Abstract
The mammalian liver is a central hub for systemic metabolic homeostasis. Liver tissue is spatially structured, with hepatocytes operating in repeating lobules, and sub-lobule zones performing distinct functions. The liver is also subject to extensive temporal regulation, orchestrated by the interplay of the circadian clock, systemic signals and feeding rhythms. However, liver zonation was previously analyzed as a static phenomenon, and liver chronobiology at tissue level resolution. Here, we use single-cell RNA-seq to investigate the interplay between gene regulation in space and time. Using mixed-effect models of mRNA expression and smFISH validations, we find that many genes in the liver are both zonated and rhythmic, most of them showing multiplicative space-time effects. Such dually regulated genes cover key hepatic functions such as lipid, carbohydrate and amino acid metabolism, but also previously unassociated genes, such as protein chaperones. Our data also suggest that rhythmic and localized expression of Wnt targets could be explained by rhythmically expressed Wnt ligands from non-parenchymal cells near the central vein. Core circadian clock genes are expressed in a non-zonated manner, indicating that the liver clock is robust to zonation. Together, our scRNA-seq analysis reveals how liver function is compartmentalized spatio-temporally at the sub-lobular scale.
Introduction
The liver is a vital organ maintaining body physiology and energy homeostasis. The liver carries out a broad range of functions related to carbohydrate and lipid metabolism, detoxification, bile acid biosynthesis and transport, cholesterol processing, xenobiotics biotransformation, and carrier proteins secretion. Notably, the liver performs catabolic and anabolic processing of lipids and amino acids and produces the majority of plasma proteins1. Liver tissue is highly structured on the cellular scale, being heterogeneous in both cell-type composition and microenvironment2. In fact, liver tissue is made up of millions of repeating anatomical and functional subunits, called lobules, which in mice contain hepatocytes arranged in about 15 concentric layers with a diameter of about 0.5mm3,4. On the portal side of the lobule, blood from the portal vein and the hepatic arteriole enters small capillaries called sinusoids and flows to the central vein. This is accompanied with gradients in oxygen concentration, nutrients and signaling along the portal-central axis, with the latter notably involving the Wnt pathway5,6. Due to this polarization, hepatocytes in different layers perform separate functions. This is accompanied with gradients of gene expression along the portal-central axis, with some genes expressed more strongly near the central vein, and vice versa for portally expressed genes. This phenomenon is termed liver zonation 1,7.
Recently, Halpern et al. combined single-cell RNA-sequencing (scRNA-seq) of dissociated hepatocytes and single-molecule RNA fluorescence in situ hybridization (smFISH) to reconstruct spatial mRNA expression profiles along the portal-central axis8. This analysis revealed an unexpected breadth of spatial heterogeneity, with ~50% of genes showing spatially non-uniform patterns. Among them, functions related to ammonia clearance, carbohydrate catabolic and anabolic processes, xenobiotics detoxification, bile acid and cholesterol synthesis, fatty acid metabolism, targets of the Wnt and Ras pathways, and hypoxia-induced genes were strongly zonated.
In addition to its spatial heterogeneity, liver physiology is also highly temporally dynamic. Chronobiology studies showed that temporally gated physiological and metabolic programs in the liver result from the complex interplay between the endogenous circadian liver oscillator, rhythmic systemic signals, and feeding/fasting cycles9,10,11. An intact circadian clock has repeatedly been demonstrated as key for healthy metabolism, also in humans12. In addition, the hepatocyte clock has specifically been shown to play a major role in the physiological coordination of nutritional signals and cell-cell communication (including non-hepatocytic cells) controlling rhythmic metabolism13. Temporal compartmentalization can prevent two opposite and incompatible processes from simultaneously occurring, for example, glucose is stored as glycogen following a meal and is later released into the blood circulation during fasting period to maintain homeostasis in plasma glucose levels. Functional genomics studies of the circadian liver were typically performed on bulk liver tissue14. In particular, several studies showed how both the circadian clock and the feeding fasting cycles pervasively drive rhythms of gene expression in bulk, impacting key sectors of liver physiology such as glucose homeostasis, lipid and steroid metabolism15–18.
Here, we asked how these spatial and temporal regulatory programs interact on the level of individual genes and liver functions more generally. In particular, can zonated gene expression patterns be temporally modulated on a 24 h time scale? And conversely, can rhythmic gene expression patterns observed in bulk samples exhibit sub-lobular structure? More complex situations may also be envisaged, such as time-dependent zonation patterns of mRNA expression (or, equivalently, zone-dependent rhythmic patterns), or sublobular oscillations that would escape detection on the bulk level due to cancelations. On the physiological level, it is of interest to establish how hepatic functions might be compartmentalized both in space and time. To study both the spatial and temporal axes, we performed scRNA-seq of hepatocytes at four different times along the 24 h day, extending a previous approach8,19 to reconstruct spatial profiles at each time point. The resulting space-time patterns were statistically classified using a mixed-effect model describing both spatial and temporal variations in mRNA levels. In total, ~5000 liver genes were classified based on their spatio-temporal expression profiles, and a few representative profiles were further analyzed with smFISH. Overall, this approach revealed the richness of space-time gene expression dynamics of the liver and provides a comprehensive view on how spatio-temporal compartmentalization is utilized at the sub-lobular scale in the mammalian liver.
Results
Single-cell RNA-seq captures space-time gene expression patterns in mouse liver
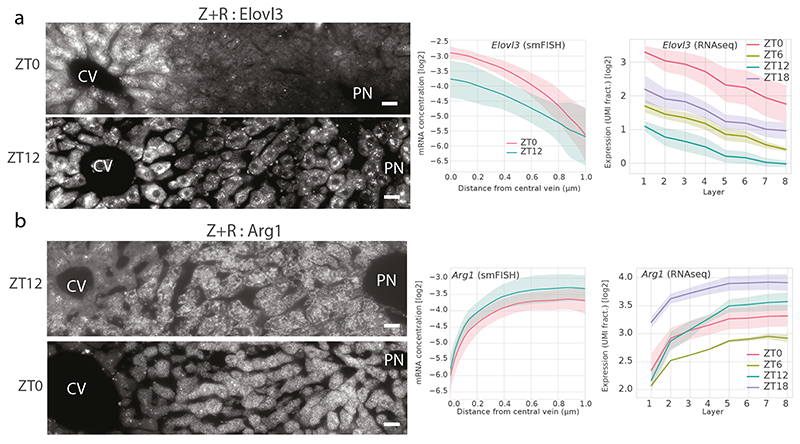
To investigate spatio-temporal gene expression patterns in mouse liver, we sequenced mRNA from individual liver cells obtained via perfusion from 10 ad libitum fed mice at 4 different times of the day (ZT = 0h, 6h, 12h and 18h, two to three replicates per time point). The interval between ZT0 and ZT12 has the light on and corresponds to the fasting period in mice, while feeding happens predominantly between ZT12 and ZT0. We here focused on hepatocytes by enrichment of cells according to size and in silico filtering, yielding a total of 19663 cells (Methods). To validate that the obtained scRNA-seq data captured the expected variability in both spatial and temporal mRNA levels, we generated a clustering analysis of all cells using a standard 2-dimensional t-SNE dimensionality reduction (Methods) and colored cells either by their positions along the central-portal axis (the a posteriori assigned layers, see below) (Figure 1a) or time (Figure 1b). The clustering revealed that portally and centrally expressed landmark transcripts, such as the cytochrome P450 oxygenases Cyp2f2 and Cyp2e1 involved in xenobiotics metabolism, mark cells in opposite regions of the projections (Figure 1c-d). Likewise, time-of-day gene expression varied along an orthogonal direction (Figure 1b), as shown for the fatty acid elongase Elovl3 peaking at ZT0 (Figure 1e).
Figure 1. A scRNA-seq approach to space-time gene expression in mouse liver.
(a-e) Global gene expression varies in both space and time, as shown using t-SNE visualizations of the scRNA-seq (n=19663 hepatocytes examined over ten independent animals). Each dot represents one cell. Individual cells are colored by the (a posteriori assigned) lobule layer (a), Zeitgeber time (b), expression levels of the zonated genes Cyp2f2 and Cyp2e1 (c-d), or the temporally-regulated and centrally zonated gene Elovl3 (e). (f-h) Reconstructed spatial profiles (lobule layers 1-8) of selected zonated genes (f, top: Glul pericentrally (PC) expressed, bottom: Ass1 periportally (PP) expressed); rhythmic but non-zonated genes (g, top: core-clock gene Bmal1 peaking at ZT18-0, bottom: clock-controlled Dbp, peaking at ZT6-12); zonated and rhythmic genes (h, Top: Elovl3, bottom: Pck1). Expression levels correspond to fraction of total UMI per cell in linear scale. Log-transformed profiles are in Extended Data Figure 1. Dots in f-h represent data points from the individual mice. Lines represent the mean expression per time point. Shaded areas represent one standard deviation (SD) across the mice. For the scRNA-seq we used n=2 (ZT6, ZT18) or n=3 mice (ZT0, ZT12) (Methods).
To obtain spatial mRNA expression profiles for each gene along the central-portal axis, we here introduced eight lobule layers, to which we assigned each individual cell. For this, we adapted a previous method that uses expression levels of landmark zonated genes to define a central-portal coordinate19, with the modification that only landmark transcripts that were sufficiently expressed and that did not vary across mice and time points were used (27 central and 28 portal landmark genes, Methods). The resulting reconstructed (binned) mRNA expression profiles yielded 80 (8 layers over 10 mice) data points for each transcript. Although our resolution is lower compared with the typically 12-15 hepatocyte layers found in the liver3,4, these reconstructions faithfully captured reference zonated genes, with both central, and portal, expression (Figure 1f). Two examples of such genes are the centrally expressed glutamine synthethase (Glul), and the portally expressed urea cycle gene argininosuccinate synthetase (Ass1), showing mutually exclusive expression along the lobule8. The reconstruction also successfully identified transcripts of the core circadian clock, such as the master transcription factor Bmal1 (also named Arntl), whose mRNA peaked between ZT18-ZT0 (Figure 1g)20. In addition to core clock genes, important clock outputs such as the PAR bZip transcription factor Dbp, which is a direct transcriptional target of BMAL1 regulating detoxification enzymes, peaked between ZT6-12 (Figure 1g)21. Finally, genes showing both zonated and rhythmic mRNA accumulation were found (Figure 1h), for example elongation of very long chain fatty acids 3 (Elovl3) is centrally expressed and peaks near ZT0, while phosphoenolpyruvate carboxykinase 1 (Pck1) regulating gluconeogenesis during fasting is expressed portally and peaks shortly before ZT12. Since most of the zonated profiles showed exponential shapes, and gene expression changes typically occur on a log scale22, we log-transformed the data for further analysis (Methods, Figure 1f and Extended Data Figure 2a-b-c). Together, these examples indicate that the obtained gene expression profiles reliably capture spatial and temporal regulation of hepatocyte gene expression.
mRNA expression profiles categorized according to zonation and rhythmicity
To gain a systematic understanding of the space-time gene expression profiles, we next investigated if zonated gene expression patterns could be dynamic along the day, or conversely whether temporal expression patterns might be zone-dependent. To select a reliable set of reconstructed mRNA expression profiles for subsequent analyses, we filtered out lowly expressed genes as well as genes with significant biological variability across replicate liver samples, although this may be at the expense of a potentially decreased sensitivity (Methods). This yielded 5058 spatio-temporal gene expression profiles (Extended Data Figure 2d). An exploratory analysis of variance clearly identified zonated genes, rhythmic genes, and fewer genes showing variability along both axes, with known zonated and rhythmic genes distributed as expected (Figure 2a).
