Abstract
Nucleic acid aptamers are single-stranded DNA or RNA molecules identified to recognize with high affinity specific targets including proteins, small molecules, ions, whole cells and even entire organisms, such as viruses or bacteria. They can be identified from combinatorial libraries of DNA or RNA oligonucleotides by SELEX technology, an in vitro iterative selection procedure consisting of binding (capture), partitioning and amplification steps. Remarkably, many of the aptamers selected against biologically relevant protein targets are G-rich sequences that can fold into stable G-quadruplex (G4) structures.
Aiming at disseminating novel inspiring ideas within the scientific community in the field of G4-structures, the emphasis of this review is placed on: 1) recent advancements in SELEX technology for the efficient and rapid identification of new candidate aptamers (introduction of microfluidic systems and next generation sequencing); 2) recurrence of G4 structures in aptamers selected by SELEX against biologically relevant protein targets; 3) discovery of several G4-forming motifs in important regulatory regions of the human or viral genome bound by endogenous proteins, which per se can result into potential aptamers; 4) an updated overview of G4-based aptamers with therapeutic potential and 5) a discussion on the most attractive G4-based aptamers for diagnostic applications. This article is part of a Special Issue entitled "G-quadruplex" Guest Editor: Dr. Concetta Giancola and Dr. Daniela Montesarchio.
Abbreviations: ABTS, 2,2′-azino-bis(3-ethylbenzothiozoline)-6-sulfonic acid; AGR2, Anterior Gradient Homolog 2; ALP, Alkaline Phosphatase; AMC, 7-amino-4-methylcoumarin; AuNP, Gold Nanoparticle; CND, Carbon Nanodot; ECL, Electrochemiluminescence; FAM, Carboxyfluorescein; G4, G-quadruplex; GCE, Glassy Carbon Electrode; GO, Graphene Oxide; GOx, Glucose Oxidase; HIV, Human Immunodeficiency Virus; KCE, Kinetic Capillary Electrophoresis; NECEEM, Non-equilibrium Capillary Electrophoresis of Equilibrium Mixtures; ON, Oligonucleotide; PDDA, Poly(diallyldimethylammonium) chloride; PET, Phosphorescence Energy Transfer; PPK, Polyphosphate Kinase; QCM, Quartz Crystal Microbalance; QDs, Quantum Dots; RT, Reverse Transcriptase; SA, Streptavidin; SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; SELEX, Systematic Evolution of Ligands by EXponential enrichment; SOMAmers, Slow Off-rate Modified Aptamers; SPR, Surface Plasmon Resonance; TBA, Thrombin Binding Aptamer; TBDPS, tert-butyldiphenylsilyl; TEL, Tetra-end-linker; TMB, 3,3′,5,5′-tetramethylbenzidine; TMPG, 3,4,5-trimethoxylphenylglyoxal; TMPyP4, 5,10,15,20-tetrakis(1-methyl-4-pyridinio)porphyrin; TPA, tetra-n-propylammonium hydroxide; VEGF, Vascular Endothelial Growth Factor
Keywords: Aptamer, G-quadruplex, Protein target, Therapy, Diagnostics, SELEX
Highlights
-
•
Oligonucleotide aptamers and methods for their selection against specific targets
-
•
The recurrence of G-quadruplex (G4) motifs in oligonucleotide aptamers
-
•
G4-forming aptamers in therapeutic applications as drugs and drug delivery systems
-
•
G4-forming aptamers in detection and diagnostic applications
1. Description and selection of nucleic acid aptamers
Nucleic acid aptamers are single-stranded DNA or RNA molecules, generally 25 to 60 bases in length, identified from pools of random-sequence oligonucleotides (ONs) by an in vitro selection process called SELEX (Systematic Evolution of Ligands by EXponential enrichment) [1], introduced in the early 90s independently by the research groups of Gold and Szostak [2], [3], [4]. Thanks to their unique three-dimensional folding, aptamers can recognize a wide range of molecular targets including proteins, small molecules, ions, whole cells and even entire organisms, such as viruses or bacteria, with affinities, expressed in terms of dissociation constants (K d), ranging from picomolar (pM) to micromolar (μM) [5], [6], [7]. Each ON aptamer can adopt a unique subset of 3D structures defined by the combination of base-pairing, π-π stacking, sugar puckering, as well as electrostatic, hydrogen bonding and non-canonical intra-molecular interactions. This structural complexity results in a high probability of selecting an aptamer that can strongly and specifically interact with the target of interest through hydrogen bonds, salt bridges, van der Waals, hydrophobic and/or electrostatic interactions. The high affinity and selectivity for the target, as well as the multiplicity of possible target types, make aptamers a valid alternative to antibodies in a wide variety of applications, from therapeutics (as drugs or drug-delivery systems) to diagnostic and sensing devices [8], [9], [10], [11], [12], [13], [14], [15]. Nucleic acid aptamers exhibit significant advantages over antibodies, such as smaller size, lower immunogenicity, remarkable stability in a wide range of pH (≈ 4–9) and temperature; in addition, aptamers allow an easy control of their folding processes, which, even after prolonged denaturation, are typically reversible upon restoring the native conditions. In addition, once selected, oligonucleotide-based aptamers are easily obtained in animal-free systems by chemical synthesis. This allows also easily performing ad hoc tailored chemical modifications that can increase aptamer chemical and biological stability and bring the production costs well below those of antibodies [16], [17].
1.1. SELEX technology
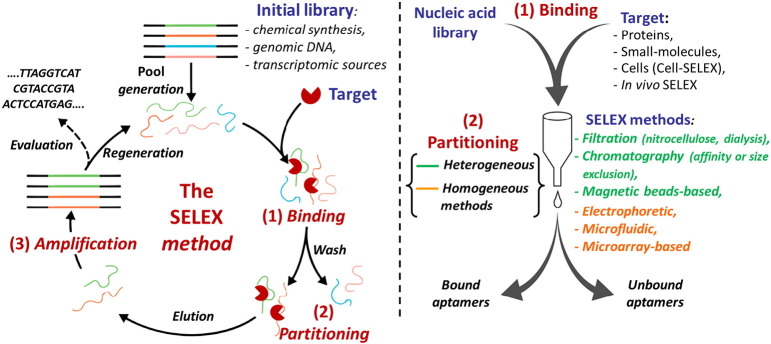
As mentioned before, aptamer development generally exploits the SELEX methodology which involves an in vitro iterative selection procedure consisting of three main steps: (i) binding (capture), where a random ON library is incubated with the desired target molecule under defined conditions; (ii) partitioning, in which the target-bound ON aptamers are separated from unbound ones; and (iii) amplification, where the enriched pool of selected aptamers is amplified before being used in the successive round of selection. During these processes, the best aptamers for the selected target are enriched and the initial ON library complexity is decreased (Fig. 1 , left).
Fig. 1.
Schematic representation of the SELEX process.
The starting pool, whose appropriate choice represents a crucial point for the success of the SELEX method, can contain 1012–1016 DNA or RNA single strands, each with an internal randomized sequence of usually 30–80 nucleotides and terminal fixed sequences, complementary to pre-defined primers used in the amplification step. Aptamer libraries can be derived from chemical synthesis, genomic DNA [18] or transcriptomic sources [19], and can be realized with deoxyribo-, ribo-, or modified nucleotides [20], [21], [22], [23]. Modified nucleotides are primarily used to increase the chemical and enzymatic stability of aptamers, but can also contribute to greatly broaden their chemical diversity, improving their binding affinity and specificity for a selected target. Among the methods developed for expanding the repertoire of nucleobase-modified aptamers, recently Mayer et al. introduced click chemistry protocols to generate a novel class of modified aptamers, termed “clickmers”, fully compatible with the common steps of established in vitro selection procedures (click-SELEX) [23]. Many enzymes that can “read” or “write” aptamer sequences containing unnatural nucleotides are available [20], [21], [22], [23], [24], [25], [26] and methods that identify the precise location of these nucleotides within an aptamer sequence are known as well [27], [28], [29].
Valuable examples of modified nucleic acids used for aptamer production are: 1) locked nucleic acids (LNAs) [16], [30]; 2) ONs with sugar backbones containing 2′-fluoro or 2′-O-methyl-ribonucleotides [16], [17], [31]; 3) spiegelmers (from the German word spiegel, “mirror”), high-affinity RNA-like ligands composed of l-ribose units, highly resistant to nuclease degradation due to their chirally-inverted backbone (developed by NOXXON Pharma; some spiegelmers are currently in advanced clinical trials) [32]; and 4) SOMAmers (Slow Off-rate Modified Aptamers, developed by SomaLogic, Inc.) which are single-stranded DNA aptamers bearing 5-methyl-dC (MedCTP) and base-modified dU (5-benzylaminocarbonyl-dU, BndU; 5-naphthylmethylaminocarbonyl-dU, NapdU; 5-tryptaminocarbonyl-dU, TrpdU; and 5-isobutylaminocarbonyl-dU, iBudU) residues, which produce aptamers with significantly increased chemical diversity, allowing higher chances of selective interactions with proteins. The development of SOMAmers constituted a highly multiplexed proteomic platform used for simultaneous identification and quantification of target proteins in complex biological samples (biomarker discovery) [20], [33].
After incubation of the library with the selected target, partition of bound from unbound sequences can be obtained in different ways (Fig. 1, right side). Typically, heterogeneous methods have been applied for this purpose: (i) filtration, based on differential retention of protein–DNA complexes and free DNA on a membrane; (ii) affinity chromatography, in which the protein is immobilized on a surface and retains oligonucleotides on the basis of their binding affinity, while the unbound sequences are discarded; or (iii) magnetic beads-based separation, in which a magnetic separator is employed for partitioning the unbound ONs from the beads-bound target-aptamer complex [34]. These surface-based techniques suffer from non-specific adsorption of free ONs to the surface of the matrix, reducing the efficiency of partitioning and leading to the necessity of multiple rounds as well as negative selection steps. Furthermore, the immobilization process used in heterogeneous partitioning techniques can negatively affect protein structure, so potentially reducing the biological relevance of the selected oligonucleotide [34]. Thus, homogeneous methods are highly desirable for the selection of aptamers for protein targets, especially in case of aptamers developed for therapeutic applications. As an alternative to surface-based methods, kinetic capillary electrophoresis (KCE) techniques have been described as very efficient systems in aptamer selection. These are based on the separation of the aptamer–target complexes from unbound ONs according to their different electrophoretic mobility in an electric field [35]. In particular, non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) is the most widely used KCE method for the highly efficient selection of aptamers, allowing also to accurately calculate the binding parameters (K d, k on and k off) of the aptamer–target interaction [36], [37]. The efficiency of partitioning by NECEEM is at least 2 orders of magnitude higher than those of conventional surface-based selection techniques [36]. As a result, library enrichment can be completed in as few as 2–4 rounds of NECEEM-based SELEX, in contrast to the 10–30 rounds typically required in SELEX based on heterogeneous partitioning. Ideally, in an electric field, the zones of free DNA, DNA–target complex, and free target are separated. The lower the K d value, the more time the aptamers spend within the target, and the closer is their migration time to that of the free target. In contrast, the higher the K d value, the more time the aptamers spend as free DNA molecules, and the more similar is their migration time with respect to that of free DNA [37]. The mobility of DNA–protein complexes is typically intermediate between those of the DNA and protein alone, and thus the whole electropherogram displays the various components in the following order: (1) free protein; (2) specific DNA–protein complex (with low K d values); (3) non-specific DNA–protein complex (with high K d values); and (4) free DNA. A fraction collection approach in capillary electrophoresis SELEX (FCE-SELEX) allows partition and collection of the aptamer–target complexes from the original DNA library, as illustrated in Fig. 2 , ensuring no aptamer is missing [38].
Fig. 2.
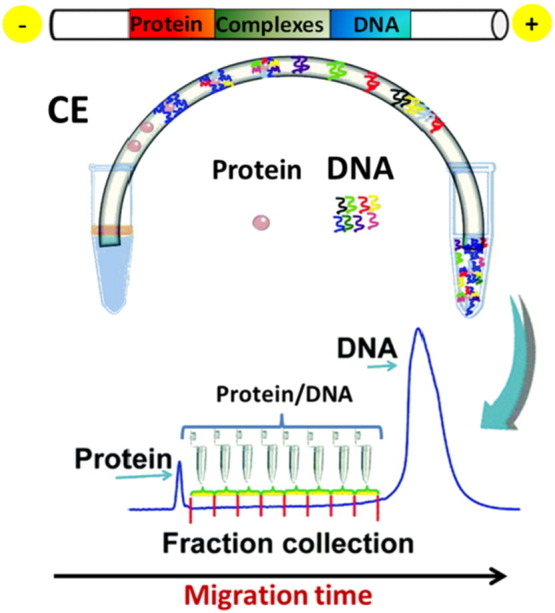
Illustration of FCE-SELEX selection for aptamers.
(Adapted from ref [38]. by permission of The Royal Society of Chemistry)
Following partitioning, the target-bound oligonucleotides are eluted from the target and amplified by PCR (DNA SELEX) or reverse transcription PCR (RNA SELEX) to give an enriched pool of selected oligonucleotides, which is used in the successive SELEX round. With increasingly stringent conditions (shorter incubation times, higher temperatures, higher ON/target ratios) in successive rounds of SELEX, the high-affinity oligonucleotides are enriched. For a number of years, the classical PCR amplification protocol has been used. However, the conventional PCR amplification method was found to quickly degrade the products and rapidly form by-products through the formation of heterodimers [39]. Additionally, DNA amplification by conventional PCR is susceptible of nonspecific primer hybridization, primer dimers, amplification bias [40], and potential aptamer loss due to their adhesion on the walls of PCR tubes [41]. A number of approaches have been proposed over the years to mend the aforementioned inefficiencies of PCR amplification in SELEX [34]. More recently, a SELEX method which combines the NECEEM partitioning with a highly efficient amplification procedure, termed Emulsion PCR (ePCR), has been described [42]. ePCR allows compartmentalizing and miniaturizing the PCR reaction in vitro, by vigorously mixing mineral oil and surfactants with a conventional PCR mix, creating a local homogeneous-like PCR amplification environment. These conditions reduce primer-dimer formation, nonspecific primer hybridization and amplification bias, i.e. the major causes of PCR inefficiencies in early amplification cycles [39], [40], [41], allowing increasing the product yield due to the close proximity in micelles of the template DNA, the primers and polymerase, each in high local concentration.
Historically, enriched aptamer pools from the SELEX experiments are cloned into a plasmid and a few hundred individual clones, at most, have to be sequenced to identify high affinity aptamers. However, the recent application of next-generation sequencing (NGS) combined with bioinformatic analysis of the growing aptamer populations is a powerful improvement in the SELEX procedure [43]. The analysis of evolving aptamer populations during SELEX, from which many sequence families can be obtained, has been approached with proper algorithms to search for structural motifs that might be critical for the aptamer–target interaction [44], [45], [46].
Next steps for obtaining functional aptamers are structure- and sequence-guided molecular engineering that include truncations and/or mutations of the sequence(s) to develop the shortest aptamer(s) with the highest affinity and specificity [1]. Once identified, the putative aptamers can be synthesized and analyzed to determine the target affinity and specificity of binding vs. other possible targets. Structural characterization is important for optimizing the aptamer features after its selection from a pool. Then, computational analysis can predict the nucleotide bases important for ligand binding and biophysical studies by NMR or X-ray crystallography can identify the molecular details of the aptamer–target interaction. These biophysical analyses, by far the most definitive and informative techniques for resolving tertiary structures, are however limited to buffer conditions compatible with data collection. The interaction sites between an aptamer and its target can be established by biochemical assays such as nuclease protection [47] and UV-cross-linking assays [48], performed on the selected ON in the presence or absence of the target [49]. Information on the binding pocket and the bases critical for target contacts are essential for aptamer optimization. This provides a map that guides the insertion of modifications to retain or increase the binding affinity while altering aptamer properties such as size or structural stability.
