Abstract
Morphological characteristics, ITS sequences, and active compounds have been used to differentiate between species of Lonicera used in the traditional Chinese medicines Flos Lonicerae Japonicae (FLJ) and Flos Lonicerae (FL). FLJ includes L. japonica whereas FJ includes Lonicera macranthoides, Lonicera hypoglauca, Lonicera confusa and Lonicera fulvotomentosa. FLJ could be distinguished from FL using four quantitative and 10 qualitative characters, ITS sequences, chlorogenic acid and luteoloside contents. Analyses revealed that L. japonica was very different from the other species. The results have implications for the identification and quality control of species of Lonicera used medicinally, suggesting that species should not be interchanged in medicinal preparations.
Keywords: Lonicera japonica, Flos Lonicerae Japonicae (FLJ), Flos Lonicerae (FL), Cfr I, Identification
Highlights
-
•
Morphological characteristics, ITS sequences and active compounds of 30 Lonicera germplasm were compared.
-
•
Flos Lonicerae Japonicae (FLJ) and Flos Lonicerae (FJ) could be distinguished.
-
•
FLJs are easy to identify from FL by Cfr I digested reaction of the ITS PCR products.
1. Introduction
Flos Lonicerae Japonicae (FLJ), the flower buds of Lonicera japonica Thunb. (Caprifoliaceae), have been used as a common traditional Chinese medicine for the treatment of exopathogenic wind-heat, epidemic febrile diseases, sores, carbuncles, furuncles, and some infectious diseases (Committee for the Pharmacopoeia of PR China, 2010, Shang et al., 2011). Recently, FLJ has been used extensively to prevent and treat some serious human and veterinary viral diseases including SARS coronavirus and H1N1 (Swine) flu virus. Therefore, FLJ has been called the ‘Plant antibiotic’ (Jiao, 2009). It has also been used as a food and as a healthy beverage, e.g. the famous Chinese herbal tea of ‘Wang Lao Ji’, all over the world (Wang, 2010). The flower buds from several species of Lonicera, such as L. macranthoides Hand. Mazz., L. fulvotomentosa Hsu., L. hypoglauca Miq., L. confusa DC., and L. similis Hemsl., are also considered as alternatives to FLJ in folk medicines and are described as ‘Jin Yin Hua’ (alternative name of FLJ) in the Chinese Pharmacopoeia (Xu et al., 1988, Committee for the Pharmacopoeia of PR China, 2000). Before 2005, L. japonica and some related species were described as the original plants of FLJ, and they were not distinguished in practical applications. Since 2005, Flos Lonicerae (FL), the flower buds of L. macranthoides Hand. Mazz., L. hypoglauca Miq., and L. confusa DC. have been listed in the Pharmacopoeia of the People's Republic of China (PPRC) (Committee for the Pharmacopoeia of PR China, 2005). In the 2010 PPRC, L. fulvotomentosa was also recorded as one of the original plants of FL. However, to date the original plants of FL and other Lonicera species have continued to be misused as FLJ in medicines, foods, and healthcare products in China.
The commercial value of FLJ in herbal medicine trading markets has increased over 400% in recent years, and over 30% of current traditional Chinese medicine prescriptions contain FLJ (Yuan et al., 2012). This increasing demand has resulted in large areas of L. japonica and related Lonicera species being cultivated with various cultivars and landraces (Hu et al., 2011), and many wild Lonicera species are also harvested as medicines or foods. However, FLJ and FL from different species, cultivars, landraces, and wild germplasm have entered the market, causing a confusing array of medicinal materials with very different qualities. This is detrimental to the development of the Lonicera-related industry. Therefore, there is an urgent need to establish a simple and effective approach to differentiating the complex original plants of FLJ and FJ.
Previous studies on the identification of L. japonica and related species have mainly focused on their morphology and histology (Xu, 1979, Xu et al., 1979, Pu et al., 2002), chemical components (Hu et al., 2011, Shang et al., 2011, Yuan et al., 2012), and various genetic molecular markers, such as RFLP (Wang et al., 2007), diagnostic PCR (Peng et al., 2009), and the psbA-trnH intergenic spacer (Sun et al., 2011). Xu (1979) described (with 15 illustrations) the botanical origin of 14 species and one subspecies of Lonicera. Glandular and non-glandular hairs were discovered as the distinguishing pharmacognostic characteristics of 20 Lonicera species (Xu et al., 1979, Xu et al., 1981). Floral morphology of L. japonica varied from that of L. hypoglauca, but not within L. japonica (Pu et al., 2002). HPLC analysis showed that ‘Nanjiang’, a variation of L. similis, had the highest content of chlorogenic acid of six different Lonicera species (Hu et al., 2011). However, the chemical components and contents of FLJ are often influenced by different habitat, harvesting time, medicinal parts, extraction methods, and whether the flowers are fresh or dry (Shang et al., 2011). Molecular markers mainly including intergenic spacers of ITS and psbA-trnH were found to identify L. japonica and related species (Wang et al., 2007, Peng et al., 2009, Chen et al., 2010, Sun et al., 2011).
The genetic relationships among L. japonica and related species are unclear when using individual morphological, chemical and molecular markers, especially for cultivars, landraces and wild germplasm within species. The effective identification and quality control of L. japonica and its related species or various germplasm within L. japonica still remain to be explored.
The objectives of this study were to (i) reveal the genetic relationships among 30 Lonicera germplasm samples through morphological characteristics, ITS sequences and active compounds; (ii) analyze the correlations among the morphological characteristics, ITS sequences and active compounds of these Lonicera germplasm samples; (iii) illustrate the differences between medicinal species of Lonicera, and confirm an effective and exercisable protocol to identify FLJ and FJ. Understanding the systematic differences between L. japonica and related species could provide valuable information to ensure their quality and therapeutic effects.
2. Materials and methods
2.1. Plant materials
Thirty Lonicera germplasm samples including 22 L. japonica, four L. macranthoides, three L. hypoglauca and one L. fulvotomentosa collected from 11 provinces of China were investigated (Table 1 ; Fig. 1 ). Among L. japonica germplasm, 10 cultivars with yellow flowers (Lj_JZ–Lj_PS2), five cultivars with red flowers (Ljv_PS1–Ljv_XXH), and seven wild germplasm (Ljw_PZ–Ljw_QZ) were included. All Lonicera germplasm are planted and stored in the Nursery of Chinese Herbal Medicine in the Lishui Institute of Forestal Sciences, Zhejiang Province.
