Abstract
G protein-coupled receptors (GPCRs) are divided phylogenetically into six classes, A-F1,2. Over 370 structures of vertebrate GPCRs (classes A, B, C and F) have been determined, leading to a substantial understanding of their function3. In contrast, there are no structures of Class D GPCRs, which are found exclusively in fungi where they regulate survival and reproduction. We have determined the first structure of a family D GPCR, the Saccharomyces cerevisiae pheromone receptor Ste2, in an active state coupled to the heterotrimeric G protein Gpa1-Ste4-Ste18. Ste2 was purified as a homodimer that coupled to two G proteins. The dimer interface of Ste2 is formed by the N-terminus, transmembrane helices H1, H2, H7 and the first extracellular loop ECL1. We established a Class D1 generic residue numbering system (CD1) to enable comparisons with orthologues and other GPCR classes. The structure of Ste2 bears similarities in overall topology to Class A GPCRs, but H4 is shifted by over 20 Å and the G protein binding site is a shallow groove rather than a cleft. The structure provides a template for the design of novel drugs targeting fungal GPCRs that could be utilised to treat a number of intractable fungal diseases4.
GPCRs comprise the largest family of receptors in fungi and they are critical for the metabolism, virulence, development, and survival of fungal species4,5. Within Class D, the fungal GPCRs are classified further into ten classes based on sequence homology, and the pheromone receptor Ste2 of S. cerevisiae is the prototypical class I fungal GPCR4. Ste2 was the first ligand binding GPCR to be sequenced6 and the well-conserved GPCR-mediated signal transduction cascade has been studied extensively in yeast7. The tridecapeptide pheromone α-factor (WHWLQLKPGQPMY) secreted by S. cerevisiaeMATα cells is the native high-affinity agonist for Ste28. Upon stimulation with α-factor, Ste2 in MATa cells facilitates GDP-GTP exchange in Gpa1 (heterotrimeric G proteinα-subunit), resulting in Ste4-Ste18 (the βγ dimer) release, which activates the Ste20 p21-activated protein kinase. Subsequent activation of the Ste11-Ste7-Fus3 MAPK cascade leads to cell cycle arrest and fusion with the opposite mating type MATα4,7.
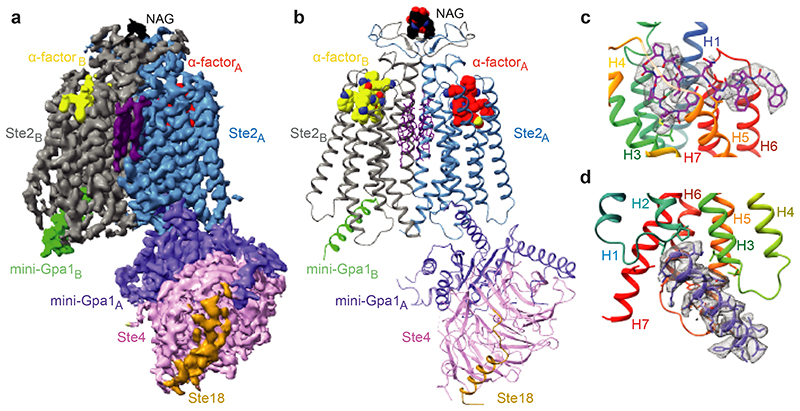
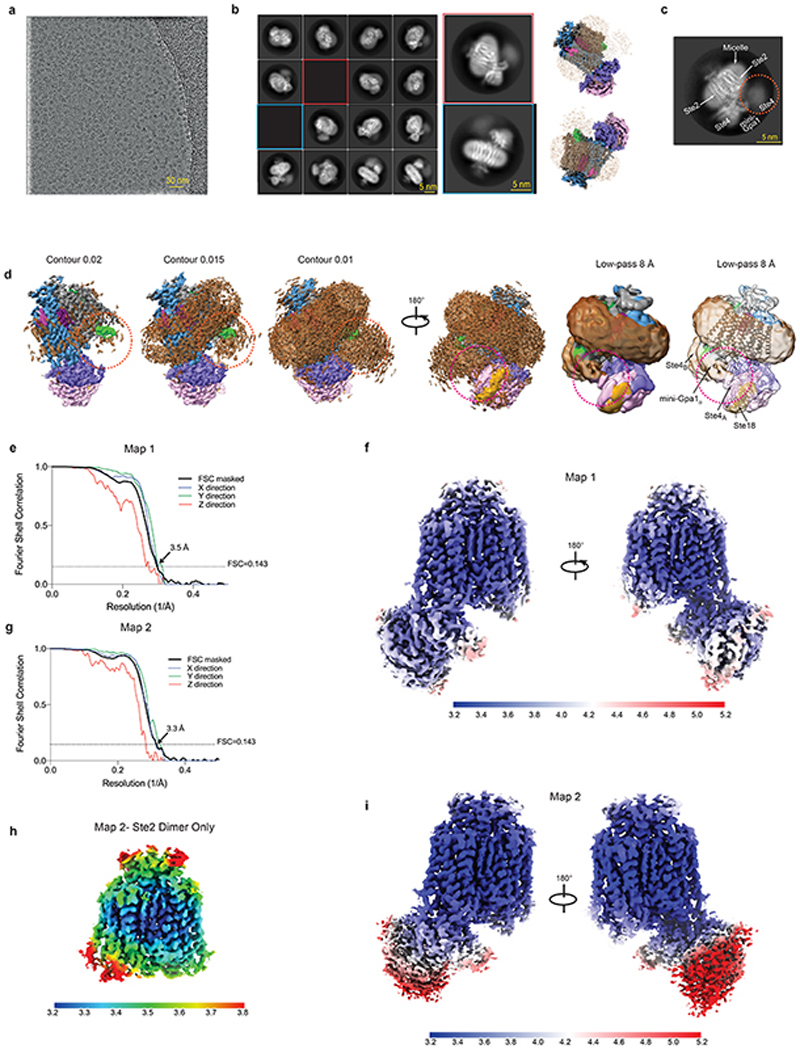
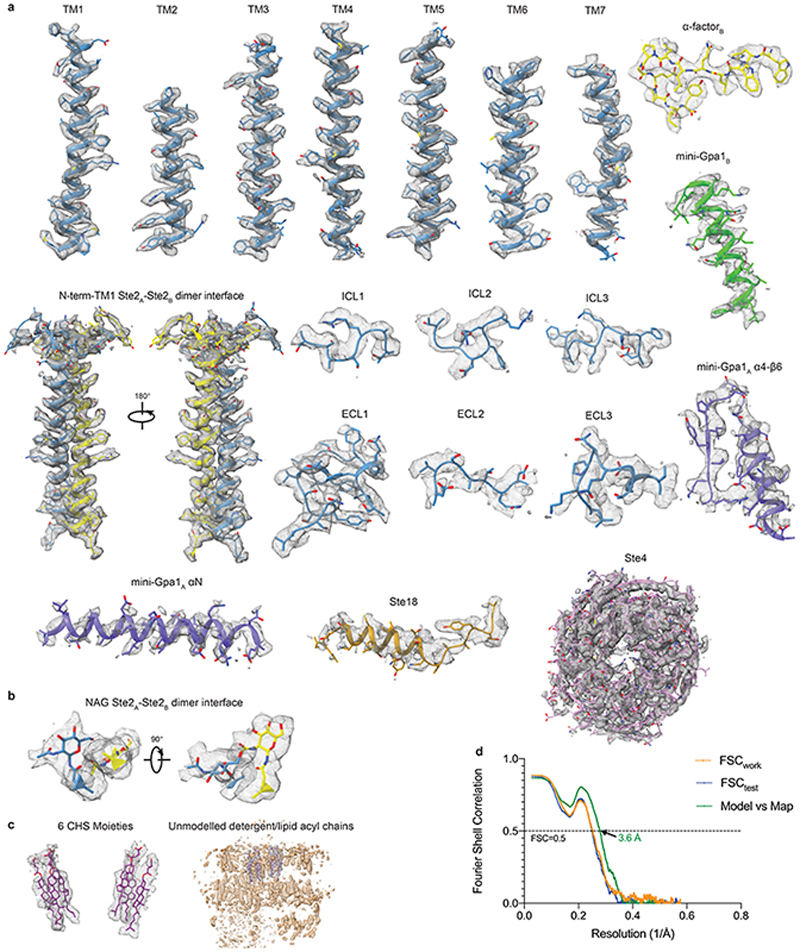
The structure of Ste2 was determined in its active state coupled to a heterotrimeric G protein and bound to α-factor. To stabilize the active state complex, an engineered version of the yeast G protein α-subunit Gpa1, called ‘mini-Gpa1’, was developed by incorporating six mutations (G50D, E51N, A345D, E348D, M450A, V453I) and two deletions based on prior work on mammalian G proteins9 (see Methods). Full-length wild type Ste2 was expressed and purified in the presence of α-factor, which was then assembled with mini-Gpa1 (α) and Ste4-Ste18 (βγ) subunits to form a stable, monodisperse complex (Extended Data Fig. 1a-f). The structure was determined by cryo-EM to an overall resolution of 3.5 Å (Map 1, Extended Data Figs. 2a-f and 3, Extended Data Table 1) exhibiting clear density for the majority of the side chains (Fig. 1d and Extended Data Fig. 4a) and α-factor (Fig. 1c). Another map (Map 2) was obtained by focussed classification of the α-factor-Ste2 portion, with the best regions resolved to 3.2 Å (Extended Data Fig. 2g-i). Two-fold symmetry was not imposed during structure determination. Ste2 was a homodimer with unambiguous occupancy of α-factor in both orthosteric binding pockets (Fig. 1a-c and Extended Data Fig. 4a) and coupled to two heterotrimeric G proteins (Extended Data Figs. 1h and 2b-d). The density for one G protein heterotrimer was well resolved to about 3.5 Å, with the best density in Gpa1 (Extended Data Fig. 2f), whilst the resolution of the other G protein was limited to about 8 Å (Extended Data Fig. 2d) except for the C-terminal α5 helix bound to the receptor (Fig. 1a and Extended Data Fig. 4a). We therefore modelled the heterotrimeric G protein coupled to Ste2 protomer A (Ste2A) and only the α5 helix of mini-Gpa1 coupled to Ste2B (Fig. 1b). Unless noted otherwise, all discussions are on the α-factor-bound Ste2A-G protein complex.
Fig. 1. Overall cryo-EM reconstruction of the Ste2-G protein heterotrimer complex.
a, Overall structure of the Ste2-G protein heterotrimer complex shown as a cryo-EM density map (sharpened with a B factor of -112 Å2) consisting of a Ste2 dimer (blue, grey), two bound α-factor ligands (red, yellow), two coupled G proteins, 6 putative CHS molecules (purple) and two NAG molecules (black). b, Cartoon of the Ste2-G protein heterotrimer complex coloured according to the polypeptides as shown in a. c, The orthosteric binding pocket of Ste2A (rainbow) with α-factorA (magenta). Residues 183 to 189 of transmembrane helix 4 are not shown for clarity. d, Binding mode of the α5 helix of mini-Gpa1A (purple) to Ste2A (rainbow).
The map in panel a was contoured at 0.02 (2 Å carve radius) and maps in panels c and d were contoured at 0.03 (2 Å carve radius) and visualised in ChimeraX35.
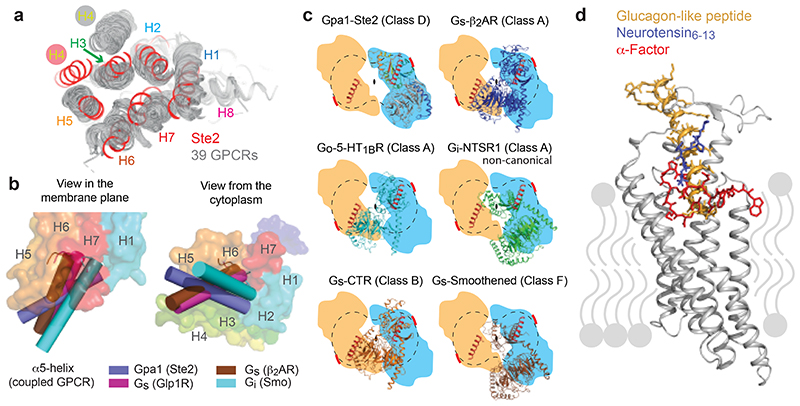
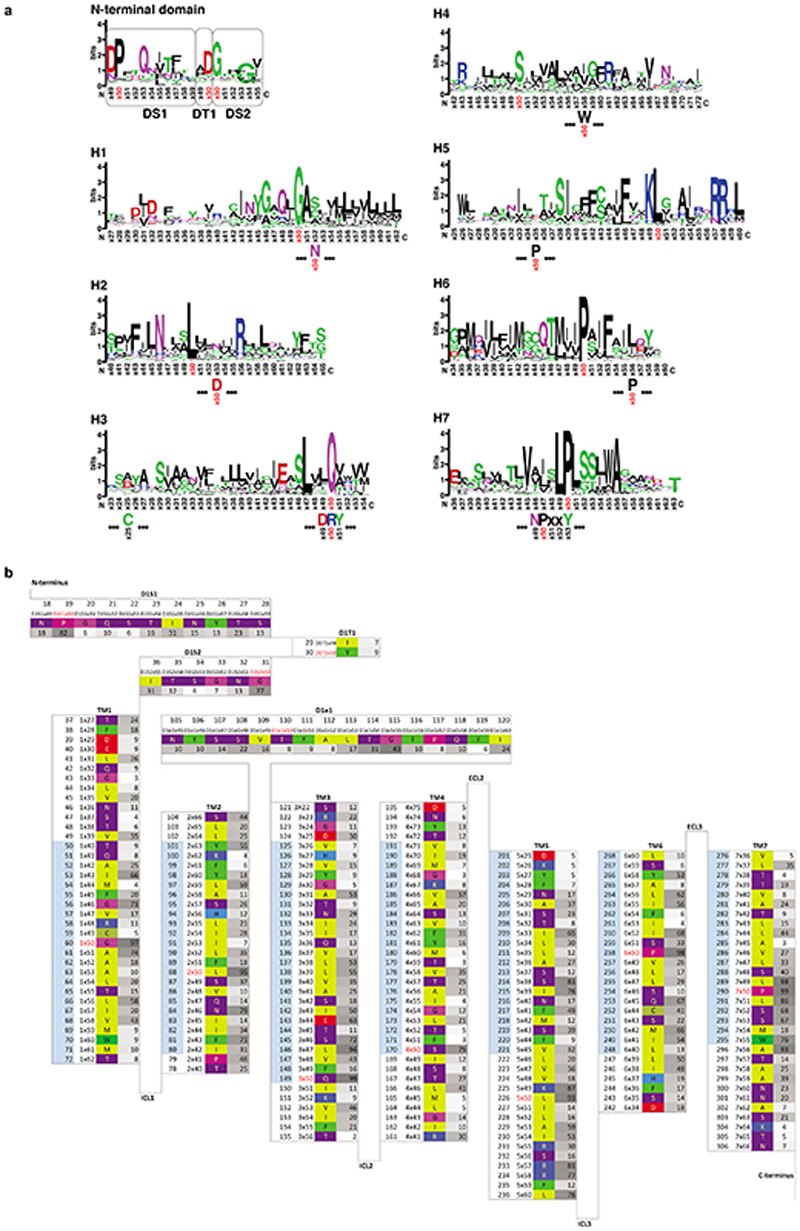
Global structural alignments of 39 GPCR structures coupled to either a G protein or arrestin showed similarities between Ste2 and receptors in Classes A, B, C and F, but also significant differences such as in the position of H4 (Extended Data Fig. 5). A generic residue numbering system was developed analogous to the Ballesteros-Weinstein, Wootten, Pin and Wang systems for Class A, B, C and F GPCRs10, respectively, where the most well conserved amino acid residue in each transmembrane α-helix is given the number Yx50 where Y is the helix number (Extended Data Fig. 6). This Class D1 numbering system (CD1) was then used to facilitate comparisons.
