Abstract
By investigating the topology of neuronal coactivity, we found that mnemonic information spans multiple operational axes in the mouse hippocampus network. High activity principal cells form the core of each memory along a first axis, segregating spatial contexts and novelty. Low activity cells join coactivity motifs across behavioural events and enable their crosstalk along two other axes. This reveals an organizational principle for continuous integration and interaction of hippocampal memories.
Assimilating new knowledge without corrupting acquired memories is critical. However, learning and memory interact: prior knowledge can proactively influence ongoing learning, and new information can retroactively modify pre-existing memories. The hippocampus is a brain region supporting memory1, yet the network-level operations that continuously incorporate new experiences, segregating them as discrete traces while enabling their interaction, are unknown. The discovery that hippocampal principal cells are tuned to the animal’s position and surrounding cues provides an important mechanistic foundation for the role of the hippocampus in memory, with each environment recruiting a discrete combination of neurons expressing a map-like representation of that space1,2. While this suggests how the hippocampus disentangles the spatial contexts of different memories, these representations further involve the fine-grained temporal coordination of neuronal spiking3,4. Notably, sets of jointly-active neurons organize motifs of coactivity (co-firing patterns), some of which underpin spatially-selective assemblies5. Here, we hypothesize that an adaptive topological reorganization of coactivity motifs enables embedding of new memory items in the hippocampal network.
We monitored dorsal CA1 (dCA1) ensembles from mice exploring a familiar environment before and after associating a novel environment with reward (sucrose), using a 1-day conditioned place preference (CPP) task (Fig. 1a and Extended Data Fig. 1a). Each day, mice first explored the familiar enclosure (exposure). Next, we identified the preference of each mouse for one of the two novel compartments connected by a bridge to form the CPP enclosure that day (pre-test). We subsequently removed that bridge in conditioning sessions where each mouse explored its non-preferred compartment baited with drops of a sucrose solution (+Suc.); and then the preferred compartment with drops of water (+Wat.). One hour after the last conditioning session, we re-inserted the bridge and tested CPP memory (CPP test; Extended Data Fig. 1b). To assess the effect of new CPP memory on prior representations, we finished each recording day by re-exposing mice to the familiar enclosure (re-exposure).
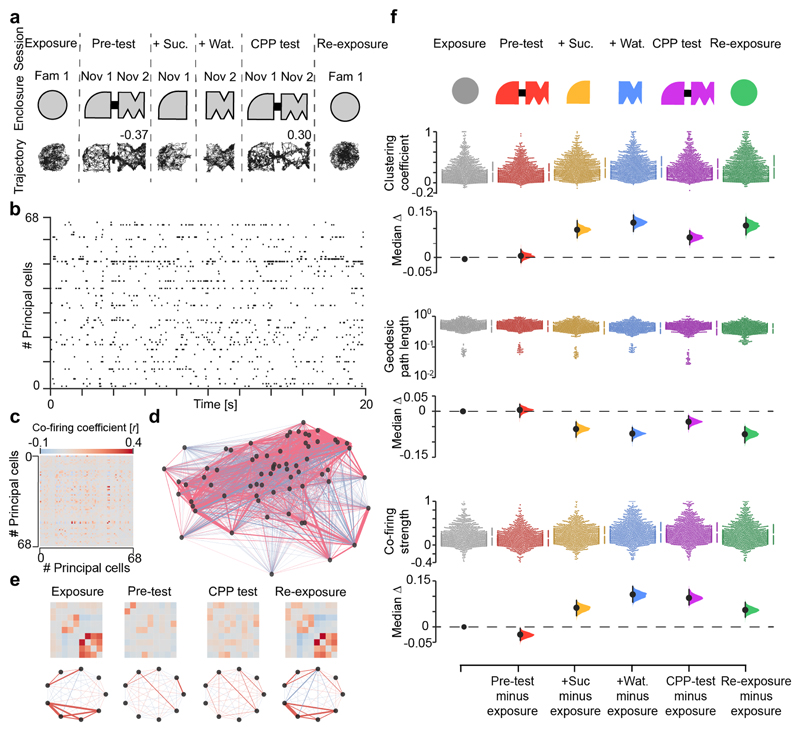
Figure 1. New CPP memory reorganizes pre-existing hippocampal co-firing topology.
(a) CPP task layout. Each day, mice explored the same familiar enclosure before and after a 4-session CPP task. Mouse trajectory in each session from one day is shown below the schematic of in-use enclosures. Numbers indicate place preference scores for pre-test and test, as the time in sucrose-paired compartment (+Suc.) minus that in water-paired compartment (+Wat.) over the sum. During CPP test, the mouse successfully changed its initial preference for the sucrose-paired compartment, as indicated by the positive score. (b) Example raster plot showing the spike trains of 68 simultaneously-recorded dCA1 principal cells (one cell per row) for the day shown in (a). For clarity, a 20-second sample is shown. (c) The corresponding adjacency matrix of the pairwise correlation coefficients measuring principal cells’ co-firing. (d) The corresponding co-firing graph. Each node represents one cell. Each edge represents the co-firing association of one cell pair, color-coded according to their correlation’s sign and width proportional to the edge’s absolute value. (e) Example adjacency matrices (top-row) and corresponding neuronal motifs (bottom-row) extracted from the graph shown in (d) to visualize some co-firing changes across sessions. (f) Changes in topological clustering (top), geodesic path length (middle) and single-neuron cumulative co-firing strength (bottom) of co-firing graphs. For each measure, the entire dataset is presented using a Cumming estimation plot to visualize the effect size; each upper panel shows the distribution of raw data points (each point represents one cell) for each color-coded session (with the gapped lines on the right as mean (gap) ± SD (vertical ends) for each session); each lower panel displays the difference between a given session and the exposure, computed from 5,000 bootstrapped resamples and with difference-axis origin aligned to the median of the exposure distribution. For each task-session: black-dot, median; black-ticks, 99% confidence interval; filled-curve: sampling-error distribution.
On each day we recorded neuronal spiking throughout the six CPP task sessions (Fig. 1a; n=1,083 total principal cells; 63.7±33.2 principal cells per day; 17 days from 7 mice). Using the spike trains recorded during active exploration (i.e. excluding immobility epochs and sharp-wave/ripples), we computed weighted graphs6,7 to explore the firing relationships between sets of coactive neurons in each task session (Fig. 1b-e; n=102 graphs; 260,058 co-firing pairs). In these co-firing graphs, each node represents one cell; the edge linking any two nodes represents the coactivity of that cell pair, with a weight computed as the Pearson correlation coefficient between their spike trains. For each graph, this procedure yielded an adjacency matrix of pairwise co-firing coefficients, with dimensions equal to the number of nodes (Fig. 1c and Extended Data Fig. 1c).
We observed that co-firing graphs include more triads of coactive nodes during CPP learning (Fig. 1f; Clustering coefficient), showing that associating a novel place with reward changes the coactivation structure of the network. Concomitantly, the average geodesic path length, calculated as the mean shortest path between any two nodes, decreases (Fig. 1f; Geodesic path length), indicating greater functional connectivity between sets of coactive neurons. By calculating for each node, the summed weight of all its edges in the adjacency matrix, we also found that the average single-neuron cumulative co-firing increases (Fig. 1f; Co-firing strength), reporting heightened firing associations among neurons during CPP learning. Similar topological deviations from the co-firing network featuring the familiar context during exposure occurred during exploration of a novel context (without reward), spontaneous preference for a novel place and reward experience in an otherwise familiar context (without CPP) (Extended Data Fig. 2), suggesting a general mechanism for integrating new information. Importantly however, these topological deviations did not reset during re-exposure one hour after CPP (Fig. 1f) while they did in the other tasks (Extended Data Fig. 2). These sustained changes following CPP neither reflected differences in exploration nor simple fluctuations in co-firing (Extended Data Fig. 3). Thus, the mnemonic operation of updating a recently-encountered place with reward caused an enduring topological reorganisation (“hysteresis”) in the coactivity structure of the network.
We next asked whether the topological hysteresis caused by CPP on the co-firing motifs expressed in the familiar enclosure (Fig. 1f) affected its spatial representation. By computing the firing maps of individual neurons in each task session (Fig. 2a), we found that both single-neuron and population-level maps featuring exposure reorganized in the CPP enclosure to then largely re-emerge during re-exposure (Extended Data Fig. 4a,b). Yet, despite their reinstatement during re-exposure after CPP, familiar maps seemed edited beyond mere fluctuations in neuronal activity (Extended Data Fig. 4c-g).
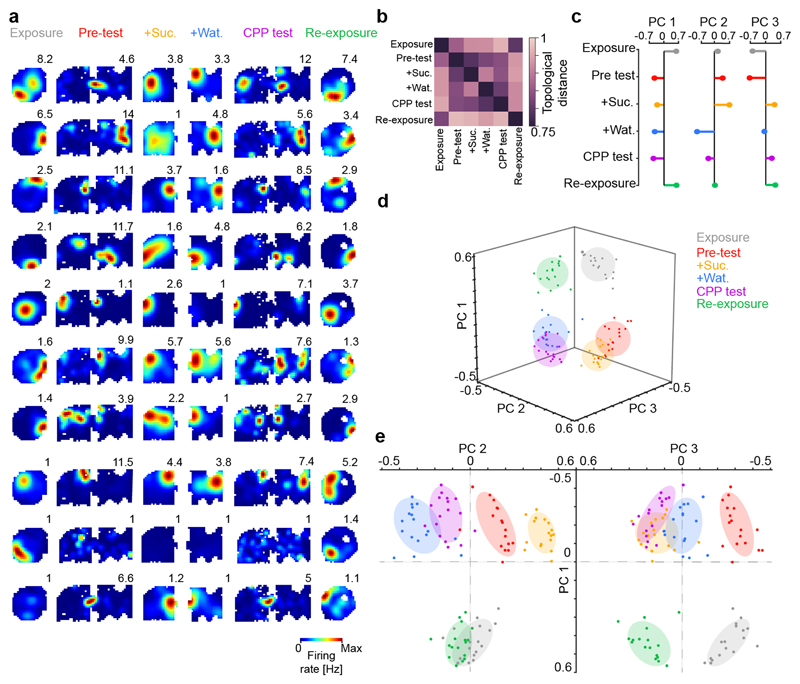
Figure 2. Mnemonic information spans multiple network co-firing axes.
(a) Example firing maps across CPP task-sessions (one cell per row; numbers indicate peak rate). (b) Average matrix of the topological distances separating the co-firing graphs across the six CPP task-sessions. (c-e) Principal Components (PCs) Analysis applied to matrices of topological distances unfolded multiple axes explaining across-session variance in co-firing. (c) Weight vectors representing the contribution of individual sessions to the variance in topological distances along the first three PCs. (d) Projection of the topological distances between co-firing motifs onto the first three PCs. Each data point represents one (color-coded) session of a given mouse day. (e) Same topological distances projected on the PC1 versus PC2 (left) and PC3 (right) planes. Note the segregation of co-firing motifs along PC1 for sessions in familiar versus CPP enclosures; for the four CPP sessions along PC2; and for exposure/re-exposure sessions along PC3.
These results indicate that new CPP memory re-structured the prior representation of the familiar environment. To discern the effect of this cross-talk, we analysed the transformation of co-firing graphs within the “network activity space”. We computed the topological distances separating graphs across the six CPP task-sessions (Fig. 1a), using the Riemannian Log-Euclidean metric. For the co-firing adjacency matrix of each session, this procedure yielded a vector of distances to the adjacency matrices of the other sessions recorded that day (n=6 task-session pairwise distance vectors, together forming one 6x6 matrix each day; Fig. 2b and Extended Data Fig. 5a). Principal components analysis of the distance matrices revealed three axes explaining ~80% of the variance between co-firing graphs across CPP task-sessions (Fig. 2c-e; see also Extended Data Fig. 5b-e). Along the first principal component, the co-firing patterns of a given session overlapped with those of the other sessions in the same environment but not across (Fig. 2c-e), thus discriminating the familiar enclosure from the novel CPP apparatus and reporting the hippocampal remapping between these contexts (Extended Data Fig. 4a-d). Coactivity along the next two components disentangled the distinct behavioural experiences within each enclosure. Namely, the four CPP sessions on the second axis and the sessions before from those after reward on the third axis, notably separating exposure and re-exposure to the familiar enclosure and thus reporting the topological hysteresis following CPP (Fig. 1f). These results held when considering separately the two CPP compartments during pre-test and test (Extended Data Fig. 5f,g). Across CPP task-sessions, co-firing motifs therefore spanned different directions of the network activity space, segregating spatial contexts while discretizing events within them. Likewise, multiple axes explained hippocampal co-firing in the other tasks (Extended Data Fig. 5h-j) as a general principle to organise information in the network.
