Abstract
Telomeres which are present at the ends of eukaryotic chromosomes mediate genome stability and determine cellular lifespan1. Telomeric repeat containing RNAs (TERRA) are long noncoding RNAs transcribed from chromosome ends2,3, which regulate telomeric chromatin structure and telomere maintenance via telomerase and homology directed repair (HDR)4,5. The mechanisms by which TERRA is recruited to chromosome ends remain poorly defined. Here we develop a reporter system to dissect the underlying mechanisms and demonstrate that the UUAGGG-repeats of TERRA are both necessary and sufficient to target TERRA to chromosome ends. TERRA preferentially associates with short telomeres through the formation of telomeric DNA:RNA hybrid (R-loop) structures that can form in trans. Telomere association and R-loop formation triggers telomere fragility and is promoted by the RAD51 recombinase and its interacting partner BRCA2 but counteracted by RNA surveillance factors, RNaseH1 and TRF1. RAD51 physically interacts with TERRA and catalyzes R-loop formation with TERRA in vitro supporting a direct involvement of this DNA recombinase in TERRA recruitment by strand invasion. Together, our findings reveal a RAD51-dependent pathway that governs TERRA mediated R-loop formation post transcription providing a mechanism of how lncRNAs can be recruited to new loci in trans.
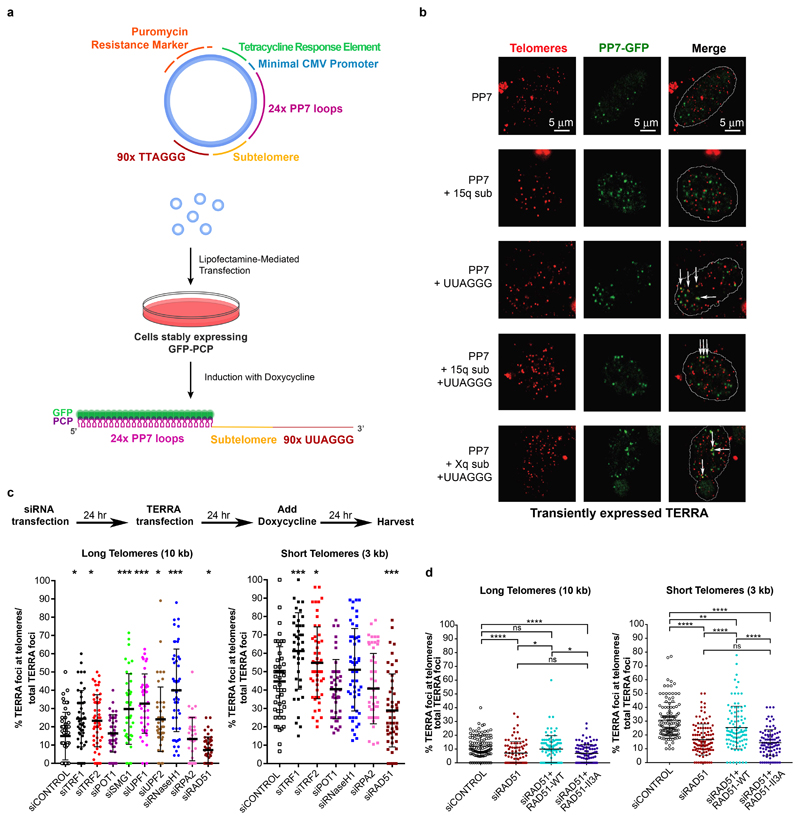
TERRA is transcribed from multiple chromosome ends and comprised of both subtelomeric sequences and telomeric repeats. More than 50% of TERRA is associated with chromatin6. To investigate how TERRA is recruited or retained at telomeres, we generated a plasmid with 24 copies of the PP7 stem-loops7 under the control of the TET promoter followed by 90 TTAGGG repeats (Fig. 1a). For full-length TERRA transcripts, the Xq and 15q subtelomeric regions containing the TERRA start sites were also cloned between the PP7 stem-loops and the TTAGGG repeats. The constructs were transiently transfected in HeLa clones constitutively expressing the PP7-coat protein fused to GFP (PCP-GFP) and a nuclear localization signal. PCP-GFP exhibited a diffuse signal in the nucleus but formed nuclear foci upon expression of the PP7 stem-loops, which are bound by PCP and can gather up to 48 PCP-GFP molecules per RNA. These foci did not co-localize with telomeres (Fig. 1b). The fusion of the subtelomeric region of 15q or Xq TERRA to the stem loops did not promote significant trafficking of the PP7 foci to telomeres. However, upon fusing the telomeric TTAGGG repeats downstream of PP7, it now co-localized with telomeres as analyzed by conventional and confocal imaging (Fig. 1b) indicating that the 5’-UUAGGG-3’ repeats of TERRA drive telomere association. The full-length PP7-tagged 15q and Xq chimeric TERRA also showed significant co-localization with telomeres (Fig. 1b and Extended Data Fig. 1a). Therefore, chimeric TERRAs originated from a plasmid were directed to telomeres in trans.
Fig. 1. Transgenic TERRA associates with telomeres and association is dependent on RAD51.
a, Schematic of chimeric TERRA construct and assay. The plasmids were transfected into HeLa cells constitutively expressing the GFP-PCP protein. After 24 hours TERRA transcription was induced with doxycycline. Chimeric TERRA, that is recognized and bo und by the GFP-PCP protein, was analyzed 24 hours upon induction. b, Fluorescence in situ hybridization (FISH) for telomeric DNA (red) and immunofluorescence (IF) for GFP (green) was employed to assess the co-localization of transiently expressed PP7 constructs with telomeres. Confocal images are shown. White arrows indicate co-localization of PP7 foci with telomeric signals. c, Schematic representation of siRNA transfection and TERRA transfection sequence. HeLa clones with long (10 kb average) and short (3 kb) telomeres were transfected with the corresponding siRNA pools, followed by chimeric TERRA transfection. The percentage of co-localizing TERRA foci was assessed by telomeric FISH combined with GFP IF. n=2 biologically independent experiments, > 40 nuclei were analyzed per condition, data are mean ± s.d. One-way ANOVA with Dunnett’s multiple comparisons test was used, comparing all conditions to siControl (*P < 0.05; **P < 0.01; ***P < 0.001). d, RAD51 enzymatic activity is required for TERRA recruitment. Endogenous RAD51 was depleted with siRNA and WT RAD51 or RAD51 II3A mutant was expressed from plasmid containing cDNA. TERRA colocalization with telomeres was assessed as in c. n=3 biologically independent experiments, > 80 nuclei were analyzed per condition, data are mean ± s.d. Two-tailed unpaired t test was used to calculate the P values. (*P < 0.05; **P < 0.01; ***P < 0.001).
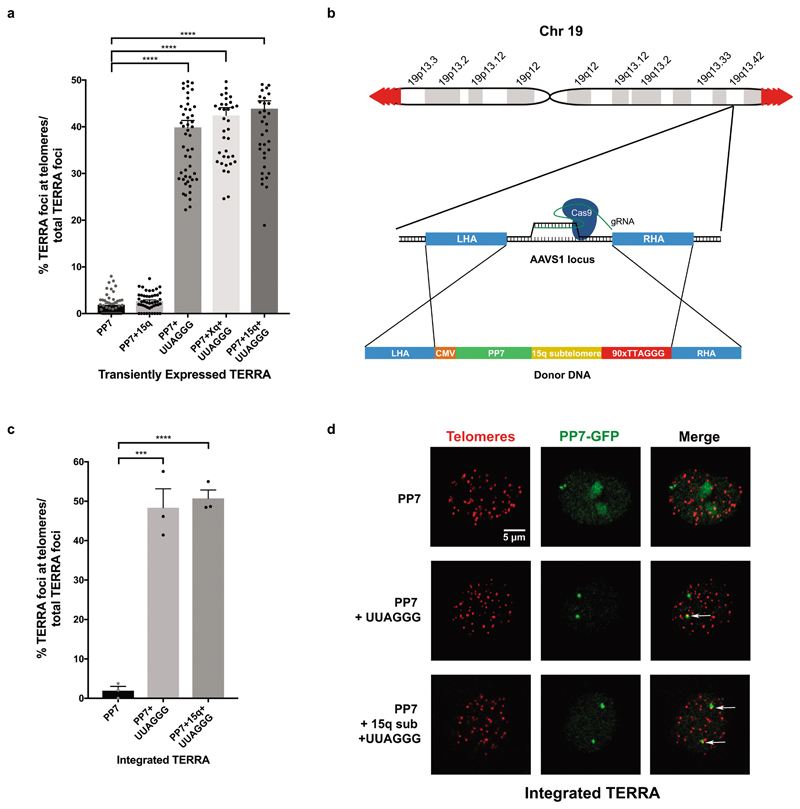
To eliminate possible confounding effects due to the high plasmid copy number or elevated levels of transgenic TERRA, using CRISPR/Cas9 technology we integrated the chimeric TERRA constructs into the genome at the Adeno-associated virus integration site 1 (AAVS1) on chromosome 19, which represents a safe harbor for transgene expression8 (Extended Data Fig. 1b). Following isolation of clones, we confirmed monoallelic site-specific integration of the full constructs by PCR and sequencing. The TERRA expression levels were lower giving 1-3 foci indicative of displacement from the transcription site, but similar to the results obtained upon transient transfection, the PP7 loops formed nuclear foci and only when fused to 5’-UUAGGG-3’ repeats, the chimeric RNAs co-localized with telomeres (Extended Data Fig. 1c,d).
