Abstract
The development of intestinal organoids from single adult intestinal stem cells in vitro recapitulates the regenerative capacity of the intestinal epithelium1,2. Here we unravel the mechanisms that orchestrate both organoid formation and the regeneration of intestinal tissue, using an image-based screen to assay an annotated library of compounds. We generate multivariate feature profiles for hundreds of thousands of organoids to quantitatively describe their phenotypic landscape. We then use these phenotypic fingerprints to infer regulatory genetic interactions, establishing a new approach to the mapping of genetic interactions in an emergent system. This allows us to identify genes that regulate cell-fate transitions and maintain the balance between regeneration and homeostasis, unravelling previously unknown roles for several pathways, among them retinoic acid signalling. We then characterize a crucial role for retinoic acid nuclear receptors in controlling exit from the regenerative state and driving enterocyte differentiation. By combining quantitative imaging with RNA sequencing, we show the role of endogenous retinoic acid metabolism in initiating transcriptional programs that guide the cell-fate transitions of intestinal epithelium, and we identify an inhibitor of the retinoid X receptor that improves intestinal regeneration in vivo.
The intestinal epithelium consists of a single layer of polarized cells, which are arranged into regular protruding units, or villi, that host differentiated cells, interspaced by crypts composed of stem cells and secretory Paneth cells3,4. At the crypt periphery, cells divide rapidly before migrating upward along the villus5 and terminally differentiating6. Intestinal epithelial cells feature unique plasticity7 that allows them to dedifferentiate and replenish the pool of cycling cells lost upon damage8,9. During regeneration, tissue pattern and homeostasis are restored through numerous signalling pathways10,11. It is, however, poorly understood how the spatial organization is regulated and coordinated, especially in the adult intestine after regeneration.
Intestinal organoids recapitulate the ability of intestinal tissue to regenerate and return to homeostasis following damage1,2,4: an entire organoid develops from a single cell, forming a self-organized structure1,12. Initially, single cells enter a regenerative state that is dependent on the transcriptional regulator YAP, and form a symmetric cyst1. Subsequently, symmetry is broken by the emergence of Paneth cells that define and maintain the crypt4, followed by the differentiation of absorptive enterocytes distal from the crypt12. Organoids thus recapitulate the regeneration of the epithelium and subsequent re-establishment of homeostasis13.
Here we have developed an image-based screening platform in organoids cultured from single cells in order to characterize the phenotypic landscape of organoid development and infer functional genetic interactions. We then focused on conditions that improve the regeneration potential, and discovered a role for nuclear retinoic acid receptors in the response to intestinal damage and homeostasis both in vitro and in vivo.
Phenotypic landscape
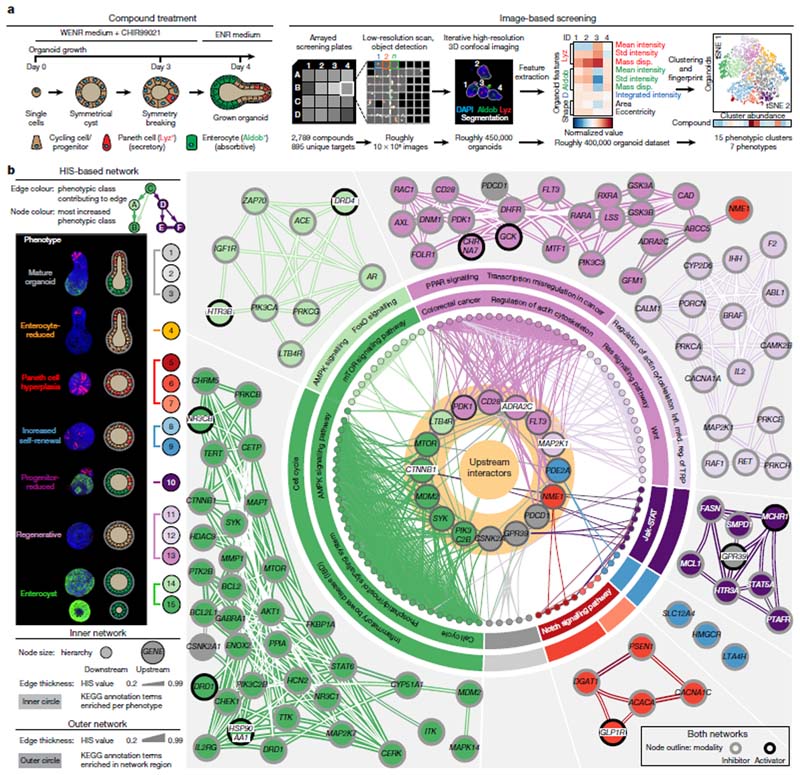
We devised an image-based screen with a chemical genetics approach, generating intestinal organoids from single cells over the course of four days in the continuous presence of 2,789 selected compounds, including inhibitors of kinases, nuclear hormone receptors, transcriptional regulators and other target classes (Supplementary Table 1). Organoids were stained for markers of enterocytes and Paneth cells (using antibodies specific to the enzyme aldolase B14,15 and lysozyme16, respectively), for DNA (using 4’,6-diamidino-2-phenylindole, DAPI) and for total protein content, and imaged with a high-throughput microscope (generating roughly 107 images). We profiled approximately 450,000 organoids using a multivariate feature set that showed reproducible, significant and condition-specific effects in organoids treated with active controls, namely the γ-secretase inhibitor17 DAPT and the glycogen synthase kinase-3β (GSK-3β) inhibitor18 CHIR99021 (Fig. 1a and Extended Data Fig. 1a–i).
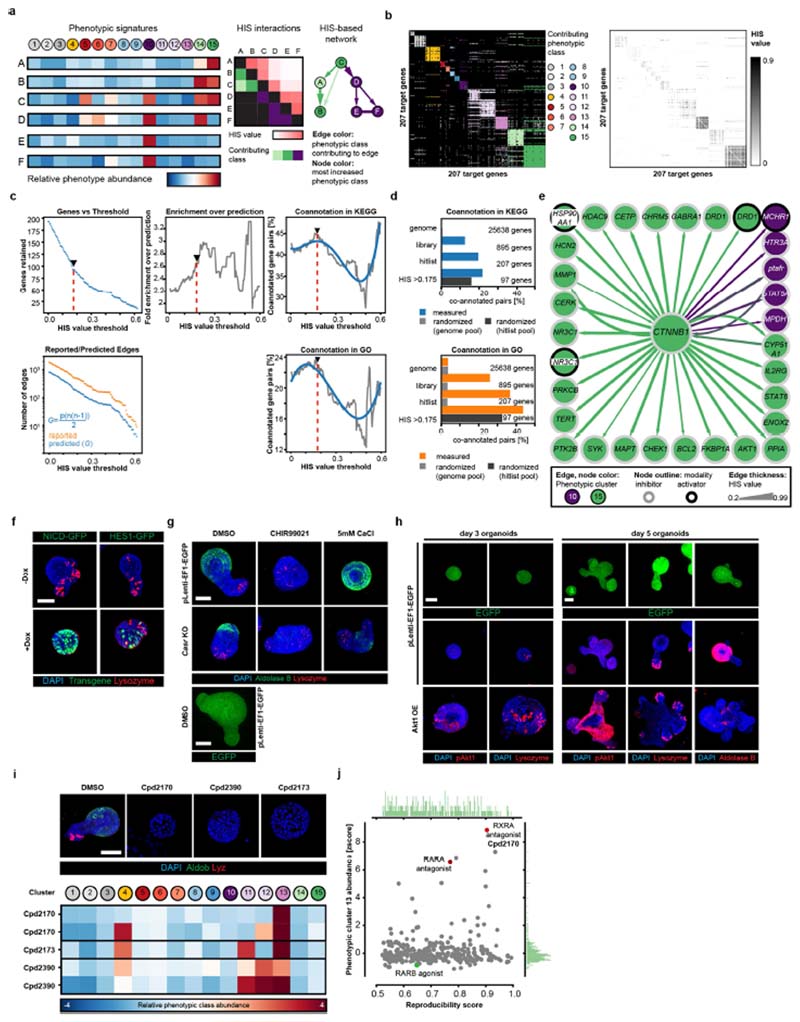
Fig. 1. Phenotypic landscape and map of functional genetic interactions in intestinal organoids.
a, Screening workflow: organoid-level multivariate feature array, clustering by similarity and generation of phenotypic fingerprints for each condition. D, DAPI; Aldob, aldolase B; Lyz, lysozyme; mass disp., mass displacement for the indicated immunofluorescence channels; tSNE, t-distributed stochastic neighbour embedding. b, Left, main panel: identified phenotypic clusters, representative images and schematic representation of the phenotypes. Right, network of HIS interactions (for HIS values of at least 0.175) between 101 genes. For the inner network, node colour and edge colour are explained in the key on the left. Highlighted nodes show off-diagonal interactors. The outer networks show detected subnetworks. Edge thicknesses show HIS values; edges with the highest HIS value per node are shown. Node outlines show compound modality (inhibitor or activator). KEGG enrichments are shown per phenotype (inner circle) and subnetwork (outer circle). Infl. med. reg., inflammation-mediated regulation. a, b, Images are maximum intensity projections (MIPs) of confocal z-stacks, showing nuclear staining (DAPI) and antibody staining for lysozyme and aldolase B.
We clustered organoids by phenotypic similarity19, assigning every organoid to one of the 15 identified phenotypic classes. To improve interpretability, we assigned the classes to seven major phenotypes, reducing the original phenotypic resolution (Extended Data Fig. 2a–c). The most frequent phenotype was the ‘wild-type’ phenotype (classes 1–3, ‘mature organoids’); others included ‘Paneth cell hyperplasia’ (classes 5–7), ‘Wnt hyperactivation’ (classes 8 and 9), ‘progenitor-reduced’ (class 10), and organoids consisting only of enterocytes (classes 14 and 15, ‘enterocysts’) or lacking both differentiated cell types (classes 11–13, ‘regenerative’) (Fig. 1b). A subpopulation (class 13) of the latter phenotype is reminiscent of the YAP1-overexpression and regenerative-state phenotype1, whereas classes 11 and 12 are more similar to an undifferentiated state and might be primed to become enterocysts over time.
To avoid any loss of fidelity, we generated a 15-element phenotypic fingerprint for every compound. We identified 301 compounds from the primary and validation screens that produced strong phenotypes and high reproducibility scores, corresponding to 207 unique target genes (Extended Data Fig. 2d-h and Supplementary Table 1).
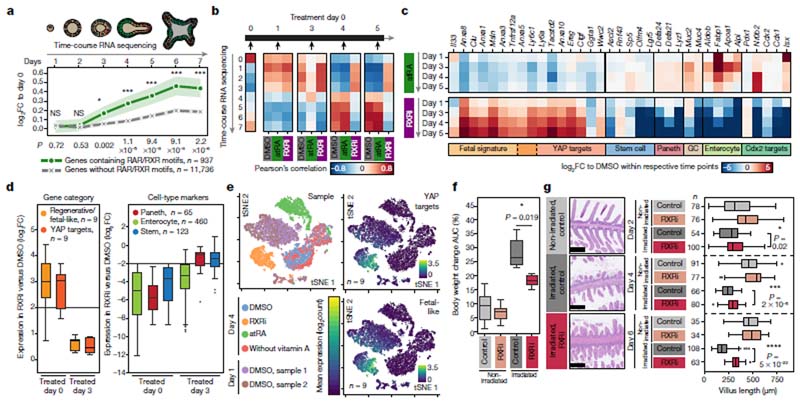
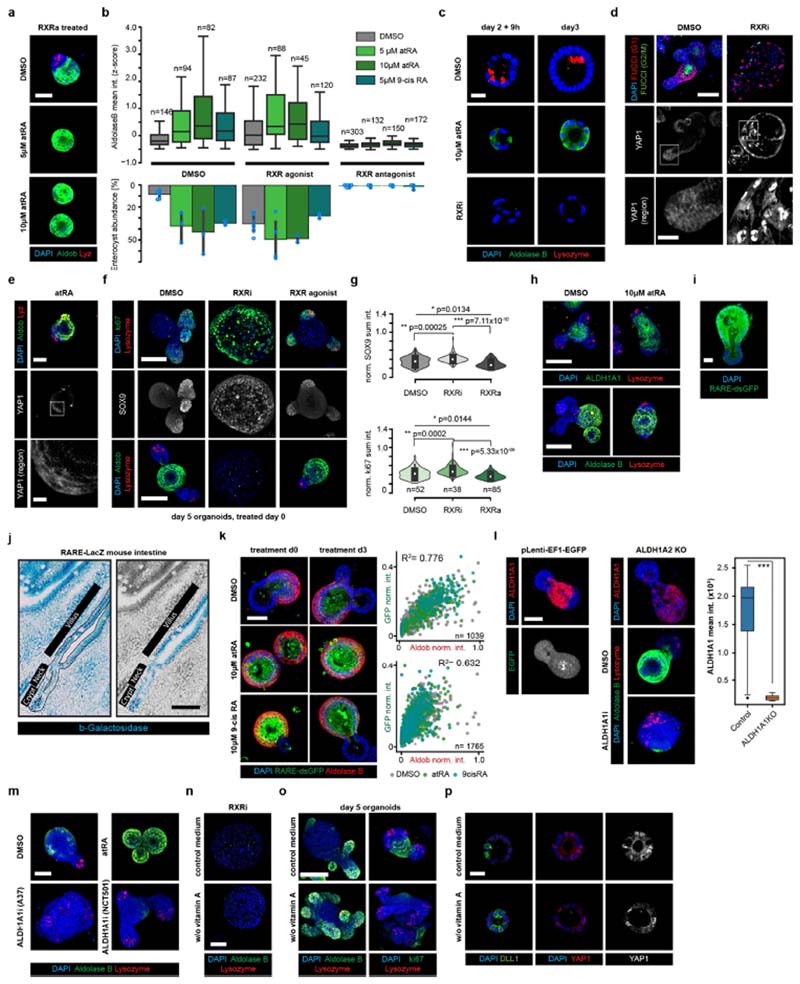
Fig. 2. Retinol metabolism and RXR activity in enterocyte differentiation.
