al. Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc Natl Acad Sci USA. 2010;107:15117–15122. doi: 10.1073/pnas.1001045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sexton T, et al. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 8.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 9.Freitas-Junior LH, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum . Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 10.Thompson J. In situ detection of RNA in blood- and mosquito-stage malaria parasites. Methods Mol Med. 2002;72:225–233. doi: 10.1385/1-59259-271-6:225. [DOI] [PubMed] [Google Scholar]
- 11.Li F, et al. Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J Biol Chem. 2008;283:5692–5698. doi: 10.1074/jbc.M707344200. [DOI] [PubMed] [Google Scholar]
- 12.Issar N, et al. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum . Microbes Infect. 2009;11:403–407. doi: 10.1016/j.micinf.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Mancio-Silva L, et al. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum . J Cell Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- 14.Contreras-Dominguez M, et al. A modified fluorescence in situ hybridization protocol for Plasmodium falciparum greatly improves nuclear architecture conservation. Mol Biochem Parasitol. 2010;173:48–52. doi: 10.1016/j.molbiopara.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Hediger F, et al. Methods for visualizing chromatin dynamics in living yeast. Methods Enzymol. 2004;375:345–365. doi: 10.1016/s0076-6879(03)75022-8. [DOI] [PubMed] [Google Scholar]
- 16.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei . Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]