Abstract
Muscle regeneration is sustained by infiltrating macrophages and consequent satellite cell (SC) activation1–4. Macrophages and SC communicate in different ways1–5 but their metabolic interplay was never investigated so far. Here, we found that muscle injuries and aging are characterized by intratissutal glutamine restriction. Low glutamine levels endow macrophages with the metabolic ability to secrete glutamine via enhanced glutamine synthetase (GS) activity at the expense of glutamate dehydrogenase-1 (GLUD1)-mediated glutamine oxidation. Glud1 knockout (KO) macrophages display constitutively high GS activity which prevents glutamine shortage. Import of macrophage-derived glutamine by SC through the glutamine-transporter SLC1A5 activates mTOR and promotes SC proliferation and differentiation. Consequently, macrophage-specific deletion or pharmacological inhibition of GLUD1 improves muscle regeneration and functional recovery in response to acute injury, ischemia, or aging. Conversely, SLC1A5 blockade in SC or GS inactivation in macrophages negatively affects SC functions and muscle regeneration. These results highlight a metabolic cross-talk between SC and macrophages whereby macrophage-derived glutamine sustains SC functions. Thus, GLUD1 targeting offers new therapeutic opportunities for the regeneration of injured or aged muscles.
Macrophages contribute to the repair of damaged skeletal muscle3,5. These cells clear tissue debris and release cytokines as well as growth factors that stimulate SC proliferation4–6. Later, macrophages promote SC differentiation4–7, and tissue revascularization7. The positive involvement of inflammatory cells in the acute phase of muscle healing is supported by the evidence that macrophage depletion impairs muscle regenerative capacity8.
Given the important role of glutamine in muscle homeostasis9–11, and our observation that glutamine production by macrophages remodels the composition of the extracellular tumormilieu12, 13, we hypothesized a function of glutamine in a yet unidentified, metabolic crosstalk between macrophages and SC.
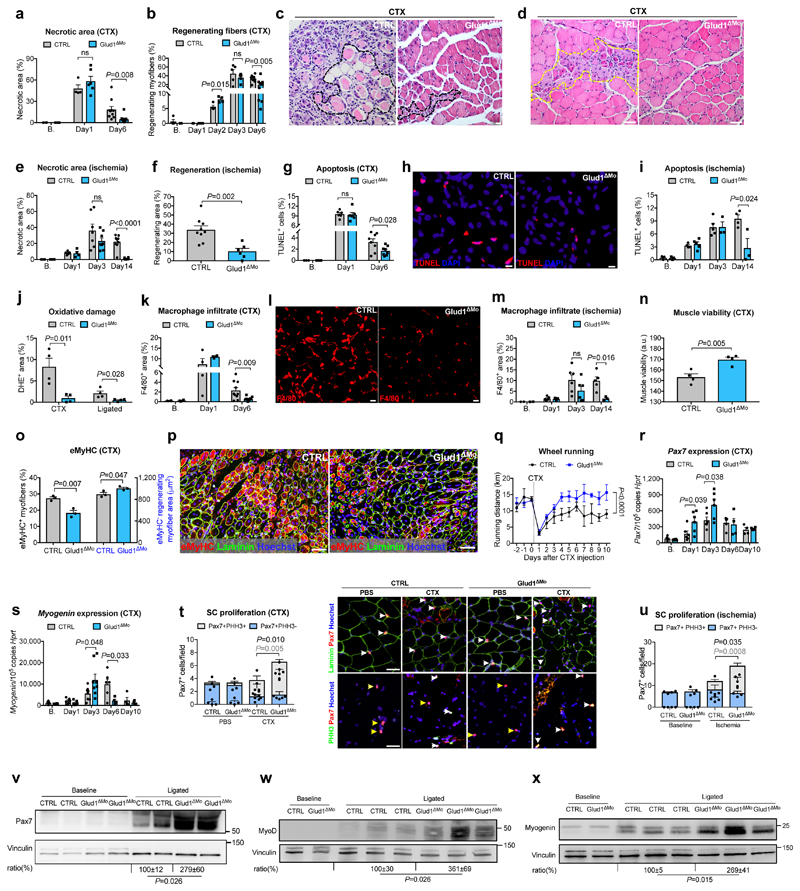
To induce myofiber death, inflammation and muscle regeneration, we injected cardiotoxin (CTX) in the tibialis anterior (TA)14 or provoked ischemia of the crural muscles15 in control (CTRL) and Glud1ΔMo mice, with complete GLUD1 deletion in macrophages and only 38% knockdown in neutrophils (Extended Data Fig. 1a-c). CTRL and Glud1ΔMo mice revealed similar muscle histology in healthy conditions and early after damage, i.e. 1 day post-CTX or 3 days post-femoral-artery-ligation (Fig. 1a-e). However, compared to CTRL, Glud1ΔMo mice displayed an earlier peak in the number of regenerating myofibers and a quicker resolution of muscle necrosis, cell death, oxidative damage, and inflammation (Fig. 1a-m). Six days post-CTX, muscle viability was higher in Glud1ΔMo vs. CTRL mice. Yet, the early regenerating myofibers (expressing embryonic myosin heavy chain) were fewer, but the late ones (negative for embryonic myosin heavy chain) were larger in Glud1ΔMo mice, pointing to a faster and more advanced regeneration (Fig. 1n-p). This phenotype was due to monocyte-derived macrophages, rather than tissue-resident macrophages (Extended Data Fig. 1d,e). Inducible deletion of Glud1 in macrophages only led to improved muscle recovery as well (Extended Data Fig. 1f-i).
Figure 1. GLUD1 loss in macrophages boosts SC activation and muscle regeneration.
a-d, Post-CTX muscle necrosis (Baseline,B. n=4; Day1 n=4,6 CTRL,Glud1ΔMo, respectively; Day6 n=10) (a) and regeneration (B. n=4,5; Day1 n=5,6; Day2 n=4; Day3 n=5; Day6 n=10) (b), with micrographs of H&E-stainings at Day6 showing necrotic (black-dotted line) (c) or regenerating (yellow-dotted line) fibers (d). e,f, Post-ligation necrosis (e) and regenerating area (f) 14 days post-ligation (B. n=4; Day1 n=5,4; Day3 n=7,8; Day14 n=9,6). g-i, Post-CTX muscle apoptosis by TUNEL staining (B. n=3; Day1 n=6; Day6 n=8) (g), with micrographs of Day6 (h), or post-ligation (B. n=3; Day1 n=4,5; Day3 n=5,3; Day14 n=4,3) (i). j, Oxidative stress by DHE stainings 6 days post-CTX or 14 days post-ligation (n=4). k-m, F/480+ macrophage infiltration post-CTX (B. n=4; Day1 n=4; Day6 n=10) (k), with micrographs of Day6 (I), or post-ligation (B. n=2; Day1 n=5; Day3 n=6; Day14 n=5,3) (m). n, Muscle viability (TTC staining) 6 days post-CTX (n=4). o,p, eMyHC+ myofibers (left) and eMyHC- regenerating myofibers area (right) over cross-section area 6 days post-CTX (n=3) (o) and representative micrographs (p). q, Voluntary running (n=5). r,s, RT-qPCR on muscle extracts for Pax7 (B./Day1/Day3 n=6; Day6 n=4; Day10 n=4) (r) and Myogenin (B./Day1 n=6; Day3 n=8; Day6 n=6,4; Day10 n=4) (s). t,u Quiescent (PHH3-) and proliferating (PHH3+) SC at baseline and 1 day post-CTX (n=4,6), with representative micrographs (t), or 3 days post-ligation (n=4) (u). White arrows indicate Pax7+ or Pax7+/PHH3+ cells; yellow arrows, Pax7+/PHH3- cells, v-x, WB on muscle extracts and densitometry for Pax7 (v), MyoD (w), Myogenin (x). A representative (a-u,w,x) or a pool (v) of at least two independent experiments is shown. Unpaired two-tailed t-test everywhere applied except in q (two-way ANOVA); ns, not significant. Bars: 10 μm (h), 20 μm (c, I), 50 μm (d, p, t). Graphs: mean ± SEM.
In voluntary wheel running tests, the baseline physical activity and its drop 1 day post-CTX were comparable in both genotypes. However, Glud1ΔMo mice re-gained the pre-injured physical capabilities earlier than CTRL mice (Fig. 1q).
Basal numbers of SC, assessed by Pax7 expression16, was comparable between genotypes (Fig. 1r-v). However, after injury, induction of SC proliferation (assessed by PHH3, or Ki67) and differentiation (assessed by the early and late differentiation markers MyoD and Myogenin, respectively17) were stronger and quicker in Glud1ΔMo than in CTRL mice (Fig. 1r-x; Extended Data Fig. 1j-n). Thus, muscle regeneration in Glud1ΔMo mice is more efficient.
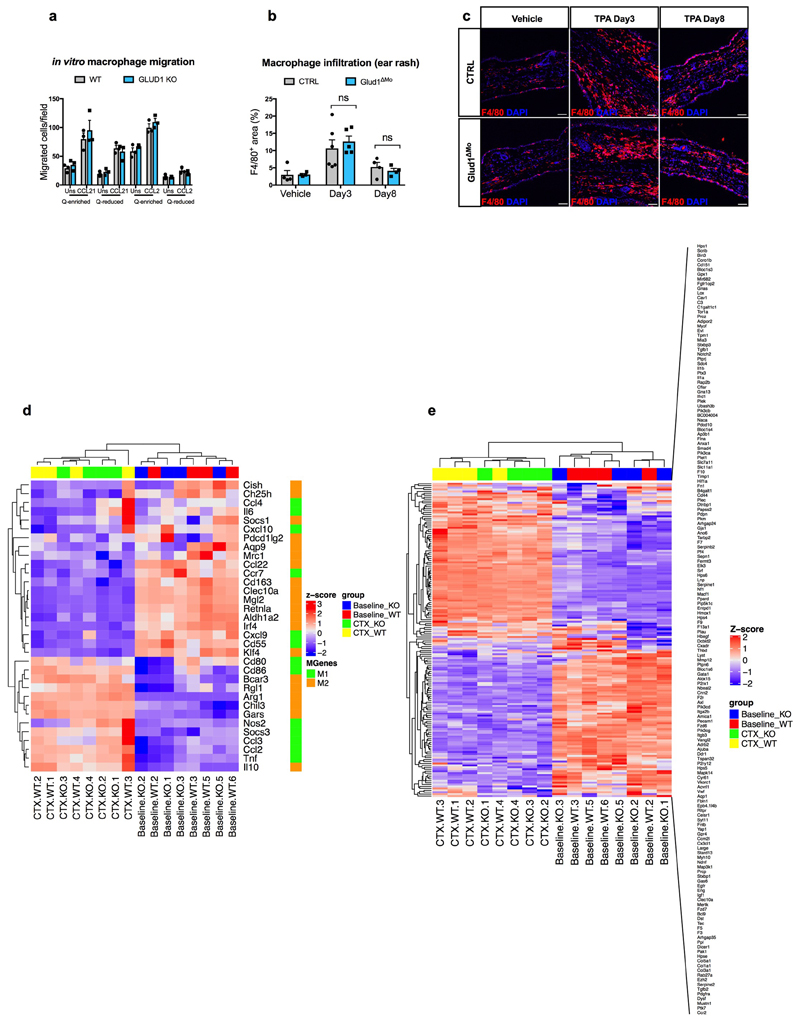
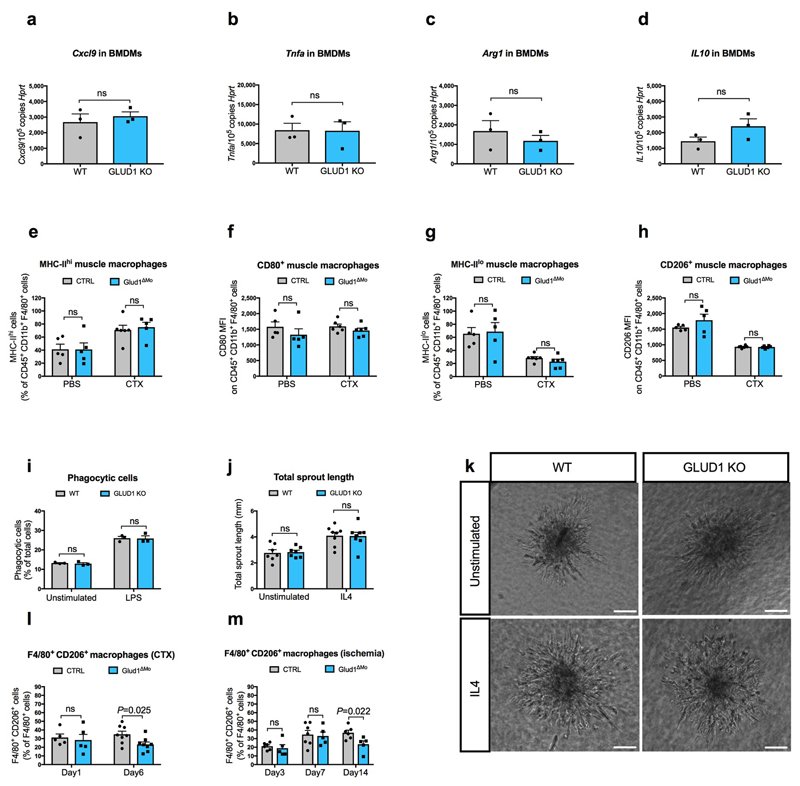
We then assessed how GLUD1-deficiency affects macrophage-mediated immunomodulation and vessel growth. Blood count, immune landscapes and vascular features of muscles at baseline and early after damage were similar in Glud1ΔMo vs. CTRL mice (Extended Data Table1; Extended Data Fig. 1o-w). In vitro and in vivo recruitment assays, macrophage polarization, as well as wound healing and angiogenic functions did not change (Extended Data Fig. 2a-e; Extended Data Fig. 3a-k). However, later after damage, total and M2-like macrophages were fewer in Glud1ΔMo vs. CTRL mice (Extended Data Fig. 3l,m), arguing that the faster resolution of inflammation is consequent to a more efficient muscle repair in Glud1ΔMo mice.
