Abstract
Peptide-based subunit vaccines are attractive in view of personalized cancer vaccination with neo-antigens, as well as for the design of the newest generation of vaccines against infectious diseases. Key to mounting robust antigen specific immunity is delivery of antigen to antigen presenting (innate immune) cells in lymphoid tissue with concomitant innate immune activation to promote antigen presentation to T cells and to shape the amplitude and nature of the immune response. Nanoparticles that co-deliver both peptide antigen and molecular adjuvants are well suited for this task. However, in the context of peptide-based antigen, an unmet need exists for a generic strategy that allows for co-encapsulation of peptide and molecular adjuvants due to the stark variation in physicochemical properties based on the amino acid sequence of the peptide. These properties also strongly differ from those of many molecular adjuvants. Here we devise a lipid nanoparticle (LNP) platform that addresses these issues. Key in our concept is poly-L-glutamic acid (PGA) which serves as a hydrophilic backbone for conjugation of respectively peptide antigen (Ag) and an imidazoquinoline (IMDQ) TLR7/8 agonist as a molecular adjuvant. Making use of the PGA’s polyanionic nature, we condensate PGA-Ag and PGA-IMDQ into LNP by electrostatic interaction with an ionizable lipid. We show in vitro and in vivo in mouse models that LNP encapsulation favors uptake by innate immune cells in lymphoid tissue and promotes to the induction of Ag-specific T cells responses both after subcutaneous and intravenous administration.
Keywords: Vaccine, lipid nanoparticles, peptides, TLR agonists
Introduction
Harnessing the immune system to recognize and kill cancer cells is a highly attractive therapeutic avenue which relies on the generation of CD8+ T cells that can recognize by their T cell receptor over-expressed (in case of self-) or uniquely expressed (in case of neo-) antigens on the cancer cell surface.1 Moreover, this approach is also of relevance for the design of vaccines against infectious diseases for which the immune response involves an important contribution by T cells. Active immunization requires the delivery of subunits of these antigens in combination with potent immune-stimulatory molecules (called molecular adjuvants) that shape the type and magnitude of the immune response.2 In the context of cancer vaccines, subunit antigens are typically peptides3 comprising the minimal amino acid sequence that can be loaded onto MHC molecules, often extended with additional amino acids to allow for conjugation, formulation, to avoid binding to randomly encountered non-immune cells or to alter the pharmacokinetic profile.
Several reports have highlighted the potential synergism between co-delivery of antigen (Ag) and molecular adjuvants to the same antigen presenting cell.4-7,8 An important challenge however relates to the stark different in physicochemical properties between such molecular adjuvants and peptide antigens and amongst peptides themselves, depending on their amino acid sequence. Hence, a need exists for generic strategies that allow for encapsulation of such compounds that are tolerant to a wide range of physicochemical properties.
Here we report on a lipid nanoparticle (LNP) platform (Figure 1) that meets the above requirements. Our choice for LNP is fostered by findings in the RNA drug field,9,10 i.e. (1 ) LNP have high potential to clinical translation/demonstrated manufacturability/scalability as demonstrated by the approval of the Onpattro siRNA drug product,11 (2) LNP have high capacity to deliver macromolecules to the cytosol; based on presence of membrane disruptive ionizable lipids and phospholipids and (3) LNP have high capacity to deliver cargo to antigen presenting cells in lymphoid tissues, as has been shown in the context of mRNA vaccines. Key in our approach is the use of the polypeptide poly-L-glutamic acid (PGA) as a scaffold for covalent attachment of peptide Ag. PGA is a hydrophilic bio-inspired synthetic polypeptide polymer12 that, owing to its abundancy of carboxylic acid groups, is well suited for the design of water-soluble small molecule and peptide-drug conjugates.13 We reasoned that the polyanionic nature of the PGA backbone could, in conjunction with cationic (ionizable) lipids, also mediate condensation into lipid nanoparticles (LNP) through electrostatic interaction in analogy to the LNP concepts developed by the RNA drug field. Such LNP would allow for co-delivery of Ag and molecular adjuvants and, when properly designed, mediate lymphatic transportation, uptake by antigen presenting cells and potentiate delivery of their payload into the cellular cytoplasm and thereby improve cross-presentation of antigen to CD8+ T cells. Furthermore, harboring electrostatic and hydrophobic binding pockets, lipid-polyglutamate LNP allows for à la carte encapsulation of a variety of TLR agonists2 with different physicochemical characteristics as molecular adjuvants. Our lab has a particular interest in small molecule imidazoquinoline TLR7/8 agonists,14-16,17 owing to their potency to activate a broad spectrum of antigen-presenting cells in mice and humans through triggering of TLR7 and TLR8 and induce robust secretion of type I interferon and IL-12 cytokines which drive anti-viral and anti-tumoral T cell immunity. Noteworthy is that imidazoquinolines are in need for a delivery agent that focuses their immune-modulatory activity to lymphoid tissue and avoids rapid systemic dissemination which causes unwanted, so called ‘wasted’ systemic inflammation.14
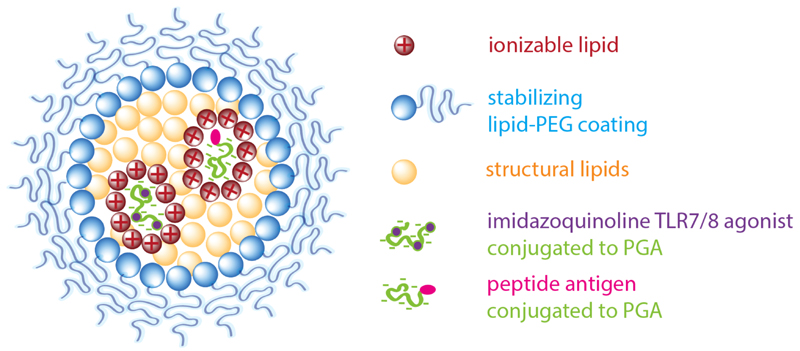
Figure 1. Design of lipid-polyglutamate vaccine nanoparticles.
Electrostatic interaction between carboxylic acid moieties of poly-L-glutamic acid (PGA) and cationic moieties of an ionizable lipid drive nanoparticle formation aided by hydrophobic interaction. A PEG-lipid provides colloidal stabilization. Antigen (Ag) and an imidazoquinoline TLR7/8 agonist (IMDQ) are separately conjugated to PGA.
Materials and methods
Materials
Poly-Glutamic Acid (PGA, 150 units) was kindly provided by Polypeptide Therapeutic Solution (Valencia, Spain). Oregon green 488 was supplied by Thermo Fisher Scientific Inc. (Madrid, Spain). Sulfo-Cyanine-5 amine was purchased from Lumiprobe (Hannover, Germany). All chemicals were reagent grade, were purchased from Sigma Aldrich (St. Louis, MO, USA) and used without further purification, unless otherwise indicated. Deuterium oxide was purchased from Deutero GmbH. Dialysis was performed in a Millipore ultrafiltration device fitted with a selected molecular weight cut-off (MWCO) regenerated cellulose membrane (Vivaspin®).
PGA synthesis and conjugation
Synthesis of PD-PGA
Synthesis of PGA-PD was performed as previously described. Briefly, PGA (acid form, 100 mg, 0.775 mmol, Glutamic Acid repeated Unit GAU, 1 eq.) was dissolved in in 5 mL of N,N’-Dimethylformamide anhydrous and DMTMM BF4 (5.08 mg, 0.0015 mmol, 0.02 eq) was added to the solution and stirred for 30 min. Pyridyldithiolethyl amine (PD, 1.44 mg, 0.075 mmol, 0.01 eq) was added and the pH of the solution was increased to 8 by adding N,N-Diisopropylethylamine. The solution was stirred for 48 hours under inert atmosphere. The product was purified by precipitation in cold ether (3 x 50 mL) and dried under vacuum overnight. Following the time, the PGA was converted into salt form by adding 5 mL of 0.5 M NaHCO3 solution and further purified using Vivaspin® MWCO 3 kDa. The final product was lyophilized and an amorphous white solid was obtained. The product was analyzed by1HNMR and UV-Vis analysis.
Yield: 90% w/w; Conjugation efficiency: 85%.
1H NMR δH (300MHz, D2O):8.60-8.37 (2H, b), 7.92-7.83 (1H, b), 7.50-7.40 (2H, b), 4.52-4.25 (1 H, b), 3.27-3.05 (4H, m), 2.60-2.30 (2H, b), 2.21-1.90 (2H, b); 1.15-1.05 (3H, t).
Synthesis of PGA-Ag
PGA-PD (salt form, 50 mg, 0.330 mmol, 1 eq.) was dissolved in 5 mL of Phosphate Buffered Saline (PBS, 10 mM phosphate, 150 mM NaCl) and CSSSIINFEKL peptide (4.92 mg, 0.0039 mmol, 0.012 eq.) was added. The mixture was stirred for 14 hours and purified by Vivaspin® MWCO 3 kDa. Then, the solution was lyophilized and an amorphous white solid was obtained. The final conjugate was characterized by1HNMR, UV-Vis and SEC-MALS analysis.
Yield: 80-90% w/w. Conjugation efficiency: 90%.
