Abstract
Gene expression programs define shared and species-specific phenotypes, but their evolution remains largely uncharacterized beyond the transcriptome layer1. Here we report an analysis of the co-evolution of translatomes and transcriptomes using ribosome-profling and matched RNA-sequencing data for three organs (brain, liver and testis) in fve mammals (human, macaque, mouse, opossum and platypus) and a bird (chicken). Our within-species analyses reveal that translational regulation is widespread in the diferent organs, in particular across the spermatogenic cell types of the testis. The between-species divergence in gene expression is around 20% lower at the translatome layer than at the transcriptome layer owing to extensive buffering between the expression layers, which especially preserved old, essential and housekeeping genes. Translational upregulation specifcally counterbalanced global dosage reductions during the evolution of sex chromosomes and the efects of meiotic sex-chromosome inactivation during spermatogenesis. Despite the overall prevalence of bufering, some genes evolved faster at the translatome layer—potentially indicating adaptive changes in expression; testis tissue shows the highest fraction of such genes. Further analyses incorporating mass spectrometry proteomics data establish that the co-evolution of transcriptomes and translatomes is refected at the proteome layer. Together, our work uncovers co-evolutionary patterns and associated selective forces across the expression layers, and provides a resource for understanding their interplay in mammalian organs.
A central goal in biology is to understand the molecular basis of phenotypic evolution, most notably that of humans and other mammals. It is thought that regulatory mutations affecting protein-coding gene expression underlie many or even most phenotypic differences between species1. Expression studies have so far focused primarily on analyses of transcriptomes and their regulation, and these studies have provided many insights into the dynamics of evolutionary changes in gene expression and associated phenotypic implications in mammals1. However, given that the expression of protein-coding genes may frequently be regulated at layers that succeed transcription2, and that it is ultimately protein abundance that is phenotypically relevant, transcriptome studies provide an incomplete picture of expression evolution. Evolutionary shifts in mRNA expression that are due to transcriptional regulatory mutations may be, for example, offset by post-transcriptional regulatory mutations that reconstitute (optimal) protein levels3. However, technologies for mass spectrometry analysis of proteins are still limited in their resolution compared to nucleic acid sequencing methodologies4.
The ribosome profiling (or ribosome-sequencing; Ribo-seq) approach provides a powerful solution to this dilemma4,5. This highly sensitive method provides a direct proxy for the rate of protein synthesis on the basis of deep sequencing of ribosome-protected mRNA fragments (“ribosome footprints”). In combination with standard RNA-sequencing (RNA-seq) for the same samples, it also enables the assessment of translational efficiency (TE) at a genome-wide scale4,5 The power and utility of Ribo-seq for comparative gene expression analyses was previously demonstrated in studies of yeast, nematodes, hybrid mouse cells and primate cell lines, providing initial insights into patterns of transcriptome versus translatome evolution6-12. However, the evolutionary comparison of translatomes across mammals and primary organs represents, as yet, uncharted territory.
To fill this gap and explore the co-evolution of regulatory processes across the transcriptome and translatome layers of gene expression, we generated Ribo-seq and matched RNA-seq data for three major mammalian organs (brain: cerebrum, liver, and testis), representing the three germ layers, from five representatives of the three main mammalian lineages: eutherian mammals (human, rhesus macaque, mouse), marsupials (grey short-tailed opossum), and egg-laying monotremes (platypus) (Fig. 1a; Supplementary Table 1). Corresponding data were generated for a bird (red junglefowl, the progenitor of domestic chicken; henceforth referred to as “chicken”), to be used as an evolutionary outgroup. To dissect patterns of gene expression regulation at the cellular level in the testis, we also generated the same types of data for spermatogenic cell types in mouse (Supplementary Table 1). Quality controls (e.g., analyses of footprint periodicity and principal component analyses (PCA)) testify to the high quality of the data (Methods, Extended Data Figs. 1-3). Notably, we observe significantly higher correlations between the translatome and proteome data than between the transcriptome and proteome data in all three organs (all P-values < 10-18, Fisher Z-transformation) (Fig. 1b). This observation is in agreement with the expectation that the rate of protein synthesis is a better predictor of protein abundance than measurements of mRNA levels5.
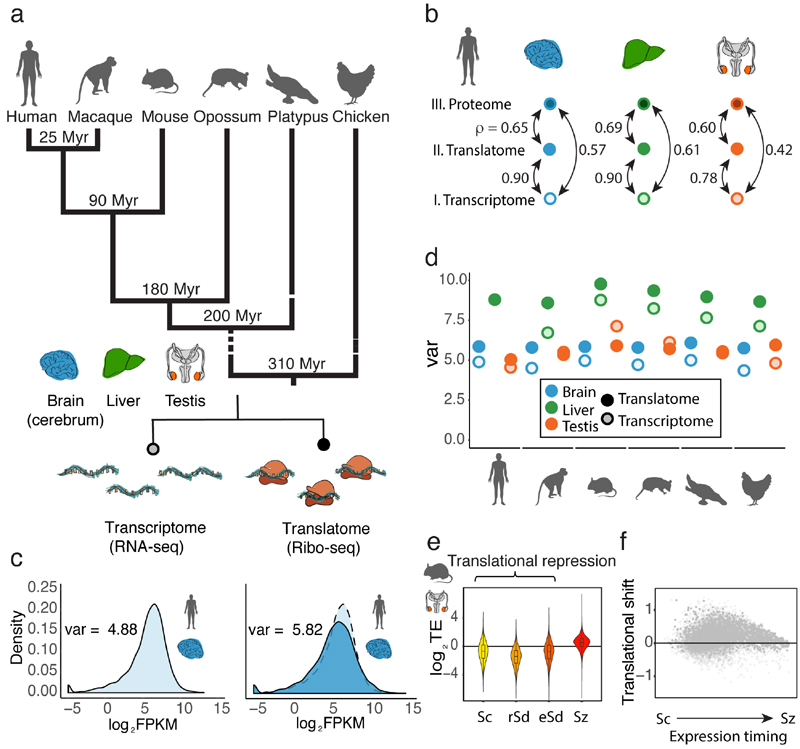
Fig. 1. Regulatory dynamics across expression layers.
a, Overview of data produced. b, Pairwise correlations (Spearman’s ρ) between transcriptomes, translatomes, and proteomes (data from ref. 22) were calculated for 9,642 genes, detected at all three expression layers in human brain, liver, and testis. c, Distribution of expression levels at the translatome layer (dark blue, measured based on Ribo-seq), compared to the transcriptome layer (light blue, measured based on RNA-seq). d, The expression variation, quantified as the variance (var) across genes of log2(FPKM+1)-transformed expression values, is calculated for expression levels at the translatome (dark colors) and transcriptome (light colors) layers. e, TE (normalized log2-transformed values) along mouse spermatogenesis was calculated for 14,979 genes detected (FPKM > 0) across all 4 stages (Sc, spermatocytes, rSd, round spermatids, eSd, elongating/elongated spermatids, Sz, spermatozoa). The zero line corresponds to the median TE of genes inferred to be expressed predominantly in somatic cells. f, Translational shift (delay) for each gene, calculated as the difference between the centers of mass for the transcriptome and translatome layers along spermatogenesis. Organ and species icons were previously used in ref. 25.
Regulation across expression layers
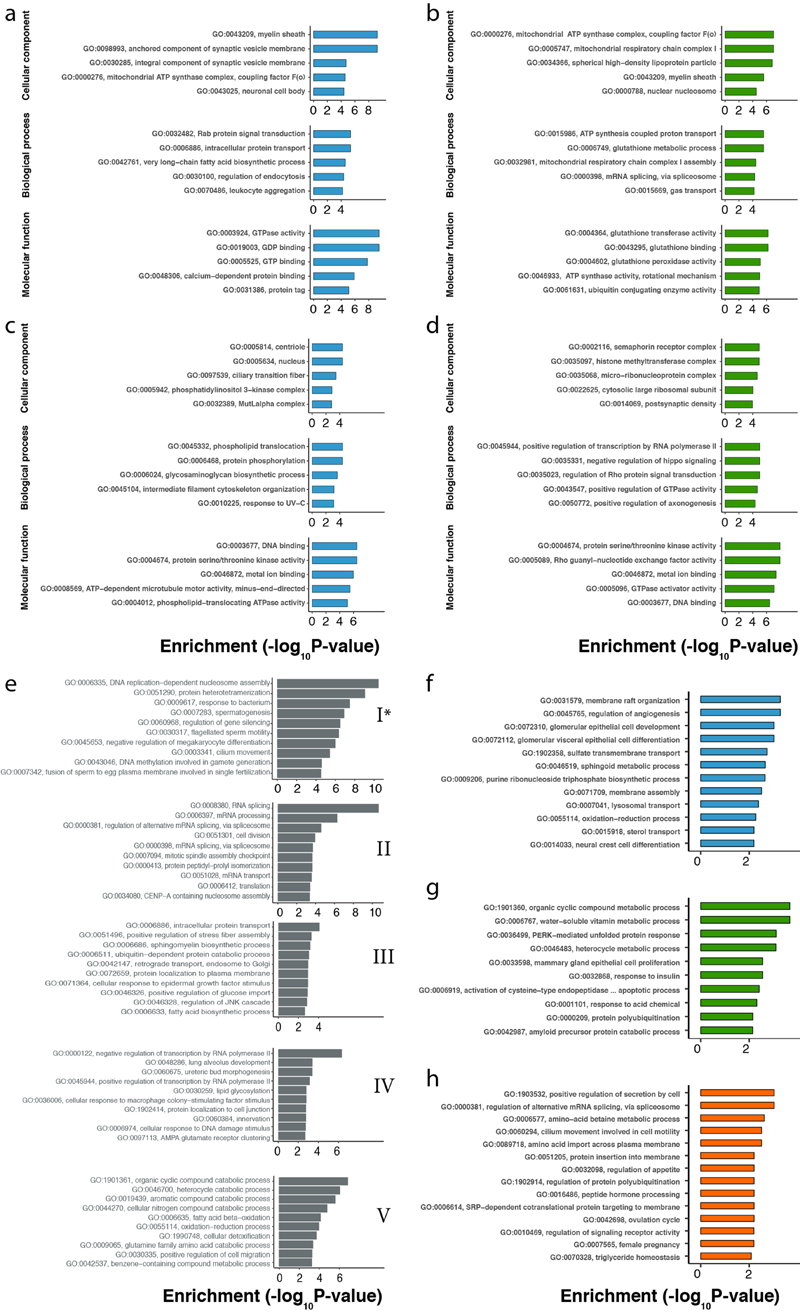
We first investigated the impact of translational regulation on gene expression in the different organs for each species. Differences in translational regulation across genes, reflected in differences in TEs, are expected to lead to an increased variation of expression levels across genes (expression variation) at the translatome compared to the transcriptome layer13 (Fig. 1c), which only reflects variation in the regulation of transcription and mRNA decay (Methods). We estimate that translational regulation increases the expression variation by 12–32% in the two somatic organs (Fig. 1d). Certain gene categories are particularly affected (i.e., potentially strongly regulated) at the translatome layer. For example, among very efficiently translated genes, there is an enrichment for membrane-associated functions, such as proton transport, signal transduction, and, in brain, also synaptic vesicle fusion (Extended Data Fig. 4a-d).
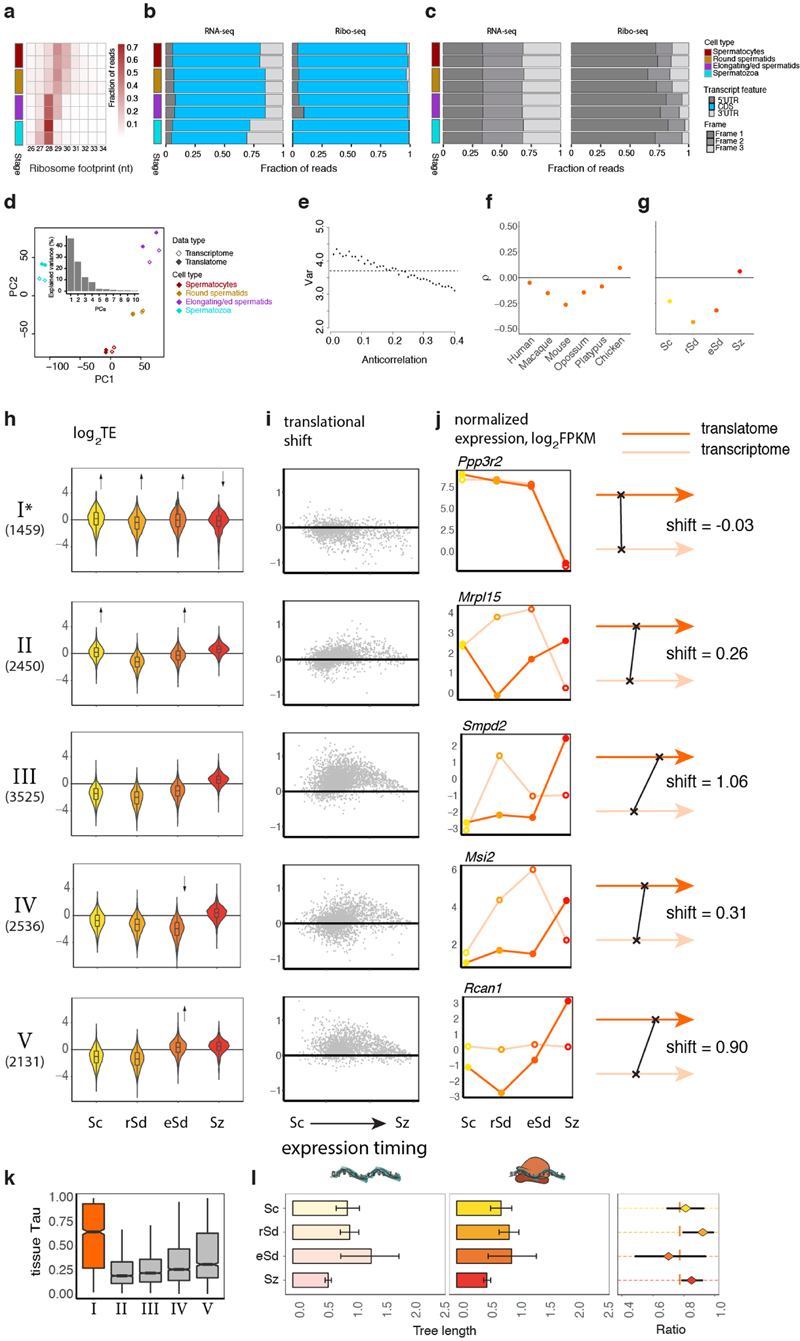
Notably, testis shows a distinct pattern, with similar or even decreased expression variation at the translatome versus transcriptome layer in four of the six species (Fig. 1d). We find this observation to be explained by an anticorrelation between transcript abundances and their TEs (Extended Data Fig. 3e, Methods). Consistently, the four species with reduced expression variation at the translatome relative to the transcriptome layer (Fig. 1d) show the strongest anticorrelations (Extended Data Fig. 3f). We hypothesized that the observed anticorrelations result from widespread translational repression of more highly expressed genes in meiotic and post-meiotic spermatogenic cells, which are abundant in the sexually mature testis14-16.