Figure 2. Space-time mRNA expression profiles categorized with mixed-effect models.
(a) Spatial and temporal variations for mRNA transcript profiles, calculated as standard deviations (SD) of log2 expression along spatial or temporal dimensions. Colored dots correspond to reference zonated genes (orange) and reference rhythmic genes (blue) (Methods). (b) Extended harmonic regression model for spatio-temporal expression profiles describing a static but zonated layer-dependent mean μ(x), as well as layer-dependent harmonic coefficients (a(x) and b(x)). All layer-dependent coefficients are modeled as second order polynomials; i denotes the biological replicates. Temporal dependency is modelled with 24h-periodic harmonic functions. μi are random effects needed due to the data structure hierarchy (Methods). (c) Schema illustrating the different categories of profiles. Depending on which coefficients are non-zero (Methods), genes are assigned to: F/N (flat or noisy, not represented), Z (zonated), R (rhythmic), Z+R (additive zonation and rhythmicity), ZxR (interacting zonation and rhythmicity). Graphs emphasize either zonation (top), with the x-axis representing layers, or rhythmicity (bottom), with the x axis representing time (ZT). Right side of the panel: two examples of fits (Elovl3 and Cyp7a1, respectively Z+R and ZxR). (d) Number of transcripts in each category. (e) Boxplot of the mean expression per category shows that zonated genes (Z, Z+R and ZxR) are more expressed than rhythmic (R) or flat/noisy (F/N). ZxR genes are the most expressed according to median expression (orange line). Box limits are lower and upper quartile, whiskers extend up to the first datum greater/lower than the upper/lower quartile plus 1.5 times the interquartile range. The number of genes per category is indicated. Remaining points are outliers. KW stands for the Kruskal-Wallis and MW for the Mann-Whitney (two-sided) tests.
To identify possible dependencies between spatial and temporal variations, we built a mixed-effect linear model23 for the space-time mRNA profiles, which extends harmonic regression to include a spatial covariate (Figure 2b). In this model, rhythms are parameterized with cosine and sine functions, while spatial profiles are represented with (up to second order) polynomials. In its most complex form, the model uses nine parameters describing spatially modulated oscillations, and one intercept per mouse (Methods). When some of the parameters are zero, the model reduces to simpler mRNA profiles, for example purely spatial or purely temporal expression profiles (Figure 2c). We then used model selection24 to identify the optimal parameterization and category for each gene (Methods). In fine, we classified each mRNA profile into one of five types of patterns (Figure 2c). If only the intercept is used, the profile will be classified as flat or noisy (F/N, Methods). If only time-independent zonation parameters are retained, the predicted profile will be purely zonated (Z). If only layer-independent rhythmic parameters are retained, the predicted profile will be purely rhythmic (R). If only layer-independent rhythmic parameters and time-independent zonation parameters are retained, the profile is classified as independent rhythmic-zonated (Z+R). If at least one layer-dependent rhythmic parameter is selected, the profile will be termed interacting (ZxR). This classification revealed that, overall, about 30% of the mRNA profiles were zonated (Z, Z+R and ZxR) and about 20% were rhythmic (R, Z+R and ZxR) (Figure 2d). The peak times of these rhythmic transcripts were highly consistent with bulk chronobiology data25 (Extended Data Figure 2e). The entire analysis can be browsed as a web-app resource along with the corresponding data (https://czviz.epfl.ch).
Interestingly, we found that 7% of the analyzed genes in the liver were both zonated and rhythmic. Such dually regulated transcripts represent 25% of all zonated transcripts, and 36% of all rhythmic transcripts, respectively. For example, the previously shown Elovl3 transcript, involved in fatty acid elongation, and Pck1, a rate limiting enzyme in gluconeogenesis, are prototypical Z+R genes (Figure 1h, Extended Data Figure 2c). Gluconeogenesis is an energetically-demanding task3. As mice are in a metabolically fasted state requiring glucose production towards the end of the light phase (~ ZT10) and oxygen needed for ATP production is most abundant portally26, this process is indeed both spatially and temporally regulated. The dual regulation of zonated-rhythmic genes may therefore ensure optimal liver function under switching metabolic conditions.
Dually regulated genes were mostly Z+R, with only a minority of ZxR patterns. The average expression across categories showed that rhythmic genes are significantly less expressed on average than genes in zonated categories, likely reflecting shorter half-lives (Figure 2e and Extended Data Figure 2f). Surprisingly, we find few highly expressed flat genes. Together, we found that mRNA expression of many zonated genes in hepatocytes is not static, and is in fact compartmentalized both in space and time.
Properties of dually zonated and rhythmic mRNA profiles
The majority of dually regulated genes are Z+R, which denotes additive (in log) space-time effects, or dynamic patterns where slopes or shapes of spatial patterns do not change with time (Figure 2c). On the other hand, interacting patterns (ZxR) are rare. Comparing the proportions of central, mid-lobular (peaking in the middle of the portal-central axis) and portal genes among the purely zonated genes (Z), and independently zonated and rhythmic genes (Z+R), did not reveal significant differences (Figure 3a), suggesting that rhythmicity is uncoupled with the direction of zonation. Similarly, comparing the phase distribution among the purely rhythmic genes (R) and the Z+R genes did not show a significant difference (Figure 3b), indicating that zonation does not bias peak expression time. Moreover, oscillatory amplitudes were uncorrelated with the zonation slopes in Z+R genes (Figure 3c). Finally, for ZxR genes with potentially more complex space-time patterns, we investigated the spreads in amplitudes and peak times across the layers (Figure 3d). For wave-like pattern (phase modulated profiles), the phase difference across the lobule was up to 3h, which corresponds to a difference in time between neighboring hepatocytes on the order of 10 minutes for lobules of about 15 cell layers. On the other hand, amplitude modulated patterns showed up to 2-fold difference in oscillatory amplitude across the lobule.
Figure 3. Properties of dually zonated and rhythmic mRNA profiles.
(a) Proportions of pericentral (green) and periportal (blue) transcripts are similar in Z and Z+R. Mid-lobular genes (orange) are rare (<2%). The number of genes are N = 484, 14, 628, and 157, 6, 184 for the portal, mid-lobular, and central genes in Z and Z+R, respectively. KS stands for the two-sided Kolmogorov-Smirnov test. (b) Peak time distributions of rhythmic transcripts are similar in R and Z+R categories (two-sample Kuiper test). (c-d) Effect sizes of zonation (slope) vs. rhythmicity (fold-change amplitude, log2 peak-to-trough) in Z+R genes (c). Magnitude of time shifts (delta phase, in hours) vs. fold-change amplitude gradient (delta amplitude, in log2) along the central-portal axis in ZxR genes (d). Genes for which the protein is also rhythmic (p<0.05, harmonic regression, F-test) in bulk data are indicated in dark red with the corresponding label (represented in Extended Data Figure 3).
To assess the potential physiological role of dually zonated and rhythmic transcripts, we asked if protein levels of the identified Z+R and ZxR genes accumulated rhythmically in a previous proteomics experiment27. In general, proteins rhythms are fewer, damped, and time-delayed compared to mRNA rhythms due to protein half-lives14,27,28 (Discussion). However, while R transcripts were twice more frequent than Z+R transcripts, the proportions were inverted for rhythmic proteins. Indeed, we found that among 65 rhythmic proteins (with q-value<0.2 in ref.27), 18 corresponded to Z+R and 10 to R transcripts. Moreover, the identified Z+R and ZxR genes with rhythmic protein accumulation cover key hepatic and zonated functions (Extended Data Figure 3, for a functional interpretation, see below) and include rate limiting enzymes. For example, for Z+R transcripts (Extended Data Figure 3a), PCK1 (rate limiting for gluconeogenesis), LPIN2 (Lipin2, catalyzes the conversion of phosphatidic acid to diacylglycerol during triglyceride, phosphatidylcholine and phosphatidylethanolamine biosynthesis), POR (cytochrome P450 oxidoreductase, required to activate P450 enzymes), DNAJA1 (HSP40 co-chaperone), ALAS1 (rate-limiting for heme biosynthesis), GNE (rate-limiting in the sialic acid biosynthetic pathway), THRSP (biosynthesis of triglycerides from medium-length fatty acid chains), show robust rhythms at the protein level. Similarly, for ZxR proteins, CYP7A1 (rate limiting enzyme in bile acid synthesis), CYP2A5 (coumarin 7-hydroxylase), SLC1A2 (high-affinity glutamate transporter), and multidrug resistance protein ABCC2 show rhythms on the protein level (Extended Data Figure 3b). Moreover, the protein rhythms accompanying those Z+R and ZxR transcripts peak with an expected delay of maximally about 6 hours28 compared to the mRNA peak times (Extended Data Figure 3c).
smFISH analysis of space-time mRNA counts
To substantiate the RNA-seq profiles, we performed RNA single molecule fluorescence in situ hybridization (smFISH) experiments on a set of selected candidate genes with diverse spatio-temporal patterns. smFISH provides a sensitive and independent, albeit low-throughput, measurement of mRNA expression. Purely zonated genes (Z) were already well studied with smFISH8. To analyze the core-clock, we measured two genes peaking at different times, Bmal1 and Per1, which were classified as R in the RNA-seq analysis. Bmal1 (~ZT0) and Per1 (~ZT12) phases were nearly identical in both experiments, and the rhythms did not depend on the lobular position consistent with R genes (Figure 4a). We analyzed three genes classified as Z+R: Pck1 was indeed both portally biased and rhythmic in RNA-seq and smFISH (Figure 4b); Elovl3 is both centrally biased and rhythmic in RNA-seq and smFISH, even though the amplitude of the oscillations was damped on the portal side in the FISH experiment (Extended Data Figure 4a); and for Arg1 (Arginase 1) the portal RNA-seq and smFISH profiles matched well (Extended Data Figure 4b). Finally, Acly showed a pattern in smFISH data which validates its classification as ZxR, with a lower amplitude on the portal side, where the transcript is more highly expressed (Figure 4c). Thus, overall, the reconstructed scRNA-seq and smFISH profiles were consistent, with minor discrepancies.
Figure 4. smFISH analysis of rhythmic and zonated transcripts.
