Abstract
The WAVE regulatory complex (WRC) is crucial for assembly of the peripheral branched actin network constituting one of the main drivers of eukaryotic cell migration. Here, we uncover an essential role of the hematopoietic-specific WRC component HEM1 for immune cell development. Germline-encoded HEM1 deficiency underlies an inborn error of immunity with systemic autoimmunity, at cellular level marked by WRC destabilization, reduced filamentous actin and failure to assemble lamellipodia. Hem1-/- mice display systemic autoimmunity, phenocopying the human disease. In the absence of Hem1, B cells become deprived of extracellular stimuli necessary to maintain the strength of B-cell receptor signaling at a level permissive for survival of non-autoreactive B cells. This shifts the balance of B-cell fate choices towards autoreactive B cells and thus autoimmunity.
Introduction
The actin cytoskeleton is fundamental to mount successful immune responses, as evident from the wide range of defects that occur in actin-related inborn errors of immunity (1–4). Studies of these conditions have revealed a pivotal role for the actin cytoskeleton and actin-binding proteins in immune system function: from hematopoiesis and immune cell development to immune cell migration to intercellular and intracellular signaling including immune cell activation (1).
The actin cytoskeleton is composed of networks and bundles of actin filaments (F-actin) that are polymerized from actin monomers e.g. underneath the plasma membrane. The polymerization is initiated by the three classes of actin nucleators, the Arp2/3 complex, the formin family, and the more recently identified Spire, cordon-bleu, and leiomodin family proteins (5). Formins nucleate and elongate linear actin filaments, while the Arp2/3 complex drives filament branch formation on pre-existing filaments (6). The Wiskott-Aldrich syndrome protein (WASP) and WASP-family verprolin-homologous (WAVE) family proteins promote F-actin-branching through the Arp2/3 complex. WAVE proteins form a 400 kDa heteropentameric (7–9) assembly called the WAVE regulatory complex (WRC). WRC-deficient cells are unable to generate lamellipodia or membrane ruffles and Wave2 knock-out mice are embryonic lethal due to impaired endothelial cell migration (10). Indeed, most studies of actin-related immunodeficiencies typically attribute abnormalities in the locomotory cell apparatus per se to the key pathogenic mechanism (1–3, 11). At the same time, recent data from non-immune cell types strongly suggest that cell-scale actin cytoskeleton morphodynamics control the spatiotemporal output and the intensity of signaling events at the molecular level (12, 13). However, the scale-bridging mechanisms linking signaling events and actin cytoskeleton-mediated cell morphogenetic behaviors remain largely unexplored in the context of immune cell physiology and pathobiology.
Here, we find that WRC deficiency caused by absence of the WRC hematopoietic cell-specific subunit HEM1 results in a previously unknown human disorder with severe immune dysregulation and recurrent infections. By studying the molecular and cellular mechanisms behind this inborn error of immunity, we uncover HEM1 as a key regulator of BCR signaling strength that is crucial for B cell development and homeostasis.
Results
Identification of human HEM1 deficiency
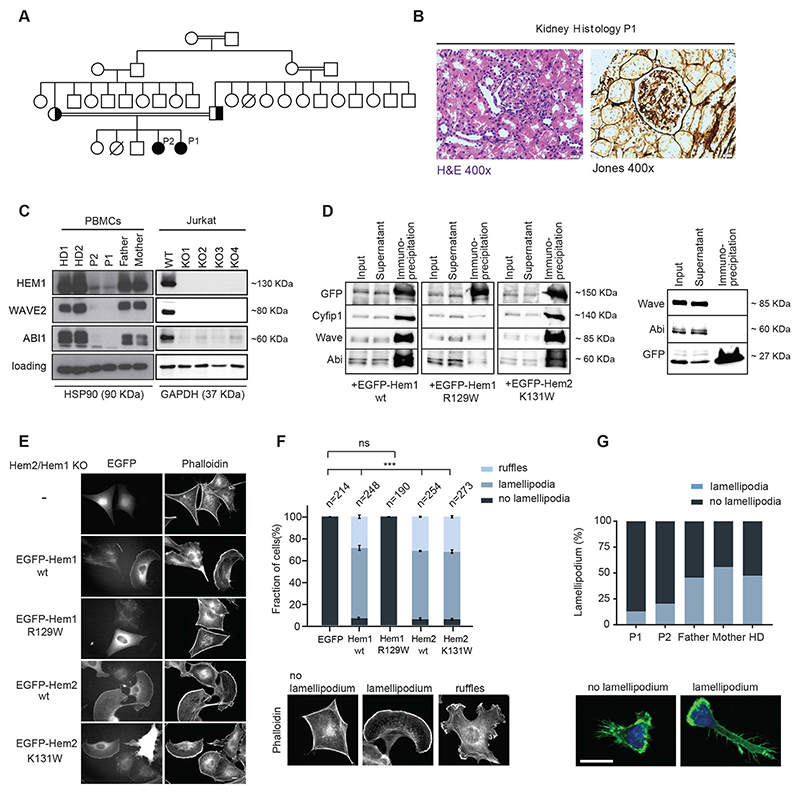
We studied 2 patients with recurrent fever with and without signs of infection from the age of 4 months (Fig. 1A). Patient 1 (P1) experienced multiple upper respiratory tract infections and later developed skin rashes, oral ulcers, photosensitivity, joint pain, fatigue, and glomerulonephritis (Fig. 1B and fig. S1A). She was tested positive for anti-dsDNA antibodies and diagnosed with systemic lupus erythematosus (SLE) according to current American College of Rheumatology (ACR) criteria (14). The patient exhibited failure to thrive with both height and weight below the third percentile. She has stable disease with long-term immunosuppression including corticosteroids and azathioprin. P1’s 8-year-old sister (P2) has experienced recurrent infections since the first year of life. Glucose-6-phosphate dehydrogenase (G6PD) deficiency (15) was diagnosed but did not result in severe hemolytic events. P2 had recurrent respiratory tract infections, repeated skin abscesses, and multiple ear infections resulting in tympanic membrane perforation and unilateral hearing loss. Recurrent lymphadenopathy and fever were evident independent of (overt) infections. Periodic fever syndromes were considered but no germline mutation in MEFV indicative of familial Mediterranean fever (16) was detected. P2 has remained negative for anti-dsDNA autoantibodies to date, does not take medications regularly but was treated with antibiotics repeatedly for assumed or proven bacterial infections. An older sister of P1 and P2 had died two hours post-partum of unknown cause without tissue samples available for molecular investigation.
Fig. 1. HEM1 deficiency results in aberrant cell morphology and defective lamellipodia formation in HEMl-deficient patients.
(A) Pedigree of the index family: double lines indicate consanguinity; black filling indicates the index patients; diagonal lines indicate deceased siblings. (B) Image of hematoxylin/eosin staining showing capillary proliferation and polymorphonuclear neutrophils in glomeruli, magnification 400x (left); Periodic Acid Schiff (PAS) staining showing mild mesangial expansion, 400x (right), consistent with Lupus nephritis, class III, active. (C) Cropped immunoblot analysis of HEM1, WAVE2, ABI1, GAPDH or HSP90 (heat shock protein 90) in peripheral blood mononuclear cells (PBMCs) of healthy donor (HD), patients (P1, P2), their parents (left) and in CRISPR/Cas9 HEM1 knock-out Jurkat T cells (right). (D) Co-immunoprecipitation of endogenous WRC subunits with ectopically expressed EGFP-tagged proteins (indicated at the bottom) in wildtype B16-F1 cells. EGFP alone was used as control (right panel). (E) Hem2/Hem1 knock-out B16-F1 clone #8 was transfected with indicated EGFP-tagged constructs. The panels display respective phalloidin stainings. Scale bar represents 10 μm. (F) Quantification of lamellipodia/ruffle formation as indicated by the color shading shows fraction of cells in % ± SEM from three independent experiments. n equals number of transfected cells analyzed. Bottom: Representative examples of cell morphologies used for cell categorization. (G) Representative images (bottom) and quantification (top) of expanded T cells spreading on fibronectin-coated surfaces stained with phalloidin and DAPI and imaged with a Zeiss LSM700 confocal microscope and a 63x oilimmersion objective. Scale bar, 10 μm.
To figure out the molecular basis of this disease, we performed exome-sequencing of P1 and identified a conserved, homozygous missense variant (c.C385T, p.R129W) in the gene NCKAP1L encoding HEM1. Genetic segregation analysis revealed perfect segregation under the assumption of autosomal-recessive inheritance including homozygosity for the affected sibling P2 (Fig. 1A and fig. S1B, C). NCKAP1L was the only gene with homozygous variants shared among the two affected siblings. The identified variant has not been reported in public databases including ExAC (17) or gnomAD (18); accession date 20.01.2020) and is predicted highly damaging using prediction tools including CADD (https://cadd.gs.washington.edu; accession date 20.01.2020). Notably, no rare homozygous premature nonsense or frameshift variants in NCKAP1L have been reported in gnomAD or other publicly available databases. The probability of cellular intolerance to a loss-of-function mutation (pLI) score of 0.85 for NCKAP1L suggests that this gene may be essential for human health. Collectively, these observations gave strong support for HEM1 as a promising candidate gene for the syndrome investigated.
Defective WRC assembly and stability
To examine the effect of the identified gene variant on protein expression, we performed immunoblotting in the following patient-derived cells: peripheral blood mononuclear cells (PBMCs) (Fig. 1C), feeder-expanded T cells, EBV-immortalized B cells (EBV-LCL) (fig. S1, D and E), and consistently observed markedly reduced HEM1 protein expression. Furthermore, the expression of the catalytic component WAVE2 and other subunits of the WRC were severely reduced in patient cells, illustrating a detrimental effect of the loss of HEM1 expression on the WRC assembly and stability. CRISPR/Cas9-engineered HEM1-deficient Jurkat T cells recapitulated the expression patterns observed in patient cells (Fig. 1C, all Western blot raw data can be found at the end of the supplementary information).
Non-hematopoietic cells express the homologous protein HEM2 instead of HEM1 (19). In contrast to previous observations (20), recent work has suggested that compensatory HEM1 expression can in principle substitute for HEM2 expression (21). HEM2 and HEM1 share approximately 60% sequence homology. However, the arginine residue at position 129 in HEM1 is not conserved in HEM2, as the respective amino acid in HEM2 is shifted by 2 residues due to an upstream insertion and constitutes a lysine (instead of arginine) at position 131. The respective murine sequences are highly conserved, including R129 in Hem1 (fig. S1C).
