Abstract
Jasmonates are known to induce the transcriptional activation of plant defense genes, which leads to the production of jasmonate-regulated proteins (JRP). We previously cloned and characterized a novel jacalin-like lectin gene (Ta-JA1) from wheat (Triticum aestivum L.), which codes a modular JRP with disease response and jacalin-related lectin (JRL) domains and is present only in the Gramineae family. The function of this protein is still unclear. Phylogenetic analysis indicated that Ta-JA1 and related proteins from cereals grouped together, which diverged from JRL with an additional N-terminal disease response domain. The recombinant Ta-JA1 proteins agglutinated rabbit erythrocytes, and this hemagglutination activity was preferentially inhibited by mannose. The Ta-JA1 protein was able to inhibit E. coli cell growth. Overexpression of Ta-JA1 in transgenic tobacco plants increased their resistance to infection by tobacco bacterial, fungal and viral pathogens. Our results suggest that Ta-JA1 belongs to a mannose-specific lectin, which may confer a basal but broad-spectrum resistance to plant pathogens. Ta-JA1 and its homologues in maize, rice, sorghum and creeping bentgrass may represent a new type of monocot lectin with a modular structure and diversity of physiological functions in biotic and abiotic stress responses.
Keywords: Broad-spectrum resistance, Jasmonates, Jasmonate-regulated proteins (JRP), Monocot jacalin-related lectins, Triticum aestivum L.
1. Introduction
Jasmonates are oxidized lipid-derived derivatives of cyclopentanone and function as plant hormones to regulate diverse developmental processes and defense responses [1]. They affect many aspects of plant growth and their adaptation to the environment. These include seed germination, pollen development, fruit ripening, wounding responses and resistance to insects and pathogens. Particularly, jasmonates play vital roles in signaling processes that link environmental changes, including abiotic and biotic stresses, to an intracellular response from plants [2].
Many jasmonate responses are mediated via changing levels of synthesis of the jasmonate-regulated proteins (JRP). Some of these JRPs with diverse physiological functions have been characterized in detail, such as proteinase-inhibitors [3], [4], thionins [5], [6], phytoalexin-synthesizing enzymes [7], [8], cell wall proteins [9], pathogenesis-related proteins and osmotin [10], and lipoxygenase [11]. In barley leaves, jasmonate treatment activates the de novo synthesis and increases the accumulation of some proteins with relative molecular mass of 23,000 [12], 32,000 and 37,000 [13]. However, the functions of the 32,000 proteins named JRP-32, are still unclear [14]. The JRP-32 mRNAs were first detected in jasmonate-treated barley leaves but these sequences are only present in the GenBank database (GenBank accession numbers AF021256, AF021257, AF021258) and no further experimental work has been documented. We have identified a cDNA encoding the wheat homologue for JRP-32 namely Ta-JA1 [14]. Ta-JA1 encoded protein together with some related proteins from Gramineae plants are modular proteins that contain the disease response and jacalin-related lectin (JRL) domains. They form a small protein family related to JRLs.
Lectins are a heterogeneous group of proteins that recognize and bind to specific carbohydrate without modifying them. Lectins function as central mediators of information transfer in biological systems by interacting with glycoproteins, glycolipids and oligosaccharides [15]. Plant lectins can be classified into 12 families according to their molecular structures, biochemical properties and sugar-binding specificities. Some lectin families, such as the legume lectins and the chitin-binding lectins composed of hevein domains, have been extensively studied [16]. The other types of lectins, namely jacalin-related lectins and the amaranthin family lectins, are less well characterized [16]. It is generally suggested that most plant lectins are directed against foreign glycans and other organisms either in recognition or in defense-related processes and that they therefore may play a crucial part in defense response against different pathogens (bacteria and fungi), phytophagous invertebrates and herbivorous animals. There is also data to suggest that some lectins may be also involved in cellular regulation and signaling [17].
Jacalin-related lectins (JRL) are structurally and evolutionally related to the jack fruit lectin [18]. This lectin family is very common among higher plants, from Musaceae [19] and Convolvulaceae [20], to Gramineae [21] and to gymnosperms [22]. Newly identified JRLs exhibit a preferential binding with mannose and are probably involved in plant defense responses. Because Ta-JA1 protein contains both disease response and JRL domains, it would be interesting to know that if it belongs to a new family of lectins and its roles in plant disease resistance. Here we report the biochemical characterization of Ta-JA1 and elucidation of its functions in transgenic tobacco.
2. Materials and methods
2.1. Similarity and evolutionary analyses
Sequence similarities of Ta-JA1 were analyzed using the SIM-Alignment Tool [23] and data from the GenBank database. Evolutionary relationships were determined using the Clustal W method with a PAM 250 residue weight table [24].
2.2. Expression and purification of recombinant Ta-JA1 protein in E. coli
For the convenient cloning of Ta-JA1 in pET32a vector (Novagen, USA), EcoRI and HindIII sites were introduced by PCR at the ATG start codon and TAG stop codon, respectively. PCR was carried out in a 50 μL volume of Gibco PCR buffer with 1 μmol/L primers, 0.4 mmol/L of each dNTP and 2.5 U Tag DNA polymerase (Gibco) using forward primer 5′-CGGAATTCATGGCCAATTTCCAGATAAC-3′, and reverse primer 5′-CCCAAGCTTTTAGAGAGGGAGCACGTAGAC-3′. The fidelity of the PCR amplification was confirmed by DNA sequencing. The PCR product was digested with EcoRI plus HindIII and ligated into pET32a vector, and then introduced into E. coli strain BL21 cells. The bacterial culture was grown at 37 °C until OD600 = 0.7, then moved to 16 °C shaken for 1 h. After that IPTG was added to the final concentration of 0.5 mM and the culture was continued to grow at 16 °C for 24 h. All purification steps were carried out at 4 °C. Induced E. coli cells were pelleted by centrifugation at 2000 g for 10 min and re-suspended in extraction buffer (50 mM Tris pH8.5, 500 mM NaCl, 10% glycerol, 1% Triton X-100, add 20 mM mercaptoethanol and 1 mM PMSF freshly). After added freshly made lysozyme to 100 μg/mL, the cells were incubated at 30 °C for 15 min. The extracts were sonicated 2 min with 20 cycles per minute and sonication was repeated 3 times, and then spun at 15,000 g for 15 min. The supernatant was used for further purification by Ni-NTA His-Bind® Spin Columns (QIAGEN, USA) according to the manufacturer's instructions. Protein concentrations were determined by the Bradford assay [25] with bovine serum albumin (BSA) as standard.