(a) smFISH (RNAscope, Methods) of the core clock genes Bmal1 and Per1 (both assigned to R) in liver slices sampled every 3 hours. Left: representative images at ZT0, ZT06, ZT12 and ZT18 for Bmal1. Central vein (CV) and a portal node (PN) are marked. Scale bar is 50μm. Endothelial cells lining the PC and cholangiocytes surrounding the PP were excluded from the quantification. mRNA transcripts and nuclei were detected in PN and PC zones (Methods). Right: temporal profiles of Bmal1 and Per1 from smFISH at 8 time points from ZT0 to ZT21, every 3 hours (the line shows the mean number of quantified mRNAs, shaded area indicate SD across 5-8 images from one animal per time point), and scRNA-seq (same representation as in Figure 1f-h, PC is layer 1, PN is layer 8), both in PN and PC regions. (b-c) smFISH (Stellaris, Methods) for Pck1 (Z+R) and Acly (ZxR) in liver slices sampled every 12 hours. Left: representative images at ZT0, ZT12 for Pck1 (b) or Acly (c). Central vein (CV) and a portal node (PN) are marked. Scale bar - 20μm. Right: quantified profiles for each gene in the two time points from smFISH (the line shows the mean number of quantified mRNAs, shaded areas indicate SD across at least 10 independent images taken from at least 2 mice per time point) and scRNA-seq (same representation as in Figure 1f-h). As above, the scRNA-seq used n=2 (ZT6, ZT18) or n=3 mice (ZT0, ZT12).
Space-time logic of hepatic functions
We next used our classification to explore the spatio-temporal dynamics of hepatic functions and signaling pathways in the liver. Given the prevalence of zonated gene expression profiles, we first analyzed if the circadian clock is sensitive to zonation. We found that profiles of reference core-clock genes (Extended Data Figure 5) were assigned to the rhythmic only category (R), except for Cry1 and Clock that were assigned to Z+R, but with high probabilities also for R (Supplementary Table 2). This suggests that the circadian clock is largely non-zonated, as also seen in the smFISH (Figure 4a), and therefore robust to the heterogenous hepatic microenvironment.
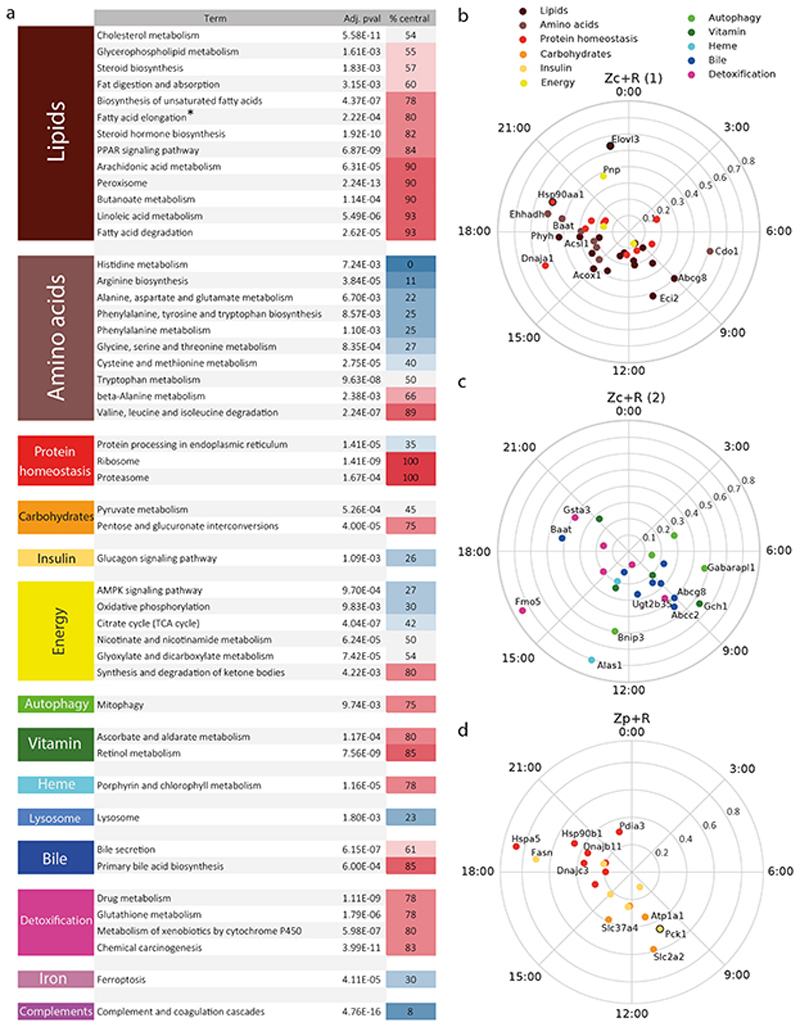
We then systematically explored enrichment of biological functions in the zonated category by querying the KEGG pathways database (Supplementary Table 3, Figure 5, Methods). In addition to recapitulating well documented zonated liver functions8,29, which we do not discuss here, this analysis highlighted Z and Z+R functions which, to our knowledge, had not been linked with liver zonation. For instance, we found that cytosolic chaperones accumulate centrally, while the endoplasmic reticulum (ER) chaperones linked with protein secretion accumulate portally (Figure 5b,d, Extended Data Figure 6). Both groups of chaperones peak during the activity/feeding phase, probably due to body temperature rhythms peaking during the active phase30,31, and likely reflect increased needs of protein folding during times of high protein synthesis. Also, we found that mRNAs of ribosomal protein genes accumulate centrally (Z), as do proteasome components (Figure 5a), the latter also containing rhythmic members (Z+R). In the liver, ribosomal proteins are rate-limiting for the synthesis of ribosomes, which themselves are rate-limiting for the synthesis of proteins32. Therefore, the overall protein synthesis rate is likely higher in these hepatocytes. Conversely, transcripts encoding components of the proteasome are involved in protein degradation. Together, these observations suggest that protein turnover is higher in centrally located hepatocytes, which are exposed to an environment with high concentrations of xenobiotics and hypoxic stress33.
Figure 5. Space-time logic of compartmentalized hepatic functions for Z+R genes.
(a) KEGG analysis of the Z and Z+R genes. For clarity, only KEGG pathways (second column) with adjusted p-value < 0.01 (standard Enrichr67 output test) are presented (Supplementary Table 3 for all enriched functions). The percentage of central genes is represented by a blue-red gradient. *: appears central due to the KEGG annotation system; however, fatty acid elongation is biased portally (see text). (b-c-d) KEGG analysis of Z+R genes. Representations of genes in central (b-c) and portal (d) enriched Z+R categories (Supplementary Table 3). Polar representation: peak expression times are arranged clockwise (ZT0 on vertical position) and amplitudes (log2, values indicated on the radial axes) increase radially. The radius coordinate of genes with an amplitude >0.9 is halved (indicated with a black circle around the colored dot).
In addition, while many mitophagy genes are expressed centrally (Z), some of those also show robust temporal rhythms peaking during the fasting period (Z+R), in particular two gamma-aminobutyric acid receptor-associated proteins (Gabarap and Gabarapl1) with an important function in autophagosome mediated autophagy34. Consistent with this temporal regulation, we had previously reported that the nuclear abundance of the fasting-dependent regulators of autophagy TFEB and ZKSCAN3 peaked near ZT635. Moreover, the centrally and synchronously accumulating Ubiquitin B mRNA (Ubb, Z+R) may contribute to triggering mitophagy36. Thus, centrally biased mitophagy may both participate in removal of damaged mitochondria in the stressed central environment.
Similarly, genes involved in bile acid synthesis and bile secretion are known to show zonated expression patterns8 (Figure 5c). Here, we found that while the rate-limiting enzyme in the bile acid biosynthetic pathway Cyp7a1 (ZxR, Figure 2c) is known to be clock-controlled, with its mRNA37,38 and protein39 (Extended Data Figure 7d) expressed maximally early during the feeding period, the ABC transporters Sterolin-1 and 2 (ABCG5/ ABCG8, both identified as Z+R) which excrete most of the biliary cholesterol40 peak towards the end of the fasting period near ZT9.
Many detoxification enzymes of the cytochromes P450 (CYPs) superfamily are known to be centrally zonated in the liver8 and several of those are found in the Z+R category. In particular, the Flavin-containing monooxygenases Fmo1, 2, and 5, which are NADPH-dependent monooxygenases involved in drug and xenobiotics detoxification, exhibit Z+R mRNA patterns with peaks near ZT16 (Figure 5c). Also, the FMO5 protein accumulates rhythmically (Figure 3c, Extended Data Figure 3). Moreover, the rate limiting enzyme ALAS1 producing the P450 cofactor heme was found as a centrally zonated Z+R transcript with peak mRNA at ZT13, showing also a robust rhythm in protein expression (Extended Data Figure 3).
To substantiate the above finding on heat shock genes, we examined the space-time behavior of temperature-regulated genes. To this end, we considered targets (bound in ChIP-seq) of the heat shock transcription factor HSF1 (Chip-Atlas41, includes our own liver data16). This showed that several targets, known to peak during the active phase30,31, are also zonated (Extended Data Figure 6). Notably, the cytoplasmic Hsp90aa1 (Hsp90A chaperone) and its interactor Dnaja1 (HSP40), respectively mitochondrial Hspd1 (HSP60), chaperones are expressed centrally where protein turnover is high. On the contrary, endoplasmic reticulum (ER) located chaperones: Hspb1 (Hsp90B) and interactor Pdia3, as well as Hspa5 (HSP70) and ER-resident Dnajc3 and Dnajb11 (DNAJ/HSP40) are expressed portally, consistent with their role in folding proteins in the secretory pathway (secretion is known to occur portally29). On the other hand, the analysis of cold induced genes (i.e. CIRBP and analogous, taken from ref.42) did not show zonated gene expression.
Finally, we note that among all KEEG pathway related to lipids, a majority show central enrichment (Figure 5a, Supplementary Table 3). Inspection of the genes involved shows that this is due to the large number of genes related to peroxisomal β-oxidation, i.e. lipid catabolism, which are incidentally also listed in biosynthesis KEGG pathways (Table S3). However, fatty acid synthesis is biased portally, as supported by key portally expressed genes such as Fasn, Srebf1, Acly, Acaca, Elovl2, Elovl5, and which is consistent with the fact that oxygen needed for mitochondrial β-oxidation is most abundant portally26. On the other hand, Elovl3, which is known to be transcriptionally controlled by the peroxisome regulator PPARα and atypically regulated among Elov family fatty acid elongases43 is expressed centrally.
