Abstract
The majority of targeted therapies for non-small-cell lung cancer (NSCLC) are directed against oncogenic drivers that are more prevalent in patients with light exposure to tobacco smoke1–3. As this group represents around 20% of all patients with lung cancer, the discovery of stratified medicine options for tobacco-associated NSCLC is a high priority. Umbrella trials seek to streamline the investigation of genotype-based treatments by screening tumours for multiple genomic alterations and triaging patients to one of several genotype-matched therapeutic agents. Here we report the current outcomes of 19 drug–biomarker cohorts from the ongoing National Lung Matrix Trial, the largest umbrella trial in NSCLC. We use next-generation sequencing to match patients to appropriate targeted therapies on the basis of their tumour genotype. The Bayesian trial design enables outcome data from open cohorts that are still recruiting to be reported alongside data from closed cohorts. Of the 5,467 patients that were screened, 2,007 were molecularly eligible for entry into the trial and 302 entered the trial to receive genotype-matched therapy, including 14 that re-registered to the trial for a sequential trial drug. Despite pre-clinical data supporting the drug–biomarker combinations, current evidence shows that a limited number of combinations demonstrate clinically relevant benefits, which remain concentrated in patients with lung cancers that are associated with minimal exposure to tobacco smoke.
In the case of cancer treatment, stratified medicine is a therapeutic strategy whereby the genotype of a tumour is used to match the patient to an appropriate targeted therapy. This strategy was first realized for the treatment of NSCLC when mutations in the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) were identified as the molecular basis of the clinical responses observed in patients treated with the EGFR tyrosine kinase inhibitor gefitinib1. In NSCLC, the majority of targetable alterations tend to occur in the cancers of patients who have never smoked or who are former light smokers. In tobacco-associated lung adenocarcinoma (LUAD) there are few actionable aberrations, and in squamous-cell lung cancer (LUSC) there are no options for targeted therapy.
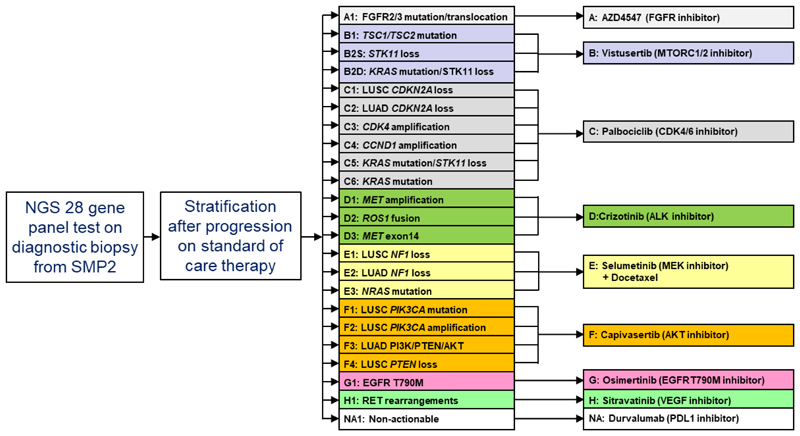
Here we report the current results of the ongoing National Lung Matrix Trial (NLMT), the largest national NSCLC umbrella study. NSCLC genotyping using next-generation sequencing (NGS) is used to stratify patients into one of 22 single-arm signal-of-activity studies, testing 8 different drugs (Fig. 1). Screening was performed on the 28-gene NGS panel from Cancer Research UK’s Stratified Medicine Programme (SMP-2) (details provided in Methods). To embed the programme into National Health Service (NHS) practice, the majority of tumours sequenced were obtained from formalin-fixed paraffin-embedded material surplus to requirements of the diagnostic work-up. Aberrations in targeted genes were tiered for oncogenic relevance using published data: tier 1 or tier 2 aberrations were eligible for inclusion4. For cases in which molecular exclusion criteria determined eligibility for the cohort (see Supplementary Information), these genes had to be read with sufficient depth to confidently call the molecularly excluded genes as wild type. Patients were eligible for entry into the trial after progression on all standard-of-care therapy; however, entry was permitted if patients refused standard treatment.
Fig. 1. Scheme of the umbrella phase II NLMT, recruiting patients with advanced NSCLC.
Patients are stratified using the 28-gene NGS panel test results from SMP2, and the trial is currently testing 8 different targeted drugs (A–H) in 22 different actionable biomarker cohorts. The trial also includes a cohort of patients with no actionable aberrations (NA).
Bayesian design for timely data release
The trial uses a Bayesian adaptive design5 to determine whether there is sufficient evidence of activity in any cohort to warrant further research. Assessment of a patient’s response to treatment is performed every 6 weeks by computerised tomography (CT) scanning and application of response evaluation criteria for solid tumours (RECIST), version 1.1. The primary outcome measures for signals of activity are either a confirmed objective response (OR) and a durable clinical benefit (DCB, defined as progression-free survival at 24 weeks, the time of the fourth on-treatment CT assessment of response) or the progression-free survival time (PFS), with the choice of outcome depending on the expected mode of action of the targeted agent. Target recruitment for each cohort is 30 patients, with futility analyses at 15 patients. Forest plots are used to display the Bayesian estimates—together with 95% credible intervals—for the true OR rate, DCB rate and median PFS, across the cohorts given the observed data and minimally informative priors. Results on closed and open cohorts are differentiated using solid and dashed lines, respectively. Waterfall plots are used to illustrate the best change in the sum of target lesion diameters. We report Bayesian posterior probabilities (PP) of a clinically relevant outcome for closed cohorts (pre-specified clinically relevant outcomes defined as: median PFS greater than 3 months, DCB rate and/or OR rate greater than 30% for single agents and greater than 40% for combination therapies) and predictive probabilities of success (PPoS) given current observed data for open cohorts—that is, the probability of a ‘go’ decision for consideration of further clinical evaluation when the cohort reaches n = 30 given the current data. Details of statistical analyses and justification of the sample sizes are provided in the Methods.
Attrition from screening to enrolment
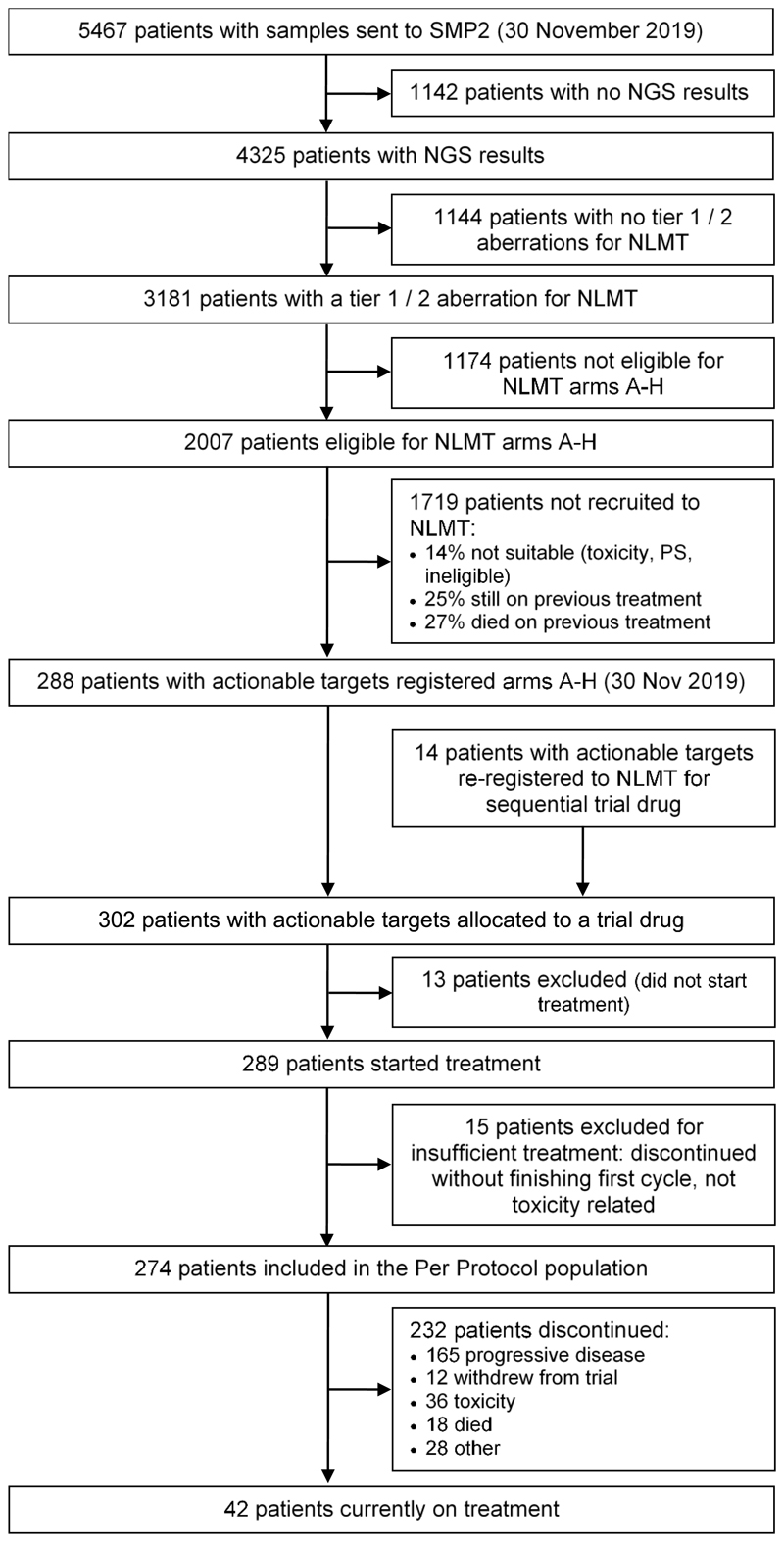
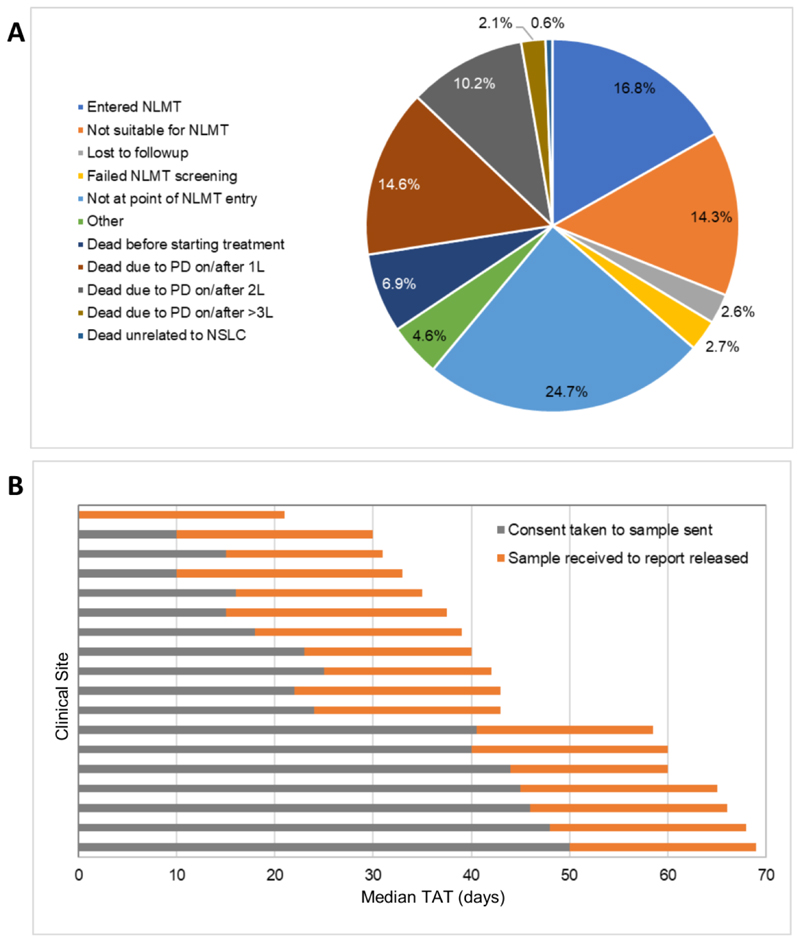
We report results from 19 cohorts (11 closed and 8 open) from the NLMT (Fig. 1; we exclude 3 cohorts E1, E3 and H1 that currently have fewer than 3 patients): each cohort represents a separate NSCLC genotype matched to its selected targeted agent. As of 30 November 2019, samples from 5,467 patients had been submitted for screening. There were 3,181 out of 5,467 patients with tier 1 or tier 2 mutations, of which 2,007 patients were molecularly eligible for entry into the trial (Fig. 2). A total of 288 out of 5,467 patients were stratified, giving a transition rate of 5.3% from samples submitted or 14% of molecularly eligible patients. To assess the reasons for attrition from screening to enrolment, a snapshot analysis of 1,433 molecularly eligible patients was undertaken (Extended Data Fig. 1a) and revealed that 25% were still on standard anticancer therapy, 7% died before starting any treatment, 15% died on or shortly after first-line standard therapy, 10% died on or shortly after second-line standard therapy, and 14% had ongoing toxicity, poor performance status or symptomatic brain metastases that precluded recruitment. The median turnaround time from the receipt of a sample to release of the results is 19 working days (interquartile range (IQR) 14–24 days) (Extended Data Fig. 1b). The median turnaround time from receiving a patient’s informed consent to be screened to the sample being sent for testing is 30 working days (range 0–50 days).
