Abstract
Cell competition allows “winner” cells to eliminate less fit “loser” cells in tissues. In Minute cell competition, cells heterozygous mutant in ribosome genes, such as RpS3 +/- cells, are eliminated by wild-type cells. How cells are primed as losers is partially understood and it has been proposed that reduced translation underpins the loser status of ribosome mutant, or Minute, cells. Here, using Drosophila, we show that reduced translation does not cause cell competition. Instead, we identify proteotoxic stress as the underlying cause of the loser status for Minute competition and competition induced by mahjong, an unrelated loser gene. RpS3 +/- cells exhibit reduced autophagic and proteasomal flux, accumulate protein aggregates, and can be rescued from competition by improving their proteostasis. Conversely, inducing proteotoxic stress is sufficient to turn otherwise wild-type cells into losers. Thus, we propose that tissues may preserve their health through a proteostasis-based mechanism of cell competition and cell selection.
Keywords: Cell competition, ribosome mutation, ribosomopathy, proteotoxic stress, autophagy, proteasome, Drosophila, aneuploidy, FOXO, Rapamycin
Introduction
Cell competition is a conserved mechanism that allows “winner” cells to eliminate viable but less fit “loser” cells in tissues1–3 This process acts as a mechanism of tissue quality control. By removing mis-specified or damaged cells, cell competition preserves tissue and organism health, potentially delaying ageing and disease onset4–6 Furthermore, an increasing body of evidence indicates that competitive interactions contribute to tissue colonisation during cancer growth7.
The first form of competition discovered was Minute cell competition, wherein cells heterozygous mutant in ribosome genes are eliminated by neighbouring wild-type cells1. Over 80 genes make up the ribosome, and most display a dominant phenotype when mutated or lost, both in Drosophila and humans8,9 Based both on phenotypic dominance and on the high number of Minute genes, spontaneously occurring Minute cell competition is likely to be a frequent event, relative to other types of cell competition. In addition, as ribosome genes are scattered across chromosomes, Minute cell competition may be frequent in diseases characterized by aneuploidy10, such as cancer, where deletions of large genomic regions often lead to single copy loss of one or more ribosome genes11.
Despite its discovery over 40 years ago1, our understanding of the mechanisms of Minute cell competition remains incomplete12. While several signals have been identified that act during cell competition4,13–19, the upstream signals priming cells as losers are mostly unknown20. It is, for instance, unclear how ribosome gene loss leads to the loser status12. Minute mutants exhibit reduced translation rate17, and it has long been assumed that this drives the loser status18,21–25 However, the actual contribution of translation has not been investigated.
Here, we investigated how ribosome mutations lead to the loser status. We find that translation is not directly linked to the loser status in Minute competition. Instead, we find that ribosome gene mutations lead to defective autophagy and proteasome flux, accumulation of protein aggregates, and proteotoxic stress. These phenotypes are causative of the loser status. In addition, inducing proteotoxic stress through overexpression of aggregate-prone proteins phenocopies these protein catabolism defects and induces the loser status. Our work identifies proteotoxic stress as the leading cause of the Minute loser status and implicates cell competition in pathologies characterized by proteotoxic stress.
Results
Reduced protein synthesis does not confer the loser status
Minute cell competition is characterized by apoptotic elimination of Minute loser cells when they are in proximity of wild-type winner cells1–3 Thus, although Minute RpS3 +/- cells display a modest increase in apoptosis compared to wild-type cells when they are in isolation (Figure 1a–b and26), apoptosis is substantially elevated during competition in RpS3 +/- cells that border wild-type cells12,27,28 (Figure 1c–d). This region-specific induction of apoptosis at clone borders is a hallmark of certain types of cell competition, including Minute competition.
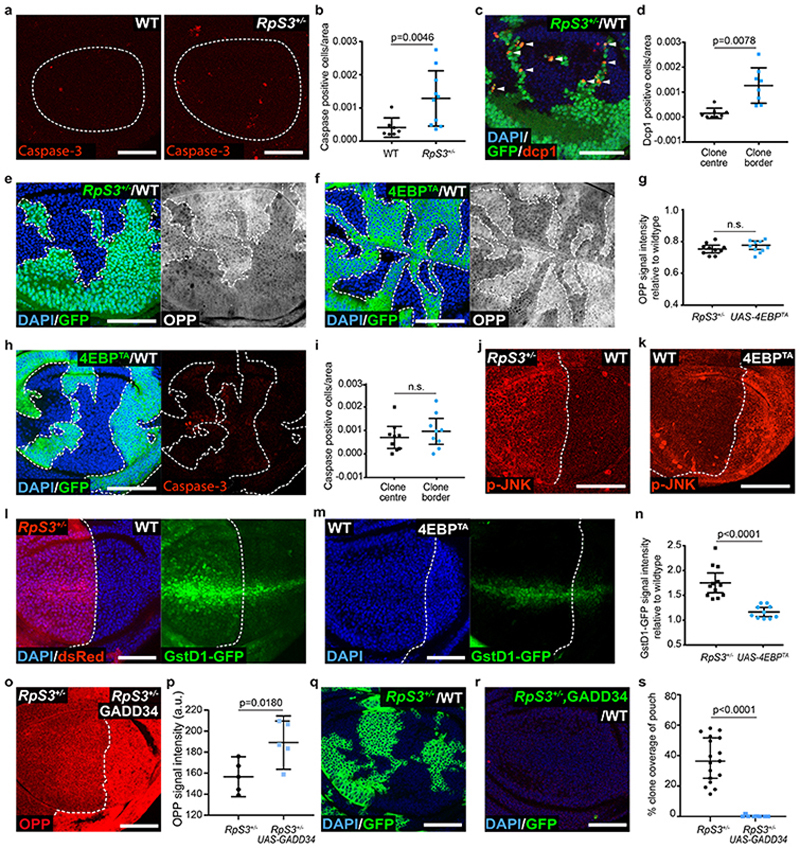
Figure 1. Reduced protein synthesis does not confer the loser status.
(a-b) Apoptosis detection by cleaved caspase-3 staining (red) in wild type or RpS3 +/- non-competing (homotypic) wing discs (a) and corresponding quantification (n=7 and 10, respectively, two-sided Mann-Whitney U Test) (b). (c-d) Apoptosis detection by dcp-1 staining (red) in competing wing discs containing RpS3 +/- cells (GFP-positive) and unlabeled wild type cells (GFP-negative) (c) and corresponding quantification (n=8, two-sided Wilcoxon signed-rank test) (d). (e-g) Translation rate measurement by OPP in wing discs containing wild-type cells and RpS3 +/- clones (GFP-positive) (e) or 4E-BPTA-expressing clones (GFP-positive) (f). Corresponding quantifications are in (g) (n=10 and 10 respectively, two-sided two sample Kolmgorov-Smirnov test). (h-i) Apoptosis detection by cleaved caspase-3 staining (red) in wing discs with mosaic expression of 4E-BPTA (GFP-positive) (h), and corresponding cell death quantifications (n=9, two-sided Wilcoxon signed-rank test) (i). (j) Wing disc harboring an RpS3 +/- Anterior (A) and a wild-type Posterior (P) compartments stained for anti-active phospho-JNK (p-JNK, red). (k) Wing disc expressing 4E-BPTA in P compartment stained for p-JNK (red). (l-n) GstD1-GFP signal (green) in wing discs harboring RpS3 +/- A cells (dsRed-positive) and wild-type P cells (dsRed-negative) (l) and in wing discs harboring 4E-BPTA-expressing P and wild-type A cells (m), and corresponding quantification (n=12 and 10 respectively, two-sided two sample Kolmgorov-Smirnov test) (n). (o-p) An RpS3 +/- wing disc over-expressing GADD34 in P cells and labelled with OPP (o), and corresponding quantification (n=5, two-sided paired t-test) (p). (q-s) Wing discs harboring wild-type cells and RpS3 +/- clones (GFP-positive) (q) or RpS3 +/- clones expressing GADD34 (GFP-positive) (r), and corresponding quantification (n=17 and 10 respectively, two-sided Mann-Whitney U test) (s). In this figure, for all micrographs, scale bars correspond to 50μm. All n numbers refer to the number of individual wing discs. In this figure and throughout: dashed lines indicate wing pouch or clonal and compartment boundaries; clone border defines cells within 2-cell diameters of the clone perimeter; Posterior is right and dorsal is up; figure panel genotypes are provided for all figures in Supplementary Table 3; each point in graphs represents one wing disc, unless otherwise indicated. For all quantifications, the horizontal line represents the mean and whiskers indicate 95% confidence intervals.
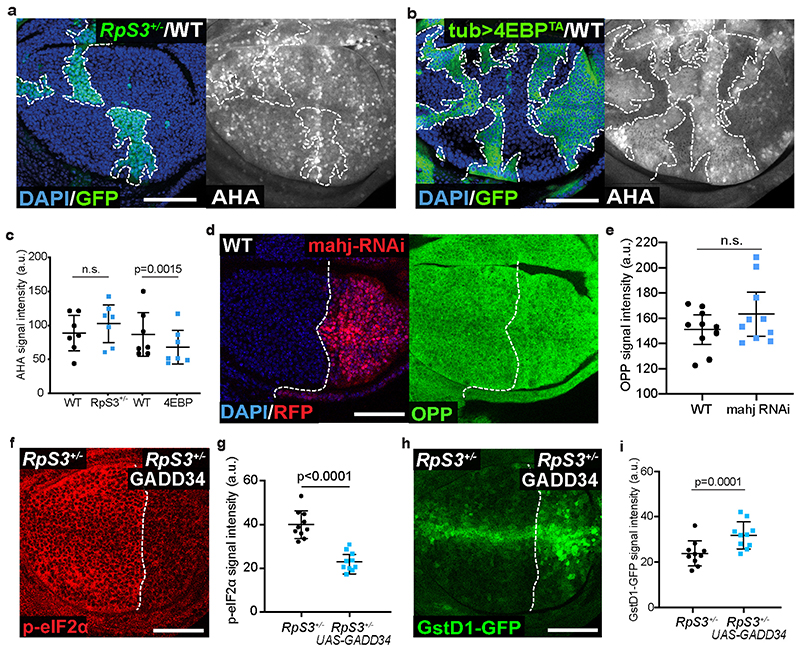
To investigate whether reduced translation triggers cell competition, we expressed a constitutively active form of the translational repressor, 4E-BP (4EBPTA)29,30, in otherwise wildtype cells. In OPP (O-propargyl-puromycin) and AHA (L-azidohomoalanine) global translation assays, 4EBPTA expression induced a reduction in protein synthesis that was comparable to (Figure 1e–g; OPP) or stronger than (Extended Data Figure 1a–c; AHA) that seen in RpS3 +/- cells. 4EBPTA expression resulted in little autonomous apoptosis (Figure 1h). Furthermore, the frequency of dying cells was similar at 4EBPTA clone borders and clone centers (Figure 1h–i). These data suggest that reducing rates of global protein synthesis alone, at levels equal to or greater than in RpS3 +/- cells, is not sufficient to trigger cell competition and indicate that additional properties induced by RpS3 +/- mutations must also play a role.
