Abstract
The formation of arteries is thought to occur by the induction of a highly conserved arterial genetic programme in a subset of vessels that will later experience an increase in oxygenated blood flow1,2. The initial steps of arterial specification require both the VEGF and Notch signalling pathways3–5. Here, we combine inducible genetic mosaics and transcriptomics to modulate and define the function of these signalling pathways in cell proliferation, arteriovenous differentiation and mobilization. We show that endothelial cells with high levels of VEGF or Notch signalling are intrinsically biased to mobilize and form arteries; however, they are not genetically pre-determined, and can also form veins. Mechanistically, we found that increased levels of VEGF and Notch signalling in pre-arterial capillaries suppresses MYC-dependent metabolic and cell-cycle activities, and promotes the incorporation of endothelial cells into arteries. Mosaic lineage-tracing studies showed that endothelial cells that lack the Notch–RBPJ transcriptional activator complex rarely form arteries; however, these cells regained the ability to form arteries when the function of MYC was suppressed. Thus, the development of arteries does not require the direct induction of a Notch-dependent arterial differentiation programme, but instead depends on the timely suppression of endothelial cell-cycle progression and metabolism, a process that precedes arterial mobilization and complete differentiation.
Angiogenesis requires the reiterative growth and remodelling of a precursor vascular network. After the formation of a rudimentary plexus, a subset of capillary branches undergo arterialization to form vessels that are specialized in the delivery of oxygen- and nutrient-rich blood. Arterialization involves numerous signalling pathways1–4 with activities that must be regulated with spatiotemporal precision. Studies in diverse model organisms have shown that the VEGF and Notch signalling pathways are essential regulators of both angiogenesis and arterialization. VEGF is a positive regulator of endothelial sprouting and proliferation, whereas Notch inhibits these processes3 in a context-dependent manner6. These pathways also have key roles in the differentiation of progenitor and capillary endothelial cells (ECs) into fully defined arterial ECs1–4. The expression of the main endothelial Notch ligand DLL4 and its receptor NOTCH1 is highest in arterial ECs, lower in capillaries, and absent from most venous ECs3,4,7. Loss of VEGF or DLL4–NOTCH signalling prevents the acquisition of molecular arterial identity markers such as Cx37 (also known as Gja4), Cx40 (Gja5) and Efnb2 5,8,9.
Activation of NOTCH receptors by their ligands triggers the release and nuclear translocation of the Notch intracellular domain (NICD), where it interacts with the transcription factor RBPJ to promote cell-type-specific and context-dependent gene expression programmes, usually related to cell proliferation or differentiation10. The current view is that the NICD–RBPJ transcriptional activator complex is a master regulator of arterial cell differentiation1,2,8. However, Notch signalling also potently inhibits the cell cycle of ECs, even before these form arteries3,4,6. Notably, arterial development coincides with the suppression of the cell cycle in ECs11,12. These observations led us to question whether the primary effect of NOTCH–RBPJ on arterialization is on the genetic differentiation of endothelial cells or on the regulation of the cell cycle.
Arteriovenous fate of Notch genetic mosaics
To map the expansion and arteriovenous fate of single ECs with normal or altered Notch signalling, we induced loss- and gain-of-function genetic mosaics by crossing ifgMosaic mice6,13 with Tie2-cre (Tie2 is also known as Tek) mice. In iChr-NotchTie2-Mosaic embryos, a mosaic of cells is induced throughout the embryonic endothelium, with normal (H2B-Cherry+), lower (dominant-negative MAML1+ and H2B-EGFP+), or higher (N1ICDP+ and HA-H2B-Cerulean+) Notch signalling (Fig. 1a, b, Extended Data Fig. 1a–d). Unlike global or EC-specific Notch1, Dll4 or Rbpj mutants, iChr-NotchTie2-Mosaic mosaic embryos developed normally (Extended Data Fig. 1e). This allowed us to compare the proliferative, migratory and differentiation dynamics of the different mutant cells with neighbouring wild-type (Cherry+) cells sharing the same environment. ECs with high Notch signalling (Cerulean+) were strongly outcompeted during the development of coronary vessels but not during endocardium development (Fig. 1c, Extended Data Fig. 1f–h). This result is consistent with the lower proliferative capacity of angiogenic ECs with higher Notch signalling6,14 and with the considerably lower proliferation activity of the Nfatc1-Cre+ endocardium (Extended Data Fig. 1i). Notably, coronary ECs with lower Notch signalling generally did not proliferate more than adjacent wild-type ECs (Extended Data Fig. 1j, k).
Fig. 1. Fate mapping of heart ECs with distinct Notch signalling levels.
a, Mice containing the iChr-Notch-Mosaic and Tie2-cre alleles undergo induction of a multispectral mosaic of ECs expressing distinct Notch signalling modulators. DN-MAML1, dominant-negative MAML1. N1ICDP, NOTCH1 intracellular domain including its native PEST sequence. b, Schematic of heart coronary development. A, artery; SV, sinus venosus; V, vein. c, Localization of ECs with distinct Notch signalling levels throughout development. Edc., endocardium; EGFP, enhanced green fluorescent protein; EMCN, endomucin. d, Contribution of ECs with distinct Notch signalling levels to the development of coronary veins or arteries. PECAM1 is used as a vascular cell marker. WT, wild type. e, Arterial fate analysis of Apln-FlpO × iFlpMTomato-cre/MYfp genetic mosaics. See also Extended Data Fig. 1. Scale bars, 100 μm. Error bars indicate s.d. NS, not significant. *P < 0.05, ***P < 0.001. For statistics, see Supplementary Data 1.
In addition to the regulation of EC proliferation, Notch also induces arterial differentiation and mobilizes ECs towards arteries1–5. In agreement with previous studies in zebrafish, we found that in iChr-NotchTie2-Mosaic hearts, ECs with reduced Notch signalling (EGFP+) were more frequently found in veins, whereas ECs with normal Notch signalling (Cherry+) were more frequently found in arteries; however, cells with low Notch signalling could be found in both types of vessel (Fig. 1d). Because the decrease in Notch signalling in ECs expressing a dominant-negative MAML1 is on average less pronounced than after the deletion of Rbpj 6, we developed an alternative Apln-FlpO inducible genetic mosaic strategy (iFlpMTomato-cre/MYfp-Mosaic) to analyse the fate of single coronary ECs with (MTomato-2A-Cre+, gene knockout) or without (MbYFP+, wild type) deletion of Rbpj (Extended Data Fig. 1l, m). The Apln-FlpO allele is located on the X chromosome and is ideal for vascular fate-mapping studies because it is mosaically expressed in female embryos at the angiogenic vascular front, which will later give rise to arteries and veins. Using this strategy, we found that coronary ECs with full Rbpj deletion (Rbpj-knockout, RbpjKO) had a more pronounced defect in artery formation than DN-MAML1+ ECs (Fig. 1d, e). However, a small fraction of RbpjKO ECs still formed coronary and retinal arteries (Fig. 1e, Extended Data Fig. 1n), which suggests that Notch activity biases ECs towards artery formation but is not essential for their differentiation. RbpjKO ECs also densely populated the peri-arterial capillary region, but most were unable to acquire the necessary phenotype to form or mobilize into arteries.
Notch activation and venous development
To investigate the phenotypic consequence of increased endothelial Notch activity, we generated a new mouse line (ROSA26LSL-MbTomato-2A-H2B-GFP-2A-N1ICDP) that allowed us to induce fluorescent labelling of the membrane (MbTomato) and nuclei (H2B-GFP) of cells that overexpress the mouse NOTCH1 intracellular domain, including its native PEST sequence (hereafter referred to as ‘N1ICDP’), that results in a moderate increase in Notch signalling (Extended Data Fig. 2a). Embryos that express this allele after Tie2-cre-mediated recombination (N1ICDPTie2-cre) can develop until embryonic day (E) 16.5 (Extended Data Fig. 2b, c). The increase in EC Notch activity markedly inhibited the proliferation of coronary vessels, thus compromising myocardial development (Extended Data Fig. 2d–h). Despite their higher Notch activity, EGFP+ N1ICDP+ ECs were frequently found in coronary (N1ICDPTie2-cre) and retinal (N1ICDPCdh5-creERT) veins, had lost the arterial marker CX40, and had normal expression of several venous markers (Extended Data Fig. 2i–w). Nonetheless, the mutant veins in hearts from N1ICDPTie2-cre mice were thinner and underdeveloped, probably owing to the lower proliferation rate and the reduced vascular density (Extended Data Fig. 2e–m).
Increases in Notch activity have been linked to an increase in arterialization1,3–5, and single-cell RNA sequencing (scRNA-seq) data revealed that pre-arterial ECs with higher Cx40 expression and Notch signalling form the main coronary arteries11. Notably, N1ICDPTie2-cre mutants did not form proper coronary arteries and instead formed small-diameter secondary collateral arteries, even though they had increased expression of arterial markers (Extended Data Fig. 2x–z).
Because N1ICDPTie2-cre mutants also had a pronounced defect in myocardial development (Extended Data Fig. 2e–h), we also induced Notch activity in coronary vessels with the Pdgfb-icreERT2 allele. This resulted in milder myocardial development defects but similar arterialization defects (Extended Data Fig. 3a–o). By contrast, activation of Notch in the endocardium (Nfatc1-cre+) induced no major defects in cardiovascular development (Extended Data Fig. 3p–u). These results suggest that ectopic Notch activation impairs the development of larger coronary arteries, presumably by promoting the arterial phenotype in ectopic pre-arterial capillary branches. Alternatively, overactivation of Notch could compromise coronary EC migration or vessel coalescence. These findings show that Notch functions during arteriovenous development are context-dependent. High Notch activity is not able to arterialize pre-venous vessels, even though it is able to promote the arterialization of pre-arterial capillary ECs.
