Graphical abstract
Keywords: Trypanosoma teixeirae sp. n, Little red flying fox (Pteropus scapulatus), Morphology, PCR, 18S ribosomal RNA (rRNA), Glycosomal glyceraldehyde phosphate dehydrogenase (gGAPDH), Phylogeny
Highlights
-
•
Characterization of Trypanosoma teixeirae sp. n. from a little red flying fox.
-
•
First trypanosome species associated with clinical disease in an Australian bat.
-
•
Morphological and molecular analyses.
-
•
T. teixeirae sp. n. clustered within the T. cruzi clade.
-
•
Evolutionary implications discussed.
Abstract
Little is known about the genetic diversity and pathogenicity of trypanosomes in Australian bats. Recently a novel trypanosome species was identified in an adult female little red flying fox (Pteropus scapulatus) with clinical and pathological evidence of trypanosomosis. The present study used morphology and molecular methods to demonstrate that this trypanosome is a distinct species and we propose the name Trypanosoma teixeirae sp. n. Morphological comparison showed that its circulating trypomastigotes were significantly different from those of Trypanosoma pteropi and Trypanosoma hipposideri, two species previously described from Australian bats. Genetic information was not available for T. pteropi and T. hipposideri but phylogenetic analyses at the 18S ribosomal RNA (rRNA) and glycosomal glyceraldehyde phosphate dehydrogenase (gGAPDH) loci indicated that T. teixeirae sp. n. was genetically distinct and clustered with other bat-derived trypanosome species within the Trypanosoma cruzi clade.
1. Introduction
Bats (order Chiroptera) are reservoirs of numerous zoonotic pathogens including rabies, Australian bat lyssavirus, severe acute respiratory syndrome (SARS), Hendra virus, Nipah virus and Ebola virus (Wood et al., 2012). Trypanosomes are blood-borne flagellate protozoan parasites that can infect a wide range of vertebrate hosts including humans. Numerous trypanosome species have been identified in bats in Asia, Africa, South America and Europe (Hoare, 1972, Baker, 1973, Marinkelle, 1976, Marinkelle, 1979, Gardner and Molyneux, 1988a, Gardner and Molyneux, 1988b, Hamanaka and Pinto Ada, 1993, Steindel et al., 1998, Barnabe et al., 2003, Grisard et al., 2003, Lisboa et al., 2008, Cottontail et al., 2009, Maia da Silva et al., 2009, Cavazzana et al., 2010, Garcia et al., 2012, Hamilton et al., 2012, Lima et al., 2012, Lima et al., 2013, Marcili et al., 2013, Silva-Iturriza et al., 2013, Cottontail et al., 2014, Ramirez et al., 2014).
In Australia, three Trypanosoma spp. have been described in bats to date: Trypanosoma pteropi from the black flying fox (Pteropus gouldii) (Breinl, 1913, Mackerras, 1959), Trypanosoma hipposideri from the dusky horseshoe bat (Hipposideros bicolor albanensis) and Trypanosoma vegrandis, in pteropid bats (Yangochiroptera) and microbats (Yinpterochiroptera) (Austen et al., 2015). None of these have been associated with clinical disease. In addition, Mackie et al. (2015) recently described the first case of trypanosomosis in a little red flying fox (Pteropus scapulatus—suborder Yinpterochiroptera) from eastern Australia, caused by an apparently novel trypanosome species.
Molecular and phylogenetic studies have suggested that bat trypanosomes are implicated in the evolutionary history of the T. cruzi clade and may potentially be the precursor of trypanosomes from Australian marsupials and several African terrestrial mammals (Hamilton et al., 2012, Lima et al., 2013). There is however very limited knowledge about the genetic diversity of Australian bat trypanosomes, where only 9 of 76 indigenous bat species have been screened for this parasite (Thompson et al., 2014).
In the present study, we describe the morphological and genetic characterisation of the novel trypanosome in the little red flying fox (Mackie et al., 2015), for which we proposed the name Trypanosoma teixeirae sp. n.
2. Material and methods
2.1. Sample collection
A venous blood sample was collected from the cephalic vein of an adult female little red flying fox that presented to the Australia Zoo Wildlife Hospital (AZWH) in April, 2014. The flying fox had been rescued from the ground at Redcliffe in south-eastern Queensland, Australia and was moribund with anaemia and icterus. Clinical and pathological evidence of disease consistent with trypanosomosis in this flying fox was described by Mackie et al. (2015).
2.2. Morphological analyses
Thin blood smears were made from a drop of fresh blood and stained with Diff Quick (Siemens, Germany). After air-drying, the slides were then cover-slipped using DePeX mounting medium Gurr (Merck Pty. Limited, Kilsyth, Victoria, Australia). Stained films were systematically examined using a BX50 microscope (Olympus, Japan) with screen views generated by a DP Controller (version 3.2.1.276, Olympus, Japan). Digital light micrograph images of any trypomastigotes observed were taken at ×1000 magnification.
Digital images of the organisms identified in the blood films were used to measure key morphological features such as total length (TL), width (W), posterior to kinetoplast (PK), kinetoplast to nucleus (KN), nucleus to anterior (NA) and free flagellum (FF), according to parameters described by Hoare (1972) and Mackerras (1959). Means and standard errors were calculated. The morphological measurements were taken using the software Image J (Abramoff et al., 2004).
As two trypanosome species have previously been described in Australian bats based on morphological analysis only (Breinl, 1913, Mackerras, 1959), morphometrics of the novel trypanosome was compared statistically with available measurements for T. pteropi and T. hipposideri. Mean values for each morphological feature were calculated for T. teixeirae sp. n. whilst median values of reported ranges were used as input data for T. pteropi and T. hipposideri, as means were not available in the bibliographical references. Statistical analyses were conducted using the one sample t-test, in the software PAST 1.43 (Hammer et al., 2001).
2.3. DNA extraction
Genomic DNA was extracted from 200 μl of whole blood, using the MasterPure Purification Kit (Epicentre Biotechnologies, USA). A DNA extraction blank (with sterile molecular-grade water instead of blood) was included in the extraction to exclude the contamination of reagents and consumables with DNA.
