Abstract
As the enzyme toolbox for biocatalysis has expanded, so has the potential for the construction of powerful enzymatic cascades for efficient and selective synthesis of target molecules. Additionally, recent advances in computer-aided synthesis planning are revolutionising synthesis design in both synthetic biology and organic chemistry. However, the potential for biocatalysis is not well captured by tools currently available in either field. Here we present RetroBioCat, an intuitive and accessible tool for computer-aided design of biocatalytic cascades, freely available at retrobiocat.com. Our approach uses a set of expertly encoded reaction rules encompassing the enzyme toolbox for biocatalysis, and a system for identifying literature precedent for enzymes with the correct substrate specificity where this is available. Applying these rules for automated biocatalytic retrosynthesis, we show our tool to be capable of identifying promising biocatalytic pathways to target molecules, validated using a test-set of recent cascades described in the literature.
Introduction
Biocatalysis is at the nexus of rapidly expanding sequence data, cheaper DNA synthesis, advances in enzyme engineering and a strong need for more sustainable manufacturing processes 1. Increasingly this means biocatalysis is an attractive option for organic synthesis, particularly where exquisite selectivity is required 2,3. Mild operating conditions afford enzymes further advantages, in that they can be combined easily into multi-step cascades without costly purification steps, often in a single reactor 4. Recent industrial examples include cascades for the production of the investigational HIV treatment drug islatravir, as well as a directed evolution campaign towards the synthesis of the Phase II clinical trial drug LSD1 inhibitor GSK2879552 5,6.
In both organic chemistry and synthetic biology, computer-aided synthesis planning (CASP) tools are increasingly used to plan synthesis routes. Tools such as RetroPath have been deployed to develop new metabolic routes to molecules of interest in synthetic biology 7, whilst in chemistry, tools such as Chematica or ASKCOS have been shown to make useful suggestions for synthetic routes to a number of target molecules 8–10. Despite successes in both of these fields, which biocatalysis spans, computer aided synthesis planning of biocatalytic cascades remains underdeveloped. Enzymatic steps are not well represented in chemical CASP tools, if they appear at all. In contrast, biological CASP tools predominantly feature biosynthetic enzymes, yet the objective of reaching a metabolic starting point, and the use of reaction rules describing transformations in metabolism, does not align well the use of enzymes for organic synthesis (Figure 1A). Indeed, enzymes in the toolbox for biocatalysis generally have a proven track-record for showing promiscuous substrate specificity, which may not be the case for all enzymes in metabolism.
Figure 1. An overview of the requirements for a biocatalysis CASP tool.
A. A CASP tool for biocatalysis requires elements of both Synthetic Biology and Chemistry. B. In the process of generating a pathway, manual and automated processes can be used synergistically for maximum benefit. CAR: Carboxylic acid reductase, ATP: Adenosine triphosphate, NADPH: Nicotinamide adenine dinucleotide phosphate.
Here we present RetroBioCat (available at retrobiocat.com), a tool for computer-aided synthesis planning of biocatalytic cascades, which builds upon CASP elements developed in both organic chemistry and synthetic biology (Figure 1A). We began the development of RetroBioCat by considering the process a scientist undergoes when planning a new biocatalytic pathway, which typically follows three stages (Figure 1B). In the first step, pathways are generated by biocatalytic retrosynthesis. Importantly, this should incorporate some scope for what enzymes could do with the application of enzyme engineering. Secondly, specific enzymes are identified for each step, before finally, potential pathways are evaluated based on factors such as the availability of starting materials or the number of steps required. In this work we describe tools which attempt to automate parts of this workflow, seeking to augment the abilities of chemists wishing to exploit the power of biocatalysis.
Results
Reaction rules for biocatalysis
For the automated generation of pathways to a target molecule, CASP tools typically rely on the use of reaction rules or templates to describe potential retrosynthetic steps, which are iteratively applied until a suitable stopping point is reached (Figure 1B, Step 1) 11,12. Rules can be manually entered 9,13,14, or automatically extracted from a database of known reactions 15,16. Critical to the success of this approach is the level of generalisation applied to the reaction rules. Rules which are too specific limit the potential to predict new routes, whilst rules which are too general can lead to unrealistic suggestions 17,18.
Rules to describe biosynthetic reactions have previously been automatically extracted from databases of metabolic reactions, with the option to specify the level of generalisation through the selection of a diameter from the reaction centre 16. Integrating these rules into RetroBioCat, we found that whilst this method has clearly been successful in generating new biosynthetic routes 19, literature examples of biocatalytic cascades were either not represented, or required the most extreme promiscuity setting in order to be captured. Crucially, treating all enzymes in metabolism as very promiscuous is unrealistic, yielding a large number of unhelpful results. Additionally, such algorithmically extracted rules typically lack the nomenclature associated with enzymes for biocatalysis. Algorithmic extraction of reaction rules from a thorough database of synthetic biotransformations relevant to biocatalysis may be an option in the future, pending the development of such a database.
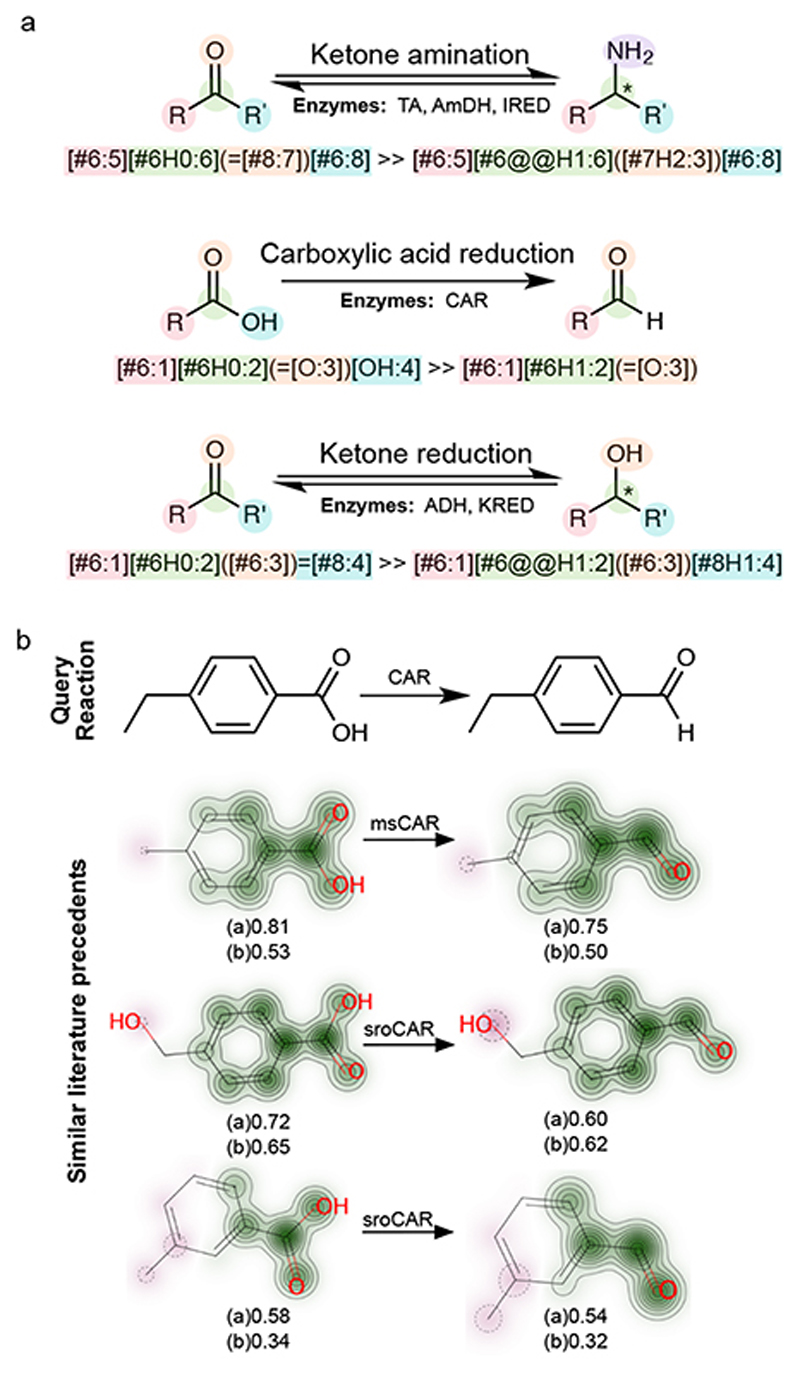
As an alternative, we developed a smaller set of expertly encoded reaction rules to describe the enzyme toolbox for biocatalysis (Figure 2A) 20–22. These rules were made relatively general in most cases to reflect established substrate promiscuity, and to highlight the scope for enzyme engineering. Importantly, the enzymes that these rules represent have been shown to be amenable to enzyme engineering in many cases, for the acceptance of highly synthetic substrates 2,23–26. In other instances, stricter limits on substrate specificity are known, and here greater context was incorporated into the reaction SMARTS. In addition, the rules include some necessary spontaneous chemical steps. An indication of whether a suggested transformation is immediately applicable, or may require enzyme engineering, is provided by the automatic identification of literature precedents by RetroBioCat (Figure 2B).
