Summary
Skin cancer risk varies substantially across the body, yet how this relates to the mutations found in normal skin is unknown. Here we mapped mutant clones in skin from high and low risk sites. The density of mutations varied by location. The prevalence of NOTCH1 and FAT1 mutations in forearm, trunk and leg skin was similar to that in keratinocyte cancers. Most mutations were caused by ultraviolet (UV) light, but mutational signature analysis suggested differences in DNA repair processes between sites. 11 mutant genes were under positive selection, with TP53 preferentially selected in the head and FAT1 in the leg. Fine scale mapping revealed 10% of clones had copy number alterations. Analysis of hair follicles showed mutations in the upper follicle resembled adjacent skin, but the lower follicle was sparsely mutated. Normal skin is dense patchwork of mutant clones arising from competitive selection that varies by location.
Introduction
Normal facial skin from middle aged humans has a substantial burden of clones carrying cancer associated mutations generated by UV light (1). Several mutant genes are under strong positive genetic selection, evidenced by an excess of protein altering relative to silent mutations (the dN/dS ratio), suggesting these mutations promote clonal expansion rather than being neutral ‘passengers’ (2). The positively selected mutant genes include those commonly mutated in basal cell carcinoma (BCC) and squamous cell carcinoma (SCC) of the skin, which are among the commonest cancers worldwide. Both cancers contain frequent mutations in TP53, NOTCH1, NOTCH2 and FAT1, all of which are positively selected in facial skin (1). These findings are consistent with cancer emerging from a subset of clones within normal tissue.
However, as the risk per unit area of developing BCC or SCC differs substantially between body sites (Figure 1a) a more systematic investigation across different parts of the skin beyond the face is warranted to understand the influence of frequency of exposure to UV light, sensitivity to UV injury (sunburn) and also the histological structure of the skin (3). These observations motivated us to use deep targeted and whole genome sequencing to perform broad and fine scale mapping of mutations across different locations. Clonal burden, mutational processes and competitive selection varied both across body sites and within millimeters in individual skin samples. The comprehensive clonal map of the skin at different locations, enables evolutionary characteristics to be compared with UV exposure and cancer risk.
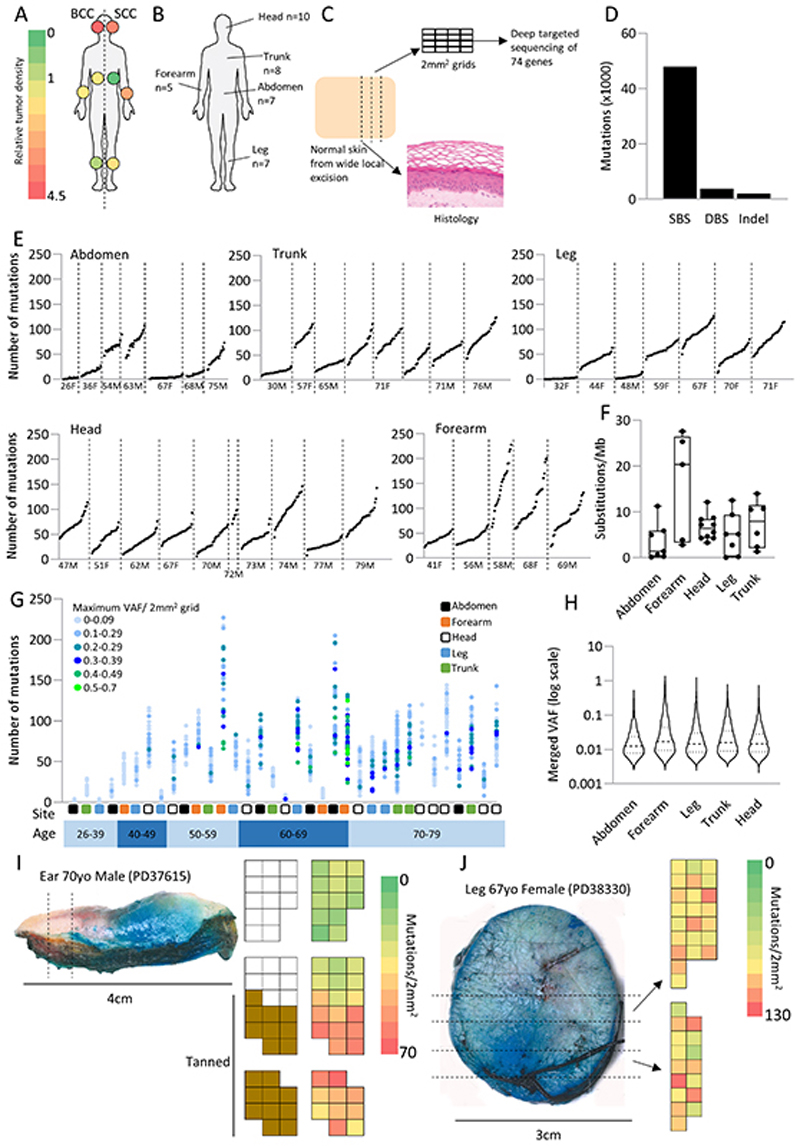
Figure 1. Human epidermis is a patchwork of small competing mutant clones.
a. Tumor density (cancers/unit area) for BCC and SCC across different body sites, whole body = 1.0. Data from (3).
b. Samples were collected from a variety of body sites with traditionally differing levels of sun exposure.
c. Experimental outline: Normal peeled epidermis was cut into 2mm2 grids and sequenced at high depth using a 74 gene panel bait set. Mutations were called using the ShearwaterML algorithm. A total of 35 patients were sequenced across 37 positions clustered into five main sites; abdomen, head, forearm, leg, and trunk. An average of 39 2mm2 grids were sequenced per position.
d. Total number of single base (SBS), double base (DBS) substitutions and insertion/deletion (indel) events across all 2mm2 grid samples from all body sites.
e. Number of mutations per 2mm2 grid from all 1261 samples from the 35 patients. Each point represents a single 2mm2 grid. Patients are ordered by age and divided by body site. M = male, F = female.
f. Average mutational burden (substitutions/Mb) per patient per site. Identical mutations in adjacent samples of a patient are merged (methods).
g. Distribution of the number of mutations and maximum variant allele frequency of all 2mm2 samples per patient. Each point represents a single 2mm2 sample and point colours represent the maximum variant allele frequency in that 2mm2 sample. Patients are ordered by age.
h. Variant allele frequency (VAF) distribution ordered by body site. Where adjacent grids share the same mutation this is assumed to be the same clone so the allele frequencies are summed as described in the methods.
i and j. Heatmaps showing the variation in the number of mutations across the epidermis. i shows an example, from a contiguous strip of skin divided into three blocks from an ear, where 2mm2 samples carrying a high number of mutations cluster together. This is not the case for the leg sample shown in j. In j the two blocks of samples are approximately 1cm apart. In i the increase in the number of mutations correlates with an increase in the pigmentation seen in the epithelia.
Results
Mapping mutations in normal skin
Normal skin from chronically and intermittently sun exposed sites was collected from 35 Caucasian donors whose ages ranged from 26-79 with a balance of males and females (Figure 1b, Table S1). Information on skin type was available for 24 donors, 19 of whom had pale skin, a history of severe sunburn and limited ability to tan (Fitzpatrick type 1 and 2) (Table S1). Eight of 28 donors with an occupational history had worked outside for 5 years or more, while of the indoor workers six had lived in subtropical or tropical countries.
Samples collected for sequencing appeared normal under a dissecting microscope and the adjacent skin had normal histology as assessed by a specialist skin pathologist. Sheets of epidermis were detached from the underlying dermis and cut into a gridded array of 2mm2 pieces, each containing approximately 105 nucleated cells of which ca. 3×104 were in the proliferative basal cell layer (4). A total of 1,261 2mm2 pieces were sequenced at an average coverage of 690x using a bait set of 74 cancer associated genes (Figure 1c, Table S2, Table S3) (1). Mutations were called using the ShearwaterML algorithm which detects mutations present in 1% or less of nucleated cells in the sample (5,6). In a total area of 25.2cm2 of skin sampled across all donors we identified 47,977 single base substitutions (SBS), 3,824 double base substitutions (DBS) and 2,090 small (<200 bp) insertion or deletion events (indels) after merging mutations shared between adjacent samples (Figure 1d, Table S4, methods,).
Variation in the density of mutant clones
The large difference in SCC and BCC risk and the variation in UV exposure across the body led us to hypothesize there would be substantial differences in the mutational landscape of different sites. However, while some samples from intermittently sun-exposed sites (e.g. abdomen and leg) did show a very low number of mutations, others had a similar number of mutations/2mm2 (clonal mutation density, CMD) and mutational burden (synonymous SNV per megabase) as frequently sun-exposed sites such as the head (Figure 1e, Figure 1f). Most mutations had a small variant allele frequency (VAF), indicating the skin is colonized by a large number of small clones (median VAF=0.015 Figure 1g, Figure 1h), though there is a tendency for clone size to increase with age (Figure S1a).
Although large differences were not seen between body sites, we noted intra-donor variability in the number of mutations detected per 2mm2 sample (Figure 1e, Figure S1b). The accumulation of mutations and the growth of mutant clones are both believed to be stochastic processes, which may lead to differences between samples (7). In stochastic spatial simulations (methods), we found the observed intra-donor range in CMD was consistent with such stochastic variation (Figure S1b, S1c).
We also investigated if the environment differed across skin from individual donors. If this was the case we would expect CMD in adjacent samples to be more similar than in more distant samples. We found six individuals who showed distinct gradients in the number of mutations across their epidermal tissue (Figure S1d). In one such specimen from the ear, the sun exposed, pigmented skin on the outside of the ear had 3-fold higher CMD than the unpigmented skin at the back of the ear (Figure 1i). Similar variability was seen in five other samples from forearm, breast, ear lobe skin, eyebrow and eyelid (Figure S1e). In the other 29 donors there was no significant spatial clustering of CMD (Figure 1j). Thus, the local level of mutations may vary due to both local differences in UV exposure and stochastic fluctuation.
Positive and negative genetic selection
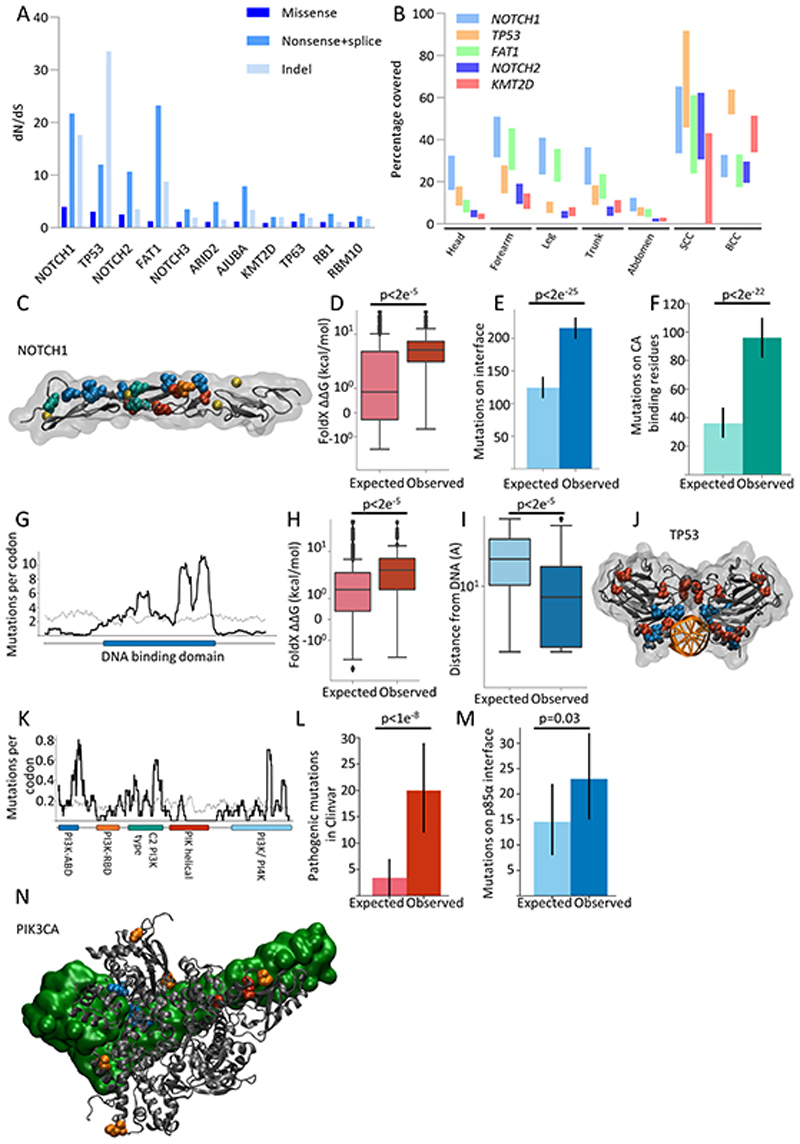
The organization of the skin allows mutant clones to spread laterally and compete for space both with wild type cells and clones carrying other mutations, a potential selective mechanism in which only the fittest mutants persist (2,7). Analysis of the dN/dS ratio, which calculates the ratio of protein altering to silent mutations, is a test for competitive selection. Genes with a dN/dS ratio >1 show that non-synonymous mutations in the gene increase cellular fitness permitting clonal expansion. 11 genes were found to be under positive selection after correcting for genomic context, mutational spectrum and multiple testing (1,5) (Figure 2a, Figure S2a, Figure S2b, Table S5). Six of these genes (NOTCH1, TP53, NOTCH2, NOTCH3, FAT1 and RBM10) have been previously identified as being positively selected in eyelid skin (1), however five are novel drivers. Of the newly identified mutant genes, the p53 family transcription factor TP63 regulates Notch signaling and keratinocyte differentiation while the KMT2D histone modifying lysine methyl transferase interacts with TP63 and promotes expression of its target genes. The SWI/SNF complex component, ARID2 and AJUBA which regulates Notch and Hippo signaling are both recurrently mutated in squamous cancers of the skin and other organs. Thus, positively selected mutant genes are predominant regulators of epidermal proliferation and/or differentiation.