2.3. Agglutinating activity and carbohydrate-binding tests
Agglutinating activity was determined using rabbit erythrocytes [26]. Freshly washed rabbit red blood cells in PSB (150 mM NaCl, 68.4 mM Na2HPO4, 31.6 mM NaH2PO4, pH 7.0) were tested for the hemagglutination by various proteins. 20 μL protein solution (1 mg/mL) was serially diluted in two-fold increments and then added to the different wells with 2% erythrocyte suspension in a 96-well plate. The mixture was kept for 1 h at room temperature and then examined visually for agglutination. The carbohydrate-binding specificity was determined by the inhibition of agglutination of rabbit erythrocytes with glucose, mannose, galactose and N-acetyl-d-glucosamine, which were serially diluted from a starting concentration of 300 mM. Solutions containing 10 μL of the purified Ta-JA1 and 10 μL saccharides were pre-incubated for 1 h, then 20 μL 2% rabbit erythrocyte suspension was added, and the agglutination was evaluated after 1 h at room temperature. The lowest concentration of saccharides that visibly decreased agglutination was defined as the minimum inhibitory concentration (MIC).
2.4. Plasmid construction and generation of transgenic tobacco
The full-length Ta-JA1 cDNA was amplified by PCR with forward primer 5′-GTCGGATCCACTAGTCACCATGGCCAATT-3′ and reverse primer 5′-CGCGAATTCAATACACACCGAAAATGAGAGC-3′. BamHI and EcoRI sites were introduced by PCR at 5′-terminus and 3′-terminus of Ta-JA1, respectively, for the convenience of subcloning. PCR was carried out with 1 μmol/L primers, 0.4 mmol/L of each dNTP and 2.5 U Taq DNA polymerase (Gibco). The PCR product was digested with BamHI plus EcoRI and ligated into pBI121 expression vector [27]. The fidelity of the construction was confirmed by enzyme digestion and DNA sequencing. The resulting binary vector was transferred by the freeze-thaw method into Agrobacterium tumefaciens strain LBA4404. Tobacco (Nicotiana tabacum cv Wisconsin 38) was transformed by the leaf disc method as previously described [28]. Rooted transformants were transferred to soil, grown in the greenhouse and allowed to self-pollinate.
Total DNA was isolated from young leaf tissues of each tobacco lines as described by Edwards et al. [29]. PCR was conducted with forward primer 5′-ACTAGTCACCATGGCCAATT-3′, and reverse primer 5′-AATACACACCGAAAATGAGAGC-3′, which were corresponding to wheat Ta-JA1 cDNA sequence. The temperature program for PCR was 5 min at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 56 °C, and 1.5 min at 72 °C, followed by 10 min at 72 °C. The amplified products were resolved on a 0.8% agarose gel and then photographed.
Total RNA was isolated from tobacco leaf tissues using the TRI reagent (Molecular Research Center Inc, Cincinnati, USA) by following the manufacturer's instructions and the expression of the Ta-JA1 gene in the transgenic tobacco plants were analyzed by RT-PCR. The first-strand cDNA was synthesized using SuperScript™ III Reverse Transcriptase Kit (Invitrogen Corporation, USA) with total RNA. Reverse transcription reactions were carried out at 50 °C for 60 min and terminated by heating at 95 °C for 10 min cDNA templates were normalized to same concentration for PCR. PCR was analyzed as described above. Primers for Ta-JA1 were: forward primer 5′-ATGCTGAAAGGTTCACAGAG-3′ and reverse primer 5′-TTAGAGAGGGAGCACGTAGAC-3′. Primers for tobacco actin were: forward primer 5′-CTATTCTCCGCTTTGGACTTGGCA-3′ and reverse primer 5′-ACCTGCTGGAAGGTGCTGAGGGAA-3′. The intensity ratio of the target gene to actin was used to estimate the relative expression level of the target.
2.5. Pathogen resistance assays
PCR-positive plants were grown in a greenhouse and allowed to self-fertilize. The segregation of the integrated gene in the progeny was tested for kanamycin resistance by germinating seeds on MS medium [30] containing kanamycin (150 μg mL−1). T2 generation plants homologous for the Ta-JA1 transgene were transplanted to potting mix and used for disease resistance analyses.
Three representative pathogens, Phytophthora parasitica var nicotianae, Pseudomonas syringae pv tabaci, and tobacco mosaic virus (TMV), which cause tobacco black shank, wildfire disease and viral disease, respectively, were used for pathogen challenges. Healthy leaves (the fourth to sixth from the top) from two month old tobacco plants were inoculated with pathogens. P. parasitica and P. syringae growth, inoculation and disease scoring were according to Guo et al. [31]. Viral infection was according to Bhargava et al. [32]. The disease lesion diameter was recorded for tobacco black shank disease, while the bacterial population inside the leaf discs was determined based on the numbers of colonies formed on King's B plates for tobacco wildfire disease. TMV infection quantitation was performed by double antibody sandwich ELISA with a commercial TMV detection kit (Agdia Inc., Indiana, USA). Absorbance at 405 nm was determined with the aid of a microplate ELISA reader (550 Microplate Reader, Bio-Rad). The Student's t-test for independent samples [33] was applied to determine the difference between the transgenic versus the control lines that were transferred with pBI121 vector alone and probability value was estimated at P 0.05 and P 0.01 levels.
3. Results
3.1. Phylogenetic analysis of Ta-JA1 and related proteins
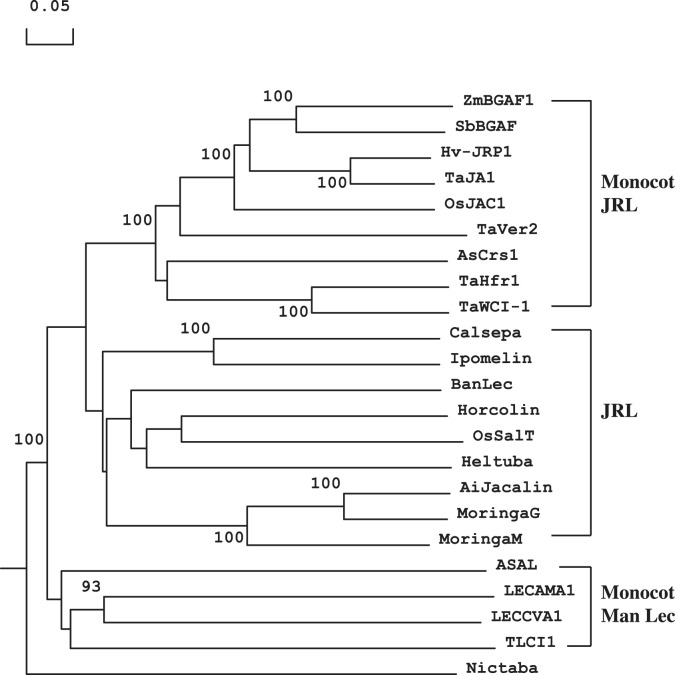
The sequence similarity search in the GenBank database revealed several proteins with high homology with Ta-JA1. The phylogenetic reconstruction showed that Ta-JA1 protein together with TaVer2 (wheat vernalization-related gene 2) [34], TaWCI-1 (wheat chemically induced gene 1) [35], TaHfr-1 (wheat Hessian fly-responsive gene 1) [36], OsJAC1 (rice JRL domain-containing protein) [37], Hv-JRP1 from barley, BGAF (β-glucosidase-aggregating factor) from maize [38] and sorghum [39], and Crs-1 from creeping bentgrass [40] clustered to one group (Monocot JRL, Fig. 1 ). All other classical jacalin-related proteins clustered to another group (JRL, Fig. 1), including AiJacalin from Artocarpus heterophyllus, Calsepa from Calystegia sepium, Ipomelin from Ipomoea batatas, MornigaG and MornigaM from Morus nigra, Heltuba from Helianthus tuberosus, Banlec from Musa paradisiaca, Horcolin from Hordeum sativum [41], and OsSalT (also called Orysata from Oryza sativa [21]. These two groups of proteins, Monocot JRL and JRL, were more closely related to each other than to the monocot mannose-binding lectins (Fig. 1; Monocot Man Lec, ASAL from Allium sativum, LECCVA1 from Crocus vernus, LECCAMA1 from Arum maculatum, and TLCI1 from Tulipa gesneriana) and were mainly differentiated due to the presence of an additional N-terminal disease response domain in Monocot JRL. Therefore, it is reasonable to believe they represent the two different types of lectins. We tentatively name Ta-JA1 and related proteins as monocot jacalin-related lectins (Monocot JRL, Fig. 1).
