Abstract
Somatic mutations in p53, which inactivate the tumour-suppressor function of p53 and often confer oncogenic gain-of-function properties, are very common in cancer1,2. Here we studied the effects of hotspot gain-of-function mutations in Trp53 (the gene that encodes p53 in mice) in mouse models of WNT-driven intestinal cancer caused by Csnk1a1 deletion3,4 or Apc Min mutation5. Cancer in these models is known to be facilitated by loss of p533,6. We found that mutant versions of p53 had contrasting effects in different segments of the gut: in the distal gut, mutant p53 had the expected oncogenic effect; however, in the proximal gut and in tumour organoids it had a pronounced tumour-suppressive effect. In the tumour-suppressive mode, mutant p53 eliminated dysplasia and tumorigenesis in Csnk1a1-deficient and Apc Min/+ mice, and promoted normal growth and differentiation of tumour organoids derived from these mice. In these settings, mutant p53 was more effective than wild-type p53 at inhibiting tumour formation. Mechanistically, the tumour-suppressive effects of mutant p53 were driven by disruption of the WNT pathway, through preventing the binding of TCF4 to chromatin. Notably, this tumour-suppressive effect was completely abolished by the gut microbiome. Moreover, a single metabolite derived from the gut microbiota–gallic acid–could reproduce the entire effect of the microbiome. Supplementing gut-sterilized p53-mutant mice and p53-mutant organoids with gallic acid reinstated the TCF4–chromatin interaction and the hyperactivation of WNT, thus conferring a malignant phenotype to the organoids and throughout the gut. Our study demonstrates the substantial plasticity of a cancer mutation and highlights the role of the microenvironment in determining its functional outcome.
TP53, the gene that encodes p53, is the most commonly mutated gene in human cancer7. Mutations in p53–including the six most frequently mutated residues, which are known as ‘hotspots’1–are mostly loss-of-function mutations in which the mutant p53 protein fails to activate the target genes of wild-type p53 that are critical for the control of homeostasis8. Mutant versions of p53 can have a dominant-negative effect over wild-type p53 and, in common with the protein products of typical dominant oncogenes, are often endowed with gain-of-function properties, thereby enhancing tumorigenesis beyond loss-of-function9. Here we tested the effects of mouse homologues of the hotspot mutations in p53 that occur most frequently in humans–R172H and R270H (R175H and R273H in humans, respectively)1. The mutations were generated by knock-in10,11, and mice were bred to homozygosity with two mouse models of WNT-driven intestinal cancer: Apc Min/+ mice (which are heterozygous for the gene encoding adenomatous polyposis coli (Apc) and develop multiple intestinal neoplasias); and mice with a gut-inducible deletion of Csnk1a1 (also known as CKIa, which encodes casein kinase 1α (CKIα))3. Whereas–as predicted–expression of mutant p53 conferred oncogenic gain-of-function properties in the distal part of the gut, notably, it exerted profound tumour-suppressive activity in the proximal part of the gut. Our findings challenge commonly held views of the effects of cancer mutations, and underscore the critical dependence of these mutant effects on the gut microbiome.
Paradoxical role of mutant p53 in the gut
To investigate the role of mutant p53 in gut tumorigenesis we first used the Csnk1a1 floxed/Vil1-Cre-ERT2 mouse model, in which Cskn1a1 (CKIa) is conditionally deleted in the gut epithelium, resulting in the activation of wild-type p533. We compared CKIa-deleted mice that have the conformational R172H hotspot mutation in Trp53 (for simplicity this gene is hereafter referred to as p53, and the mutation as p53 R172H) to CKIa-deleted mice that have either wild-type p53 or no p53 (hereafter CKIa Δgut p53 R172H, CKIa Δgut or CKIa Δgut p53 Δgut, respectively).
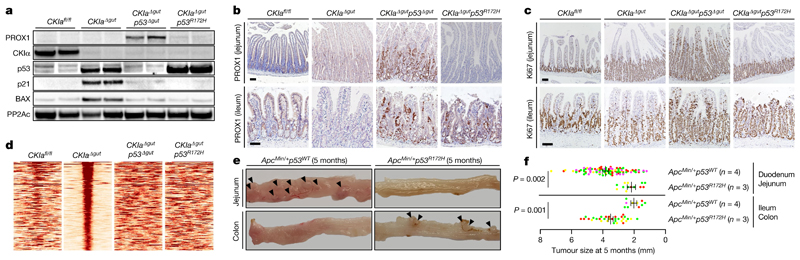
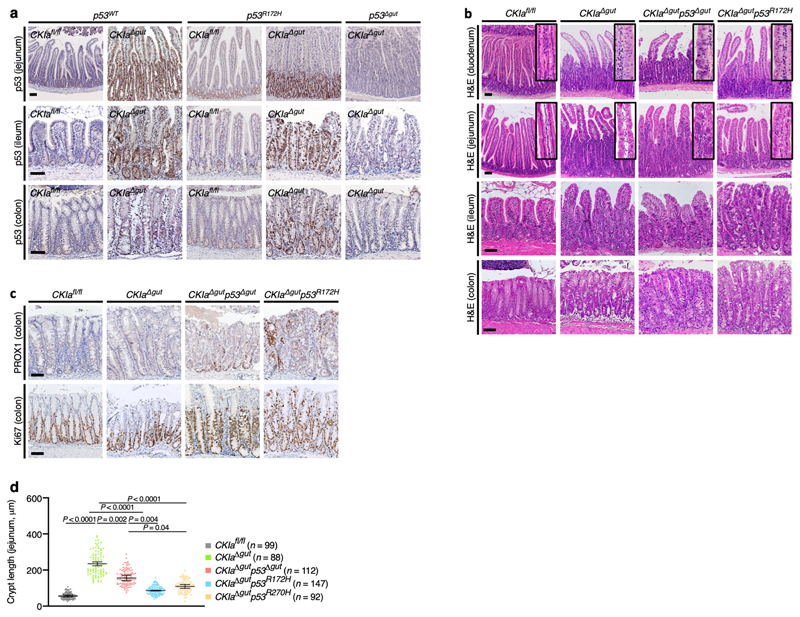
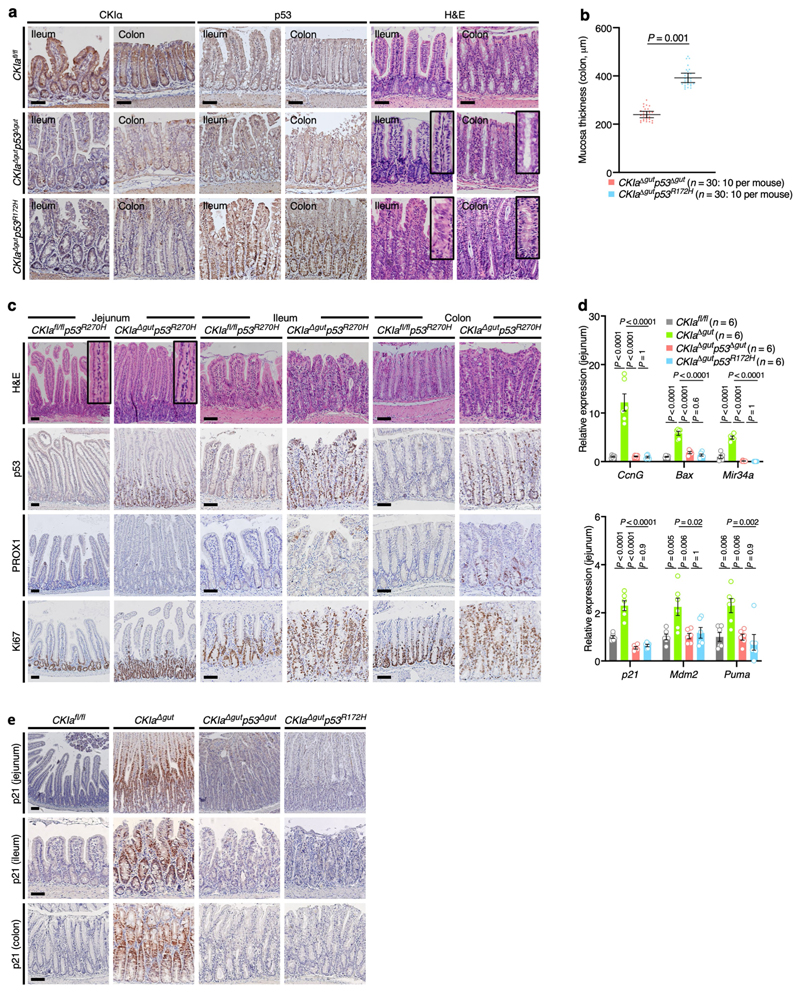
Protein stabilization is a prerequisite for efficient gain-of-function action2,8. In normal mouse gut epithelium, expression of p53 is low and restricted to the intestinal crypts. After deletion of CKIa, wild-type and mutant p53 proteins are greatly stabilized, specifically in the gut epithelium (Fig. 1a, Extended Data Fig. 1a), and show levels of expression similar to those that characterize tumours in which p53 is mutated10,11. Deleting CKIa therefore provides a means by which the effects of mutant p53 in gut carcinogenesis can be investigated. Cancer phenotype was evaluated by histological analysis of the proximal (duodenum and jejunum) and the distal (ileum and colon) gastrointestinal tract of the different mouse genotypes in comparison to CKIa fl/fl (virtually wild-type) mice. As we previously showed, gut homeostasis was maintained in CKIa Δgut mice, with moderate epithelial dysplasia, whereas CKIa Δgut p53 Δgut mice exhibited high-grade dysplasia (marked by PROX1 expression) in the entire bowel, together with enhanced proliferation12 (Fig. 1a–c, Extended Data Fig. 1b, c). However, CKIa Δgut p53 R172H mice showed an opposing phenotype at different gut segments: whereas the colonic and ileal epithelia were highly dysplastic and proliferative, the duodenum and jejunum showed normal levels of proliferation, with minimal dysplasia in the villi and significantly shorter jejunal crypts compared to CKIa Δgut and CKIa Δgut p53 Δgut mice (Fig. 1a–c, Extended Data Fig. 1b–d).
Fig. 1. Mutant p53 counteracts dysplasia and tumorigenesis in the proximal gut without regaining wild-type transcriptional activity.
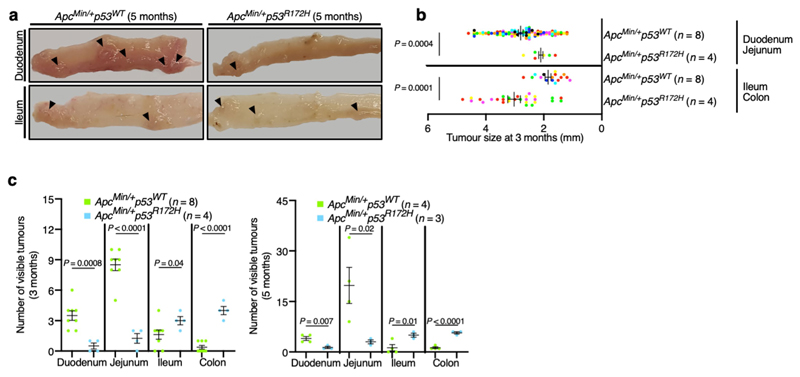
a, Immunoblot of mouse jejunal enterocytes. PP2Ac, loading control. For gel source data, see Supplementary Fig. 1. b, c, IHC of the invasion and dysplasia marker PROX1 (b) and the proliferation marker Ki67 (c) in mouse jejunum and ileum. Scale bars, 100 μm. d, p53 ChIP–seq-derived heat maps from mouse jejunal enterocytes. e, Representative images of different segments of the mouse bowel. Arrowheads indicate visible tumours. f, Tumour size (each mouse is colour-coded). Overall average of the mean values for each mouse ± s.e.m (n, number of mice), one-sided Student’s t-test. Representative data from six (a–c) or three (e) independent experiments.
