Abstract
Background: The tumor necrosis factor receptor (TNFR) associated factor 3 (TRAF3) is a key node in innate and adaptive immune signaling pathways. TRAF3 negatively regulates the activation of the canonical and non-canonical NF-κB pathways and is one of the key proteins in antiviral immunity.
Scope of Review: Here we provide a structural overview of TRAF3 signaling in terms of its competitive binding and consequences to the cellular network. For completion, we also include molecular mimicry of TRAF3 physiological partners by some viral proteins.
Major Conclusions: By out-competing host partners, viral proteins aim to subvert TRAF3 antiviral action. Mechanistically, dynamic, competitive binding by the organism's own proteins and same-site adaptive pathogen mimicry follow the same conformational selection principles.
General Significance: Our premise is that irrespective of the eliciting event – physiological or acquired pathogenic trait – pathway activation (or suppression) may embrace similar conformational principles. However, even though here we largely focus on competitive binding at a shared site, similar to physiological signaling other pathogen subversion mechanisms can also be at play. This article is part of a Special Issue entitled “System Genetics” Guest Editor: Dr. Yudong Cai and Dr. Tao Huang.
Keywords: Inflammation, Cancer, Antiviral immunity, Evolvable, Structure, Host-pathogen interactions
Highlights
-
•
TRAF3 has a key role in antiviral immune response.
-
•
TRAF3 binds to several partners through the same or overlapping interfaces.
-
•
Viruses subvert host immunity by mimicking the binding surfaces of its proteins.
-
•
Viral proteins also target the same binding site on TRAF3.
-
•
Binding of viral proteins to TRAF3 competitively displaces its host partners.
1. Introduction
The cellular communication network is ubiquitous and mobile. Signals are transferred through dynamic linkages between nodes (proteins). Nodes that transfer several signals are control switches. In principle, a control switch has a choice of which node to contact. That temporal choice or differences in avidity and state of post-translational modifications are important since they influence the consequent pathway and executed function. The multiple switches and choices along the pathways and feedback loops reflect safety checks adopted by complex systems. At each central switch the signal that is transferred (through protein-protein interaction) is determined by a number of factors, including protein concentration (determined by expression and degradation); phosphorylation (or other post-translational modification) states, the concentration of ions (particularly Ca2 +), and additional factors reflecting the cell state. In principle, a switch protein can bind to a large number of partners underscoring the many mechanisms which can be involved [1]. The principles of protein-protein interactions are fairly well understood [2], [3], [4], [5]; among them, a key feature is that proteins that bind at the same (host) site have to share a certain extent of similar molecular surface [6]. This results in competitive binding. Competitive binding is a hallmark of regulation; thus it is no wonder that pathogens have evolved to exploit it to their advantage. Virus survival relies on its adaptive evolution. Thus, its evolvability reflects a capacity to rapidly shift its genome to select proteins with states adapted to bind host switch nodes and turn them off – or on – thereby controlling their adverse (or beneficial) functions. Adapting the protein shape and shifting the conformational equilibrium may require only few residue changes to link their evolving genotype to a vital phenotype which promotes their survival.
Here we present the case of the TRAF3 switch, a master regulator at the crossroads of antiviral, anti-inflammatory and cancer pathways (Fig. 1 ). We view TRAF3 from the conformational standpoint in terms of competitive interactions on its surface. This bolsters insight into its cell signaling and evolvability of adaptive viral molecular mimicry.
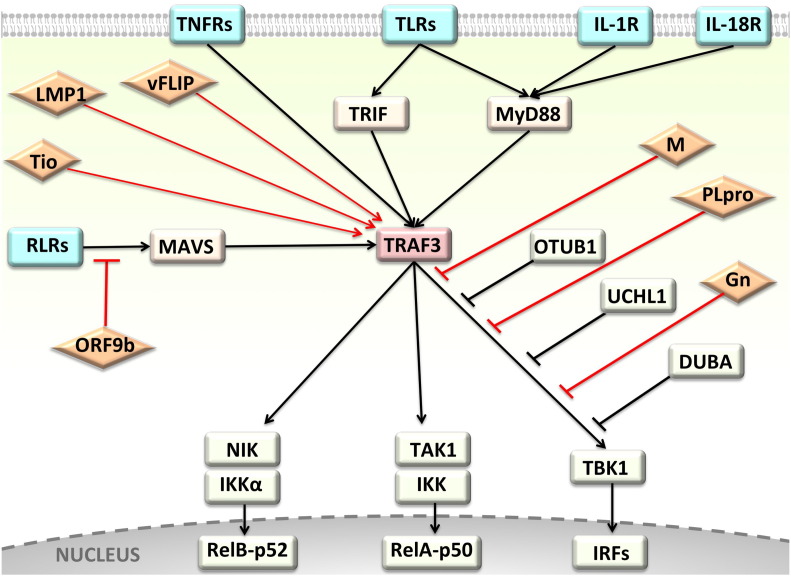
Fig. 1.
TRAF3 is a hub protein at the crossroad of immune system receptors and several viral proteins that turn its signaling on or off. TRAF3 regulates the activation of canonical and non-canonical NF-κB and the generation of anti-viral responses. Since TRAF3 controls signaling through several pathways, it is targeted by many viral proteins. Rectangular shapes correspond to cellular proteins and diamond shapes are viral proteins. Black lines show interactions through host proteins and the red lines show interactions with viral proteins.
2. TRAF3: Anti-inflammatory signaling and competitive interactions
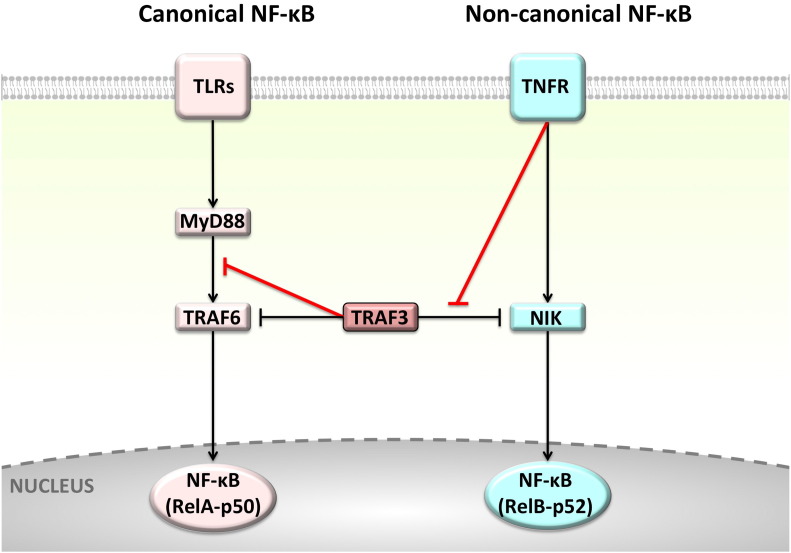
The six membered-TRAF family has important roles in pathways of the immune system, such as Toll-like receptors (TLRs), Retinoic acid inducible-gene I (RIG-I) like receptors (RLRs), NOD-like receptors (NLRs) and TNFRs [7]. Family members serve as adaptors, recruiting proteins, assembling them into large protein complexes, spatially organizing them and allosterically regulating their signaling [8]. TRAFs are non-degradative K63-specific E3 ubiquitin (Ub) ligases, ubiquitinating themselves and downstream proteins to modulate signaling [7]. Downstream kinases, such as TBK1 (TANK binding kinase 1) and IKK (IκB kinase), bind TRAFs' autoubiquitinated K63-linked Ub chain [9]. Despite the structural and sequence similarity among TRAF proteins, they have distinct preferred binding partners and perform nonredundant – sometimes opposing – signaling functions [10]. TRAF6 and TRAF3 provide a good example. Both take part in TLR signaling; however, while TRAF6 promotes signaling through the conventional downstream path which activates the classical/canonical NF-κB and results in inflammation, TRAF3 signals through interferon regulatory factors (IRFs) to produce interferons (IFNs) and anti-inflammatory cytokine IL-10 [11]. Recently, we demonstrated that TRAF6- and TRAF3-dependent downstream paths of TLRs are competitive due to overlapping binding sites on MyD88 [12]. TRAF3 negatively regulates TLR-mediated activation of classical NF-κB and MAPKs, as well as TNFR family-mediated stimulation of the alternative NF-κB pathway [7]. Canonical NF-κB signaling is activated upon stimulation of many TRAF-dependent receptors, like TNFRs and TLRs, and the non-canonical NF-κB is triggered by some of the TNFR family members, such as CD40, BAFFR, and lymphotoxin-β receptor (LTβR) [13] (Fig. 2 ). Even though TRAF3 was proposed not to play a role in the classical NF-κB, we have shown that it may restrict the activation of classical NF-κB by competing with TRAF6 to bind to MyD88 [12].