Fig. 1.
Dendrogram showing phylogenetic relationships of monocot jacalin-related lectins (Monocot JRL), jacalin-related lectins (JRL) and monocot mannose-binding lectins (Monocot Man Lec) from various plants. The tree was constructed using Clustal W method with PAM 250 residue weight table and boot-strap values were shown in each branch. AiJacalin (GenBank Accession no. L03797) from Artocarpus integrifolia, ASAL (U58947) from Allium sativum, AsCrs1 (DQ016627) from Agrostis stolonifera, Banlec (AF001527) from Musa acuminate, Calsepa (U56820) from Calystegia sepium, Heltuba (AF064029) from Helianthus tuberosus, Hv-JRP1 (AF021256) and Horcolin (AY033628) from Hordeum vulgare, Ipomelin (D89823) from Ipomoea batatas, LECCAMA1 (U12197) from Arum maculatum, LECCVA1 (AF233283) from Crocus vernus, MornigaG (AY048576) and MornigaM (AY048577) Morus nigra, OsJAC1 (DQ243708) and OsSalT (Z25811) from Oryza sativa, TaHfr-1 (AF483596), Ta-JA1 (AY372111), TaVer2 (AB012103) and TaWCI-1 (U32427) from Triticum aestivum, TLCI1 (U23041) from Tulipa hybrid and ZmBGAF1 (AF232008) from Zea mays. Nictaba (AF389848) from Nicotiana tabacum was used as the out-group.
3.2. Purified Ta-JA1 protein exhibiting lectin property with preferential binding to mannose
Previously, we showed that the levels of recombinant Ta-JA1 protein in E. coli were low and that this could only be detected by Western blotting [14]. The incubation and induction of E. coli cells at a lower temperature (16 °C instead of 37 °C) were shown to significantly increase protein levels as detected by SDS-PAGE gel. The recombinant Ta-JA1 protein, with a molecular mass about 50, 000 (Ta-JA1 plus His tag), was purified to homogeneity (Fig. 2 ).
Fig. 2.
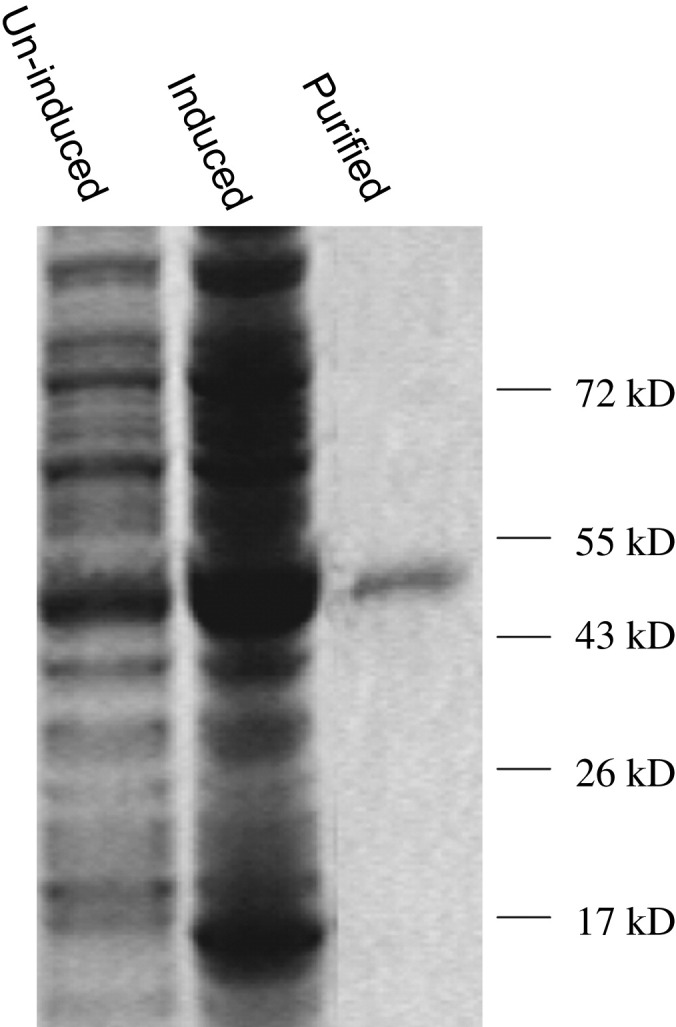
Gel electrophoretic analysis of the isolated E. coli proteins and purified Ta-JA1 protein. Proteins were separated from the total protein fractions of un-induced and IPTG-induced E. coli harbouring the Ta-JA1 expression plasmid, and after His tag resin purification of the soluble fraction from induced cells. Molecular markers are indicated on the right of the figure.
The purified recombinant Ta-JA1 protein was tested for possible lectin activity with rabbit erythrocytes. Ta-JA1 could still agglutinate rabbit red blood cells after 256 fold dilution, similar to Phaseolus vulgaris agglutinin, a typical lectin (data not shown). Carbohydrate specificity was determined by inhibition of hemagglutination using four saccharides, glucose, mannose, galactose and N-acetyl-d-glucosamine, which have been reported to have activity towards other Monocot JRL. Among the tested saccharides, d-mannose inhibited agglutination at the lowest level (MIC 6.4 mM) (Table 1 ), indicating that Ta-JA1 had lectin activity with a strong preference to d-mannose.
Table 1.