Figure 2. Critical components of RetroBioCat.
A. A selection of exemplar reaction rules for industrially relevant enzymes, written as reaction SMARTS. B. An example query for similar reactions present in a database of literature precedents. For each molecule, visualisation of the atomic contributions to the Morgan fingerprint similarity is shown. Below each molecule, the Tanimoto similarity 27 is calculated for (a) RDKit fingerprints and (b) Morgan fingerprints, each using the default settings in RDKit. The best CAR enzyme identified for each reaction is shown. TA: Transaminase, AmDH: Amine dehydrogenase, IRED: Imine reductase, CAR: Carboxylic acid reductase, ADH: Alcohol dehydrogenase, KRED: Keto reductase.
The current rule set consists of 99 reactions, described using 135 reaction SMARTS. RetroBioCat also includes the facility to accept submissions for new rules by members of the biocatalysis community, offering the potential for the community-driven development of reaction rules as the enzyme toolbox expands. Furthermore, reaction suggestions which users feel are unrealistic can be flagged for review.
Molecular similarity for identification of reaction precedents
A system for identifying specific enzyme sequences to carry out each step is also necessary for automated cascade design. Manually, scientists typically rely on extensive literature searches or enzyme screening panels to compete this step (Figure 1B, Step 2). To automate this process, a database of literature precedents for synthetic biotransformations is required. However, whilst there are many well-established enzyme databases, these tend to focus on biosynthetic reactions rather than examples of synthetic biotransformations utilised in biocatalysis. Therefore, to demonstrate this step we have begun the manual curation of a database of synthetic biotransformations, which we aim to expand upon in future work. We created a module within RetroBioCat to score reactions based on their similarity to recorded reactions 27 through the use of fingerprint similarity (Figure 2B), as has been demonstrated in both biology 7,28, and chemistry 8. Where many enzymes have been shown to catalyse a specific reaction, our approach selects the best as ranked by activity. Whilst the determinants of substrate specificity may be more complex than can be captured by fingerprint similarity alone, the selection of similar substrates allows a chemist to quickly access the relevant information to make the final decision.
Prioritising reactions by change in molecular complexity
Finally, many chemistry CASP tools feature a metric for molecular complexity to help guide the retrosynthetic search towards a simpler starting material. RetroBioCat uses the recently described SC-Score, which utilises a neural network trained on a large number of synthetic chemistry reactions to score the complexity of each molecule between 1 and 5 29. Applied to biocatalysis, this score appears to function well in guiding pathway suggestions towards synthetically useful routes (Extended Data Figure 1).
Network and pathway explorer
Having established a set of rules describing important reactions in biocatalysis (Figure 2A), a method for searching for similar reaction literature precedents (Figure 2B), and a complexity metric by which to guide retrosynthetic searches (Extended Data Figure 1), we developed two complementary approaches for exploring potential biocatalytic pathways. Firstly, a network exploration mode for human-led CASP, in which the user can explore different routes to a target molecule by expanding a network of biocatalytic disconnections (Figure 3). Alternatively, a pathway exploration mode, in which pathways are automatically generated before being ranked according to a user-defined weighted score (Figure 4). Importantly, both approaches are primarily available through an interactive web-app, but also as an open-source python package for expert users.
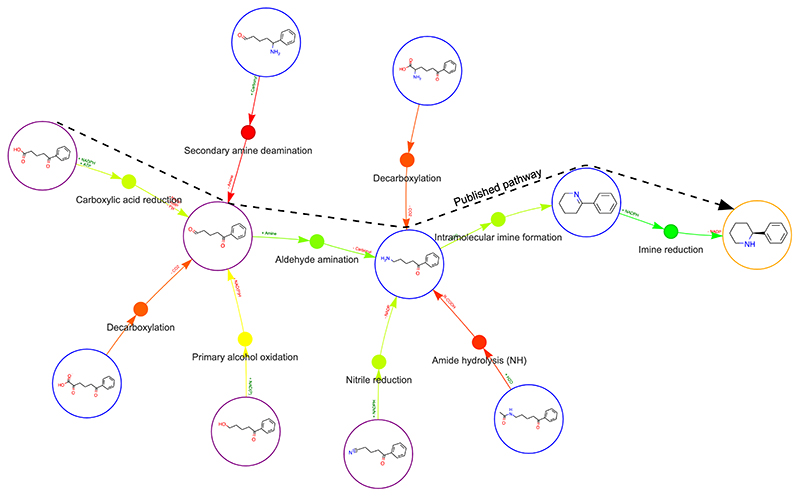
Figure 3. Human-led exploration of a network of potential biotransformations using network explorer.
Each substrate node can be iteratively expanded to reveal further possible biotransformations. Reaction nodes in green indicate high similarity to a literature reported reaction (currently a proof-of-principle dataset). The target molecule is outlined in orange, and buyable compounds outlined in purple. Interactions possible in network explorer are shown. IRED: Imine reductase, AlOx: Alcohol oxidase, ATP: Adenosine triphosphate, AMP: Adenosine monophosphate, PPi: Pyrophosphate, NAD(P): Nicotinamide adenine dinucleotide (phosphate).
