Abstract
Following the recent use of a live vaccine against porcine reproductive and respiratory syndrome virus (PRRSV) in Denmark, both American (vaccine) and European-type PRRSV now coexist in Danish herds. This situation highlighted a requirement for supplementary tests for precise virus-typing. As a result, we developed a RT-PCR assay able to detect as well as type PRRSV. To provide maximal sequence information, complete viral open reading frames (ORFs 5 and 7) were targeted for amplification. The RT-PCR test was able to amplify complete PRRSV ORFs from complex materials such as boar semen containing as little as 1 TCID50 ml−1 of PRRSV. Typing of viruses was accomplished by any one of three strategies: (i) use of type-specific PCR primers, (ii) size determination of ORF 7 amplicons, (iii) DNA sequencing. All three typing strategies showed complete concordance with the currently used method of typing with monoclonal antibodies (MAbs) when used on a panel of PRRSV field isolates covering the period 1992–1997. The ORF 7-based test had particularly desirable characteristics, namely, highly sensitive detection of PRRSV without apparent type bias, typing of the detected virus, discrimination between pure and mixed virus populations, and semi-quantitative assessment of type ratios in mixed populations, all in a single PCR reaction. In addition, the obtained sequence data were used to predict two simple and rapid strategies (single-enzyme restriction length polymorphy analysis and oligonucleotide hybridization) for confirmation of the specificity of ORF 7 RT-PCR reactions. As such, the RT-PCR assay provides a new, powerful diagnostic tool to study the population dynamics between present and emerging PRRSV-types.
Keywords: RT-PCR, PRRSV, RFLP, Vaccinations, Pig-viruses, Diagnosis-viruses
1. Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) emerged in the late 1980s as a global, major cause of reproductive disorders and pneumonia in swine. The virus, which was first isolated in the Netherlands in 1991, is an enveloped single-stranded RNA virus with a 15 kb plus-sense genome (Meulenberg et al., 1993). PRRSV belongs to the family Arteriviridae, which also includes equine arteritis virus (EAV), lactate dehydrogenase elevating virus of mice (LDHV), and simian haemorrhagic fever virus (SHFV) (Meulenberg et al., 1993).
PRRSV isolates exhibit a pronounced degree of genetic heterogeneity. The two main PRRSV types, European and American, are only 55–80% identical at the nucleotide level (Meng et al., 1995; Murtaugh et al., 1995; Suarez et al., 1996; Meulenberg et al., 1997), and the viruses can be accurately grouped in these two categories using MAbs (Yoon et al., 1995). In addition, significant genetic heterogeneity exists even within the two main groupings of `American' and `European,' so that sequence analysis, but currently not MAb-typing, allows sub-typing of PRRSV to even finer geographical levels (Meng et al., 1995; Kapur et al., 1996; Suarez et al., 1996).
While PRRSV isolates made in Denmark during the period 1992–1996 were invariably of the European-type, an increasing number of American-type PRRSV isolates has been made from aborted and stillborn piglets since the use of an attenuated American-type PRRSV vaccine starting in 1996 (Bøtner et al., 1994, Bøtner et al., 1997; Sørensen et al., 1997, Sørensen et al., 1998; Madsen et al., 1998; Nielsen et al., 1998). The emergence of an apparently pathogenic, hitherto unrecognized PRRSV-type in Denmark increased the demand for accurate typing of virus isolates. Isolation of PRRSV on cell cultures and typing with MAbs offer a combination of sturdiness, sensitivity and accuracy that, in our opinion, is unlikely to be greatly improved upon. However, in selected diagnostic cases, as well as in many research situations, the sensitivity and accurate typing afforded by RT-PCR techniques would be highly desirable. Also, cell culture methods may not be well suited to examine seminal shedding of PRRSV, due to a high content of cytotoxic compounds in this material.
It follows from the above that a RT-PCR test should be able to sensitively detect as well as type PRRSV from relevant biological material, and provide the maximal amount of sequence information, preferably by amplification of whole viral open reading frames. None of the published RT-PCR tests seems to completely fulfil this combination of requirements (Mardassi et al., 1994; Suárez et al., 1994; Van Woensel et al., 1994; Christopher-Hennings et al., 1995b; Kono et al., 1996; Gilbert et al., 1997; Larochelle and Magar, 1997; Legeay et al., 1997; Shin et al., 1997). In the present study, we report the development and characterization of a RT-PCR test which meets the above criteria, and thus should provide a valuable tool for the epidemiological surveillance of emerging PRRSV-types.
2. Materials and methods
2.1. Cells and viruses
Porcine pulmonary alveolar macrophages (PPAM) were obtained by lung lavage from specific pathogen-free piglets. All PPAM batches were tested for PRRSV susceptibility, and only highly PRRSV-susceptible PPAM cultures were used for virus isolation, propagation and titration (Bøtner et al., 1994; Nielsen et al., 1997). MARC-145 cells, which constitute the in vitro system of choice for isolating American-type PRRSV (Kim et al., 1993), were propagated as described (Nielsen et al., 1997).
Immunoperoxidase staining of PRRSV-infected PPAM or MARC monolayers with MAbs WBE4 (kindly provided by T. Drew) and VO17 (kindly provided by E. Nelson and D. Benfield) was used to characterize isolates as European- or American-type, respectively (Yoon et al., 1995).
PRRSV 111/92 is a Danish isolate of the European-type from 1992, extensively characterized by MAb reactivity as well as sequence analysis (Madsen et al., 1998). 111/92 was propagated and titrated on PPAM cultures. The stock used for the present work had a titer of 105 TCID50 ml−1. VR2332 is an American-type PRRSV isolate, and was obtained from ATCC. VR2332 was propagated and titrated on MARC cells, and had a titer of 106 TCID50 ml−1. In-house isolates of a number of other viruses (swine influenza virus, porcine circovirus, porcine respiratory coronavirus, bovine virus diarrhoea virus, Aujeszky's disease virus, porcine adenovirus and porcine enterovirus) were used to confirm the specificity of the RT-PCR test.