To dissect the functional consequences of the R129W mutation in Hem1 and potential effects of the corresponding mutation in Hem2, we employed the murine B16-F1 melanoma cell line as an established model system for actin remodeling and lamellipodium protrusion (22, 23). We first performed co-immunoprecipitation experiments with EGFP-tagged, murine wildtype (wt) Hem1, Hem1-R129W and the analogous mutation in Hem2, Hem2-K131W, using B16-F1 wildtype cells. Hem1-R129W showed strongly reduced co-precipitation of its interactors Cyfip1, Abi and WAVE as compared to Hem1-wt or Hem2-K131W (Fig. 1D), already suggesting compromised interaction of Hem1-R129W with endogenous WRC subunits. However, degradation of mutant protein was not obvious under these conditions, likely because of excess production of transient, ectopic protein, as opposed to steady-state expression of the endogenous mutant variant in mouse or human patient tissue. Lamellipodia formation relies on a functional WRC (24, 25). We thus hypothesized that the reduction of HEM1 and WAVE2 expression abrogates lamellipodia formation in Hem1- R129W-expressing patient cells. To test this, we engineered CRISPR/Cas9-edited Hem1/Hem2- deficient B16-F1 clones, which are completely devoid of lamellipodia (Fig. 1E, top panel). As opposed to EGFP expression alone, re-expression of EGFP-tagged Hem1-wt or Hem2-wt rescued lamellipodia (Fig. 1, E and F). Hem1-R129W expression failed to restore these structures, while the Hem2-K131W expression phenotype was virtually identical to that of Hem1-wt or Hem2-wt (Fig. 1, E and F). In contrast to Hem2-K131W, the Hem1-R129W protein was also unable to localize at subcellular sites normally occupied by Hem1-wt in B16-F1 wildtype cells, suggesting that the mutant Hem1 fails entirely to incorporate into functional WRCs (fig. S1F). Moreover, Hem1-R129W did not rescue lamellipodia formation in a second, independently generated Hem1/2 double-KO B16-F1 clone (fig. S1G). Consistently, cellular morphology in patient lymphocytes was aberrant, as they were unable to form stable lamellipodia and displayed reduced F-actin upon fibronectin stimulation (Fig. 1G and fig. S1, H to J). Instead, the cells showed signs of formin- and/or WASP-mediated F-actin structures with aberrant membrane spikes. Quantification of cell shape descriptors and subsequent principal component analysis of the data showed a significant difference between patient and healthy donor T cells with reduced cell spreading area being the most discriminative feature (fig. S1J; list of distinguishing features table S2).
Impaired T-cell function in HEM1 deficiency
As HEM1 deficiency resulted in loss of lamellipodia and decreased cell spreading on the extracellular matrix (ECM), we investigated the physiological consequences of these abnormalities in the context of immune cell behavior. Hem1/2 and other subunits of the WRC are classically implicated in cell migration (26, 27). However, when we examined immune cell migration efficiency using standard chemotaxis assays, we did not observe any remarkable differences between normal donor- and patient-derived cells (fig. S2A).
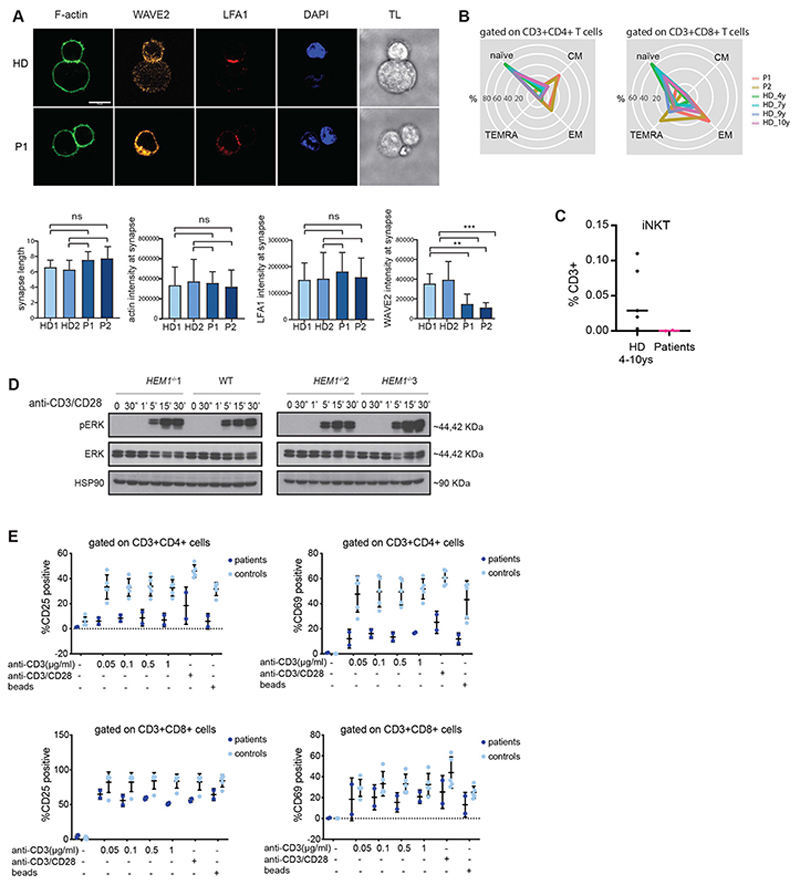
F-actin-mediated cell morphodynamics are also known to be crucially important for the effector/target cell conjugation and killing activity of cytolytic T cells (28). Nevertheless, our experiments showed that patient CD8 cells undergo normal activation (as mirrored by upregulation of LFA-1 in both patient and CRISPR/Cas9-engineered HEM1-deficient Jurkat T cells) as well as intact conjugate formation towards OKT3-coated P815 cells (Fig. 2A and fig. S2B).
Fig. 2. Impaired T-cell function in HEM1 deficiency.
(A) Representative images of immune conjugate formation in healthy donor (upper row) and patient (lower row) expanded CD8+ T cells with anti-CD3 coated P815 target cells stained with WAVE2 (orange), phalloidin (green), DAPI (blue) and LFA-1 (red), from left to right, respectively. Scale bar, 10 μm. Below: quantifications of synapse features including synapse length, F-actin, LFA-1 and WAVE2 intensities at the synapse area in both patients (P1 and P2) and two healthy donors (HD1 and HD2) TL, transmission light. (B) Radar plots showing the distribution of main T cell subpopulations of CD4+ and CD8+ T cells (naïve, central memory [CM], effector memory [EM], effector memory re-expressing CD45RA [TEMRA]) in both patients and age-matched controls. Age-matched controls are for easier illustration and comparison. Age-dependent reference values for these subsets have been published earlier (for details see Supplementary Table 4). (C) Quantification plot based on immunophenotyping data of patient iNKT cells (CD3+TCR-Vβ11+, TCRVα24+) in P1, P2, and 4 different age-matched and one adult healthy donors. (D) ERK phosphorylation in CRISPR/Cas9 HEM1 knock-out Jurkat T cells, stimulated with anti-CD3 (OKT3)/anti-CD28 for different time points. (E) Percentages of CD25- and CD69-positive cells upon stimulation with different concentrations of anti-CD3, anti-CD28 and stimulatory beads in CD4+ and CD8+ T-cell populations from both patients and 2 healthy controls.
We nonetheless hypothesized that the cytoskeletal abnormalities observed in HEM1-deficient cells may perturb immune cell functions associated with cell proliferation, survival, and differentiation. Detailed T-cell evaluation revealed normal total numbers of T lymphocytes including CD4+ and CD8+ cells and T follicular helper (TfH) cells (fig. S2, C and D, and table S3) compared to published age-dependent reference values (29). However, subset analysis showed elevated relative percentages of central memory cells and exhausted effector memory cells (table S3; Fig. 2B). Furthermore, HEM1-deficient CD4 T-helper memory subsets exhibited a hypersecretory Th1- biased phenotype compared to age-matched controls (fig. S2E). Invariant natural killer T (iNKT) cells were very low to absent in both patients (Fig. 2C), while the proportion of TCRγδ+ cells compared to TCRαβ cells showed a relative increase within the CD8+ T-cell subset (table S3), both as seen in other immunodeficiencies with marked immune dysregulation (30). Patients had only slightly elevated double-negative T (DNT) cells compared to published age-dependent reference values (29) (table S3). Increased DNTs are characteristic of auto-immune lymphoproliferative syndrome (ALPS) (31). However, expression of FAS, FAS-ligand mediated apoptosis (fig. S2F) and ALPS biomarkers were unremarkable, inconsistent with classical ALPS. Similarly, haploinsufficiency of the regulatory cytotoxic T-lymphocyte antigen (CTLA)-4 is known to cause severe systemic autoimmunity and immunodeficiency (32). Nevertheless, we found that expression of PD-1/PD-L1 and cycling of the immune checkpoint CTLA-4 were unaffected (fig. S2, G and H), and no substantial reduction of regulatory T-cell numbers was detected in the patients (table S3). We did however observe that both CD4+ T cells and CD8+ T cells were defective in antigen-induced proliferation independent of ERK activation as observed in CRISPR/Cas9-engineered HEM1 KO jurkat T cells (Fig. 2D and figS2i) after stimulation with anti-CD3 and anti-CD28 antibodies. This phenotype was accompanied by impaired expression of the activation markers CD25 and CD69 (Fig. 2E).
Aberrant B-cell phenotype in HEM1 deficiency
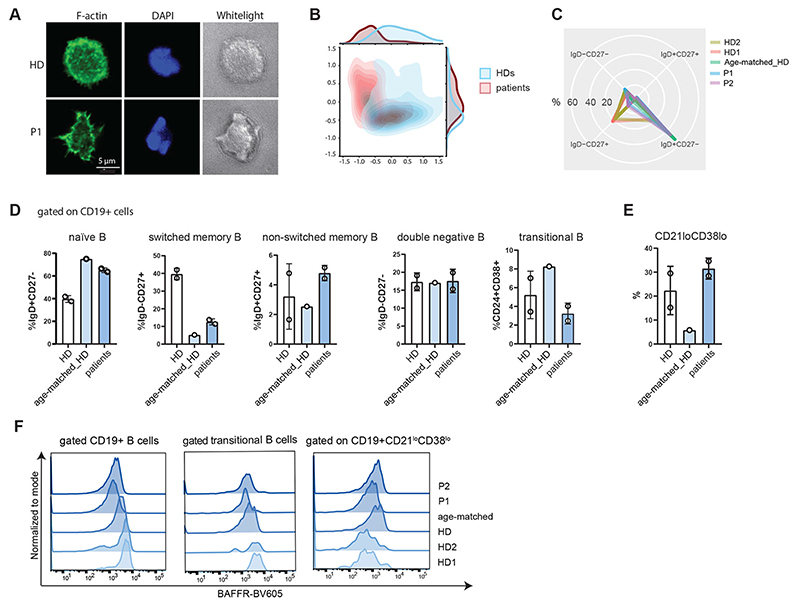
Loss of immune tolerance in SLE is supposedly multifactorial and includes a breakdown of T- and B-cell tolerance (33, 34). The presence of autoantibodies despite normal total immunoglobulin G, M or A levels in P1 suggested aberrant B-cell selection at the germinal center stage. Consistent with the observations in T cells, patient B cells also displayed decreased spreading and lack of lamellipodia upon IgM stimulation (Fig. 3, A and B, and table S4). These data suggested that in addition to processes associated with antigen-induced proliferation of T cells, HEM1 deficiency might affect cell proliferation and/or differentiation in the B-cell compartment. Therefore, we decided to evaluate patient B-cell subsets. Absolute counts of CD19+ cells, and proportions of naïve-, memory- and transitional B cells were within the aged-matched normal range compared to previously published data (35) (Fig. 3, C and D; fig. S3, A and B; table S5), but patients showed an increase in cell subsets, such as innate-like CD19+CD38loCD21lo (CD21low) memory B cells, which were previously reported to be associated with autoimmunity (35–37) (Fig. 3E). CD21low memory-like B cells expand in conditions characterized by chronic immune stimulation (38), and studies using chronic infection models showed that activity of these cells is inversely correlated with BAFFR expression due to increased cleavage of BAFFR upon BAFF stimulation (38). Therefore, we decided to evaluate BAFF levels in patients’ serum. However, surface measurements of BAFFR on HEM1-deficient patient B-cell subsets revealed reduced levels of BAFFR on CD19+ cells and B-cell subsets, indicating constant activation of these cells (Fig. 3F).