1H NMR δH (300MHz, D2O): 8.49-8.38 (2H, m), 7.53-7.16 (3H, m), 4.53-4.26 (1H, b), 4.04-3.87 (6H, m), 3.67-3.47 (4H, m), 3.11-2.88 (6H, m),2.43-2.18 (2H, m), 2.15-1.72 (2H, m), 1.61-1.41 (4H, m), 1.38-1.02 (7H, m), 1.02-0.82 (12H, m),0.79-0.69 (3H, m).
Synthesis of PGA-IMDQ
PGA (acid form, 100 mg, 0.775 mmol,1 eq.) was dissolved in 5 mL of N,N’-Dimethylformamide anhydrous. The carboxylic groups of PGA were activated by adding DMTMM BF4 (16.30 mg, 0.04967 mmol, 0.075 eq) as previously reported19 and the mixture was stirred for 30 min. Following the time, IMDQ (13.06 mg, 0.03311 mmol, 0.05 eq) was added and the pH of the solution was increased to 8 using N,N-Diisopropylethylamine. The solution was stirred for 24 hours under inert atmosphere. The product was purified by precipitation in cold ether (3 x 50 mL) and dried under vacuum overnight. The PGA was converted into salt form by adding 5 mLof 0.5 M NaHCO3 solution and further purified using Vivaspin® MWCO 3 kDa. The final product was lyophilized and an amorphous white solid was obtained. The product was quantified by1H NMR, UV-Vis and SEC-MALS analysis.
Yield: 90% w/w. Conjugation Efficiency: 80%.
1HNMR δH (300MHz, D2O): 8.02-6.87 (7H, m), 6.02-5.93 (2H, s), 4.41-4.15 (1H, b), 3.12-3.02 (2H, b), 2.58-2.16 (2H, m), 2.12-1.84 (2H, m), 1.44-1.05 (4H, m), 0.93-0.71 (3H, m).
Oregon green 488 - PGA (OG-PGA)
PGA or PGA-IMDQ (acid form, 50 mg, 0.330 mmol, 1 eq.) was dissolved in 5 mL of N,N’-Dimethylformamide anhydrous and DMTMM BF4 (3.48 mg, 0.0088 mmol, 0.02 eq) was added to the solution and stirred for 30 min. Oregon green 488-cadaverine (OG, 1.80 mg, 0.00363 mmol, 0.011 eq) was added and the pH of the solution was increased to 8 by adding N,N-Diisopropylethylamine. The solution was stirred for 48 hours under inert atmosphere. The product was purified though a LH-20 column in DMF. The solvent was evaporated under vacuum and the polymer was converted into salt form by adding 5 mLof 0.5 M NaHCO3 solution and further purified using Vivaspin® MWCO 3 kDa a. The final product was lyophilized and an amorphous yellowish solid was obtained. The product was analyzed by1HNMR and UV-Vis analysis.
Yield: 90% w/w; Conjugation efficiency > 85%.
1HNMR δH (300MHz, D2O): 8.65-6.67 (7H, b), 4.43-4.11 (1H, b), 2.37-2.06 (2H, m), 2.05-1.74 (2H, m), 1.37-1.05 (12H, m), 0.93-0.71 (3H, m).
Sulfo-Cyanine 5-PGA (Cy5 - PGA)
PGA or PGA-PD (acid form, 50 mg, 0.330 mmol, 1 eq.) was dissolved in 5 mL of Phosphate Buffered Saline (PBS, 10 mM phosphate, 150 mM NaCl) and N-hydroxysuccinimide (NHS, 1.14 mg, 0.03 eq.) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, 1.90, 0.03 eq.) were added and the solution was stirred for 30 min at room temperature. Following the time, the mixture was added of Sulfo-Cy5 amine (Cy5, 2.45 mg, 0.011 eq.) and the reaction was left under magnetic stirring for 48 hours and then purified by Vivaspin® MWCO 3 kDa. A blue amorphous solid was obtained after freeze-drying. The Cy5 loading was assessed by1 HNMR and UV-Vis (λmax=646 nm, ε= 271000 L·mol−1·cm−1) analysis.
Yield: 90% w/w; Conjugation efficiency: 70%.
1HNMR δH (300MHz, D2O): 8.60-6.50 (6H, b), 4.52-4.21 (1H, b), 3.27-3.09 (3H, m), 2.41-2.09 (2H, m), 2.07-1.80 (2H, m), 1.52-1.25 (10H, m), 0.93-0.71 (12H, m), 0.98-0.87 (3H, m).
Synthesis of DIPADS
The synthesis of DIPADS was performed in 2 steps. First, a round-bottom flask was filled with 1,1’-carbonyldiimidazole (CDI) (2.5 g, 15.2 mmol) and 10 mL anhydrous dichloromethane under a nitrogen atmosphere. Subsequently, (diisopropylamino)ethanol (1 g, 6.89 mmol) was added after which a clear solution was obtained. After 2 h stirring at room temperature, the reaction mixture was transferred to a separatory funnel and washed with water. The dichloromethane layer was dried over MgSO4 and filtered. Finally, the solvent was removed under reduced pressure, yielding the activated monomer as yellow oil (yield: quantitively).1H-NMR (CDCl3, 400 MHz), δ (ppm): 8.09 (s, 1H, imi-H), 7.39 (s, 1H, imi-H), 7.00 (s, 1H, imi-H), 4.31 - 4.22 (m, 2H, O-CH2-CH2), 3.06 - 2.89 (m, 2H, N-CH-(CH3)2), 2.78 - 2.68 (m, 2H, O-CH2-CH2), 0.95 (s, 12H, N-CH-(CH3)2).
In the second step, the activated (diisopropylamino)ethanol (0.500 g, 2.09 mmol) was dissolved in 2 mL of chloroform in a round bottom flask. To this vigorous stirring solution, dioctadecylamine (1.2 g, 2.51 mmol) dissolved in 20 mL of chloroform, was added dropwise and the reaction was stirred overnight at 50°C under a nitrogen atmosphere. Afterwards the solvent was evaporated under reduced pressure and the residual reaction mixture could be purified using silicagel column chromatography (eluent hexane/EtOAc - 8/2 (v/v)) (yield: 30%).1H-NMR (CDCl3, 300 MHz), δ (ppm): 3.95 (t, J=7.2, 2H, CH2-CH2-O), 3.14 (br s, 2H, CH2-(CH2)16-CH3), 3.05 - 2.89 (m, 2H, N-CH-(CH3)2), 2.61 (t, J=7.2, 2H, CH2-CH2-O), 1.59 - 1.40 (m, 4H, CH2-CH2-(CH2)15-CH3), 1.23 (s, 60H, CH2-CH2,(CH2)15-CH3), 0.98 (d, J=6.4, 12H, N-CH-(CH3)2), 0.88 - 0.81 (m, 6H, CH3).
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR spectra were recorded at 27°C (300K) on a 300 Ultrashield™ from Bruker (Billerica MA, USA). Data were processed with the software Mestrenova (Bruker GmbH, Karlsruhe, Germany). Samples were prepared at the desired concentration in D2O.
UV-Vis
UV-VIS analysis were performed using JASCO V-630 spectrophotometer at 25°C with a 1.0 cm quartz cells and with spectral bandwidth of 0.5 nm. Spectra analysis were recorded in the range of 200 - 700 nm.
Size Exclusion Chromatography
PGA, PGA-Ag and PGA-IMDQ conjugates were measured in an AF2000 system from Postnova Analytics (Landsberg, Germany). The system was configured to work on SEC mode with an isocratic pump (PN1130), an autosampler (PN5300) and refractive index (R.I., PN3150)/UV-visible (PN3211) detectors. A working flow rate of 0.7 mL/min at 30 °C was employed with a TSKgel G3000PWXL column.
LNP formulation
Aqueous solutions of PGA were made by adding 100 uL of a PGA stock solution (1 mg/mL) to 1.233 mL 5 mM citrate buffer (pH 4). Ethanol solutions (0.667 mL) consisted of DIPADS, DOPE, Cholesterol, DSG-PEG with a molar ratio of 50/49:10:38.5:1.5/2.5. LNP were fabricated by solvent displacement, mixing rapidly with a vortex the ethanolic solution of lipids to the aqueous solution of PGA. To remove ethanol, the formed LNP suspensions were centrifuged thrice by using an Amicon® Ultra 10K Centrifugal Filter and 5 mL of deionized water was added each time.
LNP analysis
Dynamic light scattering (DLS) and electrophoretic mobility measurements
Dynamic light scattering (DLS) and electrophoretic mobility measurements were performed on a Zetasizer Nano ZS (Malvern Instruments Ltd., Malvern, U.K.) equipped with a HeNe laser (λ. = 633 nm) and detection at scattering angle of 173°. Cumulants analysis of the data gave the z-average and polydispersity index and data fitting by CONTIN the particle size distribution. Zeta-potential values were calculated from the electrophoretic mobility based on the Smoluchowski equation.