The loss of CKIα when combined with the expression of mutant p53 is lethal within 10 days, and therefore does not allow the development of discrete tumours (polyps or carcinomas)13. To assess the gain-of-function properties of mutant p53, we compared the gut phe-notype of CKIa Δgut p53 R172H mice to that of CKIa Δgut p53 Δgut mice three days after CKIa deletion. At this time point, CKIa Δgut p53 R172H mice already showed high-grade dysplasia in the distal gut, whereas CKIa Δgut p53 Δgut mice had minimal gut pathology (Extended Data Fig. 2a, b). Notably, the gut phenotype of CKIa Δgut mice with the p53 R270H contact mutation (CKIa Δgut p53 R270H) was similar to that of mice with the p53 R172H structural mutation; CKIa Δgut p53 R270H mice showed no evidence of invasion or hyperproliferation in the proximal gut, but the distal gastrointestinal tract was highly dysplastic and proliferative (Extended Data Figs. 1d, 2c). These data indicate that in the proximal gut, mutant p53 overturns the pro-carcinogenic effects of p53 loss, and attenuates the mild proliferative and dysplastic phenotype of CKIa Δgut mice.
Wild-type p53 has a major anti-carcinogenic effect partly through the transcriptional activation of anti-proliferative and pro-apoptotic genes such as p21 (also known as Cdkn1a) and Bax 14. Therefore, one possibility to explain the tumour-suppressive effect of mutant p53 in the jejunum would be the regaining of this transcriptional activation15. However, although mutant p53 was markedly stabilized after loss of CKIα (Fig. 1a, Extended Data Fig. 1a), it was transcriptionally inactive with respect to canonical p53 target genes in the gut epithelium; we observed the robust expression of pro-apoptotic and cell-cycle-inhibitory target genes of p53 in CKIa Δgut mice, but not in CKIa Δgut p53 R172H mice, which–similarly to CKIa Δgut p53 Δgut mice–barely expressed p53 target genes (Extended Data Fig. 2d, e). Moreover, chromatin immunoprecipitation followed by sequencing (ChIP–seq) analysis revealed that the interaction of p53(R172H) with chromatin was almost completely abrogated compared to wild-type p53 (Fig. 1d), confirming that the tumour-suppressive effect of mutant p53 in the jejunum was independent of canonical p53 transcription.
To test the effects of mutant p53 in a mouse model with discrete intestinal tumours, we bred p53 R172H mutant mice with the Apc Min/+ mouse model. In this commonly used mouse model, typical adenoma tumours are produced after Apc loss-of-heterozygosity and WNT hyperactivation, and the tumours occur mostly in the proximal gut, with few or no colonic tumours5. As in CKIa Δgut p53 R172H mice, introduction of the p53 R172H mutation to Apc Min/+ mice resulted in enhanced tumorigenesis in the colon, but attenuated the tumour burden in the proximal gastrointestinal tract (Fig. 1e, f, Extended Data Fig. 3). Together, our results show that mutant p53 has a dichotomous effect in two mouse models of WNT-driven cancer: counteracting tumorigenesis in the proximal gut, but enhancing tumorigenesis in the distal gut.
Mutant p53 blocks the TCF–WNT pathway
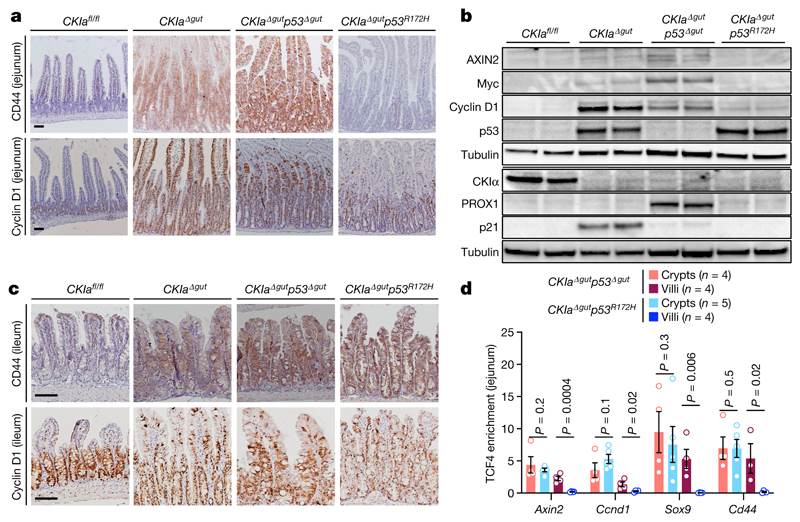
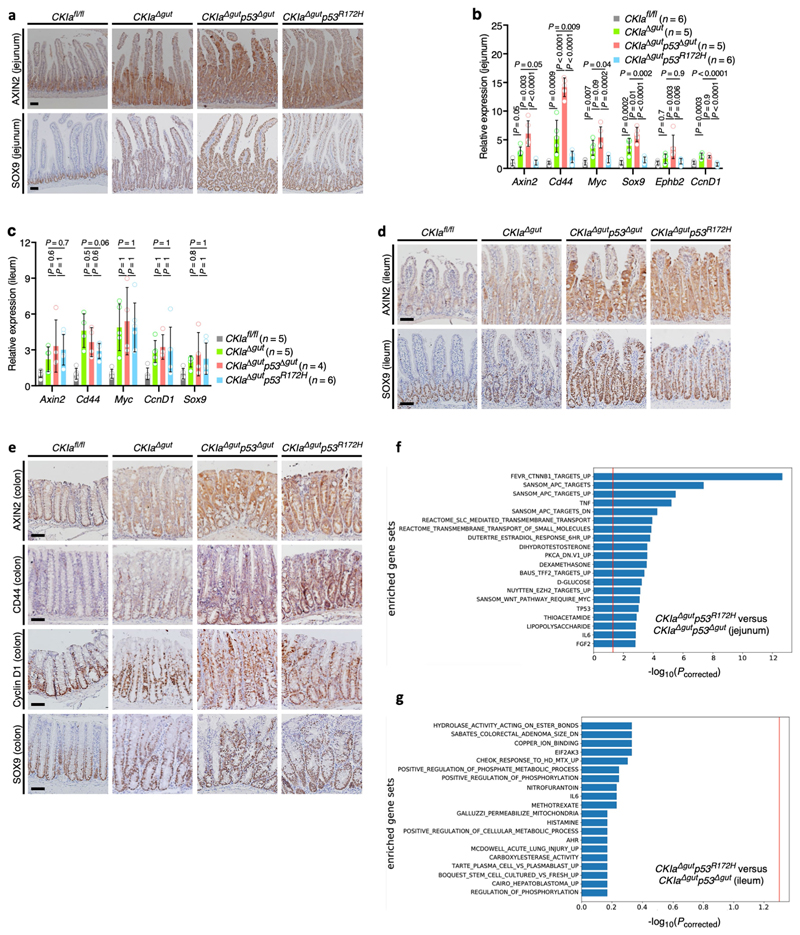
Wild-type p53 suppresses the expression of a set of genes known as the ‘p53-suppressed invasiveness signature’ (PSIS). PSIS genes are associated with a variety of invasiveness functions, and many of these genes are targets of WNT signalling3,4. To examine whether WNT signalling is also suppressed by mutant p53, we examined the expression levels of several WNT target genes in the jejunum and ileum of CKIa Δgut p53 R172H mice. As expected, a moderate activation of WNT target genes was evident in the jejunum of CKIa Δgut mice, and a far more robust activation was observed in that of CKIa Δgut p53 Δgut mice. By contrast, CKIa Δgut p53 R172H mice showed lower expression levels of all tested WNT target genes in the jejunum, in line with the tumour-suppressive activity of mutant p53 (Fig. 2a, b, Extended Data Fig. 4a, b). Notably, the expression levels of cyclin D1 and Myc–prominent WNT-driven proto-oncogenes that are critical for gut tumorigenesis16–were reduced to CKIa fl/fl levels in the jejunal enterocytes of CKIa Δgut p53 R172H mice (Fig. 2a, b, Extended Data Fig. 4b). By contrast, the expression of WNT target genes was high in the ileum and colon of CKIa Δgut p53 R172H mice, with levels of expression similar to those observed in CKIa Δgut and CKIa Δgut p53 Δgut mice (Fig. 2c, Extended Data Fig. 4c–e). To confirm the preferential influence of mutant p53 on WNT-controlled gene expression, we compared the transcriptomes of jejunal and ileal enterocytes of CKIa Δgut p53 R172H mice to those of CKIa Δgut p53 Δgut mice; this analysis showed that the effect of mutant p53 was mostly evident in WNT-dominating gene sets, and only in jejunal enterocytes (Extended Data Fig. 4f, g). No other pathway showed a similar degree of variation between the two mouse genotypes, in either the proximal or the distal gastrointestinal tract, indicating that the suppression of WNT by mutant p53 is a specific rather than a global genomic phenomenon.
Fig. 2. Mutant p53 suppresses the expression of WNT targets in the proximal gut.
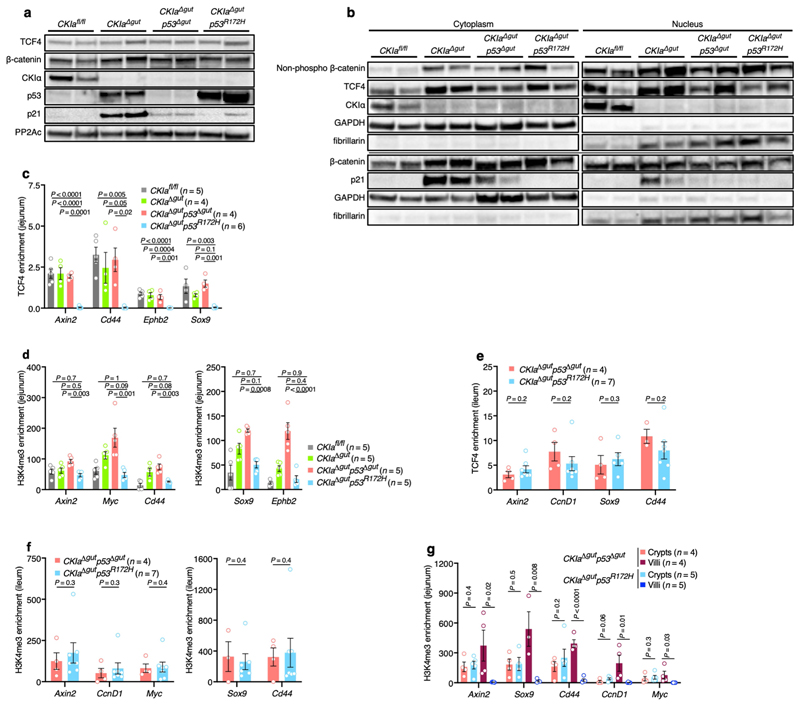
a, c, IHC of WNT targets in mouse jejunum (a) and ileum (c). Scale bars, 100 μm. b, Immunoblot of mouse jejunal enterocytes. Tubulin, loading control. For gel source data, see Supplementary Fig. 1. d, TCF4 ChIP of WNT target promoters in crypt and villus fractions of mouse jejunum. Mean enrichment ± s.e.m. (n, number of mice), one-sided Student’s t-test. Representative data from six independent experiments (a–c).