a, Top, images of organoids treated at day 0 as indicated (DMSO, dimethyl sulfoxide; control; RXRi, treatment with RXR antagonist Cpd2170). Bottom, quantified aldolase B intensity. b, Colour composite images show antibody staining for DLL1 and YAP1 under different treatments at the indicated time points. Scale bars, 20 μm. D, DAPI. Greyscale images show YAP1 localization in the highlighted regions. Scale bars, 10 μm. c, Upper four images show antibody staining for lysozyme, aldolase B and YAP1 under different treatments at day 3. Scale bar, 50 μm. Bottom, YAP1 localization in highlighted regions. Scale bar, 10 μm. d, Retinol metabolism. Cyp26, cytochrome P450; RE, retinyl ester. e, Effect of knocking out Aldhlal on organoid development. Top, images show antibody staining for ALDH1A1; bottom, antibody staining for lysozyme and aldolase B. ALDH1A1i, ALDH1A1 inhibition with A37; pLenti-EF1-EGFP, infection and selection control. f, Left, day 4 organoids under different treatments (ALDH1A1i, day 0; atRA, day 3), in medium with (control) and without vitamin A. Right, quantified aldolase B intensity. Asterisks show statistical significance for the same treatment with/without vitamin A; two-sided t-test; NS, not significant; P values shown in the plot. a-c, e, f, Representative images of organoids (n = 6 independent replicates from n = 3 independent experiments), cultured from single cells. b, Day 2 plus 9 h: organoid middle plane, day 3 and 4: MIP of confocal z-stacks. Nuclei are stained with DAPI; scale bar, 50 μm. a, f, Violin ranges extended by standard deviations; box plots show quartile ranges; whiskers show value intervals; white dots show median values; n values refer to the number of individual organoids in conditions.
Twenty-nine genes were targeted by two or more compounds, resulting in highly similar phenotypic fingerprints. Some of these genes belong to signalling pathways that have previously been implicated in organoid development and homeostasis (such as Wnt, Notch and transforming growth factor-β (T GF-β) pathways), but others contribute to pathways that have not hitherto been ascribed to organoid development (such as nuclear receptor signalling; Extended Data Fig. 3a–f and Supplementary Table 1).
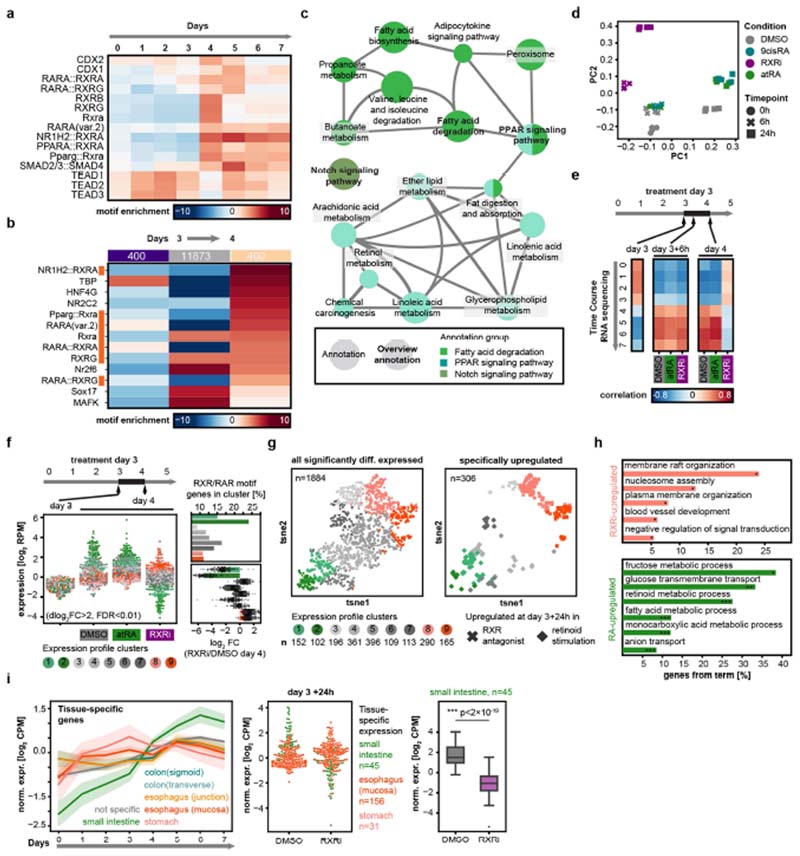
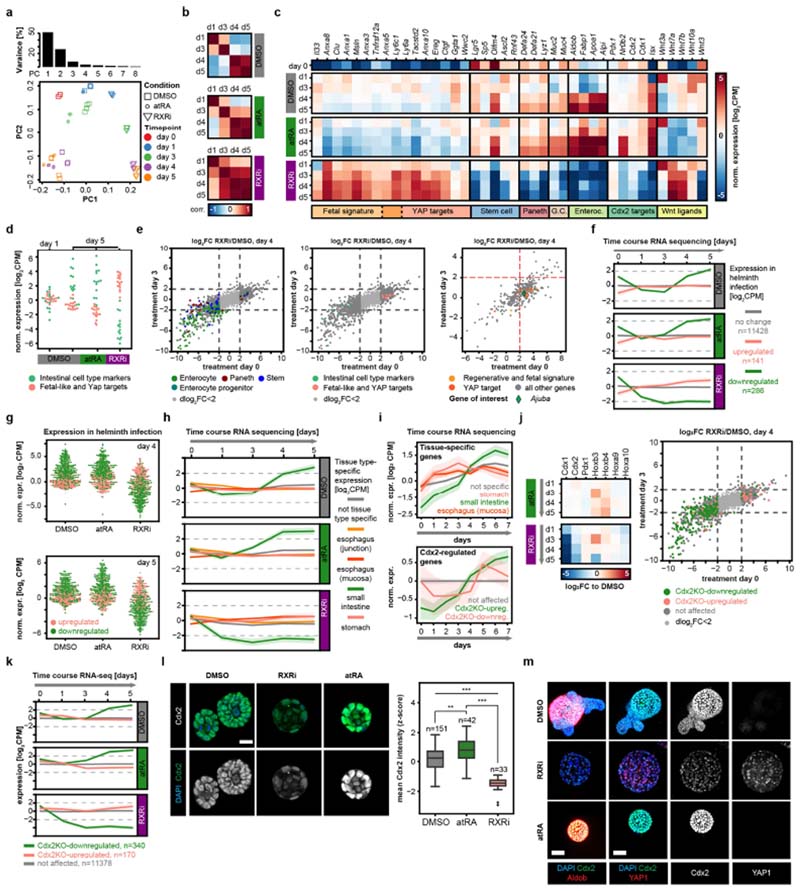
Fig. 3. RXRi treatment prevents exit from regenerative state and improves regeneration in vivo.
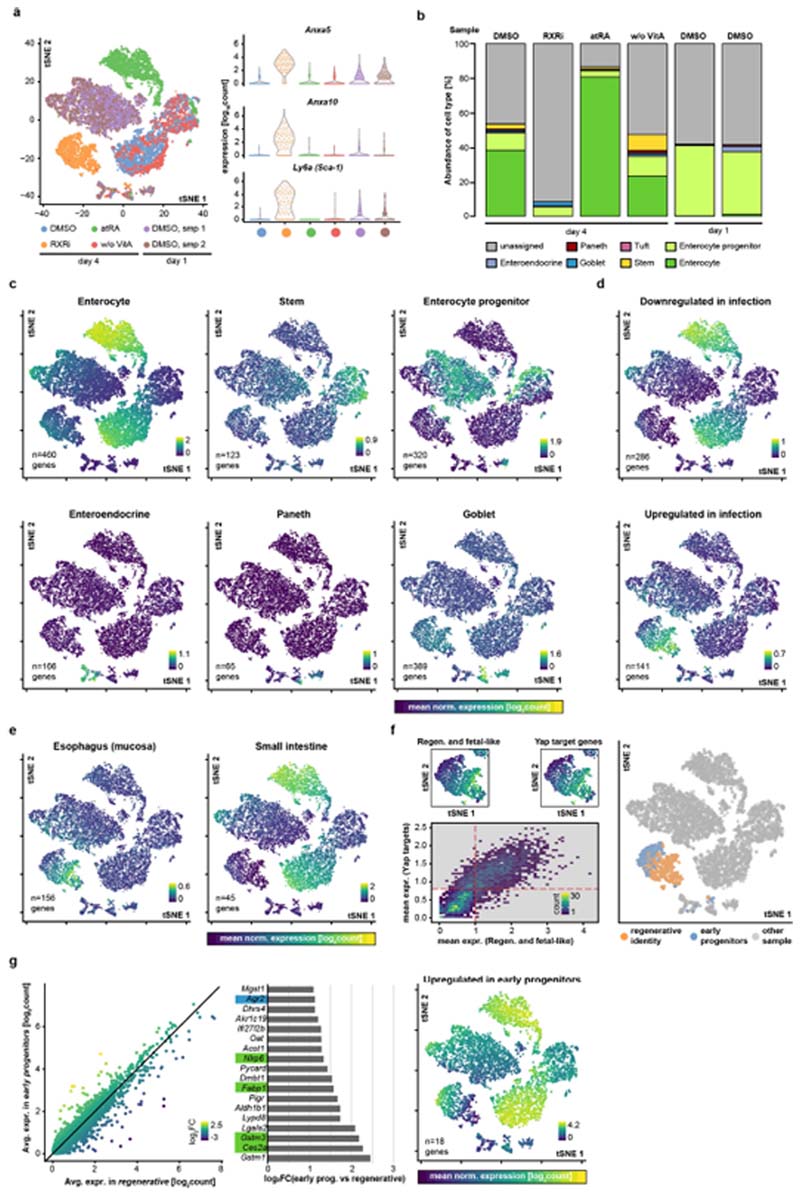
a, Temporal expression of genes containing RAR or RXR motifs, normalized to expression at day 0. Solid lines show mean values; opaque interval show standard deviation; n values refer to the number of genes per group. b, Pearson’s correlations between gene-expression levels in time-course and compound experiments. c, Differential gene expression in organoids relative to DMSO at indicated time points. GC, goblet cell. d, Fold change relative to DMSO (day 4) for specific gene categories and cell-type markers at days 0 and 3. Data points represent individual genes; horizontal lines show fourfold enrichment or depletion; n, number of genes. e, tSNE maps generated from single-cell RNA sequencing data, colour-coded by sample (left) and by mean normalized expression of gene groups (right). Data points show single cells; sampling of 5,000; n, number of genes; colour map ranges are shown in plots. f, Change in body weight (AUC, area under curve) in treatment cohorts; n = 6 mice per cohort. g, Quantification of villus length; n, number of villi; measured in n = 3 independent samples per condition. In the images, tissues are stained with haematoxylin and eosin. a, c, d, log2-transformed fold change (log2(FC)). b–d, n = 4 independent cultures from 2 independent mice. d, f, g, Boxes show quartile range; whiskers show value intervals; solid lines show median values. Two-sided t-test; asterisks indicate statistical significance; individual P values shown in plots.
Mapping functional interactions
Using the hierarchical interaction score (HIS)20,21 to investigate the phenotypic fingerprints of the 207 gene hits identified above, we inferred roughly 6,800 HIS interactions that are enriched in interactions reported by the Search Tool for Retrieving Interacting Genes/Proteins (STRING) database, and co-annotated gene pairs. We next determined the optimal HIS threshold in order to create a map of functional genetic interactions that underlie the development of intestinal organoids (Fig. 1b, Extended Data Fig. 4a–d and Supplementary Table 1).
Network connectivity identified upstream regulators and key players, including β-catenin (CTNNB1; Extended Data Fig. 4e), consistent with the crucial role of canonical Wnt signalling22,23. In addition, we identified six highly interconnected subnetworks (Fig. 1b) showing specific fingerprints and functional annotation enrichments. Interestingly, compounds that activate or inhibit Wnt signalling (GSK3B and PORCN inhibitors, respectively) both led to an increase in the abundance of symmetry-breaking-deficient phenotypes (Fig. 1b), but were assigned to different specific clusters, consistent with different phenotypic outcomes over time (Extended Data Fig. 2b).
