Abstract
Brain transcriptome and connectome maps are being generated, but an equivalent effort on the proteome is currently lacking. We performed high-resolution mass spectrometry-based proteomics for in-depth analysis of the mouse brain and its major brain regions and cell types. Comparisons of the 12,934 identified proteins in oligodendrocytes, astrocytes, microglia and cortical neurons with deep sequencing data of the transcriptome indicated deep coverage of the proteome. Cell type-specific proteins defined as tenfold more abundant than average expression represented about a tenth of the proteome, with an over representation of cell surface proteins. To demonstrate the utility of our resource, we focused on this class of proteins and identified Lsamp, an adhesion molecule of the IgLON family, as a negative regulator of myelination. Our findings provide a framework for a system-level understanding of cell-type diversity in the CNS and serves as a rich resource for analyses of brain development and function.
The complex structural and functional organization of the mammalian CNS with its enormous diversity of cell types with different morphology, connectivity and function warrants the application of global and systematic approaches. Several large-scale, collaborative initiatives have been launched that aim to establish functional connectomes and develop large-scale simulations of the brain. In addition, the Allen and the GENSAT Brain Atlas projects have taken global genetic approaches by generating genome-scale collections of gene expression profiles using in situ hybridization and BAC-EGFP transgenic mouse lines to resolve the expression pattern of genes during development and in the adult mouse brain1,2. Furthermore, gene profiling and transcriptome analyses of specific CNS cell populations have generated large collections of mRNA expression patterns across a variety of brain regions and cell types3–10. Given the enormous amount of genome and transcriptome data that has been generated, the question arises of whether these extensive databases may already represent a sufficient measure of gene expression. Several studies have reported that quantities of messenger RNA and proteins often correlate poorly, emphasizing the importance of post-transcriptional processes that control protein synthesis and degradation11. Thus, protein expression analyses are required, but a concerted effort to resolve the brain proteome is missing12. Until recently, mass spectrometry-based proteomics was limited to incomplete proteome analysis, but this situation has changed with the development of more sensitive and powerful high-resolution mass spectrometry (MS) technologies that allow in-depth coverage of nearly complete proteomes of mammalian cells 13–17.
Proteins are the main functional components in all cells, but how the proteomes differ in specialized cells in tissue is not known. Previous studies of different human cancer cell lines have found that proteins tend to be ubiquitously expressed with differences in the level of expression of the proteins rather than in their absence or presence18–20. Given the marked cell specialization that occurs in the CNS, we asked whether these general conclusions also apply to the brain by analyzing the major glial subtypes and neurons. Glia are not only abundant (constituting 50–90% of the cells in the human brain), but are also highly specialized toward cell type-selective tasks, and therefore ideally suited for these analyses21. Oligodendrocytes are primarily engaged in membrane synthesis, as their function is to insulate axons by enwrapping them with a multilamellar myelin sheath22,23.Astrocytes, in contrast, provide nutrients to the nervous tissue, maintain the extracellular ion balance, recycle neurotransmitters, shape synaptic circuits and regulate the blood-brain barrier24. Microglia cells constitute the third class of glial cells with functions mainly related to immune response and maintaining brain homeostasis25.
To resolve the mouse brain proteome and to determine the basis for cellular specialization in the CNS, we performed a global analysis of protein levels by liquid chromatography coupled to high-resolution MS of the adult mouse brain with its major brain regions and cell types (cultured and acutely isolated neuronal and glial cells).We applied deep sequencing-based transcriptome analysis for an integrative transcripts and protein expression analysis. The combined in-depth database of CNS cells represents a new resource to the neuroscience community for better understanding brain development and function. We illustrate its power by identifying previously unknown adhesion molecules in oligodendrocyte and neuron interaction.
Results
Adult mouse brain proteome
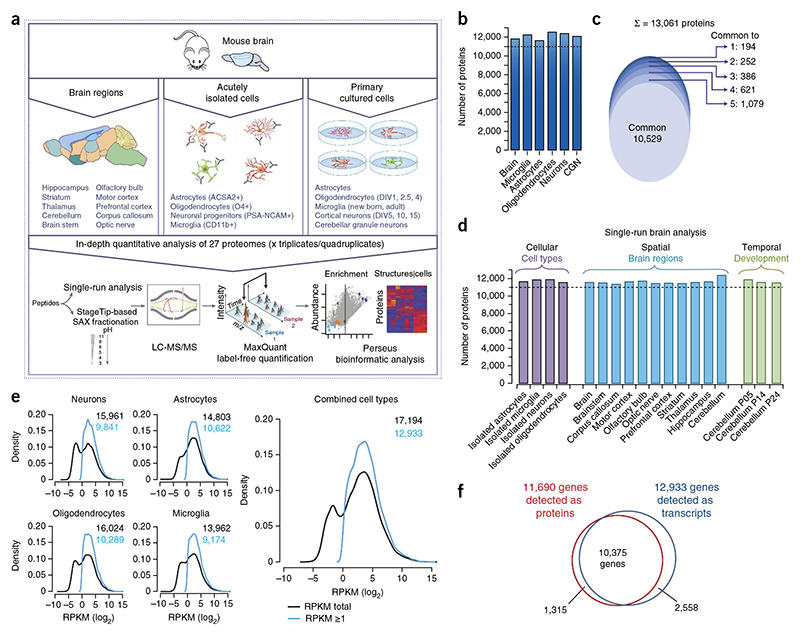
To resolve the adult mouse brain proteome, we analyzed four replicates of 9-week-old whole mouse brains after tryptic digestion of lysates and fractionation of resulting peptides into six fractions using strong anion exchange chromatography. We performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis with 4-h runs per fraction and higher energy collisional dissociation (HCD) in a quadrupole Orbitrap mass spectrometer equipped with a high-field analyzer (Online Methods). Combined analysis of the replicates using MaxQuant software26 and applying a peptide and protein false discovery rate (FDR) of 1% identified 13,214 protein groups (Fig.1a,b and Supplementary Table 1). In addition, we analyzed lysates from mouse brain in biological quadruplicates with our ‘single-shot analysis‘ approach27 using 4-h gradients. We detected more than 11,500 different proteins when we matched them against the ‘library’ of the deep brain proteome (Fig. 1c,d). To resolve the proteome of the major brain regions, we used this approach to analyze adult mouse cerebellum, hippocampus, thalamus, striatum, brain-stem, olfactory bulb, motor cortex, prefrontal cortex, corpus callosum and the optic nerve in 3–4 biological replicates. In addition, we investigated the changes in the proteome during postnatal development by determining cerebellar tissue at three different time points (∼postnatal day (P) 5, 14 and 24) (Fig. 1a,c and Supplementary Table 2). Notably, in each of the single-shot analyses, we identified more than 11,500 proteins. These results illustrate the power of proteomics technology for in-depth profiling of complex tissue in a rapid, reproducible and a quantitative fashion.
Figure 1. Comparison of proteome and RNA-Seq data.
(a) Graphical illustration of the workflow for the cell type– and brain structure-resolved mouse brain proteome. DIV, days in vitro. (b) A bar chart showing number of proteins identified in adult mouse brain and each of the cultured cell type with FDR of 1% when analyzed as 6x SAX fractions. CGN, cerebellar granule neurons. (c) A total of 10,529 proteins were identified in all cell types and brain (∼84% of all identified proteins) and an average of ∼99% protein identifications were shared between at least two proteomes. (d) Single run analysis of the mouse brain, the different brain structures and the developing cerebellum; four biological replicates (triplicates for optic nerve and corpus callosum) were measured by single 4-h LC MS/MS runs.Numbers of identified proteins are indicated after matching between runs with fractionated brain and cell-type proteome runs in the MaxQuant environment. (e) RNA-Seq analysis of cultured cells and resulting density plots of gene expression levels. The density estimates of gene expression levels are shown for each cell type and for a combination over all cell types as indicated. The combined cell type was derived by extracting the largest expression value over all cell types. In all cell types, gene expression levels followed a binomial distribution (black). Geneexpression filtered for RPKM > 1 values are shown in blue. (f) Venn diagram of the number of expressed genes on the mRNA level and on the protein level.
CNS cell-type proteome depth and quantification
To resolve cell type-specific protein expression in CNS, we wished to carry out in-depth proteomic analysis of the neurons and glia. In general, isolation of single cell types can be performed by laser microdissection or fluorescence-activated cell sorting of labeled cells in a tissue, but given that CNS-derived cells are highly branched and interconnected, these isolations lead to the loss of the most distant and connected cellular structures. Primary cell culture allows the isolation of entire cells, but may affect the expression pattern of some of the proteins. Thus,we performed an in-depth analysis of both acutely isolated and primary cultured cells to obtain a complete proteome inventory of major CNS cell types. We started with primary cell culture and confirmed the intact morphology and purity of cultured oligodendrocytes, astrocytes, microglia, cortical and cerebellar granule neurons by antibody labeling using established markers (GFAP,P-III tubulin, Ibal, O1 and MBP; Supplementary Fig. 1). Three independent preparations with a purity >95% were used for the analysis. Lysates were prepared from neurons and oligodendrocytes at different stages of development (1, 2.5 and 4 d in vitro for oligodendrocytes, and 5, 10 and 15 d in vitro for cortical neurons). We used microglia from newborn and adult mice for comparison. Combined analysis of the replicates of all cell preparations, mouse brain and major brain structures identified 294,037 sequence unique and fully tryptic peptides, which assembled into ∼13,200 protein groups (Supplementary Table 1 and protein groups table uploaded to the PRIDE repository). This corresponds to an average of 11,970 different proteins in each of these systems, of which 10,529 proteins were identified in all five cell types and brain (Fig. 1b,c).
To compare the proteome with the transcriptome and to judge the comprehensiveness of our results, we carried out RNA sequencing (RNA-Seq) of the cultured oligodendrocytes, astrocytes, microglia and cortical neurons. RNA sequencing (RNA-Seq) using the Illumina platform resulted in 19,501 unique gene symbols that were expressed in at least one cell type (Fig. 1e and Supplementary Table 3). As observed previously, the abundance of the non-filtered data, expressed as reads per kilobase of exon per million fragments mapped (RPKM), revealed a bimodal distribution (Fig. 1e), where one third of the transcripts have abundances lower than 1 RPKM15,28. These low abundance transcripts appear to be the result of rare stochastic transcription and are thought to often result in non-functional mRNAs28. There was evidence of protein translation in only about a quarter of the transcripts with RPKM values between 1 and 0.1 (Supplementary Fig. 2). When we excluded these transcripts from our analysis, a standard procedure in RNA-Seq analysis28, the number of genes was reduced to 12,933 (Fig. 1f). The 12,934 proteins identified in different CNS cell types correspond to 11,690 ENSEMBL genes, which represents ∽60% of all protein coding genes in the genome. When systematically analyzing the transcripts for which no protein was detected, the main categories that were enriched in Gene Ontology GO)-based analysis were G protein-coupled receptors (GPCRs), including olfactory receptors, which are not expected to be functionally releant in glia or cortical neurons (Supplementary Table 4). Integral membrane proteins in general were not enriched in the transcripts for whsich no protein was detected (Supplementary Table 4). Taken together with previous estimates of the size of cellular proteomes14,15,29, our data indicate that we have achieved a great depth of coverage of the proteome of glia and neurons.