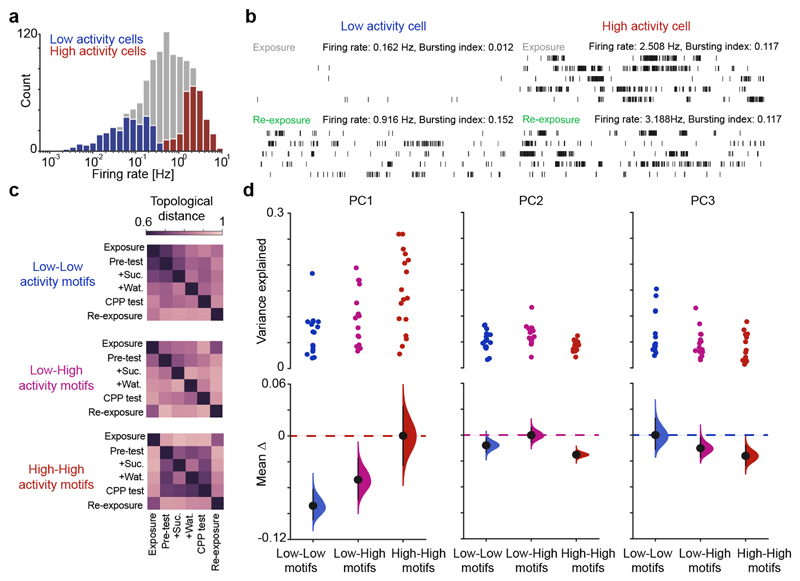
Recent findings suggest that the heterogeneity of the principal cell population, marked by the skewed (log-normal) distribution of firing rates, is central to the network operations underlying hippocampal function8–16. We thus identified principal cells in the top and bottom quartiles of the firing rate distribution during the exposure session (Fig. 3a) to examine the contribution of these two subpopulations to co-firing graphs. High and low-rate principal cells were biased towards deep and superficial pyramidal sublayers, respectively (Extended Data Fig. 6)8,13,15. We noted that low – but not high – activity cells discharged more bursts of spikes (spike packets with inter-spike intervals within 6ms) during re-exposure compared to exposure (Fig. 3b and Extended Data Fig. 7a,b), sustaining increased firing rate one hour after CPP (Extended Data Fig. 7c). These enduring activity changes were not observed in the other tasks (Extended Data Figs. 7d-i). Moreover, low activity cells with higher bursting during re-exposure showed increased spatial coherence and information content (Extended Data Fig. 8). Remarkably, these cells initially had the lowest place-field coherence during exposure and became spatially informative following CPP learning (Extended Data Fig. 9). In addition, low activity cells with the most spatially tuned activity during exposure also showed increased spatial information during re-exposure, without altered burst spiking (Extended Data Fig. 9). These results suggested that the cross-talk between the new CPP memory and the prior familiar representation involved the heightened network contribution of low activity cells following their recruitment during CPP learning. Indeed, the low – but not high – activity subpopulation exhibited sustained topological changes throughout CPP sessions (Extended Data Fig. 10a), explaining whole-network hysteresis during re-exposure (Fig. 1f). Moreover, reward-related firing modulation of low activity cells from pre-test to sucrose conditioning in the CPP enclosure predicted changes in co-firing from exposure to re-exposure in the familiar enclosure (Extended Data Fig. 10b,c), constituting another instance of the impact of reward on hippocampal activity13,17.
Figure 3. High and low activity cells contribute differentially to network co-firing axes.
(a) Firing rate (log-normal) distribution of principal cells during exposure. The highest and lowest quartiles formed the high and low activity subpopulations, respectively. (b) Example spike trains of a low and a high activity cell during exposure and re-exposure. Two raster plots shown for each cell (60-s rows; vertical ticks representing spike times) during exposure (top) and re-exposure (bottom). For clarity, the first 5-min of each session is represented. (c) Matrices of topological distances separating co-firing graphs across sessions, for low-low, low-high and high-high activity cell pairs. (d) Cumming estimation plots showing the contribution of low-low, low-high and high-high activity cell pairs to co-firing variance along the first three axes of the CPP network. Top: each data point represents the variance explained by a given co-firing motif along one axis for one day. Bottom: for each axis, the co-firing motif with the strongest contribution (color-coded dashed line) is compared with the other two motifs’ contribution. Black-dot, mean difference; black-ticks, 99% confidence interval; filled-curve: sampling-error distribution.
We finally evaluated the contribution of high and low activity subpopulations to network co-firing motifs, leveraging the Riemannian Log-Euclidean framework (Fig. 2b-e). Co-firing solely involving high activity cells segregated the two task enclosures at the onset of pre-test (Fig. 3c and Extended Data Fig. 10d). Coactivity motifs including low activity cells did not distinguish familiar exposure and novel CPP pre-test (Fig. 3c and Extended Data Fig. 10d). Rather, this subpopulation integrated co-firing patterns as mice experienced each task event, staying engaged thereafter during re-exposure where their contribution to network motifs reached that of high activity cells. Accordingly, in the CPP task the high and low activity cells contributed more to the first and third co-firing network axis, respectively; while their combination explained more the second axis (Fig. 3d). Moreover, the topological changes affecting the co-firing structure of the network during active exploration were associated with changes in sharp-wave/ripple co-firing of low activity cells during awake rest (Extended Data Fig. 10e,f), in line with recent work showing that these two subpopulations exhibit distinct sharp-wave/ripple response12,18.
In conclusion, by exploring the hippocampal network activity space from a graph-theoretical perspective, we show that new associative memories (place-reward) restructure the neural patterns representing prior memory for an unrelated environment. During this transformation of the hippocampal co-firing structure, high activity cells organize motifs that rapidly discriminate spatial contexts, robust to perturbation by subsequent experience. This contribution might instantiate a spatio-contextual backbone for memory schemas19,20. Low activity cells integrate coactivity motifs on-demand, throughout behavioural events. Their heightened engagement continues after new place-reward learning and across contexts, affecting the network representation of an otherwise familiar environment. This effect could involve low-rate principal cell plasticity12 leveraged by high computational load or neuro-modulatory processes when novel contextual information and reward experience are related in memory. The newly-acquired topology of the hippocampal firing output, shaped by past inputs, may not be permanent, perhaps slowly returning to a baseline configuration as memories consolidate to other cortical circuits. The observed hysteresis could represent a network response protecting existing memories from catastrophic interference by adjusting the co-firing structure of learnt associations or forming redundant ones. This could also reflect retroactive encoding of recent experience, juxtaposing the current spatial reference frame with those encountered before. Together, these findings support the view of a division in computational labour within the log-normally distributed principal cell population for the hippocampal discretization of memories of space and events, allowing adaptive insertion and interaction of new information within a larger network of prior knowledge.
Methods
Animals
These experiments used adult male C57BL/6J mice (Charles River Laboratories, UK) or transgenic heterozygous CamKIIa-Cre mice (Jackson Laboratories; CamKIIa-Cre B6.Cg-Tg(Camk2a-cre)T29-1Stl/J, stock number 005359, RRID: IMSR_JAX:005359; maintained on a C57BL/6J background). Mice were housed with their littermates until the surgical procedure with free access to food and water in a room with a 12/12h light/dark cycle, 19–23°C ambient temperature and 40–70% humidity. All mice held in IVC's, with wooden chew stick and nestlets. Mice were 4-7 months old at the time of testing. Experimental procedures performed on mice in accordance with the Animals (Scientific Procedures) Act, 1986 (United Kingdom), with final ethical review by the Animals in Science Regulation Unit of the UK Home Office.
Surgical procedure
Mice were implanted with a microdrive during a surgical procedure performed under deep anaesthesia using isoflurane (0.5–2 %) and oxygen (2 l/min), with analgesia (0.1 mg/kg vetergesic) provided before and after surgery. Microdrives contained 10–12 tetrodes, targeting the stratum pyramidale of the dorsal CA1 hippocampus 21. Tetrodes were constructed by twisting together four insulated tungsten wires (12 µm diameter, California Fine Wire) and heating them to fuse them into a single bundle. Each tetrode was attached to a M1.0 screw to enable their independent movement. The drive was implanted under stereotaxic control in reference to bregma21. Tetrodes were initially implanted above the CA1 pyramidal layer and their exposed parts were covered with paraffin wax. The drive was then secured to the skull using dental cement and stainless-steel anchor screws inserted into the skull. Two of the anchor screws, both above the cerebellum, were attached to a 50 µm tungsten wire (California Fine Wire) and served as ground. Tetrode placement was confirmed by the electrophysiological profile of the local field potentials in the hippocampal ripple frequency band and anatomical electrode tracks 21,22. In one mouse, a single-shank silicon probe (Neuronexus, model A1x32-5mm-25-177-H32_21mm) was implanted following the same surgical procedure to span the somato-dendritic axis of dCA1 principal cells and establish the laminar profile of the sharp-wave/ripples (SWRs) detected in the local field potentials. These silicon probe recordings allowed estimating the position (depth) of individual tetrode-recorded principal cell soma (Extended Data Fig. 6).
Recording procedure
After at least one-week post-operative recovery, mice were handled for 3-4 days and then daily familiarized to the experimental paradigm, including connection to the electrophysiological recording system and exploration of a circular-walled open-field enclosure (42 cm diameter; the familiar enclosure). Mice were food restricted (to ~90% body weight) and the various experimental conditions were randomly allocated across mice and recording days.
The 1-day CPP task included four sessions: pre-test, place conditioning to sucrose (+Suc.), to water (+Wat.) and test23. The enclosure consisted of two square-walled (46 cm × 46 cm × 38 cm) compartments with distinct inside building block configurations on each day. A bridge (8 cm length, 7 cm width) connected the two compartments during pre-test and CPP test. A linear locomotion assistant (Imetronic, Pessac, France) held the recording cable while sensing its movement using infrared light beam detectors, allowing the connected animal to move freely within and across compartments. During pre-test, mice explored the entire CPP enclosure for 15min and their baseline (spontaneous) preference for one of the two compartments was determined. Next, the bridge was removed for the conditioning sessions, and mice explored their non-preferred compartment containing drops of 20% sucrose diluted in water (2x10min sessions, 10x10μl drops per session). Next, mice explored their preferred compartment containing drops of water (2x10min sessions, 10x10μl drops per session). One hour later, CPP memory was assessed by allowing the mice to explore the entire apparatus for 15min (CPP test). We calculated a place preference score for each mouse during both pre-test and test sessions as the difference between the time spent in the compartment paired with sucrose minus that paired with water during conditioning over their sum. On the morning of each recording day, the local field potential (LFP) signals obtained from each tetrode was used to guide its optimal positioning within the dCA1 pyramidal layer in search of multi-unit spiking activity 21. Tetrodes were left in position for ~1.5-2h before recordings started. On each day, ensemble recordings were performed continuously while mice explored the familiar circular-walled enclosure before and after (“exposure” and “re-exposure”; 15 minutes each) the CPP task. Mice were subjected to a novel CPP enclosure on each recording day (i.e., novel spatial configurations and wall cue cards).
Three other behavioural tasks had a 6-session layout similar to that of the CPP task, in order to evaluate whether the topological deviations observed during CPP reflected a general network response for integrating new information. In these three tasks, each day contained 6 sessions that matched the timeline of the CPP task. Mice explored the familiar enclosure before (exposure) and one hour after (re-exposure) either: (i) four sessions of spatial exploration in a novel enclosure without reward (the “Novel context only” task; Extended Data Fig. 2a), (ii) four sessions testing spontaneous preference for one of two novel enclosures, without reward (the “Spontaneous Place Preference” task; Extended Data Fig. 2c), or (iii) four sessions of spatial exploration in a second familiar enclosure with sucrose reward and water provided in the second and third session, respectively (the “Familiar context with reward” condition; Extended Data Fig. 2f), as in the CPP task (Fig. 1a). A fifth task further allowed testing whether network topological deviations occurred during repeated exploration of a familiar enclosure (the “Familiar context only” task; four exploration sessions in the same familiar enclosure on each day; Extended Data Fig. 2h).
On each recording day, mice were returned to their homecage between task sessions, having access to water and food while the experimenter prepared the open-field arena for the next session. Data collection could not be performed blind to the conditions of the experiments since the experimenter had to be aware as to which condition they had to expose each mouse on a given day (which behavioural task) and on a given session (which open-field arena). At the end of each day, tetrodes were raised to avoid possible mechanical damage overnight.
Multichannel data acquisition and position tracking
The extracellular signals from the electrodes were amplified, multiplexed, and digitized using a single integrated circuit located on the head of the animal (RHD2164, Intan Technologies; gain x1000). The amplified and filtered (0.09Hz to 7.60kHz) electrophysiological signals were digitized at 20kHz and saved to disk along with the synchronization signals from the position tracking. To track the location of the animal, three LED clusters were attached to the electrode casing and captured at 25 frames per second by an overhead colour camera.
