Abstract
The nature of the first genetic polymer is the subject of major debate1. Although the common ‘RNA world’ theory suggests RNA as the first replicable information carrier at the dawn of life2,3, other evidence implies that life may have started with a heterogeneous nucleic acid genetic system including both RNA and DNA4. Such a theory streamlines the eventual ‘genetic takeover’ of homogeneous DNA from RNA as the principal information storage molecule in the central dogma, but requires a selective abiotic synthesis of both RNA and DNA building blocks in the same local primordial geochemical scenario. Herein, we demonstrate a high-yielding, completely stereo-, regio-, and furanosyl-selective prebiotic synthesis of the purine deoxyribonucleosides, deoxyadenosine and deoxyinosine. Our synthesis utilizes key intermediates in the prebiotic synthesis of the canonical pyrimidine ribonucleosides, and we show that, once generated, the pyrimidines persist throughout the synthesis of the purine deoxyribonucleosides, ultimately leading to a mixture of deoxyadenosine, deoxyinosine, cytidine, and uridine. These results support the notion that purine deoxyribonucleosides and pyrimidine ribonucleosides may have coexisted before the emergence of life5.
Introduction
Considerable progress in the prebiotic synthesis of the pyrimidine ribonucleosides of RNA, cytidine (C) 1 and uridine (U) 2, and their 2-thio derivatives, 3 and 4 6, 7, together with recent advances in non-enzymatic RNA replication8–10 have given credence to the RNA world theory. Progress towards the abiotic synthesis of purine nucleosides has been made, but only using routes that employ as starting materials chemically and enantiomerically pure sugars11–15, which are not likely to be have been found on the primordial earth. Additionally, no prebiotically plausible route has been shown to provide a mixture containing a competent set of nucleosides for information storage at the polymeric level.
Extant biology, in contrast to the proposed RNA world, utilizes DNA as the central information-carrying molecule. This discrepancy between the RNA world and modern biology requires a ‘genetic takeover’ that invokes the power of primitive biosynthetic machinery and natural selection operating over millions of years, ultimately resulting in an ancestral biosynthetic route to DNA16. The superior hydrolytic stability and replication fidelity17 of DNA could have resulted in selection of primitive organisms capable of synthesizing DNA, and thus its rise to prominence in the central dogma, but the feasibility of this evolutionary process in a pre-DNA world is debated1. To circumvent this potentially problematic transition, an R/DNA world has been proposed, in which nascent biology had access to both RNA and DNA building blocks from the outset, without requiring elaborate biosynthesis18–20. In such a world, heterogeneous polymers would have initially been most common, but polymers with increased homogeneity, and hence properties closer to either that of RNA or DNA, would have been selected for over their mixed counterparts4. For the R/DNA world to be plausible, an efficient prebiotic synthesis of DNA building blocks is required, and one that provides building blocks for both RNA and DNA in the same localized geochemical scenario is preferable. We recently demonstrated proof of this principle by showing that 2'-deoxy-2-thiouridine 5 – a non-canonical deoxynucleoside – can be synthesized from thioanhydrouridine 6 – an RNA derivative – by way of a prebiotically plausible, hydrogen sulfide-mediated photoreduction5. Although this finding provides an important prebiotic link between RNA and DNA building blocks, the lability of 5 to hydrolysis may limit its phosphorylation and subsequent oligomerization21, 22. Additionally, the synthesis of canonical deoxyadenosine (dA) 7 from 5 and adenine 8 was low yielding (4%), and generated a more abundant undesired side product, the α-anomer of 7 (6%). Using guidance from a geochemical scenario23, we now demonstrate a synthesis of purine deoxynucleosides that is based on prebiotically plausible reactions and substrates. We then evaluate our route at a systems level by enacting the synthesis on mixtures of materials likely to arise in a primordial environment, culminating in the demonstration of multiple reaction sequences able to selectively furnish a mixture of U (1), C (2), dA (7) and deoxyinosine (dI, 9).
Results and Discussion
Prebiotic Route to Purine Deoxyribonucleosides
A route to purine nucleosides that diverges from a prebiotic RNA synthesis is attractive because it implies that the constituents of a set of nucleosides capable of storing information – pyrimidines and purines – may have formed in the same location on a primordial Earth, rather than having been necessarily brought together by environmental processes after their separate formation. To develop such a route, we evaluated intermediates in the prebiotic RNA pyrimidine nucleoside synthesis6, 7 as ribosyl donors (Fig. 1). The RNA synthesis proceeds from RAO 10 which reacts with cyanoacetylene 11 to provide α-anhydrocytidine 12. Thiolysis of 12 in formamide produces α-2-thiocytidine 13 which undergoes efficient UV-mediated photoanomerisation to 2-thiocytidine 3, which hydrolyses to the canonical pyrimidines cytidine 1 and uridine 2, and biologically important non-canonical pyrimidine 4. Alternatively, in the dark, 13 is hydrolysed to α-2-thiouridine 14 7. Whilst 14 appeared initially only a by-product that would be produced in the dark on the early Earth, it is readily cyclised to anhydrouridine 15 at 80 °C (63% yield in water or 89% yield in formamide, Extended Data Fig. 2). We recognised α-anhydropyrimidines 12 and 15 as ideal glycosyl donors for 1',2'-cis tethered glycosylation24. Since the sugar of 12 and 15 is fixed in its furanosyl form, the formation of pyranosyl nucleosides – one of the critical downfalls of previous strategies – should be excluded. Additionally, the α-stereochemistries of C1' and C2' of 12 and 15 led us to expect transglycosylation to provide only β-anomers, the correct stereochemistry at C1' for all natural (deoxy)ribonucleosides. Finally, since 12 and 15 are ultimately derived from ribo-aminooxazoline (RAO) 10, which crystallizes enantiopure from solutions of minimally enantioenriched carbohydrates or amino acids25, 26, this route offered the so-far unmet potential to deliver enantio- and diastereomerically pure furanosyl-nucleosides by glycosylation.
