Abstract
Glioblastoma (GBM) is a devastating human malignancy. GBM stem-like cells (GSCs) drive tumor initiation and progression. Yet, the molecular determinants defining GSCs in their native state in patients remain poorly understood. Here we used single cell datasets and identified GSCs at the apex of the differentiation hierarchy of GBM. By reconstructing the GSCs’ regulatory network, we identified the YAP/TAZ coactivators as master regulators of this cell state, irrespectively of GBM subtypes. YAP/TAZ are required to install GSC properties in primary cells downstream of multiple oncogenic lesions, and required for tumor initiation and maintenance in vivo in different mouse and human GBM models. YAP/TAZ act as main roadblock of GSC differentiation and their inhibition irreversibly lock differentiated GBM cells into a non-tumorigenic state, preventing plasticity and regeneration of GSC-like cells. Thus, GSC identity is linked to a key molecular hub integrating genetics and microenvironmental inputs within the multifaceted biology of GBM.
Glioblastoma (GBM) is the most frequent and lethal form of brain cancer. GBM is characterized by high degree of intratumoral cellular heterogeneity and plasticity, contributing to therapeutic resistance and recurrence1. At the cellular level, many of the malignant traits of GBM have been interpreted through the biology of a stem-like cell population (glioblastoma stem cells, GSCs), endowed with the ability to self-renew, to initiate tumors in vivo, and to give rise to a hierarchy of more differentiated progeny2-4. These attributes establish operational definitions and corresponding bioassays for GSCs, retrospectively identifying GSCs in primary GBM5. However, although these operational definitions are effective at revealing bona fide GSCs and allowing their experimental manipulation, they also necessarily introduce experimental biases that are hardly compatible with the need of molecularly characterizing GSCs as they exist in vivo. For example, GSCs are typically sorted from other GBM cell populations through cell surface markers of disputed and incomplete specificity5; moreover, GSCs need to be expanded ex-vivo under the confounding influence of artificial culturing conditions, clearly different from those existing in vivo (e.g. in term of growth factors availability, oxygen or mechanical gradients, and more). Then, GSCs are typically cultured ex-vivo as neurospheres, where they co-exist with their more differentiated progeny6. Thus, the molecular portrait of GSCs one can obtain through these procedures may differ substantially from that one of tumor-resident, native GSCs. In turn, these confounding caveats have so far ultimately limited our understanding of the molecular underpinning of the native GSC state, its key determinants and associated vulnerabilities. Addressing this gap is essential for the design of more effective therapies.
Single-cell (sc) analyses have the potential to overcome the above limitations, allowing for a less biased identification of GSC-like cells in their native environments. Identifying the molecular features of native GSCs in single-cell data is a open quest, with some studies focusing on plasticity, some on hierarchies, some on subtype-specific stem cells7,8,9. Here we started from single-cell data to identify gene-regulatory networks of native GSCs, leading to the discovery of YAP/TAZ as key molecular engines at the heart of GSC biology.
Results
A gene expression program identifying native GSCs
To identify native GSC-like cells we aimed at visualizing the natural trajectory of differentiation of GBM cells. For this, we used scRNA-seq profiles from 32 IDH-wild type GBM patients7,8, and then validated our conclusions in large patients’ cohorts of the TCGA and REMBRANDT datasets.
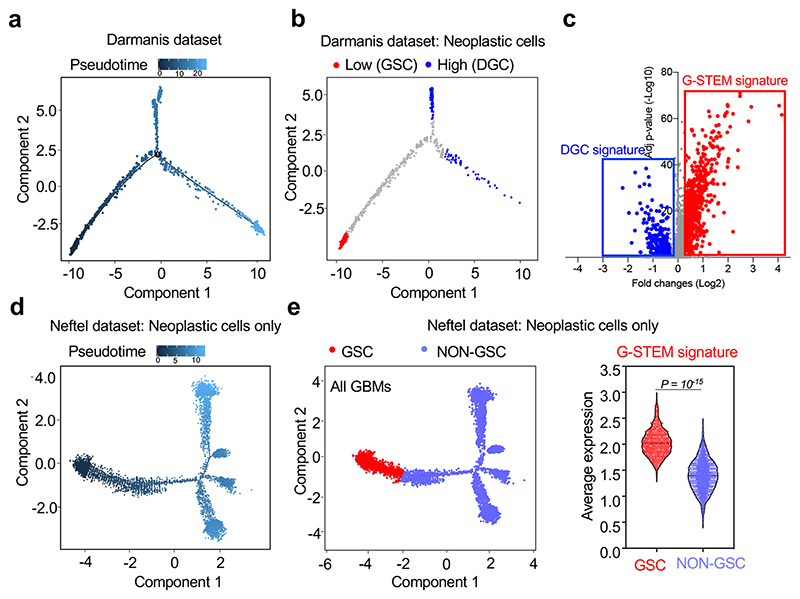
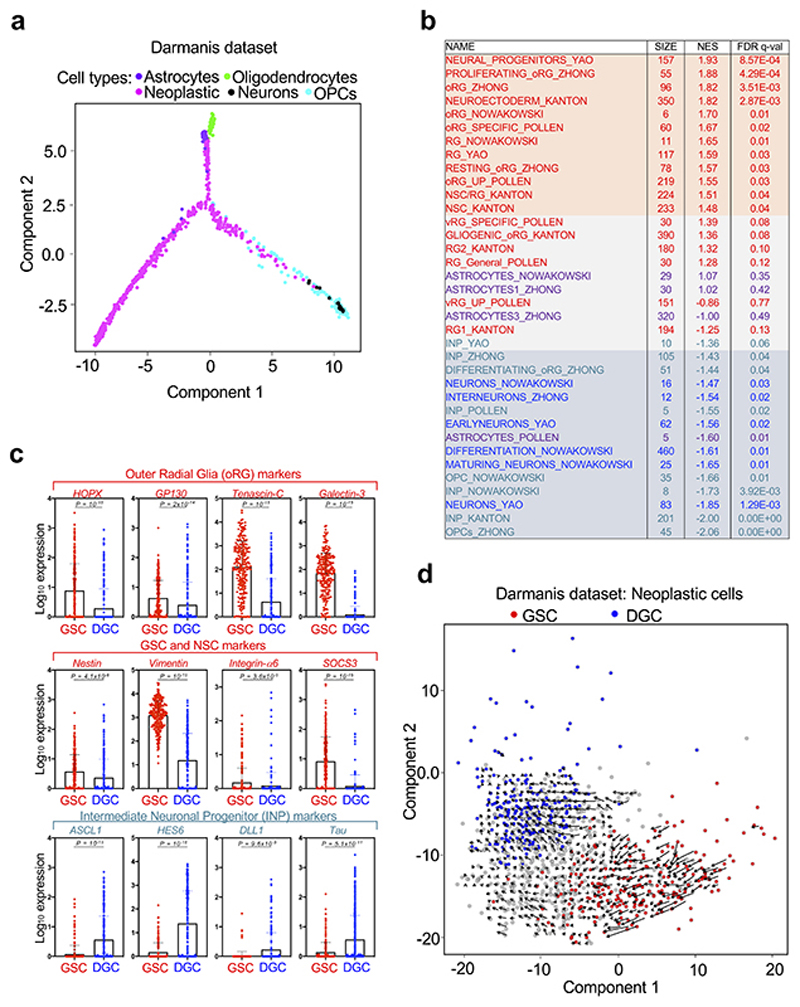
We started our investigation using Monocle10, an algorithm allowing ordering cells in a trajectory based on a “pseudotime”, a quantitative measure of the progress along a biological process defined by changes in cellular transcriptional programs. By applying this method to a scRNA-seq study of primary GBM samples (Darmanis dataset)7, we found that neoplastic cells organized along a tripartite trajectory (Fig. 1a and Extended Data Fig. 1a). To gain more insights on the nature of the cells that are at the opposite ends of this pseudotime trajectory (Fig. 1b), we compared their transcriptomes. Taking advantage of signatures derived from scRNA-seq studies of human neural development (Supplementary Table 1), we first found that neoplastic cells endowed with the highest pseudotime value were enriched for markers of differentiated neural cells (astrocytes and neurons) or of committed precursors (both OPCs and Intermediate Neuronal Precursors, INPs), as defined by Gene Set Enrichment Analysis (GSEA) (Extended Data Fig. 1b). Thus, these neoplastic cells likely represent a population of differentiated glioblastoma cells (DGCs). Conversely, cells with the lowest pseudotime value were enriched for markers of neural stem cells (NSCs) and of early neural progenitors, such as the outer Radial Glia (oRG), a primate-specific neural progenitor cell type endowed with migratory properties11 (Extended Data Fig. 1b-c). These cells expressed higher levels of markers previously associated to GSCs and NSCs, such as Nestin12, Vimentin11, Integrin-α613 and SOCS314, and lower levels of INP markers in comparison to DGCs (Extended Data Fig. 1c). All together, these data support the identification of the cell population at the start of the pseudotime trajectory as prospective native GSCs.
Fig. 1. A gene expression program identifying native GSCs.
(a) Single-cell differentiation trajectory of GBM cells reconstructed by Monocle2 using single-cell RNA-seq data from primary GBM samples of the Darmanis dataset, using the cell populations indicated in Extended Data Fig. 1a.
(b) Single-cell differentiation trajectory of the GBM cells of the Darmanis dataset hilighting the putative GSC and DGC cell populations, identified as neoplastic cell populations displaying Low (< first quartile) or High (> third quartile) pseudotime values, respectively.
(c) Volcano plot of the gene expression changes between the GSC and DGC populations of the Darmanis dataset, with indicated the genes composing the G-STEM and the DGC signatures.
(d) Single-cell trajectory of GBM cells reconstructed by Monocle2 using single-cell RNA-seq data from the sole neoplastic cells of primary GBM samples of the Neftel dataset.
(e) Violin plots showing the expression of the G-STEM signature (right panel) on the cells at the start of the pseudotime trajectory of the Neftel dataset (GSC; red dots in the left panel) vs. all the other neoplastic cells (NON GSC; light blue dots in the left panel). The p-value was determined by two-tailed Mann-Whitney test.
Although Monocle is one of the commonly used tools for delineating cell differentiation trajectories, it should be noted that such analyses do not provide a sense of direction. For this, we complemented the above analyses with “RNA velocity”15, a tool allowing the reconstruction of the differentiation trajectory of the sole neoplastic cells. Based on the dynamic of mRNA processing and degradation, this algorithm returns, for each cell, a vector (visualized as an arrow) that ultimately indicates the direction of the differentiation. As shown in Extended Data Fig. 1d, these vectors identify a differentiation trajectory that starts from GSCs (red dots) and gradually transits into DGCs (blue dots), as such solidifying the results previously obtained with Monocle.
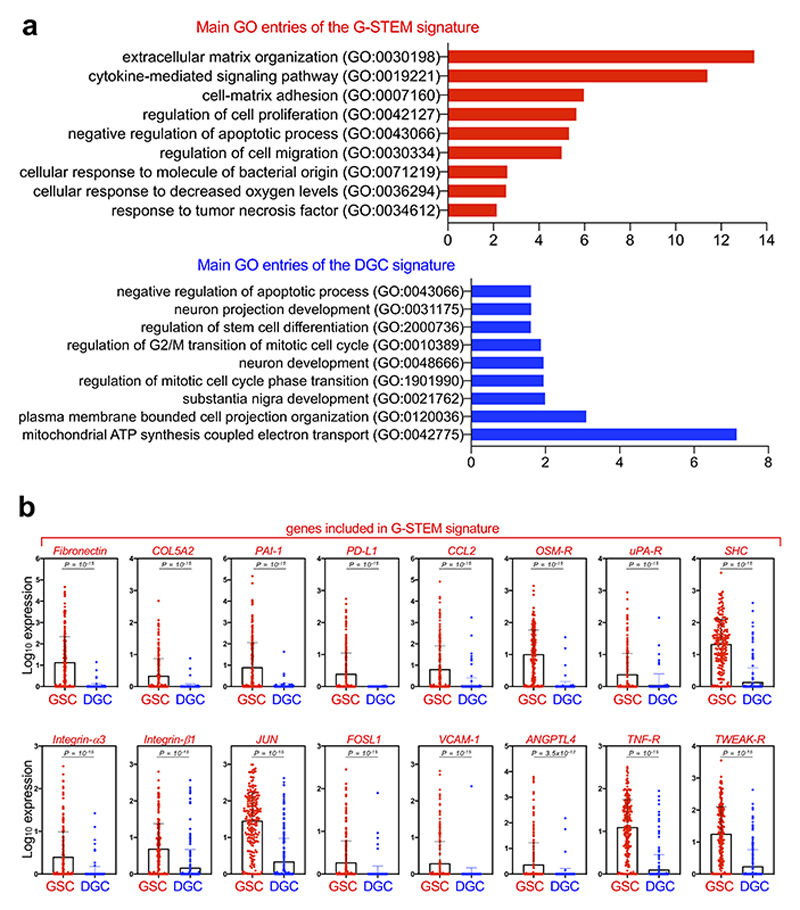
At this point, we characterized the gene expression program typifying this GSC-like cell population. We derived a gene signature, hereafter G-STEM, consisting of the genes more significantly upregulated in this cell population when compared to DGCs (Fig. 1c; Supplementary Table 2). By Monocle analyses, the G-STEM signature identifies cells at the start of the pseudotime trajectory in each individual patient (Extended Data Fig. 2a and 2b). Moreover, in order to avoid the risk of any polarization in cell trajectory caused by the presence of scRNA of normal cells, we repeated the Monocle analyses using only neoplastic cells (Darmanis dataset), finding that, also in these conditions, G-STEM identifies cells at the start of the differentiation trajectory (Extended Data Fig. 2c).
As revealed by gene ontology analysis (Extended Data Fig. 3a and 3b; Supplementary Table 3), G-STEM contains genes coding for proteins involved in ECM organization, cell-ECM adhesion, promotion of cell migration, control of cell proliferation and survival, recruitment of innate immune cells, protection against immune responses, and transduction of various extracellular signals. Overall, this molecular profile is consistent with the view that native GSCs entertain mutual relationships with their microenvironement16-18. In contrast, the gene expression program of DGCs (DGC-signature, Supplementary Table 2) mostly contains genes coding for factors involved in the early steps of neuronal differentiation (Extended Data Fig. 1c), indicating that DGCs encompass neuronal precursor-like cell states.