Space-time logic of activity of signaling pathways
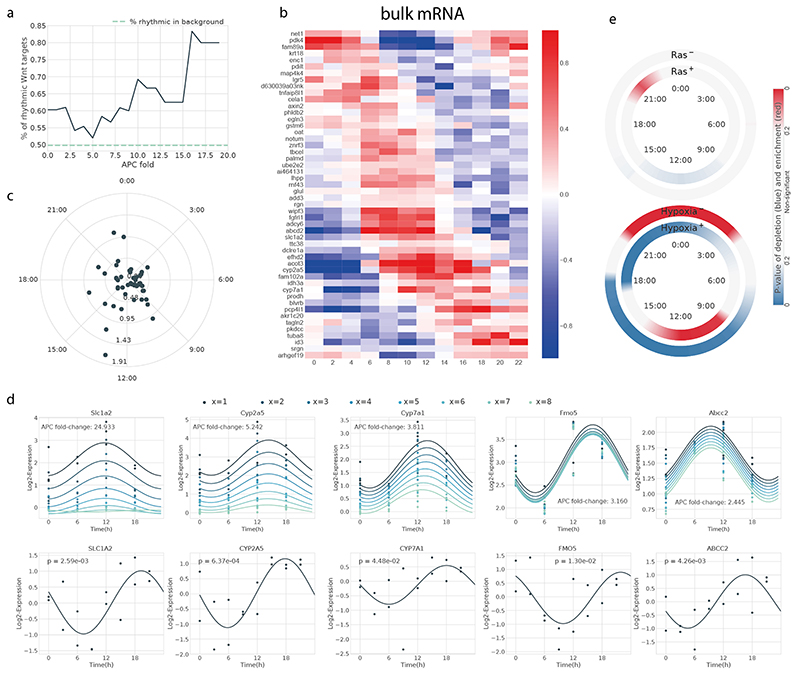
Signaling pathways that include Wnt, Ras and hypoxia have been shown to shape hepatocyte zonation8. We therefore examined the space-time activities of these pathways, extracted from the behavior of canonical target genes. We mainly focused on Wnt, as it is often considered the master regulator of liver zonation44. To systematically investigate spatio-temporal WNT/β-Catenin activity in liver, we extracted a set of Wnt targets derived from an APC KO in mouse liver8,45. We found that rhythmic transcripts (in the R, Z+R and ZxR categories) are enriched among targets of the Wnt pathway, showing a proportion that increases with the strength of the targets, with the strongest Wnt targets containing 80% of rhythmic transcripts (Extended Data Figure 7a). Positive Wnt targets were pericentral8 and peaked between ZT9 and ZT12, whereas negative Wnt targets were periportal8 and peaked between ZT21 and ZT3 (Figure 6a).
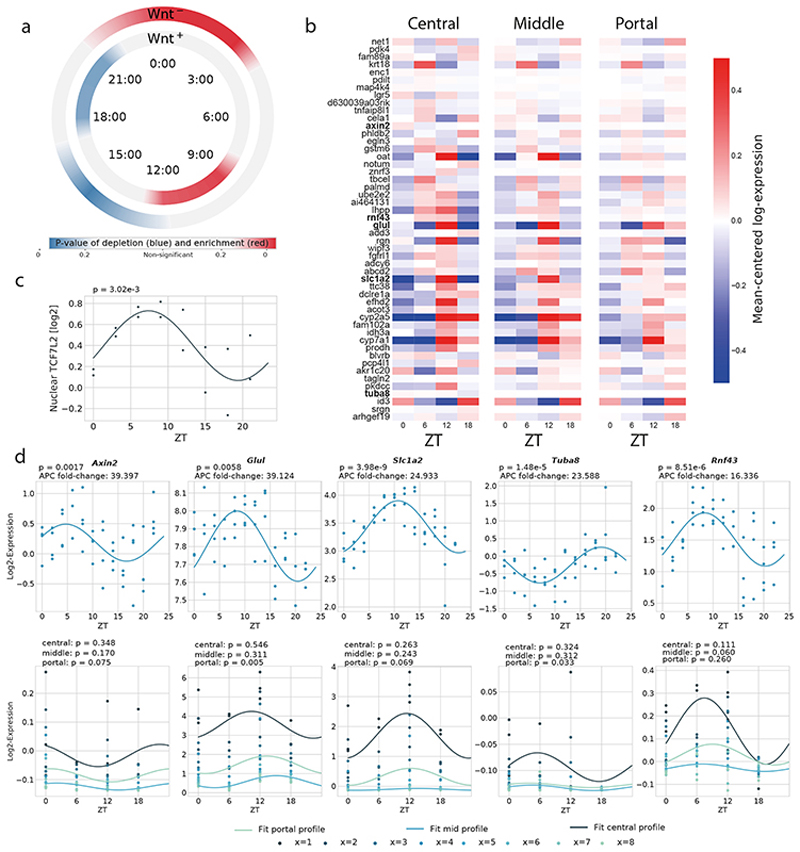
Figure 6. Rhythmic activity of Wnt signaling.
(a) Enrichment/depletion at different times (window size: 3h), of both positive (N=471) and negative (N=149) Wnt targets (background: all R and Z+R genes). Colormap shows p-values (two-tailed hypergeometric test): red (blue) indicates enrichment (depletion). (b) Heatmaps representing scRNA-seq profiles of the top 50 Wnt targets (according to the Apc-KO fold change, Lgr5 was also added) showing rhythmic mRNA in bulk (p<0.01, harmonic regression, F-test, data from ref.25). The profiles are computed in three different zones of the central-portal axis: central (layers 1-2), mid-lobular (layers 3-4-5) and portal (layers 6-7-8). Gene profiles (log2) are mean-centered in each zone. An enrichment of the phases around ZT8-14 can be observed, in agreement with Figure 6a. (c) Nuclear protein abundance from ref.35 of the Wnt effector TCF4 (encoded by the Tcf7l2 gene) in mouse liver shows a rhythm (p=0.003, harmonic regression, n=2, sampled every 3h) peaking at ZT7.5, consistent with the accumulation of mRNA targets a few hours later (panel a). (d) mRNA profiles from bulk RNA-seq (top) and scRNA-seq (bottom). Top five targets with the highest Apc-KO fold change, and rhythmicity in the bulk data (p<0.01, harmonic regression, F-test, data from ref.25). Rhythmicity (indicated above each panel) is also computed in three different zones for the scRNA-seq data. As above, the scRNA-seq used n=2 (ZT6, ZT18) or n=3 mice (ZT0, ZT12).
To obtain a temporal view of Wnt activity, we considered the top 50 Wnt pathway targets (according to the liver APC KO data) in the liver and analyzed the temporal profiles from high temporal resolution bulk liver mRNA and from our scRNA-seq binned in three different zones: central (layers 1-2), mid-lobular (layers 3-4-5) and portal (layers 6-7-8)(Figure 6b,d, Extended Data Figure 7b-c). This analysis confirmed that the peak times of rhythmic Wnt targets is preferentially between ZT9 and ZT12, and that the bulk and single cell data are consistent with each other, despite of the lower temporal sampling of the scRNA-seq. Further evidence of rhythmic WNT/β-Catenin activity was provided by our previous proteomics data35 showing that the potent Wnt effector TCF4 (encoded by the Tcf7l2 gene) has rhythmic nuclear abundance in mouse liver with a peak phase at ZT7.5 (Figure 6c), and hence explains the accumulation of its mRNA targets a few hours later. Among the rhythmic genes detected in bulk RNA-seq, the five strongest Wnt targets were, in decreasing order: Axin2, Glul, Slc1a2, Tuba8, Rnf43, with the latter showing the largest amplitude (Figure 6d); all but Tuba8 peaked in the morning. Note that Glul, an important marker of central zonation and canonical Wnt target (just like Axin2 and Rnf43), was assigned to the Z category, but with second highest probability for ZxR (Supplementary Table 2), peaking at ZT12.
In addition to the mRNA rhythms, we found that several of the Z+R or ZxR Wnt targets showed clear rhythms in bulk proteomics with the characteristic phase delays. The strongest five targets with rhythmic proteins (Extended Data Figure 7d) included the rate-limiting enzyme in the bile acid biosynthetic pathway CYP7A1, the NADPH-dependent monooxygenases involved in drug and xenobiotics detoxification FMO5, the P450 detoxification enzymes coumarin 7-hydroxylase (Cyp2a5), which may protect mice from dietary coumarin-induced toxicity46, the high-affinity glutamate transporter Slc1a2 (Eaat2), and the multidrug resistance protein ABCC2. All these proteins showed high amplitude protein rhythms peaking during the feeding phase between ZT12 and ZT18. Thus, Wnt transcription activity is clearly rhythmic in the liver, and this rhythm can propagate to protein expression.
We next asked whether the temporal oscillations in the expression of Wnt-activated genes might correlate with temporal oscillations in Wnt morphogens produced by pericentral non-parenchymal liver cells. To this end, we performed smFISH experiments and quantified the expression of the Wnt ligand Wnt2 5 and of Rspo3 6,47, a critical facilitator of Wnt signaling, as well as the Wnt antagonist Dkk3 19 (Figure 7a-b). We found that both Wnt2 and Rspo3 in liver non-parenchymal cells (NPCs) exhibit non-uniform expression around the clock, with significantly higher mRNA levels at ZT0 (p=4x10-5 for Wnt2, and p=2x10-8 for Rspo3, Kruskal-Wallis, Figure 7b). Given various delays between mRNA accumulation of ligands and expression of the Wnt targets, this timing is compatible with the peak nuclear accumulation of the TCF4 (Tcf7l2 gene) transcription factor observed at ZT7.5 (Figure 6c) and with the peaks in Wnt-activated genes between ZT6-12 (Figure 6a-b). Differences in Dkk3 expression were not significant (p=0.053). Thus, production of Wnt morphogens by central non-parenchymal liver cells might underlie the observed rhythmic Wnt pathway activity.
Figure 7. Wnt targets could be explained by rhythmically expressed Wnt ligands from non-parenchymal cells.
(a) Representative smFISH images of Wnt2, Dkk3, and Rspo3 expression at ZT0 (left) and ZT18 (right), shown in green. Markers of non-parenchymal cells (NPCs) are shown in red (Methods). Nuclei are stained in blue (DAPI). Scale bars, 2 μm. (b) Violin plots representing quantitative analysis of smFISH images (n=1420 cells from 189 central veins of at least two mice per time point). Wnt2, Dkk3, and Rspo3 transcripts are quantified in NPCs lining the central vein (Methods). mRNA expression is in smFISH dots per μm3. P-values are obtained from Kruskal-Wallis test.
Ras signalling and hypoxia are two additional pathways that have been implicated in shaping hepatocyte zonation8. In agreement with ref.8, we found that the negative targets of Ras were enriched in central genes, whereas the positive Ras targets were enriched in portal genes (not shown). The rhythmic targets (R and Z+R) of hypoxia showed a pattern of temporal compartmentalization similar with those of Wnt (Extended Data Figure 7e): the negative targets were enriched around ZT0 (dark-light transition) and underrepresented around ZT14, while the positive targets were enriched around ZT10 and underrepresented around ZT3. Ras targets, positive or negative, did not exhibit significant temporal bias.