To better understand the mechanism by which mutant p53 impedes WNT activation, we analysed the major steps that lead to the transcriptional activation of WNT target genes. Activation of the WNT pathway requires the accumulation of β-catenin and its nuclear translocation16. We found that both non-phosphorylated (active) and total nuclear levels of β-catenin were increased in mouse jejuna that lack CKIa, with no significant differences among the different CKIa-deficient genotypes (Extended Data Fig. 5a, b). β-catenin is a transcriptional coactivator that relies on T cell factor (TCF) and lymphoid enhancer factor (LEF) transcription factors16. TCF4–the main TCF in the gut epithelium–is constitutively bound to the chromatin, and requires association with β-catenin to promote WNT transcription16. Notably, ChIP analyses of jejunal enterocytes from the different mouse genotypes demonstrated the selective disengagement of TCF4 and reduced levels of the histone modification H3K4me3 at WNT-responsive elements in CKIa Δgut p53 R172H mice, although the total levels of TCF4 were comparable between genotypes (Extended Data Fig. 5a–d). Conversely, and in line with the observed phenotype, ChIP assays from ileal enterocytes showed no alteration in either TCF4 occupancy or H3K4me3 modification at WNT promoters in CKIa Δgut p53 R172H mice compared to CKIa Δgut p53 Δgut mice (Extended Data Fig. 5e, f). Despite the near-complete disengagement of TCF4, the mRNA expression of many WNT target genes was preserved at near-CKIa fl/fl levels (Extended Data Fig. 4b). Immunohistochemistry (IHC) analysis revealed that this occurred mostly in intestinal crypts (Fig. 2a, Extended Data Fig. 4a), which comprise roughly 10% of the jejunal gut epithelium. ChIP analyses of jejunal villus and crypt epithelial cells showed that in CKIa Δgut p53 R172H–but not in CKIa Δgut p53 Δgut– mice, the association of TCF4 with chromatin was limited to the crypt compartment (Fig. 2d). H3K4me3 ChIP confirmed that WNT activity was present specifically in the crypts of CKIa Δgut p53 R172H mice (Extended Data Fig. 5g). This crypt-spared effect of mutant p53 may explain the maintenance of homeostasis in the proximal CKIa Δgut p53 R172H gut.
Cell-autonomous WNT suppression by mutant p53
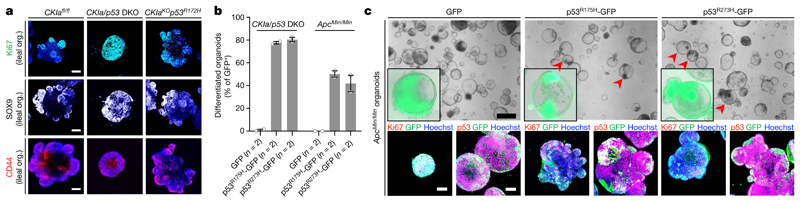
Although mutant p53 is stabilized similarly in the jejunum and the ileum, it only suppresses WNT activity in the jejunum. To ascertain the source of this discrepancy, we prepared and tested CKIa-deleted jejunal and ileal organoids that express the wild-type p53 gene, p53 R172H or no p53. Stem-cell-derived intestinal organoids are composed of a round core of differentiated cells and proliferating outpockets that surround the core; these resemble the villus and crypt compartments, respectively16. As previously reported, organoids with knockout of CKIa died shortly after the induction of the knockout, owing to the expression of p53 target genes4. By contrast, organoids with double knockout of CKIa and p53 (CKIa/p53 DKO) were highly proliferative; these organoids exhibited WNT hyperactivation, failed to differentiate and resembled highly dysplastic spheroids with no outpockets4 (Fig. 3a, Extended Data Fig. 6a–e). Expression of mutant p53 rescued both jejunal and ileal CKIa-deleted organoids, probably owing to the loss of canonical p53 target genes that induce organoid apoptosis4 (Extended Data Fig. 6a–f). However, unlike CKIa/p53 DKO organoids, CKIa-knockout organoids that express p53 R172H (CKIa KO p53 R172H) exhibited normal organoid differentiation, moderate WNT activity and proliferation and reduced dysplasia irrespective of the gut origin of the organoid, highlighting the cell-autonomous anti-proliferative and differentiation-promoting effect of mutant p53 (Fig. 3a, Extended Data Fig. 6b–e, g). It therefore appears that mutant p53 also has the potential to exert anti-tumorigenic effects in the ileum, but this is switched into a pro-tumorigenic activity in the in vivo setting.
Fig. 3. Mutant p53 promotes normally balanced growth and differentiation of intestinal tumour organoids.
a, Immunofluorescent staining of Ki67 and WNT targets in ileal organoids (org.). Nuclear counterstain, Hoechst (blue). Scale bars, 100 μm. b, Differentiated organoids as a percentage of the total number of GFP-positive organoids, transduced with the indicated lentiviruses (n, number of independent experiments). c, Bright-field imaging and merged immunofluorescence of GFP with p53 (endogenous and transduced) or with Ki67 in organoids that were transduced with the indicated lentiviruses. Nuclear counterstain (immunofluorescent images), Hoechst (blue). Inserts (bright-field images) show a GFP and bright-field merged image of a representative organoid. Scale bars, 500 μm (top); 100 μm (bottom). Representative data from three (a) or two (c) independent experiments.
To test whether human mutant p53 has a similar tumour-suppressive capacity to that found in mice, we overexpressed the R175H and R273H human homologues of mutant p53 together with GFP in CKIa/p53 DKO organoids. Organoids expressing human mutant p53 and GFP, or expressing only GFP, were identified by GFP imaging and underwent phenotypic assessment and immunofluorescent staining for p53, GFP and Ki67. Consistent with the tumour-suppressive effect of the mouse homologues in organoids and in the proximal mouse gut, human mutant p53 abolished the transformed phenotype of CKIa/p53 DKO organoids; organoids expressing human mutant p53 continued growing, yet underwent differentiation with typical crypt outpockets and a CKIa fl/fl-like redistribution of Ki67 (Fig. 3b, Extended Data Fig. 6h). Of note, given sufficient time, cancer cells might gradually rewire their signalling pathways to maximize the gain-of-function effects of their endogenous mutant p53, eventually becoming ‘addicted’ to it17. Hence, exogenous mutant p53 expression in freshly made p53-deficient orga-noids might not necessarily phenocopy the endogenous mutant effect.
Similarly to CKIa/p53 DKO organoids, Apc Min/Min adenoma organoids are highly proliferative spheroids with robust Ki67 staining, and this is despite stabilization of wild-type p53 (Fig. 3c, Extended Data Fig. 6i). Upon overexpression of human mutant p53–GFP, 40–50% of the adenoma organoids underwent a phenotypic differentiation switch during their growth, whereas none of the adenoma organoids that expressed only GFP switched phenotypes (Fig. 3b, c). Together, these data show that the tumour-suppressive power of p53 mutants is well beyond that of wild-type p53 and is sufficient to revoke an established tumorigenic phenotype in organoids that are derived from different models of intestinal cancer.
The gut microbiome turns mutant p53 oncogenic
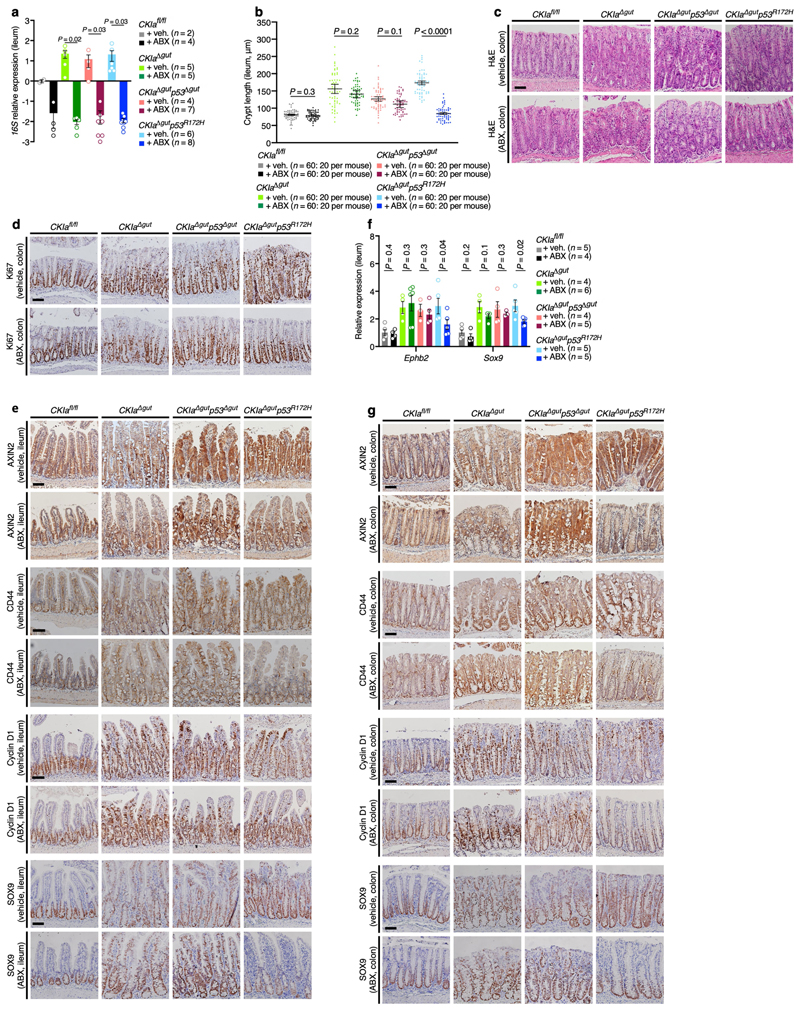
The gastrointestinal tract is a complex habitat that contains an abundance of microorganisms, the number of which increases progressively along the gastrointestinal tract: microorganisms are sparse in the proximal region and dense at the distal region of the gut18. A continuous crosstalk between the microbiome and the host cells, which is often carried out by metabolites that are derived from the microbiota, is crucial to maintain tissue homeostasis19. Dysbiosis–a perturbation in either the load or the composition of microflora–is closely associated with various illnesses; for example, obesity, inflammation and cancer18. Having shown that mutant p53 suppresses WNT and tumorigenesis only in the proximal gut and in sterile ileal organoids, we hypothesized that the gut microflora may hinder p53-mediated suppression of WNT in the distal intestine. To test this theory, we eliminated the gut microflora by treating the mice with a cocktail of antibiotics and confirmed the eradication of the gut bacteria by 16S ribosomal RNA (rRNA) analysis (Extended Data Fig. 7a). Notably, the antibiotics affected only CKIa Δgut p53 R172H mice: in these mice, treatment with antibiotics counteracted the dysplasia that was observed in the colon and ileum, and the mice had shorter crypts and better organized villi in these regions (Fig. 4a, Extended Data Fig. 7b, c). Likewise, the eradication of the gut microflora resulted in reduced WNT activation and decreased cell proliferation specifically in the ileum and colon of CKIa Δgut p53 R172H mice (Fig. 4b, c, Extended Data Fig. 7d–g). Together, our data suggest that the gut microbiota oppose the suppression of WNT that is mediated by mutant p53, and thereby promote tumorigenesis in the distal bowel.
Fig. 4. Treatment with antibiotics unleashes tumour-suppressive effects of mutant p53 in the distal gut.
a, b, Haemotoxylin and eosin (H&E)-stained sections (a) and IHC of Ki67 (b) in the ileum of mice that were treated with vehicle or a cocktail of antibiotics (ABX). Inserts (H&E) show a villus at high magnification, to demonstrate the grade of dysplasia. Scale bars, 100 μm. c, Quantitative PCR with reverse transcription (RT–qPCR) of WNT target genes in ileal enterocytes from mice that were treated with vehicle or antibiotics. Mean expression ± s.e.m. (n, number of mice) relative to vehicle-treated CKIa fl/fl mice (normalized to 1), one-sided Student’s t-test. Representative data from four independent experiments (a, b).