Ste2 dimer interface
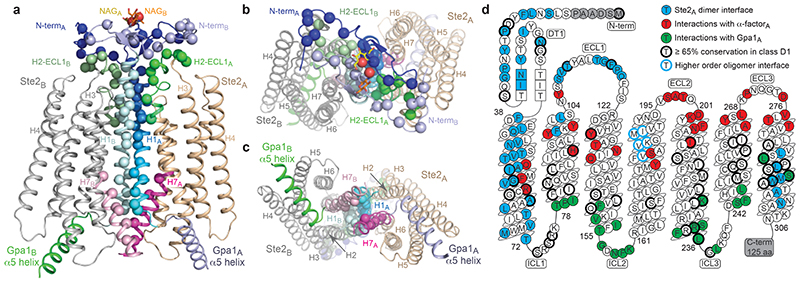
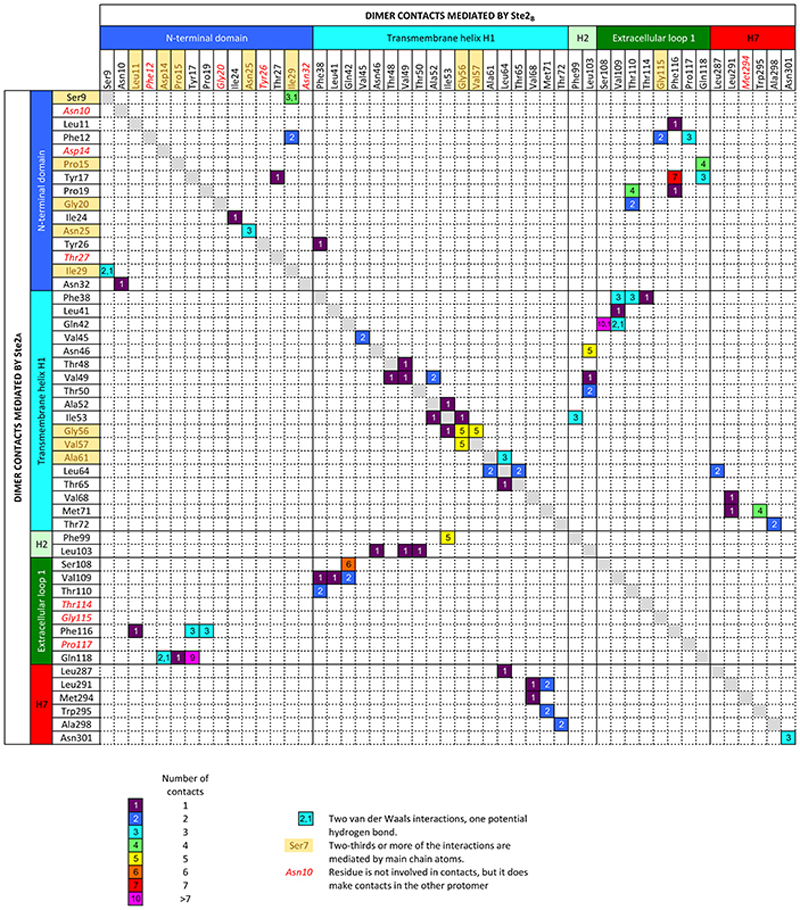
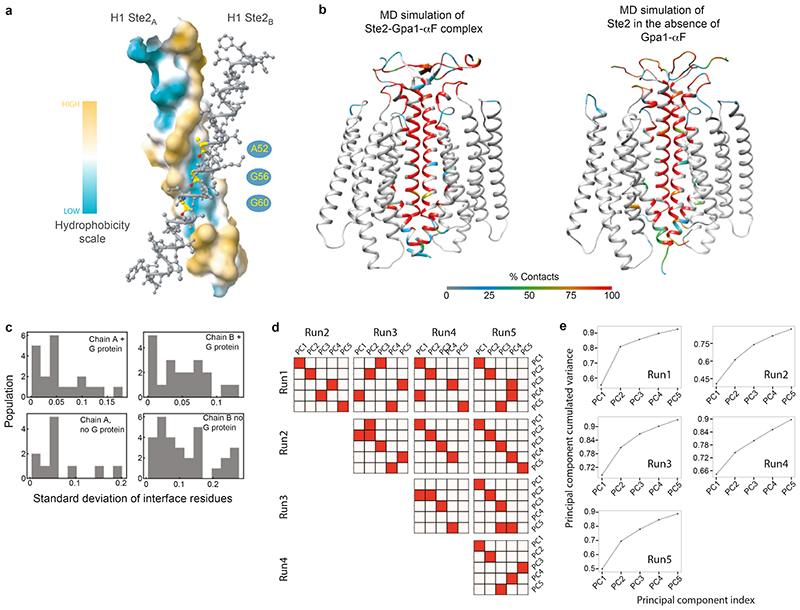
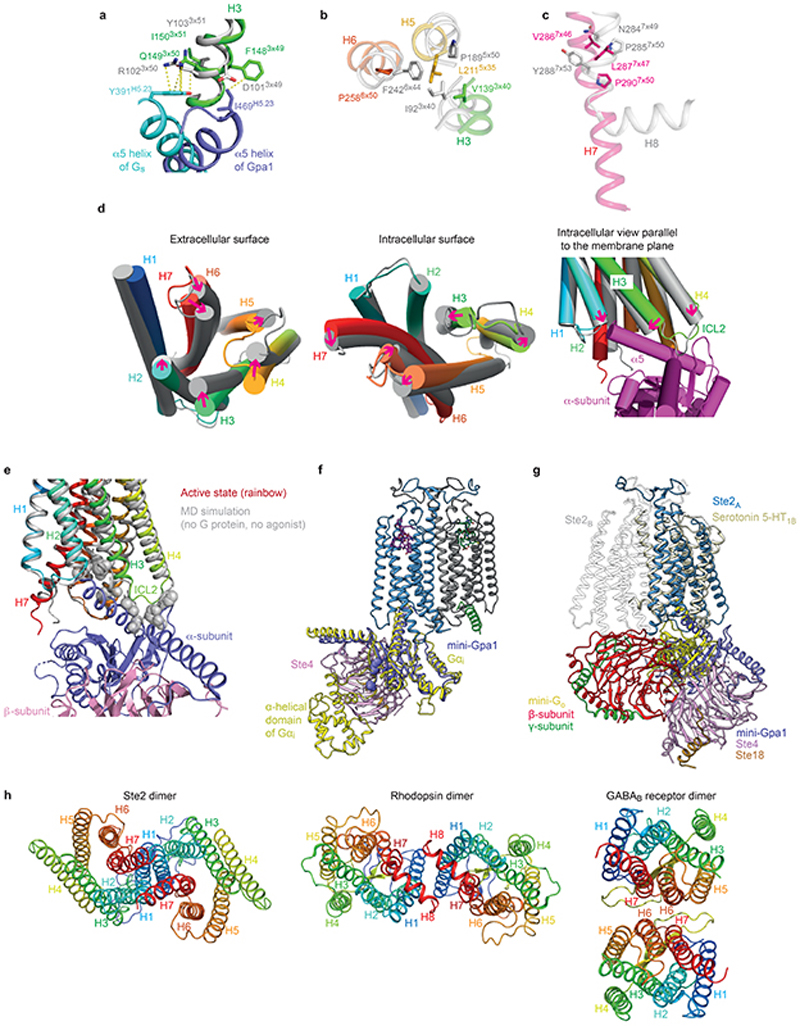
Ste2 forms a stable dimer through an extensive dimer interface (2519 Å2) that is over twice the area of the Ste2-G protein interface and involves the domain swapped N-terminus and ECL1 (44% of contacts) and transmembrane regions H1-H2-H7 (56% contacts) (Fig. 2a-c, Extended Data Fig. 7). The dimer interface is stabilised by 171 van der Waals contacts and 6 hydrogen bonds formed by 43 amino acid residues from Ste2A and 44 from Ste2B. Thirty-two of the amino acid residues at the dimer interface have been mutated previously and 27 residues shown to have a major effect on dimerisation and/or signalling when mutated, both in the presence and absence of α-factor (Supplementary Table 1). The interface is stable in molecular dynamics simulations either with or without α-factor and coupled G proteins, with many of the contacts existing 100% of the time throughout the simulation (Extended Data Fig. 8b,c; contact frequencies were calculated for 1.4 μs and 5 μs for the Ste2 dimer in the presence or the absence of α-factor-G protein heterotrimer, respectively). Ste2 was solubilized from insect cell membranes as a dimer irrespective of the detergent used (Extended Data Fig. 1a), and the dimeric nature of Ste2 before coupling to G proteins was confirmed using SEC-MALS and cryo-EM 2D class averages (Extended Data Figs. 1h and 1g, respectively). Ste2 is known to exist as a dimer in yeast cells and dimerisation is essential for biogenesis, signalling, and functional endocytosis11-14, with higher oligomeric states also being proposed15. The N-terminus of Ste2A is situated over Ste2B where it makes contacts with ECL1B. It then loops back to Ste2A, where it makes contacts with the N-terminus of Ste2B running in an antiparallel direction, before it turns into H1 (Fig. 2a). The two most conserved residues in the N-terminus, Pro19D1S1x50 and Gly31D1S2x50, contribute to the dimer interface together with the antiparallel β-sheets in the N-terminus stabilized by the conserved aliphatic residues Ile24D1S1x55 and Ile36D1S2x55 (superscripts refer to CD1 numbering system; see Methods). The dimer interface runs the length of H1A and H1B, with the helices forming a right-handed interaction (crossing angle 37°) at the highly conserved sequence I1x43xxGxxxGA1x51 (residues 53-61; Fig. 2d, Supplementary Table 2). This is a glycine zipper motif16 where shallow grooves in the α-helices interact with one another (Extended Data Fig. 8a). Mutation of Gly561x46 and Gly601x50 significantly reduced dimerisation and signalling without affecting agonist binding11,14. At the intracellular surface both H1 helices then interact with the intracellular half of H7 from the other protomer (Fig. 2a,c) and H7 crosses beneath H1 to form H7A-H7B contacts. In the detergent micelle surrounding Ste2 there were six ordered densities adjacent to the dimer interface (Fig. 1a,b and Extended Data Fig. 4c) that were assigned to the sterol cholesteryl hemisuccinate (CHS), which was present during receptor purification, although we cannot exclude the possibility they could be other sterols. Density adjacent to the N-glycosylation site17 Asn25D1S1x56 was modelled as N-acetylglucosamine and also mediated dimer contacts (Fig. 1a,b and Extended Data Fig. 4b). H4 has been suggested by mutagenesis experiments18 to be involved in the formation of higher order oligomers of Ste2, and the structure is compatible with the proposed interface, as H4 is on the opposite side of Ste2 to the H1 dimer interface (Fig. 2).
Fig. 2. The Ste2 dimer interface.
a-c The Ste2 dimer is shown as a cartoon (chain A, brown, chain B, grey) and structural elements forming the dimer interface labelled. The Cα atoms of residues forming contacts in the interface are shown as spheres. a, View parallel to the membrane; b, extracellular surface; c, intracellular surface. d, Snake plot of the amino acid sequence of S. cerevisiae Ste2 showing residues forming the dimer interface (blue), residues interacting with α-factor (red) and residues interacting with the G protein (green). Residues with ≥ 65% conservation in class D1 receptors are circled in black and residues proposed to form a higher order oligomer interface18 are circled in blue. β-Strands are shown by residues in squares. Panels a-c were prepared using PyMOL (Schrödinger Inc).