Discussion
Recent genome-wide analyses of zonated gene expression in mouse and human liver8,48,49 uncovered a rich organization of liver functions in space at the sublobular scale, while chronobiology studies of bulk liver tissue revealed a complex landscape of rhythmic regulatory layers orchestrated by a circadian clock interacting with feeding-fasting cycles and systemic signals35,50–52. Here, we established how these two regulatory programs combine to shape the daily space-time dynamics of gene expression patterns and physiology in adult liver by extending our previous scRNA-seq approach8. We found that liver uses gene expression programs with many genes exhibiting compartmentalization in both space and time.
In this study, we chose to focus on the parenchymal cells in the liver, the hepatocytes, for which smFISH data on landmark zonated genes was readily available, which enabled reconstructing spatio-temporal mRNA profiles from scRNA-seq8. Zonation profiles of other cell types in the liver may be obtained as well; in fact, static zonation mRNA expression profiles have been obtained for liver endothelial cells, using a paired-cell approach19 in which mRNA from pairs of attached mouse cells were sequenced and gene expression from one cell type was used to infer the pairs’ tissue coordinates. In addition, ab initio reconstruction methods such as diffusion pseudo time48 or novoSpaRc53, in which a zonation coordinate is inferred by assuming that the major axis of variability for a cell type reflects transcriptome-wide gene expression changes associated with zonation, could be used for spatially sparse cell types with no available zonated marker genes, e.g. stellate or resident immune Kupffer cells. Moreover, it was recently found that rhythmic gene expression and metabolism in non-hepatocyte cells can be driven both by clocks in hepatocytes via cell-cell communication as well as feeding cycles13. Our computational framework for analyzing space-time logic of gene expression could be widely applicable in such future studies.
To study whether the observed space-time expression profiles may be regulated by either liver zonation, 24h rhythms in liver physiology, or both, we developed a mixed-effect model, combined with model selection. This enabled classifying gene profiles into five categories representing different modes of spatio-temporal regulation, from flat to wave-like. To validate these, we performed smFISH in intact liver tissue, which showed largely compatible profiles although some quantitative differences were observed. These differences most likely reflect the lower sensitivity of RNA-seq, uncertainties in the spatial analysis of smFISH in tissues, as well as known inter-animal variability in the physiologic states of individual livers, notably related to the animal-specific feeding patterns25.
Together, this temporal analysis confirms that a large proportion of gene expression in hepatocytes is zonated8 or rhythmic17, and in addition reveals marked spatio-temporal regulation of mRNA levels in mouse liver (Z+R and ZxR genes, comprising 7% of all detected genes according to our criteria). This means that zonated gene expression patterns can be temporally modulated on a circadian scale, or equivalently, that rhythmic gene expression profiles can exhibit sub-lobular structure. The dominant pattern for dually regulated gene was Z+R, which corresponds to additive effects of space and time in log, or multiplicate effects of gene expression levels, and describes genes expression profiles that are compartmentalized in both space and time. In other words, such patterns are characterized by shapes (in space) that remain invariant with time, but whose magnitudes are rhythmically rescaled in time. Or equivalently, the oscillatory amplitude (fold change) and phases are constant along the lobular coordinate, but the mean expression is patterned along the lobule. Such multiplicative effects could reflect the combined actions of transcriptional regulators for the zone and rhythm on promoters and enhancers of Z+R genes. Indeed, gene expression changes induced by several regulators combine multiplicatively22. Note that though the (relative) shape of Z+R patterns is invariant in time, threshold-dependent responses that would lie downstream of such genes would then acquire domain boundaries which can shift in time. Similar phenomena are expected for interacting profiles (phase and amplitude modulated) (ZxR) that we observed for a smaller number of genes.
As shown by us and others27,28, rhythms at the protein level are typically damped and phase-delayed compared the cognate mRNA rhythms, depending on the protein half-lives. Indeed, longer protein half-lives imply smaller oscillatory amplitudes and longer delays between mRNA and protein accumulation14. Analysis of liver proteomes in bulk showed that the number of cyclic proteins is significantly lower than the number of cyclic mRNAs, with delays approaching the predicted maximum of six hours. Here, we found genes from both Z+R and ZxR that exhibited rhythmic accumulation in bulk proteomics experiments, including genes encoding rate-limiting enzymes, suggesting that dually space-time regulated patterns have a physiological role in the liver. Moreover, phase delays between the mRNA and protein profiles were as expected. Future studies will utilize emerging spatial proteomics approaches to reconstruct a space-time liver proteomic atlas54.
In addition to previously discussed zonated liver functions8, a systematic querying of KEGG pathways highlighted Z+R functions not previously associated with rhythmic liver zonation. The roles and profiles of the corresponding genes allowed us to better understand the spatiotemporal logic of the identified pathways. For instance, we found that the expression levels of both ribosome protein genes (rate-limiting for protein synthesis32) and proteasome components (involved in protein degradation) were higher in central hepatocytes. Since the central environment is subject to high concentrations of xenobiotics and hypoxic stress, this could indicate an elevated protein turnover in this region, which would ensure that damaged proteins are rapidly exchanged with new, undamaged proteins. This interpretation is corroborated by the observed increased levels of cytosolic and ER chaperones during the feeding phase, to assist protein synthesis and secretion, thereby counteracting such protein stress.
It was previously shown that Wnt signaling can explain the zonation of up to a third of the zonated mRNAs7. Wnt ligands are secreted by pericentral non-parenchymal cells, mostly endothelial cells5,6, forming a graded spatial morphogenetic field. As a result, and as observed in our enrichment analysis, Wnt-activated genes were pericentrally-zonated. Moreover, both the scRNA-seq data and previous bulk mRNA and protein measurements showed that Wnt activity is rhythmic in the liver. Our smFISH analysis suggested that temporal fluctuations in the expression of Wnt2 and Rspo3, two key Wnt ligands, secreted by pericentral non-parenchymal cells might underlie oscillatory and zonated expression of Wnt targets at times near the fasting/feeding transition.
In summary, we demonstrate how liver gene expression can be quantitatively investigated with spatial and temporal resolution and how liver functions are compartmentalized along these two axes. Our approach could be used to reconstruct spatio-temporal gene expression patterns in other zonated tissues such as the intestine and kidney3.
Methods
Animals and ethics statement
All animal care and handling were approved by the Institutional Animal Care and Use Committee of WIS and by the Canton de Vaud laws for animal protection (authorization VD3197.b). Male (C57BL/6JOlaHsd) mice aged of 6 weeks, housed under reverse-phase cycle and under ad libitum feeding (Teklad Global 18% Protein Rodent Diet) were used to generate sc-RNA-seq data of hepatocytes and single-molecule RNA-FISH (smFISH). Littermate controls were used for the ZT6 and ZT18 time points. Male mice between 8 to 10 weeks old (C57BL/6J), housed under 12:12 light-dark cycle, and having access to food only during the night (Kliba Nafag 3242 Breeding, Vitamin-fortified, irradiated > 25 kGy) were used for smFISH of circadian clock genes (Reporting Summary).
Hepatocytes isolation and single-cell RNA-seq
Liver cells were isolated using a modified version of the two-step collagenase perfusion method of Seglen55. The tissue was digested with Liberase Blendzyme 3 recombinant collagenase (Roche Diagnostics) according to the manufacturer instructions. To enrich for hepatocytes, we applied a centrifuge step at 30g for 3 min to pull down all hepatocytes while discarding most of the non-parenchymal cells that remained in the sup. We next enriched for live hepatocytes by 2 cycles of percoll gradient, hepatocytes pellet was resuspended in 25 ml of PBS, percoll was added for a final concentration of 45% and mixed with the hepatocytes. Dead cells were discarded after a centrifuge step (70g for 10min) cells were resuspended in 10x cells buffer (1x PBS, 0.04% BSA) and proceeded directly to the 10x pipeline. The cDNA library was prepared with the V2 chemistry of 10X genomics Chromium system according to manufactures instructions and sequencing was done with Illumina Nextseq 500 at estimated depth of 40,000 reads per cell. Overall, independent libraries were prepared at ZT0 (n=3 biological replicates from individual mice), ZT6 (n=2), ZT12 (n=3) and ZT18 (n=2).
Conceivably, the dissociation of liver tissue into individual cells and the purification of hepatocytes are relatively lengthy and may thus lead to alterations in mRNA expression. While it has been shown that mRNA levels do not change much during the purification and 24-hour cultivation of hepatocytes56, transcription rates on the other hand can be diminished by 8-fold to nearly 100-fold during this process57. This difference between nascent transcript and mature mRNA levels can be explained by the relatively long half-lives of liver-specific RNAs. In our case, the time needed from the dissociation of the tissue until the cell lysis is approximately 1 hour and the cell are not placed in culture. Since we are measuring mature transcripts, with half-lives in the range of typically 1-5 hours (Extended Data Figure 2f), the changes in mRNA levels due to the protocol will remain contained. In particular, the scRNA-seq of cells carry the typical hepatocyte gene expression signatures, for example, genes such as Alb or Apoa2 rank at 2nd and 5th position genome-wide. As further validation, we compared our reconstructed gene expression zonation profiles with the zonation profiles from the massively validated zonation study of ref.8, revealing a near-perfect agreement (Extended Data Figure 1f).
Filtering of raw scRNA-seq data
The initial data analysis was done in R v3.4.2 using Seurat v2.1.058. Each expression matrix was filtered separately to remove dead, dying and low-quality cells. We firstly only kept genes that were expressed in at least 5 cells in any of the ten samples. We then defined a set of valid cells with more than 500 expressed genes and between 1000 and 10000 unique molecular identifiers (UMIs) and secondly an additional expression matrix with cells having between 100 and 300 UMIs which was used for background estimation (Extended Data Figure 1a). Other UMI-filters have been tried, but yielded equal or less reliable profiles. The mean expression of each gene was then calculated for the background dataset and subtracted from the set of valid cells. This was subsequently filtered to only include hepatocytes by removing cells with expression of non-parenchymal liver cell genes. Next, the cells were filtered based on the fraction of mitochondrial gene expression. First, expression levels in each cell were normalized by the sum of all genes excluding mitochondrial and major urinary protein (Mup) genes. Indeed, as mitochondria are more abundant in periportal hepatocytes, the expression of mitochondrial genes is higher in this area59; and since these genes are very highly expressed, including them would reduce the relative expression of all other genes based on the cell’s lobular location. Mup genes are also highly abundant and mapping their reads to a reference sequence is unreliable due to their high sequence homology60. Moreover, Mup genes encode for pheromones that vary greatly between individuals to facilitate individual recognition61.