Another important consideration for aptamer development is that the target used during the selection process should be identical to the “natural” one, i.e., in the case of a protein target, for example, very similar to the protein found in vivo. This requirement is frequently fulfilled using, as SELEX targets, proteins that might only be modified with a short tag. It is often more difficult to ensure identity when the “natural target” is a membrane protein or a small molecule that has to be modified to allow its immobilization for the SELEX capture protocols. Indeed, isolated membrane proteins used for SELEX may lack the normal glycosylation pattern or the association with natural partners such as other proteins, lipids or carbohydrates, that influence/modulate their structure, the availability of surface docking sites and the affinity for aptamers preventing the access to the natural target [50]. To address some of these problems, selection techniques like cell-SELEX and tissue-SELEX have been developed [51], [52], [53], [54]. These methods provide an effective approach for the identification of new biomarkers as disease signals in diagnosis and therapy. However, the cell-SELEX system provides low aptamer enrichment efficiency because many off-target surface biomarkers are co-expressed on the cells of interest [55]. To overcome this challenge, a modified SELEX method that combines the cell-based SELEX with purified protein-based SELEX techniques was introduced (hybrid SELEX) [55], [56]. In addition to the hybrid-SELEX approach, other modified cell-SELEX technologies have been developed, such as internalized cell-SELEX, designed to select functional aptamers that can be internalized by human cells [55], [57], [58], [59], [60] and FACS-SELEX, that is used to eliminate dead cells that non-specifically bind nucleic acids and affect subsequent aptamer selection results [55], [61].
Despite the advances and huge body of literature documenting the success of aptamers, their commercial application remains relatively undeveloped, probably because of the overwhelming antibody-based market and a certain degree of hesitation to move to a new type of products. However, with the many advances made to improve the efficiency of selection and to increase the affinity, specificity, and bioavailability of aptamers, the future of therapeutic aptamers looks extremely promising. To date, Macugen (pegaptanib sodium), selected against the vascular endothelial growth factor (VEGF), is the only existing aptamer-based drug, approved by the US Food and Drug Administration for the treatment of neovascular age-related macular degeneration (AMD) in humans, also undergoing clinical trials for the treatment of diabetic macular oedema [62]. Nevertheless, new aptamer-based candidate drugs are undertaking clinical trials [9], [32], and this gives us hope that increasingly more aptamers and aptamer-like compounds will be approved as drugs in the next future.
2. G-quadruplex (G4) motifs in oligonucleotide aptamers
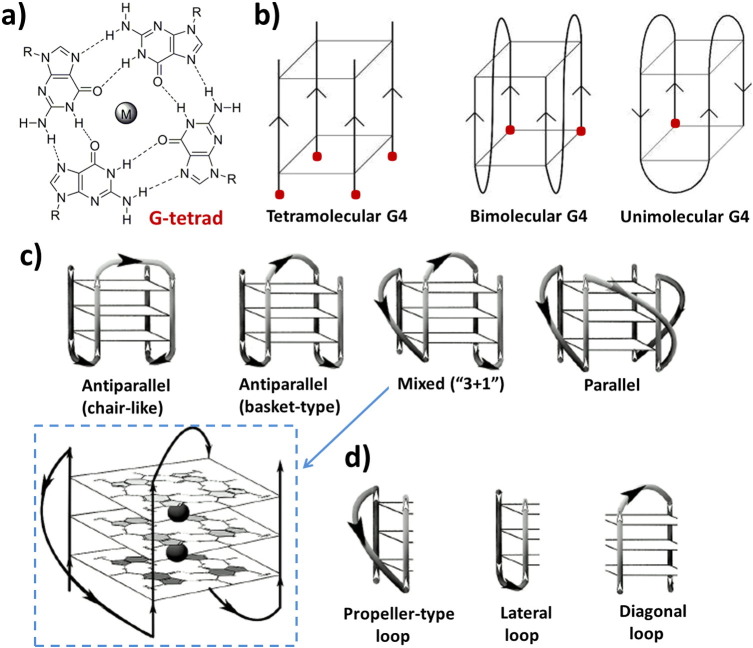
Among the combinatorially selected aptamers endowed with significant bioactivity, many are G-rich oligomers sharing a common structural feature, i.e. the ability to fold into stable G-quadruplex (G4) conformations under physiological conditions and recognize very different protein targets [63], [64]. This apparent contrast can be easily solved considering the extremely high polymorphism of G-quadruplex structures. In fact G-quadruplexes are non-canonical nucleic acid conformations adopted by guanine-rich oligonucleotides. The core G-quadruplex unit is the guanine tetrad, a cyclic planar arrangement of four guanines linked through Hoogsteen-type hydrogen bonds. Stacking of two or more guanine tetrads generates a G-quadruplex motif, which is further stabilized by cations hosted in the central cavity of the G4 [65], [66] (Fig. 3 ).
Fig. 3.
Schematic representations of: a) a G-tetrad stabilized by a metal ion in the central cavity; b) some possible topologies for simple tetramolecular, bimolecular and unimolecular G-quadruplexes (strand polarities are indicated by arrows); c) examples of unimolecular G4s that differ in strand orientation within the G4 core; and d) types of loops that join G-rich tracts in the G4 structure.
The discovery of G4-forming aptamers demonstrated that unusual DNA structures with even very similar sequences can result into a large variety of different conformations, thus embodying sufficient elements for the recognition of a wide range of unrelated protein systems, and accounting for the diversity of biological effects produced. The apparently rigid G4 structure is instead rather plastic and can fit into widely different nucleic acid architectures. Multiple elements participate in conferring G4 structures a high level of adaptability: 1) the intramolecular or intermolecular character of the system; 2) the parallel or antiparallel direction of the guanine-rich tracts in the G4 assembly; 3) the length, sequence and direction of the loops; 4) the syn or anti guanine orientation around the N-glycosidic bonds; and 5) the number of consecutive G-quartets formed. The body of evidence thus far gained indicates that intramolecular G4 conformations are generally favoured, with loops consisting of 2–4 bases, often adopting an antiparallel arrangement with alternated syn/anti orientations. The preferred number of consecutive G-quartets is 2 or 3, to avoid excessive conformational stiffness. Furthermore, other individual differentiating elements can be additionally present in the G4-based aptamers, such as bulges [67], mixed G:A:G:A, G:T:G:T and G:C:G:C tetrads [68], [69], G-vacancies [70], A-tetrads sandwiched between G-tetrads [71] or A:G4 pentads [72]. An additional level of structural diversity is related to the possibility of raising both DNA and RNA G4-based aptamers [73], [74].
One possible explanation for the high number of G4-forming sequences found in the aptamer selection processes is the significant charge density of G-quadruplex DNA — twice the negative charge per unit length compared to duplex DNA [11]. The consequent high electrostatic potential provides for strong binding to the positively charged surfaces of proteins, although hydrophobic interactions can be also critical for strong and specific binding to the target.
3. G4-forming aptamers in therapeutic applications
In the last two decades, aptamers have proved to be very attractive as therapeutic agents mainly for their enhanced cellular uptake, non-immunogenicity and lower production costs [75]. Several aptamers against different human pathologies, such as neurodegenerative diseases [76], age-related macular degeneration [77], inflammation [78], thrombosis [79], diabetes [63] etc., have been discovered and evaluated.
The ability of G4-forming aptamers to recognize various proteins playing fundamental roles in different pathologies opened the way to the development of DNA or RNA G4-based therapeutic agents. Here we will focus on these particular aptamers, the most popular of which proved to be potential drugs against cancer, human immunodeficiency virus (HIV) and coagulation (Table 1 ).
Table 1.
State-of-the-art of known G4-forming aptamers.
| Disease | Aptamer | Sequence (5′ → 3′) | Target | Current status | References |
|---|---|---|---|---|---|
| Cancer | AS1411 | d(GGTGGTGGTGGTTGTGGTGGTGGTGG) | Nucleolin | Phase II (completed) |
NCT01034410, NCT00740441, [134] |
| T40214 | d(GGGCGGGCGGGCGGGC) | STAT3 | Phase 0 | [86], [87] | |
| HJ24 | d(AGCGTCGAATACCACACGGGGGTTTTGGTGGGGGGGGCTGGGTTGTCTTGGGGGTGGGCTAATGGAGCTCGTGGTCAT) | Shp2 |
In vitro studies (IC50 = 29 nM) |
[89], [90] | |
| 3R02 | d(TGTGGGGGTGGACTGGGTGGGTACC) | VEGF |
In vitro studies (Kd = 0.30 nM) |
[92], [93], [94] | |
| Human immunodeficiency virus (HIV) | ISIS 5320 | d(T*T*G*G*G*G*T*T) | HIV gp120 |
In vitro studies (IC50 = 0.30 μM) |
[98], [99], [100] |
| Hotoda sequence | DBB-d(TGGGAG) and TBDPS-d(TGGGAG) | HIV gp120 |
In vitro studies (IC50 = 0.37 μM and 0.88 μM, resp.) |
[101], [103] | |
| Modified Hotoda sequence | (4-benzyloxy)phenylphosphate-d(TGGGAG) | HIV gp120 |
In vitro studies (EC50 = 61 nM) |
[104] | |
| Modified Hotoda sequence | [TBDPS-d(TGGGCG)]4-TEL | HIV gp120 |
In vitro studies (IC50 = 39 nM) |
[102] | |
| AS1411 | d(GGTGGTGGTGGTTGTGGTGGTGGTGG) | Nucleolin |
In vitro studies (EC50 = 15.4 nM) |
[105], [106], [107] | |
| 93del | d(GGGGTGGGAGGAGGGT) | HIV Integrase |
In vitro studies (IC50 = 42 nM) |
[108], [109] | |
| Zintevir | d(G*TGGTGGGTGGGTGGG*T) | HIV Integrase | Phase I (completed) |
NCT00002403, [134] | |
| RT6 | d(ATCCGCCTGATTAGCGATACTCAGGCGTTAGGGAAGGGCGTCGAAAGCAGGGTGGGACTTGAGCAAAATCACCTGCAGGGG) | HIV Reverse Transcriptase |
In vitro studies (IC50 = 16 nM) |
[114] | |
| ODN 93 | d(GGGGGTGGGAGGAGGGTAGGCCTTAGGTTTCTGA) | HIV RNase H |
In vitro studies (IC50 = 0.50 μM) |
[115] | |
| ODN 112 | d(CCAGTGGCGGGTGGGTGGGTGGTGGGGGGACTTGG) | HIV RNase H |
In vitro studies (IC50 = 0.50 μM) |
[115] | |
| Coagulation | TBA | d(GGTTGGTGTGGTTGG) | Thrombin | Phase I (completed) |
[117], [118], [119] |
| HD22 | d(AGTCCGTGGTAGGGCAGGTTGGGGTGACT) | Thrombin |
In vitro studies (Kd = 3.2 nM) |
[120], [121], [122] | |
| HD1–22 | d(GGTTGGTGTGGTTGGAAAAAAAAAAAAAAAAGTCCGTGGTAGGGCAGGTTGGGGTGACT) | Thrombin |
In vitro studies (Kd = 0.65 nM) |
[123], [124] | |
| RA-36 | d(GGTTGGTGTGGTTGGTGGTTGGTGTGGTTGG) | Thrombin | Phase 0 | [125], [126] | |
| NU172 | d(CGCCTAGGTTGGGTAGGGTGGTGGCG) | Thrombin | Phase II (unknown) |
NCT00808964, [134] | |
| Osteoporosis | Scl 2 | d(TTGCGCGTTAATTGGGGGGGTGGGTGGGTT) | Sclerostin |
In vitro studies (IC50 = 0.90 μM) |
[129] |
| Prion diseases | R12 | r(GGAGGAGGAGGA) | PrPc |
In vitro studies (Kd = 8.5 nM) |
[131], [132] |
| Tuberculosis | PPK2 G9 | d(AACACATAGGTTTGGTTAGGTTGGTTGGTTGAATTA) | PPK2 |
In vitro studies (IC50 = 39.3 nM) |
[133] |
* = phosphorothioate bond; DBB = 3,4-dibenzyloxybenzyl; TBDPS = tert-butyldiphenylsilyl; TEL = tetra-end-linker.
3.1. G4-based aptamers as drugs
3.1.1. Anticancer G4-aptamers
AS1411. Discovered by coincidence and not by conventional SELEX approaches [80], AS1411 (Advanced Cancer Therapeutics, formally named as ACT-GRO-777) is the most advanced and first-in-class anticancer aptamer having entered clinical trials. AS1411 is a G4-forming 26-mer DNA aptamer, highly stable and resistant to nuclease degradation, targeting nucleolin. Nucleolin is a ubiquitous and multifunctional protein playing essential roles in cell survival, growth and proliferation [78]. AS1411 binds to the external domain of nucleolin, which is overexpressed on the surface of cancer cells. Therefore, AS1411 is able to specifically recognize and then be taken up by cancer cells [81]. The internalized aptamer-nucleolin complex inhibits DNA replication, resulting in accumulation of cells in S phase and cytotoxicity against cancer cells [82]. Recently, a novel mechanism of action for AS1411 has been proposed: AS1411 is internalized via nucleolin-mediated macropinocytosis, the internalized aptamer-nucleolin complex hyperstimulates macropinocytosis, which then causes non-apoptotic cell death, known as methuosis [83].
In preclinical studies, AS1411 has shown antiproliferative activity in several cell lines, including lung, prostate and breast cancer cells. The activity of AS1411 has been also proved in nude mice, resulting in a significant growth reduction in xenograft models of prostate cancer. In Phase I clinical trials, AS1411 was well tolerated by patients with advanced solid tumors without serious toxicity [80]. In Phase II clinical trials, AS1411 has shown promising activity against metastatic renal cell carcinoma and acute myeloid leukemia with minimal toxicity [84].
T40214. STAT3 protein is a signal transducer and activator of transcription involved in cellular differentiation, proliferation and apoptosis, over-activated in many types of human cancer [63]. T40214 is a 16-mer oligonucleotide which forms a stable G-quadruplex and targets STAT3 dimer, inhibiting its DNA binding activity [85]. Injection of T40214 caused a significant reduction in STAT3 expression, increased apoptosis processes and suppressed the growth of prostate and breast xenografts tumors in nude mice [86], [87].
HJ24. Shp2 is a protein of the family of tyrosine phosphatases involved in numerous essential cellular processes, such as cell growth, differentiation and oncogenic transformation. Thus, it is considered as a key target in different human diseases and mainly in cancer pathologies [88]. SELEX methods allowed identifying the DNA aptamer HJ24, selectively recognizing Shp2: it binds Shp2 phosphatase with affinity in the nanomolar range and strongly inhibits its activity (IC50 = 29 nM) [89]. HJ24 adopts a G4 structure, which is a prerequisite for stable binding to Shp2 [90].
3R02. VEGF is a protein that acts as a key regulator in angiogenesis, thus playing an important role in several diseases, such as age-related macular degeneration and cancer. VEGF has different isoforms, the prevalent being VEGF-165 [91]. Vap7 was identified as an aptamer against VEGF after three rounds of SELEX [92]. Its sequence is d(TGTGGGGGTGGACGGGCCGGGTAGA), which folds into a stable G-quadruplex structure. On this basis, a truncated mutant of Vap7 was designed (V7t1) in which the guanine tracts necessary to form the G-quartets were conserved. V7t1 folds into several G4 structures [93] and strongly binds VEGF-165 with low nanomolar K d values. Using in silico maturation to further increase the aptamer binding affinity, a class of mutants of V7t1 was designed; from these studies, the best aptamer selected was 3R02, having a 16-fold higher affinity for the target than V7t1 (K d = 300 pM) [94].
3.1.2. Antiviral G4-aptamers
Many G-rich sequences have been identified as aptamers active against infective agents, such as the Hepatitis C Virus (HCV) and the severe acute respiratory syndrome coronavirus (SARS-CoV) [95], [96], but the class of antiviral G4-based aptamers by far most represented is certainly that against HIV. Indeed, among the anti-HIV agents, many are G4-based aptamers able to interact with: a) the HIV envelope glycoprotein gp120 (which binds the cellular receptors CD4 or CCR5/CXCR4 favouring virus entry), b) the HIV integrase, or c) the HIV reverse transcriptase, inhibiting, respectively, virus binding and entry into the target cell, viral genome integration and reverse transcription. A detailed overview of anti-HIV agents based on G-quadruplex forming oligonucleotides developed in the last two decades was recently reported by our research group [97].
Virus attachment inhibitors. Upon screening a library of phosphorothioate octanucleotides containing all possible sequences, the first anti-HIV G4-aptamer identified was d(TTGGGGTT) (ISIS 5320) [98]. It forms a tetramolecular parallel G-quadruplex which inhibits HIV infection at sub-micromolar concentrations (IC50 = 0.3 μM). Due to its highly anionic character, it strongly interacts with the cationic V3 loop of the envelope glycoprotein gp120 and inhibits virus adsorption and cell fusion [99], [100].
