Abstract
Infectious bronchitis virus (IBV) was isolated from trachea and kidney tissues of eight broiler farms in Kurdistan region of North Iraq from 2008 to 2010. The birds were suffering from respiratory and nephropathological symptoms and lesions. A 1116 bp hyper mutable spike glycoprotein (S1) gene was amplified and sequenced using conventional RT-PCR. Sequence analysis and BLAST homology search in GenBank data base indicate that two of the farms were infected with the 4/91 strain, one with an unidentified IBV and five were infected with Sul/01/09. The birds in the latter five farms were suffering from nephropathogenic lesions, however, the virus was isolated from kidney but not from trachea in these cases. The birds were vaccinated regularly with 4/91 or MA5 vaccine. The deduced amino acid sequence of the isolated and amplified S1 subunit (372 aa) of Sul/01/09 was differed in 27–28% from that of all three vaccine strains (4/91, MA5, and H120) used in the region. This dissimilarity is most likely the cause of poor efficacy of vaccines used in the region, at least in five of these farms. Amino acid sequence comparison and phylogenetic tree analysis with other published IBV genotypes indicate that this newly isolated virus together with other regionally related and recently published isolates from Israel (IS/720/99, IS/885) and Egypt (egypt/Benisuef/01) belong to a new genotype. This is the first report of identification and genotyping of IBV isolate in Iraq, which indicate the circulation of 4/91 along with a new variant (Sul/01/09) of IBV in vaccinated broiler farms.
Keywords: Avian infectious bronchitis, Coronavirus, Virus isolation, RT-PCR
1. Introduction
Avian infectious bronchitis (AIB) is an acute and highly contagious viral disease of the respiratory system in chickens. It is of significant economic importance because it results in high mortality and poor weight gain in broilers as well as decreasing egg production and quality in layers. In addition to the respiratory and reproductive system, the virus has been associated with nephritis (Dhinakar Raj and Jones, 1997, Butcher et al., 2002, Cavanagh and Naqi, 2003).
The disease is caused by infectious bronchitis virus (IBV), a member of the group 3 of coronavirus species. cDNA representing the entire IBV genome has been cloned and sequenced (Boursnell et al., 1987), it comprises of approximately 27.6 kb, a single stranded RNA virus of positive sense, encoding several proteins which are associated with RNA replication and transcription. All coronaviruses have four structural proteins, spike protein (S), small membrane envelope protein (E), membrane protein (M), and nucleoprotein (N). The spike protein of IBV undergoes post translational cleavage to form S1 and S2 subunits (Boursnell et al., 1984, Cavanagh et al., 1986). Serotype-specific determinant of IBV is thought to be located in the hypervariable regions (HVRs) of S1 glycoprotein (Cavanagh et al., 1988), and it is the S1 subunit of the spike protein which induces neutralizing antibody (Cavanagh et al., 1988, Koch et al., 1990).
In addition to the well-known Massachusetts (Mass) serotype of IBV, many other serotypes, distinct from Mass, have been isolated in Africa (El Houadfi et al., 1986, Abdel-Moneim et al., 2006), Asia (Rajeswar et al., 1998, Bochkov et al., 2006), Australia (Ignjatovic et al., 2006) and Europe (Capua et al., 1994, Worthington et al., 2008). The majority of these strains are endemic for certain geographic regions. Three closely related variants of IBV isolates in Israel (IS/720/99 and IS/885) and Egypt (Egypt/Beniseuf/01) have been genotyped by sequencing of the S1 subunit (Meir et al., 2004, Abdel-Moneim et al., 2006).
In spite of the use of three different vaccines (H120, MA5, attenuated 4/91) in poultry farms in Iraq, outbreaks have been observed with high mortality in broiler farms having nephropathogenic lesions. Since outbreaks of IBV still occur in vaccinated flocks and the virus strains isolated are frequently different from serotypes of the vaccine strains used (Liu and Kong, 2004, Cavanagh et al., 2005, Li et al., 2010), continuous identification of the genotype and production of new generations of vaccines are crucial.
In order to investigate IBV genotypes in Iraq, where the disease is endemic and widely spread in vaccinated and unvaccinated poultry farms mainly associated with kidney damage and urolethiasis, identification and molecular characterization of IBV (Sul/01/09) isolated from eight infected broiler farms was conducted and the deduced amino acid sequence of the S1 subunit of the virus was compared with geographically related isolates.