For validation, we carried out functional studies of several identified target genes from different phenotypes: Psen1 (from ‘Paneth cell hyperplasia’), Casr (encoding a calcium-sensing receptor; from ‘Wnt hyperactivation’), Akt1 (from the ‘enterocyst’ phenotype) (Extended Data Fig. 4f–h) and Rxra (from the ‘regenerative’ phenotype; Fig. 2).
RXR controls exit from regenerative state
We focused on the ‘regenerative’ phenotype, observing the highest penetrance when using Cpd2170 (RXRi), an antagonist24 of retinoid X receptor (RXR)-α (Extended Data Fig. 4i, j and Supplementary Table 1). We confirmed that RXR inhibition induced a near-complete absence of enterocytes, whereas an RXR agonist (NRX 194204), all-trans retinoic acid (atRA) or 9-cis retinoic acid (9cis-RA), individually and in combination, increased enterocyte differentiation but could not rescue the RXRi-induced differentiation defect, hinting at a retinoic acid-independent role of RXR (Fig. 2a and Extended Data Fig. 5a, b). RXRi treatment at day 0 also resulted in a lack of Paneth cells, suggesting a symmetry-breaking defect (Extended Data Fig. 5c). To investigate the initial differentiation of Paneth cells, we turned to Notch signalling1 and YAP1 localization. In RXRi-treated organoids at day 2.5, we observed an absence of cells expressing the Notch ligand DLL1 and a strong homogenous nuclear retention of YAP1; by contrast, in atRA-treated organoids YAP1 was invariably localized to the cytoplasm (Fig. 2b). Therefore, both RXRi- and atRA-treated organoids exhibit a defect in symmetry breaking, accompanied by homogenous YAP1 localization. However, RXRi-treated organoids with nuclear YAP1 retained a regenerative YAP-dependent state1,25,26 and maintained an active cell cycle (Extended Data Fig. 5d), whereas atRA-treated organoids with cytoplasmic YAP1 underwent differentiation to enterocytes.
We then treated organoids after symmetry breaking, when YAP1 is predominantly inactive (day 3). RXRi-treated organoids lacked enterocytes, were larger and had more SOX9+ and Ki67+ cells, but displayed no change in Paneth cells (Fig. 2c). By contrast, atRA treatment resulted in an increase in the number of enterocytes (Extended Data Fig. 5e–g), but again no deficiency in Paneth cells. Interestingly, RXRi treatment at day 3 also did not result in nuclear translocation of YAP1 (Fig. 2c), suggesting that RXR is not involved in direct activation of YAP1, but rather controls its nuclear export, when the latter is already active.
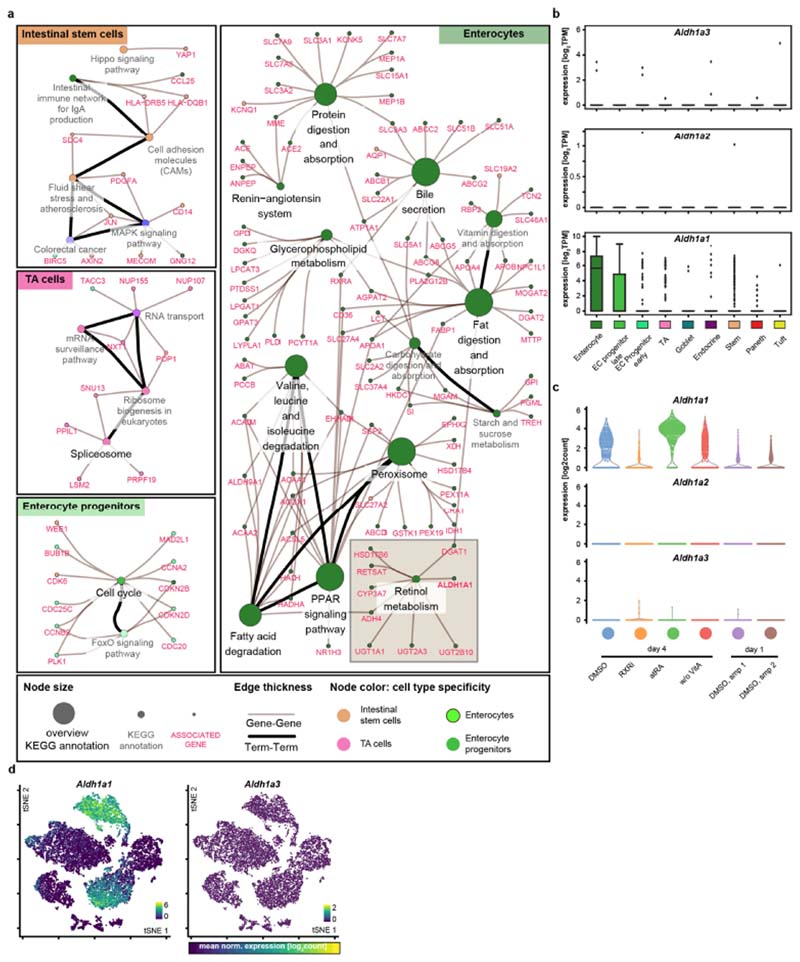
We then investigated vitamin A metabolism27,28 (Fig. 2d) and found that genes involved in retinoic acid metabolism were specifically expressed in enterocytes29,30, in particular the gene encoding the retinal- dehyde dehydrogenase ALDH1A1 (refs. 29,31,32; Extended Data Fig. 6a–d). In organoids, a higher ALDH1A1 abundance and upregulation of retinoic acid response element (RARE) motifs were exclusive to enterocytes; however, in vivo a subset of crypt cells also displayed endogenous retinoic acid activity (Extended Data Fig. 5h–k). Aldh1a1-knockout organoids and organoids cultured from single cells under ALDH1A1 inhibition or vitamin A depletion had fewer enterocytes and more cycling cells—characteristics that could be reversed by treatment with atRA (Fig. 2e, f and Extended Data Fig. 5k–n). Interestingly, organoids cultured in medium lacking vitamin A did not display perturbed YAP1 localization or symmetry breaking (Extended Data Fig. 5o). This suggests that retinoic acid metabolism is necessary for differentiation of enterocytes but not of Paneth cells (nor does it affect YAP1-expressing regenerative cells), and that enterocytes obtain regional specificity through ALDH1A1-controlled intracellular synthesis of atRA.
RXRi imposes a regenerative state
Retinoic acid receptor (RAR)-dependent signalling is important for enterocyte differentiation33, while RXR activation maintains the balance between regeneration and enterocyte differentiation (Fig. 2). We confirmed that predicted RXR/RAR target genes were induced at day 3 and were associated with enterocyte functions, and that organoids treated with RXRi at day 3 retained the day 3 transcriptional signature, whereas treatment with atRA upregulated enterocyte-specific genes (Fig. 3a, Extended Data Fig. 7a–h and Supplementary Table 2). Time-course RNA sequencing starting from single cells (Supplementary Fig. 1) showed that, whereas control organoids transition from an early regenerative state (at days 1–3) to a mature state (from day 4 onwards), RXRi-treated organoids retained the regenerative transcriptome and atRA-treated organoids matured more quickly. At the level of individual genes, in the RXRi condition, YAP targets and genes associated with a fetal-like regenerative state25 were strongly and progressively upregulated, whereas treatment with atRA caused a decrease in the regenerative signature and rapid enterocyte maturation (Fig. 3b, c and Extended Data Fig. 8a–d).
The regenerative signature and targets of YAP were significantly upregulated only in organoids treated from day 0 (Fig. 3d and Extended Data Fig. 8e), confirming the role of RXRi in maintaining the active state of YAP1, rather than in activating it. Interestingly, fetal and YAP target genes include Clu (expressed in regenerative cells), Anxa1 (an oesophageal gene) and Anxa10 (a gastric gene), suggesting that the cells have multiple regional identities (Fig. 3c). Consistent with this, enterocyte genes were downregulated in both cases, whereas marker genes of Paneth cells were downregulated only in early treatment (Fig. 3d and Extended Data Fig. 8e). Comparing RXRi treatment with helminth infection34, we observed suppression of the intestinal signature in both. By contrast, in the atRA condition, tissue specificity was acquired earlier (Extended Data Fig. 8f–h). Intestinal specification depends on tissue-specific transcription factors, such as Cdx2 and Pdx135,36. Cdx2-regulated genes decreased in expression at early time points and were re-expressed after day 3, as were intestine-specific genes. Cdx1 and Cdx2 expression was progressively downregulated in RXRi-treated organoids, whereas in the atRA condition and in enterocytes Cdx2 expression was increased (Extended Data Fig. 8i–m).
Finally, single-cell analysis revealed that day 4 organoids treated with RXRi resembled day 1 organoids, featuring upregulation of YAP targets and regenerative signature genes, such as Anxa5, Anxa10 and Sca-1 (Fig. 3e and Extended Data Fig. 9a), albeit at higher levels. Under RXRi treatment, most cells did not express intestinal markers and resembled the fetal-like state that follows helminth infection; by contrast, treatment with atRA promoted enterocyte differentiation (Extended Data Fig. 9b–e).
RXRi improves regeneration in vivo
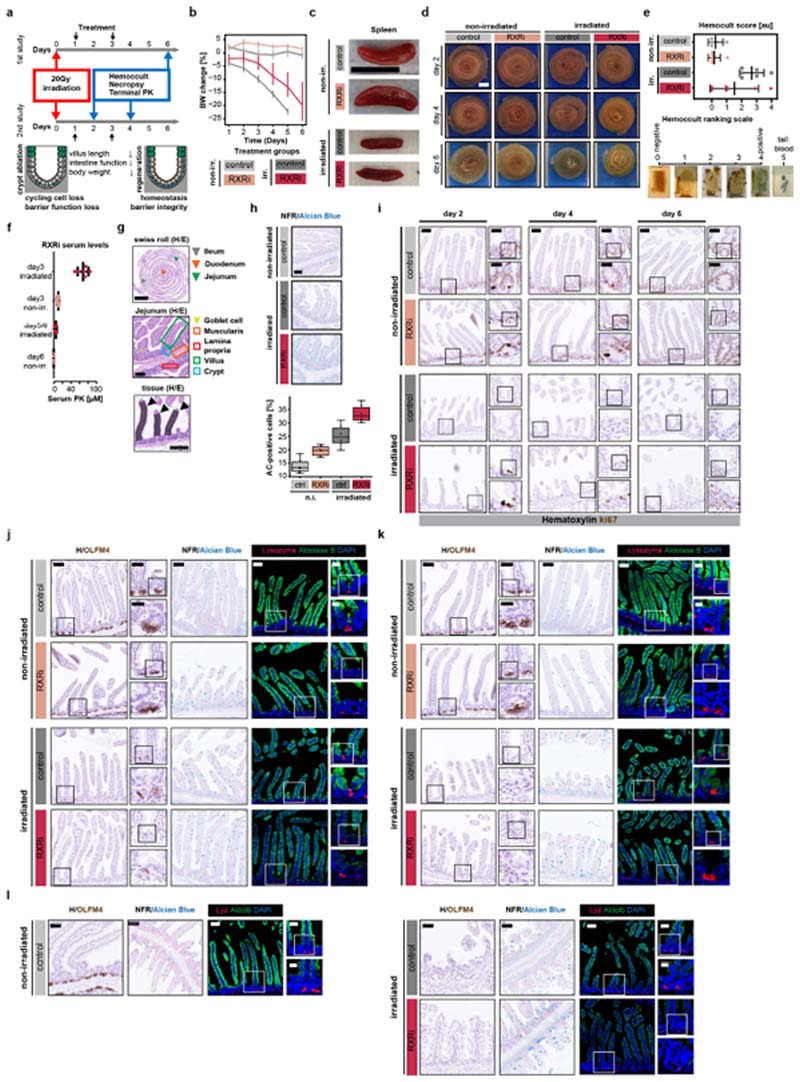
To validate the regeneration-promoting effect of RXRi, we devised an in vivo study that used a mouse model of irradiation-induced colitis, treating mice with RXRi at days 1, 2 and 3 and collecting tissue at days 2, 4 and 6 after γ-irradiation. Loss of cycling cells resulted in shorter villi and loss of barrier function, ultimately leading to weight loss. Treatment with RXRi significantly improved regeneration, resulting in reduced weight loss with no significant systemic effect on spleen size (Fig. 3f and Extended Data Fig. 10a–c). In irradiated mice, RXRi treatment resulted in less decellularization, improved barrier function and longer villi, particularly at later time points (Fig. 3g and Extended Data Fig. 10d–f). Analysis of proliferating, stem, Paneth and goblet cells and enterocytes (Extended Data Fig. 10g–l) showed an irradiation-induced increase in goblet cells. RXRi increased the abundance of goblet cells, drove a progressive accumulation of proliferating cells at the bottom of crypts, and improved crypt morphology. We did not, however, detect OLFM4+ stem cells (Extended Data Fig. 10j–l), probably owing to the longer time frames of crypt regeneration. These results validated the regenerative phenotype in vivo, suggesting transient inhibition of RXR signalling with RXR antagonists as a potentially useful therapy to improve regeneration of the intestine.