Fig. 2. Flow diagram showing the progress of patients through SMP2 and NLMT arms A–H as of 30 November 2019.
Of 302 patients recruited to the trial to receive a targeted treatment at data snapshot taken on 30 November 2019 —including 14 who were re-registered to the trial for a sequential trial drug—276 patients received sufficient treatment to be analysed for primary outcome measures (Fig. 2). The baseline characteristics are shown in Extended Data Table 1: well-documented smoking status was available for 84% of the trial patients. The molecular profile of recruited patients is shown in Extended Data Fig. 2.
Outcomes from 19 drug–biomarker cohorts
Results for the 19 cohorts are grouped into 4 modules of genomic aberrations on the basis of pathways identified from the analyses of lung cancers undertaken by The Cancer Genome Atlas (TCGA)6,7 (1) a cell cycle progression gene module, encompassing alterations in CDKN2A, CCND1 and CDK4; (2) an activated RAS module, including mutation of KRAS and loss of NF1; (3) an altered receptor tyrosine kinase (RTK) module, encompassing genomic aberrations in FGFR2 and FGFR3 (mutation and translocation), MET (amplification and mutation), ROS1 (fusion) and EGFR T790M mutation; and (4) a PI3K/PTEN/AKT/mTOR module encompassing mutation or amplification of PIK3CA, loss of PTEN, and mutations in AKT1-3 and in TSC1 and TSC2. The matching of drug to genomic aberration was based on a thorough review of all the available in vitro and in vivo lung-cancer-specific and other relevant pre-clinical data pertinent to the therapeutic targeting of each genomic aberration (Supplementary Information). Dosages and schedules are provided in Extended Data Table 2.
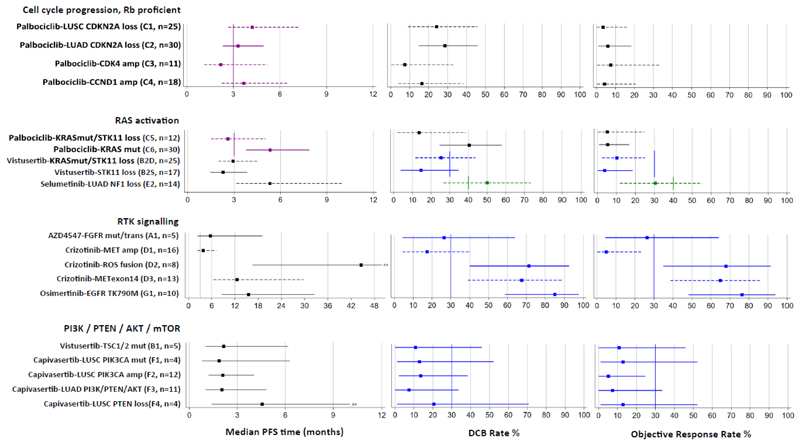
Bayesian estimates for the primary outcome measures—PFS, DCB and OR—are illustrated in Fig. 3 and tabulated in Extended Data Table 3, including the PP for closed cohorts and the PPoS for cohorts that are still open to recruitment. Plots of posterior probability distributions for median PFS, DCB rate and OR rate are shown in Extended Data Fig. 3, depth of response is illustrated by waterfall plots in Extended Data Fig. 4, and adverse reactions by drug are documented in Extended Data Table 4.
Fig. 3. Estimates of primary outcome measures for 19 drug-biomarker cohorts in NLMT grouped according to 4 modules of genomic aberrations.
Forest plots show Bayesian estimates and 95% credible intervals for true values of median PFS, DCB rate and OR rate. Purple is used to highlight estimates for which PFS is the primary outcome measure, with vertical lines showing a pre-specified clinically relevant target of a median PFS of 3 months. Green or blue is used to highlight estimates for which DCB and OR rates are co-primary outcome measures, with vertical lines showing pre-specified clinically relevant target rates of 40% or 30% respectively. Cohorts that are closed to recruitment are represented by solid lines and those still open are represented by dashed lines. Bayesian estimates are the medians of the posterior probability distributions derived from the current data and minimally informative priors. Because the trial is ongoing and the follow-up is not complete (including in some closed cohorts), these estimates are subject to change as the trial continues.
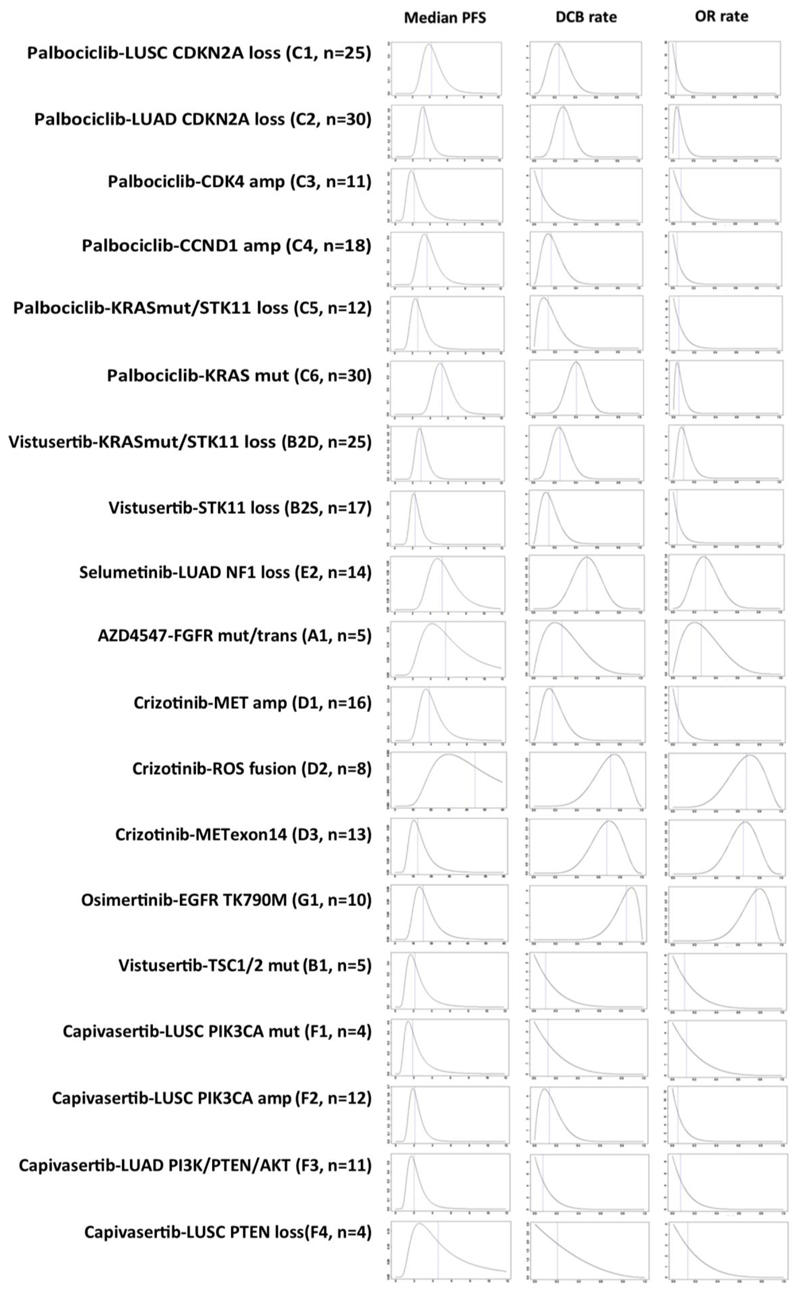
In patients with cancers that harboured aberrations in cell cycle progression genes, we assessed the effect of inhibiting CDK4 and CDK6 using palbociclib. Palbociclib is an approved agent for the treatment of breast cancer, thus its clinical efficacy has been already demonstrated. Cohorts are annotated as follows: ‘drug name - genomic aberration targeted (trial cohort label)’. The cohorts in this module are: palbociclib - LUSC CDKN2A loss (C1), palbociclib - LUAD CDKN2A loss (C2), palbociclib - CDK4 amplification (C3) and palbociclib - CCND1 amplification (C4). The current Bayesian estimates for median PFS (the primary outcome measure) range from 2.2 months (95% credible interval: 1.1–5.2) for patients with CDK4 amplification to 4.2 months (95% credible interval: 2.7–7.2) for patients with LUSC with CDKN2A loss, wherein 4 out of 18 patients obtained DCB. Recruitment continues in these cohorts, with PPoS of 0.18 and greater than 0.99, respectively. The closed cohort of patients with LUAD with CDKN2A loss achieved its primary outcome, with a median PFS of 3.3 months (95% credible interval: 2.3–5.0, PP = 0.69). There is only one confirmed objective response in 69 patients evaluable for response to palbociclib across all 4 cohorts.