Fig. 1. Previous synthesis of RNA pyrimidine nucleosides 1 (C), 2 (U) and a deoxypyrimidine nucleoside 5, and the present work.
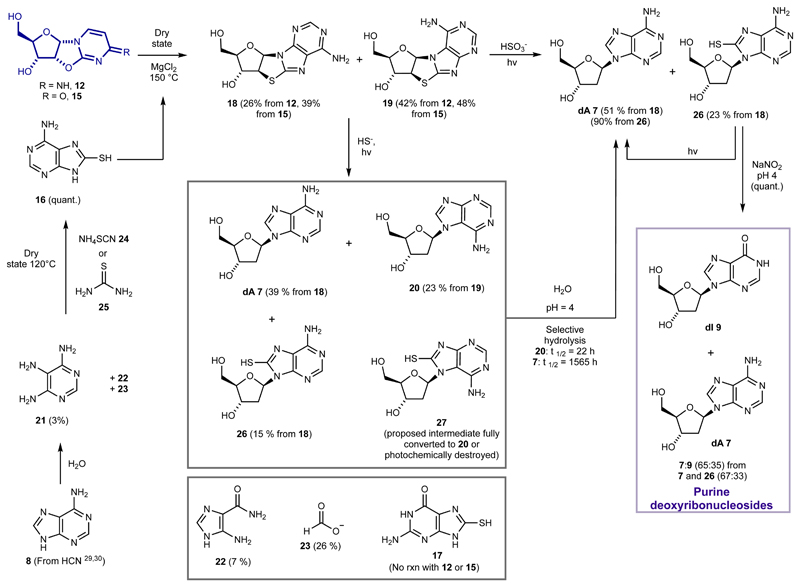
RAO 10 is a starting point in the network since it crystallises in enantiopure form from minimally enantio-enriched solutions25, 26. It can be elaborated via 12 and 3 to the pyrimidine nucleosides7. Although we had developed a low-yielding route to deoxyadenosine 7 (dA) from 6 via 5 5, we recognized that 12 and 15 are ideal candidates for tethered glycosylation with 16. The products, thioanhydropurines 18 and 19, are reduced photochemically in a similar way to 6, providing an efficient route to deoxynucleosides. Critically, once produced, pyrimidines 1 (C) and 2 (U) survive the sequence that produces purines 7 (dA) and 9 (dI), and we show that the four nucleosides 1 (C), 2 (U), 7 (dA) and 9 (dI) can be produced alongside one another.
Accordingly, we evaluated 8-mercaptoadenine 16 and 8-mercaptoguanine 17 as potential nucleophiles to participate in transglycosylation with 12 and 15 (Fig 2). Although 17 proved unreactive, 16 reacts with 12 and 15 at 150 °C in the dry state (Fig. 2), to provide two new β-configured nucleoside products in moderate yields (14% and 16% respectively from 15, trace amounts from 12). The minor product was determined to be N 9-8, 2'-anhydro-thioadenosine 18 by X-ray crystallography and 1H-NMR spiking experiments with a synthetic standard. The major product was inferred to be N 7-8,2'-anhydro-thioadenosine 19, the regioisomer of 18, by its subsequent conversion to 2'-deoxy-N 7-adenosine 20. The presence of magnesium chloride in the reaction, presumably acting as a Lewis acid27, dramatically improved the yield of 18 and 19 to 39% and 48% respectively from 15 (combined yield 87%) and 26% and 42% respectively from 12 (combined yield 68%). Thus, in a prebiotic environment where 12 or 15 and 16 are brought together, perhaps by converging streams that then undergo evaporation, 18 and 19 could be readily generated, especially in the presence of magnesium ions28.
Fig. 2. Prebiotic route to purine deoxyribonucleosides, 7 (dA) and 9 (dI).
The route starts with α-anhydropyrimidines 12 and 15, which are intermediates in the RNA pyrimidine synthesis, and 8-mercaptoadenine 16, which is available from adenine 8 via hydrolysis and reaction with ammonium thiocyanate or thiourea. Dry state tethered glycosylation of 16 and 12 or 15 provides thioanhydropurines 18 and 19, which can be photochemically reduced by two routes. If bisulfite is the reductant, only N 9-configured products 7 (dA) and 26 are formed. 26 can be converted to 7 by further irradiation, or by nitrosation. If hydrosulfide is used as the reductant, both N 9-configured 7 (dA) and 26 as well as N 7-configured 20 is formed. 20 has a half-time of hydrolysis nearly two orders of magnitude lower than 7 (dA) and so is selectively degraded. To generate deoxyinosine 9 (dI) alongside deoxyadenosine 7 (dA), the products of either photoreduction are treated with nitrous acid at pH 4.
