Abstract
Endothelial lipase (EL) changes structural and functional properties of high-density lipoprotein (HDL). HDL is a relevant modulator of endothelial nitric oxide synthase (eNOS) activity, but the effect of EL on HDL induced eNOS-activation has not yet been investigated. Here, we examined the impact of EL-modified HDL (EL-HDL) on eNOS activity, subcellular trafficking, and eNOS- dependent vasorelaxation. EL-HDL and empty virus (EV)-HDL as control were isolated from human serum incubated with EL-overexpressing or EV infected HepG2 cells. EL-HDL exhibited higher capacity to induce eNOS phosphorylation at Ser1177 and eNOS activity in EA.hy 926 cells, as well as eNOS-dependent vasorelaxation of mouse aortic rings compared to control HDL. As revealed by confocal and structured illumination-microscopy EL-HDL-driven induction of eNOS was accompanied by an increased eNOS-GFP targeting to the plasma membrane and a lower eNOS-GFP colocalization with Golgi and mitochondria. Widefield microscopy of filipin stained cells revealed that EL-HDL lowered cellular free cholesterol (FC) and as found by thin-layer chromatography increased cellular cholesterol ester (CE) content. Additionally, cholesterol efflux capacity, acyl-coenzyme A: cholesterol acyltransferase activity, and HDL particle uptake were comparable between EL-HDL and control HDL. In conclusion, EL increases eNOS activating capacity of HDL, a phenomenon accompanied by an enrichment of the plasma membrane eNOS pool, a decreased cell membrane FC and increased cellular CE content.
Keywords: eNOS, HDL, Myography, Microscopy, Endothelial lipase
1. Introduction
Maintenance of the endothelial homeostasis is largely dependent on the activity of endothelial nitric oxide synthase (eNOS) and the bioa-vailability of nitric oxide (NO). eNOS is localized in caveolae, plasma membrane microdomains rich in caveolin-1 protein, which when bound to eNOS attenuates its enzymatic activity [1]. Increase in cytosolic calcium augments formation of calcium-calmodulin complexes, which facilitate eNOS translocation to the cytosol by displacing calmodulin-1 from eNOS. In the cytosol, eNOS undergoes post-translational modifications, primarily phosphorylation [1]. eNOS phosphorylation markedly affects its enzyme activity. While phosphorylation of eNOS on serine (Ser) 1177 facilitates interaction of eNOS with calmodulin and in turn augments enzyme activity, the phosphorylation on threonine (Thr) 495 interferes with the binding of calmodulin to the enzyme resulting in decreased eNOS activity [1].
High-density lipoprotein (HDL) exerts numerous beneficial effects on the cardiovascular system [2,3]. The best studied atheroprotective function of HDL is its role in the reverse cholesterol transport, a process by which HDL prevents accumulation of cholesterol in blood vessel walls [2,3]. Not less important is the role of HDL in the maintenance of endothelial function, which is accomplished by the induction of eNOS activity and consequently increased vascular NO bioavailability [2–4]. This process involves interaction of HDL with endothelial receptors, the scavenger receptor class B type 1 (SR-BI) and sphingosine-1 phosphate (S1P) receptors, leading to eNOS phosphorylation at Ser1177 and its activation [3,5–7].
Endothelial lipase (EL) is a strong negative regulator of HDL plasma levels [8] and a potent modulator of HDL structure and composition [9,10]. EL is mainly synthesized and secreted by vascular endothelial cells [11,12]. By its pronounced phospholipase activity, EL hydrolyzes HDL phospholipids (PL) thereby producing lysophospholipids (LysoPL) and fatty acids (FA). EL-mediated depletion of HDL-PL alters structural and functional properties of HDL [9,13].
Considering pronounced EL-mediated alterations in HDL composition, structure, and function [9,10,13], we hypothesized that the eNOS activating capacity of HDL is also altered by EL. Therefore, we studied the impact of EL-modified HDL (EL-HDL) on eNOS phosphorylation, activity, and subcellular localization as well as on eNOS-dependent vascular reactivity.
2. Materials and methods
2.1. Cell culture
HepG2 cells (ATCC®, HB-8065™) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS).The human endothelial cell line EA.hy 926 [14] kindly provided by Dr. C.J.S. Edgell (University of North Carolina, Chapel Hill, NC, USA) was cultured in DMEM containing 10% FCS and 1% HAT Media Supplement (all Gibco, Life Technologies, Vienna, Austria). The immortalized human umbilical vein endothelial cell line, ECV 304 stably expressing eNOS-GFP [15] kindly provided by Dr. W.C. Sessa (Yale University School of Medicine, New Haven, CT, US) was cultured in DMEM containing 10% FCS in the presence of G418 (200 μg/mL). DMEM used for culturing of all cell lines used in the present study was supplemented with penicillin G sodium sulfate (100 units/mL), streptomycin sulfate (100 mg/mL), and amphotericin B (2.5 mg/mL) (all Gibco, Life Technologies). Cells were cultured in humidified atmosphere of 5% CO2/95% air at 37 °C and were sub-cultured using 0.025% trypsin/0.01% EDTA.
2.2. Overexpression of SR-BI in EA.hy 926 cells
Cells were plated in 6-well dishes (35,000 cells/well) and incubated under standard conditions as described above. After 24 h, the cells were washed with DMEM without FCS and transduced with multiplicity of infection of 800, using recombinant adenoviruses (Ad) encoding human SR-BI [16] or empty adenovirus containing no recombinant DNA (EV-Ad) [13] in 1 mL DMEM without FCS for 1 h. Thereafter, 1 mL DMEM containing 20% FCS was added into 1 mL infection media. After 24 h, the cells were either lysed for Western blotting analysis of SR-BI over-expression or incubated with EL-HDL or control empty virus (EV)-HDL followed by determination of eNOS activity as described below.
2.3. Human serum
Serum was obtained from healthy volunteers after overnight fasting. The local Ethics Committee of the Medical University of Graz approved all experimental protocols related to human volunteers (28–186 ex 15/ 16). Written informed consent was obtained from each subject in compliance with Good Clinical Practice.
2.4. Preparation of EL-HDL and control empty virus (EV)-HDL
HepG2 cells (2×106) plated onto 60mm dishes were transduced at a multiplicity of infection of 20 using recombinant Ad encoding human EL (EL-Ad) or EV-Ad [13]. Transduced HepG2 cells were incubated under cell culture conditions with 1.8 mL of 50% pooled human serum in DMEM without FCS for 8 h. EV-HDL and EL-HDL were isolated from modified serum as described [13]. Briefly, the density-adjusted serum (1.24 g/mL with potassium bromide) was layered underneath a potassium bromide-density solution (1.063 g/mL). Samples were centrifuged at 330,000×g for 6 h (centrifuge: Beckman Optima L-80 ultracentrifuge, rotor: Sorvall T-1270). Thereafter, the collected HDL was concentrated by Viva Spin Tubes (Sartorius, Vienna, Austria), desalted by gel filtration on Sephadex PD-10 columns (GE Healthcare, Munich, Germany) and either used directly or stored at – 80 °C. HDL protein concentration was determined with a NanoDrop ND-1000 (PeqLab Biotechnologie, Erlangen, Germany). Experiments were performed with 100 μg/mL of EV-HDL protein or EL-HDL protein.
2.5. FPLC
ÄKTA pure FPLC System (GE Healthcare, Munich, Germany) equipped with a Superdex 200 Increase 10/300 column (GE Healthcare, Munich, Germany) was used with phosphate buffered saline (PBS) as running buffer. After loading, HDL samples were separated with a constant flow of 0.5 mL/min and fractionated in a range of 9 to 13.5 mL elution volume with 0.5 mL per fraction. The software UNICORN (GE Healthcare, Munich, Germany) was used for analysis.
2.6. Western blot analyses
EA.hy 926 cells were plated either onto 24 (76,000 cells/well)- or 6 (350,000 cells/well)- well plates. Confluent cells were transduced with SR-BI-Ad or EV-Ad or treated with 100 μg/mL EV-HDL or EL-HDL in DMEM without FCS for 16 h. Thereafter, cells were placed on ice, washed once with ice cold PBS and lysed in RIPA buffer (Thermo Fisher Scientific, Schwerte, Germany) supplemented with a protease inhibitor cocktail (1 μL/million cells) (Sigma-Aldrich, Vienna, Austria) and 100 μM of the phosphatase inhibitor sodium orthovanadate (Sigma- Aldrich, Vienna, Austria).
Ten μg of cell lysate protein were supplemented with loading buffer, boiled for 10 min and subjected to gel electrophoresis using 10% SDS-polyacrylamide gels followed by transfer to PVDF or nitrocellulose membrane. PeqGOLD protein Marker IV (Peqlab, Erlangen, Germany) was used as a standard. Proteins were detected with antibodies specific against total eNOS, eNOS phosphorylated at Ser 1177 (both BD Transduction Laboratories, Biocompare, San Francisco, CA, USA), SR-BI (Abcam, ab 36,970, Cambridge, UK) or α-tubulin (Cell signaling technology, 11H10, Leiden, Netherlands), followed by appropriate HRP-conjugated secondary antibodies (Dako, Vienna, Austria). Signals were visualized using Millipore Western Blotting Substrate (Millipore Corporation, Billerica, USA). Densitometric analyses were performed using Image Lab software (Bio-Rad Laboratories, Vienna, Austria).
2.7. eNOS activity assay
EA.hy 926 cells were plated (250,000 cells/well) onto 6 well plates. After 24 h, confluent cells were treated with 100 μg/mL EV-HDL or EL-HDL in DMEM without FCS for 5 or 16 h. Intracellular conversion of L- [3H]arginine into L-[3H]citrulline was measured as previously described [17]. Briefly, after incubation with EV-HDL or EL-HDL, cells were washed and incubated at 37 °C with 50 mmol/L Tris buffer, pH 7.4, containing 100 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L MgCl2, 3 mmol/L CaCl2, 5% (vol/vol) FCS, and L-[2,3-3H]arginine (~106 dpm) in the absence or presence of 0.5 μmol/L calcium iono- phore A23187. Reactions were terminated after 5 min by washing the cells with chilled Tris buffer (50 mmol/L, pH 7.4), containing 100 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L MgCl2 and 0.1 mmol/L EGTA. Following cell lysis with 0.01 N HCl, an aliquot was removed for determination of incorporated radioactivity. To the remaining sample, 200 mmol/L sodium acetate buffer (pH 13.0) containing 10 mmol/L L-citrulline was added (final pH~ 5.0), and L-[3H]citrulline was separated from L-[3H]arginine by cation exchange chromatography. eNOS activity was expressed as the L-[3H]citrulline radioactivity relative to total radioactivity in cell lysates.
