Abstract
Organisms respond to mitochondrial stress by activating multiple defense pathways including the mitochondrial unfolded protein response (UPRmt). However, how UPRmt regulators are orchestrated to transcriptionally activate stress responses remains largely unknown. Here we identified CBP-1, the worm ortholog of the mammalian acetyltransferases CBP/p300, as an essential regulator of the UPRmt, as well as mitochondrial stress-induced immune response, reduction of amyloid-β aggregation and lifespan extension in Caenorhabditis elegans. Mechanistically, CBP-1 acts downstream of histone demethylases, JMJD-1.2/JMJD-3.1, and upstream of UPRmt transcription factors including ATFS-1, to systematically induce a broad spectrum of UPRmt genes and execute multiple beneficial functions. In mouse and human populations, transcript levels of CBP/p300 positively correlate with UPRmt transcripts and longevity. Furthermore, CBP/p300 inhibition disrupts, while forced expression of p300 is sufficient to activate, the UPRmt in mammalian cells. These results highlight an evolutionarily conserved mechanism that determines mitochondrial stress response, and promotes health and longevity through CBP/p300.
Mitochondria not only contribute to the harvesting of energy, but also serve as key signaling hubs connecting numerous metabolic processes to essential cellular and organismal functions1–3. It is therefore not surprising that the dysfunction of mitochondria is tightly associated with ageing, as well as diverse human age-related diseases, including those affecting metabolic, cardiovascular and neuromuscular systems, as well as cancer2,4–7. Moreover, mitochondria function as platforms to regulate programmed cell death and innate immune responses1,8,9. Multiple mitochondrial stress response (MSR) pathways have evolved to adapt mitochondrial function to the ever-changing cellular milieu and to a variety of extra-cellular cues10,11. However, aberrant activation of these MSR pathways may also be maladaptive and contribute to disease and ageing2, underscoring the importance of the tight control of these regulatory circuits.
The mitochondrial unfolded protein response (UPRmt), one of these MSR pathways, is triggered by mitochondrial-to-nuclear communication, leading to an adaptive transcriptional response that promotes repair and recovery of the cell or organism from transient mitochondrial dysfunction10–13. It recently emerged that the activation of the UPRmt provides resistance to pathogen infections (e.g. Pseudomonas aeruginosa) in C. elegans, and animals that failed to activate UPRmt during P. aeruginosa infection died earlier, suggesting that the UPRmt is a bona fide component of the innate immune response14,15. In mammalian cells, mitochondrial perturbations also lead to cellular stress responses closely associated with innate immunity9,16; e.g., herpesvirus infections induce a mitochondrial DNA (mtDNA) stress response, which enhances antiviral signaling and type I interferon responses and thereby confer viral resistance17.
The regulation of the UPRmt is complex and pleiotropic, and includes control at the level of transcription and chromatin organization. At the transcriptional level, the transcription factor ATFS-1 in C. elegans 18, and its functional orthologues, ATF4, ATF5 and CHOP in mammals19–21, seem to be key regulators of the UPRmt. Two recent studies suggest that an OMA1–DELE1–HRI signaling pathway relays the mitochondrial stress from mitochondria to the cytosol in mammals22,23. On the epigenetic level, the MET-2/LIN-65 histone methyltransferase complex and two histone demethylases JMJD-1.2/PHF8 and JMJD-3.1/KDM6B, regulate the UPRmt and mitochondrial stress-induced longevity in both C. elegans and mammals24,25, whereas in yeast the histone demethylase, Rph1p, is the key modulator26. However, how these different layers of UPRmt regulators are systematically coordinated to induce the expression of various UPRmt genes and execute different biological functions is still poorly understood.
Here, we demonstrate that CBP-1 acts as an essential link to translate the mitochondrial stress signal from the demethylases, JMJD-1.2/JMJD-3.1, to the UPRmt transcription factors (e.g. ATFS-1), into the coordinated transcriptional induction of a wide panel of UPRmt genes in C. elegans. Importantly, the beneficial effects induced by mitochondrial perturbations, such as resistance to pathogen infection, improved proteostasis against amyloid-β aggregation, and lifespan extension are almost completely blocked by cbp-1 silencing. Moreover, systematic correlation analysis in mouse and human populations, as well as genetic and pharmacological loss-of-function studies in mammalian cells, strongly suggest that the function of CBP/p300 in the regulation of the UPRmt, health and lifespan are also conserved in mammals. Collectively, these results highlighted that CBP/p300 is an evolutionarily conserved node for mitochondrial stress signaling that defends mitochondrial function, and promotes health and longevity.
Results
CBP-1 controls UPRmt activation in C. elegans
We used a UPRmt activation model by knocking down cco-1 (cytochrome c oxidase-1) in the UPRmt reporter hsp-6p::gfp strain27,28, and performed an RNA interference (RNAi) screen by feeding RNAi targeting all putative lysine acetyltransferases (KATs) in C. elegans (Extended Data Fig. 1a)29–32. Only RNAi of cbp-1 (R10E11.1)33, the ortholog of human CBP/p300 34–37, attenuated UPRmt activation to a similar extent as the silencing of the key UPRmt transcription factor atfs-1 18 (Fig. 1a and Extended Data Fig. 1b). The effect of cbp-1 RNAi on UPRmt activation induced by RNAi-mediated loss-of-function (LOF) of cco-1 and mrps-5 (mitochondrial ribosomal protein S5)13 was furthermore dose-dependent (Fig. 1b and Extended Data Fig. 1c, d). Moreover, another RNAi clone (cbp-1_RNAi_2), targeting a different region of the cbp-1 mRNA, compared with the one used in the RNAi screening (cbp-1_RNAi_1), also impaired UPRmt activation (Extended Data Fig. 1e, f).
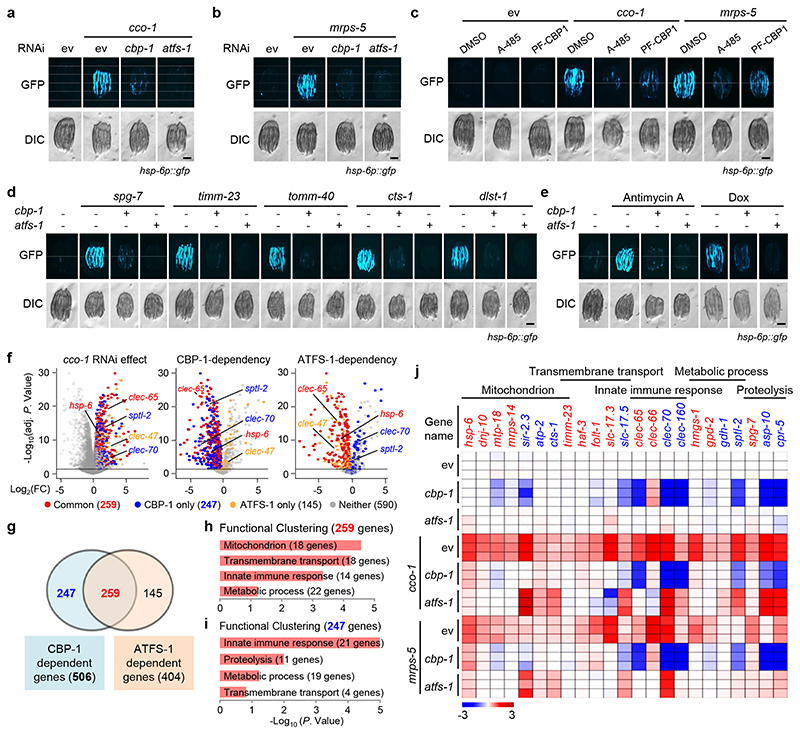
Fig. 1. CBP-1 controls the activation of UPRmt in C. elegans.
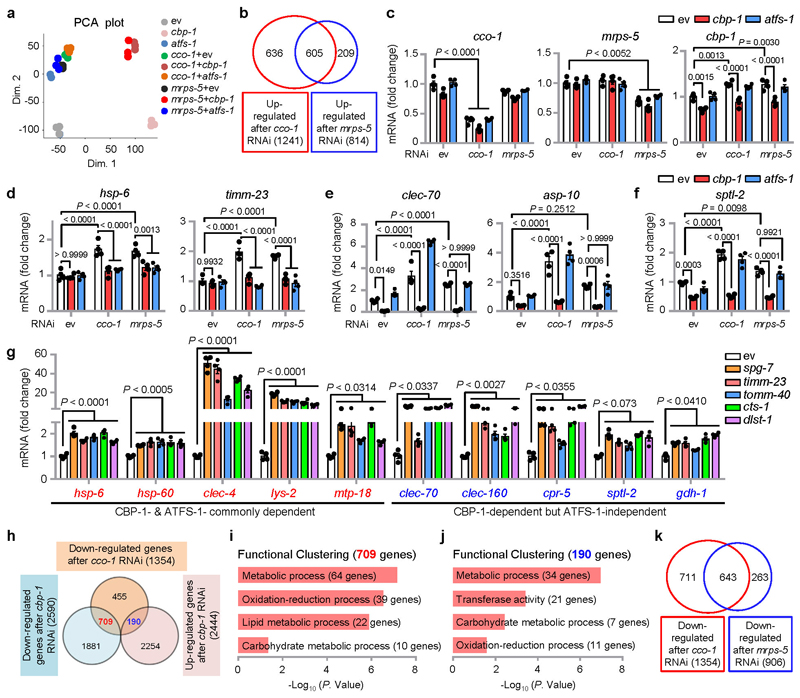
a, b, cbp-1 RNAi attenuates UPRmt activation induced by cco-1 (a) or mrps-5 (b) knockdown in hsp-6p::gfp worms. For RNAi treatment, RNAi targeting cco-1 or mrps-5 occupies 40%, cbp-1 RNAi occupies 25%, atfs-1 RNAi occupies 60%, control RNAi (ev) was used to supply to a final 100% of RNAi for all conditions. DIC, differential interference contrast image. c, CBP/p300 small-molecule inhibitors A-485 and PF-CBP1 attenuate UPRmt activation in worms. hsp-6p::gfp worms were fed with control or the UPRmt-inducing cco-1 (40%) RNAi, in combination with A-485 (10 μM) or PF-CBP1 (80 μM) treatment. d, e, cbp-1 RNAi abolishes UPRmt activation induced by spg-7, timm-23, tomm-40, cts-1 or dlst-1 RNAi (d), and by antimycin A (2.5 μM) or doxycycline (Dox) (30 μg/ml) (e), in hsp-6p::gfp worms. The atfs-1 RNAi was used as a positive control. RNAi targeting tomm-40, cts-1 or dlst-1 occupies 40%, cbp-1 RNAi occupies 25%, atfs-1 RNAi occupies 60%. f, Volcano plots showing the effects on gene expression of cco-1 RNAi compared to control RNAi (ev) (left), and of cbp-1 (middle) or atfs-1 (right) knockdown in a cco-1 RNAi background. FC, fold change. Genes dependent on both CBP-1 and ATFS-1 for induction after cco-1 RNAi are highlighted in red (common). Genes dependent on CBP-1, but not ATFS-1, for induction are in blue (CBP-1 only). Genes dependent on ATFS-1, but not CBP-1, for induction are highlighted in yellow (ATFS-1 only). g, Venn diagram of CBP-1-dependent UPRmt genes, that are in common with ATFS-1-dependent UPRmt genes in response to cco-1 RNAi, according to the RNA-seq data. h, i, Functional clustering of the 259 (h) and 247 (i) UPRmt genes as indicated in (g). j. Heat-map of the representative UPRmt genes dependent on CBP-1 for induction during cco-1 knockdown. The color-coded heat-map represents gene expression differences in log2FC relatively to control RNAi (ev) condition. Genes dependent on both CBP-1 and ATFS-1 are highlighted in red. Genes dependent on CBP-1, but not ATFS-1, are highlighted in blue. Scale bars, 0.3 mm.
As an alternative approach to inhibit CBP-1 activity, we used two mechanistically different small-molecule inhibitors, a highly specific CBP/p300 catalytic inhibitor, A-48538; and a CBP/p300 bromodomain inhibitor, PF-CBP-1 (Extended Data Fig. 1e)39. Both inhibitors suppressed UPRmt activation induced by cco-1 or mrps-5 RNAi, with A-485 having effects at a lower concentration (10 μM) compared to PF-CBP-1 (80 μM) (Fig. 1c and Extended Data Fig. 1g). Likewise, genetic or pharmacological inductions of the UPRmt by LOF of spg-7, timm-23, tomm-40, cts-1 and dlst-1, or by administering antimycin A and doxycycline (Dox)13,14,40, were abolished by cbp-1 RNAi (Fig. 1d, e). Of note, UPRmt activation is not affected by RNAi that specifically targets the two probable pseudogenes of cbp-1, cbp-2 and cbp-3 (Extended Data Fig. 1h, i), both of which lack most of the functional domains compared to CBP-1, including the histone acetyltransferase domain (Extended Data Fig. 1j)33. cbp-1 RNAi also attenuates the activation of the endoplasmic reticulum UPR (UPRER), but not the cytosolic UPR (UPRCYT)/heat shock response in C. elegans, suggesting some activity in cross-modal stress response pathways (Extended Data Fig. 1k–n).