Fig. 3. Aberrant B-cell phenotype in HEM1 deficiency.
(A) Representative images and of magnetic-activated cell sorting (MACS)-sorted B cells spreading on IgM-coated surfaces stained with phalloidin and DAPI and imaged with a Zeiss LSM700 confocal microscope and a 63x oil-immersion objective. Scale bar, 5 μm. TL, transmission light (B) Principal component analysis of cell morphology parameters extracted from immunofluorescence images using CellProfiler™. Patient (blue contour) and healthy donor (red contour) B cells can be distinguished on the basis of these parameters (list of parameters in Supplementary Table 5). (C) Radar plot showing the distribution of naïve, memory and double negative B cell subpopulations among the patients and healthy controls. (D) B-cell immunophenotyping showing relative proportions of naïve (IgD+CD27-), memory non-switched (IgD+CD27+) and memory-switched (IgD-CD27+), (IgD-CD27-) double negative B cells, relative proportions of transitional (CD19+CD24+CD38+) and (E) memory-like (CD19+CD27-IgD+CD21lowCD38low) B cells (gating strategy as outlined in Supplementary Figure 3A). (F) Expression of BAFFR (BV605) on the surface of CD19+, transitional and CD21lowCD38lowB cells of P1, P2, age-matched and adult healthy donors (gating strategy as explained in Figure S3A).
Hem1-/- mice replicate aberrant B-cell development and multifaceted autoimmunity
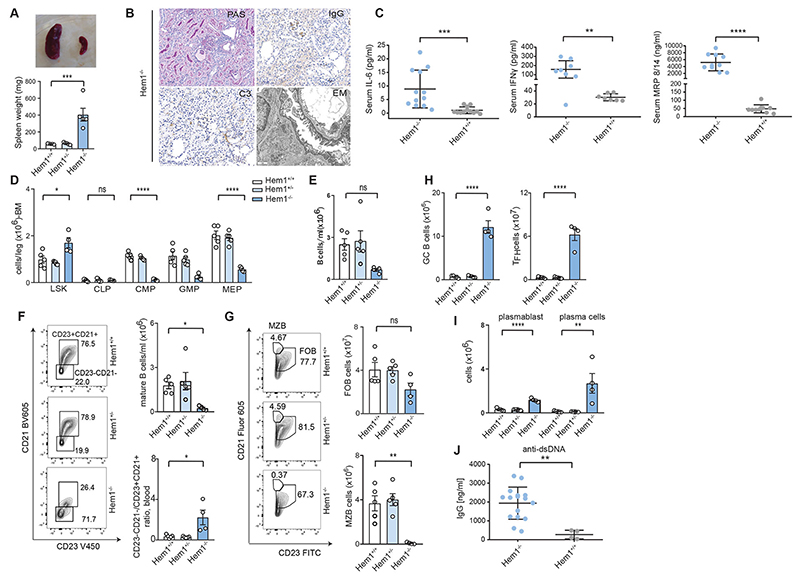
To gain a mechanistic understanding of how HEM1 deficiency affects B-cell development and germinal center maturation, we took advantage of a recently generated Hem1-/- mouse model (25) (fig. S4, A to C). We discovered that Hem1-/- mice suffered from severe systemic autoimmunity manifesting in splenomegaly, immune complex/lupus glomerulonephritis with mesangial enlargement, hypercellularity, sclerosis (Fig. 4, A and B), and IgG and C3 deposits (Fig. 4B). Further, we detected massively increased levels of pro-inflammatory markers, including IL6, IFNγ and MRP8/14 (Fig. 4C), likely underlying the observed neutrophilia and monocytosis in the peripheral blood and the spleen (fig. S4, D and E) as well as granulomatous lesions in multiple organs (fig. S4F). We also found aberrant leukocyte development in the bone marrow (Fig. 4D) and extramedullary hematopoiesis in the spleen as previously described for Hem1-/- mice (fig. S4G) (39). While T-cell numbers were unaltered in the spleen (fig. S4H), Hem1-/- mice had an expansion of the memory T-cell compartment, phenocopying the HEM1-deficient patients (Fig. 2B). Blood B-cell counts were rather reduced (Fig. 4E) and mature recirculating follicular B cells were almost absent both in the periphery (Fig. 4F) and bone marrow (fig. S4I), resulting in an increased ratio of immature/mature B cells (Fig. 4F). Intriguingly, total splenic B-cell numbers were unaltered (fig. S4J). In the spleen, Hem1-/- mice accumulated immature and transitional B cells, gated using IgD, CD19 and CD93, as previously described (40) (fig. S4k), and showed almost undetectable marginal zone and decreased follicular and mature B-cells (Fig. 4G), explaining the lack of recirculating B cells (Fig. 4E). Numbers of both GL-7hi germinal center B cells and PD-1+ TfH cells (Fig. 4H) were drastically elevated in Hem1-deficient mice. Consistent with findings in HEM1-deficient individuals, Hem1-/- mice produced manifold more potentially auto-reactive antibody-secreting plasmablasts (Fig. 4I) and showed accumulation of serum dsDNA autoantibodies as early as 3-4 weeks of age (Fig. 4J) similar to findings in HEM1-deficient patients. In sum, these data suggest that the B-cell compartment undergoes substantial changes in response to Hem1 deficiency in mice.
Fig. 4. Hem1-/- mice replicate aberrant B-cell development and autoantibody generation.
(A) Representative image and comparison of spleen size of the Hem1-/- vs. Hem1+/+ mice. (B) Immunohistochemistry and electron microscopy of kidneys from Hem1-/- mice illustrating severe systemic autoimmunity by glomerulonephritis (mesangial enlargement, hypercellularity and sclerosis and C3 deposits). (C) IL-6, IFNg and MRP8/14 cytokine levels in Hem1+/+ and Hem1-/- mice. Each dot represents one mouse. (D) LSK (Lin-Sca1+Kit-) stem cells in the bone marrow of Hem1+/+, Hem1+/- or Hem1-/- mice. (E) Quantification of total blood B cells (B220). (F) Quantification of mature blood B cells (CD21+CD23+) (top, right) and ratio of mature (CD21+CD23+) and immature B cells (CD21low CD23-) of Hem1+/+, Hem1+/+ or Hem1-/- mice in blood (bottom, right); representative FACS plots (left). (G) Representative flow cytometry plots of marginal zone (B220+CD21hiCD23-) and follicular (B220+CD21intCD23+) B cells in the spleen of Hem1+/+, Hem1+/- or Hem1-/- mice (left), quantification (right). (H) Representative flow cytometry quantification of splenic GL-7hi germinal center B cells in Hem1+/+, Hem1+/- or knockout Hem1-/- mice. Representative flow cytometry quantification of T-follicular helper cells (CD3+CD4+PD1+CXCR5+). (I) Quantification of representative flow cytometry plots along with the enumeration of splenic plasmablast (CD138+, B220+) and plasma cell (CD138+B220-) counts in Hem1+/+, Hem1+/- or Hem1-/- mice. (J) Anti-dsDNA-specific serum IgG titers of Hem1+/+, and Hem1-/- mice quantified by ELISA. FITC, fluorescein isothiocyanate; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte-erythroid progenitor; GMP, granulocyte-macrophage progenitor.
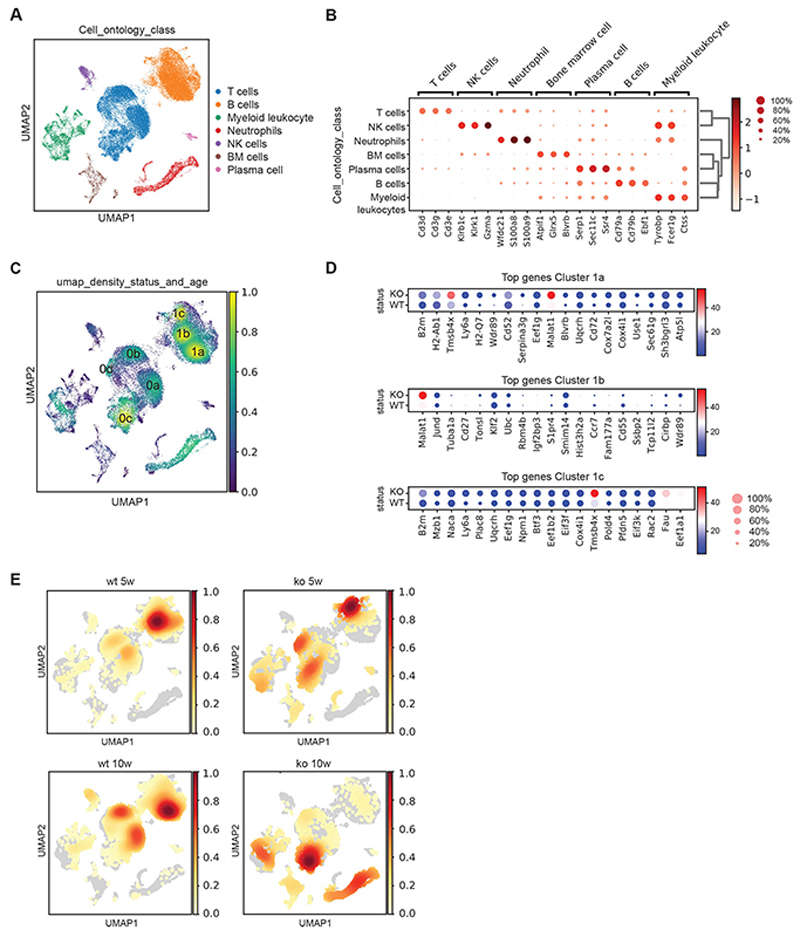
Single-cell RNA sequencing uncovers dysregulated B-cell development
To investigate the dynamics of these changes in a systematic fashion, we decided to characterize splenic cell subsets via 10X single-cell RNA sequencing in total homogenized erythrocyte-depleted spleen suspension of 5- and 10-week old Hem1+/+ and Hem1-/- mice. We were able to resolve the main leukocyte subsets using the Leiden clustering algorithm (Fig. 5, A and B). To identify hotspots of clustering, we evaluated cellular density and found 3 main B-cell (1a, 1b and 1c) and 4 main T-cell clusters (0a, 0b, 0c and 0d) (Fig. 5C; fig. S5, A and B; table S6 and S7). Based on the differential expression analysis of B-cell sub-clusters, we attributed cluster 1a to follicular B cells and 1b to germinal center B cells present in wildtype conditions. However, cluster 1c mainly present in the Hem1-/- situation could not be attributed to a specific stage of B-cell development (Fig. 5D). We speculated that this cluster may emerge from dysregulation of the B-cell developmental program in the absence of Hem1. Indeed, evaluation of cell density maps over time revealed substantial differences in B-cell subclusters in 5-week vs. 10-week-old mice suggesting a disturbed B-cell developmental program that consequently leads to alterations in T cell subsets (Fig. 5E). While in Hem1+/+ mice, the majority of B cells shifted from the germinal center cluster (1b) to the follicular B-cell cluster (1a), Hem1-/- mice showed the highest density in cluster 1c at 5 weeks but a global reduction in total B cells at 10 weeks, indicating a substantial loss of splenic B cells (Fig. 5E). Furthermore, we observed that T-cell clusters also show aberrant differentiation in Hem1-/- vs. Hem1+/+ mice calculated using entropy analyses (41, 42) (fig. S5C), further extending findings from an early study in an ENU-mutant mouse model (43).