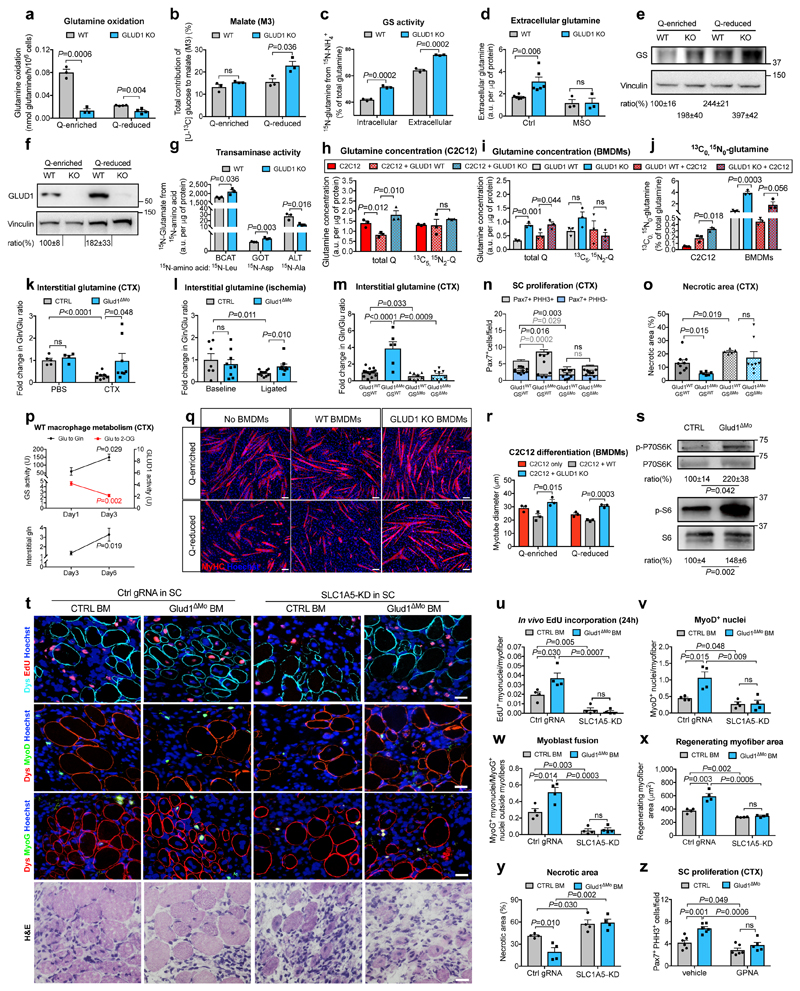
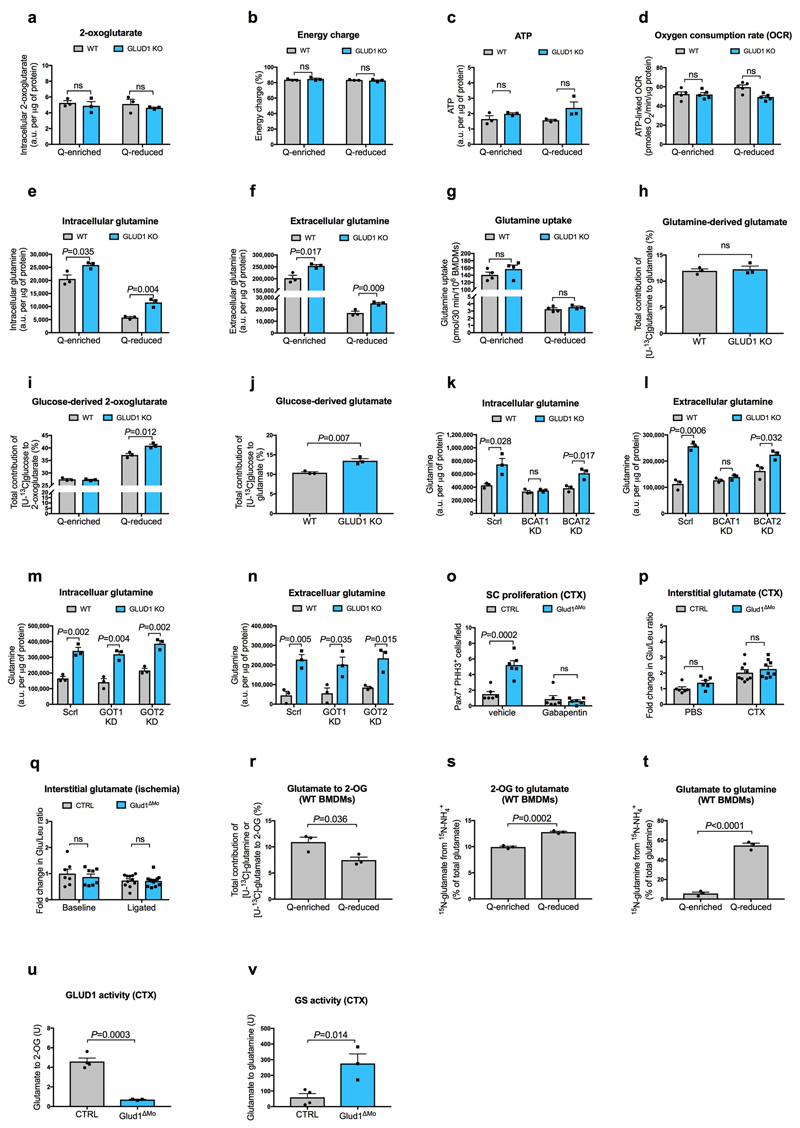
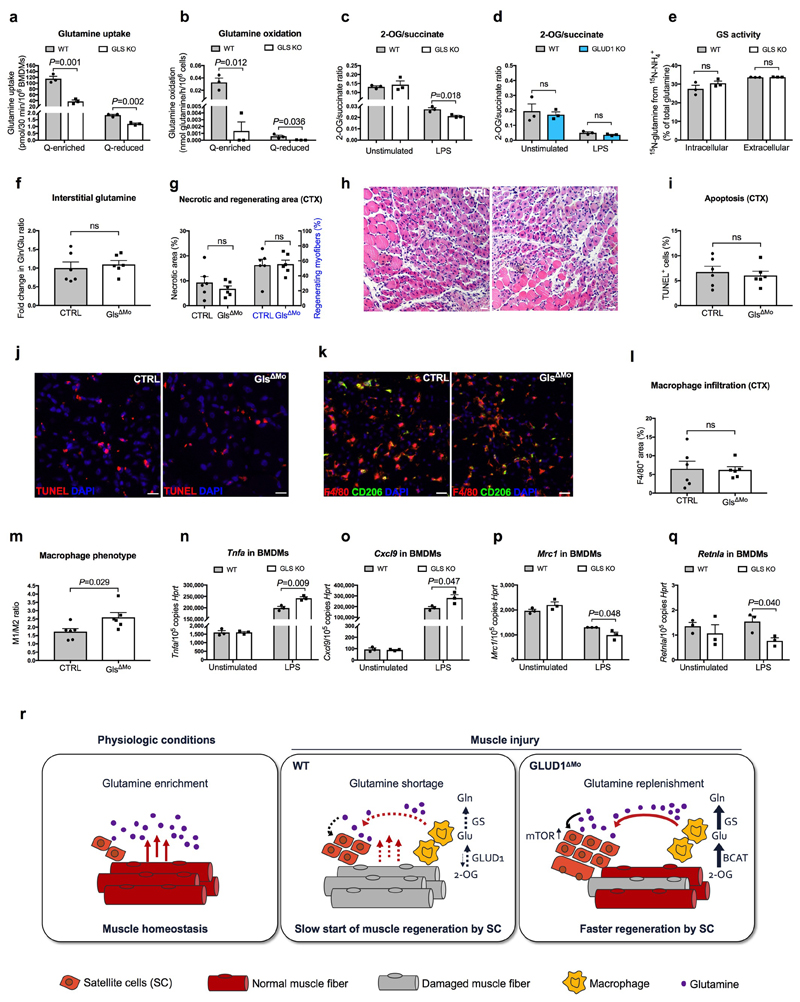
Metabolic changes within a cell affect the biology of neighbouring cells18. In WT macrophages, glutamine oxidation was 73% lower in glutamine-reduced vs. glutamine-enriched conditions (Fig. 2a). Compared to WT cells, glutamine oxidation in GLUD1 KO macrophages was lower in both culture conditions (Fig. 2a). However, total 2-oxoglutarate (2-OG) was comparable (Extended Data Fig. 4a), likely due to enhanced pyruvate carboxylase (PC)-dependent TCA-cycle anaplerosis (Fig. 2b), which compensates for the loss of glutamine-derived carbons19. Total energy charge, ATP production, ATP-linked oxygen consumption rate (OCR) were comparable in both genotypes (Extended Data Fig. 4b-d).
Figure 2. Uptake of macrophage-derived glutamine by SC boosts muscle regeneration.
a,b, Glutamine oxidation (n=3,4 WT,KO, respectively) (a), pyruvate-carboxylase activity (n=3) (b) in BMDMs under glutamine (Q)-enriched and Q-reduced conditions (n=4,3). c,d, Intra- and extracellular glutamine production (n=3) (c) and glutamine release under MSO-mediated GS inhibition (n=3) (d) in Q-starved BMDMs. e,f, WB and densitometry for GS (e) and GLUD1 (f) in BMDMs. g, BCAT, GOT, ALT activities in Q-starved BMDMs (n=3). h-j, Total glutamine, [13C5 15N2]-glutamine, [13C0 15N0]-glutamine in C2C12 cells (h,j) and BMDMs (i,j), seeded alone or co-cultured in [13C5,15N2]-glutamine-containing medium (n=3). k,l, Interstitial glutamine 1 day post-CTX (Baseline,B. n=5,4 CTRL,Glud1ΔMo, respectively; CTX n=9,8) (k) or 3 days post-ligation (B. n=7,9; ischemia n=12) (I), m-o, Interstitial glutamine and SC proliferation 1 day post-CTX (m,n), necrosis 6 days post-CTX (o) upon macrophagic GS deletion (Glud1WTGSWT n=14,6,9 in m, n, o, respectively; Glud1ΔMoGSWT n=6,4,6; Glud1WTGSΔMo n=8,5,5; Glud1ΔMoGSΔMo n=8,5,8). p, Glutamate-to-2-OG (GLUD1 activity) and glutamate-to-glutamine (GS activity) conversion in muscle-infiltrating WT macrophages (upper panel) and interstitial glutamine in WT muscles (lower panel) (n=4). q,r, C2C12 myotubes in co-culture with BMDMs (n=3). s, WB and densitometry for phospho-P70S6K (1 day post-CTX) and phospho-S6 (3 days post-CTX) in isolated SC. t-y, Micrographs (t) and quantifications (n=4) of EdU+ myonuclei (u), MyoD+ nuclei (v), myoblast fusion (w), regenerating myofiber area (x), necrosis (y) 6-day post-CTX in mice reconstituted with GLUD1-WT (CTRL BM) or GLUD1-KO (Glud1ΔMo) bone marrows (BM), and knocked-down (KD) or not (Ctrl gRNA) for SLC1A5 in SC. z, PHH3+ SC 1 day post-CTX after GPNA-mediated SLC1A5 inhibition (n=6,6,6,5 from right to left). A representative (a-d,g-j,p-r,t-z) or a pool (e,f,k-o,s) of at least two independent experiments is shown. Unpaired two-tailed t-test everywhere applied; ns, not significant; a.u., arbitrary units. Bars: 20 μm (t), 50 μm (q). Graphs: mean ± SEM.
Though glutamine oxidation in GLUD1 KO macrophages was diminished, intracellular and extracellular glutamine production was higher under both glutamine-replete and, to a greater extent, glutamine-restricted conditions (Extended Data Fig. 4e,f). This was due to enhanced GS activity (Fig. 2c,d). At the protein level, GS was induced in WT macrophages under low glutamine but this induction was stronger in GLUD1 KO macrophages (Fig. 2e). Glutamine uptake and conversion into glutamate were similar in WT and GLUD1 KO macrophages (Extended Data Fig. 4g,h).
GLUD1 converts glutamate into 2-OG but also 2-OG into glutamate, the latter used by GS to generate glutamine12. Consistent with this function, GLUD1 protein levels in WT macrophages were also upregulated under low glutamine (while undetectable in GLUD1 KO macrophages) (Fig. 2f). Under glutamine starvation, glucose utilization for glutamate production was enhanced in WT macrophages and even more in GLUD1 KO macrophages (Extended Data Fig. 4i,j). In absence of GLUD1, 2-OG into glutamate conversion was possibly taken-over by the increased activity of branched-chain amino acid aminotransferase (BCAT), utilizing branched-chain amino acids (leucine, isoleucine, and valine) as amino-group donors, or glutamate-oxaloacetate transaminase (GOT), utilizing aspartate (Fig. 2g). However, only the silencing of cytosolic BCAT (BCAT1) restored glutamine production by GLUD1 KO macrophages back to the WT levels (Extended Data Fig. 4k-n). Consistently, the BCAT1 inhibitor gabapentin prevented SC expansion in CTX-treated Glud1ΔMo mice (Extended Data Fig. 4o).
To assess the fate of macrophage-derived glutamine, we used a two-chamber co-culture of C2C12 myoblasts and macrophages in medium containing exclusively [13C5,15N2]-glutamine, i.e. glutamine labelled in the two nitrogen groups and in the five carbons. Uptake of [13C515N2]-glutamine by myoblasts did not change in all the conditions (Fig. 2h). However, compared to myoblasts alone, intracellular total glutamine (labelled and unlabelled) was lower in myoblasts cultured with WT macrophages but higher in myoblasts cultured with GLUD1 KO macrophages (Fig. 2h). In macrophages, total glutamine levels were always higher in GLUD1 KO macrophages (Fig. 2i) though a comparable uptake in all the conditions (Fig. 2i). Since in myoblasts, the contribution to the total glutamine pool of [13C0 15N0]-glutamine (derived from different sources than [13C5 15N2]-glutamine) was higher in co-culture with macrophages and peaked with GLUD1 KO macrophages (Fig. 2j), we argued that glutamine production by GLUD1 KO macrophages (Fig. 2j) outcompete glutamine consumption, increasing glutamine availability for myoblasts.