Inhibition of hemagglutination activity of recombinant Ta-JA1 protein by saccharides. Inhibition of hemagglutination was determined by serially diluting saccharides to inhibit agglutination of rabbit erythrocytes. The purified Ta-JA1 and simple saccharides solutions were pre-incubated for 1 h and rabbit erythrocytes were added. The agglutination was evaluated after 1 h at room temperature. The lowest concentration of saccharides that visibly inhibit agglutination was defined as MIC (minimum inhibitory concentration).
| Saccharides | MIC (mM) |
|---|---|
| Mannose | 6.40 |
| Galactose | 51.3 |
| N-Acetyl-d-glucosamine | 62.5 |
| Glucose | 25.7 |
3.3. Inhibition of E. coli cell growth by Ta-JA1
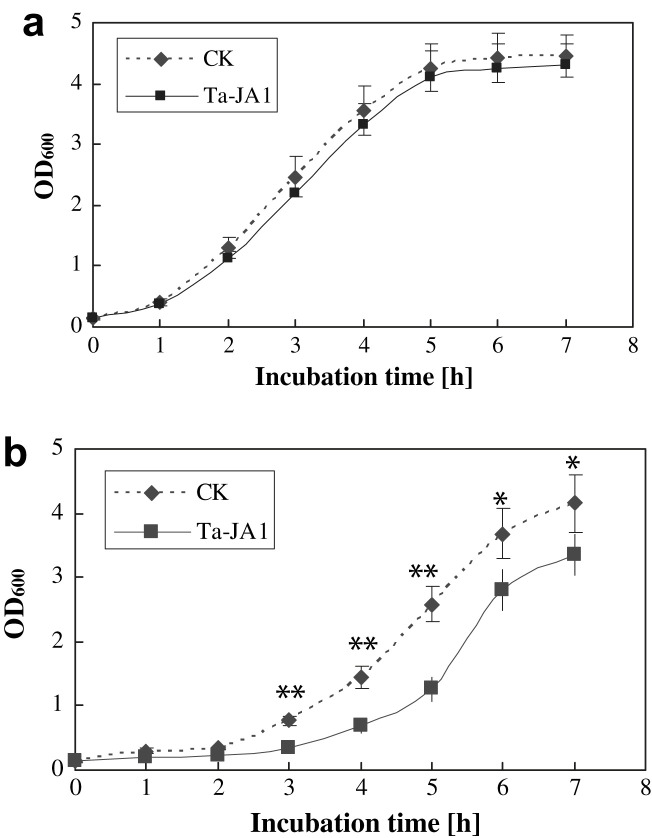
The growth curve of E. coli cells that contained Ta-JA1 expressing plasmid was determined to show the effects of Ta-JA1 on E. coli. Without IPTG-induction, the kinetic growth of cells containing Ta-JA1 plasmid was similar to that of control cells containing empty pET32a vector (Fig. 3 a). After induction with IPTG, the cell density with Ta-JA1 plasmid was remarkably lower than that of control, both in logarithmic and plateau phases (Fig. 3b). Particularly, the inhibition effects of Ta-JA1 compared with control were statistically significant at P0.01 level (by Student's t-test) from incubation time 3–6 h, which was corresponding to logarithmic growth. The cell density with Ta-JA1 plasmid was reduced to 55.8%–51.2% of control during this period.
Fig. 3.
The growth curve of E. coli cells harbouring the Ta-JA1 expressing plasmid versus empty pET32a vector plasmid (CK). a: without IPTG-induction; b: with IPTG-induction. Values are the means of three different experiments, and bars are standard errors. Probability values between CK and Ta-JA1 were estimated by the Student's t-test and significant differences at P0.05 and P0.01 levels are marked by * and **, respectively.
3.4. Improved disease resistance in the transgenic tobacco plants over-expressing Ta-JA1
In order to study the biological functions of Ta-JA1 in vivo, we generated transgenic tobacco plants with over-expressing Ta-JA1 gene. Different lines of transformed tobacco plants were obtained. Positive insertion of transgene Ta-JA1 in tobacco genome was confirmed by PCR (data not shown). Kanamycin resistance provided by the pBI121 vector was used to analyze the inheritance of the Ta-JA1 gene in self-fertilized progeny of the transgenic tobacco. Resistance versus sensitivity to kanamycin segregated in a ratio close to 3:1 in the T1 progeny of lines C2, C3, C6, and A6, and the nearly complete kanamycin resistance in homozygous T2 plants of these lines, indicating that probably one copy of T-DNA was integrated into the genome of these plants (Table 2 ). Compared with the controls, the growth, morphology, and flowering of transgenic tobacco plants did not appear significantly different.
Table 2.
Assessment of transgenic zygosity and segregation of kanamycin resistance phenotype in the T1 and T2 progenies of the transgenic tobacco.
| Plants | Progeny | KmR:KmS (seed number) | Transgene copy number | Transgenic zygosity |
|---|---|---|---|---|
| C2 | T1 | 106:28 | 1 | hemizygous |
| T2 | 99:1 | 1 | homozygous | |
| C3 | T1 | 72:20 | 1 | hemizygous |
| T2 | 95:0 | 1 | homozygous | |
| C6 | T1 | 63:17 | 1 | hemizygous |
| T2 | 85:2 | 1 | homozygous | |
| A6 | T1 | 110:24 | 1 | hemizygous |
| T2 | 112:0 | 1 | homozygous | |
To examine the expression of Ta-JA1 gene in the transgenic tobacco plants, RT-PCR was performed using total RNA from tobacco leaf tissues. A transcript of the Ta-JA1 gene with 459 bp in size was detected in all transgenic lines C2, C3, C6, and A6, but not in control line with empty pBI121 vector (Fig. 4 ). C2, C6 and A6 lines had a similar expression level, while C3 line showed the lower level than the other three lines.
Fig. 4.
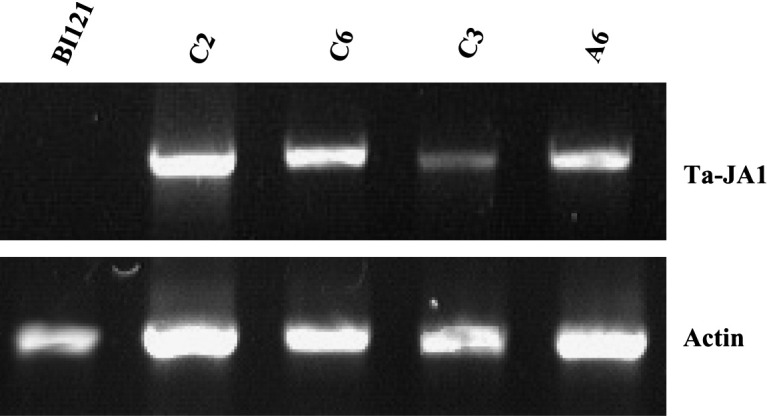
RT-PCR analysis of transgenic tobacco plants over-expressing the Ta-JA1 gene. The different transgenic lines (C2, C3, C6 and A6) and control line (BI121) that was transferred with pBI121 vector alone are shown. Actin RT-PCR was included as a control in RT-PCR analysis.