To determine the footprints of CBP-1 on the regulation of the UPRmt, we performed RNA sequencing (RNA-seq) on total RNA isolated from hsp-6p::gfp worms fed with cco-1 or mrps-5 RNAi, in combination with cbp-1 or atfs-1 RNAi (Extended Data Fig. 2a and Supplementary Table 1). The majority of transcripts induced by mrps-5 RNAi were also induced by cco-1 RNAi, but not the other way around (Extended Data Fig. 2b), which might due to the superior knockdown efficiency of cco-1 than that of mrps-5 (Extended Data Fig. 2c). We thus focused on the genes affected by cco-1 RNAi. 1,241 transcripts were significantly up-regulated after cco-1 RNAi (log2FC > 0.5, adjusted P < 0.05; defined here as UPRmt genes), among which 506 (40.8%) were CBP-1-dependent and 404 (32.6%) required ATFS-1 (Fig. 1f, g). The number of ATFS-1-dependent transcripts was similar to that found in a previous study18. Up to 259 genes induced by cco-1 RNAi were dependent on both CBP-1 and ATFS-1 (Fig. 1g). Gene ontology (GO) analysis revealed that a large number of “mitochondrion”, “transmembrane transport” and “metabolic process”-related genes including hsp-6, timm-23 and gpd-2, required both CBP-1 and ATFS-1 for induction (Fig. 1h, j and Extended Data Fig. 2d). In addition, many innate immune genes, such as the C-type lectin clec-65, were also included in this gene set (Fig. 1f, h, j), supporting a role of UPRmt in regulating the immune response14,15.
Among the 506 UPRmt transcripts regulated by CBP-1, 247 (48.8%) were only dependent on CBP-1, but not ATFS-1 (Fig. 1g), and were enriched for “innate immune response” (e.g. clec-70), “proteolysis” (e.g. asp-10), and “metabolic processes” (e.g. gdh-1) (Fig. 1i, j and Extended Data Fig. 2e). Consistent with the role of ceramide biosynthesis in mitochondrial surveillance14,41, sptl-2, which encodes a serine palmitoyltransferase, was robustly induced after cco-1 or mrps-5 knockdown in a CBP-1-, but not ATFS-1-, dependent fashion (Extended Data Fig. 2f). Moreover, other UPRmt inducers, including LOF of spg-7, timm-23, tomm-40, cts-1 and dlst-1, not only led to the induction of CBP-1-and ATFS-1-commonly dependent UPRmt transcripts (e.g. hsp-6), but also up-regulated UPRmt transcripts that were only dependent on CBP-1 but not ATFS-1 (e.g. clec-70) (Extended Data Fig. 2g).
In response to cco-1 RNAi, 1,354 transcripts were significantly down-regulated (log2FC < - 0.5, adjusted P < 0.05) (Extended Data Fig. 2h and Supplementary Table 2); among these transcripts, 709 (52.4%) were also down-regulated, and 190 (14.0%) were conversely up-regulated after cbp-1 RNAi (Extended Data Fig. 2h). Interestingly, both gene clusters were enriched for “metabolic process”, “oxidation-reduction process” and “carbohydrate metabolic process” (Extended Data Fig. 2i, j), indicating a global rewiring of metabolism during mitochondrial stress19,21,42,43, and the vital role of cbp-1 in this reprogramming. Finally, most transcripts down-regulated after mrps-5 RNAi were also down-regulated during cco-1 silencing (Extended Data Fig. 2k). Taken together, these data suggest that CBP-1 controls the induction of a broad spectrum of UPRmt genes upon various mitochondrial stresses in C. elegans.
Mitochondrial stress increases CBP-1-mediated histone acetylation at the loci of UPRmt genes
We next sought to explore the molecular mechanism on how CBP-1 regulates UPRmt activation. CBP/p300, the mammalian homologues of CBP-1, are acetyltransferases involved in histone acetylation36,37. In worms fed with cco-1 and mrps-5 RNAi, the global histone 3 acetylation at K18 (H3K18Ac) was increased by 60% and 40%, and H3K27Ac by 90% and 20%, respectively, compared with worms fed with control RNAi (Fig. 2a). This increase was remarkably attenuated by cbp-1 RNAi. Similar results were also found for H4K5Ac (Extended Data Fig. 3a), while H3K4Ac and H3K9Ac levels were not affected by cbp-1 knockdown. RNAi for atfs-1 did not alter the levels of any of the histone modifications examined (Fig. 2a and Extended Data Fig. 3a). Moreover, the CBP/p300 catalytic inhibitor A-485 also impaired H3K18Ac and H3K27Ac, but not H3K9Ac, under cco-1 knockdown condition (Fig. 2b). Additionally, other UPRmt inducers, including LOF of spg-7, timm-23, tomm-40, cts-1 and dlst-1, increased H3K18Ac by 90% to 170%, and H3K27Ac by 30% to 150% (Fig. 2c).
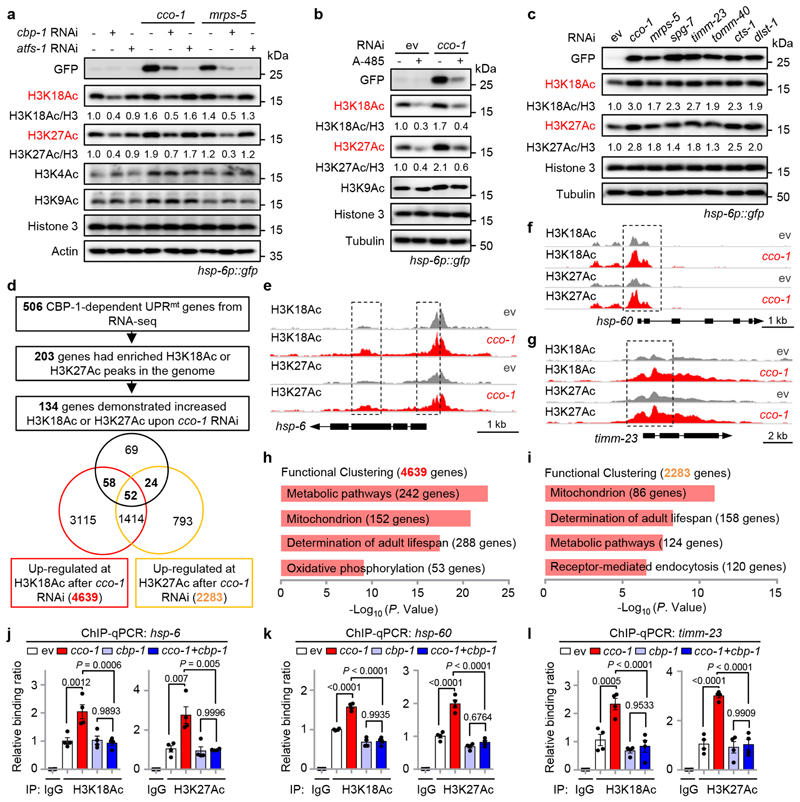
Fig. 2. Mitochondrial stress increases CBP-1-dependent histone acetylation at the loci of a large set of UPRmt genes.
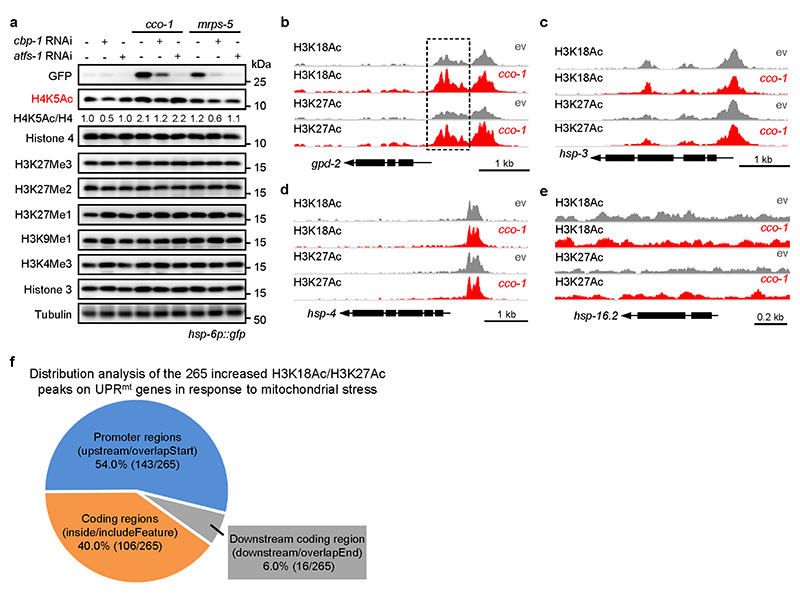
a, H3K18 and H3K27 acetylation increases in a CBP-1-dependent manner during UPRmt activation induced by cco-1 or mrps-5 RNAi. Western blots of hsp-6p::gfp worms fed with control, cbp-1, atfs-1, cco-1 or mrps-5 RNAi. RNAi targeting cbp-1 occupies 25%, atfs-1, cco-1 or mrps-5 occupies 50%, control RNAi was used to supply to a final 100% of RNAi for all conditions. b, The increase of H3K18Ac and H3K27Ac upon cco-1 RNAi is strongly blocked by chemical inhibition of CBP/p300 with A-485. Western blots of hsp-6p::gfp worms fed with control or cco-1 (50%) RNAi, in combination with DMSO or A-485 (10 μM) treatment. c, Western blots of hsp-6p::gfp worms showing increased H3K18Ac and H3K27Ac during UPRmt activation induced by RNAi targeting different mitochondrial genes as indicated. d, Summary of the H3K27Ac and H3K18Ac ChIP-seq results in worms fed with control or cco-1 RNAi. e-g, Treatment of cco-1 RNAi increases H3K18Ac and H3K27Ac at the genomic loci of representative UPRmt genes including hsp-6, hsp-60 and timm-23. Genome tracks showing the ChIP-seq analysis for H3K27Ac and H3K18Ac over the genomic loci of hsp-6 (e), hsp-60 (f) and timm-23 (g) in worms fed with control (ev) or cco-1 RNAi. The two tracks were shown with the same total count range between basal and mitochondrial stress condition for each gene. h, i, Functional clustering of the 4639 (h) and 2283 (i) genes as indicated in (d). j-l, cco-1 RNAi-induced increase of H3K18Ac and H3K27Ac at the loci of representative UPRmt genes is completely blocked by cbp-1 RNAi. ChIP-qPCR analysis of hsp-6 (j), hsp-60 (k) and timm-23 (l) in worms fed with RNAi targeting cco-1 and/or cbp-1 (n = 4 biologically independent samples). RNAi targeting cco-1 occupies 50%, cbp-1 occupies 25%. ChIP was performed by using antibodies to H3K18Ac or H3K27Ac. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test. For uncropped gel source data, see Source Data Fig. 2.
It has been known that acetylation of H3K18 and H3K27, which transforms the condensed chromatin is into a more relaxed structure, is generally linked to active transcription29,30,32,44. To examine if CBP-1-mediated histone acetylation contributes to the transcriptional activation of UPRmt genes, we performed chromatin immunoprecipitation sequencing (ChIP-seq) with antibodies against H3K18Ac and H3K27Ac in worms fed with control or cco-1 RNAi. Among the 506 UPRmt genes regulated by CBP-1 (Fig. 1g), 203 had enriched H3K18Ac or H3K27Ac peaks in the genome (Fig. 2d). Importantly, 66.0% (134/203) of these UPRmt genes (e.g. hsp-6, timm-23, hsp-60 and gpd-2) demonstrated significantly increased levels of H3K18Ac or H3K27Ac (110 genes for H3K18Ac, P < 1.9 ×10−10, Fisher’s exact test; and 76 genes for H3K27Ac, P < 1.5 ×10−12, Fisher’s exact test) after cco- 1 RNAi (FDR < 0.05) (Fig. 2d–g, Extended Data Fig. 3b and Supplementary Table 3). In contrast, no differences in H3K18Ac or H3K27Ac marks were observed for the UPRER markers hsp-3 and hsp-4, or the UPRCYT/heat shock response marker hsp-16.2, upon cco-1 RNAi treatment (Extended Data Fig. 3c–e). By analyzing the distribution of the 265 increased H3K18Ac/H3K27Ac peaks on the 134 UPRmt genes, we found that 54.0% (143/265) of them were located in promoter regions, 40.0% (106/265) were in coding regions, and 6.0% (16/265) were downstream of the coding region (Extended Data Fig. 3f and Supplementary Table 3). Indeed, for some genes (e.g. hsp-60), acetylation peaks are restricted to the promoter (Fig. 2f); whereas for other genes (e.g. hsp-6 and timm-23), acetylation marks exist in both promoter and coding regions (Fig. 2e, g). It is also noteworthy that both gene sets up-regulated for the acetylation marks in response to cco-1 RNAi (4,639 genes for H3K18Ac, and 2,283 genes for H3K27Ac) were highly enriched for GO terms including “metabolic pathways”, “mitochondrion” and “determination of adult lifespan” (Fig. 2h, i and Supplementary Table 3), supporting a critical role of these epigenetic adaptations in the control of mitochondrial metabolism and lifespan upon mitochondrial stress. Using ChIP-qPCR, we detected that the increased enrichment of H3K18Ac and H3K27Ac at the loci of UPRmt genes (e.g. hsp-6, hsp-60 and timm-23) in response to cco-1 knockdown was completely blocked by cbp-1 RNAi (Fig. 2j–l). These results indicate that increased CBP-1-dependent histone acetylation upon mitochondrial stress is closely associated with the transcriptional activation of a large set of UPRmt genes.