Fig. 5. Single-cell RNA sequencing reveals dysregulated B-cell development.
(A) Leiden clustering of pooled Hem1+/+ and Hem1-/- spleen cells identifies major leukocyte subsets. Cell clustering of single cell data using pseudo-timeline analysis. Major cell types are shown in different colors. (B) Expression levels of marker genes for identifying hotspots of clustering. (C) Cellular density map identifying 3 main B-cell (1a, 1b, 1c) and 4 main T-cell clusters (0a,0b,0c,0d) (yellow: high density area, blue low-density area). (D) Top genes identified through differential expression analysis for high density B-cell sub-clusters (1a, 1b and 1c) are shown for Hem1+/+ and Hem1-/-. (E) Cell density maps over time, in 5 versus 10-week-old mice depicting substantial differences in B-cell subclusters in 5-week vs. 10-week-old Hem1+/+ and Hem1-/- mice (red: high density area, yellow: low density area, grey: no cells).
Defective B-cell intrinsic survival in murine Hem1-/- B cells
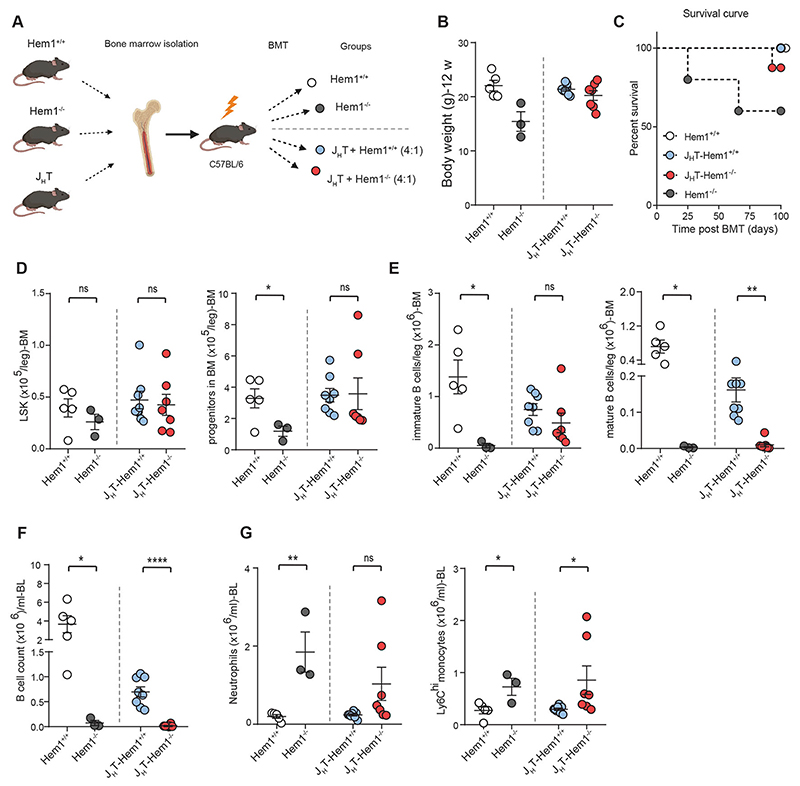
Considering the aberrations in both T- and B-cell sub clusters, we wanted to determine whether there is a B-cell intrinsic defect in the context of Hem1 deficiency. For this purpose, we made use of the JHT mouse strain (44). JHT mice harbor a deletion of the JH mini-genes generated through the Cre-loxP system and are truly devoid of B cells and immunoglobulins in all tested genetic backgrounds. Notably, the non-B cell compartment in JHT mice cannot be considered mixed, but is predominantly wildtype and therefore non-B cells are fully functional using this approach (45). We generated bone marrow chimeras of JHT and Hem1-/- vs. Hem1+/+ bone marrow, in a ratio of 80% (JHT bone marrow) to 20% (either Hem1+/+ or Hem1-/- bone marrow) and transplanted them into sub-lethally irradiated C57BL/6J mice (referred to as JHT Hem1+/+ and JHT Hem1-/- bone marrow; Fig. 6A). In recent work, Shao et al. demonstrated that Hem1-deficient hematopoietic stem cells may fail to engraft the bone marrow (39). To control for a potential engraftment issue, we also generated sub-lethally irradiated C57BL/6J mice transplanted with either Hem1+/+ or Hem1-/- bone marrow alone (referred to as C57BL/6J Hem1+/+ and C57BL/6J Hem1-/-) and monitored all mice during a recovery period. While as expected control C57BL/6J Hem1-/- mice fell severely ill and showed similar disease as seen in full-body Hem1-/- mice, C57BL/6J Hem1+/+ mice remained healthy. JHT Hem1+/+ mice appeared healthy as well, while one JHT-Heml-/- mouse died and additional three mice showed considerable weight loss (Fig. 6, B and C), forcing us to terminate the experiment. Although reconstitution of hematopoietic cells was successful, as judged by the comparable numbers of stem cells (LSKs) in both groups (Fig. 6D), JHT-Hem1-/- mice displayed a reduction in immature, and even to a greater extent mature, B-cell numbers in the bone marrow (Fig. 6E). Moreover, in the context of this bone marrow transplantation strategy, B-cell-specific Hem1 deficiency even resulted in the absence of detectable B cells both in peripheral blood (Fig. 6F), accompanied by increased numbers of circulating monocytes and neutrophils (Fig. 6G). As in other diseases, clinical phenotypes are complex and may originate from the interplay of different cell populations, however these experiments clearly indicate a critical role of HEM1 deficiency on B-cell homeostasis and hence a role for B cells in disease pathogenesis. In support, depmap analysis showed that the gene expression profiles of Hem1-/- B cells are surprisingly similar to those in B cells deficient of Pax5 (46) or Irf8 (47), well-known master regulators of B-cell development (fig. S6A).
Fig. 6. Defective B-cell intrinsic survival in murine Hem1-/- B cells.
(A) Scheme for the transplantation experiment, mixed bone marrow chimeras of JHT and Hem1-/- vs. Hem1+/+ bone marrow were transplanted into sub-lethally irradiated Bl6 mice (Hem1+/+). Control groups consisted of mice receiving Hem1+/+ or Hem1-/- bone marrow (shown in white and gray dots). The 4 transplanted grouped are depicted in the Figure. (B) Body weight of the 4 different groups of transplanted mice at the termination point of experiment (12 weeks). (C) Kaplan-Meier survival curve of bone marrow chimera experiment (C57BL/6J mice with mixed JHT and WT BM (4:1) and C57BL/6J mice with mixed JHT and KO BM (4:1). (D) Absolute number of LSK (left) and progenitor cells (right) in transplanted mice with JHT-Hem1+/+ and JHT-Hem1-/- chimeras and control groups. (E) Absolute number of immature (left) and mature (right) B cells in the bone marrow of transplanted mice. (F) Absolute number of blood B220+ B cells in transplanted mice with JHT-Hem1+/+ and JHT-Hem1-/- chimeras and the control groups. (G) Absolute number of neutrophils in blood of transplanted mice with JHT-Hem1+/+ and JHT-Hem1-/- chimeras and the control groups (left). Absolute number of blood Ly6Chi monocytes of JHT-Hem1+/+ and JHT-Hem1-/- chimeras and the control groups (right). BMT, bone marrow transplantation.
Our findings prompted us to further investigate how a cytoskeletal protein such as HEM1 may influence B-cell development. To this end, we first performed differential gene expression and pathway enrichment analyses in B cells. We found antigen presentation, leukocyte differentiation, and cell adhesion as the most enriched pathways in Hem1-/- compared to Hem1+/+ B cells (tables S8 and S9). As all these cellular pathways ensure the development of normal, non-autoreactive B cells, we decided to examine whether patient B cells display any abnormalities associated with cell spreading and interactions with antigens. Indeed, we found that patient B cells were unable to spread, as evidenced from a significantly decreased cell spreading area on ligand-coated substrates (Fig. 3, A and B, 7A and fig. S6B). Instead of assembling polarized lamellipodia and membrane ruffles, cells assumed a rather rounded blebbing or spiky morphology normally associated with high levels of cortical actomyosin tension and the formation of long linear F-actin assemblies (fig. S6C). As a result of the decreased cell spreading, B cells exhibited a reduced capacity to recruit IgM and displayed a disorganized B-cell immunological synapse (IS) required to efficiently engage and properly organize IgM molecules within the IS (Fig. 7, B and C; fig. S6, B and D). This phenotype is specific to HEM1 deficiency and was not observed to a similar extent in WASPdeficient B cells (fig. S6E).
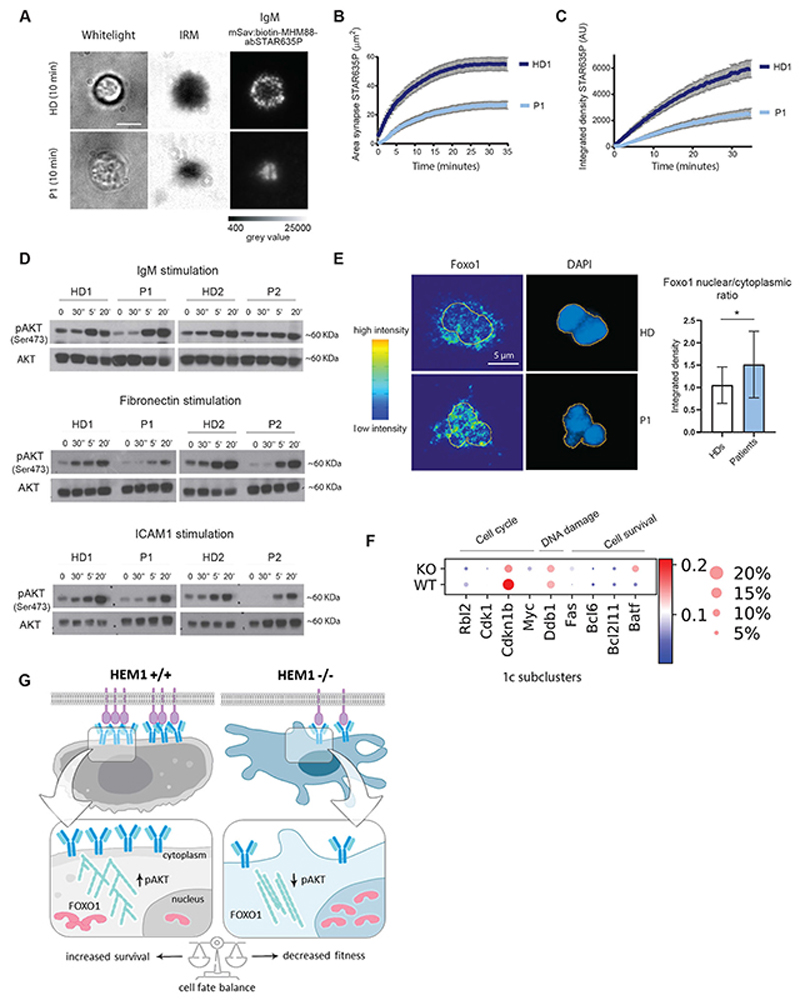
Fig. 7. Disrupted BCR signaling in HEM1 deficiency.