Mitochondrial content is often used to remove non-viable cells62. The mitochondrial content of sequenced hepatocytes exhibited a bi-modal behavior (Extended Data Figure 1b). To identify the range of mitochondrial fractions that included viable hepatocytes we used an intrinsic property of hepatocytes, which is the anti-correlation of the pericentral landmark gene Cyp2e1 and the periportal landmark gene Cyp2f2 7 (Extended Data Figure 1c). We found that hepatocytes with mitochondrial fraction in the range of 9-35% exhibited an almost perfect anticorrelation between Cyp2e1 and Cyp2f2 (Extended Data Figure 1d,e), suggesting that these are the best quality, and we consequently kept hepatocytes within this range of mitochondrial content for further analysis.
t-SNE clustering
To validate that the expected spatial and temporal axes of variation are present in the scRNA-seq data, we generated a low-dimensional representation of all cells using the standard t-SNE (t-distributed stochastic neighbor embedding)63, a nonlinear dimensionality reduction technique that embeds high-dimensional data on a 2-dimensional plane such that points that are similar in high-dimensional space are close together on the 2-dimensional representation. We then colored cells either by their position along the central-portal axis, or by time of day.
Spatial reconstruction of zonation profiles from scRNA-seq data
Choice of landmark genes
The reconstruction algorithm relies on a priori knowledge about the zonation of a small set of landmark genes to infer the location of the cells. Ref.8 used smFISH to determine the zonation pattern in situ of 6 such landmark genes and used them to reconstruct the spatial profiles of all other genes at a single time point. Since we here aimed at reconstructing zonation profiles at different time points, we could not rely on those landmark genes, which might be subject to temporal regulation. Therefore, we used an alternative strategy where we selected landmark zonated genes from ref.8 (q < 0.2), with the additional constraints that those should be highly expressed (mean expression in fraction UMI of more than 0.01% and less than 0.1%), and importantly vary little across mice and time. Specifically, we calculated the variability in the mean expression (across all layers) between all mice for every gene and removed genes with >= 10% variability. This yielded 27 central (Akr1c6, Alad, Blvrb, C6, Car3, Ccdc107, Cml2, Cyp2c68, Cyp2d9, Cyp3a11, Entpd5, Fmo1, Gsta3, Gstm1, Gstm6, Gstt1, Hpd, Hsd17b10, Inmt, Iqgap2, Mgst1, Nrn1, Pex11a, Pon1, Psmd4, Slc22a1, Tex264); and 28 portal (Afm, Aldh1l1, Asl, Ass1, Atp5a1, Atp5g1, C8a, C8b, Ces3b, Cyp2f2, Elovl2, Fads1, Fbp1, Ftcd, Gm2a, Hpx, Hsd17b13, Ifitm3, Igf1, Igfals, Khk, Mug2, Pygl, Sepp1, Serpina1c, Serpina1e, Serpind1, Vtn) landmark genes.
Reconstruction algorithm
The reconstruction algorithm is based on the algorithm in ref.8 and was used in the modified version from ref.19. Briefly, the expression of each landmark gene was normalized to its maximal expression over all cells. Then, for each cell, we divided the summed expression of all portal landmark genes by the summed expression of all portal and central landmark genes, resulting in a value μi between 0-1, which we used as the cell location in the liver lobule. We then compared the obtained μi with the distributions of cell locations from each of the layers from ref.8, which yielded a matrix in which every cell has a given probability to belong to a given layer. These weights were then used to compute the mean expression of all genes in a particular layer. The procedure was applied independently on each mouse, yielding ten spatial gene expression profiles for each gene, given as fraction of UMI per cell.
Spatiotemporal analysis of liver gene expression profiles
Data
Each profile for the 14678 genes includes 8 layers from the pericentral to the periportal zone and 4 time points: ZT0 (n=3 biological replicates from individual mice), ZT6 (n=2), ZT12 (n=3) and ZT18 (n=2). The expression levels (noted as x) are then log-transformed as follows:
| (i) |
The offset Δ = 10−4 buffers variability in lowly expressed genes, while the shift B = −log 2(11 × 10−5) changes the scale so that y = 0 corresponds to about 10 mRNA copies per cell (we expect on the order of 1M mRNA transcripts per liver cell).
Reference genes
For ease of interpretation (Figure 2 and Extended Data Figure 2), we used a set of reference circadian genes and a set of reference zonated genes, highlighted in several figures.
The reference core circadian clock and clock output genes are the following: Bmal1, Clock, Npas2, Nr1d1, Nr1d2, Per1, Per2, Cry1, Cry2, Dbp, Tef, Hlf, Elovl3, Rora, Rorc. The reference zonated genes are the following: Glul, Ass1, Asl, Cyp2f2, Cyp1a2, Pck1, Cyp2e1, Cdh2, Cdh1, Cyp7a1, Acly, Alb, Oat, Aldob, Cps1.
Gene expression variance in space and time
To analyze variability in space and time (Figure 2a) we computed, for each gene, the spatial variance Vx and the temporal variance VT. Let yx,t,j represent the expression profile, with j the replicate index, t ∈ {1,2,…, Nt} the time index, and x ∈ {1,2,…, Nx} the layer index. Then, Vx and VT are computed as follows:
| (ii) |
| (iii) |
Thus, the spatial variance Vx is computed along the space (and averaged over the replicates) for each time condition, and then averaged over time. The procedure is similar, symmetrically, for Vt.
Gene filtering
For the analyses in Figure 2, we selected transcripts that were reproducible between replicates, as well as sufficiently highly expressed (see scatterplot in Extended Data Figure 2d). To assess reproducibility across replicates, we computed the average relative variance of the spatiotemporal profiles over the replicates:
| (iv) |
We considered genes with values below 50% (Extended Data Figure 2). To filter lowly expressed genes, we required the maximum expression level across layers and time points to exceed 10-5 (fraction of UMIs) which corresponds to y = 0 or about 10 copies of mRNA per cell. While this was quite more permissive than previous scRNA-seq studies it allowed to keep most reference circadian and zonated genes. However, scRNA-seq has still limited sensitivity and some potentially important genes may have been removed in the filtering process. In the end, our filters kept 5085 genes (1437 were removed due to low expression, 4733 due to high variance, and 3543 due to both), which were then used for subsequent analyses.
Mixed-effect model for spatiotemporal mRNA profiles
Since the data is longitudinal is space (8 layers measured in each animal), modelling the space-time profiles require the use of mixed-effect models. To systematically analyze the spatiotemporal mRNA profiles, we used a parameterized function. Specifically, the model uses sine and cosine functions for the time, and polynomials (up to degree 2) for space. Possible interaction between space and time are described as space-dependent oscillatory functions, or equivalently, time-dependent polynomial parameters. Our model for the transformed mRNA expression y reads:
| (v) |
Here t is the time, x the spatial position along the liver layers, and i ∈ {1,2,…,10} the animal index. This function naturally generalizes harmonic regression, often used for analysis of circadian gene expression25, by introducing space-dependent coefficients:
Here, P 1 and P 2 are the Legendre polynomials of degrees 1 and 2, respectively; μ 0, μ 1 and μ 2 represent the static zonation profile, a 0 and b 0 represent the global (space-independent) rhythmicity of the gene, while a 1, a 2, b 1, b 2 represent layer-dependent rhythmicity. εx,t,j is a Gaussian noise term with standard deviation σ. In addition to the fixed-effect parameters described so far, we also introduced a mouse-specific random-effect μi (with zero mean). This parameter groups the dependent layer measurements (obtained in the same animal) and thereby properly adjusts the biological sample size for the rhythmicity analysis.
Phases φ (related to peak times t through t = φ * 24/2π) and amplitudes A, for each profile can then be computed for any layer from the coefficients a(x) and b(x):
| (vi) |
Note that peak-to-trough difference is 2A(x). The peak-to-trough ratio or fold change of the original expression levels is then 22A(x). We also note that an equivalent writing of the model formulates the problem in terms of time-dependent zonation parameters instead of space-dependent rhythmicity:
| (vii) |
where:
In this study, we fixed since the animals were entrained in a 24 h light-dark cycle and the low time resolution would prevent us from studying ultradian rhythms. The model parameters, including the variance of the random effects and Gaussian noise strength σ, are estimated for each gene using the fit function from the Python library StatsModels (version 0.9.0). Nelder-Mead was chosen as the optimization method, and the use of a standard likelihood was favored over the REML likelihood to allow for model comparison64. To prevent overfitting of the gene profiles, we added a noise offset σ 0 = 0.15 [log 2] to the estimated noise σ, in the expression of the likelihood function used in the mixed-effect model optimization.
Depending on the gene, the model presented in (v) and (vii) may be simplified by setting all or some of the (fixed) parameters to 0. For example, a non-oscillatory gene profile would normally have non-significant aj and bj parameters. In practice, considering the fixed effects, 29 sub-models of various complexity can be generated. However, we added a few reasonable requirements to reduce the number of models. First, the intercept μ 0 must be present in every model. Similarly, the parameters a 0 and b 0, providing a global rhythm, must be present in every rhythmic model. Finally, the parameters aj and bj for j=0,1,2 must be paired to ensure a proper phase definition (vi).
The models can then be classified in different categories, depending on the retained (non-zero) parameters (Figure 2c):
The model comprising only the intercepts μ 0 and μi, termed flat or noisy (F/N).
The models comprising only the intercepts and zonation parameters: μ 1 and/or μ 2, termed purely zonated (Z).
The models comprising only the intercepts and rhythmic parameters: a 0 and b 0, termed purely rhythmic (R).
The models comprising only the intercepts, zonated parameters and rhythmic parameters: μ 1 and/or μ 2, and a 0, b 0, termed independent (Z+R).
The models comprising interaction parameters: aj and bj for j=1,2, termed interacting (ZxR).
Note that we only plot the fixed effects in the predicted gene profiles. The Bayesian Information Criterion (BIC) is then used for model selection, enabling to choose the most parsimonious model for each gene. Consequently, the F/N class also contains noisy profiles, since genes that are not well fitted with any complex model will then be assigned to the simplest model. Additionally, it appears that, for some profiles, several competing models can result in close BIC values (see e.g. the discussion on Clock and Cry1 in the Results). Therefore, when assigning hard classes, if some models have a relative difference of less than 1% in their BIC, we systematically keep the most complex model. Moreover, we also assigned probabilities to the different categories (F, Z, R, Z+R and ZxR), computed as Schwartz BIC weights65, which is useful in case of ambiguous classification (Supplementary Table 2). All best fits with their parameter values are listed in Supplementary Table 1.
Bulk RNAseq dataset
A bulk liver RNA-seq dataset was obtained from ref.25. These data was obtained from a sampling every 2 hours for 24 hours, with 4 replicates per time condition. For Extended data Figure 2e, we only compared genes that for which rhythmicity is not changing across layers, viz. the R and Z+R categories. Note that since the scRNA-seq data has a lower temporal resolution and fewer replicates per time point, we found overall less rhythmic genes.
To assess gene rhythmicity, we used harmonic regression on the log-transformed profiles. Using the same notation as above, we define the two following models:
We then fit eq. (viii) and eq. (ix) to every transcript. Depending on the figure, we either keep the model with the lowest BIC (Extended data Figure 2e, for which we also compute the circular correlation coefficient66) or the ones having a significant rhythmicity according to a F-test (Figure 6).