2.8. Imaging of NO signals
150,000 Ea.hy 926 cells per well were seeded in 6-well plates 72 h before measurements. Infection of cells with adeno-associated virus encoding green genetically encoded fluorescent NO probe (G-geNOp) (NGFI, Graz, Austria) [18,19] was performed 48 h prior to imaging according to the manufacturer instruction. After 3 washing steps with PBS the cells were treated with the iron booster solution (NGFI, Graz, Austria) for 20 min followed by 3 washing steps with PBS and incubation with 100 μg/mL EV-HDL or EL-HDL in DMEM without FCS for 16 h. Cells were washed once with loading-buffer (2 mmol/L CaCl2, 135 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L MgCl2, 1 mmol/L HEPES, 2.6 mmol/L NaHCO3, 0.44 mmol/L KH2PO4, 0.34 mmol/L Na2HPO4, 10 mmol/L D-glucose (Roth), 0.1% vitamins, 0.2% essential amino acids and 1% penicillin/streptomycin (Gibco) at pH 7.4) and the G-geNOp fluorescence was acquired during gravity-driven perfusion of cells with Ca-Buffer (CaB) (2 mmol/L CaCl2, 140 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L MgCl2, 1 mmol/L HEPES and 10 mmol/L d-Glucose, pH adjusted to 7.4).Thereafter, cells were challenged with 100 μmol/L his-tamine, followed by a wash out and 300 μmol/L eNOS inhibitor Nω- nitro-L-arginine (L-NNA). Finally, 10 μmol/L NO donor sodium nitroprusside (SNP) freshly dissolved and prepared for each run was added as an internal control (all buffer salts were obtained from Roth, Graz, Austria).
NO measurements were performed on an Olympus IX73 equipped with a UApoN340 40× oil immol/Lersion objective (Olympus, Japan) and a CCD Retiga R1 camera (Q-imaging, Canada). For illumination a LedHUB® (Omnicron, Germany) equipped with a 470 nm LEDs in combination with a standard GFP filter set (GFP-3035D, Semrock, USA) was used for acquiring G-geNOp fluorescence. During the measurements cells were continuously perfused by a gravity-based perfusion system (NGFI, Graz, Austria). Data acquisition and control of the fluorescence microscope was performed using Visiview 4.2.01 (Visitron, Germany).
Recordings were background subtracted and bleaching corrected using an exponential decay function. To obtain suitable NO readings from the raw data three analysis steps were performed:
-
(1)
As geNOp signals are negatively correlated with the NO levels, the acquired G-geNOp signals were normalized by calculating:
N(t) = 1-Ft/F0 [18].
N: normalized geNOp signal; Ft: geNOp fluorescence over time; F0: geNOp fluorescence at time point 0.
-
(2)
The measurement was normalized to the minimal signal obtained during L-NNA perfusion to obtain basal NO values:
N1(t) = Nt-NL-NNA; min.
N1: geNOp signal normalized to the minimal response obtained in the presence of L-NNA; Nt: normalized geNOp signal over time (i.e. N). NL-NNA;min: minimal geNOp signal in the presence of L-NNA.
-
(3)
To increase the accuracy of the geNOp measurements 10 μM SNP-evoked signals were taken for every measurement as a reference to normalize histamine-induced NO signals:
N2(t) = N1t/N1snp; max.
N2: geNOp fluorescence signal over time normalized to both the minimal geNOp signal in response to L-NNA and the individual maximal response to SNP; N1t: geNOp signal over time normalized to the minimum obtained in the presence of L-NNA (i.e. N1(t)): N1snp;max: individual maximal geNOp signal induced by SNP.
Using the experimental internal references L-NNA and SNP the basal and histamine-induced maximum delta values of NO were determined as fold change of the G-geNOp signal relative to the internal reference SNP and used as a direct measure of cytosolic NO concentrations.
2.9. ATTO-HDL labeling
FPLC-purified EV-HDL and EL-HDL were labeled with ATTO-594- NHS-ester dye (ATTO-TEC GmbH, Germany) according to the manu-facturer's instructions. Labeled HDL (ATTO-HDL) was subsequently separated from unlabeled HDL and free ATTO-594 by gel filtration on Sephadex PD-10 columns (GE Healthcare, Munich, Germany) and either used directly or stored at – 80 °C for further experiments. The degree of HDL ATTO-labeling (DOL) was determined according to the manufacturer's instruction and HDL with comparable DOLs were used for experiments.
2.10. Overexpression of eNOS-GFP and G-geNOp
EA.hy 926 cells (250,000 cells/well) were plated on 30 mm- or 1.5H high-precision- glass coverslips (Marienfeld-Superior, Lauda Königshofen, Germany) and grown until 70% confluency. Cells were then transfected with a mixture of 1.5 μg of plasmids encoding eNOS-GFP [20] or G-geNOp [19] and 3 μL of Transfast™ transfection reagent (Promega, Madison, WI, USA) in serum- and antibiotic-free DMEM. After 4 h, the medium was replaced by a standard culture medium. In order to increase the transfection rate, the transfection procedure was repeated after 24 h.
2.11. Overexpression of Golgi-red fluorescence protein (RFP) and plasma membrane-RFP
ECV 304 (150,000 cells/well) or EA.hy 926 cells (250,000 cells/ well) were plated on 30 mm- or 1.5H high-precision- glass coverslips (Marienfeld-Superior, Lauda Königshofen, Germany) and grown until 70% confluency. Cells were infected with 30 particles per cell (ppc) of CellLight® Golgi-RFP, BacMam 2.0 or 30 ppc of CellLight® plasma membrane-RFP, BacMam 2.0 (ThermoFisher Scientific, Vienna, Austria) in culture medium containing antibiotics without serum. The cells were either imaged directly after l6h incubation with EV-HDL or EL-HDL or after a second round of infection.
2.12. Labelling of mitochondria with MitoTracker® Red FM
Cells were incubated with a loading buffer (135 mmol/L NaCl, 5 mmol/L KCl, 2 mmol/L CaCl2, 1 mmol/L MgCl2, 10 mmol/L Hepes, 2.6 mmol/L NaHCO3, 440 mmol/L KH2PO4, 340 mmol/L Na2HPO4, 10 mmol/L d-glucose, 0.1% vitamins, 0.2% essential amino acids, and 1% penicillin–streptomycin, pH adjusted to 7.4) containing 2.5 μmol/L MitoTracker® Red FM (MTR) (ThermoFisher Scientific, Vienna, Austria) for 10 min at 37 °C. Cells were washed twice with loading buffer and imaged afterwards.
2.13. Confocal, widefield and structured illumination microscopy (SIM)
Confocal images were acquired with a Zeiss Observer Z.1 inverted microscope equipped with a Yokogawa CSU-X1 Nipkow spinning disk system, a piezoelectric z-axis motorized stage (CRWG3-200; Nippon Thompson Co., Ltd., Tokyo, Japan), and a CoolSNAP HQ2 CCD Camera (Photometrics, Tucson, USA). The microscope was equipped with 405-, 488-, 515-, 532-, and 561-nm laser lines (Visitron Systems) and an alpha Plan-Fluar 100x/1.45 Oil M27 (Zeiss, Jena, Germany) objective was used.
The N-SIM-setup (Nikon Austria, Vienna) was equipped with 405-, 488-, 515-, 532-, and 561-nm excitation lasers introduced at the back focal plane inside the SIM box with a multimodal optical fiber. For super-resolution, a CFI SR Apochromat TIRF 100×-oil (NA 1.49) objective was mounted on a Nikon-Structured Illumination Microscope (N-SIM®).
The system with standard wide field and SIM filter sets was equipped with a dual-SIM Andor iXon3® EMCCD camera. SIM images were calibrated and reconstructed using the Nikon software Nis- Elements. General resolution (full width at half maximum; FWHM) measured with fluorescent 100-nm Tetraspec beads (Invitrogen™, Thermo Fisher Scientific, Vienna, Austria) was approximately 120 to 130 nm.
Widefield measurements were performed on an inverted and advanced fluorescent microscope with a motorized sample stage (Till Photonics, Graefling, Germany). The probes were excited via a polychrome V (Till Photonics), and emission was visualized using a 40× objective (alpha Plan Fluar 40, Zeiss, Goettingen, Germany) and a charge-coupled device camera (AVT Stringray F145B, Allied Vision Technologies, Stadtroda, Germany).
CaB was used as a general imaging buffer (all buffer salts were from Roth, Graz, Austria).
2.14. Quantification of eNOS-GFP fluorescence in perinuclear region
ECV304 cells stably expressing eNOS-GFP were treated with 100 μg/mL EV-HDL or EL-HDL in DMEM without FCS for 16 h. Confocal microscopy of eNOS-GFP fluorescence in the regions of interest (ROIs) was performed immediately after incubation with HDLs and during subsequent incubation of the cells with 0.25 μmol/L A23187 for 500 s. ROIs were set manually to contain the whole perinuclear region and the size equivalent to the cytosolic region. After background subtraction with the tool included in the Mosaic suite (Mosaic group) the ratio of perinuclear to cytosolic fluorescence intensity was taken as a measure of eNOS-GFP abundance in the perinuclear region. Images were analyzed using ImageJ software.
2.15. Colocalization of eNOS-GFP with Golgi and mitochondria
eNOS-GFP-overexpressing ECV 304 and EA.hy 926 cells were infected with CellLight® Golgi-RFP or labeled with 2.5 μmol/L MitoTracker™ Red FM (Thermo Fisher) for 10 min and incubated with 100 μg/mL EV-HDL or EL-HDL in DMEM without FCS for 5 or 16 h prior to imaging. 5×5 fields of view on the Nikon N-SIM device were imaged with the dual-cam setup. After background subtraction with the Mosaic suite (Mosaic group) included tool, the 5×5 images of each channel were stitched together with the Grid/Collection stitching imageJ plugin [21]. Cells were selected manually within the stitched images and Fiji's coloc2 was used to determine the Pearson's co-localization coefficient.