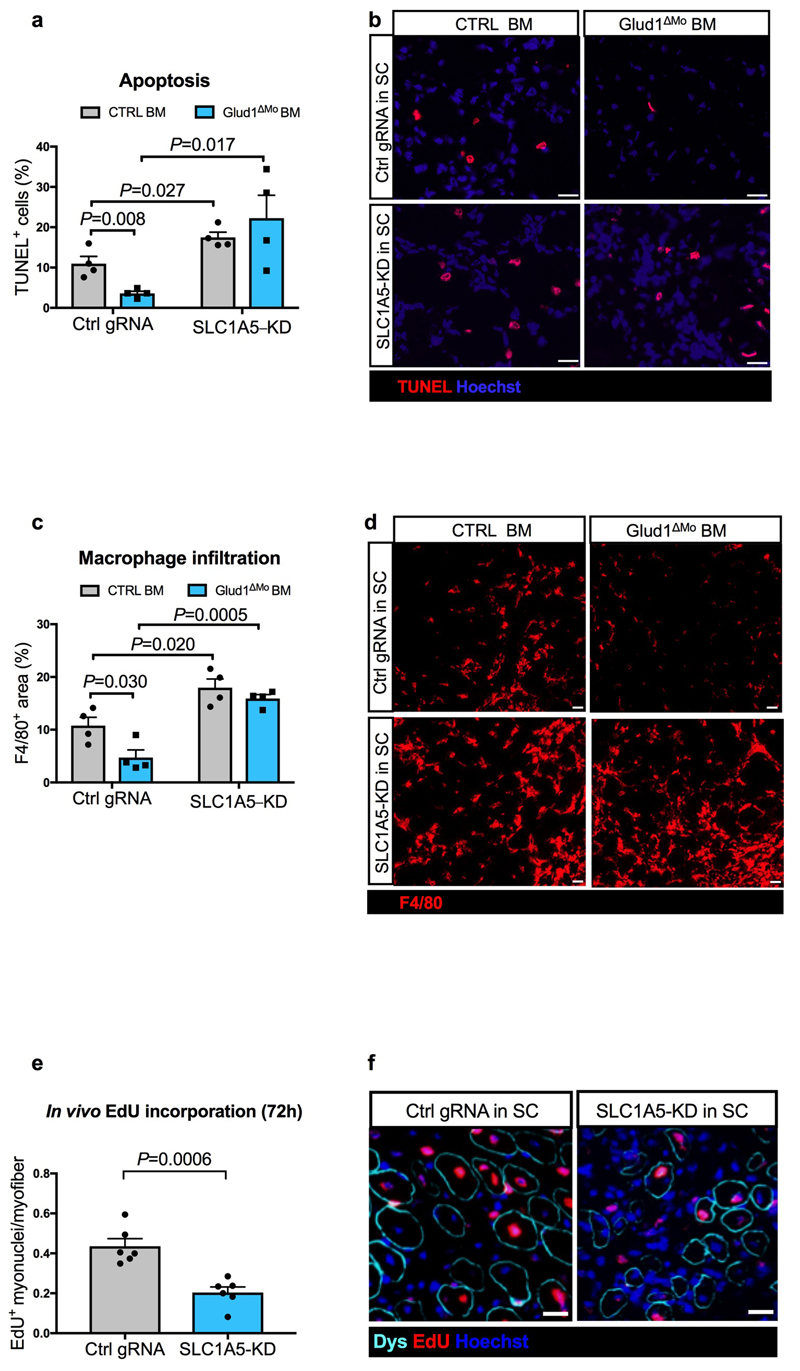
In vivo, interstitial glutamine levels in CTRL and Glud1ΔMo muscles were similar at baseline and dropped in injured CTRL but not in Glud1ΔMo muscles (Fig. 2k,l), whereas glutamate availability did not change (Extended Data Fig. 4p,q). GS depletion in macrophages worsened this post-injury glutamine shortage observed in control muscles, and also, disabled the preservation in glutamine levels seen in Glud1ΔMo muscles (Fig. 2m). Similarly, macrophage-specific depletion of GS worsened SC proliferation and muscle healing after injury, and it impeded the faster SC activation and damage resolution seen in Glud1ΔMo mice (Fig. 2n,o). Thus, macrophage GS is instrumental to replenish glutamine and promote SC activation in response to muscle damage (i.e. a glutamine-restricted condition).
Consistent with the above data, WT BMDMs placed in low glutamine showed reduced conversion of glutamate into 2-OG, a readout of oxidative GLUD1 activity (Extended Data Fig. 4r), whereas glutamine production involving the conversion of 2-OG into glutamate and glutamate into glutamine were both enhanced (Extended Data Fig. 4s,t). In muscle-infiltrating WT macrophages too, GS and oxidative GLUD1 activities, respectively, were gradually increasing and decreasing over time upon damage (Fig. 2p, upper panel), resulting in a slow increase of interstitial glutamine (Fig. 2p, lower panel). Muscle-infiltrating GLUD1 KO macrophages had constitutively inactive GLUD1 and higher GS activity (Extended Data Fig. 4u,v) which prevented the post-damage drop in interstitial glutamine (Fig. 2k).
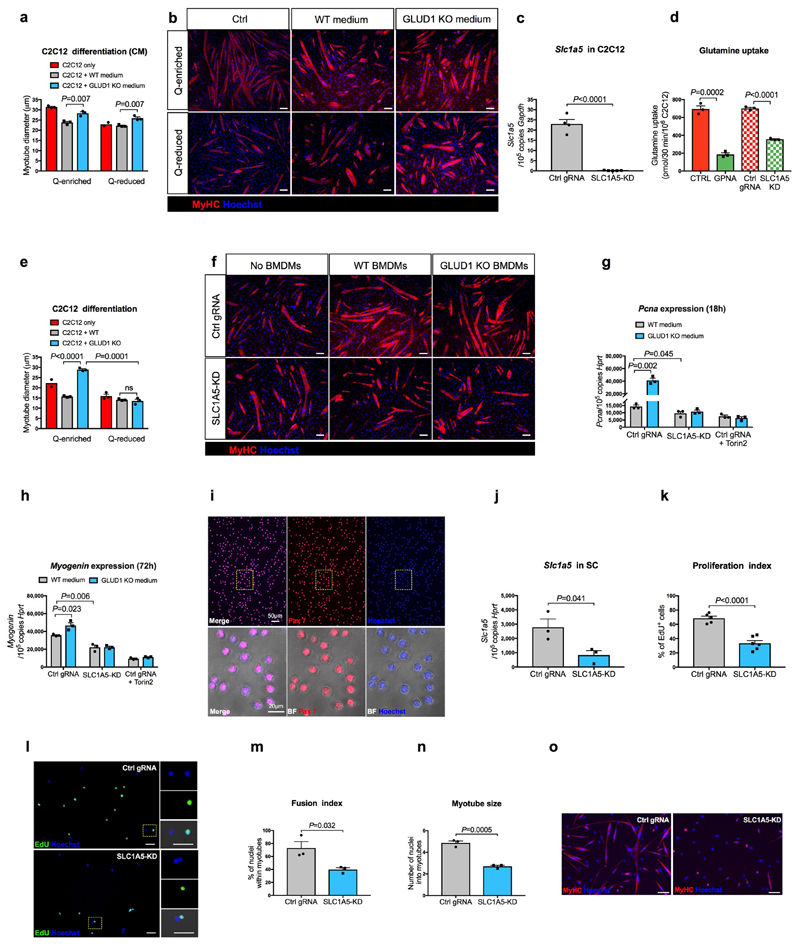
We then linked macrophage-derived glutamine and myogenic potential. Myotube formation was promoted when differentiating C2C12 myoblasts were cultured in a glutamine-rich medium and reduced under glutamine restriction (Fig. 2q,r). Co-culture of C2C12 cells with WT macrophages impaired myogenic differentiation in glutamine-enriched conditions, phenocopying a condition of low glutamine. Instead, co-culture of C2C12 cells with GLUD1 KO macrophages resulted in larger myotubes, regardless of glutamine availability in the medium (Fig. 2q,r). Similar results were obtained with macrophage-conditioned media (Extended Data Fig. 5a,b). SLC1A5 knockdown (KD) in C2C12 cells impaired glutamine uptake (Extended Data Fig. 5c,d) and abrogated the advantage offered by GLUD1 KO macrophages on myoblast differentiation (Extended Data Fig. 5e,f). Conditioned media from GLUD1 KO macrophages enhanced the expression of both the proliferation marker Pcna and the differentiation marker Myogenin and activated mTOR pathway (an important player for SC proliferation/differentiation20,21) in a glutamine-uptake-dependent manner22 (Extended Data Fig. 5g,h). The mTOR inhibitor Torin2 abolished Pcna and Myogenin induction in C2C12 cells stimulated with conditioned media from GLUD1 KO macrophages (Extended Data Fig. 5g,h). Likewise, mTOR pathway was enhanced in SC isolated from CTX-injected Glud1ΔMo vs. CTRL muscles (Fig. 2s).
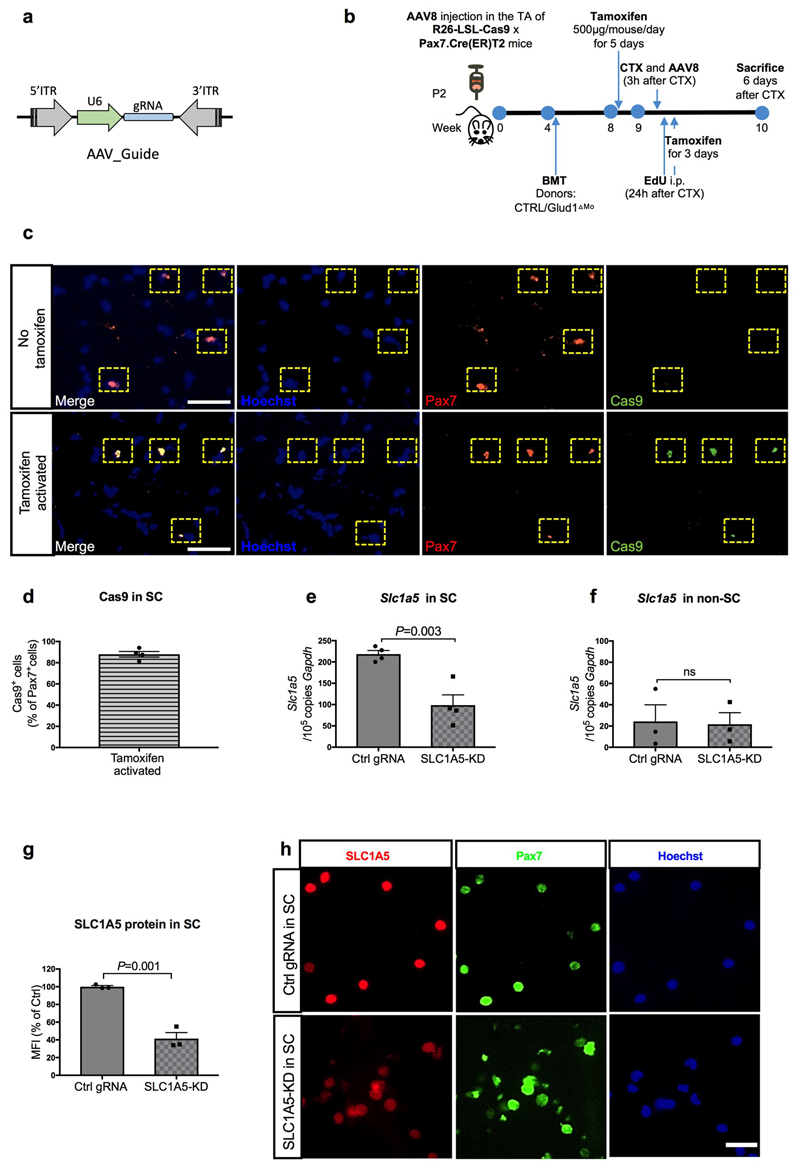
In glutamine-replete conditions, SLC1A5-KD SC had impaired in vitro proliferation/differentiation (Extended Data Fig. 5i-o). Therefore, to disrupt SLC1A5-mediated glutamine import specifically in SC in vivo, AAV8 particles containing the gRNA against Slc1a5 were injected intramuscularly in LSL-Cas9/PAX7:Cre-ERT mice, carrying a tamoxifen-inducible Cas9 in Pax7+ cells (Extended Data Fig. 6a-d). In this way SLC1A5 was selectively knocked-down by 60% in about all the SC (Extended Data Fig. 6e-h). Using bone marrow (BM) transplantation experiments, we confirmed that GLUD1 KO macrophages ameliorated muscle regeneration in mice with SC proficient in glutamine uptake (non-targeting control gRNA). Specifically, we administered EdU within 24h post-CTX and collected the injured TA muscles 6 days post-CTX. Compared to muscles in Glud1WT BM (CTRL BM), those from Glud1ΔMo BM mice treated with the non-targeting gRNA displayed a higher number of EdU+ terminally-differentiating myonuclei, increased MyoD+ myoblasts, higher number of Myogenin+ cells incorporated into the myofibers, as well as an increased area of regenerating myofibers (Fig. 2t-x). Conversely, when SC were perturbed in their capacity to import glutamine (Slc1a5-targeting gRNA), the superior myogenic potential of Glud1ΔMo BM mice was abrogated but also severely affected in Glud1WT BM (CTRL BM) mice (Fig. 2t-x).
Mirroring muscle regeneration, tissue damage was milder in Glud1ΔMo BM mice treated with the non-targeting gRNA, but worsened by the Slc1a5 gRNA in both CTRL and Glud1ΔMo BM mice (Fig. 2y, Extended Data Fig. 7a-d). As the maximal SC expansion in a WT context occurs between day 2 and day 4 post-CTX23 (Fig. 1r), we administered EdU until day 3. In this setting, the number of EdU+ myonuclei was much higher compared to a 24h EdU pulse and anyhow strongly reduced when SC were deleted for the glutamine transporter Slc1a5 (Extended Data Fig. 7e,f). Next to this approach, the SLC1A5 inhibitor gamma-L-Glutamyl-p-Nitroanilide (GPNA) impaired SC proliferation in both CTX-treated Glud1ΔMo and WT mice (Fig. 2z; Extended Data Fig. 5d). In sum, macrophage-derived glutamine uptake by SC plays a relevant role in the control of myogenic functions. Disrupting this cross-talk precludes the faster muscle repair in Glud1ΔMo mice but also delays muscle regeneration in CTRL mice.