Spike detection and unit isolation
For the offline detection of spikes, the recorded signals were first band-pass filtered (800 Hz to 5 kHz). Spikes were then detected based on the power (root-mean-square) of the filtered signal calculated in 0.2-ms sliding windows. Detected spikes of the individual electrodes were combined per tetrode. To isolate spikes belonging to the same neuron, spike waveforms were first up-sampled to 40 kHz and aligned to their maximal trough 24. Principal component analysis was applied to these waveforms ±0.5 ms from the trough to extract the first three or four principal components per channel, such that each individual spike was represented by 12 waveform parameters. An automatic clustering program (KlustaKwik, http://klusta-team.github.io) was run on this principal component space and the resulting clusters were manually recombined and further isolated based on cloud shape in the principal component space, cross-channels spike waveforms, auto-correlation histograms and cross-correlation histograms25,26. All sessions recorded on the same day were concatenated and clustered together. Each cluster used for further analysis showed throughout the entire recording day stable cross-channels spike waveforms, a clear refractory period in its auto-correlation histogram, well-defined cluster boundaries and an absence of refractory period in its cross-correlation histograms with the other clusters. Hippocampal principal cells were identified by the shape of their auto-correlation histogram, their firing rate and their spike waveform24. We further used an automated clustering pipeline using Kilosort (https://github.com/cortex-lab/KiloSort)27 via the SpikeForest sorting framework (https://github.com/flatironinstitute/spikeforest)28. To apply KiloSort to data acquired using tetrodes, the algorithm restricted templates to channels within a given tetrode bundle, while masking all other recording channels. The resulting clusters were manually curated to check all clusters and remove spurious units using metrics derived from the waveforms and spike times, and then verified by the operator. This procedure was cross-validated using several datasets and verified against manual curation, by computing confusion matrices to validate that clusters obtained automatically were also obtained with the previous method.
In total, this study includes n=3,483 principal cells from 63 recordings days: n=1,083 principal cells in the “Conditioned Place Preference” task (63.7±33.2 principal cells per day; 17 CPP 6-session days from 7 mice; 5 CamKIIa-Cre and 2 C57BL/6J; yielding 102 graphs; 260,058 co-firing pairs), n=585 principal cells in the “Novel context only” task (45.0±13.7 principal cells per day; 13 “Novel only” 6-session days from 5 mice; 3 CamKIIa-Cre and 2 C57BL/6J; yielding 78 graphs; 28,192 co-firing pairs), n=640 principal cells in the “Spontaneous Place Preference” task (49.2±17.5 principal cells per day; 13 SPP 6-session days from 6 mice; 3 CamKIIa-Cre and 3 C57BL/6J; yielding 78 graphs; 34,838 co-firing pairs), n=517 principal cells in the “Familiar context with reward” task (57.4±12.2 principal cells per day; 9 “Familiar with reward” 6-session days from 3 mice; 2 CamKIIa-Cre and 1 C57BL/6J; yielding 54 graphs; 30,512 co-firing pairs) and n=658 principal cells in the “Familiar context only” task (59.3±15.8 principal cells per day; 11 “Familial only” 4-session days from 3 mice; 2 CamKIIa-Cre and 1 C57BL/6J; yielding 44 graphs; 40,744 co-firing pairs).
Sharp-wave/ripples (SWRs)
Local field potentials (LFPs) of each pyramidal CA1 channel (for tetrode recordings) or recording site (for linear silicon probe recordings) were subtracted by the mean across all channels/sites (common average reference). These re-referenced signals were then filtered for the ripple band (110 to 250 Hz; 4th order Butterworth filter) and their envelopes (instantaneous amplitudes) were computed by means of the Hilbert transform. The peaks (local maxima) of the ripple band envelope signals above a threshold (5 times the median of the envelope values of that channel) were regarded as candidate events. Further, the onset and offset of each event were determined as the time points at which the ripple envelope decayed below half of the detection threshold. Candidate events passing the following criteria were determined as SWR events: (i) ripple band power in the event channel was at least 2 times the ripple band power in the common average reference (to eliminate common high frequency noise); (ii) an event had at least four ripple cycles (to eliminate events that were too brief); (iii) ripple band power was at least 2 times higher than the supra-ripple band defined as 200-500 Hz (to eliminate high frequency noise, not spectrally compact at the ripple band, such as spike leakage artefacts). We classified tetrodes as being in the deep or superficial sublayer of the CA1 pyramidal layer based on the mean peak amplitude of detected SWRs (Extended Data Fig. 6). Positive values indicated that the tetrode was in the deep sublayer (i.e. closest to stratum oriens) while negative values indicated tetrode was in the superficial sublayer (i.e. closest to stratum radiatum)8,15,29. SWRs were also used as time bins to calculate SWR firing response of low and high activity cells (Extended Data Fig. 10e,f).
Weighted graphs of neuronal co-firing
We constructed weighted graphs that represent the spike relationships between dCA1 principal cells recorded in a given task session, calculating for that session the set of pairwise Pearson correlation coefficients between all pairs of spike trains. These co-firing graphs were computed using time bins during active exploratory behavior (with speed>2 cm.s-1), discarding periods of immobility and further excluding sharp-wave/ripples (SWRs) in each task session. The recorded neurons (and their co-firing associations) are therefore the nodes (and their edges) in the co-firing graph of each task event. We described each graph by its adjacency matrix, A, as an N x N square matrix:
where N is the number of nodes in the graph; and each element, w ij, is a continuous weight value that defines the edge (i.e., the co-firing coefficient) between two nodes i and j. To compute each co-firing association value we used a bin-less approach by convolving the spike trains of i and j with a Gaussian kernel (SD=40ms) and then calculating their correlation coefficient r (thus, -1 ≤ r ≤ 1). As a result, A is symmetric, w ij = w ji, and the graph is undirected.
Clustering coefficient
We computed a clustering coefficient to characterize the local synchronization of network activity by quantifying the number of three-node motifs. In each graph, for any neuron i, we obtained its clustering coefficient C i using the formula proposed by Onnela et al. to quantify the strength of each triad30–32:
where j and q are neighbors of neuron i, all edge weights are normalized by the maximum edge weight in the network ŵ = w/max(w), and k i is the degree of neuron i, which in these weighted graphs with no self-connection is equal to the number of neurons minus one. Note that this formula accounts for negative edges, yielding a negative value when there is an odd number due to the negative edges in the triad; it is positive otherwise.
Geodesic path length
We measured the geodesic (i.e., shortest) path length to estimate the coordination efficacy between the activity of any two nodes in the graph. In a binary graph, this would represent the smallest number of edges connecting two nodes. Here, we embedded each weighted graph in a lattice and defined the length between two nodes i and j as: discarding all negative edges. We then identified the shortest path length between any two nodes in the graph using the Floyd-Warshall algorithm33–35.
Single-neuron cumulative co-firing strength
We defined the single-neuron cumulative co-firing strength as the total pairwise activity correlation strength of a given node in a weighted graph. As a reference, the strength in a weighted graph can be compared to the degree in a binary graph, which accounts for the number of the node’s neighbours. Here, the strength S i of a node i is the sum of all the weights w ij of the edges projected from that node:
where N is the number of neurons j that node i projects to.
Spatial rate map analyses
We divided the horizontal plane of the recording enclosures into spatial bins of approximately 2×2 cm to generate the spike count map (number of spikes fired in each bin) for each neuron and the occupancy map (time spent by the animal in each spatial bin) in each task session. All maps were then smoothed by convolution with a two-dimensional Gaussian kernel having a standard deviation equal to two bin widths. Finally, spatial rate maps were generated by normalizing the smoothed spike count maps by the smoothed occupancy map.
The single-neuron map similarity was calculated by the 2D Pearson correlation coefficient between the place maps of a given neuron across two task sessions. Here, this task session pairwise measure compares for each neuron the spatial relationship between its place map in the exposure session and that computed for another task session. For the pre-test and test sessions, two place maps were extracted (one per arena) and the single-neuron map similarity was obtained taking the maximum similarity value between either of the two CPP sessions’ maps and that of the exposure session.
The population map similarity5,36,37 compares the spatial relationships of the set of place maps computed for one task session with those from another task session. Here, this task session pairwise measure represents the extent to which sets of cells that fired in similar regions of space (that is, overlapping place fields) during the exposure session still fire together in similar regions of space later during another task session. For this measure, we used cells with a spatial coherence value (see below) above 0.2 during the exposure session. The population map similarity was calculated by first computing the place field similarity (PFS) value for each cell pair during the exposure session as the Pearson correlation coefficient from the direct binwise comparisons between the spatial rate maps of the two cells, limited to valid bins (occupancy greater than zero). This procedure yields a vector storing the PFS values for all cell pairs during the exposure session. We repeated this procedure to obtain the PFS vectors of the other task sessions. Finally, the population map similarity between two task sessions was calculated as the Pearson correlation coefficient between the PFS vector of the exposure session versus the PFS vector of the chosen comparison session.
Spatial coherence
To measure the spatial coherence (i.e. the similarity of a cell’s firing rate over spatial bins), we computed a boxcar-averaged version of its unsmoothed spatial rate map, with each bin replaced by the arithmetic mean of itself and its eight adjacent neighbours. The smoothed spatial rate map was then correlated to its unsmoothed version to yield a Pearson correlation value38.
Spatial mutual information
To estimate the amount of spatial information conveyed by the spike train of a given cell, we used the mutual information measure I(R;S ):
where S is the discrete random variable formed by the set of the animal’s spatial locations s, and R is the discrete random variable formed by the set of possible spike count responses r 39–41. This was corrected for estimation bias by subtracting an analytical estimate of the bias42.
Across-graph topological distance analyses
Co-firing graphs have symmetric and positive semi-definite adjacency matrices since their elements are computed using the Pearson correlation coefficient between pairs of spike-trains. Thus, their topologies lie on a Riemannian manifold43 and the distance between them is most accurately measured by the geodesic distance between them44. Accordingly, we employed the log-Euclidean metric45 to compute the topological distance between constellations of co-firing patterns for pairs of task session graphs. From a Euclidean norm on symmetric matrices ∥·∥, the distance D LE between two matrices is given by:
where A 1 and A 2 are the two adjacency matrices to be compared and log(·) is the matrix logarithm, the inverse of the matrix exponential defined as:
This procedure projects the adjacency matrices to a flat (zero curvature) Riemannian space, allowing applying Euclidean computations. This way, we obtained for each task session s (from exposure to re-exposure) a vector of six topological distances D s separating the co-firing graph of that session to the co-firing graph of each of the other five task sessions, as well as to itself (by adding one SD of white noise to avoid exact 0 results). We normalized each of these vectors by their maximum distance value, to compare them across days. We thus obtained a 6 x 6 matrix of the topological distances between the co-firing graphs of the six task sessions for each recording day.
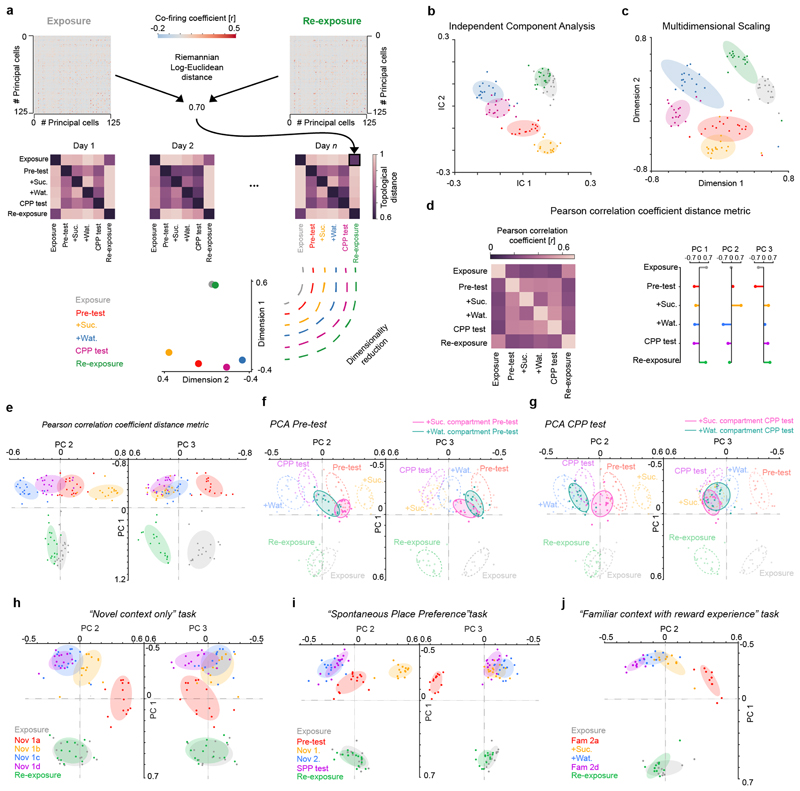
Principal Components Analysis (PCA) was used as a dimensionality reduction method to visualize the topological distances separating all co-firing graphs in the activity space of the hippocampal network. To do so, we stacked all topological distances vectors in a 6 x N matrix, where N is the total number of co-firing graphs (N=n_d x n_s, one graph per task session, n_s, per recording day, n_d). We next used PCA on the recordings with more than 40 principal cells to obtain the set of principal components that indicate trajectories in the network activity space along which co-firing graphs evolved from exposure to re-exposure task sessions. We then projected the first principal components that explained 80% of the variance in co-firing patterns onto the topological distances separating the co-firing graphs of all recordings from a given task. Independent Component Analysis (ICA) and Multidimensional Scaling (MDS) were also used as additional methods for such a visualization.