CBP-1 acts downstream of JMJD-3.1/1.2 and upstream of ATFS-1
Two histone demethylases, JMJD-3.1 and JMJD-1.2, have been reported to remove the repressive H3K27me3 mark from the promoter/coding regions of UPRmt genes, poising them for transcription, and overexpressing (OE) of jmjd-3.1 in worms is sufficient to activate the UPRmt 25. RNAi for cbp-1 abolished the activation of the UPRmt in two independently generated jmjd-3.1 OE strains (Fig. 3a). Moreover, increased levels of H3K18Ac and H3K27Ac, but not H3K9Ac, were detected in jmjd-3.1 OE worms, which was attenuated by cbp-1 RNAi (Fig. 3b). In addition, 177 (35.0%) of the 506 CBP-1-dependent UPRmt transcripts, were also induced in jmjd-3.1 OE worms (GSE78990) (Fig. 3c, d and Supplementary Table 4); and 129 (72.9%) of these 177 UPRmt transcripts were up-regulated as well upon jmjd-1.2 OE (Supplementary Table 4), underscoring a positive role of CBP-1 in the regulation of the MSR. Notably, cbp-1 stood out as the most up-regulated transcript among all the 13 putative KATs in jmjd-3.1 OE worms (Fig. 3e).
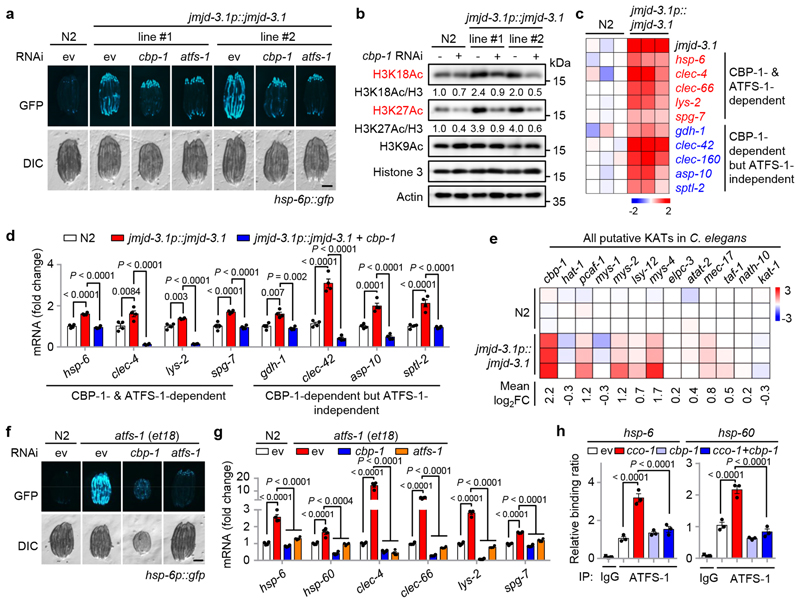
Fig. 3. CBP-1 acts downstream of JMJD-3.1/1.2, and upstream of ATFS-1, to support the expression of UPRmt genes.
a, cbp-1 RNAi attenuates the UPRmt activation induced by jmjd-3.1 overexpression. Fluorescent micrographs of wild-type (N2) or two independent hsp- 6p::gfp worm lines carrying the integrated jmjd-3.1p::jmjd-3.1 transgene fed with control, cbp-1 (25%) or atfs-1 (100%) RNAi. b, jmjd-3.1 overexpression increases H3K27Ac and H3K18Ac in a CBP-1-dependent manner. Western blots of wild-type or jmjd-3.1 transgenic worms fed with control or cbp-1 (25%) RNAi. c, Heat-map showing strong upregulation of a representative set of CBP-1-dependent UPRmt transcripts (log2FC values) in jmjd-3.1 transgenic worms compared to wild-type (N2) worms25. d, jmjd-3.1 overexpression increases the expression of a representative set of CBP-1-dependent UPRmt genes. qRT-PCR-results of wild-type or jmjd-3.1 transgenic worms fed with control or cbp-1 (25%) RNAi (n = 4 biologically independent samples). e, Heat-map (log2FC values) showing the relative transcript levels of all putative KATs in wild-type and jmjd-3.1p::jmjd-3.1 transgenic worms25. f, cbp-1 RNAi attenuates UPRmt activation in atfs-1(et18) mutant. Fluorescent micrographs of wild-type or hsp-6p::gfp worms carrying the atfs-1(et18) mutant fed with control, cbp-1 (25%) or atfs-1 (100%) RNAi. g, qRT-PCR-results of worms as indicated in (F) (n = 4 biologically independent samples). h, cbp-1 RNAi blocks the binding of ATFS-1 to the loci of UPRmt genes. ChIP-qPCR analysis of hsp-6 and hsp-60 in atfs-1p::atfs-1::flag worms fed with control, cco-1 (50%) and/or cbp-1 (25%) RNAi (n = 3 biologically independent samples). ChIP was performed by using anti-Flag M2 beads. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test. Scale bars, 0.3 mm. For uncropped gel source data, see Source Data Fig. 3.
To further explore how CBP-1 affects the transcriptional activation of UPRmt genes, we took advantage of the atfs-1(et18) mutant45, which carries a mutation in the mitochondrial targeting sequence of the transcription factor ATFS-1, leading to its nuclear accumulation and the constitutive activation of the UPRmt. Silencing of cbp-1 blocked UPRmt activation in the atfs-1(et18) mutant (Fig. 3f, g). Moreover, ATFS-1 failed to bind to the promoters of UPRmt genes (e.g. hsp-6, hsp-60) in cbp-1 RNAi-fed atfs-1p::atfs-1::flag worms even during mitochondrial stress induced by cco-1 RNAi (Fig. 3h). Collectively, these data indicate that jmjd-3.1 OE-mediated UPRmt activation requires CBP-1, and CBP-1-dependent histone acetylation acts downstream of JMJD-3.1/1.2, and meanwhile upstream of ATFS-1, leading to the transcriptional induction of UPRmt genes.
Beneficial effects of UPRmt requires CBP-1
We then explored the physiological functions of CBP-1 on MSR regulation. In line with the fact that mild mitochondrial stress protects against infection by pathogens, such as P. aeruginosa 14,15, cco-1 or mrps-5 knockdown increased the survival rate of worms exposed to P. aeruginosa, an effect that was completely abolished by cbp-1 knockdown (Fig. 4a, b). To further examine the vital role of CBP-1 in mitochondrial surveillance, we raised wild-type (N2) and the mitochondrial respiration mutants that have disruptions in one of the mitochondrial electron transport chain (ETC) components, isp-1(qm150) and clk-1(qm30) 46,47, on control or cbp-1 RNAi. Compared to C. elegans fed with control RNAi, cbp-1 RNAi even at 10% led to severe synthetic growth defects of the isp-1(qm150) and clk-1(qm30) mutants, whereas the development of wild-type worms was only slightly delayed (Fig. 4c). Similar effects were also observed in A-485 treated worms (Fig. 4c), indicating that mitochondrial mutants strongly rely on CBP-1 activity to maintain growth.
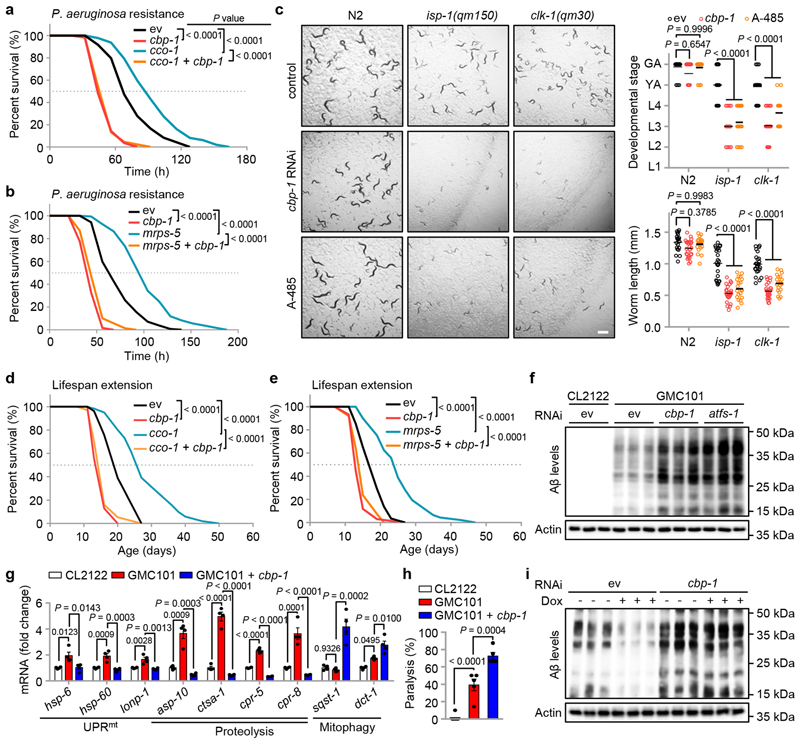
Fig. 4. CBP-1 is essential for mitochondrial surveillance, MSR-associated immune response, lifespan extension and amyloid-β proteotoxicity reduction.
a, b, The impact of cbp-1 knockdown on mitochondrial stress-induced P. aeruginosa resistance. Survival of worms fed with control (ev) or cbp-1 (20%) RNAi, in combination with cco-1 (50%) (a) or mrps-5 (80%) (b) RNAi, and exposed to P. aeruginosa. c, Representative photos of wild-type (N2), isp-1(qm150) or clk-1(qm30) worms fed with control, cbp-1 (10%) RNAi or treated with A-485 (10 μM) since maternal L4 stage. The developmental stage and body length of the F1 progeny were quantified at Day 4 after hatching (n = 25 worms for each condition). Scale bar, 1 mm. GA, gravid adult; YA, young adult; L1-4, larval stage 1-4. d, e, The impact of cbp-1 knockdown on mitochondrial stress-induced lifespan extension. cbp-1 RNAi attenuates lifespan extension induced by cco-1 (50%) (D) or mrps-5 (50%) (E) RNAi. RNAi targeting cbp-1 occupies 20% (d) or 10% (e). f, g, estern blots of Aβ aggregation and Actin levels (f), and transcript levels of selected genes measured by qRT-PCR (n = 4 biologically independent samples) (g) of CL2122 or GMC101 worms fed with control, cbp-1 (10%) or atfs-1 RNAi. h, Percentage of paralysis of CL2122 or GMC101 worms fed with control or cbp-1 (10%) RNAi at Day 5 of adulthood (n = 5 independent experiments). i, The impact of cbp-1 knockdown on mitochondrial stress-induced Amyloid-β proteotoxicity reduction. estern blots of Aβ aggregation and Actin in GMC101 worms fed with control or cbp-1 (10%) RNAi with or without Dox (15 μg/ml) treatment. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test in (c, g, h), or log-rank test in (a, b, d, e). For uncropped gel source data, see Source Data Fig. 4.
We then questioned whether cbp-1 is required for mitochondrial stress-induced lifespan extension in C. elegans 13,48. RNAi for cbp-1 at 20%, which was enough to suppress the UPRmt activation induced by cco-1 knockdown (Extended Data Fig. 1c), completely blocked the lifespan extension induced by cco-1 RNAi (Fig. 4d). Likewise, cbp-1 RNAi at 10% fully abolished mrps-5 knockdown-induced lifespan extension (Fig. 4e), in line with its capacity to block mrps-5 RNAi-induced UPRmt activation (Extended Data Fig. 1d). Meanwhile, consistent with the finding in another study49, cbp-1 knockdown alone shortened the lifespan of C. elegans (Fig. 4d, e), potentially due to the attenuated basal expression of diverse nuclear-encoded MSR transcripts (Fig. 1j).