Table 1.
Species, code, locality, latitude and longitude of 30 Lonicera germplasm samples used in this study.
| Species | Germplasm | Code | Locality (county/city, province) | Latitude/longitude | GenBank accession number |
|---|---|---|---|---|---|
| Lonicera japonica Thunb. | Cultivar with yellow flowers | Lj_JZ | Jingning, Zhejiang | N27°58′/E119°37′ | KF160894 |
| Lj_PS1 | Pingyi, Shandong | N35°30′/E117°38′ | KF160897 | ||
| Lj_BA | Bozhou, Anhui | N33°50′/E115°46′ | KF160899 | ||
| Lj_SJ | Shuyang, Jiangsu | N34°06′/E118°45′ | KF160901 | ||
| Lj_JJ | Jiujiang, Jiangxi | N29°42′/E115°59′ | KF160903 | ||
| Lj_XXH | Xinxiang, Henan | N35°18′/E113°55′ | KF160905 | ||
| Lj_QG | Qingyuan, Guangdong | N23°41′/E113°03′ | KF160907 | ||
| Lj_TH | Tianmen, Hubei | N30°39′/E113°09′ | KF160908 | ||
| Lj_XTH | Xingtai, Hebei | N37°04′/E114°29′ | KF160909 | ||
| Lj_PS2 | Pingyi, Shandong | N35°30′/E117°38′ | KF160895 | ||
| Wild | Ljw_PZ | Panan, Zhejiang | N29°03′/E120°26′ | KF160912 | |
| Ljw_QZ | Qingyuan, Zhejiang | N27°37′/E119°03′ | KF160890 | ||
| Ljw_SF | Shouning, Fujiang | N27°27′/E119°30′ | KF160892 | ||
| Ljw_LZ | Liandu, Zhejiang | N28°26′/E119°54′ | KF160893 | ||
| Ljw_JZ1 | Jingning, Zhejiang | N27°58′/E119°37′ | KF160896 | ||
| Ljw_JZ2 | Jingning, Zhejiang | N27°58′/E119°37′ | KF160898 | ||
| Ljw_QZ | Qingtian, Zhejiang | N28°08′/E120°17′ | KF160904 | ||
| Lonicera japonica Thunb. var. chinensis | Cultivars with red flowers | Ljv_PS1 | Pingyi, Shandong | N35°30′/E117°38′ | KF160886 |
| Ljv_PS2 | Pingyi, Shandong | N35°30′/E117°38′ | KF160891 | ||
| Ljv_BA | Bozhou, Anhui | N33°50′/E115°46′ | KF160900 | ||
| Ljv_JJ | Jiujiang, Jiangxi | N29°42′/E115°59′ | KF160902 | ||
| Ljv_XXH | Xinxiang, Henan | N35°18′/E113°55′ | KF160906 | ||
| Lonicera macranthoides Hand. Mazz. | Cultivars | Lm_LH1 | Longhui, Hunan | N27°07′/E111°01′ | KF160887 |
| Lm_LH2 | Longhui, Hunan | N27°07′/E111°01′ | KF160888 | ||
| Lm_LH3 | Longhui, Hunan | N27°07′/E111°01′ | KF160889 | ||
| Wild | Lmw_LH | Longhui, Hunan | N27°07′/E111°01′ | KF160915 | |
| Lonicera hypoglauca Miq. | Wild | Lhw_LZ1 | Liandu, Zhejiang | N28°26′/E119°54′ | KF160910 |
| Lhw_YZ | Yongjia, Zhejiang | N28°09′/E120°41′ | KF160911 | ||
| Lhw_LZ2 | Liandu, Zhejiang | N28°26′/E119°54′ | KF160914 | ||
| Lonicera fulvotomentosa Hsu. | Wild | Lfw_QG | Qianxinan, Guizhou | N25°05′/E104°54′ | KF160913 |
Fig. 1.
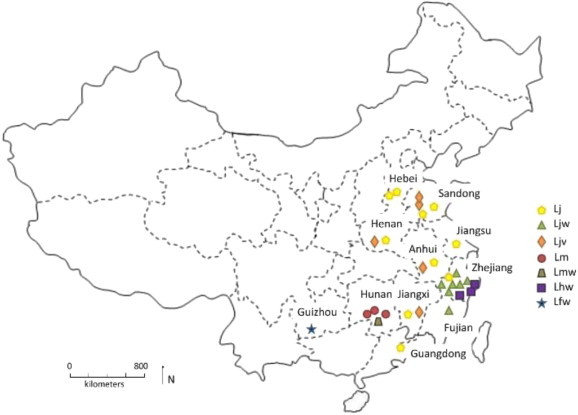
Geographic distribution of the 30 Lonicera germplasm samples. Germplasm codes refer to Table 1.
2.2. Determination of morphological characteristics
Fourteen morphological characteristics including four quantitative characters (leaf length, leaf width, leaf length/width ratio, petiole length), and 10 qualitative characters (leaf type, leaf surface coat, abaxial leaf surface coat, leaf margin coat, leaf vein coat, leaf glandular spots, branch type, branch color, branch coat, branch glandular spots) were determined for all selected Lonicera germplasm samples (Table 2 ). Five to seven plants were selected from each germplasm sample and the quantitative characters of 10 fully developed leaves were measured for each plant. Qualitative characters were observed and described as numerical values from 0–4 (Table 2).
Table 2.