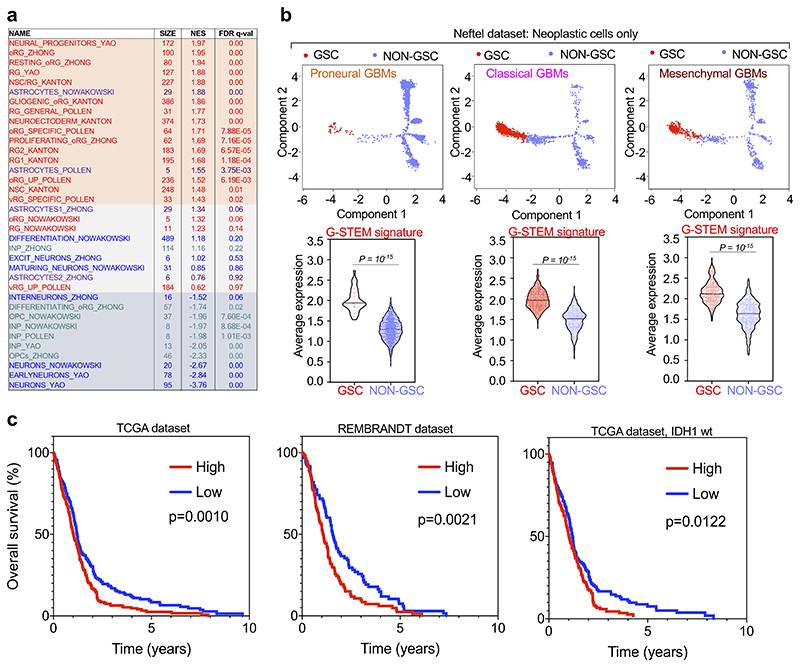
GBMs can be classified in at least three transcriptional subtypes, defined as Proneural, Classical and Mesenchymal; Proneural and Mesenchymal GBMs represent two extremes in terms of molecular marker expression and patients’ survival1. Subsequent studies have shown that the GSCs of Proneural and Mesenchymal GBMs are characterized by expression of subtype-specific cell surface markers19, raising the possibility that different GBM subtypes may originate from biologically distinct GSCs. To tackle this hypothesis, we used an independent scRNA-seq dataset from Neftel et al.8, containing 28 tumors including all GBM subtypes. Applying Monocle to the sole neoplastic cells of the Neftel dataset retrieved a complex pseudotime trajectory with many branches (Fig. 1d), consistently with the previously reported heterogeneity of GBM cell and associated plasticity. Nonetheless, at the start of the pseudotime trajectory we could identify tumor cells characterized by high-level of the G-STEM signature (Fig. 1e). Intriguingly, by GSEA, this cell population resembled human neural stem cells (NSCs) and early neural progenitors (Extended Data Fig. 4a). Collectively, these analyses identified ostensible GSC-like cells in the largest GBM sc-RNA-seq collection to date. Remarkably, when we focused on the distinct Proneural, Classical or Mesenchymal GBM subtypes, the G-STEM was invariably enriched in the cells at the start of the pseudotime trajectory (Extended Data Fig. 4b). Together, these data indicate that the GSC-like cells across all GBM subtypes share the G-STEM signature.
Data presented so far provided a molecular characterization of GSCs; next, we tested whether this correlated with the biological properties of prospective GSCs within GBM. Consistently with the role of GSCs in tumor initiation, aggressiveness and relapse, a number of studies highlighted that the GSC representation in tumor samples represents a considerable prognostic factor for poor clinical outcome in GBM patients20-22, raising the possibility that the G-STEM signature may correlate with outcome in patients. To test this prediction, we applied the GSTEM for Kaplan-Maier survival analysis of GBM patients from two large datasets, the TCGA and the REMBRANDT projects23,24. Patients were stratified in two groups, according to High or Low expression of the G-STEM signature. As shown in Extended Data Fig. 4c, high levels of G-STEM expression are indeed predictive of worse outcome in both datasets. We conclude from this collective set of results that the G-STEM signature represents a novel transcriptional program that identifies native GSCs at the top of the differentiation hierarchy in GBM patients.
Identification of candidate master Transcriptional Regulators of the GSC state
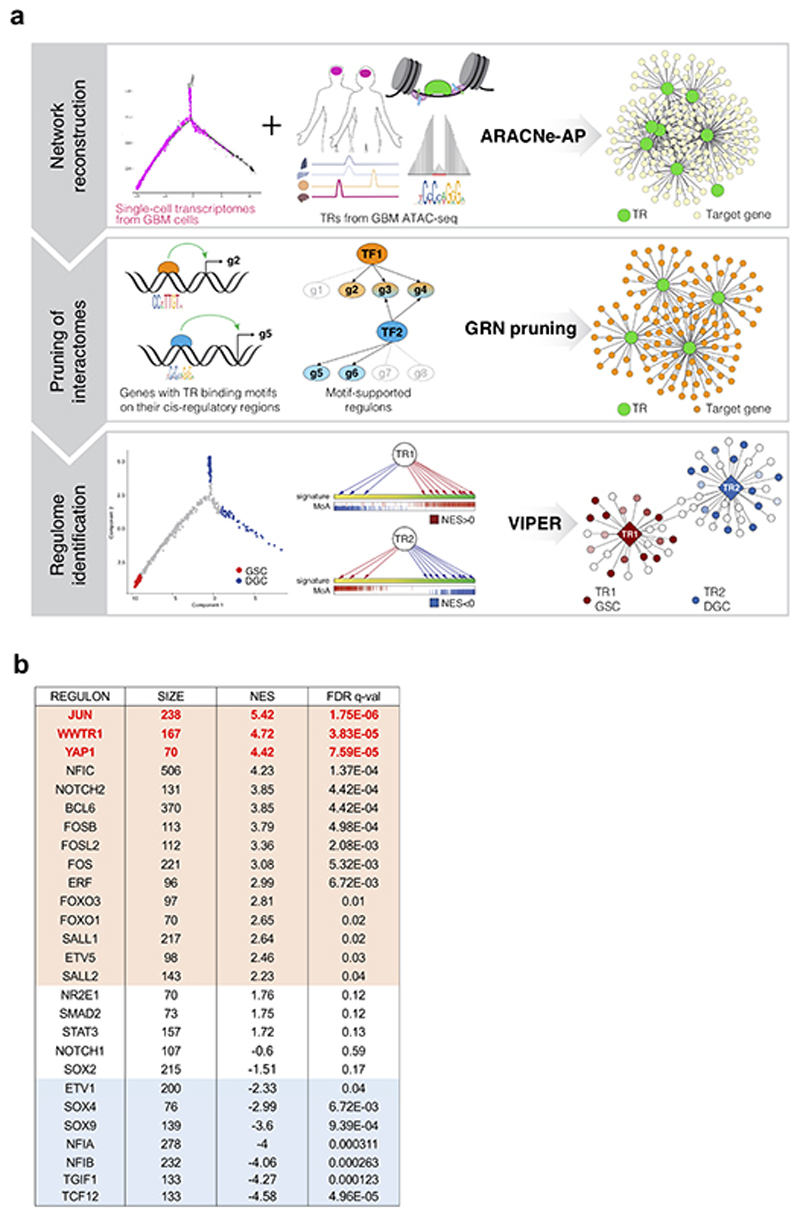
Next, we aimed to attain a deeper understanding of the molecular nature of native GSCs, whose gene-expression and functional attributes ultimately depend on transcriptional regulators (TRs) regulating each other and their downstream target genes (i.e., “regulons”), as such defining Gene regulatory networks (GRNs)25. Here we aimed at the identification of the master TRs of the GSC state. This quest was challenged by the fact that computational tools able to unbiasedly infer master TRs from single cell RNA-seq data are currently underdeveloped; indeed, the intrinsic characteristics of scRNA data - i.e., high variability per gene detected between cells, and high rates of zero values (i.e., dropouts) inherent to mRNA undersampling26,27 - present great technical challenges for TR inference. To overcome these limitations, we combined the established algorithms of ARACNE and VIPER in a multistep computational pipeline, named “Rhabdomant” (see scheme and details in Extended Data Fig. 5a and Methods), that first reconstructs a coarse gene regulatory network (GRN) from single cell gene expression profiles and transcription factors active in GBM (step 1), then “prunes” this GRN by anchoring inferred regulatory interactions to putative direct target genes of TRs (step 2), and finally prioritizes master TRs from the differential enrichment of their regulons in different cell states (step 3; Extended Data Fig. 5a).
Of the list of TRs we obtained from the Rhabdomant pipeline (Supplementary Table 4), we decided to focus our attention on the 27 TRs with the largest regulons (Extended Data Fig. 5b), controlling more than 95% of the Gene Regulatory Network (1409 out of 1465 genes). Of these TRs, 15 were candidate “master” TRs of the GSC state, indeed cumulatively controlling a large part (96%) of the G-STEM signature. Several of these factors (SALL2, NOTCH2, ETV5, FOXO1/3) are known regulators of stemness properties in both GSCs and NSCs alike28-31. As for the DGC state, we retrieved 7 TRs, most of which (TCF12, NFIA, NFIB, SOX9, SOX4) are in fact known to be involved in NSC differentiation toward neuronal or glial fates32-34. Together these results nicely validate the ability of our computational approach to identify biologically meaningful transcriptional regulators of cell states out of single-cell transcriptomic data.
Scoring the list of the most significant (FDR <0.0001) candidate master TRs of the GSC state, we focused our attention on the transcriptional coactivators YAP1 and WWTR1 (also known as TAZ). In epithelial tumors, YAP/TAZ are essential for tumor initiation and progression by inducing stemness, proliferation and chemoresistance35. Intriguingly, YAP/TAZ activation has recently emerged as hub for tumor-stromal interaction35, integrating multiple inputs, including mechanical signaling and hypoxia, that are indeed profoundly dysregulated in the GBM microenvironment, and associated to GBM recurrence1,16,18. YAP/TAZ have been reported to be regulated by CD10919, a marker of mesenchymal GBM, although the functional significance of this regulation remains undefined. Moreover, elevated YAP/TAZ expression levels have been noted in GBM, and correlated with shorter survival of glioma patients36,37, but their functional involvement in GSCs remains unexplored. Our interest on YAP/TAZ was further motivated by the fact that AP-1 family members also scored at the top of the candidate master TRs of the GSC state in our analyses. Indeed, a series of recent findings have revealed that AP-1 is a pervasive transcriptional partner of YAP/TAZ on vast number of cis-regulatory elements, and that AP-1 is functionally required for YAP/TAZ responses38,39,40. Collectively, these considerations prompted us to focus on YAP/TAZ activity as candidate overarching factor in defining the GSC state.
YAP/TAZ activity is associated to the GSC state
We addressed more directly whether YAP/TAZ are indeed specifically active in native GSCs. We thus zoomed into the GRN architecture and asked to what extent the GSC state, as defined by G-STEM, could be explained by placing YAP/TAZ at the center of a gene-regulatory cascade controlling progressive layers of downstream TRs and their targets (Fig. 2a). The YAP/TAZ-controlled GRN structure contains FOXO1 as immediate YAP/TAZ regulated downstream TR; FOXO1 regulates FOS and SALL1 that in turn regulate FOXO3, BCL6 and ERF. Collectively the target genes of these TRs accounts for a remarkable 73% of the whole G-STEM gene list, with YAP/TAZ alone directly controlling one third of this network (Supplementary Table 5).
Fig. 2. YAP/TAZ are master Transcriptional Regulators of the GSC state.
(a) Depiction of the part of the GRN of GBM controlled by YAP1, TAZ (WWTR1) and their downstream TRs. TRs are represented as diamonds; genes composing the G-STEM signature are represented as yellow dots with red borders; all the other genes composing the regulons of YAP/TAZ and of downstream TRs are depicted as small grey dots. Black edges identify the regulatory interactions between YAP/TAZ and their downstream TRs.
(b, c) Gene Set Enrichment Analysis (GSEA) enrichment score curves of the G-STEM signature (b) and the list of TRs found downstream to YAP/TAZ in the GRN of GBM (FOXO1, SALL1, FOS, FOXO3, BCL6 and ERF) (c) in YAP/TAZ knockout vs. YAP/TAZ wild-type subcutaneous KRASG12V/shp53 GBM-like tumors obtained as described in Extended Data Fig. 6. Signatures are available in Supplementary Table 7.
(d) Right panel: Heatmap showing the percentage of cells showing nuclear TAZ as deduced by immunohistochemistry (IHC) of samples from different tumor areas of 67 GBMs. Left panels: pictures of TAZ IHC in GBM samples from three different tumor areas (representative of n=25 peripheral samples, top; n=38 tumor bulk samples, middle; n=28 perinecrotic samples, bottom). ‘N’ indicates necrotic areas. Scale bars, 100 μm.
(e) Right panel: Heatmap showing standardized gene expression of the G-STEM and the DGC signatures in different histologically-defined tumor domains (the pseudopalisading cells located around necrotic areas, the “cellular tumor”, representing the bulk of tumor cells, the “infiltrating tumors” areas, where tumor cells insinuate themselves into the normal tissue, and the tumor cell-free margin, called the“leading edge”) of six different GBMs from the Ivy Atlas. Left panel: Schematics of the histologically-defined tumor domains of GBM.
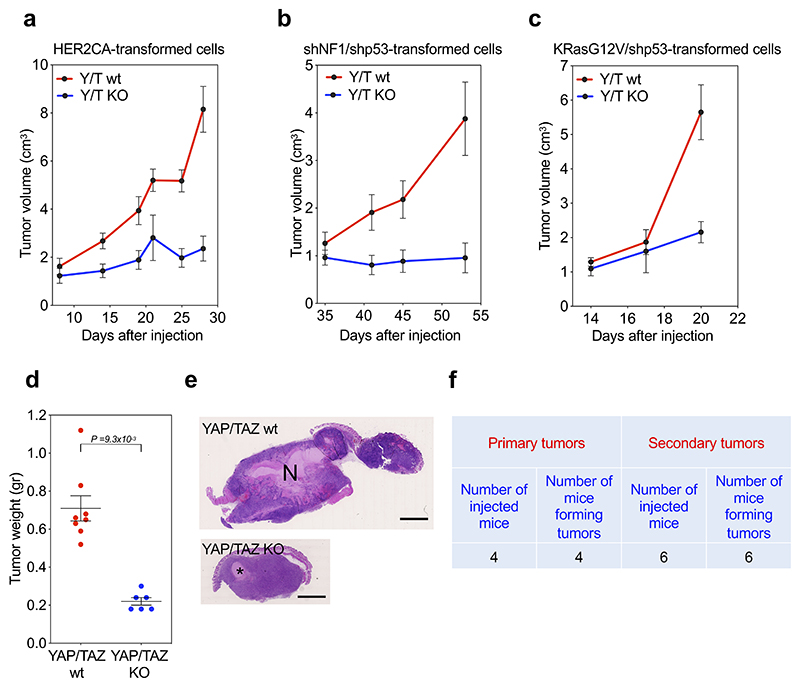
Next, we addressed experimentally to what extent YAP/TAZ are in fact required for G-STEM expression and, more broadly, for GSC biology in vivo. For this, we triggered YAP/TAZ knockout in pre-established, full blown GBM-like neoplasms derived from transformed cells bearing different oncogenic insults (KRasG12V/shp53, HER2CA or shNF1/shp53); as detailed below, these transformed cells display GSC-like properties, including ability to generate orthotopic and subcutaneous tumors driven by YAP/TAZ and displaying several features of human GBM. To assess the effects of YAP/TAZ depletion in pre-established tumors, we opted for sub-cutaneous injection12, as this set up allows careful monitoring of tumor growth and unambiguous retrieval of neoplastic cells. While YAP/TAZ wild-type tumors kept expanding, YAP/TAZ knockout halted tumor growth (Extended Data Fig. 6a-e), a result consistent with loss of GSCs in tumorigenic cell populations12,41. From YAP/TAZ wild-type tumors (4 mice out of 4), we could retrieve gliomasphere-forming cells that could be passaged ex-vivo, and that are able to re-initiate tumorigenesis in vivo (Extended Data Fig. 6f). In contrast, no gliomasphere-forming cells could be retrieved from YAP/TAZ knockout tumors (n=5). Importantly, at the molecular level, YAP/TAZ inactivation in tumors caused loss of G-STEM expression (Fig. 2b), and also led to the collapse of the TR architecture identified in our GRN as operating downstream of YAP/TAZ (i.e., with loss of FOXO1, SALL1, FOS, FOXO3, BCL6 and ERF expression; Fig. 2c). Collectively the findings indicate that YAP/TAZ are required to preserve the GSC state, that is identified by a YAP/TAZ-dependent G-STEM signature in vivo.