The orthosteric binding site
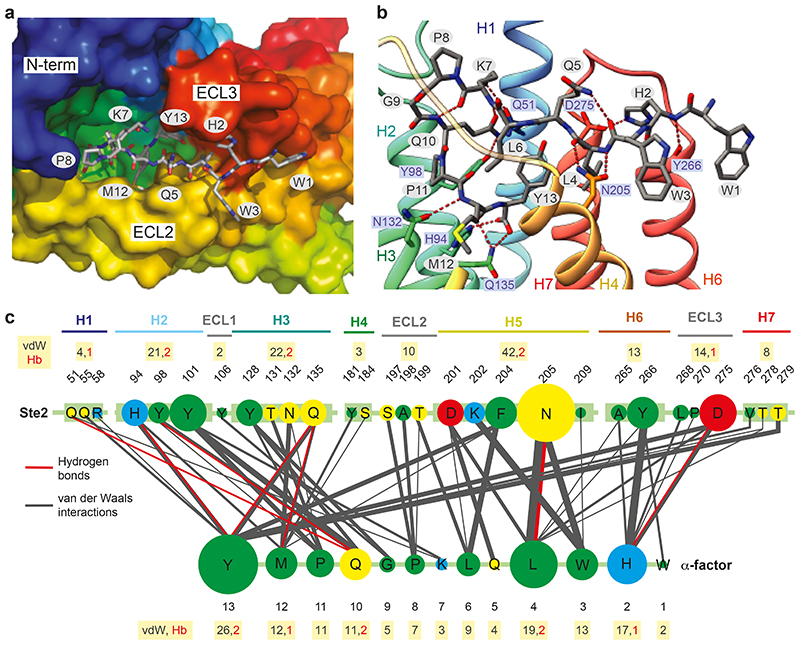
Ste2 has an extensive orthosteric binding pocket (1126 Å3) that involves residues throughout the extracellular half of the receptor (Figs. 2d and Figs. 3a-c). The N-terminal Trp1 of α-factor resides mainly outside the orthosteric binding pocket (Fig. 3b,c). In contrast, His2-Trp3-Leu4 make 38% of ligand-receptor contacts, mainly to residues in H5, H6 and ECL3. The C-terminal domain Pro11-Met12-Tyr13 contributes a further 35% of the ligand-receptor contacts, mainly to residues in H1, H2, ECL1, H3 and H4 (Fig. 3c). Mutations in 23 out of the 31 residues in Ste2 making contacts to α-factor result in major effects on ligand binding and/or signalling (Supplementary Table 3). The molecular basis for a number of key interactions between α-factor and Ste2 previously characterised by extensive mutagenesis studies19,20 of either Ste2 (Supplementary Table 3) or α-factor (Supplementary Table 4) can be rationalised by the structure (see Supplementary Tables for full mutational data and references). Amidation of the α-factor C-terminus causes a 160-fold decrease in affinity suggesting the negative charge of the carboxylate is important. In the Ste2 structure, the C-terminus of α-factor forms a hydrogen bond with His942x56 (Fig. 3b); a positively charged residue at this position is conserved in 90% of Ste2 sequences (Supplementary Table 2) and a salt bridge may form under acidic conditions in vivo. An additional positive charge (Arg581x48) adjacent to His942x56 may increase the positive electrostatic field that would be a favourable environment for α-factor’s C-terminal carboxyl group. Tyr13 in α-factor makes the most contacts to Ste2 and mutations that reduce its aromaticity (F204C and F204S) decreased ligand binding and signalling. Tyr13 also engages in polar contacts with the side chain of Gln1353x36 (Fig. 3b), and the mutations Q135A and Q135P result in decreased activity and dominant-negative phenotypes, respectively. The bend domain (Lys7-Gln10) contains a type II β-turn (Fig. 3b) that is thought to be found only in the biologically active conformation of α-factor21 and destabilization of the β-turn significantly decreases affinity19. In the N-terminal signalling domain (Trp1-Leu4), most contacts (34 van der Waals, 4 hydrogen bonds) are mediated by His2-Trp3-Leu4 (Fig. 3c). Both Asn2055x29 and Tyr2666x58 make extensive contacts to this region of α-factor and are critical for Ste2 signalling22,23. The His2 side chain of α-factor forms a hydrogen bond with Asp275ECL3 (Fig. 3b); this could possibly be a salt bridge under acidic conditions in vivo as the protonation of His2 is considered to be essential for ligand recognition and stabilization of the appropriate conformation for binding20,24. His2 also engages in multiple polar contacts with the backbone carbonyl oxygen of Tyr2666x58 and Ala2656x57 (Fig. 3b) and replacement of His2 with L-Ala results in a 100-fold less affinity19.
Fig. 3. α-Factor binding site.
a,b view of the orthosteric binding pocket; Ste2A (rainbow colouration); α-factor (sticks; carbon, grey; oxygen, red; nitrogen, blue). a, view from the extracellular surface. b, view in the membrane plane. c, Interactions between α-factor and Ste2. The circle size and line thickness are proportional to the number of interactions. Panels a-b were prepared using PyMOL (Schrödinger Inc).
The heterotrimeric G protein
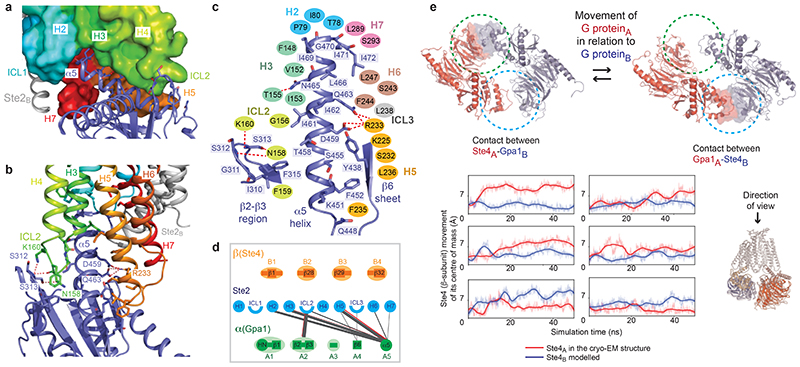
The structure of the heterotrimeric G protein Gpa1-Ste4-Ste18 is similar to the structure of Go coupled to the serotonin receptor 5-HT1B (rmsd 1.7 Å over 2832 atoms). No density was observed in the nucleotide binding pocket of Gpa1, so GDP was not bound. The interface between Ste2 and the heterotrimeric G protein (1206 Å2) is formed solely by the α-subunit mini-Gpa1 (Fig. 1a-b) and comprises 76 van der Waals contacts and 8 hydrogen bonds between 21 residues in mini-Gpa1 (Fig. 4c) and 23 residues in Ste2 (Fig. 2d). There are two main regions of Gpa1 (Fig. 4a-d) that contact the receptor, the β2-loop-β3 region (24% of all contacts) and the C-terminal α5 helix (71% of all contacts). The loop between the two β-strands β2 and β3 packs against ICL2 of Ste2 (Fig. 4a,b). The α5 helix packs against the cytoplasmic ends of H5, H6 and H7 and underneath H3-ICL2-H4, with side chains from α5 sitting in a slight groove formed between H3-ICL2 and H5-H6 (Fig. 4a,b). The C-terminal end of the α5 helix does not penetrate deeply into the receptor, but juxtaposes against the cytoplasmic end of H2 (Fig. 4a). Mutagenesis indicated that 12 out of 22 residues in Ste2 and 11 out of 21 residues in Gpa1 making direct contacts have noticeable effects on function (see Supplementary Table 5 for full mutational data and references). The C-terminal residues Ile469H5.23, Gly470H5.24, Ile471H5.25, Ile472H5.26 (superscripts refer to the residue position in the common Gα numbering scheme for G proteins25) form a ‘wavy hook’ (Fig. 4c) like in the mammalian G proteins3 and are important for G protein specificity and coupling, as mutations of Gly470-Ile472 led to strong signalling defects and loss-of-function phenotypes. Mutations of Leu2476x39, Leu2897x49 and Ser2937x53 of Ste2 that directly interact with the wavy hook of mini-Gpa1 also impact receptor dimerisation and signalling demonstrating the importance of H7 in both these processes. Most of the contacts made between the α5 helix and Ste2 are hydrophobic (Fig. 4c) although a salt bridge occurs between the conserved residue Arg2335x57 of Ste2 and Asp459H5.13 of mini-Gpa1, and Arg2335x57 also forms a polar contact with the side chain of Gln463H5.17 of mini-Gpa1 (Fig. 4b,c).
Fig. 4. G protein binding site.
a,b, Details of interactions between the α-subunit (mini-Gpa1, purple) and Ste2 (Ste2A, rainbow coloured surface; Ste2B, grey cartoon). Interacting side chains are shown as sticks and hydrogen bonds as red dashed lines. Only residues forming hydrogen bonds have been labelled. c, Schematic of the interactions between Gpa1 (purple structure) and residues in Ste2 (ovals) coloured according to the structural element in the receptor. Hydrogen bonds are depicted as red dashed lines. d, interactions between structural elements in Ste2A and G protein, with the line thickness proportional to the number of contacts (grey, van der Waals contacts; red, hydrogen bonds). No contacts are made between Ste2 and Ste4. Mammalian G proteins utilise structural elements that do not make contact with Ste2 (see ref 3). e, MD simulations of the Ste2 dimer containing the Gpa1-Ste4-Ste18 G protein determined by cryo-EM (red) and the modelled G protein (blue). Snapshots of two different states where the G proteins have interacted (circled) are shown as viewed from the extracellular surface (Ste2 removed for clarity). The degree of movement of the centre of masses of the two different β-subunits (Ste4) is indicated by the line traces as a function of MD simulation time. The line traces are the moving average of the instantaneous distances between the centres of mass. Panels a-c were prepared using PyMOL (Schrödinger Inc).
In the cryo-EM structure of the Ste2 dimer, one G protein was well ordered and could be modelled whilst the other G protein showed poor density. Molecular dynamics (MD) simulations were performed on the Ste2 dimer with two G proteins coupled (see Methods), to study the range of motions of the G proteins. Multiple separate simulations of 50-75 ns were used (total aggregate time of 1.4 μsec in 25 separate simulations; see Methods). Comparison of the movements of the β-subunit showed opposite behaviours of the two G proteins, with one showing lower dynamics than the other (Fig. 4e). When the highly dynamic G protein became more static, the other G protein transitioned into a more dynamic state. This transition seemed to occur as the β-subunit became juxtaposed to the α-subunit in the other protomer (Fig. 4e), similar to the interaction between mini-Gpa1A and Ste4B in the 8 Å filtered cryo-EM map (Extended Data Fig. 2d). In no instance did we observe a stable structure with two ordered G proteins, which is consistent with experimental data showing that the G protein was monomeric after purification (Extended Data Fig. 1h) and the cryo-EM data showing that only one G protein was ordered. However, we cannot exclude the possibility that longer simulation times and different experimental conditions could lead to both G proteins coupled to Ste2 becoming ordered. Another caveat is that the deletion of the α-helical domain in mini-Gpa1, which also contains an unstructured 109-amino acid unique ubiquitination domain insert26, could have affected the relative dynamics of the coupled G protein heterotrimers. However, SEC-MALS data show that two wild type G proteins can couple to the Ste2 dimer (Extended Data Fig. 1h). In addition, the structure of G proteins is highly conserved and alignment of mammalian Gi with the yeast G protein shows that the α-helical domain is on the opposite side to any potential interface between the yeast G proteins in the Ste2 dimer (Extended Data Fig. 9f). The significance of simultaneous coupling to two G protein heterotrimers and potential cooperativity needs to be elucidated by further experimentation.
Comparison of Ste2 with other GPCRs
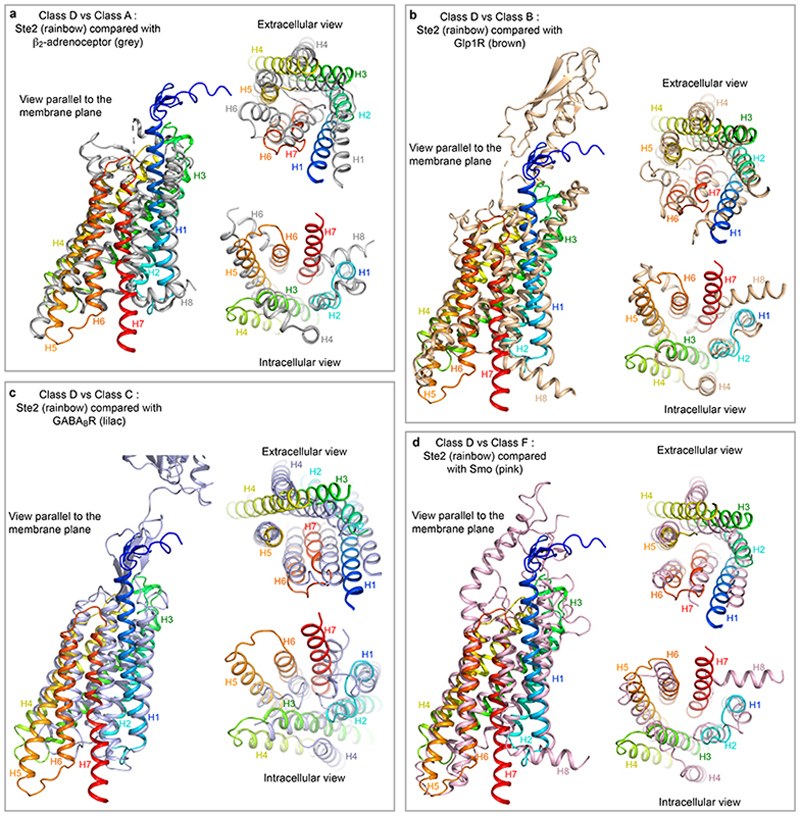
The overall arrangement of the transmembrane helix bundle in Ste2 is similar to the active state structures of Class A, B and F GPCRs (Fig. 5a), except for the position of H4, which differed by over 20 Å at the intracellular face of the receptor (comparison with the 5-HT1B receptor-Go complex). The position of the α5 helix of Gpa1 coupled to Ste2 also differs considerably from the positions of α5 helices coupled to Class A, B and F receptors, as observed upon receptor alignment by the different angles at which they insert (Fig. 5b). The α5 helices coupled to Class A, B and F all insert to approximately the same depth3, whereas the α5 helix of Gpa1 inserts less deeply (by 12 Å compared to Go-coupled 5-HT1BR). In this regard, Ste2 appears to be more similar to Class C receptors where the G protein also couples in a shallow binding cleft27. A consequence of the different insertion angle of the α5 helix into Ste2 is that the position of the G protein relative to the receptor also differs from mammalian GPCRs (Fig. 5c and Extended Data Fig. 9g) resulting in the βγ subunits not interacting with either Ste2 protomers. If Class A, B or F GPCRs dimerised in an analogous fashion to Ste2, for two G proteins to couple simultaneously it would be necessary to adjust the orientation of the G protein with respect to the receptor (Fig. 5c). An analysis of all mammalian GPCRs for a GxxxG motif in H1 identified a few candidates that could dimerise in a similar fashion to Ste2, namely the class B1 VPAC1 receptor and the class A orphan receptors MRGPRE and MRGPRG, all of which are also known to form homodimers28,29. The dimeric arrangement of Ste2 shows superficial resemblance to the low-resolution native rhodopsin (Class A) dimer in nanodiscs30, although major interactions are mediated by H8 and not H1 (Extended Data Fig. 9h). The constitutive GABAB dimer (Class C) in an active state has a different interface formed by H6 and H731.
