Abstract
Upon DNA damage, the ALC1/CHD1L nucleosome remodeling enzyme (remodeler) is activated by binding to poly(ADP-ribose). How activated ALC1 recognizes the nucleosome, as well as how this recognition is coupled to remodeling, is unknown. Here, we show that remodeling by ALC1 requires a wild-type acidic patch on the entry side of the nucleosome. The cryo-electron microscopy structure of a nucleosome-ALC1 linker complex reveals a regulatory linker segment that binds to the acidic patch. Mutations within this interface alter the dynamics of ALC1 recruitment to DNA damage and impede the ATPase and remodeling activities of ALC1. Full activation requires acidic patch-linker segment interactions that tether the remodeler to the nucleosome and couple ATP hydrolysis to nucleosome mobilization. Upon DNA damage, such a requirement may be used to modulate ALC1 activity via changes in the nucleosome acidic patches.
Introduction
Packaging into chromatin creates a barrier to DNA transactions (Lorch and Kornberg, 2017). The repeat unit of chromatin, the nucleosome, contains ~150 base pairs (bp) of DNA wrapped around a histone octamer (Luger et al., 1997). ATP-dependent nucleosome remodeling enzymes (remodelers) feature a conserved Snf2 (sucrose non-fermenter 2) family ATPase (Flaus et al., 2006) and play vital roles in regulating accessibility to DNA (Bartholomew, 2014; Becker and Workman, 2013; Bowman, 2010; Clapieretal.,2017; Narlikaret al., 2013; Lusserand Kado-naga, 2003).
Many proteins interact with a negatively charged “acidic patch” (AP), formed by histones H2A and H2B on both faces of the nucleosome (Figure 1A), by inserting an arginine (Arg) side chain into a pocket defined predominantly by H2A E61, E64, D90, and E92 (McGinty and Tan, 2016; Zhou et al., 2019). Previous studies have identified interactions between the AP and several remodelers, including SWR1 (Willhoft et al., 2018), INO80 (Eustermann et al., 2018; Ayala et al., 2018), BAF (He et al., 2020), and RSC (Valencia et al., 2019; Wagner et al., 2020; Ye et al., 2019). These structures suggested that the re-modeler-AP interaction acts as a physical tether in the assembly of the nucleosome-remodeler complex. Moreover, disrupting the AP interaction by the deletion of entire remodeler subunits impaired remodeling (Eustermann et al., 2018; Willhoft et al., 2018). Recently, the AP has been implicated in the activation of imitation-switch (ISWI)-family remodelers and chromodomain helicase DNA-binding protein 1 (Chd1) (Dann et al., 2017; Dao et al., 2020; Gamarra et al., 2018; Goldman et al., 2010; Levendosky and Bowman, 2019), and an AP-interacting basic motif was shown to be essential for remodeling by the ISWI remodeler SNF2h (Dao et al., 2020).
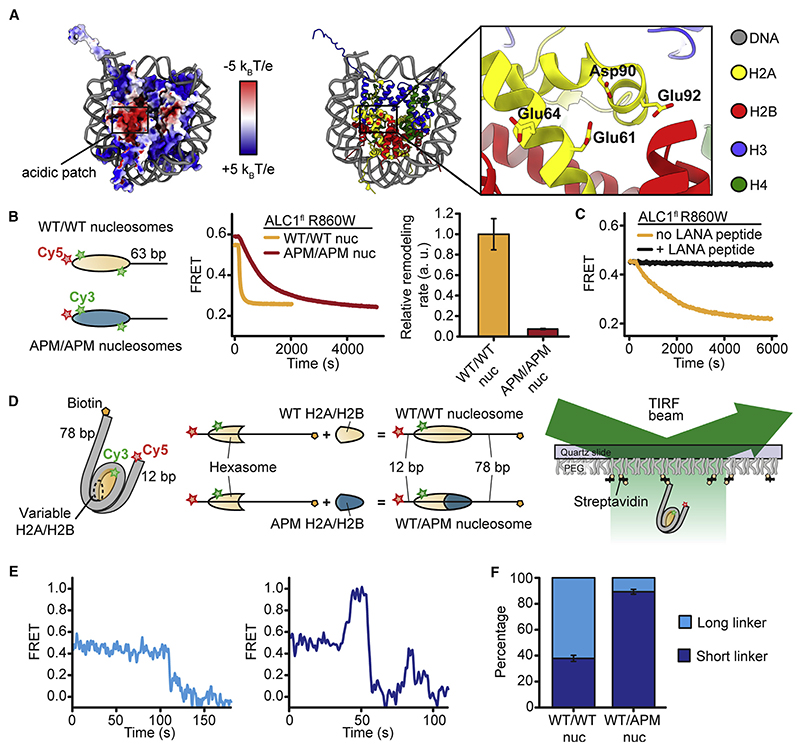
Figure 1. The AP Is Important for Remodeling by ALC1.
(A) Left: octamer surface colored by electrostatic potential (from –5 kB T/e, indicated in red, to +5 kB T/e, indicated in blue). Right: key residues of the H2A AP. Based on PDB: 1AOI.
(B) Left: schematic of FRET-labeled wild-type (WT/WT) nucleosomes and APM/APM nucleosomes with AP mutations on both faces of the octamer. Middle: ensemble remodeling of WT/WT and APM/APM nucleosomes (10 nM) in the presence of 20 μM ALC1fl R860W. Right: relative remodeling rates. Error bars represent SD (n = 3 independent experiments).
(C) Ensemble remodeling of WT/WT nucleosomes (10 nM) by 80 nM ALC1fl R860W with or without LANA peptide.
(D) smFRET labeling and detection.
(E) ALC1fl R860W-catalyzed remodeling of individual WT/WT nucleosomes toward the longer (left) or shorter (right) linker DNA.
(F) Percentages oftraceswith the initial remodeling direction toward longerorshorter linker DNAforWT/WT and WT/APM nucleosomes. Error bars indicate SEM (n > 100 traces).
See also Figure S1.
The ALC1 remodeler (Amplified in liver cancer 1) is encoded on a chromosome region frequently amplified in hepatocarcinomas (Ma et al., 2008; Marchio et al., 1997; Wong et al., 2003). ALC1 has been implicated in DNA damage response (Ahel et al., 2009; Chen et al., 2009; Gottschalk et al., 2009, 2012; Pines et al., 2012; Sellou et al., 2016) and oncogenesis (Chen et al., 2010, 2009; Ma et al., 2008; Marchio et al., 1997; Mu et al., 2015; Pines et al., 2012; Sellou et al., 2016) and differs from other remodelers by virtue of its macro domain, which binds poly(ADP-ribose) (PAR) chains (Ahel et al., 2009; Gottschalk et al., 2009) at sites of DNA damage (Hassaetal., 2006; Lindahl etal., 1995; Sa-tohand Lindahl, 1992). PARylation is linked with chromatin relaxation (Frechette et al., 1985; Leduc et al., 1986; de Murcia et al., 1986; Poirier et al., 1982), which is thought to promote DNA repair (D’Amours et al., 1999; El-Khamisy et al., 2003; Malanga and Althaus, 2005; Realini and Althaus, 1992). Initial chromatin relaxation involves PAR polymerase 1 (PARP1) and ALC1 and enables the PAR-dependent recruitment of additional proteins (Smith et al., 2018). Early PAR-dependent remodeling promotes DNA exposure but does not affect histone accessibility (Smith et al., 2019).
In the absence of DNA damage, the juxtaposition of the macro domain of ALC1 against its ATPase maintains an inactive conformation (Lehmann et al., 2017; Singh et al., 2017). PAR binding to the macro domain relieves this autoinhibition upon recruitment to DNA damage (Lehmann et al., 2017; Singh et al., 2017). However, the mechanism by which ALC1 recognizes the nucleosome, and how that recognition is coupled to remodeling, is unknown.
Here, we establish that nucleosome repositioning by ALC1 requires a wild-type (WT) AP on the entry side of the nucleosome. Our cryo-electron microscopy (cryo-EM) structure of a nucleo-some-ALC1 linker complex reveals a regulatory segment that engages the AP via an Arg anchor. Mutation of this segment or of the AP perturbs ATPase and remodeling activities of ALC1. The regulatory segment thus activates remodeling by ALC1 upon recognition of its bona fide nucleosome substrate with intact AP. We anticipate that other remodelers may use a similar mechanism to regulate their activities by analogous interactions between the AP and family-specific regulatory sequence motifs.
Results
Nucleosome Remodeling by ALC1 Requires the Entry-Side AP
To probe whether the nucleosome AP (Figure 1A) might regulate ALC1, we constructed end-positioned nucleosomes with AP mutations (APMs; alanine substitutions on H2A E61A, E64A, D90A, and E92A) on both faces of the octamer (APM/APM nucleosomes). We also reconstituted end-positioned WT nucleosomes with intact APs (WT/WT). We detected nucleosome repositioning by a constitutively active mutant of ALC1, ALC1fl R860W (Lehmann et al., 2017; Figures S1A and S1B), using fluorescence resonance energy transfer (FRET) between a Cy3 donor on H2A and a Cy5 acceptor on the octamer-proximal DNA end (Blosser et al., 2009; Deindl et al., 2013) (Figure 1B). To compare the remodeling of WT/WT and APM/APM nucleosomes under saturating concentrations of ALC1fl R860W and ATP, we first used an ensemble FRET assay (Yang et al., 2006). Upon addition of ALC1fl R860W and ATP to WT/WT nucleosomes, FRET decreased, consistent with DNA moving toward the shorter linker side. The remodeling rate decreased substantially (~13.7-fold) for APM/APM nucleosomes (Figure 1B), indicating an important role of the AP in ALC1-induced remodeling. We next determined the remodeling rate for WT/WT nucleosomes in the presence of sub-saturating concentrations of ALC1fl R860W and saturating concentrations of a peptide derived from the latency-associated nuclear antigen (LANA) of Kaposi’s sarcoma-associated herpesvirus. This LANA peptide binds to the AP of the nucleosome (Barbera et al., 2006) and has been used as a competitor to establish AP binding (England et al., 2010; Gamarra et al., 2018). Strikingly, the LANA peptide completely abrogated ALC1-catalyzed remodeling (Figures 1C and S1C).