Gallic acid mimics the microbiota effect
An analysis of the microflora composition by bacterial 16S rRNA sequencing showed that there were no major differences in species between CKIa fl/fl and CKIa fl/fl p53 R172H mice (data not shown). It is therefore unlikely that mutant p53 contributes to tumorigenesis indirectly through dysbiosis. Commensal gut bacteria–and in particular their metabolic products–strongly influence the predisposition to develop colorectal cancer18. We thus speculated that specific bacteria-derived metabolites that are present in the distal but not in the proximal gut might have the potential to counteract the WNT-suppressive effect of mutant p53. To test this hypothesis, we performed a coarse metabolite screen in CKIa KO p53 R172H jejunal organoids. Organoids were treated with metabolites from four major subgroups that have been documented to exert epigenetic effects often associated with tumorigenesis19: short-chain fatty acids (for example, butyrate and caproate); lipid derivatives (for example, deoxycholate); isothiocyanates (for example, sulforaphane); and polyphenols (for example, urolithin B, ellagic acid and gallic acid). Organoid morphology, proliferation capacity and WNT activity were tested in treated versus non-treated organoids. Whereas treatment with short-chain fatty acids, lipid derivatives or isothiocyanates did not have a marked effect on morphology, polyphenols caused organoid rounding, with polyphenol-treated organoids resembling the WNT-active CKIa/p53 DKO organoids in their morphology (Extended Data Fig. 8a). Of the polyphenols that were tested, only gallic acid significantly increased the proliferation capacity and WNT activity of CKIa KO p53 R172H organoids (Extended Data Fig. 8a, b). The morphology of CKIa fl/fl and CKIa fl/fl p53 R172H organoids was not altered after treatment with gallic acid (Extended Data Fig. 8c), suggesting that gallic acid specifically impairs the ability of mutant p53 to suppress WNT hyperactivation. Likewise, administration of gallic acid to CKIa/p53 DKO and Apc Min/Min organoids that express human mutant p53 reversed the mutant-p53-induced differentiation phenotype to a transformed phenotype characterized by spheroids with no outpockets (Extended Data Fig. 8d). Finally, we tested whether the presence of gallic acid is necessary for maintaining the suppression of the anti-tumorigenic effect of mutant p53 by removing gallic acid from the organoid culture medium. Four days after the removal of gallic acid, CKIa KO p53 R172H organoids lost their hyper-WNT-proliferative properties and reverted to the normal organoid appearance, in which WNT expression is limited to the organoid outpockets (Extended Data Fig. 8e). This indicates that the continuous presence of gallic acid is required to prevent the tumour-suppressive property of mutant p53.
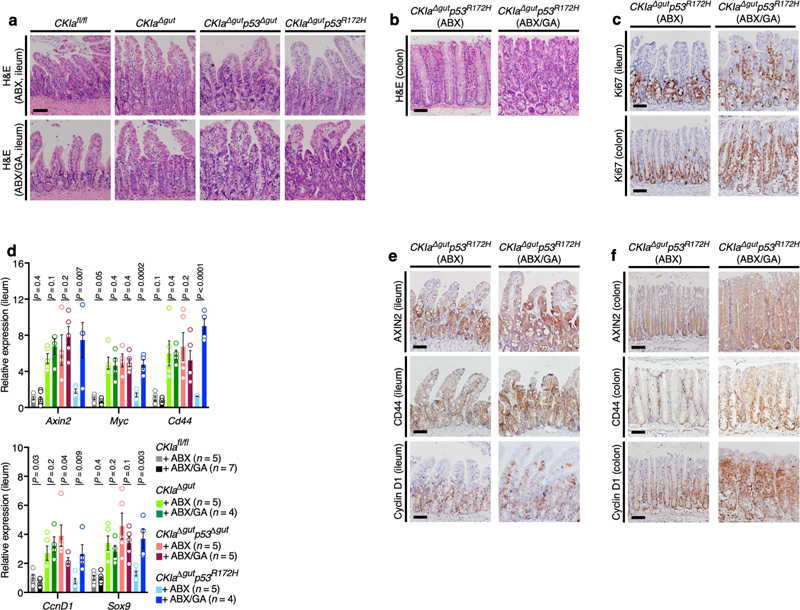
To study the effect of gallic acid on WNT suppression and gut tumo-rigenesis in vivo, we first measured the content of gallic acid in the jejunum (which is sparsely populated by bacteria) and in the ileum (which is densely populated),18 and found markedly higher levels in the distal small intestine (Extended Data Fig. 9a). Gallic acid is formed from 3-dehydroshikimate by the action of the bacterial enzyme shikimate dehydrogenase (SDH)20. Two bacterial strains–Lactobacillus plantarum and Bacillus subtilis–have specifically been identified as producers of gallic acid in humans21. We identified the SDH-coding genes of these strains (aroE of B. subtilis and aroc2 of L. plantarum) in the stool and the mucosa of CKIa fl/fl and CKIa fl/fl p53 R172H mice, with no significant differences between the two mouse genotypes (Extended Data Fig. 9b, c). Notably, whereas the jejunum contains minimal copies of the SDH genes, abundant copies were present in both the ileum and the colon of CKIa fl/fl and CKIa fl/fl p53 R172H mice (Extended Data Fig. 9c). Antibiotic treatment that eliminated nearly all of the copies of the SDH genes in mouse stools (Extended Data Fig. 9b) ameliorated tumorigenesis in CKIa Δgut p53 R172H mice (Fig. 4 and Extended Data Fig. 7). We then treated the mice with gallic acid, which increased the levels of gallic acid in the jejunum to nearly half of the endogenous levels in the ileum (Extended Data Fig. 9a). Treatment with gallic acid affected the properties of the jejunum specifically in CKIa Δgut p53 R172H mice; it became highly proliferative, with abundant high-grade dysplastic foci (Fig. 5a, Extended Data Fig. 9d). Consistent with this, the average nuclear size and overall dysplasia score were significantly higher in gallic-acid-treated versus non-treated CKIa Δgut p53 R172H mice (Extended Data Fig. 9e, f). The effect of gallic acid was then tested in the Apc Min/+ mouse models. Treatment with gallic acid completely abolished the mutant-p53-mediated suppression of tumorigenesis in the proximal gut of Apc Min/+ p53 R172H mice, but had no effect in Apc Min/+ mice that express wild-type p53 (Apc Min/+ p53 WT), indicating that–similar to our findings in the CKIa Δgut-based mouse models–the effect of this microbial metabolite is mediated through mutant p53 (Fig. 5b, c, Extended Data Fig. 9g–j).
Fig. 5. Gallic acid induces WNT-driven dysplasia and tumorigenesis when p53 is mutated.
a, IHC of Ki67 in the jejunum of mice that were treated with gallic acid (GA) or not treated (NT). Scale bar, 100 μm. b, Representative images of different segments of the mouse bowel. Arrowheads indicate visible tumours. c, Quantification of visible tumours per mouse. Mean ± s.e.m. (n, number of mice), one-sided Student’s t-test. d, TCF4 ChIP of WNT target promoters in mouse jejunal enterocytes. Mean enrichment ± s.e.m. (n, number of mice), one-sided Student’s t-test. e, Over-representation analysis of enriched gene sets differentially expressed in the jejunum of gallic-acid-treated versus not-treated CKIa Δgut p53 R172H mice; negative logarithm of Benjamini–Hochberg-corrected P values (data derived from three mice of each group). The red line indicates the significance threshold for α = 0.05. f, Immunoblot of mouse ileal enterocytes. PP2Ac, loading control. For gel source data, see Supplementary Fig. 1. Representative data from two or three independent experiments (a, b, f).
Notably, in line with the organoid results, treating mice with gallic acid reversed the WNT-suppressive effect of mutant p53. Treatment with gallic acid enabled TCF4–chromatin association and H3K4me3 modification at genomic WNT promoters exclusively in jejunal enterocytes of CKIa Δgut p53 R172H mice (Fig. 5d, Extended Data Fig. 9k), thereby inducing the expression of WNT target genes (Extended Data Fig. 9l–o). Moreover, transcriptome analysis highlighted WNT as the most differentially affected pathway in the jejunum of gallic-acid-treated CKIa Δgut p53 R172H mice (Fig. 5e). Gene sets associated with DNA replication and strand elongation were also highly enriched, possibly secondary to WNT activation.
Supplementation with gallic acid abolished the tumour-suppressive activity of mutant p53 in the distal gut of CKIa Δgut p53 R172H mice that were treated with antibiotics; the ileum and colon of the mice became hyperproliferative, with abundant high-grade dysplastic foci, and WNT target genes were highly induced throughout the ileum and colon (Fig. 5f, Extended Data Fig. 10). Together, our data indicate that gallic acid–which is produced by bacterial species that are present in both humans and mice, and is naturally abundant only in the distal gut–fully reproduces the effect of the gut microbiome in abolishing the tumour-suppressive activity of mutant p53.
Discussion
Our study reveals a remarkable functional plasticity of mutant p53 and shows the instructive role of the microbiome in moulding this plasticity. Two of the most common mutant p53 isoforms exert robust tumour-suppressive effects in the proximal mouse gut that exceed the effects of wild-type p53: mutant p53 suppresses WNT-driven hyperpro-liferation and dysplasia in CKIa Δgut mice and tumorigenesis in Apc Min/+ mice. These tumour-suppressive effects were completely abolished and instead converted to oncogenic gain-of-function activity upon interfacing with the native gut microbiota, or specifically by supplementation with gallic acid. How treatment with gallic acid overrides the WNT-blocking effect of mutant p53 remains unknown, but its reversible effect opens up possible preventive and therapeutic options for cancer; for example, through dietary management, inhibitors of gallic acid synthesis20,22 and gallic acid antagonists. It is also possible that other microbiota-derived metabolites might act as similar modulators of mutant p53, or that there are some indirect metabolite effects through the tumour stroma and the immune system.
Gain-of-function mutations in TP53 are also prevalent in tumour tissues that are inherently devoid of any substantial microbiome2, and these mutations may thus be favoured over loss-of-function mutations by mechanisms that do not involve the microbiota. Of note, TP53 and CTNNB1 mutations, which are the most frequent drivers of hepatocellular carcinoma in humans, are mutually exclusive23 (Supplementary Table 1), suggesting that WNT might be suppressed by mutant p53 in hepatocytes. Certain gain-of-function mutations in TP53 may thus show a tumour-suppressive function preferentially in WNT-driven cancers. Assuming that p53 mutations act similarly in humans, it is possible that selecting p53 mutations as tumour-suppressive entities could be of value to the maintenance of tissue homeostasis. Somatic p53 mutations are fairly prevalent and seem to promote clonal selection in healthy human skin24 and in the ageing oesophagus25,26. Why p53 mutations are associated with clonal expansion far more than other oncogenic cancer mutations is unclear. It is tempting to speculate that mutant p53 has a dual role in ageing or tissue damage, in promoting clonal expansion but also facilitating normal differentiation. Tissue damage and loss may trigger compensatory growth mechanisms, which unless supported by a differentiation capacity will not be functional. Positive selection for p53 mutations could promote functional clonal expansion, possibly in cooperation with alterations in WNT signalling–similarly to the effects of mutant p53 that we observed in gut organoids. Notable in this respect is the hypermethylation of the WNT inhibitors SFRP2 and SFRP1 that occurs in 73% of the normal oesophagus and 96% of the damaged Barrett’s oesophagus, respectively27. It is therefore possible that whereas germline TP53 mutations are detrimental and predispose to the early onset of multiple types of cancer, localized somatic TP53 mutations–especially at an increased age–could be beneficial as a protective means. This programmed reversed antagonistic pleiotropy28 may explain the plasticity of p53 mutations and why p53 is so prone to undergo mutations rather than ablation, as is typical of most other tumour-suppressor genes.