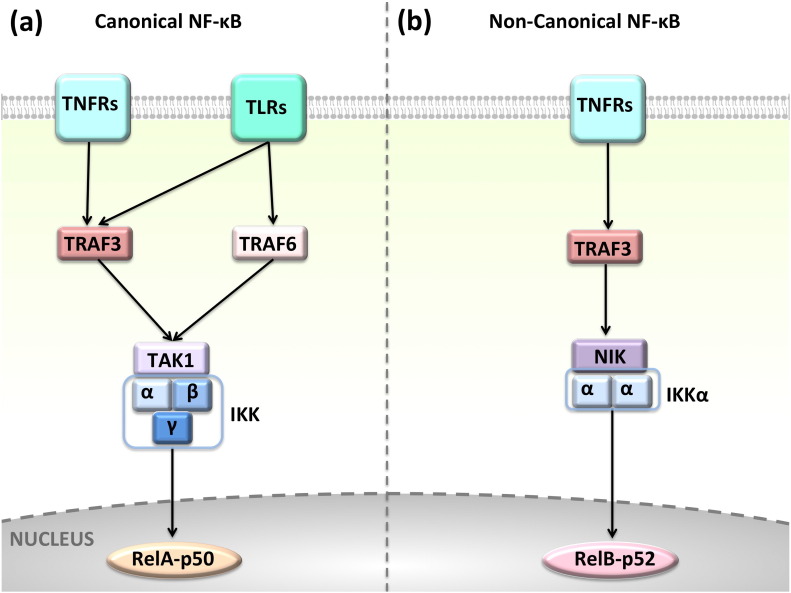
Fig. 2.
Canonical and non-canonical NF-κB pathways. TLRs and TNFRs activate the (a) classical NF-κB signaling, whereas only some subtypes of TNFRs, such as CD40, BAFFR activate (b) the alternative NF-κB. NF-κB has different subunits: RelA (p65), p50, RelB, and p52. They form heterodimers and translocate to the nucleus to initiate transcription when IKK phosphorylates and inactivates IκB (inhibitor of NF-κB). The distinct combinations of subunits trigger the transcription of different effector genes.
Current observations suggest that the antiviral function of TRAF3 cannot be replaced by other TRAF family members [10] and this function requires a structurally intact TRAF3 molecule: N-terminal or C-terminal truncated mutants cannot initiate antiviral responses in TRAF3 knock-down cells [14]. TRAF3 serves in anti-inflammatory signaling and its deletion in myeloid cells leads to inflammatory diseases and cancer in mice [15]. TRAF3 functions as a tumor suppressor in human multiple myeloma [16], [17]. Its deficiency [18] or overexpression [16] promote autoimmunity and predisposition to cancer. The outcome may depend on the microenvironment, including concentrations of the binding partners, viruses that infected the cell and pathogen-activated cell receptors. Protein expression, degradation, stability, posttranslational modifications, pH and presence of other proteins in the environment affect the affinity of a protein toward its partners. Thus, different cellular outcomes can be observed under different conditions since the preference of a protein to bind to a particular partner changes.
3. Pathogen evolvability and molecular mimicry from the structural standpoint
Pathogens developed strategies to subvert normal functions that hinder their proliferation. One way through which they accomplish this is by targeting a key node in the cellular network whose role is to shutoff (or turn on) a pathway essential for their survival. Fig. 1 demonstrates an example for this. Several viral proteins target a central protein, TRAF3 at the crossroads of immune system pathways. Viral evolvability acquired immunomodulatory activities that suppress the anti-viral immunity by encoding homologues of cytokines, their receptors or their downstream signal components [19], [20]. Here we focus on achieving this aim by analyzing conformational mimicry of TRAF3 partner proteins. TRAF3 deficiency can result in enhanced activation and proliferative canonical and non-canonical NF-κB signaling. Even though there may not be overall sequence or structural similarity of the viral proteins to those of the host, they may have evolved to adopt similar binding site surfaces, allowing them to out-compete cellular proteins. Global structural similarity is not necessary; mimicking the interacting surface can be sufficient [21], [22]. Viruses may also promote the expression of cellular proteins [23] or exploit viral proteins [24], [25] to inhibit the signaling. Structural similarity between host and virus proteins may also induce host response to pathogenic and host self-components [26], eliciting autoimmunity.
Traditionally, molecular mimicry has been viewed as sequence similarities between foreign and self-peptides that may be sufficient to induce cross-activation of autoreactive T or B cells by pathogen-derived peptides. Mechanistically, mimicry can take place through similar protein interaction sites competing to interact with a shared host surface. Similar interaction sites indicate similar interface conformations. Notably, the molecular mimicry mechanism applies not only to pathogen subversion; it simply denotes conformational similarity at the surface [27]. Surface, or interface [21], [22] mimicry implies competitive binding, a hallmark of regulation of cellular networks. Finally, interaction sites may also become similar through allosteric conformational changes. Proteins exist as dynamic ensembles of states [28]; not as rigid statues [29]. If a certain state within the ensemble is complementary to the shared binding site and has a sufficiently-high population it can act in molecular mimicry.
Hosts evolved and shaped the immune system together with the pathogens. Both acquired strategies to subvert actions of one another to their own benefit [19]. Molecular mimicry is one of the subversion mechanisms. Pathogens mimic the host proteins or interfaces and bind to their partners. Host proteins in turn alter themselves to avoid pathogenic protein binding. This phenomenon is also known as “evolutionary arms race” [22]. Evolvability is the ability of organisms to evolve and adapt themselves to become fitter to their surrounding environment. The host interfaces that participate in exogenous (inter-species, i.e. host-pathogen interactions) interactions seem to have evolved faster than those involved only in endogenous (intra-species) interactions, since they are less conserved [21], [22]. Faster evolutionary changes at the host-pathogen interfaces support the notion of host-pathogen survival competition. Changes in a few residues or even a single residue at the interface may have a large impact on the affinity toward certain partners and hence rewire pathways [30]. Pathogens are evolutionarily more plastic; that is they have higher mutation rates, particularly RNA viruses due to a lack of a proofreading capacity in RNA replicases [31]. This property allows them to quickly adapt to the host environment and develop strategies to evade the host immune surveillance.