A series of hexadeoxyribonucleotides, carrying various modifications at either the 5′ or the 3′ ends, based on the so called d(TGGGAG) Hotoda's sequence, was also synthesized and evaluated in vitro for their anti-HIV activity, proving to be potent gp120 inhibitors, with IC50 values from sub-micromolar to nanomolar concentrations [101], [102], [103], [104].
Besides the main cellular receptors for HIV entry (CD4, and the two chemokine receptors CXCR4 and CCR5), virus attachment can also occur through the interactions with heparan sulfate proteoglycans and nucleolin on the host cell-surface [105]. Therefore, also targeting nucleolin can interfere with HIV attachment, as demonstrated using AS1411, which showed a marked antiviral activity [106]. In vitro tests have shown that AS1411 is effective against HIV at low nanomolar concentrations (EC50 = 15.4 nM), thus proving to be more potent as an antiviral than as an anticancer agent [107].
Virus integration inhibitors. Anti-HIV integrase activity has been found for 93del, an aptamer with a sequence of d(GGGGTGGGAGGAGGGT) which adopts a peculiar dimeric interlocked G4 structure and exhibits in vitro inhibitory activity in the nanomolar range [108], [109]. A remarkable inhibition of the HIV integrase, with IC50 values in the nanomolar range, was observed also for T30177, an aptamer with a sequence of d(GTGGTGGGTGGGTGGGT), containing single phosphorothioate internucleoside linkages at its 5′- and 3′-ends. This oligonucleotide showed an unusual high nuclease resistance in physiological environments, due to its capacity to form a stable dimeric structure involving two subunits of propeller-type parallel-stranded G-quadruplexes [110]. T30177 was the first HIV integrase inhibitor tested in clinical trials (Zintevir, Aronex Pharmaceuticals) [111].
Reverse transcription inhibitors. The HIV reverse transcriptase (RT) is a multifunctional enzyme composed of two domains: the DNA polymerase domain converts the viral RNA genome into DNA in the cytoplasm of the host cell, while the RNase H domain specifically cleaves the RNA strand in the RNA/DNA heteroduplex [112], [113]. The first HIV RT inhibitor identified was RT6, an aptamer with a bimodular structure comprising a 5′-stem-loop module connected to a 3′-G4 module [114]. DNA aptamers for the RNase H domain of RT were then identified by SELEX procedures, using recombinant RT with or without the RNase H domain. Two aptamers, ODNs 93 and 112, selectively inhibited the HIV RNase H with IC50 values in the sub-micromolar range, not showing activity on the human enzyme [115].
3.1.3. Anticoagulant G4-aptamers
Thrombin is a serine protease that plays a key role in the last step of blood clotting process, converting soluble fibrinogen into insoluble fibrin [116]. The first anticoagulant aptamer targeting thrombin was described in 1992 by Bock et al. [117]. Subsequently, other DNA aptamers were also discovered as thrombin inhibitors, most of them adopting G-quadruplex structures.
TBA. Thrombin-binding aptamer (TBA, also known as HD1 or ARC183, Archemix and Nuvelo) is a DNA aptamer targeting thrombin exosite I with nanomolar affinity (range 25–200 nM) [117]. It folds into a chair-like G-quadruplex structure consisting of two G-quartets, a TGT loop and two TT loops [118]. In Phase I clinical trials, TBA has been tested as putative anticoagulant in coronary artery bypass graft surgery. The preliminary results confirmed the desired dose-related anticoagulation activity. However, the high amount needed to obtain the desired anticoagulation effect resulted in a sub-optimal dosing profile. Therefore, Archemix and Nuvelo interrupted Phase I clinical trials on TBA in 2005, claiming to focus on improved second generation aptamers [119].
HD22. HD22 is a DNA aptamer with a duplex/G-quadruplex mixed structure [120], [121]. HD22 recognizes the heparin-binding exosite II of thrombin, inhibiting thrombin-mediated activation of platelets, with a binding constant in the nanomolar range, showing higher affinity for thrombin than TBA [122].
HD1–22. HD1–22 is a bivalent DNA aptamer consisting of exosite I binding aptamer HD1 (TBA) and exosite II binding aptamer HD22, connected through a poly-dA linker [123]. It binds thrombin with higher affinity than the precursor aptamers alone and is able to prolong thrombin clotting times in a dose-dependent manner, proving to be a strong inhibitor of thrombin activity [124].
RA-36. RA-36 is a bivalent DNA aptamer consisting of two covalently bound G-quadruplexes, each containing the TBA motif, connected via a thymine linker. It binds thrombin exosite I and blocks fibrinogen binding to the enzyme with a dose-dependent effect, thus resulting into an efficient thrombin inhibitor. Animal tests have shown that this aptamer shows a high species-specificity [125], [126].
NU172. Discovered through SELEX and subsequently truncated to 26 nucleotides, NU172 (ARCA Biopharma) is a DNA aptamer targeting thrombin exosite I [127], able to fold into a G4 structure [128]. In preclinical studies, NU172 has shown the ability to prolong thrombin clotting time. Furthermore, due to rapid nuclease degradation, the anticoagulant activity can be halted without using antidotes. A Phase II clinical trial to test its anticoagulant efficacy in coronary artery bypass graft surgery is in unknown status [78].
3.1.4. G4-aptamers against other diseases
Skeletal diseases. Sclerostin is an extracellular protein involved in the negative regulation of bone growth and, for this reason, is a key target for the treatment of osteoporosis. A parallel G4-forming aptamer has been identified and evaluated against sclerostin. In vitro studies have shown that the G4-based aptamer specifically binds sclerostin with affinities in the nanomolar range [129].
Prion diseases. Prion diseases are neurodegenerative disorders of mammals due to the conversion of soluble normal cellular prion protein PrPc in its insoluble abnormal isoform PrPsc that accumulates in the brain [130]. Targeting PrPc can be a strategy to stabilize the normal prion protein and avoid its conversion to the abnormal PrPsc form. Mashima et al. discovered an RNA aptamer against bovine PrPc with a sequence of r(GGAGGAGGAGGA) (R12). It folds into a dimeric G-quadruplex structure [131] and exhibits anti-prion activity in mouse as a result of the PrPsc level reduction [132].
Tuberculosis. Inorganic polyphosphates act as key regulators of mycobacterials stress and virulence. Polyphosphate kinases (PPKs) are the major enzymes involved in polyphosphates metabolism proving to be key targets in Mycobacterium tuberculosis. Shum et al. identified G4 and non G4-forming aptamers against PPK2, one of the two classes of M. tuberculosis PPKs, but only the G4-forming aptamer PPK2 G9 proved to effectively bind the protein and inhibit its catalytic activity (IC50 = 39.3 nM). Mutational analysis provided further evidence of the key role played by the G-quadruplex architectures in the specific and strong binding to PPK2 [133].
3.2. G4-based aptamers as drug delivery systems
Taking into account that most of the known anticancer/antiviral drugs and candidate drugs are associated with heavy side effects due to their intrinsically poor selectivity, efficient targeted drug-delivery systems are urgently required. Due to their ability to strongly and selectively recognize protein targets, aptamers can be helpful also for specific targeting. Indeed, aptamers can be conjugated with the drug of choice in order to deliver it towards specific receptors (usually cell surface proteins) overexpressed on target cells and underexpressed on non-target healthy cells [82].
A very interesting case is the above-discussed G4-forming aptamer AS1411, which acts both as a potential drug, having promising antitumor [80], [84] and anti-HIV [106], [107] activity, and as a drug-delivery system, being efficiently and selectively internalized through nucleolin, overexpressed on cancer cells, by macropinocytosis [135]. In particular, several research groups have focused their attention on AS1411-based drug-delivery approaches, developing essentially three different kinds of systems: AS1411-drug, AS1411-drug-liposomes and AS1411-drug-nanoparticle conjugates. For each of these systems some valuable examples are reported here [136], [137], [138], [139], [140].
Shieh et al. have described an AS1411-TMPyP4 conjugate useful in photodynamic therapy [136]. The hypothesized mechanism of action is the following: the complex is efficiently taken up by cells exploiting the binding to nucleolin and, after light exposure, the activation of TMPyP4 destroys only the cells carrying the internalized complex. In vitro studies have shown that TMPyP4, when conjugated with AS1411, preferentially accumulates in breast cancer cells than in epithelium normal cells. Furthermore, the photodamage produced by this conjugate is overall more extensive in cancer cells than in normal cells. These results proved that the delivery and uptake of TMPyP4 are effectively mediated by the specific interaction of the AS1411-TMPyP4 complex with nucleolin.
Le Trinh et al. have crosslinked AS1411 with doxorubicin, a known chemotherapeutic agent [137]. In vivo tests in a murine xenograft model have shown that injection of the AS1411-doxorubicin adduct resulted in a reduced antitumor activity compared with free doxorubicin, but also in a significant reduction of the typical side effects associated to doxorubicin treatments, such as cardiotoxicity, nephrotoxicity and weight loss [137]. Another AS1411-doxorubicin adduct has been developed by Liu et al., who constructed a dimeric nanocarrier consisting of the AS1411 domain, responsible for the selective targeting of cancer cells, and a DNA duplex domain, necessary to capture and transport doxorubicin. This nanosystem showed efficient in vitro and in vivo antitumor activity on drug-resistant breast cancer cells and reduced toxicity compared to free doxorubicin [138].
In order to selectively deliver higher amounts of chemotherapeutics into cancer cells, Liao et al. have proposed the use of doxorubicin-containing liposomes functionalized with AS1411. A thiol-derivatized AS1411 was first conjugated to maleimide-PEG 2000-DSPE via formation of a thioether linkage; the purified conjugates self-assembled into spherical micelles, which were then incubated with liposomes, allowing the micelle components to be exchanged into the liposome bilayers. In vivo studies in nude mice have shown that, when delivered by liposomes, doxorubicin preferentially accumulates in tumour tissues with respect to free doxorubicin, inhibiting the tumour growth with minimal toxicity compared to the free drug [139].
Finally, Guo et al. have developed AS1411-functionalized nanoparticles to deliver paclitaxel, a potent chemotherapeutic agent. In vivo studies on these nanoparticles have shown enhancement of paclitaxel accumulation in tumor cells and higher reduction of glioma weight in nude mice compared with free paclitaxel [140].
Another G4-forming aptamer potentially useful in drug delivery, named AIR-3A, has been discovered by Meyer et al. AIR-3A is an RNA aptamer of sequence r(GGGGAGGCUGUGGUGAGGG) that specifically recognizes the receptor of interleukin-6 (IL-6), a cytokine that plays a key role in inflammatory diseases and cancer. AIR-3A was proved to specifically deliver proteins towards cells expressing IL-6R (Interleukin-6 Receptor) on their surface, such as hepatocytes, monocytes, macrophages, lymphocytes and HIV-infected cells [141], [142], further stimulating the research on novel drug delivery systems useful not only in anticancer, but also in anti-HIV treatments.
3.3. State-of-the-art on therapeutic G4-forming aptamers
The overall analysis of the known G4-forming therapeutic aptamers provides an interesting picture, from which some general conclusions can be drawn [143]. First of all, a significant number of aptamers selective against cancer cells adopt a G4 structure as a preferential conformation. The presence of a G4 folding appears to be an essential structural motif for aptamers endowed with cancer-selective antiproliferative activity. Upon screening 11 oligonucleotide combinatorial libraries, differing in nucleobase composition and length, Bates et al. found that only the G-rich libraries showing features consistent with G4 formation strongly inhibited cancer cell growth, in contrast not affecting healthy cells [144]. Then, the parallel G4 topology seems to be largely prevailing among these bioactive aptamers. One remarkable example is the nucleolin-binding aptamer AS1411, advanced to Phase II clinical trials against renal cell carcinoma [80], which, even if present as a mixture of different monomeric and even dimeric G4 conformations, essentially prefers parallel structures [145]. In another study, investigating the antiproliferative effects of G4-forming aptamers, Shangguan et al. suggested that the intramolecular parallel G4 structure may be a key feature for cellular protein recognition, allowing efficient cell uptake through endosome/lysosome pathways [146]. This study was carried out on a number of parallel G4 structures, including putative G4-forming sequences from the human genome. These findings support the idea that structure-activity relationship analyses can speed up the identification of G4 scaffolds useful for the design of optimized therapeutically active aptamers. Thus, aptamer selection can be performed using partially randomized G-rich oligonucleotide libraries combining SELEX with rational design methods. However, in some cases, even minor mutations in G4 sequences may have dramatic effects on aptamer stability and/or activity [147], [148], [149], [150], inducing marked alterations in the G4 topology (e.g., an intramolecular antiparallel G4 can be converted into an intermolecular parallel one [151]) or causing the aptamer to target different epitopes of the protein [120]. Thus, the randomization process is always a compromise between the need for the highest sequence permutation and for a stable G4-folding [152]. In the case of an intermolecular scaffold, the combinatorial libraries usually contain several consecutive guanines (G-runs) with randomized flanks [98]. In the case of an intramolecular scaffold, the G4 core is typically kept intact, while the loops are randomized.
A promising strategy to increase the molecular diversity of the aptamers within a combinatorial drug discovery approach is to exploit their modular assembly. An active G4 module can be linked to a duplex motif to get a hybrid structure with improved thermodynamic stability [153], [154]. Alternatively, polyvalent aptamers [118] can be obtained by conjugating similar or different G4 modules (homo-polyvalent or hetero-polyvalent aptamers, respectively), able to recognize distinct sites on the same target protein [123], [155], [156] or different targets [157]. A synergistic effect between the linked domains is typically expected with polyvalent systems [158]; in this case, however, the success of the overall strategy critically depends on the choice of the linker between the modules. In this context, combinatorial libraries with randomized linkers and fixed active modules have been fruitfully used [159].
Another central issue is that, due to the abundance of G4 motifs in the human genome [160], all playing diverse regulatory roles [161], a variety of genomic G4-forming motifs (particularly those located in gene promoters) can be regarded as potential candidate aptamers. In other words, G4-forming aptamers can be designed taking inspiration from natural nucleic acid substrates and then used as their competitors for endogenous G4-binding regulatory proteins, ultimately affecting gene expression and normal cell functions. In this regard, a valuable example is the G4-forming oligonucleotide carrying the sequence located in the insulin promoter, found to recognize insulin and insulin-like growth factor 2 [162], [163]. A promoter-derived aptamer selection method (G4PAS) has been successfully used to identify G4s with good affinities for VEGF, platelet-derived growth factor-AA and retinoblastoma protein RB1 [164]. Native and modified ON mimics of G4s from RAS proto-oncogenes are able to compete with DNA-protein complexes that regulate transcription and therefore can be used as decoys for transcription factors [165], [166]. Rational design of G4 decoys, first described by Xodo et al., appears to be a very promising strategy in anticancer research [167]. In parallel, it can be expected that new, more effective G4-forming antiviral agents will be developed as a consequence of the rapid progress in the studies on the G4-forming sequences of HIV-1, Epstein-Barr virus and Papilloma Virus genomes and their participation in viral life cycles [96], [97]. Hopefully, more and more gene-inspired G4-forming aptamers will be discovered in the near future.
4. G4-forming aptamers in detection and diagnostic applications
Considering all the advantages discussed before, such as the wide target library, high affinity, stability, selectivity, and capability to form specific Watson-Crick and/or Hoogsteen H-bonds, it is not surprising that nucleic acid aptamers have emerged as key recognition elements also in the design of novel biosensors (i.e., the so called “aptasensors”), in combination with various sensing strategies compatible with almost all kinds of targets, detection requirements and readout techniques [168]. The sensing process of aptasensors is similar to other biosensors: the aptamer recognizes its target, and the signal transducer transfers the recognition information into measurable signals. The aptamers can be immobilized (heterogeneous) or used in solution (homogenous phase) [169]. Once immobilized, the aptamers are able to release targets upon washing with different buffers, thus allowing in principle to reuse an aptasensor several times. Multiple sensing strategies have been developed till now, combined with various transducers, such as quartz crystal microbalance (QCM), surface plasmon resonance (SPR), fluorescence, colorimetry, electrophoresis, electrochemistry, electrochemiluminescence (ECL), field-effect transistor, and so on. These aptasensors can be generally divided into two classes: “labeled” and “label-free” sensors [170]. The most recent developments in the use of aptamers in diagnostics and imaging have been recently reviewed by Neerathilingam et al., who have discussed in detail the applications of aptamers for the detection of infective agents, antigens/toxins (bacteria) and biomarkers (cancer) [171].