To examine how ALC1 responds to AP asymmetry, we produced asymmetric nucleosomes with distinct H2A/H2B dimers (Levendosky et al., 2016). We first reconstituted FRET-labeled hexasomes with 12 bp of linker DNA on one side and 78 bp on the other side of the histone core (Figure 1D). The single H2A/ H2B dimer in these hexasomes was homogeneously incorporated on the more bendable side of the asymmetric positioning sequence (Levendosky et al., 2016; Lowary and Widom, 1998; Ngo et al., 2015), the shorter linker side. We then combined these oriented hexasomes with WT or APM H2A/H2B dimerto produce either WT/WT or WT/APM nucleosomes (Figure 1D). These nucleosomes exhibited an intermediate starting FRET value (~0.4), providing dynamic range for movements in both directions. To monitor ALC1-catalyzed remodeling of individual WT/ WT and WT/APM nucleosomes using single-molecule FRET (smFRET) (Blosser et al., 2009; Deindl and Zhuang, 2012; Deindl et al., 2013), we immobilized them and detected their fluorescence emissions (Figure 1D). Upon addition of ALC1fl R860W and ATP (but not ATPγS; Figure S1D) to WT/WT nucleosomes, a large fraction (62%) of single-nucleosome remodeling traces featured an initial decrease in FRET, consistent with the ability of ALC1 to mobilize the octamer toward the longer linker (Figures 1E, 1F, and S1 E). A smaller fraction (38%) displayed the opposite directionality, consistent with Cy5 on the shorter linker moving closer to the octamer. Our data, therefore, indicate a modest preference of ALC1 to center the histone octamer by moving it toward longer linker DNA. In stark contrast, WT/APM nucleosomes were almost exclusively (89%) moved toward the shorter linker end (Figures 1F and S1 E), implying that nucleosome repositioning by ALC1 critically depends on an entry-side WT AP.
A Regulatory Segment of the ALC1 Linker Binds to the AP
To probe ALC1-histone interactions, we carried out cross-linking coupled to mass spectrometry (XL-MS). A substantial fraction of the cross-links observed between histones and ALC1 involved its linker region (residues 570–703) (Figure 2A; Table S1). Consistent with an as yet undefined functional role of the linker region, a truncated fragment of ALC1 lacking ~60 amino acids of the linker in addition to the macro domain lost the ability to decompact a LacO array in vivo (Singh et al., 2017).
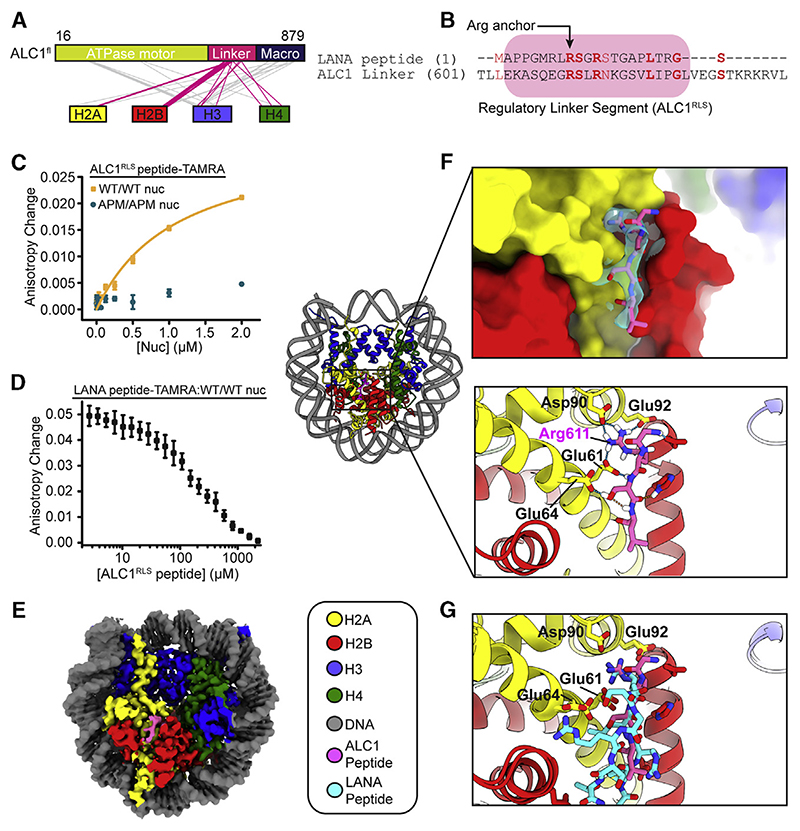
Figure 2. The ALC1 Linker Region Binds to the AP.
(A) ALC1fl-histone cross-links. Magenta lines indicate linker-histone cross-links: gray lines indicate other cross-links.
(B) Sequence alignment of LANA (1–22) and ALC1 linker(601-635).
(C) Fluorescence anisotropy of TAMRA-labeled ALC1RLS in the presence ofvarious amounts ofWT/ WT or APM/APM nucleosomes. Error bars for WT/WT nucleosome curve represent SEM (n = 3 independent experiments). APM/APM nucleosomes: one representative curve out of two independent experiments is indicated. Error bars represent SD (n = 10 technical replicates).
(D) Competition between TAMRA-labeled LANA peptide pre-bound to WT/WT nucleosomes and unlabeled ALC1RLS. See also Figure S2A. Error bars represent SEM (n = 3 independent experiments).
(E) Cryo-EM map.
(F) Left: cartoon representation. Top right inset: cryo-EM density around the ALC1 linker (cyan). Bottom right inset: hydrogen-bonding interactions.
(G) Superposition of PDB: 1ZLA onto the linkernucleosome complex.
See also Figure S2 and Tables S1 and S2.
We next aligned the sequence of the LANA peptide (Barbera et al., 2006) to the linker of ALC1 (Figure 2B). Strikingly, an N-ter-minal regulatory linker segment (ALC1RLS, residues 604–624) exhibited sequence similarity with LANA, including the conserved R611 that aligns with the Arg anchor of LANA (Figure 2B). To directly test whether ALC1RLS binds to the nucleosome, we monitored the fluorescence anisotropy of a tetramethylrhod-amine-labeled ALC1RLS peptide (ALC1RLS-TAMRA) in the presence of increasing concentrations of WT/WT nucleosomes. WT/WT nucleosomes exhibited specific affinity for the ALC1RLS-TAMRA peptide (Figure 2C; dissociation constant [KD], ~1.1 μM). Conversely, nucleosomes with AP mutations on both H2A/H2B dimers displayed substantially reduced affinity for the ALC1RLS-TAMRA peptide (Figure 2C), consistent with its binding to the WT/WT AP. Next, we probed whether the LANA peptide could compete with the ALC1RLS peptide for binding to the nucleosome (Figure 2D). Indeed, the addition of unlabeled ALC1RLS peptide to a pre-formed LANA-TAMRA peptide-nucleosome complex caused a decrease in its fluorescence anisotropy in a dose-dependent manner (Figure 2D; see also Figure S2A), demonstrating that ALC1RLS interacts with the AP.
Cryo-EM Structure of ALC1RLS Bound to the Nucleosome
We next determined a cryo-EM structure of the cross-linked complex between a linker peptide and the nucleosome (Figures 2E–2G, S2B, and S2C). The map exhibited an overall resolution of 2.5 Å (Figure S2C) with visible side chain features for central residues of ALC1RLS (Figure 2F; mean local resolution around the linker peptide: 2.5 Å). With an overall root-mean-square deviation (RMSD) of 0.46 Å between 739 Cα pairs when compared with the accession number PDB: 1ZLA, the nucleosome in our model is largely undisturbed relative to previous structures (Barbera et al., 2006). The ALC1RLS-AP interaction is stabilized by nine hydrogen bonds as well as van der Waals interactions, and ALC1RLS inserts residue R611 into a pocket formed by the α1-α2 helices of H2A (Figure 2F). Overall, ALC1RLS engages the AP in a manner that bears marked similarity to the binding mode observed for LANA (Barbera et al., 2006), albeit with reverse N-to C-terminal polarity (Figure 2G; RMSD of 1.78 Å between 11 atom pairs).
ALC1 RLS Plays a Crucial Role in Activating ATP Hydrolysis and Remodeling
In order to disrupt the AP-ALC1RLS interaction, we mutated the ALC1RLS Arg anchor (R611) and the adjacent residue (S612) to alanines. ALC1fl R611A/S612A/R860W displayed a ~2.5-fold (~2-fold under saturating conditions; see STAR Methods) reduced ATPase rate (Figure 3A). In contrast, ALC1fl R860W and ALC1fl R611A/S612A/R860W exhibited essentially identical DNA-stimulated ATPase activity (Figure S3A). The ALC1RLS mutation also increased the Michaelis constant by a factor of ~5.3 and decreased the maximum rate for remodeling by a factor of ~6.4 (Figure 3B). Taken together, our data indicate that ALC1RLS interactions play a role in nucleosome binding and in the regulation of maximum ATPase and remodeling activities of ALC1.
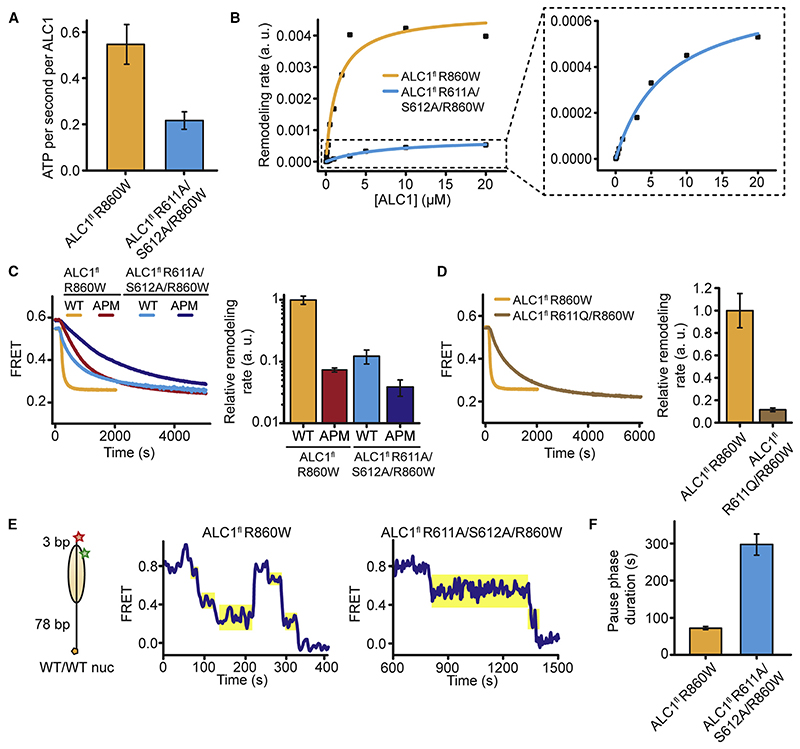
Figure 3. The Linker-AP Interaction Is Important for ALC1 Activity.