To further validate the connection between YAP/TAZ activity and GSCs, we quantified by immunohistochemistry active TAZ (i.e., nuclear and stabilized) in human GBM sections, revealing massive activation in the perinecrotic areas and a progressively more salt-and-pepper, heterogeneous staining toward the tumor periphery (Fig. 2d); these findings are nicely consistent with prior reports suggesting that the GBM perinecrotic areas are indeed enriched in GSC representation1,18. Moreover, we also detected the highest levels of G-STEM in perinecrotic areas of the Ivy Atlas42, a molecular pathology atlas providing gene expression data from human GBM after laser microdissection and RNA-seq of different histologically-defined tumor areas (Fig. 2e). Thus, G-STEM expression peaks in the same tumor areas where active TAZ peaks.
YAP/TAZ are required for oncogenes to confer GSC properties to normal neural cells
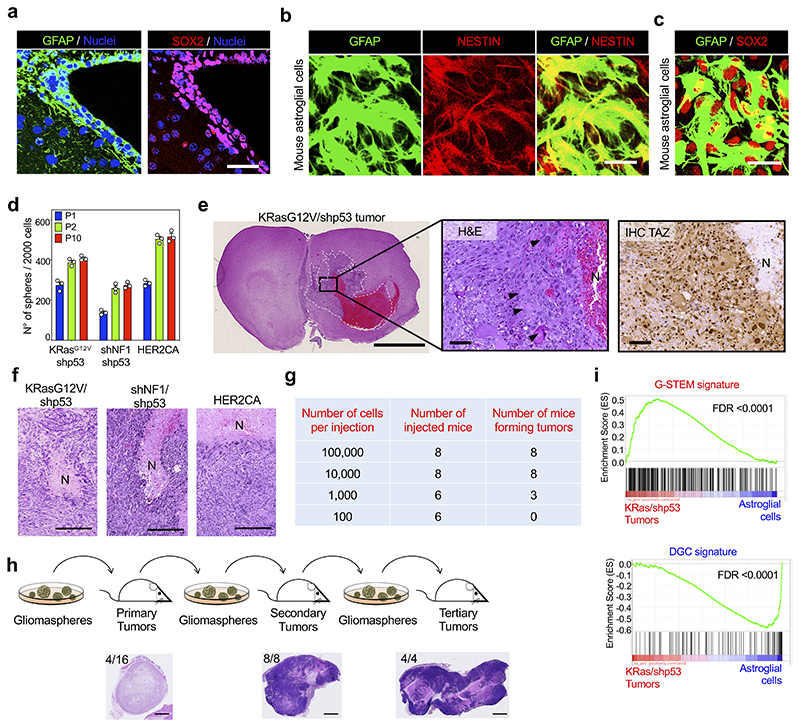
Given the activation of YAP/TAZ transcriptional programs in prospective GSCs, we next asked whether YAP/TAZ activity may represent an early addiction in gliomagenesis. Compelling evidence indicate that the cells of origin of GBM include various cell types, including astrocytes43,44, OPCs45-47 and, as recently described in human GBMs, cells of the subventricular zone (SVZ)48 that display mixed astrocyte-like and NSC-like features, such as expression of GFAP, NESTIN and SOX2 (Ref.48 and Extended Data Fig. 7a). Typical oncogenic lesions in GBM entail overactivation of the RTK/RAS pathway through genetic amplification or activating mutations of RTK-coding genes (PDGFRα, EGFR, and HER2), or mutation of the RAS inhibitor NF149.
To recapitulate, at least in part, the early step of gliomagenesis, we introduced activated oncogenes in astroglial cells from newborn mice that are highly similar to the SVZ cell population50, expressing mixed astrocyte-like (GFAP) and NSCs markers (NESTIN, SOX2) (Extended Data Fig. 7b, c). Newborn astroglial cells were transduced with lentiviral vectors coding for the activated forms of PDGFRα, EGFR or HER2, with vectors coding for oncogenic KRasG12V plus shRNA against p53, or shRNAs against NF1 and p53; this list includes established drivers of human GBM49. After approximately 3 weeks, gliomaspheres emerged from all oncogene-expressing primary astroglial monolayers, and never from controls (Fig. 3a). Gliomaspheres could be dissociated as single cells and expanded over several passages in suspension (Extended Data Fig. 7d); when transplanted orthotopically (intracranially) in immunocompromised mice, dissociated gliomasphere cells gave rise to tumors well-recapitulating key histological features of human GBM, also resembling Giant Cell Glioblastoma51, and displaying elevated levels of active TAZ in the perinecrotic regions (Extended Data Fig. 7e). Similar tumors emerged after subcutaneous cell transplantation, where they also included areas of necrosis surrounded by pseudopalisading cells, also common features of human GBM (Extended Data Fig. 7f).
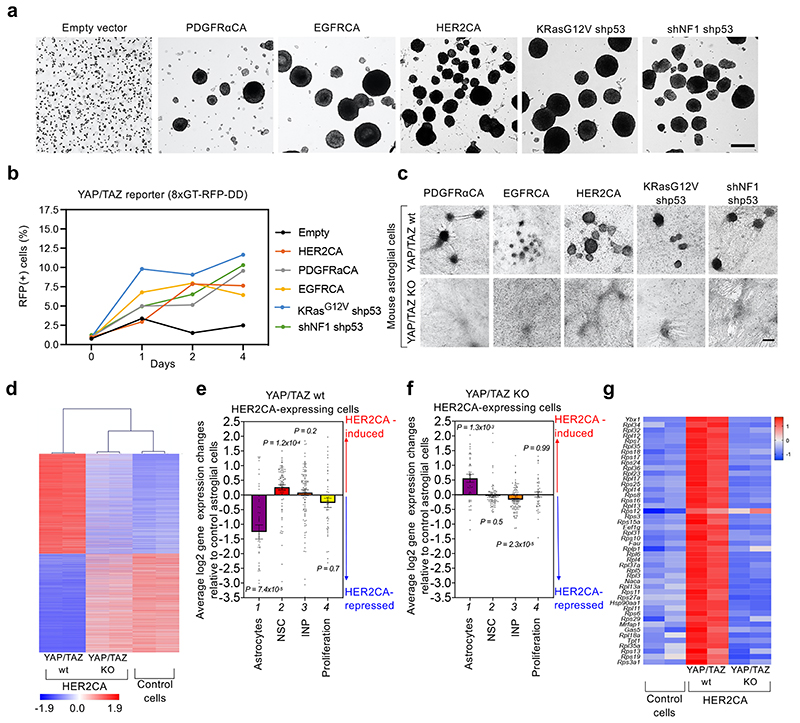
Fig. 3. YAP/TAZ are required for oncogene-dependent transformation of primary normal neural cells.
(a) Mouse newborn astroglial cells were transduced either with empty vector or with lentiviral vectors encoding for the indicated oncogenes. Bright field images of gliomaspheres (representative of n=3 experiments each) are shown. As negative control, newborn astroglial cells transduced with empty vector were not able to form any gliomasphere in suspension. Scale bar, 100 μm.
(b) Mesurement of YAP/TAZ activity in mouse newborn astroglial cells during the first days of oncogenic reprogramming. Cells were transduced with with lentiviral vectors encoding for the YAP/TAZ reporter 8xGTIIC-RFP-DD52, and with lentiviral vectors encoding for the indicated oncogenes or, as negative control, with empty vector. The graph represents the percentage of RFP-positive cells detected in newborn astroglial cell cultures at the indicated time points after the start of oncogenic reprogramming in NSC medium. The number of cells counted for each sample is reported in the corresponding Source Data file.
(c) YAP/TAZ are required for oncogene-induced transformation of mouse newborn astroglial cells. Cells from R26CAG-CreERT2; Yapfl/fl; Tazfl/fl animals were transduced with lentiviral vectors encoding for the indicated oncogenes and then cultured in NSC medium to form gliomaspheres. When 4OH-Tamoxifen (4OH-TAM) was added to the NSC medium to induce YAP/TAZ depletion, no gliomaspheres arising from the cell monolayer were observed. Images are representative of n=3 experiments each. Scale bar, 100 μm. See also Extended Data Fig. 8b for efficiency of Yap/Taz depletion.
(d) Hierarchical clustering of gene expression profiles from RNA-seq data of control (right), or HER2CA-expressing mouse newborn astroglial cells, either in the presence of endogenous YAP/TAZ (YAP/TAZ wt, left) or in YAP/TAZ-knockout setting (YAP/TAZ KO, middle). The heatmap shows standardized expression of genes significantly upregulated or downregulated in YAP/TAZ wt astroglial cells expressing HER2CA, compared to control cells. Genes are ordered according to decreasing average expression in HER2CA-transduced YAP/TAZ wt astroglial cells.
(e, f) Average log2 gene-expression changes of signatures for the indicated cell types (Astrocytes n=45 genes; NSC n=89 genes; INP n=106 genes) or for proliferating neural progenitors (Proliferation n=45 genes) in HER2CA-expressing YAP/TAZ wt newborn astroglial cells (e) or HER2CA-expressing YAP/TAZ KO newborn astroglial cells (f), compared to control newborn astroglial cells. NSC: Neural Stem Cells; INP: Intermediate Neuronal Progenitors. Signatures are derived from Ref53 and listed in Supplementary Table 6. Data are shown as mean and standard error of the mean (s.e.m.). Positive and negative values indicate, respectively, upregulation and downregulation of the indicated signatures by expression of HER2CA in newborn astroglial cells. p-values were determined by Brown-Forsythe and Welch one-way ANOVA test with Dunnett’s T3 multiple comparisons of the distribution of log2 gene-expression changes of each signature with the distribution of log2 gene-expression changes for all expressed genes (n=11,946).
(g) Heatmap showing standardized gene expression of NSCs marker genes upregulated by HER2CA in YAP/TAZ wt newborn astroglial cells (middle) compared to control cells (Co.; left) and HER2CA-expressing YAP/TAZ-KO cells (right). Genes are ordered according to the decreasing average scores in HER2CA-expressing YAP/TAZ wt newborn astroglial cells. The only gene that is upregulated by HER2CA expression irrespectively of YAP/TAZ knockout is Rps12.
Upon limiting-dilution transplantation, we found that 1,000 dissociated gliomasphere cells were sufficient to seed tumor formation (Extended Data Fig. 7g). When explanted and re-cultured ex-vivo, tumor cells remained able to form gliomaspheres that could be expanded again over several passages in culture, retaining the capacity of tumor-initiation upon serial transplantation in vivo (Extended Data Fig. 7h). As control, we could not detect any formation of spheroids from control newborn astroglial cells ex vivo (Fig. 3a); then, no outgrowth whatsoever could be detected when these parental cells were injected in recipient mice. In other words, overexpression of RAS/RTK oncogenes in otherwise normal primary cells specifically converts them into cells endowed with typical properties of GSCs, such as tumorigenic potential and the ability to undergo self-renewal, in vitro and in vivo. In accordance, by RNA-seq, tumors displayed elevated expression of the G-STEM-signature and low expression of the DGC-signature when compared with the original astroglial cell cultures (Extended Data Fig. 7i).
Are YAP/TAZ activated by oncogenic mutations during acquisition of a GSC-like state? To address this question, we first verified that YAP/TAZ were activated during the early phases of reprogramming of newborn astroglial cells into GSC-like cells by oncogenes. For this, we transduced early postnatal astroglial cells with 8xGTIIC-RFPDD, a YAP/TAZ-responsive lentiviral reporter52, and then monitored the expression of RFP during oncogene-mediated reprogramming. As shown in Fig. 3b, oncogenes specifically induced activation of the reporter within 24 hours of reprogramming; RFP-positive cells acquired elongated shapes reminiscent of that of the radial glia, spreading and migrating, typically converging toward one cell cluster (Extended Data Fig. 8a). Overall, these data indicate that YAP/TAZ activation is an early event during reprogramming of normal astroglial cells into tumorigenic GSC-like cells.
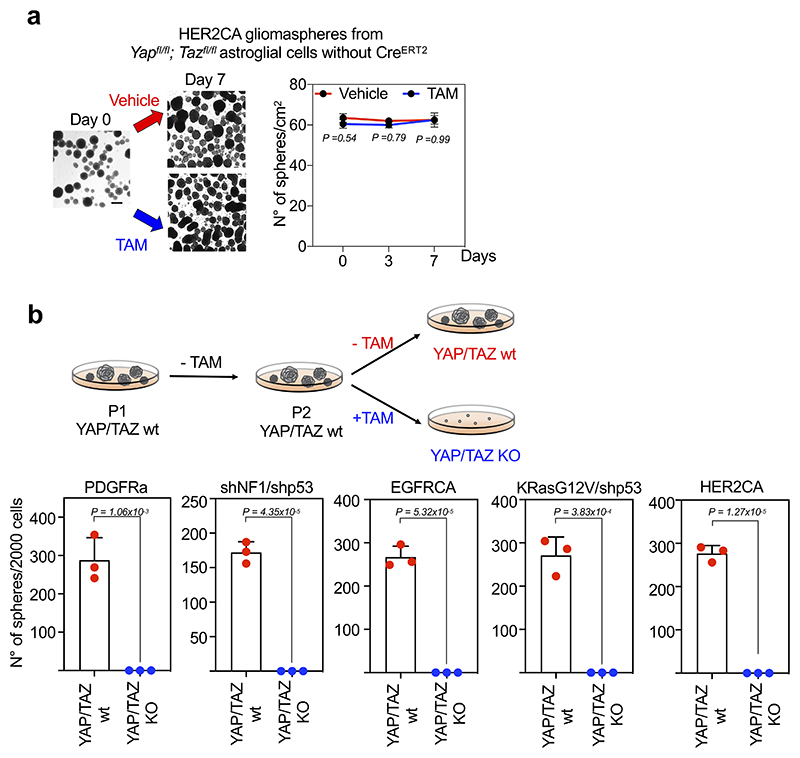
We next asked whether YAP/TAZ are required for oncogene-mediated reprogramming of normal cells into GSC-like cells. To this end, we derived astroglial cells from newborn R26 CAGCreERT2; Yap fl/fl; Taz fl/fl mice, in which genetic ablation of YAP/TAZ was induced after CRE activation by 4OH-Tamoxifen (4OH-TAM) treatment. As shown in Fig. 3c, gliomasphere formation by various RTK oncogenes, either transduced alone or in combination with shRNA targeting tumor suppressors, was completely abolished by YAP/TAZ deletion.
Next, we investigated at the molecular level how YAP/TAZ contribute to oncogenic reprogramming of normal neural cells into GSCs. We performed transcriptomic analyses by RNA-seq of astrocytes from Yap fl/fl;Taz fl/fl mice transduced with lentiviral vectors encoding CreERT2 and a doxycycline-inducible HER2CA, in absence or presence of 4OH-TAM at the incipit of cell transformation. By hierarchical clustering of samples using genes whose expression is significantly altered by HER2CA, we found that the transcriptome of YAP/TAZ-knockout cells remained remarkably similar to that of control/non-transformed cells in spite of HER2-overexpression (Fig. 3d). Specifically, HER2CA expression in newborn astroglial cells induces YAP/TAZ-dependent downregulation of astrocyte markers and upregulation of NSCs markers53 (compare lanes 1 and 2 of Fig. 3e with lanes 1 and 2 of Fig. 3f). Strikingly, 45 out of 46 NSCs marker genes upregulated by HER2CA in control cells are dependent on YAP/TAZ (Fig. 3g). Surprisingly, however, at the incipit of oncogenic transformation, YAP/TAZ appear dispensable for the expression of genes involved in proliferation (Fig. 3f, lane 4). Overall, these results indicate that the main function of YAP/TAZ during GBM initiation is to repress differentiation and promote the acquisition of NSC-like properties.
