Abstract
Small non-coding RNAs, which are 20–25 nucleotide ribonucleic acids, have emerged as an important transformation in the biological evolution over almost three decades. microRNAs (miRNAs) and short interfering RNAs (siRNAs) are two significant categories of the small RNAs that exert important effects on bone endocrinology and skeletology. Therefore, clarifying the expression and function of these important molecules in bone endocrine physiology and pathology is of great significance for improving their potential therapeutic value for metabolism-associated bone diseases. In the present review, we highlight the recent advances made in understanding the function and molecular mechanism of these small non-coding RNAs in bone metabolism, especially their potentially therapeutic values in bone-related diseases.
Keywords: miRNA, siRNA, Bone metabolism, Targeting therapeutics
Highlights
-
•
This review systematically investigated the expression and function of miRNA in bone endocrine physiology and pathology.
-
•
Comprehensive summary of recent advances made in understanding the potential therapeutic of miRNA and siRNA both in vivo and in vitro.
-
•
This study extensively generalized the delivery systems, clinical status and challenges in small non-coding RNAs-based therapeutics for bone-related disorders, together with suggested corresponding countermeasures.
1. Introduction
The discovery of small non-coding RNAs, including microRNAs (miRNAs) and short interfering RNAs (siRNAs) (Hamilton and Baulcombe, 1999, Lee et al., 1993, Reinhart et al., 2000, Aravin et al., 2007) has revolutionized our understanding of the regulation of gene expression. The effects of small RNAs on gene expression and control are generally inhibitory, and the corresponding regulatory mechanisms are therefore collectively subsumed under the heading of RNA silencing. The central theme throughout is that the small non-coding RNAs mainly serve as specific factors that combine effector proteins with target messenger RNAs (mRNAs) through complementary base pairing, and the core component of the effector machinery is a member of the argonaute protein superfamily. Compared with conventional small therapeutic molecules, miRNAs and siRNAs offer the advantages of being highly potent and able to act on “non-druggable” targets (for instance, proteins that lack an enzymatic function or that have a conformation that is inaccessible to traditional drug molecules) because they can be designed to affect virtually any gene of interest (Daka and Peer, 2012).
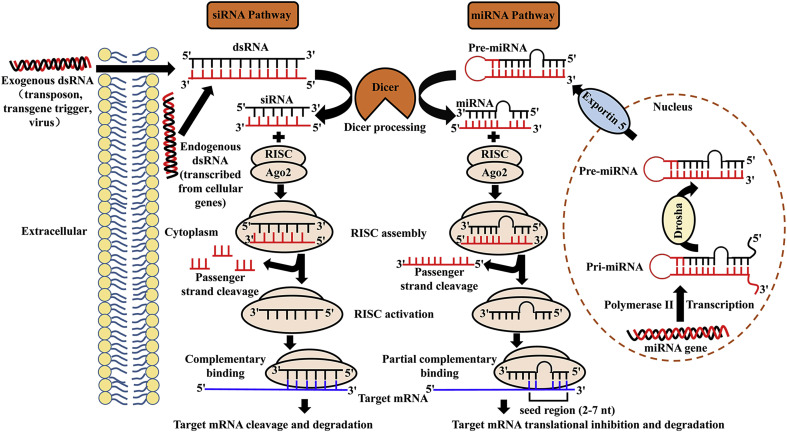
The regulation of genes and genomes mediated by miRNAs and siRNAs can occur at the levels of chromosome segregation, chromatin structure, RNA processing, RNA stability, transcription or translation (Carthew and Sontheimer, 2009). miRNAs, which are 20–25 nucleotides RNAs that regulate gene expression by targeting mRNAs at the post-transcriptional level, are purposefully expressed products of the genome originating from the organism itself (Liu and Paroo, 2010, Ha and Kim, 2014). miRNA gene transcription is executed by RNA polymerase II in the nucleus, leading to the formation of primary miRNA (pri-miRNA) with a double-stranded stem-loop structure. Then, the pri-miRNA is cleaved by a microprocessor complex (comprised of Drosha and the microprocessor complex subunit DCGR8) to form precursor miRNA (pre-miRNA, 10-100 nt). Subsequently, the pre-miRNA is transported by Exportin 5 from the nucleus to the cytoplasm. siRNAs, which are derived from the transposon, transgene trigger and virus or are transcribed from cellular genes, usually consist of 20–25 base pairs, and they regulate gene expression via a highly precise mechanism of sequence-directed gene silencing. After transporting to the cytoplasm, pre-miRNA and long double-stranded RNA (dsRNA) are processed by Dicer (known as a specialized ribonuclease (RNase) III-like enzyme) to form miRNA and siRNA, respectively. Then, both the passenger strand (sense strand) and the guide strand (antisense strand) of miRNA and siRNA are cleaved by the Argonaute 2 (AGO2) component, which belongs to RNA-induced silencing complexes (RISCs). Subsequently, the guide strand (antisense strand) of miRNA and siRNA remains associated with the RISC, whereas the passenger strand is detached from the active RISC. Finally, the guide strand of the miRNA guide the active RISC to its target mRNA with partial complementarity. The complementary pairing between mRNA and the mature miRNA typically occurs at the 3′ untranslated region (UTR) of the former and the seed region (nucleotides 2–7 from the 5′ end) of the latter, and other miRNA binding sites, such as the centred sites, 3’ supplementary sites and bulged sites are considered to be atypical. In contrast, the siRNA must be fully complementary to its target mRNA (Meister and Tuschl, 2004, Hammond et al., 2000). Different combination modes between miRNA and siRNA with target mRNA produce different target gene silencing effects, which are closely related to common events (such as Dicer processing and RISC formation). The regulation of miRNA or siRNA degradation can occur at either the transcriptional or post-transcriptional level. Transcription is a major point of regulation in miRNA biogenesis, and it is always closely associated with the activity of polymerase II (Kim et al., 2009). Drosha processing confers another important point of biogenesis for both miRNA and siRNA. For instance, a group of exoribonucleases called small RNA-degrading nuclease (SDN) proteins were reported to affect the stability of miRNA in plants (Ramachandran and Chen, 2008), however, which nucleases are responsible for miRNA degradation in animals remains unknown. There are two prominent questions closely related to the degradation of miRNA or siRNA, which are, how they are specifically marked for destruction and which ribonucleases catalyse the degradation reaction. The major difference between miRNAs and siRNAs is that the former have multiple mRNA targets, whereas the latter are highly specific with only one target. Since the discovery of siRNA, dsRNAs have been used as research tools to study the gene functions of various cell types. However, in mammalian cells, it has been verified that the delivery of long dsRNAs is closely related to the activation of the interferon (IFN) pathway, which is part of the defence mechanism against viral infection (Gantier and Williams, 2007). Because this effect can be either sequence-dependent or -independent, special care should be taken when designing siRNA therapeutics. The mechanism and pathway of miRNA and siRNA gene silencing are summarized in Fig. 1 .
Fig. 1.
Schematic gene silencing mechanism of miRNA and siRNA. miRNA gene transcription is executed by RNA polymerase II in the nucleus, leading to the formation of pri-miRNA, and then cleaved by Drosha to form pre-miRNA. Subsequently, the pre-miRNA is transported by Exportin 5 from the nucleus to the cytoplasm. siRNAs, which are exogenous products in origin, are derived directly from the transposon, transgene trigger or virus. After transporting to the cytoplasm, pre-miRNA and long dsRNA are both processed by Dicer to form miRNA and siRNA, separately. Then, both the sense strand of miRNA and siRNA are cleaved by AGO2, a component of RISC. The remaining anti-sense strand of miRNA and siRNA guide the active RISC to their target mRNA with partial complementarity and complementarity, respectively, leading to different target gene silencing effects. AGO2, Argonaute 2. RISC, RNA-induced silencing complexes.
Bone is a remarkable tissue playing key roles in critical functions in human physiology, consisting of fortification, movement and support of other vital organs, mineral storage, homeostasis and housing multiple progenitor cells, such as hematopoietic mesenchymal cells. Many cell subsets, including osteoblasts, osteoclasts and adipocytes, are involved in bone development, and the balance of bone formation, remodelling and reabsorption is extremely important to normal bone metabolism (Caplan and Dennis, 2006, Gimble et al., 1996, Gimble et al., 2006). Osteoblasts are bone-forming cells derived from bone marrow stromal cells, and they produce dense, cross-linked collagen and specialized proteins. Osteoclasts are large, multinucleated, bone-resorbing cells that arise from a monocyte-macrophage lineage, and they secrete hydrogen ions to dissolve the mineral component of bone matrix and a lytic enzyme, cathepsin K, to digest the bone matrix (mainly type I collagen). Osteocytes are star-shaped cells that represent terminally differentiated osteoblasts, they are commonly found in mature bone, and they can permeate the mineralized bone matrix. Bone is remodelled continuously during adulthood through the resorption of old bone by osteoclasts and the subsequent formation of new bone by osteoblasts (Martin, 2014).
A number of bone-related diseases, such as bone defects, osteoporosis, osteolysis, osteonecrosis, osteoarthritis and bone cancer, lead to a serious burden on the quality of life worldwide. Bone destruction begins to exceed bone formation after the age of 40 years old, leading to a local or systemic reduction of bone mass, which is well known as osteoporosis. For individuals with osteoporosis, bone fractures represent a life-threatening event, and the risk of fracture doubles for every 10% of bone that is lost (Rodan and Martin, 2000), while bone metastasis occurs in approximately 70% of patients with advanced prostate or breast cancer (Suva et al., 2011). Bone metastases leading to excess bone loss are generally classified as osteolytic (bone destruction), and those leading to bone deposition are regarded as osteoblastic (bone formation). Both bone degradation and deposition have been verified to occur in the early metastatic process, and they are always incurable once tumours metastasize to bones (Suva et al., 2011). Our increasing molecular understanding of disease-causing or disease-promoting genes of bone pathology has led to the discovery of numerous new molecular targets for the prevention and treatment of various of bone-related disorders. As a flourishing scientific field, the molecular mechanisms and therapeutic applications of the small non-coding RNAs (miRNAs and siRNAs) involved in bone-related diseases have been widely investigated in numerous studies. However, to date, no miRNA or siRNA targeting therapy-based clinical trials for musculoskeletal disorders have been initiated. This current review will focus on the roles of miRNAs in the regulation of bone formation and metabolism with an emphasis on the potential therapeutic strategies meidiated by miRNAs and siRNAs for the treatment of bone-related diseases.
2. Participation of miRNAs in bone formation
2.1. Osteogenesis-related miRNAs and target transcription factors
Bone mesenchymal stem cells (BMSCs), originating from the bone marrow possess multiple differentiation potentials. They can differentiate into osteoblasts, adipocytes and chondrocytes (Tencerova and Kassem, 2016). Osteoblasts are the major cell types in bone development and tissue engineering (Noronha-Matos et al., 2014). Multiple transcription factors participate in osteogenic differentiation, and the production and mineralization of extracellular matrix (ECM) synthesized by osteoblasts are finely regulated by different miRNAs. Among them, Runx2 and its downstream molecule Osterix are the most pivotal osteoblast-specific transcription factors that activate the relevant genes involved in the differentiation of preosteoblasts into mature osteoblasts and osteocytes, and the molecular mechanisms of the two bone -specific factors in the regulation of osteoblastogenesis were extensively described in our previous review (Fang et al., 2015).
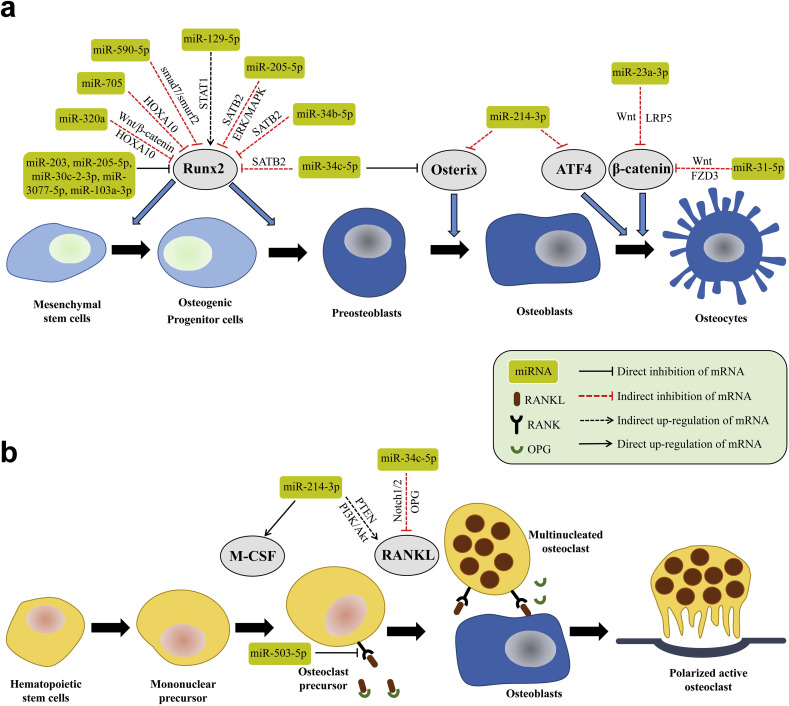
Many miRNAs are expressed during the osteogenic process induced in BMSCs, including cell proliferation, growth, matrix maturation and mineralization during the new bone formation (Fig. 2 a). Several miRNA-profiling tools, such as high throughput sequencing of small RNA libraries, quantitative real time-polymerase chain reaction (RT-PCR) with locked nucleic acid primers, and microarray analysis, have been used to screen the osteogenesis-associated miRNAs (Hu et al., 2010, Li et al., 2009, Baglìo et al., 2013). Microarray analysis, which has been widely used for the investigation of genome-wide miRNA expression, can provide the fluctuating information about various miRNAs during osteogenic differentiation, while miRNA array profiling methods have been applied for the identification of miRNAs related to osteogenic differentiation in different cell lines. Although many miRNAs have been confirmed either to descend or to ascend during the osteogenic differentiation (Li et al., 2009, Baglìo et al., 2013, Inose et al., 2009, Li et al., 2008, Eskildsen et al., 2011), few of them, including hsa-miR-15b-5p, mmu-miR-29b-2-5p, hsa-miR-34a-3p, mmu-miR- 93-5p, hsa-miR-138-5p, hsa-miR-140-5p, mmu-miR-181a-5p, hsa-miR-196a-5p, mmu-miR-210-3p, hsa-miR-218-5p, mmu-miR-335-5p, hsa-miR-338-3p, hsa-miR-346, mmu-miR-433-3p, hsa-miR-2861 and mmu-miR-3960 (Fang et al., 2015), are actually concerned with the gene expression related to skeletal formation. The biological, dynamic characteristics of several vital osteo-related miRNAs were discussed in our previous review (Fang et al., 2015). To clarify the impact on the expression profiling of miRNAs during bone metabolism, Laxman et al. performed next-generation sequencing of miRNA derived from primary human osteoblast (HOB) cells treated with dexamethasone (DEX) and parathyroid hormone (PTH). They discovered that several miRNAs such as hsa-miR-205-5p, hsa-miR-203 and hsa-miR-30c-2-3p targeting runt-related transcription factor 2 (Runx2), and hsa-miR- 320a targeting β-catenin, exhibited substantially different targeting gene-expression levels between DEX and PTH treatment (Laxman et al., 2016). Huang and colleagues reported that hsa-miR-320a negatively regulated the osteogenic differentiation of hBMSCs by repressing homeobox a10 (HOXA10) (Huang et al., 2016). HOXA10 is a bone morphogenetic protein 2 (BMP-2)-inducible gene, and it promotes osteogenic differentiation by activating Runx2 and its target genes (Hassan et al., 2007). Additionally, mmu-miR-705 and mmu-miR-3077-5p were also observed to be negative regulators of osteoblast differentiation by targeting HOXA10 (Liao et al., 2013a). Vishal et al. displayed that hsa-miR-590-5p enhanced osteoblast differentiation by indirectly stabilizing and protecting Runx2 from Smad-specific E3 ubiquitin protein ligase 2 (Smurf2)-mediated degradation by targeting Smad7, which is antagonistic to osteoblast function (Vishal et al., 2017). Intriguingly, mechanical stimulation has been confirmed to trigger osteoblast differentiation via several mechanosignal transduction pathways, playing a momentous role in bone homeostasis (Harada and Rodan, 2003, Zaidi, 2007, Hsieh and Turner, 2001, Reijnders et al., 2007). Zuo et al. reported a mechanosensitive miRNA, hsa-miR-103a-3p, that exhibited negative effects on Runx2 during cyclic mechanical stretch (CMS)-induced osteoblastogenesis, leading to decreased bone formation both in vitro and in vivo (Zuo et al., 2015). This innovative research has provided us with a feasible anabolic strategy for the treatment of skeletal disorders resulting from pathological mechanical loading with therapeutic inhibition of has-miR-103a-3p. It is well known that the skeleton is notably affected by the reduced differentiation capacity of stem cells with age (Liu and Rando, 2011). Weilner and colleagues reported for the first time that the microRNA content of microvesicles from senescent cells contributes to an aged systemic environment for compromised osteoblastogenesis (Weilner et al., 2016). They indicated that endothelial hsa-miR-31-5p, which is secreted within senescent cell-derived microvesicles, is taken up by mesenchymal stem cells, in which it suppresses osteogenic differentiation by knocking down Frizzled-3. In addition, it has been pre-clinically and successfully tested in critical size defects models (Deng et al., 2013, Deng et al., 2014), making hsa-miR-31-5p a very exciting target for bone tissue therapeutics in the near the future. Taken together, different miRNAs involved in bone formation exhibit diverse dynamic patterns, which could provide us with abundant information about the regulation of osteogenesis. However, various in vitro observations remain to be investigate further pre-clinically in vivo, and then some potential and promising candidates can be screened out after deliberative exploration.
Fig. 2.
Schematic summary of miRNA role in osteoblast and osteoclast differentiation. (a) miRNAs regulating osteoblast differentiation, proliferation and function. A cohort of transcription factors tightly regulate osteoblast commitment, such as Runx2, Osterix, ATF4 and β-catenin, from osteoprogenitors during skeletal development. (b) miRNAs effect various of molecules related to osteoclast commitment, such as RANKL and M-CSF, leading to changes in osteoclast activity in vitro as well as alterations in bone resorption in vivo. ATF4, activating transcription factor 4. HOXA10, homeobox a10. FZD3, Frizzled-3. LRP5, lipoprotein-receptor-related protein 5. M-CSF, macrophage colony-stimulating factor. Runx2, runt-related transcription factor 2. OPG, osteopontegrin. PTEN, phosphatase and tensin homologue. RANKL, receptor activator NFκB ligand. STAT1, signal transducer and activator of transcription 1. SATB2, special AT-rich sequence-binding protein 2.