(A) ATPase activity for 20 μM ALC1fl R860W or ALC1fl R611A/S612A/R860W in the presence of 2 μM WT/WT nucleosomes. Error bars represent SD (n = 3 independent experiments).
(B) Michaelis-Menten curves of remodeling rates for WT/WT nucleosomes (10 nM) and various ALC1fl R860W or R611A/S612A/R860W concentrations. Inset: enlarged view for ALC1fl R611A/S612A/R860W.
(C) Left: remodeling of10nM nucleosomes by 20 μM remodeler. Right: relative remodeling rates. Error bars represent SD(n = 3 independent experiments). Data for ALC1fl R860W and WT/WT nucleosomes as well as for ALC1fl R860W and APM/APM nucleosomes: same as indicated in Figure 1B.
(D) Left: remodeling of WT/WT nucleosomes (10 nM) in the presence of 20 μM ALC1fl R860W or ALC1fl R611Q. Right: relative rates for ALC1fl R860W or ALC1fl R611Q. Error bars represent SD (n = 3 independent experiments). Data for ALC1fl R860W and WT/WT nucleosomes: same as indicated in Figure 1B.
(E) Representative smFRET traces indicating the remodeling of individual WT/WT nucleosomeswith 3 bp of linker DNA(left) by ALC1fl R860W (middle) orALC1fl R611A/S612A/R860W (right). Only pauses immediately following translocation phases with a FRET decrease were analyzed (shaded yellow).
(F) Mean pause durations for ALC1fl R860W or ALC1fl R611A/S612A/R860W as described in (E). Error bars represent SEM (n > 170 events).
See also Figure S3.
The AP mutation should not have a dramatic additional effect on remodeling by ALC1 with a mutated ALC1RLS if both types of perturbations involved the same interface. Indeed, ALC1fl R611A/S612A/R860W remodeled APM/APM nucleosomes with only moderately (by ~3.2-fold) reduced rates when compared to WT/WT nucleosomes (Figure 3C). In contrast, a ~13.7-fold change was observed when comparing the remodeling of WT/WT versus APM/APM nucleosomes by ALC1fl R860W or a ~8.2-fold change for the remodeling of WT/WT nucleosomes by ALC1fl R860W versus ALC1fl R611A/R612A/R860W (Figure 3C). These observations are consistent with the ALC1RLS and the AP contributing to the same interface. The observed residual sensitivity of ALC1fl R611A/S612A/R860W to AP mutations most likely indicates that the R611A/S612A mutation of the regulatory linker does not completely abrogate its interaction with the AP. We cannot formally rule out an involvement of additional AP residues beyond the ones that we mutated or a contribution of additional elements of ALC1 to AP recognition. Interestingly, mutation of the Arg anchor (R611Q) alone reduced the remodeling activity of ALC1 by ~8.7-fold, close to the effect of the double-linker mutation (R611A/S612A) (Figure 3D). Consistent with our structure, the Arg anchor, therefore, plays a particularly important role in the linker-mediated regulation of ALC1.
To probe which phase of the remodeling cycle is affected by disrupting the ALC1 RLS-AP interaction, we monitored the remodeling of individual nucleosomes featuring only 3 bp of linker DNA on the acceptor-labeled side (Figure 3E). As previously observed for other remodelers (Blosser et al., 2009; Deindl et al., 2013; Gamarra et al., 2018; Hwang et al., 2014; Qiu et al., 2017), time traces exhibited phases of FRET change as well as translocation pauses. Mutation of the regulatory linker segment increased the average pause duration by over 4-fold (Figures 3E, 3F, and S3B–S3D), suggesting that interaction with the AP is important for exiting a paused state.
To probe the impact of the ALC1RLS mutation on ALC1 function in vivo, we monitored recruitment kinetics at sites of DNA damage (Ahel et al., 2009) (Figures 4A–4D). Interestingly, both R611A/S612A and R611Q mutations notably compromised the overall extent of ALC1 recruitment (Figures 4A and 4B) but only modestly delayed recruitment kinetics (Figures 4C and 4D).
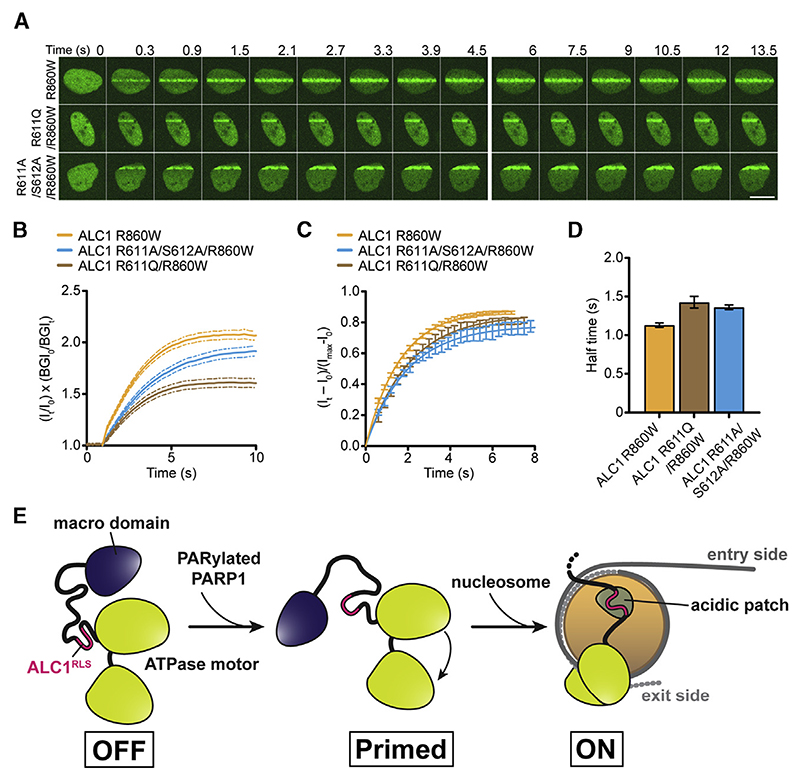
Figure 4. ALC1RLS Mutations Alter Recruitment Dynamics.
(A) U2OS cells expressing R860W, R611A/S612A/ R860W, or R611Q/R860W YFP-ALC1, imaged upon laser damage. Scale bar, 10 μm.
(B) Kinetics of R860W, R611A/S612A/R860W, and R611Q/R860W YFP-ALC1 association with DNA breaks. Error bars represent SEM (n R 131 traces from 3 independent experiments). See also Figure S4.
(C) Fractional recruitment of R860W, R611A/ S612A/R860W, and R611Q/R860W YFP-ALC1 to DNA breaks. Error bars represent SEM (n R 131 traces from 3 independent experiments).
(D) Half times from (C). Data are means ± 95% confidence intervals.
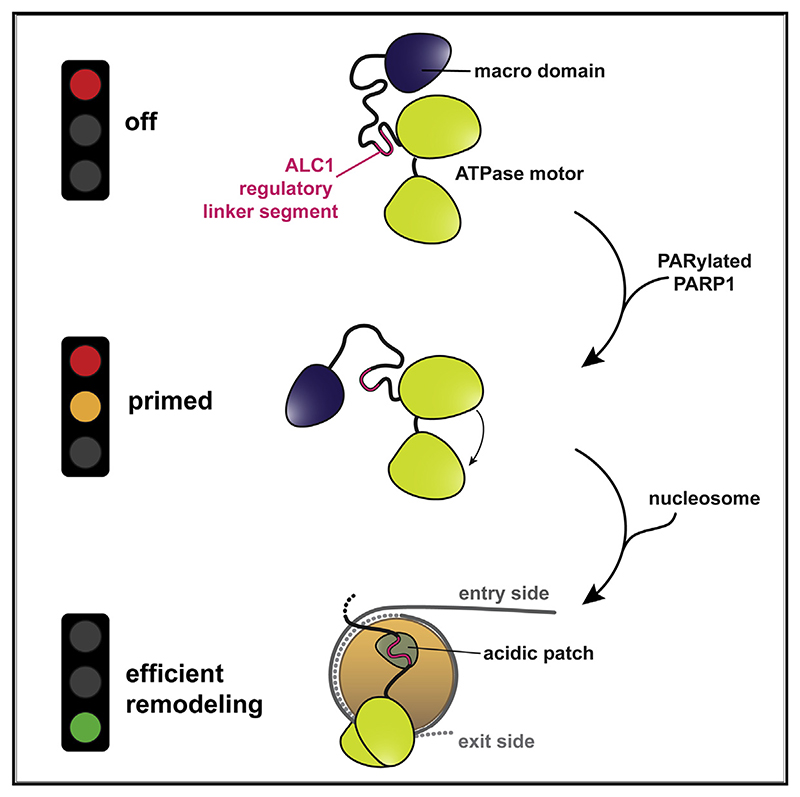
(E) In the absence of DNA damage, association of the macro domain of ALC1 with its C-terminal ATPase lobe stabilizes an ATPase “OFF” state. PAR binding to the macro domain displaces it from the ATPase. ALC1RLS then tethers the remodeler to the AP, which stabilizes an active conformation (macro domain not shown) and promotes efficient coupling of ATP hydrolysis to nucleosome mobilization.
See also Figure S4.
Discussion
We have previously described the auto-inhibited conformation of ALC1, but how it is activated upon nucleosome binding remained unclear. In this work, we have discovered a regulatory motif within the ALC1 linker that promotes activation upon recognition of an intact AP.
An important role for the AP was suggested by ensemble experiments, where the mutation of both APs or the addition of LANA peptide compromised remodeling by ALC1. Our smFRET experiments revealed its overall ability to center nucleosomes, which was lost in the absence of an entry-side WT AP. Our cryo-EM structure of a nucleosome-ALC1 linker complex shows that the ALC1RLS engages the AP via a conserved Arg anchor in a binding mode that closely resembles that of LANA, albeit with reverse N-to C-terminal polarity. Importantly, the ALC1RLS mutation affected both the remodeling and ATPase rates under saturating conditions. Although the effect was modest (~2-fold), the R611A/S612A linker mutation did not compromise ATPase activity indiscriminately but rather in a manner specific to nucleosomes. ALC1RLS thus appears to contribute to a modest extent to the activation of the ALC1 ATPase, although our data do not explain how ALC1RLS-AP interactions may promote an active ATPase conformation. Consistent with previous observations (Ahel et al., 2009; Gottschalk et al., 2009), the nucleosome-stimulated ATPase activity of ALC1fl R860W was ~1.6-fold higher than its DNA-stimulated ATPase activity. Mutation of ALC1RLS affected the remodeling rate of ALC1 more strongly than its ATPase activity (~6.4-fold versus ~2-fold, respectively), suggesting a role of the ALC1RLS-AP interaction in the coupling of ATP hydrolysis to nucleosome mobilization. Live-cell imaging showed that disrupting the ALC1-AP interaction impacts ALC1 recruitment to sites of DNA damage in vivo.