Asymmetric field flow fractionation
Asymmetric field flow fractionation (AFFF) was measured using a Shimadzu Prominence UFLC connected to a Wyatt Eclipse AFFF equipped with a Frit-inlet Channel and connected to a Wyatt Dawn Helios II multiangle light scattering (MALS) detector and fluorescence detector (Agilent Technologies, 1260 Infinity). To measure the particle size of OG-PGA LNP and encapsulation efficiency of OG-PGA, freshly prepared LNP were loaded in an Amicon Centrifugal filter unit with a 100 kDa regenerated cellulose membrane (Millipore) and the particles were centrifuged and washed with water at 2500 g to remove the ethanol from the nanoparticle solution. From the final nanoparticle solution with a volume of 300 μL, a 10x dilution was prepared in PBS. Thereof, 5 μL was injected in the AFFF channel (350 μm W spacer, 10 kDa PES membrane). To measure OG-PGA not encapsulated in the particles, the filtrate of the spinfilter was freeze dried and redissolved in 400 μL of PBS. An amount of 350 μL was injected in the AFFF channel. To quantify the free OG-PGA content in the LNP formulation, a calibration curve was prepared by injecting 10 μL, 5 μL, 2 μL and 1 μL of a 0.1 mg/mL OG-PGA stock solution in the AFFF channel. The following method was used: a cross flow of 2 mL/min for 5 min, an linear crossflow gradient from 2 to 0.3 mL/min over 10 min, a second gradient from 0.3 mL/min to 0.1 mL/min over 10 min, a cross flow of 0.1 mL/min for 10 min, a cross flow of 0 mL/min for 10 min and then a cross flow of 2 mL/min for 2 min using a detector flow of 1 mL/min and inject flow of 0.2 mL/min. A running buffer of 4.25 mM PBS was used. The particle size was determined based on the MALS measurement using the Astra 7.3.1 software and applying the Berry model to fit the data. The Sphere model was used to calculate the particle concentration. Analysis were performed in triplicate.
Fluorescence correlation spectroscopy (FSC)
Fluorescence correlation spectroscopy (FCS) experiments were performed on a commercial setup LSM 880 (Carl Zeiss), with a Zeiss C-Apochromat 40×/1.2W water immersion objective. The excitation of the OG and Cy5 labeled species was done by Argon (488 nm) and HeNe (633 nm) lasers. The respective fluorescent emissions were detected in the ranges 500 - 550 nm and 640-700 nm using a Quasar spectral detection unit (Carl Zeiss) operating in single-photon counting mode. Eight-well polystyrene-chambered coverglass (Nunc™ Lab-Tek™, Thermo Fisher Scientific) was used as a sample cell. For each solution, a series of 10 measurements with a total duration of 5 min were performed at room temperature (23°C). A calibration of the size of the confocal volumes was performed using a reference standards with known diffusion coefficients, namely Alexa Fluor 488 and Alexa Fluor 647.
In vitro experiments
MTT cytotoxicity assay
DC 2.4 cells were plated seeded in 96 well plates at a density of 8 000 cells per well in 200 μL culture medium. 50 μL of sample (dilution series in PBS), PBS (negative control, 100 % viability) and DMSO (positive control, 0 % viability) were added to the wells. After 24 h, the medium was aspirated and cells were washed with 200 μL PBS followed by addition of 100 μL of diluted MTT stock solution. After 1 h, the solution was removed and the formed formazan crystals were dissolved in 50 μL DMSO. Quantification was done by measuring the absorbance at 590 nm using a microplate reader. Note, thiazolyl blue tetrazolium bromide (MTT, 50 mg) was dissolved in 10 mL sterile PBS, filtrated (membrane 0.22 μm) and 1/5 diluted in culture medium prior to use in this assay.
Flow cytometry analysis
DC 2.4 cells were seeded out in 24 well plate at a concentration of 200 000 cells per well in 450 μL of culture medium. Cells were pulsed with samples and incubated overnight at 37 °C. Afterwards, the supernatants were removed, cells were washed with PBS and detached with cell dissociation buffer (0.5 mL, 15 min, 37 °C). The content of the wells was transferred to an Eppendorf and centrifuged (5 min, 300 G, 4 °C). After aspiration of the supernatant, the cell pellets was resuspended in PBS and analyzed using a BD Accuri Flow Cytometer. Data were processed using the FlowJo software package.
Confocal microscopy
DC 2.4 cells were seeded in Willco-Dish glass bottom at a concentration of 10 000 cells in 180 μL culture medium and allowed to adhere overnight. Cells were pulsed overnight with 10 μL of a 1 mg/mL LNP dispersion in PBS. Next, the culture medium was aspirated and cells zere fixated zith 4% PFA for 30 min followed by washing with PBS and confocal imaging using a Leica DMI6000B microscope (63x 1.40 NA objective) coupled to an AndorDSD2 confocal scanner and a Zyla5.5 CMOS camera. Image processing was done using the ImageJ software package.
RAW blue innate immune activation assay
RAW blue 264.7 macrophages were seeded in flat-bottom 96 well plate at a density of 70 000 cells per well, suspended in 180 μL culture medium and pulsed with 20 μL of sample for 24h at 37 °C at a concentration of respectively 0.8, 0.2, 0.08, 0.02, 0.08, 0.02, 0.008, 0.002, 0.0008, 0.0002, 0.00008 μg/mL. Subsequently, 50 μL of supernatant was transfer to a new flat-bottom 96 well plate followed by addition of 150 μL of QUANTI-Blue™ reagent solution, prepared according to the manufacturer’s instruction (Invivogen). After 30 minutes at 37 °C, the SEAP levels were determined by UV-Vis spectrophotometry at 620 nm using a microplate reader. Note, the colorimetric quantification of the samples was obtained relative to the negative control and each concentration was performed in fivefold.
OVA-Kb antigen presentation assay
DC 2.4 cells were seeded out in 24 well plate at a concentration of 200 000 cells per well in 450 μL of culture medium. Cells were pulsed with samples and incubated for 48 h at 37 °C. Afterwards, cells were washed and stained with 25D1.16-PE (Ebioscience) followed by flow cytometry analysis on a BD Accuri flow cytometer. Data were processed using teh FlowJo spoftware package.
In vivo experiments
IFNb luciferase reporter bioluminescence imaging
Luciferase reporter mice (IFNβ+/Δβ-luc) with a BALB/c background, aged 7-9 weeks, were housed in individual ventilated cages and given ad libitum access to food and water. 20 μL of LNP(PGA-IMDQ) or PGA-IMDQ (2 μg equivqlent IMDQ dose) were injected subcutaneously in the footpad. For in vivo imaging at the given time points, mice were injected subcutaneously with 200 μL D-luciferin and in vivo luminescence imaging was recorded 12 min later using the IVIS Lumina II imaging system.
Analysis of in vivo lymphatic drainage
20 μL (1 mg/mL in PBS, containing an equivqlent Cy5 dose of 2μg) of LNP(Cy5-PGA)or Cy5-PGA was injected into the footpad of female C57BL/6 WT mice. At the designated time point, mice where sacrificed and popliteal lymph nodes where isolated and frozen in OCT(Sakura, 4583). Frozen sections (8-μm) were cut using cryostat. These sections where fixed for 4 min in PFA 2%, washed with PBS. Images were acquired on a Zeiss LSM710 confocal microscope equipped with 488-nm, 561-nm and 633-nm lasers and with a tunable two-photon laser. Image processing was done using the ImageJ software package.
Analysis of in vivo lymphocyte targeting and activation
20 μL (1 mg/mL in PBS, containing an equivqlent Cy5 or IMDQ dose of 2μg) of LNP(Cy5-PGA), Cy5-PGA or LNP(PGA-IMDQ) was injected into the footpad of female C57BL/6 WT mice. At the designated time point, mice where sacrificed and popliteal lymph nodes where isolated. A single cell suspensions was prepared form the dissected popliteal lymph nodes for analysis by flow cytometery. Isolated lymph nodes were collected in ice cold PBS, smashed through 70 μm cell strainers, washed with PBS and stained for 30min at 4°C with following primary labeled antobibodies: CD3, CD20, CD11c, MHCII, CD86, CD40. Live dead ratio’s where determined by staining with fixable dead/live- staining and 123count ebeads were added to determine cellularity prior to acquiring them on 123count ebeads were added to determine cellularity prior to Analysis by a BD FACS Quanto flow cytometer. Data were processed using the FlowJo software package.
Immunization and tetramer staining
6 female C57BL/6 WT mice per group were injected 1 00μl either SC at the tail base with AG, PGA-Ag, LNP(PGA-AF), Ag+IMDQ,LNP(PGA-AG + PGA-IMDQ) or 100μl IV in the tail vain with (concentrations?)AG+IMDQ or LNP(PGA-AG + PGA-IMDQ). The dose as set at equivalent doses of 10 ug of peptide antigen and 10 ug of IMDQ Priming of the mice happened at day 0 and boosting at day 14 and day 28. Mice were bled at day 5, 19 and 33 to obtain 100μl of blood for flow cytometry staining. Antibodys used for flow cytometry are as followed: CD45(30-F11), CD3(145-2C11), CD4(RM4-5) by eBioscience. CD8(53-6,7) by BD biosciences. Fixable viability dye Live/Dead Efluor 506 from Invitrogen and MHC tetramer H-2Kb OVA SINFEKL by MBL. 123count ebeads (eBioscience) were added to 100μl of blood prior to staining to determine cellularity. Acquisition of the samples was done on a BD Fortessa flow cytometer. Data were processed using the FlowJo software package.