2.4. 18S rRNA and GAPDH amplification and sequencing
A nested PCR protocol using generic Trypanosoma sp. primers SLF, S762R, S823F and S662R (Maslov et al., 1996, McInnes et al., 2009) was performed to amplify an approximately 900 bp fragment of the 18S rRNA gene, as previously described by McInnes et al. (2009). The DNA sample was also amplified at the glycosomal glyceraldehyde phosphate dehydrogenase (gGAPDH) gene using a heminested PCR protocol (McInnes et al., 2009).
PCR products were run on a 2% agarose gel containing SYBR Safe Gel Stain (Invitrogen, USA), and visualized with a dark reader trans-illuminator (Clare Chemical Research, USA). The gel bands were purified using an in-house filter tip method as previously described (Yang et al., 2013). All controls (positive, negative and DNA extraction blank) produced appropriate PCR results.
The purified PCR products were sequenced using the corresponding internal reverse primers diluted at 3.2 picomoles with an ABI PrismTM Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, California, USA) on an Applied Biosystem 3730 DNA Analyzer.
2.5. Phylogenetic analysis
Nucleotide sequences obtained at both 18S rRNA and GAPDH loci were aligned with additional trypanosome sequences retrieved from GenBank (Table 1 ) by MUSCLE (Edgar, 2004) using the default settings. Ambiguous regions containing gaps or poorly aligned were removed by Gblocks (Castresana, 2000), available on the Phylogeny.fr platform (Dereeper et al., 2008), using low stringency parameters. The curated alignments were imported into MEGA 6 (Tamura et al., 2013) and the most appropriate nucleotide substitution model was selected using the dedicated function.
Table 1.
Genbank accession numbers and sources (where known) of trypanosome isolates included in the phylogenetic analyses.
| Trypanosome species | Host origin | Geographic origin | GenBank accession numbers |
|
|---|---|---|---|---|
| 18S rDNA | gGAPDH | |||
| T. teixeirae sp. n. | Bat (Pteropus scapulatus) | Australia | KT907061 | KT907062 |
| T. rangeli | Bat (Platyrrhinus lineatus) | Brazil | FJ900242 | GQ140364 |
| T. minasense | Tamarin (Saguinus midas) | Japan | AJ012413 | AB362561 |
| T. dionisii | Bat (Eptesicus brasiliensis) | Brazil | FJ001666 | GQ140362 |
| T. erneyi | Bat (Tadarida sp.) | Mozambique | JN040987 | JN040964 |
| T. vespertilionis | Bat (Pipistrellus pipistrellus) | England | AJ009166 | AJ620283 |
| T. sp. bat | Bat (Rousettus aegyptiacus) | Gabon | AJ012418 | GQ140365 |
| T. livingstonei TCC1270 | Bat (Rhinolophus landeri) | Mozambique | KF192979 | KF192958 |
| T. livingstonei TCC1953 | Bat (Hipposideros caffer) | Mozambique | – | KF192969 |
| T. cruzi Tcbat | Bat (Myotis levis) | Brazil | FJ900241 | GQ140358 |
| T. cruzi marinkellei | Bat (Carollia perspicillata) | Brazil | FJ001664 | GQ140360 |
| T. cruzi Y | Human (Homo sapiens) | Brazil | AF301912 | GQ140353 |
| T. conorhini | Rat (Rattus rattus) | Brazil | AJ012411 | AJ620267 |
| T. sp. NanDoum1 | Palm civet (Nandinia binotata) | Cameroon | FM202492 | FM164793 |
| T. sp. HochNdi1 | Monkey (Cercopithecus nictitans) | Cameroon | FM202493 | FM164794 |
| T. sp. H25 | Kangaroo (Macropus giganteus) | Australia | AJ009168 | AJ620276 |
| T. sp. AP-2011-64 | Possum (Trichosurus vulpecular) | Australia | JN315383 | AJ620276 |
| T. sp. AB-2013-G8 | Woylie (Bettongia penicillata) | Australia | KC753537 | KC812988 |
| T. avium | Eagle (Aquila pomarina) | Slovakia | AF416559 | – |
| T. sp. AAT | Currawong (Strepera sp.) | Australia | AJ620557 | AJ620264 |
| T. bennetti | American kestrel (Falco sparverius) | Germany | AJ223562 | FJ649486 |
| T. irwini | Koala (Phascolarctos cinereus) | Australia | FJ649479 | FJ649485 |
| T. lewisi | Rat (Rattus rattus) | England | AJ009156 | AJ620272 |
| T. microti | Vole (Microtis agrestis) | England | AJ009158 | AJ620273 |
| T. vivax | Cattle | EU477537 | AF053744 | |
| T. brucei brucei | – | X59955 | ||
| T. brucei rhodesiense | Human (Homo sapiens) | Uganda | AJ009142 | – |
| T. brucei gambiense | Human (Homo sapiens) | Nigeria | AJ009141 | – |
| T. evansi | Capybara (H. hydrochaeris) | Brazil | AJ009154 | AF053743 |
| T. copemani Charlton | Koala (Phascolarctos cinereus) | Australia | GU966588 | – |
| T. copemani Mika | Koala (Phascolarctos cinereus) | Australia | – | GU966585 |
| T. copemani G1 | Woylie (Bettongia penicillata) | Australia | KC753530 | KC812982 |
| T. copemani G2 | Woylie (Bettongia penicillata) | Australia | KC753531 | KC812983 |
| T. gilletti | Koala (Phascolarctos cinereus) | Australia | GU966589 | GU966587 |
| T. vegrandis G3 | Woylie (Bettongia penicillata) | Australia | KC753533 | KC812984 |
| T. vegrandis G4 | Woylie (Bettongia penicillata) | Australia | KC753532 | KC812985 |
| T. vegrandis G5 | Woylie (Bettongia penicillata) | Australia | KC753534 | KC812986 |
| T. vegrandis G6 | Woylie (Bettongia penicillata) | Australia | KC753535 | – |
| T. vegrandis G7 | Woylie (Bettongia penicillata) | Australia | KC753536 | KC812987 |
| T. mega | African toad (Bufo regularis) | Africa | AJ009157 | AJ620253 |
| T. rotatorium | Bullfrog (Rana catesbeiana) | Canada | AJ009161 | AJ620256 |
| T. binneyi | Platypus (Ornithorhynchus anatinus) | Australia | AJ132351 | AJ620266 |
| T. granulosum | Eel (Anguilla anguilla) | Portugal | AJ620552 | – |
| T. sp. CLAR | Catfish (Clarias angolensis) | Africa | AJ620555 | AJ620251 |
The evolutionary histories at both 18S rRNA and GAPDH genes were inferred by using the Maximum Likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993). The gamma shape parameter was estimated directly from the data. Reliability for internal branch was assessed using the bootstrapping method (500 bootstrap replicates) and support values (>60%) indicated at the left of each node. The phylogenetic trees were drawn to scale, with branch lengths measured in the number of substitutions per site.