Discussion
Here we established an image-based screening platform with which to characterize the phenotypic landscape of organoid development from a single screen, generating what is, to our knowledge, the first map of functional genetic interactions that govern intestinal organoid development and self-organization.
We then described two phenotypes that emerge following RXRi treatment, depending on the organoid stage. In early organoids, inhibition of RXR maintains the regenerative state (with nuclear YAP1)1,37 and prevents symmetry breaking, possibly by controlling the nuclear export of YAP1. After symmetry breaking, RXR activity is restricted to canonical retinoic acid signalling in enterocytes and is linked to ALDHlAl-dependent production of atRA. Retinol metabolism is important for maintaining the balance of cell types between enterocytes and undifferentiated progenitors, in agreement with its role in restricting proliferation38.
During organoid formation, cells undergo regenerative reprogramming, and not only lose cell-type specificity but also transiently acquire a more ‘generic’ identity, with a tendency to misexpress genes from more anterior gastrointestinal tissues, similar to the anteriorization observed in ulcerative colitis39,40. RXR inhibition results in organoids that retain the expression of fetal-like and YAP target genes, and are unable to acquire mature cell types and undergo intestinal specification. We thus propose that RXR-mediated signalling acts as a ‘homing device’ upon exit from the regenerative state, ensuring activation of intestine-specific networks of transcription factors, including Cdx2 and its downstream targets. Indeed, Cdx2 knockout in intestinal organoids induces a transformation to gastric cells41. Later in organoid development, when organoids recapitulate homeostasis, RXR is crucial for retinoic acid signalling and enterocyte differentiation. The differences between the early regenerative and later differentiation-related phenotypes probably arise from the numerous heterodimers that RXR can form42.
Finally, we demonstrated the regenerative potential of transient RXRi treatment in vivo, observing an accumulation of cycling cells. Clearly, in vivo RXRi could have broader effects, affecting other components of RXR heterodimers (such as PPAR, FXR or LXR) or non-epithelial cells (such as immune cells43). In the future, this regeneration potential could be explored by combining the inhibitor with different diets44, such as vitamin A restriction. In summary, our work illustrates how a multivariate phenotypic screening approach in an emergent organoid system can be used to identify physiologically relevant targets that can be investigated in vivo, in this case unravelling the mechanisms of intestinal regeneration.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-020-2776-9.
Methods
Animal experiments and intestinal organoid lines
Experiments involving the irradiation-induced injury model were approved by the Internal Animal Care and Use Committee (IACUC) and conducted according to IACUC guidelines at the Genomics Institute of the Novartis Research Foundation. For the irradiation study, eight-week-old male C57BL/6J mice (Jackson Labs) were used in groups of six, as this is the smallest sample size needed to achieve a significant difference, based on prior model validation. Only males were used, as they are predicted to be more resilient in this model than females, with a steadier loss of body weight and less reproductive tissue covering the intestine; use of females could affect the penetrance of the γ-irradiation dose, introducing more variability. All mice were randomized or assigned groups on a cage-by-cage basis, based on the animals’ body weight on the first day of study. It was not possible to randomize on a per-mouse basis (that is, individually) as males will fight if not housed with brothers. The animals were eight weeks old (approximately 25 g) at the start of the experiments, as this size and weight provide the best fit to the dimensions of the shielding device used for irradiation to induce injury. No blinding was performed; the researchers knew which mice were in which group during data analysis. All other animal experiments were approved by the Basel Cantonal Veterinary Authorities and conducted in accordance with the Guide for Care and Use of Laboratory Animals. Male and female outbred mice between 7 and 15 weeks old were used for all other experiments. Mouse lines used were wild-type C57BL/6 (Charles River Laboratories and Jackson Labs), Fucci2 (a gift from J. Skotheim, Stanford University, CA) and Cdx2–EGFP (a gift from K. McDole, MRC LMB, Cambridge, UK).
Organoid culture
Organoids were generated from isolated crypts of mouse small intestine as described12. Organoids were kept in IntestiCult organoid growth medium (Stem Cell Technologies) with 100 μg ml-1 penicillin–streptomycin for amplification and maintenance. For a detailed description, see Supplementary Methods.
Image-based screening assays
For a detailed description, see Supplementary Methods. In brief, organoids were collected 5–7 days after passaging and digested with TripLE (Invitrogen) for 20 min at 37 °C. Isolated cells were collected in ENR medium, composed of advanced DMEM/F-12 with 15 mM HEPES (Stem Cell Technologies) supplemented with 100 μg ml-1 penicillin–streptomycin, 1x Glutamax (Thermo Scientific), 1x B27 (Thermo Scientific), 1x N2 (Thermo Scientific), 1 mM N-acetylcysteine (Sigma), 500 ng ml-1R-Spondin (a gift from Novartis), 100 ng ml-1 Noggin (PeproTech) and 100 ng ml-1 murine epidermal growth factor (EGF; Thermo Scientific). Resuspended cells were mixed with Matrigel (Corning) in a medium to a Matrigel ratio of 1:1. After 10 min of solidification at room temperature, 40 μl of medium containing 5,000 cells was overlaid per well. From day 0 to day 3, WENRC medium (ENR medium supplemented with 20% Wnt3a-conditioned medium (Wnt3a-CM), 10 μM ROCK inhibitor Y-27632 (Stem Cell Technologies) and 3 μM GSK-3β inhibitor CHIR99021 (Stem Cell Technologies, catalogue number 72054)) was used for organoid culture. From days 3 to 5, ENR medium was used for organoid culture.
Our compound library (a gift from Novartis) was composed of 2,789 compounds (Supplementary Table 1) in the form of 1 mM solutions in DMSO. Every library plate contained 12 active controls (6 wells containing the γ-secretase inhibitor DAPT and 6 wells containing the GSK-3β inhibitor CHIR99021 (Stem Cell Technologies, catalogue number 72054) and 12 vehicle controls (DMSO). Compound treatments were performed from day 0 by adding 10 μl of 25 μM intermediate dilutions of the compound library in WENRC medium to achieve a final concentration of 5 μM. Compound treatment was repeated at day 3 by using compound library dilutions in ENR, prepared as above.
Treatment of intestinal organoids with compounds
In the indicated experiments, we used the following compounds: ALDH1A1 inhibitors A37 (Tocris, catalogue number 5802) and NCT- 501 (Tocris, catalogue number 5934); RXRA agonists NRX 194204 (Axon Med Chem, catalogue number 2408) and all-trans retinoic acid (Sigma, catalogue number R2625); RXR-α/RAR-α (RXRA/RARA) agonist 9-cis-retinoic acid (Sigma, catalogue number R4643); RXR antagonists Cpd2170 (ref. 24; a gift from Novartis) and HX 531 (Tocris, catalogue number 3912).
RXR antagonist
The RXR antagonist (Cpd2170) identified in the screen and used for follow-up studies (Figs. 2, 3 and Supplementary Table 2) was a gift from Novartis. The molecule is a diazepinylbenzoic acid derivative (International Union of Pure and Applied Chemistry (IUPAC) name 4-(2-acetyl-5-ethyl-7,7,10,10-tetramethyl-8,9-dihydronaphtho(2,3-b)(1,5) benzodiazepin-12-yl)-3-fluorobenzoic acid; ChEMBL ID CHEMBL395962). It was synthesized using hydroxy-diazepinylbenzoic ester as a precursor for an acetyl analogue by palladium coupling24. The compound was tested for its ability to inhibit the transactivation of human RXR-α in order to determine its half maximal inhibitory concentration (IC50) values24, as follows: inhibition of 9-cis-retinoic acid-induced RXR-α transactivation, 3 μM (BioAssay AID 292628); inhibition of LG-100268-induced RXR-a transactivation, 3.3 μM (BioAssay AID 292627).
Experiments in fixed organoid samples
Organoids were collected 5–7 days after passaging and digested with TripLE (Invitrogen) for 20 min at 37 °C. Dissociated cells were passed through a cell strainer with a pore size of 20 μm. For indicated experiments, single living cells were sorted by fluorescence-activated cell sorting (FACS; Becton Dickinson FACSAria cell sorter). Forward scatter and side scatter properties were used to remove cell doublets and dead cells. Single cells were derived from wild-type C57BL/6 organoids unless indicated otherwise. Resuspended cells were mixed with Matrigel (Corning) in a medium to Matrigel ratio of 1:1, plated in 96-well plates (Greiner, catalogue number 655090) in 3.5 μl droplets and exposed to the indicated compounds. All compound stocks were prepared in DMSO, and DMSO was used as a vehicle control. Organoids were treated and fixed at the indicated time points to generate samples for immunofluorescence imaging.
Organoid culture for RNA sequencing
Wild-type C57BL/6 organoids were collected 5 days after passaging and digested with TripLE (Invitrogen) for 20 min at 37 °C. Experiments were performed in duplicate for two independent biological samples (organoid cultures produced from two C57BL/6 male littermate mice). Dissociated cells were passed through a cell strainer with a pore size of 20 μm. Single living cells were sorted by FACS (Becton Dickinson FACSAria cell sorter). Forward scatter and side scatter properties were used to remove cell doublets and dead cells (Supplementary Fig. 1). Resuspended cells were mixed with Matrigel (Corning) in medium to a Matrigel ratio of 1:1, and plated in 24-well plates in 50 μl droplets. From day 0 to day 3, WENRC medium (described above in the section ‘Image-based screening assays’) was used for organoid culture. Organoids were exposed to the indicated compounds either at day 0 or at day 3 in respective assays, concomitant with the switch to ENR medium. Organoids were lysed at the indicated time points and used for RNA extraction and sequencing as described in Supplementary Methods.
Generation of organoid lines
To generate organoids expressing retinoic acid response elements (RAREs) tagged with green fluorescent protein (GFP), we infected wild-type C57BL/6 organoids with in-house-produced pGreenFire 1–RARE viral particles (System Biosciences, catalogue number TR037PA-1) or, as a control, with pEGIP (Addgene, plasmid number 26777) at 0 h. For a detailed explanation of single guide RNA (sgRNA) design in CRISPR–Cas9-mediated gene-knockout experiments and the production of lentiviral particles, see Supplementary Methods.
Organoid culture in medium without vitamin A
In the indicated experiments, organoids were cultured in medium with or without vitamin A. To prepare the medium, 1x B27 minus vitamin A (Thermo Scientific) was used instead of 1x B27 (Thermo Scientific) to prepare both ENR and WENRC media. To allow proper comparison, in these experiments, B27 minus vitamin A (Thermo Scientific) was supplemented with retinyl acetate (Merck, catalogue number 46958) corresponding to the standard B27 supplement in order to produce medium for control conditions.
Preparation and imaging of fixed samples
To allow imaging of all organoids within a similar z-range, we centrifuged the contents of each well plate at 3,000 rpm for 10 min in a pre-cooled centrifuge at 10 °C before fixation. Organoids were fixed at indicated time points in 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in phosphate-buffered saline (PBS) for 45 min at room temperature.
For image-based screening assays, organoids were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 1 h and blocked with 3% fetal calf serum (FCS; Sigma-Aldrich) in PBS with 0.1% Triton X-100 for 1 h. Primary and secondary antibodies were diluted in blocking buffer and applied as described in Supplementary Table 3. For detailed procedures, see Supplementary Methods.
In all samples, high-throughput imaging was carried out using an automated spinning disk microscope from Yokogawa (CellVoyager 7000S). For imaging, an intelligent imaging approach was used (‘search first’ module of Wako Software Suite, Fujifilm Wako Automation Corporation). For detailed procedures, see Supplementary Methods.
Primary antibody labelling
Anti-lysozyme antibody (EC3.2.1.17, Dako) was labelled with CF568 fluorophore using a labelling kit, Mix-N-Stain (Biotium), according to the manufacturer’s instructions.
Image analysis and extraction of features
Organoid segmentation in maximum intensity projections (MIPs).
For each acquired confocal z-stack field, MIPs and sum intensity projections (SIPs) were generated from all acquired z-planes per field. All MIP fields belonging to the same well were stitched together to obtain MIP well overviews for each channel. The high-resolution well overviews were used for organoid segmentation and feature extraction. Each individual organoid was automatically segmented on the basis of either its CellTrace signal (image-based screening assay) or its DAPI signal (all other imaging assays). To allow precise segmentation, clumped objects were separated using edge information (Canny edge detection algorithm) and subsequent watershedding with imposed minima.
Features in MIPs. For each segmented organoid, a total of 34 features, describing shape and intensities for each acquired channel, were extracted. A further 60 features describing Zernike polynomials were extracted and used for object filtering as described below. MIPs were used to describe fluorescence distribution and the morphological features of organoids. A subset of features was selected for multivariate feature analysis on the basis of information content and covariance as described below.
Features in SIPs. SIPs were used to extract features relating to total intensity per object to allow accurate quantification. In the image-based screen, SIP projections were used to extract all features for the segmented objects.
Feature selection for image analysis
Extracted features were analysed for covariance to exclude highly correlated (and anticorrelated) features; for details, see Supplementary Methods. In all imaging experiments, extracted features were normalized using z-score normalization within respective assay plates.
