Abstract
The repair of inflamed, demyelinated lesions as in multiple sclerosis necessitates the clearance of cholesterol-rich myelin debris by microglia/macrophages and the switch from a pro-inflammatory to an anti-inflammatory lesion environment. Subsequently, oligodendrocytes increase cholesterol levels as a prerequisite for synthesizing new myelin membranes. We hypothesized that lesion resolution is regulated by the fate of cholesterol from damaged myelin combined with oligodendroglial sterol synthesis. By integrating expression profiling, genetics, and comprehensive phenotyping, we found that paradoxically sterol synthesis in myelin-phagocytosing microglia/macrophages determines repair of acutely demyelinated lesions. Rather than producing cholesterol, microglia/macrophages synthesized desmosterol, the immediate cholesterol precursor. Desmosterol activated LXR-signaling to resolve inflammation, creating a permissive environment for oligodendrocyte differentiation. Moreover, LXR-target gene products facilitated the efflux of lipid/cholesterol from lipid-laden microglia/macrophages to support remyelination by oligodendrocytes. Consequently, pharmacological stimulation of sterol synthesis boosted repair of demyelinated lesions, suggesting novel therapeutic strategies for myelin repair in multiple sclerosis.
Introduction
Approximately 70% of the brain cholesterol in adults is associated with myelin1, a lipid-rich membrane stack that insulates axons. During brain development, oligodendrocytes synthesize most of the cholesterol for myelin membrane expansion, and correspondingly, oligodendroglial cholesterol synthesis is rate-limiting for myelination2. In the adult brain, all cell types contribute to CNS cholesterol homeostasis, which is independent of peripheral sources1. Cholesterol synthesis is counterbalanced by liver X receptor (LXR)-mediated release of cellular cholesterol3. As mammals cannot degrade cholesterol, it is either excreted from the brain or locally recycled.
Functional repair of demyelinated lesions in patients with multiple sclerosis (MS), an inflammatory demyelinating disease of the CNS4,5, is inevitably linked to local cholesterol metabolism. Myelin degenerates and releases the myelin-associated cholesterol, which decreases local sterol synthesis by feedback inhibition6–9. To phagocytose and clear myelin debris, microglia/macrophages adopt a pro-inflammatory signature10. Chronic neuroinflammation involving also T lymphocytes in MS, can cause permanent brain damage. As immunomodulatory treatments of MS ameliorate neuroinflammation but cannot restore neurological disabilities4,5, novel treatment strategies should also support remyelination.
Spontaneous repair of demyelinated lesions occurs in animal models and likely also in early MS, but necessitates the transition to a regenerative environment4. Then, oligodendrocytes synthesize myelin membranes for functional repair11,12. In atherosclerosis, a lipid-driven inflammatory disease of the vasculature, LXR-signaling is critically involved in the resolution of inflammation13. However, whether LXR-signaling links local sterol metabolism to inflammation in demyelinated CNS lesions and which cell types mediate the endogenous repair remains poorly understood.
In this study, we investigated cell type-specific sterol synthesis in the microenvironment of demyelinated lesions and its role in inflammation and remyelination. We made the surprising observation that sterol synthesis in microglia/macrophages was essential for repair after acute demyelination. Not cholesterol itself, but desmosterol, an LXR ligand, orchestrated the resolution of inflammation and facilitated lipid recycling for remyelination. Our findings highlight pharmacological support of sterol synthesis as a potential therapy in demyelinating diseases.
Results
The origin of cholesterol for remyelination depends on the disease phase
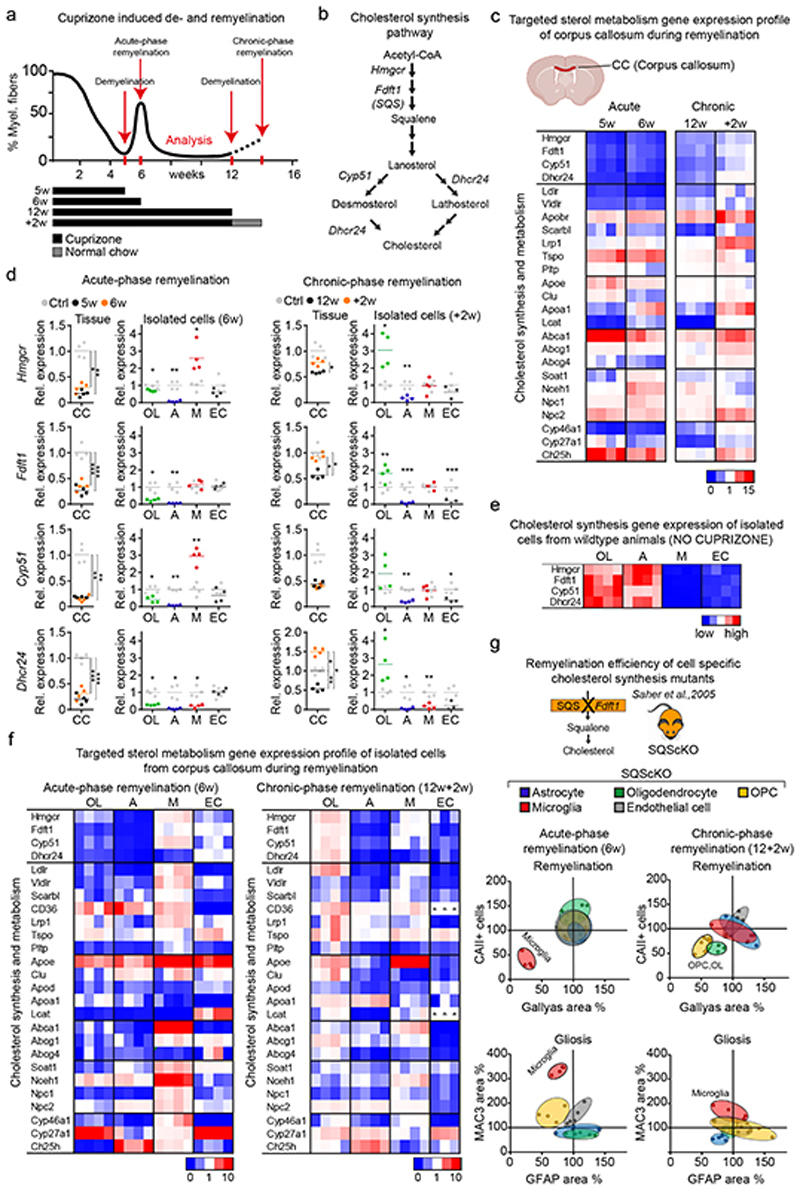
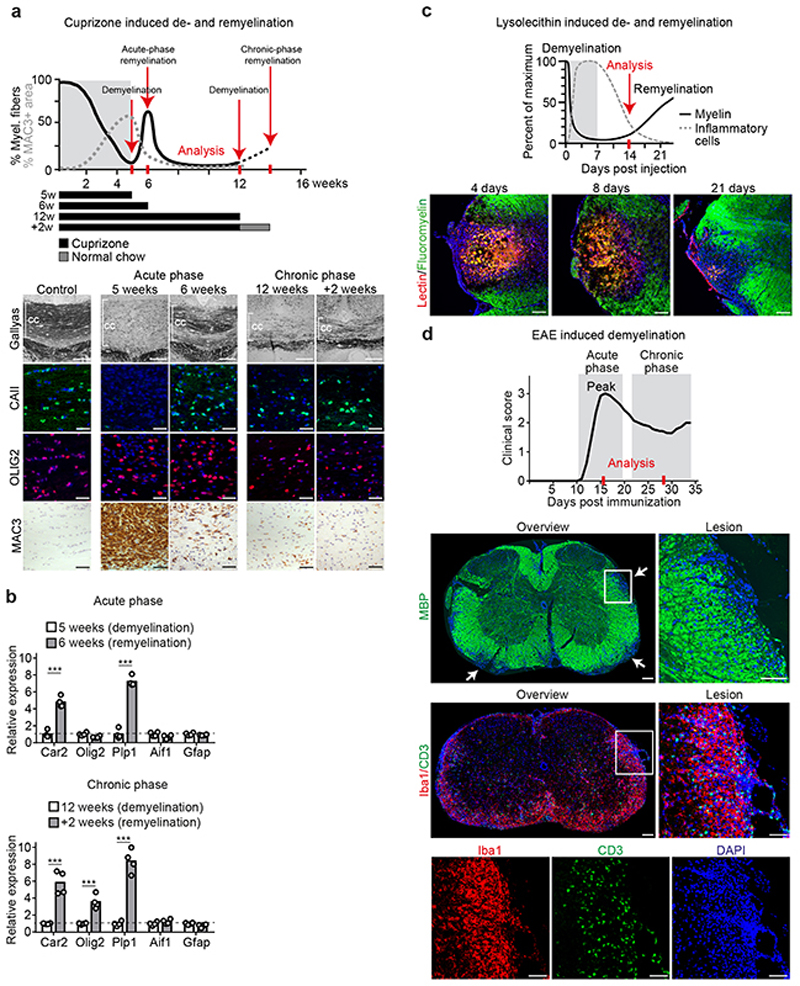
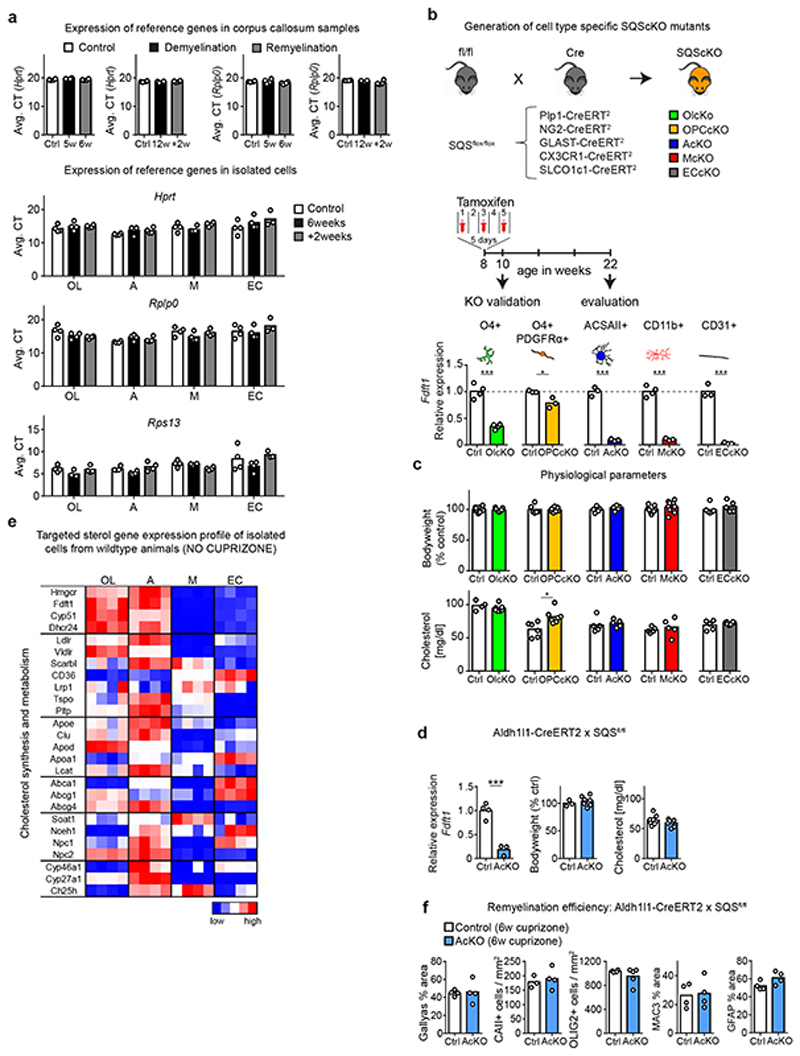
In demyelinating models, and likely also in MS, spontaneous remyelination initially occurs, but slows with disease chronicity4. To determine which cells synthesize cholesterol for remyelination in vivo, we analyzed the corpus callosum in cuprizone-treated mice at two points of demyelination (acute demyelination after 5 weeks and chronic demyelination after 12 weeks) and during two distinct phases of remyelination (Fig. 1a, Extended Data Fig. 1a-b). We chose a spontaneous transient repair phase after 6 weeks cuprizone (‘acute-phase remyelination’) and a repair phase after chronic demyelination following two weeks of cuprizone withdrawal at 12+2 weeks (‘chronic-phase remyelination’). The spontaneous transient remission phase in the 6th week intersects ongoing oligodendrocyte depletion and demyelination whilst cuprizone withdrawal triggers repair after chronic demyelination.
Figure 1. Cholesterol metabolism during de- and remyelination.
(a) Time course of demyelination/remyelination in the cuprizone model12 with treatment paradigms. See also Extended Data Fig. 1a-b.
(b) Cholesterol synthesis pathway with major enzymes and sterol intermediates.
(c) Gene expression profile in the corpus callosum following cuprizone treatment (see selected genes in (d), Supplementary Table 1 shows numeric data).
(d) Expression of sterol synthesis genes in corpus callosum and isolated cells from cuprizone-treated animals in comparison to untreated controls (16 weeks). Data points represent individual mice. Only significant changes are indicated (one-way ANOVA with Tukey’s post test; A astrocytes, CC corpus callosum, EC endothelial cells, M microglia/macrophages, OL oligodendrocytes).
(e) Gene expression profile of sterol synthesis genes in cells isolated from the corpus callosum of untreated controls.
(f) Gene expression profile in isolated cells from cuprizone-fed animals (selected genes in (d)).
(g) Remyelination efficiency and gliosis in conditional cholesterol synthesis mutants (Acute: AcKO n=4, OLcKO n=5, OPCcKO n=4, McKO n=4, ECcKO n=5; Chronic: AcKO n=4, OLcKO n=4, OPCcKO n=7, McKO n=5, ECcKO n=5) relative to controls (Acute: AcKO-Ctrl n=4, OLcKO-Ctrl n=6, OPCcKO-Ctrl n=6, McKO-Ctrl n=7, ECcKO-Ctrl n=3; Chronic: AcKO-Ctrl n=5, OLcKO-Ctrl n=4, OPCcKO-Ctrl n=4, McKO-Ctrl n=5, ECcKO-Ctrl n=8, set to 100%). Quantified histochemical stainings for myelin (Gallyas), oligodendrocytes (CAII), microgliosis (MAC3) and astrogliosis (GFAP) are shown as 95% confidence ellipses with data points of individual animals (n).
Heat maps show fold expression normalized to controls. Each square represents data obtained from one mouse (n=4). ***p<0.001, **p<0.01, *p<0.05
First, we examined expression of genes related to cholesterol synthesis (‘sterol genes’) and metabolism by RT-qPCR profiling of dissected corpus callosum (Fig. 1b-d, Supplementary Table 1, Extended Data Fig. 2a). Concomitant with demyelination, we found decreased expression of all sterol genes including Hmgcr (HMG-CoA reductase), the rate-limiting enzyme in this pathway (Fig. 1b-d, see 5w, 12w). Surprisingly, sterol genes remained downregulated during spontaneous remyelination at 6 weeks, but were restored during remyelination after chronic demyelination (Fig. 1c-d, see 6w, +2w). These findings indicate that different endogenous repair strategies occur following acute versus chronic demyelination.
This prompted us to dissect the cellular contribution to local cholesterol synthesis during remyelination following acute versus chronic demyelination. We integrated expression profiles of genes related to cholesterol synthesis and metabolism in tissue and isolated cells from corpus callosum with phenotyping of cell type-specific cholesterol synthesis mutants. In adult mice, conditional gene targeting of the squalene synthase (SQS) gene Fdft1 2 that catalyzes the first committed step of cholesterol synthesis, was efficient (up to 97 ± 0.7% loss of Fdft1 transcript levels). Loss of cholesterol synthesis in oligodendrocyte precursor cells (OPC), oligodendroglia, astrocytes, microglia, and endothelial cells was well tolerated (Extended Data Fig. 2b-d).
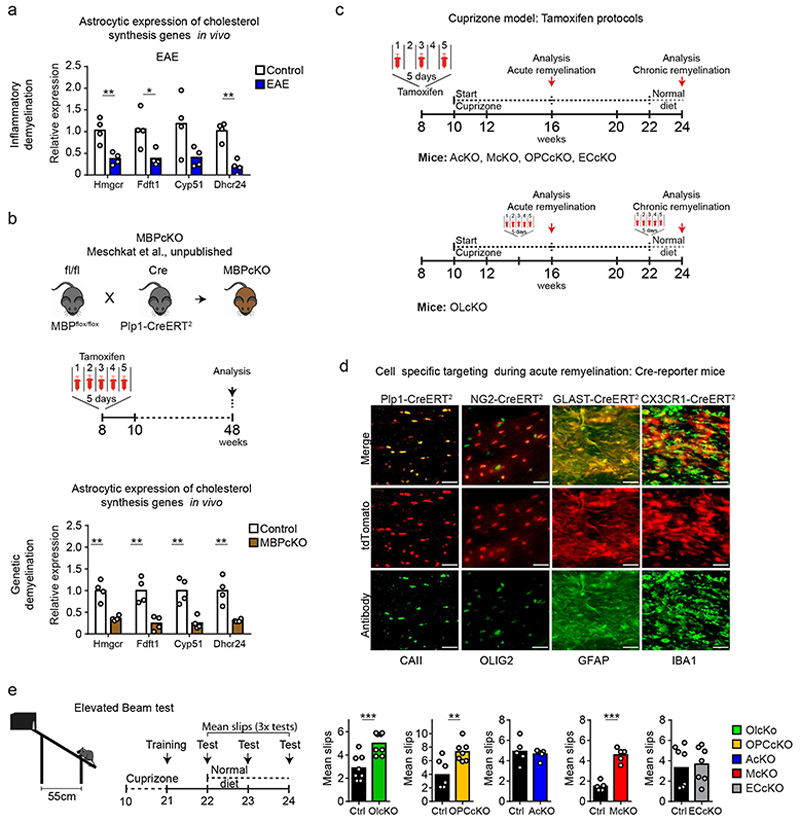
In cell populations isolated from untreated wild type (WT) mice, expression profiles of sterol genes were highest in astrocytes and oligodendrocytes, implying that homeostatic cholesterol synthesis occurs predominantly in these cells (Fig. 1e, Extended Data Fig. 2e, Supplementary Table 2). Myelin disease, whether induced pharmacologically (6w and 12+2w cuprizone), genetically (MBPcKO, inducible myelin basic protein (MBP) mutant14) or by autoimmunity (EAE, experimental autoimmune encephalomyelitis), was always associated with downregulation of sterol gene transcripts in astrocytes (Fig. 1d, 1f, Extended Data Fig. 3a-b, Supplementary Table 3), in agreement with a previous study6. Moreover, in astrocyte-specific cholesterol mutants at 6 or 12+2 weeks of cuprizone treatment, remyelination (Gallyas) and numbers of mature oligodendrocytes (CAII) were comparable to those in cuprizone-treated controls (Fig. 1g, Extended Data Fig. 2f, 3c-e). These data suggest that cholesterol synthesis in astrocytes, although part of normal cholesterol homeostasis, is not required for remyelination.
During remyelination following chronic demyelination, oligodendrocytes increased expression of sterol genes (+2w, Fig. 1d, 1f), likely explaining the restored tissue levels at this stage (+2w, Fig. 1c). Here, both OPC- and oligodendrocyte-specific cholesterol mutants showed reduced oligodendrocyte numbers, diminished remyelination and reduced motor performance (Fig. 1g, Extended Data Fig. 3e), demonstrating that cholesterol synthesis by the entire lineage is important for remyelination. In contrast, during acute-phase remyelination, oligodendroglial expression of sterol genes remained downregulated in WT oligodendrocytes, and oligodendroglial cholesterol mutants remyelinated at control levels (Fig. 1d,g). This raises the possibility that early in the disease (following acute demyelination), oligodendroglial cholesterol synthesis is dispensable and oligodendrocytes can utilize cholesterol from locally available lipoproteins for remyelination.