Fourteen morphological characteristics of 30 Lonicera germplasm samples.
| Code | Leaf traits |
Branch traits |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf length (cm) | Leaf width (cm) | Leaf length/width ratio | Petiole length (cm) | Leaf typea | Leaf surface coatb | Abaxial leaf surface coatc | Leaf margin coatd | Leaf vein coate | Leaf gland spotsf | Branch typeg | Branch colourh | Branch coati | Branch glandsj | |
| Lj_JZ | 5.1 ± 0.55 k–n* | 3.0 ± 0.20 g–i | 1.7 ± 0.22 j–o | 0.6 ± 0.11 e–k | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_PS1 | 5.0 ± 0.47 l–n | 3.5 ± 0.36 e–g | 1.4 ± 0.15 o | 0.5 ± 0.05 g–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_BA | 6.0 ± 0.60 h–k | 3.6 ± 0.22 d–f | 1.7 ± 0.17 k–o | 0.6 ± 0.09 g–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_SJ | 7.3 ± 0.55 fg | 4.5 ± 0.37 ab | 1.6 ± 0.09 l–o | 0.7 ± 0.06 d–j | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_JJ | 5.8 ± 0.51 h–l | 3.7 ± 0.64 de | 1.6 ± 0.23 m–o | 0.6 ± 0.19 f–k | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_XXH | 5.3 ± 0.66 i–n | 3.6 ± 0.32 d–f | 1.5 ± 0.20 o | 0.5 ± 0.03 i–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 |
| Lj_QG | 4.6 ± 0.81 n | 3.0 ± 0.41 g–i | 1.5 ± 0.12 no | 0.5 ± 0.06 i–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_TH | 6.3 ± 0.55 g–i | 3.9 ± 0.44 c–e | 1.6 ± 0.05 l–o | 0.5 ± 0.10 j–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_XTH | 5.4 ± 0.35 i–n | 3.5 ± 0.39 e–g | 1.6 ± 0.16 m–o | 0.5 ± 0.11 g–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Lj_PS2 | 4.7 ± 1.18 mn | 3.1 ± 0.70 f–h | 1.5 ± 0.16 no | 0.6 ± 0.11 g–l | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Ljv_PS1 | 5.0 ± 0.22 k–n | 2.8 ± 0.20 hi | 1.8 ± 0.09 i–m | 0.5 ± 0.04 kl | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 3 | 2 | 0 |
| Ljv_PS2 | 5.3 ± 0.41 j–n | 2.8 ± 0.21 hi | 1.8 ± 0.03 h–l | 0.6 ± 0.08 g–l | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 3 | 2 | 0 |
| Ljv_BA | 5.4 ± 0.47 i–n | 2.8 ± 0.31 hi | 1.9 ± 0.07 g–j | 0.4 ± 0.03 l | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 3 | 2 | 0 |
| Ljv_JJ | 5.4 ± 0.32 i–n | 2.8 ± 0.10 hi | 1.9 ± 0.12 g–k | 0.5 ± 0.12 kl | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 3 | 2 | 0 |
| Ljv_XXH | 5.5 ± 1.35 i–n | 2.8 ± 0.28 hi | 2.0 ± 0.52 e–i | 0.6 ± 0.13 g–l | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 3 | 1 | 0 |
| Ljw_PZ | 5.5 ± 1.10 i–n | 3.1 ± 0.83 f–i | 1.9 ± 0.21 g–l | 0.7 ± 0.34 c–g | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 |
| Ljw_QZ | 5.4 ± 0.37 i–n | 2.8 ± 0.24 hi | 2.0 ± 0.08 f–i | 0.5 ± 0.05 kl | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 3 | 0 |
| Ljw_SF | 5.7 ± 0.47 i–m | 2.9 ± 0.13 hi | 1.9 ± 0.12 g–j | 0.8 ± 0.16 c–f | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 0 |
| Ljw_LZ | 5.6 ± 0.19 i–n | 2.7 ± 0.15 hi | 2.1 ± 0.09 e–h | 0.7 ± 0.11 c–h | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 0 |
| Ljw_JZ1 | 6.2 ± 0.68 h–j | 3.5 ± 0.57 e–g | 1.8 ± 0.11 i–n | 0.7 ± 0.28 d–i | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 3 | 0 |
| Ljw_JZ2 | 6.6 ± 0.54 f–h | 2.8 ± 0.24 hi | 2.4 ± 0.21 b–d | 0.7 ± 0.32 c–g | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 3 | 0 |
| Ljw_QZ | 5.6 ± 1.13 i–m | 2.6 ± 0.48 i | 2.2 ± 0.41 de | 0.8 ± 0.15 b–d | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| Lmw_LH | 7.4 ± 0.44 ef | 2.9 ± 0.38 hi | 2.6 ± 0.24 ab | 0.6 ± 0.05 g–l | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 3 | 0 |
| Lfw_QG | 9.7 ± 1.13 bc | 3.5 ± 0.63 e–g | 2.8 ± 0.27 a | 0.8 ± 0.11 c–e | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 0 |
| Lhw_YZ | 8.4 ± 0.76 de | 3.8 ± 0.52 c–e | 2.2 ± 0.19 d–f | 0.9 ± 0.11 a–d | 1 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 3 | 0 |
| Lhw_LZ1 | 8.6 ± 1.01 d | 4.0 ± 0.61 b–e | 2.2 ± 0.17 d–f | 0.7 ± 0.15 c–g | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 1 | 1 |
| Lhw_LZ2 | 9.6 ± 1.26 c | 4.3 ± 0.40 bc | 2.3 ± 0.20 c–e | 0.8 ± 0.14 c–e | 1 | 0 | 2 | 1 | 2 | 1 | 0 | 1 | 1 | 0 |
| Lm_LH1 | 11.2 ± 1.16 a | 4.5 ± 0.50 ab | 2.5 ± 0.20 a–c | 1.0 ± 0.29 a | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 2 | 0 | 0 |
| Lm_LH2 | 10.0 ± 0.35 bc | 4.8 ± 0.36 a | 2.1 ± 0.09 e–g | 0.9 ± 0.08 a–c | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 2 | 0 | 0 |
| Lm_LH3 | 10.5 ± 0.89 ab | 4.1 ± 0.23 b–d | 2.6 ± 0.14 ab | 1.0 ± 0.03 ab | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 2 | 0 | 0 |
a: 0 papery, 1 stiffly papery. b, c, d, e: 0 glabrous, 1 strigose, 2 tomentose. f: 0 no gland spots, 1 white gland spots, 2 orange glandular spots. g: 0 hollow, 1 solid. h: 0 green turn to reddish brown, 1 reddish brown, 2 green, 3 purplish red. i: 0 glabrous, 1 strigose, 2 densely hispid, 3 down to glabrous, 4 coated with fulvous hair. j: 0 no glandular spots, 1 with glandular spots. Germplasm codes refer to Table 1 k–n*: klmn.