Shorter telomeres recruit more TERRA
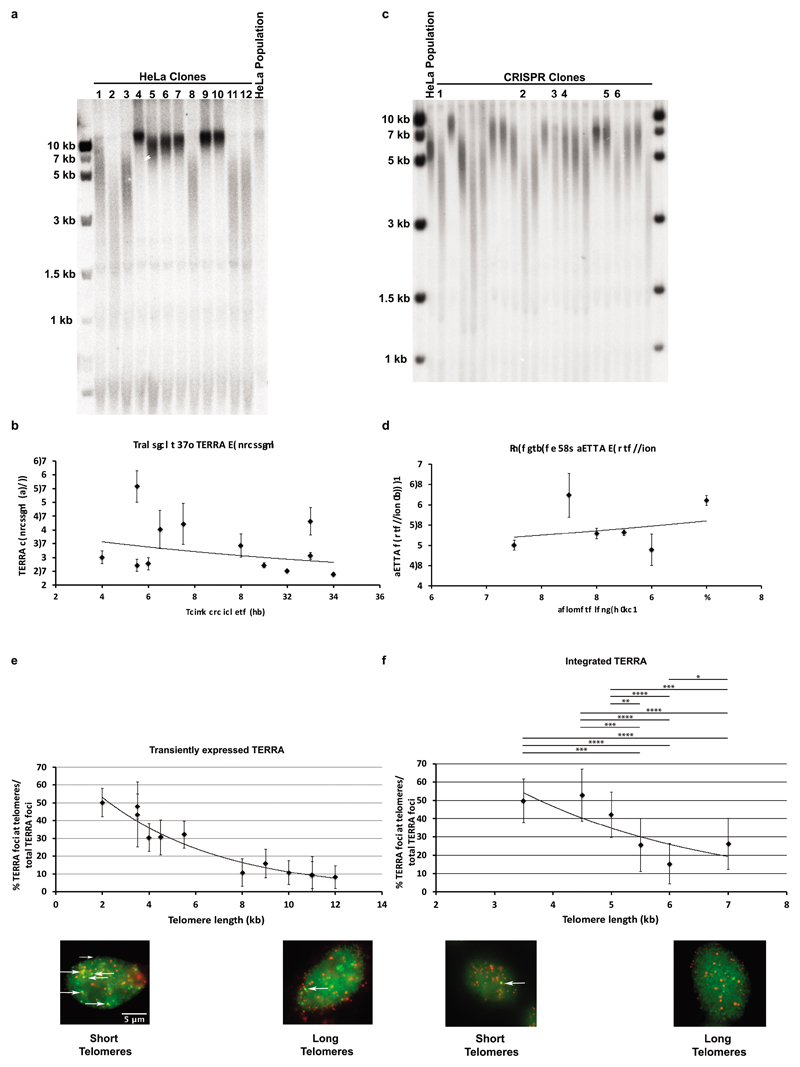
In S. cerevisiae and S. pombe, short telomeres recruit more TERRA, possibly to facilitate telomere maintenance through recombination or telomerase recruitment5,9,10. To explore the putative roles of telomere length in TERRA recruitment in human cells, we isolated individual HeLa clones that constitutively expressed the PCP-GFP and measured telomere lengths by Telomere Restriction Fragment length (TRF) analysis (Extended Data Fig. 2). Cells carrying short telomeres recruited TERRA much more efficiently than cells with long telomeres as seen upon transient or stable expression of TERRA (Extended Data Fig. 2). Therefore, short telomeres are more accessible for recruitment or retention of TERRA or long telomeres contain active systems to expel TERRA. In both experiments the overall chimeric TERRA expression levels varied in individual clones but this did not correlate with telomere length and telomere recruitment of TERRA (Extended Data Fig. 2b,d).
Recombination factors promote TERRA association with telomeres
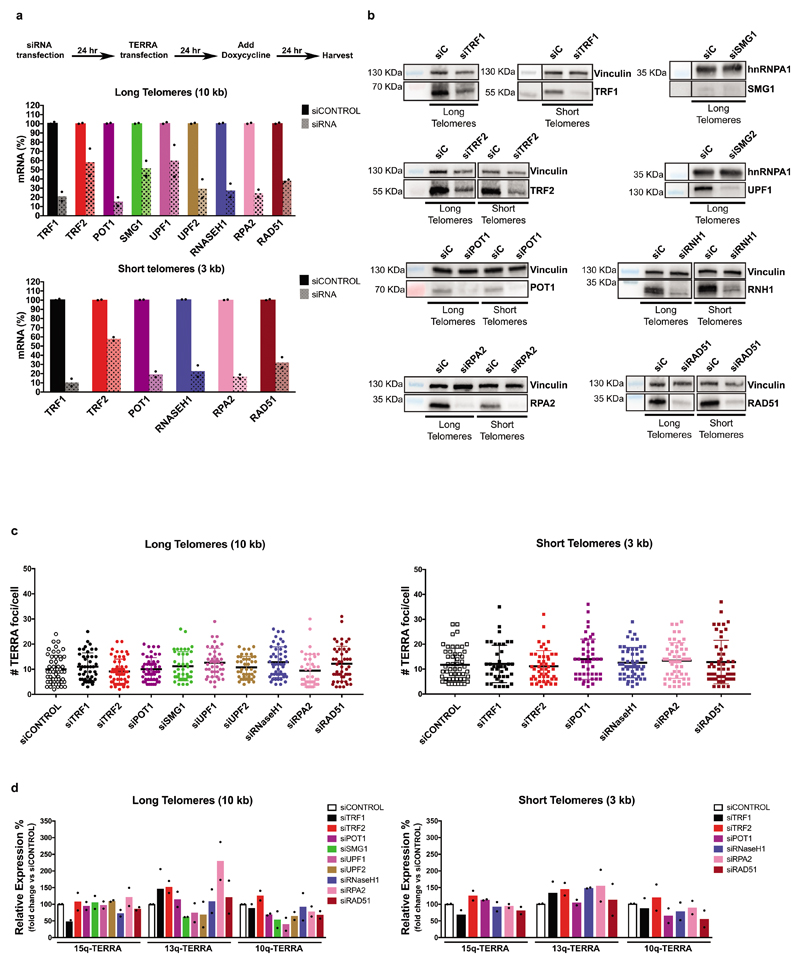
To identify proteins involved in TERRA localization at telomeres we performed siRNA screens on selected factors implicated in TERRA and telomere biology. Cell lines with long and short telomeres were transfected with siRNA pools, followed by transfection of the chimeric 15q TERRA construct. Upon induction of TERRA expression with doxycycline the cells were analyzed (Fig. 1c) and the level of depletion for each factor evaluated with RT-qPCR or Western blot (Extended Data Fig. 3a, b). The number of transgenic TERRA foci was not affected by individual depletions and no striking effects on levels of selected endogenous TERRA molecules were observed (Extended Data Fig. 3c, d). Among tested factors, depletion of TRF1 significantly increased TERRA co-localization at short and long telomeres, while removal of TRF2 led to a milder increase of recruitment (Fig. 1c). Depletion of the NMD factors also stimulated co-localization of TERRA at long telomeres supporting their crucial role in the displacement of TERRA from chromosome ends2. Similarly, removal of RNaseH1 resulted in high accumulation of TERRA at chromosome ends (~42%) in cells with long telomeres (Fig. 1c). This result indicated that TERRA recruitment or retention at long telomeres involves formation of DNA:RNA hybrids. In cells with short telomeres, depletion of RNaseH1 only marginally increased the co-localization of TERRA with telomeres. The roles of NMD factors in cells with short telomeres could not be analyzed as their depletion caused cell death. Importantly, depletion of RAD51, which facilitates strand invasion of DNA molecules during HDR, led to a significant decrease of TERRA recruitment at both long and short telomeres (Fig. 1c, d).
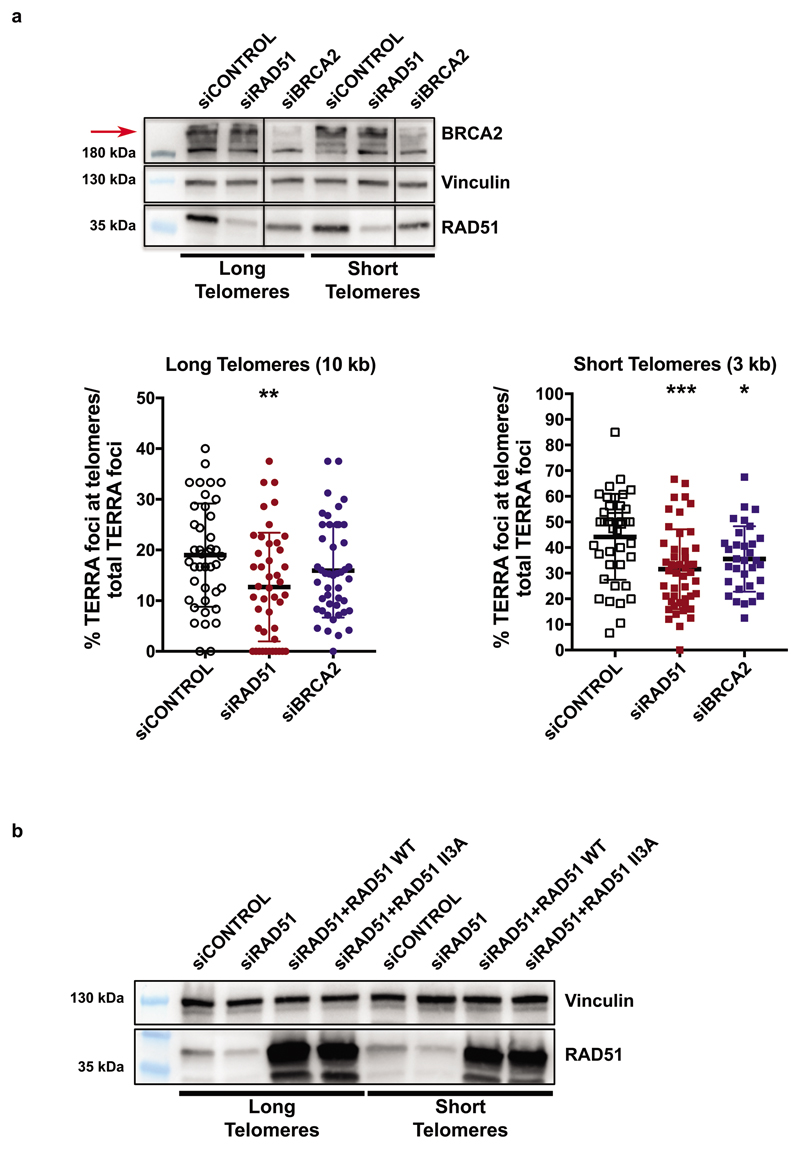
The involvement of RAD51 in TERRA recruitment prompted us to interrogate the role of BRCA2 in TERRA trafficking. BRCA2 loads RAD51 and promotes RPA displacement to form stable RAD51-ssDNA filaments capable of homology search to facilitate HDR during double strand break repair and protecting stalled replication forks11,12. Depletion of BRCA2 led to a marginal decrease in TERRA recruitment at long but more prominent reduction at short telomeres (Extended Data Fig. 4a). However, removal of BRCA2 also diminished total RAD51 protein levels in both cell lines (Extended Data Fig. 4a). Finally, we tested if RAD51 enzymatic activity is required for TERRA recruitment taking advantage of a mutation (RAD51 II3A) that retains DNA binding but not strand invasion activity13. In siRAD51 depleted cells, expression of wild type RAD51 from cDNA largely rescued TERRA colocalization with telomeres in contrast to RAD51 II3A (Fig. 1d, Extended Data Fig. 4b). Therefore, RAD51 enzymatic activity is required for TERRA association with telomeres. Overall, these data suggest that the HDR machinery promotes TERRA recruitment to telomeres.