2.2. miRNAs control bone specific signalling pathways
It has been proved that miRNAs target mRNAs through complementary base pairing to different sites in the 3’-untranslated regions, and they exert their biological functions through translational repression and/or being followed by deadenylation and degradation of their mRNA targets (Bazzini et al., 2012, Djuranovic et al., 2012). Multiple signalling pathways have been verified to participate in skeletal development and bone formation, and a good understanding of the diversity of these signals is of great importance to the manipulation of osteoblastogenesis mediated by miRNAs. A number of signalling pathways have been proved to govern the osteoblast-ogenic differentiation of BMSCs, including BMP/TGF-β and Wnt transduction, MAPK signalling, Notch signalling, IGF signalling, RANK-OPG-RANKL signalling, etc (Fang et al., 2015, Zhao et al., 2014).
Recently, Li et al. indicated that hsa-miR-23a-3p could impede the osteogenic differentiation of hBMSCs by targeting low-density lipoprotein-receptor-related protein 5 (LRP5) via inactivation of Wnt/β-catenin signalling (Li et al., 2016a). LRP5 is a single-pass transmembrane protein, and absence of or variation of in LRP5 has been shown to result in decreased bone mass and osteoporosis (Cui et al., 2011, Saarinen et al., 2007). Xiao's group demonstrated that hsa-miR-129-5p played a positive role in osteoblast differentiation by reducing signal transducer and activator of transcription 1 (STAT1) levels, leading to a dramatic increase in Runx2 expression (Xiao et al., 2016). STAT1 was originally regarded as a signalling molecule participating in immune regulation via the interferon pathway (Meraz et al., 1996), and several studies have also indicated that STAT1 served as an important negative regulator of osteogenic differentiation by suppressing the expression of Runx2 and Osterix both in vitro and in vivo (Kim et al., 2003, Tajima et al., 2010). Apart from targeting Runx2, hsa-miR-205-5p also negatively affects osteoblast differentiation by targeting special AT-rich sequence-binding protein 2 (SATB2) via the extracellular signal-regulated kinase (ERK) and p38 mitogen-activated protein kinase (MAPK) pathways (Hu et al., 2015). In addition, Wei and colleagues indicated that mmu-miR-34b-5p and mmu-miR-34c-5p inhibited osteoblast proliferation by decreasing Cyclin D1, CDK4 and CDK6 accumulation while simultaneously suppressing osteoblast differentiation by decreasing SATB2 accumulation during embryogenesis and postnatally (Wei et al., 2012). However, this study could not confirm whether other putative targets, such as Notch 1 or other genes, might additionally account for the capability of these two miRNAs to modulate osteoblastogenesis. Around the same time, Bae et al. verified that mmu-miR-34c-5p targeted multiple components of the Notch signalling pathway, including Notch1/2 and Jag 1, to regulate osteoblastogenesis in bone homeostasis (Bae et al., 2012). Considering the significant role of the Notch signalling pathway and abnormal osteoblastogenesis in osteosarcomas (Engin et al., 2009), further investigations of the functional interaction between the miR-34 family and Notch signalling could potentially lead to efficient therapeutics in bone cancer.
2.3. miRNAs control bone remodelling factors
The equilibrium of bone formation by osteoblasts and bone resorption by osteoclasts is subject to fine and tight regulation in normal bone metabolism (Karsenty and Wagner, 2002, Tanaka et al., 2005). Disruption of this balance, that is, decreased bone formation and/or excessive bone resorption, will lead to the development of osteoporosis (Zaidi, 2007, Rachner et al., 2011). The cross-talk between osteoblasts and osteoclasts is critical to the maintaining the equilibrium governed by osteoblast-directed osteoclastic bone resorption and osteoclast-directed osteoblastic bone formation (Zaidi, 2007, Martin and Sims, 2005). As a monocyte-macrophage derived cell type, osteoclasts mainly locate near the bone formation surface (Tanaka et al., 2005, Vaananen, 2005). The survival, proliferation, function and differentiation of osteoclasts are modulated by various cytokines and exogenous hormones (Del Fattore et al., 2008). It has been confirmed that receptor activator NFκB ligand (RANKL) and macrophage colony-stimulating factor-1 (M-CSF), produced by activated T cells or osteoclasts, are two key cytokines involved in osteoclastogenesis (Karsenty and Wagner, 2002, Tanaka et al., 2005, Del Fattore et al., 2008). Furthermore, immunoreceptor tyrosine-based activation motif (ITAM)-dependent costimulatory signals are also essential for osteoclastogenesis (Koga et al., 2004), and many transcription factors, especially c-Fos and NF-κB (p50 and p52), are fundamental to the normal differentiation of monocyte precursors into osteoclasts (Griqoriadis et al., 1994, Iotsova et al., 1997, Franzoso et al., 1997).
miRNAs also control osteoclastogenesis by targeting relevant gene expression involved in this delicate process (Fig. 2b). Studies of the expression patterns of miRNAs revealed that 44 miRNAs were highly expressed during the maturation of osteoclast precursors (Kagiya and Nakamura, 2013). The functions of hsa-miR-21-5p, miR-29 family, mmu-miR-31-5p, mmu-miR-124-3p, hsa-miR-133a-3p, mmu-miR-146a-5p, mmu-miR-223-3p, hsa-miR-503-5p, mmu-miR- 378-3p, hsa-miR-125a-5p, hsa-miR-148a-3p, hsa-miR-155-5p and hsa-miR-422a were systematically summarized in an excellent review (Tang et al., 2014). It has been well documented that osteoblast-to-osteoclast communication occurs via direct cell-cell contact (Irie et al., 2009, Negishi-Koga et al., 2011). To understand whether other effective paracrine pathways exist in the interaction between the two cell types, Li and colleagues conducted miRNA-mediated osteoclast-directed osteoblastic bone formation in ovariectomized (OVX) mice, indicating that inhibition of osteoclast-derived exosomal mmu-miR-214-3p induced significantly suppressed osteoclastogenesis (Li et al., 2016b). Additionally, mmu-miR-214-3p also negatively controlled osteogenic differentiation by targeting Osterix and activating transcription factor 4 (ATF4) (Shi et al., 2013, Wang et al., 2013a), promoting osteoclastogenesis via the PI3K/Akt pathway by targeting phosphatase and tensin homologue (PTEN) (Zhao et al., 2015a). Based on the above research on miRNA-modulated osteoblastogenesis and osteoclastogenesis, miRNA-targeting therapeutics for metabolic bone disorders, such as osteoporosis, osteopenia and rheumatoid arthritis (RA)-related bone destruction, are anticipated approaches for clinical application.
3. miRNA-related bone disorders and potential targeting therapeutics
3.1. Osteoporosis
Osteoporosis, resulting from an imbalance between osteoclastic bone resorption and osteoblastic bone formation, is characterized by tissue structural deterioration and low bone mineral density (Teitelbaum, 2000). It has been reported that there are approximately 75 million people suffering from osteoporosis, and osteoporotic fracture is a common complication, leading to high healthcare expenditures and loss of productivity (Muruganandan et al., 2013, Seeliger et al., 2014). Glucocorticoids (GCs), as immunosuppressive and anti-inflammatory drugs, are extensively used to treat inflammation, autoimmune disorders and infection (Tait et al., 2008). However, prolonged use of GCs has been closely associated with osteoporosis, and GC-induced osteoporosis (GCOP) is the second most common aetiology of all types of osteoporosis (Kondo et al., 2008, Weinstein et al., 1998). The suppression of osteoblast activity, differentiation, maturation and lifespan are regarded as the major mechanisms involved in GC-induced inhibition of bone formation (Weinstein, 2001, Sato et al., 2015). Moreover, capacious marrow adipogenesis is observed in glucocorticoid-mediated bone deterioration as well (Tamura et al., 2015). As the most common degenerative bone disease in elderly women, postmenopausal osteoporosis is caused by a remodelling imbalance between bone formation and bone resorption due to oestrogen deficiency. (Holroyd et al., 2008, Sambrook and Cooper, 2006) Shi and colleagues discovered that mmu-miR-17-5p and −20a-5p suppressed GC-induced osteoclast differentiation and targeted RANKL in osteoblast cells (Shi et al., 2014). This is the first report regarding microRNA-targeting RANKL in osteoclastogenesis, however, they did not explore how dexamethasone (Dex) regulated mmu-miR-17/20a-5p expression in osteoblast, which remains to be further investigated. Wang et al. observed that mmu-miR-29a-3p protected rats from GC-induced bone loss and fragility by regulating Wnt and its inhibitor Dkk-1, making miR-29a-3p a potential candidate for alleviating GCOP (Wang et al., 2013b). The inhibition of histone deacetylase (HDAC) 4 with miR-29a-3p restored the acetylation state of β-catenin and Runx2, ameliorating the harmful effects of glucocorticoids on osteoblastic mineralization (Ko et al., 2015). However, the long term effect of exogenous miR-29a-3p signaling on immune response, tissue integrity, or animal physiology warrants further evaluation.
Various of investigations on the role microRNAs in in vivo human circulating monocytes in the etiology of osteoporosis have been extensively established in order to figure out potential biomarkers or targeting therapeutics. Wang et al. illustrated that hsa-miR-133a-3p negatively regulated three potential osteoclast-related genes, chemokine (C-X-C motif) ligand 11 (CXCL-11), chemokine (C-X-C motif) receptor 3 (CXCR-3) and solute carrier family 39 A1 (SLC-39A1), indicating that hsa-miR-133a-3p in circulating monocytes might serve as a potential biomarker for postmenopausal osteoporosis (Wang et al., 2012a). In addition, hsa-miR-442a could down-regulate certain gene expression levels related to osteoclastogenesis inhibition, including cluster of differentiation 226 (CD226), phosphoprotein associated with glycosphingo-lipid microdomains 1 (PAG-1), insulin-like growth factor 1 (IGF-1) and transducer of ERBB2 (TOB-2), making hsa-miR-442a another possible biomarker underlying post-menopausal osteoporosis (Cao et al., 2014). Furthermore, hsa-miR-194-5p was identified as a potential biomarker of postmenopausal osteoporosis according to Meng et al.’s microarray analysis (Meng et al., 2015). Meanwhile, You et al. observed that hsa-miR-27a-3p-targeting myocyte enhancer factor 2c (Mef2c) was markedly decreased in osteoporotic patients through microarray analysis, and it was necessary for the shift of BMSCs from osteogenic into adipogenic differentiation during the development of osteoporosis (You et al., 2016). Overexpression of hsa-miR-133 was shown to induce postmenopausal osteoporosis by impeding osteogenic differentiation of hMSCs through the down-regulation of SLC39A1, which is positively correlated with Runx2 and Osterix (Lv et al., 2015). Weilner and colleagues performed an explorative analysis of numerous miRNAs in serum samples obtained from patients with recent osteoporotic fractures, and they discovered that hsa-miR-10a-5p, hsa-miR-10b-5p, hsa-miR-22-3p were significantly up-regulated, whereas hsa-miR-133b, hsa-miR-328-3p and hsa-let-7g-5p were down-regulated, and they also demonstrated regulatory effects on the osteogenic differentiation of BMSCs in vitro (Weilner et al., 2015). Moreover, this team also investigated the expression levels of circulating miRNAs in patients with diabetic bone disease and postmenopausal osteoporosis. They identified hsa-miR-550a-5p and hsa-miR-382-3p as the most promising circulating miRNAs for diabetic bone diseases, and hsa-miR-382-3p and hsa-miR-188-3p as the most promising circulating miRNAs for postmenopausal osteoporosis (Heilmeier et al., 2016). Identifying and evaluating patients at relatively high risk for osteoporosis-associated fragility fracture are of great significance to disease prevention and management, and further in-depth exploration of these differentially expressed miRNAs in bone tissue and in larger validation cohorts should be well established to confirm whether miRNAs can serve as potential pharmaceutically biomarkers and clinically applicable therapeutic targets for osteoporosis.
Down-regulation of mmu-miR-34a-5p-targeting cell cycle factors (CDK4, CDK6 and Cyclin D1) led to impaired BMSC osteogenic differentiation and aggravated osteoporosis in dexamethasone-induced rodent models through the Notch signalling pathway. Therefore, mmu-miR-34a-5p might be a conceivable therapeutic target for GCOP (Kang et al., 2016a). Li et al. reported that mmu-miR-140-3p and mmu-miR-214-3p impeded sponges via constructed Cre/loxP-based hybrid baculovirus vectors, demonstrating improved bone formation and mitigated excessive bone resorption in an osteoporotic bone defect rat model, thus paving a new avenue to the treatment of osteoporotic bone defects through combination of osteogenic factors and functional miRNAs (Li et al., 2016c). In addition, Krzeszinski and colleagues found that mmu-miR-34a-5p knockout and heterozygous mice exhibited elevated bone resorption and reduced bone mass by targeting transforming growth factor-β-induced factor 2 (Tgif2), indicating that miR-34a-5p was a critical osteoclast suppressor and a potential therapeutic strategy to combat osteoporosis, and could exert both anti-catabolic and anabolic effects compared to current drugs that are solely anti-catabolic (Krzeszinski et al., 2014). This innovative study paves the way for future epidemiological and clinical studies to investigate the pathological and therapeutic roles of the miR-34a-5p-Tgif2 pathway in human. It has been well established that oxidative stress plays a critical role in the development of postmenopausal osteoporosis, which is attributed largely to the decline of the enzymatic antioxidant system (Manolagas, 2010). Liao et al. illustrated that the expression of mmu-miR-705 was significantly enhanced in ovariectomized (OVX) mice through the TNF-α activated NFκB pathway, leading to the suppression of forkhead box O1 (FoxO1) protein, which is essential for defending against oxidative damage caused by oestrogen deficiency during osteoporosis (Liao et al., 2016). This study indicated that NFκB pathway is a major target for the treatment of osteoporosis, however, global inhibition of NFκB pathway may be not an ideal strategy in clinic because NFκB signaling is of great importance for immnue system and other physiological. Thus, modulation of its downstream signalling will be a promising strategy. In addition, this group also reported that mmu-miR-705 and mmu-miR-3077-5p could mediate the shift of BMSC cell lineages into adipocytes in osteoporosis by targeting HOXA10 and Runx2, respectively (Liao et al., 2013b). Furthermore, TNF-α was reported to impede bone formation by down-regulating the expression of hsa-miR-21-5p in oestrogen deficiency-induced osteoporosis, with enhanced expression of Spry1 (Yang et al., 2013), which negatively regulates the FGF and ERK-MAPK signalling pathways.
3.2. Bone defects
The aetiologies of bone defects mainly include osteoporosis, trauma and bone tumour resection. Minor bone defects can be self-rehabilitated; however, bone restoration can be difficult in cases of critically sized bone defects. Traditionally, grafting (including autografts, allografts and xenografts) and the introduction of metal implants were the gold standard treatment for these defects. Nevertheless, several common complications, such as tissue function loss, implant-related infection and implant rejection, have restricted the therapeutic effects of these methods. More recently, bone tissue engineering, which provides a relatively disease-free implant for damaged bones without imposing any obvious adverse effects on the skeleton and its environment, has emerged as an alternative strategy to resolve these problems originating from traditional therapy for bone defects. Bioactive and biogradable scaffolds with a similar micro-architecture to that of bone tissues, which can be fabricated by various methods such as electrospinning (Meka et al., 2016), freeze drying (Cholas et al., 2016), low-temperature rapid prototyping (Qin et al., 2016), 3D printing (Zhao et al., 2015b), etc., have been widely used to accelerate and enhance bone repair for critically sized bone defects. Biomaterial-mediated miRNA delivery for bone regeneration is being extensively exploited based on the molecular mechanisms discussed above. The miRNA expression profiles of BMSCs at the defect site can be differentially altered by osteoinductive and osteoconductive biomaterials. Thus, genetically modified scaffolds incorporating with various miRNAs could pave the way for new manipulation of osteoblastogenesis and osteoclastogenesis during bone restoration.
Palmieri et al. first effort to investigated the regulation of miRNAs in osteoblast-like cells via calcium sulphate (CaS) by using microarray techniques, providing a better understanding of the molecular mechanism of interactions between biomaterials and bone regeneration (Palmieri et al., 2008). The nano-structure interface of biomaterials was shown to promote bone mineralization through the regulation of relevant miRNA expression. Mahmood et al. observed that many osteogenic genes were differentially regulated by silver nano-particles (AgNPs) (Mahmood et al., 2011). Apart from investigating the effects of different biomaterials on whole miRNAs profiles, numerous miRNA-targeting strategies with varied biomaterials to treat bone defects have been thoroughly studied. Anti-miRNA oligonucleotides (AMOs), which are complementary to target miRNAs, were used to suppress the expression of certain critical miRNAs in osteoblast differentiation. Yan et al. reported BMSC sheets incorporating with non-viral oligonucleotide anti-mmu-miR-138-5p by a vitamin C-induced method, which displayed significantly improved bone regeneration in vivo by activating the ERK1/2 pathway via targeting Runx2 (Yan et al., 2014). It was verified that the miR-29 family could negatively regulate the extracellular matrix (ECM) production during the early stage of osteogenesis (Kapinas et al., 2009), and inhibition of the miR-29 family could promote ECM synthesis in osteoblastogenesis. James et al. determined that mmu-miR- 29a-3p inhibitor-loaded gelatine nanofibre scaffolds prepared by electrospinning could increase ECM deposition, which might serve as a novel platform for exploiting miRNA-based therapeutics in bone defects (James et al., 2014). It was identified that miR-133a served as a direct negative regulator of the master transcription factor of osteogenesis, Runx2 (Fang et al., 2015). Castaño and colleagues fabricated porous collagen/nano-hydroxyapatite scaffolds with miR-133a-inhibiting complexes, demonstrating significantly promoted osteogenic differentiation in vitro, thus offering potential applications for bone repair and tissue engineering (Castaño et al., 2016). However, complete inhibition of such functional miRNAs may trigger unwanted side-effects, thus, systematic investigations of the stability of the delivery system and in vivo validation remain to be performed to verify the therapeutic efficacy of such an antago-miRNAs-based osteoconductive scaffolds for bone engineering.