2.16. Colocalization of eNOS-GFP with plasma membrane
EA.hy 926 cells co-overexpressing plasma membrane-RFP and eNOS-GFP were incubated with 100 μg/mL EV-HDL or EL-HDL for 16 h followed by acquiring confocal dual color images. Background was subtracted using the histogram based Mosaic suite (Mosaic group) [21]. Internalized RFP signal was cropped out of the image manually using ImageJ to obtain a clear plasma membrane-staining. Afterwards the Pearson's co-localization coefficient between the eNOS-GFP and plasma membrane-RFP channel was determined using Fijis coloc2 plugin.
2.17. Internalization of ATTO-labeled EV-HDL and EL-HDL
EA.hy 926 cells (250,000/well) overexpressing the cytosolic marker geNOP-GFP [19] were incubated with 100 μg/mL EV-ATTO-HDL or EL-ATTO-HDL for 1 or 16 h followed by acquiring of confocal z-stacks with a stepping of 0.2 μm. Total ATTO-HDL fluorescence intensity per cell was determined after blind deconvolution of the acquired dual color z-stacks using NIS-Elements. ATTO-HDL vesicles were identified by auto Otsu thresholding using the stack histogram. Count, volume, surface, and total fluorescence intensity of the vesicles containing ATTO-HDL were measured with the ImageJ 3D Manager using the cytosolic geNOP-GFP as a mask to analyze exclusively ATTO-HDL internalized by cells.
2.18. Determination of cellular free cholesterol (FC) content using filipin staining
EA.hy 926 cells were incubated with 100 μg/mL of ATTO-EV-HDL or ATTO-EL-HDL in DMEM without FCS for 1, 5 or 16 h, respectively. After washing with warm PBS, cells were fixed with 4% paraformaldehyde for 15 min. FC was visualized by staining with 0.05 mg/ mL filipin (Sigma Aldrich, Vienna, Austria) (diluted in PBS) for 2 h. Imaging was performed immediately after staining on a wide field microscope using 361 ± 10 and 580 ± 10 nm excitation and 510 nm (495dcxru; Omega Optical, Brattleboro, VT, USA) and 635 nm (59,004; Chroma, Bellows Falls, VT, USA) emission for capturing the fluorescence of filipin and ATTO-HDL, respectively. The illumination intensity was kept constant for all experiments. Cells in the field of view were counted manually and the cell area was measured using auto Li threshold binary image. The mean filipin fluorescence intensity per cell was normalized to the cell area. To obtain filipin fluorescence associated exclusively with cell membranes i.e. without the filipin fluorescence associated with ATTO-HDL-containing vesicles the ATTO-HDL channel thresholded by an auto Otsu threshold was used as a mask to subtract filipin fluorescence colocalized with ATTO-HDL.
2.19. Cholesterol efflux capacity of EV-HDL and EL-HDL
Cholesterol efflux capacity was assessed using an established assay with minor modifications [13,22]. Briefly, EA.hy 926 cells were plated on 48-well plates (40,000 cells/well). Cells were labeled with 1 μCi/mL [3H]-cholesterol for 24 h (Perkin Elmer, Boston, MA, USA). After labeling, cells were washed twice with serum free DMEM and subsequently equilibrated in serum-free DMEM containing 0.2% BSA for 2 h. After additional two washing steps, [3H]-cholesterol efflux was determined by incubating cells for 1, 3, 5, or 16 h with serum- free DMEM containing 100 μg/mL EV-HDL (protein) or EL-HDL (protein), respectively. Cholesterol efflux was expressed as the radioactivity in the medium relative to total radioactivity in medium and cells. All steps were performed in the presence of 2 μg/mL of the acyl-coenzyme A: cholesterol acyltransferase (ACAT) inhibitor Sandoz 58-035 (Sigma, Darmstadt, Germany).
2.20. Thin-layer chromatography (TLC) analysis of cell lipids
EA.hy 926 cells plated in 6-well plates were incubated with 100 μg/ mL of EV-HDL or EL-HDL for 16 h. Thereafter, the cells were washed with PBS and lipids were extracted with hexane/isopropanol (3:2, v:v), extracts evaporated in the SpeedVac and redissolved in chloroform before TLC using hexane-diethyl ether-glacial acetic acid (70:29:1, v:v:v) as mobile phase. The signals corresponding to PL, FC, FA, triglycerides, and cholesterol ester (CE) were visualized by primulin and the signal intensity evaluated by densitometry. Intra-plate-variations of the FC and CE signals in EV-HDL samples were 8 and 5% and in EL-HDL samples 8 and 6%, respectively. The inter-plate-variations of the FC and CE signals in EV-HDL samples were 26 and 23% and in EL-HDL samples 13 and 17%, respectively. The delipidated cell layers were lysed with 1 mL of 0.3 mol/L NaOH/0.1% SDS and the protein content was determined with Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Schwerte, Germany).
2.21. Analysis of ACAT activity by [3H] oleate pulse
Albumin-oleate complex was prepared according to [23] with slight modifications. In brief, radiolabeled oleic acid [9,10-3H]; 50 Ci/mmol; (Hartmann Analytic, Braunschweig, Germany) was dried in a glass tube under a stream of nitrogen. 10 μL of non-radiolabeled 20 mmol/L sodium oleate (Merck, Vienna, Austria) was added per 1 μCi dried [3H] oleic acid followed by vortexing. Finally, an equal volume of 20% fatty acid-free BSA solution (Merck, Vienna, Austria) was added and vortexed. EA.hy 926 cells were incubated with 100 μg/mL EV-HDL or EL-HDL for 5 or 16 h, washed with PBS and pulsed with [3H] oleic acid/ BSA for 30 min as described [24]. Cells were washed twice with ice-cold PBS and extracted with hexane/isopropanol (3:2, v/v), extracts evaporated in the SpeedVac and redissolved in chloroform before TLC using Silica Gel 60 plastic sheets (20×20 cm; Merck, Austria) and a solvent system for the separation of neutral lipids (hexane/diethyl ether/acetic acid, 70:29:1, vol/vol). After primuline staining lipids were visualized using an UV lamp (366 nm) and [3H]CE and [3H]tria-cylglycerol (TAG) spots were cut out for scintillation counting. Following lipid extraction, cells were lysed with 0.3 mol/L NaOH/0.1% SDS, and the protein concentrations were determined according to [25].
2.22. Vasorelaxing capacity of EV-HDL and EL-HDL
Aortic rings, approximately 2mm in length were cut from descending thoracic aorta from 14 weeks old male C57BL/6 mice. The arterial rings were positioned in small wire myograph chambers (Danish MyoTechnology, Aarhus, Denmark), which contained physiological salt solution (PSS) (119 mmol/L NaCl, 4.7 mmol/L KCl, 1.18 mmol/L KH2PO4, 1.2 mmol/L MgCl2, 2.5 mmol/L CaCl2, 25 mmol/ L NaHCO3, 0.03 mmol/L EDTA-Na2·2H2O, and 5.5 mmol/L d-glucose pH 7.4) aerated with 5% CO2/95% O2 at 37 °C. The myograph chambers were connected to force transducers for isometric tension recording (PowerLab, ADInstruments). The rings were kept in PSS buffer at 37 °C. A normalization procedure was performed in order to estimate the luminal diameter (passive tension) for each aortic ring according to the manufacturer's instructions (Multiwire myograph system – model 620M; Danish MyoTechnology, Aarhus, Denmark) followed by stabilization period for 20 min. Modified PSS containing high concentrations of KCl (58.88 mmol/L KCl, 1.17 mmol/L MgSO4·7H2O, 1.18 g/L KH2PO4, 2.5 mmol/L CaCl2·2H2O, 25 mmol/L NaHCO3, 0.03 mmol/L EDTA-Na2·2H2O, 5.5 mmol/L D-glucose) was used to determine maximum contractility of the tissue. When the developed tension attained its peak value, the rings were relaxed by rinsing with the buffer. Next, the rings were contracted with increasing concentrations of norepinephrine (NE) (1 nmol/L – 0.3 μmol/L) (Sigma-Aldrich) to produce 80% of the maximum contraction achieved by modified PSS followed by endothelium-dependent relaxation to cumulatively increasing concentrations of acetylcholine chloride (1 nmol/L – 0.3 μmol/L) (Sigma- Aldrich). After washout and equilibration, the rings were constricted with one dose of NE in the presence or absence of L-NNA (300 μmol/L) to 80% of the maximal constriction achieved by modified PSS [17]. Thereafter, 100 μg/mL EV-HDL protein or EL-HDL protein were added to the rings in the absence or presence of L-NNA (300 μmol/L). HDL-induced relaxation was expressed as a percentage of the initial NE-induced constriction.
2.23. Impact of EV-HDL and EL-HDL on contractile response to L-NNA
Aortic rings were isolated as described above. Each ring was incubated in a separate well of a 96-well plate with 150 μL DMEM without FCS containing 100 μg/mL EV-HDL or EL-HDL in a cell culture incubator at 37 °C for 16 h. Thereafter, the rings were transferred to the myograph, followed by stabilization in PSS, determination of passive tension and exposure to modified PSS, containing high concentrations of KCl for maximal constriction. After relaxation achieved by switching from modified PSS to PSS, the rings were exposed to the required NE concentrations for 10% of the maximal KCl-induced constriction, followed by addition of L-NNA (300 μmol/L). The ratio of constriction observed after and before addition of L-NNA is indicative of NO availability, whereby a higher ratio indicates a higher NO availability [17].
All animals received care in accordance with the Austrian law on experimentation with laboratory animals, which is based on the US National Institutes of Health guidelines. Experiments were approved by the Austrian Federal Ministry for Education, Science, and Research (BMWF-66.010/0020-WF/V/3b/2016).