2.3. PCR amplification and ITS sequence analysis
The total genomic DNA was extracted from gel-dried leaves using a modified CTAB method (Wang et al., 2008). The entire ITS sequences, including ITS1, 5.8SrDNA gene and ITS2, were amplified using universal primers ITS1 ‘AGAAGTCGTAACAAGGTTTCCGTAGG’ and ITS4 ‘TCCTCCGCTTATTGATATGC’ for all Lonicera germplasm samples (Zhu et al., 2010). The reaction mixture (50 μL) for PCR consisted of 25 ng total genomic DNA, 0.25 mM each of dNTP, 0.2 mM MgCl2, 1 μM of each forward and reverse primer, 1× buffer, 10% (v/v) DMSO, and 0.03 U Ex Taq polymerase (TaKaRa). The ITS sequence was amplified at 94 °C for 4 min followed by 30 cycles of 94 °C for 1 min, 50 °C for 2 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min in a Mastercycler Gradient PCR (Eppendorf, Germany). All amplified products were detected by electrophoresis on 1.5% agarose gels in 1× TAE buffer (100 V for 35 min), and purified and sequenced by the Shanghai Invitrogen Company. After manual correction, the nucleotide sequences were aligned using MEGA 5.0 software (Koichiro et al., 2011). Genetic distances among the 30 germplasm samples were calculated based on Kimura's two-parameter model using MEGA 5.0. Gaps were treated as missing data. Phylogenetic trees were constructed using the UPGMA method, and the bootstrap values for the interior nodes were performed with 1000 replicates. The ITS sequence of L. japonica, L. macranthoides (JQ731716), L. confusa (FJ774986), L. fulvotomentosa (FJ774988), L. hypoglauca (FJ372916) and L. similis (FJ774987) from GenBank were also analyzed for constructing the phylogenetic trees.
2.4. Restriction endonuclease digestion reaction
Restriction endonuclease analysis was performed by DNAman to find appropriate endonucleases for the digestion reaction. The selected restriction endonuclease Cfr I was used to digest the amplified ITS sequence of FLJ and FL. The digestion reaction was performed in a 20 μL reaction mixture containing 2 μL 10× buffer, 10 μL PCR products, 5 U restriction endonuclease Cfr I, and ddH2O. Then the mixture was placed in a 37 °C water bath overnight. The enzyme-digested product was detected by 2% agarose gel electrophoresis. The electrophoretogram was used to identify FLJ and FL.
2.5. Determination of active components
The chlorogenic acid and luteoloside contents were determined by the high-performance liquid chromatography (HPLC) method according to the current PPRC (Committee for the Pharmacopoeia of PR China, 2010). HPLC was conducted by an Agilent 1100 system with an Elit C18 chromatographic column (250 mm × 4.6 mm, 5 μm). To determine the chlorogenic acid content, 0.5 g of each sample powder was accurately weighed and extracted with 50 mL of 50% methanol by ultrasonication for 30 min (250 W, 35 kHz). The extract was filtered, and 5 mL of filtrate was supplemented with 50% methanol to 25 mL. Ten μL of the filtrate was injected into the HPLC system for chlorogenic acid analysis at 327 nm with a flow rate of 1.0 mL/min. The mobile phase contained acetonitrile/0.4% phosphate (13:87, V/V). To determine the luteoloside content, 2 g of each sample powder was accurately weighed and extracted with 50 mL of 70% alcohol by ultrasonication for 1 h (250 W, 35 kHz). Ten μL of the filtrate was injected into the HPLC system for luteoloside analysis at 350 nm with a flow rate of 1.0 mL/min. The mobile phase contained acetonitrile (A) and 0.5% glacial acetic acid (B).
2.6. Statistical analysis
The mean values, standard deviations of morphological characteristics, chlorogenic acid and luteoloside contents were calculated using the statistical package SPSS v17.0 (SPSS Inc., USA). The differences among the germplasm samples were compared by the Minimum Significant Difference method (LSD). Cluster analysis of morphological characteristics and active components were conducted using the average linkage method in SAS 9.1 and SPSS v17.0 software, respectively. Principal component analysis (PCA) was performed using SPSS v17.0.
Distance matrixes of morphological and componential data were carried out by SPSS v17.0. The correlation analysis among the morphological characteristics, ITS sequences and active components of Lonicera germplasm samples were performed by a Mantel test using GenAlEx 6.5 software (Peaktall and Smouse, 2012).
3. Results
3.1. Morphological analysis
The morphological characteristics were significantly different among the 30 Lonicera germplasm samples (Table 2). Cluster analysis showed that they were divided into five clusters: I) 22 L. japonica, II) a wild L. hypoglauca of Lhw_YZ, III) a wild L. fulvotomentosa of Lfw_QG and a wild L. macranthoides of Lmw_LH, IV) another wild L. hypoglauca of Lhw_LZ1, V) three Lonicera macranthoides cultivars of Lm_LH1-3 and the other wild L. hypoglauca of Lhw_LZ2 (Fig. 2 ).
Fig. 2.
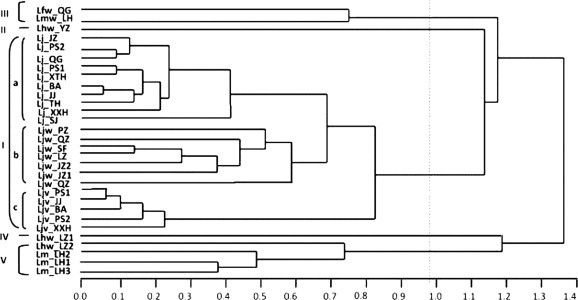
Dendrogram of 30 Lonicera germplasm samples based on morphological characteristics. Germplasm codes refer to Table 1.
All the 22 L. japonica germplasm samples formed cluster I with three subgroups: a) yellow flower cultivars, b) wild germplasm, c) red flower cultivars. Germplasm in subgroup a were characterized by papery and oval leaves with a length/width ratio of about 1.5, and green hollow branches. Germplasm in subgroup b were characterized by stiffly papery, glabrous and strigose leaves, and green and reddish-brown hollow branches. Germplasm in subgroup c were characterized by stiffly papery and glabrous leaves, and purplish red and solid branches. The inter-germplasm differences of wild L. japonica were higher than those of cultivars with red flowers. Details are described in Table 2. The PCA analysis of morphological characteristics also showed the same pattern: the 22 L. japonica germplasm samples were separated into three groups of A, B and C (Fig. 3 L), corresponding to the subgroups of a, b and c in Fig. 2.