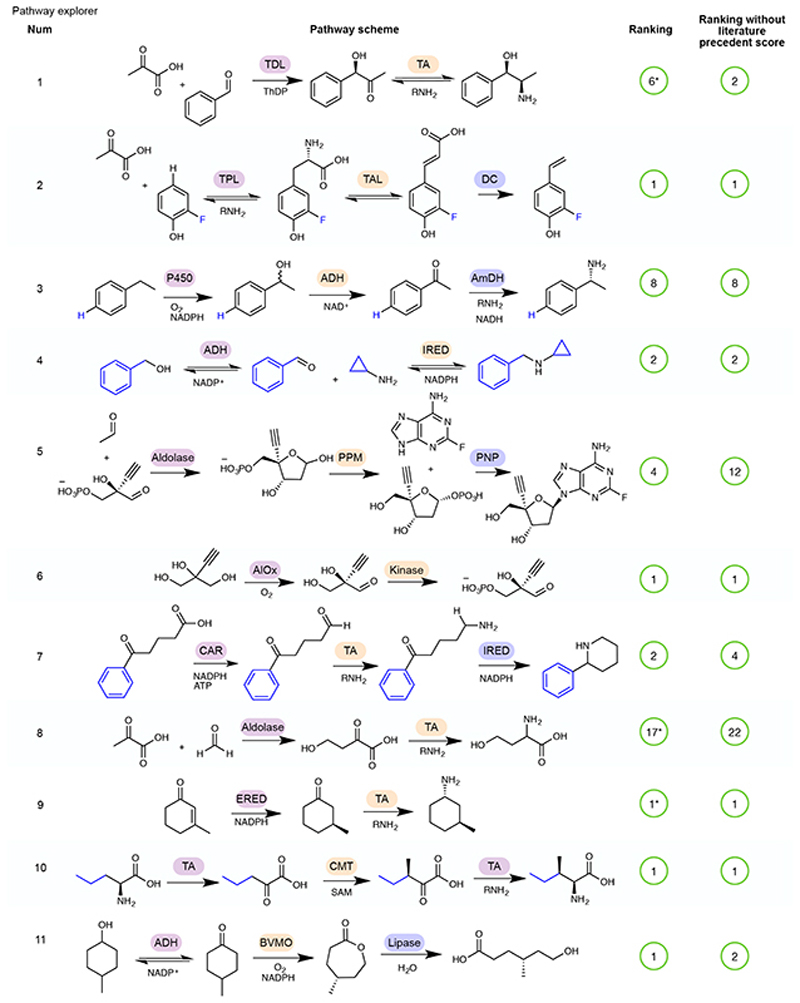
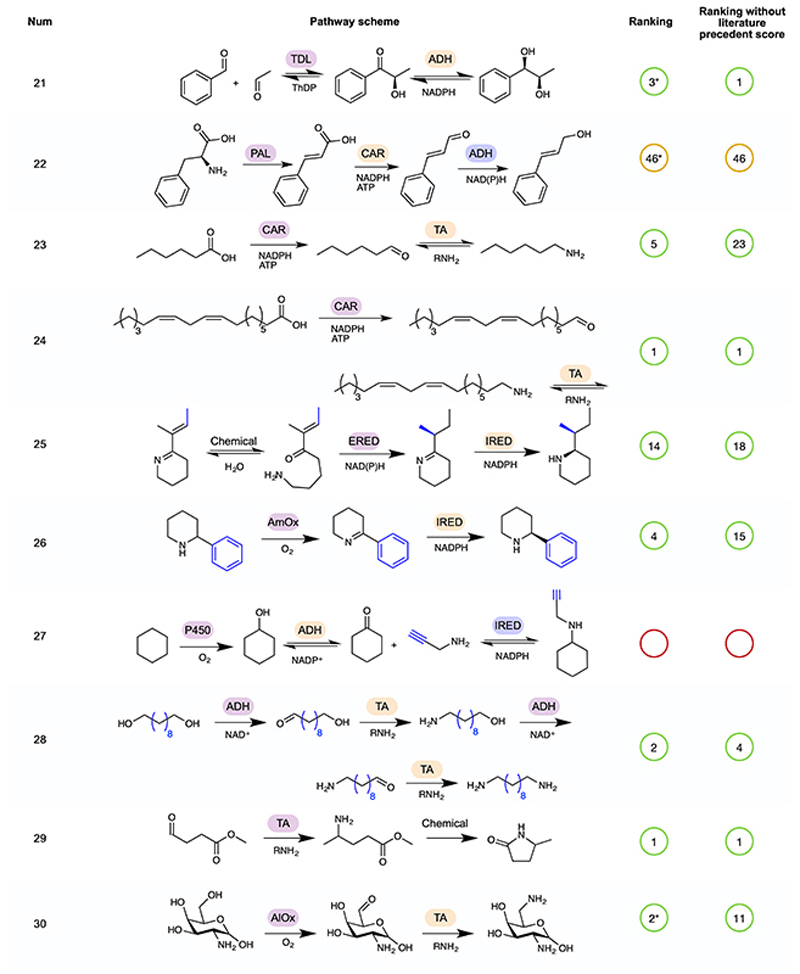
Figure 4. - An example selection of some of the biocatalytic cascades identified in the literature and used as a test-set for Pathway explorer.
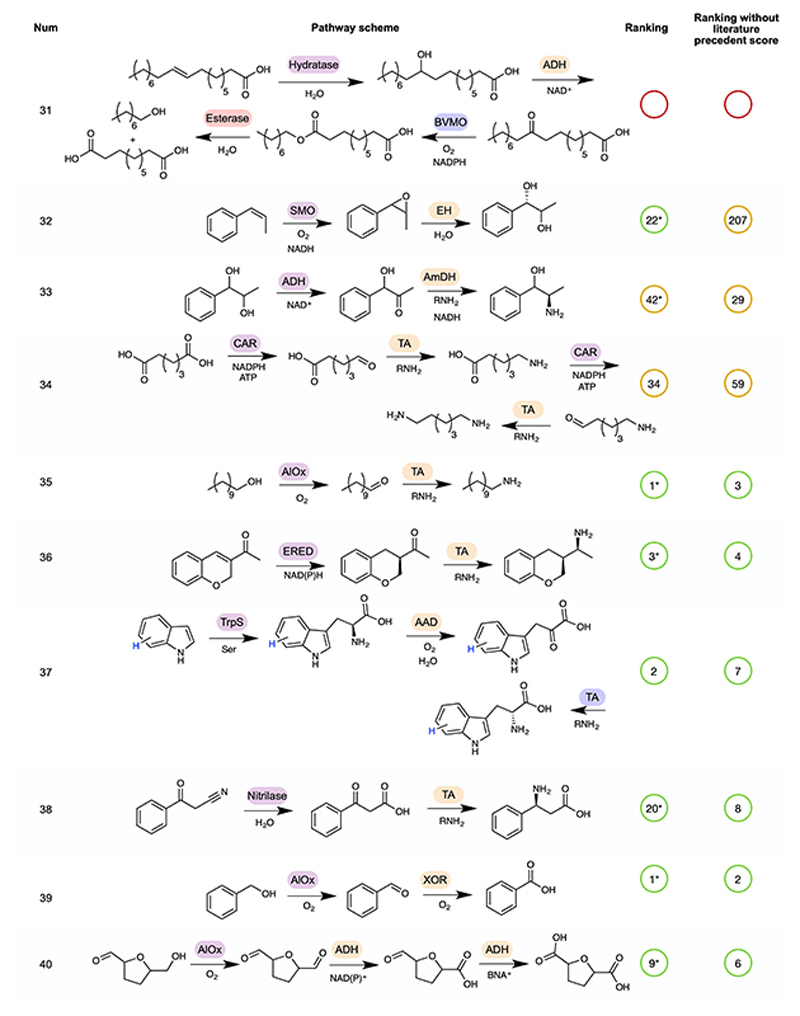
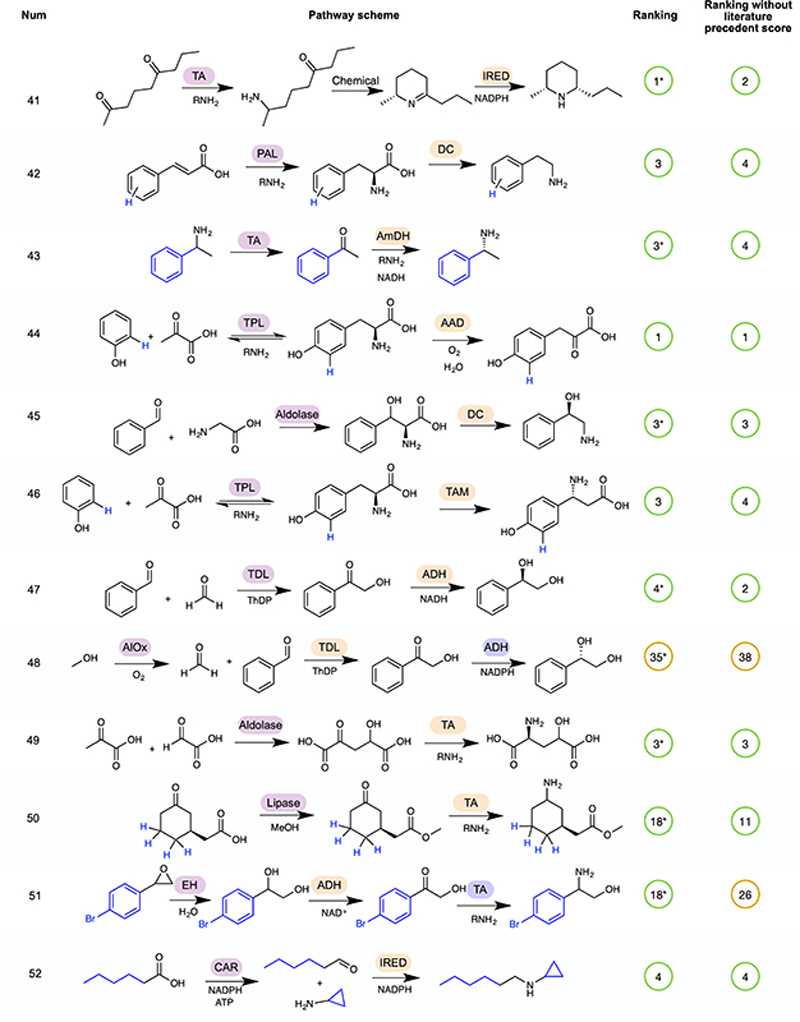
The remainder of the 52 cascades are available in Extended Data Figures 2–5 5,31–72. In some cases, a number of cascades were demonstrated with different R groups, for which we have chosen a single example, highlighted in blue. Pathway rankings by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TDL: thiamine-dependent lyase, TA: transaminase, TPL: tyrosine phenol lyase, TAL: tyrosine ammonia lyase, DC: decarboxylase, P450: cytochrome P450, ADH: alcohol dehydrogenase, AmDH: amine dehydrogenase, IRED: imine reductase, PPM: phosphopentomutase, PNP: purine nucleoside phosphorylase, AlOx: alcohol oxidase, BVMO: Baeyer-Villiger monooxygenase, CAR: carboxylic acid reductase, ERED: ene reductase, CMT: C-methyltransferase, AAD: amino acid deaminase, AADH: amino acid dehydrogenase, PAL: phenylalanine ammonia lyase, AmOx: amine oxidase, TrpS: tryptophan synthase, ThDP: thiamine diphosphate, ATP: Adenosine triphosphate, NADP: Nicotinamide adenine dinucleotide phosphate.