Figure 2. Positive selection of genes linked with cancer in normal skin.
a. dN/dS ratios for missense, nonsense/splice substitutions and insertions/deletions (indel) across all body sites for genes under significant (global q <0.01 and dN/dS>2) positive selection.
b. Estimated percentage of cells carrying mutations in the most strongly positively selected genes (NOTCH1, FAT1, TP53 and NOTCH2) for each body site as well as for basal and squamous cell carcinomas (see methods). Upper and lower bound range allows for uncertainty in copy number and biallelic mutations. Upper bound represents no CNA and one mutant allele per gene.
c-f: Positive selection of categories of missense mutations in NOTCH1 EGF repeats 11-12 that form part of the ligand binding domain:
c. Structure of NOTCH1 EGF11-13 (PDB 2VJ3). Residues containing missense mutations that occur >10 times are highlighted. Ligand binding interface residues, blue; calcium binding residues, green; destabilising residues, red; D464N, orange, does not fit into the previous categories. Calcium ions shown in yellow.
d. Missense mutations that are not on ligand-interface or calcium binding residues are significantly more destabilising than would be expected under neutral selection (p<2e−5, n=452, two-tailed Monte Carlo test, methods).
e. Non-calcium binding missense mutations with ΔΔG < 2kcal/mol (i.e. are not highly destabilising) occur on the ligand-binding interface significantly more than would be expected under neutral selection (p=2e−25, n=315, two-tailed binomial test, error bars show 95% confidence intervals, methods).
f. Missense mutations with ΔΔG < 2kcal/mol (i.e. are not highly destabilising) and that are not on the ligand-binding interface occur on calcium binding residues significantly more than would be expected under neutral selection (p=2e−22, n=195, two-tailed binomial test, error bars show 95% confidence intervals, methods).
g-h: Positive selection of missense mutations in TP53
g. Sliding window plot of missense mutations per codon in TP53. Observed counts shown by the black line. Expected counts assuming that missense mutations were distributed across the gene according to the mutational spectrum (methods) shown in grey. DNA-binding domain (DBD) of TP53 shown in blue below the x-axis.
h. Missense mutations in the TP53 DBD that are more than 5Å from the DNA are significantly more destabilising than would be expected under neutral selection (p<2e−5, n=760, two-tailed Monte Carlo test, methods).
i. Missense mutations with ΔΔG < 2kcal/mol (not highly destabilising) in the TP53 DBD are significantly closer to the DNA than would be expected under neutral selection (p<2e−5, n=395, two-tailed Monte Carlo test, methods).
j. Structure of the TP53 DNA-binding domain (PDB 2AC0) bound to DNA (orange). Residues containing missense mutations that occur at least 10 times are highlighted. Highly destabilising mutations (ΔΔG >= 2 kcal/mol) shown in red. Non-destabilising mutations shown in blue.
k-n: Positive selection of missense mutations in PIK3CA
k. Sliding window plot of missense mutations per codon in PIK3CA. Observed counts shown by the black line. Expected counts assuming that missense mutations were distributed across the gene according to the mutational spectrum (methods) shown in grey. Domains of PIK3CA encoded protein are shown below the x-axis.
l. Significantly more single nucleotide substitutions in PIK3CA are annotated as pathogenic/likely pathogenic in the Clinvar database than would be expected under neutral selection (q=1e−8, n=216, two-tailed binomial test, error bars show 95% confidence intervals, methods).
m. Significantly more missense mutations in PIK3CA occur in codons at the interface binding PIK3R1 (defined as PIK3CA residues with atoms within 5Å of PIK3R1 in PDB 4L1B) than would be expected under neutral selection (p=0.03, n=157, two-tailed binomial test, error bars show 95% confidence intervals, methods).
n. Structure of PIK3CA protein, grey, bound to PIK3R1, green (PDB 4L1B). Residues with mutations occurring at least 3 times are highlighted. Mutations close to PIK3R1 shown in blue, other mutations that are annotated as pathogenic/likely pathogenic shown in red, all others shown in orange.
The proportion of skin colonized by mutant clones in each sample may be estimated by summing their variant allele frequencies for each mutant gene, within upper and lower bounds that allow for uncertainty over copy number (5). Five positively selected mutant genes, NOTCH1, TP53, FAT1, NOTCH2 and KMT2D occupied a substantial proportion of normal skin (Figure 2b; Figure S2c). At several sites the prevalence of mutant NOTCH1 and FAT1 was similar to that found in BCC and SCC, hinting that these genes may appear as cancer drivers by virtue of colonizing normal skin rather than promoting tumor formation (Figure 2b). In contrast, TP53, NOTCH2 and KMT2D mutants are enriched in both SCC and BCC compared with normal tissue.
The large number of missense mutations in NOTCH1 and TP53 allowed us to explore mutant selection in more detail. Consistent with previous data sets, missense mutations in NOTCH1 are concentrated in the ligand-binding region (Figure S2d) (5). Missense mutations that destabilise the protein, likely to be functionally equivalent to nonsense mutations, or mutations that disrupt the ligand-binding interface or calcium binding sites, which are expected to interfere with ligand-mediated NOTCH activation, are significantly selected (Figure 2c–f). Most missense mutations in TP53 occur in the DNA binding domain (Figure 2g), and were either protein destabilising or clustered close to the DNA binding surface (Figure 2h–j).
Alongside missense mutations that inactivate proteins, there are also activating point mutations. These may not be detected by dN/dS analysis if they are outnumbered by neutral or inactivating mutations in a gene. We found examples of canonical ‘hot spot’ activating mutations in receptor tyrosine kinases (EGFR, ERBB2, ERBB3 and FGFR3), signalling intermediates (KRAS, HRAS, AKT1, PIK3CA) and the redox response regulatory transcription factor NFE2L2. The most frequent activating mutations were in PIK3CA, which encodes the catalytic p110a subunit of phosphatidylinositol 3-kinase (PI3K) (Figure 2k). The dN/dS results showed that nonsense mutations in PIK3CA were negatively selected, but that missense selection was not significantly different from neutral. However, there was a significant enrichment of specific PIK3CA mutations associated with various cancers or overgrowth diseases that are known to be driven by PIK3CA activating mutations (Figure 2l) (8). This suggests that there is negative selection of inactivating PIK3CA mutations and positive selection of activating PIK3CA mutations. To test this hypothesis, we assessed the selection of mutations in PIK3CA predicted to alter the binding of p110α protein to the inhibitory p85a PI3K subunit, finding a small, but statistically significant, enrichment of mutations at the interface of p110α with p85α (p=0.03, two-tailed binomial test Figure 2m–n). We conclude that activating PIK3CA mutants are positively selected but mutations that inhibit its function are under negative selection.
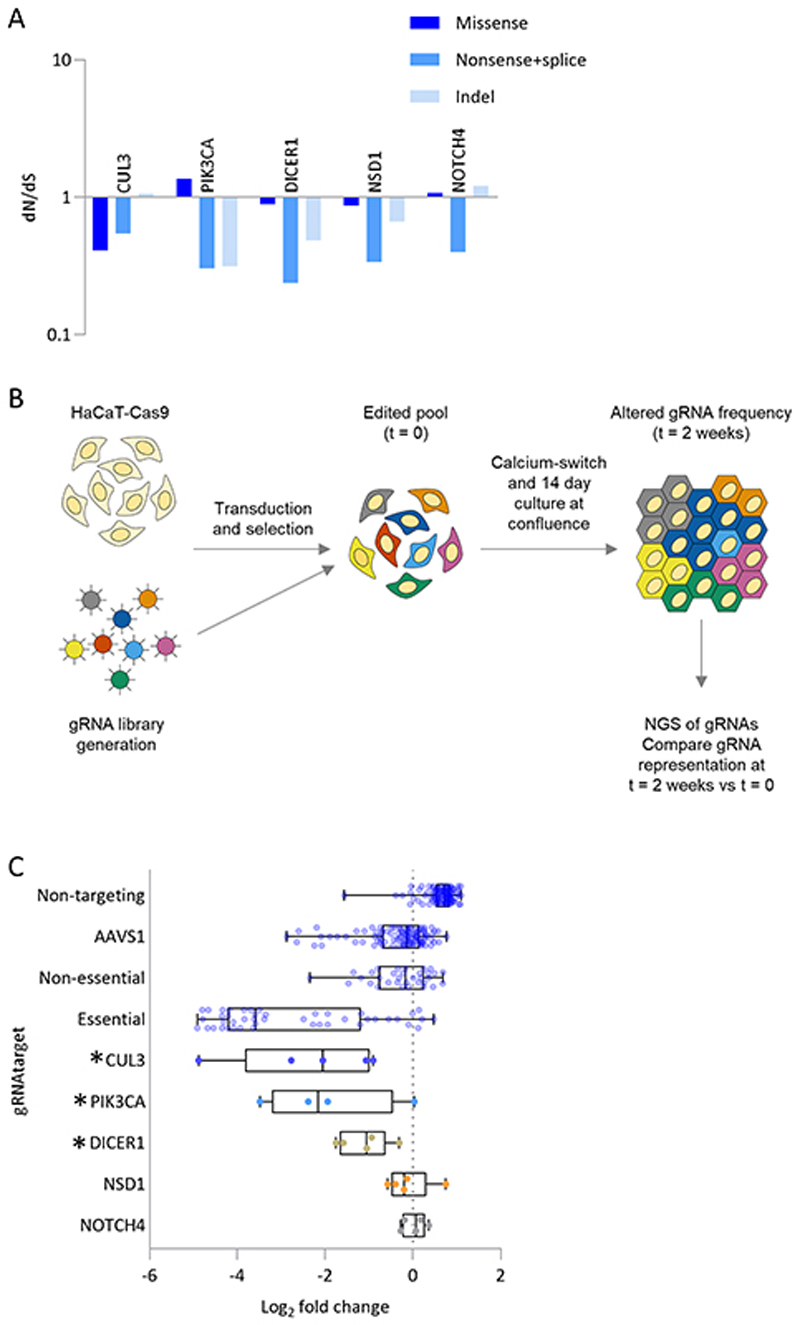
In addition to PIK3CA we also found a further four mutant genes that appeared to be under negative selection, with a dN/dS ratio below 1 (Figure 3a). PIK3CA, DICER1, CUL3, NSD1 and NOTCH4 were all depleted for protein truncating mutations and indels relative to the number of silent mutations. This was not due to variations in sequencing depth at the gene level (Figure S3a, methods). Synonymous mutations are generally distributed evenly across protein coding genes. However, in the case of CUL3 two synonymous mutations within the first exon were highly enriched (Figure S3b). This may reflect altered translational start sites or transcription factor binding due to the nucleotide changes (Figure S3c). Such synonymous mutations with the potential to alter cellular fitness may render the dN/dS ratio an unreliable metric of selection.
Figure 3. Human skin shows evidence of negative selection.
a. dN/dS ratios for missense, nonsense/ splice substitutions and insertions/deletions (indels) across all body sites for genes under significant negative selection. Only genes with global q<0.01 are shown.
b. Experimental outline: HaCaT cells (an immortalised keratinocyte cell line), were infected with lentivirus encoding Cas9 and guide RNAs (gRNA) targeting negatively selected genes or controls of non-targeting, AAVS1 safe harbour site, known essential and non-essential genes. Following puromycin selection, cells were cultured for a further two weeks at confluence in a high calcium media, which permits differentiation. Sequencing of gRNAs immediately after puromycin selection (t=0) and after two weeks at confluence (t=2) allows monitoring of changes in gRNA representation.
c. Log fold change of gRNA representation between t=0 and t=2. Each dot represents a gRNA Genes chosen are those predicted to be under negative selection according to dN/dS ratios. *MAGeCK FDR<0.1.