Fig. 3.
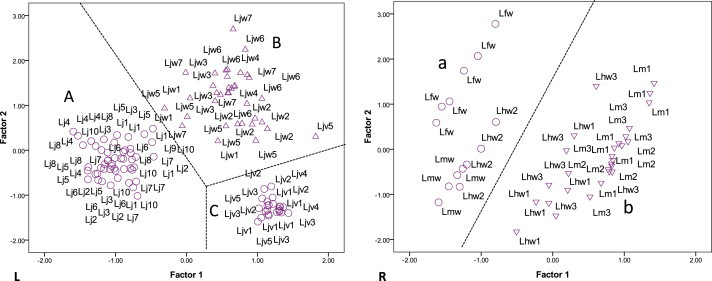
Principal component analysis of morphological characteristics variation of 22 L. japonica germplasm (L), and eight Lonicera germplasm samples except for L. japonica (R). Germplasm codes refer to Table 1. Lj1–Lj10 represent Lj_JZ–Lj_PS2, Ljw1–Ljw7 represent Ljw_PZ–Ljw_QZ, Ljv1–Ljv5 represent Ljv_PS1–Ljv_XXH, Lm1–Lm3 represent Lm_LH1–Lm_LH3, Lmw represent Lmw_LH, Lhw1–Lhw3 represent Lhw_LZ1–Lhw_LZ2, Lfw represent Lfw_QG. Each triangle or cycle represents an individual.
Apart from the 22 L. japonica germplasm samples, another eight Lonicera germplasm samples formed four distantly related clusters in the dendrogram based on morphological characteristics (Fig. 2). PCA analysis demonstrated that the eight Lonicera germplasm samples were separated into two groups (Fig. 3R), corresponding to cluster II, III, and cluster V, IV in Fig. 2. The germplasm in cluster II and III showed similar morphological characteristics such as large, oblong leaves, strigose leaf margins and veins, and specificities such as the coating of fulvous hair of Lfw_QG, papery and strigose leaves of Lmw_LH, and orange glandular spots of the abaxial leaf surface of Lhw_YZ. The germplasm in clusters IV and V were specialized with a dense tomentose leaf with a length of 9–12 cm. However, Lm_LH1–Lm_LH3 with green and glabrous branches were distinguished from Lhw_LZ1 and Lhw_LZ2.
3.2. ITS sequence analysis
The total length of ITS sequence of the 30 Lonicera germplasm samples was 722–723 bp, with one insertion or deletion of base pairs, of which 16 nucleotide sites were variable sites, i.e. the average divergence rate of the ITS sequence was 2.2%. The phylogenetic tree constructed based on ITS sequences of the 30 Lonicera germplasm samples showed that two large, distinct clusters matched with the FLJ and FL, respectively (Fig. 4 ). In the FLJ cluster, five L. japonica cultivars with red flowers (Ljv) were closer to the wild L. japonica germplasm (Ljw) than cultivars with yellow flowers (Lj), although the genetic relationships among all 22 L. japonica germplasm samples were very close. The FJ cluster was composed of the eight germplasm of FL and five Lonicera species (L. macranthoides, L. confusa, L. fulvotomentosa, L. hypoglauca and L. similis). Three L. macranthoides cultivars (Lm_LH1–Lm_LH3) were very different from their wild germplasm (Lmw_LH). However, they had a close genetic relationship with the wild germplasm of L. hypoglauca (Lhw_YZ, Lhw_LZ1, Lhw_LZ2), which was similar to that according to the morphological characteristics. The correlation analysis also showed a significant correlation (r = 0.754, P < 0.01) between ITS sequences and morphological characteristics of all the 30 Lonicera germplasm samples, suggesting consistency of morphological characteristics and the ITS sequences.
Fig. 4.
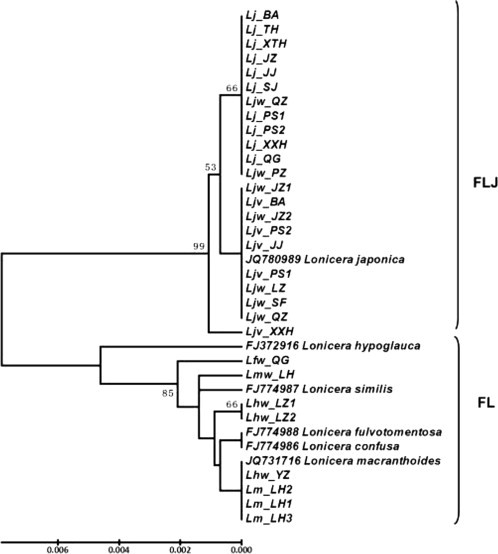
UPGMA dendrogram showing the genetic relationships of Lonicera japonica and related species based on ITS sequences. The numbers between nodes indicate percentages of bootstrap support. Germplasm codes refer to Table 1.
3.3. Active components analysis
Chlorogenic acid contents were significantly different between FLJ and FL (Table 3 ). The chlorogenic acid content of the L. macranthoides cultivar was significantly higher than that of L. japonica. The highest chlorogenic acid contents were detected in L. macranthoides cultivars of Lm_LH2 (5.55%) and Lm_LH1 (4.36%), while L. japonica cultivars with red flowers of Ljv_PS2 (0.95%) and Ljv_PS1 (0.99%) had the lowest content. The luteoloside contents were also significantly different between FLJ and FL, ranging from 0.039% (Ljw_LZ) to 0.187% (Lj_SJ). The order in luteoloside content was L. japonica cultivars with yellow flowers (e.g., Lj_SJ) > L. japonica cultivars with red flowers (e.g., Ljv_PS1) > L. macranthoides cultivars (e.g., Lm_LH1) > L. japonica wild germplasm (e.g., Ljw_LZ).
Table 3.