Unexpectedly, microglia that express only low levels of sterol genes under normal physiological conditions, upregulated the majority of these genes following acute demyelination (Fig. 1d-f). Moreover, ablation of cholesterol synthesis in CNS microglia/macrophages (collectively designated phagocytes) resulted in severely impaired remyelination following acute demyelination (Fig. 1g). Together, these data demonstrate that astroglial cholesterol is dispensable for remyelination. Oligodendroglial cholesterol synthesis promotes remyelination after chronic demyelination, and remyelination after acute demyelination requires an intact sterol synthesis pathway in CNS phagocytes. We therefore focused our analysis on repair early in the disease (acutely demyelinating states).
Loss of sterol synthesis in microglia/macrophages induces persistent activation and impairs lipid export during remyelination
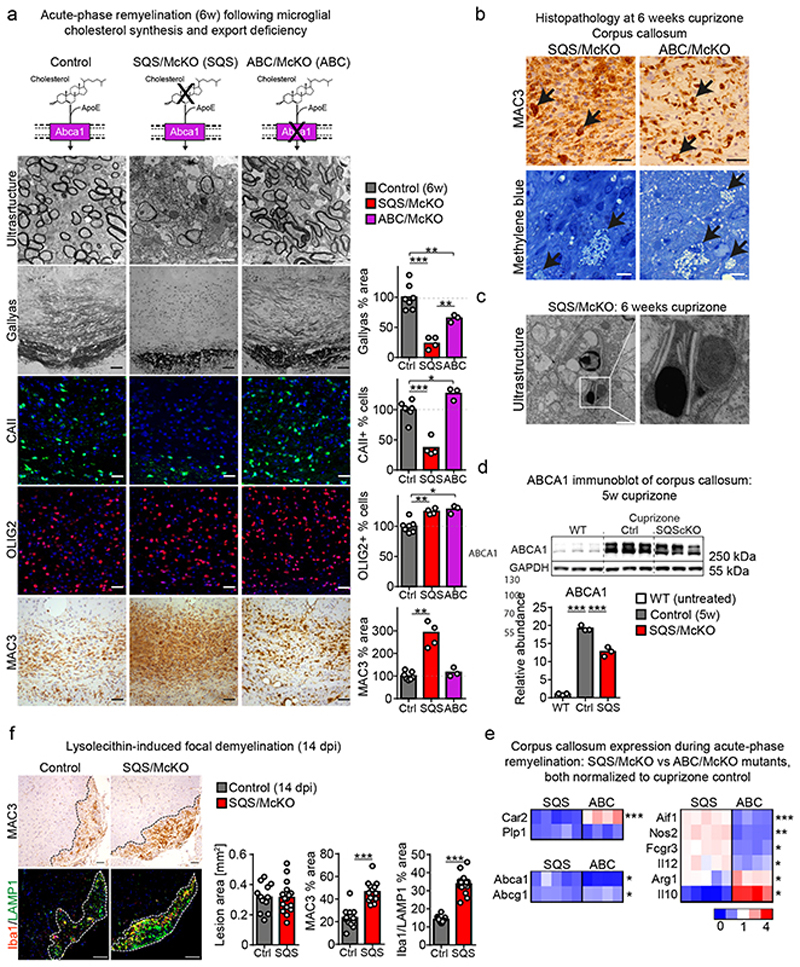
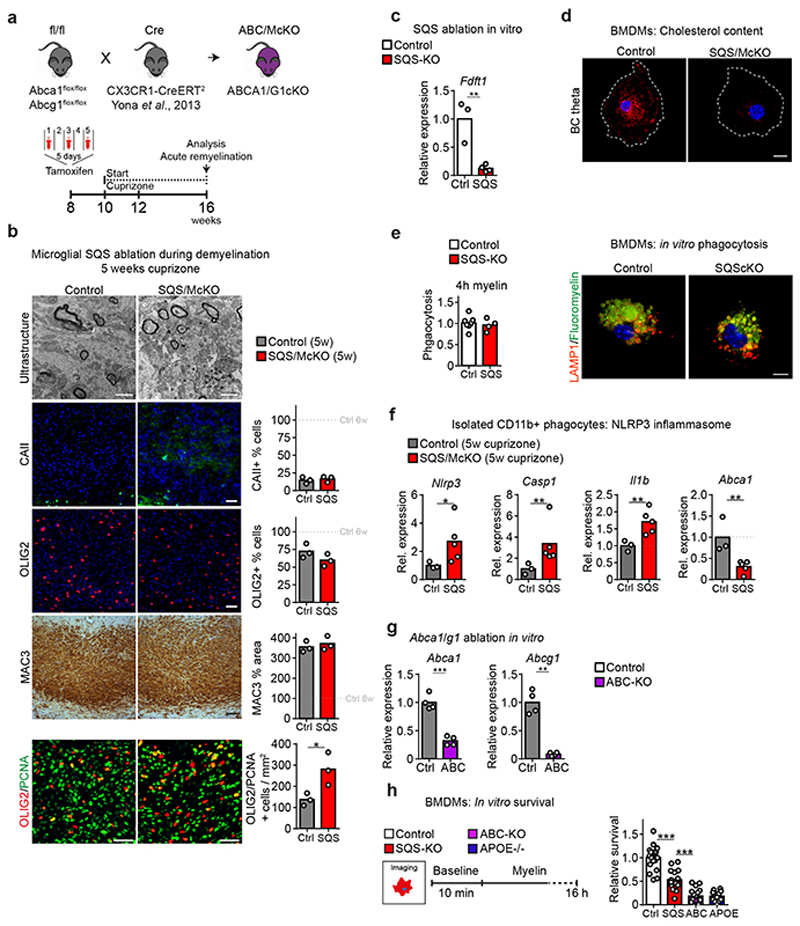
In microglia/macrophages, perturbed cholesterol efflux has been linked to impaired remyelination15 but the relationship of phagocyte sterol synthesis versus cholesterol export for the repair of demyelinated lesions is unclear. Therefore, we directly compared microglial sterol synthesis mutants (CX3CR1-CreERT2::Fdft1 flox/flox, termed SQS/McKO) with microglial lipid efflux (Abca1 [adenosine triphosphate–binding cassette A1] and Abcg1) mutant mice (CX3CR1-CreERT2::Abca1 flox/flox Acbg1 flox/flox, termed ABC/McKO) in the acute-phase remyelination paradigm. As expected, histological analysis revealed remyelination deficits in both mutants (Fig. 2a, Extended Data Fig. 4a). However, SQS/McKO mutants were more strongly affected (~25% remyelination in the corpus callosum of control levels, compared to ~65% in ABC/McKO animals). Thus, in phagocytes both sterol synthesis and sterol efflux are required to fully restore myelin following acute demyelination.
Figure 2. Impaired remyelination in sterol metabolism mutants.
(a) Histopathology of microglial cholesterol synthesis (SQS/McKO, n=4 animals) and cholesterol export (ABC/McKO, n=3 animals) mutants, showing myelin loss (electron microscopic images, Gallyas), mature oligodendrocytes (CAII), oligodendrocyte linage cells (OLIG2) and microgliosis (MAC3) (Scale 50 μm) with quantification normalized to cuprizone controls (n=7 animals). Asterisks mark significant changes (one-way ANOVA with Tukey’s post test).
(b) Foamy phagocytes following demyelination, visualized by MAC3 (Scale 50 μm) and methylene blue (Scale 10 μm) staining. Representative image of stainings from SQS/McKO (n=4 animals) and ABC/McKO (n=3) animals.
(c) Cholesterol crystals in SQScKO cuprizone mice (n=4 animals, Scale 1 μm).
(d) Cropped ABCA1 immunoblot following demyelination (5w cuprizone) of corpus callosum samples from untreated WT, cuprizone control and cuprizone SQS/McKO (n=3 animals). Reprobing for GAPDH confirms comparable protein loading. Asterisks mark significant changes (one-way ANOVA with Tukey’s post test).
(e) Gene expression related to oligodendrocytes (Car2, Olig2), sterol efflux (Abca1, Abcg1) and inflammation during acute remyelination in corpus callosum from SQS/McKO (n=5) and ABC/McKO (n=4) mice normalized to cuprizone control (n=4 mice). Asterisks mark significant changes between SQS/McKO and ABC/McKO mutants (Student’s t-test, two-sided).
(f) Microgliosis (MAC3, Iba1/LAMP1) in the DAPI-positive lesion area (dotted line) at 14 dpi after lysolecithin-mediated demyelination with quantification (Scale 100 μm). Asterisks mark significant changes between control (n= 12 lesions from 6 animals) and SQS/McKO (n=14 lesions from 7 animals) mutants (Student’s t-test, two-sided).
***p<0.001, **p<0.01, *p<0.05
During lesion repair, OPC proliferation precedes oligodendrocyte differentiation and remyelination16. To test whether impaired OPC proliferation or differentiation accounted for the poor remyelination in SQS conditional mutants, we quantified OLIG2/PCNA double-positive proliferating OPCs at demyelination (5w of cuprizone), and OLIG2+ oligodendrocyte lineage cells and CAII+ mature oligodendrocytes during spontaneous remyelination (6w cuprizone) by cell counting. Proliferating OPCs densities were approximately doubled in SQS/McKO mice compared to controls (Extended Data Fig. 4b). Oligodendroglial lineage cell densities were slightly above control levels in either conditional mutant, whereas oligodendrocyte maturation was strongly inhibited only in SQS/McKO mutants (Fig. 2a).
Potentially, oligodendrocyte maturation could be impeded by defective clearance of myelin debris16 by SQS mutant myeloid cells. However, myelin phagocytosis by SQS mutant bone marrow-derived macrophages (BMDM) was at control levels in vitro (Extended Data Fig. 4c-e). Five weeks of cuprizone resulted in comparable demyelination and microgliosis in SQS/McKO mice and controls (Extended Data Fig. 4b). However, in contrast to cuprizone-treated WT mice, we observed numerous lipid-laden foamy phagocytes containing cholesterol-like crystals in SQS/McKO mice (Fig. 2b-c), similar to lipid efflux-deficient ABC/McKO mutants15. SQS mutant phagocytes isolated at this peak of demyelination, significantly upregulated gene expression associated with NLRP3 inflammasome activation (Extended Data Fig. 4f). Accordingly, myelin phagocytosis in vitro decreased survival of SQS mutant phagocytes with a morphology attributable to pyroptosis (Extended Data Fig. 4g-h, Video S1), reminiscent of the lipid overload in atherosclerosis-associated macrophages17. We therefore tested whether loss of sterol synthesis in phagocytes affects lipid turnover. In cuprizone-treated SQS/McKO mutant mice, mRNA and protein levels of ABCA1 and ABCG1 were strongly reduced compared to controls (Fig. 2d-e, Supplementary Table 5), likely caused by diminished expression in SQS mutant phagocytes (Extended Data Fig. 4f).
After clearance of myelin debris, phagocytes in MS lesions can adopt an anti-inflammatory phenotype characterized by downregulation of pro-inflammatory mediators such as NOS2 and up-regulation of anti-inflammatory cytokines e.g. IL1018. Similarly, in controls during acute-phase remyelination, microgliosis largely resolved exhibiting an anti-inflammatory / pro-regenerative gene expression signature, the latter even more pronounced in ABC/McKO mice (Fig. 2a, e). In contrast, SQS/McKO mice showed persistent microgliosis, increased mRNA expression of pro-inflammatory mediators such as NOS2 and IL12 as well as decreased expression of the anti-inflammatory mediators IL10 and arginase-1. Persistent microgliosis was also observed in SQS/McKO mice focally demyelinated with lysolecithin (Fig. 2f, Extended Data Fig. 1c), suggesting prolonged microglial activation as a general feature of myelin-phagocytosing microglia that lack sterol synthesis. These findings revealed overlapping mechanisms underlying impaired repair in the sterol metabolism mutants (pyroptotic cell death and lipid flux deficits in both) that are nevertheless distinct (disparity in resolution of microgliosis). Independent of phagocytic lipid processing, sterol synthesis appeared critical for inducing an anti-inflammatory signature and for resolving microgliosis after clearing myelin debris. Thus, it is conceivable that the persistent pro-inflammatory activation of SQS mutant microglia caused the arrest of oligodendrocyte maturation and poor remyelination.
Defective LXR-signaling underlies the non-resolving inflammation in cholesterol mutants
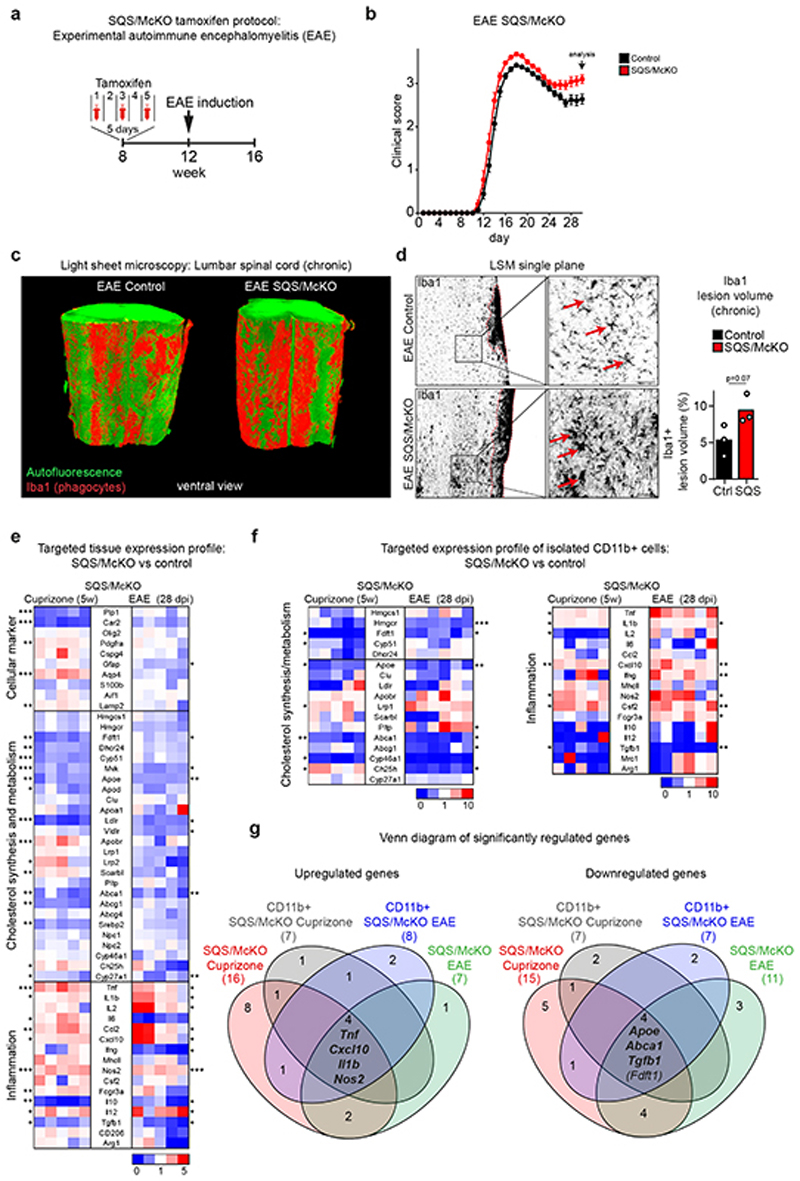
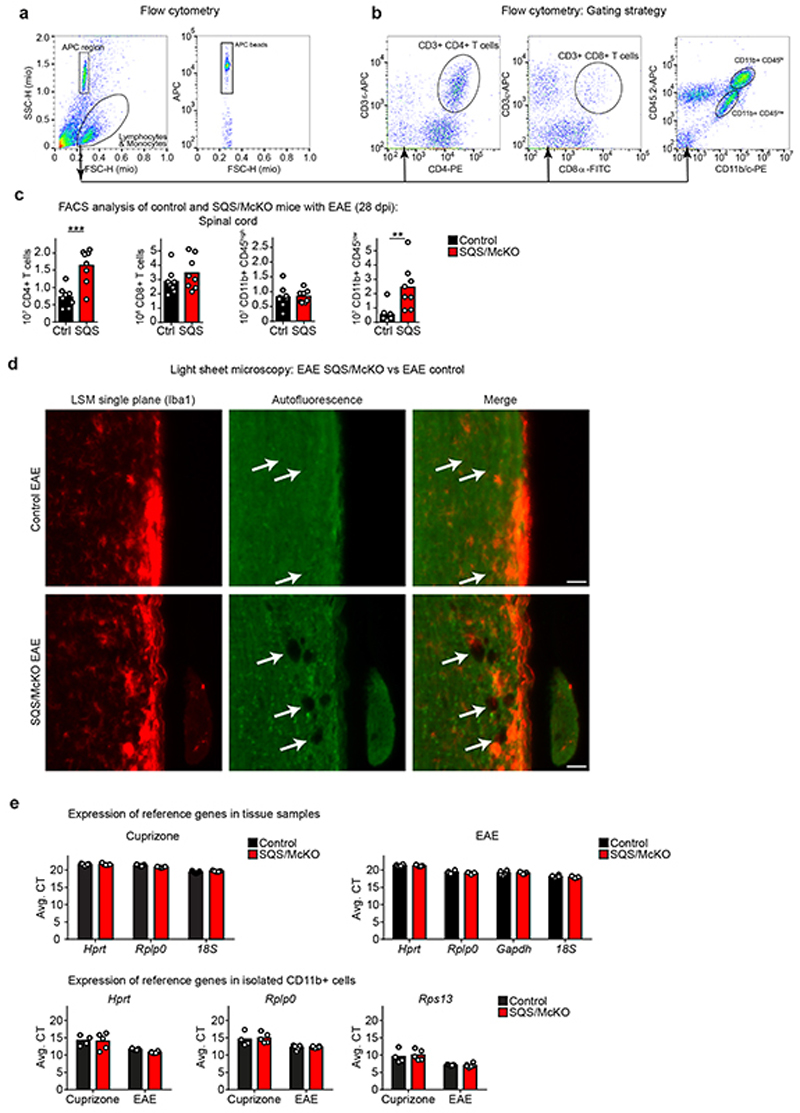
In EAE (Extended Data Fig. 1d), an autoreactive T cell-driven model of MS, microglia/macrophages clear myelin debris. We asked whether also in EAE, sterol synthesis in phagocytes is critical to resolve inflammation. SQS/McKO mice developed aggravated disease (Fig. 3a-b) with increased density of CD4+ T cells and CD11b+/CD45low microglia by FACS of spinal cord (Extended Data Fig. 5a-c). By light sheet microscopy, we detected an almost twofold increase in the total volume of SQS mutant phagocytes, exhibiting a more activated, less ramified morphology (Fig. 3c-d, Extended Data Fig. 5d, Video S2-S3).
Figure 3. Non-resolving inflammation in demyelinated microglia-specific cholesterol mutants.
(a) Recombination strategy in EAE. Inducing EAE 4 weeks after tamoxifen injections allowed renewal of peripheral monocytes such that only microglia but not monocyte derived macrophages lacked sterol synthesis.
(b) Mean clinical score ± SEM of control (n=34 animals) and SQS/McKO (n=27 animals) mice.
(c) Representative light sheet microcopy of lumbar spinal cord samples (~2.25 mm length) from SQS/McKO and controls 28d after EAE induction, stained for Iba1+ phagocytes (n=3 animals).
(d) Single plane from (c) with detail (Scale 50 μm) and quantification of percent Iba1+ lesion volume of control and SQS/McKO mutants (n=3 animals, Student’s t-test, two-sided).
(e) Fold expression levels in corpus callosum of cuprizone (5w) treated SQS/McKO mice (n=5 animals) normalized to control mice (n= 7 animals, set to 1) and lumbar spinal cord of EAE (28 dpi) treated SQS/McKO mice (n=6 animals) normalized to control mice (n=6 animals, set to 1). Each square represents data obtained from one mouse. Asterisks mark significant changes (Student’s t-test, two-sided). Numerical data with statistics is shown in Supplementary Table 6-7.
(f) Fold gene expression in isolated CD11b+ cells from SQS/McKO mice normalized (Cuprizone n=5 animals, EAE n=6 animals) to controls (Cuprizone n=4 animals, EAE n=6 animals, set to 1) treated as in (e). Asterisks mark significant changes (Student’s t-test, two-sided).
(g) Venn diagram of differentially expressed genes in (e) and (f).
