Summary
Cancers arise through the acquisition of oncogenic mutations and grow through clonal expansion1,2. Here we reveal that most mutagenic DNA lesions are not resolved as mutations within a single cell-cycle. Instead, DNA lesions segregate unrepaired into daughter cells for multiple cell generations, resulting in the chromosome-scale phasing of subsequent mutations. We characterise this process in mutagen-induced mouse liver tumours and show that DNA replication across persisting lesions can produce multiple alternative alleles in successive cell divisions, thereby generating both multi-allelic and combinatorial genetic diversity. The phasing of lesions enables the accurate measurement of strand biased repair processes, the quantification of oncogenic selection, and the fine mapping of sister chromatid exchange events. Finally, we demonstrate that lesion segregation is a unifying property of exogenous mutagens, including UV light and chemotherapy agents in human cells and tumours, which has profound implications for the evolution and adaptation of cancer genomes.
Sequencing and analysis of cancer genomes have identified a wealth of driver mutations and mutation signatures1,3, illustrating how environmental mutagens cause genetic damage and elevate cancer risk4,5. The diversity of mutation patterns identified from cancer genome sequencing is testament to the temporal and spatial heterogeneity of exogenous and endogenous exposures, mutational processes, and germline variation amongst patients. A recent study of diverse human cancers identified 49 distinct single base substitution signatures, with almost all tumours demonstrating evidence of at least three signatures3.
Such intrinsic heterogeneity leads to overlapping mutation signatures that confound our ability to accurately disentangle the biases of DNA damage and repair, or to interpret the dynamics of clonal expansion. We reasoned that a more controlled and genetically uniform cancer model system would overcome some of these limitations and complement human cancer studies. By effectively re-running cancer evolution hundreds of times, we aimed to explore oncogenesis and mutation patterns at high resolution and with good statistical power.
We chemically induced liver tumours in fifteen-day-old (P15) male C3H/HeOuJ inbred mice (Fig. 1a; subsequently C3H, n=104) using a single dose of diethylnitrosamine (DEN), thus greatly extending our previous study6. To provide a genetic comparison and a validation dataset in a divergent mouse strain7, we treated a cohort of CAST/EiJ mice with DEN (subsequently CAST, n=54).
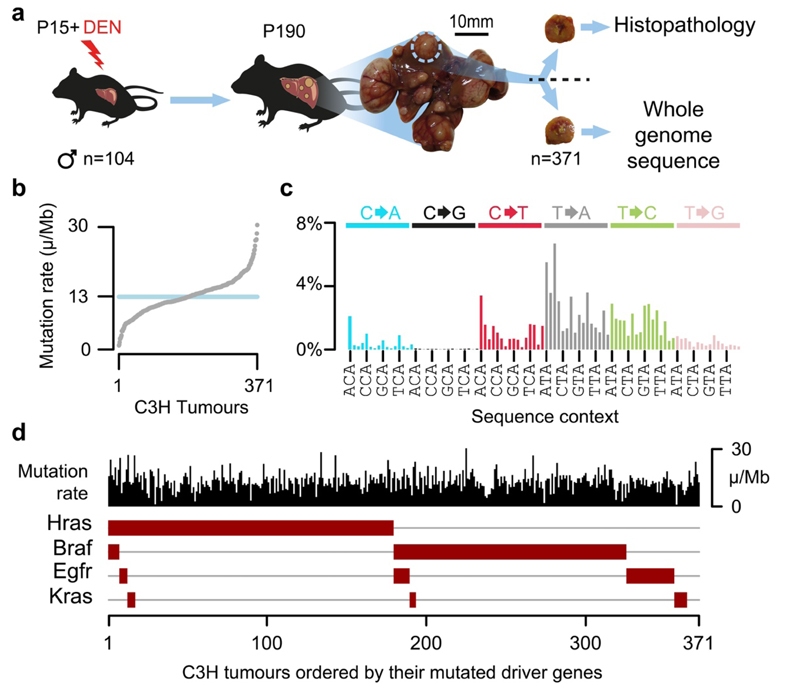
Fig.1. DEN-initiated tumours have a high burden of point mutations with a distinct mutation signature and driver mutations in the EGFR/RAS/RAF pathway.
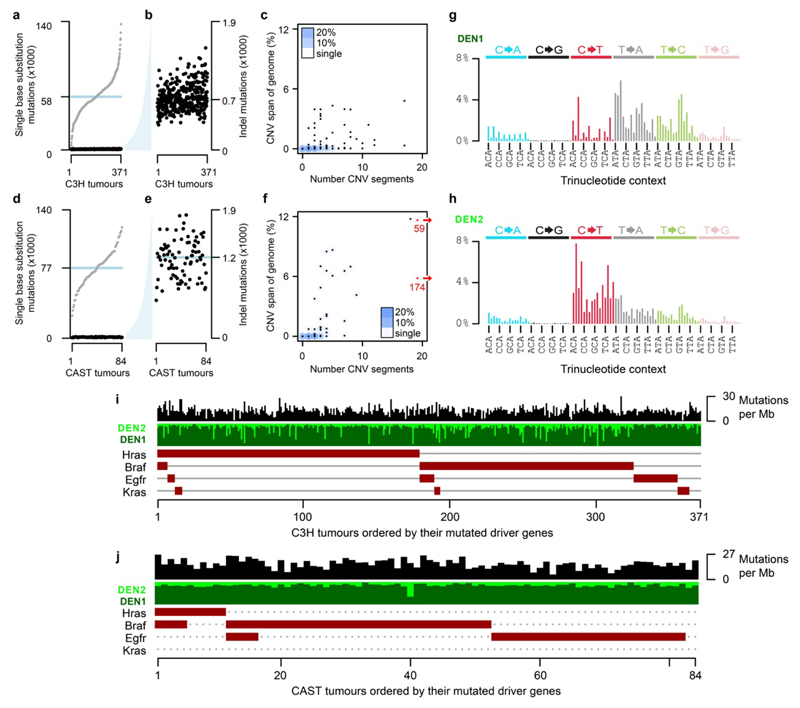
a, Fifteen-day-old (P15) male C3H/HeOuJ mice received a single dose of diethylnitrosamine (DEN); tumours were isolated 25 weeks after DEN treatment (P190), histologically analysed and subjected to whole genome sequencing. b, DEN-induced tumours displayed a median mutation rate of 13 mutations per million base pairs (μ/Mb). c, Mutation spectra histogram for the aggregated mutations of 371 C3H tumours showing the distribution of nucleotide substitutions, stratified by flanking nucleotide sequence context (96 categories). Sequence context for every fourth trinucleotide context is annotated (x-axis). d, Oncoplot summarising each tumour as a column with its mutation rate (black) and the presence of driver mutations in known driver genes (brown boxes). Tumours are ordered by the driver mutations identified.
Whole genome sequencing (WGS) of 371 independently-evolved tumours from 104 C3H mice (Supplementary Table 1) revealed that each genome harboured ~60,000 somatic point mutations, which equates to 13 mutations per megabase (Fig. 1b) and is comparable to human cancers caused by exogenous mutagen exposure such as tobacco smoking and UV exposure8,9. Insertion-deletion mutations, larger segmental changes, and aneuploidies were rare (Extended Data Fig. 1a-f). The tumour genomes were dominated (76%) by T→N/A→N mutations (where N represents any alternate nucleotide, Fig. 1c), consistent with previous studies implicating the long-lived thymine adduct O4-ethyl-deoxythymine as one of the principal mutagenic lesions generated through bioactivation of DEN by cytochrome P450 (CYP2E1)10. In addition to the predominantly T→N signature (subsequently DEN1), deconvolution of mutation signatures revealed a second signature prominent in a minority of tumours (DEN2) but typically present at a low level (Extended Data Fig. 1g-j). DEN2 is mainly composed of C→T/G→A substitutions, likely representing O6-ethyl-2-deoxyguanosine10, which can be repaired by the enzyme MGMT11. Known driver mutations were identified in the EGFR/RAS/RAF pathway6,12,13 (Fig. 1d). These exhibited a strong propensity to be mutually exclusive: 82% of C3H tumours had only a single known driver mutation. Similar results were replicated in CAST mice (Extended Data Fig. 1i,j).
Chromosome-scale segregation of lesions
Strikingly, in each tumour genome we observed multi-megabase segments with pronounced Watson versus Crick strand asymmetries of mutation spectra (Fig. 2). We define Watson strand bias as an excess of T→N over A→N mutations, when called on the forward strand of the reference genome, and the opposite as Crick strand bias. The asymmetrically mutated segments often encompass an entire chromosome, and have a median span of 55Mb (Fig. 2a-d). The scale of these segmental asymmetries is orders of magnitude greater than those generated by transcription coupled repair (TCR)14, APOBEC mutagenesis15,16, or produced by replication strand asymmetries14,17. Despite segmental strand asymmetry, mutation load remains approximately uniform across the genome (Fig. 2e) and asymmetric segments do not correspond to changes in DNA copy-number (Fig. 2f).
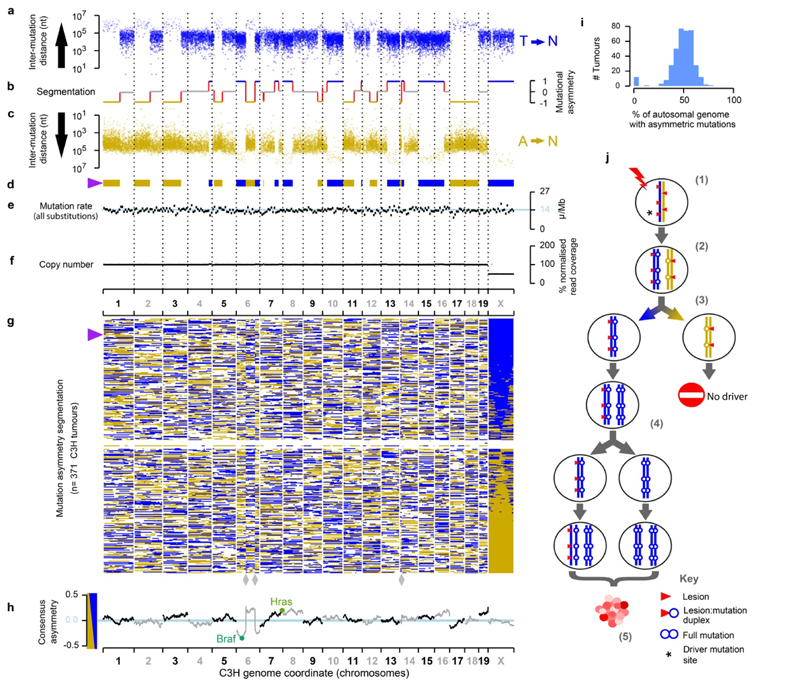
Fig. 2. Chromosome-scale and strand asymmetric segregation of DNA lesions.
a-f, An example DEN-induced C3H tumour (identifier: 94315_N8) with the genome shown over the x-axis. a-c, Mutational asymmetry. Individual T→N mutations shown as points, blue (T on the Watson strand, a) and gold (T on the Crick strand, c), the y-axis representing the distance to the nearest neighbouring T→N mutation on the same strand. b, Segmentation of mutation strand asymmetry patterns. Y-axis position shows the degree of asymmetry (no bias: grey); mutational symmetry switches indicated as red lines. d, Segmentation profile summarised as ribbon showing only the asymmetric segments. e, Mutation rate in 10Mb windows, blue line shows the genome wide rate for this tumour. f, DNA copy number in 10Mb windows (grey) and for each asymmetry segment (black). g, Summary ribbon plots (as in d) for all 371 C3H tumours, ranked by chromosome X asymmetry. Purple triangle indicates tumour shown in panels a-f. Reference genome mis-assembly points marked (grey diamonds). h, Balance of Watson versus Crick asymmetry amongst tumours, showing deviations at driver genes. i, Tumours consistently show segmental mutational asymmetry across 50% of their autosomal genome. j, Model for DNA lesion segregation as a mechanism to generate mutational asymmetries. The exposure of a mutagen generates lesions (red triangles) on both strands of the DNA duplex (1). If not removed before or during replication (2) those lesions will segregate into two sister chromatids, one (blue) carrying only Watson strand lesions and subsequent templated errors, and the second (gold) only Crick strand lesions and their induced errors. Following mitosis, the daughter cells will have a non-overlapping complement of mutagen-induced lesions and resulting replication errors (3), which are resolved into full mutations in the next round of replication (4). The lesion containing strands segregate, becoming a progressively diminishing fraction of the lineage, yet continue as a template for replication. Only cell lineages containing cancer driver changes (* in step (1)) will expand into substantial clonal populations (5).