Indeed, translation of transcripts in spermatocytes and in particular in spermatids is on average strongly downregulated compared to translation of transcripts inferred to be expressed in somatic cells and is shifted towards later spermatogenic stages (Fig. 1e, f). Similarly to the whole mouse testis, we observe reduced expression variation (by 4–22%) at the translatome relative to the transcriptome layer and an anticorrelation between transcript abundances and their TEs in spermatocytes and spermatids (Extended Data Fig. 3g).
Despite the overall repression of translation in spermatocytes and spermatids and its delay towards later spermatogenic stages, different groups of genes have distinct patterns of TE dynamics, as revealed by a clustering analysis (Methods, Extended Data Fig. 3h-j, Supplementary Table 2). Notably, genes in cluster I, enriched for spermatogenesis and sperm-related functions (Extended Data Fig. 4e) and largely testis-specific (Extended Data Fig. 3k), are efficiently translated in spermatocytes and spermatids and therefore escape the overall translational repression and delay (Extended Data Fig. 3h-j).
Co-evolution of expression layers
To obtain global view of rates of gene expression evolution, we reconstructed expression trees for the trancriptome and translatome layers in the three organs (Fig. 2a-c; Methods, Extended Data Fig. 5a). Trees for both expression layers recapitulate the known mammalian phylogeny and are consistent with previous transcriptome analyses17. For example, the longer branches in the testis trees, compared to those in the brain and liver trees, reflect the rapid gene expression evolution of this organ, which is thought to reflect strong positive selection related to male reproductive success as well as the overall leaky transcription during the massive chromatin remodeling during spermatogenesis1,17. However, differences in cell type abundances across species, which are pronounced for the testis18, may also have contributed to its rapid evolutionary divergence.
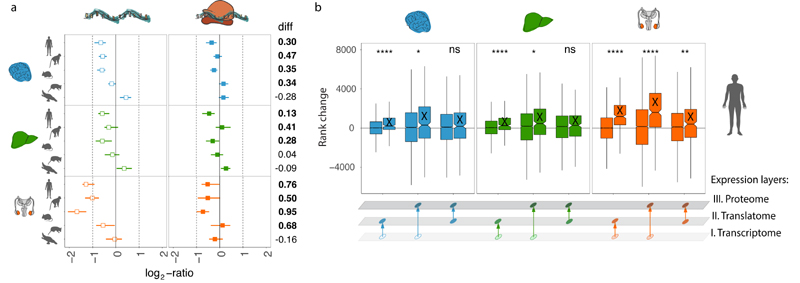
Fig. 2. Evolution of gene expression across expression layers.
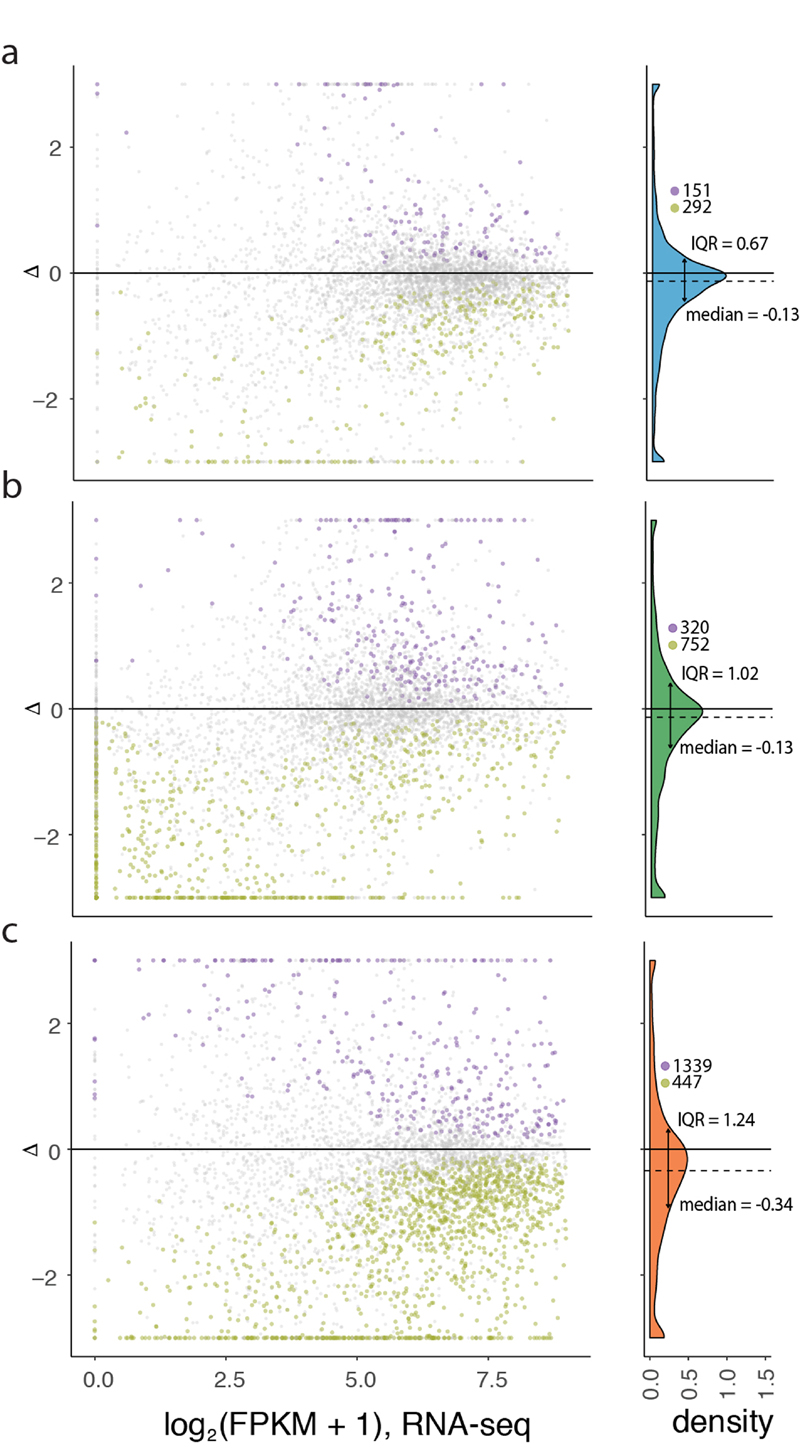
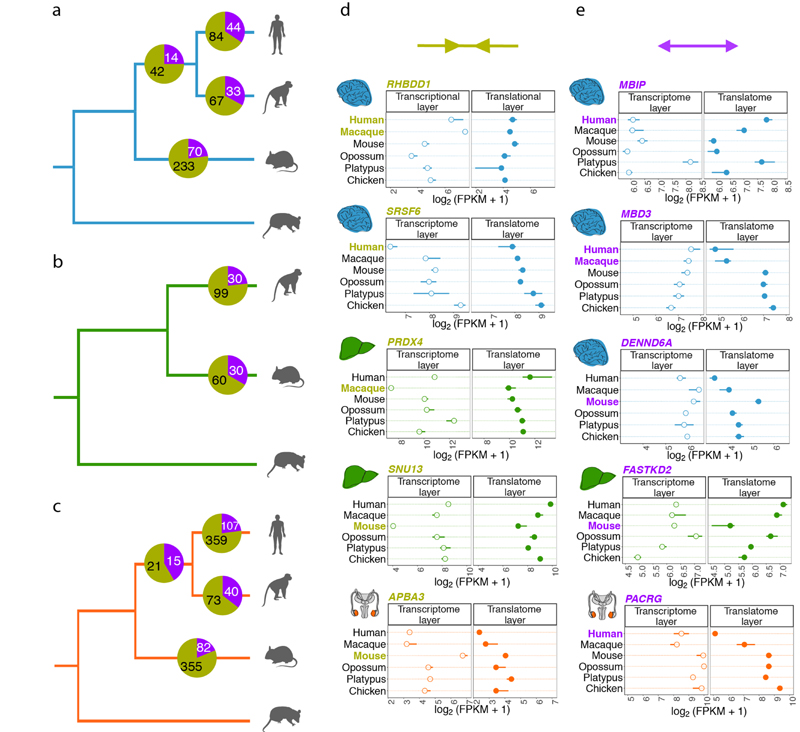
a-c, Gene expression phylogenies of 5,060 robustly expressed (FPKM > 1 across all libraries) 1:1 orthologues at the transcriptome (light and thick branches) and translatome (dark and thin) layers for brain (a), liver (b), and testis (c). Branch lengths represent the fractions of expression variation, which correspond to evolutionary changes in expression levels (Extended Data Fig. 5a). Due to the lack of a biological replicate, the branch leading to human was omitted in the liver phylogeny for the transcriptome layer. Proportions of bootstrapped trees supporting branching patterns are indicated next to the respective nodes. d-f, Differences in the evolution between transcriptome and translatome layers for individual genes in brain (d), liver (e), and testis (f). Density distribution, median Δ, interquartile range (IQR) of Δ, and number of cases with Δ significantly higher (potentially driven by directional selection) or lower (stabilizing selection) than zero are shown to the right of each panel. All genes in graphs d-f can be interactively explored in our Ex2plorer database (https://ex2plorer.kaessmannlab.org/). g, Similarity of gene expression (rank) changes between human and mouse brains at the proteome layer compared to the changes at the underlying translatome and transcriptome expression layers, respectively, as assessed by Spearman’s correlation coefficients (ρ). Proteomics data were retrieved from previous studies21,22. Organ and species icons were previously used in ref. 25.
Notably, the lengths of branches in the translatome data trees are overall 20-22% shorter than those defined by the transcriptome data, indicating that expression levels are more conserved at the translatome layer, which reflects the joint effects of evolutionary changes at both expression layers (Fig. 2a-c). These estimates are robust to read downsampling (Supplementary Table 3), and we obtain similar estimates using an alternative rank correlation-based approach17 (Methods; Supplementary Table 4). The observed pattern therefore likely reflects an overall scenario of compensatory evolution, in which regulatory changes at both expression layers counterbalance (buffer) each other (Extended Data Fig. 6a and Methods). Additionally, unproductive transcript isoforms19, whose expression levels likely evolve under relaxed selective constraints, may contribute to the overall greater divergence at the transcriptome layer.
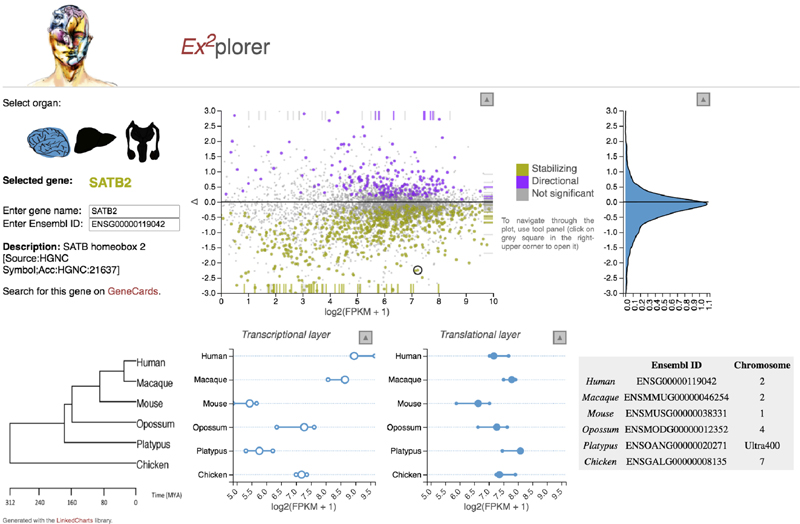
We also estimated differences in rates of evolution between the two expression layers for individual (1:1 orthologous) genes based on the difference (Δ) in expression variance between these layers across all studied species (Fig. 2d-f, Extended Data Fig. 6b, Supplementary Table 5, Methods,). Δ = 0 indicates equal evolutionary rates at both expression layers, Δ > 0 indicates a higher evolutionary rate at the translatome layer, and Δ < 0 indicates a lower evolutionary rate at the translatome layer. Our analysis reveals overall lower rates of evolution at the translatome layer compared to the transcriptome layer in all three organs (i.e., median Δ < 0; Fig. 2d-f), a result that is robust to read downsampling (Extended Data Fig. 7) and is consistent with the tree analysis (Fig. 2a-c). Overall, our observations bolster the notion that gene expression evolution has been shaped predominantly by stabilizing selection1,17. An illustrative example of a strongly buffered gene is SATB2, a gene whose encoded protein is highly conserved across vertebrates and is linked to developmental delay/intellectual disability in humans20; SATB2 displays high variation in expression across species at the transcriptome layer, but only small changes at the translatome layer (Extended Data Fig. 8). We developed a resource that allows the interactive exploration of the evolution at both expression layers for all 1:1 orthologous genes (Ex2plorer: https://ex2plorer.kaessmannlab.org/) (Extended Data Fig. 8).
Despite the overall buffering pattern, there is a high diversity among individual genes, with many of them changing faster at the translatome layer than at the transcriptome layer (Fig. 2d-f); these genes are enriched for specific biological processes (Extended Data Fig. 4f-h). The testis displays the biggest contrast in the evolution of individual genes between the two expression layers, with a substantially larger spread of Δ values (interquartile range, IQR of Δ = 1.08) compared to liver (IQR of Δ = 0.86) and especially brain (IQR of Δ = 0.58) (Fig. 2d-f). This pattern likely reflects differential selective forces shaping the testis and its peculiar patterns of transcription and translational control (see above).
Analyses that incorporated previous human and mouse proteomics data21,22 illustrate that the aforementioned co-evolution of regulatory changes at the first two expression layers, which determines the evolutionary dynamics of protein synthesis rates, is overall reflected at the proteome layer. First, we find that evolutionary expression (rank) changes between human and mouse brains are significantly more similar (correlated) between the proteome and translatome than between the proteome and transcriptome (Fig. 2g; P < 10-8, Fisher Z-transformation). Second, a rank-based comparison of protein expression levels between human and mouse brains revealed that genes with a lower rate of evolution at the translatome than transcriptome layer (corresponding to genes with Δ < 0 in the Δ analyses described above) evolved significantly more slowly at the proteome layer than genes with higher rates of evolution at the translatome layer (corresponding to genes with Δ > 0) (P < 10-5, Mann-Whitney U test)) (Extended Data Fig. 9).