In the specific case of G4-forming aptamers, the versatility and plasticity of these structures can be fruitfully exploited in various types of detection assays [64].
4.1. Thrombin detection by TBA and its variants: a biologically relevant case and/or a proof-of-concept
The most studied G4-forming aptamers in many diagnostic applications are the thrombin binding aptamers (TBAs), two short DNA sequences of 15 (TBA15) and 29 (TBA29) nucleotides in length, relatively easy and inexpensive to synthesize and able to recognize with high affinity and specificity the fibrinogen- and heparin-recognition exosite on thrombin, respectively (exosites I and II) [117]. Despite the high biological significance of thrombin as a target, both TBA15 and TBA29, properly modified to fulfill the desired detection process, have been widely used essentially as a proof-of-concept to verify the feasibility of the proposed analytical method. The success of TBAs as model systems in the development of detection assays is due to several factors, such as: 1) the high affinity and specificity for the target; 2) the availability of two G4-based thrombin aptamers, each targeting a specific exosite on the protein in a non-interferring manner, giving the possibility to perform sandwich-type assays; 3) the rapid and unambiguous switching of the aptamer structures triggered by thrombin binding, and 4) the catalytic activity of the protein, still present after the binding event for both aptamers. The scientific literature has plenty of research studies for the detection of thrombin. They are based either on very complicated multi-recognition systems or on rapid and easy methods, and can be grouped in homogenous- or heterogeneous-phase assays, using labeled or label-free, as well as enzyme-assisted assays. For a detailed recent review, illustrating the development of more than one hundred analytical techniques centered on the TBA-thrombin system, see also ref. [172]. We report here an update on recent TBA-based sensing systems, discussing in parallel other, less recent assays, selected on the basis of their apparent efficiency and simplicity.
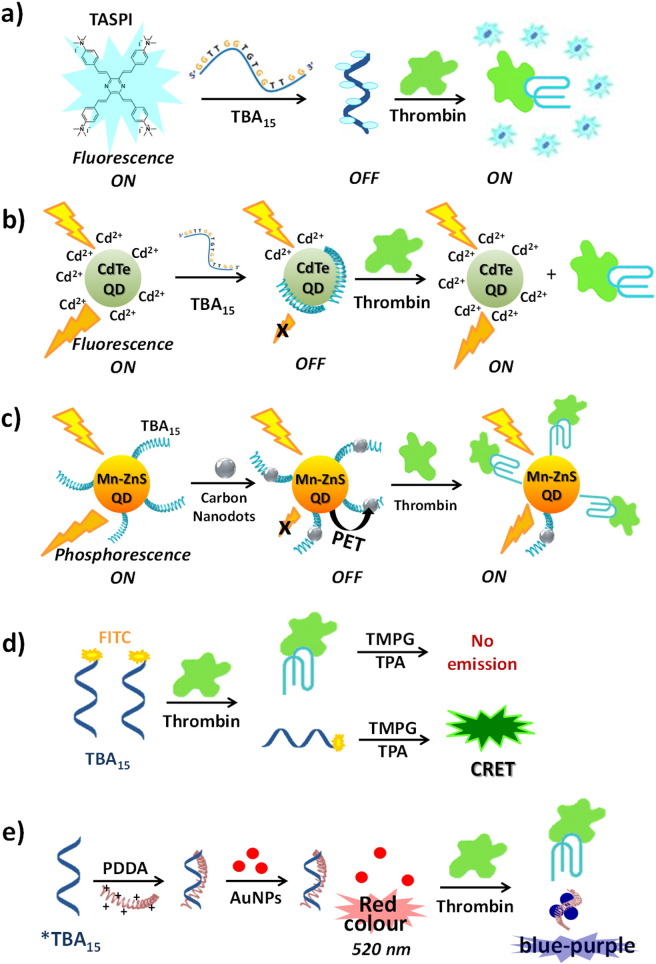
In 2011 Yan et al. developed a probe for thrombin sensing based on TBA15 and a multi-branched fluorescent pyrazine derivative (TASPI) which, upon strongly interacting with the single strand DNA, turns off its bright fluorescence. In the presence of thrombin, which specifically binds to TBA inducing its folding into an antiparallel G-quadruplex structure, TASPI molecules are released from the aptamer, resulting into vivid and facile fluorescence recovery (“turn ON” system, Fig. 4a). Interestingly, this approach can be employed also in real plasma samples during the coagulation process [173].
Fig. 4.
Schematic representation of thrombin detection using various TBA-based sensing systems and different detection techniques employing fluorescence, phosphorescence, chemiluminescence or colorimetry.
Another similar example for the detection of thrombin has been reported in 2013 as a facile and sensitive label-free homogeneous assay. In this approach, Zhang et al. used CdTe quantum dots (QDs), Cd2 + ions and the TBA15 [174]. The QDs can be “activated” with fluorescence enhancement by adding extra Cd2 + to the solution in a basic medium; the positively charged Cd2 +-activated CdTe QDs interact with the negatively charged TBA, leading to fluorescence quenching; when thrombin is added, TBA forms a G-quadruplex structure through specific interaction with its target, releasing the QDs with a recovery of the fluorescence intensity (Fig. 4b). A linear response for thrombin has been observed in the range from 1.4 nM to 21 nM, with a detection limit of 0.70 nM. This method is easy to perform since covalent modifications or immobilizations of the aptamer are avoided, can reduce background signal and improve sensitivity due to the turn-on sensing mode, and can be performed directly on real serum samples, not requiring pre-treatments of the serum matrix.
A remarkable fluorescence enhancement upon thrombin recognition has been obtained also using a dansyl/cyclodextrin-tagged TBA15: in this case, thrombin binding to the TBA produces a fluorescence enhancement due to encapsulation of dansyl, attached at the 3′-end of the TBA, into the apolar cavity of the β-cyclodextrin at the 5′-end of the aptamer sequence [175].
Similarly, another aptamer-based turn-on thrombin biosensor was based on Mn-doped ZnS quantum dots, whose phosphorescence was restored after thrombin-aptamer recognition [176]. The system involved an efficient phosphorescence energy transfer (PET) donor-acceptor pair: Mn-doped ZnS quantum dots labeled with thrombin-binding aptamers (TBA QDs) as donors, and carbon nanodots (CNDs) as acceptors. Due to the π-π stacking interactions between the aptamer and CNDs, the energy donor and acceptor are taken into close proximity, leading to the phosphorescence quenching of TBA QD-donors. With the introduction of thrombin onto the TBA-QDs-CNDs system in “off state”, the phosphorescence is “turned on” due to the formation of G4-thrombin complexes, which release the energy acceptor CNDs from the energy donors (Fig. 4c). The sensor displays a linear range of 0–40 nM for thrombin, with a detection limit of 0.013 nM in pure buffers. The proposed aptasensor has also been used to monitor thrombin in complex biological fluids, including serum and plasma, with satisfactory recovery ranging from 96.8 to 104.3%. These results can be explained because the interference to phosphorescence deriving from auto-fluorescence and scattering light in complex matrixes can be easily avoided.
Also chemiluminescence was employed in a turn-off solution assay for thrombin detection. In this case TBA15, conjugated to 6-carboxyfluorescein (6-FAM), was reacted with 3,4,5-trimethoxylphenylglyoxal (TMPG) in the presence of tetra-n-propylammonium hydroxide (TPA): the high-energy intermediate formed transfers energy to 6-FAM, emitting bright light based on chemiluminescence energy transfer (CRET). In the presence of thrombin, the brightness of guanine chemiluminescence was quenched due to the formation of G-quadruplex TBA-conjugated 6-FAM bound with the target (Fig. 4d). The detection limit of G4-TBA aptasensor with good linear calibration curves, accuracy, precision, and recovery was 12.3 nM in 5% human serum [177].
In another study, a colorimetric ultrasensitive thrombin-detection assay has employed an elongated variant of TBA15 (21-mer), the cationic polymer poly(diallyldimethylammonium) chloride (PDDA) and gold nanoparticles as sensing elements [178]. Initially, the cationic polymer interacts with the TBA and the dispersed gold nanoparticles confer a wine-red color to the solution. Upon thrombin addition, the target strongly interacts with the TBA probe displacing the cationic polymer, which in turn induces gold nanoparticle (AuNP) aggregation; this event produces a visible change in color from red-wine to blue-purple, detectable also by naked eye (Fig. 4e). This colorimetric sensor could detect thrombin up to 1 pM with high selectivity also in the presence of other interfering proteins and was successfully applied to determine thrombin levels in human serum samples.
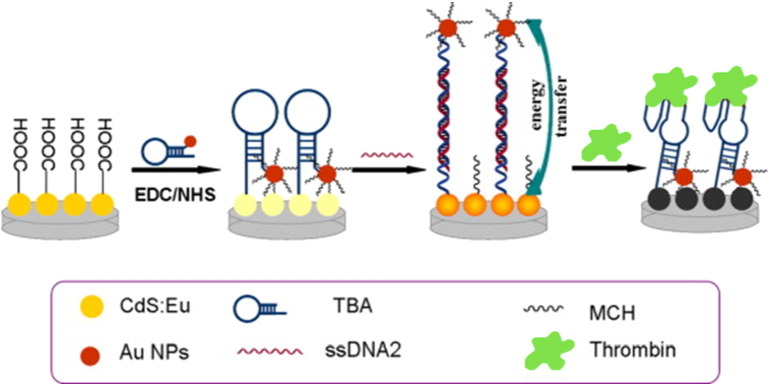
A more complicated but ultrasensitive assembled system for thrombin detection was based on an off-on-off electrochemiluminescence (ECL) approach [179]. The system is composed of an Eu3 +-doped CdS nanocrystal (CdS:Eu NC) film on a glassy carbon electrode (GCE) as ECL emitter. AuNPs, labeled with a hairpin-DNA probe (ssDNA1, Fig. 5 ) containing TBA15, are linked on the NC film: an ECL quenching effect (off) is produced as a result of Förster-resonance energy transfer (FRET) between the CdS:Eu NC film and the proximal AuNPs. Upon hybridization with a free ssDNA (ssDNA2) complementary to the hairpin, a duplex structure is formed and the AuNPs are pulled away from the CdS:Eu NC film, causing an ECL enhancement (on) due to the interactions of the excited CdS:Eu NCs with ECL-induced surface plasmon resonance (SPR) in AuNPs at large separation. The addition of thrombin induces ssDNA1 to form a G-quadruplex structure, displacing the ssDNA2 and allowing the AuNPs to get close to the CdS:Eu NC film again, eventually producing an enhanced ECL quenching (off). This “off-on-off” system showed a maximum 7.4-fold change of ECL intensity, due to the configuration transformation of ssDNA1, providing great sensitivity in thrombin detection in a wide detection range, from 50 aM to 1 pM.
Fig. 5.
TBA-related ECL “off–on–off” platform based on energy transfer between CdS:Eu NC film and AuNPs. EDC = N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride; NHS = N-hydroxysuccinimide; MCH = 6-mercapto-1-hexanol.
(Adapted with permission from ref. [179]. Copyright 2014, Elsevier B.V.)
Very recently, a novel label-free electrochemical aptasensing platform for rapid and facile detection of thrombin, using a graphene oxide (GO)-modified pencil graphite electrode and TBA15 has been designed [180]. The strategy relies on the preferential adsorption of single-stranded DNA (ssDNA) to GO over aptamer-target complexes. The TBA-thrombin complex formation was monitored by differential pulse voltammetry (DPV) using the guanine oxidation signal. In the absence of thrombin, the aptamers are adsorbed onto the surface of GO, leading to a strong background guanine oxidation signal. Conversely, in the presence of thrombin, the conformational transformation of TBA after incubation with the thrombin solution and formation of the aptamer-thrombin complexes, which have weak binding affinity for GO, leads to the desorption of TBA-thrombin complex from the electrode surface and significant oxidation signal decrease (Fig. 6 ). The selectivity of the biosensor was studied using other biological substances. The biosensor signal was proportional to the thrombin concentration from 0.1 to 10 nM with a detection limit of 0.07 nM.
Fig. 6.
Schematic illustration of the label-free electrochemical aptasensor for thrombin detection.
In order to evaluate the analytical reliability and application potential of this biosensor in clinical analysis, the proposed biosensor was used for the detection of thrombin in healthy human blood serum samples under optimal experimental conditions. Similar responses were found for both buffer and serum samples, and proved the applicability of the proposed sensor for real sample analysis. Moreover, this biosensor was used for the determination of thrombin concentration in a spiked diluted serum sample by the standard addition method. The obtained results for measurements at four different concentrations showed good recovery values (98–103%), indicating that the proposed biosensor has high accuracy in practical applications and can be used for the thrombin detection in real biological samples.
Many other sensing systems for the detection of thrombin, or of other protein targets of therapeutic relevance, require an enzymatic activity to obtain the signal transduction producing fluorescence, colorimetric or electrochemical changes.
Very important advances in the field of G4 aptasensors came from the discovery of a peroxidase activity associated with the G4-hemin complex (DNAzyme). Initially, in 1998, a specific aptamer binding iron (III)-protoporphyrin IX (hemin) was selected (PS2.M), which was a G-rich DNA sequence able to form a stable G-quadruplex structure [181]. It was demonstrated that the aptamer-hemin complex had significantly higher peroxidase activity than hemin alone, under physiological conditions, catalyzing (DNAzyme) the oxidation of 2,2′-azino-bis(3-ethylbenzothiozoline)-6-sulfonic acid (ABTS) by hydrogen peroxide (H2O2), with concomitant formation of the colored radical cation product ABTS• + [181]. Its peroxidase activity was also significantly higher than that of a previously reported hemin-catalytic-antibody complex [182]. It was later realized that the RNA version of PS2.M (rPS2.M) also showed enhanced peroxidase activity, demonstrating that RNA-based aptamers can have a similar structure and function [183]. The intramolecular G-quadruplex structure was shown to play an important role in the peroxidase activity [184]. As a general DNA ligand with the porphyrin ring, hemin should bind also to other G-quadruplex families. This point was indeed investigated exploring the interactions between hemin and other G-quadruplex aptamers, discovering that most G4-forming aptamers were able to bind hemin, with different affinities depending on the type of G4 structure, and had peroxidase activity (DNAzyme) [185]. Regarding the sensing system, in addition to ABTS, many other compounds can also act as substrates being oxidized by H2O2 upon G4–hemin complex catalysis, such as luminol to produce chemiluminescence [186] or chemiluminescence resonance energy transfer [187], 3,3′-diaminobenzidine (DAB) [188], etc. The peroxidase-mimicking activity of G4 DNAzyme is related to the topology and conformation of the G4 structure [189]: remarkably, parallel G-quadruplexes display much higher peroxidase activity than those displayed by hemin and antiparallel G-quadruplexes.
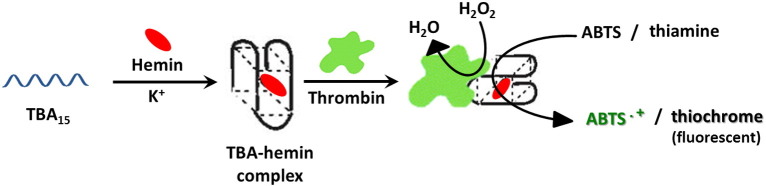
Also TBA15 binds hemin; this occurs when the G4-forming aptamer is in the folded state, thus forming a stable complex with weak peroxidase-like activity. After incubation with thrombin, the TBA–hemin complex is recognized by the protein target, forming a supramolecular DNAzyme with significantly increased catalytic activity. This recognition event produces an absorbance change in the ABTS–H2O2 system which can be easily monitored using UV–vis absorption spectroscopy (Fig. 7 ), thus providing a facile approach to reveal thrombin with a detection limit of 20 nM [190].
Fig. 7.
Colorimetric and fluorimetric approaches for thrombin detection based on the DNAzyme formation between hemin, TBA and the target protein.
Improved sensitivity with respect to the colorimetric assay with ABTS was obtained combining the TBA G-quadruplex-based DNAzyme (TBA-hemin-thrombin) with the H2O2–thiamine fluorescent system. The H2O2-mediated oxidation of thiamine, a cheap fluorescent substrate, gives rise to fluorescence emission (Fig. 7) allowing the quantitative analysis of thrombin with a detection limit of 1 pM [191].