Pervasive, strand-asymmetric mutagenesis can be explained as the consequence of DEN-induced lesions remaining unrepaired prior to genome replication. The first round of replication after DEN exposure results in two sister chromatids with independent lesions on their parent strands (Fig. 2j). The daughter strand is produced using a lesion-containing template whose complement is synthesised with reduced replication fidelity over damaged nucleotides, resulting in nucleotide misincorporation errors complementary to lesions. These two sister chromatids necessarily segregate into separate daughter cells during mitosis. The heteroduplexes of lesions with paired mismatches are resolved into full mutations by later replication cycles (Fig. 2j). We subsequently refer to this phenomenon as “lesion segregation”.
The haploid X chromosome always contains segments with a strong strand bias (Fig. 2g). On autosomal chromosomes, we also observe an unbiased state, which we interpret as the aggregated biases of the two allelic autosomal chromosomes with opposing strand asymmetries. More explicitly, when both copies of a chromosome have Watson bias, the genome shows a Watson bias (e.g. chromosome 15 in Fig. 2a-d); when one copy has Watson bias and the other a Crick bias the two will cancel each other out and appear unbiased (e.g. chromosome 19 in Fig. 2a-d). Under lesion segregation, these asymmetries represent the random retention of Watson or Crick biased segments over the whole genome, and are essentially the output of two independent 1:1 Bernoulli processes, analogous to two fair coin flips. In such a model, we expect (1) 50% of the autosomal genome and (2) 100% of the haploid X chromosome to show mutational asymmetry; both predictions are supported by the observed data (Fig. 2g,i). A small fraction of tumours (3.5%) are outliers (Fig. 2g,i), with absent or muted mutational asymmetry; these features are associated with atypically low variant allele frequency distributions, indicating they may be polyclonal or polyploid (Supplementary Table 1). The asymmetric regions show a 23-fold (median) excess of their preferred mutation over its reverse complement, suggesting that >95% of those lesions that go on to produce a mutation, segregate for at least one mitosis.
Resolving sister chromatid exchange
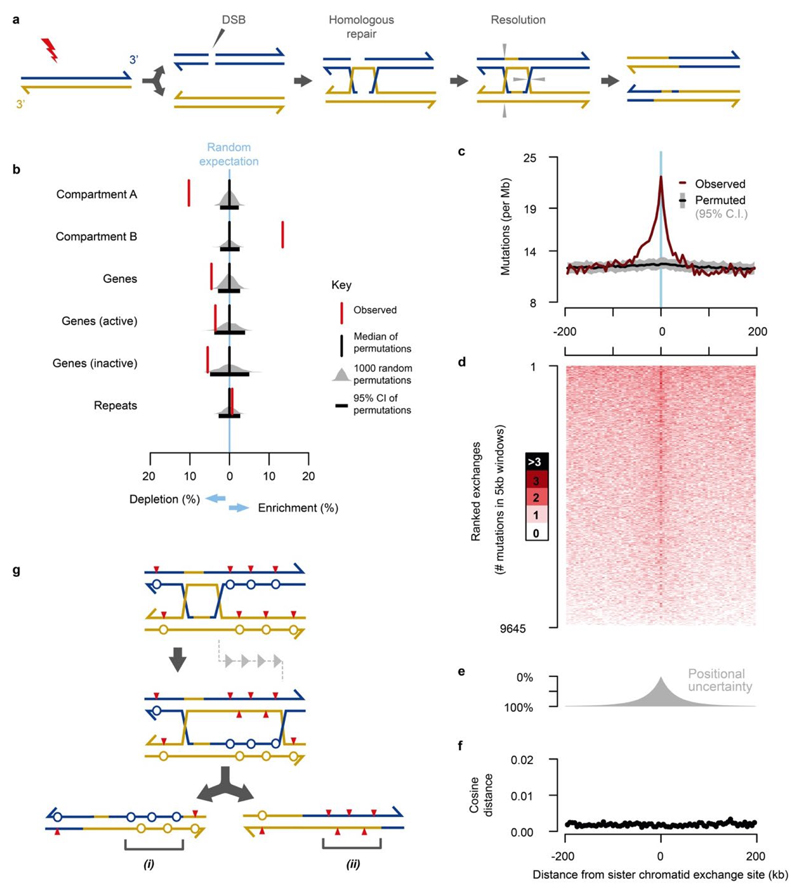
The lesion segregation model predicts mutational asymmetries should span whole chromosomes, yet we commonly observe discrete switches between multi-megabase segments of Watson and Crick bias within a chromosome (Fig. 2a-d,g). Such switches likely represent sister chromatid exchanges (SCEs) resulting from homologous recombination (HR) mediated DNA repair events18 (Extended Data Fig. 3a) that are typically invisible to sequencing technologies because HR between sister chromatids is generally thought to be error-free19.
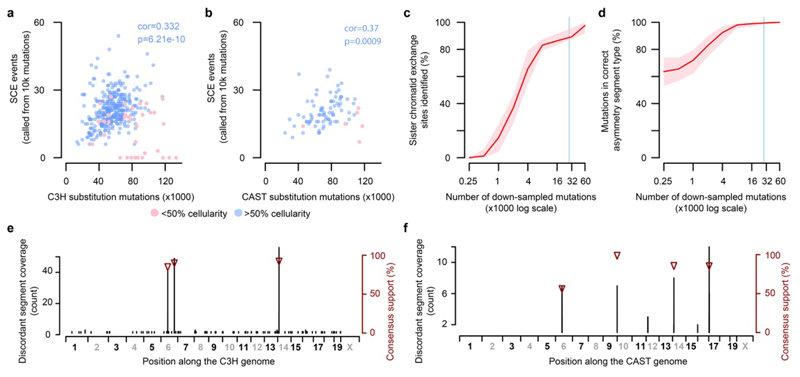
The observed rate of sister-chromatid exchange positively correlates with the genome-wide load of point mutations (Extended Data Fig. 2a,b). The presence of ~27 (median) SCEs in each of 371 diploid tumour genomes meant we were well-powered to detect recurrent exchange sites and biases in genomic context (Extended Data Fig. 2c,d). After filtering out three reference genome mis-assemblies (Fig. 2g; Extended Data Fig. 2e,f), we find that SCEs occur throughout the genome, with modest enrichment in transcriptionally inactive, late replicating regions (Extended Data Fig. 3b).
The fine mapping (~20kb resolution) of SCEs allowed us to test the fidelity of HR between sister chromatids. The mutation rate appears locally elevated at SCEs, but the spectrum of mutations matches the rest of the genome (Extended Data Fig. 3c-f). We propose that a model of Holliday intermediate branch migration could explain these observations (Extended Data Fig. 3g).
Lesion segregation reveals oncogenic selection
The random segregation of sister chromatids into daughter cells would result in 50% Watson and 50% Crick strand lesion retention on average across tumours, and the majority of the genome conforms to this prediction (Fig. 2h). We observe striking deviations at loci spanning known murine hepatocellular carcinoma driver genes (Fig. 2h). For example, the Braf T→A mutation at codon 584 is a known oncogenic driver 6 and is observed in 153/371 C3H tumours. Presuming that the Braf mutation was DEN induced, we would expect the mutation to have occurred in a chromosomal segment that retained T-lesions on the same strand as the driver T→A change. Indeed this is the case (94%; 144/153 tumours retain lesions on the expected strand, Fisher’s exact test p=3.6x10-19, rejecting the 50:50 null expectation). In contrast, tumours lacking the Braf mutation do not show a systematic bias (47% Crick bias, 53% Watson bias, p=0.88, not rejecting the 50:50 null expectation). We applied this general test for oncogenic selection at sites with sufficient recurrent mutations to have statistical power. Our results confirmed significant oncogenic selection of previously identified driver mutations in Hras, Braf and Egfr (Fig. 1d; Extended Data Table 1).
DNA repair with lesion strand resolution
Resolving DNA lesions to specific strands in a single mutagenised cell cycle presents a unique opportunity to investigate strand-specific interactions with DNA damage and repair in vivo. In expressed genes, transcription-coupled nucleotide excision repair specifically removes DNA lesions from the mRNA template strand rather than from the non-template strand (Fig. 3a)20,21. To explore this, we generated total transcriptomes of liver tissue from P15 C3H and CAST mice, corresponding to the known tissue of origin as well as the exact developmental timing of DEN mutagenesis.