***p<0.001, **p<0.01, *p<0.05
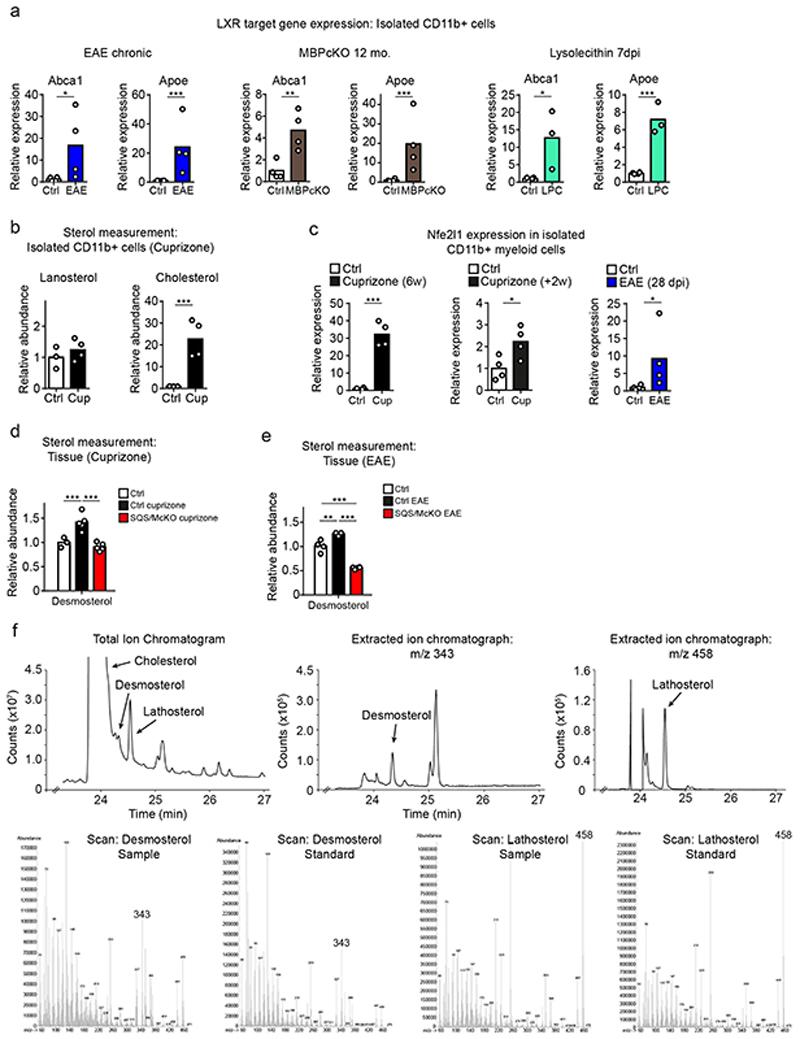
To identify factors that could link sterol metabolism with inflammation, we determined expression profiles in tissue samples and isolated CD11b+ myeloid cells in demyelinating conditions (5w cuprizone and 28d post immunization EAE). By setting a threshold of a 1.5-fold change (p<0.05), we found 18 differentially expressed genes in EAE, and 32 in cuprizone-treated SQS/McKO mutants compared to controls (Fig. 3e-g, Extended Data Fig. 5e, Supplementary Table 6-7). In conditional mutants of both disease models, the pro-inflammatory genes Tnf, Il1b, Nos2 and Cxcl10 were upregulated, while the anti-inflammatory gene Tgfb1 and the cholesterol efflux genes Abca1 and Apoe were downregulated. All these genes are linked to LXR signaling and most of them (Tnf, Il1b, Nos2, Cxcl10, Abca1, Apoe) are direct transcriptional targets3. In addition to increasing lipid efflux, LXR activation can dampen inflammation. These data raise the possibility that sterol synthesis in phagocytes is necessary to activate LXR-signaling for promoting cholesterol efflux and resolving inflammation.
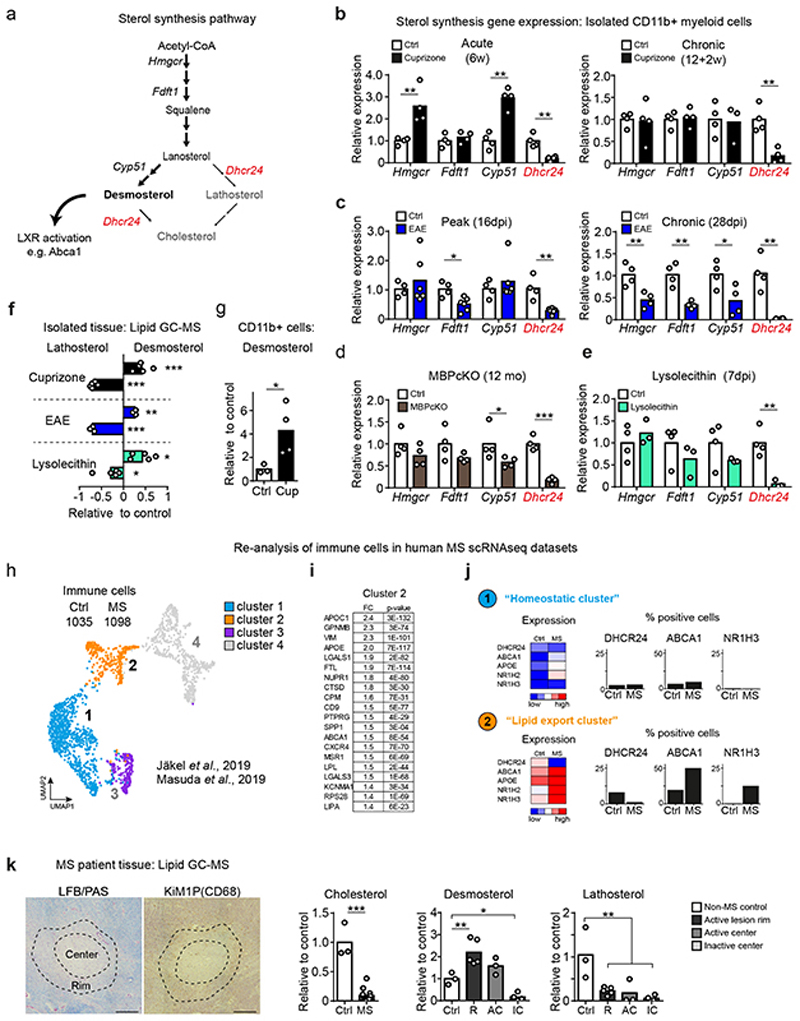
Stalling sterol synthesis to increase desmosterol is a common mechanism in demyelinating disease
The potential perturbation of LXR-signaling in sterol synthesis mutants prompted us to test whether demyelination in general induces LXR target genes in WT phagocytes. Indeed, similar to phagocytes from (i) cuprizone-treated mice (Fig. 1f), we also found increased expression of the LXR-dependent cholesterol efflux genes Abca1 and Apoe in phagocytes isolated from (ii) mice with EAE, (iii) genetic myelin mutants (MBPcKO) and (iv) lysolecithin-treated mice (Extended Data Fig. 6a). These findings suggest that LXR activation is a general response of myelin-clearing microglia/macrophages.
The major endogenous LXR agonists are oxysterols and certain cholesterol synthesis intermediates3. Inferred from the failure to activate LXR-signaling in our sterol synthesis mutants, we surmised that sterol intermediates accounted for the observed LXR activation in our disease models (Fig. 4a). We first determined the expression levels of key genes of sterol synthesis in acutely isolated CD11b+ myeloid cells from mice after cuprizone treatment (Fig. 4b), EAE (Fig. 4c), genetic myelin mutants (Fig. 4d), and lysolecithin treatment (Fig. 4e). We found a consistent and drastic downregulation only of Dhcr24 (24-dehydrocholesterol reductase). As reduced levels of DHCR24 leads to the accumulation of the LXR agonist desmosterol, we next quantified sterol intermediates by mass spectrometry. In agreement with the exclusive downregulation of Dhcr24 expression, we found increased abundance of desmosterol, both in tissue samples (Fig. 4f), and in isolated phagocytes (Fig. 4g, Extended Data Fig. 6b). The reduced abundance of lathosterol, whose synthesis requires DHCR24 activity, further supports a prominent role of desmosterol in these experimental models. Expression of the NRF1/NFE2L1 transcriptional regulator that links sterol content to LXR-signaling19, was strongly upregulated in phagocytes of mice following demyelination (Extended Data Fig. 6c), supporting a central role of LXR activation in the repair of demyelinated lesions. If phagocytes are responsible for the increased tissue levels of desmosterol in WT animals, such increase of this cholesterol precursor should not occur in SQS/McKO mutants. Indeed, cuprizone and EAE treatment did not raise desmosterol levels in SQS/McKO mutants (Extended Data Fig. 6d-e). In summary, myelin clearance is accompanied by downregulation of Dhcr24, leading to the accumulation of the LXR agonist, desmosterol, in phagocytes. Enhanced LXR-signaling contributes to intracellular cholesterol homeostasis and the return of phagocytes to an anti-inflammatory phenotype.
Figure 4. Desmosterol synthesis is a common mechanism in demyelinating disease.
(a) Scheme depicting the sterol synthesis pathway with the link to LXR activation via the agonist function of desmosterol.
(b-e) Expression of cholesterol synthesis genes in isolated CD11b+ cells from individual mice (n) during cuprizone (b; n=4), EAE (c; control n=4, EAE n=6) MBPcKO (d; n=4) and lysolecithin (e; n=3) treatment (Student’s t-test, two-sided).
(f) Mean relative abundance of desmosterol and lathosterol in tissue from cuprizone (5w, n=5 animals), EAE (n=3) and lysolecithin-treated mice (n=5) measured by GC-MS (one-way ANOVA, Tukey’s post test).
(g) Mean relative abundance of desmosterol in isolated CD11b+ cells from 5w cuprizone-treated mice (n=4 animals) compared to untreated controls (n=3), measured by GC-MS (Student’s t-test, two-sided).
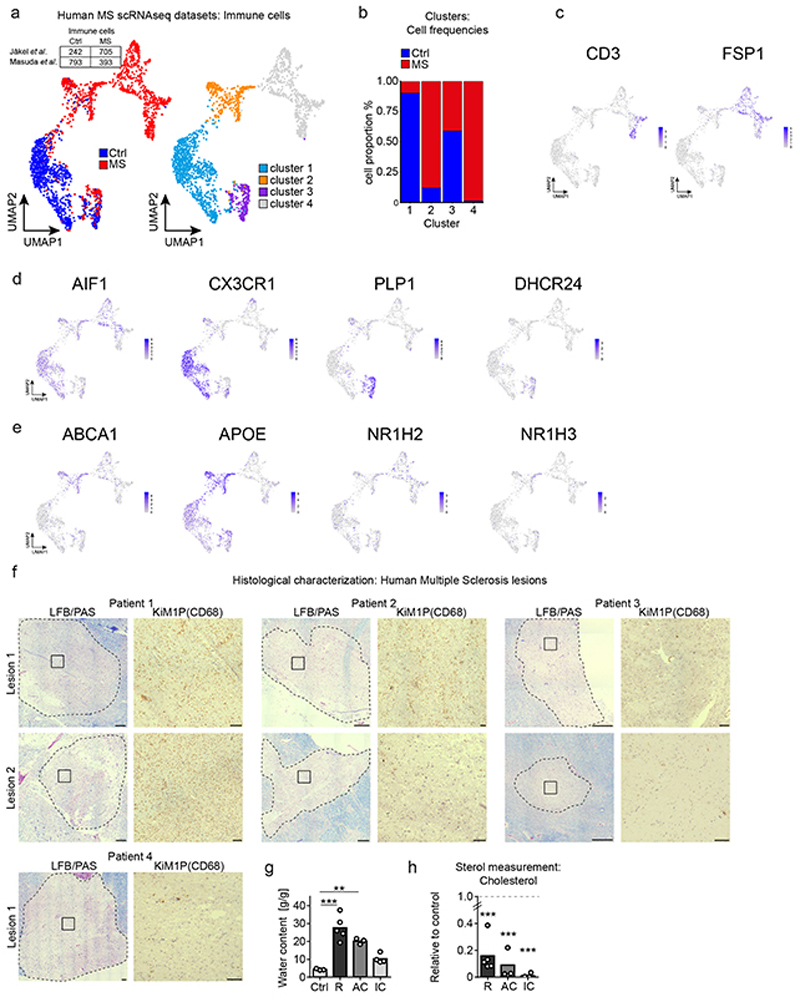
(h) UMAP and unsupervised clustering of immune cell clusters from human MS scRNAseq datasets (GSE118257, GSE124335).
(i) Top 20 upregulated genes of cluster 2 compared to cluster 1 showing fold change (FC) and p-value (Wilcoxon Rank Sum test, two sided).
(j) Mean expression (related to the mean of cluster 1-3) and expression frequency (% positive nuclei/cells).
(k) MS lesions (scale 500 μm) stained for myelin (LFB/PAS) and phagocytes (CD68/KiM1P) to illustrate lesion center and rim (dotted lines). Mean relative abundance of lipids (as indicated) in tissue samples from 4 MS patients, normalized to 3 control individuals. Ctrl: control white matter (n=3 samples); R: Active lesion rim (n=5 lesions); AC: Active lesion center (n=3 lesions); IC: Inactive lesion center (n=4 lesions) (one-way ANOVA, Holm-Sidak post test).
Significant changes are marked by asterisks (***p<0.001, **p<0.01, *p<0.05).
To examine whether desmosterol-activated LXR-signaling in myeloid cells is relevant in human MS, we analyzed single-cell gene expression profiles from MS patients of two recent studies (GSE118257 and GSE12433520,21). Datasets include healthy control white matter, normal appearing white matter, active lesions and chronic active lesions. We merged designated immunological cell subpopulations, and unsupervised clustering gave rise to four clusters (Fig. 4h, Extended Data Fig. 7a-b). Myeloid cells (microglia/macrophages) allocated to clusters 1 and 2 (Extended Data Fig. 7c-d). Cluster 1 largely contained homeostatic myeloid cells that were more abundant in control subjects (‘homeostatic cluster’). Cells from MS patients dominated cluster 2. We designated this cluster as ‘lipid export cluster’ because its transcription profile related to lipid export and mobilization, as defined by the most differentially expressed genes and gene set enrichment analysis (Fig. 4i, Supplementary Table 8). Importantly, the cells from MS patients upregulated expression of LXR alpha and beta (NR1H3 and NR1H2 genes) and LXR targets ABCA1 and APOE, but strongly downregulated DHCR24 (Fig. 4j, Extended Data Fig. 7e, Supplementary Table 9).
To explore whether this altered gene regulation translated into increased desmosterol abundance, we performed mass spectrometry on human MS tissue (Supplementary Table 10). Human samples were histochemically stained for the inflammatory marker CD68/KiM1P and for myelination (LFB/PAS) (Extended Data Fig. 7f) to discriminate between active lesion rim (R), active lesion center (AC), and inactive lesion center (IC). Increased tissue water content confirmed active inflammatory processes in the rim and active lesions centers, but not in inactive lesion centers (Extended Data Fig. 7g). Irrespective of the lesion type, MS samples contained <20% of cholesterol levels found in non-MS white matter, likely reflecting myelin loss (Fig. 4k, Extended Data Fig. 7h). Strikingly, we detected significantly elevated levels of desmosterol but not lathosterol in the rim and center of active lesions but not in inactive lesions (Fig. 4k). These data support the model that in microglia/macrophages at sites of active myelin/lipid remodeling, the sterol synthesis pathway up to the level of DHCR24 is essential to produce desmosterol that activates LXR-signaling, which can then restrict inflammation and facilitate cholesterol efflux and recycling.
Squalene administration restores LXR-signaling in SQS mutant phagocytes in vitro
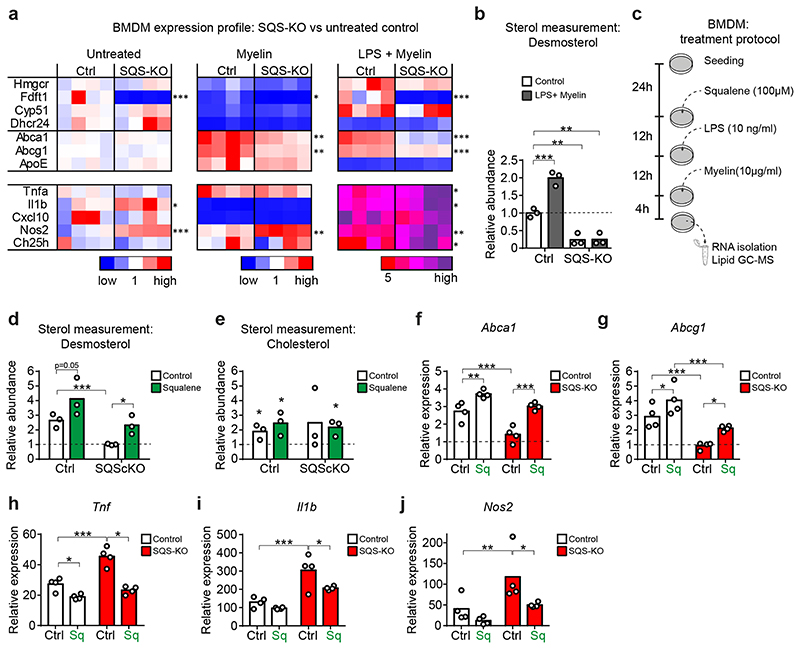
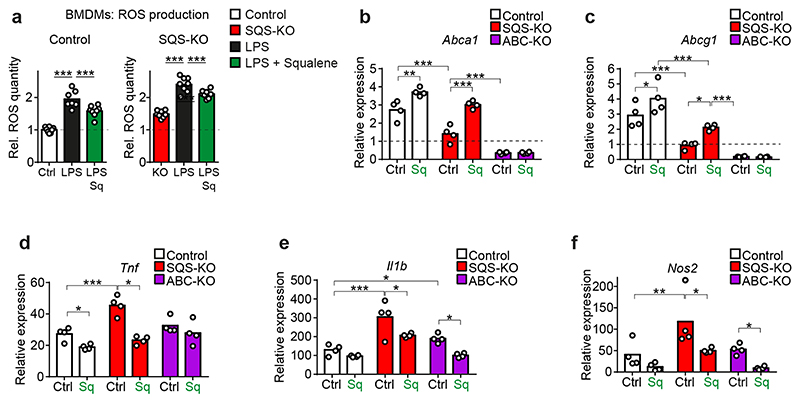
To investigate this regulatory system mechanistically, we turned to in vitro experiments using WT and SQS knockout BMDMs. We exposed cells to purified myelin or myelin+LPS to simulate an inflammatory environment during acute demyelination, and determined expression of sterol synthesis genes, LXR targets and inflammatory genes. As expected from a homeostatic response, WT BMDMs treated with myelin alone downregulated expression of the entire cholesterol synthesis pathway (Fig. 5a). The myelin+LPS treatment induced sterol genes apart from downregulating Dhcr24, leading to a 2-fold increased cellular desmosterol content (Fig. 5b). In contrast, SQS knockout BMDMs failed to increase desmosterol, which attenuated LXR target gene regulation (reduced efflux gene expression, increased inflammatory gene expression) (Fig. 5a), recapitulating our in vivo findings. Concordantly, steady state levels of reactive oxygen species (ROS) were elevated in SQS mutant BMDM compared to controls (Extended Data Fig. 8a).
Figure 5. Squalene administration restores LXR-signaling in SQS mutant phagocytes in vitro .
(a) Relative expression of genes related to cholesterol synthesis, cholesterol export and inflammation in control and SQS-KO BMDMs treated with myelin (10 ng/ml) or LPS (10 ng/ml) + myelin for 16h normalized to untreated control cultures (n=4 cultures set to 1; one-way ANOVA with Tukey’s post test).
(b) Desmosterol abundance in control and SQS-KO BMDMs (n=3 cultures) treated as in (a).
(c) Scheme of squalene treatment of BMDM cultures. 24h after seeding, cells were treated with squalene or left untreated, followed by administration of LPS and myelin.
(d-e) Abundance of desmosterol (d,) and cholesterol (e) in LPS + myelin-treated BMDM cultures with or without squalene administration (as in c), normalized to untreated WT controls (n=3 cultures). Asterisks mark significant changes (Student’s t-test, two-sided).