We have previously shown that RpS3 +/- cells and cells mutant in the loser gene and ubiquitin ligase mahjong 31 (mahj), share what we have termed the ‘prospective loser status’ – a cellular state which predisposes cells to act as losers when confronted with wildtype winners20. This state is characterized by activation of a range of stress response pathways, even in the absence of cell competition20. For example, RpS3 +/- and mahj -/- cells display chronic activation of JNK signaling20,32 and of the Nrf2-mediated oxidative stress response20. Furthermore, Nrf2 activation is sufficient to induce the loser status in competition with wild-type cells20. To determine whether a reduction in protein synthesis is sufficient to activate these pathways, we examined the levels of phospho-JNK and the activation of an Nrf2 reporter, GstD1-GFP, in the absence of competition33. As Minute cell competition does not occur across compartment boundaries, we are able to use compartment-specific transcriptional drivers to generate wing discs with two distinct but non-competing cell populations, one in the anterior compartment and one in the posterior. Similarly to RpS3 +/- cells, the levels of phospho-JNK were higher in wing disc cells expressing 4EBPTA than in the wild-type compartment (Figure 1j–k). However, GstD1-GFP levels were only minimally affected in 4EBPTA cells (Figure 1l–n). Thus, a reduction in protein synthesis can produce some aspects of the prospective loser status (JNK activation) but is insufficient to induce oxidative stress response activity or provoke cell competition.
We next asked whether reduced protein synthesis is necessary for mahj -/- cells or RpS3 +/- cells to behave as losers. Knock-down of Mahj did not affect protein translation rate (Extended Data Figure 1d–e), indicating that translation inhibition does not play a role in priming mahj -/- cells as losers. Next, we sought to boost rates of translation in RpS3 +/- cells and assess the resulting effect on the prospective loser status and on Minute competition. GADD34 can stimulate translation via dephosphorylation of the translation initiation factor, eIF2α34 Indeed, GADD34 overexpression in RpS3 +/- cells caused a reduction in phospho-eIF2α (Extended Data Figure 1f–g) and a corresponding rescue of translation, as assessed by OPP incorporation (Figure 1o–p). Surprisingly, GADD34-expressing RpS3 +/- cells displayed higher levels of the GstD1-GFP oxidative stress reporter (Extended Data Figure 1h–i) and performed worse than RpS3 +/- cells in competition, with hardly any surviving at the point of dissection (Figure 1q–s). Thus, translation inhibition seems to counter the loser status rather than contribute to it, in RpS3 +/- cells.
Prospective losers display dependence on autophagy and defective autophagic flux
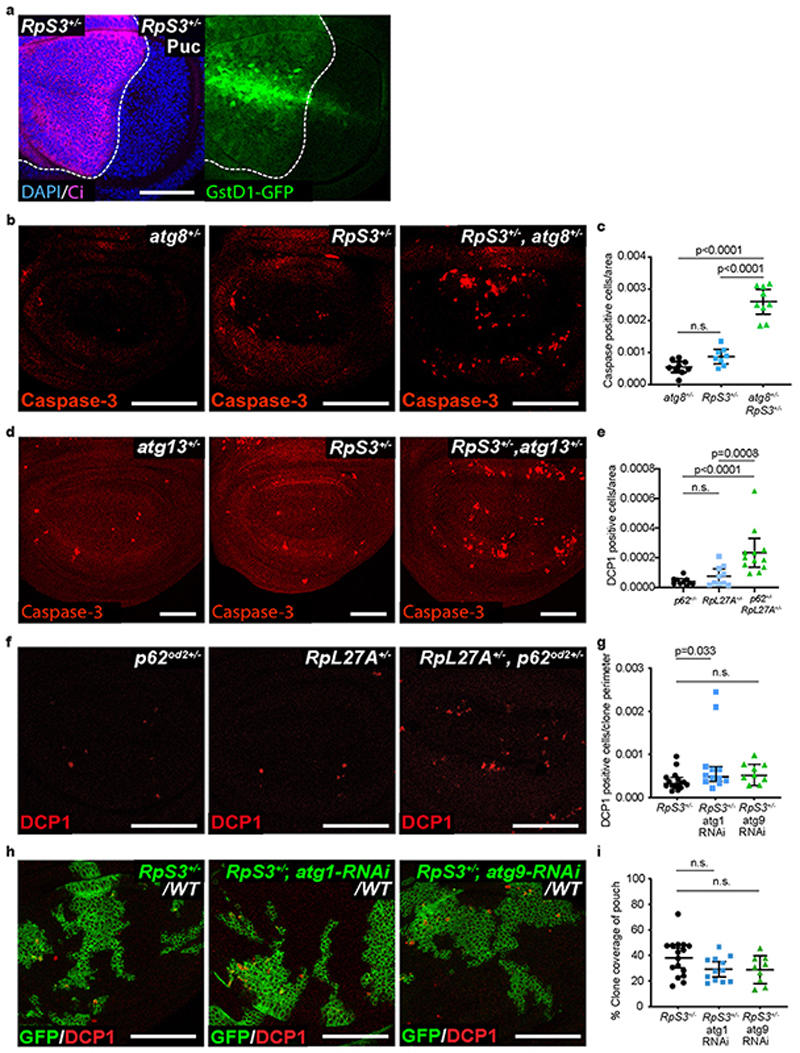
In order to seek out an alternative cause of the prospective loser status, we turned to a known rescue of Minute competition: inhibition of JNK signaling. In addition to rescuing RpS3 +/- cells from competition, JNK inhibition partially reverses activation of the transcriptional signature associated with prospective losers20, Furthermore, it reduces GstD1-GFP reporter activation in RpS3 +/- cells (Extended Data Figure 2a). Thus, we compared the transcriptional profiles of RpS3 +/- wing discs with or without JNK signaling inhibition20, to identify pathways associated with JNK inhibition and with a rescue of the loser status. This revealed differential expression of genes involved in protein catabolism, the proteasome, autophagy, and the unfolded protein response (Supplementary Table 1). These pathways have all been implicated in Nrf2 regulation35,36, supporting a potential role in cell competition.
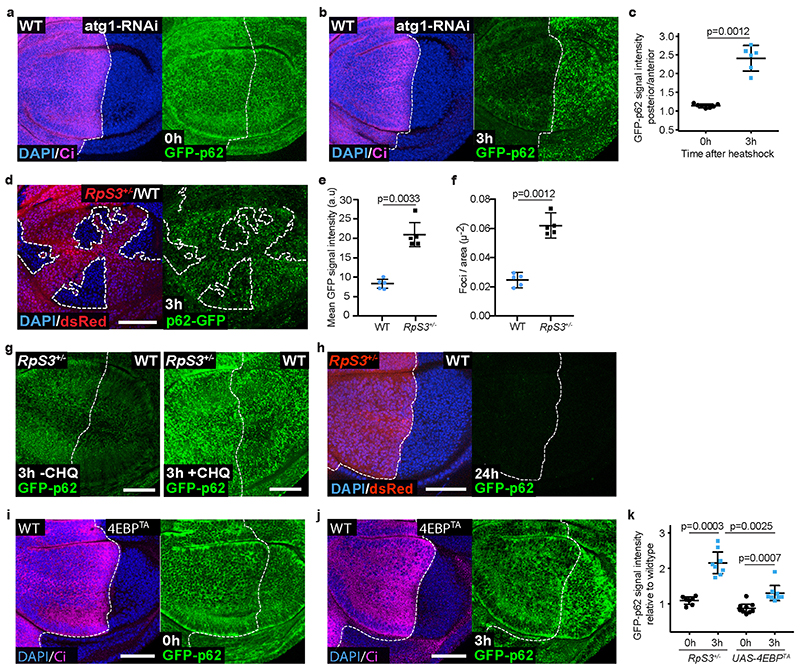
In order to examine the role of autophagy in RpS3 +/- cells, we obtained wing discs from larvae carrying heterozygous mutations for both RpS3 and one of several autophagy-related genes: p62 (ref(2)P in Drosophila), atg8 or atg13 37. We found that all three autophagy mutations caused a cell-autonomous increase in apoptotic events in an RpS3 +/- background, as compared to RpS3 +/- or autophagy mutations alone (Figure 2a–b, Extended Data Figure 2b–d). Heterozygous mutations in another ribosome loser mutation, RpL27A, also caused increased apoptosis in combination with heterozygous mutations in the autophagy gene p62 (Extended Data Figure 2e–f). Thus, Minute cells are acutely reliant on autophagy. However, autophagy inhibition did not impact the competitive status of RpS3 +/- cells, as knockdown of autophagy genes atg1 or atg9 by RNAi did not affect clone coverage or competition-induced cell death in competing RpS3 +/- cells (except for a mild increase in competitive death in the case of atg1 RNAi; Extended Data Figure 2g–i). This contrasts with data from Nagata et al.,18, who have instead shown that inhibiting autophagy rescues Minute cells from competition. Non-competing RpS3 +/- cells also appeared to have more atg8-positive foci (Figure 2c) and had more p62-positive foci (Figure 2d–e) than wild-type cells.
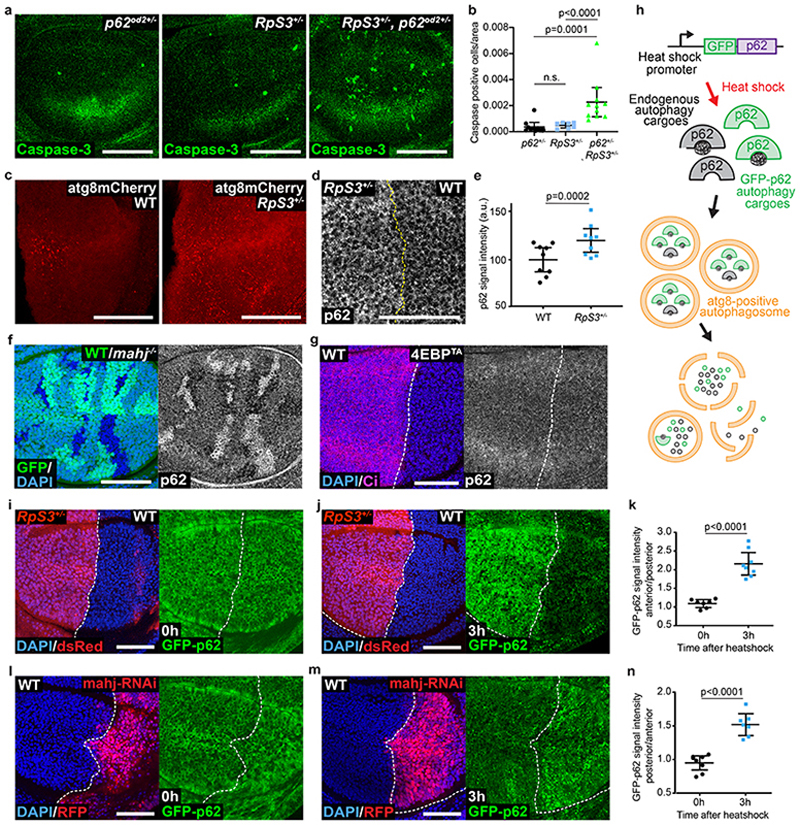
Figure 2. Prospective losers display defective autophagic flux.