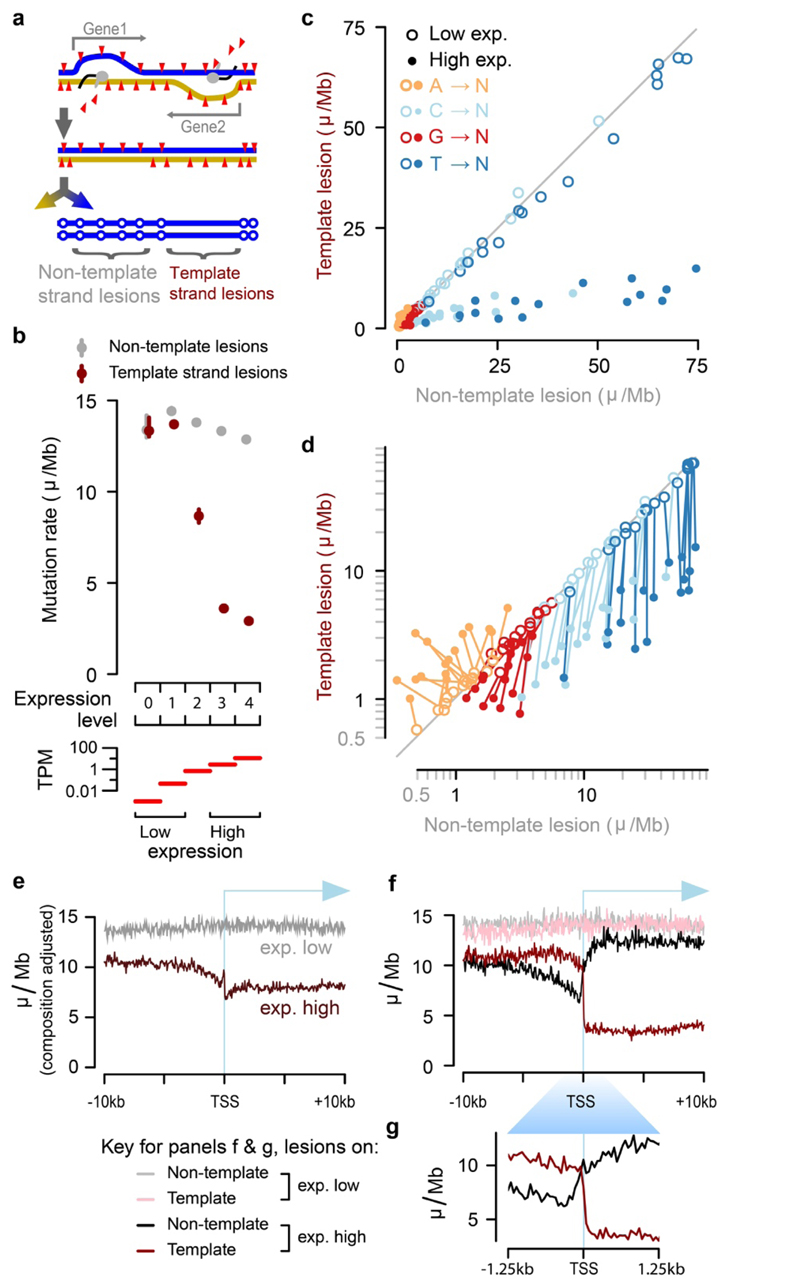
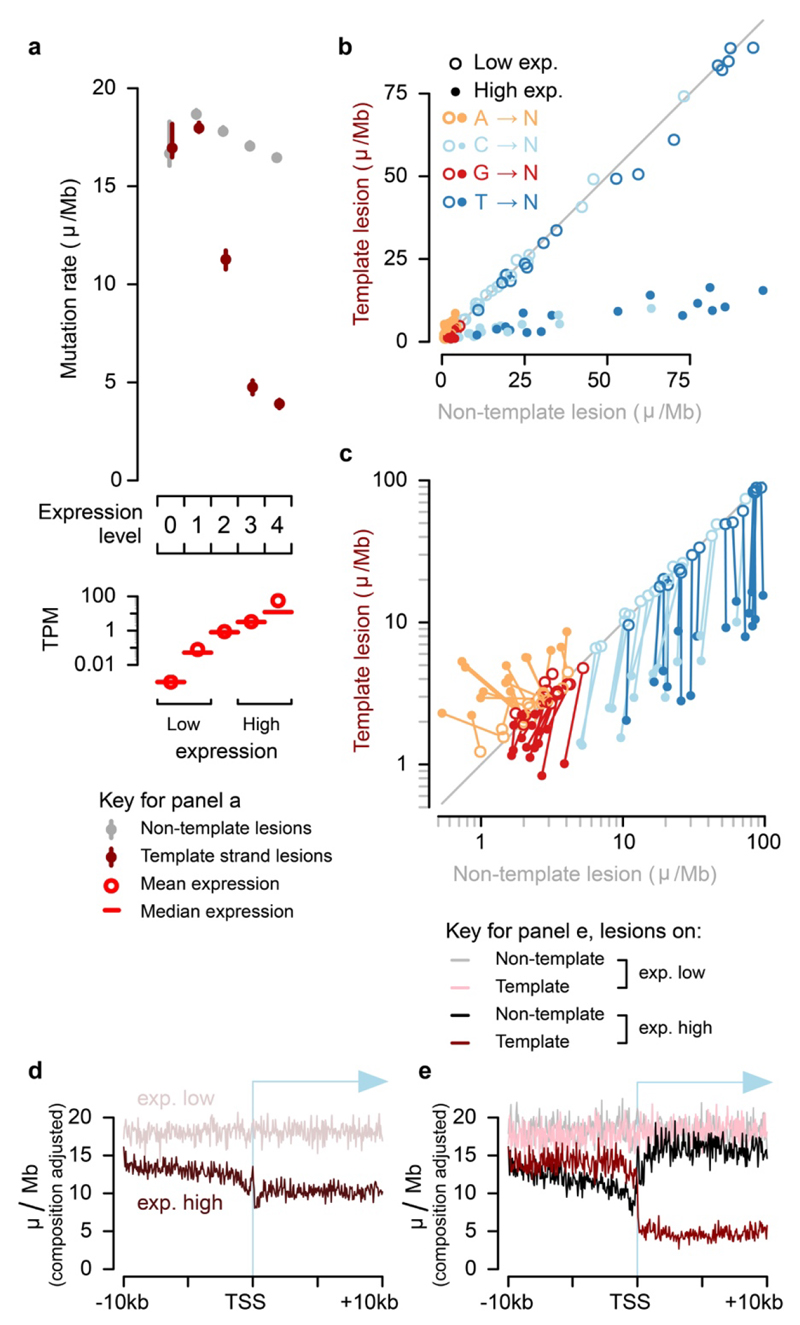
Fig.3. Identification of the lesion containing DNA strand allows processes such as transcription coupled repair (TCR) to be quantified with strand specificity.
a, TCR of DNA lesions is expected to reduce the mutation rate only when lesions are on the template strand of an expressed gene. b, TCR of template strand lesions is dependent on transcription level (P15 liver, transcripts per million (TPM)). Confidence intervals (99%) are shown as whiskers. c, Comparison of mutation rates for the 64 trinucleotide contexts: each context has one point for low and one point for high expression. d, Data as in panel c plotted on log scale; there is a line linking low and high expression for the same trinucleotide context. e, Sequence composition normalised profiles of mutation rate around transcription start sites (TSS). f, Stratifying the data plotted in e by lesion strand reveals much greater detail, including the pronounced net influence of bidirectional transcription initiation on the observed mutation patterns. g, TSS region detail from panel above, f.
For each gene in each tumour, we resolved whether the lesion-containing strand was the mRNA template or not, and calculated mutation rates stratified by both expression level and lesion strand (Fig. 3b). As expected, TCR was highly specific to the template strand and correlated closely with gene expression. Among genes without detectable expression, there was no reduction or observable transcription strand-bias in the mutation rate. In contrast, mutation in the most highly expressed genes was reduced by 79.8±1.0%, if the tumour inherited lesions on the template strand. We also detected a small transcription-associated decrease of 10.7±1.4% in mutation rate for lesions on the non-template strand, relative to lowly-expressed genes.
We next considered the specificity of TCR, comparing the rates of mutation for each trinucleotide context between template and non-template strands, stratified by expression level (Fig. 3c,d). The most common mutations (T→N), have an 82% (s.d. 6.8% across sequence contexts) lower rate on the template strand than the non-template strand for highly expressed genes; the non-template mutation rate is the same regardless of expression level (Fig. 3d, dark-blue lines are close to vertical), as expected20.
Mutations from C and G on the template strand show a high efficiency of TCR (70% (s.d. 7.8%) and 34% (s.d. 21%) respectively, Fig. 3d), but there is a consistent transcription-dependent reduction of mutation rate when these lesions are on the non-template strand (lines are deflected from vertical), possibly revealing activity of non-TCR repair processes in accessible genic regions. Though comparatively rare, mutations from adenine on the lesion containing strand are increased with transcription (Fig. 3d). This unexpected observation could be due to the activity of error-prone translesion DNA polymerase Pol-η which targets transcribed regions, where it specifically mutates A:T base-pairs22.
Prior analyses of TCR could not resolve the lesion containing strand14,20,23. Consistent with these previous findings, we observe reduced mutation rates broadly across the transcription start site (TSS) region and into active gene bodies (Fig. 3e). A notable feature of this profile is the relative increase in mutation rate for the core promoter located in the 200 nucleotides immediately upstream of the TSS24. Including lesion strand information in the analysis (Fig. 3f,g) shows the relative increase in mutation rate over the core promoter to be a result of high rates of TCR upstream and downstream, but a relative depletion of TCR activity over the promoter itself, results that are replicated in CAST mice (Extended Data Fig. 4a-e). The ability to resolve the lesion strand newly reveals the striking and distinct contributions of bidirectional transcription from active promoters25 in shaping the observed mutation patterns.
Lesion segregation generates genetic diversity
The lesion segregation model (Fig. 2j) predicts that a segregating lesion may template multiple rounds of replication in successive cell cycles. In such a scenario, each replication across the lesion could incorporate different incorrectly paired nucleotides - or even the correctly paired nucleotide - opposite a persistent lesion. Consistent with this notion, hundreds of multi-allelic mutations have recently been reported from single cell sequencing of human cancer samples26 and a well-controlled cell lineage tracking system27.
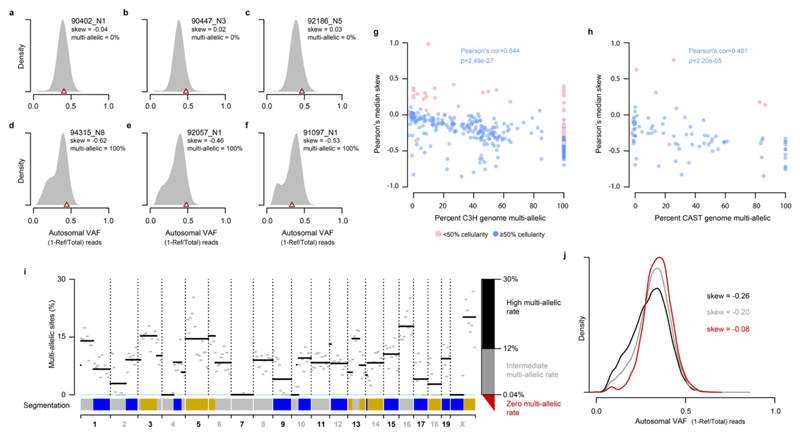
We investigated the extent of multi-allelic variation within each liver tumour genome by analysing the sequencing reads overlapping identified mutations. For example, a nucleotide position with multiple high-confidence reads supporting a reference T, mutation to A and also mutation to C would be considered multi-allelic. On average, 8% of mutated sites in DEN induced tumours exhibit evidence of multi-allelic variants (n=1.8 million sites in C3H tumours), though this value ranges from <1% to 25.7% between tumours (Fig. 4a). As a control we performed equivalent analysis on sites that had been called as mutated in a randomly selected proxy tumour; on average, only 0.098% (95% CI: 0.043-0.25%) show evidence of non-reference nucleotides.
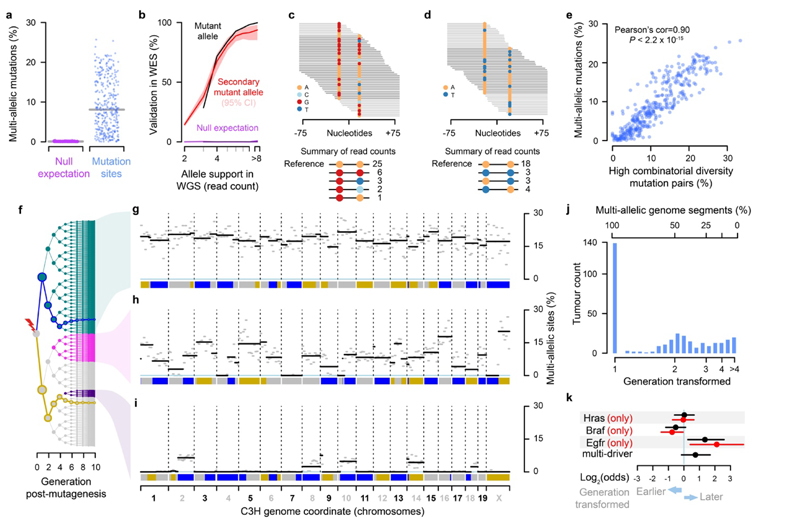
Fig.4. Lesion segregation generates multi-allelic and combinatorial genetic diversity.
a, Percent of mutation sites with robust support for multi-allelic variation, one point per tumour. Grey line indicates median. Null expectation (magenta) from permutation between tumours. b, Validation rate for whole genome sequence (WGS) mutation calls in replication whole exome sequencing (WES). Null expectation from permuting tumour identity between WGS and WES. c, Sequence reads spanning proximal mutations, showing nucleotide calls per read. d, As c, showing combinatorial diversity between a pair of biallelic sites. e, Correlation between per-tumour multi-allelic rate and high combinatorial diversity mutation pairs (as in c, d), one point per tumour. f, Tree showing all possible progeny of a DEN mutagenised cell for the subsequent 10 generations. Blue and gold lines trace the simulated segregation of lesion-containing strands from a single haploid chromosome. Coloured nodes show hypothetical transformation events and their daughter lineages that would give rise to the multi-allelic patterns in tumours shown to the right. g-i, Mutation asymmetry summary ribbons for example C3H tumours that show high g, variable h, or low i rates of genetic diversity; genome on the x-axis. The percent of mutation sites with robust support for multi-allelic variation calculated in 10Mb windows (grey) and for each asymmetric segment (black). j, Histogram of the estimated cell generation post-DEN exposure from which C3H tumours developed based on the proportion of multi-allelic segments. k, Enrichment of specific driver gene mutations in earlier (generation 1) and later (generation >1) developing tumours. All tumours containing the indicated driver mutation (black); the subset of tumours with just the indicated driver and no other driver mutation (red); multi-driver denotes all tumours that contain multiple identified driver genes in the EGFR/RAS/RAF pathway.
We further validated the multi-allelic variant calls from whole genome sequencing within independently performed exome sequencing of the same tumours6. The second and subsequent alternate alleles show the same profile of read depth-dependent validation rate as the called mutant allele, and clear separation from control analyses with mis-paired exome and genome sequence (Fig. 4b).
