Abstract
Recently, a coronavirus strain (179/07-11) was isolated from water buffalo (Bubalus bubalis) and the virus which displayed a strict genetic and biological relatedness with bovine coronavirus (BCoV) was referred to as bubaline coronavirus (BuCoV). Here, we report the characterisation of four BuCoVs strains identified in the faeces or intestinal contents of water buffalo calves with acute gastroenteritis. Single BuCoV infections were detected in all but one cases from which two clostridia species were also isolated. Sequence and phylogenetic analyses of the 5′ end of the spike-protein gene showed that three BuCoVs were closely related to the prototype strain 179/07-11, whereas the fourth isolate (339/08-C) displayed a higher genetic identity to recent BCoV reference strains. Three strains adapted to the in vitro grow on human rectal tumour cells were also evaluated for their ability to replicate in a bovine cell line (Madin Darby bovine kidney) and to cause haemagglutination of chicken erythrocytes and all displayed biological properties similar to those already described for the prototype BuCoV. The present report shows that albeit genetically heterogeneous, the different BuCoV strains possess a common biological pattern which is different from most BCoV and BCoV-like isolates.
Keywords: Coronavirus, Bubalus bubalis, Gastroenteritis, Virus characterisation
1. Introduction
Coronaviruses (CoVs) are enveloped, positive-stranded RNA viruses that cause respiratory and/or enteric disease in mammals and birds (Enjuanes et al., 2000). With the largest known genomic RNA, CoVs are prone to dramatic shifts of the tissue tropism or host range through accumulation of point mutations or deletions/insertions or recombination events affecting the structural and non-structural proteins (Decaro and Buonavoglia, 2008). Currently, CoVs are classified into three different genetic and antigenic groups with group 2 being further divided into subgroups 2a and 2b. Subgroup 2a consists of mouse hepatitis virus, rat sialodacryadenitis virus, human coronavirus (HCoV) HKU1 (Woo et al., 2005) and several bovine-like CoVs, including bovine coronavirus (BCoV), porcine haemagglutinating encephalomyelitis virus (PHEV), HCoV-OC43, human enteric coronavirus (HECoV) 4408 (Enjuanes et al., 2000), and the newly recognized equine coronavirus (ECoV) (Guy et al., 2000) and canine respiratory coronavirus (CRCoV) (Erles et al., 2003, Decaro et al., 2007). There are multiple genetic and antigenic evidences that several subgroup 2a CoVs, such as HCoV-OC43, HECoV-4408, PHEV and CRCoV, have arisen as consequence of trans-species infections caused by BCoV (Zhang et al., 1994, Vijgen et al., 2005, Vijgen et al., 2006, Erles et al., 2007, Lorusso et al., 2009). According to the proposal of the Coronavirus Study Group of the International Committee of Taxonomy of Viruses, given their close genetic relatedness, all BCoV-like CoVs are now considered not as separate viruses but rather as host range variants of the same species Betacoronavirus 1 within genus Betacoronavirus (formerly group 2) (de Groot et al., 2008).
Recently, another bovine-like CoV, which was referred to as bubaline coronavirus (BuCoV), was isolated from water buffalo (Bubalus bubalis) calves with fatal gastroenteritis in Italy (Decaro et al., 2008d). The virus was characterised at the genetic level, displaying intact structural and non-structural proteins with respect to BCoV, whereas biologically it presented unique characteristics including the inability to grow on bovine derived cell cultures and to agglutinate chicken erythrocytes.
Here, we report the clinical history and virus characterisation of four outbreaks of gastroenteric disease caused by BuCoV in Italian buffalo herds.
2. Material and methods
2.1. Clinical outbreaks
The first outbreak was registered in July, 2007 in a buffalo herd of the Caserta province (Campania region) and involved 7–15-day-old calves with the occurrence of depression, anorexia and haemorrhagic diarrhoea. The affected calves recovered progressively after treatment with oral rehydrants and antibiotics (sulfadimidin + trimetoprim). The laboratory analyses were carried out on faecal samples collected from six diarrhoeic calves (339/07-A-F).