To test if the negatively selected mutant genes altered keratinocyte proliferative potential or survival, we performed a pooled lentiviral CRISPR/Cas9 knockout screen with a targeted library of 317 gRNAs in HaCaT epidermal keratinocyte cells. Whole genome sequencing of HaCaT cells showed that PIK3CA, CUL3, NOTCH1, NSD1 and NOTCH4 were wild type and there was a p.S1631A (c.4891T>G) mutation with a VAF of 0.49 in DICER1, that represents a conservative change in a poorly conserved region of the protein and therefore seems likely to be non-pathogenic (https://www.ncbi.nlm.nih.gov/clinvar/variation/221072/). gRNA abundance was determined following lentiviral infection and after 14 days in culture (Figure 3b). The specific depletion of gRNAs targeting known essential genes compared to those targeting known non-essential genes, or the safe-harbour AAVS1 locus, confirmed the ability of this assay to detect the dropout of gRNAs affecting cell growth or survival (Figure 3c). Of the genes under negative selection, gRNAs targeting CUL3, PIK3CA and DICER1 were significantly depleted arguing knockout of these genes is detrimental to keratinocyte growth or survival in vitro (Figure 3c, Table S6). gRNAs targeting NSD1 and NOTCH4 were not significantly depleted, which may reflect differences between the cultured HaCaT cells and keratinocytes in the epidermis. Although a Crispr screen will, for any given sgRNA, generate a cell population containing inactivating indels in either one or both alleles and therefore imperfectly models the monoallelic loss we see in human epidermis, it does provide evidence that these genes are detrimental to keratinocyte fitness. In addition, developmental mutations argue these genes are haploinsufficient (Decipher database haploinsufficiency scores: PIK3CA - 0.26%, DICER1 - 1.99% and CUL3 - 4.57%, https://decipher.sanger.ac.uk), suggesting that a single wild type allele is insufficient to maintain cellular fitness.
Negative selection has not been observed other epithelial tissues including oesophagus and colon (5,9). To show a mutant gene is negatively selected, a large number of silent mutations must be detected for reduction in the dN/dS ratio below 1 to reach statistical significance. It is possible that in normal tissues other than skin, the density of mutations not been high enough to reveal negative selection.
Mutant gene selection and mutational signature vary across the body
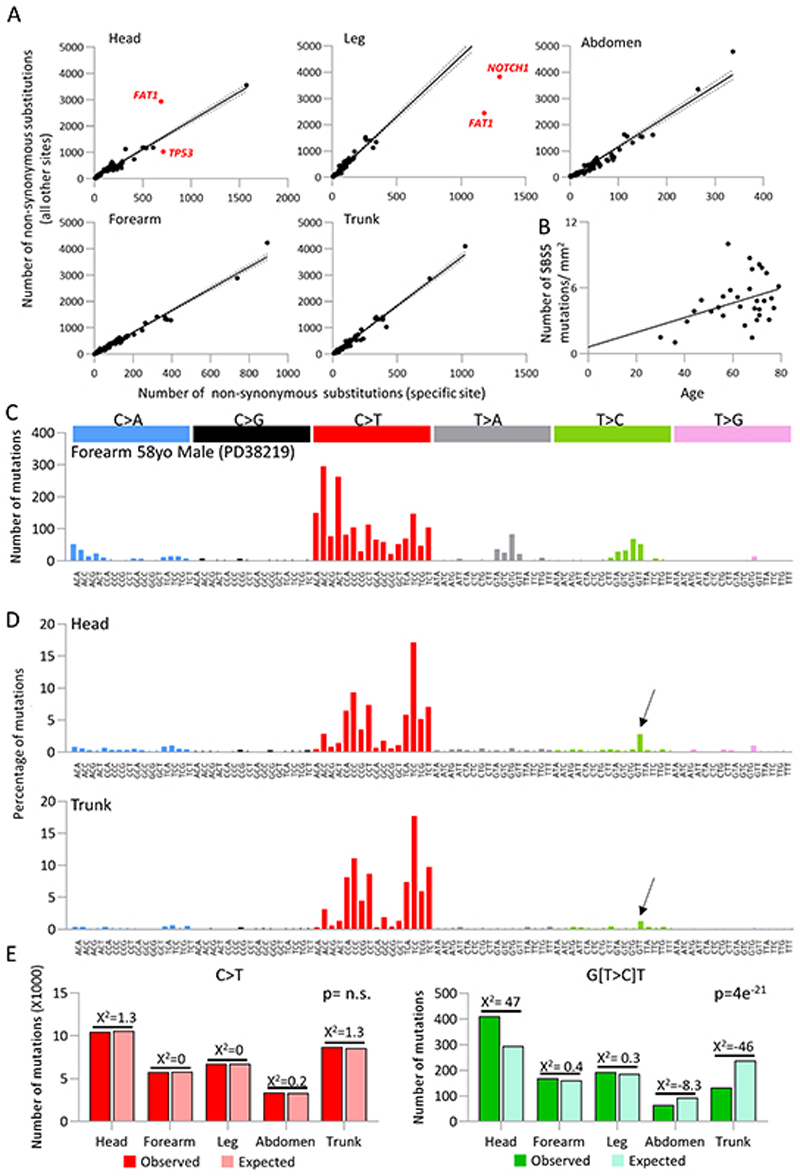
To investigate gene selection between body sites we compared the number of non- synonymous mutations in each gene at each site with that found at all other sites (Figure 4a). This revealed differential selection with protein altering FAT1 and NOTCH1 mutants over represented in the leg. In contrast FAT1 mutants were depleted in the head and TP53 mutants enriched over other sites. This possibly reflects environmental differences between sites such as the frequency of UV exposure.
Figure 4. Human epidermis shows selection and signature variation between body sites.
a. Differential selection of TP53, NOTCH1 and FAT1 across different body sites. Number of non-synonymous mutations per gene from indicated body site versus all non-synonymous mutations in that gene from all other body sites. Each dot represents a gene from the 74 gene targeted bait panel. Solid line represents trendline with 95% CI marked (dotted line). For points highlighted in red, q<1×10−6 using a likelihood ratio test of dN/dS ratios (methods).
b. Correlation between patient age and number of mutations attributed to signature SBS5. Linear regression p=0.0299, slope 0.067, intercept 0.608
c. Trinucleotide spectrum for single base substitutions of donor PD38219 which shows considerable differences to all other donors. This signature is consistent with SBS32 (cosine similarity 0.95) linked with azathioprine treatment.
d. Combined trinucleotide spectra for single base substitutions in all samples from the head (top panel) and trunk (bottom panel) from targeted sequencing. Arrow indicates G(T>C)T peak which contributes to a higher proportion of overall burden in the head. Head = 14561 mutations, Trunk = 10515 mutations.
e. Chi-squared test for all C>T mutations versus G(T>C)T mutations by body site (chi-squared 105, df=4, p=0). A higher proportion of G(T>C)T mutations are observed at sites with higher relative risk of cSCC.
The majority of mutations observed are consistent with damage induced by UV light, with C>T (or G>A) changes accounting for 77% of all single base substitutions and CC>TT (or GG>AA) changes accounting for 88% of all double base substitutions (Figure S2b). Although C>T changes can be linked with the ageing signature SBS1 as well as the SBS7 UV signatures, once the trinucleotide context was taken into account, only a small fraction of the C>T changes can be attributed to SBS1. In addition, both C>T and CC>TT substitutions were more frequently found on the un-transcribed strands of genes, suggesting a link with transcription-coupled nucleotide excision repair, an observation again consistent with SBS7a and SBS7b rather than SBS1 (10). In common with other epithelial tissues, there was a positive correlation between SBS5, a signature associated with cellular ageing, and patient age (Figure 4b) (linear regression p=0.0299, slope 0.067, intercept 0.608) (5,9). However, the contribution of SBS5 is relatively modest compared to that of SBS7 (average 19.8% and 76% respectively).
Using the trinucleotide context of each mutation, four single-base substitution reference signatures associated with damage by UV light (SBS7a-d) have recently been characterised in cancers of the skin (10). SBS7a and SBS7b are likely to arise from UV induced cyclobutane pyrimidine dimers or 6-4 photoproducts, with recent work showing that SBS7b is enriched in open chromatin and SBS7a in quiescent regions, perhaps caused by differences in repair due to genomic context (11). It has been hypothesised that SBS7c and SBS7d may be due to translesion DNA synthesis by error-prone polymerases inserting T rather than A, or G rather than A, opposite UV damage respectively (10). In all but one donor (Figure 4c), trinucleotide spectra were characteristic of SBS7a, SBS7b and SBS7d composition (Figure 4d), however mutations consistent with SBS7c (T[T>A]T) were not observed at a frequency any greater than other T>A mutation contexts.
Forearm skin from 58 year male (PD38219) was an outlier in terms of mutational spectrum (Figure 4c) and also the donor with the highest mutation burden (Figure 1e). This spectrum has a cosine similarity of 0.95 with the reference signature SBS32 (10). SBS32 was first identified in patients treated with azathioprine to induce immunosuppression and is characterised by a predominance of C>T mutations (75%) with a very strong transcriptional strand bias favouring higher rates of C>T mutations on the transcribed strand. The strand bias is consistent with azathioprine interfering with purine synthesis leading to incorporation of guanine derivates into DNA, which lead to C>T (G>A) mutations, especially with guanosine alterations on the coding strand, that escape nucleotide excision repair. This strand bias is in contrast to that caused by UV damage, which induces cyclobutane pyrimidine dimers, which are more efficiently repaired by transcription coupled-nucleotide excision repair on the transcribed strand leading to higher rates of C>T in template strand orientation. Here, this donor was on medication for immunosuppression since age 24 following a kidney transplant. Despite the high cosine similarity of this donor’s spectrum to SBS32, it is likely that SBS32 itself is contaminated by the SBS7 (UV) signatures as it was first characterised in cutaneous SCCs (12). The majority (58%) of DBS in this donor were CC>TT substitutions but with the opposite transcriptional strand bias to UV induced DBS. Long term azathioprine treatment may generate most of the mutations in the normal skin of transplant recipients.
In all other donors, we noticed a visible difference in the frequency of G[T>C]T mutations (Figure 4d). A chi-square test revealed a significant difference in the proportions of G[T>C]T mutations and all C>T mutations by body site (chi-square = 105, d.f. = 4, p = 0). In fact, the proportion of G[T>C]T mutations was found to increase with the increased relative risk of cSCC at that site (Figure 4e) (3). G[T>C]T mutations make up the predominant peak of SBS7d, suggesting a lower proficiency for DNA repair after UV damage at sites at higher risk of cSCC. We conclude that exposure to UV light dominates over aging in generating mutations in normal skin and that the nature of UV induced DNA damage and repair differ between high and low cancer risk sites.
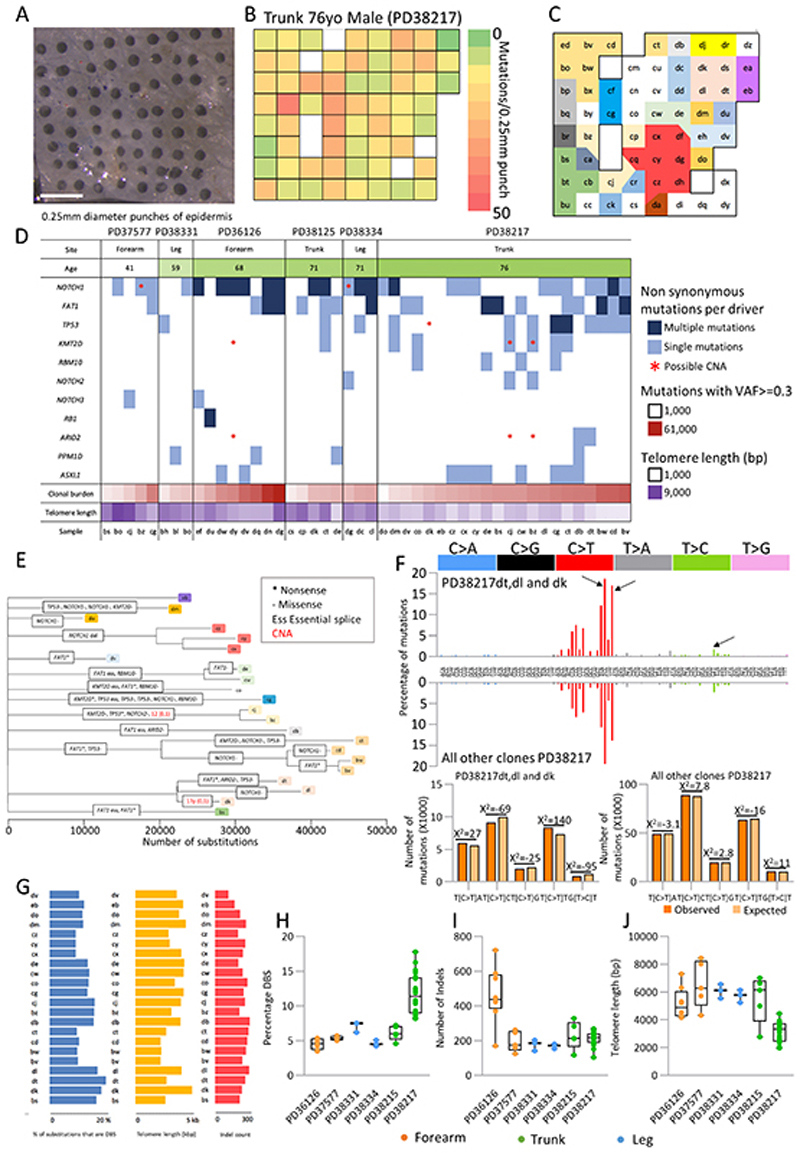
Higher resolution mapping and clonal genomes
To map mutant clones at increased spatial resolution 232 additional samples were taken from 6 donors with a 250 μm diameter circular punch, collected in a rectangular array with approximately a punch diameter between each sample (Figure 5a). Each sample corresponds to ca. 2400 nucleated cells of which 750 were in the basal layer. While 23 samples could not be processed due to low DNA yield, the remaining samples were sequenced for 324 cancer associated genes and the mutations called with ShearwaterML as described above (Figure 1c, Table S2, Table S3). A total of 2805 mutations were identified with a similar mutational spectrum and distribution of single-base substitutions, double-base substitutions and indels to the larger skin samples (Figure S4a–c, Table S7). Two additional genes in the expanded gene set, PPM1D and ASLX1, were under positive selection (Figure S4d, Figure S4e, Figure S4f). PPM1D is a regulator of TP53 under positive selection in normal esophagus while mutations of the epigenetic regulator ASLX1 are selected in squamous head and neck cancer. The number of mutations per sample was highly variable within each individual site (Figure 5b, Figure S4b, Figure S5a). Two additional genes in the expanded gene set, PPM1D and ASLX1, were under positive selection (Figure S4d, Figure S4e). PPM1D is a regulator of TP53 under positive selection in normal esophagus while mutations of the epigenetic regulator ASLX1 are selected in squamous head and neck cancer. A filtered set of mutations with a variant allele fraction of at least 0.2 was used to spatially map the clones present in the tissue (Figure 5c, Figure S5b). The density of clones varied substantially between individuals and within sites; for example, trunk samples from PD38217 showed the highest density of clones (Figure 5c) whereas the trunk skin from donor PD38215 was lightly mutated (Figure S5b).