A panel of 52 PRRSV field isolates (comprising 36 Danish isolates and 16 isolates from other European countries distributed to diagnostic laboratories under the European Union concerted action programme on PRRSV) were used to verify the typing and detection accuracy of the RT-PCR test.
2.2. RNA isolation
Boar semen and swine serum were used without any pretreatment, fractionation or dilution. Cell culture material was freeze-thawed and pre-cleared by centrifugation. Lung tissue was prepared by standard methods as for virus isolation: a 10% homogenate was pre-cleared by centrifugation, and sterile-filtered through a 0.45 μm filter.
30 μl of material to be assayed for PRRSV was added to 1 ml of 5 M guanidine isothiocyanate (GnSCN) − 31 mM citrate, pH 5.2. After a 5 min incubation at room temperature, residual cellular debris was cleared by centrifugation. RNA was recovered from GnSCN using acid-treated silica particles (Boom et al., 1990; Legeay et al., 1997). In our hands, the silica method yielded superior results to acid phenol/chloroform extraction, particularly for PRRSV RNA extraction from semen. 5 μl silica suspension was added to the cleared sample in GnSCN buffer, and incubated for 20 min on a rotating spear. The silica was washed twice with 1 ml GnSCN buffer, twice with 1 ml 70% ethanol and once with 1 ml absolute ethanol. Following air-drying, the silica was re-suspended in 45 μl nuclease-free water containing 0.03 U μl−1 of a heat-stable, redox-independent RNase inhibitor (Murphy et al., 1995) (5′-3′, Boulder, CO, USA), and nucleic acid was eluted by heating the samples for 5 min at 65°C. Finally, the silica was pelleted by spinning samples for 5 min at 11 g in a microcentrifuge, and 28 μl of the supernatant (referred to as RNA below) was used for cDNA synthesis.
2.3. Primers
PCR primers were chosen flanking the ORF 5 and ORF 7 genes (see Table 1 ). Inosines were utilized at wobble base positions. ORF 7 encodes the 15 kDa viral nucleocapsid protein, and ORF 5 encodes a 25 kDa glycosylated membrane protein, which is a major constituent of the viral envelope (Meulenberg et al., 1995). Two pairs of ORF 5 primers were chosen so that one pair was specific for European-type (EU), and the other type specific for American-type (US) PRRSV. To afford type characterization, the cDNA was examined in two PCR reactions, with EU ORF 5 primers being used in one PCR reaction, and US ORF 5 primers in the other.
Table 1.
Description of the primers used for reverse transcription and PCR
| RT-PCR type | Primer sequencea | Primer specificity | Size of RT-PCR productb |
|---|---|---|---|
| ORF 7 | RT primer | All known PRRSV-types | American-type PRRSV: 660 bp |
| 5′ TCG CCC TAA T 3′ | |||
| Forward PCR primer | European-type PRRSV: 637 bp | ||
| 5′ GCC CCT GCC CAI CAC G 3′ | |||
| Reverse PCR primer | |||
| 5′ TCG CCC TAA TTG AAT AGG TGA 3′ | |||
| EU ORF 5 PCR | RT primer | European-type PRRSV | 719 bp |
| 5′ TAT GTI ATG C 3′ | |||
| Forward PCR primer | |||
| 5′ CAA TGA GGT GGG CIA CAA CC 3′ | |||
| Reverse PCR primer | |||
| 5′ TAT GTI ATG CTA AAG GCT AGC AC 3′ | |||
| US ORF 5 PCR | RT primer | American-type PRRSV | 818 bp |
| 5′ AAA GGT GCA G 3′ | |||
| Forward PCR primer | |||
| 5′ GCT CCA TTT CAT GAC ACC TG 3′ | |||
| Reverse PCR primer | |||
| 5′ AAA GGT GCA GAA GCC CTA GC 3′ |
PCR primer pairs were used in singleplex PCR reactions on cDNA primed with a cocktail of the three RT primers. Note that RT primers are 3′-end truncated versions of the corresponding reverse PCR primer, lending a semi-nested effect to the RT-PCR test. `I' denotes inosines.
b All RT-PCR products extend from 50–150bp upstream from the start codon to 60–70 downstream from the stop codon, thus containing the whole coding sequence for the indicated gene. The combined size of the ORF 5 and 7 amplicons corresponds to 9% of the whole viral sequence.
A different strategy was used for amplification of ORF 7: the chosen ORF 7 primer-binding sites were highly conserved between all PRRSV-types (European as well as American), so that the single pair of ORF 7 primers was expected to amplify all known PRRSV-types. In this case, the known length polymorphism of the ORF 7 amplicon (US-types are approximately 4% longer than EU-types) allowed type-calling following sieving agarose gel electrophoresis (Mardassi et al., 1994).
The relatively high genetic variability of PRRSV made primer selection a non-trivial task. Therefore, it was highly desirable to utilize the anti-sense PCR primers to also prime the reverse transcription reaction. Yet, mispriming by full-length anti-sense PCR primers during the low-stringency conditions of the RT reaction would increase the risk of generating an unspecific product in the subsequent PCR reaction. To overcome this problem, we used 3′-end truncated versions of the anti-sense PCR primers to prime the RT reaction. Use of these specific RT primers increased sensitivity of the test about 10-fold compared to alternative RT priming methods such as random hexamers or poly dT.
2.4. cDNA synthesis
Following elution from silica, RNA was immediately subjected to reverse transcription (RT) with Moloney murine virus reverse transcriptase using `Ready to Go' tubes, according to the manufacturer's instructions (Pharmacia, Allerød, Denmark). The reactions (total volume 33 μl, containing 28 μl RNA and 1.25 μM each RT primer) were incubated for 45 min at 45°C and heat-inactivated for 5 min at 90°C. Although not specifically recommended by the manufacturer, heat inactivation of the RT reactions increased the sensitivity of the RT-PCR test, probably reflecting the known ability of non-denatured reverse transcriptases to inhibit Taq DNA polymerase activity (Sellner et al., 1992). RT reactions were stored at −20°C.