The independent generation of multi-allelic variation across the genome produces combinatorial genetic diversity not expected under purely clonal expansion. This combinatorial diversity can be directly visualised in pairs of mutated sites close enough to be spanned by individual sequencing reads (Fig. 4c,d). These reads report allele combinations that required lesions to have been replicated over without generating a mutation in some cell divisions (Fig. 4d). Allele frequency distributions indicate that non-mutagenic synthesis over DNA lesions is common (Extended Data Fig. 5). As expected for orthogonal measures of the generated genetic diversity, the tumour-wide level of combinatorial diversity from proximal mutation pairs closely correlates with the multi-allelic rate (Fig. 4e), and highlights the consistently high variance of these measures between tumours.
The explanation for this inter-tumour variance becomes evident when plotting the distribution of multi-allelism along each tumour genome (Fig. 4f-i). Tumours with high rates of genetic diversity typically have uniformly high rates of multi-allelism across their genome (Fig. 4g). They likely developed from a first generation daughter of the original DEN mutagenised cell, in which all DNA is a duplex of a lesion containing and non-lesion containing strand. Replication over lesion containing strands in subsequent generations produces multi-allelic variation at a uniform rate throughout the genome.
Tumours with lower total levels of genetic diversity exhibit discrete genomic segments of high and low multi-allelism (Fig. 4h,i). These tumours can be explained as having developed from a cell a few generations subsequent to DEN treatment. Each mitosis following DEN exposure is expected to dilute the lesion containing strands present in each daughter cell by approximately 50%, assuming random segregation. Only lesion-retaining fractions of the genome generate multi-allelic and combinatorial genetic diversity in the daughter lineages. As expected from the lesion segregation model with SCE, the multi-allelic patterns mirror the mutational asymmetry segmentation pattern.
By estimating the fraction of multi-allelic chromosomal segments, we can infer the cell generation post-DEN exposure that the tumour grew from (Fig. 4j). 67% of C3H and 21% of CAST tumours developed from first generation daughter cells following DEN exposure, indicating the single large burst of mutations was instantly transformative. For the remainder, the observed fractions of multi-allelic segments cluster around expectations for second and subsequent cell generations, suggesting that the production of a specific mutant allele combination, an additional mutation, or an external trigger was required for transformation. Intriguingly, Egfr driven tumours appear to transform significantly later after DEN treatment, suggesting that driver gene identity may influence the timing of tumour inception (Fig. 4k).
Lesion segregation is ubiquitous
Lesion segregation is a major feature of DEN mutagenesis in mouse liver. This immediately raises two important questions: are other DNA damaging agents also characterised by lesion segregation? And does lesion segregation occur in human cells and cancers? A recent study exposed human induced pluripotent stem cells (iPSCs) to 79 known or suspected environmental mutagens and found 41 of them produced nucleotide substitutions above background expectations5. Although not previously noted in these data, we found that many of these exposures generated chromosome-scale lesion segregation patterns (Extended Data Fig. 6a-d) similar to our in vivo DEN model.
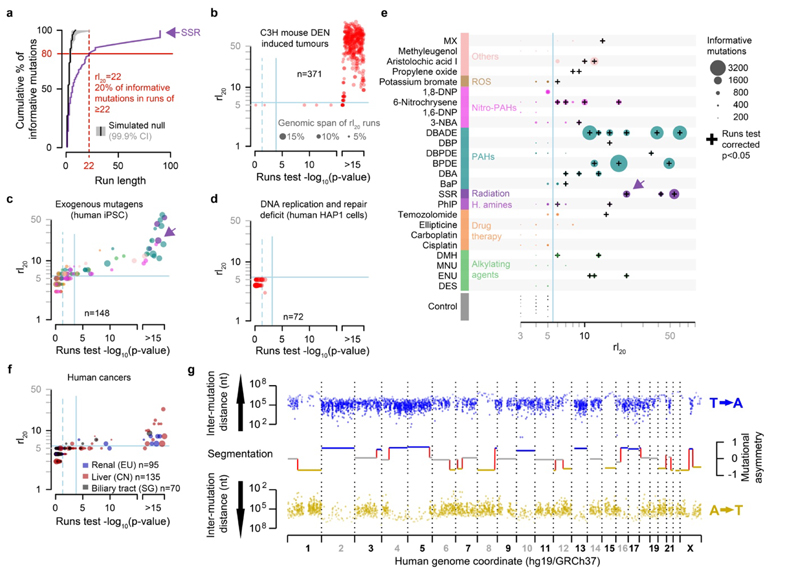
For each mutagenic agent, we identified the most common nucleotide substitution type in each sample (e.g. C→T/G→A in simulated solar radiation exposure, Extended Data Fig. 6a) and applied runs-based tests to quantify their segmental asymmetry. The application of runs-based tests (e.g. the rl20 metric) (Fig. 5a-f) revealed that segmental mutational asymmetry is a common feature of DNA damaging mutagens in human cells (Fig. 5c). We detect significant mutational asymmetry in every sample with good statistical power (>1,000 informative mutations, Fig. 5e), including clinically relevant insults, such as sunlight (simulated solar radiation, SSR), tobacco smoke (BPDE) and chemotherapeutic agents (temozolomide). We conclude that the chromosome-scale segregation of lesions and the resulting strand asymmetry of mutation patterns is a general feature of all tested DNA damaging mutagens.
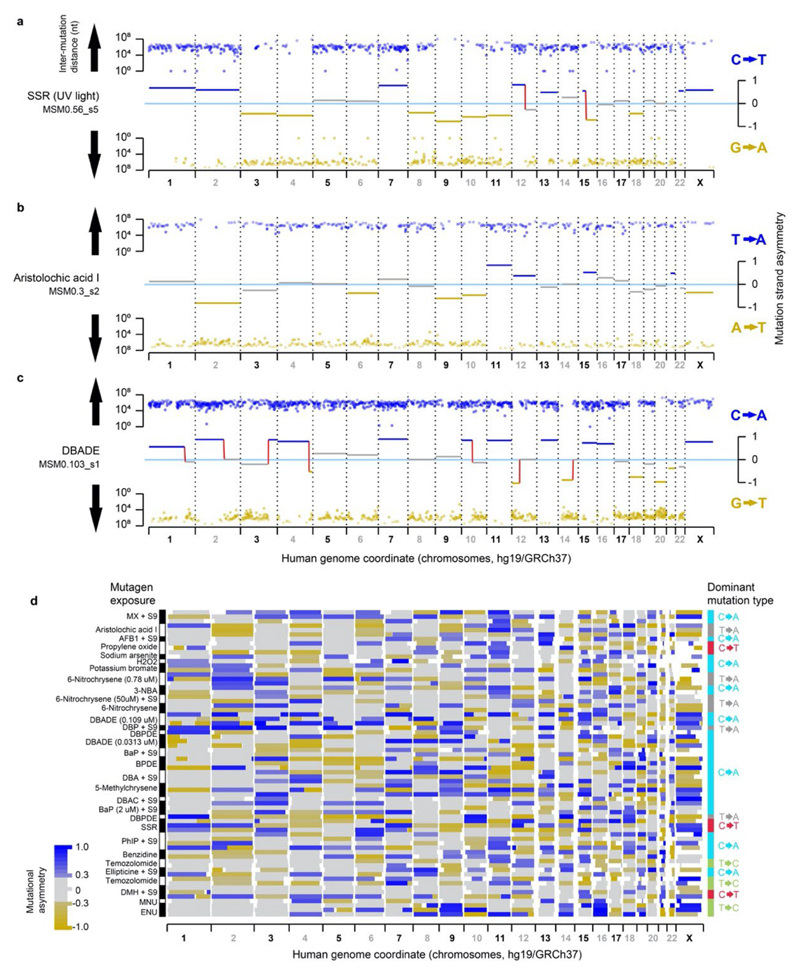
Fig.5. Lesion segregation is a pervasive feature of exogenous mutagens and is evident in human cancers.
a, The runs-based rl20 metric, calculated for the simulated solar radiation (SSR) clone MSMO_56.s5 (Extended Data Fig. 6a); here, 20% of informative mutations (C→T/G→A) are in strand asymmetric runs of 22 consecutive mutations or longer (e.g. ≥22 C→T without an intervening G→A). b-d, The rl20 metric and runs tests. Solid blue lines show Bonferroni adjusted p=0.05 thresholds, p-values < 1x10-15 are rank-ordered. b, DEN-induced C3H tumours (this study). c, Mutagen exposed human cells5, colour corresponds to the mutagen key in panel g. d, Cell-lines with genetically perturbed genome replication and maintenance machinery28. e, All 25 mutagens identified as producing robust mutation spectra when human induced pluripotent stem cells are exposed5, grouped by type of agent. See Supplementary Table 2 for the details of abbreviated mutagen exposures. The rl20 metric (x-axis) is plotted for each replicate clone, the size of each data point is scaled to the number of informative mutations. f, The rl20 metric and runs tests for human cancers from International Cancer Genome Consortium projects. g, Mutational asymmetry in an example human hepatocellular carcinoma, donor DO231953, which shows a single dominant mutation signature for aristolochic acid exposure (43.3%).
In an analogous experiment, similar numbers of mutations were induced by perturbation of replication and repair pathways28. These mutator phenotypes are independent of DNA lesions and correspondingly significant asymmetry was neither expected nor detected (Fig. 5d).
A common feature of our DEN mutagenesis experiment and mutagen exposure in human iPSCs5 is the striking pattern of mutation asymmetry that occurs as a consequence of a single mutagenic insult. By contrast, most human cancers are subject to multiple damaging events over their history. Our lesion segregation model predicts that such tumours will acquire new waves of segregating lesions after each exposure, thus progressively masking and mutually confounding their mutation patterns. Therefore, even though we have shown that UV exposure does cause striking lesion segregation in human cells (Fig. 5a; Extended Data Fig. 6a), it is unlikely that the mutational asymmetry diagnostic of lesion segregation would be detected in skin cancers.
Despite the low prior expectation of detecting lesion segregation patterns in human cancers, we used the same algorithm as for human iPSCs to search for such patterns in human cancer genomes29 (n=18,965 cancers from 22 primary sites). This identified multiple cancers that clearly show mutational asymmetry characteristic of lesion segregation (Fig. 5f,g). The majority of these tumours are renal, hepatic or biliary in origin, and show a high mutation rate and strand asymmetry of T→A/A→T mutations, consistent with known aristolochic acid exposure3 (Supplementary Table 2). We conclude that while visualised most clearly in tumours subjected to a single dose of a mutagen, lesion segregation has likely shaped all genomes that have suffered DNA damage, which has important implications for tumour evolution and heterogeneity.
Discussion
Here we have shown that most mutation-causing DNA lesions are not resolved as mutations within a single cell-cycle. Instead, lesions segregate unrepaired into daughter cells for multiple cellular generations, resulting in chromosome-scale strand asymmetry of subsequent mutations. This suggests that lesion removal prior to replication is high fidelity, rarely resulting in mutations. Low fidelity replication over persistent lesions implicates the involvement of DNA damage tolerance mechanisms30 over genomic perfection31. Initially discovered in a well-powered in vivo mammalian model of oncogenesis, we also demonstrate that lesion segregation is ubiquitous to all tested mutagens, occurs in human cells, and is evident in human cancers. Similar patterns of asymmetry in bacterial mutagenesis posit that the underlying mechanisms are deeply conserved32,33.
Our discovery of pervasive lesion segregation challenges long standing assumptions in the analysis of cancer evolution34. For example, the widely used infinite sites model35 does not allow for recurrent rounds of mutation at the same site. These findings also provide new opportunities for understanding cancer evolution, through the use of the mutational asymmetry and multi-allelic rate patterns to track events during oncogenesis and to quantify selection. A far-reaching implication of lesion segregation is that it may provide a window of opportunity for a cancer to sample the fitness of mutation combinations within the lineage, circumventing Muller’s ratchet36 and Hill-Robertson interference: low efficiency of selection due to the inability to separate mutations of opposing fitness effects37,38. Consequently, DNA damaging chemotherapeutics, particularly large or closely spaced doses generating persistent lesions, could inadvertently provide an opportunity for efficient selection of the resulting mutations. This insight may guide the development of more effective chemotherapeutic regimens.
