Summary
Inflammasomes are signaling platforms that are assembled in response to infection or sterile inflammation by cytosolic pattern recognition receptors (PRRs). The consequent inflammasome-triggered Caspase-1 activation is critical for the host defense against pathogens. During infection, NLRP3, a PRR also called Cryopyrin, triggers the assembly of an inflammasome activating Caspase-1 via the recruitment of ASC and Nek7. The NLRP3 inflammasome activation is tightly controlled both transcriptionally and post-translationally. Despite the importance of the NLRP3 inflammasome regulation in autoinflammatory and infectious diseases, little is known about the mechanism controlling the NLRP3 activation and the upstream signaling that regulates the NLRP3 inflammasome assembly. We have previously shown that the RhoGTPases-activating toxin from Escherichia coli, CNF1, activates Caspase-1, but the upstream mechanism is unclear. Here we provide evidence of the role of the NLRP3 inflammasome in sensing the activity of bacterial toxins and virulence factors that activate host RhoGTPases. We demonstrate that this activation relies on monitoring of the toxin’s activity on the RhoGTPase Rac2. We also show that the NLRP3 inflammasome is activated by a signaling cascade involving the P21 activated kinases (Pak) 1/2 and the Pak1-mediated phosphorylation of Threonine 659 of NLRP3, which is necessary for the NLRP3-Nek7 interaction, the inflammasome activation and the IL-1ß cytokine maturation. Furthermore, inhibition of the Pak-NLRP3 axis diminishes the bacterial clearance of CNF1-expressing UTI89 E. coli during bacteremia in mice. Altogether, our results establish Pak1/2 as critical regulators of the NLRP3 inflammasome and reveal the role of the Pak-NLRP3 signaling axis in vivo during bacteremia in mice.
Introduction
Uropathogenic Escherichia coli (UPEC) is the leading causative agent of bacteremia1. Therefore, it is fundamental to decipher the mechanisms that determine the fate of this pathogen in blood. The innate immune sensing of E. coli is mediated by Pattern Recognition Receptors (PRR), mainly by Toll-Like Receptor (TLR)-4 that detects bacterial lipopolysaccharides (LPS). LPS is the principal component of the external membrane of both pathogenic and non-pathogenic E. coli, thus Pattern-Triggered Immunity (PTI) does not appear to be sufficient to gauge the pathogenic potential of microbes. TLR4 being activated by both live and dead bacteria, PTI is certainly important to monitor the quantity of bacteria but not sufficient to determine their quality2. One strategy to determine microbial pathogenicity is the detection of virulence factors activities that are specific to pathogens3. Virulence factors of uropathogenic E. coli include CNF1, a RhoGTPases targeting toxin. The CNF1 toxin bears an enzymatic activity responsible for the post-translational deamidation of a specific glutamine residue on a subset of Rho GTPases, namely Rac, Cdc42 and RhoA4–6. This modification destroys their intrinsic and GAP-regulated ability to hydrolyze GTP, conferring to Rho proteins dominant-positive mutant characteristics4–6. This modification increases GTP bound activated-Rho proteins and the activation of their downstream signaling pathways6. By modulating the host cytoskeleton, these virulence factors confer to bacteria invasion properties and modulation of inflammatory responses7–10. Among virulence factors, more than 30 target RhoGTPases. They are either activators or inhibitors of RhoGTPases and both activate Caspase-111,12.
Inflammasomes are signaling platforms assembled by cytosolic PRRs that activate Caspase-1. NLRP3 oligomerizes upon infection or cellular damage and recruits ASC, Nek7, and Caspase-1 to form the NLRP3 inflammasome. This assembly results in ASC speck formation, cleavage of pro-Caspase-1 into active Caspase-1 and the maturation of pro-IL-1ß into IL-1ß. The NLRP3 assembly is controlled by both the priming by TLR ligands and activation signals. In addition, the NLRP3 assembly is regulated by phosphorylation and ubiquitination events13,14. Despite variety of identified NLRP3 activators, both upstream signaling pathways controlling NLRP3 post-translational modifications and activation mechanisms are still unclear14. Interestingly, toxins that inactivate RhoGTPases activate the Pyrin inflammasome via the modification of its phosphorylation status by PKN1/2 kinases. The Pyrin inflammasome has been shown to detect toxins inhibiting Rho GTPases but nothing is known about the sensing of toxins activating RhoGTPases by inflammasomes15,16. Here, we used the CNF1 toxin as a model of the RhoGTPase-activating virulence factor to demonstrate the role of the Pak-NLRP3 axis in sensing CNF1 activity and controlling the bacterial clearance during bacteremia.
Results
The CNF1-triggered immunity requires NLRP3
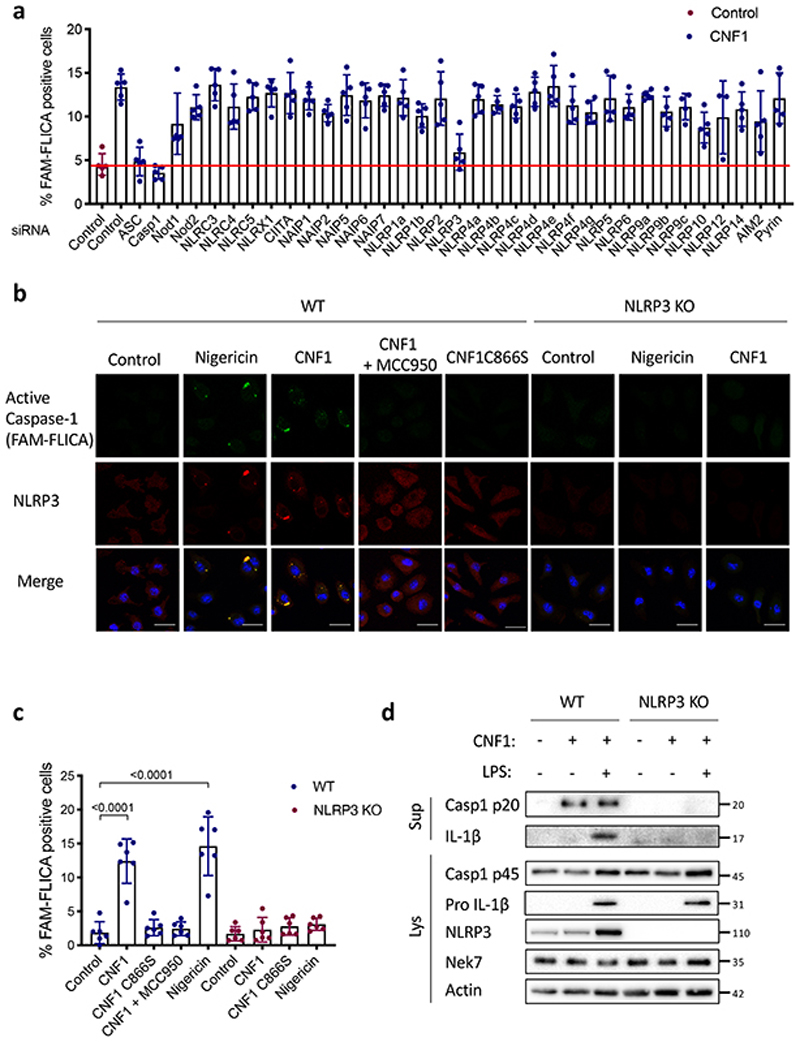
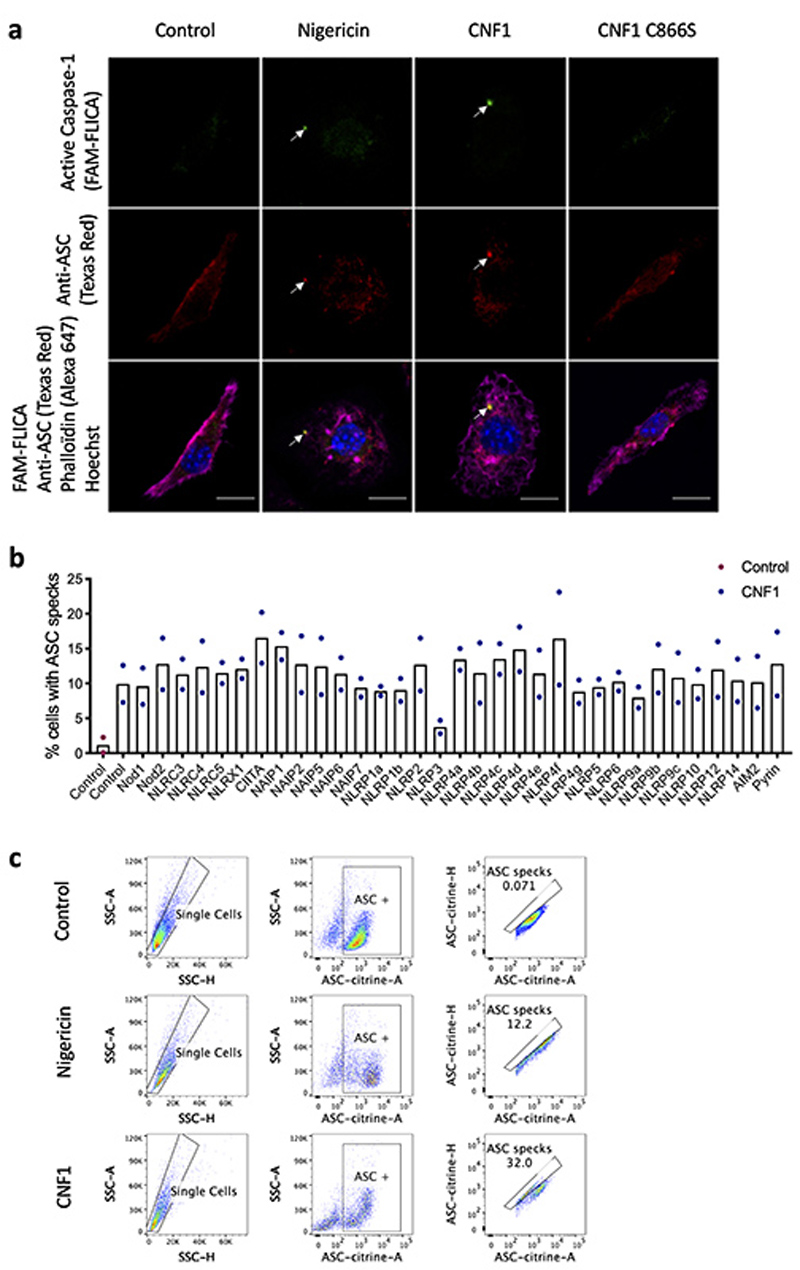
We set-up an assay to monitor the CNF1-triggered Caspase-1 activation using a FAM-YVAD-FMK (FAM-FLICA) probe. Primary bone marrow-derived macrophages (BMDMs) isolated from BALB/c mice were treated with CNF1 and analyzed using confocal microscopy. Cells with dots of FAM-FLICA staining corresponding to ASC specks were counted (Extended Data 1a). This unbiased screen revealed NLRP3 as the major NLR involved in the CNF1-triggered Caspase-1 activation (Figure 1a). The role of NLRP3 in this pathway was confirmed in BMDMs isolated from C57BL/6J mice bearing ASC-citrine knock-in using flow cytometry (Extended Data 1b–c). In this assay, we quantified the percentage of cells with ASC specks as previously described17,18. These data revealed the conserved role of NLRP3 in the response to CNF1 in macrophages isolated from both BALB/c and C57BL/6J background.
Figure 1. CNF1-triggered caspase-1 activation and IL-1ß maturation requires NLRP3.