Signs of enteritis and tympanism occurred in February, 2008 in 10–60-day-old calves of a buffalo herd of the province of Latina (Lazio region). In this case, the mortality rates did not exceed 20%. Necropsy carried out on a dead buffalo calf (82/08) revealed the presence of severe gastroenteritis, with enlargement of the mesenteric lymph nodes and gall bladder.
Another gastroenteritis outbreak was observed in April, 2008 in the Caserta province. The affected buffalo calves, 15–20 days of age, showed anorexia, apathy, yellow or bloody diarrhoea and dehydration, which led to the death of the affected animals within 48–72 h after the onset of clinical signs. The mortality rate was about 30%. The carcasses of two calves (153/08-A, 153/08-B) were submitted to necropsy, showing congestion and haemorrhages in the small and large intestines that were filled with abundant liquid content.
In the same period, clinical signs of coronavirosis (loss of appetite, liquid diarrhoea) were observed in 10–20-day-old calves form a buffalo herd of the Foggia province (Apulia region). In this case, the dams had been vaccinated against rotavirus, BCoV and Escherichia coli before calving in order to transfer the maternally derived immunity to their offspring. At necropsy on a dead buffalo calf (155/08), the typical gross lesions of coronaviral enteritis were observed and the intestinal content was collected for routine analyses.
2.2. Bacteriological and parasitological investigations
For bacteriological investigations, tissue samples and faeces or intestinal contents were plated out on 5% sheep blood agar and cultured aerobically at 37 °C for 24 h for detection of aerobic pathogens. Anaerobic pathogens were searched for by obtaining ten-fold dilutions of the samples in fluid thioglycolate medium and subsequent plating each sample dilution onto 5% sheep blood agar and egg yolk agar with d-cycloserine 400 μg/ml. Bacteria were allowed to grow overnight at 37 °C in anaerobic condition. Bacteria identification was achieved by standard biochemical procedures and analytical profile index (API, BioMérieux Italia S.p.A., Rome, Italy). To detect intestinal parasites in the faeces or intestinal contents, zinc sulphate flotation was used, whereas the Ziehl Nielsen staining was performed on the faecal samples or intestinal sections for identification of Cryptosporidium spp.
2.3. Screening for viral pathogens
DNA extraction from tissue samples and faeces or intestinal contents was carried out by means of the DNeasy Tissue Kit (QIAGEN S.p.A., Milan, Italy), whereas RNA was extracted from the fecal samples or intestinal contents and from tissues by using the QIAamp® Viral RNA Mini Kit and QIAamp® RNeasy Mini Kit (QIAGEN S.p.A.), respectively. DNA extracts were subjected to PCR assays for detection of bovine herpesvirus types 1 (BoHV-1) (Vilcek, 1993) and 4 (BoHV-4) (Boerner et al., 1999) using the kit LA PCR Kit Ver. 2.1 (TaKaRa Bio Inc., Shiga, Japan). RNA extracts were used for detection of BCoV and BCoV-like viruses (Erles et al., 2003), toroviruses (Hoet et al., 2002), rotaviruses (Gouvea et al., 1994), caliciviruses (Jiang et al., 1999), bovine viral diarrhea virus (BVDV) (Sullivan and Akkina, 1995) and bovine respiratory syncytial virus (Valarcher et al., 1999). RT-PCR assays were performed using SuperScript™ One-Step RT-PCR for Long Templates (Life Technologies, Invitrogen srl, Milan, Italy).
2.4. Quantification of BCoV-like CoV RNA
The RNA of BCoV-like CoVs was quantified in the RT-PCR positive samples by means of a real-time RT-PCR assay recently established (Decaro et al., 2008b). Briefly, viral RNAs were reverse-transcribed using GeneAmp® RNA PCR (Applied Biosystems, Applera Italia, Monza, Italy) and the c-DNAs were real-time PCR-amplified in a 50 μl-reaction mixture containing 25 μl of IQ™ Supermix (Bio-Rad Laboratories Srl), 600 nM of primers BCoV-F (CCTTCATATCTATACACATCAAGTTGTT) and BCoV-R (ACCAGCCATTTTAAATCCTTCA), 200 nM of probe BCoV-Pb (6FAM - CCTTCATATCTATACACATCAAGTTGTT-BHQ1) and 20 μl of c-DNA. The thermal profile consisted of activation of iTaq DNA polymerase at 95° C for 10 min, followed by 45 cycles of denaturation at 95° C for 15 s and annealing-extension at 60° C for 1 min. BCoV-like RNA copy numbers were calculated on the basis of the standard curves generated by 10-fold dilutions of a synthetic RNA obtained by in vitro transcription of a plasmid containing the M gene of BCoV strain 339/06 (Decaro et al., 2008c).