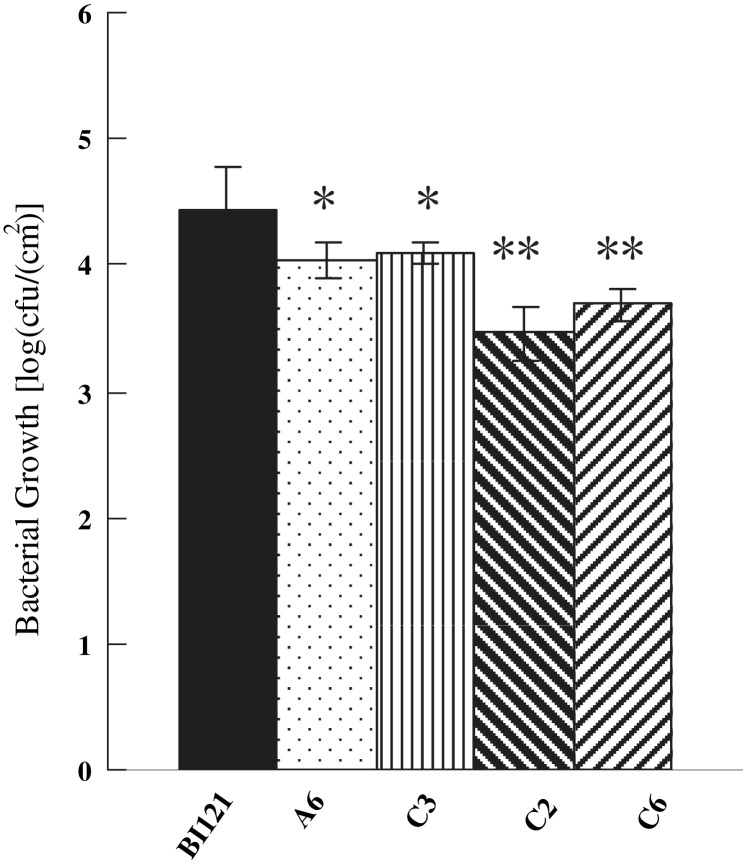
To examine whether the Ta-JA1 gene was involved in disease resistance, tobacco bacterial, fungal and viral pathogens, P. syringae pv tabaci, P. parasitica var nicotianae and TMV were used to inoculate the transgenic plants. As illustrated in Fig. 5 , the growth of P. syringae in tobacco cells was inhibited by one order of magnitude in the T2 progeny of C2 and C6 transgenic lines compared to the control line with only pBI121 vector. The inhibition of pathogen growth in C3 and A6 lines was less than that in C2 and C6, but was still statistically significant compared with that in the control tobacco.
Fig. 5.
Analysis of bacterial resistance to Pseudomonas syringae tabaci in Ta-JA1 transgenic tobacco plants. P. syringae (107 colony forming units/ml) was inoculated into the fourth to sixth upper leaves of Ta-JA1 transgenic plants of T2 progeny and the control (BI121) that was transferred with pBI121 vector alone. The infected leaves were collected and the bacterial populations were determined seven days after inoculation. Values are the means of three different experiments, and bars are standard errors. Probability values between the control and transgenic tobacco were estimated by the Student's t-test and significant differences at P0.05 and P0.01 levels are marked by * and **, respectively.
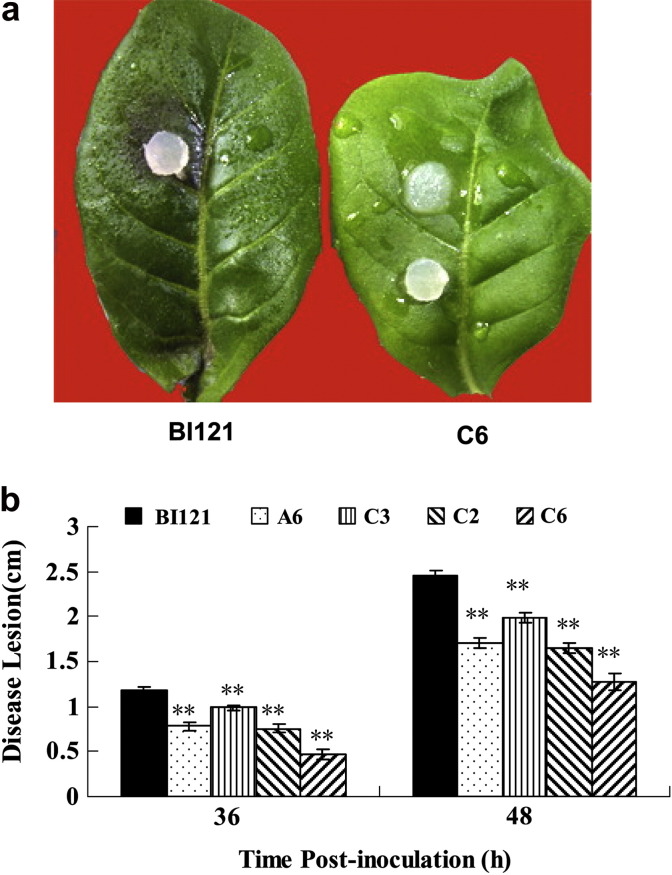
Similarly, the transgenic C2, C3, C6 and A6 lines exhibited less symptoms or significantly delayed development of the disease than the control after being infected by P. parasitica var nicotianae (Fig. 6 ). However, C3 line showed less resistance than the other three lines, which was probably associated with a low expression level of the transferred gene (Fig. 4). Similar results with remarkable reduction in TMV infectivity in transgenic plants were seen by ELISA of leaf extracts (Fig. 7 ). Taken together, the data indicate that the overexpression of Ta-JA1 gene in tobacco plants enhances the disease resistance to bacterial, fungal and viral pathogens.
Fig. 6.
Analysis of resistance to fungal pathogen Phytophthora parasitica nicotianae in Ta-JA1 transgenic tobacco plants. The fourth to sixth leaves from the top of Ta-JA1 transgenic plants of T2 progeny and the control (BI121) that was transferred with pBI121 vector alone were inoculated with mycelium-agar plugs of P. parasitica. a: The disease lesion symptoms on control (BI121) and transgenic C6 line; b: The disease lesion diameter was measured as an average of the inoculated leaves 36 and 48 h post inoculation, respectively. Values are the means of three different experiments, and bars are standard errors. Probability values between the control and transgenic tobacco were estimated by the Student's t-test and significant differences at P0.05 and P0.01 levels are marked by * and **, respectively.
Fig. 7.