Fig. 5. Comparison of Ste2 with mammalian GPCRs.
a, Alignment of 39 mammalian GPCRs in an active state (grey) with Ste2 (red), viewed from the extracellular surface. b, Positions of the α5 helices (cylinders) of coupled G proteins after global alignment of the receptors used in a. The positions of the α5 helices are shown relative to Ste2 (rainbow colouration) shown as a transparent surface. c, Relative positions of coupled G proteins after global alignment of receptors in relation to Ste2: blue outline, position of Gpa1-Ste4-Ste18 coupled to Ste2A from the cryo-EM structure; orange outline, theoretical position of disordered G protein coupled to Ste2B if it were related by a 2-fold axis perpendicular to the membrane to the ordered G protein. A thick red line denotes where the deleted α-helical domain and ubiquitination domain would be expected to protrude. The position of Ste2 is indicated by a dumbbell (dashed line) and the positions of the two α5 helices are shown as a cartoon (red). d, Alignment of Ste2, neurotensin receptor (NTSR1; Class A) and GLP-1 receptor (GLP1R; Class B) and comparison of the binding mode of the native ligands superimposed onto the structure of Ste2 (grey). Panels a, b and d were prepared using PyMOL (Schrödinger Inc).
The overall similarity of the Ste2 structure with mammalian GPCRs and a similar mode of G protein coupling perhaps suggest similar mechanisms of activation. Indeed, the depth of the orthosteric binding pockets of a Class B receptor such as GLP1R is very similar to Ste2, although both are deeper than that of a Class A receptor such as NTSR1 (Fig. 5d). However, the canonical motifs in Class A receptors such as the PIF, DRY and NPxxY motifs32 are not present in Ste2. Comparing a structural alignment of GPCRs alongside sequence conservation shows that Gln1493x50 of Ste2 (99% conservation in Class D1) aligns well with Arg3x50 of the DRY motif in Class A receptors and could be functionally equivalent in stabilising the inactive state (Extended Data Fig. 9a). Similarly, the PIF motif analogues in Ste2 form a cluster (Extended Data Fig. 9b). The NPxxY motif in H7 corresponds with the highly conserved motif LPLSSxWA in Ste2, although Pro2907x50 in Ste2 does not correspond with the kink in H7 present in Class A GPCRs (Extended Data Fig. 9c). To study the dynamics of the receptor and its G protein coupling interface in the absence of the G protein and α factor, five independent simulations of 1 μsec each were performed and they all converged to a stable state (Extended Data Fig. 8c-e). We cannot state whether an inactive state was reached, as was achieved in MD simulations of the β2-adrenoceptor (sometimes within 1 μsec)33, as there is no inactive state structure of Ste2 for comparison. However, it seems likely that the simulated receptor is tending towards the inactive state based on the decrease in the volume of the G protein binding site by 70% and increase in the volume of the orthosteric binding pocket of Ste2A by 20% (see Methods). Inactive state β1-adrenoceptor showed a consistently larger orthosteric binding pocket volume compared to the active state with the same ligand bound34. On the intracellular face of Ste2A, the change in tilt of H3 and H4 (11° and 14°, respectively) and the new disposition of ICL2 occlude the binding site for the α5 helix of Gpa1, which would thus prevent G protein coupling (Extended Data Fig. 9d,e). This is different from the changes observed in analogous simulations of β2AR, where H5-H6 moves to block the G protein binding site33. These data suggest that the dynamics of the helices in Ste2 differ from those in Class A GPCRs, which hints at a different mechanism of receptor activation.
Materials and Methods
Cloning, expression and purification of Ste2
A construct encoding wild-type Saccharomyces cerevisiae Ste2 (residues 1-431) followed by a TEV cleavage site, EGFP, and a decahistidine tag (wtSte2-TEV-EGFP-His10) was synthesized as a gBlocks® gene fragment (IDT) and cloned into pAKGP67-B by in vivo assembly in E. coli XL10-Gold36. High-titre recombinant baculoviruses expressing Ste2 were prepared using the flashBAC ULTRA system (Oxford Expression Technologies). Trichoplusia ni High Five™ cells (Thermo Fischer Scientific) were maintained in log phase growth in suspension in ESF 921 medium (Expression Systems). Insect cells at 2-3 x 106 cells/mL were infected with 3% (v/v) Ste2 baculovirus and cultured for 48 h. The cells were harvested by centrifugation, flash-frozen in liquid nitrogen, and stored at -80°C until further use. All purification steps described below were carried out at 4°C. Insect cell membranes from 2 L of culture were prepared by two iterations of homogenization using an Ultra-Turrax disperser (IKA) and centrifugation at 125,000 xg for 90 min in 10 mM HEPES pH 7.5, 1 mM EDTA, 2 mM PMSF, 25 U/mL Benzonase®, 10 mM MgCl2, supplemented with EDTA-free Protease Inhibitor Cocktail tablets (PIC, Roche). Membrane suspensions were supplemented with 5 μM α-factor (GenScript) and incubated overnight. The membranes were solubilized in 20 mM HEPES pH 7.5, 100 mM NaCl, 20% glycerol, 2 mM PMSF, 25 U/mL Benzonase®, 10 mM MgCl2, 1% (w/v) Lauryl Maltose Neopentyl Glycol (LMNG, Anatrace), 0.1% (w/v) Cholesteryl Hemisuccinate (CHS; Anatrace) and PIC. The sample was clarified by ultracentrifugation, supplemented with 10 mM imidazole, and mixed in batch with 10 mL Super Ni-NTA Agarose resin (Generon) for 2 h. The resin was packed by gravity-flow and washed with 4 x 10 CV wash buffer [20 mM HEPES pH 7.5, 300 mM NaCl, 20% glycerol, 10 mM MgCl2, 0.02% (w/v) LMNG, 0.01% (w/v) CHS, 1 μM α-factor, 40 mM imidazole]. The bound species was eluted with elution buffer [20 mM HEPES pH 7.5, 100 mM NaCl, 20% glycerol, 10 mM MgCl2, 0.02% (w/v) LMNG, 0.01% (w/v) CHS, 1 μM α-factor, 300 mM imidazole]. The eluate was incubated overnight with 2.5 mg TEV protease and 1 mM DTT. The sample was subsequently exchanged into desalting buffer [20 mM HEPES pH 7.5, 100 mM NaCl, 20% glycerol, 10 mM MgCl2, 0.02% (w/v) LMNG, 0.01% (w/v) CHS, 1 μM α-factor, 5 mM imidazole] using Sephadex G-25 PD-10 desalting columns (GE Healthcare). TEV protease and free GFP were removed by negative purification on TALON® resin (Takara Bio). The flow-through fraction was concentrated using a 100 kDa MWCO Amicon® Ultra centrifugal concentrator (Merck) and loaded at 0.35 mL/min onto an Agilent Bio SEC-5 500 Å column, pre-equilibrated in 20 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.001% (w/v) LMNG, 0.0005% (w/v) CHS, 2 μM α-factor. The peak fractions containing Ste2 were pooled and concentrated. Protein concentration was estimated by NanoDrop 2000 (Thermo Scientific) at 280 nm. A typical yield was ~1-2 mg pure Ste2/L cell culture (using an extinction co-efficient of 1.157).
Engineering, cloning, expression and purification of mini-Gpa1
Engineering of mini-Gpa1 was based in part on the design of mini-Go 9. Human Go and S. cerevisiae Gpa1 share amino acid sequence identity and similarity of 51% and 67%, respectively. S. cerevisiae Gpa1 was mutated to incorporate: (1) 6 point mutations (G50D, E51N, A345D, E348D, M450A, V453I); (2) deletion of the switch III region (residues 350-359), ubiquitination domain and α-helical domain (residues 66-299) and palmitoylation site (Cys3)37; (3) addition of a GGSGGSGG linker between residues 65 and 300; and (4) introduction of a His6 tag and TEV cleavage site between N-terminal residues 2 and 4. The His6-TEV-miniGpa1 cDNA was synthesized as a gBlocks® gene fragment (IDT) and cloned into pET15b by in vivo assembly in E. coli XL10-Gold36. The plasmid was transformed into E. coli strain BL21-CodonPlus(DE3)-RIL cells (Agilent). To express mini-Gpa1, a 200 mL LB pre-culture (supplemented with 0.2% glucose, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol) was inoculated with a single bacterial colony and grown for 18 h at 30°C. For expression and purification, 2 L TB (supplemented with 0.2% glucose, 5 mM MgSO4, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol) was inoculated and cultured from an initial OD600 ~0.15. Expression was induced at OD600 ~0.6 with 50 μM IPTG. The cells were subsequently cultured at 17°C for 18-20 h, harvested, flash-frozen in liquid nitrogen, and stored at -80°C until further use. All purification steps described below were carried out at 4°C. Cells from 2 L of culture were resuspended in lysis buffer (40 mM HEPES pH 7.5, 100 mM NaCl, 10 mM imidazole, 10% glycerol, 5 mM MgCl2, 0.05 mM GDP, 250 units/μL Benzonase®, 2 mM PMSF, PIC (Roche), 50 μg/mL lysozyme, 0.1 mM DTT) and disrupted by sonication. The cell lysate was clarified by centrifugation and mixed in batch with 10 mL Super Ni-NTA agarose resin (Generon) for 2 h. The resin was packed by gravity-flow and washed with 4 x 10 CV wash buffer (20 mM HEPES pH 7.5, 200 mM NaCl, 10% glycerol, 1 mM MgCl2, 40 mM imidazole, 0.05 mM GDP). The bound species was eluted with elution buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM MgCl2, 250 mM imidazole, 0.05 mM GDP). The eluate was supplemented with 2.5 mg TEV protease and 1 mM DTT, transferred to a 10 kDa MWCO dialysis bag, and dialyzed overnight against SEC buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM MgCl2, 0.01 mM GDP). TEV protease was removed by negative purification on Super Ni-NTA agarose resin (Generon). The flow-through was concentrated using a 10 kDa MWCO Amicon® Ultra centrifugal concentrator (Merck) and loaded onto a Superdex 200 HiLoad 26/60 column (GE Healthcare) pre-equilibrated in SEC buffer with 0.1 mM TCEP. The peak fractions containing mini-Gpa1 were pooled and concentrated. Protein concentration was estimated by NanoDrop 2000 (Thermo Scientific) at 280 nm. A typical yield was ~4 mg pure mini-Gpa1/L cell culture (using an extinction coefficient of 0.702).
Cloning, expression and purification of wild type Gpa1
The construct encoding a wild type S. cerevisiae Gpa1 containing an N-terminal His6-TEV tag was synthesized as a gBlocks® gene fragment (IDT) and cloned into pET15b by in vivo assembly in E. coli XL10-Gold36. The plasmid was then transformed into E. coli strain BL21-CodonPlus(DE3)-RIL cells (Agilent). To express full-length Gpa1, a 5 mL LB starter culture (supplemented with 0.2% glucose, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol) was inoculated with a single bacterial colony and grown for 6-8 h at 37°C shaking at 220 rpm. 5 mL from this starter culture was used to inoculate 150 mL LB media (supplemented with 0.2% glucose, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol) and the culture was incubated for 16-20 h at 30°C. 2L TB media (supplemented with 0.2% glucose, 5 mM MgSO4, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol) was inoculated with the overnight culture at an OD600 of 0.15 and expression was induced with 50 μM IPTG when the cells reached an OD600 ~0.6. The cells were subsequently cultured at 25°C for 18-20 h, harvested, flash-frozen in liquid nitrogen, and stored at -80°C until further use. All purification steps described below were carried out at 4°C. Cells from 2 L of culture were resuspended in lysis buffer (40 mM HEPES pH 7.5, 100 mM NaCl, 10 mM imidazole, 10% glycerol, 5 mM MgCl2, 0.05 mM GDP, 250 units/μL Benzonase®, 2 mM PMSF, PIC (Roche), 50 μg/mL lysozyme, 0.1 mM DTT) and disrupted by sonication. The cell lysate was clarified by centrifugation and loaded onto HisTrap-FF column (GE Healthcare). The column was washed with 4 x 10 CV wash buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 1 mM MgCl2, 40 mM imidazole, 0.05 mM GDP). The bound species was eluted with elution buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM MgCl2, 250 mM imidazole, 0.05 mM GDP). The eluate was supplemented with 4 mg TEV protease and 1 mM DTT, transferred to a 10 kDa MWCO dialysis bag, and dialyzed overnight against SEC buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM MgCl2, 0.01 mM GDP). TEV protease was removed by negative purification on Super Ni-NTA agarose resin (Generon). The flow-through was concentrated using a 10 kDa MWCO Amicon® Ultra centrifugal concentrator (Merck) and loaded onto a Superdex 200 HiLoad 26/60 column (GE Healthcare) pre-equilibrated in SEC buffer with 0.1 mM TCEP. The peak fractions containing wild type Gpa1 were pooled and concentrated. Protein concentration was estimated using a NanoDrop 2000 (Thermo Scientific) at 280 nm. A typical yield was ~15 mg pure wild type Gpa1/L cell culture (using an extinction coefficient of 0.769).
Cloning, expression and purification of Ste4 and Ste18
Constructs encoding wild-type S. cerevisiae Ste4 (residues 1-423) containing an N-terminal His6-TEV tag, and full-length Ste18 (residues 1-110) containing two mutations (C106S, C107S) were synthesized as gBlocks® gene fragments (IDT). Ste18 residues C106 and C107 are known to be palmitoylation and prenylation sites, respectively38. The gene fragments were cloned separately into pBacPAK8 by in vivo assembly in E. coli XL10-Gold36. Separate high-titre recombinant baculoviruses for the expression of either Ste4 or Ste18 were prepared using the flashBAC ULTRA system (Oxford Expression Technologies). Trichoplusia ni cells (Expression Systems) were maintained in log phase growth in suspension in ESF 921 medium (Expression Systems). For large-scale production, 4 L of cells at 2-3 x 106 cells/mL were supplemented with 5% serum, co-infected with 3% (v/v) Ste4 and 3% (v/v) Ste18 baculoviruses, and cultured for 48 h. The cells were harvested by centrifugation, flash-frozen in liquid nitrogen, and stored at -80°C until further use. All purification steps described below were performed at 4°C. Cells were resuspended in lysis buffer (30 mM Tris pH 8.0, 100 mM NaCl, 5 mM Imidazole, 5 mM MgCl2, 250 units/μL Benzonase®, 2 mM PMSF, PIC) and disrupted by sonication. The cell lysate was clarified by centrifugation and mixed in batch with 10 mL Super Ni-NTA agarose resin (Generon) overnight. The resin was packed by gravity-flow and washed sequentially with 2 x 10 CV wash buffer 1 (20 mM Tris pH 8.0, 250 mM NaCl, 10% glycerol, 10 mM imidazole), 2 x 10 CV wash buffer 2 (20 mM Tris pH 8.0, 250 mM NaCl, 10% glycerol, 20 mM imidazole). The bound species was eluted with elution buffer (20 mM Tris pH 8.0, 25 mM NaCl, 10% glycerol, 300 mM imidazole). The eluate was supplemented with 6 mg TEV protease and 1 mM DTT, transferred to a 10 kDa MWCO dialysis bag, and dialyzed overnight against SEC buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol). TEV protease was removed by negative purification on Super Ni-NTA agarose resin (Generon). The flow through was concentrated in a 10 kDa MWCO Amicon® Ultra concentrator (Merck) and loaded onto a Superdex 200 HiLoad 26/60 column (GE Healthcare) pre-equilibrated in SEC buffer with 0.1 mM TCEP. The peak fractions containing Ste4-Ste18 dimer were pooled and concentrated.