In particular, the network exploration mode can be useful for scientists who may not be familiar with biocatalysis, allowing them to visualise potential biocatalytic disconnections to their target molecule. Integrated is the enzyme identification module, which colours reaction nodes green where a similar literature precedent is identified, or red where only negative data has been reported. Further data on substrate specificity, buy-ability or molecular complexity is also available through the interactive graph. For example, hovering over a green node displays further data on the activity and literature source for that enzyme. For more suggestions and detail on a particular reaction, clicking and holding a reaction node launches a pop-up window with further information (Figure 3). In addition, custom reactions may be added to the graph, allowing custom chemical steps to be included by the user. Alternatively, the reaction rules which are applied can be switched over to suggestions which make use of the recently described chemistry CASP tool AIZynthfinder 30, for the creation of powerful chemo-enzymatic cascades.
Alternatively, the pathway exploration mode seeks to automatically generate useful suggestions for possible pathways to a target molecule. To do this, a network is first generated by applying reaction rules iteratively up to a user-defined maximum length. Networks which reach a user-defined limit to the number of nodes are reduced in size by removing the outer-most worst reactions, as scored by the change in molecular complexity. Pathways are then generated by a best first search approach, which prioritises steps with higher changes in molecular complexity until all possible pathways have been generated, or a limit to number of pathways is reached. A weighted score is used to evaluate and rank each pathway, taking into account the change in molecular complexity, the number of steps, whether the starting material appears in a catalogue of buyable building blocks, and the number of steps with a similar literature precedent.
Most chemistry CASP tools utilise a stopping criterion (other than maximum pathway length), such as the commercial availability of starting materials 8–10. Whilst starting material availability is clearly of relevance to the design of biocatalytic cascades, often experimenters are also interested in demonstrating enzymatic cascades with commercially available intermediates, inhibiting the use of starting material availability as a stopping criterion for RetroBioCat. Instead, RetroBioCat generates pathways of all lengths from a network, relying on the weighted score to determine which pathways are the most promising. Changing the weighted score might result in longer or shorter pathways being suggested more highly. For example, shorter pathways to buyable starting materials can be favoured by increasing both the weight for the number of steps, and the weight for whether a buyable starting material is available. However, in general, the default weights are a good starting point.
Evaluation using a test-set of 52 literature cascades
To test both network and pathway explorer, we carried out a thorough review of the biocatalytic cascades reported in the literature, generating a test-set of 52 pathways (Figure 4, Extended Data Figures 2–5) 5,31–72. Except for C-H oxidation by P450 enzymes, all of the reactions in the test-set were correctly predicted by RetroBioCat. Importantly, the majority of pathways were suggested within the top few suggestions using pathway explorer with only the default settings for the weighted score, validating this as a useful approach for the automated design of biocatalytic cascades.
Discussion
CASP tools should strive to augment the abilities of scientists seeking to design new routes to a target molecule. An intuitive and easy to use user-interface, as we have developed for RetroBioCat, is therefore crucial. Furthermore, the manually curated reaction rules utilised by RetroBioCat strike a balance between being general enough, so that the potential for enzyme engineering or discovery is captured, whilst providing context where necessary so as to be realistic.
Suggestions for potential biocatalytic transformations even without literature precedent are themselves a valuable resource, as in many cases enzyme screening panels can be employed to find the right enzyme for a specific reaction. However, where there is literature precedent for a reaction, suggestions are more robust and easier to implement if these are automatically identified. Here, we have demonstrated the use of molecular similarity to automate this process and have begun the construction of a database of synthetic biotransformations described in the literature, with further contributions to be reported in future work.
Pathway explorer offers automated ranking of suggested pathways using a selection of metrics. We have shown that this functions well in suggesting previously reported pathways early in the ranking system. Future improvements could seek to provide further information on the suitability of each suggested pathway. For example, thermodynamics, cofactor usage, substrate and product solubility or stability 74, toxicity, reaction conditions, starting material price and predicted pathway kinetics 75, could all offer more insight into which pathway is the most promising for experimental characterisation. Substantial advances are being made in the pathway searching algorithms utilised in organic chemistry. As we seek to incorporate organic chemistry or biosynthetic steps into RetroBioCat, or simply as we expand the reactions rules for biocatalysis, it may become necessary to exploit more advanced algorithms for pathway generation, such as the Monte Carlo tree search (MCTS) 7,10,76,77.
Several challenges still remain for the refinement of RetroBioCat. For example, at present enzymatic C-H activations such as hydroxylations and halogenations are currently not fully included, as the context in these reactions rules requires more careful consideration. Additionally, larger, more complex target molecules are sometimes handled inadequately by RetroBioCat, possibly highlighting the need for increased research into bond forming enzymes in the biocatalysis field as a whole. Indeed, with exceptions, most biocatalytic transformations are performed on small molecules of typically less than 500 Da. To help mitigate this issue, RetroBioCat features an option to fragment a molecule along synthetically accessible bonds prior to the generation of pathways or networks. Additionally, chemical steps can be suggested in network explorer 30. Future work to include chemistry steps in pathway explorer will allow better automated suggestions to be made where some chemical steps are necessary, although care must be taken that enzymatic steps are well represented amongst the more numerous chemical options. Additionally, incorporating the reaction rules developed for metabolic engineering could further blur the lines between biocatalysis and biosynthesis, and open up access to a broad pool of renewable resources for use as substrates. Crucially, many recent CASP tools are written in python using open-source libraries such as RDKit 7,8,11,30. Furthermore, the use of reaction SMARTS to describe reaction templates is relatively common across many tools, which should facilitate the combination of approaches from different fields into a single solution in the future.