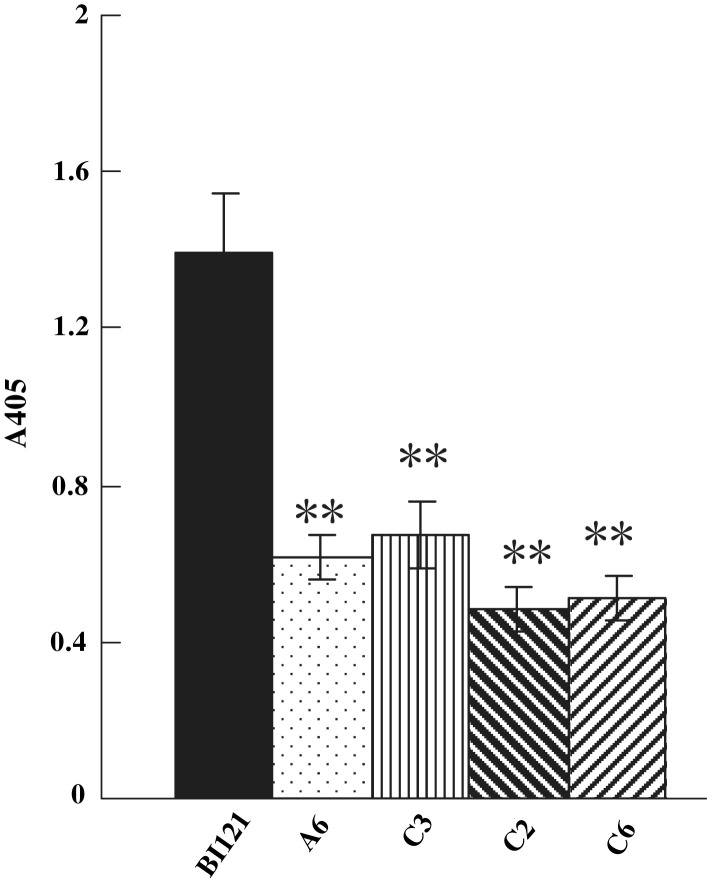
Analysis of resistance to tobacco mosaic virus (TMV) in Ta-JA1 transgenic tobacco plants. The fourth to sixth leaves from the top of Ta-JA1 transgenic plants of T2 progeny and the control (BI121) that was transferred with pBI121 vector alone were infected with TMV. TMV resistance shown by transgenic tobacco plants was measured by ELISA absorbance values at 405 nm (A405) 10 day post inoculation. Values are the means of three different experiments, and bars are standard errors. Probability values between the control and transgenic tobacco were estimated by the Student's t-test and significant differences at P0.05 and P0.01 levels are marked by * and **, respectively.
4. Discussion
4.1. Ta-JA1 and related proteins may consist of a new type of lectins
Classical plant lectins including legume and chitin-binding lectins are constitutively expressed in plant tissues, but there is increasing evidence that some newly identified lectins are induced by biotic or abiotic stimuli [17]. The jacalin-related lectins (JRL) may be involved in plant defense responses, particularly for the sub-family of mannose-specific homologues (mJRL). It has been shown that treatment with methyl jasmonate can induce lectin activity in the leaves of rice, barley, wheat, maize and rye. Purification and characterization of the cDNA clones revealed that these lectins all belong to cytoplasmic mJRL [42]. This suggests that the induction of mJRL by jasmonates is a common phenomenon in cereals. Ta-JA1 is similar in protein structure to mJRL, as it contains a jacalin-related domain and has the beta-prism architecture [14]. Furthermore, an N-terminal domain equivalent to disease response protein is found in Ta-JA1. This inducible JRL with modular structure has been proposed as a different type of JRL [17]. Here the phylogenetic analyses of JRLs and monocot mannose-binding lectins confirmed that Ta-JA1 together with other related proteins might form a new type of lectins, namely monocot jacalin-related lectins (Monocot JRL). Monocot JRLs have two features that are distinct from classical JRLs. (1) All Monocot JRLs are found in the Poaceae family. The evolutional analysis of Crs-1 indicated that Monocot JRL homologues were present in tribe Triticeae (barley and wheat), Oryzeae (rice), and Andropogoneae (maize and sorghum), while being lost in the Aveneae [40]. (2) Monocot JRL proteins contain two functional domains. A disease response domain is located in their N-termini, while a JRL domain is located in their C-termini, in contrast with the classical JRL proteins that just have a JRL domain. Hence, Monocot JRL proteins can be regarded as modular proteins composed of an N-terminal disease response domain followed by a JRL domain. Physiological functions of Monocot JRL proteins remain largely open to be investigated. Their insect resistant properties have been demonstrated by feeding Drosophila melanogaster larvae with TaHfr-1 protein, a similar Monocot JRL protein that can be induced by wheat Hessian fly [43]. Crs-1 is a Ta-JA1 homologous gene in creeping bentgrass with up-regulation upon inoculation of dollar spot pathogen, implying a possible role in pathogen reactions [40]. There is currently no evidence for the involvement of Monocot JRL proteins in disease resistance.
4.2. Ta-JA1 is a unique lectin protein and confers pathogen resistance
Our data showed that Ta-JA1 was a mannose-specific lectin protein (Table 1). A homologous protein from rice, OsJAC1, has also been shown to be a mannose-specific lectin [37]. Therefore, previously named jasmonate-regulated proteins JIP-32 may belong to a lectin family namely Monocot JRL (Fig. 1).
The levels of Ta-JA1 protein in E. coli were quite low (Fig. 2). Generally, the low levels of protein may be due to two reasons. Firstly it may be that some membrane proteins cannot be synthesized in E. coli cells [44]. Ta-JA1 protein could be induced at low temperature (Fig. 2), therefore this was not case for Ta-JA1. Second, it may be that the induced protein is toxic to E. coli cells as is the case with the viscotoxins from European mistletoe (Viscum album L.) [45]. As shown in Fig. 3b, induction of Ta-JA1 protein markedly inhibited E. coli growth compared to control cells with just empty pET32a vector, with 55.8%–51.2% reduced cell density in logarithmic growth period. This indicated that the inhibition on cell growth of Ta-JA1 protein was caused by its toxicity to the cell. Direct supplement of the purified Ta-JA1 protein in LB medium, however, did not inhibit the growth of E. coli (unpublished data). This suggests that Ta-JA1 protein is active after entering the cell. Furthermore, the overexpression of Ta-JA1 gene in transgenic tobacco increased the disease resistance to bacterial (wildfire disease), fungal (black shank disease) and viral (TMV) pathogens. Therefore, Ta-JA1 has a broadened role in the plant defense response, which can act on general bacteria (E. coli) and harmful pathogens (wildfire, black shank and TMV pathogens). Inhibition of E. coli growth has been reported for a lectin that contains chitinase domain, BjCHI1 [46]. Transplastomic tobacco expressing BjCHI1 have also shown anti-fungal activities against different pathogens, including Colletotrichum truncatum, C. acutatum, Botrytis cinerea, and Ascochyta rabiei [47]. The actions of Ta-JA1 on disease resistance look similar with that of BjCHI1. Compared with specific disease resistant genes, the actions of Ta-JA1 on pathogens are not very strong, suggesting Ta-JA1 confers a basal but broad-spectrum resistance to pathogens. Ta-JA1 is the first Monocot JRL protein that has been shown to increase disease resistance. Considering Ta-JA1 as a jasmonate-regulated protein, which is in downstream of jasmonate-induced processes, Ta-JA1 may interact with microbes instead of inducing the other gene actions. In contrast, overexpression of an AP2/EREBP transcription factor, OPBP1, has been shown to enhance both disease resistance and salt tolerance of tobacco through regulating expression in sets of stress-related genes [31]. The mechanism of Ta-JA1 with microbes is interesting and warrants further investigation.