2.5. Virus isolation, haemagglutination and receptor-destroying enzyme activity
BCoV-like-positive samples representative of each outbreak (339/07-C, 82/08, 153/08-B, 155/08) were subjected to virus isolation attempts on human rectal tumour (HRT-18) and Madin Darby bovine kidney (MDBK) cells, grown in Dulbecco's minimal essential medium (D-MEM) added with 10% foetal calf serum, as previously described (Decaro et al., 2008d). Viral growth was monitored by visual inspection of cytopathic effect and by an immunofluorescence assay using a BCoV-positive bovine serum and a rabbit anti-bovine IgG conjugated with fluorescein isothiocyanate (Sigma Aldrich srl, Milan, Italy).
Viral isolates were tested as previously described (Hasoksuz et al., 1999, Decaro et al., 2008d) for their haemagglutination and receptor-destroying enzyme activities in comparison with the BuCoV prototype isolate 179/07-11 and BCoV enteric isolates 339/06 (Decaro et al., 2008c) and 9WBL7 (kindly provided by Dr Paolo Cordioli, Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna, Brescia, Italy) and nasal isolate 438/06-2 (Decaro et al., 2008a).
2.6. RT-PCR amplification, sequence analysis and phylogeny of the spike-protein gene
The 5′ end of the spike-protein (S) gene of the detected BuCoVs was amplified using primers BCV-23510F (TATGATCCGCTACCAATTATTTTGCTTGGCA) and BCV-24326R (ACAACACCAGTGTCTGTAAAATATGCA) and SuperScript™ One-Step RT-PCR for Long Templates (Invitrogen srl, Milan, Italy), according to the manufacturer's instructions and a previous report (Decaro et al., 2008d). The PCR-amplified products (817 bp) were cloned into pCR®2.1-TOPO® vectors (TOPO TA Cloning®, Invitrogen srl) and the recombinant clones individualized by blue/white screening. Plasmid DNAs were sequenced by Genome Express (Meylan, France) and the obtained sequences were assembled and analysed using the BioEdit software package (Hall, 1999) and the NCBI's (http://www.ncbi.nlm.nih.gov) and EMBL's (http://www.ebi.ac.uk) analysis tools. Phylogenetic and molecular evolutionary analyses were conducted using Mega3 (Kumar et al., 2004). Phylogenetic trees based on 686 nucleotides (nt) of the S-gene 5′ end were elaborated using both parsimony and neighbor-joining methods, supplying a statistical support with bootstrapping over 1000 replicates.
2.7. Nucleotide sequence accession number
The nt sequences of the partial S gene sequences of the four bubaline strains have been deposited in GenBank under accession numbers GU001944–GU001947.
3. Results
3.1. Identification of BuCoV in clinical samples
All collected faecal samples and intestinal contents tested positive for bovine-like CoVs by a classical, gel-based RT-PCR targeting the S gene. A TaqMan assay confirmed the presence of the nucleic acid of BCoV-like CoVs in all tested samples but intestinal content of calf 82/08, with viral RNA titres ranging from 3.31 × 102 to 7.16 × 106 viral RNA copies μl−1 template (Table 1 ). Electrophoretic analysis of the real-time RT-PCR products showed the presence of the band of the expected size also for sample 82/08, thus proving the presence of the viral RNA in the tested sample as obtained by conventional RT-PCR. The band was excised form the gel, purified and sequenced, displaying a mismatch in the probe binding region, that could have prevented the annealing of the specific probe (data not shown).
Table 1.