2.5. PCR
5 μl of the RT reaction was used as template in a 50 μl PCR reaction containing (including the contribution from the RT reaction) 15 mM Tris (pH 8.3), 58 mM KCl, 750 μM DTT, 2.2 mM MgCl2, 640 μM each dNTP, 500 nM each PCR primer and 2.5 U Ampli Taq Gold (Perkin Elmer, Birkerod, Denmark). Depending on the PCR primer pair used, ORF 7, EU ORF 5 or US ORF 5 was amplified per PCR reaction.
PCR reactions were prepared in thin-walled `Gene Amp' tubes (Perkin Elmer), covered with a mineral oil overlay, and cycled in a Perkin Elmer `Thermal Cycler' under the following conditions: [94°C for 10 min], 55 × [95°C for 20 s – annealing temperature for 20 s – 72°C for 20 s], [72°C for 5 min]. The Ampli Taq Gold enzyme was only activated during the 10 min incubation at 94°C, thus ensuring `hot start' conditions. The annealing temperature was 55°C for the ORF 7 primers, and 60°C for the ORF 5 primers. PCR sensitivity was not improved by using longer denaturation, annealing and extension times. On the other hand, use of short segment times made it possible to take urgent samples through the RT-PCR test in a single day.
2.6. Agarose gel electrophoresis, gel purification and sequencing
For low resolution electrophoresis, 10 μl of the PCR reactions was electrophoresed in 1–2% GTG agarose (FMC, Medinova, Denmark) in 1 × TBE at 8 V cm−1, followed by staining in 1 μg ml−1 ethidium bromide, and photography under 254 nm epi-illumination in a standard Polaroid setup.
To resolve ORF 7 amplicons from US (660 bp) and EU (637 bp) PRRSV-types, we used 1.9% highly sieving Metaphor agarose (FMC) in 0.8 × TBE with 0.5 μg ml−1 ethidium bromide. The gels were run for 5–6 h in 0.4 × TBE at 17 V cm−1, with constant cooling and recirculation of the running buffer (Hoefer SuperSub gel apparatus, Pharmacia), and were photographed as above. Diagnostically adequate resolution of ORF 7 bands could also be obtained without the use of buffer cooling and recirculation, by means of slower electrophoretic runs (6 V cm−1, 18 h) in 4% `3 : 1' agarose (FMC) in 1 × TBE.
In selected cases, bands were excised from 1% agarose gels, and the purified DNA sequenced using the corresponding PCR primers and Perkin Elmer fluorescent chain terminator chemistry.
3. Results
3.1. The RT-PCR test specifically detects as well as types field PRRSV isolates
To investigate the specificity of the test, we first used RNA extracted from our reference viruses 111/92 (EU-type) and VR2332 (US-type). The RNA was reverse-transcribed in a non-type-specific fashion using a mix of three anti-sense primers binding downstream of the ORF 5 and ORF 7 genes of EU- as well as US-types. Then, aliquots of single RT reactions were examined in three singleplex PCR reactions using ORF 7, US ORF 5 and DK ORF 5 PCR primer pairs. As shown in Fig. 1 , none of the three PCR primer pairs produced bands from negative controls. As expected, the ORF 7 primers reacted with 111/92 as well as VR2332 cDNA, and yielded bands of the expected size. The size difference between ORF 7 amplicons from 111/92 (637 bp) and VR2332 (660 bp) was only poorly resolved under the electrophoretic conditions used for Fig. 1. In contrast to the ORF 7 PCR primers, the ORF 5 PCR primers were type-specific: the EU ORF 5 PCR primers reacted only with 111/92, to give a specific band of 719 bp, while the US ORF 5 PCR primers reacted only with VR2332, to give a specific band of 818 bp (Fig. 1).
Fig. 1.
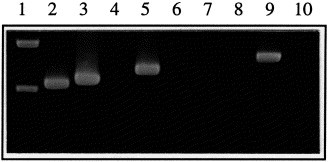
Specificity of the test on EU-type (111/92) and US-type (VR2332) reference viruses. RNA was extracted from 111/92 stock, VR2332 stock and water, followed by reverse transcription with a cocktail of 3 RT primers designed to prime cDNA synthesis from ORFs 5 and 7 of all known types of PRRSV. cDNA aliquots were examined by singleplex PCR with primers specific for all known PRRSV-types (ORF 7 PCR) or specific for EU- or US-type PRRSV (EU ORF 5 or US ORF 5 PCR, respectively). Lane 1, Phi-X HaeIII digest. Lanes 2–4, ORF 7 PCR on the following templates: 2, 111/92. 3, VR2332. 4, H2O. Lanes 5–7, EU ORF 5 PCR on the following templates: 5, 111/92. 6, VR2332. 7, H2O. Lanes 8–10, US ORF 5 PCR on the following templates: 8, 111/92. 9, VR2332. 10, H2O.
Next, we examined the RT-PCR test for false-positive reactivity on a panel of eight in-house isolates of relevant porcine and bovine viruses (see Section 2). None of the tested viruses gave a false-positive result in the RT-PCR test with any of the three PCR primer pairs (not shown).
Finally, we examined the typing accuracy of the RT-PCR test on a panel of 52 field PRRSV isolates. The results are presented in Table 2 . To provide a basis for comparison, 40 of the isolates were typed as European or American by MAbs. The 36 Danish isolates were selected to cover the period 1992 (when PRRSV was first diagnosed in Denmark) to 1997. Note that some Danish isolates were of the American-type, reflecting spread of an American-type live PRRSV vaccine virus in Danish herds (Madsen et al., 1998; Nielsen et al., 1998; Sørensen et al., 1998). Sixteen isolates distributed among European diagnostic laboratories under the EU concerted action programme against PRRSV were included to represent `non-Danish' European-types. The 12 isolates for which MAb-typing was not done were from before the licensing of the live American-type PRRSV vaccine, and were thus all expected to be of European-type.