Of note, there was a possibility that using the Pearson correlation to evaluate distances between each pair of co-firing matrices could alter the eigenvalues and the positive semi-definiteness of the co-firing matrices. Since the eigenvalues are the basis for computing geodesic distances, the Pearson correlation coefficient could distort the similarity evaluation between co-firing matrices44. Yet, we additionally applied the Pearson correlation coefficient to measure the relationships between co-firing matrices across CPP task sessions: the first three PCs extracted this way corresponded to those revealed by the Riemannian log-Euclidean distance metric (Extended Data Fig. 5d,e), notably showing that the co-firing network axes reported in this study are not mere outputs of a specific analysis method.
Bursting index
For the spike train of each neuron, we defined spike bursts as transient packets of spikes with inter-spike intervals less than 6ms 46,47. We used the inter-spike interval t versus inter-spike interval t + 1 plot to identify the first, mid and last spikes of each burst candidate. The bursting index was then defined by the ratio of bursting spikes out of all the spikes fired by the neuron.
High and low activity cells contribution to network co-firing patterns
For each task session s, we quantified the contributions of the high and low activity sub networks G to network-level co-firing patterns by computing the Euclidean distance between the six-dimensional topological distance vector representing the entire network D s and those representing the two sub-networks (see section above):
Data and statistical analyses
Data were analysed in Python 3.6 and using the packages DABEST v0.3.048, scikit-learn v0.23.249, NetworkX v2.450, pyentropy v0.5.051, Numpy v1.18.1, Scipy v1.4.1, Matplotlib v3.1.2, Pandas v0.25.3 and Seaborn v0.11.0. All statistical tests related to a symmetric distribution were performed two-sided using Gardner-Altman plots (to compare 2 groups) and Cumming plots (for more groups) from the Data Analysis with Bootstrap-coupled ESTimation (DABEST) framework48. These DABEST plots allow visualizing the effect size by plotting the data as the mean or median difference between one of the groups (the left-most group of each plot, used as group-reference) and the other groups (to the right, along the x-axis of each plot). For each estimation plot: (i) the upper panel shows the distribution of raw data points for the entire dataset, superimposed on bar-plots reporting group mean±SEM, unless stated otherwise; and (ii) the lower panel displays the difference between a given group and the (left-most) group-reference, computed from 5,000 bootstrapped resamples and with difference-axis origin aligned to the mean or the median of the group-reference distribution. For each estimation plot: black-dot, mean (for normal distributions) or median (for skewed distributions) as indicated; black-ticks, 95% or 99% confidence interval as indicated; filled-curve: bootstrapped sampling-error distribution. Data distributions were assumed to be normal but this was not formally tested. We also used the t-test to compare two conditions; the Wald test for assessing the significance of regression lines; and the Kolmogorov–Smirnov test for comparing probability distributions. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications (e.g., 5,8,10,12,13,15–19). Neural and behavioural data analyses were conducted in an identical way regardless of the identity of the experimental condition from which the data were collected, with the investigator blind to group allocation during data collection and/or analysis. See also the corresponding Life Sciences Reporting Summary.
Extended Data
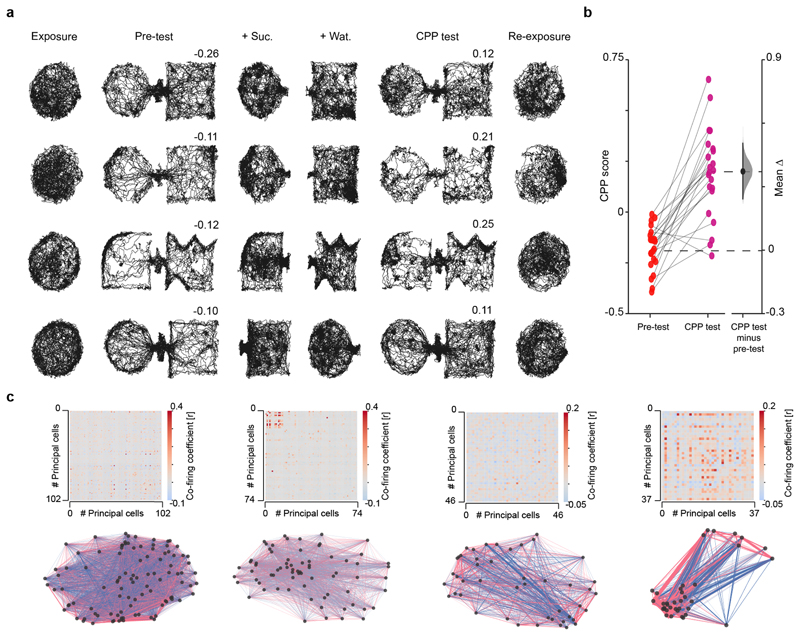
Extended Data Fig. 1. Behavioural performance and hippocampal co-firing graphs in the CPP task.
(a) Additional example trajectories for four mice. Each day (one day per row), mice explored the same (circular-walled) familiar enclosure before (exposure) and after (re-exposure) a 1 day 4-session CPP task (Fig 1a). The CPP apparatus was formed by two novel compartments on each day. Numbers indicate place preference scores for pre-test and CPP test sessions, as the time in sucrose-paired compartment (+Suc.) minus that in water-paired compartment (+Wat.) divided by the sum. (b) Place preference scores. A negative score indicates that mice spent less time in the compartment paired with sucrose during conditioning. Note that during CPP test, mice successfully changed their preference for the compartment recently paired with sucrose, as indicated by the positive score. Scores are presented using a Gardner-Altman estimation plot to visualise the effect size. The left panel shows the distribution of raw data points for the pre-test and test sessions. The right panel displays the difference between CPP test and pre-test, computed from 5,000 bootstrapped resamples and with difference-axis origin aligned to the mean of the pre-test session distribution. Black-dot, mean difference; black-ticks, 99% confidence interval; gray-filled-curve: sampling-error distribution. (c) The adjacency matrices (top row) of the pairwise correlation coefficients measuring principal cells’ co-firing for days shown in (a), with the corresponding co-firing graphs (bottom row). Each node represents one principal cell. Each edge represents the co-firing association of one cell pair, color-coded according to their correlation’s sign and width proportional to the edge’s absolute value.
Extended Data Fig. 2. Graph topology during exploration of a novel enclosure, spontaneous novel place preference and exploration of a familiar enclosure with or without reward.
(a,b) “Novel context only” task. Behavioural protocol (a) and topology of hippocampal co-firing graphs (b) for unrewarded exploration of a novel enclosure (n=585 total principal cells; 45.0±13.7 principal cells per day; 13 days from 5 mice yielding n=78 graphs; 28,192 co-firing pairs). Each day, mice explored the familiar enclosure before (exposure) and after (re-exposure) exploring a novel enclosure over four sessions, without sucrose reward. The session timeline matches that of the CPP task. (a) An example mouse trajectory for each session is shown for one day below the schematic of in-use enclosures. Additional examples shown below for two other days (one day per row). (b) The topological changes in clustering (top), geodesic path length (middle) and single-neuron cumulative co-firing strength (bottom) of co-firing graphs. For each measure, the entire “Novel context only” dataset is presented using a Cumming estimation plot to visualize the effect size. Each upper panel shows the distribution of raw data points (one point represents one principal cell) for each color-coded session (with the gapped lines on the right as mean (gap) ± SD (vertical ends) for each session). Each lower panel displays the difference between a given session and exposure, computed from 5,000 bootstrapped resamples and with difference-axis origin aligned to the median of the exposure distribution. For each session: black-dot, median; black-ticks, 99% confidence interval; filled-curve: sampling-error distribution. Note that exploring a novel context causes topological deviations from the co-firing graph that had featured exposure, indicating that the hippocampal network “learns” about the novel spatial layout. These deviations no longer occurred during re-exposure. (c-e) “Spontaneous Place Preference” (SPP) task. Behavioural protocol (c) and performance (d) along with the graph topology (e) (n=640 total principal cells; 49.2±17.5 principal cells per day; 13 days from 6 mice yielding n=78 graphs; 34,838 co-firing pairs). Each day, mice explored the familiar arena before (exposure) and after (re-exposure) exploring a CPP-like apparatus formed by two novel compartments (Nov 1 and Nov 2) connected with a bridge (pre-test). Having identified the preference of each mouse for one of the two novel compartments, the bridge was next removed; each mouse explored its non-preferred compartment, and then its preferred compartment, as in the CPP task but without sucrose nor water. One hour after, the bridge was re-inserted to test place preference (SPP test). The session timeline matches that of the CPP task. (c) Example mouse trajectories as in (a). Numbers indicate place preference scores for pre-test and SPP test, as the time in the non-preferred compartment minus that in the preferred compartment over the sum. Note that mice did not changed their preference, as indicated by the negative scores. (d) Scores presented using a Gardner-Altman estimation plot (as in Extended Data Fig. 1b). Note that co-firing topology also deviates during SPP sessions from that featuring exposure. Topological deviations no longer marked re-exposure. (f, g) “Familiar context with reward” task. Behavioural protocol (f) and graph topology (g) (n=517 total principal cells; 57.4±12.2 principal cells per day; 9 days from 3 mice yielding n=54 graphs; 30,512 co-firing pairs). Each day, mice explored the first familiar arena before (exposure) and after (re-exposure) exploring a second familiar enclosure during four sessions. The timeline matches that of the CPP task. Drops of sucrose (+Suc.) and water (+Wat.) were provided during Fam 2b and Fam 2c sessions, respectively. Example mouse trajectories from three days (f) shown as in (a). (g) Similar to the unrewarded exploration of novel enclosures (b,e), co-firing graph topology deviated during exploration of the second familiar arena when paired with reward. These deviations no longer marked re-exposure to the first familiar enclosure. (h, i) “Familiar context only” task. Behavioural protocol (h) and network topology measured by co-firing strength (i) during exploration of a familiar enclosure without reward (n=658 total principal cells; 59.3±15.8 principal cells per day; 11 days from 3 mice yielding n=44 graphs; 40,744 co-firing pairs). Each day, mice explored a familiar enclosure (Fam 1 or Fam 2) over four sessions, to match the four sessions used between exposure and re-exposure in the other tasks (Fig. 1a and Extended Data Fig. 2a-g). (h) Example mouse trajectories shown as in (a). (i) Co-firing strength did not deviate during repeated familiar explorations. (j) Topological changes between exposure and re-exposure are compared across tasks. Note the sustained deviations (topological “hysteresis”) following CPP.
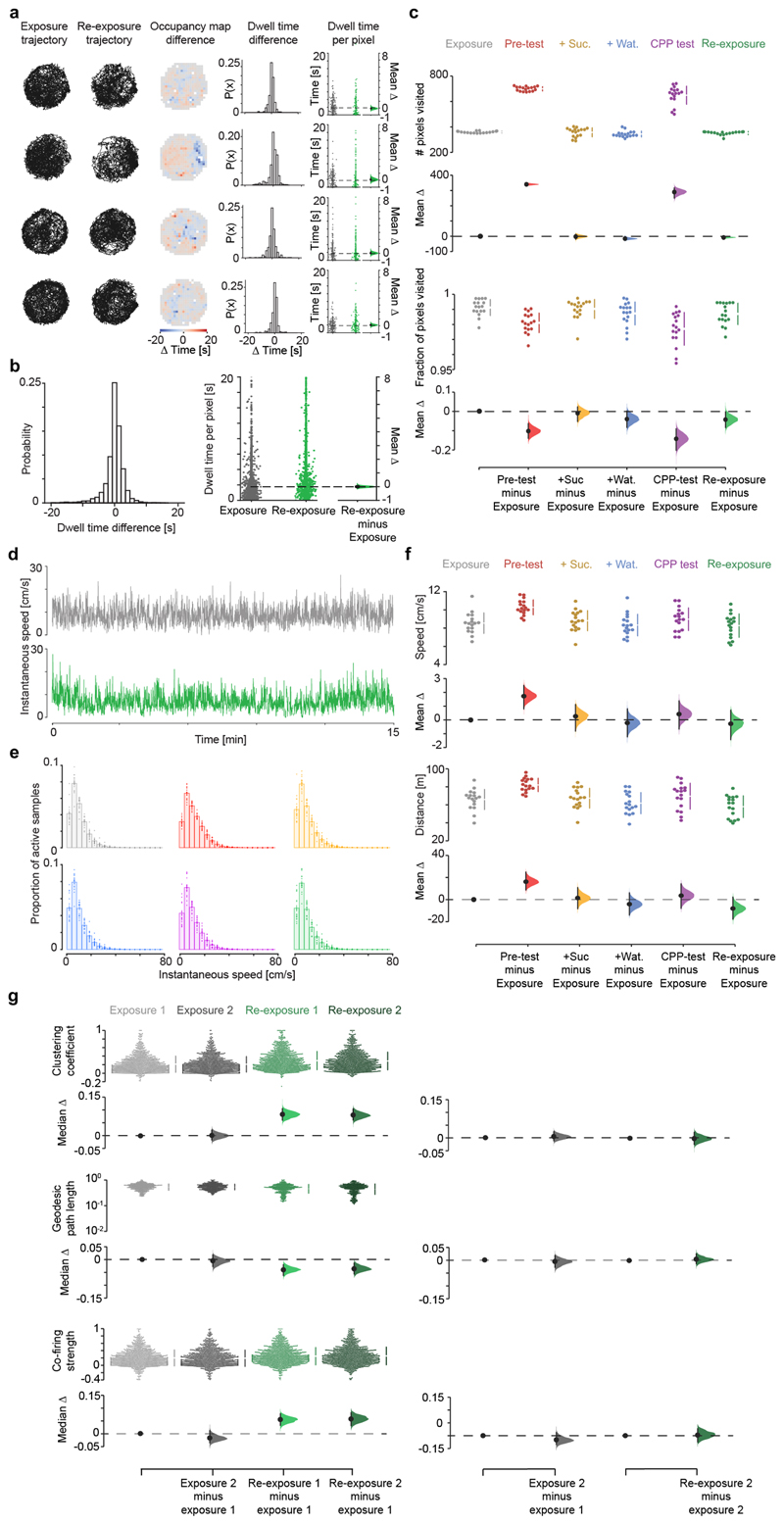
Extended Data Fig. 3. Topological changes between exposure and re-exposure in the CPP task do not relate to differences in spatial exploration or mere fluctuations in co-firing.