(a) BMDMs isolated from BALB/c mice were transfected with the indicated siRNA for 72 h prior to 6 h of CNF1 treatment (500 ng/mL). The active Caspase-1 was detected using the FAM-FLICA probe. Cells harboring FAM-FLICA dots were counted positive using Fiji Software. Each dot represents 200 cells (n=1800 cells). Data are expressed as the mean ± SEM. (b-c) BMDMs extracted from wild-type or NLRP3 knock-out C57BL/6J mice were pretreated 45 min or not with MCC950 (1 μM) prior to treatment for 6 h with CNF1 (500 ng/mL), or the CNF1 catalytic inactive mutant CNF1 C866S (500ng/mL) or Nigericin (5 μM). Cells were analyzed by immunofluorescence and confocal imaging. Active Caspase-1 (FAM-FLICA) is shown in green, NLRP3 in red and nuclei in blue. Scale bar 20 μm. (c) Quantification of FAM-FLICA positive cells in wild-type (blue) or NLRP3 knock-out BMDMs (red). Each dot represents 100 cells (n=600 cells). Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. (d) wild-type or NLRP3 knock-out BMDMs were treated with CNF1 (500 ng/mL) and/or LPS (100 ng/mL) for 8 h prior to supernatant and cell lysates collection and immunoblot analysis. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
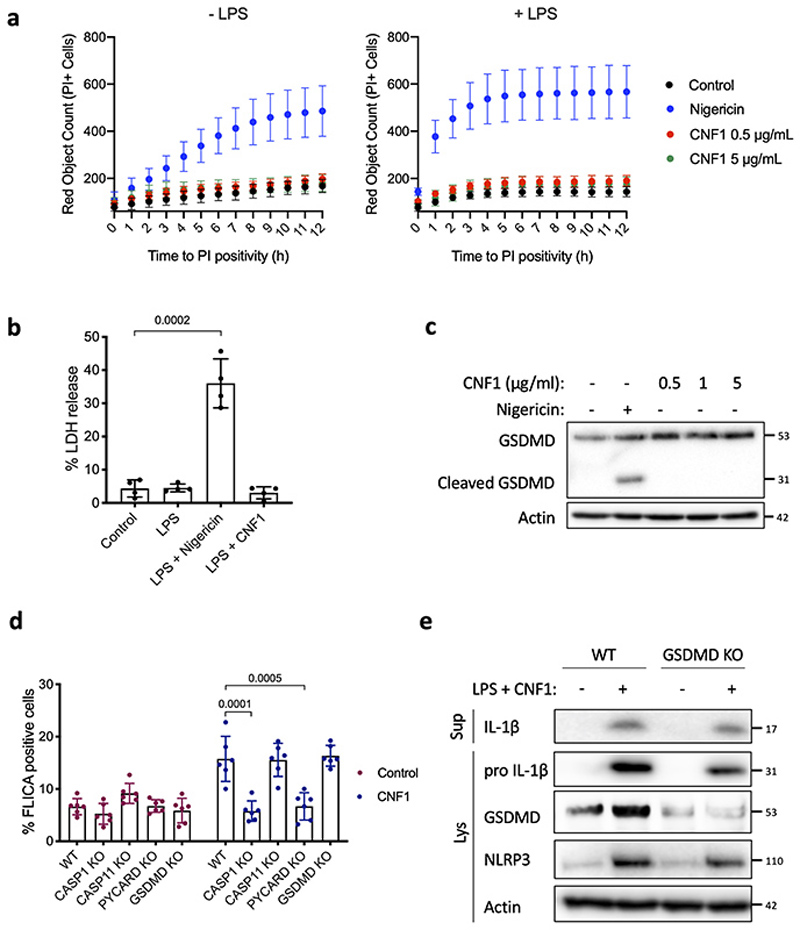
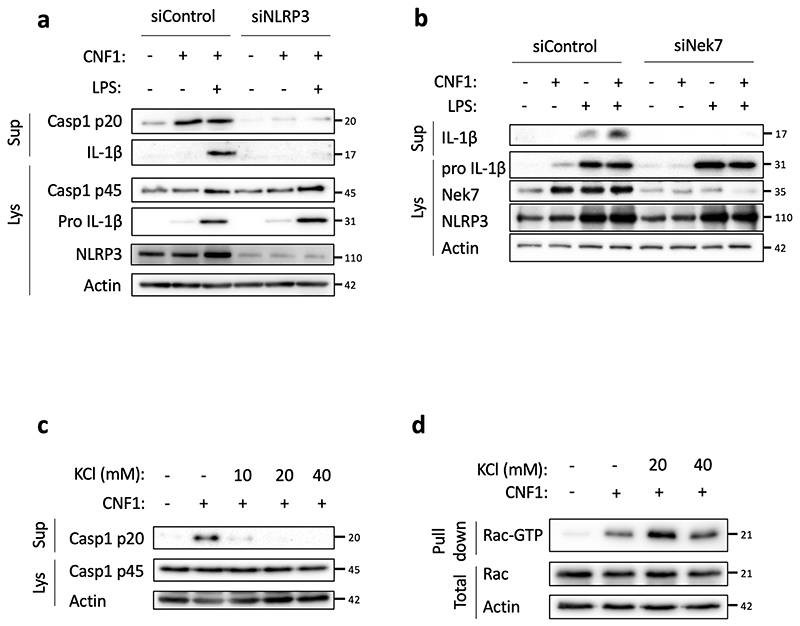
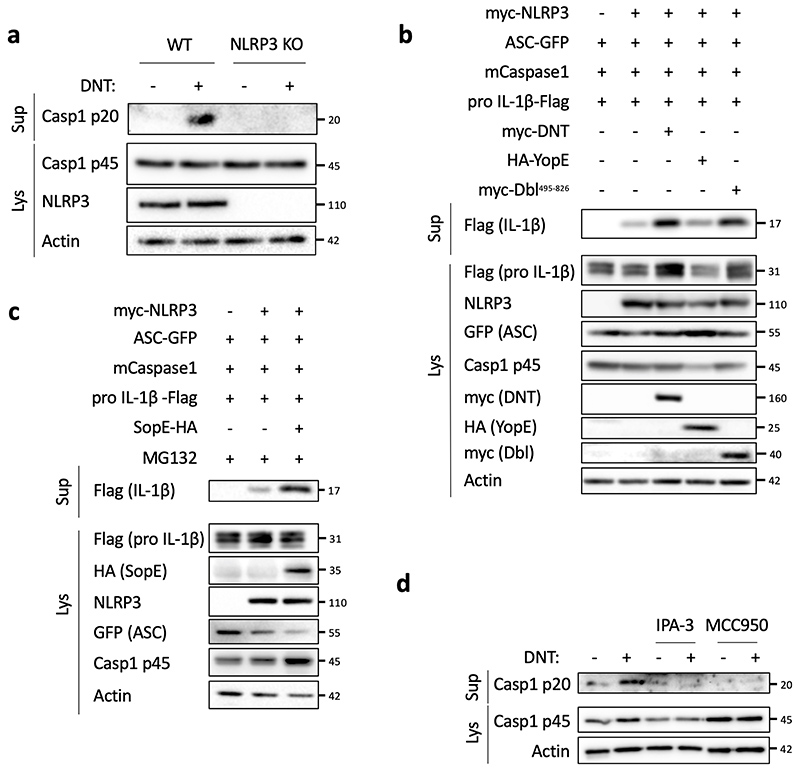
Next, we investigated the role of NLRP3 in the CNF1-triggered immunity. The co-treatment of BMDMs isolated from wild-type mice with the CNF1 toxin together with the NLRP3 inhibitor MCC950 was sufficient to block Caspase-1 activity demonstrating the CNF1 toxin as a NLRP3 activator (Figure 1b–c). Importantly, the number of FAM-FLICA positive cells was dramatically reduced in BMDMs treated with CNF1 catalytic inactive mutant C866S. This result provided the first evidence that CNF1 toxin activity is monitored by NLRP3, rather than the pattern of the toxin3. Furthermore, the CNF1-triggered IL-1ß maturation/secretion and Caspase-1 activation was impaired in NLRP3 knock-out BMDMs (Figure 1d and Supplementary figure 1). In contrast, the CNF1 treatment did not affect the secretion of the IL-6 or TNF-α, two cytokines not regulated by inflammasomes (Figure 1d and Supplementary figure 1). The NLRP3 inflammasome activation is often associated to pyroptosis19. To investigate whether CNF1 was triggering pyroptosis we measured propidium iodide (PI) incorporation (Extended Data 2a), lactate dehydrogenase (LDH) release (Extended Data 2b), as well as Gasdermin D (GSDMD) cleavage (Extended Data 2c). In contrast to Nigericin, we did not observe any of these pyroptosis markers following CNF1 treatment and we observed similar level of CNF1-triggered Caspase-1 activation and IL-1ß maturation/secretion in wild-type and GSDMD knock-out macrophages (Extended Data 2d–e). We subsequently tested the role of the NLRP3 inflammasome regulator Nek7 in the CNF1-triggered immunity. The NLRP3 or Nek7 siRNA transfection in BMDMs inhibited the IL-1ß maturation triggered by CNF1 (Extended Data 3a–b). K+ efflux is an upstream event for NLRP3 inflammasome activation and Nek7 requires K+ efflux for NLRP3 inflammasome assembly14,20. We observed that KCl treatment was sufficient to inhibit the CNF1-triggered Caspase-1 cleavage (Extended Data 3c). Importantly, we confirmed that the KCl treatment was not inhibiting the CNF1 toxin activity toward RhoGTPases activation by using a Pak effector pull-down (Extended Data 3d). Next, we investigated whether other toxins targeting RhoGTPases were able to activate the NLRP3 inflammasome. Dermonecrotic toxin (DNT) from Bordetella has a transglutaminase activity toward RhoGTPases that allows RhoGTPases constitutive activation6,10. We observed that purified recombinant DNT triggered the activation of Caspase-1 in wild-type but not in NLRP3 knock-out macrophages (Extended Data 4a). We next tested whether the NLRP3 activation was triggered specifically by virulence factors activating RhoGTPases using a NLRP3 inflammasome reconstitution in HEK 293T cells21. Cells were transfected with plasmids encoding the DNT toxin or the injected bacterial virulence factors YopE from Yersinia that contains a GAP domain allowing inactivation of the RhoGTPases6. We observed the NLRP3 dependent IL-1ß maturation when cells were transfected with the RhoGTPases activating toxin DNT, but not with the RhoGTPases inhibitor YopE (Extended Data 4b). The expression of the virulence factor SopE from Salmonella containing a GEF domain activating Rac and Cdc426,22 or the expression of the GEF domain of the Dbl exchange factor (Dbl495-826)23 were sufficient to trigger NLRP3 dependent IL-1ß maturation (Extended Data 4b–c). Altogether, we showed that toxins and virulence factors activating RhoGTPases triggered the NLRP3 inflammasome activation and suggested a major role for Rac in this pathway.
The NLRP3 inflammasome activation by CNF1 relies on Rac2 and the Pak serine/threonine kinases
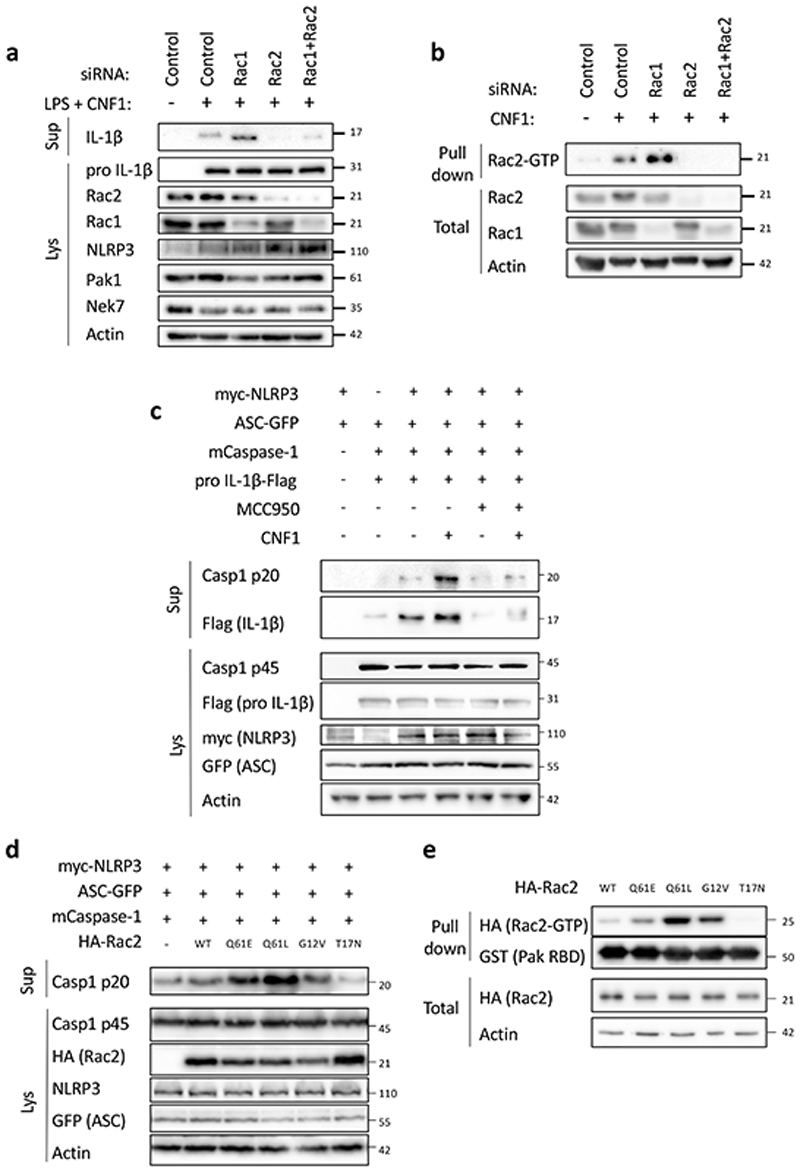
Although the CNF1 toxin is a RhoGTPase activator and Rac2 is a hematopoietic-specific RhoGTPase involved in the innate immune response to the CNF1 toxin24, the contribution of Rac1 and Rac2 in this process is still unknown. To investigate the role of Rac in the CNF1-triggered NLRP3 inflammasome activation, we knocked-down Rac1 and/or Rac2 using siRNA in BMDMs. Interestingly, Rac1 knock-down resulted in an increased level of CNF1-triggered IL-1ß maturation whereas Rac2 knock-down was sufficient to block it (Figure 2a). We tested whether activated GTP-bound Rac2 levels would increase when Rac1 was targeted by siRNA. The GST-Pak-RBD pull-down analysis indicated an increase in activated Rac2 when Rac1 was knocked-down by siRNA (Figure 2b). These data demonstrated the critical role of Rac2 in the CNF1-triggered IL-1ß maturation (Figure 2a and 2b). To determine the molecular mechanism of the Caspase-1 activation, we used the system of NLRP3 inflammasome reconstitution in HEK 293T cells. This analysis showed that CNF1 is sufficient for the NLRP3 inflammasome activation-triggered Caspase-1 cleavage and that the co-treatment of CNF1 with MCC950 inhibited this Caspase-1 activation (Figure 2c). The transfection of Rac2GTPase or activated mutant forms of Rac2 (including Q61E mimicking the CNF1 modification or Q61L and G12V) were sufficient to activate Caspase-1 in contrast to the inactive mutant Rac2T17N (Figure 2d). Interestingly, the strength of Caspase-1 activation observed with the activated forms of Rac2GTPase correlated with the amount of RhoGTPases bound to GST-Pak-RBD (Figure 2e). These data indicated that NLRP3 senses the activation level of the RhoGTPase Rac2 proportionally to the strength of activation rather than by detecting the structural modification made by the toxin as it would be predicted for a classical PRR.