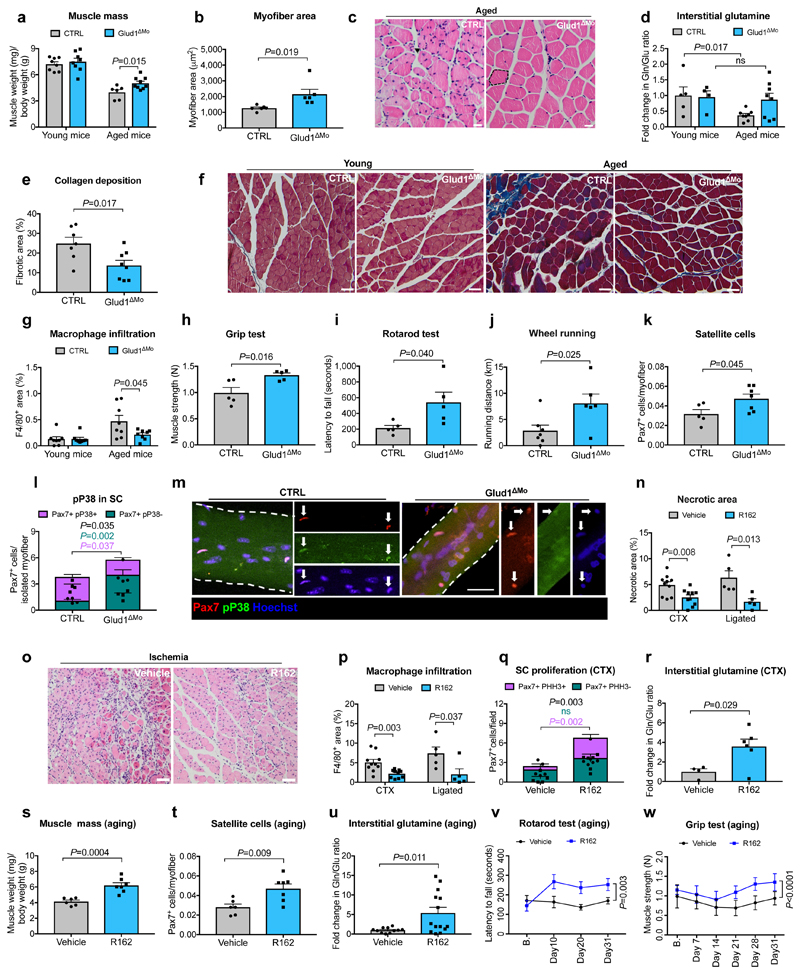
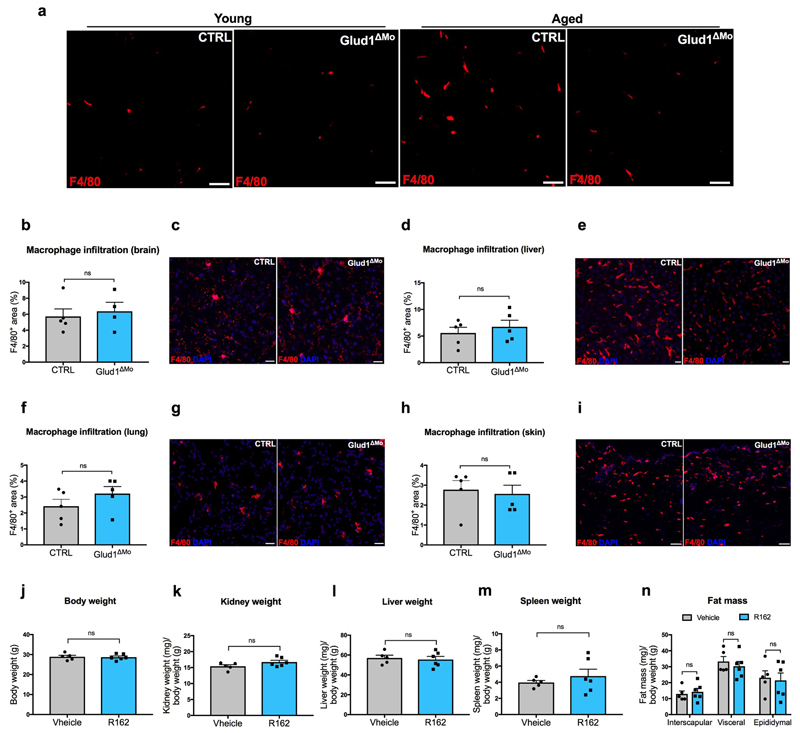
Regenerative functions are known to decline with aging24. Muscle weight index in 18 month-old CTRL mice was reduced when compared to young mice, but this drop was less severe in Glud1ΔMo mice (Fig. 3a), as also indicated by the presence of larger myofibers (Fig. 3b,c). Interstitial glutamine followed the same trends (Fig. 3d). Fibrosis and macrophage infiltration were exacerbated in old vs. young CTRL mice, but to a lesser extent in Glud1ΔMo mice (Fig. 3e-g; Extended Data Fig. 8a), arguing that chronic inflammation inversely correlates with muscle health. Macrophage density in other tissues did not change (Extended Data Fig. 8b-i). Overall, muscle performance of 18 month-old Glud1ΔMo mice was better than age-matched CTRL mice (Fig. 3h-j). In line with this, Glud1ΔMo muscles displayed increased numbers but decreased ratio of phospho-p38 positive vs. negative SC (Fig. 3k-m), suggesting that SC self-renewal was better preserved in Glud1ΔMo mice25.
Figure 3. GLUD1 loss or inhibition in macrophages benefits damaged and aged muscles.
a, Gastrocnemius weight (young mice n=8; aged mice n=6,9 CTRL,Glud1ΔMo, respectively). b, c, Quantification (b) and micrographs (c) of fiber area in H&E-stained gastrocnemius sections (aged mice n=6). d, Intra-TA interstitial glutamine (young mice n=5,4; aged mice n=8). e,f, Quantification (e) for collagen deposition (in blue) in crural muscles of aged mice (n=7,8) on Masson’s trichrome stainings (f). g, Macrophage infiltration in crural muscles of young or aged mice (n=8). h-j, Grip strength (n=5) (h), rotarod test (n=5) (i), voluntary running (n=7,6) (j) in aged mice, k, Intra-TA SC density (n=5,7). l-m Pax7 and phospho-P38 in SC, associated to myofibers isolated from extensor digitorum longus muscles of aged mice (n=4). n,o, Muscle necrosis 6 days post-CTX (n=10) or 14 days post-ligation (n=5) upon R162-mediated GLUD1 inhibition (n), and micrographs showing ischemic necrosis (o). p, Macrophage infiltration 6 days post-CTX (n=10) or 14 days post-ligation (n=5). q, PHH3+/- SC 1 day post-CTX (n=5,6). r, Interstitial glutamine 1 day post-CTX (n=4,6). s-w, Gastrocnemius weight (n=6,7) (s), SC number per isolated TA myofiber (n=6,7) (t), interstitial glutamine (n=11,14) (u), rotarod (n=7) (v) and grip strength test (n=7,8) (w) in vehicle and R162-treated aged mice. A representative of at least two independent experiments is shown in a-r. Unpaired two-tailed t-test everywhere applied except in v and w (two-way ANOVA); ns, not significant. Scale bars: 20 μm (c, f, m, o). Graphs: mean ± SEM.
We finally assessed the potential therapeutic effect of the GLUD1 inhibitor R16 226. In response to CTX or ischemia, R162 treatment reduced muscle necrosis and inflammation (Fig. 3n-p), and boosted SC proliferation and interstitial glutamine concentration (Fig. 3q,r). In 18 month-old mice as well, administration of R162 for a month increased muscle mass, SC numbers, and interstitial glutamine, and improved physical performance (Fig. 3s-w). R162 did not affect body and organs’ weights (Extended Data Fig. 8j-n).
This therapeutic benefit was selective for GLUD1 targeting. Unlike GLUD1 deletion (Extended Data Fig. 4g), macrophage-specific knockout of glutaminase (GLS), converting glutamine into glutamate, resulted in reduced glutamine uptake, glutamine oxidation, and 2-OG-to-succinate ratio but did not promote GS activity (Extended Data Fig. 9a-e). Six days post-CTX, interstitial glutamine levels, muscle damage, and macrophage infiltration were comparable in CTRL and GlsΔMo mice, but macrophages were more M1-like in GlsΔMo mice (Extended Data Fig. 9f-m), consistent with in vitro data (Extended Data Fig. 9n-q) and previous findings27.
Muscle tissue is a major site for glutamine synthesis in the body9,10,11. We show here that muscle damage and aging restrain glutamine availability. Muscle-infiltrating macrophages sense this shortage and tilt down glutamine oxidation in favor of glutamine production. Macrophage-released glutamine is uptaken by SC promoting their proliferation and differentiation through mTOR activation. Unlike WT macrophages, GLUD1 KO macrophages are pre-adapted to glutamine starvation, which results in improved regeneration (Extended Data Fig. 9r). Thus, a metabolic rewiring in macrophages re-establishes muscle homeostasis in response to damage.
The risk of vascular occlusion and skeletal muscle impairment is increased in case of diabetes, hypercholesterolemia, and obesity, which are recurrent in today’s society28. Similarly, sarcopenic patients are arising together with the population age29. However, little can be done to improve these conditions28,30. Our data suggest a pharmacologic approach to treat damage and age-related skeletal muscle decline.
Methods
Mouse models
GLUD1 (Glud1tm1.1Pma, MGI:3835667)31, GS (Glultm3Whla, MGI:4462791)32, GLS (Glstm2.1Sray, MGI:95752)33 floxed mouse lines, all in a C57BL/6 background, were obtained respectively from Dr. Pierre Maechler (University of Geneva, Switzerland), Dr. Wouter H. Lamers (Academic Medical Center, Amsterdam, Netherlands) and Dr. Stephen Rayport (Columbia University, NY, US). Glud1 L/L;CSF1R:Cre-ERT transgenic mice were generated by intercrossing Glud1 floxed mice with the tamoxifen-inducible, macrophage-specific CSF1R:Cre-ERT deleter mouse line (a gift of Dr. Jeffrey Dr. W. Pollard, University of Edinburgh, UK). PAX7:Cre-ERT transgenic mice34, harboring a tamoxifen-inducible Cre under the Pax7 promoter for SC-specific expression, were provided by Dr. Katrien De Bock (ETH, Switzerland). LoxP-STOP-LoxP Cas9 mice (B6J.129(B6N)- Gt(ROSA) 26Sortm1(CAG-cas9*,-EGFP)Fezh/J)35 were purchased from Jackson Laboratory. LSL-Cas9 x PAX7:Cre-ERT transgenic mice, a strain that has inducible Cas9 expression in germline under the SC-specific Pax7 promoter, were generated by intercrossing LoxP-STOP-LoxP Cas9 mice with PAX7:Cre-ERT transgenic mice line. Acute deletion of Glud1 in macrophages was obtained by daily intraperitoneal (i.p.) injection of tamoxifen (0.05 mg per gram of body weight) for 5 days before and during cardiotoxin (CTX) (Latoxan) induced injury. Control mice were treated with tamoxifen according to the same protocol. All mice used for ischemia and CTX experiments were on a C57BL/6 background between 8 and 15 weeks old, while 18-month old mice were used for the aging experiments. Mice were used without specific gender selection. In all experiments, littermate controls were used. Housing and all experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven.
Cardiotoxin muscle injury
Mice were anaesthetized with isoflurane and 50 μl of 10 μM CTX was injected in the tibialis anterior (TA) muscle14. Control mice (denoted as baseline) were subjected to PBS injection. For SC isolation, mice were injected with 50 μl of 10 μM CTX in TA and 100 μl of 10 μM CTX in gastrocnemius muscles. Muscles were harvested for analysis at different time points post-injury. In vivo GLUD1 inhibition was achieved by bi-daily gavage of R162 (Focus Biomolecules) at 0.6 mg/mouse. Mice were pretreated 1 day before CTX injection. Afterwards, mice continued to receive bi-daily treatment until their sacrifice. To inhibit SLC1A5, mice were treated by oral gavage 3 times per day with a solution (200 μl/mouse) containing 500 mM GPNA (Sigma-Aldrich). CTX was injected 1 h after the first gavage, and mice were sacrificed 24 h afterwards. In vivo BCAT1 inhibition was achieved by bi-daily i.p. injection of gabapentin (Sigma-Aldrich) at 2 mg/mouse. Mice were pretreated once before CTX injection, and mice were sacrificed 24 h afterwards.
Hindlimb ischemia
To induce acute hindlimb ischemia and greatly prevent flow redirection into the collateral circulation, which leads to severe muscle necrosis, unilateral or bilateral ligations of the high femoral artery were performed without damaging the nervus femoralis as previously described15. Control mice (denoted as baseline) were subjected to a sham operation that did not involve the ligation of the femoral artery. Functional perfusion measurements were performed using a Lisca PIM II camera (Gambro)15. GLUD1 inhibition was achieved by bi-daily gavage of R162 (Focus Biomolecules) at 0.6 mg/mouse. Mice were pretreated 1 day before femoral artery ligation. Afterwards, mice continued to receive bi-daily treatment until their sacrifice.
Wheel running test
Physical activity was assessed with voluntary wheel running test. Mice were individually housed in cages equipped with 12 cm diameter wheels for rodents, wheels were connected with cycle computers (SunDing SD-568AE). After 1 week of acclimatization, physical activity (i.e. duration, running speed, and distance) was recorded daily.
Rotarod test
Whole body mobility and coordination was assessed by rotarod performance. Following a 5 min acclimatization in the test room, mice were placed on the rod (Biological research apparatus), which was rotating at an initial speed of 4 rpm. The speed was increased gradually from 4 rpm to 40 rpm within 5 min and latency to fall on to a soft pad was recorded. The test was repeated twice more, with 15 min between tests. After 3 days training, latency to fall was calculated over 3 trials.
Grip test
Muscle strength was measured by pulling backwards the mice with a continuous movement when the mice were hold firmly to the grip of strength meter (Chatillon, DFE II Digital Force Gauge). The test was repeated twice more, with 15 min between tests. Results was calculated over the 3 trials.
TTC staining
TA muscles were collected 6 days after CTX injection. After scarification muscles were washed in PBS and cut in transversal sections. Sections were incubated with a buffered (pH 7.4) 1.5% 2,3,5- triphenyl-tetrazolium chloride (TTC) solution at a temperature of 37°C for 40 min. Pictures were taken with NIKON camera. The quantification was performed with Image J software. Images were converted in a grey scale and it was calculated the mean grey intensity of each section.