(a) Spatial occupancy during exposure and re-exposure for example CPP days from four mice (one mouse day per row; using examples shown in Extended Data Fig. 1). Shown for each day, from left to right: (i) animal’s trajectory for each familiar session, (ii) map of pixel-wise dwell time difference across the two sessions (for each pixel, the time spent in that pixel during exposure minus re-exposure), (iii) distribution of pixel-wise dwell time differences for all pixels covering the familiar enclosure, showing no significant difference of the mean from 0 (all Ps>0.05), and (iv) corresponding Gardner-Altman estimation plot to visualize the effect size of the pixel-wise dwell time difference across the two sessions. For each Gardner-Altman plot: left panels show the raw data points for exposure (grey) and re-exposure (green), with each point representing the dwell time in a given pixel; right panel: mean (black-dot), 95% confidence interval (black-ticks) and sampling-error distribution (filled-curve) of the difference between re-exposure and exposure, computed from 5,000 bootstrapped resamples and with the difference-axis (dashed-line) origin aligned to the mean of the exposure distribution. (b) Pixel-wise dwell time difference in spatial occupancy across the two sessions for all CPP days, as in (a). Average time difference distribution (left) not significantly different from 0 (1-sample t-test, p=0.53, t=0.62, df=1461; in sec per spatial bin: mean difference=0.05±0.08; 95% confidence interval=[0.106, -0.205]; interquartile range=[1.306, -0.870]), as also shown in the corresponding Gardner-Altman plot (right). (c) Cumming estimation plots showing the absolute number of pixels visited in each CPP task session (top; each dot representing one mouse CPP day) and the fraction of visited pixels in each enclosure (bottom). Note that the animal’s coverage was not significantly different between exposure and re-exposure. The higher number of pixels visited during pre-test and CPP test merely reflects the higher dimension of the whole-CPP apparatus. (d) Example time course of a mouse instantaneous speed during exposure (top) and re-exposure (bottom) in one CPP day. (e) Instantaneous speed across the six sessions for all CPP days (bar charts: mean±SEM; with each superimposed dot representing one mouse CPP day). No significant differences across sessions with respect to exposure (P values: pre-text=0.25; +Suc.=0.38; +Wat.=0.76; CPP test=0.82; Re-exposure=0.98; all 2-sample Kolmogorov-Smirnov tests). (f) Average speed across the six CPP task sessions (top) along with the total distance travelled (bottom; calculated for the first 15 min of each session). Note that the significant increased speed and distance travelled during pre-test (when the mouse is exposed for the first time to the novel CPP apparatus) do not translate in topological differences (Fig. 1f). These analyses (a-f) show that the topological hysteresis during re-exposure compared to exposure (Fig. 1f) does not reflect non-specific changes in spatial exploration. (g) Topology alterations of hippocampal graphs in re-exposure (Fig. 1f) do not reflect mere fluctuations in co-firing. To control for natural variations in co-firing graphs, we split both exposure and re-exposure in two sections (1 and 2) with equal duration of active exploration (speed>2cm/sec; exposure: 6.89±0.13 versus 6.97±0.10 min; re-exposure: 6.99±0.30 versus 6.85±0.31 min, all Ps>0.05; Wilcoxon signed-rank test) and quantified topological changes across. As for all topological analyses, sharp-wave/ripples were excluded (though their occurrence did not differ between exposure and re-exposure; t=-1.55, p=0.14, paired t-test). For each measure: the top panel shows the raw data points for each (color-coded) section (with the gapped lines on the right as mean (gap) ± SD (vertical ends)); the bottom-left panel shows the difference between the bootstrapped distribution with respect to first section of exposure; the bottom-right panel shows the difference between the distribution of the second compared to the first section within each familiar session. Note that co-firing topology did not significantly change during each individual familiar exploration. For each section: black-dot, median; black-ticks, 95% confidence interval; filled-curve: bootstrapped sampling-error distribution.
Extended Data Fig. 4. Familiar map-representations are largely reinstated during re-exposure after CPP but include edits not explained by mere fluctuations.
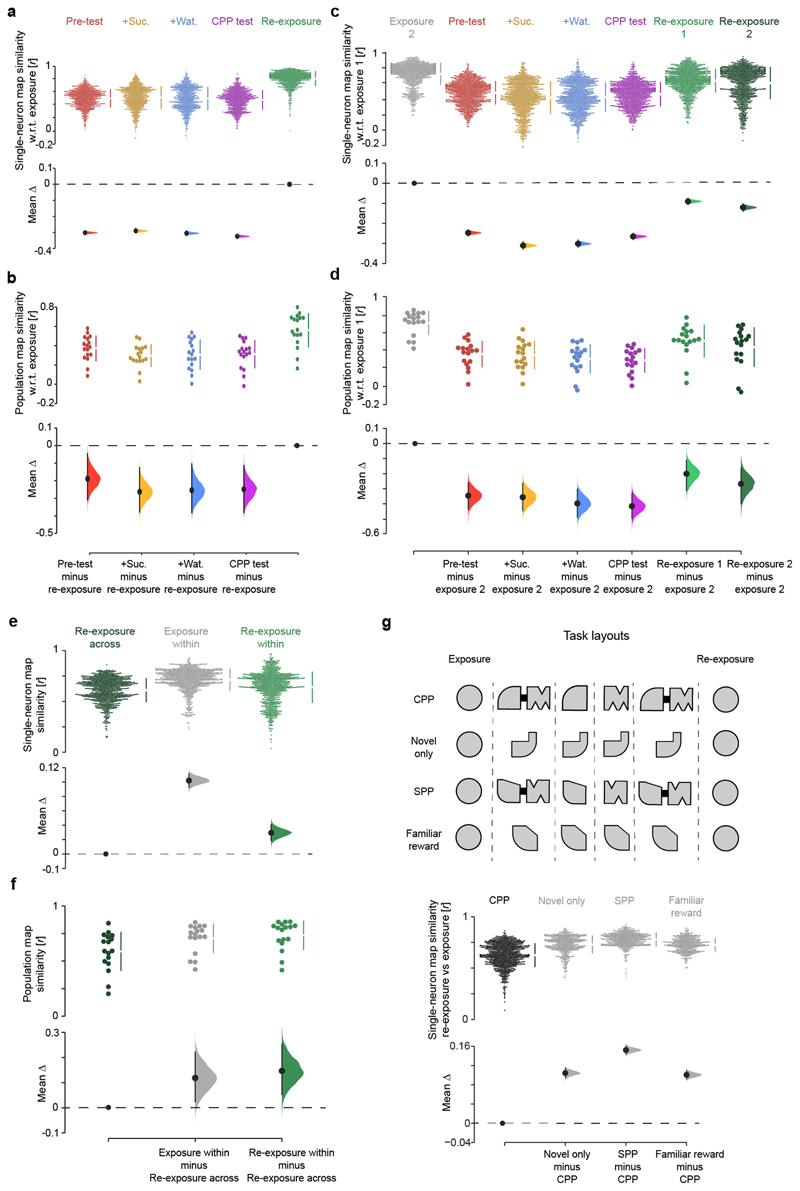
(a,b) Cumming estimation plots showing the effect size for changes in the similarity of both single-neuron (a) and population (b) spatial maps across CPP task sessions, with respect to (w.r.t.) exposure. For the single-neuron map similarity analysis (a), each data point represents the Pearson correlation using the firing rate of an individual neuron between the spatial bins of its map during exposure matched to those in a subsequent session. Note that single-neuron familiar maps are well reinstated during re-exposure following their reorganization during CPP sessions. For the population-level map similarity analysis (b), each data point represents the extent to which on a given CPP day pairs of cells that jointly represented a location during exposure continued to show spatially overlapping firing fields in subsequent sessions. This indicates that the combination of cell pairs sharing place fields during exposure largely re-emerges during re-exposure after their re-organization in the CPP enclosure, consistent with the remapping of hippocampal maps across spatial contexts (e.g., Muller, R. U. & Kubie, J. L., J. Neurosci. 1987; Wilson, M. A. & McNaughton, B. L., Science 1993; Leutgeb, S. et al, Science 2004; Wills, T. J. et al, Science 2005; Colgin, L. L. et al, Trends in Neurosciences 2008). (c-g) The strength of familiar map reinstatement from exposure to re-exposure was compared to non-specific fluctuations in firing activity over time. (c) With respect to the first section (half) of exposure, shown is the effect size for changes in single-neuron map similarity during the second section of the exposure, the 4 CPP sessions and the two (first and second) sections of re-exposure. (d) Same as (c) but for the population map similarity. (e,f) To contrast the effect of CPP on familiar map reinstatement against within-session variations in single-neuron (c) and population (d) map similarity, we compared the spatial correlation of hippocampal maps between exposure and re-exposure (Re-exposure across) with that between the two sections of the exposure (Exposure within) and of the re-exposure (Re-exposure within). The across-session fluctuations were quantified by comparing maps of each of the two re-exposure half sections (computed as in (c,d)) with those of the two exposure half sections, taking the mean of the four resulting similarity scores. The within-session fluctuations were obtained by comparing maps of the two sections of exposure or re-exposure, as indicated. Note that both single-neuron (e) and population-level (f) map fluctuations are significantly smaller within an exploration session of the familiar enclosure than across, even though single-neuron map variations (e) within re-exposure are markedly larger than those within exposure before CPP. (g) In addition, the effect of CPP on familiar map reinstatement was compared to familiar map reinstatement between exposure and re-exposure in the other tasks: unrewarded exploration of a novel enclosure (Novel only), spontaneous place preference for a novel place (SPP) and reward experience in another familiar enclosure (Familiar with reward). Altogether, these analyses show that while spatial maps expressed during re-exposure following CPP are strongly correlated with those initially seen during exposure before CPP (a,b), these reinstated maps nevertheless differ across these two sessions in the familiar enclosure of the CPP task more than changes expected from non-specific fluctuations occurring within a given exploration of the familiar enclosure or those due to the temporal gap between exposure and re-exposure (c-g). These results indicate a crosstalk between the new CPP memory and the prior hippocampal representation of the familiar enclosure. Subsequent analyses in this study relate such a crosstalk to changes in firing activity of low rate principal cells (see Extended Data Figs. 7-10). For each Cumming estimation plot: black-dot, median or mean as indicated; black-ticks, 99% confidence interval; filled-curve: bootstrapped sampling-error distribution.
Extended Data Fig. 5. Visualizing directions of hippocampal graph transformations in the network co-firing space.