Estimates of genetic divergence between sequences were generated in MEGA 6 based on the Tamura-Nei algorithm, using uniform rates and a partial deletion of 95%.
3. Results
3.1. Microscopy and morphometric analysis of T. teixeirae sp. n.
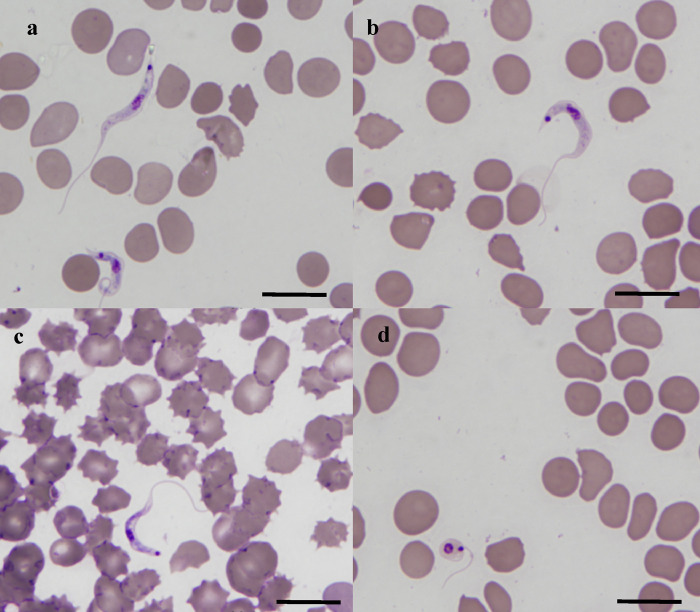
A total of nine organisms morphologically consistent with a trypanosome were detected by light microscopy in blood films from the little red flying fox. The extracellular organisms were slender with tapered ends, with long free flagellums and either an undeveloped or absent undulating membrane. A nearly central nucleus and a terminal small round deeply staining internal structure consistent with a kinetoplast were also observed (Fig. 1 a–c). The trypomastigotes varied in length from 20.4 to 30.8 μm (average 25.9 μm) and in width from 1.3 to 2.3 μm (average 1.9 μm) (Table 2 ).
Fig. 1.
Light photomicrographs of diff quick stained blood film showing Trypanosoma teixeirae sp. n. trypomastigotes in the blood of a red flying fox (Pteropus scapulatus) (a–c) and (d) Round epimastigote form. Scale bars represent 10 μm.
Table 2.
Mean dimensions and standard errors (S.E.) of morphological features of Trypanosoma teixeirae sp.n. isolated from a little red flying fox’s blood.
| Featurea | No. of organisms measured | Observed range (μm) | Mean ± S.E. (μm) |
|---|---|---|---|
| Total length | 8 | 20.4–30.8 | 25.9 ± 1.2 |
| Width | 8 | 1.3–2.3 | 1.9 ± 0.1 |
| PK | 7 | 1.5–2.4 | 2 ± 0.15 |
| KN | 9 | 3.3–6.2 | 4.9 ± 0.3 |
| NA | 9 | 5.1–9.8 | 7.8 ± 0.5 |
| FF | 8 | 10.0–12.9 | 11.3 ± 0.4 |
aTotal length: total body length measured along mid-line including free-flagellum.
Width: maximum width measured at nucleus level (undulating membrane included).
PK: distance between the posterior end and the kinetoplast.
KN: distance between the kinetoplast and posterior edge of the nucleus.
NA: distance between the anterior edge of the nucleus and the anterior end of the body.
FF: length of the free flagellum.
Among the nine long slender organisms observed, two were not true trypomastigotes as their kinetoplast was located at the very end of the posterior, what made it impossible to calculate the PK distance. In another instance, the trypomastigote’s free flagellum was apparently under a red blood cell, hence any measurements taken of FF or TL would have been inaccurate. We have therefore only measured what was feasible, which explains the divergence in the number of organisms measured for each morphological feature (Table 2).
Three flagellate round forms with a flagellum running round the organism about 90 degrees were also observed (Fig. 1d). Their body shape was consistent with a sphaeromastigote or a round epimastigote if their flagellar position was considered.
Morphometric analysis revealed that although the reported length and width ranges for the T. teixeirae sp. n. and T. pteropi overlap, the former was significantly longer and thinner than the latter (p < 0.01) (Table 3 ). There was no significant difference between KN, NA and FF dimensions between T. teixeirae sp. n. and T. pteropi. In addition, T. hipposideri was significantly smaller than T. teixeirae sp. n. for TL and FF dimensions (p < 0.01) (Table 3). No significant difference was observed for B, PK and KN between T. teixeirae sp. n. and T. hipposideri.
Table 3.