Assessment of proteome quality and comparison to the transcriptome
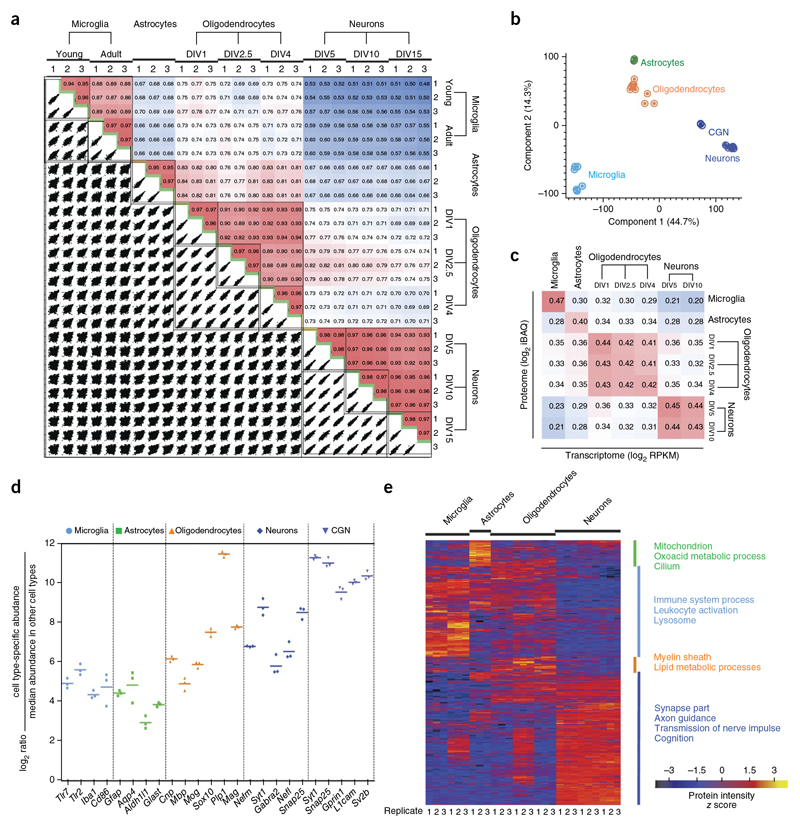
To further determine the quality of our proteome analysis, we quantitatively compared protein abundances across the different cell types. For this analysis, we analyzed a total of 162 raw files (nine cell preparations x six fractions x three replicates) with MaxQuant using its label-free quantification algorithm21,25, in which the MS signals detected from the same peptides in the different cell types are quantified relative to each other Supplementary Table 5). Between biological replicates of the same cell types, Pearson’s correlation coefficients were about 0.96, demonstrating excellent technical and biological reproducibility (Fig. 2a). We also observed high correlations of the biological replicates for the RNA-Seq data (Pearsons correlation of ∼0.98; Supplementary Fig. 3). Between different cell types, correlations were much lower, from 0.48 to 0.84 for the proteome (Fig. 2a) and from 0.25 to 0.86 for the transcriptome (Supplementary Fig. 3). Consistent with their hematopoietic origin, microglia were the most diverging cell type; and, as expected from their shared lineage, astrocytes and oligodendrocytes were closest to each other (Fig. 2b). As the correlation between the proteomes and the corresponding transcriptomes was only 0.4 to 0.45,our analysis captured important differences between transcriptome and proteome30,31 (Fig. 2c).
Figure 2. Comparative analysis of cell proteomes.
(a) The matrix of 162 correlation plots revealed very high correlations between LFQ intensities in triplicates (Pearson correlation coefficient 0.94–0.98 between cell types). The color code follows the indicated values of correlation coefficient. (b) PCE. The proteome of all cell types and their differentiation states measured in triplicates segregated into major cell types based on component 1 and component 2, which account for 44.7% and 14.3% of variability, respectively. (c) Correlation plots of iBAQ intensities (proteome) versus RPKM values (transcriptome).The color follows the indicated values of correlation coefficient. (d) Fold expression of the indicated marker proteins in individual replicates is shown on a log2 scale as points with mean in the specified cell type in comparison with other cell types. (e) Heat map of proteins differentially expressed across different cell types (n = 3 for each cell type). The top categories enriched for clusters are shown. Heat map of z-scored LFQ intensities of the significantly differentially expressed proteins after unsupervised hierarchical clustering. Proteins are divided into four clusters showing the top categorical annotations enriched after a Fisher’s exact test (P ═0.02).
The expression levels of well-described markers for astrocytes (Aquaporin 4, Gfap, Glast and Aldh1l1), oligodendrocytes (Cnp1, Sox10, Mbp, Mag, Mog, Plp1), cortical neurons (neurofilament, Snap25, synaptotagmin I, Gabra 2), cerebellar granule neurons (Synaptotagmin I, Snap25, Sv2b, Gprin1, L1cam) and microglia (Iba1, Tlr2, Tlr7, CD86) in our triplicate proteome analysis revealed that almost all of these markers were reproducibly enriched at least 16-fold, thereby confirming the quality of the proteome analysis (Fig. 2d). In addition, our analysis revealed several previously unknown cell type-specific proteins (see below; Supplementary Tables 6 and 7).
Clustering and cluster-specific enrichment analysis using Gene Ontology Biological Processes (GOBP), Gene Ontology Cellular Components (GOCC), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Comprehensive Resource of Mammalian Protein Complexes (CORUM) clearly recalled the knownbiological processes, components and pathways of each cell type from unbiased analysis (Fig. 2e and Supplementary Tables 8 and 9). In an orthogonal approach, we assessed the enrichment of biological pathways in a cell type-specific manner instead of across a selected cluster by relating proteome expression levels to any protein annotation category32. Close to 1,000 annotation terms, including about 60 KEGG pathways, were significantly enriched in at least one cell type proteome (annotation matrix resolved to individual replicate; Supplementary Table 10).
Pathways and biological functions enriched in oligodendrocytes include the known highly specific processes such as myelination, ensheathment of axons, lipid metabolism and membrane-associated processes. In the oligodendrocyte precursor cells population, we observed a strong enrichment in RNA related to metabolic processes, which would be expected for proliferating cells. We also found accumulation of components of the myelin sheath during oligodendrocyte differentiation. The pathways enriched in microglia were mostly those involving regulation of immune processes, antigen processing and presentation, defense response, or leukocyte activation. For cortical and cerebellar granule neurons, we found the expected pathways involving the synapse and pathways, including dendrite development and morphogenesis, axonogenesis, calcium-mediated signaling, and post-and presynaptic membrane potential processes. Among the cellular components, a range of neuron-specific components such as axon terminus, axons, synaptosome, and neuronal projection were enriched. Processes related to cGMP metabolism and semaphoring receptor activity were more abundant in cortical neurons as compared with CGN Supplementary Fig. 4).Pathways enriched in astrocytes were mainly metabolic in nature, including cellular ketone, organic acid, carboxylic, oxidation-reduction processes, amino acid amine and small molecule catabolic processes. Notably, cilium assembly was prominently enriched in cultured astrocytes (Supplementary Table 8).
In the analysis of the developing cerebellum (P5, P14, P24) we observed that pathways related to synaptogenesis, myelination and axonal growth increased at P14 and P24(Supplementary Fig. 5). Together these analyses of our deep primary cell proteomes faithfully reproduce known and characteristic cellular functions as well as their markers, providing a solid foundation for their global characterization and for new discoveries.
Definition of global differences between CNS cell proteomes
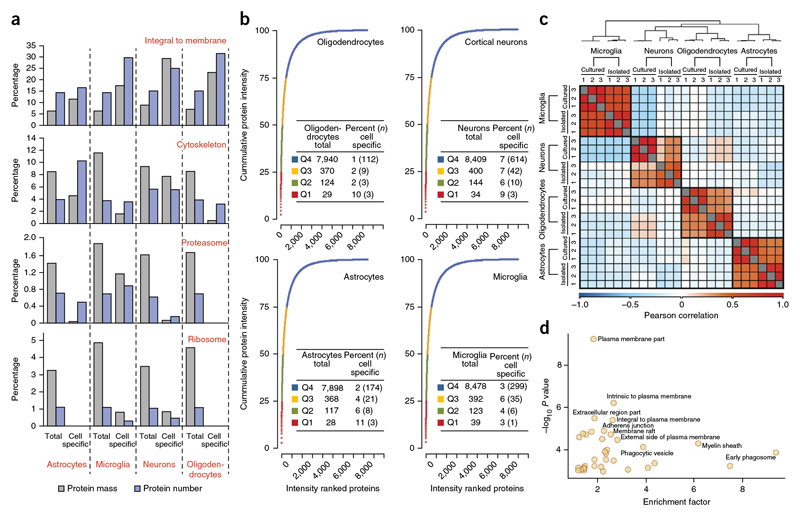
The large majority of proteins were expressed in all four cell types, whereas only about 15% of the quantified proteins could be considered to be cell type specific (>10-fold more abundant in one cell compared with all others; Supplementary Table 6). GO-based enrichment analysis revealed that cell type-specific proteins are highly enriched for cell surface and extracellular proteins (Supplementary Table 11). A similar analysis for the RNA-Seq data set revealed that cell type-specific transcripts were also significantly enriched for functionsrelated to the cell surface and extracellular space (Supplementary Table 12). The MS signal of peptides identifying each protein can also be used to estimate its absolute abundance using the iBAQ algorithm, which normalizes the summed peptide intensities by the number of theoretically observable peptides of the protein. These iBAQ values yield protein copy number estimates of each protein in the proteome33. They determine the proportional contribution of any protein to the total proteome, as well as of every pathway or GO annotation. We performed a quantitative comparison of the proteins identified as similarly or differentially expressed, which revealed marked differences for proteins related to the cell surface or being integral to the plasma membrane (Fig. 3a).Whereas integral membrane proteins contributed to only ∼6–8% of the protein mass of the total proteome, they accounted for ∼12–29% of the protein mass of the cell type-specific proteins (Fig. 3a).