Any prebiotic synthesis requires a viable route to all reagents from plausible early-Earth feedstocks. We were drawn towards adenine 8 as a starting point for the provision of 8-mercaptoadenine 16, due to its widely accepted prebiotic plausibility as a relatively stable pentamer of hydrogen cyanide29, 30. Remarkably, despite the reactivity of related purines31, adenine did not react with elemental sulfur at temperatures up to 300 °C. However, adenine does undergo (slow) hydrolysis in aqueous media. Miller et. al. reported a half-time for hydrolysis of adenine of about 1 year at 100 °C, and identified (but did not quantify) 4,5,6-triaminopyrimidine 21 (TAP) among the products of hydrolysis32. We reinvestigated this hydrolysis of adenine 8, under conditions more suited to a laboratory time-scale (138 °C, phosphate buffer pH 8), and at partial conversion after 10–12 days confirmed the presence of TAP in yields of 2–3% (8–9% based on recovered adenine) (Fig. 2). Due to the differential solubilities of adenine and TAP, the supernatants of adenine hydrolysis reactions are enriched in TAP after cooling. A typical supernatant contains 5-aminoimidazole-4-carboxamide 22, TAP 21, and adenine 8 in a 4:2:1 ratio, and formate 23 as the only other major component (See Fig. S1–S5 for full details). We found that TAP (either commercially supplied or that in the crude adenine hydrolysate) is converted to 8-mercaptoadenine 16 by heating in the dry state with either ammonium thiocyanate 24 or thiourea 25. 24 is an inevitable by-product of the photochemistry of hydrogen cyanide and hydrogen sulfide33, two precursors likely to have been abundant on the primordial earth, and heavily implicated in the origin of life by our cyanosulfidic chemical network23. Thiourea 25 has also been widely invoked as a prebiotically plausible reagent34. Thus, we envision that a primordial environment supplied with adenine and water would continuously generate TAP, which can be enriched in aqueous solution by moving down a thermal gradient. Ammonium thiocyanate 24 can be mixed with the TAP at any stage, and eventual evaporation and dry state reaction leads to 8-mercaptoadenine 16. This method of accumulation of TAP also improves the plausibility of some aspects of other prebiotic syntheses12.
With thioanhydropurine nucleosides 18 and 19 in hand, we moved on to evaluate their photoreduction chemistry to see if we might directly generate deoxyadenosine. Our previous synthesis of a deoxypyrimidine via a thioanhydropyrimidine 6 (Fig. 1) proceeded by the reduction of a C–S to a C–H bond mediated by a hydrated electron, generated by UV irradiation of hydrosulfide5, 33. 18 and 19 were separately subjected to UV irradiation at 254 nm in water with hydrogen sulfide (H2S) as the reductant (Fig. 2). In the photoreduction of 18, the natural regioisomer N 9-deoxyadenosine 7 (dA) was detected in 39% yield, along with 15% of 8-mercapto-deoxyadenosine 26. 26 was demonstrated to be a competent intermediate in the reaction by desulfurization to give 7 (dA) either by UV irradiation35, or treatment with nitrous acid, which is produced from common atmospheric gases, nitrogen and carbon dioxide36. Nucleobase loss was also apparent (8-mecaptoadenine 16 in 10% yield and adenine 8 in 17% yield). The same reaction starting with 19 gave N 7-deoxyadenosine 20 in 23% yield with no other nucleoside products. Our proposed intermediate in this process, 8-mercapto-N 7-deoxyadenosine 27, is either fully converted to 20 or photochemically destroyed. Photoreduction was also carried out on a mixture of 18 and 19 compatible with our synthesis by tethered transglycosylation. The ratio of N 9:N 7 regioisomers was increased from 38:62 of 18:19 in the starting mixture to 56:44 of 7:20 after photoreduction (31% yield for 7, 17% yield for 20), indicating an enhanced stability of intermediates or products bearing the natural N 9 glycosidic linkage, compared to N 7 isomers. Replacing hydrosulfide as the electron donor with bisulfite (HSO3 -, pH 7)37, which is readily formed by the dissolution of atmospheric SO2 in water38, improved both the yield and selectivity of photoreduction. Photoreduction with bisulfite of 18 alone provided deoxyadenosine 7 (dA) in 51% yield and 8-mercapto-deoxyadenosine 26 in 23% yield, while a similar reaction with the N 7-regio-isomer 19 led only to its photochemical destruction. Photoreduction of a mixture of 18 and 19 with bisulfite led only to N 9-linked products, 7 and 26 in 44% and 18% yield respectively (Extended Data Fig. 3). Separate experiments probing the stability of starting materials and products under the reaction conditions indicated that the relative stabilities of intermediates are the cause of this selectivity. This strikingly selective destruction is highly suggestive of a potential mechanism by which primordial nucleosides were restricted to a near-canonical set39, 40. We found further evidence for such restriction in the hydrolysis rates of the N 9 and N 7 isomers of deoxyadenosine. In acetate buffer (pH 4, room temperature), the natural isomer 7 (dA) is more than 70 times more stable than 20 ( half-lives of 1565 and 22 hours respectively), which is consistent with the reported difference in stabilities towards acid hydrolysis between the corresponding isomers of adenosine41, 42.