Results and Discusison
Polyglutamate conjugation of peptide Ag and TLR7/8 agonist
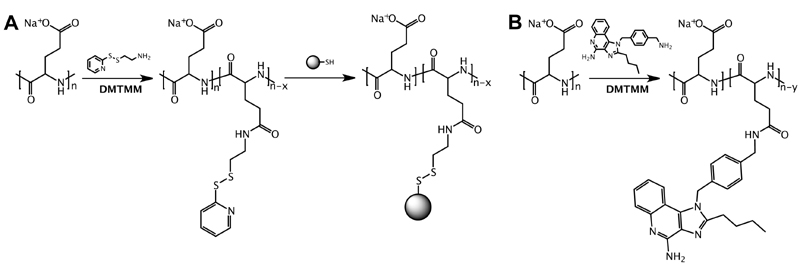
PGA with a molecular weight of 22.6 kDa (corresponding to 150 glutamate repeating units on average) was used as a platform for Ag and imidazoquinoline conjugation. Our strategy for conjugation of respectively Ag and imidazoquinoline TLR7/8 agonist to PGA is depicted in Scheme 1A. For conjugation of peptide Ag, first a piridyldisulfide moiety was introduced onto the PGA backbone by reacting carboxylic acid moieties of PGA with pyridyldithiolethyl amine mediated by 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium tetrafluoroborate (DMTMM BF4) as amidation reagent, yielding a 1% mol/mol PGA- pyridyldithiolethyl amide (PGA-PD, Figure S1), corresponding to an average of 1.5 PD moiety per chain. We selected this linking chemistry due to the need of the antigen peptide to be released in the intracellular milieu.20 As a model peptide Ag, we opted in this study for the CD8 epitope of ovalbumin (OVA). For conjugation purpose, the minimal epitope sequence SIINFEKL was extended at the C-terminus with two serines and a cysteine, resulting in CSSSIINFEKL.21 Subsequently, this peptide was conjugated to the PGA-PD by disulfide bond formation targeting one peptide per PGA chain. The conjugate was synthesized with high conjugation yield and characterized by1HNMR, showing an average of one peptide per chain (Figure S2A). The UV-Vis analysis showed the pyridyl moiety displacement and the effective conjugation of the antigen. Peptide antigen-conjugated PGA will further on be denoted as PGA-Ag (Figure S2B).
Scheme 1. Poly-L-glutamic acid (PGA) conjugation.
(A) For conjugation of peptide antigen (Ag), PGA is first functionalized with a pyridyldisulfide moiety followed by conjugation of the Ag to a terminal cysteine residue via a disulfide bond. (B) For conjugation of the imidazoquinoline TLR7/8 agonist IMDQ, aliphatic amine of IMDQ is conjugated to a carboxylic acid moiety of PGA through an amide bond.
With regard to the molecular adjuvant, we chose the imidazoquinoline TLR7/8 agonist 1-(4-(aminomethyl)benzyl)-2-butyl-1Himidazo[4,5-c]quinolin-4-amine (IMDQ)22 which combines high potency with the availability of a primary aliphatic amine. The latter was used for conjugation to carboxylic acid moieties of PGA using 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium tetrafluoroborate (DMTMM) and leading to an IMDQ content of 10 wt% (Scheme 1B). The final conjugate was characterized with1HNMR and UV-Vis analysis showing a good yield and the chemical identity of the desired product (Figure S3). The IMDQ-conjugated PGA will further on be denoted as PGA-IMDQ. The MW of the conjugates was calculated by Size Exclusion Chromatography- Multiangle light scattering (SEC-MALS) and is in fair agreement with the theoretical MW, showing an increment of MW with the conjugation of the antigen or IMDQ. The zeta potential of the conjugates was measured, and the PGA derivatization do not affect the negative charge of the polymers, permitting the complexation into the SLN (Table S1).
It could be argued that being a small molecule, IMDQ could also be conjugation to a lipid for subsequent encapsulation into LNP. And indeed, lipid-conjugation has been reported by others to alter the pharmacokinetic behavior of imidazoquinolines and promote loading into liposomes.23 However, direct conjugation to lipids often results in a dramatic loss in TLR-agonistic activity. In previous work we have shown that conjugation of IMDQ to nanogels and lipid-polymer amphiphiles, which are both much more hydrated, is a more viable approach to combine a favorable pharmacokinetic profile, i.e. delivery to lymph nodes, with only a moderate loss in activity. Hence, we reasoned in the context of the present work that conjugation of IMDQ to PGA could be more advantageous.
Lipid-polyglutamate nanoparticle fabrication
LNP were fabricated by rapid mixing under vigorous stirring of an aqueous solution containing PGA and an ethanolic solution containing an ionizable lipid in addition to cholesterol and dioleoylphosphatidylethanolamine (DOPE) as structural helper lipids to aid in nanoparticle formation24 and PGA encapsulation, and possibly improve cytosolic delivery. Also, distearoyl-rac-glycerol-poly(ethylene gylcol) (DSG-PEG; PEG length of 2 kDa) was added to the ethanolic solution to provide sterical stabilization against aggregation of the resulting LNP. A screening experiment was conducted, aiming at elucidating formulation parameters that yielded colloidally stable LNP suspension with a mean nanoparticle diameter below 200 nm and a zeta-potential close to neutral or negative (Figure 1 ). The choice of the latter two parameters was fostered by earlier work by our lab and others with regard to optimizing nanoparticle properties in view of maximizing lymphatic translocation upon subcutaneous administration.14,25
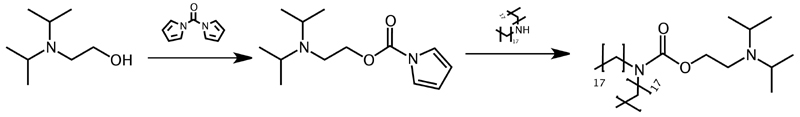
In the context of the present work, a novel ionizable lipid was synthesized according to Scheme 2, by conjugating 2-(diisopropylamino)ethanol to distearylamine through a carbamate linkage which we further abbreviate as DIPADS. Diisopropylamino moieties have a pKa below 6.5 and hence have a low degree of ionization at physiological pH but become strongly ionized at the acidic pH that is sensed by nanoparticles upon endocytosis into endosomal vesicles. In the nanomedicine field, diisopropylamino moieties have extensively been reported to promote endosomal escape through a combination of effects.26-28 Recently, we also observed that the hydrophobicity of the diisopropyl group has a potent membrane destabilizing activity.29
Scheme 2. Synthesis of the ionizable lipid DIPADS through carbamate conjugation of distearylamine to 2-(diisopropylamino)ethanol.
To formulate the PGA LNPs, a molar ratio of ionizable lipid (N) to glutamic acid repeating units (COOH) of respectively 10:1 and 5:1 were chosen. Note that this corresponds to the well-known N/P (P: molar fraction of negatively charged phosphates) ratio from the RNA drug field.9 An initial molar ratio of 50:10: 38.5: 1.5 between ionizable lipid: DOPE : cholesterol: DSG-PEG was selected, identical to the ratio commonly used for LNP mediated delivery of siRNA and mRNA.
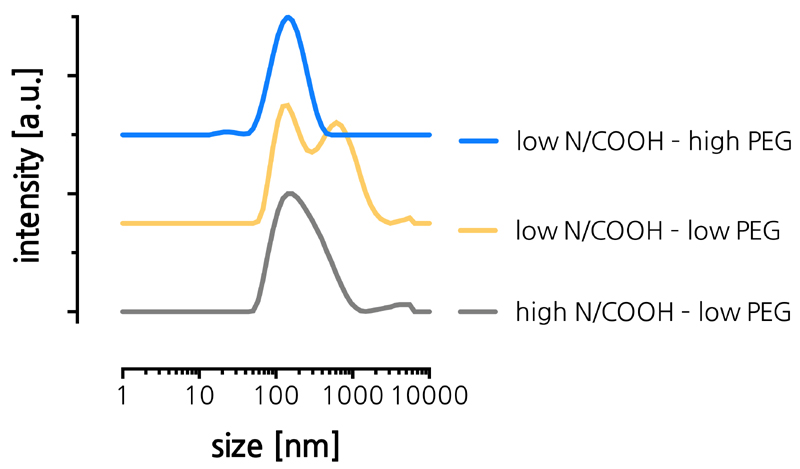
PGA and lipids were mixed at a 2:1 ratio of aqueous buffer and ethanolic solution. The pH of the aqueous solution was set at 4 (5 mM citrate buffer) to ensure a high degree of ionization of DIPADS to drive electrostatic interaction with the PGA. Immediately after mixing of the aqueous and ethanolic solutions, the formed suspensions were purified by spin filtration to remove ethanol. Table 1 summarizes the composition of the respective batches and their characteristics after spin filtration, including particle size and dispersity as measured by dynamic light scattering (DLS) and the zeta-potential calculated from measuring the electrophoretic mobility (in HEPES buffer, pH of 7.4). Figure 2A shows the corresponding DLS-based size distributions of the LNP. From these data it could be concluded that at a DSG-PEG content of 1.5%, both for a N/COOH ratio of 10:1 and 5:1, no stable LNP were formed. To improve on the stability of the LNPs, we decided to augment the mol% of DSG-PEG to 2.5%. This increase in PEG-lipid content now enabled stable LNP formation, yielding LNP with slightly negative zeta-potential and a sub 100 nm diameter. We attribute the need for a higher degree of PEGylation to the low degree of ionization of DIPADs which we assume is responsible for lower charge repulsion between the LNP and a larger extent of hydrophobic domains on the LNP surface which could promote nanoparticle aggregation. These findings meet the initial expectation and give a first proof that PGA can become encapsulated by LNP using solvent displacement from ethanol into water as fabrication method for LNP.