Detection of segmentation artefacts by SVMs
A linear support vector machine (SVM) was trained on a dataset of 100 randomly picked individual organoids with correct segmentation and 100 organoids with observed segmentation artefacts. Features used for SVM prediction consisted of the 9 features used for PhenoGraph analysis, and also included 60 Zernike polynomials.
Filtering of sparse conditions
For image-based screening assays, conditions with fewer than ten organoids detected per well were discarded from the analysis. In other assays, the threshold level for sparse conditions was assessed on an assay-to-assay basis; in general, all conditions with less than 20% of the mean organoid count in the given assay were discarded.
Generation of phenotypic signatures
Phenotypic clustering was carried out using the entire dataset (402,930 organoids) with the feature set defined in Extended Data Fig. 1, using the software package PhenoGraph (MATLAB implementation; https://github.com/dpeerlab/cyt3) as above. The abundance of every phenotypic class was calculated as the fraction of organoids belonging to the class in every individual condition. Abundance was z-score-transformed within assay plates to minimize plate effects. For detailed procedures, see Supplementary Methods.
Hit selection
Individual treatment conditions were ranked by reproducibility between replicates. The reproducibility score was defined as the correlation coefficient for 15-element phenotypic fingerprints of respective conditions. Conditions with a reproducibility score of more than 0.5 and a z-score value for phenotypic class abundance of more than 1.5 or less than -1.5 for any of the classes were included in the hit list. Conditions with a z-score value for phenotypic class abundance of more than 4 or below -4 for any of the classes were selected for validation.
Hit validation with randomization trials
To calculate statistical parameters, we carried out a randomization trial, reshuffling the phenotypic class labels randomly between the 400,000 organoids in the dataset. Cluster cardinalities were kept, as the pool of class labels has not been altered. The z-scored abundances of phenotypic classes in every condition were then calculated from 1,000 trials to estimate the probability that a given condition would reproduc-ibly (with correlation between replicas used as the cutoff parameter) present a significant phenotypic change after level permutation. For detailed procedures, see Supplementary Methods.
Target-gene-enrichment score
To ensure that the hit list was not prone to contain genes targeted by a high number of compounds in the initial library, we calculated a target-gene-enrichment score (5enr) as described in Supplementary Methods.
HIS calculations
Hierarchical interaction scores were calculated as described21 using the 15-element phenotypic fingerprints as inputs. The resulting interaction matrix was used to infer edges for generation of a network of phenotypic interactions. For further analysis, only those genes that were connected by edges with HIS values of more than 0.2 were kept.
Subnetworks were identified using the Cytoscape implementation of the ClusterOne algorithm45. For visualization purposes, only edges with the highest HIS value per node were kept.
STRING validation of HIS predictive power
We used a prediction model, calculating the predicted number of edges for a set of genes that are retained when a sliding HIS threshold is applied (Extended Data Fig. 4b). For the detailed procedure, see Supplementary Methods.
Co-annotation analysis
A progressive HIS threshold with steps of 0.005 was applied to the list of inferred HIS interactions, to eliminate those genes that are not connected by HIS interactions at a given threshold. The optimal value for the HIS threshold was calculated by using the percentage of gene pairs connected by retained HIS interactions that are co-annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG) or Gene Ontology (GO) terms. The resulting first infliction point (defined as the absence of a gene-pair dropout between two subsequent steps of the sliding threshold) was used as the HIS threshold for generating the functional interaction network (Fig. 1b).
Annotation enrichment analysis
Annotation enrichment analysis was performed using the ClueGo plugin for Cytoscape46. The enrichment of KEGG functional annotations (Homo sapiens and Mus musculus KEGG pathways, version of 1 March 2017) was calculated against the corresponding background (all detected genes for RNA-sequencing (RNA-seq) experiments, and a list of the unique targets of the compound library (Supplementary Table 1) for the image-based screen).
Annotation enrichment analysis in HIS network
For annotation enrichment analysis in the network of HIS interactions (Fig. 1), the minimal number of genes was adjusted according to subnetwork size in the range two to four genes.
Annotation enrichment analysis in RNA-seq data
For annotation enrichment analysis using the RNA-seq dataset (Fig. 3), a minimal number of three genes was used, and only those annotations with an enrichment P-value (two-tailed hypergeometric test with Bon-ferroni correction) of less than 0.05 were included.
RNA purification for bulk RNA sequencing
RNA was isolated using a single-cell RNA purification kit (Norgen Biotek Corporation, catalogue number 51800), pooling 3 wells of organoid culture from 24-well plates (wild-type C57BL/6 background). Organoids were seeded as single cells at 50,000 cells per well and cultured as described above. Organoids were treated with compounds at either day 0 or day 3 in the relevant experiments. RNA purification was performed in duplicate for two organoid cultures (wild-type C57BL/6, male littermate mice) for all treatment conditions. A step of DNase treatment was included (RNase-free DNase I kit, Norgen Biotek, catalogue number 25710) for all samples.
Bulk RNA sequencing and raw data processing
RNA-seq libraries were prepared using the TruSeq Illumina messenger RNA library preparation kit, and sequenced using the Illumina HiSeq2500 platform. For detailed procedures and differential expression analysis, see Supplementary Methods.
Single-cell RNA-seq and raw data processing
Single cells were isolated from organoids (wild-type C57BL/6 background, male littermate mice) at the indicated time points, passed through a cell strainer with a pore size of 30 μm and used for FACS to discard debris and dead cells. Cellular suspensions were loaded on a 10x Genomics Chromium single-cell instrument to generate single-cell gel beads in emulsion (GEMs). For detailed procedures and differential expression analysis, see Supplementary Methods.
Analysis of single-cell RNA-seq
tSNE maps were generated from normalized read counts for dimensionality reduction and used to display the expression levels of gene categories in single cells from the samples included in the single-cell RNA-seq experiment. Cell-type identities of single cells were defined on the basis of mean expression levels of cell-type marker genes29.
Analysis of transcription-factor-binding motifs
Analysis of transcription-factor-binding sites was performed using HOMER (version 4.8; http://homer.ucsd.edu/homer/motif/) as described47. For detailed procedures, see Supplementary Methods.
Correlation with published datasets
For cross-correlation studies with published RNA-seq datasets, raw data were obtained from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) repository: helminth infection in mouse intestine34, GEO accession number GSE97405; time course of organoid development1, GEO accession number GSE115955; Cdx2-knockout organoids41, GEO accession number GSE62784. Data were processed from read counts as above.
To identify tissue-specific genes, we selected all genes from the GTEx repository (https://gtexportal.org/), mapping the human genes to their respective mouse orthologues. Analysis was performed as in the Supplementary Information.
Irradiation-induced injury and RXRi dosing
Eight-week-old male C57BL/6J mice (Jackson Labs) were weighed and divided into groups on day 0. Mice were exposed to 20 Gy γ-irradiation. For detailed procedures, see Supplementary Methods.
Tissue collection and histology
Mice were euthanized using an overdose of isoflurane and cervical dislocation on either day 2 (3 h post final dose of RXR antagonist, 3 total doses), day 4 (16 h post final dose, 6 total doses), day 5 (40 h post final dose, 6 total doses) or day 6 (65 h post final dose, 6 total doses). Small intestines were collected and cleaned using cold Hank’s balanced salt solution (HBSS; Hyclone) and fixed in a Swiss roll conformation with 10% neutral buffered formalin in PBS (Avantik) for 48 h at room temperature before transfer into 70% ethanol. Small-intestine Swiss rolls were embedded in paraffin (Sakura Tissue Tek) and 5-μm longitudinal serial sections were cut using a Leica RM2255. For detailed procedures, see Supplementary Methods.
Immunohistochemical imaging of tissue samples
The 5-μm longitudinal serial sections of small-intestine Swiss rolls embedded in paraffin (described above) were mounted on glass slides and used for deparaffinizing with UltraClear reagent. Deparaffinized samples were used for sodium-citrate-based antigen retrieval. Stained slides were mounted with precision coverslips using ibidi mounting medium (ibidi) and used for imaging with a high-throughput confocal microscope as above to generate MIPs from confocal z-stacks. For detailed procedures, see Supplementary Methods.
Quantification of villus length
Individual villi were measured in ImageJ in jejunal regions of the histologically stained longitudinal serial sections of small-intestine Swiss rolls. For every condition, at least three regions from at least two mice were used per time point and per treatment.
Statistics and reproducibility
The investigators were not blinded to allocation during experiments and data analysis. The allocation of wells for experimental conditions was randomly assigned. All statistics were calculated on the basis of independent replicates, unless stated otherwise in figure legends. Statistical significance was determined by two-sided t-test, unless stated otherwise in figure legends. No statistical methods were used to predetermine sample size. Experiments were repeated at least twice, except the following experiments, which were carried out once: the image-based screen was performed once in two independent replicates from two independent mice; bulk and RNA-seq time courses were performed in n = 4 individual replicates from n = 2 independent mice; the in vivo irradiation time course was performed in cohorts of n = 6 independent mice per treatment condition. In vivo irradiation study, performed with a single terminal time point (day 6), was repeated twice with similar results.
Extended Data
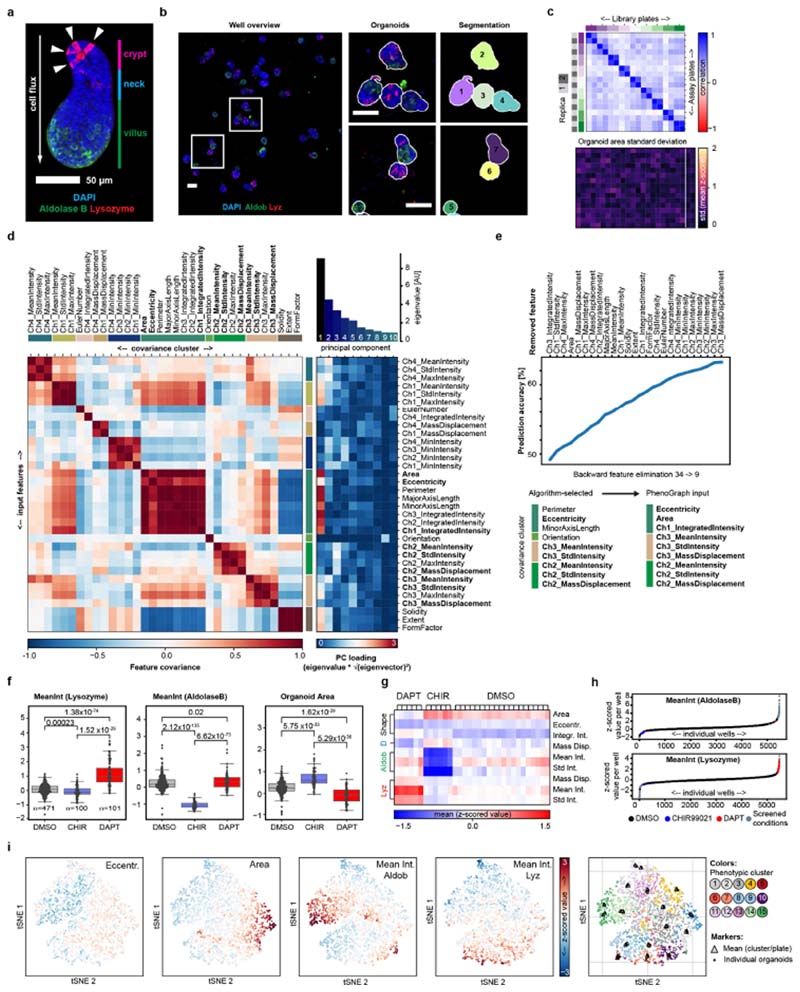
Extended Data Fig. 1. Reproducibility of the image-based screen.
a, An example image (MIP of a confocal z-stack) of an organoid, with annotated regions along the major axis, and showing antibody staining for lysozyme (Lyz) and aldolase B (Aldob). Lysozyme-positive Paneth cells are indicated by white arrowheads. Scale bar, 50 μm. b, Left, an example image of a stitched well-level overview. Scale bar, 100 μm. Middle, enlarged regions from the left panel. Scale bar, 100 μm. Right, segmentation masks, with object unique identifier numbers overlaid. a, b, Representative images of the DMSO control condition, repeated in n = 471 independent replicates with similar results. c, Top, reproducibility of detected organoid counts in replicas of the library plates. Heat map of Pearson’s correlation of organoid counts in screen replicates, with plate replicates grouped by parent library plates as indicated; n = 384 wells per plate. Bottom, heat map of standard deviation of organoid size (z-scored organoid area) for respective well positions. Passive and active controls are in wells outlined by the white box. n = 18 independent plates. d, Left, covariance matrix of the 34 extracted features, clustered by covariance (c-means clustering, ten clusters). Ch, channel; Stdlntensity, standard deviation of intensity. Right, heat map showing principal component loading of the extracted features; bar chart above, eigenvalues of first ten principal components. e, Top, changes in the accuracy of the naive Bayes predictor throughout the backwards-feature- elimination test. Bottom, final set of nine features (‘Algorithm-selected’) and list of features used as input for clustering with PhenoGraph. f, Distribution of mean values per well of the indicated features in active and passive control conditions. CHIR is the GSK3B inhibitor CHIR99021; DAPT is a γ-secretase inhibitor. Centre line, median; boxes, quartile range; individual data points overlaid. Two-sided t-test, with P-values shown in the plot. MeanInt, mean intensity. n = 471, 100 and 101 independent samples for the DMSO, CHIR99021 and DAPT conditions respectively. g, Z-scored organoid features used for clustering, shown for individual well positions of in-plate replicates of active and passive control conditions. n = 6, 6 and 20 wells per plate for DAPT, CHIR and DMSO respectively; shown are mean values over n = 18 assay plates. D, DAPI; Eccentr., eccentricity, Integr. Int., integrated intensity; Mass disp., mass displacement for the indicated immunofluorescence channels; Std Int., standard deviation of intensity. h, Mean z-scored values for the indicated features per well from the entire dataset, sorted in ascending order. Every data point is an average value per well, with conditions colour-coded as in the key. n = 5,250 independent samples measured in the screen. i, tSNE maps colour coded for z-scored values of indicated features (left to right: eccentricity, organoid area, mean intensity of aldolase B and mean intensity of lysozyme antibody staining) and PhenoGraph classification. Every data point represents an individual organoid, with sampling stratified by PhenoGraph assignment. The sample size is 5,000 individual organoids.