Figure 3. Quantitative analysis of expressed genes.
(a) Analysis of specific GO annotation terms (indicated in red above the bar graph) is shown as the percent of the genes corresponding to the annotation term and the percent of the protein mass that was attributed to these annotations.The analysis was performed separately for all proteins identified in indicated cell type or for those proteins that were specific to the indicated cell type. (b) Cumulative protein mass from the highest to the lowest abundance proteins for the indicated cell type. The table liststotal number of proteins constituting different quantiles (Q1-Q4) and the percent of these proteins that showed cell type-specific expression. (c,d) Comparison of acutely isolated cells with cultured cells. (c) Cells were isolated using MACS microbeads coupled with antibodies to 04 for oligodendrocytes, PSA-NCAM for neuronal progenitors, CD11b for microglia and ACSA-2 for astrocytes. The heat map shows the Pearson correlation coefficientsbetween acutely isolated and cultured cell types. The color code indicates the values of the correlation coefficients. (d) Plot of GOCC enrichment of proteins > 10-fold enriched is shown. –log10 P value is plotted against enrichment factor of the GOCC terms.
For the analysis of the cell type-specific proteins in the total proteome, we ranked the proteins according to their abundance in the proteome. As previously observed in ultra-deep proteome analysis, a relatively small number of proteins make up for a high proportion of the total protein mass15,34. In fact, in all four cell types, ∼ 30–40 of the most abundant proteins accounted for ∼25% of the total protein molecules, and some of these were cell type specific (for oligodendrocytes Mbp, Plp, Cnp1; for cortical neurons Atp1b1, Crmp1, Gpm6a; for astrocytes Gfap, Gstm1, Prdx6; and for microglia Gm5483; Fig. 3b).
Next, we compared the differentially expressed proteins in each cell type individually.The top 50 cell type-specific proteins (the most enriched and abundant proteins) included many well-established cell type-specific markers, but, based on PubMed search, also a relatively large number of proteins that have previously not been studied in the context of the respective cell type (more than one-third of the 50 proteins in the various glial types). In oligodendrocytes, several of these proteins had functions related to the extracelluar matrix (Col11a1, Col1a1, Leprl1, Bgn, Plod2), the cytoskeleton (Afap1l2, Tagln, Kank1, Rtkn, Tns3) and vesicular trafficking (Dnm3, Rtkn). The cell type-specific proteins in microglia had functions in membrane trafficking (Rab32, Myo1f), lipid catabolism (Apobr, Lipa) and signaling at the cell surface (CD180, Dock2, Nckap1l,Gpnmb, Fermt3), whereas proteins with designated functions in actin-related processes (Cnn1, Fmn2, Nebl, Pdlim7, Synpo2) and cilia (Rsph1, Ccdc39) were enriched in astrocytes.
We found that oligodendrocytes expressed the smallest number (341 proteins) of cell type-specific proteins, defined by the greater than tenfold fold enrichment criterion, and cortical neurons the largest (669 proteins). Similar results were obtained with at least fourfold (and with statistical significance) difference between the different cells. This selection resulted in 411 oligodendrocyte-enriched and 1,388 neuron-enriched gene products (Supplementary Table 6). Transcription factors (for example, Sox8, Sox10, Myt1, Myrf, Olig2, Nkx2.2 in oligodendrocytes; Tbr1, Bcl11b, Myt1l, Emx1, Bhlhe22, Arx in neurons; Sfpi1, Irf5, Irf8, Runx1, Ikzf1 in microglia; Sox9 and Pax6 in astrocytes) were also among these cell type-specific proteins.
Proteome analysis of acutely isolated CNS cell types
Given that the results obtained thus far have been based on ex vivo cultured cells, we asked how they compare with data derived from acutely isolated cells. We used antibody-coupled microbeads for magnetic affinity cell sorting to isolate CD11b+ microglia, O4+ oligodendrocytes, ACSA-2+ astrocytes and PSA-NCAM+ neuronal progenitors from P8 mice. We achieved highly purified populations of cells with mean purity in excess of 95% in all three biological replicates (Supplementary Fig. 1), in each of which we identified and quantified more than 11,500 proteins (Fig. 1d and Supplementary Tables 2 and 13). The expression levels of cell type-specific markers for astrocytes (Aqp4, Gfap, Aldh1l1), oligodendrocytes (Cnp1, Mbp, Plp1), microglia (Iba1, Tlr2, Tlr7) and neuronal progenitors (Bcl11a, Bcl11b, Ebf3) showed that almost all of these markers were reproducibly enriched at least 16-fold, also confirming the quality of the proteome analysis (Supplementary Fig. 6a). We observed high correlation in the fold enrichments of proteins when comparing the related cell types in culture and acutely isolated (Fig. 3c). The significantly enriched annotation terms revealed very similar functional proteomics portraits in the corresponding cell types (Supplementary Table 14). Given that microglia and astrocytes are thought to undergo the largest changes in cell culture, we compared significantly enriched annotation terms between cultured and acutely isolated preparations of these cells (Supplementary Fig. 6b,c). The most substantial increase was observed for pathways related to antigen-presentation and extracellular matrix in primary cultures of microglia and astrocytes, respectively (Supplementary Fig. 6b,c). To extract globally unique proteins from the proteome data, we determined the number of cell type-specific proteins (>10-fold more abundant in one cell type compared to all others). As in cultured cells, their total number was relatively small 768 proteins). Again, as observed in cultured CNS cell types, our GO-based enrichment analysis of cell type-specific proteins showed a high enrichment for cell surface proteins (Fig. 3d).
The most abundant and enriched proteins inthe brain proteome
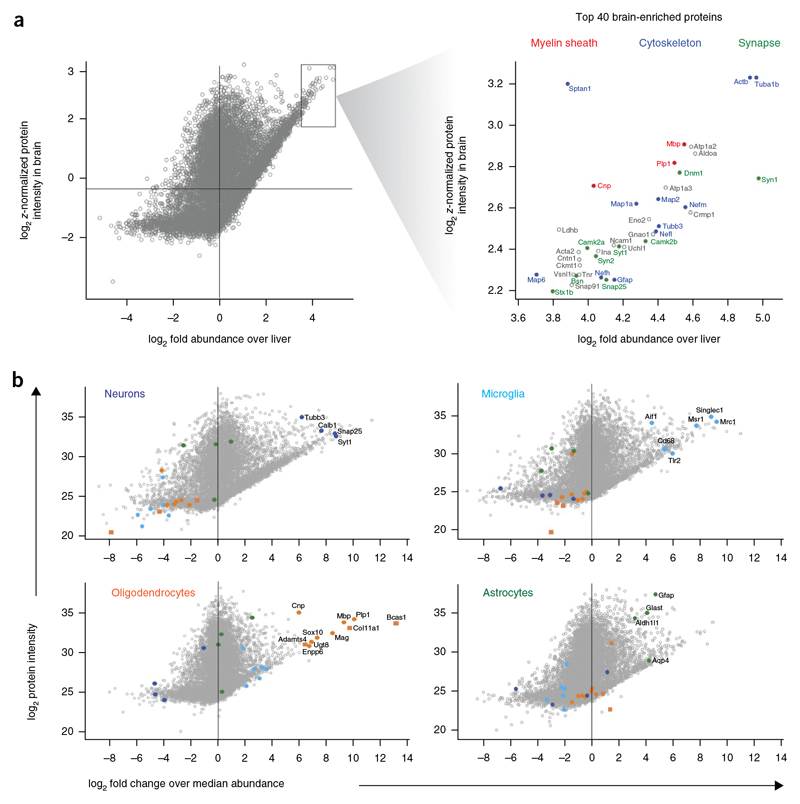
As our analysis demonstrated that the different CNS cell types had a large fraction of the proteome in common, but differed in their expression of cell surface proteins, we asked whether this conclusion could be extended to the entirebrain proteome when compared with an unrelated tissue proteome. To this end, we performed a quantitative comparison of the adult mouse brain (Fig.4 and Supplementary Table 2) and liver proteomes and provide a list of the 147 most abundant brain-enriched (>10-fold enrichment as compared to the liver) proteins (Supplementary Table 15). We plotted the proteins of the adult mouse brain proteome against abundance and enrichment to highlight the top proteins of this category (Fig. 4a).Among the top 40 most abundant and enriched ones were proteins of the myelin sheath (Mbp, Plp1 and Cnp), the cytoskeleton (Tuba1b, Actb, Sptan1, Map2, Map1a, Tubb3, Nefl, Map6, Nefh, Gfap), synapses (Dnm1, Syn1, Camk2b, Syt1, Camk2a, Syn2, Bsn, Snap25, Stx1b) and glycolysis, as well as energy pathways (Aldoa, Eno2, Ldhb). Similar to the comparison between the different cell types, GO-based enrichment analysis revealed that cell surface proteins were highly overrepresented in the group of the most abundant brain-versus liver-enriched proteins and four out of the top five GOCC terms were related to membranes (Supplementary Table 16).
Figure 4. Abundant and enriched proteins in the mouse brain and its cell types.
(a) Scatter plot of log2 fold expression versus log2 LFQ intensity in the adult mouse brain in comparison to the mouse liver proteome. Among the top 40 most abundant and enriched proteins of the adult mouse brain are proteins of the myelin sheath (red), the cytoskeleton (blue) and synapses (green). (b) Scatter plot of log2 fold expression versus log2 LFQ intensity in the indicated cell type in comparison with other cell types with highlighting of known and previously unknown cell type-specific markers.