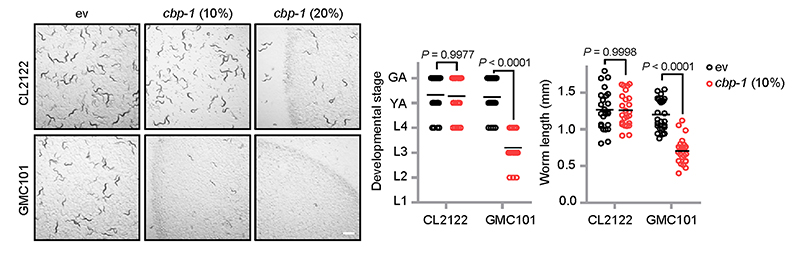
We have previously shown that humans with Alzheimer’s disease (AD), as well as mouse and C. elegans models of AD, are all typified by the induction of a cross-species conserved MSR transcript signature50. Strikingly, further activation of these MSR pathways reduced amyloid-β (Aβ) proteotoxicity in cells, worms and in transgenic mouse models of AD50. The GMC101 strain is a worm AD model that expresses the human Aβ 1–42 peptide in body wall muscle cells51. Adults of GMC101 develop age-progressive paralysis and amyloid deposition after a temperature shift from 20 to 25 °C. In these worms, cbp-1 RNAi at 10% caused a severe developmental delay even in the absence of the disease-inducing temperature shift, phenocopying mitochondrial respiration mutants that rely on cbp-1 for adaption, whereas the control CL2122 strain was not affected (Extended Data Fig. 4). Similar to atfs-1 RNAi, cbp-1 RNAi exacerbated Aβ aggregation in the GMC101 strain (Fig. 4f). In addition, cbp-1 knockdown in GMC101 worms prominently repressed not only the classical UPRmt transcripts (e.g. hsp-6), but also many UPRmt genes involved in “proteolysis” that only depend on CBP-1, but not ATFS-1 (e.g. asp-10) (Figs. 1i, j and 4g). Interestingly, the transcripts of another branch of the MSR, i.e. autophagy/mitophagy (e.g. sqst-1, dct-1), were conversely increased during cbp-1 RNAi, suggesting a specific role of CBP-1 in regulating the UPRmt branch of the MSR. Finally, cbp-1 RNAi worsened the paralysis and completely blocked the beneficial effect of Dox, an antibiotic that inhibits mitochondrial translation and activates the MSR13, on the reduction of Aβ aggregates in GMC101 worms50 (Fig. 4h, i).
Together, these results indicate that CBP-1 is essential for mitochondrial stress-induced immune response, lifespan extension and amyloid-β aggregation reduction in C. elegans.
CBP/p300 expression correlates with UPRmt transcripts and lifespan
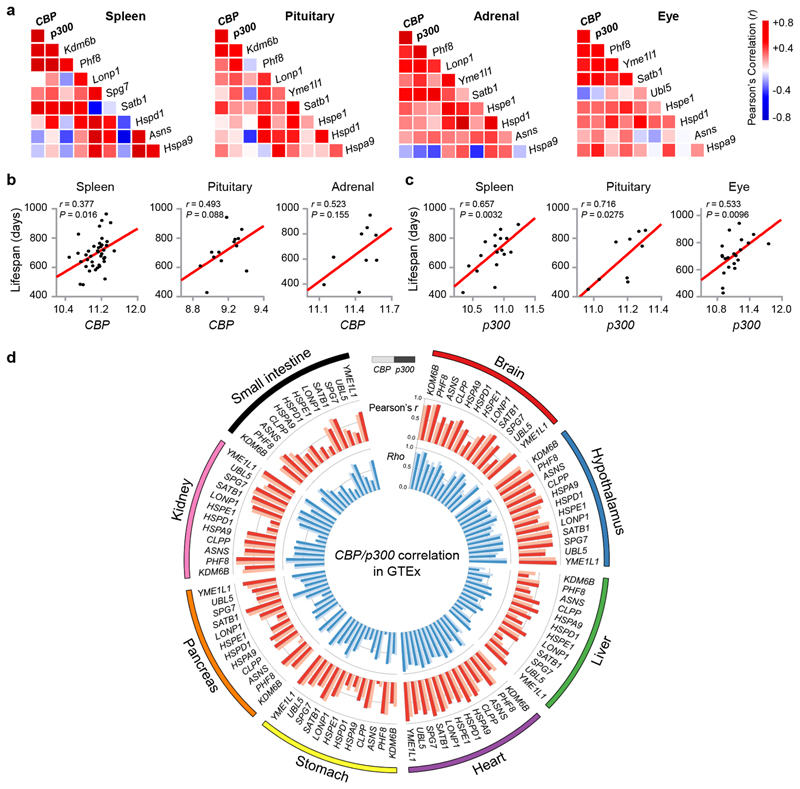
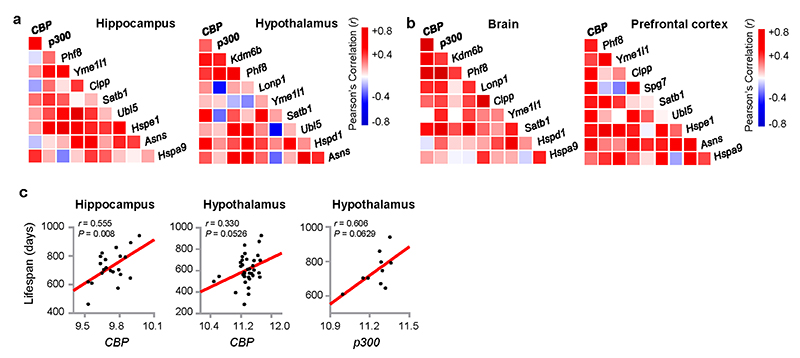
Next, we examined if the role of CBP-1 in UPRmt activation and MSR-associated beneficial effects is conserved in mammals. CBP expression in spleen, pituitary, adrenal and eye positively correlated with p300 expression in the BXD mouse genetic reference population (GRP)43,52 (www.genenetwork.org, Fig. 5a), confirming a complementary function of the two acetyltransferases35–37. Their expression levels also correlate with transcript levels of Kdm6b and Phf8, the murine homologs of jmjd-3.1 and jmjd-1.2 (Fig. 5a). Moreover, in these tissues, CBP/p300 expression overall positively correlated with transcripts of UPRmt-related genes10–12, including the mitochondrial proteases (Lonp1, Yme1l1 and Spg7), the DNA-binding proteins (Satb1 and Ubl5), the mitochondrial chaperones (Hspe1, Hspd1 and Hspa9), and asparagine synthetase Asns (Fig. 5a). Similar correlation networks were also found in the hippocampus and hypothalamus of BXD mice (Extended Data Fig. 5a), and in the brain and prefrontal cortex of mice from a different GRP, the LXS cohort53 (Extended Data Fig. 5b). In accordance with the indispensable role of CBP-1 in MSR-associated health and lifespan regulation in C. elegans (Fig. 4), we observed positive correlations between lifespan and CBP/p300 expression in spleen, pituitary, adrenal, eye, hippocampus and hypothalamus of the BXD strains (Fig. 5b, c and Extended Data Fig. 5c).
Fig. 5. Expression of CBP/p300 positively correlates with UPRmt transcripts and longevity in mouse and human populations.
a, Pearson’s correlation co-expression heat-map for CBP, p300, Kdm6b, Phf8 and UPRmt genes in spleen, pituitary, adrenal and eye of the BXD mouse genetic reference population43, 52. Positive and negative correlations are indicated in red and blue, respectively. The intensity of the colors corresponds to correlation coefficients. b, Positive correlations between lifespan and CBP transcript levels in spleen, pituitary and adrenal of BXD mice (Pearson’s r, two-sided). c, Positive correlations between lifespan and p300 transcript levels in spleen, pituitary and eye of BXD mice (Pearson’s r, two-sided). d, Circos plot of the expression correlations between CBP/p300 transcripts and the UPRmt gene transcripts in the brain (cerebellar hemisphere), hypothalamus, liver, heart (left ventricle), stomach, pancreas, kidney and small intestine of human samples derived from the Genotype-Tissue Expression (GTEx) database (v8)54. Red bar ring: the Pearson’s correlation coefficients (r) between UPRmt gene transcripts and CBP (light red) or p300 (dark red). Blue bar ring: the Spearman Rank correlation coefficients (Rho) between UPRmt gene transcripts and CBP (light blue) or p300 (dark blue). All correlations were found to be positive.
Finally, in the human Genotype-Tissue Expression (GTEx) database54, mRNA levels of CBP and p300 positively correlated with KDM6B, PHF8 and UPRmt transcripts in many tissues including brain, hypothalamus, liver, heart, stomach, pancreas, kidney and small intestine, forming a systematic network (Fig. 5d). These observations suggest that CBP/p300 likely play an evolutionarily conserved role in MSR regulation across species from worms to human.
A conserved role of CBP/p300 in MSR
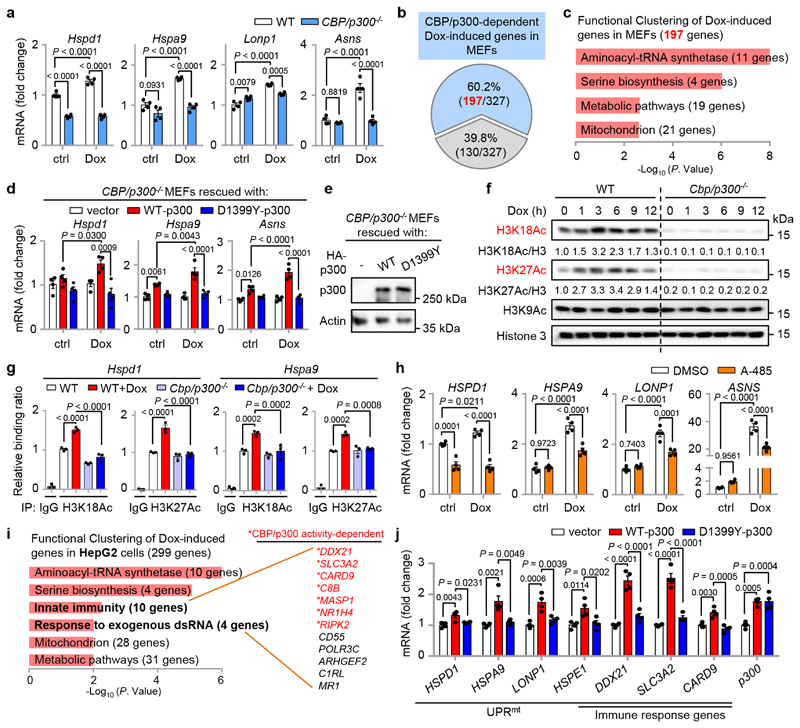
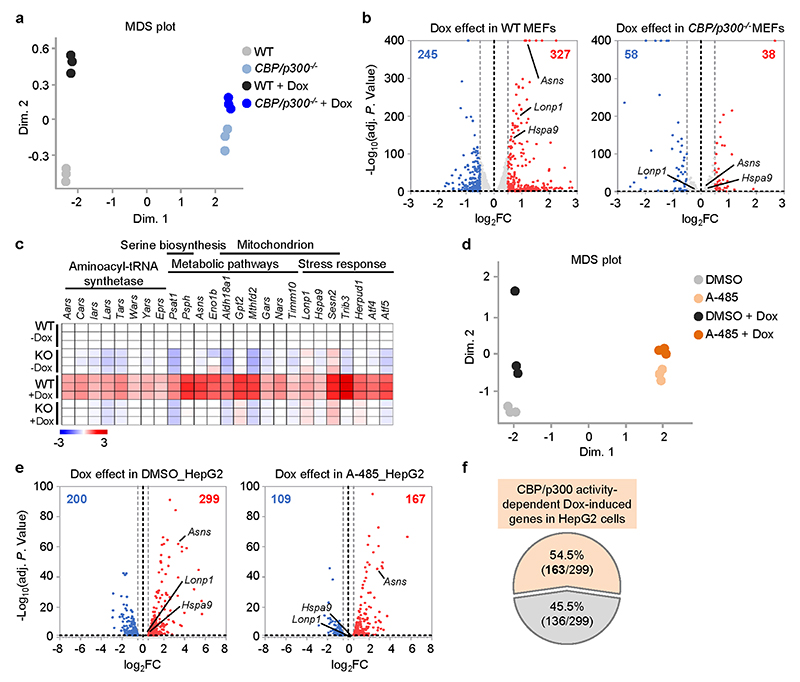
To validate the strong connections between CBP/p300 and UPRmt activation in mammals, we challenged wild-type (WT) and CBP/p300 knockout (KO) mouse embryonic fibroblasts (MEFs) with the mitochondrial stress inducer Dox13,36. Dox induced many UPRmt transcripts such as Hspd1, Hspa9, Lonp1 and Asns, a response that was remarkably blocked by CBP/p300 KO (Fig. 6a). RNA-seq analysis revealed that 327 transcripts were up-regulated (log2FC > 0.5, adjusted P < 0.05), and 245 transcripts were down-regulated (log2FC < -0.5, adjusted P < 0.05) in wild-type MEFs upon Dox treatment (Extended Data Fig. 6a, b and Supplementary Table 5). In contrast, only 38 up-regulated and 58 down-regulated transcripts were detected in CBP/p300 -/- MEFs (Extended Data Fig. 6b). Importantly, up to 197 (60.2%) of the 327 Dox-induced transcripts in wild-type MEFs were dependent on CBP/p300 for induction (Fig. 6b). These Dox-induced and CBP/p300-dependent transcripts were enriched for “aminoacyl-tRNA synthetase”, confirming a close link between mRNA translation and the UPRmt 21,55; for “serine biosynthesis”, including Phgdh, Psat1, Psph and Shmt2 19,56; and for metabolic and mitochondrial pathways (e.g. Eno1b, Timm10) (Fig. 6c and Extended Data Fig. 6c). Similar gene sets were also recently reported to be induced by other MSR inducers, such as CCCP (carbonyl cyanide m-chlorophenyl hydrazone) and oligomycin in different mammalian cells22,23. It is also noteworthy that Dox-induced expression of both Atf4 and Atf5, two key transcriptional regulators of the UPRmt 19,20, was heavily dependent on CBP/p300 (Extended Data Fig. 6c), suggesting a commanding role of CBP/p300 in UPRmt activation. In addition, reconstitution of WT-p300, but not a p300 acetyltransferase activity-defective mutant, restored Dox-induced UPRmt activation in CBP/p300 -/- MEFs (Fig. 6d, e), confirming that the catalytic activity of CBP/p300 is indispensable for this stress response.