(A) Healthy donor (top) and patient (bottom) B cells following IgM stimulation using a planar-supported lipid bilayer functionalized with a monovalent streptavidin platform presenting biotinylated and abSTAR635P-conjugated MHM-88 mAbs, transmission light, TL (left), Interference Reflection Microscopy, IRM (middle) and IgM (right). Scale bar: 5μM. (B) Area of IgM (abSTAR635P)/μm2 over time illustrates accumulation of IgM at the immune synapse. (C) Integrated density of IgM [Arbitrary Unit (AU)] over time illustrates intensity of IgM at the immune synapse (HD1-dark blue, patient 1 (light blue) (D) Representative EBV-LCL transformed B-cells Western blots of P1, P2 and 2 healthy donors of pAKT (Ser473) and total AKT including ratios of phospho/total AKT by IgM, fibronectin and ICAM1 stimulation. (E) Immunofluorescence staining of patient and healthy donor-derived primary B cells for Foxo1 and DAPI following IgM stimulation illustrating increased levels of Foxo1 in the nucleus of P1. Quantification of nuclear/cytoplasmic ratio of both P1 and p2 (pooled) and two healthy donors. The yellow line indicates the nuclear border, intensity map (left). (F) Representative graphs for Foxo1 target genes regulating cell cycle progression (Rbl2, Cdk1, Cdknlb, Myc), DNA damage (Ddb1) and apoptosis (Bcl6, Bcl2lll, Batf). (G) Scheme illustrating proposed mechanism.
Since actin cytoskeleton-mediated organization of the B-cell synapse is key to integrate stimulatory cues and activate downstream signaling pathways (48), we hypothesized that in the absence of HEM1, B cells fail to activate critical signaling cascades to a degree sufficient to maintain cell fitness and proper development. Such signaling cascades classically include the kinases ERK and PI3K ultimately activating AKT, whose functions are critical to skew the balance of cell fate choices towards growth and proliferation. Hence, when we biochemically probed the activation status of the kinase AKT, we found a significant decrease in the amount of phosphorylated AKT (pAKT Ser473) in patient cells compared to normal donor B cells (Fig. 7D). To execute its cell fate-controlling function, AKT operates through transcriptional regulators such as the evolutionarily conserved Daf-16-related Forkhead O (Foxo) family. Phosphorylation of the Foxo proteins by AKT is established to induce their nuclear export and subsequent inactivation of transcriptional activity indispensable for B-cell development. Thus, we used nuclear levels of Foxo proteins as a readout of its transcriptional activity (49, 50). Indeed, we found that IgM-stimulated normal donor-derived B cells display significantly decreased nuclear levels of Foxo1 (Fig. 7E). By contrast, patient cells retained Foxo1 in the nucleus upon IgM stimulation, indicating that the AKT-mediated control of Foxo1 nuclear-cytoplasmic shuttling and thus transcriptional activity is attenuated in HEM1-deficient B cells (Fig. 7E) (50).
Foxo1 also operates as a transcriptional repressor of genes promoting B-cell survival, proliferation and proper differentiation. Indeed, consistent with the described above accumulation of Foxo1 in the nucleus of patient HEM1-deficient B cells, our analysis of single-cell RNAseq data in mouse splenic B cells revealed Foxo1 targets (51) among top transcriptionally deregulated genes in Hem1-/- vs. Hem1+/+ B cells. For instance, we found pro-proliferative genes (Myc, Cdkn1b, Cdk1, Rbl2), normally repressed by Foxo1, were reduced (Fig. 7F), while genes promoting controlled DNA damage (Ddb1) and controlling apoptosis (Fas, Bcl2, Bcl6, and Batf) via transcriptional activation by Foxo1 were upregulated in Hem1-/- B cells (Fig. 7F and fig. S6F). This transcriptional pattern was most evident in the cluster 1c (Fig. 5D) that did not appear to correspond to any specific stage of B-cell development. Therefore, we concluded that this cell cluster most probably emerges from the dysregulation of B-cell developmental program resulting from Hem1 deficiency.
Discussion
The multicomponent WAVE regulatory complex (WRC) is traditionally considered as one of the key molecular machineries enabling actin-driven migration of a variety of animal tissue cell types (7, 10, 26). More recent studies of the WRC in immune cell migration reveal that its subunits are not necessarily essential for basal cell migration but mainly contribute to the ability of rapidly crawling amoeboid leukocytes to navigate extracellular spaces with increasing geometrical complexity (25). Given the conditional nature of the role of the WRC in cell migration and the systemic severity of knock-out phenotypes for WRC subunits (e.g., Wave2) at an organismal level (10), it is unlikely that WRC has evolved to regulate cell locomotion only. Here, by dissecting the pathobiology underlying a previously unknown inborn error of human immunity caused by deficiency in the hematopoietic-specific WRC component HEM1, we uncover a role for the WRC in the context of B-cell development and homeostasis (scheme Fig. 7G).
Over the last years with the discoveries of key regulators of immune homeostasis it became clear that there is considerable variability as the clinical presentation of even patients with the same genetic disorder may vary from sub-clinical to severe systemic autoimmunity, as exemplified in patients with germline mutations in PIK3CD, CTLA4 or DEF6 (32, 52, 53). Reminiscent of such observations, we detected phenotypic variability in HEM1-deficient individuals, with the more severely affected patient P1 showing an SLE-like autoimmune disease phenocopied by a Hem1- deficient mouse model, while the clinical phenotype of P2 – to date – has remained milder despite similar B-cell phenotypic aberrations including increased CD21loCD38lo B cells. Notably, aberrant B-cell phenotypes similarly to our observations for HEM1 deficiency are seen in other SLE and CVID (Common Variable Immunodeficiency) patients, including reduced switched memory B cells and expanded CD21loCD38lo B cells. Although clonally related to memory B cells and plasmablasts, these cells form distinct clades within phylogenetic trees based on accumulated variable gene mutations, supporting exit from active GCs (37). CD21loCD38lo B cells express a transcriptional program suggesting they are primed for plasma cell differentiation and are refractory to GC differentiation(37). During GC maturation, B cells repetitively test and adapt their B-cell receptor to ensure the development of a tolerant, non-autoreactive BCR (54). B cells constantly take up soluble antigens predominantly via clathrin-coated pits (55), but uptake of cellbound antigens requires additional, larger-scale processes involving F-actin-mediated cell shape reorganization. Encountering antigen (Ag)-presenting cells (APCs) bearing cognate antigen, B cells begin establishing the immune synapse (IS) (56) by spreading across the surface of the APC to scan for additional antigens. This process is followed by membrane contraction and the formation of antigen receptor microclusters initiating the signaling reactions necessary for lymphocyte activation (47).
We found that in the absence of HEM1, B cells fail to spread properly and exhibit a lower coverage of antigen-presented surfaces due to the lack of lamellipodia. This in turn results in significantly less efficient aggregation of membrane bilayer-bound IgM molecules within the B-cell IS. These morphodynamic abberations in B-cell IS formation and function decrease the strength of the extracellular signal required to tune BCR signaling toward normal B-cell development. We hypothesize that in HEM1 deficient GCs the control of developmental and survival programs is hijacked by auto-reactive antigens as they elicit stronger signals, shifting the balance of B-cell fate choices towards survival of autoreactive B cells and the development of autoimmunity. Similar defects in T-cell IS formation may further promote the disease progression as cognate interactions between T and B lymphocytes lead to a bidirectional signal exchange(28, 47, 55). Additionally, we speculate that when HEM1-deficient B cells extract antigens from live APCs, lack of Arp2/3- mediated actin branching reduces membrane flexibility hindering BCR-dependent membrane invagination and as a result antigen extraction. At the same time, formins known to take over the actin polymerase activity in the absence of Arp2/3-mediated actin branching, would promote unbranched linear actin filament generation, which can sterically impede dynamic signal extraction. Collectively, these processes explain a reduced amount of perceived cell-bound signal. Mutations in genes encoding multiple actin cytoskeleton regulators have now been characterized as causing distinct inborn error of immunity (IEI) entities (57). In addition to combined impairments in innate and adaptive immunity, autoimmunity emerges as a common feature of some actin associated IEIs. Studies in the prototypical Wiskott-Aldrich syndrome have highlighted the pivotal role of the Arp2/3 activator WASP in controlling BCR and TLR signaling in a B-cell differentiation stage dependent manner, with elevated responsiveness in immature B cells and reduced responsiveness in mature B cells (58, 59). Compared to WASP deficiency, DOCK8 deficiency is associated to accumulation of auto-reactive B cells (60). As suggested by studies in murine models, WIP deficiency, still poorly investigated in humans, might also be associated with autoimmunity due to aberrant B-cell activation (61). In parallel to WAVE, also WASP, DOCK8 and WIP, which have been shown to interact (62), control actin branching via the ARP2/3 complex. The overlapping function of these proteins in tuning B-cell activation and tolerance may therefore be due to the fact that they converge in controlling spatio-temporal activation of the ARP2/3 complex. A recent study that pharmacological inhibition of ARP2/3 impairs BCR microcluster mobility promoting APC-induced B-cell activation at the IS (63). Our study highlights that beyond these overlapping aspects, the WAVE versus WASP-mediated activation of Arp2/3 in B cells may differ, both in terms of molecular control and associated phenotype.
Collectively, we here characterize a loss of function mutation in WRC actin regulatory complex, which results in B-cell developmental dysregulation and autoimmunity. We demonstrate that the WRC critically regulates extracellular signal perception and signal-bridging, identifying a major and previously unrecognized role for the WRC in B-cell-development and immune homeostasis across species.
Materials and Methods
Study design
The objective of this study was to investigate the role of HEM1 in human and mouse immune system and the consequence of HEM1 deficiency in immune dysregulation and autoimmunity. For this purpose, we made use of primary material from two HEM1 deficient patients, which were genetically analyzed in our lab primarily and also we used Hem1-/- mouse as an animal model. To study the cellular functions in this genetic defect we also produced cell line models which is described at the following sections.