KEGG pathway Enrichment analysis
Functional annotation clustering from Enrichr67 for the categories F/N, Z and Z+R (which is then subdivided in central and portal), Zc+R (central zonated and rhythmic), Zp+R (portal zonated and rhythmic) and finally R was ran (https://maayanlab.cloud/Enrichr/) with standard parameters, using the standard KEGG 2019 Mouse set of pathways. The enriched pathways (adjusted p-value < 0.1, standard Enrichr output test) were then further annotated to compute, e.g. the number of central/portal genes in each category and the phase of each gene. This analysis is available Supplementary Table 3.
smFISH
Analysis of Z+R and ZxR genes (Stellaris smFIH probes)
Preparation of probe libraries, hybridization procedure and imaging conditions were previously described19. Briefly, smFISH probe libraries were coupled to TMR, Alexa594 or Cy5. Cell membranes were stained with alexa fluor 488 conjugated phalloidin (Rhenium A12379) that was added to GLOX buffer68. Portal node was identified morphologically on DAPI images based on bile ductile, central vein was identified using smFISH for Glul in TMR, included in all hybridizations. Images were taken as scans spanning the portal node to the central vein. Images were analyzed using ImageM68. Quantification of zonation profiles in different circadian time points were generated by counting dots and dividing the number of dots in radial layers spanning the portal-central axis by the layer volume.
Temporal analysis of circadian genes (RNA scope smFISH probes)
smFISH of R genes were done on fresh-frozen liver cryosections (8μm) embedded in O.C.T Compound (Tissue-Tek; Sakura-Finetek USA), sampled every three hours (ZT0 to ZT21). RNAscope® probes for Bma1l mRNA (Mm-Arntl, catalog #: 438748-C3) and Per1 mRNA (Mm-Per1, catalog #: 438751) were used, according to the manufacturer’s instructions for the RNAscope Fluorescent Multiplex V1 Assay (Advanced Cell Diagnostics). To detect the central vein, an immunofluorescence of Glutamine Synthetase (ab49873, Abcam, diluted 1:2000 in PBS/BSA 0.5%/Triton-X0.01%) was done together with smFISH. Nuclei were counterstained with DAPI and sections were mounted with ProLong™ Gold Antifade Mountant. Liver sections were imaged with a Leica DM5500 widefield microscope and an oil-immersion x63 objective. Z-stacks were acquired (0.2μm between each Z position) and mRNA transcripts were quantified using ImageJ, as described previously in ref.50. Pericentral (PC) and Periportal (PP) veins were manually detected based on Glutamine Synthetase IF or on bile ducts (DAPI staining). The Euclidean distance between two veins and the distance from the vein of each mRNA transcript were calculated. mRNA transcripts were assigned to a PP or PC zone if the distance from the corresponding vein was smaller than one-third of the distance between the PP and PC veins (ranging from 50 to 130μm).
Wnt2, Rspo3 and Dkk3 expression in non-parenchymal cells (NPCs) (Stellaris smFIH probes)
Preparation of probe libraries, hybridization protocol and imaging conditions were previously described19. The Aqp1, Igfbp7 and Ptprb probe libraries were coupled to TMR, the Wnt2 library was coupled to Alexa594 and the Dkk3 or Rspo3 library were coupled to Cy5. Cell membranes were stained with alexa fluor 488 coupled to phalloidin (Rhenium A12379) that was added to GLOX buffer68. The central vein was identified based on morphological features inspected in the DAPI and Phalloidin channels and presence of Wnt2-mRNA (detected by smFISH). Central vein niche NPCs were identified by co-staining of Aqp1, Igfbp7 and Ptprb. The central vein area was imaged and the images were analyzed using ImageM68. We counted dots of Wnt2, Rspo3 and Dkk3 expression (corresponding to single mRNA molecules) in NPCs lining the central vein and removed background dots larger than 25 pixels. We then divided the dot count by the segmented cell volume. In total 1420 NPCs from 189 central veins of at least 2 mice per time point (ZT0,6,12,18) were imaged and a Kruskal-Wallis test based on the mean mRNA dot concentration in each cell was performed to compare the ZT0 and ZT18 time points.
Extended Data
Extended Data Fig. 1. scRNA-seq pre-processing.
(a) Histogram of number of UMIs per cell barcode for each mouse. Red patches mark the cells used for background estimation (100-300 UMI/cell barcode), gray patches mark the cells used for downstream analysis (1000-10000 UMI/cell barcode).
(b) Histogram of fraction of all UMIs mapping to mitochondrial genes. Filter used for downstream analysis in grey (0.09-0.35).
(c) smFISH staining of a liver lobule with probes against Cyp2e1 (red) and Cyp2f2 (green). CV = central vein, PN = portal node. Overall, the data combine 10 images from from two mice.
(d) Expression of Cyp2e1 and Cyp2f2 in cells with different fraction of mitochondrial expression. Three different filters for the fraction of UMIs mapping to mitochondrial genes (0-0.09, 0.09-0.35, 0.35-1) were applied, the data of all mice merged and the resulting datasets visualized as t-SNE plots.
(e) Violin plots for the correlations between Cyp2e1 and Cyp2f2 expression in single hepatocyte populations with different filters for fractions of mitochondrial expression. Each dot represents one mouse (n=10 mice for each distribution) and the shape of the violin represents the density of points.
(f) Comparison of the zonation profiles of Z and Z+R genes obtained in our current study and the previous reconstruction from Halpern et al. 2017. Profiles were interpolated to fit 15 layers, where 1 is pericentral and 8 is periportal. Dots indicate the center of mass (expression-weighted lobule layer) of the Z and Z+R genes computed in both datasets, for gene having an average expression of at least 10-5 in Halpern et al. r is the correlation coefficient, p the corresponding p-value from a standard linear regression.
Extended Data Fig. 2. Log-transformed reconstructed profiles, pre-filtering of the genes and comparison with external datasets.
(a-c) Expression levels of the reconstructed profiles for the genes from Figure 1F-H after log-transformation (Methods). Dots in represent data points from the individual mice. Lines represent mean expression per time point. Shaded areas represent one standard deviation (SD) across the mice (n=2 or n=3 depending on the time point, Methods).
(d) Biological variability of gene profiles across independent replicate liver samples, quantified in terms of the average relative replicate variance. 0 shows perfectly reproducible profiles while 1 the most variable genes (Methods). Genes inside the bottom-right box (x-cutoff at 10-5; y-cutoff at 0.5) are selected and contain all but one of the reference genes. Colored dots show reference zonated genes (blue) and reference rhythmic genes (orange).
(e) Comparison of the peak times for rhythmic genes in R and Z+R, with the dataset from Atger et al, PNAS 2015. Circular correlation coefficient is 0.746 (Methods).
(f) Boxplot of the mRNA half-lives (data from Wang, J. et al, 2017) shows that R genes as a group (median, orange line) are the shortest-lived. Box limits are lower and upper quartiles, whiskers extend up to the first datum greater/lower than the upper/lower quartile plus 1.5 times the interquartile range. Remaining points are marked. MW stands for the two-sided Mann-Whitney test, and KW stands for Kruskal-Wallis test.
Extended Data Fig. 3. Z+R and ZxR transcripts with corresponding rhythmic protein accumulation in bulk mass spectrometry data.
(a-b) Rhythmic proteins corresponding to Z+R (a) and ZxR (b) transcripts were selected from Robles et al., 2014 (from original Table S2), and fitted with harmonic regression (p-value of rhythmicity from F-tests are indicated above the plot). Only proteins having a p-value<0.01 are shown.
(c) Scatter plot of the phase of the fits from the transcripts (x-axis) against the phase of the fits from the proteins (y-axis). The diagonal is indicated with a dashed grey line, the theoretical upper bound (6h) for the delay between mRNA and protein is indicated with a dashed red line. All rhythmic proteins (q<0.2 in the original analysis) are represented.}
Extended Data Fig. 4. Additional validations for the Z+R category.
(a-b) smFISH (Stellaris, Methods) for Elovl3 (Z+R) and Arg1 (Z+R). smFISH quantifications were made for ZT0 and ZT12 (Methods). Left: representative images at ZT0, ZT12 for Elovl3 (a) or Arg1 (b). Pericentral veins (CV) and a periportal node (PN) are marked. Scale bar - 20μm. Right: quantified profiles for each gene at the two time points from smFISH (top, line plot is the mean number of mRNAs, shaded area indicate SD across twelve images), and scRNA-seq data (bottom, line plot is mean expression, shaded area is SD across mice, n=2 or n=3 depending on the time point, Methods).
Extended Data Fig. 5. The core circadian-clock is not zonated.
(a) Spatial and temporal profiles and fits for circadian core-clock genes. Peak times are indicated on the temporal representation. For the genes Cry1 and Clock, additional dashed lines represent fits for the R model, as the Schwartz BIC weights from the R and Z+R models were close (Supplementary Table 2).
(b) Amplitudes and peak times of the core-clock circadian genes in a polar coordinate representation (clock-wise ZT times are indicated, distance from the center corresponds to the amplitude) show the expected organization of core clock transcript in the liver.
Extended Data Fig. 6. Spatio-temporal mRNA expression profiles for heat-responsive genes. Represented genes correspond to the top 200 targets from the ChIP-Atlas list for mouse HSF1.
(a) Polar plot representation of the transcripts that are R, Z+R, or ZxR among the HSF1 targets from the ChIP-Atlas (http://chip-atlas.org/). Genes involved in chaperone functions (chaperones, co-chaperones, or chaperone facilitators) are named. Color indicates zonation, while grey dots show purely rhythmic genes.
(b) Spatial representation of the transcripts corresponding to proteins involved in chaperone functions, separated in central (left, cytoplasmic function) and portal (right, endoplasmic reticulum function) zonation.
Extended Data Fig. 7. Rhythmicity of Wnt targets in bulk RNA-seq, and proteomics liver time series data.
(a) Enrichment of rhythmic genes (R, Z+R and ZxR) among the targets of the Wnt pathway, computed on the bulk dataset (Atger et al., 2015). Targets above a given percentile (x-axis) of Apc-KO fold change are considered. The percentage of rhythmic genes in the whole Atger et al. dataset is indicated by a dashed blue line.
(b) Bulk mRNA (coming from Atger et al. dataset) rhythmicity profiles of Wnt targets among the top-50 targets with highest Apc-KO fold change. Gene profiles are centered around their mean. An enrichment of the phases around ZT8-14 is observed, in agreement with Figure 6a.
(c) Polar plot representation of the individual gene phases and amplitudes represented in panel b (bulk data).
(d) Temporal representation of selected genes profiles from the scRNA-seq (top, n=2 or n=3 animals depending on the time point, Methods) and bulk proteomics (bottom, data from Robles et al, 2014, n=2 replicates per time point sampled every 3h) data. Represented profiles are ones with (1) the highest Apc-KO fold change, (2) a significantly rhythmic protein (p < 0.05, standard harmonic regression, F-test), and (3) belonging to the Z+R or ZxR category.