Table 1. Composition and properties of LNP measured by dynamic light scattering (DLS) and electrophoretic mobility analysis.
| high N/COOH - low PEG | low N/COOH - low PEG | low N/COOH - high PEG | |
|---|---|---|---|
| N/COOH ratio | 10:1 | 5:1 | 5:1 |
| DIPADS (mol%) | 50 | 50 | 49 |
| DOPE (mol%) | 10 | 10 | 10 |
| cholesterol (mol%) | 38.5 | 38.5 | 38.5 |
| DSG-PEG (mol%) | 1.5 | 1.5 | 2.5 |
| size [nm] | / | / | 87 |
| PDI | 0.3 | 0.43 | 0.2 |
| zeta potential [mv] | -3 | -3 | -7 |
Figure 2. (A) Size distribution curves measured by DLS of LNP formulations listed in Table 1.
Physicochemical characterization of LNP
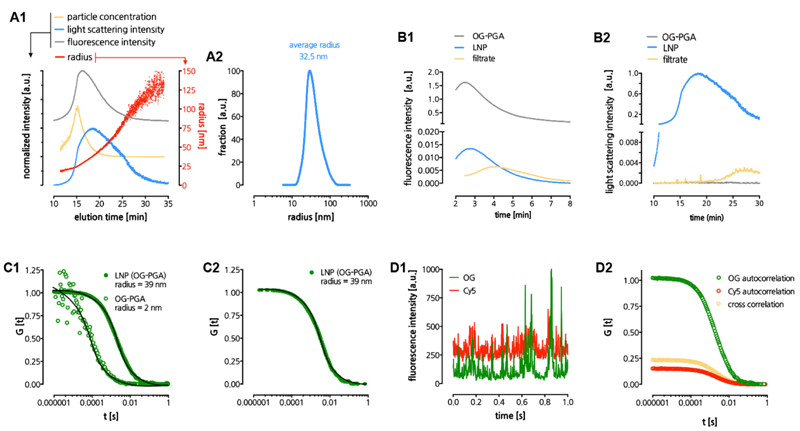
Having selected suitable parameters for lipid-polyglutamate LNP formulation, we performed a more thorough characterization of the LNP using a number of complimentary techniques. Asymmetric field flow fractionation (AFFF) coupled to fluorescence, UV-vis and multi angle light scattering detection was used to analyze the LNP size distribution in greater detail and provide evidence on PGA encapsulation (Figure 3A-B). LNP loaded with Oregon Green (OG) labeled PGA (OG-PGA) (cfr. Experimental sections for details on synthesis) showed an excellent overlay between the fluorescence and light scattering signal (Figure 3A1), hinting at a single homogenous population of OG-PGA-loaded LNP. The number average LNP radius was calculated to be 32.5 nm (Figure 3A2). The corresponding diameter of 63 nm is slightly smaller than the hydrodynamic diameter obtained by DLS. This can be attributed to DLS being much more affected by a low fraction of larger LNP due to the non-linear increase in light scattering intensity with size.
Figure 3. Physicochemical characterization of LNP.
(A) Asymmetric field flow fractionation (AFFF) analysis of LNP. Panel A1 depicts multiple parameters as function of the elution time, panel A2 depicts the calculated size distribution based on the light scattering signal. (B) AFFF analysis used to determine encapsulation of OG-PGA in LNP. Panel B1 depicts the fluorescence signal in the time interval where OG-PGA elutes. Panel B2 depicts the light scattering signal in the time interval where the LNP elute. (C) LNP analysis by fluorescence correlation spectroscopy (FCS). Panel C1 depicts normalized autocorrelation curves (evaluated from measuring fluorescence fluctuations in the OG(-PGA) signal) and their fits (solid lines) that yielded the radii of PGA and LNP in aqueous medium. Panel C2 depicts normalized autocorrelation curves (calculated from measuring fluorescence fluctuations in the OG(-PGA) signal) and the corresponding fit (solid line) yielding the radius of LNP in human blood plasma. Panel C3 depicts time fluorescence fluctuations (time traces) of the OG(-PGA) and Cy5(-DOPE) signals. Panel C4 depicts auto- and cross-correlation curves the OG(-PGA) and Cy5(-DOPE) signals from LNP in aqueous medium.
To assess encapsulation of OG-PGA in LNP, we used AFFF to measure freely soluble OG-PGA in the LNP formulation and in the filtrate, obtained after the spin filtration step during purification of the LNP. Based on the initial amount of OG-PGA that was used for LNP formulation and a calibration curve of OG-PGA, an encapsulation efficiency of 91+1 % was calculated (Figure 3B1). Furthermore, by analyzing the light scattering signal of the filtrate, no LNP could be detected (Figure 3B2), indicating that the latter were well retained in the designated compartment during the spin filtration purification process.
Additional confirmation regarding encapsulation of PGA in LNP and whether PGA remains encapsulated in presence of serum was sought by fluorescence correlation spectroscopy (FCS).30 Hereto, double fluorescently labeled LNP were prepared containing (fluorescent green) OG-PGA and (fluorescent red) Cy5-DOPE. For FCS, a confocal microscope equipped with sensitive detectors is used to measure fluctuations in fluorescence in the confocal volume of a solution containing the specimen of interest, being LNP in the present case. Based on the autocorrelation of OG with itself as a function of a shift time, a radius of 2 nm for OG-PGA and 39 nm for the LNP was calculated. Off note, this value corresponds well with the value obtained by AFFF analysis with multi angle light scattering detection. When the experiment was repeated with prior incubation of 2 h at 37 °C in 10% fetal bovine serum (FBS), an identical radium of 39 nm was measured, indicating at a good stability of the LNP in a complex, serum rich medium.
Additional confirmation regarding encapsulation of PGA in LNP and whether PGA remains encapsulated in presence of serum was sought by fluorescence correlation spectroscopy (FCS). Hereto, double fluorescently labeled LNP were prepared containing (fluorescent green) OG-PGA and (fluorescent red) Cy5-DOPE. For FCS, a confocal microscope equipped with sensitive detectors was used to study a solution containing the fluorescent specimen of interest, being LNP in the present case. The fluctuations in detected fluorescence intensity, caused by the diffusion of fluorescent species through the confocal volume were recorded and used to calculate experimental autocorrelation curves. A fit of these curves with a model function for freely diffusing species30 yielded the hydrodynamic radii of 2 nm for OG-PGA and 39 nm for the LNP dispersed in aqueous medium (Figure 3 C1). Off note, this value corresponds well with the value obtained by AFFF analysis with multi angle light scattering detection. When the FCS experiment was repeated with LNP incubated for 2 hours in human blood plasma (Figure 3 C2), an identical radius of 39 nm was measured after accounting for plasma viscosity.31 These results indicate a good stability with neither decomposition no aggregation of the LNP in a complex, application relevant, medium.
We also performed dual color fluorescence cross correlation (DC-FCCS) experiments by creating two overlapping confocal volumes (“green” and “red”) and monitoring independently the fluctuations caused by the diffusion of OG-PGA and Cy5-DOPE containing species. The simultaneous detection of both dyes as indicated by the time traces (Figure 3 D1) and the strong cross-correlation with amplitude laying between the amplitudes of the two autocorrelations (Figure 3 D2), confirmed that all LNP successfully carried both OG-PGA and Cy5-DOPE, with absence of freely soluble OG-PGA.
In vitro LNP evaluation
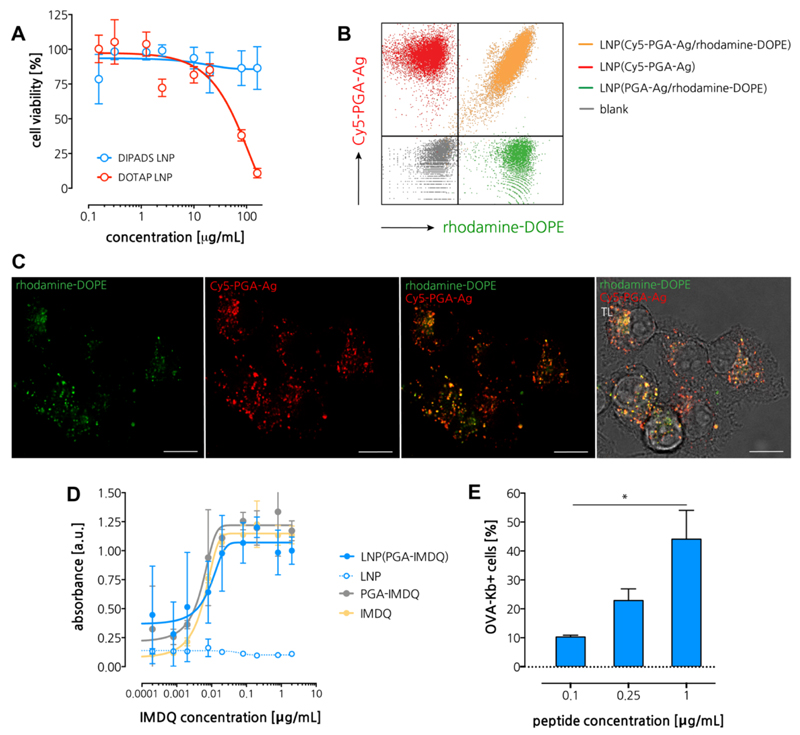
First, we measured the cytotoxicity on an in vitro cell culture. Hereto we made use of DC2.4,32 an immortalized mouse dendritic cells (DC), as model antigen presenting cells (APC). As shown in Figure 4A, no sign of cytotoxicity was detected within the tested experimental window. By means of comparison, we also tested LNP prepared with DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), an often-used cationic lipid for gene transfection. Interestingly, DOTAP-based LNP (size (DLS): 81 nm and zeta-potential: 12 mV) were found to be significantly toxic to DC2.4 cells, which we hypothesize to be due to the strongly cationic nature of DOTAP, which induces extensive destabilization of cellular membranes. These findings further advocate the translational value of ionizable lipids with moderate pKa.