Once identified, lesion segregation is a deeply intuitive concept. Its practical applications provide new vistas for the exploration of genome maintenance and fundamental molecular biology. The discovery of pervasive lesion segregation profoundly revises our understanding of how the architecture of DNA repair and clonal proliferation can conspire to shape the cancer genome.
Methods
Mouse colony management
Animal experimentation was carried out in accordance with the Animals (Scientific Procedures) Act 1986 (United Kingdom) and with the approval of the Cancer Research UK Cambridge Institute Animal Welfare and Ethical Review Body (AWERB). Animals were maintained using standard husbandry: mice were group housed in Tecniplast GM500 IVC cages with a 12-hour light / 12-hour dark cycle and ad libitum access to water, food (LabDiet 5058), and environmental enrichments.
Chemical model of hepatocarcinogenesis
15-day-old (P15) male C3H and CAST mice were treated with a single intraperitoneal (IP) injection of N-Nitrosodiethylamine (DEN; Sigma-Aldrich N0258; 20 mg/kg body weight) diluted in 0.85% saline. Liver tumour samples were collected from DEN-treated mice 25 weeks (C3H) or 38 weeks (CAST) after treatment. All macroscopically identified tumours were isolated and processed in parallel for DNA extraction and histopathological examination. Non-tumour tissue from untreated P15 mice (ear, tail, and background liver) was sampled for control experiments.
Tissue collection and processing
Liver tumours of sufficient size (≥2 mm diameter) were bisected; one half was flash frozen in liquid nitrogen and stored at -80°C for DNA extraction, and the other half was processed for histology. Tissue samples for histology were fixed in 10% neutral buffered formalin for 24 h, transferred to 70% ethanol, machine processed (Leica ASP300 Tissue Processor; Leica, Wetzlar, Germany), and paraffin embedded. All formalin-fixed paraffin-embedded (FFPE) sections were 3 μm in thickness.
Histochemical staining
FFPE tissue sections were haematoxylin and eosin (H&E) stained using standard laboratory techniques. Histochemical staining was performed using the automated Leica ST5020; mounting was performed on the Leica CV5030.
Imaging
Tissue sections were digitised using the Aperio XT system (Leica Biosystems) at 20x resolution; all H&E images are available in the BioStudies archive at EMBL-EBI under accession S-BSST129.
Tumour histopathology
H&E sections of liver tumours were blinded and assessed twice by a pathologist (S.J.A); discordant results were reviewed by an independent hepatobiliary pathologist (S.E.D). Tumours were classified according to the International Harmonization of Nomenclature and Diagnostic Criteria for Lesions in Rats and Mice (INHAND) guidelines39. In addition, tumour grade, size, morphological subtype, nature of steatosis, and mitotic index were assessed (Supplementary Table 1), as well as the presence of cystic change, haemorrhage, necrosis, or vascular invasion.
Sample selection for WGS
Tumours which met the following histological criteria were selected for whole genome sequencing (C3H n=371, CAST n=84): (i) diagnosis of either dysplastic nodule (DN) or hepatocellular carcinoma (HCC), (ii) homogenous tumour morphology, (iii) tumour cell percentage >70%, and (iv) adequate tissue for DNA extraction. Neoplasms with extensive necrosis, mixed tumour types, a nodule-in-nodule appearance (indicative of an HCC arising within a DN), or contamination by normal liver tissue were excluded. Since carcinogen-induced tumours arising in the same liver are independent6, multiple tumours were selected from each mouse to minimise the number of animals used. A subset of normal (non-tumour) samples from untreated mice were also sequenced (C3H n=13, CAST n=7).
Whole genome sequencing
Genomic DNA was isolated from liver tissue and liver tumours using the AllPrep 96 DNA/RNA Kit (Qiagen, 80311) according to the manufacturer's instructions. DNA quality was assessed on a 1% agarose gel and quantified using the Quant-IT dsDNA Broad Range Kit (Thermo Fisher Scientific). Genomic DNA was sheared using a Covaris LE220 focused-ultrasonicator to a 450 bp mean insert size.
WGS libraries were generated from 1 μg of 50 ng/ul high molecular weight gDNA using the TruSeq PCR-free Library Prep Kit (Illumina), according to the manufacturer's instructions. Library fragment size was determined using a Caliper GX Touch with a HT DNA 1k/12K/Hi Sensitivity LabChip and HT DNA Hi Sensitivity Reagent Kit to ensure 300-800 bp (target ~450 bp).
Libraries were quantified by real-time PCR using the Kapa library quantification kit (Kapa Biosystems) on a Roche LightCycler 480. 0.75 nM libraries were pooled in 6-plex and sequenced on a HiSeq X Ten (Illumina) to produce paired-end 150 bp reads. Each pool of 6 libraries was sequenced over eight lanes (minimum of 40x coverage).
Variant calling and somatic mutation filtering
Sequencing reads were aligned to respective genome assemblies (C3H = C3H_HeJ_v1; CAST = CAST_EiJ_v1)40 with bwa-mem (v.0.7.12)41 using default parameters. Reads were annotated to read groups using the picard (v.1.124)42 tool AddOrReplaceReadGroups, and minor annotation inconsistencies corrected using the picard CleanSam and FixMateInformation tools. Bam files were merged as necessary, and duplicate reads were annotated using the picard tool MarkDuplicates.
Single nucleotide variants were called using Strelka2 (v.2.8.4)43 implementing default parameters. Initial variant annotation was performed with the GATK (v.3.8.0)44 walker CalculateSNVMetrics45. Genotype calls with a variant allele frequency < 0.025 were removed. Although inbred strains were used, fixed genetic differences between the colonies and the reference genome, as well as small numbers of germline variants segregating within the colonies were identified. For each strain, fixed differences identified as homozygous changes present in 100% of genotyped samples were filtered out. Segregating variants were filtered based on the excess clustering of mutations to animals with shared mothers. To generate a null expectation taking into account the family structure of the colonies, the parent-offspring relationships were randomly permuted 1,000 times. For each count of recurrent mutation (range 5 to 371 inclusive), we determined the null distribution of expected distinct mothers. Comparing this to the observed count of distinct mothers for each recurrent (n>4) mutation, those with a low probability (p<1x10-4, pnorm function from R (v.3.5.1)46) under the null were excluded from analyses.
Copy number variation between tumours within strains was called using CNVkit (v.0.9.6)47. Non-tumour reference coverage was provided from non-tumour control WGS data (C3H n=11, CAST n=7) and per tumour cellularity estimates (see below) were provided.
RNA-sequencing
Total RNA was extracted from P15 liver tissue (n=4 biological replicates per strain) using QIAzol Lysis Reagent (Qiagen), according to the manufacturer’s instructions. DNase treatment and removal were performed using the TURBO DNA-freeTM Kit (Ambion, Life Technologies), according to the manufacturer’s instructions. RNA concentration was measured using a NanoDrop spectrophotometer (Thermo Fisher); RNA integrity was assessed on a Total RNA Nano Chip Bioanalyzer (Agilent)
Total RNA (1 μg) was used to generate sequencing libraries using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina), according to the manufacturer’s instructions. Library fragment size was determined using a 2100 Bioanalyzer (Agilent). Libraries were quantified by qPCR (Kapa Biosystems). Pooled libraries were sequenced on a HiSeq4000 to produce ≥40 million paired-end 150 bp reads per library.
RNA-seq data processing and analysis
Transcript abundances were quantified with Kallisto (v.0.43.1)48 (using the flag --bias) and a transcriptome index compiled from coding and non-coding cDNA sequences defined in Ensembl v9149. Transcripts per million (TPM) estimates were generated for each annotated transcript and summed across alternate transcripts of the same gene for gene-level analysis. Transcription start sites (TSS) for each gene were annotated with Ensembl v91 and based upon the most abundantly expressed transcript. RNA-seq data are available at Array Express at EMBL-EBI under accession E-MTAB-8518.
Genomic annotation data
Mouse liver proximity ligation sequencing (HiC) data were downloaded from GEO (GSE65126)50, replicates were combined, then aligned to GRCm3851 and processed using the Juicebox (v.7.5) and Juicer scripts52 to obtain the HiC matrix. Eigenvectors were obtained for 500kb consecutive genomic windows over each chromosome from the HiC matrix using Juicebox and subsequently oriented (to distinguish compartment A from B) using GC content per 500kb bin. We used progressiveCactus53 to project the 500kb windows into the C3H reference genome and Bedtools (v.2.28.0) to merge syntenic loci between 450 and 550 kb in size, removing the second instance where we observed overlaps.
Genic annotation was obtained from Ensembl v9149 for the corresponding C3H and CAST reference genome assemblies (C3H_HeJ_v1, CAST_EiJ_v1). Genomic repeat elements were annotated using RepeatMasker (v.20170127)54 with the default parameters and libraries for mouse annotation.
The analysable fraction of the genome
Analysis and sequence composition calculations were confined to the main chromosome assemblies of the reference genome (chromosomes 1-19 and X). Using WGS of non-tumour liver, ear and tail samples (C3H n=11, CAST n=7) collected and sequenced contemporaneously with tumour samples, genome sequencing coverage was calculated for 1kb windows using multicov in Bedtools (v.2.28.0)55. Windows with read coverage >2 s.d. from the autosomal mean were flagged as suspect in each tumour. Read coverage over the X chromosome was doubled in these calculations to account for the expected hemizygosity in these male mice. Any 1kb window identified as suspect in >90% of these non-tumour samples was flagged as “abnormal read coverage” (ARC) and masked from subsequent analysis. This masked 12.7% of the C3H and 11.5% of the CAST reference genomes yielding analysable haploid genome sizes of C3H = 2,333,783,789 nt and CAST = 2,331,370,397 nt.
Mutation rate calculations
Mutation rates were calculated as 192 category vectors representing every possible single nucleotide substitution conditioned on the identity of the upstream and downstream nucleotides. Each rate being the observed count of a mutation category divided by the count of the trinucleotide context in the analysed sequence. To report a single aggregate mutation rate, the three rates for each trinucleotide context were summed to give a 64 category vector and the weighted mean of that vector reported as the mutation rate. The vector of weights being the trinucleotide sequence frequency of a reference sequence, for example the composition of the whole genome. In the case of whole genome analysis, the same trinucleotide counts are used in (1) the individual category rates calculation and (2) the weighted mean of the rates, cancelling out. For windowed comparisons of mutation rates, the weighted mean is calculated using the genome wide composition of trinucleotides rather than the local sequence composition, providing a compositionally adjusted mutation rate estimate. For mutation rates in TCR analysis, the same compositional adjustment was carried out but using the trinucleotide composition of the aggregate genic spans of genome (minus ARC regions) for normalisation.
Mutation signatures
The 96 category “folded” mutation counts for each of the 371 C3H tumours were deconvolved into the best fitting number (K) of component signatures using sigFit (v.2.0)56 with 1,000 iterations and K set to integers 2 to 8 inclusive. A heuristic goodness of fit score based on cosine similarity favoured instances where K=2. The DEN1 and DEN2 signatures reported were obtained by running sigFit with 30,000 iterations for K=2. Analysis of CAST tumours gave less distinct separation of signatures so the C3H derived DEN1 and DEN2 were used for both strains. To fit signatures to each tumour we used sigFit provided with the DEN signatures and additional SPONT1 and SPONT2 signatures that were derived from equivalent WGS analysis of spontaneous (non-DEN induced) C3H tumours.
Driver mutation identification
Candidate driver mutations were identified by applying oncodriveFML (v.2.2.0 using the SIFT scoring scheme)57 and oncodriveCLUSTL (v.1.1.1)58 to mutations identified in C3H tumours. The only genes convincingly identified as significantly enriched for functionally impactful or clustered mutations were Hras, Braf, and Egfr. Kras appeared as marginally significant. These four genes were identified for C3H6. Protein altering mutations in those genes were annotated as driver mutations in C3H and CAST tumours.