Photoreduction Mechanism
To provide mechanistic rationale for the observed photochemical selectivity, we performed quantum chemical calculations using density functional theory and algebraic diagrammatic construction to the second order [ADC(2)] methods43, 44. These calculations revealed, in the case of bisulfite, two possible competing mechanisms that explain the difference in reactivity of the two regioisomers. 18 and 19 can both undergo photoexcitation, but generate dissimilar biradical species (Fig. 3a). Photoexcitation of 18 leads to rupture of the C2'–S bond on the surface of the lowest excited singlet (S1) state, generating biradical 28 (Fig 3a, N 9; Extended Data Fig. 4a). Reduction of this species by intermolecular hydrogen atom transfer (HAT) or proton-coupled electron transfer (PCET) is likely to lead to C2'-reduced species 29, and ultimately, via a second HAT or PCET and subsequent photolysis of the C8–S bond of 26 35, deoxyadenosine 7 (dA) (Fig. 3a, N 9). In contrast, photoexcitation of 19 leads to N7–C8 bond rupture through the S1/S0 state crossing (Fig. 3a, N 7; Extended Data Fig. 4b), generating 30, which is likely to undergo decomposition without C2'–S reduction. Since bisulfite is well-known to provide a hydrated electron upon irradiation45, a second possibility is the reduction by hydrated electrons of 18 and 19 in the ground state. Again, calculations suggest different fates of 18 and 19 upon reduction. Reduction of 18 is predicted to proceed with concomitant barrierless C2'–S bond rupture to give radical anion intermediate 31 (Fig. 3b, N 9; Extended Data Fig. 5) whereas reduction of 19 is predicted to lead to formation of a C8, N9 radical anion 32 which also is likely to undergo decomposition rather than C2' reduction (Fig. 3b N 7, Extended Data Fig. 5). In the absence of any reducing agent, both 18 and 19 undergo (equally) slow photochemical decomposition, presumably via the calculated biradical structures 28 and 30, but in the presence of bisulfite, reduction of the ground state or photochemically generated intermediates results in remarkably different fates.
Fig. 3. Proposed mechanism of photoreduction of N 7-8,2'-anhydro-thioadenosine 18 and N 9-8,2'-anhydro-thioadenosine 19 nucleosides.
a) Potential mechanism involving bisulfite proceeding with initial photoexcitation of the thioanhydronucleosides to 28, followed by reduction of C2', sulfur, and C8. Photoexcitation of the N 7 isomer 19 to 30 leads to decomposition. b) Potential mechanism involving bisulfite proceeding via intial reduction of ground state thioanhydronucleosides, followed by desulfurisation of 26. Reduction of 19 gives 32 which leads to decomposition. c) Distinct mechanism involving reduction of thioanhydronucleoside–hydrosulfide encounter complexes, 33 and 34, which both undergo charge transfer and concomitant C–S bond cleavage to produce 31 and 35. 31 and 35 undergo reduction at C2' and desulfurisation to furnish 7 (dA) and 20.
The successful reduction of 19 alongside 18 when using hydrosulfide as the reducing agent is explained by a distinct mechanism. Calculations located stable encounter complexes, 33 and 34, between HS- and thioanhydronucleosides 18 and 19, respectively (Fig. 3c, Extended Data Fig. 4c and 4d). This interaction is predominantly stabilized by electrostatic and dispersion interactions and our interaction energy decomposition demonstrates its stability in aqueous solution (see the SI for detail). Similar S…S interactions were recently identified in intramolecular complexes and were classified as chalcogen bonds46. Such an encounter complex facilitates charge transfer (CT) from the hydrosulfide anion to the thioanhydropurine fragment almost immediately after UV absorption by the complex to the S1 state. Subsequent relaxation on the S1 surface enables practically barrierless C2'–S bond breaking completed by a peaked S1/S0 state crossing for both intermediates 31 and 35, thus facilitating C2'–S reduction of both 18 and 19 (Extended Data Fig. 4c and 4d). The products of this photochemical transformation, 26 and 27, may further undergo photochemical sulfur cleavage through the mechanism described by Roberts et al.35 (Fig. 3c). Thus, a HS-thioanhydropurine encounter complex facilitates C–S bond cleavage and partially protects N 7 isomer 19 from the photodestruction observed in the presence of bisulfite. This finding not only explains the distinctive outcomes of photoreduction between the two reducing agents, but also points towards a potentially important stabilising role for hydrosulfide in prebiotic chemistry and photochemistry in general.
Prebiotic Route to A Purine/Pyrimidine Genetic System
Whilst our attempts to glycosylate 8-mercaptoguanine 17 to provide thioanhydroguanosine (and ultimately deoxyguanosine) failed, the triple selectivity and high yield of our route to deoxyadenosine combined with recent results from the Szostak group47 suggest a possible alternative genetic alphabet that does not include (deoxy)guanosine. Guanosine is yet to succumb to a plausible prebiotic synthesis, but Szostak et al. have recently shown that inosine (I), which is capable of base-pairing with cytosine, can replace guanosine in non-enzymatic RNA replication systems with no loss of rate or fidelity. (Deoxy)adenosine 7 (dA) is readily converted to (deoxy)inosine 9 (dI) (Fig. 2) by deaminative hydrolysis, which spontaneously occurs very slowly in nucleic acid polymers48, and is greatly accelerated by the presence of nitrous acid49. To demonstrate that this conversion can occur under mild conditions consistent with our primordial geochemical scenario50, we treated deoxyadenosine 7 (dA) with nitrous acid at pH 4 (the same conditions by which we could effect desulfurization of 26). After four days at room temperature, approximately 40% of 7 (dA) had been converted to 9 (dI), providing a 60:40 mixture of 7 (dA) and 9 (dI) (Fig 2). A control experiment monitoring the decomposition of deoxyadenosine 7 (dA) at pH 4, without nitrous acid, showed only a trace of depurination (t1/2 = 1600 h). When a 67:33 mixture of 7 and 26, representative of the outcome of photoreduction, was submitted to the reaction conditions, 26 underwent relatively rapid desulfurization first, with deoxyadenosine 7 (dA) undergoing slower deaminative hydrolysis to ultimately provide a 65:35 mixture of 7 (dA) and 9 (dI). Thus, mixtures of deoxyadenosine 7 (dA) and deoxyinosine 7 (dA) are readily obtainable from partial deaminative hydrolysis of deoxyadenosine 7 (dA) or its precursor 26, thereby supplying half of a potential primordial alphabet. Despite the potential for a mismatch in reactivity between deoxypurines and pyrimidines, a 47:53 mixture of deoxyadenosine 7 (dA) and cytidine 1 (C) underwent nitrous acid-promoted deamination to provide all four (deoxy)nucleosides deoxyadenosine 7 (dA), deoxyinosine 9 (dI), cytidine 1 (C), and uridine 2 (U) (30:17:42:11 ratio) (Extended Data Fig. 6). A similar primordial mixture may have been a starting point for genetic information storage. Furthermore, in the absence of significant geochemically plausible sources of pyrimidine deoxynucleotides and purine ribonucleotides, heteropolymers made from a mixture of purine deoxyribonucleotides and pyrimidine ribonucleotides should possess heritable backbone heterogeneity and thus a 1:1 phenotype to genotype correspondence, which is potentially advantageous in the evolution of catalytic activity18.