TERRA forms R-loops in trans promoting telomere fragility
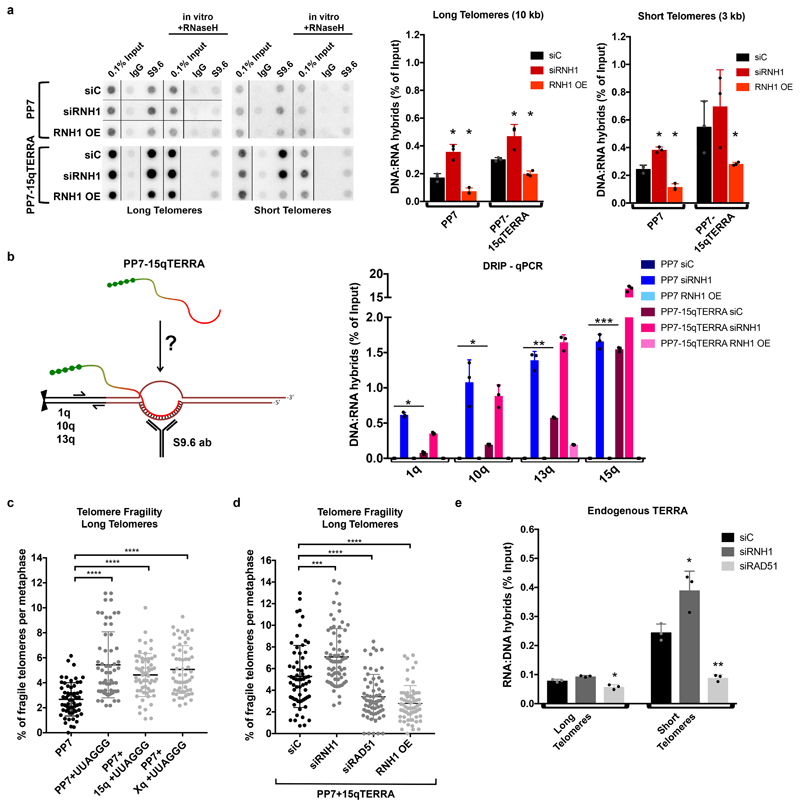
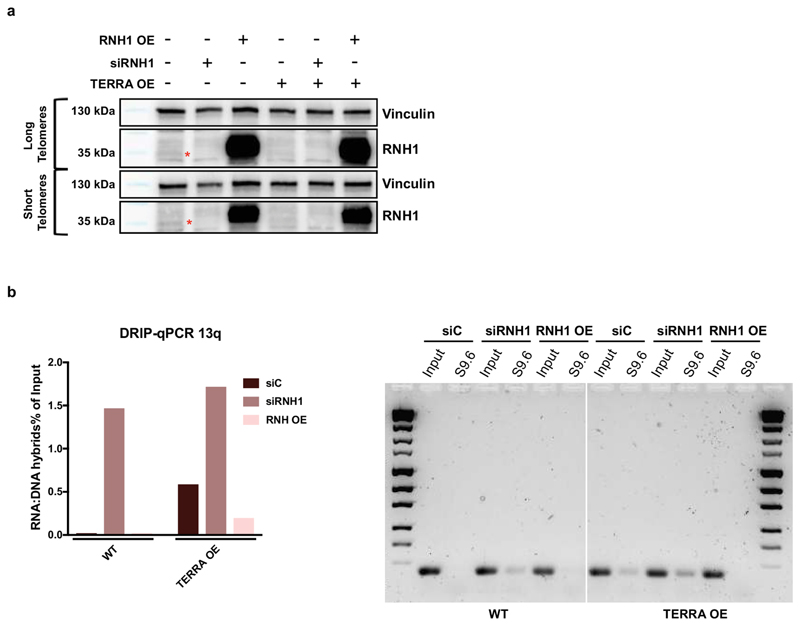
Since TERRA association with long telomeres was increased upon depletion of RNaseH1, we hypothesized that the transgenic TERRA may form R-loops with telomeres in trans. To explore this possibility, we applied the DNA:RNA immunoprecipitation protocol (DRIP) in which the specificity of the S9.6 monoclonal antibody to recognize 8-9 bp of DNA-RNA hybrids is exploited14. Precipitated nucleic acids were probed for telomeric repeats and as a control for specificity, isolated nucleic acids were treated in vitro with RNaseH1 prior to immunoprecipitation. Abolishment of the signal upon RNaseH1 pre-treatment confirmed the specificity of the assay for detection of telomeric R-loops (Fig. 2a). As expected, we detected R-loops at telomeres in wild type cells (Fig. 2a). Depletion of RNaseH1 (RNH1) led to R-loop increase while its overexpression to R-loop decrease at both long and short telomeres (Fig 2a, Extended Data Fig. 5a). Overexpression of chimeric TERRA further increased R-loop frequency in cells with both long and short telomeres (Fig. 2a), which again increased upon RNaseH1 depletion and decreased upon overexpression (Fig. 2a and Extended Data Fig. 5a) indicating that the transgenic TERRA formed DNA:RNA hybrids.
Fig. 2. TERRA forms R-loops in trans inducing telomere fragility in dependency of RAD51.
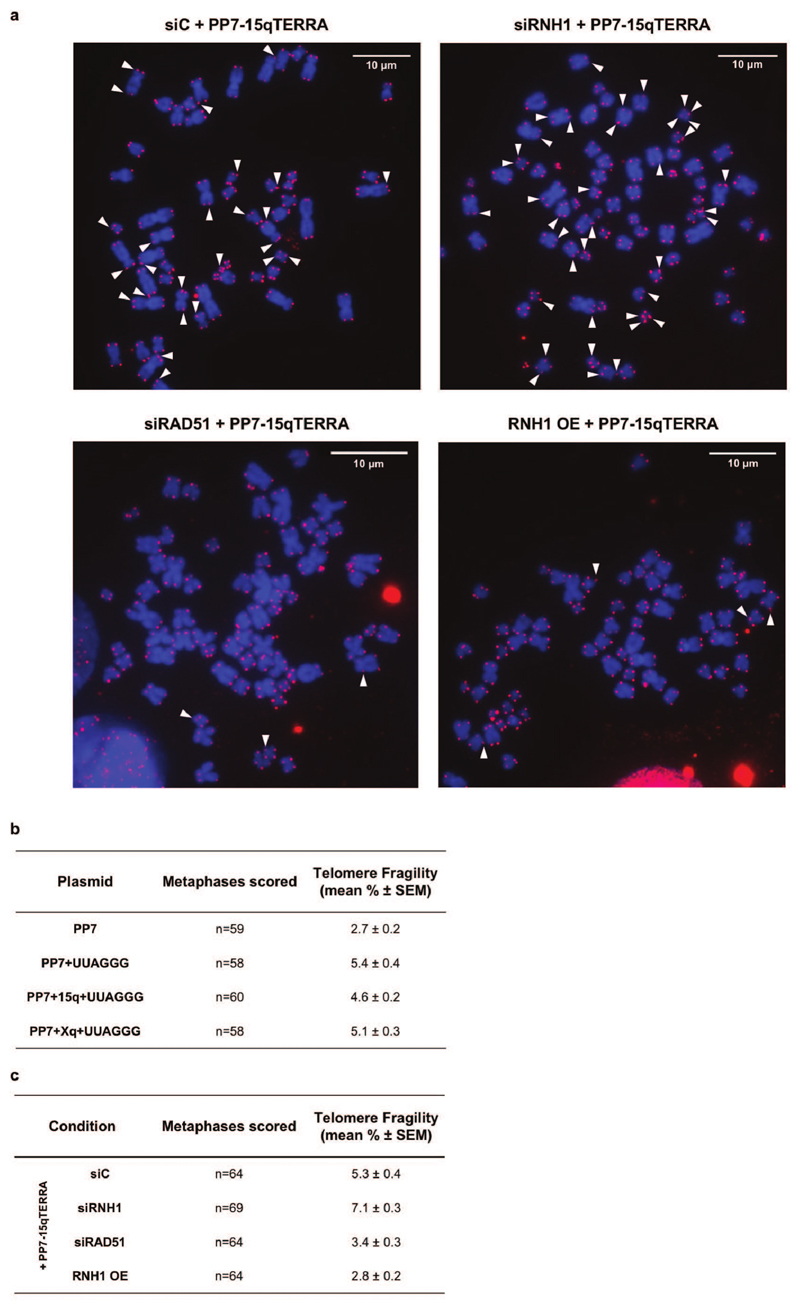
a, Detection of telomeric hybrids on dot-blots. Telomeric hybrids were isolated by DNA-RNA immunoprecipitation with S9.6 antibody from cells with long or short telomeres. Cells expressed either the PP7 stem loops or the PP7-15q TERRA and had been transfected with control siRNA (siC), siRNAs against RNaseH1 (RNH1), or with a plasmid that overexpressed RNH1. In vitro treatment with RNaseH1 served as a negative control. Quantification of signals is shown on the right. b, Schematic representation of PP7-15q TERRA expressed from a plasmid and forming R-loops in trans at telomeres 1q, 10q and 13q. Telomeric hybrids arising at the indicated chromosome ends were quantified by qPCR with primers amplifying specific subtelomeric DNA next to the telomeric tracts in cells with long telomeres. c, Telomere fragility induced upon expression of TERRA from a plasmid. The fraction of fragile telomeres was quantified on metaphase chromosomes stained with a telomeric FISH probe. d, Effects of RNaseH1 and RAD51 on telomere fragility. In cells expressing PP7+15qTERRA from a plasmid, telomere fragility was quantified upon depletion (si) or overexpression (OE) of RNaseH1 (RNH1), or depletion of RAD51. e, Detection of endogenous telomeric R-loops in siC, siRNH1 and siRAD51 transfected cells. For all data n=3 biologically independent experiments, data are mean ± s.d. Two-tailed unpaired t test was used to calculate the P values. (*P < 0.05; **P < 0.01; ***P < 0.001). For Fig. c and d the number of metaphases scored are indicated for each condition in Extended Data Fig. 6 b and c.