We assessed the effect of three different treatments for targeting aberrations activating KRAS. The cohorts in this module are as follows: palbociclib - KRAS mutation with no concomitant aberration activating AKT (AKT activation abrogates RAS-induced senescence mediated by CDK4 loss (Supplementary Information)) (C6), palbociclib -KRAS mutation/dual STK11 loss (C5), vistusertib (inhibitor of mTORC1 and mTORC2) -KRAS mutation/dual STK11 loss (B2D), vistusertib – STK11 loss only (B2S) and docetaxel + selumetinib (MEK inhibitor) - LUAD NF1 loss (E2). The Food and Drug Administration have recently approved selumetinib for paediatric patients with germline loss of NF1 who develop symptomatic inoperable plexiform neurofibromas8. Numerically, the highest median PFS of all the palbociclib-treated cohorts is the closed RAS mutant cohort (C6), at 5.3 months (95% credible interval: 3.8–7.9, PP > 0.99) with DCB rate 40% (95% credible interval: 25–58%). The dual STK11 loss/RAS mutant palbociclib cohort (C5) currently has a median PFS of 2.6 months (95% credible interval: 1.5–5.0) and continues to recruit with PPoS of 0.27. A second dual STK11 loss/RAS mutant cohort (B2D) was treated with vistusertib—pre-clinical data strongly suggested the need to target mTORC2 as well as mTORC1 when trying to reverse the metabolic reprogramming in cancers with STK11 loss/KRAS mutation (Supplementary Information). Currently, 2 out of 25 patients have shown confirmed responses, and 6 out of 25 patients obtained DCB in this cohort, which continues to recruit with a PPoS for DCB of 0.13. The single STK11-loss cohort (B2S) was closed at interim for futility. There is encouraging preliminary data for the docetaxel + selumetinib combination in the NF1 loss LUAD cohort (E2): 4 out of 14 patients currently have confirmed OR, and the estimated DCB rate is 50% (95% credible interval: 27–73%; PPoS = 0.89).
We assessed the effect of targeting mutations, amplifications and fusions of genes encoding several receptor tyrosine kinases. The cohorts in this module are as follows: AZD4547 (FGFR inhibitor) – FGFR2 and FGFR3 mutations and translocations (A1), crizotinib (Met inhibitor) - MET amplification (D1), crizotinib (also ROS inhibitor) – ROS1 fusion (D2), crizotinib - MET exon 14 skipping mutations (D3) and osimertinib (EGFR inhibitor) - EGFR T790M mutation (G1). One out of five patients had a confirmed response to FGFR inhibition using AZD4547, with the response ongoing after more than 20 months. The tumour from this patient harbours an FGFR translocation (FGFR2-MBIP_F19:M1)—none of the patients with FGFR mutations (FGFR2(V515L), FGFR3(P575S), FGFR3(S758P), FGFR3(S294C)) responded or obtained a DCB. This cohort is now closed owing to drug availability issues. For patients with cancers harbouring MET amplifications (six or more gene copies, D1), 0 out of 13 patients currently show a response to crizotinib, and the estimated DCB rate is 17% (95% credible interval: 4–41%). In the MET exon 14 skipping mutation cohort (D3), the estimated confirmed OR rate is currently 65% (8 out of 12 patients responded, 95% credible interval: 39–86%) and the DCB rate is 68% (95% credible interval: 39–89%). Both MET alteration cohorts continue to recruit, with PPoS for DCB of 0.07 and greater than 0.99, respectively. The crizotinib-ROS1 fusion (D2) and osimertinib-EGFR mutation (T790M) (G1) cohorts were closed early owing to the licensing of these drugs in these indications; however, even with small numbers (n = 8 and 10, respectively) the trial clearly confirms substantial efficacy, with OR rates of 68% (95% credible interval: 35–92%; PP = 0.99) and 76% (95% credible interval: 48–94%; PP > 0.99), respectively.
We assessed the effect of inhibiting mTOR (activated via a TSC1 or TSC2 mutation mutation) using vistusertib or inhibiting AKT using capivasertib in patients with cancers harbouring genomic aberrations that activated AKT. In patients with PI3K/PTEN/AKT-altered triple-negative breast cancer, capivasertib significantly improved PFS in combination with paclitaxel when compared with paclitaxel alone9; this provides clinical proof of the relevance of targeting activated AKT with this drug. The cohorts in this module are as follows: vistusertib - TSC1 and TSC2 mutation (B1), capivasertib - LUSC PIK3CA mutation (F1), capivasertib - LUSC PIK3CA amplification (F2), capivasertib - LUSC PTEN loss (F4) and capivasertib - LUAD with aberrations of PI3K/PTEN or AKT genes (F3). Zero out of five patients with cancers harbouring TSC1 and TSC2 mutations responded or obtained DCB using vistusertib. Of 28 patients across 4 cohorts with cancers harbouring PIK3CA or PTEN aberrations, no patient responded to AKT inhibition using capivasertib, and only one patient obtained a DCB. These cohorts have been closed owing to futility.
Histology, smoking history and outcome
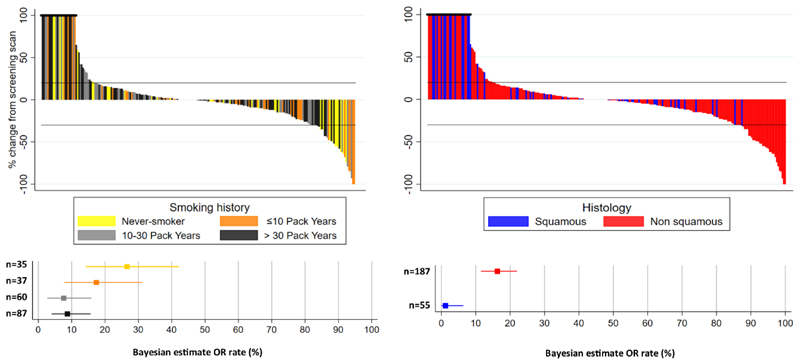
Data on smoking history was available for 253 (84%) of the 302 patients in the trial (Extended Data Table 1). A waterfall plot including the smoking-history data for all trial patients (Fig. 4) shows that decreases in the sum of target-lesion diameters are mainly seen for patients that have never smoked or those with a low cumulative smoking duration, and these are largely patients that have known actionable genomic aberrations. A total of 30 out of 187 patients with non-squamous NSCLC had a confirmed OR to genotype-matched targeted therapy, whereas 0 out of 55 LUSC patients had a confirmed OR to stratified therapy. Excluding actionable aberrations that have become apparent during the timeframe of NLMT, there were 9 confirmed ORs out of all patients screened across the entire study, 4 of which were in the NF1 LUAD cohort treated with docetaxel + selumetinib.
Fig. 4. OR rates and best percentage change in sum of target lesion diameters across 19 reported cohorts in NLMT according to smoking history and histology.
a, Top, waterfall plot shows, for each patient, the best percentage change in sum of target lesion diameters according to RECIST v.1.1. Bars are coloured according to the patient’s smoking history. Patients who discontinued before assessment, or had a value greater than 100% were capped at 100%. Bottom, forest plots show Bayesian estimates (with 95% credible intervals) of OR rates, grouped according to the smoking history of the patients. b, As for a, but coloured and grouped according to the histology of the tumour.
Optimising precision medicine outcomes
Precision medicine has transformed the outcomes of many patients with NSCLC, and the continued search for new genotype-directed stratified therapies remains a high priority. Whilst uncertainty remains regarding the patient benefit of some drug–biomarker cohorts in the NLMT, recruitment to the trial will continue. In addition, the trial provides an ongoing platform on which to test potential new drug–biomarker combinations as they emerge. There are four essential elements that a precision medicine study needs in order to deliver on its goal of discovering new targeted therapy options: the right genomic targets, the right drugs to target those genomic alterations, the right infrastructure to run the study and the right patient population.
Pre-clinical work is essential to identify promising biomarker–drug combinations, but the models used must recapitulate the genomic context and evolutionary trajectory of the targeted genomic alteration. Tobacco-associated NSCLCs harbour many more clonal mutations compared with NSCLCs in non-smokers10, which increases the chance that other oncogenic drivers could co-exist with the targeted genomic aberration. Ongoing genomic instability driven largely by tobacco exposure, APOBEC and mitotic clock signatures11—combined with extensive somatic copy-number alterations (SCNAs) — may also lead to the rapid evolution of resistance. A reduction in clinical benefit with increasing cumulative smoking duration12 and higher complexity of the mutational landscape13 has been demonstrated in patients with EGFR mutations treated with EGFR tyrosine kinase inhibitors. Although we relied on extensive pre-clinical data to inform our biomarker–drug selections, a substantial amount of data was generated using models that lacked other genomic aberrations besides the targeted alteration. Genetically engineered mouse models of NSCLC have mutational burdens more than 100-fold lower than that of human disease14. Carcinogen-induced models have few SCNAs15. Non-malignant cells that are engineered to harbour single genomic aberrations (as used to select mutations treated with AZD454716) fail to replicate genomic complexity. The results of targeting common SCNAs were disappointing. There was a stark difference in the activity of crizotinib in patients with cancers that harbour MET amplification compared with MET exon 14 mutations: the former is characterized by greater genomic instability and can be heterogeneous, the latter often occurs in non-smokers in cancers without concurrent driver mutations17. Chromosomal copy-number amplicons can encode multiple genes, all of which might subtly affect the phenotype. Using two genomic datasets—TCGA and TRACERx (tracking non-small cell lung cancer evolution through therapy)—we investigated whether this was the case with PIK3CA amplification. We found that there are indeed a substantial number of potential drivers that are co-amplified with PIK3CA and, furthermore, that there is considerable heterogeneity of amplicon size across individual tumours (Extended Data Figs. 5, 6).