We next identified genes with lineage-specific differences of evolutionary change between the expression layers; that is, genes with a significantly faster or slower evolution at the translatome than the transcriptome layer on the primate or rodent lineages, potentially driven by directional or stabilizing selection, respectively (Extended Data Fig. 10 and Supplementary Table 6). Most of these cases (73% in brain, 73% in liver, 77% in testis) correspond to instances of compensatory evolution. However, a subset of genes (161 in brain, 60 in liver, and 244 in testis) changed significantly more or almost exclusively at the translatome layer (Extended Data Fig. 10).
Factors of gene expression evolution
To dissect and understand the genomic sources of the differential evolutionary conservation patterns at the two expression layers, we investigated the effect of different gene characteristics on rates of expression divergence and strength of buffering, based on branch lengths of expression trees reconstructed for different gene categories. First, we considered the phenotypic impact of a gene (i.e., how essential its function is for organismal fitness) by leveraging several metrics that assess the extent of mutational tolerance (typically within coding sequences) across the genome23. Our analyses revealed a strong relationship between gene essentiality and expression evolution in the three organs; that is, genes that are highly sensitive to mutations (essential genes) show lower expression divergence at both layers together with stronger buffering compared to genes that are relatively tolerant to mutations (Fig. 3a). The impact of essentiality is higher in somatic tissues compared to testis, and is particularly strong in brain. We also observe similar but overall weaker effects for dosage sensitivity (Fig. 3a) 23.
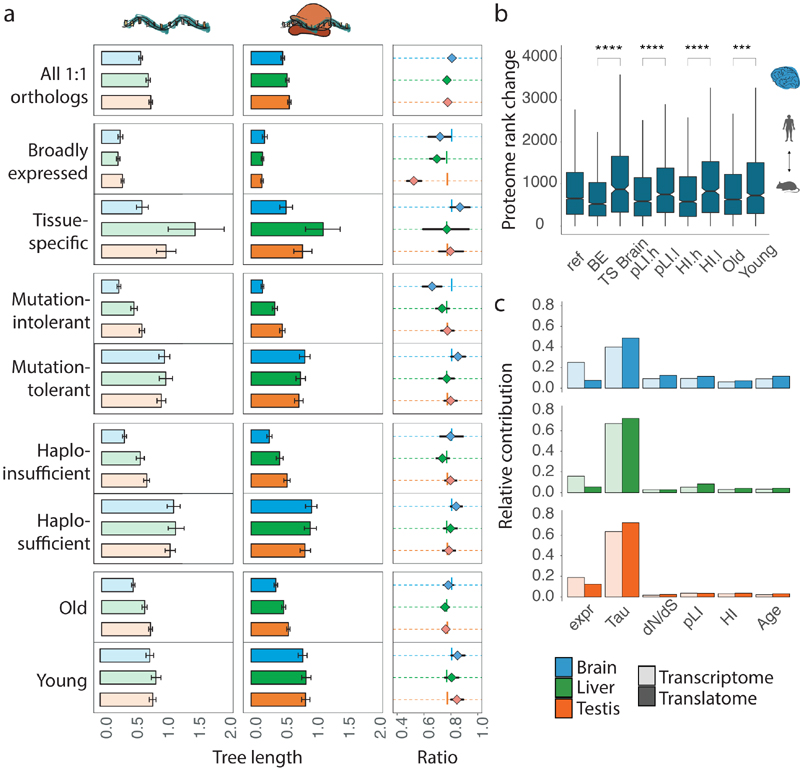
Fig. 3. Co-evolution of expression layers across gene classes.
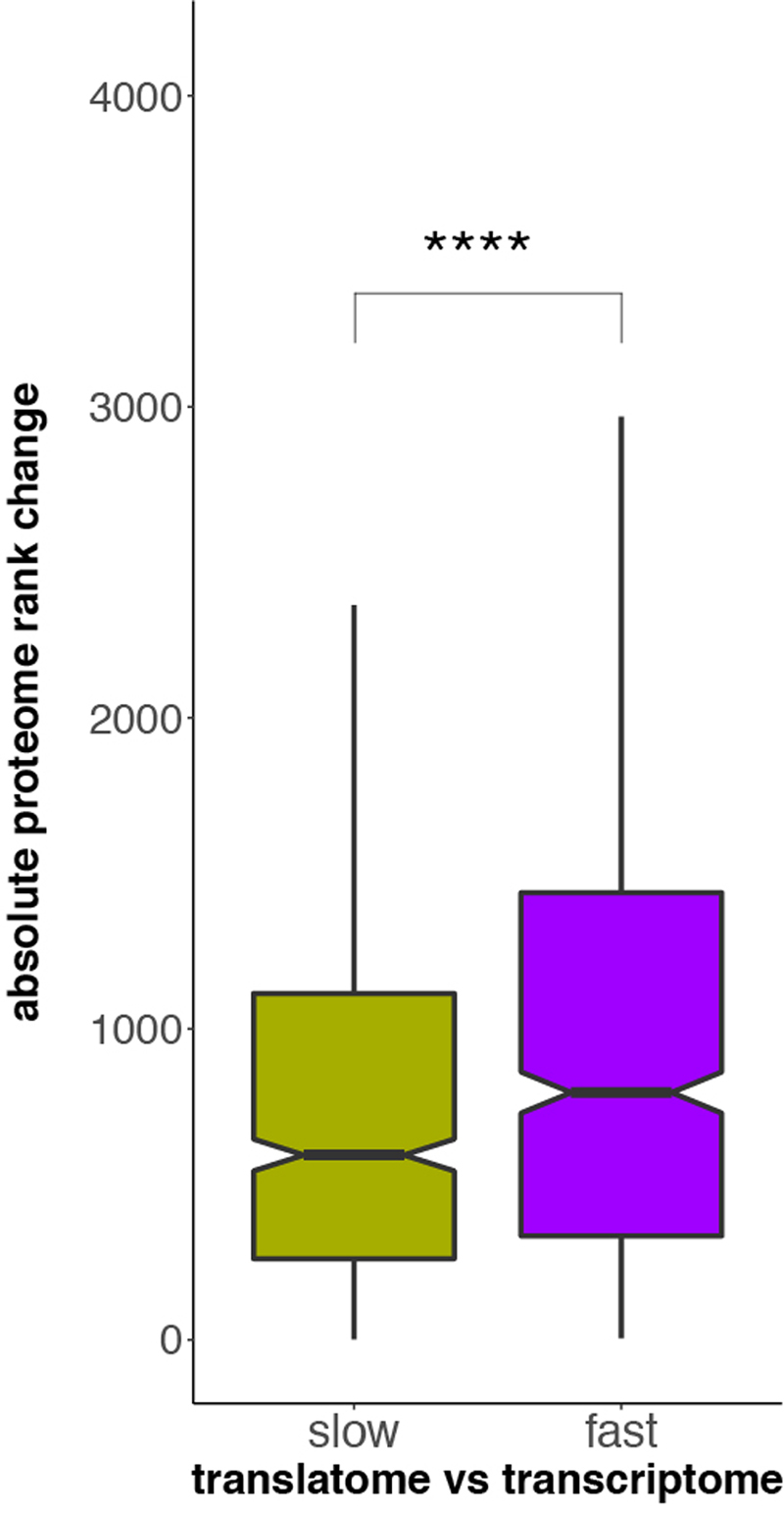
a, Gene-expression divergence at the two expression layers was calculated for all 8,109 1:1 orthologues robustly expressed (FPKM > 1) in macaque, mouse and opossum (Ref), and for specific gene sets: genes with particular spatial expression patterns (broadly expressed (BE) or tissue-specific (TS)); genes with a high (mutation-intolerant, pLI.h) or low (mutation-tolerant, pLI.l) probability of being loss-of-function-intolerant; genes with high (haploinsufficient, HI.h) or low (haplosufficient, HI.l) sensitivity to copy number reductions; and genes that duplicated in the common bony vertebrate ancestor (old) or that have duplication origins in tetrapods (young). Analysis was restricted to three species to increase the number of available 1:1 orthologues. Ratios between rates of gene-expression divergence (translational to transcriptional) are shown to the right of each set of bar plots (vertical lines indicate ratios obtained for the complete set of orthologues). Error bars correspond to 95% confidence intervals, calculated based on 1,000 bootstrap replicates. b, Absolute rank changes of proteome gene expression levels were calculated across the same categories for 6,972 1:1 orthologues, detected in human and mouse brains at all three expression layers; Mann-Whitney U tests (two-sided) were performed for statistical comparisons (***P = 0.00034, ****P < 0.0001). Box plots represent the median ± 25th and 75th percentiles, whiskers are at 1.5 times the interquartile range. c, Contribution of different factors to gene expression divergence rates at the transcriptional and translational layers. Expr, gene expression level (as log2(FPKM+1)); Tau, tissue-specificity, measured as τ; dN/dS, ratio of substitutions in non-synonymous to synonymous sites; pLI – loss of function intolerance; HI – haploinsufficiency; Age – age of the last duplication. Organ and species icons were previously used in ref. 25.
We next assessed the relationship between spatial expression characteristics and expression evolution. Housekeeping genes are broadly expressed genes that are required for the maintenance of fundamental cellular functions across all tissues in the body24. We observed very low and overall similar rates of expression evolution for broadly expressed genes across organs (Fig. 3a). Extensive buffering leads to even higher conservation at the translatome layer, with a particularly strong effect in testis. Depending on the organ, tissue-specific genes evolve two to six times faster and display much weaker buffering compared to broadly expressed genes (Fig. 3a). Contrary to broadly expressed genes, tissue-specific genes evolve with different rates in different organs, with brain-specific genes evolving considerably more slowly than genes specific to liver or testis. Genes specifically expressed at a particular stage of spermatogenesis (i.e., cell type-specific genes) (Extended Data Fig. 3l), in turn, evolve faster than genes on average in testis (Fig. 3a, all 1:1 orthologues), with the exception of genes specifically expressed in mature spermatozoa, which evolve approximately two times slower at both expression layers than genes specific to earlier spermatogenic stages.
Finally, we hypothesized that genes of different ages may show differential divergence dynamics across the two expression layers. For instance, given that new (duplicate) genes are typically functional in later, less constrained developmental stages25 and are less likely to be essential compared to older genes26, they could be expected to show less constrained and less buffered gene expression change. Indeed, we find that older genes show overall lower gene expression divergence and also stronger buffering than genes in the younger category (Fig. 3a).
Notably, all of the aforementioned contrasts between categories at the transcriptome and translatome layers are also reflected at the proteome layer. That is, comparisons of human and mouse proteome data for the brain21,22 reveal higher rank preservation for broadly-expressed, essential, haploinsufficient, and old genes than for tissue-specific, mutationally tolerant, haplosufficient, and young genes, respectively (Fig. 3b).
To assess the relative contribution of the different gene characteristics to expression divergence rates, we implemented a multiple regression analysis (Methods). This analysis reveals that spatial expression has the highest impact on evolutionary rates among the considered characteristics in all three tissues, followed by transcript abundance (Fig. 3c). The contributions of gene sequence conservation and age, as well as functional characteristics, such as gene essentiality and dosage sensitivity, are higher in brain than in liver and testis. Importantly, at the translatome layer, there is a higher contribution of all characteristics but transcript abundance, consistent with a higher functional relevance of expression output at this layer than at the transcriptome layer.
Translational X upregulation
Motivated by the pronounced buffering of expression changes across organs and gene classes, we explored whether this mechanism mitigated the consequences of the massive remodeling of gene contents during sex chromosome evolution. The differentiation of mammalian sex chromosomes, which are derived from an ordinary pair of ancestral autosomes, entailed the loss of most genes on the Y chromosome, leaving males with a single copy for nearly all X chromosomal genes27. It was hypothesized that a twofold transcriptional upregulation of the single remaining gene copies on the X restored ancestral expression outputs in males. In this model, the overabundance of X-linked transcripts in females, resulting from the combined activity of the two upregulated X chromosomes, was then secondarily compensated by the well-known process of X chromosome inactivation27. However, previous evolutionary transcriptome studies revealed that, in eutherians, X-linked genes in males and females lack full global transcriptional compensation, whereas there may be full transcriptional upregulation of the X in marsupials1,27-29. Furthermore, sex chromosome differentiation also triggered the emergence of complete meiotic silencing of sex chromosomes30 (MSCI) in the male germline of eutherians and marsupials, and alternative mechanisms must have evolved to compensate for this lack of active X-linked gene transcription in the testis during meiosis30. So far, one mechanism – the generation of autosomal substitute gene copies – has been discovered, which, however, only compensates for the silencing of a limited number of key X-linked genes1,30.
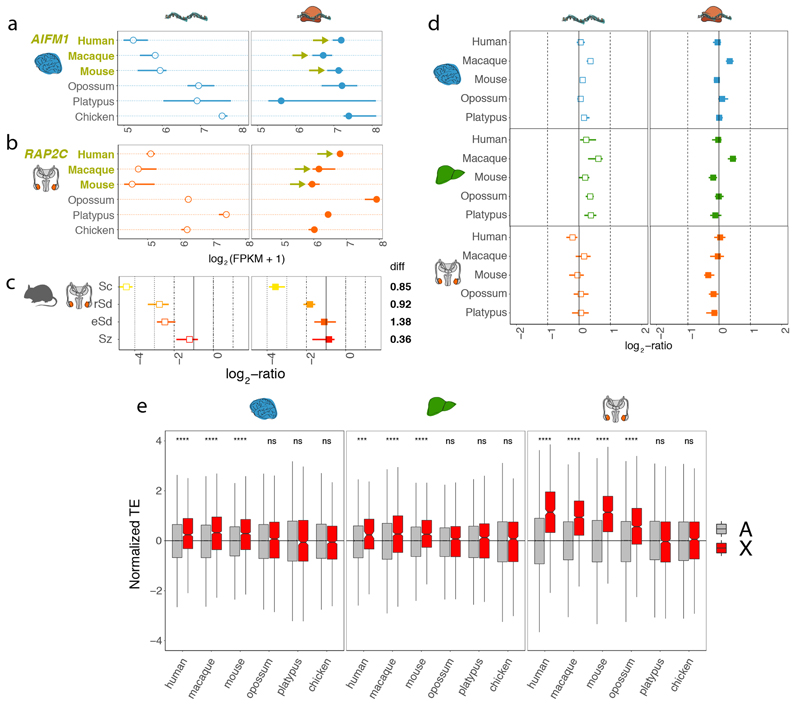
To evaluate to what extent buffering through translational upregulation might have attenuated consequences of the X dosage reduction and/or the complete silencing through MSCI in meiotic cells, we compared, for both expression layers, current X expression levels with ancestral (proto-sex chromosomal) expression levels (Methods). Ancestral X expression levels were inferred from expression levels of autosomal orthologues (mostly located on chromosome 4) in the chicken outgroup, which has a different sex chromosome system and no MSCI, an approach that was previously shown to reliably approximate global ancestral expression patterns1,28. Our analyses revealed that current X expression levels are significantly more similar to ancestral levels at the translatome layer than at the transcriptome layer in eutherian organs (Fig. 4a, Extended Data Fig. 11a, b). Notably, the strongest translational upregulation occurs in the testis (Fig. 4a), which is dominated by meiotic spermatocytes and post-meiotic spermatid cells, where MSCI exerts its effect30,31. Indeed, a dissection of this pattern at the level of individual cell types reveals it to be driven by spermatocytes and spermatids (Extended Data Fig. 11c).