Chlorogenic acid and luteoloside contents of FLJ (L. japonica) and FL (L. macranthoides).
| Medicinal name | Code | Chlorogenic acid (%) | Galuteolin (%) |
|---|---|---|---|
| Flos Lonicerae Japonicae (FLJ) |
Lj_JZ | 1.04 ± 0.075 h | 0.075 ± 0.0077 ef |
| Lj_PS1 | 1.93 ± 0.017 fg | 0.074 ± 0.0013 ef | |
| Lj_BA | 2.07 ± 0.009 fg | 0.176 ± 0.0127 ab | |
| Lj_SJ | 2.60 ± 0.035 e | 0.187 ± 0.0063 a | |
| Lj_JJ | 2.73 ± 0.086 e | 0.167 ± 0.0027 abc | |
| Lj_XXH | 2.25 ± 0.011 f | 0.063 ± 0.0023 fg | |
| Lj_TH | 2.09 ± 0.025 fg | 0.071 ± 0.0032 ef | |
| Lj_PS2 | 3.18 ± 0.02 cd | 0.165 ± 0.0666 abc | |
| Ljv_PS1 | 0.99 ± 0.008 h | 0.137 ± 0.0063 d | |
| Ljv_PS2 | 0.95 ± 0.012 h | 0.145 ± 0.0027 cd | |
| Ljv_BA | 1.23 ± 0.035 h | 0.181 ± 0.0061 ab | |
| Ljv_JJ | 2.23 ± 0.051 f | 0.156 ± 0.0083 bcd | |
| Ljw_QZ | 1.26 ± 0.011 h | 0.088 ± 0.0013 ef | |
| Ljw_SF | 3.26 ± 0.02 c | 0.041 ± 0.0007 g | |
| Ljw_LZ | 2.89 ± 0.012 de | 0.039 ± 0.0010 g | |
| Ljw_JZ1 | 3.27 ± 0.011 c | 0.042 ± 0.0020 g | |
| Ljw_JZ2 | 1.87 ± 0.115 g | 0.088 ± 0.0202 ef | |
| Ljw_QZ | 2.16 ± 0.005 fg | 0.095 ± 0.0103 e | |
| Flos Lonicerae (FL) | Lm_LH1 | 4.36 ± 0.292 b | 0.072 ± 0.0117 ef |
| Lm_LH2 | 5.55 ± 0.802 a | 0.098 ± 0.0086 e |
Mean values with alphabetical suffices within the same column are statistically different at the significance level of 0.05 based on the protected least significant difference test. Germplasm codes refer to Table 1.
FL and FLJ could be divided into two clusters based on the contents of chlorogenic acid and luteoloside. Two L. macranthoides cultivars, Lm_LH1 and Lm_LH2, formed FL cluster II and were obviously distant from the FLJ cluster that contained L. japonica germplasm. The distinction in contents of active components between FLJ and FL was similar to those in the morphological characteristics and ITS sequence. Correlation analysis demonstrated that chlorogenic acid and luteoloside contents among different Lonicera germplasm samples was significantly correlated with morphological characteristics (r = 0.619, P < 0.01) and the ITS sequence (r = 0.727, P < 0.01).
3.4. Molecular identification of FLJ and FL
A mutation that appeared around site 541 bp of the ITS sequence between FLJ and FL was used for the digestion reaction. Restriction endonuclease analysis revealed that endonuclease Cfr I could be used for discrimination of FLJ and FL. The endonuclease recognized site C∧GGCCG existed in FLJ and FL owned CGGCTG in homologous sites. The digested reaction resulted in the 722–723 bp ITS PCR products of FLJ being digested into two fragments around 500 bp and 200 bp using endonuclease Cfr I; meanwhile, the FL remained unchanged. Consequently, the FLJ can be identified from FL.
4. Discussion
In 1963 the PPRC recorded that the origin plant of FLJ was L. japonica Thunb. (Wang et al., 2009). Three Lonicera species, L. hypoglauca Miq., L. confusa DC. and L. dasystyla Rhed., were also included in the PPRC with L. japonica as the origin plant of FLJ since 1977. In 2005, the PPRC changed the rule once more, and although L. japonica was retained as the origin plant of FLJ, new medicinal material of FL from the flower buds of L. macranthoides, L. hypoglauca and L. confusa were recorded. Lonicera fulvotomentosa was included as one of the origin plants of FL in the latest PPRC (Committee for the Pharmacopoeia of PR China, 2010). The repeated changes in the rules of FLJ caused confusion about the origin plants of FLJ and FJ. The morphological characteristics of several Lonicera species were described in the Flora of China (Xu et al., 1988). However, it is usually difficult to identify closely related species of Lonicera based on traditional classification owing to their similar morphological characteristics (Sun et al., 2011). Generally, the morphological characteristics were only qualitatively described with illustrations (Xu, 1979, Xu et al., 1979). In this study, multi-character joint analysis showed that L. japonica could be distinguished from L. macranthoides, L. hypoglauca and L. fulvotomentosa based on four quantitative and 10 qualitative characters. L. japonica has been domesticated from wild species and cultivated for hundreds of years mainly in the Shandong and Henan provinces of China. Many L. japonica germplasm with obvious different morphological characteristics, yield and quality were formed after long-term natural and artificial selection (Zhang, 2005). In this study, three kinds of L. japonica germplasm, yellow flowers cultivars, wild germplasm, and red flowers cultivars were also identified with morphological characteristics (see Table 2).
Previous research found a suitable DNA marker (psbA-trnH intergenic spacer) for the identification of the botanical origins of FLJ and FL by testing the genetic differences of L. japonica and its closely related species (Sun et al., 2011). Chen et al. (2010) proposed that the ITS2 region was the most promising universal DNA barcode for authenticating medicinal plants. A cleavage rate of ITS PCR products by EcoN I can be used to classify geo-authentic L. japonica from different geographical origins (Wang et al., 2007). However, the discrimination power of these sequences was not enough if different cultivars and wild germplasm of L. Japonica were included. We clarified the genetic relationships among FLJs (from different L. japonica germplasm samples) and FL (L. macranthoides, L. hypoglauca, L. fulvotomentosa and L. confusa), and distinguished the origins of FLJ and FL based on an ITS sequence. Furthermore, FLJs can be identified from FL by Cfr I digested reaction of the ITS PCR products.