A similar Monocot JRL protein TaHfr-1, which is up-regulated by Hessian fly, has detrimental effects on insect growth and development when feeding D. melanogaster larvae directly with the recombinant protein [43]. It is interesting to note that both Ta-JA1 and TaHfr-1 show the broad-spectrum actions individually in disease resistance and insecticidal activity, as Ta-JA1 exhibited not only to inhibit E. coli cell growth, but also to resist bacterial, fungal and viral pathogens in plant, while TaHfr-1 appeared to be virulent to both Hessian fly and D. melanogaster larvae (belonging to the Cecidomyiidae and Drosophilidae families, respectively). These broad-spectrum actions resemble the behavior of other lectins such as BjCHI1 [47] but are distinguishing to typical disease resistance gene (R gene) as gene-to-gene action [48]. The strong anti-coronavirus activity was reported for the mannose-binding lectins when tested with a severe acute respiratory syndrome coronavirus (SARS-CoV) and feline infectious peritonitis virus (FIPV) in vitro [49]. RTM1 (Restricted TEV Movement 1) protein from Arabidopsis thaliana was documented to be similar to the alpha-chain of the Artocarpus integrifolia lectin, jacalin. The RTM1 was also shown to restrict long-distance movement of tobacco etch virus without causing a hypersensitive response or inducing systemic acquired resistance [50], [51]. These data further support the wide-spectrum of pathogen resistance for mannose-binding lectins, including Ta-JA1.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No.30671043, No.30671134, No. 30970261), the Natural Science Foundation of Beijing (No. 6082018), the National High Technology and Research Development Program of P. R. China (“863” project, No. 2007AA10Z101), the Chinese National Special Foundation for Transgenic Plant Research and Commercialization (No. 2008ZX08002-003), and Innovation Project of Chinese Academy of Sciences.
References
- 1.Wasternack C. Jasmonates: an update on biosynthesis, signal, transduction and action in plant stress response, growth and development. Ann. Bot. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pena-Cortes H., Barrios P., Dorta F., Polanco V., Sanchez C., Sanchez E., Ramırez I. Involvement of jasmonic acid and derivatives in plant responses to pathogens and insects and in fruit ripening. J. Plant Growth Regul. 2005;23:246–260. [Google Scholar]
- 3.Farmer E.E., Johnson R., Ryan C.A. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 1992;98:995–1002. doi: 10.1104/pp.98.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zavala J.A., Patankar A.G., Gase K., Hui D., Baldwin I.T. Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol. 2004;134:1181–1190. doi: 10.1104/pp.103.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andresen I., Becker W., Schlüter K., Burges J., Parthier B., Apel K. The identification of leaf thionin as one of the main jasmonate-induced proteins of barley (Hordeum vulgare) Plant Mol. Biol. 1992;19:193–204. doi: 10.1007/BF00027341. [DOI] [PubMed] [Google Scholar]
- 6.Kitanaga Y., Jian C., Hasegawa M., Yazaki J., Kishimoto N., Kikuchi S., Nakamura H., Ichikawa H., Asami T., Yoshida S., Yamaguchi I., Suzuki Y. Sequential regulation of gibberellin, brassinosteroid, and jasmonic acid biosynthesis occurs in rice coleoptiles to control the transcript levels of anti-microbial thionin genes. Biosci. Biotechnol. Biochem. 2006;70:2410–2419. doi: 10.1271/bbb.60145. [DOI] [PubMed] [Google Scholar]
- 7.Gundlach H., Müller M.J., Kutchan T.M., Zenk M.H. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. USA. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J.W., Wu J.Y. Nitric oxide is involved in methyl jasmonate-induced defense responses and secondary metabolism activities of taxus cells. Plant Cell Physiol. 2005;46:923–930. doi: 10.1093/pcp/pci098. [DOI] [PubMed] [Google Scholar]
- 9.Ellis C., Karafyllidis I., Wasternack C., Turner J.G. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Chang P.F.L., Liu D., Narasimhan M.L., Raghothama K.G., Hasegawa P.M., Bressan R.A. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feussner I., Hause B., Vörös K., Parthier B., Wasternack C. Jasmonate-induced lipoxygenase forms are localized in chloroplasts of barley leaves (Hordeum vulgare cv. Salome) Plant J. 1995;7:949–957. [Google Scholar]
- 12.Hause B., Hertel S.C., Klaus D., Wasternack C. Cultivar-specific expression of the jasmonate-induced protein of 23 kDa (JIP-23) occurs in Hordeum vulgare L. by jasmonates but not during seed germination. Plant Biol. 1999;1:83–89. [Google Scholar]
- 13.Leopold J., Hause B., Lehmann J., Graner A., Parthier B., Wasternack C. Isolation, characterization and expression of a cDNA coding for a jasmonate-inducible protein of 37 kDa in barley leaves. Plant Cell Environ. 1996;19:675–684. [Google Scholar]
- 14.Wang X.M., Ma Q.H. Characterization of a jasmonate-regulated wheat protein related to a beta-glucosidase-aggregating factor. Plant Physiol. Biochem. 2005;43:185–192. doi: 10.1016/j.plaphy.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Sharon N., Lis H., editors. Lectins. Kluwer Academic Publishers; 2003. [Google Scholar]
- 16.Van Damme E.J.M., Lannoo N., Peumans W.J. Plant Lectins, Adv. Bot. Res. 2008;48:107–209. [Google Scholar]
- 17.Van Damme E.J.M., Barre A., Rouge P., Peumans W.J. Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci. 2004;9:484–489. doi: 10.1016/j.tplants.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Moreira R.A., Ainouz I.L. Lectins from seeds of jack fruit (Artocarpus integrifolia L.): isolation and purification of two isolectins from the albumin fraction. Biol. Plant. 1981;23:186–192. [Google Scholar]
- 19.Clendennen S.K., May G.D. Differential gene expression in ripening banana fruit. Plant Physiol. 1997;115:463–469. doi: 10.1104/pp.115.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peumans W.J., Winter H.C., Bemer V., Van Leuven F., Goldstein I.J., Truffa-Bachi P., Van Damme E.J.M. Isolation of a novel plant lectin with an unusual specificity from Calystegia sepium. Glycoconjugate J. 1997;14:259–265. doi: 10.1023/a:1018502107707. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W., Peumans W.J., Barre A., Astoul C.H., Rovira P., Rouge P., Proost P., Truffa-Bachi P., Jalali A.A., Van Damme E.J.M. Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta. 2000;210:970–978. doi: 10.1007/s004250050705. [DOI] [PubMed] [Google Scholar]