In addition, the osteogenesis of BMSC sheets was greatly ameliorated by hsa-miR-21-5p-loaded chitosan/hyaluronic acid nanoparticles (Wang et al., 2016a). Therefore, such miRNA-based cell sheet engineering might serve as a novel approach to more rapid and robust bone regeneration clinically. Deng and colleagues reported poly(glycerol sebacate) (PGS) scaffolds loaded with anti-miR-31-expressing BMSCs, showing better biocompatibility and bone regeneration when repairing critical-sized calvarial defects (CSDs) in rats (Deng et al., 2014). They also systematically evaluated the roles of mmu-miR-31-5p-modified adipose tissue-originated stem cells (ASCs) both in vitro and in vivo, indicating that knockdown of miR-31-5p by lentivirus (antisense) in ASCs exhibited dramatically improved repair of CSDs by targeting Runx2 and SATB2 (Deng et al., 2013). Silver nanoparticle (SNP) compounds conjugated with hsa-miR-148b-3p, a positive regulator of osteogenesis (Schoolmeesters et al., 2009), enhanced osteogenic differentiation by improving ALP activity and calcium mineralization (Qureshi et al., 2013). Thus, this study provided the possibility of 3D scaffolds combined with miR-148b-3p-SNP conjugates as a light activated miRNA delivery vehicle and the capability to modulate the osteogenic differentiation using light as a non-invasive triggering modality. Moreover, the incorporation of mmu-miR-26a-5p-transfected ASCs into porous hydroxyapatite scaffolds also showed satisfactory bone repair in a rat tibial defect model (Wang et al., 2015a). The osseointegration between titanium implants and surrounding bone tissues is of great importance to the immobilization of large bone defects. To improve the loading capability of titanium-based implants, the microarc-oxidation method was introduced to form micropore structures. For instance, hsa-miR-21-5p-loaded chitosan/hyaluronic acid nanoparticles coated over the surface of titanium using cross-linking methods could promote the osteogenic differentiation of hBMSCs (Wang et al., 2015b), providing a unique cross-linking-mediated therapeutic approach to prepare miRNA-loaded metal implant surfaces for bone repair.
3.3. Primary bone tumor and secondary metastasis
miRNA expression was first linked to cancer in 2002, initially with chronic lymphocytic leukaemia and then with other different types of malignancies (Mirnezami et al., 2009). It has been generally recognized that some miRNAs function as tumour-inhibitor genes, whereas others alternatively behave as oncogenes (Nugent, 2014). The development of cancer can be attributed to the dysregulation of miRNA expression to a certain extent, making miRNA-targeted interventions a promising approach for the diagnosis, as well as the treatment, of different malignancies. Primary bone tumours encompass a broad range of types, including osteosarcoma (OS), Ewing sarcoma (ES), chondrosarcoma (CS), multiple osteochondromatosis (MOC), and giant cell tumour of bone (GCTB), etc., among which osteosarcoma and Ewing sarcoma are the two most common malignant primary tumours of bone (Nugent and Santulli, 2015).
The roles and target genes of miRNAs in primary bone tumours and metastatic bone tumours were elucidated in an elaborative review (Nugent, 2014). Currently, there appear to be a few new miRNAs and concomitant targets associated with bone neoplasms. Sun and colleagues observed that hsa-miR-646 was a suppressor of OS cell metastasis by down-regulating fibroblast growth factor 2 (FGF-2) (Sun et al., 2015). Shi et al. found that the proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) of OS cells were effectively prevented by overexpression of hsa-miR-204-5p directly targeting Sirtuin 1 (Sirt 1) (Shi et al., 2015). Sirt 1 is a type of class III histone deacetylase that promotes tumourigenesis and metastasis (Lin et al., 2014). Moreover, Sirt 1 is also a direct target of hsa-miR-126-5p that suppresses the proliferation of OS cells by down-regulating Sirt 1 expression (Xu et al., 2013). Wu et al. reported that the expression of hsa-miR-409-3p in OS tissue was significantly lower than in adjacent non-tumour tissue, and it inhibited OS cell migration and invasion by targeting catenin-δ1, which is encoded by CTNND1 and serves as a tumour-suppressing or oncogenic factor, depending on the cancer type (Wu et al., 2016a). This study for the first time discovered a unique function of catenin-δ1 in osteosarcoma progression, and further studies by exploring the interconnected pathway in relevant models will be beneficial to fully elucidate the function of miR-409-3p/catenin-δ1 in osteosarcoma development. Zhou et al. observed that hsa-miR-20a-5p exerted oncogenic effects on OS cells via direct inhibition of early growth response 2 (EGR-2) (Zhou et al., 2015), which negatively regulates cell proliferation and cell cycle (Unoki and Nakamura, 2001), indicating that miR-20a-5p may serve as a promising prognostic and therapeutic target for OS. He and colleagues discovered that up-regulation of hsa-miR-106a-5p could inhibit OS proliferation, migration and invasion by targeting high-mobility group AT-hook 2 (HMGA2) (He et al., 2016), which is critical to the chromosomal aberrations and tumourigenesis (Fusco and Fedele, 2007).
The 5-year survival was reported at approximately 20% in osteosarcoma patients with lung metastasis (Siegel and Pressey, 2008), and this relatively poor survival might be caused by the difficulty of eliminating cancer stem cells (CSCs), a small subset of tumour cells and also the possible primary cause of tumour metastasis and relapse (Mitra et al., 2015). hsa-miR-26a-5p was shown to play a positive role in osteoblastogenesis, and it could suppress the CSC phenotype and tumour growth of OS by inhibiting Jagged1/Notch pathway, which plays a pivot role in maintaining CSCs function (Lu et al., 2016). Thus, the metastasis of OS might be effectively prevented using certain miRNAs to suppress CSC function. The reprogramming of metabolic pathways, such as increased de novo lipid synthesis, represents one of the most critical and common features of cancer cells. As the precursor of de novo lipogenesis, ATP citrate lyase (ACLY) is a cross-link between the mevalonate pathways and fatty acid synthesis, and it is one of the most significant components of ACLY metabolic post-transcriptional regulation (Zaidi et al., 2012). The tumour growth and metastasis of OS could be inhibited by hsa-miR-22-3p targeting ACLY, yielding an effective therapeutic approach to different cancers (Xin et al., 2016). Apart from inhibition of the Notch-1 pathway (Li et al., 2013), hsa-miR-145-5p also acted as a tumour inhibitor of OS via direct suppression of friend leukaemia virus integration 1 (FLI-1) that enhances oncogenesis (Wu et al., 2016b). This is the first observation of miR-145-5p/FLI-1 interaction in OS cells, and the direct interaction between this miRNA and proliferation of OS cells remains to be further investigated.
Extensive bone resorption is one of the most featured characteristics of GTCB, causing regional pain and high risk of pathological fractures (Cowan and Singh, 2013). RANK-expressing monocytic cells are regarded as the neoplastic component of GTCB (Atkins et al., 2006), thus, RANKL-targeted intervention by miRNAs could be a meaningful method to restrain the development of GTCB. As a member of the miR-106b-25 cluster, hsa-miR-106b-5p was verified to be a novel inhibitor of osteoclastogenesis and osteolysis by targeting RANKL and other cytokines (IL-8, MMP2 and TWIST) in GTCB (Wang et al., 2015c), while hsa-miR-126- 5p also restrained osteoclast differentiation and bone resorption in GTCB through inhibition of matrix metalloproteinase-13 (MMP-13) and parathyroid hormone-related protein (PTHrP) (Wu et al., 2014a, Zhou et al., 2014). Meanwhile, the osteoclast differentiation and bone resorption in GCTB involve various factors, therefore, miR-126-5p might influence some other factors expression in the process via either a direct or indirect way, which requires further study in the near future. Some hypoxia-induced factors, such as vascular endothelial growth factor (VEGF) and hypoxia-inducible factor 1α (HIF-1α), were overexpressed in sacral GCTB samples, and they are closely related to tumour microvessel density (Fu et al., 2014). Guo et al. elucidated for the first time that hsa-miR-210-5p was up-regulated by HIF-1α in GCTB stromal cells, suggesting an effective strategy to suppress the hypoxic adaptation of GCTBs (Guo et al., 2015).
The most common metastatic bone tumours are the carcinomas derived from the prostate, breast and lung. Ma et al. first proposed the involvement of miRNAs in metastasis in 2007, indicating that hsa-miR-10b-5p initiated the invasion and metastasis of breast cancer (Ma et al., 2007). This significant study opened the door for the investigations of the role of miRNAs in mediating cancer metastasis. Bone metastasis is one of the most typical characteristics of prostate cancer (PC) clinically, and it is usually treated with androgen deprivation therapy to relieve symptoms and restrain tumour growth. Comprehending the molecular mechanisms of the miRNAs that regulate metastasis is essential to improve the therapeutic outcomes of patients (Shah et al., 2004). Liu et al. reported that low levels of androgen-regulated hsa-mir-1-2 could promote PC bone metastasis by activating SRC through the ERK signalling pathway, suggesting that SRC activity might be regarded as a potential biomarker in patients receiving androgen deprivation treatment (Liu et al., 2015). Aberrant activation of Ras and Wnt signallings were critical events during the development of PC bone metastasis (Carter et al., 1990, Kypta and Waxman, 2012). hsa-miR-34a-5p was reported to restrain the bone metastasis of Ras-activated PC via the Wnt signalling pathway by targeting transcription factor 7 (TCF7). TCF7 plays a vital role in enhancing the oncogenic activity of the Wnt signalling pathway, and the up-regulation of TCF7 depends on the suppression of miR-34a (Chen et al., 2015). It has been well established that transforming growth factor-β (TGF-β) signalling promotes malignancy in tumourigenic tumour cells while impeding tumour growth in benign/normal tissues (Ao et al., 2006). TGF-β serves as a momentous factor in facilitating bone metastasis by activating mammalian target of rapamycin (mTOR) function. mmu-mir-96 was observed to be up-regulated by TGF-β through Smad-dependent transcription in the progression of PC bone metastasis, and AKT1S1, the translated protein of which negatively regulates mTOR kinase, was identified as a downstream target of the TGF-β-miR-96 pathway (Siu et al., 2015). Endothelial nitric oxide synthase (eNOS) has been detected in clinical PC tissues and is positively correlated with the invasion, metastasis and antiandrogen resistance of PC (Nanni et al., 2009, Yu et al., 2013). Fu and colleagues found that hsa-miR-335-5p and hsa-miR-543 restrained bone metastasis in PC by targeting eNOS, indicating that these two miRNAs might be used clinically for the prediction of bone metastasis in PC patients (Fu et al., 2015). Even though the regulatory role of miRNAs in the control of metastatic bone tumors has been well described in numerous investigations, the specific network and interaction remains to be further explored in the future.
3.4. Osteoarthritis
Osteoarthritis (OA) is the most prevalent degenerative bone joint disease characterized by progressive deterioration of articular cartilage, sclerosis of subchondral bone and synovial hyperplasia (Martel-Pelletier, 2004). Over the past decades, there has been an increasing concern about the functional and therapeutic roles of miRNAs in OA, and some elegant reviews have systematically elucidated miRNA-mediated homeostasis and pathogenesis in OA (Miyaki and Asahara, 2012, Wu et al., 2014b, Sondag and Haggi, 2016). Here, we only focus on several of the most recently reported miRNAs involved in the pathogenesis and targeting therapeutics of OA. Hu et al. reported that hsa-miR-139-5p promoted the inhibition of chondrocyte proliferation and migration in human chondrocyte CHON-001 cells by suppressing insulin-like growth factor 1 receptor (IGF1R) and eukaryotic translation initiation factor 4 gamma 2 (EIF4G2) (Hu et al., 2016), both of which play pivotal roles in facilitating the translation initiation and proliferation of cancer cells (Coldwell et al., 2013, Maloney et al., 2003). Kang et al. observed that hsa-miR-23a-3p expression was significantly higher in OA cartilage than in normal tissues and it decreased chondrocyte ECM synthesis by targeting Smad3, a regulator of TGF-β in the homeostasis and restoration of cartilage (Kang et al., 2016b). In addition, Smad3 was also a target of hsa-miR-16-5p, which might contribute to the deterioration of OA (Li et al., 2015). As a proinflammatory cytokine, IL-1β is an inhibitor of chondrocytes proliferation and collagen biosynthesis (Goldring et al., 1994). IL-1β-mediated overexpression of inducible nitric oxide synthase (iNOS) strongly contributes to cartilage deterioration and inflammation (Abramson et al., 2001). Rasheed and colleagues indicated that up-regulation of hsa-miR-26a-5p in human OA chondrocytes could effectively restrain the expression of iNOS via activation of the NF-κB pathway (Rasheed et al., 2016). In addition, IL-1β inhibited the expression of miR-30a by recruitment of activator protein 1 (AP-1) and overexpression of disintegrin-like and metalloproteinase with thrombospondin-5 (ADAMTS-5), leading to aggravated degradation of cartilage matrix in OA (Ji et al., 2016a). Similarly, hsa-miR-142-3p and mmu-miR-210-5p could suppress inflammation and chondrocyte apoptosis in OA via the NF-κB pathway by targeting HMGB-1 and death receptor 6 (DR6), respectively (Wang et al., 2016b, Zhang et al., 2015a). Inhibited production of proteoglycans and an imbalance between collagens I and II induced by fibroblast growth factor 2 (FGF2) are among the important pathogenic factors in OA (Ellman et al., 2013). FGF2-induced ADAMTS expression inhibited the transcription of hsa-miR-105-5p by targeting Runx2, thus accelerating the progression of OA (Ji et al., 2016b). In addition to the inflammatory factor-mediated cartilage deterioration, mechanical stress is also a significant biochemical factor involved in the initiation and progression of OA, especially traumatic OA. Yang et al. reported that mechanically responsive miR-365 was significantly increased after cyclic loading and IL-1β stimulation by targeting histone deacetylase 4 (HDAC4), which acts as a potent suppressor of chondrocyte hypertrophy. Thus, inhibition of hsa-miR-365a-3p might be an effective therapy for the prevention and treatment of traumatic OA (Yang et al., 2016). In short, inflammation-mediated degeneration and apoptosis of cartilage are the most important causes of the progression of OA, and regulation of the miRNAs involved in the pathophysiological process in OA could pave new and anticipated therapeutic avenues to treat this degenerative and inflammatory disorder.
4. Potential targeting therapeutics of siRNAs in bone disorders
4.1. siRNA-mediated bone formation and resorption
Silencing of gene expression in a sequence-specific manner at post-transcriptional levels can be achieved by small-interfering RNAs (siRNAs), which were initially proposed by Fire and colleagues in 1998 and were developed rapidly to treat musculoskeletal disorders over the last two decades (Fire et al., 1998). The discovery of this process has opened the doors to the development of siRNA-based therapeutics. Recently, siRNAs have been fully exploited to regulate the expression of various proteins involved in many therapeutic conditions, such as osteoporosis, osteoarthritis, and bone regeneration, etc. However, siRNA-based therapy is still gaining ground in musculoskeletal tissue repair, bone regeneration and tumour suppression. With advancement in the understanding of the targeting biological pathways involved in bone regeneration and also the rapid progress in siRNA technologies, application of siRNAs for bone-related disorders has great therapeutic potential. siRNA therapeutics targeting the osteoclast-mediated bone resorption involved in osteoporosis and osteoarthritis have been well elucidated in a few excellent reviews (Shi et al., 2011, Wang and Grainger, 2012, Wang and Grainger, 2013). In this review, we mainly focus on siRNA-mediated bone regeneration and several newly released investigations of the treatment of bone resorption using siRNA-targeting technologies.
Two major categories of DDSs used for siRNA delivery in therapeutic applications for bone regeneration are lipid-based and polymer-based systems. Currently, the majority of commercial transfection reagents are modified cationic liposome-based systems. Farnesyl pyrophosphate synthase (FPPS), an enzyme in the mevalonate pathway, is the inhibition target of alendronate, which is a potent FDA-approved nitrogen-containing bisphosphonate drug. Knockdown of FPPS activity mediated by siRNA exhibited selectively inhibited osteoclast-mediated bone resorption and promoted bone mass maintenance without negative effects on the proliferation of preosteoblasts (Wang et al., 2011). This is the first report to knock down the molecular target of clinically common nitrogen-containing bisphosphonates through siRNA. However, the direct effect of FPPS changes on preosteoblasts mineralization is still unclear, and further investigation is required to confirm whether down-regulation of FPPS via siRNA influence the osteoblast-osteoclast communication. Ríos and colleagues administered siRNA silencing prolyl hydroxylase domain-containing protein 2 (siPHD2) and guanine nucleotide-binding protein alpha-stimulating activity polypeptide 1 (siGNAS1) via Ambion (A commercially purchased polyamine-based transfection agent), inducing enhanced osteogenesis in vitro and bone formation in vivo (Ríos et al., 2012). Nevertheless, the complex and different intercellular mechanisms of siPHD2 and siGNAS1 during the regulation of bone formation remains to be further determined, and the prefabricated bone defect model with no mechanical loading could impair the clinical application potential of this siRNA-based targeting therapeutic. Jia et al. fabricated a siRNAs-incorporating chitosan sponge, within which two types of siRNA-Lipofectamine 2000 complexes targeting soluble vascular endothelial growth factor receptor 1 (siFlt-1) and casein kinase 2 interaction protein 1 (Ckip-1) were delivered together. This composite siRNAs-delivered scaffold showed satisfactory angiogenesis and osteogenesis in BMSCs and bone repair efficacy in vivo (Jia et al., 2014). Furthermore, Guo and colleagues also reported a similar Ckip-1-siRNA-Lipofectamine 2000 complex, demonstrating increased osteogenic differentiation in vitro and reversal of bone loss in osteoporotic mice (Guo et al., 2014). Zhang and colleagues developed a bone-formation surface-targeting system based on dioleoyl trimethylammonium propane (DOTAP) cationic liposomes, which displayed high affinity for poorly crystallized hydroxyapatite (HA) due to an (AspSerSer)6 moiety. An osteogenic siRNA targeting casein kinase-2 interacting protein-1 (Plekho1) was encompassed into the liposomes, producing significantly increased bone micro-architecture and bone formation in both healthy and osteoporotic rats (Zhang et al., 2012a). Subsequently, Liang et al. encapsulated the same siRNA into a more efficient osteoblast-specific aptamer-functionalized lipid nanoparticle (LNPs-siRNA) to promote the selective uptake of Plekho1 siRNA, which might act as a novel and desirable siRNA-based bone anabolic strategy to improve bone mass in osteoporosis (Liang et al., 2015). Bone-specific targeting delivery system as discussed above is a promising approach to specially delivering siRNA drugs to bone-formation surfaces and the osteogenic cells that reside there, thus, providing a potential solution to the bottleneck in clinical translation of siRNA-based bone anabolic therapeutics in the near future. Moreover, lipidoids, which possess a similar chemical structure to liposomes, are another new type of lipid-based nanoparticle. Lipidoids gained significantly higher transfection efficiency over Lipofectamine 2000 in human umbilical vein endothelial cells (HUVECs) at a low serum level (2%) (Fulmer, 2010). Ramasubramanian et al. performed delivery of BMP-2 DNA along with Noggin and guanine nucleotide binding protein alpha-stimulating activity polypeptide (GNAS) targeting siRNA in an immortalized human foetal osteoblast cell line (hFOB1.19) using a lipidoid called NA114, which was previously reported to be an efficient siRNA carrier (Cho et al., 2009). This study determined that this osteoinductive genes delivery caused decreased cell viability and promoted cell apoptosis in a dose-dependent manner, emphasizing the significance of distinguishing the biological effects of osteoinductive signals on various cell types in bone regeneration (Ramasubramanian et al., 2015). Targeting the component of the triad OPG/RANKL/RANK by siRNA technique seems to be a promising approach to regulate the osteoclastogenesis. Córdova and colleagues reported local administration of a RANK-targeting siRNA, delivered by the cationic liposome DMAPAP/DOPE, which revealed effectively impeded osteolysis and increased bone formation in a polyethylene (PE) particle-induced osteolysis model in mice (Córdova et al., 2015). This study demonstrated for the first time an effective inhibition of particle-induced osteolysis in vivo using a RANK-directed siRNA-sequence. Interestingly, the unexpected bone formation observed in siRNA-treated mice indicated that the injection of this cationic liposome might produce a favourable local anabolic microenvironment, and further study remains to be established to understand the biological effects of delivery system on the targeting cells or tissues, which may be useful to improve the biocompatibility of the siRNA delivery system.