Mutational asymmetry segmentation and scoring
For each tumour a focal subset of “informative” mutation types were defined, T→N/A→N mutations, in the case of DEN-induced tumours. The order of focal mutations along each chromosome was represented as a binary vector (e.g. 0 for T→N, 1 for A→N). Vectors corresponding to each chromosome of each tumour were processed with the cpt.mean function of the R Changepoint (v.2.2.2)59 package run with an Akaike information criterion (AIC) penalty function, maximum number of changepoints set to 12 (Q=12), and implementing the PELT algorithm for optimal changepoint detection. Following segmentation, the defined segments were scored for strand asymmetry, taking into account the sequence composition of the segment. For example in tumours with T→N/A→N informative mutations the number of Ts on the forward strand is the count of Watson sites G W and the number of T→N mutations is μ W which together give the Watson strand rate R W=μ W/G W. The forward strand count of As and mutations from A likewise give the Crick strand rate R C=μ C/G C. From these two rates we calculate a relative difference metric, the mutational asymmetry score S=(R W-R C)/(R W+R C).
The parameter S scales from 1 all Watson (e.g. DEN T→N mutations) through 0 (50:50 T→N:A→N) to -1 for all Crick (e.g. DEN A→N). For the categorical assignment, S ≥ 0.3 is Watson strand asymmetric, S ≤- 0.3 Crick strand asymmetric and in the range -0.3 < S < 0.3 symmetric, though more stringent filtering was applied where noted. Segments containing <20 informative mutations were discarded from subsequent analyses. To test for oncogenic selection at sites with recurrent mutations, mutational asymmetry segments overlapping the focal mutation were categorised based on their asymmetry score S, as above. The test was implemented as a Fisher’s exact test with the 2x2 contingency table comprising the counts of chromosomes (two autosomes per cell) stratified by Watson versus Crick asymmetry and the presence of the focal mutation in the tumour. Tumours containing another known driver gene or recurrent mutation within the focal asymmetry segment were discarded from the analysis. We estimated the minimum recurrence of a mutation necessary to reliably detect oncogenic selection through simulation. Biased segregation of chromosomes containing drivers was modelled using the observed median excess of T→N over A→N lesions (23 fold), and random segregation of non-driver containing strands (1:1 ratio). Our model predicted >33 C3H recurrences or >41 CAST recurrences would give 80% power to detect oncogenic selection if present.
Tumour cellularity estimates
We calculated tumour cellularity as a function of the non-reference read count in autosomal chromosomes (1-R/d)*2 where R is the reference read count at a mutated site and d is the total read depth at the site. For each tumour these values were binned in percentiles and the midpoint of the most populated (modal) percentile taken as the estimated cellularity of the tumour. Given the low rate of copy number variation across the DEN induced tumours, no correction was made for copy-number distortion. Skew in the variant allele frequency (VAF=(1-R/d)) distribution was calculated using Pearson’s median skewness coefficient implemented in R as (3(mean-median))/sd of the VAF distribution.
Identifying and filtering reference genome mis-assemblies
Since lesion segregation, mutation asymmetry patterns allow the long-range phasing of chromosome strands, they can detect discrepancies in sequence order and orientation between the sequenced genomes and the reference. We identified autosomal asymmetry segments that immediately transitioned from Watson bias (S > 0.3) to Crick (S < -0.3) or vice versa without occupying the intermediate unbiased state (-0.3 < S < 0.3); such “discordant segments” are unexpected. Allowing for ±100kb uncertainty in the position of each exchange site we produced the discordant segment coverage metric. At sites with discordant segment coverage >1 we calculated percentage consensus for mis-assembly M=ds/(ds+cs) where ds is the number of discordant segments over the exchange site and cs the number of concordant: where either Watson or Crick mutational asymmetry extends at least 1x106 nucleotides on both sides of the exchange site. The approximate genomic coordinates for a C3H strain specific inversion on chromosome 6 were previously reported60.
Sister chromatid exchange site analysis
Identified SCE sites were aggregated across tumours from each strain. Exchange sites within 1x106 nt of known and proposed reference genome mis-assembly sites were excluded from analysis. The mid-point between the flanking informative mutations was taken as the reference genome position of the exchange event, and the distance between those flanking mutations as the positional uncertainty of the estimate. To generate null expectations for mutation rate measures, the coordinate of an exchange was projected into the genome of a proxy tumour and the mutation rates and patterns measured from that proxy tumour (repeated 100 times). The permutation of tumour identifiers for the selection of proxy tumours was a shuffle without replacement that preserved the total number of exchange sites measured in each tumour.
The comparison of mutation spectra between windows was calculated as the cosine distance between the 96 category trinucleotide context mutation spectra for the whole genome and that calculated for the aggregated 5kb window. The 96 categories were equally weighted for this comparison.
Exchange site enrichment analysis used Bedtools55 shuffle to permute the genomic positions of exchange sites into the analysable fraction of the genome (defined above). Observed rates of annotation overlap were compared to the distribution of values from 1,000 permuted exchange sites. For genic overlaps we used Ensembl v9149 coordinates for genic spans; gene expression status was based on the summed expression over all annotated transcripts for the gene from P15 liver from the matched mouse strain. Expression thresholds were defined as >50th centile for active and <50th centile for inactive genes.
A higher count of informative mutations provides greater power to identify shorter mutational asymmetry segments. To fairly test for correlation between nucleotide substitution rate and SCE rate we randomly down-sampled informative mutations to 10,000 per tumour genome and recomputed the mutational asymmetry segmentation patterns from the sampled data. Tumours with <10,000 informative mutations were excluded. We then correlated the total (not down sampled) nucleotide substitution load to the count of SCE events inferred from the down-sampled data.
Transcription coupled repair calculations
For each protein coding gene, the maximally expressed transcript isoform was identified from P15 liver in the matched strain (TPM expression), subsequently the primary transcripts. In the case of ties, transcript selection was arbitrary. Genes were partitioned into five categories based on the expression of the primary transcript: expression level 0 (<0.0001 TPM) and four quartiles of detected expression.
Using the segmental asymmetry patterns of each tumour and the annotated coordinates (Ensembl v91) of the selected transcripts, we identified transcripts completely contained in a single Watson or Crick asymmetric segment and located at least 200kb from the segment boundary at both ends. We also applied strict asymmetry criteria of mutational asymmetry scores S > 0.8 for Watson and S < -0.8 for Crick asymmetry segments, though analysis with the standard asymmetry thresholds and no segment boundary margin give similar results and identical conclusions. For each transcript in each tumour we then used both the transcriptional orientation of the gene and the mutational asymmetry of the segment containing it to resolve the segregated lesions to either the template (anti-sense) or non-template (sense) strand of the gene. Transcripts contained in mutationally symmetric regions or not meeting the strict filtering criteria were excluded from analysis.
We then analysed mutation rates stratifying by gene expression level and the template/non-template strand of the lesions but aggregating between tumours within the same strain. The transcription start site coordinates used correspond to the annotated 5’ end of the primary transcripts.
Multi-allelic variation
Aligned reads spanning genomic positions of somatic mutations were re-genotyped using Samtools mpileup (v.1.9)61. Genotypes supported by ≥2 reads with a nucleotide quality score of ≥20 were reported, considering sites with two alleles as biallelic, those with three or four alleles as multi-allelic. The fraction of called mutations exhibiting multi-allelic variation was calculated for the analysable fraction of the genome, across 10Mb consecutive windows and also for each of the mutational asymmetry segments calculated for each tumour.
A null expectation for the multi-allelic rate estimate was generated per C3H tumour; genomic positions identified as mutated across the other 370 tumours were down-sampled to match the mutation count in the focal tumour. Any of these proxy mutation sites with a non-reference genotype supported by ≥2 reads and nucleotide quality score ≥20 at the focal site were referred to as “multi-allelic” for the purposes of defining a background expectation for the calling of multi-allelic variation. For each tumour, this was repeated 100 times and the mean reported.
We used whole exome sequencing (WES) of fifteen C3H tumours from prior work6 that have subsequently been used to generate WGS data in this study as a basis for validating multi-allelic calls. Multi-allelic variant positions derived from WGS were genotyped in WES using Samtools mpileup, as described above. Only sites with ≥30x WES coverage were considered and alleles were found to be concordant if a WGS genotype was supported by ≥1 read in the WES data. To provide a null expectation, the analysis was repeated using WES data from a different tumour and validation rates reported for all versus all combinations of mismatched WGS-WES pairs (152-15=210).
To quantify combinatorial genetic diversity for each tumour, pairs of mutations located between 3-150nt apart were phased using sequencing reads that traversed both mutation sites. Distinct allelic combinations were counted after extraction with Samtools mpileup using only reads with nucleotide quality score ≥20 over both mutation sites.
Estimating the cell generation of transformation
Knowing the faction of lesion segregation segments that generated multi-allelic variation across a tumour genome allows the inference of the generation time post-mutagenesis of the cell from which the tumour developed, because each successive cell generation is expected to retain only 50% of the lesion containing segments. We estimate this fraction as follows. Let p denote the fraction of multi-allelic segments and let q be its complement, i.e. the fraction of non-multi-allelic segments, for each tumour genome. Segment boundaries being SCE sites or chromosome boundaries. In order to determine p, we re-purpose the quadratic Hardy-Weinberg equation: p+q=p 2+2pq+q 2 =1, which holds since the two possible fractions need to sum to unity. Given an asymmetric segment of interest in the diploid genome, there are 3 distinct scenarios: (i) both chromosomes are multi-allelic (p 2), (ii) One of the chromosomes is multi-allelic and the other is not (pq+qp) and (iii) both chromosomes are non-multi-allelic (q 2). The first two scenarios are not distinguishable from the data as both appear multi-allelic (m). However, in the third scenario, for a segment to be non-multi-allelic (biallelic, b), both chromosomal copies have to be non-multi-allelic. As described below, q 2 can be estimated directly from the data and is subsequently used to estimate p=1-sqrt(q 2 ) and hence the cell generation number of transformation post-mutagenesis.
The estimation of q 2 requires computing the ratio q 2=b/(b+m). We can directly observe the counts of b as non-multi-allelic segments. The number of autosomal chromosome pairs (n=19) and count of sister chromatid exchange events (x) give the total number of segments in the genome b+m=n+x. Exchange events are not expected to align between allelic chromosomes which will result in the partial overlap of segments between allelic copies. Although this increases the number of observed segments (b and m) relative to actual segments, assuming the independent behaviour of allelic chromosomes and that segment length is independent of multi-allelic state, this partial overlap does not systematically distort the quantification of b or the estimation of q 2.
To call a non-multi-allelic segment (b) we require less than 0.04% multi-allelic sites. The threshold based on the tri-modal frequency distribution of multi-allelic rates per-segment, aggregated over all 371 C3H tumours. The 0.04% threshold separates the lower distribution of multi-allelic rates from the mid and higher distributions.
To test for the enrichment of specific driver gene mutations in early generation versus late generation transformation post-DEN treatment, we applied Fisher’s exact test (fisher.test function in R) to compare the generation 1 ratio of tumours with, versus those without a focal mutation, to the same ratio for tumours inferred to have transformed in a later generation. We additionally report the same odds ratios, but requiring that the “with focal mutation” tumours had a driver mutation in only one of the driver genes: Hras, Braf, or Egfr.
Cell-line and human cancer mutation analysis
Somatic mutation calls were obtained from DNA maintenance and repair pathway perturbed human cells28. Of the 128,054 reported single nucleotide variants, 6,587 unique mutations (genomic site and specific change) were shared between two or more sister clones, so likely represent mutations present but not detected in the parental clone. All occurrences of the shared mutations were filtered out leaving 106,688 mutations for analysis, although the inclusion of these filtered mutations does not alter any conclusions drawn. Somatic mutation calls from mutagen exposed cells5 were obtained, no additional filtering was applied to these sub-clone mutations.