Methods
Data reporting
No statistical methods were used to predetermine sample size. Unless otherwise stated, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Mouse breeding and genotyping
All mice were of the species Mus musculus (C57BL/6J). Csnk1a1 fl/fl; Villin-Cre-ERT2 (CKIa fl/fl) and Csnk1a1 fl/fl;Trp53 fl/fl;Villin-Cre-ERT2 (CKIa fl/fl p53 fl/fl) mice were generated as previously described3. Mice with a germline knock-in Trp53 point mutation R172H or R270H were purchased from The Jackson Laboratory (B6.129S4(Cg)-Trp53 tm2.1Tyj/J or B6.129S4-Trp53 tm3.1Tyj/J, respectively)10,11. Mice were backcrossed for seven generations with C57BL/6 mice to generate a pure genetic background. CKIa fl/fl p53 R172H and CKIa fl/fl p53 R270H mice were generated by crossing mice with a knock-in Trp53R172H or Trp53 R270H mutation with CKIa fl/fl mice. Mice with a heterozygous germline nonsense mutation at codon 850 of Apc (Apc Min/+) were purchased from The Jackson Laboratory (C57BL/6J-Apc Min/J)5, and Apc Min/+ p53 R172H mice were generated by crossing mice with a knock-in Trp53 R172H mutation with Apc Min/+ mice. All mice were kept under specific-pathogen-free (SPF) conditions in the Hadassah Medical School animal facility unit of the Hebrew University. All experiments were performed in accordance with the guidelines of the Hebrew University’s ethics committee according to AAALAC standards. The study is in compliance with all relevant ethical regulations. To reduce the suffering of the mice, mice were euthanized and excluded from experiments if they lost 20% of their initial bodyweight during the progression of the experiment or 10% within 2–3 days. All mice were from the same genetic background (C57BL/6) and all experiments were performed on male or female mice of similar ages and in similar numbers, as our initial data did not indicate any notable differences between mouse genders. For mouse genotyping, DNA from the tail or ear of 4-week-old pups was extracted using standard protocols and the following primers were used: Csnk1a1 fl forward: TCCACAGTTAACCGTAATCGT, reverse: AACTGCAAATGAAAGCCCTG; Trp53 fl forward: CACAAAAAACAGGTTAAACCCAG, reverse: AGCACATAGG AGGCAGAGAC; Trp53 R172H forward: ACCTGTAGCTCCAGCACTGG, reverse: ACAAGCCGAGTAACGATCAGG; Trp53 R270H forward: AGCCTGCCTAGCTTCCTCAGG, reverse: CTTGGAGACATAGCCACACTG; Apc WT forward: GCCATCCCTTCACGTTAG, reverse: TTCCACTTTGGCATAAGGC; Apc Min forward: TTCTGAGAAAGACAGAAGTTA, reverse: TTCCACTTTGGCATAAGGC; Cre forward: CAAGCCTGGCTCGACGGCC, reverse: CGCGAACATCTTCAGGTTCT. For all experiments, sample size was chosen on the basis of previous experiments involving the same mouse models in our laboratory, and pilot studies. These data were also used to estimate the number of samples needed for conclusive and statistically significant results.
Tamoxifen administration, antibiotic and gallic acid treatments and tissue preparation
Mice were randomly separated from breeding cages 21 days after birth and allotted to different experimental groups. Knockout of CKIa and p53 was induced in 6–8-week-old mice by four subcutaneous tamoxifen injections (150 mg/kg) (Sigma-Aldrich) every alternate day and mice were euthanized afterwards. For antibiotic treatment, a combination of vancomycin (250 mg/l, BioAvenir), imipenem (250 mg/l, MSD) and metronidazole (250 mg/l, B. Braun) was administered in the drinking water, supplemented with sucrose (20 g/l, Sigma-Aldrich) for two weeks before the first tamoxifen injection and throughout the eight days of knockout induction for a total period of three weeks. Vehicle-treated mice were administered sucrose. For gallic acid treatment, gallic acid (1 g/l, Sigma-Aldrich) was administered in the drinking water for four weeks before the first tamoxifen injection and throughout the eight days of knockout induction for a total period of five weeks. Mice co-treated with gallic acid and antibiotics were subjected to treatment with gallic acid for two weeks, followed by treatment with gallic acid and a cocktail of antibiotics for an additional two weeks before the first tamoxifen injection and throughout the eight days of knockout induction for a total period of five weeks. Apc Min/+ p53 WT and Apc Min/+ p53 R172H mice were euthanized at 3 or 5 months of age. Representative gut images were positioned on a virtual background. For gallic acid treatment of Apc Min/+ mice, gallic acid (1 g/l) was continuously administered in the drinking water of four-week-old mice for three months. Tumour burden was evaluated by quantification of visible tumours in different regions of the gut and measurement of tumours size was done using a caliper (Fine Science Tools). The intestine, liver, spleen, kidney and lung were subjected to fixation in 4% formaldehyde and paraffin embedding in formalin-fixed paraffin-embedded (FFPE) blocks. The duodenum, jejunum, ileum and colon were flushed with ice-cold phosphate-buffered saline (PBS) and cut open longitudinally before the fixation. Intestinal epithelial cells from different gut sections were isolated as described previously3. For villus and crypt fractions, jejuna were shaken in 2 mM EDTA in PBS for 30 min, followed by a gentle shaking and centrifugation (villus fraction). The crypt fraction was prepared by a vigorous shaking of the remaining tissue, followed by 70-μm filtration and centrifugation.
Histology and immunohistochemistry
For H&E staining and IHC analysis, FFPE blocks were cut into 5-μm sections. For IHC, sections were blocked with normal horse or goat serum (Vector) for 1 h and then incubated overnight at 4 °C with the following antibodies in CAS-Block (Invitrogen): AXIN2 (1:200, Abcam), CD44 (1:200, eBioscience), CKIα (1:1,000, Abcam) Cyclin D1 (1:125, Thermo Fisher Scientific), Ki67 (1:200, Neomarkers), p21 (1:50, Santa Cruz Biotechnology), p53 (1:200, Novocastra), PROX1 (1:200, R&D Systems) and SOX9 (1:1,000, Abcam). Secondary antibodies were horseradish peroxidase (HRP) polymer-conjugated goat anti-rat, horse anti-rabbit, horse anti-goat (Vector) or anti-mouse (mouse-on-mouse; Nichirei). Diaminobenzidine (DAB) chromogen (Thermo Fisher Scientific) was used for detection and haematoxylin was used as a counterstain. Images were taken with a BX51 Olympus microscope. Measurement of the number and size of tumours and the degree of dysplasia in histology sections was performed blindly.
Intestinal crypt cultures (organoids)
Intestinal crypts from the jejunum or ileum of the different mouse genotypes were isolated on the basis of previously published methods4,29. To ascertain the preservation of jejunal and ileal properties of the respective organoid cultures, the levels of Lct and Fabp6 were measured, demonstrating higher levels of the former in jejunal organoids and higher levels of the latter in ileal organoids30. Crypts were collected as described and embedded onto Matrigel (BD Biosciences) at 50 μl per well in 24-well plates. The culture medium was Dulbecco’s modified Eagle medium (DMEM)/F12 (Gibco), containing GlutaMAX (1:100, Gibco) and penicillin–streptomycin (1:100, Biological Industries). The medium was supplemented with B27 (1:50, Gibco), mouse Noggin (100 ng/ml, Peprotech), mouse EGF (20 ng/ml, Peprotech), human basic FGF (10 ng/ml, Peprotech) and human R-spondin-1 (500 ng /ml, Peprotech). For Apc Min/Min organoids (derived from adenomas after Apc loss-of-heterozygosity in Apc Min/+ mice) the medium was DMEM/F12 supplemented with mouse EGF and B27 alone. Organoid cultures were split 1:4 and embedded onto new Matrigel every five to seven days. Knockout induction in the crypt cultures was achieved by incubation with 300 nM 4-hydroxytamoxifen (4OHT; Sigma-Aldrich) for 72 h. Selection for organoids with an active WNT–β-catenin pathway was done by splitting the 4OHT-induced organoids and culturing them without R-spondin-1 in the culture medium. For the screen of bacterial metabolites, organoids were first treated with several increasing concentrations of the different bacterial metabolites in the medium, and then for each metabolite the maximum non-toxic concentration was chosen: butyric acid (100 μM, Sigma-Aldrich), caproic acid (100 μM, Sigma-Aldrich), sulforaphane (500 nM, Sigma-Aldrich), sodium deoxy-cholate (50 μM, Sigma-Aldrich), ellagic acid (50 μM, Sigma-Aldrich), gallic acid (50 μM, Sigma-Aldrich) and urolithin B (50 μM, Sigma-Aldrich). Organoids were also screened and treated with propionic acid (100 μM, Sigma-Aldrich), acetic acid (100 μM, Sigma-Aldrich) and sodium litho-cholate (50 μM, Sigma-Aldrich); all of these three had no effect and therefore are not included. To test whether the effect of gallic acid on organoids is reversible, organoids were either grown with gallic acid for nine days continuously, or five days with and then four days without gallic acid.
Histology, immunofluorescent staining and imaging of intestinal crypt organoids
Organoids were cultured on round cover slips in 24-well plates. Fixation was performed with 4% paraformaldehyde for 15 min, and this was followed by permeabilization with 0.5% Triton X-100 in PBS for 30 min. Blocking was performed with CAS-Block (Invitrogen) for 30 min and then organoids were incubated with the following primary antibodies for 3 h: AXIN2 (1:200, Abcam), CD44 (1:200, eBioscience), Cyclin D1 (1:125, Thermo Fisher Scientific), GFP (1:500, Abcam), Ki67 (1:200, Neomarkers), p53 (1:200, Novocastra), p53 (1:1,000, Cell Signaling Technology) and SOX9 (1:1,000, Abcam). Secondary antibodies, Alexa Fluor 488-conjugated donkey anti-mouse and donkey anti-rabbit and Alexa Fluor 647-conjugated donkey anti-goat and goat anti-rat (1:1,000, Thermo Fisher Scientific), were applied for 3 h. Hoechst (1 mg/ml, Molecular Probes) was used as a counterstain for nuclei. The orga-noids were mounted in mounting medium (Immco Diagnostics) and imaged with a Olympus IX81 confocal microscope. For H&E staining, Matrigel-embedded organoids from 6 wells of a 24-well plate were fixed with 100 μl plasma and 50 μl thrombin (20 IU/ml, Siemens), followed by overnight incubation in 4% formaldehyde (Bio-Lab). Fixed organoids were embedded in paraffin and blocks were cut into 5-μm sections and stained with H&E. Bright-field images of infected organoids were taken using a fluorescent microscope (Observer Z1, Zeiss).
Virus preparation
Authenticated 293T cells purchased from ATCC (CRL-3216) were grown in DMEM medium (Gibco) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific), glutamine (1:100, Biological Industries) and penicillin–streptomycin (1:100, Biological Industries). Cells were routinely tested for mycoplasma contamination and were negative. The 293T cells were transfected with lentiviral vectors expressing either GFP, p53R175H-GFP or p53R270H-GFP, using a TransIT-LT1 transfection reagent (MirusBio). Virus-containing medium was collected three days after the infection and concentrated (30×) by ultracentrifugation (Beckman Coulter Diagnostics, 18 h, 70,000 RCF) and virus pellets were resuspended in 500 μl organoid infection medium.
Organoid infection and quantification of outpocket budding
Freshly split CKIa/p53 DKO and Apc Min/Min organoids were treated with 80 ng WNT3A (Peprotech) and 10 mM nicotinamide (Sigma-Aldrich) for three days before infection. On the day of infection, organoids were incubated in 200 μl of concentrated virus in infection medium (DMEM/F12 supplemented with WNT3A (80 ng/ml, Peprotech), 10 mM nicotinamide, B27 (1:50, Gibco), mouse Noggin (100 ng/ml, Peprotech), mouse EGF (20 ng/ml, Peprotech), human basic FGF (10 ng/ml, Peprotech) human R-spondin-1 (500 ng/ml, Peprotech), ROCK inhibitor (Sigma-Aldrich) and polybrene (8 μg /ml, Sigma-Aldrich) for 4 h. Infected organoids were collected, Matrigel-embedded and grown in DMEM/F12 medium for five days. Immunofluorescent double staining for p53–GFP and Ki67–GFP was performed and the percentage of organoids with outpocket budding was calculated out of the total GFP-expressing population of organoids. Two independent orga-noid infection cultures were analysed in Fig. 3b. Bright-field and GFP imaging were performed using a fluorescent microscope (Observer Z1, Zeiss).