(a-b) Apoptotic cell death, as detected by anti-cleaved Caspase-3 reactivity (green), in wing discs of a p62 +/- heterozygote (a, left), RpS3 +/- heterozygote (a, middle), or p62 +/-, RpS3 +/- transheterozygote (a, right) and corresponding quantification (n=10, 7, and 11 respectively, two-sided Mann-Whitney U test without p-adjustment for multiple comparisons) (b). (c) Staining of autophagosomes and autolysosomes, as detected by atg8-GFP-mCherry expression (red) in the P-compartment of wild type (c, left), or Rps3 +/- (c, right) wing discs. (d-e) Immunostaining for p62 in wing discs harboring RpS3 +/- A cells and wild-type P cells (d) and corresponding fluorescence intensity quantification (n=9, two-sided paired t-test) (e). (f) Immunostaining of p62 in a wing disc with mahj -/- clones (GFP-negative) induced in a mahj +/- heterozygous background (1XGFP). Wild-type twin spots are 2XGFP. (g) Immunostaining for p62 in wing discs harboring wild-type A cells and 4E-BPTA-expressing P cells (labelled by the absence of Ci, magenta). (h) Schematic representation of ReFLUX: the autophagy cargo p62 is fused to GFP and driven by a hs promoter for pulse-chase expression. (i-k) GFP-p62 ReFlux signal (green) in wing discs harboring RpS3 +/- A cells (dsRed-positive) and wildtype P cells (dsRed-negative) immediately after heat shock (i), or three hours later (j) and corresponding signal quantifications (n= 7 and 8 respectively, two-sided student’s t-test) (k). (l-n) GFP-p62 ReFlux signal (green) in wing discs expressing mahj-RNAi in the P compartment (RFP-positive), immediately after heat shock (l) or three hours later (m) and corresponding signal quantifications (n=8 and 7 respectively, two-sided student’s t-test) (n). For all micrographs, scale bars correspond to 50μm. For all quantification, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Cells with reduced function of the loser gene and ubiquitin ligase mahj share with RpS3 +/- cells a cell-autonomous signature of hundreds of differentially expressed genes relative to wild-type cells, as well as a cell-autonomous activation of the oxidative stress response20. This suggests that mutations in mahj and RpS3 lead to cell competition using a convergent mechanism Thus, we examined the autophagic state in mahj -/- cells. mahj -/- homozygous clones in a background of mahj +/- and wild type cells also accumulated p62 foci (Figure 2f), whereas 4EBPTA had no effect on the number of p62 foci (Figure 2g). Thus, deregulated autophagy is associated with the prospective loser status of two functionally unrelated mutants, and this is not a consequence of reduced protein synthesis.
Accumulation of Atg8- and p62-positive autophagosomes can reflect either decreased or increased autophagic flux38. To measure autophagic flux in prospective losers, we designed the reporter “ReFlux” (Ref(2)P autophagy Flux) that measures the rate of p62 degradation38,39. p62 is both an autophagy adaptor and an autophagy cargo that is degraded upon autophagosome degradation by the lysosome38. Thus, measuring the rate of p62 degradation provides a direct measure of autophagic flux38. In ReFlux, p62 is fused to GFP and driven by a heat-shock (hs) promoter for pulse-chase expression40 (Figure 2h). As a control, we confirmed that ReFlux reports reduced autophagic flux upon depletion of the autophagy gene atg1 (Extended Data Figure 3a–c). Then, we expressed ReFlux across wing discs containing RpS3 +/- anterior and wild-type posterior compartments. We found that RpS3 +/- and wild-type cells show similar GFP-p62 ReFlux signal intensity immediately following pulse expression. However, after a chase period, GFP-p62 ReFlux signal perdures in RpS3 +/- cells compared to wild-type cells, indicating reduced autophagic flux (Figure 2i–k). A reduced autophagic flux was also seen in competing RpS3 +/- cells, relative to competing wild-type cells (Extended Data Figure 3d–f). Treatment with the autophagy inhibitor chloroquine led to persistence of the GFP-p62 ReFlux signal, confirming that GFP-p62 ReFlux loss is due to autophagic degradation (Extended Data Figure 3g). ReFlux was eventually cleared from the RpS3 +/- compartment (Extended Data Figure 3h), indicating that autophagic degradation is delayed but not blocked. Knockdown of Mahj also reduced autophagic flux (Figure 2l–n). Overexpression of 4EBPTA also reduced autophagic flux, albeit with a substantially smaller effect size than RpS3 +/- mutations (Extended Data Figure 3i–k).
Defective autophagy does not cause the loser status
Defective autophagy has been associated with the loser status in mouse embryonic stem cells41. Having observed reduced autophagic flux in both RpS3 +/- and mahj -/- prospective losers, we next investigated whether reduced autophagy is sufficient to induce the loser status in these epithelia. Clones of cells expressing atg1 RNAi within wild-type imaginal discs did not show cell death enrichment at the clone borders (Figure 3a–b), even though they accumulated p62 foci (Figure 3c), indicative of impaired autophagy. atg1-depleted cells also failed to activate the oxidative stress response in a non-competitive context (Figure 3d, right), despite confirmation of autophagy impairment from p62 accumulation (Figure 3d, left). Similarly, inhibiting autophagy in clones by mutating atg13 caused accumulation of p62 foci (Figure 3e), but did not result in cell competition with wild-type cells, as neither cell death nor clonal disadvantage were observed (Figure 3f–h). Therefore, reduced autophagic flux is observed in RpS3 +/- cells both in the absence of and during competition but is not sufficient to cause cell competition.
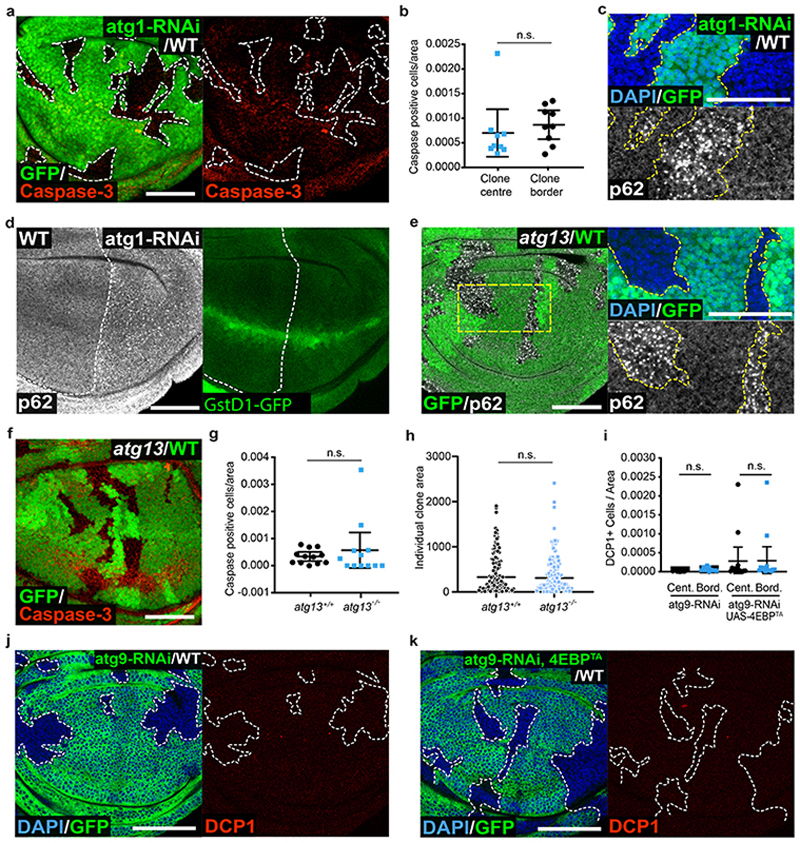
Figure 3. Autophagy impairment does not confer the loser status.
(a-b) Apoptosis detection by cleaved caspase-3 staining (red) in wing discs with mosaic expression of atg1-RNAi (GFP-positive cells) (a) and corresponding quantifications (n=9, two-sided Wilcoxon signed-rank test) (b). Cell death is classed as border death or center death, as described in Figure 1. (c) p62 staining in wing discs of the same genotype as in (a). (d) p62 staining (left) and GstD1-GFP signal (right) in wing discs harboring atg1-RNAi expressing P cells and wild-type A cells. (e-h) p62 staining (e) and apoptosis detection by cleaved caspase-3 staining (red) (f) in wing discs with atg13 -/- clones (GFP-negative) induced in an atg13 +/- heterozygous background (1XGFP), and corresponding cell death (g, n=12, two-sided Wilcoxon signed-rank test) and clone size (h, n=95 and 105, respectively, two-sided Mann-Whitney U test) quantifications for atg13 -/- clones and wild-type atg13 +/+ twin spots (2XGFP). Each dot or square on the graph in (h) represents one clone, and the horizontal line represents the median and whiskers indicate the 95% confidence interval. (i-k) Wing discs harboring GFP-positive clones expressing atg9-RNAi (j) or expressing atg9-RNAi and 4E-BPTA (k) and stained for cleaved-dcp1 (red) and corresponding cell death quantification in clone centers (Cent.) versus borders (Bord.) (n=11 and 14 respectively, two-sided Wilcoxon signed-rank test) (i). For all micrographs, scale bars correspond to 50μm. For all quantifications provided other than (h), the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs, except in (h) wherein n numbers refer to the number of individual twin-spot clones.
As reduced protein synthesis and autophagy flux are observed in RpS3 +/- losers but neither is sufficient to confer the loser status, we asked whether they might do so in concert. However, co-expressing atg9 RNAi and 4EBPTA in clones of cells in a wild-type wing disc did not result in border cell death, indicating that reduced protein synthesis and defective autophagy together are not sufficient to induce the competitive elimination of losers (Figure 3i–k).