Notch suppresses cell growth pathways
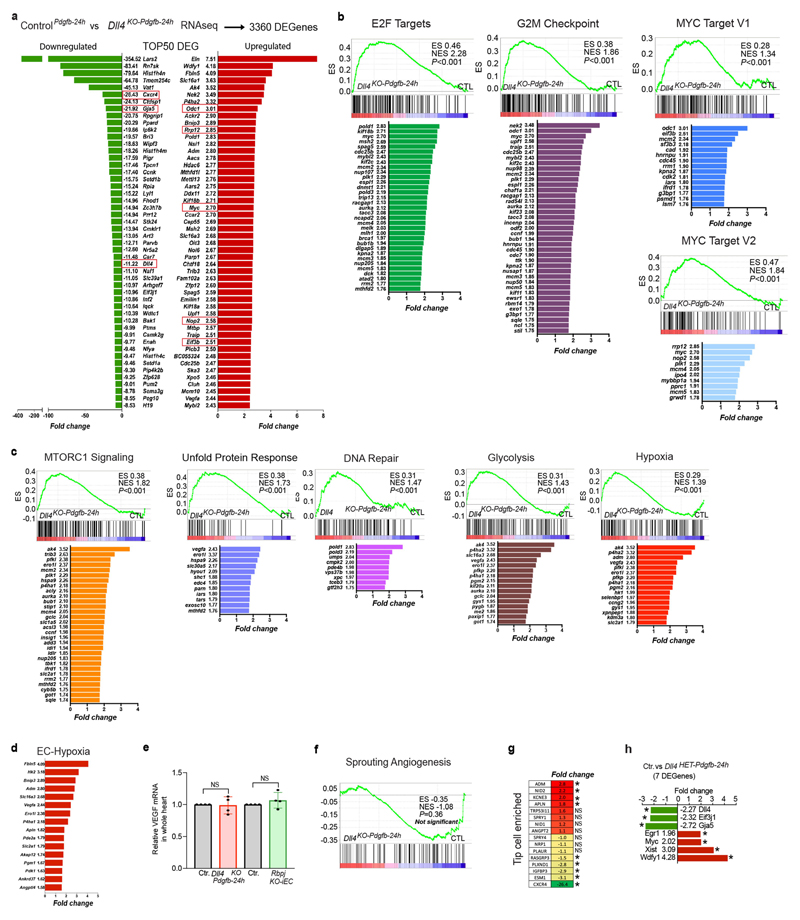
To investigate the molecular mechanisms that are regulated by DLL4–NOTCH signalling, we isolated Pdgfb-icreERT2-ires-egfp+ coronary ECs from wild-type control and Dll4KO-Pdgfb-24h E14.5 hearts (Fig. 2a, b). RNA-seq analysis revealed that loss of DLL4–NOTCH signalling from E13.5 to E14.5 (24 h) markedly altered the expression of 3,360 genes (Extended Data Fig. 4a). The set of differentially expressed genes included downregulation of most coronary arterial-enriched genes11, whereas few coronary-vein-enriched genes were upregulated (Fig. 2c). This confirmed that DLL4–NOTCH signalling is essential for arterialization, as previously shown5,8,9.
Fig. 2. Transcriptional and phenotypic changes after loss of DLL4 or Notch signalling in coronary ECs.
a, The Pdgfb-icreERT2-ires-egfp allele is expressed only in myocardium vessels. b, Validation of Dll4 deletion. c, Fold change in expression of arterial and venous genes. d, Top upregulated gene sets in Dll4KO-Pdgfb-24h samples. Adj. FDR, adjusted false discovery rate; NES, normalized enrichment score. e, Fold change of MYC target genes in Dll4KO-Pdgfb-24h samples. f, Differentially expressed genes (DEG) after heterozygous and full loss of DLL4. Ctrl, control. g, ICAM2 immunostaining showing lack of coronary artery (CA) in Dll4 mutants (Dll4Het and Dll4KO). h, Arterial gene expression in Dll4 mutants. i, Endothelial MYC pathway gene expression in Dll4 mutants. Scale bars, 100 μm. *P < 0.05, Benjamin–Hochberg adjusted FDR. ‘#’ denotes not significant.
Notably, the downregulation of arterial-enriched genes coincided with a profound upregulation of genes related to cell proliferation and metabolism (E2F targets, G2–M checkpoint genes, MYC targets, mTORC1, glycolysis and hypoxia genes), all of which are linked to a hyperactivated or hyperproliferative endothelial state (Fig. 2d, e, Extended Data Fig. 4b–e). DLL4–NOTCH signalling has also been implicated in the negative regulation of endothelial sprouting of retinal ECs3. However, angiogenic coronary vessels with loss of DLL4–NOTCH signalling did not show increased sprouting or vascular density, or increased expression of sprouting or tip-cell-enriched genes (Extended Data Figs. 4f, g, 5a). In coronary vessels, DLL4–NOTCH signalling thus seems to control endothelial arteriovenous differentiation and proliferation but not sprouting.
We also performed RNA-seq analysis on coronary ECs with a two-fold decrease in Dll4 expression for 24 h (Dll4Het-Pdgfb-24h). In contrast to coronary ECs with a complete loss of Dll4, a twofold decrease in Dll4 expression resulted in only seven differentially expressed genes with a Benjamin–Hochberg adjusted P value < 0.05 (Fig. 2f, Extended Data Fig. 4h). In these hemizygous mutant hearts, pre-arterial ECs were also unable to form arteries (Fig. 2g, Extended Data Fig. 5b–f), even though only Dll4 itself and Cx40 were significantly downregulated (twofold), and the expression of all other artery-enriched genes did not change (Fig. 2h). Thus, major coronary arteries can fail to develop in Dll4 hemizygous mutants even when acquisition of the pre-arterial molecular identity is not markedly compromised. Notably, ECs with hemi- or homozygous loss of Dll4 showed a similar increase in the expression of Myc and in many of its cell cycle- and metabolism-related targets (Fig. 2i), which suggests that increased MYC activity is a shared feature of the arterialization defects in the Dll4Het and Dll4KO mutants.
Notch has arteriovenous context-dependent effects
The data above show that ECs with full or hemizygous loss of DLL4–NOTCH signalling upregulate pathways that are linked to cell metabolism and proliferation, which should result in increased EC proliferation. Paradoxically, however, sub-epicardial venous ECs proliferated significantly less when they lost DLL4–NOTCH signalling or the transcription factor RBPJ (Extended Data Fig. 5g, h), even though the expression of mitogenic markers such as phosphorylated ERK (p-ERK) increased considerably (Extended Data Fig. 5i). However, the high levels of p-ERK correlated with increased expression of the cell-cycle inhibitor p21 (Extended Data Fig. 5j, k) in coronaries adjacent to the VEGF-rich myocardium15. This result is consistent with a bell-shaped response of proliferating ECs to mitogenic stimulation, according to which ECs arrest when exposed to a highly mitogenic stimulus as a result of a loss of Notch signalling in a VEGF-high environment6.
Unlike the highly proliferative sub-epicardial venous vessels, arterial zone capillaries, located deeper in the myocardium, have higher Notch signalling and proliferate considerably less12,16. EC proliferation in these arterial capillaries was increased after the loss of DLL4 or RBPJ (Extended Data Fig. 5l–m). This dual and location-dependent role of Notch is highly conserved, and also occurs in the developing retinal vasculature, where angiogenic ECs become arrested after the inhibition of DLL4–NOTCH signalling, whereas periarterial and more mature ECs re-enter the cell cycle and proliferate more6.
Arteriovenous fate of VEGFR2 genetic mosaics
Our results suggest that the transcriptional Notch programme induced in pre-arterial capillaries is associated with exit from the cell cycle as a result of lower levels of p-ERK or MYC. However, VEGF, the most important endothelial pro-mitogenic factor, is known to be required for arterialization in embryos and coronary artery development5,15. To investigate the influence of VEGFR2 and ERK signalling levels on arterialization, we analysed coronary arterialization in iMb-Vegfr2-Mosaic mice13 (Fig. 3a). MbYFP+ (VEGFR2low) ECs were rarely found in the intramyocardial pre-arterial coronary plexus and did not form arteries; however, they did form the endocardium and heart-surface venous vessels, probably owing to genetic compensation by VEGFC–VEGFR3 signalling in this heart region17. By contrast, MbTomato+ (VEGFR2high) ECs frequently formed arteries but tended not to incorporate into veins (Fig. 3b–e). These results show that ECs with very high VEGFR2–ERK signalling are more likely to form arterial vessels, which is in contrast to our finding that Notch activation induces arterialization by suppressing ERK signalling and mitogenic activity in pre-arterial ECs. However, both overactivation of VEGFR2–ERK and Notch activation induced exit from the cell cycle during coronary angiogenesis (Fig. 3f, Extended Data Fig. 2f), through a bell-shaped dose-dependent-response to mitogenic stimuli6. Low VEGFR2–ERK signalling also induced frequent exit from the cell cycle, but this was insufficient to induce effective arterial differentiation (Fig. 3e, f). This probably reflects the requirement of VEGFR2 signalling for cell mobilization or adequate activation of DLL4 and NOTCH signalling in pre-arterial ECs3,5, and this activity may be needed for the adequate suppression of the downstream MYC programme. Supporting this interpretation, MbTomato+ (VEGFR2high) cells with high levels of ERK signalling had low levels of MYC (Fig. 3g), whereas ECs with loss of DLL4 or RBPJ signalling had high activity of both ERK and MYC (Extended Data Figs. 5i, 6a, b). These results therefore suggest that the low MYC levels induced by high arterial VEGF and Notch activity set the cell-cycle or metabolic suppression that enables subsequent arterial development. The tight regulation of MYC expression by DLL4, NOTCH and RBPJ during arterialization in vivo was not observed after overactivation of Notch signalling in EC lines in vitro (Extended Data Fig. 6c–h). This suggests that MYC is not a direct target of the NICD–RBPJ complex or of the downstream HEY transcription factors, and that its regulation by Notch probably involves additional genetic or biophysical elements present in ECs undergoing arterialization in vivo.