2. Materials and methods
Samples: Tissue samples from eight suspected IBV outbreaks in Sulaimani broiler farms were collected. Trachea, lung and kidneys were obtained from 3 to 4 birds in each farm and transferred to the laboratory on ice. The birds suffered from respiratory symptoms and lesions as well as kidney damage (enlargement, congestion, and uroletheasis). The flocks were vaccinated with 4/91 or MA5 vaccine. Three different strains of IBV vaccines (H120, CEVA), (MA5, Intervet) and (4/91, Intervet) were used as positive control.
Serological test: Blood samples were collected from 10 birds of each of the eight broiler farms and repeated one week later. The serum was separated; enzyme linked immunosorbent assay (ELISA) was conducted for the detection of IBV antibody in clinically suspected farms using IBV antibody test kit (Synbiotics, USA). This ELISA kit is highly specific in which a titer of (>5 × 103) indicates the present of IBV infection.
RNA extraction: 50–100 mg tissue from trachea, lung and kidney of the birds were homogenized in liquid nitrogen. TRIZOL® reagent was used to extract the RNA from tissue samples according to the manufacturer's instructions. The RNAs were dissolved in 40 μl RNase-DNase free distilled water and directly used for subsequent RT-RCR or stored at (−20 °C).
Virus isolation: Tissue homogenate (200 μl) from trachea and kidney of each PCR positive sample was inoculated in the allantoic cavities of 3 SPF chicken egg embryos (9–11 days) and incubated for 3 further days at 37 °C and candled daily. After 5 passages, allantoic fluids were collected and RNA was extracted for RT-PCR (Momayez et al., 2002).
Oligonucleotides: Oligonucleotides used in this project are illustrated in Table 1 .
Table 1.
Sequences, genome location and the references of primers used in this study.
| Primers | Sequences 5′–3′ | Gene | Position in the sequenceψ | Reference | |
|---|---|---|---|---|---|
| 1 | r-N1221 | CATTCTCTCCTAGAGCTGCA | N | 27,075–27,094 | Designed by researcher |
| 2 | f-N784 | AATTTTGGTGATGACAAGATGA | N | 26,626–26,647 | (Farsang et al., 2002) |
| 3 | f-XCE1+ | CACTGGTAATTTTTCAGATGG | S1 | 21,069–21,089 | (Adzhar et al., 1997) |
| 4 | r-XCE2− | CTCTATAAACACCCTTACA | S1 | 21,507–21,526 | (Adzhar et al., 1997) |
| 5 | f-S1Uni2+ | (CCC)aAATTTAAAACTGAACA | S1 | 20,298–20,314 | (Adzhar et al., 1996) |
| 7 | rS1982 | ACCAGCCGGTTTAGTAGAAG | S1 | 22,331–22,350 | Designed by researcher |
| 6 | f-col VI a2 | ACACGCGAGGCGCTGCCCGGGAC | Col. VI α 2 | 1978–2000 | Designed by researcher |
| 8 | r-col VI a2 | ACAGGAGGTAACAGGTTCATA | Col. VI α 2 | 5311–5332 | Designed by researcher |
cDNA synthesis: SuperScript ™ II RT protocol (Invitrogen) was used according to the manufacturer's instruction, briefly, 1 μl total RNA (1 μg/μl) was mixed with 1 μl reverse primer (10 pmole/ul), r-N1221 for N gene or r-S1982 for S gene (Table 1), 1 μl dNTPs (10 mM each) and RNase free distilled water in a total volume of 12.5 μl mixture. The mixture heated to 65 °C for 5 min then directly chilled on ice. 4 μl from 5× First-strand buffer and 2 μl Dithiotheritol (0.1 mM), 0.5 μl (40 units/μl) RNase inhibitor were added to the mixture. After incubating the mixture at 42 °C for 2 min, 1 μl (200 units/μl) Superscript™ II Reverse Transcriptase was added and further incubated at 42 °C for 50 min, and finally, the enzyme was inactivated at 70 °C for 15 min. The cDNA was stored at 20 °C for the following PCR amplifications.