2.24. RNA isolation and quantitative real-time PCR analysis
Total RNA was isolated using TriFast™ reagent according to the manufacturer's protocol (Peqlab, Erlangen, Germany). One μg of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). Quantitative real-time PCR was performed using the GoTaq® qPCR MasterMix (Promega, Madison, WI) on a Bio-Rad CFX96 Touch™ Real-Time PCR (Bio-Rad, Hercules, CA). Expression profiles and associated statistical parameters were determined using the 2–ΔΔCT method. Following primers were used: abca1 (forward: acatcctgaagccaatcctga; reverse: ctcctgtcgcatgtcactcc), abcg1 (forward: attcagggacctttcctattcgg; reverse: ctcaccactattgaacttcccg), scarb1 (forward: acttctggcattccgatcagt; reverse: acgaagcgataggtggggat).
2.25. Statistical analysis
Data were expressed as mean ± standard error of mean (SEM). Statistical significance was determined by unpaired, two-tailed t-test, one-way ANOVA with the Tukey's post-hoc test, or multiple t-test with a Sidak-Bonferroni correction for multiple comparison; p-values< 0.05 were considered significant. Statistical analyses were performed using GraphPadPrism (version 6.01).
3. Results
3.1. EL-HDL exhibits increased eNOS activating and vasorelaxing capacity
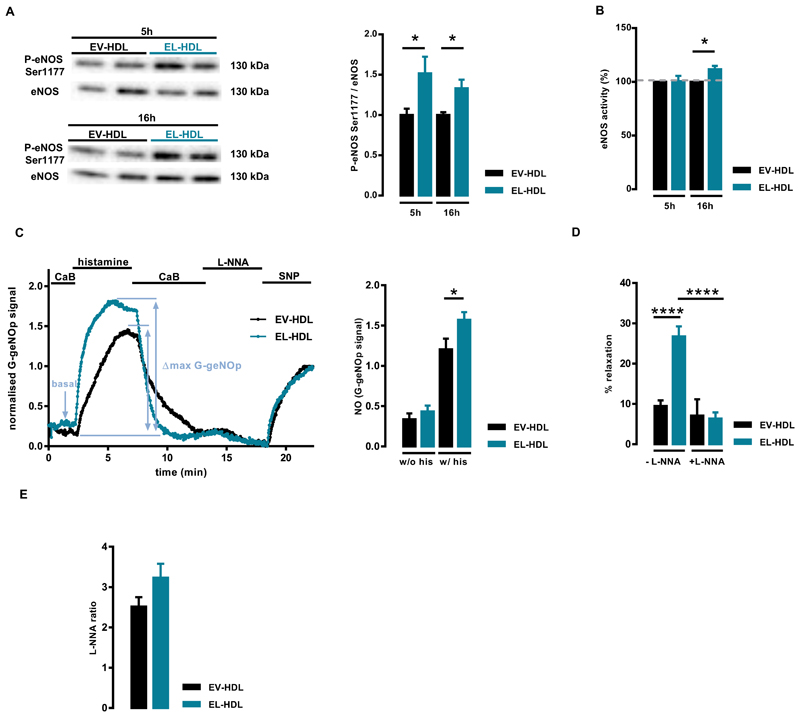
To assess eNOS activating capacity of EL-HDL, we examined the rate of eNOS phosphorylation at Ser1177 and eNOS activity in EA.hy 926 cells incubated with EV-HDL or EL-HDL for 5 and 16 h, respectively. These time points were selected due to the results of the initial time course experiments. The strongest induction of eNOS phosphorylation by EL-HDL was observed after 5 and 16 h is the latest time-point at which EL-HDL-induced eNOS phosphorylation was still significantly higher compared to EV-HDL (Suppl. Fig. S1). The impact of EV-HDL and EL-HDL on eNOS phosphorylation was not studied at time points earlier than 1 h of exposure because mechanical forces induced by changing of cell culture medium and/or washing of cells strongly and rapidly induced eNOS phosphorylation (Suppl. Fig. S2). As shown in Fig. 1, eNOS phosphorylation at Ser1177 was significantly higher at both time points in the cells incubated with EL-HDL compared to EV-HDL. However, eNOS activity was significantly higher in EL-HDL treated cells only after 16 but not 5 h when compared to EV-HDL (Fig. 1B). Comparable results were obtained when we measured eNOS activity in the presence of the calcium ionophore A23187 (Suppl. Fig. 3). Furthermore, histamine-induced cytosolic NO levels were significantly higher in EL-HDL treated cells, whereas basal NO levels were only slightly and not significantly higher in these compared to EV-HDL treated cells (Fig. 1C). Of note, relatively long incubation times of EV-HDL and EL-HDL with EA.hy 926 cells might have affected structural and functional properties of HDLs. Accordingly, it is possible that the observed augmented eNOS-inducing capacity of EL-HDL was not solely due to the EL-induced changes in HDL functionality but also, to a lesser extent, due to the modifications of EL-HDL entailed by incubation with EA.hy 926 cells.
Fig. 1.
EL-HDL exhibits increased eNOS activating and vasorelaxing capacity. (A) EA.hy 926 cells were incubated with 100 μg/mL EV-HDL protein or EL-HDL protein for 5 and 16 h, respectively. Ten μg of cell lysate protein were separated by 10% SDS-PAGE followed by Western blotting analyses of eNOS and eNOS phosphorylated at Ser1177. Protein size annotations refer to protein marker bands on the membrane. (B) Cells were treated with 100 μg/mL EV-HDL protein or EL-HDL protein for 5 and 16 h followed by incubation with L-[3H]arginine for 5 min and subsequent quantification of L-[3H]citrulline after its separation from L-[3H]arginine by exchange chromatography. eNOS activity is presented as % conversion of L-[3H]arginine to L-[3H]citrulline; the values in cells incubated with EV-HDL were set to 100%. The values in (A) and (B) are mean ± SEM of 3 or 4 independent experiments, respectively, performed in duplicate (A) or triplicate (B) and analyzed by two-tailed unpaired t-test; *p < 0.05. (C) Left panel: Representative normalized curves of G-geNOP signals obtained in the G-geNOp-expressing Ea.hy 926 cells incubated with 100 μg/mL EV-HDL protein or EL-HDL protein for 16 h. The G-geNOp fluorescence was acquired during perfusion of the cells with a calcium-buffer (CaB), followed by 100 μmol/L histamine, wash out and eNOS inhibitor Nω-nitro-L-arginine (L-NNA, 300 μmol/L). Finally, 10 μmol/L NO donor sodium nitroprusside (SNP) was added as an internal reference for normalization. Right panel: Basal and histamine-induced G-geNOp signals reflecting respective cytosolic NO levels/signals normalized to 10 μmol/L SNP. The values are mean ± SEM of 3 independent experiments with a total of 31 (EV-HDL) and 27 (EL-HDL) imaged cells analyzed by twotailed unpaired t-test; *p < 0.05. (D) Mouse aortic rings were precontracted with norepinephrine in the absence or presence of the eNOS inhibitor Nω-nitro-L-arginine (L-NNA, 300 μmol/L) followed by the addition of 100 μg/mL EV-HDL protein or EL-HDL protein onto precontracted rings in the absence or presence of L-NNA. HDL-induced relaxation was expressed as a percentage of the initial norepinephrine-induced contraction. The values are mean ± SEM obtained with 11–16 rings for each condition analyzed by two-tailed unpaired t-test; ****p < 0.0001. (E) Aortic rings were incubated in DMEM without FCS containing 100 μg/mL EV-HDL or EL-HDL in cell culture incubator at 37 °C for 16 h. Thereafter, the rings were transferred into the myograph and precontracted to 10% of the maximal KCl contraction followed by addition of 300 μmol/L L-NNA. The L-NNA ratio of contraction achieved after and before addition of L-NNA is shown. The values are presented as mean ± SEM of 16 rings for each condition analyzed by two-tailed unpaired t-test.
Functional implication of EL-HDL-induced eNOS activity was ex-amined ex vivo using mouse aortic rings and wire myography, a method suitable for testing the impact of various compounds, including HDL, on vascular constriction and relaxation. These experiments showed, that EL-HDL-induced vasorelaxation of mouse aortic rings constricted with noradrenaline was significantly stronger compared to EV-HDL and, importantly, could be completely inhibited with the eNOS inhibitor, L-NNA (Fig. 1D). In addition, the contractile response of mouse aortic rings to L-NNA (indicative of NO bioavailability), tended to be higher in rings which were incubated with EL-HDL prior to myography compared to those incubated with EV-HDL (Fig. 1E).
3.2. EL-HDL alters subcellular localization of eNOS
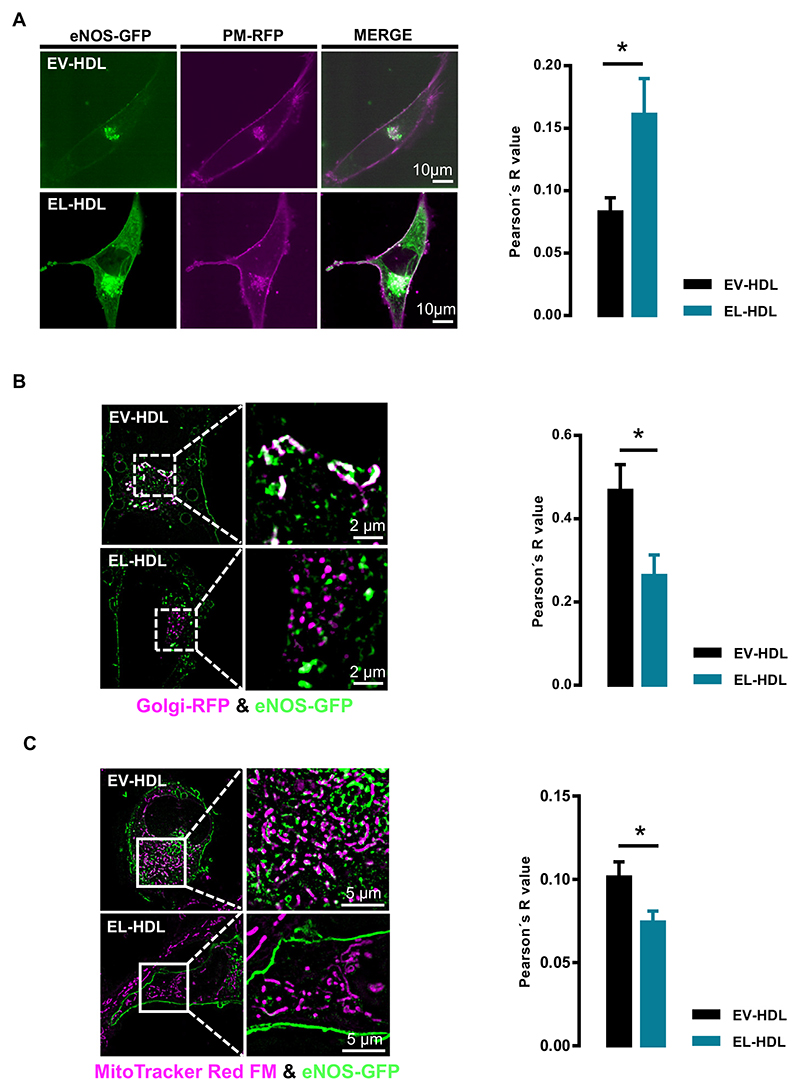
Since eNOS activity is largely dependent on its subcellular localization [26–29], we expected the increased eNOS activity observed after 16 h incubation with EL-HDL being accompanied by an altered subcellular localization of eNOS. Indeed, the colocalization of eNOS-GFP with plasma membrane-RFP was significantly higher after 16 h (Fig. 2A), whereas it was significantly lower with Golgi and mitochondria (Fig. 2B, C and Suppl. Fig. 4A, B) in EA.hy 926 cells incubated with EL-HDL compared to EV-HDL. Similar results were obtained in ECV 304 cells (Suppl. Fig. 5A, B and Suppl. Fig. 6A, B). In line with the decreased A23187-induced eNOS-GFP colocalization with Golgi (Suppl. Fig. 5A), the kinetics experiments revealed a slower A23187-induced eNOS-GFP shuttling to the perinuclear region in EL- HDL compared to EV-HDL treated cells (Suppl. Fig. 7). In contrast to what we observed after 16 h of incubation, the impact of EV-HDL and EL-HDL on eNOS-GFP colocalization with Golgi (Suppl. Fig. 8 A,B) and mitochondria (Suppl. Fig. 9 A,B) did not differ significantly after 5 h of incubation.