Comparison between morphological dimensions of blood trypomastigotes of Trypanosoma teixeirae sp. n. with Trypanosoma pteropi and Trypanosoma hipposideri.
| Morphological Feature (μm) |
||||||
|---|---|---|---|---|---|---|
| Total length | Width | PK | KN | NA | FF | |
| Trypanosoma teixeirae sp.n. | 20.4–30.8 25.9 | 1.3–2.3 | 1.5–2.4 | 3.3–6.2 | 5.1–9.8 | 10.0–12.9 |
| 1.9 | 2.0 | 4.9 | 7.8 | 11.3 | ||
| Trypanosoma pteropi | 18.0–22.0 | 2.0–4.0 | 1.5–4.0 | 4.0–5.0 | 8.0–10.0 | 8.0–12.0 |
| 20.0* | 3.0* | 2.75* | 4.5 | 9.0 | 10.0 | |
| Trypanosoma hipposideri | 10.5–13.0 | 1.5–2.0 | 1.0–2.5 | 4.0–6.0 | 1.5–5.0 | 4.0–8.0 |
| 11.65* | 1.75 | 1.75 | 5.0 | 3.25* | 6.0* | |
Total length: total body length measured along mid-line including free-flagellum; Width: maximum width measured at nucleus level (undulating membrane included); PK: distance between the posterior end and the kinetoplast; KN: distance between the kinetoplast and posterior edge of the nucleus; NA: distance between the anterior edge of the nucleus and the anterior end of the body; FF: length of the free flagellum. Ranges given with mean for Trypanosoma teixeirae sp. n. As the mean dimensions were not available for Trypanosoma pteropi or Trypanosoma hipposideri the median value of the range is presented in the table and was used for statistical analysis. For each column, values followed by an asterisk are significantly different to the T. teixeirae sp.n. value (p < 0.01).
3.2. Sequence and phylogenetic analysis
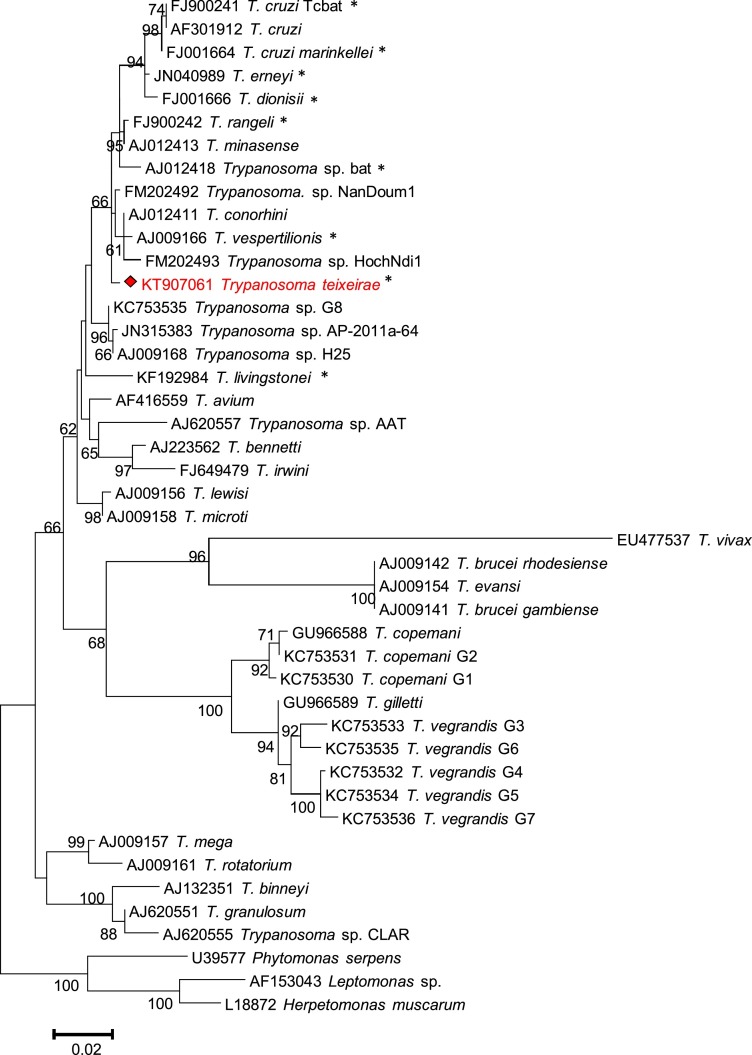
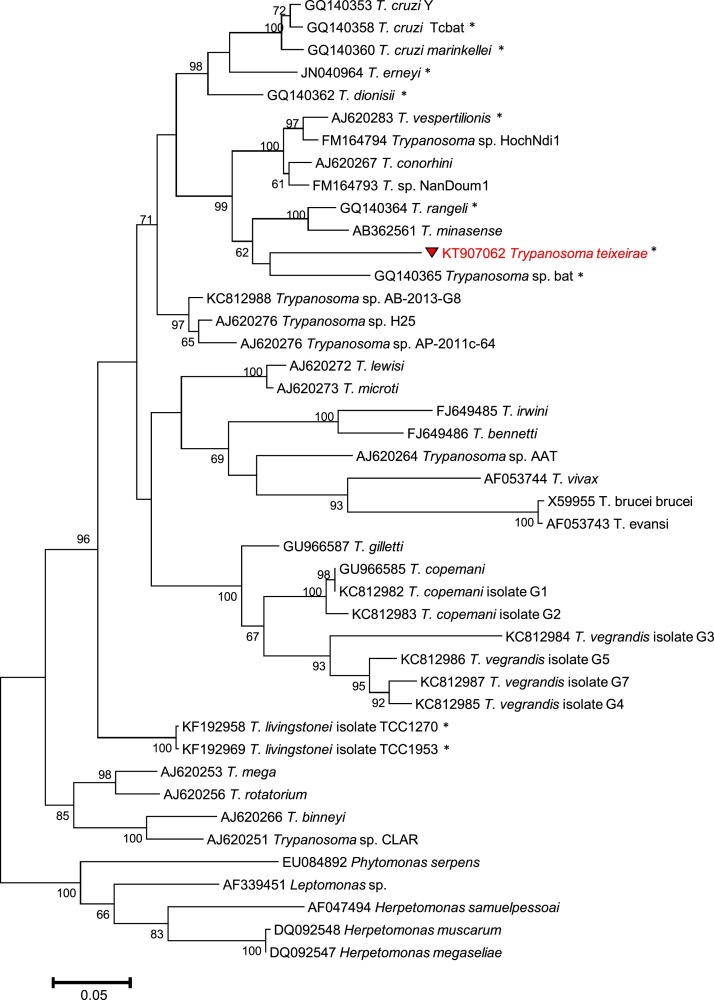
Maximum Likelihood analysis at both the 18S rDNA and GAPDH loci (Fig. 2, Fig. 3 , respectively) produced concordant tree topologies and revealed that T. teixeirae sp. n. grouped with other trypanosomes belonging to the T. cruzi clade, including 7 bat-derived isolates (T. cruzi Tcbat, T. cruzi marinkellei, T. erneyi, T. dionisii, T. rangeli, T. vespertilionis and T. sp. AJ012418/GQ140365) and three isolates from Australian marsupials (T. sp. H25 from a kangaroo- AJ009168/AJ620276; T. sp. AB-2003-G8 from a woylie- KC753537/KC812988; and T. sp. AP-2011-64 from a brush-tailed possum − JN315383/AJ620276). The phylogenetic trees also corroborated the evolutionary relationships among all major trypanosome clades described in previous broader analyses. Nucleotide sequences were obtained at both loci for T. teixeirae sp. n. were submitted to GenBank under the following accession numbers: KT907061 and KT907062.