In summary, RetroBioCat offers an accessible set of tools for computer-aided design of biocatalytic cascades. These tools should be useful in highlighting the potential of enzymes for organic synthesis, and for the design of de novo biocatalytic pathways.
Methods
Overview
Both network explorer and pathway explorer utilise the creation of a bipartite directional graph using the NetworkX Python package, to hold all the possible transformations to the target molecule. The first node in the network is the target molecule as a SMILES string. On applying the reaction rules, reactions are added as nodes with edges between the new reaction nodes and the molecule the rules were applied to. The products of the reactions are then also added as nodes, in the form of SMILES strings, with edges between these new molecules and the reaction node that produced them. Applying the reaction rules iteratively creates a network of potential routes leading back to the target molecule. A maximum number of nodes can be set to limit combinatorial explosion, above which the outer-most reactions are deleted according to which has the worst change in molecule complexity. Nodes in the network are scored as described below, with the results held in a dictionary for each node. The web interface offers a network exploration mode, in which double clicking on a molecule applies reaction rules to that molecule to expand the network in this location. Alternatively, pathway explorer automatically generates pathways by automatically expanding a network out to a specified number of steps, before generating the possible pathways present in the network and ranking them, as described below.
Chemistry
The RDKit chemoinformatics library is used to implement all chemistry-related methods, such as reaction transformations or calculating molecular similarity. Molecules are stored in pathways or networks as SMILES strings, as generated by RDKit using the default settings. For processing, SMILES are first translated into a mol object as defined in RDKit.
Reaction rules are defined using reaction SMARTS. Reaction SMARTS are developed manually, often making use of the capability of Marvin JS (https://chemaxon.com/products/marvin-js) to draw the proposed reaction and extracting it as a reaction SMARTS. Positive and negative tests, in the form of SMILES strings which should or should not be transformed by the rules, are defined to ensure reaction SMARTS act as planned. Reaction rules are applied using a modified version of rdChiral 78 (https://github.com/connorcoley/rdchiral).
Node scoring
In both network and pathway explorer, molecule and reaction nodes are scored to allow scoring of individual steps or entire pathways, as detailed below.
Whether a particular molecule is available as a buyable building block is identified by querying a database of buyable SMILES strings. We used a combination of the ‘in-stock building blocks’ list in the ZINC database, the building blocks listed by emolecules, and the building blocks available from molport to construct this database. All SMILES strings were pre-processed to be in the form generated by RDKit using the default settings. If a SMILES string is in the database, the is_buyable attribute is marked as 1.
To calculate molecular complexity, we use the SC-Score 29. Code for this module is taken from https://github.com/connorcoley/scscore/. We use the standalone numpy version of the SC-Scorer, utilising Boolean fingerprints with a length of 1024. Every molecule is scored using the SC-Scorer, with the result saved in an attribute on the node as ‘complexity’. For every molecule, a ‘relative complexity’ is also calculated, by taking the difference between the complexity of the current molecule and the complexity of the target molecule. The difference in complexity between a reaction substrate and product is used to calculate ‘change in complexity’ for every reaction node. Where a reaction has multiple substrates, the substrate with the highest complexity is used.
A module for comparing molecule similarity is available within RetroBioCat for comparing suggested reactions against a database of literature precedent. To do this, fingerprints are constructed for every molecule in the database, and for the molecules in the query reaction. RDKit fingerprints are used with the default settings as implemented in RDKit. Fingerprints are compared by calculating Tanimoto similarity, again implemented using RDKit. Molecules with similarity below a cut-off value are discarded. Similarity is scored using either only the similarity of the products, or the average of similarities for both the products and the substrates. The highest scoring reaction is used as a suggestion in network or pathway explorer, with the enzyme with the highest activity chosen. Optionally, negative data can be included in this search. A number of alternative fingerprints are available within RDKit, such as Morgan fingerprints, Avalon Fingerprints, or Atom-Pair and Topological-Torsion Fingerprints. Each of these fingerprints extracts features of a molecule in a different way, and could be used to calculate similarity. In our hands, the RDKit fingerprint functions well in identifying similar molecules in our dataset.
Pathway generation
To automatically generate pathways to a target molecule, a network is first generated by iteratively applying the reaction rules up a specified number of steps. Pathways are generated by applying a best first search on the network, using molecular complexity as the selection criteria, adjusted to only include positive values. At each step, the option to stop the search is also available. Once all the possible pathways in the network have been generated, or the maximum number of pathways reached, pathways are scored and ranked. Pathways are scored on their total change in complexity, the number of enzymatic steps in the pathway, the percentage of starting material which is marked as ‘buyable’, and the number of steps which have been identified with similar literature precedent. Each score is normalised to between 0 and 1, to which a user-specified weight can be applied. The pathways are ranked in order of the total of the weighted scores, presenting the user with the highest scoring pathways first. In addition, a diversity score is applied which penalises reactions which have appeared in the prior suggestions.
Pathway explorer test set evaluation
The test set of 52 pathways consists of the target molecules, the starting materials used and the enzymes in the published pathway. For each test, pathways are automatically generated and ranked. Each generated pathway is then compared with the published version. Pathways which contain the starting molecules and include the recorded enzymes for the published pathway are marked as a match, with the ranking of the pathway recorded.
Extended Data
Extended Data Fig. 1. An example generated using Network Explorer to illustrate changes in molecular complexity.
Arrows and reactions are coloured by the change in molecular complexity, determined using the SC-Score. Green indicates a negative change in molecule complexity, which in most cases corresponds to a synthetically useful transformation. Red indicates a positive change in molecular complexity. Colours are determined relative to the other transformations leading to a specific molecule. Some reactions have been removed for clarity. Pathway published in reference 68.
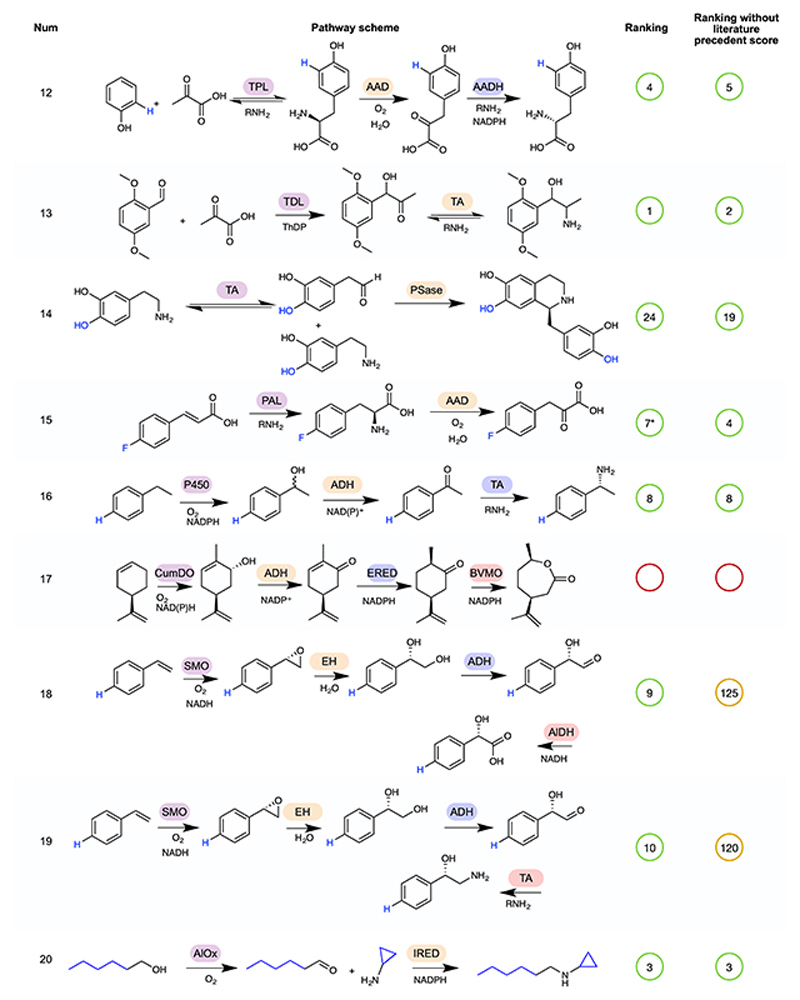
Extended Data Fig. 2. Rankings for pathways 12 to 20 of the test-set for Pathway explorer.