Detection of the virus by N gene amplification: A primer pair (f-N784 and r-N1221) was used to detect IBV in clinical samples amplifying 437 bp of N gene. This pair of primers was designed specific to a conserved region of nucleocapsid (N) gene to ensure a wide detection range. The PCR amplification reaction was carried out in 25 μl mixture containing 2.5 μl of 10× PCR reaction buffer, 0.7 μl of 10 mM dNTPs, 0.5 μl of each of 10 pmol forward (f-N784) and reverse (r-N1221) primers, 0.1 μl of Red Hot Taq DNA polymerase (5 unit/μl) (ThermoScientific, UK), 1 μl cDNA, the mixture is finalized to 25 μl by the addition of 19.7 μl DNase-RNase free distilled water. Amplification was performed with a thermo cycler (Eppindorf, USA) at 94 °C for 5 min initial denaturation step, 35 cycles (94 °C for 1 min, 52 °C for 30 s and 72 °C for 1 min) and a final extension at 72 °C for 10 min.
PCR amplification of S1 gene: RT-PCR was applied for the first 1230 base of S1 gene which contains the 3 hyper variable regions (HVRs) of S gene (Dolz et al., 2006, Cavanagh et al., 2005). The amplification reaction was carried out in 25 μl reaction mixture containing 2.5 μl of 10× PCR reaction buffer, 1 μl of 10 mM dNTPs, 0.5 μl of 10 pmol forward (f-S1 Uni2+) and reverse (r-XCE2−) primers, 0.3 μl of 5 U/μl Thermoprime Taq DNA polymerase, and 1 μl cDNA. Amplification was carried out with a thermal profile at 94 °C for 5 min initial denaturation then 35 cycles (94 °C for 1 min, 50 °C for 30 s and 72 °C for 2 min) amplification with a final extension of 72 °C for 10 min. The primer pair (f-col VIα2 and r-col VIα2) (Table 1) was used to amplify housekeeping gene of chicken collagen VIα2 as control for cDNA synthesis.
Nucleotide sequencing: About 50 μl of 1230 bp PCR product of amplified S1 subunit from 8 broiler farms were purified using PCR purification kit (Qiagen, Germany), and sequenced with two forward (f-S1Uni2+ and f-XCE1+) and one reverse (r-XCE2−) primers in Innovations Biochemical Laboratory (IBL), Vienna-Austria.
GenBank accession numbers: GenBank accession number of the nucleotide and amino acid sequence of Sul/01/09 isolate S1 gene reported in this study is (GQ281656).
GenBank accession numbers of IBV sequences used in the analysis are: Israel/720/99 (AY091552), IS/885 (AY279533), Egypt/beniSeuf/01 (AF395531), IR-1061-PH (AY544778), IR-1062GA (AY544777), 4/91 (AF093794), Spain/00/338 (DQ064814), Italy-02(AJ457137), QXIBV (AF193423), H120 (M21970), MA5 (AY561713), M41 (AY561711), Connecticut (LI8990), and Australia (AY775779).
3. Results
Clinical signs and virus isolation: All eight clinically suspected farms were serologically positive, even vaccinated farms had a very high titer of antibody ab to 1.4 × 104, in most of the farms the ELISA titer was increased in the second sampling, which indicate that reaction is not due to the vaccination. The IBV-Ab-test kit used allows differentiating between vaccinated and non-vaccinated farms, depending on the Antibody titer. The virus has been detected in all 8 poultry farms. The mortality in these farms was approximately 30% in the first 5 days of the onset of the disease and the birds suffered mainly from kidney damage and urolitheasis. In addition to kidney damage, respiratory signs and lesions were found in two broiler farms, where the disease was further complicated with other infections such as Airsacculitis due to E. coli infection, in which 4/91 was isolated, with a mortality rate of approximately 50–60%. These two farms lucked the hygienic measures and the birds had been vaccinated with 4/91 vaccine two weeks before the onset of the disease. The sequences of the isolated viruses from these two vaccinated broiler farms were identical with vaccine strain 4/91. A local isolate Sul/01/09 has been detected in five broiler farms, where the birds were suffering from nephropathologic lesions. On culturing the virus in egg embryos, one third of the embryos died after each passage of 4/91 strain. Dwarfism has been observed in both 4/91 and Sul/01/09 infected embryos.