Figure 5. Variation in mutational load, mutational signatures and telomere length at fine scale resolution.
a. 0.25mm diameter punches were taken in a gridded array format from peeled epidermis. Samples were sequenced using a 324 gene bait panel and mutations identified using ShearwaterML as previously described. 232 punches from 6 individuals from leg, trunk and forearm were sequenced. A subset of samples were subsequently whole genome sequenced. Scale bar = 1mm.
b. Heat map of a single individual (Trunk 76yo male PD38217; shown in A) showing the number of mutations per 0.25mm punch detected from targeted sequencing.
c. Clonal map of the same individual as b. A filtered set of mutations with a variant allele fraction >= 0.2 was used to spatially map the clones, letters indicate individual samples, each color denotes a separate clone. White is used for polyclonal samples. Samples with too low DNA yield for sequencing have been removed from the map.
d. Plot summarising the mutations (VAF>=0.3) and copy number aberrations for genes identified as being under positive selection in targeted sequencing data for 46 wholegenome punch samples. Age of donor, number of clonal (VAF>=0.3) mutations and telomere length for each sample is shown. Not all events are independent as some samples are part of the same clone (Figure 5e, Figure S5).
e. Maximum parsimony tree of clonal substitutions detected in 32 whole-genome punch samples of trunk skin from a 76yo male (PD38217). Branch lengths are equivalent to the number of clonal single and double base substitutions and are annotated with clonal non-synonymous mutations detected in the 13 genes found to be under positive selection. Within each branch, driver mutations are arbitrarily ordered. Copy number alterations are shown in red.
f. Combined trinucleotide spectra for single base substitutions assigned to branches of the dt/dl/dk clade (top panel) versus those assigned to all other branches (bottom panel) from same individual as E (PD38217). Arrows show trinucleotide contexts found to differ the most between these two groups (chi-squared =397, d.f. =4, p=0). Clone dt, dl, dk = 49144 mutations, all other clones = 458479 mutations.
g. Variation in a single individual (PD38217) in the percentage of substitutions that are double base (DBS), telomere length and number of clonal (VAF of at least 0.3) insertion/deletions (indel) per whole genome sample.
h-j. Variation in the percentage of substitutions that are double base (h), number of insertions/ deletions (indel) (i) and telomere length (j) per whole genome sample, by donor. Three different body sites are shown: forearm (orange), trunk (green) and leg (blue).
In order to better understand the genome-wide mutational burden, mutational processes and copy number aberrations, 46 samples from 34 different clones were submitted for wholegenome sequencing (WGS). All but three clones harboured one or more of the mutant genes under positive selection in the skin, no additional genes under selection were detected (Figure 2a, Figure 5d, Figure S4d). The majority of clones (58%) carried 2 or more positively selected mutant genes (Figure S4f). In terms of copy number alteration, we found two genomes with clonal non-synonymous mutations in NOTCH1 that displayed loss of heterozygosity at 9q, the NOTCH1 locus. In addition, a clone spanning samples PD38217cj and bz had a clonal deletion of one whole allele of chromosome 12. This clone harboured a missense mutation in KMT2D, a positively selected driver of clonal expansion located on chromosome 12. In contrast, PD36126dy had an amplification for one whole allele of chromosome 12 (this sample had clonal non-synonymous mutations in 18 genes located on this chromosome, but none were under selection or seem to be relevant to skin biology or carcinogenesis). Finally, one sample, PD38217dk, has a deletion of one allele at 17p, the TP53 locus (Figure 5d).
Phylogenetic trees were constructed using shared clonal single-base and double-base substitutions across all whole-genome samples for each donor (Figure 5e, Figure S5c). We observed a 2 to 3-fold variation in the number of clonal substitutions per genome in the same donor, despite the clones being almost adjacent in the skin.
Comparison of the proportion of the mutations in each trinucleotide context revealed differences between the branches. The spectrum of the clone which covers samples dt, dl and dk in PD38217 showed several differences when compared with all other clones sequenced in this donor (chi-square = 397, d.f. = 4, p = 0, Figure 5f). Each branch of this clade was found to have an increase of T[C>T]T mutations and fewer T[C>T]C and G[T>C]T mutations, with the G[T>C]T mutations being characteristic of SBS7d. Since the differences we observe in the frequency of mutations in these trinucleotide contexts were common to branches of the same clone, it is likely they are caused by biological differences in the cells of that clone (see Methods). In the whole genome data, we do detect an increased frequency of T[T>A]T mutations, characterised as SBS7c, that was not observed in the targeted data. However, the contribution of these mutations to the overall burden is small and less than that previously described in skin cancers (10). We conclude that genuine differences in UV damage or repair exist between clones in very close spatial proximity.
We found double base substitutions, indels and telomere length also varied between clones, from the same individual in some cases by nearly 2-fold (Figure 5g). The intra-individual range in DBS and indels was as large as that between donors (Figure 5h, Figure 5i respectively). Telomere length varied in a similar manner, indeed the variation seen in individuals was similar to that across cancers derived from different tissues (Figure 5j).
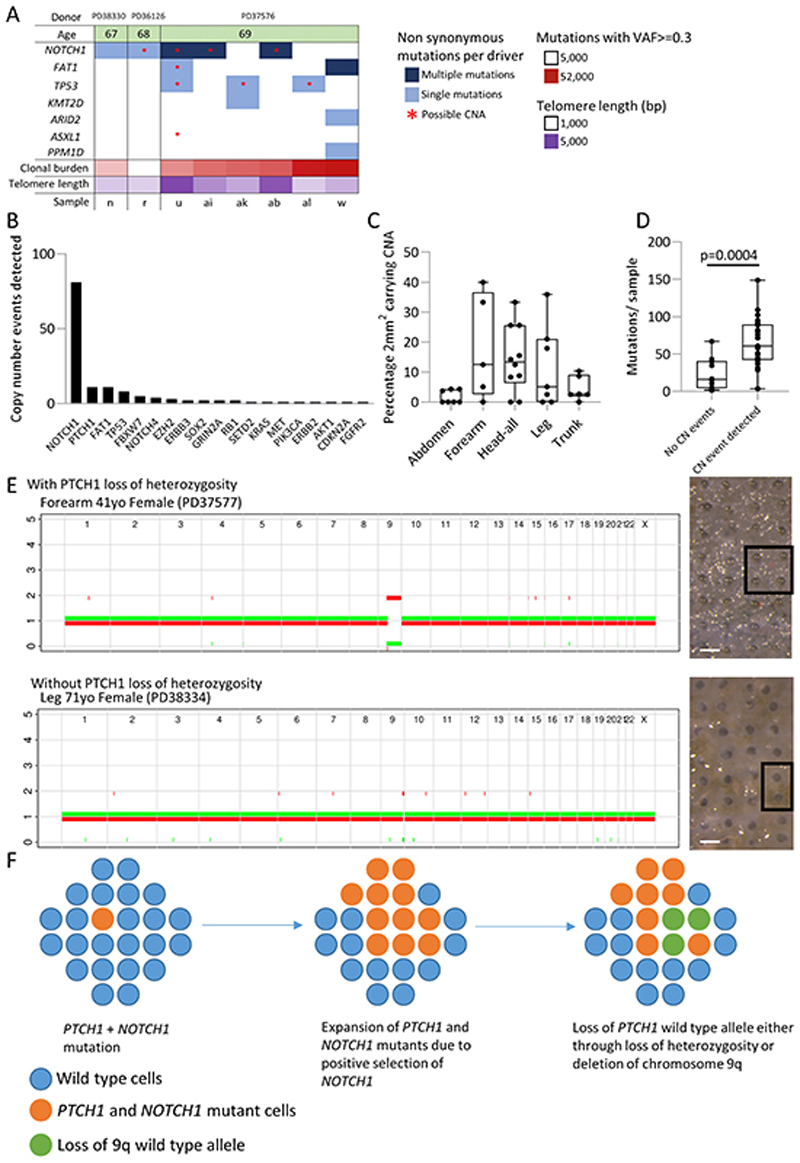
Copy Number Alteration (CNA) in normal skin
Given the presence of CNA in 6/46 genomes from micro punch biopsies, we explored other samples for evidence of genomic instability. Eight (0.4%) of 2mm2 samples were also clonal and were submitted for WGS. We found non-synonymous mutations in at least one driver gene in all samples, and CNA of one or more of NOTCH1, FAT1, ASXL1 and TP53 in six of the eight samples (Figure 6a).
Figure 6. Normal human skin shows frequent copy number changes including loss of heterozygosity of PTCH1 .
a. Plot summarising the mutations (VAF>=0.3) and copy number alterations for genes identified as being under positive selection in targeted sequencing data for eight clonal whole-genome 2 mm2 grid samples. Age of donor, number of clonal (VAF>=0.3) mutations and telomere length for each sample is also shown. Not all events are independent since some samples are part of the same clone. Sample PD37576u shows multiple changes with loss of heterozygosity at 4q (FAT1), 9q (NOTCH1), 17p (TP53) and 20q (ASXL1).
b. Number of copy number events per gene detected by SNP phasing of targeted sequence data in all 1261 2mm2 grid samples.
c. Percentage of grids carrying a copy number event detectable by SNP phasing segregated by body site. Each dot represents an individual.
d. Average number of mutations per 2mm2 grid in patients either carrying or not carrying a copy number event p=3.8×10−4, Student’s two tailed t-test
e. Copy number profile of whole genome sequenced punch samples showing chromosome 9 loss of heterozygosity. The top example shows loss of heterozygosity for both NOTCH1 and PTCH1; the bottom example shows just NOTCH1 LOH. Scale bar = 0.5mm
f. Possible origin of basal cell carcinoma. In a wild type population of cells (blue circles) a single cell acquires PTCH1 and then NOTCH1 non-synonymous mutations (marked orange), the clone size expands and persists due to NOTCH1 positive selection. At a later time point the wild type PTCH1 allele is lost either through deletion or loss of heterozygosity (marked green), thus leaving a clone lacking functional PTCH1 expression and primed for BCC transformation.
To get further insight into the extent of CNA we used a heterozygous SNP phasing approach to detect sub-clonal allele loss of the genes in the targeted sequencing of 2mm2 samples (Figure 6b) (5). This revealed recurrent allele loss of NOTCH1, NOTCH1 and PTCH1, TP53, FAT1 and FAT1 and FBXW7. There was a higher prevalence of CNA in frequently sun exposed head and forearm skin which reached borderline significance (p=0.06 one-way ANOVA), though changes were seen at all body sites (Figure 6c). Across all samples there were significantly more mutations in samples containing copy number alterations (Figure 6d, Student’s 2 tailed t-test, p=3.8×10−4).