A continuation of Figure 4, showing rankings for pathways 12 to 20 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TPL: tyrosine phenol lyase, AAD: amino acid deaminase, AADH: amino acid dehydrogenase, TDL: thiamine-dependent lyase, TA: transaminase, PSase: Pictet-Spenglerase, PAL: phenylalanine ammonia lyase, P450: cytochrome P450, ADH: alcohol dehydrogenase, CumDO: cumene dioxygenase, ERED: ene reductase, BVMO: Baeyer-Villiger monooxygenase, SMO: styrene monooxygenase, AlDH: aldehyde dehydrogenase, AlOx: alcohol oxidase.
Extended Data Fig. 3. Rankings for pathways 21 to 30 of the test-set for Pathway explorer.
A continuation of Figure 4, showing rankings for pathways 21 to 30 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TDL: thiamine-dependent lyase, ADH: alcohol dehydrogenase, PAL: phenylalanine ammonia lyase, CAR: carboxylic acid reductase, ERED: ene reductase, IRED: imine reductase, AmOx: amine oxidase, P450: cytochrome P450, ATP: Adenosine triphosphate, NADP: Nicotinamide adenine dinucleotide phosphate.
Extended Data Fig. 4. Rankings for pathways 31 to 40 of the test-set for Pathway explorer.
A continuation of Figure 4, showing rankings for pathways 31 to 40 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. ADH: alcohol dehydrogenase, BVMO: Baeyer-Villiger monooxygenase, SMO: styrene monooxygenase, EH: epoxide hydrolase, AmDH: amine dehydrogenase, CAR: carboxylic acid reductase, TA: transaminase, AlOx: alcohol oxidase, TA: transaminase, ERED: ene reductase, TrpS: tryptophan synthase, XOR: xanthine oxidoreductase, AAD: amino acid deaminase, ATP: Adenosine triphosphate, NADP: Nicotinamide adenine dinucleotide phosphate.
Extended Data Fig. 5. Rankings for pathways 41 to 52 of the test-set for Pathway explorer.
A continuation of Figure 4, showing rankings for pathways 41 to 52 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TA: transaminase, IRED: imine reductase, PAL: phenylalanine ammonia lyase, DC: decarboxylase, AmDH: amine dehydrogenase, TPL: tyrosine phenol lyase, AAD: amino acid deaminase, TAM: tyrosine aminomutase, TDL: thiamine-dependent lyase, ADH: alcohol dehydrogenase, AlOx: alcohol oxidase, EH: epoxide hydrolase, CAR: carboxylic acid reductase, ATP: Adenosine triphosphate, NADP: Nicotinamide adenine dinucleotide phosphate.
Supplementary Material
Acknowledgements
We kindly acknowledge financial support from the European Research Council (788231-ProgrES-ERC-2017-ADG to SLF; BIO-HBORROW: Grant No. 742987 to NJT). The authors also thank all the beta-testers of RetroBioCat, particularly Sebastian Cosgrove and Robert Speight.
Footnotes
Author Contributions
WF, LJH, SLF and NJT planned the work. Code for RetroBioCat written by WF. Pathway test-set generated by LJH. Initial draft of the manuscript written by WF, with subsequent contributions from LJH, SLF and NJT. All authors have given approval to the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
Other than literature precedent data which can currently only be accessed at https://retrobiocat.com pending future publications, the database files for RetroBiocat at the time of publication are available at 10.6084/m9.figshare.12696482.v4. The 52 pathway test-set is available along with the source code at 10.6084/m9.figshare.12698072.v7 or https://github.com/willfinnigan/retrobiocat
Code availability
RetroBioCat is freely available as a web-app at https://retrobiocat.com. We have also made the source code freely available under the MIT license, available at https://github.com/willfinnigan/retrobiocat, or specifically for the version described here, at 10.6084/m9.figshare.12698072.v7.
References
- 1.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 2.Sheldon RA, Brady D. The limits to biocatalysis: Pushing the envelope. Chem Commun. 2018;54:6088–6104. doi: 10.1039/c8cc02463d. [DOI] [PubMed] [Google Scholar]
- 3.Hönig M, Sondermann P, Turner NJ, Carreira EM. Enantioselective Chemo-and Biocatalysis: Partners in Retrosynthesis. Angew Chem Int Ed. 2017;56:8942–8973. doi: 10.1002/anie.201612462. [DOI] [PubMed] [Google Scholar]
- 4.France SP, Hepworth LJ, Turner NJ, Flitsch SL. Constructing Biocatalytic Cascades: In Vitro and In Vivo Approaches to De Novo Multi-Enzyme Pathways. ACS Catal. 2017;7:710–724. [Google Scholar]
- 5.Huffman MA, et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science. 2019;366:1255–1259. doi: 10.1126/science.aay8484. [DOI] [PubMed] [Google Scholar]
- 6.Schober M, et al. Chiral synthesis of LSD1 inhibitor GSK2879552 enabled by directed evolution of an imine reductase. Nat Catal. 2019;2:909–915. [Google Scholar]
- 7.Koch M, Duigou T, Faulon JL. Reinforcement learning for bioretrosynthesis. ACS Synth Biol. 2020;9:157–168. doi: 10.1021/acssynbio.9b00447. [DOI] [PubMed] [Google Scholar]
- 8.Coley CW, Rogers L, Green WH, Jensen KF. Computer-assisted retrosynthesis based on molecular similarity. ACS Cent Sci. 2017;3:1237–1245. doi: 10.1021/acscentsci.7b00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szymkuć S, et al. Computer-Assisted Synthetic Planning: The End of the Beginning. Angew Chem Int Ed. 2016:55. doi: 10.1002/anie.201506101. [DOI] [PubMed] [Google Scholar]
- 10.Segler MHS, Preuss M, Waller MP. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 11.Landrum G. RDKit: Open-cource cheminformatics software. 2016 [Google Scholar]
- 12.Coley CW, Green WH, Jensen KF. Machine learning in computer-aided synthesis planning. Acc Chem Res. 2018;51:1281–1289. doi: 10.1021/acs.accounts.8b00087. [DOI] [PubMed] [Google Scholar]
- 13.Grzybowski BA, et al. Chematica: A story of computer code that started to think like a chemist. Chem. 2018;4:390–398. [Google Scholar]
- 14.Hartenfeller M, et al. A collection of robust organic synthesis reactions for in silico molecule design. J Chem Inf Model. 2011;51:3093–3098. doi: 10.1021/ci200379p. [DOI] [PubMed] [Google Scholar]
- 15.Plehiers PP, Marin GB, Stevens CV, Van Geem KM. Automated reaction database and reaction network analysis: extraction of reaction templates using cheminformatics. J Cheminform. 2018;10:1–18. doi: 10.1186/s13321-018-0269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duigou T, Du Lac M, Carbonell P, Faulon JL. Retrorules: A database of reaction rules for engineering biology. Nucleic Acids Res. 2019;47:D1229–D1235. doi: 10.1093/nar/gky940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molga K, Gajewska EP, Szymkuć S, Grzybowski BA. The logic of translating chemical knowledge into machine-processable forms: A modern playground for physical-organic chemistry. React Chem Eng. 2019;4:1506–1521. [Google Scholar]