Some biomarker–drug combinations may fail because the selected drug fails to adequately inhibit the selected target. The differences in outcome for patients with RET-fusion NSCLC treated with multi-kinase inhibitors18 or with highly selective RET inhibitors19 is a paradigm example of the importance of selecting optimal best-in-class agents to match to selected targets: we abandoned a RET-fusion cohort treated with the multi-kinase inhibitor sitravatinib on the basis of these data. It is essential that robust pharmacodynamics in the relevant cancer are obtained during the early phases of testing20. Although treatment with vistusertib reduced tumour pS6 levels in all patients, the effects on p4EBP1 were modest21, and these pharmacodynamic data were generated with daily dosing rather than with the higher pulsatile doses that were used in this study.
The screening platform is an essential infrastructural element in precision medicine studies. Our screening turnaround time was clearly slower than that of some commercial providers. Although this is unlikely to have substantially affected the entry of participants into this trial—because the majority of screening was performed at diagnosis and patients were enrolled after standard therapy—prolonged waits for biopsy slots and slow recovery of blocks for testing suggest that testing for circulating tumour DNA at the time of trial entry might increase the number of participants that enter the trial22. Finally, we revealed very large attrition rates, which highlight the scale of the screening effort required to run studies that treat patients after the completion of standard-of-care therapies in lung cancer. Analysis of circulating tumour DNA in the minimal residual disease setting might be a viable way of performing precision-medicine studies in diseases for which rapid progression and deterioration of performance status when treated with standard anticancer therapies is still common. It is hoped that the lessons learnt from the current data from the NLMT will help to inform the next wave of precision medicine studies.
Methods
SMP2 study design and patient eligibility
The SMP2, funded by Cancer Research UK (CRUK), is an observational pre-screening study for advanced lung cancer that was launched in 2014. Patients with locally advanced or late-stage metastatic NSCLC (stage III or IV) that were not eligible for primary surgery or radical radiotherapy and with a performance status of 0–2 were eligible for the study. All patients were requested to give signed informed consent for the trial and genomic analyses.
Patients were recruited to SMP2 through an extended network of 23 hospitals, called clinical hubs (CHs), which include the 18 Experimental Cancer Medicine Centres (ECMCs) and 5 non-ECMC centres. Through a hub-and-spoke model, patients from local feeder hospitals were also referred via the CHs for enrolment in the study, with a total of over 50 hospitals involved in the study.
The clinical sites obtained patients’ consent for screening on SMP2 at either primary diagnosis or at relapse using a local consent form or the specific CRUK SMP2 consent form. After consenting, a sample from a diagnostic biopsy together with matched blood was sent to one of the three technology hubs (THs) for molecular testing. The THs are either ISO 15189 or CPA-accredited NHS Molecular Genetics Laboratories located at Birmingham (BMH; West Midlands Regional Genetics Service), Cardiff (All Wales Medical Genetics Service) and at the Royal Marsden Hospital London (RMH; The Centre for Molecular Pathology) and are paired evenly across the CHs.
Samples required for molecular testing included sections from formalin-fixed paraffin-embedded (FFPE) samples from various sample types (prepared as 8 × 6 μm, 7 × 10 μm or 11 × 5 μm sections depending on the specific TH requirements) with ≥20% tumour content and medium–high cellularity (>4,000 cells), or locally extracted DNA from tumour biopsies (70 ng of tumour DNA at a minimum concentration of 2 ng μl−1) together with a marked haematoxylin and eosin (H&E) slide and matched blood sample for germline comparison (minimum 4 ml EDTA). The tumour cellularity was assessed by a senior pathologist on an H&E slide from the same biopsy core as part of NHS routine care.
Matched tumour and blood samples or extracted DNA were sent from the CHs to their paired THs either frozen or at room temperature with accompanying paperwork.
DNA extraction
Tumour DNA and germline DNA from whole-blood samples were extracted at the THs using either the Maxwell 16 FFPE Plus LEV DNA Purification Kit (Promega; Cardiff and BMH THs) and Qiagen DNA extraction kit (Qiagen; RMH) according to the manufacturer’s instructions. The extracted DNA was quantified using Qubit broad range blood or high-sensitivity FFPE assay according to the manufacturer’s instructions. Samples with DNA concentrations lower than 50 ng were failed.
Panel design
The custom SMP2 v.01 panel was designed using illumina DesignStudio, a web-based design tool that converts target regions to capture probes. Probes were designed at the maximum allowed density (Probe Spacing = Overlapping). SMP2 targets in 28 genes were identified by pharma partners and CRUK. The extent of the region targeted varied across the 28 genes. Some genes required only an individual exon to give a readout of a particular hotspot (for example, AKT1), whereas others required all exons plus intronic regions to assist with somatic copy number alteration (SCNAs) calling or complete tiling of specific introns for structural variant detection.
Genetic variants were identified through a custom Nextera Rapid Capture sequencing assay (Illumina). Illumina sequencing libraries were generated from 50 ng of DNA samples (FFPE and blood) using Nextera transposons which simultaneously fragment and add sequencing adapters to the DNA. Libraries from 5 tumour normal pairs were pooled (500 ng tumour, 250 ng matched normal) and enriched for regions of interest using a custom SMP2 panel following the standard protocol. Enriched libraries were diluted to 13 pM and sequenced on the Illumina MiSeq system using paired 75-bp reads according to the manufacturer’s recommendation.
In March 2017, the SMP2 v.01 panel was updated to SMP2 v.02. Although the majority of the desired target regions remained the same, some changes were made to improve the performance of the panel. Specifically, for highly repetitive intronic regions required to detect fusions and alternative splicing events, the number of probes in the least unique regions were reduced to lower the coverage in off-target regions. Conversely, more probes in regions where more coverage was desired—including the target region around MET exon 14 to cover all previously characterized deletion events in this region—were included. Furthermore, the coverage of genes with high failure rates in panel v.01 was improved (RB1 and FGFR3).
There were also substantial changes to the target regions used for copy-number calling, in which extra target regions were included to increase their resolution; in addition, a more even scattering of target regions across the genome was provided to increase the information available to make a copy-number call. Finally, we added targets for single-nucleotide polymorphisms (SNPs) common in the population23 to confirm that tumour and normal samples were derived from the same individual.
Pipeline for analysis and variant classification
Sequencing reads were aligned to hg19 using iSAAC (Illumina). Variant calling was performed using Strelka24 for single-nucleotide variants, CRAFT (Illumina) for SCNAs and Manta25 for structural variants. Variant call files were gathered into an Excel report to assist with summarizing and reporting the data. A bespoke SMP2 app was created on Illumina BaseSpace to automate the process.
The panel can detect single-nucleotide variants and indels at >5% frequency (10/200 reads). SCNA calling was attempted on all tumours that were NGS-tested, although SCNAs could be confidently detected by NGS in samples with a high tumour percentage (>60% tumour content) and if the SCNA was large. Low-level or suspected SCNAs were confirmed by fluorescence in-situ hybridization (FISH) before reporting to the CHs. Similarly, FISH was used to confirm deletions identified by NGS and determine whether the deletion was homozygous or heterozygous.
Observed aberrations were tiered on the basis of a lab manual compiled and updated by the Pharma partners (Pfizer and AstraZeneca) from published data. Aberrations were classified as tier 1, tier 2 and tier 3, where tiers 1 and 2 denote aberrations that confer eligibility for one or more NLMT cohorts and tier 3 contains non-NLMT genes.
FISH analyses
FISH analyses were performed for the following genes to confirm SCNAs and deletions identified by NGS and were divided between the 3 THs: MET and ROS1 (performed by RMH TH), PIK3CA, PTEN and CCND1 (performed by Cardiff TH), and CDK4 and CDKN2A (performed by BMH TH). All probes were purchased from Cytocell and used according to the manufacturers’ instructions.
NLMT design and procedures
NLMT is a multi-centre, multi-arm, umbrella phase II platform trial in advanced NSCLC, recruiting from 24 approved hospitals across the UK (ClinicalTrials.Gov NCT02664935, ISRCTN38344105, EudraCT 2014-000814-73). Each arm is testing an experimental targeted drug intervention in a population stratified by multiple pre-specified target biomarkers. A Bayesian adaptive design5 is used to screen each experimental targeted drug for signals of efficacy in each selected molecularly-defined cohort. In addition, patients with no actionable genetic change (NA) are included and treated with the latest drug from a sequential pipeline of experimental agents (data not reported here).
As a platform trial with an adaptive design, the number of treatment arms and molecularly defined cohorts is dynamic. The trial design reported here includes 8 targeted drugs (initially 7) being tested in 22 molecularly-defined cohorts (initially 20) (Fig. 1) and results from 19 are reported here. The protocol is currently being developed for the inclusion of a further 2 treatment arms and 3 new cohorts, and further arms and cohorts may be added subject to funder and regulatory approval. Details of the delivery of each treatment according to the protocol are specified in Extended Data Table 2.
The trial opened to recruitment in March 2015 and is ongoing. This interim report of results relates to a data freeze on 16 December 2019 and included only patients recruited to the trial by 30 November 2019. At this time, 12 cohorts were closed to recruitment (all reported here except H1) and 10 continue to recruit (8 reported here, with E1 and E3 excluded).
Eligible patients had received previous anticancer treatment or refused standard-of-care first-line therapy; had provided an adequate specimen to adequately characterize the molecular genotype of the tumour in the molecular pre-screening according to the molecular exclusion rules; had histologically or cytologically confirmed NSCLC stage III (not suitable for radical radiotherapy or surgery) or stage IV; had CT or magnetic resonance imaging (MRI) scanning of head, chest, abdomen demonstrating measurable disease according to response evaluation criteria in solid tumours (RECIST) v.1.1; had adequate haematological, hepatic and renal function; were at least 18 years of age and using adequate contraceptive measures. Patients had no known: major surgery in the 4 weeks before trial treatment; nausea, vomiting, chronic gastrointestinal diseases or other issues that would preclude adequate drug absorption; psychological, familial, sociological or geographical condition hampering protocol compliance; concurrent malignancies diagnosed in the last 3 years (except for adequately treated basal cell carcinoma of the skin and in situ carcinoma of the uterine cervix); unresolved toxicity of grade 2, 3 or 4 from previous treatment; evidence of severe or uncontrolled systemic diseases, including active bleeding diatheses; active infection including hepatitis B, hepatitis C and human immunodeficiency virus; pregnancy or were lactating. Patients that were judged to be unlikely to comply with study procedures, restrictions and requirements were also excluded. Specific eligibility criteria is applicable for each treatment arm on the basis of the investigator brochure or summary of product characteristics requirements. All patients gave written informed consent in accordance with good clinical practice guidelines and the Declaration of Helsinki’s ethical principles for medical research.