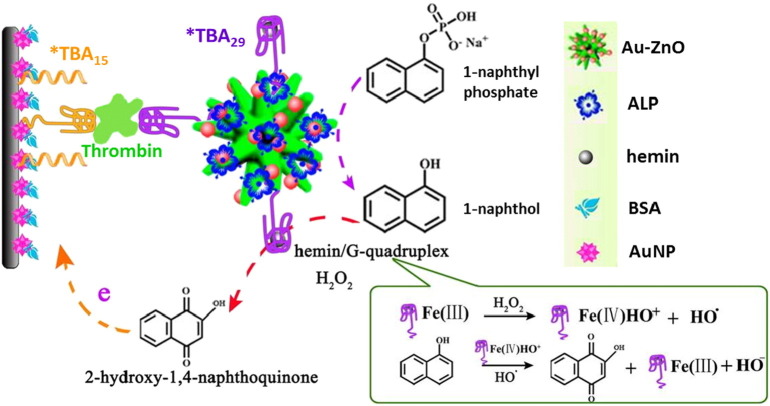
Employing the synergistic effect of the hemin/G-quadruplex peroxidase activity, of the enzymatic activity of an alkaline phosphatase (ALP) as well as of the sandwich-type recognition of thrombin by the two TBAs (TBA15 and TBA29) specific for the exosites I and II of the target protein, respectively, an ingeniously-designed and very sensitive electrochemical aptasensor for thrombin has been obtained [192]. This thrombin aptasensor is composed of two systems: 1) gold-nanoparticle-decorated zinc oxide nanoflowers (Au-ZnO) as the matrix for immobilizing ALP and TBA labeled with hemin to form hemin/G-quadruplex/ALP/Au-ZnO bioconjugates, and 2) a solid platform as the sensing interface, constituted by a GCE on which a thin layer of AuNPs, functionalized with TBA15, is deposited. Thrombin detection was obtained exploiting a sandwich-type recognition, which led in close proximity the TBA29-Au-ZnO bioconjugates and the TBA15-AuNP modified electrode. In these conditions, upon addition of 1-naphthyl phosphate, ALP efficiently catalyzed its oxidation to 1-naphthol on the surface of the electrode, on the other hand, 1-naphthol could be oxidized by the hemin/TBA/thrombin system in the presence of H2O2, resulting in the final promotion of electron transfer to the electrode and amplification of the electrochemical signal. An enzyme-assisted signal amplification strategy was incorporated into this assay to increase the sensitivity of the electrochemical aptasensor (Fig. 8 ). The proposed thrombin aptasensor yielded a linear response in the range 1 pM–30 nM with a detection limit of 0.37 pM.
Fig. 8.
Schematic illustration of ALP and hemin/G-quadruplex as catalysts for an in situ amplified electrochemical detection signal.
(Adapted with permission from ref. [192]. Copyright 2015, American Chemical Society)
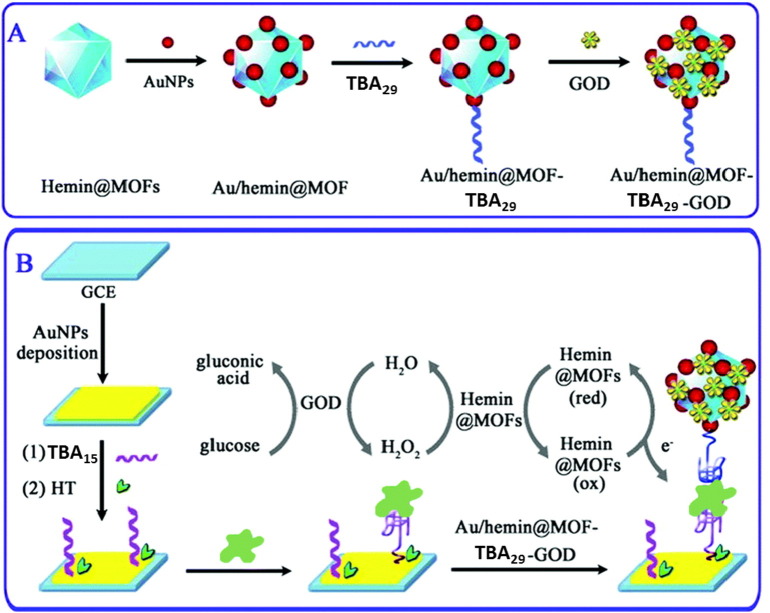
In a similar system, the same authors used another enzyme, glucose oxidase (GOx), to construct a sensitive electrochemical aptasensor for thrombin, exploiting the enzyme-assisted signal amplification strategy. They employed the synergistic effect of hemin as a peroxidase mimic, of the enzymatic activity of GOx, and of the sandwich-type recognition of thrombin by the two TBAs (TBA15 and TBA29) [193]. The system has been obtained combining two components: 1) a multifunctional bionanocomposite in solution composed of nano-sized Fe-MIL-88 MOFs encapsulating hemin and functionalized with gold nanoparticles, glucose oxidase and TBA29 (Fig. 9A); and 2) a solid platform as the sensing interface, constituted by a GCE on which a thin layer of AuNPs functionalized with TBA15 was deposited (Fig. 9B). The sandwich-type recognition involving the Au/hemin@MOF–TBA29–GOx bioconjugates and the TBA15-AuNP modified electrode allowed sensitive thrombin detection (Fig. 9B).
Fig. 9.
(A) The preparation of Au/hemin@MOF–TBA29–GOx bioconjugates. (B) Schematic representation of the stepwise assembly procedure and electrocatalysis detection principle of the proposed electrochemical aptasensor. HT = hexanethiol.
(Adapted with permission from ref. [193]. Copyright 2015, The Royal Society of Chemistry)
Upon addition of glucose, GOx efficiently catalyzed the oxidation of glucose to gluconolactone, coupled with the local formation of H2O2 in the presence of dissolved O2; the generated H2O2 on the electrode surface was reduced by the hemin@MOF system, with the final promotion of electron transfer to the electrode and signal amplification. Thus, the enzyme-assisted signal amplification strategy increased the sensitivity of the constructed electrochemical aptasensor employing hemin@MOFs as redox mediators for signal generation based on the reversible redox of Fe(III)/Fe(II) within hemin. The so-obtained sandwich-type aptasensor showed good sensitivity, stability and satisfactory reproducibility, as well as good precision in thrombin determination in serum, indicating that the method has the potential to be used for the detection of this protein in complex samples.
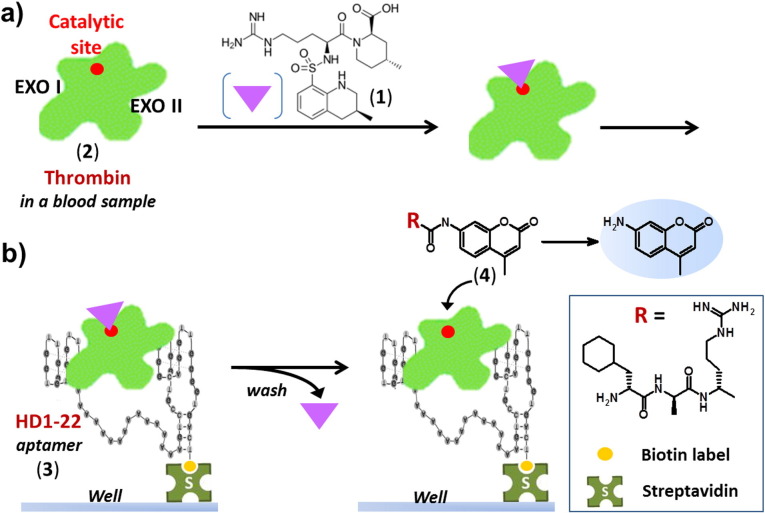
In another detection method based on an enzymatically-assisted assay, the catalytic activity of the target, i.e. thrombin, was exploited [194]. Indeed, thrombin is a protease enzyme that can specifically catalyze the cleavage of proteins and peptides and the binding sites of aptamers on the protein are different from the enzyme catalytic sites. In the absence of aptamer binding to thrombin, the enzymatic activity of thrombin can be maintained and used for the development of detection methods. During the blood sampling process, an anticoagulant buffer (citrate) containing the reversible active-site inhibitor argatroban (1, Fig. 10 ) is added to the blood sample containing thrombin (2, PDB 1DWC). Complex formation between argatroban and thrombin efficiently prevents irreversible inhibition of thrombin by endogenous thrombin inhibitors. Wells of streptavidin-coated microtiter modules, previously loaded with the 3′-biotinylated anti-thrombin aptamer HD1–22 (3), that simultaneously targets exosites I and II of thrombin, are overlaid with plasma. After incubation and capturing of the argatroban-thrombin complex by HD1–22, the wells are washed to remove plasma remains and reversibly bound argatroban. Subsequently, a thrombin-specific peptide substrate, bearing an AMC (7-amino-4-methylcoumarin) fluorogenic probe (H-D-CHA-Ala-Arg-AMC, 4), is added to quantitatively determine the amount of functional active thrombin captured in the wells. Thus a supramolecular oligonucleotide-based enzyme capture assay for the sensitive detection of thrombin was developed in plasma, under routine clinical conditions [194].
Fig. 10.
General principle of the oligonucleotide-based enzyme capture assay for thrombin detection developed by Pötzsch et al. [194]. a) During the blood sampling process, an anticoagulant buffer (citrate) containing the reversible active-site inhibitor argatroban (1) is added to the blood sample containing thrombin (2, PDB 1DWC). Complex formation between argatroban and thrombin efficiently prevents irreversible inhibition of thrombin by endogenous thrombin inhibitors. b) Wells of streptavidin-coated microtiter modules previously loaded with the 3′-biotinylated anti-thrombin aptamer HD1–22 (3) that simultaneously targets exosites I and II of thrombin are overlaid with plasma. After incubation and capturing of the argatroban-thrombin complex by HD1–22, wells are washed to remove plasma remains and reversibly bound argatroban. Subsequently, a thrombin-specific peptide substrate, bearing an AMC (7-amino-4-methylcoumarin) fluorogenic probe (H-D-CHA-Ala-Arg-AMC, 4), is added to quantitatively determine the amount of functional active thrombin captured in the wells.
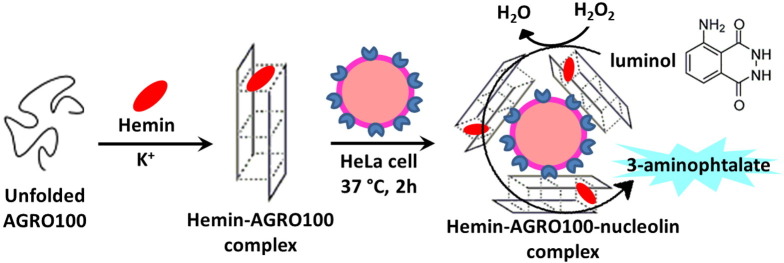
In addition to thrombin, two other well-known G4-forming aptamers named AGRO100 (AS1411) and T30695 (a Zintevir analogue), identified, as discussed before, as high-affinity binders of the proteins nucleolin and HIV-1 integrase, respectively, with effective anticancer and anti-HIV biological activity, exhibit high hemin-binding affinities comparable to that of the known aptamer (termed PS2M) selected by the in vitro evolution process. They are capable of simultaneously binding their protein target and hemin through two different modes. Most importantly, their corresponding hemin-DNA complexes reveal excellent peroxidase-like activities, higher than that of the reported hemin-PS2M DNAzyme. The potential of these systems in bioanalysis was demonstrated for AGRO100, which was applied to the chemiluminescence detection of nucleolin expressed at the surface of HeLa cells. Based on the specific AGRO100-nucleolin interaction, the surface-expressed nucleolin of HeLa cells is labeled in situ with the hemin-AGRO100 DNAzyme, and then determined in the luminol-H2O2 system (Fig. 11 ). Through this approach, the sensitive detection of total nucleolin expressed on the surface of about 6000 HeLa cells was accomplished [195].
Fig. 11.
Multifunctional aptamer AGRO100 (better known as AS1411) as a DNAzyme-based platform for the chemiluminescence detection of nucleolin expressed at the surface of HeLa cells. After the binding of hemin, the folded AGRO100 can label the cell-surface-expressed nucleolin in situ, providing an approach to sensing this protein marker in the luminol–H2O2 system.
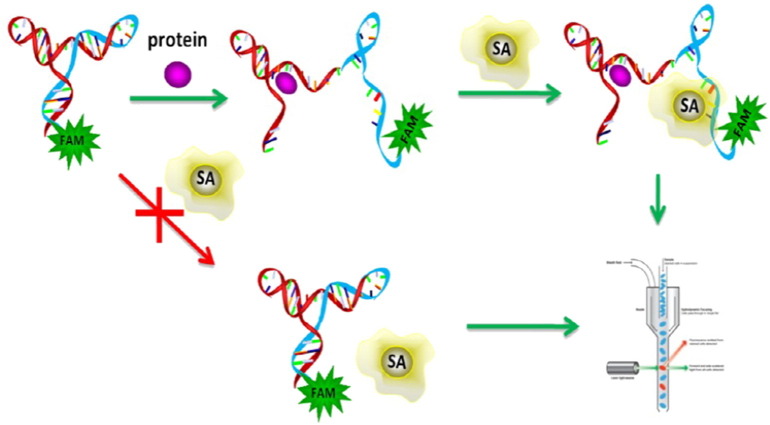
Another G4-forming aptamer probe with great potential as a useful tool for early diagnosis and prognosis of cancer is C14B1, identified by SELEX and subsequently optimized for the specific binding to anterior gradient homolog 2 (AGR2), a good cancer biomarker [196]. Structural studies have identified that the binding motif of the aptamer is an intramolecular parallel G-quadruplex structure and its structure and binding affinity are strongly dependent on the nature of the monovalent ion. For AGR2 detection, an allosteric molecular beacon, named AGR2-aMB, coupled with a fluorescence flow cytometry sensing system, was developed (Fig. 12 ). The AGR2-aMB is a single strand DNA consisting of a streptavidin (SA) aptamer sequence [197], a C14B1 sequence, a short sequence complementary to a small tract of the SA aptamer sequence, and a fluorophore. A stable hairpin structure is formed by intramolecular hybridization between the SA aptamer sequence and the complementary sequence, temporarily disabling the probe ability to bind with SA beads. Consequently, when incubated with SA beads, no probe can bind to SA beads, and the beads display very low fluorescence. In the presence of AGR2, however, the C14B1 sequence in the loop of aMB binds to the target sequence, in turn disrupting the hairpin structure to release the SA binding sequence, and thereby activating the binding affinity of the probe to SA beads. Thus, the AGR2-bound probe will bind to SA beads, which will strongly fluoresce due to FAM labeling of the probe. Target molecules can be detected and quantified by reading the fluorescence intensity of SA beads through a flow cytometer (Fig. 12).
Fig. 12.
Working principle of an allosteric molecular beacon probe for sensitive and selective detection of AGR2 based on fluorescent flow cytometry analysis.
(reproduced from ref. [196])
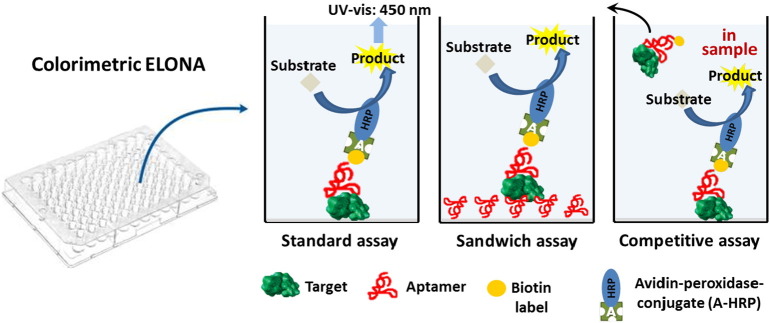
Recently, the possibility to detect bacterial pathogens by a G4-forming aptamer, selected for Protein A by the FluMag-SELEX process [198], was evaluated in an integrated assay called ELONA (Enzyme-Linked OligoNucleotide Assay) [199], for the recognition of intact bacterial cells of Staphylococcus aureus presenting Protein A on their surface (Fig. 13 ). The protein target is a typical component of the cell wall of this gram-positive bacterium and also represents one of its virulence factors, rendering Protein A a suitable target for its detection. Even if the functionality of the aptamer was based on a very complex and highly variable structure, potentially forming dimeric and multimeric G-quadruplex assembly, the full-length aptamer and one of its truncated variants were proved to specifically bind to Protein A-expressing intact cells of S. aureus, giving a positive signal in the assay.
Fig. 13.
Schematic representation of different ELONA (Enzyme-Linked OligoNucleotide Assay) formats used for aptamer-based protein detection.