Figure 4. In vitro immuno-biological characterization of LNP.
(A) Cell viability measured by MTT assay. (n=6) (B) Flow cytometry analysis if DC2.4 cells pulsed with single-dye or double-dye labeled LNP. (C) Confocal microscopy of DC2.4 cells pulsed with LNP. Scale bar represents 15 microns. (D) Dose-response innate activation measured by the RAW Blue reporter cell assay (n=3). (E) Flow cytometry analysis of DC2.4 cells pulsed with different concentration of antigen loaded LNP followed by immunostaining with the OVA-Kb antibody (n=3; t-test *: p<0.05).
Next, we assessed uptake of LNP by antigen presenting cells (APC) in vitro. For detection purpose, we encapsulated Cy5-labeled PGA-Ag into LNP and made used of rhodamine-labeled DOPE (rhodamine-DOPE) to obtain double-fluorescent labeled LNP. DCs were pulsed with these LNP followed by flow cytometry analysis and confocal microscopy imaging. As a control, we also tested LNP containing only a single fluorophore (either rhodamine or Cy5). Flow cytometry analysis (Figure 4B) showed that single dye-labeled controls were located in the corresponding quadrant. By contrast, DCs pulsed with LNP containing both Cy5-PGA-Ag and rhodamine-DOPE, were located in the double-positive quadrant, thereby hinting at successful PGA-Ag delivery mediated by LNP. Moreover, these data provide evidence that despite the presence of a PEG-coating and negative zeta-potential, LNP can still to associate with cells. Confocal microscopy (Figure 4C) provided further confirmation of cellular uptake, showing co-localization between rhodamine-DOPE (green fluorescence) and Cy5-PGA-Ag (red fluorescence). These findings allowed us to conclude that our LNP formulations is well suited for encapsulation and intracellular delivery of PGA-Ag. In vitro antigen presentation was assessed by pulsing DCs cells with different concentration of PGA-Ag loaded LNP followed by counterstaining with a PE-labeled monoclonal antibody recognizing MHC class I molecule Kb bound to the peptide SIINFEKL, and flow cytometry analysis (Figure 4D). A clear dose-response binding of the antibody was observed, suggesting that LNP allow for efficient processing and presentation of Ag on the surface of antigen presenting cells.
To engineer the LNP with immune-stimulatory properties, PGA-IMDQ was formulated into LNP similarly as described above for encapsulation of PGA-Ag. To test whether PGA-conjugation and subsequent LNP formulation alters the TLR agonistic activity of IMDQ, we used the in vitro RAW Blue NF‐kB activation reporter cell assay (Figure 4E). The latter engineered cell line expresses a spectrum of TLRs (including TLR7/8a) and produces secreted embryonic alkaline phosphatase (SEAP) in response to TLR-triggered NF‐kB activation, which can be measured by an enzymatic colorimetric assay. Interestingly, contrary to our earlier iterations on IMDQ conjugation, no loss in activity was observed for PGA-IMDQ relative to IMDQ in native soluble state. These findings hint at either intracellular processing of the PGA-IMDQ through proteases thereby liberating small PGA-IMDQ oligomers that witness reduced steric hindrance compared to macromolecular IMDQ-conjugates, or a favorable binding to the TLR7/8 receptor either as a monovalent interaction or through multivalency by multiple IMDQ motifs on a single PGA backbone. This will be subject to further investigation in upcoming work. Interestingly, also encapsulation into LNP did not induce a reduction in TLR agonistic activity, which could potentially be attributed to the efficient cellular uptake of the LNP. Whereas the employed RAW Blue assay is highly sensitive to TLR7 triggering as well as to triggering of various other patter-recognition receptors, it cannot be excluded that PGA and LNP (PGA), although quiescent in this assay, are capable of triggering other innate pathways. This will be subject of future endeavors.
In vivo LNP evaluation
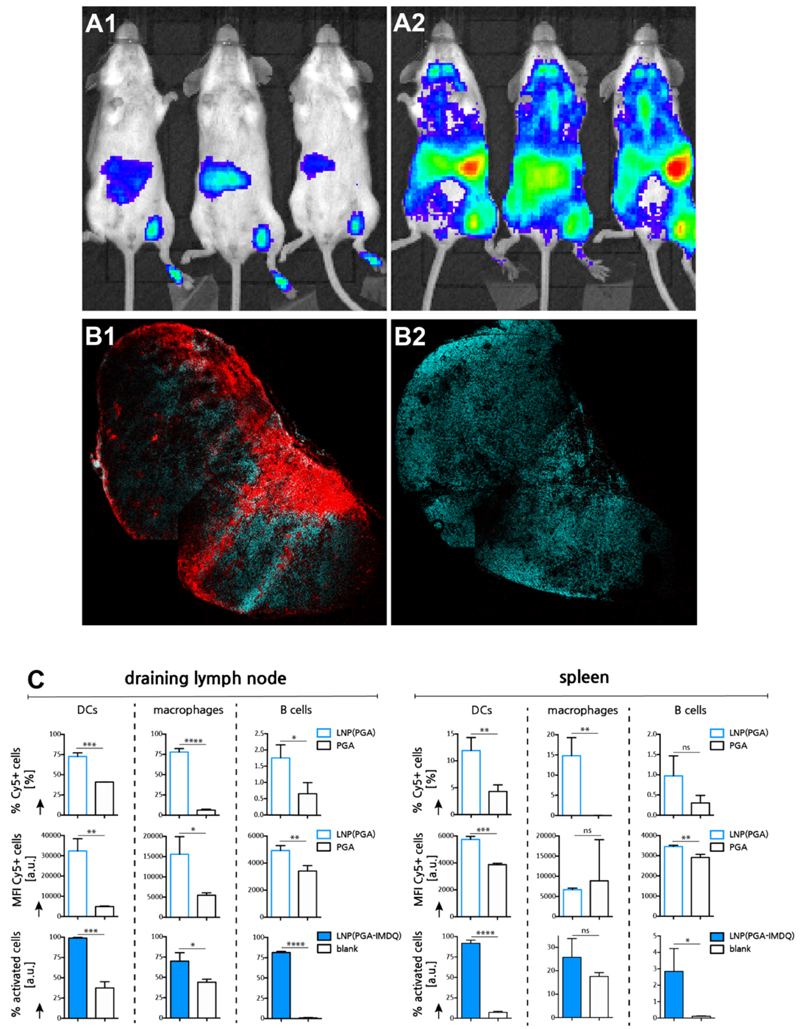
Next, we aimed at investigating the im mu no-biological behavior of the LNPin vivoin mouse models. Type I interferons represent the signature cytokines induced by TLR7 activation. To assess the intensity and biodistribution of TLR7 mediated innate activation, we thereby imaged the type I interferon response using an IFN‐β‐luciferase reporter mouse model, in which a firefly luciferase encoding sequence had been placed under the control of the IFN-β promoter.33 This model enables spatiotemporal analysis of IFN‐β induction, thereby offering semi-quantitative insights in tissue disposition and systemic exposure of IMDQ formulations. Soluble PGA-IMDQ and PGA-IMDQ loaded LNP were injected into the footpad of mice followed by bioluminescence imaging 4 h post injection. As presented in Figure 5A, PGA-IMDQ provokes mild innate immune activation at the site of administration, in the draining popliteal lymph node and in the liver. By contrast, PGA-IMDQ loaded LNP induces potent innate activation in the draining popliteal lymph node and in the spleen and to a smaller extent also in the liver. These findings are unlike any of our previous observation on IMDQ-conjugated materials which only showed translocation to draining lymph nodes but induced no or little activation in distal lymphoid tissue. In particular, the potent innate immune activation in the spleen is of striking interest due to the vast numbers of antigen presenting cells and T cells which makes it a primary organ for inducing adaptive immune responses. Of note, empty LNP, that do not contain IMDQ did not yield a detectable response in this experimental setting (Figure S8 in Supporting Information). Confocal microscopy imaging of the draining lymph node (Figure 5B) of mice injected with respectively Cy5-PGA loaded LNP and soluble Cy5-PGA gave proof that the LNP indeed distributed through lymphoid tissue, and this to a much higher extent than Cy5-PGA.
Figure 5. In vivo lymphatic transportation and innate immune activation by LNP.