Analysis of protein expression in brain regions
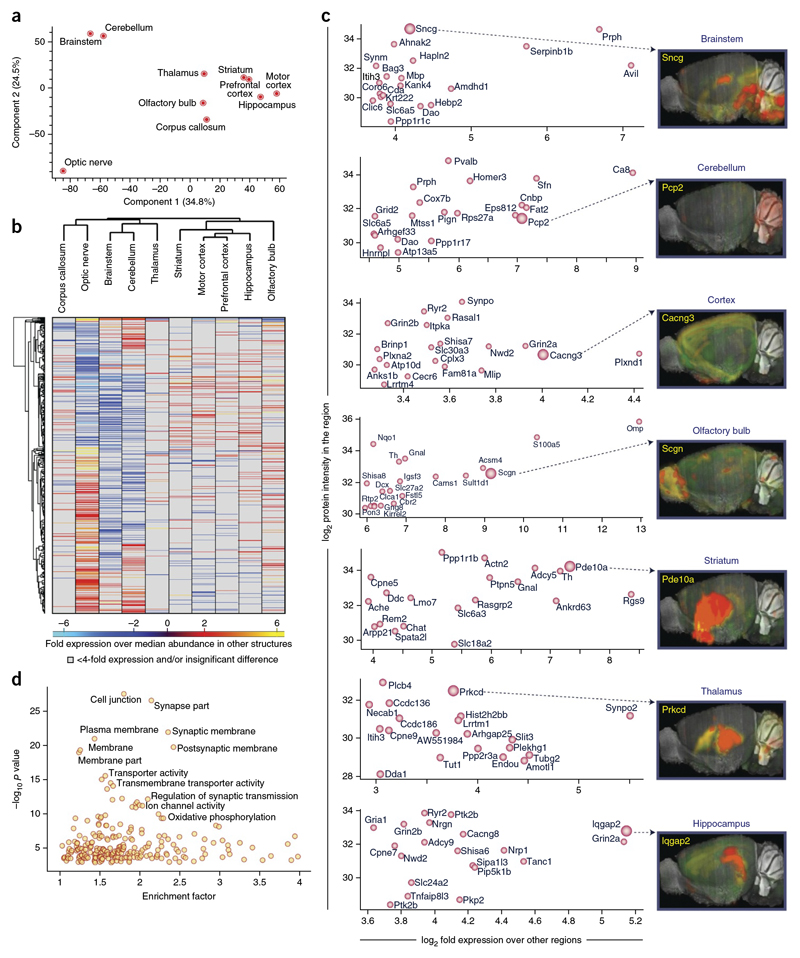
To directly determine the quantitative landscape of the brain proteome and to identify the most specific proteins for each of the major brain regions, we undertook an in-depth analysis of ten major brain regions. We performed a quantitative comparison of the expression of proteins in the cerebellum, hippocampus, thalamus, striatum, brainstem, olfactory bulb, motor corex, prefrontal cortex, corpus callosum and the optic nerve. As judged by principal component analysis (PCA) and hierarchical clustering, the optic nerve, the cerebellum and the brain stem were the most diverging brain regions, whereas the remaining seven regions clustered more tightly (Fig. 5a,b). In total, we identified 2,901 brain region-enriched proteins (>4-fold difference) and found that the thalamus expressed the smallest and the optic nerve the largest numbers of brain region-enriched proteins (154 and 1,179, respectively; Supplementary Tables 17 and 18). We plotted the abundance versus the enrichment of proteins in the different brain regions to highlight the top 20 most abundant, yet brain region-enriched, proteins. Prph, Sncg and Serpinb1b were most abundant and enriched in the brain stem, Omp and S100a5 in the olfactory bulb, Ppp1r1b, Actn2 and Pde10a in the striatum, and Plcb4, Prkcd and Synpo2 in the thalamus (Fig. 5c). These proteomic findings can be compared and contrasted with in situ data from the Allen Brain Atlas1, 2, which allows one to immediately compare and contrast gene expression at the transcriptome and proteome levels (Fig 5c). GO enrichment analysis of all the enriched proteins together again highlighted the cell surface as one of the most significant components (P < 0.02; Fig. 5d).
Figure 5. Brain region-resolved proteome.
(a) PCA. The proteomes of major mouse brain regions (P60) were measured in quadruplicates (triplicates for optic nerve and corpus callosum) and segregated based on component 1 and component 2, which accounted for 34.8% and 24.5% of variability, respectively. (b) Heat map of proteins differentially expressed across the different brain regions. The heat map is based on the z-scored LFQ intensities of the significantly differentially expressed proteins after unsupervised hierarchical clustering. Proteins with more than fourfold expression differences are shown. (c) Scatter plot of log2 fold expression versus log2 LFQ intensity of the top 20 proteins in the indicated brain region in comparison with other brain region. The larger red circles indicate proteins chosen for the comparison with the corresponding transcripts analyzed by in situ hybridization in the Allen Brain Atlas project. Images are taken from the Allen Brain Atlas (http://mouse.brain-map.org). Image credit: Allen Institute for Brain Science. (d) Plot of GOCC enrichment of proteins > 10-fold enriched is shown. -log10 P valueis plotted against enrichment factor of the GOCC terms.
Identification of Lsamp as a negative regulator of myelination
Having demonstrated that the different cell types can be distinguished by a relatively small number of proteins associated with the cell surface and extracellular matrix, we asked whether we can identify previously unknown cell type-specific markers. We reasoned that the most enriched and abundant proteins should be preferred candidates for such an analysis and plotted them against abundance and enrichment for each cell type (Fig. 4b).For the three pooled oligodendrocyte stages, we found three thus far unstudied proteins just among the top ten proteins. We overlaid cell type-specific proteins for oligodendrocytes with reported protein-protein interaction network based on physical interaction and known functions (Supplementary Fig. 7). These analyses identified proteins of the extracellular matrix (Col11A1, Col1A1, Col1A2, Col4A1, Col3A1, Col5A2), of which Col11A1 was the most highly expressed and enriched protein. It constitutes one of the two alpha chains of type XI collagen and belongs to the fibrillar class of collagens. Mutations in the gene are associated with a type of Stickler syndrome, a connective tissue disorder characterized by ocular, skeletal, orofacial and auditory defects35. Using western blotting of purified CNS cell types, we confirmed that Col11A1 is enriched in oligodendrocytes at an early stage of development (Supplementary Fig. 8). Immunofluorescence staining of cultured cells showed that Col11A1 can be used as a new marker for immature oligodendrocytes and immunohistochemistry of brain sections revealed a weak association of Col11A1 with white matter tracts (Supplementary Fig. 8).
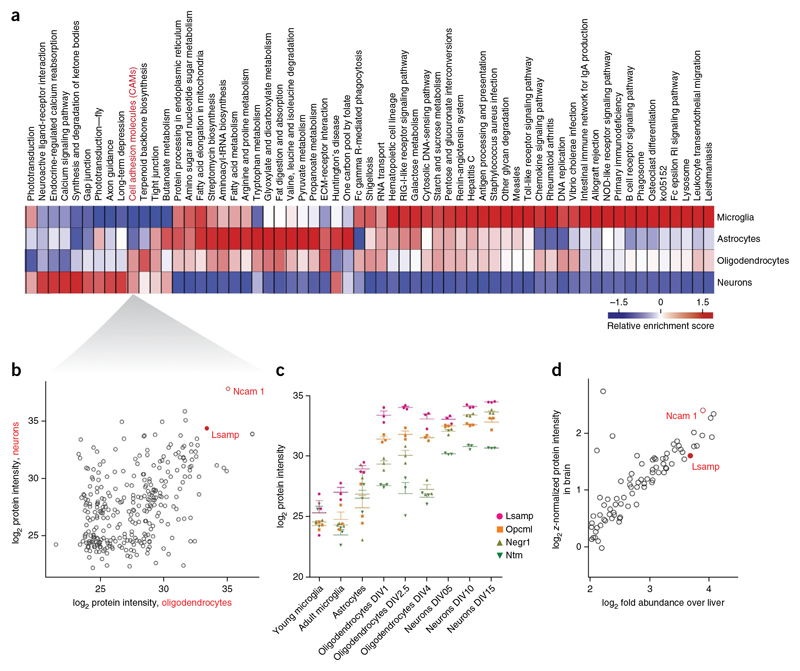
To further illustrate the power of the resource, we investigated whether we could discover proteins that are functionally shared between two cell types. As there is a high degree of functional coupling between neurons and glia, we performed comparative pathway enrichment analysis to detect KEGG pathways and gene ontology terms that were common to both. Cell adhesion was among the KEGG pathways that were shared between neurons and oligodendrocytes (Fig. 6a). As little is known about the adhesion molecules that mediate neuron and oligodendrocyte interaction, we asked whether our proteome analysis was able to identify new candidate molecules. We focused on abundantly and differentially expressed adhesion molecules and plotted the 282 proteins of this category. The most abundant and enriched adhesion molecule shared between oligodendrocytes and neurons was Ncam1 (Fig. 6b),a protein that negatively regulates myelination36. The limbic system-associated membrane protein (Lsamp) was the second-most enriched adhesion molecule in neurons and oligodendrocytes. Lsamp belongs to the IgLON family of adhesion molecules, which also comprises Ntm (neurotrimin), Opcml (opiod-binding cell adhesion molecule) and Negr1 (neuronal growth regulator1)37–39.Notably, most family members were enriched in neurons and oligodendrocytes (except of Ntm, which was only enriched in neurons; Fig. 6c). All four IgLONs are glycosylphosphati-dylinositol (GPI)-anchored glycoproteins with three Ig domains that have been implicated in cell adhesion and growth guidance in neurons, but not in glia. By plotting the most abundant and enriched (as compared with the liver) adhesion molecules of the adult mouse brain, we identified Lsamp among the top proteins (Fig. 6d), further supporting the importance of this still poorly described molecule in brain function.
Figure 6. Comparative pathway enrichment analysis identifies cell adhesion molecules enriched In oligodendrocytes and neurons.
(a) Annotation matrix of KEGG pathways enriched in different cell types shown as a heat map (red indicating KEGG pathways higher abundance and blue indicating lower abundance) after clustering of score differences from one-dimensional annotation testing (Online Methods). (b) Scatter plot for LFQ intensities of proteins corresponding to KEGG pathway cell adhesion molecules (CAM) in oligondendrocytes versus neurons. (c) Label-free quantification of individual triplicates is shown as points with mean ± s.e.m. for the IgLON family proteins in the different CNS cells types. (d) Scatter plot of log2 fold expression versus log2 z-normalized protein intensity of proteins corresponding to KEGG pathway cell adhesion molecules in brain versus liver.