ELONA was performed by immobilization of native or recombinant Protein A in microtiter plates followed by addition of 3′-biotinylated aptamer PA#2/8 for binding and subsequent washing of the unbound aptamer (Fig. 13a). It was demonstrated that PA#2/8 strongly binds to native and recombinant Protein A and clearly distinguishes its target from functional related proteins like Protein G and Protein L, which were not recognized. Remarkably, the aptamer does not bind to human serum albumin (HSA) used as unspecific control protein. ExtrAvidin-peroxidase conjugate was then applied to each well, following a washing step (Fig. 13a). Successively, a 3,3′,5,5′-tetramethylbenzidine (TMB) solution was added to each well and the optical density at 450 nm was measured. Similar experiments were performed by coating the wells with bacterial cells of Staphylococcus aureus to assess the binding ability of aptamer PA#2/8 and truncated variants to Protein A in an intact cell context. The results of the ELONA assay clearly demonstrate the specific recognition and binding of the aptamer to the cells via Protein A. PA#2/8 and its truncated variant PA#2/8[S1–58] were able to differentiate between the Protein A-producing S. aureus Cowan strain and the Protein A-deficient Wood46 strain (both as formaldehyde-fixed cells) or living E. coli as the negative control. Improvements of the ELONA assay in terms of binding results will be obtained by development of a sandwich or competitive assays (Fig. 13b and c), which could avoid the immobilization of the target cells and improve the assay handling.
5. Conclusions and perspectives
In conclusion, G4-based aptamers have been identified against a wide variety of protein targets. The discovery of many G4-forming aptamers, able to recognize even very different proteins, fully demonstrated that such an unusual nucleic acid structure, either DNA- or RNA-based, may contain efficient recognition elements for several unrelated protein systems, spanning from the coagulation cascade to spongiform encephalopathy. Despite having an overall similar scaffold, a large variety of different structural arrangements are available for G-quadruplexes, accounting for their remarkable diversity in the target recognition events, also ensuring high specificities and binding affinities.
The first aptamer identified was the thrombin binding aptamer (TBA15), forming an intramolecular antiparallel chair-like G-quadruplex with two planes of guanine tetrads, able to inhibit the thrombin-catalyzed polymerization of fibrinogen [117]. Although TBA – in its unmodified version – did not prove to be clinically acceptable as an anticoagulant, it is still extensively used as a model G-quadruplex structure in fundamental, methodological and ‘proof-of-concept’ works [117], [118], [172]. Since these original studies, the G-quadruplex structure has been observed as main conformational feature in a variety of aptamer selection experiments, clear evidence that this structural motif is among the most informative and plastic single-stranded nucleic acid structures which can be identified by in vitro selection.
A significant number of selective cancer cell-binding aptamers adopt the parallel G4 topology. Due to the high polymorphism of G4 structures, on one side, and the relative paucity of structural studies on biologically relevant G-rich oligonucleotides, on the other side, no clear structure-activity relationship can be drawn to correlate a given conformation with a specific bioactivity. However, further advances in the knowledge of the structural features of the most biologically relevant G4-forming oligonucleotides will allow identifying G4 scaffolds essential for the design of optimized, specific therapeutic aptamers. To this purpose, an integrated approach for novel aptamer design, combining combinatorial methods, i.e. SELEX, with various rational design methods, including the modular assembly approach, is highly desirable.
It is noteworthy that, due to the G4 motif abundance in the human genome [160] and the diverse regulatory roles of G4s [161], several oligonucleotide sequences taken from genomic G4 motifs (particularly those located in gene promoters) can be used as potential aptamers. Thus, with the aid of computational methods [such as the Quadruplex forming G-rich Sequences (QGRS) mapper] able to identify G4-forming sequences in the genomes on the basis of the primary sequence input [200], [201], many potential G4-based aptamers acting as decoy molecules can be developed and tested for a specific protein target of biological relevance. Rational design of G4-forming decoys appears to be a promising approach in anticancer studies [167]. Likewise, rapid progress in the studies of the G4-forming sites of HIV-1, Epstein-Barr Virus and Papilloma Virus genomes and their participation in viral life cycles will facilitate the development of new G4-based antiviral agents [96], [97]. More genomic G4-inspired aptamers will be hopefully tested in the near future.
The relatively low number of aptamers presently approved or in clinical trials might leave the reader with the impression that these compounds represent interesting biochemical tools, but hold poor promise for real therapeutic applications. We should, however, bear in mind that aptamer technology for biomedical use is still relatively young, since the underlying basic knowledge is little over 25 years old [5]. Hence, much more has to be learned to fully master the tools that will eventually allow a mature development of aptamer-based drugs. Nonetheless, the existing data, starting from Macugen's approval as a drug, indicate that the research in the field (particularly the G4 area) is properly addressed and will soon bear a number of relevant and concrete applications.
Furthermore, the versatility and plasticity of aptamers in G-quadruplex form can be fruitfully exploited in various detection assays [179], [190], [192], [193], [194], [195]. Many analytical methods make use of thrombin as target and TBA as a proof-of-principle of the proposed analytical method. The literature has plenty of research studies developing both very complicated multi-recognition systems and rapid, easy systems for the detection of thrombin. Indeed, in almost all the research works describing a new TBA-based sensing system, the authors claim that the realized assay can be potentially extended to other G4-forming aptamers for whatever target. Therefore, assays for the selective detection of other molecular targets than thrombin, using G4-forming aptamers, can be expected. However, we critically need high-resolution structures of other G4-forming aptamers in complex with their targets to fully answer the question whether TBA is typical or atypical, and how (and if) other G4-forming aptamers can afford the same high target specificity and binding affinity displayed by TBAs. New structures will help G4-forming aptamers to meet their therapeutic promise and analytical potential, leading to a new chapter in DNA G-quadruplex research.
Transparency document
Transparency document
Acknowledgements
Funding: This work was supported by the Italian Association for Cancer Research (AIRC) (IG 2015 n. 17037 to D.M.)
Footnotes
This article is part of a Special Issue entitled "G-quadruplex" Guest Editor: Dr. Concetta Giancola and Dr. Daniela Montesarchio.
The Transparency document associated with this article can be found, in the online version.
References
- 1.Stoltenburg R., Reinemann C., Strehlitz B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Ellington A.D., Szostak J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 5.Sullenger A.B. Aptamers coming of age at twenty-five. Nucleic Acid Ther. 2016;26:119. doi: 10.1089/nat.2016.29001.sul. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee J., Nilsen-Hamilton M. Aptamers: multifunctional molecules for biomedical research. J. Mol. Med. 2013;91:1333–1342. doi: 10.1007/s00109-013-1085-2. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez L.I., Machado I., Schafer T., Hernandez F.J. Aptamers overview: selection, features and applications. Curr. Top. Med. Chem. 2015;15:1066–1081. doi: 10.2174/1568026615666150413153717. [DOI] [PubMed] [Google Scholar]
- 8.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier K.E., Levy M. From selection hits to clinical leads: progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016;5:16014–16023. doi: 10.1038/mtm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan W., Wang H., Chen Y., Zhang X., Zhu H., Yang C., Yang R., Liu C. Molecular aptamers for drug delivery. Trends Biotechnol. 2011;29:634–640. doi: 10.1016/j.tibtech.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho E.J., Lee J.W., Ellington A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y.S., Raston N.H., Gu M.B. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016;76:2–19. doi: 10.1016/j.bios.2015.06.040. [DOI] [PubMed] [Google Scholar]
- 14.Sun H., Tan W., Zu Y. Aptamers: versatile molecular recognition probes for cancer detection. Analyst. 2016;141:403–415. doi: 10.1039/c5an01995h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang D., Shigdar S., Qiao G., Wang T., Kouzani A.Z., Zhou S.F., Kong L., Li Y., Pu C., Duan W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine. Theranostics. 2015;5:23–42. doi: 10.7150/thno.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuwahara M. Progress in chemically modified nucleic acid aptamers. In: Erdmann V.A., Volker A., Markiewicz W.T., Barciszewski J., editors. Chemical Biology of Nucleic Acids. RNA Technologies; 2014. [Google Scholar]
- 17.Wang R.E., Wu H., Niu Y., Cai J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011;18:4126–4138. doi: 10.2174/092986711797189565. [DOI] [PubMed] [Google Scholar]
- 18.Vu M.M., Jameson N.E., Masuda S.J., Lin D., Larralde-Ridaura R., Lupták A. Convergent evolution of adenosine aptamers spanning bacterial, human, and random sequences revealed by structure-based bioinformatics and genomic SELEX. Chem. Biol. 2012;19:1247–1254. doi: 10.1016/j.chembiol.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto Y., Nakamura Y., Ohuchi S. HEXIM1-binding elements on mRNAs identified through transcriptomic SELEX and computational screening. Biochimie. 2012;94:1900–1909. doi: 10.1016/j.biochi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E.N. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keefe A.D., Cload S.T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 22.Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.Y., McLaughlin S.H., Herdewijn P., Holliger P. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolle F., Brändle G.M., Matzner D., Mayer G. A versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed. Eng. 2015;54:10971–10974. doi: 10.1002/anie.201503652. [DOI] [PubMed] [Google Scholar]
- 24.Meek K.N., Rangel A.E., Heemstra J.M. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods. 2016;106:29–36. doi: 10.1016/j.ymeth.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Laos R., Thomson J.M., Benner S.A. DNA polymerases engineered by directed evolution to incorporate non-standard nucleotides. Front. Microbiol. 2014;5:565. doi: 10.3389/fmicb.2014.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz H.J., Gochi A.M., Chia H.E., Ogonowsky A.L., Chiang S., Filipovic N., Weiden A.G., Hadley E.E., Gabriel S.E., Leconte A.M. Taq DNA polymerase mutants and 2′-modified sugar recognition. Biochemistry. 2015;54:5999–6008. doi: 10.1021/acs.biochem.5b00689. [DOI] [PubMed] [Google Scholar]
- 27.Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 28.Sefah K., Yang Z., Bradley K.M., Hoshika S., Jiménez E., Zhang L., Zhu G., Shanker S., Yu F., Turek D., Tan W., Benner S.A. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z., Durante M., Glushakova L.G., Sharma N., Leal N.A., Bradley K.M., Chen F., Benner S.A. Conversion strategy using an expanded genetic alphabet to assay nucleic acids. Anal. Chem. 2013;85:4705–4712. doi: 10.1021/ac400422r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt K.S., Borkowski S., Kurreck J., Stephens A.W., Bald R., Hecht M., Friebe M., Dinkelborg L., Erdmann V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egli M., Herdewijn P. Wiley; 2012. Chemistry and Biology of Artificial Nucleic Acids. [Google Scholar]
- 32.Vater A., Klussmann S. Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics. Drug Discov. Today. 2015;20:147–155. doi: 10.1016/j.drudis.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Rohloff J.C., Gelinas A.D., Jarvis T.C., Ochsner U.A., Schneider D.J., Gold L., Janjic N. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopinath S.C. Methods developed for SELEX. Anal. Bioanal. Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- 35.Mendonsa S.D., Bowser M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2003;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 36.Berezovski M., Drabovich A., Krylova S.M., Musheev M., Okhonin V., Petrov A., Krylov S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures: a universal tool for development of aptamers. J. Am. Chem. Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 37.Drabovich A., Berezovski M., Krylov S.N. Selection of smart aptamers by equilibrium capillary electrophoresis of equilibrium mixtures (ECEEM) J. Am. Chem. Soc. 2005;127:11224–11225. doi: 10.1021/ja0530016. [DOI] [PubMed] [Google Scholar]
- 38.Zhaofeng L., Hongmin Z., Jiang H., Ou H., Li X., Zhang L. Development of a fraction collection approach in capillary electrophoresis SELEX for aptamer selection. Analyst. 2015;140:2664–2670. doi: 10.1039/c5an00183h. [DOI] [PubMed] [Google Scholar]
- 39.Musheev M.U., Krylov S.N. Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue. Anal. Chim. Acta. 2006;564:91–96. doi: 10.1016/j.aca.2005.09.069. [DOI] [PubMed] [Google Scholar]
- 40.Kanagawa T. Bias and artifacts in multitemplate polymerase chain reactions (PCR) J. Biosci. Bioeng. 2003;96:317–323. doi: 10.1016/S1389-1723(03)90130-7. [DOI] [PubMed] [Google Scholar]
- 41.Kanoatov M., Krylov S. DNA adsorption to the reservoir walls causing irreproducibility in studies of protein-DNA interactions by methods of kinetic capillary electrophoresis. Anal. Chem. 2011;83:8041–8045. doi: 10.1021/ac202048y. [DOI] [PubMed] [Google Scholar]
- 42.Yufa R., Krylova S.M., Bruce C., Bagg E.A., Schofield C.J., Krylov S.N. Emulsion PCR significantly improves nonequilibrium capillary electrophoresis of equilibrium mixtures-based aptamer selection: allowing for efficient and rapid selection of aptamer to unmodified ABH2 protein. Anal. Chem. 2015;87:1411–1419. doi: 10.1021/ac5044187. [DOI] [PubMed] [Google Scholar]
- 43.Schutze T., Wilhelm B., Greiner N., Braun H., Peter F., Morl M., Erdmann V.A., Lehrach H., Konthur Z., Menger M., Arndt P.F., Glokler J. Probing the SELEX process with next-generation sequencing. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoinka J., Zotenko E., Friedman A., Sauna Z.E., Przytycka T.M. Identification of sequence-structure RNA binding motifs for SELEX-derived aptamers. Bioinformatics. 2012;28:215–223. doi: 10.1093/bioinformatics/bts210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoinka J., Berezhnoy A., Sauna Z.E., Gilboa E., Przytycka T.M. AptaCluster — a method to cluster HT-SELEX aptamer pools and lessons from its application. Res. Comput. Mol. Biol. 2014;8394:115–128. doi: 10.1007/978-3-319-05269-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alam K.K., Chang J.L., Burke D.H. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papavassiliou A.G. Determination of a transcription factor-binding site by nuclease protection footprinting onto southwestern blots. Methods Mol. Biol. 2009;543:201–218. doi: 10.1007/978-1-60327-015-1_14. [DOI] [PubMed] [Google Scholar]
- 48.Chodosh L.A. UV crosslinking of proteins to nucleic acids. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb1205s36. [DOI] [PubMed] [Google Scholar]