Figure 2. Rac2 activation triggers NLRP3 inflammasome activation.
(a) BMDMs extracted from BALB/c mice were transfected with siRNA targeting the indicated isoform of RacGTPase for 72 h and treated with CNF1 (500 ng/mL) and LPS (100 ng/mL) for 8 h. Supernatants and cell lysates were analyzed by immunoblot. (b) iBMDM were transfected with the indicated siRNA for 72 h before being treated or not with CNF1 (500 ng/mL) for 6 h prior being analyzed using a GST-Pak Effector pull-down assay. The Rac2 associated with the GST-Pak-RBD beads is indicated as Rac2-GTP. (c-d) HEK293T cells were transfected for 16 h with plasmids encoding NLRP3 inflammasome components: myc-NLRP3, ASC-GFP, mCaspase-1 and pro-IL-1β-Flag as indicated, prior to Caspase-1 cleavage or pro-IL-1β maturation analysis by immunoblot. (c) Cells were pretreated or not with 1 μM MCC950 for 45 min prior to 6 h of CNF1 treatment (500 ng/mL). (d) Cells were transfected with HA-tagged mutants of Rac2: the constitutively active mutant mimicking CNF1-induced deamidation Rac2Q61E (Q61E), the constitutively active mutants Rac2Q61L (Q61L), Rac2G12V (G12V) or the dominant negative mutant Rac2T17N (T17N). Supernatants and cell lysates were analyzed by immunoblot. (e) HEK293T cells were transfected 16 h with HA-tagged active mutants Rac2Q61E, Rac2Q61L, Rac2G12V or the dominant negative mutant Rac2T17N prior to be analyzed using a GST-Pak Effector pull-down assay. The HA-Rac2 associated to the GST-Pak-RBD beads is indicated as Rac2-GTP. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
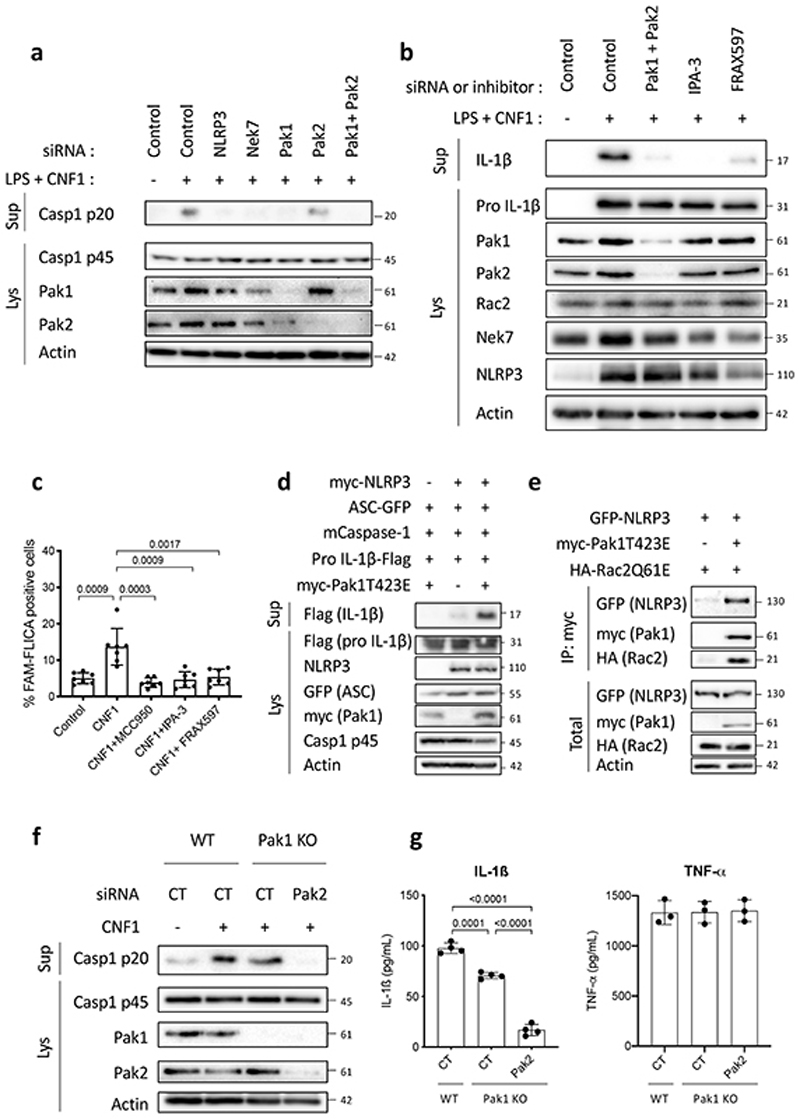
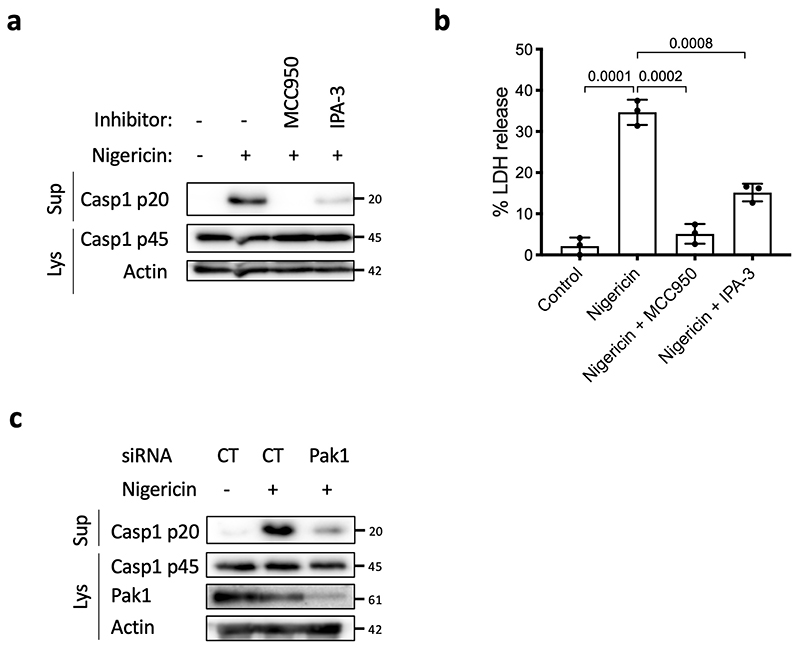
The correlation between Caspase-1 activation and the amount of Rac2 bound to GST-Pak-RBD suggested a potential role of Pak kinases in the CNF1-triggered NLRP3 inflammasome activation. We thus knocked-down Pak1 and/or Pak2 in BMDMs by transfecting siRNA (Pak3 is predominantly expressed in the brain25,26). We observed a major reduction of Caspase-1 cleavage in Pak1 siRNA treated cells but a moderate impact with the siRNA targeting Pak2 indicating a main role for Pak1 (Figure 3a). However, we could not exclude that the total inhibition of Caspase-1 cleavage observed with the Pak1 siRNA treatment might be due to a limit in the detection level of cleaved Caspase-1 or to Pak1 siRNA that may affect Pak2 suggesting a partial Pak1/2 redundancy. The treatment with Pak1 inhibitors (IPA-3 or FRAX597) was sufficient to block the CNF1-triggered IL-1ß maturation observed in macrophages treated with LPS and the Caspase-1 activation (Figure 3b–c). The inhibition of the Caspase-1 cleavage upon IPA-3 treatment was similarly observed in macrophages treated with DNT (Extended Data 4d). Interestingly, IPA-3 was shown to inhibit the binding of activated forms of Rac and Cdc42 to Pak1 thereby inhibiting the Threonine 423 autophosphorylation, whereas the FRAX597 is an ATP-competitive inhibitor27. We expressed in the inflammasome reconstitution system in HEK293T the activated form of Pak1 (T423E) together with Caspase-1, ASC and pro-IL-1β and observed no IL-1β maturation. In contrast, when NLRP3 was transfected together with ASC and Caspase-1, the expression of the activated form of Pak1 was sufficient to trigger the IL-1β maturation (Figure 3d). In addition, phosphorylated forms of Pak co-localized in dot-like structures with NLRP3 and active Caspase-1 (Supplementary Figure 2). We next investigated whether Rac2, Pak1 and NLRP3 proteins formed a complex. We found NLRP3 interacting with activated Rac2 when activated Pak1 was expressed (Figure 3e). We next investigated whether Pak1 was involved in the Nigericin-triggered NLRP3 inflammasome activation and observed that IPA-3 treatment was sufficient to inhibit both the Caspase-1 cleavage and the LDH release (Extended Data 5a–b). Furthermore, siRNA targeting of Pak1 was found to reduce the Nigericin triggered Caspase-1 maturation (Extended Data 5c).
Figure 3. Rac2-NLRP3 signaling depends on Pak1 kinase.
(a) BMDMs isolated from BALB/c mice were transfected for 72 h with siRNA targeting either NLRP3, Nek7, Pak1, Pak2 as indicated and non-targeting siRNA used as control. Cells were treated CNF1 (500 ng/mL) and LPS (100 ng/mL) for 8 h, as indicated. Supernatants and cell lysates were analyzed by immunoblots. (b) BMDMs isolated from BALB/c mice were transfected with Pak1 and Pak2 targeting siRNA or with non-targeting siRNA for 72 h and treated either with 5 μM IPA-3, 1 μM FRAX597 or vehicle for 45 min prior to treatment with CNF1 (500 ng/mL) and LPS (100 ng/mL) for 8 h. Supernatants and cell lysates were analyzed by immunoblot. (c) BMDMs isolated from BALB/c mice were treated with vehicle (Control) or treated either with 1 μM MCC950, 5 μM IPA-3 or 1 μM FRAX597 for 45 min prior to treatment with CNF1 (500 ng/mL) for 6 h. Active Caspase-1 was stained with FAM-FLICA, analyzed by microscopy and FAM-FLICA positive cells were counted. Each dot represents 100 cells (n=800 cells). Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. (d) HEK293T cells were transfected as indicated with plasmids encoding NLRP3 inflammasome component (myc-NLRP3, ASC-GFP, mCaspase-1) and pro-IL-1β-Flag together with myc-Pak1T423E and pro-IL-1β-Flag cleavage was analyzed by immunoblot. (e) HEK293T cells were transfected with plasmids encoding GFP-NLRP3, myc-Pak1T423E and HA-Rac2Q61E. Cell lysates were processed for anti-myc immunoprecipitation. (f) BMDMs isolated from wild-type or Pak1 knock out C57BL/6J mice were transfected 72 h with non-targeting or Pak2-targeting siRNA prior to CNF1 (500 ng/mL) treatment for 8h. Supernatants and cell lysates were analyzed by immunoblots. (g) BMDMs isolated from wild-type or Pak1 knock out C57BL/6J mice were transfected 72 h with non-targeting or Pak2-targeting siRNA prior to CNF1 (500 ng/mL) and LPS (100 ng/mL) treatment for 8 h. Supernatants were analyzed by ELISA for IL-1β (n=4 biologically independent samples) and TNF-α (n=3 biologically independent samples). Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
To genetically prove the involvement of Pak1 in the NLRP3 inflammasome activation, we took advantage of Pak1 knock-out mice. We observed a reduction of the Caspase-1 cleavage triggered by CNF1 in Pak1 knock-out macrophages and a reduction of the IL-1β secretion (Figure 3f and 3g). In contrast, the secretion of TNFα was unaffected (Figure 3g). Both Caspase-1 cleavage and IL-1β secretion triggered by CNF1 were drastically reduced when the Pak1 knock-out macrophages were treated with the Pak2 siRNA suggesting a partial compensation in Pak1 knock-out macrophages (Figure 3f and 3g).