Protein concentration was estimated by NanoDrop 2000 (Thermo Scientific) at 280 nm. A typical yield was ~4 mg pure Ste4-Ste18/L of culture (using an extinction co-efficient of 1.321).
Formation of Ste2-G protein heterotrimer complex
Mini-Gpa1 heterotrimer or wild type Gpa1 heterotrimer was formed by mixing purified mini-Gpa1 or wild type Gpa1, respectively, with Ste4-Ste18 dimer at 1:1 molar ratio and the complex was loaded and separated on Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated with 10 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM MgCl2, and 0.01 mM GDP. Peak fractions were pooled and the complex was concentrated in a 10K MWCO Amicon® Ultra centrifugal concentrator (Merck). Ste2 was mixed with mini-Gpa1 heterotrimer or wild type Gpa1 heterotrimer at 1.1:1 molar ratio, at a final Ste2 concentration of 2 mg/mL. The mixture was supplemented with 40 μM α-factor and incubated overnight on ice. Apyrase (2 U/mL) and lambda protein phosphatase (400 units) (NEB) were added and allowed to incubate at 4°C for 1 h. The sample was loaded onto an Agilent Bio SEC-5 500 Å column pre-equilibrated in 20 mM HEPES pH 7.5, 100 mM NaCl, 2 mM MgCl2, 0.001% (w/v) LMNG, and 1 μM μ-factor. Peak fractions were pooled and the complex was concentrated in a 100K MWCO Amicon® Ultra centrifugal concentrator (Merck).
Ste2-G protein complex cryo-grid preparation and data collection
Cryo-EM grids were prepared by applying 3 μL α-factor-bound Ste2-mini-Gpa1-Ste4-Ste18 (~0.7 mg/mL) onto glow-discharged holey gold (Quantifoil Au 1.2/1.3 300 mesh) and copper grids (Quantifoil Cu 1.2/1.3 300 mesh). Excess sample was removed by blotting with filter paper for 2.5 s before plunge-freezing in liquid ethane (at -181°C) using a FEI Vitrobot Mark IV at 100% relative humidity and 4°C. Data collection was carried out on grids made from a single preparation of Ste2-G protein heterotrimer complex. Images were collected on a FEI Titan Krios microscope operating at 300 kV using a GIF Quantum K2 detector (Gatan) in counting mode. Data were collected in three independent sessions (eBIC Krios1, LMB Krios1, LMB Krios2) producing a total of 7,660 movies. On eBIC Krios1 and LMB Krios1, data collection was performed using EPU (Thermo Fisher Scientific). Individual grid holes were centred with stage shift for data acquisition. On LMB Krios2, automated data acquisition was performed using SerialEM39. In this case, beam tilt-compensated image shift with calibrated coma correction was used to acquire images from 3 x 3 grid hole arrays39,40. Two non-overlapping exposures were collected per grid hole in all sessions. Micrographs were collected with a fluence of ~40-48 e−/Å2. Each micrograph was collected as dose-fractionated movie frames (~1.0 e−/Å2/frame) at fluxes of 5 e−/pix/s (eBIC Krios1), 5.7 e−/pix/s (LMB Krios1), and 5.2 e−/pix/sec (LMB Krios2), with an energy selection slit width of 20 eV. The datasets were collected at a magnification of 130,000× (1.047 Å/pix, eBIC Krios1), 105,00× (1.145 Å/pix, LMB Krios1) and 105,000× (1.1 Å/pix, LMB Krios2).
Data processing and model building
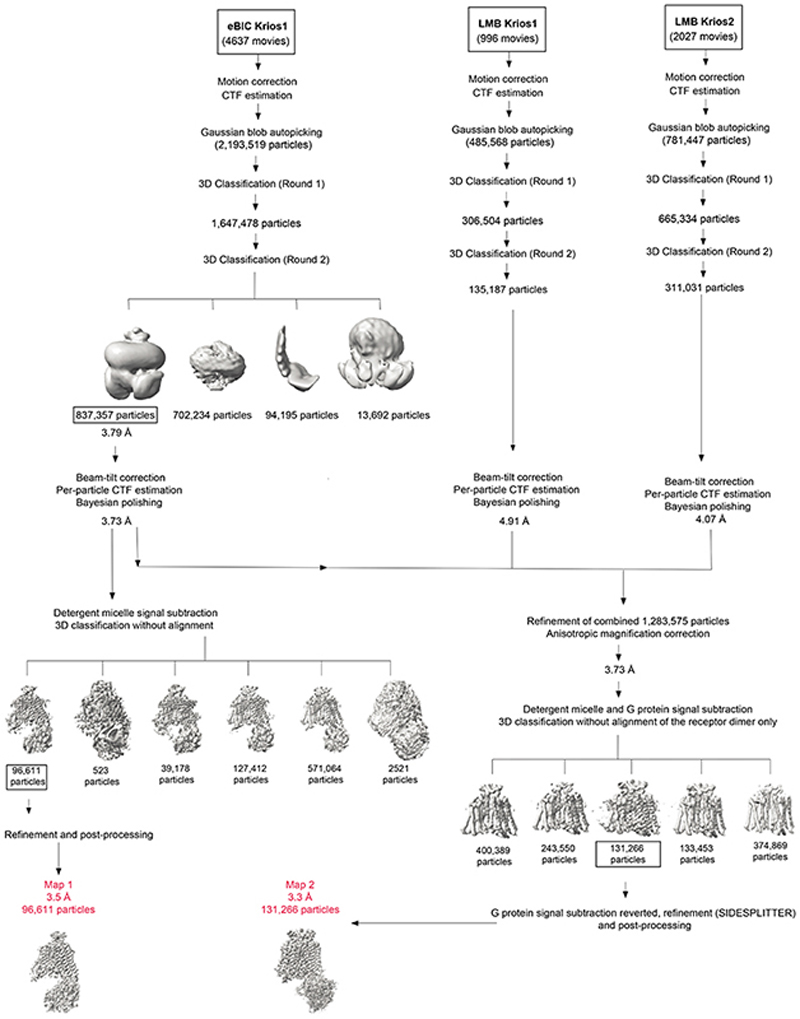
RELION-3.1 was used for all data processing unless specified otherwise41. Due to the beam-image shift strategy used in the collection of LMB Krios2 dataset, the micrographs from this session were grouped into one of 18 optics groups based on their positions within the 3 x 3 (x2 exposures) array. Micrographs from eBIC Krios1 and LMB Krios1 were assigned to one optics group each. Micrographs were subjected to beam-induced motion correction using MotionCor242 by dividing each frame into 5 x 5 patches. CTF parameters were estimated from non-dose-weighted micrographs in GCTF43 with equiphase averaging. Autopicking was performed with a Gaussian blob as template, which led to an initial selection of 2,193,519 particles, 485,568 particles, and 781,447 particles from eBIC Krios1, LMB Krios1 and LMB Krios2 datasets, respectively. Particles were extracted in a box-size equivalent to 220 Å and down-scaled initially to 4.1 Å/pix. The extracted particles were subjected to two rounds of 3D classification using an ab initio model as reference. This resulted in a major class with features consistent with a Ste2 homodimer coupled to two G protein heterotrimers. This major class comprised 837,357 particles, 135,187 particles, and 311,031 particles from the eBIC Krios1, LMB Krios1 and LMB Krios2 datasets, respectively. The eBIC Krios1 dataset also produced a class that is consistent with a G protein-free Ste2 homodimer. This class accounted for 702,234 particles but was not observed in the other two datasets. No reliable classes consistent with either a Ste2 monomer or monomer-G protein complex were identified from the datasets despite extensive 2D and 3D classifications. The particles belonging to the major class of each dataset were re-extracted at a pixel size of 1.6 Å/pix and refined iteratively with beam-tilt correction, per-particle CTF estimation, and Bayesian polishing procedures. At this stage, the reconstructions consistently exhibited clear transmembrane features as well as unambiguous occupancy of the two putative orthosteric ligand binding sites. The density for one G protein was relatively well defined. However, in comparison, the second G protein was poorly resolved, with reasonable alignment limited to the α5-helical segment of mini-Gpa1. The particles were re-extracted at the corresponding pixel sizes of data collection with another round of Bayesian polishing.
Map 1 was generated from the eBIC dataset. The initial stack of 837,357 particles was subjected to signal subtraction in order to exclude the detergent micelle and most of the poorly resolved G protein, but retaining the interfacial α-helical segment. Following consensus refinement, subtracted particles were characterised by 3D classification without alignment into 6 classes (regularisation parameter, T=20), masked to encompass signal from the entire post-subtraction component. This identified a final stack of 96,611 particles that refined to an overall resolution of 3.5 Å according to the gold standard FSC criterion of 0.143. It includes density for the α-factor-bound Ste2 homodimer, one G protein heterotrimer, and the remaining α□-helical segment of the second G protein. The α-factor-bound Ste2 and mini-Gpa1 regions feature clear density for side chains, which facilitated de novo model building. The Ste4-Ste18 region is less well resolved, but allowed reasonable model building in the resolved regions (Extended Data Fig. 2f).
To improve confidence in the de novo modelling of Ste2 and its interface with G protein, a second reconstruction (Map 2) was performed with specific emphasis on the receptor region. The initial stack of 837,357 particles from the eBIC dataset was combined with the equivalent stacks from LMB Krios1 (135,187 particles) and LMB Krios2 (311,031 particles) datasets. The merged particle stack was corrected for anisotropic magnification and subjected to signal subtraction to exclude, in this case, the detergent micelle and both G proteins. Subtracted particles were refined to produce a consensus alignment on the Ste2 homodimer region. Newly refined orientations were characterised by 3D classification without alignment into 5 classes (T=20), masked to encompass signal from the receptor region. This identified a subset of 131,266 particles that aligned well. To this final set of particles, signal for the G protein and α5-helical segment was restored. During subsequent refinement of these particles, half maps were locally filtered between refinement iterations using SIDESPLITTER44, which is adapted from the LAFTER45 algorithm and maintains gold-standard independence between the two half maps. Map 2 reached an overall resolution of 3.2 Å and showed improved features and local resolution in the Ste2 region, while resolution in the G protein regions was comparatively limited (Extended Data Fig. 2h,i).
Throughout data processing, one G protein remained less well resolved than the other. Some secondary structural features become more apparent when the reconstruction is low-pass filtered to 8 Å (Extended Data Fig. 2d). Attempts to improve this region of the map by focused classification and refinement proved intractable, reflecting the dynamic nature of this G protein. All classifications and refinements were performed with C1 symmetry. The application of C2 symmetry did not improve the map. Local resolution estimates for Map 1 and Map 2 were calculated with RELION-3.1. The directional FSC estimates were calculated using 3DFSC46. With previous datasets, reconstruction was hindered by moderately severe preferential orientation of the particles. Treatment of the pre-formed complex with phosphatase gave rise to an improvement in orientation distribution. Map 1 was assessed by 3DFSC to have sphericity of 0.958. The mechanism underlying this improvement was not investigated further. The C-terminal region of Ste2 is known to be extensively phosphorylated47 in vivo and it is possible that the removal of existing phosphoryl groups facilitates the reorientation of particles at the air-water interface. The overall stability of the complex and the asymmetry apparent in the dynamics between the two G proteins were unaffected by the phosphatase treatment.
Map 1 was used for model building and refinement, and Map 2 was used for visualization of some less well resolved receptor regions in Map 1. Map densities depicted in this study are of Map 1 unless otherwise specified. Model building and refinement were performed using the CCP-EM48 and PHENIX49 software suites. Buccaneer50 was used for chain tracing to generate a partial ab initio model of Ste2, which was rebuilt manually in COOT16. The cryo-EM model of mini-Go-β1-γ2 (PDB ID 6G79) was used as a starting model for mini-Gpa1-Ste4-Ste18. In referring to the polypeptides that exist as two copies, molecules A (in subscript) denote the components associated with the well-resolved G protein; molecules B denote those associated with the more dynamic G protein. The following residues were not modelled due to poor map density: (1) Ste2A and Ste2B, N-terminal residues 1 to 7 and C-terminal residues 307 to 431); (2) mini-Gpa1A, N-terminal residues 4-6 and residues 360-361; (3) Ste4, N-terminal residues 2-13 and C-terminal residues 347-423; and (4) Ste18, N-terminal residues 1-49 and C-terminal residues 75-110. Mini-Gpa1B has been modelled from residues 450-472, corresponding to its α5 helix. The model was built by iterative remodelling in COOT and real space refinement in PHENIX. Restraints included standard geometric, secondary structure, Ramachandran, and rotamer. Restraints for N-acetylglucosamine (NAG) and CHS (monomer library ID Y01) were derived in eLBOW51 and AceDRG52, respectively. The final model achieved good geometry (Extended Data Table 1). Model validation was performed in PHENIX, Molprobity53 and EMRinger54. The goodness of fit of the model to the map was assessed in PHENIX using a global model-vs-map FSC (Extended Data Fig. 4d). FSCwork/FSCtest validation was performed to monitor over fitting during refinement by refining a ‘shaken’ model55 against half map-1 and then calculating the FSC of the resulting refined model against half map-2.
Data collection and processing of Ste2 with a Volta Phase Plate
To analyse the oligomeric state of Ste2 in the absence of coupled G proteins, cryo-EM grids were prepared as described above with α-factor-bound Ste2 (~0.32 mg/mL). Samples were imaged on a Titan Krios operating at 300 kV using a GIF Quantum K2 detector (Gatan) and a Volta Phase Plate56 (VPP) to improve contrast. 623 micrographs were collected at a magnification of 105,000× (1.1 Å/pix) using SerialEM. Each micrograph was collected as dose-fractionated movie frames (~1.0 e−/Å2/frame) at a flux of 4.3 e−/pix/s with an energy selection slit width of 20 eV, resulting in fluence of 35 e−/Å2. Micrographs were subjected to beam-induced motion correction using MotionCor242 by dividing each frame into 5x5 patches. CTF parameters were estimated from non-dose-weighted micrographs in GCTF43 with equiphase averaging. Autopicking was performed with a Gaussian blob as template, which led to an initial selection of 390,123 particles. Particles were extracted in a box-size equivalent to 184 Å and down-scaled to 2.2 Å/pix. Representative 2D class averages from the processing of this dataset (Extended Data Fig. 1g) indicate that α-factor-bound Ste2 exists as a dimer in the absence of G protein. No classes of monomeric Ste2 were obtained. This dataset did not yield a high-resolution 3D reconstruction.