Immunoblot analysis
Whole-cell protein extracts were prepared from intestinal organoids and enterocytes isolated from jejunal and ileal epithelia using lysis buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA and 0.5% NP40) containing protease inhibitor cocktail (1:200, Calbiochem), PMSF (1 mM, Sigma-Aldrich) and phosphatase inhibitors: β-glycerophosphate (20 mM, Sigma-Aldrich), PNPP (20 mM, Sigma-Aldrich) and okadaic acid (100 nM, Calbiochem). For preparation of the cytoplasmic and nuclear protein fractions, enterocyte pellets were incubated in hypotonic buffer (10 mM HEPES, 150 μM spermine, 750 μM spermidine, 100 μM EDTA, 100 μM EGTA, 2 mM DTT and 10 mM KCl with protease and phosphatase inhibitors). After centrifugation, the cytoplasmic fraction (supernatant) was collected and the nuclear pellet was suspended in nuclear extraction buffer (20 mM HEPES, 150 μM spermine, 750 μM spermi-dine, 200 μM EDTA, 2 mM EGTA, 500 μM DTT, 42 mM NaCl and 25% (v/v) glycerol with protease and phosphatase inhibitors) and sonicated (VC750, Sonics). The nuclear fraction (supernatant) was collected after centrifugation. Immunoblot analysis was performed using standard techniques. Blots were incubated with the following primary antibodies in 3% BSA in TBST: AXIN2 (1:1,000, Abcam), BAX (1:1,000, Cell Signaling Technology), β-catenin (1:750, BD Transduction), non-phospho (active) β-catenin (1:1,000, Cell Signaling Technology), CKIα (1:1,000, Santa Cruz Biotechnology), Cyclin D1 (1:500, Thermo Fisher Scientific), fibrillarin (1:500, Abcam), GAPDH (1:10,000, Merck), Myc (1:1,000, Cell Signaling Technology), p21 (1:200, Santa Cruz Biotechnology), p53 (1:1,000, Novocastra), PP2Ac (1:1,000, Merck), PROX1 (1:200, R&D Systems), TCF4 (1:1,000, Abcam) and tubulin (1:5,000, Sigma-Aldrich). Secondary antibodies were HRP-linked goat anti-mouse, goat anti-rabbit and rabbit anti-goat (1:10,000; Jackson). Blots were developed using enhanced chemiluminescence (GE Healthcare).
RNA expression analysis
Total RNA was extracted from cell pellets using the miRNeasy extraction kit (Qiagen) and subjected to reverse transcription using M-MLV RT (Invitrogen). For quantification of miR34a, 0.5 μg RNA was poly-A-polymerized (Ambion) before reverse transcription. mRNA expression levels were measured by RT–qPCR using SYBR Green (Invitrogen) in a QuantStudio 12K Flex Real-Time PCR system (ABI). Relative quantities of gene transcripts were analysed in ExpressionSuite v.1.1 software and normalized to the Ubc (mRNA) or U6 (Mir34a) transcripts. The sequences of RT–qPCR primers are as follows: 16S forward: AGTGTTTGATCCTGGCTC, reverse: TGCTGCCTCCCGTAGGAGT; Axin2 forward: TAGGCGGAATGAAGATGGAC, reverse: CTGGTCACCCAACAAGGAGT; Bax forward: ATGCGTCCACCAAGAAGCTGA, reverse: AGCAATCATCCTCTGCAGCTCC; Ccnd1 forward: TTGACTGCCGAGAAGTTGTG, reverse: CCACTTGAGCTTGTTCACCA; Ccng1 forward: GCTGGCGCTATCTATCCTTG, reverse: GGTCAAATCTCGGCCACTTA; Csnk1a1 forward: GGTGCGGAAGATCGGATCT, reverse: TTCACTGCCACTTCCTGCC; Ephb2 forward: TGGCAAGTTCAGTGGCAAGA, reverse: GGCCACCAGAGACACAACAA; Fabp6 forward: TTCTTCCTTAGCTGCTCGC, reverse: CTGCTGGACCTCTGTGATG; Lct forward: AGATCCAGGGCACCTTTGAC, reverse: GAAGCAACTCCCCTATCGGC; Mdm2 forward: TGTGTGAGCTGAGGGAGATG, reverse: CACTTACGCCATCGTCAAGA; Mir34a forward: TGGCAGTGTCTTAGCTGGTTGTT, reverse: GCGAGCACAGAATTAATACGAC ; Myc forward: TGAGCCCCTAGTGCTGCAT, reverse: AGCCCGACTCCGACCTCTT; p21 forward: TCCACAGCGATATCC AGACA, reverse: AGACAACGGCACACTTTGCT; Puma (Bbc3) forward: CAAGAAGAGCAGCATCGACA, reverse: TAGTTGGGCTCCATTTCTGG; Sox9 forward: GGAGCTAFGCAAGACTCTGG, reverse: TGTAATCGGGGTGGTCTTTCT; Trp53 forward: TGAAACGCCGACCTATCCTTA, reverse: GGCACAAACACGAACCTCAAA; U6 forward: GATGACACGCAAATTCGTGAA, reverse: GCGAGCACAGAATTAATACGAC; Ubc forward: CAGCCGTATATCTTCCCAGAC, reverse: CTCAGAGGGATGCCAGTAATC.
RNA sequencing
For quality control of RNA extraction yield and library synthesis products, the RNA ScreenTape kit (Agilent Technologies), D1000 Screen-Tape kit (Agilent Technologies), Qubit RNA HS Assay kit (Invitrogen) and Qubit DNA HS Assay kit (Invitrogen) were used for each specific purpose. For mRNA library preparation, the KAPA Stranded mRNA-seq kit with mRNA capture beads (Kapa Biosystems) was used. In brief, 1 μg was used for the library construction, and the library was eluted in 20 μl of elution buffer. Libraries were adjusted to 10 mM, then 10 μl from each sample was collected and pooled in one tube. A pool of multiplex samples (1.5 pM including PhiX 1.5%) was loaded into the NextSeq 500/550 High Output v2 kit (75 cycles) cartridge (Illumina) and loaded on a NextSeq 500 System (Illumina), with 75 cycles and single-read-sequencing conditions. Obtained sequencing data were aligned to the Ensembl Mus musculus reference genome, revision 92 (mm10), and read counts per gene were normalized to sequencing depth and gene length as fragments per kilobase per million reads mapped (FPKM). Differential expression was assessed using DESeq231. Genes were considered differentially expressed if they had an FPKM > 1 in at least one sample and showed an absolute log-transformed fold change ≥ 1 together with a false discovery rate (FDR) > 0.05 (Benjamini– Hochberg-corrected P value). Gene sets for the over-representation analysis were obtained from the Ingenuity database and the Molecular Signatures Database (MsigDB; version 5.0, collections H, C2, C5, C6 and C7). The significance of over-representation was assessed using a hypergeometric test and corrected for multiple hypothesis testing using the Benjamini–Hochberg correction. RNA-sequencing (RNA-seq) results and gene expression data are available at Array Express under E-MTAB-7859.
Chromatin immunoprecipitation
Freshly extracted enterocytes were fixed and cross-linked using 1% paraformaldehyde solution for 10 min and neutralized with 125 mM glycine for 5 min. Cells were resuspended and lysed in RIPA buffer containing 0.5% SDS. To solubilize and shear cross-linked DNA molecules, lysates were sonicated with a Covaris M220 Sonicator at 75W with 20% duty factor and 200 cycles for 20 min. Immunobeads were prepared by incubation of 80 μL Dynabeads Protein G magnetic beads (Life Technologies) for 6 h with 2 μg of either anti-TCF4 (Millipore) or anti-H3K4me3 (Active Motif) antibodies. For p53 ChIP 80 μl packed antibody-conjugated agarose beads (Santa Cruz Biotechnology) were used. Sonicated lysates were centrifuged at 13,000 rpm for 5 min and the chromatin was incubated overnight at 4 °C with the anti-TCF4, anti-H3K4me3 or anti-p53 immune-beads. After ChIP, samples were washed and bound complexes were eluted by Direct Elution Buffer (10 mM Tris-HCl pH 8, 0.3 M NaCl, 5 mM EDTA and 0.5% SDS), followed by treatment with RNase A (Thermo Fisher Scientific) and Proteinase K (Invitrogen). Chromatin was purified with the QIAquick Gel Extraction Kit (Qiagen). For p53 ChIP–seq, libraries were prepared from purified chromatin using the Microplex Library Preparation Kit (Diagenode) according to the manufacturer’s instructions. The quality of sequencing libraries was controlled using a Bioanalyzer 2100 with the Agilent High Sensitivity DNA Kit (Agilent). Pooled sequencing libraries were quantified with digital PCR (QuantStudio 3D, Thermo Fisher Scientific) and sequenced on an Illumina HiSeq 1500 platform in rapid-run mode with 50-base single reads. For TCF4 and H3K4me3, ChIP products were tested by qPCR using SYBR Green (Invitrogen) in a QuantStudio 12K Flex Real-Time PCR system (ABI). Enrichment values were calculated compared to the Cryaa gene and were analysed in ExpressionSuite 1.1 software with the following primers: TCF4 ChIP: Axin2 forward: CTCCAG TAGAACATGGGGGAC, reverse: GCCCAGGGCAAAGTAATCC; Ccnd1 forward: GAAGGGTTTGGGCTTGC, reverse: CCTCTGGAGGCTGCAGG; Cd44 forward: GAATTGCAATTTTACATACGA, reverse: CTGTTCTTTCTTGT TTACATTTTAT; Cryaa forward: CCATCAGCTCATGACCCATA, reverse: CTTCAGGACATCCCAACCC; Ephb2 forward: GGGAGAGACCACTGCTGC, reverse: GGTTGGACTGCCAATCCTG; Myc forward: CAGCCGTATATCTTCCCAGAC, reverse: CTCAGAGGGATGCCAGTAATC; Ubc forward: GGAAACTGGGAAATTAATGT, reverse: GGACAAACCCAAGCTTTC; Sox9 forward: GAGGAAAACGAGCTTTTCAA, reverse: CATT-TAAACATCCCCTTTCTG. H3K4me3 ChIP: Axin2 forward: ACAGCCCAAGAACCGG, reverse: TTCTTAGAAACGGCTGCGA; Ccnd1 forward: GGTGTCAGGGTACGCGC, reverse: GGAGACCGGCAGTACAGC; Cd44 forward: CACTACAGTCCCTCCCCAG, reverse: TCCGTTTCATCCAGCACG; Cr yaa forward: CCATCAGCTCATGACCCATA, reverse: CTTCAGGACATCCCAACCC; Ephb2 forward: GGTGCAACCCGTGGG, reverse: CGCACACACACACGAGC; Myc forward: GGAAAAAAAAATAGAGAGAGG TGGG, reverse: GAGTGAGGCGAGTCGGAC; Sox9 forward: CTCAAGAGCTAGCCGTGATTG, reverse: GGACTGAAACTGGTAAAGTTGTCG;
ChIP–seq data analysis
Reads were aligned to the Ensembl Mus musculus reference genome, revision 92 (mm10), using Bowtie2 (v.2.0.0) using the default parameter settings32. The expected number of duplicates based on sequencing depth was estimated using a binomial distribution and lanes were de-duplicated to this estimate, keeping only these effective reads for further analysis. Peak calling was performed using MACS2 (v.2.1.0) with default parameters for all samples. To additionally reduce the number of false positive peaks, obtained peak lists were filtered to those peaks that showed at least threefold enrichment over input and a minimum of 50 effective foreground reads. To enable comparisons between samples, peak signals were normalized to the sequencing depth and expressed as counts per million (CPM). A heat map was produced showing the CPM signal in a 2,000-base-pair window centred on the peak summits for all filtered peaks obtained for the CKIa Δgut sample. Three ‘hyper-ChIPable’ regions contained in all samples including negative controls were considered artefacts and subsequently removed. CPMs were normalized to the 90th quantile per lane for a set of 166 peaks found in CKIa Δgut mice and ordered according to signal strength in descending order. ChIP–seq results and enrichment data are available at Array Express under E-MTAB-7858.