The DRIP assay (Fig. 2a) could not distinguish to what extent the DRIP signals for transgenic TERRA were derived from R-loops forming within the transgenic plasmid during transcription or R-loops forming post transcription in trans with telomeres. To measure R-loops specifically at telomeres, we used the DRIP samples derived from HeLa cells with long telomeres (Fig. 2a) and determined the presence of four specific chromosome ends by quantitative PCR (qPCR) using subtelomere-specific primers residing in immediate proximity to the terminal 5’-TTAGGG-3’ repeats (Fig. 2b). Telomeric R-loops became detectable at all four chromosome ends upon depletion of RNaseH1 (Fig. 2b). Specifically, 1q, 10q and 13q subtelomeric DNA strongly increased in abundance upon expression of transgenic PP7-15qTERRA indicating that R-loops had formed at these telomeres with PP7-15qTERRA. The 15q signal was enhanced even more extensively presumably due to R-loops forming with plasmid DNA containing the 15q sequence. As for wild type TERRA, RNaseH1 overexpression almost completely abolished the signals whereas RNAseH1 depletion increased R-loop abundance. The qPCR products (Extended Data Fig. 5b) were sequenced verifying the identity of products for each chromosome end. Together, these data confirmed that transgenic PP7-15qTERRA associated with telomeres in trans through the formation of R-loop structures.
TERRA R-loops have been implicated in interfering with telomere replication4,15–17 which is manifested in telomere fragility18 being characterized by the accumulation of telomeric signals in metaphase chromosomes with a smeary or discontinuous appearance (Extended Data Fig. 6a). Transgenic TERRA increased telomere fragility (Fig. 2c and Extended Data Fig. 6b) which was suppressed by RAD51 depletion or RNaseH1 overexpression but increased by RNaseH1 depletion (Fig. 2d and Extended Fig. 6c, d). These results confirm that post transcription from plasmids, TERRA forms R-loops at telomeres in trans.
RAD51 promotes telomeric R-loop formation
We next tested by DRIP assay the role of RAD51 in hybrid formation of endogenous TERRA at telomeres in WT cell lines (Fig. 2e, Extended Data Fig. 7a). While depletion of RNaseH1 led to expected mild increase of R-loops, depletion of RAD51 caused a significant decrease in hybrid accumulation. Even stronger reduction of R-loops was observed in RAD51-depleted cells with short telomeres, which are characterized by higher levels of DNA:RNA hybrids. Therefore, RAD51 promotes association of endogenous TERRA with telomeres through R-loop formation.
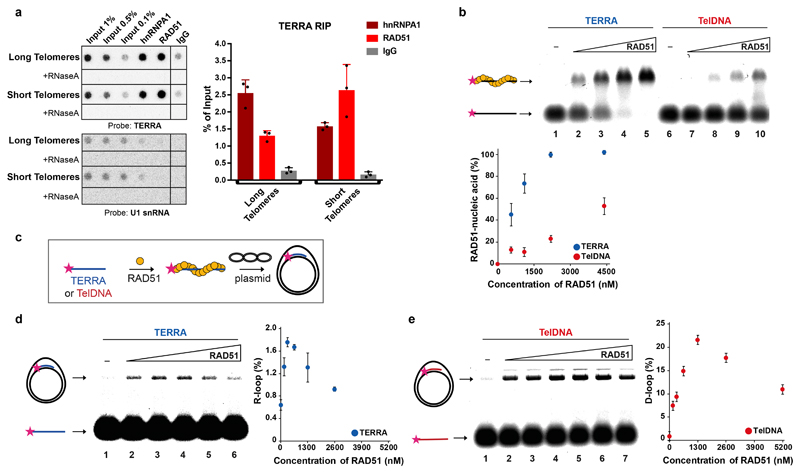
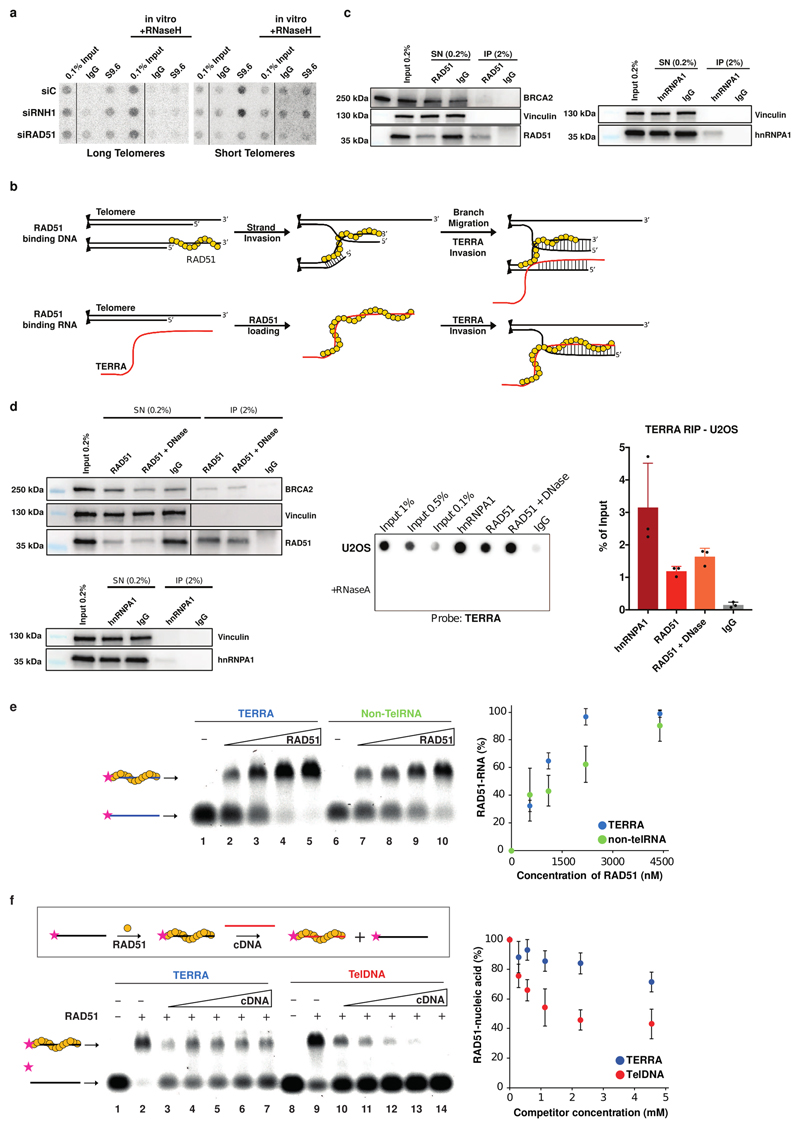
We hypothesized that RAD51 either binds TERRA to catalyze its strand invasion reaction into the telomeric repeats (Extended Data Fig. 7b lower panel) or TERRA might hybridize to exposed single stranded telomeric DNA during RAD51-mediated HDR between telomeric DNA molecules even in the absence of a physical interaction between TERRA with RAD51 (Extended Data Fig. 7b upper panel). To explore these hypotheses, we performed native RNA immunoprecipitations (RNA-IP) with anti-RAD51 antibodies in HeLa cells. We also included anti-hnRNPA1 antibodies as a control, as this protein binds TERRA19. Immunoprecipitation of endogenous RAD51 specifically retrieved TERRA and not the nuclear U1 snRNA (Fig. 3a and Extended Data Fig. 7c) and similar results were observed in the U2OS cell line (Extended Data Fig. 7d). The TERRA signal was sensitive to RNaseA treatment showing specific recovery of RNA, but the RNA signal was insensitive to DNaseI treatment. Together these experiments indicate that TERRA associates with RAD51 in vivo.
Fig. 3. RAD51 associates with TERRA and catalyzes R-loop formation.
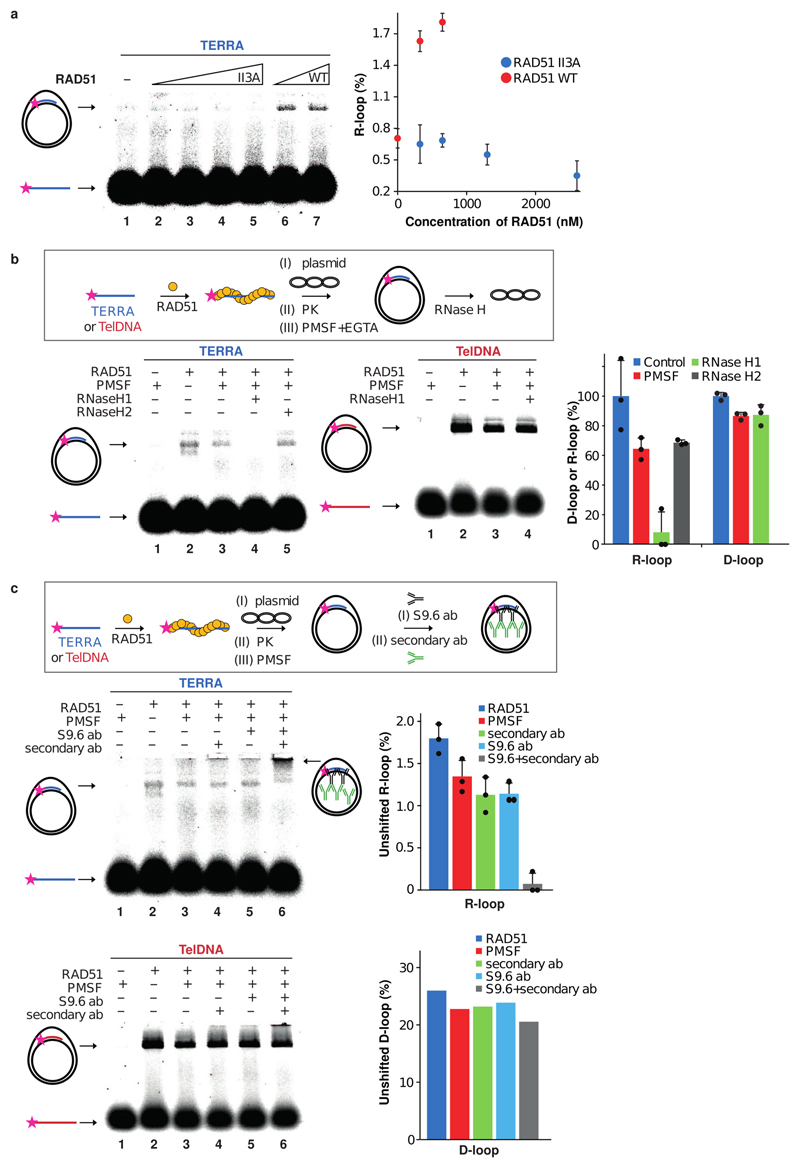
a, Native RNA-immunoprecipitation (RNA-IP) assay with anti-RAD51 and anti-hnRNPA1 antibodies performed in extracts from HeLa cell clones with long or short telomeres. Western blotting was used to evaluate the IP efficiency of RAD51 and hnRNPA1, and co-IP of BRCA2 (see Extended data Fig. 7c, d). IP-recovered RNA was analyzed for TERRA and U1 snRNA. n=3 biologically independent experiments, data are mean ± s.d. Two-tailed unpaired t test was used to calculate the P values. (*P < 0.05; **P < 0.01; ***P < 0.001). b, Affinity of RAD51 for TERRA (lanes 1 – 5) and TelDNA (lanes 6 – 10) oligonucleotides analyzed by EMSA. Fluorescently labelled RNA or DNA substrates (20 nM) were incubated with increasing concentration of RAD51 protein (550, 1100, 2200, and 4400 nM). Quantification is shown on the bottom. n=3 independent experiments, data are mean ± s.d. c, Schematics for R-loop and D-loop formation assay. d, RAD51 concentration dependent formation of R-loops, its detection on native gel and quantification. n=3 independent experiments, data are mean ± s.d. e, Detection of D-loops on native gel and quantification. n=3 independent experiments, data are mean ± s.d.