FSEC Assays
Fluorescence-detection size-exclusion chromatography (FSEC) assays were performed following established procedures57. Insect cells (500 μL, 3 x 106 cells/mL) expressing wtSte2-TEV-EGFP-His10 were resuspended in 20 mM HEPES pH 7.5, 150 mM NaCl, 20% glycerol, 2 mM PMSF, 25 U/mL Benzonase®, 10 mM MgCl2, PIC. The cells were split into aliquots and solubilized with different detergents (final concentration 1%; CHS final concentration 0.1%, where applicable): dodecyl-β-D-maltoside (DDM)+CHS, LMNG+CHS, LMNG, digitonin and GDN. The cells were solubilized for 1 h and clarified by ultracentrifugation. The solubilisate was loaded onto an Agilent Bio SEC-5 500 Å column attached to a Shimadzu HPLC system. UV absorbance was measured at 280 nm. GFP fluorescence was measured at excitation and emission wavelengths of 475 nm and 515 nm, respectively. The running buffer contained 20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, and 0.03% (w/v) DDM. The column was calibrated with the following molecular weight standards: thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa). The data was plotted and analysed in GraphPad Prism 8.
SEC-MALS
Size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) experiments were performed by injecting 100 μL samples onto an Agilent Bio SEC-5 500 Å column. Light scattering was measured in-line using a Wyatt HELEOS-II 18-angle photometer coupled to a Wyatt Optilab rEX differential refractometer (Wyatt Technology Corp). Ste2-mini Gpa1 heterotrimer complex, Ste2-wild type Gpa1 heterotrimer complex, Ste2, and mini-Gpa1 heterotrimer were injected at 0.36 mg/mL, 0.2 mg/mL, 0.18 mg/mL and 0.47 mg/mL, respectively. Analytical SEC of Ste2-mini Gpa1 complex, Ste2-wild type Gpa1 complex, and Ste2 were performed at 25°C in a buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM α-factor, 0.001% (w/v) LMNG, 0.0005% (w/v) CHS. Mini-Gpa1 heterotrimer was characterised in 20 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.01 mM GDP. BSA was used for calibration. Molecular masses were determined using the three-detector methodology58 implemented in the ASTRA software package (Wyatt Technology). Extinction coefficients of 1.218, 1.154, 1.157 and 1.129 for Ste2-mini G protein heterotrimer, Ste2-wild type G protein heterotrimer, Ste2, and mini-Gpa1 heterotrimer, respectively, were calculated in PROTPARAM. A dn/dc value of 0.139 was used for the detergent micelle59. The data was plotted in GraphPad Prism 8.
Thermostability assay
Thermostability measurements were performed on a Prometheus NanoTemper NT.48 and the data was analysed in PR.ThermControl v2.3.1 software package. Ste2-G protein heterotrimer complex and Ste2 were resuspended in 20 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM α-factor, 0.001% (w/v) LMNG, 0.0005% (w/v) CHS. The temperature was increased from 15.0°C to 95.0°C at 2°C/min. The melting curve was computed from the first derivative of the ratio of intrinsic fluorescence intensities (350/330 nm). The data was plotted in GraphPad Prism 8.
Molecular dynamics simulations
The purpose of the MD simulations was to examine the local dynamics (local to the EM structure starting structure) of the Ste2 dimer with and without G protein coupled. Multiple short simulations were performed rather than one long simulation as the random initial velocity assignments allowed the two G proteins to explore more possible motions60,61. The structure of mini-Gpa1B-Ste4B-Ste18B was not determined by cryo-EM, except for the α5 helix in the mini-Gpa1B subunit. We generated a starting model of a Ste2 dimer coupled to two G proteins by using the cryo-EM structure mini-Gpa1-Ste4-Ste18 unit (resolved G protein). The resolved subunit of Gpa1 was overlaid on the unresolved mini-Gpa1B peptide, to generate the full Ste2-G protein dimer complex, as shown in Fig. 4e. Here we performed all-atom MD simulations on the Ste2-G protein dimer complex in explicit lipid POPC bilayer and water. The total number of atoms including lipid and water in this system was 366,667. The modelled full dimer complex was energy minimized using GROMACS package62 with CHARMM36m63 force field, with restraints of 5 kcal/mol-Å2 applied on all backbone heavy atoms during the minimization process. The minimized structure of the Ste2-G protein dimer complex was placed in a 13*13 nm2 POPC bilayer environment using CHARMM-GUI64, and then solvated with a water box with margins of 14 Å from the protein. The system was then neutralized with 150 mM NaCl. This system was then slowly heated from 0 K to 310 K using a NVT ensemble and Nosé-Hoover thermostat65. This heating process was followed by a 30 ns equilibration protocol performed using NPT ensemble. Throughout the heating and equilibration processes we used a harmonic position restraint on the proteins, ligand and the POPC lipid heavy atoms. The constraints force constant was set to 5 kcal/mol-Å2 initially, and gradually reduced to 0 kcal/mol-Å2 with a 1 kcal/mol-Å2 per 5 ns window. The pressure was controlled using the Parrinello-Rahman method66 and the simulation system was coupled to a 1 bar pressure bath. The last frame from the equilibrium process was taken as the starting conformation for 25 unconstrained NPT simulation runs at 310K each lasting for 50 ns with 6 of the runs extended to 75 ns. We performed 25 short MD simulations to refine the fit of the second unit of mini-Gpa1B in the Ste2 dimer structure. In all simulations the LINCS algorithm is applied on all bonds and angles of waters with 2 fs time step used for integration. We use a cut-off of 12 Å for non-bond interaction and used the particle mesh Ewald method67 to treat long range L-J interactions. The MD snapshots were stored at every 20 ps interval.
To calculate the movement of the Ste4 subunit, snapshots from the MD trajectories were aligned to the starting EM structure based on backbone atoms of the transmembrane regions of Ste2. We calculated the distance between the centre of mass of both Gβ Ste4 units and the centre of mass from the initial position in the EM structure. The distance moved by each unit from the starting positions were plotted as a time series and we observed a significant shift away from the initial position when the distance between the centres of mass with respect to the EM structure was greater than 7 Å. Within one simulation run, we observed the movement of either of the two monomers but not both the monomers at the same time. Three scenarios were observed in all the 25 simulations: (i) the modelled Ste4 unit shifted away; (ii) the EM resolved Ste4 unit shifted away; (iii) one of the monomer shifts away and swings back completing one period within 40 ns, soon after which the other unit began to shift away. We speculate that this periodic motion could be a consequence of interaction between Ste4 unit from one G-protein and mini-Gpa1 unit of the other G-protein (Fig. 4e).
We performed MD simulations on the Ste2 dimers in the absence of G protein and α-factor by removing the peptide ligand and G protein from the starting cryo-EM structure. The Ste2 dimer was put into POPC bilayer with dimensions of 13*13 nm2. The minimisation-heating-equilibration protocol described above was used for this system as well. We performed five production runs each 1000 ns long, giving a total of 5000 ns of simulation time. We tested the convergence of these 5 simulations by calculating the dot product of the top 5 principal components across the 5 runs. As shown in Extended Data Figs. 8d,e the 5 MD simulation runs have converged a stable, possibly tending towards inactive state.
To calculate the inter-residue contact frequency, the residue non-bond contacts (van der Waals interactions between heavy atoms) within 4.5Å between the two Ste2 monomers in the dimer interface was calculated. The percentage of the MD snapshots that contain these contacts is the frequency of the contacts shown in Extended Data Fig. 8c. The contact frequencies were calculated for both 1400 ns of Ste2-mini-Gpa1-Ste4-Ste18 complex simulation and 5000 ns of Ste2 dimer only simulation. The contact frequencies are coloured as b-factor on Ste2 structure (Extended Data Fig. 8b). There was no significant difference between the dimer interface after 1000 ns simulation without ligand and G-protein, indicating the dimer interface remains stable during the simulations. The standard deviation in the contact frequency for the dimer interface residues among the five MD simulation runs for Ste2 dimer without the G protein and α factor and among the 25 simulation runs for Ste2 dimer with G protein are shown in Extended Data Fig. 8c.
To better understand the structural changes in the Ste2 conformation after the MD simulations, we calculated the tilt angles of the transmembrane α-helices before and after the MD simulations of the Ste2 dimer without the G protein. The tilt angle differences were calculated for the representative structures extracted from the most populated conformation cluster from the MD simulations. We carried out clustering of the 5 μs MD trajectories of Ste2 dimer without the G protein, by the root mean square deviation (RMSD) in the coordinates of the backbone atoms in the transmembrane α-helices. We used a cut-off of 1.5Å in RMSD for the clustering method. The clustering was done using the g_cluster script from GROMACS62. The most occupied cluster had a population of 66% of the MD snapshots in this cluster. We extracted the representative conformation from this most occupied conformation cluster to calculate the tilt angles. An in-house software68 was used to calculate the tilt angle of the transmembrane α-helices on this representative structure. We also calculated the tilts of the α-helices in the cryo-EM structure. The difference in the tilt angles of each transmembrane α-helix in the MD structure compared to the starting structure are: H1 9°; H2 5°; H3 11°; H4 14°; H5 6°; H6 9°; and H7 9°. The volume of the orthosteric binding pocket and G protein binding pocket was calculated for the cryo-EM structure and a representative structure of the 5 μs simulation (no G protein or ligand) from clustering analysis using trj_cavity69. The grid space for pocket searching was set to 1.7 Å. The respective volumes calculated were the following: orthosteric binding site, 1066 Å3 for Ste2A in the cryo-EM structure and 1125 Å3 for the simulated structure; G protein binding pocket, 712 Å3 for Ste2A in the cryo-EM structure and 176 Å3 for the simulated structure.
Ste2 sequence conservation analysis
All sequences with a Ste2 PFAM domain (PF02116) were retrieved from UniProt (accessed on May 14th, 2020). This comprised 4 Swiss-Prot reviewed sequences and 829 TrEMBL unreviewed sequences, 380 of which are preliminary data obtained from whole genome shotgun entries. All partial sequences, defined as tagged as “Fragment” by UniProt or having fewer than 7 predicted transmembrane helices, were removed which left 479 entries. Entries with the same sequence were merged and for each species/strain the entry with the highest number of residues aligning with residues in the STE2_YEAST sequence in a pairwise alignment was kept, resulting in a final dataset of 395 unique sequences (Supplementary Table 2). These sequences were aligned with Motif-Aware PRALINE and subsequently manually refined using JalView70 (version 2.11.1.0) and AliView71 (version 1.26).
Class nomenclature ‘D1’
In the GPCR nomenclature system, fungal receptors have been placed in a single class, namely class D, but they are exceedingly diverse in amino acid sequence. For example, there is no amino acid sequence similarity between Ste2 and Ste3 receptors. Consequently, previous studies that classified fungal GPCRs separated them into ten classes (I-X)4. As all GPCR classes have been defined by amino acid sequence similarity, and not by species (for example, there are no GPCR classes specific to humans), we have followed the categorisation of fungal receptors and named the nomenclature ‘class D1’, as Ste2 was previously categorised as a ‘class I’ receptor in fungi. Hopefully this system will be expanded to include all the other classes of fungal GPCRs once their structures have been determined, but this will inevitably require further classes as we were unable to include Ste3 in class D1.
Class D1 and cross-class generic residue numbering
Consistent with the generic residue numbering schemes of other GPCR classes72, the most conserved residue in each predicted transmembrane helix was identified and annotated as the numbering reference position with the number 50. Other residues in each of the helices, indexed 1-7, were numbered relative to this reference position. ECL1 has an ordered secondary structure (an α-helix) and was assigned generic residue numbers in the same way. This segment was named D1e1 (class D1 ECL1), as its secondary structure differs from the class A ECL1. Furthermore, the β-sheets in the N-terminus were assigned generic residue numbers distributed over three segments: D1S1 (class D1 Sheet 1), D1T1 (class D1 Turn 1), and D1S2 (class D1 Sheet 2) (Extended Data Fig. 6). Based on a structure superposition, a sequence alignment was generated between classes D1 and A. Only the 7-transmembrane helix bundle of Ste2 was structurally superposed to the heptahelical transmembrane bundle of all class A GPCR structures available in the PDB with TM-align73. The match with the lowest RMSD to Ste2 was obtained with a structure of the angiotensin II type 2 receptor (5XJM). Reference x50 positions from this class A reference structure were transferred to the closest residue in Ste2 and all other residues numbered relative to this. For H3, which acts as a hub in the GPCR fold, the class D1 and class A reference (number 50 positions) were found to be in an equivalent position and thus assigned the same number (Extended Data Fig.6). The Ste2 sequence was integrated along with 10 orthologues into the GPCRdb. This enables the comparison of the many species orthologues – currently 829 STE2 sequences from 691 unique species/strains in UniProt – and of the corresponding positions in the classes A, B, C and F.
In addition to classifying Ste2 sequences, the sequence of Gpa1 (GPA1_YEAST) was annotated according to the CGN scheme25 and integrated into the GPCRdb resources. Gpa1 is most similar to the human Gi/o subtypes with key differences in 3 loop regions, namely a 69 residue long insertion in loop hahb, an insertion of 40 residues in loop hbhc, and a single extra residue in loop hchd.
Extended Data
Extended Data Fig. 1. Purification, formation, and biophysical characterization of the Ste2-G protein heterotrimer complex.