Systems-level Prebiotic Plausibility
Having demonstrated the potential of a divergent route to yield a local mixture of 7 (dA), 9 (dI), 1 (C) and 2 (U), we sought to evaluate the key question of whether all four nucleosides could persist after divergence in the sequence. We chose a 1:1 mixture of α– and β–2-thiocytidines 13 and 3 as our starting point, which could be obtained from the partial photoanomerisation of 13, and evaluated two particular combinations of reactions as representative permutations of a primordial geochemical process (Fig. 4, Route A and B). In route A, exposure of the mixture to nitrous acid (pH 4) generates a mixture of 12 and 1 (100% yield for 12 from 13, 54% yield for 1 from 3). 12 is potentially formed from 13 by intramolecular addition of the C2' hydroxyl to C2 of an S-nitrosyl intermediate, and subsequent elimination of SNO-. Dry state glycosylation of 16 and a 1:1 mixture of 12 and 1 (C), in the presence of MgCl2, leads to a mixture of 18 and 19 as described in our route development above, however, critically, 95% of 1 persists in this mixture. Subsequent photoreduction in the presence of ferrocyanide and bisulfite generates the expected mixture of purine nucleosides 7 (dA), 26, 20 and 27 alongside 1 (C). Finally, a second exposure to nitrous acid converts this mixture into the components of a competent genetic system, 7 (dA), 9 (dI) (10% and 9% yield respectively from 12 for 3 steps), 1 (C) and 2 (U) (84% combined persistence after 3 steps) with no significant nucleoside impurities. Products derived from 19 – with the wrong N 7 regiochemistry – are hydrolysed in the last step. It is noteworthy that this route is only viable from a systems level approach – for instance, the pyrimidines are fairly rapidly destroyed in the photoreduction step in the absence of the thioanhydropurines (Extended Data Fig. 7). Route B presents an alternative in which initial hydrolysis of the mixture of 13 and 3 generates glycosyl donor 15 (26% yield) alongside pyrimidine nucleosides (4% of 1 (C), 2% of 2 (U), 92% 3 remaining). 3 has previously been shown to hydrolyse to 1 (C) and 2 (U) in greater yields (44%) over longer periods7. A representative mixture of 15, 1 (C) and 2 (U) (2:1:1) was then subjected to tethered glycosylation, resulting in 18 and 19 as above (30% and 50% yield respectively) with 80% and 95% persistence of 1 (C) and 2 (U). Photoreduction of the mixture, this time with hydrogen sulfide, provides purine products 7, 26, 20 and 27 alongside the pyrimidines 1 (C) and 2 (U). Finally, nitrosation furnished the key mixture of 7 (dA) and 9 (dI) (6% for each from 15 for three steps) alongside pyrimidine nucleosides (43% persistence over 3 steps, final ratio of dA:dI:C:U in the mixture is 14:13:45:28, Extended Data Fig. 8). Thus, sequences comprised of various orders of operations and various photoreduction conditions, which might plausibly emulate a terrestrial geochemical scenario, generate the components of a mixed genetic system alongside one another. The exact ratio of 1 (C) and 2 (U) (ribosylpyrimidines) to 7 (dA) and 9 (dI) (deoxyribosylpurines) in the final mixture will depend on the ratio of α– (anhydro)pyrimidines (13, 12, and 15) to β–(thio)pyrimidines (1, 2 and 3) earlier in the sequence, which will vary based on environmental conditions.
Fig. 4. A systems-level approach to a potential primordial genetic alphabet composed of 1 (C), 2 (U), 7 (dA) and 9 (dI).
A mixture of the α– and β–epimers of 2-thiocytidine 13 and 3, which interconvert in UV light, can generate a mixture containing 1 (C), 2 (U), 7 (dA) and 9 (dI). A general route is shown at left. The thiopyrimidines are initially converted into the canonical pyrimidines (cytidine 1 and uridine 2) and the α-anhydropyrimidines 12 and 15. The latter undergo tethered glycosylation and then photoreduction to selectively provide purine deoxyribonucleosides 7 (dA) and 9 (dI) as depicted in Fig. 2. The pyrimidines 1 (C) and 2 (U) persist through each step of this sequence, ultimately generating a mixture of all four nucleosides. Specific conditions and yields for two possible particular routes (Routes A and B) are shown at right.
In conclusion, a highly efficient synthesis of both deoxyadenosine 7 (dA) and deoxyinosine 9 (dI), requiring only prebiotically plausible reagents and conditions, is reported. In contrast to all previous attempts to synthesize purine nucleosides, our synthesis is both prebiotically plausible and strictly stereo-, regio-, and furanosyl-selective for the only isomer of the deoxypurine nucleosides used in modern biology. The pathway proceeds mostly via simple hydrolysis or dry state processes, with a key reduction step promoted by UV irradiation supported by distinct mechanisms. The (photo)chemical selection exhibited by this route hints at an explanation for Nature’s choice of one isomer of nucleic acid from the many that are conceivable. Critically, we have demonstrated that sequences leading selectively to both RNA pyrimidine and DNA purine nucleosides can occur together simultaneously, providing mixtures which could conceivably complete a genetic alphabet. The fact that DNA building blocks can be co-produced with the RNA pyrimidine nucleosides is consistent with and perhaps evidence for the coexistence of RNA and DNA building blocks at the dawn of life.