4. Available structures and structural models for TRAF3 interactions
TRAF3 has a TRAF domain, five Zinc finger (ZF) motifs and a RING domain (Fig. 3 ). The TRAF domain is divided into a β-sandwich TRAF-C lobe at the C-terminal, through which it interacts with upstream proteins and a coiled-coil TRAF-N lobe at its N-terminal through which it homotrimerizes [7]. The TRAF domain is responsible for the varying specificities of TRAF proteins [10]. The N-terminal RING domain harbors the E3 Ub-ligase activity. The structures of some TRAF-C interactions have been resolved, such as with LTβR [32], Caspase activation and recruitment domain adaptor-inducing IFN-β (Cardif, also known as MAVS) [10], CD40 [33], TRAF-associated NF-κB activator (TANK) [34], the Epstein Barr virus latent membrane protein-1 (LMP1) [24], and BAFFR [35]. All recognize the same TRAF-C β-sandwich binding crevice, thus are competitive binders. To enrich the TRAF3 structural space, we modeled additional TRAF-C and RING domains' interactions by a template-based protein-protein interaction prediction tool, PRISM (PRotein Interactions by Structural Matching) [36], [37], [38]. The rationale behind PRISM is also interface mimicking [39]. Although global structures of proteins are diverse, they use similar interface architectures to bind to their partners. The interfaces of resolved structures of protein complexes can serve as templates to predict new interactions. Table 1 lists TRAF3-binding proteins. Almost all interactions are competitive. Below, we present examples of individual interactions of TRAF3 and their available or predicted structural interaction models.
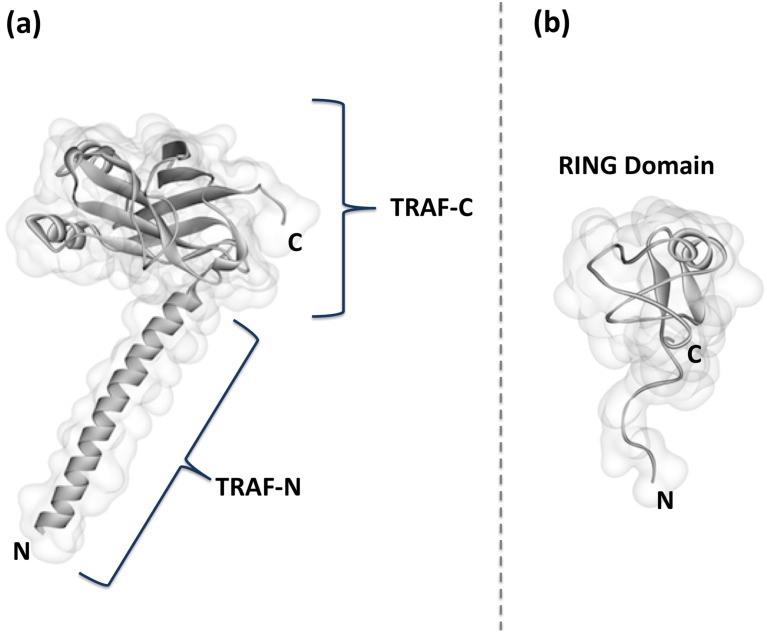
Fig. 3.
Domain structure of TRAF3. TRAF3 has two domains: (a) a TRAF domain at its C terminus – consisting of β-sandwich TRAF-C and coiled coil TRAF-N –, and (b) a RING domain at its N terminus.
Table 1.
TRAF3 interactions through its TRAF-C and RING domains. All of the interactions through the same domain are competitive due to overlapping interfaces.
| TRAF3 interactions | Domain of TRAF3 |
|---|---|
| LTBR | Beta-sandwich of TRAF-C |
| CD40 | Beta-sandwich of TRAF-C |
| BAFFR | Beta-sandwich of TRAF-C |
| MAVS | Beta-sandwich of TRAF-C |
| TANK | Beta-sandwich of TRAF-C |
| LMP1 | Beta-sandwich of TRAF-C |
| cIAP1 | Beta-sandwich of TRAF-C |
| cIAP2 | Beta-sandwich of TRAF-C |
| TRADD | Beta-sandwich of TRAF-C |
| MyD88 | Beta-sandwich of TRAF-C |
| NIK | Beta-sandwich of TRAF-C |
| DUBA | Beta-sandwich of TRAF-C |
| UCHL1 | Beta-sandwich of TRAF-C |
| ORF-9b | Beta-sandwich of TRAF-C |
| vFLIP | Beta-sandwich of TRAF-C |
| Plpro | Beta-sandwich of TRAF-C |
| OTUB1 | RING domain |
| SRC | RING domain |
4.1. TRAF3-NIK interaction
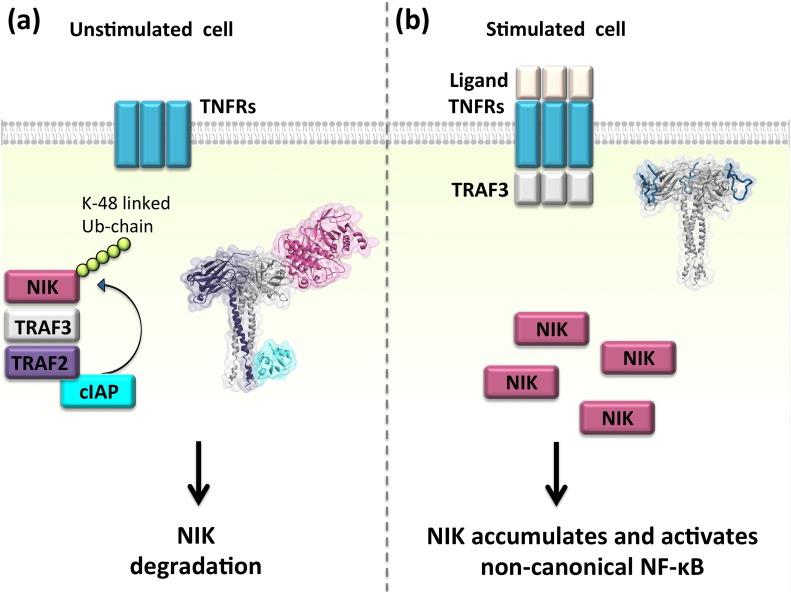
Fig. 4 displays the binary competitive interactions through the TRAF-C domain in the TRAF3 protein. The straight lines that converge to the same site on TRAF3 bind to fully/partially overlapping surface on TRAF3. For example, NF-κB inducing kinase (NIK) and some TNFRs such as LTβR, BAFFR, and CD40 bind to overlapping surfaces on TRAF3. Normally, in unstimulated cells, when the receptor is not bound to its ligand, NIK is bound to TRAF3 and is ubiquitinated by cellular inhibitors of apoptosis (cIAPs). The Ub chain labels NIK for degradation, keeping its concentration below a certain threshold (Fig. 5 ) [40]. On the other hand, if the receptor is bound to its ligand, NIK-TRAF3 interaction is abrogated and NIK is not degraded; it accumulates. Accumulation of NIK leads to activation of the alternative NF-κB pathway [40]. The structure of TRAF3-NIK interaction is not available. We modeled it by exploiting PRISM [36], [37], [38]. All details regarding the models and interface and hotspot residues are presented in the supplementary materials (Table S1 through Table S19). Our structural model supports earlier experimental findings that show competitive NIK displacement from TRAF3 upon LTβR binding [41]. This reveals how/why NIK is not degraded and accumulates since the cytoplasmic part of the receptor interacts with TRAF3 and prevents TRAF3-NIK interaction (Fig. 4). Although Liao et al. [42] suggested that the N-terminal of NIK, residues 78ISIIAQA84, binds to TRAF3, in our model the middle part of NIK (residues 464–539) interacts. The F474E mutation on TRAF3 leads to constitutive activation of the non-canonical NF-κB, since NIK cannot bind to this mutant form of TRAF3 and be degraded [41], suggesting that this residue is likely to be in the interface. Our model also has F474 in the interface of TRAF3-NIK. If NIK cannot bind to TRAF3, it cannot get ubiquitinated by cIAPs and targeted for degradation. For instance, Tio oncoprotein of Herpesvirus ateles displaces NIK when it binds to TRAF3 and activates non-canonical NF-KB constitutively [43]. Structural details of the interactions elucidate how and why distinct downstream outcomes such as accumulation or degradation of NIK are observed under different conditions – stimulated or unstimulated cells – as in our case here.