- 49.Wilkinson K.A., Merino E.J., Weeks K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 50.Lis H., Sharon N. Protein glycosylation: structural and functional aspects. Eur. J. Biochem. 1993;218:1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- 51.Lyu Y., Chen G., Shangguan D., Zhang L., Wan S., Wu Y., Zhang H., Duan L., Liu C., You M., Wang J., Tan W. Generating cell targeting aptamers for nanotheranostics using cell-SELEX. Theranostics. 2015;6:1440–1452. doi: 10.7150/thno.15666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dua P., Kim S., Lee D.K. Nucleic acid aptamers targeting cell-surface proteins. Methods. 2011;54:215–225. doi: 10.1016/j.ymeth.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Cerchia L., Esposito C.L., Jacobs A.H., Tavitian B., de Franciscis V. Differential SELEX in human glioma cell lines. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Li S., Xu H., Ding H., Huang Y., Cao X., Yang G., Li J., Xie Z., Meng Y., Li X., Zhao Q., Shen B., Shao N. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J. Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- 55.Sun H., Zhu X., Lu P.Y., Rosato R.R., Tan W., Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hicke B.J., Marion C., Chang Y.F., Gould T., Lynott C.K., Parma D., Schmidt P.G., Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 57.Yan A., Levy M. Cell internalization SELEX: in vitro selection for molecules that internalize into cells. Methods Mol. Biol. 2014;1103:241–265. doi: 10.1007/978-1-62703-730-3_18. [DOI] [PubMed] [Google Scholar]
- 58.Gourronc F.A., Rockey W.M., Thiel W.H., Giangrande P.H., Klingelhutz A.J. Identification of RNA aptamers that internalize into HPV-16 E6/E7 transformed tonsillar epithelial cells. Virology. 2013;446:325–333. doi: 10.1016/j.virol.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camorani S., Esposito C.L., Rienzo A., Catuogno S., Iaboni M., Condorelli G., de Franciscis V., Cerchia L. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRß aptamer. Mol. Ther. 2014;22:828–841. doi: 10.1038/mt.2013.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iaboni M., Fontanella R., Rienzo A., Capuozzo M., Nuzzo S., Santamaria G., Catuogno S., Condorelli G., de Franciscis V., Esposito C.L. Targeting insulin receptor with a novel internalizing aptamer. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mayer G., Ahmed M.S., Dolf A., Endl E., Knolle P.A., Famulok M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 62.Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 63.Gatto B., Palumbo M., Sissi C. Nucleic acid aptamers based on the G-quadruplex structure: therapeutic and diagnostic potential. Curr. Med. Chem. 2009;16:1248–1265. doi: 10.2174/092986709787846640. [DOI] [PubMed] [Google Scholar]
- 64.Tucker W.O., Shum K.T., Tanner J.A. G-quadruplex DNA aptamers and their ligands: structure, function and application. Curr. Pharm. Des. 2012;18:2014–2026. doi: 10.2174/138161212799958477. [DOI] [PubMed] [Google Scholar]
- 65.Neidle S., Balasubramanian S. The Royal Society of Chemistry; Cambridge: 2006. Quadruplex Nucleic Acids. [Google Scholar]
- 66.Spindler L., Fritzsche W. The Royal Society of Chemistry; Cambridge: 2012. Guanine Quartets: Structure and Application. [Google Scholar]
- 67.Mukundan V.T., Phan A.T. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013;135:5017–5028. doi: 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- 68.Tran P.L., De Cian A., Gros J., Moriyama R., Mergny J.L. Tetramolecular quadruplex stability and assembly. Top. Curr. Chem. 2013;330:243–273. doi: 10.1007/128_2012_334. [DOI] [PubMed] [Google Scholar]
- 69.Malgowska M., Gudanis D., Kierzek R., Wyszko E., Gabelica V., Gdaniec Z. Distinctive structural motifs of RNA G-quadruplexes composed of AGG, CGG and UGG trinucleotide repeats. Nucleic Acids Res. 2014;42:10196–10207. doi: 10.1093/nar/gku710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X.M., Zheng K.W., Zhang J.Y., Liu H.H., He Y.D., Yuan B.F., Hao Y.H., Tan Z. Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc. Natl. Acad. Sci. U. S. A. 2015;112:14581–14586. doi: 10.1073/pnas.1516925112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Virgilio A., Esposito V., Citarella G., Mayol L., Galeone A. Structural investigations on the anti-HIV G-quadruplex-forming oligonucleotide TGGGAG and its analogues: evidence for the presence of an A-tetrad. Chem. Biochem. 2012;13:2219–2224. doi: 10.1002/cbic.201200481. [DOI] [PubMed] [Google Scholar]
- 72.Phan A.T., Kuryavyi V., Ma J.B., Faure A., Andreola M.L., Patel J.D. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji X., Sun H., Zhou H., Xiang J., Tang Y., Zhao C. Research progress of RNA quadruplex. Nucleic Acid Ther. 2011;21:185–200. doi: 10.1089/nat.2010.0272. [DOI] [PubMed] [Google Scholar]
- 74.Collie G.W., Parkinson G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 75.Santosh B., Yadava P.K. Nucleic acid aptamers: research tools in disease diagnostics and therapeutics. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/540451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qu J., Yu S., Zheng Y., Zheng Y., Yang H., Zhang J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2016 doi: 10.1007/s00018-016-2345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drolet D.W., Green L.S., Gold L., Janjic N. Fit for the eye: aptamers in ocular disorders. Nucleic Acid Ther. 2016;26:127–146. doi: 10.1089/nat.2015.0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu Y., Liang C., Lv Q., Li D., Xu X., Liu B., Lu A., Zhang G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016;17:358–376. doi: 10.3390/ijms17030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Becker R.C., Povsic T., Cohen M.G., Rusconi C.P., Sullenger B. Nucleic acid aptamers as antithrombotic agents: opportunities in extracellular therapeutics. Thromb. Haemost. 2010;103:586–595. doi: 10.1160/TH09-10-0716. [DOI] [PubMed] [Google Scholar]
- 80.Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soundararajan S., Wang L., Sridharan V., Chen W., Courtenay-Luck N., Jones D., Spicer E.K., Fernandes D.J. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 2009;76:984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu X., Chen J., Wu M., Zhao J.X. Aptamers: active targeting ligands for cancer diagnosis and therapy. Theranostics. 2015;5:322–344. doi: 10.7150/thno.10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reyes-Reyes E.M., Šalipur F.R., Shams M., Forsthoefel M.K., Bates P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015;9:1392–1405. doi: 10.1016/j.molonc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosenberg J.E., Bambury R.M., Van Allen E.M., Drabkin H.A., Lara P.N., Jr., Harzstark A.L., Wagle N., Figlin R.A., Smith G.W., Choueiri T., Erlandsson F., Laber D.A. A phase II trial of the nucleolin-targeted DNA aptamer AS1411 in metastatic refractory renal cell carcinoma. Investig. New Drugs. 2014;32:178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jing N., Li Y., Xu X., Sha W., Li P., Feng L., Tweardy D.J. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003;22:685–696. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- 86.Weerasinghe P., Garcia G.E., Zhu Q., Yuan P., Feng L., Mao L., Jing N. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int. J. Oncol. 2007;31:129–136. [PubMed] [Google Scholar]
- 87.Weerasinghe P., Li Y., Guan Y., Zhang R., Tweardy D.J., Jing N. T40214/PEI complex, a potent therapeutics for prostate cancer that targets STAT3 signaling. Prostate. 2008;68:1430–1442. doi: 10.1002/pros.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neel B.G., Gu H., Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signalling. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 89.Hu J., Wu J., Li C., Zhu L., Zhang W.Y., Kong G., Lu Z., Yang C.J. A G-quadruplex aptamer inhibits the phosphatase activity of oncogenic protein Shp2 in vitro. Chembiochem. 2011;12:424–430. doi: 10.1002/cbic.201000470. [DOI] [PubMed] [Google Scholar]
- 90.Zhao X.Q., Wu J., Liang J.H., Yan J.W., Zhu Z., Yang C.J., Mao B.W. Single-molecule force spectroscopic studies on intra- and intermolecular interactions of G-quadruplex aptamer with target Shp2 protein. J. Phys. Chem. B. 2012;116:11397–11404. doi: 10.1021/jp303518b. [DOI] [PubMed] [Google Scholar]
- 91.Ferrara N., Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr. Rev. 1997;18:4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- 92.Nosaka Y., Sode K., Ikebukuro K. Screening and improvement of an anti-VEGF DNA aptamer. Molecules. 2010;15:215–225. doi: 10.3390/molecules15010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marusic M., Veedu R.N., Wengel J., Plavec J. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013;41:9524–9536. doi: 10.1093/nar/gkt697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nonaka Y., Yoshida W., Abe K., Ferri S., Schulze H., Bachmann T.T., Ikebukuro K. Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal. Chem. 2012;85:1132–1137. doi: 10.1021/ac303023d. [DOI] [PubMed] [Google Scholar]
- 95.Wandtke T., Wozniak J., Kopinski P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7:751–780. doi: 10.3390/v7020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Metifiot M., Amrane S., Litvak S., Andreola M.L. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014;42:12352–12366. doi: 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Musumeci D., Riccardi C., Montesarchio D. G-Quadruplex forming oligonucleotides as anti-HIV agents. Molecules. 2015;20:17511–17532. doi: 10.3390/molecules200917511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wyatt J.R., Vickers T.A., Roberson J.L., Buckheit R.W., Jr., Klimkait T., DeBaets E., Davis P.W., Rayner B., Imbach J.L., Ecker D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buckheit R.W., Jr., Roberson J.L., Lackman-Smith C., Wyatt J.R., Vickers T.A., Ecker D.J. Potent and specific inhibition of HIV envelope-mediated cell fusion and virus binding by G quartet-forming oligonucleotide (ISIS 5320) AIDS Res. Hum. Retrovir. 1994;10:1497–1506. doi: 10.1089/aid.1994.10.1497. [DOI] [PubMed] [Google Scholar]
- 100.Stoddart C.A., Rabin L., Hincenbergs M., Moreno M., Linquist-Stepps V., Leeds J.M., Truong L.A., Wyatt J.R., Ecker D.J., McCune J.M. Inhibition of human immunodeficiency virus type 1 infection in SCID-hu Thy/Liv mice by the G-quartet-forming oligonucleotide, ISIS 5320. Antimicrob. Agents Chemother. 1998;42:2113–2115. doi: 10.1128/aac.42.8.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hotoda H., Koizumi M., Koga R., Kaneko M., Momota K., Ohmine T., Furukawa H., Agatsuma T., Nishigaki T., Sone J., Tsutumi S., Kosaka T., Abe K., Kimura S., Shimada K. Biologically active oligodeoxyribonucleotides. 5. 5′-End-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 1998;41:3655–3663. doi: 10.1021/jm970658w. [DOI] [PubMed] [Google Scholar]
- 102.D'Atri V., Oliviero G., Amato J., Borbone N., D'Errico S., Mayol L., Piccialli V., Haider S., Hoorelbeke B., Balzarini J., Piccialli G. New anti-HIV aptamers based on tetra-end-linked DNA G-quadruplexes: effect of the base sequence on anti-HIV activity. Chem. Commun. 2012;48:9516–9518. doi: 10.1039/c2cc34399a. [DOI] [PubMed] [Google Scholar]
- 103.D'Onofrio J., Petraccone L., Erra E., Martino L., Di Fabio G., De Napoli L., Giancola C., Montesarchio D. 5′-Modified G-quadruplex forming oligonucleotides endowed with anti-HIV activity: synthesis and biophysical properties. Bioconjug. Chem. 2007;18:1194–1204. doi: 10.1021/bc070062f. [DOI] [PubMed] [Google Scholar]
- 104.Di Fabio G., D'Onofrio J., Chiapparelli M., Hoorelbeke B., Montesarchio D., Balzarinib J., De Napoli L. Discovery of novel anti-HIV active G-quadruplex-forming oligonucleotides. Chem. Commun. 2011;47:2363–2365. doi: 10.1039/c0cc04751a. [DOI] [PubMed] [Google Scholar]
- 105.Nisole S., Said E.A., Mische C., Prevost M.C., Krust B., Bouvet P., Bianco A., Briand J.P., Hovanessian A.G. The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J. Biol. Chem. 2002;277:20877–20886. doi: 10.1074/jbc.M110024200. [DOI] [PubMed] [Google Scholar]
- 106.Perrone R., Butovskaya E., Lago S., Garzino-Demo A., Pannecouque C., Palù G., Richter S.N. The G-quadruplex-forming aptamer AS1411 potently inhibits HIV-1attachment to the host cell. Int. J. Antimicrob. Agents. 2016;47:311–316. doi: 10.1016/j.ijantimicag.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Metifiot M., Amrane S., Mergny J.L., Andreola M.-L. Anticancer molecule AS1411 exhibits low nanomolar antiviral activity against HIV-1. Biochimie. 2015;118:173–175. doi: 10.1016/j.biochi.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 108.Metifiot M., Leon O., Tarrago-Litvak L., Litvak S., Andreola M.L. Targeting HIV-1 integrase with aptamers selected against the purified RNase H domain of HIV-1 RT. Biochimie. 2005;87:911–919. doi: 10.1016/j.biochi.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 109.De Soultrait V.R., Lozach P.Y., Altmeyer R., Tarrago-Litvak L., Litvak S., Andreola M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- 110.Mukundan V.T., Do N.Q., Phan A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011;39:8984–8991. doi: 10.1093/nar/gkr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marchand C., Maddali K., Metifiot M., Pommier Y. HIV-1 IN inhibitors: 2010 update and perspectives. Curr. Top. Med. Chem. 2009;9:1016–1037. doi: 10.2174/156802609789630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu S., Abbondanzieri E.A., Rausch J.W., le Grice S.F.J., Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abbondanzieri E.A., Bokinsky G., Rausch J.W., Zhang J.X., le Grice S.F.J., Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Michalowski D., Chitima-Matsiga R., Held D.M., Burke D.H. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008;36:7124–7135. doi: 10.1093/nar/gkn891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Andreola M.L., Pileur F., Calmels C., Ventura M., Tarrago-Litvak L., Toulme J.J., Litvak S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry. 2001;40:10087–10094. doi: 10.1021/bi0108599. [DOI] [PubMed] [Google Scholar]
- 116.Stubbs M.T., Bode W. The clot thickens: clues provided by thrombin structure. Trends Biochem. Sci. 1995;20:23–28. doi: 10.1016/s0968-0004(00)88945-8. [DOI] [PubMed] [Google Scholar]
- 117.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 118.Musumeci D., Montesarchio D. Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer. Pharmacol. Ther. 2012;136:202–215. doi: 10.1016/j.pharmthera.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 119.2005. http://www.hcp.com/content5788.html (accessed 18.09.2016)
- 120.Tasset D.M., Kubik M.F., Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 121.Russo Krauss I., Pica A., Merlino A., Mazzarella L., Sica F. Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta Crystallogr. D Biol. Crystallogr. 2013;69:2403–2411. doi: 10.1107/S0907444913022269. [DOI] [PubMed] [Google Scholar]
- 122.Trapaidze A., Hérault J.P., Herbert J.M., Bancaud A., Gué A.M. Investigation of the selectivity of thrombin-binding aptamers for thrombin titration in murine plasma. Biosens. Bioelectron. 2016;78:58–66. doi: 10.1016/j.bios.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 123.Muller J., Wulffen B., Potzsch B., Mayer G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. Chembiochem. 2007;8:2223–2226. doi: 10.1002/cbic.200700535. [DOI] [PubMed] [Google Scholar]
- 124.Muller J., Freitag D., Mayer G., Potzsch B. Anticoagulant characteristics of HD1-22, a bivalent aptamer that specifically inhibits thrombin and prothrombinase. J. Thromb. Haemost. 2008;6:2105–2112. doi: 10.1111/j.1538-7836.2008.03162.x. [DOI] [PubMed] [Google Scholar]
- 125.Zavyalova E., Samoylenkova N., Revishchin A., Golovin A., Pavlova G., Kopylov A. Evaluation of antithrombotic activity of thrombin DNA aptamers by a murine thrombosis model. PLoS One. 2014;9:107–113. doi: 10.1371/journal.pone.0107113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zavyalova E., Golovin A., Timoshenko T., Babiy A., Pavlova G., Kopylov A. DNA aptamers for human thrombin with high anticoagulant activity demonstrate target- and species-specificity. Curr. Med. Chem. 2012;19:5232–5237. doi: 10.2174/092986712803530575. [DOI] [PubMed] [Google Scholar]
- 127.Buff M.C., Schäfer F., Wulffen B., Müller J., Pötzsch B., Heckel A., Mayer G. Dependence of aptamer activity on opposed terminal extensions: improvement of light-regulation efficiency. Nucleic Acids Res. 2010;38:2111–2118. doi: 10.1093/nar/gkp1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zavyalova E., Tagiltsev G., Reshetnikov R., Arutyunyan A., Kopylov A. Cation coordination alters the conformation of a thrombin-binding G-quadruplex DNA aptamer that affects inhibition of thrombin. Nucleic Acid Ther. 2016;26:299–308. doi: 10.1089/nat.2016.0606. [DOI] [PubMed] [Google Scholar]