- 18.Segler MHS, Waller MP. Neural-symbolic machine learning for retrosynthesis and reaction prediction. Chem - A Eur J. 2017;23:5966–5971. doi: 10.1002/chem.201605499. [DOI] [PubMed] [Google Scholar]
- 19.Fehér T, et al. Validation of RetroPath, a computer-aided design tool for metabolic pathway engineering. Biotechnol J. 2014;9:1446–1457. doi: 10.1002/biot.201400055. [DOI] [PubMed] [Google Scholar]
- 20.Turner NJ, Humphreys L. Biocatalysis in organic synthesis: The retrosynthesis approach. Royal Society of Chemistry; 2018. [Google Scholar]
- 21.Turner NJ, O’Reilly E. Biocatalytic retrosynthesis. Nat Chem Biol. 2013;9:285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- 22.de Souza ROM, Miranda LSMA, Bornscheuer UT. A Retrosynthesis Approach for Biocatalysis in Organic Synthesis. Chem - A Eur J. 2017;23:12040–12063. doi: 10.1002/chem.201702235. [DOI] [PubMed] [Google Scholar]
- 23.Heath RS, et al. An engineered alcohol oxidase for the oxidation of primary alcohols. ChemBioChem. 2019;20:276–281. doi: 10.1002/cbic.201800556. [DOI] [PubMed] [Google Scholar]
- 24.Batista VF, Galman JL, Pinto DC, Silva AMS, Turner NJ. Monoamine oxidase: Tunable activity for amine resolution and functionalization. ACS Catal. 2018;8:11889–11907. [Google Scholar]
- 25.Devine PN, et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat Rev Chem. 2018;2:409–421. [Google Scholar]
- 26.Arnold FH. Directed Evolution: Bringing New Chemistry to Life. Angew Chem Int Ed. 2018;57:4143–4148. doi: 10.1002/anie.201708408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rácz A, Bajusz D, Héberger K. Life beyond the Tanimoto coefficient: Similarity measures for interaction fingerprints. J Cheminform. 2018;10:1–12. doi: 10.1186/s13321-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breitling R, et al. Selenzyme: enzyme selection tool for pathway design. Bioinformatics. 2018;34:2153–2154. doi: 10.1093/bioinformatics/bty065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coley CW, Rogers L, Green WH, Jensen KF. SCScore: Synthetic Complexity Learned from a Reaction Corpus. J Chem Inf Model. 2018;58:252–261. doi: 10.1021/acs.jcim.7b00622. [DOI] [PubMed] [Google Scholar]
- 30.Genheden S, et al. AiZynthFinder: A Fast Robust and Flexible Open-Source Software for Retrosynthetic Planning. 2020:1–14. doi: 10.26434/chemrxiv.12465371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sehl T, et al. Two steps in one pot: Enzyme cascade for the synthesis of nor(pseudo)ephedrine from inexpensive starting materials. Angew Chem Int Ed. 2013;52:6772–6775. doi: 10.1002/anie.201300718. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, et al. Efficient production of phenylpropionic acids by an amino-group-transformation biocatalytic cascade. Biotechnol Bioeng. 2020;117:614–625. doi: 10.1002/bit.27241. [DOI] [PubMed] [Google Scholar]
- 33.Erdmann V, et al. Methoxamine Synthesis in a Biocatalytic 1-Pot 2-Step Cascade Approach. ACS Catal. 2019;9:7380–7388. [Google Scholar]
- 34.Lichman BR, et al. One-pot triangular chemoenzymatic cascades for the syntheses of chiral alkaloids from dopamine. Green Chem. 2015;17:852–855. [Google Scholar]
- 35.Parmeggiani F, Lovelock SL, Weise NJ, Ahmed ST, Turner NJ. Synthesis of D-and L-Phenylalanine Derivatives by Phenylalanine Ammonia Lyases: A Multienzymatic Cascade Process. Angew Chem Int Ed. 2015;54:4608–4611. doi: 10.1002/anie.201410670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Both P, et al. Whole-Cell Biocatalysts for Stereoselective C-H Amination Reactions. Angew Chem Int Ed. 2016;55:1511–1513. doi: 10.1002/anie.201510028. [DOI] [PubMed] [Google Scholar]
- 37.Oberleitner N, et al. From waste to value - Direct utilization of limonene from orange peel in a biocatalytic cascade reaction towards chiral carvolactone. Green Chem. 2017;19:367–371. [Google Scholar]
- 38.Wu S, et al. Highly regio-and enantioselective multiple oxy-and amino-functionalizations of alkenes by modular cascade biocatalysis. Nat Commun. 2016;7:11917. doi: 10.1038/ncomms11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsden JI, et al. Biocatalytic N-Alkylation of Amines Using Either Primary Alcohols or Carboxylic Acids via Reductive Aminase Cascades. J Am Chem Soc. 2019;141:1201–1206. doi: 10.1021/jacs.8b11561. [DOI] [PubMed] [Google Scholar]
- 40.Jakoblinnert A, Rother D. A two-step biocatalytic cascade in micro-aqueous medium: Using whole cells to obtain high concentrations of a vicinal diol. Green Chem. 2014;16:3472–3482. [Google Scholar]
- 41.Klumbys E, Zebec Z, Weise NJ, Turner NJ, Scrutton NS. Bio-derived production of cinnamyl alcohol via a three step biocatalytic cascade and metabolic engineering. Green Chem. 2018;20:658–663. doi: 10.1039/C7GC03325G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busto E, Simon RC, Kroutil W. Vinylation of Unprotected Phenols Using a Biocatalytic System. Angew Chem Int Ed. 2015;54:10899–10902. doi: 10.1002/anie.201505696. [DOI] [PubMed] [Google Scholar]
- 43.Citoler J, Derrington SR, Galman JL, Bevinakatti H, Turner NJ. A biocatalytic cascade for the conversion of fatty acids to fatty amines. Green Chem. 2019;21:4932–4935. [Google Scholar]
- 44.Thorpe TW, et al. One-Pot Biocatalytic Cascade Reduction of Cyclic Enimines for the Preparation of Diastereomerically Enriched N-Heterocycles. J Am Chem Soc. 2019;141:19208–19213. doi: 10.1021/jacs.9b10053. [DOI] [PubMed] [Google Scholar]
- 45.Heath RS, Pontini M, Hussain S, Turner NJ. Combined Imine Reductase and Amine Oxidase Catalyzed Deracemization of Nitrogen Heterocycles. ChemCatChem. 2016;8:117–120. [Google Scholar]
- 46.Tavanti M, Mangas-Sanchez J, Montgomery SL, Thompson MP, Turner NJ. A biocatalytic cascade for the amination of unfunctionalised cycloalkanes. Org Biomol Chem. 2017;15:9790–9793. doi: 10.1039/c7ob02569f. [DOI] [PubMed] [Google Scholar]
- 47.Sattler JH, et al. Redox Self-Sufficient Biocatalyst Network for the Amination of Primary Alcohols. Angew Chem Int Ed. 2012;51:9156–9159. doi: 10.1002/anie.201204683. [DOI] [PubMed] [Google Scholar]
- 48.Mourelle-Insua Á, Zampieri LA, Lavandera I, Gotor-Fernández V. Conversion of γ-and δ-Keto Esters into Optically Active Lactams. Transaminases in Cascade Processes. Adv Synth Catal. 2018;360:686–695. [Google Scholar]
- 49.Aumala V, et al. Biocatalytic Production of Amino Carbohydrates through Oxidoreductase and Transaminase Cascades. ChemSusChem. 2019;12:848–857. doi: 10.1002/cssc.201802580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song J-W, et al. Multistep Enzymatic Synthesis of Long-Chain α,ω-Dicarboxylic and ω-Hydroxycarboxylic Acids from Renewable Fatty Acids and Plant Oils. Angew Chem Int Ed. 2013;52:2534–2537. doi: 10.1002/anie.201209187. [DOI] [PubMed] [Google Scholar]