YAP/TAZ are required for the intrinsic differentiation plasticity of GBM cells
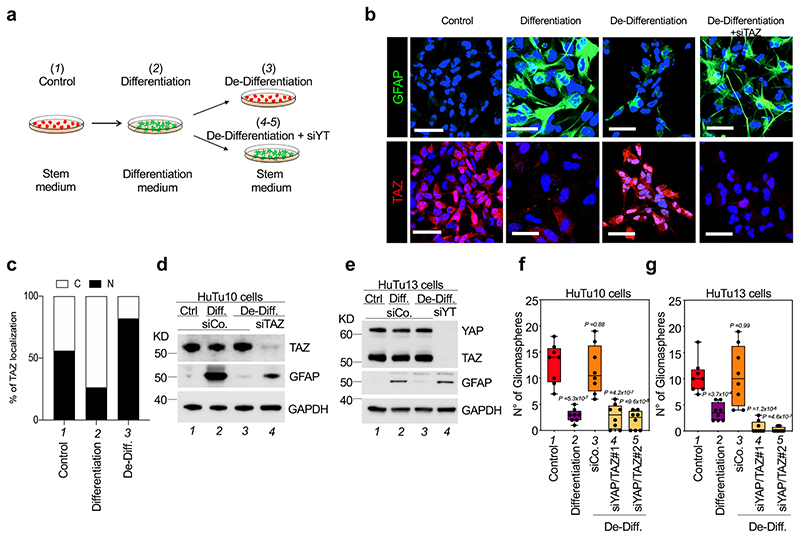
One of the most lethal properties of GBM cells is their intrinsic plasticity54, allowing for interconversion between GSC and non-GSC states depending on a number of factors, as such promoting tumor relapse55. We thus tested the role of YAP/TAZ in GBM plasticity. For this, we took advantage of a previously reported differentiation/de-differentiation protocol for human GBM cells54,56 (Fig. 4a). Specifically, we used two independent primary GMB cell lines, HuTu10 and HuTu1357, corresponding, by transcriptomics, to proneural and mesenchymal subtypes, respectively. When cultured in serum-free, growth-factor rich conditions, both cell types display no expression of the astrocyte marker GFAP; however, after exposure to serum and BMP2-containing media, cells en mass differentiate into GFAP-positive cells with a stellate morphology, reminiscent of normal astrocytes (Fig. 4b, and Fig. 4d, e, compare lanes 1 and 2). This event is accompanied by a dramatic decrease in the ability to form gliomaspheres (Fig. 4f, g, compare lanes 1 and 2), indicating that acquisition of astrocyte-like properties is associated with loss of stemness properties. Of note, this process is also accompanied by relocalization of TAZ in the cytoplasm, indicating that YAP/TAZ are inactivated during differentiation (Fig. 4b and Fig. 4c, compare lanes 1 and 2). The return to a GSC-like, less differentiated phenotype can be induced by placing cells back into serum-free, growth-factor rich culture conditions (de-differentiation), promoting YAP/TAZ re-entry in the nucleus, progressive disappearance of GFAP and re-acquisition of gliomasphere-forming abilities (Fig. 4b and Fig. 4c-g, compare lanes 2 and 3). Remarkably, under these conditions, YAP/TAZ depletion prevented de-differentiation, as cells remained as differentiated astrocyte-like cells in spite of being exposed to GSC-inducing medium, as revealed by their retaining high levels of GFAP (Fig. 4b, and Fig. 4d, e, compare lanes 3 and 4), and permanent loss of gliomasphere-forming ability (Fig. 4f, g, compare lane 3 with lanes 4 and 5).
Fig. 4. YAP/TAZ control GBM cell plasticity.
(a) Schematic representation of the experimental setup used to promote differentiation of HuTu cells, and then revert them back to a dedifferentiated state.
(b-e) Effects of TAZ depletion on the plasticity of HuTu10 and HuTu13 cells subjected to the differentiation/de-differentiation protocol depicted in (a). (b) Representative GFAP and TAZ stainings (scale bars, 100 μm). (c) Quantifications of the percentage of cells showing predominantly nuclear ‘N’ or predominantly cytoplasmic ‘C’ TAZ localization. Data are representative of at least 200 cells for each condition. (d, e) Western blot analysis for GFAP and YAP/TAZ; GAPDH serves as loading control. Uncropped images are in Source Data. Experiments were independently repeated three (d) and two (e) times, with similar results.
(f, g) HuTu10 (f) and HuTu13 (g) cells were subjected to the differentiation/de-differentiation protocol depicted in (a) and plated for sphere-forming assays. Panels are quantifications of gliomaspheres (n=8), presented as box and whisker plots: the box extends from the 25th to the 75th percentile, the line within the box represents the median, whiskers extend to show the highest and lowest values. For each condition, experiments were repeated four times with two independent replicas. All data are plotted. p-values were determined by one-way ANOVA with Dunnett’s T3 multiple comparisons.
YAP/TAZ preserve the GSC state by preventing differentiation
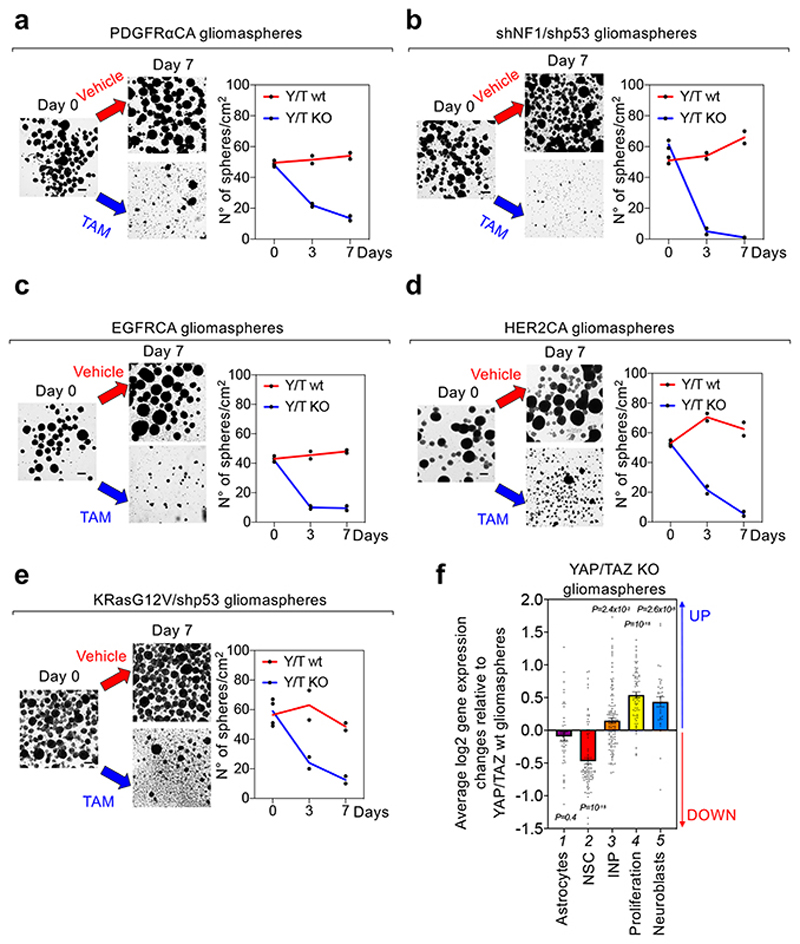
Having shown above the role of YAP/TAZ at inducing GSC-like properties in otherwise normal cells at the incipit of oncogenic transformation, or in differentiated tumor cells, we next asked whether YAP/TAZ are required to preserve GSC-like cells after their establishment. For this, we first monitored the effects of YAP/TAZ ablation within pre-established gliomaspheres. YAP/TAZ depletion induced progressive disaggregation and demise of gliomaspheres, irrespectively of the type of oncogenic lesion that drove gliomasphere emergence (Fig. 5a-e). After single cell replating, YAP/TAZ knockout cells were unable to sustain any outgrowth/passaging (Extended Data Fig. 9b), indicating that YAP/TAZ are essential for self-renewal.
Fig. 5. YAP/TAZ are required to prevent GSC differentiation.
(a-e) Gliomaspheres derived from PDGFRαCA (a)-, shNF1/shp53 (b)-, EGFRCA- (c), HER2CA- (d), or KRasG12V/shp53- (e) transformed R26CAG-CreERT2; Yapfl/fl; Tazfl/fl newborn astroglial cells were treated with either ethanol (YAP/TAZ wt) or 4OH-Tamoxifen (YAP/TAZ KO). Shown are representative images (left; scale bar, 100 μm) and quantifications (right; mean and individual data points of two independent experiments, each performed with two replicas) of the number of gliomaspheres/cm2 in vehicle versus TAM-treated samples. See also Extended Data Fig. 9a for a specificity control, showing that, in absence of CREER-expression, 4OH-Tamoxifen does not induce gliomasphere disaggregation.
(f) Analysis of RNA-seq data from gliomaspheres derived from KRasG12V/shp53-transformed R26CAG-CreERT2; Yapfl/fl; Tazfl/fl newborn astroglial cells and treated either with vehicle or with 4OH-TAM as described above. The graph shows average log2 gene-expression changes of signatures for the indicated cell types (Astrocytes n=44 genes; NSC n=89 genes; INP n=119 genes; Neuroblasts n=37 genes) or for proliferating neural progenitors (Proliferation n=66 genes) in 4OH-TAM-treated (YAP/TAZ KO) KRasG12V/shp53 gliomaspheres, compared to vehicle-treated (YAP/TAZ wt) KRasG12V/shp53 gliomaspheres. Abbreviations are as in Fig. 3e, f. Data are shown as mean and standard error of the mean (s.e.m.). Positive and negative values indicate, respectively, upregulation and downregulation of the indicated signatures after YAP/TAZ knockout. p-values were determined by Brown-Forsythe and Welch one-way ANOVA test with Dunnett’s T3 multiple comparisons of the distribution of log2 gene-expression changes of each signature with the distribution of log2 gene-expression changes for all expressed genes (n=12,211).
To investigate at the molecular level the consequences of YAP/TAZ ablation, we first compared the transcriptomes of gliomasphere cultures arising from KRasG12V/shp53-transformed R26CAGCreERT2; Yapfl/fl; Tazfl/fl astroglial cells, either treated with vehicle-control or with 4OH-TAM. Surprisingly, loss of YAP/TAZ in gliomaspheres was accompanied by upregulation of proliferation markers (Fig. 5f, lane 4), indicating that processes other than loss of proliferation are relevant for gliomasphere demise after YAP/TAZ inactivation. As shown in Fig. 5f, upon YAP/TAZ ablation, NSCs markers were strongly downregulated whereas markers of neuroblasts were induced (including Dlx2, Stmn1, TUJ1 and many others, see Supplementary Table 6), suggesting that YAP/TAZ is primarily required in GSCs to prevent their differentiation along the neuronal lineage. Consistently, YAP/TAZ ablation in gliomaspheres also caused upregulation of markers of intermediate neuronal progenitors (INP, Fig. 5f, lane 3), that is, proliferating neuroblasts’ precursors.
YAP/TAZ control GBM initiation and differentiation in vivo
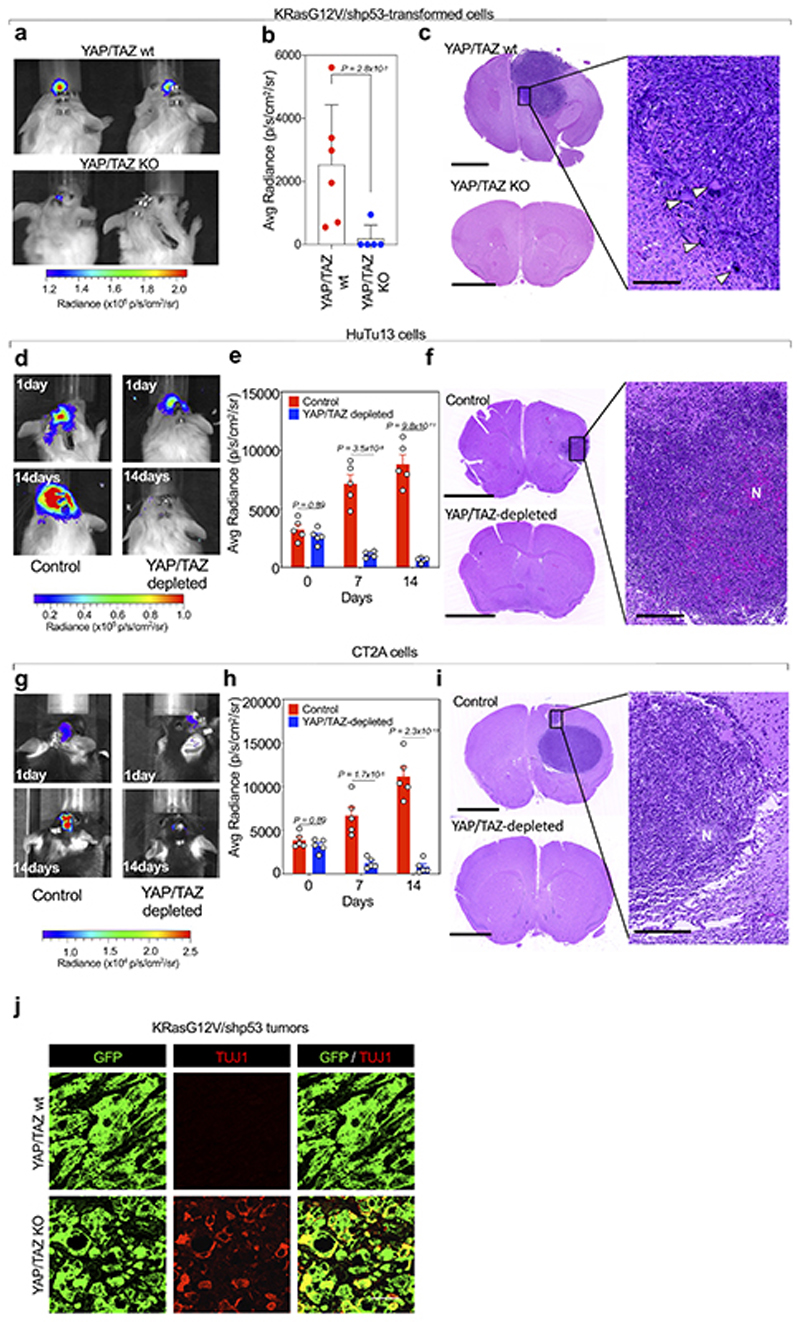
In keeping with the relevance of YAP/TAZ at preserving GSCs-like cell population in vitro, we next verified if YAP/TAZ are required for one of the cardinal features of GSCs in vivo, that is, tumor initiation. We tested this idea by injecting GBM cells orthotopically, in the brain of immunocompromised mice. We first used transformed Yapfl/fl; Tazfl/fl cells carrying either shNF1/shp53 or KRasG12V/shp53, and dual luciferase-GFP expression vectors, allowing for a non-invasive readout of tumor growth. Mice injected intracranially with parental GBM cells invariably formed large tumor masses with invasive behavior, as revealed by bioluminescence and by histological analyses (Fig. 6a-d and Extended Data Fig. 10a-c). In contrast, YAP/TAZ knockout cells failed to form any outgrowths (Fig. 6a-c and Extended Data Fig.10a-c). Similar results were obtained in immunocompromised mice by orthotopically-injected control vs. YAP/TAZ-depleted human GBM cells (HuTu13 cells, Extended Data Fig. 10d-f).
Fig. 6. YAP/TAZ are required for GBM initiation by preventing GSC differentiation.
(a-d) Immunocompromised mice were injected intracranially with shNF1/shp53-transformed cells derived from Yapfl/fl; Tazfl/fl newborn astroglial cells, also transduced with dual luciferase-GFP expression vectors. Control (YAP/TAZ wt) animals (n=5) were injected with cells transduced with Ad-GFP, whereas YAP/TAZ KO refers to animals (n=5) injected with cells transduced with Ad-Cre. (a) Representative images of brain bioluminescence. (b) Bioluminescence quantification shown as scatter dot plots and bar graphs showing mean with s.d.; p-value was calculated by unpaired two-tailed t-test. (c) Representative H&E staining; scale bar, 1 mm. (d) Magnification of the tumor generated by YAP/TAZ wt cells; scale bar, 250 μm. Arrowheads point to polynucleated giant cells, a characteristic trait of giant cells Glioblastoma.