BuCoV detection in buffalo calves with diarrhoea by virus isolation, conventional and real-time RT-PCR.
| Prot. no. | Age of the animals | Sample type | Virus isolation | RT-PCR | Real-time RT-PCRa |
|---|---|---|---|---|---|
| 339/07-A | 15 days | Faeces | ND | + | 2.45 × 104 |
| 339/07-B | 11 days | Faeces | ND | + | 6.54 × 103 |
| 339/07-C | 11 days | Faeces | + | + | 8.17 × 104 |
| 339/07-D | 10 days | Faeces | ND | + | 3.31 × 102 |
| 339/07-E | 11 days | Faeces | ND | + | 5.08 × 103 |
| 339/07-F | 7 days | Faeces | ND | + | 1.13 × 104 |
| 82/08 | 23 days | Intestinal content | − | + | − |
| 153/08-A | 15 days | Intestinal content | ND | + | 3.26 × 105 |
| 153/08-B | 18 days | Intestinal content | + | + | 7.16 × 106 |
| 155/08 | 20 days | Intestinal content | + | + | 4.30 × 106 |
+, positive; −, negative; ND, not done.
Real-time RT-PCR results are expressed as RNA copies μl−1 of template.
Common parasites of ruminants were not recovered from any gastroenteritis outbreaks, whereas only the intestinal content of calf 82/08 was found to contain high amounts of clostridia, including Clostridium perfringens type A and Clostridium butyrricum.
3.2. Biological characterisation
All BuCoVs inoculated on cell cultures but strain 82/08 were successfully isolated on HRT-18 cells (Table 1), reaching at the 3rd passage viral titres higher than 107 TCID50 ml−1. In order to evaluate the growth characteristics of the BuCoV strains, prototype isolates from each outbreak (3rd passages on HRT-18 cells) were subjected to ten consecutive passages on MDBK cells and the cryolysates of the last passage were submitted to real-time RT-PCR. As shown in Fig. 1 , the bubaline strains were not able to replicate efficiently on the bovine cell line. In fact, by using real-time RT-PCR quantification, viral replication of strains 339/07-C and 155/08 was no longer observed after the fifth and eighth passages, respectively, whereas the RNA of strain 153/08-B was detected at the 10th passage but at a low titre.
Fig. 1.
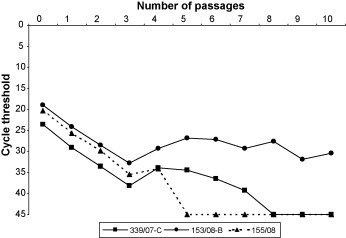
Kinetic of the growth of the bubaline coronavirus isolates on Madin Darby bovine kidney cells. The number of passages is indicated on the x-axis, whereas the corresponding cycle threshold (CT) values are presented on the y-axis. Passage 0 is the inoculum used to infect the bovine cells and corresponding to the cryolysate of the 3rd passage on HRT-18 cells.
The three BuCoV isolates as well as the BuCoV and BCoV reference strains agglutinated mouse erythrocytes at high titres, whereas only the BuCoV isolate 155/08 was able to cause low-titre HA of chicken erythrocytes. In contrast, all BCoV reference strains agglutinated chicken erythrocytes with high efficiency. As previously observed (Decaro et al., 2008d), HA titres were higher at 4 °C than at 37 °C. Using mouse erythrocytes, RDE activity (loss of the HA pattern) was observed for the bovine and bubaline isolates (Table 2 ).
Table 2.
Haemagglutination (HA) and receptor-destroying enzyme (RDE) titers of BuCoV and BCoV isolates.
| CoV isolate | Origin | HA titrea |
RDE titrea |
||||
|---|---|---|---|---|---|---|---|
| 4 °C |
37 °C |
Mouse | Chicken | ||||
| Mouse | Chicken | Mouse | Chicken | ||||
| BuCoV-339/07 | Faeces | 512 | <2 | 128 | <2 | <2 | <2 |
| BuCoV-153/08 | Intestinal content | 512 | <2 | 256 | <2 | 256 | <2 |
| BuCoV-155/08 | Intestinal content | 128 | 16 | 64 | 4 | 128 | <2 |
| BuCoV-179/07-11 | Faeces | 512 | <2 | 128 | <2 | 128 | <2 |
| BCoV-339/06 | Faeces | 128 | 64 | 64 | 32 | 64 | 16 |
| BCoV-9WBL7 | Faeces | 1024 | 256 | 1024 | 64 | 256 | <2 |
| BCoV-438/06-2 | Nasal swab | 512 | 256 | 128 | 16 | 128 | 64 |
HA and RDE titres are referred to 50 μl of viral suspension.