(a) Computing the topological distances that separate co-firing graphs across the six task sessions. The co-firing graph of each task session was used to define a 6-dimensional vector of topological Riemannian Log-Euclidean distances to the other co-firing graphs obtained that day (e.g. here illustrated re-exposure versus exposure), including itself (see methods). For each 6-session task day, this procedure thus gives a 6 x 6 matrix of topological distances. All distance matrices were then stacked together to form a 6 x N matrix (with N the number of total graphs, i.e. N = 17 days x 6 sessions = 102) onto which we apply a dimensionality reduction technique (PCA, ICA or MDS). (b, c) Segregation of co-firing graphs using Independent Components Analysis (ICA; b) or Multidimensional Scaling (MDS; c). (b) ICA applied to the same matrices of topological distances used with PCA in the CPP dataset (Fig. 2) as another dimensionality reduction method to visualize axes explaining across-session variance in co-firing motifs. Note that co-firing graphs computed for the exposure and the re-exposure overlap on the first two independent components (IC) as they do along the first principal component (Fig. 2c-e); co-firing graphs are separated across the 4 CPP task events, as they are along the second principal component (Fig. 2c-e). (c) MDS also applied to the same matrices of topological distances used with PCA in the CPP dataset (Fig. 2); this method preserves the six-dimensional distances between co-firing graphs and maps them onto a 2D plane. Note that the co-firing graphs computed for each task session of each CPP day are well separated, indicating the existence of multiple axes along which co-firing patterns change across CPP task events. (d, e) Here the Pearson correlation coefficient is used instead of the Riemannian Log-Euclidean distance to compute the topological distance between co-firing graphs across the six CPP task sessions. In (d) the average topological distance matrix (left) and its first three PCs (right) are shown. In (e) the segregation of the six CPP task sessions is shown using the PCs shown in (d). Note that for both the Riemannian Log-Euclidean distance (Fig. 2c-e) and the Pearson correlation coefficient (d,e) approaches, the PCA of the CPP dataset reveals that the variance in hippocampal co-firing segregated the familiar enclosure from the whole CPP test apparatus along PC1. Surprisingly, PC1 did not segregate the two compartments (Nov1 and Nov2) that formed the CPP apparatus on each day. This suggests that these compartments were treated together as one spatial continuum along PC1 because when the animal first encountered the CPP apparatus, these two compartments were equally novel and physically connected by the bridge during the pre-test. Therefore, a refined interpretation why along PC1 the two CPP compartments are clustered together (and different from the familiar enclosure) is that neuronal co-firing along this axis also accounts for spatial familiarity versus novelty. (f, g) PCA with the two compartments of the CPP apparatus considered separately during both pre-test (f) and CPP test (g) sessions. Here, we computed a co-firing graph using the spike trains associated with the visits of each CPP compartment during both test sessions, thus obtaining one co-firing graph per individual compartment (Nov1 versus Nov2) during each test session. Projecting the resulting four co-firing graphs (two for pre-test and two for CPP test) onto the PCA axes obtained when considering the CPP apparatus as a whole entity (Fig. 2e) shows that PC2 segregates co-firing patterns related to each CPP compartment during test compared to pre-test. Likewise, PC2 segregated the two CPP compartments when explored separately during the conditioning sessions where we removed the bridge (Fig. 2e). (h-j) The topological distance projections of the co-firing graphs from the other 6-session tasks. As for the CPP dataset, multiple PCs accounted for the variance in co-firing across the 6 sessions of each task. Note, however, that co-firing variance relates to the specifics of each task, and so is each set of PCs and their interpretation. For each task, we used the PCs explaining at least 80% of the total variance to project the co-firing graphs topological distance. The variance explained by these PCs is: “Novel context only” PC1=57%, PC2=19% and PC3=8%; “Spontaneous Place Preference” PC1=52%, PC2=18% and PC3=16%; “Familiar context with reward experience” PC1=69%, PC2=15%; and “CPP” PC1=48%, PC2=19% and PC3=14% (Fig. 2e).
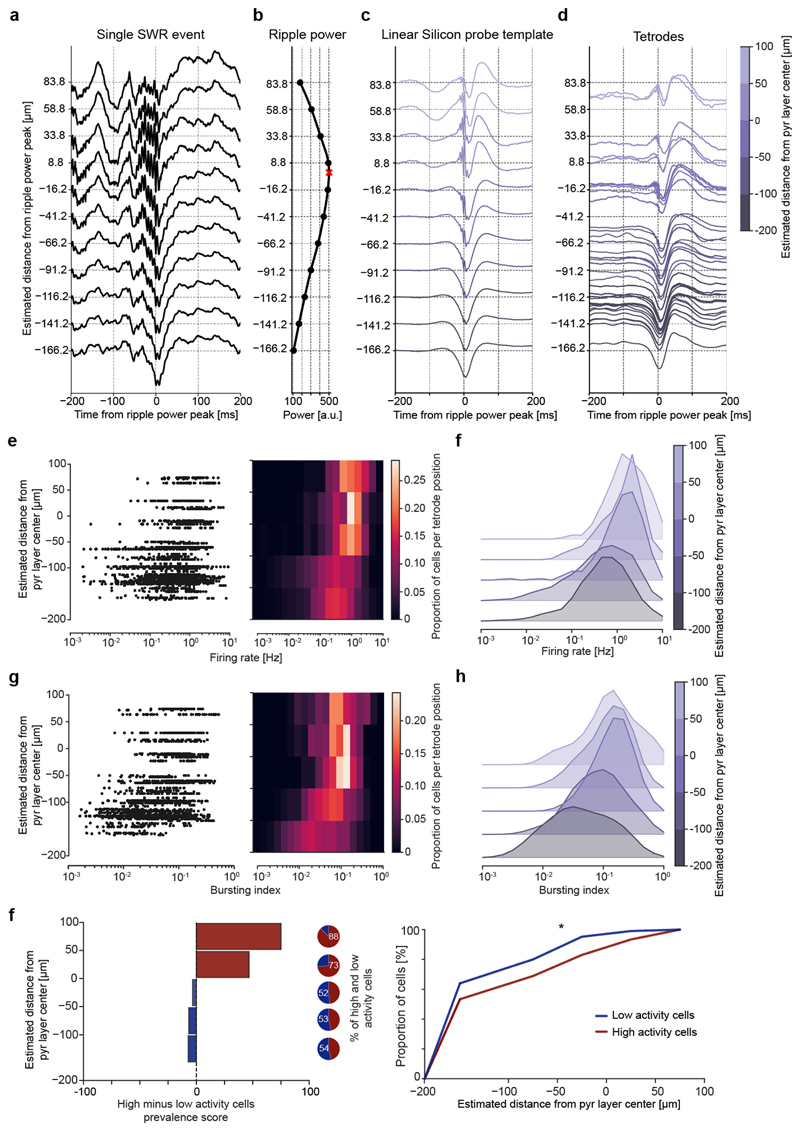
Extended Data Fig. 6. High and low rate CA1 principal cells are skewed towards deep and superficial pyramidal sublayers, respectively.
(a-d) For each CPP task day, we estimated the position (depth) of individual tetrode-recorded principal cell soma by leveraging silicon probe recordings with known spacing between the recording sites along a linear shank (25-μm steps). From these silicon probe recordings, we first computed the laminar profile of sharp-wave/ripples (SWRs) detected in the local field potentials (LFPs) along the radial axis of the dCA1 hippocampus (a). We used the peak of the corresponding depth profile of the ripple-band (110-250Hz) power to estimate the centre (middle) of the pyramidal (pyr.) layer (b; red cross). Using the average LFP waveform of the SWR events detected in these silicone probe recordings, we then established a SWR template where we reported the distance relative to the estimated centre of the pyramidal layer, knowing the precise distance between the recording sites on the linear shank (c). Next, we computed the individual SWR profile of each tetrode for CPP recording days finishing with a sleep session, this way estimating the depth of each individual tetrode (and thus that of the somas of its recorded neurons) by positioning its SWR profile within the silicone probe SWR template (the ground-truth vertical depth; c). Shown in (d) are examples of single-tetrode SWR profiles and their estimated depth. (e) Left: the estimated depth of principal cells as a function of the average firing rate measured in the CPP task. Each data point represents one principal cell. Right: the same data plotted as a firing rate probability distribution per estimated depth. (f) Same data as in (e) but plotted as a ridge plot to better visualise the relation between firing rate of principal cells and depth of their recording tetrodes. Note that principal cells in the deep sublayer of the dCA1 pyramidal layer (closer to stratum oriens) show higher firing rates than those in the superficial sublayer (closer to stratum radiatum) (line of best fit: y=23.34log(x)-74.7; p=10-30; two-sided Wald Test). (g,h) The same analyses shown in (e,f) but performed for the bursting index. Note that principal cells of the deep dCA1 sublayer show higher spike bursting compared to those of the superficial sublayer (line of best fit: y=47.43log(x)-15.0; p=10-64; two-sided Wald Test). (i) Relative prevalence (left) and cumulative distribution (right) of low (blue) and high (red) rate principal cells along the dCA1 radial axis. Low and high activity cells are skewed towards superficial and deep dCA1 pyramidal sublayers, respectively (p=0.016, Kolmogorov-Smirnov 2-sample test).
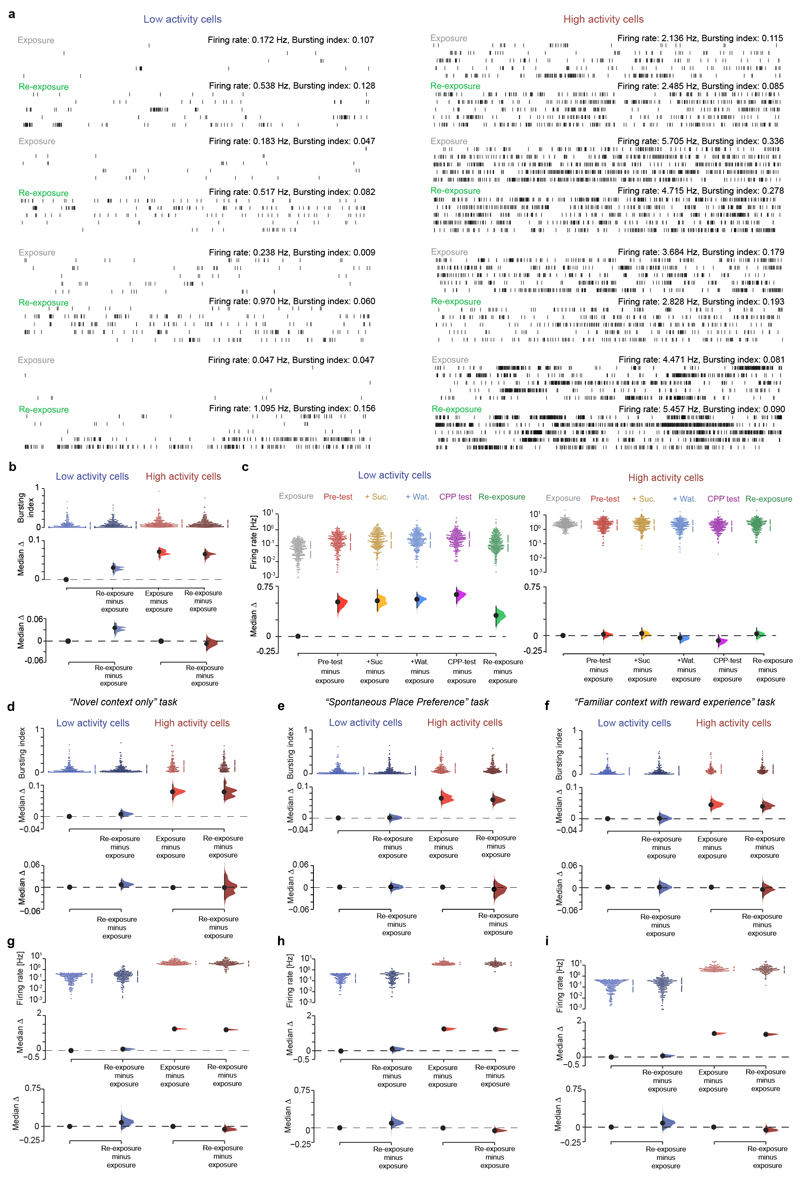
Extended Data Fig. 7. Low activity principal cells show increased firing activity during re-exposure to the familiar enclosure following CPP learning.