S1 sequence analysis: The first 1116 nucleotides encoding S1 spike glycoprotein subunit including the hyper variable regions were sequenced. Comparative analysis of the nucleotides and the deduced amino acid sequence of S1 subunit were performed to assess the relation of Sulaimani (Sul/01/09) isolate with other sequences and to find similarities with vaccine strains H120, MA5 and 4/91. It has been found that Sul/01/09 isolate is 27–28% different from all vaccine strains. However, it only deviates in 6% from Egypt/Benisuef/01 isolate and 5% from IS/885 and Israel/720/99 isolates from Israel (Table 2 ). Nucleotide sequences of these four isolates are identical with about 96–97% similarity (Table 2). Only a few restriction enzymes can be used to differentiate between these closely related isolates (Fig. 1 ), these enzymes can be used for restriction fragment length polymorphism (RFLP) to identify and differentiate between them. Multiple sequence alignment of 14 different strains and isolates have been performed using CLUSTAL W version 1.83 (www.genome.jp) (Fig. 2 ), which indicate that the regions at the positions 27–36, 95–114, 164–178, 207–247 and 335–363 of S1 subunit based on 4/91 strain (accession no. AF093794) are significantly conserved regions in almost all compared sequences. Hyper variable regions (HVRs) of 14 selected S1 amino acid sequences from geographically different regions are located in positions 50–78, 115–148, and 284–292 (Fig. 2). The phylogenetic tree of the aligned amino acids was also produced using MEGA5 Beta version 6.1 online software (www.megasoftware.net) which shows a typical relatedness between sul/01/09 isolate and reference IBV strains from Israel, but not with isolates from our neighbor country Iran (Fig. 3 ).
Table 2.
Nucleotide and deduced amino acid identities of IBV S1 gene sequences.
| Nucleotide identity | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| 1 | 97 | 96 | 96 | 76 | 76 | 78 | 77 | 77 | 76 | 77 | 79 | 78 | 76 | 77 | 1 Sul/01/09 isolate | |
| 2 | 95 | 98 | 98 | 77 | 77 | 78 | 78 | 78 | 77 | 77 | 78 | 78 | 76 | 77 | 2 IS/720/99 | |
| 3 | 95 | 97 | 97 | 77 | 77 | 78 | 77 | 78 | 77 | 77 | 79 | 78 | 77 | 77 | 3 IS/885 | |
| 4 | 94 | 97 | 96 | 74 | 74 | 77 | 74 | 76 | 73 | 74 | 77 | 76 | 75 | 75 | 4 Egypt/BeniSeuf/01 | |
| 5 | 72 | 73 | 73 | 71 | 99 | 80 | 93 | 77 | 76 | 76 | 77 | 77 | 78 | 97 | 5 H 120 (vaccine) | |
| 6 | 72 | 73 | 73 | 72 | 99 | 80 | 93 | 77 | 76 | 76 | 77 | 77 | 78 | 97 | 6 MA5 (vaccine) | |
| 7 | 76 | 76 | 77 | 72 | 78 | 78 | 80 | 77 | 76 | 77 | 80 | 80 | 77 | 81 | 7 Australian T strain | |
| 8 | 73 | 73 | 73 | 70 | 90 | 90 | 79 | 76 | 76 | 76 | 77 | 77 | 77 | 94 | 8 Connecticut | |
| 9 | 73 | 75 | 75 | 72 | 73 | 73 | 74 | 73 | 93 | 94 | 81 | 81 | 78 | 77 | 9 4/91 | |
| 10 | 72 | 74 | 73 | 68 | 72 | 72 | 74 | 72 | 90 | 97 | 80 | 80 | 76 | 76 | 10 IR-1061-PH (Iran) | |
| 11 | 73 | 74 | 74 | 70 | 72 | 72 | 74 | 72 | 90 | 96 | 81 | 81 | 78 | 77 | 11 IR-1062-GA (Iran) | |
| 12 | 77 | 79 | 79 | 75 | 75 | 75 | 80 | 75 | 82 | 81 | 81 | 98 | 77 | 77 | 12 Spain/00/338 | |
| 13 | 77 | 78 | 78 | 74 | 75 | 75 | 80 | 75 | 82 | 81 | 81 | 97 | 77 | 78 | 13 Italy-02 | |
| 14 | 74 | 75 | 76 | 72 | 76 | 76 | 74 | 75 | 77 | 75 | 76 | 76 | 76 | 78 | 14 QXIBV | |
| 15 | 70 | 72 | 71 | 69 | 95 | 95 | 78 | 89 | 73 | 72 | 72 | 74 | 74 | 75 | 15 M41 | |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| Amino acids identity | ||||||||||||||||
Fig. 1.