- 129.Shum K.T., Chan C., Leung C.M., Tanner J.A. Identification of a DNA aptamer that inhibits sclerostin's antagonistic effect on Wnt signaling. Biochem. J. 2011;434:493–501. doi: 10.1042/BJ20101096. [DOI] [PubMed] [Google Scholar]
- 130.Linden R., Martins V.R., Prado M.A., Cammarota M., Izquierdo I., Brentani R.R. Physiology of the prion protein. Physiol. Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 131.Mashima T., Matsugami A., Nishikawa F., Nishikawa S., Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mashima T., Nishikawa F., Kamatari Y.O., Fujiwara H., Saimura M., Nagata T., Kodaki T., Nishikawa S., Kuwata K., Katahira M. Anti-prion activity of an RNA aptamer and its structural basis. Nucleic Acids Res. 2013;41:1355–1362. doi: 10.1093/nar/gks1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shum K.T., Lui E.L.H., Wong S.C.K., Yeung P., Sam L., Wang Y., Watt R.M., Tanner J.A. Aptamer-mediated inhibition of mycobacterium tuberculosis polyphosphate kinase 2. Biochemist. 2011;50:3261–3271. doi: 10.1021/bi2001455. [DOI] [PubMed] [Google Scholar]
- 134.2016. https://clinicaltrials.gov/ (accessed 15.09.2016)
- 135.Reyes-Reyes E.M., Teng Y., Bates P.J. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shieh Y.A., Yang S.J., Wei M.F., Shieh M.J. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano. 2010;4:1433–1442. doi: 10.1021/nn901374b. [DOI] [PubMed] [Google Scholar]
- 137.Le Trinh T., Zhu G., Xiao X., Puszyk W., Sefah K., Wu Q., Tan W., Liu C. A synthetic aptamer-drug adduct for targeted liver cancer therapy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu J., Wei T., Zhao J., Huang Y., Deng H., Kumar A., Wang C., Liang Z., Ma X., Liang X.J. Multifunctional aptamer-based nanoparticles for targeted drug delivery to circumvent cancer resistance. Biomaterials. 2016;91:44–56. doi: 10.1016/j.biomaterials.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 139.Liao Z.X., Chuang E.Y., Lin C.C., Hoc Y.C., Lin K.J., Cheng P.Y., Chen K.J., Wei H.J., Sung H.W. An AS1411 aptamer-conjugated liposomal system containing a bubble-generating agent for tumor-specific chemotherapy that overcomes multidrug resistance. J. Control. Release. 2015;208:42–51. doi: 10.1016/j.jconrel.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 140.Guo J., Gao X., Su L., Xia H., Gu G., Pang Z., Jiang X., Yao L., Chen J., Chen H. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32:8010–8020. doi: 10.1016/j.biomaterials.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 141.Meyer C., Eydeler K., Magbanua E., Zivkovic T., Piganeau N., Lorenzen I., Grötzinger J., Mayer G., Rose-John S., Hahn U. Interleukin-6 receptor specific RNA aptamers for cargo delivery into target cells. RNA Biol. 2012;9:67–80. doi: 10.4161/rna.9.1.18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Szameit K., Berg K., Kruspe S., Valentini E., Magbanua E., Kwiatkowski M., Chauvot de Beauchêne I., Krichel B., Schamoni K., Uetrecht C., Svergun D.I., Schlüter H., Zacharias M., Hahn U. Structure and target interaction of a G-quadruplex RNA-aptamer. RNA Biol. 2016;13:973–987. doi: 10.1080/15476286.2016.1212151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Varizhuk A., Ilyinsky N., Smirnov I., Pozmogova G. G4 aptamers: trends in structural design. Mini-Rev. Med. Chem. 2016;16:1321–1329. doi: 10.2174/1389557516666160321114715. [DOI] [PubMed] [Google Scholar]
- 144.Choi E.W., Nayak L.V., Bates P.J. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010;38:1623–1635. doi: 10.1093/nar/gkp1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dailey M.M., Miller M.C., Bates P.J., Lane A.N., Trent J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chang T., Qi C., Meng J., Zhang N., Bing T., Yang X., Cao Z., Shangguan D. General cell-binding activity of intramolecular G-quadruplexes with parallel structure. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fujita H., Imaizumi Y., Kasahara Y., Kitadume S., Ozaki H., Kuwahara M., Sugimoto N. Structural and affinity analyses of G-quadruplex DNA aptamers for camptothecin derivatives. Pharmaceuticals (Basel) 2013;6:1082–1093. doi: 10.3390/ph6091082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Arora A., Nair D.R., Maiti S. Effect of flanking bases on quadruplex stability and Watson-Crick duplex competition. FEBS J. 2009;276:3628–3640. doi: 10.1111/j.1742-4658.2009.07082.x. [DOI] [PubMed] [Google Scholar]
- 149.Smirnov I., Shafer R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 150.Bugaut A., Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Avino A., Portella G., Ferreira R., Gargallo R., Mazzini S., Gabelica V., Orozco M., Eritja R. Specific loop modifications of the thrombin-binding aptamer trigger the formation of parallel structures. FEBS J. 2014;281:1085–1099. doi: 10.1111/febs.12670. [DOI] [PubMed] [Google Scholar]
- 152.McManus S.A., Li Y. Assessing the amount of quadruplex structures present within G(2)-tract synthetic random-sequence DNA libraries. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tatarinova O., Tsvetkov V., Basmanov D., Barinov N., Smirnov I., Timofeev E., Kaluzhny D., Chuvilin A., Klinov D., Varizhuk A., Pozmogova G. Comparison of the ‘chemical’ and ‘structural’ approaches to the optimization of the thrombin-binding aptamer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dolinnaya N.G., Yuminova A.V., Spiridonova V.A., Arutyunyan A.M., Kopylov A.M. Coexistence of G-quadruplex and duplex domains within the secondary structure of 31-mer DNA thrombin-binding aptamer. J. Biomol. Struct. Dyn. 2012;30:524–531. doi: 10.1080/07391102.2012.687518. [DOI] [PubMed] [Google Scholar]
- 155.Mayer G., Muller J., Mack T., Freitag D.F., Hover T., Potzsch B., Heckel A. Differential regulation of protein subdomain activity with caged bivalent ligands. Chem. Biochem. 2009;10:654–657. doi: 10.1002/cbic.200800814. [DOI] [PubMed] [Google Scholar]
- 156.Rohrbach F., Fatthalla M.I., Kupper T., Potzsch B., Muller J., Petersen M., Pedersen E.B., Mayer G. Chemical maturation of a bivalent aptamer by single domain variation. Chem. Biochem. 2012;13:631–634. doi: 10.1002/cbic.201200015. [DOI] [PubMed] [Google Scholar]
- 157.Opazo F., Eiden L., Hansen L., Rohrbach F., Wengel J., Kjems J., Mayer G. Modular assembly of cell-targeting devices based on an uncommon G-quadruplex aptamer. Mol. Ther. Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mendelboum Raviv S., Horvath A., Aradi J., Bagoly Z., Fazakas F., Batta Z., Muszbek L., Harsfalvi J. 4-Thiodeoxyuridylate-modified thrombin aptamer and its inhibitory effect on fibrin clot formation, platelet aggregation and thrombus growth on subendothelial matrix. J. Thromb. Haemost. 2008;6:1764–1771. doi: 10.1111/j.1538-7836.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 159.Ahmad K.M., Xiao Y., Soh H.T. Selection is more intelligent than design: improving the affinity of a bivalent ligand through directed evolution. Nucleic Acids Res. 2012;40:11777–11783. doi: 10.1093/nar/gks899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Saini N., Zhang Y., Usdin K., Lobachev K.S. When secondary comes first-the importance of non-canonical DNA structures. Biochimie. 2013;95:117–123. doi: 10.1016/j.biochi.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Connor A.C., Frederick K.A., Morgan E.J., McGown L.B. Insulin capture by an insulin-linked polymorphic region G-quadruplex DNA oligonucleotide. J. Am. Chem. Soc. 2006;128:4986–4991. doi: 10.1021/ja056097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiao J., Carter J.A., Frederick K.A., McGown L.B. A genome inspired DNA ligand for the affinity capture of insulin and insulin like growth factor-2. J. Sep. Sci. 2009;32:1654–1664. doi: 10.1002/jssc.200900060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yoshida W., Saito T., Yokoyama T., Ferri S., Ikebukuro K. Aptamer selection based on G4-forming promoter region. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Membrino A., Cogoi S., Pedersen E.B., Xodo L.E. G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cogoi S., Zorzet S., Rapozzi V., Geci I., Pedersen E.B., Xodo L.E. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013;41:4049–4064. doi: 10.1093/nar/gkt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Miglietta G., Gouda A.S., Cogoi S., Pedersen E.B., Xodo L.E. Nucleic acid targeted therapy: G4 oligonucleotides downregulate HRAS in bladder cancer cells through a decoy mechanism. ACS Med. Chem. Lett. 2015;6:1179–1183. doi: 10.1021/acsmedchemlett.5b00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ilgu M., Nilsen-Hamilton M. Aptamers in analytics. Analyst. 2016;141:1551–1568. doi: 10.1039/c5an01824b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sassolas A., Blum L.J., Leca-Bouvier B.D. Homogeneous assays using aptamers. Analyst. 2011;136:257–274. doi: 10.1039/c0an00281j. [DOI] [PubMed] [Google Scholar]
- 170.Du Y., Li B., Wang E. “Fitting” makes “sensing” simple: label-free detection strategies based on nucleic acid aptamers. Acc. Chem. Res. 2013;46:203–213. doi: 10.1021/ar300011g. [DOI] [PubMed] [Google Scholar]
- 171.Chandola C., Kalme S., Casteleijn M.G., Urtti A., Neerathilingam M. Application of aptamers in diagnostics, drug-delivery and imaging. J. Biosci. 2016;41:535–561. doi: 10.1007/s12038-016-9632-y. [DOI] [PubMed] [Google Scholar]
- 172.Deng B., Lin Y., Wang C., Li F., Wang Z., Zhang H., Li X.F., Le X.C. Aptamer binding assays for proteins: the thrombin example—a review. Anal. Chim. Acta. 2014;837:1–15. doi: 10.1016/j.aca.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 173.Yan S., Huang R., Zhou Y., Zhang M., Deng M., Wang X., Weng X., Zhou X. Aptamer-based turn-on fluorescent four-branched quaternary ammonium pyrazine probe for selective thrombin detection. Chem. Commun. 2011;47:1273–1275. doi: 10.1039/c0cc02792h. [DOI] [PubMed] [Google Scholar]
- 174.Zhang X., Hu R., Shao N. Label-free sensing of thrombin based on quantum dots and thrombin binding aptamer. Talanta. 2013;107:140–145. doi: 10.1016/j.talanta.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 175.De Tito S., Morvan F., Meyer A., Vasseur J.J., Cummaro A., Petraccone L., Pagano B., Novellino E., Randazzo A., Giancola C., Montesarchio D. Fluorescence enhancement upon G-quadruplex folding: synthesis, structure, and biophysical characterization of a dansyl/cyclodextrin-tagged thrombin binding aptamer. Bioconjug. Chem. 2013;24:1917–1927. doi: 10.1021/bc400352s. [DOI] [PubMed] [Google Scholar]
- 176.Zhang L., Cui P., Zhang B., Gao F. Aptamer-based turn-on detection of thrombin in biological fluids based on efficient phosphorescence energy transfer from Mn-doped ZnS quantum dots to carbon nanodots. Chemistry. 2013;19:9242–9250. doi: 10.1002/chem.201300588. [DOI] [PubMed] [Google Scholar]
- 177.Cho S., Park L., Chong R., Kim Y.T., Lee J.H. Rapid and simple G-quadruplex DNA aptasensor with guanine chemiluminescence detection. Biosens. Bioelectron. 2014;52:310–316. doi: 10.1016/j.bios.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhengbo C., Tan Y., Zhang C., Yin L., Ma H., Nengsheng Y., Qiang H., Lin Y. A colorimetric aptamer biosensor based on cationic polymer and gold nanoparticles for the ultrasensitive detection of thrombin. Biosens. Bioelectron. 2014;56:46–50. doi: 10.1016/j.bios.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 179.Deng L., Du Y., Xu J.J., Chen H.Y. An off-on-off electrochemiluminescence approach for ultrasensitive detection of thrombin. Biosens. Bioelectron. 2014;59:58–63. doi: 10.1016/j.bios.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 180.Ahour F., Ahsani M.K. An electrochemical label-free and sensitive thrombin aptasensor based on graphene oxide modified pencil graphite electrode. Biosens. Bioelectron. 2016;86:764–769. doi: 10.1016/j.bios.2016.07.053. [DOI] [PubMed] [Google Scholar]
- 181.Travascio P., Li Y., Sen D. DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem. Biol. 1998;5:505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- 182.Cochran A.G., Schultz P.G. Peroxidase activity of an antibody-heme complex. J. Am. Chem. Soc. 1990;112:9414–9415. [Google Scholar]
- 183.Travascio P., Bennet A.J., Wang D.Y., Sen D. A ribozyme and a catalytic DNA with peroxidase activity: active sites versus cofactor-binding sites. Chem. Biol. 1999;6:779–787. doi: 10.1016/s1074-5521(99)80125-2. [DOI] [PubMed] [Google Scholar]
- 184.Travascio P., Witting P.K., Mauk A.G., Sen D. The peroxidase activity of a hemin-DNA oligonucleotide complex: free radical damage to specific guanine bases of the DNA. J. Am. Chem. Soc. 2001;123:1337–1348. doi: 10.1021/ja0023534. [DOI] [PubMed] [Google Scholar]
- 185.Kosman J., Juskowiak B. Peroxidase-mimicking DNAzymes for biosensing applications: a review. Anal. Chim. Acta. 2011;707:7–17. doi: 10.1016/j.aca.2011.08.050. [DOI] [PubMed] [Google Scholar]
- 186.Xiao Y., Pavlov V., Gill R., Bourenko T., Willner I. Lighting up biochemiluminescence by the surface self-assembly of DNA-hemin complexes. Chembiochem. 2004;5:374–379. doi: 10.1002/cbic.200300794. [DOI] [PubMed] [Google Scholar]
- 187.Liu X., Freeman R., Golub E., Willner I. Chemiluminescent and chemiluminescence resonance energy transfer (CRET) detection of DNA, metal ions, and aptamer–substrate complexes using hemin/G-quadruplexes and CdSe/ZnS quantum dots. ACS Nano. 2011;5:7648–7655. [Google Scholar]
- 188.Thirstrup D., Baird G.S. Histochemical application of a peroxidase DNAzyme with a covalently attached hemin cofactor. Anal. Chem. 2010;82:2498–2504. doi: 10.1021/ac902887j. [DOI] [PubMed] [Google Scholar]
- 189.Kong D.M., Yang W., Wu J., Li C.X., Shen H.X. Structure–function study of peroxidase-like G-quadruplex-hemin complexes. Analyst. 2010;135:321–326. doi: 10.1039/b920293e. [DOI] [PubMed] [Google Scholar]
- 190.Li T., Wang E., Dong S. G-quadruplex-based DNAzyme for facile colorimetric detection of thrombin. Chem. Commun. 2008;9:3654–3656. doi: 10.1039/b805565c. [DOI] [PubMed] [Google Scholar]
- 191.Zhang Y., Li B., Jin Y. Label-free fluorescent detection of thrombin using G-quadruplex-based DNAzyme as sensing platform. Analyst. 2011;136:3268–3273. doi: 10.1039/c1an00002k. [DOI] [PubMed] [Google Scholar]
- 192.Yang Z.H., Zhuo Y., Yuan R., Chai Y.Q. Amplified thrombin aptasensor based on alkaline phosphatase and hemin/G-quadruplex-catalyzed oxidation of 1-naphthol. ACS Appl. Mater. Interfaces. 2015;7:10308–10315. doi: 10.1021/acsami.5b00988. [DOI] [PubMed] [Google Scholar]
- 193.Xie S., Ye J., Yuan Y., Chai Y., Yuan R.A. A multifunctional hemin@metal-organic framework and its application to construct an electrochemical aptasensor for thrombin detection. Nanoscale. 2015;7:18232–18238. doi: 10.1039/c5nr04532k. [DOI] [PubMed] [Google Scholar]
- 194.Müller J., Becher T., Braunstein J., Berdel P., Gravius S., Rohrbach F., Oldenburg J., Mayer G., Potzsch B. Profiling of active thrombin in human blood by supramolecular complexes. Angew. Chem. Int. Ed. 2011;50:6075–6078. doi: 10.1002/anie.201007032. [DOI] [PubMed] [Google Scholar]
- 195.Li T., Shi L., Wang E., Dong S. Multifunctional G-quadruplex aptamers and their application to protein detection. Chemistry. 2009;15:1036–1042. doi: 10.1002/chem.200801282. [DOI] [PubMed] [Google Scholar]
- 196.Wu J., Wang C., Li X., Song Y., Wang W., Li C., Hu J., Zhu Z., Li J., Zhang W., Lu Z., Yang C.J. Identification, characterization and application of a G-quadruplex structured DNA aptamer against cancer biomarker protein anterior gradient homolog 2. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bing T., Yang X., Mei H., Cao Z., Shangguan D. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg. Med. Chem. 2010;18:1798–1805. doi: 10.1016/j.bmc.2010.01.054. [DOI] [PubMed] [Google Scholar]
- 198.Stoltenburg R., Schubert T., Strehlitz B. In vitro selection and interaction studies of a DNA aptamer targeting Protein A. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Stoltenburg R., Krafčiková P., Víglaský V., Strehlitz B. G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep. 2016;6:33812. doi: 10.1038/srep33812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kikin O., D'Antonio L., Bagga P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bedrat A., Lacroix L., Mergny J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document