As other siRNA DDSs, synthetic and natural polymers have been widely studied and modified to improve intracellular uptake, and they were elaborately analysed in a previous review (Whitehead and Langer, 2009). Wang and colleagues reported a local siRNA delivery strategy to target selectively and to suppress osteoclastogenesis at bone sites with osteoporosis. They fabricated passive phagocyte-targeting poly(lactic-co-glycolic acid) (PLGA) microparticles and incorporated them within biomedical grade calcium-based injectable bone cement to deliver RANK siRNAs to both osteoclasts and their precursors, providing a proof of concept to use a locally delivered siRNA therapeutic strategy for direct targeting of osteoporosis sites (Wang et al., 2012b). We have discussed the role of glucocorticoids (GCs) in osteoblastogenesis and osteoclastogenesis in this review; therefore, designing siRNA targeting GC receptor (GCR) could be a feasible approach for regulating the function of hBMSCs, which was explored in an in vitro study (Wei et al., 2013). Moreover, Hong et al. reported that GCR-siRNA-delivered PLGA microparticles showed effectively inhibited, detrimental effects of endogenous GC on hBMSCs, with improved osteogenic proliferation and differentiation (Hong et al., 2012). Wang et al. fabricated a site-specific bone-targeting system based on an octa-aspartic acid sequence (Asp8) and bisphosphonates, which was demonstrated to favorable recognize and strongly bind to biomineral (e.g. hydroxyapatite) surfaces (Wang et al., 2006). In their studies, a highly hydrophilic synthetic polymer, N-(2-hydroxypropyl) methacryl-amide (HPMA), was coupled with Asp8 and alendronate (a bisphosphonate) to deliver semaphorin 4d (Sema4d) siRNAs to specific bone sites, exhibiting partial counteraction of bone loss originating from osteoporosis (Zhang et al., 2014, Zhang et al., 2015b). Recently, this research team fabricated a PLLA scaffold incorporating siRNA-Sema4d to improve bone formation in osteoporosis-related bone defects (Zhang et al., 2016). Sema4d, also known as CD100, was proved to be expressed by osteoclasts and negatively regulated osteoblastogenesis (Negishi-Koga et al., 2011). It has been reported that the physical chemistry of bone-formation surfaces covered with osteoblasts is characterized by lowly crystallized hydroxyapatite, while the physical chemistry of bone-resorption surfaces covered with osteoclasts is characterized by highly crystallized hydroxyapatite (Wang et al., 2007). As two bone-targeted molecules, (AspSerSer)6 and Asp8 both demonstrated effective bone targeting functions (Zhang et al., 2012a, Wang et al., 2006). The stronger affinity of (AspSerSer)6 to lowly crystallized hydroxyapatite rather than highly crystallized hydroxyapatite provided an explanation for the preferential binding of (AspSerSer)6 to bone-formation surfaces, whereas the stronger affinity of Asp8 to highly crystallized hydroxyapatite rather than lowly crystallized hydroxyapatite produced a preferential binding of Asp8 to bone-resorption surfaces (Zhang et al., 2012a). The different affinity of these two molecules to the bone-formation or resorption surfaces deserves great attention in the siRNA-based bone anabolic therapy.
Except for polymer-based particles, polymeric hydrogels with controllable three dimensional networks have also been used for siRNA delivery in bone regeneration. Nguyen and colleagues described a localized and sustained Noggin siRNA DDS, based on engineered poly(ethylene glycol) (PEG) hydrogels scaffolds, which exhibited a controlled releasing course of 3–6 weeks and well-guided osteogenic differentiation of hBMSCs (Nquyen et al., 2014). Their work provided an efficient methodology for realizing controlled stem cell fate in bone tissue engineering in the future. Furthermore, Manaka and colleagues examined the efficacy of Noggin-siRNA delivered by a biodegradable hydrogel (poly-D,L-lactic acid-p-dioxanone-polyethylene glycol block co-polymer, PLA-DX-PEG), which displayed enhanced BMP-induced ectopic bone formation (Manaka et al., 2011). Noggin is a major extracellular antagonist of BMPs, and Noggin silencing by siRNA exhibited significantly accelerated osteoblastic differentiation in vitro and promoted new bone formation in vivo (Wan et al., 2007, Takyama et al., 2009). However, the effect of local inhibition of Noggin expression by targeting siRNA transfection in vivo was not as dramatic as expected from the in vitro findings, which may due to the inadequate dose of Noggin-targeted siRNA and/or insufficient efficacy of the delivery system for siRNA into cells. Even though plenty of siRNA-based targeting systems have been effectively confirmed in vitro as mentioned before, a number of in vivo investigations are badly required to further determine the application efficacy of those siRNA-based targeting therapeutic in bone disorders.
4.2. siRNA-mediated therapy for bone tumour
In addition to the investigations of the potent application of siRNAs to combat bone defects and resorption, siRNAs could be also efficient and anticipated tools to restrain tumour development and metastasis to the skeletal system. As a sequence-specific gene -silencing tool at the post-transcriptional level, siRNAs have been widely explored for targeting specific oncogenes, leading to effectively impeded bone tumour growth and ameliorated chemosensitivity.
It is widely acknowledged that VEGF regulates tumour angiogenesis by directly increasing neovascularization, vascular permeability and remodelling (Bradshaw et al., 2013). Silencing of VEGF via siRNAs was confirmed to suppress osteosarcoma angiogenesis and to promote tumour cell apoptosis via activation of phosphoinositide 3-kinase (PI3K) and the protein kinase B (AKT) signalling pathway in SaOS-2 cells (Peng et al., 2016). Moreover, combined application of VEGF-siRNA and neoadjuvant chemotherapy manifested preferable synergistic therapeutic effect to simple biological therapy or VEGF-siRNA plus anti-angiogenesis chemotherapy in OS-bearing rats, suggesting that the addition of neoadjuvant chemotherapy might be beneficial for enhancing the therapeutic efficiency of VEGF-siRNA (Zhu and Mei, 2015). Translationally controlled tumour proteins (TCTPs), such as Mcl-2 (known as a Bcl-2 family member), have been described as stabilizing and promoting the anti-apoptotic activity of several oncogenes, thus facilitating the progression of tumour cells (Liu et al., 2005). siRNA-mediated silencing of TCTP displayed obviously promoted apoptosis of the Saos-2 and U2OS cell lines, together with inhibited development of xenografts in nude mice (Shen et al., 2016). The L1 cell adhesion molecule (L1CAM), belonging to the immunoglobulin superfamily, has been identified as a critical mediator of tumour progression. Based on the discovery of high expression of L1CAM in androgen-insensitive and metastatic prostate cancer (PC) cell lines, Sung et al. investigated the therapeutic effects of liposome-encapsulated L1CAM-siRNAs exhibiting admirable suppression of PC proliferation and bone metastasis through the inhibition of matrix metalloproteinase-2 (MMP-2) and MMP-9 and activation of the NF-κB pathway (Sung et al., 2014). However, the mechanism by which the L1CAM regulates mesenchymal-to-epithelial transition (EMT)-related gene expression and confers enhanced motility and metastatic capabilities to PC remains to be further determined.
Multidrug resistance (MDR) represents an enormous barrier in the treatment of osteosarcoma. The recovery rate is dramatically decreased in patients with recurrence caused by MDR (Schwartz et al., 2007). Overexpression of membrane-bound drug transporter proteins, including MDR-related proteins (ABCC1, ABCC2, MRP1 and MRP2), P-glycoprotein (P-ag, ABCB1) and breast cancer resistance proteins (ABCG2 and BCRP), is one of the considered causes of MDR (Lourda et al., 2007). To overcome MDR in treating OS, Susa and colleagues fabricated MDR1-siRNA-loaded, lipid-modified, dextran-based polymeric nanoparticles, suggesting that this approach could reverse drug resistance of MDR cell lines via effective suppression of P-ag expression (Susa et al., 2010). Similarly, siRNA-mediated down-regulation of P-ag also displayed restored chemosensitivity in chemoresistant carcinoma and sarcoma cell lines (Perez et al., 2011). To improve further therapeutic efficiency against MDR, Chen and colleagues studied multifunctional nanoparticles with modified delivery properties (Chen et al., 2010). In their work, liposome-polycation-DNA (LPD) was sequentially modified with cationic lipid, VEGF-siRNA and doxorubicin (Dox) to improve drug uptake and penetration and to suppress angiogenesis. For the sake of inhibition of the drug efflux mediated by P-glycoprotein, they finally designed an anionic LPD, which exhibited significantly improved entrapment efficiency of Dox over cationic LPD. This exploration was the first to co-deliver the chemotherapy agent and oncogene-targeting siRNAs to overcome MDR in different carriers, providing a novel insight into the designing of multi-functional gene-delivery systems to solve MDR in future clinical practice.
Furthermore, siRNA-based gene delivery systems have also been applied in other bone neoplasms. Cheng et al. observed that CD147 expression affected the recurrence rate of GCTB after a clinicopathological analysis of 80 cases and CD147 knockdown by siRNA revealed dramatically inhibited proliferation, migration and invasion of GCTB cells in vitro (Cheng et al., 2015). Lau and colleagues found that p63 was an important factor facilitating GCT tumourigenesis via suppression of p53 by binding to the CDC2 and CDC25C p53-REs, and knockdown of p63 by siRNA manifested greatly decreased proliferation and promoted the apopotsis of GCT stromal cells (Lau et al., 2013). Bertrand and colleagues described that silencing of EWS/FLI-1 (one of the critical oncogenes in Ewing sarcoma) by siRNAs delivered by hydrogenated cationic nanodiamonds (ND-H) had enhanced the cytotoxic impact of vincristine on Ewing sarcoma (ES) cells (Bertrand et al., 2015). Nanodiamonds (NDs), which are suitable for surface modification with therapies, targeting, or labelling of moiety due to their carbon-related surface chemistry, were reported to deliver siRNAs to ES cells by targeting the oncogene EWS/FLI-1 (Alhaddad et al., 2011). This siRNA delivery system not only exhibited strongly suppression oncogene expression, but also enhanced the cytotoxic effect of vincristine, a chemio-therapeutic agent already used in clinics to treat Ewing sarcoma. Therefore, ND-H constitutes a great promising platform for siRNA delivery in anti-bone cancer therapy.
Cancer-induced bone pain is a severe clinical issue affecting the quality of life of patients and even therapeutic outcomes. Recently, siRNA-based gene treatment emerged as an anticipated method to attenuate bone cancer pain (BCP). Huang and colleagues reported that silencing of phosphatidylinositol 3-kinase cb (PI3Kcb) alleviated BCP by increasing glial excitability (Huang and Zhang, 2014). Toll-like receptor 4 (TLR4) has been preliminarily corroborated to be an important synergistic factor involved in proinflammatory cytokine-mediated neuron-to-glial signal activation (Lan et al., 2010), and siRNA-mediated down-regulation of TLR4 yielded obviously attenuated nociception in rats with BCP (Pan et al., 2015). It was reported that hyper-active platelet-derived growth factor (PDGF) could induce tactile, allodynia and selective inhibition of PDGF receptor (PDGFR) produced suppressed tactile allodynia and thermal hyperalgesia (Masuda et al., 2009). Based on this discovery, Xu and colleagues observed that siRNA-mediated PDGF knockdown effectively relieved BCP by blocking the AKT/ERK signalling pathway (Xu et al., 2016). Central sensitization promotes the development of BCP and cAMP response element binding protein (CREB), which is positively regulated by CREB-regulated transcription coactivator1 (CRTC1), which in turn plays a critical role in central sensitization (Bourtchuladze et al., 1994). Liang and colleagues observed that suppression of spinal CRTC1 by siRNA resulted in effectively reduced BCP in mice orthotopically implanted with OS cells (Liang et al., 2016). These studies discussed above may provide further insight into the mechanisms of cancer pain, and contribute to the development of therapeutic strategies for clinical treatment of bone cancer pain.
4.3. siRNA-mediated therapy for other non-lethal diseases
Some other non-lethal diseases, such as osteoarthritis, osteonecrosis and heterotopic ossification, also exert many adverse effects on the quality of life of patients. Recently, siRNA-based targeting gene technologies have offered potential benefits for the treatment of these bone-related diseases. Corticosteroids have been widely used in some infectious and rheumatoid diseases such as severe acute respiratory syndrome (SARS) and rheumatoid arthritis. However, steroid-associated osteonecrosis (SAON) usually occurs after pulsed steroid application for these diseases (Xie et al., 2015). Destructive bone repair is one of the most important pathological changes involved in SAON, which is characterized by obvious bone resorption and vascular hyperpermeability. VEGF mainly contributes to vascular hyperpermeability and it potentially mediates osteoclastic bone resorption (Kim et al., 2004). Src, which is a downstream molecule of VEGF signalling encoded by the c-src gene, participates in osteoclastic bone resorption and VEGF-induced vascular permeability (Boyce et al., 1992). Zheng and colleagues performed systematical evaluation of the therapeutic efficiency of Src blockage by siRNA with regard to SAON both in vitro and in vivo, and they revealed that siRNA-mediated Src suppression significantly suppressed VEGF-induced osteoclastic bone resorption and vascular hyperpermeability by destroying osteoclast cytoskeletons and maintaining endothelial junctional integrity (Zheng et al., 2015, Zheng et al., 2016, Cao et al., 2015a). Moreover, they determined that up-regulation of VEGF expression in chondrocytes derived from femoral head osteonecrosis in pigs was mediated partially by HIF-1α, and silencing of HIF-1α by siRNA transfection led to obviously decreased VEGF expression (Zhang et al., 2012b). Therefore, suppression of VEGF expression using siRNA technology might gain desirable therapeutic value in preventing destructive bone repair due to SAON. Li and colleagues further reported that blockage of peroxisome proliferator-activated receptor-γ (PPAR-γ) by siRNA inhibited adipogenesis and enhanced osteogenesis in BMSCs, thus providing a novel avenue for impeding the development of SAON (Li et al., 2014). PPAR-γ is a specific transcriptional factor correlated with adipogenesis, and knockout of PPAR-γ in BMSCs led to increased osteoblastogenesis (Cao et al., 2015b). This evidence indicates that PPAR-γ is an significant target gene in the development of SAON.
Analogous to the miRNA-mediated targeting therapy for OA, siRNA-based gene-specific therapeutics have displayed potential treatment value in degenerative diseases as well. MMP-13 was reported to facilitate the progression of OA due to its preferential digestion of type II collagen in the early stage of cartilage degradation (Takaishi et al., 2008). Akagi and colleagues demonstrated that intra-articular injection of MMP-13 siRNA in the early stage of a surgically-induced murine OA model showed delayed cartilage degradation (Akagi et al., 2014). However, it remains to be determined whether the duration of MMP-13 siRNA retention is altered in joints with experimental OA, and the knockdown effect mediated by siRNA in other tissues such as cartilage should also be investigated. Autologous chondrocyte implantation (ACI) is a cell-based therapy and was first reported in 1994 (Brittberg et al., 1994). Since then, it has been widely performed on more than 12,000 patients with OA. However, the success of ACI always depends on the phenotypic stabilization of chondrocytes. Legendre et al. observed that the gene blockage of collagen type I alpha 1 (COL1A1) and high temperature requirement factor A1 (HtrA1) via siRNA induced an distinctly enhanced differentiation index of chondrocytes cultured with BMP-2 under hypoxic conditions, thus offering the potential opportunity to explore novel ACI-based treatment in the future (Legendre et al., 2013, Ollitrault et al., 2015). It was corroborated that overexpression of ADAMTS-5 mediated by IL-1β induced aggravated degradation of cartilage matrix in OA and significantly inhibited miR-30a expression levels (Ji et al., 2016a). siRNA-mediated blockage of ADAMTS-5 demonstrated protective effects on cartilage degradation (Chu et al., 2013). Moreover, intra-articular delivery of hypoxia-inducible factor-2α (HIF-2α) siRNA by chondrocyte-homing nanoparticles resulted in considerably decreased expression of some catabolic proteins involved in the progression of OA, such as MMP-13, VEGF, and ADAMTS-5, thus providing better prevention of cartilage degeneration in OA patients (Pi et al., 2015). The hypoxia signalling pathway involved in bone and cartilage regeneration was comprehensively summarized in a detailed review (Stiers et al., 2016), and further research is badly needed to determine hypoxia-associated gene targeting strategies to improve the therapeutic potential regarding to OA.
Additionally, siRNAs have also been used to explore potential therapies for autosomal dominant diseases in bone, which are characterized by severe morbidity and lack of effective treatments. It has been verified that Albers-Schönberg disease, also called autosomal dominant osteopetrosis type 2 (ADO2), is characterized by significantly inhibited osteoclast activity, mainly due to heterozygous missense mutations of the CLCN7 gene. Capulli et al. reported a CLCN7-specific siRNA-based exploration using a cationic polymer transfection reagent both in vitro and in vivo, leading to overall improvement in the osteopetrotic bone phenotype (Capulli et al., 2015). This study provided a feasible siRNA-based therapeutic strategy for CLCN7-dependent ADO2, and it emphasizes the translational impact of future treatment on the therapeutically neglected osteopetrosis.
5. Summary of delivery systems and clinical status in small non-coding RNAs-based therapeutics for bone-related disorders
The primary role of a delivery system is to facilitate the cellular uptake of miRNAs or siRNAs to their target sites. A delivery system can also protect nucleic acids from premature nuclease degradation, which can affect the specificity and functionality of the RNA molecules. It has been well documented that miRNAs or siRNAs themselves are prone to nuclease degradation with low cell membrane penetration capability due to their highly charged backbones. In addition, inflammation-related cell toxicity and unintended gene knockdown caused by off-target effects derived from the application of miRNAs or siRNAs should be paid great attention when delivering them in vivo. Thus, establishing rational and effective miRNAs- or siRNAs-based drug delivery systems (DDSs) is of great importance to their targeting therapeutics in bone disorders. Since miRNAs and siRNAs have similar physicochemical properties (double-stranded RNAs with 20–25 nucleotides) and the same intracellular sites of actions (both require enzymatic functions of the RISC to be active against target mRNAs), similar delivery technologies can be applied to both types of RNA molecules.