3.3. Genetic characterisation
The 5′ end of the S gene of the four prototype BuCoV strains was PCR-amplified and sequenced as previously described (Decaro et al., 2008d) and the obtained sequences were compared to reference BuCoV strain 179/07-11 and BCoV strains Mebus (old strain) and 339/06 (recently isolated in Italy, Decaro et al., 2008c). By sequence analysis of the 5′ end 686 nt, three out of four BuCoV strains were found to be closely related to each other (98.7–99.3% of nt identity) and to the prototype isolate 179/07-11 (99.0–99.6%), whereas strain 339/07-C displayed the highest identity (97.5%) to the recent Italian BCoV isolate 339/06 and only 96.2–97.2% identity to prototype and new BuCoVs (Table 3 ).
Table 3.
Nucleotide identity (%) of BuCoV strains to BuCoV and BCoV reference isolates in the 5′ end of the S gene.
| BuCoV | BuCoV | BuCoV | BuCoV | BuCoV | BCoV | BCoV | |
|---|---|---|---|---|---|---|---|
| 179/07-11 | 155/08 | 153/08-B | 339/07-C | 82/08 | 339/06 | Mebus | |
| 179/07-11 | – | 99.6 | 99.3 | 97.2 | 99.0 | 98.5 | 96.7 |
| 155/08 | – | 99.3 | 97.2 | 99.0 | 98.2 | 96.4 | |
| 153/08 | – | 96.9 | 98.7 | 97.9 | 96.1 | ||
| 339/07 | – | 96.2 | 97.5 | 96.2 | |||
| 82/08 | – | 97.5 | 96.4 | ||||
| 339/06 | – | 97.2 | |||||
| Mebus | – |
Phylogeny inferred with the neighbor-joining method showed that three BuCoV strains formed a unique cluster together with the prototype virus 179/07-11, which was separate from BCoVs albeit distantly related to the recent Italian BCoV isolate 339/06 (Fig. 2 ). In contrast, strain 339/07-C was intermingled with classical BCoV isolates, particularly with reference strains OK-0514-3 and LSU-94LSS-051-2 that had been isolated in USA from calves with respiratory disease (Chouljenko et al., 1998). This pattern of segregation was confirmed by the tree constructed with the maximum parsimony method (data not shown).
Fig. 2.
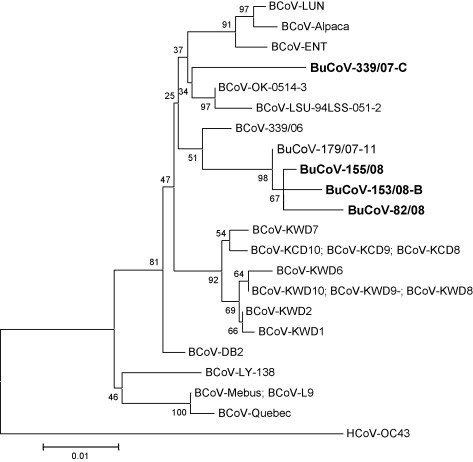
Phylogenetic analysis of bubaline coronavirus (BuCoV). Neighbor-joining tree based on partial 5′ end of the spike-protein gene of bovine and bubaline CoVs. For phylogenetic tree construction, the following reference strains were used (GenBank accession numbers are reported in parentheses): BuCoV strain 179/07-11 (EU019216); BCoV strains Mebus (U00735), Quebec (AF220295), DB2 (DQ811784), ENT (AF391541), LUN (AF391542), Alpaca (DQ915164), KWD1 (AY935637), KWD2 (AY935638), KWD7 (AY935643), KWD8 (AY935644), KWD9 (AY935645), KWD10 (AY935646), KCD8 (DQ389639), KCD9 (DQ389640), KCD10 (DQ389641), L9 (M64667), OK-0514-3 (AF058944), LSU-94LSS-051-2 (AF058943), LY-138 (AF058942), 339/06 (EF445634). The tree is rooted on the group-2 human coronavirus HCoV-OC43 (NC_005147). A statistical support was provided by bootstrapping over 1000 replicates. The scale bars indicate the estimated numbers of nucleotide substitutions per site.