Extended Data
Extended Data Fig. 1. A summary of the main findings of the work.
Previously, a prebiotically plausible synthesis of beta-ribopyrimdines C and U has been identified using α–-thiocytidine. Herein, we demonstrate that the same intermediate can undergo a distinct prebiotically plausible process that could have happened in a similar, or the same, environment. The new process furnishes β–D-N 9-deoxyribopurine nucleosides, dA and dI, alongside the pyrimidines. Remarkable selectivity enforced by UV irradiation and hydrolysis operates throughout the reported ribosylpyrimidine synthesis and the newly discovered deoxyribosylpurine synthesis, resulting in a set of nucleosides with only the canonical regio- and stereochemistry. The coexistence in one location of a set of nucleosides similar to this is thought by many to be a precondition for the spontaneous emergence of life on Earth.
Extended Data Fig. 2. 1H NMR spectra of conversion of α-anhydrouridine 15 from α-thiouridine 14.
a) 1H NMR spectrum of α-anhydrouridine 15; b) 1H NMR spectrum of the reaction mixture after heating α-thiouridine 14 in H2O; c) 1H NMR spectrum of the reaction mixture after heating α-thiouridine 14 in formamide.
Extended Data Fig. 3. 1H NMR spectra of photoreduction of N 7-8,2'-anhydro-thioadenosine 18 and N 9-8,2'-anhydro-thioadenosine 19 mixture with bisulfite.
a) 1H NMR spectrum of the crude mixture before irradiation (the ratio of N 7 : N 9 isomer was 4 : 5); b) 1H NMR spectrum of the mixture after irradiation for 7 hrs (the N 9 isomers dA 7 and 26 are the only detectable products).
Extended Data Fig. 4. Potential energy surfaces and S1/S0 state crossings of the key photochemical steps in deoxyadenosine synthesis calculated at the ADC(2)/madef2-TZVP level (see the SI for more details).
a) C-S bond opening may spontaneously occur in 18 leading to a peaked S1/S0 state crossing, however, a reducing agent is necessary to maintain that geometry after reaching the S0 state; b) N7-C8 bond rupture is the lowest energy photochemical process in 19 and results in destruction of the purine ring; c) and d) encounter complexes of 18 and 19 with HS-, which readily undergo photochemical C–S bond rupture induced by charge transfer from HS- to chromophore.
Extended Data Fig. 5. Equilibrium geometries of C2, S8 radical anion 31 and C8, N9 radical anion 32 radical anions which may be formed after accepting a hydrated electron from the environment and the adiabatic electron affinities calculated at the ωB97X-D/IEFPCM/ma-def2-TZVP.
Extended Data Fig. 6. 1H NMR spectra for the reactions of deoxyadenosine 7 and cytidine 1 with nitrous acid.
a) 1H NMR spectrum of the mixture of deoxyadenosine 7 and cytidine 1; b) 1H NMR spectrum of the reaction mixture after 4 days, showing the ratio of all four (deoxy)nucleosides deoxyadenosine 7, deoxyinosine 9, cytidine 1, and uridine 2 is 30:17:42:11.
Extended Data Fig. 7. 1H NMR spectra for stability study of cytidine 1 and uridine 2 at 254 nm irradiation with bisulfite.
a) 1H NMR spectrum of the mixture of cytidine 1, bisulfite and K4Fe(CN)6 in the dark; b) as a), 1H NMR spectrum after 10 hours of irradiation; c) 1H NMR spectrum of the mixture of uridine 2, bisulfite and K4Fe(CN)6 in the dark; d) as c), 1H NMR spectrum after 10 hours of irradiation; e) 1H NMR spectrum of the mixture of cytidine 1, uridine 2, N 9-thioanhydroadenosine 18, bisulfite and K4Fe(CN)6 in the dark; f) as e), 1H NMR spectrum after 10 hours of irradiation.
Extended Data Fig. 8. 1H NMR spectra for sequential reactions with the mixture of α-anhydrouridine 15, cytidine 1 and uridine 2.
a) 1H NMR spectrum of the mixture after heating with 8-mercaptoadenine 16 and magnesium chloride at 150 °C for 1.5 days; b) 1H NMR spectrum of the same mixture after irradiation with hydrogen sulfide at 254 nm; c) 1H NMR spectrum of the same mixture after reacting with nitrous acid for 2 days (dA 7: dI 9: C 1: U 2= 14: 14: 44: 28).
Supplementary Material
Acknowledgments
The authors thank all JDS group members for fruitful discussions. This research was supported by the Medical Research Council (MC_UP_A024_1009), the Simons Foundation (290362 to JDS, 494188 to RS), and a grant from the National Science Centre Poland (2016/23/B/ST4/01048 to RWG). MJJ acknowledges support within the “Diamond Grant” (0144/DIA/2017/46) from the Polish Ministry of Science and Higher Education and a computational grant from Wroclaw Centre of Networking and Supercomputing (WCSS).
Footnotes
Author contributions: Experimental: J. X., V. C., N. J. G., D. A. R., A. D. B.; Theoretical: M. J. J., R. W. G., R. S.; Crystallography: A. D. B.; Supervision: J. D. S.; all authors co-wrote the manuscript.