(a) Example spike trains of low and high activity cells during exposure and re-exposure to the familiar environment. For each of the eight example cells, shown are two raster plots (60-s rows; vertical ticks representing spike times) for exposure (upper plot) and re-exposure (lower plot). For clarity, only the first 5 min of each session are presented. Note the increased spike burst discharge by low activity cells during re-exposure compared to exposure. (b) Cumming estimation plot used to visualize for low (blue colours) and high (red colours) activity cells the effect size for changes in spike bursting between re-exposure and exposure. Upper panel: distributions of raw data points (each color-coded point represents one cell from the CPP task: n=272 low activity cells and 272 high activity cells) during exposure and re-exposure in light and dark colours, respectively; with the gapped lines on the right as mean (gap) ± SD (vertical ends) for a given subpopulation in a given session. Middle panel: the median (black-dot), 95% confidence interval (black-ticks) and sampling-error distribution (filled-curve) of the difference between (low or high activity) cells in a given session and the low activity cells in exposure, computed from 5,000 bias-corrected bootstrapped resamples and with the difference-axis (dashed-line) origin aligned to the median of low activity cells in exposure. Lower panel: similarly, compares the distribution in re-exposure to that in exposure within each subpopulation. Note that low, but not high, activity cells significantly discharge more bursts of spikes during re-exposure following CPP compared to exposure. The lack of increased bursting of the high activity cells during re-exposure compared to exposure does not reflect a ceiling effect given that the burst spiking distribution of this subpopulation had an inter-quartile range=0.07–0.18, a median=0.12 and a mean=0.15 throughout the CPP task sessions (see also the corresponding raw data points in Extended Data Fig. S9d and the coherence-percentiles' mean in Extended Data Fig. S9h), showing that this subpopulation could have discharged more bursts during re-exposure. (c) Cumming estimation plots to visualize the change in firing rate across CPP task sessions for the low (left panel) and the high (right panel) activity cell subpopulations. Note that firing rate of low activity cells increased during CPP sessions and stayed at higher values during re-exposure, not resetting to the values seen in exposure. (d-i) Bursting index (d-f) and firing rate (g-i) of high and low activity principal cells in the exposure and re-exposure to the familiar enclosure during the other tasks: exploration of a novel enclosure only (d,g), spontaneous place preference for a novel place (e,h) and exploration of another familiar enclosure with reward (f,i). Each (color-coded) data point represents one cell (“Novel context only” task: n=214 low activity cells and 128 high activity cells; “Spontaneous Place Preference” task: n=188 low activity cells and 152 high activity cells; “Familiar context with reward experience” task: n=148 low activity cells and 96 high activity cells). All differences presented using Cumming estimation plots as in (b). Note that in these three other tasks, low activity cells did not show significant changes in burst spiking and firing rate during re-exposure compared to exposure.
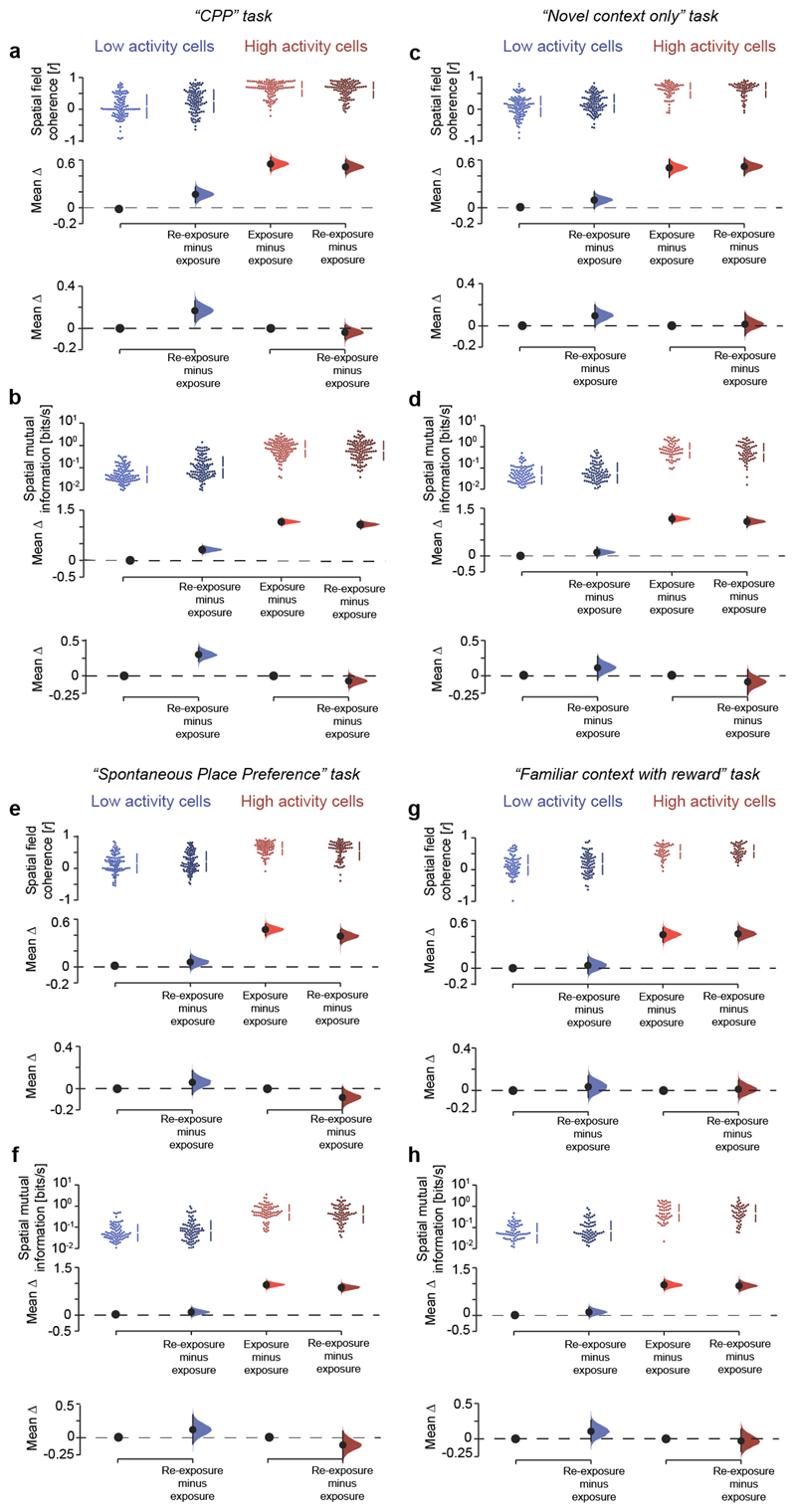
Extended Data Fig. 8. Low activity cells with increased burst spiking following CPP exhibited stronger place field coherence and spatial information content.
(a,b) Cumming estimation plots showing for low (blue colours) and high (red colours) activity cells the effect size in changes of place field coherence (a) and spatial mutual information (b) between exposure and re-exposure in the CPP task. The most burst spiking cells of both subpopulation (top 40% of bursting index distribution) in the re-exposure session are considered. Note that low, but not high, activity cells have significantly higher spatial coherence (a) and mutual information (b) during re-exposure after CPP. (c-h) Similarly, the place field coherence (c,e,g) and the spatial mutual information (d,f,h) of low and high activity principal cells is shown for the other tasks including exposure and re-exposure sessions. Low and high activity cells selected in the same way as in the CPP task dataset (a,b) to allow for comparison. For all Cumming estimation plots: black-dot, median (for skewed distributions) or mean (for normal distributions) as indicated; black-ticks, 95% confidence interval; filled-curve: bootstrapped sampling-error distribution.
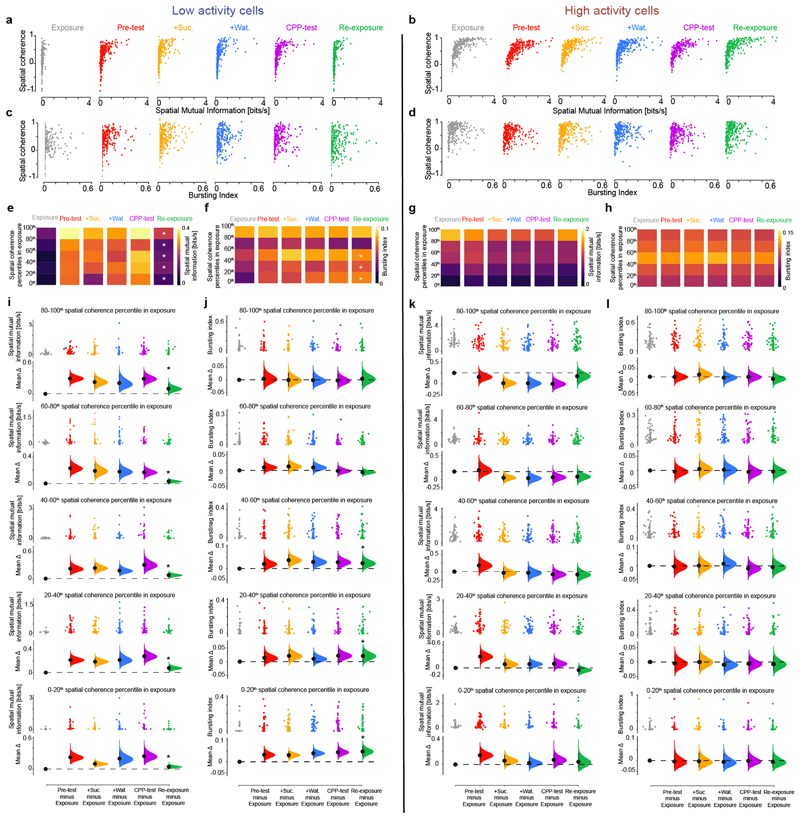
Extended Data Fig. 9. Relation between spatial field coherence during exposure and subsequent changes in both burstiness and spatial information for low and high activity cells in the CPP task.
(a,b) The spatial mutual information of low and high activity cells is plotted against their respective spatial coherence for each of the six CPP task sessions. Each data point represents one cell. Comparing with exposure, note the right-shift towards higher spatial information for a set of low activity cells during the CPP sessions, which then remained at higher values during re-exposure (a). (c,d) Similarly, the bursting index of low and high activity cells is plotted against their spatial coherence for each CPP task session. Also comparing with exposure, note the right-shift towards higher spike bursting in the distributions of low activity cells during CPP sessions, remaining higher during re-exposure (c). (e-h) For each of the six CPP task sessions, both low (e,f) and high (g,h) activity cell subpopulations were binned in 20th-percentiles according to the spatial coherence of each cell’s place field during the exposure session. Then, the average spatial mutual information (e,g) and bursting index (f,h) were computed for each 20th-percentile bins across all task sessions. For clarity, the bins of the re-exposure session marked with a white star (*) have significant changes in either measure (spatial information or bursting index) compared to the corresponding bins in the exposure session. Note that during re-exposure, low activity cells show significant increase in spatial information following CPP compared to exposure (e). This was not the case for high activity cells (g). Moreover, low activity cells with the least (0th–60th percentiles) spatial coherence during exposure increased their spike bursting during the CPP sessions and thereafter during re-exposure (f; with statistically different percentile bins marked with a white star compared to their corresponding bins in exposure). This was not seen for high activity cells (h). (i-l) Cumming estimation plots showing the effect size for changes in spatial information (i,k) and bursting index (j,l) for the low (i,j) and the high (k,l) activity cell subpopulations, binned as 20th-percentiles according to the spatial coherence of each cell’s place field during exposure. For further clarity, the black stars above the green filled-curves in re-exposure indicate significant changes compared to exposure. Note that across all percentile bins, low activity cells show significant increase in spatial information in re-exposure compared to exposure (i). This was not seen for high activity cells (k). Moreover, low activity cells with the least (0th–60th percentiles) spatial coherence during exposure thereafter discharged more spike bursts during the CPP sessions and continued to exhibit significant higher burstiness during re-exposure compared to exposure (f,j; see stars). High activity cells maintained the same burstiness throughout all task sessions (h,l). For each Cumming estimation plot: black-dot, mean; black-ticks, 95% confidence interval; filled-curve: bootstrapped sampling-error distribution. Altogether, these analyses show that CPP learning subsequently affected low activity cells during re-exposure to the familiar environment. Notably, these results show that the dCA1 network gained in spatial information content during re-exposure (compared to exposure) from both: (1) low activity cells that were already spatially tuned during exposure and then exhibited higher spatial informativeness following CPP (e,i), and (2) low activity cells that were not spatially tuned during exposure but were de-novo recruited during CPP, after which they stayed more active during re-exposure (f,j). These results are in line with the increase in spatial coherence observed for the most burst spiking of the low, but not high, activity cells during re-exposure compared to exposure (Extended Data Fig. 8a), and the gradual engagement of low activity cells in whole-network co-firing motifs (see Fig. 3c and Extended Data Fig. 10d).
Extended Data Fig. 10. High and low activity principal cells make distinct contributions to network co-firing motifs.