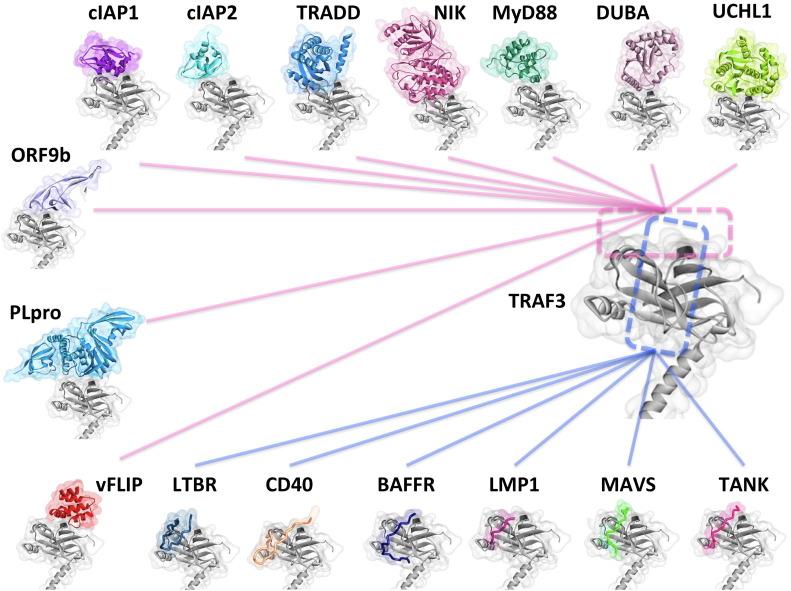
Fig. 4.
Interactions of TRAF3 through its TRAF-C region. TRAF3 interacts with many proteins through its TRAF-C region. Since the surface on this region is limited, TRAF3 uses same or overlapping surface patches (interfaces) to interact with different partners. Some of the interactions shown here have already resolved structures (LTβR [32], CD40 [33], BAFFR [35], LMP1 [24], MAVS [10], and TANK [34]). We modeled the rest. The dashed rectangles show the interfaces on TRAF3 and the solid lines show which interactions share these interfaces. Since the two dashed rectangles overlap, all interactions shown in this figure have overlapping interfaces with all other interactions. Thus, all are potentially competitive.
Fig. 5.
Distinct cellular outcomes are observed when TRAF3 interacts with NIK and TNFRs. Our structural model supports the experimental finding [41] of NIK's displacement upon receptor binding and explains why NIK is not degraded in stimulated cells. Some TNFRs form trimers upon stimulation, but others preassemble into trimers before stimulation [44]. They recruit trimeric TRAFs. TRAFs trimerize through their TRAF-C region and dimerize through their RING domain [45], [46]. (a) In the unstimulated cell, NIK is bound to TRAF3-TRAF2 heterotimer, gets ubiquitinated by cIAPs, and gets degraded. The structure of TRAF2-cIAP complex is available (PDB ID: 3M0A). We obtained the heterotrimer of TRAF3 and TRAF2 by superimposing TRAF2 onto TRAF3 unit cell trimer. TRAF3-NIK interaction is modeled by PRISM [36], [37], [38]. (b) When TNFRs recognize their ligands they recruit TRAF3 homotrimers to their cytoplasmic domain. Since TNFRs and NIK bind to the same site on TRAF3, TNFR-TRAF3 interaction liberates NIK and prevents its ubiquitination. NIK accumulates and activates non-canonical NF-κB. Here, the structure of TRAF3-LTβR (PDB ID: 1RF3) is shown as an example of TNFR activation.
Without structural information, we surmised that TRAF3 inhibits the canonical NF-κB through TRAF6 in the TLR pathway, and the non-canonical NF-κB through NIK [18], [42] in the TNFR pathway. However, atomic details of TRAF3 interactions corrected our presumption: TRAF3 does not inhibit TRAF6; instead, it competes with TRAF6 to bind to MyD88 (Fig. 6 ), which explains why TRAF3 restricts TRAF6-dependent NF-κB activation. Similarly, TRAF3 does not block NIK; TNFRs and NIK compete to bind to TRAF3 (Fig. 6).
Fig. 6.
Structural details of TRAF3 interactions revealed how TRAF3 inhibits both the canonical and non-canonical NF-κB pathways. Structural details of TRAF3 interactions corrected critical misinterpretation and unravel what may happen in actual scenario. In TLR signaling, it was assumed that TRAF3 inhibits directly TRAF6, but structural data show that it inhibits the MyD88-TRAF6 interaction and hence prevents activation of the canonical NF-κB pathway. In TNFR signaling, the presumption was that TRAF3 blocks NIK; however structural data suggest that TRAF3-TNFR interaction prevents TRAF3-NIK association. Black lines show what was assumed before and red lines show what is revealed by the structural details of our models.
4.2. TRAF3-cIAPs interaction
cIAPs are important players in TNFR pathways. They have baculoviral IAP repeats (BIR domains) and a RING domain, through which they function as E3 Ub-ligase enzymes [47]. In the unstimulated cell, TRAF3 requires TRAF2 to recruit cIAPs. NIK-bound TRAF3 recruits TRAF2, which is bound to cIAPs through its coiled-coil TRAF-N [7], [48]. When NIK interacts with TRAF2-bound TRAF3, cIAPs (cIAP1 and cIAP2) conjugate the degradative K48-linked Ub-chain to NIK, which targets it to proteosomal degradation [7], [49]. TRAF3 cannot associate with cIAPs in the unstimulated cell [48], consequently it cannot catalyze this Ub-chain conjugation to NIK; it requires TRAF2 to recruit cIAPs. Thus, even though TRAF2 and TRAF3 are structurally very similar (RMSD of 1.31 Å over 187 residues), they have nonredundant functions [49], which might reflect cell/tissue specificity. Viral stimulation induces cIAP1-TRAF3 interaction, which results in ubiquitination and degradation of TRAF3 [50]. This interaction is detected only after viral infection. Upon stimulation, not only NIK accumulation is observed, but also TRAF3 degradation, probably due to TRAF3-cIAP1 interaction. Similarly, cIAP2 was also suggested not to interact with TRAF3 [43]. However, it may follow a scenario similar to that of cIAP1 where viral stimulation triggers its interaction with TRAF3. We modeled TRAF3 interactions with both cIAP1 and cIAP2 (Fig. 4). In the TRAF3-cIAP1 interaction model, BIR3 domain of cIAP1 interacts with the TRAF-C region of TRAF3, where LTβR and NIK bind (F474E mutation is close to interface). In the TRAF3-cIAP2 complex that we modeled, BIR1 domain of cIAP2 interacts with TRAF3 and this complex is very similar to the TRAF3-cIAP1 interaction.