Fig. 2.
Impact of EL-HDL on subcellular localization of eNOS-GFP. EA.hy 926 cells overexpressing eNOS-GFP and (A) plasma membrane-RFP (PM-RFP) or (B) Golgi-RFP, or (C) stained with MitoTracker® Red were incubated with 100 μg/mL EV-HDL or EL-HDL for 16 h followed by (A) confocal microscopy or (B, C) structured illumination microscopy. Representative images and the Pearson's co-localization coefficients are shown. The colocalizations of eNOS-GFP (green) with (A) PM-RFP (magenta), (B) Golgi-RFP (magenta) or (C) MitoTracker® Red FM (magenta) are indicated in white. The values are expressed as mean ± SEM of 4 independent experiments performed in duplicate and analyzed by two-tailed unpaired t-test; *p < 0.05.
3.3. EL-HDL decreases cellular FC content
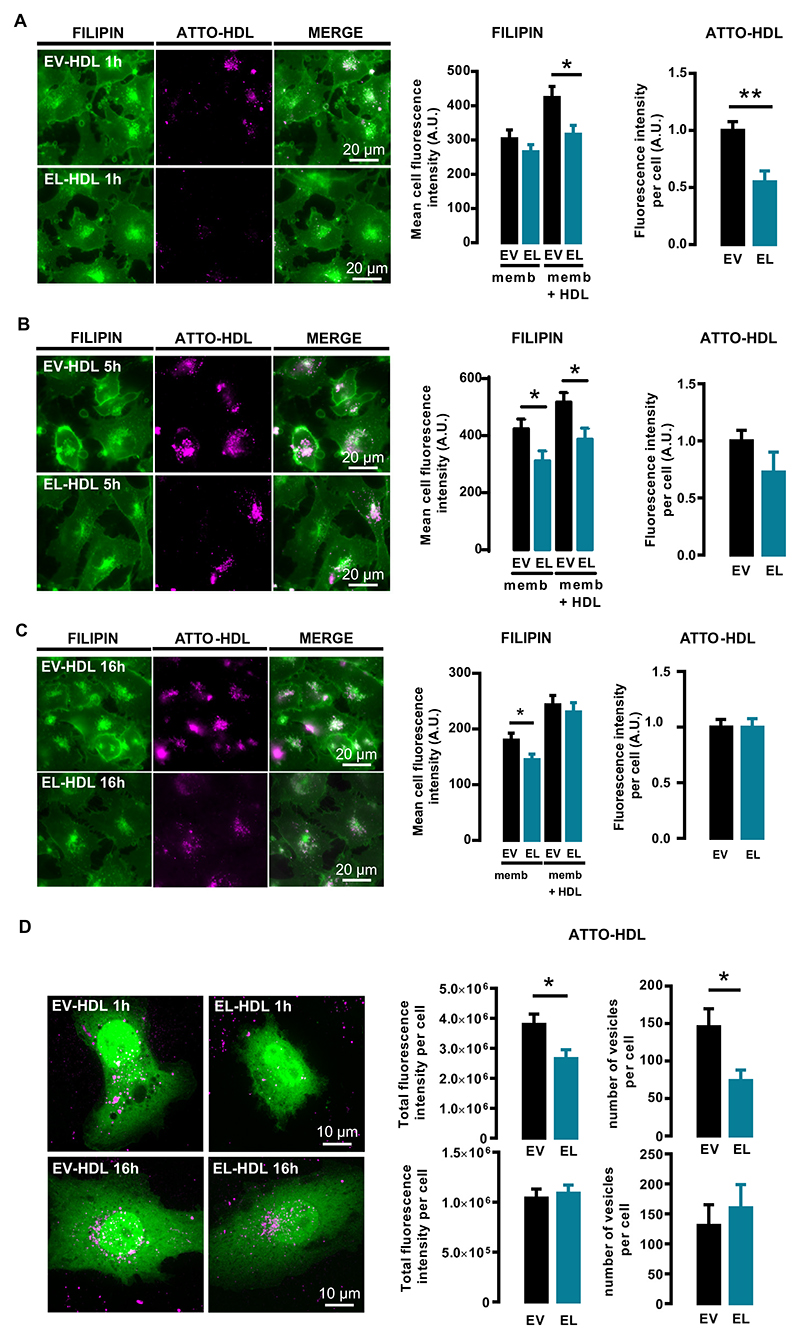
The FC content of cellular membranes affects eNOS activity and subcellular trafficking [29,30]. We therefore examined the cellular FC content by quantifying filipin fluorescence in filipin-labeled cells exposed to ATTO-EV-HDL or ATTO-EL-HDL for 1, 5 or 16 h, respectively. After 1 h, the total filipin fluorescence (memb + HDL), which is the sum of the filipin fluorescence associated with cellular membrane structures and the filipin fluorescence associated with ATTO-HDL-containing vesicles, was significantly lower in the ATTO-EL-HDL compared to EV-HDL-ATTO treated cells (Fig. 3A). In contrast, the filipin fluorescence associated with cellular membrane structures (memb) was comparable between ATTO-EV-HDL and ATTO-EL-HDL treated cells (Fig. 3A). In addition, the total ATTO-fluorescence and the number of ATTO-HDL containing vesicles were significantly lower in ATTO-EL-HDL compared to ATTO-EV-HDL treated cells (Fig. 3A, D). After 5 h, both the total filipin fluorescence and the fluorescence associated with cellular membranes were significantly lower in the ATTO-EL-HDL compared to ATTO-EV-HDL treated cells, whereas the total ATTO-fluorescence was not significantly different in cells treated with ATTO-EL-HDL or ATTO-EV-HDL (Fig. 3B). While after 16 h the total filipin fluorescence was not significantly different between ATTO-EV-HDL and ATTO-EL-HDL treated cells, the filipin fluorescence associated with cellular membranes was significantly lower in the ATTO-EL-HDL compared to ATTO-EV-HDL treated cells (Fig. 3C). This was accompanied by similar total ATTO-fluorescence and similar number of ATTO-HDL vesicles in ATTO-EV-HDL and ATTO-EL-HDL treated cells (Fig. 3C, D). From these results we concluded that after 1 and 5 h, primarily the different rate of HDL uptake, whereas after 16 h the extent of cholesterol supply, cholesterol efflux, or cholesterol esterification determine the difference in the filipin fluorescence (i.e. FC content) observed in cells treated with ATTO-EV-HDL or ATTO-EL-HDL.
Fig. 3.
ATTO-EL-HDL uptake and impact on cellular FC content. EA.hy 926 cells were incubated with 100 μg/mL ATTO-EV-HDL protein or ATTO-EL-HDL protein for (A) 1, (B) 5 or (C) 16 h followed by washing, fixation, filipin staining, and imaging of filipin (green)- and ATTO-fluorescence (magenta) by wide-field microscopy. Representative images and corresponding analyses are shown. Total filipin-fluorescence (memb. + HDL) is the sum of the filipin fluorescence associated with cell membranes (memb.) and the filipin fluorescence of ATTO-HDL-vesicles (HDL), the latter determined by using the ATTO-fluorescence as a mask for the filipin fluorescence. The values are presented as mean ± SEM of 3 independent experiments analyzed by two-tailed unpaired t-test; *p < 0.05, **p < 0.01. (D) EA.hy 926 cells overexpressing the cytosolic marker geNOP-GFP were incubated with 100 μg/mL ATTO-EV-HDL protein or ATTO-EL-HDL protein for 1 or 16 h, followed by 3D spinning disk confocal microscopy to analyze the total ATTO-fluorescence and the number of ATTO-HDL-containing vesicles taken up per cell. Representative images and corresponding analyses are shown. The values are expressed as mean ± SEM of 4 independent experiments performed in duplicate and analyzed by twotailed unpaired t-test; *p < 0.05.