Patient registration into the trial by the treating clinician was by telephone to the central registration service at the Cancer Research UK Clinical Trials Unit at the University of Birmingham, where the appropriate experimental treatment was allocated to the patient according to the molecular stratification.
The trial complied with all regulatory requirements; ethical approval for the trial protocol (currently v.8.0, dated 18 October 2019) was obtained from South Central - Oxford C Research Ethics Committee and clinical trial authorisation was granted by UK Medicines and Healthcare products Regulatory Agency (MHRA). The trial was sponsored by the University of Birmingham and run by the Cancer Research UK Clinical Trials Unit located there. Funding for the trial came from Cancer Research UK with drugs provided by pharmaceutical partners. The trial was initiated and conducted independently by the trial investigators in collaboration with Cancer Research UK. The corresponding author had full access to all the data in the trial and had final responsibility for the decision to submit for publication. The independent Trial Steering Committee reviewed the interim data every 6 months to ensure patient safety and are responsible for making decisions to close cohorts early and to share interim data with investigators and pharmaceutical industry partners. In particular, they endorsed the decision to publish the interim data.
Patient history of smoking was taken at baseline and self-reported measures of smoking status were taken at baseline, clinic visits and treatment discontinuation using a smoking questionnaire. Exhaled carbon monoxide (CO) measurements were also recorded at clinic visits using a MicroMedical Micro CO Monitor.
NLMT statistical analysis and justification for sample size
For treatment arms A, B, D, E, F, G and H, co-primary outcome measures are: (i) OR, defined as the incidence of a confirmed complete or partial response according to RECIST v.1.126 and (ii) DCB, defined as the incidence of remaining free of disease progression at the fourth scan at approximately 24 weeks from the start of treatment. For treatment arm C the primary outcome measure is PFS, defined as the time from commencement of trial treatment to the date of the CT scan at which progressive disease is first recorded, or date of death without previously recorded progression. The choice of primary outcome was based on the expected mode of action of the targeted agent.
Statistical analysis uses a Bayesian conjugate analysis to generate the posterior probability distribution for the primary outcome measure summary statistic to represent the signal of activity for each drug–biomarker cohort. For OR and DCB the analysis uses a beta-binomial conjugate analysis5 with a minimally informative Beta(1,1) prior distribution and for PFS an exponential-inverse-gamma conjugate analysis27 with a minimally informative inverse gamma prior distribution IG(0.001,0.001). From these posterior probability distributions, estimates (using the median of the posterior probability distributions) and 95% credible intervals for the true OR rate, DCB rate and median PFS are derived together with relevant probabilities (PP and PPoS) on which decisions for further recruitment or research are based. For any cohort, the influence of the minimally informative priors on estimates and probabilities will decrease as the sample size increases.
Target recruitment for each drug–biomarker cohort is 30 patients, with interim analyses after 15 patients to allow early termination for futility. Pre-specified guidelines for decision-making at interim and final analyses are specific to the treatment arms. For treatment arms A, B, D, F and G, if the interim PP shows a high chance (>0.9) that the true OR or DCB rate is <30% then the cohort is recommended for early closure. Further, if the final PP shows a moderate chance (>0.5) that the true OR and/or DCB rate >30% then the signal in that cohort is considered worthy of further investigation. The guidelines are similar for treatment arm E, but as it is testing a combination of agents, it has a higher clinically relevant cut-off of 40%. For treatment arm C, if the interim PP shows a high chance (>0.8) that the true median PFS is less than 3 months then the cohort is recommended for early closure. Further, if the final PP shows a moderate chance (>0.5) that the true median PFS is greater than 3 months then the signal in that cohort is considered worthy of further investigation. We report PP of a clinically relevant outcome as specified above for all closed cohorts.
For open cohorts that are continuing to recruit, we report interim results for primary outcomes using the PPoS. This is the probability of a ‘go’ decision for consideration of further clinical evaluation when the cohort reaches the target of 30 patients, given the minimally informative prior and the trial data observed at that point. This new approach provides insight, while the trial is ongoing, into the drug–biomarker combinations that have the strongest potential for further research.
Operating characteristics of the trial design were evaluated for the decision criteria specified above for a range of interim and final sample sizes to determine the appropriate number. The selected sample sizes of 15 patients for interim and 30 patients for final were selected because they gave an acceptable balance of error rates for this early phase trial. In summary, the operating characteristics for co-primary outcomes of DCB and OR and for PFS outcome (which depends on recruitment rate) were as follows: the chance of correctly stopping early at interim was greater than 0.60, the chance of incorrectly stopping early at interim was less than 0.05, the chance of a correct ‘go’ decision at final was greater than 0.80, and the chance of an incorrect ‘go’ decision at final was less than 0.15.
TCGA and TRACERx data processing
Analysis of whole exome sequencing (n = 327 samples from 100 patients) from TRACERx was performed as in Jamal-Hanjani et al.11. Copy number segmentation, tumour purity and ploidy for each sample were estimated using ASCAT28 as in Jamal-Hanjani et al.11. These data were used as input to a multi-sample SCNA estimation approach to produce genome wide estimates of the presence of loss of heterozygosity as well as loss, neutral, gain, and amplification copy number states relative to sample ploidy. The log ratio (LogR) values present in each copy number segment with ≥ 5 LogR values in all samples of a tumour were examined relative to three sample-ploidy adjusted LogR thresholds using one-tailed ttests with a P < 0.01 threshold. These LogR thresholds were equivalent to < log2[1.5/2] for losses, > log2[2.5/2] for gains and > twice sample ploidy for amplifications in a diploid tumour. Any segment not classified as a loss, gain or amplification was classed as neutral. For each segment, these relative to ploidy definitions were combined with loss of heterozygosity detection across all samples from a single tumour. SCNA segments demonstrating amplification/gain involving PIK3CA in any sample of an individual tumour were isolated for analysis, and oncogenes29 within them annotated. Affymetrix SNP6 profiles were obtained for paired tumour-normal samples from the TCGA [dataset ID: phs000178.v10.p8] and processed by using PennCNV libraries30 to obtain BAFs and LogR from each tumour-normal pair. LogR values were GC corrected using a wave-pattern GC correction method31. LogR and BAFs were processed with ASCAT 2.4.228 to identify SCNAs. To determine genome-wide copy-number gain, total copy number values in copy-number segmentation data for each sample were divided by the sample mean ploidy, then log2 transformed. Gains were defined as greater than log2[2.5/2]. Amplifications were defined as a total copy number greater than twice the sample ploidy plus an additional single copy. TCGA SCNA segments harbouring an amplification/gain involving PIK3CA were isolated for analysis, and oncogenes29 within them annotated.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
Extended Data Fig. 1. Reasons for attrition and median testing turnaround time in the SMP2 study.
a, Reasons why patients enrolled in SMP2 did not enter the NLMT were collected for a subset of patients (N=1433). PD, progressive disease; 1L, first-line treatment; 2L, second-line treatment; 3L, third-line treatment. b, Median turnaround time (TAT) of SMP2 testing. Turnaround time was measured in days from the 18 SMP2 clinical sites that recruited patients. This consists of the median time from when informed consent was received from the patient to enter SMP2 to the tissue sample being sent for testing (grey bars) and from receipt of the tissue sample at the SMP2 technical hubs to the release of the SMP2 screening report (orange bars).
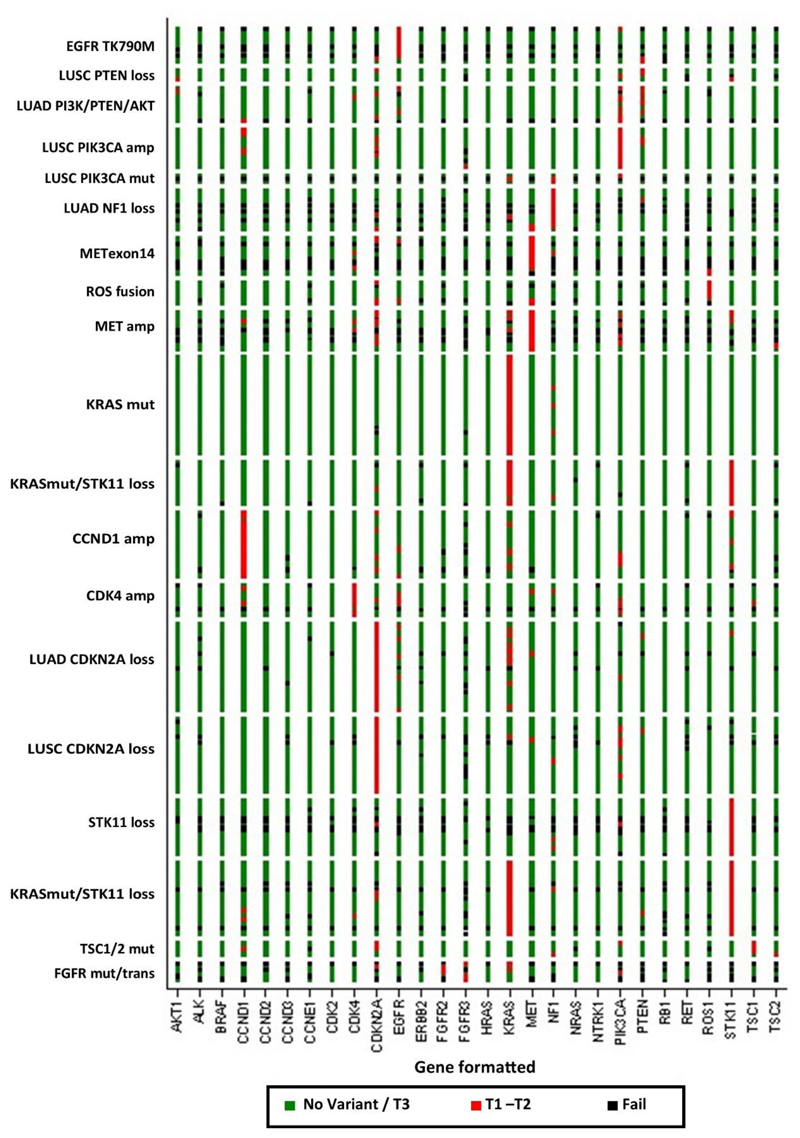
Extended Data Fig. 2. Heat map of all 28 genes for patients registered to the 19 reported cohorts in NLMT.
Detailed 28-gene NGS panel results were available for 283 patients included in the reported analysis, organized by molecular cohort and drug treatment. Green elements indicate wild-type or tier 3 aberration, red indicates a tier 1 or tier 2 aberration, and black a fail.