Fig. 6. Functions of CBP/p300 in UPRmt activation are conserved in mammals.
a, Knockout of CBP/p300 attenuates Dox-induced transcription of key UPRmt genes in MEFs. qRT-PCR-results of wild-type (WT) and CBP/p300 -/- MEFs treated with or without Dox (30 μg/ml) for 24 h (n = 4 biologically independent samples). b, Diagram of the UPRmt genes that are dependent (blue) or independent (grey) on CBP/p300 for induction upon Dox treatment in MEFs, according to the RNA-seq data. c, Functional clustering of the 197 genes as indicated in (b). d, e, Transcripts (n = 4 biologically independent samples) (d) and protein levels (e) of CBP/p300 -/- MEFs reconstituted with empty vector, WT-p300 or the D1399Y acetyltransferase activity-defective mutant of p300, after control or Dox (30 μg/ml) treatment for 24 h. f, Western blots showing time-dependent changes of H3K18Ac and H3K27Ac in WT and CBP/p300 -/- MEFs treated with Dox (30 μg/ml) for 0-12 h. g, CBP/p300 is essential for Dox-induced H3K18Ac and H3K27Ac at the promoters of UPRmt genes (e.g. Hspd1 and Hspa9). ChIP-qPCR analysis of Hspd1 and Hspa9 in wild-type and CBP/p300 -/- MEFs treated with or without Dox (30 μg/ml) for 3 h (n = 4 biologically independent samples). ChIP was performed by using antibodies to H3K18Ac or H3K27Ac. h, CBP/p300 acetyltransferase activity inhibitor A-485 strongly suppresses Dox-induced transcription of key UPRmt genes in HepG2 cells. qRT-PCR-results of HepG2 cells pretreated with DMSO or A-485 (5 μM) for 1 h, and then co-treated with or without Dox (30 μg/ml) for 24 h (n = 4 biologically independent samples). i, Functional clustering of the 299 genes up-regulated upon Dox treatment (30 μg/ml, 24 h) and the CBP/p300 activity-dependency in human HepG2 cells, according to the RNA-seq data with A-485. CBP/p300 activity-dependent genes are indicated in red. j, qRT-PCR-results of HepG2 cells overexpressing empty vector, WT-p300 or the acetyltransferase activity-defective mutant D1399Y-p300 (n = 4 biologically independent samples). Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test. For uncropped gel source data, see Source Data Fig. 6.
In line with increased CBP/p300-mediated histone acetylation during mitochondrial perturbations in C. elegans (Fig. 2a–c), H3K18Ac and H3K27Ac levels peaked at 3-6 hour of Dox treatment in WT MEFs, which was significantly attenuated in CBP/p300 -/- MEFs (Fig. 6f). Meanwhile, H3K9Ac was not affected by the KO of CBP/p300 (Fig. 6f). ChIP-qPCR analysis further revealed that CBP/p300 is essential for Dox-induced increases in H3K18Ac and H3K27Ac levels at the promoters of prototypical UPRmt genes (e.g. Hspd1 and Hspa9) (Fig. 6g).
Liver is the central hub for metabolism and we have previously found that hepatocytes respond robustly to Dox treatment13. We thus further tested the impact of Dox treatment in the human hepatoma cell line HepG2. Similar to the effect of CBP/p300 KO, the induction of multiple prototypical UPRmt transcripts upon Dox treatment were abolished by the CBP/p300 KAT activity inhibitor A-485 (Fig. 6h). RNA-seq analysis revealed that the expressions of a much smaller number of transcripts were altered upon Dox treatment in A-485-treated cells, compared to that in control cells (Extended Data Fig. 6d, e). Moreover, Dox treatment induced 299 transcripts (log2FC > 0.5, adjusted P < 0.05), and the induction of 163 (54.5%) of them were abrogated by A-485 (Extended Data Fig. 6f and Supplementary Table 6). Notably, in addition to the GO terms found in MEFs (e.g. “aminoacyl-tRNA synthetase” and “Mitochondrion”), the Dox-induced transcripts in HepG2 cells were also enriched for “Innate immunity” and “Response to exogenous dsRNA”, containing 12 genes (two genes belonged to both terms) and 7 of them were dependent on CBP/p300 activity for induction (Fig. 6i and Supplementary Table 6). Finally, forced expression of WT-p300, but not the KAT activity-defective mutant of p300, is sufficient to induce the expression of UPRmt and Dox-induced immune response genes (e.g. DDX21, SLC3A2) in HepG2 cells (Fig. 6j). Taken together, these results point to a conserved and central role of CBP/p300 in MSR regulation in mammals.
Discussion
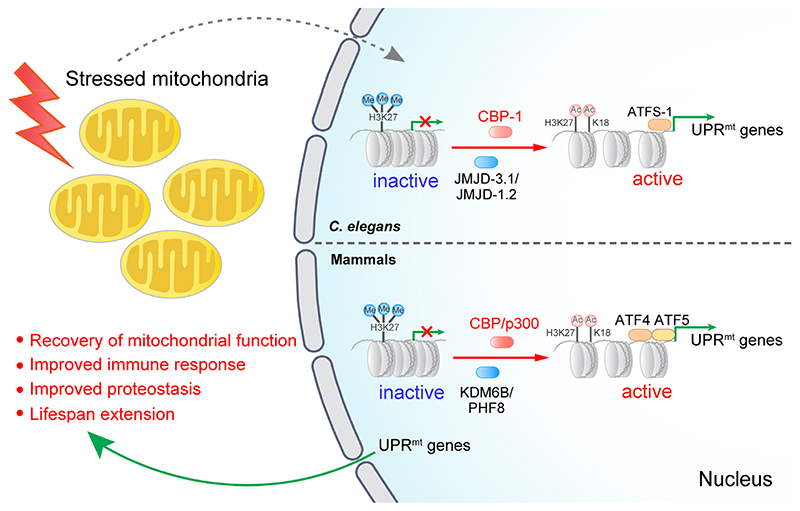
Here, by employing multilayered genetic and pharmacological approaches applied to C. elegans, mouse and human populations and cell lines, we provided strong evidence that CBP-1 or the mammalian CBP/p300 act downstream of demethylases JMJD-3.1/JMJD-1.2 or mammalian KDM6B/PHF8, switching the transcription-repressive histone methylation marks (e.g. H3K27Me3) to the transcription-active acetylation marks (e.g. H3K27Ac), and thereby relays the mitochondrial stress signal to the transcriptional induction of diverse UPRmt genes in C. elegans as well as in mammals (Fig. 7). Notably, many of the CBP-1-or CBP/p300-dependent UPRmt effectors positively contribute to mitochondrial function recovery, improved immune response, enhanced proteostasis against amyloid-β aggregation, and lifespan extension. In support of these findings, changes in CBP/p300 function tightly associate with multiple ageing/mitochondrial-related diseases, including Alzheimer’s and Huntington’s diseases57–59, and forced expression or pharmacological activation of CBP/p300 is sufficient to ameliorate neurodegenerative phenotypes in both mice and Drosophila AD models60–62.
Fig. 7. Model for CBP-1- or CBP/p300-mediated regulation of mitochondrial stress response and longevity.
When the mitochondria are stressed in response to various intracellular or extracellular stimuli, CBP-1 or the mammalian CBP/p300 act downstream of demethylases JMJD-3.1/JMJD-1.2 or mammalian KDM6B/PHF8, switching the transcription-repressive histone methylation marks (e.g. H3K27Me3) to the transcription-active acetylation marks (e.g. H3K27Ac), and thereby relays the mitochondrial stress signal to the transcriptional induction of diverse UPRmt genes in C. elegans as well as in mammals. Many of these UPRmt effectors play positive roles in the recovery of mitochondrial function, improved immune response and proteostasis, and lifespan extension.
How CBP-1 or CBP/p300 as well as the histone demethylases, sense mitochondrial stress remains an important direction for future work. One possibility is that CBP-1 itself is a downstream target that is activated in response to mitochondrial stress, as evidenced by increased cbp-1 expression after cco-1 and mrps-5 silencing (Extended Data Fig. 2c), and after jmjd-3.1 OE (Fig. 3e). Changes in mitochondrial metabolism may also modulate the levels of acetyl-CoA, which acts as a substrate for the acetyltransferase activity of KATs including CBP/p30030,32,63.
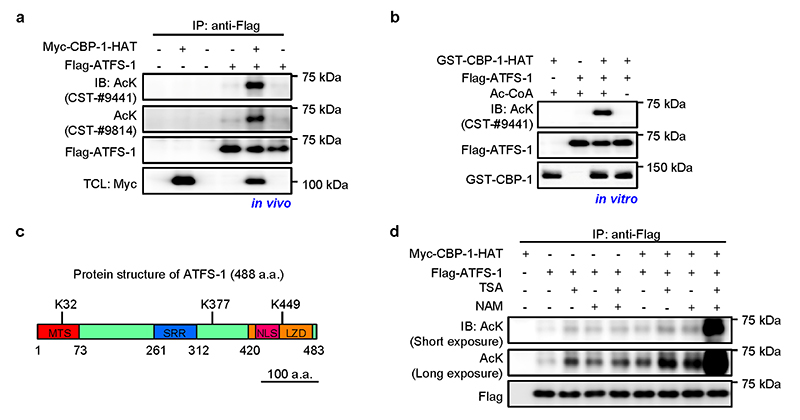
Of note, despite that we mainly focused on the regulation of H3K27Ac and H3K18Ac upon mitochondrial stress due to the availability of the reagents, it is very likely that similar regulatory mechanism exists as well for other CBP-1- or CBP/p300-mediated histone acetylation sites32,37, e.g. H4K5Ac (Extended Data Fig. 3a), which could also positively contribute to chromatin decompaction and transcriptional reactivation44. Moreover, CBP/p300 may also affect mitochondrial function and stress resistance by targeting proteins besides histones. As a first attempt in this direction, we investigated whether ATFS-1 could be acetylated by CBP-1. CBP-1 could indeed acetylate ATFS-1 both in vivo and in vitro (Extended Data Fig. 7a, b). Through mass spectrometry, we identified three acetylation sites in ATFS-1 (Extended Data Fig. 7c). Additionally, we also investigated which class of deacetylase (HDAC) is responsible for the deacetylation of ATFS-1. By using Trichostatin A (TSA, class I/II HDACs inhibitor), and nicotinamide (NAM, class III HDACs inhibitor), we found that HDACs belonging to at least two different classes participate in the deacetylation of ATFS-1 (Extended Data Fig. 7d). Furthermore, it has been reported that the PPARγ coactivator-1 (PGC-1α) can be acetylated by p300 and deacetylated by Sirt1, serving as an important switch controlling mitochondrial biogenesis and function64,65. In another study, p300 was identified as a binding partner for ATF4, and could enhance ATF4-mediated transcriptional activation through a mechanism independent of its acetyltransferase activity66.
In addition to the indispensable role of CBP-1 or CBP/p300 in MSR, we have noticed that the basal expression of some UPRmt transcripts also decreased after cbp-1 silencing, CBP/p300 KO or CBP/p300 activity inhibition (Figs. 1j, 6a, h; Supplementary Tables 1 and 5), suggesting that CBP/p300 functions in maintaining “basal UPRmt activity” as well. Nevertheless, the distinction between “basal” and “stress” conditions is somehow artificial, especially considering that organisms and cells are constantly exposed to multiple cues, and different wild C. elegans strains differ with respect to the level of UPRmt activation at “basal” condition67. Moreover, it is likely that some UPRmt genes controlled by CBP-1 or CBP/p300 may also contribute to basal mitochondrial function. For example, the chaperone hsp-60 or its mouse ortholog Hspd1, which demonstrated decreased “basal” H3K18Ac/H3K27Ac enrichment, ATFS-1 binding and mRNA expression upon cbp-1 RNAi or CBP/p300 suppression (Figs. 2k, 3h, 6a, g, h), have been reported to be essential for mitochondrial homeostasis even at basal state27,68. It is also noteworthy that we detected increased CBP-1 or CBP/p300-mediated acetylation marks during mitochondrial stress in both promoter and coding regions for a large set of genes (Extended Data Fig. 3f and Supplementary Table 3). According to a systematic study on mapping the global histone acetylation patterns to gene expression in yeast69, hyperacetylation of both intergenic and coding regions genome-wide at histone H3K18/H3K27 are significantly correlated with active transcription, it is hence likely that acetylation marks in both the promoter and coding regions in our context may contribute to transforming the condensed chromatin into a more relaxed structure and thus facilitate transcription29,30,32,44.