(A) Bioluminescence imaging of IFNb-luciferase reporter mouse 4h post injection. (A1: PGA-IMDQ; A2: LNP(PGA-IMDQ)) (B) Confoca microscopy image of a sectioned popliteal lymph node, 24h post injection of LNP in the foot pad. (cyan: DAPI, red:Cy5-PGA loaded LNP). Note that some irregularities in the image are due to stitching. (B1 : LNP(Cy5-PGA); B2: Cy5-PGA) (C) Flow cytometry analysis of LNP uptake by innate immune cells and innate immune cell activation in the draining inguinal lymph node) 24h post injection of LNP in the tail base (n=3; t-test *: p<0.05, **:p<0.01, ***:p<0.001, ****:p<0.0001).
To gain a more detailed understanding of the distribution of the LNP over different immune cell subsets in the draining lymph nodes and spleen, Cy5-PGA loaded LNP were injected subcutaneously in the tail base followed by dissection and flow cytometry analysis of the draining inguinal lymph node and the spleen (Figure 5C). The reason for choosing the tail base was that this will also be the site of administration for later immunization experiments, whereas footpad injection and popliteal lymph node imaging is better suited for rapid screening of lymphatic translocation. At the level of the draining lymph node, vast numbers of DCs and macrophages became positive for LNP. B cells too received LNP, albeit to a lower extent. In all cases, Cy5-PGA loaded LNP resulted in a stronger cellular association compared with an equivalent dose of unformulated soluble Cy5-PGA. At the level of the spleen, a similar trend is witnessed, with large fractions of DCs and macrophages that became positive for LNP, in addition to a smaller fraction of B cells. Moreover, when taking into account the cellular mean fluorescence intensity (MFI) values, it is clear that LNP-mediated delivery not only allows for reaching more cells, but also delivers a higher payload to these cells. Note that the aberrant values of spleen macrophages are due to the insignificant numbers of these cells that received Cy5-PGA. To measure activation of innate immune cell subsets by PGA-IMDQ loaded LNP, the draining lymph node and the spleen were analyzed by flow cytometry for the upregulation of maturation and activation markers (Figure 5C). In the draining lymph node, a very significant increase in mature (CD86+) DCs and macrophages was observed in addition to a vast activation (CD69+) of B cells. In the spleen, a vast increase in maturation of the DC population was observed, in addition to a more moderate maturation/activation, of the macrophage and B cell populations. Taken together, our data suggest that, upon local subcutaneous injection, LNP can enter systemic circulation and predominantly accumulate in the spleen, thereby provoking activation of a broad range of immune cell subsets.
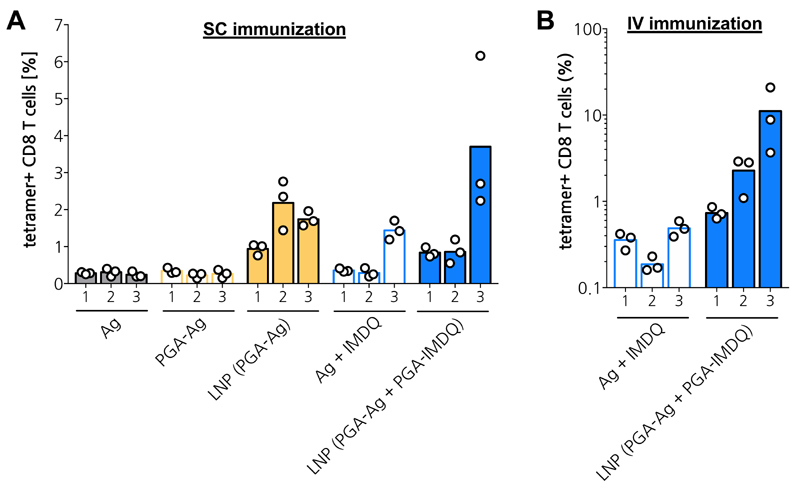
Encouraged by having observed potent innate activation in vivo by LNP, we set out to further investigate the potential of the LNP approach to drive the induction of adaptive T cell mediated immune responses against encapsulated peptide antigen. Hereto, mice were immunized subcutaneously at the tail base following a prime and double boost scheme, with a standard 3-week interval between each immunization. 8 days post immunization, the presence of antigen-specific CD8 T cells in the blood was detected by measuring OVA-tetramer positive CD8 T cells in blood samples (Figure 6A). Unformulated antigen in soluble form and PGA-conjugated antigen (PGA-Ag) yielded detectable although very low responses. LNP-encapsulated antigen provoked considerably higher responses. The exact reason for the latter, being either solely the effect of enhanced delivery of antigen to antigen presenting cells in lymphoid tissue, or a combination of delivery and activation of pathways that were not detected during the in vitro innate immune activation assay, remains elusive and will be subject of future research.
Figure 6. In vivo adaptive immune response.
(A) Subcutaneous immunization in tail base. (n=3) (B) Intravenous immunization in tail vain. (n=3)
Also, soluble antigen, adjuvanted with IMDQ in soluble form, induced a considerable response, but only after the second boost immunization. Importantly, overall, LNP encapsulating both antigen and IMDQ yielded consistently the highest percentages of antigen-specific CD8 T cells after each immunization and the highest percentage after the second boost. To further explore the potential of the LNP vaccine platform, we also investigated the response to intravenous administration (Figure 6B). We found that soluble antigen, adjuvanted with soluble IMDQ, induced very low responses, similar to those measured upon subcutaneous immunization. In stark contrast, LNP encapsulating antigen and IMDQ induced robust fractions of antigen-specific CD8 T cells that consistently increased upon each immunization and mounted up to 10% on average after the second boost.
Conclusions
In conclusion, we reported in this paper on an elegant and efficient route for co-encapsulation of peptide Ag and a molecular adjuvant using a lipid-polyglutamate nanoparticle (LNP) platform that favors lymphatic and splenic delivery upon local subcutaneous administration. LNP encapsulating an imidazoquinoline TLR7/8 agonist (IMDQ) induce potent innate immune activation in lymph nodes and in the spleen. Immunization with LNP encapsulating antigen and IMDQ induced antigen-specific T cell responses in vivo in mice. In addition, we also highlighted the potential of LNP to induce robust responses after intravenous immunization.
Our approach holds the potential to co-encapsulate multiple antigens and well as varying the type of molecular adjuvants. Indeed, harboring electrostatic and hydrophobic binding pockets, lipid-polyglutamate LNP allows for à la carte encapsulation of TLR agonists as molecular adjuvants. Indeed, electrostatic interaction could be used for co-encapsulation of RNA- or DNA- based TLR agonists such as CpG (TLR9 agonist), polyI:C (TLR3 agonist), and ssRNA (TLR7/8 agonist) could be encapsulated through electrostatic interaction. Lipophilic adjuvants such as MPLA (TLR4 agonist) or LTA and PamCySK (TLR2 agonists) could be encapsulated through hydrophobic interaction. Moreover, besides co-delivery of TLR agonist and antigen, TLR agonist loaded LNP could also serve as stand-alone adjuvant as shown by Swaminathan et al. for the case of oligonucleotide TLR9 agonist loaded LNP.34
Furthermore, we aim at screening libraries of ionizable lipids to identify formulations that allow for optimal cytosolic delivery in combination with an acceptable safety profile and explore these for the delivery of clinically relevant tumor antigens. Finally, in view of the high response to intravenous immunization, also this route will receive additional attention, in particular in view of biodistribution and safety aspects. In this context, producing LNP with different, and more discrete, size distributions could also be of great relevance to elucidate, depending on the route of administration, the influence of LNP size on biodistribution and further downstream the resulting immune response.
Supplementary Material
Supporting Information containing additional characterization data is available free of charge.
Acknowledgements
The authors acknowledge Snezana Dordevic for the Size Exclusion Chromatography measurements. This work was supported by the Spanish Ministry of Science and Innovation (SAF2016-80427-R and PID2019-108806RB-I00) and the European Research Council (ERC-CoG-2014-648831 ‘MyNano’ and ERC-PoC-2018-825798 ‘Polymmune’) (MJV). Part of the equipment employed in this work has been funded by Generalitat Valenciana and co-financed with FEDER funds (PO FEDER of Comunitat Valenciana 2014-2020). BGDG acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 817938).