Fig. 3. High VEGF-ERK signalling induces arterialization by suppressing EC proliferation.
a, Mice containing an iMb-Vegfr2-Mosaic allele were crossed with Tie2-cre mice to generate a mosaic of coronary ECs with distinct levels of VEGFR2 signalling (expressing constitutively active VEGFR2 (VEGFR2Ac) or a tyrosine kinase mutant (VEGFR2TK-). b–e, Relative p-ERK levels and localization of MbTomato+ (VEGFR2high) and MbYFP+ (VEGFR2low) in coronary vessels and endocardium. f, Analysis of proliferation (EdU+) of ECs with distinct VEGFR2–ERK signalling levels. g, Comparison of MYC expression in wild-type (Tomato-) and VEGFR2high (Tomato+) ERG+ ECs. Scale bars, 100 μm. Data are mean and s.d. *P < 0.05, ***P < 0.001. For statistics, see Supplementary Data 1.
Arterialization without Notch and MYC
To determine the cell-autonomous role of MYC in arterial differentiation, without affecting the whole embryo and heart vascular development (Extended Data Fig. 6i–k), we used the inducible genetic mosaic system (iFlpMTomato-cre/MYfp) presented above. Although ECs with loss of Myc (MTomato-2A-Cre+) proliferated less, they formed arteries at a much higher rate than the control (MYFP+) cells (Fig. 4a–c). This suggests that MYC negatively regulates pre-arterial specification by enhancing the proliferative and metabolic activity of coronary ECs. These results are consistent with recent single-cell RNA-seq data showing a considerable decrease in Myc expression during coronary arterial differentiation11 and with the absence of a Myc-dependent proliferation effect in the Notch-high arterial zone (Fig. 4b).
Fig. 4. Arterial development occurs in the absence of NOTCH–RBPJ signalling when MYC function is suppressed.
a, b, Reduced EC proliferation in MycKO-iEC coronary vessels. PA, periarterial; PV, perivenous. c, Arterial fate analysis of Myc loss-of-function genetic mosaics. d, Loss of Myc rescues the defects in arterial differentiation (CX40) and proliferation (EdU+) induced by full loss of Rbpj (see also Extended Data Figs. 7a–c, 8, 9). e, Arterial fate analysis of indicated genetic mosaics in adult heart coronary arteries. f, g, Arterial, capillary and venous frequency analysis of the indicated genetic mosaics in P6 retinas. The log2-transformed fold change indicates comparisons between arteries and capillaries of the same genotype. Only Rbpj-knockout (RbpjKO) cells are underrepresented in arteries. Scale bars, 100 μm. Data are mean and s.d. *P < 0.05, **P < 0.01, ***P < 0.001. For statistics, see Supplementary Data 1.
The Dll4Het and Dll4KO data presented above suggested that Notch inhibition interferes with arterial development not by directly inhibiting arterial differentiation, but by inducing MYC upregulation and cell-cycle progression in pre-arterial vessels. To test this hypothesis, we analysed coronary and retina EC proliferation and arterial differentiation in a series of mutants with single or compound deletion of the genes Rbpj, Dll4 and Myc. Notably, Myc deletion was sufficient to revert the increase in EC proliferation and lack of arterialization observed in Rbpj or Dll4 mutants (Fig. 4d, Extended Data Fig. 7). Endothelial cells with double-knockout (DKO) of both Dll4 and Myc or of both Rbpj and Myc regained the expression of arterial genes and had reduced expression of E2F, cell-cycle and metabolic genes (Extended Data Fig. 8)
We next used the iFlpMTomato-cre/MYfp mosaic genetic system to investigate whether single cells with full loss of RBPJ and MYC could form functional arteries for long periods of time in several organs. In contrast to RbpjKO ECs, Rbpj/MycDKO ECs formed a high proportion of the aorta, heart, retina and liver arteries, even though they were relatively less frequent in capillaries owing to the loss of MYC (Fig. 4e–g, Extended Data Fig. 9).
These results suggest that Notch is a master regulator of arterialization not because it directly induces the arterial genetic program, but because it suppresses MYC and its downstream metabolic and cell-cycle responses, a prerequisite for subsequent acquisition of arterial cell fate.
Discussion
Activation of Notch signalling culminates in the transcriptional activation of several artery-enriched genes and is required for capillary ECs to form arteries5,8,9. This led to the model that Notch induces the direct genetic differentiation or pre-determination of pre-arterial ECs. However, we are not aware of any studies that show the direct binding of the Notch downstream effector transcription factors RBPJ or HEY to the promoters or enhancers of arterial or venous genes. Moreover, overactivation of Notch in vitro is unable to induce the expression of most arterial-specific genes18.
Here, we show that single ECs with high Notch activity can form veins and that ECs with full loss of Notch activity can form arteries, although at a much lower frequency than wild-type cells. This would not be possible if a Notch-driven, hard-wired transcriptional pre-specification existed. In addition, the complete loss of Notch–RBPJ in already developed and mature arteries does not compromise their arterial phenotype or identity19. Thus, the evidence suggests that Notch function is important only in the capillary-to-arterial transition, a process associated with a timely reduction in metabolic and cell-cycle activity11,12,16. We show that this transition can occur almost normally in the absence of Notch signalling, as long as MYC function is suppressed. The ability of ECs to differentiate and form arterial vessels without the activity of the NICD–RBPJ transcriptional activator complex and MYC suggests that Notch does not directly induces arterial differentiation, but instead reduces their MYC-dependent metabolic and cell-cycle activities.
Our results also explain how VEGF, a potent inducer of ERK and mitogenic activity, can induce arterial differentiation. A relatively mild increase in ERK activity is known to induce MYC activity20; however, strong VEGF–ERK stimulation induces high DLL4–Notch signalling, and this reduces MYC levels and EC proliferation, thereby promoting arterialization.
Recent studies using refined technologies in lineage tracing and imaging suggested that ECs with high Notch expression are particularly able to migrate against flow towards arteries, a process that involves a combination of CXCL12–CXCR4 chemokine signalling and an arterial fate-inductive stimulus21,22. Given the interaction and often mutually exclusive nature of cell proliferation and migration6, it is reasonable to think that a timely inhibition of MYC and the cell cycle by the higher Notch activity in pre-arterial ECs will render them more responsive to chemokine signalling or other migratory cues. This in turn may facilitate their assembly and incorporation into developing arteries, where they subsequently adapt to their inductive microenvironment and take on their final arterial identity. The model proposed here (Extended Data Fig. 10) also fits with the recently published observation that overexpression of the venous-determining transcription factor COUP-TFII (also known as NR2F2) in pre-arterial ECs compromises arterial development by inducing the expression of cell-cycle genes11,23.
From a translational standpoint, the ability to modulate or reprogramme the arterial or venous identities of vessels is of interest for the treatment of coronary artery disease24,25. The mechanistic insights presented here help to advance our knowledge of the process of arterialization and may enable better induction of this process during tissue growth, regeneration, or ischaemic cardiovascular disease.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-020-3018-x.
Methods
Mice
The following published mouse alleles were used: iChr2-Control-Mosaic 13; iMb2-Control-Mosaic 13; iChr2-Notch-Mosaic 13; iMb-Vegfr2-Mosaic 13; Tie2-cre 26; Cdh5(PAC)-creERT2 27; Pdgfb(BAC)-icreERT2-ires-egfp 28; Nfact1-cre 15; Dll4 flox/flox 29; Rbpjflox/flox 30; Mycflox/flox 31; GFP-Myc 32; and iSuRe-cre 33. ROSA26LSL-MbTomato-2A-H2B-GFP-2A-N1ICDP mice were generated by targeting the construct indicated in Extended Data Fig. 2a (Addgene 159220) to the Rosa26 locus of mouse ES cells with G4 background following standard procedures13. The iFlpMTomato-cre/MYfp mice were generated by using CRISPR–Cas9 assisted gene targeting of the construct indicated in Extended Data Fig. 1l to the Rosa26 locus using previously published methods42 in mouse G4 ES cells. Apln-FlpO mice were also generated by using CRISPR–Cas9 assisted gene targeting (guide RNA target sequence GAATCTGAGGCTCTGCGTGCAGG) in mouse G4 ES cells to insert the HA-NLS-FlpO-WPRE-Sv40-pA-loxP-PGK-Neo-polyA-loxP cassette (Addgene 159222) in frame with the ATG of the endogenous Apln gene. All primer sequences required to genotype these mice are provided in Supplementary Table 1.
To induce CreERT2 activity in embryos, 20 mg tamoxifen (Sigma-Aldrich, T5648) and 10 mg progesterone (Sigma-Aldrich, P0130) were first dissolved in 140 μl absolute ethanol and after 860 μl corn oil (20 mg ml−1 tamoxifen and 10 mg ml−1 progesterone). This solution was given via oral gavage (200 μl, total dose of 4 mg tamoxifen per adult mouse). To induce CreERT2 activity in pups, 3 mg of 4-OH tamoxifen (Sigma-Aldrich, H6278) was first dissolved in 75 μl absolute ethanol and after 465 μl Kolliphor EL (Sigma-Aldrich, C5135), then added 935 μl PBS. Each pup received 20 μg g−1 of 4-OH tamoxifen. Injection and dissection stages for individual embryo and pup experiments are mentioned in Supplementary Table 2.