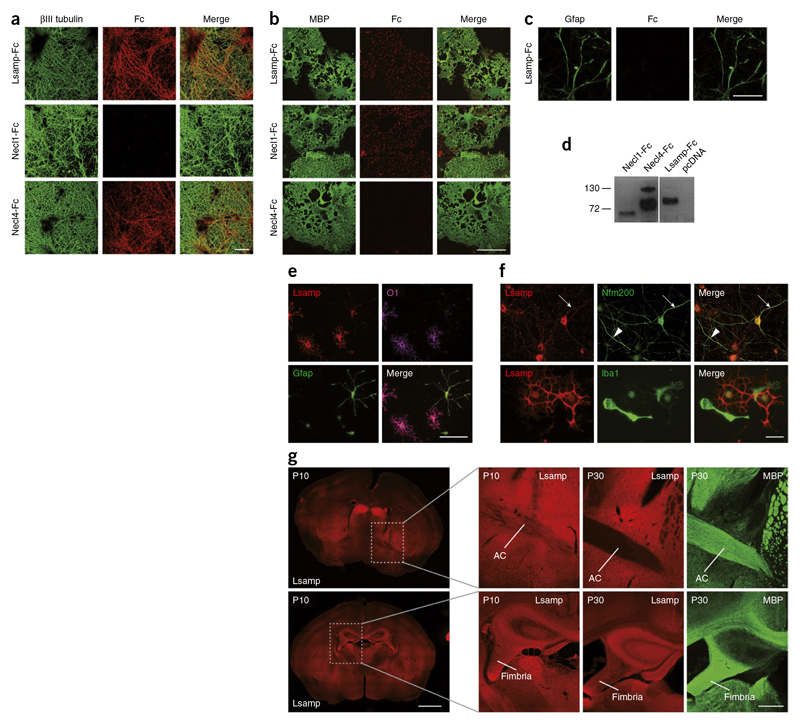
Given that proteins of the IgLON family promote cell-to-cell adhesion by homo- and heterophilic interactions40,41, they represent ideal candidates for axon and oligodendrocyte interaction. We generated a soluble version of the proteins by cloning the predicted extracellular domain and tagging it with a human Fc fragment for detection and used the soluble and purified extracellular domains of the proteins in binding studies (Fig.7). The assay was performed by adding the soluble extracellular part of proteins to either neuronal or oligodendrocyte cultures. As positive controls, we used Necl1-Fc and Necl4-Fc42,43, which, as expected, bound to oligodendrocytes and axons, respectively. All of the members of the IgLON family that we tested (Lsamp-Fc, Neurotrimin-Fc, Opcml-Fc) bound to both neurons and oligodendrocytes (Fig. 7a,b ,d and Supplementary Fig. 9). The interaction was specific, as none of the proteins interacted with astrocytes or HEK293T cells (Fig. 7c and Supplementary Fig.10).
Figure 7. Lsamp interacts with and is expressed on oligodendrocytes and neurons.
(a) Binding of Lsamp-Fc to neurons labeled with an antibody against βMI tubulin. Necl4-Fc served as a positive and Necl 1-Fc as a negative control.The Fc fragments were visualized with Cy3-conjugated anti-Fc antibodies. (b) Binding of Lsamp-Fc to oligodendrocytes labeled with an antibody against MBP. Necl 1-Fc served as a positive and Necl4-Fc as a negative control. (c) Lsamp-Fc did not bind to astrocytes labeled with an antibody against Gfap. (d) Immunoblot of secreted Fc fusion proteins containing the extracellular domains of the indicated proteins. (e) Immunofluorescence of mixed glial cultures shows staining of oligodendrocytes (O1; magenta), but not astrocytes (Gfap; green) with an antibody against Lsamp (red). (f) Top, Lsamp was present on a subpopulation of neurons (neurofilament 200 kDa) (top right). Arrow indicates toward an Lsamp-positive neuronal process, arrowhead indicates an Lsamp-negative process. Bottom, Lsamp was absent from microglia (Iba1), as shown by staining of a mixed glial culture. (g) Brain sections of wild-type mice were immunostained with antibodies against Lsamp (red) at P10, and Lsamp and MBP (green) at P30. Lsamp staining was enriched along the axonal tracts of the fimbria and anterior commissure (AC) at P10, but not at P30. Scale bars represent 10 μm.
Next, we transfected plasmids encoding for full-length Lsamp, Neurotrimin and Opcml tagged with EGFP into HEK293T cells, added soluble Fc fusion proteins and observed homo- and heterophilic interactions as noted previously41 (Supplementary Fig. 10). We continued our analysis by characterizing the function of Lsamp, the most abundant and enriched IgLON family member in neurons and oligodendrocytes. Using antibody staining, we confirmed the expression of Lsamp in oligodendrocytes and neurons, but not in astrocytes or microglia (Fig. 7e,f). In primary cortical neuronal culture, we found Lsamp in the cell body and processes of a subpopulation of neurons, consistent with the previously described regional specific expression pattern of Lsamp in the brain44 (Fig. 7f).
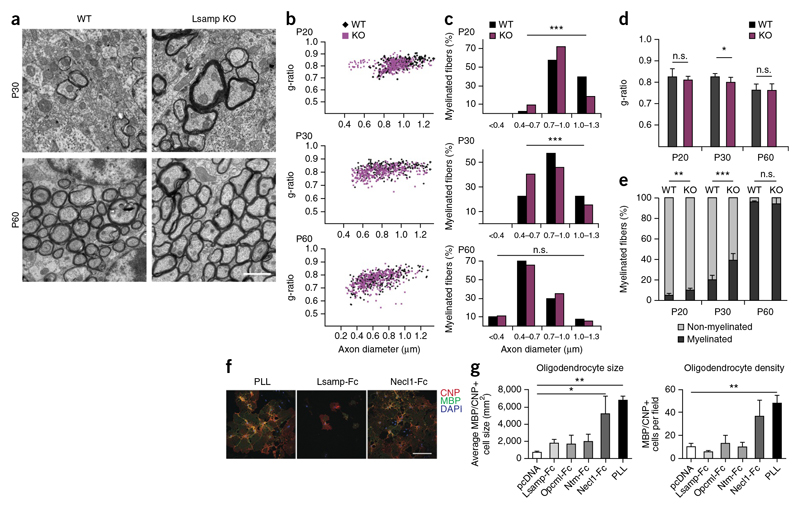
In brain sections, Lsamp was highly enriched in limbic structures. Early in development, before myelination had started (up to P20), Lsamp appeared along the axonal tracts connecting limbic areas, including the frontal corpus callosum, fimbria, stria terminalis and on fibers of the anterior commissure (Fig. 7g). In adult mice, the labeling of Lsamp in white matter was reduced. We confirmed specificity of the staining using brain tissue from Lsamp knockout mice (Supplementary Fig. 11). Comparing the overall architecture of fiber tracts in knockout and wild-type mice on brain sections stained for MBP revealed no major differences. To determine whether Lsamp could, nevertheless, be responsible for more subtle differences in CNS myelination, we used electron microscopy to study myelination in the corpus callosum and fimbrial axons of knockout and wild-type mice starting at P20, a time point at which fiber tracts are still stained for Lsamp. Myelin thickness was determined by calculating the g-ratio (the ratio of axon diameter to the diameter of axon and myelin sheath) at P20, P30 and P60. We observed that g-ratios were significantly different only at P30 in the fimbria of Lsamp knockout (thicker myelin) as compared with wild-type mice (P = 0.0105; Fig. 8a,b,d and Supplementary Fig. 12). When we analyzed the axonal size distribution of myelinated fibers, we observed that thinner axons started to be myelinated earlier in Lsamp knockout mice than in wild-type animals (Fig. 8c). Given that myelination starts in the thickest axons and progresses to small diameter axons with time, such a shift is an indication for premature myelination. Indeed, we observed a higher number of myelinated axons in Lsamp-deficient animals at P20 and P30 (Fig. 8e). Together, these data point to a role of Lsamp as a negative regulator of myelination in specific fiber tracts. To directly test whether Lsamp acts as a repulsive molecule for oligodendrocytes, we performed cell-spreading assays. Coverslips were coated with the respective Fc-fusion proteins and growth and spreading of oligodendrocytes was determined. Whereas Necl1-Fc, used as a positive control, promoted growth and spreading of oligodendrocytes, cells did not attach and grow on Lsamp-Fc coated coverslips (Fig. 8f,g). Similar results were obtained for Ntm-Fc and Opcml-Fc, suggesting that members of the IgLON family are repulsive to oligodendrocytes. Apart from identifying a previously unknown negative regulator of myelination in specific fiber tracts of the CNS, these results exemplify the utility of our proteome resource.
Figure 8. Lsamp is a negative regulator of myelination in the fiber tracts of the fimbria-fornix.
(a) Electron microscopy images of the fimbria/fornix at P30 and P60 from controls (wild type, WT) and Lsamp knockout (KO) mice. Scale bar represents 1 |μm (b) Scatter plots of g-ratios of individual fibers of the fimbria/fornix at P20,P30 and P60 from control (black)and Lsamp KO mice (magenta). (c) The histogram shows the percentage of myelinated axons with respect to axon diameter at 0.3-μm intervals at P20, P30 and P60 for wild-type and Lsamp KO mice. There was a shift toward myelination of low-caliber axons in the mutant as compared with the control (chi-square test; from top to bottom: P ═ 4.5 x 10-10, P ═ 2.8 x 10-5, P ═ 0.36, ***P <0.0001).More than ∼250 axons for each genotype were counted (three animals per genotype) (d) Average g-ratio at P20, P30 and P60 for wild-type and Lsamp KO mice (Student's t test, P ═ 0.0105; n ═ 3 mice per genotype). Error bars represent s.d. (e) Percentage of myelinated and unmyelinated axons counted at P20, P30 and P60. More than 1,500 axons were counted for each time point (n ═ 5 mice for P20 and P30, n ═ 4 for P60) per genotype (bars show mean ± s.d.; Student's t-test; **P ═ 0.0055, ***P ═ 0006). (f,g) Coverslips were coated with 10 μg ml-1 Fc-fusion proteins (IgLON family proteins and control), and oligodendrocyte precursor cells were plated and allowed to adhere and grow for 4 d. PLL and Necl1-Fc coating were used as positive controls. The purified supernatant of HEK 293T cells transfected with an empty vector (pcDNA) was used as negative control (bars show mean ± s.d.; ANOVA, P ⋖ 0.05, Dunnet post hoc test with pcDNA as control; n ═ 3 experiments; *P ═ 0.0087, **P ═ 0.0025). Scale bar represents 20 μm.
Discussion
To obtain a global understanding of brain function, large-scale neuroscience projects are underway that seek to generate anatomical maps and molecular information of cells in the CNS. Although proteins are the functional units of cells, concerted, large-scale initiatives trying to generateproteome maps of the brain and its components have not yet been successful, mainly because the required technology has only very recently been developed. To provide a starting point for a comprehensive brain proteome map, we performed in-depth proteome analysis of the adult mouse brain, its major brain-region and CNS cell types that resulted in the largest collection thus far of cell type- and brain region-resolved protein expression data of the brain. To estimate the coverage and completeness of our proteomes, we included transcriptome analyses of the same samples. The identified 17,634 transcripts from the RNA sequencing showed a bimodal distribution, where one-third of the transcripts were of low abundance, possibly as a result of stochastic transcription28. Commonly used filtering criteria forRNA-Seq reduced the number to 12,933, indicating that our proteome coverage is close to completion, at least in terms of detection of each functionally expressed gene. This analysis is supported by data from several large scale, high-resolution, FDR-controlled cellular proteome projects, where the coverage saturated at approximately 11,000 (refs. 15,18,45,46).