- 22.Yagi F., Iwaya T., Haraguchi T., Goldstein I.J. The lectin from leaves of Japanese cycad, Cycas revoluta Thunb. (gymnosperm) is a member of the jacalin-related family. Eur. J. Biochem. 2002;269:4335–4341. doi: 10.1046/j.1432-1033.2002.03127.x. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman J.D. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J.D., Higgins D.G., Gibson T.J., Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choices. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 26.Debray H., Daecourt D., Strecker G., Spik G., Montruil J. Specificity of twelve lectins towards oligosaccharides and glycopeptides related to N-glycosylproteins. Eur. J. Biochem. 1981;117:41–55. doi: 10.1111/j.1432-1033.1981.tb06300.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen P.Y., Wang C.K., Soong S.C., To K.Y. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 2003;11:287–293. [Google Scholar]
- 28.Ma Q.H., Wang X.M., Wang Z.M. Expression of isopentenyl transferase gene controlled by seed-specific lectin promoter in transgenic tobacco influences seed development. J. Plant Growth Regul. 2008;27:68–76. [Google Scholar]
- 29.Edwards K., Johnstone C., Thompson C. A simple rapid method for the preparation of plant genomic DNA for PCR analysis. Nucl. Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murashige T., Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 31.Guo Z.J., Chen X.J., Wu X.L., Ling J.Q., Xu P. Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol. Biol. 2004;55:607–618. doi: 10.1007/s11103-004-1521-3. [DOI] [PubMed] [Google Scholar]
- 32.Bhargava A., Osusky M., Hancock R.E., Forward B.S., Kay W.W., Misra S. Antiviral indolicidin variant peptides: evaluation for broad-spectrum disease resistance in transgenic Nicotiana tabacum. Plant Sci. 2007;172:515–523. [Google Scholar]
- 33.Geng S., Hills F.G. Kendell Hunt Publisher; Dubuque, Iowa: 1989. Biometrics in Agriculture Science. [Google Scholar]
- 34.Yong W.D., Xu Y.Y., Xu W.Z., Wang X., Li N., Wu J.S., Liang T.B., Chong K., Xu Z.H., Tan K.H., Zhu Z.Q. Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta. 2003;217:261–270. doi: 10.1007/s00425-003-0994-7. [DOI] [PubMed] [Google Scholar]
- 35.Gorlach J., Volrath S., Knauf-Beiter G., Hengy G., Beckhove U., Kogel K., Oostendorp M., Staub T., Ward E., Kessmann H., Ryals J. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams C.E., Collier C.C., Nemacheck J.A., Liang C., Cambron S.E. A lectin-like gene responds systemically to attempted feeding by avirulent first-instar Hessian fly larvae. J. Chem. Ecol. 2002;28:1411–1428. doi: 10.1023/a:1016200619766. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J.F., Han Y., Xing L.J., Xu Y.Y., Xu Z.H., Chong K. Cloning and expression of a novel cDNA encoding a mannose-specific jacalin-related lectin from Oryza sativa. Toxicon. 2006;47:133–139. doi: 10.1016/j.toxicon.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Esen A., Blanchard D.J. A specific beta-glucosidase-aggregating factor is responsible for the beta-glucosidase null phenotype in maize. Plant Physiol. 2000;122:563–572. doi: 10.1104/pp.122.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kittur F.S., Yu H.Y., Bevan D.R., Esen A. Homolog of the maize β-glucosidase aggregating factor from sorghum is a jacalin-related GalNAc-specific lectin but lacks protein aggregating activity. Glycobiology. 2009;19:277–287. doi: 10.1093/glycob/cwn132. [DOI] [PubMed] [Google Scholar]
- 40.Li H.M., Rotter D., Bonos S.A., Meyer W.A., Belanger F.C. Identification of a gene in the process of being lost from the genus Agrostis. Plant Physiol. 2005;138:2386–2395. doi: 10.1104/pp.105.063297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grunwald I., Heinig I., Thole H.H., Neumann D., Kahmann U., Kloppstech K., Gau A.E. Purification and characterisation of a jacalin-related, coleoptile specific lectin from Hordeum vulgare. Planta. 2007;226:225–234. doi: 10.1007/s00425-006-0467-x. [DOI] [PubMed] [Google Scholar]
- 42.Van Damme E.J.M., Zhang W., Peumans W.J. Induction of cytoplasmic mannose-binding jacalin-related lectins is a common phenomenon in cereals treated with jasmonate methyl ester. Comm. Appl. Biol. Sci. 2004;69:23–31. Ghent University. [PubMed] [Google Scholar]
- 43.Subramanyam S., Smith D.F., Clemens J.C., Webb M.A., Sardesai N., Williams C.E. Functional characterization of HFR1, a high-mannose N-glycan-specific wheat lectin induced by Hessian fly larvae. Plant Physiol. 2008;147:1412–1426. doi: 10.1104/pp.108.116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonander N., Hedfalk K., Larsson C., Mostad P., Chang C., Gustafsson L., Bill R.M. Design of improved membrane protein production experiments: quantitation of the host response. Protein Sci. 2005;14:1729–1740. doi: 10.1110/ps.051435705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bogomolovas J., Simon B., Sattler M., Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 2009;64:16–23. doi: 10.1016/j.pep.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Zhao K.J., Chye M.L. Methyl jasmonate induces expression of a novel Brassica juncea chitinase with two chitin-binding domains. Plant Mol. Biol. 1999;40:1009–1018. doi: 10.1023/a:1006266407368. [DOI] [PubMed] [Google Scholar]
- 47.Guan Y.F., Ramalingam S., Nagegowda D., Taylor P.W.J., Chye M.L. Brassica juncea chitinase BjCHI1 inhibits growth of fungal phytopathogens and agglutinates Gram-negative bacteria. J. Exp. Bot. 2008;59:3475–3484. doi: 10.1093/jxb/ern197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayliffe M.A., Lagudah E.S. Molecular genetics of disease resistance in cereals. Ann. Bot. 2004;94:765–773. doi: 10.1093/aob/mch207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keyaerts E., Vijgen L., Pannecouque C., Van Damme E.J.M., Peumans W., Egberink H., Balzarini J., Van Ranst M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007;75:179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahajan S.K., Chisholm S.T., Whitham S.A., Carrington J.C. Identification and characterization of a locus (RTM1) that restricts long-distance movement of tobacco etch virus in Arabidopsis thaliana. Plant J. 1998;14:177–186. doi: 10.1046/j.1365-313x.1998.00105.x. [DOI] [PubMed] [Google Scholar]
- 51.Chisholm S.T., Mahajan S.K., Whitham S.A., Yamamoto M.L., Carrington J.C. Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc. Natl. Acad. Sci. USA. 2000;97:489–494. doi: 10.1073/pnas.97.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]