Our results demonstrate a critical role for the interaction between ALC1RLS and the AP in tethering the remodeler to the nucleosome and in stimulating both remodeling and, to a modest extent, the ATPase activities of ALC1. Taken together, our data are consistent with the following model for activation (Figure 4E): upon recruitment to DNA damage and displacement of the macro domain, ALC1 associates with the nucleosome (Lehmann et al., 2017). In the fully activated state of ALC1, the AP engages the ALC1RLS, which tethers the remodeler to the nucleosome and promotes efficient coupling of ATP hydrolysis to remodeling.
Our work has uncovered an unexpected function of the ALC1RLS that enables full activation only upon specific recognition of a nucleosome with an intact AP. Our experiments reveal that, with both APs intact, ALC1 exhibits a preference to center the octamer, albeit to a lesser extent than ISWI remodelers such as ACF and ISW2 (Ito et al., 1997; Längst et al., 1999; Tsukiyama et al., 1999; Varga-Weisz et al., 1997). Our data further reveal an unexpected divergence between ALC1 and the related Chd1 in their response to AP asymmetry. Unlike ALC1, Chd1 is only modestly affected by AP mutations (Levendosky and Bowman, 2019). Moreover, Chd1 responds similarly to AP mutations on entry-side and exit-side H2A/H2B dimers. In stark contrast, ALC1 depends much more strongly on a WT AP on the entry side. For ALC1-catalyzed remodeling of nucleosomes with AP asymmetry, net movement occurs toward the WT AP-containing side. Our smFRET analyses suggest that the pause phase of the remodeling cycle might serve to probe for an intact AP. Although future studies with histone variants are required, we speculate that, in vivo, the selective removal or blocking of one of the two APs might locally redirect ALC1 remodeling away from spacing and toward the disorganization of nucleosomes. Such remodeling may facilitate chromatin relaxation in a DNA-damage-specific chromatin context.
Star★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| hnRNPAI | Sant Cruz Biotechnology | Cat#SC-32301; RRID: AB_627729 |
| CHD1L | Cell Signaling Technology | Cat#13460; RRID: AB_2798225 |
| Bacterial and Virus Strains | ||
| E. coli Rosetta 2 (DE3) | Novagen | Cat#71400 |
| E. coli BL21 (DE3) pLysS | Novagen | Cat#69451 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BrdU | Sigma-Aldrich | Cat#B5002 |
| Doxycycline | Sigma-Aldrich | Cat#M0503-5X2MG |
| Blasticidin | ThemoFisher Scientific | Cat#A1113903 |
| Hygromycin B | ThemoFisher Scientific | Cat#10687010 |
| Zeocin | ThemoFisher Scientific | Cat#R25005 |
| Lipofectamine 2000 | ThemoFisher Scientific | Cat#11668019 |
| EDTA-free Complete protease inhibitor cocktail | Roche | Cat#COEDTAF-RO |
| PhosSTOP phosphatase inhibitor cocktail | Roche | Cat#PHOSS-RO |
| 4x NuPAGE LDS sample buffer | ThemoFisher Scientific | Cat#NP0008 |
| LANA peptide | Peptide 2.0 | N/A |
| LANA peptide-TAMRA | Peptide 2.0 | N/A |
| ALC1RLS peptide | Peptide 2.0 | N/A |
| ALC1RLS peptide-TAMRA | Peptide 2.0 | N/A |
| ALC1 linker peptide-Biotin | Peptide 2.0 | N/A |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#5056489001 |
| Benzonase Nuclease | Sigma-Aldrich | Cat#E1014 |
| Cy3-maleimide | GE Healthcare | Cat#PA23031 |
| Pyruvate Kinase/Lactic Dehydrogenase | Sigma-Aldrich | Cat#P0294 |
| Trypsin | Promega UK | Cat#V5111 |
| Glu-C | Promega UK | Cat#V165A |
| Deposited Data | ||
| Raw cryo-EM videos and final particle images (deposited on EMPIAR) | This study | EMPIAR-10465 |
| Nucleosome cryo-EM maps (deposited on EMDB) | This study | EMD-11220 |
| Hexasome cryo-EM maps (deposited on EMDB) | This study | EMD-11221 |
| Nucleosome atomic model (deposited in PDB) | This study | 6ZHX |
| Hexasome atomic model (deposited in PDB) | This study | 6ZHY |
| Experimental Models: Cell Lines | ||
| U2OS Flp-In T-Rex HOST | Durocher lab | N/A |
| ALCI –/– U2OS Flp-In T-REx | This paper | N/A |
| ALCI –/– U2OS Flp-In YFP-ALC1 | This paper | N/A |
| ALCI –/– U2OS Flp-In YFP-ALC1 R860W | This paper | N/A |
| ALCI –/– U2OS Flp-In YFP-ALC1 R611A/ R612AR860W | This paper | N/A |
| ALC1 –/– U2OS Flp-In YFP-ALC1 R611Q R860W | This paper | N/A |
| Oligonucleotides | ||
| DNA oligonucleotides | This paper | Table S3 |
| Recombinant DNA | ||
| Human ALC1fl (16-879) pNIC-CH2 | Lehmann et al., 2017 | N/A |
| Human PARP1 (1-1014) pET-28 | Langelier et al., 2017 | N/A |
| px459 | Addgene | #62988 |
| px459 ALC1 EX3 | This paper | N/A |
| pOG44 Flp-Recombinase expression vector | ThemoFisher Scientific | Cat#V600520 |
| ALC1 WT YFP-ALC1 pDEST-YFP/FRT/TO | Ahel et al., 2009 | N/A |
| ALC1 R860W YFP-ALC1 pDEST-YFP/FRT/TO | Lehmann et al., 2017 | N/A |
| ALC1 R611A/R612AR860W YFP-ALC1 pDEST-YFP/ | ||
| FRT/TO | This paper | N/A |
| ALC1 R611Q/R860W YFP-ALC1 pDEST-YFP/FRT/TO | This paper | N/A |
| X. laevis Pet3a_H2A | Geeta Narlikar Lab | N/A |
| X. laevis Pet3a_H2B | Geeta Narlikar Lab | N/A |
| X. laevis Pet3a_H3 | Geeta Narlikar Lab | N/A |
| X. laevis Pet3a_H4 | Geeta Narlikar Lab | N/A |
| Software and Algorithms | ||
| Warp | Tegunov and Cramer, 2019 | http://www.warpem.eom/warp/# |
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| RELION-3.1 | Scheres, 2012; Zivanov et al., 2018, 2020 | https://www3.mre-lmb.eam.ae.uk/relion/index.php/Main_Page |
| UCSF ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| Coot | Casahal et al., 2020 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| ISOLDE | Croll, 2018 | https://isolde.cimr.cam.ac.uk/ |
| Prism 8 | GraphPad Software | https://www.graphpad.com/ |
| Fiji | NIH | https://imagej.net/Fiji/Downloads |
| Image Lab 5.2.1 | Bio-Rad Laboratories | https://www.bio-rad.com/en-uk/product/image-lab-%20software?ID%20=%20KRE6P5E8Z |
| Andor iQ. | Oxford Instruments | https://andor.oxinst.com/products/iq-live-cell-imaging-software/ |
| Adobe Illustrator 23.11 | Adobe | https://www.adobe.com/uk/products/illustrator.html |
| Phenix suite | Liebschner et al., 2019 | https://www.phenix-online.org/ |
| Other | ||
| 5 mL HisTrap FF | GE Healthcare | Cat#17525501 |
| HiPrep 26/10 Desalting | GE Healthcare | Cat#17508701 |
| 5 mL HiTrap Q HP | GE Healthcare | Cat#17115401 |
| 5 mL HiTrap SP HP | GE Healthcare | Cat#17115201 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat#28989335 |
| 5 mL HiTrap Heparin HP | GE Healthcare | Cat#17040703 |
| HiPrep 16/60 Sephacryl S-200 HR | GE Healthcare | Cat#17116601 |
| Superdex Peptide 3.2/300 (discontinued) | GE Healthcare | Cat# 29036231 |
| Acquity UPLC CSH C18 1.7 mm, 1.0 x 100 mm column | Waters | Cat#176002137 |
| C18 Acclaim PepMap100 5 mm, 100 mm x 20 mm nanoViper | Thermo Scientific | Cat#164564 |
| C18 Acclaim PepMap100 3 mm, 75 mm x 250 mm nanoViper | Thermo Scientific | Cat#164569 |
| Pierce Monomeric Avidin Agarose resin | Thermo Scientific | Cat#20228 |
| Quantifoil R 2/2 Cu 300 grids | Electron Microscopy Sciences | Cat#Q3100CR2 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sebastian Deindl (sebastian.deindl@icm.uu.se).
Materials Availability
This study did not generate new unique reagents.
Experimental Model and Subject Details
cDNAs for ALC1 and PARP1 were of Homo sapiens origin. Histone proteins (recombinantly expressed in E. coli) were of Xenopus laevis origin. Cell lines as well as growth conditions are described in the Method Details.
Method Details
Cloning, plasmids, and cell lines
Oligonucleotides used in this study were ordered from IDT (see Table S3). The ALC1fl (16-879) construct contains the human ALC1 sequence and a 6xHis-tag, as previously published (Lehmann et al., 2017). All point mutations were generated by PCR-based site-directed mutagenesis. The plasmid for human PARP1 (1-1014) with an N-terminal 6xHis-tag was a kind gift from John M. Pascal (Langelier et al., 2017).