Genome-wide transcriptome analyses have been performed extensively with CNS cell types,but the correlations between mRNA and protein levels are too weak and variable for each protein to allow for reliable quantitative predictions on protein abundance. We observed differences in the proteome and transcriptome with Pearson correlation coefficients between 0.40 and 0.45, somewhat poorer than those previously reported for cell lines15, but in the range of recent comparisons of human tissues16.
The brain proteome presented here represents an incomplete snapshot. It will be necessary to identify the missing proteins by further improvements in technology and to obtain time-dependent expression patterns47. In addition, the analyses of post-translational modifications and isoforms will continue to represent a major challenge and opportunity toward completion of a truly comprehensive brain proteome.
In the context of the enormous cellular diversity and specialization of cells in the brain, one interesting question is how proteomes differ between highly specialized cells. An unexpected finding of our study was the high qualitative degree of similarity of the proteomes, even in the context of very dedicated cell types in a highly complex organ. So far, in-depth proteome studies analyzing the shared and distinct features among different cells had primarily been carried out in cultured fibroblast-like or cancer cell lines15, 18–20. An interesting aspect of our analysis is that cell type-specific proteins represent many relatively abundantintegral membrane proteins associated with the cell surface and the extracellular matrix. Even when comparing the different brain regions, the cell surface emerged as the cellular component containing the proteins that contribute the most to brain-region specificity. Considering the highly specialized tasks that the glia and neuronal cells need to carry out, the small number of cell type-specific proteins was unexpected. Could the strong similarity be a result of cell culture conditions? Although intercellular interactions of the various CNS cells in vivo are known to be important, this is unlikely to be the reason. Using acutely purified cell populations, we found highly overlapping protein expression patterns between cultured and isolated CNS cell preparations. This indicates that these cultured primary cells can serve as an alternative for cells in vivo in many contexts (see Supplementary Fig. 13 for RNA-Seq comparison).
The in-depth analysis of proteins that are abundantly and differentially expressed in CNS cells is a promising strategy for understanding cell-specific functions. We provide proof of concept by applying this strategy to identify adhesion molecules in neuron and oligodendrocyte interaction. Given that the molecularnature of axon and glia interaction is unknown, we applied bioinformatic tools and found that cell adhesion was among the KEGG pathways that were shared between neurons and oligodendrocytes. Based on this information, we plotted all 282 proteins of this category and found that the most abundant and enriched adhesion molecule shared between oli-godendrocytes and neurons was Ncam1, a protein that negatively regulates myelination36. Lsamp wasthe second most enriched adhesion molecule in neurons and oligodendrocytes. Lsamp is known to mediate homophilic, as well as heterophilic, interactions with other members of the IgLON family48. Myelin is critical for information processing and contributes to learning and cognition, but little is knownof how axons are selected for myelination49. Notably, subtle changes in myelin have been observed in a wide range of psychiatric disorders, including schizophrenia, depression and bipolar disorders49. Thus,such minor alterations may already be relevant and could well have an effect on higher brain functions. Given the association between single-nucleotide polymorphisms in the LSAMP gene and psychiatric diseases 50,it would be interesting to investigate whether differences in myelination are involved. Up to now,behavioral abnormalities with hyperactivity in novel environment have been reported in Lsamp-deficient mice, but structural analysis of brain anatomy has been lacking50. Future studies with various IgLON compound mutants will be needed to investigate their possible functional redundancies in axon-glial interaction and axonal myelination.
In summary, our proof-of-concept study found that proteome analysis may serve as a rich resource for the understanding of brain development and function, complementing existing transcriptome databases. The entire data set and the MaxQuant output tables are available on proteomeXchange (see Supplementary Fig. 14 for explanation of data visualization in MaxQB database, http://maxqb.biochem.mpg.de/mxdb/project/show/P009); and, for simpler access to key quantitative features, see http://www.mousebrainproteome.com/ Quantitative proteomic data sets are required, for example, for systems biology, which try to describe and simulate biological processes by integrating comprehensive data from various sources. As the analysis of proteomes is substantially more challenging than that of genomes, previous efforts have been biased toward transcriptome analyses. However, cost-effective, rapid, accurate and sensitive proteomics have now become possible. We further optimized this technology by extending our single-shot analysis27, which resulted in the detection of more than 11,500 different proteins in only 4-h replicate analysis. Such a deep proteome coverage in a single run was made possible by matching to the brain proteome library composed of the fractionated analysis of mouse brain and its primary cell types that we generated.We believe that these technological advances can be used broadly in the proteomic study of diverse and rare brain cell types. In a more global view, our data and the rapidity with which it can now be generated, suggest that MS-based proteomics can be implemented more broadly into neuroscience to help resolve the full molecular complexity of the brain at the protein level, with cellular or even subcellular resolution.
Methods
Methods and any associated references are available in the online version of the paper.
Accession codes. All proteomics data for each individual proteome sample from different cell types and brain regions are available at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository with the data set identifier PXD001250.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper
Online Methods
Tissue preparation, cell culture and cell isolation
All animal experiments were performed according to the Lower Saxony State regulations for animal experimentation. Brain regions were dissected from P60 male C57BL/6 mice using a ‘rodent brain matrix’ 1-mm coronal slicer (ASI Instruments) according to coordinates obtained from the Mouse Brain Library (http://www.mbl.org/) using the C57BL/6J atlas as reference. The preparation of primary cell cultures was performed according to previously described protocol with modifications51,52. Briefly, brains were removed from newborn mice (P1) were separated by midline incision and transferred into Hanks Buffered saline solution (HBSS). Under a dissection microscope, the meninges were removed and the hindbrains were discarded. A total of five brains were pooled and incubated with 0.25% trypsin/EDTA (wt/vol) at 37 °C for 10 min. The tissues were washed twice with BME growth medium (supplemented with 10% fetal calf serum (FCS, vol/vol), GlutaMAX, penicillin and streptomcyin) and further dissociated by trituration (ten times) using a glass pipette. The cell suspension was passed through a cell sieve and subsequently plated onto 5 PLL-coated cell culture flasks with BME growth medium. Two thirds of the growth medium was exchanged twice a week. 10 d after plating the glial mixed cultures, microglia were removed by gentle, manual shaking, leaving a culture that consisting of OPCs on top of a confluent layer of astrocytes. The next day, OPCs were shaken off by hand and pelleted by centrifugation. The OPC-pellet was re-suspended in Super Sato differentiation medium (high-glucose DMEM with B27-supplement, 1% horse serum (vol/vol), 110 ag ml-1 sodium pyruvate, 0.5 nM tri-iodo-thyronin, 0.5 μM L-thyroxin, GlutaMAX and penicillin/streptomycin). The OPC suspension was transferred into an untreated cell culture dish and incubated at 37 °C for 3 min. During this time, contaminating microglia and astrocytes adhered to the culture dish, while OPCs remained in suspension. This step was repeated a second time, to yield highly pure OPC cultures. These cells were plated in an appropriate cell number onto PLL-coated cell culture dishes or glass cover slips with Super Sato. The day of OPC plating was defined as DIV-0 and cells were harvested for experiments at the subsequent days. After shaking off the OPCs, fresh DMEM (supplemented with 10% FCS, GlutaMAX, penicillin and streptomycin) was added to the remaining astrocytes and the cells were allowed to recover for 72 h. The astrocytes were then harvested for further experiments. The purity of these cells was determined separately by trypsination, seeding onto PLL-coated cover-slips and immunostaining after 48 h. The preparation of microglia was performed as described previously53. In brief, glial mixed cultures were treated with microglia colony stimulating factor (MCSF) to enhance microglia proliferation. After 3 d, microglia were harvested by gentle shaking and seeded onto PLL-coated cell culture dishes with DMEM (supplemented with 10% FCS, GlutaMAX, penicillin and streptomycin). After another 3 d, cells were used for experiments. Cortical neuron cultures were prepared from embryonic mice at E16.5 as described52 with minor modifications. Pregnant mice were killed by cervical dislocation and opened to reveal the embryos. The embryos were decapitated; the brains were exposed by a midline incision and transferred into HBSS. Meninges were stripped from the surface of the brain and hindbrain was discarded. Three brains were pooled, treated with 0.25% trypsin/EDTA for 10 min and washed with neuronal growth medium (MEM with B27-supplement, 0.6% glucose (wt/vol), 0.22% bicarbonate (wt/vol), pyruvate, GlutaMAX, penicillin and streptomycin). Cells were plated onto PLL-coated cell culture dishes with neuronal medium. After 16 h, cytosine arabinoside (AraC) was added in a final concentration of 4 μM. The addition was repeated the second day after plating. The third day, AraC was washed out and cells were cultivated in neuronal growth medium until day 5, 10 and 16, respectively.
For acute cell isolation, cells were isolated using the MACS cell selection kit (Miltenyi Biotec). Briefly, brains from mice at P8 were isolated, minced and mechanically dissociated with the gentleMACS dissociator according to the manufacturer instructions. The cells were then passed through a 45-μm strainer and incubated with the cell-specific beads for 15 min. The cells were then loaded on LS columns and separated on a quadroMACS magnet. The target cells were washed once in phosphate-buffered saline (PBS), then pelleted and snap-frozen. The remaining cells were collected for the isolation of the other cell population.
Sample preparation for mass spectrometric analysis
Cells were lysed in a buffer consisting of 0.1 M Tris-HCl, pH 8.0, 0.1 M DTT, and 2% SDS (wt/vol) at 99 °C for 5 min. The lysates were sonicated using a Branson type sonicator and protein content was determined using a Fluorescence Spectrometer. 100 ag of protein was subjected to chloroform methanol precipitation and the proteins were digested with LysC and trypsin according to the standard protocol54. After desalting on StageTips55, digested peptides were subjected to either fractionation or single-shot analysis as indicated in Figure 1. 25 μg of the digested peptides were separated into six fractions using strong anion exchange (SAX) according to the previously described pipette tip protocol56. For single-shot analysis digested peptide samples were directly analyzed without any fractionation.
Lc MS/MS analysis
MS analysis was performed using Q Exactive mass spectrometers (Thermo Fisher Scientific) coupled on-line to a nanoflow UHPLC instrument (Easy nLC, Thermo Fisher Scientific). Eluted peptides were separated over a 240-min gradient on a reverse phase 50-cm-long C18 column (75-μm inner diameter, ReproSil-Pur C18-AQ 1.8 ¡am beads (GmbH)). The survey scans (300-1,700 m/z, target value 3E6, maximum ion injection times 20 ms) were acquired and followed by higher energy collisional dissociation (HCD) based fragmentation (normalized collision energy 25). For measurements in Q Exactive mass spectrometers, a resolution of 70,000 at m/z 400 was used for survey scans and up to ten dynamically chosen most abundant precursor ions were fragmented (isolation window 1.6 m/z). The MS/MS scans were acquired at a resolution of 17,000 at m/z 400 (target value 1E6, maximum ion injection times 60 ms). For survey scans in Q Exactive HF, a resolution of 60,000 at m/z 200 was used and up to 15 dynamically chosen most abundant precursor ions were fragmented (isolation window 1.6 m/z). The MS/MS scans were acquired at a resolution of 15,000 at m/z 200 (target value 1E5, maximum ion injection times 25 ms).