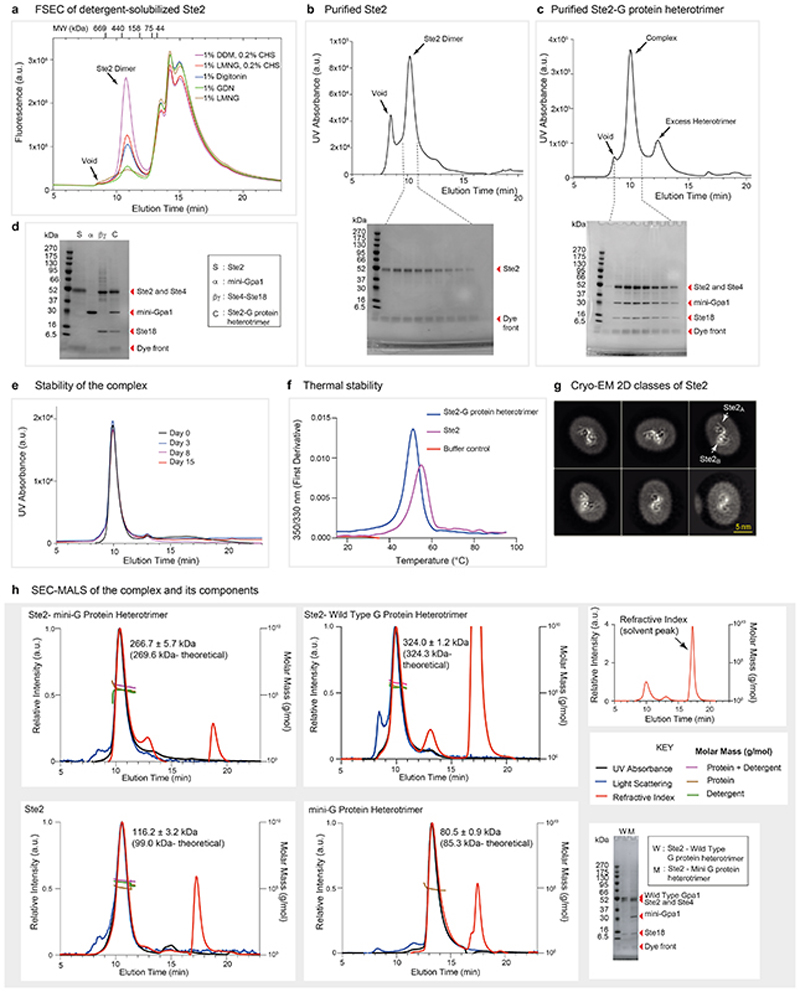
a, FSEC trace of insect cells expressing wtSte2-TEV-eGFP-His10 solubilised in different detergents. Ste2 migrated as a dimer in all tested detergents with an apparent molecular weight of ~380 kDa. The molecular weight standards used for calibration of the column are shown for reference. Results are representative of three independent experiments. b, Size-exclusion chromatography (SEC) profiles of Ste2 and SDS-PAGE analysis of the eluted fractions. c, SEC profiles of Ste2-G protein heterotrimer complex and SDS-PAGE analysis of the eluted fractions from the complex formation reaction (see methods). d, SDS-PAGE analysis of the pooled and concentrated Ste2-G protein complex along with the individual components used to form the complex. Experiments in b, c, and d are representative of three independent purifications. e, Complex stability as analysed by SEC after incubation of the purified complex on ice for up to 15 days. This experiment was carried out once. f, Thermal stability as analysed by nanoDSF, which measures changes in the intrinsic fluorescence of the protein when subjected to thermal denaturation. The melting temperatures (Tm) of Ste2-G protein complex and Ste2 dimer were calculated to be 51.2 °C and 55.0 °C, respectively. This experiment was performed once in two replicates. g, Representative 2D class averages of Ste2 imaged on a Titan Krios with the GIF Quantum K2 detector and a Volta Phase Plate56 (VPP). All classes are consistent with a dimer and no monomer classes were observed. h, SEC-MALS analysis of the Ste2-mini-G protein heterotrimer complex, Ste2-wild type G protein heterotrimer complex, Ste2 dimer and mini-G protein heterotrimer complex. A large change in refractive index that occurs at the end of column volume due to the elution of salts and/or detergents present in the solvent is indicated as solvent peak. Where indicated, chromatograms show signal from UV absorbance, light scattering and refractive index. The traces were normalized to the peak maxima. Calculated molar masses are depicted as traces. The molar masses of the protein components across the peak and the theoretical molar masses are indicated. SDS-PAGE analysis of the Ste2-mini-G protein heterotrimer and Ste2-wild type G protein heterotrimer complexes are shown. The results are representative of three independent experiments, except the experiment with Ste2 dimer, which was performed once. For gel source data (panels b, c, d and h), see Supplementary Figure 1
Extended Data Fig. 2. Cryo-EM single-particle reconstruction of the Ste2-G protein heterotrimer complex.
a, A representative micrograph (defocus −2.0 μm) from the eBIC Krios1 dataset. b, Representative 2D class averages of the Ste2-G protein heterotrimer complex determined using the initial set of particles from eBIC Krios1 dataset following 3D classification. Reconstructions with the detergent micelle are juxtaposed to indicate relative orientations. c, A representative 2D class average is shown from the final set of particles (eBIC Krios1 dataset) used to determine the model. While one G protein is well-resolved, the second heterotrimeric G protein manifests as a poorly-resolved density in the reference-free 2D averages (circled). d, The poorly-resolved G protein heterotrimer (orange circle) becomes apparent at lower contour levels and certain secondary structural features become visible when the map is low-pass filtered to 8 Å. The contact between mini-Gpa1B and Ste4A of the adjacent mini-Gpa1A heterotrimer is indicated (red circle) e, g, FSC curves of the reconstructions Map 1 and Map 2 (black lines) shows an overall resolution of 3.5 Å and 3.3 Å, respectively, using the gold standard FSC of 0.143. The directional 3D-FSC curves computed from the two halfmaps are shown in colour. h, Local resolution estimation of the Ste2 dimer portion from Map 2 as calculated by RELION. f, i, Local resolution estimation of the Ste2-G protein heterotrimer complex from Map 1 and Map 2 as calculated by RELION. Map 2 shows a higher local resolution in the Ste2 dimer, but the resolution is limited in the G protein regions.
Extended Data Fig. 3. Flow chart of single particle cryo-EM data processing.
Micrographs were collected from three independent sessions (between 24 h and 72 h long) on a Titan Krios with GIF Quantum K2. Each dataset was corrected separately for drift, beam-induced motion and radiation damage. After estimation of CTF parameters, particles were picked using a Gaussian blob and subjected to two rounds of 3D classification using an ab initio model as reference. The number of final particles from each round of 3D classification is shown. A representative 3D classification scheme (round 2) from the eBIC Krios1 dataset is shown. The initial set of best particles from the eBIC Krios1 dataset was refined and corrected for beam-tilt. Per-particle CTF estimation and Bayesian polishing was performed, detergent micelle signal was subtracted, and a 3D classification without alignment was performed. A model (Map 1) with 96,611 particles was refined and achieved a global resolution of 3.5 Å. In parallel, the final set of particles from LMB Krios1 and LMB Krios2 were refined and corrected for beam-tilt, which was followed by per-particle CTF estimation and Bayesian polishing. The initial set of particles from eBIC Krios1 was combined with the final set of particles from LMB Krios1 and LMB Krios2. The combined set of particles was refined, anisotropic magnification correction was performed, and the particles were subjected to a 3D classification without alignment after subtraction of the signal from the detergent micelle and the G protein. After restoring the G protein signal, a set of 131,266 particles was refined to a global resolution of 3.3 Å (Map 2) with improved features in the Ste2 dimer region. Resolution of the models after refinements was calculated with the gold-standard FSC of 0.143 in RELION.
Extended Data Fig. 4. Cryo-EM map quality of the Ste2-mini-Gpa1-Ste4-Ste18 heterotrimer complex and model validation.
Unless stated otherwise, densities from Map 1 were visualised using UCSF ChimeraX35 (contour level 0.03) and encompass a carve radius of 2 Å around the indicated region. a, Densities of transmembrane helices of Ste2A (contour level 0.045), α-factorB, mini-Gpa1B, dimer interface between Ste2A and Ste2B, intracellular loops ICL1-3, and extracellular loops ECL1-3 are shown as a mesh. Densities of αN and α4-β6 are shown as representative densities from mini-Gpa1A. Densities of Ste4 and Ste18 (contour 0.025) are shown as a mesh at a contour level of 0.01 and a carve radius of 2 Å. b, Two NAG molecules were modelled attached to Asn25 of SteA and Ste2B and they contribute to the dimer interface (contour level 0.01 from Map 2). Asn25 is a well characterized glycosylation site in Ste217. c, Densities for six ordered putative CHS molecules surrounding the dimer interface were visible and were modelled (contour level 0.01). Several other detergent or lipid acyl chain densities were visible in the map before detergent micelle signal subtraction, but were left unmodelled. d, FSC of the refined model versus the map (green curve) and FSCwork/FSCfree validation curves (orange and blue curves, respectively).
Extended Data Fig. 5. Comparison of Ste2 to other classes of GPCRs.
Thirty-nine receptors in the active state were aligned using GESAMT74 and then prototypical members of different classes were compared with Ste2. Both GESAMT74 and TM-align73 provided similar results. In all panels, Ste2 is shown in rainbow colouration and is compared with: a, β2-adrenoceptor (Class A; PDB code 3SN6); b, glucagon-like peptide receptor 1 (Class B; PDB code 6B3J); c, GABAB receptor (Class C, PDB code 7C7Q); d, smoothened (Class F; PDB code 6D32).
Extended Data Fig. 6. Amino acid conservation of Ste2 and the Class D1 (CD1) numbering system.
a, The size of the letter is related to the degree of conservation from the aligned Ste2 orthologues (395 sequences). The CD1 numbering system is shown alongside the positions of the defining residue and Ballesteros-Weinstein number for Class A GPCRs.
Generic residue numbers use the GPCRdb scheme, which uses the same reference positions as the Ballesteros-Weinstein scheme but corrects offsets due to helix bulges or constrictions. Two structurally important motifs in Class A GPCRs (DRY and NPxxY) are depicted. b, The amino acid sequence of S. cerevisiae Ste2 is shown (coloured boxes) in relation to secondary structural elements. The CD1 number (e.g. 5×50) is given adjacent to the amino acid residue number. Numbers in grey boxes represent the sequence conservation of the S. cerevisiae Ste2 residue at that position amongst Ste2 orthologues from 395 unique sequences. The light blue colour on the residue sequence numbers indicates residues predicted to be positioned in the lipid bilayer based on structural superposition with the Class B receptor GLP1R (6B3J).
Extended Data Fig. 7. Amino acid residues forming the Ste2 dimer interface.
Contacts are shown as coloured squares with a digit representing the number of van der Waals interactions (≤ 3.9 Å) and hydrogen bonds.
Extended Data Fig. 8. MD simulations of the Ste2 dimer in the absence of G proteins and ligands.
a, Groove in the H1-H1 dimer interface. b, Heat map of the percentage of MD snapshots that show residue contacts in the dimer interface in the EM structure and in the representative structure from MD simulations in the absence of the ligands and G proteins. c, Top row, histograms of the standard deviation among 25 simulation runs (total 1.4 μs) for Ste2 dimer with G protein and α factor. Bottom row, histograms of the standard deviation in Ste2A and Ste2B interface residue contacts among 5 simulation runs (total 5 μs) of the Ste2 dimer without G protein. d, Convergence of the five 1 μs simulations was tested by calculating dot product among the top 5 weighted principal components (PC) for the 5 simulations of Ste2 dimer without G protein or α factor. The top weighted principal components PC1 to PC5 from each run all show strong overlap (red squares) with at least one of the PCs from other runs. e, Cumulative variance of PCs show that the top five PCs occupy more than 90% of the populations and therefore they are sufficient to describe the motion of the complex.
Extended Data Fig. 9. Structural features of Ste2 and comparison with mammalian GPCRs.
a-c, Alignment of GPCRs was performed by GESAMT74 (CCP4 suite of programs) in conjunction with 39 other G protein or arrestin-coupled GPCR structures (Fig. 5a). Ste2 is depicted in rainbow colouration and the adenosine A2A receptor (A2AR) in grey. a, DRY motif showing the stacking (van der Waals contacts, yellow dashed lines) of the highly conserved Arg3×50 residue with PheH5.23 of the α-subunit of Gs. b, Pro5×50-Ile3×40-Phe6×44 motif provides a conserved packing of residues in the active state at the core of Class A receptors. A similar role could be played by Leu5×35 and Val3×40 in Ste2. c, the NPxxY motif in Class A GPCRs aligns with a conserved region in Ste2 receptors (see also Extended Data Fig. 6a), but Pro7×50 in Ste2 is not associated with a kink in H7 as it does in Class A GPCRs. d, Conformational changes in the transmembrane α-helices observed during MD simulations of the dimer without the G proteins or the ligands. e, Alignment of the active state Ste2 (rainbow coloration) with the Ste2 conformation (grey) reached after MD simulations in the absence of G protein and ligand. Gpa1 would not be able to couple to this new conformation due to clashes with atoms in H2 (Ile80), H3 (Phe148, Gln149, Lys151, Val152, Thr155) and ICL2 (Gly156, Asp157, Asn158, Phe159). Clashes are defined as atoms (grey spheres) within 2 Å of atoms in Gpa1 coupled to the active state of Ste2. f, The position of the α-helical domain in Gpa1 distal to the dimer interface can be inferred from comparison with the structure of Gi (yellow) coupled to rhodopsin (PDB code 6CMO) after alignment with mini-Gpa1 (purple). The position in Gpa1 of the deletion of the α-helical domain (which also includes the ubiquitination domain) is indicated by the amino acid residues immediately before and after the deletion being shown as spheres. g, superposition of Go-coupled serotonin 5-HT1B receptor (yellow) with Ste2A (blue). The position of heterotrimeric Go does not align with the yeast G protein and is sited below Ste2B (grey). h, Views of the intracellular surfaces of dimers from Class D (Ste2), Class A (rhodopsin; PDB code 6OFJ) and Class C (GABAB receptor; PDB code 7C7Q). All receptors are in rainbow coloration.