4. Discussion
Recently, a coronavirus strain was isolated from an outbreak of gastroenteritis in an Italian water buffalo herd. The bubaline virus (BuCoV-179/07-11) was found to be strictly related to BCoV at the genetic level, but displayed some unique biological properties such as poor growth on MDBK cells and lack of HA activity using chicken erythrocytes (Decaro et al., 2008d). In this paper, we have reported the genetic characterisation of four additional BuCoV strains detected in buffalos with gastroenteritis. While three strains were strictly related to each other and to the prototype BuCoV isolate, strain 339/07-C was more similar to BCoVs by both sequence analysis and phylogeny of the 5′ end of the S gene. These findings seem to suggest a distinct origin of the two clusters of BuCoVs from BCoV. However, as other genomic regions were not analysed in this study, a potential origin of strain 339/07-C as recombinant virus between BuCoV and BCoV cannot be ruled out definitively.
The BCoV origin or, alternatively, the presence of a common ancestor, has been hypothesised for several group 2a CoVs, including HCoV-OC43 (Vijgen et al., 2005), PHEV (Vijgen et al., 2006), CRCoV (Erles et al., 2007, Lorusso et al., 2009) and some ruminant CoVs (Hasoksuz et al., 2007, Jin et al., 2007, Alekseev et al., 2008, Decaro et al., 2008d). We have already shown that more than one BCoV strain or ancestor virus was likely involved in the origin of CRCoV, thus leading to the emergence of different canine strains (Lorusso et al., 2009). Accordingly, a similar mechanism may have been involved in the emergence of genetically distinct BuCoV strains.
Three BuCoV strains were adapted to grow on HRT-18 cells but only one of those was able to replicate with low efficiency on MDBK cells and none displayed evident HA activity using chicken erythrocytes. These findings are in agreement with those already obtained with the prototype strain 179/07-11 (Decaro et al., 2008d), the giraffe isolate GiCoV-OH3 and recent BCoVs (Hasoksuz et al., 2007). In the HA test with mouse erythrocytes, the results for the GiCoV-OH3 strain were similar to those for BCoV strains at both 4 °C and 37 °C, whereas the ability to agglutinate chicken erythrocytes by GiCoV-OH3 was observed only at 4 °C. Similar results were also obtained for one enteric BCoV (BCoV-TS) and two respiratory BCoVs (67NS and 220NS) (Hasoksuz et al., 2007). The absent or poorly efficient adaptation to MDBK cells and the lack of HA activity with chicken red blood cells could be considered typical features of BuCoV. In fact, although other ruminant CoVs and even some BCoV strains displayed similar characteristics, those were never combined (Benfield and Saif, 1990, Hasoksuz et al., 2007). Two recent BCoV isolates from southern Italy (the enteric strain 339/06 and the respiratory strain 438/06-2) exhibited HA and growth patterns very different from those of BuCoV isolates. However, more studies of contemporary BCoV isolates in the same geographic area should be done to decide if these attributes apply only to BuCoV or if such differences are consistent among CoVs from different ruminant species.
The BuCoV strains identified so far have displayed similar biological but not genetic features, thus requiring further studies aiming to isolate and characterise additional strains.