MS data analysis
Mass spectra were processed using the MaxQuant computational platform26 versions 1.5.2.1, 1.5.0.8 and 1.3.9.21. The spectra were searched by the Andromeda search engine57 against the mouse Uniprot sequence database. The search included cysteine carbamidomethylation as a fixed modification and N-acetylation of protein and oxidation of methionine as variable modifications. Peptides with minimum of seven amino-acid length were considered and the required FDR was set to 1% at the peptide and protein level. Where possible, peptide identification were transferred based on accurate mass and retention times across liquid chromatography (LC)-MS runs to obtain peptide identity in cases where the precursor peptides were present in MS1 but not selected for fragmentation and identification by MS2 in given run. This matching between runs is a feature in the MaxQuant software21,58. For single run analysis of brain and cerebellum, matching was done against the library of the deep brain proteome composed of fractionated and single shot runs (Supplementary Table 2). Otherwise fractionated samples and single shot runs were matched within themselves (Supplementary Table 1). Protein identification required at least one unique or razor peptide per protein group. Quantification in MaxQuant was performed using the label free quantification (LFQ) algorithm32. Briefly, MaxLFQ is an intensity determination and normalization procedure that is fully compatible with any peptide or protein separation before LC-MS analysis. Given that the presence of quantifiable pep-tides varies from sample to sample, the algorithm uses only common peptides for pair-wise ratio determination for each protein and calculates a median ratio to protect against outliers. It then determines all pair-wise protein ratios and requires a minimal number of two peptide ratios for a given protein ratio to be considered valid. A least-squares analysis is then performed to reconstruct the abundance profile before rescaling the whole profile to the cumulative intensity across samples. This step preserves the total summed intensity for a protein over all samples. This procedure is repeated for all proteins, resulting in an accurate abundance profile for each protein across the samples.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001250. The data set can also be accessed through MaxQB database (http://maxqb.biochem.mpg.de/mxdb/project/show/P009) and for simpler access to key quantitative features: http://www.mousebrainproteome.com/.
Downstream bioinformatics analyses
Bioinformatic analysis was performed in the Perseus software environment. All MS data were mapped to gene symbols for comparison with the RNA-Seq data. We performed two-sample welch t-tests to identify proteins with a significant differential expression in a cell type in comparison to others employing a 2% permutation-based FDR filter. For each cell type, fold expression values were calculated from the individual median averaged protein LFQ intensities and the corresponding median LFQ intensities of the other cell types and their differentiation stages. PCA was performed on the LFQ intensity data.
The Illumina raw reads were binned according to the barcodes and the barcodes and adaptors were trimmed away. The pre-processed reads were then subjected to quality control using FastQC. The quality filtered reads were further aligned to the mouse transcriptome Mus musculus (UCSC version mm10) using Tophat read mapper58. RPKM for gene features were then calculated using GenomicRanges and GenomicFeatures packages59 from the R Bioconductor tool set.
Pathway membership information was obtained from KEGG pathways and categorical annotation was supplied as GO biological process (BP), molecular function (MF), and cellular component (CC). We filtered for 8,147 protein groups that were quantified in all replicates for at least one cell type or its differentiation stage. Differentially expressed proteins were identified by an ANOVA based test at permutation-based FDR cutoff of 0.05. Hierarchical clustering was performed after z-score normalization of the regulated data set and enrichment for annotation categories was evaluated by Fisher exact test to obtain a P value.
The one-dimensional annotation matrix algorithm ranks the protein abundance and tests if the proteins corresponding to every annotation term tend to be ranked higher or lower than the ranking of all proteins in the data set. In this way, it tests and computes the difference of every protein annotation from the overall intensity distribution using the two-sided Wilcoxon-Mann-Whitney test where the multiple hypothesis testing is adjusted by applying a Benjamini-Hochberg FDR threshold of 0.02. For significant cat-egories, a position score (termed s; a number between -1 and 1) is calculated, indicating where the center of the distribution of protein abundance for the annotation category is located relative to the overall distribution of values. We used this algorithm to create an annotation matrix of ‘position score’ for GO annotation terms and KEGG pathways separately for all 27 proteomes applying a P value threshold of 0.005. The replicates were grouped based on cell type and differentiation stage before applying an ANOVA test on the annotation matrix with a permutation-based FDR cutoff of 0.001 to identify annotation terms that are significantly different between at least two groups. The z-scored annotation matrix is supplied as Supplementary Table 10.
Immunocytochemistry
In order to determine the purity of the primary neural cells, every cell culture dish contained two 8-mm round glass cover slips. Before lysis of the cells for RNA isolation or protein analysis, the coverslips were removed and fixed with 4% paraformaldehyde (PFA, wt/vol) in PBS. Cells on the coverslips were immunostained as described. We used antibodies directed against O1 (1:50) and MBP (1:300) (Millipore, AB5864) to stain oligodendrocytes, against GFAP (1:200, GA5, Novocastra and G5601, Promega) for astrocytes, Iba1 (1:1,000, 019-19741, Wako) to label microglia and against p-III-tubulin (1:1,000, T4026, Sigma-Aldrich) or neurofilament 200 kDa (1:80, N0142, Sigma-Aldrich) to visualize neurons. Col11a1 was detected using the polyclonal ab64883 antibody (Abcam) Appropriate fluorescent-labeled antibodies were purchased from Dianova and used in 1:300 dilutions. Samples were analyzed using a Leica DMI 6000 fluorescence microscope with a 40x NA 1.10 plan-apochromat objective. The number of contaminating cells was counted and only cultures of at least 95% purity were included in further experiments.
Immunohistochemistry
The following primary antibodies were used in respective concentrations: Col11a1 (ab64883, Abcam), MBP (SMI94, Covance; DakoCytomation), Lsamp (2G9 DSHD, University of Iowa). As secondary antibodies, appropriate fluorescent-labeled antibodies from Dianova were employed. For tissue processing mice were killed, the tissue was fixed with 4% PFA in PBS, saturated with 30% sucrose (wt/vol) and frozen. Tissue slices of 30 μM were prepared using a Leica CM 1900 Cryostat and mounted onto superfrost slides. Antigen retrieval with 10 mM citrate buffer was performed, sections were permeabilized with 0.5% Triton X-100 (vol/vol) in PBS for 30 min, washed and blocked in blocking solution (5% horse serum and 0.5% Triton X-100 in PBS) for 1 h at 20 °C. Sections were incubated with primary antibodies in 3% horse serum and 0.5% Triton X-100 in PBS at respective concentrations overnight. Next, sections were washed with PBS and incubated with secondary antibodies for 1 h at RT. After washing with PBS, the sections were mounted in fluorescence mounting medium (Dako). For staining with antibodies against Lsamp, Triton X-100 was omitted as it affected the quality of the stainings.
RNA extraction, generation of the cDNA libraries and Illumina RNA-Seq
Cell cultures were washed three times with PBS, approximately 1 million cells were lysed in 750 μl of RLT buffer (Qiagen) and passed through a 27G needle. Samples were stored at −80 °C until RNA preparation. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s guidelines. RNA concentration was determined using a NanoDrop ND1000 spectrophotometer and RNA integrity was assessed with an Agilent 2100 Bioanalyzer.
For the first round of amplification, cDNA synthesis was performed using 300 ng of RNA and the Superscript III first strand synthesis kit (Life Technologies). A mixture of T7-B primers was used in this reaction. Afterwards, second strand synthesis was performed using DNA Polymerase I and second-strand buffer (Life Technologies). The double-stranded cDNA was purified using a CyScribe GFX Purification kit (GE Healthcare) and used for in-vitro-transcription (MEGAscript T7 Kit, Ambion). The resulting aRNA was purified using a RNeasy Micro column kit (Qiagen).
For the second round of amplification, between 300 and 800 ng of aRNA were mixed with a Dec1-hairpin-N9 primer and first strand synthesis was performed using the Superscript III first strand synthesis kit (Invitrogen). Second strand synthesis was done with a B-short primer and Taq-Polymerase. cDNA was purified with the NuleoSpin Extract II Kit (Macherey & Nagl).
Sequencing codes were introduced by step-out PCR. Therefore, cDNA was mixed with the appropriate Code_Cis40 primers and amplified by PCR using the PWO Master Mix (Roche).
The Illumina sequencing adaptors were introduced in a second PCR using the appropriate primers. Samples were analyzed using Illumina2000 sequencing60.
RT-qPCR analysis
Expression of Col11a1 on an mRNA level was confirmed by RT-qPCR analysis as described previously. In brief, 1,000,000 cells were lysed in 1 ml of TRIzol (Life Technologies) and total RNA was isolated according to the manufacturer’s guidelines. cDNA synthesis was performed using 2 μg of total RNA, Oligio(dT)12-18 primer and the Superscript III first strand synthesis kit (life technologies). Quantitative real-time PCR was performed in triplicates using the Power SYBR Green PCR Master Mix (Roche) according to the manufacturer’s recommendations on a Roche LightCycler 480 instrument. For Col11a1, the primers 5’-CAGATTGTGACTTAACATCCAAGG-3’ and 5’-CTCGATTATATCCTCAGGTGCAT-3’ were used, GAPDH was amplified with primers 5’-CAATGAATACGGCTACAGCAAC-3’ and 5’-TTACTCCTTGGAGGCCATGT-3’. Quantification was performed using the ACt method (2-ΔΔt). Data were normalized to GAPDH.
Western blot analysis
Cell cultures were lysed in lysis buffer (1% NP40, 0.1% SDS (vol/vol), 0.25 mM EDTA in PBS) with complete protease inhibitor cocktail (Roche). Proteins were resolved in 7.5% SDS-PAGE gels and transferred to a nitrocellulose membrane. Antibodies against Col11a1 (Abcam) and Calnexin (Stressgen) were used in 1:2,000 dilution, detected with appropriate HRP-conjugated antibodies and visualized by ECL (Amersham).