Fig. 2.
Phylogenetic relationships of Trypanosoma teixeirae sp. n. with other trypanosomes, based on 18S rDNA partial sequences (∼730 bp). Evolutionary relationships were determined by Maximum Likelihood, based on the Tamura-Nei model (Tamura et al., 2013). Bootstrap values (>60%) based on 500 replicates are indicated at the left of each supported node. The scale bar is the proportion of base substitutions per site. Trypanosome species from bats are shown with an asterisk.
Fig. 3.
Phylogenetic relationships of Trypanosoma teixeirae sp. n. with other trypanosomes, based on gGAPDH partial sequences (∼775 bp). Evolutionary relationships were determined by Maximum Likelihood, based on the Tamura-Nei model (Tamura et al., 2013). Bootstrap values (>60%) based on 500 replicates are indicated at the left of each supported node. The scale bar is the proportion of base substitutions per site. Trypanosome species from bats are shown with an asterisk.
Estimates of evolutionary divergence between nucleotide sequences revealed that T. teixeirae sp. n. was genetically distinct but most closely related to T. minasense and T. rangeli (genetic distances of 1% at the 18S rDNA locus and 14%–15% at the gGAPDH, respectively) (Table 4 ).
Table 4.
Genetic distances between Trypanosoma teixeirae sp. n. and other trypanosome species at 18S rRNA and gGAPDH loci.
| Trypanosome species/isolate |
Genetic distances (%) |
|
|---|---|---|
| 18S rDNA | gGAPDH | |
| T. minasense (AJ012413/AB362561) | 1% | 14% |
| T. rangeli (FJ900242/GQ140364) | 1% | 15% |
| T. sp. bat (AJ012418/GQ140365) | 1% | 16% |
| T. conorhini (AJ012411/AJ620267) | 1% | 16% |
| T. sp. NanDoum1 (FM202492/FM164793) | 1% | 17% |
| T. vespertilionis (AJ009166/AJ620283) | 1% | 17% |
| T. sp. HochNdi1 (FM202493/FM164794) | 1% | 17% |
| T. sp. H25 (AJ009168/AJ620276) | 2% | 17% |
| T. sp. AB-2013-G8 (KC753537/KC812988) | 1% | 18% |
Genetic distances were calculated in MEGA 6 (Tamura et al., 2013) using the Tamura-Nei model (Tamura and Nei 1993).
4. Species description
Species Name: Trypanosoma teixeirae sp. n. (Fig. 1)
Type host: Little red flying fox (Pteropus scapulatus).
Other hosts: Unknown
Type Locality: Redcliffe peninsula, Queensland, Australia.
Prevalence: Unknown
4.1. Morphology
T. teixeirae sp. n. trypomastigotes are on average 25.9 μm long and 1.9 μm wide, exhibiting a slender shape with tapered ends, a nearly central nucleus and a small terminal kinetoplast. Undulating membrane either absent or under-developed.
4.2. Etymology
The species is named T. teixeirae sp. n. in honour of Prof. Marta Maria Geraldes Teixeira, from the University of Sao Paulo, who has greatly contributed to the biology and phylogeny of trypanosome species.
5. Discussion
In the present study, we have described T. teixeirae sp. n., the causative agent of trypanosomosis in an Australian little red flying fox, using morphological and molecular techniques. This is the fourth trypanosome species to be reported in indigenous Australian bats and the first one associated with clinical disease.
Unfortunately no genetic data was available for two of the previously reported bat-derived trypanosomes in Australia (T. pteropi and T. hipposideri). Trypansosoma pteropi was described as having a slender body (total length 18–22 μm; width 2–4 μm), an under-developed undulating membrane and a long free flagellum whereas T. hipposideri is very small and slender (total length 10.5–13 μm; width 1.5–2 μm), with a delicate short free flagellum at the anterior end (Breinl, 1913, Mackerras, 1959). Statistical analysis however, revealed that T. teixeirae sp. n. was significantly larger than both T. pteropi and T. hipposideri, even though they had several other overlapping morphometric features. However, morphology alone is not a reliable tool to delimit trypanosome species due to the interspecific similarities and intraspecific variability (Dunn et al., 1963, Marinkelle, 1966, Dunn, 1968).
Besides the typical trypomastigotes, two round forms (sphaeromastigotes or round epimastigotes) were also observed. The term ‘sphaeromastigote’ (Brack, 1968) refers to the parasite body shape only and has been applied without reference to the flagellar development. However, as these forms may occur within different stages of the parasite’s development, it is more appropriate to characterise the round organisms observed in the present study as round epimastigotes, considering both their body form and flagellar features (Elliott et al., 1974). This stage normally occurs in the interior of the cell, in vessels or in the insect gut.