There are mainly two types of DDSs for transferring miRNAs or siRNAs: viral-mediated and non-viral vectors. Viruses, such as lentiviruses, retroviruses, adenoviruses, and herpesviruses, appear to be the first choices for miRNA or siRNA delivery, and they are extremely efficient in transferring RNA-encoding vectors into the nuclei of mammalian cells to ensure high expression of RNA. Almost 70% of gene therapy clinical trials have involved the application of viral vectors to deliver nucleic acids due to their high transduction efficiency (Yin et al., 2014). In addition, long-term expression can be achieved using viruses. However, the major limitations of viral vectors are biological safety issues, including possible carcinogenicity, high viral toxicity and verified immunogenicity (Wang et al., 2008). Therefore, despite the inferior transfection efficiency of viral vectors, non-viral transfer systems with low acute toxicity, lower production cost, non-infection and non-immunogenicity are emerging as a desirable approach to deliver miRNAs or siRNAs in vivo (Castanotto et al., 2009). Two major categories of non-viral DDSs used for miRNAs or siRNAs delivery in therapeutic applications are lipid-based and polymer-based systems. Apart from the advantages mentioned above (i.e., relatively favourable safety profiles and low production costs), nonviral vectors are highly versatile. They can be easily modified to promote their delivery efficiency, for example, to improve serum stability and to extend the circulation time by PEGylation (Buyens et al., 2012), or to achieve site-specific delivery by incorporating targeting ligands (i.e., (AspSerSer)6 and Asp8) (Zhang et al., 2012a, Liang et al., 2015). Despite efforts to develop suitable miRNA or siRNA delivery systems for clinical application, a poor correlation between in vitro and in vivo efficacies has also been observed, and it has often been reported that a delivery system functioned effectively in vitro but failed in vivo because of poor pharmacokinetic profiles, toxicity issues, immune responses or nonspecific uptake (Lv et al., 2006). Therefore, the success of therapeutic miRNAs and siRNAs is highly dependent on the availability of an efficient and safe delivery system. Selected examples of viral and non-viral delivery systems that have been explored to deliver therapeutic miRNAs and siRNAs both in vivo and in vitro are summarized in Table 1 and Table 2 , respectively. For a better understanding of the nucleic acids and gene delivery, please refer to the chapter summarized by Akita et al. (2011)
Table 1.
Summary of preclinical investigations on miRNAs-mediated targeting therapeutics in bone-related disorders.
| Disorders | miRNA | Delivery system | Regulation | Gene of interest | In vitro/In vivo | Outcome summary | Reference |
|---|---|---|---|---|---|---|---|
| Osteoporosis (OP) | mmu-miR-17/20a-5p | Lipofectamine 2000 complexes | ↑ | RANKL | MC3T3-E1, C57BL/6 mice | 窗体顶端 | (Shi et al., 2014) |
| Attenuated Dexamethasone- induced osteoclast differentiation and function | |||||||
| mmu-miR-29a-3p | Lentivirus | ↑ | Dkk-1 | SD rat | Protects against GC-induced bone loss and fragility | (Wang et al., 2013b) | |
| mmu-miR-34a-5p | X-tremeGENE Transfection Reagent | ↓ | CDK4, CDK6, | C57BL/6 mice | 窗体顶端 | (Kang et al., 2016a) | |
| Cyclin D1 | Impaired BMSC osteogenic differentiation and aggravated osteoporsis | ||||||
| mmu-miR-140/214-3p | Baculovirus vectors | ↓ | JAG1, Tmem119, ATF-4, Osterix | SD rat | Promoted bone formation and mitigated excessive bone resorption | (Li et al., 2016c) | |
| mmu-miR-34a-5p | Lipofectamine RNAi MAX |
↑ | Tgif2 | C57BL/6J mice | Elevated bone resorption and reduced bone mass | (Krzeszinski et al., 2014) | |
| Bone defect | hsa-miR-21-5p | Chitosan/hyaluronic acid nanoparticles | ↑ | Runx2 | hBMSC | Significantly improved osteogenesis | (Schoolmeesters et al., 2009, Wang et al., 2015b) |
| mmu-miR-26a-5p | Lipofectamine 2000 complexes | ↑ | OCN, COL1 | 窗体顶端 | Satisfactory bone repair in tibial defect | (Wang et al., 2015a) | |
| Rabbit ASC | |||||||
| mmu-miR-29a-3p | Gelatin solutions | ↓ | IGF1, tfgb1 | MC3T3-E1 | Significantly enhanced ECM deposition | (James et al., 2014) | |
| mmu-miR-31-5p | Lipofectamine 2000 complexes | ↓ | Runx2, Satb2 | SD rat | Good biocompatibility and bone regeneration | (Deng et al., 2014, Deng et al., 2013) | |
| mmu-miR-138-5p | Lipofectamine 2000 complexes | ↓ | Runx2 | C57BL/6 mice, | 窗体顶端 | (Yan et al., 2014) | |
| rat BMSC | Massive bone with good vascularization | ||||||
| hsa-miR-148b-3p | Silver nanoparticle complexes | ↑ | Runx2, OCN | hASC | Promoted osteogenic differentiation | (Qureshi et al., 2013) | |
| Primary bone tumor | hsa-miR-20a-5p | Lipofectamine 2000 complexes | ↓ | EGR | MG63, SaSO2, SW1353 | Inhibited cell proliferation and cycle | (Zhou et al., 2015) |
| hsa-miR-22-3p | Lipofectamine 2000 complexes | ↑ | ACLY | SaSO2, PC3, Hela, A549 | Inhibited growth and metastasis of various tumors | (Xin et al., 2016) | |
| hsa-miR-26a-5p | Lentivirus vector | ↑ | Jagged1 | MG63, SaSO2, 143B, U2OS, BALA/c nude mice | Suppressed CSCs phenotype and tumor growth | (Lu et al., 2016) | |
| hsa-miR-106a-5p | Lipofectamine 2000 complexes | ↑ | HMGA2 | MG63, SaSO2, HOS, U2OS | Inhibited proliferation, migration and invasion | (He et al., 2016) | |
| hsa-miR-106b-5p | FuGENE HD transfection agent | ↑ | RANKL, IL-8, MMP2, TWIST | mice BMM, GCTSC, OVX mouse | Inhibited bone resorption and osteolysis | (Wang et al., 2015c) | |
| hsa-miR-126-5p | FuGENE HD transfection agent | ↑ | MMP-13, PTHrP | mice BMM, GCTSC | Suppression of osteoclast differentiation and bone resorption | (Wu et al., 2014a, Zhou et al., 2014) | |
| hsa-miR-126-5p | Lipofectamine 2000 complexes | ↑ | Sirt 1 | SaSO2 | 窗体顶端 | (Shi et al., 2015, Xu et al., 2013) | |
| inhibited proliferation, migration, invasion and epithelial-mesenchymal transition | |||||||
| hsa-miR-204-5p | |||||||
| hsa-miR-145-5p | Lipofectamine 3000 complexes | ↓ | FLI-1 | MG63, SaSO2 | Inhibited tumor progression | (Wu et al., 2016b) | |
| hsa-miR-409-3p | Lipofectamine 2000 complexes | ↑ | Catenin-δ1 | U2OS | Inhibited migration and invasion | (Wu et al., 2016a) | |
| hsa-miR-646 | Lipofectamine 2000 complexes | ↑ | FGF-2 | MG63, SaSO2, HOS, U2OS | Suppression of metastasis | (Sun et al., 2015) | |
| Secondary metastasis | hsa-miR-34a-5p | Lentivirus vector | ↑ | TCF-7 | PC3, RasB1 | Inhibited bone metastasis of Ras-activated PC | (Chen et al., 2015) |
| hsa-miR-335-5p, | Lentivirus vector | ↑ | eNOS | PC3 | Inhibited bone metastasis of PC | (Fu et al., 2015) | |
| hsa-miR-543 | |||||||
| Osteoarthritis (OA) | hsa-miR-23a-3p, | Lipofectamine 2000 complexes | ↓ | Samd3 | SW1353 | Promoted type II collagen and aggrecan expression | (Kang et al., 2016b, Li et al., 2015) |
| hsa-miR-16-5p | |||||||
| hsa-miR-139-5p | Lipofectamine RNAi MAX |
↓ | EIF4G2, IGF1R | CHON-001 | Protective effects on chondrocyte proliferation and migration | (Hu et al., 2016) | |
| hsa-miR-142-3p | Lipofectamine 2000 complexes | ↑ | HMGB-1 | C57BL/6 mice | Suppressed inflammation and chondrocyte apoptosis | (Wang et al., 2016b) | |
| mmu-miR-210-5p | Geneporter 2 Transfection Reagent |
↑ | DR6 | SD rat | Suppressed inflammation and chondrocyte apoptosis | (Zhang et al., 2015a) | |
| hsa-miR-365a-3p | Lipofectamine 2000 complexes | ↓ | HDAC4 | Primary chondrocyte from OA patient | Inhibited expression of MMP13 and Col X | (Yang et al., 2016) |
Table 2.
Summary of preclinical investigations on siRNAs-mediated targeting therapeutics in bone-related disorders.
| Disorders | Targeting gene | Delivery system | In vitro/In vivo | Outcome summary | Reference |
|---|---|---|---|---|---|
| Bone formation and resorption | FPPS | DharmaFECT4 (cationic lipid transfection reagent) | rat BMSC, MC3T3-E1 | Selectively inhibiting osteoclast-mediated bone resorption and improving osteoblast differentiation without effects on osteoblast mineralization | (Wang et al., 2011) |
| GNAS1, PHD2 | Ambion (polyamine-based transfection agent) | hBMSC, Sheep (local administration) | Both support bone regeneration in vivo, whereas only siGNAS1 promoted collagen I and osteopontin expression in vitro | (Ríos et al., 2012) | |
| siFlt-1, Ckip-1 | Lipofectamine 2000 complexes | rat BMSC, SD rat (local administration) | Satisfactory angiogenesis and osteogenesis in BMSCs and bone repair efficacy in vivo | (Jia et al., 2014) | |
| Ckip-1 | Lipofectamine 2000 complexes | MC3T3-E1,, C57/BL mice, SD rat (tail vein injection) | Increased osteogenic differentiation in vitro and reversed bone loss in osteoporotic mice | (Guo et al., 2014) | |
| Plekho1 | DOTAP cationic liposomes | SD rat (tail vein injection) | Increased bone microarchitecture and bone formation in both healthy and osteoporotic rats. | (Zhang et al., 2012a) | |
| Plekho1 | LNPs | SD rat (tail vein injection) | Improved bone mass in osteoporotic rats | (Liang et al., 2015) | |
| RANK | Cationic liposome DMAPAP/DOPE | C57BL/6 mice (local administration) | Effectively impeded osteolysis and increased bone formation | (Córdova et al., 2015) | |
| RANK | PLGA microparticles | mice BMSC | Inhibited osteoclastic phenotype | (Wang et al., 2012b) | |
| GCR | PLGA microparticles | hBMSC | Improved osteogenic proliferation and differentiation | (Hong et al., 2012) | |
| Sema4d | HPMA copolymer | Balb/c; Kunming mice (intravenous injection) | Partly counteracted bone loss originated from osteoporosis | (Zhang et al., 2014, Zhang et al., 2015b) | |
| Bone formation and resorption | Sema4d | PLLA scaffold | Wistar rat (local administration) | Improved bone formation in osteoporosis-related bone defects | (Zhang et al., 2016) |
| Noggin | PEG hydrogels scaffolds | hBMSC | Well guided osteogenic differentiation | (Nquyen et al., 2014) | |
| Noggin | PLA-DX-PEG | ICR mice (local administration) | Enhanced BMP-induced ectopic bone formation | (Manaka et al., 2011) | |
| Bone tumor | VEGF | Adenoviral vector | SaSO2 | Suppressed osteosarcoma angiogenesis and promoted apoptosis | (Peng et al., 2016) |
| VEGF | Adenoviral vector | SD rat (intratumoral injection) | Inhibited tumor growth | (Zhu and Mei, 2015) | |
| TCTP | Lentiviral particles | SaSO2, U2OS, MG63,nude mice (subcutaneous injection) | Promoted cell apoptosis and inhibited development of xenografts | (Shen et al., 2016) | |
| L1CAM | Liposome SN | PC3, nude mice (intratumoral injection) | Admirable suppression of PC proliferation and bone metastasis | (Sung et al., 2014) | |
| ABCB1 | Lipid-modified dextran polymer | KHOSR2, U2OSR2 | Reversed drug resistance of MDR cell lines | (Susa et al., 2010) | |
| ABCB1 | Lipofectant | hMNNG/HOS, 293T | Restored chemosensitivity in MDR cell lines | (Perez et al., 2011) | |
| VEGF | liposome-polycation-DNA (LPD) | NCI/ADR-RES,OVCAR-8, nude mice (tail vein injection) | Reversed MDR by multi-functional gene delivery systems | (Chen et al., 2010) | |
| CD147 | Adenoviral vector | Primary GCTB from patient | Dramatically inhibited proliferation, migration and invasion of GCTB cell | (Cheng et al., 2015) | |
| P63 | Lipofectamine RNAiMAX | Primary GCTSCs from patient | Dramatically inhibited proliferation of GCTB cell | (Lau et al., 2013) | |
| Bone tumor | EWS/FLI-1 | hydrogenated cationic nanodiamonds | A673 human; Ewing sarcoma cell line | Strongly inhibited oncogene expression and enhanced cytotoxic effect of vincristine | (Bertrand et al., 2015) |
| PI3Kcb | Adenoviral vector | MADB-106, SD rat (local administration) | Alleviated bone cancer pain (BCP) by increasing glia excitability | (Huang and Zhang, 2014) | |
| TLR4 | Adenoviral vector | Walker256, SD rat (intrathecal injection) | Attenuated nociception in rats with BCP | (Pan et al., 2015) | |
| PDGFR | HSV vector | SD rat (subarachnoid administration) | Effectively relieved BCPin the spinal cord | (Xu et al., 2016) | |
| CRTC1 | Adenoviral vector | NCTC2472, C3H/HeNCrl/Vr mice (intrathecal injection) | Reduced BCP in mice orthotopically implanted with OS cells | (Liang et al., 2016) | |
| SAON | Src | Lipofectamine 2000 complexes | New-Zealand white Rabbit (local administration) | Inhibited destructive repair of osteonecrosis | (Zheng et al., 2015, Cao et al., 2015a, Zheng et al., 2016) |
| PPAR-γ | Adenovirus shuttle vector | New-Zealand white Rabbit (local administration) | Inhibited adipogenesis of BMSCs and prevention of SAON in rabbit | (Li et al., 2014) | |
| MMP-13 | Dharmacon Accell | C57BL/6 mice (intra-articular injection) | Delayed cartilage degradation at the early stage of OA development | (Akagi et al., 2014) | |
| COL1A1, HtrA1 | Type I collagen sponges | Human articular chondrocyte (HAC), rabbit articular chondrocyte (RAC) | Enhanced differentiation index of chondrocytes cultured with BMP-2 under hypoxia condition | (Legendre et al., 2013, Ollitrault et al., 2015) | |
| ADAMTS-5 | Lentivirus vector | SD rat (intra-articular injection) | Protective effect on cartilage degradation | (Chu et al., 2013) | |
| HIF-2α | Chondrocyte- homing nanoparticles | Mouse (intra-articular injection) | Inhibited catabolic proteins expression in OA, and maintained cartilage integrity | (Pi et al., 2015) | |
| ADO2 | CLCN7 | Lipofectamine | RAW264.7, Human osteoclasts, mice (subcutaneous injection) | Inhibited increase of bone mass in growing ADO2 mice | (Capulli et al., 2015) |
The first clinical trial of a miRNA therapy began in 2013, with the second one starting in early 2015. In contrast, the first clinical trial of a siRNA therapeutic was initiated in 2004 (Ozcan et al., 2015). Through searching on ClinicalTrials.gov, to date, we found approximately 50 siRNA candidates that have reached various stages of clinical trials for the treatment of different diseases, and approximately 50% of these trials are focused on tumour-related targeting therapeutics. However, the clinical development of miRNAs as therapeutics is lagging significantly, with only the two miRNA therapeutics mentioned above, both of which are indicated for the treatment of cancers. The relatively slow progress of miRNA therapeutics is mainly due to their uncertain mechanisms and complex roles of action and specificity during the early years of their discovery. In addition, the diverse potential applications (e.g., as drug targets and biomarkers) of miRNAs, which have been extensively discussed before, might also have detracted from their development as therapeutic agents. However, all of the therapeutic miRNAs or siRNAs in clinical trials are either for topical administration (i.e., ocular or dermal), or they systemically targeting the liver, where most injected RNAs end up non-specifically anyway. Unfortunately, there have been no clinical trials initiated for bone-related disorders at the time of our writing. Therefore, many efforts still must be exerted for the improvement and better understanding of miRNA and siRNA therapeutics, which will be discussed latter.
6. Challenges facing small non-coding RNAs-based therapeutics in bone-related disorders
Synthetic miRNAs and siRNAs hold great promise as new classes of therapeutic agents by silencing genes of interest, and they have been studied for the treatment of various human diseases, such as cancers, genetic disorders, cardiovascular diseases and bone-related disorders. It has been well documented that miRNAs and siRNAs have similar physicochemical properties that are involved in the regulation of gene silencing, yet their mechanisms of action are different; thus, the requirements for sequence design and therapeutic application are distinct. The major difference between them is that the former has multiple mRNA targets, whereas the latter is highly specific, with only one target. Therefore, in some complex multi-genic diseases, miRNAs might have predominance over siRNAs. The design of miRNAs is more straightforward than that of siRNA when used for bone therapeutics because the sequence of the former should be almost, if not entirely, identical to the endogenous miRNA of interest. Moreover, miRNAs have an advantage over siRNAs as therapeutics for complex multigenic bone-related diseases, such as congenital dislocation of the hip (CDH) and rachischisis, which require modulation of multiple pathways for effective treatment. With the ability to inhibit the expression of a number of target genes, which often work together as a network within the same cellular pathway, a whole disease phenotype can potentially be altered through a single miRNA sequence. In contrast, the therapeutic potential of siRNAs is sometimes limited by their ability to target only one specific gene. Therefore, it could be challenging to employ siRNAs to regulate complex bone-related diseases.