(e-g) GL261 cells, an established mouse model of GBM, were injected intracranially in syngeneic (C57BL/6) mice. Control animals (n=5) were injected with cells transduced with lentiviral vectors coding for control shRNA, whereas YAP/TAZ-depleted refers to animals (n=5) injected with cells transduced with lentiviral vectors coding for doxycycline-inducible YAP and TAZ shRNAs and exposed to doxycycline prior to injection to induce YAP/TAZ depletion. Cells were also transduced with dual luciferase-GFP expression vectors. To sustain YAP/TAZ depletion after injection, doxycycline was added to the drinking water of all mice. (e) Representative images of brain bioluminescence at one day and 14 days after injection. (f) Bioluminescence quantification at three different time points shown as scatter dot plots and bar graphs showing mean with s.d.; unpaired two-tailed t-test p-values are shown. (g) Representative H&E stainings of brain sections from mice injected with control (upper panel and corresponding magnification) or with YAP/TAZ-depleted GL261 cells (middle and lower panels), the latters displaying either no remaining tumor cells (middle panel, representative of n=3 mice), or a residual amount of injected cells converging toward the right ventricle (lower panel and corresponding magnification, representative of n=2 mice). Scale bars, 2.5 mm in left panels and 250 μm in the magnifications shown on the right. ‘N’ indicates necrotic areas.
(h) Representative GFP and TUJ1 stainings (scale bars, 50 μm) in sections from the same mouse brains injected with control (upper panel) or YAP/TAZ depleted GL261 cells (lower panel) shown in the upper and lower panels of (g), respectively. The arrowhead point to a single TUJ1-positive cell in the control tumor.
(i) Gene Set Enrichment Analysis (GSEA) enrichment score curve of known markers of neuronal differentiation of NSC in YAP/TAZ KO vs. YAP/TAZ wt subcutaneous tumors from KRasG12V/shp53-transformed cells, following the experimental setup indicated in Extended Data Fig. 6. Signatures are available in Supplementary Table 7.
(j) GFP and TUJ1 stainings (scale bars, 50 μm) in sections from YAP/TAZ wt and YAP/TAZ KO tumors (representative of n=3 independent tumor samples each) derived from shNF1/shp53-transformed cells, following the experimental setup indicated in Extended Data Fig. 6.
Next, we validated these findings using some of the most established models of mouse GBM, that is, the mouse glioma cell lines GL261 and CT2A. After orthotopic transplantation in syngeneic mice, these cells form tumors displaying several characteristics of human GBMs, including intra-tumoral heterogeneity, pseudopalisading necrosis, radio-resistance, and chemo-resistance58. To study the role of YAP/TAZ in these models, we injected GL261 or CT2A expressing anti-YAP/TAZ doxycycline-inducible shRNAs in the brain of immunocompetent syngeneic mice. Control cells formed large tumor masses, while, upon doxycycline treatment, YAP/TAZ-depleted cells did not (Fig. 6e-g and Extended Data Fig. 10g-i). However, in some brain sections we could still detect residual YAP/TAZ-depleted cells tumor cells as such allowing in vivo investigation of their differentiation state by immunofluorescence. As shown in Fig. 6h for GL261 cells, YAP/TAZ depletion causes a strikingly wholesale differentiation toward the neuronal lineage, being essentially all these cells positive for TUJ1. In contrast, tumors generated by parental GL261 cells are almost invariably negative for TUJ1, with the exception of a minority of cells (about 3% of tumor cells typically found as small clusters).
We then confirmed that tumor cell differentiation is also at the roots of the halted tumor growth after YAP/TAZ inactivation in pre-established lesions in vivo (as in Extended Data Fig. 6). As mentioned above, loss of YAP/TAZ causes depletion of GSCs and downregulation of G-STEM signature. Remarkably, this is accompanied by massive upregulation in the expression of early markers of neuronal differentiation (e.g., of Ascl1, Tau, Tuj1, NCAM, Stathmin, SOX11) (Fig. 6i). By IF, YAP/TAZ knockout tumor cells acquired the expression of TUJ1, confirming that YAP/TAZ prevents GSCs differentiation, with their ablation skewing the fate of GBM cells towards neuronal-like fate (Fig. 6j and Extended Data Fig. 10j). We conclude from this collective set of results that YAP/TAZ are required for tumorigenesis and to prevent differentiation in vivo in multiple cellular and experimental contexts, in primary astroglial/SVZ-like cells transformed with different activated oncogenes, and in classic GBM models growing in a syngeneic context.
Discussion
In this work, we advance on the molecular foundations of the GSC state, identifying YAP/TAZ as the transcriptional determinants that define GSC populations in their native state. YAP/TAZ activation occurs downstream of classic oncogenic drivers of GBM to induce GSC-containing tumorigenic cell populations; within such populations, YAP/TAZ remain key for self-renewal of gliomaspheres in vitro, and for both tumor initiation and maintenance in vivo, as shown in different mouse and human GBM cellular contexts. Consistently, YAP/TAZ activity, as monitored by the G-STEM signature, can identify prospective GSCs in the heterogeneous GBM cell populations, at least in the IDH-wild type GBMs.
A recent seminal study of Suvà and colleagues, Neftel et al.8, presented a model in which cellular heterogeneity of IDH-wild type GBM reflects the coexistence, within each individual tumor, of four cellular states, able to intercovert into each other. Still unclear is whether these cell subtypes are connected to a shared GSC-state, or whether different stem cell populations exist in different GBM subtypes. The present identification of the G-STEM only in part advances on these open issues. Our analyses of Neftel et al., scRNA-seq data reveal GSC populations earmarked by the G-STEM transcriptional program in different GBM subtypes. However, projecting a shared molecular signature into cell populations should be interpreted with caution, as this may imply either of two scenarios: one in which the G-STEM indeed identifies a specific GSC population common to all GBM subypes, or, alternatively, that distinct stem cells in different GBM may share part of their anti-differentiation mechanisms, and that one of this overlapping program may be highlighted by elevated YAP/TAZ activity as denoted by G-STEM.
Notably, we found that these native GSCs display a hybrid phenotype between the Astrocyte-like and Mesenchymal-like cell states of Neftel et al. For example, we found that native GSCs are enriched of molecular markers typical of both Astrocyte-like state (e.g., HOPX, GFAP, MLC1) and of the Mesenchymal-like state (e.g., VIM, CD44, LGALS3) (see Supplementary Table 2). This conclusion is apparently in contrast with our observation that GSCs acquire astrocyte-like features after in vitro differentiation induced by serum. However, normal astrocytes do not express mesenchymal markers11, and appear transcriptionally distinct from GSCs in our analyses. Rather, it is tempting to speculate that native GSCs may resemble fetal-like cells, such as early neural progenitors which also co-express markers of astrocyte (e.g., HOPX, GFAP) and mesenchymal-like states (e.g., VIM, LGALS3)11.
Restoring differentiation capacity of GBM might represent a therapeutic option56, although this is complicated by the ability of differentiated cells to revert back to a GSC state54. Here we advance in these directions by showing that YAP/TAZ activity peaks in GSCs and that targeting YAP/TAZ is instrumental to cause their irreversible conversion into committed neural progenitors and more differentiated neural cell types. Of note, this occurs independently of proliferation control, providing a departure from current models envisioning a central role of YAP/TAZ as regulators of cell cycle progression in cancer38. We propose that an anti-YAP/TAZ therapy has the potential to be more effective than current chemotherapeutic regimens targeting cell proliferation, whose efficacy in GBM patients is in fact very limited. Strategies aimed at blunting YAP/TAZ activity in vivo have been recently proposed, including inhibitors of Brd4, a YAP/TAZ co-activator59 that represents an addiction of GBM although only in vivo60, consistently with the identification of YAP/TAZ activity at the core of the native GSC state.
In conclusion, we have here advanced on a key molecular underpinning of the GSC native state, as such unveiling a core vulnerability and addiction of GBM, all in all hinting to new perspectives to ameliorate treatment of a devastating malignancy.
Methods
Analysis of single-cell RNA-seq data from primary glioblastomas
Darmanis dataset
We analyzed single-cell RNA-seq data of primary glioblastoma samples from Darmanis et al.7, retaining the cell annotation provided by the authors. Raw reads were downloaded from GEO (GSE84465) and mapped to the human reference genome GRCh38 using STAR61. Raw gene counts were obtained using the featureCounts function of the Rsubread R package62 and the GENCODE release 25 (GRCh38.p7) basic gene annotation. Quality controls and normalization were carried out using Seurat63 (version 2.3.1) with default parameters. We retained for subsequent analyses 3,188 cells (out of 3,588 cells) with i) number of unique detected genes between 500 and 8000; ii) total number of detected molecules between 1×105 and 1.5×106; iii) fraction of reads mapping to the mitochondrial genome ≤0.2. Pseudotime trajectories on neoplastic and normal neural cells were constructed using Monocle210 (version 2.8.0); we used the unsupervised “dpFeature” procedure to order cells based on genes that differ between clusters and the “DDRTree” algorithm for the dimensionality reduction step, as recommended by the authors. Based on the distribution of pseudotime values, we selected two populations of neoplastic cells, with pseudotime values below the first quartile (Low), and above the third quartile (High), respectively. We compared the transcriptomes of these two populations using the FindMarkers function of Seurat, setting the following parameters: only.pos=F, min.pct=0.01, logfc.threshold=0.01, min.cells.gene=1, min.cells.group=1. Results from the analysis of differential expression were functionally annotated using GSEA and gene sets derived from previously published gene signatures11,64-67 (Supplementary Table 1). The GSEA software (http://software.broadinstitute.org/gsea/index.jsp) was applied in preranked mode to the gene list ranked on log2 fold change. Gene sets were considered significantly enriched at FDR ≤0.05 when using classic enrichment statistics and 1,000 permutations of gene sets.
The same pseudotime trajectory analysis was applied to the neoplastic and normal cells of each single patient (with the exclusion of patient BT-S6 that was characterized by a very limited number of sequenced cells) and to the sole neoplastic cells of all patients. In both cases, we defined two populations of neoplastic cells, one with pseudotime values below the first quartile (Q1-cells, Low) and the other with pseudotime values above the third quartile (Q3-cells, High) of the distribution of pseudotime values, and compared the transcriptomes of these two populations (Low versus High) as described above. In the case of the pseudotime trajectory analyses of single patients, we set the root on the end that is opposite to those containing normal cells.
To reconstruct the differentiation trajectory of the neoplastic cells, we estimated the RNA velocities of neoplastic cells using velocyto.R, a package for the analysis of expression dynamics in single cell RNA data15. In velocyto.R, we estimated the RNA velocity with the gene-relative model, which combines cell kNN pooling with the gamma fit based on an extreme quantiles, and set the parameter kCells=25 and the parameter fit.quantile=0.02.
G-STEM signature
From the differentially expressed genes between Low- and High-cells, we selected the 895 genes with fold-change ≥1.25 and adjusted p-value ≤0.05 to define the G-STEM signature (Supplementary Table 2). We annotated the G-STEM signature on the Biological Process gene ontologies using the Enrichr website (https://maayanlab.cloud/Enrichr/).
Neftel dataset
We analyzed single-cell RNA-seq data of primary glioblastoma samples from Neftel et al.8, retaining the cell annotation provided by the authors. Expression matrix and metadata were downloaded from the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP393/single-cell-rna-seq-of-adult-and-pediatric-glioblastoma#study-summary). Pseudotime trajectories on the sole neoplastic cells were constructed using Monocle210 (version 2.8.0); we used the unsupervised “dpFeature” procedure to order cells based on genes that differ between clusters and the “DDRTree” algorithm for the dimensionality reduction step. Based on the distribution of pseudotime values, we select two populations of neoplastic cells, the first with pseudotime values below the first quartile (<Q1), and the second with pseudotime values above the first quartile (>Q1). We compared the transcriptomes of the two populations of neoplastic cells calculating the log2 fold change of the expression level for each gene. We applied the GSEA software in preranked mode to the gene list ranked on log2 fold change to evaluate the functional enrichment for the same gene sets tested in the Darmanis dataset (Supplementary Table 1). Gene sets were considered significantly enriched at FDR ≤ 0.05 when using classic enrichment statistics and 1,000 permutations of gene sets.
Signature scores
Signature scores have been calculated as the average expression of the genes comprised in each signature. All analyses have been performed in R 3.5.0.
Collection and processing of GBM transcriptomes from the TCGA and REMBRANDT studies
Gene expression data of the TCGA and REMBRANDT were obtained from Gene Expression Omnibus (see Data Availability and Supplementary Tables 8, 9). For the TCGA dataset, related clinical and molecular subclass data have been obtained from Table S7 of Ref.23 and NCI Genomic Data Commons (https://gdc.cancer.gov/about-data/publications/lgggbm_2016), whereas clinical data for the REMBRANDT dataset were downloaded from GEO GSE108474.
For the TCGA dataset, expression values were generated from intensity signals using a custom definition file (CDF) for Affymetrix HT HG-U133A arrays based on Entrez genes (hthgu133ahsentrezgcdf version 21.0.0; http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/21.0.0/entrezg.asp). Intensity values for 552 samples (n=542 brain glioblastomas and n=10 brain tissues; Supplementary Table 8) have been background-adjusted, normalized using quantile normalization, and gene expression levels calculated using median polish summarization of Bioconductor affy package (multi-array average procedure, RMA 68). For the REMBRANDT dataset, after removing low grade gliomas, probe level signals for a total of 248 samples (n=220 glioblastomas and n=28 non tumor brain tissues; Supplementary Table 9) were converted to expression values using RMA and a custom CDF for Affymetrix HG-U133Plus2 arrays based on Entrez genes (hgu133plus2hsentrezgcdf version 21.0.0; http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/21.0.0/entrezg.asp). All analyses were performed in R 3.5.0.
Kaplan-Meier survival analysis of human GBM datasets
To identify two groups of tumors with either high or low G-STEM signature we applied the following classification rule. Briefly, each tumor was classified as G-STEM signature high if the average standardized expression level of G-STEM signature genes (fold-change ≥2; Supplementary Table 2) was larger than the mean of average standardized expression signals of all samples, and as G-STEM signature low vice versa. This classification was applied to expression values of TCGA and REMBRANDT glioblastomas. To evaluate the prognostic value of the G-STEM signature, we applied the Kaplan-Meier method on the patients’ survival data to estimate the probabilities that patients classified as “G-STEM high” and “G-STEM low” would survive. To confirm these findings, the Kaplan-Supplemental Meier curves were compared using the log-rank (Mantel-Cox) test. P values were calculated according to the standard normal asymptotic distribution. Survival analysis was performed in GraphPad Prism.
Identification of candidate master Transcriptional Regulators
To identify candidate master transcriptional regulators we assembled a 3-step computational workflow named “Rhabdomant”.
In the first step, to reconstruct the GRN, we initially defined a list of transcription factors active in GBM, defined as TRs potentially associated to chromatin in GBM tumors. For this, we took advantage of the epigenetic analyses provided by a recent large-scale ATAC-seq profiling of several human tumor types, including GBM69. We carried out a DNA binding motif enrichment analysis using the HOMER algorithm on the open chromatin regions of GBMs, and then selected the list of transcription factors whose DNA-binding motifs were highly enriched (FDR<0.0001) in these genomic regions. We manually implemented this list with partner transcriptional co-factors (see Supplementary Table 10) for a total list of 151 TRs.