Extended Data Fig. 2. Robust phenotypic signatures.
a, Top, phenotypic clusters and phenotypic groups (see Fig. 1b) identified using the multivariate feature array, and heat map of mean z-scored values per class for the indicated feature array. Aldob, aldolase B; Ar, area; D, DAPI; Ecc, eccentricity; II, integrated intensity; Lyz, lysozyme; MD, mass displacement; MI, mean intensity; SI, standard deviation of intensity; Sh, shape. n = 40,624, 34,265, 20,134, 31,687, 2,549, 702, 23,643, 48,992, 30,791, 21,342, 23,610, 32,773, 19,172, 41,931 and 29,939 organoids for classes 1-15 respectively. Bottom, phenotypic class assignment for organoids in control conditions; outer circle colour, phenotypic group; inner circle colour, assigned phenotypic cluster; n = 7367, 7982 and 33395 individual organoids for DAPT, CHIR99021 (CHIR) and DMSO conditions respectively. b, Potential loss of phenotypic resolution owing to phenotype grouping. Three phenotypic clusters are assigned to the ‘regenerative’ phenotype, but the exact 15-element phenotypic signature differs depending on whether PORCN or GSK3β is inhibited. Mean of n = 100, 4, 2 and 2 independent replicates for CHIR99021, Cpd2170, Cpd420 and Cpd1772 respectively. c, Top, relative z-score transformed abundances of 15 phenotypic clusters, shown as means ± s.d. Bottom, heat maps showing individual and mean phenotypic signatures for 50 replicates of the indicated control conditions. n = 50 sampled independent replicates for all 3 conditions. d, Left, randomization trial for reshuffling of phenotypic class labels in the entire dataset. Right, scatter plots and histograms showing the distribution of replica correlation and maximum class variance in permuted and non-permuted data for the selected and filtered conditions. Red dashed lines show the original selection stringency. e, Distribution of calculated P-values in the indicated groups of conditions. Top, scatter plot and kernel density estimation; bottom, box plots. Boxes, quartile range; whiskers, value interval with excluded outliers; solid lines, median values. n = 2,242 unique conditions corresponding to 5,204 individual wells. n = 279, 15 and 1,948 conditions for ‘hit’, ‘hit (one replica)’ and ‘not selected’ groups respectively. f, Top left, mean z-scored values per identified phenotypic class for the indicated feature array. Bottom left, heat map representation of the phenotypic signature for the indicated compounds, grouped by the most abundant phenotype in the original screen. Top right, tSNE map colour coded for the highest enriched phenotypic clusters. Every data point represents a compound. tSNEs were calculated from phenotypic signatures. Bottom, calculated reproducibility values per compound and numbers of replicates used for analysis of the indicated compounds. The replica count is the number of non-sparse (n > 10 organoids) independent wells recorded per condition. Assay performed in n = 8 independent replica from 2 independent organoid cultures. g, Flowchart depicting filtering of the screened conditions. h, tSNE maps colour coded for the highest enriched phenotypic clusters as indicated; every data point represents either a compound condition (top; n = 301 compound conditions) or a unique target gene-modality pair (bottom; n = 207 target genes); tSNEs calculated from 15-dimensional phenotypic signatures.
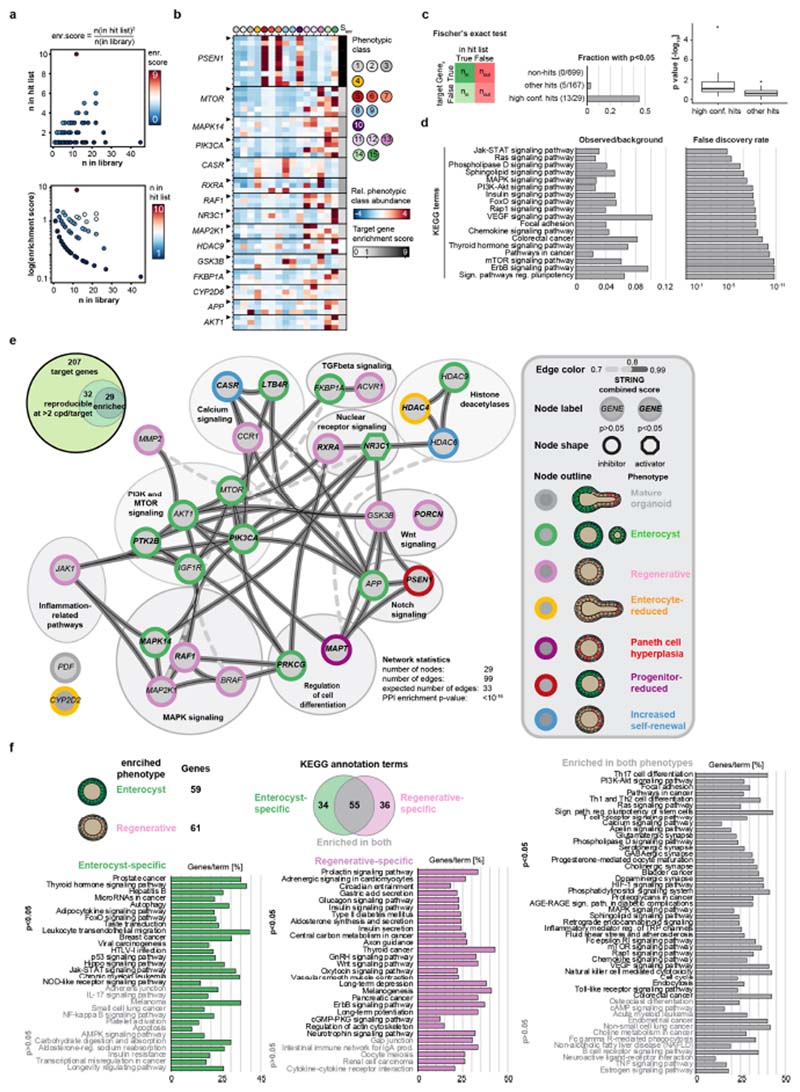
Extended Data Fig. 3. Identification of hit compounds and target genes.
a, Top, compound abundance in the library and hit list. Bottom, distribution of enrichment scores for targets of the screen hits. Every data point represents a unique target-gene–modality pair. b, Heat map showing phenotypic signatures for genes with a compound coverage of at least three compounds per gene. Heat map rows marked by arrowheads show compounds used as representative conditions per target. Senr, target enrichment score (see a). c, Left, contingency table for two-sided Fischer’s exact test; right, bar plot and box plot of P-value distributions in the indicated compound condition groups. High conf. hits, high-confidence hits. In the box plot, boxes show quartile ranges; whiskers show value intervals with excluded outliers; centre lines show median values. n = 699, 167 and 29 unique target genes for non-hits, other hits and high-confidence hits respectively. d, Enrichment and false discovery rate for functional annotation enrichments reported by the STRING database tool (https://string-db.org/) for the list of 29 target genes (e). e, Top left, Venn diagram showing the distribution of target genes by compound coverage. Main panel, network of functional interactions for the set of genes with highcompound coverage and hitlist specific enrichment. Edges show interactions known in the STRING database; thickness and line type represent the STRING combined score (see network key); nodes show unique target genes; node outlines show phenotypic clusters with the highest enrichment; circles indicate groups of similar functional annotations. f, Enrichment of KEGG terms (percentage of genes annotated with KEGG terms) in conditions from the hit list where the highest enriched phenotype is either ‘regenerative’ or ‘enterocyst’. Values are sorted by corresponding P-values (two-sided test based on hypergeometric distribution with Bonferroni correction). n = 59 and 61 genes for the enterocyst and regenerative phenotypes, respectively.
Extended Data Fig. 4. Validation of HIS predictive power and screen hits.
a, Principle of the hierarchical interaction score (HIS). HIS is calculated pairwise for a set of genes described by multivariate phenotypic signatures (left), to generate a matrix of graded interactions (middle) in which every interaction contains information on which readout contributed the most. In the resulting HIS-based network (right), the strength of the interactions (the thickness of the edges) reflects the similarity of the observed enrichment or depletion, and directionality is from the gene with the broader set of effects to the gene with the narrower set of effects. b, Heat maps of the HIS pairs for n = 207 target genes; interactions with non-zero HIS values are shown colour-coded for the phenotypic class that contributes the most to HIS inference (left) or for the HIS value (right). c, Top left, number of retained nodes; bottom left, number of STRING-reported and predicted edges (see Supplementary Methods); centre, fold enrichment of reported STRING interactions over prediction; right, percentage of co-annotated gene pairs with increasing HIS threshold cutoff (top right, KEGG co-annotations; bottom right, GO co-annotations (levels 5–15)). Red dashed line, HIS threshold (0.175); arrowheads, inflection points. Top and bottom right plots: blue line, polynomial fit. d, Percentage of coannotated gene pairs in indicated sets of genes in measured and randomized data (top, KEGG co-annotations; bottom, GO co-annotations (levels 5–15)). The corresponding number of genes is drawn either from the genome or the hitlist pool, as indicated. e, HIS-based interaction network for β-catenin. Node colour shows the highest enriched phenotypic class; edge colours show the phenotypic class contributing the most to HIS value inferences (see key); edge arrow shapes show HIS-inferred directionality. All edges directed to/from β-catenin are shown. f, Inducible NICD–GFP and HES1–GFP knock-in organoids with and without doxycycline (dox) treatment from day 0, showing loss of Paneth cells in NICD-overexpressing organoids. Nuclei are stained with DAPI (blue); lysozyme is stained with antibody (red). Scale bar, 50 μm. g, Day 5 puromycin-selected lentiCRISPRv2-mediated Casr knockout (CasrKO) organoids, with corresponding infection and selection controls (pLenti–EF1–EGFP organoids). Organoids were cultured from single cells in the indicated conditions. The upper six images show the perturbed crypt phenotype and lack of phenotypic response to CaCl2 treatment in CasrKO organoids. Bottom, lentivirally introduced EGFP expression after puromycin selection. Scale bars, 30 μm. h, Top two rows, day 3 and day 5 puromycin-selected control (pLenti–EF1–EGFP) organoids; bottom row, lentiviral Akt1-overexpressing (Akt1 OE) organoids cultured from single cells, showing perturbed phenotypes and lack of enterocyte differentiation. Scale bars, 30 μm. i, Top, organoids from DMSO control and RXR antagonist conditions analysed in the screen. Scale bar, 50 μm. Bottom, corresponding phenotypic signatures. n = 2, 2 and 1 non-sparse (n > 10 organoids) independent replica for Cpd2170, Cpd2390 and Cpd2173, respectively. j, Abundance of phenotypic cluster 13 in hit conditions with a reproducibility of more than 0.5. Compounds targeting RXR and RAR are depicted with coloured dots. Histograms show distributions of respective values. n = 301 independent conditions. Microscopy images shown in f-i are MIPs of confocal z-stacks. f-h, Assays performed in n = 12 independent replica from 2 independent organoid cultures with similar results.