The onward industrialisation of the bubaline farming has led to the emergence of diseases that until few years ago were considered typical of cattle herds but were rarely observed in water buffaloes. Among the emergent pathologies, viral gastroenteritis, mainly associated with coronavirus (Muniiappa et al., 1985, Decaro et al., 2008d) and rotavirus (Sunil-Chandra and Mahalingam, 1994, Martella et al., 2003) infections, is playing an increasing role in the economical losses related to the calf mortality, especially when female calves are affected. The changes in the hygienic conditions highlight the need for a more in-depth investigation of these viral infections that are frequently associated to different clinical pictures in comparison with cattle. For instance, coronavirus infections can cause enteric as well as respiratory disease in cattle (Lathrop et al., 2000, Storz et al., 2000, Decaro et al., 2008a), whereas respiratory distress has not been associated with BuCoV so far. In addition, the impact of these emerging infections in water buffalo herds should be evaluated as for the extensive use of vaccines.
In conclusion, based on the present report, a potential role as enteric pathogen should be assigned to BuCoV and this virus should be included in the diagnostic panel for the infectious agents involved in the occurrence of acute gastroenteritis in buffaloes. To what extent BuCoV impacts on the buffalo productions should be assessed in the next future through extensive epidemiological and clinical survey in buffalo herds.
Acknowledgement
This work was supported by grants from the Italian Ministry of Health, Ricerca corrente IZSME 08/07 “Studio della epidemiologia e delle caratteristiche genetiche del coronavirus del bufalo”.
References
- Alekseev K.P., Vlasova A.N., Jung K., Hasoksuz M., Zhang X., Halpin R., Wang S., Ghedin E., Spiro D., Saif L.J. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J. Virol. 2008;82:12422–12431. doi: 10.1128/JVI.01586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield D.A., Saif L.J. Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J. Clin. Microbiol. 1990;28:1454–1457. doi: 10.1128/jcm.28.6.1454-1457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner B., Weigelt W., Buhk H.J., Castrucci G., Ludwig H. A sensitive and specific PCR/Southern blot assay for detection of bovine herpesvirus 4 in calves infected experimentally. J. Virol. Methods. 1999;83:169–180. doi: 10.1016/s0166-0934(99)00117-2. [DOI] [PubMed] [Google Scholar]
- Chouljenko V.N., Kousoulas K.G., Lin X., Storz J. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes. 1998;17:3–42. doi: 10.1023/A:1008048916808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Ziebuhr J., Poon L.L., Woo P.C., Talbot P., Rottier P.J.M., Holmes K.V., Baric R., Perlman S., Enjuanes L. 2008. Revision of the Family Coronaviridae. Taxonomic Proposal of the Coronavirus Study Group to the ICTV Executive Committee. Available from http://talk.ictvonline.org/media/p/1230.aspx (accessed 20.10.09) [Google Scholar]
- Decaro N., Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet. Microbiol. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Elia G., Mari V., Lucente M.S., Cordioli P., Colaianni M.L., Martella V., Buonavoglia C. Serological and molecular evidence that canine respiratory coronavirus is circulating in Italy. Vet. Microbiol. 2007;121:225–230. doi: 10.1016/j.vetmic.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Campolo M., Desario C., Cirone F., D’abramo M., Lorusso E., Greco G., Mari V., Colaianni M.L., Elia G., Martella V., Buonavoglia C. Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy. J. Vet. Diagn. Invest. 2008;20:28–32. doi: 10.1177/104063870802000105. [DOI] [PubMed] [Google Scholar]