Phytochemical and pharmacological research showed that more than 140 chemical compounds have been isolated from L. japonica (Shang et al., 2011). Among these compounds, chlorogenic acid and luteoloside have been chosen as biomarkers to characterize the quality of FLJ by the PPRC (Chinese Pharmacopoeia Commission, 2010). However, the active components of FLJ varied in contents along with different L. japonica germplasm. Chlorogenic acid and luteoloside contents were very different among the L. japonica germplasm samples of the current study, and only 10 were up to a grade of ≥1.5% chlorogenic acid and ≥0.05% luteoloside as decided by the 2010 PPRC. The FLJ from wild L. japonica germplasm had a high chlorogenic acid content and relatively low luteoloside content. Yuan et al. (2012) reported that the chlorogenic acid, luteoloside, quercitin, and isopropyl laurate contents are higher overall in red FLJ flower buds compared with those of FLJ. We detected that the chlorogenic acid and luteoloside contents were different in red FLJ from the four cultivars with red flowers, and two red flower cultivars had the lowest chlorogenic acid contents. The active components of FLJ have been shown to vary with habitat (Xing et al., 2003), harvesting time (Xu et al., 2010), processing (Huang et al., 2009), and so on. Whether the labile and inconsistent chemical components are appropriate biomarkers for identifying and quality control of FLJ should be studied further.
The active components, which are secondary metabolites produced in the secondary metabolism process, differ between species of Lonicera (Xu et al., 1988, Hu et al., 2011). The chlorogenic acid contents of L. japonica, L. confusa, L. hypoglauca, L. macranthoides, L. acuminate and L. similis were 5.06%, 3.25%, 2.11%, 5.72%, 3.06% and 7.66%, respectively (Hu et al., 2011). Chlorogenic acid content (2.80%–5.08%) of L. macranthoides is higher than that of FLJ, and luteoloside is relatively low (0.04%–0.06%) in L. macranthoides (Li et al., 2007). Recently, chlorogenic acid and luteolin were found in other medicinal plants, where the contents were higher than L. japonica (Committee for the Pharmacopoeia of PR China, 2010, Shang et al., 2011). In this study, two cultivars of L. macranthoides are up to the grade of chlorogenic acid and luteoloside contents for FLJ as decided by the PPRC. The results also confirm the conclusion that employing chlorogenic acid and luteolin to control the quality of FLJ lacks specificity (Shang et al., 2011). The specific effective component Macranthoidin B of FJ may be more suitable for identifying and evaluating the FLJ and related preparations blended with FJ (Lin et al., 2013).
Pharmacological studies have demonstrated that both FLJ and FL possess wide efficacies as anti-inflammatory (Cui et al., 2007, Yang et al., 2009), anti-viral (Wang et al., 2011), anti-bacterial (Li and Zhao, 2010, Liu and Li, 2012), anti-oxidant (Wang et al., 2010), hepatoprotective (Hu et al., 2008) and anti-tumor (Zhang et al., 2007) medicines. The similar pharmacological actions of FLJ and FL might be due to their similar chemical components, such as phenolic acids (e.g. chlorogenic acid), flavonoids (e.g. luteoloside), saponins and iridoids (Shang et al., 2011, Liu et al., 2013). However, a couple of chemical components differ between FLJ and FL, for example macranthoidin B is present in L. macranthoides, L. hypoglauca, L. fulvotomentosa and L. confusa, but not in L. japonica (Lin et al., 2013). The distinct components and contents of FLJ and FL might affect their efficacies by the interactions of chemical components.
In conclusion, the morphological characteristics, ITS sequences and active components have advantages in clarifying the genetic relationships among L. japonica and closely related species of L. macranthoides, L. hypoglauca, L. fulvotomentosa and L. confusa, and in the identification of FLJ (L. japonica germplasm) from FL (L. macranthoides, L. hypoglauca, L. fulvotomentosa and L. confusa). FLJs are easy to identify from FL by the Cfr I digested reaction of the ITS PCR products. All of the analyses revealed that L. japonica differed from its related species, implying caution in replacing or blending the traditional Chinese medicines of FLJ and FL.
Acknowledgements
We thank Mr. Zhao-Cheng TANG for the technical assistance of identification of morphological characteristics. This study was supported by the Natural Science Foundation of Shanghai (No. 12ZR1432900) and the Program of Lishui Administration of Science and Technology, Zhejiang Province, China (No. 20110427).
References
- Chen S.L., Yao H., Han J.P., Liu C., Song J.Y., Shi L.C., Zhu Y.J., Ma T.G., Pang X.H., Luo K., Li Y., Li X.W., Jia X.C., Lin Y.L., Leon C. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. Plos One. 2010;5:1–7. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee for the Pharmacopoeia of PR China . Chemistry Industry Publishing House; Beijing: 2000. Pharmacopoeia of the People's Republic of China. [Google Scholar]
- Committee for the Pharmacopoeia of PR China . Chemistry Industry Publishing House; Beijing: 2005. Pharmacopoeia of the People's Republic of China. [Google Scholar]
- Committee for the Pharmacopoeia of PR China . Chemistry Industry Publishing House; Beijing: 2010. Pharmacopoeia of the People's Republic of China. [Google Scholar]
- Cui X.Y., Wang S.X., Hou Y.L. Antiinflammatory mechanism of the Lonicera japonica Thunb. extract. Chin. Parm. 2007;18:1861–1863. [Google Scholar]
- Hu C.M., Jiang H., Liu H.F., Li R., Li J. Effects of LJTF on immunological liver injury in mice. Anhui. Med. Pharm. J. 2008;12:295–296. [Google Scholar]