We determined whether purified RAD51 can bind TERRA directly carrying out electrophoretic mobility shift assays (EMSA). Recombinant RAD51 was incubated with fluorescently labelled TERRA or telomeric DNA oligonucleotides and the reactions were resolved by PAGE (Fig. 3b). RAD51 bound the TERRA oligonucleotide with 3-fold higher affinity than the corresponding telomeric DNA sequence, as lower protein concentration was required to shift the migration of the labelled oligonucleotide-RAD51 complexes with TERRA (Fig. 3b). RAD51 binding to TERRA oligonucleotide was tighter than to an unrelated RNA (Extended Data Fig. 7e) and also more stable when compared to telomeric DNA as demonstrated by the competition assay in which pre-formed TERRA- or TelDNA-RAD51 complexes were challenged with excess of unlabeled 49mer competitor ssDNA (cDNA) (Extended Data Fig. 7f).
To test if RAD51 promotes R-loop formation in vitro, we carried a strand invasion assay with recombinant RAD51 and telomeric RNA and DNA (Fig. 3c, d, e). RAD51 catalyzed both R-loop and D-loop formation in concentration dependent manner in contrast to the catalytically dead RAD51 II3A (Extended Data Fig. 8a). As expected, canonical R-loops were sensitive to treatment with RNaseH1 and were supershifted with the R-loop recognizing S9.6 antibody when combined with anti-mouse IgG (Extended Data Fig. 8b,c). Taken together this data supports a direct role of RAD51 protein in TERRA localization to initiate strand invasion and promote the formation of DNA:RNA hybrids at telomeres.
Discussion
Our data reveal the mechanism of how TERRA is recruited to chromosome ends through RNA strand invasion. The recruitment and R-loop formation depends on the RAD51 recombinase and the 5’-UUAGGG-3’ repeats of TERRA. Though the subtelomeric TERRA sequences were not required in our assay for telomere recruitment, we do not exclude that they may contribute in targeting endogenous TERRA molecules to their native chromosome ends. The observed base pairing between TERRA and telomeric DNA provides a mechanism of how this long noncoding RNA can encounter its major site of action at chromosome ends. Therefore, TERRA recruitment appears to occur in a process that strongly resembles the strand invasion and homology search mechanism exploited in all living organisms during DNA repair by HDR and which is also characteristic for ALT-mediated telomere stabilization in cancer cells. A direct involvement of RAD51 in the strand invasion reaction is supported by our finding that RAD51 strongly binds TERRA and catalyzes strand invasion into telomeric sequence in vitro. However, it is likely that important differences between DNA- and RNA-mediated strand invasion exist with the regard to the mechanism and the requirement of accessory factors, which must be explored in the future.
Increased R-loop formation and RAD51 association with short telomeres was previously shown in yeast5. Our findings in human cells are fully consistent with these observations. Furthermore, we reveal that TERRA can form R-loops post transcriptionally at telomeres in trans. R-loops were generally assumed to form only during transcription through unsuccessful removal of the native RNA from its DNA template20. However, previous studies in yeast suggested that R-loops may also form at loci that are distinct from their site of synthesis21. Our work here suggests that R-loop formation in trans plays a major role for telomere homeostasis. Through its mechanism of recruitment, TERRA may represent a scaffold to guide and regulate telomerase, HDR proteins and chromatin modifiers specifically at the chromosome ends that may need their attention. It will be important to determine what other long noncoding RNAs are recruited to their sites of action through similar mechanisms.
Methods
Cell culture and transfections
The telomerase positive cell line HeLa (cervical cancer) was from ATCC and the ALT cell line U2OS (osteosarcoma) from ECACC. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml of penicillin/streptomycin (Thermo Fisher Scientific). The cell lines were maintained in a 5% CO2 incubator at 37°C.
HeLa cells were transfected with Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). To induce TERRA transcription, doxycycline (1μg/ml) was added to the medium 24 hours after transfection and the cells were harvested 48 hours post transfection. To isolate clones containing integrated TERRA trans-genes, puromycin (1μg/ml) was added to the medium 24 hours following transfection with the pSpCas9(BB)-2A-Puro plasmid. Selection was maintained for 5 days. SiRNAs were purchased as pools from Dharmacon (siGENOME SMARTpool). Cells were transfected with 20 pmol siRNAs using calcium phosphate transfection in 6-well plates in DMEM supplemented with 10% fetal bovine serum (FBS). Cells were harvested 72 hours post transfection. WT RAD51 or RAD51 II3A mutant was expressed from plasmid containing cDNA13.
Lentivirus production and cell transduction
Plasmids pMD2.G (1μg) and pCMVR8.74 (3μg) (kind gifts from Didier Trono, EPFL) were mixed with PP7_eGFP (3μg) (kind gift from Daniel Larson, NIH) or pLenti CMV rtTA3 Hygro (addgene #26730) for transfection of HEK293T in OPTIMEM medium (Thermo Fisher Scientific) using Lipofectamine 2000. The transfection mix was incubated overnight and the medium replaced the next day. The supernatants were collected the next two consecutive days upon centrifugation and cleared through a 0.22 μm filter unit (Stericup, Millipore), before aliquoting the viruses and freezing them at -80°C. HeLa cells were transduced with 1 ml of viral particles of pLenti CMV rtTA3 Hygro. Following infection, the cells were selected with hygromycin (200 μg/ml) for 5 days, before they were infected again with the PP7_GFP viruses. GFP positive clones with similar GFP intensity were isolated with a FACSAria Fusion sorter by EPFL’s Flow Cytometry Core Facility.
Cloning of TERRA constructs
The TERRA constructs were cloned in the pTRE2puro vector of Clontech’s TET On system without the corresponding polyA region. The PP7 stem loops were amplified from Addgene plasmid # 61762 (kind gift from Daniel Larson) and introduced into the pTRE2puro through In-Fusion cloning. To amplify the TTAGGG sequence we performed a modified PCR protocol using the Phusion Green Hot Start II High-Fidelity DNA polymerase (F537S) with no template and the following reaction: 1x GC Phusion Buffer, 0.2 mM dNTPs, 0.4 μM primer 5’-TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG-3’, 0.4 μM primer 5’-CCCTAACCCTAACCCTAACCCTAACCCTAA-3’, and 2 U of polymerase. The PCR consisted of 30 sec at 98°C followed by 10 cycles at 98°C for 5 sec, 60°C for 10 sec, and at 72°C for 15 sec. The DNA products of variable size were fractionated on a 1.2% agarose gel and the desired size was excised, extracted with the QIAquick gel extraction kit, and cloned into the pTRE2puro vector containing the 24 copies of PP7 stem loops. The subtelomeric sequences were amplified with a high fidelity polymerase from phenol-chloroform extracted genomic DNA from HeLa and introduced into the corresponding vectors through In-Fusion cloning. All constructs were verified by restriction digestion and sequencing. The plasmids were amplified in HR-deficient E. coli strains (i.e. Stbl3) at 30°C. All the primers are listed in Extended Data Table 1.
Generation of integrated TERRA cell lines
The integration of TERRA constructs into the AAVS1 locus was performed as previously described22. The gRNA for AAVS1 (5’-ACCCCACAGTGGGGCCACTA-3’) and its complementary strand were annealed, and cloned into the pSpCas9(BB)-2A-Puro plasmid acquired from Addgene (#48139). The donor template encompassed ~800bp of homology arms for AAVS1, which were amplified from HeLa genomic DNA, and cloned into the plasmids containing the control PP7 stem loops construct and the different TERRA transgenes. HeLa cells were transfected with both the gRNA/Cas9 and the donor template vectors. Individual clones were screened by PCR for the presence of the transgene in the AAVS1 locus.