Based on the overall analysis of miRNA- and siRNA-based therapeutics mentioned above, we can know that these therapies have emerged as excellent genetic tools in bone biology, holding great therapeutic promise in the field of gene therapy. However, there are several universal challenges facing the application of miRNA and siRNA therapeutics, although one of the distinctive features that differentiate siRNAs from miRNAs is that siRNAs are designed to silence the expression of a specific target mRNA, leading to the downregulation of unintended, unpredicted targets, which are also called off-target effects. Three main off-target effects of siRNAs were discovered with potentially unwanted side effects almost a decade ago (Jackson and Linsley, 2010). Firstly, silencing of many unintended genes through partial sequence complementarity always produces adverse effects on the normal biology. In addition, there was an inflammatory response caused by either the siRNA itself or the DDS used. Finally, the saturation of endogenous RNAi machinery could inevitably affect normal miRNA functionality. Thus, one of the major challenges of siRNA therapy is to reduce off-target effects, which have been verified to compromise the therapeutic effects and specificity and can even lead to cell death (Fedorov et al., 2006). It is also important to have a clear overview of the barriers to in vivo treatment with miRNAs and siRNAs. Obstacles to miRNA or siRNA delivery depend on the target organ and route of administration. Different in vivo administration routes will encounter different physiological barriers. For instance, intravenous administration is the most commonly used technique, and the endothelial wall in the vasculature serves as the primary barrier to RNA delivery. Moreover, the mononuclear phagocytes and reticulo-endothelial system are responsible for the physiological barriers that hinder miRNAs or siRNAs from reaching their targets, leading to reduced therapeutic efficacy. Furthermore, both miRNA and siRNA therapeutics face the same major challenges of prolonged and stable delivery, improved potency and minimized cell and tissue toxicity. Finally, several current in vitro studies with therapeutic RNAi have provided results that are largely not comparable with in vivo results with the same reagents. This disconnect is a current barrier to predictability and reliability for clinical translation.
7. Conclusion and future expectation
miRNAs and siRNAs, both of which exert gene silencing effects on physiological and pathological processes at the post-transcriptional level by targeting relevant and specific mRNA, represent innovative and promising breakthroughs in the field of genetic engineering. Remarkable progress has been made in the biological, diagnostic and therapeutic potential of these two small non-coding RNAs for bone metabolism and bone-related disorders. The experience from antisense and gene therapy has contributed to the rapid progress of miRNAs and siRNAs in clinical studies and in particular, the technology of the chemical modification and delivery of nucleic acids developed previously can be applied to both miRNAs and siRNAs. Thorough investigation and full understanding of the function of miRNAs involved in various bone-related physiological and pathological activities hold great significance for the design of specific and accurate gene-silencing therapeutics mediated via siRNAs.
Currently, siRNAs have demonstrated obvious superiority over miRNAs in the progress of advancing into clinical trials, as discussed before. Orthopaedic applications for therapeutics RNAs have the advantage of the surgeon entering the site of therapy surgically for repair, allowing RNA to be administered locally to these sites (topically or in combination with a device). The most attractive aspects of miRNA and siRNA therapeutics are their abilities to target virtually any genes, which might not be possible with protein-based drugs or small molecules. While the therapeutic efficacy of miRNAs and siRNAs has been successfully shown both in vitro and in vivo, several technical barriers still must be overcome for these RNA molecules to be used clinically. While the majority of the literature focuses on miRNA or siRNA expression and gene targeting, there is a need to assess their processing and architecture because they are closely related to the targeting effects of MSC differentiation. There remains a need for more studies evaluating the regulation of miRNA or siRNA processing and biogenesis during osteogenesis and their related proteins, which could lead to novel mechanisms for miRNA- or siRNA-based interventions in bone disorders. To avoid silencing of unintended genes, lowering the concentration of miRNAs or siRNAs used could be an effective approach for reducing these unwanted side-effects mentioned above, while decreasing the concentration to the minimally effective limit can also significantly alleviate the burden on RNAi machinery and avoid disrupting physiological processes. For proper therapeutic use, the sequences of RNA must be carefully designed to avoid any specific or nonspecific unwanted side effects and immune responses. To avoid the off-target effects induced by siRNAs, one feasible approach is to reduce the complementarity between the seed region (2–7 nucleotides of 5′ end) of the siRNA and the 3′UTR of the mRNA. In addition, the siRNA should have a low thermodynamic stability of the duplex between the seed region of the guide strand of the siRNA and its target mRNA since a low seed-target duplex stability reduces the ability of the siRNA to trigger seed-dependent, off-target effects (Naito and Ui-Tei, 2013).
The use of nonviral vectors, including polymer-based or lipid-based delivery systems, offers the advantages of better safety profiles and lower production costs compared to viral vectors, despite inferior delivery efficiency. In addition, there are also a few additional design criteria that can promote the efficacy of miRNA and siRNA therapeutics in bone. Incorporation of miRNA-or siRNA-loaded particles inside injectable biodegradable matrices might be highly favorable in localizing the delivery of miRNAs or siRNAs to the sites of bone defects, osteoporosis or tumour, thus, maximizing the therapeutic effect. Meanwhile, such an approach would also circumvent many physiological barriers in the case of systemic administration. The addition of targeting moieties is considered to be another significant approach to improving the efficacy of siRNA delivery. The three most commonly used moieties for bone-related disorders are bisphosphonates, bone-specific oligopeptides and aptamers, which have been discussed before (Zhang et al., 2012a, Liang et al., 2015, Wang et al., 2006, Zhang et al., 2014, Zhang et al., 2015b). The discovery of molecules with high affinity for bone surfaces will improve the therapeutic effect and clinical relevance of miRNA- or siRNA-based delivery systems. In addition to improving the therapeutic effects of any miRNA or siRNA therapies analysed above, there are also safety issues to be considered to move the technology into a clinical setting. It is very important to fabricate the DDS and injectable matrices with biocompatible and degradable biomaterials, with degradation byproducts that are noncytotoxic, and with materials can be cleared rapidly and readily from the body without accumulation in the liver or other organs. Cationic polymers have been extensively used for oligonucleotide delivery due to their capability to complex readily with RNA or DNA as mentioned above (Wang et al., 2011, Zhang et al., 2012a, Córdova et al., 2015). However, upon degradation some cationic polymers can induce cytotoxic effects on surrounding cells and can cause red blood cell lysis (Fischer et al., 2003). Therefore, hydrolysable polymers, such as cyclodextrin-containing polymers, which can degrade more readily and rapidly, could gain greater attention when designing the DDS involved in miRNA-or siRNA-based therapeutics. In addition, the duration of silencing effect after a single dose of miRNAs or siRNAs depends on several factors, such as the stability of the RNA molecules, the rate of RNA release from the delivery system, the type of target tissues, together with the half-life and turnover rate of the target proteins. A good understanding of this area could also contribute to the rational design of therapeutic strategies to promote clinical outcomes of miRNA and siRNA treatments. For instance, PEGylated nanoparticles incorporating targeting ligands might be effective for avoiding reticulo-endothelial system (RES) clearance, thus prolonging the circulation time and the stability of miRNA and siRNA delivery systems.
To improve the stability of miRNAs and siRNAs as therapeutic agents, relevant chemical modifications of RNA were developed initially to address this issue. Successful chemical modification should not compromise the gene-silencing efficiency of these RNA molecules. The major types of chemical modification that are commonly investigated in miRNA and siRNA design including locked and unlocked nucleic acids, ribose 2′-OH groups and phosphorothioate modifications (Allerson et al., 2005). Different RNA modifications can be used to serve various functions, and proper chemical modification is also an effective approach for reducing the off-target effects analysed above. Efficient gene silencing can also be achieved with naked RNA; however, this approach might be limited to local administration to organs such as the lungs and the eyes. Additionally, proper chemical modification (such as incorporation of specific nuclease inhibitors) of the RNA molecule could be beneficial for improving its stability against nuclease activity in the airway fluid of the lungs or in the vitreous humour of the eye. Although miRNA-and siRNA-based therapy remains a nascent field in bones, it is expected that with advances in RNA design, specific biochemical modifications and better understand-ing of the effects and duration of gene silencing, miRNAs and siRNAs will realize a gradual and progressive translation of preclinical experiments into clinical trials in the near future.
Disclosure
The authors report no conflicts of interest in this work.
References
- Abramson S.B., Attur M., Amin A.R. Nitric oxide and inflammatory mediators in the perpetuation of osteoarthritis. Curr. Rheumatol. Rep. 2001;3:535–541. doi: 10.1007/s11926-001-0069-3. [DOI] [PubMed] [Google Scholar]
- Akagi R., Sasho T., Saito M. Effective knock down of matrix Metalloproteinase-13 by an intra-articular injection of small interfering RNA (siRNA) in a murine surgically-induced osteoarthritis model. J. Orthop. Res. 2014;32:1175–1180. doi: 10.1002/jor.22654. [DOI] [PubMed] [Google Scholar]
- Akita H., Hatakeyama H., Khalil I.A. Chapter 4.425 delivery of nucleic acids and gene delivery. Compr. Biomater. 2011;23:411–444. [Google Scholar]
- Alhaddad A., Adam M.P., Botsoa J. Nanodiamond as a vector for siRNA delivery to Ewing sarcoma cells. Small. 2011;7:3087–3095. doi: 10.1002/smll.201101193. [DOI] [PubMed] [Google Scholar]
- Allerson C.R., Sioufi N., Jarres R. Fully 2’-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- Ao M., Williams K., Bhowmick N.A. Transforming growth factor- beta promotes invasion in tumorigenic but not in nontumorigenic human prostatic epithelial cells. Cancer Res. 2006;66:8007–8016. doi: 10.1158/0008-5472.CAN-05-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Atkins G.J., Kostakis P., Vincent C. RANK expression as a cell surface marker of human osteoclast precursors in peripheral blood, bone marrow, and giant celltumors of bone. J. Bone. Min. Res. 2006;21:1339–1349. doi: 10.1359/jbmr.060604. [DOI] [PubMed] [Google Scholar]
- Bae Y.J., Yang T., Zeng H.C. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglìo S.R., Devescovi V., Granchi D. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene. 2013;527:321–331. doi: 10.1016/j.gene.2013.06.021. [DOI] [PubMed] [Google Scholar]
- Bazzini A.A., Lee M.T., Giraldez A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.R., Pioche-Durieu C., Ayala J. Plasma hydrogenated cationic detonation nanodiamonds efficiently deliver to human cells in culture functional siRNA targeting the Ewing sarcoma junction oncogene. Biomaterials. 2015;45:93–98. doi: 10.1016/j.biomaterials.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Bourtchuladze R., Frenguelli B., Blendy J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Boyce B.F., Yoneda T., Lowe C. Requirement of pp60c-src expression for osteoclasts to form ruffled borders and resorb bone in mice. J. Clin. Invest. 1992;90:1622–1627. doi: 10.1172/JCI116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw M., Mansfield A., Peikert T. The role of vascular endothelial growth factor in the pathogenesis, diagnosis and treatment of malignant pleural effusion. Curr. Oncol. Rep. 2013;15:207–216. doi: 10.1007/s11912-013-0315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittberg M., Lindahl A., Nilsson A. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N. Engl. J. Med. 1994;331:889–895. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- Buyens K., De Smedt S.C., Braeckmans K. Liposome based systems for systemic siRNA delivery : stability in blood sets the requirements for optimal carrier design. J. Control. Release. 2012;158:362–370. doi: 10.1016/j.jconrel.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Cao Z., Moore B.T., Wang Y. MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One. 2014;9:e97098. doi: 10.1371/journal.pone.0097098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H.J., Zheng L.Z., Wang N. Src blockage by siRNA inhibits VEGF-induced vascular hyperpermeability and osteoclast activity-an in vitro mechanism study for preventing destructive repair of osteonecrosis. Bone. 2015;74:58–68. doi: 10.1016/j.bone.2014.12.060. [DOI] [PubMed] [Google Scholar]
- Cao J., Ou G., Yang N. Impact of targeted PPARγ disruption on bone remodeling. Mol. Cell. Endocrinol. 2015;410:27–34. doi: 10.1016/j.mce.2015.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A.I., Dennis J.E. Mesenchymal stem cells as trophic mediators. J. Cell. Biochem. 2006;98:1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- Capulli M., Maurizi A., Ventura L. Effective small interfering RNA therapy to treat CLCN7-dependent autosomal dominant osteopetrosis type 2. Mol. Ther-Nucl Acids. 2015;4:e248. doi: 10.1038/mtna.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B.S., Epstein J.I., Isaacs W.B. Ras gene mutations in human prostate cancer. Cancer Res. 1990;50:6830–6832. [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño I.M., Curtin C.M., Duffy G.P. Next generation bone tissue engineering: non-viral miR-133a inhibition using collagen-nanohydroxyapatite scaffolds rapidly enhances osteogenesis. Sci. Rep. 2016;6:27941. doi: 10.1038/srep27941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., Rossi J.J. The promises and pitfalls of RNA- interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bathula S.R., Li J. Multifunctional nanoparticles delivering small interfering RNA and doxorubicin overcome drug resistance in cancer. J. Biol. Chem. 2010;285:22639–22650. doi: 10.1074/jbc.M110.125906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Y., Liu S.Y., Chang Y.S. MicroRNA-34a regulates WNT/TCF7 signaling and inhibits bone metastasis in Ras-activated prostate cancer. Oncotarget. 2015;6:441–457. doi: 10.18632/oncotarget.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D.D., Hu T., Zhang H.Z. Factors affecting the recurrence of giant cell tumor of bone after surgery: a clinicopathological study of 80 cases from a single center. Cell Physiol. Biochem. 2015;36:1961–1970. doi: 10.1159/000430164. [DOI] [PubMed] [Google Scholar]
- Cho S.W., Goldberg M., Son S.M. Lipid-like nanoparticles for small interfering RNA delivery to endothelial cells. Adv. Funct. Mater. 2009;19:3112–3118. doi: 10.1002/adfm.200900519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholas R., Kunjalukkal Padmanabhan S., Gervaso F. Scaffolds for bone regeneration made of hydroxyapatite microspheres in a collagen matrix. Mater. Sci. Eng C. Mater. Biol. Appl. 2016;63:499–505. doi: 10.1016/j.msec.2016.03.022. [DOI] [PubMed] [Google Scholar]
- Chu X., You H., Yuan X. Protective effect of lentivirus-mediated siRNA targeting ADAMTS-5 on cartilage degradation in a rat model of osteoarthritis. Int. J. Mol. Med. 2013;31:1222–1228. doi: 10.3892/ijmm.2013.1318. [DOI] [PubMed] [Google Scholar]
- Coldwell M.J., Cowan J.L., Vlasak M. Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: a reappraisal of translation initiation during mitosis. Cell Cycle. 2013;12:3615–3628. doi: 10.4161/cc.26588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdova L.A., Trichet V., Escriou V. Inhibition of osteolysis and increase of bone formation after local administration of siRNA-targeting RANK in a polyethylene particle-induced osteolysis model. Acta Biomater. 2015;13:150–158. doi: 10.1016/j.actbio.2014.10.042. [DOI] [PubMed] [Google Scholar]
- Cowan R.W., Singh G. Giant cell tumour of bone: a basic science perspective. Bone. 2013;52:238–246. doi: 10.1016/j.bone.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Cui Y., Niziolek P.J., MacDonald B.T. Lrp5 functions in bone to regulate bone mass. Nat. Med. 2011;17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daka A., Peer D. RNAi-based nanomedicines for targeted personalized therapy. Adv. Drug. Deliv. Rev. 2012;64:1508–1521. doi: 10.1016/j.addr.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Del Fattore A., Teti A., Rucci N. Osteoclast receptors and signaling. Arch. Biochem. Biophys. 2008;473:147–160. doi: 10.1016/j.abb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Deng Y., Zhou H., Zou D. The role of miR-31-modified adipose tissue-derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials. 2013;34:6717–6728. doi: 10.1016/j.biomaterials.2013.05.042. [DOI] [PubMed] [Google Scholar]
- Deng Y., Bi X., Zhou H. Repair of critical-sized bone defects with anti-miR-31-expressing bone marrow stromal stem cells and poly(glycerol sebacate) scaffolds. Eur. Cell. Mater. 2014;27:13–24. doi: 10.22203/ecm.v027a02. [DOI] [PubMed] [Google Scholar]
- Djuranovic S., Nahvi A., Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman M.B., Yan D., Ahmadinia K. Fibroblast growth factor control of cartilage homeostasis. J. Cell. Biochem. 2013;114:735–742. doi: 10.1002/jcb.24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F., Bertin T., Ma O. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum. Mol. Genet. 2009;18:1464–1470. doi: 10.1093/hmg/ddp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen T., Taipaleenmäki H., Stenvang J. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.J., Deng Y., Gu P. MicroRNAs regulate bone development and regeneration. Int. J. Mol. Sci. 2015;16:8227–8253. doi: 10.3390/ijms16048227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y., Anderson E.M., Birmingham A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischer D., Li Y., Ahlemeyer B. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- Franzoso G., Carlson L., Xing L. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Bai R., Zhao Z. Overexpression of hypoxia-inducible factor -1α and vascular endothelial growth factor in sacral giant cell tumors and the correlation with tumor microvessel density. Exp. Ther. Med. 2014;8:1453–1458. doi: 10.3892/etm.2014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Liu X., Liu Y. MicroRNA-335 and -543 suppress bone metastasis in prostate cancer via targeting endothelial nitric oxide synthase. Int. J. Mol. Med. 2015;36:1417–1425. doi: 10.3892/ijmm.2015.2355. [DOI] [PubMed] [Google Scholar]