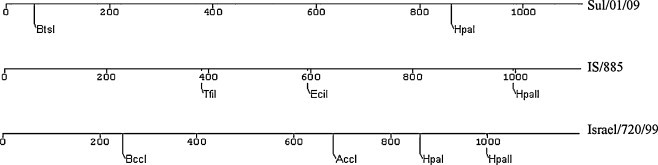
Restriction map of Sul/01/09 IBV and two closely related Israeli isolates (IS/885 and Israel/720/99) that show the restriction enzymes which can be used to differentiate between these isolates. All positions of other known restriction enzymes are identical. The process was done using ChromasPro (version 1.5) online software.
Fig. 2.
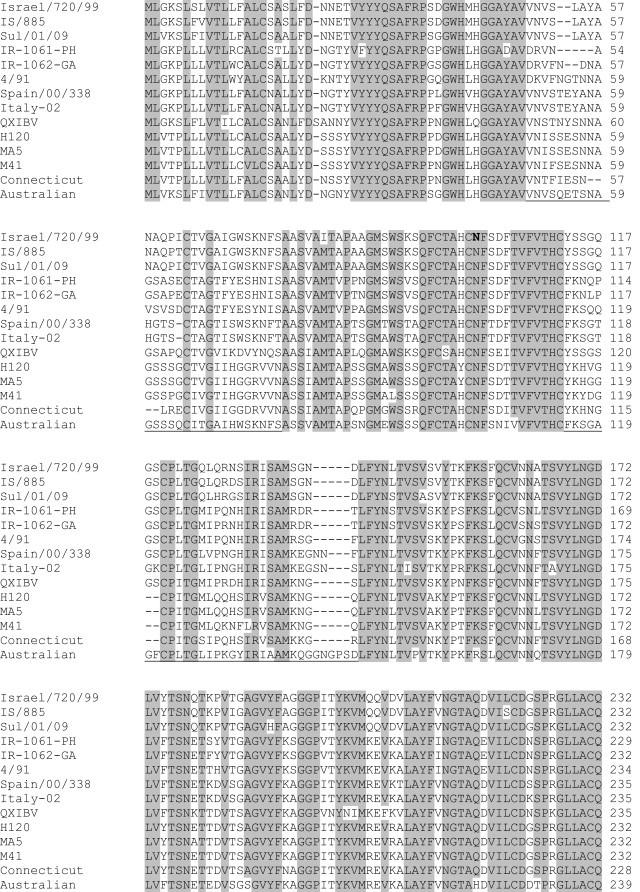

Multiple sequence alignment of the amino acid of Sulaimani isolate (Sul/01/09) with other selected isolates from GenBank. Egypt/Benisuef/01 is not included since the published sequence is too short. Conserved amino acids are indicated in Gray boxes. The hypervariable regions are underlined. Gaps indicate in (−)(CLUSTAL 2.0.12 multiple sequence alignment is used for the alignment).
Fig. 3.
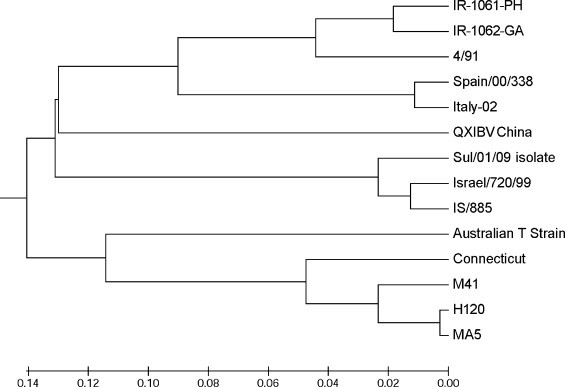
Phylogenetic tree of Sul/01/09 isolate and other selected strains and isolates from GenBank representing different geographic regions show the relationship among the deduced S1 amino acid sequence of isolates from the Middle East (IS/885, Israel/720/99, and Sul/01/09), reference strains from USA (H120, MA5, M41, and connecticut), isolates from Europe (Spain/00/338, and Italy-02), isolates from neighbor country Iran (IR-1061-PH, IR-1062-GA), UK strain (4/91), Australian T strain, and QXIBV from China.