Prospective losers have defective proteasome flux
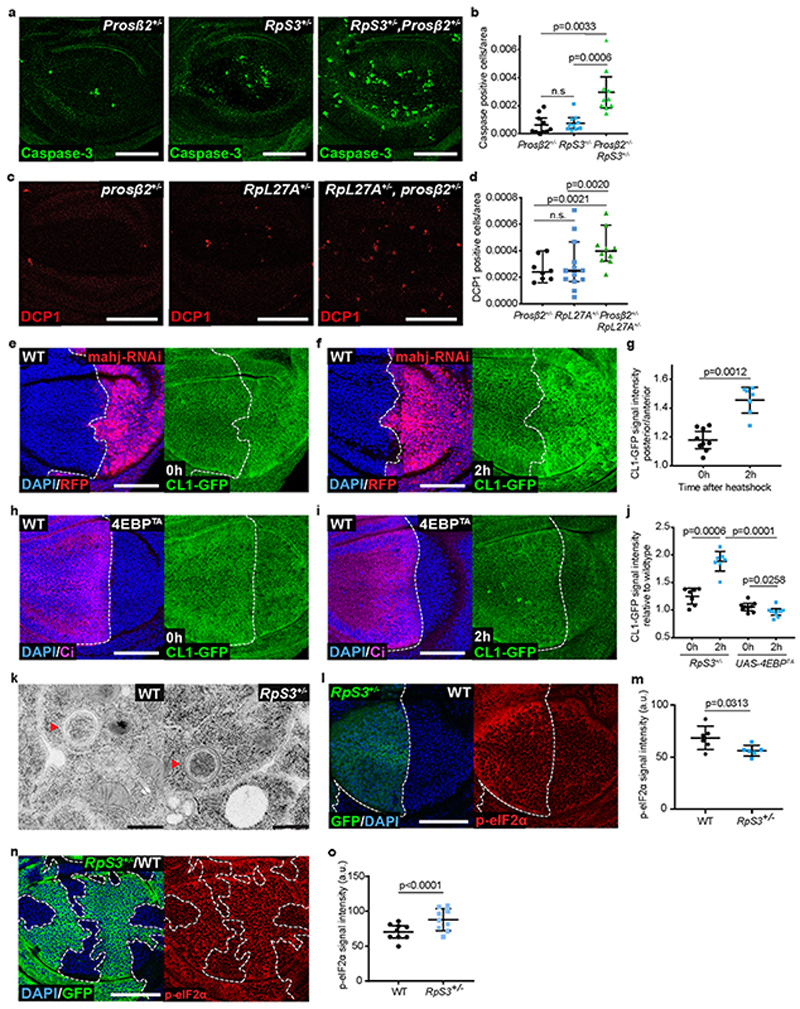
Proteasome genes were also differentially expressed in RpS3 +/- cells upon JNK signaling inhibition (Supplementary Table 1), prompting us to investigate the role of the proteasome in Minute cells. Heterozygosity of a proteasomal core subunit gene caused increased apoptosis in RpS3 +/- cells and in RpL27A +/- cells (Extended Data Figure 4a–d). Similarly, feeding flies the proteasome inhibitor bortezomib42 increased the number of dying cells in RpS3 +/- but not wild-type wing discs (Figure 4a–c). Thus, ribosome mutant cells are cell-autonomously reliant on proteasome function in addition to autophagy.
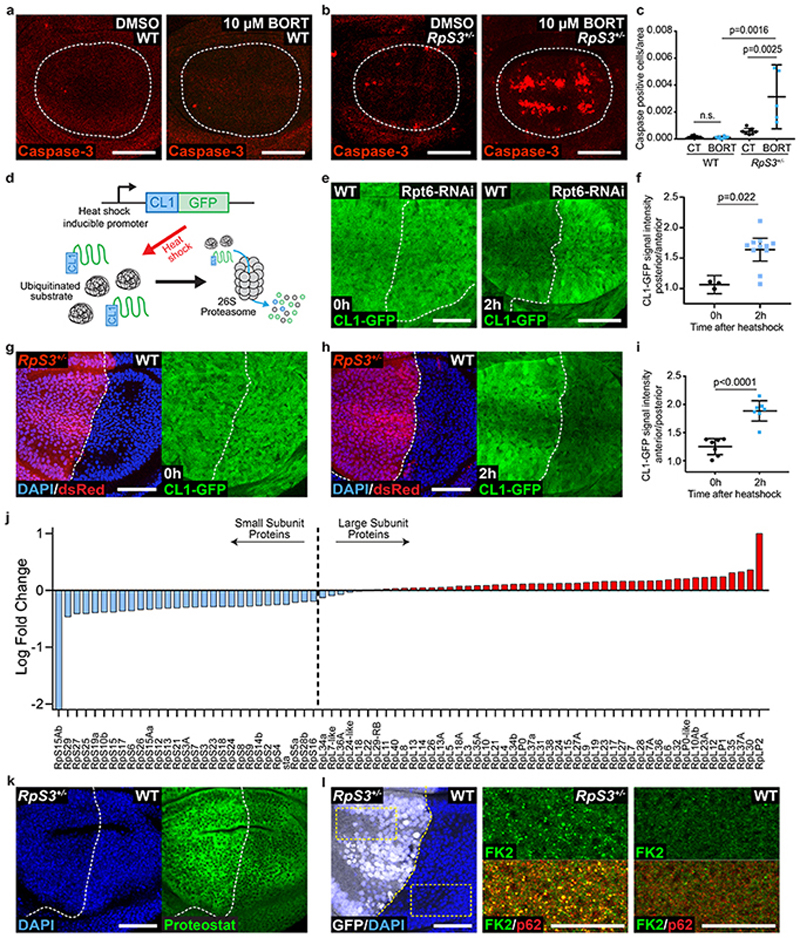
Figure 4. Prospective losers display proteotoxic stress.
(a-b) Apoptosis detection by cleaved caspase-3 staining (red) in wild type (a) or RpS3 +/- (b) wing discs fed DMSO or 10 μM bortezomib, as indicated. (c) Quantification of dying cell numbers within the pouch region of wing discs from the conditions indicated in (a-b) (n=8, 8, 7, and 5, respectively, two-sided Mann-Whitney U test without p-adjustment for multiple comparisons). (d) Schematic representation of ProteoFLUX: a fusion of GFP with the proteasome degradation signal CL1, driven by a hs promoter for pulse-chase expression. (e-f) ProteoFLUX CL1-GFP signal (green) in wing discs expressing RNAi against the proteasomal subunit Rpt6 specifically in P cells, immediately after heat shock or two hours later, as indicated (e), and corresponding signal quantifications (n=3 and 11 respectively, two-sided Mann-Whitney U test) (f). (g-i) ProteoFLUX CL1-GFP signal (green) in wing discs harboring RpS3 +/- A cells (dsRed-positive) and wild-type P cells (dsRed-negative), immediately after heat shock (g), or two hours later (h), and corresponding signal quantifications (n=7 and 7 respectively, two-sided student’s t-test) (i). (j) Abundance of Ribosomal subunit proteins in RpS3 +/- wing discs relative to wild-type wing discs by TMT Mass Spectrometry. Bars indicate average log fold change values across two independent biological replicates. (k) Proteostat protein aggregate staining (green) in wing discs harboring RpS3 +/- A cells and wild-type P cells. (l) FK2 anti-conjugated ubiquitin (green) and anti-p62 (red) staining in a wing disc harboring an RpS3 +/- A compartment and a wild-type P compartment, as indicated. Yellow boxes mark inset locations. For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
To determine whether proteasome function is dysregulated in RpS3 +/- cells, we examined proteasome activity with CL1-GFP, a fusion of GFP with the proteasome degradation signal CL1, which targets GFP for efficient proteasomal degradation43. To enhance reporter sensitivity, we designed the reporter ProteoFlux, a hs-driven CL1-GFP, to enable pulse-chase measurements of proteasome flux (Figure 4d). We confirmed that ProteoFlux CL1-GFP detects reduced proteasome flux when we interfere with proteasome function by knockdown of the proteasome subunit Rpt6 (Figure 4e–f). We then expressed ProteoFLUX CL1-GFP in wing discs harboring RpS3 +/- anterior and wild-type posterior compartments, so that we could compare directly their proteasome flux in the absence of cell competition. RpS3 +/- and wild-type cells showed similar ProteoFLUX CL1-GFP signal intensity immediately after pulse expression. After a chase period, however, we observed higher GFP intensity in RpS3 +/- than in wild-type cells, indicating slower proteasome flux in RpS3 +/- cells (Figure 4g–i). ProteoFlux CL1-GFP degradation was also delayed in cells depleted for Mahj (Extended Data Figure 4e–g), but not in 4EBPTA-expressing cells (Extended Data Figure 4h–j). Therefore, like reduced autophagic flux, reduced proteasomal flux is a common feature of genetically distinct prospective losers.
RpS3 +/- mutations induce protein aggregates and stoichiometric imbalance in ribosome proteins
Ribosomal proteins are degraded by the proteasome44 and by autophagy45,46 Indeed, electron microscopy analysis showed phago-lysosomal structures containing ribosomes both in wild-type and in RpS3 +/- wing disc cells (Extended Data Figure 4k). We reasoned that RpS3 +/- mutations could lead to a stoichiometric imbalance in ribosome proteins, which could in turn cause proteotoxic stress and overload the proteasome and autophagy machineries47,48 To test this, we measured relative levels of ribosome proteins, by Tandem Mass Tag (TMT) Spectrometry of RpS3 +/- and wild-type wing discs. TMT successfully identified 78 ribosome proteins of the 93 reported on Flybase (of the missing 15, 8 are not expected to be expressed in wing discs). This showed that the RpS3 +/- mutation causes a reduction in RpS3 protein of 0.291 log-fold relative to wild-type levels. Interestingly, a reduction was observed for all small ribosome subunit proteins detected (Figure 4j), indicating coordinated regulation, but this was not seen for components of the large subunit, whose levels were, with few exceptions, equal to or higher than in wild-type cells (Figure 4j). Thus, at steady state, RpS3 +/- cells have a stoichiometric excess of ribosome proteins from the large subunit relative to small subunit ribosome proteins. This could contribute to proteasome and autophagy overload.
When they are not efficiently cleared by degradation, ribosome proteins can form protein aggregates44,47,48 To test this, we used Proteostat, a dye which fluoresces upon intercalation with protein aggregate-associated quaternary structures. Indeed, Proteostat staining detected accumulation of protein aggregates in RpS3 +/- cells relative to wild-type cells, in the absence of cell competition (Figure 4k). Protein aggregates are often ubiquitin-positive49,50, and immunostaining with the FK2 antibody, which detects mono- and poly-ubiquitin conjugates, revealed that RpS3 +/- cells, but not wild-type cells, accumulate large, ubiquitin-positive foci in the cytoplasm (Figure 4l). Many of these foci were also positive for the autophagy adapter/cargo p62 (Figure 4l), which is often recruited to cytosolic protein aggregates50. Furthermore, phospho-eIF2α, a marker of proteotoxic stress and of the integrated stress response34, was upregulated in RpS3 +/- cells, both in homotypic conditions (Extended Data Figure 4l–m) and during cell competition (Extended Data Figure 4n–o). Collectively, RpS3 +/- cells show reduced autophagy flux, reduced proteasome flux, accumulation of ubiquitinated protein aggregates, and markers of proteotoxic stress.