(f-j) Relative expression of cholesterol efflux (f, g) and inflammatory (h-j) genes in control and SQS-KO BMDMs treated with myelin+LPS with or without squalene (as in c), normalized to untreated WT cultures (n=4 cultures, two-way ANOVA with Holm-Sidak post test).
Bars represent the mean of individual cultures. ***p<0.001, **p<0.01, *p<0.05.
Next, we explored whether squalene, the product of squalene synthase (SQS), could rescue impaired LXR-signaling in SQS-deficient BMDM challenged with myelin+LPS (Fig. 5c). Indeed, squalene increased the desmosterol content in mutant BMDM to control levels (Fig. 5d), independent of the elevated cellular cholesterol that likely reflected uptake of cholesterol-rich myelin (Fig. 5e). This led to normalized expression of cholesterol efflux genes (Fig. 5f-g) and attenuated expression of inflammatory genes in mutant BMDM (Fig. 5h-j), demonstrating that LXR-signaling depended on post-squalene sterol synthesis. In addition to fueling sterol synthesis, squalene itself has anti-inflammatory properties as a ROS scavenger22, which potentially contributed to reducing ROS levels in LPS-activated BMDMs (Extended Data Fig. 8a). Interestingly, squalene treatment also increased desmosterol levels and repressed inflammatory genes in WT BMDM (Fig. 5f-j) and cholesterol export-deficient BMDMs (Extended Data Fig. 8b-f). These data support the hypothesis that under our experimental conditions, endogenous squalene availability became limiting and squalene supplementation enhanced LXR-signaling via desmosterol synthesis, stimulating cholesterol efflux and attenuating inflammation (Extended Data Fig. 8g).
Exogenous squalene ameliorates non-resolving inflammation in SQS/McKO mice in vivo and promotes remyelination
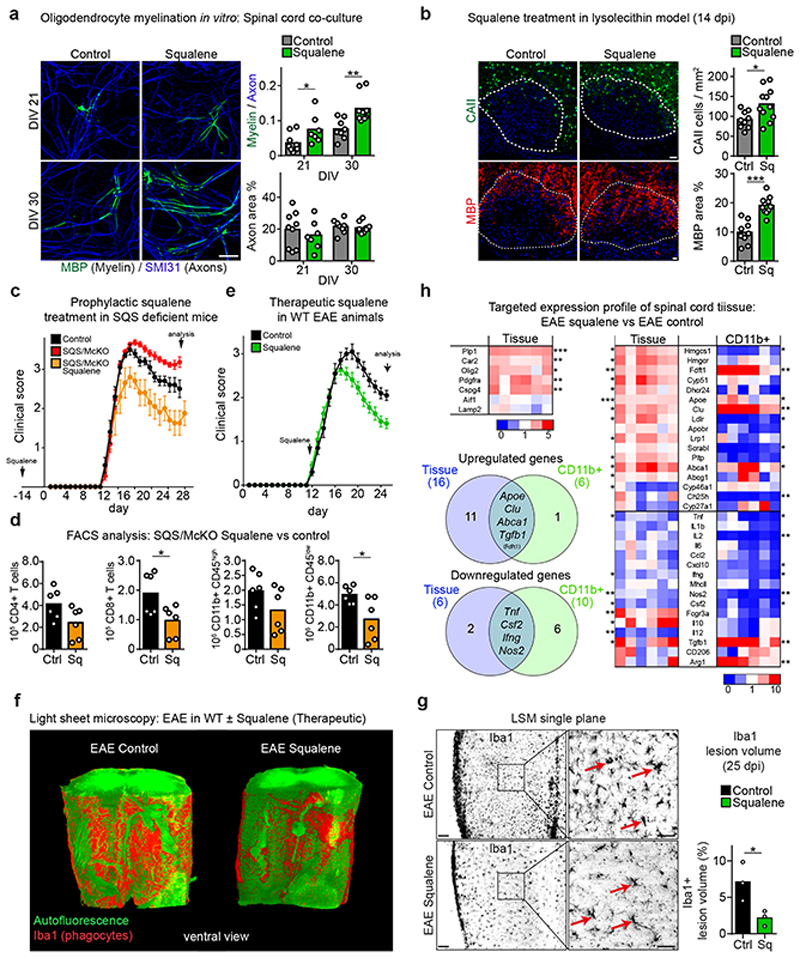
Besides serving as a precursor for desmosterol in phagocytes, exogenous squalene could facilitate cholesterol synthesis in oligodendrocytes and thus directly support myelination, as cholesterol is essential for myelination and directly enhances remyelination2,12,23. When we supplemented spinal cord cell cultures with squalene, axon densities remained unaffected but the rate of myelination, measured by MBP-positive myelin internodes, increased significantly compared to vehicle-treated controls (Fig. 6a). This suggests a direct pro-myelinating effect of squalene on oligodendrocytes, distinct from its homeostatic and anti-inflammatory effects in phagocytes.
Figure 6. Exogenous squalene rescues non-resolving inflammation in SQS/McKO mice in vivo and promotes remyelination.
(a) Mean myelination in spinal cord cultures at 21-30 days in vitro (DIV) with (DIV21, n=7 cultures; DIV30, n=9) or without squalene (DIV21, n=9; DIV30, n=8) by quantification of MBP+ myelin segments relative to SMI31+ axons (two-way ANOVA, Holm-Sidak post test). Scale, 50 μm.
(b) Mean CAII+ oligodendrocytes and MBP+ myelin in lesions from control (n=9 lesions from 5 animals) and squalene-treated animals (n=11 lesions from 6 animals, two-sided Student’s t-test) at 14dpi lysolecithin. Scale, 50 μm.
(c) Mean clinical score ± SEM of EAE, comparing controls (n=6), SQS/McKO (n=11) and SQS/McKO treated with prophylactic squalene.
(d) Mean density inflammatory cells as in (c) from controls (n=6) and squalene-treated SQS/McKO mice (n=6) by flow cytometry (Student’s t-test, two-sided).
(e) Mean clinical score ± SEM of WT EAE animals therapeutically treated with (n=11) or without squalene (n=10).
(f) Representative light sheet microcopy of spinal cord samples (~2.25 mm length) from WT EAE mice treated with or without squalene (n=3 animals), stained for Iba1+ phagocytes (red) and autofluorescence imaging.
(g) Single plane from (f) with detail and quantification of % Iba1+ spinal cord volume (n=3 animals, Student’s t-test, two-sided). Scale, 50 μm.
(h) Fold expression profiles in tissue and CD11b+ cells from EAE animals treated with or without prophylactic squalene (normalized to vehicle-treated EAE mice, n=6), showing cellular markers (upper left) and genes related to sterol metabolism and inflammation (right). Venn diagrams indicate significantly differentially expressed genes (Student’s t-test, two-sided).
***p<0.001, **p<0.01, *p<0.05.
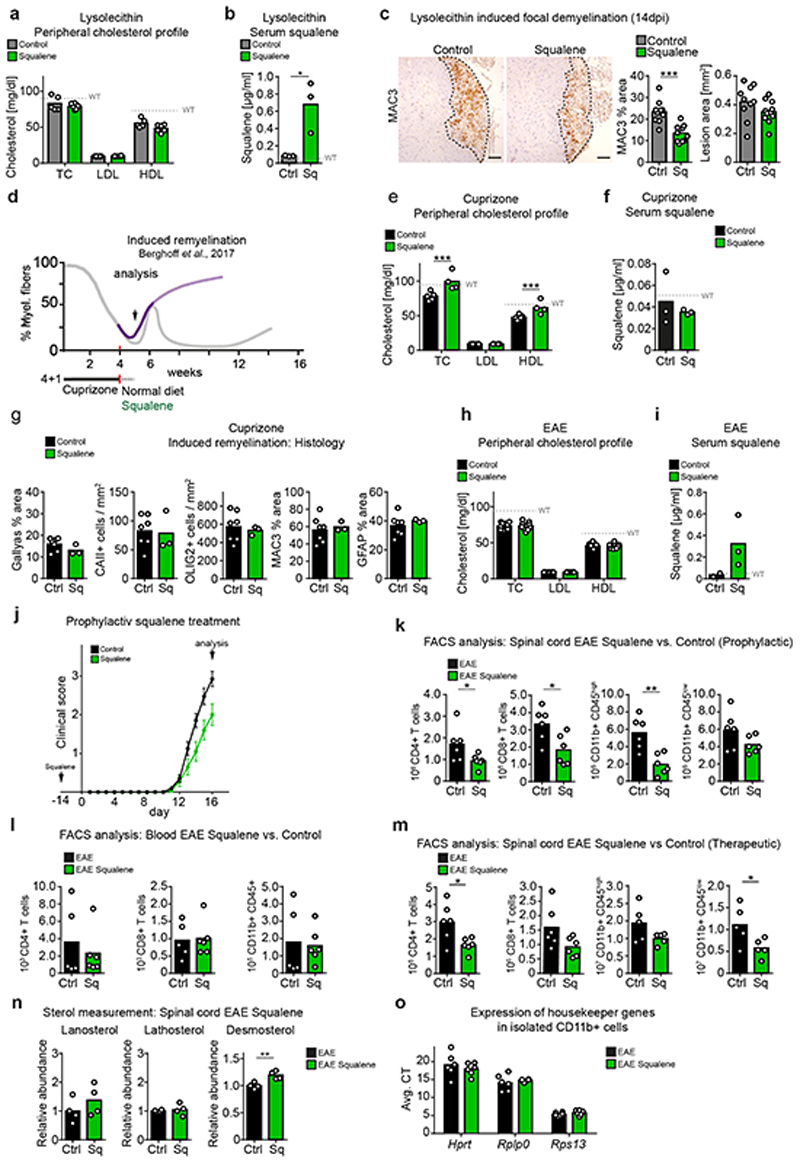
To investigate whether squalene supplementation supports regenerative processes in vivo, we demyelinated mouse spinal cord by lysolecithin and fed a chow with 0.5% squalene. Squalene feeding strongly increased serum squalene levels, while serum cholesterol remained unaltered (Extended Data Fig. 9a-b), as expected from human subjects24. Squalene administration reduced the density of MAC3+ microglia, increased the density of CAII+ oligodendrocytes and the MBP+ area in lesions, indicating attenuated inflammation and enhanced remyelination (Fig. 6b, Extended Data Fig. 9c). Of note, squalene supplementation did not increase serum squalene levels in the cuprizone model, possibly because of hepatic defects in this model25, and failed to facilitate remyelination (Extended Data Fig. 9d-g).
In WT mice with EAE, squalene supplementation selectively increased serum squalene levels by ~10-fold (Extended Data Fig. 9h-i). To test whether squalene could rescue the worsened disease course of EAE in SQS/McKO mice (Fig. 3a-d), we applied a prophylactic squalene treatment. Unexpectedly, exogenous squalene ameliorated clinical disabilities and reduced inflammatory cell number in conditional mutants to levels lower than in WT EAE controls (Fig. 6c-d). This overcompensation of the genetic defect suggests that squalene is limiting in WT microglia/macrophages, and squalene administration could enhance desmosterol-mediated LXR-signaling in WT mice with EAE.
Therefore, we administered prophylactic squalene to WT mice with EAE. Squalene-treated animals showed reduced clinical scores at the peak of disease (Extended Data Fig. 9j), and reduced numbers of inflammatory cells in the spinal cord (Extended Data Fig. 9k). Peripheral disease processes appeared largely unaffected by squalene, as evidenced by comparable numbers of peripheral T cells and myeloid cells in blood samples (Extended Data Fig. 9l). To exclude interference with the immunological priming phase of EAE, we applied squalene therapeutically commencing at disease onset. Again, squalene fed animals showed reduced clinical disease severity and reduced density of infiltrating CD4+ T cells and CD11b+/CD45+ phagocytes (Fig. 6e, Extended Data Fig. 9m). By light sheet microscopy, the total volume of Iba1+ phagocytes in squalene-treated mice was only about a third of control volumes. Further, phagocytes more frequently resembled homeostatic ramified microglia (Fig. 6f-g, Video S4-S5). Analysis in spinal cord tissue and isolated CD11b+ myeloid cells revealed that squalene ameliorated disease by reducing expression of pro-inflammatory markers such as Tnf and Nos2, and enhancing expression of anti-inflammatory markers such as Tgfb1 and Arg1 (Fig. 6h, Supplementary Table 11, Extended Data Fig. 9o). Increased expression of cholesterol efflux genes (Abca1, Apoe) in myeloid cells was accompanied by increased desmosterol tissue levels (Extended Data Fig. 9n), implicating enhanced LXR activation in squalene-treated mice. In squalene-treated EAE animals, expression of genes related to oligodendrocyte differentiation and remyelination (Plp1, Car2) increased, paralleled by upregulation of cholesterol synthesis genes including Dhcr24 (Fig. 6h). These in vitro and in vivo data support the concept that squalene supplementation ameliorates inflammatory demyelination by activating LXR-signaling to resolve inflammation and enhance repair.
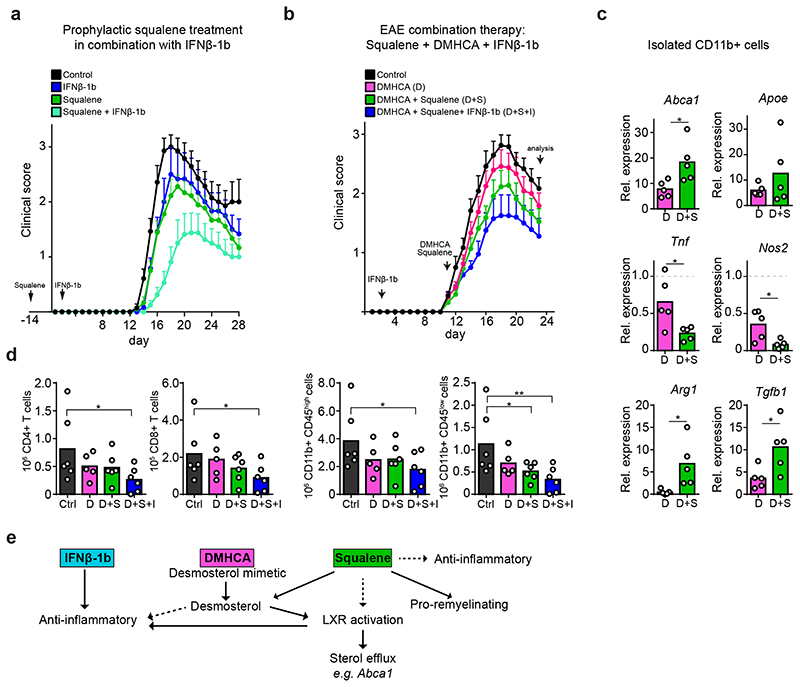
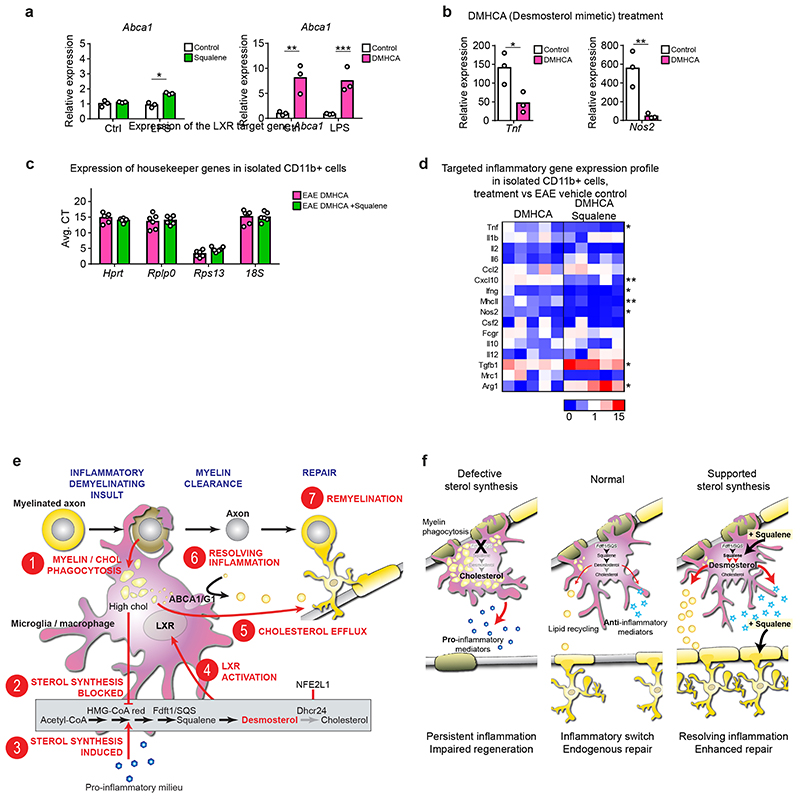
We next explored the relationship between squalene treatment and the anti-inflammatory standard first-line therapeutic IFNβ-1b. Single treatments ameliorated clinical scores to comparable degrees. However, in combination, squalene and daily injections of IFNβ-1b further reduced clinical scores. This shows the feasibility and safety of this treatment paradigm (Fig. 7a). We next tested whether squalene led to an additional benefit, when LXR-signaling was activated by the desmosterol mimetic and potent LXR agonist DMHCA (N,N-dimethyl-3ß-hydroxycholenamide, Extended Data Fig. 10 a-b)26. Therapeutic co-administration of DMHCA+squalene further ameliorated disease scores over DMHCA alone (Fig. 7b), potentially because of enhanced bioavailability of squalene compared to DMHCA under these experimental demyelinating conditions or LXR-independent anti-inflammatory properties of squalene. Accordingly, the effect of DHMCA on gene expression in CD11b+ phagocytes (upregulation of LXR target genes and anti-inflammatory mediators, repression of pro-inflammatory genes) was at least doubled in DMHCA+squalene-treated EAE mice (Fig. 7c, Extended Data Fig. 10c-d). Finally, the triple treatment with squalene, DMHCA and IFNβ-1b ameliorated EAE clinical disability most strongly; reflected also in attenuated densities of inflammatory cells (Fig. 7d). Taken together, in our MS models of demyelination, squalene administration mitigated disease severity at the level of microglia/macrophages by LXR-mediated suppression of inflammation and support of lipid recycling. In addition, squalene directly facilitated remyelination by oligodendroglia (Fig. 7e-f). Furthermore, squalene treatment can be combined with immunomodulatory drugs, suggesting profound implications for therapeutic intervention in MS.
Figure 7. Combination therapy of squalene and interferone ameliorates EAE.
(a) Mean clinical scores ± SEM of WT control (n=7), IFNβ-1b-treated (n=6), prophylactically squalene-treated (n=9) and combination-treated (squalene + IFNβ-1b, n=8) EAE animals.
(b) Mean clinical scores ± SEM of EAE animals treated therapeutically with DMHCA (n=12), DMHCA + Squalene (n=11), DMHCA + Squalene in combination with IFNβ-1b (n=10) or left untreated animals.
(c) Relative expression of LXR target genes and inflammatory genes in isolated CD11b+ cells from treatment groups in (b, n=5 animals), normalized to EAE control (n=6 animals). Asterisks mark significant changes (Student’s t-test, two-sided).
(d) Flow cytometric quantification of inflammatory cells at chronic EAE of animals in (b, n=5 animals, one-way ANOVA with Holm-Sidak).
(e) Scheme depicting suggested treatment effects of IFNβ-1b, DMHCA and squalene on inflammation, LXR-signaling and remyelination.
Data represent means (data points) of individual animals. **p<0.01, *p<0.05.
Discussion
Oligodendroglial synthesis of cholesterol, or access to cholesterol, is essential for myelin membrane growth in development2 and myelin disease12,23, and compounds linked to cholesterol homeostasis promote OPC differentiation27. Here, we could confirm this concept in animal models of myelin repair. Importantly, the cell type-specific sterol synthesis and horizontal cholesterol flux in the microenvironment of demyelinated lesions likely contributes to efficacy of repair. The analysis of mice with genetically blocked sterol synthesis at the level of squalene synthase (SQS) in different brain cell types led to the unexpected discovery that efficient myelin repair of acute lesions requires sterol synthesis by microglia/macrophages but not oligodendrocytes. Since in acutely demyelinated CNS lesions, microglia/macrophages internalize (cholesterol-rich) myelin debris, it is particularly difficult to understand why these cells have to synthesize sterols to reinforce remyelination by oligodendrocytes.