4.3. TRAF3-TRADD interaction
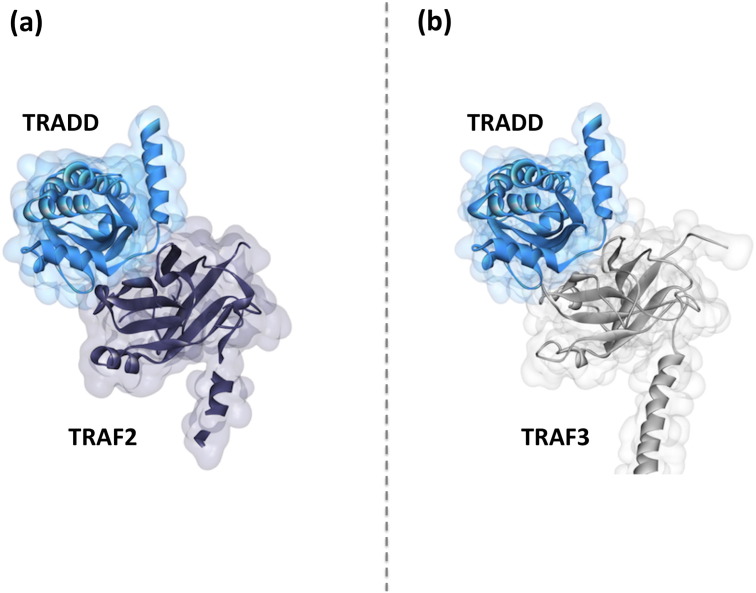
Some TNFRs have death domain (DD) in their cytoplasmic domains – and some TNFRs do not. The ones with DD do not interact with TRAFs directly; instead, they employ a bridging protein, either TRADD or RIPK1 to recruit TRAFs [34]. TRADD was suggested to interact specifically with TRAF1 and TRAF2, interactions vital for suppressing apoptosis [51]. Michallet et al. experimentally showed that TRADD also interacts with MAVS, TRAF3, and TANK in the RIG-I induced pathway [52]. They also suggested that TRADD functions downstream of MAVS but upstream of TRAF3 and TANK. PRISM [36], [37], [38] identified TRADD-TRAF3 interaction, which is very similar to TRAF2-TRADD crystal structure (PDB ID: 1F3V) [51] (Fig. 7 ). TRAF3-MAVS (PDB ID: 4GHU) [10], and TRAF3-TANK (PDB ID: 1KZZ) [34] complexes have resolved structures in which TANK and MAVS bind at exactly the same site on TRAF3 and according to our model this interface also overlaps the TRAF3-TRADD interaction (Fig. 4). Although TRADD is experimentally shown to bind to MAVS, TANK and TRAF3 [52], these interactions cannot take place simultaneously with a single TRAF3 molecule. TRAF3 functions as a trimer. It is possible that even though one TRAF3 molecule in the trimer is in contact with TRADD, others associate with MAVS and TANK.
Fig. 7.
TRADD binds to TRAF2 and TRAF3 in a similar fashion. (a) While TRAF2-TRADD complex structure is available on PDB (PDB ID: 1F3V) [51], (b) we modeled TRAF3-TRADD by PRISM [36], [37], [38]. Our structural model for TRADD-TRAF3 is very similar to TRADD-TRAF2 crystal structure.
4.4. TRAF3-DUBA interaction
Ubiquitination is one of the most common post-translational modifications in the TLR and NF-κB pathways [53], [54], [55]. Linkage of different Ub chain types has distinct consequences. While K48-linked Ub chain targets proteins for degradation, K63-linked Ub does not: instead it modifies the interaction modes by serving as an anchor [7]. Several Ub-dependent regulation mechanisms have evolved to modulate these signaling pathways and dampen the production of excess inflammatory cytokines and IFNs [56], [57]. Some regulators are ubiquitinases, like TRIAD3A, that add K48-linked chains and target the proteins for proteosomal degradation [53], [58], whereas others are deubiquitinating enzymes, such as CYLD (Cylindromatosis D), DUBA (Deubiquitynating enzyme A), and OTUB1 (Otubain 1) removing the K63-linked Ub-chains from proteins, thereby preventing interactions with downstream partners and turning off signaling [53], [55], [57]. TRAF6 is a substrate of CYLD [53], [59] and TRAF3 of DUBA [60] and OTUB1 [9], [57]. Our recent study provided structural insight into how TRAF6 and TRAF3 are negatively regulated by these deubiquitinases [55]. In addition to removing Ub-chains, these deubiquitinating enzymes also abolish the interactions of TRAF6 and TRAF3 with their partners due to overlapping binding site.
Fig. 4 shows the TRAF3-DUBA interaction model that we obtained by PRISM [36], [37], [38], in which the concave crevice on TRAF-C of TRAF3 is in contact with DUBA. Although the RING domain of TRAF3 was anticipated to interact with DUBA since there is less Ub removal in RING-domain deleted TRAF3 [60], PRISM finds interaction only with the TRAF-C region. If DUBA binds to both TRAF-C and the catalytic RING domain of TRAF3, it impedes TRAF3 signaling not only by removing the Ub chains, but also by preventing its interactions.
4.5. TRAF3-OTUB1 interaction
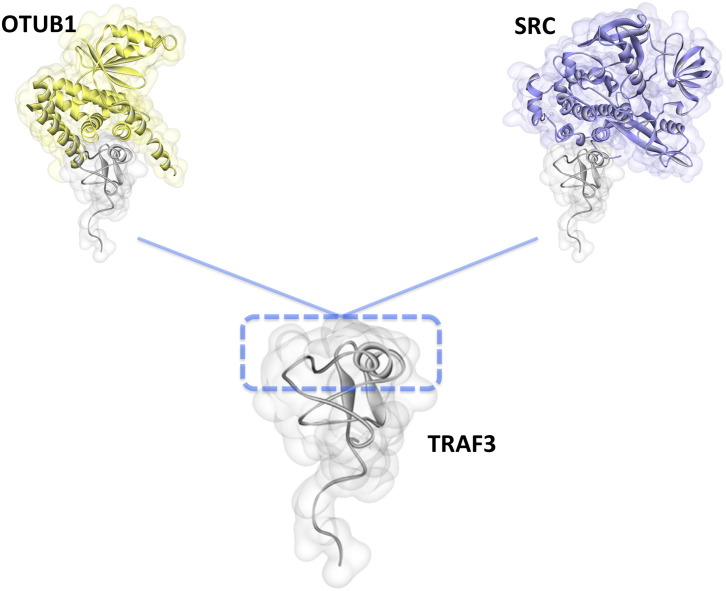
OTUB1 is also a debubiqutinase that targets TRAF3 after viral infection and attenuates virus-triggered IFN production [57]. We modeled this interaction and observed that the RING domain of TRAF3 can be in contact with OTUB1 (Fig. 8 ). Some proteins, like HSCARG facilitate the deubiquitination of TRAF3 by recruiting OTUB1 in order to negatively regulate IFN expression upon Sendai virus infection to prevent excess immune response, which could be devastating for the cell [9].
Fig. 8.
Modeled TRAF3 interactions through its RING domain. PRISM [36], [37], [38] predicted TRAF3 interactions with OTUB1 and SRC through its RING domain. These interactions have fully or partially overlapping interfaces, therefore they cannot co-exist. OTUB1 is a negative regulator of TRAF3-dependent IFN production [57]. However, SRC promotes TRAF3-dependent IFN production since its siRNA leads to decreased IFN production [61]. Competitive displacement of OTUB1 from TRAF3 may be the underlying cause for the increased IFN production in SRC recruitment to TRAF3, since TRAF3 signaling is not restricted to one negative regulator.