Improving proteostasis in RpS3 +/- cells rescues their loser status
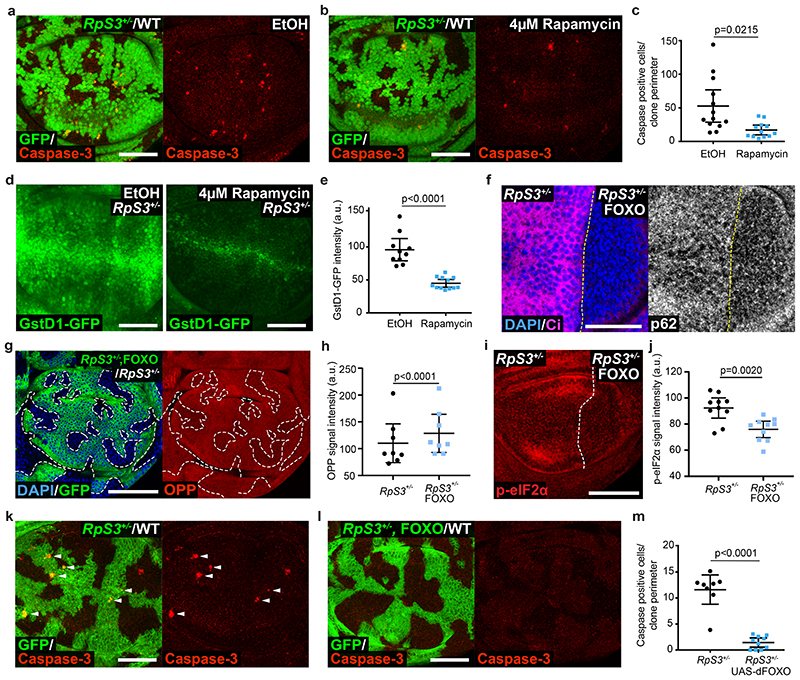
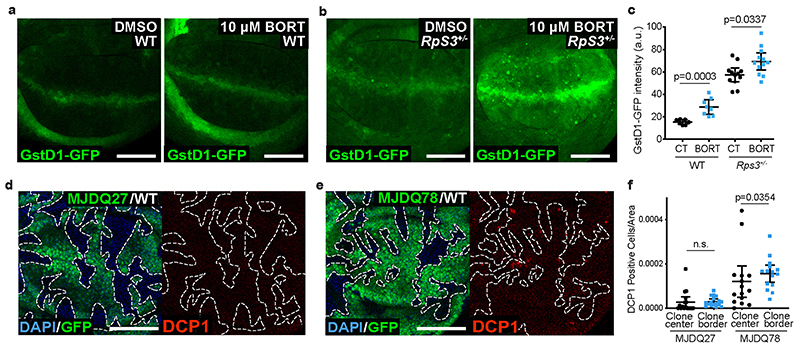
Proteotoxic stress can induce Nrf2 activation51, and this in turn is linked to the loser status20, suggesting a link between proteotoxic stress and the prospective loser status. Consistent with this, inhibiting the proteasome with bortezomib was sufficient to elevate GstD1-GFP signal in non-competing wild-type and RpS3 +/- wing disc cells (Extended Data Figure 5a–c). We therefore asked whether alleviating proteotoxic stress would rescue loser cells from competition. Rapamycin inhibits TOR signaling and promotes proteostasis via multiple mechanisms, including inhibiting translation and activating autophagy and proteasome functions52,53. We found that rapamycin feeding reduced the frequency of competition-induced apoptosis in RpS3 +/- cells bordering wild-type cells (Figure 5a–c). Rapamycin feeding also reduced the cell-autonomous activation of the oxidative stress reporter GstD1-GFP in RpS3 +/- cells (Figure 5d–e). As rapamycin was fed systemically, the observed rescue of competition-induced cell death could in part arise from the effects of rapamycin on wild-type cells. We therefore sought to improve proteostasis specifically in RpS3 +/- cells. To this end, we overexpressed, in RpS3 +/- cells, the transcription factor FOXO, which is inhibited by TOR signaling54,55 and promotes both autophagy and proteasome functions55. FOXO overexpression reduced the number of p62-positive aggregates (Figure 5f), increased protein synthesis (Figure 5g–h) and reduced mildly the levels of phospho-eIF2α (Figure 5i–j) in RpS3 +/- cells, indicating overall improved proteostasis. Strikingly, FOXO overexpression in RpS3 +/- cells abolished competition-induced cell death, as very few apoptotic bodies could be detected in competition with wild-type cells (Figure 5k–m). These data indicate that reducing proteotoxic stress inhibits the competitive elimination of RpS3 +/- cells.
Figure 5. Alleviating proteotoxic stress rescues the loser status.
(a-b) Apoptosis detection by cleaved caspase-3 staining (red) in competing wing discs containing RpS3 +/- cells (GFP-positive) and unlabeled wild type cells (GFP-negative) from larvae fed ethanol carrier (a) or 4 μM rapamycin (b). (c) Quantification of cell death at RpS3 +/- clone boundaries for the experiments in (a-b) (n=13 and 12 respectively, two-sided two sample Kolmgorov-Smirnov test). (d-e) GstD1-GFP signal (green) in RpS3 +/- wing discs fed EtOH control or 4μM Rapamycin, as indicated (d), and corresponding quantification (n=10 and 12 respectively, two-sided student’s t-test) (e). (f) p62 staining in RpS3 +/- wing discs expressing FOXO in P cells (labelled by the absence of Ci, magenta). (g-h) An RpS3 +/- wing disc harboring FOXO expressing clones (GFP-positive) and labelled with OPP (red) (g) with corresponding quantification in (h) (n=8, two-sided paired t-test). (i-j) Phospho-eIF2α staining (red) in RpS3 +/- wing discs expressing FOXO in P cells (i) and corresponding quantification (n=10, two-sided Wilcoxon signed-rank test. Due to low genetic frequency and the presence of an internal control, samples from multiple experiments were pooled together) (j). (k-l) Apoptosis detection by cleaved caspase-3 staining (red) in competing wild-type/RpS3 +/- mosaic wing discs without (k) or with (l) additional expression of dFOXO specifically in RpS3 +/- cells. (m) Quantification of cell death at RpS3 +/- clone boundaries for the experiments in (k-l) (n=8 and 10, respectively, two-sided two sample Kolmgorov-Smirnov test). For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Proteotoxic stress in sufficient to cause the loser status
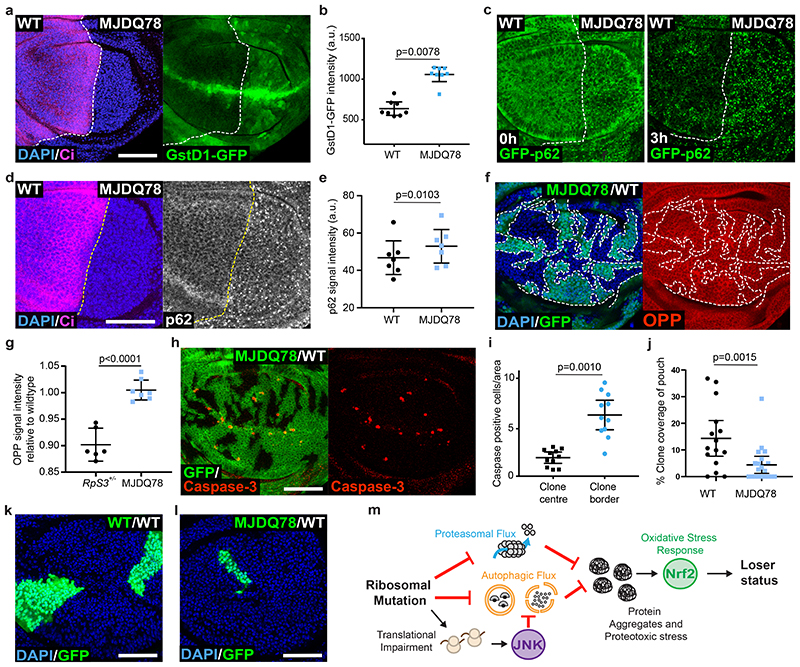
We considered that protein aggregation and proteotoxic stress could be sufficient to cause the loser status in competitive contexts. To test this hypothesis, we ectopically expressed the human aggregate-prone polyQ protein ataxin-3 (SCA3/MJDQ78), which is responsible for the human neurodegenerative disorder Machado Joseph Disease56 and has been used in Drosophila to model this neurodegenerative condition57. MJDQ78 expression was sufficient to recapitulate many features shared by RpS3 +/- and mahj -/- prospective losers, namely up-regulation of GstD1-GFP (Figure 6a–b), reduced autophagic flux (Figure 6c), and accumulation of p62-positive structures (Figure 6d–e). MJDQ78 however, did not perceptibly impact on rates of translation, as measured by OPP incorporation (Figure 6f–g). Importantly, clones overexpressing MJDQ78 in wild-type wing disc showed a local induction of apoptosis, specifically at their borders with wild-type cells (Figure 6h–i), and grew poorly relatively to wild-type clones (Figure 6j–l), indicating that these cells are eliminated by cell competition. This was specifically induced by proteotoxic stress, as clones expressing the wild-type version of Ataxin-3 (MJDQ27)57 did not show induction of border death (Extended Data Figure 5d–f). Thus, proteotoxic stress is sufficient to turn otherwise wild-type cells into losers (Figure 6m).
Figure 6. Proteotoxic stress is sufficient to confer the loser status.
(a-b) GstD1-GFP signal (green) in a wing disc expressing MJDQ78 in P cells (labelled by the absence of Ci, magenta) (a) and corresponding quantification (n=8, two-sided Wilcoxon signed-rank test) (b). (c) GFP-p62 ReFlux signal (green) in wing discs expressing MJDQ78 in P cells, immediately after heat shock or three hours later, as indicated. (d-e) p62 staining in a wing disc expressing MJDQ78 in P cells (labelled by the absence of Ci, magenta) (d), and corresponding quantification in (e) (n=7, two-sided paired t-test). (f-g) Wing discs harboring GFP-positive clones expressing MJDQ78 labelled with OPP (red) (f) with corresponding quantification relative to wing discs containing competing RpS3 +/- clones and wildtype winners (image not shown) in (g) (n=6 and 7 respectively, two-sided student’s t-test). (h-i) Mosaic wing disc containing GFP-positive clones overexpressing MJDQ78, immuno-stained for cleaved Caspase-3 (red) (h), and corresponding cell death quantification (n= 11, two-sided Wilcoxon signed-rank test) (i). (j-l) Wing discs harboring wild-type cells and wildtype control clones (GFP-positive) (k) or clones expressing MJDQ78 (GFP-positive) (l), and corresponding quantification (n=15 and 20 respectively, two-sided Mann-Whitney U test) (j). (m) Model summarizing how ribosome gene loss leads to proteotoxic stress and to the loser status. For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Discussion
Our work shows that single copy loss of ribosome genes leads to major defects in cellular proteostasis, as also shown in the accompanying paper from Recanses-Alvarez et al.,58 Heterozygosity of ribosome genes in humans leads to genetic disorders collectively known as ribosomopathies, characterized by severe malformations and pathologies9 The mechanisms through which ribosomal mutations lead to these defects are only partially understood9 Our work suggests that proteotoxic stress may be an underlying cause for some such defects and that they might be improved by drugs that promote proteostasis, such as the FDA-approved compound rapamycin53 that we have used in this study.
Our work shows that proteotoxic stress is sufficient to confer the loser status. This finding broadens the scope of cell competition and suggests it may be an active mechanism in physiological and pathological contexts characterized by proteotoxic stress. This may help explain the competitive elimination of neurons in Drosophila models of neurodegenerative diseases59. It may be especially relevant to cancer, where proteotoxic stress is often observed60. Our findings suggest that cancer cells might represent concealed losers that have escaped proteotoxic stress-induced cell competition through masking mutations. Understanding how Minute mutations and proteotoxic stress lead to cell competition may help unmask the loser status in cancer cells in ways that could be exploited therapeutically7.
Healthy proteostasis is a driver of organism fitness61 and contributes to organism longevity62, whereas impaired proteostasis is associated with aging and with age-related pathologies62, 63. We propose that tissues preserve their health and youth through a proteostasis-based mechanism of cell elimination. By measuring cell fitness on the basis of proteostasis and converting it into the loser status through the activation of the oxidative stress response, proteostasis-based cell competition could act as a general mechanism of cell selection in adult homeostasis. How proteotoxic stress induces the loser status remains to be established.