Patient and ethics
Patients and other family member samples were included after collecting informed written consent and with approval from the ethics committee of Tehran University of Medical Sciences. The patients were evaluated, followed up and treated at the Departments of Rheumatology and Immunology at Children’s medical center hospital, Tehran University of Medical Sciences, Tehran, Iran. The study was approved by the local ethics committee of the Medical University of Vienna, Austria.
Animal Experiments Ethics
All experimental studies were approved by the Animal Ethics Committee of the Medical University of Vienna, Austria and were performed according to the guidelines for Good Scientific Practice of the Medical University of Vienna, Austria.
Exome sequencing (ES)
For ES, a TrueSeq Rapid Exome kit as well as the Illumina HiSeq3000 system and the cBot cluster generation instruments were used as previously described (64, 65), with minor changes. Briefly, reads were aligned to the human genome version 19 by means of the Burrows-Wheeler Aligner (BWA). Variants effect predictor (VEP) was used for annotating single nucleotide variants (SNVs) and insertions/deletions lists. The obtained list was then filtered according to the presence of variants with a minor allele frequency (MAF) >0.01 in 1,000 Genomes, gnomAD, and dbSNP build 149. After further filtering steps for nonsense, missense, and splice-site variants using VCF. Filter software (66), an internal database was used to filter for recurrent variants. Moreover, variants are prioritized using tools, such as SIFT, Polyphen-2 and the combined annotation dependent depletion (CADD) score as well as HGCS, that predict the deleteriousness of a present variant.
Preprocessing and analysis of single-cell RNA-seq data
For details regarding sample preparation please see supplementary methods. Preprocessing of the single-cell RNA-seq data was performed using Single-Cell Software Suite (version 3.0.2, 10x Genomics) (67). Raw sequencing files were demultiplexed using the Cell Ranger command ‘mkfastq’. Each sample was aligned to the human reference genome assembly ‘refdata-cellranger-mm10-1.2.0’ using the Cell Ranger command ‘count’, and all samples were aggregated using the Cell Ranger command ‘aggr’ without depth normalization. Raw expression data were then loaded into python 3.7.3 and analyzed using the scanpy (1.4.5.post1) package (68).
Filtering and preparation of single-cell RNA-seq data
Cells with fewer than 700 detected genes, fewer than 1,200 counts or exceeded a total mitochondrial gene expression of 20% were removed. Genes expressed in fewer than 20 cells or fewer than 3 counts were also removed. Count data were normalized (excluding highly expressed genes), batch effect corrected and unwanted variation (number of counts per unique molecular identifier [UMI], mitochondrial fraction and ribosomal fraction) were regressed out.
Clustering, differential expression and further analyses
Highly variable genes (4000) were identified using the Cell Ranger (67) implementation in Scanpy (Single-Cell Analysis in Python) (68). Dimensionality reduction was performed using uniform manifold approximation and projection (UMAP) (69) and principal component analysis (PCA). The PCA was based on the identified variable genes and limited to the first 50 principal components. The neighborhood graph was computed taking the UMAP and PCA into account. The Leiden (70) algorithm with a resolution of 0.1 grouped cells into clusters. By quantifying the connectivity of partitions with partition-based graph abstraction (PAGA) (71), the UMAP was recalculated and sub-clusters within the B-cell and T-cell identified using the Leiden algorithm with a resolution of 0.6 and 0.4 respectively. Differential expression analysis was performed based on the t-test, which overestimates variance of each group. The P values were adjusted for multiple testing using the Benjamini-Hochberg correction. Entropy analysis was performed using the SLICE (72) package following the standard workflow.
Flow cytometry analyses
All flow cytometric analyses were recorded on a BD LSR Fortessa. All data were analyzed using FlowJo X (TreeStar) and data was graphed with Prism 8.0 (GraphPad Software). For detailed experimental procedures and antibodies see supplementary information.
Cell line generation and editing
For details regarding Jurkat, B16-F1 EBV-LCL generation and editing as well as feeder T-cell expansion see supplementary information.
Rescue experiments and microscopy
Cells were transfected as described above and seeded on the following day onto 15 mm-diameter glass coverslips coated with 25 μg/ml laminin (Sigma-Aldrich, L2020) in laminin-coating buffer: 50 mM Tris, 150 mM NaCl. Cells were allowed to adhere overnight and then fixed with prewarmed, 4% paraformaldehyde (PFA; Sigma-Aldrich P6148) in PBS for 20 min. Permeabilization was carried out using 0.05% Triton-X100 in PBS for 1 min, subsequently followed by 3 washing steps with 1xPBS. In order to visualize the actin cytoskeleton, cells were subjected to phalloidin staining (ATTO-594-conjugated phalloidin, ATTO-TEC AD594-81, 1:200) and finally mounted onto glass slides using ProLong Gold antifade reagent (Thermo Fisher Scientific, P36934). PFA-fixed samples of cells expressing certain EGFP-tagged proteins of interest were analyzed on an inverted microscope (Axiovert 100TV, Zeiss) equipped with an HXP 120 lamp for epifluorescence illumination, a Coolsnap-HQ2 camera (Photometrics), a filter wheel (LUDL Electronic Products LTD) and electronic shutters driven by MetaMorph software (Molecular Devices Corp., Sunnyvale, CA, USA). Images were taken using either 63x/1.4NA or 100x/1.4NA Plan apochromatic oil objectives. Cells devoid of Hem1/Hem2 expression and thus incapable of lamellipodia formation served as tool to address and compare the effects of certain point mutations in either Hem1 or Hem2 on lamellipodia formation. Cells expressing respective construct (positive for EGFP-signal) were categorized according to their cellular morphology as exemplified in “final figure C”. Fractions of cells in % were plotted as stacked bar charts using Excel 2010 (Microsoft).
Video microscopy and immunofluorescence studies of patient and healthy controls
For details regarding IF, TCR, BCR signaling analyses and video microscopy please refer to the supplementary information.
Generation of Ilem1-/- mice
To generate conditional Hem1-knockout mice, a targeting vector was used in which exons 4 and 5 of the Hem1 gene were flanked by loxP sites. A Neo-cassette flanked by frt sites was inserted into intron 3 allowing for G418 selection. This vector was electroporated into murine embryonic stem cells (ES line IGD3.2)(25) Homologous recombinant clones were identified by genomic Southern hybridization and PCR. A targeted ES cell clone was injected into C57BL/6 blastocysts and gave rise to germ line chimeric animals. The Neo-cassette was removed from the hem1 locus by mating the resulting animals (fl_neo/wt), with transgenic mice expressing flip recombinase, resulting in conditional Hem1 mice (fl/wt) that were backcrossed to a C57BL/6 background. Finally, male Hem1 fl/wt animals were crossed with female K14-cre mice to obtain animals being heterozygously deleted for Hem1 (del/wt). Unless otherwise indicated, age-matched female and male animals were used at 4-8 weeks of age. All mice were group housed on a 12:12-h light-dark cycles at 22°C with food and water provided ad libitum.
Bone marrow transplantation
Bone marrow transplantation studies were performed as previously described (74). In short, 8- week-old C57BL/6J mice were sublethally irradiated (2×6Gy) and were subsequently injected intravenously via the retro-orbital plexus with 3×106 mixed bone marrow from 8-week-old JHT and Hem1+/+ versus Hem1-/- donors (4:1 ratio). The recipient mice were given a 14-week recovery period before intervention studies.
Microscopy
We further operated an Eclipse Ti-E (Nikon) inverted microscope system (setup #2) that was equipped with a high NA objective (100x magnification, NA=1.49, Nikon SR APO TIRF) and a low NA objective (20x magnification, NA=0.75, Nikon S Fluor). For excitation, we employed five diode lasers with the wavelengths 405 nm, 488 nm, 514 nm, 642 nm (iBeam smart Toptica) and 532 nm and 561 nm (Obis). The excitation light was cleaned up with clean up filters that matched the corresponding wavelengths (Chroma). Setup 2 was also coupled to a xenon lamp (Lambda LS lamp, Sutter Instrument Company), which was equipped with a filter wheel (Sutter Instrument Company) with the following filters installed: 340/26, 387/11, 370/36, 474/27, 554/23, 635/18 (all Chroma). This setup was further operated with the beam splitters zt405/488/532/640rpc and ET-Dualb. Sedat CFP/YFP sbxm + HC510/20 (all Chroma), and a Notch filter blocking 405, 488, 532 and 640 nm light (Chroma). Emission light was cleaned up with the use of a filter wheel (Sutter Instrument Company) which was equipped with the bandpass filters ET450/50, ET510/20, ET525/50, ET605/52 and ET700/75 (all Chroma). Emission light was recorded with a backside-illuminated electron-multiplying charge-coupled device (EM-CCD) camera (Andor iXon Ultra 897). An 8 channel DAQ-card (National Instruments) and the microscopy automation and image analysis software Metamorph (Molecular Devices) were used to program and apply timing protocols and control all hardware components. Microscopy setups were covered by a box and temperature sensitive experiments were conducted under the control of a temperature control system from Leica (setup #1) and Pecon (setup #2). For details regarding bilayer preparation and antibody labelling please see supplementary information.
B-cell synapse formation
Primary human B cells were washed once in imaging buffer [HBSS (Life technologies) supplemented with 0.4 mg ml-1 Ovalbumin (Merck), 2 mM CaCl2 and 2 mM MgCl2 (Merck)] and seeded on supported lipid bilayers (SLBs) functionalized with mSav:STAR635P-MHM88-biotin (20 – 100 molecules per μm2) and ICAM-1-12His (~100 molecules per μm2) to record B cell synapse formation. Images were recorded in TIRF mode using an 100x TIRF objective (Nikon) and at 25°C. We recorded B cell synapses only from adherent cells, a condition that was verified by Interference Reflection Microscopy (IRM) using 554/23 nm lamp excitation and a CFP/YFP +HC510/20 cube. To visualize B cell synapse formation in TIRF, we recorded a red image (640 nm laser excitation, 705/72 emission filter) of mSav:STAR635P-MHM88-biotin followed by a blue image (488 nm laser excitation, 510/20 emission filter) of ICAM-1-AF488 with an illumination time of 10 ms. We used a laser power density of 0.16 kW cm-2 and 0.06 kW cm-2 to excite STAR635P and AF488, respectively. Videos of B cell synapse formation were recorded with an 20x objective in epi fluorescence mode using 635/18 nm lamp illumination to visualize mSav:STAR635P-MHM88-biotin, 474/27 nm lamp illumination to excite ICAM-1-AF488 and 554/23 lamp illumination to excite 7AAD (7-aminoactinomycin D) (live/dead cell discrimination). We recorded an image stack (whitelight, GFP, Cy3, Cy5 illumination) every 10 s with an illumination time of 100 ms for all channels.