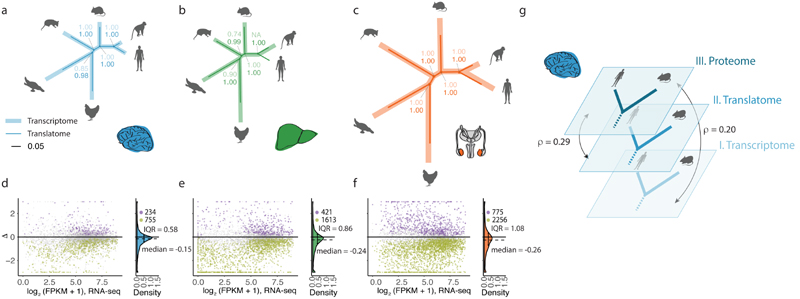
Fig. 4. Compensatory evolution of X-linked genes.
a, Median current to ancestral gene expression ratios at the two expression layers for 1:1 orthologous X-linked genes in eutherians for brain, liver, and testis. Expression levels of chicken orthologues were used as proxy for ancestral expression levels. Platypus genes that are 1:1 orthologous to human X-linked genes and that are present and expressed in chicken were used as a control (i.e., they lack evolutionary dosage reduction and MSCI). Differences in log2-ratios between expression layers are shown to the right of each plot. Cases where the log2-ratio at the translatome layer is significantly (P < 0.05, Mann-Whitney U test, two-sided) higher than at the transcriptome layer are marked in bold. Solid vertical lines correspond to expression levels expected under no dosage reduction. b, Changes in gene expression ranks between transcriptome, translatome, and proteome expression layers for X-linked (X) and autosomal (A) genes for human brain, liver, and testis, respectively. Mann-Whitney U tests (two-sided) were performed for statistical comparisons (non-significant, ns: brain P = 0.416, liver P = 0.399; * P < 0.05, ** P = 0.0067, **** P < 0.0001). Box plots represent the median ± 25th and 75th percentiles, whiskers are at 1.5 times the interquartile range. Organ and species icons were previously used in ref. 25.
As a control for these analyses, we assessed current/ancestral expression of platypus genes that are orthologous to human X-linked genes; these genes are autosomal in platypus, given that sex chromosomes originated independently in monotremes from different autosomes than those that gave rise to sex chromosomes in eutherians/marsupials. As expected, we observe neither a reduction of current transcript abundances nor translational upregulation for these platypus genes compared to the chicken reference (Fig. 4a). A second control, in which we compared expression patterns of eutherian autosomal genes with those of orthologues on chicken chromosome 4, showed similar patterns (Extended Data Fig. 11d).
Moreover, in full agreement with these evolutionary inferences of translational X upregulation, we find that TEs are significantly higher for eutherian X-linked than autosomal genes, consistent with previous work32, especially in the testis, whereas they are not higher for 1:1 orthologues of eutherian X-linked genes compared to other autosomal genes in platypus and chicken (Extended Data Fig. 11e). We then sought to assess whether the observed translational upregulation of X-linked genes relative to autosomal genes has actually led to higher protein abundances. Rank-based analyses across the three expression layers revealed higher expression levels of X-linked genes at both the proteome and translatome layer compared to the transcriptome layer in all three organs, with the strongest upregulation in testis (Fig. 4b). The only marginal increase of expression ranks from the translatome to proteome layer for X-linked genes suggests that X upregulation is mostly confined to the translatome layer.
Discussion
In our study, we contrasted transcriptomes and translatomes across multiple primary organs and representative mammals and a bird, based on extensive RNA-seq and Ribo-seq datasets. Our analyses uncovered detailed patterns of variation and co-evolution that we confirmed to be overall reflected at the level of the proteome, on the basis of state-of-the-art proteomics data for human and mouse21,22, and therefore to be of phenotypic relevance. First, we compared the regulatory dynamics across the two expression layers between the different organs within each species, which revealed a unique pattern in the testis that is explained by strong differential regulation of translation across spermatogenic cell types. Second, our evolutionary analyses unveiled differential and fine-tuned patterns of compensatory evolution between the two expression layers across organs and gene classes, where regulatory changes between expression layers compensate each other. We also show that translational upregulation specifically counteracts consequences of sex chromosome differentiation. Together with other mechanisms1,28, translational compensation in somatic tissues may therefore have contributed to the emergence of X inactivation in eutherian females. The evolution of this mechanism has remained enigmatic, given the lack of full global transcriptional X upregulation1,28. We note that a previous human proteome study did not detect dosage compensation33, presumably because of the limited resolution of available data. Despite the overall buffering pattern, we observed substantial variation in expression divergence rates across individual genes. Notably, some genes changed significantly more or even almost exclusively at the translatome layer, which potentially contributed to lineage- or species-specific organ adaptations.
Initial yeast hybrid work revealed a dominant role of buffering6,7, although this conclusion was subsequently challenged based on analytical considerations9,34, which also apply to a study reporting an excess of compensatory change in hybrid fibroblasts from two mouse strains12. The only previous between-species translatome comparison in mammals, a comparative study of lymphoblastoid cell lines between human, chimpanzee, and macaque across the three main expression layers (transcriptome, translatome, proteome), found very little evidence of buffering11. The short evolutionary divergence time of the species covered in that study may have limited the emergence of compensatory mutations and/or the power to detect (potentially subtle) translational changes. Previous yeast work is consistent with this notion, given that there is less evidence of buffering in hybrids of yeast strains from the same species compared to hybrid work based on different species35.
Altogether, our work identified strong and important differences in patterns of gene expression divergence between the transcriptome and translatome layers in mammalian organs, with an overall lower rate of expression divergence at the translatome layer due to widespread compensatory co-evolution between the layers. Our data and results provide a major resource for future explorations of gene expression evolution across the different layers and its underlying regulatory mechanisms. We therefore developed an online resource that allows for the interactive and integrated exploration of the transcriptome and translatome layers across mammals (Ex2plorer: https://ex2plorer.kaessmannlab.org/).
Methods
Biological samples
We generated Ribo-seq and matched RNA-seq data for the following samples: brain (cerebrum), liver, and testis from human (Homo sapiens), rhesus macaque (Macaca mulatta), mouse (Mus musculus, strain: CD-1, RjOrl:SWISS), grey short-tailed opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), and chicken (red junglefowl, Gallus gallus) (Supplementary Table 1). Additionally we generated Ribo-seq and matched RNA-seq for cells corresponding to four different stages of mouse spermatogenesis: spermatocytes, round spermatids, elongating/elongated spermatids, and spermatozoa (Supplementary Table 1). Isolation of spermatogenic cells was conducted as described in our previous study31. The purity of the isolated cell type pools was ~82–88% for spermatocytes, ~88–90% for round spermatids, ~84-95%, for elongating/elongated spermatids, and ~95% for spermatozoa. In our study, we also retrieved published mass spectrometry proteomics data for human brain, liver, and testis samples from Wang et al. 2019 (ref. 22) and mouse brain from Sharma et al. 2015 (ref. 21).
Our study complies with all relevant ethical regulations with respect to both human samples and samples for the other mammals. Human samples were obtained from scientific tissue banks or dedicated companies; informed consent was obtained by these sources from donors prior to death or from next-of-kin. The use of all human samples for the type of work described in this study was approved by an Ethics Screening panel from the European Research Council (ERC) (associated with H.K.’s ERC Consolidator Grant 615253, OntoTransEvol) and local ethics committees; from the Cantonal Ethics Commission in Lausanne (authorization 504/12) and the Ethics Commission of the Medical Faculty of Heidelberg University (authorization S-220/2017). The use of all other mammalian samples for the type of work described in this study was approved by ERC Ethics Screening panels (ERC Starting Grant 242597, SexGenTransEvolution, and ERC Consolidator Grant 615253, OntoTransEvol).
Ribo-seq and matched RNA-seq data production
The Ribo-seq data was generated based on the ribosome profiling method established by Ingolia et al. 36, but differs from the original protocol5 in that it includes an additional rRNA depletion step (see below for details). The Ingolia 2012 protocol36 has been implemented in the TruSeq Ribo Profile (Mammalian) Library Prep Kit (Illumina) (formerly ARTseq) used in our study. Specifically, frozen tissues were treated in 3 volumes of ice-cold lysis buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.4, 5 mM MgCl2, 5 mM DTT, 100 μg/ml cycloheximide, 1% Triton X-100, 0.5% Sodium deoxycholate, complete EDTA-free protease inhibitors (Roche) and 40 U/ml RNasin plus (Promega)) using a Teflon homogenizer. Lysates were incubated for 10 min on ice and cleared by centrifugation at 3,000 x g and 4 °C for 3 min. Supernatants were flash-frozen and stored in liquid nitrogen. For absorbance measurements, lysates were gently thawed on ice and the OD260 was determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific). From the lysate pool, 15 OD260 were incubated with 650 U RNase I (Ambion) and 5 U Turbo DNase (Ambion) for 45 min at room temperature and gentle agitation. Nuclease digestion was stopped through addition of 8.7 μl SUPERase In RNase Inhibitor (Ambion). Subsequently, lysates were applied to MicroSpin S-400 HRcolumns (GE Healthcare Life Sciences); pre-washed 3 times with 700 μl polysome buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.4, 5 mM MgCl2, 5 mM DTT, 100 μg/ml cycloheximide, 40 U/ml RNasin plus (Promega), complete EDTA-free protease inhibitors (Roche)) for 1 min at 450 x g; and centrifuged for 2 min at 650 x g at 4 °C. The flow-through was immediately mixed with 1 ml Qiazol (Qiagen) and ribosome-protected mRNA fragments were purified using the miRNeasy Micro kit (Qiagen) according to the manufacturer’s instructions, and the concentration of the RNA was determined using a NanoDrop spectrophotometer.
Prior to library preparation, a total of 5 μg RNA from each sample was subjected to ribosomal RNA depletion (Ribo-Zero rRNA Removal kit, Illumina) and subsequently purified using the RNA Clean & Concentrator-5 kit (Zymo Research) according to the manufacturer’s protocol. The rRNA-depleted RNA was separated on a denaturing 15% urea polyacrylamide gel (Thermo Fisher Scientific) and stained with SYBR-Gold (Thermo Fisher Scientific). Gel slices between 26-34 nt were excised and the RNA was extracted using a 450 μl gel extraction buffer (0.5 M Ammonium acetate and 0.05% SDS) for 2 hours at room temperature and with gentle agitation. Gel pieces were removed by centrifugation over Spin-X filter tubes (Corning) for 2 min at 15,000 x g. RNA was precipitated over night at -20 °C in the presence of 1 ml 100% ethanol and 3 μl glycogen. RNA was pelleted for 25 min and washed with 80% ethanol in a tabletop centrifuge at maximum speed and 4 °C. Sequencing libraries were generated using the TruSeq Ribo Profile (Mammalian) Library Prep Kit (Illumina). End-repair, 3’ adapter, reverse transcription, cDNA purification, and circularization were done according to the manufacturer’s instructions.
For opossum, platypus, and chicken samples, an additional rRNA depletion step was implemented, given that the standard depletion step (see above) is based on human, mouse and rat sequence information and that it was determined to be inefficient for these non-model species (which are evolutionarily highly diverged from human/mouse) in test experiments. Specifically, to reduce rRNA contamination for these species (and thus reduce the number of sequencing reads needed for in-depth analyses), first strand cDNAs derived from species-specific rRNA contaminants were further depleted after circularization by hybridization to 5’-biotinylated sense strand oligonucleotides followed by removal of the duplexes through streptavidin affinity as previously described36. Supplementary Table 7 lists the subtractive hybridization oligonucleotides, which correspond to the most abundant rRNA contaminants that were determined in a pilot ribosome profiling experiment (not shown). PCR amplification of the circularized cDNA product was done using the TruSeq Ribo Profile (Mammalian) Library Prep Kit (Illumina) according to the manufacturer’s instructions. The final library of 150-200 bp was gel-purified on a 10% polyacrylamide non-denaturing gel (Thermo Fisher Scientific), excised and recovered with 330 μl gel extraction buffer for 1 hour at 37 °C and with gentle agitation. Gel pieces were removed by centrifugation over Spin-X filter tubes (Corning) for 2 min at 15,000 x g. Libraries were precipitated at -20°C for 1 hour in the presence of 525 μl 100% isopropanol and 2 μl glycogen, pelleted for 25 min at 4 °C and 15,000 x g, washed with 80% ethanol and resuspended in water. Libraries were sequenced on Illumina HiSeq 2500 or, for tests, Illumina MiSeq machines (read lengths: 50 or 100 nucleotides, nt). After data processing and initial quality controls (see below), analyses revealed that the additional depletion step reduced rRNA contamination from 48% to 13%, from 49% to 38%, and from 54% to 28%, for opossum, platypus and chicken, respectively.
In parallel to the Ribo-seq library preparation, matched RNA-seq libraries were prepared from the same lysates based on Ingolia et al.36 and using TruSeq Ribo Profile (Mammalian) Library Prep Kit (Illumina). There was no extra rRNA depletion step during the preparation of RNA-seq libraries. The concentration and the quality of both the Ribo-seq and RNA-seq libraries were determined using Qubit (Thermo Fisher Scientific) and Fragment Analyzer (Advanced Analytical) platforms.