Interstitial fluid preparation
Intact TA muscles for the CTX model, or crural muscles (gastrocnemius and soleus muscles) for the ischemia model, were placed into test tubes with perforated bottom. 20 μL of 0.9% NaCl solution (pH 7.4) was added to the sample. Interstitial fluid was collected by centrifugation (110 g, 10 min, 4°C). Protein within theinterstitial fluid was precipitated using -20°C cold methanol/water-mix (5:3) and centrifuged (20000 x g, 5 min, 4°C). The supernatant was dried using a vacuum centrifuge and derivatized for mass spectrometry analysis.
Bone marrow transplantation
5-6 weeks old recipient mice were irradiated with 9.2 Gy. Subsequently, 10x10s bone marrow cells from the appropriate genotype were injected intravenously via the tail vein. Muscle injury experiments were initiated 5 weeks after bone marrow reconstitution. Red and white blood cell count was determined using a hemocytometer on peripheral blood collected in heparin with capillary pipettes by retro-orbital bleeding.
In vitro and in vivo targeting of the Slcla5 locus
To generate in vitro a stable C2C12 mouse myoblast cell line and primary SC, deficient for SLC1A5, lentiCRISPRv2 vectors expressing the Cas9 along with a gRNA targeting the Slcla5 locus (AATCCCTATCGATTCCTGTG) or a non-taregting control gRNA (GAACAGTCGCGTTTGCGACT), were used. 2xl04 C2C12 cells/well were seeded in 12-well plates and transduced with purified lentiviral vectors in the presence of 8 Mg/ml polybrene for 48 h. After adding virus, the plate was centrifuged for 30 min at 800x g, 25-32°C. Transduced cells were selected with puromycin (4 μg/ml) during a period of 5 to 7 days. The lentiCRISPRv2 was a gift from Dr. Feng Zhang (Addgene plasmid # 52961)36. The gRNAs were cloned as described previously36. For an optimal transduction of SCs cells in vivo and specifc targeting of the Slcla5 locus exclusively in SCs, the gRNAs described above were cloned in the AAV_Guide vector (see also Extended Data Fig. 6a). AAV8 production and titration was outsourced to Vigene Biosciences (Rockville, MD 208050, USA). Two-day-old (P2) neonatal LSL-Cas9 x PAX7:Cre-ERT transgenic mice were then administered locally into the TA muscles with 6.7 X 1010 vector genomes (vg) per injection (5 μl/leg). Bone marrow cells from CTRL and Glud1ΔMo mice were transplanted at the age of five weeks. Five weeksafter bone marrow reconstitution, tamoxifen was given by daily oral gavage for five days before and three days after CTX induced injury. 180 μg EdU was given by i.p. injection 24 h after CTX injection. 3 hours after CTX injection mice also received a second AAV injection into the TA muscles (4 X 1011 vg per injection, see shematic in Extended Data Fig. 6b).
SC isolation and culture conditions
Hindlimb muscles were dissected and minced in small fragments by scissors and digested in 0.1% collagenase/0.25% trypsin solution at 37°C with gentle shaking for 20 min as previously described37. Supernatants were collected in fetal bovine serum (FBS) at 37°C and passed through 40 μm filters. The digestion step was repeted 3 times until the complete dissociation of samples. The fraction of freshly isolated SC was enriched by using the SC isolation kit (Miltenyi Biotec, 130-104-268). SC were collected on glass slides by cytospin for immunofluorescence analyses, or snap frozen for protein extraction. The fraction of non-SC (Miltenyi Biotec, 130-104-268) was used as control where indicated.
SC were seeded on collagen-coated dishes (Sigma) and maintained in growth medium (GM), that, for SC, is DMEM (Gibco) supplemented with 20% FBS (Gibco), 10% Horse Serum (HS) (Gibco), 1% Chicken embryo extract (Seralab), 2 mM glutamine, 110 mg/ml sodium pyruvate (Gibco), 100 units/ml penicillin and 100 μg/mL streptomycin (Gibco), or in differentiation medium (DM), that is DMEM supplemented with 2% HS, 2 mM glutamine, 110 mg/ml sodium pyruvate, 100 units/ml penicillin and 100 μg/mL streptomycin.
EdU-based SC proliferation assays were performed by seeding in 6 well plates 3.0 x 104 cells/well in GM containing 10 μM of 5-ethylnyl-2’-deoxyuridine (EdU) (Thermo Fisher, C10337) for 24 h prior the fixation. Click-iT® reaction and staining were performed according to the manufacturer’s instructions.
Differentiation assays were performed by seeding 1.0 x 104/cm2 SC in GM. After 24 h, GM was replaced with DM for 5 days. Myotubes-derived SC were fixed in 4% paraformaldehyde and stained for myosin heavy chain. Fusion index (percentage of nuclei within myotubes: myotube=nuclei>2) and myotube size (mean of number of nuclei into myotubes) were analyzed.
Single fiber isolation and immunostaining
Single myofibers were isolated from extensor digitorum longus (EDL) muscles of aged mice as previously described37. Briefly, intact muscles were dissected from tendon to tendon and digested with a solution containing 0.2% collagenase type I (Sigma) in DMEM (Dulbecco’s modified Eagle’s medium; high glucose, L-glutamine with 110 mg/ml sodium pyruvate) at 37°C for 3h. Afterward, individual fibers were separated from each others by pipetting under a dissecting microscope, and then washed in DMEM.
Fresh isolated fibers were fixed in 4% paraformaldehyde, 6min at RT, permeabilized by a solution containing 0.5% Triton, 8 min at RT, blocked with 20% HS lh at RT and incubated overnight at 4°C with anti mouse pax7 (DHSB, 1:20) and anti rabbit Phospho-p38 MAPK (Cell Signaling Technology, 1:200) followed by incubation with anti rabbit Alexa 488, and anti mouse 568 conjugated secondary antibodies (Invitrogen, 1:1000). 28-30 fibers per point were analysed.
Cell lines
C2C12 murine myoblast cells were obtained from the American Type Culture Collection (ATCC). C2C12 authentication was confirmed by testing their myogenic differentiation capacity. Specifically, RT-qPCR and immunofluorescence analyses of the expression of specific myogenic markers (e.g. myogenin and myosin heavy chain) confirmed their myogenic origin. Cells were regularly tested for mycoplasma via PCR.
Glutamine-enriched and glutamine-reduced media preparation
When specified, the glutamine (Q)-enriched medium (4mM glutamine) was obtained by supplementing M199 or DMEM with 10% FBS. A glutamine (Q)-reduced medium (0.03 mM glutamine) was obtained by supplementing M199 or DMEM (with no glutamine) with 10% FBS, dialyzed for glutamine. Dyalized FBS was obtained by slide A lyzer dialysis cassette (Thermo Scientific 66130).
In vitro C2C12 co-cultures with BMDMs
C2C12 cells were cultured in growth medium (GM), that for C2C12 cells, is DMEM (Gibco) supplemented with 10% FBS (Gibco), 2 mM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. C2C12 myoblasts were cultured for no more than 6 passages in a humidified incubator in 5% CO2 and 95% air at 37°C. To obtain BMDM-conditioned media (CM), 2 x 106 BMDMs (WT or GLUD1 KO) were cultured in a 6 cm dish for 48h in Q-enriched or Q-reduced medium.
In the proliferation assay, 6 x 104 C2C12 cells/well were seeded in 6-well plates in GM. After 24 h, regular GM was replaced with a Q-reduced GM that was previously conditioned by WT or GLUD1 BMDMs.
In the differentiation assay, 6 x 104 C2C12 cells/well were seeded in a 6-well plates in GM. After 24 h, GM was replaced with Q-enriched or Q-reduced differentiation medium (DM), that is the same medium as described above for SC. At day 2 of myoblast differentiation, 1.5 x 105 BMDMs or BMDM-conditioned DM were added to the differentiating myotubes for additional 4 days. Afterwards, samples were collected for RNA or fixed in 4% formaldehyde for 10 min at RT and permeabilized in PBS with 1% BSA, 0.2% TritonX for 30 min and blocked in 10% Donkey serum lh at RT. Finally, samples were incubated overnight at 4°C with mouse anti-MF20 (DSHB, 2μg/ml) and subsequently incubated with AlexaFluor 568-conjugateddonkey anti-mouse (Invitrogen 1:1000). When indicated, mTOR inhibition was achieved by adding 500nM Torin2 (Selleck Chemicals) to the medium.
In vitro BMDM-C2C12 co-cultures with [13C515N2]-glutamine
2.5 x 105 BMDMs were seeded in the top chambers of a 24-well transwell plate (0.4 μm polycarbonate membrane) whereas 1.5 x 105 C2C12 cells were seeded in the lower wells, in a medium where the only glutamine present was labelled in the two nitrogen groups and in the five carbons ([13C515N2]-glutamine). After 48h, cells were scraped in 80% methanol and phase separation was achieved by centrifugation at 4°C. Methanol-water phase containing polar metabolites was separated and dried using a vacuum concentrator. The dried metabolite samples were stored at -80°C. Isotopomer distributions and metabolite levels were measured with a 7890A GC system (Agilent Technologies) combined with a 5975C Inert MS system (Agilent Technologies).
Glutamine oxidation
Each M199 media (Q-enriched or Q-reduced) was supplemented with 0.5 μCi/ml [U-14C]-glutamine. After incubating 1 x 106 BMDMs in a 12-well plate for 6h, 250 μl of 2 M perchloric acid was added to each well to stop cellular metabolism. Each well was immediately covered with a 1 x hyamine hydroxide-saturated Whatman paper. Overnight absorption of14CC>2 released during the oxidation of glutamine into the paper was performed at RT and radioactivity in the paper was determined by liquid scintillation counting.
Glutamine uptake
2 x 106 BMDMs were seeded in a 6-well plate and cultured in M199 medium (Gibco) and 10% FBS, supplemented with 0.5 μCi/ml [U-14C]-glutamine for 30 min at 37°C. Cells were lysed in IN NaOH and the radioactivity was measured by liquid scintillation counting.
A(X)P detection by LC-MS
2 x 106 BMDMs were lysed in 300 μl extraction buffer (50:30:20 mix of methanol:acetonitrile:10 mM Tris pH 9.3). Following extraction, samples were centrifuged for 10 min at 20000 x g (at 4°C). The supernatant was transferred to a vial. 35 μl was loaded onto an Ultimate 3000 UPLC (Thermo Scientific, Bremen, Germany) equipped with a ZIC-pHILIC column (2.1 x 150 mm, 5 μm particle size, cat# 1.50460.0001, Merck, Darmstadt, Germany) in line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). A linear gradient was carried out starting with 90% solvent A and 10% solvent B. From 2 to 20 min the gradient changed to 80% B and was kept at 80% until 23 min. Next a decrease to 40% B was carried out to 25 min, further decreasing to 10% B at 27 min. Finally 10% B was maintained until 35 min. The solvent was used at a flow rate of 200 ul/min, the columns temperature was kept constant at 25°C. The mass spectrometer operated in negative ion mode, settings of the HESI probe were as follows: sheath gas flow rate at 35, auxiliary gas flow rate at 10 (at a temperature of 260°C). Spray voltage was set at 4.8 kV, temperature of the capillary at 300°C and S-lens RF level at 50. A full scan (resolution of 140.000 and scan range of m/z 70-1050) was applied. For the data analysis we used an in-house library and metabolites of interest were quantified (area under the curve) using the XCalibur 4.0 (Thermo Scientific) software platform. The energy charge was calculated as ([ATP]+1/2[ADP])/ ([ATP]+ [ADP] +[AMP]).
Oxygen consumption
1.5 x 104 BMDMs were incubated overnight on Seahorse XF24 tissue culture plates (Agilent). During the assay, the medium was replaced by unbuffered DMEM supplemented with 5 mM D-glucose and 2 mM L-glutamine, pH 7.4. The measurement of oxygen consumption was performed at 6 min intervals (2min mixing, 2min recovery, 2min measuring) using the Seahorse XF24 analyzer. Inhibitors were serially injected at thefollowing concentrations: oligomycin (1 μM), FCCP (fluoro-carbonyl cyanide phenylhydrazone, 1.5 μM), antimycin A (1 μM) (all from Sigma-Aldrich).
13C and 15N tracing experiments
For13C and 15N tracing experiments, cells were incubated with [U-13C]-L-glutamine (2 mM), [U-13C]-L-glutamate (0.25 rnM), [U-13C]-D-glucose (5 rnM), 15NH4CI (2 mM), [15N,13C4]-aspartate (1 mM), [15N,13C4]-alanine (1 mM) or [15N]-leucine (0.8 mM) for 48 h (confirmation of steady state) respectively (all from Cambridge Isotope Laboratories).
Metabolites quantification by LC-MS/MS
For mass spectrometry analysis of glutamate and glutamine, 2 x 106 cell pellets were washed twice in PBS and extracted in 500 μl of 80% methanol. Upon extraction, samples were centrifuged at 20000 x g for 15 min and the supernatant was dried using a vacuum centrifuge. 25 μl of a 2% methoxyamine hydrochloride solution were added to the dried pellet and the tubes were then placed at 37°C for 90 min. Then 75 μl of N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1% N-tert-Butyldimethyl-chlorosilane (Sigma-Aldrich, Bornem, Belgium) was added and the reaction was carried out for 30 min at 60°C. Reaction mixtures were then centrifuged for 15 min at 20000 g at 4°C in order to remove insolubilities, the supernatant was transferred to a glass vial with conical insert (Agilent). GC-MS analyses were performed using an Agilent 7890A GC equipped with a HP-5 ms 5% Phenyl Methyl Silox (30 m - 0.25 mm i.d. - 0.25 μm; Agilent Technologies, Santa Clara, California, USA) capillary column, interfaced with a triple quadruple tandem mass spectrometer (Agilent 7000B, Agilent Technologies) operating under ionization by electron impact at 70 eV. The injection port, interface and ion source temperatures were kept at 230°C. Temperature of the quadrupoles was maintained at 150°C. The injection volume was 1 μl, and samples were injected at 1:25 split ratio. Helium flow was kept constant at 1 ml/min. The GC oven temperature was held at 60°C for 3 min, increased to 300°C at 9°C/min, and kept for 2 min. The mass spectrometer operated in SIM mode, glutamine and glutamate were determined from the m/z 341.2 and 342.2 respectively. For quantifications, the MassHunter Workstation Software B.06.00 SP01 (Agilent Technologies) was used.