Methods
Fly husbandry
Fly lines were maintained at 25°C on a flour-based food supplemented with yeast. Our standard recipe contains 7.5g/L agar powder, 50g/L baker’s yeast, 55g/L glucose, 35g/L wheat flour, 2.5 % nipagin, 0.4 % propionic acid and 1.0% penicillin/streptomycin. For some chemical feeding experiments, drugs were diluted in Nutrifly GF food (Scientific Laboratory Supplies) made to manufacturer’s instructions. Sexes were not differentiated for any experiments, except in cases where transgenes were X-linked. Eggs were collected for 24 hours and wing discs were dissected from wandering third instar larvae. For each dataset, including across different vials or genotypes, egg collections, heat-shocks and harvesting of wandering stage larvae for dissections were done in parallel. All Drosophila strains used in this study are provided in Supplemental Table 2, and genotypes for all experimental crosses are provided in Supplemental Table 3.
Immunostaining
Wing discs were dissected in phosphate-buffered saline (PBS) before fixation in 4% formaldehyde/PBS solution for 20 minutes at room temperature. Dissected hemi-larvae were subsequently washed three times in PBS (30 seconds each), before permeabilisation in PBS containing 0.25% Triton X-100 (PBS-T). Samples were next incubated in blocking buffer (PBS-T supplemented with 4% fetal calf serum) for 30 minutes at room temperature. Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. Samples were washed three times in PBS-T (10 minutes each) before incubation in secondary antibody (diluted in blocking buffer) for 1 hour at room temperature. The secondary antibodies used were conjugated with Alexa 488, Alexa 555 or Alexa 633 dyes (Molecular probes). Nuclei were counterstained with DAPI (0.5 μg/ml). After three 5-minute washes in PBS-T, wing discs were mounted in Vectashield (Vector laboratories) on a borosilicate glass side (no 1.5, VWR international). For anti-FK-2 staining, the blocking buffer was substituted with a 3% BSA in PBS solution. Details and sources of all antibodies are provided in Supplemental Table 2. Dilutions for primary antibodies used are as follows: 1 in 500 for anti-pJNK, 1 in 1000 for anti-Ci, 1 in 2000 for anti-Ref(2)P, 1:25000 for anti-cleaved Caspase-3, 1 in 2500 for anti-DCP1, 1 in 500 for anti-p-eIF2α, and 1 in 5000 for anti-FK2.
Clonal analysis
Mosaic wing discs were generated using the FLP/FRT system employing hs-FLP or en-Gal4-UAS-FLP transgenic strains. For clone induction, heat shocks were carried out 2-4 days after egg laying (depending on experiment), in a 37°C water bath before returning flies to a 25°C incubator, or for experiments employing a temperature sensitive Gal80 (Gal80TS), to a water bath at the indicated temperature. The exact temperature for Gal80TS experiments together with heat shock conditions and clone age, which were optimized for each experiment individually, are listed in Supplemental Table 3.
Translation Assays
AHA and OPP assays were carried out using the Click-iT™ Plus OPP Protein Synthesis Assay kit and Click-iT Plus™ AHA Protein Synthesis Assay kit, respectively. For the AHA assay, wing discs were dissected and inverted in a glass dish before incubation in methionine free Schneider’s medium at 25 °C for 45 min. Hemi-larvae were then incubated for a further 45 min in methionine free medium supplemented with 2 mM AHA reagent. Samples were subsequently washed in PBS before fixation in 4% formaldehyde/PBS solution. For OPP assays, larvae were dissected in normal Schneider’s medium before transfer to a 1.5 ml Eppendorf containing 5 μM OPP reagent in Schneider’s medium and incubation for 15 min at 25 °C. Samples were subsequently washed in PBS before fixation. For both assays, fixed tissues were subsequently stained using the standard Click-iT protocol according to manufacturer’s instructions. Details for reagents are provided in Supplemental Table 2.
Identification of proteostasis genes
The full list of genes differentially expressed in RpS3+/- cells plus/minus expression of the JNK inhibitor puc was reported previously 20. To identify differentially expressed proteostasis genes from this list we selected genes associated with the following GO terms: autophagy, response to unfolded proteins, proteasome complex, proteasome catabolic process.
Re-Flux and Proteo-Flux Assays
Re-Flux and Proteo-Flux assays were carried out as pulse-chase experiments. Third instar wandering larvae were heat-shocked for 40 to 45 minutes, to induce a pulse of GFP-p62 or CL1-GFP, respectively. Larvae were incubated at 25 degrees for the indicated times to chase protein levels before dissection.
Proteostat assay
For PROTEOSTAT® Protein Aggregation Assay larvae were dissected and inverted in PBS before transfer to a 1.5 ml Eppendorf tube containing 4% formaldehyde diluted in 1X PROTEOSTAT assay buffer (PAB). The samples were subsequently permeabilized in 0.5% Triton X-100, 3 mM EDTA, pH 8.0 diluted in 1X PAB, before staining with PROTEOSTAT detection reagent diluted 1 in 20,000 together with Hoechst 33342 at 1 μg/ml in PAB. Hemi-larvae were subsequently washed three times in PBS before separating wing discs from the larval body and mounting in PBS under our standard cover slips. Wing discs were imaged immediately. Details for reagents are provided in Supplemental Table 2.
Transmission electron microscopy
Larvae were washed and dissected in Schneider’s Insect Medium and imaginal wing discs were dissected out and subjected to high-pressure freezing in a 20% BSA solution followed by an osmium tetroxide freeze substitution and Epon embedding. The resulting blocks were sectioned onto grids using an ultramicrotome and stained with uranyl acetate and lead citrate. Sections were then imaged on a Tecnai 12 transmission electron microscope.
Chemical feeding
For bortezomib feeding, eggs were collected for 24 hours and larvae grown on normal food for 72 hours before being floated in a 20% sucrose solution. Floated larvae were thoroughly washed with PBS before transferring to Nutri-Fly™ GF Premixed food containing 10 μM bortezomib or the equivalent volume of DMSO (as a carrier control). Larvae were grown until they were at third instar wandering stages. For rapamycin feeding, 4 μM rapamycin was diluted in standard wheat-based food and floated larvae were maintained on the drug (or equivalent carrier control of ethanol) until wandering stage. For chloroquine incubation, dissected larvae were incubated in 50 μM chloroquine diluted in normal Schneider’s medium (or the equivalent volume of water as a carrier control) for three hours at 25 °C, before washing in PBS and fixation. Details for reagents are provided in Supplemental Table 2.
Proteomics
Third instar larvae raised on normal food were dissected in ice-cold PBS containing 1X Phos-STOP phosphatase inhibitor and 1X Halt Protease Inhibitor cocktail. Wing discs were then centrifugated in an Eppendorf containing PBS/inhibitor cocktail for 30 seconds at 6,000 rcf at 4 °C before being lysed in ice-cold RIPA lysis buffer. Lysed samples were centrifugated at 12,500 rcf at 4 °C for ten minutes. Aliquots of 50µg of each sample were digested with trypsin (1.25µg trypsin; 37°C, overnight), and labelled with Tandem Mass Tag (TMT) ten plex reagents according to the manufacturer’s protocol (Thermo Fisher Scientific, Loughborough, LE11 5RG, UK) before samples were pooled. 40ug of the pooled sample was desalted using a SepPak cartridge according to the manufacturer’s instructions (Waters, Milford, Massachusetts, USA). Eluate from the SepPak cartridge was evaporated to dryness and resuspended in buffer A (20 mM ammonium hydroxide, pH 10) prior to fractionation by high pH reversed-phase chromatography using an Ultimate 3000 liquid chromatography system (Thermo Fisher Scientific). In brief, the sample was loaded onto an XBridge BEH C18 Column (130Å, 3.5 µm, 2.1 mm X 150 mm, Waters, UK) in buffer A and peptides eluted with an increasing gradient of buffer B (20 mM Ammonium Hydroxide in acetonitrile, pH 10) from 0-95% over 60 minutes. The resulting fractions were evaporated to dryness and resuspended in 1% formic acid prior to analysis by nano-LC MSMS using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific).
High pH reversed-phase fractions were further fractionated using an Ultimate 3000 nano-LC system in line with an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). All spectra were acquired using an Orbitrap Fusion Lumos mass spectrometer controlled by Xcalibur 3.0 software (Thermo Scientific) and operated in data-dependent acquisition mode using an SPS-MS3 workflow. FTMS1 spectra were collected at a resolution of 120 000, with an automatic gain control (AGC) target of 400 000 and a max injection time of 100ms. Precursors were filtered with an intensity threshold of 5000, according to charge state (to include charge states 2-7) and with monoisotopic peak determination set to Peptide. Previously interrogated precursors were excluded using a dynamic window (60s +/-10ppm). The MS2 precursors were isolated with a quadrupole isolation window of 0.7m/z. ITMS2 spectra were collected with an AGC target of 10 000, max injection time of 70ms and CID collision energy of 35%.
For FTMS3 analysis, the Orbitrap was operated at 30 000 resolution with an AGC target of 50 000 and a max injection time of 105ms. Precursors were fragmented by high energy collision dissociation (HCD) at a normalised collision energy of 60% to ensure maximal TMT reporter ion yield. Synchronous Precursor Selection (SPS) was enabled to include up to 5 MS2 fragment ions in the FTMS3 scan.
The raw data files were processed and quantified using Proteome Discoverer software v2.1 (Thermo Scientific) and searched against the UniProt Drosophila melanogaster database (downloaded March 2020: 41311 entries) using the SEQUEST HT algorithm. Peptide precursor mass tolerance was set at 10ppm, and MS/MS tolerance was set at 0.6Da. Searches were performed with full tryptic digestion and a maximum of 2 missed cleavages were allowed. The reverse database search option was enabled and all data was filtered to satisfy false discovery rate (FDR) of 5%. Ribosomal proteins were identified by cross referencing the proteomic results against the ‘Ribosomal Protein’ category in FlyBase using R statistical software. Average fold changes were obtained for Ribosomal Proteins which exhibited a consistent change in relative abundance across both biological replicates. Two biological replicates were performed.
Cloning and transgenics
To isolate genomic DNA, a single fly was homogenized in 50 μl extraction buffer containing 10 mM Tris HCl pH 8.2, 2 mM EDTA pH 8.0, 0.1% Triton X-100 and 200 μg/ml proteinase K. Samples were then heated to 55 °C for 30 min in a Thermoshaker with occasional vortexing, before increasing the temperature to 95 °C for 15 min to inhibit protease activity. Samples were then cooled to 4 °C and centrifuged at 5,000 x g for 5 min at 4 °C. The supernatant was subsequently transferred to a fresh 0.5 ml Eppendorf tube and stored at 4 °C. Alternatively, DNA was isolated from 10-15 flies using a Gentra Puregene Tissue Kit using the following protocol: flies were homogenized using a motorized pestle in 200 μl cell lysis buffer and incubated at 65 °C in a Thermoshaker for 15 min. Then, 1 μl RNAase A solution was added, before incubation at 37 °C for a further 15 min. A volume of 100 μl of protein precipitation buffer was subsequently added and samples were thoroughly mixed and incubated on ice for 5 min. Samples were centrifuged for 10 min at 4 °C, at max speed before adding 300 μl isopropanol to the supernatant, mixing well and a further 15 min in the centrifuge. The resulting pellet was washed twice with 70 % ethanol before re-suspending in 50 μl of DNAse free water.