4.6. TRAF3-UCHL1 interaction
Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) is another deubiquitinase that cleaves the K63-linked poly-Ub chain from TRAF3 [23]. High risk human Papilloma virus (HPV) induces the overexpression of UCHL1 in keratinocytes. HPV is seen in 99% of cervical cancers and also in head and neck squamous cell carcinoma (HNSCC) [62], [63]. However, the exact molecular mechanism by which the virus promotes tumor development is not fully understood. HPV infection alone is not sufficient for malignant transformation; genetic impairments are also necessary [62]. HPV escapes from host immune surveillance by altering the ubiquitination state of TRAF3 which results in decrease in IFNs and cytokines [23]. From the viral standpoint this is a good strategy: it allows persistent virus infection without getting recognized by the host. Since TRAF3 is indispensable for antiviral immunity, its signaling is frequently targeted by proteins of viruses [24], [43], [64], [65], [66]. Table 2 lists viral proteins interacting with TRAF3. We modeled the TRAF3-UCHL1 interaction (Fig. 4 ). As observed with DUBA, UCHL1 also binds to the TRAF-C. Like DUBA, UCHL1 modifies the ubiqutination state and hinders the interactions of TRAF3. Thus, here rather than directly acting to mimic cellular interaction, HPV evolved a strategy that acts by promoting mimicry through UCHL1, a cellular protein that assists in viral survival. That is, HPV acts by manipulating a cellular messenger.
Table 2.
TRAF3 interactions with viral proteins and the cellular outcome observed.
| Viral proteins that disrupt TRAF3 interactions | Virus | Interactions abolished | Structure | Domain of TRAF3 | Cellular outcome | Reference |
|---|---|---|---|---|---|---|
| LMP1 | EBV | TRAF3-CD40 | 1zms.pdb | TRAF-C | Constitutive activation of NF-KB | [24] |
| vFLIP | Kaposi's sarcoma herpes virus | TRAF3-NIK | PRISM model | TRAF-C | Activation of NF-KB | [65] |
| ORF9b | SARS Coronavirus | TRAF3-MAVS | PRISM model | TRAF-C | Inhibition of IFN production | [66] |
| Plpro | SARS Coronavirus | TRAF3-MAVS | PRISM model | TRAF-C (our model) | Inhibition of IFN production | [64] |
| M | SARS Coronavirus | TRAF3-TBK1 | Unknown | Unknown | Inhibition of IFN production | [67] |
| Gn | NY-1 Hantavirus | TRAF3-TBK1 | Unknown | RING | Inhibition of IFN production | [68], [69] |
| Tio | Herpesvirus ateles | TRAF3-NIK | Unknown | TRAF-C | Constitutive activation of NF-KB | [43] |
Why are there redundant deubiquitinases that target TRAF3? The recruited deubiquitinase may be signaling pathway – e.g. RLR or TLR – specific. Alternatively, different viruses may employ distinct deubiquitinases to halt the IFN production. Spatial and temporal concentrations may also influence which enzyme is exploited.
4.7. TRAF3-PLpro interaction
Viruses can also evade the host immune system by exploiting their own proteins, besides inducing the expression of cellular proteins. Papain-like protease (PLpro) protein of SARS Coronavirus is also a deubiquitinase. It inhibits IFN production by abolishing the formation of the TRAF3-TBK1 complex [64]. We modeled its interaction with TRAF3 and found that PLpro binds to the crevice on TRAF-C (Fig. 4). As TBK1 associates with TRAF3 – not through direct physical contact, but through the Ub-chain that is attached to TRAF3 [9] – PLpro binding to TRAF-C does not compete with TBK1. There may be some allosteric events causing the abrogation of TBK1 interaction upon PLpro binding.
Gn protein (also known as G1) of NY-1 Hantavirus [68], [69] and M protein of SARS Coronavirus [67] also disrupts the formation of TRAF3-TBK1 complex and inhibits IFN production. Structures of these proteins are not available.
4.8. TRAF3-LMP1 interaction
The LMP1 protein is encoded by the Epstein-Barr virus (EBV, a human herpes virus) genome. It transforms B-lymphocytes to malignant cells by mimicking the constitutively active TNFR CD40 and serving as a CD40 decoy [24]. LMP1 enables NIK accumulation and activates the non-canonical NF-κB signaling in a ligand-independent manner [70], which helps the virus to replicate in the host [19] and promotes adenotonsillar B lymphocyte transformation and nasopharyngeal carcinoma [24], [71]. The structure of TRAF3-LMP1 is already resolved: LMP1 recognizes the same site on TRAF3 that CD40 does (Fig. 4), and makes additional H-bonds with TRAF3, which stabilize the complex and may allow LMP1 to outcompete CD40 [24].
4.9. TRAF3-vFLIP interaction
Like LMP1, other viral proteins such as vFLIP (viral FADD-like interleukin-1-b-converting enzyme (FLICE)/caspase-8-inhibitory protein) of Kaposi's sarcoma herpesvirus activate NF-κB to ensure their survival by binding to TRAF2 and TRAF3 [65]. vFLIP is the viral substitute of the cellular FLIP proteins that inhibit death receptor-mediated apoptosis. vFLIP has two DEDs. The second DED was suggested to interact with TRAF3 because it possesses the conserved TRAF-interacting motif (93PYQLT97). The structure of vFLIP of Kaposi's sarcoma herpesvirus has not yet been resolved. We modeled its structure by the I-Tasser server [72] and its interaction by PRISM [36], [37], [38]. In our model (Fig. 4), vFLIP binds to the β-sandwich of TRAF3 where DD of MyD88 binds, but the TRAF-interacting motif of vFLIP is not at the interface. NIK and vFLIP bind to overlapping surfaces on TRAF3. Therefore, vFLIP binding liberates NIK and allows its accumulation and resulting NF-κB activation.
4.10. TRAF3-SRC interaction
The proto-oncogene non-receptor tyrosine kinase c-Src plays a key role in antiviral TLR3 and RIG-I responses, and in type-I IFN production upon Sendai virus infection [61]. Knockdown of c-Src by siRNA resulted in decreased production of antiviral proteins [61]. TRAF3 is the common intermediate in TLR3 and RIG-I pathways. c-Src associates with the RING domain of TRAF3 and this interaction promotes the production of IFNs through RIG-I signaling [61]. The crystal structure of TRAF3-SRC is not yet available, but we have modeled it as shown in Fig. 8. It has overlapping interfaces with OTUB1. Competitive displacement of a negative regulator, OTUB1, from TRAF3 may be the reason for increased IFN production in Src recruitment to TRAF3.
4.11. TRAF3-MAVS interaction
Some receptors, like TNFRs physically associate with TRAF3, but some cytoplasmic receptors, like RIG-I and melanoma differentiation-associated protein 5 (MDA5) employ a bridging MAVS protein to recruit TRAF3. MAVS acts as a scaffolding protein to facilitate the assembly of large protein complexes. The structure of TRAF3- MAVS complex is available in the PDB (PDB ID: 4GHU) [10]. In this structure (Fig. 4), MAVS binds to the same binding pocket in the β-sandwich of TRAF3.
4.12. TRAF3-ORF9b interaction
ORF-9b of SARS coronavirus localizes to the mitochondria. It interacts with MAVS and triggers degradation of MAVS, TRAF3 and TRAF6 [66], thereby profoundly constraining IFN production. Its crystal structure is available [73]. We built its complex structure with TRAF3 (Fig. 4). TRAF3-ORF9b interaction is mutually exclusive with TRAF3-MAVS interaction.
5. Conclusions
TRAF3 is a critical control switch that negatively regulates the activation of the canonical and non-canonical NF-κB pathways; it is also a key protein in antiviral immunity. It is no wonder that it is targeted by pathogens in multiple ways. Here we took steps toward unraveling its mechanism of action in the cell and under viral onslaught on the molecular level. Exploiting available crystal structures and supplementing them by modeling, our structural analysis observes that TRAF3 binds many proteins at the same or partially overlapping binding sites. The signaling output can then be determined by the specific temporal interaction – which results in pro- or anti-proliferative cell output. TRAF3 selection of a specific partner among the many interacting at the same site is influenced by the cell state and many factors are at play.