Extended Data Table 1. Cryo-EM data collection and refinement statistics.
| (α-factor-bound Ste2 coupled to mini-Gpa1-Ste4-Ste18) | |||
|---|---|---|---|
| (EMDB-XXXXX) | |||
| (PDB XXXX) | |||
| Session | eBIC Krios1 | LMB Krios1 | LMB Krios2 |
| Data collection and processing | |||
| Magnification | 130,000x | 105,000x | 105,000x |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e-/Å2) | 40 | 40 | 40 |
| Defocus range (μm) | -0.9 to -2.7 | -0.9 to -2.7 | -0.9 to -2.7 |
| Pixel size (Å) | 1.047 | 1.145 | 1.1 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images*(no.) | 837,357 | 135,187 | 311,031 |
| Contribution to final particle images (no.) | 96,611 (Map 1), 110,300 (Map 2) | 16,418 (Map 2) | 4,556 (Map 2) |
| Final particle images (no.) | 96,611 (Map 1) | 131,274 (Map 2) | |
| Map resolution (Å) | 3.5 | 3.3 | |
| FSC threshold | 0.143 | 0.143 | |
| Map resolution range† (Å) | ~3.4 to ~4.7 | ~3.2 to ~6.5 | |
| Refinement | |||
| Initial model used (PDB code) | 6G79 (for G protein heterotrimer) | ||
| Model resolution‡ (Å) | 3.6 | ||
| FSC threshold | 0.5 | ||
| Map sharpening B factor (Å2) | -112 | ||
| Model composition | |||
| Non-hydrogen atoms | 9420 | ||
| Protein residues | 1202 | ||
| Ligands | 8 | ||
| B factors (Å2) | |||
| Protein | 47.44 | ||
| Ligand | 25.73 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.005 | ||
| Bond angles (°) | 0.895 | ||
| Validation | |||
| Molprobity score | 1.47 | ||
| Clashscore | 4.98 | ||
| Poor rotamers (%) | 0.00 | ||
| EMRinger score | 1.89 | ||
| Ramachandran plot | |||
| Favored (%) | 96.69 | ||
| Allowed (%) | 3.31 | ||
| Disallowed (%) | 0.00 | ||
After 3D classification.
Local resolution range.
Resolution at which FSC between map and model is 0.5.
Supplementary Material
Acknowledgements
V.V. was funded by a Gates Cambridge Scholarship. The work in C.G.T.’s laboratory was funded by a grant from the European Research Council (EMPSI 339995), Sosei Heptares and core funding from the Medical Research Council [MRC U105197215]. The work in N.V.’s lab was funded by grants from the National Institutes of Health (R01-GM117923, R01-GM097261). The work in D.E.G.’s laboratory was funded by Independent Research Fund Denmark | Natural Sciences (8021-00173B), Lundbeck Foundation (R163-2013-16327) and Novo Nordisk Foundation (NNF17OC0031226). D.E.G. is a member of the Integrative Structural Biology at the University of Copenhagen (ISBUC). We thank T. Nakane, P. Emsley, A.G.W. Leslie, S. Scheres, E. Miller, E.R.S. Kunji, P. Edwards, T. Warne and V. Chandrasekaran for helpful discussions, and S. McLaughlin and C. Johnson from the LMB biophysics facility, G. Cannone and G. Sharov from the LMB EM facility and J. Grimmett and T. Darling from the LMB scientific computing for technical support during this work.
Footnotes
Author contributions
V.V. performed receptor and G protein engineering, expression, purification, complex formation, biophysical characterisation, preparation of cryo-EM grids, cryo-EM data collection, data processing, structure determination and model building. N.M. performed the molecular dynamics simulations, and N.M. and N.V. did the analysis of the MD simulation trajectories. G.P.S. developed the generic residue numbers and GPCRdb resources. A.J.K. analysed sequence conservation, structural mechanisms and co-supervised G.P.S. Y.L. advised on cryo-EM data collection, data processing and model building. D.E.G. supervised G.P.S. and A.J.K. and contributed to the design of residue numbering and GPCRdb resource. V.V. and C.G.T. carried out structure analysis and manuscript preparation. C.G.T. managed the overall project. The manuscript was written by V.V. and C.G.T., and included contributions from all the authors.
Author information. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare the following competing interests: CGT is a shareholder, consultant and member of the Scientific Advisory Board of Sosei Heptares, who also partly funded this work.
Data availability statement
The structure of the Ste2 dimer coupled to G proteins has been deposited in the Protein Data Bank (https://www.rcsb.org/) with accession number 7AD3 and the associated cryo-EM data has been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) with accession number EMDB-11720. Cryo-EM data eBIC Krios1, LMB Krios1 and LMB Krios2 are available from the authors. There are no restrictions on data availability. Ste2 orthologue sequences and the generic residue numbering are available at https://gpcrdb.org/protein/ste2_yeast and the structure-based sequence alignments are at https://gpcrdb.org/alignment/targetselection. Structures with PDB codes 6G79, 3SN6, 6B3J, 7C7Q, 6D32, 6CMO, 6OFJ are available at https://www.rcsb.org/.
References
- 1.Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- 2.Kolakowski LF., Jr GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- 3.Garcia-Nafria J, Tate CG. Cryo-EM structures of GPCRs coupled to Gs, Gi and Go. Mol Cell Endocrinol. 2019;488:1–13. doi: 10.1016/j.mce.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Brown NA, Schrevens S, van Dijck P, Goldman GH. Fungal G-protein-coupled receptors: mediators of pathogenesis and targets for disease control. Nat Microbiol. 2018;3:402–414. doi: 10.1038/s41564-018-0127-5. [DOI] [PubMed] [Google Scholar]
- 5.Van Dijck P, et al. In: The Fungal Kingdom. Heitman J, et al., editors. Wiley; 2017. pp. 417–439. [Google Scholar]
- 6.Burkholder AC, Hartwell LH. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985;13:8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue C, Hsueh YP, Heitman J. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev. 2008;32:1010–1032. doi: 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee BK, Khare S, Naider F, Becker JM. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to alpha-factor pheromone binding. J Biol chem. 2001;276:37950–37961. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- 9.Nehme R, et al. Mini-G proteins: Novel tools for studying GPCRs in their active conformation. PloS one. 2017;12 doi: 10.1371/journal.pone.0175642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isberg V, et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overton MC, Chinault SL, Blumer KJ. Oligomerization, biogenesis, and signaling is promoted by a glycophorin A-like dimerization motif in transmembrane domain 1 of a yeast G protein-coupled receptor. J Biol Chem. 2003;278:49369–49377. doi: 10.1074/jbc.M308654200. [DOI] [PubMed] [Google Scholar]
- 12.Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Current Biology. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 13.Yesilaltay A, Jenness DD. Homo-oligomeric complexes of the yeast alpha-factor pheromone receptor are functional units of endocytosis. Mol Biol Cell. 2000;11:2873–2884. doi: 10.1091/mbc.11.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehret AU, Bajaj A, Naider F, Dumont ME. Oligomerization of the yeast alpha-factor receptor: implications for dominant negative effects of mutant receptors. J Biol Chem. 2006;281:20698–20714. doi: 10.1074/jbc.M513642200. [DOI] [PubMed] [Google Scholar]
- 15.Stoneman MR, et al. Quaternary structure of the yeast pheromone receptor Ste2 in living cells. Biochim Biophys Acta Biomembr. 2017;1859:1456–1464. doi: 10.1016/j.bbamem.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Kim S, et al. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci U S A. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mentesana PE, Konopka JB. Mutational analysis of the role of N-glycosylation in alpha-factor receptor function. Biochemistry. 2001;40:9685–9694. doi: 10.1021/bi0108507. [DOI] [PubMed] [Google Scholar]
- 18.Wang HX, Konopka JB. Identification of amino acids at two dimer interface regions of the alpha-factor receptor (Ste2) Biochemistry. 2009;48:7132–7139. doi: 10.1021/bi900424h. [DOI] [PubMed] [Google Scholar]
- 19.Abel M, Zhang Y, Lu HF, Naider F, Becker J. Structure - function analysis of the Saccharomyces cerevisiae tridecapeptide pheromone using alanine - scanned analogs. J Peptide Res. 1998;52:95–106. doi: 10.1111/j.1399-3011.1998.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 20.Naider F, Becker JM. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G proteincoupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Shenbagamurthi P, Kundu B, Raths S, Becker JM, Naider F. Biological activity and conformational isomerism in position 9 analogs of the des-1-tryptophan, 3. beta-cyclohexylalanine-alpha-factor from Saccharomyces cerevisiae Biochemistry. 1985;24:7070–7076. doi: 10.1021/bi00346a008. [DOI] [PubMed] [Google Scholar]
- 22.Lee YH, Naider F, Becker JM. Interacting residues in an activated state of a G protein-coupled receptor. J Biol Chem. 2006;281:2263–2272. doi: 10.1074/jbc.M509987200. [DOI] [PubMed] [Google Scholar]
- 23.Lee BK, et al. Tyr266 in the sixth transmembrane domain of the yeast alpha-factor receptor plays key roles in receptor activation and ligand specificity. Biochemistry. 2002;41:13681–13689. doi: 10.1021/bi026100u. [DOI] [PubMed] [Google Scholar]
- 24.Blumer KJ, Thorner J. Beta and gamma subunits of a yeast guanine nucleotide-binding protein are not essential for membrane association of the alpha subunit but are required for receptor coupling. Proc Natl Acad Sci U S A. 1990;87:4363–4367. doi: 10.1073/pnas.87.11.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flock T, et al. Universal allosteric mechanism for Galpha activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixit G, Baker R, Sacks CM, Torres MP, Dohlman HG. Guanine nucleotide-binding protein (Ga) endocytosis by a cascade of ubiquitin binding domain proteins is required for sustained morphogenesis and proper mating in yeast. Journal of Biological Chemistry. 2014;289:15052–15063. doi: 10.1074/jbc.M114.566117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao C, et al. Cryo-EM structures of inactive and active GABA B receptor. Cell Res. 2020:1–10. doi: 10.1038/s41422-020-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SY, Lee LT, Chow BK. Receptor oligomerization: from early evidence to current understanding in class B GPCRs. Front Endocrinol (Lausanne) 2012;3:175. doi: 10.3389/fendo.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milasta S, et al. Interactions between the Mas-related receptors MrgD and MrgE alter signalling and trafficking of MrgD. Mol Pharmacol. 2006;69:479–491. doi: 10.1124/mol.105.018788. [DOI] [PubMed] [Google Scholar]
- 30.Zhao DY, et al. Cryo-EM structure of the native rhodopsin dimer in nanodiscs. J Biol Chem. 2019;294:14215–14230. doi: 10.1074/jbc.RA119.010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao C, et al. Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 2020;30:564–573. doi: 10.1038/s41422-020-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weis WI, Kobilka BK. The Molecular Basis of G Protein-Coupled Receptor Activation. Annu Rev Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dror RO, et al. Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warne T, Edwards PC, Dore AS, Leslie AGW, Tate CG. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019;364:775–778. doi: 10.1126/science.aau5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Extended Data References
- 36.García-Nafría J, Watson JF, Greger IH. IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci Rep. 2016;6:27459. doi: 10.1038/srep27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manahan CL, Patnana M, Blumer KJ, Linder ME. Dual lipid modification motifs in Gα and Gγ subunits are required for full activity of the pheromone response pathway in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:957–968. doi: 10.1091/mbc.11.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirschman JE, Jenness DD. Dual lipid modification of the yeast ggamma subunit Ste18p determines membrane localization of Gbetagamma. Mol Cell Biol. 1999;19:7705–7711. doi: 10.1128/mcb.19.11.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Mastronarde DN. Advanced data acquisition from electron microscopes with serialEM. Microscopy Microanalysis. 2018;24:864–865. [Google Scholar]
- 41.Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramlaul K, Palmer CM, Nakane T, C HSA. Mitigating Local Over-fitting During Single Particle Reconstruction with SIDESPLITTER. J Struct Biol. 2020:107545. doi: 10.1016/j.jsb.2020.107545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramlaul K, Palmer CM, Aylett CHS. A Local Agreement Filtering Algorithm for Transmission EM Reconstructions. J Struct Biol. 2019;205:30–40. doi: 10.1016/j.jsb.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods. 2017;14:793. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KM, et al. Multiple regulatory roles of the carboxy terminus of Ste2p a yeast GPCR. Pharmacol Res. 2012;65:31–40. doi: 10.1016/j.phrs.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burnley T, Palmer CM, Winn M. Recent developments in the CCP-EM software suite. Acta Crystallogr D Struct Biol. 2017;73:469–477. doi: 10.1107/S2059798317007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 51.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long F, et al. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr D Struct Biol. 2017;73:112–122. doi: 10.1107/S2059798317000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danev R, Baumeister W. Cryo-EM single particle analysis with the Volta phase plate. Elife. 2016;5:e13046. doi: 10.7554/eLife.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Hayashi Y, Matsui H, Takagi T. Method Enzymol. Vol. 172. Elsevier; 1989. pp. 514–528. [DOI] [PubMed] [Google Scholar]
- 59.Cooley RB, O’Donnell JP, Sondermann H. Coincidence detection and bidirectional transmembrane signaling control a bacterial second messenger receptor. Elife. 2016;5:e21848. doi: 10.7554/eLife.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grossfield A, Zuckerman DM. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu Rep Comput Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monticelli L, Sorin EJ, Tieleman DP, Pande VS, Colombo G. Molecular simulation of multistate peptide dynamics: a comparison between microsecond timescale sampling and multiple shorter trajectories. J Comput Chem. 2008;29:1740–1752. doi: 10.1002/jcc.20935. [DOI] [PubMed] [Google Scholar]
- 62.Berendsen HJ, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Computer Phys Com. 1995;91:43–56. [Google Scholar]
- 63.Lee J, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comp. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 65.Martyna GJ, Klein ML, Tuckerman M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J Chem Phys. 1992;97:2635–2643. [Google Scholar]
- 66.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 67.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 68.Bhattacharya S, et al. Critical analysis of the successes and failures of homology models of G protein-coupled receptors. Proteins. 2013;81:729–739. doi: 10.1002/prot.24195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paramo T, East A, Garzon D, Ulmschneider MB, Bond PJ. Efficient Characterization of Protein Cavities within Molecular Simulation Trajectories: trj_cavity. J Chem Theory Comput. 2014;10:2151–2164. doi: 10.1021/ct401098b. [DOI] [PubMed] [Google Scholar]
- 70.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Isberg V, et al. Generic GPCR residue numbers-aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucl Acid Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J Mol Biol. 2012;1:76. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure of the Ste2 dimer coupled to G proteins has been deposited in the Protein Data Bank (https://www.rcsb.org/) with accession number 7AD3 and the associated cryo-EM data has been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) with accession number EMDB-11720. Cryo-EM data eBIC Krios1, LMB Krios1 and LMB Krios2 are available from the authors. There are no restrictions on data availability. Ste2 orthologue sequences and the generic residue numbering are available at https://gpcrdb.org/protein/ste2_yeast and the structure-based sequence alignments are at https://gpcrdb.org/alignment/targetselection. Structures with PDB codes 6G79, 3SN6, 6B3J, 7C7Q, 6D32, 6CMO, 6OFJ are available at https://www.rcsb.org/.