Somatic mutation calls from the International Cancer Genome Consortium (ICGC)62 were obtained as simple_somatic_mutation.open.* files from release 28 of the consortium, one file for each project. These somatic mutations have been called from a mixture of whole genome and whole exome sequencing. Of the 18,965 patients represented (and not embargoed in the release 28 dataset), 116 were excluded from analysis; these represent a distinct whole exome sequenced subset of the LICA-CN project that appear to show a processing artefact in the distribution of specific mutation subsets. ICGC mutations were filtered to remove insertion and deletion mutations and also filtered for redundancy so that each mutation was only reported once for each patient. Mutation signatures deconvolution was performed using the R MutationPatterns (v.1.4.2)63 package and COSMIC signature 22 was interpreted as aristolochic acid3.
The rl20 metric and runs tests
Amongst only the informative mutations (e.g. T→N/A→N in DEN) three consecutive T→N without an intervening A→N is a run of three. The R function rle was used to encode the run-lengths for binary vectors of informative mutations along the genome of a focal tumour.
Ranking them from the longest to the shortest run, we find the set of longest runs that encompass 20% of all informative mutations in the tumour. The run-length of the shortest of those is reported as the rl20 metric. The threshold percent of mutations was defined as having to be less than 50%, as on average only 50% of the autosomal genomes are expected to show mutational asymmetry patterns. On testing with randomised data, the value of 20% gave a stable null expectation (maximum observed value of a run of five) and still encompassed a large fraction of the informative mutations. All rl20 results reported were implemented so that runs were broken when crossing chromosome boundaries.
The Wald-Wolfowitz runs test was performed using the runs.test function of the R randtests (v.1.0)64 library. It was applied to binary vectors of informative changes as described above, with threshold=0.5.
The Wald-Wolfowitz runs test significance is inflated by coordinated dinucleotide changes, such as those produced by UV light exposure and also other local mutational asymmetries such as replication asymmetry14 and kataegis events15,65. The rl20 metric appears robust to most such distortions but we find it efficiently detects kataegis events that are in an otherwise mutationally quiet background, as is often the case for breast cancer. For this reason we also indicate the total genomic span of mutations in the rl20 subset of mutation runs: kataegis events typically span a tiny (<5%) fraction of the whole genome.
Computational analysis environment
Primary data processing was performed in shell-scripted environments calling the software indicated. Except where otherwise noted, analysis processing post-variant calling was performed in a Conda environment and choreographed with Snakemake running in an LSF batch control system (Supplementary Table3). The analysis pipeline including Conda and Snakemake configuration files can be obtained from the repository https://git.ecdf.ed.ac.uk/taylor-lab/lce-ls.
Key resources
The key reagents and resources required to replicate our study are listed in Supplementary Table 3. For externally sourced data, where applicable, URLs that we used can be found in the Git repository https://git.ecdf.ed.ac.uk/taylor-lab/lce-ls.
Extended Data
Extended Data Fig. 1. Summary mutation metrics for both C3H and CAST tumours.
a, Single nucleotide substitution rates per C3H tumour, rank ordered over x-axis (grey points, median blue line). Insertion/deletion (indel, <11 nt) rates show as black. b, Y-axis from a, expanded to show distribution of indel rates with preserved tumour order. c, Number of C3H copy number variant (CNV) segments and their total span as a percent of the haploid genome. Blue shading shows intensity of overlapping points as a percent of all tumours in the plot. d-f, Corresponding plots for CAST derived tumours, f, two extreme x-axis outliers relocated (red) and x-axis value shown. g-h, Mutation spectra deconvolved from the aggregate spectra of 371 C3H tumours, subsequently referred to as the DEN1 and DEN2 signatures. i, Oncoplot summarising mutation load, mutation spectra, and driver gene mutation complement of C3H tumours. j, Oncoplot of CAST derived tumours as i.
Extended Data Fig. 2. The frequency of sister chromatid exchanges correlates with mutation rate, and localising reference genome assembly errors.
a, The relationship between single nucleotide substitution mutation load and detected sister chromatid exchange (SCE) events in C3H tumours. Counts of SCE (y-axis) are based on down-sampling to 10,000 informative mutations per tumour to ensure equal power to detect SCE in each tumour. Tumours with <50% cellularity (pink) have high mutation load and form a sub-group with few detected sister chromatid exchange events; these are suspected to be polyclonal tumours and were excluded from the Pearson’s correlation reported. b, As for a but showing CAST derived tumours. c, Evaluation of the relationship between mutation load and ability to detect sister chromatid exchange events. Mutations from C3H tumour 94315_N8 (shown in Fig. 2) randomly down-sampled and segmentation analysis applied. Y-axis shows the percentage of sister chromatid exchange events detected (100 replicates, 95% C.I. pink). X-axis is on a log-scale: 95% of C3H and >95% of CAST tumours have mutation counts to the right of the blue vertical line. Down-sampling other tumours gave comparable results. d, The same down-sampling data as shown in panel c but the y-axis shows the percent of mutations with the correct (same as full data) mutational asymmetry assignment. e, Candidate C3H reference genome assembly errors. Genome coordinates shown on the x-axis. Immediate switches between Watson and Crick asymmetry are not expected on autosomes unless both copies of the chromosome have a SCE event at equivalent sites. However, inversions and translocations between the sequenced genomes and the reference assembly are expected to produce immediate asymmetry switches. The discordant segment coverage (DSC) count (black y-axis) shows the number of informative tumours (those with either Watson or Crick strand asymmetry at the corresponding genome position) that suggest a tumour genome to reference genome discrepancy. Consensus support (brown y-axis) plotted as triangles shows the percentage of informative tumours that support a genomic discrepancy at the indicated position (only shown for values >50% support). The two sites on chromosome 6 in C3H correspond to a previously identified C3H strain specific inversion that is known to be incorrectly oriented in the C3H reference assembly60. f, As for e, but showing CAST tumours. The candidate mis-assembly on chromosome 14 in both strains at an approximately orthologous position suggesting a rearrangement shared between strains or a missassembly in the BL6 GRCm38 reference assembly against which other mouse reference genome assemblies have been scaffolded.
Extended Data Fig. 3. Locally elevated mutation load is driven by sister-chromatid exchange.
a, Double strand breaks (DSBs) and other DNA damage can trigger homologous recombination (HR) mediated DNA repair between sister chromatids. The repair intermediate resolves into separate chromatids through cleavage and ligation; grey triangles denote cleavage sites for one of the possible resolutions that would result in a large-scale sister-chromatid exchange event. Although illustrated for double-ended DNA breaks, single ended breaks from collapsed replication forks can be repaired through HR and could similarly lead to the formation of repair intermediate structures that can be resolved as SCEs. b, Enrichment analysis of sister chromatid exchanges sites (red) compared with null expectations from randomly permuting locations into the analysable fraction of the genome (grey distributions), the black boxes denote 95% of 1,000 permutations. Sister chromatid exchange events are enriched in later replicating and transcriptionally less active genomic regions (Hi-C defined compartment B), and correspondingly depleted from early replicating active regions. c, Aggregating across n=9,645 sister chromatid exchange sites, the observed mutation rate approximately doubles at the inferred site of exchange (x=0). Aggregate mutation rates (brown) were calculated in consecutive 5kb windows. Compositionally matched null expectation was generated by permuting each exchange site into 100 proxy tumours and calculating median (black) and 95% confidence intervals (grey) while preserving the total number of projected sites per proxy tumour. d, The elevated mutation count is not the result of a high mutation density in a subset of exchange sites, rather it is a subtle increase in mutations across most exchange sites. Heatmap showing mutation counts calculated in consecutive 5kb windows across each exchange site. Rows represent each exchange site, rank-ordered by total mutation count across each 400kb interval. e, The distribution of positional uncertainty in exchange site location approximately mirrors the decay profile of elevated mutation frequency. f, Divergence of mutation rate spectra is shown as cosine distance between the analysed window and the genome wide mutation rate spectrum aggregated over all C3H tumours. Despite the elevated mutation frequency, there is no detected distortion of the mutation spectrum. g, A model based on HR repair intermediate, branch migration that produces heteroduplex segments of (i) mismatch:mismatch (circles) and (ii) lesion:lesion (red triangles) strands. Subsequent strand segregation would increase the mutational diversity of a descendant cell population but not the mutation count per cell (key as per Fig. 2).
Extended Data Fig. 4. Replication of transcription coupled repair with lesion strand resolution in Mus musculus castaneus.
a, Transcription coupled repair of template strand lesions is dependent on transcription level (P15 liver, transcripts per million, TPM). Confidence intervals (99%) are shown as whiskers, where broad enough to be visible. b, Comparison of mutation rates for the 64 trinucleotide contexts: each context has one point for low and one point for high expression. c, Data as in panel b plotted on log scale; there is a line linking low and high expression for the same trinucleotide context. d, Sequence composition normalised profiles of mutation rate around transcription start sites (TSS). e, Stratifying the data plotted in d by lesion strand reveals much greater detail on the observed mutation patterns, including the pronounced influence of bidirectional transcription initiation.
Extended Data Fig. 5. Variant allele frequency distributions demonstrate high rates of non-mutagenic replication over segregating lesions.
a-f, Variant allele frequency (VAF) distributions shown as probability density functions (total area under curve=1) for example tumours, calculated taking into account observed multi-allelic variation. The VAF for identified driver mutations is indicated (brown triangle). Tumour identifiers are shown top right along with the percent of genomic segments (based on mutation asymmetry segmentation) that are multi-allelic. Skew shows Pearson’s median skewness coefficient for the VAF distributions. Panels a-c show tumours with no multi-allelic segments and exhibit a symmetric VAF distribution showing minimal sub-clonal structure; d-f tumours with all segments multi-allelic, illustrating the sub-clonal structure generated by segregating lesions. g, Tumours with a high proportion of multi-allelic segments have a left-skewed VAF distribution indicating frequent non-mutagenic replication over segregating lesions. Percent of genome segments that are multi-allelic (x-axis) plotted against VAF distribution skew for 371 C3H tumours. Tumours with low estimated cellularity indicated in pink and excluded from correlation analysis. h, As for g but showing 84 CAST tumours. i, Mutation asymmetry summary ribbon for example C3H tumour 90797_N2; genome on the x-axis. The percent of mutation sites with robust support for multi-allelic variation (y-axis) calculated in 10Mb windows (grey) and for each asymmetric segment (black). Thresholds for high (black), intermediate (grey) and zero (red) rates of multi-allelic sites shown on the right axis. j, VAF density plots for the example tumour 90797_N2 (shown in i) mutations in asymmetry segments stratified by the multi-allelic rate thresholds defined in panel i. As with whole tumour based analysis (a-h), high multi-allelic rates correspond to a leftward skew of the VAF (black, grey) whereas segments without multi-allelic variation (red) show a minimally skewed distribution.
Extended Data Fig. 6. Examples of mutation patterns generated by lesion segregation from a diverse range of clinically relevant mutagens.
a, Genome wide mutation asymmetry plot (as per Fig. 2a-c) for simulated solar radiation (SSR) exposed human iPSCs5 illustrating lesion segregation for ultraviolet damage. Immediately adjacent mutations (inter-mutation distance 100) indicate CC->TT dinucleotide changes. Despite a low total mutation load (1,308 nucleotide substitutions, 842 informative T→A changes), the mutational asymmetry of lesion segregation is evident for the aristolochic acid exposed clone5 b, and the polycyclic aromatic hydrocarbon DBADE, c that is found in tobacco smoke. d, Summary mutation asymmetry ribbons (as per Fig. 2d) for all mutagen exposed clones with rl20 >5, which illustrates the independence of asymmetry pattern between replicate clones, almost universal asymmetry on chromosome X, and approximately 50% of the autosomal genome with asymmetry over autosomal chromosomes. The dominant mutation type is indicated for each mutagen. In those clones with low mutation rates, some sister exchange sites are likely to have been missed leading to reduced asymmetry signal (e.g. on the X chromosome). Segments with <20 informative mutations are shown in white.