PTCH1 is of particular interest as it mutated in almost all cases of BCC but is not under selection in the epidermis (13). We identified examples of clones that had undergone LOH of 9q, some including PTCH1, a gene frequently showing CNA in BCC (Figure 6e). In addition, the relatively close proximity of PTCH1 to NOTCH1 suggest a potential mechanism for the persistence of PTCH1 mutations, despite their not being under positive selection (Figure 6f). Should a PTCH1 mutant cell acquire a NOTCH1 mutation, it will be carried as a passenger in a clonal expansion driven by NOTCH1. Subsequent loss of the wild type 9q allele involving PTCH1 as well as NOTCH1 would result in biallelic disruption of PTCH1 in a highly competitive clone, with the potential to generate a BCC. We conclude that normal skin harbors substantial numbers of clones with CNA, with recurrent LOH of positively selected mutants linked to cancer, particularly in frequently sun exposed sites.
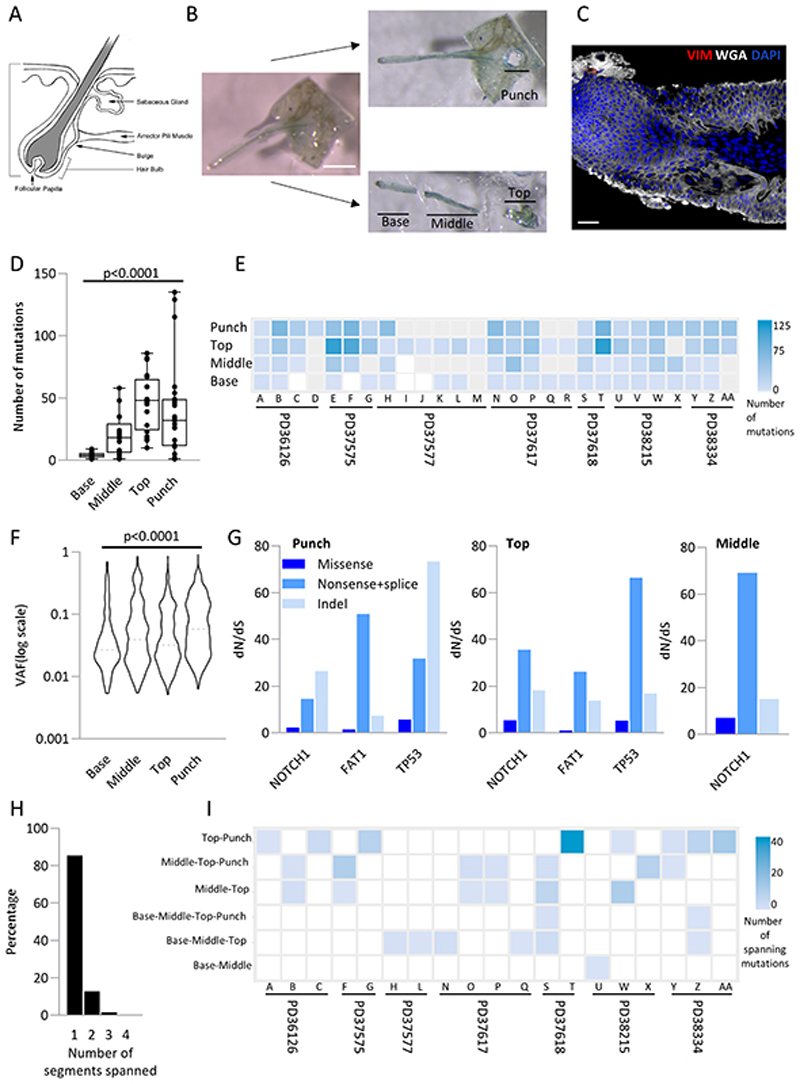
Mutations within hair follicles
Finally, we assayed mutations in hair follicles, appendages which are discrete self-maintaining units and have been proposed as a site of origin of BCC and SCC (Figure 7a)(14). Each follicle undergoes cyclical expansion and regression throughout life. Coarse (intermediate type) follicles at the largest (anagen) stage of the hair cycle were isolated from forearm, leg and trunk skin of 7 donors. We took a 250μm diameter micro-biopsy from adjacent epidermis and then dissected each follicle into thirds which were sequenced as described for the punch biopsies (Figure 7b). The upper section included the funnel shaped infundibulum which is in continuity with the epidermis. The mid follicle included the bulge region that contains stem cells that maintain the follicle base as it undergoes cyclical expansion and regression. Confocal imaging of adjacent follicles suggested there was minimal contamination with underlying dermis (Figure 7c).
Figure 7. Human hair follicles are polyclonal with the base of the follicle showing differences in mutational load compared to the top.
a. Structure of the human hair follicle (https://emedicine.medscape.com/article/835470-overview)
b. Experimental outline: Intact hair follicles are dissected from the epidermis and cut into three (designated base, middle and top). A 0.25mm punch of epidermis was taken adjacent to the follicle (designated punch). All samples were sequenced at high depth using a 324 gene bait panel and mutations identified using ShearwaterML as previously. Scale bar = 0.5mm.
c. Example confocal image of hair follicle stained with WGA (white), Vimentin (red), and dapi (blue). Scale bar = 38μm
d. Distribution of the number of mutations in different parts of the follicle. Each dot represents a sample. Solid line indicates the median. p<0.0001 Kruskal-Wallis test.
e. Number of exonic mutations per follicle across 324 genes. Each column represents a follicle with patient noted below. Each row is either the punch, top, middle or base. Samples that had insufficient DNA for sequencing are shown as grey.
f. Violin plot of the variant allele frequency at each part of the follicle. Dashed line indicates the median. p<0.0001 Kruskal-Wallis test.
g. dN/dS ratios for missense, nonsense/ splice substitutions and insertions/deletions (indel) across different parts of the follicle. Only genes with globalq<0.01 are shown.
h. Percentage of mutations spanning more than one location (n=2009 mutations)
i. Heatmap of number of mutations spanning different segments of the hair follicle. Each column is a follicle with the patient indicated below. Only follicles with spanning mutations are shown (methods). Spanning segments are detailed on the left.
The total number of mutations varied along the follicle; the number of mutations in the upper follicle was similar to that in the adjacent epidermis, but lower in the mid-follicle and base, consistent with the limited depth penetration of UV light into the skin (Figure 7d, Figure 7e, Table S8). The type of mutations and the mutational spectrum were similar to that of the epidermis (Figure S2a, Figure S2d, Figure S4c, Figure S4e Figure S6a, Figure S6b,). There was considerable variation in the number of mutations in each segment of the different follicles ranging from 3-6 fold in the middle and top areas (Figure 7d, Figure 7e).
The maximum VAF across all mutations indicates the clonality of each sample. In all three regions VAFs above 0.4 (or 0.8 in mutations with LOH) were seen, indicating it is possible for a clone to take over all or most of each region of the follicle (Figure S6c). The distribution of all VAFs was similar in the upper and mid follicle, but VAFs were significantly lower in the follicle base, indicating the bulk of clones were relatively small in this region (p<0.0001, Kruskal Wallis) (Figure 7f). dN/dS analysis showed NOTCH1, TP53 and FAT1 were positively selected both in the upper follicle and the adjacent epidermal punch, while NOTCH1 was selected in the mid follicle (Figure 7g). There were too few mutations to analyze selection in the base, but no TP53 and only 3 NOTCH1 mutations were detected.
Most mutations were private to each region (Figure 7h) however some mutations spanned multiple segments (Figure 7i, methods). One mutation in the non-selected ACVR gene was present with a VAF of over 0.4 in the base and mid follicle. Several follicles had one or more mutations spanning the mid and top follicle and the top follicle to the epidermal sample. Seven follicles had clones spreading from mid follicle to the epidermis and two clones spanned from the base to the epidermis. These clones reveal major migrations of cells perhaps in response to local injury.
Discussion
This study reveals normal appearing human epidermis as a competitive battleground between mutant clones generated by UV light. We find multiple lines of evidence supporting strong competitive selection of mutant genes, both positive and negative. There is strong genetic selection and marked enrichment of protein destabilizing or activating missense mutations of a subset of mutant genes. Using fine scale spatial mapping we observe multiple tightly packed clones indicating the skin is a dense patchwork of mutant clones. These findings are consistent with the hypothesis that selected mutant clones expand laterally within the proliferative layer of the epidermis as they outcompete neighboring cells. Mutant clones then collide and compete at clonal boundaries, resulting in the elimination of less fit mutants (7). That this all happens within a normal appearing tissue argues even the fittest mutant clones are constrained eventually and revert towards homeostatic behavior (2).
Whilst competitive selection is strong it is not uniform over the body, illustrated by the different prevalence of TP53 and FAT1 mutants in head and leg skin. Long term low dose UV exposure promotes the expansion of pre-existing TP53 mutant clones as well as generating new mutations, but the impact of UV on FAT1 clonal fitness is unknown (7). We also see variation in the UV signature 7d between frequently sun exposed and intermittently sun exposed sites, hinting at differences in UV lesion repair processes.
Hair follicles have been the subject of intense study in mice, but far less is known about human hair dynamics. The upper follicle resembles the adjacent epidermis in terms of gene selection and the distribution of mutant VAFs. The follicle base is maintained by a small population of cells in the bulge region of the mid follicle, but remains highly polyclonal, arguing multiple clones sustain the hair cycle over several decades. This differs from other tissues with spatially confined niches containing small numbers of stem cells, such as colonic crypts which become monoclonal with age (9).
What insights does this study give into carcinogenesis? Firstly, the differences we see in mutational landscape across sites are small relative to those in cancer risk. One possible explanation for this apparent discrepancy may be variations in environment between body sites, indeed this is hinted at by differences seen in mutational signatures and the strength of selection of some mutant genes. If environmental factors that promote transformation can be defined they may represent opportunities to intervene to reduce cancer risk.
In addition to these broad patterns, whole genome sequencing revealed differences in mutation burden, indels, double base substitution and telomere length in individual clones from the same donor in very close proximity. In addition, differences in mutational signatures point to variation in DNA damage and repair in clones with near identical UV exposure. What may cause such heterogeneity? It may reflect stochastic fluctuations, ‘neutral drift’, as it is unclear if the extent to which the burden of mutations, indels, DBS and telomere length, or indeed the differences in DNA damage/repair alter clonal fitness. Another source of variation is time. WGS reports the genome of the common ancestor of the clone, which reflects the history of that cell up until the time the clone was founded. A clone originating in the donors’ 20’s may differ in parameters such as mutational burden and telomere length from a clone founded decades later. Finally, the fitness of clones and their likelihood of surviving to expand to a detectable size may also not merely depend on the genome/epigenome of the clone itself but on the fitness of its immediate neighbours during clone expansion (15).
The high prevalence of some positively selected mutant genes in normal skin, similar to that in cancers, suggests that FAT1 and NOTCH1 may have little role in transformation, whereas TP53 and NOTCH2, which are enriched in cancers, play a part in transformation. One mutant gene that is not selected and striking by its low prevalence is PTCH1, mutated in almost all BCC. If the vast majority of mutations that have no effect on competitive fitness are eliminated by neutral drift, as is the case in mouse epidermis, heterozygous PTCH1 mutant clones are unlikely to persist for long enough to undergo biallelic inactivation (7). A potential mechanism that would allow heterozygous PTCH1 mutants to expand and persist follows from the proximity of PTCH1 and NOTCH1 on chromosome 9q. If a PTCH1 mutant cell acquires a NOTCH1 mutation it will have a strong fitness advantage and will expand and persist. Subsequent LOH affecting both genes would result in biallelic PTCH1 inactivation (Figure 6e, Figure 6f). The similarities between the upper hair follicle and the epidermis make it possible that some cancers may arise from follicles, but the sparsity of clonal mutations in the base of larger follicles suggest cancers arising from this area are likely to be rare.
In conclusion, the skin carries a far higher burden of mutant clones than other epithelial tissues, due to the effects of UV exposure that both generates mutations and promotes clonal expansion (7). In the light of this, the high incidence of BCC and SCC compared with other epithelial cancers seems unsurprising but the resilience of the tissue to mutation is remarkable (5,9). The selected mutant genes in skin partially overlap with those in esophageal epithelium, another squamous tissue, but there are key differences, for example NOTCH1 mutant clones are unable to colonize the majority of the skin as they do in the esophagus, perhaps because their expansion is restricted by the presence of other strongly competitive mutants. Understanding the environmental differences that promote the transformation of mutants in high and low risk sites may give insights that guide skin cancer prevention.
Methods
Ethics
Written informed consent was obtained in all cases under ethically approved protocols (Research Ethics Committee references 15/EE/0152 NRES Committee East of England – Cambridge South and 15/EE/0218 NRES Committee East of England - Cambridge East). The study was conducted in accordance with the Declaration of Helsinki.