Pak1 phosphorylates NLRP3 and triggers inflammasome activation
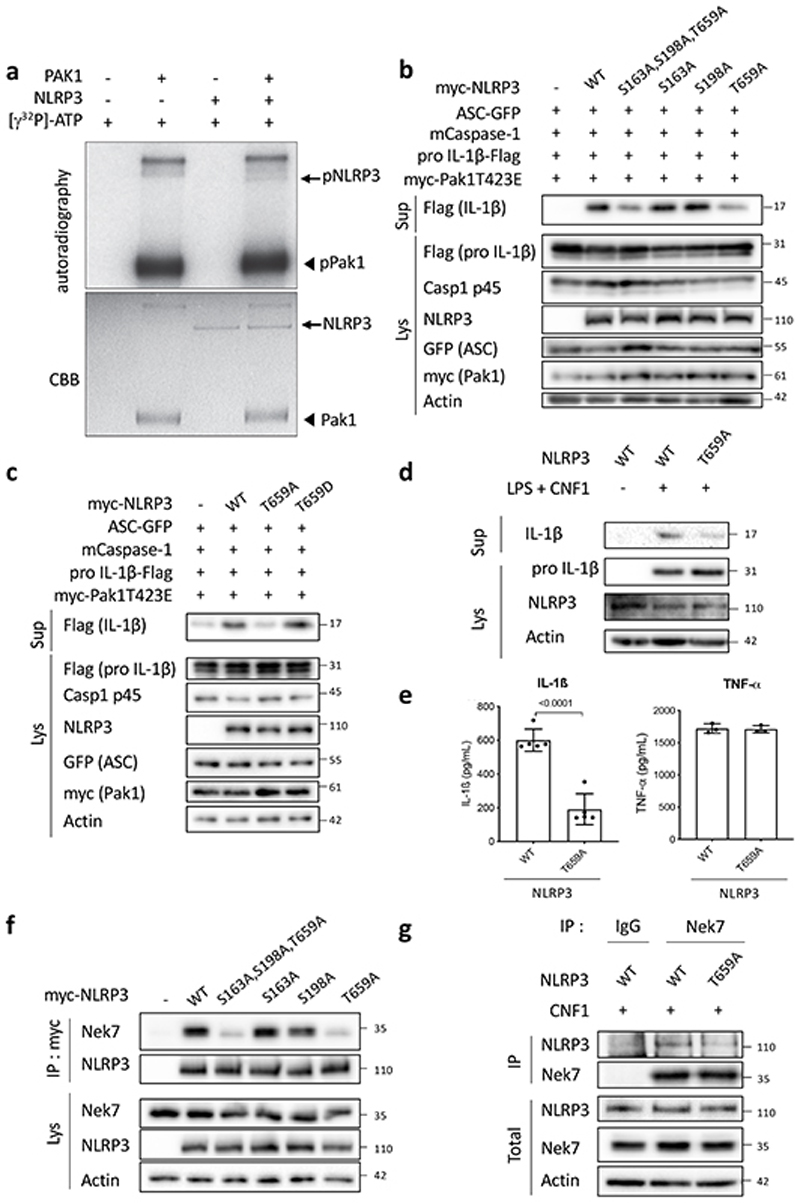
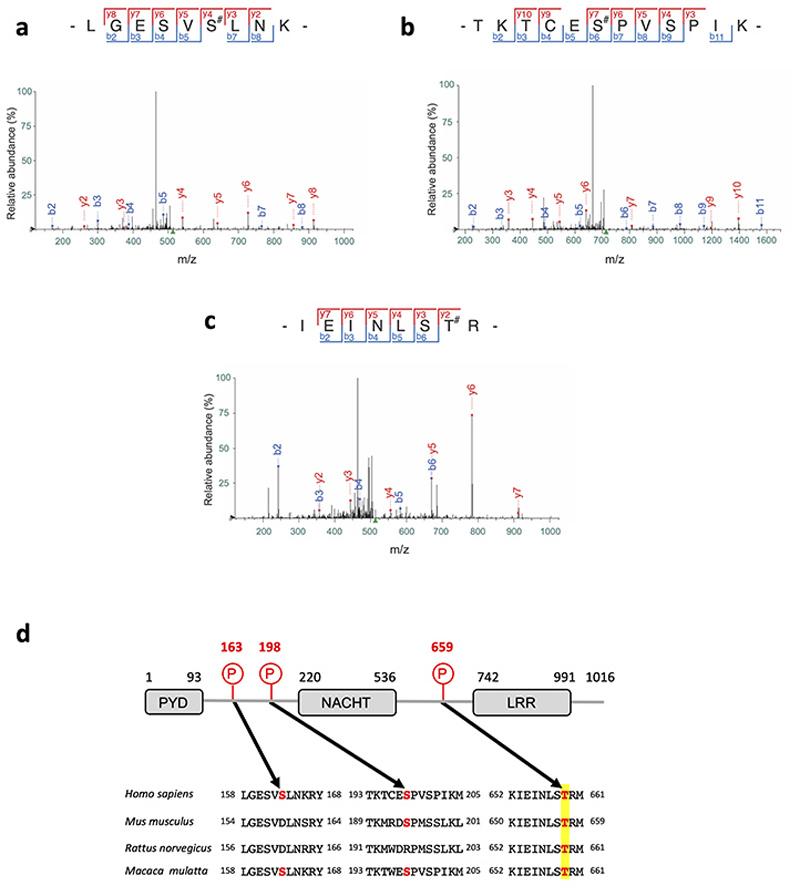
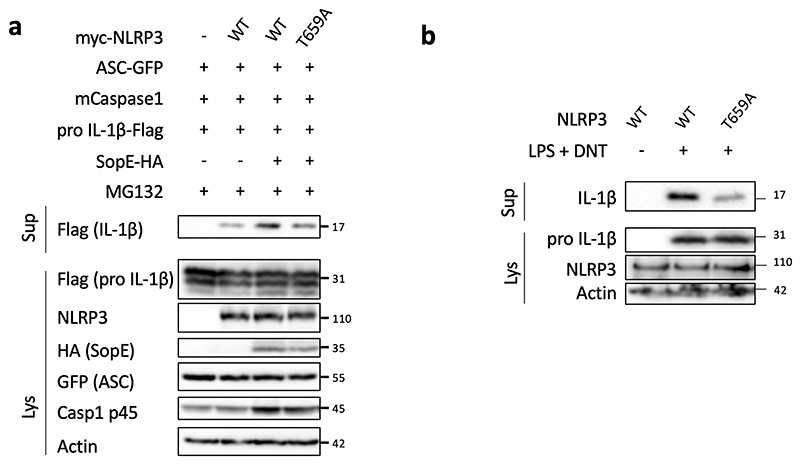
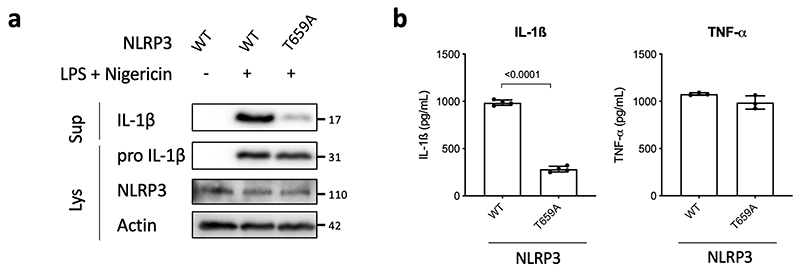
To further investigate whether NLRP3 is a substrate for the Pak1 serine/threonine kinase, we set-up an in vitro kinase assay. When both Pak1 and NLRP3 proteins were incubated with ATPγ32P, we observed a band at the size of NLRP3 indicating the direct phosphorylation of NLRP3 by Pak1 in vitro (Figure 4a). The in vitro kinase assay was then used to identify the phosphorylated sites of NLRP3 by analyzing the band corresponding to NLRP3 using mass spectrometry. The analysis revealed that Pak1 phosphorylates NLRP3 at 3 independent positions corresponding to S163, S198 and T659 in the human NLRP3 (Extended Data 6a–c and Supplementary Table1). Interestingly, the S163 and S198 were previously reported to be phosphorylated and S198 as important for NLRP3 priming28. The T659 was not reported to be phosphorylated and, interestingly, the identified peptide appears to be conserved between human and mouse (Extended Data 6d). Reinforcing the potential conservation of the Pak-NLRP3 axis, CNF1-triggered Caspase-1 activation was observed in primary human macrophages and was inhibited by treatment with NLRP3 inhibitor or Pak1 inhibitor (Extended Data 7a–b). We next expressed in the inflammasome reconstitution system the activated form of Pak1 (T423E) and compared the effect of the expression of NLRP3 wild-type to the NLRP3 triple mutant S163A, S198A, T659A or single mutant NLRP3 S163A, NLRP3 S198A and NLRP3 T659A in which the phosphorylated residue has been replaced by an Alanine that is not sensitive to phosphorylation. The results show that the NLRP3 triple mutant S163A, S198A, T659A or the single mutant T659A is impaired in IL-1β maturation triggered by the activated Pak1 T423E, indicating an important role for the T659 residue in the Pak1-triggered NLRP3 inflammasome activation (Figure 4b). In addition, we generated a T659D phosphomimetic NLRP3 mutant and we observed that, compared to the T659A mutant, the NLRP3 T659D had increased capacity to trigger the pro-IL-1β maturation (Figure 4c). Importantly, similar results were obtained when the RhoGTPases activating virulence factor SopE was transfected to activate the pathway, highlighting the involvement of this NLRP3 post translational regulation for the sensing of other virulence factors activating RhoGTPases (Extended Data 8a). Next, immortalized macrophages knocked-out for NLRP3 were stably reconstituted with plasmids encoding either NLRP3 or NLRP3 T659A. Confirming our results, we observed an impaired CNF1-triggered IL-1β maturation/secretion in macrophages expressing NLRP3 T659A compared to NLRP3 wild-type expressing macrophages (Figure 4d and 4e). In contrast, TNFα was similarly secreted by macrophages expressing either NLRP3 or NLRP3 T659A (Figure 4e). We also observed with the DNT toxin an impaired CNF1-triggered IL-1β maturation in macrophages expressing the T659A mutant compared to NLRP3 wild-type expressing macrophages (Extended Data 8b). Reinforcing the importance of this phosphorylation site of NLRP3 in the inflammasome activation process, the Nigericin-triggered IL-1β maturation and secretion were reduced in macrophages expressing the NLRP3 T659A compared to the macrophages expressing NLRP3 whereas TNFα secretion was unaffected (Extended Data 9). Altogether, these results show a functional role for the T659 NLRP3 phosphorylation and determine Pak1 as a regulator of the NLRP3 inflammasome. The structural analysis of the NLRP3-Nek7 interaction revealed a putative interaction domain at the level of the T659 of NLRP329. Using co-IP experiments, we tested whether S163A, S198A, T659A NLRP3 triple mutant or single mutants NLRP3 S163A, NLRP3 S198A and NLRP3 T659A affected the interaction with endogenous Nek7. The interaction of NLRP3 and Nek7 was impaired in the NLRP3 triple mutant S163A, S198A, T659A and in the NLRP3 T659A mutant indicating that Threonine 659 is a critical site for NLRP3-Nek7 interaction and suggesting the importance of Threonine 659 phosphorylation for the NLRP3-Nek7 interaction (Figure 4f). This observation was confirmed using an anti-Nek7 immunoprecipitation in macrophages in which we found a reduced NLRP3 T659A binding to Nek7 compared to wild-type NLRP3 (Figure 4g).
Figure 4. Pak1 phosphorylates NLRP3 to promote inflammasome activation.