- Hu S.Q., Dong G.L., Chen X.M., Huang L.L., Yang X., Tong W. ITS sequence-based identification and utilization evaluation of “Nanjiang” (Lonicera similis Hemsl.), a local cultivar in Sichuan, China. Genet. Resour. Crop. Evol. 2011;59:547–555. [Google Scholar]
- Huang J.J., Song B.W., Zhu X.Y., Fu Z.L. Analysis of components of volatile oils of Lonicera and heated vol and comparison of their anti-inflammatory activities. J. Zhejiang Univ. Tec. 2009;37:126–130. [Google Scholar]
- Jiao S.G. Research and comprehensive utilization of honeysuckle. Qilu Pharm. Aff. 2009;28:487–489. [Google Scholar]
- Koichiro T., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tao Z.M., Lei H.Q., Yan T.L., Wu Z.G. Survey and evaluation of Flos Lonicerae grown in Zhejiang province. Acta. Agric. Zhejianggensis. 2007;19:431–434. [Google Scholar]
- Li P., Zhao C. Antibacteria test of aqueous extract and ethanol extract of Flos Lonicerae Japonicae. Chin. Mod. Med. 2010;17:48–50. [Google Scholar]
- Lin Y.Q., Wang S.H., Xu L.H., Wang B., Lin L. Study on identification method for Flos Lonicerae japonicae and Flos Lonicerae. J. Pharm. Res. 2013;32:69–71. [Google Scholar]
- Liu L., Li R. Extract of ingredients for medicine in Lonicera Confusa DC. and study on antibiotic activity. J. Med. Sci. Centr. South. Chin. 2012;40:298–330. [Google Scholar]
- Liu W.J., Chen Y., Ma X., Feng X., Liang J.Y. Progress in the research on chemical constituents of Lonicera macranthoides Hand.-Mazz. Chin. Wild. Plant. Res. 2013;32:6–10. [Google Scholar]
- Peaktall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X.X., Li W.D., Wang W.Q., Bai G.B. Identification of Lonicera japonica by PCR-RFLP and allele-specific diagnostic PCR based on sequences of internal transcribed spacer regions. Planta. Med. 2009;75:1–3. doi: 10.1055/s-0029-1186235. [DOI] [PubMed] [Google Scholar]
- Pu Z.M., Xing J.B., Li P., Liu T., Wang Z.H. Study on floral morphology of Flos Lonicerae. Chin. Tradit. Herb. Drugs. 2002;25:854–859. [PubMed] [Google Scholar]
- Shang X.F., Pan H., Li M.X., Miao X.L., Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2011;138:1–21. doi: 10.1016/j.jep.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.Y., Gao T., Yao H., Shi L.C., Zhu Y.Z., Chen S.L. Identification of Lonicera japonica and its related species using the DNA barcoding method. Planta. Med. 2011;77:301–306. doi: 10.1055/s-0030-1250324. [DOI] [PubMed] [Google Scholar]
- Wang C.Z., Li P., Ding J.Y., Fishbein A., Yunb C.S. Discrimination of Lonicera japonica Thunb. from different geographical origins using restriction fragment length polymorphism analysis. Biol. Pharm. Bull. 2007;30:779–782. doi: 10.1248/bpb.30.779. [DOI] [PubMed] [Google Scholar]
- Wang L.J. The study progress of Lonicera japonica. Mediators. Inflamm. 2010;8:2293–2296. [Google Scholar]
- Wang L.P., Xin N.Q., Qiu Y., Yang B. Antioxidative effects of total flavonoids from Guangxi Lonicera hypoglauca Miq. and Shandong Lonicera Japonica in vivo. J. Guangxi. Med. Univ. 2010;27:681–683. [Google Scholar]
- Wang L.Q., Cui B.A., Zhang H.Y. Antivirai effect of Flos Lonicerae japonica and Flos Lonicerae flavonoids ingredients to PRV in vitro. Chin. Anim. Husb. Veter. Med. 2011;38:183–187. [Google Scholar]
- Wang T., Zhou F.Q., Li J., Zhang Y.Q., Guo Q.M., Xia Y. A study on methods for extraction of genomic DNA from Lonicera japonica Thunb. dry leaves. Chin. Arch. Tradit. Chin. Med. 2008;26:496–498. [Google Scholar]
- Wang Z., Wen H.L., Mei S.M., Xiao C.Y., Zheng L. Discussion on separation of Flos Lonicerae japonicae and Flos Lonicerae in pharmacopoeia commission of the People's Republic of China 2005 edition. Lishizhen Med. Mater. Med. Res. 2009;20:150–151. [Google Scholar]
- Xing J.B., Li P., Liu Y. Study on variation of chlorogenic acid in Flos Lonicerae with phenological phases and localities. Chin. Pharm. 2003;38:19–21. [Google Scholar]
- Xu B.S. Study on origin plants of Jin-Yin-Hua. Acta. Pharmacol. Sin. 1979;14:23–33. [Google Scholar]
- Xu B.S., Hu J.Q., Wang H.J. vol. 72. Science Press; Beijing: 1988. pp. 143–259. (Flora of China). [Google Scholar]
- Xu G.J., Xu L.S., Niu Y.Z., Xu B.S. Studies on the identification of the Chinese drug Jin-Yin-Hua I. J. Nanjing. Coll. Pharm. 1979;1:82–87. [Google Scholar]
- Xu G.J., Xu L.S., Zu Y.Q., Wu W.Y., Zheng X.Z. Studies on the identification of the Chinese drug Jin-Yin-Hua II. J. Nanjing. Coll. Pharm. 1981;1:1–5. [Google Scholar]
- Xu L., Zeng Z.L., Xu X.Y., Luo Y.J. Effects of different collecting time, flowering phase and drying method on the content of chlorogenic acid in Xiushan Honeysuckle. J. Anhui. Agri. Sci. 2010;38:2938–2939. [Google Scholar]
- Yang B., Qiu Y., Wang L.P., Zhang X.L. Studies on the anti-inflammatory molecular mechanism of chlorogenic acid extracted from Lonicera confusa DC. in vitro. Chin. Pharm. Bull. 2009;25:542–545. [Google Scholar]
- Yuan Y., Song L.P., Li M.H., Liu G.M., Chu Y.N., Ma L.Y. Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica thunb. BMC Genomics. 2012;13:1–17. doi: 10.1186/1471-2164-13-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Shandong Traditional Chinese Medicine University; 2005. Studies on the Germplasm Resources of FLOS Lonicera. Master thesis. [Google Scholar]
- Zhang Y.N., Pei Q., Li S., Liu L.H. Anti-tumor experiment research of total saponins from Flos Lonicerae. Chin. Tradit. Herb. Drugs. 2007;38:181–183. [Google Scholar]
- Zhu Y., Dong Y.Z., Zan S.P., Chen H. Phylogenetic relationships of Lonicera based on sequences of nuclear ribosomal DNA. Aeta Bot. Boreal. Occident. Sin. 2010;30:250–254. [Google Scholar]