Grasping function in the complex cellular milieu and relating the atomic-scale conformational behavior of molecules to the regulation of the protein in the cell is a formidable aim [74]. At the fundamental level, proteins (and RNA and DNA) exist as ensembles of conformational states and work through dynamic shifts of their conformational distributions. At the cellular level, it presents a challenge because of the intricacy of the cellular network and the heterogeneity of the regulatory mechanisms. Living organisms evolved a winning strategy: merging the fundamental with the cellular needs through ‘evolvability’. Evolvability strives to bridge the nuanced functional spectrum based on principles of physics and evolution. Evolvability optimizes and adapts available molecular cellular building blocks and molds them for enhanced and more robust cellular function.
Viral molecular mimicry is a common way of inhibiting (or activating) the host signaling pathways [19], [25]. Taken together, competitive binding and the evolvability of adaptive viral molecular mimicry appear key players in cell function and its hijacking by pathogens. We believe that one way to grasp the interplay between pathogen and commensal microbiota and the human host is to fuse the respective networks, including structural data; structural data is the basis for in-depth mechanistic understanding.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Transparency document
Transparency document.
Acknowledgements
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E, and NIDCD intramural projects ZIA-DC-000016, 73 and 74. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. O.K. and A.G. acknowledge Science Academy (of Turkey).
Footnotes
This article is part of a Special Issue entitled “System Genetics” Guest Editor: Dr. Yudong Cai and Dr. Tao Huang.
The Transparency document associated with this article can be found, in online version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2016.05.021.
Contributor Information
Emine Guven-Maiorov, Email: emine.guven-maiorov@nih.gov.
Ozlem Keskin, Email: okeskin@ku.edu.tr.
Attila Gursoy, Email: agursoy@ku.edu.tr.
Carter VanWaes, Email: vanwaesc@nidcd.nih.gov.
Zhong Chen, Email: chenz@nidcd.nih.gov.
Chung-Jung Tsai, Email: tsaic@mail.nih.gov.
Ruth Nussinov, Email: nussinor@helix.nih.gov.
Appendix A. Supplementary data
Supplementary material
References
- 1.Tsai C.J., Ma B., Nussinov R. Protein-protein interaction networks: how can a hub protein bind so many different partners? Trends Biochem. Sci. 2009;34:594–600. doi: 10.1016/j.tibs.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai C.J., Lin S.L., Wolfson H.J., Nussinov R. Protein-protein interfaces: architectures and interactions in protein-protein interfaces and in protein cores. Their similarities and differences. Crit. Rev. Biochem. Mol. Biol. 1996;31:127–152. doi: 10.3109/10409239609106582. [DOI] [PubMed] [Google Scholar]
- 3.Keskin O., Ma B., Rogale K., Gunasekaran K., Nussinov R. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up systems biology approach. Phys. Biol. 2005;2:S24–S35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- 4.Snider J., Kotlyar M., Saraon P., Yao Z., Jurisica I., Stagljar I. Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 2015;11:848. doi: 10.15252/msb.20156351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiva E., Babbitt P.C. Evolutionary reprograming of protein-protein interaction specificity. Cell. 2015;163:535–537. doi: 10.1016/j.cell.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Keskin O., Nussinov R. Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure. 2007;15:341–354. doi: 10.1016/j.str.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Hacker H., Tseng P.H., Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 8.Nussinov R., Ma B., Tsai C.J. A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochim. Biophys. Acta. 2013;1834:820–829. doi: 10.1016/j.bbapap.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Peng Y., Xu R., Zheng X. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P., Reichardt A., Liang H., Aliyari R., Cheng D., Wang Y., Xu F., Cheng G., Liu Y. Single amino acid substitutions confer the antiviral activity of the TRAF3 adaptor protein onto TRAF5. Sci. Signal. 2012;5:ra81. doi: 10.1126/scisignal.2003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider K., Benedict C.A., Ware C.F. A TRAFfic cop for host defense. Nat. Immunol. 2006;7:15–16. doi: 10.1038/ni0106-15. [DOI] [PubMed] [Google Scholar]
- 12.Guven-Maiorov E., Keskin O., Gursoy A., VanWaes C., Chen Z., Tsai C.J., Nussinov R. Scientific Reports. 2015. The architecture of the TIR domain signalosome in the Toll-like Receptor-4 signaling pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun S.C. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha S.K., Pietras E.M., He J.Q., Kang J.R., Liu S.Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T.L., Wang Y., Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25:3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalani A.I., Luo C., Han Y., Xie P. TRAF3: a novel tumor suppressor gene in macrophages. Macrophage (Houst) 2015;2 doi: 10.14800/macrophage.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zapata J.M., Llobet D., Krajewska M., Lefebvre S., Kress C.L., Reed J.C. Lymphocyte-specific TRAF3 transgenic mice have enhanced humoral responses and develop plasmacytosis, autoimmunity, inflammation, and cancer. Blood. 2009;113:4595–4603. doi: 10.1182/blood-2008-07-165456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keats J.J., Fonseca R., Chesi M., Schop R., Baker A., Chng W.J., Van Wier S., Tiedemann R., Shi C.X., Sebag M., Braggio E., Henry T., Zhu Y.X., Fogle H., Price-Troska T., Ahmann G., Mancini C., Brents L.A., Kumar S., Greipp P., Dispenzieri A., Bryant B., Mulligan G., Bruhn L., Barrett M., Valdez R., Trent J., Stewart A.K., Carpten J., Bergsagel P.L. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annunziata C.M., Davis R.E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., Dave S., Hurt E.M., Tan B., Zhao H., Stephens O., Santra M., Williams D.R., Dang L., Barlogie B., Shaughnessy J.D., Jr., Kuehl W.M., Staudt L.M. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 20.Standfuss J. Structural biology. Viral chemokine mimicry. Science. 2015;347:1071–1072. doi: 10.1126/science.aaa7998. [DOI] [PubMed] [Google Scholar]
- 21.Franzosa E.A., Garamszegi S., Xia Y. Toward a three-dimensional view of protein networks between species. Front. Microbiol. 2012;3:428. doi: 10.3389/fmicb.2012.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzosa E.A., Xia Y. Structural principles within the human-virus protein-protein interaction network. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10538–10543. doi: 10.1073/pnas.1101440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karim R., Tummers B., Meyers C., Biryukov J.L., Alam S., Backendorf C., Jha V., Offringa R., van Ommen G.J., Melief C.J., Guardavaccaro D., Boer J.M., van der Burg S.H. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S., Xie P., Welsh K., Li C., Ni C.Z., Zhu X., Reed J.C., Satterthwait A.C., Bishop G.A., Ely K.R. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J. Biol. Chem. 2005;280:33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- 25.Stebbins C.E., Galan J.E. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 26.Christen U., Hintermann E., Holdener M., von Herrath M.G. Viral triggers for autoimmunity: is the ‘glass of molecular mimicry’ half full or half empty? J. Autoimmun. 2010;34:38–44. doi: 10.1016/j.jaut.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada K., Gherasim C., Banerjee R., Koutmos M. Structure of human B12 trafficking protein CblD reveals molecular mimicry and identifies a new subfamily of nitro-FMN reductases. J. Biol. Chem. 2015;290:29155–29166. doi: 10.1074/jbc.M115.682435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei G., Xi W., Nussinov R., Ma B. Protein ensembles: how does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem. Rev. 2016 doi: 10.1021/acs.chemrev.5b00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nussinov R., Wolynes P.G. A second molecular biology revolution? The energy landscapes of biomolecular function. Phys. Chem. Chem. Phys. 2014;16:6321–6322. doi: 10.1039/c4cp90027h. [DOI] [PubMed] [Google Scholar]