U2OS Flp-In T-REx cells were a kind gift from Durocher lab and maintained in DMEM medium (GIBCO/Thermo Fisher) supplemented with 10% FBS and Pen/Strep with 15.5 μg/ml zeocin (Invitrogen) and 4 μg/ml blasticidin. CRISPR inactivation of ALC1 was carried out in Flp-In T-REx U2OS cells by transiently expressing Cas9 and gRNA targeting Exon3 of ALC1 F-oligo: CACCGCAG GAAGATTAAATGATGAA R-oligo: AAACTTCATCATTTAATCTTCCTGC with px459 (addgene) (Ran et al., 2013). Single cell clones were grown by limiting dilution and then screened for ALC1 expression by western blot. For in vivo experiments, an YFP-labeled construct of ALC1 (YFP-ALC1 pDEST-YFP/FRT/TO) was used, previously generated by amplification from a human HeLa cDNA library and subsequent cloning by Gateway LR reaction (Ahel et al., 2009). ALC1 linker-mutations R611Q and R611A/S612A were introduced into ALC1 R860W YFP-ALC1 pDEST-YFP/FRT/TO (from Lehmann et al., 2017) by quick-change mutagenesis. Inducible YFP-ALC1 cell lines were generated using the Flp-In T-REx system (Invitrogen) as described in the manufacturer’s protocol. Each YFP/FRT/TO construct was co-transfection with the pOG44 vector (Flp recombinase) into ALC1 –/– U2OS host cell lines. Recombination events were selected with 250 μg/ml hygromycin B (ThermoScientific).
DNA constructs
The DNA constructs used in the nucleosome assembly for single-molecule and ensemble remodeling studies comprised the Widom 601 nucleosome positioning sequence (Lowary and Widom, 1998) as well as either 3 bp or 12 bp of linker DNA on one side and 78 bp on the other side (for the single-molecule remodeling construct) or 63 bp of linker DNA on one side and no linker DNA on the other side (for the ensemble remodeling construct). All constructs were 5’-labeled with Cy5 on the short linker end. The constructs for single-molecule experiments additionally featured a biotin on the long linker end. DNA constructs were generated by PCR using HPLC-purified labeled oligonucleotide primers (IDT), followed by PAGE purification of the PCR product using a BioRad MiniPrep Cell or Prep Cell apparatus. The DNA constructs for the assembly of nucleosomes used in fluorescence anisotropy experiments and ATPase assays comprised the Widom 601 positioning sequence without additional linker DNA (for the fluorescence anisotropy construct) or 63 bp of linker DNA on one side and no linker DNA on the other side (for the ATPase assay construct). Both constructs were purified as described above.
Peptides
The following peptides were purchased from Peptide 2.0:
LANA peptide: MAPPGMRLRSGRSTGAPLTRGS
LANA peptide-TAMRA: MAPPGMRLRSGRSTGAPLTRGSK-/TAMRA/
ALC1RLS peptide: EKASQEGRSLRNKGSVLIPGL
ALC1RLS peptide-TAMRA: EKASQEGRSLRNKGSVLIPGLK-/TAMRA/
ALC1 linker peptide-Biotin: EKASQEGRSLRNKGSVLIPGLVEGSTKRKRVLSPEEK-/Biotin/
ALC1 expression and purification
All ALC1 constructs were expressed and purified as previously described (Lehmann et al., 2017). In brief, proteins were expressed in Rosetta 2 (DE3) cells (Novagen) and induced at 18°C overnight with 0.5 mM IPTG at OD600 = 2.0. For the purification, cell pellets were resuspended in Nickel column buffer A (20 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole, 10% glycerol, 5 mM β-mercaptoe-thanol), containing a protease inhibitor cocktail (Roche) and Benzonase Nuclease (Sigma). The cells were lysed using a sonicator, centrifuged, and the resulting supernatant was filtered through a 0.45 μm filter and purified over a 5 mL HisTrap FF column (GE Healthcare). Peak fractions were pooled and desalted using a HiPrep 26/10 desalting column (GE Healthcare) into S column buffer (20 mM MES pH 6.0,300 mM NaCl, 10% glycerol, 1 mM DTT). The protein was loaded onto a 5 mLSP HP column (GE Healthcare), with a 5 mL Q HP column (GE Healthcare) attached in tandem to trap contaminating DNA, and eluted with a linear salt gradient. Peak fractions were injected onto a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) equilibrated with 20 mM MES pH 6.5,300 mM NaCl, 5% glycerol, and 0.5 mM TCEP. The final protein was concentrated to typically 10 mg/ml, flash frozen in liquid nitrogen, and stored at –80°C.
PARP1 expression and purification
Full-length human PARP1 was expressed and purified as previously described (Langelier et al., 2017). In brief, the PARP1 construct was introduced in Rosetta 2 (DE3) cells (Novagen) that were grown in the presence of 10 mM benzamide to OD600 = 0.8-1.0 at 37°C and after the addition of ZnSO4 (final conc. 0.1 mM), expression was induced at 16°C overnight with 0.2 mM IPTG. For the purification, cell pellets were resuspended in resuspension buffer (25 mM HEPES pH 8,500 mM NaCl, 0.5 mM TCEP, 10 mM benzamide and 0.1% NP-40), containing a protease inhibitor cocktail (Roche). The cells were lysed using a sonicator, centrifuged, and the resulting supernatant was filtered through a 0.22 μm filter and loaded on to a 5 mL HisTrap FF column (GE Healthcare) equilibrated with equilibration buffer (25 mM HEPES pH 8, 500 mM NaCl, 0.5 mM TCEP). After consecutive washes with Low Salt and High Salt Wash buffers (25 mM HEPES pH 8, 500 mM (low)/1 M (high) NaCl; 0.5 mM TCEP, 20 mM imidazole and protease inhibitor cocktail) protein was eluted using elution buffer supplemented with 400 mM imidazole. Peak fractions were pooled and diluted with No Salt Heparin buffer (50 mM Tris pH 7.0,1 mM EDTA, 0.1 mM TCEP) to a final salt concentration of 375 mM NaCl prior to loading the protein onto a 5 mL HiTrap Heparin HP column (GE Healthcare) equilibrated with 375 mM NaCl containing Low Salt Heparin buffer. Protein was eluted using a linear salt gradient. Peak fraction were pooled, filtered through a 0.22 μm filter and loaded onto a HiPrep 16/60 Sephacryl S-200 HR (GE Healthcare) column equilibrated with gel filtration buffer (25 mM HEPES pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1 mM TCEP). The final protein was concentrated to 30 mg/ml and after flash freezing in liquid nitrogen stored at –80°C.
Histone expression and purification
Recombinant histones (H2A, H2A K120C, H2AAPM, H2A APM K120C, H3 C110Aand H4) from Xenopus laevis were purified essentially as described previously (Klinkeret al., 2014). In brief, proteins were expressed in BL21 (DE3) pLysS (Novagen) cellsand induced at OD600 = 0.6-0.8 for 2 h at 37°C with 1 mM IPTG. The cell pellets were resuspended in 40 mM NaOAc pH 5.2,1 mM EDTA, 10 mM lysine, 200 mM NaCl, 5 mM β-mercaptoethanol, 6 M urea, and a protease inhibitor cocktail (Roche). Once homogenized, the cells were lysed using a sonicator, centrifuged, and the resulting supernatant was filtered through a 0.45 μm filter and purified over a 5 mL SP HP column (GE Healthcare), with a 5 mL Q HP column (GE Healthcare) attached in tandem to trap contaminating DNA and eluted with a salt gradient. Peak fractions were dialyzed overnight against cold water. The dialysate was mixed with Tris pH 8.0 to a final concentration of 15 mM and then passed over a 5 mLQ HP column (GE Healthcare). The final protein was concentrated to 5 mg/ml, flash frozen in liquid nitrogen, and stored at –80°C.
Histone labeling
Histones H2A K120C and H2AAPM K120C were site-specifically labeled at cysteine residue 120 with Cy3-maleimide (GE Healthcare) as previously described (Yang et al., 2006; Blosser et al., 2009). In brief, one mg of histone protein was diluted in labeling buffer (20 mM Tris pH 7.0, 7 M guanidine-HCl, 5 mM EDTA, 1.25 mM TCEP) and incubated for 2 h at room temperature in the dark. Cy3-maleimide was dissolved in DMSO and added to the protein at a final concentration of 0.75 mM. After 3 h in the dark at room temperature, the reaction was quenched with a final concentration of 80 mM β-mercaptoethanol. The labeled protein was dialyzed nine times against dialysis buffer (20 mM Tris pH 7.0, 7 M guanidine-HCl, 1 mM DTT) and then used directly in an octamer assembly.
Histone octamer assembly
The four histones were combined at a molar ratio 1.2:1.2:1:1 (H2A:H2B:H3:H4), unfolded in 400 mM Tris pH 7.5, 7 M guanidine-HCl, 200 mM DTT and the histone octamer was assembled by dialyzing three times against refolding buffer (10 mM Tris-HCl pH 7.5, 2 M NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol). The assembly reaction was filtered through a 0.22 μm filter and loaded onto a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) equilibrated with refolding buffer. Peakfractions were pooled, concentrated to 3 mg/ml, flash frozen in liquid nitrogen, and stored at –80°C.
Nucleosome assembly
Mononucleosomes were reconstituted from the DNA constructs and histone octamer (detailed above) by salt dialysis (Dyer et al., 2004). Nucleosomes for FRET-based assays were additionally subjected to PAGE purification using a BioRad MiniPrep Cell or Prep Cell apparatus.
ATPase activity measurements
ATPase activity was measured using a coupled assay that measures ADP production as described previously, (Willhoft et al., 2016). Final concentrations of 0.45 mM NADH, 1.0 mM phosphoenolpyruvate, 54 U/ml pyruvate kinase (Sigma), and 78 U/ml lactic dehydrogenase (Sigma) were used. For measurements of nucleosome-stimulated ATPase activity, reactions contained 20 μM ALC1,2 μM WT/WT nucleosomes, and 1 mM ATP in a volume of 30 μl. Fluorescence was monitored (excitation wavelength: 340 nm; emission wavelength: 430 nm) at 37°C using a CLARIOstar microplate reader (BMG Labtech) and black low-volume 384-well microplates (Corning). For measurements of DNA-stimulated ATPase activity, the absorbance was monitored at 340 nm for reactions containing a final of 1 μM ALC1 and either 20 μM or 50 μM 145 bp nucleosomal double-stranded (ds) DNA in 100 μl. Reaction rates were determined using mean linear slopes. For measurements of nucleosome-stimulated ATPase activity, we estimated Vmax for both ALC1fl R860W and ALC1fl R611A/S612A/R860W using the Michaelis-Menten equation and the Km values that we obtained from nucleosome sliding kinetics in Figure 3B. Based on Vmax, the difference in nucleosome-stimulated ATPase activity is ~2-fold lower for ALC1fl R611A/S612A/R860W compared to ALC1fl R860W (rather than ~2.5-fold).