Human samples
Normal skin samples were collected from patients undergoing wide local excision after initial melanoma excision, patients undergoing browplexy or from deceased organ donors from whom organs were being retrieved for transplantation. For every sample as much underlying fat and dermis as possible was removed and samples were cut into approximately 0.5cm× 0.5cm pieces. Samples were incubated in 20mM EDTA for 2 hours at 37°C. The epidermis was then peeled away from the dermis using fine forceps and a dissecting microscope. The epidermis was fixed for 30mins with 4% paraformaldehyde (FD Neurotechnologies) before being washed three times in 1xPBS. For sequencing the epidermis was cut into 2×1mm grid samples and DNA extracted using the QIAamp micro DNA extraction kit (Qiagen) by digesting overnight and following manufacturer’s instruction. DNA was eluted using pre-warmed AE buffer where the first eluent was passed through the column two further times. DNA was extracted from flash-frozen fat and dermis as for the epidermal samples and was used as the germline control. Patient PD38218 contributed three independent trunk samples. For the purpose of mutational burden, signature and selection these were treated as a single site. 0.25mm diameter epidermal samples were collected using a brain punch biopsy (Stoelting Europe).
Hair follicles were dissected away from the peeled epidermis and cut into equal thirds. DNA from both the punches and hair follicles was extracted using the Arcturus Picopure kit (Applied Biosystems) following manufacturer’s instructions.
Immunofluorescence of hair follicles
PFA-fixed wholemounts were blocked for 2 hours in blocking buffer (0.5% bovine serum albumin, 0.25% fish skin gelatin, 0.5% Triton X-100 and 10% donkey serum) dissolved in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 4 mM MgSO4-7H20). Follicles were stained with Vimentin (ab92547, Abcam RRID:AB_10562134) diluted 1:500 in blocking buffer for 24 hours at room temperature with continuous rocking. Samples were washed for a minimum of 24 hours with 0.2% Tween-20 in PHEM buffer changing daytime washes every 2-3 hours. Follicles were then stained with donkey anti-rabbit Alexa-conjugated 555 (1:500 dilution), wheat germ agglutinin 657 (1:500) and 1 μg/ml DAPI. All secondary antibodies were diluted in blocking buffer and samples were incubated for 24 hours at room temperature with continuous rocking. Samples were washed for a minimum of 24 hours with 0.2% Tween-20 in PHEM buffer changing daytime washes every 2-3 hours before imaging on a Leica SP8 confocal microscope.
CRISPR-Cas9 knockout screening
The plasmids used are listed below:
Plasmids.
| Plasmid name | Description | Supplier/Source |
|---|---|---|
| pKLV2-EF1a-Cas9Bsd-W | Cas9 expression vector | Addgene #68343 RRID:Addgene_68343 |
| pKLV2-U6gRNA5(BbsI)- ccdb-PGKpuro2ABFP-w | gRNA vector, modified to contain ccdB gene between Bbs1 sites for gRNA subcloning. | Gene Editing, Cellular operations, Wellcome Sanger Institute |
| psPAX2 | VSV-G envelope expressing plasmid. | Addgene #12260 RRID:Addgene_12260 |
| pMD2.G | 2nd generation lentiviral packaging plasmid. | Addgene #12259 RRID:Addgene_ 12259 |
| pKLV2-U6gRNA5(gGFP)-PGKBFP2AGFP-W | Cas9 activity reporter plasmid. | Addgene #67980 RRID:Addgene_ 67980 |
| pKLV2-U6gRNA5(Empty)-PGKGFP2ABFP-W | Cas9 activity reporter plasmid (control). | Addgene #67983 RRID:Addgene_ 67983 |
Cell culture
HaCaT keratinocytes are a spontaneously immortalised but non-tumorigenic cell line obtained from DKFZ (now distributed by CLS, Eppelheim Germany). Short tandem repeat profiling by the Eurofins Cell Line Authentication Service confirmed the cells had the correct marker profile as listed in Cellosaurus (https://web.expasy.org/cellosaurus/CVCL_0038). HaCaT cells were maintained in calcium-free DMEM (Invitrogen) with 10% FBS (calcium-chelated using 5% chelex resin) and supplemented to 0.03mM CaCl2 (“low calcium DMEM”). HaCaT cells maintained in low calcium DMEM were passaged twice weekly to maintain cells in an undifferentiated, proliferating state. Following lentiviral delivery of Cas9 and the gRNA library, cells were switched to DMEM with 10% FBS and supplemented with 1.8mM CaCl2, a concentration permissive to keratinocyte differentiation. Cells tested negative for mycoplasma on completion of experiments using a LookOut Mycoplasma Detection Kit (Sigma-Aldrich Cat# MP0035).
gRNA library design and synthesis
A minipool gRNA library was designed to include gRNAs targeting five genes under apparent negative selection (globalq<0.01 see dN/dS analysis). gRNA sequences were selected from the previously described Human CRISPR Library v.1.0 (16). Control gRNAs targeting known essential and nonessential genes (17) and the AAVS1 safe-harbour locus were also included. An 89-mer oligo pool (oPool) was purchased from IDT in the format ATATATCTTGTGGAAAGGACGAAACACCGN19GTTTAAGAGCTATGCTGGAAACAGCATAGCAAGT TTAAATA, where N19 specifies the variable gRNA sequence. gRNAs were PCR-amplified from the oPool under the following conditions:
| Component | 25 μl Reaction | Final concentration |
|---|---|---|
| 2X Q5 Mastermix (NEB | 12.5 μl | 1X |
| 10 μM Forward Primer | 1.25 μl | 0.5 μM |
| 10 μM Reverse Primer | 1.25 μl | 0.5 μM |
| Template DNA (1ng/pl) | 1.0 μl | 0.04ng/ul |
| Nuclease-Free Water | 9.0 μl | Up to 25 μl |
| Step | Temperature | Time |
| Initial Denaturation | 98°C | 30s |
| 98°C | 10s | |
| 14 Cycles | 67°C | 10s |
| 72°C | 15s | |
| Final Extension | 72°C | 2mins |
| Hold | 4°C | Hold |
| Forward | Primer: |
| ATCATATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGA AACACC | |
| Reverse | Primer: |
| TGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCATAGCTCT TAAAC | |
To avoid the need for gel extraction of digested fragments, we utilised a modified version of pKLV2-U6gRNA5(BbsI)-PGKpuro2ABFP-W in which the ccdB cassette was inserted between the BbsI sites. pKLV2-U6gRNA5(BbsI)-ccdb-PGKpuro2ABFP-W was digested with Fastdigest BbsI (Fermentas) for 1h at 37°C. The vector digest and PCR products (4 pooled 25μl PCR reactions) were purified using the DNA clean and concentrator kit (Zymo Research) according to the manufacturer’s instructions.
Gibson assembly reactions were set up using 100ng each of the PCR-amplified oligo library and the BbsI-digested gRNA vector, and incubated at 50°C for 1 hour. Gibson assembly products were purified and concentrated by ethanol precipitation (1h at 4°C) and resuspended in RNAse-free water to a final concentration of 100ng/μl. 100μl of Lucigen Endura electrocompetent cells were combined with 1μl of Gibson assembly product and electroporated using the BTX ECM630 (Harvard Apparatus) (2000V, 200Ω, 25μF). Cells were recovered in 1ml of Lucigen recovery medium for 1 hour at 37°C. Bacterial plates were prepared using a serial dilution of recovered cells to estimate the total number of transformants and calculate library coverage. The remaining cells were grown overnight in a 250ml low-salt 2xLB + ampicillin liquid culture at 37°C. Plasmid DNA was extracted using the Endofree Maxi Kit (Qiagen) according to the manufacturer’s instructions. The 317 gRNA library was transformed at an estimated coverage of >100,000X. Control Gibson assembly reactions without oligo insert yielded a negligible number of transformants, indicating a low background due to undigested vector and efficient suppression of growth by the ccdB cassette.
For quality control of the generated plasmid library, PCR amplification, Illumina sequencing (19-bp single-end sequencing with custom primers on the MiSeq platform) and sgRNA counting were performed as described previously (18). 97.1% (309/317) of gRNAs were recovered, with a skew ratio of 2.2, indicating a favourable gRNA distribution when compared to broadly used genome-wide gRNA libraries (19).
The ‘ineq’ R package was used to generate Lorenz curves and to calculate a Gini index for the generated library and widely used genome-wide libraries, as shown below. The Gini index is a measurement of statistical dispersion and reflects the evenness of read count distribution. A lower Gini index is indicative of a more even distribution (Figure S7).
| gRNA library | Skew Ratio | Gini Index |
|---|---|---|
| SMS (this study) | 2.2 | 0.19 |
| Avana | 4.9 | 0.36 |
| Brunello | 4.0 | 0.29 |
| Yusa_V1 | 7.9 | 0.34 |
| Yusa_V1.1 | 3.2 | 0.23 |
Lentivirus production and transduction
Lentiviruses were generated by transfection of HEK293FT cells using Lipofectamine 3000 (Invitrogen). One day before transfection, 4 million HEK293FT cells were plated in a 10cm dish to reach 80 – 90% confluence after 24h. On the day of transfection, 6μg of lentiviral vector, 7.5μg of the 2nd generation lentiviral packaging plasmid psPAX2, 2.5μg of VSV-G envelope expressing plasmid pMD2.G and 35μl of P3000 Enhancer were added to 1.5ml of serum-free OptiMEM (Invitrogen). 41μl of Lipofectamine 3000 was diluted in 1.5ml serum-free OptiMEM and incubated at room temperature for 5 minutes, before combining with the prepared DNA mixture and incubating at room temperature for a further 30 minutes. HEK293FT media was replaced with 6ml serum-free OptiMEM, before adding the transfection complexes. After 6 hours, the transfection medium was replaced with complete DMEM. Lentiviral supernatants were collected 24 and 48 hours post-transfection. Harvests were pooled together, centrifuged to remove cellular debris and filtered (0.45μm pore, PES)(Merck). Virus aliquots were stored at −80°C.
Generation and validation of Cas9-expressing HaCaT keratinocytes
HaCaT cells were transduced with Cas9-Bsd lentivirus at a low MOI and expanded in the presence of blasticidin (10μg/ml) for 7 days. Cas9 activity was confirmed in >90% of cells using a previously described dual fluorescence reporter vector system2. Briefly, HaCaT or HaCaT-Cas9 keratinocytes were transduced with a lentiviral vector to express GFP and BFP in addition to a gRNA targeting GFP. Cells were also transduced with vector lacking a targeting gRNA in parallel to serve as a control. >90% of transduced (BFP+) cells were efficiently edited (GFP-), specifically in HaCaT-Cas9 cells. No GFP loss was detected in parental HaCaT cells, or in either cell population transduced with the control vector.
Generation of genome-wide mutant libraries and screening
1.5 × 106 Cas9-expressing HaCaT keratinocytes were transduced in a total volume of 15ml low calcium DMEM containing polybrene (8ug/ml) and gRNA lentivirus at a 1/30 dilution. The cell/virus suspension was plated at 20,000 cells/cm2 in a T75 cell culture flask, After 6 hours, cells had adhered and the transduction medium was replaced with low-calcium DMEM. Transduction efficiencies were measured 72h post-transduction by cytometric analysis of the BFP+ fraction. Transduction efficiencies varied between 10 – 15% between replicates, corresponding to an initial library coverage of ~500 – 700X. gRNA-expressing cells were enriched to >95% by 72 hour puromycin selection (2ug/ml).
Puromycin-selected cells were replated at 10,000 cells/cm2 and grown for 72h under low-calcium conditions until confluent. Cells were then switched to DMEM + 1.8mM CaCl2 and maintained for a further 14 days. Media was refreshed every 3 days. Cells were trypsinized and genomic DNA was extracted from ~1.7 million cells (5000X coverage) using a previously described method using salt precipitation and purification by isopropanol precipitation.
PCR amplification, Illumina sequencing (19-bp single-end sequencing with custom primers on the MiSeq platform) and sgRNA counting were performed as described previously (18).
Identification of positively and negatively selected guide RNAs
Read counts generated from samples collected 2 weeks post-library transduction were compared to those from samples collected immediately after puromycin selection (t = 0) using the MAGeCK package to identify positively and negatively selected gRNAs and genes (20).
Sequencing
The two sequence capture bait sets used in this study have been described previously (5,9). The ‘grid’ bait set contains a set of 74 genes recurrently mutated in SCC and BCC as well as genes commonly mutated in other epithelial cancers. The ‘punches + follicles’ bait set is broader range of genes frequently mutated in a range of cancers based on the COSMIC cancer gene census (https://cancer.sanger.ac.uk/census). Samples were sequenced with each bait set as detailed below using fat/dermis from the same patient as a germline control. A list of all genes covered by the bait sets can be found in Table S3, and metrics of the bait sets are summarized below:
| Bait set name | Size (Mb) | Size (bases) | Number of Genes | Usage |
|---|---|---|---|---|
| Grid | 0.39 | 391,028 | 74 | 2mm2 samples |
| Punches+ follicles | 2.00 | 1,995,559 | 324 | 0.25 mm diameter punch samples and hair follicles |
Target-enriched samples were multiplexed and sequenced on HiSeq 2000 (Illumina) to generate 75bp paired-end reads. Whole genome sequencing was performed on either HiSeq X Ten or NovaSeq 6000 machines (Illumina) to generate 150bp paired end reads.