3.4. Cholesterol efflux and cholesterol ester content in cells exposed to EL- HDL or EV-HDL
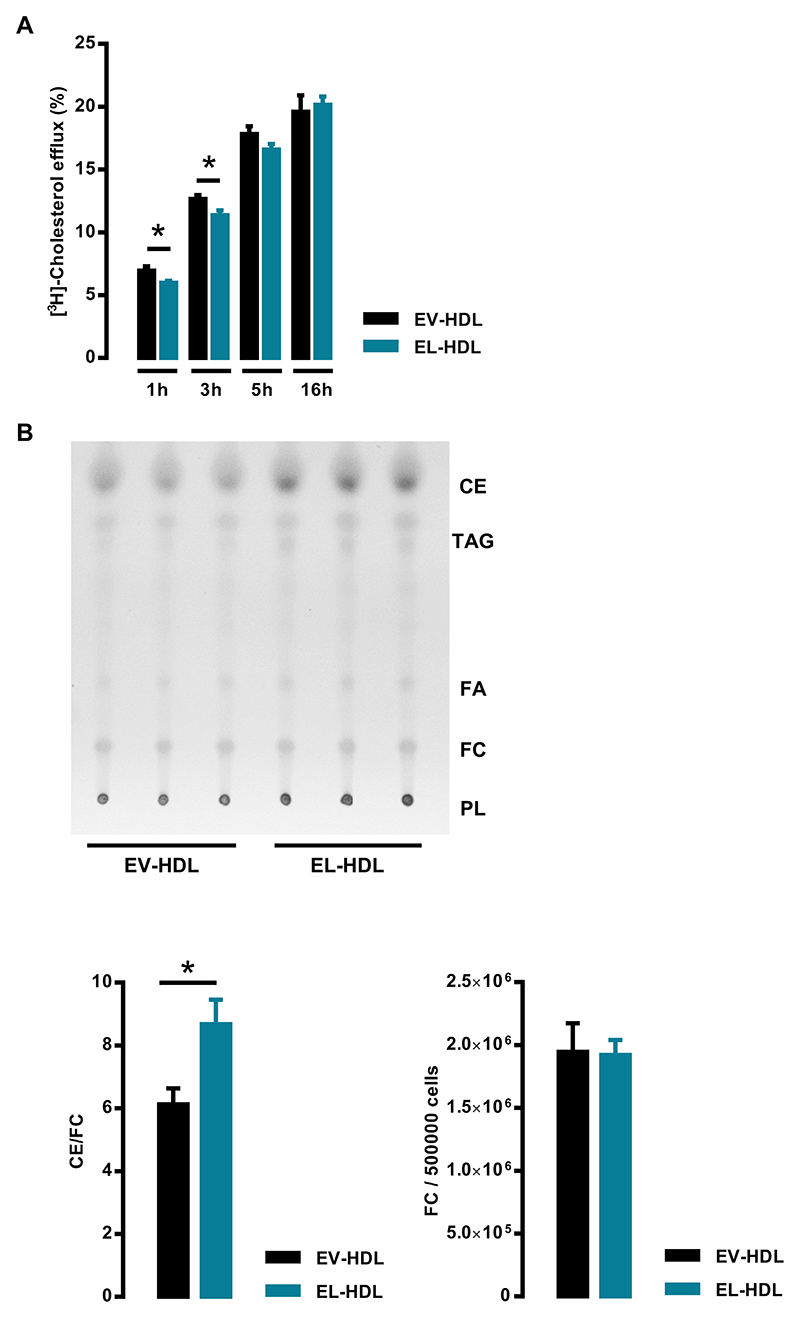
To examine whether a higher cholesterol efflux capacity of EL-HDL contributes to the lower FC content of cells incubated with EL-HDL compared to EV-HDL, we measured [3H]-cholesterol efflux from EA.hy 926 cells incubated with EL-HDL or EV-HDL for different time periods. While after 1 and 3 h cholesterol efflux was significantly lower in the presence of EL-HDL compared to EV-HDL, we observed no differences after 5 and 16 h (Fig. 4A). Importantly, incubation of EA.hy 926 cells with EL-HDL for 16 h resulted in significantly higher ratio of CE to FC compared to EV-HDL treated cells (Fig. 4B and Suppl. Fig. S10). In accordance with comparable total filipin fluorescence (Fig. 3C) the FC levels determined by TLC were comparable in EL-HDL and EV-HDL treated cells (Fig. 4B and Suppl. Fig. S10).
Fig. 4.
Impact of EL-HDL on cholesterol efflux as well as FC and CE content in EA.hy 926 cells. (A) Cells labeled with [3H]-cholesterol for 24 h were incubated with 100 μg/mL EV-HDL protein or EL-HDL protein for indicated time periods. Cholesterol efflux was expressed as the radioactivity in the medium relative to total radioactivity in medium and cells. The values are presented as mean ± SEM of 4 independent experiments performed in duplicate analyzed by two-tailed unpaired t-test; *p < 0.05. (B) EA.hy 926 cells plated in 6-well plates were incubated with 100 μg/mL of EV-HDL protein or EL-HDL protein for 16 h. Thereafter, the cells were washed with PBS and lipids were extracted with hexane/isopropanol (3:2, v:v). Extracts were evaporated and dissolved in chloroform before thin layer chromatography using hexane-diethyl ether-glacial acetic acid (70:29:1, v:v:v) as a mobile phase. The signals corresponding to phospholipids (PL), free cholesterol (FC), triacylglycerols (TAG), fatty acids (FA), and cholesterol ester (CE) were visualized by primulin and the signal intensity determined by densitometry. Annotations of the lipid species refer to the lipid standards on the plate. Representative TLC plate and the ratio of CE and FC signals as well as arbitrary units of FC per 500,000 cells both normalized to background are shown. The values are presented as mean ± SEM of 4 independent experiments performed in triplicate and analyzed by two-tailed unpaired t-test; *p < 0.05.
4. Discussion
Considering the profound effect of EL on the structure and composition of HDL [9,13], we hypothesized that EL modulates the eNOS activating capacity of HDL. To test our hypothesis, we generated EL- modified HDL by in vitro incubation of human serum with EL-over-expressing cells and tested the capacity of the isolated modified HDL to induce eNOS activity and vasorelaxation. Experiments were performed in the endothelial cell line EA.hy 926 as well as in the ECV 304 cells stably overexpressing eNOS-GFP. Although the latter cells are not endothelial cells, they have been widely used as a model for studying the relationship between subcellular eNOS targeting and activity [15,27,31]. In addition, the eNOS activating capacity of EL-HDL was also tested ex vivo by wire myography in mouse aortic rings.
Here, we show for the first time that the EL modification augments the capacity of HDL to induce eNOS activity and eNOS-dependent va-sorelaxation. The EL-HDL-driven induction of eNOS activity was ac-companied by an increased eNOS targeting to the plasma membrane and concomitantly lower eNOS colocalization with Golgi and mitochondria. The fact that the SR-BI-overexpression in EA.hy 926 cells increased the EV-HDL- and EL-HDL-induced eNOS activity to a similar extent (Suppl. Fig. S11) indicates that SR-BI-mediated eNOS activation is not responsible for the stronger eNOS-activating capacity of EL-HDL.
Cultured endothelial cells have two major cellular pools of eNOS, a plasma membrane pool and a perinuclear pool, the latter one being associated primarily with the Golgi apparatus [32,33]. Previous studies have shown that the plasma membrane-associated eNOS is constitutively active and highly sensitive to transmembrane Ca2+ flux, whereas the perinuclear pool is less responsive to Ca2+ but highly responsive to phosphoinositide-3-kinase (PI3K)/protein kinase-B (AKT)- mediated phosphorylation [27,29]. Accordingly, the EL-HDL-driven enrichment of eNOS in the plasma membrane pool might be responsible for the increased eNOS activating capacity of EL-HDL observed in the present study under both basal and A23187-induced conditions. In contrast to eNOS activity, the NO levels in EL-HDL treated cells were significantly higher only under histamine-induced but not basal conditions. This is consistent with the myography data showing no significant difference in contractile response to L-NNA (indicative of NO levels) in EL-HDL- versus EV-HDL-treated mouse aortic rings under basal conditions.
It is generally accepted that the phosphorylation of eNOS at Ser1177 increases eNOS activity. In accordance with a previous study [26], we, however, observed a dissociation between eNOS activity and the degree of phosphorylation at Ser1177 upon 5 h incubation of EA.hy 926 cells with modified HDL. More specifically, despite significantly higher Ser1177 phosphorylation, eNOS activity was comparable between EL-HDL- and EV-HDL-treated cells as was the eNOS colocalization with Golgi and mitochondria. This finding is in contrast to 16 h of incubation, which resulted in higher eNOS activity and Ser1177-phosphorylation but lower colocalization of eNOS with Golgi and mitochondria in EL-HDL-compared to EV-HDL-treated cells. We conclude that the EL-HDL-driven subcellular eNOS trafficking plays an important role in the EL-HDL-driven induction of eNOS activity.
Subcellular targeting of eNOS is largely determined by palmitoylation, which facilitates the hydrophobic interaction of eNOS with the plasma membrane, particularly with caveolae [1,32]. It is well known that biophysical features of the plasma membrane are determined by the FC content, which in turn may affect subcellular eNOS localization and activity [29,30,34]. Accordingly, the decreased cell membrane FC content observed in the EL-HDL-treated cells, is a likely determinant of the EL-HDL-driven subcellular eNOS trafficking and enrichment of eNOS in the plasma membrane pool.
The lower cell membrane FC content accompanied by an increased cellular CE content in EL-HDL treated cells suggested increased FC esterification rate in these cells. However, we found that incorporation of [3H]oleic acid into the cellular [3H]CE pool, indicative of ACAT activity, was low and comparable in EL-HDL and EV-HDL treated cells (Suppl. Fig. S12A). Markedly higher incorporation of [3H]oleic acid into [3H]TAG in EV-HDL and EL-HDL treated cells (Suppl. Fig. S12B) excluded the possibility that impaired uptake of [3H]oleic acid was a cause for its decreased incorporation into [3H]CE. The low ACAT ac-tivities measured in the present study are most probably a consequence of the incubation of cells with HDLs in the absence of serum, and accordingly absence of low-density lipoprotein, which in contrast to HDL, is crucial for induction of ACAT activity in cultured cells [35]. All in all, higher ACAT activity [36] and cholesterol esterification rate do not explain lower FC levels in cellular membranes and higher CE content in EL-HDL-treated cells. Further studies are needed to examine whether differences between EL-HDL and EV-HDL in the FC/CE supply and/or the rate of degradation of the delivered CE contribute to the lower membrane FC and higher CE content in EL-HDL treated cells.