Genome and transcript isoform annotation
Given that the quality of genome annotation differs substantially between the studied species and that we aimed for optimal transcript isoform reconstructions for each tissue as a foundation for all analyses in this study, we refined previous annotations from Ensembl37 for each tissue using our extensive stranded poly(A)-selected RNA-seq datasets25,29. Specifically, for each species we downloaded the reference genome from Ensembl release 87 (ref. 37): hg38 (human), rheMac8 (rhesus macaque), mm10 (mouse), monDom5 (opossum), ornAna1 (platypus), and galGal5 (chicken). For every species-organ combination, the Ensembl annotation was extended using our previous stranded (100 nt, single-end) RNA-seq data25,29. Raw reads were first trimmed with cutadapt v1.8.3 (ref. 38) to remove adapter sequences and low-quality (Phred score < 20) nucleotides, then reads shorter than 50 nt were filtered out (parameters: --adapter=AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC --match-read-wildcards --minimum-length=50 -q 20). Processed reads were then mapped to the reference transcriptome and genome using Tophat2 v2.1.1 (ref. 39) (parameters: --bowtie1 --read-mismatches 6 --read-gap-length 6 --read-edit-dist 6 --read-realign-edit-dist 0 --segment-length 50 --min-intron-length 50 --library-type fr-firststrand --max-insertion-length 6 --max-deletion-length 6).
We then assembled models of transcripts expressed in each tissue using StringTie v1.3.3 (ref. 40) (parameters: -f 0.1 -m 200 -a 10 -j 3 -c 0.1 -v -g 10 -M 0.5). Stringent requirements on the number of reads supporting a junction (-j 3), minimum gap between alignments to be considered as a new transcript (-g 10), and fraction covered by multi-hit reads (-M 0.5) were used to avoid merging independent transcripts and to reduce noise caused by unspliced or incompletely spliced transcripts. We compared the assembled transcript models to the corresponding reference Ensembl annotations using the cuffcompare program v2.2.1 from the cufflinks package41. We then combined the newly identified transcripts with the respective Ensembl gene annotation into a single gtf file. We extended the original Ensembl transcriptome annotation by 4.1-18.9 Mbp with novel transcripts and by 26.8-42.0 Mbp with new splice isoforms, providing, as expected, longer total extension for rhesus macaque, opossum, platypus, and chicken than for the well-studied species (i.e., human and mouse) (Supplementary Table 8).
Selecting the dominant splice isoform
Gene expression level estimates may strongly depend on the proper choice of splice isoforms. A previous study based on proteome data suggested that the vast majority of genes have a single dominant splice isoform42, which is not necessarily the longest. In this study, we focused on the dominant isoform, which was identified by taking into account transcript abundances and coding sequence (CDS) lengths according to the following criteria. For genes with a single annotated isoform, this isoform is by definition the dominant isoform. For genes with multiple isoforms, we proceeded as follows. If the most abundant isoform (i.e., with largest FPKM - fragments per kilobase of transcript per million reads mapped - value based on RNA-seq data) has a more than 5 times higher expression level than the second most abundant isoform, then the most abundant isoform is chosen as the dominant isoform, akin to previous work43. Else, we examined whether the most abundant isoform has a more than 5 times higher expression level than the third most abundant isoform. If so (or if there is no third isoform), we considered the two most abundant isoforms for the final selection step. If not, the final selection was made among the three most abundant isoforms. In the final selection step, the dominant isoform was defined as that with the longest CDS, or, if CDS lengths were the same, the longest transcript.
Orthologous gene sets
Gene expression comparisons between species were made based on genes with a 1:1 orthologous relationship across the species investigated in a given analysis (Supplementary Table 9). Orthology relationships were extracted from Ensembl release 87 (ref. 37). In cases where the dominant splice isoforms of two neighboring genes overlapped in the genome of a species, both genes and their 1:1 orthologues in the other species were removed from all subsequent analyses to avoid read assignment ambiguities.
To ensure that our results and inferences are not affected by potential differences in gene structures between species, we restricted the analyses to the coding regions of the longest protein-coding isoform of 1:1 orthologues that perfectly align across species (i.e., same length, without any gaps). Multiple species alignments to human (hg38) obtained from the UCSC site (http://hgdownload.soe.ucsc.edu/downloads.html) were used to extract genomic coordinates for sequences that aligned without gaps across all 6 species.
Compiling structural RNA sequences for each species
To assess how much each library was contaminated by unusable reads generated from structural RNAs, we first collected for each type of major structural RNAs the annotated sequences from multiple public databases. rRNA sequences for each species were retrieved from several sources: Ensembl release 87, SILVA rRNA database v128 (ref. 44), and NCBI. Transfer RNA (tRNA) sequences were obtained from Ensembl release 87, the genomic tRNA database (gtRNAdb) (ref. 45), and NCBI. Small nucleolar RNAs (snoRNAs) were downloaded from Ensembl release 87 via BioMart.
Read mapping and processing
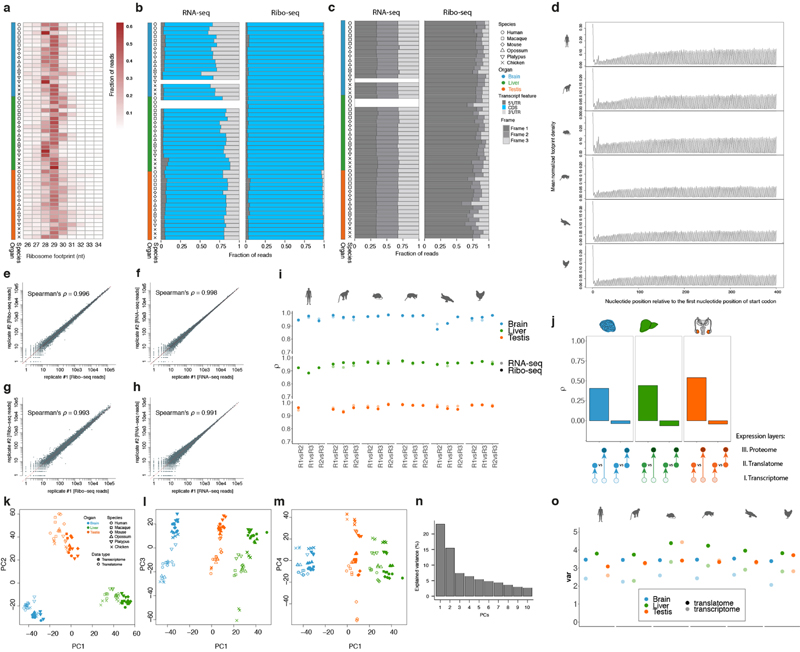
Initial quality assessment of the sequencing reads (e.g., average GC content, base composition, and variability between clusters) was conducted based on the preliminary quality values produced by the Illumina Casava 1.82 software. Raw reads with known 3’ adaptor and low quality bases (Phred score < 20) were trimmed with cutadapt v1.8.3 (ref. 38) (parameters: --adapter=AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC --minimum-length=6 --maximum-length=60 -q 20), and then the trimmed reads were sequentially mapped to the index libraries of species-specific rRNAs, human/mouse/rat rRNAs, species-specific tRNAs, and species-specific snoRNAs using Bowtie2 v2.3.1 (ref. 46) (parameters: --phred33 -L 20 -N 1 -t --no-unal). We discarded the alignments in each step and kept the unaligned reads. Only reads with specific lengths (26-34 and 20-50 nt for Ribo-seq and RNA-seq reads, respectively) were used in downstream analyses. Consistent with biological expectations, we observe Ribo-seq read length distribution peaking around 28-30 nt and predominant mapping to coding regions in all samples (Extended Data Figs. 1a, b, 3a, b).
Overall, the RNA-seq reads (median at ~37 nt) are longer than the Ribo-seq reads (median at ~29 nt). To avoid differences in the mappability of reads spanning the exon-exon junction due to read length differences between Ribo-seq and RNA-seq data, reads longer than 29 nt were trimmed down to 29 nt. Subsequently, the reads were first aligned against organ transcriptomes and then mapped to their respective reference genome with Tophat2 v2.1.1 (ref. 39) (parameters: --no-novel-juncs --library-type fr-firststrand --read-realign-edit-dist 0 --segment-length 20 --min-anchor-length 5 --min-intron-length 50). Uniquely aligned reads with up to a single mismatch between the query sequence and the reference sequence were accepted. For each gene, only reads with A-site (aminoacyl tRNA site, defined and calibrated as in ref. 5) mapped inside the coding region of its dominant splice isoform were quantified and used. Alignment statistics for each filtering and mapping step are provided in Supplementary Table 1.
Triplet periodicity analysis
We used triplet periodicity to evaluate the quality of Ribo-seq experiments, given that it reflects the pattern of genuine translation. Footprint profiles within CDSs were generated by assigning ribosomal A-sites to each nucleotide position of each codon (reading frames 1, 2, and 3). The number of reads mapped to each of the three reading frames was normalized by the total number of reads within the CDS. In sharp contrast to the RNA reads, which mapped evenly to the three codon positions, the ribosome footprints mapped mostly to the first nucleotide of the codon; i.e., to the canonical reading frame (Extended Data Figs. 1c, 3c). The average footprint density of metagene profiles along the CDS faithfully reflects mRNA translocation by codon as translation occurs (Extended Data Fig. 1d).
Assessment of reproducibility for both data types
To assess the reproducibility of the Ribo-seq and RNA-seq datasets and its similarity between the two data types, we calculated the Spearman’s correlation coefficient (ρ) between the read counts of protein-coding genes for each pair of biological replicates and technical replicates (generated for mouse and chicken livers) for the Ribo-seq and RNA-seq data, respectively. The high correlation coefficients observed across technical replicates (ρ > 0.99) and biological replicates (ρ: 0.95-0.99) for both the Ribo-seq and RNA-seq datasets indicate high technical and biological reproducibility (i.e., low technical/biological variation) (Extended Data Fig. 1e-i). Importantly, the correlation coefficients and hence the reproducibility are similar between the two data types (Extended Data Fig. 1e-i). These observations, together with the observation that the estimates in our study are robust to downsampling to equal amount of reads across samples (main text, Extended Data Figs. 1o and 7 and Supplementary Table 3), rule out that observed patterns in downstream biological analyses are explained by technical differences between the Ribo-seq and RNA-seq data (e.g., higher technical variation in the Ribo-seq data than in the RNA-seq data).
Expression levels and normalization
Gene expression levels at the transcriptome and translatome layers for each gene were measured in fragments per kilobase of CDS per million uniquely CDS-aligning reads (FPKM), a unit which corrects for both feature length and sequencing depth. We calculated FPKM based only on the coding region of each locus (i.e., the dominant splice isoform — see above) for both Ribo-seq and RNA-seq libraries, to exclude biased measurements due to heterogeneous quality of annotations for UTRs across species/organs and the fact that Ribo-seq reads, contrary to RNA-seq reads, predominantly map to the main coding region. To render the data comparable across species and organs, translatome and transcriptome FPKMs were separately normalized based on our published approach17. Specifically, among the genes with median expression ranks in the interquartile range, we identified the 1,000 genes that have the most conserved ranks among samples and calculated their median expression levels in each sample. We then derived scaling factors that adjusted these medians to a common value (i.e., dividing each individual median value by the mean of all median values). Finally, these factors were used to scale expression values of all genes in the corresponding samples.
Principal component analysis (PCA)
The PCA of normalized expression (log2(FPKM+1)-transformed) in amniote organs was based on 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologues (Extended Data Fig. 1k-n). The same genes were used for Ribo-seq data without any further filtering. PCA was performed using the prcomcp function in the stats package of R environment for statistical computing (R) (ref. 47). The PCA of gene expression during mouse spermatogenesis was based on 11,057 genes robustly expressed (median FPKM > 1) across mouse spermatogenesis libraries (Extended Data Fig. 3d).
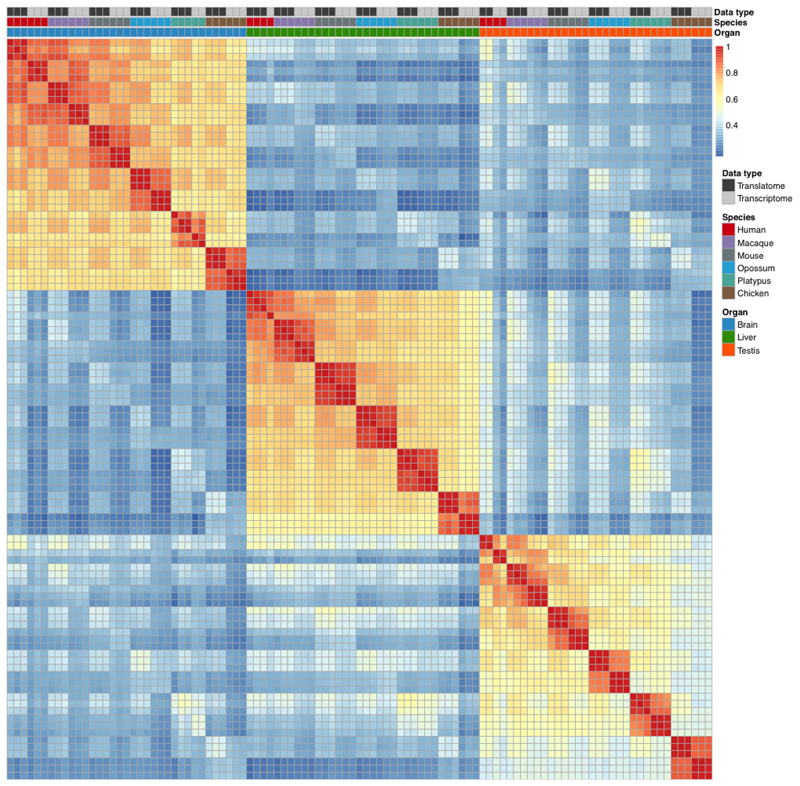
The PCA shows overall highly consistent clustering of the different aspects of the translatome and transcriptome data (Extended Data Fig. 1k-n, see also correlation heatmap, Extended Data Fig. 2). The first principal component (PC1), explaining most gene expression variance, separates the samples by organs, while PC2 separates the germline (testis) and somatic (brain and liver) data (Extended Data Fig. 1k), in agreement with previous work1,17,25. PC1, PC2 and in particular PC3 represent the distinct clustering of the translatome and transcriptome data (Extended Data Fig. 1k, l), whereas PC4 separates the data according to the different species/lineages (Extended Data Fig. 1m).