- Fulmer T. Curiously strong lipidoids. SciBX. 2010;3:1–2. [Google Scholar]
- Fusco A., Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- Gantier M.P., Williams B.R. The response of mammalian cells to doublestranded RNA. Cytokine. Growth. Factor. Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble J.M., Robinson C.E., Wu X. The function of adipocytes in the bone marrow stroma: an update. Bone. 1996;19:421–428. doi: 10.1016/s8756-3282(96)00258-x. [DOI] [PubMed] [Google Scholar]
- Gimble J.M., Zvonic S., Floyd Z.E. Playing with bone and fat. J. Cell. Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- Goldring M.B., Birkhead J.R., Suen L.F. Interleukin-1 beta- modulated gene expression in immortalized human chondrocytes. J. Clin. Invest. 1994;94:2307–2316. doi: 10.1172/JCI117595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griqoriadis A.E., Wang Z.Q., Cecchini M.G. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- Guo B., Zhang B., Zheng L. Therapeutic RNA interference targeting CKIP-1 with a cross-species sequence to stimulate bone formation. Bone. 2014;59:76–88. doi: 10.1016/j.bone.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Guo S., Bai R., Liu W. MicroRNA-210 is upregulated by hypoxia- inducible factor-1 in the stromal cells of giant cell tumors of bone. Mol.Med. Rep. 2015;12:6185–6192. doi: 10.3892/mmr.2015.4170. [DOI] [PubMed] [Google Scholar]
- Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein E., Beach D. An RNA directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Harada S., Rodan G.A. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- Hassan M.Q., Tare R., Lee S.H. Hoxa10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.Y., Wang G.C., Zhang H. miR-106a-5p suppresses the proliferation, migration, and invasion of osteosarcoma cells by targeting HMGA2. DNA. Cell. Biol. 2016;35:506–520. doi: 10.1089/dna.2015.3121. [DOI] [PubMed] [Google Scholar]
- Heilmeier U., Hackl M., Skalicky S. Serum miRNA signatures are indicative of skeletal fractures in postmenopausal women with and without type 2 diabetes and influence osteogenic and adipogenic differentiation of adipose tissue-derived mesenchymal stem cells in vitro. J. Bone Min. Res. 2016 doi: 10.1002/jbmr.2897. [DOI] [PubMed] [Google Scholar]
- Holroyd C., Cooper C., Dennison E. Epidemiology of osteoporosis. Best. Pract. Res. Clin. Endocrinol. Metab. 2008;22:671–685. doi: 10.1016/j.beem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Hong L., Wei N., Joshi V. Effects of glucocorticoid receptor small interfering RNA delivered using poly lactic-co-glycolic acid microparticles on proliferation and differentiation capabilities of human mesenchymal stromal cells. Tissue. Eng. 2012;18(Part A):775–784. doi: 10.1089/ten.tea.2011.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.F., Turner C.H. Effects of loading frequency on mechanically induced bone formation. J. Bone. Min. Res. 2001;16:918–924. doi: 10.1359/jbmr.2001.16.5.918. [DOI] [PubMed] [Google Scholar]
- Hu R., Li H., Liu W. Targeting miRNAs in osteoblast differentiation and bone formation. Expert. Opin. Ther. Targets. 2010;14:1109–1120. doi: 10.1517/14728222.2010.512916. [DOI] [PubMed] [Google Scholar]
- Hu N., Feng C., Jiang Y. Regulative effect of Mir-205 on osteogenic differentiation of bone mesenchymal stem cells (BMSCs): possible role of SATB2/runx2 and ERK/MAPK pathway. Int. J. Mol. Sci. 2015;16:10491–10506. doi: 10.3390/ijms160510491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zhang W., Li F. miR-139 is up-regulated in osteoarthritis and inhibits chondrocyte proliferation and migration possibly via suppressing EIF4G2 and IGF1R. Biochem. Biophys. Res. Commun. 2016;474:296–302. doi: 10.1016/j.bbrc.2016.03.164. [DOI] [PubMed] [Google Scholar]
- Huang H.J., Zhang M. Downregulation of PI3Kcb utilizing adenovirus- mediated transfer of siRNA attenuates bone cancer pain. Int. J. Clin. Exp. Pathol. 2014;7:8127–8135. [PMC free article] [PubMed] [Google Scholar]
- Huang J., Meng Y., Liu Y. MicroRNA-320a regulates the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting HOXA10. Cell. Physiol. Biochem. 2016;38:40–48. doi: 10.1159/000438607. [DOI] [PubMed] [Google Scholar]
- Inose H., Ochi H., Kimura A. A microRNA regulatory mechanism of osteoblast differentiation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iotsova V., Caamaño J., Loy J. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat. Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- Irie N., Takada Y., Watanabe Y. Bidirectional signaling through ephrinA2-EphA2 enhances osteoclastogenesis and suppresses osteoblastogenesis. J. Biol. Chem. 2009;284:14637–14644. doi: 10.1074/jbc.M807598200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug. Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- James E.N., Delany A.M., Nair L.S. Post-transcriptional regulation in osteoblasts using localized delivery of miR-29a inhibitor from nanofibers to enhance extracellular matrix deposition. Acta Biomater. 2014;10:3571–3580. doi: 10.1016/j.actbio.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q., Xu X., Zhang Q. The IL-1β/AP-1/miR-30a/ADAMTS-5 axis regulates cartilage matrix degradation in human osteoarthritis. J. Mol. Med. 2016;94:771–785. doi: 10.1007/s00109-016-1418-z. [DOI] [PubMed] [Google Scholar]
- Ji Q., Xu X., Xu Y. miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J. Mol. Med. 2016;94:681–694. doi: 10.1007/s00109-016-1380-9. [DOI] [PubMed] [Google Scholar]
- Jia S., Yang X., Song W. Incorporation of osteogenic and angiogenic small interfering RNAs into chitosan sponge for bone tissue engineering. Int. J. Nanomedicine. 2014;9:5307–5316. doi: 10.2147/IJN.S70457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiya T., Nakamura S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor alpha and RANKL during osteoclast differentiation. J. Periodontal. Res. 2013;48:373–385. doi: 10.1111/jre.12017. [DOI] [PubMed] [Google Scholar]
- Kang H., Chen H., Huang P. Glucocorticoids impair bone formation of bone marrow stromal stem cells by reciprocally regulating microRNA-34a-5p. Osteoporos. Int. 2016;27:1493–1505. doi: 10.1007/s00198-015-3381-x. [DOI] [PubMed] [Google Scholar]
- Kang L., Yang C., Song Y. MicroRNA-23a-3p promotes the development of osteoarthritis by directly targeting SMAD3 in chondrocytes. Biochem. Biophys. Res. Commun. 2016;478:467–473. doi: 10.1016/j.bbrc.2016.06.071. [DOI] [PubMed] [Google Scholar]
- Kapinas K., Kessler C.B., Delany A.M. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 2009;108:216–224. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Wagner E.F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kim S., Koga T., Isobe M. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Gene. Dev. 2003;17:1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Bian H., Randall T. Increased VEGF expression in the epiphyseal cartilage after ischemic necrosis of the capital femoral epiphysis. J. Bone. Min. Res. 2004;19:2041–2048. doi: 10.1359/JBMR.040911. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Ko J.Y., Chuang P.C., Ke H.J. MicroRNA-29a mitigates glucocorticoid induction of bone loss and fatty marrow by rescuing Runx2 acetylation. Bone. 2015;81:80–88. doi: 10.1016/j.bone.2015.06.022. [DOI] [PubMed] [Google Scholar]
- Koga T., Inui M., Inoue K. Costimulatory signals mediated by the itam motif cooperate with rankl for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- Kondo T., Kitazawa R., Yamaguchi A. Dexamethasone promotes osteoclastogenesis by inhibiting osteoprotegerin through multiple levels. J. Cell. Biochem. 2008;103:335–345. doi: 10.1002/jcb.21414. [DOI] [PubMed] [Google Scholar]
- Krzeszinski J.Y., Wei W., Huynh H. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431–435. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kypta R.M., Waxman J. Wnt/β-catenin signalling in prostate cancer. 2012;9:418–428. doi: 10.1038/nrurol.2012.116. [DOI] [PubMed] [Google Scholar]
- Lan L.S., Ping Y.J., Na W.L. Down-regulation of Toll-like receptor 4 gene expression by short interfering RNA attenuates bone cancer pain in a rat model. Mol. Pain. 2010;6:2. doi: 10.1186/1744-8069-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.P., Ng P.K., Li M.S. p63 regulates cell proliferation and cell cycle progression-associated genes in stromal cells of giant cell tumor of the bone. Int. J. Oncol. 2013;42:437–443. doi: 10.3892/ijo.2012.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman N., Rubin C.J., Mallmin H. Second generation sequencing of microRNA in human bone cells treated with parathyroid hormone or dexamethasone. Bone. 2016;84:181–188. doi: 10.1016/j.bone.2015.12.053. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Legendre F., Ollitrault D., Hervieu M. Enhanced hyaline cartilage matrix synthesis in collagen sponge scaffolds by using siRNA to stabilize chondrocytes phenotype cultured with bone morphogenetic Protein-2 under hypoxia. Tissue. Eng. PartC. Methods. 2013;19:550–567. doi: 10.1089/ten.TEC.2012.0508. [DOI] [PubMed] [Google Scholar]
- Li Z., Hassan M.Q., Volinia S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Hassan M.Q., Jafferji M. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang J., Zhang L. Diallyl trisulfide inhibits proliferation, invasion and angiogenesis of osteosarcoma cells by switching on suppressor microRNAs and inactivating of Notch-1 signaling. Carcinogenesis. 2013;34:1601–1610. doi: 10.1093/carcin/bgt065. [DOI] [PubMed] [Google Scholar]
- Li J., Li Y., Wang Y. Preventive effects of siRNA targeting PPARγ gene on steroid-induced osteonecrosis in rabbits. Connect. Tissue. Res. 2014;55:322–330. doi: 10.3109/03008207.2014.941106. [DOI] [PubMed] [Google Scholar]
- Li J., Jia L., Liu X. MicroRNA-16-5p controls development of osteoarthritis by targeting SMAD3 in chondrocytes. Curr. Pham. Des. 2015;21:5160–5167. doi: 10.2174/1381612821666150909094712. [DOI] [PubMed] [Google Scholar]
- Li T., Li H., Wang Y. microRNA-23a inhibits osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting LRP5. Int. J. Biochem. Cell. Biol. 2016;72:55–62. doi: 10.1016/j.biocel.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Li D., Liu J., Guo B. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat. Commun. 2016;7:10872. doi: 10.1038/ncomms10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.C., Chang Y.H., Yeh C.L. Healing of osteoporotic bone defects by baculovirus-engineered bone marrow-derived MSCs expressing MicroRNA sponges. Biomaterials. 2016;74:155–166. doi: 10.1016/j.biomaterials.2015.09.046. [DOI] [PubMed] [Google Scholar]
- Liang C., Guo B., Wu H. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference based bone anabolic strategy. Nat. Med. 2015;21:288–294. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Liu Y., Hou B. CREB-regulated transcription coactivator 1 enhances CREB-dependent gene expression in spinal cord to maintain the bone cancer pain in mice. Mol. Pain. 2016;12:1–11. doi: 10.1177/1744806916641679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L., Yang X., Su X. Redundant mir-3077-5p and mir-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell. death. Dis. 2013;4:e600. doi: 10.1038/cddis.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L., Yang X., Su X. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell. death. Dis. 2013;4:e600. doi: 10.1038/cddis.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L., Su X., Yang X. TNF-α inhibits FoxO1 by upregulating miR-705 to aggravate oxidative damage in bone marrow-derived mesenchymal stem cells during osteoporosis. Stem Cells. 2016;34:1054–1067. doi: 10.1002/stem.2274. [DOI] [PubMed] [Google Scholar]
- Lin L., Zheng X., Qiu C. SIRT1 promotes endometrial tumor growth by targeting SREBP1 and lipogenesis. Oncol. Rep. 2014;32:2831–2835. doi: 10.3892/or.2014.3521. [DOI] [PubMed] [Google Scholar]
- Liu Q., Paroo Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- Liu L., Rando T.A. Manifestations and mechanisms of stem cell aging. J. Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Peng H.W., Cheng Y.S. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell. Biol. 2005;25:3117–3126. doi: 10.1128/MCB.25.8.3117-3126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.N., Yin J., Barrett B. Loss of androgen-regulated MicroRNA 1 activates SRC and promotes prostate Cancer bone metastasis. Mol. Cell. Biol. 2015;35:1940–1951. doi: 10.1128/MCB.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourda M., Trougakos I.P., Gonos E.S. Development of resistance to chemotherapeutic drugs in human osteosarcoma cell lines largely depends on up- regulation of Clusterin/Apolipoprotein. Int. J. Cancer. 2007;120:611–622. doi: 10.1002/ijc.22327. [DOI] [PubMed] [Google Scholar]
- Lu J., Song G., Tang Q. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2016 doi: 10.1038/onc.2016.194. [DOI] [PubMed] [Google Scholar]
- Lv H., Zhang S., Wang B. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Lv H., Sun Y., Zhang Y. MiR-133 is involved in estrogen deficiency- induced osteoporosis through modulating osteogenic differentiation of mesenchymal stem cells. Med. Sci. Monit. 2015;21:1527–1534. doi: 10.12659/MSM.894323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Mahmood M., Li Z., Casciano D. Nanostructural materials increase mineralization in bone cells and affect gene expression through miRNA regulation. J. Cell. Mol. Med. 2011;15:2297–2306. doi: 10.1111/j.1582-4934.2010.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney E.K., McLaughlin J.L., Dagdigian N.E. An anti-insulin-like growth factor I receptor antibody that is a potent inhibitor of cancer cell proliferation. Cancer Res. 2003;63:5073–5083. [PubMed] [Google Scholar]
- Manaka T., Suzuki A., Takyama K. Local delivery of siRNA using a biodegradable polymer application to enhance BMP-induced bone formation. Biomaterials. 2011;32:9642–9648. doi: 10.1016/j.biomaterials.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Manolagas S.C. From estrogen-centric to aging and oxidative stress: a revised perspective of the pathogenesis of osteoporosis. Endocr. Rev. 2010;31:266–300. doi: 10.1210/er.2009-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel-Pelletier J. Pathophysiology of osteoarthritis. Osteoarthr. Cartil. 2004;12(Suppl. A):S31–S33. doi: 10.1016/j.joca.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Martin T.J. Bone biology and anabolic therapies for bone: current status and future prospects. J. Bone. Metab. 2014;21:8–20. doi: 10.11005/jbm.2014.21.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.J., Sims N.A. Osteoclast-derived activity in the coupling of bone formation to resorption. Trends. Mol. Med. 2005;11:76–81. doi: 10.1016/j.molmed.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Masuda J., Tsuda M., Tozaki-Saitoh H. Intrathecal delivery of PDGF produces tactile allodynia through its receptors in spinal microglia. Mol. Pain. 2009;5:23. doi: 10.1186/1744-8069-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meka S.R., Jain S., Chatterjee K. Strontium eluting nanofibers augment stem cell osteogenesis for bone tissue regeneration. Colloids. Surf B. Biointerfaces. 2016;146:649–656. doi: 10.1016/j.colsurfb.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Meng J., Zhang D., Pan N. Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. Peer J. 2015;3:e971. doi: 10.7717/peerj.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz M.A., White J.M., Sheehan K.C. Targeted disruption of the Stat1 gene in mice reveals unexpected physiological specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Mirnezami A.H., Pickard K., Zhang L. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur. J. Surg. Oncol. 2009;35:339–347. doi: 10.1016/j.ejso.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Mitra A., Mishra L., Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6:10697–10711. doi: 10.18632/oncotarget.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki S., Asahara H. Macro view of microRNA function in osteoarthritis. Nat. Rev. Rheumatol. 2012;8:543–552. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruganandan S., Dranse H.J., Rourke J.L. Chemerin neutralization blocks hematopoietic stem cell osteoclastogenesis. Stem Cells. 2013;31:2172–2182. doi: 10.1002/stem.1450. [DOI] [PubMed] [Google Scholar]
- Naito Y., Ui-Tei K. Designing functional siRNA with reduced off-target effects. Methods. Mol. Biol. 2013;942:57–68. doi: 10.1007/978-1-62703-119-6_3. [DOI] [PubMed] [Google Scholar]
- Nanni S., Benvenuti A., Grasselli C. Endothelial NOS, estrogen receptor β, and HIFs cooperate in the activation of a prognostic transcriptional pattern in aggressive human prostate cancer. J. Clin. Invest. 2009;119:1093–1108. doi: 10.1172/JCI35079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi-Koga T., Shinohara M., Komatsu N. Suppression of bone formation by osteoclastic expression of semaphorin 4D. Nat. Med. 2011;17:1473–1480. doi: 10.1038/nm.2489. [DOI] [PubMed] [Google Scholar]
- Noronha-Matos J.B., Coimbra J., Sa-E-Sousa A. P2X7-induced zeiosis promotes osteogenic differentiation and mineralization of postmenopausal bone marrow-derived mesenchymal stem cells. FASEB J. 2014;28:5208–5222. doi: 10.1096/fj.14-257923. [DOI] [PubMed] [Google Scholar]
- Nquyen M.K., Jeon O., Krebs M.D. Sustained localized presentation of RNA interfering molecules from in situ forming hydrogels to guide stem cell osteogenic differentiation. Biomaterials. 2014;35:6278–6286. doi: 10.1016/j.biomaterials.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer. Manag. Res. 2014;6:15–25. doi: 10.2147/CMAR.S53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. microRNA and Bone Cancer. In: Santulli G., editor. microRNA: Cancer. From Molecular Biology to Clinical Practice. vol. 11. 2015. pp. 201–230. (Adv. Exp. Med. Biol. Chapter). [DOI] [PubMed] [Google Scholar]
- Ollitrault D., Legendre F., Drougard C. BMP-2, hypoxia, and COL1A1/HtrA1 siRNAs favor neo-cartilage hyaline matrix formation in chondrocytes. Tissue. Eng. PartC. Methods. 2015;21:133–147. doi: 10.1089/ten.tec.2013.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan G., Ozpolat B., Coleman R.L. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug. Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri A., Pezzetti F., Brunelli G. Calcium sulfate acts on the miRNA of MG63E osteoblast-like cells. J. Biomed. Mater. Res. B. Appl. Biomater. 2008;84:369–374. doi: 10.1002/jbm.b.30880. [DOI] [PubMed] [Google Scholar]
- Pan R., Di H., Zhang J. Inducible Lentivirus-Mediated siRNA against TLR4 Reduces Nociception in a Rat Model of Bone Cancer Pain. 2015;2015:523896. doi: 10.1155/2015/523896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng N., Gao S., Guo X. Silencing of VEGF inhibits human osteosarcoma angiogenesis and promotes cell apoptosis via VEGF/PI3K/AKT signaling pathway. Am. J. Transl. Res. 2016;8:1005–1015. [PMC free article] [PubMed] [Google Scholar]
- Perez J., Bardin C., Rigal C. Anti-MDR1 siRNA restores chemosensitivity in chemoresistant breast carcinoma and osteosarcoma cell lines. Anticancer Res. 2011;31:2813–2820. [PubMed] [Google Scholar]
- Pi Y., Zhang X., Shao Z. Intra-articular delivery of anti-Hif-2α siRNA by chondrocyte-homing nanoparticles to prevent cartilage degeneration in arthritic mice. Gene Ther. 2015;22:439–448. doi: 10.1038/gt.2015.16. [DOI] [PubMed] [Google Scholar]