Microglia in CNS remyelination
Remyelination of experimentally demyelinated lesions requires the pro-inflammatory activation of microglia/macrophages16,28 to clear myelin debris. Then, pro-inflammatory destructive phagocytes undergo active cell death or adopt an anti-inflammatory pro-regenerative signature10,29–31 followed by lipid remodeling.
In non-repairing MS lesions, the pro-inflammatory milieu and microgliosis persist32, and also that sterol synthesis-deficient microglia/macrophages remain chronically activated in demyelinated lesions. In contrast, accumulating sterol precursors due to the genetic defect should rather decrease pro-inflammatory cell activation33. We hypothesize that the inability of SQS-mutant phagocytes to adopt an anti-inflammatory signature reflects a critical absence of desmosterol. Desmosterol, the immediate cholesterol precursor, is a well-known activator of LXR-signaling that, outside the nervous system, facilitates the resolution of the pro-inflammatory phenotype13,34–36. Our mutant microglia/macrophages did not upregulate classical LXR target genes of cholesterol export and hence failed to secrete phagocytosed lipids. In our experiments, desmosterol dictated the efficiency of resolving gliosis, cholesterol recycling and remyelination. Together with our findings of genetic and pharmacological manipulations of post-squalene sterol synthesis in vitro and in vivo, we can propose a working model of endogenous repair of demyelinated lesions (Fig. 7f, Extended Data Fig. 10e).
Cholesterol, sterol synthesis and remyelination – a working model
When myelin degenerates, microglia/macrophages take up large amounts of cholesterol from phagocytosed myelin debris (Fig. 7f, Extended Data Fig. 10e). While this inhibits their cholesterol synthesizing machinery by feedback control37, the pro-inflammatory milieu promotes cholesterol synthesis38. Indeed, we observed upregulated transcripts of sterol synthesizing enzymes apart from Dhcr24 mRNA, encoding the last enzyme of cholesterol biosynthesis. Dhcr24 downregulation could be mediated by NFE2L1/NRF1, an inflammation-responsive transcriptional repressor whose activity depends on cellular cholesterol excess19. The selective loss of Dhcr24 in microglia/macrophages at sites of inflammatory demyelination will transiently increase desmosterol levels and activate LXR-signaling. This stimulates transcriptional upregulation of genes for cholesterol export (recycling) and importantly suppresses the pro-inflammatory phenotype, which collectively facilitates remyelination. The cholesterol for remyelination of acute lesions likely originates from recycled myelin debris as oligodendrocytes as well as astrocytes decrease transcripts of cholesterol synthesis.
In addition to desmosterol, oxysterols could induce LXR-signaling3. Oxysterols contributed to LXR activation in our mice but failed to compensate the lack of desmosterol synthesis in SQS mutant phagocytes. In patients with MS, the role of oxysterols is unclear, and the increased oxysterol concentrations found in some individuals are rather considered biomarker of disease progression39,40.
We speculate that this sterol-regulated process helps emerging lesions to repair efficiently, as long as the microglia/macrophages respond properly, which is the case in juvenile or young adult wild type brains. However, in the aging white matter, microglia/macrophage functions are increasingly compromised, giving rise to a pro-inflammatory phenotype and perturbed lipid recycling15,41,42, paralleled by a gradual decrease of sterol synthesis in the aging brain43. Thus, in diseases involving acute demyelination and severe inflammation, such as in mouse EAE or human MS, aging microglia/macrophages constitute a critical risk factor for remyelination to fail.
Relevance to Multiple Sclerosis
Our data are relevant for human MS, also showing downregulation of DHCR24 and upregulation of LXR target genes in repairing lesion areas18,44,45. It is tempting to presume that the persistent MS lesions are exactly the sites where the local repair mechanisms involving active sterol synthesis, lipid recycling and switching to an anti-inflammatory phenotype in microglia/macrophages have failed, leading to a vicious circle of inflammation and demyelination. We speculate the local coincidence of extensive demyelination (cholesterol uptake blocking sterol synthesis), auto-aggressive immune cells (activating microglia/macrophages), and “above-threshold” microglial perturbation with the combined effects of aging (lipofuscin deposits, cholesterol crystals, epigenetic changes) causes lesion pathology. In contrast, normal appearing white matter would not exceed this "threshold" of microglia/macrophage impairment. Here, inflammation would largely resolve and lesions would quickly remyelinate, possibly even without leaving a visible pathology, as suggested by gene expression profiles and C14 carbon dating11,20.
Therapeutic implications
In MS, there is urgent need for therapeutic strategies targeting both, inflammation and remyelination4. To show proof of principle for the critical role of desmosterol in microglia, we have successfully applied the desmosterol mimetic DMHCA (a synthetic LXR agonist), which efficiently targets traumatic brain injury in mice26,46. In models of inflammatory demyelination, the combination of this synthetic LXR agonist with the first-line anti-inflammatory MS medication and squalene achieved best therapeutic benefits. The natural compound squalene feeds into sterol synthesis and enhances both, i) the synthesis of desmosterol in microglia/macrophages (anti-inflammatory mode of action), and ii) cholesterol synthesis in oligodendrocytes for myelination (remyelinating mode of action). Importantly, squalene administration was well tolerated in mice, in agreement with studies in humans24. Interestingly, Mediterranean diets may reduce disease severity in MS47,48. The Mediterranean life-style can result in squalene consumption levels of 200-400 mg/day (compared to 30 mg/day in the US)49,50. Thus, squalene supplementation should be further investigated as a therapeutic strategy in human MS.
Materials and Methods
Animals
All animal studies were performed in compliance with the animal policies of the Max Planck Institute of Experimental Medicine, and were approved by the German Federal State of Lower Saxony. Animals were group-housed (3-5 mice) in a controlled environment (temperature of 22°C, 30-70% humidity, 12 hour dark/light cycle) and had access to food and water ad libitum. Adult male and female C57BL/6N mice (8-10 weeks of age) or genetic mouse mutants (8–10 weeks of age) were taken for all experiments. Male mice were subjected to cuprizone, EAE and lysolecithin experiments. Female mice were used for non-pathology induced experiments. Animals of same gender were randomly assigned to experimental groups (3-12 mice). Conditional cholesterol synthesis mutants in this study were generated by crossbreeding cell type-specific Cre lines (Supplementary Table 12) with mice harboring squalene synthase with two loxP sites flanking exon 5, referred to as SQS-flox mice (SQScKO). Conditional mutant were compared with the respective Cre control, i.e. PLP1-CreERT2::SSFXflox/flox mutants and PLP1-CreERT2 controls, Cspg4/NG2CreERT2/+::SSFXflox/flox and Cspg4/NG2CreERT2/+ controls, GLASTCreERT2/+::SSFXflox/flox mutants and GLASTCreERT2/+ controls, CX3CR1CreERT2/+::SSFXflox/flox mutants (SQS/McKO) and CX3CR1CreERT2/+ controls, SLCO1c1-CreERT2::SSFXflox/flox mutants and SLCO1c1-CreERT2 controls, Aldh1l1-CreERT2::SSFXflox/flox mutants and Aldh1l1-CreERT2 controls. To compensate for the prominent liver recombination by Aldh1l1-CreERT2, experimental animals were kept on chow supplemented with 0.5% cholesterol. Adult genetic demyelination was induced by establishing conditional MBP mutants, i.e. PLP1-CreERT2::MBPflox/flox (MBPcKO) and PLP1-CreERT2 controls. To analyses cholesterol export deficiency in phagocytes, CX3CR1CreERT2/+::ABCA1flox/flox ABCG1flox/flox experimental animals (ABC/McKO) and CX3CR1CreERT2/+ controls as well as APOE-/- experimental animals (ApoEKO) and APOE+/+(WT) controls were used. Cell type-specific Cre expression was verified by crossbreeding different Cre-driver lines to Rosa26tdTomato reporter mice.
Tamoxifen induced recombination
Transgenic mice received tamoxifen either by oral administration, three times every second day at a concentration of 0.4 mg/g body weight dissolved in corn oil:ethanol (1:9) or by intraperitoneal injections on 5 consecutive days at a concentration of 75 μg/g body weight.
Experimental autoimmune encephalomyelitis (EAE)
MOG-EAE was induced in mice purchased from Charles River by immunizing subcutaneously with 200 mg myelin oligodendrocyte glycoprotein peptide 35–55 (MOG35–55) in complete Freund’s adjuvant (M. tuberculosis at 3.75 mg ml-1, BD) and i.p. injection twice with 500 ng pertussis toxin (Sigma) as described12. Animals were examined daily and scored for clinical signs of the disease. If disease did not start within 15 days after induction or the clinical score rose above 4, animals were excluded from the analysis. The clinical score was: 0 normal; 0.5 loss of tail tip tone; 1 loss of tail tone; 1.5 ataxia, mild walking deficits (slip off the grid); 2 mild hind limb weakness, severe gait ataxia, twist of the tail causes rotation of the whole body; 2.5 moderate hind limb weakness, cannot grip the grid with hind paw, but able to stay on a upright tilted grid; 3 mild paraparesis, falls down from a upright tiled grid; 3.5 paraparesis of hind limbs (legs strongly affected, but move clearly); 4 paralysis of hind limbs, weakness in forelimbs; 4.5 forelimbs paralyzed; 5 moribund/dead. Treatment commencing either two weeks before immunization was defined as prophylactic regimen or at the first appearance of EAE symptoms defined as therapeutic regimen. IFNβ1b (Bayer) was administrated by daily intraperitoneal (i.p.) injections 30.000 U per animal.
Cuprizone
Cuprizone pathology was induced by feeding mice with 0.2% w/w cuprizone (Sigma-Aldrich) in powder chow. Mice received cuprizone for ‘demyelination’ (5 weeks), ‘acute remyelination’ (6 weeks) and ‘chronic remyelination’ (12 weeks followed by 2 weeks normal chow) paradigms. For ‘induced remyelination’ experiments, mice were fed cuprizone in standard chow for 4 weeks, followed by cuprizone withdrawal and feeding mice standard chow with or without therapeutic dietary supplementation (e.g. Squalene). Mice were fed three times a week an exceeding amount of chow by dispenser. Food intake and animal weight was monitored. Age-matched untreated controls were fed standard powder chow.
Focal spinal cord demyelinating lesions
Focal spinal cord demyelinating lesions were induced under anaesthesia (10 mg/ml ketamine and 1 mg/ml xylazine) by stereotactic injection of 1 μl lysolecithin (1%, from egg yolk, alphalysophosphatidylcholine, Sigma-Aldrich) into the left and right ventro-lateral funiculus between Th3 and Th4 of 10-week old animals, as previously described15. Midline dorsal blood vessel was used as reference and capillary was positioned 0.5 mm left and right. The injection was performed at a rate of 150 nl/minute. Injection site was marker by addition of Monastral blue (Sigma-Aldrich). 5 minutes after the delivery of the lysolecithin, the capillary was slowly retracted. At the day of injection, mice were randomly assigned to treatment groups.
Therapeutic diets
Therapeutic diets were prepared in powder chow (V1124 ssniff Spezialdiäten GmbH, Germany). Mice received either 0.5 % squalene (Sigma, oil) or 0.04% DMHCA51 (Avanti, in corn oil) chow. Controls received either normal powder chow or in case of DMHCA, corn oil supplemented chow. Mice were fed three times a week an exceeding amount of chow by dispenser. Therapeutic dose was calculated according to mean daily food intake per gram mouse52.
Serum Analysis
Animals were anesthetized by intraperitoneal injection of Avertin (tribromethanol), blood was collected by cardiac puncture, and serum was prepared after 4h clotting by centrifugation. Cholesterol measurements were done with the architectII system (Abbott Diagnostics). For squalene measurement, lipids were extracted from serum samples according to Bligh and Dyer53. The chloroform phase containing the lipids was evaporated at 40°C in a vacuum concentrator (Qiagen) and extracted lipids redissolved and saponified in glass tubes with 0.5 M KOH in EtOH for 30 min. Non-saponifiable lipids were extracted with n-hexane. After evaporation of the n-hexane phase at 40°C in a vacuum concentrator, non-saponifiable lipids were redissolved in MetOH and subjected to reverse phase HPLC (250/4 Nucleoshell RP C18 (Macherey and Nagel) connected to an Äkta FPLC (GE Healthcare, Unicorn 1.0), flow rate 0.25 ml/min, liquid phase acetonitril:EtOH 70:30 (v/v), online UV detection at 205 nm. Squalene retention and peak amplitude were related to injections of a range of concentration standards in MetOH. Amplitudes were normalized to the neutral lipid recovery as assessed via online detection of fluorescence of dehydroergosterol (excitation 325 nm, emission 375 nm, Quantamaster (PTI Felix 32 1.2) that was spiked into the serum sample.
Elevated Beam Test
To assess bilateral sensorimotor coordination that involves connecting white matter structures like the corpus callosum, elevated beam test (in-house made, MPI of experimental medicine) was applied. Mice were trained to run toward a hiding box on a beam (width 1.5 cm) one week before testing. In one testing session number of slips in a defined 55 cm distance over three repeats was counted. Mean number of slips after three testing sessions was used as read out.
Flow cytometry
Single-cell suspensions from spinal cords were obtained via mechanical dissociation on a cell strainer. Immune cells were separated over a two-phase Percoll-density gradient by centrifugation. Blood was collected by cardiac puncture in EDTA (80mM) and single-cell suspension was obtained by centrifugation over lymphocyte separation medium (PAA). Staining of CD3+/CD4+ T cells, CD3+/CD8+ T cells and CD11b/CD45 cells (macrophages/microglia) was performed using the following antibodies in a 1:200 dilution: anti-CD3e (Biolegend Cat # 100312, clone 145-2C11), anti-CD4 (Becton Dickinson Cat # 553730, clone GK 1.5), anti-CD8 (Becton Dickinson Cat # 100706, clone 53–6.7), anti-CD11b (Biolegend Cat # 101207, clone M1/70), anti-CD45.2 (Biolegend Cat # 109814, clone 104). The addition of Calibrite APC beads54 allowed for cell quantification. Flow cytometry was performed using a FACSCalibur operated by Cell Quest software c0.0 (Becton Dickinson) and a CytoFLEX S operated by CytExpert v2.4 software (Beckman Coulter v2.3). Small debris was removed with the preliminary FSC/SSC gating. Single, living cells were obtained by doublet exclusion. Analyzed cells were defined by marker expression and positive and negative staining populations were defined using istopye controls if necessary. Calibrite APC beads (Alizadeh and Karimi-Abdolrezaee) were used for cell quantification (For gating strategy see Extended Data Figures 5a, 5a).
Magnetic cell isolation (MACS)
Glial cells and neurons were isolated according to the adult brain dissociation protocol (Miltenyi biotec). Corpus callosum and/or cortex were isolated from Bregma +1.10 to −2.46. Antibody labeling was done according to the Microbead kit protocols (Miltenyi biotec): oligodendrocytes (O4, 1:40, Cat #130–096-670); astrocytes (ACSA-2, 1:10. Cat #130–097-679), microglia (CD11b, 1:10, Cat #130–093-636), and endothelial cells (CD31, 1:10, Cat #130–091-935). Purity of cell populations was routinely determined by qPCR on extracted and reverse transcribed RNA. To account for increased microglial cell populations in diseased corpus callosum, we pooled the corpus callosum from two control mice (n=1 sample) for the isolation of microglia and compared this with microglia isolated from one diseased mouse (n=1 sample). Expression levels of housekeeper genes in these cell fractions were in a comparable range (Extended Data Figures 2a, 5e, 9o, 10a).
Histochemistry
Mice were anesthetized and perfused with 4% formaldehyde (PFA). In case of cuprizone-treated animals, brain samples were cut at Bregma 1.58 to account for regional specificity of cuprizone pathology. Tissue was postfixed overnight, embedded in paraffin and cut into 5 mm sections (HMP 110, MICROM). Gallyas silver impregnation was done as described23. Immunohistological stainings were done on deparaffinized sections followed by antigen-retrieval in sodium citrate buffer (0.01 M, pH 6.0). For immunofluorescence detection, blocking was performed with serum-free protein block (Dako). Primary antibodies were anti-CAII (provided by S. Ghandour (no commercial antibody), dilution 1:150), anti-Iba1 (Wako #019-19741, dilution 1:1000), anti-Iba1 (Abcam Cat # ab107159, dilution 1:200), anti-CD3 (Serotec #MCA1477, 1:150), anti-LAMP1 (BD Pharmingen Cat #09671, 1:200), anti-PCNA (Abcam #ab29, dilution 1:1000), anti-OLIG2 (C. Stiles / J. Alberta, DF308 (no commercial antibody), dilution 1:200), diluted in 2% bovine serum albumin (BSA)/PBS and incubated for 48 h followed by incubation with fluorophore-coupled secondary antibodies (Alexa488 donkey anti-mouse Invitrogen Cat #A-21202, dilution 1:1000; Alexa488 donkey anti-rabbit Invitrogen Cat #A-21206, dilution 1:1000; Alexa488 donkey anti-rat Invitrogen Cat #A-21208, dilution 1:1000; Alexa594 donkey anti-rat Invitrogen Cat #A-21209, dilution 1:1000; Alexa555 donkey anti-rabbit Invitrogen Cat #A-31572, dilution 1:1000; Cy3 donkey anti-goat Jackson Immuno Research #705-165-147, dilution 1:500; Dylight633 donkey anti-rabbit Invitrogen #A-35562, dilution 1:1000). For chromogenic stainings, blocking of endogenous peroxidase activity with 3% hydrogen peroxide was performed followed by 20% goat serum (BSA/PBS)block and incubation with primary antibodies (anti-MAC3 Pharmigen Cat #01781D, dilution 1:400, anti-GFAP Chemicon #MAB3402, dilution 1:200). Detection was carried out with the LSAB®2 System-HRP (Dako, Hamburg, Germany, Anti-rabbit/mouse LSAB2 Kit Dako Cat#K0679, dilution 1:100) or the VECTASTAIN® Elite® ABC HRP Kit (Vector Labs, Anti-Rat IgG Vector Cat#BA-9400, dilution 1:100). HRP substrate 3,30-Diaminobenzidine (DAB) was applied by using the DAB Zytomed Kit (Zytomed Systems GmbH). Nuclear labeling was done by Haematoxylin stain followed by mounting (Eukitt). Samples were analyzed on an Axio Imager.Z1 (Zeiss) equipped with an AxioCam MRc3, x0.63 Camera Adaptor and the ZEN 2012 blue edition software using 10x objective (Plan Apochromat 10x/0.45 M27) or 20x objective (Plan-Apochromat x20/0.8) and evaluated with Image J software. Quantification of areas (Gallyas, GFAP, MAC3) were done by applying semi-automated ImageJ 1.52p software macro including thresholding (variable threshold in case of Gallyas and fixed threshold for antibody stainings) and color deconvolution. Two to four sections per animal were analyzed.
Multiple sclerosis tissue
Human cryoconserved material of MS diagnosed and non-MS patients was purchased from Netherland Brain Bank after being approved by the Ethics Tissue Advisory Board, Nederlands Herseninstituut (Project no. 1206). Four MS patients (three males, one female) and three non-MS control samples (two males, one female) were analyzed (Supplementary Table 10). Age of patients ranged from 47 to 61 years, with a median age of 49 years. Post mortem delay ranged between 5:15 to 11 hours. Non-MS material was selected by the absence of histopathological hallmarks. MS diagnosed patients were selected according to the presence of active, chronic active and chronic inactive lesions which was confirmed by histopathological examination. Frozen human material was cut on a cryotome to determine lesion localization and verify classification. Cryosections were fixed with 4% PFA for 5 days. For LFB staining section were incubated in working solution over night at 60°C following differentiation with 0.05% lithium carbonate and 70% isopropyl alcohol. PAS staining was achieved by 1% periodic acid incubation for 5 min. For KiM1p55 staining (anti-KiM1P antibody provided by HJ Radzun (no commercial antibody) dilution 1:50), antigen retrieval was performed with 10 mM citric acid buffer (pH 6.0) followed by blocking of endogenous peroxidase with 0.3% H2O2 in PBS for 10 min at 4°C and incubation and blocking with 10% FCS. First antibody was incubated overnight diluted in blocking buffer following biotinylated secondary antibody (GE Healthcare, Jackson ImmunoResearch and DCS Innovative Diagnostic Systems) incubation and peroxidase-conjugated avidin and DAB (Sigma-Aldrich) visualization. For sterol analysis by mass spectrometry, center and lesion rim as well as white matter of non-MS patients were dissected and stored at -80°C.