(a) In vitro [γ-32P]-ATP kinase assay using human recombinant NLRP3 (arrows) and human recombinant Pak1 (arrowheads) analyzed by autoradiography and Coomassie Brilliant Blue (CBB) staining. (b) HEK293T cells were transfected with plasmids encoding NLRP3 inflammasome component (ASC-GFP, mCaspase-1), pro-IL-1β-Flag, myc-Pak1T423E, with either myc-NLRP3 or myc-NLRP3 S163A or myc-NLRP3 S198A or myc-NLRP3 T659A or myc-NLRP3 S163A, S198A, T659A and IL-1β maturation was analyzed by immunoblot. (c) HEK293T cells were transfected with plasmids encoding NLRP3 inflammasome components (ASC-GFP, mCaspase-1), myc-Pak1T423E, with either myc-NLRP3 or myc-NLRP3 T659A or myc-NLRP3 T659D and IL-1β maturation was analyzed by immunoblot. (d-e) NLRP3 knock-out iBMDMs reconstituted either with NLRP3 or NLRP3 T659A were treated with vehicle or LPS (100 ng/mL) and CNF1 (500ng/mL) for 8h, (d) supernatants and cell lysates were analyzed by immunoblot, (e) supernatants were analyzed by ELISA for IL-1β (n=4 biologically independent samples) and TNF-α (n=3 biologically independent samples). Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. (f) HEK293T cells were transfected with plasmids encoding either myc-NLRP3 or myc-NLRP3 S163A or myc-NLRP3 S198A or myc-NLRP3 T659A or myc-NLRP3 S163A, S198A, T659A. Cell lysates were processed for anti-myc immunoprecipitation and endogenous Nek7 was revealed using an anti-Nek7 antibody. (g) NLRP3 knock-out iBMDMs reconstituted either with NLRP3 or NLRP3 T659A were treated with CNF1 (500 ng/mL) for 6 h. Cell lysates were processed for anti-Nek7 or isotypic IgG immunoprecipitation. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
Clearance of CNF1-expressing E. coli during bacteremia requires the Pak-NLRP3 signaling axis
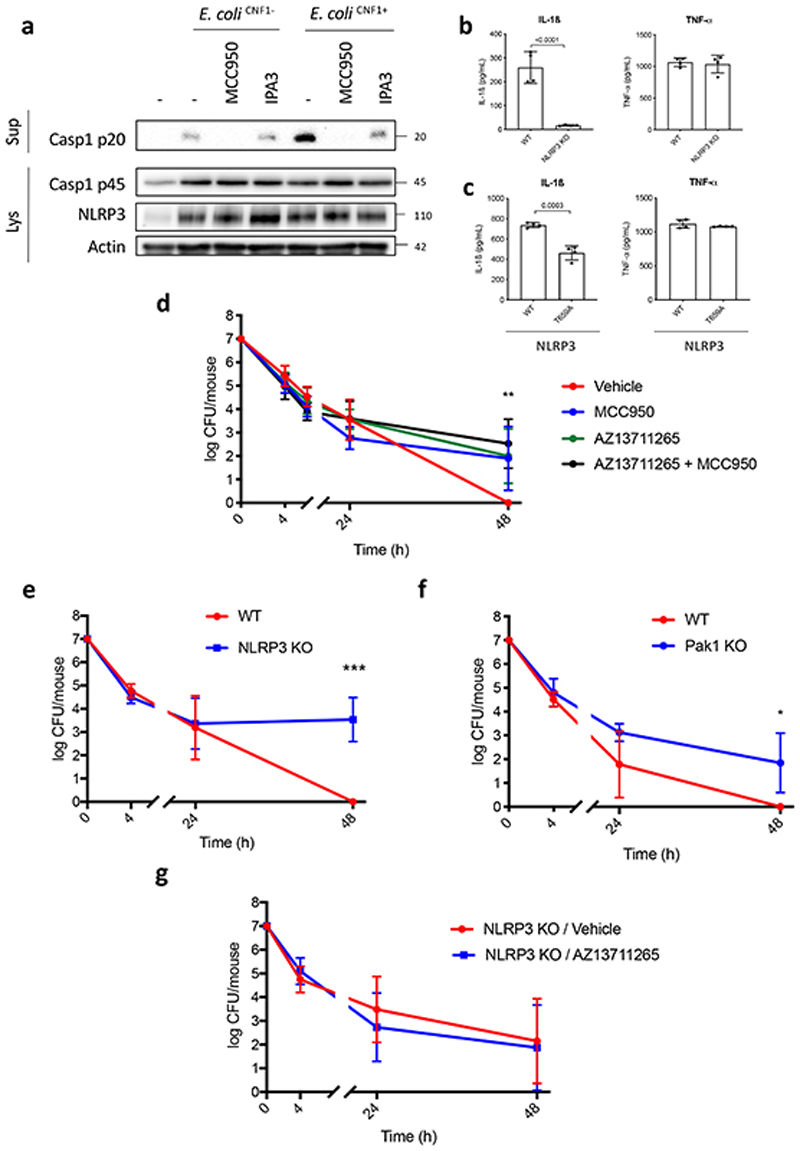
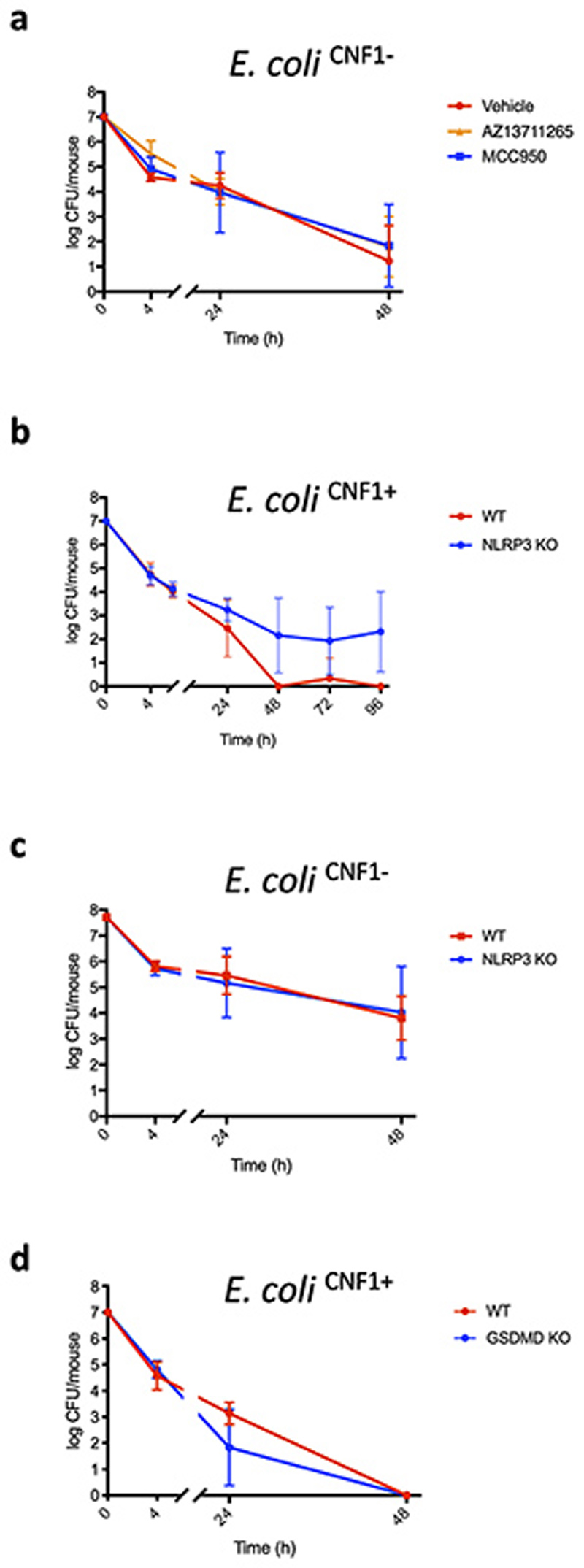
We next addressed the relevance of the CNF1-triggered Pak-NLRP3 signaling axis during infection We observed an increased Caspase-1 maturation when macrophages were infected with CNF1-expressing E. coli compared to the isogenic E. coli CNF1 knock-out strain and treatment with NLRP3 or Pak1 inhibitors decreased the Caspase-1 cleavage triggered by the E. coli expressing CNF1 (Figure 5a). Furthermore, IL-1ß secretion triggered by the CNF1-expressing E. coli was reduced when bacteria were added to NLRP3 knock-out macrophages (Figure 5b). In addition, the CNF1-expressing E. coli triggered the IL-1ß secretion in macrophages complemented with the NLRP3 T659A mutant was reduced compared to control macrophages expressing NLRP3 (Figure 5c). The TNFα secretion measured during CNF1-expressing E. coli infection was not affected in macrophages isolated from NLRP3 knock-out mice or macrophages expressing the NLRP3 T659A mutant (Figure 5b and 5c). We previously demonstrated that the CNF1 toxin expressed by E. coli triggered both an immune response in vivo and the bacterial clearance during bacteremia11. To investigate the role of Pak1 during CNF1-expressing E. coli bacteremia, we used the Pak1 inhibitor AZ13711265 which blocks the CNF1-triggered IL-1ß maturation (Supplementary Figure 3) and was associated with good in vivo pharmacokinetic properties27. We monitored the bacterial burden during bacteremia in control or mice injected with AZ13711265. Mice bacteremia was measured for each mouse at 4 h, 24 h and 48 h post-infection (Figure 5d). The bacterial clearance of CNF1-expressing E. coli was observed with no bacteria detectable at 48 h in all control animals (Figure 5d). We measured a statistically significant higher bacterial load at 48 h and 77 % of animals found to be positive for bacteremia in mice injected with the Pak1 inhibitor indicating that in vivo the inhibition of Pak1 is sufficient to inhibit the CNF1-expressing E. coli clearance (Figure 5d). Next, we used an NLRP3 inhibitor, MCC950, which has been shown to be efficient in vivo 30. We monitored bacteremia in mice injected with MCC950 compared to control. The bacterial clearance of CNF1-expressing E. coli in mice injected with the NLRP3 inhibitor was significantly higher at 48 h and 70% of animals found to be positive for bacteremia (Figure 5d). Consistent with our model, we observed no significant effect of both AZ13711265 and MCC950 toward the bacterial clearance when we infected mice with the isogenic E. coli CNF1 knock-out strain (Extended Data 10a). To genetically prove this point, we infected wild-type mice, NLRP3 knock-out mice or Pak1 knock-out mice and compared the CNF1-expressing E. coli burden. Consistent with the previous results obtained with the NLRP3 and Pak1 inhibitors, we did not detect any bacteria in the blood of infected wild-type mice at 48 h while we measured a mean of 1.5×104 and 2.5×102 bacteria per mouse in the blood of NLRP3 knock-out and Pak1 knock-out mice, respectively (Figures 5e and 5f). The lower effect observed in Pak1 knock-out mice compared to NLRP3 knock-out mice could be explained by the redundancy observed between Pak1 and Pak2 at the cellular level. The difference of CNF1-expressing E. coli clearance measured at 48 h between wild-type and NLRP3 knock-out mice was still observable at later time points and was not observed when mice were infected with the isogenic E. coli CNF1 knock-out strain indicating the specificity of the CNF1 response toward the NLRP3 pathway in vivo (Extended Data 10b–c). In addition, we measured a similar trend in the clearance of the CNF1-expressing E. coli strain in wild-type and GSDMD knock-out mice (Extended Data 10d). We next monitored the bacterial burden in mice infected with CNF1-expressing E. coli and treated with AZ13711265, MCC950, or both. We observed no differences in bacterial clearance between the three groups (Figure 5d). NLRP3 knock-out mice injected with vehicle or with AZ13711265 demonstrated no differences in bacterial clearance suggesting that Pak1 and NLRP3 are acting within the same signaling pathway during bacteremia (Figure 5g). Altogether these results unravel the critical role of Pak1 and NLRP3 in the clearance of CNF1-expressing bacteria and their importance in the innate immune response during bacteremia.
Figure 5. Pak1 and NLRP3 control the burden of CNF1-expressing E. coli during bacteremia.
(a) BMDMs isolated from C57BL/6 mice were pretreated for 45 min with 1 μM MCC950 or 5 μM IPA-3 and were uninfected or infected (MOI: 5) either with E. coli CNF1+ or isogenic CNF1-deleted mutant E. coli CNF1-. Supernatants and cell lysates were analyzed by immunoblot. (b) BMDMs isolated from C57BL/6 or C57BL/6 NLRP3 knock-out mice or (c) iBMDMs expressing NLRP3 or NLRP3 T659A were infected (MOI: 5) with E. coli CNF1+. Supernatants were analyzed by ELISA (n=4 biologically independent samples per group). Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. (d-g) Mice were intravenously infected with 107 CFU of E. coli CNF1+, prior to the collection of peripheral blood at 4 h, 24 h and 48 h for the measurement of bacteremia. (d) C57BL/6J mice were injected intraperitoneally either with vehicle, or 50 mg/kg MCC950 (n=10 mice), 10 mg/kg AZ13711265 (n=10 mice) or both once a day (n=9 mice). p-value: **<0.01, for each individual inhibitor-treated group compared to the group injected with vehicle. (e) wild-type (n=7 mice) or NLRP3 knock-out C57BL/6J mice (n=6 mice) were analyzed p-value: ***<0.001. (f) wild-type (n=4 mice) or Pak1 knock-out C57BL/6J mice were analyzed (n=4 mice) p-value: *<0.05. (g) NLRP3 knock-out C57BL/6J mice were injected intraperitoneally with 10 mg/kg AZ13711265 (n=7 mice) or vehicle (n=9 mice) once a day. Experiments were repeated at least two times, and representative data are shown. The numbers on the side of the immunoblots indicate molecular weight (kDa). Data are expressed as the geometric mean ± 95 CI. Statistical analyses were performed using a two-tailed nonparametric Mann Whitney test. p-value: *<0.05, **<0.01, ***<0.001.
Discussion
Our results shed light on a regulatory mechanism for NLRP3 following the activation of Rac2 by the CNF1 bacterial toxin. The level of NLRP3 inflammasome activation correlates with the strength of interaction between activated Rac2 and the Pak1-RBD, indicating that the innate immune system can adapt its response to the Rac2 activity level. This appears as an elegant strategy to deliver a commensurate response to the level of CNF1 toxin activity. Remarkably, the phosphorylated peptide containing the Threonine 659 of human NLRP3 isolated by mass spectrometry is highly conserved between species and the Pak-NLRP3 axis is conserved in human macrophages and is involved in the Nigericin-triggered NLRP3 inflammasome activation. Complementary studies will be necessary to determine the precise molecular mechanism in other species or other contexts and whether the NLRP3 Threonine 659 phosphorylation is a consensus site used by other kinases. Nevertheless, our results show that T659 NLRP3 phosphorylation is important for NLRP3 inflammasome activation and suggest its implication in NLRP3-related inflammatory disorders or susceptibility to infection.
Our results suggest a redundancy between Pak1 and Pak2 unraveled by a partial compensation by Pak2 in Pak1 knock-out macrophages and mice. Further studies would be required to clarify the importance of each Group-1 Pak proteins in the NLRP3 inflammasome activation. Unfortunately, the Pak2 knock-out mice are embryonic lethal (E8.0) and these studies would require the generation of conditional transgenic mice31.