Table 2.
Verification of the RT-PCR test on a panel of 52 field PRRSV isolates from different European countries
| Country of origin a | Period covered by material | Material used for RT-PCR | Number of isolates or animals | MAb-type of virusb | ORF 7 RT-PCR resultc | US ORF 5 RT-PCR resultc | EU ORF 5 RT-PCR resultc |
|---|---|---|---|---|---|---|---|
| Denmark | 1992–1997 | Cell culture isolates | 15 | European | + | − | + |
| '' | 16 | American (vaccine) | + | + | − | ||
| Lung homogenate | 2 | American (vaccine) | + | + | − | ||
| Pleural fluid | 3 | American (vaccine) | + | + | − | ||
| Spain | 1991–1992 | Cell culture isolates | 1 | European | + | − | + |
| '' | 2 | Not done | + | − | + | ||
| Belgium | 1992 | Cell culture isolates | 1 | European | + | − | + |
| '' | 1 | Not done | + | − | + | ||
| United Kingdom | 1991 | Cell culture isolates | 1 | European | + | − | + |
| ” | 2 | Not done | + | − | + | ||
| Germany | Before 1993 | Cell culture isolates | 5 | Not done | + | − | + |
| France | 1991–1993 | Cell culture isolates | 1 | European | + | − | + |
| '' | 2 | Not done | + | − | + |
Danish isolates were selected to cover the period from 1992 (when PRRSV was first isolated in Denmark) to the present day. Isolates were from aborted or weak born piglets from sows without any history of PRRSV vaccination. Isolates from other European countries were sent out under the EU concerted action on PRRSV programme.
b Isolates were typed by reactivity of infected cell monolayers with MAbs VO17 (specific for American-type PRRSV) and WBE 4 (specific for European-type PRRSV).
c RT-PCR test result was determined by absence (negative result, indicated by `−' in the table) or presence (positive result, indicated by `+' in the table) of bands of the expected size following agarose gel electrophoresis. Identity of bands was confirmed by excision from agarose gels and subsequent DNA sequencing for all data shown in the table. Negative controls (uninfected PPAM and MARC cells, as well as serum, semen and lung homogenate from uninfected animals) were negative in RT-PCR.
RT-PCR with ORF 7 primers amplified a specific band from all field PRRSV isolates. Negative controls (uninfected PPAM and MARC cells as well as semen, serum and lung homogenate from uninfected animals) were always negative. The size of the ORF 7 amplicon of selected Danish field isolates was compared to the size of the ORF 7 amplicon of the reference EU- (111/92) and US- (VR2332) type viruses (Fig. 2 ). As expected, field isolates MAb-typed as European, produced a 111/92-size (637 bp) ORF 7 amplicon, whereas field isolates MAb-typed as American, produced a VR2332-size (660 bp) ORF 7 amplicon. Since the examined isolates covered the period from 1992 to 1997, the results confirmed that ORF 7 amplicon size is a temporally stable marker of PRRSV-type.
Fig. 2.
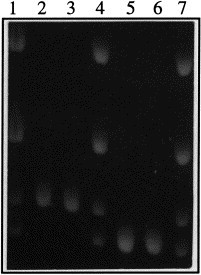
Typing of Danish field PRRSV isolates by ORF 7 amplicon size. ORF 7 RT-PCR reactions from 4 MAb-typed Danish field PRRSV isolates (part of the dataset presented in Table 2) were examined by highly sieving agarose gel electrophoresis. To facilitate the precise comparison of ORF 7 amplicon size, a marker (`5 + 7 marker') containing pooled RT-PCR product from ORF 5 and ORF 7 PCR on US- (VR2332) and EU- (111/92) type virus was utilized in lanes across the gel. The 4 bands in the 5 + 7 marker are: 818 bp (US ORF 5), 719 bp (DK ORF 5), 660 bp (ORF 7 from US-type virus), 637 bp (ORF 7 from EU-type virus). Lanes 1, 4 and 7, 5 + 7 marker. Lanes 2 and 3, two isolates from 1997 MAb-typed as American-type. Lanes 5 and 6, two isolates from 1993 and 1995 MAb-typed as European-type.
In the 40 cases where MAb-typing was performed, all PRRSV isolates reacted with either DK ORF 5 primers or US ORF 5 primers in a manner fully consistent with the MAb-type of the virus. The 12 isolates for which MAb-typing was not performed all reacted as European-type by ORF 5 PCR, in full concordance with their geographical/chronological origin.
It should also be mentioned that for all the data shown in Table 2, the bands were confirmed as PRRSV-specific by sequencing the whole coding sequence of ORFs 5 and 7. In all cases, the sequences were in full accordance with the MAb-type of isolate (the sequence data will be presented elsewhere).
Taken together, the results above confirmed the PRRSV specificity of the RT-PCR primers. Also, the results confirmed that field isolates of PRRSV could be rapidly and precisely typed as European or American based on either one of two strategies: generic ORF 7 PCR combined with the size polymorphy of the ORF 7 amplicon, or type-specific ORF 5 PCR. Sequencing was a slower technique, but provided detailed sub-typing of PRRSV, which could not be achieved by the aforementioned methods (Storgaard, 1998).
Finally, although the bulk of the field isolates listed in Table 2 were grown in cell culture prior to RT-PCR, the detection of PRRSV in 5/5 samples of primary material (lung homogenate and pleural fluid) indicated that the RT-PCR test was amply sensitive to detect PRRSV directly in the tissues of clinically affected animals. The sensitivity of the test is investigated below.