In the present study, we observed a reduced endocytosis of ATTO-EL-HDL after 1 h of incubation, exemplified by a lower total ATTO fluorescence, and a lower number of ATTO-vesicles, whose size was not different between ATTO-EV-HDL and ATTO-EL-HDL (Suppl. Fig. S13). This might reflect a diminished affinity of EL-HDL for SR-BI [9], which, together with the decreased phospholipid content of EL-HDL [13,37] most likely underlies the determined reduced EL-HDL-mediated cholesterol efflux observed after 1 and 3 h in the present as well as in our previous studies [9,13]. Since in the present study cholesterol efflux was studied under basal conditions (without pre-treatment of cells with cyclic AMP) the expression of SR-BI in EA.hy 926 cells was markedly higher compared to the levels of ABCA1 and ABCG1 (Suppl. Fig. S14). This suggests that EL reduces cholesterol efflux capacity of HDL, which is mediated mainly by aqueous diffusion and SR-BI [38] and only slightly by ABCG1 and ABCA1. This is in accordance with our previous findings showing that EL-HDL shows diminished ABCA1-independent but unaltered ABCA1-dependent efflux [13]. Furthermore, in our previous study we found that in contrast to EL-HDL, the in vitro generated EL-modified serum exhibits an increased total and ABCA-dependent cholesterol efflux capacity [13]. This is presumably due to the EL-mediated generation of lipid-free/-poor apoA-I which accumulates in the EL-modified serum. However, as a result of its higher density, lipid- free/-poor apoA-I is not co-isolated with EL-HDL upon ultracentrifugation and accordingly has no impact on the cholesterol efflux capacity of isolated EL-HDL. Of note, in the present study, we performed cholesterol efflux experiments in the presence of an ACAT inhibitor. Accordingly, our results reflect the cholesterol efflux capacity of HDL and not the cellular FC availability.
It is conceivable that the EL-induced changes in HDL size, structure, and composition [9,13] may attenuate the interaction of HDL with cells independently of SR-BI. Along these lines and in analogy with a diminished endocytosis efficiency of negatively charged nanoparticles [39], the negative charge of EL-HDL particles, a consequence of the HDL enrichment with EL-generated non-esterified fatty acids [9], might (at least in part) explain the slower endocytosis of EL-HDL observed in the present study.
It has been shown that an increased membrane fluidity caused by low membrane cholesterol content increases, whereas a decreased membrane fluidity caused by cholesterol enrichment, reduces eNOS activity [40]. Similarly, enrichment of the plasma membrane with unsaturated PL has been shown to increase plasma membrane fluidity and in consequence activities of some plasma membrane associated receptors and enzymes [41,42]. We have previously shown that EL-generated unsaturated lysoPL, which are enriched in EL-HDL [13,43], are taken up and converted into unsaturated PL by cells [44]. Accordingly, EL-HDL-mediated enrichment of the plasma membrane with unsaturated lysoPL and their subsequent conversion to unsaturated PL together with a reduced FC content may contribute to the EL-HDL-driven changes in plasma membrane fluidity and in turn altered subcellular eNOS-targeting and -activity.
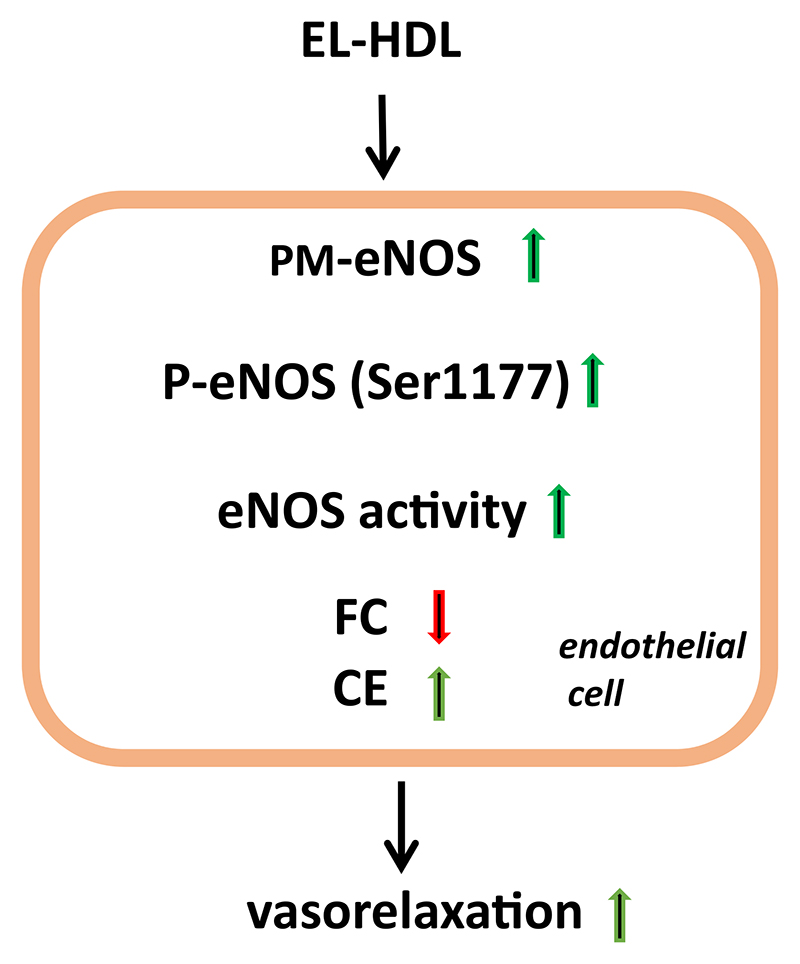
5. Conclusions
Based on our results, we conclude that EL increases eNOS activating capacity of HDL, a phenomenon accompanied by an enrichment of the plasma membrane eNOS pool as well as a decreased cell membrane FC and increased cellular CE content (Fig. 5).
Fig. 5.
Impact of EL-HDL on eNOS and vasorelaxation. EL-HDL promotes enrichment of the plasma membrane (PM) eNOS pool, increases eNOS phosphorylation at Ser1177, eNOS activity and vasorelaxation as well as decreases cellular free cholesterol (FC) and increases cellular cholesterol ester (CE) content.
Supplementary Material
Acknowledgements
This research was supported by the Austrian Science Fund FWF (P27166-B23 to SF, DK-MOLIN W1241 to G.M., DK-MCD W1226 to W.F.G. and D.K., SFB F73 to D.K.) and the PhD program “Molecular Metabolism” of the Medical University of Graz. The funders had no roles in the design of the study, data collection, analysis, and interpretation, report writing or article submission.
The authors wish to thank Margarete Lechleitner, Luca Schmid, Anton Ibovnik, Christoph Russ, and Evamaria Imp for their expert technical assistance.
Abbreviations:
- EL
endothelial lipase
- eNOS
endothelial nitric oxide synthase
- NO
nitric oxide
- GFP
green fluorescence protein
- RFP
red fluorescence protein
- CE
cholesterol ester
- FC
free cholesterol
- LysoPL
lysophospholipids
- FA
fatty acids
- EL-HDL
EL-modified HDL
- EL-HDL
EL-modified HDL
- SR-BI
scavenger receptor class B type 1
- S1P
sphingosine-1 phosphate
- FCS
fetal calf serum
- DMEM
Dulbecco's modified Eagle medium
- Ad
adenoviruses
- EV
empty virus
- HDL
high-density lipoprotein
- FPLC
fast performance liquid chromatography
- geNOP
genetically encoded fluorescent NO probe
- MTR
MitoTracker® Red FM
- ROIs
regions of interest
- L-NNA
Nrn-nitro-L-arginine
- ACAT
acyl-coenzyme A: cholesterol acyltransferase
- G-geNOp
green genetically encoded fluorescent NO probe
Footnotes
Declaration of competing interest
The authors have no conflict of interest to disclose.