Extended Data Table 1. A lesion segregation based test for oncogenic selection.
| Strain | Gene | Mutation | Mutation count | Odds ratio | P-value | Known driver |
|---|---|---|---|---|---|---|
| C3H | Braf | 6:37548568_A/T | 151 | 2.13 | 5.77x10-6 | Yes |
| C3H | Hras | 7:145859242_T/C | 81 | 2.67 | 6.88x10-6 | Yes |
| C3H | Hras | 7:145859242_T/A | 65 | 1.02 | 1 | Yes |
| C3H | Intronic Fmnl1 | 11:105081902_A/C | 44 | 1.03 | 1 | No |
| C3H | Intergenic | 9:73125689_G/C | 42 | 1.13 | 1 | No |
| C3H | Egfr | 11:14185624_T/A | 34 | 3.87 | 1.23x10-4 | Yes |
| CAST | Braf | 6:37451282_A/T | 42 | 1.41 | 0.338 | Yes |
Recurrently mutated sites in both C3H and CAST with sufficient estimated power to detect oncogenic selection through biased strand retention analysis (required >33 C3H recurrences or >41 CAST recurrences). Odds ratio values >1 indicate the predicted correlation of driver mutation and Watson/Crick strand retention in tumours with the candidate driver mutation, but not for those without the mutation. The Fisher’s exact test P-value is shown after Bonferroni correction. Known driver indicates the mutation or its orthologous change has previously been implicated as a driver of hepatocellular carcinoma6. The CAST 6:37451282_A/T mutation is orthologous to the C3H 6:37548568_A/T mutation.
Supplementary Material
Acknowledgements and funding
We gratefully acknowledge Maša Roller and Florian Markowetz for supervision, and Loris Mularoni and Graham Ritchie for software support. Also, the valuable contributions by the CRUK Cambridge Institute Core facilities: CRUK Biological Resources (Angela Mowbray), Preclinical Genome Editing (Lisa Young, Steven Kupczak, Maureen Cronshaw, Paul Mackin, Yi Cheng, Lena Hughes-Hallett), Genomics (James Hadfield, Fatimah Bowater), Bioinformatics (Gord Brown, Matthew Eldridge, Richard Bowers), Histopathology and ISH, Research Instrumentation, and Biorepository; Edinburgh Genomics (Clinical) Facility; and the EMBL-EBI technical services cluster (Zander Mears, Andrea Cristofori, Tomasz Nowak, Sundeep Nanuwa, Vahit Tabak, Alessio Checcucci). We thank Wendy Bickmore and Chris Ponting for comments on the manuscript.
This work was supported by: Cancer Research UK (20412, 22398), the European Research Council (615584, 682398), the Wellcome Trust (WT108749/Z/15/Z, WT106563/Z/14/A, WT202878/B/16/Z), the European Molecular Biology Laboratory, the MRC Human Genetics Unit core funding programme grants (MC_UU_00007/11, MC_UU_00007/16), and the ERDF/Spanish Ministry of Science, Innovation and Universities-Spanish State Research Agency/DamReMap Project (RTI2018-094095-B-I00). S.J.A. received a Wellcome Trust PhD Training Fellowship for Clinicians (WT106563/Z/14/Z) and is now funded by a National Institute for Health Research (NIHR) Clinical Lectureship. O.P. is funded by a BIST PhD fellowship supported by the Secretariat for Universities and Research of the Ministry of Business and Knowledge of the Government of Catalonia and the Barcelona Institute of Science and Technology. V.S. is supported by an EMBL Interdisciplinary Postdoc (EIPOD) fellowship under Marie Skłodowska-Curie actions COFUND (664726). E.K. is supported by the EMBL International PhD Programme. C.A-P. is supported by La Caixa Foundation fellowship (ID 100010434; LCF/BQ/ES18/11670011). S.V.B. is supported by ERC Starter Grant 759967. A.E. is supported by a UKRI Innovation Fellowship (MR/RO26017/1). A.K. is a cross-disciplinary postdoctoral fellow supported by funding from the University of Edinburgh and Medical Research Council (core grant to the MRC Institute of Genetics and Molecular Medicine).
Footnotes
Author contributions
S.J.A., F.C., C.F., D.T.O. conceived the project and designed the experiments. S.J.A., F.C., C.F., performed the mutagenesis experiments and sequencing experiments. E.L-A, A.M.R. performed supporting experiments. J.S-L provided contract sequencing. S.J.A. performed the histopathological analyses with advice from S.E.D.. C.J.A., M.S.T. designed and implemented computational analysis. M.S.T. discovered lesion segregation. O.P., V.S., T.F.R., M.L., S.A., E.K., J.L. performed supporting computational analysis. C.A-P., S.V.B., R.M.D., A.E., V.B.K., A.K., I.S., L.T. contributed to the computational analyses. T.F.R., M.L., S.A., A.D.Y. curated data. S.J.A., C.A.S., N.L.B., P.F., D.T.O., M.S.T. supervised the work. S.J.A., C.A.S., N.L.B., P.F., D.T.O., M.S.T. lead the Liver Cancer Evolution Consortium. S.J.A. and P.F. provided scientific and administrative organisation. S.J.A., C.A.S., N.L.B., P.F., D.T.O., M.S.T. funded the work. S.J.A., D.T.O., M.S.T. wrote the manuscript. All authors had the opportunity to edit the manuscript. All authors approved the final manuscript.
Competing interests
P.F. is a member of the Scientific Advisory Boards of Fabric Genomics, Inc., and Eagle Genomics, Ltd.
Data availability
The WGS FASTQ files are available from the European Nucleotide Archive (ENA) under accession number PRJEB37808. RNA-seq files are available from Array Express under experiment number E-MTAB-8518. Digitised histology images are available from Biostudies under accession S-BSST383.
References
- 1.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20:404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrov LB, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kucab JE, et al. A Compendium of Mutational Signatures of Environmental Agents. Cell. 2019;177:821–836. doi: 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor F, et al. Mutational landscape of a chemically-induced mouse model of liver cancer. J Hepatol. 2018;69:840–850. doi: 10.1016/j.jhep.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maronpot RR. Biological Basis of Differential Susceptibility to Hepatocarcinogenesis among Mouse Strains. J Toxicol Pathol. 2009;22:11–33. doi: 10.1293/tox.22.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, et al. Whole-genome sequencing reveals genomic signatures associated with the inflammatory microenvironments in Chinese NSCLC patients. Nat Commun. 2018;9:2054. doi: 10.1038/s41467-018-04492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verna L, Whysner J, Williams GM. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol Ther. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 11.Kondo N, Takahashi A, Ono K, Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/543531. 543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maronpot RR, Fox T, Malarkey DE, Goldsworthy TL. Mutations in the ras proto-oncogene: clues to etiology and molecular pathogenesis of mouse liver tumors. Toxicology. 1995;101:125–156. doi: 10.1016/0300-483x(95)03112-s. [DOI] [PubMed] [Google Scholar]
- 13.Buchmann A, Karcier Z, Schmid B, Strathmann J, Schwarz M. Differential selection for B-raf and Ha-ras mutated liver tumors in mice with high and low susceptibility to hepatocarcinogenesis. Mutat Res. 2008;638:66–74. doi: 10.1016/j.mrfmmm.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Haradhvala NJ, et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petljak M, et al. Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell. 2019;176:1282–1294.e20. doi: 10.1016/j.cell.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomkova M, Tomek J, Kriaucionis S, Schuster-Böckler B. Mutational signature distribution varies with DNA replication timing and strand asymmetry. Genome Biol. 2018;19:129. doi: 10.1186/s13059-018-1509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry P, Evans HJ. Cytological detection of mutagen-carcinogen exposure by sister chromatid exchange. Nature. 1975;258:121–125. doi: 10.1038/258121a0. [DOI] [PubMed] [Google Scholar]
- 19.Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez BS. Is homologous recombination really an error-free process? Front Genet. 2014;5:175. doi: 10.3389/fgene.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strick TR, Portman JR. Transcription-Coupled Repair: From Cells to Single Molecules and Back Again. J Mol Biol. 2019;431:4093–4102. doi: 10.1016/j.jmb.2019.05.040. [DOI] [PubMed] [Google Scholar]
- 21.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supek F, Lehner B. Clustered Mutation Signatures Reveal that Error-Prone DNA Repair Targets Mutations to Active Genes. Cell. 2017;170:534–547.e23. doi: 10.1016/j.cell.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Huang MN, et al. Genome-scale mutational signatures of aflatoxin in cells, mice, and human tumors. Genome Res. 2017;27:1475–1486. doi: 10.1101/gr.220038.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forrest AR. The FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers J, Jahn K, Raphael BJ, Beerenwinkel N. Single-cell sequencing data reveal widespread recurrence and loss of mutational hits in the life histories of tumors. Genome Res. 2017;27:1885–1894. doi: 10.1101/gr.220707.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brody Y, et al. Quantification of somatic mutation flow across individual cell division events by lineage sequencing. Genome Res. 2018;28:1901–1918. doi: 10.1101/gr.238543.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou X, et al. Validating the concept of mutational signatures with isogenic cell models. Nat Commun. 2018;9:1744. doi: 10.1038/s41467-018-04052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nik-Zainal S, Hall BA. Cellular survival over genomic perfection. Science. 2019;366:802–803. doi: 10.1126/science.aax8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhomchuk D, Amstislavskiy V, Soldatov A, Ogryzko V. Use of high throughput sequencing to observe genome dynamics at a single cell level. Proceedings of the National Academy of Sciences. 2009;106:20830–20835. doi: 10.1073/pnas.0906681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan K, Gordenin DA. Clusters of Multiple Mutations: Incidence and Molecular Mechanisms. Annu Rev Genet. 2015;49:243–267. doi: 10.1146/annurev-genet-112414-054714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz R, Schäffer AA. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet. 2017;18:213–229. doi: 10.1038/nrg.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics. 1969;61:893–903. doi: 10.1093/genetics/61.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Genetic Load and Potential Mutational Meltdown in Cancer Cell Populations. Mol Biol Evol. 2019;36:541–552. doi: 10.1093/molbev/msy231. [DOI] [PubMed] [Google Scholar]
- 37.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 38.Tilk S, Curtis C, Petrov D, McFarland CD. Most cancers carry a substantial deleterious load due to Hill-Robertson interference. bioRxiv. 2019 doi: 10.1101/764340. 764340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thoolen B, et al. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol. 2010;38:5S–81S. doi: 10.1177/0192623310386499. [DOI] [PubMed] [Google Scholar]
- 40.Lilue J, et al. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat Genet. 2018;50:1574–1583. doi: 10.1038/s41588-018-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broad Institute. Picard Tools. Broad Institute, GitHub Repository. 2019 http://broadinstitute.github.io/picard.
- 43.Kim S, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 44.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eldridge M. gatk-tools: Utilities for processing sequencing data and genomic variants using GATK. https://github.com/crukci-bioinformatics/gatk-tools. [Google Scholar]
- 46.R Core Team. R: A Language and Environment for Statistical Computing. 2017 [Google Scholar]
- 47.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray N, Pimentel H, Melsted P, Pachter L. Near-optimal RNA-Seq quantification with kallisto. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 49.Cunningham F, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vietri Rudan M, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durand NC, et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armstrong J, et al. Progressive alignment with Cactus: a multiple-genome aligner for the thousand-genome era. bioRxiv. 2019 doi: 10.1101/730531. 730531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2013-2015 http://www.repeatmasker.org. [Google Scholar]
- 55.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gori K, Baez-Ortega A. sigfit: flexible Bayesian inference of mutational signatures. bioRxiv. 2018 doi: 10.1101/372896. 372896. [DOI] [Google Scholar]
- 57.Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnedo-Pac C, Mularoni L, Muiños F, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUSTL: a sequence-based clustering method to identify cancer drivers. Bioinformatics. 2019;35:5396. doi: 10.1093/bioinformatics/btz588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Killick R, Eckley IA. changepoint: An R Package for Changepoint Analysis. J Stat Softw. 2014;58 [Google Scholar]
- 60.Akeson EC, et al. Chromosomal inversion discovered in C3H/HeJ mice. Genomics. 2006;87:311–313. doi: 10.1016/j.ygeno.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 61.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.International Cancer Genome Consortium et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caeiro F, Mateus A. randtests: Testing randomness in R. 2014 [Google Scholar]
- 65.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WGS FASTQ files are available from the European Nucleotide Archive (ENA) under accession number PRJEB37808. RNA-seq files are available from Array Express under experiment number E-MTAB-8518. Digitised histology images are available from Biostudies under accession S-BSST383.