All mouse husbandry and experimentation was conducted using protocols approved by local animal ethics committees and authorities (Comunidad Autónoma de Madrid and Universidad Autónoma de Madrid CAM-PROEX 177/14 and CAM-PROEX 167/17). The CNIC mouse colony (Mus musculus) is maintained in racks with individual ventilation cages according to current Spanish and European legislation (RD 53/2013 and EU Directive 63/2010, respectively). Mice have a dust/pathogen-free bedding, sufficient nesting and environmental enrichment material for the development of species-specific behaviour. All mice have food and water ‘ad libitum’ in environmental conditions of 45–65% relative humidity, temperatures of 21–24 °C and a 12 h/12 h light/dark cycle. In addition and to preserve animal welfare, an animal health surveillance program is applied for health monitoring, which follows FELASA recommendations for specific pathogen-free facilities.
We used M. musculus with C57BL6 or C57BL6×129SV genetic backgrounds. To generate embryos for analysis, we intercrossed mice with an age range between 7 and 30 weeks. All adult mice analysed were less than 12 weeks old. We do not expect our data to be influenced by mouse age or sex, with the exception of experiments conducted with the Apln-FlpO X chromosome-linked allele that is mosaically inactivated in female embryos as explained in the text.
Whole-mount immunofluorescence and imaging of embryonic hearts, retinas and aortas
Whole embryos were dissected between E12.5 and E17.5 and their chests fixed in 4% paraformaldehyde (PFA, EMS 15714) on ice for 2 h (E12.5) or 5 h (E14.5–E17.5) with shaking. Chests were washed with shaking for 2 × 10 min in PBS at room temperature. Hearts were removed and cut on the ventral side (for artery morphological analysis) and the dorsal side (for venous morphological analysis), and the endocardium was trimmed with microdissection scissors (FST 15001-08, FST 11210-10). For immunostaining, hearts were first blocked and permeabilized in PBBT solution (PBS with 0.5% Triton X-100 and 10% donkey serum) for 2 h at room temperature with shaking. Subsequently, primary antibodies were added to fresh PBBT solution (Supplementary Table 3) and hearts were incubated at 4 °C overnight with shaking. The next day, hearts were washed in PBT (PBS with 0.5% Triton X-100) for 6 × 15 min to remove unbound primary antibody. Secondary antibodies (Supplementary Table 3) were diluted in PBT, and samples were incubated at 4 °C overnight with shaking, followed the next day by 6 × 15 min washes in PBT. Heart samples were placed in the middle of a 100-μm spacer (Invitrogen, S24737) and mounted in Fluoromount-G (SouthernBiotech, 0100-01). Large Z-volumes of the samples were imaged using different objectives and tile scanning on a Leica SP5, Leica SP8 STED or Leica SP8 Navigator microscopes.
For mouse retina immunostaining, eyes were collected at the indicated time points and fixed in 4% PFA in PBS for 20 min at room temperature. After microdissection, retinas were fixed in 4% PFA for an additional 45 min, followed by two PBS washes of 5 min each. Retinas were blocked and permeabilized with PBTS (0.3% Triton X-100, 3% fetal bovine serum (FBS) and 3% donkey serum) for 1 h. Samples were then incubated overnight at 4 °C in Biotinylated isolectinB4 (Vector Labs, B-1205, diluted 1:50) and primary antibodies (Supplementary Table 3) diluted in PBTS buffer. After five washes (20 min each) in PBTS buffer diluted 1:2, samples were incubated for 2 h at room temperature with Alexa conjugated secondary antibodies (Thermo Fisher). After two washes in PBS, retinas were mounted with Fluoromount-G (SouthernBiotech).
For whole-mount immunofluorescence of mouse aortas, adult mice were euthanized with CO2 and intially perfused with 4 ml of KCl 50 mM, followed by perfusion-fixation with 4% formaldehyde (PanReac Apply-Chem, 2529311212) injected in the left ventricle during 5 min. Aortas were obtained from the spine and collected in 4% PFA to complete fixation for 2 h at 4 °C. After three 10 min washes in PBS, aortas were cleaned from perivascular fat tissue and opened longitudinally. Aortas were than incubated in permeabilization-blocking buffer (0.5% Triton X-100 and 20% donkey serum in PBS) for 2 h at room temperature with shaking. Subsequently the aortas were incubated in PBST (10% Tween 20, 10% FBS, 10% donkey serum in PBS) for 2 h at room temperature and after incubated in the same solution containing primary antibodies (Supplementary Table 3) overnight at 4 °C. The next day, antibodies were washed four times (1 h each) in PBT at 4 °C. After washing, aortas were incubated in PBT containing fluorochrome-conjugated secondary antibodies (Supplementary Table 3) at 4 °C overnight. After five washes (1 h each) in PBT at 4 °C, aorta samples were flat-mounted on a microscope slide, with the luminal endothelium facing up, with Fluoromount-G (SouthernBiotech, 0100-01).
Immunofluorescence on cryosections
Embryonic, postnatal or adult hearts and livers were isolated and fixed in 4% PFA for 2 h on ice and then washed with PBS 3 × 5 min. After washing, organs were incubated in 30% sucrose in PBS overnight at 4 °C with shaking. Organs were then embedded in OCT (Sakura), snap frozen on dry ice and stored at –80 °C. Cryosections (25 μm) were cut with a Leica Cryostat (Leica CM1950). For immunofluorescence, slides were dried at room temperature for 10 min followed by washing with PBS 3 × 5 min, then blocked with PBBT (0.5% Triton X-100 and 10% donkey serum in PBS) for 1 h at room temperature and incubated with primary antibodies (Supplementary Table 3) diluted in PBBT overnight at 4 °C in a humidified chamber. The following day, slides were washed in PBS for 3 × 5 min and then incubated with secondary antibodies (Supplementary Table 3) diluted in PBS for 2 h at room temperature in humidified chamber. Next, slides were washed in PBS at room temperature for 3 × 5 min and mounted in Fluoromount-G (SouthernBiotech, 0100-01) before confocal imaging.
For phosphoextracellular signal-regulated kinase (p-ERK) staining, we used a different protocol with the tyramide signal amplification kit (TSA Plus Fluorescein System; PerkinElmer Life, NEL774). In brief, cryosections were dried at room temperature for 30 min followed by washes in PBS for 3 × 5 min. Cryosections were then incubated in sub-boiling sodium citrate buffer (10 mM, pH 6.0) for 30 min, allowed to cool at room temperature for 30 min, and incubated for 30 min in 3% H2O2 in methanol to quench endogenous peroxidase activity. Slides were rinsed in double-distilled H2O and washed in PBS for 3 × 5 min, followed by blocking in PBBT for 1 h and incubation with the primary antibody (Supplementary Table 3) in PBBT overnight at 4 °C. Slides were then washed and incubated for 2 h with anti-rabbit-HRP secondary antibody at room temperature; after washes, the signal was amplified using the TSA fluorescein kit (NEL774).
Microscopy
We used different laser-scanning confocal microscopes depending on the complexity of the immunostainings and the combination of signals or fluorescent proteins detected. We used the inverted Leica TCS SP5 confocal or the Leica TCS SP8 STED or Navigator Confocal microscopes containing a 405 nm laser and a white laser that allows excitation at any wavelength from 470 to 670 nm. Confocal Zeiss LSM780 was also used sporadically. We acquired either individual fields or tiles of large areas with 10×, 20× and 40× lens. All images shown are representative of the results obtained for each group and experiment. Littermates were dissected and processed under exactly the same conditions. Comparisons of phenotypes or signal intensity were made with pictures obtained using the same laser excitation and confocal scanner detection settings. Images were processed and analysed using Leica SP5, SP8, Zeiss LSM780, ImageJ/Fiji v1.53c and Adobe Photoshop CC 19.1.5 software.
Cell culture and in vitro cell treatments
Pooled human umbilical vein endothelial cells (HUVECs) were purchased from Lonza (CC-2519) and cultured in endothelial basal medium (EBM, Lonza) supplemented with hydrocortisone (1 μg ml−1), bovine brain extract (12 μg ml−1), gentamicin (50 μg ml−1), amphotericin B (50 ng ml−1), human recombinant epidermal growth factor (10 ng ml−1), and 10% fetal bovine serum (FBS, Life Technologies). Human embryonic kidney cells (HEK293FT) were used for lentivirus production and purchased from Life Technologies (R70007). These cells were cultured in DMEM supplemented with 10% FBS (Life Technologies) and geneticin (500 μg ml−1, Invitrogen). All cells used were from commercial sources and therefore were not authenticated but they had the reported characteristics and gene expression profiles. They were tested negative for mycoplasma and maintained at 37 °C in a humidified atmosphere with 5% CO2. Notch activity was induced with DLL4 ligands as previously described34. In brief, recombinant human DLL4 (R&D Systems) was reconstituted at 100 μg ml−1 in sterile PBS containing 0.1% bovine serum albumin. DLL4 was immobilized by coating culture dishes with 500 ng ml−1 DLL4 in PBS for 1 h at room temperature or overnight at 4 °C. To inhibit Notch signalling (by γ-secretase inhibition), HUVECs were treated with 0.1 μM DBZ (CalBiochem) in DMSO. Control groups were treated with the respective vehicles. Induction of HUVECs stably transduced with doxycycline-inducible N1ICD-V5 encoding lentiviruses or controls was performed with 200 ng ml−1 of doxycycline (Sigma).