Extended Data Fig. 3. Posterior probability distribution plots by cohort for median PFS in months, DCB rate and OR rate.
Plots show the posterior probability distribution for true values of the relevant outcome measure, given the prior probability distribution and the observed data. The blue dotted line indicates the median of the posterior distribution. Given the prior and the observed data, there is an equal probability that the true value is greater than or less than the median value. Median PFS uses an inverse-gamma distribution with a prior of IG(0.001, 0.001), which provides minimal information and hence the posterior is dominated by the observed data in the trial. DCB and OR rate use a beta distribution, with a prior of Beta(1,1), which attributes equal probability to all possible rates of response from 0%–100%, and contributes data to the posterior equivalent to two trial patients. This will therefore be more influential at early stages of recruitment, but as more patients contribute their results the posterior will be dominated by the trial data. Bayesian estimates and 95% credible intervals for the true median PFS, DCB rate and OR rate are generated from these posterior probability distributions, together with PP and PPoS.
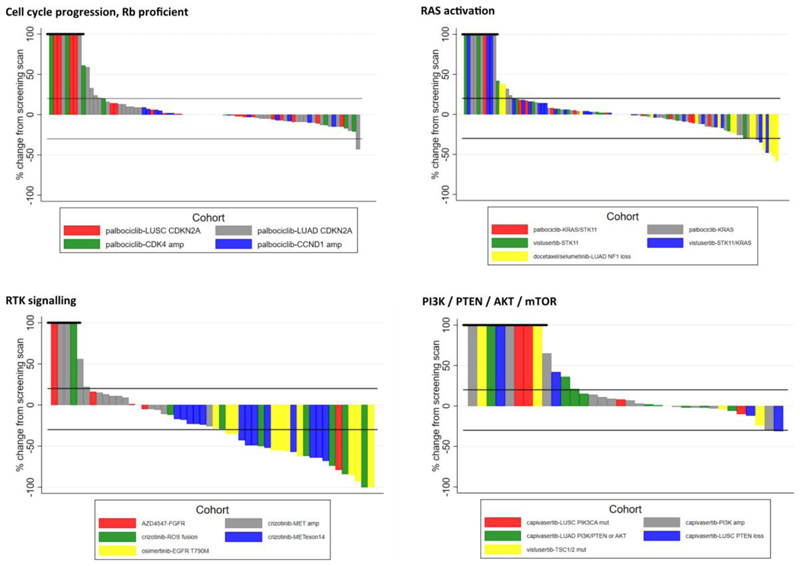
Extended Data Fig. 4. Waterfall plots for 19 drug–biomarker cohorts from NLMT.
Plots are grouped according to 4 genomic modules of genomic aberrations showing, for each patient, the best percentage change in sum of target lesion diameters according to RECIST.
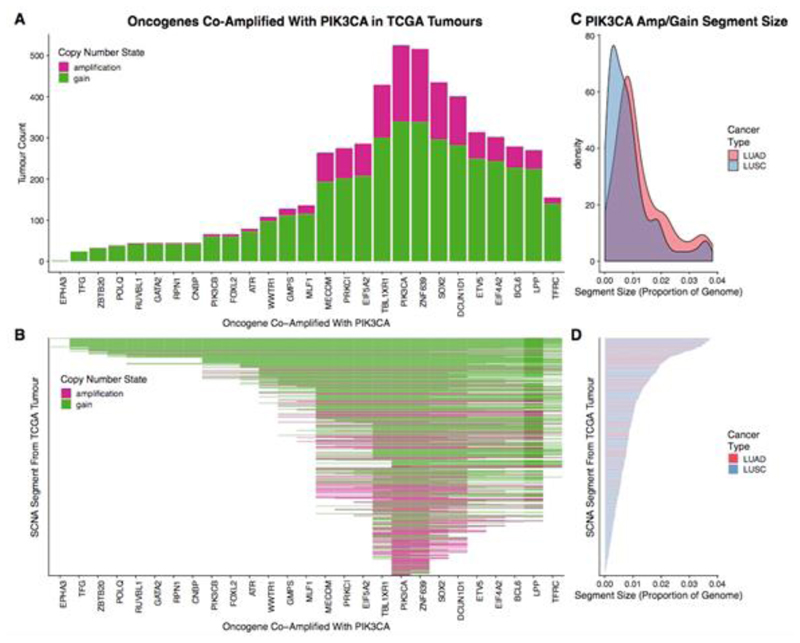
Extended Data Fig. 5. PIK3CA amplifications in the TCGA LUAD and LUSC cohort.
a, Bar plot of oncogenes that are in close genomic proximity to PIK3CA and are gained or amplified (co-amplified) on the same SCNA segment with PIK3CA. The height of the bars represents the number of tumours that have the particular oncogene co-amplified with PIK3CA. b, Heat map indicating whether oncogenes are co-amplified on the same SCNA segment with PIK3CA. In a, b, genes are ordered on the basis of genomic location. Dark pink shading indicates that the corresponding SCNA segment was amplified, whereas green shading indicates that the corresponding SCNA segment was gained. c, Density plot indicating the frequency of the sizes of SCNA segments harbouring an amplification or gain involving PIK3CA. The distribution representing LUAD cases is indicated in red, while the distribution representing LUSC cases is indicated in blue. d, Bar plot indicating the sizes of the SCNA segments harbouring the PIK3CA amplification/gain for each TCGA case. Bars are coloured according to cancer type (red, LUAD; blue, LUSC). SCNA segments in b, d are in the same order. Only TCGA cases with a PIK3CA gain or amplification (n = 524 of 1,010) are included in this plot.
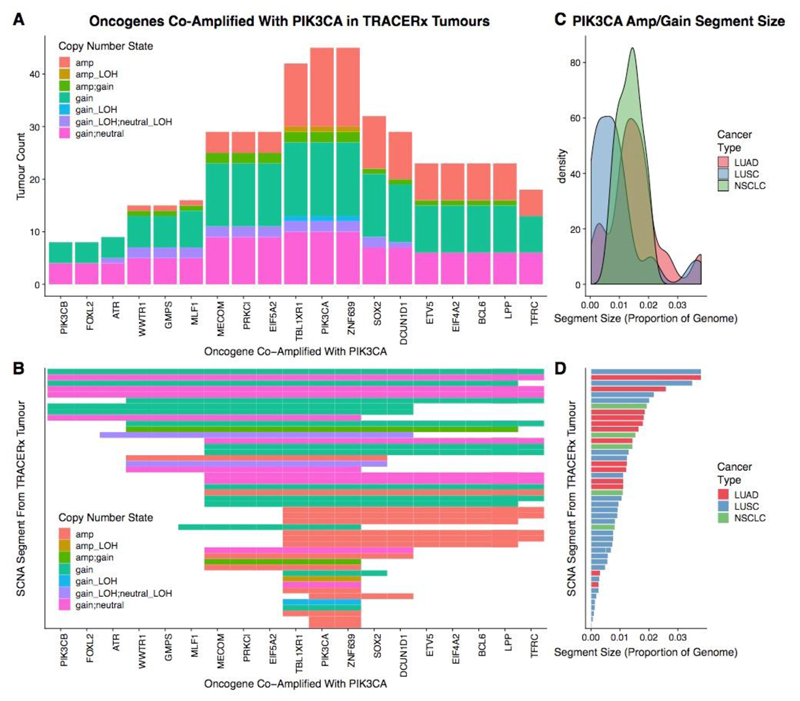
Extended Data Fig. 6. PIK3CA amplifications in the TRACERx 100 cohort.
a, Bar plot of oncogenes which are in close genomic proximity to PIK3CA and are gained or amplified (co-amplified) on the same SCNA segment with PIK3CA. The height of the bars represented the number of tumours that have the particular oncogene co-amplified with PIK3CA. b, Heat map indicating whether oncogenes are co-amplified on the same SCNA segment with PIK3CA. In a, b, genes are ordered on the basis of genomic location. The shading in the heat map indicates the type of SCNA affecting the genomic segment encompassing PIK3CA. As TRACERx data are multi-regional, some segments are assigned two different SCNAs (for example, “gain_neutral” indicates that this case harboured a subclonal gain in PIK3CA, where the gain was observed in some regions of that tumour, whereas other regions of that tumour were copy-number neutral at the same locus). c, Density plot indicating the frequency of the sizes of SCNA segments harbouring an amplification or gain involving PIK3CA. The distribution representing LUAD cases is indicated in red, the distribution representing LUSC cases is indicated in blue, and the distribution representing other NSCLC is indicated in green. d, Bar plot indicating the sizes of the SCNA segments harbouring the PIK3CA gain or amplification for each TRACERx case. Bars are coloured according to cancer type (red, LUAD; blue, LUSC; green, other NSCLC). SCNA segments in b, d are in the same order. Only TRACERx cases with a PIK3CA gain or amplification (n = 45 of 100) are included in this plot.
Extended Data Table 1. Baseline characteristics for patients recruited to targeted treatment arms in NLMT (all patients registered up to 30 November 2019).
| All (n=302) | |
|---|---|
| Age (years) | |
| Median | 65 |
| Inter-quartile range | 59-70 |
| Range | 26-86 |
| Performance Status | |
| 0 | 64 (21%) |
| 1 | 203 (67%) |
| 2 | 32 (11%) |
| Not known | 3(1%) |
| Histology | |
| Adenocarcinoma | 212 (70%) |
| Squamous cell carcinoma | 72 (24%) |
| Carcinoma NOS | 9 (3%) |
| Not known | 9 (3%) |
| Smoking History (pack years) | |
| Never-smoker | 40 (13%) |
| <10 | 46 (15%) |
| 10-30 | 67 (22%) |
| >30 | 100 (33%) |
| Not known | 49 (16%) |
| Metastases | |
| No | 100 (33%) |
| Yes | 197 (65%) |
| Not known | 5 (2%) |
| Previous lines (systemic anticancer treatment) | |
| One | 96 (32%) |
| Two | 104 (34%) |
| Three | 47 (16%) |
| More than three | 35 (12%) |
| Not known | 20 (7%) |
Extended Data Table 2. Details of targeted drugs included in the NLMT.