Competing interests: The authors declare no competing financial interests.
Reprints and permission information is available at http://www.nature.com/reprints.
Data and materials availability
Supplementary Information is available containing all procedures, characterization data, NMR spectra, HPLC traces, X-Ray data and CCDC numbers, and theoretical methods and data. Any additional data are available from the corresponding author upon reasonable request.
Code availability
All custom code used to generate the data in this study is available upon reasonable request.
References
- 1.Samanta B, Joyce GF. A reverse transcriptase ribozyme. Elife. 2017;6 doi: 10.7554/eLife.31153. e31153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert W. Origin of life: The RNA world. Nature. 1986;319:618. [Google Scholar]
- 3.Joyce GF. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 4.Bhowmik S, Krishnamurthy R. The role of sugar-backbone heterogeneity and chimeras in the simultaneous emergence of RNA and DNA. Nat Chem. 2019;11:1009–1018. doi: 10.1038/s41557-019-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Green NJ, Gibard C, Krishnamurthy R, Sutherland JD. Prebiotic phosphorylation of 2-thiouridine provides either nucleotides or DNA building blocks via photoreduction. Nat Chem. 2019;11:457–462. doi: 10.1038/s41557-019-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powner MW, Gerland B, Sutherland JD. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, et al. A prebiotically plausible synthesis of pyrimidine beta-ribonucleosides and their phosphate derivatives involving photoanomerization. Nat Chem. 2017;9:303–309. doi: 10.1038/nchem.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heuberger BD, Pal A, Del Frate F, Topkar VV, Szostak JW. Replacing uridine with 2-thiouridine enhances the rate and fidelity of nonenzymatic RNA primer extension. J Am Chem Soc. 2015;137:2769–2775. doi: 10.1021/jacs.5b00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walton T, Szostak JW. A highly reactive imidazolium-bridged dinucleotide intermediate in nonenzymatic RNA primer extension. J Am Chem Soc. 2015;138:11996–12002. doi: 10.1021/jacs.6b07977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, et al. Enhanced nonenzymatic RNA copying with 2-aminoimidazole activated nucleotides. J Am Chem Soc. 2017;139:1810–1813. doi: 10.1021/jacs.6b13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller WD, Orgel LE, Sanchez RA. Studies in Prebiotic Synthesis: VI. Solid-State Synthesis of Purine Nucleosides. J Mol Evol. 1972;1:249–257. doi: 10.1007/BF01660244. [DOI] [PubMed] [Google Scholar]
- 12.Becker S, et al. A high-yielding, strictly regioselective prebiotic purine nucleoside formation pathway. Science. 2016;352:833–836. doi: 10.1126/science.aad2808. [DOI] [PubMed] [Google Scholar]
- 13.Kim H, Benner SA. Prebiotic stereoselective synthesis of purine and noncanonical pyrimidine nucleotides from nucleobases and phosphorylated carbohydrates. Proc Nat Acad Sci USA. 2017;114:11315–11320. doi: 10.1073/pnas.1710778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker S, et al. Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides. Science. 2019;366:76–82. doi: 10.1126/science.aax2747. [DOI] [PubMed] [Google Scholar]
- 15.Teichert JS, Kruse FM, Trapp O. Direct prebiotic pathway to DNA nucleosides. Angew Chem Int Ed. 2019;55:9944–9947. doi: 10.1002/anie.201903400. [DOI] [PubMed] [Google Scholar]
- 16.Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 17.Leu K, Obermayer B, Rajamani S, Gerland U, Chen IA. The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res. 2011;39:8135–8147. doi: 10.1093/nar/gkr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutherland JD, Whitfield JN. Prebiotic chemistry: a bioorganic perspective. Tetrahedron. 1997;53:11493–11527. [Google Scholar]
- 19.Trevino SG, Zhang N, Elenko MP, Lupták A, Szostak JW. Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc Nat Acad Sci USA. 2011;108:13492–13497. doi: 10.1073/pnas.1107113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavette JV, Stoop M, Hud NV, Krishnamurthy R. RNA–DNA chimeras in the context of an RNA world transition to an RNA/DNA world. Angew Chem Int Ed. 2016;55:13204–13209. doi: 10.1002/anie.201607919. [DOI] [PubMed] [Google Scholar]
- 21.Schoffstall AM. Prebiotic phosphorylation of nucleosides in formamide. Orig Life. 1976;7:399–412. doi: 10.1007/BF00927935. [DOI] [PubMed] [Google Scholar]
- 22.Lohrmann R, Orgel LE. Urea-Inorganic Phosphate Mixtures as Prebiotic Phosphorylating Agents. Science. 1971;171:490–494. doi: 10.1126/science.171.3970.490. [DOI] [PubMed] [Google Scholar]
- 23.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat Chem. 2015;7:301–307. doi: 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishiwata A, Lee YJ, Ito Y. Recent advances in stereoselective glycosylation through intramolecular aglycon delivery. Org Biomol Chem. 2010;8:3596–3608. doi: 10.1039/c004281a. [DOI] [PubMed] [Google Scholar]
- 25.Springsteen G, Joyce GF. Selective derivatization and sequestration of ribose from a prebiotic mix. J Am Chem Soc. 2004;126:9578–9583. doi: 10.1021/ja0483692. [DOI] [PubMed] [Google Scholar]