Electron microscopy
Electron microscopic analysis was done as described12. 4% PFA, 2.5% Glutaraldehyde, 0.1 M Phosphate buffer fixed sagittal sections were cut on a vibratome (Leica VT1200, 300 mm). The corpus callosum with adjacent tissue (-0.04mm lateral) was punched with a 2mm diameter punching tool and embedded in epon (EMTP, Leica) following region verification with methylene blue staining. Representative pictures (x12000 magnification, TRS, Moorenweis) of uranyl acetate contrasted ultrathin sections were taken with the Zeiss EM900.
Expression analyses
For expression analyses of tissue samples, mice were killed by cervical dislocation. Samples were quickly cooled and region of interest prepared. RNA was extracted using RNeasy Mini (Qiagen). cDNA was synthesized with Superscript III (Invitrogen). Concentration and quality of RNA was evaluated using a NanoDrop spectrophotometer and RNA Nano (Agilent). RNA from MACS-purified cells was extracted using QIAshredder and RNeasy protocols (Qiagen). cDNA was amplified by Single Primer Isothermal Amplification (Ribo-SPIA® technology) using Ovation PicoSL WTA System V2 (NuGEN) following the manufactures protocol. Quantitative PCRs were done in triplicates on 384-well plates using the GoTaq® qPCR Master Mix (Promega, A6002) and the LightCycler® 480 Instrument. Background subtraction and thresholding was performed using the LightCycler® 480 software 1.5 (Roche)56. Expression values were normalized to the mean of at least 2 out of the housekeeping genes Hprt, Rps13, Rplp0, Gapdh, 18S (Extended Data Fig. 2a, 5e, 9o, 10a). Quantification was done by applying the ΔΔCt method, normalized to experimental controls (set to 1). All primers (Supplementary Table 4) were designed to fulfill optimal criteria e.g. primer length (18-22 bp), melting temperature (52-58°C), GC content (40-60%), low number of repeats, amplicon length (<220 bp). All primers were intron-spanning.
Myelin preparation
Myelin was isolated from 12 weeks old mouse brains as described23. Brains were homogenized in 0.32M saccharose solution and added gently on top of 0.85M saccharose solution in an ultracentrifugation bucket and centrifuged for 30 minutes at 75.000 x g. Interphase, with roughly purified myelin, was removed washed with water, and centrifuged for 15 min at 75,000 x g. Osmotic shock was performed by water incubation for 15 minutes followed by centrifugation for 15 minutes at 12,000 x g. Pellet was resuspended in 0.35M saccharose solution added on top of 0.85m saccharose solution and centrifuged for 30 minutes at 75,000 x g. Purified myelin was washed with water, and centrifuged for 15 minutes at 75,000 x g. The pellet resuspended in PBS, homogenized and stored at -80°C. Protein concentration of myelin was measured with Lowry DC Protein Assay (Bio-Rad).
Myelinating Co-cultures
Myelinating cell cultures were established from embryonic day 13 (E13) mouse embryo (C57BL6/J Charles River) spinal cords as described previously57. Briefly, spinal cords were dissected and dissociated in trypsin, then plated at 150,000 cells per coverslip (3 coverslips per 35 mm Petri dish) in serum-containing plating medium (50% DMEM, 25% HBSS, 25% horse serum). Two hours later, differentiation medium (DMEM with 4.5 mg/ml glucose, hydrocortisone, insulin, biotin and N1) was added58. Two to three days later, half the medium was removed and replaced with differentiation medium plus HBSS (vehicle) or squalene (final concentration 100 mM). Cells were fed every 2-3 days. Differentiation medium minus insulin was used from day in vitro (DIV) 12. On DIV 21 and DIV 30 (both +/- 2 days), cells were fixed for 10 minutes in 4% paraformaldehyde then stored in PBS until immunostained. Treatments were randomised across 8 x 35 mm Petri dishes, and individual coverslips from 3 separate Petri dishes were fixed at each time point. Four independent experiments (embryos derived from 4 pregnant dams) were conducted. Cells were permeabilized in 0.5% Triton X100 in PBS for 10 minutes, blocked in 10% goat serum for 30 minutes and incubated overnight at 4°C in primary antibody (anti-MBP, Serotec Cat #MCA 4095, dilution 1:400; anti-SMI31 Covance Cat #SMI31P, dilution 1:400) in blocking solution. Secondary antibody (Alexa 488, Invitrogen Cat #A-21208, dilution 1:100; Alexa 594 Invitrogen Cat #A-21209, dilution 1:100) was applied in blocking solution for 1 hour after thorough washing. Cells were washed in PBS and briefly in tap water then coverslips were mounted on glass slides in Mowiol with DAPI. Axonal (SMI31) area and the area with myelin sheaths (MBP) of seven randomly chosen visual fields of myelinating co-cultures (x 20 magnification) was measured by automated threshold with Fiji Software (SMI31 Otsu, MBP Triangle). Specimens were analyzed on an Axiophot observer.Z1 (Zeiss) equipped with an AxioCam MRm and the ZEN 2012 blue edition software and evaluated with Image J software.
Bone marrow derived macrophages (BMDM)
BMDMs were isolated from femora bones of adult mice. 5x105 cells were plated 10 cm-diameter tissue culture dish in L929-CM (20%). Cells were cultivated (DMEM, 1% P/S, 10% fetal calf serum) at 37 °C with 7.5% CO2 v/v. In vitro recombination was achieved by 4-hydroxytamoxifen (Sigma-Aldrich) administration at a final dose of 4 μM for 10 days (dissolved in EtOH) starting 24h after plating. For experimental seeding, cells were harvested following accutase (Innovative Cell Technologies Inc.) induced detachment. Cells were either seeded into cell culture grade plasticware, onto Poly-L-lysine (PLL) coated, HCl-washed 1.5# 12 mm diameter glass coverslips, or into PLL coated glass bottom chamber slides (Ibidi) for live imaging. Cells were seeded (6-well: 250k cells; 96-well: 18k cells; 12mm coverslip: 18k cells; 8-chamber slide: 18k cells) in cultivation media. For activation, cells were treated with LPS (10 ng/mL) and/or myelin (10 μg/mL) if not otherwise stated. Squalene (100 μM in HBSS) or DMHCA (10 μM in DMF) were added 24h before activation.
In vitro phagocytosis
In vitro phagocytosis was analyzed by Fluoromyelin Green pre-labeling of myelin and fixation with 4% PFA. For fluorescence stainings, cell were permeabilized for 5 minutes with 0.1% TritonX-100, blocked for 30 minutes with 4% HS, 2% BSA in PBS and incubated with primary antibody (anti-Iba1, Wako Cat #019-19741, dilution 1:1000; anti-LAMP1, BD Pharmingen Cat #09671D, dilution 1:400) overnight. Cells were incubated with secondary antibodies in blocking solution for 1h followed by nuclear DAPI labeling. For analysis mean fluorescence intensity of Fluoromyelin-positive accumulations were quantified using CellProfiler 4.0, defining cells by DAPI and Iba1-positive staining. For phagocyte survival assay, live imaging was performed on an Axio Imager.Z1 (Zeiss) equipped with an AxioCam MRc3, x0.63 Camera Adaptor and the ZEN 2012 blue edition software using 20x objective (Plan-Apochromat x20/0.8) at 37°C and 5% CO2 (v/v). Cells were imaged over 16h with a frame rate of 5-8 minutes and Definite Focus (Zeiss). Analysis was performed with ImageJ 1.52p to determine survival rate.
Cholesterol staining
To visualize cholesterol-rich cellular domains BCtheta live cell staining was performed. Cell were washed on ice (DPBS), blocked for 10 minutes with 1% BSA in PBS and incubated with BCtheta (0.015 mg/mL) for 30 minutes following fixation with 4% PFA. Cells were imaged with a Leica SP8 Lightning microscope and LAS X software ver 1.0.0 at 63x magnification and stacks were processed as maximum intensity projection.
Measurement of Reactive Oxygen Species (ROS)
ROS production was measured by 2’,7’ dichlorofluoresceindiacetate (DCFDA) detection according to published protocols59 with minor modifications. For measurement, cells were incubated for 25 minutes at 37 °C with 5 μM DCFDA in 1x DPBS with 4.5 g/L D-glucose. DPBS was aspirated and cells were incubated for 10 - 20 minutes in DMEM (without PhenolRed, 4.5 g/L D-glucose and 3.7 g/L sodium bicarbonate), to allow deacetylation and oxidation. 96-well flat bottom plates were then measured with Eon plate reader (Biotek, Gen5 2.0) at excitation / emission of 495 nm / 529 nm. Relative ROS production was calculated after blank substraction and normalization to untreated controls (set to 1).
Quantification of sterols
Sterol abundance was quantified by lipid gas chromatography coupled to mass spectrometry (GC-MS) in corpus callosum (5w cuprizone), in lumbar spinal cord (28 days post immunization [dpi] EAE, 7 dpi lysolecithin), in acutely isolated cells (4-5 animals grouped for each replicate) or in cultured cells (700.000 to 3.500.000 cells). Samples were lyophilized at a shelf temperature of -56 °C for 24h under vacuum of 0.2 mBar (Christ LMC-1 BETA 1-16) and weighed for calculation of water content and normalization as described53. For the sterol analysis by GC-MS analysis, lyophilized tissue was ground to a fine powder using a shaking mill and glass balls (5 mm). Metabolites were extracted in a two-phase system of Methyl-tert-butyl ether: Methanol 3:1 (v/v) and H2O, and pentadecanoic acid was added as an internal standard. 10-200 μl of organic phase was dried under a stream of nitrogen, dissolved in 10-15 μl pyridine and derivatised with twice the volume of N-Methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) to transform the sterols and the standard to their trimethylsilyl (TMS)-derivatives. Each sample was analyzed twice, with a higher split to quantify cholesterol and with a lower split to measure all other sterols. The samples were analyzed on an Agilent 5977N mass selective detector connected to an Agilent 7890B gas chromatograph equipped with a capillary HP5-MS column (30 m x 0.25 mm; 0.25 μm coating thickness; J&W Scientific, Agilent). Helium was used as carrier gas (1 ml/min). The inlet temperature was set to 280 °C and the temperature gradient applied was 180 °C for 1 min, 180 – 320 °C at 5 K/min and 320°C for 5 min. Electron energy of 70 eV, an ion source temperature of 230°C, and a transfer line temperature of 280°C was used. Spectra were recorded in the range of 70-600 Da/e (ChemStation Software D.01.02.16). Sterols were identified by the use of external standards.
Immunoblot
Animals were sacrificed by cervical dislocation, corpus callosum samples were dissected on ice and snap frozen on dry ice. For protein analyses tissue samples were homogenized in pre-cooled sucrose buffer containing protease and phosphatase inhibitors (PhosSTOPTM, Roche) using precellys 24 (Bertin technologies) at 5.000rpm for 3x10 seconds. Lysates were centrifuged for 5min at 13.000 rpm to remove nuclei. Protein concentrations were measured with Lowry method (LOWRY et al., 1951) and Bio-Rad DC Protein Assay kit following manufacturer´s instructions. Protein separation was performed by SDS-PAGE (12 %) using Mini-PROTEANTM Handcast System (BIO-RAD). Samples were diluted in 1x SDS sample buffer and 8% 2-Mercaptoethanol or Dithiothreitol (0,5M). Protein lysates were denaturated on a shaker for 10 min at 40°C or 70°C corresponding to the used antibody. 10-25 μg sample was loaded at as well as 3 μl of pre-stained protein ladder (PageRulerTM, ThermoFisher Scientific). Proteins were transferred to PVDF membranes (GE Healthcare, 10600023) using a tank blot system (XCell SureLockTM Mini-Cell Electrophoresis System, ThermoFisher Scientific). Membranes were incubated in 1x TBS containing 0.05% Tween-20 (Promega, H5151, 1x TBS-T) and 5% w/v skim milk powder (blocking buffer) for 60 min under constant shaking at RT. Primary Antibodies (anti-ABCA1, Santa Cruz Cat#sc-58219, dilution 1:500; anti-GAPDH, Enzo Cat #ADI-CSA-335-E, dilution 1:500) were diluted in blocking buffer and incubated overnight at 4°Cunder constant shaking. Immunodetection was archieved by horseradish peroxidase (HRP)-coupled secondary antibodies (1:5000) diluted in blocking buffer for 1h at RT following incubation in Enhanced Chemiluminescence solution (Western Lightning® Plus, Perkin Elmer) and signal detection using the ChemoStar ECL Imager HR 6.0 (ChemoStar Touch v.0.5.68)60. Relative protein abundance (controls set to 1) was calculated using Image J Gel analyzer and normalized to loading controls (Actin or GAPDH).
Light sheet microscopy
PFA immersion fixed spinal cord segments were processed for whole mount immune-labelling and tissue clearing following a modified iDISCO protocol61. Briefly, samples were dehydrated in ascending methanol (MeOH)/PBS series followed by overnight bleaching/permeabilization in a mix of 5% H2O2/20% DMSO/MeOH at 4°C. Samples were further washed in MeOH and incubated in 20% DMSO/MeOH at RT for 2h. Then, samples were rehydrated using a descending methanol/PBS series and further washed with in PBS/0.2% TritonX-100 for 2h. The samples were then incubated overnight in 0.2% TritonX-100, 20% DMSO, and 0.3 M glycine in PBS at 37°C and blocked using PBS containing 6% goat serum, 10% DMSO and 0.2% Triton-X100 for 2 days at 37°C. Samples were retrieved, washed twice in PBS containing 0.2% Tween20 and 10μg/ml heparin (PTwH) and incubated with primary antibody solution (anti-Iba1, Wako Cat #019-19741, dilution 1:500; PTwH/5%DMSO/3% goat serum) for 7 days at 37°C. After several washes, samples were incubated with secondary antibody solution (Alexa 555 Invitrogen Cat #A-31572 dilution 1:500 in PTwH/3% goat serum) for 4 days at 37°C. Prior to clearing, the samples were washed in PTwH and embedded in 2% Phytagel (Sigma Aldrich #P8169) in water. The embedded tissue was then dehydrated using an ascending series of Methanol/PBS and incubated overnight incubation in a mixture of 33% dichloromethan (DCM) and 66% MeOH at RT. Samples were further delipidated by incubation in 100% DCM for 40min and transferred to pure ethyl cinnamate (Eci; Sigma Aldrich #112372) as clearing reagent. Tissues became transparent after 15min in Eci and were stored at RT until imaging. Light sheet microscopy was performed using a LaVision Ultramicroscope II equipped with 2x objective, corrected dipping cap and zoom body. Spinal cords were mounted onto the sample holder with the dorsal/ventral axis facing down (z imaging axis = Dorsoventral axis spinal cord). The holder was placed into the imaging chamber filled with Eci. Images were acquired in 3D multicolour mode with the following specifications: 5μm sheet thickness; 40% sheet width; 2x zoom; 4μm z-step size; one site sheet illumination; 100ms camera exposure time; full field of view. Autofluorescence was recorded using 488nm laser excitation (80% laser power) and a 525/40 emmision filter and red fluorescence was recorded using 561nm laser excitation (30% laser power) and 585/40 emission filters. Images were loaded into Vision4D 3.0 (Arivis) and the imageset was cropped to 500 - 2000 pixels corresponding to 2,2 mm of spinal cord length. The volume of the spinal cord was determined by performing an automatic intensity thresholding on the autofluorescence channel. Phagocytes were detected by running a manual intensity thresholding on the 561nm channel and Iba1 cell accumulation with a volume of <1000μm3 was considered lesion positive. Then total lesion volume as well as the lesion volume fraction in respect to the total spinal cord volume were calculated. For 3D rendering, the autofluorescence and Iba1 channel were depicted in green and red pseudocolor, respectively. High-resolution images as well as videos were created using the Arivis 4D viewer.
Human single-cell/nuclei transcriptome sequencing datasets
Human single-cell/nuclei RNA sequencing profile were obtained from two available datasets, GSE118257 and GSE124335, and analyzed by R 4.0.2 using R package Seurat v3.1.0 and R package ggplot2 v3.2.062,63. Subset gene counts of 990 nucleus which are identified as microglia, macrophages or immunological oligodendrocytes from GSE118257, were filtered by genes per nuclei with a UMI cut-off of >200 and <3,000. Raw counts matrix recovered from GSE124335 was filtered with number of genes per cell >200 and <3,500. After quality control, both expression matrices were log-normalized with a scale factor of 10,000 and further centered with Seurat analytical pipeline default setting. For merging the two matrices, Canonical Correlation Analysis eliminated the batch effect. The union of top 2,000 variable features from each matrix, and the first 30 canonical correlation components were chosen for further analysis. The resulting four major clusters were detected under resolution 0.1 and two dimensional visualization was performed by Uniform Manifold Approximation and Projection (UMAP)64. Subsequently, we analyzed gene expression by subgrouping cells into control (Ctrl; non-MS control/normal appearing white matter) or multiple sclerosis (MS; Active lesions). Proportion and average gene expression level of cells in each cluster were calculated and then visualized using R package ggplot2 v3.2.065. Cluster 4 was annotated as ‘non-phagocyte cluster’, containing mostly cells from MS patients, especially CD3 expressing T cells and FSP1 expressing non-microglia/macrophages (Extended Data Fig. 7). The expression profile of cells in cluster 3 that contained similar cell numbers from control individuals and MS patients, is in agreement with the recently designated ‘immune oligodendroglia cluster’ exemplified by expression of canonical oligodendrocyte genes such as PLP1 (Extended Data Fig. 7)20. Cluster 1 and 2 were annotated as myeloid cells (Microglia/Macrophages) expressing typical marker genes such as CX3CR1, CD68 and AIF1. Cells in cluster 2 (annotated as lipid export cluster) were compared to cells in cluster 1 (annotated as homeostatic cluster) to detect Differentially Expressed Genes (DEGs). Gene ontology enrichment analysis of DEGs were performed by GSEA 4.1.066,67.
Quantification and statistical analysis
Number of animals and in vitro replicates for each experiment is shown in the figure legends. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications2,12,23. No inclusion or exclusion criteria were used if not otherwise stated. Animals/cultures were randomly allocated to treatment groups. Studies were conducted blinded to investigators. In EAE experiments, we excluded animals from the analysis, if disease did not start within 15 days after induction or the clinical score rose above 4. No other animals or data points were excluded from the analysis. Genes not detected in the expression analysis are indicated as NA. Data are expressed as mean ±SEM unless otherwise indicated. For statistical analysis, unpaired two-sided Student’s t-test, one-way ANOVA with Tukey correction for multiple comparisons, two-way ANOVA followed by Tukey's post hoc test to compare interactions or Holm-Sidak method when appropriate, were applied. Normality was tested by using the Kolmogorov-Smirnov test. In case of n numbers were below 5, we assumed normal distribution. Nonparametric Mann-Whitney test was used when data did not pass the normality test. Wilcoxon Rans Sum test was used for analysis of scRNA-seq data material. Data analysis was performed using GraphPad Prism Software Version 6 and the R software. A value of p<0.05 was considered statistically significant. Asterisks depict statistically significant differences (* p<0.05, ** p<0.01, *** p<0.001). Statistical source data with exact P values for each comparison are provided in Supplementary Table 13.
Extended Data
Extended Data Fig. 1. Mouse models of demyelination and remyelination.