BAM files were mapped to the GRCh37d5 reference genome using BWA-mem (version 0.7.17) (21) and targeted sequencing was aligned using the GATK tool IndelRealigner (version 3.6.0) (22). Duplicate reads were marked using Biobambam2 (Biobambam2 version 2.0.86. https://gitlab.com/german.tischler/biobambam2, https://www.sanger.ac.uk/science/tools/biobambam). Depth of coverage was calculated using Samtools (version 0.1.18) to exclude reads which were: unmapped, not in the primary alignment, failing platform/vendor quality checks or were PCR/Optical duplicates. BEDTools (version 2.23.0) coverage program was then used to calculate the depth of coverage per base across samples (Table S2).
To determine which samples were suitable for WGS, we used the VAF from targeted sequencing to determine which samples were clonal. For the 0.25mm samples, WGS was performed on all clonal samples in the individuals studied. For the 2mm2 samples only eight out of 1261 samples were clonal and all of these were sequenced.
Variant calling
For targeted sequencing data sub-clonal mutation variant calling was made using the deepSNV R package (also commonly referred to as ShearwaterML), version 1.21.3, available at https://github.com/gerstung-lab/deepSNV, used in conjunction with R version 3.3.0 (2016-05-03) (5).
deepSNV makes use of statistical testing to differentiate sequencing errors from true low-frequency mutations, and has been shown to be reliable down to a detection limit of 1/10,000 alleles (23). The statistical tests compare by position and strand between skin samples and a panel of control samples to estimate how likely an observed nucleotide is a sequencing error or a true variant. Combining the information for each strand generates a single value used for filtering false positive variants. It was noted in development that the performance of deepSNV was not strongly dependent upon p, q-values or PCR amplifications, and its sensitivity can be increased through higher sequencing depths. A q-value of 0.01 was used to filter the variant calls.
Fat and dermis was used as the germline sample for each donor, and sequenced as outlined previously. Aligned germline BAM files for each corresponding sample type (grids, punches and follicles) were provided to deepSNV, excluding the germline for the sample being analysed, to form a normal sample panel used for statistical testing and false-positive variant identification. Variants called from a donor’s germline sample were subtracted from the list of variants called from non-germline samples belonging to the donor.
Mutations called by ShearwaterML were filtered using the following criteria
-
○
Positions of called SNVs must have a coverage of at least 100 reads (10 reads for 0.25mm diameter punch biopsy samples).
-
○
Germline variants called from the same individual were removed from the list of called variants.
-
○
The p-values of the putative mutations were adjusted with FDR and filtered with a q-value threshold of 0.01
-
○
Mutations not present in at least one read from both strands were removed.
-
○
Pairs of SNVs on adjacent nucleotides within the same sample are merged into a dinucleotide variant if at least 90% of the mapped DNA reads containing at least one of the SNV pair, contained both SNVs.
-
○
Identical mutations found in multiple contiguous tissue biopsies are merged and considered as a single clone in order to prevent duplicate clone counting.
For the hair follicles samples due to the very low input quantity of DNA, particularly towards the middle and base of the follicle, false positive variant calls attributed to sequencing artefacts are more likely. We therefore applied a strict method of variant calling with ShearwaterML in order to be as conservative as possible. This involved, for all samples within each follicle, calling variants against a custom normal panel consisting of all other follicle samples from all donors, plus the dermis/fat samples from all other donors. The corresponding dermis/fat sample for the donor of that follicle was then used to remove germline variants. For follicle spanning mutations only follicles where adjacent segments were successfully sequenced or where the same mutation is present within the same follicle, ie base and top, are shown Shearwater was run with a normal panel of >24,000, >31,000, and >12,000× mean coverage depth for the 2mm2 grid samples, hair follicles and punch biopsy samples respectively.
For whole genome sequencing data variants were called using the CaVEMan and Pindel algorithms (24) (25). For SNVs CaVEMan was run with the major copy number set 10 and the minor copy number set to 2. Only SNVs which passed all CaVEMan filters were kept. Additional filtering to remove mapping artefacts associated with BWA-MEM were: the median alignment score of reads supporting a variant had to be at least 140 and the number of clipped reads equal to zero. In addition, the proportion of mutant reads present in the matched sample also had to be zero. Variants with at least one mutant read present in the matched sample were also removed. Two SNVs called at adjacent positions within the same sample were merged to form a doublet-base substitution if at least 90% of the mapped DNA reads containing at least one of the SNV pair contained both SNVs. Small (<200bp) insertions and de1letions were called using Pindel. Only indels which passed all Pindel filters were kept. For the punch samples only, variants were filtered to remove a large excess of single base pair insertions at homopolymers of length five or more, an artefact likely caused by PCR amplification of low-input DNA concentrations during whole-genome sequencing. Indels were then classed as clonal if the VAF was at least 0.3.
Variants were annotated using VAGrENT (26). A full list of called variants from 2mm2 gridded samples, 0.25mm diameter punch samples and hair follicles are shown in Table S4, Table S7 and Table S8 respectively.
Clonal maps
Clonal maps were plotted using exonic mutations called from the targeted sequencing data. Coloured squares indicate samples dominated by a clone, after correcting for copy number. Polygons are used where more than two dominant clones occupy a sample, the clone with the larger VAF is shown as the larger polygon. White squares indicate polyclonal samples not dominated by a large clone.
Mutational signature analysis
Mutational spectra and signatures are described using the notation employed by the PCAWG Mutational Signatures working group (10). The frequency of mutations within each trinucleotide context was calculated using deconstructSigs (https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0893-4). SigProfiler, a method of non-negative matrix factorisation, was used to calculate transcriptional strand bias and was also used to estimate the contribution of 49 single-basesubstitution and 11 doublet-base-substitution reference signatures (originally characterised in human cancers) to the combined mutational spectrum of each donor (10).
Copy number calling
For whole genome sequencing data the copy number was inferred using ascatNgs using dermis/ fat from the same donor as a germline control (27).
SNP phasing
For targeted sequenced samples the copy number was estimated on a per gene basis by phasing heterozygous SNPs, as described previously (1,5). For each sample, a panel of all other samples from the same donor was used to call SNP loci.
Telomere length estimation
Telomere length was estimated using the Telomerecat software package (28). The telomere length given is a median of all chromosomes for all cells in that sample.
Phylogeny construction of whole genome sequenced punches
Substitutions were genotyped prior to phylogeny construction. For each sample in a donor, a pile-up of all quality reads was constructed and the number of mutant and wild-type reads counted for every locus that had a mutation called in any sample of that donor. Only reads with a mapping quality of at least 30 and bases with a base quality of at least 25 were included. Mutations with a VAF less than 0.2 were given a genotype of 0 and those with a VAF greater than or equal to 0.3 a genotype of 1. For mutations with a VAF between 0.2 and 0.3, the genotype was set to NA (not applicable) for the purposes of phylogeny construction. Maximum parsimony trees were drawn for each donor after 1000 iterations with MPBoot (29). Phylogenies for each donor were drawn with single-base and double-base substitutions combined and with single-base substitutions only. The addition of double-base substitutions did not alter the structure of the tree, only the branch lengths. Branch lengths of the final phylogenies were adjusted so that each double-base change counted as a single substitution. Substitutions were then re-assigned to each branch of a phylogeny. Each branch was annotated with exonic non-synonymous (missense, nonsense, essential splice, insertion or deletion) mutations in genes previously identified as being under positive selection by dNdScv in the targeted grid dataset. To reduce the number of false negatives, each phylogeny was annotated with a driver mutation in these genes if it was present in either the filtered wholegenome CaVEMan and Pindel calls, the targeted ShearwaterML calls and/or the targeted CaVEMan and Pindel calls with a VAF of at least 0.3. Copy number changes were assigned to each phylogeny manually.
Trinucleotide spectra were drawn for all single base substitutions that had been assigned to each branch of a phylogeny. A visible difference was noticeable in the frequency of T[C>T]A, T[C>T]T and G[T>C]T peaks in branches of the same donor. There are four single-base substitution reference signatures associated with damage by UV light (SBS7a-d) that have been characterised in cancers of the skin (10) We used a chi-square test to quantitatively compare the proportions of mutations in the most prominent peaks of these reference signatures (SBS7a and b: C[C>T]A, C[C>T]C, C[C>T]T, T[C>T]A, T[C>T]C and T[C>T]T; SBS7c: T[T>A]T; SBS7d: G[T>C]T) for each branch. Any branches with less than five mutations in any of these categories was removed from the analysis. The differences observed between clones of the same donor are unlikely to be due to sequencing artefact as they are common to branches of samples that share thousands of substitutions attributed to UV damage and are spatially adjacent in the tissue (i.e. are of the same clone). Since each branch is treated as an independent sample in the analysis, we can be confident that the differences observed in mutational spectra are due to biological differences of the cells within each clone.
Calculations of percentage mutant epithelia
The prevalence of BCC and SCC mutations were assessed using variant calls from previously published studies. Variant calls for BCC samples were taken from the publication by Bonilla et al (13). 126 whole exome sequenced samples, Agilent SureSelect Human All Exon V5 capture kit (50Mb), and 157 targeted samples capturing 387 cancer genes were used for BCCs. Variant calls from a total of 103 whole exome sequenced SCC samples were collated from five studies (12,30-33). Two studies used Agilent SureSelect Human All Exon V5 (50Mb) (12,31). The remaining studies each used: NimbleGen HGSCVCR EXome 2.1 design1 (42Mb) (30). Illumina Nextera Rapid Capture Exome Library Prep (37Mb) (32) and unspecified version of the Agilent SureSelect Human All Exon capture kit (33). Variant calls were filtered to remove low confidence variants below a coverage depth of 10x. Variant annotation was performed using VAGrENT to enable the calculation of the percentage of samples containing non-synonymous mutations for genes of interest. A weighted mean and the confidence interval (95%) were calculated for each gene.
For the current study percentage of mutant tissue and dN/dS analysis was performed as described previously (5). Mutation burden was calculated as per section 5.1 of the supplementary methods of (5). This method accounts for the synonymous footprint (Number of possible synonymous mutations) over the baitset.
To test for variation in selection between body sites, dN/dS ratios were compared using likelihood ratio tests, as used previously to compare selection between individuals (5).
Simulations of mutation number per biopsy
Simulations were run using a Wright-Fisher process (34) constrained to a 2-D hexagonal grid with periodic boundary conditions. At the start of a simulation, all cells in the grid are assigned a fitness of 1. In each step of the simulation, a new generation of cells pick their parents” from neighbouring cells in the previous generation (35). Each cell inherits the fitness of its parent unless a mutation occurs. Mutations are introduced at random to a number of cells in each generation. Most mutations are “neutral” and do not alter the cell fitness inherited from the parent cell, but a small proportion of mutations introduce a fitness advantage drawn from an exponential distribution. The new cell fitness is then the product of the parental cell fitness and the fitness advantage of the mutation. The fitter a cell compared to its neighbours, the more likely it is to be picked as a parent by cells in the next generation, and hence fitter cells produce more offspring (35). In this way, a fit cell is more likely to expand into a larger clone. Aside from the use of a Wright-Fisher style model to simulate one generation of cells in a single step, the simulations follow the same principles as a Moran-style model (single pairs of cell birth/stratification events per step) described previously (2). New generations of cells are assumed to occur every two weeks. Biopsies are then taken from the simulated grids after 3000 or 4000 weeks (~58, ~77 years), with a simulated coverage of 690 reads, a minimum of 5 mutant reads required for a clone to be observed, and 3·104 cells per 2mm2 biopsy (4). Simulations were run for grids of 40 adjacent biopsies (8×5 grid).
The simulations were run for a wide range of mutation rates and mutation fitness advantages. All combinations of the following parameters were run. The mutation rates used were 0.0000001, 0.000001, 0.00001, 0.0001, 0.001 or 0.01 mutations per cell per generation. The proportion of mutations which were non-neutral (changed the cell fitness) was 0.00001, 0.0001, 0.001, 0.01 or 0.1. The fitness advantages of the non-neutral mutations were drawn from exponential distributions with mean 0.01, 0.05, 0.1, 0.2, 0.5, 1 or 4. If the pre-mutation fitness of the cell was f and the fitness advantage drawn was s, then the post-mutation cell fitness was f × (1 + s).
Testing clustering of mutation counts
We used the Moran’s I test of spatial autocorrelation (36) to investigate whether environmental variation across each sampled region might be affecting mutation density. Clones which spread between multiple samples can create local clusters of higher mutation density if they contain many passenger mutations, but these clusters could be unrelated to environmental factors. To limit the effect of these clusters due to clonal spread, all mutations spanning multiple samples were excluded from this test.
We used the python package PySAL (37) to run Moran’s I. We used the p-value under the randomization assumption and multiple test correction was applied using the Benjamini-Hochberg method (38).
Protein structure analysis
ΔΔG calculation
We used FoldX 5 (39) to calculate ΔΔG, a measure of the change in protein folding energy caused by a mutation.
For each pdb file, the FoldX command RepairPDB was run to minimise steric clashes and optimise residue orientation. Then the FoldX command PositionScan was run for every residue of the proteins of interest in the structure, which mutates each residue to every other amino acid and calculates the ΔΔG value. Default FoldX settings were used for both the RepairPDB and PositionScan commands.