For experiments involving chemical fixation, cell samples were fixed with 4% paraformaldehyde (Thermo Fisher Scientific) in PBS for 10 minutes at room temperature and subsequently washed in HBSS (Life technologies) supplemented with 0.4 mg ml-1 ovalbumin (Merck), 2 mM CaCl2 and 2 mM MgCl2 (Merck) to terminate the fixation reaction. Microscopy images were processed and analyzed with the open-source image processing package Fiji (75). B cell synapse dynamics was assessed by measuring the ability of B cells to collect stimulatory anti-IgM antibodies (mSav:STAR635P-MHM88-biotin) upon interaction with the bilayer. For that purpose, the Fiji software was used to extract the area and the integrated density of fluorescently labeled anti-IgM (mSav:STAR635P-MHM88-biotin) associated with each cell. This analysis was applied to a sequence of 100 images covering an acquisition time of 34 minutes, in order to capture both the spreading of individual B cell synapses and the progressive enrichment in collected anti-IgM antibodies
Supplementary Material
One Sentence Summary.
HEM1 deficiency causes heritable autoimmunity and immunodeficiency interface required for B-cell receptor complex assembly.
Acknowledgments
The authors gratefully acknowledge the patients and parents for their participation in the study. Meinrad Busslinger provided JHT knockout mice to Christoph Binder for further breeding and bone marrow transplantation experiments. We thank Sophie Allart and Astrid Canivet from the CPTP cell imaging facility in Toulouse. We would also like to thank Meinrad Busslinger for providing JHT knockout mice for bone marrow transplantation experiments. We would like to thank Bernhard Ransmayr for assistance with Figure preparation. We Thank Tatjana Hirschmugl for graphical adaptation of the scheme shown in Fig. 7G.
Funding
The research leading to these results has received funding from the European Research Council Consolidator Grant iDysChart (ERC grant agreement number: 820074) (to K.B.), the FWF through project FWF I2250-B28 (to K.B. and E.S.), a Science Award of the Austrian Society of Pediatrics and Adolescent Medicine (to E.S.), grants by the Deutsche Forschungsgemeinschaft (to T.E.S. and K.R.) and intramural funding from the Helmholtz Society (to T.E.S.). Parts of Figure 3 and 4 were created using the Biorender software.
Footnotes
Author contributions: ES, SZ and CR performed most experiments, analyzed data and interpreted results. ES supervised CR. SZ identified the mutation in HEM1. MGK, ES and SZ designed, performed and analyzed murine immune phenotyping and transplantation experiments. PH and TES provided Hem1-/- mice originally generated in her lab. SSt performed initial characterizations of Hem1-/- mice, was responsible for genotyping, and together with PH supervised breeding and delivery of Hem1-/- mice for further analyses. FK generated Nap1/Hem1 double knock-out B16-F1 melanoma cells and performed all experiments in this cell type. SSt performed dsDNA and cytokine analyses in Hem1-/- mice. MC together with JM performed CellProfiler analyses. DM provided critical input and performed initial mouse experiments. LD performed video microscopy analyses and provided critical intellectual input. BH, RCA and JB performed selected experiments. RP performed lipid bilayer experiments with SZ, supervised by JBH. RK performed histological analyses and electron microscopy of Hem1-/- mice. KR and TES supervised FK and SSt and provided critical input to experiments and manuscript. NR and SZ coordinated the study in Tehran. NR, VZ and SSh provided local infrastructure in Tehran, cared for the patients and identified the family. CJB provided laboratory resources for mouse experiments, supervised MGK and provided critical intellectual input. AL and MS provided critical intellectual input. MH analyzed 10x scRNAseq data together with CB, SZ, ES with input from CS. KB, ES, SZ and AL wrote the manuscript together with input from CR and MGK. KB coordinated the study, provided laboratory resources, interpreted data, and took overall responsibility for the study. All of the authors provided critical input and agreed to this publication
Competing interests: The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to kaan.boztug@ccri.at.
Data availability
All single cell RNA-Seq data included in this manuscript have been deposited with the functional genomics data store ArrayExpress (73) archive and accessible under the accession number E-MTAB-8706.
Data and materials availability
All single cell RNA-Seq data included in this manuscript have been deposited with the functional genomics data store ArrayExpress archive and are accessible under the accession number E-MTAB-8706.
References and Notes
- 1.Moulding DA, Record J, Malinova D, Thrasher AJ. Actin cytoskeletal defects in immunodeficiency. Immunol Rev. 2013;256:282–99. doi: 10.1111/imr.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahr WHA, Pluthero FG, Elkadri A, Warner N, Drobac M, Chen CH, Lo RW, Li L, Li R, Li Q, Thoeni C, et al. Loss of the Arp2/3 complex component ARPC1B causes platelet abnormalities and predisposes to inflammatory disease. Nat Commun. 2017;8:14816. doi: 10.1038/ncomms14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanzi G, Moratto D, Vairo D, Masneri S, Delmonte O, Paganini T, Parolini S, Tabellini G, Mazza C, Savoldi G, Montin D, et al. A novel primary human immunodeficiency due to deficiency in the WASPinteracting protein WIP. J Exp Med. 2012;209:29–34. doi: 10.1084/jem.20110896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suetsugu S, Miki H, Takenawa T. Identification of Two Human WAVE/SCAR Homologues as General Actin Regulatory Molecules Which Associate with the Arp2/3 Complex. Biochem Biophys Research Commun. 1999;260:296–302. doi: 10.1006/bbrc.1999.0894. [DOI] [PubMed] [Google Scholar]
- 5.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 7.Litschko C, Linkner J, Brühmann S, Stradal TEB, Reinl T, Jänsch L, Rottner K, Faix J. Differential functions of WAVE regulatory complex subunits in the regulation of actin-driven processes. Eur J Cell Biol. 2017;96:715–727. doi: 10.1016/j.ejcb.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. The WAVE Regulatory Complex Links Diverse Receptors to the Actin Cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan C. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiskott A. Familiärer, angeborener Morbus Werlhofii? Monatsschriftfür Kinderheilkunde. 1937:212–216. [Google Scholar]
- 12.Mohan AS, Dean KM, Isogai T, Kasitinon SY, Murali VS, Roudot P, Groisman A, Reed DK, Welf ES, Han SJ, Noh J, et al. Enhanced Dendritic Actin Network Formation in Extended Lamellipodia Drives Proliferation in Growth-Challenged Rac1P29S Melanoma Cells. Dev Cell. 2019;49:444–460.e9. doi: 10.1016/j.devcel.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moujaber O, Stochaj U. The Cytoskeleton as Regulator of Cell Signaling Pathways. Trends Biochem Sci. 2019 doi: 10.1016/j.tibs.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Brunner HI, Holland M, Beresford MW, Ardoin SP, Appenzeller S, Silva CA, Flores F, Goilav B, Wenderfer SE, Levy DM, Ravelli A, et al. the PRCSG and PRINTO Investigators, American College of Rheumatology Provisional Criteria for Global Flares in Childhood-Onset Systemic Lupus Erythematosus. Arthritis Care Res. 2018;70:813–822. doi: 10.1002/acr.23557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beutler E. G6PD deficiency. Blood. 1994;84:3613–3636. [PubMed] [Google Scholar]
- 16.Özen S, Batu ED, Demir S. Familial Mediterranean Fever: Recent Developments in Pathogenesis and New Recommendations for Management. Front Immunol. 2017;8:253. doi: 10.3389/fimmu.2017.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Exome Aggregation Consortium. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. Genomics. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 19.Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TEB. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakeman AS, Anderson KV. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development. 2006;133:3075–3083. doi: 10.1242/dev.02473. [DOI] [PubMed] [Google Scholar]
- 21.Dolati S, Kage F, Mueller J, Müsken M, Kirchner M, Dittmar G, Sixt M, Rottner K, Falcke M. On the relation between filament density, force generation, and protrusion rate in mesenchymal cell motility. Mol Biol Cell. 2018;29:2674–2686. doi: 10.1091/mbc.E18-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dürre K, Keber FC, Bleicher P, Brauns F, Cyron CJ, Faix J, Bausch AR. Capping protein-controlled actin polymerization shapes lipid membranes. Nat Commun. 2018;9:1630. doi: 10.1038/s41467-018-03918-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kage F, Winterhoff M, Dimchev V, Mueller J, Thalheim T, Freise A, Brühmann S, Kollasser J, Block J, Dimchev G, Geyer M, et al. FMNL formins boost lamellipodial force generation. Nat Commun. 2017;8:14832. doi: 10.1038/ncomms14832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaks M, Singh SP, Kage F, Thomason P, Klünemann T, Steffen A, Blankenfeldt W, Stradal TE, Insall RH, Rottner K. Distinct Interaction Sites of Rac GTPase with WAVE Regulatory Complex Have Non-redundant Functions in Vivo. Current Biology. 2018;28:3674–3684.e6. doi: 10.1016/j.cub.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leithner A, Eichner A, Müller J, Reversat A, Brown M, Schwarz J, Merrin J, de Gorter DJJ, Schur F, Bayerl J, de Vries I, et al. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat Cell Biol. 2016;18:1253–1259. doi: 10.1038/ncb3426. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Z, Chai Y, Jiang Y, Li W, Hu H, Li W, Wu J-W, Wang Z-X, Huang S, Ou G. Functional Coordination of WAVE and WASP in C. elegans Neuroblast Migration. Developmental Cell. 2016;39:224–238. doi: 10.1016/j.devcel.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 27.Squarr AJ, Brinkmann K, Chen B, Steinbacher T, Ebnet K, Rosen MK, Bogdan S. Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J Cell Biol. 2016;212:591–603. doi: 10.1083/jcb.201508081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanzavecchia A, Sallusto F. Antigen decoding by T lymphocytes: from synapses to fate determination. Nat Immunol. 2001;2:487–492. doi: 10.1038/88678. [DOI] [PubMed] [Google Scholar]
- 29.Schatorjé EJH, Gemen EFA, Driessen GJA, Leuvenink J, van Hout RWNM, de Vries E. Paediatric Reference Values for the Peripheral T cell Compartment: T Lymphocyte Subsets in Children. Scand J Immunol. 2012;75:436–444. doi: 10.1111/j.1365-3083.2012.02671.x. [DOI] [PubMed] [Google Scholar]
- 30.Minton K. Innate lymphoid cells: Human ILCs face redundancy. Nat Rev Immunol. 2016;16:596–597. doi: 10.1038/nri.2016.106. [DOI] [PubMed] [Google Scholar]