Average nuclear size and dysplasia score
The dysplasia score was calculated on the basis of five criteria: the number of high-grade dysplasia foci; nuclear size; nuclear morphology; deviation of nuclei from the villus axis; and number of nucleoli in a nucleus. In each group, a total of 30 villi were microscopically scanned: 5 villi per field, 2 fields per mouse, 3 mice. For each villus, high-grade dysplasia foci were counted, and 10 nuclei were examined in terms of size, shape, number of nucleoli and nucleus deviation from the villus axis. The average nuclear size was calculated for each field. Each criterion was given a rank ranging from 0 (normal, as seen in CKIa fl/fl mice) to 3 (highly dysplastic, as seen in CKIa Δgut p53 Δgut mice) and the total dysplasia score was calculated by summing up the 5 ranks of all mentioned criteria. Measurements were done with a BX51 Olympus microscope using a 60× objective lens.
Extraction of DNA from stool and gut mucosa, PCR of SDH-coding genes and sequencing of 16S rRNA
Stool samples (around 100 mg) and mucosa pieces were freshly collected. For antibiotic-treated mice, samples were collected after two weeks of treatment. DNA was extracted using the QIAamp Fast DNA stool Mini kit (Qiagen). Relative copy numbers of SDH-coding genes were determined using qPCR with the following primers: aroC2 forward: TTAAGCGCCTTACTAGTATGGTCG, reverse: AAACAAGAGCCGAACACTTCAC; aroE forward: ATGAAACGAGATAAGGTGCAGAC, reverse: TGCTGCTTTGCTCAATGTG. For 16S rRNA sequencing, the V3 and V4 variable regions of the 16S gene were amplified using the following primer sequences: forward: CTACGGGNGGCWGCAG, reverse: GACTAC HVGGGTATCTAATCC. PCR products were cleaned from primers using AMPure XP beads, barcoded and cleaned again with AMPure XP beads. Concentrations were normalized to 10 nM and samples were pooled and paired-end reads (2 × 250 bp) were sequenced on a MiSeq platform (Illumina). The taxonomic composition of the samples was explored using the QIIME2 v.2019.4. pipeline33 implemented by the workflow platform NeatSeq-Flow34. In brief, the feature table and 16S sequences were generated by implementing the dada2 package35 using only the forward (R1) reads of each sequenced amplicon. A rooted phylogenetic tree was generated using the ‘align-to-tree-mafft-fasttree’ pipeline. The phylogenetic tree was used for alpha and beta diversity analyses using ‘core-metrics-phylogenetic’, ‘alpha-rarefaction’ (sampling depth was 5,000) and ‘alpha-group-significance’ methods. Taxonomic assignment of the sequences to deduce the composition of the samples was performed using a pre-trained naive Bayes classifier and the q2-feature-classifier plug-in. Differential abundance testing was performed using ANCOM36 and Gnesis37. An alpha group significance test and ANCOM and Gnesis differential abundance testing showed that there are no significant differences in taxonomic composition between the two bacterial populations
High-performance liquid chromatography–mass spectrometry
For jejunal and ileal samples, 5–7-cm segments from the jejunum and ileum of four non-treated and four gallic-acid-treated mice were taken, cleaned of stool and weighed. Samples were chopped, homogenized (PT 2100, Polytron) and centrifuged, and supernatants collected from every two mice of respective treatment groups were pooled. Samples were treated with 1% (v/v) formic acid to improve the recovery of gallic acid38, and methanol (4:1) was added for protein precipitation. Samples were vigorously vortexed and centrifuged, and supernatants were dried completely using a vacuum concentrator (Speed-Vac). Samples were resuspended in ultra-pure water and the concentration of gallic acid was quantified using a Quantum Access MAX Triple-Stage Quadrupole mass spectrometer.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 8 software. For all in vivo and ex vivo experiments, an unpaired one-tailed Student’s t-test was applied for comparisons of two groups. A one-way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis was applied when comparing three or more groups. A Kruskal– Wallis one-way analysis followed by Conover’s test was used to compare relative copy numbers of SDH genes in different segments of the gut. Data in graphs are presented as mean ± s.e.m. The threshold of significance was 0.05 for both P and q values. Exact P values, sample sizes (n) and the statistical test used are provided in the figures and the corresponding figure legends. For ex vivo organoid experiments, technical triplicates as well as independent experiments were performed on separate days to ensure reproducibility. For in vivo experiments, biological replicates as well as independent cohorts of mice were used as stated in the respective figure legends. RNA-seq and ChIP–seq experiments were performed with biological replicates. Biological and technical replicates from independent experiments were used for all other data. All technically sound replication attempts were successful.
Extended Data
Extended Data Fig. 1. Mutant p53 counteracts dysplasia and proliferation in the proximal mouse gut.
a, IHC of p53 in the jejunum, ileum and colon of p53 WT and p53 R172H mice before and after CKIa deletion, and in CKIa Δgut p53 Δgut mice. Scale bars, 100 μm. b, H&E-stained sections of different mouse gut segments. Inserts show high-magnification images of duodenal and jejunal villi, demonstrating the grade of dysplasia. Scale bars, 100 μm. c, IHC of PROX1 and Ki67 in mouse colon. Scale bars, 100 μm. d, Jejunal crypt length. Overall average of the mean values for each mouse ± s.e.m (n, number of crypts pooled from three mice), one-way ANOVA with Tukey’s test. Representative data from six independent experiments (a–c).
Extended Data Fig. 2. Mutant p53 exerts oncogenic GOF at the distal gut and tumour-suppressive effects at the proximal gut.
a, H&E-stained sections and IHC of CKIα and p53 in mouse ileum and colon, three days after knockout induction. Inserts (H&E) show high-magnification images of colon and ileum, demonstrating the grade of dysplasia. Scale bars, 100 μm. b, Colon thickness of mice described in (a). Overall average of the mean values for each mouse ± s.e.m (n, number of colon thickness measurements), one-sided Student’s t-test. c, H&E-stained sections and IHC of p53, PROX1 and Ki67 in indicated mouse gut segments. Inserts (H&E) show high-magnification images of jejunal villi, demonstrating the grade of dysplasia. Scale bars, 100 μm. d, RT–qPCR of p53 targets in mouse jejunal enterocytes indicating that mutant p53 has not regained wild-type transcriptional activity. Mean ± s.e.m. (n, number of mice) relative to CKIa fl/fl (normalized to 1), one-way ANOVA with Tukey’s test. e, IHC of p21 in indicated mouse gut segments. Scale bars, 100 μm. Representative data from two (a) or six (c, e) independent experiments.
Extended Data Fig. 3. Mutant p53 counteracts tumorigenesis in the proximal mouse gut.
a, Representative images of different mouse bowel segments. Arrowheads indicate visible tumours. b, Tumour size (each mouse is colour-coded). Overall average of the mean values for each mouse ± s.e.m (n, number of mice), one-sided Student’s t-test. c, Quantification of visible tumours per mouse. Mean ± s.e.m. (n, number of mice), one-sided Student’s t-test. Representative data from three independent experiments (a).
Extended Data Fig. 4. Mutant p53 suppresses the expression of WNT targets exclusively in the proximal gut.
a, IHC of WNT targets in mouse jejunum. Scale bars, 100 μm. b, c, RT–qPCR of WNT targets in mouse jejunal (b) and ileal (c) enterocytes. Mean ± s.e.m. (n, number of mice) relative to CKIa fl/fl (normalized to 1), one-way ANOVA with Tukey’s test. d, e, IHC of WNT targets in mouse ileum (d) and colon (e). Scale bars, 100 μm. f, g, Over-representation analysis of enriched gene sets differentially expressed in jejunum (f) and ileum (g) of CKIa Δgut p53 R172H versus CKIa Δgut p53 Δgut mice; negative logarithm of Benjamini–Hochberg-corrected P values (data derived from three mice of each group). Red lines indicate the significance threshold for α = 0.05. Representative data from four independent experiments (a, d, e).
Extended Data Fig. 5. Suppression of WNT target gene expression by mutant p53 is mediated by preventing the association of TCF4 with WNT promoters.
a, Immunoblot of mouse jejunal enterocytes. PP2Ac, loading control. For gel source data, see Supplementary Fig. 1. b, Immunoblot of cytoplasmic and nuclear extracts from mouse jejunal enterocytes. GAPDH and fibrillarin, cytoplasmic and nuclear markers, respectively. For gel source data, see Supplementary Fig. 1. c, d, TCF4 (c) and H3K4me3 (d) ChIP of WNT target promoters in mouse jejunal enterocytes. Mean enrichment ± s.e.m. (n, number of mice), one-way ANOVA with Tukey’s test. e, f, TCF4 (e) and H3K4me3 (f) ChIP of WNT target promoters in mouse ileal enterocytes. Mean enrichment ± s.e.m. (n, number of mice), one-sided Student’s t-test. g, H3K4me3 ChIP of WNT target promoters in crypt and villus fractions of mouse jejunum. Mean enrichment ± s.e.m. (n, number of mice), one-sided Student’s t-test. Representative data from three independent experiments (a, b).
Extended Data Fig. 6. Mutant p53 promotes balanced growth and differentiation of intestinal organoids.
a, RT–qPCR of Csnk1a1 and Trp53 in jejunal organoids. Mean ± s.e.m. (n, number of mice that were used as a source for organoid cultures) relative to CKIa fl/fl (normalized to 1), one-way ANOVA with Tukey’s test. b, e, Immunofluorescent staining of p53 (b) and H&E (e) of jejunal organoids; different levels of dysplasia are evident in e. Nuclear counterstain (immunofluorescence), Hoechst (blue). Scale bars, 100 μm (b) and 50 μm (e). c, d, Bright-field imaging of jejunal (c) and ileal (d) organoids that express p53 WT or p53 R172H, with or without CKIa knockout induction, and of CKIa/p53 DKO organoids. Inserts show a high-magnification image of a representative organoid. Scale bars, 500 μm. f, RT–qPCR of p53 targets in jejunal organoids. Mean ± s.e.m. (n, number of independent experiments performed with organoid cultures of two different mice), relative to CKIa fl/fl (normalized to 1), one-way ANOVA with Tukey’s test. g, Immunofluorescent staining of Ki67 and WNT targets in jejunal organoids. Nuclear counterstain, Hoechst (blue). Scale bars, 100 μm. h, Bright-field imaging and merged immunofluorescence of GFP with p53 or Ki67 in jejunal CKIa/p53 DKO organoids transduced with the indicated lentiviruses. Inserts (bright-field) show a GFP and bright-field merged image of a representative organoid. Nuclear counterstain (immunofluorescence), Hoechst (blue). Scale bars, 500 μm (top); 100 μm (bottom). i, Immunoblot of wild-type and Apc Min/Min organoids. Tubulin, loading control. For gel source data, see Supplementary Fig. 1. Representative data from two or three (b, g–i) or five (c–e) independent experiments.
Extended Data Fig. 7. Treatment with antibiotics unleashes tumour-suppressive effects of mutant p53 in the distal gut.
a, RT–qPCR of the 16S subunit of microbial rRNA in the ileum of vehicle- or antibiotic-treated mice. Mean ± s.e.m. (n, number of mice) relative to vehicle-treated CKIa fl/fl (normalized to 1), one-sided Student’s t-test. The y axis is represented as log10-transformed expression. b, Length of crypts in mouse ileum. Overall average of the mean values for each mouse ± s.e.m (n, number of crypts), one-sided Student’s t-test. c, d, g, H&E-stained sections (c) and IHC of Ki67 (d) and WNT targets (g) in mouse colon. Scale bars, 100 μm. e, IHC of WNT targets in mouse ileum. Scale bars, 100 μm. f, RT–qPCR of WNT targets in ileal enterocytes from vehicle- or antibiotic-treated mice. Mean ± s.e.m. (n, number of mice) relative to vehicle-treated CKIa fl/fl mice (normalized to 1), one-sided Student’s t-test. Representative data from four independent experiments (c, d, e, g).