3.2. The test detects PRRSV at concentrations two orders of magnitude below those found in clinical material
From the outset, a main goal was that the test should be sufficiently sensitive to detect PRRSV directly in clinical samples, without resorting to impractical techniques such as nested PCR or Southern blot detection of PCR product. PRRSV infection results in transient viraemia (reaching 1.000 TCID50 ml−1 serum), with replication of virus occurring preferentially in lung macrophages (up to 1.000 TCID50 g−1 lung tissue at peak viral replication 7–14 days post-infection) (Duan et al., 1997; Nielsen et al., 1997). Also, characteristically for the Arteriviruses to which PRRSV belongs, the virus is intermittently excreted in semen for long periods of time, in currently unknown quantities (Swenson et al., 1994; Christopher-Hennings et al., 1995a; Gradil et al., 1996; Nielsen et al., 1997; Sur et al., 1997). Therefore, the sensitivity of the RT-PCR test was evaluated on 10-fold serial dilutions of the reference viruses 111/92 (EU-type) and VR2332 (US-type) in clinically relevant materials such as whole boar semen (Fig. 3 ).
Fig. 3.
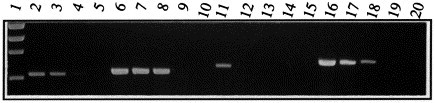
Sensitivity of the test on EU-type (111/92) and US-type (VR2332) reference viruses. The two reference viruses were diluted in whole (undiluted and unfractionated) boar semen to the concentrations given below. 30 μl of each dilution was examined by RT-PCR. Lane 1, Phi-X HaeIII marker. Lanes 2–5, ORF 7 PCR on the following dilutions of 111/92 (EU-type virus): (2) 10 TCID50 ml−1. (3) 1 TCID50 ml−1. (4) 0.1 TCID50 ml−1. (5) 0.01 TCID50 ml−1. Lanes 6–9, ORF 7 PCR on the following dilutions of VR2332 (US-type virus): (6) 100 TCID50 ml−1. (7) 10 TCID50 ml−1. (8) 1 TCID50 ml−1. (9) 0.1 TCID50 ml−1. Lane 10, negative control: ORF 7 PCR on boar semen without PRRSV. Lanes 11–14, EU ORF 5 PCR on the following dilutions of 111/92 (EU-type virus): 11, 10 TCID50 ml−1. 12, 1 TCID50 ml−1. 13, 0,1 TCID50 ml−1. 14, 0,01 TCID50 ml−1. Lane 15, negative control: EU ORF 5 PCR on 100 TCID50 ml−1 of VR2332 (US-type virus). Lanes 16–19, US ORF 5 PCR on the following dilutions of VR2332 (US-type virus): 16, 100 TCID50 ml−1. 17, 10 TCID50 ml−1. 18, 1 TCID50 ml−1. 19, 0,1 TCID50 ml−1. Lane 20, negative control: US ORF 5 PCR on 10 TCID50 ml−1 111/92 (DK-type virus).
Since the inter-type size difference of the ORF 7 amplicon is only 4%, and the GC content is similar (51% for the slightly larger VR2332 amplicon, 54% for the smaller 111/92 amplicon), detection sensitivity of the ORF 7 PCR was expected to be similar for US- and EU-type viruses. In fact, the US-type reference virus was detected at 1 TCID50 ml−1, while the EU-type reference virus produced a strong band at 1 TCID50 ml−1, and a very faint one at 0.1 TCID50 ml−1. Given the inherent uncertainties in titering and diluting the reference viruses, we consider the results to be in exceedingly good accordance with the theoretical prediction of bias-less detection.
The EU ORF 5 PCR detected virus in a 10 TCID50 ml−1 dilution, and the US ORF 5 PCR detected virus in a 1 TCID50 ml−1 dilution. Thus, these PCRs exhibited a high sensitivity, which was comparable to the ORF 7 PCR. Despite the high sensitivity of ORF 5 PCRs on the cognate virus-type, no reaction was seen with the reciprocal virus-type (Fig. 3, Lanes 15 and 20).
Based on at least three independent experiments with each PCR on virus dilutions in semen and serum, our experience was that in dilutions below 10 TCID50 ml−1, detection of PRRSV was indeed often possible, but became more variable (not shown); therefore, 10 TCID50 ml−1 was set as the nominal sensitivity for all three PCR tests.
3.3. The ORF 7 PCR is able to identify the concomitant presence of US- and EU-type virus in ratios spanning two orders of magnitude
The power of the ORF 7 PCR for bias-less detection and typing of both EU and US PRRSV in a single reaction makes this test highly suited for countries where European and American PRRSV-types coexist, such as Denmark. Yet, the value of the test would be greatly enhanced if ORF 7 PCR could also distinguish between infection with a single PRRSV-type, and concomitant infection in the same animal with US- and EU-types. The use of size polymorphy in an ORF 7 amplicon to type PRRSV viruses was first described by Mardassi et al., but these authors did not investigate the applicability of the technique to double infection. The results in Fig. 4 shows that ORF 7 PCR in fact correctly identified mixed-infection situations by displaying both size variants of the ORF 7 amplicon (compare lanes 2, 4 and 6). In the experiment shown in Fig. 4, 3% of a minority-type was detected in a mixed population (see Lanes 3 and 5. The US-type band in Lane 3 was clearly visible on the original photo). Further experiments indicated that a 1 : 100 ratio between viruses (measured in TCID50) resulted in the simultaneous appearance of both size variants of the ORF 7 amplicon, whereas a 1 : 1.000 ratio between viruses lay below the resolving ability of the ORF 7 PCR. Importantly, judged by eye, the ratio between viruses was reflected in relative intensities of ORF 7 bands (compare Lanes 3, 4 and 5). The difference in intensities of ORF 7 bands from the minor virus population in Lanes 3 (very faint band from 3% US-type PRRSV) and 5 (somewhat stronger band from 3% EU-type PRRSV) most likely reflected the uncertainty in titrating stock viruses, which would be expected to figure very prominently in this type of analysis. Thus, the ORF 7 PCR was able not only to identify a mixed population, but also to provide semi-quantitative information about the population composition in the sample.