Immunofluorescence (IF) and Fluorescence In Situ Hybridization (FISH)
The cells were grown on glass coverslips following the culture conditions mentioned above. The coverslips were washed twice with 1xPBS, fixed with 4% paraformaldehyde for 10 min at room temperature, and washed twice with 1xPBS. The cells were permeabilized in 1xdetergent solution (0.1% Triton X-100, 0.02% SDS in 1 × PBS), followed by pre-blocking with 2%BSA in 1xPBS. The cells were blocked with 10% normal goat serum in 2%BSA/1xPBS. Coverslips were incubated with primary and subsequently with secondary antibodies in blocking solution, fixed with 4% paraformaldehyde for 5 min at room temperature, and the samples were dehydrated with a serial concentration of ethanol. For FISH staining the cells were hybridized with the hybridization solution containing 10 mM Tris–HCl pH 7.4, 70% formamide, 0.5% blocking reagent and 1/1,000 Cy3 probe, denatured at 80 °C for 3 min, and hybridized for 3 h at room temperature. The cells were washed with Wash 1 (10 mM Tris–HCl pH 7.4, 70% formamide) and Wash 2 (0.1M Tris–HCl pH 7.4, 0.15M NaCl, 0.08% Tween-20) twice, stained with DAPI, and dehydrated with ethanol. Images were acquired on an Upright Zeiss Axioplan or on a Zeiss LSM700 equipped with an AxioCam MRm B/W. Images were processed and analyzed with ImageJ and Adobe Photoshop. All statistical analysis was performed in GraphPad Prism. All the figures were created in Adobe Illustrator
Telomeric FISH on metaphase spreads
Cells were treated with 0.05 μg/ml demecolcine for 2 h, collected and incubated in hypotonic solution (0.075M KCl) at 37 °C for 8 min. Swollen cells were collected and fixed in ice-cold methanol:acetic acid (3:1) overnight at 4 °C. The next day 100 μl of cell suspension was dropped onto slides, incubated at 70 °C for 1 min and air-dried overnight at room temperature. FISH staining was performed as described above.
Telomere length analysis
HeLa genomic DNA was isolated with the Wizard Genomic DNA purification according to the manufacturer’s instructions (Promega). 6 μg of genomic DNA was digested with 30U of HinfI and RsaI overnight at 37°C. The digested DNA was mixed with 6xMassRuler DNA Loading Dye (Thermo Fisher Scientific), loaded on a 0.8% agarose gel in 1xTBE and fractionated by gel electrophoresis at 2V/cm for 20 hours. The gels were dried for two hours at 50°C in vacuum, treated with denaturation buffer (0.5 M NaOH, 1.5 M NaCl) and neutralization buffer (0.5 M Tris-HCl pH 7.5, 1.5 M NaCl), followed by pre-hybridization with Church buffer for 1 hour at 50°C. The gels were hybridized overnight at 50°C with a randomly labeled TeloC probe as described previously23. The gels were washed for 1 hour at 50°C with 4xSSC, 4xSSC 0.1%SDS, and 2xSSC 0.1%SDS, exposed to a phosphorimager screen and analyzed on a Typhoon phosphorimager (GE).
RT-qPCR
For RT-qPCR of TERRA, 3x106 cells were harvested following transfection of the chimeric TERRA constructs. RNA was isolated with the NuceloSpin RNA kit (Macherey-Nagel). RT-qPCR for 15q TERRA was performed using our previously described protocol24. To assess mRNA levels post siRNA transfections, we used ThermoFisher Scientific’s SuperScript III Reverse transcriptase and Power SYBR Green PCR Master Mix on an Applied Biosystems 7900HT Fast Real-Time System according to the manufacturer’s instructions.
Western blotting
Antibodies are listed in Extended Data Table 2. Protein samples were mixed with 2xLaemmli buffer, boiled for 5 min at 95°C, run on 4-15% SDS-PAGE precast gels (Mini-PROTEAN TGX Gels, BioRad), transferred to nitrocellulose blotting membranes (Amersham), blocked with blocking solution (3%BSA in 1xPBS, 0.1% Tween-20), and incubated overnight at 4°C with the corresponding primary antibody. Membranes were washed three times for 5 min with 1xPBST followed by incubation for 1 hour at RT with HRP-conjugated secondary antibodies (Promega) in blocking solution. Membranes were washed again three times for 5 min with 1xPBST before revealing them with a chemiluminescence detection kit (Western bright ECL, Advansta) and analyzing them on a Vilber Fusion FX imaging system.
DNA-RNA Immunoprecipitation
Cells at ~40% confluence in 6-well plates were transfected with siRNAs, followed by plasmid transfection the next day. The cells were harvested on ice 48 hours later, counted on a CASY Cell Counter, washed with 1xPBS, and samples were taken for DRIP and Western blot analysis. For DRIP, 107 cells were dissolved in 175 μl of ice-cold RLN buffer (50 mM Tris-HCl pH 8, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM DTT, and 100 U/ml RNasIN PLUS), incubated on ice for 5 min, and centrifuged (300xg, 2 min, 4°C). The nuclei were lysed in 500 μl RA1 buffer (NucleoSpin RNA, Macherey-Nagel) containing 5 μl of β-mercaptoethanol and homogenized by passing them through a 20G x 11/2 syringe (0.9mm x 40 mm). The nucleic acids (NAs) containing extracts were mixed with 250 μl H2O and 750 μl phenol:chloroform:isoamylalcohol (25:24:1) in a Phase Lock Gel heavy (5PRIME), centrifuged (13,000xg, 5 min, RT), and the upper aqueous phase was mixed with 750 μl of ice-cold isopropanol, NaCl was added to 50 mM, incubated on ice for 30 min and precipitated NAs were collected by centrifugation at 10,000xg for 30 min at 4°C. The pellets were washed twice with 70 % ice-cold ethanol, air-dried, dissolved in 130 μl of H2O, and sonicated on a Covaris system (10% Duty Factor, 200 Cycles per Burst, for 180 seconds, with an AFA intensifier) to obtain 10-500 bp long fragments. 90 μg of sonicated NAs were mixed with RNaseH1 or H2O in RNaseH1 buffer (20 mM HEPES-KOH pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT) and incubated at 37°C for 90 min. The samples were diluted 10 times in DIP-1 buffer (10 mM HEPES-KOH pH 7.5, 275 mM NaCl, 0.1% Na-deoxycholate, 0.1% SDS, 1% Triton X-100) and pre-cleared with 80 μl of Sepharose Protein G beads for 1 hour, on a rotating wheel, at 4°C. 1% of NAs was kept as input. Half of the samples (~45 μg of NAs) were incubated with 3 μg of S9.6 antibody or mouse IgG and 40 μl of Sepharose Protein G beads on a rotating wheel at 4°C overnight. The next day the samples were washed 5 min on a rotating wheel at 4°C with DIP-2 (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton X-100, 0.1% Na-deoxycholate), DIP-3 (50 mM HEPES-KOH pH 7.5, 500 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton-X100, 0.1% Na-deoxycholate), DIP-4 (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 250 mM LiCl, 1% NP-40, 1% Na-deoxycholate), and TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0). The samples were incubated with Cross-link reversal buffer (1% SDS, 0.1 M NaHCO3, 0.5 mM EDTA pH 8.0, 20 mM Tris-HCl pH 8.0, 10 μg/ml RNase, DNase free (Roche)) at 65°C overnight. The DNA was isolated using the QIAGEN PCR Clean-up kit. The DNA was pipetted onto a positively charged nylon membrane (Amersham Hybond N+) and telomeric DNA was detected with a telomeric probe as previously described25.
RNA Immunoprecipitation
The cells were grown to ~70% confluence in 15 cm dishes, harvested on ice, counted on a CASY Cell Counter, washed with 1xPBS, and lysed in RNA-IP RLN buffer (50 mM Tris-HCl pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM DTT 400 U/ml RNasIN PLUS, and protease inhibitors (Complete, Roche)). The lysates were pre-cleared with Dynabeads Protein G for 1 hour on a rotating wheel at 4°C, and incubated overnight with 6 μg of the corresponding antibody on a rotating wheel at 4°C. 35 μl of with yeast tRNA pre-blocked Dynabeads Protein G (ThermoFisher Scientific) per IP were added to the mixtures and incubated for 2 hours on a rotating wheel at 4°C, followed by 5 washes with RLN buffer supplemented with 6 mM EDTA pH 8.0. The RNA was eluted in 1% SDS, 5 mM EDTA pH 8.0, and 5 mM β-mercaptoethanol at 42°C for 30 min, followed by 30 min at 65°C. The RNA was purified using the RNA clean-up protocol of the NucleoSpin RNA kit (Macherey-Nagel). The RNA was denatured at 65°C for three minutes and loaded on a positively charged nylon membrane (Amersham Hybond N+). TERRA and U1 snRNA were detected with the corresponding probe as previously described26.
EMSA
RAD51 protein was purified27 and diluted in Dilution Buffer (25 mM Tris-HCl pH 7.5, 10% (v/v) glycerol, 0.5 mM EDTA, pH 7.5, 50 mM KCl, 1 mM DTT and 0.01% NP40,) supplemented with 2 mM MgCl2 and 2 mM ATP. Increasing concentration of RAD51 was incubated with 20 nM Cy3 labelled 41mer TERRA (5’-UUAGGGUUAGGGUUAGGGUUAGGGUUAGGGUUAGGGUUAGG-3’), 41mer TelDNA (5’-TTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGG-3’), 41mer non-TelRNA (5’-AGUAUAUAUGAGUAAACUUGGUCUGACAGUUACCAAUGCUU-3’) or 40mer non-TelDNA (5’-AAATTAACAAGTATAATAAGAAATAGAAACAAGAAATAGA-3’) substrate at 37 °C for 10 minutes in 10 μl of buffer D (50 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM MgCl2, 1 mM ATP). Reaction mixtures were then crosslinked with 0.01% glutaraldehyde for 10 min. Products were resolved using 0.8 % TBE agarose gels at 4°C for 50 min (6.5 V/cm). Gels were imaged on a FLA-9000 scanner (Fujifilm) and quantified with Multi Gauge V3.2 (Fujifilm).
Stability EMSA
Fluorescently labelled 41mer TERRA or 41mer TelDNA substrates were incubated with indicated concentration of RAD51 at 37 °C for 10 minutes in 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 1mM MgCl2 and 1 mM ATP. To challenge assembled RAD51-ssDNA complexes, increasing concentration of unlabeled 49mer competitor ssDNA (5’-CCTGTTCAAACGCACATATTAAGCATTTCCTGTCATTGGCGGCTAATTC-3’) was added and incubated for another 10 minutes at 37 °C ssDNA. Products were crosslinked with 0.005% glutaraldehyde for further 10 minutes and resolved using 0.8% TBE agarose gel at 4 °C for 50 min (6.5 V/cm). Gels were imaged on a FLA-9000 scanner (Fujifilm) and quantified with Multi Gauge V3.2 (Fujifilm).