For cloning of both ReFLUX (hs-GFP-p62) and ProteoFLUX (hs-CL1-GFP) constructs, gDNA was isolated from 10-15 flies of the genotypes UAS-GFP-p62 or UAS-CL1-GFP respectively. The resulting gDNA was used as template for a PCR using primers designed to amplify constructs introduced in the common pUAST vector. To generate pCaSper-hs-GFP-p62 three different pairs of primers were used to generate a PCR product that could be inserted into the pCR™4-TOPO™ vector. The resulting pTOPO-GFP-p62 together with pCaSper-hs were digested with XbaI and NotI restriction enzymes (New England Biosciences Ltd) to produce a fragment containing GFP-p62 that could be ligated into the pCaSper-hs backbone. For the hs-CL1-GFP, a protocol using Infusion® HD Cloning Plus Kit was designed to infuse a PCR product containing the CL1-GFP sequencing into the pCasper-hs-GFP-p62 plasmid.
For cloning of the act>RpS3>Gal4 construct, the Infusion® HD Cloning Plus Kit (Clontech, 638909) was used to linearize an extant pCaSper2-act>CD2>Gal4 vector 64, by digestion with the Acc65I restriction enzyme (NEB). Two PCR products from a plasmid encoding RpS3 together with Hsp70 terminator sequences, were then infused. The resulting plasmid was transformed into Stellar™ competent cells (Clontech, 636766).
Plasmids for all constructs were sent for injection into a w118 line by Genetics Services, University of Cambridge or BestGene Drosophila embryo injection services. Exact primers used are provided in Supplemental Table 2.
Image acquisition and processing
Confocal images were acquired using Leica SP5 and SP8 confocal microscopes using a 40x 1.3 NA P Apo Oil objective. All wing discs were imaged as z-stacks with each section corresponding to 0.5-1 μm. Images were subsequently analysed and processed using Fiji2 and Photoshop (Adobe Version CS6). Clonal areas were determined using a custom script built in Fiji. For cell death quantifications, caspase-3 or DCP1 positive cells were counted in the region specified in each experiment (as reported in the figure legend). All counts were normalized to their respective area as measured in Fiji. For signal intensity, mean grey value was measured in Fiji for the specified genotypes within the pouch region of the wing disc.
Quantifications
For immunofluorescence and fluorescent reporter microscopy-based assays, all measurements were derived from the pouch region of the wing disc. For cell death assays, death counts were normalized to the area of the wing pouch or to the specified region of the clones within the pouch. For all scatter dot plots, unless otherwise specified, the horizontal line represents the mean and whiskers indicate 95% confidence intervals.
Statistics and reproducibility
All data used for statistical tests along with the specific test used for each experiment are shown in the Statistics Source Data table. Statistical tests were performed using GraphPad Prism 7.0a and Rstudio software. P-values were determined using univariate statistics. We consider not significant (n.s.) p-values >0.05. Parametric tests were used in cases where assumptions of normality and equivalence of variance were met. Non-parametric tests were used otherwise. The parametric tests used were Student’s T-Test and paired T-Test for matched data. The non-parametric tests used were either a Kolmogorov-Smirnov test or Mann Whitney U-test, or Wilcoxon matched-pairs signed rank test for matched data. P-value corrections for multiple comparisons were not considered due to the low number of comparisons. All statistical tests were two-sided. A minimum of three biological repeats were used for experiments comparing across separate wing discs. For matched experiments containing an internal control, a minimum of two biological repeats were performed. Functional validation of reagents and Drosophila stocks (e.g. RNAi) was carried out at least once. All data points for all replicates for specific quantifications are provided in the ‘Statistics Source Data’ supplemental file.
Extended Data
Extended Data Fig. 1. Protein synthesis and its regulation in Rps3 +/- cells.
(a-c) AHA (grey) protein synthesis assay in wing discs harboring either Rps3 +/- clones (GFP-positive) (a) or clones overexpressing 4EBPTA (GFP-positive) (b), and corresponding quantification (n=7 and 7, respectively, two-sided paired t-test without p-adjustment for multiple comparisons) (c). (d-e) OPP (green) protein synthesis assay in a wing disc expressing mahj-RNAi in the P compartment (positively labelled with RFP) (d) and corresponding quantification (n=10, two-sided Wilcoxon signed-rank test) (e). (f-g) An RpS3 +/- wing disc expressing GADD34 in the P compartment and labelled with phospho-eIF2α (red) (f), and corresponding quantification (n=10, two-sided paired t-test) (g). (h-i) GST-GFP reporter (green) activation in an RpS3 +/- wing disc expressing GADD34 in the P compartment (h), and corresponding quantification (n=10, two-sided paired t-test) (i). For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Extended Data Fig. 2. The role of autophagy in prospective losers.
(a) GstD1-GFP signal (green) in a RpS3 +/- wing disc expressing Puc in P cells (labelled by the absence of Ci, magenta). (b-c) Apoptotic cell death, as detected by anti-cleaved Caspase-3 reactivity (red), in wing discs of an atg8 +/- heterozygote (b, left), RpS3 +/- heterozygote (b, middle), or atg8 +/-, RpS3 +/- transheterozygote (b, right) and corresponding quantification (n=9, 8, and 9 respectively, two-sided two sample Kolmgorov-Smirnov test without p-adjustment for multiple comparisons) (c). (d) Apoptotic cell death, as detected by anti-cleaved Caspase-3 reactivity (red), in wing discs of an atg13 +/- heterozygote (d, left), RpS3 +/- heterozygote (d, middle), or atg13 +/-, RpS3 +/- transheterozygote (d, right). (e-f) Apoptotic cell death, as detected by anti-cleaved dcp1 antibody staining (red), in wing discs of a p62 +/- heterozygote (f, left), RpL27A +/- heterozygote (f, middle), or RpL27A +/-, p62 +/- transheterozygote (f, right) and corresponding quantification (n=10, 10, and 12 respectively, two-sided Mann-Whitney U test without p-adjustment for multiple comparisons) (e). (g-i) Wing discs harboring RpS3 +/- clones (GFP-positive) (h, left), RpS3 +/- clones expressing atg1-RNAi (GFP-positive) (h, middle), or RpS3 +/- clones expressing atg9-RNAi (GFP-positive) (h, right) stained with cleaved-dcp1 (red) and corresponding quantification of border cell death (n=16, 12, and 9 respectively, two-sided Mann-Whitney U test without p-adjustment for multiple comparisons) (g) and clone coverage (n=16, 12, and 9 respectively, two-sided student’s t-test without p-adjustment for multiple comparisons) (i). For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Extended Data Fig. 3. Autophagy flux in ribosome mutants and upon translation inhibition.
(a-c) GFP-p62 ReFlux signal (green) in wing discs expressing RNAi against the autophagy gene atg1 specifically in P cells (labelled by the absence of Ci, magenta), immediately after heat shock (a) or three hours later (b), and corresponding signal quantifications (n=7 and 6 respectively, two-sided two sample Kolmgorov-Smirnov test) (c). (d-f) GFP-p62 ReFlux signal (green) in a wing disc harboring RpS3 +/- clones (dsRed-positive) three hours after heat-shock (d) and corresponding quantification of GFP-p62 signal intensity (e) and number of GFP-p62 foci per area (f) (for both measurements, n=5, two-sided paired t-test). (g) GFP-p62 ReFlux signal (green) in wing discs harboring RpS3 +/- A cells and wild-type P cells, three hours after heat-shock, with or without addition of chloroquine, as indicated. (h) GFP-p62 ReFlux signal (green) in wing discs harboring RpS3 +/- A cells (dsRed-positive) and wild-type P cells (dsRed-negative) twenty-four hours after heat-shock. (i-k) GFP-p62 ReFlux signal (green) in wing discs harboring wild-type A cells and 4E-BPTA -expressing P cells (labelled by the absence of Ci, magenta), immediately after heat shock (i) or three hours later (j), and corresponding signal quantifications relative to wing discs containing an RpS3 +/- A compartment and wildtype P compartment (images not shown) (n=9 and 8 for 0 and 3 hour 4E-BPTA, and n=7 and 8 for 0 and 3 hour RpS3 +/-, respectively; two-sided two-sample Kolmgorov-Smirnov test without p-adjustment for multiple comparisons) (k). For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Extended Data Fig. 4. Proteasome defects are linked to the prospective loser status but not to translation inhibition.
(a-b) Apoptosis as detected by anti-cleaved caspase-3 reactivity (green), in Prosβ2 +/- (a, left), RpS3 +/- (a, middle), or Prosβ2+/-, RpS3 +/- transheterozygote (a, right) wing discs and corresponding quantification (n=10, 10, and 10 respectively, two-sided two sample Kolmgorov-Smirnov test without p-adjustment for multiple comparisons) (b). (c-d) Apoptotic cell death as detected by cleaved-dcp1 (red) in Prosβ2 +/- (c, left), a RpL27A +/- (c, middle), or a RpL27A +/-, prosβ2 +/- transheterozygote (c, right) wing discs, and corresponding quantification (n=8, 13, and 10 respectively, two-sided Mann-Whitney U test without p-adjustment for multiple comparisons) (d). (e-g) ProteoFLUX CL1-GFP signal (green) in wing discs expressing mahj-RNAi in the P compartment (RFP-positive), immediately after heat shock (e) or two hours later (f) and corresponding signal quantifications (n=9 and 7 respectively, two-sided two sample Kolmgorov-Smirnov test) (g). (h-j) ProteoFLUX CL1-GFP signal (green) in wing discs harboring wild-type A cells and 4E-BPTA-expressing P cells (labelled by the absence of Ci, magenta), immediately after heat shock (h) or two hours later (i), and corresponding signal quantifications relative to wing discs containing an RpS3 +/- A compartment and wildtype P compartment (images not shown) (n=9 and 10 for 0 and 2 hour 4E-BPTA, and n=7 and 7 for 0 and 2 hour RpS3 +/-, respectively; two-sided two-sample Kolmgorov-Smirnov test without p-adjustment for multiple comparisons) (j). (k) Transmission Electron microscopy images of a wing disc with wildtype P (left panel) and RpS3 +/- A compartments (right panel). Red arrows indicate phago-lysosomal structures containing ribosomes. The scale bar is 500 nm. (l-m) Phospho-eIF2α staining (red) in wing discs harboring RpS3 +/- A cells (GFP-positive) and wild-type P cells (GFP-negative) (l) and corresponding signal quantifications (n=6, two-sided Wilcoxon ranked-sum test) (m). (n-o) A wing disc harboring RpS3 +/- clones (GFP-positive) and stained for phospho-elF2α (red) (n) and corresponding signal quantification (n=9, two-sided paired t-test) (o). For all micrographs other than those in (k), scale bars correspond to 50μm. For all quantifications, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Extended Data Fig. 5. Proteostasis and the oxidative stress response.