Calcium binding sites
The calcium binding residues in EGF11-12 of NOTCH1 were identified defined using MetalPDB (40) and the 2VJ3 (41).
NOTCH1 ligand binding interface
We used the residues previously identified as on the ligand binding interface (42).
Distance measurements in protein structures
Distances from residues in p53 to the DNA molecule were measured using the python package MDanalysis (43) and PDB 2AC0 (44).
Enrichment of pathogenic variants
We used the variant summary file of Clinvar annotations, https://ftp.ncbi.nlm.nih.gov/pub/clinvar/tab_delimited/variant_summary.txt.gz - downloaded 7th April 2020) to identify missense variants annotated as pathogenic or likely pathogenic.
The proportions of variants annotated as pathogenic/likely pathogenic in each gene were tested using the method described in Methods - Statistical testing for bias in mutation composition and a binomial test. Multiple test correction was applied using the Benjamini-Hochberg method (38).
Many pathogenic mutations are likely to be missing from the Clinvar database so the numbers of annotated pathogenic mutations we find per gene may be an underestimate of the true number of pathogenic mutations.
Statistical testing for bias in mutation composition
If certain types of mutations convey a fitness advantage to their cell of origin, then they will be more likely to form clones large enough to detect through sequencing. The observed mutations can be compared to the mutations expected under a neutral assumption (that all mutations in a gene have an equivalent impact) to test whether certain mutation features are selected for.
The trinucleotide mutational spectrum was used to estimate how frequently each possible single nucleotide mutation in a gene would appear in the data if there was no bias due to selection. Each possible mutation was scored for a particular metric and weighted using the spectrum to form the “neutral” null distribution. The observed distribution was then compared to the null to test for a significant bias in the metric scores. For example, if destabilising mutations provided a fitness advantage, we would expect the observed distribution of a destabilisation metric (like FoldX ΔΔG) to be skewed towards higher values than would expected under the neutral null hypothesis.
The test detects differences in relative selection in a gene or section of a gene. Therefore, if there are multiple selected features of mutations in the same region it becomes harder to detect selection of each individual feature. This can be mitigated by testing for a one feature after segmenting the mutations using the other features.
Full details of the method have been described previously (45).
For continuous metrics (ΔΔG, distance between the mutated residue and a particular set of atoms in the protein structure), a two-tailed Monte Carlo test was used to compare the null and observed distributions. 100000 random samples were taken from the null distribution, meaning the minimum p-value possible in this test was slightly less than 2e−5.
For true/false metrics (is the mutation annotated as pathogenic/likely pathogenic in Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/); is the mutation on an interface or on a calcium-binding site), a two-tailed binomial test was used. Where 95% confidence intervals are shown for the true/false distributions, the intervals were calculated by taking 10000 random samples from the null distribution or by bootstrapping 10000 random samples with replacement from the observed data.
Visualization of protein structures
Images of protein structures were generated using VMD (46).
Supplementary Material
Statement of significance.
Mapping mutant clones across the body reveals normal skin is a dense patchwork of mutant cells. The variation in cancer risk between sites substantially exceeds that in mutant clone density. More generally, mutant genes cannot be assigned as cancer drivers until their prevalence in normal tissue is known.
Acknowledgements
This work was supported by the Wellcome Trust to the Wellcome Sanger Institute (098051 and 296194) and Cancer Research UK Programme Grants to P.H.J. (C609/A17257 and C609/A27326). B.A.H. and M.W.J.H. are supported by Medical Research Council grants MC_UU_12022/9 and MR/S000216/1. M.W.J.H. was supported by Clare College, Cambridge. B.A.H. acknowledges support from the Royal Society (grant no. UF130039). S.D. benefited from an ESPOD fellowship, 2018-21, from the Wellcome Sanger and European Bioinformatics Institutes EMBL-EBI. AR is supported by a Cancer Research UK Clinician Scientist Fellowship (C64667/A27958). K.M. is supported by the Chan Zuckerberg Initiative. We thank the Cambridge Biorepository for Translational Medicine for access to human tissue.
Financial Support
This study was supported by Cancer Research UK, the Wellcome Trust, the Medical Research Council, the Royal Society, the Chan Zuckerberg Initiative, Clare College, Cambridge University, and the European Bioinformatics Institute EMBL-EBI.
Footnotes
Author Contributions
PJ and JF conceived study and devised experiment strategy with input from KF and AD. JF, EE and DFA performed the experiments, apart from the CRISPR-Cas9 knockout screen which was designed, performed and analysed by CB and JK. Data was analysed by CK, MH, RS, JF, SO, DS, with supervision from MG and BH. Clinical aspects of the study were performed by AD, KF, DM, AR, KM, K S-P. ER performed histological review and processed samples. Manuscript was written by PJ, JF, CK, MH, CB, SD and MG.
Declaration of Interests
The authors declare no competing interests.
Conflict of Interest: The authors declare no potential conflicts of interest.
Data and code availability statement
The sequencing datasets generated during study are available at the European Genome-phemone Archive (EGA). 2mm2 grid samples (Accession number EGAS00001001933, EGAS00001002165), 0.25mm diameter punch and hair follicle samples (Accession number EGAS00001002708) and whole genome sequencing (Accession number EGAS00001002416) Code for the simulations of mutations counts per sample https://github.com/michaelhall28/Fowler_et_al_scripts https://github.com/michaelhall28/darwinian_shift https://github.com/michaelhall28/clone-competition-simulation
References
- 1.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348(6237):880–6. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall MWJ, Jones PH, Hall BA. Relating evolutionary selection and mutant clonal dynamics in normal epithelia. Journal of the Royal Society Interface. 2019;16(156):20190230. doi: 10.1101/480756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramaniam P, Olsen CM, Thompson BS, Whiteman DC, Neale RE. Anatomical Distributions of Basal Cell Carcinoma and Squamous Cell Carcinoma in a PopulationBased Study in Queensland, Australia. JAMA Dermatol. 2017;153(2):175–82. doi: 10.1001/jamadermatol.2016.4070. [DOI] [PubMed] [Google Scholar]
- 4.Bergstresser PR, Pariser RJ, Taylor JR. Counting and sizing of epidermal cells in normal human skin. The Journal of investigative dermatology. 1978;70(5):280–4. doi: 10.1111/1523-1747.ep12541516. [DOI] [PubMed] [Google Scholar]
- 5.Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362(6417):911–7. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerstung M, Papaemmanuil E, Campbell PJ. Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics (Oxford, England) 2014;30(9):1198–204. doi: 10.1093/bioinformatics/btt750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murai K, Skrupskelyte G, Piedrafita G, Hall M, Kostiou V, Ong SH, et al. Epidermal Tissue Adapts to Restrain Progenitors Carrying Clonal p53 Mutations. Cell stem cell. 2018;23(5):e8. doi: 10.1016/j.stem.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madsen RR, Vanhaesebroeck B, Semple RK. Cancer-Associated PIK3CA Mutations in Overgrowth Disorders. Trends in molecular medicine. 2018;24(10):856–70. doi: 10.1016/j.molmed.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574(7779):532–7. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vöhringer H, Gerstung M. Learning mutational signatures and their multidimensional genomic properties with TensorSignatures. bioRxiv. 2019:850453. doi: 10.1101/850453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inman GJ, Wang J, Nagano A, Alexandrov LB, Purdie KJ, Taylor RG, et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nature communications. 2018;9(1):3667. doi: 10.1038/s41467-018-06027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. doi: 10.1038/ng.3525. http://www.nature.com/ng/journal/v48/n4/abs/ng.3525.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 14.Page ME, Lombard P, Ng F, Gottgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell stem cell. 2013;13(4):471–82. doi: 10.1016/j.stem.2013.07.010. S1934-5909(13)00318-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colom B, Alcolea MP, Piedrafita G, Hall MWJ, Wabik A, Dentro SC, et al. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat Genet. 2020;52(6):604–14. doi: 10.1038/s41588-020-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019;568(7753):511–6. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 17.Chen C-H, Li W, Xiao T, Xu H, Jiang P, Meyer CA, et al. Integrative analysis and refined design of CRISPR knockout screens. bioRxiv. 2017 [Google Scholar]
- 18.Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey Oliver M, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell reports. 2016;17(4):1193–205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nature protocols. 2017;12(4):828–63. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biology. 2014;15(12):554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. 2013 1303.3997. [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstung M, Beisel C, Rechsteiner M, Wild P, Schraml P, Moch H, et al. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nature communications. 2012;3(1):811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
- 24.Jones D, Raine KM, Davies H, Tarpey PS, Butler AP, Teague JW, et al. cgpCaVEManWrapper: Simple Execution of CaVEMan in Order to Detect Somatic Single Nucleotide Variants in NGS Data. Current protocols in bioinformatics. 562016:0.1-.0.8. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raine KM, Hinton J, Butler AP, Teague JW, Davies H, Tarpey P, et al. cgpPindel: Identifying Somatically Acquired Insertion and Deletion Events from Paired End Sequencing. Current protocols in bioinformatics. 522015:7.1-.7.2. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menzies A, Teague JW, Butler AP, Davies H, Tarpey P, Nik-Zainal S, et al. VAGrENT: Variation Annotation Generator. Current protocols in bioinformatics. 2015;52(1):8.1-.8.1. doi: 10.1002/0471250953.bi1508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raine KM, Van Loo P, Wedge DC, Jones D, Menzies A, Butler AP, et al. ascatNgs: Identifying Somatically Acquired Copy-Number Alterations from Whole-Genome Sequencing Data. Current protocols in bioinformatics. 2016;56:9.1-.9.7. doi: 10.1002/cpbi.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farmery JHR, Smith ML, Lynch AG. Telomerecat: A ploidy-agnostic method for estimating telomere length from whole genome sequencing data. Scientific reports. 2018;8(1):1300. doi: 10.1038/s41598-017-14403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang DT, Vinh LS, Flouri T, Stamatakis A, von Haeseler A, Minh BQ. MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC evolutionary biology. 2018;18(1):11. doi: 10.1186/s12862-018-1131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20(24):6582–92. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.South AP, Purdie KJ, Watt SA, Haldenby S, den Breems N, Dimon M, et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. The Journal of investigative dermatology. 2014;134(10):2630–8. doi: 10.1038/jid.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yilmaz AS, Ozer HG, Gillespie JL, Allain DC, Bernhardt MN, Furlan KC, et al. Differential mutation frequencies in metastatic cutaneous squamous cell carcinomas versus primary tumors. Cancer. 2017;123(7):1184–93. doi: 10.1002/cncr.30459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durinck S, Ho C, Wang NJ, Liao W, Jakkula LR, Collisson EA, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer discovery. 2011;1(2):137–43. doi: 10.1158/2159-8290.Cd-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright S. Evolution in Mendelian Populations. Genetics. 1931;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etheridge A. Springer Science & Business Media; 2011. Some Mathematical Models from Population Genetics; p. 119. [Google Scholar]
- 36.Moran PAP. Notes on Continuous Stochastic Phenomena. Biometrika. 1950;37(1/2):17–23. doi: 10.2307/2332142. [DOI] [PubMed] [Google Scholar]
- 37.Rey SJ, Anselin L. PySAL: A Python Library of Spatial Analytical Methods. In: Fischer MM, Getis A, editors. Handbook of Applied Spatial Analysis: Software Tools, Methods and Applications. Springer Berlin Heidelberg; Berlin, Heidelberg: 2010. pp. 175–93. [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 39.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Research. 2005;33(2):W382–W8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putignano V, Rosato A, Banci L, Andreini C. MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Research. 2017;46(D1):D459–D64. doi: 10.1093/nar/gkx989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cisinhibition. Nature structural & molecular biology. 2008;15(8):849–57. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355(6331):1320–4. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gowers RJ, Linke M, Barnoud J, Reddy TJE, Melo MN, Seyler SL, et al. MDAnalysis: A python package for the rapid analysis of molecular dynamics simulations. In: Benthall S, Rostrup S, editors. Proceedings of the 15th Python in Science Conference; Austin, TX. 2016. pp. 98–105. SciPy. [DOI] [Google Scholar]
- 44.Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, et al. Structural Basis of DNA Recognition by p53 Tetramers. Molecular Cell. 2006;22(6):741–53. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Hall MWJ, Shorthouse D, Jones PH, Hall BA. Investigating structure function relationships in the NOTCH family through large-scale somatic DNA sequencing studies. bioRxiv. 2020 [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing datasets generated during study are available at the European Genome-phemone Archive (EGA). 2mm2 grid samples (Accession number EGAS00001001933, EGAS00001002165), 0.25mm diameter punch and hair follicle samples (Accession number EGAS00001002708) and whole genome sequencing (Accession number EGAS00001002416) Code for the simulations of mutations counts per sample https://github.com/michaelhall28/Fowler_et_al_scripts https://github.com/michaelhall28/darwinian_shift https://github.com/michaelhall28/clone-competition-simulation