Extended Data Fig. 5. Profiling the effects of treatment with an RXR antagonist.
a, Images of organoids cultured in the presence of an RXR agonist (RXRa) in the indicated treatment conditions. Scale bar, 50 μm. Assay performed in n = 12 (DMSO) or n = 6 (other conditions) independent replica from 2 independent organoid cultures. b, Top, distribution of mean aldolase B staining intensity in control organoids and those treated at day 4. Bottom, abundance of the ‘enterocyst’ phenotype in the indicated conditions. Error bars show standard deviations between n = 6 (DMSO) or n = 3 (other conditions) independent replica. Assays were performed with the same number of replicas from two independent organoid cultures with similar results. c, Images of organoids fixed at indicated time points, showing antibody staining for lysozyme. Scale bar, 20 μm. d, Top two rows, images of organoids from Fucci2 mice, expressing a dual-colour cell-cycle reporter (mVenus-hGem(1/110), G2/M-phase reporter; mCherry–hCdt1(30/120), G1-phase reporter) cultured with or without RXR antagonist (RXRi), fixed at day 4. Top row, expression of mVenus–hGem(1/110) (FUCCI (G2/M)), mCherry–hCdt1(30/120)(FUCCI (G1)); bottom row, antibody staining for YAP1. Scale bar, 100 μm. Bottom row, enlargements of those areas outlined in the middle row. Scale bar, 20 μm. e, Images of organoids following 10 μM atRA treatment at day 3. Top row, scale bar, 50 μm. Bottom row, scale bar, 10 μm. f, Organoids fixed at day 5, 48 h after switch to ENR medium. Scale bar, 100 μm. g, Top, distribution of mean SOX9, and bottom, ki67 staining intensity in control and treated organoids. Organoids were fixed at day 5, 48 h after switching to ENR medium. Violin ranges extended by one standard deviation. Two-sided t-test; P-values shown in the plot. h, Images of wild-type organoids, treated as indicated at day 3, and fixed at day 5. Scale bar, 100 μm. i, Images of organoids expressing RARE–dsGFP at day 4. dsGFP, destabilized GFP; scale bar, 10 μm. Assay performed in n = 12 independent replicates from n = 3 independent organoid cultures with similar results. j, Histological images of mouse small intestinal epithelium from RARE-LacZ mouse. Regions along the crypt–villus axis are indicated. A single crypt flanked by villus regions, outlined in the left panel, is shown in blue at the right. Scale bar, 50 μm. Experiment performed in n = 4 independent histological samples from n = 2 independent mice with similar results. k, Left, RARE–dsGFP organoids in the indicated treatment conditions. Scale bar, 60 μm. Right, DAPI-normalized sum intensity (norm. int.) of RARE–dsGFP reporter and aldolase B in individual organoids. Data points represent individual organoids, colour-coded according to treatment as indicated. R 2, Pearson’s correlation coefficient. l, Left, puromycin-selected ALDH1A2- knockout organoids. Scale bar, 50 μm. Right, ALDH1A1 staining intensity in control and ALDH1A1-knockout organoids. n = 36 and 17 organoids for control and ALDH1A1-knockout conditions respectively. ***P = 5 x 10-18, two-tailed t-test. m, Images of organoids treated as indicated from day 0. ALDH1A1i, 5 μM ALDH1A1 inhibitor; atRA, 10 μM all-trans retinoic acid. Scale bar, 50 μm. Assay performed in n = 8 independent replica. n, Images of organoids treated with RXR antagonist from day 0 in medium with or without vitamin A. Organoids were fixed at day 5, 48 h after switch to ENR medium. Scale bar, 50 μm. o, Wild-type organoids cultured in medium with or without vitamin A. Scale bar, 100 μm. p, Organoids cultured in medium with or without vitamin A and fixed at the indicated time points. Scale bar, 20 μm. a, d–f, h, k–o, Microscopy images are composite MIPs of confocal z-stacks. c, i, p, Microscopy images are composite individual z-planes from the organoid middle plane. b, g, k, n indicates the number of individual organoids used as data points. b, l, Boxes show quartile ranges; whiskers show value intervals with excluded outliers; middle lines or white dots show median values. c–f, h, Assays performed in at least four independent replicates from at least two independent organoid cultures with similar results. a, c–f, h, i, k–p, Organoids were cultured in the indicated conditions from single cells.
Extended Data Fig. 6. Retinol metabolism in enterocytes.
a, Network representation of annotation enrichment analysis for a published RNA-seq dataset describing marker genes of intestinal epithelium cell types29. Small nodes represent genes; large nodes represent functional annotations; edge thickness show gene-annotation assignments (grey) or term–term relations (black). Colours indicates cell-type specificities of annotations (see key). b, Box plots showing expression (in log2-normalized transcripts per kilobase million (TPM) values) of Aldhlal, Aldh1a2 and Aldh1a3 across cell types in the published single-cell RNA-seq (scRNA-seq) dataset29. Boxes show quartile ranges; whiskers represent value intervals with excluded outliers; middle lines show median values. Each data point represents a single cell. EC, enterocyte; TA, transient-amplifying. n = 1,522 single cells. c, Expression (in log2 normalized transcript counts) of Aldhlal, Aldh1a2 and Aldh1a3 across indicated treatment conditions in scRNA-seq (Fig. 3 and Extended Data Fig. 9). Sampling of n = 3,000 single cells. d, tSNE maps of the scRNA-seq experiment. Each data point represents a single cell, colour-coded by expression (log2 normalized transcript counts) of indicated genes in samples of the scRNA-seq experiment (Fig. 3). Ranges of the colour mapping are indicated in each plot. Sampling of n = 5,000 single cells. b–d, Each data point represents a single cell. atRA, 10 μM all-trans retinoic acid; DMSO, DMSO control, RXRi, 5 μM RXR antagonist; w/o VitA, vitamin-A-depleted medium.
Extended Data Fig. 7. Transcriptional signature of organoids treated with RXR antagonist.
a, Motif enrichments in the 1,000-base-pair regions that are centred on promoters of genes that are more highly expressed at a given day compared with the average from the whole period of organoid development. b, Enrichment heat map of transcription-factor-binding motifs at the transition from day 3 to day 4. Genes are binned by expression pattern. c, Network of functional annotation (KEGG) terms specifically enriched in genes containing RXR- and RAR-containing transcription-factor-binding motifs. Nodes show annotation terms; edges represent term–term interactions; node colours show annotation groups (see key). d, Principal component (PC) analysis of RNA-seq samples from the indicated treatment conditions and time points. The scatter plot shows the first two principal components. n = 4 (2 independent organoid cultures each from 2 independent mice) for all individual conditions. e, Pearson’s correlation values between samples of the time-course RNA-seq experiment and compound-stimulation RNA-seq experiment. n = 12,025 genes detected in both datasets. f, Left, expression of selected genes in untreated organoids at day 3 and in the indicated treatment conditions 24 h after treatment. Every data point represents a gene, colour-coded according to assigned cluster label (k-means clustering). Stratified sampling of 1,884 genes; n = 50 genes per cluster. FDR, false discovery rate. Top right, abundance of genes containing RXR/RAR-binding motifs in assigned clusters; bottom right, mean expression of genes from respective clusters in the RXR-antagonist condition compared with the DMSO control (log2 fold change, log2FC). Data shown as means ± s.d. n = 152, 102, 196, 361, 396, 109, 113, 290 and 165 for expression-profile clusters 1–9 respectively. g, tSNE maps colour-coded according to assigned cluster labels (k-means clustering). Left, tSNEs calculated from expression values measured in the bulk RNA-seq experiment (e). Right, genes showing treatment-specific upregulation and downregulation in the orthogonal compound condition. Every data point represents a gene. h, Annotation enrichment analysis (GO terms) for gene categories described in g. Asterisks depict statistical significance determined with two-tailed hypergeometric test with Bonferroni correction. *P < 0.05; **P < 2 x 10-3; ***P < 2 x 10-6. i, Left, expression of genes reported to have biased expression in the indicated gastrointestinal-tract tissues during unperturbed intestinal organoid development. log2-transformed count per million (CPM) values are normalized to mean expression over all samples. Solid lines show mean expression per time point; opaque intervals show standard deviation. n = 83, 20, 63, 156, 259, 45 and 31 genes for colon (sigmoid), colon (transverse), oesophagus (junction), oesophagus (mucosa), not specific, small intestine and stomach respectively. Middle, expression of genes specific to the indicated gastrointestinal tissues in RXRi-treated organoids and DMSO controls after 24 h of treatment. Right, expression of small-intestine-specific genes in DMSO- and RXRi-treated organoids. Boxes show quartile ranges; whiskers denote value intervals with excluded outliers; middle lines show median values. ***P < 2 x 10-19 (two-tailed t-test). d–i, atRA, 10 μM all-trans retinoic acid; 9cisRA, 10 μM 9-cis-retinoic acid; DMSO, DMSO control; RXRi, 5 μM RXR antagonist. g, i, log2-transformed CPM values, normalized to mean expression over all samples.
Extended Data Fig. 8. Organoids treated with an RXR antagonist lose intestinal identity.
a, Principal component analysis of RNA-seq samples (treated at day 0) from different treatment conditions and time points. The scatter plot shows the first two principal components. n = 4 (2 technical replicates of 2 biological replicates) for all individual conditions. The bar chart above depicts the percentage of variance explained by the first eight principal components. b, Pearson’s correlation values between samples of the time- course RNA-seq experiment. n = 13,502 genes detected in all samples. c, Normalized gene expression (in log2CPM) of the indicated genes in samples of the time-course RNA-seq experiment (Fig. 3b). Enteroc., enterocyte specific; G.C., goblet cell specific. d, Expression of genes from the indicated categories in treatment conditions at days 1 and 5 of organoid development. Data points represent genes. n = 14 and 16 genes for intestinal cell type markers and fetal- like and YAP target genes respectively (gene names are in b). e, Normalized gene expression in contrast (RXRi-treated over DMSO control, at day 4) for samples treated at days 0 and 3. Left and middle, colours show cell-type markers (left) or indicated gene categories (middle); dashed lines are at fourfold enrichment or depletion; opaque areas cover genes not significantly perturbed in either contrast; data points represent genes; colours represent indicated gene categories; marker types highlight the genes of interest. Right, only those genes that were upregulated on transition from day 0 to day 1 in the DMSO control are shown. Red dashed lines are at fourfold enrichment. Marker types highlight the genes of interest. f, Expression of genes grouped by response to intestinal helminth infection34. Expression is quantified by log2CPM, normalized to the mean expression over all samples in the respective datasets. Solid lines show mean values per time points; opaque intervals show standard deviations; dashed lines show fourfold enrichment or depletion. g, Normalized expression (log2CPM) of genes differentially regulated in helminth infection34 at day 4 (top) and day 5 (bottom). Each data point represents a gene; n = 141 and 286 genes for infection-upregulated and -downregulated genes respectively. h, Expression of tissue-specific genes over time points and treatments (DMSO, atRA and RXRi). log2CPM, normalized to mean expression over all samples; dashed lines show fourfold enrichment or depletion. n = 63, 156, 259, 45 and 31 genes for oesophagus (junction), oesophagus (mucosa), not specific, small intestine and stomach respectively. i, Top, expression of tissue-specific genes (n = 156, 259, 45 and 31 genes for oesophagus (mucosa), not specific, small intestine and stomach respectively), and bottom, expression of genes regulated by Cdx2 (n = 340, 170 and 11,378 genes for Cdx2-upregulated, Cdx2-downregulated and not affected respectively) in unperturbed organoid development. log2CPM, normalized to mean expression over all samples. j, Left, fold changes for the indicated genes in the indicated contrasts over the time-course RNA-seq experiment (Fig. 3b). Right, normalized gene expression in the indicated contrasts for samples treated at days 0 and 3; dashed lines show fourfold enrichment or depletion; the opaque area covers genes not significantly perturbed in either contrasts; colours show the indicated gene categories; every data point represents a gene, n = 340, 170 and 11,378 genes for Cdx2-upregulated, Cdx2- downregulated and not affected respectively. k, Expression of genes regulated by Cdx2 in organoids over the time course of the experiment. Solid lines show mean values per time point; opaque intervals show standard deviations; dashed lines show fourfold enrichment or depletion. l, Left panels, wild-type organoids at day 3 cultured from single cells; images are composite MIPs; scale bar, 10 μm. Right, intensity of Cdx2 staining; boxes show quartile ranges; whiskers show value intervals with excluded outliers; solid lines show median values. Asterisks indicate statistical significance; two-sided t-test; P-values are as follows: DMSO versus RXRi, 2.04 x 10-23; DMSO versus atRA, 8.17 x 10-7; atRA versus RXRi, 5.84 x 10-24; n, number of organoids analysed in each condition. m, Cdx2-EGFP organoids at day 4, cultured from single cells. Images are composite MIPs. Scale bars, 50 μm. The right two columns show YAP1 and Cdx2 staining from respective composite images. Assay performed in n = 16 independent replicates from n = 2 independent organoid cultures with similar results. f, h, i, k, Solid lines show mean values per time points; opaque intervals show standar deviations. atRA, 10 μM all-trans retinoic acid; DMSO, DMSO control; RXRi, 5 μM RXR antagonist.