Lentivirus production and HUVEC transductions
Lentiviruses were used to infect HUVECs. Viruses were collected from HEK293FT cells co-transfected with psPAX2 (Addgene, 12260), pMD2.G (Addgene, 12259) and human N1ICD-V5 inserted into the doxycycline-inducible transfer vector pLVX-TetOne-Puro (Clontech), using the Lipofectamine 2000 transfection reagent (Life Technologies). Viruses were collected 48 and 72 h after transfection, and incubated with HUVECs for 16 h in presence of 8 μg ml−1 polybrene (Santa Cruz). Fresh EBM media containing 1 μg ml−1 puromycin (InvivoGen) was added to select transduced cells.
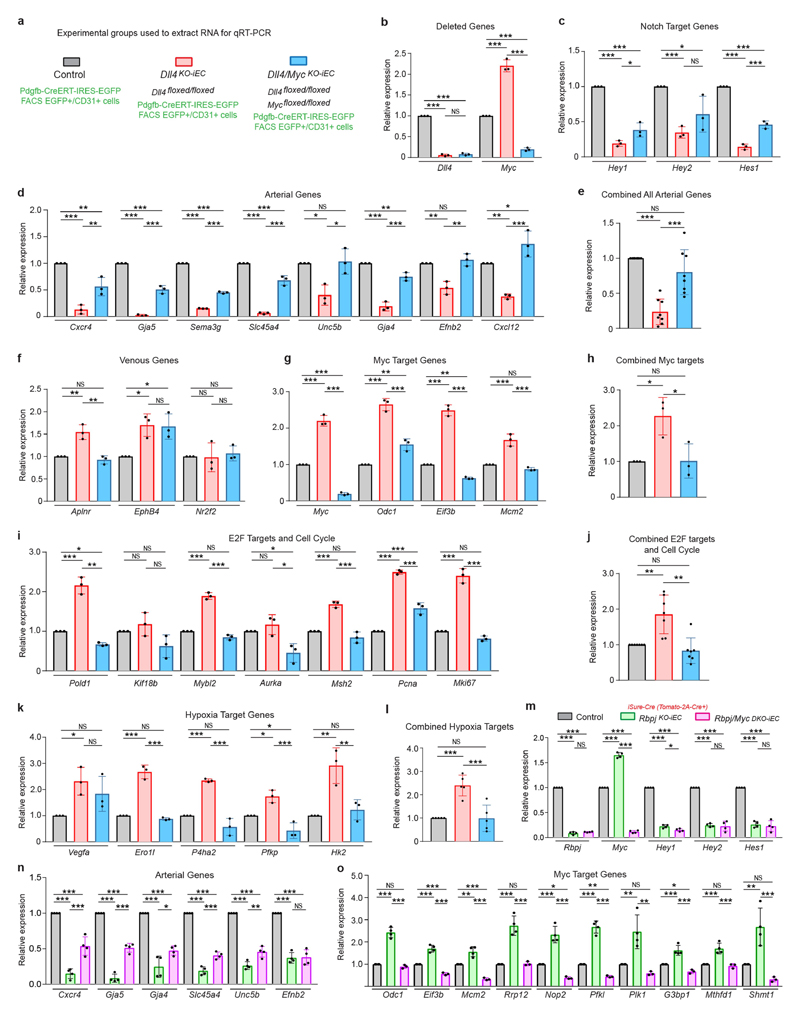
Endothelial cell isolation, RNA extraction and qRT–PCR
Coronary ECs with distinct Notch signalling levels were isolated according to their fluorescence from iChr2-Notch-Mosaic or ROSA26LSL-MbTomato-2A-H2B-GFP-2A-N1ICDP mice after crossing with Tie2-cre mice. Fluorescent (mutant) and non-fluorescent (wild type/control) cells were isolated as indicated further below. To isolate and profile coronary ECs with unaltered or half or full loss of DLL4 signalling, we inter-crossed Pdgfb-icreERT2-ires-egfp mice with wild-type females or Dll4flox/flox Pdgfb-icreERT2-ires-egfp male mice with wild-type or Dll4flox/flox females. Tamoxifen was administered at E13.5 and embryos were collected at E14.5. Thousands of GFP+ cells (from several hearts per litter) were isolated by FACS to obtain three independent samples per group: controls, Dll4 hemizygous mutants, and Dll4 homozygous mutants (Supplementary Table 4). To isolate and profile coronary ECs with unaltered or full loss of Rbpj or Rbpj/Myc we intercrossed mice containing Rbpjflox/flox;Cdh5-creERT2;iSuRe-cre alleles or Rbpjflox/flox/Mycflox/flox;Cdh5-creERT2;iSuRe-cre alleles. Tamoxifen was administered at E12.5 and embryos collected at E15.5. Note that RBPJ turnover after inducing its deletion is slower than DLL4, and a longer window of genetic deletion needed to be used. Thousands of MbTomato+ (mutant) and MbTomato- (control) cells were isolated by FACS from several independent litters per experimental group (Supplementary Table 4). To isolate and profile coronary ECs from iFlpMTomato-cre/MYfp genetic mosaics with unaltered or full loss of Rbpj or Rbpj/Myc, we intercrossed mice containing the Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjflox/flox alleles or Apln-FlpO;iFlpMTomato-cre/MYfp; Rbpjflox/flox;Mycflox/flox alleles. Thousands of MbTomato-2A-Cre (mutant) and MbYFP (control) expressing cells were isolated by FACS from several independent litters containing E15.5 hearts (Supplementary Table 4).
Embryonic hearts were dissected, minced and digested for 20 min at 37 °C with 2.5 mg ml−1 collagenase type I (Thermo Fisher), 2.5 mg ml−1 dispase II (Thermo Fisher) and 50 μg ml−1 DNaseI (Roche). Dissociated samples were filtered through a 70-μm cell strainer, and cells were centrifuged (300g, 4 °C for 5 min). Cell pellets were gently resuspended in blood lysis buffer (0.15 M NH4Cl, 0.01 M KHCO3 and 0.01 M EDTA in distilled water) and incubated for 10 min on ice to remove erythroid cells. Cells were centrifuged (300g at, 4 °C for 5 min), and cell pellets were gently resuspended in blocking solution (DPBS without Ca2+ or Mg2+ and containing 3% dialysed FBS (Thermo Fisher)) and incubated at 4 °C with shaking for 20 min. Cells were centrifuged (300g at 4 °C for 5 min), resuspended and incubated for 30 min at 4 °C with APC rat-anti-mouse CD31 (BD Pharmigen, 551262, 1:200). Cells were then centrifuged (300g, 4 °C for 5 min), resuspended and washed in DPBS without Ca2+ or Mg2+ and centrifuged again, and cell pellets were resuspended in blocking solution. Cells were kept on ice until used for FACS. DAPI (5 mg ml−1) was added to the cells immediately before FACS. Cells were sorted in a FACS Aria Cell Sorter (BD Biosciences). For each group, approximately 2,000–6,000 DAPI negative APC-CD31+ cells without or with fluorescence (GFP or Tomato or Cherry or Cerulean) were sorted directly to buffer RLT (RNAeasy Micro kit, Qiagen) and RNA extracted according to the manufacturer instructions and stored at –80 °C. This RNA was later used for cDNA production and quantitative PCR (qPCR) or RNA-seq analysis.
For quantitative reverse transcription PCR (qRT–RCR), total RNA was retrotranscribed with the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Thermo Fisher, 4368814). cDNA was preamplified with Taqman PreAmp Master Mix containing a Taqman Assay-based pre-amplification pool containing a mix of the Taqman assays indicated in Supplementary Table 5. Preamplified cDNA was used for qRT–PCR with the same gene specific Taqman Assays and Taqman Universal Master Mix in a AB7900 thermocycler (Applied Biosystems). AB7900 software was used to retrieve and analyse data.
In the case of the experiments with HUVECs, RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer protocol. For cDNA synthesis, 1 μg of total RNA was reverse transcribed with the M-MLV reverse transcriptase (Thermo Fisher). qRT–PCR was performed using the human Taqman assays indicated in Supplementary Table 5, with the StepOnePlus real-time PCR system (Applied Biosystems). Gene expression was calculated using the ΔΔC t method and Actb used as reference gene for data normalization.
Western blot analysis
Protein extracts from HUVECs were collected by lysis in RIPA buffer (Sigma; 150 mM NaCl, 1.0% (v/v) IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) supplemented with 1 × EDTA-Free Complete Protease Inhibitor Cocktail (Roche) and 1 mM phenylmethylsulfonyl fluoride (Carl Roth). Proteins were separated by SDS–PAGE (Tris-glycine gels with Tris/glycine/SDS buffer, Bio-Rad) and transferred onto nitrocellulose membranes using the Trans Turbo Blot system (Bio-Rad). The following primary antibodies were used: anti-c-MYC (Cell Signaling Technology, 9402, 1:1,000), anti-N1ICD (Cell Signaling Technology, 4147, 1:1,000), anti-V5 (Cell Signaling Technology, 13202, 1:2,500) and anti-TUBULIN (Cell Signaling Technology, 2148, 1:5,000). Peroxidase-conjugated Goat IgGs (1:5,000) secondary antibodies were purchased from Jackson Immuno Research Labs. Target proteins were visualized by chemiluminescence using the ECL detection kit (Clarity Western ECL Substrate, Bio-Rad) and the ChemiDoc MP Imaging System (Bio-Rad).
Next-generation sequencing
Next-generation sequencing services were provided by the Genomics Unit of CNIC. One microlitre of stored RNA per sample was quantified using the Agilent 2100 Bioanalyzer RNA6000 pico total RNA chip. After quantification, 700 pg of total RNA were used for cDNA amplification with the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech-Takara). 1 ng of amplified cDNA per sample was used to generate barcoded libraries using the Nextera XT DNA library preparation kit (Illumina). In brief, cDNA was fragmented and adapters are added in a single reaction followed by amplification and clean-up steps. Library size was checked using the Agilent 2100 Bioanalyzer High Sensitivity DNA chip, and library concentration was determined with a Qubit fluorometer (Thermo Fisher Scientific). Libraries were sequenced on a HiSeq2500 (Illumina) to generate 100 bases paired-end reads.