(e) Enrichment/depletion at different times (window size: 3h), of both positive and negative Ras (N=31 and N=33, respectively) and Hypoxia (N=73 and N=41, respectively) targets (background: all R and Z+R genes). Colormap shows p-values (two-tailed hypergeometric test): red (blue) indicates enrichment (depletion).
Supplementary Material
Acknowledgments
We thank Daniel Mauvoisin for assistance with the animal work, Cédric Gobet for bioinformatics advice, and the EPFL BIOP facility for advice with microscopy. This work was supported by the Rothschild Caesarea Foundation’ fund managed by Weizmann Institute and EPFL, a Swiss National Science Foundation Grant 310030_173079 (to F. Naef), and the EPFL. S. Itzkovitz is supported by the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics, The Leir Charitable Foundations, Richard Jakubskind Laboratory of Systems Biology, Cymerman-Jakubskind Prize, The Lord Sieff of Brimpton Memorial Fund, the Wolfson Foundation SCG, the Wolfson Family Charitable Trust, Edmond de Rothschild Foundations, the I-CORE program of the Planning and Budgeting Committee and the Israel Science Foundation (grants 1902/12 and 1796/12, the Israel Science Foundation grant no. 1486/16, the Chan Zuckerberg Initiative grant no. CZF2019-002434, the Broad Institute-Israel Science Foundation grant no. 2615/18, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 768956, the Bert L. and N. Kuggie Vallee Foundation, and the Howard Hughes Medical Institute (HHMI) international research scholar award.
Footnotes
Author contributions
F.N. and S.I. conceived the study. K.B.H., J.E.K. and C.H. prepared all the samples and performed the experiments. C.D. and F.N designed the modeling. C.D., J.E.K., K.B.H. and C.H. analyzed the data. M.R. and S.M. assisted with the scRNA-seq and smFISH experiments. F.N and S.I. supervised the study. C.D., J.E.K. and F.N. wrote the manuscript. All authors reviewed the manuscript and provided input.
Competing interests
The authors declare no competing interests.
Data availability
scRNA-seq data
All scRNA-seq data is deposited in GEO with accession code GSE145197.
Reconstructed gene profiles
Reconstructed spatio-temporal gene profiles are available as Matlab files at https://github.com/naef-lab/Circadian-zonation
Web-application
The whole dataset of gene profiles along with the analysis is available online as a web-application at the URL https://czviz.epfl.ch/. The application was built in Python using the library Dash by Plotly (version 1.0).
Code availability
The code for fitting the mixed-effects models and generating the main figures is available at https://github.com/naef-lab/Circadian-zonation
Details regarding the statistics, software and data are provided in the Reporting Summary.
References
- 1.Gebhardt R. Metabolic zonation of the liver: Regulation and implications for liver function. Pharmacol Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 2.Hoehme S, et al. Prediction and validation of cell alignment along microvessels as order principle to restore tissue architecture in liver regeneration. Proc Natl Acad Sci. 2010;107:10371–10376. doi: 10.1073/pnas.0909374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Moshe S, Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol. 2019;16:395–410. doi: 10.1038/s41575-019-0134-x. [DOI] [PubMed] [Google Scholar]
- 4.Colnot S, Perret C. Liver Zonation. In: Monga SPS, editor. Molecular Pathology of Liver Diseases. Vol. 5. Springer US; 2011. pp. 7–16. [Google Scholar]
- 5.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Planas-Paz L, et al. The RSPO–LGR4/5–ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 7.Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 8.Halpern KB, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dibner C, Schibler U, Albrecht U. The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 10.Green CB, Takahashi JS, Bass J. The Meter of Metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung J, Naef F. Rhythms of the Genome: Circadian Dynamics from Chromatin Topology, Tissue-Specific Gene Expression, to Behavior. Trends Genet TIG. 2018;34:915–926. doi: 10.1016/j.tig.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Maury E, Ramsey KM, Bass J. Circadian Rhythms and Metabolic Syndrome: From Experimental Genetics to Human Disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan D, et al. The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science. 2020;369:1388–1394. doi: 10.1126/science.aba8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermet J, Yeung J, Naef F. Systems Chronobiology: Global Analysis of Gene Regulation in a 24-Hour Periodic World. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028720. a028720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobel JA, et al. Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLOS Biol. 2017;15 doi: 10.1371/journal.pbio.2001069. e2001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung J, et al. Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res. 2018;28:182–191. doi: 10.1101/gr.222430.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halpern KB, et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 2018;36:962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunger MK, et al. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 22.Beal J. Biochemical complexity drives log-normal variation in genetic expression. Eng Biol. 2017;1:55–60. [Google Scholar]
- 23.McLean RA, Sanders WL, Stroup WW. A Unified Approach to Mixed Linear Models. Am Stat. 1991;45:54. [Google Scholar]
- 24.Wagenmakers E-J, Farrell S. AIC model selection using Akaike weights. Psychon Bull Rev. 2004;11:192–196. doi: 10.3758/bf03206482. [DOI] [PubMed] [Google Scholar]
- 25.Atger F, et al. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci. 2015;112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kietzmann T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robles MS, Cox J, Mann M. In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004047. e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauvoisin D, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gebhardt R. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J Gastroenterol. 2014;20:8491. doi: 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-Driven and Oscillator-Dependent Circadian Transcription in Mice with a Conditionally Active Liver Clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinke H, et al. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinturel F, et al. Diurnal Oscillations in Liver Mass and Cell Size Accompany Ribosome Assembly Cycles. Cell. 2017;169:651–663.e14. doi: 10.1016/j.cell.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kietzmann T, Cornesse Y, Brechtel K, Modaressi S, Jungermann K. Perivenous expression of the mRNA of the three hypoxia-inducible factor alpha-subunits, HIF1alpha, HIF2alpha and HIF3alpha, in rat liver. Biochem J. 2001;354:531–537. doi: 10.1042/0264-6021:3540531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ: C/EBPβ regulates circadian autophagy rhythm. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab. 2017;25:102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harper JW, Ordureau A, Heo J-M. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 2018;19:93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 37.Le Martelot G, et al. REV-ERBα Participates in Circadian SREBP Signaling and Bile Acid Homeostasis. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000181. e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrell JM, Chiang JYL. Short-Term Circadian Disruption Impairs Bile Acid and Lipid Homeostasis in Mice. Cell Mol Gastroenterol Hepatol. 2015;1:664–677. doi: 10.1016/j.jcmgh.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma K, et al. Circadian Dysregulation Disrupts Bile Acid Homeostasis. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0006843. e6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu L, et al. Disruption of Abcg5 and Abcg8 in mice reveals their crucial role in biliary cholesterol secretion. Proc Natl Acad Sci. 2002;99:16237–16242. doi: 10.1073/pnas.252582399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oki S, et al. Ch IP -Atlas: a data-mining suite powered by full integration of public Ch IP -seq data. EMBO Rep. 2018;19 doi: 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gotic I, et al. Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp. Genes Dev. 2016;30:2005–2017. doi: 10.1101/gad.287094.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakobsson A, Jörgensen JA, Jacobsson A. Differential regulation of fatty acid elongation enzymes in brown adipocytes implies a unique role for Elovl3 during increased fatty acid oxidation. Am J Physiol-Endocrinol Metab. 2005;289:E517–E526. doi: 10.1152/ajpendo.00045.2005. [DOI] [PubMed] [Google Scholar]
- 44.Burke ZD, Tosh D. The Wnt/β-catenin pathway: master regulator of liver zonation? BioEssays. 2006;28:1072–1077. doi: 10.1002/bies.20485. [DOI] [PubMed] [Google Scholar]
- 45.Gougelet A, et al. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 46.Lavery DJ, et al. Circadian Expression of the Steroid 15 α-Hydroxylase (Cyp2a4) and Coumarin 7-Hydroxylase (Cyp2a5) Genes in Mouse Liver Is Regulated by the PAR Leucine Zipper Transcription Factor DBP. Mol Cell Biol. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brosch M, et al. Epigenomic map of human liver reveals principles of zonated morphogenic and metabolic control. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mermet J, et al. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev. 2018;32:347–358. doi: 10.1101/gad.312397.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Martelot G, et al. Genome-Wide RNA Polymerase II Profiles and RNA Accumulation Reveal Kinetics of Transcription and Associated Epigenetic Changes During Diurnal Cycles. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001442. e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, et al. Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc Natl Acad Sci. 2018;115:E1916–E1925. doi: 10.1073/pnas.1715225115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nitzan M, Karaiskos N, Friedman N, Rajewsky N. Gene expression cartography. Nature. 2019 doi: 10.1038/s41586-019-1773-3. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Moshe S, et al. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab. 2019;1:899–911. doi: 10.1038/s42255-019-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seglen PO. Methods in Cell Biology. Vol. 13. Elsevier; 1976. Chapter 4 Preparation of Isolated Rat Liver Cells; pp. 29–83. [DOI] [PubMed] [Google Scholar]
- 56.Darnell AM, Subramaniam AR, O’Shea EK. Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol Cell. 2018;71:229–243.e11. doi: 10.1016/j.molcel.2018.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clayton DF, Harrelson AL, Darnell JE. Dependence of liver-specific transcription on tissue organization. Mol Cell Biol. 1985;5:2623–2632. doi: 10.1128/mcb.5.10.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loud AV. QUANTITATIVE STEREOLOGICAL DESCRIPTION OF THE ULTRASTRUCTURE OF NORMAL RAT LIVER PARENCHYMAL CELLS. J Cell Biol. 1968;37:27–46. doi: 10.1083/jcb.37.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuhn NJ, Woodworth-Gutai M, Gross KW, Held WA. Subfamilies of the mouse major urinary protein (MUP) multi-gene family: sequence analysis of cDNA clones and differential regulation in the liver. Nucleic Acids Res. 1984;12:6073–6090. doi: 10.1093/nar/12.15.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hurst JL, et al. Individual recognition in mice mediated by major urinary proteins. Nature. 2001;414:631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 62.Ilicic T, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maaten L, van der Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 64.Müller S, Scealy JL, Welsh AH. Model Selection in Linear Mixed Models. Stat Sci. 2013;28:135–167. [Google Scholar]
- 65.Schwarz G. Estimating the Dimension of a Model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 66.Jammalamadaka SR, SenGupta A. Topics in Circular Statistics. Vol. 5 WORLD SCIENTIFIC; 2001. [Google Scholar]
- 67.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyubimova A, et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat Protoc. 2013;8:1743–1758. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data
All scRNA-seq data is deposited in GEO with accession code GSE145197.
Reconstructed gene profiles
Reconstructed spatio-temporal gene profiles are available as Matlab files at https://github.com/naef-lab/Circadian-zonation
Web-application
The whole dataset of gene profiles along with the analysis is available online as a web-application at the URL https://czviz.epfl.ch/. The application was built in Python using the library Dash by Plotly (version 1.0).
The code for fitting the mixed-effects models and generating the main figures is available at https://github.com/naef-lab/Circadian-zonation
Details regarding the statistics, software and data are provided in the Reporting Summary.