- 30.Davey N.E., Trave G., Gibson T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Finlay B.B., McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 32.Li C., Norris P.S., Ni C.Z., Havert M.L., Chiong E.M., Tran B.R., Cabezas E., Reed J.C., Satterthwait A.C., Ware C.F., Ely K.R. Structurally distinct recognition motifs in lymphotoxin-beta receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling. J. Biol. Chem. 2003;278:50523–50529. doi: 10.1074/jbc.M309381200. [DOI] [PubMed] [Google Scholar]
- 33.Ni C.Z., Welsh K., Leo E., Chiou C.K., Wu H., Reed J.C., Ely K.R. Molecular basis for CD40 signaling mediated by TRAF3. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10395–10399. doi: 10.1073/pnas.97.19.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., Ni C.Z., Havert M.L., Cabezas E., He J., Kaiser D., Reed J.C., Satterthwait A.C., Cheng G., Ely K.R. Downstream regulator TANK binds to the CD40 recognition site on TRAF3. Structure. 2002;10:403–411. doi: 10.1016/s0969-2126(02)00733-5. [DOI] [PubMed] [Google Scholar]
- 35.Ni C.Z., Oganesyan G., Welsh K., Zhu X., Reed J.C., Satterthwait A.C., Cheng G., Ely K.R. Key molecular contacts promote recognition of the BAFF receptor by TNF receptor-associated factor 3: implications for intracellular signaling regulation. J. Immunol. 2004;173:7394–7400. doi: 10.4049/jimmunol.173.12.7394. [DOI] [PubMed] [Google Scholar]
- 36.Baspinar A., Cukuroglu E., Nussinov R., Keskin O., Gursoy A. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285–W289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuncbag N., Gursoy A., Nussinov R., Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat. Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogmen U., Keskin O., Aytuna A.S., Nussinov R., Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muratcioglu S., Guven-Maiorov E., Keskin O., Gursoy A. Advances in template-based protein docking by utilizing interfaces towards completing structural interactome. Curr. Opin. Struct. Biol. 2015;35:87–92. doi: 10.1016/j.sbi.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Sun S.C. Controlling the fate of NIK: a central stage in noncanonical NF-kappaB signaling. Sci. Signal. 2010;3:e18. doi: 10.1126/scisignal.3123pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanjo H., Zajonc D.M., Braden R., Norris P.S., Ware C.F. Allosteric regulation of the ubiquitin:NIK and ubiquitin:TRAF3 E3 ligases by the lymphotoxin-beta receptor. J. Biol. Chem. 2010;285:17148–17155. doi: 10.1074/jbc.M110.105874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao G., Zhang M., Harhaj E.W., Sun S.C. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 43.de Jong S.J., Albrecht J.C., Giehler F., Kieser A., Sticht H., Biesinger B. Noncanonical NF-kappaB activation by the oncoprotein Tio occurs through a nonconserved TRAF3-binding motif. Sci. Signal. 2013;6:ra27. doi: 10.1126/scisignal.2003309. [DOI] [PubMed] [Google Scholar]
- 44.Naude P.J., den Boer J.A., Luiten P.G., Eisel U.L. Tumor necrosis factor receptor cross-talk. FEBS J. 2011;278:888–898. doi: 10.1111/j.1742-4658.2011.08017.x. [DOI] [PubMed] [Google Scholar]
- 45.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin Q., Lamothe B., Darnay B.G., Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vince J.E., Pantaki D., Feltham R., Mace P.D., Cordier S.M., Schmukle A.C., Davidson A.J., Callus B.A., Wong W.W., Gentle I.E., Carter H., Lee E.F., Walczak H., Day C.L., Vaux D.L., Silke J. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (tnf) to efficiently activate nf-{kappa}b and to prevent tnf-induced apoptosis. J. Biol. Chem. 2009;284:35906–35915. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zarnegar B.J., Wang Y., Mahoney D.J., Dempsey P.W., Cheung H.H., He J., Shiba T., Yang X., Yeh W.C., Mak T.W., Korneluk R.G., Cheng G. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P.H., Keats J.J., Wang H., Vignali D.A., Bergsagel P.L., Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao A.P., Li S., Zhong B., Li Y., Yan J., Li Q., Teng C., Shu H.B. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-beta) and cellular antiviral response. J. Biol. Chem. 2010;285:9470–9476. doi: 10.1074/jbc.M109.071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park Y.C., Ye H., Hsia C., Segal D., Rich R.L., Liou H.C., Myszka D.G., Wu H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell. 2000;101:777–787. doi: 10.1016/s0092-8674(00)80889-2. [DOI] [PubMed] [Google Scholar]
- 52.Michallet M.C., Meylan E., Ermolaeva M.A., Vazquez J., Rebsamen M., Curran J., Poeck H., Bscheider M., Hartmann G., Konig M., Kalinke U., Pasparakis M., Tschopp J. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–661. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Bibeau-Poirier A., Servant M.J. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine. 2008;43:359–367. doi: 10.1016/j.cyto.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Liu S., Chen Z.J. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guven-Maiorov E., Keskin O., Gursoy A., Nussinov R. A structural view of negative regulation of the toll-like receptor-mediated inflammatory pathway. Biophys J. 2015 doi: 10.1016/j.bpj.2015.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wullaert A., Heyninck K., Janssens S., Beyaert R. Ubiquitin: tool and target for intracellular NF-kappaB inhibitors. Trends Immunol. 2006;27:533–540. doi: 10.1016/j.it.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Li S., Zheng H., Mao A.P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., Shu H.B. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 2010;285:4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida H., Jono H., Kai H., Li J.D. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J. Biol. Chem. 2005;280:41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 60.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K.M., Eby M., Pietras E., Cheng G., Bazan J.F., Zhang Z., Arnott D., Dixit V.M. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 61.Johnsen I.B., Nguyen T.T., Bergstroem B., Fitzgerald K.A., Anthonsen M.W. The tyrosine kinase c-Src enhances RIG-I (retinoic acid-inducible gene I)-elicited antiviral signaling. J. Biol. Chem. 2009;284:19122–19131. doi: 10.1074/jbc.M808233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 63.Tummers B., Burg S.H. High-risk human papillomavirus targets crossroads in immune signaling. Viruses. 2015;7:2485–2506. doi: 10.3390/v7052485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guasparri I., Wu H., Cesarman E. The KSHV oncoprotein vFLIP contains a TRAF-interacting motif and requires TRAF2 and TRAF3 for signalling. EMBO Rep. 2006;7:114–119. doi: 10.1038/sj.embor.7400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siu K.L., Kok K.H., Ng M.H., Poon V.K., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alff P.J., Sen N., Gorbunova E., Gavrilovskaya I.N., Mackow E.R. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J. Virol. 2008;82:9115–9122. doi: 10.1128/JVI.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muyangwa M., Martynova E.V., Khaiboullina S.F., Morzunov S.P., Rizvanov A.A. Hantaviral proteins: structure, functions, and role in hantavirus infection. Front. Microbiol. 2015;6:1326. doi: 10.3389/fmicb.2015.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Repic A.M., Shi M., Scott R.S., Sixbey J.W. Augmented latent membrane protein 1 expression from Epstein-Barr virus episomes with minimal terminal repeats. J. Virol. 2010;84:2236–2244. doi: 10.1128/JVI.01972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung G.T., Lou W.P., Chow C., K.F. To, Choy K.W., Leung A.W., Tong C.Y., Yuen J.W., Ko C.W., Yip T.T., Busson P., Lo K.W. Constitutive activation of distinct NF-kappaB signals in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2013;231:311–322. doi: 10.1002/path.4239. [DOI] [PubMed] [Google Scholar]
- 72.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nussinov R., Tsai C.J. Free energy diagrams for protein function. Chem. Biol. 2014;21:311–318. doi: 10.1016/j.chembiol.2013.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.
Supplementary material