Estimating the impact of translational regulation
The impact of translational regulation on gene expression can be estimated as an increase in variance of gene expression levels at the translatome layer compared to the transcriptome layer. For both expression layers, we estimated the expression variation across genes, as the variance of the log2(FPKM+1)-transformed median expression values (ėg) across biological replicates in a particular species and organ across 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologous genes (g). The obtained values were corrected for the sampling variance (ϑ), stemming from variation in expression between individuals within species and measurement error. The corrected values were calculated for every species and organ as:
| (1) |
where n corresponds to the number of replicates for a particular species-organ combination, and ϑ is calculated as the average variance of expression levels of a gene (eg,r) across biological replicates (r):
| (2) |
Although the variance in general is expected to be higher at the translatome layer compared to the transcriptome layer due to additional regulatory steps, any anticorrelation between transcript abundance and TE would reduce the variance of the sum of the two expression layers and would thus lead to the variance at the translatome layer being lower than that at the transcriptome layer, as is demonstrated in simulated scenarios (Extended Data Fig. 3e). We modeled expression at the translatome layer as ~(N(σtr2 = 3.61) + N(σtl2 = 0.59) + N(σe_tl2 = 0.10)), with anticorrelation between transcript abundance and TE taking values from 0 to -0.4 with step -0.01; σtr2 and σtl2 correspond to variances of median expression levels for transcript abundances and translation efficiencies, and σe_tr2 and σe_tl2 correspond to the variation in expression levels between individuals within species and measurement errors for transcriptome and translatome layers, respectively. We observe anticorrelation signals of different strengths in all mammalian testis samples, except mouse spermatozoa, (Extended Data Fig. 3f, g), which we believe is caused by the widespread translational repression in certain germ cells (see main text), which highly outnumber somatic cells in the adult testis (ref. 14).
Expression across mouse spermatogenesis
We applied calculations of the measure tau (τ) (ref. 48) across spermatogenesis stages (spermatocytes, round spermatids, elongating/elongated spermatids, and spermatozoa) to determine cell type-specific genes. A gene was considered to be specifically (i.e., predominantly) expressed in a particular cell type, if τ > 0.8, its FPKM expression is greater than 1 and maximal in samples corresponding to that cell type. We then used the sets of cell type-specific genes to normalize expression values of spermatogenesis samples at both expression layers, assuming that their expression outside of the specific stage is negligible. Namely, we multiplied gene expression level values in spermatogenesis samples by the ratio of the median expression level value of cell type-specific genes in full testis to the median expression of cell type-specific genes in that cell type for the transcriptome and translatome layers and for the four cell types, respectively.
Normalized log2-transformed median expression values across replicates at the transcriptome (ėtr) and translatome (ėtl) layers were used to calculate translation efficiency (TE) across spermatogenesis samples as:
| (3) |
Finally, we subtracted from the obtained TE values the TE calculated for genes expressed predominantly in the somatic cells of testis, so that TE = 0 would correspond to the median TE of genes expressed predominantly in the somatic cells. Genes were considered as predominantly expressed in the somatic cells, if FPKM expression values were < 1 in all spermatogenesis samples, and > 1 in the full testis sample.
To investigate patterns of translational regulation during spermatogenesis we clustered TE trajectories using the unsupervised soft clustering method from Mfuzz package49 (v.2.42.0) (number of clusters = 5, fuzzification = 2.5). The fuzzification parameter was estimated with mestimate function from Mfuzz package. 12,101 genes with FPKM > 0 across all stages were used in the analysis.
For every gene, we also assessed potential shifts in expression between the transcriptome and the translatome layers. For each expression layer we calculated a center of mass of gene expression along spermatogenesis (see also schematic illustration in Extended Data Fig. 3j) based on normalized log2-transformed expression values, and used the difference between the centers of mass as a measure of shift in expression at the translatome relative to the transcriptome layer (translational shift), so that shift = 0 indicates synchronous expression at the two layers, shift > 0 indicates a delay in translation, whereas shift < 0 indicates less efficient translation in the later stages of spermatogenesis (Extended Data Fig. 3i-j).
GO enrichment analyses
GO enrichment analyses were carried out with topGO50 (v.2.34.0, R). We used the weight01 algorithm and fisher statistics for the enrichment test and limited the analysis to categories with at least 5 genes (nodeSize = 5). In the analysis of TE in somatic organs, we selected the genes with log2-fold change > 1 between transcriptome and translatome layers of expression and padj < 0.05, calculated in DESeq2 (v.1.20.0, R, ref. 51). Genes with FPKM > 0 in the respective organs were used as the universe sets. In the analysis of TE during spermatogenesis, we used Mfuzz clusters as gene sets of interest and genes with FPKM > 0 across all stages as the universe gene set. In the analysis of genes that changed faster at the translatome than at the transcriptome layer, we selected genes with Δ significantly higher than 0 for every organ, respectively, and used 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologous genes as the gene universe.
Gene expression phylogenies
We estimated expression divergence at the transcriptome and translatome layers based on the assumption that gene expression evolution represents the succession of independent changes in gene expression levels, consistent with Brownian motion-based models of gene expression evolution52. Therefore, differences in population-average expression between, for example, human (ėH) and macaque (ėMa) can be quantified as:
| (4) |
which captures changes in gene expression between 1:1 orthologues that occurred in the human and macaque lineages since their last common ancestor (corrected for sampling variance similarly to (1)), as illustrated in Extended Data Fig. 5a (left column).
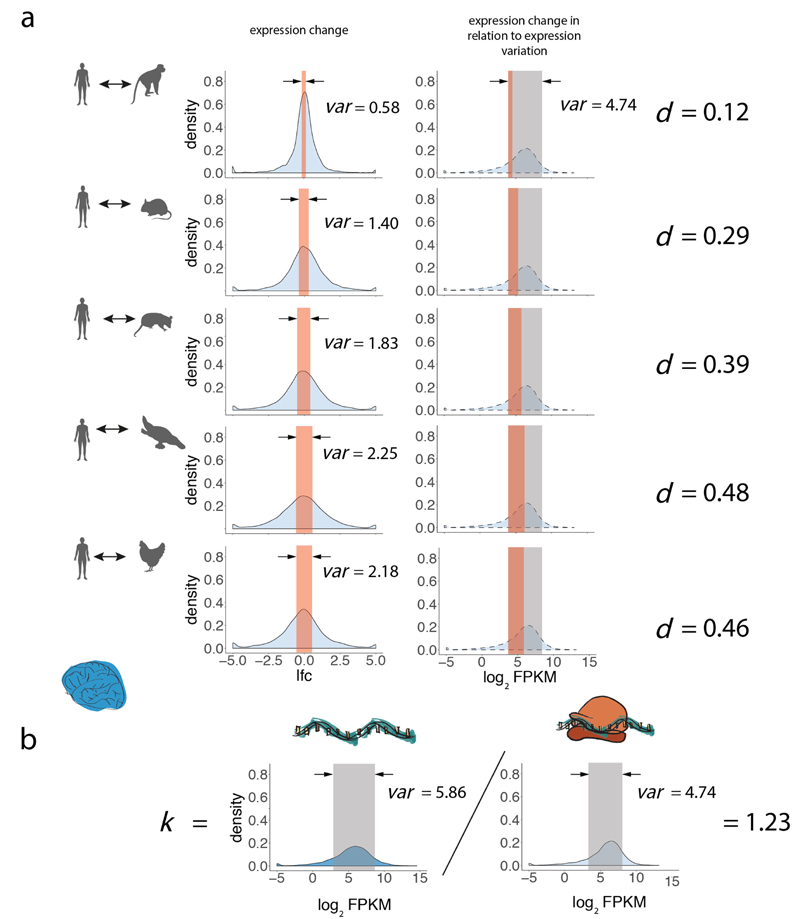
To scale the expression change, i.e. to understand how much it contributes to expression variation, we divide the metrics obtained in (4) by the variance of expression levels across genes, averaged across all 6 species. We use the normalized metric as an estimate of the expression divergence between species (in this example between human and macaque) (Extended Data Fig. 5a, right column):
| (5) |
de was calculated for every pair of species in each organ for the transcriptome and translatome layers. It is important to note that the normalization (i.e., the term in the denominator) accounts for the differences in the expression variation across genes between expression layers and organs (Fig. 1d).
Based on pairwise distance matrices between species, for each organ and both expression layers, we constructed gene expression trees using the neighbor-joining (NJ) approach, akin to our previous procedure17. 5,060 robustly expressed (median FPKM > 1 across all libraries) 1:1 orthologues among the set of 6,327 1:1 orthologues were considered in the analysis. The NJ trees were constructed using functions in the ‘ape’ package in R (ref. 53). The reliability of branching patterns was assessed with bootstrap analyses (1:1 orthologues were randomly sampled with replacement 100 times). The bootstrap values are the proportions of replicate trees that share the branching pattern of the majority-rule consensus tree and are shown in Fig. 2a-c.
We estimated robustness of expression divergence estimates to the amount of sampled reads. We sampled 0.5, 1, 1.5, 2, and 2.5 million reads mapped to perfectly aligned positions in coding regions of 6,327 1:1 orthologues from each library, and estimated overall divergences (i.e., the sum of lengths of all branches in the tree; human branch was not included for liver trees due to a lack of replicates) based on these downsampled datasets. Downsampling suggests that the estimates are almost indistinguishable from the full dataset at already 2 million reads (Supplementary Table 4). Ratios of translatome tree lengths to transcriptome tree lengths, corresponding to the amount of buffering, are also robust to downsampling (Supplementary Table 4).
Compared to a Spearman rank correlation-based approach, previously used to calculate expression divergence rates (ref. 14), our method provides a unit-based metrics to estimate divergence. Also, unlike correlation-based approaches, our method allows for comparison of evolutionary rates between different gene categories (see Fig. 3a and corresponding Method section below). Reassuringly, however, the Spearman rank correlation-based approach gives very similar estimates of buffering (i.e., 19-21%) across organs (Supplementary Table 4) to what we observe in our variance-based approach (20-22%) (Fig. 2a-c).
Modeling gene expression divergence
The overall lower expression divergence observed at the translatome layer compared to transcriptome layer (Fig. 2a-c) may be due to different factors. First, this pattern may result from compensatory changes; that is, when evolutionary changes in transcript abundances and their translational regulation (reflected in their TEs) compensate each other. Second, even without any compensatory changes, divergence at the translatome layer can be lower than at the transcriptome layer due to lower rates of TE changes compared to changes in transcript abundance. Indeed, in a scenario with no changes in TEs, only changes in transcript abundances would contribute to the divergence at both layers, but their relative impact would be less at the translatome layer, due to higher expression variation across genes at the translatome layer (Fig. 1c).
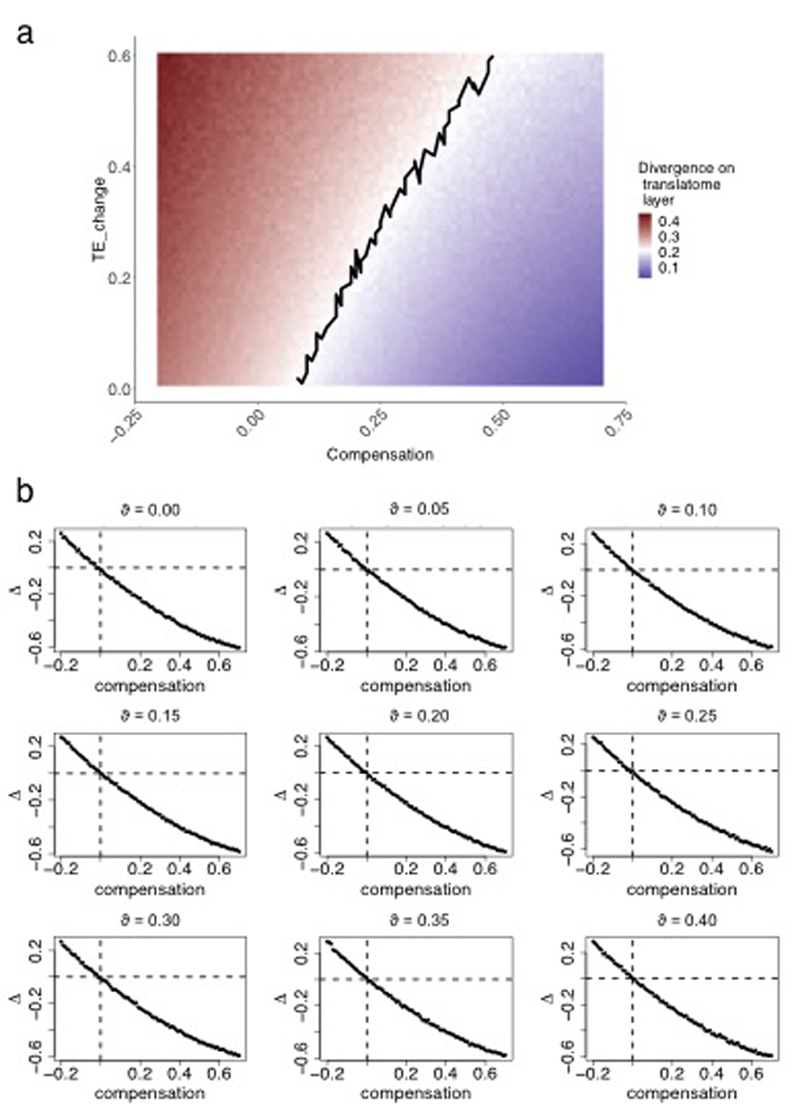
To dissect the aforementioned factors that may shape the evolution of gene expression at the translatome layer, we modeled the divergence of expression levels at the translatome layer between macaque and mouse brain over parameters corresponding to the rates of TE divergence and extent of compensatory evolution (Extended Data Fig. 6a). Modeled variables appear as , to discriminate from real data observations, which are used for parameter estimations. The simulations were done for 5,060 genes according to the following steps:
- Gene expression of the last common ancestor between macaque (Ma) and mouse (Mo) at the transcriptome layer was modeled using the normal distribution generator function rnorm in R as:
(6) - Gene expression divergences from the ancestor at the transcriptome layer was modeled for macaque and for mouse separately as:
(7) - Gene expression levels at the transcriptome layer in present-day macaque and mouse were modeled as:
(8) - TE of the last common ancestor (A) between macaque and mouse was modeled as:
(9) - Divergences in TE rates from the ancestor were modeled for macaque and mouse as:
where c is a parameter that varies from -0.25 to 0.75, with steps of 0.01, and indicates the amount of compensatory evolution between the two layers, where = 0 corresponds to a scenario with no compensation, c > 0 corresponds to scenarios where transcriptome and translational changes are cancelling each other to different degrees, and c < 0 corresponds to scenarios where transcriptome and translational changes are acting in the same direction; and d is a parameter, which varies from 0.01 to 0.60, with steps of 0.01, and indicates the change in TE relative to its estimated ancestral value.(10) - Finally, gene expression at the translatome layer in modern macaque and mouse were modeled as:
(11)
Expression divergences for every simulated scenario were calculated as described in the previous section and are shown in Extended Data Fig. 6a.