Next, we applied the reverse-engineering algorithm ARACNe-AP70 to the gene expression signals of the neoplastic cells of the Darmanis scRNA-seq dataset, allowing to map the interactions between GBM-specific transcription factors and their coregulated genes. To limit the effects of scRNA-seq data sparsity, we removed genes expressed only in a limited number of cells (normalized counts >0 in less than 100 cells).
In the second step, in order overcome spurious TR-target gene association intrinsic to the noise generated by scRNA-seq data, we pruned the GRN by retaining only candidate direct targets genes of each TR. For this, we took advantage of the association map of between each ATAC-seq peak and its target genes, as provided by Corces et al.69 (Supplementary Table 11). We then intersected this map with the gene interactomes obtained in step 1, in so doing retaining in the GRN only the lists of target genes (regulons) associated to a binding motif for a candidate TR in their cis-regulatory elements.
In the third step, we interrogated the GRN with the VIPER algorithm71 to identify candidate master TRs of GSCs and DGCs, namely, transcriptional determinants whose regulons were enriched of genes activated in one of the two opposite cell populations of the pseudotime trajectory of neoplastic cells, that is the G-STEM and DGC signatures. Based on VIPER analysis, we defined as candidate master TRs those regulomes (i.e., the TR and its regulon) with an FDR ≤0.05 and a number of target genes >70. This resulted in 27 candidate MRs, of which 15 were candidate master TRs of the GSC state and 7 were candidate master TRs of the DGC state.
An extended version of these procedures is provided as Protocol Exchange, at DOI....
Comparison of G-STEM signature and master TR regulomes
To compare the G-STEM signature with the GBM regulomes, we calculated the enrichment of G-STEM genes in the regulomes of YAP/TAZ and of their downstream TRs. Specifically, we considered the first 3-layers of the YAP/TAZ gene-regulatory network and identified 6 TRs (FOXO1, FOS, SALL1, FOXO3, BCL6 and ERF); then we calculated the intersection between the 313 G-STEM signature genes comprised in the GBM regulons resulting from Step 2 and the targets positively interacting (Mode of Action >0) with TAP/TAZ or one of their downstream TRs (Supplementary Table 5). The statistical significance of the overlaps was calculated in R using the fisher.test function of the stat package.
Analysis of samples from the Ivy Glioblastoma Atlas
RNA-seq data from 6 GBM were downloaded from the Anatomic Structures RNA-Seq repository of the Ivy Glioblastoma Atlas Project42 (see Supplementary Table 12 and Data Availability). Gene expression was quantified in R 3.3.1 using the featureCounts function of the Rsubread R package62 and the UCSC gene annotation (GRCh37/hg19). Data normalization has been performed using the edgeR package72 (version 3.20.0); briefly, raw counts were normalized to counts per million mapped reads (CPM) and to fragments per kilobase per million mapped reads (FPKM). Gene expression data were then standardized tumor-wise. Signature scores have been calculated as the average expression of the genes comprised in each signature. For each area of each tumor, the signature scores were calculated as the average of the signature scores of the different samples of same area from the same tumor.
TAZ immunohistochemistry
Archival frozen GBM specimens were collected at the Azienda Ospedaliera, Padua, Italy. For IHC, 4 μm thick sections were obtained from tumor samples. IHC was performed with rabbit polyclonal anti-TAZ (Sigma, HPA007415;1:50 diluted) as previously described73. For mouse tissues, IHC was performed using a fully automated system (Bond-maX; Leica).
Slide images were captured using the D-Sight-F system for digital pathology (Menarini Diagnostics) and the percentage of TAZ-positive nuclei was determined using the Nuclei Analysis module of the D-Sight Viewer software.
Reagents and plasmids
Doxycycline hyclate, Tamoxifen, 4-Hydroxytamoxifen, Hygromycin and Puromycin were from Sigma. Fibronectin was from Santa Cruz Biotechnologies. Recombinant human/mouse/rat BMP2, hEGF, hbFGF, mEGF, mbFGF were from Peprotech. BIT9500 Serum Substitute was from StemCell Technologies. Growth-factor-reduced Matrigel (Phenol Red-free) was from Corning. XenoLight D-Luciferin-K+ Salt Bioluminescent Substrate was from Perkin Elmer. Cre- and GFP-expressing adenoviruses were from University of Iowa, Gene Transfer Vector Core.
For inducible expression of HER2CA, HER2CA cDNA (from pcDNA3-HER2-CA (Addgene#16259) was subcloned in FUW-tetO-MCS (Addgene#84008). Empty vector (FUW-tetO-MCS) was used as negative control. Inducible lentiviral vectors were used in combination with FUdeltaGW-rtTA (Addgene#19780).
For constitutive expression of RTKs, cDNAs of HER2CA (from FUW-tetO-HER2-CA), PDGFRαCA (from pcDNA5FRT-EF-Pdgfrα-CA-EGFPN, Addgene#66789) and EGFRCA (from pBabe EGFR-L858R/T790M, Addgene#32073) were subcloned in CSII-CMV-MCS-IRES-puro empty vector backbone (obtained by substituting the blasticidin (bsd) resistance of CSII-CMV-MCS-IRES-bsd (a gift of H. Miyoshi) with the puromycin resistance).
For constitutive depletion of Nf1 and p53 in mouse cells, we used pTomo-shNF1-shp53 (a gift from Inder Verma43). For constitutive lentiviral expression of mutant KRas, the pTomo KRasG12V was generated by subcloning the KRasG12V from the pBabe KRasG12V (Addgene#46746) to the pTomo-MCS Empty vector, obtained by substituting the loxP-RFP-loxP cassette from the pTomo vector (Addgene #26291) with a synthetic multiple cloning sites (MCS). For constitutive expression of mutant KRas and shp53, pTomo-KRasG12V-shp53 was created by inserting the shp53-containing cassette (from the pTomo-HRas-shp53, a gift of Inder Verma43) into the pTomo KRas G12V with SalI/SfiI restriction sites. The 8xGTIIC-RFP-DD lentiviral vector is a gift from Joan Massagué52.
For doxycycline-inducible downregulation of mouse YAP and TAZ in GL261 and CT2A cell lines, we used Tet-pLKO-puro (a kind gift of Giannino Del Sal) lentiviral vectors expressing doxycycline-inducible sh-mouseTAZ in combination with Tet-pLKO-hygro lentiviral vectors (obtained by substituting the puromycin resistance cassette in Tet-pLKO-puro with the hygromycin resistance cassette from pBABEhygro) expressing doxycycline-inducible sh-mouseYAP. Tet-pLKO-puro shControl lentiviral vectors were used as control.
LV-CreERT2 was obtained by substituting the Cre coding sequence of the LV-Cre-SD (Addgene#12106) with CreERT2 of the pCAG-CreERT2 (Addgene#14797).
All constructs were confirmed by sequencing.
Cell cultures
HuTu10 and HuTu13 patient-derived GBM cell lines were gently donated by Giuseppe Basso57, and were cultured on fibronectincoated dishes in DMEM/F12 (Gibco), 10% BIT9500, 25 ng/ml hEGF, 25 ng/ml hbFGF, glutamine and antibiotics.
HEK293T cells were from ATCC and were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS), glutamine and antibiotics. HEK293T cells were authenticated by DSMZ/Eurofins Genomics. CT2A and GL261 mouse glioma cell lines were purchased from Millipore (Catalog # SCC194) and from DSMZ (Catalog # ACC 802), respectively, and cultured as described in manufacturer’s instructions. All cells were routinely tested negative for mycoplasma.
For experiments with inducible transgenes, cells were treated with 2 μg/ml doxycycline for the whole duration of the experiments. siRNA transfections were done with Lipofectamine RNAi-MAX (Thermo Fisher Scientific) in antibiotics-free medium according to manufacturer instructions. Sequences of siRNAs are provided in Supplementary Table 13. Lentiviral particles preparation and cell culture infections were as in Ref.74.
Mice
Animal experiments were performed adhering to our institutional and national guidelines as approved by OPBA (Padova) and the Ministry of Health of Italy. The housing conditions comprised a diet with 28% protein. A maximum of five adult mice weighing up to 20 g were homed in a single cage, maintaining the ambient temperature at 19–23 °C, the humidity at 55%±10% and a 12-h light/12-h dark cycle.
6-8 week-old female NOD-SCID mice (Charles River) were used for subcutaneous injections. 6-8 week-old female NSG or C57BL/6 mice (Charles River) were used for intracranial injections.
Transgenic lines used in the experiments were gently provided by: Duojia Pan (Yapfl/fl mice75); Dieter Saur and Jens Siveke (R26CAG FSF-CreER mice76) and Paolo Bonaldo (CMV-Flp mice). Double Yapfl/fl;Tazfl/fl conditional knock-out mice were as described in Ref.77. To obtain R26CAG-CreER;Yapfl/fl;Tazfl/fl mice, first we obtained the R26CAG-CreER line by crossing R26CAG-FSF-CreER mice with CMV-Flp mice, R26CAG-CreER mice were then intercrossed with Yapfl/fl;Tazfl/fl mice.
Gliomasphere preparation
Primary newborn astroglial cells were isolated and maintained as previously described78. For gliomasphere preparation, astroglial cells were plated at 20-30% confluence in 6-well plates in 2 ml astrocyte medium (DMEM medium supplemented with 10% FBS, glutamine and antibiotics). The next day cells were transduced with lentiviral vectors coding for oncogenes, or with empty vector as negative controls. After 24 hours (day 3), transduced astrocytes were switched to NSC medium (DMEM/F12 supplemented with 100X N2, 20 ng/ml mEGF, 20 ng/ml mbFGF, glutamine, and antibiotics). Spheres arising from the cell monolayer were evident after approximately 3 weeks. Sphere passaging was performed as described previously74.
To evaluate self-renewal properties (Extended Data Fig. 7d and 9b), gliomaspheres were dissociated to single cells and replated in Ultra Low Attachment 24-wells plates (Corning), at the concentration of 2,000 cells per well; fully gliomaspheres were counted by visual inspection.
Monitoring YAP/TAZ activity during gliomagenesis
Newborn mouse astroglial cells were plated at 20-30% confluence in 6-well plates in 2 ml of astrocyte medium. 24 hours after seeding, cells were infected with the 8xGTIIC-RFP-DD lentiviral vector52. Transduced cells were infected with oncogene-expressing or empty lentiviral vectors and, the day after, switched to NSC medium supplemented with trimethoprim (TMP, 10μM). Bright-field and fluorescent images were acquired daily with a Leica DMIL LED microscope equipped with a Leica DFC 3000G camera using LAS AF version X software.
For the experiments depicted in Fig. 3d-g, astroglial cells from Yapfl/fl;Tazfl/fl mice were transduced with LV-CreERT2 and then with FUdeltaGW-rtTA and Doxy-inducible HER2CA lentiviruses. After 24 hours, infected newborn astroglial cells were switched to NSC medium containing 2 μg/ml doxycycline to induce HER2CA minus/plus 1 μM 4OH-Tamoxifen to induce YAP/TAZ knockout. Media were replaced every 3-4 days, till day 19, when cells were harvested for RNA extraction. Negative controls were provided by astroglial cells transduced with empty vector.
Testing YAP/TAZ requirement for gliomasphere maintenance
Single glioblastoma cells obtained from R26CAG-CreERT2;Yapfl/fl;Tazfl/fl P2 gliomaspheres were seeded to form P3 gliomaspheres. Fully-formed P3 gliomaspheres were treated with either vehicle (Ethanol) or 1 μM 4OH-Tamoxifen. Freshly 4OH-Tamoxifen was added every 3 days and sphere morphology and size was evaluated 3 days and 1 week after the first treatment. Bright-field images were acquired with a Leica DMIL LED microscope equipped with a Leica DFC 3000G camera using LAS version X software.
Subcutaneous tumor experiments
Oncogene-induced P2/P3 gliomaspheres were dissociated into single cells with Tryple, resuspended in ice-cold Matrigel and injected into the flank of NOD/SCID mice (750,000 cells/200 μl of Matrigel). Tumor growth was followed during time, and masses were harvested for histological analyses when they reached 1-2 cm of diameter, before any apparent ulceration of the skin.
To test the requirement of YAP/TAZ on the maintenance of subcutaneous tumors, we injected in the flank of NOD/SCID mice single cells dissociated from oncogene-induced R26CAG-CreERT2;Yapfl/fl;Tazfl/fl P2-P3 gliomaspheres (750,000 cells/200 μL of Matrigel). At the appearance of palpable masses, a group of mice received a TAM400/CreER diet (Envigo) to promote YAP/TAZ knockout in tumor cells (YAP/TAZ KO), whereas a second group, serving as control, continued to be fed with normal diet (YAP/TAZ wt). Tumor growth was followed during time, and masses were harvested for histological analyses when YAP/TAZ wt tumors reached 1-2 cm of diameter. For the serial passaging experiments, cell cultures were obtained from primary tumor masses and expanded in vitro as gliomaspheres in NSC medium for three passages. Gliomaspheres were then dissociated into single cells and subcutaneously transplanted (10,000 cells/200 μl of Matrigel) for secondary tumor formation. This procedure was repeated again for tertiary tumor formation of Extended Data Fig. 7h.
For the experiments in Extended Data Fig. 7g, P3 KRasG12V/shp53-gliomaspheres were dissociated with Tryple, resuspended at different cell dilutions in 200 μl ice-cold Matrigel (100,000; 10,000; 1000; 100 cells) and injected into the flank of NOD/SCID mice. Tumor growth was followed during time, and masses were harvested for histological analyses when they reached 1-2 cm of diameter, before any apparent ulceration of the skin.
Brain tumor experiments
For experiments depicted in Fig. 6a-d and Extended Data Fig. 8a-c, single cells obtained from oncogene-induced Yapfl/fl;Tazfl/fl gliomaspheres were transduced with adenoviruses encoding for CRE recombinase (Ad-Cre) to induce YAP/TAZ knockout, or for GFP (Ad-GFP) as negative controls. Cells were then orthotopically injected into NSG mice (300,000 cells/2 μl PBS).
For the experiments in Extended Data Fig. 10d-f, HuTu13 cells were transfected with siCo. or siYAP/TAZ for 48 hours and then orthotopically injected into 6-8 weeks NSG mice (300,000 cells/2 μl PBS).
For the experiments in Fig. 6e-h and in Extended Data Fig. 10g-i, CT2A or GL261 shControl and shYAP/TAZ lines were generated by infecting glioma cells either with the pLKO-hygro-puro-Tet-On-shControl or with pLKO-hygro-Tet-On-shYap and pLKO-puro-Tet-On-shTaz lentiviral particles respectively; cells were expanded in hygromycin/puromycin-containing medium to select for transduced cells. GL261 and CT2A cells were then treated for 48 hours with 2 μg/ml doxycycline and then orthotopically injected into the brain of 6-8 weeks C57BL/6J mice (300,000 cells/2 μl PBS). To sustain YAP/TAZ depletion after injection, doxycycline was added to the drinking water of all mice.
Prior to injection, all cells were transduced with a lentiviral construct coding for eGFP and firefly luciferase (GFP/Luc)79. The procedure and the coordinates used for the injection were as described previously79. Brain tumor growth was monitored by in vivo Luciferase assay, by intraperitoneal injection of 150 μg/g of XenoLight D-luciferin in PBS (PerkinElmer), and detecting brain luminescence with a Xenogen IVIS Lumina II System (Xenogen Corporation). Data analysis was performed using Living Image software version 4.7.2 (PerkinElmer), and the intensity of the signal was quantified in the regions of interest. Mouse brains were harvested as described previously79, fixed overnight in 4%PFA and then processed for H&E staining and immunofluorescence. NDPscan3.1 was used to acquire H&E images.