Fluorescence anisotropy binding assay
For binding studies, we used nucleosomes without linker DNA (WT/WT or APM/APM) as well as the following synthetic peptides (as detailed above): LANA peptide-TAMRA, ALC1RLS peptide-TAMRA, and ALC1RLS peptide. Fluorescence anisotropy measurements were carried out at 30°C using a CLARIOstar microplate reader (BMG Labtech). Binding curves were recorded in black low-volume 384-well assay plates (Corning). Individual wells contained 10 nM peptide and the indicated concentration of nucleosomes in 15 mM HEPES pH 7.5,1 mM EDTA, 5% glycerol, and 1 mM DTT. Dissociation constants were determined by fitting the fluorescence anisotropy data to the solution of a quadratic equation derived from the binding isotherm, which took into account depletion of peptide: , where r is the fluorescence anisotropy, A represents the total concentration of peptide, a is a scaling factor, KD is the dissociation constant, and B is the total concentration of titrated ligand. For competition binding experiments, WT/WT nucleosomes (100 nM) were incubated with LANA-TAMRA peptide (10 nM) prior to the addition of varying concentrations of unlabeled ALC1RLS peptide.
Ensemble FRET assay for nucleosome remodeling
Nucleosome ensemble remodeling kinetics were measured by monitoring the Cy5 (under 620 nm and 520 nm excitation) and Cy3 (under 520 nm excitation) fluorescence emission signals of a solution of FRET-labeled nucleosomes using a Spark (Tecan) or CLARIOstar (BMG Labtech) multimode microplate reader. Ensemble nucleosome remodeling assays were performed with 10 nM nucleosomes, varying concentrations of ALC1 as indicated, 1 mM MgCl2 and 1.5 mM ATP in remodeling buffer (20 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 5% sucrose, 0.1 mg/ml BSA, 1 mM DTT). Where indicated, PARP1 was PARylated prior to nucleosome remodeling by incubating (in remodeling buffer at 37°C for 5 min) 80 nM or 5 μM PARP1 with 120 nM or 7.5 μM nucleosomal DNA as well as 50 μM or 100 μM NAD+, for reactions with 80 nM or 5 μM ALC1, respectively. For remodeling assays in the presence of LANA peptide, 10 nM nucleosomes were incubated with 80 μM LANA peptide at room temperature for 5 min prior to the addition of MgCl2 and ALC1. Remodeling rates were obtained as initial slopes of the remodeling curves. Reaction kinetics were analyzed assuming a standard Michaelis-Menten mechanism.
Single-molecule FRET remodeling
Using the DNA constructs for single-molecule FRET described above, we first generated oriented hexasomes with H2A-Cy3 on the short linker side using a salt dialysis method as described previously (Dyer et al., 2004; Levendosky et al., 2016; Sabantsev et al., 2019). Nucleosomes were then reconstituted by adding a 3-fold excess of WT or APM H2A/H2B (final concentration of dimer 1.2 μM) to purified hexasomes and incubating the mixture at 37°C for 10 min (Levendosky et al., 2016). This allowed for the controlled incorporation of the Cy3 label and AP mutations in a specific orientation (Figure 1D). Labeled nucleosomes were immobilized on a PEG (poly[ethylene glycol])-coated quartz slide using biotin-streptavidin interactions (Deindl et al., 2013). Cy3 and Cy5 fluorophores were excited with 532 nm Nd:YAG and 638 nm diode lasers, respectively, and fluorescence emissions from Cy3 and Cy5 were detected using a custom-built prism-based TIRF microscope. To check the presence of an intact donor fluorophore, the sample was alternately excited with 532 nm and 638 nm lasers during the experiment. Data acquisition was controlled using MicroManager (Edelstein et al., 2010). Data were analyzed using custom scripts for the Fiji distribution of ImageJ (Schindelin et al., 2012; Schneider et al., 2012), IDL, and MATLAB (Sabantsev et al., 2019). The initial remodeling direction was determined by visual inspection of the FRET traces. Remodeling experiments were carried out in the imaging buffer containing 40 mM Tris pH 7.5,12 mM HEPES pH 7.9,60 mM KCl, 0.32 mM EDTA, 3 mM MgCl2, 100 μg/mL acetylated BSA (Promega), 10% (v/v) glycerol, 10% (w/v) glucose, 2 mM Trolox to reduce photoblinking of the dyes (Rasnik et al., 2006), as well as an enzymatic oxygen scavenging system (composed of 800 μg/ ml glucose oxidase and 50 μg/ml catalase). Using a syringe pump (Harvard Apparatus), remodeling was initiated by infusing the sample chamber with imaging buffer supplemented with ALC1fl R860W (2.5 μM), ATP (1 mM) and MgCl2 (1 mM). The nucleosomes used for the quantification of remodeling directionality (Figure 1D) exhibited an intermediate starting FRET value of ~0.4. Upon remodeling, a subset of time traces displayed an intial FRET increase, followed by a subsequent FRET decrease after reaching the maximum FRET value. Such a FRET signature could be caused by unidirectional remodeling that proceeds beyond the DNA end rather than by a change in remodeling direction. As soon as the end of the FRET dynamic range is reached (a maximum FRET value in this case), we cannot reliably distinguish between these two cases. For this reason, we limited our analyses to the initial portions of the time traces (before the edges of the FRET dynamic range are reached) that can be unambiguously interpreted.
Most traces exhibit repeated directional switching, which might be caused by two ALC1 molecules bound at opposing sides of the same nucleosome taking turns in remodeling it. Alternatively, such non-monotonous FRET changes could also be due to the consecutive binding of ALC1 molecules in different orientations, orstemfrom the same ALC1 molecule switching between the two opposing binding sites within a single binding event, as has been observed for Chd1 (Qiu et al., 2017). We note that our data do not allow us to distinguish between these scenarios.
The unusually long pauses observed for ALC1fl R611A/S612A/R860W predominantly represent pauses in translocation by the same remodeler as opposed to separate remodeler binding events, based on the following considerations. First, the average pause duration was 2.8-fold shorter than the average binding time for the remodeler at a concentration of 20 μM. Second, lowering the remodeler concentration by a factor of 4 produced only a minor (30%) increase in the observed pause duration.
Cross-linking coupled to mass spectrometry
For cross-linking coupled to mass spectrometry, instead of ALC1fl R860W we used ALC1fl R857Q because that mutant could be produced in the required larger quantities. Importantly, biochemical and cell-based approaches have previously shown that both the R857Q and R860W mutations affect the same electrostatic interface and can be used interchangeably to render ALC1 constitutively active independent of PARP1 activation (Lehmann et al., 2017). ALC1fl R857Q-nucleosome complexes were cross-linked by incubation with a 100-fold molar excess of N-hydroxysuccinimide (NHS) ester disuccinimidyl dibutyric urea (DSBU, formerly BuUrBu) for 45 min at room temperature. The reactions were quenched by adding NH4HCO3 to a final concentration of 50 mM and incubating for an additional 15 min. The cross-linked samples were freeze-dried and resuspended in 50 mM NH4HCO3, reduced with 10 mM DTT, and alkylated with 50 mM iodoacetamide. Following alkylation, proteins were digested with trypsin (Promega, UK) at an enzyme-to-substrate ratio of 1:20 overnight at 37°C or sequentially with trypsin and Glu-C (Promega, UK) at an enzyme-to-substrate ratio of 1:20 and 1:50 at 37°Cand 25°C, respectively. The samples were acidified with formic acid to a final concentration of 2% (v/v), split into two equal amounts for peptide fractionation by peptide size exclusion chromatography (SEC) and reverse phase C18 high pH chromatography (C18-Hi-pH). For SEC, a Superdex Peptide 3.2/300 column (GE Healthcare) with 30% (v/v) acetonitrile/0.1% (v/v) TFA as the mobile phase and a flow rate of 50 μl/min were used, and fractions were collected every 2 min over an elution volume of 1.0 ml to 1.7 ml. C18-Hi-pH fractionation was carried out on an Acquity UPLC CSH C18 1.7 μm, 1.0 × 100 mm column (Waters) over a gradient of acetonitrile 2%-40% (v/v) and 100 mM NH4HCO3. Fractions were lyophilized and resuspended in 2% (v/v) acetonitrile and 2% (v/v) formic acid and analyzed by nano-scale capillary LC-MS/MS using an Ultimate U3000 HPLC (ThermoScientific Dionex, USA) to deliver a flow of approximately 300 nl/min. A C18 Acclaim PepMap100 5 μm, 100 μm × 20 mm nanoViper (Thermo-Scientific Dionex, USA) trapped the peptides before separation on a C18 Acclaim PepMap100 3 μm, 75 μm × 250 mm nanoViper (ThermoScientific Dionex, USA). Peptides were eluted with an acetonitrile gradient. The analytical column outlet was directly interfaced via a nano-flow electrospray ionisation source, with a hybrid quadrupole orbitrap mass spectrometer (Q-Exactive HF-X, ThermoScientific, USA). MS data were acquired in data-dependent mode. High-resolution full scans (R = 120,000, m/z 350-2000) were recorded in the Orbitrap followed by higher energy collision dissociation (HCD) (stepped collision energy 30 ± 3) of the 10 most intense MS peaks. MS/MS scans (R = 45,000) were acquired with a dynamic exclusion window of 20 s.
For data analysis, Xcalibur raw files were converted into the MGF format through MSConvert and used directly as input files for MeroX2. Searches were performed against an ad hoc protein database containing the sequences of the complexes and a set of randomized decoy sequences generated by the software. The following parameters were set for the searches: maximum number of missed cleavages 3; targeted residues K, S, Y and T; minimum peptide length 5 amino acids; variable modifications: carbamido-methyl-Cys (mass shift 57.02146 Da), Met-oxidation (mass shift 15.99491 Da); DSBU modification fragments: 85.05276 Da and 111.03203 (precision: 5 ppm MS1 and 10 ppm MS2); False Discovery Rate cut-off: 5%. Finally, fragmentation spectra were manually inspected and validated.