4. Discussion
Poultry farms in Iraq are suffering from huge economic losses caused by Avian infectious bronchitis disease. In spite of the use of three different vaccine strains (H120, CEVA), (MA5, Intervet) and (4/91, Intervet) the nephropathogenic type of the disease and even 4/91 infection is appearing in vaccinated farms. Since Iraq suffered from three wars and long time embargos in the last three decades, little has been done to investigate the spread of the disease and the genotype of the causative agent of IBV.
A nephropathogenic IBV (Sul/01/09) isolate was identified from the tissue of kidney of vaccinated broiler flocks with a mortality rate of approximately 30% at the time of sampling. Sequence analysis of Sulaimani isolate S1 gene revealed its close relatedness only to three field isolates in Israel and Egypt (Table 2). Since high rate of antiserum immunized with different homologous field genotype could neutralize each other (Shimazaki et al., 2009, Wang and Huang, 2000), which suggest genotypes based on S1 sequence analysis to some extent correlate with serotypes of IBV strains, therefore Isolates from Egypt, Iraq and Israel can be considered as the same novel Middle East genotype, and most likely to be the same serotype.
In spite of the long border and a large trade relation with Iran, especially import of chicken and chicken products since 2003, it is not clear why Iraq's isolate (Sul/01/09) differs from isolates from Iran (IR-1061-PH, and IR-1062-GH) in approximately 27% amino acids. Further investigation is necessary to exclude the existence of Iranian isolates in other parts of Iraq. Since diverse coronaviruses have been detected recently in wild birds (Hughes et al., 2009) and it is common among mallard ducks (Muradrasoli et al., 2009), it is very likely that such identical isolates transmitted between Iraq, Israel and Egypt through wild migrated birds, but not through poultry product trade, which does not existing between these countries. Further investigation of IBV in migrated wild birds is necessary to elucidate this measure.
The low nucleotide and amino acid similarities between the (H120, MA5) Massachusetts and 4/91 vaccine with Sulaimani isolate may account for the occurrence of the IBV outbreak in vaccinated flocks. Production of a new vaccine based on the Middle East genotype is necessary to protect the poultry industry in Iraq. Isolation of 4/91 from vaccinated broiler farms may be due to the involvement of the vaccine strain, which has been previously reported (Farsang et al., 2002).
Multiple sequence alignment and phylogenetic tree analysis separate the isolates into different groups which are genotypically related to each other. Most of the serotypes are different from each other in the first HVR (positions 50-78) of S1 spike glycoprotein by more than 20%. This region seems to be conserved in each geographically isolated virus, which can be used for genotyping of the virus. It is most likely that this region is a serotype-specific determinants of IBV and contains antigenic epitope, which is serologically important for serotyping of IBV. Amino acid sequences of European, North American or Middle East groups are highly homologous in the first hyper variable region of S1 subunit of each group, but differ with other groups (Fig. 2). This data indicates that in addition to 4/91 a new nephropathogenic IBV is circulating in Iraq, which belongs to the IBV variant previously isolated in Middle East.
Acknowledgments
This work was carried out with financial support from Kurdistan Institute for strategic studies and scientific researches, University of Sulaimani, and Directorate of Veterinary in Sulaimani, Iraq.