Evolutionary reconstructions at both 18S rDNA and gGAPDH revealed that T. teixeirae sp. n. was genetically distinct from all known trypanosomes. The use of these two genes is recommended for taxonomic analysis of trypanosomatids and validation of new trypanosome species (Hamilton et al., 2004, Viola et al., 2009, Teixeira et al., 2011, Lima et al., 2012, Lima et al., 2013, Borghesan et al., 2013).
Phylogenetic analyses at both 18S rRNA and gGAPDH loci revealed that T. teixeirae sp. n. clustered within the T. cruzi clade together with all other bat-derived trypanosome species described to date, except T. livingstonei (which was positioned basal to the T. cruzi clade), T. evansi (which belongs to the T. brucei clade) and T. vegrandis (which forms a separate group associated with other marsupial-derived trypanosomes found in Australia) (Hamilton et al., 2007, Botero et al., 2013, Lima et al., 2013, Austen et al., 2015, Carnes et al., 2015). At the gGAPDH locus, T. teixeirae sp. n. was closest to T. minasense and T. rangeli and exhibited 14% and 15% genetic distance from these two species respectively. T. minasense has been found in neotropical non-human primates from South America (Ziccardi and Lourenco-de-Oliveira, 1999) whilst T. rangeli has been reported in a range of mammalian hosts including Brazilian bats (Maia da Silva et al., 2009). Although T. teixeirae sp. n. exhibited a relatively low (1%) genetic distance from its closest related species at the 18S rRNA locus, a similar pattern was observed when comparing other previously described species among each other. For instance, genetic distances between T. minasense and T. vespertilionis were 1% and 12% at the 18S rRNA and gGAPDH loci respectively. Trypanosomes have few morphological features detectable using light microscopy which can adequately delimit species (Gibson, 2009). Previous studies have reported that a genetic distance of 3.75% at the GAPDH gene is sufficient to delimit a new trypanosome species (McInnes et al., 2011). By this criterion, T. teixeirae sp. n. is clearly a separate species.
Bat trypanosomes have been implicated in the evolutionary origin of T. cruzi, the causative agent of Chagas disease, one of the most important public health issues in South America (Hamilton et al., 2012, Bonney, 2014). The ‘bat-seeding’ theory suggests that T. cruzi evolved from within a broad clade of bat-derived species, which have made the switch into terrestrial mammals (Hamilton et al., 2012, Lima et al., 2013). The theory also implies that these arboreal trypanosomes species could potentially be evolutionary precursors for the terrestrial trypanosome lineage within Australian mammals (Hamilton et al., 2012, Lima et al., 2013, Thompson et al., 2014).
It is therefore possible that T. teixeirae sp. n. could be the precursor of three marsupial-derived trypanosomes belonging to the T. cruzi clade: T. sp. H25 (Averis et al., 2009), T. sp. AP-2011-64 (Paparini et al., 2011) and T. sp. AB-2013-G8 (Botero et al., 2013). As most native bat species remain unsampled (Thompson et al., 2014), future studies are required to provide more evidence to support the ‘bat-seeding’ theory in Australia and elucidate evolutionary relationships between trypanosomes.
Similarly to most bat trypanosomes described worldwide, the prevalence, distribution, vectors, life cycle and zoonotic potential of T. teixeirae sp. n. remain unclear. Therefore, more studies comprising a larger sample size are required to better understand the prevalence and clinical impacts of T. teixeirae sp. n. on bat populations, taking into account ecological and stress factors that could play a role in the expression of clinical disease.
References
- Abramoff M.D., Magalhaes P.J., Ram S.J. Image processing with image. J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Austen J.M., O'Dea M., Jackson B., Ryan U. High prevalence of Trypanosoma vegrandis in bats from Western Australia. Vet. Parasitol. 2015;142:1443–1452. doi: 10.1016/j.vetpar.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averis S., Thompson R.C., Lymbery A.J., Wayne A.F., Morris K.D., Smith A. The diversity, distribution and host-parasite associations of trypanosomes in Western Australian wildlife. Parasitology. 2009;136:1269–1279. doi: 10.1017/S0031182009990801. [DOI] [PubMed] [Google Scholar]
- Baker J.R. First european record of Trypanosoma (Megatrypanum) sp. of bats. Nat. New Biol. 1973;241:96. doi: 10.1038/newbio241096a0. [DOI] [PubMed] [Google Scholar]
- Barnabe C., Brisse S., Tibayrenc M. Phylogenetic diversity of bat trypanosomes of subgenus Schizotrypanum based on multilocus enzyme electrophoresis, random amplified polymorphic DNA, and cytochrome b nucleotide sequence analyses. Infect. Genet. Evol. 2003;2:201–208. doi: 10.1016/s1567-1348(02)00130-2. [DOI] [PubMed] [Google Scholar]
- Bonney K.M. Chagas disease in the 21st century: a public health success or an emerging threat? Parasite. 2014;21:11. doi: 10.1051/parasite/2014012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesan T.C., Ferreira R.C., Takata C.S., Campaner M., Borda C.C., Paiva F., Milder R.V., Teixeira M.M., Camargo E.P. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164:129–152. doi: 10.1016/j.protis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Botero A., Thompson C.K., Peacock C.S., Clode P.L., Nicholls P.K., Wayne A.F., Lymbery A.J., Thompson R.C. Trypanosomes genetic diversity, polyparasitism and the population decline of the critically endangered Australian marsupial, the brush tailed bettong or woylie (Bettongia penicillata) Int. J. Parasitol. Parasites Wildl. 2013;2:77–89. doi: 10.1016/j.ijppaw.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack C. Elektronenmikroskopische untersuchungen zum lebenszyklus von Trypanosoma cruzi unter besonderer beruckischtigung der entwicklungsformen im ubertrager Rhodnium prolixus. Acta Trop. 1968;25 [PubMed] [Google Scholar]