Fig. 4.

Examination of mixed-type PRRSV populations by ORF 7 PCR. Reference viruses VR2332 (American-type) and 111/92 (European-type) were mixed in different ratios (based on TCID50), but maintaining a constant PRRSV concentration of 100 TCID50 ml−1 (thus ensuring that even a 3% minority-type would be above the detection sensitivity of the RT-PCR), and the mixed populations were analyzed by ORF 7 RT-PCR. Lanes 1 and 7, `5 + 7' size marker, as described in Fig. 2. Lane 2, EU-type virus only. Lane 3, EU : US-type ratio of 29 : 1. Lane 4, EU : US-type ratio of 1 : 1. Lane 5, EU : US-type ratio of 1 : 29. Lane 6, US-type virus only.
4. Discussion
European as well as American-types of PRRSV are currently associated with pathogenicity in Danish pigs; the former type entered Denmark in 1992 as part of the general appearance of PRRSV in Western Europe, and the latter type was introduced in 1996, due to the use of an attenuated, live PRRSV vaccine based on American-type virus. This situation requires that diagnostic tests be able to detect as well as precisely type the PRRSV virus. Virus isolation on PPAM and MARC monolayers is currently the gold standard of virus detection; yet, some PRRSV-containing materials (such as semen) contain too high levels of cytotoxic compounds to be reliably screened by tissue culture methods (Christopher-Hennings et al., 1995b; Nielsen et al., 1997; Shin et al., 1997). Typing PRRSV isolates with EU-type specific and US-type-specific MAbs is a rapid and accurate method; however, finer sub-typing of virus is not currently possible with this method, excluding detailed epidemiological studies. Although a number of RT-PCR tests for PRRSV have been published by others, the current situation in Denmark (co-existence of European and American-type PRRSV) poses unique requirements, particularly for precise differentiation of European, wildtype-American and vaccine-American PRRSV. Thus, the RT-PCR test described above was tailored to specifically address these diagnostic concerns.
By optimizing RNA extraction as well as crucial steps of the RT-PCR procedure, we routinely detected PRRSV diluted in clinically relevant materials such as serum and boar semen at a concentration of 1–10 TCID50 ml−1. In previous studies, detection of PRRSV at these low levels in boar semen required fractionation of semen and nested PCR (Christopher-Hennings et al., 1995b; Shin et al., 1997), making the test cumbersome for routine use, or was accompanied by a very small PCR amplicon size (Legeay et al., 1997), making the test of limited value for virus-typing/sequencing. Thus, the ability to sensitively amplify PRRSV sequences of up to 820 bp from `difficult' material such as boar semen represents a significant and non-trivial technical advantage of our assay.
The nominal sensitivity of the RT-PCR test (10 TCID50 ml−1) is very similar to the sensitivity of routine cell culture methods. In fact, we detected virus by RT-PCR in 5/5 samples of lung and pleural fluid from which the virus was also isolated by cell culture (Table 2). Thus, RT-PCR detection is mainly expected to be advantageous on material where culture is considered difficult, such as boar semen. It should be mentioned that since clinical material such as serum and lung homogenate contain up to 1000 TCID50 ml−1 during the peak of viral replication at 10–14 days post-infection, the RT-PCR test is expected to have up to a 100-fold excess of sensitivity for direct detection of PRRSV in the tissues of infected pigs, without any need for nested PCR. These calculations were supported by preliminary semi-quantitative RT-PCR analysis: sera, lung homogenates and semen from PRRSV-infected pigs at 10–14 days post-infection could be diluted 10–100-fold, and still be detected positive by ORF 7 RT-PCR. Furthermore, our ORF 7 RT-PCR detected PRRSV nucleic acid in the semen of experimentally infected boars from 4 to 42 days post-infection, without need for nested PCR (results not shown). Further experiments are planned to examine the duration and quantity of PRRSV excretion in boar semen.
Robust RT-PCR detection requires that primer-binding sites in the viral genome should be conserved over time. Although primers (Table 1) were chosen in what appeared to be conserved regions by sequence analysis, experimental confirmation of this seemed important, because of the pronounced genetic heterogeneity between PRRSV isolates. When applied to a panel of 52 PRRSV field isolates from the period 1991–1997, the test in fact correctly detected PRRSV in all the samples (Table 2). Thus, primer-binding sites did not significantly change over a 6-year period, and, by inference, the risk of PRRSV evolution interfering with detection in our test would appear to be small.
Differential sensitivity of the RT-PCR for various PRRSV-types is an important characteristic, determining the usefulness of the test for epidemiological studies. Experimental data (Fig. 3) suggest that the ORF 7 PCR exhibits equal sensitivity for European- and American-types, making it a highly suited test for estimating type prevalence. Theoretical considerations strongly support that ORF 7 PCR should exhibit bias-less detection: detection of all types is accomplished by the same RT-PCR primers, and the ORF 7 amplicons from EU- and US-types exhibit similar GC content and only a slight (4%) size difference, suggesting similar amplification efficiencies in RT-PCR. For the two ORF 5 PCR, the situation was more complex: since both ORF 5 PCR detected the cognate virus-type at 10 TCID50 ml−1, we conclude that no type-bias existed at virus concentrations well below those normally found in clinical material. However, we cannot exclude that detection with ORF 5 PCR may be biased at extremely low virus concentrations.
Typing of PRRSV in the RT-PCR assay was accomplished by three different strategies (direct sequencing of the PCR product, type-specific PCR primers, and size determination of the ORF 7 amplicon), which all showed complete concordance with the currently used method of MAb-typing. Sequencing is cumbersome, but allows unrivalled precision in typing, with sub-typing of PRRSV even within the main categories of `European' and `American' being possible. On the other hand, use of type-specific primers or size determination of ORF 7 amplicons are rapid methods, that offer broad classification of virus as `European' or `American' only. Together, the flexibility of the described RT-PCR assay should thus cover most diagnostic eventualities.