- Decaro N., Elia G., Campolo M., Desario C., Mari V., Radogna A., Colaianni M.L., Cirone F., Tempesta M., Buonavoglia C. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J. Virol. Methods. 2008;151:167–171. doi: 10.1016/j.jviromet.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Desario C., Campolo M., Elia G., Martella V., Greco G., Cirone F., Colaianni M.L., Cordioli P., Buonavoglia C. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet. Microbiol. 2008;126:30–39. doi: 10.1016/j.vetmic.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Elia G., Campolo M., Mari V., Desario C., Lucente M.S., Lorusso A., Greco G., Corrente M., Tempesta M., Buonavoglia C. Biological and genetic analysis of a bovine-like coronavirus isolated from water buffalo (Bubalus bubalis) calves. Virology. 2008;370:213–222. doi: 10.1016/j.virol.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S., Spaan W.J.M., Taguchi F., Talbot P. Family Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy, Classification and Nomenclature of Viruses. Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310:216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Shiu K.B., Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 2007;124:78–87. doi: 10.1016/j.virusres.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V., Santos N., Timenetsky, Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.S., Breslin J.J., Breuhaus B., Vivrette S., Smith L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hasoksuz M., Lathrop S.L., Gadfield K.L., Saif L.J. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 1999;60:1227–1233. [PubMed] [Google Scholar]
- Hasoksuz M., Alekseev K., Vlasova A., Zhang X., Spiro D., Halpin R., Wang S., Ghedin E., Saif L.J. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 2007;81:4981–4990. doi: 10.1128/JVI.02361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet A.E., Cho K.O., Chang K.O., Loerch S.C., Wittum T.E., Saif L.J. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 2002;63:342–348. doi: 10.2460/ajvr.2002.63.342. [DOI] [PubMed] [Google Scholar]
- Jiang X., Huang P.W., Zhong W.M., Farkas T., Cubitt D.W., Matson D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods. 1999;83:145–154. doi: 10.1016/s0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- Jin L., Cebra C.K., Baker R.J., Mattson D.E., Cohen S.A., Alvarado D.E., Rohrmann F. Analysis of the genome sequence of an alpaca coronavirus. Virology. 2007;365:198–203. doi: 10.1016/j.virol.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lathrop S.L., Wittum T.E., Brock K.V., Loerch S.C., Perino L.J., Bingham H.R., McCollum F.T., Saif L.J. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am. J. Vet. Res. 2000;61:1062–1066. doi: 10.2460/ajvr.2000.61.1062. [DOI] [PubMed] [Google Scholar]
- Lorusso A., Desario C., Mari V., Campolo M., Lo russo E., Elia G., Martella V., Buonavoglia C., Decaro N. Molecular characterization of a canine respiratory coronavirus strain detected in Italy. Virus Res. 2009;141:96–100. doi: 10.1016/j.virusres.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Ciarlet M., Pratelli A., Arista S., Terio V., Elia G., Cavalli A., Gentile M., Decaro N., Greco G., Cafiero M.A., Tempesta M., Buonavoglia C. Molecular analysis of the VP7, VP4, VP6, NSP4, and NSP5/6 genes of a buffalo rotavirus strain: identification of the rare P[3] rhesus rotavirus-like VP4 gene allele. J. Clin. Microbiol. 2003;41:5665–5675. doi: 10.1128/JCM.41.12.5665-5675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniiappa L., Mitov B.K., Kharalambiev Kh.E. Demonstration of coronavirus infection in buffaloes. Vet. Med. Nauki. 1985;22:27–32. [PubMed] [Google Scholar]
- Storz J., Purdy C.W., Lin X., Burrell M., Truax R.E., Briggs R.E., Frank G.H., Loan R.W. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp. from cattle involved in two natural outbreaks of shipping fever. J. Am. Vet. Med. Assoc. 2000;216:1599–1604. doi: 10.2460/javma.2000.216.1599. [DOI] [PubMed] [Google Scholar]
- Sullivan D.G., Akkina R.K. A nested polymerase chain reaction assay to differentiate pestiviruses. Virus Res. 1995;38:231–239. doi: 10.1016/0168-1702(95)00065-x. [DOI] [PubMed] [Google Scholar]
- Sunil-Chandra N.P., Mahalingam S. Rotavirus-associated diarrhoea in buffalo calves in Sri Lanka. Res. Vet. Sci. 1994;56:393–396. doi: 10.1016/0034-5288(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Valarcher J.F., Bourhy H., Gelfi J., Schelcher F. Evaluation of a nested reverse transcription-PCR assay based on the nucleoprotein gene for diagnosis of spontaneous and experimental bovine respiratory syncytial virus infections. J. Clin. Microbiol. 1999;37:1858–1862. doi: 10.1128/jcm.37.6.1858-1862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek S. Detection of the bovine herpesvirus-1 (BHV-1) genome by PCR. J. Virol. Methods. 1993;41:245–247. doi: 10.1016/0166-0934(93)90132-b. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]