- 31.Bride K, Teachey D. Autoimmune lymphoproliferative syndrome: more than a FAScinating disease. F1000Res. 2017;6:1928. doi: 10.12688/f1000research.11545.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab C, Gabrysch A, Olbrich P, Patiño V, Warnatz K, Wolff D, Hoshino A, Kobayashi M, Imai K, Takagi M, Dybedal I, et al. Phenotype, penetrance, and treatment of 133 cytotoxic T-lymphocyte antigen 4-insufficient subjects. J Allergy Clin Immunol. 2018;142:1932–1946. doi: 10.1016/j.jaci.2018.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pernis AB, Ivashkiv LB. ‘-Omics’ shed light on B cells in lupus. Nat Immunol. 2019;20:946–948. doi: 10.1038/s41590-019-0446-6. [DOI] [PubMed] [Google Scholar]
- 34.Sharabi A, Tsokos GC. T cell metabolism: new insights in systemic lupus erythematosus pathogenesis and therapy. Nat Rev Rheumatol. 2020 doi: 10.1038/s41584-019-0356-x. [DOI] [PubMed] [Google Scholar]
- 35.Piątosa B, Wolska-Kuśnierz B, Pac M, Siewiera K, Gałkowska E, Bernatowska E. B cell subsets in healthy children: Reference values for evaluation of B cell maturation process in peripheral blood. Cytometry. 2010;78B:372–381. doi: 10.1002/cyto.b.20536. [DOI] [PubMed] [Google Scholar]
- 36.Lau D, Lan LY-L, Andrews SF, Henry C, Rojas KT, Neu KE, Huang M, Huang Y, DeKosky B, Palm A-KE, Ippolito GC, et al. Low CD21 expression defines a population of recent germinal center graduates primed for plasma cell differentiation. Sci Immunol. 2017;2:eaai8153. doi: 10.1126/sciimmunol.aai8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorarinsdottir K, Camponeschi A, Gjertsson I, Mårtensson I-L. CD21low B cells: A Snapshot of a Unique B Cell Subset in Health and Disease. Scand J Immunol. 2015;82:254–261. doi: 10.1111/sji.12339. [DOI] [PubMed] [Google Scholar]
- 38.Shao L, Chang J, Feng W, Wang X, Williamson EA, Li Y, Schajnovitz A, Scadden D, Mortensen LJ, Lin CP, Li L, et al. The Wave2 scaffold Hem-1 is required for transition of fetal liver hematopoiesis to bone marrow. Nat Commun. 2018;9:2377. doi: 10.1038/s41467-018-04716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shikatani EA, Besla R, Ensan S, Upadhye A, Khyzha N, Li A, Emoto T, Chiu F, Degousee N, Moreau JM, Perry HM, et al. c-Myb Exacerbates Atherosclerosis through Regulation of Protective IgM-Producing Antibody-Secreting Cells. Cell Reports. 2019;27:2304–2312.e6. doi: 10.1016/j.celrep.2019.04.090. [DOI] [PubMed] [Google Scholar]
- 40.Teschendorff AE, Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat Commun. 2017;8:15599. doi: 10.1038/ncomms15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teschendorff AE, Sollich P, Kuehn R. Signalling entropy: A novel network-theoretical framework for systems analysis and interpretation of functional omic data. Methods. 2014;67:282–293. doi: 10.1016/j.ymeth.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Park H, Staehling-Hampton K, Appleby MW, Brunkow ME, Habib T, Zhang Y, Ramsdell F, Liggitt HD, Freie B, Tsang M, Carlson G, et al. A point mutation in the murine Hem1 gene reveals an essential role for Hematopoietic Protein 1 in lymphopoiesis and innate immunity. J Exp Med. 2008;205:2899–2913. doi: 10.1084/jem.20080340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Trounstine M, Alt FW, Young F, Kurahara C, Loring JF, Huszar D. Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int Immunol. 1993;5:647–656. doi: 10.1093/intimm/5.6.647. [DOI] [PubMed] [Google Scholar]
- 44.Shen P, Roch T, Lampropoulou V, O’Connor RA, Stervbo U, Hilgenberg E, Ries S, Dang VD, Jaimes Y, Daridon C, Li R, et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature. 2014;507:366–370. doi: 10.1038/nature12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minton K. Recipe for a B cell. Nat Rev Immunol. 2004;4:840–840. [Google Scholar]
- 46.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Yin W, Jing Y, Kang D, Yang L, Cheng J, Yu Z, Peng Z, Li X, Wen Y, Sun X, et al. The Coordination Between B Cell Receptor Signaling and the Actin Cytoskeleton During B Cell Activation. Front Immunol. 2019;9:3096. doi: 10.3389/fimmu.2018.03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, Dalla-Favera R. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity. 2015;43:1064–1074. doi: 10.1016/j.immuni.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 49.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochimica et Biophysica. Acta (BBA) - Molecular Cell Research. 2011;1813:1978–1986. doi: 10.1016/j.bbamcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Serwas NK, Hoeger B, Ardy RC, Stulz SV, Sui Z, Memaran N, Meeths M, Krolo A, Yüce Petronczki Ö, Pfajfer L, Hou TZ, et al. Human DEF6 deficiency underlies an immunodeficiency syndrome with systemic autoimmunity and aberrant CTLA-4 homeostasis. Nat Commun. 2019;10:3106. doi: 10.1038/s41467-019-10812-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh A, Joshi V, Jindal AK, Mathew B, Rawat A. An updated review on activated PI3 kinase delta syndrome (APDS) Genes & Diseases. 2020;7:67–74. doi: 10.1016/j.gendis.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leavy O. Mediators of central tolerance. Nat Rev Immunol. 2015;15:404–404. doi: 10.1038/nri3879. [DOI] [PubMed] [Google Scholar]
- 54.Stoddart A, Jackson AP, Brodsky FM. Plasticity of B Cell Receptor Internalization upon Conditional Depletion of Clathrin. MBoC. 2005;16:2339–2348. doi: 10.1091/mbc.E05-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batista FD, Iber D, Neuberger MS. B cells acquire antigen from target cells after synapse formation. Nature. 2001;411:489–494. doi: 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- 56.Janssen E, Geha RS. Primary immunodeficiencies caused by mutations in actin regulatory proteins. Immunol Rev. 2019;287:121–134. doi: 10.1111/imr.12716. [DOI] [PubMed] [Google Scholar]
- 57.Kolhatkar NS, Brahmandam A, Thouvenel CD, Becker-Herman S, Jacobs HM, Schwartz MA, Allenspach EJ, Khim S, Panigrahi AK, Luning Prak ET, Thrasher AJ, Notarangelo LD, et al. Altered BCR and TLR signals promote enhanced positive selection of autoreactive transitional B cells in Wiskott-Aldrich syndrome. J Exp Med. 2015;212:1663–1677. doi: 10.1084/jem.20150585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becker-Herman S, Meyer-Bahlburg A, Schwartz MA, Jackson SW, Hudkins KL, Liu C, Sather BD, Khim S, Liggitt D, Song W, Silverman GJ, et al. WASp-deficient B cells play a critical, cell-intrinsic role in triggering autoimmunity. The Journal of Experimental Medicine. 2011;208:2033–2042. doi: 10.1084/jem.20110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janssen E, Morbach H, Ullas S, Bannock JM, Massad C, Menard L, Barlan I, Lefranc G, Su H, Dasouki M, Al-Herz W, et al. Dedicator of cytokinesis 8-deficient patients have a breakdown in peripheral B-cell tolerance and defective regulatory T cells. Journal of Allergy and Clinical Immunology. 2014;134:1365–1374. doi: 10.1016/j.jaci.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keppler SJ, Gasparrini F, Burbage M, Aggarwal S, Frederico B, Geha RS, Way M, Bruckbauer A, Batista FD. Wiskott-Aldrich Syndrome Interacting Protein Deficiency Uncovers the Role of the Co-receptor CD19 as a Generic Hub for PI3 Kinase Signaling in B Cells. Immunity. 2015;43:660–673. doi: 10.1016/j.immuni.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janssen E, Tohme M, Hedayat M, Leick M, Kumari S, Ramesh N, Massaad MJ, Ullas S, Azcutia V, Goodnow CC, Randall KL, et al. A DOCK8-WIP-WASp complex links T cell receptors to the actin cytoskeleton. JCI. 2016;126:3837–3851. doi: 10.1172/JCI85774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger-Munro M, Choi K, Scurll JM, Abraham L, Chappell RS, Sheen D, Dang-Lawson M, Wu X, Priatel JJ, Coombs D, Hammer JA, et al. Arp2/3 complex-driven spatial patterning of the BCR enhances immune synapse formation, BCR signaling and B cell activation. eLife. 2019;8:e44574. doi: 10.7554/eLife.44574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozen A, Comrie WA, Ardy RC, Dominguez Conde C, Dalgic B, Beser OF, Morawski AR, Karakoc-Aydiner E, Tutar E, Baris S, Ozcay F, et al. CD55 Deficiency, Early-Onset Protein-Losing Enteropathy, and Thrombosis. N Engl J Med. 2017;377:52–61. doi: 10.1056/NEJMoa1615887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salzer E, Cagdas D, Hons M, Mace EM, Garncarz W, Petronczki OY, Platzer R, Pfajfer L, Bilic I, Ban SA, Willmann KL, et al. RASGRP1 deficiency causes immunodeficiency with impaired cytoskeletal dynamics. Nat Immunol. 2016;17:1352–1360. doi: 10.1038/ni.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller H, Jimenez-Heredia R, Krolo A, Hirschmugl T, Dmytrus J, Boztug K, Bock C. VCF.Filter: interactive prioritization of disease-linked genetic variants from sequencing data. Nucleic Acids Research. 2017;45:W567–W572. doi: 10.1093/nar/gkx425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2018 available at http://arxiv.org/abs/1802.03426 1802.03426 [cs, stat] [Google Scholar]
- 69.Traag V, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, Theis FJ. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo M, Bao EL, Wagner M, Whitsett JA, Xu Y. SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1278. gkw1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, Megy K, et al. ArrayExpress update—simplifying data submissions. Nucleic Acids Research. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li AC, Binder CJ, Gutierrez A, et al. Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J Clin Invest. 2004;114(11):1564–1576. doi: 10.1172/JCI18730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Houmadi R, Guipouy D, Rey-Barroso J, Vasconcelos Z, Cornet J, Manghi M, Destainville N, Valitutti S, Allart S, Dupré L. The Wiskott-Aldrich Syndrome Protein Contributes to the Assembly of the LFA-1 Nanocluster Belt at the Lytic Synapse. Cell Reports. 2018;22:979–991. doi: 10.1016/j.celrep.2017.12.088. [DOI] [PubMed] [Google Scholar]
- 77.Fairhead M, Krndija D, Lowe ED, Howarth M. Plug-and-play pairing via defined divalent streptavidins. J Mol Biol. 2014;426:199–214. doi: 10.1016/j.jmb.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, Ting AY. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–73. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossboth BK, Platzer R, Sevcsik E, Baumgart F, Stockinger H, Schütz GJ, Huppa JB, Brameshuber M. Unscrambling Fluorophore Blinking for Comprehensive Cluster Detection via PALM. Biophysics. 2019 doi: 10.1101/545152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gudipati V, Rydzek J, Perez ID, Scharf L, Königsberger S, Einsele H, Stockinger H, Hudecek M, Huppa JB. Inefficient ZAP70-Signaling Blunts Antigen Detection by CAR-T-Cells. Immunology. 2019 doi: 10.1101/720417. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All single cell RNA-Seq data included in this manuscript have been deposited with the functional genomics data store ArrayExpress (73) archive and accessible under the accession number E-MTAB-8706.
All single cell RNA-Seq data included in this manuscript have been deposited with the functional genomics data store ArrayExpress archive and are accessible under the accession number E-MTAB-8706.