Extended Data Fig. 9. scRNA-seq analysis of phenotypes produced by RXRi treatment.
a, Left, tSNE maps from scRNA-seq. Each data point represents a single cell, colour-coded by sample (see key). n = 4,787, 3,570, 5,365, 4,963, 5,576 and 5,304 single cells for day 4 DMSO, RXRi, atRA and vitamin A-free, and day 1 DMSO control samples 1 and 2 respectively. Right, expression (log 2normalized transcript counts) of indicated genes in samples of the scRNA-seq experiment (Fig. 3); sampling of n = 3,000 cells. b, Cell-type composition of scRNA-seq samples. Confidence cutoffs for cell-type assignment were set individually for each cell type on the basis of the mean expression levels of marker genes from ref. 29; n = 29,565 single cells. c-e, tSNE maps of our scRNA- seq experiment, colour coded by mean normalized expression (log2-transformed counts) of gene groups. c, Expression of cell-type signature genes from ref. 29. The numbers of genes per cell-type signature are shown in individual plots. d, Expression of genes that are differentially regulated following intestinal helminth infection34. e, Expression of genes with biased expression in tissues (GTEX database; https://gtexportal.org/home/). f, Left, partial tSNE maps (top) and density map (bottom) of single cells from the RXR-antagonist-treated sample of the scRNA-seq experiment. In the density map, the x- and y-axes show the mean expression (mean of log2-transformed counts) of genes from the indicated categories. Colours show cell counts in respective bins. Red dashed lines show cutoffs used to assign populations. Right, tSNE maps of the scRNA- seq experiment, showing assigned identities in the RXR-antagonist-treated sample. g, Left, average expression of genes in populations of cells as assigned in f. Colours depict the log2-transformed fold change (log2FC) in the ‘early progenitor’ population compared with the ‘regenerative identity’ population. Every data point represents a gene. Centre, log2FC values of the indicated genes. Green, absorptive-lineage-specific genes; blue, secretory-lineage- specific genes. Right, tSNE maps of the scRNA-seq experiment colour-coded by mean normalized expression (log2-transformed counts) of genes with log2FC values of greater than 0.5 between ‘early progenitors’ and ‘regenerative’ populations. a, b, atRA, 10 μM all-trans retinoic acid; DMSO, DMSO control; RXRi, 5 μM RXR antagonist; w/o VitA, vitamin-A-depleted medium. a–f, In tSNE maps each data point represents a single cell; sampling of 5,000 single cells. c–e, g, Ranges of colour mapping and numbers of genes per category (n) are indicated in each plot.
Extended Data Fig. 10. Treatment with an RXR antagonist improves intestinal regeneration in an irradiation-induced mouse model of colitis.
a, irradiation-induced colitis model. PK, pharmacokinetic analysis. b, Change in body weight in the indicated treatment cohorts over the time frame of the study. Data shown are mean ± s.d values for cohorts of n = 6 independent mice per cohort. Non-irr., non-irradiated vehicle-treated cohort. c, Images of spleen extracted from mice from the indicated treatment cohorts. Scale bar, 1 cm. d, Small-intestine’ Swiss rolls’ from mice belonging to the indicated treatment cohorts and time points. Scale bar, 5 mm. e, Top, haemoccult score of stool samples from the indicated treatment cohorts; middle lines show mean; error bars show ± s.d.; n = 6 independent animals per cohort. Bottom, representative images of the haemoccult samples corresponding to the quantitative haemoccult scores. f, Serum levels of the RXRi compound at 7 h post-treatment at the indicated time points in RXRi-treated mice. Middle lines show means; error bars show ± s.d.; n = 6 independent animals per cohort. g, Top, small-intestine Swiss roll, showing the ileum, duodenum and jejunum. Scale bar, 5 mm. Middle, jejunum part of the intestine. Scale bar, 250 μm. Bottom, enlarged view of villi units; individual are villi indicated with arrowheads; the shaded area indicates measured villi length. Scale bar, 250 μm. h, Top, abundance of goblet cells, measured by staining with alcian blue, in histological samples at day 6 in mice from the indicated groups. Bottom, quantified percentage of alcian blue (AC)-positive cells in the entire intestine of mice from the indicated treatment cohorts. Boxes show, quartile ranges; whiskers show value intervals; solid lines show median values; individual data points are overlaid. n = 6 independent animals per cohort. i, Histological profiling of the abundance of Ki67+ cells in the intestinal epithelium at days 2, 4 and 6 in the indicated treatment cohorts. j–l, Histological profiling of the intestinal epithelium at days 2 (j), 4 (k) and 6 (l) extracted from mice from the indicated treatment cohorts. Left panels, haematoxylin and antibody staining for OLFM4; scale bars, 50 μm. Enlarged regions: upper insets, scale bars, 25 μm; lower insets, scale bars, 12 μm. Middle panels, nuclear fast red (NFR) and alcian blue staining; scale bars, 50 μm. Right panels, confocal imaging of histological sections (composite MIPs ofz-stacks); scale bars, 50 μm. Enlarged regions: upper insets, scale bars, 25 μm; lower insets, scale bars, 12 μm. In all panels, experiments were carried out in n = 6 independent mice per treatment cohort with similar results. c, d, Representative images from respective treatment cohort animals. g–l, Representative images from n = 24 histological samples from n = 6 independent mice per treatment cohort. b–h, Day 6 (terminal time point) mouse study repeated twice with n = 6 mice per cohort with similar results.
Supplementary Material
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Acknowledgements
We thank D. Vischi, S. Iftikhar and E. Tagliavini for IT support; L. Gelman for assistance and training; H. Kohler for sorting; S. Smallwood for sequencing; and L. Pelkmans, J. Betschinger, C. Tsiairis, S. Gasser and laboratory members for reading the manuscript. This work received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 758617) and from the Swiss National Foundation (POOP3_157531 to P.L.).
Footnotes
Author contributions
P.L. conceived and supervised the study. P.L. and I.L. designed the experiments. J.J. designed the annotated compound library. I.L. and K.V. performed the image-based screen. I.L. performed the image analysis. L.C.M. designed CRISPR-Cas9 guides and carried out cloning. F.M. and I.L. performed lentivirus production and organoid preparations. I.L. performed imaging and transcriptomics experiments. M.B.S. designed the analysis of transcription-factor motifs. I.L., D.S. and M.B.S. analysed RNA-seq data. J.B., R.Z., K.C. and S.M. designed, performed and analysed the mouse irradiation study. P.L. and I.L. wrote the paper.
Competing interests
P.L. and I.L. are inventors on the patent application EP19182782.3, filed on 27 June 2019, with the title ‘Promoting tissue regeneration’, pertaining to the use of RXR antagonists as therapeutic agents in tissue regeneration. The patent applicant is the Friedrich Miescher Institute for Biomedical Research. Other authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-2776-9.
Peer review information Nature thanks Hans Clevers, Francesco Ioro, Bon-Kyoung Koo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at http://www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
All RNA-seq data generated here (bulk RNA-seq data (compound treatment at day 3), bulk RNA-seq data (compound treatment at day 0) and single-cell RNA-seq data) are available at the GEO under accession number GSE147136. The RXR antagonist used here (Cpd2170) is available through a material transfer agreement (MTA) with Novartis. The source data for following figures are available in the manuscript files: Figs. 1a, b, 2a, f, 3a–d, f, g and Extended data Figs. 1c–i, 2a–h, 3a–f, 4b–d, i, j, 5b, j–l, 6b, c, 7b, d–g, i, 8a–l, 9a, b, f, g, 10b, e, f, h. Gene sets used for visualization in Fig. 3e and Extended Data Fig. 9c–e, g are provided in Supplementary Table 2. Source data are provided with this paper.
Code availability
Code used for image analysis was developed in the Liberali laboratory using MATLAB and Python 3. Segmentation for the image-based screen was performed using code developed in the Liberali laboratory in MATLAB, and is available at https://github.com/fmi-basel/glib-lukonin-et-al-2020. The code for organoid two-dimensional segmentation and feature extraction in other assays is available at https://github.com/fmi-basel/glib-nature2018-materials.
References
- 1.Serra D, et al. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grün D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 3.Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J Biol Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 4.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. BioEssays. 2002;24:91–98. doi: 10.1002/bies.10028. [DOI] [PubMed] [Google Scholar]
- 6.Bullen TF, et al. Characterization of epithelial cell shedding from human small intestine. Lab Invest. 2006;86:1052–1063. doi: 10.1038/labinvest.3700464. [DOI] [PubMed] [Google Scholar]
- 7.de Sousa e Melo F, de Sauvage FJ. Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell. 2019;24:54–64. doi: 10.1016/j.stem.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Tetteh PW, et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 9.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian H, et al. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joosten SPJ, et al. MET signaling mediates intestinal crypt-villus development, regeneration, and adenoma formation and is promoted by stem cell CD44 isoforms. Gastroenterology. 2017;153:1040–1053. doi: 10.1053/j.gastro.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 13.Chacón-Martínez CA, Koester J, Wickstrom SA. Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development. 2018;145 doi: 10.1242/dev.165399. dev165399. [DOI] [PubMed] [Google Scholar]
- 14.Chang J, et al. Proteomic changes during intestinal cell maturation in vivo. J Proteomics. 2008;71:530–546. doi: 10.1016/j.jprot.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carithers LJ, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peeters T, Vantrappen G. The Paneth cell: a source of intestinal lysozyme. Gut. 1975;16:553–558. doi: 10.1136/gut.16.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanDussen KL, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin X, et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine JH, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberali P, Snijder B, Pelkmans L. A hierarchical map of regulatory genetic interactions in membrane trafficking. Cell. 2014;157:1473–1487. doi: 10.1016/j.cell.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 21.Snijder B, Liberali P, Frechin M, Stoeger T, Pelkmans L. Predicting functional gene interactions with the hierarchical interaction score. Nat Methods. 2013;10:1089–1092. doi: 10.1038/nmeth.2655. [DOI] [PubMed] [Google Scholar]
- 22.Andreu P, et al. A genetic study of the role of the Wnt/beta-catenin signalling in Paneth cell differentiation. Dev Biol. 2008;324:288–296. doi: 10.1016/j.ydbio.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaki J, et al. Synthesis and structure-activity relationship of novel RXR antagonists: orally active anti-diabetic and anti-obesity agents. Bioorg Med Chem Lett. 2007;17:4804–4807. doi: 10.1016/j.bmcl.2007.06.080. [DOI] [PubMed] [Google Scholar]
- 25.Yui S, et al. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell. 2018;22:35–49. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 27.Borel P, et al. Processing of vitamin A and E in the human gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2001;280:G95–G103. doi: 10.1152/ajpgi.2001.280.1.G95. [DOI] [PubMed] [Google Scholar]
- 28.Lampen A, Meyer S, Arnhold T, Nau H. Metabolism of vitamin A and its active metabolite all-trans-retinoic acid in small intestinal enterocytes. J Pharmacol Exp Ther. 2000;295:979–985. [PubMed] [Google Scholar]
- 29.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goverse G, et al. Diet-derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J Immunol. 2017;198:2172–2181. doi: 10.4049/jimmunol.1600165. [DOI] [PubMed] [Google Scholar]
- 31.Molotkov A, Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem. 2003;278:36085–36090. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- 32.Bowles J, et al. ALDH1A1 provides a source of meiosis-inducing retinoic acid in mouse fetal ovaries. Nat Commun. 2016;7 doi: 10.1038/ncomms10845. 10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ordonez-Moran P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell. 2015;28:815–829. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Nusse YM, et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 2018;559:109–113. doi: 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim TH, Shivdasani RA. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J Biol Chem. 2011;286:11427–11433. doi: 10.1074/jbc.M110.188797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharya N, et al. Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8+ T cell-mediated immunity in colorectal cancer. Immunity. 2016;45:641–655. doi: 10.1016/j.immuni.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dotti I, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. doi: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serafini EP, Kirk AP, Chambers TJ. Rate and pattern of epithelial cell proliferation in ulcerative colitis. Gut. 1981;22:648–652. doi: 10.1136/gut.22.8.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmini S, et al. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat Commun. 2014;5 doi: 10.1038/ncomms6728. 5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassani B, Villablanca EJ, De Calisto J, Wang S, Mora JR. Vitamin A and immune regulation: role of retinoic acid in gut-associated dendritic cell education, immune protection and tolerance. Mol Aspects Med. 2012;33:63–76. doi: 10.1016/j.mam.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rangan P, et al. Fasting-mimicking diet modulates microbiota and promotes intestinal regeneration to reduce inflammatory bowel disease pathology. Cell Rep. 2019;26:2704–2719. doi: 10.1016/j.celrep.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barisic D, Stadler MB, lurlaro M, Schubeler D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature. 2019;569:136–140. doi: 10.1038/s41586-019-1115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data generated here (bulk RNA-seq data (compound treatment at day 3), bulk RNA-seq data (compound treatment at day 0) and single-cell RNA-seq data) are available at the GEO under accession number GSE147136. The RXR antagonist used here (Cpd2170) is available through a material transfer agreement (MTA) with Novartis. The source data for following figures are available in the manuscript files: Figs. 1a, b, 2a, f, 3a–d, f, g and Extended data Figs. 1c–i, 2a–h, 3a–f, 4b–d, i, j, 5b, j–l, 6b, c, 7b, d–g, i, 8a–l, 9a, b, f, g, 10b, e, f, h. Gene sets used for visualization in Fig. 3e and Extended Data Fig. 9c–e, g are provided in Supplementary Table 2. Source data are provided with this paper.
Code used for image analysis was developed in the Liberali laboratory using MATLAB and Python 3. Segmentation for the image-based screen was performed using code developed in the Liberali laboratory in MATLAB, and is available at https://github.com/fmi-basel/glib-lukonin-et-al-2020. The code for organoid two-dimensional segmentation and feature extraction in other assays is available at https://github.com/fmi-basel/glib-nature2018-materials.