(a) Cumming estimation plots showing CPP task-related changes in topological clustering (top), geodesic path length (middle) and single-neuron cumulative co-firing strength (bottom) of co-firing graphs (with respect to exposure) for low and high activity cells. Black-dots, median; black-ticks, 99% confidence interval; filled-curve: sampling-error distribution. (b) Top, schematic showing the location of the containers for the sucrose and water drops in an example CPP enclosure. Bottom, distribution of firing rate changes (scores) between the +Suc and pre-test sessions for low and high activity cells (low activity cells: p=2.68x10-5, t=4.279, df=251; high activity cells: p=0.25, t=1.160, df=271; 1-sample t-tests against 0 mean). For every cell that fired at least 100 spikes in either session, a score is obtained by taking the difference between its mean firing rate at the containers during +Suc and pre-test sessions, dividing by the sum. (c) The change in firing rate at the containers from pre-test to +Suc. session correlated with the change in co-firing strength from exposure to re-exposure for the low (regression line y=0.21x-0.11; p=0.018, Wald test) but not the high (regression line y=0.12x-0.23; p=0.51, Wald test) activity cells. Together with the topological deviations that feature the low activity cell co-firing graphs (a), this result supports the idea that during the mnemonic update of a newly encountered place with reward experience, a change in the firing activity of low activity cells allows a cross-talk between the new CPP memory and the prior representation of the familiar enclosure, as reported along PC3 (Fig. 3d). (d) Contribution of low and high activity cells to network co-firing motifs, as measured by the proximity between the high (right) and low (left) activity sub-networks to the whole network (i.e., containing the full distribution of all recorded neurons) in the topological distance space across the six CPP task events (w.r.t. low activity cells in exposure). Black-dot, median or mean as indicated; black-ticks, 99% confidence interval; filled-curve: sampling-error distribution. (e) For low and high activity cells, firing rate changes during sharp-wave/ripples (SWRs) detected in periods of immobility (speed<2cm/sec) of exposure and re-exposure sessions. For every cell, the change in SWR firing is measured as the difference between its mean firing rate during SWRs in the exposure and re-exposure sessions divided by the subpopulation’s average firing rate during SWRs of exposure. (f) Change in co-firing during SWRs between exposure and re-exposure for low-low, low-high and high-high activity cell pairs. SWR co-firing computed during SWRs detected in periods of immobility (speed<2cm/sec) of a given exploration session in the familiar enclosure. The change in co-firing between re-exposure and exposure was then divided by the average subpopulation’s co-firing. Note the increased SWR co-firing between low-low and low-high activity cells during the re-exposure session following CPP learning.
Acknowledgements
we would like to thank J. Csicsvari and R. Lambiotte for commenting on a previous version of the manuscript; J. Janson for technical assistance; H.C. Barron and all members of the Dupret and Schultz labs for discussions and feedback during the course of this project. This work was supported by the Engineering and Physical Sciences Research Council UK Centre for Doctoral Training in Neurotechnology (EP/L016737/1 to S.R.S.), the Biotechnology and Biological Sciences Research Council UK (Award BB/N002547/1 to D.D.) and the Medical Research Council UK (Programmes MC_UU_12024/3 and MC_UU_00003/4 to D.D.).
Footnotes
Author contributions: all authors contributed to the preparation of the manuscript. G.P.G., S.R.S. and D.D. designed the study, developed the methodology, and analyzed the data. S.B.Mc., L.L., V.L-d-S, M.E. S.T. and D.D. acquired the data and helped with data analysis. S.R.S. and D.D. acquired funding.
Competing interests: The authors declare no competing interests.
Code availability
The software used for data acquisition and analysis are available using the web links mentioned in the methods.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Andersen P, Morris RGM, Amaral D, Bliss T, O’Keefe J. The Hippocampus Book. Oxford University Press; 2006. [Google Scholar]
- 2.Moser EI, Moser M-B, McNaughton BL. Spatial representation in the hippocampal formation: a history. Nat Neurosci. 2017;20:1448–1464. doi: 10.1038/nn.4653. [DOI] [PubMed] [Google Scholar]
- 3.Buzsáki G, Llinás R. Space and time in the brain. Science. 2017;358:482–485. doi: 10.1126/science.aan8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubie JL, Levy ERJ, Fenton AA. Is hippocampal remapping the physiological basis for context? Hippocampus. 2020;30:851–864. doi: 10.1002/hipo.23160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill J, Senior TJ, Allen K, Huxter JR, Csicsvari J. Reactivation of experience-dependent cell assembly patterns in the hippocampus. Nat Neurosci. 2008;11:209–215. doi: 10.1038/nn2037. [DOI] [PubMed] [Google Scholar]
- 6.Bassett DS, Sporns O. Network neuroscience. Nature Neuroscience. 2017;20:353–364. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries MD. Dynamical networks: Finding, measuring, and tracking neural population activity using network science. Netw Neurosci. 2017;1:324–338. doi: 10.1162/NETN_a_00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuseki K, Diba K, Pastalkova E, Buzsáki G. Hippocampal CA1 pyramidal cells form functionally distinct sublayers. Nat Neurosci. 2011;14:1174–1181. doi: 10.1038/nn.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzsáki G, Mizuseki K. The log-dynamic brain: how skewed distributions affect network operations. Nature Reviews Neuroscience. 2014;15:264–278. doi: 10.1038/nrn3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rich PD, Liaw H-P, Lee AK. Large environments reveal the statistical structure governing hippocampal representations. Science. 2014;345:814–817. doi: 10.1126/science.1255635. [DOI] [PubMed] [Google Scholar]
- 11.Soltesz I, Losonczy A. CA1 pyramidal cell diversity enabling parallel information processing in the hippocampus. Nat Neurosci. 2018;21:484–493. doi: 10.1038/s41593-018-0118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosmark AD, Buzsáki G. Diversity in neural firing dynamics supports both rigid and learned hippocampal sequences. Science. 2016;351:1440–1443. doi: 10.1126/science.aad1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danielson NB, et al. Sublayer-Specific Coding Dynamics during Spatial Navigation and Learning in Hippocampal Area CA1. Neuron. 2016;91:652–665. doi: 10.1016/j.neuron.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cembrowski MS, Spruston N. Heterogeneity within classical cell types is the rule: lessons from hippocampal pyramidal neurons. Nature Reviews Neuroscience. 2019;20:193–204. doi: 10.1038/s41583-019-0125-5. [DOI] [PubMed] [Google Scholar]
- 15.Oliva A, Fernández-Ruiz A, Buzsáki G, Berényi A. Spatial coding and physiological properties of hippocampal neurons in the Cornu Ammonis subregions. Hippocampus. 2016;26:1593–1607. doi: 10.1002/hipo.22659. [DOI] [PubMed] [Google Scholar]
- 16.Navas-Olive A, et al. Multimodal determinants of phase-locked dynamics across deep-superficial hippocampal sublayers during theta oscillations. Nature Communications. 2020;11:2217. doi: 10.1038/s41467-020-15840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauthier JL, Tank DW. A Dedicated Population for Reward Coding in the Hippocampus. Neuron. 2018;99:179–193.e7. doi: 10.1016/j.neuron.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valero M, et al. Determinants of different deep and superficial CA1 pyramidal cell dynamics during sharp-wave ripples. Nature Neuroscience. 2015;18:1281–1290. doi: 10.1038/nn.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenzie S, et al. Hippocampal Representation of Related and Opposing Memories Develop within Distinct, Hierarchically Organized Neural Schemas. Neuron. 2014;83:202–215. doi: 10.1016/j.neuron.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tse D, et al. Schemas and Memory Consolidation. Science. 2007;316:76–82. doi: 10.1126/science.1135935. [DOI] [PubMed] [Google Scholar]
- 21.van de Ven GM, Trouche S, McNamara CG, Allen K, Dupret D. Hippocampal Offline Reactivation Consolidates Recently Formed Cell Assembly Patterns during Sharp Wave-Ripples. Neuron. 2016;92:968–974. doi: 10.1016/j.neuron.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Csicsvari J, Hirase H, Czurkó A, Mamiya A, Buzsáki G. Oscillatory coupling of hippocampal pyramidal cells and interneurons in the behaving Rat. J Neurosci. 1999;19:274–287. doi: 10.1523/JNEUROSCI.19-01-00274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trouche S, et al. A Hippocampus-Accumbens Tripartite Neuronal Motif Guides Appetitive Memory in Space. Cell. 2019;176:1393–1406.e16. doi: 10.1016/j.cell.2018.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csicsvari J, Hirase H, Czurko A, Buzsáki G. Reliability and State Dependence of Pyramidal Cell–Interneuron Synapses in the Hippocampus. Neuron. 1998;21:179–189. doi: 10.1016/s0896-6273(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 25.Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J Neurophysiol. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- 26.Kadir SN, Goodman DFM, Harris KD. High-Dimensional Cluster Analysis with the Masked EM Algorithm. Neural Computation. 2014;26:2379–2394. doi: 10.1162/NECO_a_00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pachitariu M, Steinmetz N, Kadir S, Carandini M, D HK. Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. bioRxiv 061481. 2016 doi: 10.1101/061481. [DOI] [Google Scholar]
- 28.Magland JF, et al. SpikeForest: reproducible web-facing ground-truth validation of automated neural spike sorters. bioRxiv 2020.01.14.900688. 2020 doi: 10.1101/2020.01.14.900688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Csicsvari J, Hirase H, Czurko A, Mamiya A, Buzsáki G. Fast network oscillations in the hippocampal CA1 region of the behaving rat. Journal of neuroscience. 1999;19:1–4. doi: 10.1523/JNEUROSCI.19-16-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onnela J-P, Saramäki J, Kertész J, Kaski K. Intensity and coherence of motifs in weighted complex networks. Physical review. E, Statistical, nonlinear, and soft matter physics. 2005;71 doi: 10.1103/PhysRevE.71.065103. 065103. [DOI] [PubMed] [Google Scholar]
- 31.Costantini G, Perugini M. Generalization of Clustering Coefficients to Signed Correlation Networks. PLOS ONE. 2014;9 doi: 10.1371/journal.pone.0088669. e88669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saramäki J, Kivelä M, Onnela J-P, Kaski K, Kertész J. Generalizations of the clustering coefficient to weighted complex networks. Phys Rev E. 2007;75 doi: 10.1103/PhysRevE.75.027105. 027105. [DOI] [PubMed] [Google Scholar]
- 33.Floyd RW. Algorithm 97: Shortest Path Commun. Vol. 5. ACM; 1962. p. 345. [Google Scholar]
- 34.Roy B. Transitivité et connexité. C R Acad Sci Paris. 1959;249:216–218. [Google Scholar]
- 35.Warshall S. A Theorem on Boolean Matrices. J ACM. 1962;9:11–12. [Google Scholar]
- 36.Dupret D, O’Neill J, Pleydell-Bouverie B, Csicsvari J. The reorganization and reactivation of hippocampal maps predict spatial memory performance. Nat Neurosci. 2010;13:995–1002. doi: 10.1038/nn.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNamara CG, Tejero-Cantero Á, Trouche S, Campo-Urriza N, Dupret D. Dopaminergic neurons promote hippocampal reactivation and spatial memory persistence. Nat Neurosci. 2014;17:1658–1660. doi: 10.1038/nn.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Schönfeld F, Wiskott L, Manahan-Vaughan D. Spatial representations of place cells in darkness are supported by path integration and border information. Front Behav Neurosci. 2014;8:222. doi: 10.3389/fnbeh.2014.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butts DA. How much information is associated with a particular stimulus? Network. 2003;14:177–187. [PubMed] [Google Scholar]
- 40.Cover TM, Thomas JA. Elements of Information Theory. John Wiley & Sons; 1991. [Google Scholar]
- 41.Shannon C, Weaver W. University of Illinois; Urban, Illinois: 1949. The Mathematical Theory of Communication; p. 131. [Google Scholar]
- 42.Panzeri S, Treves A. Analytical estimates of limited sampling biases in different information measures. Network: Computation in Neural Systems. 1996;7:87–107. doi: 10.1080/0954898X.1996.11978656. [DOI] [PubMed] [Google Scholar]
- 43.Moakher M. A Differential Geometric Approach to the Geometric Mean of Symmetric Positive-Definite Matrices. SIAM J Matrix Anal & Appl. 2005;26:735–747. [Google Scholar]
- 44.Venkatesh M, Jaja J, Pessoa L. Comparing functional connectivity matrices: A geometry-aware approach applied to participant identification. NeuroImage. 2020;207 doi: 10.1016/j.neuroimage.2019.116398. 116398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Magnetic Resonance in Medicine. 2006;56:411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- 46.Harris KD, Hirase H, Leinekugel X, Henze DA, Buzsáki G. Temporal Interaction between Single Spikes and Complex Spike Bursts in Hippocampal Pyramidal Cells. Neuron. 2001;32:141–149. doi: 10.1016/s0896-6273(01)00447-0. [DOI] [PubMed] [Google Scholar]
- 47.Ranck JB. Studies on single neurons in dorsal hippocampal formation and septum in unrestrained rats. I. Behavioral correlates and firing repertoires. Exp Neurol. 1973;41:461–531. doi: 10.1016/0014-4886(73)90290-2. [DOI] [PubMed] [Google Scholar]
- 48.Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nature Methods. 2019;16:565–566. doi: 10.1038/s41592-019-0470-3. [DOI] [PubMed] [Google Scholar]
- 49.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 50.Hagberg AA, Schult DA, Swart PJ. Exploring Network Structure, Dynamics, and Function using NetworkX. Proceedings of the 7th Python in Science Conference; 2008. pp. 11–15. [Google Scholar]
- 51.Ince RAA, Petersen RS, Swan DC, Panzeri S. Python for information theoretic analysis of neural data. Front Neuroinform. 2009:3. doi: 10.3389/neuro.11.004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The software used for data acquisition and analysis are available using the web links mentioned in the methods.
The data that support the findings of this study are available from the corresponding author upon reasonable request.