(a) The cuprizone model of demyelination and remyelination exhibits some similarities to pattern III MS lesions25. Although the exact molecular cause of the cuprizone induced pathology remains elusive, copper chelation induced dysfunction of mitochondrial enzymes could selectively lead to apoptosis in oligodendrocytes (Praet et al., 2014). Cuprizone (bis-cyclohexanone oxaldihydrazone) is administered to adult mice as a 0.2% dietary supplement, which leads to a highly reproducible oligodendropathy followed by demyelination of distinct brain regions, predominantly the corpus callosum (CC). The graphical representation of myelination and microgliosis is based on histochemical stainings as shown in lower panels (further details to this model12,25 and Matsushima and Morell, 2001). Feeding mice with cuprizone induces progressive loss of mature oligodendrocytes (CAII) that is already detectable two to three days following cuprizone administration and accompanied by gliosis and blood-brain barrier impairment (Berghoff et al., 2017, Stumpf et al., 2019). Loss of oligodendrocytes induces demyelination (Gallyas) in the corpus callosum starting after 2-3 weeks of cuprizone treatment. Loss of mature oligodendrocytes to less than 10% of control numbers and maximum demyelination of the corpus callosum is achieved after 5 weeks of continuous cuprizone supplementation (5w, acute demyelination). During the 5 weeks, OPCs proliferate (OLIG2) and remain largely undifferentiated. Whether OPCs do not differentiate during this disease period or differentiate, attempt to remyelinate and subsequently die is unclear. Demyelination is accompanied by progressive microgliosis (MAC3, dashed gray line). Following the peak at 5w, microgliosis quickly resolves within the following week despite ongoing cuprizone administration, leading to moderate microgliosis at 6 weeks cuprizone treatment (about 30% of MAC3 levels at 5w). This coincides with a brief remission phase characterized by oligodendrocyte maturation. The corpus callosum remyelinates reaching levels of about 50% myelin content of untreated controls (6w, acute-phase remyelination). In case of ongoing cuprizone treatment, remyelinated myelin sheaths subsequently degenerate. Microglial numbers progressively decrease despite ongoing degeneration leading to residual numbers of microglia at 12 weeks cuprizone. At this stage, the corpus callosum is chronically demyelinated (12w, chronic demyelination). CAII numbers of ~30% of untreated controls might also reflect a second spontaneous but minor remyelination phase. When cuprizone is withdrawn at this point and mice are fed normal chow, remyelination commences and reaches ~20% myelin content of untreated controls after 2 weeks recovery (12+2 weeks, chronic-phase remyelination) (Scale 50 μm).
(b) Quantitative RT-PCR analysis determining the expression of oligodendrocyte and myelin related genes (Car2, Plp1, Olig2), and marker genes for microglia (Aif1) and astrocytes (Gfap) in corpus callosum samples during acute and chronic remyelination. Bars represent the means with individual data points normalized to the demyelination time point 5w cuprizone (n=4 animals, set to 1, dotted line). Acute-phase (6w, n=4 animals) and chronic-phase (12+2w, n=4 animals) remyelination is reflected in increased transcripts of genes related to oligodendrocyte maturation and myelination (Car2, Plp1) in corpus callosum samples. The resolving microgliosis between 5w and 6w of cuprizone feeding is reflected in an about 40% drop of Aif1 transcripts. Asterisks mark significant changes, ***p<0.001 (Student’s t-test, two-sided).
(c) The lysolecithin model of demyelination and remyelination in the spinal cord15 (Bjelobaba et al., 2018). Focal spinal cord demyelinating lesions are induced by stereotactic injection of 1 μl lysolecithin (1%) into the left and right ventro-lateral funiculus between Th3 and Th4. Demyelination starts within hours and proceeds until about 5-7 days post injection, which is paralleled by microgliosis and OPC proliferation. Then phagocyte activity progressively resolves resulting in low levels of activated microglia at 3 weeks post induction. After clearance of myelin debris, oligodendrocytes differentiate and remyelination starts after about 10 days post induction. Like other toxic models, pathology in the lysolecithin is not mediated by inflammatory immune cell infiltration, although some invasion of T- and B-lymphocytes occur and the blood-brain barrier is disrupted by the experimental procedure (Yazdi et al., 2015) (Scale 100 μm). In contrast to the cuprizone model, repeated demyelination does not result in limited remyelination and reduced number of OPCs (Penderis et al., 2003).
(d) MOG-EAE (experimental autoimmune encephalomyelitis) causes a monophasic inflammatory demyelinating disease and models predominantly inflammatory aspects of MS pathology (Constantinescu et al., 2011; Ransohoff, 2012). Mice are immunized with the MOG35-55 peptide (amino acid 35-55 of myelin oligodendrocyte protein) that induces expansion of autoreactive T lymphocytes during a preclinical phase. Already during this disease period, the blood-brain barrier is compromised (Paul et al., 2013) and some CNS immune cell entry and cytokine production occur. After 10-13 days post immunization clinical disabilities become evident characterized by ascending paralysis. Histopathology is more pronounced in spinal cord tissue than in brain. Lesions are characterized by marked infiltration of encephalitogenic effector T cells (CD3), as well as recruitment and activation of microglia/macrophages cells (Iba1) and demyelination (MBP). Tissue damage during this acute disease phase of EAE reaches a peak after several days followed by a chronic phase with milder but persistent disability. Remyelination events that are considered to be minor. Scale 50 μm.
Berghoff, S.A., Düking, T., Spieth, L., Winchenbach, J., Stumpf, S.K., Gerndt, N., Kusch, K., Ruhwedel, T., Möbius, W., and Saher, G., Acta neuropathologica communications, 2017, 5, 94.
Bjelobaba, I., Begovic-Kupresanin, V., Pekovic, S., and Lavrnja, I., J Neurosci Res, 2018, 96, 1021-1042.
Constantinescu, C.S., Farooqi, N., O'Brien, K., and Gran, B., Br J Pharmacol, 2011, 164, 1079-1106.
Matsushima, G.K., and Morell, P., Brain pathology, 2001, 11, 107-116.
Paul, D., Cowan, A.E., Ge, S., and Pachter, J.S., Microvascular research, 2013, 86, 1-10.
Penderis, J., Shields, S.A., and Franklin, R.J., Brain : a journal of neurology, 2003, 126, 1382-1391.
Praet, J., Guglielmetti, C., Berneman, Z., Van der Linden, A., and Ponsaerts, P., Neurosci Biobehav Rev, 2014, 47, 485-505.
Ransohoff, R.M., Nature neuroscience, 2012, 15, 1074-1077.
Stumpf, S.K., Berghoff, S.A., Trevisiol, A., Spieth, L., Düking, T., Schneider, L.V., Schlaphoff, L., Dreha-Kulaczewski, S., Bley, A., Burfeind, D., Kusch, K., Mitkovski, M., Ruhwedel, T., Guder, P., Rohse, H., Denecke, J., Gärtner, J., Möbius, W., Nave, K.A., and Saher, G., Acta Neuropathol, 2019, 138, 147-161.
Yazdi, A., Baharvand, H., and Javan, M., Neuroscience, 2015, 311, 34-44.
Extended Data Fig. 2. Generation and validation of cell specific cholesterol synthesis mutants.
(a) Quantitative RT-PCR analysis determining the expression of housekeeping reference genes in corpus callosum or isolated cells from mice (n=4 animals) treated with cuprizone (A astrocytes, EC endothelial cells, M microglia/macrophages, OL oligodendrocytes).
(b) Conditional mutants were generated by crossing SQSflox/flox mice to Cre driver lines. Fdft1 was inactivated by tamoxifen injections, and mice were analyzed at time points as indicated (middle panel). Fdft1 (SQS) expression in acutely isolated cells targeted by the conditional mutation shown as mean of fold values (OLcKO n=4, OPCcKO n=3, AcKO n=3, McKO n=3, ECcKO n=3) normalized to cells from control mice (OLcKO-Ctrl n=4, OPCcKO-Ctrl n=3, AcKO-Ctrl n=3, McKO-Ctrl n=4, ECcKO-Ctrl n=3; set to 1). Asterisks mark significant changes (Student’s t-test, two-sided).
(c) Body weight (OLcKO n=15, OPCcKO n=7, AcKO n=5, McKO n=10, ECcKO n=5; OLcKO-Ctrl n=11, OPCcKO-Ctrl n=6, AcKO-Ctrl n=5, McKO-Ctrl n=9, ECcKO-Ctrl n=6) and total serum cholesterol (OLcKO n=7, OPCcKO n=7, AcKO n=5, McKO n=5, ECcKO n=8; OLcKO-Ctrl n=4, OPCcKO-Ctrl n=6, AcKO-Ctrl n=5, McKO-Ctrl n=5, ECcKO-Ctrl n=5) of analyzed cell-specific cholesterol synthesis mutants in (b) at the age of 22 weeks. Bars represent mean of individual animals. Asterisks mark significant changes (Student’s t-test, two-sided).
(d) Evaluation of astrocyte-specific cholesterol synthesis mutants (Aldh1l1-CreERT2 driver line, Winchenbach et al., 2016), determining Fdft1 expression in isolated astrocytes, body weight and total serum cholesterol in mutants (isolated astrocytes n=3 animals; body weight n=9 animals, cholesterol n=7 animals and Cre controls (isolated astrocytes n=4 animals; body weight n=3 animals, cholesterol n=8 animals). Bars represent the means with individual data points normalized to controls. Asterisks mark significant changes (Student’s t-test, two-sided).
(e) Targeted sterol metabolism gene expression profile of isolated cells from the corpus callosum of n=4 untreated WT animals. The relative expression heat map shows low transcript levels (blue) and high transcript levels (red) for each gene normalized to the mean of all samples.
(f) Quantification of histological stainings for myelination (Gallyas), mature oligodendrocytes (CAII), oligodendrocyte linage cells (Olig2), microgliosis (MAC3) and astrogliosis (GFAP) in Aldh1l1-CreERT2 astrocyte cholesterol synthesis mutants (n=4 animals) as in (d) at 6 weeks cuprizone. Bars represent the means with individual data points normalized to controls (n=4 animals) set to 100).
***p < 0.001, **p < 0.01, *p < 0.05
Winchenbach, J., et al. Inducible targeting of CNS astrocytes in Aldh1l1-CreERT2 BAC transgenic mice. F1000Research 5, 2934 (2016).
Extended Data Fig. 3. Remyelination efficiency in conditional mutants.
(a) Expression of cholesterol synthesis genes in isolated astrocytes from EAE lesions. Bars represent the means with individual data points (n=4 animals) normalized to isolated cells from controls (n=4 animals, set to 1). Asterisks mark significant changes (Student’s t-test, two-sided).
(b) (Upper panel) Generation of MBP conditional mutants (MBPcKO) and (Lower panel) expression of cholesterol synthesis genes in isolated astrocyte 12 month post tamoxifen administration. Bars represent the means with individual data points (n=4 animals) normalized to isolated cells from control (n=4 animals, set to 1). Asterisks mark significant changes (Student’s t-test, two-sided).
(c) Tamoxifen protocol to induced recombination during cuprizone paradigm. Tamoxifen was given either before cuprizone application to target SQS in astrocytes, microglia, OPCs and endothelial cells or before remyelination to target oligodendrocytes.
(d) TDTO reporter expression during remyelination to evaluate cell targeting in cuprizone fed mice by co-labeling with CAII for oligodendrocytes, OLIG2 for oligodendrocyte linage cells, GFAP for astrocytes and Iba1 for microglia (Scale 50 μm).
(e) Elevated beam testing of cholesterol synthesis mutants during chronic remyelination (OLcKO n=8, OPCcKO n=7, AcKO n=5, McKO n=5, ECcKO n=7; OLcKO-Ctrl n=8, OPCcKO-Ctrl n=7, AcKO-Ctrl n=5, McKO-Ctrl n=5, ECcKO-Ctrl n=8). Bars represent the means with individual data points of three testing sessions. Asterisks mark significant changes (Student’s t-test, two-sided).
***p < 0.001, **p < 0.01.
Extended Data Fig. 4. Ablation of microglial cholesterol synthesis does not affect demyelination in the cuprizone model.
(a) Generation and cuprizone treatment paradigm of phagocyte-specific Abca1/Abcg1 cholesterol export mutants (ABC/McKO).
(b) Representative images from control (n=3 animals) and SQS/McKO mice (n=3 animals) after 5w cuprizone (acute demyelination), showing myelination (ultrastructure, scale 1 μm), oligodendrocytes (CAII), oligodendrocyte lineage cells (OLIG2) and microgliosis (MAC3) (Scale 50 μm) with corresponding quantification on the right. Asterisks mark significant changes (Student’s t-test, two-sided).
(c) Fdft1 expression in SQS-KO BMDMs cultures that were recombined in vitro (n=4 cultures). Data normalized to control cells (n=3 cultures, set to 1). Asterisks mark significant changes (Student’s t-test, two-sided).
(d) Cholesterol labeling in SQS-KO BMDMs (Ctrl n=3, SQS-KO n=4 cultures) cultured in lipid-depleted media by live staining with BCtheta (Scale 10μm).
(e) In vitro Fluoromyelin phagocytosis in control and SQS-KO BMDM cultures after a 4h challenge with myelin (Scale 10μm). Internalized myelin (fluoromyelin) was quantified by co-labeling for Lamp1-positive endo/lysosomes (Ctrl n= 7 cultures, SQS-KO n=4 cultures; 600-1050 cells).
(f) Mean gene expression related to NLRP3 inflammasome and cholesterol efflux (Abca1) in isolated phagocytes from SQS/McKO mice (n=5 animals) normalized to cuprizone control (n=3 animals). Asterisks mark significant changes (Student’s t-test, two-sided).
(g) Abca1 and Abcg1 expression in ABC-KO BMDMs cultures that were recombined in vitro (n=4 cultures). Data normalized to control cells (n=4 cultures, set to 1). Asterisks mark significant changes (Student’s t-test, two-sided).
(h) In vitro survival of BMDM following myelin (10 μg/ml) treatment (n=17 cultures from control mice; n=16 from SQS mutants and ABC mutants, n=12 from APO knockout mice; each with 251-351 cells). About 50% of SQS mutant phagocytes died within 16h of incubation with myelin, with morphology attributable to pyroptosis (Video S1). Of note, in two lipid export mutants, i.e. ABCA1/ABCG1 and ApoE-deficient phagocytes, pyroptotic cell death was even more pronounced, whereas control phagocyte numbers did not change. Asterisks mark significant changes (one-way ANOVA with Tukey’s post test).
***p < 0.001, **p < 0.01, *p < 0.05
Extended Data Fig. 5. EAE pathology in SQS/McKO animals.
(a) Representative pictures showing gating strategy to analyze lymphocytes and monocytes by flow cytometry.
(b) Flow cytometry plots depicting CD4+ T cells (left), CD8+ T cells (middle) and myeloid subpopulations (right) consisting of macrophages (CD11b+/CD45high) and microglia (CD11b+/CD45low).
(c) Flow cytometric quantification of inflammatory cell abundance at chronic EAE of SQS/McKO (n=8 animals) and control (n=8 animals) spinal cord samples. Asterisks mark significant changes (Student’s t-test, two-sided).
(d) Light sheet microcopy of SQS/McKO (n=3 animals) and control (n=3 animals) spinal cord samples stained for Iba1+ phagocytes (red) and autofluorescence imaging. Arrows point to circular areas with low optical density suggestive of focal edema that was only occasionally observed in EAE controls (Scale 100μm).
(e) Quantitative RT-PCR analysis determining the expression housekeeping reference genes in tissue and isolated CD11b+ cells from cuprizone treated SQS/McKO (n=5 animals), cuprizone control mice (n=4 animals), EAE (n=6 animals) and EAE control mice (n=5 animals).
***p < 0.001, **p < 0.01, *p < 0.05
Extended Data Fig. 6. LXR target gene upregulation and desmosterol accumulation in demyelinating mouse models.
(a) Quantitative RT-PCR analysis determining expression of Abca1 and Apoe in isolated CD11b+ cells from chronic EAE (left panel, n=4 animals), MBPcKO (middle panel, n=4 animals) and lysolecithin (right panel, n=3 animals) lesions. Bars represent the means with individual data points normalized to isolated cells from control (EAE n= 4 animals, MBPcKO-Ctrl n=4 animals, Lysolecithin Ctrl n=4 animals, set to 1) (Student’s t-test, two-sided).
(b) Relative abundance of lanosterol and cholesterol normalized to cell number in isolated CD11b+ myeloid cells during acute cuprizone-mediated demyelination (5w cuprizone), measured by lipid mass spectroscopy (Ctrl n=3 animals, cuprizone n=4 animals) (Student’s t-test, two-sided).
(c) Relative expression of Nrf1/Nfe2l1 in isolated microglia isolated from control mice and diseased mice (n=4 mice per group) after feeding cuprizone for 6 weeks (left), 12+2 weeks (middle) and 28d after induction of EAE (right) (Student’s t-test, two-sided).
(d) Relative abundance of desmosterol in corpus callosum during acute demyelination (5w cuprizone) of SQS/McKO animals (n=5 animals) and cuprizone controls (n=5 animals) measured by lipid mass spectrometry. Data normalized to non-disease controls (n=3 animals, set to 1) (one-way ANOVA with Tukey’s post test).
(e) Relative abundance of desmosterol in spinal cord tissue during chronic EAE (28 dpi) of SQS/McKO animals (n=4 animals) and EAE controls (n=3 animals) normalized to untreated controls (n=4 animals, set to 1) measured by lipid mass spectroscopy (one-way ANOVA with Tukey’s post test).
(f) Total ion chromatogram, selected ion mass chromatograms and spectra from representative human tissue sample and commercial standards analyzed by GC-MS. Time axes of chromatograms are broken to show only the elution of sterols. Lower panel shows background subtracted (adjacent to peak) mass spectra of N-Methyl-N (trimethylsilyl) trifluoroacetamide (MSTFA)-derivated metabolites.
Asterisks mark significant changes, ***p < 0.001, **p < 0.01, *p < 0.05
Extended Data Fig. 7. Identity of immune cell cluster in human MS scRNA-seq datasets and characterization of analyzed human MS lesions.
(a) Distribution of immune cells from control and MS patients (left) and k-means clustering (right) of cells (resolution 0.1) on UMAP plot of merged external scRNA-seq datasets.
(b) Frequency distribution of cells from control and MS patients in each cluster.
(c) Expression of selected marker genes in T cells (CD3) and non-microglia/macrophages (FSP1, Fibroblast-Specific Protein-1) in cluster 4 of sc-RNAseq datasets.
(d) Expression of selected marker genes in myeloid cells (AIF1, CX3CR1), oligodendrocytes (PLP1) and DHCR24 expressing cells in analyzed immune cells of sc-RNAseq datasets.
(e) Expression of genes related to cholesterol export in analyzed immune cells of sc-RNAseq datasets.
(f) Histological characterization of isolated lesions from individual patients. Dotted line indicates lesion rim defined by myelin staining (LFB/PAS).Center activity (square) was determined by presents of phagocytic cells (KiM1P) (Scale 500μm).
(g) Tissue water (g/g dry tissue) of control white (n=3 patients) matter and MS lesion material (n=4 patients). R: Active lesion rim (n=5 lesions); AC: Active lesion center (n=3 lesions); IC: Inactive lesion center (n=4 lesions). Asterisks mark significant changes (one-way ANOVA with Holm-Sidak post test). **p < 0.01,*p < 0.05.
(h) Relative abundance of cholesterol normalized to input tissue weight and standard (controls set to 1) of control white (n=3 patients) matter and MS lesion material (n=4 patients). R: Active lesion rim (n=5 lesions); AC: Active lesion center (n=3 lesions); IC: Inactive lesion center (n=4 lesions). Asterisks mark significant changes (one-way ANOVA with Holm-Sidak post test). ***p < 0.001, **p < 0.01, *p < 0.05
Extended Data Fig. 8. Inflammatory response in wild type and SQS-KO BMDMs challenged with LPS.
(a) Mean ROS levels in control (left) and SQS-KO BMDMs (right) stimulated with or without LPS and treated with or without squalene (100μM) (n=8 cultures per condition). Data normalized to untreated control (set to 1). Asterisks mark significant changes (two-way ANOVA with Holm-Sidak post test).
(b-f) Relative expression of cholesterol efflux (b, c) and inflammatory (d-f) genes in control and SQS-KO BMDMs treated with myelin+LPS with or without squalene (as depicted in Fig. 5c), normalized to untreated WT cultures(n=4 cultures) (two-way ANOVA with Holm-Sidak post test).
Asterisks mark significant changes, ***p < 0.001, **p < 0.01, *p < 0.05
Extended Data Fig. 9. Squalene affects serum squalene levels in lysolecithin and EAE models of MS but not in cuprizone-treated mice.
(a) Peripheral serum cholesterol profile of control (n=5 animals) and squalene-treated WT (n=6 animals) animals 14 days post injection with lysolecithin. Measurement shows total cholesterol (TC), low-density lipoprotein cholesterol (LDL) and high-density lipoprotein cholesterol (HDL). Dotted lines indicate values of non-diseased WT mice (Student’s t-test, two-sided).