R-loop/D-loop formation assay
Fluorescently labelled 41mer TERRA or 41mer TelDNA (50 nM) were pre-incubated for 10 minutes at 37 °C with increasing concentration of RAD51 in 50 mM Tris-HCl, pH 7.5, 1 mM MgCl2 supplemented with 1 mM CaCl2 and 1 mM AMP-PNP. Reaction was started by addition of 600 ng of pCR4-TOPO vector containing the 15q subtelomeric sequence followed by 15 copies of TTAGGG repeats. The mixture was incubated for another 10 minutes at 37 °C and then stopped by addition of SDS (0,1%) and proteinase K (0.1 mg/ml) followed by 3 minutes incubations at 37 °C. Reaction mixtures were separated on 0.8% agarose gel and analyzed as described for EMSA. For digestion of R-loops by RNaseH, the mixtures were inhibited for 1 minute with PMSF (2 mM) and with EGTA (1.6 mM) at 37°C to inhibit proteinase K and chelate calcium ions, respectively. The products were digested by RNaseH1 (6.8 mU/μl, Thermo Scientific™) or RNaseH2 (6.8 mU/μl, New England Biolabs) for 1 minute at 37 °C and resolved as described above. For antibody-specific supershift of R-loops, the products were formed as described for RNaseH digestion followed by incubation with S9.6 antibodies (0.02 μg/μl) for 2 minutes at 37°C and/or anti-mouse horseradish peroxidase conjugated antibody
Telomeric DNA:RNA hybrid formation and RNaseH1 digestion
Fluorescently labelled TERRA or non-telomeric RNA were mixed with their corresponding complementary ssDNA in the annealing buffer (10 mM Tris at pH 8.5, 50 mM NaCl, 1 mM EDTA). To form the DNA:RNA hybrid the mixture was heated for 5 min at 95 °C, followed by gradual cooling down at room temperature. The DNA:RNA hybrid (40 nM) was cleaved by RNaseH1 (6.8 mU/μl) in the presence or absence 1 mM CaCl2 for 10 minutes at 37 °C in 50 mM Tris (7.5), 1 mM MgCl2 and 1 mM AMP-PNP. Reaction was stopped by addition of SDS/PK mixture and subsequent incubation for 10 minutes at 37 °C. Reaction mixtures were loaded on 10% PAGE gel, separated by electrophoresis (90 V for 60 minutes at 4 °C), scanned by Image Reader FLA-9000 scanner and quantified by MultiGauge V3.2 software.
Supershift of DNA:RNA hybrid with S9.6 antibody
Fluorescently labelled TERRA and non-telomeric DNA:RNA hybrids (40 nM) were incubated with 0.015 μg/μl of S9.6 antibody for 10 minutes at 37 °C and then crosslinked with 0.01% glutaraldehyde for 10 minutes at 37 °C. Products were crosslinked with 0.015% glutaraldehyde for further 10 minutes and resolved using 0.8% TBE agarose gel at 4 °C for 50 min (6.5 V/cm). Gels were imaged on a FLA-9000 scanner (Fujifilm) and quantified with Multi Gauge V3.2 (Fujifilm).
Extended Data
Extended Data Figure 1. Colocalization of transgenic TERRA with telomeres.
a, Quantification of colocalization of transgenic RNA foci expressed from plasmids with telomeres. b, Strategy for targeting the AAVS1 locus with CRISPR/CAS9 to integrate the chimeric TERRA constructs into the genome. c, Quantification of colocalization of transgenic RNA foci with telomeres when TERRA was expressed from the AAVS1 locus. d, Confocal images with TERRA expressed from the AAVS1 locus. For Fig. a and c, n=3 biologically independent experiments, data are mean ± s.d. Two-tailed unpaired t test was used to calculate the P values. (***P < 0.001; ****P<0.0001).
Extended Data Figure 2. TERRA associates preferentially with short telomeres.
a, TRF analysis of HeLa clones used for transient expression of TERRA b, d, TERRA expression levels measured by RT-qPCR and compared to the levels of TERRA in the clone with the shortest telomeres. n=3 biologically independent experiments, data are mean ± s.d. c, TRF analysis of HeLa clones expressing transgenic TERRA from the AAVS1 locus. e, HeLa clones of different telomere length were transiently transfected with the PP7-15q TERRA construct and co-localization was assessed by IF-FISH. The percentage of TERRA foci that co-localized with telomeres was plotted as a function of telomere length. White arrows indicate co-localization of PP7 foci with the telomeric signal. Random colocalization events (~3%) were not subtracted. f, Co-localization events as in e but for PP7-15q TERRA expressed from the AAVS locus on chromosome 19. n=3 biologically independent experiments, data are mean ± s.d. Significant differences are indicated. Two-tailed unpaired t test was used to calculate the P values. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P<0.0001).
Extended Data Figure 3. Depletion of factors regulating TERRA trafficking.
a, Timeline of transfections and cell harvest. mRNA levels were determined by RT-qPCR for both long and short telomere cell lines. b, For the second replicate of the siRNA screen the functional level of depletion was also evaluated on Western blots. Vinculin and hnRNPA1 were used as loading controls. c, Number of TERRA foci per cell were plotted for each depleted factor. n=2 biologically independent experiments, ≥ 40 nuclei were analyzed per condition. d, Quantification of endogenous TERRA stemming from chromosome ends 15q, 13q and 10q upon depletion of the indicated factors relative to the negative control (siCONTROL). n=2 biologically independent experiments.
Extended Data Figure 4. Depletion of RAD51 and BRCA2 which regulate TERRA association with telomeres.
a, HeLa clones with long and short telomeres were transfected with siRAD51 and siBRCA2, followed by chimeric TERRA transfection. Representative immunoblots show RAD51 and BRCA2 depletion and Vinculin as a loading control. Upon depletion, co-localization of TERRA with telomeres was assessed with IF-FISH. n=2 biologically independent experiments, ≥ 54 nuclei were analyzed per condition, data are mean ± s.d. One-way ANOVA with Dunnett’s multiple comparisons test was used, comparing all conditions to siCONTROL. (*P < 0.05; **P < 0.01; ***P < 0.001). b, Detection of endogenous and transgenic RAD51 on a Western blot. Endogenous RAD51 was depleted with siRNA and WT RAD51 or RAD51 II3A mutant was expressed from plasmid containing cDNA.
Extended Data Figure 5. RNaseH1 regulated formation of telomeric R-loops in trans.
a, RNH1 depletion and overexpression was assessed by Western blotting in cell lines with long and short telomeres and upon TERRA overexpression. b, Representative example of one DRIP assay followed by qPCR for chromosome 13q. The hybrids are expressed as fraction of Input. The amplified DNA was run on a gel, isolated, and sequenced.
Extended Data Figure 6. Transgenic TERRA expressed from plasmids induces telomere fragility.
a, Representative examples of metaphase chromosomes stained with FISH to visualize telomeres. DNA is stained with DAPI. Fragile telomeres are indicated by white arrowheads. b, Quantification of telomere fragility. c, Quantification of telomere fragility in cells expressing PP7-15qTERRA in which expression of RNaseH1 (RNH1) and RAD51 was manipulated as indicated. For Fig. b and c, the number of metaphases scored over 3 biologically independent experiments are indicated for each condition as n.
Extended Data Figure 7. RAD51 binds TERRA and promotes R-loop formation.
a, Detection of endogenous telomeric R-loops on dot-blots as in Fig. 2a. Cells were transfected with siCONTROL (siC), siRNH1 and siRAD51. b, Western blot analysis of RAD51 and hnRNPA1 upon native RNA-IP (see Fig. 3a for RNA analysis). c, Possible models for the roles of RAD51 in mediating TERRA-telomere associations. Upper row: RAD51 binds telomeric ssDNA inducing strand invasion of another telomere. TERRA hybridizes to the displaced strand upon branch migration. Lower row: RAD51 binds TERRA directly and initiates homology search and strand invasion of TERRA at a telomere. d, Native RNA-IP performed in U2OS cell extracts demonstrating association of TERRA with RAD51 and hnRNPA1. n=3 biologically independent experiments, data are mean ± s.d. e, Affinity of RAD51 for TERRA and non-telomeric RNA oligonucleotides. Quantification is shown on the right. n=3 independent experiments, data are mean ± s.d. f, Schematic representation of EMSA-competition assay (upper). Pre-formed RAD51-nucleic acid complex with 20 nM TERRA (lanes 2 – 7) or TelDNA oligonucleotide (lanes 9 – 14) was challenged with increasing concentration of unlabelled competitor ssDNA (cDNA; 0.28, 0.56, 1.13, 2.27 and 4.54 μM). Quantification is shown on the right. n=3 independent experiments, data are mean ± s.d.
Extended Data Figure 8. RAD51 catalyzes formation of canonical R-loops.
a, RAD51 II3A mutant (lines 2 – 5; 325, 650, 1300, and 2600 nM) or RAD51 WT (lines 6 – 7; 325 and 650 nM) was incubated with TERRA substrate (50 nM) followed by addition of plasmid containing homologous region. Quantification of R-loop in the presence of RAD51 WT or RAD51 II3A (right panel). n=2 independent experiments, data are mean ± s.d. b, Schematic of the R-loop assay (upper). After RAD51-mediated R-loop or D-loop formation, proteins were digested with proteinase K (PK), which was then inactivated with PMSF and EGTA. R-loops/D-loops are detected on the native gel as indicated. Treatment with RNaseH1 which degrades the RNA moiety in DNA:RNA hybrid structures eliminates the R-loops but has no effect on D-loop. RNaseH2 which cleaves ribonucleotides in DNA had no effect as expected. Quantification is shown on the right. n=3 independent experiments, data are mean ± s.d. c, Schematic of the R-loop assay (upper). RAD51 mediated R-loops (middle, n=3 independent experiments, data are mean ± s.d.), in contrast to D-loops (bottom), are recognized by the S9.6 antibody and supershifted in presence of both, S9.6 and anti-mouse IgG. Quantification is shown on the right. n=1 experiment.