Electron microscopy
Lsamp knockout mice (C57/BL6J) and wild-type littermates were perfused with 4% PFA and 2.5% glutaraldehyde (vol/vol) in PBS and brains were stored in the fixative. Electron microscopy was performed as described previously61. Brains were cut into 300-μm sections using a vibratome and 2-mm punches of the genu of the corpus callosum and the fimbria were prepared. Tissue was embedded in Epon, ultrathin sections were prepared and stained with 1% uranyl acetate (wt/vol) and lead citrate. Electron micrographs were taken on a LEO 912AB electron microscope (Zeiss) using an on-axis 2,048 x 2,048 charge-coupled device camera (Proscan). Blinded quantification was performed by determining areas of the myelinated and unmyelinated axons. The diameter of a circle with that area calculated and g-ratio was determined by dividing axonal diameter by myelinated diameter.
Fc-fusion protein generation and purification
Necl1-Fc and Necl4 -Fc plasmids were kindly provided by E. Peles (Weizmann Institute). The Fc fragment was amplified and inserted into a pcDNA3.1+ plasmid using the primers 5'-AAAAGAGCT CGGAGGAGGAGGAGAT CCCCGT CGTGCAT CTAT C -3’ and 5’-AAA AGGGCCCTCTAGATCATTTACCC -3'. The extracellular domain from Lsamp, Ntm, Opcml and Mcam were amplified and inserted into the Fc plasmid using NheI restriction sites. Soluble Fc-fusion proteins were purified using Protein A HP Spin Trap columns (GE Healthcare), following manufacturer’s instructions. Briefly, HEK cells were transfected as described in the previous section. After 2 -3 d, the supernatant was collected, and centrifuged for 15 min at 4,000g at 4 °C. 1x Complete Protease Inhibitor Cocktail (Roche) was added to the supernatant and then concentrated using Amicon Ultra-15 Centrifugal Filter Units (Millipore) according to manufacturer’s recommendations. The final volume of 2 ml was diluted in equal volume of binding buffer (20 mM sodium phosphate, pH 7.0). Briefly, the storage solution from the column was removed by centrifugation for 30 s at 100g. The column was equilibrated adding 600 μl of binding buffer, centrifuged for 30 s at 100g. Then, 600 μl of the antibody solution were added, incubated for 4 min while gently mixing and then centrifuged for 30 s at 100g. This procedure was repeated until all the volume was loaded on the column. Then the column was washed twice adding 600 μl binding buffer and centrifuging for 30 s at 100g. Two collection tubes per sample were prepared for eluted fractions, each one containing 30 ¡al of neutralizing buffer. The proteins were eluted twice by adding 400 ¡al of elution buffer (0.1 M glycine-HCl, pH 2.7), mixing by inversion, placing the column in a 2 -ml microcentrifuge tube containing 30-jal neutralizing buffer (1 M Tris-HCl, pH 9.0) and centrifuged for 1 min at 50g. The proteins were aliquoted and kept at −20 °C.
Binding assay
For binding assays, supernatant from transfected HEK cells was retrieved and centrifuged 10 min at 5,000g. Per each 18 mm coverslip of neuronal or oligodendroglial culture, 150 μl of media were mixed with 1.5 μl of Cy3-conjugated anti-human Fc antibody (Dianova) and incubated for 30 min at 20 °C. Then, the mix was added to each coverslip and incubated for 20 min at 20 °C in a humid chamber. Finally, the coverslips were washed 3 times with PBS and the cells were fixed with 4% PFA for 15 min at 20 °C.
Adhesion assay
Glass coverslips in a 24-well plate were coated with 500 μl of a solution with 10 ag ml-1 of donkey anti-human Fc antibody (Dianova) in 50 mM Tris-HCl (pH 9.0) and left overnight at 4 °C. Then, the coverslips were washed three times with supplement-free DMEM and 500 μl of a solution containing 10 ag ml-1 of Fc-fusion protein in 0.2% BSA/PBS (wt/vol). After 1 h of incubation at 37 °C, the coverslips were washed threetimes with DMEM and fresh Super Sato media was added. PLL coating (100 μg ml-1) was used as a positive control to verify the quality of the primary oligodendrocyte preparation. After washing and placing new media, 25,000 OPC were plated and allowed to grow for four days, and fixed with 4% PFA.
Statistical analyses
All statistical analyses were performed on at least three different biological replicates. The sample size was similar or higher to those generally employed in the field. Data distribution was assumed to be normal but this was not formally tested. Statistical details are reported in each figure legend. Two sided Fischer’s exact test, Chi-square test, one way ANOVA Dunnett’s multiple comparison, Wilcoxon-Mann-Whitney test and two tailed unpaired t test were performed using the Prism Software package (GraphPad).
A Supplementary Methods checklist is available.
Supplementary Material
Acknowledgmentsts
We would like to acknowledge the PRIDE Team for upload of proteomics raw data. We thank J. Cox, M.Y. Hein and K. Mayr for helpful discussions, and C. Velte, N. Schwedhelm-Dornmeyer, S.Safaiyan, G. Sowa and M. Dodel for technical assistance. Illumina sequencing was performed by R. Reinhardt at the Max-Planck Genome Center Cologne.The work was supported grants from the German Research Foundation (SI 746/9-1; 10-1;TRR43; RO 4076/3-1), the Tschira- Stiftung, and grants from the Estonian Research Council (IUT20-41 and PUT129).The research leading to these results has received funding from the European Commission under FP7 GA n°ERC-2012-SyG_318987-ToPAG and MC-ITN IN-SENS (#607616). S.S. received a PhD scholarship from the Boeringer-Ingelheim Fonds and N.K. this recipient of a Marie-Curie fellowship from the INSENS/FP7-PEOPLE-2013 (607616) framework.
Footnotes
Author Contribution
K.S., M.M. and M.S. designed the experiments. K.S., S.S., C.G.B., N.K., N.M.-H., L.C. and U.-K.H. performed the experiments. K.S., S.S., S.T., N.K., M.J.R., M.M. and M.S. analyzed the data. K.K. and M.-A.P. provided materials. K.S., M.M. and M.S. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 2.Thompson CL, et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–323. doi: 10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitchen RR, Rozowsky JS, Gerstein MB, Nairn AC. Decoding neuroproteomics: integrating the genome, translatome and functional anatomy. Nat Neurosci. 2014;17:1491–1499. doi: 10.1038/nn.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck M, Claassen M, Aebersold R. Comprehensive proteomics. Curr Opin wBiotechnol. 2011;22:3–8. doi: 10.1016/j.copbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013;49:583–590. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 15.Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 17.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. M111014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundberg E, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pontén F, et al. A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol. 2009;5:337. doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Sherman DL, Brophy PJ. Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci. 2005;6:683–690. doi: 10.1038/nrn1743. [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal S, Yurlova L, Simons M. Central nervous system myelin: structure, synthesis and assembly. Trends Cell Biol. 2011;21:585–593. doi: 10.1016/j.tcb.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Molofsky AV, et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Nagaraj N, et al. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013722. M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hebenstreit D, et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beck M, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, et al. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski JR, et al. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol Syst Biol. 2012;8:611. doi: 10.1038/msb.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snead MP, Yates JR. Clinical and molecular genetics of Stickler syndrome. J Med Genet. 1999;36:353–359. [PMC free article] [PubMed] [Google Scholar]
- 36.Charles P, et al. Negative regulation of central nervous system myelination by polysialylated-neural cell adhesion molecule. Proc Natl Acad Sci USA. 2000;97:7585–7590. doi: 10.1073/pnas.100076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimenta AF, et al. The limbic system-associated membrane protein is an Ig superfamily member that mediates selective neuronal growth and axon targeting. Neuron. 1995;15:287–297. doi: 10.1016/0896-6273(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 38.Schofield PR, et al. Molecular characterization of a new immunoglobulin superfamily protein with potential roles in opioid binding and cell contact. EMBO J. 1989;8:489–495. doi: 10.1002/j.1460-2075.1989.tb03402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struyk AF, et al. Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. J Neurosci. 1995;15:2141–2156. doi: 10.1523/JNEUROSCI.15-03-02141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed J, McNamee C, Rackstraw S, Jenkins J, Moss D. Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J Cell Sci. 2004;117:3961–3973. doi: 10.1242/jcs.01261. [DOI] [PubMed] [Google Scholar]
- 41.Gil OD, Zanazzi G, Struyk AF, Salzer JL. Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J Neurosci. 1998;18:9312–9325. doi: 10.1523/JNEUROSCI.18-22-09312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiegel I, et al. A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat Neurosci. 2007;10:861–869. doi: 10.1038/nn1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurel P, et al. Nectin-like proteins mediate axon Schwann cell interactions along the internode and are essential for myelination. J Cell Biol. 2007;178:861–874. doi: 10.1083/jcb.200705132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed JE, et al. Expression of cellular adhesion molecule ‘OPCML’ is down-regulated in gliomas and other brain tumours. Neuropathol Appl Neurobiol. 2007;33:77–85. doi: 10.1111/j.1365-2990.2006.00786.x. [DOI] [PubMed] [Google Scholar]
- 45.Moghaddas Gholami A, et al. Global proteome analysis of the NCI-60 cell linepanel. Cell Rep. 2013;4:609–620. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Azimifar SB, Nagaraj N, Cox J, Mann M. Cell type-resolved quantitative proteomics of murine liver. Cell Metab. 2014;20:1076–1087. doi: 10.1016/j.cmet.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Bayés A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gil OD, et al. Complementary expression and heterophilic interactions between IgLON family members neurotrimin and LAMP. J Neurobiol. 2002;51:190–204. doi: 10.1002/neu.10050. [DOI] [PubMed] [Google Scholar]
- 49.Fields RD. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Innos J, Koido K, Philips MA, Vasar E. limbic system associated membrane protein as a potential target for neuropsychiatric disorders. Front Pharmacol. 2013;4:32. doi: 10.3389/fphar.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trajkovic K, et al. Neuron to glia signaling triggers myelin membrane exocytosis from endosomal storage sites. J Cell Biol. 2006;172:937–948. doi: 10.1083/jcb.200509022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fitzner D, et al. Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO J. 2006;25:5037–5048. doi: 10.1038/sj.emboj.7601376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regen T, et al. CD14 and TRIF govern distinct responsiveness and responses in mouse microglial TLR4 challenges by structural variants of LPS. Brain Behav Immun. 2011;25:957–970. doi: 10.1016/j.bbi.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 55.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 56.Wiániewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 57.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J.Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 58.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003118. e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnell C, et al. The multispecific thyroid hormone transporter OATP1C1 mediates cell-specific sulforhodamine 101-labeling of hippocampal astrocytes. Brain Struct Funct. 2015;220:193–203. doi: 10.1007/s00429-013-0645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snaidero N, et al. Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell. 2014;156:277–290. doi: 10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.