Cryo-EM sample preparation
A biotinylated ALC1 linker peptide was added to core nucleosomes (without any linker DNA on either side of the histone octamer) at 10-fold molar excess in the reaction buffer (15 mM HEPES pH 7.5,1 mM EDTA, 1 mM DTT). After 10 min of incubation on ice, the complex was cross-linked with 0.05% glutaraldehyde for 5 min on ice. The cross-linking reaction was quenched by adding 1 M Tris-HCl pH 7.5 and the sample was dialyzed against purification buffer (15 mM HEPES 7.5,150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.01% NP-40, 1 mM DTT) for one hour at 4°C. After dialysis, the sample was applied onto a column custom-packed with Pierce Monomeric Avidin Agarose resin (Thermo Scientific) and incubated for 30 min to allow binding to the column. Washing and elution steps were performed with a controlled flow rate of 0.1 ml/min. The cross-linked complex between the nucleosome and biotinylated ALC1 linker peptide was eluted with the same purification buffer supplemented with 2 mM biotin. Fractions containing the complex were identified by native PAGE, pooled, concentrated, and buffer-exchanged using a 30 kDa cut-off spin concentrator in cryo-EM buffer (15 mM HEPES pH 7.5, 50 mM NaCl, 1 mM DTT). Quantifoil R 2/2 Cu 300 grids (Electron microscopy sciences) were glow-discharged at 20 mA for 60 s (PELCO easiGlow). 3 μl of the sample at 6.4 μM were applied onto grids and immediately blotted for 2.5 s. Sample application and blotting was repeated once more before rapid plunge-freezing into liquid ethane, using a Vitrobot Mark IV (Thermo Scientific) operated at 100% relative humidity and 4°C.
Cryo-EM data collection and processing
Cryo-EM data were collected at the Karolinska Institutet 3D-EM facility (Stockholm, Sweden) using a Titan Krios G3 microscope operated at 300 kV and equipped with a Gatan K3 Bioquantum detector and a GIF Quantum LS energy filter (slit width 20 eV). Movies were recorded in counting mode, gain-corrected, at a calibrated pixel size of 0.654 Å/pixel, with a total exposure of 50.4 electrons/Å2 fractionated over 60 movie frames (resulting in an exposure per frame of 0.84 electrons/Å2/frame). Motion correction, CTF estimation and particle picking were performed during data collection using Warp (Tegunov and Cramer, 2019). A subset of the particles extracted by Warp were imported into cryoSPARC (Punjani et al., 2017) for evaluation by 2D and 3D classification. The entire dataset was then reprocessed in RELION-3.1.0 (Scheres, 2012; Zivanov et al., 2018, 2020). Motion correction was performed using Motion-Cor2 version 1.3.1 (Zheng et al., 2017) and CTF estimation was performed using Gctf version 1.18b2 (Zhang, 2016), both from within RELION. Particle coordinates from Warp were imported into RELION. Unsuitable particles were excluded after one round of reference-free 2D classification. The initial 3D model was generated from the data using the InitialModel (stochastic gradient descent) procedure in RELION. Unsuitable particles were further excluded in six rounds of 3D classification, without any symmetry constraints applied (C1). Round 3 of 3D classification identified two sets of particles corresponding to a nucleosome with both APs occupied by a peptide and a hexasome with its single AP occupied by a peptide. These two classes were independently subjected to 3D refinement, CTF refinement (in three successive steps: beam tilt and higher-order aberrations, anisotropic magnification, and per-particle defocus plus per-micrograph astigmatism), 3D refinement, Bayesian polishing and a final 3D refinement (Figure S2; Table S2). Consistent with complex preparation by first saturating the nucleosome with ALC1 linker peptide, followed by crosslinking and purification of nucleosomes-peptide complexes, all nucleosome particles had two peptides bound, occupying the APs on both sides of the nucleosome. The dataset contains neither unbound nucleosomes, nor nucleosomes with only one AP occupied. Our dataset also contains hexasomes (nucleosomes lacking one H2A-H2B dimer), since partial nucleosome disassembly upon vitrification is a common phenomenon (Bilokapic et al., 2018). Interestingly, the hexasome class also displayed density from the bound ALC1 linker peptide on the remaining H2A/H2B dimer, supporting our in vitro characterization of cross-linking efficiency: because the affinity of the peptide for the AP is much lower than that of the H2A/H2B dimer for the hexasome, we would not expect to see density for the peptide under conditions that lead to dissociation of one H2A/H2B dimer, yet we observe such density here, consistent with efficient cross-linking. The local resolution was estimated with RELION’s local Fourier Shell Correlation procedure.
Model building and refinement
We built a model of a nucleosome with both APs bound by the ALC1 linker peptide from PDB entries 3LZ0 (nucleosome with Xenopus laevis histones and Widom 601 DNA) (Vasudevan et al., 2010) and 1ZLA (LANA peptide) (Barbera et al., 2006) and performed rigid-body fitting of this model into our cryo-EM map using UCSF ChimeraX version 0.93 (Goddard et al., 2018). The resolution of our maps allowed us to distinguish between the two asymmetric sides of the 601 sequence, so we oriented the nucleosome model properly into the map in the orientation that gave the best model-to-map real-space correlation coefficient. We edited the LANA peptide sequence to match the ALC1 linker peptide and deleted residues not supported by density in our cryo-EM map, using Coot version 0.9-pre-855 (Casañal et al., 2020). We finally performed flexible fitting of the model into the cryo-EM map using ISOLDE version 1.0b5 (Croll, 2018). The resulting refined model was then rigid-body fit into the hexasome map, the H2A, H2B and ALC1 linker peptide chains were deleted from the model on the side of the hexasome map devoid of density for these chains, DNA nucleotides not supported by density were also deleted from the model (corresponding to the more easily unwrapped side of the 601 sequence), and the resulting hexasome model was subjected to flexible fitting in ISOLDE. For more details on the model building, see also Supplemental Methods. Model-to-map real space correlation coefficients and model geometry statistics were calculated with phenix.validation_-cryoem, from the Phenix suite version 1.18.2-3874 (Liebschner et al., 2019). Model validation statistics are presented in Table S2.
Whole-cell extracts and immunoblotting
For whole cell lysates, PBS washed cells were lysed in RIPA Buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl, 1x phosphatase mix (Phos-Stop, Roche) and complete EDTA-free protease inhibitor mix (Roche)) on ice for 20 min. Lysates were sonicated with a probe at medium intensity for 5 s in a Soniprep 150 instrument and clarified by centrifugation at 13000 xg for 10 min at 4°C. Proteins were denatured in 2X NuPAGE LDS sample buffer (Invitrogen) and 1% 2-metcaptoethanol (Sigma-Aldrich) for 5 min at 95°C. Proteins were separated by SDS-PAGE using Nu-PAGE mini gels (Invitrogen) and transferred onto 0.4 PVDF (Millipore). Membranes were blocked with 5% skim milk/PBST (PBS/0.1% Tween-20) for 1 h at room temperature and probed with ALC1 1:1000 (Cell Signaling Technology), hnRNPA1 1:3000 (Sant Cruz Biotechnology) primary antibodies overnight at 4°C. Membranes were then washed 3 times for 10 min with PBST, incubated with appropriate secondary antibodies conjugated to a horseradish peroxidase (HRP) for 1 h at room temperature and washed again 3 times for 10 min with PBST. Immunoblots were developed using Clarity or Clarity Max Western ECL Substrate (Bio-Rad).
Laser damage
Cells were seeded on 8 well Lab-Tek chamber slides (Thermo Fisher Scientific). Cells were pre-sensitized with 10 μM BrdU and treated with 1 μg/ml doxycycline 24 hours prior to imaging. Cells were transferred to an Nikon Ti-E with: Andor FRAPPA unit, Yoko-gawa CSU-X spinning disk scanhead, Andor iXon 897 EM-CCD camera with a heat and atmosphere controlled incubator from OKO labs. Laser micro-irradiation was performed with a 405 nm laser focused through a 60x 1.45 NA TIRF lens. Laser power was set to 50% and regions of interest (ROIs) were bleached for 20 iterations. In order to monitor the association kineticsof YFP-tagged ALC1 at sites of laser micro-irradiation, the time evolution of YFP fluorescence in the damaged region of interest was recorded upon excitation with a 488 nm laser. Recruitment of YFP to bleached ROIs was analyzed in FIJI (Schindelin et al., 2012) and data were processed in R. In brief, nuclear masks were segmented by local thresholding. FRAPped nuclei were identified by overlap of the nuclear mask with a bleached ROI. Nuclei with a circularity score less of than 0.4 were excluded from the analysis or if they contained multiple ROIs. In order to quantify recruitment, the mean YFP fluorescence intensities at a given time point t (It) were normalized to the mean initial fluorescence intensities I0 of the same region of interest. Normalized intensities were additionally multiplied with BGI0/BGIt to correct for photobleaching, where BGI represents the background fluorescence intensity. In order to compare association rates, the time course of fractional recruitment was determined as (It-I0)/(Imax-I0). Association kinetics were fit with a single-exponential (one-phase association) to obtain half-time constants.
Quantification and Statistical Analysis
For fluorescence anisotropy binding experiments, the reported values represent the mean ± SEM from 3 independent experiments. For ATPase activity measurements, the reported values represent the mean ± standard deviation from 3 independent experiments. For ensemble nucleosome remodeling time courses, each experimental condition was recorded in 3 independent experiments, and representative remodeling time courses are shown. Relative remodeling rates derived from these curves represent the mean ± standard deviation from 3 independent experiments. For smFRET experiments, values are reported as mean ± SEM from several experiments as specified in the corresponding figure legends. For association kinetics from live-cell imaging, error bars represent SEM from N ≥ 131 traces from 3 independent experiments.
Supplementary Material
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108529.
Highlight.
Remodeling by ALC1 requires an acidic patch on the nucleosome entry side
A cryo-EM structure reveals how a regulatory linker segment binds to the acidic patch
Interfacial mutations compromise recruitment to DNA damage and catalytic activities
Graphical Abstract.
Acknowledgments
We thank S.D. Knight for insightful comments; M. Hällberg, M. Carroni, and J. Conrad for assistance with cryo-EM data collection; J. Locke for early cryo-EM work; and A. Emmerich for preliminary smFRET experiments. This work was supported by the European Research Council (StG, 714068 to S.D.; AdG, Tel-Metab to S.J.B.; and CoG, 820102 to A.C.), the KAW Foundation (grant KAW 019.0306), the Swedish Research Council (VR grant 2019-03534), and Cancer-fonden (grant 19 0055 Pj). Work at the Francis Crick Institute was funded by CRUK, Wellcome Trust, and MRC (FC001065 to A.C. and FC0010048 to S.J.B.). S.J.B. acknowledges support from Wellcome Trust Senior Investigator and collaborative grants, and S.T. acknowledges support from NIH NIGMS grant R35 GM127034. S.D. is an EMBO Young Investigator.