- 26.Anastasi C, Crowe MA, Powner MW, Sutherland JD. Direct Assembly of Nucleoside Precursors from Two- and Three-Carbon Units. Angew Chem Int Ed. 2006;45:6176–6179. doi: 10.1002/anie.200601267. [DOI] [PubMed] [Google Scholar]
- 27.Vorbrüggen H, Ruh-Pohlenz C. Handbook of nucleoside synthesis. Wiley; 2001. [Google Scholar]
- 28.Holm NG, Oze C, Mousis O, Waite JH, Guilbert-Lepoutre A. Serpentinization and the formation of H2 and CH4 on celestial bodies (planets, moons, comets) Astrobiology. 2015;15:587–600. doi: 10.1089/ast.2014.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez RA, Ferris JP, Orgel LE. Studies in prebiotic synthesis II: Synthesis of purine precursors and amino acids from aqueous hydrogen cyanide. J Mol Biol. 1967;80:223–253. [PubMed] [Google Scholar]
- 30.Hudson JS, et al. A unified mechanism for abiotic adenine and purine synthesis in formamide. Angew Chem Int Ed. 2012;51:5134–5137. doi: 10.1002/anie.201108907. [DOI] [PubMed] [Google Scholar]
- 31.Giner-Sorolla A, Thom E, Bendich A. Studies on the Thiation of Purines. J Org Chem. 1964;29:3209–3212. [Google Scholar]
- 32.Levy M, Miller SL. The stability of the RNA bases: implications for the origin of life. Proc Natl Acad Sci USA. 1998;95:7933–7938. doi: 10.1073/pnas.95.14.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritson DJ, Sutherland JD. Synthesis of aldehydic ribonucleotide and amino acid precursors by photoredox chemistry. Angew Chem Int Ed. 2013;52:5845–5847. doi: 10.1002/anie.201300321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson MP, Levy M, Miller SL. Prebiotic synthesis of diaminopyrimidine and thiocytosine. J Mol Evol. 1996;43:543–550. doi: 10.1007/BF02202102. [DOI] [PubMed] [Google Scholar]
- 35.Roberts SJ, et al. Selective prebiotic conversion of pyrimidine and purine anhydronucleosides into Watson-Crick base-pairing arabino-furanosyl nucleosides in water. Nat Commun. 2018;9:4073–4082. doi: 10.1038/s41467-018-06374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranjan S, Todd ZR, Rimmer PB, Sasselov DD, Babbin AR. Nitrogen oxide concentrations in natural waters on early Earth. Geochem Geophy Geosy. 2019;20:2021–2039. [Google Scholar]
- 37.Xu J, et al. Photochemical reductive homologation of hydrogen cyanide using sulfite and ferrocyanide. Chem Commun. 2018;54:5566–5569. doi: 10.1039/c8cc01499j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marion GM, Kargel JS, Crowley JK, Catling DC. Sulfite–sulfide– sulfate–carbonate equilibria with applications to Mars. Icarus. 2013;225:342–351. [Google Scholar]
- 39.Rios AC, Tor Y. On the origin of the canonical nucleobases: an assessment of selection pressures across chemical and early biological evolution. Isr J Chem. 2013;53:469–483. doi: 10.1002/ijch.201300009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rios AC, Yu HT, Tor Y. Hydrolytic fitness of N-glycosyl bonds: comparing the deglycosylation kinetics of modified, alternative, and native nucleosides. J Phys Org Chem. 2014;28:173–180. doi: 10.1002/poc.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panzica RP, Rousseau RJ, Robins RK, Townsend LB. Relative stability and a quantitative approach to the reaction mechanism of the acid-catalyzed hydrolysis of certain 7-and 9-β-D-ribofuranosylpurines. J Am Chem Soc. 1972;94:4708–4714. doi: 10.1021/ja00768a045. [DOI] [PubMed] [Google Scholar]
- 42.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 43.Hättig C. Structure Optimizations for Excited States with Correlated Second-Order Methods: CC2 and ADC(2) Adv Quantum Chem. 2005;50:37–60. [Google Scholar]
- 44.Dreuw A, Wormit M. The algebraic diagrammatic construction scheme for the polarization propagator for the calculation of excited states. Wiley Interdiscip Rev Comput Mol Sci. 2015;5:82–95. [Google Scholar]
- 45.Sauer MC, Crowell RA, Shkrob IA. Electron Photodetachment from Aqueous Anions. 1. Quantum Yields for Generation of Hydrated Electron by 193 and 248 nm Laser Photoexcitation of Miscellaneous Inorganic Anions. The Journal of Physical Chemistry A. 2004;108:5490–5502. [Google Scholar]
- 46.Pascoe DJ, Ling KB, Cockroft SL. The origin of chalcogen-bonding interactions. J Am Chem Soc. 2017;139:15160–15167. doi: 10.1021/jacs.7b08511. [DOI] [PubMed] [Google Scholar]
- 47.Kim SC, O’Flaherty DK, Zhou L, Lelyveld VS, Szostak JW. Inosine, but none of the 8-oxo-purines, is a plausible component of a primordial version of RNA. Proc Natl Acad Sci USA. 2018;115:13318–13323. doi: 10.1073/pnas.1814367115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karran P, Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 49.Shapiro R, Pohl SH. Reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry. 1968;7:448–455. doi: 10.1021/bi00841a057. [DOI] [PubMed] [Google Scholar]
- 50.Mariani A, Russell DA, Javelle T, Sutherland JD. A light-releasable potentially prebiotic nucleotide activating agent. J Am Chem Soc. 2018;140:8657–8661. doi: 10.1021/jacs.8b05189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Information is available containing all procedures, characterization data, NMR spectra, HPLC traces, X-Ray data and CCDC numbers, and theoretical methods and data. Any additional data are available from the corresponding author upon reasonable request.
All custom code used to generate the data in this study is available upon reasonable request.