Altogether, by applying genetic and pharmacological LOF approaches, combined with bioinformatic and mechanistic studies, we identified the acetyltransferase CBP-1, as an essential regulator for the activation of the MSR and in particular the UPRmt. The beneficial effects on pathogen infection resistance, protein aggregation reduction and lifespan extension caused by mitochondrial perturbations are almost completely dependent on CBP-1 in C. elegans. Furthermore, systematic correlation analysis in mouse and human populations, as well as LOF studies in mammalian cells, indicate that functions of CBP/p300 in UPRmt regulation and longevity are also conserved in mammals. Our results thus reveal an evolutionarily conserved mechanism that coordinates the multiple layers of UPRmt regulators to systematically activate the stress responses, defend mitochondrial function, and promote health and longevity. Further studies will have to define whether genetically or pharmacologically targeting these CBP/p300-driven MSR pathways can have therapeutic applications against mitochondrial-related diseases, pathogen infections as well as ageing.
Methods
C. elegans strains
The Bristol strain (N2) was used as the wild-type strain. SJ4100 (zcIs13[hsp-6p::GFP]), MQ887 (isp-1(qm150)IV), MQ130 (clk-1(qm30) III), QC118 (atfs-1(et18)), OP675 (atfs-1::TY1::EGFP::3xFLAG), GMC101 (dvIs100 [unc-54p::A-beta-1-42::unc-54 3’-UTR + mtl-2p::GFP]) and CL2122 (dvIs15 [(pPD30.38) unc-54(vector) + (pCL26) mtl-2::GFP]) were obtained from the Caenorhabditis Genetics Center (CGC; Minneapolis, MN). Strains with jmjd-3.1 overexpression line #1 AUW3 (N2, epfIs3[myo-2p::cfp, jmjd-3.1p::jmjd-3.1]; zcIs13[hsp-6p::gfp]V) and line #2 AUW4 (N2, epfIs4[myo-2p::cfp, jmjd-3.1p::jmjd-3.1]; zcIs13[hsp-6p::gfp]V) were described previously25. The strain atfs-1(et18); zcIs13[hsp-6p::GFP] was generated by crossing the SJ4100 (zcIs13[hsp- 6p::GFP]) males with the QC118 (atfs-1(et18)) early 4 hermaphrodites. orms were cultured at 20 °C and fed with E. coli OP50 on Nematode Growth Media (NGM) plates unless otherwise indicated.
RNA interference
Bacterial feeding RNAi experiments were performed as described13. RNAi clones were used from either the Ahringer or Vidal libraries and verified by sequencing. Double RNAi experiments were carried out by mixing bacterial cultures normalized to their optical densities (OD600) before seeding onto NGM plates.
The alternative cbp-1 RNAi clone (cbp-1 RNAi_2) was constructed by PCR amplification of cbp-1 cDNA from total RNA with the following primers: cbp-1_RNAi2_632_Fw: 5’-CTCGAGGGTGTGGAAGGTGGACGTAG-3’, cbp-1_RNAi2_632_Rv: 5’-AGATCTTCCATTGGGCGCTTGATGAT-3’. The PCR product was then ligated into the L4440 empty vector and transformed into E. coli HT115 competent cells. The cbp-1 RNAi clone from Ahringer library (cbp-1 RNAi_1) was used for all experiments related to cbp-1 RNAi unless otherwise indicated.
Lifespan experiments were performed at 20 °C as described previously70. Briefly, 75-100 animals were used per condition and scored every other day, and those disappeared or exploded at the vulva were censored. All RNAi treatment for lifespan started since the maternal L4 stage.
Induction of the UPRmt
For RNAi-induced UPRmt, RNAi bacteria were grown in B containing 25 mg/ml carbenicillin at 37 °C overnight. The bacteria were then seeded onto 6 cm NGM plates with 2 mM IPTG. Dried plates were ept at room temperature overnight to allow IPTG induction of dsRNA expression. 4 worms or synchronized worm eggs were raised on the RNAi plates at 20 °C. The F0 worms were then removed the next day if L4 worms were seeded the day before. Fluorescent images with the same exposure time for each condition were taken after 2-3 days. For antimycin A or doxycycline induced UPRmt, antimycin A (Cat. A8674, Sigma) with a final concentration of 2.5 μM, or doxycycline (Cat. D9891, Sigma) with a final concentration of 30 μg/ml were added into the NGM just before pouring the plates.
RNA extraction and RNA-seq analysis
For worm samples, worms were synchronized by bleaching. Synchronized worm eggs were plated in NGM plates under the described conditions and raised at 20 °C. Worms were harvested after 2 days (at L4/young adult stage), washed with M9 buffer for three times to remove the bacteria, then snap frozen in liquid nitrogen. On the day of the extraction, 1 ml of TriPure Isolation Reagent (Cat. 11667165001, Roche) was added to each tube. The samples were then frozen and thawed quickly 8 times with liquid nitrogen and water bath to rupture cell membranes. RNA was then extracted by using a column-based kit from Macherey-Nagel (Cat. 740955.250). For mammalian cell samples, cells were directly dissolved in 1 ml of the TriPure Isolation Reagent and extracted by using the kit from Macherey-Nagel (Cat. 740955.250). RNA-seq was performed by BGI with the BGISEQ-500 platform.
RNA-seq data analysis for worm samples was performed using the R version 3.6.3 (https://www.r-project.org/). Briefly, after sequencing on the BGIseq-500 platform, the raw reads were filtered by removing adaptor sequences, contamination and low-quality (phred quality < 20) reads. FastQC71 was used to verify the quality of the sequence data. Sequenced reads were mapped to the worm genome “Caenorhabditis_elegans. Bcel235.89” with STAR aligner version 2.6.0a72. Reads were counted using htseq-count version 0.10.073, using these flags: -f bam -r pos -s no -m union -t exon -i gene_id. Differential expression of genes was calculated by using Limma-Voom74,75. The genes with a Benjamini-Hochberg adjusted P-value of less than 0.05 were defined as statistically significant. Genes whose expressions were significantly up-regulated with log2FC > 0.5 (adjusted P < 0.05) in cco-1 RNAi condition; and were then down-regulated by more than 25% of the log2FC after cbp-1 or atfs-1 RNAi co-treatment, compared to the log2FC of cco-1 RNAi condition, were considered as CBP-1- or ATFS-1-dependent. Genes whose expressions were significantly down-regulated with log2FC < -0.5 (adjusted P < 0.05) were defined as the down-regulated genes. For MEFs or human HepG2 cell samples, similar analysis procedure was used, except that the “Mus_musculus.GRCm38.95” genome or the “Homo_sapiens.GRCh38.95” was used for mapping. Genes whose expressions were up-regulated with log2FC > 0.5 (adjusted P < 0.05) in Dox treatment condition; and were then down-regulated by more than 25% of the log2FC after CBP/p300 KO or A-485 treatment, compared to the log2FC of WT-Dox condition, were considered as CBP/p300-dependent or CBP/p300 activity-dependent. Functional clustering was performed by using the DAVID (Database for Annotation, Visualization and Integrated Discovery) database76. Heat-maps were generated by using Morpheus (https://software.broadinstitute.org/morpheus).
Quantitative RT-PCR
Worms were raised and total RNA was isolated as described for the RNA-seq studies. cDNA was then synthesized from total RNA using the Reverse Transcription Kit (Cat. 205314, Qiagen). qRT-PCR was performed by using the LightCycler 480 SYBR Green I Master kit (Cat. 04887352001, Roche). The primers used for qRT-PCR are listed in Supplementary Table 7. Primers for worm pmp-3, mouse Actin and human ACTB were used as normalization controls.
Western blotting
For worm samples, proteins were extracted as described previously13. Western blotting was performed with antibodies against GFP (1:1000, Cat. 2956, CST), Actin (1:2000, Cat. A5441, Sigma), H3K18Ac (1:1000, Cat. 07-354, Merck), H3K27Ac (1:1000, Cat. ab4729, abcam), H3K9Ac (1:1000, Cat. 06-942, Merck), H3K4Ac (1:1000, Cat. Ab176799, abcam), Histone 3 (1:1000, Cat. 9715, CST), Tubulin (1:2000, Cat. T5168, Sigma), H3K27Me3 (1:1000, Cat. 07-449, millipore), H3K27Me2 (1:1000, Cat. ab24684, abcam), H3K27Me1 (1:1000, Cat. 07-448, millipore), H3K9Me1 (1:1000, Cat. 07-450, millipore), H3K4Me3 (1:1000, Cat. 07-473, millipore), Histone 4 (1:1000, Cat. sc-10810, Santa Cruz), H4K5Ac (1:1000, Cat. ab51997, abcam), β-amyloid 1–16 (6E10) (1:1000, Cat. 803001, BioLegend), HA-tag (1:2000, Cat. 3724, CST), Flag-tag (1:1000, Cat. F7425, Sigma), Myc-tag (1:2000, Cat. sc-40, Santa Cruz), GST-tag (1:1000, Cat. 2625, CST), AcK (1:1000, Cat. 9441, CST), AcK (1:1000, Cat. 9814, CST) and HRP-labelled anti-rabbit (Cat. 7074, CST) and anti-mouse (Cat. 7076, CST) secondary antibodies.
Chromatin immunoprecipitation (ChIP) and ChIP-seq of worms
The ChIP of worms was performed as described77, with slight modifications. Briefly, worms were synchronized by bleaching. Synchronized worm eggs were plated in NGM plates under the described conditions and raised at 20 C. Worms were harvested after 2 days (at L4/young adult stage) and washed with M9 buffer for three times. Worms were then fixed with 1% formaldehyde in PBS for 30 min, and quenched by glycine. Immunoprecipitations were carried out by using antibodies against H3K18Ac (1:100, Cat. 07-354, Merck) or H3K27Ac (1:100, Cat. ab4729, abcam). For ChIP of ATFS-1, the OP675 (atfs-1::TY1::EGFP::3xFLAG) worm strain and anti-Flag M2 beads (Cat. A2220, Sigma) were used. The primers used for ChIP-qPCR were listed in Supplementary Table 7.
For ChIP-seq, DNA fragments were sequenced using the BGISEQ-500 platform. The data analysis was performed using the R version 3.6.3 (https://www.r-project.org/). FastQC71 was used to verify the quality of the sequence data. Alignment was performed against the C. elegans genome “Caenorhabditis_elegans. Bcel235.89” following the Bowtie2 (version 2.3.5)78 manual guidelines with default parameters. SAMtools (version 1.4.1)79 was used to sort, filter and index the obtained alignments. Peak calling was then performed using MACS2 (version 2.1.2)80 against default Poisson distribution to generate raw counts for each sample, or between samples of interest for comparison. The peak scores between treatment and control for each histone modification were generated with an associated FDR value (default value of FDR 0.05). Quality of alignment and peaks were assessed using the ChIPQC (version 1.18.2)81 before proceeding with the analysis. Read counts per peak were obtained using BEDTools (version 2.26.0)82 and SAMtools (version 1.4.1)79 packages. Intersection between sets and their associated p-values were computed using the SuperExactTest7 (version 1.0.6)83 package. Genome tracks were revealed by Integrative Genomics Viewer IGV (version 2.8.0)84, the two tracks were shown with the same total count range between basal and mitochondrial stress conditions for each gene.
Pseudomonas aeruginosa infection assay
The P. aeruginosa PA14 slow killing assay was performed as described85. Briefly, P. aeruginosa overnight cultures were seeded onto slow-killing NGM agar plates with 0.35% peptone. Plates were allowed to dry for 20 min at room temperature, and then incubated at 37 °C for 24 h and allowed to equilibrate at 25 °C for another 24 h. Synchronized worm eggs were raised on RNAi bacteria as indicated in the Figure legends, until they reach L4 stage. The worms were then transferred to P. aeruginosa slow-killing plates and were counted every 12 h. Animals were scored as dead if they failed to respond when gently punched with a worm picker. 80-100 worms were used for each condition, and those disappeared or exploded at the vulva were censored. Each experiment was performed at least two times, and the log rank (Mantel-Cox) statistical test was used to calculate P values.