In vivo GS activity and glutamate to 2-OG conversion
GS activity and glutamate to 2-OG conversion in muscle-infiltrating macrophages, sorted 1 and 3 days after CTX injection, were measured respectively with the glutamine synthetase microplate assay kit (ABIN2704091, Cohesion Biosciences) and the glutamate dehydrogenase activity assay kit (MAK099-1KT, Sigma-Aldrich).
TPA model of acute skin inflammation
Phorbol ester TPA was used to induce acute skin inflammation as described before38. Briefly, TPA (2.5 μg in acetone, 20 μl total volume/site) was topically applied on the ear skin of anaesthetized mice. As vehicle control, the ear was painted with acetone alone. After the indicated time points, tissue was harvested for further analysis.
FACS analysis of muscle macrophages
TA muscles were dissected, dissociated mechanically, digested using 800U/ml collagenase II (10 ml per sample) for 1 h at 37°C, centrifuged and resuspended with lOOOU/ml collagenase II (1 ml per sample) and llU/ml Dispase (1 ml per sample) solution and incubated for 30 min at 37°C. The digested tissue was filtered using a 40 μm pore sized mesh and cells were centrifuged 5 min at 500 g. Cells were resuspended in FACS buffer (PBS containing 2% FBS and 2 mM EDTA), incubated for 15 min with Mouse BD Fc Block purified anti-mouse CD16/CD32 mAb (BD-pharmingen) and stained with the following antibodies for 30 min at 4°C: viability dye (Invitrogen), anti-CD45 (Biolegend), anti-CDllb (Invitrogen), anti-F4/80 (Invitrogen), anti-MHCII (Invitrogen), anti-CD80 (Invitrogen) and anti-CD206 (Biorad), anti-CD45R (BD Biosciences), anti-Ly6G (BD Biosciences,), anti-TCR(3 (BD Biosciences), anti-CD4 (BD Biosciences) and anti-CD8 (Invitrogen,). Cells were subsequently, washed and resuspended in cold FACS buffer before FACS analysis or flow sorting by a FACS Verse or FACS Aria (BD Biosciences), respectively. Fluorescence minus one (FMO) controls were performed in all the stainings and used for the proper gating in all the analysis.
Histology and immunostainings
7 μm-thick cryosections were obtained by using a Leica cryostat from frozen muscles collected in optimal cutting temperature compound (OCT) and fixed in 4% formaldehyde for 10 min at RT. Alternatively, TA and crural muscle were fixed in 2% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned at 7 μm thickness. Necrotic muscle area was detected by H&E staining as the area which includes necrotic myocytes, inflammatory cells and interstitial cells. After deparaffinization and rehydration, muscle sections were permeabilized by a solution containing 1% BSA, 0.2% Triton-X in PBS 30 min at RT, blocked with 10% Donkey serum (Sigma) 1 h at RT and treated with target retrieval solution at pH 6.1 (Dako, S1699) at 92°C for 20 min. To reduce the immune background sections were blocked with 10% donkey serum in PBS, lh at RT, followed by blocking with FAB fragment anti-mouse IgG (Jackson Immunoresearch, 1:5-1:10), lh at RT and 0.3% H202 in PBS for 30 min at RT to block endogenous peroxidase activity. Slides were incubated overnight at 4°C with primary antibodies: rat anti-F4/80 (Bio-Rad, 1:100), rabbit anti-laminin (Sigma-Aldrich, 1:300), rabbit anti-phospho-histone H3 (Millipore, 1:1000), mouse anti-MF20 (DSHB, 2μg/ml), rabbit anti-Dystrophin (Abeam, 1:50), mouse anti-MyoD (Santa Cruz, 1:10), mouse anti-embryonic Myosin (DSHB, 1:20), mouse anti-Myogenin (DSHB, 1:1), mouse anti-Pax7 (DSHB, 1:20), rat anti-CD34 (BD Biosciences), mouse anti-MMR/CD206 (R&D Systems, 1:100), rabbit anti-Ki67 (Abeam, 1:100), rabbit anti-CRISPR-Cas9 (Abeam, 1:500). Appropriate secondary antibodies were used: Alexa 488, or 568 conjugatedsecondary antibodies (Invitrogen, 1:1000), biotin-labeled antibodies (Jackson Immunoresearch, 1:500-1:2000) and, when necessary, TSA fluorescein tyramide, TSA Plus Cyanine 3 or Cyanine 5 System amplification (Perkin Elmer, Life Sciences, 1:500-1:2000) were performed according to the manufacturer’s instructions. Immunofluorescence of SC cytospinned on glass slides was performed by fixation in 4% formaldehyde for 10 min at RT, followed by incubation with 0.5% Triton-X in PBS for 20 min at RT and blocking with with 10% donkey serum for 1h at RT. Samples were then probed with mouse anti-Pax7 (DSHB, 1:20) alone or incombination with rabbit anti-SLC1A5 (Alomone Labs, 1:50) for 2h at RT followed by incubation with mouse Alexa 568 (Invitrogen, 1:10000), or a combination of mouse Alexa 488 and rabbit 568 conjugated secondary antibodies (Invitrogen, 1:1000), respectively. In vivo cell proliferation and differentiation was detected by DNA incorporation of thymidine analogue EdU (ThermoFisher, Click-iT C10337) in combination with Dystrophin staining on muscle cryosections. Oxidative damage was detected by dihydroethidium (DHE) (Life Technology). Samples were incubated with 10 ^M DHE at 37°C for 30 min. Apoptosis was detected by TUNEL assay kit (Sigma-Aldrich) according to the manufacturer’s instructions. Nuclei were counterstained with Hoechst-33342 (Invitrogen, 1:1000). Whenever sections were stained in fluorescence, ProLong Gold mounting medium with or without DAPI (Invitrogen) was used.
Imaging and Morphometry
Images were acquired by an Olympus BX41 microscope equipped with the CellSense Dimension imaging software. CellSense Dimension software was used for the morphometric analyses of cell cultures and muscle tissue. Myotubes diameter was measured as the average from three independent measurements per myotube.
Bone marrow-derived macrophages (BMDMs)
Macrophages were derived from bone marrow precursors as described before38. Briefly, bone marrow cells (1.6 x 106 cells/ml) were cultured in a volume of 6 ml in a 10 cm Petri dish in DMEM supplemented with 20% FBS and 30% L929 conditioned medium as a source of M-CSF. After 3 days of culture, an additional 3 ml of differentiation medium was added. At day 7, macrophages were harvested with ice cold Ca2+ and Mg2+-free PBS. The cells obtained were uniformly macrophages as assessed by FACS, using the pan-macrophage marker F4/80. When indicated, GS inhibition in cultured BMDMs was achieved by adding 1 mM L-methionine-SR-sulfoximine (MSO; Sigma) to the medium for 48 h. Silencing of Bcatl, Bcat2, Gotl, Got2 and Alt in BMDMs was achieved by electroporation with specific siRNAs (IDT). Briefly, 5.6 xlO6 BMDMs were resuspended in 750 μl of Opti-MEM and were electroporated (250V, 950 μF, ∞ Ω) with 120 pmol of total siRNA IDT. Control BMDMs were electroporated with scrambled siRNA sequences.
BMDM migration assay
Migration of BMDMs was assessed by using a 8-μm-pore Transwell permeable plate (Corning Life Science). The bottom chambers contained DMEM with specific chemoattractants or controls (specified in each Figure), BMDMs were harvested and then seeded in the upper chamber (2.5 x 105 cells in 200 μl of DMEM at 2% HS). After 4 h incubation, migrated cells were fixed with 4% paraformaldehyde, stained with 5mg/ml crystal violet/20% methanol and counted under the microscope.
Macrophage phagocytosis
BMDMs were treated with 50 ng/ml of IL-4 in DMEM complete, non-treated condition was used as control. 6 h after treatment, cells were harvested and incubated 40 min at 37°C with latex beads (1:5000) (Polysciences Ref. # 17152-10), negative controls were incubated 40 min at 4°C. After incubation, cells were washed with 2ml MACS buffer, and read by FACS in the FITC channel.
Endothelial sprouts
BMDMs were treated with 10 ng of IL-4 in DMEM complete, non-treated condition was used as control. 24 h after treatment, macrophages were harvested and incubated overnight with HUVECs (1400 BMDMs and 1400 HUVECs) in hanging drops of 25 μL in EGM-2 medium containing methylcellulose (methylcellulose 4000 cP, Sigma-Aldrich, Bornem, Belgium) to form spheroids. Then, spheroids were harvested and embedded in collagen type I gel in a 24-well plate and cultured for 20 h to induce sprouting. Spheroids were fixed with 4% PFA and images were captured with a LEICA DM1600B inverted light microscope. The total sprout length per spheroid (cumulative length of primary sprouts and branches) was done manually using the Image J software.
Protein extraction and immunoblot
Whole cell protein extraction was performed using extraction Buffer (20 mM Tris HCI, 150 mM NaCI, 1% Triton X-100, 10% glycerol, 5 mM EDTA) supplemented with Complete Mini protease inhibitor (Roche) and PhosSTOP Phosphatase Inhibitor (Roche). A protein load (from 15-40 μg) was separated by NuPAGE® 4-12% Bis-Tris (Thermofisher) or Any kD™ Mini-PROTEAN® TGX™ (Biorad) Precast Gels and transferred electrophoretically to nitrocellulose membrane by iBIot System (Thermofisher). Nonspecific binding was blocked in Tris-CI Buffered Saline Solution with 0.05% Tween-20 (TBST) containing 10% non-fat dry milk or 5% of bovine serum albumin. The following antibodies were used: GLUD1 (Abeam, 1:3000), GS (Sigma-Aldrich, 1:5000), Pax7 (DSHB, 0.5μg/ml), MyoD (Santa Cruz, 1:500), Myogenin (DSHB, 0.2μg/ml), phospho-p70S6K (Cell signaling technology), p70S6K (Cell signaling technology, 1:1000), phospho-S6 ribosomal protein (Cell signaling technology, 1:1000), S6 ribosomal protein (Cell signaling technology, 1:1000), Vinculin (Sigma-Aldrich, 1:1000), and appropriate HRP-conjugated secondary antibodies (Cell signaling technology, 1:3000-1:10000). Signal was visualized by Enhanced Chemiluminescent Reagents (ECL, Invitrogen) or West Femto by Thermo Scientific accordingto the manufacturer’s instructions and acquired by a LAS 4000 CCD camera with ImageQuant software (GE Healthcare). Densitometry was performed by using Image J software, expressing the data as percentage of the signal for the indicated protein vs. an house-keeping control or the phosphorylated form vs. its unphosphorylated total protein.
RT-qPCR
Cells were washed in PBS, collected in RLT buffer (Qiagen) and kept at -80°C. RNA was extracted with the RNeasy Micro kit (Qiagen) according to manufacturer’s instructions. Reverse transcription to cDNA was performed with the SuperScript® III First Strand cDNA Synthesis Kit (Life Technologies) according to manufacturer’s protocol. Pre-made assays were purchased from IDT (Pax7, Mm.PT.58.12398641; Myogenin, Mm.PT.58.6732917; Pcna, Mm.PT.58.33207367; Cxcl9, Mm.PT.58.5726745; Tnfa, Mm.PT.58.12575861; Arg1, Mm.PT.58.8651372; Il10, Mm.PT.58.13531087, Mrc1, Mm.PT.47.7673017; Retnla, Mm.PT.58.43062398; Glud1, Mm.PT.58.43368019; Hprt, Mm.PT.58.32092191; Slc1a5, Mm.PT.58.33492914). cDNA, primer/probe mix and TaqMan Fast Universal PCR Master Mix were prepared in 10 μl according to manufacturer’s instructions (Applied Biosystems). Samples were loaded into an optical 96-well Fast Thermal Cycling plate (Applied Biosystems) and RT-qPCR were performed using an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems). To detect genome editing (indel) of the Slc1a5 target locus at transcriptional level, we designed a spefic primer set. The forward primer 5’-AATCCCTATCGATTCCTGTGG - 3’ anneals to the gRNA cutting site whereas, the reverse primer 5’- GAACCGGCTGATGTGTTTGG -3’ anneals to a non-targeted coding region. Thus, mutations of the gRNA target site will disrupt the amplification of the target region. cDNA, primers and PowerUp SYBR Green Master Mix were prepared in a volume of 20 μl according to manufacturer’s instructions (Applied Biosystems).
RNA sequencing
RNA concentration and purity were determined spectrophotometrically using the Nanodrop ND-1000 (Nanodrop Technologies) and RNA integrity was assessed using a Bioanalyser 2100 (Agilent). Per sample, an amount of 4 ng of total RNA was used as input for the SMART-Seq v4 Ultra Low Input RNA protocol (version “091817”) from Takara Bio USA, Inc. Subsequently, 5 ng of purified cDNA was sheared to 300bp using the Covaris M220. From the sheared material, sequencing libraries were prepared with the NEBNext Ultra DNA Library Prep Kit for Illumina (version 6.0 -2/18), according to the manufacturer’s protocol including a size selection to 250bp insert size. Sequence-libraries of each sample were finally equimolarly pooled and sequenced on 1 NextSeq500 v2 flow-cell at 1x75 bp (76-6-0-0).
Statistics
Data entry and all analyses were performed in a blinded fashion. All statistical analyses were performed using GraphPad Prism software on mean values calculated from the averages of technical replicates. Statistical significance was calculated by two-tailed unpaired t-test on two experimental conditions or two-way ANOVA when repeated measures were compared, with P<0.05 considered statistically significant. The exact P values are always reported except when P<0.0001. No values were excluded from the analyses. Sample sizes for all experiments were chosen based on previous experiences. Independent experiments were pooled and analyzed together whenever possible as detailed in figure legends. All graphs show mean values ± standard error of the mean (SEM).