We unravel a CNF1-triggered secretion of IL-1β that is not linked to an increased cell death and independent of GSDMD. Studies of NLRP3 inflammasome canonical triggers have demonstrated different IL-1β secretion scenarios. In the conventional scenario, Caspase-1 cleaves the inflammasome related cytokines and GSDMD to generate active N-GSDMD32,33. N-GSDMD relocate in the plasma membrane to form pores allowing IL-1β secretion32,33. GSDMD pores are associated with pyroptosis in the case of classical inflammasome activation or are controlled during inflammasome hyperactivation, leading to secretion without pyroptosis34–36. The CNF1-triggered IL-1β secretion appears to fall in another category, independent of both GSDMD cleavage and cell death that may share similarities with the unconventional IL-1β secretion37. This unconventional secretion relies on the IL-1β affinity with plasma membrane ruffles that are characteristic of the CNF1-triggered RacGTPases activation. The mechanism explaining how CNF1-triggered Caspase-1 activation without GSDMD cleavage remains to be elucidated and may be unique to toxins activating RhoGTPases. An hypothesis would be that the activation of Rac2, in parallel to the Pak1-NLRP3 pathway activation, inhibits the cleavage of GSDMD. Similar to Toxoplasma gondii, the NF-κB activation triggered by Rac2 might be another mechanism explaining the IL-1β secretion independent of cell death and GSDMD38. A Rac2 signaling may also regulate a potassium channel explaining the inhibition by KCl of the CNF1-triggered IL-1β secretion. Favoring this hypothesis, RacGTPases have been found to modulate Kir2.1, a Kir family potassium channel39.
Our study shows NLRP3 as a major sensor of toxins activating RhoGTPases whereas previous work has shown the sensing of RhoGTPase-inactivating toxins by Pyrin12. These studies highlight that mammalian innate immune system has evolved strategies that share similarities with the Effector Triggered Immunity (ETI) to detect abnormal activation of RhoGTPases3,40,41. Interestingly, both inactivation of RhoA and activation of Rac2 by bacterial toxins are monitored by Pyrin and NLRP3, respectively. More precisely, here we show that RacGTPases activating bacterial factors are sensed by NLRP3 independently of the type of modification made. Similarly, bacterial factors inactivating RhoA activate Pyrin independently of the type of modifications12,42,43. These results suggest that the host guarding of RhoGTPases signaling integrity relies on two sensors that monitor the abnormal RhoGTPases cycling rather than toxin-triggered post-translational modifications of host proteins or virulence factors enzymatic activities. Interestingly, both Pyrin and NLRP3 require the regulation by serine/threonine kinases respectively PKN1/2 and Pak1/2. The fact that two different inflammasomes have been evolutionarily selected to detect bacterial toxins that modify RhoGTPases highlights the importance of RhoGTPases in innate immunity.
Further studies would be necessary to determine the in vivo conservation of the Pak-NLRP3 axis and whether the sensing by the NLRP3 inflammasome of other RhoGTPases-activating virulence factors impact the bacterial burden during infection. Similarly, we would expect that further studies will determine the importance of other inflammasomes in detecting bacterial virulence factors endowed with enzymatic activities.
Our results reveal the importance of the Pak1 and NLRP3 in controlling the bacterial burden during bacteremia in mice. Even though additional studies will be required to determine the role of the Pak-NLRP3 signaling axis in bacteremic patients, our results showing increased bacterial burden in MCC950-treated mice suggest that caution will be necessary for the use of NLRP3 inhibitors in clinical settings. This is consistent with clinical data showing an increased risk of infections associated with IL-1 signaling inhibition44,45. An option would be to consider combining inflammasome inhibitors with antibiotherapies or with an enhanced surveillance for a potential for bacteremia risk.
Methods
Ethics statement
This study was carried out in strict accordance with the guidelines of the Council of the European Union (Directive 86/609/EEC) regarding the protection of animals used for experimental and other scientific purposes. The protocol was approved by the Institutional Animal Care and Use Committee on the Ethics of Animal Experiments of Nice, France (reference: APAFIS#18322-20181218099427035 v2 and APAFIS#24906-2020031614223228 v2).
Bacterial strains and toxins
The E. coli UTI89 clinical isolate was originally obtained from a patient with cystitis46 and the isogenic UTI89 CNF1+ (E. coli CNF1+) or UTI89 CNF1- (E. coli CNF1-) streptomycin-resistant strain generation and culture conditions were previously described11. For the infections, a 1/100 dilution of an overnight culture was inoculated and grown in up to OD600 = 1.2 using a Luria-Bertani (LB) medium supplemented with streptomycin (200 μg/mL). Bacteria were harvested by centrifugation and washed twice in PBS before dilution in PBS to obtain the desired bacterial concentrations for the mouse infection experiments. Recombinant wild-type Cytotoxic Necrotizing Factor-1 (CNF1) and its catalytically inactive form (CNF1-C866S) were produced and purified as previously reported47,48. The recombinant DNT toxin was purified from the pQEDNTwt using the same protocol49. The recombinant proteins were passed through a polymyxin B column (Affinity Detoxi-Gel, Pierce). The endotoxin removal was verified using a colorimetric LAL assay (LAL QCL-1000, Cambrex). Each stock of the CNF1 preparation (2 mg/mL) was shown to contain less than 0.5 endotoxin units/mL. Plasmid expressing the virulence factor pCMV-SopE-HA was previously reported22 and SopE expression was stabilized by adding MG132 (10 μM) to the cells in order to block its proteasomal degradation as previously described50. Plasmids expressing the pRK5-myc-DNT was obtained by PCR amplification of the pQEDNTwt and the pCMV-HA-YopE was obtained by PCR amplification and subcloning from the pACY184-YopE-GSK (kind gift of Igor Brodsky). All the plasmids were verified by sequencing (Eurofins).
Cell culture, transfection and inhibitors
HEK 293T cells were obtained from ATCC (CRL-3216) and maintained according to ATCC instructions. Bone marrow-derived macrophages (BMDMs) were extracted from femurs of 6-10 weeks-old BALB/c, C57BL/6J, C57BL/6J knock-in or C57BL/6J knock-out mice as indicated in the legends and were cultured in RPMI GlutaMax medium (Life Technologies) supplemented with 100 ng/mL M-CSF (Premium grade, Miltenyi Biotec), 10% heat-inactivated FBS (Biowest) and 50 μg/mL gentamycin (Life Technologies) at 37°C in an atmosphere containing 5% CO2. The cells were seeded at a concentration of 106 cells per well in a 6-well plate. After 6 days of differentiation, BMDMs were used for experiments. HEK 293T cells were transfected with plasmids using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. SiRNAs were transfected in BMDMs for 72 h using Lipofectamine RNAiMAX Reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. Cells were transfected as indicated in figure legends with siRNAs (Dharmacon) targeting NLRP3 (L-053455-00), Rac1 (L-041170-00), Rac2 (L-041171-01), Nek7 (J-063266-09), Pak1 (L-048101-00), Pak2 (L-040615-00) or non-targeting control siRNA (D-001810-10). For the siRNA screen, the BMDM were transfected with siRNA (Dharmacon) targeting Nod1 (L-055182-00), Nod2 (L-052735-00), NLRC3 (L-052823-01), NLRC4 (L-055000-00), NLRC5 (L-067620-01), NLRX1 (L-057712-01), CIITA (L-043166-02), NAIP1 (L-047682-00), NAIP2 (L-044151-01), NAIP5 (L-044142-01), NAIP6 (L-044145-01), NAIP7 (L-065757-00), NLRP1a (L-066229-00), NLRP1b (L-161107-01), NLRP2 (L-053528-01), NLRP3 (L-053455-01), NLRP4a (L-052395-01), NLRP4b (L-058181-01), NLRP4c (L-049416-01), NLRP4d (L-067051-01), NLRP4e (L-068064-01), NLRP4f (L-052668-01), NLRP4g (L-066364-01), NLRP5 (L-045315-01), NLRP6 (L-066157-01), NLRP9a (L-058269-01), NLRP9b (L-066417-01), NLRP9c (L-057344-01), NLRP10 (L-056559-01), NLRP12 (L-060234-01), NLRP14 (L-066093-01), Pycard (L-051439-01), MEFV (L-048693-01), AIM2 (L-044968-01). BMDMs were pre-treated with inhibitors for 45 min: 1 μM CP-456773 or MCC950 (Sigma), 5 μM IPA-3 (Tocris), 1 μM FRAX597 (Tocris), the indicated concentration of AZ13711265 (AGV Discovery) in 2% FBS containing RPMI followed by the addition of CNF1 500 ng/mL and/or ultrapure LPS 100 ng/mL (Invivogen) as indicated in the figure legends. Cells treated with Nigericin 5 μM (Invivogen) or ATP 5 mM (Invivogen) for 30 min were used as positive control for NLRP3 inflammasome activation. For K+ efflux preventing experiments, BMDMs were treated with 10 mM, 20 mM or 40 mM KCl. Primary macrophages were infected with E. coli CNF1+ or the isogenic E. coli CNF1- (MOI=5) for 16 h. Immortalized BMDM knock-out for NLRP3 were stably complemented with pINDUCER21 plasmids encoding human NLPR3 wild-type or NLRP3 T659A under a doxycycline inducible promoter as previously described51. NLRP3 expression was induced by adding 2 μg/mL doxycycline for 16 h (Takara Bio). All cell lines were authenticated by using PCR assays with species-specific primers. Mycoplasma testing was negative.
Mouse model of infection
7 weeks old female C57BL/6J mice (Charles River Laboratory) were injected intraperitoneally with MCC950 (Sigma) 50 mg/kg/24 h or AZ13711265 (AGV Discovery) 10 mg/kg/24 h or both. NLRP3 knock-out mice were kindly provided by V. Petrilli and have been previously described52. Pak1 knock-out, GSDMD knock-out, and ASC-citrine knock-in mice used in this study have been previously reported17,32,53. Female NLRP3 knock-out or Pak1 knock-out and female congenic wild-type C57BL/6J littermate mice were injected intravenously with 107 CFUs of E. coli and the determination of bacteremia was monitored as previously described11. Mice were housed with their littermates and kept on a regular 12-h light/dark cycle, room temperature: 20–25 °C; relative humidity: 50–70%. Food and water were available ad libitum. Experiments were carried out under pathogen-free conditions with randomly chosen animals (same sex, matched by age and body weight). Investigators were blinded for in vivo experiments. Sample size was determined based on our previous work11 and G*Power software.
Reconstituted NLRP3 inflammasome in HEK293T cell system
HEK293T cells were transfected with plasmids encoding the NLRP3 inflammasome components as previously described21, 28. HEK293T cells were transfected with plasmids encoding either myc-NLRP3 or NLRP3 mutants, ASC-GFP, mpro-Caspase1, and pro-IL-1ß-Flag. When indicated in the legend, cells were co-transfected with HA-Rac2, the constitutively active mutant of Rac2 mimicking CNF1-induced deamidation Rac2Q61E, Rac2Q61L, Rac2G12V, or Rac2T17N a dominant negative mutant of Rac2 for 16 h. The monitoring of Caspase-1 or IL-1ß cleavage was performed using supernatant immunoblotting.
Immunoprecipitation
HEK293T cells were transfected with plasmids encoding myc-NLRP3, myc-NLRP3 S163A S198A T659A, myc-NLRP3 S163A, and myc-NLRP3 T659A GFP-NLRP3, myc-Pak1 T423E and HA-Rac2 Q61E or NLRP3 expression was induced by adding 2 μg/mL doxycycline for 16 h to iBMDMs stably expressing NLPR3 or NLRP3 T659A. Cells were lysed and processed for immunoprecipitation using 2 μg of anti-myc or 3 μg of anti-Nek7 antibodies per conditions as previously described54. The NLRP3 expression and endogenous Nek7 levels were monitored in the cell lysate as well as in the IP fraction.
LDH release
The supernatant of stimulated macrophages was collected and centrifuged at 300 g for 5 min to remove cellular debris. LDH measurement was performed with the LDH Cytotoxicity Assay Kit (Thermo Scientific) according to the manufacturers’ instructions, in samples diluted 1:5 in PBS. Data were plotted as % of LDH release considering a Triton X-100 treated well as 100%.
Cell permeabilization kinetics
BMDMs were plated and stimulated in a 96-well plate in media containing propidium iodide (0.1 μg/ml) and data were acquired with a 10× objective using the IncuCyte Zoom system v6.2.9200.0 (Essen BioScience) in a CO2 and temperature-controlled environment. Each condition was run in quadruplicate. The number of fluorescent objects was counted with Incucyte Zoom (Essen BioScience) software.
In vitro kinase assay
500 ng of recombinant purified Pak1 were incubated with 1 μg of recombinant human NLRP3 protein (Abcam, ab165022), and with 50 μM ATP and 4 μCi of [γ 32 P]-ATP in kinase buffer (50mM HEPES pH 7.3, 50 mM NaCl, 0.05% Triton X-100, 10 mM β-glycerophosphate, 5 mM NaF, 10 mM MgCl2, and 0.2 mM MnCl2) at 30 °C for 30 min in a final volume of 39 μL. Reaction was stopped by adding 15 μL of LDS (ThermoFisher Scientific) and 6 μL of DTT 500 mM. Samples were analyzed by electrophoresis using Bolt 4-12% Bis-Tris Plus gels (ThermoFisher Scientific) followed by Coomassie blue staining and autoradiography.