HuTu cell plasticity assay
HuTu cells have been classified based on their transcriptional profiles (centroids are reported in Supplementary Table 14). Details of the procedure are provided as Protocol Exchange at DOI…
HuTu cells were plates on fibronectin-coated 6-well plates and cultured in stem medium (DMEM/F12, 10% BIT9500, 25 ng/ml h-EGF, 25 ng/ml h-bFGF, glutamine and antibiotics). Differentiation was established by switching to differentiation medium (DMEM/F12, 10% FBS, BMP2 50 ng/ml, glutamine and antibiotics) for 15 days. De-differentiation was induced through medium switch to stem medium. When indicated, siRNA transfections were performed the last day of differentiation, before starting dedifferentiation. De-differentiation was maintained for 5 days before harvesting for western blot. For immunofluorescence, at the end of the de-differentiation process cells were plated on fibronectin-coated glass slides for 24 hours and then fixed with 4% PFA.
For experiments in Fig. 4f, g, cells were transfected with control siRNA or with two independent YAP/TAZ siRNA mixes after 15 days of differentiation. Cells were then plated in low-attachment 24-well (2,000 cells per well) in HuTu stem medium; growing spheres were counted after 4 days.
Gene expression analyses by RNA-seq
RNA extraction from cells was performed with NucleoSpin 8 RNA Core Kit (Macherey-Nagel) according to the manufacturer’s instructions, using an automated system (Freedom EVO, Tecan). Preparation and sequencing of RNA-seq libraries were as in Ref.59. Raw reads were mapped to the mouse reference genome (GRCm38) using STAR61. Raw gene counts were obtained using the featureCounts function of the Rsubread R package62 and the UCSC gene annotation (GRCm38/mm10). Raw counts were normalized to counts per million mapped reads (CPM) and to fragments per kilobase per million mapped reads (FPKM) using the edgeR package72; only genes with a CPM greater than 1 in at least 1 sample (or 2 samples when replicates are available) were further retained for differential analysis. Differential gene expression analysis was performed using the exactTest function of the edgeR package72.
Hierarchical clustering of Fig. 3d was performed using the Hierarchical Clustering of the MultiExperiment Viewer (MeV 4.8) package with Pearson correlation as distance metric and average linkage clustering, using the row-wise standardized FPKM of genes significantly (FDR ≤0.05) upregulated (fold change ≥1.33; FPKM ≥1 in YAP/TAZ wt HER2CA-expressing astrocytes) or downregulated (fold change ≤0.75; FPKM ≥1 in control newborn astroglial cells) in YAP/TAZ wt astrocytes expressing HER2CA compared to control newborn astroglial cells. Gene expression heatmaps have been generated in GraphPad Prism 8.0.2 software using row-wise standardization of the expression values.
Average log2 gene expression changes have been calculated as the standardized average log2 fold change of signature genes in all samples and plotted as mean and standard error of the mean (SEM). For Fig. 3e, f, displaying log2 gene expression changes between HER2CA-expressing and control astrocytes, we considered only genes expressed either in HER2CA-expressing astrocytes for upregulated genes or in control astrocytes for downregulated genes, i.e., genes displaying FPKM ≥1 in HER2CA astrocytes for genes with log2 fold change >0, and genes displaying FPKM ≥1 in control astrocytes for genes with log2 fold change <0. For Fig. 5f, displaying log2 gene expression changes between YAP/TAZ KO and YAP/TAZ wt gliomaspheres, we considered only genes expressed either in YAP/TAZ KO gliomaspheres for upregulated genes or in YAP/TAZ wt gliomaspheres for downregulated genes, i.e., genes displaying FPKM ≥1 in YAP/TAZ KO gliomaspheres for genes with log2 fold change >0, and genes displaying FPKM ≥1 in YAP/TAZ wt gliomaspheres for genes with log2 fold change <0.
Quantitative Real-Time PCR
Real-Time PCR was performed as described previously59, using System thermal cycler and analyzed with QuantStudio™ Software (ThermoFisher) (version 1.4.3). Expression levels are normalized to GAPDH. PCR oligo sequences are listed in Supplementary Table 15.
Immunofluorescence
Immunofluorescence on PFA-fixed cells and tissue samples was performed as previously described80. Primary and secondary antibodies and their working dilutions are described in Supplementary Table 16. Slides were mounted with Fluoroshield Mounting Medium with DAPI (F6057, Sigma). Images were acquired with Leica TCS SP5II confocal microscope equipped with a CCD camera using LAS AF 2.7.3.9723 software, and analyzed using Volocity software 6.0 (PerkinElmer).
Western Blot
Immunoblots were performed as previously described73. Chemiluminescence was digitally acquired by ImageQuant LAS 4000 1.2 (GE healthcare). Primary and secondary antibodies and their working dilutions are described in Supplementary Table 16.
Statistics and reproducibility
Data are mean ± s.d. or s.e.m., as indicated in the figure legends. Statistical tests (Student’s t-test, ANOVA and Kaplan-Maier survival analyses) are indicated in the figure legends and were performed with GraphPad Prism8.0.2 software. Sample sizes for each experiment are stated in the corresponding figure legends. No statistical method was used to predetermine sample size.
All tested animals were included in the analysis. All experiments were reproducible. Every figure states how many times each experiment was performed with similar results. Mice were randomly allocated to experimental or treatment groups. Investigators were not blinded to mouse grouping. Pathological examination of histological section was carried out by M. Fassan (a professional pathologist), who was blind to animal treatments.
Extended Data
Extended Data Fig. 1. Identification of the gene expression program of GSCs.
(a) Single-cell differentiation trajectory of GBM cells reconstructed by Monocle2 using single-cell RNA-seq data of the indicated cell populations from primary GBM samples of the Darmanis dataset.
(b) Gene set enrichment analysis (GSEA) for association between the cell populations at the start and at the end of the pseudotime trajectory of the neoplastic cells of the Darmanis datasets (as depicted in Fig. 1c), and gene sets denoting the identity of specific cell types. Gene lists denoting early neural progenitor cells (RG: Radial Glia; oRG: outer Radial Glia; vRG: ventricular Radial Glia) or neural stem cells (NSC) are indicated in red; those identifying neurons, astrocytes or committed neuronal progenitors (OPC: Oligodendrocyte Progenitor Cells; INP: Intermediate Neuronal Progenitors) are, respectively, in blue, purple and blue-green colors; gene lists enriched in the putative GSC and DGC populations are highlighted in orange and in light blue, respectively. Signatures are available in Supplementary Table 1. GSEA calculated FDR adjusting for multiple comparisons; details of p-value and FDR calculation are described in the GSEA website (http://software.broadinstitute.org/gsea/index.jsp). Related to Fig. 1c.
(c) Log2 expression levels of the indicated oRG (top graphs), NSC and GSC (middle graphs) and INP markers (bottom graphs) in the subpopulations of neoplastic cells of the Darmanis dataset that are at the start (GSC, n=221 cells) and at the end (DGC, n=221 cells) of the pseudotime trajectory depicted in Fig. 1c. Data are presented as mean + s.d. p-values were determined by unpaired two-tailed t test.
(d) RNA velocities (arrows) of neoplastic cells of the Darmanis dataset projected in the space of the first two principal components. Red and blue dots are the cells that are at the start (GSC) and at the end (DGC) of the pseudotime trajectory depicted in Fig. 1c.
Extended Data Fig. 2. Validation of the G-STEM signature.
(a-b) Violin plots showing the expression of the G-STEM signature (right panels in (b)) on the cells at the start (Low; red dots in the left panels in (b)) of the pseudotime trajectories (a) of patient-specific cohorts of the Darmanis dataset, vs. the neoplastic cells that are on the opposite ends of the same trajectories (High; blue dots in the left panels in (b)). The p-values were determined by two-tailed Mann-Whitney test.
(c) Violin plots showing the expression of the G-STEM signature (right panel) on the cells at the start (Low; red dots in the middle panel) of the pseudotime trajectory (left panel) of the sole neoplastic cells of the Darmanis dataset, vs. the cells that are on the opposite ends of the same trajectory (High; blue dots in the middle panel). The p-values were determined by two-tailed Mann-Whitney test.
Extended Data Fig. 3. Characterization of the G-STEM signature.
(a) Graphs depicting the most significant GO terms emerging from the Gene Ontology analyses of the genes composing the G-STEM and the DGC signatures. The full lists of significant GO terms of both signatures are in Supplementary Table 3.
(b) Log2 expression levels of the indicated components of the G-STEM signature in in the subpopulations of neoplastic cells of the Darmanis dataset that are at the start (GSC, n=221 cells) and at the end (DGC, n=221 cells) of the pseudotime trajectory depicted in Fig. 1c. Data are presented as mean + s.d. p-values were determined by unpaired two-tailed t test.
Extended Data Fig. 4. Validation of the G-STEM signature in large datasets of GBM patients.
(a) Gene set enrichment analysis (GSEA) for association between the cell population at the start of the pseudotime trajectory of the neoplastic cells of the Neftel datasets (as depicted in Fig. 1e) vs. all the other neoplastic cells and gene sets denoting the identity of specific cell types. Abbreviations and color codes are as in Extended Data Fig. 1b. Signatures are available in Supplementary Table 1. GSEA calculated FDR adjusting for multiple comparisons; details of p-value and FDR calculation are described in the GSEA website (http://software.broadinstitute.org/gsea/index.jsp). Related to Fig. 1e.
(b) Violin plots showing the expression of the G-STEM signature (bottom panels) on the cells at the start of the pseudotime trajectory (GSC; red dots in the top panels) of small tumor cohorts of the Neftel dataset, pre-sorted according to the Proneural, Classical or Mesenchymal classification of GBMs, vs. all the other neoplastic cells of the same cohorts (NON GSC; light blue dots in the top panels) of the same dataset. The p-values were determined by two-tailed Mann-Whitney test.
(c) Kaplan–Meier analysis representing the probability of survival in n=541 GBM patients from the TCGA dataset (left panel), n=210 GBM patients from the REMBRANDT dataset (middle panel), and n=390 GBM patients carrying wild-type IDH1 from the TCGA dataset (right panel), stratified according to high or low GSC-signature. The p-value of the Log-rank (Mantel-Cox) test reflects the significance of the association between GSC-signature “low” and longer survival. G-STEM expression is prognostic for the vast majority of GBM, that is IDH1-wild type tumors (93%, of those annotated in the TGCA dataset; n=390 out of 419 IDH1-annotated samples).
Extended Data Fig. 5. A computational procedure to identify candidate TRs controlling the gene expression program of GSCs.
(a) Overview of the experimental flow for inference of the master Transcriptional Regulators (TRs) of the GSC state using the Rhabdomant pipeline on the Darmanis sc-RNA-seq dataset of primary GBM samples. See Methods for details.
(b) List of candidate master Transcriptional Regulators (TRs) emerging from the analysis of the Darmanis dataset of scRNA-seq dataset with the Rhabdomant pipeline, ordered on the base of their normalized enrichment signal (NES). The Rhabdomant pipeline calculated FDR adjusting for multiple comparisons; see Methods for details about p-value and FDR calculation. The lists of candidate master TRs of the GSC and of the DGC state are highlighted in orange and in light blue, respectively. The most significant candidate master TRs of the GSC state are indicated in red.
Extended Data Fig. 6. YAP/TAZ are required for GSC maintenance in vivo.
(a-c) Effects of YAP/TAZ knockout on the growth of established subcuteaneous GBM-like lesions. Transformed cells were obtained by dissociation of gliomaspheres obtained from HER2CA- (a), shNF1/shp53- (b) or KRasG12V/shp53- (c) transformed R26CAGCreERT2; Yapfl/fl; Tazfl/fl newborn mouse astroglial cells (as in Fig. 3), and then injected in NOD-SCID mice. When subcutaneous tumors reached approximately 0.5 cm of diameter, mice were either fed with Tamoxifen food to induce YAP/TAZ knockout (YAP/TAZ KO), or maintained under normal diet (YAP/TAZ wt). Graphs are growth curves of YAP/TAZ wt (KRasG12V/shp53-, n=4 mice; HER2CA, n=6 mice; shNF1/shp53, n=5 mice) and YAP/TAZ KO (KRasG12V/shp53-, n=4 mice; HER2CA, n=4; shNF1/shp53, n=8 mice) tumors (average volume ± s.e.m.).
(d, e) Effects of YAP/TAZ knockout in tumors derived from KRasG12V/shp53 gliomaspheres, following the experimental setup described in a-c. (d) Dot plot for tumor weight at sacrifice (YAP/TAZ wt, n=8; YAP/TAZ KO, n=6). Mean ± s.e.m. of the distribution are also shown. p-value was calculated by unpaired two-tail t-test. (e) Representative H&E stainings. Scale bar, 2.5 mm. N, necrotic area; *, Matrigel residue.
(f) Tabular results showing the number of NOD/SCID mice displaying subcutaneous tumor formation after injection of cells dissociated either from gliomaspheres derived from HER2CA-transformed primary newborn astroglial cells (Primary tumors), or from HER2CA-gliomaspheres derived from one of the Primary tumors (Secondary tumors).
Extended Data Fig. 7. Ex-vivo reprogramming of normal neural cells into GSC-like cells.
(a) GFAP and SOX2 stainings (scale bars, 50 μm) of the mouse SVZ, representative of n=3 mice. Nuclei were counterstained with DAPI.
(b, c) GFAP, NESTIN and SOX2 stainings (scale bars, 50 μm) in mouse newborn astroglial cells, representative of two independent experiments.
(d) Gliomaspheres emerging from newborn astroglial cell cultures transformed by the indicated oncogenes (P0 spheres) were dissociated to single cells and replated at clonal density for gliomasphere formation (P1 to P10 spheres). Results are representative of three experiments with n=3 replicates each. Data are presented as scatter dot plots and bar graphs showing mean with s.d.
(e) Left panel: H&E staining of a lesion obtained after intracranial transplantation of shNf1/shp53-transformed astroglial cells. N, necrotic area. Scale bar, 2.5 mm. Middle panel: High magnification of the same tumor, showing large polynucleated cells (arrowheads). Right panel: TAZ IHC on the same tumor. Scale bars, 100 μm. Experiments were independently repeated on n=10 mice, with similar results.
(f) H E staining of subcutaneous tumors obtained by injecting cells dissociated from gliomaspheres carrying the indicated oncogenic lesions, representative of: KRasG12V/shp53, n=4 tumors; HER2CA, n=6 tumors; shNf1/shp53, n=5 tumors. N, necrotic areas. Scale bars, 250 μm.
(g) Number of mice displaying tumor formation after injection of cells dissociated from KRasG12V/shp53-gliomaspheres at the indicated cell dilutions.
(h) Top, Schematic representation of the serial transplantation assay performed with HER2CA-transformed cells (see Methods for details). Bottom, H&E staining (scale bars, 2.5 mm) of tumors obtained after each round of transplantation, representative of n=4 primary tumors, n=8 secondary tumors and n=4 tertiary tumors, respectively. Numbers of mice developing tumors per numbers of transplanted mice are indicated in each picture.
(i) GSEA curves of the G-STEM and the DGC signatures in KRasG12V/shp53-tumors compared to the astroglial cells from which they derive. Signatures are available in Supplementary Table 7.
Extended Data Fig. 8. Oncogenic insults activate YAP/TAZ in transformed primary astroglial cells.
(a) Bright-field and fluorescent pictures (representative of n=5 independent samples each) of newborn astroglial cells transduced with lentiviral vectors encoding for the YAP/TAZ reporter 8xGTIIC-RFP-DD52, and with lentiviral vectors encoding for the indicated oncogenes or, as negative control, with empty vector, as in Fig. 3b. Images were taken 4 days after inducing oncogenic reprogramming by incubating cells in NSC medium. Scale bars, 50 μm.