Extended Data
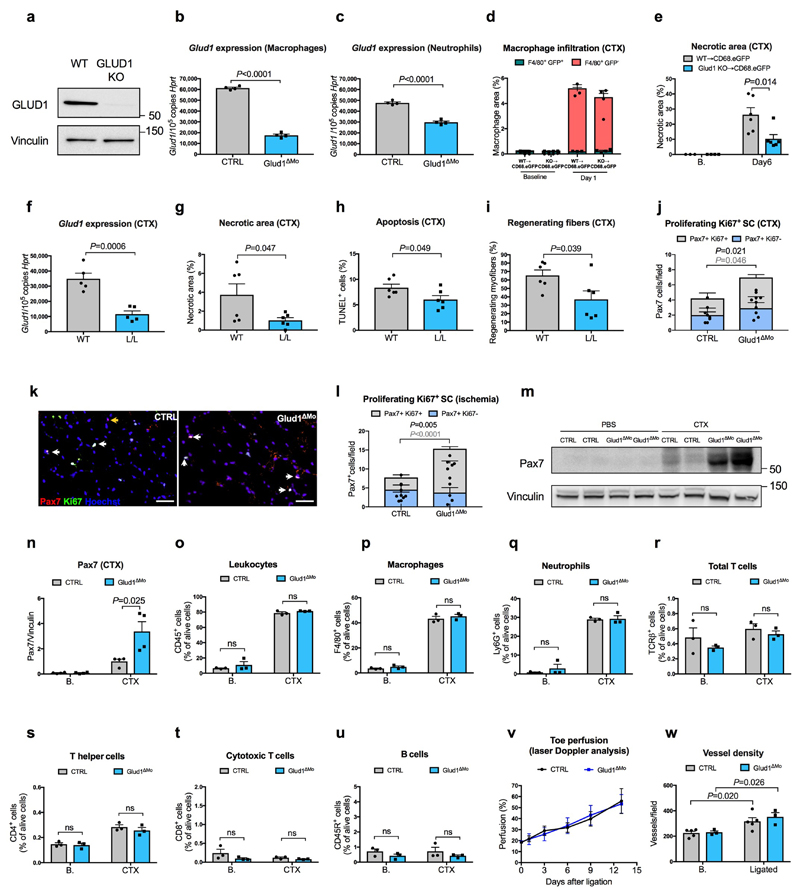
Extended Data Figure 1. Infiltrating GLUD1-deficient macrophages improve muscle repair.
a, WB for GLUD1 in BMDMs from CTRL and Glud1ΔMo mice. Vinculin was used as loading control. Representative image of 3 independent blots.
b,c, RT-qPCR of Glud1 in F4/80+ macrophages (b), and Glud1 in Ly6G+ neutrophils (c), sorted from TA muscles 1 day post-CTX (n=4).
d, Monocyte-derived macrophages (F4/80+ GFP-) and tissue-resident macrophages (F4/80+ GFP+) in TA muscles 1day post-CTX. Injured mice were CD68.eGFP transgenic mice reconstituted with WT (WT→CD68.eGFP) (n=3) or Glud1ΔMo bone marrow cells (Glud1 KO→CD68.eGFP) (n=4).
e, Necrotic area on H&E stained sections from TA muscles 6 days post-CTX. Injured mice were CD68.eGFP transgenic mice reconstituted with WT (WT→CD68.eGFP) or Glud1ΔMo bone marrow cells (KO→CD68.eGFP) (n=6). Baseline: WT→CD68.eGFP (n=3); KO→CD68.eGFP (n=4).
f, RT-qPCR of Glud1 in F4/80+ macrophages, sorted from spleens upon tamoxifen-induced macrophage-specific Glud1 deletion in Glud1L/L;CSF1R:Cre-ERT mice (L/L in short); tamoxifen injected littermates (Glud1L/L and negative for CSF1R:Cre-ERT; WT in short) were used as controls (n=5).
g-i, Quantification of necrosis (g), apoptosis (h), and regenerating fibers (i), from TA muscles 6 days post-CTX in tamoxifen-injected Glud1 L/L;CSF1R:Cre-ERT (L/L) mice and littermate controls (Glud1 L/L and negative for CSF1R:Cre-ERT; WT in short) (n=6).
j-l, Quantification of proliferating (Ki67-expressing) SC in TA muscles (j) 1 day post-CTX injury (CTR n=4, Glud1ΔMo n=5), with representative images (k), or in crural muscles (l) 3 days post-ligation (n=5). The yellow arrows indicate Pax7+ Ki67- cells, and the white arrows indicate Pax7+ Ki67+ cells.
m,n, WB for Pax7 in TA muscles lysates (m) from CTRL or Glud1ΔMo mice 1 day post-CTX (n=4), and densitometric quantification (n). Vinculin was used as loading control. Numbers represent fold change vs. Vinculin.
o-u, FACS quantification of total CD45+ leukocytes (o), F4/80+ macrophages (p), Ly6G+ neutrophils (q), TCR)+ total T cells (r), CD4+ T cells (s), CD8+ cytotoxic T cells (t), and CD45R+ B cells (u), in TA muscles at baseline or 1 day post-CTX (n=3).
v, Laser Doppler analysis 1, 3, 6, 9 and 13 days post-ligation (CTRL n=5 for all the time points; Glud1ΔMo Day0/1/3/6 n=4, Day9/13 n=3). Toe perfusion of non-ligated control was defined as 100%.
w, Quantification of vessel density in crural muscles 14 days post-ligation (CTRL n=5; Glud1ΔMo n=3).
A representative (everywhere except for n) or a pool (n) of at least two independent experiments is shown. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05). Scale bars: 50 μm (k). Graphs show mean ± SEM.
Extended Data Figure 2. GLUD1 loss in macrophages does not alter either their recruitment or M1/M2/wound-healing gene expression patterns.
a, Crystal-violet-stained bone-marrow derived macrophages (BMDMs), migrating towards CCL21, CCL2 or PBS (Uns) in glutamine (Q)-enriched or Q-reduced media (n=3).
b,c, Quantification (b) and representative images (c) of F4/80 staining in ear-sections with acetone (vehicle) or upon phorbol ester (TPA)-induced cutaneous rash, 3 days and 8 days after TPA applying (Vehicle n=4; TPA Day3 n=6,5 CTRL and Glud1ΔMo, respectively; TPA Day8 n=4).
d, Heatmap analysis of M1 and M2 macrophage gene expression in CD45+ F4/80+ macrophages sorted from TA muscles at baseline and 1 day post-CTX (n=4).
e, Heatmap analysis of wound healing gene expression in CD45+ F4/80+ macrophages sorted from TA muscles at baseline and 1 day post-CTX (n=4).
a-c experiments show representative values of 2 independent experiments, d-e show values from one single experiment. Unpaired two-tailed t-test was applied in b; ns, not significant (P>0.05). Scale bars: 50 μm (c). Graphs show mean ± SEM.
Extended Data Figure 3. GLUD1 loss in macrophages does not alter either M1/M2 polarization or their related functions.
a-d, RT-qPCR of Cxcl9 (a), Tnfa (b), Arg1 (c), and Il10 (d) in BMDMs isolated from CTRL and Glud1ΔMo mice (n=3).
e-h, FACS analysis of different M1 (e, f) or M2 (g, h) polarization states in CD45+ CD11b+ F4/80+ macrophages isolated from TA muscles at baseline (n=5) or 1 day post-CTX (n=6).
i, Quantification of macrophage phagocytosis. BMDMs were treated with LPS or PBS (unstimulated) prior to the assay (n=3).
j,k, Quantification (j), and representative images (k) of total endothelial sprout length of spheroid containing HUVEC and WT or Glud1ΔMo BMDMs. BMDMs were treated with IL4 prior to the assay; unstimulated BMDMs were used as control (Unstimulated n=7; IL4 n=8).
l-m, CD206+ F4/80+ area in TA muscles 1 day (n=5) and 6 days (n=8) post-CTX (l) or in crural muscles 3 days (CTRL n=6; Glud1ΔMo n=5), 7 days (CTRL n=7; Glud1ΔMo n=5) and 14 days (CTRL n=6; Glud1ΔMo n=5) post-ligation (m).
All experiments show representative values of at least 2 independent experiments. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05). Scale bars: 50 μm (k). Graphs show mean ± SEM.
Extended Data Figure 4. GLUD1 loss in macrophages enhances GS-mediated glutamine release.
a, Quantification (by GC-MS) of intracellular 2-oxoglutarate content in BMDMs cultured in Q-enriched or Q-reduced media (n=3).
b,c, LC-MS measurement of total cellular energy charge ([ATP + 1/2ADP]/[ATP + ADP + AMP]) (b) and ATP content (c) in BMDMs (n=3).
d, Oxygen consumption rate (OCR) in BMDMs (n=5).
e-f, Quantification of intracellular (e) and extracellular (f) glutamine content in BMDMs cultured in Q-enriched or Q-reduced media (n=3).
g, [U-14C]-glutamine uptake in BMDMs cultured in Q-enriched (n=4) or Q-reduced (WT n=4; Glud1ΔMo n=3) media.
h, Evaluation of [U-13C]-glutamine-derived carbon incorporation into glutamate in BMDMs (n=3).
i-j, Evaluation of [U-13C]-glucose-derived carbon incorporation levels into 2-oxoglutarate (i) and glutamate (j) in BMDMs (n=3).
k-l, Quantification of intracellular (k) and extracellular (l) glutamine content in BMDMs upon silencing of BCAT1 or BCAT2 (n=3).
m-n, Quantification of intracellular (m) and extracellular (n) glutamine content in BMDMs upon silencing of GOT1 or GOT2 (n=3).
o, Quantification of SC on TA muscles 1 day post-CTX injury, stained for PHH3 and Pax7. CTRL and Glud1ΔMo mice were treated 2 times per day with the BCAT1 inhibitor Gabapentin, or vehicle as control (n=6).
p, Fold change in glutamate to leucine ratio in the interstitial fluid of TA muscles 1day post-CTX, relative to PBS-injected CTRL muscle (PBS n=6; CTX n=9).
q, Fold change in glutamate to leucine ratio in the interstitial fluid of crural muscles 3 days post-ligation, relative to CTRL baseline muscle (Baseline n=7,8 CTRL, Glud1ΔMo, respectively; ligated n=11,12 CTRL, Glud1ΔMo, respectively).
r, Evaluation of the conversion of glutamate to 2-OG by analyzing [U-13C]-glutamine (Q-enriched condition) or [U-13C]-glutamate (Q-reduced condition) incorporation into 2-OG in WT BMDMs (n=3).
s, Evaluation of the conversion of 2-OG to glutamate by analyzing 15NH4 + incorporation into glutamate in WT BMDMs (n=3).
t, Evaluation of glutamine synthetase (GS) activity by analyzing 15NH4 + incorporation into glutamine in BMDMs (n=3).
u,v, Evaluation of the conversion of GLUD1 activity (u), and glutamine synthetase (GS) activity (v), in muscle-infiltrating macrophages, sorted 1 day post-CTX. One unit for the conversion of glutamate to 2-OG is the amount of enzyme that will generate 1 μmole of NADH per minute at pH 7.6 at 37°C. One unit of GS activity is defined as the enzyme producing 1 nmole of gamma-glutamyl hydroxamic acid per minute (CTRL n=4; Glud1ΔMo n=3). The control condition (CTRL) in u,v is the same one displayed in Fig. 2p at day 1.
All experiments (except for o) show representative values of at least 2 independent experiments, o shows values from one single experiment. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05); a.u., arbitrary unit. Graphs show mean ± SEM.
Extended Data Figure 5. Harnessing glutamine uptake in vitro.
a,b, Quantifications (a) and representative images (b) of myotube diameter in C2C12 cells cultured in BMDM-conditioned media (CM) (n=3 except for Q-reduced C2C12 where n=2).
c, RT-qPCR of SLC1A5 knockdown efficiency in C2C12 cells. Cells were transduced with a LV co-expressing Cas9 and a gRNA targeting the Slc1a5 locus (SLC1A5-KD) (n=5) or a non-targeting control gRNA (Ctrl gRNA) (n=4).
d, [U-14C]-glutamine uptake in SLC1A5-deficient C2C12 cells (SLC1A5 KD) generated by coexpressing Cas9 along with a gRNA targeting the Slc1a5 locus. Parental cells (CTRL) and cells transduced with a non-targeting control gRNA (Ctrl gRNA) were used as negative controls. C2C12 cells treated with SLC1A5 inhibitor gamma-L-Glutamyl-p-Nitroanilide (GPNA) were used as a positive control (n=3).
e-f, Quantification (e) and representative images (f) of myotube diameter in control or SLC1A5-KD C2C12 cells co-cultured with BMDMs under glutamine deprivation (n=3 except Ctrl C2C12 n=2).
g, RT-qPCR analysis of the proliferation marker Pcna in control or SLC1A5-KD C2C12 cells, or control C2C12 treated with the mTOR inhibitor Torin2, cultured for 18 hours in BMDM-conditioned, Q-reduced growth media, where the only glutamine present comes from WT or GLUD1 KO BMDMs. A non-targeting control gRNA (Ctrl gRNA) was used as control (n=3).
h, RT-qPCR analysis of the differentiation marker Myogenin in control or SLC1A5-KD C2C12 cells, or control C2C12 treated with the mTOR inhibitor Torin2, cultured for 72 hours in BMDM-conditioned, Q-reduced differentiation media, where the only glutamine present comes from WT or GLUD1 KO BMDMs. A non-targeting control gRNA (Ctrl gRNA) was used as control (n=3).
i, Representative images of an immunofluorescence for Pax7 on a pure SC population, freshly isolated from hindlimb muscles of WT mice.
j, RT-qPCR for Slc1a5 in SC, transduced with the same LV as above. The graph shows values of 3 biological repetitions per condition.
k-l, Quantification (k) and representative images (l) of EdU by immunofluorescence in control or SLC1A5-KD SC. A non-targeting control gRNA (Ctrl gRNA) was used as a control (Ctrl gRNA n=5; SLC1A5-KD n=6).
m-o, Quantification (m,n) and representative images (o) of fusion index and myotube size in control or SLC1A5-KD SC 5 days of culture in differentiation media. A non-targeting control gRNA (Ctrl gRNA) was used as a control. The graph shows values of 3 biological repetitions per condition.
All experiments show representative values of at least 2 independent experiments. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05). Scale bars: 50 μm (b,f,l); 100 μm (o). Graphs show mean ± SEM.