Differences in evolutionary rates between expression layers for individual genes
To understand how changes at the translatome layer affect the expression evolution of individual genes, we calculated the difference in evolutionary rates between the two expression layers across 6 species for each of 5,060 1:1 orthologues, for brain, liver, and testis as:
| (12) |
where and are medians of log2(FPKM+1)-transformed expression levels across replicates for gene g in 6 species at the translatome and transcriptome layers, respectively; k is a coefficient set to normalize for the differences in the within-species expression variation across genes between expression layers (seeformula (1) and associated explanation), as illustrated in Extended Data Fig. 5b:
| (13) |
Therefore, Δ represents the difference in the amount of evolutionary (across species) gene expression change between the translatome layer ( term) and transcriptome layer ( term, where k adjusts for differences in expression variation across genes between the two expression layers). Δ = 0 indicates equal evolutionary rates at both expression layers, Δ > 0 indicates a faster evolutionary rate at the translatome layer, and Δ < 0 indicates a slower evolutionary rate at the translatome layer. To judge the statistical significance of the observed sign of Δ, we estimate its standard error by repeatedly replacing median expression values across replicates with expression values in individual replicates (rs) for every species and calculating standard deviation over the bootstrapped Δ as:
| (14) |
and then calculate a Z-score for every gene as:
| (15) |
A gene was considered as evolving significantly faster or slower at the translatome layer compared to the transcriptome layer, if the corresponding P-value is less than 0.1 after multiple test correction using the Benjamini-Hochberg method54.
To estimate the variation of Δ and to compare it between organs, we calculated the interquartile range (IQR) of Δ across all 5,060 1:1 orthologues.
Calculating Δ on simulated data for the brain of 6 species (performed similarly to as described in the previous section) shows that the metric is indeed centered at 0 when evolutionary rates are the same at both expression layers, and that it decreases with increasing amount of compensation (Extended Data Fig. 6b). The metric remains unbiased with increasing amount of biological/technical variation across individuals (Extended Data Fig. 6b).
Lineage-specific changes
To identify orthologues that changed significantly more at one expression layer compared to the other, we calculated Z-scores based on log2-fold changes and standard errors obtained with DESeq2 R-package (v 1.20.0, ref. 51). For a species pair and a particular organ, Z-scores were calculated for pairs of orthologues as
| (16) |
where and correspond to log2-fold changes between two species at the transcriptome and translatome layer for gene g, respectively; and correspond to standard errors for gene g, estimated by DESeq2 based on biological replicates and the RNA-seq and Ribo-seq data, respectively; k – normalization coefficient, calculated as:
| (17) |
where spA and spB correspond to different species in a pair (e.g., human and macaque).
Z > 0 indicates greater change at the translatome layer, whereas Z < 0 indicates greater change at the transcriptome layer. The difference was considered significant, if the corresponding P-value is less than 0.05 after Benjamini-Hochberg correction for multiple testing54. Changes were assigned to specific lineages according to maximum parsimony. That is, a change was considered specific to the human lineage, if the Z-score was significant and of the same sign between human and mouse and human and opossum; a change was considered specific to the macaque lineage, if the Z-score was significant and of the same sign between macaque and mouse and macaque and opossum; an overlap between human and macaque lineages was attributed to their common ancestor; a change was considered specific to the mouse lineage, if the Z-score was significant and of the same sign between mouse and macaque and mouse and opossum.
Expression divergence across gene classes
The gene sets underlying the different classes analyzed in this paper were retrieved from various sources. Gene essentiality was defined based on the probability of being loss-of-function intolerant, that is, the pLI score55; the score data were obtained from ExAC release 0.3.1 (http://exac.broadinstitute.org/). Haploinsufficiency (HI) scores from Shihab et al. 56 were used as proxies of the extent of haploinsufficiency for human genes. We projected human HI scores to mouse 1:1 orthologues. Mouse HI scores for 11,828 1:1 orthologues among the three representative species (i.e., macaque (Ma), mouse (Mo), and opossum (O)) were first ranked from the largest to the smallest values and then used to define two gene subsets (first/last quartile) that were defined as sensitive/insensitive to haploinsufficiency, respectively.
To define the sets of both broadly expressed genes and tissue-specific genes for mouse, we relied on the RNA-seq data for five adult mouse tissues (i.e., brain, heart, liver, kidney and testis) obtained from our previous study25. That study also included cerebellum, but we did not include it in this analysis, because considering both brain (prefrontal cortex) and cerebellum would reduce the number of brain-specific genes due to the frequently shared gene expression profiles in these two tissues25. We assessed gene expression breadth using the tau (τ) tissue specificity index48. Genes with τ ≤ 0.2 and τ ≥ 0.7 were defined as broadly expressed and tissue-specifically expressed, respectively (Supplementary Table 10).
Finally, the phylogenetic age of each mouse gene was retrieved from the GenTree database57 (http://gentree.ioz.ac.cn/). Given that the analyses that consider gene age are based on shared 1:1 therian orthologues, we focused the age analyses on two sets of orthologues: 1) those that emerged in therian, mammalian, amniote or tetrapod ancestors (genes defined as relatively young); and 2) orthologues that emerged before (i.e., ancestors of jawed vertebrates) (genes defined as relatively old). The set of 11,828 1:1 orthologues among macaque, mouse, and opossum were considered for total branch length analyses. Specific gene class information for every orthologous gene is provided in Supplementary Table 10.
Expression divergence for a particular gene set (e.g., housekeeping genes) between a pair of species (e.g., macaque and mouse) was calculated for the transcriptome and translatome layers as:
| (18) |
where indicates restriction to a particular gene set, with genes robustly expressed (median FPKM > 1) across all libraries, or, in the case of tissue-specific genes, robustly expressed (median FPKM > 1) in the tissue of interest.
Natural selection acting to preserve the encoded amino-acid sequence was estimated as ω = dN/dS, the ratio of nonsynonymous/synonymous substitution rates, using the basic model implemented in the codeml function of the PAML package58 (v.4.9f). Calculations of ω were done based on coding sequences, extracted from multiple species alignments to human (hg38) from the UCSC site (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/multiz100way/), according to annotation coordinates of the dominant isoform in human.
To estimate relative contributions of different factors (i.e., gene expression level, tissue-specificity, conservation of the coding sequence, loss of function intolerance, haploinsufficiency, and gene age) to rates of evolution at the two expression layers, measured as variance in expression levels across species for each gene g, we built a multiple linear regression model separately for the transcriptome and translatome layers as:
| (19) |
where ėg,s corresponds to the median log2(FPKM+1)-transformed expression levels across replicates for gene g in 3 species, and mediang[ėg,s] corresponds to a median value across 3 species; τ – tissue specificity index of mouse gene (calculated as above); ω – dN/dS ratio (calculated as above); pLI – loss of function intolerance (projected from human orthologues); HI – haploinsufficiency (projected from human orthologues); t – phylogenetic duplication age of mouse gene.
Assessment of X to proto-X expression levels
To assess the occurrence of X upregulation as a response to sex chromosome differentiation, current X expression levels were compared to ancestral X (proto-X) expression levels for the RNA-seq and Ribo-seq data, respectively, following our previous procedures1,28,29. Ancestral expression levels of proto-sex chromosomal genes were inferred from expression levels of autosomal 1:1 orthologues in the chicken (the evolutionary outgroup), which has a different sex chromosome system1,28,29. Prior to any direct comparisons, raw X expression levels in each of the focal eutherian species (i.e., human, macaque, or mouse) and chicken were normalized relative to their autosomal backgrounds. Briefly, for each library, X expression levels were normalized on the basis of a scaling factor that was derived from adjusting the median expression levels of robustly expressed (FPKM > 1 across RNA-seq libraries for the respective organ) autosomal 1:1 orthologues across all RNA-seq and Ribo-seq libraries to a common value (i.e., each individual median value was divided by the mean of all median values). We next computed current X to proto-X expression log2-ratios (i.e., log2-transformed median across biological replicates expression values of focal species divided by that of chicken replicates) for every orthologous pair for the transcriptome and translatome layers, respectively. Finally, the median ratio across the 1:1 orthologues was calculated, with 95% confidence intervals estimated by 100 resamplings with replacement. Statistically significant differences in log2-ratios between expression layers were assessed using Mann-Whitney U tests.
We also compared TEs of human X-linked and autosomal genes and their orthologues in the other five species for the three studied organs, filtering for robustly expressed genes (FPKM > 1 across RNA-seq libraries for the respective organ from a given species) (Extended Data Fig. 10e). Statistically significant differences between gene sets were assessed using Mann-Whitney U tests.
Proteome data analyses
To assess the extent to which patterns of variation and co-evolution of transcriptomes and translatomes are reflected at the level of the proteome, we performed various rank-based analyses, given that the mass spectrometry-based data are not directly quantitatively comparable to the RNA-seq/Ribo-seq data and across species. Mass spectrometry data were retrieved from published work21,22 (see also section: Biological samples). First, we assessed correlations across the three expression layers in the three studied organs in human based on Spearman’s rank correlation coefficients for 9,642 genes with detectable expression at all three expression layers (Fig. 1b). In the framework of the cross-layer comparisons, we also more specifically investigated the similarity of expression (rank) changes between the different pairs of layers using Spearman correlation analyses (Extended Data Fig. 1j). In concordance with the correlation analysis across expression layers discussed in the main text (Fig. 1b), we find that expression (rank) changes between the transcriptome and translatome are overall similar to those between the transcriptome and proteome, whereas changes between the transcriptome and translatome are completely different from those between the translatome and proteome (Extended Data Fig. 1j).
Second, focusing on 6,972 1:1 orthologous genes with detectable expression in the brain in human and mouse at all three expression layers, we determined changes in expression ranks between the two species for each of the three expression layers. We then assessed the correlation of these changes between: 1) the proteome and translatome; 2) the proteome and transcriptome (Fig. 2g).
Third, for the same set of 6,972 1:1 orthologues, we performed rank-based comparisons of protein expression levels between human and mouse. Specifically, for each gene, we assessed rank changes between human and mouse for the translatome and transcriptome layers, respectively. These served as proxies of evolutionary divergence rates at these expression layers. Next, we calculated differences of the inferred rates between the two expression layers and selected two sets of genes: 1) the 10% of genes with the most decelerated rate of evolution at the translatome compared to the transcriptome layer; 2) the 10% of genes with the most accelerated rate of evolution at the translatome compared to the transcriptome layer. Finally, we calculated the absolute amount of rank change at the proteome layer for these two sets of genes (Extended Data Fig. 9).
Fourth, the same rank-change approach and set of 6,972 1:1 orthologues were used to assess the evolution at the proteome layer across the different gene classes (Fig. 3b).
Fifth and finally, to assess whether the observed translational upregulation of X-linked genes has actually led to higher protein abundances, we assessed the amount of rank changes and their directionality (i.e., expression rank increases or decreases relative to preceding layers) across the three expression layers for X-linked and autosomal genes in human brain, liver, and testis (Fig. 4b).
General statistics and plots
All statistical analyses and graphical representations were done in R 3.3.3 (ref. 47) using the R packages DESeq2 (1.20.0) (ref. 51), ggplot2 (2.2.1) (ref. 47), ape (5.0) (ref. 53), pheatmap (1.0.10) (ref. 47), gridExtra (2.3) (ref. 47), plyr (1.8.4) (ref. 47), dplyr (0.7.4) (ref. 47), Mfuzz (2.42.0) (ref. 49), topGO (2.34.0) (ref. 50), cowplot (0.9.4) (ref. 47), org.Hs.eg.db (3.7.0) (ref. 47), org.Mm.eg.db (3.7.0) (ref. 47), tidyverse (1.2.1) (ref. 47), and plotly (4.8.0) (ref. 47).
Extended Data
Extended Data Fig. 1. Information on generated RNA-seq and Ribo-seq data.
a, Ribosome footprint length distributions across Ribo-seq libraries (nt, nucleotides). b, Fractions of Ribo-seq and RNA-seq reads mapped to 5’ untranslated regions (5’UTRs), coding sequences (CDS), and 3’ untranslated regions (3’UTRs), respectively. c, Distribution of Ribo-seq and RNA-seq reads across the three reading frames in the coding sequence (CDS) of dominant splicing isoforms (Frame 1: canonical reading frame). d, Mean normalized density of footprints along the coding region of the dominant isoforms of protein-coding genes for the brain Ribo-seq data. The Ribo-seq read (A-site) density for each position is plotted relative to the first nucleotide position of the start codon. e-h, Spearman’s correlation coefficient (ρ) of read counts for protein-coding genes with a mean read count > 1 between the two technical replicates for mouse liver Ribo-seq (e) and RNA-seq (f) data, and for chicken liver Ribo-seq (g) and RNA-seq (h) data. i, Correlations between biological replicates for Ribo-seq and RNA-seq data. Each dot corresponds to Spearman’s correlation coefficient (ρ) in pairs of biological replicates for every species-organ combination. Only 1 replicate (therefore no pairs) is available for the human liver transcriptome, only 2 replicates (1 pair) are available for the human testis transcriptome and translatome, and only 2 replicates (1 pair) are available for the platypus brain transcriptome. The correlation coefficients between the replicates are similar for the two data types and statistically indistinguishable (P = 0.159) in a Mann-Whitney U test (two-sided). j, Comparisons of gene expression (rank) changes between the three expression layers. Changes in gene expression ranks were calculated between expression layers (i.e., from transcriptome to translatome, from transcriptome to proteome, and from translatome to proteome), and Spearman’s ρ was calculated to estimate the similarity of rank changes between the different pairs of expression layers. k-n, PCA based on 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologues. Factorial maps represent the relations of PC2 versus PC1 (k), PC3 versus PC1 (l), and PC4 versus PC1 (m). The scree plot (n) indicates the percentage of variance explained by each of the first 10 PCs. (o), Variance at the two expression layers across mammalian organs for downsampled data. For this analysis data were downsampled to 2.5 million reads in each library. See Fig. 1d for the analysis of the full dataset. Organ and species icons were previously used in ref. 25.
Extended Data Fig. 2. Correlations of gene expression levels between sequenced libraries.
The heatmap of the pairwise Spearman’s correlation coefficient (ρ) is based on the set of 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologues for perfectly aligned regions (see Methods). It represents the degree of similarity of gene expression profiles between data types (translatome, transcriptome), species (human, macaque, mouse, opossum, platypus, chicken) and tissues (brain, liver, testis).