- Breinl A. Parasite protozoa encountered in the blood of Australian native animals. Aust. Inst. Trop. Med. 1913:30–38. [Google Scholar]
- Carnes J., Anupama A., Balmer O., Jackson A., Lewis M., Brown R., Cestari I., Desquesnes M., Gendrin C., Hertz-Fowler C., Imamura H., Ivens A., Koreny L., Lai D.H., MacLeod A., McDermott S.M., Merritt C., Monnerat S., Moon W., Myler P., Phan I., Ramasamy G., Sivam D., Lun Z.R., Lukes J., Stuart K., Schnaufer A. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl. Trop. Dis. 2015;9:e3404. doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavazzana M., Jr., Marcili A., Lima L., da Silva F.M., Junqueira A.C., Veludo H.H., Viola L.B., Campaner M., Nunes V.L., Paiva F., Coura J.R., Camargo E.P., Teixeira M.M. Phylogeographical, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotrypanum parasitic in Brazilian bats. Int. J. Parasitol. 2010;40:345–355. doi: 10.1016/j.ijpara.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Cottontail V.M., Wellinghausen N., Kalko E.K. Habitat fragmentation and haemoparasites in the common fruit bat, Artibeus jamaicensis (Phyllostomidae) in a tropical lowland forest in Panama. Parasitology. 2009;136:1133–1145. doi: 10.1017/S0031182009990485. [DOI] [PubMed] [Google Scholar]
- Cottontail V.M., Kalko E.K., Cottontail I., Wellinghausen N., Tschapka M., Perkins S.L., Pinto C.M. High local diversity of Trypanosoma in a common bat species, and implications for the biogeography and taxonomy of the T. cruzi clade. PLoS One. 2014;9:e108603. doi: 10.1371/journal.pone.0108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the nonspecialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn F.L., Lambrecht F.L., Duplessis R. Trypanosomes of south american monkeys and marmosets. Am. J. Trop. Med. Hyg. 1963;12:524–534. doi: 10.4269/ajtmh.1963.12.524. [DOI] [PubMed] [Google Scholar]
- Dunn F.L. The TIF direct smear as an epidemiological tool; with special reference to counting helminth eggs. Bull. World Health Organ. 1968;39:439–449. [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott K., O'Connor M., Wolstenholme G.E.W. John Wiley & Sons, Ltd.; Chichester, UK: 1974. Trypanosomiasis and Leishmaniasis (with Special Reference to Chagas’ Disease) [Google Scholar]
- Garcia L., Ortiz S., Osorio G., Torrico M.C., Torrico F., Solari A. Phylogenetic analysis of Bolivian bat trypanosomes of the subgenus schizotrypanum based on cytochrome B sequence and minicircle analyses. PLoS One. 2012;7:e36578. doi: 10.1371/journal.pone.0036578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.A., Molyneux D.H. Schizotrypanum in british bats. Parasitology. 1988;97:43–50. doi: 10.1017/s0031182000066725. Pt 1. [DOI] [PubMed] [Google Scholar]
- Gardner R.A., Molyneux D.H. Trypanosoma (Megatrypanum) incertum from Pipistrellus pipistrellus: development and transmission by cimicid bugs. Parasitology. 1988;96:433–447. doi: 10.1017/s0031182000080082. Pt 3. [DOI] [PubMed] [Google Scholar]
- Gibson W. Species-specific probes for the identification of the African tsetse-transmitted trypanosomes. Parasitology. 2009;136:1501–1507. doi: 10.1017/S0031182009006179. [DOI] [PubMed] [Google Scholar]
- Grisard E.C., Sturm N.R., Campbell D.A. A new species of trypanosome Trypanosoma desterrensis sp. n., isolated from South American bats. Parasitology. 2003;127:265–271. doi: 10.1017/s0031182003003536. [DOI] [PubMed] [Google Scholar]
- Hamanaka S.I., Pinto Ada S. Growth and differentiation on a trypanosome of the subgenus Schizotrypanum from the bat Phyllostomus hastatus. Rev. Soc. Bras. Med. Trop. 1993;26:225–230. doi: 10.1590/s0037-86821993000400005. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Stevens J.R., Gaunt M.W., Gidley J., Gibson W.C. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int. J. Parasitol. 2004;34:1393–1404. doi: 10.1016/j.ijpara.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Gibson W.C., Stevens J.R. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol. Phylogenet. Evol. 2007;44:15–25. doi: 10.1016/j.ympev.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Teixeira M.M., Stevens J.R. The evolution of Trypanosoma cruzi: the ‘bat seeding’ hypothesis. Trends Parasitol. 2012;28:136–141. doi: 10.1016/j.pt.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Hammer Ø., Harper D.A.T., Ryan P.D. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:9. [Google Scholar]
- Hoare C.A. Blackwell Scientific Publishing; Oxford, England: 1972. The Trypanosomes of Mammals: A Zoological Monograph. [Google Scholar]
- Lima L., Silva F.M., Neves L., Attias M., Takata C.S., Campaner M., de Souza W., Hamilton P.B., Teixeira M.M. Evolutionary insights from bat trypanosomes: morphological, developmental and phylogenetic evidence of a new species Trypanosoma (Schizotrypanum) erneyi sp. nov., in African bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012;163:856–872. doi: 10.1016/j.protis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Lima L., Espinosa-Alvarez O., Hamilton P.B., Neves L., Takata C.S., Campaner M., Attias M., de Souza W., Camargo E.P., Teixeira M.M. Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasites Vectors. 2013;6:221. doi: 10.1186/1756-3305-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa C.V., Pinho A.P., Herrera H.M., Gerhardt M., Cupolillo E., Jansen A.M. Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) genotypes in neotropical bats in Brazil. Vet. Parasitol. 2008;156:314–318. doi: 10.1016/j.vetpar.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Mackerras M.J. The haematozoa of Australian mammals. Aust. J. Zool. 1959;7:105–135. [Google Scholar]