References
- (1).Melief CJM, van Hall T, Arens R, Ossendorp F, van der Burg SH. Therapeutic Cancer Vaccines. J Clin Invest. 2015;125(9):1–12. doi: 10.1172/JCI80009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mancini RJ, Stutts L, Ryu KA, Tom JK, Esser-Kahn AP. Directing the Immune System with Chemical Compounds. ACSChem Biol. 2014;9(5):1075–1085. doi: 10.1021/cb500079s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schumacher TN, Schreiber RD. Neoantigens in Cancer Immunotherapy. Science (80-. ) 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- (4).Irvine DJ, Hanson MC, Rakhra K, Tokatlian T. Synthetic Nanoparticles for Vaccines and Immunotherapy. Chem Rev. 2015;115(19):11109–11146. doi: 10.1021/acs.chemrev.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Moon JJ, Suh H, Bershteyn A, Stephan MT, Liu H, Huang B, Sohail M, Luo S, Ho Um S, Khant H, Goodwin JT, et al. Interbilayer-Crosslinked Multilamellar Vesicles as Synthetic Vaccines for Potent Humoral and Cellular Immune Responses. Nat Mater. 2011;10(3):243–251. doi: 10.1038/nmat2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nembrini C, Stano A, Dane KY, Ballester M, van der Vlies AJ, Marsland BJ, Swartz MA, Hubbell JA. Nanoparticle Conjugation of Antigen Enhances Cytotoxic T-Cell Responses in Pulmonary Vaccination. Proc Natl Acad Sci. 2011;108(44):E989–E997. doi: 10.1073/pnas.1104264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lynn GM, Laga R, Darrah PA, Ishizuka AS, Balaci AJ, Dulcey AE, Pechar M, Pola R, Gerner MY, Yamamoto A, Buechler CR, et al. In Vivo Characterization of the Physicochemical Properties of Polymer-Linked TLR Agonists That Enhance Vaccine Immunogenicity. Nat Biotechnol. 2015;33(11):1201–1210. doi: 10.1038/nbt.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wei L, Zhao Y, Hu X, Tang L. Redox-Responsive Polycondensate Neoepitope for Enhanced Personalized Cancer Vaccine. ACS Cent Sci. 2020;6(3):404–412. doi: 10.1021/acscentsci.9b01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kauffman KJ, Webber MJ, Anderson DG. Materials for Non-Viral Intracellular Delivery of Messenger RNA Therapeutics. J Control Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- (10).Kulkarni JA, Witzigmann D, Chen S, Cullis PR, Van Der Meel R. Lipid Nanoparticle Technology for Clinical Translation of SiRNA Therapeutics. Acc Chem Res. 2019;52(9):2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- (11).A Triumph of Perseverance over Interference. Nature Biotechnology. 2018;36:775. doi: 10.1038/nbt.4255. [DOI] [PubMed] [Google Scholar]
- (12).Singer JW. Paclitaxel Poliglumex (XYOTAX, CT-2103): A Macromolecular Taxane. J Control Release. 2005;109(1–3):120–126. doi: 10.1016/j.jconrel.2005.09.033. [DOI] [PubMed] [Google Scholar]
- (13).Eldar-Boock A, Miller K, Sanchis J, Lupu R, Vicent MJ, Satchi-Fainaro R. Integrin-Assisted Drug Delivery of Nano-Scaled Polymer Therapeutics Bearing Paclitaxel. Biomaterials. 2011;32(15):3862–3874. doi: 10.1016/j.biomaterials.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Nuhn L, Vanparijs N, De Beuckelaer A, Lybaert L, Verstraete G, Deswarte K, Lienenklaus S, Shukla NM, Salyer ACD, Lambrecht BN, Grooten J, et al. PH-Degradable Imidazoquinoline-Ligated Nanogels for Lymph Node-Focused Immune Activation. Proc Natl Acad Sci U S A. 2016;113(29):8098–8103. doi: 10.1073/pnas.1600816113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nuhn L, De Koker S, Van Lint S, Zhong Z, Portela Catani JP, Combes F, Deswarte K, Li Y, Lambrecht BN, Lienenklaus S, Sanders NN, et al. Nanoparticle-Conjugate TLR7/8 Agonist Localized Immunotherapy Provokes Safe Antitumoral Responses. Adv Mater. 2018;30(45):e1803397. doi: 10.1002/adma.201803397. [DOI] [PubMed] [Google Scholar]
- (16).Van Herck S, Deswarte K, Nuhn L, Zhong Z, Portela Catani JP, Li Y, Sanders NN, Lienenklaus S, De Koker S, Lambrecht BN, David SA, et al. Lymph-Node-Targeted Immune Activation by Engineered Block Copolymer Amphiphiles-TLR7/8 Agonist Conjugates. J Am Chem Soc. 2018;140(43):14300–14307. doi: 10.1021/jacs.8b08595. [DOI] [PubMed] [Google Scholar]
- (17).De Vrieze J, Louage B, Deswarte K, Zhong Z, De Coen R, Van Herck S, Nuhn L, Kaas Frich C, Zelikin AN, Lienenklaus S, Sanders NN, et al. Potent Lymphatic Translocation and Spatial Control Over Innate Immune Activation by Polymer-Lipid Amphiphile Conjugates of Small‐Molecule TLR7/8 Agonists. Angew Chemie Int Ed. 2019;58(43):15390–15395. doi: 10.1002/anie.201905687. [DOI] [PubMed] [Google Scholar]
- (18).Talelli M, Vicent MJ. Reduction Sensitive Poly(L-Glutamic Acid) (PGA)-Protein Conjugates Designed for Polymer Masked-Unmasked Protein Therapy. Biomacromolecules. 2014;15(11):4168–4177. doi: 10.1021/bm5011883. [DOI] [PubMed] [Google Scholar]
- (19).Barz M, Duro-Castano A, Vicent MJ. A Versatile Post-Polymerization Modification Method for Polyglutamic Acid: Synthesis of Orthogonal Reactive Polyglutamates and Their Use in “Click Chemistry.”. Polym Chem. 2013;4(10):2989–2994. [Google Scholar]
- (20).Kalam MA, Khan AA, Alshamsan A. Non-Invasive Administration of Biodegradable Nano-Carrier Vaccines. Am J Transl Res. 2017;9(1):15–35. [PMC free article] [PubMed] [Google Scholar]
- (21).Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ. Designer Vaccine Nanodiscs for Personalized Cancer Immunotherapy. Nat Mater. 2017;16(4):489–496. doi: 10.1038/nmat4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shukla NM, Malladi SS, Mutz Ca, Balakrishna R, David Sa. Structure-Activity Relationships in Human Toll-like Receptor 7-Active Imidazoquinoline Analogues. J Med Chem. 2010;53(11):4450–4465. doi: 10.1021/jm100358c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Smirnov D, Schmidt JJ, Capecchi JT, Wightman PD. Vaccine Adjuvant Activity of 3m-052: An Imidazoquinoline Designed for Local Activity without Systemic Cytokine Induction. Vaccine. 2011;29(33):5343–5442. doi: 10.1016/j.vaccine.2011.05.061. [DOI] [PubMed] [Google Scholar]
- (24).Kulkarni JA, Witzigmann D, Leung J, Tam YYC, Cullis PR. On the Role of Helper Lipids in Lipid Nanoparticle Formulations of SiRNA. Nanoscale. 2019;11(45):21733–21739. doi: 10.1039/c9nr09347h. [DOI] [PubMed] [Google Scholar]
- (25).De Koker S, Cui J, Vanparijs N, Albertazzi L, Grooten J, Caruso F, De Geest BG. Engineering Polymer Hydrogel Nanoparticles for Lymph Node-Targeted Delivery. Angew Chem Int Ed Engl. 2016;55(4):1334–1339. doi: 10.1002/anie.201508626. [DOI] [PubMed] [Google Scholar]
- (26).Lomas H, Canton I, MacNeil S, Du J, Armes SP, Ryan AJ, Lewis AL, Battaglia G. Biomimetic PH Sensitive Polymersomes for Efficient DNA Encapsulation and Delivery. Adv Mater. 2007;19(23):4238–4243. [Google Scholar]
- (27).Liang K, Such GK, Zhu Z, Yan Y, Lomas H, Caruso F. Charge-Shifting Click Capsules with Dual-Responsive Cargo Release Mechanisms. Adv Mater. 2011;23(36):H273–H277. doi: 10.1002/adma.201101690. [DOI] [PubMed] [Google Scholar]
- (28).Yu H, Zou Y, Wang Y, Huang X, Huang G, Sumer BD, Boothman DA, Gao J. Overcoming Endosomal Barrier by Amphotericin B-Loaded Dual PH-Responsive PDMA-b-PDPA Micelleplexes for SiRNA Delivery. ACSNano. 2011;5(11):9246–9255. doi: 10.1021/nn203503h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).De Coen R, Nuhn L, De Geest BG. Engineering Mannosylated Nanogels with Membrane-Disrupting Properties. Polym Chem. 2019;10(31):4297–4304. [Google Scholar]
- (30).Koynov K, Butt HJ. Fluorescence Correlation Spectroscopy in Colloid and Interface Science. Current Opinion in Colloid and Interface Science. 2012;17(6):377–387. [Google Scholar]
- (31).Negwer I, Best A, Schinnerer M, Schäfer O, Capeloa L, Wagner M, Schmidt M, Mailänder V, Helm M, Barz M, Butt H-J, et al. Monitoring Drug Nanocarriers in Human Blood by Near-Infrared Fluorescence Correlation Spectroscopy. Nat Commun. 2018;9(1):5306. doi: 10.1038/s41467-018-07755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Shen Z, Reznikoff G, Dranoff G, Rock K. Cloned Dendritic Cells Can Present Exogenous Antigens on Both MHC Class I and Class II Molecules. J Immunol. 1997;158(6):2723–2730. [PubMed] [Google Scholar]
- (33).Lienenklaus S, Cornitescu M, Zietara N, Łyszkiewicz M, Gekara N, Jabłónska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, et al. Novel Reporter Mouse Reveals Constitutive and Inflammatory Expression of IFN-Beta in Vivo. J Immunol. 2009;183(5):3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- (34).Swaminathan G, Thoryk EA, Cox KS, Meschino S, Dubey SA, Vora KA, Celano R, Gindy M, Casimiro DR, Bett AJ. A novel lipid nanoparticle adjuvant significantly enhances B and T cell respones to sub-unit vaccine antigens. Vaccine. 2016;34:110–119. doi: 10.1016/j.vaccine.2015.10.132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.