(b) Serum squalene concentration measured by HPLC in control and squalene-treated lysolecithin animals treated as in (a) (n=3 animals each group) (Student’s t-test, two-sided).
(c) Histopathological quantification of the lesion size and MAC3+ microglia/macrophages in the lesions from control (n=9 lesions from 5 animals) and squalene treated animals (n=11 lesions from 6 animals) at 14 dpi lysolecithin (Scale 100μm) (Student’s t-test, two-sided).
(d) Scheme depicting induced remyelination paradigm in the cuprizone model12. Mice were fed cuprizone for 4 weeks after which mice received chow supplemented with vehicle or squalene for one week (dotted line) followed by analysis. The gray line depicts the degree of myelination in case of continued cuprizone feeding.
(e) Peripheral serum cholesterol profile determining total cholesterol (TC), low-density lipoprotein cholesterol (LDL) and high-density lipoprotein cholesterol (HDL) in control (n=5 animals) and cuprizone (n=4 animals) mice treated as in (d). Dotted lines indicate values of non-disease WT mice (Student’s t-test, two-sided).
(f) Serum squalene concentration measured by HPLC in control and squalene-treated lysolecithin animals treated as in (d) (n=3 animals each group). Dotted line indicates values of non-disease controls.
(g) Histological quantification of myelination (Gallyas), mature oligodendrocytes (CAII), oligodendrocyte linage cells (OLIG2), microgliosis (MAC3) and astrogliosis (GFAP) at induced remyelination with squalene supplementation (n=3 animals) compared to control (n=7 animals) in mice treated as in (d).
(h) Peripheral serum cholesterol profile of control (n=10 animals) and squalene-treated (n=11 animals) EAE animals during peak of disease (16 dpi). Measurement shows total cholesterol (TC), low-density lipoprotein cholesterol (LDL) and high-density lipoprotein cholesterol (HDL). Dotted lines indicate values of non-disease WT mice.
(i) Serum squalene concentration of control (n=2 animals) and squalene-treated (n=3 animals) EAE animals during peak of disease (16 dpi) measured by HPLC.
(j) Clinical score ± SEM of control (n=6) and prophylactically squalene-treated (n=6) EAE animals analyzed at peak of disease (16 dpi).
(k) Number of CD4+ and CD8+ T cells, CD11b/CD45high macrophages and CD11b/CD45low microglia per gram spinal cord determined by flow cytometry at peak of EAE (16 dpi) in control (n=6 animals) and squalene-treated animals (n=6 animals) (Student’s t-test, two-sided).
(l) Quantification of blood inflammatory cells by flow cytometry from animals as in (j) during peak of EAE disease (n=6 animals each group).
(m) Analysis of inflammatory cells in spinal cord tissue by flow cytometry from control (n=6 animals) and therapeutically squalene-treated (n=5 animals) EAE animals (Fig. 6e) (Student’s t-test, two-sided).
(n) Relative abundance of sterol intermediates in spinal cord of squalene-treated EAE animals (n=4 animals) normalized to untreated EAE controls (n=4 animals, set to 1) (Student’s t-test, two-sided).
(o) Quantitative RT-PCR analysis determining the expression housekeeping reference genes in isolated CD11b+ cells from spinal cord from EAE control (n=6 animals) and EAE mice treated with squalene (n=6 animals).
Bar graphs represent means with individual data points. Asterisks mark significant changes, ***p < 0.001, **p < 0.01, *p < 0.05
Extended Data Fig. 10. Working model.
(a) Abca1 expression following squalene (100 μM, left) and DMHCA (10 μM, right) treatment in LPS activated BMDMs (n=3 cultures) (Student’s t-test, two-sided).
(b) Expression of inflammatory genes (Tnf, Nos2) in LPS (10ng/ml) activated WT BMDMs (n=3 cultures) 24h after DMHCA (10μM) treatment. Data normalized to untreated control (n=3 cultures, set to 1) (two-way ANOVA with Holm-Sidak post test).
(c) Quantitative RT-PCR analysis determining the expression housekeeping reference genes in spinal cord from EAE mice treated with DMHCA (n=6 animals) and squalene (n=6 animals).
(d) Fold expression profiles of inflammatory genes in CD11b+ cells isolated from spinal cord of EAE animals treated with DMHCA (n=6 animals) and squalene (n=6 animals) (normalized to vehicle-treated EAE mice, n=6 animals). Asterisks indicate significant differences between DMHCA and DMHCA/squalene co-treatment (Student’s t-test, two-sided).
(e) Resulting working model of the relationship between sterol metabolism in microglia/macrophages in an inflammatory demyelinating lesion and myelin repair. (1) Phagocytosis of lipid-rich myelin increases the cellular cholesterol content in microglia/macrophages, which (2) inhibits sterol synthesis by negative feedback regulation. (3) In demyelinating lesions, pro-inflammatory mediators counteract this inhibition and induce sterol synthesis. NRF1/NFE2L1 likely senses intracellular cholesterol excess and represses Dhcr24, leading to the synthesis of the sterol intermediate desmosterol. (4) Increasing desmosterol levels activate LXR-signaling. (5) This leads to LXR-mediated export of cholesterol via ABC transporters (cholesterol recycling) and (6) dampening of the pro-inflammatory phenotype of microglia/macrophages. (7) Resolved inflammation and increased disposition of cholesterol facilitates remyelination by oligodendrocytes.
(f) In case of normal repair of demyelinated lesions, microglia/macrophages are capable of lipid/cholesterol recycling and switching to an anti-inflammatory signature that depends on desmosterol synthesis. We speculate that this conserved mechanism mediates “silent” regeneration in asymptomatic individuals and in the preclinical phase of MS. When endogenous sterol synthesis is impaired as in our SQS conditional mutants or presumably also during aging or advanced disease, these repair strategies fail leading to cholesterol overloading of microglia/macrophages, persistent inflammation and impaired regeneration. Dietary squalene supplementation supports sterol synthesis. This increases desmosterol levels in microglia/macrophages that facilitates to resolve the local inflammation and to recycle lipid/cholesterol. In addition, squalene promotes cholesterol synthesis by oligodendrocytes for remyelination that collectively enhances repair.
Asterisks mark significant changes, ***p < 0.001, **p < 0.01, *p < 0.05.
Supplementary Material
Acknowledgments
We cordially thank A. Fahrenholz, T. Freerck, B. Sadowski, L. Fernandez Garcia-Agudo, and M. Lim for technical support. We thank C. Stiles, J. Alberta, S. Ghandour and HJ. Radzun for generous gifts of antibodies. We thank M. Schwaninger for providing SLCO1c1-CreERT2 mice. This work was funded by the Deutsche Forschungsgemeinschaft (SA2014/2-1 to GS). GS is supported by the Wilhelm-Sander Stiftung and the Alzheimer Forschung Initiative. Myelinating cell culture work was funded by the UK MS Society (Grant 38 to JME). KAN is supported by a European Research Council Advanced Grant. The Adelson Medical Research Foundation supports MSD and KAN. MWS is supported by the BMBF (CMT-NET01GM1511C, CMT-NRG 01GM1605). PS is supported by the Studienstiftung des deutschen Volkes. MP is supported by the Sobek Foundation, the Ernst-Jung Foundation, the German Research Foundation (SFB992, SFB1160, Reinhart-Koselleck-Grant, TRR167) and the Ministry of Science, Research and Arts, Baden-Wuerttemberg (Sonderlinie “Neuroinflammation”) and by the BMBF-funded competence network of multiple sclerosis (KKNMS). FO is supported by the Deutsche Forschungsgemeinschaft (OD87/1-1, OD87/3-1), FO and LH are supported by the Deutsche Forschungsgemeinschaft (TRR274/1).
Footnotes
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Author Contributions
S.A.B. and G.S. planned and designed the study. S.A.B. and L.Sp. were involved in all experiments. T.S. performed reanalysis of human sc-RNAseq datasets. L.Sc. performed cell culture experiments. L.H. and F.O. did flow cytometry with subsequent expression analyses. T.D. was involved in MACS cell isolation. D.E., M.W.S. and performed HPLC serum analysis. J.N. did western blot analysis. T.R. performed electron microscopy analysis. T.I and P.S. performed lipid mass spectrometry. I.H. provided human tissue. C.S.N. and F.v.d.M. characterized and isolated human MS tissue lesion. R.S. and M.P. provided RNAseq datasets. L.C. and M.S. were in involved in lysolecithin experiments. J.W., W.M. and M.M. characterized mouse pathology. J.M.E performed myelinating co-culture experiments. C.D. and A.O.S. did light sheet microscopy. S.A.B., J.M.E, K.A.N. and G.S. wrote and edited the manuscript.
Competing interests
SAB and GS are listed as inventors on pending patent claims (PCT###) filed by MPG covering the application of squalene in demyelinating disorders.
Data availability
Human single-cell/nuclei RNA sequencing data were obtained from two previously published datasets, GSE118257 and GSE124335. All the data supporting the conclusions of the current study are presented in the figures (Figure 4h-j contains visualizations of RNA-seq data; Extended data figure 6a-e contains visualizations of RNA-seq data). If necessary, the data that support the findings of this study are available from the corresponding author Gesine Saher upon reasonable request. There are no restrictions on data availability. Source data are provided with this paper.
Code availability
Code for data integration and analysis can be found at https://github.com/TSun-tech/MS_scRNAseq.
References
- 1.Dietschy JM. Central nervous system: cholesterol turnover, brain development and neurodegeneration. Biological chemistry. 2009;390:287–293. doi: 10.1515/BC.2009.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saher G, et al. High cholesterol level is essential for myelin membrane growth. Nature neuroscience. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- 3.Courtney R, Landreth GE. LXR Regulation of Brain Cholesterol: From Development to Disease. Trends in endocrinology and metabolism: TEM. 2016;27:404–414. doi: 10.1016/j.tem.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich DS, Lucchinetti CF, Calabresi PA. Multiple Sclerosis. The New England journal of medicine. 2018;378:169–180. doi: 10.1056/NEJMra1401483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plemel JR, Liu WQ, Yong VW. Remyelination therapies: a new direction and challenge in multiple sclerosis. Nature reviews Drug discovery. 2017;16:617–634. doi: 10.1038/nrd.2017.115. [DOI] [PubMed] [Google Scholar]
- 6.Itoh N, et al. Cell-specific and region-specific transcriptomics in the multiple sclerosis model: Focus on astrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E302–E309. doi: 10.1073/pnas.1716032115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voskuhl RR, et al. Gene expression in oligodendrocytes during remyelination reveals cholesterol homeostasis as a therapeutic target in multiple sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:10130–10139. doi: 10.1073/pnas.1821306116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurevics H, et al. Alterations in metabolism and gene expression in brain regions during cuprizone-induced demyelination and remyelination. Journal of neurochemistry. 2002;82:126–136. doi: 10.1046/j.1471-4159.2002.00954.x. [DOI] [PubMed] [Google Scholar]
- 9.Lavrnja I, et al. Expression profiles of cholesterol metabolism-related genes are altered during development of experimental autoimmune encephalomyelitis in the rat spinal cord. Scientific reports. 2017;7:2702. doi: 10.1038/s41598-017-02638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunha MI, et al. Pro-inflammatory activation following demyelination is required for myelin clearance and oligodendrogenesis. J Exp Med. 2020;217 doi: 10.1084/jem.20191390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung MSY, et al. Dynamics of oligodendrocyte generation in multiple sclerosis. Nature. 2019;566:538–542. doi: 10.1038/s41586-018-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berghoff SA, et al. Dietary cholesterol promotes repair of demyelinated lesions in the adult brain. Nature Communications. 2017;8 doi: 10.1038/ncomms14241. 14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spann NJ, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meschkat M, et al. White matter integrity requires continuous myelin synthesis at the inner tongue. bioRxiv. 2020 doi: 10.1038/s41467-022-28720-y. 2020.2009.2002.279612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantuti-Castelvetri L, et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science. 2018;359:684–688. doi: 10.1126/science.aan4183. [DOI] [PubMed] [Google Scholar]
- 16.Kotter MR, Li WW, Zhao C, Franklin RJ. Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:328–332. doi: 10.1523/JNEUROSCI.2615-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boven LA, et al. Myelin-laden macrophages are anti-inflammatory, consistent with foam cells in multiple sclerosis. Brain : a journal of neurology. 2006;129:517–526. doi: 10.1093/brain/awh707. [DOI] [PubMed] [Google Scholar]
- 19.Widenmaier SB, et al. NRF1 Is an ER Membrane Sensor that Is Central to Cholesterol Homeostasis. Cell. 2017;171:1094–1109 e1015. doi: 10.1016/j.cell.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Jakel S, et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566:543–547. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda T, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 22.Cardeno A, et al. Squalene targets pro- and anti-inflammatory mediators and pathways to modulate over-activation of neutrophils, monocytes and macrophages. J Funct Foods. 2015;14:779–790. [Google Scholar]
- 23.Saher G, et al. Therapy of Pelizaeus-Merzbacher disease in mice by feeding a cholesterol-enriched diet. Nat Med. 2012;18:1130–1135. doi: 10.1038/nm.2833. [DOI] [PubMed] [Google Scholar]
- 24.Gylling H, Miettinen TA. Postabsorptive metabolism of dietary squalene. Atherosclerosis. 1994;106:169–178. doi: 10.1016/0021-9150(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 25.Gudi V, Gingele S, Skripuletz T, Stangel M. Glial response during cuprizone-induced de- and remyelination in the CNS: lessons learned. Front Cell Neurosci. 2014;8:73. doi: 10.3389/fncel.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muse ED, et al. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E4680–E4689. doi: 10.1073/pnas.1714518115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubler Z, et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature. 2018;560:372–376. doi: 10.1038/s41586-018-0360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miron VE, et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nature neuroscience. 2013;16:1211–1218. doi: 10.1038/nn.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogie JF, et al. Myelin-derived lipids modulate macrophage activity by liver X receptor activation. PloS one. 2012;7:e44998. doi: 10.1371/journal.pone.0044998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd AF, et al. Central nervous system regeneration is driven by microglia necroptosis and repopulation. Nature neuroscience. 2019;22:1046–1052. doi: 10.1038/s41593-019-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olah M, et al. Identification of a microglia phenotype supportive of remyelination. Glia. 2012;60:306–321. doi: 10.1002/glia.21266. [DOI] [PubMed] [Google Scholar]
- 32.Kuhlmann T, et al. An updated histological classification system for multiple sclerosis lesions. Acta neuropathologica. 2017;133:13–24. doi: 10.1007/s00401-016-1653-y. [DOI] [PubMed] [Google Scholar]
- 33.Akula MK, et al. Protein prenylation restrains innate immunity by inhibiting Rac1 effector interactions. Nat Commun. 2019;10:3975. doi: 10.1038/s41467-019-11606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araldi E, et al. Lanosterol Modulates TLR4-Mediated Innate Immune Responses in Macrophages. Cell reports. 2017;19:2743–2755. doi: 10.1016/j.celrep.2017.05.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korner A, et al. Inhibition of Delta24-dehydrocholesterol reductase activates pro-resolving lipid mediator biosynthesis and inflammation resolution. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:20623–20634. doi: 10.1073/pnas.1911992116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C, et al. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. The Journal of biological chemistry. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- 37.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 38.Liebergall SR, et al. Inflammation Triggers Liver X Receptor-Dependent Lipogenesis. Mol Cell Biol. 2020;40 doi: 10.1128/MCB.00364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fellows Maxwell K, et al. Oxysterols and apolipoproteins in multiple sclerosis: a 5 year follow-up study. J Lipid Res. 2019;60:1190–1198. doi: 10.1194/jlr.M089664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zmyslowski A, Szterk A. Oxysterols as a biomarker in diseases. Clin Chim Acta. 2019;491:103–113. doi: 10.1016/j.cca.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Safaiyan S, et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nature neuroscience. 2016;19:995–998. doi: 10.1038/nn.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marschallinger J, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nature neuroscience. 2020;23:194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thelen KM, Falkai P, Bayer TA, Lutjohann D. Cholesterol synthesis rate in human hippocampus declines with aging. Neurosci Lett. 2006;403:15–19. doi: 10.1016/j.neulet.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 44.Hendrickx DAE, et al. Gene Expression Profiling of Multiple Sclerosis Pathology Identifies Early Patterns of Demyelination Surrounding Chronic Active Lesions. Frontiers in immunology. 2017;8:1810. doi: 10.3389/fimmu.2017.01810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mailleux J, et al. Active liver X receptor signaling in phagocytes in multiple sclerosis lesions. Multiple sclerosis. 2018;24:279–289. doi: 10.1177/1352458517696595. [DOI] [PubMed] [Google Scholar]
- 46.Yu S, et al. Dissociated sterol-based liver X receptor agonists as therapeutics for chronic inflammatory diseases. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2016;30:2570–2579. doi: 10.1096/fj.201600244R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katz Sand I. The Role of Diet in Multiple Sclerosis: Mechanistic Connections and Current Evidence. Current nutrition reports. 2018;7:150–160. doi: 10.1007/s13668-018-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sedaghat F, Jessri M, Behrooz M, Mirghotbi M, Rashidkhani B. Mediterranean diet adherence and risk of multiple sclerosis: a case-control study. Asia Pacific journal of clinical nutrition. 2016;25:377–384. doi: 10.6133/apjcn.2016.25.2.12. [DOI] [PubMed] [Google Scholar]
- 49.Beltrán G, Bucheli ME, Aguilera MP, Belaj A, Jimenez A. Squalene in virgin olive oil: Screening of variability in olive cultivars. European Journal of Lipid Science and Technology. 2016 [Google Scholar]
- 50.Smith TJ. Squalene: potential chemopreventive agent. Expert opinion on investigational drugs. 2000;9:1841–1848. doi: 10.1517/13543784.9.8.1841. [DOI] [PubMed] [Google Scholar]
- 51.Quinet EM, et al. Gene-selective modulation by a synthetic oxysterol ligand of the liver X receptor. J Lipid Res. 2004;45:1929–1942. doi: 10.1194/jlr.M400257-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Bachmanov AA, Reed DR, Beauchamp GK, Tordoff MG. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behavior genetics. 2002;32:435–443. doi: 10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian journal of biochemistry and physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 54.Alizadeh A, Karimi-Abdolrezaee S. Microenvironmental regulation of oligodendrocyte replacement and remyelination in spinal cord injury. The Journal of physiology. 2016;594:3539–3552. doi: 10.1113/JP270895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radzun HJ, et al. Detection of a monocyte/macrophage differentiation antigen in routinely processed paraffin-embedded tissues by monoclonal antibody Ki-M1P. Laboratory investigation; a journal of technical methods and pathology. 1991;65:306–315. [PubMed] [Google Scholar]
- 56.Lampron A, et al. Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J Exp Med. 2015;212:481–495. doi: 10.1084/jem.20141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bijland S, et al. An in vitro model for studying CNS white matter: functional properties and experimental approaches. F1000Research. 2019;8:117. doi: 10.12688/f1000research.16802.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:514–517. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Roper MG. Measurement of DCF fluorescence as a measure of reactive oxygen species in murine islets of Langerhans. Analytical methods : advancing methods and applications. 2014;6:3019–3024. doi: 10.1039/C4AY00288A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lucchinetti CF, et al. Clinical and radiographic spectrum of pathologically confirmed tumefactive multiple sclerosis. Brain : a journal of neurology. 2008;131:1759–1775. doi: 10.1093/brain/awn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Renier N, et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 62.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McInnes Leland, H J, Melville James. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv.org. 2018 1802.03426. [Google Scholar]
- 65.Wickham H. ggplot2 - Elegant Graphics for Data Analysis. Springer; 2009. [Google Scholar]
- 66.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Human single-cell/nuclei RNA sequencing data were obtained from two previously published datasets, GSE118257 and GSE124335. All the data supporting the conclusions of the current study are presented in the figures (Figure 4h-j contains visualizations of RNA-seq data; Extended data figure 6a-e contains visualizations of RNA-seq data). If necessary, the data that support the findings of this study are available from the corresponding author Gesine Saher upon reasonable request. There are no restrictions on data availability. Source data are provided with this paper.
Code for data integration and analysis can be found at https://github.com/TSun-tech/MS_scRNAseq.