Cell culture and drug treatment
HepG2 cells were obtained from ATCC. Cells were validated to be free of mycoplasma contamination and maintained in DMEM medium containing 4.5 g glucose per liter and 10% FBS. Immortalized Crebbpfl/fl; Ep300fl/fl mouse embryonic fibroblasts (MEFs) stably expressing Cre-ERT2 were kindly provided by Dr. P. K. Brindle36. The floxed CBP and p300 alleles were deleted as previously described37. Briefly, Crebbpfl/fl; Ep300fl/fl MEFs were treated with 2 μM 4-hyroxy-tamoxifen (Cat. H7904, Sigma), media were changed and fresh 4-OHT added every 12 h for 2 days. Cells were cultured for an additional 1 day to allow for complete depletion of CBP and p300 protein. For transfection, plasmids expressing human full-length WT-p300 (Cat. 89094, addgene), and the p300 acetyltransferase activity-defective mutant (Cat. 89095, addgene)86, were purchased from addgene and transfected with the Lipofectamine 3000 Reagent (Cat. L3000015, ThermoFisher). CBP/p300 acetyltransferase inhibitors A-485 (Cat. 6387, TOCRIS) and bromodomain inhibitor PF-CBP1 (Cat. S8180, Selleck Chemicals) were dissolved in DMSO and treated with final concentrations as described in the Figure legends. For the treatment of worms with CBP/p300 inhibitors, A-485 or PF-CBP1 was added into the NGM agar medium with final concentrations as indicated just before pouring the plates.
ATFS-1 acetylation analysis
For in vivo analysis, plasmids expressing full length ATFS-1 or the HAT domain of CBP-1 (aa 803-1620) were created by PCR amplification from total worm cDNA and verified by sequencing. Transfection were performed with the Lipofectamine 3000 Reagent (Cat. L3000015, ThermoFisher) in HEK293T cells, 5 μM Trichostatin A (TSA, Cat. T8552 Sigma), and/or 10 mM nicotinamide (NAM, Cat. N0636, Sigma) were added to the culture medium 8 h before harvesting. For in vitro analysis, the HAT domain of CBP-1 (aa 803-1620) were subcloned to the GST-tag-containing pGEX-4T-1 vector, and purified from the BL21 bacteria. In vitro acetylation assay was carried using 50 μl reactions contained 50 mM HEPES (pH 8.0), 10% glycerol, 1 mM DTT, 1 mM PMSF, 5 μM TSA, 10 mM NAM, 100 μM acetyl-CoA, immune-purified ATFS-1 from HEK293T cells and GST-HAT-CBP-1. After incubation at 30 °C for 1 h, the reaction was stopped by addition of 10 μl of 5 × SDS sample buffer. The samples were then subjected to SDS–PAGE and western blotting.
Bioinformatic analyses
All BXD, LXS and GTEx transcriptome data sets for bioinformatic analyses were downloaded from GeneNetwork (http://www.genenetwork.org) and performed as described in previous studies43,52,53. The BXD transcriptome datasets used to establish genetic correlations were UTHSC Affy MoGene 1.0 ST Spleen (Dec10) RMA Males, INIA_Pituitary_RMA_M_0612, INIA Adrenal Affy MoGene 1.0ST (Jun12) RMA Males, Eye M430v2 (Sep08) RMA, Hippocampus Consortium M430v2 (Jun06) RMA, and INIA Hypothalamus Affy MoGene 1.0 ST (Nov10) Male. The IDs for lifespan datasets were 12564, 17475, 18435, 19422 and 19424. The LXS transcriptome datasets used were UCAMC LXS Whole Brain Saline RNA Sequence (Feb16) FPKM, and VCU LXS PFC Sal M430A 2.0 (Aug06) RMA. For human genetic correlation analyses, the “GTEx v8 All Tissues” dataset was used54. Pearson’s r was used for measuring the correlations. Correlation heat-maps were generated with Morpheus (https://software.broadinstitute.org/morpheus). The Circos plot was generated using Circos (http://www.circos.ca)87.
Statistics and reproducibility
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications13,50,70. Samples were allocated to groups/treatments randomly, and steps were taken to avoid batch effects. Experimental conditions were not blinded. However, data analysis was performed blind whenever possible. No data were excluded from the analysis, except for the C. elegans lifespan or survival experiments (the reasons for censoring were the “exploded vulva” phenotype or worms that crawled off the plate. These reasons were pre-established before the beginning of the experiment13,70). All individual data points have been shown in the figures, data distribution was assumed to be normal but this was not formally tested. All the experiments, particularly the representative micrographs shown in Fig. 1a–e, 3a, 3f; Extended Data Fig. 1b–d, 1f–h, 1k–n, were repeated at least twice and similar results were found. Survival analyses were performed using the Kaplan-Meier method and the significance of differences between survival curves calculated using the log rank (Mantel-Cox) test. Differences between two groups were assessed using two-tailed unpaired Student’s t-tests. Analysis of variance (ANOVA) followed by Tukey post-hoc test (one-way ANOVA for comparisons between groups, two-way ANOVA for comparisons of magnitude of changes between different groups from different cell lines or treatments) was used when comparing more than two groups, P values were adjusted for multiple comparisons. GraphPad Prism 6 program was used for all statistical analyses. Fiji (version 1.47b) was used to quantify the western blots as indicated in Fig. 2a–c and Extended Data Fig. 3a.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
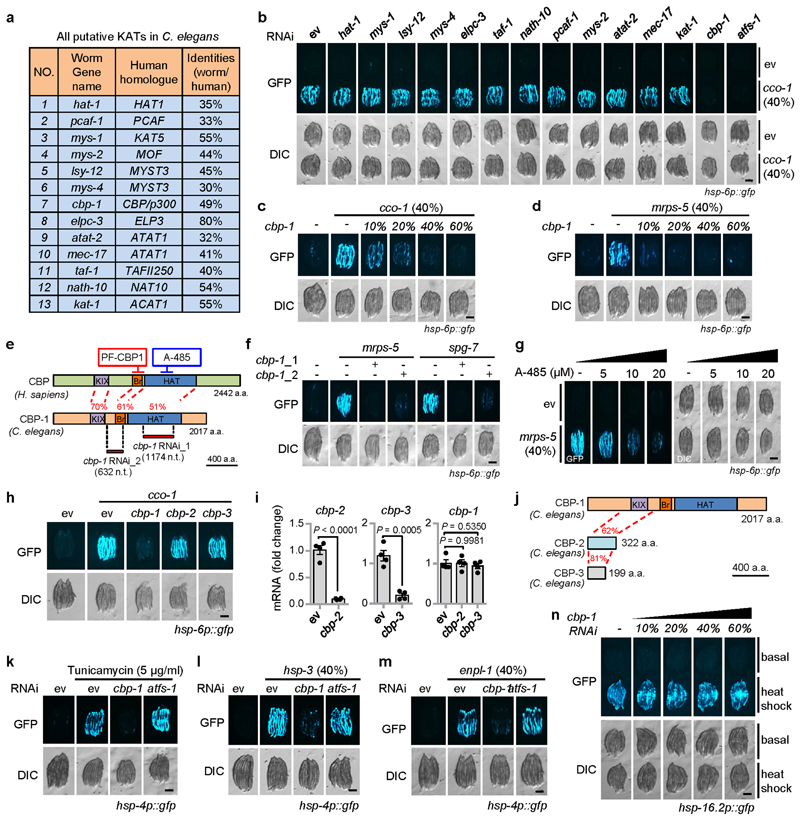
Extended Data Fig. 1. Inhibition of CBP-1 by RNAi or pharmacological inhibitors attenuates UPRmt activation in C. elegans.
a, All putative lysine acetyltransferases (KATs) in C. elegans and their human homologues. The worm KATs were validated/identified by searching the C. elegans protein database for proteins with a conserved acetyltransferase domain, and high amino acid sequences identities of the known human KATs30. b, Role of KATs in UPRmt activation in C. elegans. hsp-6p::gfp worms were fed with control (ev) or cco-1 (40%) RNAi in combination with RNAi targeting different KATs (60%). c, d, cbp-1 RNAi attenuates the UPRmt activation induced by cco-1 (c) or mrps-5 (d) RNAi in a dose-dependent manner. RNAi targeting cco-1 or mrps-5 occupied 40%. cbp-1 RNAi occupied 10-60%. Control RNAi was used to supply to a final 100% of RNAi for all conditions. e, Schematic diagram showing the different regions regulated by the two different cbp-1 RNAi, and the two CBP/p300 inhibitors (PF-CBP1 and A-485). KIX, kinase-inducible domain interacting domain; Br, bromodomain; HAT, histone acetyltransferase domain; a.a., amino acids; n.t., nucleotides. f, The alternative cbp-1 RNAi (cbp-1_2) also inhibits UPRmt activation. RNAi targeting mrps-5 or spg-7 occupies 40%, cbp-1 RNAi occupies 25%. g, A-485 attenuates UPRmt activation induced by mrps-5 RNAi in a dose-dependent manner. hsp-6p::gfp worms were fed with control or mrps-5 RNAi (40%), in combination with 0-20 μM A-485. h, i, RNAi that specifically targets cbp-2 or cbp-3 failed to abolish UPRmt activation in hsp-6p::gfp worms. Photos (h) and qRT-PCR-results (n = 4 biologically independent samples) (i) of hsp-6p::gfp worms fed with control, cco-1 (40%), cbp-1 (25%), cbp-2 (Ahringer library) or cbp-3 RNAi. Error bars denote SEM. Statistical analysis was performed by two-tailed unpaired Student’s t-test. j, Schematic diagram showing the protein structure of CBP-1, CBP-2 and CBP-3. The numbers in red indicate the amino acid sequence identities between two proteins in comparison. k-m, cbp-1 RNAi attenuates the UPRER activation induced by tunicamycin (5 μg/ml) (k), hsp-3 (l) or enpl-1 (m) RNAi in hsp-4p::gfp worms. RNAi targeting hsp-3 or enpl-1 occupies 40%, cbp-1 RNAi occupies 25%, atfs-1 RNAi occupies 60%. n, cbp-1 RNAi does not affect the cytosolic UPR (UPRCYT)/heat shock response activation induced by heat shock. hsp16.2p::gfp reporter worms were fed with different percentages of cbp-1 RNAi as indicated. As positive control, heat shock for 8 h at 31°C could induce the UPRCYT and cbp-1 RNAi did not block this response. Scale bars, 0.3 mm.
Extended Data Fig. 2. UPRmt genes dependent or independent of CBP-1 and ATFS-1 for expression.
a, Principal-component analysis (PCA) of the RNA-seq profiles of worms treated with the indicated RNAi. b, Diagram of genes up-regulated after cco-1 RNAi, in common with genes up-regulated after mrps-5 RNAi according to the RNA-seq data. c-f, qRT-PCR-results of indicated genes in hsp-6p::gfp worms fed with control (ev), cco-1, mrps-5, cbp-1 or atfs-1 RNAi (n = 4 biologically independent samples). RNAi targeting cco-1 or mrps-5 occupies 50%, cbp-1 occupies 25%, atfs-1 occupies 50%. g, qRT-PCR-results of hsp-6p::gfp worms fed with control (ev), spg-7, timm-23, tomm-40, cts-1 or dlst-1 RNAi (n = 4 biologically independent samples). UPRmt genes dependent on both CBP-1 and ATFS-1 for induction according to the RNA-seq dataset (as summarized in Fig. 1g) are highlighted in red. Genes only dependent on CBP-1, but not ATFS-1, are highlighted in blue. h, Diagram of the down-regulated genes after single cco-1 RNAi (orange), in common with down-regulated (blue) or up-regulated (pink) genes after single cbp-1 RNAi according to the RNA-seq data. i, j, Functional clustering of the 709 (i) and 190 (j) genes as indicated in (h). k, Diagram of genes down-regulated after cco-1 RNAi, in common with genes down-regulated after mrps-5 RNAi according to the RNA-seq data. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test.
Extended Data Fig. 3. Mitochondrial stress increases CBP-1-mediated histone acetylation at the loci of UPRmt, but not UPRER or UPRCYT genes.
a, Western blots of hsp-6p::gfp worms fed with control, cbp-1, atfs-1, cco-1 or mrps-5 RNAi. RNAi targeting cbp-1 occupies 25%, atfs-1, cco-1 or mrps-5 occupies 50%. b-e, Genome tracks showing the ChIP-seq analysis for H3K27Ac and H3K18Ac over the genomic loci of gpd-2 (b), hsp-3 (c), hsp-4 (d) and hsp-16.2 (e) in worms fed with control or cco-1 RNAi. The two tracks were shown with the same total count range between basal and mitochondrial stress condition for each gene. f, Summary of the distribution analysis of the 265 increased H3K18Ac/H3K27Ac peaks on the 134 UPRmt genes (as indicated in Fig. 2d) in response to mitochondrial stress. For uncropped gel source data, see Source Extended Data Fig. 3.
Extended Data Fig. 4. RNAi of cbp-1 caused a severe developmental delay in the worm Alzheimer’s disease model GMC101, but not in the control CL2122 strain.
Representative photos of CL2122 or GMC101 worms fed with control or cbp-1 (10% or 20%) RNAi since maternal L4 stage. The developmental stage and body length of the F1 progeny were quantified at Day 4 after hatching (n = 25 worms for each condition). Conditions with 20% cbp-1 RNAi were not quantified as most of the eggs failed to hatch in GMC101 worms fed with 20% cbp-1 RNAi. Scale bar, 1 mm. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test.