(a-c) GstD1-GFP signal (green) in wild type (a) or RpS3 +/- wing discs (b) fed DMSO control or 10μM bortezomib, as indicated, and corresponding quantification (n=7, 8, 12, and 12, two-sided Mann-Whitney U test without p-adjustment for multiple comparisons) (c). (d-f) Wing discs harboring GFP-positive clones expressing MJDQ27 (d) or MJDQ78 (e) and stained with cleaved-dcp1 (red) and corresponding quantification of cell death (n=17 and 15 respectively, two-sided Wilcoxon signed-rank test without p-adjustment for multiple comparisons) (f). For all micrographs, scale bars correspond to 50μm. For all quantifications provided, the horizontal line represents the mean and whiskers indicate 95% confidence intervals. All n numbers refer to the number of individual wing discs.
Supplementary Material
Acknowledgments
We thank the Piddini group for input on the project and manuscript, Rafael Carazo Salas for feedback and discussions on the data and Life Science Editors for editorial assistance. We thank the Wolfson Bioimaging Facility for access to microscopes and for assistance in performing electron microscopy. We also thank the University of Bristol Proteomics Facility for performing the TMT proteomic experiments and for proteomics bioinformatics support. We are grateful to Tor Erik Rusten for the generous gift of the p62 antibody. This work was supported by Wellcome Trust PhD studentships to MPD and to IK, a Cancer Research UK Programme Grant to E.P (A12460), a Cancer Research UK Programme Foundation Award to E.P. (Grant C38607/A26831) and a Royal Society University Research fellowship to E.P. (UF0905080). E.P. is a Wellcome Trust Senior Research Fellow (205010/Z/, 16/Z).
Footnotes
Author contributions:
E.P. led the project. All authors conceived the experiments. M.P.D, M.B, I.K. and P.F.L performed and analysed the experiments. M.P.D, M.B, P.F.L and E.P. wrote the manuscript.
Financial and non-financial competing interests: The authors declare no competing interests.
Code availability
The Fiji-based custom-made script can be made available to individuals upon reasonable request, while we seek to publish it independently of this study.
Data availability
All source numerical data are provided in the Statistics Source Data table. All other data supporting the findings of this study are available upon reasonable request. The following publicly available databases were used in this study: Flybase (https://flybase.org); Uniprot D. melanogaster proteome (https://www.uniprot.org/proteomes/UP000000803).
References
- 1.Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Developmental Biology. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 2.Baker NE. Mechanisms of cell competition emerging from Drosophila studies. Curr Opin Cell Biol. 2017;48:40–46. doi: 10.1016/j.ceb.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maruyama T, Fujita Y. Cell competition in mammals - novel homeostatic machinery for embryonic development and cancer prevention. Curr Opin Cell Biol. 2017;48:106–112. doi: 10.1016/j.ceb.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Merino MM, et al. Elimination of unfit cells maintains tissue health and prolongs lifespan. Cell. 2015;160:461–476. doi: 10.1016/j.cell.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S, et al. Correction of aberrant growth preserves tissue homeostasis. Nature. 2017;548:334–337. doi: 10.1038/nature23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N, et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature. 2019;568:344–350. doi: 10.1038/s41586-019-1085-7. [DOI] [PubMed] [Google Scholar]
- 7.Vishwakarma M, Piddini E. Outcompeting cancer. Nature Reviews Cancer. 2020;20:187–198. doi: 10.1038/s41568-019-0231-8. [DOI] [PubMed] [Google Scholar]
- 8.Marygold SJ, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills EW, Green R. Ribosomopathies: There’s strength in numbers. Science. 2017;358 doi: 10.1126/science.aan2755. eaan2755. [DOI] [PubMed] [Google Scholar]
- 10.Baker NE. Cell competition. Curr Biol. 2011;21:R11–5. doi: 10.1016/j.cub.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 11.Ajore R, et al. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol Med. 2017;9:498–507. doi: 10.15252/emmm.201606660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker NE. Emerging mechanisms of cell competition. Nat Rev Genet. 2020;29:1–15. doi: 10.1038/s41576-020-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhiner C, et al. Flower Forms an Extracellular Code that Reveals the Fitness of a Cell to its Neighbors in Drosophila. Dev Cell. 2010;18:985–998. doi: 10.1016/j.devcel.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Meyer SN, et al. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science. 2014;346 doi: 10.1126/science.1258236. 1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baillon L, Germani F, Rockel C, Hilchenbach J, Basler K. Xrp1 is a transcription factor required for cell competition-driven elimination of loser cells. Sci Rep. 2018;8:17712–10. doi: 10.1038/s41598-018-36277-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kale A, Li W, Lee C-H, Baker NE. Apoptotic mechanisms during competition of ribosomal protein mutant cells: roles of the initiator caspases Dronc and Dream/Strica. Cell Death Differ. 2015;22:1300–1312. doi: 10.1038/cdd.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C-H, et al. A Regulatory Response to Ribosomal Protein Mutations Controls Translation, Growth, and Cell Competition. Dev Cell. 2018;46:456–469.e4. doi: 10.1016/j.devcel.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata R, Nakamura M, Sanaki Y, Igaki T. Cell Competition Is Driven by Autophagy. Dev Cell. 2019;51:99–112.e4. doi: 10.1016/j.devcel.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Blanco J, Cooper JC, Baker NE. Roles of C/EBP class bZip proteins in the growth and cell competition of Rp (‘Minute’) mutants in Drosophila. Elife. 2020;9:5184. doi: 10.7554/eLife.50535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kucinski I, Dinan M, Kolahgar G, Piddini E. Chronic activation of JNK JAK/STAT and oxidative stress signalling causes the loser cell status. Nature Communications. 2017;8:136. doi: 10.1038/s41467-017-00145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kale A, et al. Ribosomal Protein S12e Has a Distinct Function in Cell Competition. Dev Cell. 2018;44:42–55.e4. doi: 10.1016/j.devcel.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C-H, et al. A Regulatory Response to Ribosomal Protein Mutations Controls Translation, Growth, and Cell Competition. Dev Cell. 2018;46:807. doi: 10.1016/j.devcel.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milán M. Survival of the fittest. Cell competition in the Drosophila wing. EMBO reports. 2002;3:724–725. doi: 10.1093/embo-reports/kvf151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 25.Amoyel M, Bach EA. Cell competition: how to eliminate your neighbours. Development. 2014;141:988–1000. doi: 10.1242/dev.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho CMA. Growth and cell survival are unevenly impaired in pixie mutant wing discs. Development. 2005;132:5411–5424. doi: 10.1242/dev.02148. [DOI] [PubMed] [Google Scholar]
- 27.Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Baker NE. Engulfment is required for cell competition. Cell. 2007;129:1215–1225. doi: 10.1016/j.cell.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 29.Imai Y, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. The EMBO Journal. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Molecular and Cellular Biology. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamori Y, et al. Involvement of Lgl and Mahjong/VprBP in Cell Competition. Plos Biol. 2010;8 doi: 10.1371/journal.pbio.1000422. e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamori Y, Deng W-M. Cell competition and its implications for development and cancer. Journal of Genetics and Genomics. 2011;38:483–495. doi: 10.1016/j.jgg.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pakos-Zebrucka K, et al. The integrated stress response. EMBO reports. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain A, et al. p62/Sequestosome-1, Autophagy-related Gene 8, and Autophagy in Drosophila Are Regulated by Nuclear Factor Erythroid 2-related Factor 2 (NRF2), Independent of Transcription Factor TFEB. Journal of Biological Chemistry. 2015;290:14945–14962. doi: 10.1074/jbc.M115.656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva-Islas CA, Maldonado PD. Canonical and non-canonical mechanisms of Nrf2 activation. Pharmacol Res. 2018;134:92–99. doi: 10.1016/j.phrs.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 38.Mauvezin C, Ayala C, Braden CR, Kim J, Neufeld TP. Assays to monitor autophagy in Drosophila. Methods. 2014;68:134–139. doi: 10.1016/j.ymeth.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang Y-Y, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piddini E, Marshall F, Dubois L, Hirst E, Vincent J-P. Arrow (LRP6) and Frizzled2 cooperate to degrade Wingless in Drosophila imaginal discs. Development. 2005;132:5479–5489. doi: 10.1242/dev.02145. [DOI] [PubMed] [Google Scholar]
- 41.Sancho M, et al. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev Cell. 2013;26:19–30. doi: 10.1016/j.devcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paramore A, Frantz S. Bortezomib. Nat Rev Drug Discov. 2003;2:611–612. doi: 10.1038/nrd1159. [DOI] [PubMed] [Google Scholar]
- 43.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 44.Sung M-K, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol Biol Cell. 2016;27:2642–2652. doi: 10.1091/mbc.E16-05-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nature Publishing Group. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 46.Wyant GA, et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360:751–758. doi: 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tye BW, et al. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. Elife. 2019;8:3429. doi: 10.7554/eLife.43002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albert B, et al. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife. 2019;8:720. doi: 10.7554/eLife.45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 50.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p 62 is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 52.Bjedov I, et al. Mechanisms of Life Span Extension by Rapamycin in the Fruit Fly Drosophila melanogaster. Cell Metabolism. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metabolism. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harbor Perspectives in Biology. 2012;4 doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends in Biochemical Sciences. 2014 doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klockgether T, Mariotti C, Paulson HL. Spinocerebellar ataxia. Nat Rev Dis Primers. 2019;5:24–21. doi: 10.1038/s41572-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 57.Bonini NM. A genetic model for human polyglutamine-repeat disease in Drosophila melanogaster. Philos Trans R Soc Lond, B, Biol Sci. 1999;354:1057–1060. doi: 10.1098/rstb.1999.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Recasens-Alvarez C, Alexandre C, Kirkpatrick J, Nojima H, Huels DJ, Snijders AP, Vincent J-P. Ribosomopathy-associated mutations cause proteotoxic stress that is alleviated by TOR inhibition. Nature Cell Biology. 2020 doi: 10.1038/s41556-020-00626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coelho DS, et al. Culling Less Fit Neurons Protects against Amyloid-β-Induced Brain Damage and Cognitive and Motor Decline. CellReports. 2018;25:3661–3673.e3. doi: 10.1016/j.celrep.2018.11.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guang MHZ, et al. Targeting Proteotoxic Stress in Cancer: A Review of the Role that Protein Quality Control Pathways Play in Oncogenesis. Cancers. 2019;11:66. doi: 10.3390/cancers11010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu Rev Genet. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- 62.Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 63.Taylor RC, Dillin A. Aging as an Event of Proteostasis Collapse. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a004440. a004440–a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Q, Neal SJ, Pignoni F. Mutant analysis by rescue gene excision: New tools for mosaic studies in Drosophila. Genesis. 2016;54:589–592. doi: 10.1002/dvg.22984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katheder NS, et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541:417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gay P, Contamine D. Study of the ref(2)P locus of Drosophila melanogaster. II. Genetic studies of the 37DF region. Mol Gen Genet. 1993;239:361–370. doi: 10.1007/BF00276934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Fiji-based custom-made script can be made available to individuals upon reasonable request, while we seek to publish it independently of this study.
All source numerical data are provided in the Statistics Source Data table. All other data supporting the findings of this study are available upon reasonable request. The following publicly available databases were used in this study: Flybase (https://flybase.org); Uniprot D. melanogaster proteome (https://www.uniprot.org/proteomes/UP000000803).