- 51.Corrado ML, Knaus T, Mutti FG. Regio-and stereoselective multi-enzymatic aminohydroxylation of β-methylstyrene using dioxygen, ammonia and formate. Green Chem. 2019;21:6246–6251. doi: 10.1039/C9GC03161H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fedorchuk TP, et al. One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases. J Am Chem Soc. 2020;142:1038–1048. doi: 10.1021/jacs.9b11761. [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Zheng Y-C, Chen F-F, Xu J-H, Yu H-L. Enantioselective Bioamination of Aromatic Alkanes Using Ammonia: A Multienzymatic Cascade Approach. ChemCatChem. 2020;12:2077–2082. [Google Scholar]
- 54.Pickl M, Fuchs M, Glueck SM, Faber K. Amination of ω-Functionalized Aliphatic Primary Alcohols by a Biocatalytic Oxidation-Transamination Cascade. ChemCatChem. 2015;7:3121–3124. doi: 10.1002/cctc.201500589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parmeggiani F, et al. One-Pot Biocatalytic Synthesis of Substituted d -Tryptophans from Indoles Enabled by an Engineered Aminotransferase. ACS Catal. 2019;9:3482–3486. [Google Scholar]
- 56.Zhang Z-J, Cai R-F, Xu J-H. Characterization of a new nitrilase from Hoeflea phototrophica DFL-43 for a two-step one-pot synthesis of (S)-β-amino acids. Appl Microbiol Biotechnol. 2018;102:6047–6056. doi: 10.1007/s00253-018-9057-7. [DOI] [PubMed] [Google Scholar]
- 57.Bechi B, et al. Catalytic bio-chemo and bio-bio tandem oxidation reactions for amide and carboxylic acid synthesis. Green Chem. 2014;16:4524–4529. [Google Scholar]
- 58.Jia H-Y, Zong M-H, Zheng G-W, Li N. One-Pot Enzyme Cascade for Controlled Synthesis of Furancarboxylic Acids from 5-Hydroxymethylfurfural by H2O2 Internal Recycling. ChemSusChem. 2019;12:4764–4768. doi: 10.1002/cssc.201902199. [DOI] [PubMed] [Google Scholar]
- 59.Alvarenga N, et al. Asymmetric Synthesis of Dihydropinidine Enabled by Concurrent Multienzyme Catalysis and a Biocatalytic Alternative to Krapcho Dealkoxycarbonylation. ACS Catal. 2020;10:1607–1620. [Google Scholar]
- 60.Weise NJ, et al. Bi-enzymatic Conversion of Cinnamic Acids to 2-Arylethylamines. ChemCatChem. 2020;12:995–998. [Google Scholar]
- 61.Yoon S, et al. Deracemization of Racemic Amines to Enantiopure (R)-and (S)-amines by Biocatalytic Cascade Employing ω-Transaminase and Amine Dehydrogenase. ChemCatChem. 2019;11:1898–1902. [Google Scholar]
- 62.Steinreiber J, et al. Overcoming thermodynamic and kinetic limitations of aldolase-catalyzed reactions by applying multienzymatic dynamic kinetic asymmetric transformations. Angew Chem Int Ed. 2007;46:1624–1626. doi: 10.1002/anie.200604142. [DOI] [PubMed] [Google Scholar]
- 63.Shanmuganathan S, Natalia D, Greiner L, Domínguez de María P. Oxidation-hydroxymethylation-reduction: a one-pot three-step biocatalytic synthesis of optically active α-aryl vicinal diols. Green Chem. 2012;14:94–97. [Google Scholar]
- 64.Montgomery SL, et al. Direct Alkylation of Amines with Primary and Secondary Alcohols through Biocatalytic Hydrogen Borrowing. Angew Chem Int Ed. 2017;129:10627–10630. doi: 10.1002/anie.201705848. [DOI] [PubMed] [Google Scholar]
- 65.Guérard-Hélaine C, et al. Stereoselective synthesis of γ-hydroxy-α-amino acids through aldolase-transaminase recycling cascades. Chem Commun. 2017;53:5465–5468. doi: 10.1039/c7cc00742f. [DOI] [PubMed] [Google Scholar]
- 66.Siirola E, et al. Asymmetric Synthesis of 3-Substituted Cyclohexylamine Derivatives from Prochiral Diketones via Three Biocatalytic Steps. Adv Synth Catal. 2013;355:1703–1708. [Google Scholar]
- 67.Zhang J-D, et al. Asymmetric ring opening of racemic epoxides for enantioselective synthesis of (S)-β-amino alcohols by a cofactor self-sufficient cascade biocatalysis system. Catal Sci Technol. 2019;9:70–74. [Google Scholar]
- 68.France SP, et al. One-Pot Cascade Synthesis of Mono-and Disubstituted Piperidines and Pyrrolidines using Carboxylic Acid Reductase (CAR), ω -Transaminase (ω-TA), and Imine Reductase (IRED) Biocatalysts. ACS Catal. 2016;6:3753–3759. [Google Scholar]
- 69.Hernandez K, et al. Combining Aldolases and Transaminases for the Synthesis of 2-Amino-4-hydroxybutanoic Acid. ACS Catal. 2017;7:1707–1711. [Google Scholar]
- 70.Monti D, et al. Cascade Coupling of Ene-Reductases and ω-Transaminases for the Stereoselective Synthesis of Diastereomerically Enriched Amines. ChemCatChem. 2015;7:3106–3109. [Google Scholar]
- 71.Liao C, Seebeck FP. Asymmetric β-Methylation of L-and D-α-Amino Acids by a Self-Contained Enzyme Cascade. Angew Chem Int Ed. 2020;59:7184–7187. doi: 10.1002/anie.201916025. [DOI] [PubMed] [Google Scholar]
- 72.Schmidt S, et al. Biocatalytic Access to Chiral Polyesters by an Artificial Enzyme Cascade Synthesis. ChemCatChem. 2015;7:3951–3955. [Google Scholar]
- 73.Koszelewski D, Tauber K, Faber K, Kroutil W. ω-Transaminases for the synthesis of non-racemic a-chiral primary amines. Trends Biotechnol. 2010;28:324–332. doi: 10.1016/j.tibtech.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 74.Li X, et al. DeepChemStable: Chemical Stability Prediction with an Attention-Based Graph Convolution Network. J Chem Inf Model. 2019;59:1044–1049. doi: 10.1021/acs.jcim.8b00672. [DOI] [PubMed] [Google Scholar]
- 75.Finnigan W, et al. Engineering a seven enzyme biotransformation using mathematical modelling and characterized enzyme parts. ChemCatChem. 2019;11:3474–3489. doi: 10.1002/cctc.201900646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen B, Li C, Dai H, Song L. Retro*: Learning Retrosynthetic Planning with Neural Guided A* Search. 2020 Preprint at https://arxiv.org/abs/2006.15820.
- 77.Kishimoto A, Buesser B, Chen B, Botea Eaton A. Depth-First Proof-Number Search with Heuristic Edge Cost and Application to Chemical Synthesis Planning. Adv Neural Inf Process Syst. 2019:7226–7236. [Google Scholar]
- 78.Coley CW, Green WH, Jensen KF. RDChiral: An RDKit Wrapper for Handling Stereochemistry in Retrosynthetic Template Extraction and Application. J Chem Inf Model. 2019;59:2529–2537. doi: 10.1021/acs.jcim.9b00286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Other than literature precedent data which can currently only be accessed at https://retrobiocat.com pending future publications, the database files for RetroBiocat at the time of publication are available at 10.6084/m9.figshare.12696482.v4. The 52 pathway test-set is available along with the source code at 10.6084/m9.figshare.12698072.v7 or https://github.com/willfinnigan/retrobiocat
RetroBioCat is freely available as a web-app at https://retrobiocat.com. We have also made the source code freely available under the MIT license, available at https://github.com/willfinnigan/retrobiocat, or specifically for the version described here, at 10.6084/m9.figshare.12698072.v7.