Extended Data Fig. 5. CBP/p300 expression positively correlates with Kdm6b/Phf8, UPRmt transcripts and lifespan in mouse populations.
a, Pearson’s correlation co-expression heat-map for CBP/p300, Kdm6b/Phf8 and UPRmt genes in hippocampus and hypothalamus of the BXD mouse genetic reference population43,52. Positive and negative correlations are indicated in red and blue, respectively. The intensity of the colors corresponds to correlation coefficients. b, Pearson’s correlation co-expression heat-map for CBP/p300, Kdm6b/Phf8 and UPRmt genes in the brain (whole brain) and prefrontal cortex of the LXS mouse genetic reference population53. c, Positive correlations between lifespan and CBP or p300 transcript levels in hippocampus and hypothalamus of BXD mice (Pearson’s r, two-sided). Each dot indicates an independent BXD strain.
Extended Data Fig. 6. An essential role of CBP/p300 and CBP/p300 acetyltransferase activity in UPRmt activation in mammalian cells.
a, Multidimensional scaling (MDS) plot of the RNA-seq profiles of wild-type (WT) and CBP/p300 -/- MEFs treated with or without Dox (30 μg/ml) for 24 h. Note the decreased distance between Dox-treated and un-treated condition in the CBP/p300 -/- background compared to that in WT background. b, CBP/p300 -/- MEFs are insensitive to the treatment of mitochondrial stress inducer Dox. Volcano plots showing the effect of Dox treatment in wild-type (WT) (left) or CBP/p300 -/- (right) MEFs on gene expression. FC, fold change. Genes that were up-regulated (log2FC > 0.5, adjusted P < 0.05) during Dox treatment were highlighted in red. Genes that were down-regulated (log2FC < -0.5, adjusted P < 0.05) were highlighted in blue. c, Heat-map of the representative UPRmt genes dependent on CBP/p300 for induction in response to Dox treatment in WT and CBP/p300 -/- (KO) MEFs, according to the RNA-seq data. The heat-map was shown in log2FC values. d, MDS plot of the RNA-seq profiles of HepG2 cells treated with or without CBP/p300 acetyltransferase activity inhibitor A-485 (5 μM) and/or Dox (30 μg/ml) for 24 h. e, The CBP/p300 catalytic inhibitor A-485 attenuates the effect of Dox on gene expression in HepG2 cells. Volcano plots showing the effect of Dox on gene expression of HepG2 cells in control (DMSO) (left) or A-485 (right) treatment background. FC, fold change. Genes that were up-regulated (log2FC > 0.5, adjusted P < 0.05) during Dox treatment were highlighted in red. Genes that were down-regulated (log2FC < -0.5, adjusted P < 0.05) were highlighted in blue. f, Diagram of the UPRmt genes that are dependent (orange) or independent (grey) on CBP/p300 activity in response to Dox treatment in HepG2 cells, according to the RNA-seq data with A-485.
Extended Data Fig. 7. ATFS-1 can be acetylated by CBP-1 and affected by both class I/II and class III HDACs.
a, ATFS-1 was acetylated by CBP-1 in vivo. Flag-tagged ATFS-1 was expressed with or without CBP-1 acetyltransferase domain (CBP-1-HAT) in HEK293T cells, immunoprecipitated with anti-Flag antibody and analyzed by western blots. TCL, total cell lysate. b, ATFS-1 was acetylated by CBP-1 in vitro. Bacterially expressed GST tagged CBP-1-HAT was incubated with Flag-ATFS-1 with or without acetyl-CoA (Ac-CoA) and immunoblotted as indicated. c, Schematic diagram showing the protein structure of ATFS-1 and the acetylated sites identified by mass spectrometry. MTS, mitochondrial targeting sequence; SRR, Serine-rich region; NLS, Nuclear localization signal; LZD, Leucine zipper domain. d, HEK293T cells transfected as indicated were treated with or without TSA (class I/II HDAC inhibitor), or NAM (class III HDAC inhibitor) 8 h before harvesting, and analyzed by western blots. For uncropped gel source data, see Source Extended Data Fig. 7.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center for providing C. elegans strains. We thank Dr. P. K. Brindle for providing CBP/p300 -/- MEFs. We thank all members of J. Auwerx and K. Schoonjans laboratories for helpful discussions. This work was supported by grants from the EPFL, the European Research Council (ERC-AdG-787702), the Swiss National Science Foundation (SNSF 31003A_179435) and GRL grant of the National Research Foundation of Korea (NRF 2017K1A1A2013124). T.Y.L. was supported by the “Human Frontier Science Program” (LT000731/2018-L). L.J.E.G. is supported by a Swiss Government Excellence Scholarship (FCS ESKAS-Nr. 2019.0009).
Footnotes
Author contributions
T.Y.L. and J.A. conceived the project. T.Y.L. performed most of the experiments. A.W.G. contributed to the C. elegans lifespan experiments. A.M. contributed to the P. aeruginosa infection experiment. T.Y.L., M.B.S., H.L., A.B., G.E.A., X.L. and L.J.E.G. performed data analysis. K.S. and J.A. supervised the study. T.Y.L. and J.A. wrote the manuscript with comments from all authors.
Competing interests
Authors declare no competing interests.
Data availability
The RNA/DNA sequencing datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with the accession numbers: GSE131611 for worm RNA-seq, GSE148328 for worm ChIP-seq, GSE131613 for MEFs RNA-seq, and GSE156830 for human HepG2 RNA-seq. Functional clustering in this study was performed by using the DAVID (Database for Annotation, Visualization and Integrated Discovery) database v6.8 (https://david.ncifcrf.gov/home.jsp). The BXD, LXS and GTEx transcriptome datasets used in this study are available in the GeneNetwork database (https://www.genenetwork.org). All data supporting the findings of this study are available from the corresponding author J.A. upon request. Source data are provided with this paper.
References
- 1.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mottis A, Herzig S, Auwerx J. Mitocellular communication: Shaping health and disease. Science. 2019;366:827–832. doi: 10.1126/science.aax3768. [DOI] [PubMed] [Google Scholar]
- 3.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nature reviews Molecular cell biology. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 5.Sun N, Youle RJ, Finkel T. The Mitochondrial Basis of Aging. Molecular cell. 2016;61:654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth LN, Brunet A. The Aging Epigenome. Molecular cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh PP, Demmitt BA, Nath RD, Brunet A. The Genetics of Aging: A Vertebrate Perspective. Cell. 2019;177:200–220. doi: 10.1016/j.cell.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nature reviews Molecular cell biology. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi-Sanabria R, Frankino PA, Paul JW, 3rd, Tronnes SU, Dillin A. A Futile Battle? Protein Quality Control and the Stress of Aging. Developmental cell. 2018;44:139–163. doi: 10.1016/j.devcel.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response - synchronizing genomes. Curr Opin Cell Biol. 2015;33:74–81. doi: 10.1016/j.ceb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrino MW, et al. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 17.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quiros PM, et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorese CJ, et al. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Current biology: CB. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munch C, Harper JW. Mitochondrial unfolded protein response controls matrix pre- RNA processing and translation. Nature. 2016;534:710–713. doi: 10.1038/nature18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fessler E, et al. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature. 2020;579:433–437. doi: 10.1038/s41586-020-2076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo X, et al. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature. 2020;579:427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Y, et al. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt) Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merkwirth C, et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell metabolism. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 28.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Bio. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 30.Menzies KJ, Zhang HB, Katsyuba E, Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol. 2016;12:43–60. doi: 10.1038/nrendo.2015.181. [DOI] [PubMed] [Google Scholar]
- 31.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nature reviews Molecular cell biology. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 32.Sheikh BN, Akhtar A. The many lives of KATs - detectors, integrators and modulators of the cellular environment. Nature reviews Genetics. 2019;20:7–23. doi: 10.1038/s41576-018-0072-4. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, Mello C. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes & development. 1998;12:943–955. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chrivia JC, et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 35.Arany Z, Sellers WR, Livingston DM, Eckner R. E1a-Associated P300 and Creb-Associated Cbp Belong to a Conserved Family of Coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 36.Kasper LH, et al. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. The EMBO journal. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinert BT, et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell. 2018;174:231–244 e212. doi: 10.1016/j.cell.2018.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasko LM, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chekler EL, et al. Transcriptional Profiling of a Selective CREB Binding Protein Bromodomain Inhibitor Highlights Therapeutic Opportunities. Chemistry & biology. 2015;22:1588–1596. doi: 10.1016/j.chembiol.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 40.Nolden M, et al. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Kim HE, et al. Lipid Biosynthesis Coordinates a Mitochondrial-to-Cytosolic Stress Response. Cell. 2016;166:1539. doi: 10.1016/j.cell.2016.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YF, Haynes CM. Metabolism and the UPR(mt) Molecular cell. 2016;61:677–682. doi: 10.1016/j.molcel.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor perspectives in biology. 2014;6 doi: 10.1101/cshperspect.a018762. a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rauthan M, Ranji P, Pradenas NA, Pitot C, Pilon M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. P Natl Acad Sci USA. 2013;110:5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 47.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 48.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, et al. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS biology. 2009;7:e1000245. doi: 10.1371/journal.pbio.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorrentino V, et al. Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature. 2017;552:187. doi: 10.1038/nature25143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McColl G, et al. Utility of an improved model of amyloid-beta (A beta(1-42)) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol Neurodegener. 2012;7 doi: 10.1186/1750-1326-7-57. doi:Artn 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreux PA, et al. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams RW, et al. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mammalian genome: official journal of the International Mammalian Genome Society. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- 54.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molenaars M, et al. A Conserved Mito-Cytosolic Translational Balance Links Two Longevity Pathways. Cell metabolism. 2020;31:549. doi: 10.1016/j.cmet.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao XR, et al. Mitochondrial dysfunction remodels one - carbon metabolism in human cells. Elife. 2016;5:e10575. doi: 10.7554/eLife.10575. doi:ARTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schueller E, et al. Dysregulation of histone acetylation pathways in hippocampus and frontal cortex of Alzheimer?s disease patients. Eur Neuropsychopharm. 2020;33:101–116. doi: 10.1016/j.euroneuro.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 58.Nucifora FC, et al. Interference by Huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 59.Nativio R, et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nature genetics. 2020 doi: 10.1038/s41588-020-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer’s disease. P Natl Acad Sci USA. 2010;107:22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chatterjee S, et al. Reinstating plasticity and memory in a tauopathy mouse model with an acetyltransferase activator. Embo Mol Med. 2018;10 doi: 10.15252/emmm.201708587. doi:ARTN e8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cutler T, et al. Drosophila Eye Model to Study Neuroprotective Role of CREB Binding Protein (CBP) in Alzheimer’s Disease. Plos One. 2015;10 doi: 10.1371/journal.pone.0137691. doi:ARTN e0137691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC- 1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 66.Lassot I, et al. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J Biol Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 67.Yin JA, et al. Genetic variation in glia-neuron signalling modulates ageing rate. Nature. 2017;551:198–203. doi: 10.1038/nature24463. [DOI] [PubMed] [Google Scholar]
- 68.Magnoni R, et al. Late onset motoneuron disorder caused by mitochondrial Hsp60 chaperone deficiency in mice. Neurobiol Dis. 2013;54:12–23. doi: 10.1016/j.nbd.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 69.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 70.Mouchiroud L, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 72.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 77.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carroll TS, Liang Z, Salama R, Stark R, de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet. 2014;5:75. doi: 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang M, Zhao Y, Zhang B. Efficient Test and Visualization of Multi-Set Intersections. Sci Rep. 2015;5:16923. doi: 10.1038/srep16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirienko NV, Cezairliyan BO, Ausubel FM, Powell JR. Pseudomonas aeruginosa PA14 pathogenesis in Caenorhabditis elegans. Methods in molecular biology. 2014;1149:653–669. doi: 10.1007/978-1-4939-0473-0_50. [DOI] [PubMed] [Google Scholar]
- 86.Askew EB, Bai S, Blackwelder AJ, Wilson EM. Transcriptional synergy between melanoma antigen gene protein-A11 (MAGE-11) and p300 in androgen receptor signaling. J Biol Chem. 2010;285:21824–21836. doi: 10.1074/jbc.M110.120600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA/DNA sequencing datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with the accession numbers: GSE131611 for worm RNA-seq, GSE148328 for worm ChIP-seq, GSE131613 for MEFs RNA-seq, and GSE156830 for human HepG2 RNA-seq. Functional clustering in this study was performed by using the DAVID (Database for Annotation, Visualization and Integrated Discovery) database v6.8 (https://david.ncifcrf.gov/home.jsp). The BXD, LXS and GTEx transcriptome datasets used in this study are available in the GeneNetwork database (https://www.genenetwork.org). All data supporting the findings of this study are available from the corresponding author J.A. upon request. Source data are provided with this paper.