References
- Abdel-Moneim A.S., El-Kady M.F., Ladman B.S., Gelb J. S1 gene sequence analysis of a nephropathogenic strain of avian infectious bronchitis virus in Egypt. Virol. J. 2006;3:78. doi: 10.1186/1743-422X-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhar A., Gough R.E., Haydon D., Shaw K., Britton P., Cavanagh D. Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian Pathol. 1997;26:625–640. doi: 10.1080/03079459708419239. [DOI] [PubMed] [Google Scholar]
- Adzhar A.B., Shaw K., Britton P., Cavanagh D. Universal oligonucleotides for the detection of infectious bronchitis virus by the polymerase chain reaction. Avian Pathol. 1996;25:817–836. doi: 10.1080/03079459608419184. [DOI] [PubMed] [Google Scholar]
- Bochkov Y.A., Batchenko G.V., Shcherbakova L.A., Borisov A.V., Drygin V.V. Molecular epizootiology of avian infectious bronchitis in Russia. Avian Pathol. 2006;35:379–393. doi: 10.1080/03079450600921008. [DOI] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Binns M.M. Sequence of the membrane protein gene from avian coronavirus IBV. Virus Res. 1984;1(4):303–313. doi: 10.1016/0168-1702(84)90019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Butcher G.D., Shapiro D.P., Miles R.D. Institute of Food and Agricultural Science, University of Florida; 2002. Infectious Bronchitis Virus: Classical and Variant Strains. [Google Scholar]
- Capua I., Gough R.E., Mancini M., Casaccia C., Weiss C. A ‘novel’ infectious bronchitis strain infecting broiler chickens in Italy. J. Vet. Med. B. 1994;41:83–89. doi: 10.1111/j.1439-0450.1994.tb00211.x. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Mockett A.P.A. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988;11:141–150. doi: 10.1016/0168-1702(88)90039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Pappin D.J., Binns M.M., Boursnell M.E., Brown T.D. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986;4:133–143. doi: 10.1016/0168-1702(86)90037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Naqi S. Infectious bronchitis. In: Barnes H.J., Fadly A.M., GlissonF J.R., McDougald L.R., Swayne D.E., editors. Diseases of Poultry. 11th edn. Iowa State University Press; Ames: 2003. pp. 101–119. [Google Scholar]
- Cavanagh D., Picault J.-P., Gough R.E., Hess M., Mawditt K., Britton P. Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 2005;34:20–25. doi: 10.1080/03079450400025414. [DOI] [PubMed] [Google Scholar]
- Dhinakar Raj G., Jones R.C. Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol. 1997;26:677–706. doi: 10.1080/03079459708419246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolz R., Pujols J., Ordonez G., Porta R., Majo N. Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype. Avian Pathol. 2006;35:77–85. doi: 10.1080/03079450600597295. [DOI] [PubMed] [Google Scholar]
- El Houadfi M.D., Jones R.C., Cook J.K.A., Ambali A.G. The isolation and characterization of six avian infectious bronchitis virus isolated in Morocco. Avian Pathol. 1986;15:93–105. doi: 10.1080/03079458608436269. [DOI] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renstro’m L.H.M., Baule C., Soós T., Belák S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31:229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L.A., Savage C., Naylor C., Bennett M., Chantrey J., Jones R. Genetically diverse coronaviruses in wild bird populations of north England. Emerg. Infectious Dis. 2009;15(7):1091–1094. doi: 10.3201/eid1507.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic J., Gould G., Sapats S. Isolation of a variant infectious bronchitis virus in Australia that further illustrates diversity among emerging strains. Arch. Virol. 2006;151:1567–1585. doi: 10.1007/s00705-006-0726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G., Hartog L., Grant A., Van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71:1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xue C., Chen F., Qin J., Xie Q., Bi Y., Cao Y. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet. Microbiol. 2010;143:145–154. doi: 10.1016/j.vetmic.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir R., Rosenblut E., Perl S., Kass N., Ayali G., Hemsani E., Perk S. Identification of a novel nephropathogenic infectious bronchitis virus in Israel. Avian Dis. 2004;48:635–641. doi: 10.1637/7107. [DOI] [PubMed] [Google Scholar]
- Momayez R., Pourbakhsh S.A., Khodashenas M., Banani M. Isolation and identification of Infectious bronchitis virus from commercial chickens. Arch. Razi Inst. 2002;53:1–9. [Google Scholar]
- Muradrasoli S., Mohamed N., Hornyak A., Fohlman J., Olsen B., Belak S., Blomberg J. Broadly targeted multiprobe QPCR for detection of coronaviruses: coronavirus is common among mallard ducks (Anas Platyrhynchos) J. Virol. Methods. 2009;159(2):277–287. doi: 10.1016/j.jviromet.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswar J.J., Dorairajan N., Paul W.M. Seroprevalence study of infectious bronchitis among poultry in Tamilnadu. Indian Vet. J. 1998;75:973–974. [Google Scholar]
- Shimazaki Y., Harada M., Horiuchi T., Yoshida K., Tanimura C., Nakamura S., Mase M., Suzuki S. Serological studies of infectious bronchitis vaccines against Japanese field isolates of homologous and heterologous genotypes. J. Vet. Med. Sci. 2009;71:891–896. doi: 10.1292/jvms.71.891. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Huang Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington K.J., Currie R.J.W., Jones R.C. A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37:247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]