RNA-seq data analysis
RNA-seq data were analysed by the CNIC Bioinformatics Unit. Sequencing reads were processed with a pipeline that used FastQC v0.11.5 (Babraham Bioinformatics, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to evaluate their quality, and cutadapt35 to trim sequencing reads, thus eliminating Illumina and SMARTer adaptor remains, and discard reads shorter than 30 bp. Resulting reads were mapped against mouse transcriptome GRCm38.76, and gene expression levels were estimated with RSEM v.1.2.3036. Expression count matrices were then processed with an analysis pipeline that used the bioconductor package limma v.3.32.1037 for normalization (using the TMM method) and differential expression testing; matrix processing considered only genes expressed with at least 1 count per million (CPM) in at least as many samples as the condition with the least number of replicates. Two pairwise contrasts were performed: Dll4f/f-iDEC-Pdgfb-24h (abbreviated in raw data file as ‘KO’) versus ControliDEC-Pdgfb-24h (abbreviated in raw data file as ‘WT’) and Dll4f/wt-iDEC-Pdgfb-24h (abbreviated in raw data file as ‘Het’) versus ControliDEC-Pdgfb-24h. Changes in gene expression were considered significant if associated to Benjamini and Hochberg adjusted P < 0.05.
Complementary enrichment analyses with GSEA38 were performed for each contrast, using the whole collection of genes detected as expressed (12,872 genes) to identify gene sets that had a tendency to be more expressed in either of the conditions being compared. Gene sets, representing pathways or functional categories from the Hallmark and Gene Ontology collections were retrieved from MsigDB39. Enriched gene sets with FDR < 0.25 were considered of interest.
Ultrasound analysis of cardiac function
Mice aged 2 to 5 months were analysed for cardiac function at the CNIC Advanced Imaging Unit. In brief, mice were anaesthetized by inhalation of isoflurane/oxygen and examined with a 30-MHz transthoracic echocardiography probe and a Vevo 2100 ultrasound system (Visual-Sonics). End-systolic and end-diastolic left ventricle volumes (LVESV and LVEDD), ejection fraction (EF) and fractional shortening (FS) were calculated from short-axis and long-axis B-mode views.
In vivo EdU labelling and EC proliferation detection
To detect EC proliferation rates in embryonic hearts, 50 μg g−1 body weight EdU (Invitrogen, A10044) was injected intraperitoneally into pregnant mice 1 h before dissection. Embryonic hearts were isolated for cryosection or wholemount analysis. EdU signals were detected with the Click-it EdU Alexa Fluor 647 or 488 Imaging Kit (Invitrogen, C10340 or C10337). In brief, after all other primary and secondary antibody incubations, samples were washed according to the immunofluorescence staining procedure and then incubated with Click-iT EdU reaction cocktails for 40 min, followed by DAPI counterstaining.
Statistical analysis
As indicated in Supplementary Data 1 file, two groups of samples with a Gaussian distribution were compared by unpaired two-tailed Student’s t-test. Comparisons among more than two groups were made by ANOVA followed by the Tukey multiple comparisons test. Data are mean and s.d. as indicated, and differences were considered significant at P < 0.05. Initial calculations were done in Excel 2016, and final data points were represented and analysed with GraphPad Prism v.7.03.
Mice were treated randomly before tissues collection and analysis. Mice or tissues were selected for a posteriori analysis based on their genotype, the detected Cre-dependent recombination frequency, and quality of multiplex immunostaining. For in vitro experiments samples were allocated randomly in each experiment. Sample size was chosen according to the observed statistical variation and published protocols. In most cases, investigators were not blinded during data collection or analysis owing to its impracticality and the need for a priori knowledge of which control and mutant samples are being handled and selected for downstream analysis. As an example, in 8 embryos of a litter, often only 1 or 2 have the desired combination of alleles, and therefore those selected mutant embryos need to be compared with few selected control embryos, so that all downstream costs and analysis are kept to the minimum necessary. ImageJ/FIJI software was used to analyse the microscopy data in an automatic and objective manner. All experiments in the paper were quantified using standardized experimental controls and quantitative methods to avoid bias.
Extended Data
Extended Data Fig. 1. Multispectral genetic mosaics to analyse the proliferation and arteriovenous fate of heart and retina ECs.
a, Schematic of Dual ifgMosaic technology to multispectrally barcode cells with different combinations of chromatin (iChr2-Mosaic) or membrane (iMb2-Mosaic) tagged fluorescent proteins. b, Combined or individual channels of multispectral confocal imaging of the heart-surface coronary vasculature and sections. c, iChr-Notch-Mosaic allele targeted to the Rosa26 locus. In cells expressing Cre, the allele undergoes stochastic recombination, generating a mosaic of cells expressing either H2B-Cherry (control wild-type cells with Cherry FP bound to chromatin/nuclei), H2B-GFP-2A-DN-MAML1 (GFP labels the nuclei of cells with induced downregulation of Notch/Rbpj signalling) or HA-H2B-Cerulean-2A-N1ICDP (Cerulean containing the HA epitope labels the nuclei of cells with induced upregulation of NOTCH1 signalling). d, qRT–PCR analysis of Notch target-genes in H2B-GFP+ and H2B-Cherry+ ECs obtained from iChr-NotchTie2-Mosaic E15.5 hearts by FACS. e, Survival rate of adult iChr-NotchTie2-Mosaic mice follows expected Mendelian frequencies indicating no lethality. f, g, Confocal images of iChr-NotchNfatc1-Mosaic embryonic hearts sections showing the localization and relative frequency of ECs with distinct Notch signalling levels. h, Quantification of experiments in f and g showing how the relative percentage of each type of cell changes throughout development according to their location. i, Proliferation (EdU+) of coronary ECs (ICAM2+/ERG+ in myocardium) and endocardium (EMCN+/ERG+) at E12.5 and E15.5. j, k, Analysis of cell proliferation in iChr-NotchTie2-Mosaic ECs. l, Illustration showing the iFlpMTomato-cre/MYfp mosaic allele and expected outcomes in cells expressing Flp/FlpO recombinase. m, qRT–PCR analysis of Rbpj mRNA levels in CD31+/MbYFP+ and CD31+/MbTomato+ ECs collected by FACS from Apln-FlpO;iFlpMTomato-cre/MYfp; Rbpjf/f E15.5 hearts. Residual Rbpj expression is due to the FACS and qRT–PCR assay as previously described33 (see Methods). n, Confocal images and quantification of postnatal day 6 (P6) retinas showing the localization and frequency of MTomato+ (RbpjKO) and MYFP+ (wild-type) cells in retina arteries (A), veins (V) and intermediate capillaries. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 2. Ectopic Notch activation is compatible with vein development and impairs coronary artery development.
a, Genetic construct used to generate mice with Cre-dependent conditional expression of MbTomato, H2B-GFP and N1ICDP in Tie2-Cre + coronary vessels which results in increased expression of canonical Notch target genes. b, Bright-field images of E12.5 and E15.5 embryos and hearts showing the appearance of microcirculatory defects at E15.5. c, Statistical analysis of mouse survival rate per stage indicates that most mutant mice do not pass embryonic stage E16.5. d, Cardiovascular development defects in N1ICDPTie2-cre hearts may result in tissue hypoxia and the observed upregulation of Vegf mRNA. e, f, Myocardial EC proliferation analysis. g, h, CD31 staining of control and N1ICDPTie2-cre heart sections showing vascular defects and thinner myocardial wall (inset) in mutant hearts. i–m, Analysis of the heart surface coronary vein diameter and plexus development and density. n, Confocal images showing immunostaining of the heart surface venous plexus for the arterial marker CX40. ECs with high Notch activity (Tomato+) do not express CX40 and are able to form veins. o, Confocal images of E14.5 heart veins showing that Tomato/N1ICDP+ cells (yellow arrowheads) express EPHB4 like wild-type cells (white arrowheads). p, Confocal images of E14.5 heart veins showing that GFP+/N1ICDP+ cells (yellow arrowheads) express COUP-TFII like GFP-/wild-type cells (white arrowheads). q, Chart showing the quantification of COUP-TFII immunosignals intensity in 280GFP+and 280GFP- cells present in the veins of 3 independent N1ICDPTie2-cre hearts (2 veins per heart were quantified). r, Schematic of the mouse postnatal retina vascular development, in which tamoxifen was induced at P1, when the vessels start to grow, and analysis was carried at P6. s–v, Cells with induced high Notch activity (MbTomato+or H2B-GFP+, yellow arrowheads) can differentiate and form veins like wild-type cells (white arrowheads). The Tomato+/N1ICDP+ cells found in veins do not express the arterial marker CX40, and instead express the venous markers VEGFR3, COUP-TFII and EPHB4. w, Quantification of signals (in pictures like shown in s–v) indicate no change in the expression of arteriovenous genes. x, Analysis of coronary artery development. y, z, Confocal images of heart sections and quantifications of immunosignals in the myocardium showing an increase in CX40, DLL4 and SM22α in N1ICDPTie2-cre myocardium vessels. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 3. Overactivation of Notch in PDGFB+ coronary ECs, but not in NFATC1+ endocardium, compromises coronary vessel and heart development.