The specificity of our RT-PCR test rests on the detection and precise sizing of amplicons by agarose gel electrophoresis. Although the test was optimized to avoid unspecific bands, and did not show cross-reactivity to a number of relevant viruses (see Section 2), it can be anticipated that additional examination of the specificity of an amplicon may be required. For this purpose, RFLP analysis offers a very simple and attractive alternative to the more impractical techniques of sequencing, Southern blotting or nested PCR. In fact, the relatively large amplicon size in our test (637–818 bp) provides an ideal target for RFLP. For example, analysis of our own as well as GeneBank ORF 7 sequences (total of 61 European- and 19 American-type) predicts that the digestion of the ORF 7 amplicon produced in our test with a single, inexpensive restriction enzyme (HaeII), could provide discrimination between US- (215 and 445 nt fragments) and EU-types (299 and 338 nt fragments). Also, the ORF 7 sequence data indicated that a 29 mer oligonucleotide (5′ AT GGC CAG CCA GTC AAT CA A/G CTG TGC AAG 3′) would hybridize to an internal, highly conserved sequence in ORF 7 amplicons from EU as well as US viruses. Preliminary experiments indicate that using an alkaline phosphatase-labelled form of this oligo in a rapid dot-blot assay is highly useful for enhancing both specificity and sensitivity of the ORF 7 RT-PCR.
Given the proclivity of RNA viruses to recombine (Lai, 1992; Lai and Banner, 1991), the presence of two radically disparate PRRSV-types in Denmark raises the question of PRRSV genomic stability, which in turn may have implications for detection and pathogenicity of viruses. Insight into these problems will require the study of animals naturally or experimentally co-infected with both PRRSV-types. For such studies, easy semi-quantitative assessment of mixed viral populations over a wide-type ratio, as is afforded by the ORF 7 PCR, is likely to prove valuable.
Acknowledgements
F. Lihme and P. Normann are thanked for excellent and invaluable technical assistance. J. Nielsen and S. Alexandersen are thanked for helpful suggestions during the project and during the writing of this manuscript.
References
- Boom R., Sol C.J.A., Salimans M.M.M., Jansen C.L., Wertheim-van Dillen P.M.E., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøtner A., Nielsen J., Bille-Hansen V. Isolation of porcine reproductive and respiratory syndrome (PRRS) virus in a Danish swine herd and experimental infection of pregnant gilts with the virus. Vet. Microbiol. 1994;40:351–360. doi: 10.1016/0378-1135(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Bøtner A., Strandbygaard B., Sørensen K.J., Have P., Madsen K.G., Smedegaard Madsen E., Alexandersen S. Appearance of acute PRRS-like symptoms in sow herds after vaccination with a modified live PRRS vaccine. Vet. Rec. 1997;141:497–499. doi: 10.1136/vr.141.19.497. [DOI] [PubMed] [Google Scholar]
- Christopher-Hennings, J., Nelson, E.A., Hines, R.J., Nelson, J.K., Swenson, S.L., Zimmerman, J.J., Chase, C.L., Yaeger, M.J., Benfield, D.A., 1995a. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of abult boars. J. Vet. Diag. Invest. 7, 456–464 [DOI] [PubMed]
- Christopher-Hennings, J., Nelson, E.A., Nelson, J.K. Hines, R.J., Swenson, S.L., Hill, H.T., Zimmerman, J.J., Katz, J.B., Yaeger, M.J., Chase, C.C., 1995b. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J. Clin. Microbiol. 33, 1730–1734 [DOI] [PMC free article] [PubMed]
- Duan X., Nauwynck H.J., Pensaert M.B. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV) Vet. Microbiol. 1997;56:9–19. doi: 10.1016/S0378-1135(96)01347-8. [DOI] [PubMed] [Google Scholar]
- Gilbert S.A., Larochelle R., Magar R., Cho H.J., Deregt D. Typing of porcine reproductive and respiratory syndrome viruses by a multiplex PCR assay. J. Clin. Microbiol. 1997;35:264–267. doi: 10.1128/jcm.35.1.264-267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradil C., Dubuc C., Eaglesome M.D. Porcine reproductive and respiratory syndrome virus: Seminal transmission. Vet. Rec. 1996;138:521–522. doi: 10.1136/vr.138.21.521. [DOI] [PubMed] [Google Scholar]
- Kapur V., Elam M.R., Pawlovich T.M., Murtaugh M.P. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the mid-western United States. J. Gen. Virol. 1996;77:1271–1276. doi: 10.1099/0022-1317-77-6-1271. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Kwang J., Yoon I.J., Joo H.S., Frey M.L. Enhanced replication of poroine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- Kono Y., Kanno T., Shimizu M., Yamada S., Ohashi S., Nakamine M., Shirai J. Nested PCR for detection and typing of porcine reproductive and respiratory syndrome (PRRS) virus in pigs. JV Medical Science. 1996;58:941–946. doi: 10.1292/jvms.58.10_941. [DOI] [PubMed] [Google Scholar]
- Lai M.C. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C., Banner L.R. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology. 1991;185:441–445. doi: 10.1016/0042-6822(91)90795-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle R., Magar R. Evaluation of the presence of porcine reproductive and respiratory syndrome virus in packaged pig meat using virus isolation and polymerase chain reaction (PCR) method. Vet. Microbiol. 1997;58:1–8. doi: 10.1016/s0378-1135(97)00150-8. [DOI] [PubMed] [Google Scholar]