References
- [1].Vanhoutte PM, Zhao Y, Xu A, Leung SW. Thirty years of saying NO: sources, fate, actions, and misfortunes of the endothelium-derived vasodilator mediator. Circ Res. 2016;119:375–396. doi: 10.1161/CIRCRESAHA.116.306531. [DOI] [PubMed] [Google Scholar]
- [2].Besler C, Luscher TF, Landmesser U. Molecular mechanisms of vascular effects of high-density lipoprotein: alterations in cardiovascular disease. EMBO Mol Med. 2012;4:251–268. doi: 10.1002/emmm.201200224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kratzer A, Giral H, Landmesser U. High-density lipoproteins as modulators of endothelial cell functions: alterations in patients with coronary artery disease. Cardiovasc Res. 2014;103:350–361. doi: 10.1093/cvr/cvu139. [DOI] [PubMed] [Google Scholar]
- [4].Calabresi L, Gomaraschi M, Franceschini G. Endothelial protection by high-den-sity lipoproteins: from bench to bedside. Arterioscler Thromb Vasc Biol. 2003;23:1724–1731. doi: 10.1161/01.ATV.0000094961.74697.54. [DOI] [PubMed] [Google Scholar]
- [5].Mineo C, Shaul PW. Novel biological functions of high-density lipoprotein cholesterol. Circ Res. 2012;111:1079–1090. doi: 10.1161/CIRCRESAHA.111.258673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mineo C, Shaul PW. Functions of scavenger receptor class B, type I in atherosclerosis. Curr Opin Lipidol. 2012;23:487–493. doi: 10.1097/MOL.0b013e328357ba61. [DOI] [PubMed] [Google Scholar]
- [7].Yuhanna IS, Zhu Y, Cox BE, Hahner LD, Osborne-Lawrence S, Lu P, Marcel YL, Anderson RG, Mendelsohn ME, Hobbs HH, Shaul PW. High-den-sity lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat Med. 2001;7:853–857. doi: 10.1038/89986. [DOI] [PubMed] [Google Scholar]
- [8].Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T. Endothelial lipase is a major determinant of HDL level. J Clin Invest. 2003;111:347–355. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gauster M, Oskolkova OV, Innerlohinger J, Glatter O, Knipping G, Frank S. Endothelial lipase-modified high-density lipoprotein exhibits diminished ability to mediate SR-BI (scavenger receptor B type I)-dependent free-cholesterol efflux. Biochem J. 2004;382:75–82. doi: 10.1042/BJ20031882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nijstad N, Wiersma H, Gautier T, van der Giet M, Maugeais C, Tietge UJ. Scavenger receptor BI-mediated selective uptake is required for the remodeling of high density lipoprotein by endothelial lipase. J Biol Chem. 2009;284:6093–6100. doi: 10.1074/jbc.M807683200. [DOI] [PubMed] [Google Scholar]
- [11].Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- [12].Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- [13].Schilcher I, Kern S, Hrzenjak A, Eichmann TO, Stojakovic T, Scharnagl H, Duta-Mare M, Kratky D, Marsche G, Frank S. Impact of endothelial lipase on cholesterol efflux capacity of serum and high-density lipoprotein. Sci Rep. 2017;7 doi: 10.1038/s41598-017-12882-7. 12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Edgell CJ, McDonald CC, Graham JB. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc Natl Acad Sci U S A. 1983;80:3734–3737. doi: 10.1073/pnas.80.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sowa G, Liu J, Papapetropoulos A, Rex-Haffner M, Hughes TE, Sessa WC. Trafficking of endothelial nitric-oxide synthase in living cells. Quantitative evidence supporting the role of palmitoylation as a kinetic trapping mechanism limiting membrane diffusion. J Biol Chem. 1999;274:22524–22531. doi: 10.1074/jbc.274.32.22524. [DOI] [PubMed] [Google Scholar]
- [16].Strauss JG, Zimmermann R, Hrzenjak A, Zhou Y, Kratky D, Levak-Frank S, Kostner GM, Zechner R, Frank S. Endothelial cell-derived lipase mediates uptake and binding of high-density lipoprotein (HDL) particles and the selective uptake of HDL-associated cholesterol esters independent of its enzymic activity. Biochem J. 2002;368:69–79. doi: 10.1042/BJ20020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kozina A, Opresnik S, Wong MS, Hallstrom S, Graier WF, Malli R, Schroder K, Schmidt K, Frank S. Oleoyl-lysophosphatidylcholine limits endothelial nitric oxide bioavailability by induction of reactive oxygen species. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113443. e113443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Eroglu E, Rost R, Bischof H, Blass S, Schreilechner A, Gottschalk B, Depaoli MR, Klec C, Charoensin S, Madreiter-Sokolowski CT, Ramadani J, Waldeck-Weiermair M, Graier WF, Malli R. Application of genetically encoded fluorescent nitric oxide (NO*) probes, the geNOps, for real-time imaging of NO* signals in single cells. J Vis Exp. 2017;121:1–10. doi: 10.3791/55486. e55486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eroglu E, Gottschalk B, Charoensin S, Blass S, Bischof H, Rost R, Madreiter-Sokolowski CT, Pelzmann B, Bernhart E, Sattler W, Hallstrom S, Malinski T, Waldeck-Weiermair M, Graier WF, Malli R. Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat Commun. 2016;7 doi: 10.1038/ncomms10623. 10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Charoensin S, Eroglu E, Opelt M, Bischof H, Madreiter-Sokolowski CT, Kirsch A, Depaoli MR, Frank S, Schrammel A, Mayer B, Waldeck-Weiermair M, Graier WF, Malli R. Intact mitochondrial Ca(2+) uniport is essential for agonist-induced activation of endothelial nitric oxide synthase (eNOS) Free Radic Biol Med. 2017;102:248–259. doi: 10.1016/j.freeradbiomed.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Khera AV, Cuchel M, de la Llera-Moya M, Rodrigues A, Burke MF, Jafri K, French BC, Phillips JA, Mucksavage ML, Wilensky RL, Mohler ER, Rothblat GH, Rader DJ. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N Engl J Med. 2011;364:127–135. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Van Harken DR, Dixon CW, Heimberg M. Hepatic lipid metabolism in experimental diabetes. V. The effect of concentration of oleate on metabolism of trigly-cerides and on ketogenesis. J Biol Chem. 1969;244:2278–2285. [PubMed] [Google Scholar]
- [24].Cadigan KM, Heider JG, Chang TY. Isolation and characterization of Chinese hamster ovary cell mutants deficient in acyl-coenzyme A:cholesterol acyltransferase activity. J Biol Chem. 1988;263:274–282. [PubMed] [Google Scholar]
- [25].Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- [26].Wang H, Wang AX, Liu Z, Chai W, Barrett EJ. The trafficking/interaction of eNOS and caveolin-1 induced by insulin modulates endothelial nitric oxide production. Mol Endocrinol. 2009;23:1613–1623. doi: 10.1210/me.2009-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fulton D, Babbitt R, Zoellner S, Fontana J, Acevedo L, McCabe TJ, Iwakiri Y, Sessa WC. Targeting of endothelial nitric-oxide synthase to the cytoplasmic face of the Golgi complex or plasma membrane regulates Akt-versus calcium-dependent mechanisms for nitric oxide release. J Biol Chem. 2004;279:30349–30357. doi: 10.1074/jbc.M402155200. [DOI] [PubMed] [Google Scholar]
- [28].Boo YC, Kim HJ, Song H, Fulton D, Sessa W, Jo H. Coordinated regulation of endothelial nitric oxide synthase activity by phosphorylation and subcellular localization. Free Radic Biol Med. 2006;41:144–153. doi: 10.1016/j.freeradbiomed.2006.03.024. [DOI] [PubMed] [Google Scholar]
- [29].Zhang Q, Church JE, Jagnandan D, Catravas JD, Sessa WC, Fulton D. Functional relevance of Golgi- and plasma membrane-localized endothelial NO synthase in reconstituted endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:1015–1021. doi: 10.1161/01.ATV.0000216044.49494.c4. [DOI] [PubMed] [Google Scholar]
- [30].Everson WV, Smart EJ. Influence of caveolin, cholesterol, and lipoproteins on nitric oxide synthase: implications for vascular disease. Trends Cardiovasc Med. 2001;11:246–250. doi: 10.1016/s1050-1738(01)00119-0. [DOI] [PubMed] [Google Scholar]
- [31].Sanchez FA, Rana R, Gonzalez FG, Iwahashi T, Duran RG, Fulton DJ, Beuve AV, Kim DD, Duran WN. Functional significance of cytosolic endothelial nitric-oxide synthase (eNOS): regulation of hyperpermeability. J Biol Chem. 2011;286:30409–30414. doi: 10.1074/jbc.M111.234294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Garcia-Cardena G, Oh P, Liu J, Schnitzer JE, Sessa WC. Targeting of nitric oxide synthase to endothelial cell caveolae via palmitoylation: implications for nitric oxide signaling. Proc Natl Acad Sci U S A. 1996;93:6448–6453. doi: 10.1073/pnas.93.13.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sessa WC, Garcia-Cardena G, Liu J, Keh A, Pollock JS, Bradley J, Thiru S, Braverman IM, Desai KM. The Golgi association of endothelial nitric oxide synthase is necessary for the efficient synthesis of nitric oxide. J Biol Chem. 1995;270:17641–17644. doi: 10.1074/jbc.270.30.17641. [DOI] [PubMed] [Google Scholar]
- [34].Blair A, Shaul PW, Yuhanna IS, Conrad PA, Smart EJ. Oxidized low density lipoprotein displaces endothelial nitric-oxide synthase (eNOS) from plasmalemmal caveolae and impairs eNOS activation. J Biol Chem. 1999;274:32512–32519. doi: 10.1074/jbc.274.45.32512. [DOI] [PubMed] [Google Scholar]
- [35].Goldstein JL, Dana SE, Brown MS. Esterification of low density lipoprotein cholesterol in human fibroblasts and its absence in homozygous familial hypercholesterolemia. Proc Natl Acad Sci U S A. 1974;71:4288–4292. doi: 10.1073/pnas.71.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Smith JL, Madden LJ, de Jersey J. Effect of exogenous cholesterol and dithio- threitol on the activity of human liver microsomal acyl-coenzyme A:cholesterol acyltransferase (ACAT) Clin Chim Acta. 1996;256:13–25. doi: 10.1016/s0009-8981(96)06408-x. [DOI] [PubMed] [Google Scholar]
- [37].Schilcher I, Ledinski G, Radulovic S, Hallstrom S, Eichmann T, Madl T, Zhang F, Leitinger G, Kolb-Lenz D, Darnhofer B, Birner-Gruenberger R, Wadsack C, Kratky D, Marsche G, Frank S, Cvirn G. Endothelial lipase increases antioxidative capacity of high-density lipoprotein. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:1363–1374. doi: 10.1016/j.bbalip.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Phillips MC. Molecular mechanisms of cellular cholesterol efflux. J Biol Chem. 2014;289:24020–24029. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomedicine. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Deliconstantinos G, Villiotou V, Stavrides JC. Modulation of particulate nitric oxide synthase activity and peroxynitrite synthesis in cholesterol enriched endothelial cell membranes. Biochem Pharmacol. 1995;49:1589–1600. doi: 10.1016/0006-2952(95)00094-g. [DOI] [PubMed] [Google Scholar]
- [41].Loesberg C, Spence S, Nijkamp FP, Houslay MD. Dietary linoleic acid-induced changes in respiratory beta-adrenergic receptor function and the form of arrhenius plots of isoprenaline- and prostaglandin E2-stimulated adenylate cyclase activity in a model for atopy. Cell Signal. 1994;6:187–199. doi: 10.1016/0898-6568(94)90076-0. [DOI] [PubMed] [Google Scholar]
- [42].McMurchie EJ, Patten GS, Charnock JS, McLennan PL. The interaction of dietary fatty acid and cholesterol on catecholamine-stimulated adenylate cyclase activity in the rat heart. Biochim Biophys Acta. 1987;898:137–153. doi: 10.1016/0005-2736(87)90032-0. [DOI] [PubMed] [Google Scholar]
- [43].Gauster M, Rechberger G, Sovic A, Horl G, Steyrer E, Sattler W, Frank S. Endothelial lipase releases saturated and unsaturated fatty acids of high density lipoprotein phosphatidylcholine. J Lipid Res. 2005;46:1517–1525. doi: 10.1194/jlr.M500054-JLR200. [DOI] [PubMed] [Google Scholar]
- [44].Riederer M, Kofeler H, Lechleitner M, Tritscher M, Frank S. Impact of endothelial lipase on cellular lipid composition. Biochim Biophys Acta. 2012;1821:1003–1011. doi: 10.1016/j.bbalip.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.