Extended Data Fig. 3. Quality assessment and analysis of mouse spermatogenesis data.
a, Ribosome footprint length distributions across Ribo-seq libraries (nt, nucleotides). b, Fractions of Ribo-seq and RNA-seq reads mapped to 5’ untranslated regions (5’UTRs), coding sequences (CDS), and 3’ untranslated regions (3’UTRs), respectively. c, Distribution of Ribo-seq and RNA-seq reads across the three reading frames in the coding sequence (CDS) of dominant splicing isoforms (Frame 1: canonical reading frame). d, PCA based on 11,057 genes robustly expressed (median FPKM > 1) across murine spermatogenesis libraries. The scree plot (inset) indicates the percentage of variance explained by each of the first 10 PCs. e, Variance at the translatome layer calculated for simulated scenarios with different amounts of translational contribution (see Methods for details). Dashed line corresponds to IQR calculated at the transcriptome layer; f,g, Spearman’s ρ between transcription abundance and TE was calculated for 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologues in bulk testis across the amniotes (f) and across spermatogenesis stages in mouse (g). h,i, TE (h) and translational shift (i) for clusters of genes (gene numbers in parentheses) with distinct TE patterns (Mfuzz clustering). Arrows indicate TE increases/decrease compared to the respective global pattern (Fig. 1e). *indicates a cluster of genes, which escape expression repression and delay at the translatome layer. j, Expression of individual genes, representing each of the five TE clusters, at the transcriptome and translatome layers (left column); shift in expression timing between expression layers for the corresponding genes (right column) with crosses representing the centers of mass of gene expression across spermatogenesis. k, Tissue-specificity (tissue Tau) across TE clusters. Cluster I, highlighted in color, is dominated by testis-specific genes. Box plots represent the median ± 25th and 75th percentiles, whiskers are at 1.5 times the interquartile range. l, Gene expression divergence at the two expression layers for genes with stage-specific expression across spermatogenesis among 8,109 1:1 orthologues robustly expressed (FPKM > 1) in macaque, mouse, and opossum. Sc, spermatocytes, rSd, round spermatids, eSd, elongating/elongated spermatids, Sz, spermatozoa.
Extended Data Fig. 4. GO enrichment analyses.
a-d, Top 5 significantly enriched GO terms among genes with high (a, b) and low (c, d) TE for brain (a, c; blue) and liver (b, d; green) in mouse. e, Top 10 significantly enriched GO terms for each of the mouse spermatogenesis TE trajectory cluster (Extended Data Fig. 3h-k). f-h, Significantly enriched GO terms (biological processes) among genes that changed significantly more at the translatome compared to the transcriptome layer in brain (f), liver (g), and testis (h). Significance was estimated in Fisher’s exact test (P < 0.05), with P values adjusted for multiple testing using Benjamini-Hochberg method.
Extended Data Fig. 5. Normalization procedures in the evolutionary expression analyses.
a, Illustration of the normalization approach used in our study to globally assess gene expression evolution. In this approach, evolutionary changes in gene expression are based on the assessment of expression differences across 1:1 orthologues between species. Specifically, we quantify the differences across orthologues as the variance (var) of their log2-fold expression changes between species (left column), which is then divided (normalized) by the expression variation, calculated as the variance (var) of expression levels across genes, averaged across all studied species (right column). This procedure provides the expression divergence estimate (d). We note that the variance is similar across species for a given organ and expression layer (Fig. 1d). The example shown illustrates changes between human and each of the other five species in brain at the transcriptome layer. b, Illustration of the normalization procedure used to assess the expression evolution of individual genes. The normalization coefficient k is calculated as the ratio of the variances (var) across genes between the translatome and the transcriptome layer. The brain is shown as an example. Organ and species icons were previously used in ref. 25.
Extended Data Fig. 6. Simulation of gene expression divergence across expression layers.
a, Simulation of gene expression divergence across different evolutionary scenarios. Expression divergence at the translatome layer between macaque and mouse brain was modeled over parameters of compensation and TE change (see “Modeling gene expression divergence” in Methods for details). Red (blue) correspond to simulated scenarios with expression divergence higher (lower) than in actual data. Black line corresponds to simulated scerarios demonstrating expression divergence values observed in actual data. b, Contrast in evolutionary rates between the two expression layers for simulated data. Δ was calculated for simulated datasets with different amounts of compensation and different amounts of, corresponding to expression variation between individuals and measurement errors (see Methods for details).
Extended Data Fig. 7. Contrast in evolution between transcriptome and translatome layers for individual genes in downsampled data.
Δ was calculated based on datasets downsampled to 0.5 million in each library for brain (a), liver (b), and testis (c). See Fig. 2e-f in the main text for the analysis of the full dataset.
Extended Data Fig. 8. Screenshot of SATB2 gene in Ex2plorer app.
SATB2 is an example of a gene that changes significantly less on translational layer compared to transcriptional layer in mammalian brain. Organ icons were previously used in ref. 25.
Extended Data Fig. 9. Evolution at the proteome layer between human and mouse brain for genes with slower/faster evolution at the translatome compared to the transcriptome layer.
Absolute rank changes of proteome expression levels were calculated for genes with slower (olive) and faster (purple) evolution at the translatome compared to the transcriptome layer. The difference of the distributions between the two gene sets is statistically significant (****P < 0.0001, Mann-Whitney U test, two-sided). Box plots represent the median ± 25th and 75th percentiles, whiskers are at 1.5 times the interquartile range.
Extended Data Fig. 10. Mammalian lineage-specific changes between expression layers.
a-c, Number of genes with lineage-specific patterns of slower (olive) or faster (purple) evolution at the translational layer, potentially driven by stabilizing and directional selection, respectively, for brain (a), liver (b), and testis (c). Due to the lack of a biological replicate, the branch leading to human was omitted in the liver phylogeny for the transcriptional layer. d, e, Examples of individual genes with potential patterns of stabilizing (d) or directional (e) evolution. Species names with significant changes are marked by corresponding colors. Organ and species icons were previously used in ref. 25.
Extended Data Fig. 11. Compensatory evolution of X-linked genes.
a, b, examples of upregulation for the dosage reduction at the transcriptome layer. Species affected by upregulation are shown in olive, with arrows representing compensatory changes at the translatome layer. c, Median ratio of X-linked gene expression values in murine spermatogenic cell types to expression values of their 1:1 orthologues in chicken testis. In all cases log2-ratio at the translatome layer is significantly (P < 0.05, Mann-Whitney U test, two-sided) higher than at the transcriptome layer (marked in bold). Solid vertical lines correspond to expression levels expected under no dosage reduction (i.e., log2-ratio = -1). d, Median current to ancestral gene expression ratios at two expression layers for 1:1 orthologous autosomal genes located on chromosome 4 in chicken for brain, liver, and testis. Chicken orthologues were used as a proxy for ancestral expression. See Fig. 4a and main text for details. e, Normalized TEs for 1:1 orthologues of eutherian X-linked and autosomal genes across amniote organs. Mann-Whitney U tests (two-sided) were performed for statistical comparisons (non-significant, ns: P > 0.05, ***P = 0.00003, ****P < 0.0001). P values were adjusted for multiple testing using Bonferroni method. Box plots represent the median ± 25th and 75th percentiles, whiskers are at 1.5 times the interquartile range. Organ and species icons were previously used in ref. 25.
Supplementary Material
Acknowledgments
We thank K. Harshman and the Lausanne Genomics Technology Facility for high-throughput sequencing support; I. Xenarios and the Vital-IT computational facility for computational support; M. Baumann and S. Richling from the Heidelberg University Computational Center (Universitätsrechenzentrum, URZ) for supporting our computational work on the bwForCluster; M. Sanchez-Delgado for figure assistance; J. VandeBerg for opossum samples; P. Khaitovich for human and macaque samples; P. Jensen and A. Fallahshahroudi for red junglefowl samples; and A.B. Arpat and the Kaessmann group for helpful discussions.
Funding
This work was primarily supported by a grant (KA 1710/3-1) to H.K. from the German Research Council (DFG) and also by a European Research Council grant (615253, OntoTransEvol) to H.K. We also acknowledge computational support by the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through grant INST 35/1134-1 FUGG. D.G. acknowledges funding through the National Centre of Competence in Research (NCCR) RNA & Disease. F.G. is supported by an Australian Research Council Fellowship. S.O. was supported by the DFG (CRC 1366). S.A. was supported by the DFG (SFB 1036). M.E.G. and A.H.F.M.P acknowledge funding from the Swiss National Science Foundation (31003A_146293) and the Novartis Research Foundation. B.dM. was funded by grants from the Centre National pour la Recherche Scientifique (CNRS) and the European Research Council (ERC) Executive Agency under the European Community’s Seventh Framework Programme (FP7/2007-2013 Grant Agreement no. [322788]).
Footnotes
Author contributions
H.K. conceived and designed the original study. The work was supervised by H.K. and E.L. Z.Y.W. and E.L. processed all data and performed the analyses; A.L., K.M., T.B. and C.R. performed all experimental work. F.G. provided platypus samples and related biological expertise. M.C.M. provided key analytical ideas and discussions. B.D., B.dM., M.E.G. and A.H.F.M.P provided purified mouse spermatogenic cell samples. P.J. and D.G. helped to establish the original ribosome profiling method for solid tissues. D.G. provided key experimental input and guidance during the data production and initial analysis phase. S.O. developed the Ex2plorer app. S.A. provided key statistical advice. Z.Y.W., E.L. and H.K. wrote the manuscript, with input from all authors.
Competing interests
The authors declare no competing interests.
Data availability
Raw and processed Ribo-seq and RNA-seq data are available from ArrayExpress (accession number: E-MTAB-7247, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-7247/). All other data are available as Supplementary information or are available upon request. We also created a publicly available data resource (Ex2plorer: https://ex2plorer.kaessmannlab.org/), which allows the interactive exploration of the evolution at both expression layers for all 1:1 orthologous genes.
Code availability
Custom R scripts used to generate the results reported in the manuscript and processed data are available at https://github.com/evgenyleushkin/translatome.
References
- 1.Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet. 2014;15:734–748. doi: 10.1038/nrg3802. [DOI] [PubMed] [Google Scholar]
- 2.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan Z, et al. Primate Transcript and Protein Expression Levels Evolve under Compensatory Selection Pressures. Science. 2013 doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015;16:651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McManus J, May G, Spealman P, Shteyman A. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2013 doi: 10.1101/gr.164996.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artieri CG, Fraser HB. Evolution at two levels of gene expression in yeast. Genome Res. 2013 doi: 10.1101/gr.165522.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadler M, Fire A. Conserved translatome remodeling in nematode species executing a shared developmental transition. PLoS Genet. 2013;9:e1003739. doi: 10.1371/journal.pgen.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albert FW, Muzzey D, Weissman JS, Kruglyak L. Genetic influences on translation in yeast. PLoS Genet. 2014;10:e1004692. doi: 10.1371/journal.pgen.1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Evolution of gene regulation during transcription and translation. Genome Biol Evol. 2015;7:1155–1167. doi: 10.1093/gbe/evv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang SH, Hsiao CJ, Khan Z, Pritchard JK. Post-translational buffering leads to convergent protein expression levels between primates. Genome Biol. 2018;19:83. doi: 10.1186/s13059-018-1451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou J, et al. Extensive allele-specific translational regulation in hybrid mice. Mol Syst Biol. 2015;11:825. doi: 10.15252/msb.156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csardi G, Franks A, Choi DS, Airoldi EM, Drummond DA. Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast. PLoS Genet. 2015;11:e1005206. doi: 10.1371/journal.pgen.1005206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech Dev. 2001;106:3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- 15.Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res. 2003;103:217–224. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- 16.Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci U S A. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 18.Ramm SA, Scharer L, Ehmcke J, Wistuba J. Sperm competition and the evolution of spermatogenesis. Mol Hum Reprod. 2014;20:1169–1179. doi: 10.1093/molehr/gau070. [DOI] [PubMed] [Google Scholar]
- 19.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 20.Zarate YA, Fish JL. SATB2-associated syndrome: Mechanisms, phenotype, and practical recommendations. Am J Med Genet A. 2017;173:327–337. doi: 10.1002/ajmg.a.38022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma K, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Syst Biol. 2019;15:e8503. doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartha I, di Iulio J, Venter JC, Telenti A. Human gene essentiality. Nat Rev Genet. 2018;19:51–62. doi: 10.1038/nrg.2017.75. [DOI] [PubMed] [Google Scholar]
- 24.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Cardoso-Moreira M, et al. Gene expression across mammalian organ development. Nature. 2019;571:505–509. doi: 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WH, Trachana K, Lercher MJ, Bork P. Younger genes are less likely to be essential than older genes, and duplicates are less likely to be essential than singletons of the same age. Mol Biol Evol. 2012;29:1703–1706. doi: 10.1093/molbev/mss014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graves JA. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet. 2016;17:33–46. doi: 10.1038/nrg.2015.2. [DOI] [PubMed] [Google Scholar]
- 28.Julien P, et al. Mechanisms and evolutionary patterns of Mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marin R, et al. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 2017;27:1974–1987. doi: 10.1101/gr.223727.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner JM. Meiotic Silencing in Mammals. Annu Rev Genet. 2015;49:395–412. doi: 10.1146/annurev-genet-112414-055145. [DOI] [PubMed] [Google Scholar]
- 31.Soumillon M, et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 32.Faucillion ML, Larsson J. Increased expression of X-linked genes in mammals is associated with a higher stability of transcripts and an increased ribosome density. Genome Biol Evol. 2015;7:1039–1052. doi: 10.1093/gbe/evv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Zhang J. No X-chromosome dosage compensation in human proteomes. Mol Biol Evol. 2015;32:1456–1460. doi: 10.1093/molbev/msv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bader DM, et al. Negative feedback buffers effects of regulatory variants. Mol Syst Biol. 2015;11:785. doi: 10.15252/msb.20145844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefke B, Sun W, Li YS, Fang L, Chen W. The evolution of posttranscriptional regulation. Wiley Interdiscip Rev RNA. 2018:e1485. doi: 10.1002/wrna.1485. [DOI] [PubMed] [Google Scholar]
References
- 36.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 39.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tress ML, Abascal F, Valencia A. Alternative Splicing May Not Be the Key to Proteome Complexity. Trends Biochem Sci. 2017;42:98–110. doi: 10.1016/j.tibs.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez-Porta M, Frankish A, Rung J, Harrow J, Brazma A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14:R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 48.Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 49.Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 50.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci U S A. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the false discivery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 55.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shihab HA, Rogers MF, Campbell C, Gaunt TR. HIPred: an integrative approach to predicting haploinsufficient genes. Bioinformatics. 2017;33:1751–1757. doi: 10.1093/bioinformatics/btx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao Y, et al. GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Res. 2019;29:682–696. doi: 10.1101/gr.238733.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed Ribo-seq and RNA-seq data are available from ArrayExpress (accession number: E-MTAB-7247, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-7247/). All other data are available as Supplementary information or are available upon request. We also created a publicly available data resource (Ex2plorer: https://ex2plorer.kaessmannlab.org/), which allows the interactive exploration of the evolution at both expression layers for all 1:1 orthologous genes.
Custom R scripts used to generate the results reported in the manuscript and processed data are available at https://github.com/evgenyleushkin/translatome.