Footnotes
Author Contributions
S.D. conceived and oversaw the study. L.C.L., L.B., G.H., K.B., A.S., G.G., S.P., G.D., H.O., S.T., A.C., J.M.S., S.J.B., and S.D. designed and/or conducted experiments. A.S. collected/analyzed smFRET data. L.B. collected cryoEM data, with help from G.G. G.G. analyzed cryo-EM data, with help from L.B. S.D. and S.J.B. wrote the manuscript, with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Data and Code Availability
Raw cryo-EM videosand final particle images: accession code EMPIAR-10465. EMDB accession codes: EMD-11220 (nucleosome) and EMD-11221 (hexasome). PDB accession codes: 6ZHX (nucleosome) and 6ZHY (hexasome).
References
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Willhoft O, Aramayo RJ, Wilkinson M, McCormack EA, Ocloo L, Wigley DB, Zhang X. Structure and regulation of the human INO80-nucleosome complex. Nature. 2018;556:391–395. doi: 10.1038/s41586-018-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a017905. a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Strauss M, Halic M. Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol. 2018;25:101–108. doi: 10.1038/s41594-017-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD. Mechanisms of ATP-dependent nucleosome sliding. Curr Opin Struct Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanal A, Lohkamp B, Emsley P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Huang J-D, Hu L, Zheng B-J, Chen L, Tsang SL, Guan X-Y. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS ONE. 2009;4:e6727. doi: 10.1371/journal.pone.0006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan THM, Yuan Y-F, Hu L, Huang J, Ma S, Wang J, Dong S-S, Tang KH, Xie D, et al. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these pro-cessesin human patients. J Clin Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll TI. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ri-bosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- Dann GP, Liszczak GP, Bagert JD, Müller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature. 2017;548:607–611. doi: 10.1038/nature23671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao HT, Dul BE, Dann GP, Liszczak GP, Muir TW. A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat Chem Biol. 2020;16:134–142. doi: 10.1038/s41589-019-0413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, Poirier GG. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J Biol Chem. 1986;26:7011–7017. [PubMed] [Google Scholar]
- Deindl S, Zhuang X. Monitoring conformational dynamics with single-molecule fluorescence energy transfer: applications in nucleosome remodeling. Methods Enzymol. 2012;513:59–86. doi: 10.1016/B978-0-12-391938-0.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell. 2013;152:442–452. doi: 10.1016/j.cell.2012.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England JR, Huang J, Jennings MJ, Makde RD, Tan S. RCC1 uses a conformationally diverse loop region to interact with the nucleosome: a model for the RCC1-nucleosome complex. J Mol Biol. 2010;398:518–529. doi: 10.1016/j.jmb.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner K-P. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018;556:386–390. doi: 10.1038/s41586-018-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DMA, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frechette A, Huletsky A, Aubin RJ, de Murcia G, Mandel P, Lord A, Grondin G, Poirier GG. Poly(ADP-ribosyl)ation of chromatin: kinetics of relaxation and its effect on chromatin solubility. Can J Biochem Cell Biol. 1985;63:764–773. doi: 10.1139/o85-096. [DOI] [PubMed] [Google Scholar]
- Gamarra N, Johnson SL, Trnka MJ, Burlingame AL, Narlikar GJ. The nucleosomal AP relieves auto-inhibition by the ISWI remodeler SNF2h. eLife. 2018;7:e35322. doi: 10.7554/eLife.35322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JA, Garlick JD, Kingston RE. Chromatin remodeling by imitation switch (ISWI) class ATP-dependent remodelers is stimulated by histone variant H2A.Z. J Biol Chem. 2010;285:4645–4651. doi: 10.1074/jbc.M109.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Trivedi RD, Conaway JW, Conaway RC. Activation of the SNF2 family ATPase ALC1 by poly(ADP-ribose) in a stable ALC1·PARP1·nucleosome intermediate. J Biol Chem. 2012;287:43527–43532. doi: 10.1074/jbc.M112.401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y. Structure of nucleosome-bound human BAF complex. Science. 2020;367:875–881. doi: 10.1126/science.aaz9761. [DOI] [PubMed] [Google Scholar]
- Hwang WL, Deindl S, Harada BT, Zhuang X. Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA. Nature. 2014;512:213–217. doi: 10.1038/nature13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Klinker H, Haas C, Harrer N, Becker PB, Mueller-Planitz F. Rapid purification of recombinant histones. PLoS ONE. 2014;9:e104029. doi: 10.1371/journal.pone.0104029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M-F, Steffen JD, Riccio AA, McCauley M, Pascal JM. Purification of DNA Damage-Dependent PARPs from E. coli for Structural and Biochemical Analysis. Methods Mol Biol. 2017;1608:431–444. doi: 10.1007/978-1-4939-6993-7_27. [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DFV, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Leduc Y, de Murcia G, Lamarre D, Poirier GG. Visualization of poly(ADP-ribose) synthetase associated with polynucleosomes by immunoelectron microscopy. Biochim Biophys Acta. 1986;885:248–255. doi: 10.1016/0167-4889(86)90239-9. [DOI] [PubMed] [Google Scholar]
- Lehmann LC, Hewitt G, Aibara S, Leitner A, Marklund E, Maslen SL, Maturi V, Chen Y, van der Spoel D, Skehel JM, et al. Mechanistic Insights into Autoinhibition of the Oncogenic Chromatin Remodeler ALC1. Mol Cell. 2017;68:847–859.e7. doi: 10.1016/j.molcel.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levendosky RF, Bowman GD. Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. eLife. 2019;8:e45472. doi: 10.7554/eLife.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levendosky RF, Sabantsev A, Deindl S, Bowman GD. The Chd1 chromatin remodeler shifts hexasomes unidirectionally. eLife. 2016;5:e21356. doi: 10.7554/eLife.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung L-W, Jain S, McCoy AJ, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Satoh MS, Dianov G. Enzymes acting at strand interruptions in DNA. Philos Trans R Soc Lond B Biol Sci. 1995;347:57–62. doi: 10.1098/rstb.1995.0009. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Kornberg RD. Chromatin-remodeling for transcription. Q Rev Biophys. 2017;50:e5. doi: 10.1017/S003358351700004X. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–2. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-depen-dent molecular machines. BioEssays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- Ma N-F, Hu L, Fung JM, Xie D, Zheng B-J, Chen L, Tang D-J, Fu L, Wu Z, Chen M, et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1 within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;18:59–65. [PubMed] [Google Scholar]
- McGinty RK, Tan S. Recognition of the nucleosome by chromatin factors and enzymes. Curr Opin Struct Biol. 2016;37:54–61. doi: 10.1016/j.sbi.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Q-J, Li H-L, Yao Y, Liu S-C, Yin C-G, Ma X-Z. Chromodomain Helicase/ATPase DNA-Binding Protein 1-Like Gene (CHD1L) Expression and Implications for Invasion and Metastasis of Breast Cancer. PLoS ONE. 2015;10:e0143030. doi: 10.1371/journal.pone.0143030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo TTM, Zhang Q, Zhou R, Yodh JG, Ha T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell. 2015;160:1135–1144. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199:235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Levendosky RF, Chakravarthy S, Patel A, Bowman GD, Myong S. The Chd1 chromatin remodeler shifts nucleosomal DNA bidirectionally as a monomer. Mol Cell. 2017;68:76–88.e6. doi: 10.1016/j.molcel.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosyla-tion) J Biol Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- Sabantsev A, Levendosky RF, Zhuang X, Bowman GD, Deindl S. Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat Commun. 2019;10:1720. doi: 10.1038/s41467-019-09657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to Im-ageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, Kozlowski M, Bultmann S, Ladurner AG, Timinszky G, Huet S. The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol Biol Cell. 2016;27:3791–3799. doi: 10.1091/mbc.E16-05-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh HR, Nardozza AP, Möller IR, Knobloch G, Kistemaker HAV, Hassler M, Harrer N, Blessing C, Eustermann S, Kotthoff C, et al. A Poly-ADP-Ribose Trigger Releases the Auto-Inhibition of a Chromatin Remodeling Oncogene. Mol Cell. 2017;68:860–871.e7. doi: 10.1016/j.molcel.2017.11.019. [DOI] [PubMed] [Google Scholar]
- Smith R, Sellou H, Chapuis C, Huet S, Timinszky G. CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res. 2018;46:6087–6098. doi: 10.1093/nar/gky334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Lebeaupin T, Juhász S, Chapuis C, D’Augustin O, Dutertre S, Burkovics P, Biertümpfel C, Timinszky G, Huet S. Poly(ADP-ribose)-dependent chromatin unfolding facilitates the association of DNA-binding proteins with DNA at sites of damage. Nucleic Acids Res. 2019;47:11250–11267. doi: 10.1093/nar/gkz820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia AM, Collings CK, Dao HT, St Pierre R, Cheng Y-C, Huang J, Sun Z-Y, Seo H-S, Mashtalir N, Comstock DE, et al. Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling. Cell. 2019;179:1342–1356.e23. doi: 10.1016/j.cell.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Wagner FR, Dienemann C, Wang H, Stützer A, Tegunov D, Urlaub H, Cramer P. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature. 2020;579:448–451. doi: 10.1038/s41586-020-2088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willhoft O, Bythell-Douglas R, McCormack EA, Wigley DB. Synergy and antagonism in regulation of recombinant human INO80 chromatin remodeling complex. Nucleic Acids Res. 2016;44:8179–8188. doi: 10.1093/nar/gkw509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willhoft O, Ghoneim M, Lin C-L, Chua EYD, Wilkinson M, Chaban Y, Ayala R, McCormack EA, Ocloo L, Rueda DS, Wigley DB. Structure and dynamics of the yeast SWR1-nucleosome complex. Science. 2018;362:eaat7716. doi: 10.1126/science.aat7716. [DOI] [PubMed] [Google Scholar]
- Wong N, Chan A, Lee S-W, Lam E, To K-F, Lai PB-S, Li X-N, Liew C-T, Johnson PJ. Positional mapping for amplified DNA sequences on 1q21-q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol. 2003;38:298–306. doi: 10.1016/s0168-8278(02)00412-9. [DOI] [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- Ye Y, Wu H, Chen K, Clapier CR, Verma N, Zhang W, Deng H, Cairns BR, Gao N, Chen Z. Structure of the RSC complex bound to the nucleosome. Science. 2019;366:838–843. doi: 10.1126/science.aay0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Gaullier G, Luger K. Nucleosome structure and dynamics are coming of age. Nat Struct Mol Biol. 2019;26:3–13. doi: 10.1038/s41594-018-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108529.
Data Availability Statement
Raw cryo-EM videosand final particle images: accession code EMPIAR-10465. EMDB accession codes: EMD-11220 (nucleosome) and EMD-11221 (hexasome). PDB accession codes: 6ZHX (nucleosome) and 6ZHY (hexasome).