Extended Data Table 1. Oligonucleotides used in this study.
| Primers used for Chimeric TERRA constructs | ||
|---|---|---|
| Name | Forward | Reverse |
| Amplification of PP7 stem loops | CCGCGGCCCCGAATTCTATCGATACTCGAGATCCTA | GCTGACTAGAGGATCCACACGCGTTCTCGATAATGAA |
| Amplification of TTAGGG repeats | TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG | CCCTAACCCTAACCCTAACCCTAACCCTAA |
| Amplification of 15q subtelomere | GAACGCGTGTGGATCATTCTCCTCAGGTCAGACCCG | CTAACCCTAAGGATCCTAACCGTGACCCTGACCCCG |
| Amplification of Xq subtelomere | GAACGCGTGTGGATCCCGAGTTGCGTTCTCG | CTAACCCTAAGGATCGCACATGAGGAATGTGGGTG |
| Amplificiation of AAVS1 left homology arm | TGACCGGTTGCTTTCTCTGACCAGCATTC | GTGTTAACCACTGTGGGGTGGAGGGGAC |
| Amplificiation of AAVS1 right homology arm | CGGATATCACTAGGGACAGGATTGGT | GTGATATCCTGTAGGAAGGGGCAGGAGA |
| Primers used for RT-qPCR | ||
| Name | Forward | Reverse |
| 15qTERRA | CAGCGAGATTCTCCCAAGCTAAG | AACCCTAACCACATGAGCAACG |
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
| TRF1 | CTTGCCAGTTGAGAACGATATACA | CATCAGGGCTGATTCCAAGG |
| TRF2 | GGGTTATGCAGTGTCTGTCGCG | CAGTGGTGTGAGCTCAGCCT |
| POT1 | CCAAGCTCTGGATCAGTATCATT | CATAGTGGTGTCCTCTCAAATAC |
| SMG1 | CCAAGCACCGTTCCAGGAACTG | CTCTCTTGCACCGCTTTCCCAG |
| UPF1 | CGCAGGGCTACATCTCCATGAG | CTCGTCACCAAGGTAACTGTCCTG |
| UPF2 | GGCTGAGTCTGCAGACACAATGC | GCAGCAAGTTGAGAGGACATGGG |
| RNaseHI | GGCCAGGCCATCCTTTAAATGTAGG | GCTTGTTCAATGGCTTTGCAGGC |
| RPA2 | GCAGGGAACTTTGGTGGGAATAGC | CCCTTCAGGTCTTGGACAAGCC |
| RAD51 | GAGGAAAGGAAGAGGGGAAACCAG | CCCACTCCATCTGCATTAATGGCG |
| 1q subtelomere | CAGCGTCGCAACTCAAATG | CCCTCACCCTCCATGAGTAATA |
| 10q subtelomere | GCATTCCTAATGCACACATGAC | TACCCGAACCTGAACCCTAA |
| 13q subtelomere | GCACTTGAACCCTGCAATACAG | CCTGCGCACCGAGATTCT |
| 15q subtelomere | AACCCTAACCACATGAGCAACG | GCTGCATTAAAGGGTCCAGT |
| siRNAs | ||
| Gene name | Catalog number | siRNA sequences |
| siControl | D-001206-13-20 | UAGCGACUAAACACAUCAA, UAAGGCUAUGAAGAGAUAC, AUGUAUUGGCCUGUAUUAG, AUGAACGUGAAUUGCUCAA |
| SiTRF1 | M-010542-02 | CAAGAUAAACCUAGUGGUA, GGUGAUCCAAAUUCUCAUA, GGAAACUGGUCUAAAAUAC, GCCAGUUGAGAACGAUAUA |
| siTRF2 | M-003546-00 | GAAGUGGACUGUAGAAGAA, GGAAGCUGCUGUCAUUAUU, GGAAUCAGCUAUCAAUGUG, GAAGACAGUACAACCAAUA |
| SiPOT1 | M-004205-01 | AGAAAGAUGUCAACAGCUA, GAGGCAAAGAAUCGAAAUA, CAGGAGUACUAGAAGCCUA, UGCAAGAUCUCCACGUUAA |
| SiSMG1 | M-005033-01 | GUGAAGAUGUUCCCUAUGA, GAGGUUAGCUGCGGAAAGA, GGUCAGACAUCCACCAGAA, UAACUUGGCUCAGCUGUAU |
| SiUPF1 | M-011763-01 | GCUCCUACCUGGUGCAGUA, UCAAGGUCCCUGAUAAUUA, GGAAGUCGACCUCCUUUGA, CAAGAUAACAUCACUGUCA |
| siUPF2 | M-012993-01 | GGAACGAGAAUUCUUAAUA, GCAUGUACCUUGUGUAGAA, GAAGAUAUUCGAUUAGGAA, GGUCUAGAGAGUUGCGAAU |
| siRNaseH1 | M-012595-00 | GCGCAGAGCCGUAUGCAAA, GAGCUAAACAAUCGGAAGA, GCCAGGCCAUCCUUUAAAU, GACAUUCAGUGGAUGCAUG |
| siRPA2 | M-017058-00 | GAUCAAUGCACACAUGGUA, CAAAAUAGAUGACAUGACA, GAGUGAAGCAGGGAACUUU, GUGGAACAGUGGAUUCGAA |
| SiRAD51 | M-003530-04 | GAAGCUAUGUUCGCCAUUA, GCAGUGAUGUCCUGGAUAA, CCAACGAUGUGAAGAAAUU, AAGCUAUGUUCGCCAUUAA |
| siBRCA2 | M-003462-01 | GAAACGGACUUGCUAUUUA, GUAAAGAAAUGCAGAAUUC, GGUAUCAGAUGCUUCAUUA. GAAGAAUGCAGGUUUAAUA |
Extended Data Table 2. Antibodies used in this study.
| Antibody | Company | Catalogue No | Dilution (Technique) |
|---|---|---|---|
| GFP | Home-Made | 1:1000 (IF) | |
| GFP | Merck Millipore | MAB3580 | 1:1000 (IF, WB) |
| RAD51 | Santa-Cruz | sc-8349 | 1:1000 (WB) |
| RAD51 | ABCAM | ab133534 | 6μgr(IP) |
| BRCA2 | Merck Millipore | MAB3580 | 1:1000 (WB) |
| Vinculin | ABCAM | ab129002 | 1:10,000 (WB) |
| hnRNPA1 | Santa-Cruz | sc-32301 | 1:1000 (WB), 6μgr(IP) |
| RNaseH1 | GeneTex | GTX-117624 | 1:1000 (WB) |
| TRF1 | Santa-Cruz | sc-6165R | 1:1000 (WB) |
| TRF2 | Merck Millipore | 05-521 | 1:1000 (WB) |
| POTI | ABCAM | ab124784 | 1:1000 (WB) |
| SMG1 | Abgent | AP8055a | 1:500 (WB) |
| UPF1 | Bethyl Laboratories | A300-037A | 1:1000 (WB) |
| RPA2 | ABCAM | ab2175 | 1:1000 (WB) |
| Anti-DNA-RNA Hybrid [S9.6] | Kerafast | ENH001 | 1 μgr / 10 μgr nucleic acids (DRIP) |
| Anti-Rabbit IgG (H+L), HRP Conjugate | Promega | W4011 | 1:10,000 (WB) |
| Anti-Mouse IgG (H+L), HRP Conjugate | Promega | W4021 | 1:10,000 (WB) |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 633 | Thermo Fisher | A-21070 | 1:1,000 (IF) |
Acknowledgements
We thank Daniel Larson (NIH), Douglas Bishop (University of Chicago), Viesturs Simanis (EPFL), Pierre Gönczy (EPFL) and Didier Trono (EPFL) for providing material. The BIOP core facility at EPFL, the Gönczy lab, and Mario Spirek is thanked for technical support and advice. M.F. was supported by European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 702824. J.L.’s laboratory was supported by the Swiss National Science Foundation (SNFS), the SNFS funded NCCR RNA and disease network and EPFL. L.K.’s laboratory was supported by the Wellcome Trust Collaborative Grant 206292/E/17/Z, the Czech Science Foundation (GACR 17-17720S), project LQ1605 from the National Program of Sustainability II (MEYS CR). Both J.L. and L.K. were also supported by an Initial Training Network (ITN) grant (aDDRess) from the European Commission’s Seventh Framework Programme.
Footnotes
Authors contributions M.F. and J.L. conceived the study. M.F. and R.V.F. executed all cell and molecular biology experiments. M.P. performed all biochemistry experiments and T.L. some EMSA experiments. L.K. conceived the RAD51 biochemistry experiments and advised on text. J.L. and M.F. wrote the paper.
Author Information The authors declare no competing financial interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nature Reviews Molecular Cell Biology. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat–Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 3.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature Cell Biology. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 4.Arora R, et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nature Communications. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graf M, et al. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell. 2017;170:72–85.e14. doi: 10.1016/j.cell.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular Dissection of Telomeric Repeat-Containing RNA Biogenesis Unveils the Presence of Distinct and Multiple Regulatory Pathways. Molecular and Cellular Biology. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-Time Observation of Transcription Initiation and Elongation on an Endogenous Yeast Gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cusanelli E, Romero CAP, Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell. 2013;51:780–791. doi: 10.1016/j.molcel.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 10.Moravec M, et al. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016;17:999–1012. doi: 10.15252/embr.201541708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorslund T, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason JM, Chan Y-L, Weichselbaum RW, Bishop DK. Non-enzymatic roles of human RAD51 at stalled replication forks. Nature Communications. 2019;10 doi: 10.1038/s41467-019-12297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boguslawski SJ, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. Journal of Immunological Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 15.Sagie S, et al. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nature Communications. 2017;8 doi: 10.1038/ncomms14015. 14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013;32:2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balk B, et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 18.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redon S, Zemp I, Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41:9117–9128. doi: 10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Molecular Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. eLife. 2013 doi: 10.7554/eLife.00505. https://elifesciences.org/articles/00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grolimund L, et al. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nature Communications. 2013;4:2848. doi: 10.1038/ncomms3848. [DOI] [PubMed] [Google Scholar]
- 24.Feretzaki M, Lingner J. A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods. 2017;114:39–45. doi: 10.1016/j.ymeth.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Porro A, et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nature Communications. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porro A, Feuerhahn S, Lingner J. TERRA-Reinforced Association of LSD1 with MRE11 Promotes Processing of Uncapped Telomeres. Cell Reports. 2014;6:765–776. doi: 10.1016/j.celrep.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Špírek M, et al. Human RAD51 rapidly forms intrinsically dynamic nucleoprotein filaments modulated by nucleotide binding state. Nucleic Acids Research. 2018;46:3967–3980. doi: 10.1093/nar/gky111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.