a, Schematic of the inducible Notch gain-of-function allele, that once crossed with the Pdgfb-icreERT2-ires-egfp allele enables the co-expression of Mbtomato, H2B-GFP and N1ICDP in PDGFB+ coronary ECs. b, Confocal picture showing the strong expression of the Pdgfb-icreERT2-ires-egfp allele in coronary vessels of the myocardium (m), but not in the EMCN+ endocardium (e). c, Stereomicroscope analysis of mutant embryos and control littermates showing growth retardation at E16.5. d, Immunostaining for the reporter MbTomato shows the induction of the allele in Pdgfb-icreERT2-ires-egfp+ myocardium vessels (ICAM2+) but not endocardium. e, g, Thinner myocardial wall and vascular defects in N1ICDPPdgfb-creERT hearts. f, h, Malformation and reduced coronary arterial diameter in N1ICDPPdgfb-creERT hearts. i–k, Reduced coronary vessel density and vein diameter in N1ICDPPdgfb-creERT hearts. l, m, Expression of the arterial proteins DLL4 and CX40 in control and N1ICDPPdgfb-creERT coronary vessels. n, o, Reduced proliferation (ERG+/EdU+) of N1ICDPPdgfb-creERT subepicardium venous vessels (whole-mount and sections). p, Induction of N1ICDP expression (MbTomato+) in the NFATC1+ endocardium and its lineages does not cause major cardiovascular development defects at E15.5. q–t, Comparative analysis of coronary artery and vein development in control and N1ICDPNfatc1-cre hearts. Note the absence of GFP+/N1ICDP+ cells in veins derived from Nfatc1-cre lineage in N1ICDPNfatc1-cre mice. u, Haematoxylin and eosin staining of 7-month-old hearts of control and N1ICDPNfatc1-cre mice. Mutant hearts are larger. Quantifications of heart weight per body weight and left ventricle (LV) fraction shortening are shown. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 4. Induction of cell-cycle and metabolic pathways in coronary ECs 24 h after Dll4 deletion.
a, List of the 50 most up- and downregulated genes (from a total of 3,360) after Dll4 deletion in coronary ECs for 24 h. b, c, List of dysregulated genes within a selected enriched gene set (gene set enrichment analysis, GSEA). d, Fold changes for a selected list of genes previously shown to be upregulated when HUVECs are exposed to hypoxia40,41. e, Vegf mRNA levels do not change in hearts from Dll4KO-Pdgfb-24h (E14.5) and RbpjKO-iEC (E15.5) embryos, suggesting the absence of cardiac hypoxia, despite the existence of endothelial hypoxia. f, The group of genes belonging to the sprouting angiogenesis GSEA pathway are not differentially regulated. g, The expression of most genes previously found to be upregulated in tip cells is not increased after the loss of DLL4–NOTCH signalling. h, Only 7 genes are significantly dysregulated in Dll4Het-Pdgfb-24h samples. Data shown as mean ± s.d. *P < 0.05, Benjamini and Hochberg adjusted.
Extended Data Fig. 5. DLL4 signalling dose is essential for coronary proliferation and arterialization but not sprouting or connection to the aorta.
a, Dll4 deletion in Pdgfb-icreERT2-ires-egfp + coronary ECs for 24 h (from E11.5–E12.5 or E13.5–E14.5) does not induce endothelial sprouting, but slightly reduces the mean density of heart subepicardial vessels at E14.5. b–e, The coronary plexus forming the coronary artery connects (yellow arrows) to the main aorta of control, Dll4Het-iEC and Dll4KO-iEC hearts. However, despite no defects in the connection, the calibre of the coronary artery stem is reduced and most Dll4Het-iEC hearts and all Dll4KO-iEC hearts do not develop proper coronary arteries with CX40+signals (pink arrows) at E14.5. f, At E17.5, the surviving Dll4Het-iEC embryos also show a major developmental defect or delay in coronary artery development. Of note, the DLL4 heterozygous haploinsufficiency is variable and more pronounced on the C57BL/6J background used in this study, as previously reported8,9. g, h, Dll4 and Rbpj deletion reduces the frequency of EdU+ ECs in subepicardium vessels. i, Dll4 deletion increases p-ERK activity in subepicardium vessels. j, k, Dll4 and Rbpj deletion increases the frequency of p21+/ERG+ ECs in subepicardium vessels. l, m, Dll4 and Rbpj deletion increases the frequency of proliferation (EdU+/ERG+) in arterial zone coronary vessels. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 6. Regulation of MYC by DLL4–NOTCH signalling in vivo and in vitro.
a, MYC–GFP fusion protein expression is upregulated in coronary vessels (ICAM2+) after inducing heterozygous Dll4 deletion. b, Comparison of MYC protein expression in wild-type and Rbpj mutant ERG+ ECs. c, Western blot analysis showing no major changes in MYC protein levels after stimulation of HUVECs with DLL4 ligands for 6 or 24 h, in the presence or absence of the Notch inhibitor DBZ. d, qRT–PCR analysis showing significant changes of Notch target genes (Dll4, Hey1, Hey2, Hes1), but not Myc and Odc1, after stimulation of HUVECs with DLL4 ligands for 8 h in the presence or absence of the Notch inhibitor DBZ. e–h, Western blot and qRT–PCR analysis showing that an even stronger activation of Notch signalling (for 24 h, e and f; and 48 h g and h) and its downstream target genes (HEY1,HEY2,NRARP and HES1) in HUVECs overexpressing Dox-inducible N1ICD-V5 has relatively minor or non-significant effects on MYC expression. ODC1 expression is 40% decreased. i, Whole-mount analysis of control and MYC mutant hearts showing that global MYC deletion compromises the development of coronary vessels and consequently arteries (CX40+).j, Sectional analysis of control and MYC mutant hearts showing a severe depletion of coronary vessels in the myocardium which causes a reduction in its thickness. k, Mosaic induction of Mycflox/flox Cdh5-creERT2 iSuRe-cre mice shows that MYC-null ECs (MbTomato-2A-Cre+) form well coronary arteries, in contrast to hearts with full induction of Myc deletion. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1. For western blot gel source data, see Supplementary Fig. 1. Western blot controls (tubulin) were run on separate gels, owing to overlapping size with MYC, as sample processing controls.
Extended Data Fig. 7. Loss of Myc rescues the proliferation and arterialization defects caused by Rbpj or Dll4 deletion.
a, Sectional analysis of control and Rbpj/MycDKO-iEC iSuRe-cre mutant hearts showing that MbTomato-2A-Cre-expressing cells (from the induced iSuRe-cre allele) form CX40+ coronary arteries. b, qRT–PCR analysis showing the efficient deletion of Rbpj and Myc in Rbpj/MycDKO-iEC mutant cells. Note residual expression detected is due to the FACS and qRT–PCR assay as previously described33 (see Methods). c, Confocal pictures of the heart deeper arterial zone and quantifications showing that the additional loss of Myc rescues the proliferation (ERG+/EdU+) defects of RbpjKO ECs, rescuing also arterial development. d–f, Confocal pictures and quantifications showing that loss of Myc rescues the proliferation and arterial development defects induced by hemizygous loss of Dll4. g, h, The additional use of the iSuRe-cre allele, ensures that Tomato-2A-Cre expressing cells have deletion of all floxed alleles (in this case Dll4 and Myc). These cells do not form arteries on a Dll4floxed background but yes on a Dll4/Mycfloxed background. Note that Tomato-cells also express Cdh5-creERT2, which also efficiently deletes Dll4 and Myc, independently of recombination of the iSuRe-cre transgene. i, Arterial development in control, RbpjKO-iEC and Rbpj/MycDKO-iEC P6 retinas. Tamoxifen was injected at P2 and P3. Data shown as mean ± s.d. ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 8. Gene expression signatures of Dll4/Myc and Rbpj/Myc mutant coronary vessels.
a–l, qRT–PCR analysis of the indicated genes in the samples mentioned in a. Each dot in a chart column bar represents the relative measure in a sample of cells obtained by FACS from an entire litter containing several Pdgfb-icreERT2-ires-egfp+ hearts. Charts in e, h, j and l represent the fold change of all genes belonging to the indicated group and shows that the additional deletion of Myc rescues the expression of all sets of genes to control sample levels. m–o, qRT–PCR analysis of the indicated genes in the samples mentioned in m. Each dot in a chart column bar represents the relative measure in a sample of cells obtained by FACS from an entire litter containing several Tomato- and Tomato+ hearts. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. For statistics, see Supplementary Data 1.