(b) Compendium of Fig. 3c. Efficiency of Yap and Taz downregulation in R26CAG-CreERT2; Yapfl/fl; Tazfl/fl mouse newborn astroglial cells treated with either vehicle (Control) or 4OH-TAM (YAP/TAZ KO), as measured by qRT-PCR (mean + s.d. of all independent samples of three experiments). p-values are calculated by two-way ANOVA with Sidak’s multiple comparisons.
Extended Data Fig. 9. YAP/TAZ are required for GSC maintenance in vitro.
(a) Control experiment of Fig. 5a-e. Gliomaspheres derived from HER2CA-transformed Yapfl/fl; Tazfl/fl newborn astroglial cells, not expressing CREERT2, were treated with either ethanol (Vehicle) or 4OH-TAM (TAM). Panels are representative images (left; scale bar, 100 μm) and quantifications (right; mean ± s.d. of two independent experiments, each performed with two replicates) of the number of gliomaspheres/cm2 in vehicle versus 4OH-TAM-treated samples. p-values were determined by two-way ANOVA with Sidak’s multiple comparisons test. In the absence of CREERT2 expression 4OH-TAM tamoxifen is inconsequential for gliomasphere formation, indicating that gliomasphere disaggregation shown in Fig. 4a-e is specifically caused by YAP/TAZ deletion.
(b) P2 gliomaspheres derived from R26CAG-CreERT2; Yapfl/fl; Tazfl/fl newborn astroglial cells transformed with the indicated oncogenes were dissociated to single cells and replated at clonal density for P3 gliomasphere formation in presence of ethanol (YAP/TAZ wt), or of 4OH-TAM to induce YAP/TAZ knockout (YAP/TAZ KO). Data are presented as scatter dot plots (n=3 replicates each) and bar graphs showing mean with s.d. The p-values were calculated by unpaired two-tailed t-test.
Extended Data Fig. 10. YAP/TAZ are required for GBM initiation in vivo.
(a-c) Immunocompromised mice were injected intracranially with KRasG12V/shp53-transformed Yapfl/fl;Tazfl/fl cells, also transduced with dual luciferase-GFP expression vectors. Control animals (n=6) were injected with cells transduced with Ad-GFP, whereas YAP/TAZ KO animals (n=5) were injected with cells transduced with Ad-Cre. (a) Representative images of brain bioluminescence. (b) Bioluminescence quantification shown as scatter dot plots and bar graphs showing mean with s.d; p-value was calculated by unpaired two-tailed t-test. (c) Representative H&E stainings. Scale bars, 2.5 mm in left panels and 250 μm in the magnification shown on the right. Arrowheads highlight the presence of large, polynucleated cells.
(d-f) Immunocompromised mice were injected intracranially with HuTu13 cells transduced with dual luciferase-GFP expression vectors, and transfected with siCo (Control; n=5) or siYAP/TAZ (YAP/TAZ depleted; n=5). (d) Representative images of brain bioluminescence. (e) Bioluminescence quantification shown as scatter dot plots and bar graphs showing mean with s.d.; unpaired two-tailed t-test p-values are shown. (f) Representative H&E stainings. Scale bars, 2.5 mm in left panels and 250 μm in the magnification shown on the right. ‘N’ indicates necrosis.
(g-i) CT2A cells were transduced with dual luciferase-GFP expression vectors and injected intracranially in syngeneic mice. Control animals (n=5) were injected with cells expressing anti-GFP shRNA, whereas YAP/TAZ-depleted animals (n=5) were injected with cells expressing doxycycline-inducible YAP and TAZ shRNAs. (g) Representative brain bioluminescences at one day and 14 days after injection. (h) Bioluminescence quantification at three different time points shown as scatter dot plots and bar graphs showing mean with s.d.; unpaired two-tailed t-test p-values are shown. (i) Representative H&E stainings. Scale bars, 2.5 mm in left panels and 250 μm in the magnification shown on the right. N, necrotic areas.
(j) GFP and TUJ1 stainings in sections from YAP/TAZ-wt and YAP/TAZ-KO subcutaneous shNF1/shp53-induced tumors (representative of n=3 independent samples each). Scale bars, 50 μm.
Acknowledgments
We thank I. Verma, J. Massagué and L. Naldini for plasmids, and colleagues sharing their plasmids through Addgene (Mien-Chie Hung, Lotte Pedersen, Christopher Counter, Connie Cepko, Konrad Hochedlinger, Matthew Meyerson). We thank D.J. Pan, D. Saur, J. Siveke, and P. Bonaldo for gifts of mice, Giuseppe Basso for HuTu cells, Gaia Zuccolotto for GFP/Luc-expressing lentiviral construct, Vito Barbieri for in vivo experiments, Vincenza Guzzardo for histology, Silvia Bresolin for microarrays, Genesio Leo for TAZ IHC analysis, and Mattia Forcato for comments. M.Ca. was supported by a FIRC-AIRC fellowship for Italy. O.R. is supported by Fondazione Umberto Veronesi (Post-Doctoral Fellowship 2020). The research leading to these results has received funding: from AIRC 5x1000 2018 “Metastasis as mechanodisease” (ID: 22759) grant to S.P.; from AIRC IG Grant 2019 (ID: 23307) to S.P.; from the Italian Ministry of Education, University and Research under a MIUR-FARE grant to S.P., and a MIUR-PRIN Bando 2017 grant to SP (cod. 2017HWTP2K); from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (DENOVOSTEM grant agreement No 670126) to S.P.
Footnotes
Author contributions M.Ca. performed most of the experiments in vitro and in vivo, and contributed to the writing. A.F. carried out the initial experiments of this study. A.Guarnieri and G.B. carried out experiments with HuTu cells. T.P. optimized mouse astrocytes isolation and infection. F.Z., contributed to manuscript preparation. F.Z., H.L.S., P.C. and A.C. optimized technical procedures critical for experiments in vivo and in vitro; M.F. performed histology and histopathological evaluations. E.P. and A.R. performed brain tumor experiments. O.R., A.Grilli and S.B. performed bioinformatic analyses. S.P. and M.Co. conceived the initial hypothesis and experimental design, organized the work and wrote the MS.
Competing interests
The authors declare no competing interests.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All RNA-seq and microarray raw data generated for the present study, along with counts matrices and metadata for each sample, are publicly available in Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession GSE133471. Single-cell RNA-seq data of primary glioblastoma samples from Darmanis et al. were downloaded as raw reads from GEO GSE84465. Expression matrix and metadata of the Neftel dataset were downloaded from the Single Cell Portal of the Broad Institute (https://singlecell.broadinstitute.org/single_cell/study/SCP393/single-cell-rna-seq-of-adult-and-pediatric-glioblastoma#study-summary). Raw gene expression data (.CEL files) of the GBM TCGA cohort were downloaded from GEO GSE83130. Raw gene expression (.CEL files) and clinical data of the REMBRANDT study were downloaded from GEO GSE108474. BAM files of Ivy Atlas GBM samples were downloaded from the Anatomic Structures RNA-Seq repository of the Ivy Glioblastoma Atlas Project (http://glioblastoma.alleninstitute.org/rnaseq/bam.csv).
Source data for Fig. 1e, 3b, 3e-f, 4c, 4f-g, 5a-f, 6b, 6f, and Extended Data Fig. 1c, 2b-c, 3b, 4b, 6a-d, 7d, 8b, 9a-b, 10b, 10e, 10f have been provided with the paper. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code availability
All code was performed using R 3.5.0 and publicly available packages explicitly cited in the manuscript. No custom functions were written for the analysis. STAR is available in GitHub (https://github.com/alexdobin/STAR). ARACNe-AP is available in GitHub (https://github.com/califano-lab/ARACNe-AP). HOMER is available at http://homer.ucsd.edu/homer/. MultiExperiment Viewer is available at http://mev.tm4.org.
References
- 1.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 3.Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer research. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 4.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 5.Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes & development. 2019;33:591–609. doi: 10.1101/gad.324301.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard SM, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell stem cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Darmanis S, et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell reports. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neftel C, et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell. 2019;178:835–849 e821. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nature methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lathia JD, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell stem cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertolini JA, et al. Mapping the Global Chromatin Connectivity Network for Sox2 Function in Neural Stem Cell Maintenance. Cell stem cell. 2019;24:462–476 e466. doi: 10.1016/j.stem.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miroshnikova YA, et al. Tissue mechanics promote IDH1-dependent HIF1alpha-tenascin C feedback to regulate glioblastoma aggression. Nature cell biology. 2016;18:1336–1345. doi: 10.1038/ncb3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nature neuroscience. 2016;19:20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes & development. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minata M, et al. Phenotypic Plasticity of Invasive Edge Glioma Stem-like Cells in Response to Ionizing Radiation. Cell reports. 2019;26:1893–1905 e1897. doi: 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laks DR, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallini R, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 22.Zeppernick F, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 23.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusev Y, et al. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Scientific data. 2018;5 doi: 10.1038/sdata.2018.158. 180158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nature methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 27.Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nature methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 28.Suva ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park NI, et al. ASCL1 Reorganizes Chromatin to Direct Neuronal Fate and Suppress Tumorigenicity of Glioblastoma Stem Cells. Cell stem cell. 2017;21:411. doi: 10.1016/j.stem.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Breunig JJ, et al. Ets Factors Regulate Neural Stem Cell Depletion and Gliogenesis in Ras Pathway Glioma. Cell reports. 2015;12:258–271. doi: 10.1016/j.celrep.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell stem cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Dreau G, et al. E proteins sharpen neurogenesis by modulating proneural bHLH transcription factors’ activity in an E-box-dependent manner. Elife. 2018:7. doi: 10.7554/eLife.37267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang P, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mu L, et al. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:3067–3080. doi: 10.1523/JNEUROSCI.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanconato F, Cordenonsi M, Piccolo S. YAP and TAZ: a signalling hub of the tumour microenvironment. Nature reviews Cancer. 2019 doi: 10.1038/s41568-019-0168-y. [DOI] [PubMed] [Google Scholar]
- 36.Orr BA, et al. Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J Neuropathol Exp Neurol. 2011;70:568–577. doi: 10.1097/NEN.0b013e31821ff8d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian T, et al. TAZ promotes temozolomide resistance by upregulating MCL-1 in human glioma cells. Biochem Biophys Res Commun. 2015;463:638–643. doi: 10.1016/j.bbrc.2015.05.115. [DOI] [PubMed] [Google Scholar]
- 38.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nature cell biology. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koo JH, et al. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes & development. 2020;34:72–86. doi: 10.1101/gad.331546.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan X, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549:227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puchalski RB, et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360:660–663. doi: 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai C, et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes & development. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Persson AI, et al. Non-stem cell origin for oligodendroglioma. Cancer cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alcantara Llaguno SR, et al. Adult Lineage-Restricted CNS Progenitors Specify Distinct Glioblastoma Subtypes. Cancer cell. 2015;28:429–440. doi: 10.1016/j.ccell.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galvao RP, et al. Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proceedings of the National Academy of Sciences of the United States of America; 2014. pp. E4214–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560:243–247. doi: 10.1038/s41586-018-0389-3. [DOI] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganat YM, et al. Early postnatal astroglial cells produce multilineage precursors and neural stem cells in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:8609–8621. doi: 10.1523/JNEUROSCI.2532-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Histological Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer; France: 2016. [Google Scholar]
- 52.Er EE, et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nature cell biology. 2018;20:966–978. doi: 10.1038/s41556-018-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zywitza V, Misios A, Bunatyan L, Willnow TE, Rajewsky N. Single-Cell Transcriptomics Characterizes Cell Types in the Subventricular Zone and Uncovers Molecular Defects Impairing Adult Neurogenesis. Cell reports. 2018;25:2457–2469 e2458. doi: 10.1016/j.celrep.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Caren H, et al. Glioblastoma Stem Cells Respond to Differentiation Cues but Fail to Undergo Commitment and Terminal Cell-Cycle Arrest. Stem cell reports. 2015;5:829–842. doi: 10.1016/j.stemcr.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Auffinger B, et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell death and differentiation. 2014;21:1119–1131. doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piccirillo SG, Vescovi AL. Bone morphogenetic proteins regulate tumorigenicity in human glioblastoma stem cells. Ernst Schering Foundation symposium proceedings; 2006. pp. 59–81. [DOI] [PubMed] [Google Scholar]
- 57.Pistollato F, et al. Molecular mechanisms of HIF-1alpha modulation induced by oxygen tension and BMP2 in glioblastoma derived cells. PloS one. 2009;4 doi: 10.1371/journal.pone.0006206. e6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh T, et al. Immunocompetent murine models for the study of glioblastoma immunotherapy. J Transl Med. 2014;12:107. doi: 10.1186/1479-5876-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanconato F, et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nature medicine. 2018;24:1599–1610. doi: 10.1038/s41591-018-0158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller TE, et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature. 2017;547:355–359. doi: 10.1038/nature23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao Z, et al. A Single-Cell Roadmap of Lineage Bifurcation in Human ESC Models of Embryonic Brain Development. Cell stem cell. 2017;20:120–134. doi: 10.1016/j.stem.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nowakowski TJ, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong S, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–528. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 67.Kanton S, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 68.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 69.Corces MR, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018:362. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lachmann A, Giorgi FM, Lopez G, Califano A. ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics. 2016;32:2233–2235. doi: 10.1093/bioinformatics/btw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alvarez MJ, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nature genetics. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 74.Panciera T, et al. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell stem cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Developmental cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schonhuber N, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nature medicine. 2014;20:1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Azzolin L, et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 78.Schildge S, Bohrer C, Beck K, Schachtrup C. Isolation and culture of mouse cortical astrocytes. Journal of visualized experiments : JoVE. 2013 doi: 10.3791/50079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Porcu E, et al. BMP9 counteracts the tumorigenic and pro-angiogenic potential of glioblastoma. Cell death and differentiation. 2018;25:1808–1822. doi: 10.1038/s41418-018-0149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Totaro A, et al. YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nature communications. 2017;8 doi: 10.1038/ncomms15206. 15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All RNA-seq and microarray raw data generated for the present study, along with counts matrices and metadata for each sample, are publicly available in Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession GSE133471. Single-cell RNA-seq data of primary glioblastoma samples from Darmanis et al. were downloaded as raw reads from GEO GSE84465. Expression matrix and metadata of the Neftel dataset were downloaded from the Single Cell Portal of the Broad Institute (https://singlecell.broadinstitute.org/single_cell/study/SCP393/single-cell-rna-seq-of-adult-and-pediatric-glioblastoma#study-summary). Raw gene expression data (.CEL files) of the GBM TCGA cohort were downloaded from GEO GSE83130. Raw gene expression (.CEL files) and clinical data of the REMBRANDT study were downloaded from GEO GSE108474. BAM files of Ivy Atlas GBM samples were downloaded from the Anatomic Structures RNA-Seq repository of the Ivy Glioblastoma Atlas Project (http://glioblastoma.alleninstitute.org/rnaseq/bam.csv).
Source data for Fig. 1e, 3b, 3e-f, 4c, 4f-g, 5a-f, 6b, 6f, and Extended Data Fig. 1c, 2b-c, 3b, 4b, 6a-d, 7d, 8b, 9a-b, 10b, 10e, 10f have been provided with the paper. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
All code was performed using R 3.5.0 and publicly available packages explicitly cited in the manuscript. No custom functions were written for the analysis. STAR is available in GitHub (https://github.com/alexdobin/STAR). ARACNe-AP is available in GitHub (https://github.com/califano-lab/ARACNe-AP). HOMER is available at http://homer.ucsd.edu/homer/. MultiExperiment Viewer is available at http://mev.tm4.org.