Immunofluorescence staining, antibodies and ELISA assays
Caspase-1 activation was detected using the fluorescent probe FAM-FLICA (ImmunoChemistry Technologies) after 6 h of treatment, according to the manufacturer’s instructions. After labelling, cells were fixed in 4% paraformaldehyde for 15 min, PFA was neutralized with 50 mM NH4Cl for 15 min, cells were permeabilized with 0.5% Triton X-100 for 5 min and blocked with 2% TBS-BSA. Cells were incubated with mouse anti-NLRP3 (clone Cryo-2, Adipogen) and/or rabbit anti-ASC (AG-25B-0006, Adipogen) or rabbit anti-Phospho-Pak (ab40795, Abcam) antibodies for 1 h followed by incubation with secondary antibodies TexasRed anti-mouse IgG (TI-2000, Vector Laboratories) or Cy5 anti-mouse IgG (715-175-151, Jackson Immunoresearch) and/or TexasRed anti-rabbit IgG (711-075-152, Jackson Immunoresearch) and/or Phalloidin Alexa Fluor 647 (ab176759, Abcam) and Hoechst 33342 (H1399, ThermoFisher Scientific) for 30 min. Cells were imaged using Nikon A1R confocal microscope. Antibodies used in this study were: rabbit anti-IL-lβ (GTX74034, Genetex), mouse anti-Caspase-1 (clone Casper-1, Adipogen), mouse anti-Rac (clone 102/Rac1, BD Biosciences), goat anti-Rac2 (ab2244, Abcam), mouse anti-NLRP3 (clone Cryo-2, Adipogen), rabbit anti-Nek7 (ab133514, Abcam), rabbit anti-Pak1 (2602, CST), rabbit anti-Pak2 (2608, CST), rabbit monoclonal anti-GSDMD (ab209845), mouse anti-β-actin (AC-74, Sigma), mouse anti-myc (9E10, Roche), mouse anti-HA (16B12, Covance), mouse anti-Flag (clone M2, Sigma), mouse anti-GFP (clone 7.1, 13.1, Roche). Cytokines secretion was determined by ELISA using the mouse Quantikine ELISA kits for mouse IL-6, IL-18, TNFα and IL-1ß (R&D Systems) according to the manufacturer’s instructions.
Flow cytometry analysis
BMDM isolated from C57BL/6J mice constitutively expressing ASC-citrine fusion protein (R26-CAG-ASC-citrine) were treated with LPS (100 ng/mL) for 16h before 6h of treatment with vehicle or CNF1 (500 ng/mL) or 30 min with Nigericin (5μM). Cells were collected and analyzed by flow cytometry using the BD FACSCanto II cytometer (BD Biosciences). Cytometry data were analyzed using the FlowJo v10.6.2 software. Doublets were excluded using SSC-A (area) and SSC-H (height) plot, cells with a high expression of ASC-citrine were gated and then analyzed for ASC-citrine signal area (ASC-citrine-A) and ASC-citrine signal height (ASC-citrine-H). Cells with ASC specks were defined with a higher ASC-H:ASC-A ratio.
Statistical analyses
Statistical analyses were performed using GraphPad Prism V8.2.1 software. Comparisons of mice’s bacterial load were performed using the Mann-Whitney nonparametric test. Statistical analyses of FAM-FLICA positive cells, cytokines secretion and LDH release were performed using an unpaired two-tailed Student’s t-test.
Source data
Source Data Fig. 1 Unprocessed western blots.
Source Data Fig. 2 Numerical data.
Extended Data
Extended Data Fig. 1. CNF1 triggers Caspase-1 activation and ASC specks formation.
(a) BMDMs isolated from BALB/c mice were either treated with vehicle (control) or CNF1 (500 ng/mL) inactive catalytic mutant CNF1 C866S (500 ng/mL) for 6 h, or Nigericin (5 μM) for 30 min. Active Caspase-1 was revealed with FAM-FLICA (green), ASC was stained using an anti-ASC antibody (Texas Red), nuclei and actin filament were stained with Hoechst and phalloidin-Alexa 647 respectively. Cells were analyzed by confocal microscopy. Arrows indicates FAM-FLICA dots that colocalize with the ASC staining. Scale bar: 10μm. (b) BMDM isolated from C57BL/6J mice constitutively expressing ASC-citrine fusion protein (R26-CAG-ASC-citrine) were transfected with the indicated siRNA for 72 h prior to 6 h of CNF1 treatment (500 ng/mL) or treated with vehicle (control). Percent of cells with ASC specks. Data are expressed as the mean ± SEM. Each dot represents 105 cells (n=2 biologically independent samples). (c) BMDM isolated from C57BL/6J mice constitutively expressing ASC-citrine fusion protein (R26-CAG-ASC-citrine) were treated 6 h with CNF1 (500 ng/mL) or Nigericin (5 μM) for 30 min or vehicle (control). Cells were analyzed for ASC speck formation by flow cytometry as indicated, doublets were excluded using SSC-A and SSC-H plot, cells with a high expression of ASC-citrine were gated and then analyzed for ASC-citrine area (ASC-citrine-A) and ASC-citrine height (ASC-citrine-H). Cells with ASC specks are defined with a higher ASC-H:ASC-A ratio. Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 2. NLRP3 inflammasome activation by CNF1 does not induce pyroptosis.
(a) BMDMs isolated from C57BL/6J mice were treated with vehicle (control), Nigericin (5 μM) or CNF1 (500 ng/mL or 5 μg/mL) with or without LPS (100 ng/mL). Propidium iodide (PI) uptake was monitored over time (red object count) by real time imaging. Data are expressed as mean ± SD. 104 cells were analyzed for each replicate (n=4 independent wells). (b) BMDMs isolated from C57BL/6J mice were treated with vehicle (control, n=6 independent experiments), LPS (100 ng/mL, n=4 independent experiments), LPS and CNF1 (500 ng/mL, n=6 independent experiments) or LPS and Nigericin (5 μM, n=4 independent experiments), and LDH release was assessed. Data are expressed as the mean ± SEM. Statistical analyses were performed using a two -tailed nonparametric Mann Whitney test. (c) BMDMs isolated from C57BL/6J mice were treated either with Nigericin (5 μM) for 30 min or CNF1 (0.5, 1 or 5 μg/mL) for 8h and GSDMD cleavage in cell lysates is shown. (d) BMDMs isolated from C57BL/6J wild-type or CASP1, CASP11, PYCARD (coding for ASC) or GSDMD knock-out mice were untreated or treated with CNF1 (500 ng/mL) for 6h and were analyzed for Caspase-1 activation using the FAM-FLICA probe. Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. Each dot represents 100 cells (n=700 cells). (e) BMDMs isolated from wild-type or GSDMD knock-out mice were treated with CNF1 (500 ng/mL) and LPS (100 ng/mL) for 8h as indicated. Supernatants and cell lysates were analyzed by immunoblot. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 3. CNF1-triggered inflammasome activation depends on NLRP3, Nek7 and K+ .
(a, b) BMDMs isolated from C57BL/6J mice were transfected with siRNA-targeting NLRP3 (a), siRNA-targeting Nek7 (b), or control non-targeting siRNA for 72 h before treatment with CNF1 (500 ng/mL) and/or LPS (100 ng/mL) for 8 h. Supernatants and cell lysates were analyzed by immunoblot. (c, d) BMDMs isolated from C57BL/6J mice (c) or iBMDMs (d) were treated with the indicated KCl concentration and CNF1 (500 ng/mL) for 8 h. (c) Supernatants and cell lysates were analyzed by immunoblot, or (d) cell lysates were analyzed using a GST-Pak effector pull-down assay. The Rac associated with the GST-Pak-RBD beads is indicated as Rac-GTP. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 4. Toxins mediated Rho GTPases activation but not inhibition trigger the NLRP3 inflammasome.
(a) BMDMs isolated from wild-type or NLRP3 knock out C57BL/6J mice were treated with DNT (1 μg/mL) for 8 h. Supernatants and cell lysates were analyzed by immunoblot. (b-c) HEK293T cells were transfected as indicated with plasmids encoding NLRP3 inflammasome components (myc-NLRP3, ASC-GFP, mCaspase-1) and pro-IL-1β-Flag together with (b) myc-DNT, HA-YopE or myc-Dbl495-826 or (c) transfected with SopE-HA and treated with MG132 to block SopE degradation (10 μM Supernatants and cell lysates were analyzed by immunoblot. (d) BMDMs isolated from C57BL/6J mice were treated with IPA-3 (5 μM) or MCC950 (1 μM) for 45 min prior to 8 h of DNT treatment (1 μg/mL). Supernatants and cell lysates were analyzed by immunoblot. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 5. Inhibition of Pak1 diminishes NLRP3 activation by Nigericin.
(a, b) BMDMs isolated from C57BL/6J mice were treated with MCC950 (1 μM) or IPA-3 (5 μM) for 45 min prior to Nigericin (5 μM) treatment for 30 min. Supernatan and cell lysates were analyzed by (a) immunoblot and (b) supernatants were analyzed for LDH release (n=3 biologically independent experiments). Statistical analyses were performed using a two-tailed nonparametric Mann Whitney test. n=3 biologically independent samples were analyzed. (c) BMDMs isolated from C57BL/6J mice were treated for 72 h with non-targeting or Pak1 -targeting siRNA before treatment with Nigericin (5 μM) for 30 min. Supernatants and cell lysates were analyzed by immunoblot. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown. Data are expressed as the mean ± SEM.
Extended Data Fig. 6. Mass spectrometry analysis of Pak1 triggered NLRP3 phosphorylation.
(a-c) Fragmentation spectra of human NLRP3 peptides showing phosphorylation of Ser-163, Ser-198 and Thr-659. (d) Representation of NLRP3 domain structure and sequence alignment of NLRP3 ortholog peptides surrounding phosphorylated residues identified by mass spectrometry. The phosphorylated residues are in bold red.
Extended Data Fig. 7. Conservation of the Pak-NLRP3 axis in Human monocyte-derived macrophages.
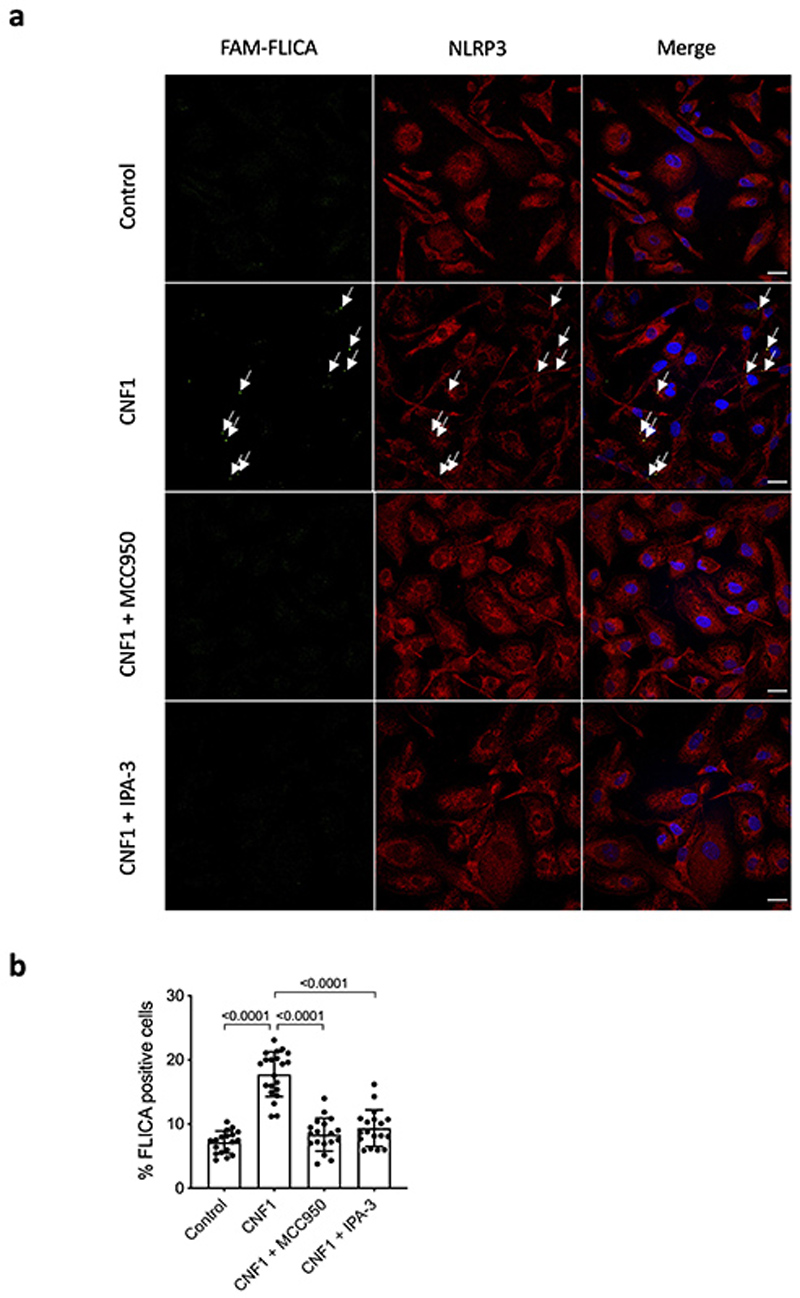
(a-b) Human monocyte-derived macrophages (hMDMs) were pretreated with vehicle, MCC950 (1 μM) or IPA-3 (5 μM) for 45 min before CNF1 (500 ng/mL) treatment for 6 h. Active Caspase-1 was stained with FAM-FLICA (green), NLRP3 (red) and nuclei (blue) were stained for immunofluorescence and confocal microscopy imaging. Arrows indicates FAM-FLICA dots that colocalize with NLRP3. Scale bar: 20 μm. (b) quantification of FAM-FLICA positive cells. Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. Each dot represents 100 cells (n=1800 cells). Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 8. The NLRP3 T659A mutant inhibit the IL-1β maturation triggered by SopE and DNT.