- Legeay O., Bounaix S., Denis M., Arnauld C., Hutet E., Cariolet R., Albina E., Jestin A. Development of a RT-PCR test coupled with a microplate colorimetric assay for the detection of a swine Arterivirus (PRRSV) in boar semen. J. Virol. Methods. 1997;68:65–80. doi: 10.1016/s0166-0934(97)00110-9. [DOI] [PubMed] [Google Scholar]
- Madsen K.G., Hansen C.M., Madsen E.S., Strandbygaard B., Bøtner A., Sørensen K.J. Detection of porcine reproductive and respiratory syndrome virus of the American-type in Danish swine herds. Arch. Virol. 1998;143:1683–1700. doi: 10.1007/s007050050409. [DOI] [PubMed] [Google Scholar]
- Mardassi H., Wilson L., Mounir S., Dea S. Detection of porcine reproductive and respiratory syndrome virus and efficient differentiation between Canadian and European strains by reverse transcription and PCR amplification. J. Clin. Microbiol. 1994;32:2197–2203. doi: 10.1128/jcm.32.9.2197-2203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Halbur P.G., Lum M.A. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): Implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch. Virol. 1995;140:745–755. doi: 10.1007/BF01309962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J.M., Hulst M.M., De Meijer E.J., Moonen P.L.J.M., Den Besten A., De Kluyver E.P., Wensvoort G., Moormann R.J.M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.M., Petersen-den B.A., Kluyver E.d., Moormann R.M., Schaaper W.M., Wensvoort G., De K.E. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J., Besten A.d., Kluyver E.d., Nieuwstadt A.v., Wensvoort G., Moormann R.J. Molecular characterization of Lelystad virus. Vet. Microbiol. 1997;55:197–202. doi: 10.1016/S0378-1135(96)01335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy N.R., Leinbach S.S., Hellwig R.J. A potent, cost-effective RNase inhibitor. BioTechniques. 1995;18:1068–1073. [PubMed] [Google Scholar]
- Murtaugh M.P., Elam M.R., Kakach L.T. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch. Virol. 1995;140:1451–1460. doi: 10.1007/BF01322671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J., Bøtner, A., Oleksiewicz, M.B., Storgaard, T., 1998. Experimental inoculation of late-term pregnant sows with a field isolate of PRRS vaccine-like virus, In: Proc. 15th Int. Pig Vet. Soc. Congr., Birmingham, UK
- Nielsen T.L., Nielsen J., Have P., Baekbo P., Hoff J.R., Botner A. Examination of virus shedding in semen from vaccinated and from previously infected boars after experimental challenge with porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 1997;54:101–112. doi: 10.1016/s0378-1135(96)01272-2. [DOI] [PubMed] [Google Scholar]
- Sellner L.N., Coelen R.J., Mackenzie J.S. Revers transcriptase inhibits Taq polymerase activity. Nucleic Acids Res. 1992;20:1487–1490. doi: 10.1093/nar/20.7.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Torrison J., Choi C.S., Gonzalez S.M., Crabo B., Molitor T.W. Monitoring of porcine reproductive and respiratory syndrome virus infection in boars. Vet. Microbiol. 1997;55:337–346. doi: 10.1016/s0378-1135(96)01336-3. [DOI] [PubMed] [Google Scholar]
- Storgaard, T., 1998. (unpub)
- Suarez P., Zardoya R., Martin M.J., Prieto C., Dopazo J., Solana A., Castro J.M. Phylogenetic relationships of european strains of porcine reproductive and respiratory syndrome virus (PRRSV) inferred from DNA sequences of putative ORF-5 and ORF-7 genes. Virus Res. 1996;42:159–165. doi: 10.1016/0168-1702(95)01305-9. [DOI] [PubMed] [Google Scholar]
- Suárez P., Zardoya R., Prieto C., Solana A., Tabarés E., Bautista J.M., Castro J.M. Direct detection of the porcine reproductive and respiratory syndrome (PRRS) virus by reverse polymerase chain reaction (RT-PCR) Arch. Virol. 1994;135:89–99. doi: 10.1007/BF01309767. [DOI] [PubMed] [Google Scholar]
- Sur J.-H., Doster A.R., Christian J.S., Galeota J.A., Wills R.W., Zimmerman J.J., Osorio F.A. Porcine reproductive and respiratory syndrome virus replicates in testicular germ cells, alters spermatogenesis, and induces germ cell death by apoptosis. J. Virol. 1997;71:9170–9179. doi: 10.1128/jvi.71.12.9170-9179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson S.L., Hill H.T., Zimmerman J.J., Evans L.E., Landgraf J.G., Wills R.W., Sanderson T.P., McGinley M.J., Brevik A.K., Ciszewski D.K. Excretion of porcine reproductive and respiratory syndrome virus in semen after experimentally induced infection in boars. J. Am. Vet. Med. Assoc. 1994;204:1943–1948. [PubMed] [Google Scholar]
- Sørensen K.J., Bøtner A., Smedegaard Madsen E., Strandbygaard B., Nielsen J. Evaluation of a blocking ELISA for screening of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus. Vet. Microbiol. 1997;56:1–8. doi: 10.1016/s0378-1135(96)01345-4. [DOI] [PubMed] [Google Scholar]
- Sørensen K.J., Strandbygaard B., Bøtner A., Madsen E.S., Nielsen J., Have P. Blocking ELISA's for the distinction between antibodies against European and American strains of porcine reproductive and respiratory syndrome (PRRS) virus. Vet. Microbiol. 1998;60:169–177. doi: 10.1016/s0378-1135(98)00159-x. [DOI] [PubMed] [Google Scholar]
- Van Woensel P., Van der Wouw J., Visser N. Detection of porcine reproductive respiratory syndrome virus by the polymerase chain reaction. J. Virol. Methods. 1994;47:273–278. doi: 10.1016/0166-0934(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Yoon K.J., Zimmerman J.J., McGinley M.J., Landgraf J., Frey M.L., Hill H.T., Platt K.B. Failure to consider the antigenic diversity of porcine reproductive and respiratory syndrome (PRRS) virus isolates may lead to misdiagnosis. J. Vet. Diagn. Invest. 1995;7:386–387. doi: 10.1177/104063879500700315. [DOI] [PubMed] [Google Scholar]