BD, twice a day; OD, once a day; NOS, Not Otherwise Specified.
| Treatment Arm | IMP | Route and formulation | Trial dose and schedule |
|---|---|---|---|
| A | AZD4547 FGFR inhibitor | Oral tablets (20 &80mg) | 80mg BD Continuous dosing 21 day cycles |
| B | Vistusertib MTORC1/2 inhibitor | Oral tablets (25 mg) | 125mg BD Intermittent dosing (2 continuous days in 7) 28 day cycles |
| C | Palbociclib CDK4/6 inhibitor | Oral capsules (75, 100 & 125 mg) | 125mg OD Intermittent dosing (21 days on, 7 days off) 28 day cycles |
| D | Crizotinib ALK inhibitor | Oral capsules (200& 250mg) | 250 mg BD Continuous dosing 21 day cycles |
| E | Selumetinib MEK inhibitor | Oral capsules (25 mg) | 75mg BD Continuous dosing 21 day cycles |
| Docetaxel chemotherapy | IV infusion over 30-60 minutes, concentrate for solution for infusion | 75mg/m2 3-weekly | |
| F | Capivasertib (AZD5363) AKT inhibitor | Oral tablets (160& 200mg) | 480 mg BD Intermittent dosing (4 days on, 3 days off) 28 day cycles |
| G | Osimertinib EGFRM+ and T790M+ inhibitor | Oral tablets (80mg) | 80 mg OD Continuous dosing 21 day cycles |
| H | Sitravatinib VEGFR Inhibitor | Oral capsules (40mg & 10mg) | 120 mg OD Continuous dosing 21 day cycles |
Extended Data Table 3. Primary outcome measures for each drug-biomarker cohort in NLMT.
Number of patients in the intention-to-treat (ITT) and per protocol populations are shown. For OR and DCB, the observed number is reported together with the denominator that shows the number of patients currently with sufficient follow-up data to be included in the analysis. Bayesian estimates of the true OR rate and DCB rate, given the current data and minimally informative priors, are reported together with 95% credible intervals (CrI). For PFS, Bayesian estimates of the true median PFS in months, given the current data and minimally informative priors, are reported together with 95% credible interval. For closed cohorts, the Bayesian PP is reported for primary outcomes, showing the probability that the true value is greater than the pre-specified clinically relevant targets as follows: OR and/or DCB rates of 30% for B2S, A1, D2, G1, B1, F1-F4; median PFS of 3 months for C2 and C6. For open cohorts, the PPoS is reported for primary outcomes, showing the probability of a go decision when the cohort reaches n = 30, given the current observed data, with pre-specified clinical relevant targets as follows: OR and/or DCB rates of 30% for B2D, D1, D3, and 40% for E2; median PFS of 3 months for C1, C3-C5.
| Cohort | ITT | Per Protocol | Median PFS (months) l95%Crlf | PPoS | PP | Observed DCB | DCB rate estimate (95%Crl) | PPoS | PP | Observed OR | OR rate eslimate (95%Crl) | PPoS | PP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell cycle progression Rb proficient | |||||||||||||
| Palbociclib-LUSC CDKN2A loss (C1) | 27 | 25 | 42 (2.7 - 7.2) |
>0.99 | (-) | 4/18 | 24% (9 - 48) |
(-) | (-) | 0/19 | 3% (0-17) |
(-) | (-) |
| Palbociclib-LUAD CDKN2A loss (C2) | 32 | 30 | 3.3 (2.3 - 5.0) |
(-) | 069 | 8/29 | 29% (15 - 46) |
(-) | (-) | 1/27 | 6% (1-18) |
(-) | (-) |
| Palboclclib-CDK4 amplification (C3) | 12 | 11 | 22 (1.1-5.2) |
0.18 | (-) | 0/8 | 7% (0-34) |
(-) | (-) | 0/8 | 7% (0 - 34) |
(-) | (-) |
| Palbociclib-CCND1 amplification (C4) | 22 | 18 | 3.7 (2.3 - 6,5) |
0.85 | (-) | 2/15 | 16% (4 - 38) |
(-) | (-) | 0/15 | 4% (0-21) |
(-) | (-) |
| RAS activation | |||||||||||||
| Palbociclib-KRAS mutation+ dual STK11 loss (C5) | 15 | 12 | 26 (1.5 - 5.0) |
0.27 | 1/11 | 14% (2 - 39) |
(-) | (-) | 0/12 | 5% (0 - 25) |
(-) | (-) | |
| Palbociclib-KRAS mutation (C6) | 33 | 30 | 5.3 (3.8 - 7.9) |
(-) | >0.99 | 12/30 | 40% (25- 53) |
(-) | (-) | 1/30 | 5% (1-17) |
(-) | (-) |
| Vistusertib-KRAS mutation + dual STK11 loss (B2D) | 28 | 25 | 2.9 (2.0 - 4.6) |
(-) | 6/25 | 25% (12 - 44) |
0.13 | (-) | 2/25 | 10% (2 - 25) |
O.01 | (-) | |
| Vistusertib-STKl l loss (B2S) | 19 | 17 | 23 (1.5 - 3.8) |
(-) | 2/17 | 15% (4 - 35) |
(-) | 006 | 0/17 | 4% (0 - 19) |
(-) | O.01 | |
| Selumetinib-LUAD NF1 loss(E2) | 14 | 14 | 5.3 (3.2 - 10.0) |
(-) | 7/14 | 50% (27 - 73) |
ose | (-) | 4/14 | 31% (12 - 55) |
0.17 | (-) | |
| RTK Signalling | |||||||||||||
| AZD4547-FGFR mutations/translocations (A1) | 5 | 5 | 5.6 (2.4 - 19.0) |
(-) | (-) | 1/5 | 26% (4 - 64) |
(-) | 0.42 | 1/5 | 26% (4 - 64) |
(-) | 0.42 |
| Crizotlnib-Met amplification (D1) | 19 | 16 | 3.8 (2.3 - 7.2) |
(-) | (-) | 2/14 | 17% (4-41) |
007 | (-) | 0/13 | 5% (0 - 23) |
<0.01 | (-) |
| Crizotlnib-ROS fusion (D2) | 8 | 8 | 44.6 (16 5- 192.7) |
(-) | (-) | 6/8 | 71% (40 - 93) |
(-) | >0.99 | 5/7 | 68% (35 - 92) |
(-) | 0.99 |
| Crizotlnib-Met eKon 14 skipping mutations (D3) | 13 | 13 | 12.5 (6 4 - 29.7) |
(-) | (-) | 7/10 | 68% (39 - 89) |
>0 99 | (-) | 8/12 | 65% (39 - 86) |
>0.99 | (-) |
| Osimertinib-EGFR T790M (G1) | 10 | 10 | 15.5 (8.5 - 32.6) |
(-) | (-) | 9/10 | 85% (59 - 98) |
(-) | >0.99 | 8/10 | 76% (48 - 94) |
(-) | =0.99 |
| PI3K / PTEN / AKT i mTOR | |||||||||||||
| Vlstuser1lb-TSC1/2 mutation (B1) | 5 | S | 2.1 (1.0 - 6.2) |
(-) | (-) | 0/5 | 11% (0 - 46) |
(-) | 0.12 | 0/5 | 11% (0-46) |
(-) | 0.12 |
| Capivasertlb-LUSC PIK3CA mutation (F1) | 5 | 4 | 1.9 (0.8 - 6.3) |
(-) | (-) | 0/4 | 13% (1 - 52) |
(-) | 0-17 | 0/4 | 13% (1 -52) |
(-) | 0.17 |
| Capivasertlb-LUSC PI3K amplification (F2) | 14 | 12 | 2.1 (1.2 - 4.1) |
(-) | (-) | 1/11 | 14% (2 - 39) |
(-> | 0-09 | 0/12 | 5% (0 - 25) |
(-) | O.01 |
| Caplvasertib-LUAD with aberrations PI3K/PTEN/AKT (F3) | 12 | 11 | 20 (1.0 - 4.8) |
(-) | (-) | 0/8 | 7% (0-34) |
(-) | 004 | 0/8 | 7% (0 - 34) |
(-) | 0.04 |
| Capivasertlb-LUSC PTEN loss (F4) | 4 | 4 | 4.6 (1.4 - 31.6) |
(-) | (-) | 0/2 | 21% (1-71) |
(-) | 0.34 | 0/4 | 13% (1 -52) |
(-) | 0.17 |
Extended Data Table 4. Adverse reactions by treatment arm in NLMT.
The table shows for each treatment arm, the number (and percentage) of patients with an adverse reaction reported at any grade (1-4), at grade 2 and above (2-4), at grade 3 and above (3-4) and at grade 4. For those patients with at least one adverse reaction reported, the table shows the median, interquartile range (IQR) and range for the number of adverse reactions per patient.
| Highest AR grade reported | ||||||
|---|---|---|---|---|---|---|
| Treatment arm | 1-4 | 2-4 | 3-4 | 4 | Median (IQR) | Range |
| AZD4547 (n = 6) | 4 (67%) | 4 (67%) | 2 (33%) | 0 (0%) | 19.5(12 - 29) | 10 - 33 |
| Vistusertib (n=53) | 42 (79%) | 33 (62%) | 18 (34%) | 0 (0%) | 9 (4 - 17) | 1 - 39 |
| Palbociclib (n=142) | 115 (81%) | 91 (64%) | 44 (31%) | 1 (1%) | 11 (6 - 16) | 1 - 59 |
| Crizotinib (n=40) | 36 (90%) | 24 (60%) | 8 (20%) | 0 (0%) | 20 (9 - 30) | 1 - 88 |
| Selumetinib/Docetaxel (n=17) | 17(100%) | 16 (94%) | 9 (53%) | 1 (6%) | 20 (12 - 27) | 3 - 50 |
| Capivasertib (n=35) | 30 (86%) | 26 (74%) | 17 (49%) | 0 (0%) | 9 (7 - 15) | 1 - 28 |
| Osimertinib (n = 10) | 10(100%) | 6 (60%) | 1 (10%) | 0 (0%) | 25.5(11 - 33) | 8 - 47 |
| Sitravatinib (n=1) | 1 (100%) | 1 (100%) | 1 (100%) | 0 (0%) | 16 (16 - 16) | 16 - 16 |
Supplementary Material
Acknowledgements
The Investigators and Sponsor thank all the patients and their families who participated in this trial, as well as the NHS Trusts and staff and the members of the Trial Steering Committee, chaired by R. Kaplan, who have supported this trial. We thank our patient and public representatives, M. Baker on the Trial Steering Committee and T. Haswell on the Trial Management Group. This is an investigator-initiated and investigator-led trial funded by CRUK grant C11497/A19363 and C11497/A22209. Trial drugs were supplied free of charge by AstraZeneca (AZD4547, vistusertib, selumetinib, capivasertib, osimertinib and durvalumab), Pfizer (palbociclib and crizotinib) and Mirati (sitravatinib), who also supported the programme through the funding of SMP2. The Cancer Research UK Clinical Trials Unit at the University of Birmingham is supported by CRUK grant C22436/A25354. The National Lung Matrix Trial was supported by Experimental Cancer Medicine Centres (ECMC) funding and by the ECMC Network. This trial has been independently peer reviewed and has been adopted by the National Institute for Health Research Clinical Research Network Portfolio.