- Mackie J.T., Stenner R., Gillett A., Barbosa A.D., Ryan U., Irwin P. Trypanosomiasis in an Australian little red flying-fox (Pteropus scapulatus) J. Vet. Diagn. Invest. 2015 doi: 10.1111/avj.12597. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia da Silva F., Marcili A., Lima L., Cavazzana M., Jr., Ortiz P.A., Campaner M., Takeda G.F., Paiva F., Nunes V.L., Camargo E.P., Teixeira M.M. Trypanosoma rangeli isolates of bats from Central Brazil: genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Trop. 2009;109:199–207. doi: 10.1016/j.actatropica.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Marcili A., da Costa A.P., Soares H.S., Acosta Ida C., de Lima J.T., Minervino A.H., Melo A.T., Aguiar D.M., Pacheco R.C., Gennari S.M. Isolation and phylogenetic relationships of bat trypanosomes from different biomes in Mato Grosso, Brazil. J. Parasitol. 2013;99:1071–1076. doi: 10.1645/12-156.1. [DOI] [PubMed] [Google Scholar]
- Marinkelle C.J. Observations on human, monkey and bat trypanosomes and their vectors in Colombia. Trans. R. Soc. Trop. Med. Hyg. 1966;60:109–116. [Google Scholar]
- Marinkelle C.J. The Biology of the Trypanosomes of Bats. Biology of the Kinetoplastida. In: Lumdsen W.H.R., Evans D.A., editors. Academic; New York: 1976. pp. 175–216. [Google Scholar]
- Marinkelle C.J. Trypanosoma (Megatrypanum) megachiropterum sp. n. from the flying fox Pteropus tonganus, Quoy, Gaimard. J. Protozool. 1979;26:352–353. [Google Scholar]
- Maslov D.A., Lukes J., Jirku M., Simpson L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: implications for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 1996;75:197–205. doi: 10.1016/0166-6851(95)02526-x. [DOI] [PubMed] [Google Scholar]
- McInnes L.M., Gillett A., Ryan U.M., Austen J., Campbell R.S., Hanger J., Reid S.A. Trypanosoma irwini n. sp. (Sarcomastigophora: trypanosomatidae) from the koala (Phascolarctos cinereus) Parasitology. 2009;136:875–885. doi: 10.1017/S0031182009006313. [DOI] [PubMed] [Google Scholar]
- McInnes L.M., Hanger J., Simmons G., Reid S.A., Ryan U.M. Novel trypanosome Trypanosoma gilletti sp. (Euglenozoa: Trypanosomatidae) and the extension of the host range of Trypanosoma copemani to include the koala (Phascolarctos cinereus) Parasitology. 2011;138:59–70. doi: 10.1017/S0031182010000971. [DOI] [PubMed] [Google Scholar]
- Paparini A., Irwin P.J., Warren K., McInnes L.M., de Tores P., Ryan U.M. Identification of novel trypanosome genotypes in native Australian marsupials. Vet. Parasitol. 2011;183:21–30. doi: 10.1016/j.vetpar.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Ramirez J.D., Tapia-Calle G., Munoz-Cruz G., Poveda C., Rendon L.M., Hincapie E., Guhl F. Trypanosome species in neo-tropical bats: biological, evolutionary and epidemiological implications. Infect. Genet. Evol. 2014;22:250–256. doi: 10.1016/j.meegid.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Iturriza A., Nassar J.M., Garcia-Rawlins A.M., Rosales R., Mijares A. Trypanosoma evansi kDNA minicircle found in the Venezuelan nectar-feeding bat Leptonycteris curasoae (Glossophaginae), supports the hypothesis of multiple origins of that parasite in South America. Parasitol. Int. 2013;62:95–99. doi: 10.1016/j.parint.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Steindel M., Grisard E.C., de Carvalho Pinto C.J., Cordeiro F.D., Ribeiro-Rodrigues R., Romanha A.J. Characterization of trypanosomes from the subgenus Schizotrypanum isolated from bats Eptesicus sp. (Chiroptera: Vespertilionidae), captured in Florianopolis, Santa Catarina State, Brazil. J. Parasitol. 1998;84:601–607. [PubMed] [Google Scholar]
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.M.G., Borghesan T.C., Ferreira R.C., Santos M.A., Takata C.S., Campaner M., Nunes V.L., Milder R.V., de Souza W., Camargo E.P. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of Proteobacterial symbionts. Protist. 2011;162:503–524. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Thompson C.K., Godfrey S.S., Thompson R.C. Trypanosomes of australian mammals: a review. Int. J. Parasitol. Parasites Wildl. 2014;3:57–66. doi: 10.1016/j.ijppaw.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola L.B., Attias M., Takata C.S., Campaner M., De Souza W., Camargo E.P., Teixeira M.M. Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake Trypanosomes: Trypanosoma serpentis n. sp. from Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. J. Eukaryot. Microbiol. 2009;56:594–602. doi: 10.1111/j.1550-7408.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- Wood J.L., Leach M., Waldman L., Macgregor H., Fooks A.R., Jones K.E., Restif O., Dechmann D., Hayman D.T., Baker K.S., Peel A.J., Kamins A.O., Fahr J., Ntiamoa-Baidu Y., Suu-Ire R., Breiman R.F., Epstein J.H., Field H.E., Cunningham A.A. A framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos. Trans. R Soc. Lond. B Biol. Sci. 2012;367:2881–2892. doi: 10.1098/rstb.2012.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Murphy C., Song Y., Ng-Hublin J., Estcourt A., Hijjawi N., Chalmers R., Hadfield S., Bath A., Gordon C., Ryan U. Specific and quantitative detection and identification of Cryptosporidium hominis and C. parvum in clinical and environmental samples. Exp. Parasitol. 2013;135:142–147. doi: 10.1016/j.exppara.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Ziccardi M., Lourenco-de-Oliveira R. Polymorphism in trypomastigotes of Trypanosoma (Megatrypanum) minasense in the blood of experimentally infected squirrel monkey and marmosets. Mem. Inst. Oswaldo Cruz. 1999;94:649–653. doi: 10.1590/s0074-02761999000500016. [DOI] [PubMed] [Google Scholar]