Extended Data Figure 6. Selective and inducible knockdown of Slc1a5 in SC.
a, Schematic representation of the AAV8 expression vector for in vivo targeting of SC. U6, Pol III promoter driving the expression of the gRNA targeting the Slc1a5 locus or a non-targeting control gRNA. Since the mice used in this experiment are LSL-Cas9 x PAX7:Cre-ERT mice, Cas9 is exclusively activated in Pax7+ cells upon tamoxifen administration and, genome editing of the Slc1a5 locus will occur selectively in SC.
b, Schematic overview of an AAV8-based CRISPR/Cas9-mediated in vivo genome editing.
c-d, Representative images (c) and quantification (d) for Pax7 and Cas9 staining on uninjured muscles before and after tamoxifen administration (n=4).
e-f, RT-qPCR for Slc1a5 in freshly isolated SC (n=4) (e) and all other mononuclear cells (non-SC) (n=3) (f) upon in vivo genome editing of the Slc1a5 locus (SLC1A5-KD) specific in SC. Non-targeting control gRNA (Ctrl gRNA) was used as a control.
g-h, Quantification (g) and representative images (h) of SLC1A5 and Pax7 stainings on freshly isolated SC, upon in vivo genome editing of the Slc1a5 locus (SLC1A5-KD) specific in SC (n=3).
All experiments show representative values of at least 2 independent experiments. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05). Scale bars: 50 μm (c); 20 μm (h). Graphs show mean ± SEM.
Extended Data Figure 7. Slc1a5 knockdown in SC impairs the recovery of the muscle from CTX-induced damage.
a-d, Quantification of TUNEL+ cells (a), F4/80+ area (c) and representative images (b,d) respectively, in TA muscle 6 days post-CTX obtained from LSL-Cas9 x PAX7:Cre-ERT mice treated with an AAV8 vector encoding for Ctrl gRNA (Ctrl gRNA) or Slc1a5 gRNA (SLC1A5-KD) (n=4).
e,f, Quantification (e) and representative images (f) of EdU+ myonuclei in TA muscle 6 days post-CTX, upon in vivo genome editing of the Slc1a5 locus (SLC1A5-KD) specific in SC. EdU was given by i.p. injection at 24h, 48h and 72h after CTX injection (n=6).
a-d show representative values of 2 independent experiments, e-f show values of 1 experiment. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05). Scale bars: 20 μm (b,d,f). Graphs show mean ± SEM.
Extended Data Figure 8. Macrophage-specific genetic deletion or pharmacologic inhibition of GLUD1 alters only the basal inflammation and weight of muscle tissue in aged mice.
a, Representative images of F4/80+ area in crural muscles of young and aged mice.
b-i, Quantification and representative images of F4/80+ area in brain (b,c), liver (d,e), lung (f,g), and skin (h,i) of aged mice (n=5 except in b for Glud1ΔMo n=4).
j-n, Body weight (j) and mass to body weight ratio of kidney (k), liver (l), spleen (m), and fat tissues (n) of aged mice upon R162 treatment (CTRL n=5; Glud1ΔMo n=6).
a-i show representative values of at least 2 independent experiments, j-n show values of 1 experiment. Unpaired two-tailed t-test was everywhere applied, ns, not significant (P>0.05). Scale bars: 50 μm (a,i); 20 μm (c,e,g). Graphs show mean ± SEM.
Extended Data Figure 9. GLS loss in macrophages is not advantageous for muscle repair.
a,b, [U-14C]-glutamine uptake (a) and glutamine oxidation (b) in WT or GLS KO BMDMs cultured with Q-enriched or Q-reduced media (n=3).
c,d, 2-oxoglutarate (2-OG) to succinate ratio in WT or GLS KO BMDMs (c) and 2-OG to succinate ratio in WT or GLUD1 KO BMDMs (d). BMDMs were treated with 50ng/mL LPS or PBS (unstimulated) prior to the assay (n=3).
e, Evaluation of GS activity by analyzing the percentage of the 15NH4 +-derived ammonia incorporation levels into glutamine in BMDMs isolated from CTRL and Gls ΔMo mice (n=3).
f, Fold change in glutamine to glutamate ratio in the interstitial fluid of TA muscle 1 day post-CTX, relative to PBS injected CTRL muscle (n= 6).
g,h, Quantification of necrotic (right side of the graph) and regenerating (left side of the graph) areas on H&E-stained sections from TA muscles 6 days post-CTX (n= 6) (g) and representative images (h).
i,j, Quantification (i) and representative images (j) of TUNEL+ cells in TA muscle 6 days post-CTX (n=6).
k-m, Representative images (k) and quantification of F4/80+ area (l), CD206- F4/80+ cells (M1) to CD206+ F4/80+ cells (M2) ratio (m) in TA muscles 6 days post-CTX (n=6).
n-q, RT-qPCR of Tnfa (n), Cxcl9 (o), Mrc1 (p) and Retnla (q) in BMDMs isolated from CTRL and GlsΔMo mice. BMDMs were treated with LPS or PBS (unstimulated) prior to the assay (n=3).
r, Scheme illustrating the physiological role of Glud1 in macrophages in response to muscle damage. During muscle disruption, ischemia or aging, interstitial glutamine drops likely because of the loss in myofibers (a major glutamine source) and poor blood supply. Infiltrating macrophages respond to glutamine starvation by reducing their oxidative GLUD1 activity in favour of GS activity. Macrophage-derived glutamine is released and progressively fills the muscle interstitium, where it is uptaken by SC promoting their proliferation and differentiation into new fibers, two processes that are favoured by glutamine-dependent mTOR activation. Towards the end of this regenerative process, the newly generated fibers will undertake glutamine production while inflammation will be progressively resolved. GLUD1-deficient macrophages are metabolically pre-adapted towards glutamine synthesis and release, thus preventing this glutamine drop. It follows that, in case of muscle damage, macrophage-specific knockout of Glud1 or pharmacologic GLUD1 blockade strengthens SC activation, ultimately leading to therapeutic muscle regeneration.
All experiments show representative values of at least 2 independent experiments. Unpaired two-tailed t-test was everywhere applied; ns, not significant (P>0.05). Scale bars: 20 μm (h); 10 μm (j,k). Graphs show mean ± SEM.
Extended Data Table 1. Blood count in CTRL and Glud1ΔMo mice.
The values show the haematological parameters in CTRL and Glud1ΔMo mice. Data are pooled from 2 independent experiments, CTRL n=8;Glud1ΔMo n=10. Abbreviations: white blood cell (WBC), neutrophil (NEU), lymphocyte (LYM), monocyte (MON), eosinophil (EOS), basophil (BAS), red blood cell (RBC), platelet (PLT). Values show mean ± SEM.
| Parameter | CTRL | Glud1ΔMo |
|---|---|---|
| WBC (K/μl) | 4.51±1.09 | 4.56±0.88 |
| NEU (%) | 11.49±2.11 | 12.30±2.80 |
| LYM (%) | 84.95±2.38 | 83.61±2.45 |
| MON (%) | 1.81±0.44 | 2.01±0.70 |
| EOS (%) | 0.88±0.32 | 0.93±0.32 |
| BAS (%) | 0.16±0.07 | 0.18±0.08 |
| RBC (M/μI) | 8.84±0.32 | 8.51±0.47 |
| PLT (K/μI) | 630.4±99.97 | 692±81.10 |
Supplementary Material
Acknowledgments
MM was supported by an ERC Consolidator-grant (ImmunoFit), FWO-SBO (ZL3C3602), Horizon 2020 (research and innovation program under the Marie Sktodowska-Curie grant agreement No 766214). We thank Vincent van Hoef for bioinformatic analyses; Prof. Sarah-Maria Fendt, Prof. Christian Frezza, Prof. Antonio Musaro, Prof. Giulio Cossu, Prof. Jean-Chrisophe Marine for advices; Sarah Trusso Cafarello and Sander Willox for technical support. PC and MM received long-term structural Methusalem funding by the Flemish Government; PC is supported by an ERC PoC [ERC-713758] and Advanced-grant [EU-ERC743074], MSh is granted by China Scholarship Council (CSC); E.B. by FWO (1525315N).
Footnotes
Author Contribution
MSh performed experimental design, all experiments, data acquisition and interpretation, and wrote the manuscript. FC and RA performed in vitro assays and histology. JS performed all the ligations and histological stainings. FV performed angiogenic and in vitro assays. MYR provided AAV vectors. GE performed Seahorse measurements. SC and PM generated GLUD1 cKO mice and provided critical suggestions. KDB provided the transgenic mice expressing Cre-ERT under the Pax7 promoter, and provided critical edits to the text. MSa provided critical edits to the text. PC helped in the experiments with GLS KO macrophages and provided the mice. BG and PC supported with metabolic assays and critical suggestions in manuscript writing. MDM designed and supervised all the in vitro and in vivo gene editing approaches, and provided critical edits to the text. EB performed performance experiments, histology, experimental design, data analysis, and wrote the manuscript. MM and EB performed the experimental design, data analysis, conducted scientific direction, and wrote the manuscript.
Competing Financial Interests
No competing financial interests to declare.
Data Availability
RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) data repository with accession number GSE123825. Source Data are provided for all the experiments. Other data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Bentzinger CF, Wang YX, Dumont NA, Rudnicki MA. Cellular dynamics in the muscle satellite cell niche. EMBO reports. 2013;14:1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costamagna D, Berardi E, Ceccarelli G, Sampaolesi M. Adult Stem Cells and Skeletal Muscle Regeneration. Current gene therapy. 2015;15:348–363. doi: 10.2174/1566523215666150630121024. [DOI] [PubMed] [Google Scholar]
- 3.Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. Febs J. 2013;280:4118–4130. doi: 10.1111/febs.12166. [DOI] [PubMed] [Google Scholar]
- 4.Saclier M, et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem cells. 2013;31 doi: 10.1002/stem.1288. 384396. [DOI] [PubMed] [Google Scholar]
- 5.Tidball JG. Regulation of muscle growth and regeneration by the immune system. Nature reviews. Immunology. 2017;17:165–178. doi: 10.1038/nri.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perdiguero E, et al. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J Cell Biol. 2011;195:307–322. doi: 10.1083/jcb.201104053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latroche C, et al. Coupling between Myogenesis and Angiogenesis during Skeletal Muscle Regeneration Is Stimulated by Restorative Macrophages. Stem cell reports. 2017;9:2018–2033. doi: 10.1016/j.stemcr.2017.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summan M, et al. Macrophages and skeletal muscle regeneration: a clodronate-containing liposome depletion study. American journal of physiology. Regulatory, integrative and comparative physiology. 2006;290:R1488–1495. doi: 10.1152/ajpregu.00465.2005. [DOI] [PubMed] [Google Scholar]
- 9.Rennie MJ, et al. Skeletal muscle glutamine transport, intramuscular glutamine concentration, and muscle-protein turnover. Metabolism: clinical and experimental. 1989;38:47–51. doi: 10.1016/0026-0495(89)90140-6. [DOI] [PubMed] [Google Scholar]
- 10.Biolo G, Fleming RY, Maggi SP, Wolfe RR. Transmembrane transport and intracellular kinetics of amino acids in human skeletal muscle. Am J Physiol. 1995;268:E75–84. doi: 10.1152/ajpendo.1995.268.1.E75. [DOI] [PubMed] [Google Scholar]
- 11.Nurjhan N, et al. Glutamine: a major gluconeogenic precursor and vehicle for interorgan carbon transport in man. J Clin Invest. 1995;95:272–277. doi: 10.1172/JCI117651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmieri EM, et al. Pharmacologic or Genetic Targeting of Glutamine Synthetase Skews Macrophages toward an M1-like Phenotype and Inhibits Tumor Metastasis. Cell Rep. 2017;20:1654–1666. doi: 10.1016/j.celrep.2017.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St Pierre BA, Tidball JG. Differential response of macrophage subpopulations to soleus muscle reloading after rat hindlimb suspension. J Appl Physiol (1985) 1994;77:290–297. doi: 10.1152/jappl.1994.77.1.290. [DOI] [PubMed] [Google Scholar]
- 14.Guardiola O, et al. Induction of Acute Skeletal Muscle Regeneration by Cardiotoxin Injection. Journal of visualized experiments : JoVE. 2017 doi: 10.3791/54515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda Y, et al. Macrophage skewing by Phd2 haplodeficiency prevents ischaemia by inducing arteriogenesis. Nature. 2011;479:122–126. doi: 10.1038/nature10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zammit PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. 2017;72:19–32. doi: 10.1016/j.semcdb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Wenes M, et al. Macrophage Metabolism Controls Tumor Blood Vessel Morphogenesis and Metastasis. Cell metabolism. 2016;24:701–715. doi: 10.1016/j.cmet.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Yang C, et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers JT, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, et al. mTOR is necessary for proper satellite cell activity and skeletal muscle regeneration. Biochem Bioph Res Co. 2015;463:102–108. doi: 10.1016/j.bbrc.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jewell JL, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347:194–198. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rayagiri SS, et al. Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03425-3. 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa-Victor P, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 25.Bernet JD, et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin L, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu PS, et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nature immunology. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 28.Obara H, Matsubara K, Kitagawa Y. Acute Limb Ischemia. Ann Vasc Dis. 2018;11:443–448. doi: 10.3400/avd.ra.18-00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayer AA, et al. New horizons in the pathogenesis, diagnosis and management of sarcopenia. Age Ageing. 2013;42:145–150. doi: 10.1093/ageing/afs191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinciguerra M, Musaro A, Rosenthal N. Regulation of muscle atrophy in aging and disease. Adv Exp Med Biol. 2010;694:211–233. doi: 10.1007/978-1-4419-7002-2_15. [DOI] [PubMed] [Google Scholar]
- 31.Carobbio S, et al. Deletion of glutamate dehydrogenase in beta-cells abolishes part of the insulin secretory response not required for glucose homeostasis. J Biol Chem. 2009;284:921–929. doi: 10.1074/jbc.M806295200. [DOI] [PubMed] [Google Scholar]
- 32.He Y, et al. Glutamine synthetase deficiency in murine astrocytes results in neonatal death. Glia. 2010;58:741–754. doi: 10.1002/glia.20960. [DOI] [PubMed] [Google Scholar]
- 33.Mingote S, et al. Genetic Pharmacotherapy as an Early CNS Drug Development Strategy: Testing Glutaminase Inhibition for Schizophrenia Treatment in Adult Mice. Front Syst Neurosci. 2015;9:165. doi: 10.3389/fnsys.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guardiola O, et al. Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc Natl Acad Sci USA. 2012;109:E3231–3240. doi: 10.1073/pnas.1204017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaFleur MW, et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09656-2. 1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasut A, Jones AE, Rudnicki MA. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J Vis Exp. 2013:e50074. doi: 10.3791/50074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casazza A, et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrpl signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) data repository with accession number GSE123825. Source Data are provided for all the experiments. Other data that support the findings of this study are available from the corresponding author upon reasonable request.