Extended Data Fig. 9. iFlpMTomato-cre/MYfp genetic mosaics reveal the long term fate of RbpjKO and Rbpj/MycDKO ECs in different organs.
a, Schematic of the Apln-FlpO and iFlpMTomato-cre/MYfp alleles used to induce full loss-of-function genetic mosaics in the mouse heart, retina, liver and aorta. b, Lineage qRT–PCR analysis of Tomato+ and YFP+ cells isolated from Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjf/f and Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjf/f;Mycf/f E15.5 hearts. Note that Apln-FlpO recombines the alleles at an early stage and that mutant cells differentially segregate throughout development, leading to gene expression changes at E15.5 that may reflect not only the regulation by Rbpj and Myc (shown in Extended Data Fig. 8) but also their segregational arteriovenous fate. c, Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjf/f;Mycf/f mouse survival rate. d, Expression of Apln-HA-FlpO in retina vessels at P6 shows the localization of HA-FlpO in the nuclei of angiogenic front ECs, and not in arteries. e, Schematic of recombination of the Apln-FlpO and iFlpMTomato-cre/MYfp alleles in the retina angiogenic front at P2 and distribution of MbTomato+ and MbYFP+ cells at P6. f–h, Representative confocal images of P6 retinas from mice with the indicated genotypes, and immunostained for isolectinB4, Tomato/Dsred and YFP. Arteries are easily distinguishable from veins by their relatively stronger IsolectinB4 signal and morphology/diameter. i, Quantifications shown are complementary to those in Fig. 4g and allow the direct comparison of significance in the differential distribution of Tomato+ cells only within arteries, or only within capillaries, or only within veins, without taking into account their distinct capillary frequencies and developmental proliferation bias. The results indicate that Rbpj/MycDKO ECs are less frequently found in capillaries, and therefore proliferate less, but have a normal ability of forming arteries, unlike RbpjKO ECs, that occur at a relatively high frequency in capillaries and at very low frequency in arteries (see also log2-transformed fold change indicated in Fig. 4g). j, k, Sectional analysis of control and mutant genetic mosaics of embryonic, newborn and adult hearts showing the very low frequency or absence of Tomato+/mutant cells in coronary arteries of Rbpjflox/flox hearts, and their presence in the coronary arteries of control and Rbpjflox/flox/Mycflox/flox hearts. For quantifications of adult hearts data, see Fig. 4e. l, Sectional analysis of control and mosaic mutant adult livers showing portal arteries (PA) and portal veins (PV) and respective quantifications in these oxygen-rich (DLL4+ and EFNB2+) liver portal vessels. m, Confocal imaging of the inner surface of aortas, stained for CD144 (VE-cadherin), showing the relative occurrence of control and mutant cells and chart with the respective quantifications. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 10. Models for the regulation of arterialization by Notch.
The initial model used to describe arterialization was based on the assumption that an increase in pulsatile and oxygenated blood flow induces the direct differentiation and development of arterial vessels. The current model is based on the evidence that pre-arterial capillary ECs have higher Notch–RBPJ activity and higher expression of arterial genes. This model assumes that arterial fate is determined before the increase in arterial blood flow, through the direct induction of arterial gene expression via the Notch–RBPJ transcriptional complex. Here, we propose a model in which Notch–RBPJ suppresses MYC-dependent cell cycle or biosynthetic activity, rendering ECs more permissive to the adoption of an arterial phenotype without the need for Notch-dependent genetic pre-determination or differentiation. This model is supported by the observed phenotypic and transcriptomic outcomes in several mouse mutant models.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-3018-x.
Acknowledgements
Research in the Benedito laboratory was supported by the European Research Council (ERC) Starting Grant AngioGenesHD (638028), the CNIC Intramural Grant Program Severo Ochoa (11-2016-IGP-SEV-2015-0505), and the Ministerio de Ciencia y Innovación (MCIN SAF2013-44329-P, RYC-2013-13209 and SAF2017-89299-P). The CNIC is currently supported by MCIN and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505). Research in the Potente laboratory was supported by the Max Planck Society, the ERC Consolidator Grant EMERGE (773047), the Deutsche Forschungsgemeinschaft (SFB 834), and the Foundation Leducq Transatlantic Network. W.L. received a Marie Curie FP7 COFUND CNIC fellowship. M.F.-C. and I.G.-G. were supported by PhD fellowships from Fundación La Caixa (CX_E-2015-01 and CX-SO-16-1, respectively) and S. M. by the Austrian Science Fund (FWF) project J4358. We thank S. Bartlett and S. Rocha for English editing; J. L. de La Pompa and D. Macgrogan for scientific input and the CNIC Transgenesis, Microscopy, Genomics and Bioinformatic units. We also thank M. Yanagisawa, F. Radtke, R. H. Adams, M. Fruttiger, F. Alt, B. Sleckman and T. Honjo for sharing the Tie2-cre, Dll4floxed, Cdh5(PAC)-creERT2, Pdgfb-icreERT2-ires-egfp, Mycfloxed, GFP-Myc and Rbpjfloxed mice, respectively.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Author contributions W.L. and R.B. designed most of the experiments, interpreted results and wrote the manuscript. I.G.-G. engineered the DNA constructs, validated the iFlpMTomato-cre/MYfp and Apln-FlpO mice, and gave input on genetic mosaics analysis. M.F.-C. supported RNA-seq, FACS, qRT–PCR and iSuRe-cre analysis. L.G.-O. performed mouse aortas dissection, immunostaining and analysis. V.C.-G. and M.S.S.-M. gave general technical assistance, managed the mouse colonies and breedings, and established some of the ES cell lines used to generate mouse models. S.M., J.A. and M.P. designed and performed experiments with endothelial cell lines, interpreted results and revised the manuscript.
Competing interests The authors declare no competing interests.
Peer review information Nature thanks Andreas Fischer, Holger Gerhardt and Kristy Red-Horse for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at http://www.nature.com/reprints.
Data availability
The RNA-seq data can be viewed at the Gene Expression Omnibus (GEO) under accession number GSE158731. We used in this study the MsigDB as indicated above. All other data supporting the study findings are available from the corresponding author upon request. This includes additional raw data such as unprocessed original pictures and independent replicates, which are not displayed in the Article but are included in the data analysis in the form of graphs. Source data are provided with this paper.
References
- 1.Red-Horse K, Siekmann AF. Veins and arteries build hierarchical branching patterns differently: bottom-up versus top-down. BioEssays. 2019;41:e1800198. doi: 10.1002/bies.201800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons M, Eichmann A. Molecular controls of arterial morphogenesis. Circ Res. 2015;116:1712–1724. doi: 10.1161/CIRCRESAHA.116.302953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Benedito R, Hellström M. Notch as a hub for signaling in angiogenesis. Exp Cell Res. 2013;319:1281–1288. doi: 10.1016/j.yexcr.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Lawson ND, Vogel AM, Weinstein BM. Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 6.Pontes-Quero S, et al. High mitogenic stimulation arrests angiogenesis. Nat Commun. 2019;10:2016. doi: 10.1038/s41467-019-09875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benedito R, Duarte A. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr Patterns. 2005;5:750–755. doi: 10.1016/j.modgep.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Duarte A, et al. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale NW, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci USA. 2004;101:15949–15954. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 11.Su T, et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature. 2018;559:356–362. doi: 10.1038/s41586-018-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang JS, et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun. 2017;8:2149. doi: 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pontes-Quero S, et al. Dual ifgMosaic: a versatile method for multispectral and combinatorial mosaic gene-function analysis. Cell. 2017;170:800–814. doi: 10.1016/j.cell.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benedito R, et al. The Notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 15.Wu B, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang AH, et al. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev. 2017;31:1308–1324. doi: 10.1101/gad.301549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen HI, et al. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development. 2014;141:4500–4512. doi: 10.1242/dev.113639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aranguren XL, et al. Unraveling a novel transcription factor code determining the human arterial-specific endothelial cell signature. Blood. 2013;122:3982–3992. doi: 10.1182/blood-2013-02-483255. [DOI] [PubMed] [Google Scholar]
- 19.Ehling M, Adams S, Benedito R, Adams RH. Notch controls retinal blood vessel maturation and quiescence. Development. 2013;140:3051–3061. doi: 10.1242/dev.093351. [DOI] [PubMed] [Google Scholar]
- 20.Lee T, Yao G, Nevins J, You L. Sensing and integration of Erk and PI3K signals by Myc. PLOS Comput Biol. 2008;4:e1000013. doi: 10.1371/journal.pcbi.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasan SS, et al. Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat Cell Biol. 2017;19:928–940. doi: 10.1038/ncb3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitulescu ME, et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat Cell Biol. 2017;19:915–927. doi: 10.1038/ncb3555. [DOI] [PubMed] [Google Scholar]
- 23.Sissaoui S, et al. Genomic characterization of endothelial enhancers reveals a multifunctional role for NR2F2 in regulation of arteriovenous gene expression. Circ Res. 2020;126:875–888. doi: 10.1161/CIRCRESAHA.119.316075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudino M, et al. Radial-artery or saphenous-vein grafts in coronary-artery bypass surgery. N Engl J Med. 2018;378:2069–2077. doi: 10.1056/NEJMoa1716026. [DOI] [PubMed] [Google Scholar]
- 25.Fujita M, Sasayama S. Coronary collateral growth and its therapeutic application to coronary artery disease. Circ J. 2010;74:1283–1289. doi: 10.1253/circj.cj-10-0376. [DOI] [PubMed] [Google Scholar]
- 26.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 28.Claxton S, et al. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 2008;46:74–80. doi: 10.1002/dvg.20367. [DOI] [PubMed] [Google Scholar]
- 29.Koch U, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 31.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 32.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Chacón M, et al. iSuRe-Cre is a genetic tool to reliably induce and report Cre-dependent genetic modifications. Nat Commun. 2019;10:2262. doi: 10.1038/s41467-019-10239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim R, et al. Deubiquitinase USP10 regulates Notch signaling in the endothelium. Science. 2019;364:188–193. doi: 10.1126/science.aat0778. [DOI] [PubMed] [Google Scholar]
- 35.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10. [Google Scholar]
- 36.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheurer SB, Rybak JN, Rösli C, Neri D, Elia G. Modulation of gene expression by hypoxia in human umbilical cord vein endothelial cells: A transcriptomic and proteomic study. Proteomics. 2004;4:1737–1760. doi: 10.1002/pmic.200300689. [DOI] [PubMed] [Google Scholar]
- 41.Weigand JE, Boeckel JN, Gellert P, Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One. 2012;7:e42697. doi: 10.1371/journal.pone.0042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu VT, et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 2016;16:4. doi: 10.1186/s12896-016-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data can be viewed at the Gene Expression Omnibus (GEO) under accession number GSE158731. We used in this study the MsigDB as indicated above. All other data supporting the study findings are available from the corresponding author upon request. This includes additional raw data such as unprocessed original pictures and independent replicates, which are not displayed in the Article but are included in the data analysis in the form of graphs. Source data are provided with this paper.