Extended Data Fig. 8. Mutant p53 induces WNT suppression and differentiation of intestinal tumour organoids, an effect which is reversibly blocked by gallic acid treatment.
a, Bright-field and immunofluorescent imaging of WNT targets and Ki67 in CKIa KO p53 R172H jejunal organoids treated with different bacterial metabolites. Nuclear counterstain (immunofluorescence), Hoechst (blue). Inserts (bright-field) show a high-magnification image of a representative organoid. Scale bars, 500 μm (bright-field); 100 μm (immunofluorescence). b, RT–qPCR of WNT targets in jejunal organoids. Mean ± s.e.m. (n, number of independent experiments performed with organoid cultures from two different mice) relative to CKIa fl/fl (normalized to 1), one-way ANOVA with Tukey’s test. c, Bright-field imaging of non-treated and gallic-acid-treated jejunal organoids. Inserts show a high-magnification image of a representative organoid. Scale bar, 500 μm. d, Merged immunofluorescence of GFP and Ki67 in non-treated and gallic-acid-treated jejunal CKIa/p53 DKO and Apc Min/Min organoids transduced with the indicated lentiviruses; three representative organoid fields. Expression of p53R175H-GFP in CKIa/p53 DKO organoids and p53R273H-GFP in Apc Min/Min organoids produced a similar effect. Nuclear counterstain, Hoechst (blue). Scale bars, 100 μm. e, Bright-field and immunofluorescent imaging of WNT targets and Ki67 in CKIa KO p53 R172H jejunal organoids grown with gallic acid for 9 days continuously, or 5 days with and then 4 days without gallic acid. Nuclear counterstain (immunofluorescence), Hoechst (blue). Inserts (bright-field) show a high-magnification image of a representative organoid. Scale bars, 500 μm (top); 100 μm (bottom). Representative data: a, c-e from two to three independent experiments.
Extended Data Fig. 9. Gallic acid reverses mutant-p53-induced WNT suppression and promotes dysplasia and tumorigenesis across the entire gut.
a, Levels of gallic acid in the jejunum and ileum of gallic-acid-treated and non-treated CKIa Δgut p53 R172H mice. Mean ± s.e.m. (n, number of samples, each pooled from two mice), one-way ANOVA with Tukey’s test. b, qPCR of SDH-coding genes and 16S rRNA genes from mouse stool. Mean ± s.e.m. (n, number of mice from which stool was collected), relative to CKIa fl/fl (normalized to 1), one-way ANOVA with Tukey’s test. c, qPCR of SDH-coding genes from the mucosa of different segments of the mouse gut. Mean ± s.e.m. (n, number of mice) relative to CKIa fl/fl jejunum (normalized to 1), Kruskal–Wallis with Conover’s test. The y axis is represented as a power of 10. d, H&E-stained sections of mouse jejunum. Inserts show high-magnification images of jejunal villi, demonstrating the grade of dysplasia. Scale bar, 100 μm. e, Average nuclear size in mouse jejunal villi. Overall average of the mean values for each mouse ± s.e.m (n, number of microscope fields), one-sided Student’s t-test. f, Dysplasia score (see Methods) of mouse jejunal villi. Overall average of the mean values for each mouse ± s.e.m (n, number of villi), one-sided Student’s t-test. g, h, Representative images of different segments of the mouse bowel. Arrowheads indicate visible tumours. i, Quantification of visible tumours per mouse. Mean ± s.e.m. (n, number of mice), one-sided Student’s t-test. j, Tumour size (each mouse is colour-coded). Overall average of the mean values for each mouse ± s.e.m (n, number of mice), one-way ANOVA with Tukey’s test. k, H3K4me3 ChIP of WNT target promoters in mouse jejunal enterocytes. Mean enrichment ± s.e.m. (n, number of mice), one-sided Student’s t-test. l, RT–qPCR of WNT targets in mouse jejunal enterocytes. Mean ± s.e.m. (n, number of mice) relative to non-treated CKIa fl/fl (normalized to 1), one-sided Student’s t-test. m, IHC of WNT targets in mouse jejunum. Scale bars, 100 μm. n, o, Immunoblot of mouse jejunal enterocytes. PP2Ac, loading control. For gel source data, see Supplementary Fig. 1. Representative data from two or three independent experiments (d, g, h, m–o).
Extended Data Fig. 10. Gallic acid reverses the effect of antibiotic treatment in the distal gut of CKIa Δgut p53 R172H mice.
a, b, H&E-stained sections of mouse ileum (a) and colon (b). Scale bars, 100 μm. c, IHC of Ki67 in mouse ileum and colon. Scale bars, 100 μm. d, RT–qPCR of WNT targets in mouse ileal enterocytes. Mean ± s.e.m. (n, number of mice) relative to antibiotic-treated CKIa fl/fl (normalized to 1), one-sided Student’s t-test. e, f, IHC of WNT targets in mouse ileum (e) and colon (f). Scale bars, 100 μm. Representative data from two or three independent experiments (a–c, e, f).
Supplementary Material
Acknowledgements
We thank N. Cohen-Saban and N. Amsalem for assistance with mouse models; M. Biton for project discussions in the initial phase of this study; the Mass Spectrometry Unit of the Institute for Drug Research, School of Pharmacy, The Hebrew University of Jerusalem (HUJI) for MS analysis; and the Genomic Applications Laboratory, Core Research Facility and Faculty of Medicine at HUJI and I. Plaschkes (Bioinformatics Unit, The Robert H. Smith Faculty of Agriculture, Food and Environment at HUJI) for assistance with 16S rRNA sequence analysis. This work was supported by the Israel Science Foundation (ISF) Centers of Excellence (2084/15) to Y.B.-N., M.O. and E.P., the ISF (3165/19) within the Israel Precision Medicine Program to Y.B.-N., the European Research Council within the FP-7 to Y.B.-N. (294390 PICHO) and E.P (281738 LIVERMICROENV) and the Israel Cancer Research Fund Professorship to Y.B.-N.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author contributions E.K., I.S.-A. and Y.B.-N. planned the study and Y.B-N. supervised the research. E.K. prepared the mouse models, performed the molecular, cellular and mouse studies and analysed the data related to Figs. 1a–f, 2a–d, 3a–c, 4a–c, 5a–f and Extended Data Figs. 1a–d, 2a–e, 3a–c, 4a–g, 5a–g, 6a–i, 7a–g, 8a–e, 9a–o, 10a–f; I.S.-A. contributed to Fig. 3b, c and Extended Data Figs. 6h, i, 8d, 9a and independently reproduced similar data to Figs. 1a, 2b, 5f and Extended Data Figs. 5a, b, 9n, o; A.V. contributed to Fig. 4a, Extended Data Figs. 7c, 9a, d, 10a, b and Supplementary Table 1 and independently reproduced similar data to Fig. 5f; S.M. contributed to Figs. 1e, 5a and Extended Data Figs. 1b, 2a, c, e, 3a, 6b, 10c, e, f and independently reproduced similar data to Figs. 2b, 3a, 4b and Extended Data Figs. 6g, 7d, e, g, 8b, 9m; A.L. contributed to Figs. 1d, 4a and Extended Data Figs. 1b, 2c, 7c; E.E. generated the original CKIa fl/fl mouse model of CKIa Δgut and CKIa Δgut p53 Δgut, contributed to Extended Data Fig. 1b and independently reproduced similar data to Fig. 1a and Extended Data Fig. 1a; A.Z. contributed to Fig. 4a and Extended Data Figs. 7c, 9d; M.S. contributed to Extended Data Fig. 9a and independently reproduced similar data to Figs. 1a, 2b, 5f and Extended Data Figs. 5a, b, 9n, o; G.V. contributed to Extended Data Fig. 9b, c; M.M. and T.S. contributed to Figs. 1d, 5e, Extended Data Fig. 4f, g and Supplementary Table 1; and T.S., E.P. and M.O. provided discussions and advice on the study. E.K., I.S.-A., A. V., T.S., E.P., M.O. and Y.B.-N. wrote the paper.
Competing interests A US provisional patent application (no. 62/987,058) entitled ‘A method for the diagnosis and treatment of cancer’ was filed on 9 March 2020. The inventors are Y.B.-N., E.K. and I.S.-A. The invention is based on the finding that gut microbiota, in particular polyphenol-producing microbiota, promote colorectal cancer and have a pro-tumorigenic effect both in vitro and in vivo. The invention concerns a method for the prevention and therapy of cancer in patients who have a tumour with an oncogenic p53 mutation. The method involves therapy with agents that reduce the levels of polyphenols, in particular the levels of gallic acid in the gut.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-2541-0.
Peer review information Nature thanks Christian Jobin, Carol Prives and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at http://www.nature.com/reprints.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The source gels for immunoblots are provided in Supplementary Fig. 1. All sequencing data of the study are available at Array Express: the ChIP–seq results and enrichment data are available with accession code E-MTAB-7858 and the RNA-seq results and gene expression data are available with accession code E-MTAB-7859.Source data are provided with this paper.
References
- 1.Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol. 2018;15:13–30. doi: 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- 2.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elyada E, et al. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 4.Pribluda A, et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell. 2013;24:242–256. doi: 10.1016/j.ccr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 6.Halberg RB, et al. Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proc Natl Acad Sci; USA. 2000. pp. 3461–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiewe T, Haran TE. How mutations shape p53 interactions with the genome to promote tumorigenesis and drug resistance. Drug Resist Updat. 2018;38:27–43. doi: 10.1016/j.drup.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Dittmer D, et al. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 10.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Petrova TV, et al. Transcription factor PROX1 induces colon cancer progression by promoting the transition from benign to highly dysplastic phenotype. Cancer Cell. 2008;13:407–419. doi: 10.1016/j.ccr.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Fearon EF, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 14.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 16.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Schulz-Heddergott R, et al. Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits Stat3-mediated tumor growth and invasion. Cancer Cell. 2018;34:298–314. doi: 10.1016/j.ccell.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 19.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muir RM, et al. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia) Plant Mol Biol. 2011;75:555–565. doi: 10.1007/s11103-011-9739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, et al. The microbiota is essential for the generation of black tea theaflavins-derived metabolites. PLoS ONE. 2012;7:e51001. doi: 10.1371/journal.pone.0051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Díaz-Quiroz DC, et al. Synthesis, biological activity and molecular modelling studies of shikimic acid derivatives as inhibitors of the shikimate dehydrogenase enzyme of Escherichia coli . J Enzyme Inhib Med Chem. 2018;33:397–404. doi: 10.1080/14756366.2017.1422125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurent-Puig P, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763–1773. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]
- 24.Yizhak K, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019;364:eaaw0726. doi: 10.1126/science.aaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martincorena I, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama A, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 27.Clément G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett’s esophagus. Oncogene. 2006;25:3084–3092. doi: 10.1038/sj.onc.1209338. [DOI] [PubMed] [Google Scholar]
- 28.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 30.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sklarz M, Levin L, Gordon M, Chalifa-Caspi V. NeatSeq-Flow: a lightweight high-throughput sequencing workflow platform for non-programmers and programmers alike. 2018 Preprint at https://www.biorxiv.org/content/10.1101/173005v3.
- 35.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandal S, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morton JT, et al. Balance trees reveal microbial niche differentiation. mSystems. 2017;2:e00162–16. doi: 10.1128/mSystems.00162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, et al. A UPLC-MS/MS method for simultaneous determination of free and total forms of a phenolic acid and two flavonoids in rat plasma and its application to comparative pharmacokinetic studies of Polygonum capitatum extract in rats. Molecules. 2017;22:353. doi: 10.3390/molecules22030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source gels for immunoblots are provided in Supplementary Fig. 1. All sequencing data of the study are available at Array Express: the ChIP–seq results and enrichment data are available with accession code E-MTAB-7858 and the RNA-seq results and gene expression data are available with accession code E-MTAB-7859.Source data are provided with this paper.