- Qin L., Yao D., Zheng L. Phytomolecule icaritin incorporated PLGA/TCP scaffold for steroidassociated osteonecrosis: proof-of-concept for prevention of hip joint collapse in bipedal emus and mechanistic study in quadrupedal rabbits. Biomaterials. 2016;59:125–143. doi: 10.1016/j.biomaterials.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi A.T., Monroe W.T., Dasa V. miR-148b-nanoparticle conjugates for light mediated osteogenesis of human adipose stromal/stem cells. Biomaterials. 2013;34:7799–7810. doi: 10.1016/j.biomaterials.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Rachner T.D., Khosla S., Hofbauer L.C. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubramanian A., Jeeawoody S., Yang F. Gene delivery of osteo- inductive signals to a human fetal osteoblast cell line induces cell death in a dose- dependent manner. Drug. Deliv. Transl. Res. 2015;5:160–167. doi: 10.1007/s13346-013-0163-x. [DOI] [PubMed] [Google Scholar]
- Rasheed Z., Al-Shobaili H.A., Rasheed N. MicroRNA-26a-5p regulates the expression of inducible nitric oxide synthase via activation of NF-κB pathway in human osteoarthritis chondrocytes. Arch. Biochem. Biophys. 2016;594:61–67. doi: 10.1016/j.abb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Reijnders C.M., Bravenboer N., Tromp A.M. Effect of mechanical loading on insulin-like growth factor-I gene expression in rat tibia. J. Endocrinol. 2007;192:131–140. doi: 10.1677/joe.1.06880. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ríos C.N., Skoracki R.J., Mathur A.B. GNAS1 and PHD2 short- interfering RNA support bone regeneration in vitro and in an in vivo sheep model. Clin. Orthop. Relat. Res. 2012;470:2541–2553. doi: 10.1007/s11999-012-2475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan G.A., Martin T.J. Therapeutic approaches to bone diseases. Science. 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- Saarinen A., Välimäki V.V., Välimäki M.J. The A1330 V polymorphism of the low-density lipoprotein receptor-related protein 5 gene (LRP5) associates with low peak bone mass in young healthy men. Bone. 2007;40:1006–1012. doi: 10.1016/j.bone.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Sambrook P., Cooper C. Osteoporosis. Lancet. 2006;367:2010–2018. doi: 10.1016/S0140-6736(06)68891-0. [DOI] [PubMed] [Google Scholar]
- Sato A.Y., Tu X., McAndrews K.A. Prevention of glucocorticoid inducedapoptosis of osteoblasts and osteocytes by protecting against endoplasmic reticulum (ER) stress in vitro and in vivo in female mice. Bone. 2015;73:60–68. doi: 10.1016/j.bone.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoolmeesters A., Eklund T., Leake D. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5605. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C.L., Gorlick R., Teot L. Multiple drug resistance in osteogenic sarcoma: INT0133 from the Children's Oncology Group. J. Clin. Oncol. 2007;25:2057–2062. doi: 10.1200/JCO.2006.07.7776. [DOI] [PubMed] [Google Scholar]
- Seeliger C., Karpinski K., Haug A.T. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Min. Res. 2014;29:1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- Shah R.B., Mehra R., Chinnaiyan A.M. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- Shen J.H., Qu C.B., Chu H.K. siRNA targeting TCTP suppresses osteosarcoma cell growth and induces apoptosis in vitro and in vivo. Biotechnol. Appl. Biochem. 2016;63:5–14. doi: 10.1002/bab.1335. [DOI] [PubMed] [Google Scholar]
- Shi Q., Zhang X.L., Dai K.R. siRNA therapy for cancer and non-lethal diseases such as arthritis and osteoporosis. Expert. Opin. Biol. Ther. 2011;11:5–16. doi: 10.1517/14712598.2010.532483. [DOI] [PubMed] [Google Scholar]
- Shi K., Lu J., Zhao Y. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone. 2013;55:487–494. doi: 10.1016/j.bone.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Shi C., Qi J., Huang P. MicroRNA-17/20a inhibits glucocorticoid-in duced osteoclast differentiation and function through targeting RANKL expression in osteoblast cells. Bone. 2014;68:67–75. doi: 10.1016/j.bone.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Shi Y., Huang J., Zhou J. MicroRNA-204 inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in osteosarcoma cells via targeting Sirtuin 1. Oncol. Rep. 2015;34:399–406. doi: 10.3892/or.2015.3986. [DOI] [PubMed] [Google Scholar]
- Siegel H.J., Pressey J.G. Current concepts on the surgical and medical management of osteosarcoma. Expert. Rev. anticancer. Ther. 2008;8:1257–1269. doi: 10.1586/14737140.8.8.1257. [DOI] [PubMed] [Google Scholar]
- Siu M.K., Tsai Y.C., Chang Y.S. Transforming growth factor-β promotes prostate bone metastasis through induction of microRNA-96 and activation of the mTOR pathway. Oncogene. 2015;34:4767–4776. doi: 10.1038/onc.2014.414. [DOI] [PubMed] [Google Scholar]
- Sondag G.R., Haggi T.M. The role of MicroRNAs and their targets in osteoarthritis. Curr. Rheumatol. Rep. 2016;18:56. doi: 10.1007/s11926-016-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiers P.J., van Gastel N., Carmeliet G. Targeting the hypoxic response in bone tissue engineering: a balance between supply and consumption to improve bone regeneration. Mol. Cell. Endocrinol. 2016;432:96–105. doi: 10.1016/j.mce.2015.12.024. [DOI] [PubMed] [Google Scholar]
- Sun X.H., Geng X.L., Zhang J. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2) Tumor Biol. 2015;36:2127–2134. doi: 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- Sung S.Y., Wu I.H., Chuang P.H. Targeting L1 cell adhesion molecule expression using liposome encapsulated sirna suppresses prostate cancer bone metastasis and growth. Oncotarget. 2014;5:9911–9929. doi: 10.18632/oncotarget.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa M., lyer A.K., Ryu K. Inhibition of ABCB1 (MDR1) expression by an siRNA nanoparticulate delivery system to overcome drug resistance in osteosarcoma. PLoS One. 2010;5:e10764. doi: 10.1371/journal.pone.0010764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva L.J., Washam C., Nicholas R.W. Bone metastasis: mechanisms and therapeutic opportunities. Nat. Rev. Endocrinol. 2011;7:208–218. doi: 10.1038/nrendo.2010.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait A.S., Butts C.L., Sternberg E.M. The role of glucocorticoids and progestins in inflammatory, autoimmune, and infectious disease. J. Leukoc. Biol. 2008;84:924–931. doi: 10.1189/jlb.0208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima K., Takaishi H., Takito J. Inhibition of STAT1 accelerates bone fracture healing. J. Orthop. Res. 2010;28:937–941. doi: 10.1002/jor.21086. [DOI] [PubMed] [Google Scholar]
- Takaishi H., Kimura T., Dalal S. Joint diseases and matrix metallo- proteinases: a role for MMP-13. Curr. Pharm. Biotechnol. 2008;9:47–54. doi: 10.2174/138920108783497659. [DOI] [PubMed] [Google Scholar]
- Takyama K., Suzuki A., Manaka T. RNA interference for noggin enhances the biological activity of bone morphogenetic proteins in vivo and in vitro. J. Bone. Min. Metab. 2009;27:402–411. doi: 10.1007/s00774-009-0054-x. [DOI] [PubMed] [Google Scholar]
- Tamura Y., Kawao N., Yano M. Role of plasminogen activator inhibitor-1 in glucocorticoid-induced diabetes and osteopenia in mice. Diabetes. 2015;64:2194–2206. doi: 10.2337/db14-1192. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Nakamura K., Takahasi N. Role of RANKL in physiological and pathological bone resorption and therapeutics targeting the RANKL-RANK signaling system. Immunol. Rev. 2005;208:30–49. doi: 10.1111/j.0105-2896.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- Tang P., Xiong Q., Ge W. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol. 2014;11:1355–1363. doi: 10.1080/15476286.2014.996462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum S.L. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- Tencerova M., Kassem M. The bone marrow-derived stromal cells: commitment and regulation of adipogenesis. Front. Endocrinol. 2016;7:127. doi: 10.3389/fendo.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M., Nakamura Y. Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene. 2001;20:4457–4465. doi: 10.1038/sj.onc.1204608. [DOI] [PubMed] [Google Scholar]
- Vaananen K. Mechanism of osteoclast mediated bone resorption-rationale for the design of new therapeutics. Adv. Drug Deliv. Rev. 2005;57:959–971. doi: 10.1016/j.addr.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Vishal M., Vimalraj S., Ajeetha R. MicroRNA-590-5p stabilizes Runx2 by targeting Smad7 during osteoblast differentiation. J. Cell. Physiol. 2017;232:371–380. doi: 10.1002/jcp.25434. [DOI] [PubMed] [Google Scholar]
- Wan D.C., Pomerantz J.H., Brunet L.J. Noggin suppression enhances in vitro osteogenesis and accelerates in vivo bone formation. 2007;282:26450–26459. doi: 10.1074/jbc.M703282200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Grainger D.W. RNA therapeutics targeting osteoclast-mediated excessive bone resorption. Adv. Drug. Deliv. Rev. 2012;64:1341–1357. doi: 10.1016/j.addr.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Grainger D.W. Developing siRNA therapies to address osteoporosis. Ther. Deliv. 2013;4:1239–1246. doi: 10.4155/tde.13.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Sima M., Mosley R.L. Pharmacokinetic and bio- distribution studies of a bone-targeting drug delivery system based on N- (2-hydroxypropyl) methacrylamide copolymers. Mol. Pham. 2006;3:717–725. doi: 10.1021/mp0600539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Miller S.C., Shlyakhtenko L.S. Osteotropic peptide that differentiates functional domains of the skeleton. Bioconjug Chem. 2007;18:1375–1378. doi: 10.1021/bc7002132. [DOI] [PubMed] [Google Scholar]
- Wang Q.Z., Lv Y.H., Diao Y. The design of vectors for RNAi delivery system. Curr. Pharm. Des. 2008;14:1327–1340. doi: 10.2174/138161208799316357. [DOI] [PubMed] [Google Scholar]
- Wang Y., Panasiuk A., Grainger D.W. Small interfering RNA knocks down the molecular target of alendronate, farnesyl pyrophosphate synthase, in osteoclast and osteoblast cultures. Mol. Pharm. 2011;8:1016–1024. doi: 10.1021/mp100374n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li L., Moore B.T. MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One. 2012;7:e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tran K.K., Shen H. Selective local delivery of RANK siRNA to bone phagocytes using bone augmentation biomaterials. Biomaterials. 2012;33:8540–8547. doi: 10.1016/j.biomaterials.2012.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guo B., Li Q. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- Wang F.S., Chuang P.C., Lin C.L. MicroRNA-29a protects against glucocorticoid-induced bone loss and fragility in rats by orchestrating bone acquisition and resorption. Arthritis Rheum. 2013;65:1530–1540. doi: 10.1002/art.37948. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang D., Hu Z. MicroRNA-26a-modified adipose- derived stem cells incorporated with a porous hydroxyapatite scaffold improve the repair of bone defects. Mol. Med. Rep. 2015;12:3345–3350. doi: 10.3892/mmr.2015.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu G., Feng Z. Microarc-oxidized titanium surfaces functionalized with microrNa-21-loaded chitosan/hyaluronic acid nanoparticles promote the osteogenic differentiation of human bone marrow mesenchymal stem cells. Int. J. Nanomedicine. 2015;10:6675–6687. doi: 10.2147/IJN.S94689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yin H., Wang J. MicroRNA-106b inhibits osteoclast- ogenesis and osteolysis by targeting RANKL in giant cell tumor of bone. Oncotarget. 2015;6:18980–18996. doi: 10.18632/oncotarget.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu G., Wei M. Improving the osteogenesis of human bone marrow mesenchymal stem cell sheets by microrna-21-loaded chitosan/hyaluronic acid nanoparticles via reverse transfection. Int. J. Nanomedicine. 2016;11:2091–2105. doi: 10.2147/IJN.S104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guo Y., Wang C. MicroRNA-142-3p inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting HMGB1. Inflammation. 2016;39:1718–1728. doi: 10.1007/s10753-016-0406-3. [DOI] [PubMed] [Google Scholar]
- Wei J.W., Shi Y., Zheng L.H. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Yu Y., Joshi V. Glucocorticoid receptor antagonist and siRNA prevent senescence of human bone marrow mesenchymal stromal cells in vitro. Cell. Tissue. Res. 2013;354:461–470. doi: 10.1007/s00441-013-1700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilner S., Skalicky S., Salzer B. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. doi: 10.1016/j.bone.2015.05.027. [DOI] [PubMed] [Google Scholar]
- Weilner S., Schraml E., Wieser M. Secreted microvesicular miR-31 inhibits osteogenic differentiation of mesenchymal stem cells. Aging Cell. 2016;15:744–754. doi: 10.1111/acel.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein R.S. Glucocorticoid-induced osteoporosis. Rev. Endocr. Metab. Disord. 2001;2:65–73. doi: 10.1023/a:1010007108155. [DOI] [PubMed] [Google Scholar]
- Weinstein R.S., Jilka R.L., Parfitt A.M. Inhibition of osteoblasto- genesis and promotion of apoptosis of osteoblasts and osteocytes by glucocorticoids. Potential mechanisms of their deleterious effects on bone. J. Clin. Invest. 1998;102:274–282. doi: 10.1172/JCI2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K.A., Langer R. Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug. Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yin H., Liu T. MiR-126-5p regulates osteoclast differentiation and bone resorption in giant cell tumor through inhibition of MMP-13. Biochem. Biophys. Res. Commun. 2014;443:944–999. doi: 10.1016/j.bbrc.2013.12.075. [DOI] [PubMed] [Google Scholar]
- Wu C., Tian B., Qu X. MicroRNAs play a role in chondrogenesis and osteoarthritis (review) Int. J. Mol. Med. 2014;34:13–23. doi: 10.3892/ijmm.2014.1743. [DOI] [PubMed] [Google Scholar]
- Wu S., Du X., Wu M. MicroRNA-409-3p inhibits osteosarcoma cell migration and invasion by targeting catenin-δ1. Gene. 2016;584:83–89. doi: 10.1016/j.gene.2016.03.021. [DOI] [PubMed] [Google Scholar]
- Wu P., Liang J., Yu F. miR-145 promotes osteosarcoma growth by reducing expression of the transcription factor friend leukemia virus integration 1. Oncotarget. 2016;7:42241–42251. doi: 10.18632/oncotarget.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W.Z., Gu X.C., Hu B. Role of microRNA-129-5p in osteoblast differentiation from bone marrow mesenchymal stem cells. Cell. Mol. Biol. 2016;62:95–99. [PubMed] [Google Scholar]
- Xie X.H., Wang X.L., Yang H.L. Steroid-associated osteonecrosis: epidemiology, pathophysiology, animal model, prevention, and potential treatments (an overview) J. Orthop. Transl. 2015;3:58–70. doi: 10.1016/j.jot.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Qiao Z., Li J. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7:44252–44265. doi: 10.18632/oncotarget.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.Q., Liu P., Si M.J. MicroRNA-126 inhibits osteosarcoma cells proliferation by targeting Sirt1. Tumor Biol. 2013;34:3871–3877. doi: 10.1007/s13277-013-0974-x. [DOI] [PubMed] [Google Scholar]
- Xu Y., Liu J., He M. Mechanisms of PDGF siRNA-mediated inhibition of bone cancer pain in the spinal cord. Sci. Rep. 2016;6:27512. doi: 10.1038/srep27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Zhao Y. Non-viral oligonucleotide antimiR-138 delivery to mesenchymal stem cell sheets and the effect on osteogenesis. Biomaterials. 2014;35:7734–7749. doi: 10.1016/j.biomaterials.2014.05.089. [DOI] [PubMed] [Google Scholar]
- Yang N., Wang G., Hu C. Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J. Bone. Min. Res. 2013;28:559–573. doi: 10.1002/jbmr.1798. [DOI] [PubMed] [Google Scholar]
- Yang X., Guan Y., Tian S. Mechanical and IL-1β responsive miR-365 contributes to osteoarthritis development by targeting histone deacetylase 4. Int. J. Mol. Sci. 2016;17:e436. doi: 10.3390/ijms17040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Kanasty R.L., Eltoukhy A.A. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- You L., Pan L., Chen L. MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell. Physiol. Biochem. 2016;39:253–265. doi: 10.1159/000445621. [DOI] [PubMed] [Google Scholar]
- Yu S., Jia L., Zhang Y. Increased expression of activated endothelial nitric oxide synthase contributes to antiandrogen resistance in prostate cancer cells by suppressing androgen receptor transactivation. Cancer Lett. 2013;328:83–94. doi: 10.1016/j.canlet.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Zaidi M. Skeletal remodeling in health and disease. Nat. Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- Zaidi N., Swinnen J.V., Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 2012;72:3709–3714. doi: 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- Zhang G., Guo B., Wu H. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat. Med. 2012;18:307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- Zhang C., Li Y., Cornelia R. Regulation of VEGF expression by HIF-1α in the femoral head cartilage following ischemia osteonecrosis. Sci. Rep. 2012;2:650. doi: 10.1038/srep00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei L., Miron R.J. Prevention of alveolar bone loss in an osteoporotic animal model via interference of semaphorin 4d. J. Dent. Res. 2014;93:1095–1100. doi: 10.1177/0022034514552676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Cao X., Li J. MiR-210 inhibits NF-κB signaling pathway by targeting DR6 in osteoarthritis. Sci. Rep. 2015;5:12775. doi: 10.1038/srep12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei L., Miron R.J. Anabolic bone formation via a site specific bone targeting delivery system by interfering with semaphorin 4D expression. J. Bone. Min. Res. 2015;30:286–296. doi: 10.1002/jbmr.2322. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wei L., Miron R.J. Bone scaffolds loaded with siRNA Semaphorin4d for the treatment of osteoporosis related bone defects. Sci. Rep. 2016;6:26925. doi: 10.1038/srep26925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Xu D., Li Y. MicroRNAs regulate bone metabolism. J. Bone. Min. Metab. 2014;32:221–231. doi: 10.1007/s00774-013-0537-7. [DOI] [PubMed] [Google Scholar]
- Zhao C., Sun W., Zhang P. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol. 2015;12:343–353. doi: 10.1080/15476286.2015.1017205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Zhang J., Zhu M. Three-dimensional printed strontium- containing mesoporous bioactive glass scaffolds for repairing rat critical-sized calvarial defects. Acta Biomater. 2015;12:270–280. doi: 10.1016/j.actbio.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Zheng L.Z., Cao H.J., Chen S.H. Blockage of src by specific siRNA as a novel therapeutic strategy to prevent destructive repair in steroid- associated osteonecrosis in rabbits. J. Bone. Min. Res. 2015;30:2044–2057. doi: 10.1002/jbmr.2542. [DOI] [PubMed] [Google Scholar]
- Zheng L.Z., Wang X.L., Cao H.J. Src siRNA prevents cortico- steroid-associated osteoporosis in a rabbit model. Bone. 2016;83:190–196. doi: 10.1016/j.bone.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Yin H., Wang T. MiR-126-5p regulates osteolysis formation and stromal cell proliferation in giant cell tumor through inhibition of PTHrP. Bone. 2014;66:267–276. doi: 10.1016/j.bone.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Zhou W., Ge W., Meng G. MicroRNA-20a promotes the proliferation and cell cycle of human osteosarcoma cells by suppressing early growth response 2 expression. Mol. Med. Rep. 2015;12:4989–4994. doi: 10.3892/mmr.2015.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.Z., Mei J. Effect and mechanism analysis of siRNA in inhibiting VEGF and its anti-angiogenesis effects in human osteosarcoma bearing rats. Eur. Rev. Med. Pharmacol. Sci. 2015;19:4362–4370. [PubMed] [Google Scholar]
- Zuo B., Zhu J.F., Li J. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone. Min. Res. 2015;30:330–345. doi: 10.1002/jbmr.2352. [DOI] [PubMed] [Google Scholar]