(a) HEK293T cells were transfected with plasmids encoding NLRP3 inflammasome components (ASC-GFP, mCaspase-1) and pro-IL-1β-Flag and either myc-NLRP3 (WT) or myc-NLRP3 T659A together with SopE-HA and treated with MG132 (10 μM) to block SopE degradation. Supernatants and cell lysates were analyzed by immunoblot. (b) NLRP3 knock-out iBMDMs reconstituted either with NLRP3 or NLRP3 T659A were treated with vehicle or LPS (100 ng/mL) and DNT (1 μg/mL) for 8 h. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 9. Macrophages expressing the NLRP3 T659A mutant have an impaired Nigericin-triggered IL-1β maturation.
(a-b) iBMDMs stably expressing either NLRP3 or NLRP3 T659A were treated with Nigericin (5 μM) for 30 min. Supernatants and cell lysates were analyzed by immunoblot and by ELISA for IL-1β (n=4 biologically independent samples) and TNF-α (n=3 biologically independent samples). Data are expressed as the mean ± SEM. Statistical analyses were performed using a two-tailed unpaired Student’s t-test. The numbers on the side of the immunoblots indicate molecular weight (kDa). Experiments were repeated at least three times, and representative data are shown.
Extended Data Fig. 10. E. coli CNF1- clearing is not affected by Pak1 or NLRP3 inhibition and E. coli CNF1+ clearing does not rely on GSDMD.
(a-d) Wild-type or knock-out mice were infected intravenously with isogenic CNF1-deleted E. coli (E. coli CNF1-) or CNF1 expressing E. coli (E. coli CNF1+). (a) Wild-type mice were injected intraperitoneally with 10 mg/kg AZ13711265 or 50 mg/kg MCC950 or vehicle once a day and were infected intravenously with isogenic CNF1-deleted E. coli (E. coli CNF1-) prior to the collection of peripheral blood at 4 h, 24 h and 48 h for measurement of bacteremia (n=5 mice per group). (b) Wild-type or NLRP3 knock-out C57BL/6J mice were infected intravenously with CNF1 expressing E. coli (E. coli CNF1+) prior to the collection of peripheral blood at 4 h, 24 h, 48 h, 72 h and 96 h for measurement of bacteremia (n=6 per group). (c) Wild-type (n=6 mice) or NLRP3 knock-out C57BL/6J mice (n=4 mice) were infected intravenously with isogenic CNF1-deleted E. coli (E. coli CNF1-) prior to the collection of peripheral blood at 4 h, 24 h and 48 h for measurement of bacteremia (n=6 per group). (d) Wild-type (n=6 mice) or GSDMD knock-out C57BL/6J mice (n=6 mice) were infected intravenously with E. coli CNF1+ prior to the collection of peripheral blood at 4 h, 24 h and 48 h for measurement of bacteremia. Experiments were repeated two times and representative data are shown. Data are expressed as the geometric mean ± 95 CI.
Supplementary Material
Acknowledgements
We thank Patrick Auberger, Andreas Baumler, Igor Brodsky, Jonathan Chernoff, Douglas Golenbock, Thomas Henry, Marijke Keestra-Gounder, Emmanuel Lemichez, Edward Manser, Etienne Meunier, Virginie Petrilli, Didier Pisani, Jean-Ehrland Ricci, Guillaume Robert, Pierre-Marie Roger, Lynda Stuart, Peter Vandenabeele, Stoyan Ivanov and Laurent Yvan-Charvet for sharing materials or fruitful discussions. We thank Anne-Sophie Dufour, Elsa Garcia, Marie Irondelle and Joseph Murdaca for technical assistance. We are grateful to the Innate Sensors Community (InnaSCo) for sharing tools. We thank Abby Cuttriss and the Office of International Scientific Visibility of Université Côte d’Azur for professional language editing. The mouse strain used for this research project, B6.129S2-Pak1tm1Cher/Mmnc, RRID:MMRRC_031838-UNC, was obtained from the Mutant Mouse Resource and Research Center (MMRRC) at University of North Carolina at Chapel Hill, an NIH-funded strain repository, and was donated to the MMRRC by Jonathan Chernoff, Ph.D., Fox Chase Cancer Center. We thank the Etablissement Français du Sang of Marseille for providing human blood from human healthy donors. We thank the C3M facilities (animal, genomic, cytometry, imaging) and the Harvard Taplin mass spectrometry core. This work was supported by grants from ANR (ANR-17-CE15-0001), “Investments for the Future” programs LABEX SIGNALIFE ANR-11-LABX-0028-01, IDEX UCAJEDI ANR-15-IDEX-01, ARC (RAC15014AAA), Université Côte d’Azur, Infectiopole sud and REDPIT. BFP is supported by ERC (ERC-2013-CoG_616986). AM supported by a fellowship from FRM, CT by a fellowship from Ville de Nice and OD is supported by a fellowship from INSERM and Université Côte d’Azur.
Footnotes
Author contributions
OD and AD designed, performed, and analyzed most of the experiments with input from CT, CL, AJ, AG, AM, PC, AR and SM. JC and RR provided advice on the mice infection model. EV, RG, DC, BFP, AR, PHVS, and ML provided tools and advice on NLRP3 inflammasome regulation. PM generated NLRP3 mutants and with GM and OD performed and analyzed most of the in vivo experiments. OV performed virulence factors and toxins subcloning, protein purifications, the in vitro kinase assay and analyzed the mass spectrometry results. LB conceived the project, designed experiments, and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Data availability
All data supporting the findings of this study are available within the article and its supplementary information or from the corresponding author upon reasonable request. Source data are provided with this paper.
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:1021. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol. 2013 doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flatau G, et al. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt G, et al. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-I. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 6.Aktories K, Barbieri J. Bacterial cytotoxins: targeting eukaryotic switches. Nat Rev Micro. 2005;3:397–410. doi: 10.1038/nrmicro1150. [DOI] [PubMed] [Google Scholar]
- 7.Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruno VM, et al. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoSPathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munro P, et al. Activation and proteasomal degradation of rho GTPases by cytotoxic necrotizing factor-1 elicit a controlled inflammatory response. J Biol Chem. 2004;279:35849–35857. doi: 10.1074/jbc.M401580200. [DOI] [PubMed] [Google Scholar]
- 10.Boquet P, Lemichez E. Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol. 2003;13:238–246. doi: 10.1016/s0962-8924(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 11.Diabate M, et al. Escherichia coli alpha-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia. PLoS Pathog. 2015;11:e1004732. doi: 10.1371/journal.ppat.1004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 13.Groslambert M, Py BF. Spotlight on the NLRP3 inflammasome pathway. J Inflamm Res. 2018;II:359–374. doi: 10.2147/JIR.S141220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Wang H, Kouadir M, Song H, Shi F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019;10:128. doi: 10.1038/s41419-019-1413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao W, Yang J, Liu W, Wang Y, Shao F. Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation. Proc Natl Acad Sci U S A. 2016;113:E4857–66. doi: 10.1073/pnas.1601700113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YH, Wood G, Kastner DL, Chae JJ. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol. 2016;17:914–921. doi: 10.1038/ni.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzeng TC, et al. A Fluorescent Reporter Mouse for Inflammasome Assembly Demonstrates an Important Role for Cell-Bound and Free ASC Specks during In Vivo Infection. Cell Rep. 2016;16:571–582. doi: 10.1016/j.celrep.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sester DP, et al. Assessment of Inflammasome Formation by Flow Cytometry. Curr Protoc Immunol. 2016;114 doi: 10.1002/cpim.13. 14.40.1-14.40.29. [DOI] [PubMed] [Google Scholar]
- 19.Lamkanfi M, Dixit VM. In Retrospect: The inflammasome turns 15. Nature. 2017;548:534–535. doi: 10.1038/548534a. [DOI] [PubMed] [Google Scholar]
- 20.He Y, Hara H, Núñez G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, Murray A, Beutler B. Reconstruction of the Mouse Inflammasome System in HEK293T Cells. Bio Protoc. 2016;6 doi: 10.21769/BioProtoc.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keestra AM, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 2013;496:233–237. doi: 10.1038/nature12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doye A, et al. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- 24.Boyer L, et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 26.Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J. 2010;425:465–473. doi: 10.1042/BJ20091173. [DOI] [PubMed] [Google Scholar]
- 27.Semenova G, Chernoff J. Targeting PAK1. Biochem Soc Trans. 2017;45:79–88. doi: 10.1042/BST20160134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song N, et al. NLRP3 Phosphorylation Is an Essential Priming Event for Inflammasome Activation. Mol Cell. 2017;68:185–197. doi: 10.1016/j.molcel.2017.08.017. e6. [DOI] [PubMed] [Google Scholar]
- 29.Sharif H, et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019;570:338–343. doi: 10.1038/s41586-019-1295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly ML, Chernoff J. Mouse models of PAK function. Cell Logist. 2012;2:84–88. doi: 10.4161/cl.21381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 33.He WT, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broz P, Pelegrín P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 35.Rühl S, et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science. 2018;362:956–960. doi: 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- 36.Evavold CL, et al. The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity. 2018;48:35–44. doi: 10.1016/j.immuni.2017.11.013. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monteleone M, et al. Interleukin-1 β Maturation Triggers Its Relocation to the Plasma Membrane for Gasdermin-D-Dependent and -Independent Secretion. Cell Rep. 2018;24:1425–1433. doi: 10.1016/j.celrep.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 38.Pandori WJ, et al. Toxoplasma gondii activates a Syk-CARD9-NF-κB signaling axis and gasdermin D-independent release of IL-1β during infection of primary human monocytes. PLoS Pathog. 2019;15:e1007923. doi: 10.1371/journal.ppat.1007923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muessel MJ, Harry GJ, Armstrong DL, Storey NM. SDF-1α and LPA modulate microglia potassium channels through rho gtpases to regulate cell morphology. Glia. 2013;61:1620–1628. doi: 10.1002/glia.22543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 41.Lopes Fischer N, Naseer N, Shin S, Brodsky IE. Effector-triggered immunity and pathogen sensing in metazoans. Nat Microbiol. 2020;5:14–26. doi: 10.1038/s41564-019-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aubert DF, et al. A Burkholderia Type VI Effector Deamidates Rho GTPases to Activate the Pyrin Inflammasome and Trigger Inflammation. Cell Host Microbe. 2016;19:664–674. doi: 10.1016/j.chom.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Medici NP, Rashid M, Bliska JB. Characterization of Pyrin Dephosphorylation and Inflammasome Activation in Macrophages as Triggered by the Yersinia Effectors YopE and YopT. Infect Immun. 2019;87 doi: 10.1128/IAI.00822-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabral VP, Andrade CA, Passos SR, Martins MF, Hökerberg YH. Severe infection in patients with rheumatoid arthritis taking anakinra, rituximab, or abatacept: a systematic review of observational studies. Rev Bras Reumatol Engl Ed. 2016;56:543–550. doi: 10.1016/j.rbre.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Ridker PM, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 46.Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buetow L, Flatau G, Chiu K, Boquet P, Ghosh P. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nat Struct Biol. 2001;8:584–588. doi: 10.1038/89610. [DOI] [PubMed] [Google Scholar]
- 48.Doye A, Boyer L, Mettouchi A, Lemichez E. Ubiquitin-mediated proteasomal degradation of Rho proteins by the CNF1 toxin. Methods Enzymol. 2006;406:447–456. doi: 10.1016/S0076-6879(06)06033-2. [DOI] [PubMed] [Google Scholar]
- 49.Matsuzawa T, Kashimoto T, Katahira J, Horiguchi Y. Identification of a receptor-binding domain of Bordetella dermonecrotic toxin. Infect Immun. 2002;70:3427–3432. doi: 10.1128/IAI.70.7.3427-3432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubori T, Galán JE. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 51.Lagrange B, et al. Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nat Commun. 2018;9 doi: 10.1038/s41467-017-02682-y. 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric ack crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 53.McDaniel AS, et al. Pak1 regulates multiple c-Kit mediated Ras-MAPK gain-in-function phenotypes in Nf1+/- mast cells. Blood. 2008;112:4646–4654. doi: 10.1182/blood-2008-04-155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stutz A, et al. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med. 2017;214:1725–1736. doi: 10.1084/jem.20160933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its supplementary information or from the corresponding author upon reasonable request. Source data are provided with this paper.