Footnotes
Author contributions
G.M., L.B. and P.F. designed the study. All analysis was performed by G.M., P.F. and L.B., with the exception of SMP2 data (Extended Data Fig. 1), which was analysed by T.C.M. and M.A.C., and TCGA and TRACERx data processing (Extended Data Figs. 5, 6), which was performed by E.L. and T.B.K.W. Interpretation of data was performed by G.M., L.B., P.F., J. Savage and C.S. The trial was managed by G.M., J. Savage, D.W., M.Mehmi., R.S., A.F. and S.P. The manuscript was written by G.M., L.B., P.F., J. Savage, T.C.M. and M.A.C. Patient recruitment was carried out by G.M., S.P., Y.S., A.G., D.G., J.C., N.O., A.B., E.T., J. Spicer, P.J., A.D., M.Mackean. and M.F. Lead Arm Investigators were G.M., S.P., T.A.Y., Y.S. and J. Spicer. All authors critically reviewed the manuscript.
Competing interests
G.M. reported receiving research funding from Bristol-Myers Squibb, Kael-Gemvax, Merck Sharp & Dome and Plexxikon, and personal fees from BioLineRx, Boehringer Ingelheim, Bristol-Myers Squibb, Merck Sharp & Dome and Roche. S.P. reported receiving research funding from Boehringer Ingelheim, Epizyme, Bristol-Myers Squibb, Clovis Oncology, Roche, Eli Lilly and Takeda, and personal fees from Boehringer Ingelheim, AstraZeneca, Roche, Takeda, Chugai Pharma, Merck Sharp & Dohme, Bristol-Myers Squibb, EMD Serono, Abbvie, Guardant Health, Pfizer and Novartis. J. Savage reported receiving personal fees from Eli Lilly. Y.S. reported personal fees from Roche, AstraZeneca, Takeda, Pfizer and Merck Sharp & Dohme. A.G. reported personal fees from Pfizer, Boehringer Ingelheim, Merck Sharp & Dohme, Novartis, AstraZeneca, Bristol-Myers Squibb, Takeda, Abbvie, Roche and Foundation Medicine. D.G. reported receiving personal fees from AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Genesis Care UK, Pfizer, Roche, Takeda and Merck Sharp & Dohme and non-financial competing interests with Roy Castle Lung Cancer Foundation. E.T. reported receiving personal fees from Roche, Pfizer, Boehringer Ingelheim and Merck Sharp & Dohme. J. Spicer reported receiving research funding from Starpharma, Taiho Pharmaceutical, Bristol-Myers Squibb, Roche, BerGenBio, Genmab and Curis, personal fees from Bristol-Myers Squibb, Lytix Biopharma and IObiotech and owning stocks and shares in IGEA. P.J. reported receiving personal fees from AstraZeneca, Boehringer Ingelheim, Eli Lilly and Pfizer. T.Y. reported research funding from AstraZeneca, Vertex, Pfizer, Bayer, Tesaro, Jounce Therapeutics, Eli Lilly, Seattle Genetics, Kyowa Hakko Kirin, Constellation Pharmaceuticals and personal fees from AstraZeneca, Pfizer, Tesaro, EMD Serono, Vertex, Seattle Genetics, Roche, Janssen, Clovis Oncology, Ignyta, Atrin Pharmaceuticals, Aduro Biotech, Merck Sharp & Dohme, Almac Group, Bayer, Bristol-Myers Squibb, Calithera Biosciences and Cybrexa Therapeutics. C.S. reports receiving research funding from Boehringer Ingelheim and personal fees from Genentech, Roche, Sarah Cannon Research Institute, Boehringer Ingelheim, GlaxoSmithKline, Eli Lilly, Celgene, Ono Pharmaceutical, SERVIER, Pfizer, Bristol-Myers Squibb, Novartis, AstraZeneca and Ilumina and owning stocks and shares in Epic Sciences, Apogen Biotechnologies, GRAIL and Achilles Therapeutics. J.C. reported receiving personal fees from Novartis, Eli Lilly and Boehringer Ingelheim. A.F. reported receiving research funding from Ethicon (Johnson and Johnson). R.S., T.C.M. and M.C. were employed by Cancer Research UK who fund the study. L.B. reports receiving personal fees from AstraZeneca, Novartis and Springer. The other authors declare no competing interests.
Data availability
For NLMT data, scientifically sound proposals from appropriately qualified Research Groups will be considered for data sharing. Requests should be made by returning a completed Data Sharing Request Form and curriculum vitae of the lead applicant and statistician to newbusiness@trials.bham.ac.uk. The Data Sharing Request Form captures information on the specific requirements of the research, the statistical analysis plan, and the intended publication schedule. The request will be reviewed independently by the Cancer Research UK Clinical Trials Unit (CRCTU) Directors at University of Birmingham in discussion with the Chief Investigator and relevant Trial Management Group and independent Trial Steering Committee. In making their decision the Director’s Committee will consider the scientific validity of the request, the qualifications of the Research Group, the views of the Chief Investigator, Trial Management Group and Trial Steering Committee, consent arrangements, the practicality of anonymising the requested data and contractual obligations. Where the CRCTU Directors and appropriate Trial Committees are supportive of the request, and where not already obtained, consent for data transfer will be sought from the Sponsor of the trial before notifying the applicant of the outcome of their request. It is anticipated that applicants will be notified of a decision within 3 months of receipt of the original request. The results published here are based in part upon data generated by The Cancer Genome Atlas pilot project established by the NCI and the National Human Genome Research Institute. The data were retrieved through database of Genotypes and Phenotypes (dbGaP) authorization (accession number : phs000178.v10.p8). TRACERx sequencing datasets used in this study are described in Hanjani et al.11 and Abbosh et al.32
Code availability
NLMT Statstical analysis code is available for download from the Github repository https://github.com/pfletchergit/NLMT_Nature2020.
References
- 1.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Bergethon K, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr TH, et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer. 2016;16:319–329. doi: 10.1038/nrc.2016.35. [DOI] [PubMed] [Google Scholar]
- 5.Berry SM, Carlin BP, Lee JJ, Muller P. CRC; 2010. Bayesian adaptive methods for clinical trials. [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koselugo (selumetinib) approved in US for paediatric patients with neurofibromatosis type 1 plexiform neurofibromas. 2020 https://www.astrazeneca.com/media-centre/press-releases/2020/koselugoselumetinib-approved-in-us-for-paediatric-patients-with-neurofibromatosis-type-1-plexiform-neurofibromas.html. [Google Scholar]
- 9.Schmid P, et al. Capivasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer: the PAKT trial. J Clin Oncol. 2020;38:423–433. doi: 10.1200/JCO.19.00368. [DOI] [PubMed] [Google Scholar]
- 10.Jamal-Hanjani M, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim IA, Lee JS, Kim HJ, Kim WS, Lee KY. Cumulative smoking dose affects the clinical outcomes of EGFR-mutated lung adenocarcinoma patients treated with EGFR-TKIs: a retrospective study. BMC Cancer. 2018;18:768. doi: 10.1186/s12885-018-4691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Offin M, et al. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer Res. 2019;25:1063–1069. doi: 10.1158/1078-0432.CCR-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFadden DG, et al. Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci USA. 2016;113:E6409–E6417. doi: 10.1073/pnas.1613601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westcott PM, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao RG, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castiglione R, et al. Comparison of the genomic background of MET-altered carcinomas of the lung: biological differences and analogies. Mod Pathol. 2019;32:627–638. doi: 10.1038/s41379-018-0182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautschi O, et al. Targeting RET in patients with RET-rearranged lung cancers: results from the global, multicenter RET registry. J Clin Oncol. 2017;35:1403–1410. doi: 10.1200/JCO.2016.70.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbiah V, et al. Precision targeted therapy with BLU-667 for RET-driven cancers. Cancer Discov. 2018;8:836–849. doi: 10.1158/2159-8290.CD-18-0338. [DOI] [PubMed] [Google Scholar]
- 20.Morgan P, et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving phase II survival. Drug Discov Today. 2012;17:419–424. doi: 10.1016/j.drudis.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Basu B, et al. First-in-human pharmacokinetic and pharmacodynamic study of the dual m-TORC 1/2 inhibitor AZD2014. Clin Cancer Res. 2015;21:3412–3419. doi: 10.1158/1078-0432.CCR-14-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothwell DG, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25:738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 23.Pengelly RJ, et al. A SNP profiling panel for sample tracking in whole-exome sequencing studies. Genome Med. 2013;5:89. doi: 10.1186/gm492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 26.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Thall PF, Wooten LH, Tannir NM. Monitoring event times in early phase clinical trials: some practical issues. Clin Trials. 2005;2:467–478. doi: 10.1191/1740774505cn121oa. [DOI] [PubMed] [Google Scholar]
- 28.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey MH, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng J, et al. Single-cell copy number variation detection. Genome Biol. 2011;12:R80. doi: 10.1186/gb-2011-12-8-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbosh C, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For NLMT data, scientifically sound proposals from appropriately qualified Research Groups will be considered for data sharing. Requests should be made by returning a completed Data Sharing Request Form and curriculum vitae of the lead applicant and statistician to newbusiness@trials.bham.ac.uk. The Data Sharing Request Form captures information on the specific requirements of the research, the statistical analysis plan, and the intended publication schedule. The request will be reviewed independently by the Cancer Research UK Clinical Trials Unit (CRCTU) Directors at University of Birmingham in discussion with the Chief Investigator and relevant Trial Management Group and independent Trial Steering Committee. In making their decision the Director’s Committee will consider the scientific validity of the request, the qualifications of the Research Group, the views of the Chief Investigator, Trial Management Group and Trial Steering Committee, consent arrangements, the practicality of anonymising the requested data and contractual obligations. Where the CRCTU Directors and appropriate Trial Committees are supportive of the request, and where not already obtained, consent for data transfer will be sought from the Sponsor of the trial before notifying the applicant of the outcome of their request. It is anticipated that applicants will be notified of a decision within 3 months of receipt of the original request. The results published here are based in part upon data generated by The Cancer Genome Atlas pilot project established by the NCI and the National Human Genome Research Institute. The data were retrieved through database of Genotypes and Phenotypes (dbGaP) authorization (accession number : phs000178.v10.p8). TRACERx sequencing datasets used in this study are described in Hanjani et al.11 and Abbosh et al.32
NLMT Statstical analysis code is available for download from the Github repository https://github.com/pfletchergit/NLMT_Nature2020.